Abstract
Background
It is challenging to differentiate air and bone on MR images of conventional sequences due to their low contrast. We propose to combine semantic feature extraction under auto-context manner into random forest to improve reasonability of the MRI segmentation for MRI-based radiotherapy treatment planning or PET attention correction.
Methods
We applied a semantic classification random forest (SCRF) method which consists of a training stage and a segmentation stage. In the training stage, patch-based MRI features were extracted from registered MRI-CT training images, and the most informative elements were selected via feature selection to train an initial random forest. The rest sequence of random forests was trained by a combination of MRI feature and semantic feature under an auto-context manner. During segmentation, the MRI patches were first fed into these random forests to derive patch-based segmentation. By using patch fusion, the final end-to-end segmentation was obtained.
Results
The Dice similarity coefficient (DSC) for air, bone and soft tissue classes obtained via proposed method were 0.976±0.007, 0.819±0.050 and 0.932±0.031, compared to 0.916±0.099, 0.673±0.151 and 0.830±0.083 with random forest (RF), and 0.942±0.086, 0.791±0.046 and 0.917±0.033 with U-Net. SCRF also outperformed the competing methods in sensitivity and specificity for all three structure types.
Conclusions
The proposed method accurately segmented bone, air and soft tissue. It is promising in facilitating advanced MR application in diagnosis and therapy.
Keywords: MRI segmentation, semantic classification random forest (SCRF), auto-context
Introduction
Magnetic resonance image (MRI) is a widely accepted modality for cancer diagnosis and radiotherapy target delineation due to its superior soft tissue contrast. Bone and air segmentations are important tasks for MRI, and facilitate several clinical applications, such as MRI-based treatment planning in radiation oncology (1), MRI-based attenuation correction for positron emission tomography (PET) (2), and MR-guided focused ultrasound surgery (FUS) (3).
MRI-only based treatment planning process is desired since it can bypass CT acquisition, which eliminates the MRI-CT registration errors and ionizing radiation, as well as reduces medical cost and clinical workflow. However, it is not feasible for current clinical practice since MRI images do not provide electron density information for dose calculation and reference images for patient setup (4-9). While ignoring inhomogeneity gives rise to 4–5% of dose errors, simply assigning three bulk densities, such as, bone, tissue and air, could reduce deviations to less than 2%, which is clinically acceptable (10). Even with the use of synthetic CTs that provides continuous electron density estimation for improved dose calculation accuracy, bone and air identification is considered key to heterogeneity correction and thus accurate dose estimation (11). The hybrid PET/MRI system has emerged as a promising imaging modality due to the unparalleled soft tissue information provided by the non-ionizing imaging modality. Though different types of MR-based attenuation correction methods have been investigated, virtually all current commercial PET/MRI systems employ segmentation-based methods due to its efficiency, robustness and simplicity (12). Accurate segmentation of different tissue types, especially of bone and air, directly impacts the estimation accuracy of attenuation map. Bone segmentation in MRI also facilitates the quickly-developing technology of image-guided FUS (3). FUS requires a refocusing of the ultrasound beams to compensate for distortion and translation caused by the attenuation and scattering of the beams through bone (13). Since many procedures utilize, and are based on MR capabilities, it would be desirable to delineate bone from MRI, potentially avoiding the additional steps of CT acquisition and subsequent CT-MRI co-registration.
In contrast to CT which provides excellent tissue-bone and tissue-air contrast, conventional MR images have weak signals at bone and air regions, which makes bone and air segmentation particularly difficult. To accurately delineate structures, a straightforward approach is to warp atlas templates to the MRI, allowing one to exploit the excellent bone and air contrast on CT images to identify the corresponding structures in MRI (14,15). Besides computational cost, atlas-based methods are prone to registration errors as well as inter-patient variability. A larger and more varied atlas dataset could help to improve registration accuracy. However, because of organ morphology and substantial variability across patients, it is difficult to satisfy all possible scenarios. Moreover, larger atlas templates are usually associated with significantly increased computational cost. To solve this issue, recently, Bourouis et al. integrated an expectation-maximization (EM) algorithm and a deformable level-set model into atlas-based framework to perform brain MRI segmentation (16). In this method, atlas dataset is used to initialize the segmentation. EM algorithm is applied to generate a global shape of the specific object. Finally, a level-set method is used to refine the segmentation. Specialized MR sequences, such as ultrashort echo time (UTE) pulse sequences, have been investigated for bone visualization and segmentation. Its performance is limited by of noise and image artefacts (17). Moreover, due to considerably long acquisition time, the application of UTE MR sequence is usually limited to brain imaging or small field-of-view images. Segmentation of the cranial bone from MRI is challenging, because compact bone is characterized by very short transverse relaxation times and typically produces no signal when using conventional MRI sequences. Recently, Krämer et al. proposed a fully automated segmentation algorithm using dual-echo, UTE MRI data (18). Machine learning-based and deep learning-based segmentation and synthetic CT (also known as pseudo CT) generation methods have been intensively studied for the last decades (19-22), among which random forest-based method is one popular machine learning approach. The popularity of random forest arises from its appealing features, such as its capability of handling a large variety of features and enabling feature sharing of a multi-class classifier, robustness to noise and efficient parallel processing (23). Random forest has been employed to generate synthetic MRIs of different sequences for improved contrast (24), synthetic CTs for MRI-only radiotherapy treatment planning (25,26), as well as PET AC (27).
In this work, we propose to integrate an auto-context model and patch-based anatomical signature into a random forest framework to iteratively segment air, soft tissue and bone on routine anatomical MRIs. This semantic classification random forest (SCRF)-based approach has three distinctive strengths: (I) in order to enhance feature sensitivities to detect structures, three types of features are chosen to characterize information of an image patch at different levels from voxel level, sub-region level, to whole-patch level; (II) to improve the classification efficiency, a feature selection strategy is introduced into random forest-based model to first identify the most discriminative features from the MRI features; (III) to enhance the reasonability of segmentation, semantic features are extracted under an auto-context manner to consider surrounding information to guide more reasonable result iteratively.
