Abstract
Background: Hypertension has been continuing to be a major contributor to the global burden of disease and to the global mortality, leading to over 10 million deaths each year. The purpose of this study was to investigate the association between Adiponectin gene polymorphism with Essential hypertension (EH). Methods: PubMed, EMbase, the Cochrane Library, and China National Knowledge Infrastructure (CNKI) were searched independently by two investigators. Pooled odds ratios and 95% confidence intervals were calculated to estimate the associations of Adiponectin polymorphism with EH. Results: Thirteen studies with 3198 cases and 3076 controls for meta-analysis (MA) were included in present study. Pooled results showed that rs2241766 polymorphism is associated with the risk of EH in the allelic model (G vs. T: OR=1.10; 95% CI, 1.01-1.21). In the <40 years subgroup, rs2241766 polymorphism is associated with the risk of EH in allele model (G vs. T: OR=1.43; 95% CI, 1.06-1.94), recessive model (GG vs. GT + TT: OR=5.26, 95% CI=1.47-18.76), homozygous model of GG (GG vs.TT: OR=5.27, 95% CI=1.47-18.95), and rs266729 in recessive model (GG vs. GT + TT: OR=2.33, 95% CI=1.33-4.08). Conclusions: Our meta-analysis results show that the rs2241766 polymorphism is associated with the risk of hypertension. There still need a larger sample with better design to verify.
Keywords: Adiponectin, polymorphisms, hypertension, meta-analysis
Introduction
Hypertension has been continuing to be a major contributor to the global burden of disease and to the global mortality, leading to over 10 million deaths each year [1,2]. The etiology of hypertension has not been fully elucidated , however, the interaction between genes and environmental factors is thought to play an important role in the pathological process of hypertension [3]. In recent years, researches have been focusing on the genetic susceptibility and genetic polymorphism of hypertension [4,5].
Adiponectin (ADIPOQ) is a recently discovered cytokine specifically secreted by adipocytes, with plasma concentrations range from 5 to 30 mg/L and accounting for approximately 0.05% of total plasma protein in healthy adults [6]. Studies have shown that adiponectin plays an vital role in improving insulin sensitivity, anti-inflammation and anti-atherosclerosis [7,8]. In recent years, there was a study have reported that the polymorphism of ADIPOQ gene is associated with plasma levels of adiponectin, and low adiponectin levels predispose essential hypertension [9]. Some studies have investigated the correlation between several polymorphisms in ADIPOQ gene and hypertension, including rs2241766 (+45T>G), rs1501299 (+276G>T), and rs266729 (-11377C>G). A study showed [10] that the association between polymorphisms of adiponectin and essential hypertension and found +45T/G and +276G/T were significantly associated with essential hypertension. Also reported by other scholars [11] that although -11377C>G in the promoter region of ADIPOQ gene was not yet suggested to be associated with essential hypertension, they confirmed that -11377C>G was associated with reduced plasma lipids. In addition, it is reported [12] that there was no significant association between rs2241766 and rs1501299 and hypertension. And other scholars reported [13,14] found that rs2241766 was associated with the risk of hypertension, but rs1501299 was not associated with the risk of hypertension. Therefore, results of these studies are inconsistent and the association between polymorphisms in ADIPOQ gene and hypertension remains to be determined. We performed this meta-analysis aimed at determining the associations of rs2241766, rs1501299, and rs266729 polymorphisms in the ADIPOQ gene with hypertension susceptibility.
The present meta-analysis aimed to analyze the relationship between three widely evaluated genetic polymorphisms in ADIPOQ gene and the risk of hypertension. The ADIPOQ gene (OMIM: 605441, Gene ID: 9370) is located in human 3q27 and has a DNA length of approximately 16 kb and contains three exons and two introns. ADIPOQ is a specific hormone protein secreted by fat cells. It is a protective factor and plays a very important role in anti-atherosclerosis, insulin resistance and anti-inflammation [15].
Adamczak et al. [16] first analyzed and reported a negative correlation between plasma levelsof adiponectin and the systolic blood pressure, diastolic blood pressure, and mean arterial pressure and concluded that low levels of adiponectin may play a role in the pathogenesis of essential hypertension. Mangge et al. [17] reported that the polymorphism of ADPIOQ gene correlated with adiponectin concentration. Therefore, the polymorphism of ADPIOQ gene was considered associating with the risk of hypertension.
Materials and methods
The meta-analysis is conducted in accordance with the guidelines of the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) statement [18].
Search strategy
To identify studies eligible for the systematic review and meta-analysis, we searched databases of PubMed, EMbase, the Cochrane Library, and China National Knowledge Infrastructure up to December 19, 2019, The following terms: “hypertension”, “adiponectin”, “ADIPOQ”, “intron”, “exon” and “polymorphism” was used for this searching. Additional potential relevant articles were identified by us through screened references of retrieved articles.
Inclusion and exclusion criteria
Inclusion criteria
Studies included must meet the following criteria: (1) Studies were case-control or cohort design; (2) Studies have evaluated the association between single nucleotide polymorphisms in ADIPOQ gene and susceptibility of hypertension; (3) Studies evaluated rs2241766, rs1501299, or/and rs266729; (4) Studies provided details of associations between genotypes and phenotypes and sufficient data for estimating odds ratios (ORs) with 95% confidence intervals (CIs).
Exclusion criteria
(1) Conference abstracts, case reports, comments, repeated researches, and systematic reviews were excluded; (2) Studies lacking data for genotype distribution; (3) Studies unable to determine whether the control population meets the Hardy-Weinberg equilibrium (HWE) balance.
Data extraction and quality assessment
Two reviewers independently screened the articles, extracted data, and performed quality evaluations. Data extracted from studies were as follows: first author, country, year of publication, average age, type of study, sample size, ethnicity, genotyping method, genotype distributions, and body mass index (BMI). Disagreements were resolved through discussion or consulting third-party experts. The quality of the included studies was assessed using the Newcastle-Ottawa (NOS) scale, with a rating of 0-9 and a maximum of 9 [19].
Statistical analysis
Stata version 12.0 (Stata Corp, College Station, TX) was used for statistical analyses. A chi-square test was performed to evaluate the HWE for the control group in each study group. Pooled ORs with corresponding 95% CIs were calculated to evaluate associations between the ADIPOQ gene and hypertension for the following genetic models: rs2241766 (allelic model: G vs. T, recessive models: GG vs. GT + TT, dominant models: GG + GT vs. TT, heterozygote model: GT vs. TT, homozygote model: GG vs. TT); rs1501299 (allelic model: T vs. G, recessive models: TT vs. TG + GG, dominant models: TT + TG vs. GG, heterozygote model: TG vs. GG, homozygote model: TT vs. GG); rs266729 (allelic model: G vs. C, recessive models: GG vs. GC + CC, dominant models: GG + GC vs. CC, heterozygote model: GC vs. CC, homozygote model: GG vs. CC). Q tests and I2 statistics were performed to assess heterogeneity between studies. If P Q<0.05 or I2>50%, a significant heterogeneity was existed among studies and then, a random effects model was used to calculate the pooled OR, otherwise (i.e., P Q>0.05 or I2<50%), a fixed effects model was performed.
For studies with significant heterogeneity, stratified analysis for samples may be performed by subgroup analysis to identify potential sources of heterogeneity. Gradually eliminating individual studies or studies deviating from HWE for sensitivity analysis were performed to assess the stability of results. Begg’s funnel plot and Egger’s test were used to justify potential publication bias.
