Abstract
The SGS1 gene of Saccharomyces cerevisiae is a homologue for the Bloom's syndrome and Werner's syndrome genes. The disruption of the SGS1 gene resulted in very poor sporulation, and the majority of the cells were arrested at the mononucleated stage. The recombination frequency measured by a return-to-growth assay was reduced considerably in sgs1 disruptants. However, double-strand break formation, which is a key event in the initiation of meiotic DNA recombination, occurred; crossover and noncrossover products were observed in the disruptants, although the amounts of these products were slightly decreased compared with those in wild-type cells. The spores produced by sgs1 disruptants showed relatively high viability. The sgs1 spo13 double disruptants sporulated poorly, like the sgs1 disruptants, but spore viability was reduced much more than with either sgs1 or spo13 single disruptants. Disruption of the RED1 or RAD17 gene partially alleviated the poor-sporulation phenotype of sgs1 disruptants, indicating that portions of the population of sgs1 disruptants are blocked by the meiotic checkpoint. The poor sporulation of sgs1 disruptants was complemented with a mutated SGS1 gene encoding a protein lacking DNA helicase activity; however, the mutated gene could suppress neither the sensitivity of sgs1 disruptants to methyl methanesulfonate and hydroxyurea nor the mitotic hyperrecombination phenotype of sgs1 disruptants.
Proteins having DNA helicase activity play important roles in many processes involving DNA, such as replication, repair, and recombination. The product of the Escherichia coli recQ gene, which has DNA helicase activity, is a member of the RecF pathway of recombination. The recF mutants lack conjugal recombination proficiency and UV resistance in the background of recBCD (lacking active endonuclease V) and sbcBC (lacking active exonuclease I), and recQ deletion mutants in the background of recBC sbcBC display UV and methyl methanesulfonate (MMS) sensitivity (22, 30).
We (38) and Puranam and Blackshear (32) cloned cDNAs encoding a RecQ homologue of human cells, DNA helicase Q1 and RECQL, respectively. Since these are the same gene, we tentatively designated this gene RECQL1. We also cloned a gene of Saccharomyces cerevisiae encoding a protein having DNA helicase motifs with high homology to those of E. coli RecQ and human ATPase RECQL1. This gene soon was found to be identical to the SGS1 (slow growth suppressor 1) gene.
A mutant allele of the SGS1 gene was identified as a suppressor of the slow-growth phenotype of top3 mutants (11). Two-hybrid experiments indicated that the yeast Sgs1 protein interacts with DNA topoisomerase III (Top3) (11) as well as DNA topoisomerase II (50). The protein encoded by the SGS1 gene has seven conserved helicase motifs, and Sgs1 was shown to actually have DNA helicase activity (3, 23). Deletion mutants of the SGS1 gene showed a reduction in the fidelity of chromosome segregation during mitosis and meiosis (50, 51); mitotic hyperrecombination phenotypes in interchromosomal homologous recombination, intrachromosomal excisional recombination, ectopic recombination (51), unequal sister chromatid recombination (31), and illegitimate recombination (53); and premature aging (44). The sgs1 mutants were shown to be moderately sensitive to MMS (10) and hydroxyurea (HU) (53) but not to ionizing radiation or UV light (51).
Four human genes encoding a RecQ homologue have been identified in addition to RECQL1. These are the Bloom's syndrome (BLM) gene (8), the Werner's syndrome (WRN) gene (54), RECQL4 (19, 20), and RECQL5 (19). The representative clinical manifestations of Bloom's syndrome (BS) are cancer predisposition, immunodeficiency, and male infertility (13, 16). In BS cells, the interchanges between homologous chromosomes are increased and an abnormally large number of sister chromatid exchanges are present (13). Werner's syndrome (WS) patients prematurely develop a variety of major age-related diseases, such as arteriosclerosis, malignant neoplasms, melituria, and cataracts (9). The cells derived from WS patients show chromosome instability and a shorter life span in cultures (28). However, it remains unclear how the dysfunction of the gene products is related to the observed phenotypes of cells derived from these patients.
To date, a number of RecQ homologues have been reported from prokaryotes, such as Bacillus subtilis (L47648 in GenBank) and Haemophilus influenzae (HI32756 in GenBank); eukaryotes, such as S. pombe (45) and S. cerevisiae; and higher eukaryotes, including humans. Although significant homology is present within the consensus helicase domains, these RecQ homologues are classified into two groups, according to size. One group includes prokaryotic RecQ homologues, E. coli RecQ, human RecQL1, and RecQL5, which consist of about 600 to 650 amino acids; the other group includes Sgs1, Hus2/Rqh1/Rad12 of S. pombe, RecQL4, BLM, and WRN, consisting of about 1,400 amino acids. The latter RecQ homologues have a highly charged N- or C-terminal domain (19). A search of the entire genome of S. cerevisiae revealed that SGS1 is the sole homologue of recQ in S. cerevisiae. Thus, it seems likely that SGS1 is a functional homologue of one or several human RECQ genes.
To clarify the functions of Sgs1 and to obtain insight into the functions of BLM and WRN, we analyzed in detail the cause of the poor-sporulation phenotype of sgs1 disruptants in relation to meiotic processes, including meiotic recombination.
MATERIALS AND METHODS
Yeast strains and plasmids.
The origins and relevant genotypes of the strains used are listed in Table 1. The strains designated MR966 and MR93-28C (33), NKY1303 and NKY1543 (46), and S754 and S756 (rad50S) (4) are SK1 derivatives. Yeast manipulations were carried out as described by Sherman et al. (42). Plasmids were constructed by standard procedures (37). The full-length SGS1 gene was isolated by PCR using a genomic DNA isolated from strain W303. PCR products were cloned into pBluescript SK(+). pYCp1305 contains the full-length SGS1 gene (nucleotides [nt] −207 to 5558, from the XhoI site to the SacI site) of the YCp vector, pRS314, which includes a centromere element, an autonomously replicating sequence, and a TRP1 marker (43).
TABLE 1.
Experimental strains of S. cerevisiae
| Strain | Genotype | Reference or source |
|---|---|---|
| W303-1A | MATa ura3-1 leu2-3,112 trp1-1 his3-11 ade2oc can1-100 Gal+ | Thomas and Rothstein (48) |
| WQ701 | MATa/MATα W303-1A but sgs1::URA3 | This study |
| YPH499 | MATa/MATα ade2-100oc leu2Δ his3Δ200 lys2-801am ura3-52 trp1Δ63 Gal+ | Sikorski and Hieter (43) |
| YQ401 | MATa/MATα YPH499 but sgs1::URA3 | This study |
| MR966 | MATa/MATα ura3-52 leu2-3,112 trp1-289 his1-7 | Resnick et al. (33) |
| MR93-28C | MATa/MATα ura3-52 leu2-3,112 trp1-289 his1-1 | Resnick et al. (33) |
| MR101 | MATa/MATα MR966/MR93-28c | This study |
| MR202 | MATa/MATα MR966 but sgs1::URA3/MR93-28c but sgs1::LEU2 | This study |
| MR301 (ds1) | MATa/MATα MR966 but sgs1Δ::AUR/MR93-28c but sgs1Δ::AUR | This study |
| ds13 | MATa/MATα MR966 but spo13::URA3/MR93-28c but spo13::URA3 | This study |
| ds1m11 | MATa/MATα MR966 but sgs1Δ::AUR mre11::URA3/MR93-28c but sgs1Δ::AUR mre11::URA3 | This study |
| ds1s13 | MATa/MATα MR966 but sgs1Δ::AUR spo13::URA3/MR93-28c but sgs1Δ::AUR spo13::URA3 | This study |
| ds13m11 | MATa/MATα MR966 but mre11::URA3 spo13::hisG/MR93-28c but mre11::URA3 spo13::hisG | This study |
| dr1 | MATa/MATα MR966 but red1::URA3/MR93-28c but red1::URA3 | This study |
| dr17 | MATa/MATα MR966 but rad17::LEU2/MR93-28c but rad17::LEU2 | This study |
| ds1r1 | MATa/MATα MR966 but sgs1Δ::AUR red1::URA3/MR93-28c but sgs1Δ::AUR red1::URA3 | This study |
| ds1r17 | MATa/MATα MR966 but sgs1Δ::AUR rad17::LEU2/MR93-28c but sgs1Δ::AUR rad17::LEU2 | This study |
| NKY1303 | MATa/MATα lys2 ho::LYS2 ura3 leu2::hisG arg4-bgl his4B::LEU2 | Storlazzi et al. (46) |
| NKY1543 | MATa/MATα lys1 ho::hisG ura3 leu2::hisG arg4-nsp his4X::LEU2 (BamHI)-URA3 | Storlazzi et al. (46) |
| RKH112 | MATa/MATα NKY1303 but sgs1::AUR | This study |
| RKH113 | MATa/MATα NKY1543 but sgs1::AUR | This study |
| NKY2846 | MATa/MATα NKY1303/NKY1543 | This study |
| RKH225 | MATa/MATα RKH112/RKH113 | This study |
| S754 | MATa/MATα ura3 lys2 ho::LYS2 leu2Δ (asp718-ecoRI) arg4Δ(eco47III-hpaI) his4::URA3-(arg4-bgl) rad50S-KI81::URA3 | Bishop et al. (4) |
| S756 | MATa/MATα ura3 lys2 ho::LYS2 leu2Δ(asp718-ecoRI) arg4Δ(eco47III-hpaI) his4::URA3-(arg4-nsp) rad50S-KI81::URA3 | Bishop et al. (4) |
| RKH110 | MATa/MATα S754 but sgs1::AUR | This study |
| RKH111 | MATa/MATα S756 but sgs1::AUR | This study |
| S1510 | MATa/MATα S754/S756 | This study |
| RKH221 | MATa/MATα RKH110/RKH11 | This study |
Gene disruption.