To demonstrate the effectiveness of the auto-context model (28), we compared the performance of the proposed method with a conventional random forest framework without the auto-context model. Deep learning-based methods show state-of-the-art performances in various medical imaging applications. Therefore, we trained a well-established deep learning-based model, U-Net (29), and compared its segmentation accuracy with the proposed method.
Methods
Method overview
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by institutional review board and informed consent was not required for this Health Insurance Portability and Accountability Act (HIPAA)-compliant retrospective analysis.
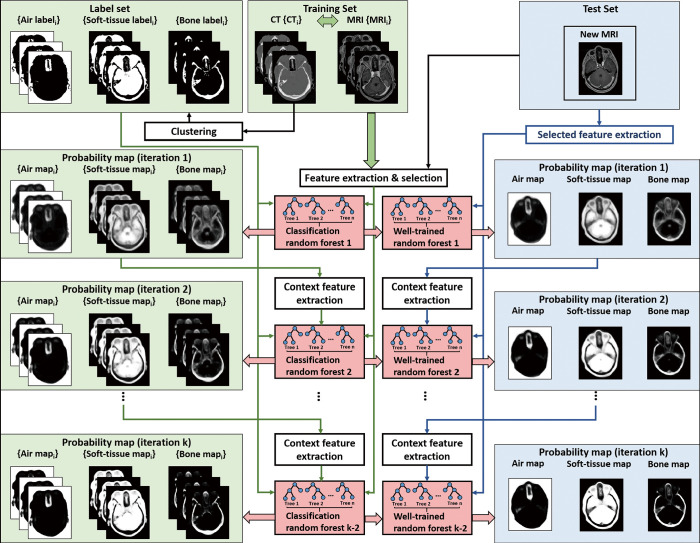
Figure 1 shows the workflow of proposed SCRF. The proposed method was trained with registered MR and CT images. During training, for a set of registered brain MR and CT images, the tissue labels, which are obtained from CT images were used as classification target for its corresponding MRI. These tissue labels include air, soft-tissue and bone. These tissue labels were derived via segmenting on CT images. Fuzz C-means was used for this segmentation. CT images were first segmented by a previously described thresholding method (30). Segmentation holes were filled by morphological erosion with a 5 voxel spherical kernel to eliminate any objects that were not physically part of the patient.
Figure 1.
The workflow of the proposed brain MRI SCRF method. The training stage is shown on the left, and the segmentation stage is shown on the right. MRI, magnetic resonance image; SCRF, semantic classification random forest.
Since image quality would affect the training and inference of the learning-based model, a noise reduction method was used to improve the feeding MRI’s image quality. The noise reduction was implemented via non-local means method (31). In addition, since MRI’s quality is often affected by inhomogeneity, nonparametric nonuniform intensity normalization (N3) algorithm was applied for MR image inhomogeneity bias correction.
For each patient, the MR and CT images were registered along with the corresponding labels. MRI data were first resampled to match the resolution of CT data. For each patient, all training MR and CT images were first rigidly registered by an intra-subject registration. Then the MR-CT pair for all the patients were fused into a same coordinate. Inter-patient registration of MR images consisted of a rigid registration followed by a B-spline deformable image registration. The transformation matrix obtained during this registration process was applied to the CT images to generate the deformable template CT. All registrations were rigidly performed on commercial software, Velocity AI 3.2.1 (Varian Medical Systems, Palo Alto, CA).
The input patch size of the MRI was [33, 33, 33]. The corresponding CT segmentation label on that patch’s central position was regarded as the learning-based classification target. Multiple features were extracted from MRI patch to represent that patch’s class. These features can be summarized as three folds. The first fold extracts feature via a voxel-wise manner, such as pairwise difference (PD) performed on two voxels. The second fold extracts feature via a sub-region manner, such as local binary pattern (LBP) and discrete cosine transformation (DCT) feature extractions which are performed on sub-region of a patch, and PD feature extraction performed on mean values of two sub-regions of a patch. The third fold extracts feature on whole patch, such as LBP and DCT feature extractions which are performed on whole patch. To further enlarge the feature variation, multiscale strategy was utilized. We performed feature extraction on both original scale MRI and on three rescaled MRIs (with scaling factor of 0.75, 0.5 and 0.25).
Since the extracted features may include some noisy and uninformative elements, which may affect the performance of classification model, feature selection was used to reduce these elements. The feature selection is implemented by logistic LASSO, which is introduced in our previous work (32). Auto-context strategy was used to iteratively improve the segmentation results, which means the first classification model is trained on extracted features only, and the rest models are trained on extracted features from both MRI and segmentation results (33). The features extracted from segmentation results are called as semantic features or auto-context features in this work.
The classification model is implemented by random forest model. The implementation details for random forest used in this work are as follows. The number of trees was set to 100. Minimum Gini impurity optimization was used to create each tree in the forest. A node in a tree will be split if this induces a decrease of the Gini impurity greater than or equal to this value. Gini impurity is a measure of how often a randomly chosen element from the set would be incorrectly labeled if it were randomly labeled according to the distribution of labels in the subset. The Gini impurity can be computed by summing the probability pi of an item with label i being chosen times the probability of a mistake in categorizing that item. It reaches its minimum (zero) when all cases in the node fall into a single target category. The weighted impurity decrease equation is calculated as follow:
| [1] |
where N is the total number of samples, Nt is the number of samples at the current node, is the number of samples in the left child node, and is the number of samples in the right child node. IGini denotes the Gini impurity of that set.
Once the sequence of random forests was trained, the segmentation of an arrival MRI can be obtained by feeding the MRI patches into the sequence of trained models to derive patch-based classification. Finally, by perform patch fusion, the segmentation result was derived.
Semantic feature
Traditional random forest feeds feature vector of a patch into the model and predict the class of central voxel or central sub-region of that patch. The challenge of this work is that MRI patches would share similar structures but would be corresponding to different central CT intensities and sub-regions, namely, different classes. The extracted feature vectors of similar structured MRI patches would be similar, whereas their learning targets are different. This would cause a potential ambiguity for the training of the random forest model, since the training of the random forest model is performed via building a serious of binary decision trees that are optimized based on extracted feature vectors (27). Thus, the features extracted from MRI-only may not be accurate to derive reasonable tissue segmentation.