Results
Literature screening
A total of 308 studies were retrieved in our work. After excluding duplicate publications, case reports, reviews, and studies that could not be combined, thirteen articles with 3198 cases and 3076 controls were included in this study. Studies by Zhang et al. [20], Leu et al. [21], Kang et al. [22], Jiang et al. [23], and Machado et al. [24] contained two studies, which were analyzed separately in the following meta-analysis. A flow chart summarizing the process of literature is depicted in Figure 1.
Figure 1.
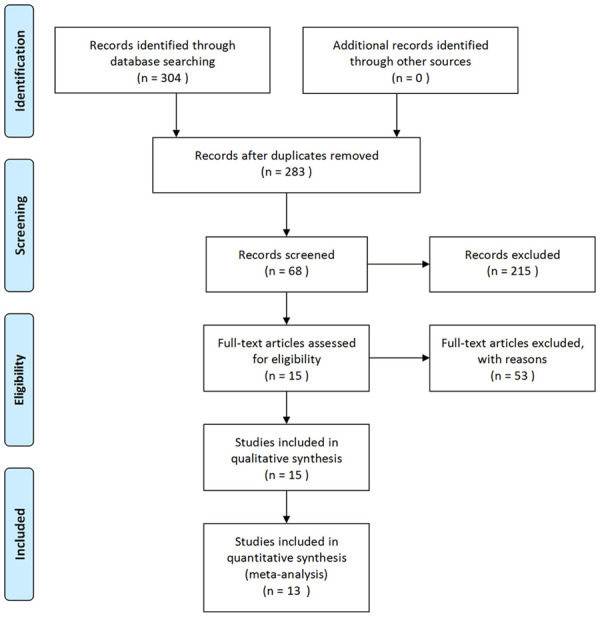
PRISMA flow diagram of studies included in the Meta-analysis.
Characteristics and quality evaluations for included studies
Table 1 summarizes characteristics of all included studies. Eleven studies were carried out in China, one in Brazil, and one in Turkey. The rs2241766 polymorphism was analyzed in 12 studies, the rs1501299 polymorphism was in 10, and the rs266729 polymorphism was in 8.
Table 1.
Principle characteristics of the studies included in the meta-analysis
| Study | Country | Year | Age | Type of study | Sample size | Ethnicity | Genotyping method | Genotype distributions (case/control) | P value of HWE | Quality score | BMI | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||||||||||
| Case | Control | Case | Control | Case | Control | |||||||||||
| rs2241766 | TT | TG | GG | |||||||||||||
| Yan [12] | China | 2006 | 49.0±9.0 | 48.0±10.0 | Case control | 482 | 497 | Asian | PCR | 201/222 | 186/203 | 95/72 | 0.024 | 8 | 30.80±2.50 | 22.40±1.80 |
| Jeng [35] | China | 2007 | 51.6±14.7 | 50.7±12.1 | Case control | 212 | 356 | Asian | PCR | 82/126 | 99/173 | 31/57 | 0.853 | 7 | 25.70±3.60 | 23.80±3.40 |
| Tang [13] | China | 2008 | 63.5±6.5 | 60.5±7.1 | Case control | 151 | 100 | Asian | PCR | 99/63 | 32/28 | 20/9 | 0.036 | 8 | NR | NR |
| Wang [14] | China | 2008 | 46.4±14.0 | 46.8±15.4 | Case control | 80 | 160 | Asian | PCR | 41/87 | 34/60 | 5/13 | 0.561 | 6 | 26.82±2.44 | 22.43±3.99 |
| Youpeng [36] | China | 2010 | 30.9±6.0 | 30.0±4.5 | Case control | 107 | 81 | Asian | PCR | 74/53 | 26/27 | 7/1 | 0.228 | 8 | 22.05±2.65 | 20.10±1.80 |
| Leu A [21] | China | 2011 | 41.4±0.7 | 46.7±0.8 | Case control | 159 | 446 | Asian | PCR | 74/233 | 70/181 | 15/32 | 0.696 | 8 | 25.40±0.30 | 23.50±0.10 |
| Leu B [21] | China | 2011 | 41.7±0.6 | 57.5±1.0 | Case control | 192 | 165 | Asian | PCR | 92/78 | 81/75 | 19/12 | 0.696 | 8 | 28.30±0.30 | 27.10±0.30 |
| Kang B [22] | China | 2013 | 52.3±9.6 | 49.8±8.7 | Case control | 153 | 126 | Asian | PCR | 63/75 | 68/40 | 11/22 | 0.105 | 7 | 22.57±2.27 | 21.53±2.17 |
| Jiang A [23] | China | 2014 | 68.8±6.7 | 67.1±7.1 | Case control | 223 | 176 | Asian | PCR | 102/91 | 86/65 | 10/14 | 0.72 | 9 | 24.50±4.30 | 23.10±3.30 |
| Jiang B [23] | China | 2014 | 68.2±6.1 | 67.1±7.2 | Case control | 181 | 176 | Asian | PCR | 90/91 | 67/65 | 12/10 | 0.72 | 9 | 26.00±3.70 | 23.10±3.30 |
| Machado A [24] | Brazil | 2014 | 26.0±4.5 | 24.0±4.0 | Case control | 113 | 161 | Caucasian | PCR | 81/139 | 29/29 | 3/1 | 0.697 | 8 | 27.53±5.15 | 22.80±2.70 |
| Machado B [24] | Brazil | 2014 | 27.0±4.5 | 24.0±5.0 | Case control | 127 | 161 | Caucasian | PCR | 97/139 | 26/29 | 4/1 | 0.697 | 8 | 26.28±4.80 | 22.80±2.70 |
| rs1501299 | GG | TG | TT | |||||||||||||
| Iwashima [9] | Japan | 2004 | 59.4±0.5 | 57.1±0.6 | Case control | 446 | 312 | Asian | PCR | 225/165 | 180/124 | 41/23 | 0.964 | 8 | 24.40±0.10 | 23.10±0.20 |
| Yan [12] | China | 2006 | 49.0±9.0 | 48.0±10.0 | Case control | 482 | 497 | Asian | PCR | 259/274 | 184/187 | 39/36 | 0.599 | 8 | 30.80±2.50 | 22.40±1.80 |
| Wang [14] | China | 2008 | 46.4±14.0 | 46.8±15.4 | Case control | 80 | 160 | Asian | PCR | 41/74 | 28/73 | 11/13 | 0.392 | 6 | 26.82±2.44 | 22.43±3.99 |
| Youpeng [36] | China | 2010 | 30.9±6.0 | 30.0±4.5 | Case control | 107 | 81 | Asian | PCR | 19/19 | 60/49 | 28/13 | 0.051 | 8 | 22.05±2.65 | 20.10±1.80 |
| Leu A [21] | China | 2011 | 41.4±0.7 | 46.7±0.8 | Case control | 159 | 446 | Asian | PCR | 85/228 | 60/178 | 14/40 | 0.536 | 8 | 25.40±0.30 | 23.50±0.10 |
| Leu B [21] | China | 2011 | 41.7±0.6 | 57.5±1.0 | Case control | 192 | 165 | Asian | PCR | 117/93 | 68/56 | 7/16 | 0.089 | 8 | 28.30±0.30 | 27.10±0.30 |
| Kang B [22] | China | 2013 | 52.3±9.6 | 49.8±8.7 | Case control | 153 | 126 | Asian | PCR | 100/82 | 46/38 | 7/6 | 0.56 | 7 | 22.57±2.27 | 21.53±2.17 |
