Abstract
Chromatin remodeling complexes help regulate the structure of chromatin to facilitate transcription. The multisubunit human (h) SWI-SNF complex has been shown to remodel mono- and polynucleosome templates in an ATP-dependent manner. The isolated hSWI-SNF ATPase subunits BRG1 and hBRM also have these activities. The intact complex has been shown to produce a stable remodeled dimer of mononucleosomes as a product. Here we show that the hSWI-SNF ATPases alone can also produce this product. In addition, we show that hSWI-SNF and its ATPases have the ability to transfer histone octamers from donor nucleosomes to acceptor DNA. These two reactions are characterized and compared. Our results are consistent with both products of SWI-SNF action being formed as alternative outcomes of a single remodeling mechanism. The ability of the isolated ATPase subunits to catalyze these reactions suggests that these subunits play a key role in determining the mechanistic capabilities of the SWI-SNF family of remodeling complexes.
Chromatin acts as a barrier to eukaryotic transcription by blocking transcription factor access and polymerase movement. To contend with this barrier, cells make use of a variety of evolutionarily conserved ATP-dependent chromatin remodeling complexes. These complexes have been shown to influence transcription and chromatin access in vivo and in vitro, but the extent of their roles and their mechanisms of action are only beginning to be understood (15, 17, 40, 43).
Human cells contain a family of SWI-SNF complexes that are closely related to the yeast (y) SWI-SNF and RSC complexes. Human (h) SWI-SNF has been implicated in transcriptional activation of several genes (1, 6, 10, 16, 20, 25), as well as in transcriptional repression and growth control through the p105Rb retinoblastoma protein (9, 26, 34, 38). To determine the function of SWI-SNF in gene regulation, we have examined its effects on chromatin in vitro. hSWI-SNF contains at least eight subunits and can be isolated in two forms, which contain either BRG1 or hBRM as a central ATPase subunit (18, 41). These complexes possess DNA- and nucleosome-stimulated ATPase activity and have an ATP-dependent ability to remodel mononucleosome core particles (referred to as nucleosomes or cores herein) (as assayed by changes in DNase I digestion patterns) and plasmid chromatin (as assayed by a reduction in nucleosome-constrained negative supercoils) (18). Each ATPase alone, when purified from insect cells, appears capable of these activities, although at reduced levels (28). Addition of three other conserved subunits—namely BAF155, BAF170, and Ini1—to the ATPase subunit generates a minimal hSWI-SNF complex that is almost as active as the intact complex.
While ATP is required for remodeling by intact hSWI-SNF, the remodeled state is stable in the absence of ATP (13) or SWI-SNF in several in vitro assays. These observations led to the discovery that SWI-SNF can act enzymatically to create a stable, altered dimer of mononucleosome cores (30). This product, which we refer to here as the remodeled dimer, could be separated from hSWI-SNF and was shown to have the proper ratio of the four core histones and DNA but an apparent molecular weight twice that of a normal nucleosome. It was determined to have distinct DNase and micrococcal nuclease digestion patterns and altered susceptibility to restriction enzyme digestion. Increased affinity of GAL4 for this product suggested that it might be more amenable to transcriptional activation. The hSWI-SNF complex also converts this product back to cores, and both the creation of the remodeled dimer and reconversion to cores requires ATP hydrolysis. In general, these activities and properties are shared by hSWI-SNF and the related yeast complexes. Notably, yeast RSC generates and reconverts a highly similar stable product (22), and the remodeling of mononucleosomes by ySWI-SNF is stable after ySWI-SNF removal (8).
Many different chromatin remodeling complexes have been identified to date. Some can be classified as SWI-SNF-like, by virtue of their subunit composition and the similarity of their ATPases to ySWI2-SNF2 (for a review, see reference 17). A second major family contains ISWI or a related protein as its central ATPase (for reviews, see references 11 and 40). The complexes from this ISWI-based family are unlike SWI-SNF-like complexes in that their ATPase activity is stimulated primarily by nucleosomes and not by DNA alone. Nevertheless, ISWI is similar to BRG1 and hBRM in that it can also perform at least some of the in vitro activities of the complex in which it is found (7, 19). A third family, which includes the nucleosome remodeling and deacetylase (NuRD) complexes, contains a CHD/Mi-2 protein(s) as the central ATPase(s) (for a review, see reference 2). All of these complexes have been shown to remodel chromatin in vitro in some of the many available assays. An exhaustive comparison across all assays has not been done, but current data indicate that each remodeling complex can perform only a subset of known remodeling activities (see, e.g., references 17 and 40 for reviews). These activities include those described above, as well as the abilities to disorder arrays of nucleosomes, space nucleosomes evenly along an array, reposition individual nucleosomes, increase restriction enzyme access to nucleosomes, increase transcriptional initiation or elongation, and transfer histone octamers to acceptor DNAs in trans. It is not known how these activities are related at a mechanistic level, nor is it known which components of the complexes are required for each specialized function.
To begin to address these questions, we have examined two activities of the hSWI-SNF complex: the ability to form a remodeled nucleosome dimer (30) and the ability to transfer histone octamers in trans (this report). Recently, the ySWI-SNF and RSC complexes have been shown to possess the ability to transfer histone octamers to acceptor DNAs in trans under certain conditions (23, 42). Here we show that hSWI-SNF has a similar activity. Analysis of this activity and comparison of it to the ability to form remodeled dimers suggest that both products may be formed through a shared mechanism. To address the protein subunit requirements for these reactions, we tested isolated BRG1 and hBRM. Each protein also possesses both activities, suggesting that the enzymatic capabilities of the complex are largely determined by its ATPase subunits.
MATERIALS AND METHODS
Protein purification.
hSWI-SNF was purified from a HeLa cell line overexpressing the FLAG-tagged Ini1 subunit of the complex, using an M2 anti-FLAG affinity resin (Sigma) as described previously (33). The purity of these fractions was ∼70%, as estimated from silver-stained gels. Molarity estimations assumed this level of purity and a molecular mass of 2 MDa. There were trace amounts (<1%, as estimated from silver-stained gels) of histones evident in the SWI-SNF preparation, which might have been in the form of nucleosomes. The FLAG epitope-tagged hSWI-SNF ATPases BRG1 and hBRM were expressed and purified from Sf9 cells by using a baculovirus system as described previously (28). These proteins were ∼80% pure as determined by silver staining. Molarity estimations assumed this level of purity and a monomeric molecular mass of 190 kDa. For the experiments presented in Fig. 3, BRG1 was further purified in monomeric form by glycerol gradient centrifugation as follows. To promote self-dissociation, immunopurified BRG1 was incubated on ice in BC buffer (20 mM HEPES [pH 7.9], 10% glycerol, 0.2 mM EDTA, 1 mM dithiothreitol [DTT], 0.2 mM phenylmethylsulfonyl fluoride [PMSF]) containing 1 M urea and approximately 1 M NaCl or KCl for 3.5 h; then it was diluted with BC buffer containing no KCl and no glycerol to reduce the density of the solution, immediately layered onto a 10 to 40% glycerol gradient (50 mM Tris [pH 8.0], 0.1 M urea, 0.1 M KCl, 0.1% NP-40, 1 mM DTT, 1 mM EDTA, 0.2 mM PMSF), and centrifuged for 17 h at 35,000 rpm (∼150,000 × g) and 4°C in an SW55 rotor (Beckman). Twenty-two 6-drop fractions were collected from the bottom of the gradient by the use of a butterfly needle. For Fig. 3A, 50 μg of a BRG1 sample was incubated in 158 μl of BC buffer containing 1 M urea, 1 M NaCl, 95 mM KCl, and 3% glycerol. The incubation mixture was then diluted 1.9-fold, and 200 μl of this dilution (containing about 34 μg of BRG1) was layered onto the glycerol gradient. For Fig. 3B, approximately 7.5 μg of BRG1 was incubated in 50 μl of BC buffer containing 1 M urea, 930 mM NaCl, 65 mM KCl, and 13% glycerol. The incubation mixture was then diluted fourfold, and 190 μl of this was layered onto the glycerol gradient.