In order to solve this problem, we aim to use the semantic information from the segmenting result to further improve the reasonability and accuracy of segmentation under an auto-context manner. First we trained a random forest model based on arrival MRI’s features. Rather than applying this model to derive segmentation, we fed MRI’s features to derive the posterior probability of tissue class via maximizing a posterior (MAP). These posterior probabilities are fused together via patch fusion to obtain an end-to-end (equal sized) probability map corresponding to MRI’s segmentation.
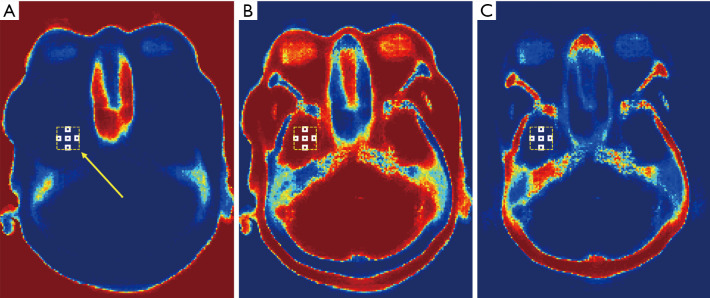
The auto-context procedure iteratively update the random forest model and probability map of MRI by feeding features from MRI and from probability map into the model (34). This procedure would improve the reasonability of probability map, since it leverages the information surrounding the object of interest (34). The features extracted from probability map are called semantic features. These semantic features are extracted by searching surrounding mean sub-regions’ probabilities corresponding to each MRI patch’s central voxel. The surrounding locations are shown in Figure 2. These locations represent each central position and its superior/inferior positions, left/right positions, and anterior/posterior positions, which are called as context locations in this work.
Figure 2.
An example illustrating the blocks within a window for semantic feature extraction. The axial view of probability map of air, soft tissue and bone classes are shown by (A), (B) and (C), respectively. The semantic features are extracted within an extracting window, which is shown as yellow dashed rectangles. For each semantic feature, it is calculated by the mean value of a white rectangle located within extracting window. The yellow arrow indicates a window position.
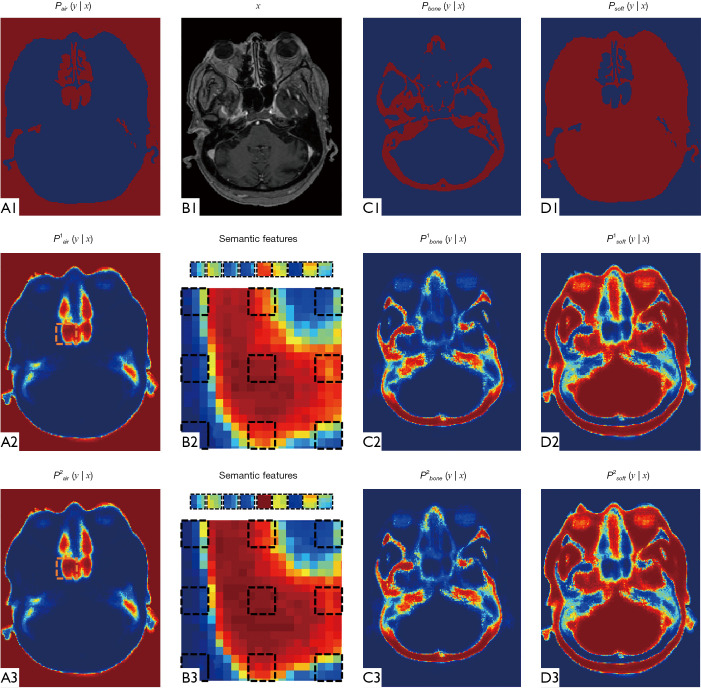
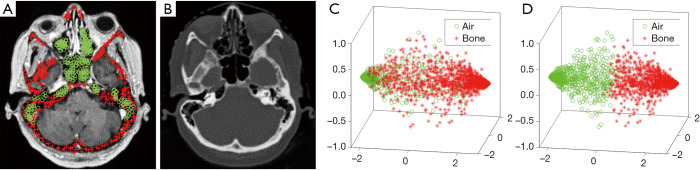
Figure 3 demonstrates the advantage of semantic feature extraction under auto-context procedure. (B1) shows the MRI (denoted by x) in axial view. (A1) shows the ground truth probability map of air, which is a binary map of air segmentation. (A2) shows the probability map derived via first random forest. (B2) shows the context locations (black dashed rectangles) for semantic feature extraction. (A3) shows the updated probability map derived under auto-context manner, i.e., feeding MRI features and previous probability map’s semantic features into a second random forest. We repeated semantic feature extraction and random forest training steps iteratively and alternatively until convergence. By using thresholding, the final air segmentation can be obtained by last air probability map. The threshold was set to 0.6, i.e., a voxel belongs to air if its air probability is larger than 0.6. This threshold was set by its best segmentation performance. (C1,C2,C3) and (D1,D2,D3) show the updating of probability map of bone and soft tissue, respectively. As compared between (A3,C3,D3) and (A2,C2,D2), it is shown that in first iteration, the central position’s probability within selected air region was ambiguous by a mixture of three tissue probabilities. In second iteration, the central position’s air probability is raised, and the rest two classes probabilities decreased, which can provide a clearer differentiation between these three classes. Figure 4 illustrates the utility of feature selection. We scatter plot feature significant elements that are belong to two kind of samples as shown in (A). The sample patches for feature extraction belong to air and bone tissues, and their central position are highlighted by green circles and red asterisks, respectively. The significant feature elements are derived by collect the top three element after principle component analysis. As can be seen from the visual comparison between before and after feature selection, the feature selection can improve the feature’s discriminative ability to be easier to differentiate the air and bone region, which is challenging in MRI brain classification.
