Abstract
Deep learning, a new branch of machine learning algorithm, has emerged as a fast growing trend in medical imaging and become the state-of-the-art method in various clinical applications such as Radiology, Histo-pathology and Radiation Oncology. Specifically in radiation oncology, deep learning has shown its power in performing automatic segmentation tasks in radiation therapy for Organs-At-Risks (OAR), given its potential in improving the efficiency of OAR contouring and reducing the inter- and intra-observer variabilities. The similar interests were shared for target volume segmentation, an essential step of radiation therapy treatment planning, where the gross tumor volume is defined and microscopic spread is encompassed. The deep learning-based automatic segmentation method has recently been expanded into target volume automatic segmentation. In this paper, the authors summarized the major deep learning architectures of supervised learning fashion related to target volume segmentation, reviewed the mechanism of each infrastructure, surveyed the use of these models in various imaging domains (including Computational Tomography with and without contrast, Magnetic Resonant Imaging and Positron Emission Tomography) and multiple clinical sites, and compared the performance of different models using standard geometric evaluation metrics. The paper concluded with a discussion of open challenges and potential paths of future research in target volume automatic segmentation and how it may benefit the clinical practice.
Keywords: Deep learning, target volume delineation, auto segmentation, radiation therapy
Introduction
Radiation therapy is one of the most common treatments for cancer, requiring patient-specific treatment planning to deliver conformal radiation doses to the target while sparing the healthy tissues. The treatment planning starts with the target volumes and Organs-At-Risks (OAR) contouring on computed tomography (CT), magnetic resonance (MR) or positron emission tomography (PET), which lays the foundation of the precision of the entire workflow moving forward. The target volume contouring is manually delineated by radiation oncologists, which is taken as the golden standard in the clinical practice but a time-consuming process and may suffer from substantial inter- and intra-observer variability (1-3). In addition, the rapid development of online adaptive radiation therapy also raises the requirement of contouring efficiency (4). Based on the failures mode analysis of chart review in radiation therapy indicated by AAPM Task Group 275 (5), the wrong or inaccurate target volume contours was ranked as the top one high-risk failure mode through the entire workflow, indicating plenty of room for improvements in safety and efficiency of radiation therapy that may be achieved through the development of target volume automatic segmentation.
Prior to the emerging trend of deep learning, classic imaging processing techniques such as atlas-based methods had shown promising performance in target volume automatic segmentation in selective clinical sites (6-9). However, the fuzzy anatomical features shown in images impose a challenge on deformable registration, which is the pre-requisite step of multi-atlas-based automatic segmentation (MABAS), thus set a ceiling for the accuracy of MABAS. In comparison, deep learning-based methods, a new branch of machine learning with greater modeling complexity and trainable to vast amount of data, are good at automatic feature extraction from image data, and are capable of directly learning from the imaging data in an end-to-end manner. Benefiting from the development of advanced computer hardware as well as accumulatively increasing clinical data for model training, deep learning-based automatic segmentation has gradually outperformed previous methods and become the state-of-the-art methods in automatic segmentation field. This trend can be observed in Figure 1, which shows how the number of deep learning-based papers for automatic segmentation in radiotherapy has increased strongly in the last years.
Figure 1.
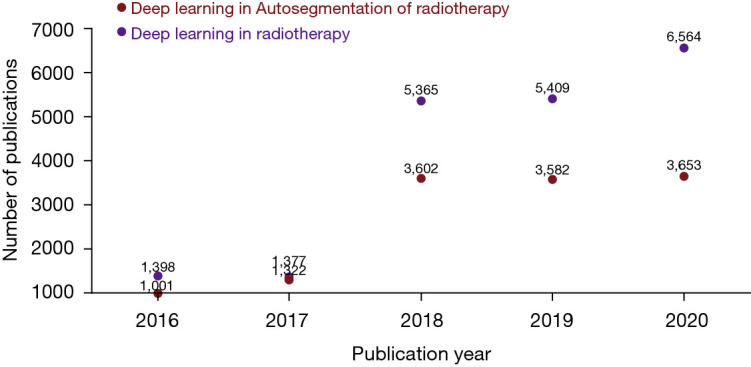
Overview of numbers of papers published from 2016 to 2020 regarding deep learning-based methods for radiotherapy applications and deep learning-based automatic segmentation applications for radiotherapy.
In this paper, we provide an overview of state-of-the-art deep learning techniques for target volume automatic segmentation in three commonly used imaging modalities in radiotherapy (i.e., CT, MRI, PET), and discuss the advantages and remaining challenges of current deep learning-based target volume segmentation methods that hinder widespread clinical deployment. To our knowledge, there have been several papers that reviewed the general applications of deep learning in radiotherapy (10-12), however, none of them has provided a systematic overview focused on the automatic segmentation of target volumes. This review paper aims at providing a comprehensive overview from the debut to the state-of-the-art of deep learning algorithms, focusing on a variety of target volume segmentation in different clinical sites and multi-modality imaging.
To identify related studies, search engines like PubMed were queried for papers containing (“deep learning”) and (“target volume”) and (“delineation”) or (“segmentation”) in the title or abstract. Additionally, for conference proceedings such as MICCAI and ISBI were searched separately based on the titles of papers. The last update to the included papers was on Feb 5th, 2021.
Fundamentals of deep learning methods
Architectures
In this section, we first introduce basic deep neural network architectures and then briefly introduce variation models with building blocks that are commonly used to boost the ability of the networks to perform automatic delineation of radiotherapy target volumes.
Convolutional neural networks (ConvNets)
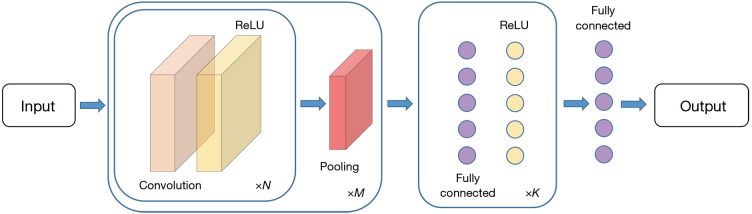
The typical structure of ConvNets usually consists of: Convolutional layers, Pooling layers and Fully Connected layers. As indicated by Karpathy et al. (13), a common pattern of ConvNets can be visualized as in Figure 2.