The SGS1 gene was disrupted by the one-step gene substitution method (36). A 0.7-kbp (nt 2258 to 3048) fragment containing URA3, LEU2, or AUR (for aureobasidin 1-C) at the StuI site in the middle of the helicase domain was introduced into desired strains by a conventional transformation method. The resultant transformants were selected on synthetic complete medium (SC) plates lacking uracil or leucine or on yeast extract-peptone-adenine-dextrose (YPAD) plates containing aureobasidin A. sgs1 null deletion disruptants (sgs1Δ::AUR) were made by replacing 4,365 bp of the SGS1 sequence (nt −207 to 4158, from the XhoI site to the EheI site) with the aureobasidin 1-C (AUR) gene. The resultant transformants were selected on YPAD plates including aureobasidin A. Plasmids pNKY58 (from N. Kleckner), HT16 (from H. Ogawa), pHSS6 (from R. E. Malone), and pWL8 (from T. Weinert) were used to generate spo13, mre11, red1, and rad17 disruptants, respectively. Gene disruption was confirmed by PCR or Southern blot analysis. Diploid sgs1 disruptants of W303-1A and YPH499 were constructed using the HO plasmid on WQ701 and YQ401, respectively. In the case of SK1 background strains, MR966, MR93-28C, NKY1303, NKY1543, S754, and S756, a and α haploid gene disruptants were constructed and then mated to make diploids.
Sporulation and return-to-growth assay.
For sporulation on plates, cells were incubated on yeast extract-peptone-dextrose (YPD) plates for 2 days and allowed to sporulate for 5 days on plates containing 1% potassium acetate (KAC plates). Sporulation was monitored with a phase-contrast microscope, and the percentage of cells forming asci was calculated. For sporulation in liquid medium, sporulation and the return-to-growth assay were performed essentially as described by Dykstra et al. (7). Cells were grown with vigorous aeration at 28°C in presporulation medium (SPS) containing 0.5% yeast extract, 1% peptone, 0.17% yeast nitrogen base without amino acids, 0.05 M potassium phthalate, 1% KAC, 0.5% ammonium sulfate, and the required amino acids. Cells were grown to a density of 2 × 107 to 5 × 107 cells/ml, washed twice with warmed 1% KAC, and suspended in warmed sporulation medium (SPM) (1% KAC, one-fifth the standard concentration of required amino acids, 0.005% Nonidet P-40) at a density of 2 × 107 cells/ml. Cells were incubated with vigorous shaking; the volume of the cell suspension was less than one-eighth the capacity of the flask to ensure good aeration. The frequency of recombination between the his1-1 and his1-7 alleles was examined at different times after the shifting to SPM by inoculating cells on SC plates lacking histidine [SC-His(−)] or containing histidine [SC-His(+)]. The recombination frequency was determined by comparing the number of colonies on SC-His(−) plates with the number of colonies on SC-His(+) plates.
Flow cytometry.
Cells were fixed with 70% ethanol, washed with 0.2 M Tris-HCl (pH 7.5), and exposed to 0.1 mg of RNase A per ml for 3 h at 37°C. Cells were washed with 0.2 M Tris-HCl (pH 7.5), stained with 0.1 mg of propidium iodide per ml for 15 min on ice, and analyzed with a FACScan/CellFIT system (Becton Dickinson).
Preparation of DNA and detection of DSBs or physical recombinants.
DNA was isolated from yeast cells using the Zymolyase method (14). For the detection of double-strand breaks (DSBs) in MR and rad50S strains, 5 μg of DNA samples was digested with XhoI and fractionated in an 0.8% agarose gel. Hybridization was performed using rapid-hyb buffer (Amersham) essentially as described by Sambrook et al. (37) and using the XhoI-NheI fragment from a plasmid having the YCR47C-YCR48W region in chromosome III (51a). The detection of physical recombinants of strains NKY2846 and RKH225 was performed essentially as described by Storlazzi et al. (46). For the detection of physical recombinants of RKH strains, 5 μg of DNA samples was digested with XhoI or XhoI-MluI and fractionated in a 0.6% agarose gel. Hybridization was performed using probe B (see Fig. 5A). Probes were labeled with 32P using a Rediprime random primer labeling kit (Amersham). Bands were visualized and quantified with a BAS-Mac system (Fuji Film).
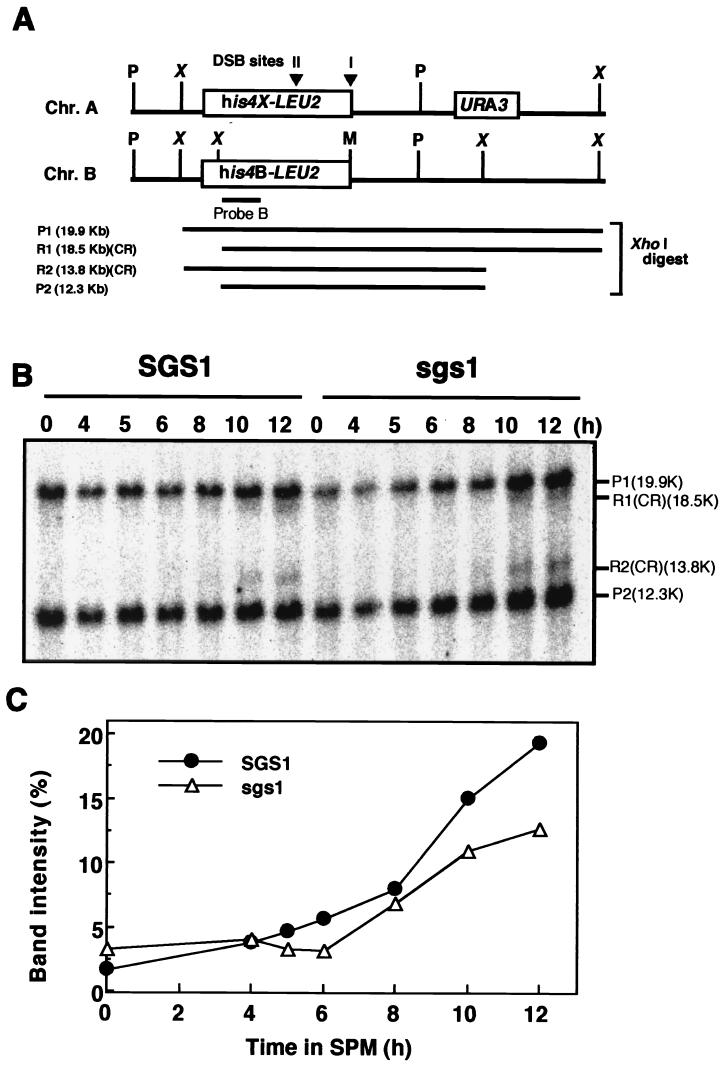
FIG. 5.