Figure 3.
An example of probability maps generation during auto-context procedure. (B1) shows the MRI in axial view. (A1), (C1) and (D1) show the ground truth probability map of tissue classes, which is a binary mask of ground truth contour. (A2), (C2) and (D2) show the estimated probability maps via first random forest. (A3), (C3) and (D3) show the estimated probability maps via second random forest. (B2,B3) show the zoomed-in regions close to the nasopharynx. The context locations are shown in black dashed rectangles. By calculating mean value, each context location generates a semantic feature. MRI, magnetic resonance image.
Figure 4.
An example illustrating the significance of feature selection. (A) and (B) show axial viewed MRI and CT image, where the sample voxels corresponding to air tissue are represented by green circles, and the sample voxels of bone tissue are highlighted by red asterisks. (C) Shows the scatter plots of first three principle components of MRI features of these samples. The principle components are obtained via principle component analysis. (D) Shows the scatter plots of first three principle components of MRI features that are derived after feature selection. MRI, magnetic resonance image.
For implementation, in our work we trained 4 random forests in total, i.e., one initial random forest trained by MRI features, and the rest three random forests trained by both MRI features and semantic features. The more details of random forest can be found in our previous work (35).
Evaluation
A total of 14 patients who had MRI and CT data acquired and underwent cranial irradiation treatment were retrospectively investigated in this study. The MRIs were acquired on a Siemens Avanto 1.5T MRI scanner with T1 magnetization-prepared rapid gradient echo (MP-RAGE) sequence and 1.0×1.0×1.4 mm3 voxel size (TR/TE: 950/13 ms, flip angle: 90°). The MP-RAGE sequence can capture whole brain in a short scan time with good tissue contrast and high spatial resolution. The main patient selection criterion was that their MRIs were acquired with the same 3D sequences, had fine spatial resolution, and the entire head was imaged. The number of slices ranges from 176–224 for different patients. The CTs were acquired by a Siemens SOMATOM Definition AS CT scanner at 120 kVp and 220 mAs with the patient in treatment position; each 0.6-mm-thick slice had a resolution of 512×512 pixels, with pixel spacing of 0.586 mm. Bone, air and soft tissue were segmented on CT images and registered to MR images, which were used as ground truth. Leave-one-out cross-validation was used for evaluation. Dice similarity coefficient (DSC) of each class was used for numerical quantification (36). The formula of DSC measurement is as follows:
| [2] |
where X and Y denotes the binary mask of ground truth contour and the segmentation derived via proposed method, respectively. We also calculated sensitivity and specificity using the overlapping ratio inside and outside the ground truth volume,
| [3] |
| [4] |
where and denotes the volumes outside binary mask of ground truth contour and segmentation derived via proposed method, respectively.
To study the effectiveness of the proposed SCRF model, we ran the random forest (RF) method without an auto-context model and patch-based anatomical signatures and compared the resulting segmentation accuracy with the proposed method. Deep learning-based methods have been intensively studied in the last decade for various medical imaging applications. To compare the performance of SCRF method with deep learning-based methods, we also trained a well-established deep learning-based model, U-Net, for air, bone and soft tissue classification. To fairly compare the performance of the proposed method and competing methods, all methods were evaluated based on same experiment settings (same input image and leave-one-out cross-validation scheme).
Results
Comparison with random forest method
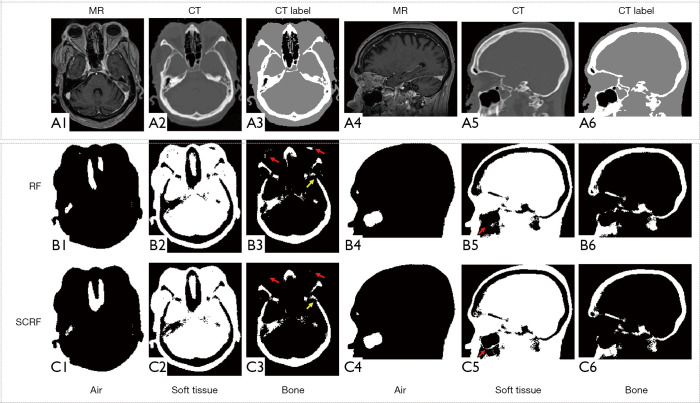
Figure 5 shows the qualitative comparison between RF and SCRF methods. Both RF and SCRF generate air, soft tissue and bone classification similar to the classification obtained with CT images. However, the left lens was mislabeled as bone with RF method, as indicated by the red arrows in Figure 5 (B3), and correctly classified as soft tissue with SCRF method. As shown by the red arrows in (B5) and (C5), the fine structure delineating maxillary sinus was better identified with SCRF.
Figure 5.
Qualitative comparison between RF and SCRF methods. (A1) and (A4) shows MRI in axial view and sagittal view, respectively. (A2), (A5) and (A3), (A6) are corresponding CT images and CT labels (black for air, gray for soft-tissue, and white for bone) respectively. Row (B) and (C) are results generated with RF and SCRF respectively. The yellow arrows indicate bone region that is challenging during classification. The red arrows indicate soft tissue and air regions that are challenging during classification. MRI, magnetic resonance image; SCRF, semantic classification random forest.
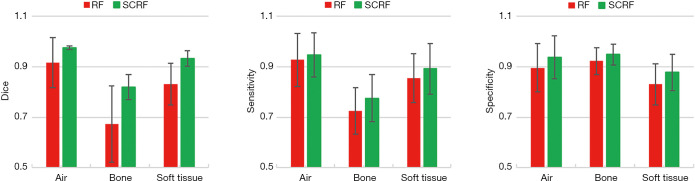
Figure 6 shows the quantitative comparisons of DSC, sensitivity and specificity between the two methods. With the integration of auto-context model and patch-based anatomical signature, the proposed SCRF method outperformed the RF method on all calculated metrics. The DSC results of air, bone and soft tissue classes were 0.976±0.007, 0.819±0.050 and 0.932±0.031, compared to 0.916±0.099, 0.673±0.151 and 0.830±0.083 with RF. Sensitivities were 0.947, 0.775, 0.892 for the three tissue types with SCRF, and 0.928, 0.725, 0.854 with RF. Specificity for the three tissue types was also improved with proposed method, which were 0.896, 0.823 and 0.830 with RF, compared to 0.938, 0.948, 0.878 with proposed SCRF.