Figure 2.
An illustration of the typical ConvNets architecture. ×N, ×M, and ×K indicate repetitions of the corresponding structure.
The convolutional layers take the original input image and extract feature maps from the image by convolving the input image with a series of kernels (also known as filters or features). For an m × n kernel K and a 2D input image I, the output unit after convolution equals to
| [1] |
In modern ConvNets architectures, element wise non-linear transformation is applied after each convolution operation, and the default recommendation is to use Rectified Linear Unit (ReLU) (14,15) defined as g(z)=max(0,z) to threshold at zero. Therefore, the (i,j) element of the output feature map, after applying a non-linear transformation ReLU and adding a bias B(i,j), equals to
| [2] |
The values of each kernel is learned by the ConvNets during the training process, and generate lower-level features focusing on local details, which aggregating together into higher-level features corresponding to complex and abstract human visual concepts. The size of the output feature map is controlled by three hyper-parameters: (I) depth d, corresponds to the number of different kernels applied to the input image; (II) stride s, indicates the number of pixels that are jumped every time the kernel slides over the input image; (III) zero-padding p, refers to the amount of zeros padded around the input image border that help control the output feature map size. Assuming the input image size is I, and the kernel size is K, the output size of the feature map stack is given by F=[(1−K+2p)/s+1]×d.
The pooling layers takes a large feature map generated by the convolutional layer and shrinks its dimension by replacing each value in the feature map with the statistics of the nearby outputs, such as the maximum, or the sum the average. One of the most commonly used pooling methods is max pooling, which moves a window in steps across the feature map and extract the largest value within the window. While introducing pooling layers to ConvNets can improve the invariance of local deviations, reduce the amount of parameters to be computed and control over fitting, it can also lead to problems such as losing pose and orientation information in deeper layers, as well as complicating the architectures that use top-down design (16). Some researchers have proposed alternative methods to replace the pooling layers in the ConvNets: Springenberg et al. (17) suggested competitive performance of object recognition can be accomplished by using a ConvNets composed solely of repeated convolutional layers with greater strides. In a latest architecture CapsNet proposed by Sabour et al. (18), the dynamic routing mechanism that generates a parent capsule and calculates the output from grouped capsules, replaces the max pooling in typical ConvNets by applying the dynamic routing on the capsule outputs to ensure the feature representations are equivariant.
Different from Convolutional layers and Pooling layers that preserve the spatial structure of an image, a Fully Connected layer can be considered as a one-dimensional list that connects with every single neuron, i.e., the activation function in the previous layer which is calculated by matrix multiplication followed by a bias offset.
Although the major applications of ConvNets is in image classification, it can also be employed in target volume delineation tasks without major revisions to the architecture. However, this requires extracting patches from the original image and then classify the pixel at the center of each patch. Major drawbacks associated with this sliding-window method include (I) redundant computation caused by repetitive convolutions of highly overlapped patches and (II) inability for ConvNets to learn global features due to the small patch size and (III) being applicable only for binary segmentation task while fully connected layers exist. As a result, an end-to-end segmentation network at pixel level, namely, fully convolutional neural network (FCN) is more commonly used, and will be discussed in the next section.
FCN
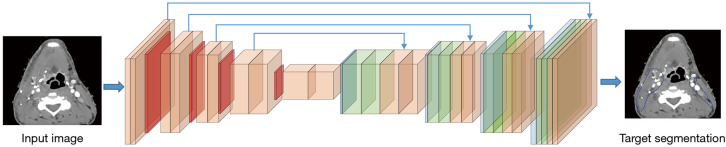
In 2014, Long et al. (19) proposed their FCN for dense predictions. With fully connected layers replaced by convolutions, the resulting FCN can use existing classification ConvNets to learn the hierarchies of features and output feature maps in a more efficient manner compared to the sliding-window method. The FCN architecture proposed by Long et al. is considered a milestone of applying ConvNets in semantic segmentation and most state-of-the-art segmentation neural networks have adopted this paradigm (20). A classic example of FCN was shown in Figure 3.
Figure 3.
A classic example of FCN-based target segmentation. The model takes the whole image as input through encoder, extracts feature maps through a series of deconvolution layers in the decoder, and eventually generates the probability maps for multiple target classes, where each pixel is assigned to the class with the highest probability.
Apart from fully connected layers, pooling layers also hinder the direct use of ConvNets in segmentation tasks. In ConvNets, pooling layers are used to increase the field of view and prevent overwhelm of system memory while reducing the feature map resolution and discarding the spatial information. From the aspect of segmentation, however, this is unfavorable since segmentation tasks require accurate alignment of classification map, hence the need to preserve spatial information. Various methods have been developed to tackle the challenge of preserving spatial information while reducing the resolution of feature maps, leading to multiple variants of the initial FCN architecture.
In general, all the FCN-based architectures employ classification ConvNets, with their fully connected layers replaced by convolutions, and produce low-resolution feature maps. This process is usually named Encoder. Decoder then refers to the process of mapping the low-resolution feature maps to the pixel-wise prediction in terms of segmentation. As above mentioned, variant architectures of FCN usually diverge at this point: Long et al. (19) utilized backward-strided convolution (or well-known as deconvolution) to up-sample the feature maps by fusing features from different coarseness and produce dense per-pixel outputs with the original image size. Ronneberger et al. (21) pushed one step further and proposed U-net architecture, which up-sampled the feature maps by connecting the encoder and the decoder at every stage. Although not the first paper proposing up-sampling method through learned deconvolutions, an important contribution by Ronneberger et al. (21) is introducing the skip-connections that directly forward the feature maps computed in each encoding stage to each decoding stage, allowing the decoder of each stage to learn relevant features that will be lost when pooled by the encoder. The original U-net architecture is 2D ConvNets, and it was later expanded to 3D architecture by Çiçek et al. (22) Furthermore, Milletari et al. (23) modified the original U-Net (known as V-Net) by introducing ResNet-like (24) residual blocks and replace the original cross-entropy objective function with dice similarity coefficients. Despite architecture variations, Isensee et al. (25) proposed “no new net” (nnU-Net) to show that the basic U-Net can still outperform many recently proposed methods by autonomous transforms to different datasets and segmentation tasks.