| Machado A [24] | Brazil | 2014 | 26.0±4.5 | 24.0±4.0 | Case control | 113 | 161 | Caucasian | PCR | 59/68 | 46/74 | 8/19 | 0.868 | 8 | 27.53±5.15 | 22.80±2.70 |
| Machado B [24] | Brazil | 2014 | 27.0±4.5 | 24.0±5.0 | Case control | 127 | 161 | Caucasian | PCR | 66/68 | 47/74 | 14/19 | 0.868 | 8 | 26.28±4.80 | 22.80±2.70 |
| Demir [30] | Turkey | 2016 | 63.6±7.1 | 64.6±7.3 | Case control | 170 | 170 | Caucasian | PCR | 78/92 | 73/72 | 19/6 | 0.071 | 8 | 27.30±3.20 | 26.10±3.20 |
| rs266729 | CC | CG | GG | |||||||||||||
| Jia [37] | China | 2008 | 61.1 | 57.0 | Case control | 182 | 58 | Asian | PCR | 96/37 | 77/19 | 9/2 | 0.817 | 8 | NR | NR |
| Zhang A [20] | China | 2011 | 52.4±9.6 | 49.3±10.8 | Case control | 112 | 128 | Asian | PCR | 72/78 | 33/46 | 7/4 | 0.368 | 7 | NR | NR |
| Zhang B [20] | China | 2011 | 53.9±10.5 | 49.8±10.3 | Case control | 108 | 140 | Asian | PCR | 66/85 | 37/38 | 5/17 | <0.01 | 7 | NR | NR |
| Leu A [21] | China | 2011 | 41.4±0.7 | 46.7±0.8 | Case control | 159 | 446 | Asian | PCR | 85/84 | 90/64 | 23/11 | 0.8 | 8 | 25.40±0.30 | 23.50±0.10 |
| Leu B [21] | China | 2011 | 41.7±0.6 | 57.5±1.0 | Case control | 192 | 165 | Asian | PCR | 93/84 | 68/64 | 12/11 | 0.8 | 8 | 28.30±0.30 | 27.10±0.30 |
| Kang A [22] | China | 2013 | 52.3±9.6 | 49.8±8.7 | Case control | 153 | 126 | Asian | PCR | 95/70 | 48/46 | 10/10 | 0.532 | 7 | 22.57±2.27 | 21.53±2.17 |
| Machado A [24] | Brazil | 2014 | 26.0±4.5 | 24.0±4.0 | Case control | 113 | 161 | Caucasian | PCR | 61/93 | 36/57 | 16/11 | 0.577 | 8 | 27.53±5.15 | 22.80±2.70 |
| Machado B [24] | Brazil | 2014 | 27.0±4.5 | 24.0±5.0 | Case control | 127 | 161 | Caucasian | PCR | 81/93 | 27/57 | 19/11 | 0.577 | 8 | 26.28±4.80 | 22.80±2.70 |
Abbreviations: PCR: polymerase chain reaction; BMI: body mass index.
According to the Newcastle-Ottawa quality assessment scale, the quality of the included literature was evaluated. The scores are shown in Table 1 and 9 studies have a score of no less than 8.
Quantitative synthesis for MA
For a comprehensive evaluation, we calculated Ors with 95% CIs to estimate associations of the three polymorphisms in ADIPOQ with the risk of hypertension under the genetic models of alleles, recessive genotypes, dominant genotypes, heterozygote, and homozygote. The genetic polymorphisms of the three loci were not heterogeneous in general, so the combined effect model was used for the combined analysis (as shown in Table 2). According to ORs and 95% CIs, there were no statistical significances between the rs1501299 polymorphism and the rs266729 polymorphism and the risk of hypertension in all of the above mentioned genetic models. In the allelic model of the rs2241766 polymorphism, Figure 2 showed the allele of G was associated with an increased risk of hypertension (G vs. T: OR=1.10; 95% CI, 1.01-1.21). However, the difference was not statistically significant in the other four genetic models for the rs2241766 polymorphism (Table 2).
Table 2.
Pooled ORs and 95% CIs of the associations of ADIPOQ polymorphsims with hypertension
| Genotype | s.s | Pooled estimate | Heterogeneity | P value for Begg’s test | ||
|---|---|---|---|---|---|---|
|
|
|
|||||
| OR, 95% CI | P Z | I 2 | P Q | |||
| rs2241766 | ||||||
| G vs. T | 12 | 1.10, 1.01-1.21 | 0.035 | 0.00% | 0.563 | 0.304 |
| GG vs. GT + TT | 12 | 1.17, 0.96-1.42 | 0.124 | 40.20% | 0.073 | 0.537 |
| GG + GT vs. TT | 12 | 1.12, 0.99-1.26 | 0.073 | 0.00% | 0.567 | 0.837 |
| GT vs. TT | 12 | 1.09, 0.96-1.23 | 0.206 | 25.00% | 0.198 | 0.837 |
| GG vs. TT | 12 | 1.19, 0.97-1.47 | 0.093 | 23.30% | 0.215 | 0.537 |
| rs1501299 | ||||||
| T vs. G | 10 | 1.01, 0.92-1.11 | 0.824 | 45.90% | 0.055 | 0.592 |
| TT vs. TG + GG | 10 | 1.13, 0.91-1.41 | 0.257 | 49.00% | 0.039 | 0.858 |
| TT + TG vs. GG | 10 | 0.98, 0.86-1.11 | 0.720 | 15.30% | 0.302 | 0.858 |
| TG vs. GG | 10 | 0.96, 0.84-1.09 | 0.522 | 0.00% | 0.633 | 0.474 |
| TT vs. GG | 10 | 1.09, 0.87-1.37 | 0.455 | 53.70% | 0.022 | 1.000 |
| rs266729 | ||||||
| G vs. C | 8 | 1.08, 0.94-1.24 | 0.263 | 25.90% | 0.223 | 0.711 |
| GG vs. GC + CC | 8 | 1.34, 0.98-1.84 | 0.063 | 45.20% | 0.078 | 0.174 |
| GG + GC vs. CC | 8 | 1.03, 0.87-1.22 | 0.708 | 17.40% | 0.293 | 1.000 |
| GC vs. CC | 8 | 0.98, 0.82-1.17 | 0.801 | 38.20% | 0.125 | 0.902 |
| GG vs. CC | 8 | 0.76, 0.55-1.05 | 0.095 | 2.70% | 0.409 | 0.266 |
Figure 2.
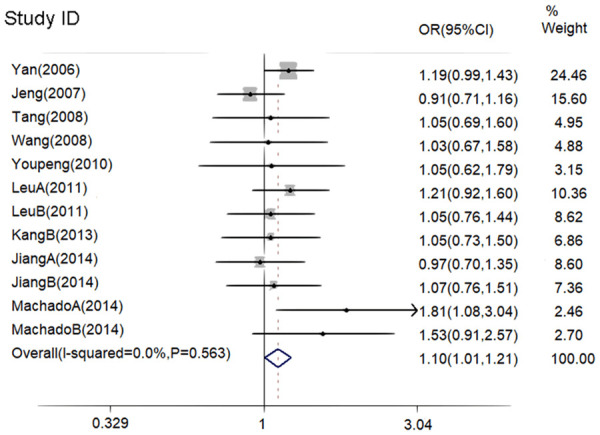
Forest plot for the risk of hypertension with allele model of rs2241766.