Detection of crossover-type recombination products in sgs1 disruptants. (A) Map of the his4 LEU2 locus on chromosome III. P, X, and M, restriction sites for PstI, XhoI, and MluI, respectively; P1 and P2, fragments derived from the parental DNAs after digestion with XhoI; R1 and R2, fragments derived from crossover-type (CR) recombination products. (B) NKY2846 (SGS1) and RKH225 (sgs1::AUR) cells treated as described in Materials and Methods were harvested at various times after transfer to SPM. DNA was isolated from the cells, and aliquots (5 μg) of XhoI-digested DNA were analyzed by Southern blotting. The probe used was derived from pNKY155. (C) The intensity of the bands corresponding to R1 and R2 was quantified and expressed as a percentage of the intensity of all bands in the lane.
Site-directed mutagenesis.
pYCp1306 was constructed by converting the PstI site (CTGCAG) of SGS1 in pRS314 to a SacII site (CCGCGG) without a change in amino acid sequence. pYCp1309 (sgs1-hd) was constructed by replacing the HindIII-PstI fragment of SGS1 in pYCp1306 with a fragment encoding alanine (GCA) instead of lysine (AAA) at amino acid position 706 in helicase motif I. The mutagenized fragment was made by PCR, and the mutation was confirmed by DNA sequencing.
RESULTS
Poor sporulation of sgs1 disruptants of various strains.
Diploid sgs1 disruptants were constructed using three yeast strains, W303, YPH, and MR, to investigate the function of SGS1 in meiosis. Sporulation on KAC plates was assessed by counting the cells containing asci. The sporulation frequencies of the W303 and YPH strains on KAC plates were 16.2 and 11.0%, respectively. Those of the MR strain, a derivative of SK1, were 52.8% on KAC plates and about 90% in SPM. The sporulation frequencies of the sgs1 disruptants derived from the W303, YPH, and MR strains were decreased by about 1/7, 1/22, and 1/13, respectively, compared with that of the corresponding wild-type cells (Table 2).
TABLE 2.
Poor sporulation of sgs1 disruptants
| Strain background | Genotype | Sporulation efficiency (% of asci formed)a |
|---|---|---|
| W303-1A | SGS1/SGS1 | 16.2 ± 3.7 |
| sgs1::URA3/sgs1::URA3 | 2.2 ± 1.4 | |
| YPH499 | SGS1/SGS1 | 11.0 ± 2.1 |
| sgs1::URA3/sgs1::URA3 | 0.5 ± 0.3 | |
| MR966/MR93-28C | SGS1/SGS1 | 52.8 ± 9.1 |
| sgs1::URA3/sgs1::LEU2 | 4.2 ± 2.2 |
Diploid cells incubated on YPD plates for 2 days were transferred to 1% KAC plates and incubated for 5 days to allow sporulation. Sporulation was monitored with a phase-contrast microscope.
Spore formation and premeiotic DNA replication in MR strains.
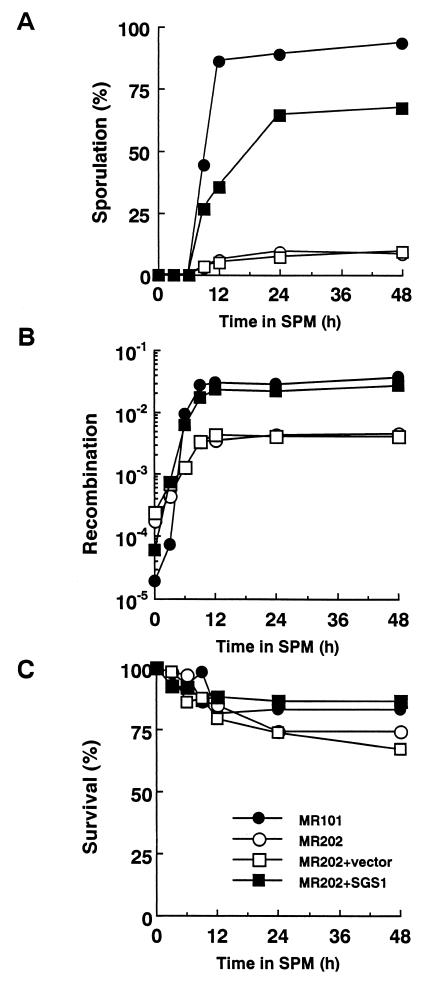
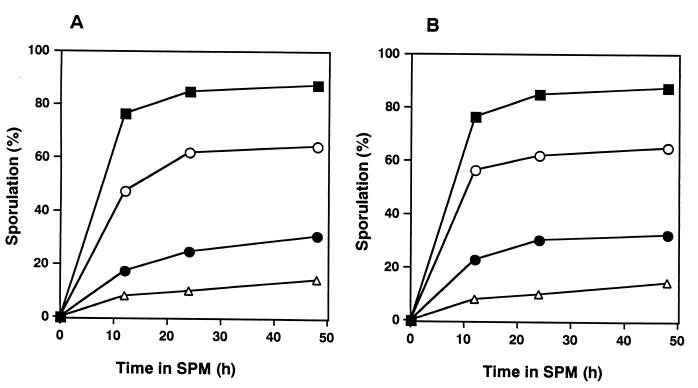
At 24 h after the change to SPM, the sporulation frequency of sgs1 disruptants (MR202) reached only 9.3%, while that of wild-type cells (MR101) attained 89% (Fig. 1A). We also examined the effect of disruption of the SGS1 gene on the frequency of recombination between heteroalleles at the HIS1 locus during meiosis. The frequency of recombination, measured by a return-to-growth assay, was reduced by 8.2-fold in sgs1 disruptants compared to that of wild-type cells (Fig. 1B). The increased recombination frequency of sgs1 disruptants compared with that of wild-type cells at time zero seems to correspond to the hyperrecombination phenotype of sgs1 disruptants during mitotic growth, as reported previously (51). The viability of sgs1 disruptants at 24 h after the transfer to SPM was 74%, while that of wild-type cells was 84% (Fig. 1C). These poor-sporulation and reduced-recombination phenotypes were complemented with SGS1 on either a single-copy plasmid (pYCp1305) (Fig. 1A and B) or a multicopy plasmid (data not shown).
FIG. 1.
Poor sporulation and reduction in the level of intergenic recombination of sgs1 disruptants. Cells treated as described in Materials and Methods were removed after transfer to SPM. Aliquots of 1 ml were removed at various times after the shift, and sporulation, recombination, and cell viability were assessed. YCp vectors, pRS314 (vector only), and pYCp1305 (containing full-length SGS1) were transfected into disruptants to confirm the effect of SGS1. Symbols: ●, MR101 (SGS1/SGS1); ○, MR202 (sgs1::URA3/sgs1::LEU2); □, MR202 plus pRS314; ■, MR202 plus pYCp1305. (A) Sporulation was monitored with a phase-contrast microscope, and the percentage of cells which formed asci containing any spores was measured. (B) The frequency of recombination between the his1-1 and his1-7 alleles was measured by a return-to-growth assay. The frequency was determined by comparing the number of colonies formed on SC-His(−) plates with the number formed on SC plates after incubation for 3 days at 28°C. (C) The viability of the cells was measured by enumerating colonies that appeared on SC plates after incubation for 3 days at 28°C, taking the viability of the cells at time zero as 100%.