As an alternative to the encoder-decoder design, another way of restoring spatial information in the image is Dilated Convolutions. The main idea of dilated convolutions is to insert spaces between each convolution filter cell; in other words, dilated convolution is like a usual convolution but with up-sampled filters, and it is useful to expand the field of view exponentially without increasing parameters or computation time. Chen et al. (26) and Yu et al. (27) firstly combined the dilated convolutions with semantic segmentation. Specifically, the dilated convolutional layers with increasing dilation rates were inserted to replace the last several pooling and convolutional layers and to output dense predictions.
Model enhancement techniques
There are several techniques in the literatures that have been used to enhance classic FCN architectures (e.g. U-Net, ResNet) for more accurate target volume segmentation: (I) multi-branch inputs: allows additional information from other imaging modalities to be fused with the planning images to propagate feature extraction (28,29); (II) advanced convolution kernels: examples include using dilated kernel that performs convolution with a wider stride to expand the receptive field in the feature extraction process (30); (III) inter-channel connections: examples include using (i) residual connections that connect outputs from previous layer to the feature map generated by the current layer to boost feature reusability and thereby increase the depth of the network (31,32), (ii) dense connections that link all the outputs from previous layers to the feature map generated by the current layer to boost feature exploration (33,34).
Deep learning model training
Common loss functions
Given the class imbalance issue is usually associated with target volume automatic segmentation, cross entropy (CE) is one of the most common loss functions being used in many studies. In particular, the cross-entropy loss for target volume segmentation summarizes pixel-wise probability errors between a predicted output and its corresponding ground truth label map for each class c through the total number of data samples N:
| [3] |
where c=0 accounts for the background class and 1≤c≤C accounts for all target volume foreground classes.
Another classic loss function that is frequently used in image segmentation tasks is soft-Dice loss, which penalizes the misalignments between a predicted probability map and its ground truth label map at the pixel-wise level:
| [4] |
Evaluation metrics
To quantitatively evaluate the performance of automatic target segmentation methods, there are three types of metrics commonly used: (I) volume-based metrics (e.g., Dice similarity coefficient, Jaccard similarity index), (II) distance-based metrics (e.g., surface distance, Hausdorff distance), (III) comparisons with expert variability (e.g., inter- and intra-observer variabilities).
The Dice similarity coefficient evaluates the ratio of overlap between the automatic segmentation and the ground truth contours:
| [5] |
where A stands for Auto-segmented contours, G represents the Ground-truth contours, TP is the true positive rate, FP is the false positive rate and FN is the false negative rate.
Amongst distance-based evaluation metrics, mean surface distance and Hausdorff distance are commonly used to find the maximum distance of all voxels on the automatically segmented target surface to the ground truth contour surface, where the mean surface distance is defined as:
| [6] |
where marks the distance from the current voxel to the closest voxel on ground truth contours first proposed by Gerig et al. (35).
Hausdorff distance is another variation of surface distance metric, in which it accounts for the maximum distance of all voxels along the segmentation surface:
| [7] |
Often the accuracy of automatic segmentation were compared with variability among expert human operators. The rational for such comparison was to provide a fair evaluation of the automatic segmentation results, because in the case that the training data is comprised of inputs from various observers, the output of the model is expected to reflect that variability as well. There are two major types of observer variability: Inter-observer variability, which is quantified by the differences among contours delineated by multiple observers on the same target volume; and intra-observer variability, which is quantified by the differences among contours delineated by a single observer on the same target volume at several different tries, usually longitudinally at different time points.
It is worth noting even though the segmentation accuracies of different methods were presented in this paper, they are not directly comparable unless these methods are evaluated on the same dataset. This is because, even for the same target volume segmentation task, the intrinsic characteristics of datasets can vary across imaging modalities, acquisition protocols, and patient populations—each of these variations can lead to significant effects on the data distribution and result in different model performances.
Deep learning for target volume delineation
CT-based target volume segmentation
CT is a routine technique that is used for target definition and treatment planning in radiotherapy. There are two main imaging modalities: non-contrast CT imaging and contrast-enhanced CT. Non-contrast CT utilizes the variation in density of tissues to generate the image, such that different densities using various attenuation values can be easily distinguished, and is generally the standard of clinical practice for treatment planning. In comparison, contrast-enhanced CT is acquired after the injection of a contrast agent, which provides better visualization of lymph nodes and vessels, has shown to be helpful in target volume definition (36,37). In the following sections, we will review some of the most commonly used deep-learning-based target delineation methodologies using CT images as input.