We then performed analyses for associations of the three single nucleotide polymorphisms in ADIPOQ gene with the risk of hypertension in subgroups stratified by age, ethnicity, BMI, and whether the control group met the HWE (Hardy-Weinberg equilibrium) or not. The results of all subgroup analyses were listed in Table 3. We found that rs2241766 SNP was significantly associated with the risk of hypertension in individuals no more than 40 years old. Increased risk of hypertension was observed for the allele of G (G vs. T: OR=1.43; 95% CI, 1.06-1.94), for the recessive model (GG v GT + TT: OR=5.26, 95% CI=1.47-18.76), and for the homozygous model of GG (GG v TT: OR=5.27, 95% CI=1.47-18.95). In the Caucasian population, the risk of hypertension still increased significantly for the allele of G (G VS T: OR=1.67, 95% CI=1.16-2.40), for the recessive model (GG v GT + TT: OR=5.04, 95% CI=1.04-24.50), for the dominant models (GG + GT v TT: OR=1.62, 95% CI=1.08-2.42), and for the homozygous model of GG (GG v TT: OR=5.46, 95% CI=1.12-26.64). In the population with BMI of more than25, allele models (G VS T: OR=1.13, 95% CI=1.02-1.26), recessive models (GG v GT + TT: OR=1.27, 95% CI=1.02-1.59), and homozygous models (GG vs. TT: OR=1.28; 95% CI, 1.01-1.62) have significant associations with the risk of hypertension. The results of the stratified study showed that the polymorphism of rs1501299 was not associated with the risk of hypertension. Although there was no significant association between the rs266729 polymorphism and the risk of hypertension for the overall group without stratification, it was found that the recessive model of GG in individuals no more than40 years old (GG v GT + TT: OR=2.33, 95% CI=1.33-4.08) and in the Caucasian population (GG v GT + TT: OR=2.33, 95% CI=1.33-4.08) was significantly associated with an increased risk for the occurrence of hypertension (Table 3).
Table 3.
Hierarchical analysis results for the associations of three locis polymorphsims with hypertension
| Subgroups (rs2241766) | s.s | G vs. T | GG v GT + TT | GG + GT v TT | GT v TT | GG v TT | |||||
|
|
|
|
|
|
|||||||
| OR, 95% CI | I 2, P Q | OR, 95% CI | I 2, P Q | OR, 95% CI | I 2, P Q | OR, 95% CI | I 2, P Q | OR, 95% CI | I 2, P Q | ||
|
| |||||||||||
| Age | |||||||||||
| ≤40 years | 3 | 1.43, 1.06-1.94 | 7.5%, 0.339 | 5.26, 1.47-18.76 | 0.0%, 0.991 | 1.33, 0.95-1.86 | 40.8%, 0.185 | 1.18, 0.84-1.67 | 53.9%, 0.114 | 5.27, 1.47-18.95 | 0.0%, 0.996 |
| >40 years | 9 | 1.07, 0.98-1.18 | 0.0%, 0.836 | 1.11, 0.91-1.35 | 37.6%, 0.118 | 1.09, 0.96-1.24 | 0.0%, 0.758 | 1.07, 0.93-1.23 | 20.3%, 0.262 | 1.13, 0.91-1.39 | 9.4%, 0.356 |
| Ethnicity | |||||||||||
| Asian | 10 | 1.07, 0.98-1.18 | 0.0%, 0.895 | 1.13, 0.93-1.38 | 40.2%, 0.090 | 1.08, 0.95-1.22 | 0.0%, 0.777 | 1.05, 0.92-1.20 | 23.4%, 0.227 | 1.15, 0.94-1.42 | 15.9%, 0.297 |
| Caucasian | 2 | 1.67, 1.16-2.40 | 0.0%, 0.648 | 5.04, 1.04-24.50 | 0.0%, 0.913 | 1.62, 1.08-2.42 | 0.0%, 0.551 | 1.49, 0.98-2.25 | 0.0%, 0.494 | 5.46, 1.12-26.64 | 0.0%, 0.947 |
| BMI | |||||||||||
| ≤25 | 3 | 1.01, 0.81-1.26 | 0.0%, 0.940 | 0.65, 0.39-1.09 | 60.4%, 0.080 | 1.16, 0.88-1.52 | 15.7%, 0.306 | 1.26, 0.94-1.68 | 70.2%, 0.035 | 0.77, 0.45-1.32 | 44.1%, 0.167 |
| >25 | 8 | 1.13, 1.02-1.26 | 19.3%, 0.277 | 1.27, 1.02-1.59 | 0.0%, 0.474 | 1.12, 0.98-1.29 | 0.0%, 0.486 | 1.07, 0.93-1.24 | 0.0%, 0.632 | 1.28, 1.01-1.62 | 2.1%, 0.413 |
| HWE | |||||||||||
| P H<0.05 | 2 | 1.17, 0.99-1.38 | 0.0%, 0.583 | 1.46, 1.07-2.00 | 0.0%, 0.890 | 1.08, 0.86-1.36 | 0.0%, 0.435 | 0.96, 0.74-1.23 | 0.0%, 0.325 | 1.45, 1.04-2.02 | 0.0%, 0.949 |
| P H>0.05 | 10 | 1.08, 0.97-1.20 | 0.0%, 0.461 | 1.00, 0.78-1.29 | 38.8%, 0.100 | 1.13, 0.98-1.30 | 0.0%, 0.449 | 1.14, 0.98-1.32 | 27.1%, 0.195 | 1.05, 0.80-1.37 | 24.3%, 0.219 |
|
| |||||||||||
| Subgroups (rs1501299) | T v G | TT v TG + GG | TT + TG v GG | TG v GG | TT v GG | ||||||
|
|
|
|
|
|
|||||||
| OR, 95% CI | I 2, P Q | OR, 95% CI | I 2, P Q | OR, 95% CI | I 2, P Q | OR, 95% CI | I 2, P Q | OR, 95% CI | I 2, P Q | ||
|
| |||||||||||
| Age | |||||||||||
| ≤40 years | 3 | 0.89, 0.71-1.10 | 67.9%, 0.044 | 1.05, 0.68-1.61 | 54.0%, 0.114 | 1.03, 0.90-1.17 | 0.0%, 0.628 | 0.76, 0.55-1.05 | 0.0%, 0.369 | 0.89, 0.55-1.44 | 63.7%, 0.064 |
| >40 years | 7 | 1.04, 0.94-1.16 | 30.8%, 0.193 | 1.16, 0.91-1.50 | 54.4%, 0.041 | 0.77, 0.57-1.04 | 41.8%, 0.179 | 1.00, 0.87-1.16 | 0.0%, 0.844 | 1.16, 0.89-1.50 | 54.4%, 0.041 |
| Ethnicity | |||||||||||
| Asian | 7 | 1.03, 0.92-1.15 | 2.6%, 0.405 | 1.13, 0.88-1.44 | 37.6%, 0.142 | 1.00, 0.87-1.16 | 0.0%, 0.795 | 0.99, 0.85-1.15 | 0.0%, 0.883 | 1.10, 0.85-1.43 | 34.7%, 0.163 |
| Caucasian | 3 | 0.96, 0.79-1.18 | 80.3%, 0.006 | 1.16, 0.74-1.83 | 75.2%, 0.018 | 0.89, 0.68-1.16 | 70.9%, 0.032 | 0.85, 0.65-1.12 | 46.9%, 0.152 | 1.06, 0.66-1.69 | 80.3%, 0.006 |
| BMI | |||||||||||
| ≤25 | 3 | 1.13, 0.95-1.35 | 0.0%, 0.522 | 1.38, 0.92-2.06 | 0.0%, 0.571 | 1.11, 0.87-1.40 | 0.0%, 0.715 | 1.06, 0.83-1.36 | 0.0%, 0.902 | 1.40, 0.90-2.15 | 0.0%, 0.510 |
| >25 | 7 | 0.97, 0.86-1.08 | 54.5%, 0.040 | 1.04, 0.80-1.35 | 60.7%, 0.018 | 0.93, 0.81-1.08 | 29.5%, 0.203 | 0.92, 0.79-1.07 | 0.0%, 0.436 | 0.99, 0.76-1.29 | 63.2%, 0.012 |
|
| |||||||||||
| Subgroups (rs266729) | G v C | GG v GC + CC | GG + GC v CC | GC v CC | GG v CC | ||||||
|
|
|
|
|
|
|||||||
| OR, 95% CI | I 2, P Q | OR, 95% CI | I 2, P Q | OR, 95% CI | I 2, P Q | OR, 95% CI | I 2, P Q | OR, 95% CI | I 2, P Q | ||
|
| |||||||||||
| Age | |||||||||||
| ≤40 years | 2 | 1.18, 0.9-1.55 | 0.0%, 0.414 | 2.33, 1.33-4.08 | 0.0%, 0.911 | 0.95, 0.68-1.33 | 26.9%, 0.242 | 0.73, 0.50-1.06 | 54.0%, 0.140 | 0.82, 0.46-1.47 | 0.0%, 0.440 |
| >40 years | 6 | 1.05, 0.89-1.23 | 38.8%, 0.147 | 1.04, 0.71-1.53 | 35.9%, 0.168 | 1.06, 0.87-1.29 | 26.4%, 0.236 | 1.07, 0.87-1.32 | 19.4%, 0.287 | 0.73, 0.49-1.08 | 22.9%, 0.262 |
| Ethnicity | |||||||||||
| Asian | 6 | 1.05, 0.89-1.23 | 38.8%, 0.147 | 1.04, 0.71-1.53 | 35.9%, 0.168 | 1.06, 0.87-1.29 | 26.4%, 0.236 | 1.07, 0.87-1.32 | 19.4%, 0.287 | 0.73, 0.49-1.08 | 22.9%, 0.262 |
| Caucasian | 2 | 1.18, 0.9-1.55 | 0.0%, 0.414 | 2.33, 1.33-4.08 | 0.0%, 0.911 | 0.95, 0.68-1.33 | 26.9%, 0.242 | 0.73, 0.50-1.06 | 54.0%, 0.140 | 0.82, 0.46-1.47 | 0.0%, 0.440 |
Sensitivity analysis and publication bias
To rule out the impact of low-quality literature on meta-analysis results, we performed a sensitivity analysis by omitting individual studies one by one. Results showed that no individual studies on the polymorphisms of the three loci affected the value of the combined OR, which proved that the MA has good stability.