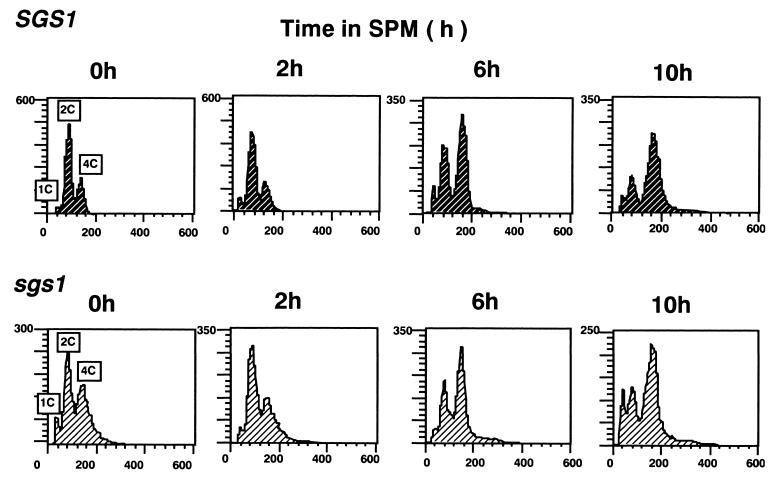
Most of the sgs1 disruptants were arrested at the mononucleated stage after the shift to SPM, raising the possibility that premeiotic DNA replication is affected in sgs1 disruptants. Thus, DNA replication in sgs1 disruptants (MR202) and wild-type cells (MR101) after transfer to SPM was monitored by flow cytometric analysis. The majority of disruptants and wild-type cells had 2C DNA before induction of sporulation (Fig. 2). At 10 h after the shift, the majority of disruptants and wild-type cells had 4C DNA, indicating that the bulk of DNA replication was completed in most cells, even disruptants. These results suggest that Sgs1 is not involved until after most of the DNA is synthesized.
FIG. 2.
Flow cytometric analysis of DNA content. MR101 (SGS1/SGS1) and MR202 (sgs1::URA3/sgs1::LEU2) cells were removed at various times after the shift to SPM and fixed with 70% ethanol. The DNA content of the samples was analyzed with a FACScan/CellFIT system.
Analysis of meiosis-specific DSBs in sgs1 disruptants.
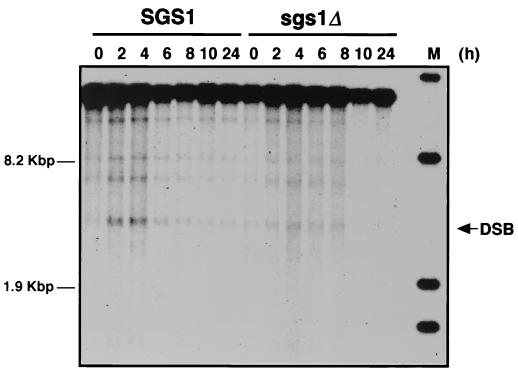
The formation of DSBs is considered the initial event in meiotic recombination (2, 5, 47). We examined DSB formation during meiosis in sgs1 disruptants. As shown in Fig. 3, transient DSB signals in the YCR47C-YCR48W region, which is the THR4 centromere-proximal region on chromosome III, were visualized by using the YCR48W probe (51a). DSB signals were observed in wild-type cells 2 h after the shift to SPM, reached a maximum level at 4 h, and gradually decreased. In sgs1 disruptants, the number of DSBs was considerably decreased.
FIG. 3.
Detection of DSB in sgs1 disruptants. MR101 (SGS1) and MR301 (sgs1Δ::AUR) cells were treated as described in Materials and Methods and harvested at various times after transfer to SPM. DNA samples were isolated from the cells, and aliquots (5 μg) of the DNA were separated on an agarose gel. DSB signals in the YCR47C-YCR48W region were analyzed by Southern blotting.
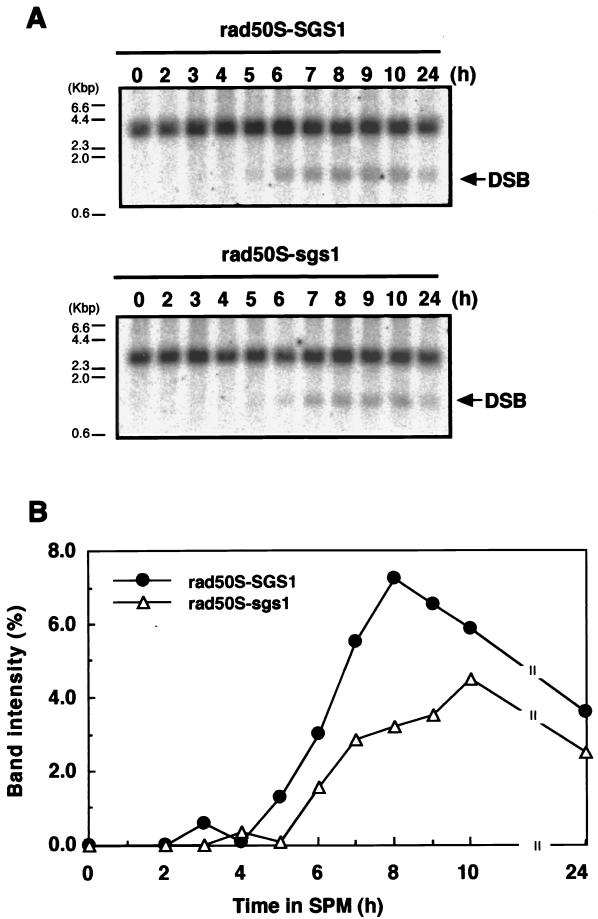
To elucidate whether the formation of DSBs was really reduced in sgs1 disruptants, we examined the accumulation of DSBs at the YCR47C-YCR48W loci in chromosome III in the rad50S (rad50S-KI81) background (4), in which the processing but not the formation of DSBs is blocked (2, 5). As shown in Fig. 4, DSBs appeared about 5 h after the shift to SPM and accumulated gradually in rad50S cells (S1510). In rad50S sgs1 disruptants (RKH221), the appearance of DSBs was delayed about 1 h and the maximum level attained was slightly lower than that in rad50S cells.
FIG. 4.
Accumulation of DSBs in rad50S sgs1 mutants. (A) Wild-type cells and sgs1 disruptants with the rad50S background, S1510 (SGS1) and RKH221 (sgs1::AUR), were treated as described in Materials and Methods and harvested at various times after transfer to SPM. DNA was isolated from the cells, and aliquots (5 μg) of the DNA were separated on an agarose gel. DSB signals in the YCR47C-YCR48W region were analyzed by Southern blotting. (B) The intensity of the band corresponding to the DSB signal was quantified and expressed as a percentage of the intensity of all bands in the lane.
Physical analysis of the recombination products in sgs1 disruptants during meiosis.
The physical recombinants at the his4-LEU2 loci were examined (Fig. 5A) (46). Bands corresponding to crossover-type recombination can be discriminated from parental fragments by restriction site polymorphism of the XhoI site between homologous chromosomes. The bands derived from crossover-type recombination were detected 8 h after the shift to SPM in wild-type cells and sgs1 disruptants. The intensities of the bands reached 20 and 13% that of the parental bands by 12 h for wild-type cells and sgs1 disruptants, respectively (Fig. 5B). Bands derived from non-crossover-type recombination (gene conversion) are detectable with the same specific probe by Southern blotting with DNA samples digested with XhoI and MluI (46). We examined non-crossover-type recombination and found that it occurred in sgs1 disruptants as well as in wild-type cells (data not shown).
Analysis of SGS1 function during sporulation by deletion of SPO13 and MRE11.
Spo13 is required for meiosis I, and deletion of the SPO13 gene results in a single meiosis II-like division, producing dyad asci, and rescues the meiotic lethality of early recombination-deficient mutants by bypassing meiosis I division (29). Mre11 is required in the early stages of meiotic recombination, including DSB formation, and mre11 mutants produce nonviable spores (1, 15). To analyze the function of SGS1 during sporulation, we constructed double disruptants, spo13 sgs1, mre11 sgs1, and spo13 mre11 (Table 1), and examined whether these cells were capable of sporulation. These double disruptants were analyzed for mitotic recombination frequency, sporulation, spore viability, and meiotic recombination frequency, as measured by the return-to-growth assay (Table 3). spo13 mre11 double disruptants produced dyad asci with 63.2% spore viability and showed a remarkably reduced meiotic recombination frequency, as reported by Ajimura et al. (1). For sgs1 spo13 double disruptants, 10% of the cells formed dyad asci at 24 h after the shift to SPM; this value was the same as that for sgs1 disruptants and was 5.2- and 3.3-fold lower than those for spo13 and spo13 mre11 cells, respectively. The spore viability of sgs1 spo13 cells was 5.0- and 3.4-fold lower than those of sgs1 and spo13 cells, respectively. Very few sgs1 mre11 double disruptants sporulated, and the spores were nonviable. All the single and double disruptants except for spo13 showed mitotic hyperrecombination.