FCN-based segmentation
Men et al. (30) were among the first ones to apply a FCN to segment the Clinical Target Volume (CTV) on non-contrast CT images of rectal cancer. Their end-to-end FCN approach enhanced by dilated convolution channels achieved competitive segmentation performance, significantly outperforming traditional non-deep-learning-based methods in terms of both speed and accuracy. In the following years, multiple studies based on FCNs have been proposed, aiming at achieving further improvements in automatic target delineation performance. In this regard, one stream of work focuses on modifying the network architecture to enhance the feature extraction capacity - for example, Men et al. (38) modified the structure of VGG-16 with reversed deconvolutions at decoder level to re-establish the feature maps to achieve pixel-wise segmentation of gross tumor volume (GTV) and CTV segmentation in nasopharyngeal cancer. Schreier et al. (39) modified the U-Net architecture by adding interconnected channels between high- and low-resolution feature maps. Cardenas et al. (40) and Wang et al. (41) investigated different loss functions, such as weighted cross-entropy and weighted dice loss to boost the target delineation performance. Initially, many FCN-based methods used 2D networks rather than 3D networks for segmentation. This is mainly due to the typical computational memory constraints on the hardware, which limits the applicability of 3D networks. Besides, training 2D networks requires less data, since each CT slice can be one sample. Li et al. (42) investigated a hybrid 2D/3D to re-use the 2D features map in a 3D architecture to take advantage of both intra- and inter-slice features extraction. As more datasets and hardware with larger memory becoming available, more 3D networks on target volume segmentation are expected.
Data pre-processing
Due to different protocols being used during patient simulation, CT images being input into FCN may have various contrasts, resolutions and image size, which make it difficult to standardize the network structure. To overcome these challenges, most studies employed data pre-processing as a pre-requisite step before the model training process. Two main concerns were addressed through the pre-processing step. (I) Homogenize the input image size. Schreier et al. (39) and Song et al. (31) chose to re-sample the breast imaging data from three institutions into a uniform size along both axial and longitudinal directions for the sake of easiness of model training and hardware memory consumption. Furthermore, Cardenas et al. (43) not only performed data re-sampling but also used anatomical landmarks in the cranial and caudal directions and body contour to help identify the region of interests. (II) Enhance contrast-to-noise ratio of the input image. Many classic image processing methods were fused with FCN through this process. For example, Men et al. (30) utilized contrast limited adaptive histogram equalization (CLAHE) (44) to improve the image contrast especially on the boundaries while eliminating the noise amplification in the homogeneous area. Pixel intensity normalization was also frequently used to remove contrast variations among images by different scanners (28,45).
Hybrid loss
Another problem that may limit the target delineation performance in both 2D and 3D FCNs is that they are typically only trained with pixel-wise loss functions, such as Cross Entropy and soft-Dice loss. These pixel-wise loss functions may not be sufficient to learn features that represent the underlying anatomical structures. Several approaches therefore focus on designing combined loss functions to address class imbalance issues and improve the predictive accuracy and robustness of the network. The anatomical constraints are represented as regularization terms to take into account the shape information (46) or contour and region information (45), encouraging the network to generate more anatomically plausible segmentations.
Other imaging modalities
Although CT is taken as the standard imaging technique during radiotherapy treatment planning, its application in target definition is sometimes constrained by the limited contrast on the boundary of tumor volumes. The pitfalls brought by the inherent characteristics of CT were usually addressed by associating with other imaging modalities such as magnetic resonance imaging (MRI), and positron emission tomography (PET). In MRI, by utilizing different sequences, it allows accurate quantification of functional and pathological tissue. PET, on the other hand, provides quantitative metabolic information by highlighting malignant regions with greater signal, thus helpful in characterizing lesions. Recently, there is an emerging trend of utilizing multi-modality imaging to train deep learning-based target delineation models. In the following sections, we will describe and discuss these methods in detail regarding different applications.
MRI-based target volume segmentation
The architectures utilizing MRI recently emerged in the target volume delineation, and are mainly focusing on the brain tumor treatment, where MRI is the dominant if not exclusive imaging modality. On other body sites, however, MRI-based target volume segmentation is only auxiliary due to MRI’s intrinsic limitations, including geometry distortion, filed of view limitation, and unstandardized intensity representation. For instance, Ermis et al. (33) adapted DenseNet (47), an encoder-decoder FCN composed of four levels connecting using skip connections, to perform resection cavity in glioblastoma patients using T1-weighted, T2-weighted and fluid-attenuated inversion recovery (FLAIR) MRI sequences. Peeken et al. (48) proposed a regression fully connected neural network that was capable of estimating tumor volume fraction based on diffusion signals, the model was then applied to T1-weighted (with and without contrast), T2-weighted and FLAIR MRIs to estimate the infiltrative GTV. More recently, one of the variations of FCN named U-net has become the method of choice for target volume segmentation: He et al. (24) modified the 3D U-Net (22) with dual contraction paths for nasopharyngeal carcinoma segmentation. Lin et al. (32) also worked on nasopharyngeal carcinoma target volume segmentation, but utilized a different 3D encoding-decoding FCN, VoxelResNet (49), to do the work.
PET-based target volume segmentation
Different from MRI that could function as an independent imaging modality in the treatment planning of radiotherapy, PET images almost always need to be registered with CT due to the limited resolution and the lack of density conversion table. Depending on whether the PET images were acquired at the same time of patient CT simulation, there may be substantial misalignments between PET and CT acquired at two different times and with different patient positioning setup. To solve this issue, Jin et al. (29) proposed an architecture that firstly performed automatic image registration between PET and CT, and then trained a two-channel encoding-decoding network, one channel with CT only and the other channel with both CT and registered PET-CT. These two results from separate channels were then fused through late fusion to generate the final prediction. The model has been developed to tackle the challenging problem of esophageal cancer tumor delineation and achieved promising performance compared to clinical contours (50). Guo et al. (28), on the other hand, used the PET scans acquired at the same time as planning CT images, and used the fused images as input into a single-channel model to perform target volume delineation in head and neck cancer. Table 1 summarized some selective deep learning studies on target volume automatic segmentation with various imaging modalities and at different clinical sites. Several studies also employed inter-observer (quantifies the variations in contours made by multiple observers) or intra-observer variabilities (quantifies the variations in contours made by a single observer longitudinally across time) as a gauge of their auto-segmentation performance, which can provide a fair comparison of auto-segmentation versus human expert contours as most models are trained in a supervised fashion.