The publication bias was analyzed using the Stata 12.0 software. We found that the funnel plot was basically symmetrical with respect to the central axis, and P values of Begg’s test were all more than 0.05 (Table 2), indicating that there was no publication bias in each genetic model.
Discussion
Our results of the present study indicate that there is a significant association between the polymorphism of rs2241766 and the risk of hypertension. The T→G mutation was observed in the allelic model and increased the risk of hypertension by 10% (OR=1.10, 95% CI=1.01-1.21), meanwhile, no statistical significances were found in other genetic models nor in any genetic models for the polymorphisms of rs1501299 and rs266729. Although no heterogeneity was identified between studies and considering the asymmetry of factors such as race, age, and BMI in the study, we performed analyses for subgroups stratified by these factors. Subgroup analysis showed that significant associations of the rs2241766 polymorphism with the risk of hypertension were observed not only in the allelic model, recessive model, dominant model, and homozygous model in the subgroup of the Caucasian population, but also in the allelic model, recessive model, and homozygous model in individuals with BMI more than 25. In subgroups stratified by age, we found that the allelic model, recessive model, and homozygous models was significantly associated with an increased risk of hypertension in individuals no more than 40 years old. In order to prevent the impact of the HWE study on the analysis, we analyzed the test value based on the control group P, and found that the recessive and homozygous models were significantly associated with the risk of onset in the group at P<0.05. The association between the rs2241766 polymorphism in ADIPOQ gene and the susceptibility to hypertension has also been discussed by Xi et al. [25] Zhaoet al. [26]. In their study, the rs2241766 was not associated with the hypertension susceptibility, whichis inconsistent with our findings. However, the rs2241766 in ADIPOQ gene is associated with risks of obesity, coronary heart disease, and type 2 diabetes. Therefore, no association between the rs2241766 polymorphism and the risk of hypertension that reported by Zhao et al. [26] and Xi et al. [25] may be related to environmental factors such as diet-induced obesity which were not considered in their studies. The level of adiponectin in the plasma of patients with hypertension is low, and low levels of adiponectin are associated with single nucleotide polymorphisms of ADIPOQ [27]. Mousavinasab et al. [28] found that the GG genotype of the rs2241766 had a higher prevalence in individuals with hypertension than TT genotype. Tang et al. [5,28] reported that GG + GT genotype rather than TT genotype was significantly associated with the low level of adiponectin. Therefore, we speculate that the association between hypertension susceptibility and rs2241766 polymorphism may be affected by environmental factors such as obesity. The rs2241766 polymorphism in the exon 2 of the ADIPOQ gene may affect the expression of ADIPOQ genein adipose tissues and thereby associate with a reduced plasma concentration of adiponectin, which may increase the probability of developing hypertension. In the present study, the significance of association identified in the subgroup of the Caucasian may indicate the genetic heterogeneity. We speculate that the linkage disequilibrium structure of human ADIPOQ gene may be different in different ethnicities, i.e., the rs2241766 polymorphism is not consistently associated with the risk of hypertension in different races. Further exploration of the genetic structure of ADIPOQ gene may improve our understanding of mechanisms by which genetic factors work in hypertension.
We found that the rs266729 polymorphism was not significantly associated with the risk of hypertension risk at an overall level for included subjects. However, after stratification, in individuals no more than 40 years old and those who were Caucasians, the recessive genetic model of GG of the rs266729 were associated with an increased probability of developing hypertension (GG v GT + TT: OR=2.33, 95% CI=1.33-4.08). Studies on rs266729 and hypertension risk included subjects from different regions. A study of the British population found that the rs266729 polymorphism was associated with the levels of systolic and diastolic blood pressure [29]. The -11377C>G mutation in the proximal promoter region is associated with lower levels ofadiponectin and with an increased risk of hypertension in Hong Kong and the mainland of China. Although our results were also statistically significant, considering the limited number of the included studies, the interpretation of our results needs to be cautious. The association between the rs266729 polymorphism and the risk of hypertension and the underlying mechanism deserve further study.
In our present study, the polymorphism of rs1501299 was not significantly associated with the risk of hypertension in either overall or subgroup analyses. In our included studies, it was reported that the rs1501299 polymorphism is a protective factor for hypertension or a risk factor for hypertension [24,30], which is inconsistent to our null conclusions. Hara et al. [31] found association between the rs1501299 polymorphism and adiponectin concentration in obese individuals with a BMI of no less than 26.7. We considered that the inconsistency may be due to differences in stratification and assumed that rs1501299 might be less able to regulate adiponectin levels than other polymorphism loci, all of which may be unrelated. Therefore, more research is needed to explore the effect of rs1501299 polymorphism on hypertension.
Nowadays, it has been proved that adiponectin can decrease blood pressure through central and peripheral mechanisms. However, experiments for these findings are performed in environment that were strictly controlled and had well designed controls, which may not directly and accurately reflect the occurrence and development of human hypertension in the real world. Hypertension is affected by a combination of environmental and genetic factors. Adiponectin levels are also affected by both the ADIPOQ polymorphism and the environment. Risk factors relating to lifestyle such as obesity may often play a more decisive role than genetic factors. Some studies have shown that after adjustment of some adipokines such as resistin and leptin, adiponectin levels are no longer associated with the risk of hypertension, meanwhile, adipocytokines secreted by perivascular adipose tissue still cause contractions of adjacent small arteries [32,33]. Under this circumstance, the level of adiponectin may not be sufficient to affect blood pressure, and other fat factors may do. However, a genome-wide association study showed that the ADIPOQ polymorphism was identified as an important genetic factor affecting blood pressure, and the ADIPOQ gene affects blood pressure by regulating adiponectin levels, rather than by regulating adipokines or other acquired risk factors identified [34].