Figure 6.
Quantitative comparison of RF and SCRF methods for DSC (left), sensitivity (middle), and specificity (right). Error bars show standard deviation. SCRF, semantic classification random forest; DSC, Dice similarity coefficient.
Comparison with U-Net model
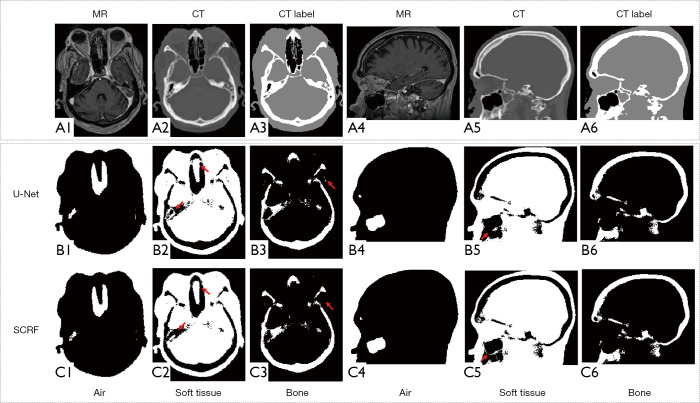
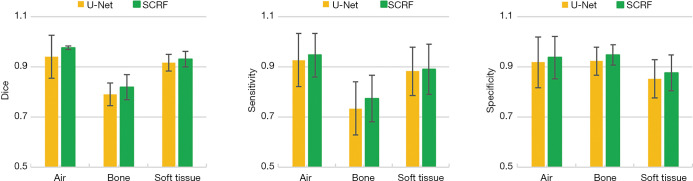
We compared the performance of the proposed method against a well-established deep learning-based model, U-Net. As indicated by the red arrows in Figure 7 (B2) and (C2), SCRF generated more accurate classification in challenging areas, such as the paranasal sinuses. U-Net failed to delineate the soft tissue around maxillary sinus [Figure 7 (B5)], while the soft tissue structure was accurately identified with SCRF [Figure 7 (C5)]. SCRF also corrected the mislabeling produced by the U-Net method in Figure 7 (B3). The performance improvement was further illustrated in the quantitative comparison. As shown in Figure 8, U-Net obtained DSCs of 0.942, 0.791 and 0.917 for air, bone and soft tissue classes, and SCRF improved DSCs by 0.034, 0.028 and 0.016 respectively. Similarly, sensitivity calculated U-Net classification results were 0.927, 0.735 and 0.883 for air, bone and soft tissue, and increased by 0.020, 0.040 and 0.009 with SCRF. Specificity was improved by 0.019, 0.025 and 0.025 on the three tissue types with the proposed SCRF method.
Figure 7.
Qualitative comparison between U-Net and SCRF methods. (A1) and (A4) shows MRI in axial view and sagittal view, respectively. (A2), (A5) and (A3), (A6) are corresponding CT images and CT labels (black for air, gray for soft-tissue, white for bone) respectively. Row (B) and (C) are results generated with RF and SCRF respectively. The red arrows indicate soft tissue and bone regions that are challenging during classification. MRI, magnetic resonance image; SCRF, semantic classification random forest.
Figure 8.
Quantitative comparison of U-Net and SCRF methods on DSC (left), sensitivity (middle) and specificity (right). Error bars show standard deviation. SCRF, semantic classification random forest; DSC, Dice similarity coefficient.
Discussion
In this paper, we have investigated a learning-based approach to classify tissue labels for arrival MRI. The novelty of our approach is the integration of semantic feature extraction under an auto-context manner, which would be helpful to iteratively improve segmentation’s reasonability via considering each voxel’s surrounding information. To reduce the effect of noisy and uninformative elements during MRI’s feature extraction, a LASSO-based feature selection is used. As can be seen from Figure 4, after feature selection, it would be easier to group MRI’ features into air and bone classes as compared to before feature selection [shown in scatter plot in (D) vs. (C)], which means the features after feature selection would be more discriminative for classification.
In this study, we demonstrated the clinical feasibility of the proposed method on 14 patients. The proposed method can be a promising tool in recently proposed advanced MR applications in diagnosis and therapy, such as MRI-based radiation therapy, attenuation correction for a hybrid PET/MRI scanner or MR-guided FUS.
Special MR sequences, such as UTE (17,37) and zero time echo (ZTE) (38,39), have been investigated for bone detection and visualization, which were also employed on commercial PET/MRI systems for segmentation-based attenuation correction. However, the segmentation accuracy with conventional methods on those special sequences are usually limited due to the high level of noise and the presence of image artifacts. Juttukonda et al. derived intermediate images from UTE and Dixon images for bone and air segmentation, which obtained Dice coefficients of 0.75 and 0.60 for the two tissue types, respectively (40). An et al. improved the UTE MR segmentation accuracy with a multiphase level-set algorithm (41). The bone and air DSC obtained on 18F-FDG datasets were 0.83 and 0.62. Baran et al. built a UTE MR template that contained manual-delineated air/bone/soft tissue contours, and used Gaussian mixture models to fit UTE images, which obtained average Dice coefficients of 0.985 and 0.737 for air and bone (42). The proposed SCRF method was implemented on MR images generated with routinely-acquired T1 sequences. Despite the limited bone-air contrast, our method demonstrated superior segmentation performance. Considering the valuable patient-specific bone extraction information provided by UTE and ZTE sequences, combining the superior detection capability of machine learning techniques with special bone-visualization sequences has the potential to generate promising results. In the future, we will explore the possibility of integrating the proposed method with UTE or ZTE MR sequences for better bone and air differentiation.