Table 1. A summary of target volume automatic segmentation accuracies in representative deep learning studies on various clinical sites and with multiple imaging modalities.
| Selected studies | Year | Network architecture | Imaging modalities | Clinical sites | No. of patients | Mean dice coefficient | |||
|---|---|---|---|---|---|---|---|---|---|
| Train/validation | Test | Auto | Observer variability | ||||||
| Men et al. (30) | 2017 | 2D DenseNet with multiple dilated convolution paths | CT | Rectal | 218 | 60 | 0.88 | N/A | |
| Cardenas et al. (42) | 2018 | 3D U-Net | CT | Oropharyngeal | 210 (3-fold CV) | 75 | 0.81 | 0.80 (1) | |
| Men et al. (38) | 2017 | 2D FCN with VGG-16 encoding structure | CT | Nasopharyngeal | 184 | 46 | 0.83 | 0.80 (1) | |
| Guo et al. (28) | 2019 | 3D DenseNet | PET/CT | Head and Neck | 175 | 75 | 0.73 | 0.80 (1) | |
| Jin et al. (50) | 2021 | Two-channel 3D progressive nested network with early and late fusions | PET/CT | Esophageal | 148 (4-fold CV) | N/A | 0.79 | N/A | |
| Ermis et al. (33) | 2020 | 2D DenseNet | MRI | GBM | 30 (6-fold CV) | N/A | 0.84 | 0.85 | |
| Lin et al. (32) | 2019 | 3D ResNet | MRI | Nasopharyngeal | 715 | 203 | 0.79 | 0.74 | |
| Cardenas et al. (45) | 2020 | 3D U-Net | CT | Head and neck | 71 | 32 | Up to 0.93 | Contours modified by physicians | |
| Men et al. (51) | 2018 | 2D ResNet | CT | Breast | 800 (5-fold CV) | N/A | 0.91 | N/A | |
| Bi et al. (52) | 2019 | 2D ResNet | CT | NSCLC | 200 | 50 | 0.75 | 0.72 | |
| Liu et al. (34) | 2020 | 2.5D U-Net | CT | Cervical | 210 (5-fold CV) | 27 | 0.86 | 0.65 (53) | |
CV, cross-validation; N/A, not applicable. FCN, fully convolutional neural network; NSCLC, non-small cell lung cancer; CT, computerized tomography.
Challenges and future work
Scarcity and inconsistency of supervised labels
One of the biggest challenges for deep learning models is the scarcity of high-quality annotated data. In this review, we found that the majority of studies used a supervised fashion to train their networks, which requires a large number of annotated images. As mentioned, annotating target volumes on treatment planning images is time consuming and often requires significant amounts of clinical expertise. To tackle this challenge, current studies either compromised the network structure with 2D fashion, or introduced data augmentation into the pipeline, which intends to boost the number of training samples and the variety of training images by artificially generating new samples from existing annotated data. Networks can be robust to simple variations, such as translation and rotation, with data augmentation. This method, however, still has its own limitations where the artificial augmentation may fail to reflect the real-world data distributions. On the other hand, previous studies have observed substantial differences in target volume delineation among multiple physicians mainly driven by clinical experience and expertise. These inconsistencies could be a major bottleneck of deploying and transferring the deep learning target delineation model from one institution to the other. In the future, building and setting up benchmark datasets at various clinical sites is essential to stimulate the research in this area and help standardize the model performance evaluation by different studies.
Incorporation of multi-modality clinical inputs
Different from OAR automatic segmentation, target volume delineation relies not only on treatment planning images, but also supportive information provided by functional imaging, microscopic involvement and clinical health record. The diversity of the input information drives the future development direction towards multi-modality imaging learning or even joint learning across heterogeneous data spaces (i.e., images and texts) (54).
Model generability across various machines and institutions
Another common limitation in deep learning-based target volume delineation is the lack of generalization power when presented with samples subject to different data distributions. In other words, deep learning models tend to be biased by their respective training data, thus limit the ability to deploy single-institution-trained models to other clinics with different delineation guidelines or protocols, patient population with different anatomies, and machines with different imaging parameters. Currently there is an emerging trend in radiology to build up large multi-vendor, multi-institution diagnostic datasets being annotated with heterogonous clinical practice (55). However, this approach may not scalable in radiation oncology, as the process of data annotation is much more involved in radiation therapy applications. Instead, some recent studies have started to investigate the use of unsupervised learning that aims at optimizing the model performance on a target dataset without additional labeling costs. Several works have successfully applied generative-adversarial learning to cross-modality OAR segmentation tasks, i.e., adapting the segmentation model trained on synthetic MR to aid segmentation tasks on CT (56,57).
Conclusions
In this review paper, we provided a comprehensive overview of deep learning-driven target volume segmentation using three common imaging modalities in radiation therapy (CT, MRI, PET), covering a wide range of existing deep learning approaches that are designed to segment CTVs and GTVs of various clinical sites. We also outlined the remaining challenges and discussed the future potential of these deep learning-based automatic segmentation methods. Regardless of the challenges facing down the road, the great potential of deep learning-based target volume automatic segmentation in efficiency and standardization enhancement of radiation therapy will keep the topic an active direction in research, product development, and clinical implementation.
Acknowledgments
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
Footnotes
Provenance and Peer Review: With the arrangement by the Guest Editors and the editorial office, this article has been reviewed by external peers.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/qims-21-168). The special issue “Artificial Intelligence for Image-guided Radiation Therapy” was commissioned by the editorial office without any funding or sponsorship. LD reports NIH grants for research in proton therapy and outcome studies, unrelated to this work. Sponsored research and honoraria from Varian Medical System. TL reports consulting fees, honoraria, and travel expenses from Varian Medical Systems unrelated to this work. Patent titled “Systems and methods for automatic, customized radiation treatment plan generation for cancer” was filed. The authors have no other conflicts of interest to declare.