TABLE 3.
Sporulation, spore viability, and recombination in sgs1 spo13 and sgs1 mre11 double disruptants
| Strain | Genotype | Mitotic recombination (His+ frequency 10−5)a | Sporulation (%) atb:
|
Spore viabilityc | Meiotic recombination (His+ frequency, 10−3) at 24 hd | |
|---|---|---|---|---|---|---|
| 24 h | 96 h | |||||
| MR101 | SGS1 SPO13 MRE11 | 5.8 | 81 | 88 | 75/84 (89.3) | 37.9 |
| ds1 | sgs1Δ | 57.6 | 8 | 10 | 72/124 (58.1) | 3.9 |
| ds13 | spo13 | 9.5 | 52 | 79 | 48/120 (40.0) | 6.5 |
| ds1s13 | sgs1Δ spo13 | 56.3 | 10 | 15 | 15/128 (11.7) | 3.9 |
| ds1m11 | sgs1Δ mre11 | 38.8 | 0.6 | 0.1 | 0/120 (0.0) | 0.2 |
| ds13m11 | spo13 mre11 | 44.6 | 33 | 55 | 86/136 (63.2) | 0.1 |
The frequency of mitotic recombination between the his1-1 and his1-7 alleles was determined by comparing the number of colonies on SC-His(−) plates with the number on SC plates.
Diploid cells were precultured in SPS and inoculated into SPM at a density of 2 × 107 cells/ml with vigorous shaking. Aliquots of 1 ml were taken at the times indicated, sporulation was monitored at 24 or 96 h after the shift to SPM with a phase-contrast microscope, and the percentage of tetrad asci or dyad asci (boldface values) for spo13 disruptants relative to total cell number was calculated.
Spore viability was determined after dissection by dividing the number of spores forming colonies by the total number dissected. Numbers in parentheses are percentages.
The frequency of meiotic recombination between the his1-1 and his1-7 alleles was determined at 24 h after the shift to SPM by comparing the number of colonies on SC-His(−) plates with the number on SC plates.
Either the red1 or the rad17 mutation partially alleviates the prophase arrest of sgs1 disruptants.
The facts that the majority of sgs1 disruptants remained in the mononucleated stage after transfer to SPM and showed relatively high viability indicate that the defect due to the dysfunction of Sgs1 produces a signal to arrest cells at or before meiosis I. Thus, we examined the possibility that mononucleated sgs1 cells were arrested by meiotic checkpoint control.
The disruption of either the RED1 or the RAD17 gene, both of which are involved in meiotic checkpoint control (25, 52), resulted in partial alleviation of the poor-sporulation phenotype of sgs1 disruptants (Fig. 6).
FIG. 6.
Partial suppression of poor sporulation of sgs1 null mutants by the deletion of RED1 or RAD17 gene function. Cells treated as described in Materials and Methods were harvested after transfer to SPM. Aliquots (1 ml) were removed at various times after the shift to SPM, and the cells were examined for sporulation. (A) Symbols: ■, wild-type cells (MR101); ▵, sgs1 cells (MR301); ○, red1 cells (dr1); ●, red1 sgs1 cells (dslr1). (B) Symbols: ■, wild-type cells (MR101); ▵, sgs1 cells (MR301); ○, rad17 cells (dr17); ●, rad17 sgs1 cells (dslr17).
Missense mutation in the ATP binding motif of SGS1 affects sensitivity to MMS and HU but not sporulation.
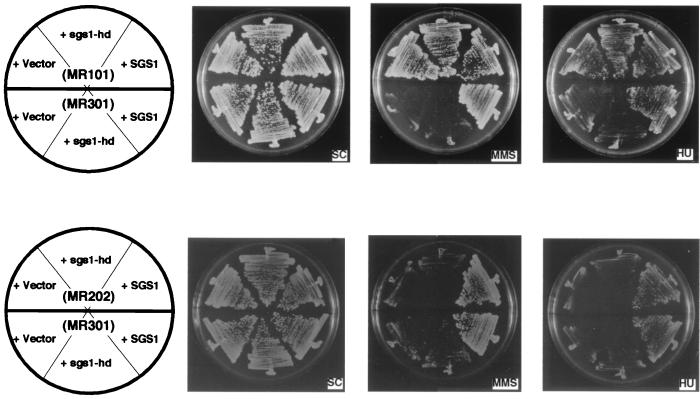
DNA helicase motif I is known to be involved in ATP binding. Lu et al. reported that a missense mutation of Sgs1 in helicase motif I at amino acid position 706, from lysine to alanine, abolished helicase activity (23). To determine the requirement of helicase activity for various functions of Sgs1, a plasmid carrying an sgs1 gene encoding Sgs1 having the same missense mutation (sgs1-hd) was transformed into sgs1 disruptants. As shown in Fig. 7, sgs1-hd was not able to complement the sensitivity to MMS or HU of the sgs1 partial disruptant (MR202) or null mutant (MR301). In contrast, the poor-sporulation phenotype and reduced frequency of meiotic recombination monitored by the return-to-growth assay were rescued by sgs1-hd in the sgs1 null mutant (MR301) as well as the partial disruptant (MR202) (Table 4). The increased recombination in sgs1 disruptants at time zero, which corresponds to mitotic recombination, was not suppressed by sgs1-hd.
FIG. 7.
An sgs1 gene coding for a protein defective in DNA helicase activity cannot complement the MMS and HU sensitivities of sgs1 disruptants. Wild-type cells (MR101), sgs1 disruptants (MR202), and sgs1 null deletion disruptants (MR301) were transformed with pRS314 (vector only), pYCp1309 (sgs1-hd), and pYCp1305 (containing full-length SGS1). Cells were streaked onto SC plates containing MMS (0.02%) or HU (100 mM) and were photographed after 3 days at 28°C.
TABLE 4.
Analysis of Sgs1 functionsa
| Strain | Plasmid | Mitotic recombination (His+ frequency, 10−5) at 0 h | Sporulation (% of asci formed) at:
|
Meiotic recombination (His+ frequency, 10−3) at 24 h | |
|---|---|---|---|---|---|
| 12 h | 24 h | ||||
| MR101 (WT) | Vector | 2.7 | 82 | 89 | 27.8 |
| SGS1 | 3.1 | 82 | 89 | 26.1 | |
| sgs1-hd | 3.5 | 81 | 89 | 24.1 | |
| MR202 (sgs1) | Vector | 23.4 | 4.9 | 7.4 | 4.2 |
| SGS1 | 6.0 | 36 | 64 | 22.3 | |
| sgs1-hd | 16.3 | 23 | 50 | 15.5 | |
| MR301 (sgs1Δ) | Vector | 20.1 | 7.0 | 9.2 | 5.5 |
| SGS1 | 2.2 | 25 | 58 | 24.8 | |
| sgs1-hd | 27.2 | 28 | 51 | 17.3 | |
Wild-type (WT) cells (MR101), sgs1 disruptants (MR202), and sgs1 null deletion disruptants (MR301) were transformed with pRS314 (vector only), pYCp1305 (containing full-length SGS1), and pYCp1309 (sgs1-hd). Sporulation was monitored with a phase-contrast microscope at 12 and 24 h after the shift to SPM. The frequency of recombination between the his1-1 and his1-7 alleles was examined at 0 h (for mitotic recombination) and 24 h (for meiotic recombination by a return-to-growth assay) after the shift to SPM.