Several machine learning- and deep learning-based MR segmentation methods have been studied in the literature. Yang et al. proposed a multiscale skull segmentation method for MR images. A multiscale bilateral filtering scheme is used to improve the robustness of the method for noise MR images (43). The average Dice coefficient of skull is 0.922. Liu et al. trained a deep convolutional auto-encoder network for soft tissue, bone and air identification (44). This deep learning-based method generated average Dice coefficients of 0.936 for soft tissue, 0.803 for bone, and 0.971 for air. Convolution encoder-decoder (CED) was also implemented on MR images acquired with both UTE and out-of-phase echo images for better bone and air differentiation, which generated mean Dice coefficient of 0.96, 0.88 and 0.76 for soft tissue, bone and air, respectively (45). Comparing to those state-of-the-art methods, the proposed method demonstrates competitive segmentation performance. In our experiment 14 patients with leave-one-out cross-validation was used to verify the proposed method and competing methods. For each experiment, we used 13 patients as training data, and the rest 1 patient as testing data. Thus, this patient is separated from the training data. We then repeated this experiment 14 times so that each patient can be used as testing data only one time. If a hold-out test with several patients are used to verify the proposed method, we may need more patient data to enlarge the data variation of training data. The small training data variation may cause overfitting issue and further affect the robustness of the performance of the proposed method. To train and test the proposed model with additional more patients’ data will be our future work.
We evaluate the performance of the proposed method on brain images, and demonstrated superior performances for bone and air segmentation even in challenging areas, such as paranasal sinuses. Bone and air segmentation are crucial for attenuation correction of brain PET images, and facilitates MR-guided FUS. Bone segmentation in whole-body MR also have important clinical implications, such as musculoskeletal applications (46) and traumatic diagnoses (47,48). Different from brain MRI where the difficulty lies in the bone/air segmentation, challenges of bone segmentation on whole-body MRI are areas where spongy bone exists, such as the vertebra. The application of special MR sequences, such as UTE, is not clinically feasible for routine whole-body imaging due to the prolonged scanning time. For future work, we will modify the proposed method to include both cortical and spongy bone segmentation in the framework and implement it on whole-body MRI. As compared to bone, air and soft-tissue segmentation, tissue structures segmentation from MRI would be more challenging. Recently, deep learning-based networks demonstrated their performance on neonatal cerebral tissue type identification (49). Ding et al. performed deep learning-based models on both dual-modality MRI (T1 and T2) and single modality MRI, and found that the average Dice coefficient of dual-modality MRI can reach at 0.94/0.95/0.92 or gray matter, white matter, and cerebrospinal fluid, respectively, compared with 0.90/0.90/0.88 of T2 MRI (49). Evaluating our method on tissue structures segmentation for multi-modality MRI will be our future work.
Conclusions
We have applied a machine learning-based automatic segmentation method that could identify soft tissue, bone and air on MR images. The proposed method has acceptable segmentation accuracy and can be promising in facilitating advanced MR applications in diagnosis and therapy.
Acknowledgments
Funding: This research is supported in part by the National Cancer Institute of the National Institutes of Health under Award Number R01CA215718 (XY).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by institutional review board and informed consent was not required for this Health Insurance Portability and Accountability Act (HIPAA)-compliant retrospective analysis.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
Footnotes
Provenance and Peer Review: With the arrangement by the Guest Editors and the editorial office, this article has been reviewed by external peers.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/qims-20-1114). The special issue “Artificial Intelligence for Image-guided Radiation Therapy” was commissioned by the editorial office without any funding or sponsorship. All authors report this research was supported in part by the National Cancer Institute of the National Institutes of Health Award Number R01CA215718. The authors have no other conflicts of interest to declare.
References
- 1.Wang T, Manohar N, Lei Y, Dhabaan A, Shu HK, Liu T, Curran WJ, Yang X. MRI-based treatment planning for brain stereotactic radiosurgery: Dosimetric validation of a learning-based pseudo-CT generation method. Med Dosim 2019;44:199-204. 10.1016/j.meddos.2018.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang X, Wang T, Lei Y, Higgins K, Liu T, Shim H, Curran WJ, Mao H, Nye JA. MRI-based attenuation correction for brain PET/MRI based on anatomic signature and machine learning. Phys Med Biol 2019;64:025001. 10.1088/1361-6560/aaf5e0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schlesinger D, Benedict S, Diederich C, Gedroyc W, Klibanov A, Larner J. MR-guided focused ultrasound surgery, present and future. Med Phys 2013;40:080901. 10.1118/1.4811136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang T, Hedrick S, Lei Y, Liu T, Jiang X, Dhabaan A, Tang X, Curran W, McDonald M, Yang X. Dosimetric Study of Cone-Beam CT in Adaptive Proton Therapy for Brain Cancer. Med Phys 2018;45:E259. [Google Scholar]