References
- 1.Wong J, Fong A, McVicar N, Smith S, Giambattista J, Wells D, Kolbeck C, Giambattista J, Gondara L, Alexander A. Comparing deep learning-based auto-segmentation of organs at risk and clinical target volumes to expert inter-observer variability in radiotherapy planning. Radiother Oncol 2020;144:152-8. 10.1016/j.radonc.2019.10.019 [DOI] [PubMed] [Google Scholar]
- 2.Reed VK, Woodward WA, Zhang L, Strom EA, Perkins GH, Tereffe W, Oh JL, Yu TK, Bedrosian I, Whitman GJ, Buchholz TA, Dong L. Automatic segmentation of whole breast using atlas approach and deformable image registration. Int J Radiat Oncol Biol Phys 2009;73:1493-500. 10.1016/j.ijrobp.2008.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou R, Liao Z, Pan T, Milgrom SA, Pinnix CC, Shi A, Tang L, Yang J, Liu Y, Gomez D, Nguyen QN, Dabaja BS, Court L, Yang J. Cardiac atlas development and validation for automatic segmentation of cardiac substructures. Radiother Oncol 2017;122:66-71. 10.1016/j.radonc.2016.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glide-Hurst CK, Lee P, Yock AD, Olsen JR, Cao M, Siddiqui F, Parker W, Doemer A, Rong Y, Kishan AU, Benedict SH, Li XA, Erickson BA, Sohn JW, Xiao Y, Wuthrick E. Adaptive Radiation Therapy (ART) Strategies and Technical Considerations: A State of the ART Review From NRG Oncology. Int J Radiat Oncol Biol Phys 2021;109:1054-75. 10.1016/j.ijrobp.2020.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ford E, Conroy L, Dong L, de Los Santos LF, Greener A, Gwe-Ya Kim G, Johnson J, Johnson P, Mechalakos JG, Napolitano B, Parker S, Schofield D, Smith K, Yorke E, Wells M. Strategies for effective physics plan and chart review in radiation therapy: Report of AAPM Task Group 275. Med Phys 2020;47:e236-72. 10.1002/mp.14030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anders LC, Stieler F, Siebenlist K, Schäfer J, Lohr F, Wenz F. Performance of an atlas-based autosegmentation software for delineation of target volumes for radiotherapy of breast and anorectal cancer. Radiother Oncol 2012;102:68-73. 10.1016/j.radonc.2011.08.043 [DOI] [PubMed] [Google Scholar]
- 7.Yang J, Beadle BM, Garden AS, Gunn B, Rosenthal D, Ang K, Frank S, Williamson R, Balter P, Court L, Dong L. Auto-segmentation of low-risk clinical target volume for head and neck radiation therapy. Pract Radiat Oncol 2014;4:e31-7. 10.1016/j.prro.2013.03.003 [DOI] [PubMed] [Google Scholar]
- 8.Stapleford LJ, Lawson JD, Perkins C, Edelman S, Davis L, McDonald MW, Waller A, Schreibmann E, Fox T. Evaluation of automatic atlas-based lymph node segmentation for head-and-neck cancer. Int J Radiat Oncol Biol Phys 2010;77:959-66. 10.1016/j.ijrobp.2009.09.023 [DOI] [PubMed] [Google Scholar]
- 9.Young AV, Wortham A, Wernick I, Evans A, Ennis RD. Atlas-based segmentation improves consistency and decreases time required for contouring postoperative endometrial cancer nodal volumes. Int J Radiat Oncol Biol Phys 2011;79:943-7. 10.1016/j.ijrobp.2010.04.063 [DOI] [PubMed] [Google Scholar]
- 10.Cardenas CE, Yang J, Anderson BM, Court LE, Brock KB. Advances in Auto-Segmentation. Semin Radiat Oncol 2019;29:185-97. 10.1016/j.semradonc.2019.02.001 [DOI] [PubMed] [Google Scholar]
- 11.Meyer P, Noblet V, Mazzara C, Lallement A. Survey on deep learning for radiotherapy. Comput Biol Med 2018;98:126-46. 10.1016/j.compbiomed.2018.05.018 [DOI] [PubMed] [Google Scholar]
- 12.Boldrini L, Bibault JE, Masciocchi C, Shen Y, Bittner MI. Deep Learning: A Review for the Radiation Oncologist. Front Oncol 2019;9:977. 10.3389/fonc.2019.00977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karpathy A. Cs231n: Convolutional neural networks for visual recognition. Available online: http://cs231n.stanford.edu/2016/
- 14.Jarrett K, Kavukcuoglu K, LeCun Y, editors. What is the best multi-stage architecture for object recognition? Kyoto: 2009 IEEE 12th International Conference on Computer Vision, 2009. [Google Scholar]
- 15.Nair V, Hinton GE, editors. Rectified linear units improve restricted boltzmann machines. Haifa, Israel: 27th International Conference on Machine Learning (ICML-10), 2010. [Google Scholar]
- 16.Goodfellow I, Bengio Y, Courville A. Deep learning. Cambridge: MIT Press, 2016. [Google Scholar]
- 17.Springenberg JT, Dosovitskiy A, Brox T, Riedmiller M. "Striving for Simplicity: The All Convolutional Net." Available online: https://arxiv.org/pdf/1412.6806.pdf
- 18.Sabour S, Frosst N, Hinton GE, editors. Dynamic routing between capsules. Proceedings of the 2017 Neural Information Processing Systems, 2017. [Google Scholar]
- 19.Long J, Shelhamer E, Darrell T, editors. Fully convolutional networks for semantic segmentation. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2015. [DOI] [PubMed] [Google Scholar]
- 20.Garcia-Garcia A, Orts-Escolano S, Oprea S, Villena-Martinez V, Garcia-Rodriguez J. A Review on Deep Learning Techniques Applied to Semantic Segmentation. arXiv preprint arXiv:170406857 2017. Available online: https://arxiv.org/pdf/1704.06857.pdf