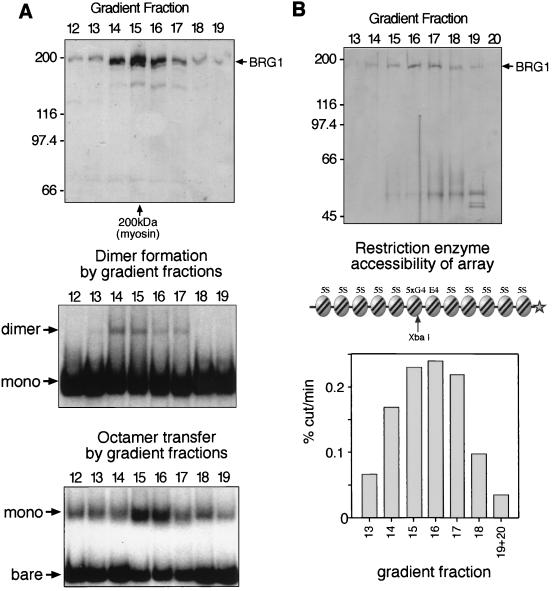
FIG. 3.
Remodeling activities coelute with monomeric BRG1. (A) Coelution of octamer transfer and remodeled-dimer activities with monomeric BRG1. BRG1 incubated in a solution containing 1 M urea, 1 M NaCl, and 95 mM KCl was centrifuged through a 10 to 40% glycerol gradient (see Materials and Methods). Fractions were analyzed by silver staining following sodium dodecyl sulfate-PAGE (top panel; 16.7 μl of each peak fraction). The relative position of myosin (200 kDa) centrifuged under identical conditions is indicated (below), as are the positions of molecular mass markers in the gel (left; in kilodaltons). The ∼150-kDa band comigrates with a BRG1 breakdown product identified by Western blotting (data not shown). The BRG1-containing fractions were analyzed for their ability to convert mononucleosome cores to a dimeric form (middle panel; EMSA) and to transfer octamers from unlabeled genomic HeLa mononucleosomes to labeled TPT DNA (bottom panel; EMSA). These reactions were performed under standard conditions, except that the final reaction conditions were 33 mM KCl, 0.3% NP-40, and 11 mM (dimer formation) or 33 mM (octamer transfer) urea. Dimer formation reactions contained 2% gradient fraction and 240 pM TPT mononucleosomes, while octamer transfer reactions contained 20% gradient fraction, 120 pM bare TPT, and 24 nM HeLa cell mononucleosomes (mono). The concentration of BRG1 in these reactions was estimated, from the silver stained gel, to range from 0.5 to 3 nM (dimer formation) or from 5 to 25 nM (octamer transfer). (B) Remodeling activity correlates with BRG1 in a quantitative assay. BRG1 incubated in a solution containing 1 M urea, 930 mM NaCl, and 65 mM KCl was centrifuged through a 10 to 40% glycerol gradient (see Materials and Methods) and analyzed by silver staining following sodium dodecyl sulfate-PAGE (top panel; 20 μl of each fraction). The remodeling activity of these fractions was assayed by measuring changes in the rate of restriction enzyme cutting of a nucleosomal array (shown; see Materials and Methods for details). The rate of cutting in the presence of 15 μl of each gradient fraction (or a 1:1 mixture of fractions 19 and 20) in a total of 25 μl is shown in the bottom panel. The concentration of BRG1 in these reactions was estimated to range from 1 to 5 nM.
Bulk HeLa cell polynucleosomes were purified as described previously (39). Mono- and dinucleosomes were created from these polynucleosomes by redigestion with MNase and purification by centrifugation at 150,000 × g on a 10 to 30% glycerol gradient as described previously (30), except that the glycerol gradient buffer (referred to hereafter as GGB) contained 20 mM HEPES (pH 7.9), 1 mM EDTA, 0.1% NP-40, and 30 mM KCl. Nucleosome concentrations (given as DNA concentrations) were determined by measuring the A260 in 2 M NaCl or by comparison in ethidium bromide-stained gels to nucleosomes whose concentration had been determined by measuring their A260.
Nucleosome assembly and labeling.
The 155-bp TPT MluI-EcoRI fragment (which contains two 20-bp nucleosome phasing sequences at the MluI end [32]) was used to assemble nucleosomes. It was purified, assembled into nucleosomes, and labeled on the MluI site end with [32P]dCTP as described previously (30). For nonlabeled TPT nucleosomes, the full-length 155-bp TPT fragment was amplified by PCR and purified by 5% polyacrylamide gel electrophoresis (PAGE) followed by elution by soaking of the excised band and ethanol precipitation. Mononucleosomes were assembled as described above or by gradient dialysis (29), using histone/DNA ratios of 0.9:1 or 1.6:1 (Geeta Narlikar, G. R. Schnitzler, and R. E. Kingston, unpublished data). They were purified by centrifugation in 10 to 30% glycerol gradients prepared with GGB or 50 mM Tris (pH 7.5)–1 mM EDTA–100 mM KCl, and concentrations were determined as described above. Bulk HeLa cell mononucleosomes (0.25 μg) were labeled by using T4 polynucleotide kinase (10 U) and [γ-32P]ATP for 30 min at 30°C and 15 min at 42°C in 26 μl of 10% gradient buffer with 7 mM MgCl2. Unreacted label was removed with a G50 TE spin column. For random-sequence mononucleosome DNA, an identical reaction product was extracted with neutralized phenol before the spin column step.
Remodeling reactions.
Experiments involving interconversion of mononucleosome cores and remodeled dimer species were performed as described previously (30) except where noted in the figure legends. In general, dimer formation and octamer transfer reactions were performed at 30°C in 25-μl reaction volumes containing in a solution containing 16 mM HEPES (pH 7.9), 10 mM Tris (pH 8.0), 60 mM KCl, and 5 mM MgCl2, with or without 2 mM ATP-MgCl2, 7 to 10% glycerol, 0.1% NP-40, 20 μg of bovine serum albumin (BSA)/ml, 0.16 mM EDTA, 0.8 mM DTT, and 0.16 mM PMSF. Significant variations on these conditions are noted in the figure legends. We have found that these reactions occur efficiently in the following solutes concentration ranges: 2 to 5 mM MgCl2, 0.1 to 2 mM ATP-MgCl2, 20 to 70 mM KCl or NaCl, and up to 0.1% NP-40 (13, 28, 30) (this report and data not shown). For gel shift analysis, plasmid DNA or KCl was added at the concentrations indicated in the figure legends to disrupt SWI-SNF interactions with the templates and allow products to be resolved by 5% polyacrylamide–0.5× Tris-borate-EDTA electrophoresis at 4°C (2 h, 200 V). For footprinting analysis, reaction mixtures were treated with DNase for 2 min at room temp and analyzed as described previously (13). Variations are detailed in the figure legends. Quantitation was performed with a PhosphorImager (Molecular Dynamics).
Purification and analysis of reaction products.
Scaled-up remodeling or transfer reactions were performed with reduced proportions of glycerol (to allow layering on the gradient). These reactions were stopped by addition of plasmid DNA (which binds SWI-SNF and its ATPases, competing them off of test templates) and separated on glycerol-GGB gradients. The large size of the plasmid DNA caused it to pellet, removing SWI-SNF proteins from nucleosome-containing gradient fractions. Alternatively, reactions were stopped by addition of KCl to 230 mM (which disrupts SWI-SNF–nucleosome interactions [30]) and purified on glycerol-GGB gradients containing 180 mM KCl. Western blot analysis showed that there was some contamination of salt-stopped remodeled dimer fractions with SWI-SNF complexes. These complexes, however, were inactive for remodeling when ATP was added.
Restriction enzyme assay for remodeling activity.