- 5.Yang X, Lei Y, Wang T, Patel P, Dhabaan A, Jiang X, Shim H, Mao H, Curran W, Jani A. Patient-Specific Synthetic CT Generation for MRI-Only Prostate Radiotherapy Treatment Planning. Med Phys 2018;45:E701-E702. [Google Scholar]
- 6.Lei Y, Shu H, Tian S, Wang T, Dhabaan A, Liu T, Shim H, Mao H, Curran W, Yang X. A Learning-Based MRI Classification for MRI-Only Radiotherapy Treatment Planning. Med Phys 2018;45:E524. [Google Scholar]
- 7.Jeong J, Wang L, Bing J, Lei Y, Liu T, Ali A, Curran W, Mao H, Yang X. Machine-Learning-Based Classification of Glioblastoma Using Dynamic Susceptibility Enhanced MR Image Derived Delta-Radiomic Features. Med Phys 2018;45:E584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shafai-Erfani G, Wang T, Lei Y, Tian S, Patel P, Jani AB, Curran WJ, Liu T, Yang X. Dose evaluation of MRI-based synthetic CT generated using a machine learning method for prostate cancer radiotherapy. Med Dosim 2019;44:e64-e70. 10.1016/j.meddos.2019.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y, Lei Y, Wang Y, Wang T, Ren L, Lin L, McDonald M, Curran WJ, Liu T, Zhou J, Yang X. MRI-based treatment planning for proton radiotherapy: dosimetric validation of a deep learning-based liver synthetic CT generation method. Phys Med Biol 2019;64:145015. 10.1088/1361-6560/ab25bc [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Owrangi AM, Greer PB, Glide-Hurst CK. MRI-only treatment planning: benefits and challenges. Phys Med Biol 2018;63:05TR1. [DOI] [PMC free article] [PubMed]
- 11.Karlsson M, Karlsson MG, Nyholm T, Amies C, Zackrisson B. Dedicated magnetic resonance imaging in the radiotherapy clinic. Int J Radiat Oncol Biol Phys 2009;74:644-51. 10.1016/j.ijrobp.2009.01.065 [DOI] [PubMed] [Google Scholar]
- 12.Mehranian A, Arabi H, Zaidi H. Vision 20/20: Magnetic resonance imaging-guided attenuation correction in PET/MRI: Challenges, solutions, and opportunities. Med Phys 2016;43:1130-55. 10.1118/1.4941014 [DOI] [PubMed] [Google Scholar]
- 13.Clement GT, Hynynen K. A non-invasive method for focusing ultrasound through the human skull. Phys Med Biol 2002;47:1219-36. 10.1088/0031-9155/47/8/301 [DOI] [PubMed] [Google Scholar]
- 14.Dowling JA, Lambert J, Parker J, Salvado O, Fripp J, Capp A, Wratten C, Denham JW, Greer PB. An atlas-based electron density mapping method for magnetic resonance imaging (MRI)-alone treatment planning and adaptive MRI-based prostate radiation therapy. Int J Radiat Oncol Biol Phys 2012;83:e5-11. 10.1016/j.ijrobp.2011.11.056 [DOI] [PubMed] [Google Scholar]
- 15.Uh J, Merchant TE, Li Y, Li X, Hua C. MRI-based treatment planning with pseudo CT generated through atlas registration. Med Phys 2014;41:051711. 10.1118/1.4873315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bourouis S, Hamrouni K, Betrouni N, editors. Automatic MRI Brain Segmentation with Combined Atlas-Based Classification and Level-Set Approach. Image Analysis and Recognition; 2008 2008//; Berlin, Heidelberg: Springer Berlin Heidelberg. [Google Scholar]
- 17.Keereman V, Fierens Y, Broux T, De Deene Y, Lonneux M, Vandenberghe S. MRI-based attenuation correction for PET/MRI using ultrashort echo time sequences. J Nucl Med 2010;51:812-8. 10.2967/jnumed.109.065425 [DOI] [PubMed] [Google Scholar]
- 18.Krämer M, Herzau B, Reichenbach JR. Segmentation and visualization of the human cranial bone by T2* approximation using ultra-short echo time (UTE) magnetic resonance imaging. Z Med Phys 2020;30:51-9. 10.1016/j.zemedi.2019.06.003 [DOI] [PubMed] [Google Scholar]
- 19.Han X. MR-based synthetic CT generation using a deep convolutional neural network method. Med Phys 2017;44:1408-19. 10.1002/mp.12155 [DOI] [PubMed] [Google Scholar]
- 20.Lei Y, Harms J, Wang T, Liu Y, Shu HK, Jani AB, Curran WJ, Mao H, Liu T, Yang X. MRI-only based synthetic CT generation using dense cycle consistent generative adversarial networks. Med Phys 2019;46:3565-81. 10.1002/mp.13617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang T, Lei Y, Fu Y, Wynne JF, Curran WJ, Liu T, Yang X. A review on medical imaging synthesis using deep learning and its clinical applications. J Appl Clin Med Phys 2021;22:11-36. 10.1002/acm2.13121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lei Y, Fu Y, Wang T, Qiu RLJ, Curran WJ, Liu T, Yang X. Deep Learning Architecture Design for Multi-Organ Segmentation. 1st Edition ed. Auto-Segmentation for Radiation Oncology: State of the Art. Boca Raton: CRC Press, 2021. [Google Scholar]
- 23.Lebedev AV, Westman E, Van Westen GJ, Kramberger MG, Lundervold A, Aarsland D, Soininen H, Kloszewska I, Mecocci P, Tsolaki M, Vellas B, Lovestone S, Simmons A, Alzheimer's Disease Neuroimaging I, the AddNeuroMed c. Random Forest ensembles for detection and prediction of Alzheimer's disease with a good between-cohort robustness. Neuroimage Clin 2014;6:115-25. 10.1016/j.nicl.2014.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jog A, Carass A, Roy S, Pham DL, Prince JL. Random forest regression for magnetic resonance image synthesis. Med Image Anal 2017;35:475-88. 10.1016/j.media.2016.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang X, Lei Y, Higgins K, Wang T, Tian S, Dhabaan A, Shim H, Mao H, Curran W, Shu H. Patient-Specific Pseudo-CT Generation Using Semantic and Contextual Information-Based Random Forest for MRI-Only Radiation Treatment Planning. Med Phys 2018;45:E702-E.