- 21.Ronneberger O, Fischer P, Brox T, editors. U-net: Convolutional networks for biomedical image segmentation. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, 2015. [Google Scholar]
- 22.Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O, editors. 3D U-Net: learning dense volumetric segmentation from sparse annotation. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, 2016. [Google Scholar]
- 23.Milletari F, Navab N, Ahmadi SA, editors. V-net: Fully convolutional neural networks for volumetric medical image segmentation. 2016 Fourth International Conference on 3D Vision (3DV), 2016:565-71. doi: 10.1109/3DV.2016.79. 10.1109/3DV.2016.79 [DOI] [Google Scholar]
- 24.He K, Zhang X, Ren S, Sun J, editors. Deep residual learning for image recognition. Available online: https://arxiv.org/abs/1512.03385 10.1109/CVPR.2016.90 [DOI]
- 25.Isensee F, Jaeger PF, Kohl SAA, Petersen J, Maier-Hein KH. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods 2021;18:203-11. 10.1038/s41592-020-01008-z [DOI] [PubMed] [Google Scholar]
- 26.Chen LC, Papandreou G, Kokkinos I, Murphy K, Yuille AL. Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. arXiv preprint arXiv:160600915 2016. Available online: https://arxiv.org/pdf/1606.00915.pdf [DOI] [PubMed]
- 27.Yu F, Koltun V. Multi-scale context aggregation by dilated convolutions. arXiv preprint arXiv:151107122 2015. Available online: https://arxiv.org/pdf/1511.07122.pdf
- 28.Guo Z, Guo N, Gong K, Zhong S, Li Q. Gross tumor volume segmentation for head and neck cancer radiotherapy using deep dense multi-modality network. Phys Med Biol 2019;64:205015. 10.1088/1361-6560/ab440d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin D, Guo D, Ho TY, Harrison AP, Xiao J, Tseng CK, Lu L, editors. Accurate esophageal gross tumor volume segmentation in pet/ct using two-stream chained 3d deep network fusion. Available online: https://arxiv.org/pdf/1909.01524.pdf
- 30.Men K, Dai J, Li Y. Automatic segmentation of the clinical target volume and organs at risk in the planning CT for rectal cancer using deep dilated convolutional neural networks. Med Phys 2017;44:6377-89. 10.1002/mp.12602 [DOI] [PubMed] [Google Scholar]
- 31.Song Y, Hu J, Wu Q, Xu F, Nie S, Zhao Y, Bai S, Yi Z. Automatic delineation of the clinical target volume and organs at risk by deep learning for rectal cancer postoperative radiotherapy. Radiother Oncol 2020;145:186-92. 10.1016/j.radonc.2020.01.020 [DOI] [PubMed] [Google Scholar]
- 32.Lin L, Dou Q, Jin YM, Zhou GQ, Tang YQ, Chen WL, Su BA, Liu F, Tao CJ, Jiang N, Li JY, Tang LL, Xie CM, Huang SM, Ma J, Heng PA, Wee JTS, Chua MLK, Chen H, Sun Y. Deep Learning for Automated Contouring of Primary Tumor Volumes by MRI for Nasopharyngeal Carcinoma. Radiology 2019;291:677-86. 10.1148/radiol.2019182012 [DOI] [PubMed] [Google Scholar]
- 33.Ermiş E, Jungo A, Poel R, Blatti-Moreno M, Meier R, Knecht U, Aebersold DM, Fix MK, Manser P, Reyes M, Herrmann E. Fully automated brain resection cavity delineation for radiation target volume definition in glioblastoma patients using deep learning. Radiat Oncol 2020;15:100. 10.1186/s13014-020-01553-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Z, Liu X, Guan H, Zhen H, Sun Y, Chen Q, Chen Y, Wang S, Qiu J. Development and validation of a deep learning algorithm for auto-delineation of clinical target volume and organs at risk in cervical cancer radiotherapy. Radiother Oncol 2020;153:172-9. 10.1016/j.radonc.2020.09.060 [DOI] [PubMed] [Google Scholar]
- 35.Gerig G, Jomier M, Chakos M, editors. Valmet: A new validation tool for assessing and improving 3D object segmentation. Utrecht, Netherlands: 4th International Conference on Medical Image Computing and Computer-Assisted Intervention, MICCAI 2001. Springer Verlag, 2001. [Google Scholar]
- 36.Rodrigues RS, Bozza FA, Christian PE, Hoffman JM, Butterfield RI, Christensen CR, Heilbrun M, Wiggins RH, 3rd, Hunt JP, Bentz BG, Hitchcock YJ, Morton KA. Comparison of whole-body PET/CT, dedicated high-resolution head and neck PET/CT, and contrast-enhanced CT in preoperative staging of clinically M0 squamous cell carcinoma of the head and neck. J Nucl Med 2009;50:1205-13. 10.2967/jnumed.109.062075 [DOI] [PubMed] [Google Scholar]
- 37.Choi Y, Kim JK, Lee HS, Hur WJ, Hong YS, Park S, Ahn K, Cho H. Influence of intravenous contrast agent on dose calculations of intensity modulated radiation therapy plans for head and neck cancer. Radiother Oncol 2006;81:158-62. 10.1016/j.radonc.2006.09.010 [DOI] [PubMed] [Google Scholar]