DISCUSSION
It has been reported that sgs1 mutants show a poor-growth phenotype in the top1 mutant background (23), suppression of top3-associated poor growth (11), hypersensitivity to HU (53) and MMS (10), defects in faithful chromosome segregation in mitosis as well as meiosis (50), mitotic hyperrecombination phenotypes (31, 51, 53), poor sporulation (12, 51), and premature aging (44). In this study, we analyzed in detail the poor-sporulation phenotype of sgs1 disruptants in relation to meiotic recombination.
Meiotic recombination in an sgs1 mutant examined by physical analysis.
We observed poor-sporulation phenotypes in sgs1 disruptants from several strains with different genetic backgrounds (Table 2), as reported previously (12, 50, 51). The apparent frequency of meiotic recombination, measured by a return-to-growth assay, was decreased severalfold in sgs1 disruptants compared with wild-type cells (Fig. 1). Thus, we examined recombination intermediates of sgs1 disruptants by physical analysis. Although the number of DSBs was reduced severalfold in sgs1 disruptants compared with wild-type cells, almost the same number of DSBs accumulated in sgs1 rad50S cells as in rad50S cells. Similar results were reported for red1 and mek1/mre4 mutants (52). In red1 and mek1/mre4 mutants, the steady-state level of DSBs was reduced, but the number of DSBs that accumulated in red1 rad50S and mek1 rad50S cells was almost the same as that in rad50S cells. Xu et al. proposed that in red1 and mek1/mre4 mutants, the kinetics of DSB formation are negatively regulated by meiosis-specific surveillance mechanisms, and the conversion of DSBs to double Holliday junctions would be one of the most important checkpoints (52). Thus, we speculated that the kinetics of DSB formation, rather than the machinery to form DSBs, are somehow affected in sgs1 disruptants.
Meiotic recombination products of either the crossover type or the gene conversion type appeared in sgs1 disruptants as well as wild-type cells. Since the appearance of recombinant molecules does not necessarily require the resolution of Holliday junctions, the possibility remains that the defect of Sgs1 function affects the recombination process itself.
The frequency of meiotic recombination measured by the return-to-growth assay was decreased severalflold in sgs1 disruptants compared with wild-type cells (Fig. 1). Watt et al. reported no decrease in the meiotic recombination frequency in spores formed by sgs1 mutants (51). This discrepancy can be explained by the differences in the populations analyzed; that is, Watt et al. dealt with only viable spores, and the return-to-growth assay dealt with the whole population, most of which was not able to form spores (Fig. 1). However, the physical assay of amounts of meiotic recombinants showed little difference between sgs1 and wild-type cells. The decrease in meiotic recombination in the return-to-growth assay was also observed in top3 disruptants, and the low level of meiotic recombination in this assay was explained to be due to the loss of viability of meiotic cells following DSB formation in top3 mutants (12). However, this explanation cannot be applied for the sgs1 disruptants in this study, because only a slight reduction in viability was observed with sgs1 disruptants in the return-to-growth assay (Fig. 1).
In most sgs1 disruptants, meiosis I is not bypassed upon disruption of SPO13.
On deletion of the SPO13 gene, meiosis I is bypassed and dyad asci are produced (29). Deletion of the SPO13 gene in a rad52, rad50S, or dmc1 background, which is defective at a point after DSB formation, produced dead or very-low-viability dyad asci (4, 27). The viability of spores formed in either rad52, rad50S, or dmc1 single mutants also was very low (4, 27). Disruptants of the genes required before or for DSB formation during meiotic recombination, such as spo11, mre11, and rad50, sporulate with almost normal efficiency but produce nonviable spores, and disruption of SPO13 in spo11, mre11, or rad50 mutants results in the formation of viable dyad asci without meiotic recombination (1, 21, 26).
In fact, spo13 mre11 mutants sporulated and produced dyad asci containing viable spores (Table 3), as reported previously (1). The results obtained with sgs1 and sgs1 spo13 disruptants were quite different from those obtained with the above mutants. The sgs1 disruptants showed low sporulation efficiency, but the spores produced showed relatively high viability, and the disruption of SPO13 in sgs1 disruptants did not alleviate the low sporulation efficiency of the sgs1 disruptants. Thus, it must be emphasized that the majority of sgs1 disruptants have a defect that renders them unable to bypass meiosis I on disruption of SPO13. A similar result was obtained with top3 disruptants (12). A small portion of the sgs1 population underwent meiosis almost normally, and the spores showed relatively high viability. However, a defect in segregation was observed in this population, since a considerable number of the asci formed by sgs1 disruptants contained odd numbers of spores (data not shown), as reported previously (51).
The rad50S mutants, unable to process DSBs, showed a poor-sporulation phenotype, and both dmc1 and top2 mutants, which are defective in reciprocal recombination and the resolution of recombination intermediates, respectively, were arrested at meiotic prophase under meiosis-specific checkpoint control. The introduction of mutations in either MRE11 or RAD50 in the above mutants to form mre11 rad50S, rad50 dmc1, and rad50 top2, which eliminates DSB formation, resulted in the restoration of sporulation and the production of dead spores (1, 4, 35). Thus, if Sgs1 is required only after DSB formation in meiotic recombination, sgs1 mre11 double mutants should show phenotypes similar to those of mre11 single mutants. However, this was not the case, because the reduced level of spore formation by sgs1 mutants could not be alleviated by introducing the MRE11 mutation, that is, eliminating DSBs. Similar results were obtained for top3 mutants by Gangloff et al. (12), who reported that disruption of SPO11, which encodes the enzyme essential to form DSBs (17), could not alleviate the top3 sporulation defect.
Arrest of sgs1 disruptants in the mononucleated stage is caused partially by meiotic checkpoint function.
The facts that the majority of sgs1 disruptants remained in the mononucleated stage after transfer to SPM and showed relatively high viability indicate that the defect due to the dysfunction of Sgs1 produces a signal to arrest cells at or before meiosis I. Gangloff et al. (12) showed that sgs1 disruptants of strain W303 could sporulate but that the process was delayed and inefficient compared to that of wild-type cells, and they argued that the Sgs1 defect generates a checkpoint signal. We also monitored the sporulation of sgs1 disruptants (MR301) for up to 12 days. The percentage of asci that were formed gradually increased but at 12 days was still only 18.5%, compared with 85% for wild-type cells. For dmc1 mutants, lacking Dmc1, which is the meiosis-specific Rad51 homologue, cells with recombination intermediates arrested in the prophase without a loss of viability (4). The disruption of RED1 in dmc1 mutants released the prophase arrest of the dmc1 mutants and restored spore formation (52). Cells lacking Red1, which is the meiosis-specific component of the axial element, failed to form a synaptonemal complex (34) and to check aberrant DNA recombination (52). In addition, the disruption of RAD17 in dmc1 mutants alleviated the prophase arrest of the dmc1 mutants (25), suggesting the existence of single-stranded DNA regions in uncompleted recombination intermediates in dmc1 cells, because Rad17 is able to sense single-stranded DNA regions (24).
The disruption of either RED1 or RAD17 in sgs1 disruptants partially alleviated the poor-sporulation phenotype of sgs1 mutants. The reason why the effect was partial is not clear. One explanation is that cells in which the phenotype was not alleviated by the disruption of either RED1 or RAD17 have defects that cause the suppression and that can be alleviated only by disruption of both RED1 and RAD17 or defects that are sensed by sensors other than Red1 and Rad17. Another is that cells in which the phenotype was alleviated by the red1 or rad17 mutation are of different populations. Thus, it seems that sgs1 disruptants are arrested at multiple points rather than at a single unique point. The partial suppression caused by the disruption of RAD17 indicates the existence of single-stranded DNA regions in a portion of the arrested population of sgs1 cells (24, 25). It seems likely that sgs1 disruptants contain incomplete homologous recombination intermediates in which single-stranded DNA regions exist. The suppression of the late S/G2 delay of top3 mutants by an SGS1 mutation suggests that both Sgs1 and Top3 are involved in the late stage of DNA replication in the mitotic cell cycle (11). Although premeiotic DNA synthesis followed a normal time course even in sgs1 disruptants (Fig. 2), the possibility cannot be excluded that DNA replication is inhibited at a late stage and single-stranded DNA regions remain in sgs1 disruptants.