End-labeled nucleosomal array 5S-G5E4 (27) (a gift from J. L. Workman) was formed by salt dialysis as described previously (24), with the following modifications. DNA and purified HeLa cell core histones were mixed at about a 1:1 ratio at a final DNA concentration of ∼0.1 mg/ml in a 100-μl volume containing 20 mM Tris (pH 7.8), 2 M NaCl, 10 mM DTT, 1 mM EDTA, 0.5 mM benzamidine-HCl, and 0.1 mg of BSA/ml. This mixture was transferred to dialysis tubing (SpectraPor) with a 6- to 8-kDa molecular mass cutoff and dialyzed against 200 ml of a solution containing 20 mM Tris (pH 7.7), 2 M NaCl, 1 mM DTT, 1 mM EDTA, and 0.5 mM benzamidine-HCl. The dialysis buffer was exchanged for a similar buffer containing 0.25 M NaCl at a rate of 12 to 13 ml/h over 48 h, and then the assembly was further dialyzed for 12 h against Tris-EDTA buffer. Assembly was verified by electrophoresis on a Tris-acetate-EDTA–1% polyacrylamide gel. To assay remodeling, 3.35 ng of this nucleosomal array was incubated in a total volume of 25 μl containing 15 μl of gradient buffer or BRG1-containing glycerol gradient fractions, 20 U of XbaI (under standard conditions but with 3.5 mM MgCl2, 0.5 mM ATP, and 60 mM urea) at 30°C. Samples were removed at three time points, deproteinized, and resolved on a Tris-acetate-EDTA–1% agarose gel to determine the extent of XbaI digestion.
RESULTS
Creation of a stably remodeled mononucleosome dimer by the ATPase subunits.
The hSWI-SNF complex, immunopurified from a HeLa cell line expressing a FLAG-tagged INI1 subunit, can generate a stable remodeled dimer from mononucleosome cores (30). To determine whether the individual ATPase subunits are also capable of creating this remodeled dimer, we examined the activity of FLAG-tagged versions of BRG1 and hBRM that had been overexpressed and immunopurified from baculovirus-infected Sf9 cells. Such preparations of BRG1 or hBRM have been shown to remodel both mononucleosomes and nucleosomal arrays (28).
Isolated BRG1 and hBRM ATPases were incubated with mononucleosomes and ATP, and the resultant reaction products were separated on a glycerol gradient. Analysis of the input to the gradient by mobility shift gel assay demonstrated that BRG1 and hBRM created a species that comigrated with the stable dimer created by intact SWI-SNF (Fig. 1A, compare lanes 1, 4, and 7). Separation of the reaction products on a gradient demonstrated that the remodeled dimer created by the isolated subunits migrated in a peak separate from that containing mononucleosomes and at the same position as the remodeled dimer created by SWI-SNF (Fig. 1A, compare lanes 2 and 3 with lanes 5 and 6 and lanes 8 and 9). The ability of BRG1 and hBRM to create these species was dependent on the presence of ATP (data not shown).
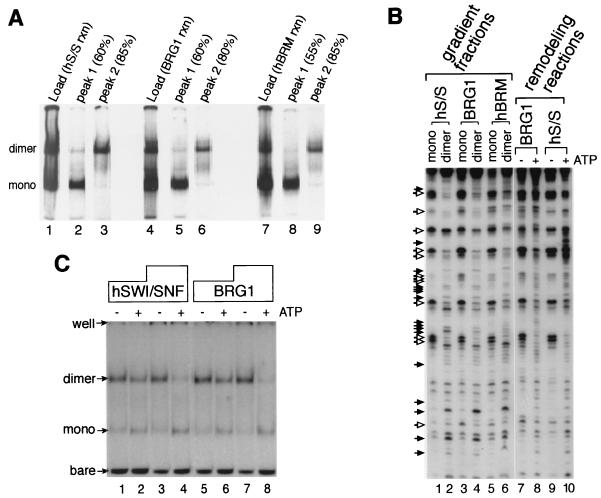
FIG. 1.
BRG1 and hBRM are capable of interconverting mononucleosome cores between a rotationally phased monomeric state and a stably remodeled dimeric state. (A) Creation of remodeled dimer product by hSWI-SNF and isolated ATPases. SWI-SNF (hS/S; 1 nM), 4 nM BRG1, or 12 nM hBRM was incubated with 390 pM labeled TPT cores and 2 nM unlabeled HeLa cell mononucleosomes in 200-μl remodeling reactions under standard conditions (see Materials and Methods), but with 30 mM KCl, 3.5 mM MgCl2, and 0.5 mM ATP, at 30°C for 40 min. The reactions were stopped by addition of KCl to 240 mM, and the products were separated on a 10 to 30% glycerol gradient by centrifugation for 19.5 h. Samples (2 μl) of the reaction products loaded on the gradients (Load) were analyzed by the gel shift method (lanes 1, 4, and 7). Peak gradient fractions (5-μl volumes) containing mononucleosome cores (mono) (peak 1; lanes 2, 5, and 8) and the novel dimer species (peak 2; lanes 3, 6, and 9) were also analyzed by the gel shift method. Percentages indicate the distances of these fractions from the top of the gradient, which is indicative of relative S values. (B) Comparison of the DNase I digestion patterns of mononucleosome dimers formed by BRG1, hBRM, and the hSWI-SNF complex. Aliquots of peak gradient fractions from the gel shown in panel A (0.075 ng of labeled DNA) were incubated with 0.25 ng of unlabeled HeLa cell mononucleosomes/μl under standard conditions, but with 36 mM KCl, 3.5 mM MgCl2, and 0.5 mM ATP, at 30°C for 30 min; this was followed by equilibration to room temperature for 5 min and digestion with 80 U of DNase I/ml for 2 min. DNase digestion was stopped with 200 μl of phenol, and DNA was isolated and separated by denaturing PAGE (lanes 1 to 6). Positions where cutting is increased (closed arrows) or decreased (open arrows) in the dimer species (lanes 2, 4, and 6) are indicated. For comparison, the DNase I patterns of unseparated BRG1 and SWI-SNF remodeling reactions, performed in the presence (+) or absence (−) of ATP, are shown (lanes 7 to 10). (C) BRG1 reconverts remodeled dimer back to mononucleosome cores. Remodeled dimers (4 pM), generated by BRG1 and isolated by glycerol gradient centrifugation (Fig. 1A, lane 6), were incubated with 2.5 nM HeLa cell mononucleosomes and with 250 pM (lanes 1 and 2) or 500 pM (lanes 3 and 4) hSWI-SNF or 4 nM (lanes 5 and 6) or 8 nM (lanes 7 and 8) and BRG1 for 35 min at 30°C in 50-μl standard reaction volumes containing 44 mM KCl and 3.5 mM MgCl2, with (+) or without (−) 0.5 mM ATP. The reactions were stopped by increasing the KCl concentration to 240 mM, and the products were resolved by EMSA. The bare DNA in these lanes resulted from dissociation of the nucleosomes in the dimer-containing gradient fraction following storage at 4 to 8°C for several weeks.
We compared the DNase I digestion pattern of the DNA within each of the isolated forms. Because the DNA template (referred to as TPT) used in these experiments contains a rotational phasing sequence, it displays a distinct 10-bp pattern of accessibility in the absence of remodeling (see, e.g., Fig. 1B, lane 7). Previously we showed that the hSWI-SNF-generated remodeled dimer formed from these mononucleosomes has an altered DNase I digestion pattern, indicative of a stably remodeled species (30) (Fig. 1B, compare lanes 1 and 2). The novel species produced by BRG1 and hBRM display a cutting pattern (Fig. 1B, lanes 4 and 6) similar to that of the hSWI-SNF-generated remodeled dimer and distinct from that of the starting mononucleosomes (Fig. 1B, lanes 3 and 5). This pattern also has many similarities to that of unfractionated reaction mixtures in which mononucleosomes were undergoing active remodeling at the time of DNase I treatment (Fig. 1B, lanes 8 and 10). Thus, the products formed by the hSWI-SNF complex and by the individual ATPase subunits displayed the same mobility in two assays and similar patterns of DNase I accessibility. We conclude that BRG1 and hBRM are capable of creating stably remodeled dimers from mononucleosome cores.