Our research has some limitations: ① the literature we included is either in Chinese or in English and may have some bias in the results; ② when we conducted subgroup analysis, limited studies were available for certain subgroup analyses and the results need to be treated with caution; ③ there were two control group in the literature that does not meet the HWE balance, which mightdue to genotyping errors or other biases in the original study; ④ our analysis was based primarily on unadjusted effect estimates, while confounding factors were not controlled and gene-environment interactions with disease were not considered; ⑤ only Asians and Caucasians were included, and the conclusions may not be suitable for all races.
In summary, our meta-analysis results show that the rs2241766 polymorphism is associated with the risk of hypertension. The polymorphism of rs1501299 is not significantly associated with the risk of hypertension. The rs266729 polymorphism is only found to be associated with the risk of hypertension in subgroups of individuals no more than 40 years old and in Caucasian population. Whether these polymorphisms may have a clinical significance in patients with hypertension remains to be determined, and further research is needed to confirm our finding in the future.
Acknowledgements
This work was supported by: (1) The new era comprehensively and thoroughly promote the study of healthy Anhui construction, Anhui Province philosophy and social science planning general project (No. AHSKY2018D55); (2) Molecular epidemiological study of cZNF609/mir-615-5p/target gene signaling pathway involved in hypertension and stroke mechanism, National Natural Science Foundation of China (No. 81874280).
Disclosure of conflict of interest
None.
References
- 1.Forouzanfar MH, Liu P, Roth GA, Ng M, Biryukov S, Marczak L, Alexander L, Estep K, Hassen Abate K, Akinyemiju TF, Ali R, Alvis-Guzman N, Azzopardi P, Banerjee A, Barnighausen T, Basu A, Bekele T, Bennett DA, Biadgilign S, Catala-Lopez F, Feigin VL, Fernandes JC, Fischer F, Gebru AA, Gona P, Gupta R, Hankey GJ, Jonas JB, Judd SE, Khang YH, Khosravi A, Kim YJ, Kimokoti RW, Kokubo Y, Kolte D, Lopez A, Lotufo PA, Malekzadeh R, Melaku YA, Mensah GA, Misganaw A, Mokdad AH, Moran AE, Nawaz H, Neal B, Ngalesoni FN, Ohkubo T, Pourmalek F, Rafay A, Rai RK, Rojas-Rueda D, Sampson UK, Santos IS, Sawhney M, Schutte AE, Sepanlou SG, Shifa GT, Shiue I, Tedla BA, Thrift AG, Tonelli M, Truelsen T, Tsilimparis N, Ukwaja KN, Uthman OA, Vasankari T, Venketasubramanian N, Vlassov VV, Vos T, Westerman R, Yan LL, Yano Y, Yonemoto N, Zaki ME, Murray CJ. Global burden of hypertension and systolic blood pressure of at least 110 to 115 mmHg, 1990-2015. JAMA. 2017;317:165–182. doi: 10.1001/jama.2016.19043. [DOI] [PubMed] [Google Scholar]
- 2.GBD 2017 Risk Factor Collaborators. Global, regional, and national comparative risk assessment of 84 behavioural, environmental and occupational, and metabolic risks or clusters of risks for 195 countries and territories, 1990-2017: a systematic analysis for the global burden of disease study 2017. Lancet. 2018;392:1923–1994. doi: 10.1016/S0140-6736(18)32225-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harrison M, Maresso K, Broeckel U. Genetic determinants of hypertension: an update. Curr Hypertens Rep. 2008;10:488–495. doi: 10.1007/s11906-008-0091-1. [DOI] [PubMed] [Google Scholar]
- 4.Rhodes CJ, Batai K, Bleda M, Haimel M, Southgate L, Germain M, Pauciulo MW, Hadinnapola C, Aman J, Girerd B, Arora A, Knight J, Hanscombe KB, Karnes JH, Kaakinen M, Gall H, Ulrich A, Harbaum L, Cebola I, Ferrer J, Lutz K, Swietlik EM, Ahmad F, Amouyel P, Archer SL, Argula R, Austin ED, Badesch D, Bakshi S, Barnett C, Benza R, Bhatt N, Bogaard HJ, Burger CD, Chakinala M, Church C, Coghlan JG, Condliffe R, Corris PA, Danesino C, Debette S, Elliott CG, Elwing J, Eyries M, Fortin T, Franke A, Frantz RP, Frost A, Garcia JGN, Ghio S, Ghofrani HA, Gibbs JSR, Harley J, He H, Hill NS, Hirsch R, Houweling AC, Howard LS, Ivy D, Kiely DG, Klinger J, Kovacs G, Lahm T, Laudes M, Machado RD, MacKenzie Ross RV, Marsolo K, Martin LJ, Moledina S, Montani D, Nathan SD, Newnham M, Olschewski A, Olschewski H, Oudiz RJ, Ouwehand WH, Peacock AJ, Pepke-Zaba J, Rehman Z, Robbins I, Roden DM, Rosenzweig EB, Saydain G, Scelsi L, Schilz R, Seeger W, Shaffer CM, Simms RW, Simon M, Sitbon O, Suntharalingam J, Tang H, Tchourbanov AY, Thenappan T, Torres F, Toshner MR, Treacy CM, Vonk Noordegraaf A, Waisfisz Q, Walsworth AK, Walter RE, Wharton J, White RJ, Wilt J, Wort SJ, Yung D, Lawrie A, Humbert M, Soubrier F, Trégouët DA, Prokopenko I, Kittles R, Gräf S, Nichols WC, Trembath RC, Desai AA, Morrell NW, Wilkins MR UK NIHR BioResource Rare Diseases Consortium; UK PAH Cohort Study Consortium; US PAH Biobank Consortium. Genetic determinants of risk in pulmonary arterial hypertension: international genome-wide association studies and meta-analysis. Lancet Respir Med. 2019;7:227–238. doi: 10.1016/S2213-2600(18)30409-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang JM, Shi N, Dong K, Brown SA, Coleman AE, Boegehold MA, Chen SY. Response gene to complement 32 maintains blood pressure homeostasis by regulating Α-adrenergic receptor expression. Circ Res. 2018;123:1080–1090. doi: 10.1161/CIRCRESAHA.118.313266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stefan N, Stumvoll M. Adiponectin-its role in metabolism and beyond. Horm Metab Res. 2002;34:469–474. doi: 10.1055/s-2002-34785. [DOI] [PubMed] [Google Scholar]
- 7.Liu M, Liu F. Transcriptional and post-translational regulation of adiponectin. Biochem J. 2010;425:41–52. doi: 10.1042/BJ20091045. [DOI] [PubMed] [Google Scholar]
- 8.Ouchi N, Kihara S, Arita Y, Maeda K, Kuriyama H, Okamoto Y, Hotta K, Nishida M, Takahashi M, Nakamura T, Yamashita S, Funahashi T, Matsuzawa Y. Novel modulator for endothelial adhesion molecules: adipocyte-derived plasma protein adiponectin. Circulation. 1999;100:2473–2476. doi: 10.1161/01.cir.100.25.2473. [DOI] [PubMed] [Google Scholar]