Relationship between Top3 and Sgs1 functions in meiosis.
The protein encoded by SGS1 was shown to have DNA helicase activity (3, 23). It has been reported that a missense mutation of lysine to alanine at amino acid position 706 abolishes the DNA helicase activity of Sgs1 in vitro (23). The plasmid carrying the gene with this missense mutation, sgs1-hd, rescued neither MMS sensitivity nor HU sensitivity in sgs1 disruptants, indicating that repair of certain types of DNA damage requires the DNA helicase activity of Sgs1 (Fig. 7). Similar results were reported recently (10). In addition, sgs1-hd could suppress neither elevated homologous recombination between heteroalleles (Table 4) nor elevated unequal sister chromatid exchanges in sgs1 mutants (31). In contrast, it was reported that the sgs1-hd allele behaves just like the wild-type allele: it decreases the rate of growth of top3 sgs1 mutants and improves the poor growth of top1 sgs1 mutants (23). In other words, DNA helicase activity is not required for complementation of top1- and top3-related sgs1 phenotypes. In addition, we found that sgs1-hd almost completely rescued the poor-sporulation phenotype and the reduced frequency of meiotic recombination of sgs1 disruptants. Thus, two different mechanisms underlie the functions of Sgs1; one requires DNA helicase activity, and the other does not. This finding should help us to analyze the molecular mechanisms producing the pleiotropic phenotypes of sgs1 disruptants.
Budding yeast Top3 has a weak ability to relax negatively supercoiled double-stranded DNA and preferentially binds to single-stranded DNA regions (18). Both genetic and physical interactions have been demonstrated for Sgs1 and Top3, indicating that these proteins function as a complex in the mitotic cell cycle (11). Gangloff et al. (11) proposed that movement of the Sgs1 helicase along the DNA melts the duplex, providing a preferential single-stranded substrate for Top3, and that the Sgs1-Top3 complex might act as a reverse gyrase (6). The observation that Sgs1 devoid of DNA helicase activity could carry out its role in sporulation indicates that the postulated reverse gyrase activity of the Sgs1-Top3 complex is dispensable from the meiotic process, because helicase activity is essential for introducing positive supercoils. Thus, it seems likely that one of the possible functions of Sgs1 in the meiotic process is to recruit Top3. In this context, it is interesting that mouse Top3α and Top3β as well as Blm mRNAs were highly expressed in the testis and that the levels of their expression in the testis were increased simultaneously and markedly by 17 days after birth, when numbers of the cells in pachytene phase increase (39, 40, 41). In addition, it was reported recently that BLM, one of the Sgs1 homologues in higher eukaryotes, was localized on mouse meiotic chromosomes during meiotic DNA recombination (49). Taken together with the report that male BS patients are infertile (16), these results suggest that BLM plays roles in meiotic recombination.
Recently, the sporulation of top3 mutants as well as sgs1 mutants has been reported (12). Although the frequency of sporulation of sgs1 mutants is low, it is higher than that of top3 mutants, which make no spore. In addition, deletion of the SGS1 gene restores sporulation in top3 mutants to a level below that in sgs1 single mutants. Thus, Sgs1 does more than simply recruit Top3; an unknown protein has the ability to recruit Top3, or Top3 itself can access the meiotic chromosome at a low efficiency.
ACKNOWLEDGMENTS
We are grateful to A. Sugino, N. Kleckner, H. Ogawa, R. E. Malone, and T. Weinert for providing plasmid DNAs and yeast strains.
This work was supported by grants-in-aid for scientific research and for scientific research on priority areas from the Ministry of Education, Science, Sports and Culture of Japan, health sciences research grants from the Ministry of Health and Welfare of Japan, a grant from the CREST of the JST and the Human Frontier Science Program, and a grant from the Mitsubishi Foundation.
REFERENCES
- 1.Ajimura M, Leem S-H, Ogawa H. Identification of new genes required for meiotic recombination in Saccharomyces cerevisiae. Genetics. 1993;133:51–66. doi: 10.1093/genetics/133.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alani N, Padmore R, Kleckner N. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990;61:419–436. doi: 10.1016/0092-8674(90)90524-i. [DOI] [PubMed] [Google Scholar]
- 3.Bennett R J, Sharp J A, Wang J C. Purification and characterization of the Sgs1 DNA helicase activity of Saccharomyces cerevisiae. J Biol Chem. 1998;273:9644–9650. doi: 10.1074/jbc.273.16.9644. [DOI] [PubMed] [Google Scholar]
- 4.Bishop D K, Park D, Xu L, Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- 5.Cao L, Alani E, Kleckner N. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell. 1990;61:1089–1101. doi: 10.1016/0092-8674(90)90072-m. [DOI] [PubMed] [Google Scholar]
- 6.Confalonieri F, Elie C, Nadal M, de la Tour C B, Forterre P, Duguet M. Reverse gyrase: a helicase-like domain and a type I topoisomerase in the same polypeptide. Proc Natl Acad Sci USA. 1993;90:4753–4757. doi: 10.1073/pnas.90.10.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dykstra C C, Kitada K, Clark A B, Hamatake R K, Sugino A. Cloning and characterization of DST2, the gene for DNA strand transfer protein β from Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:2583–2592. doi: 10.1128/mcb.11.5.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellis N A, Groden J, Ye T-Z, Straughen J, Lennon D J, Ciocci S, Proytcheva M, German J. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 9.Epstein C J, Martin G M, Schultz A L, Motulsky A G. Werner's syndrome: a review of its symptomatology, natural history, pathologic features, genetics and relationship to the natural aging process. Medicine. 1966;45:177–222. doi: 10.1097/00005792-196605000-00001. [DOI] [PubMed] [Google Scholar]
- 10.Frei C, Gasser S M. The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes Dev. 2000;14:81–96. [PMC free article] [PubMed] [Google Scholar]
- 11.Gangloff S, McDonald J P, Bendixen C, Arthur L, Rothstein R. The yeast type 1 topoisomerase Top3 interacts with Sgs1, a DNA helicase homologue: a potential eukaryotic reverse gyrase. Mol Cell Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gangloff S, deMassy B, Arthur L, Rothstein R, Fabre F. The essential role of yeast topoisomerase III in meiosis depends on recombination. EMBO J. 1999;18:1701–1711. doi: 10.1093/emboj/18.6.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.German J. Bloom syndrome: a Mendelian prototype of somatic mutational disease. Medicine. 1993;72:393–406. [PubMed] [Google Scholar]
- 14.Goyon C, Lichten M. Timing of molecular events in meiosis in Saccharomyces cerevisiae: stable heteroduplex DNA is formed late in meiotic prophase. Mol Cell Biol. 1993;13:373–382. doi: 10.1128/mcb.13.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johzuka K, Ogawa H. Interaction of Mre11 and Rad50: two proteins required for DNA repair and meiosis-specific double-strand break formation in Saccharomyces cerevisiae. Genetics. 1995;139:1521–1532. doi: 10.1093/genetics/139.4.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kauli R, Prager-Lewin R, Kaufman H, Laron Z. Gonadal function in Bloom's syndrome. Clin Endocrinol. 1977;6:285–289. doi: 10.1111/j.1365-2265.1977.tb02013.x. [DOI] [PubMed] [Google Scholar]
- 17.Keeney S, Giroux C N, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 18.Kim R A, Wang J C. Identification of the yeast TOP3 gene product as a single strand-specific DNA topoisomerase. J Biol Chem. 1992;267:17178–17185. [PubMed] [Google Scholar]