Previously, we also showed that the complete hSWI-SNF complex can convert remodeled dimers back to their original monomeric form in an ATP-dependent reaction, thus establishing a dynamic equilibrium between these two nucleosomal states (30). We directly tested BRG1 and hBRM for the ability to perform the reverse reaction by incubating them with remodeled dimer isolated on a glycerol gradient. Both BRG1 and hBRM were capable of converting these remodeled dimers back to cores in an ATP-dependent reaction (Fig. 1C and data not shown). These results indicate that the ability to create and reconvert this altered nucleosomal species from mononucleosomes is intrinsic to the ATPase subunits of hSWI-SNF complexes and does not require other subunits.
Octamer transfer by the hSWI-SNF complex and its ATPase subunits.
The yeast RSC and SWI-SNF complexes have been shown to be able to transfer histone octamers to DNA in trans or in cis around a barrier (23, 42). Little is known, however, about the mechanism involved or the requirement for different subunits in this reaction. To address these issues, we examined the ability of hSWI-SNF and its ATPases to carry out octamer transfer. Transfer from an excess of unlabeled HeLa cell-derived mononucleosome cores to a radiolabeled 155-bp nucleosomal template, TPT, was measured by the creation of a radiolabeled product with the same mobility as control mononucleosome cores on a native gel (Fig. 2A). In the absence of ATP or remodeler, a small amount of transfer appeared to occur (Fig. 2A, lanes 2 to 4, 6, and 8). Addition of ATP to reaction mixtures containing hSWI-SNF or either of the ATPases (BRG1 or hBRM) resulted in a significant increase in the amount of transfer (Fig. 2A, lanes 5, 7, and 9, and data not shown).
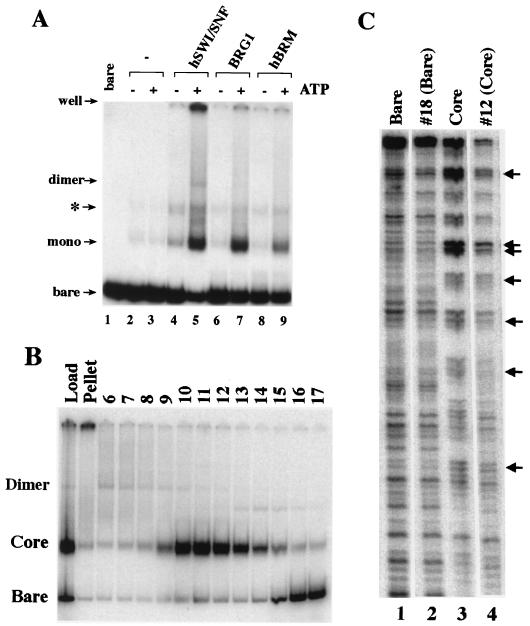
FIG. 2.
hSWI-SNF and ATPases BRG1 and hBRM can transfer histone octamers from nucleosomes to bare DNA. (A) hSWI-SNF, BRG1, and hBRM transfer histone octamers. Labeled bare TPT fragment (120 pM) was incubated for 35 min at 30°C with 24 nM bulk HeLa cell mononucleosomes (mono) in 12.5-μl standard reactions but with 30 mM KCl and 3.5 mM MgCl2 and with (+) or without (−) 0.5 mM ATP. The remodeler concentrations were 5.6 nM hSWI-SNF complex (lanes 4 and 5), 8 nM BRG1 (lanes 6 and 7), and 8 nM hBRM (lanes 8 and 9). Reactions were stopped by addition of 2 μg of plasmid DNA, and the products were separated by EMSA. The product marked with an asterisk is created by SWI-SNF and bare DNA in the absence of nucleosomes. Preliminary results suggest that its formation, which is stimulated by ATP (see, e.g., Fig. 5A), is due to protein binding (J. Guyon and R. E. Kingston, unpublished observations). However, since its formation is actually repressed by the presence of donor nucleosomes, this product was not further analyzed here. (B) The transfer product has the same gradient mobility as a nucleosome core. A scaled-up transfer reaction mixture contained 590 pM bare TPT, 280 ng of HeLa cell polynucleosomes (∼11 nM in nucleosomes), and 4 nM SWI-SNF in a 200-μl volume, and the reaction was performed under standard reaction conditions but with 30 mM KCl, 3.5 mM MgCl2, 1 mM ATP, and no glycerol. After 1 h, the reaction was stopped by addition of 22 μg of a 8-kb plasmid (pKS-BRG1), and the products were purified on a 10 to 30% GGB-glycerol gradient. Fractions were then separated by EMSA. (C) DNase I accessibility of DNA before and after octamer transfer. Gradient-isolated cores (fraction no. 12, lane 4) or bare DNA (fraction no. 18, lane 2) and control cores (lane 3) or DNA (lane 1) (0.3 ng each) were brought up to a 100-μl volume by addition of 10% GGB, adjusted to 5 mM MgCl2 and 40-μg/ml BSA, and digested for 2 min at room temp with DNase (0.03 U for bare DNA; 0.3 U for nucleosomal DNA). The DNase reaction was stopped by addition of EDTA to 15 mM prior to DNA purification and denaturing PAGE. Arrows indicate the 10-bp periodicity of DNase cuts indicative of a rotationally phased nucleosome core.
The primary transfer product migrated at the position of control mononucleosomes in a glycerol gradient (Fig. 2B and data not shown). Treatment of this isolated product with DNase I resulted in a 10-bp digestion pattern characteristic of rotationally phased TPT mononucleosomes (Fig. 2C, compare lanes 3 and 4) and distinct from that of bare TPT DNA (Fig. 2C, lanes 1 and 2). These results indicate that the ability to transfer histone octamers in trans has been conserved between the yeast RSC and SWI-SNF complexes and the hSWI-SNF complex and that, at least for hSWI-SNF, this activity is intrinsic to the ATPase subunits. This reaction is characterized further below.
Isolation of ATPase subunits under denaturing conditions.
The results reported above, combined with previous results from studies of the enzymatic activities of individual hSWI-SNF subunits (28), indicate that the isolated BRG1 and hBRM ATPases are able to perform a variety of remodeling reactions. Despite the fact that these proteins were highly overexpressed in the Sf9 cells prior to purification, there remained the formal possibility that copurifying contaminants from insect cells were partially responsible for the observed activities. From previous work this appeared unlikely; BRG1 and hBRM fractions did not contain significant amounts of additional peptides, and preparations of non-ATPase hSWI-SNF subunits purified from Sf9 cells did not exhibit remodeling activity and thus did not copurify with Sf9 ATPases.
To address this issue further, we fractionated the BRG1 preparation under conditions that inhibited its association with other subunits. Under normal salt conditions (100 mM KCl), most of the immunopurified BRG1 eluted from a gel filtration column in a broad peak from 500 kDa to 2 MDa and pelleted in a 10 to 30% glycerol gradient after 18 h (data not shown). This behavior appears to be due to self-aggregation, since these preparations were up to 90% pure. We found that if BRG1 was first incubated in a solution of 1 M urea plus 1 M salt, it subsequently migrated as an apparent monomer in gel filtration and gradient centrifugation, at the approximate position of the 200-kDa myosin marker run in parallel (Fig. 3, top panels, and data not shown). We tested these fractions for their ability to form the remodeled dimer and to carry out octamer transfer (Fig. 3A, lower panels). Both of these activities coeluted with the protein peak of BRG1. In the experiment shown, the peak fractions for each activity were not identical; however, these assays have not proven to be linear over a wide range of concentrations, and this skewing has not been reproducible.
To better distinguish whether remodeling activity precisely coeluted with the BRG1 protein, we used a sensitive and quantifiable assay which measures changes in the accessibility of restriction enzymes to sites within a nucleosomal array. A similar assay has been shown to be linear with time and enzyme concentration for the ySWI-SNF complex (21). We performed a variation of this assay, measuring XbaI access to a site in the 5S-G5E4 array, which is also linear with time and hSWI-SNF concentration (reference 27 and data not shown) (see diagram in Fig. 3B). The ability of the gradient fractions to increase the rate of digestion of a nucleosomal array correlated with the amount of BRG1 in each fraction, but not with the level of any contaminating protein (compare upper and lower panels of Fig. 3B) (the primary contaminants in these fractions are likely to be keratins). This coelution of activity with monomeric BRG1 in each assay indicates that it is BRG1 alone that is responsible for these remodeling activities, and not BRG1 in association with endogenous SF9 proteins.