- 9.Iwashima Y, Katsuya T, Ishikawa K, Ouchi N, Ohishi M, Sugimoto K, Fu Y, Motone M, Yamamoto K, Matsuo A, Ohashi K, Kihara S, Funahashi T, Rakugi H, Matsuzawa Y, Ogihara T. Hypoadiponectinemia is an independent risk factor for hypertension. Hypertension. 2004;43:1318–1323. doi: 10.1161/01.HYP.0000129281.03801.4b. [DOI] [PubMed] [Google Scholar]
- 10.Ronconi V, Turchi F, Rilli S, Di Mattia D, Agostinelli L, Boscaro M, Giacchetti G. Metabolic syndrome in primary aldosteronism and essential hypertension: relationship to adiponectin gene variants. Nutr Metab Cardiovasc Dis. 2010;20:93–100. doi: 10.1016/j.numecd.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 11.Suriyaprom K, Phonrat B, Namjuntra P, Harnroongroj T, Tungtrongchitr R. The -11377C>G adiponectin gene polymorphism alters the adiponectin concentration and the susceptibility to type 2 diabetes in thais. Int J Vitam Nutr Res. 2010;80:216–24. doi: 10.1024/0300-9831/a000020. [DOI] [PubMed] [Google Scholar]
- 12.Yan WL, Chen SF, Huang JF, Shen Y, Qiang BQ, Liu DH, Gu DF. Common SNPs of APM1 gene are not associated with hypertension or obesity in Chinese population. Biomed Environ Sci. 2006;19:179–84. [PubMed] [Google Scholar]
- 13.Tang X, Li J, Wu L, Zhuang M, Lu Z. Relationship between adiponectin+ 45 nucleotide T/G polymorphism and essential hypertension. Tianjin Medical Journal. 2008:762–764. [Google Scholar]
- 14.Wang Z, Xia B, Jiang L, Hu Z, Cao P. Association between adiponectin gene polymorphisms and hypertension. Journal of New Medicine. 2008:25–26. [Google Scholar]
- 15.Zhang M, Peng Y, Lyu SZ. Association between genetic variants in the adiponectin gene and premature myocardial infarction. Zhonghua Xin Xue Guan Bing Za Zhi. 2016;44:577–582. doi: 10.3760/cma.j.issn.0253-3758.2016.07.005. [DOI] [PubMed] [Google Scholar]
- 16.Adamczak M, Wiecek A, Funahashi T, Chudek J, Kokot F, Matsuzawa Y. Decreased plasma adiponectin concentration in patients with essential hypertension. Am J Hypertens. 2003;16:72–75. doi: 10.1016/s0895-7061(02)03197-7. [DOI] [PubMed] [Google Scholar]
- 17.Mangge H, Almer G, Truschnig-Wilders M, Schmidt A, Gasser R, Fuchs D. Inflammation, adiponectin, obesity and cardiovascular risk. Curr Med Chem. 2010;17:4511–4520. doi: 10.2174/092986710794183006. [DOI] [PubMed] [Google Scholar]
- 18.Gu C, Qu Y, Zhang G, Sun L, Zhu Y, Ye D. A single nucleotide polymorphism in ADIPOQ predicts biochemical recurrence after radical prostatectomy in localized prostate cancer. Oncotarget. 2015;6:32205–11. doi: 10.18632/oncotarget.4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wells G. The Newcastle-Ottawa Scale (NOS) for assessing the quality of non randomised studies in meta-analyses. http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp 2001.
- 20.Zhang ZB, Yu L, Yang KJ, Xu LW, Sheng T, Hao P, Wang YP, Meng F. Association between single nucleotide polymorphisms (SNPs) at the promoter of adiponectin gene and essential hypertension in Chinese Korean and Han of Yanbian region. Yi Chuan. 2011;33:54–59. doi: 10.3724/sp.j.1005.2011.00054. [DOI] [PubMed] [Google Scholar]
- 21.Leu HB, Chung CM, Lin SJ, Jong YS, Pan WH, Chen JW. Adiponectin gene polymorphism is selectively associated with the concomitant presence of metabolic syndrome and essential hypertension. PLoS One. 2011;6:e19999. doi: 10.1371/journal.pone.0019999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang Z, Zhao HZ, Heng SU, Zhang Y, Yan-Fang YU, Yang OU, Xue YM, Endocrinology DO. Genetic association analysis of variation in the adiponectin gene promoter with essential hypertension in Dali Bai populations. Chinese Journal of Clinicians. 2013 [Google Scholar]
- 23.Jiang B, Liu Y, Liu Y, Fang F, Wang X, Li B. Association of four insulin resistance genes with type 2 diabetes mellitus and hypertension in the Chinese Han population. Mol Biol Rep. 2014;41:925–933. doi: 10.1007/s11033-013-2937-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Machado JS, Palei AC, Amaral LM, Bueno AC, Antonini RS, Duarte G, Tanus-Santos JE, Sandrim VC, Cavalli RC. Polymorphisms of the adiponectin gene in gestational hypertension and pre-eclampsia. J Hum Hypertens. 2014;28:128–32. doi: 10.1038/jhh.2013.53. [DOI] [PubMed] [Google Scholar]
- 25.Xi B, He D, Wang Q, Xue J, Liu M, Li J. Common polymorphisms (rs2241766 and rs1501299) in the ADIPOQ gene are not associated with hypertension susceptibility among the Chinese. Mol Biol Rep. 2012;39:8771–8775. doi: 10.1007/s11033-012-1739-0. [DOI] [PubMed] [Google Scholar]
- 26.Zhao T, Zhao J. Genetic effects of adiponectin on blood lipids and blood pressure. Clin Endocrinol (Oxf) 2011;74:214–222. doi: 10.1111/j.1365-2265.2010.03902.x. [DOI] [PubMed] [Google Scholar]
- 27.Ong KL, Li M, Tso AW, Xu A, Cherny SS, Sham PC, Tse HF, Lam TH, Cheung BM, Lam KS Investigators of the Hong Kong Cardiovascular Risk Factor Prevalence Study. Association of genetic variants in the adiponectin gene with adiponectin level and hypertension in Hong Kong Chinese. Eur J Endocrinol. 2010;163:251–257. doi: 10.1530/EJE-10-0251. [DOI] [PubMed] [Google Scholar]
- 28.Mousavinasab F, Tähtinen T, Jokelainen J, Koskela P, Vanhala M, Oikarinen J, Keinänen-Kiukaanniemi S, Laakso M. Common polymorphisms (single-nucleotide polymorphisms SNP+ 45 and SNP+ 276) of the adiponectin gene regulate serum adiponectin concentrations and blood pressure in young finnish men. Mol Genet Metab. 2006;87:147–151. doi: 10.1016/j.ymgme.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 29.Avery P, Patel S, Ibrahim I, Walker M, Keavney B. Common variation in the adiponectin gene has an effect on systolic blood pressure. J Hum Hypertens. 2011;25:719–24. doi: 10.1038/jhh.2010.122. [DOI] [PubMed] [Google Scholar]
- 30.Demir AK, Kaya SU, Şahin Ş, Benli İ, Bütün İ, Erken E, Tasliyurt T. Single nucleotide polymorphism of adiponectin+ 276 G/T is associated with the susceptibility to essential hypertension in a Turkish population. Clin Exp Hypertens. 2016;38:686–690. doi: 10.1080/10641963.2016.1200607. [DOI] [PubMed] [Google Scholar]