- 38.Men K, Chen X, Zhang Y, Zhang T, Dai J, Yi J, Li Y. Deep Deconvolutional Neural Network for Target Segmentation of Nasopharyngeal Cancer in Planning Computed Tomography Images. Front Oncol 2017;7:315. 10.3389/fonc.2017.00315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schreier J, Attanasi F, Laaksonen H. A Full-Image Deep Segmenter for CT Images in Breast Cancer Radiotherapy Treatment. Front Oncol 2019;9:677. 10.3389/fonc.2019.00677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cardenas CE, McCarroll RE, Court LE, Elgohari BA, Elhalawani H, Fuller CD, Kamal MJ, Meheissen MAM, Mohamed ASR, Rao A, Williams B, Wong A, Yang J, Aristophanous M. Deep Learning Algorithm for Auto-Delineation of High-Risk Oropharyngeal Clinical Target Volumes With Built-In Dice Similarity Coefficient Parameter Optimization Function. Int J Radiat Oncol Biol Phys 2018;101:468-78. 10.1016/j.ijrobp.2018.01.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang T, Lei Y, Tian S, Jiang X, Zhou J, Liu T, Dresser S, Curran WJ, Shu HK, Yang X. Learning-based automatic segmentation of arteriovenous malformations on contrast CT images in brain stereotactic radiosurgery. Med Phys 2019;46:3133-41. 10.1002/mp.13560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li X, Chen H, Qi X, Dou Q, Fu CW, Heng PA. H-DenseUNet: Hybrid Densely Connected UNet for Liver and Tumor Segmentation From CT Volumes. IEEE Trans Med Imaging 2018;37:2663-74. 10.1109/TMI.2018.2845918 [DOI] [PubMed] [Google Scholar]
- 43.Cardenas CE, Anderson BM, Aristophanous M, Yang J, Rhee DJ, McCarroll RE, Mohamed ASR, Kamal M, Elgohari BA, Elhalawani HM, Fuller CD, Rao A, Garden AS, Court LE. Auto-delineation of oropharyngeal clinical target volumes using 3D convolutional neural networks. Phys Med Biol 2018;63:215026. 10.1088/1361-6560/aae8a9 [DOI] [PubMed] [Google Scholar]
- 44.Pizer SM, Amburn EP, Austin JD, Cromartie R, Geselowitz A, Greer T, ter Haar Romeny B, Zimmerman JB, Zuiderveld K. Adaptive histogram equalization and its variations. Computer vision, graphics, and image processing 1987;39:355-68. Available online: https://www.cs.unc.edu/Research/Image/MIDAG/pubs/papers/Adaptive%20Histogram%20Equalization%20and%20Its%20Variations.pdf 10.1016/S0734-189X(87)80186-X [DOI] [Google Scholar]
- 45.Cardenas CE, Beadle BM, Garden AS, Skinner HD, Yang J, Rhee DJ, McCarroll RE, Netherton TJ, Gay SS, Zhang L, Court LE. Generating High-Quality Lymph Node Clinical Target Volumes for Head and Neck Cancer Radiation Therapy Using a Fully Automated Deep Learning-Based Approach. Int J Radiat Oncol Biol Phys 2021;109:801-12. 10.1016/j.ijrobp.2020.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tong N, Gou S, Yang S, Ruan D, Sheng K. Fully automatic multi-organ segmentation for head and neck cancer radiotherapy using shape representation model constrained fully convolutional neural networks. Med Phys 2018;45:4558-67. 10.1002/mp.13147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang G, Liu Z, Van Der Maaten L, Weinberger KQ, editors. Densely connected convolutional networks. Available online: https://arxiv.org/pdf/1608.06993.pdf
- 48.Peeken JC, Molina-Romero M, Diehl C, Menze BH, Straube C, Meyer B, Zimmer C, Wiestler B, Combs SE. Deep learning derived tumor infiltration maps for personalized target definition in Glioblastoma radiotherapy. Radiother Oncol 2019;138:166-72. 10.1016/j.radonc.2019.06.031 [DOI] [PubMed] [Google Scholar]
- 49.Chen H, Dou Q, Yu L, Qin J, Heng PA. VoxResNet: Deep voxelwise residual networks for brain segmentation from 3D MR images. Neuroimage 2018;170:446-55. 10.1016/j.neuroimage.2017.04.041 [DOI] [PubMed] [Google Scholar]
- 50.Jin D, Guo D, Ho TY, Harrison AP, Xiao J, Tseng CK, Lu L. DeepTarget: Gross tumor and clinical target volume segmentation in esophageal cancer radiotherapy. Med Image Anal 2021;68:101909. 10.1016/j.media.2020.101909 [DOI] [PubMed] [Google Scholar]
- 51.Men K, Zhang T, Chen X, Chen B, Tang Y, Wang S, Li Y, Dai J. Fully automatic and robust segmentation of the clinical target volume for radiotherapy of breast cancer using big data and deep learning. Phys Med 2018;50:13-9. 10.1016/j.ejmp.2018.05.006 [DOI] [PubMed] [Google Scholar]
- 52.Bi N, Wang J, Zhang T, Chen X, Xia W, Miao J, Xu K, Wu L, Fan Q, Wang L, Li Y, Zhou Z, Dai J. Deep Learning Improved Clinical Target Volume Contouring Quality and Efficiency for Postoperative Radiation Therapy in Non-small Cell Lung Cancer. Front Oncol 2019;9:1192. 10.3389/fonc.2019.01192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eminowicz G, McCormack M. Variability of clinical target volume delineation for definitive radiotherapy in cervix cancer. Radiother Oncol 2015;117:542-7. 10.1016/j.radonc.2015.10.007 [DOI] [PubMed] [Google Scholar]
- 54.Wang X, Peng Y, Lu L, Lu Z, Summers RM, editors. Tienet: Text-image embedding network for common thorax disease classification and reporting in chest x-rays. Available online: https://arxiv.org/pdf/1801.04334.pdf
- 55.Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM, editors. Chestx-ray8: Hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. Available online: https://arxiv.org/pdf/1705.02315.pdf
- 56.Liu Y, Lei Y, Fu Y, Wang T, Zhou J, Jiang X, McDonald M, Beitler JJ, Curran WJ, Liu T, Yang X. Head and neck multi-organ auto-segmentation on CT images aided by synthetic MRI. Med Phys 2020;47:4294-302. 10.1002/mp.14378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dong X, Lei Y, Tian S, Wang T, Patel P, Curran WJ, Jani AB, Liu T, Yang X. Synthetic MRI-aided multi-organ segmentation on male pelvic CT using cycle consistent deep attention network. Radiother Oncol 2019;141:192-9. 10.1016/j.radonc.2019.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]