Role of the remodeled dimer and ATP hydrolysis in octamer transfer.
It is conceivable that octamer transfer does not proceed directly from nucleosome cores but rather proceeds from the remodeled dimer generated by SWI-SNF. Either the dimer would be the substrate on which SWI-SNF acts to facilitate transfer, or it might spontaneously transfer octamers once formed. To test these possibilities, we compared the abilities of mononucleosome cores and the remodeled dimer to act as donors in the transfer reaction (in the presence or absence of SWI-SNF). hSWI-SNF-remodeled, unlabeled HeLa cell mononucleosome dimers were isolated by glycerol gradient centrifugation and tested for their ability to donate octamers. We did not detect significant octamer transfer from either mononucleosomes or remodeled dimers in the absence of hSWI-SNF (Fig. 4A, lanes 2 and 3). This indicated that transfer from the remodeled dimer did not proceed spontaneously. The inclusion of hSWI-SNF and ATP in the reaction mixture allowed transfer from the remodeled dimer to occur (Fig. 4A, lanes 4 to 6). The rate of this transfer was comparable to the rate of transfer from mononucleosomes (Fig. 4A, lanes 7 to 9). Under these conditions, the rate of dimer formation was not significantly higher than the rate of octamer transfer (data not shown); thus, mononucleosomes need not first be converted to dimers before acting as donors.
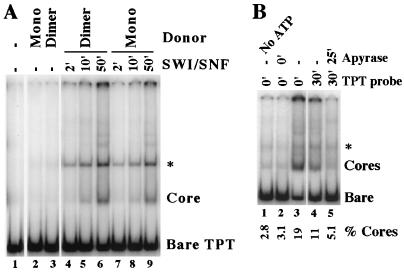
FIG. 4.
Octamer transfer requires an unstable intermediate of the hSWI-SNF remodeling reaction. (A) The remodeled dimer does not support transfer without SWI-SNF. Nonlabeled remodeled dimer was generated by incubating 100 nM HeLa cell mononucleosome cores with 3.9 nM SWI-SNF in a 200-μl volume under standard reaction conditions, but in the absence of glycerol and in the presence of 30 mM KCl and 3.5 mM MgCl2, for 2 h. The reaction was stopped by addition of KCl to 230 mM, and the products were separated on 180 mM–KCl–GGB–glycerol gradients as described in Materials and Methods. Octamer transfer reactions were performed with 80 pM labeled TPT bare DNA and 0.8 nM control mononucleosomes or gradient-purified remodeled dimers under standard conditions but with 0.1 mM ATP-MgCl2, 3 mM MgCl2, and 50 mM KCl. SWI-SNF was added at the onset of the reactions or either 10 or 2 min before the reactions were stopped (at 50 min) with 3 μg plasmid DNA. ∗, donor-independent nonnucleosomal band described in the legend to Fig. 2. (B) Transfer requires continuous ATP hydrolysis. Octamer transfer reactions with 4 nM HeLa cell cores and 120 pM labeled TPT bare DNA and 4.1 nM SWI-SNF were performed under standard conditions but with 2.5 mM MgCl2 and 0.1 mM ATP. Apyrase was added where indicated, and in the last two lanes the labeled TPT bare DNA was added after 30 min. All reactions were stopped with 3 μg of plasmid DNA at 55 min, and products were separated by EMSA. Percentages of cores formed, relative to total counts in the lane, are indicated.
Although efficient transfer from the remodeled dimer does not occur spontaneously, it might proceed from some other, as-yet-unidentified stable product of SWI-SNF. To address this, we did a study to determine whether octamer transfer requires continuous ATP hydrolysis (Fig. 4B). Donor mononucleosomes were incubated with hSWI-SNF and ATP for 25 min; this was followed by removal of ATP by apyrase for 5 min and then addition of labeled acceptor DNA. When the reaction products were separated by electrophoretic mobility shift assay (EMSA) after a total incubation of 55 min, the amount of labeled DNA converted to mononucleosomes (lane 5) was reduced relative to that of an incubated reaction mixture to which no apyrase was added (lane 4) or that resulting when the acceptor DNA was present throughout the incubation with SWI-SNF (lane 3). The amount of transfer was similar to that seen when no ATP was added to the reaction or when apyrase was added at time zero (lanes 1 and 2). A similar result was seen when the reaction was stopped by addition of excess nonhydrolyzable adenosine 5′-O-(3-thiotriphosphate) prior to addition of acceptor DNA (data not shown). These experiments indicate that efficient octamer transfer is dependent on an unstable intermediate or product of the hSWI-SNF reaction that does not last long after ATP removal. Alternatively, it may depend on a stable product of low abundance that requires continued SWI-SNF action to facilitate high levels of transfer.
Donor nucleosome and acceptor DNA requirements for octamer transfer.
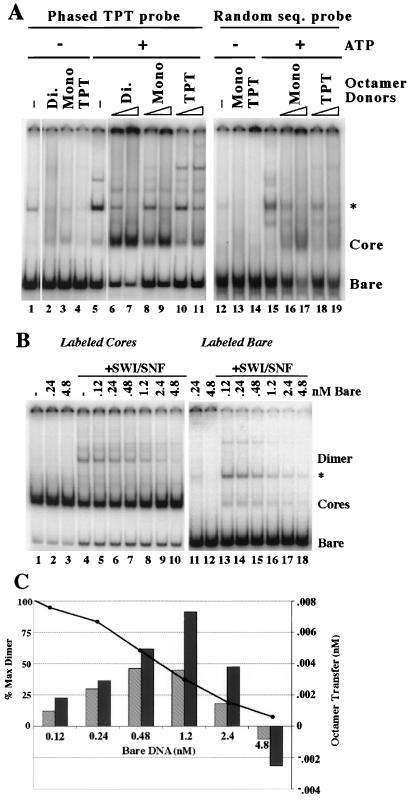
To characterize the generality and significance of octamer transfer, we further examined the donor and acceptor requirements for the reaction. We first examined the effect of using different DNA lengths and sequences as octamer donors and acceptors. hSWI-SNF was able to transfer octamers from HeLa cell mononucleosomes, dinucleosomes, and polynucleosomes to bare acceptor DNA, suggesting that the length of the donor is not important for transfer (Fig. 5A, lanes 6 to 9, and data not shown). In all cases, the percentage of labeled acceptor DNA converted to nucleosomes increased with increasing amounts of donor.
FIG. 5.
Effects of donor nucleosome and acceptor DNA type and concentration on transfer. (A) Donor and acceptor requirements for transfer. Transfer reactions (standard conditions but with 21 mM KCl) were performed with 0.12 nM labeled TPT DNA (left panel) or labeled bulk ∼150-bp DNA purified from HeLa cell mononucleosome cores (right panel), 3.8 nM hSWI-SNF, 2 mM MgCl2 with (+) or without (−) ATP, and 0.6 or 3 nM bulk HeLa cell mononucleosomes (Mono), Dinucleosomes (Di.), or TPT mononucleosomes (TPT) where indicated. At 30 min, 3 μg of plasmid DNA was added, and the reaction products were separated by EMSA. ∗, see the legend to Fig. 2. (B) Inhibition of transfer by high concentrations of bare DNA. Transfer reactions (standard conditions but with 3 mM MgCl2 and 34 mM KCl) were performed with 0.24 nM TPT cores (labeled [left panel] or unlabeled [right panel]), 3.8 nM SWI-SNF (where indicated), and the indicated amount of bare TPT DNA (80 pM of which was labeled in the panel on the right). Reactions were stopped with 3 μg of plasmid at 40 min, and products were separated by EMSA. (C) Quantitation of the results in panel B. For transfer from labeled cores (light bars), lane 4 (no added bare DNA) was set to background. For transfer from unlabeled cores (dark bars), lane 11 (no SWI-SNF) was set to background. The line represents the amount of remodeled dimer formed as a percentage of the amount formed in the absence of bare DNA (lane 4).