- 31.Hara K, Boutin P, Mori Y, Tobe K, Dina C, Yasuda K, Yamauchi T, Otabe S, Okada T, Eto K, Kadowaki H, Hagura R, Akanuma Y, Yazaki Y, Nagai R, Taniyama M, Matsubara K, Yoda M, Nakano Y, Tomita M, Kimura S, Ito C, Froguel P, Kadowaki T. Genetic variation in the gene encoding adiponectin is associated with an increased risk of type 2 diabetes in the Japanese population. Diabetes. 2002;51:536–540. doi: 10.2337/diabetes.51.2.536. [DOI] [PubMed] [Google Scholar]
- 32.Asferg C, Møgelvang R, Flyvbjerg A, Frystyk J, Jensen JS, Marott JL, Appleyard M, Jensen GB, Jeppesen J. Leptin, not adiponectin, predicts hypertension in the Copenhagen city heart study. Am J Hypertens. 2010;23:327–333. doi: 10.1038/ajh.2009.244. [DOI] [PubMed] [Google Scholar]
- 33.Patel JV, Lim HS, Dubb K, Hughes EA, Lip GY. Circulating levels of adiponectin, leptin, and tumour necrosis factor α in hypertension. Ann Med. 2009;41:291–300. doi: 10.1080/07853890802672839. [DOI] [PubMed] [Google Scholar]
- 34.International Consortium for Blood Pressure Genome-Wide Association Studies; Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, Smith AV, Tobin MD, Verwoert GC, Hwang SJ, Pihur V, Vollenweider P, O’Reilly PF, Amin N, Bragg-Gresham JL, Teumer A, Glazer NL, Launer L, Zhao JH, Aulchenko Y, Heath S, Sõber S, Parsa A, Luan J, Arora P, Dehghan A, Zhang F, Lucas G, Hicks AA, Jackson AU, Peden JF, Tanaka T, Wild SH, Rudan I, Igl W, Milaneschi Y, Parker AN, Fava C, Chambers JC, Fox ER, Kumari M, Go MJ, van der Harst P, Kao WH, Sjögren M, Vinay DG, Alexander M, Tabara Y, Shaw-Hawkins S, Whincup PH, Liu Y, Shi G, Kuusisto J, Tayo B, Seielstad M, Sim X, Nguyen KD, Lehtimäki T, Matullo G, Wu Y, Gaunt TR, Onland-Moret NC, Cooper MN, Platou CG, Org E, Hardy R, Dahgam S, Palmen J, Vitart V, Braund PS, Kuznetsova T, Uiterwaal CS, Adeyemo A, Palmas W, Campbell H, Ludwig B, Tomaszewski M, Tzoulaki I, Palmer ND CARDIoGRAM consortium; CKDGen Consortium; KidneyGen Consortium; EchoGen consortium; CHARGE-HF consortium. Aspelund T, Garcia M, Chang YP, O’Connell JR, Steinle NI, Grobbee DE, Arking DE, Kardia SL, Morrison AC, Hernandez D, Najjar S, McArdle WL, Hadley D, Brown MJ, Connell JM, Hingorani AD, Day IN, Lawlor DA, Beilby JP, Lawrence RW, Clarke R, Hopewell JC, Ongen H, Dreisbach AW, Li Y, Young JH, Bis JC, Kähönen M, Viikari J, Adair LS, Lee NR, Chen MH, Olden M, Pattaro C, Bolton JA, Köttgen A, Bergmann S, Mooser V, Chaturvedi N, Frayling TM, Islam M, Jafar TH, Erdmann J, Kulkarni SR, Bornstein SR, Grässler J, Groop L, Voight BF, Kettunen J, Howard P, Taylor A, Guarrera S, Ricceri F, Emilsson V, Plump A, Barroso I, Khaw KT, Weder AB, Hunt SC, Sun YV, Bergman RN, Collins FS, Bonnycastle LL, Scott LJ, Stringham HM, Peltonen L, Perola M, Vartiainen E, Brand SM, Staessen JA, Wang TJ, Burton PR, Soler Artigas M, Dong Y, Snieder H, Wang X, Zhu H, Lohman KK, Rudock ME, Heckbert SR, Smith NL, Wiggins KL, Doumatey A, Shriner D, Veldre G, Viigimaa M, Kinra S, Prabhakaran D, Tripathy V, Langefeld CD, Rosengren A, Thelle DS, Corsi AM, Singleton A, Forrester T, Hilton G, McKenzie CA, Salako T, Iwai N, Kita Y, Ogihara T, Ohkubo T, Okamura T, Ueshima H, Umemura S, Eyheramendy S, Meitinger T, Wichmann HE, Cho YS, Kim HL, Lee JY, Scott J, Sehmi JS, Zhang W, Hedblad B, Nilsson P, Smith GD, Wong A, Narisu N, Stančáková A, Raffel LJ, Yao J, Kathiresan S, O’Donnell CJ, Schwartz SM, Ikram MA, Longstreth WT Jr, Mosley TH, Seshadri S, Shrine NR, Wain LV, Morken MA, Swift AJ, Laitinen J, Prokopenko I, Zitting P, Cooper JA, Humphries SE, Danesh J, Rasheed A, Goel A, Hamsten A, Watkins H, Bakker SJ, van Gilst WH, Janipalli CS, Mani KR, Yajnik CS, Hofman A, Mattace-Raso FU, Oostra BA, Demirkan A, Isaacs A, Rivadeneira F, Lakatta EG, Orru M, Scuteri A, Ala-Korpela M, Kangas AJ, Lyytikäinen LP, Soininen P, Tukiainen T, Würtz P, Ong RT, Dörr M, Kroemer HK, Völker U, Völzke H, Galan P, Hercberg S, Lathrop M, Zelenika D, Deloukas P, Mangino M, Spector TD, Zhai G, Meschia JF, Nalls MA, Sharma P, Terzic J, Kumar MV, Denniff M, Zukowska-Szczechowska E, Wagenknecht LE, Fowkes FG, Charchar FJ, Schwarz PE, Hayward C, Guo X, Rotimi C, Bots ML, Brand E, Samani NJ, Polasek O, Talmud PJ, Nyberg F, Kuh D, Laan M, Hveem K, Palmer LJ, van der Schouw YT, Casas JP, Mohlke KL, Vineis P, Raitakari O, Ganesh SK, Wong TY, Tai ES, Cooper RS, Laakso M, Rao DC, Harris TB, Morris RW, Dominiczak AF, Kivimaki M, Marmot MG, Miki T, Saleheen D, Chandak GR, Coresh J, Navis G, Salomaa V, Han BG, Zhu X, Kooner JS, Melander O, Ridker PM, Bandinelli S, Gyllensten UB, Wright AF, Wilson JF, Ferrucci L, Farrall M, Tuomilehto J, Pramstaller PP, Elosua R, Soranzo N, Sijbrands EJ, Altshuler D, Loos RJ, Shuldiner AR, Gieger C, Meneton P, Uitterlinden AG, Wareham NJ, Gudnason V, Rotter JI, Rettig R, Uda M, Strachan DP, Witteman JC, Hartikainen AL, Beckmann JS, Boerwinkle E, Vasan RS, Boehnke M, Larson MG, Järvelin MR, Psaty BM, Abecasis GR, Chakravarti A, Elliott P, van Duijn CM, Newton-Cheh C, Levy D, Caulfield MJ, Johnson T. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–9. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeng JR. Plasma adiponectin, T94G gene polymorphism and PAI-1 in patients with and without hypertension. Cardiology. 2007;107:30–37. doi: 10.1159/000093610. [DOI] [PubMed] [Google Scholar]
- 36.Youpeng B, Wei X, Wei L, Jin J, Haiyan Y, Yuan Y, Rong Z. Relationships among adiponectin gene polymorphisms, proteinuria and increased blood pressure in the context of placental diseases. Hypertens Res. 2010;33:1066–70. doi: 10.1038/hr.2010.134. [DOI] [PubMed] [Google Scholar]
- 37.Jia M, Qiao BM, Zheng Z. Study on correlation of genetic polymorphism of adiponectin promoter and adiponectin levelwith hypertension and its complications. Chinese Journal of Laboratory Diagnosis. 2008;2:222–226. [Google Scholar]