- 19.Kitao S, Ohsugi I, Ichikawa K, Goto M, Furuichi Y, Shimamoto A. Cloning of two new human helicase genes of the RecQ family: biological significance of multiple species in higher eukaryotes. Genomics. 1998;54:443–452. doi: 10.1006/geno.1998.5595. [DOI] [PubMed] [Google Scholar]
- 20.Kitao S, Shimamoto A, Goto M, Miller R W, Smithson W A, Lindor N M, Furuichi Y. Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nat Genet. 1999;22:82–84. doi: 10.1038/8788. [DOI] [PubMed] [Google Scholar]
- 21.Klapholz S, Waddell C S, Esposito R E. The role of the SPO11 gene in meiotic recombination in yeast. Genetics. 1985;110:187–216. doi: 10.1093/genetics/110.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kowalczykowski S C, Dixon D A, Eggleston A K, Lauder S D, Rehrauer W M. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu J, Mullen J R, Brill S J, Kleff S, Romeo A M, Sternglanz R. Human homologues of yeast helicase. Nature. 1996;383:678–679. doi: 10.1038/383678a0. [DOI] [PubMed] [Google Scholar]
- 24.Lydall D, Weinert T. Yeast checkpoint genes in DNA damage processing: implications for repair and arrest. Science. 1995;270:1488–1491. doi: 10.1126/science.270.5241.1488. [DOI] [PubMed] [Google Scholar]
- 25.Lydall D, Nikolsky Y, Bishop D K, Weinert T. A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature. 1996;383:840–843. doi: 10.1038/383840a0. [DOI] [PubMed] [Google Scholar]
- 26.Malone R E, Esposito R E. Recombinationless meiosis in Saccharomyces cerevisiae. Mol Cell Biol. 1981;1:891–901. doi: 10.1128/mcb.1.10.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mao-Draayer Y, Galbraith A M, Pittman D L, Cool M, Malone R E. Analysis of meiotic recombination pathways in the yeast Saccharomyces cerevisiae. Genetics. 1996;144:71–86. doi: 10.1093/genetics/144.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin G M. Cellular aging-clonal senescence. Am J Pathol. 1977;89:484–511. [PMC free article] [PubMed] [Google Scholar]
- 29.McCarroll R M, Esposito R E. SPO13 negatively regulates the progression of mitotic and meiotic nuclear division in Saccharomyces cerevisiae. Genetics. 1994;138:47–60. doi: 10.1093/genetics/138.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakayama K, Irino N, Nakayama H. The recQ gene of Escherichia coli K12: molecular cloning and isolation of insertion mutants. Mol Gen Genet. 1985;200:266–271. doi: 10.1007/BF00425434. [DOI] [PubMed] [Google Scholar]
- 31.Onoda F, Seki M, Miyajima A, Enomoto T. Elevation of sister chromatid exchange in Saccharomyces cerevisiae sgs1 disruptants and the relevance of the disruptants as a system to evaluate mutations in Bloom's syndrome gene. Mutat Res. 2000;459:203–209. doi: 10.1016/s0921-8777(99)00071-3. [DOI] [PubMed] [Google Scholar]
- 32.Puranam K L, Blackshear P J. Cloning and characterization of RecQL, a potential human homologue of the Escherichia coli DNA helicase RecQ. J Biol Chem. 1994;269:29838–29845. [PubMed] [Google Scholar]
- 33.Resnick M A, Sugino A, Nitiss J, Chow T. DNA polymerase, deoxyribonuclease, and recombination during meiosis in Saccharomyces cerevisiae. Mol Cell Biol. 1984;12:2811–2817. doi: 10.1128/mcb.4.12.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rockmill B, Roeder G S. Meiosis in asynaptic yeast. Genetics. 1990;126:563–574. doi: 10.1093/genetics/126.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rose D, Thomas W, Holm C. Segregation of recombined chromosomes in meiosis I requires DNA topoisomerase II. Cell. 1990;60:1009–1017. doi: 10.1016/0092-8674(90)90349-j. [DOI] [PubMed] [Google Scholar]
- 36.Rothstein R. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Seki M, Miyazawa H, Tada S, Yanagisawa J, Yamaoka T, Hoshino S, Ozawa K, Eki T, Nogami M, Okumura K, Taguchi H, Hanaoka F, Enomoto T. Molecular cloning of cDNA encoding human DNA helicase Q1 which has homology to Escherichia coli RecQ helicase and localization of the gene at chromosome 12p12. Nucleic Acids Res. 1994;22:4566–4573. doi: 10.1093/nar/22.22.4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seki T, Seki M, Katada T, Enomoto T. Isolation of a cDNA encoding mouse DNA topoisomerase III which is highly expressed at the mRNA level in the testis. Biochim Biophys Acta. 1998;1396:127–131. doi: 10.1016/s0167-4781(97)00192-9. [DOI] [PubMed] [Google Scholar]
- 40.Seki T, Seki M, Onodera R, Katada T, Enomoto T. Cloning of cDNA encoding a novel mouse DNA topoisomerase III (Topo IIIβ) possessing negatively supercoiled DNA relaxing activity, whose message is highly expressed in the testis. J Biol Chem. 1998;273:28553–28556. doi: 10.1074/jbc.273.44.28553. [DOI] [PubMed] [Google Scholar]
- 41.Seki T, Wang W-S, Okumura N, Seki M, Katada T, Enomoto T. cDNA cloning of mouse BLM gene, the homologue to human Bloom's syndrome gene, which is highly expressed in the testis at the mRNA level. Biochim Biophys Acta. 1998;1398:377–381. doi: 10.1016/s0167-4781(98)00066-9. [DOI] [PubMed] [Google Scholar]
- 42.Sherman F, Fink G R, Hicks J B. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1983. [Google Scholar]
- 43.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sinclair D A, Mills K, Guarente L. Accelerated aging and nucleolar fragmentation in yeast sgs1 mutants. Science. 1997;277:1313–1316. doi: 10.1126/science.277.5330.1313. [DOI] [PubMed] [Google Scholar]
- 45.Stewart E, Chapman C R, Al-Khodairy F, Carr A M, Enoch T. rqh1+, a fission yeast gene related to the Bloom's and Werner's syndrome genes, is required for reversible S phase arrest. EMBO J. 1997;16:2682–2692. doi: 10.1093/emboj/16.10.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Storlazzi A, Xu L, Cao L, Kleckner N. Crossover and noncrossover recombination during meiosis: timing and pathway relationships. Proc Natl Acad Sci USA. 1995;92:8512–8516. doi: 10.1073/pnas.92.18.8512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun H, Treco D, Schultes N P, Szostak J W. Double-strand breaks at an initiation site for meiotic gene conversion. Nature. 1989;338:87–90. doi: 10.1038/338087a0. [DOI] [PubMed] [Google Scholar]
- 48.Thomas B J, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 49.Walpita D, Plug A W, Neff N F, German J, Ashley T. Bloom's syndrome protein, BLM, colocalizes with replication protein A in meiotic prophase nuclei of mammalian spermatocytes. Proc Natl Acad Sci USA. 1999;96:5622–5627. doi: 10.1073/pnas.96.10.5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Watt P M, Louis E J, Borts R H, Hickson I D. Sgs1: a eukaryotic homolog of E. coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell. 1995;81:253–260. doi: 10.1016/0092-8674(95)90335-6. [DOI] [PubMed] [Google Scholar]
- 51.Watt P M, Hickson I D, Borts R H, Louis E J. SGS1, a homologue of the Bloom's and Werner's syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics. 1996;144:935–945. doi: 10.1093/genetics/144.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51a.Wu T-C, Lichten M. Meiosis-induced double strand break sites determined by yeast chromatin structure. Science. 1994;263:515–518. doi: 10.1126/science.8290959. [DOI] [PubMed] [Google Scholar]
- 52.Xu L, Weiner B M, Kleckner N. Meiotic cells monitor the status of the interhomolog recombination complex. Genes Dev. 1997;11:106–118. doi: 10.1101/gad.11.1.106. [DOI] [PubMed] [Google Scholar]
- 53.Yamagata K, Kato J, Shimamoto A, Goto M, Furuichi Y, Ikeda H. Bloom's and Werner's syndrome genes suppress hyperrecombination in yeast sgs1 mutant: implication for genomic instability in human diseases. Proc Natl Acad Sci USA. 1998;95:8733–8738. doi: 10.1073/pnas.95.15.8733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu C-E, Oshima J, Fu Y-H, Wijsman E M, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, Martin G M, Mulligan J, Schellenberg G D. Positional cloning of the Werner's syndrome gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]