We then looked at the effect of DNA sequence on transfer. The octamer transfer experiments to this point measured transfer from genomic HeLa cell nucleosomes to 155-bp TPT DNA. TPT contains nucleosome phasing sequences that introduce a bend at one end of the DNA (32). This results in preferential wrapping of the DNA with the bent face toward the histone octamer, presumably because this creates a more energetically stable nucleosome. Theoretically, this increased stability could inhibit transfer from DNA with phasing sequences to other DNA sequences. To examine this, we used unlabeled TPT mononucleosomes as octamer donors and found that transfer still occurred, albeit about fivefold less efficiently (Fig. 5A, lanes 10 and 11). We then replaced the TPT acceptor DNA with labeled genomic HeLa cell DNA (∼145 to 155 bp) isolated from mononucleosomes. hSWI-SNF was capable of transferring octamers from both unlabeled HeLa cell and TPT mononucleosomes to this genomic DNA to generate cores with the same mobility as mononucleosome cores from HeLa cells (Fig. 5A, lanes 16 to 19; Fig. 2B; and data not shown). Again, transfer from TPT is about fivefold less efficient than that from random DNA. Note that the broader core and bare bands are reflective of the slight heterogeneity of acceptor DNA length. These results indicate that hSWI-SNF can transfer octamers between a variety of donor and acceptor DNA species and that phasing sequences do not impart absolute directionality to this reaction.
Since transfer occurred with a variety of nucleosome donors, we tested whether free histones could act as donors. We found that in reactions containing free histones, hSWI-SNF and/or ATP had no effect (data not shown). Thus, SWI-SNF appears to require nucleosomal histones for transfer. Furthermore, this result suggests that hSWI-SNF cannot act by itself as a nucleosome assembly factor utilizing free histones.
Inhibition of remodeling and transfer by bare DNA.
To further elucidate the relationship between formation of the stably remodeled dimer and transfer of histone octamers, we examined the effect of bare DNA concentration on both reactions. We found that while bare DNA is a required substrate in octamer transfer, high concentrations of bare DNA inhibited both reactions.
Reaction mixtures containing 0.24 nM TPT nucleosome cores were incubated with SWI-SNF and increasing amounts of bare DNA (Fig. 5B). The reaction was performed two ways; for lanes 1 to 10 the nucleosome cores were labeled and octamer transfer was measured as the release of labeled bare DNA, while for lanes 11 to 18 the bare DNA was labeled and transfer was determined by measuring the increase in labeled cores. The values for transfer were plotted against the concentration of bare DNA (Fig. 5C). In both cases, transfer increased with up to 1.2 nM bare DNA (fivefold excess over cores) and decreased at higher concentrations. Similar results were seen when a 1.2 or 4.8 nM concentration of cores was used as the donor (data not shown). This behavior is consistent with bare DNA being a required substrate but also a competitive inhibitor. Such a situation might arise if nucleosomes and bare DNA competed for the same binding sites.
Bare DNA is not a substrate for remodeled-dimer formation and more directly inhibits the formation of dimers (Fig. 5B, lanes 4 to 10; plotted as a line in Fig. 5C). Intriguingly, dimer formation and octamer transfer were inhibited at similar DNA concentrations (Fig. 5C and data not shown). The fact that our nucleosome core preparations were contaminated with small amounts (∼2%) of bare DNA precluded both a rigorous assessment of the Km for nucleosomes in either the octamer transfer or remodeled-dimer reactions and a rigorous measurement of the Ki of bare DNA in these reactions. By using only low nucleosome concentrations under conditions of linearity with substoicheometric amounts of SWI-SNF, however, we can estimate that the Ki for bare DNA is ∼3 nM in reactions that measure formation of the remodeled dimer (data not shown). Interestingly, this value is in the range of the Kd measured for bare DNA binding to BRG1 (G. Narlikar and R. E. Kingston, unpublished observations).
DISCUSSION
We have shown that the highly purified hSWI-SNF ATPase subunits, BRG1 and hBRM, can interconvert a stable remodeled mononucleosome dimer with a standard nucleosome core in a reaction that is indistinguishable from that involving the entire hSWI-SNF complex. In addition, hSWI-SNF and the isolated ATPases are each capable of transferring histone octamers from donor nucleosomes to acceptor DNA in trans. The dependence of both reactions on continuous ATP hydrolysis suggests that they may proceed through an unstable intermediate formed by hSWI-SNF. To date, both the formation of a stably remodeled dimer and octamer transfer are activities that have been demonstrated only for members of SWI-SNF-related complexes. The observation that the isolated ATPases of hSWI-SNF can catalyze both of these reactions suggests that it is the ATPase subunit that defines these particular capabilities of the SWI-SNF family of complexes.
A model for nucleosome remodeling and octamer transfer.
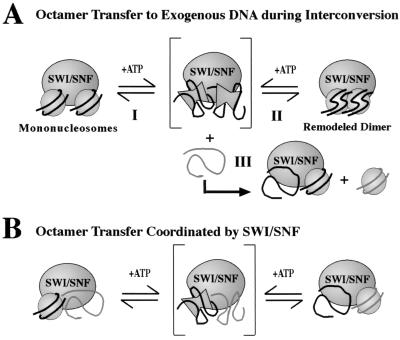
We have proposed a model for the ability of SWI-SNF to interconvert mononucleosome cores and the remodeled dimer in which SWI-SNF has two nucleosome binding sites with which to bind two mononucleosomes or one dimer (17, 30, 31) (Fig. 6A, top). The presence of two nucleosome binding sites per SWI-SNF complex is suggested by electron microscopic studies of ySWI-SNF (3) and the apparent molecular weight of hSWI-SNF (17). It is important to note, however, that the stoichiometries of the ATPase and other subunits in SWI-SNF complexes have not yet been determined. After substrate binding, the hydrolysis of ATP is hypothesized to result in the generation of an activated intermediate in which histone-DNA and perhaps interhistone interactions are dramatically loosened. Following the hydrolysis cycle, these interactions would reform stochastically to local energy minima, generating either individual mononucleosomes (Fig. 6A, reaction I) or the remodeled dimer (Fig. 6A, reaction II). In this way, hSWI-SNF might be acting analogously to ATP-dependent chaperones that unfold misfolded proteins and allow them to refold spontaneously, effectively reducing the activation barrier between folded states (for a review, see reference 12).
FIG. 6.
Model of hSWI-SNF mechanism. (A) When the SWI-SNF ATPase(s) (BRG1 or hBRM alone, or in the SWI-SNF complex) binds two mononucleosome cores (left side) or one remodeled dimer (right side), the hydrolysis of ATP is hypothesized to generate a high-energy SWI-SNF nucleosome intermediate. The loosened histone-DNA contacts in this intermediate facilitate remodeling, allowing these transiently disrupted nucleosomes to stochastically reform normal mononucleosomes (reconversion; reaction I) or remodeled dimers (remodeling; reaction II). In addition, the weakened contacts may allow the histones in this disrupted complex to be pulled away by exogenous bare DNA, resulting in octamer transfer (reaction III). (B) Alternatively, transfer may occur in a concerted reaction when one SWI-SNF site is bound by a nucleosome and the other is bound by bare DNA.