- 26.Yang XF, Lei Y, Shu HK, Rossi P, Mao H, Shim H, Curran WJ, Liu T. Pseudo CT Estimation from MRI Using Patch-based Random Forest. Proc SPIE: Medical Imaging 2017: Image Processing 2017;10133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huynh T, Gao Y, Kang J, Wang L, Zhang P, Lian J, Shen D, Alzheimer's Disease Neuroimaging I. Estimating CT Image From MRI Data Using Structured Random Forest and Auto-Context Model. IEEE Trans Med Imaging 2016;35:174-83. 10.1109/TMI.2015.2461533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang T, Lei Y, Tian S, Jiang X, Zhou J, Liu T, Dresser S, Curran WJ, Shu HK, Yang X. Learning-based automatic segmentation of arteriovenous malformations on contrast CT images in brain stereotactic radiosurgery. Med Phys 2019;46:3133-41. 10.1002/mp.13560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. arXiv:150504597 2015. 10.1007/978-3-319-24574-4_28 [DOI]
- 30.Huang LK, Wang MJJ. Image Thresholding by Minimizing the Measures of Fuzziness. Pattern Recognition 1995;28:41-51. 10.1016/0031-3203(94)E0043-K [DOI] [Google Scholar]
- 31.Yang J, Fan J, Ai D, Zhou S, Tang S, Wang Y. Brain MR image denoising for Rician noise using pre-smooth non-local means filter. Biomed Eng Online 2015;14:2. 10.1186/1475-925X-14-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang X, Lei Y, Shu HKG, Rossi PJ, Mao H, Shim H, Curran WJ, Liu T. A Learning-Based Approach to Derive Electron Density from Anatomical MRI for Radiation Therapy Treatment Planning. Int J Radiat Oncol 2017;99:S173-S4. 10.1016/j.ijrobp.2017.06.437 [DOI] [Google Scholar]
- 33.Tu Z, Bai X. Auto-Context and Its Application to High-Level Vision Tasks and 3D Brain Image Segmentation. Ieee Transactions on Pattern Analysis and Machine Intelligence 2010;32:1744-57. 10.1109/TPAMI.2009.186 [DOI] [PubMed] [Google Scholar]
- 34.Tu Z, Bai X. Auto-context and its application to high-level vision tasks and 3D brain image segmentation. IEEE Trans Pattern Anal Mach Intell 2010;32:1744-57. 10.1109/TPAMI.2009.186 [DOI] [PubMed] [Google Scholar]
- 35.Lei Y, Harms J, Wang T, Tian S, Zhou J, Shu HK, Zhong J, Mao H, Curran WJ, Liu T, Yang X. MRI-based synthetic CT generation using semantic random forest with iterative refinement. Phys Med Biol 2019;64:085001. 10.1088/1361-6560/ab0b66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang T, Lei Y, Tang H, He Z, Castillo R, Wang C, Li D, Higgins K, Liu T, Curran WJ, Zhou W, Yang X. A learning-based automatic segmentation and quantification method on left ventricle in gated myocardial perfusion SPECT imaging: A feasibility study. J Nucl Cardiol 2020;27:976-87. 10.1007/s12350-019-01594-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Catana C, van der Kouwe A, Benner T, Michel CJ, Hamm M, Fenchel M, Fischl B, Rosen B, Schmand M, Sorensen AG. Toward implementing an MRI-based PET attenuation-correction method for neurologic studies on the MR-PET brain prototype. J Nucl Med 2010;51:1431-8. 10.2967/jnumed.109.069112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weiger M, Brunner DO, Dietrich BE, Muller CF, Pruessmann KP. ZTE imaging in humans. Magn Reson Med 2013;70:328-32. 10.1002/mrm.24816 [DOI] [PubMed] [Google Scholar]
- 39.Wiesinger F, Sacolick LI, Menini A, Kaushik SS, Ahn S, Veit-Haibach P, Delso G, Shanbhag DD. Zero TE MR bone imaging in the head. Magn Reson Med 2016;75:107-14. 10.1002/mrm.25545 [DOI] [PubMed] [Google Scholar]
- 40.Juttukonda MR, Mersereau BG, Chen Y, Su Y, Rubin BG, Benzinger TLS, Lalush DS, An H. MR-based attenuation correction for PET/MRI neurological studies with continuous-valued attenuation coefficients for bone through a conversion from R2* to CT-Hounsfield units. Neuroimage 2015;112:160-8. 10.1016/j.neuroimage.2015.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.An HJ, Seo S, Kang H, Choi H, Cheon GJ, Kim HJ, Lee DS, Song IC, Kim YK, Lee JS. MRI-Based Attenuation Correction for PET/MRI Using Multiphase Level-Set Method. J Nucl Med 2016;57:587-93. 10.2967/jnumed.115.163550 [DOI] [PubMed] [Google Scholar]
- 42.Baran J, Chen Z, Sforazzini F, Ferris N, Jamadar S, Schmitt B, Faul D, Shah NJ, Cholewa M, Egan GF. Accurate hybrid template-based and MR-based attenuation correction using UTE images for simultaneous PET/MR brain imaging applications. BMC Medical Imaging 2018;18:41. 10.1186/s12880-018-0283-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang X, Fei B. Multiscale segmentation of the skull in MR images for MRI-based attenuation correction of combined MR/PET. Journal of the American Medical Informatics Association 2013;20:1037-45. 10.1136/amiajnl-2012-001544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu F, Jang H, Kijowski R, Bradshaw T, McMillan AB. Deep Learning MR Imaging-based Attenuation Correction for PET/MR Imaging. Radiology 2018;286:676-84. 10.1148/radiol.2017170700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jang H, Liu F, Zhao G, Bradshaw T, McMillan AB. Technical Note: Deep learning based MRAC using rapid ultrashort echo time imaging. Med Phys 2018. [Epub ahead of print]. doi: . 10.1002/mp.12964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blemker SS, Asakawa DS, Gold GE, Delp SL. Image-based musculoskeletal modeling: applications, advances, and future opportunities. J Magn Reson Imaging 2007;25:441-51. 10.1002/jmri.20805 [DOI] [PubMed] [Google Scholar]
- 47.Lubovsky O, Liebergall M, Mattan Y, Weil Y, Mosheiff R. Early diagnosis of occult hip fractures MRI versus CT scan. Injury 2005;36:788-92. 10.1016/j.injury.2005.01.024 [DOI] [PubMed] [Google Scholar]
- 48.Saini A, Saifuddin A. MRI of osteonecrosis. Clin Radiol 2004;59:1079-93. 10.1016/j.crad.2004.04.014 [DOI] [PubMed] [Google Scholar]
- 49.Ding Y, Acosta R, Enguix V, Suffren S, Ortmann J, Luck D, Dolz J, Lodygensky GA. Using Deep Convolutional Neural Networks for Neonatal Brain Image Segmentation. Front Neurosci 2020;14:207. 10.3389/fnins.2020.00207 [DOI] [PMC free article] [PubMed] [Google Scholar]