Our data suggest that octamer transfer may occur by a modification of the above mechanism. Dimer formation and octamer transfer require continuous ATP hydrolysis, and formation of the remodeled dimer does not appear to be required for transfer to occur. The concentrations of DNA that inhibit transfer are quite similar to those that inhibit the formation of the remodeled dimer, consistent with the same active site being used for both reactions. We propose that each binding site in SWI-SNF can bind either DNA or a nucleosome, but not both. For octamer transfer to occur, one site would be bound by a nucleosome (the donor) and the other would be bound by DNA (the acceptor) (Fig. 6B). High concentrations of bare DNA would inhibit transfer and remodeled-dimer formation similarly, since without nucleosomes bound to one site (transfer) or both sites (dimer formation) SWI-SNF will have no lasting effect. When ATP is hydrolyzed, the nucleosome will be transiently disrupted and the loosened histone-DNA contacts that would have allowed two nucleosomes to form the remodeled dimer will now allow the donor's histones to transfer to the acceptor DNA. While it is possible that octamer transfer and dimer formation use distinct intermediates, a single intermediate state provides the simplest explanation for our results. A second possibility is that the reaction is not actively coordinated by SWI-SNF; instead, histones from one of the two transiently disrupted nucleosomes may be scavenged by an exogenous piece of DNA, resulting in octamer transfer (Fig. 6A, reaction III). If the first model (Fig. 6B) were valid, very high concentrations of nucleosomes should inhibit transfer (since both binding sites would be bound by nucleosomes and not DNA). Unfortunately, contamination of our nucleosome preparations by small amounts of bare DNA has prevented us from directly testing this hypothesis.
ySWI-SNF has been shown to facilitate both transfer and the sliding of nucleosomes in cis along polynucleosome-length DNAs (42). Sliding in cis could be accomplished as in the model above, except that the acceptor DNA would be the DNA that is linked in cis. Alternatively, as an activated intermediate relaxes to form a nucleosome, the newly formed histone-DNA contacts may differ from those in the original nucleosome, resulting in shifting of the octamer position.
Donor and acceptor requirements for octamer transfer.
Random-sequence poly-, di-, and mononucleosomes isolated from HeLa cell nuclei by MNase digestion, as well as in vitro-assembled rotationally phased TPT mononucleosomes, all function as octamer donors. Both TPT DNA and nucleosome-length HeLa cell genomic DNA function as acceptors. A moderate influence of the phasing sequence can be seen as an approximately fivefold decrease in transfer from phased- to random-sequence DNA. As is seen with methods of nucleosome assembly involving salt dialysis, the rotational orientation of nucleosomes formed by octamer transfer appears to be determined by the phasing sequences (see, e.g., Fig. 2C). This is presumably because having the phasing sequences out of alignment produces a nucleosome with higher free energy. The low efficiency of transfer in this and previous work means that it is possible that transfer requires a contaminant or a certain form of the nucleosome that is present at low levels in our donor samples. However, the protein content in these HeLa and TPT nucleosome preparations is greater than 95% core histones, and other contaminants are likely to differ among these samples. Also, the most likely candidate for contamination across these samples, free histones, cannot be used by SWI-SNF to assemble nucleosomes from bare DNA (G. R. Schnitzler and R. E. Kingston, unpublished observation).
Functions of isolated BRG1 and hBRM ATPases.
In this article we have extended our earlier results to show that the central ATPases of human SWI-SNF complexes, BRG1 and hBRM, can individually perform all of the tested catalytic functions of the entire complex. Both can generate a remodeled dimer product indistinguishable from that of complete hSWI-SNF. They can also reconvert this product back to normal cores, establishing an equilibrium between the two nucleosomal states. As is true for the complete SWI-SNF complex, the altered DNase digestion pattern due to hBRM and BRG1 action can be largely explained as being a mixture of the DNase patterns for normal cores and remodeled dimers (Fig. 1B) (30). Furthermore, BRG1 and hBRM can catalyze the octamer transfer reaction. We believe that these activities are intrinsic to the ATPases, since they—and nucleosomal array remodeling activity—coelute with BRG1 under conditions in which BRG1 runs as a monomer.
These observations suggest that the central ATPases of hSWI-SNF are sufficient for many of the ATP-dependent remodeling functions of the complex. If SWI-SNF requires two nucleosome binding sites to function, these results suggest either that BRG1 and hBRM contain two nucleosome binding sites each or that they can dimerize in solution. Some of the other evolutionarily conserved subunits, namely BAF155, BAF170, and Ini1, have been shown to affect the rate of ATP hydrolysis and the efficiency of plasmid chromatin remodeling (28). Other subunits, however, may have little effect on the enzymatic activities of hSWI-SNF and may be involved in the proper regulation of the complex in vivo. Such may be the case in these assays as well, since we estimate the intact complex to be approximately 4- to 16-fold more active per mole than BRG1 for dimer formation (Fig. 1A), reconversion (Fig. 1B), and octamer transfer (Fig. 2A).
Role of octamer transfer in SWI-SNF function.
hSWI-SNF can stimulate RNA polymerase II elongation in vitro on nucleosomal templates activated by HSF or VP16 activators (5). The ability of SWI-SNF to transfer an octamer to bare DNA may be important for this effect and thus potentially for transcriptional elongation in vivo. As RNA polymerase II moves, downstream nucleosomes act as barriers and cause dramatic pauses (5, 14). The transfer of a nucleosome to upstream sequences as a polymerase passes has been observed in vitro for a bacteriophage polymerase and RNA polymerase III (35, 36). The ability of SWI-SNF to carry out octamer transfer may facilitate this process, accommodating both polymerase movement and maintenance of the nucleosomal content of the gene. An ability of SWI-SNF to reposition or alter nucleosomes to accommodate a passing polymerase is one possible explanation for yeast SWI-SNF being continuously required for transcription in vivo (4, 37). Remodeling complexes probably also facilitate transcription in vivo by helping transcription factors bind their cognate sites in promoter chromatin. An ability to transfer an octamer to surrounding DNA, either in cis or in trans, would open up DNA binding sites and allow factor access. Together with the creation of remodeled nucleosome dimers, which allows increased access of Gal4 to a site originally near the dyad of the unremodeled nucleosome (30), octamer transfer and/or mobilization may allow remodeling complexes to create highly dynamic and accessible chromatin.
ACKNOWLEDGMENTS
M.L.P. and G.R.S. contributed equally to this work; the order of their names was determined by a coin flip.
We thank Jerry Workman for the kind gift of the plasmid containing the 5S-G5E4 sequence; Geeta Narlikar (G.N.) for sharing the PCR method of assembling concentrated mononucleosomes; Andy Saurin, G.N., and Pu Zhang for aid in assembly of the 5S array; and G.N., Jeff Guyon, Jeff Aalfs, Laura Corey, Stuart Levine, Kyu-Min Lee, Kelly Sullivan, Alona Weiss, and other members of the Kingston lab for helpful comments and discussions and for help during the planning and execution of these experiments.
This work was funded by NIH grants to R.E.K. M.L.P. was a Research Fellow of the National Cancer Institute of Canada supported with funds from the Terry Fox Run.
REFERENCES
- 1.Armstrong J A, Bieker J J, Emerson B M. A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell. 1998;95:93–104. doi: 10.1016/s0092-8674(00)81785-7. [DOI] [PubMed] [Google Scholar]
- 2.Ayer D E. Histone deacetylases: transcriptional repression with SINers and NuRDs. Trends Cell Biol. 1999;9:193–198. doi: 10.1016/s0962-8924(99)01536-6. [DOI] [PubMed] [Google Scholar]
- 3.Bazett-Jones D P, Côté J, Landel C C, Peterson C L, Workman J L. The SWI/SNF complex creates loop domains in DNA and polynucleosome arrays and can disrupt DNA-histone contacts within these domains. Mol Cell Biol. 1999;19:1470–1478. doi: 10.1128/mcb.19.2.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biggar S R, Crabtree G R. Continuous and widespread roles for the Swi-Snf complex in transcription. EMBO J. 1999;18:2254–2264. doi: 10.1093/emboj/18.8.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown S A, Imbalzano A N, Kingston R E. Activator-dependent regulation of transcriptional pausing on nucleosomal templates. Genes Dev. 1996;10:1479–1490. doi: 10.1101/gad.10.12.1479. [DOI] [PubMed] [Google Scholar]
- 6.Chiba H, Muramatsu M, Nomoto A, Kato H. Two human homologues of Saccharomyces cerevisiae SWI2/SNF2 and Drosophila Brahma are transcriptional coactivators cooperating with the estrogen receptor and the retinoic acid receptor. Nucleic Acids Res. 1994;22:1815–1820. doi: 10.1093/nar/22.10.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corona D F, Langst G, Clapier C R, Bonte E J, Ferrari S, Tamkun J W, Becker P B. ISWI is an ATP-dependent nucleosome remodeling factor. Mol Cell. 1999;3:239–245. doi: 10.1016/s1097-2765(00)80314-7. [DOI] [PubMed] [Google Scholar]
- 8.Cote J, Peterson C L, Workman J L. Perturbation of nucleosome core structure by the SWI/SNF complex persists after its detachment, enhancing subsequent transcription factor binding. Proc Natl Acad Sci USA. 1998;95:4947–4952. doi: 10.1073/pnas.95.9.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunaief J L, Strober B E, Guha S, Khavari P A, Alin K, Luban J, Begemann M, Crabtree G R, Goff S P. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell. 1994;79:119–130. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- 10.Fryer C J, Archer T K. Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature. 1998;393:88–91. doi: 10.1038/30032. [DOI] [PubMed] [Google Scholar]
- 11.Guschin D, Wolffe A P. SWItched-on mobility. Curr Biol. 1999;9:R742–R746. doi: 10.1016/s0960-9822(99)80473-4. [DOI] [PubMed] [Google Scholar]
- 12.Hartl F U. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 13.Imbalzano A N, Schnitzler G R, Kingston R E. Nucleosome disruption by human SWI/SNF is maintained in the absence of continued ATP hydrolysis. J Biol Chem. 1996;271:20726–20733. doi: 10.1074/jbc.271.34.20726. [DOI] [PubMed] [Google Scholar]
- 14.Izban M G, Luse D S. Factor-stimulated RNA polymerase II transcribes at physiological elongation rates on naked DNA but very poorly on chromatin templates. J Biol Chem. 1992;267:13647–13655. [PubMed] [Google Scholar]
- 15.Kadonaga J T. Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- 16.Khavari P A, Peterson C L, Tamkun J W, Mendel D B, Crabtree G R. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- 17.Kingston R E, Narlikar G J. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 1999;13:2339–2352. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- 18.Kwon H, Imbalzano A N, Khavari P A, Kingston R E, Green M R. Nucleosome disruption and enhancement of activator binding by a human SWI/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 19.Langst G, Bonte E J, Corona D F, Becker P B. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell. 1999;97:843–852. doi: 10.1016/s0092-8674(00)80797-7. [DOI] [PubMed] [Google Scholar]
- 20.Lee C H, Murphy M R, Lee J S, Chung J H. Targeting a SWI/SNF-related chromatin remodeling complex to the beta-globin promoter in erythroid cells. Proc Natl Acad Sci USA. 1999;96:12311–12315. doi: 10.1073/pnas.96.22.12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Logie C, Peterson C L. Catalytic activity of the yeast SWI/SNF complex on reconstituted nucleosome arrays. EMBO J. 1997;16:6772–6782. doi: 10.1093/emboj/16.22.6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lorch Y, Cairns B R, Zhang M, Kornberg R D. Activated RSC-nucleosome complex and persistently altered form of the nucleosome. Cell. 1998;94:29–34. doi: 10.1016/s0092-8674(00)81218-0. [DOI] [PubMed] [Google Scholar]
- 23.Lorch Y, Zhang M, Kornberg R D. Histone octamer transfer by a chromatin-remodeling complex. Cell. 1999;96:389–392. doi: 10.1016/s0092-8674(00)80551-6. [DOI] [PubMed] [Google Scholar]
- 24.Luger K, Rechsteiner T J, Flaus A J, Waye M, Richmond T J. Characterization of nucleosome core particles containing histone proteins made in bacteria. J Mol Biol. 1997;272:301–311. doi: 10.1006/jmbi.1997.1235. [DOI] [PubMed] [Google Scholar]
- 25.Muchardt C, Yaniv M. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 1993;12:4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphy D J, Hardy S, Engel D A. Human SWI-SNF component BRG1 represses transcription of the c-fos gene. Mol Cell Biol. 1999;19:2724–2733. doi: 10.1128/mcb.19.4.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neely K, Hassan A, Wallberg A, Steger D, Cairns B, Wright A, Workman J. Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription from nucleosome arrays. Mol Cell. 1999;4:649–655. doi: 10.1016/s1097-2765(00)80216-6. [DOI] [PubMed] [Google Scholar]
- 28.Phelan M L, Sif S, Narlikar G J, Kingston R E. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol Cell. 1999;3:247–253. doi: 10.1016/s1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- 29.Richmond T, Searles M, Simpson R. Crystals of a nucleosome core particle containing defined sequence DNA. J Mol Biol. 1988;1998:161–170. doi: 10.1016/0022-2836(88)90386-5. [DOI] [PubMed] [Google Scholar]
- 30.Schnitzler G, Sif S, Kingston R E. Human SWI/SNF interconverts a nucleosome between its base state and a stable remodeled state. Cell. 1998;94:17–27. doi: 10.1016/s0092-8674(00)81217-9. [DOI] [PubMed] [Google Scholar]
- 31.Schnitzler G R, Sif S, Kingston R E. A model for chromatin remodeling by the SWI/SNF family. Cold Spring Harbor Symp Quant Biol. 1998;63:535–543. doi: 10.1101/sqb.1998.63.535. [DOI] [PubMed] [Google Scholar]
- 32.Shrader T E, Crothers D M. Artificial nucleosome positioning sequences. Proc Natl Acad Sci USA. 1989;86:7418–7422. doi: 10.1073/pnas.86.19.7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sif S, Stukenberg P T, Kirschner M W, Kingston R E. Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes Dev. 1998;12:2842–2851. doi: 10.1101/gad.12.18.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strober B E, Dunaief J L, Guha S, Goff S P. Functional interactions between the hBRM/hBRG1 transcriptional activators and the pRB family of proteins. Mol Cell Biol. 1996;16:1576–1583. doi: 10.1128/mcb.16.4.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Studitsky V M, Clark D J, Felsenfeld G. A histone octamer can step around a transcribing polymerase without leaving the template. Cell. 1994;76:371–382. doi: 10.1016/0092-8674(94)90343-3. [DOI] [PubMed] [Google Scholar]
- 36.Studitsky V M, Kassavetis G A, Geiduschek E P, Felsenfeld G. Mechanism of transcription through the nucleosome by eukaryotic RNA polymerase. Science. 1997;278:1960–1963. doi: 10.1126/science.278.5345.1960. [DOI] [PubMed] [Google Scholar]
- 37.Sudarsanam P, Iyer V R, Brown P O, Winston F. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2000;97:3364–3369. doi: 10.1073/pnas.050407197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trouche D, Le Chalony C, Muchardt C, Yaniv M, Kouzarides T. RB and hbrm cooperate to repress the activation functions of E2F1. Proc Natl Acad Sci USA. 1997;94:11268–11273. doi: 10.1073/pnas.94.21.11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Utley R T, Owen-Hughes T A, Juan L J, Cote J, Adams C C, Workman J L. In vitro analysis of transcription factor binding to nucleosomes and nucleosome disruption/displacement. Methods Enzymol. 1996;274:276–291. doi: 10.1016/s0076-6879(96)74024-7. [DOI] [PubMed] [Google Scholar]
- 40.Varga-Weisz P D, Becker P B. Chromatin-remodeling factors: machines that regulate? Curr Opin Cell Biol. 1998;10:346–353. doi: 10.1016/s0955-0674(98)80010-0. [DOI] [PubMed] [Google Scholar]
- 41.Wang W D, Cote J, Xue Y, Zhou S, Khavari P A, Biggar S R, Muchardt C, Kalpana G V, Goff S P, Yaniv M, Workman J L, Crabtree G R. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 42.Whitehouse I, Flaus A, Cairns B R, White M F, Workman J L, Owen-Hughes T. Nucleosome mobilization catalyzed by the yeast SWI/SNF complex. Nature. 1999;400:784–787. doi: 10.1038/23506. [DOI] [PubMed] [Google Scholar]
- 43.Workman J L, Kingston R E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]