Abstract
The clinical success of engineered, CD19-directed chimeric antigen receptor (CAR) T cells in relapsed, refractory B-cell acute lymphoblastic leukemia (B-ALL) has generated great enthusiasm for the use of CAR T cells in patients with cytogenetics that portend a poor prognosis with conventional cytotoxic therapies. One such group includes infants and children with mixed lineage leukemia (MLL1, KMT2A) rearrangements (MLL-r), who fare much worse than patients with low- or standard-risk B-ALL. Although early clinical trials using CD19 CAR T cells for MLL-r B-ALL produced complete remission in most patients, relapse with CD19-negative disease was a common mechanism of treatment failure. Whereas CD19neg relapse has been observed across a broad spectrum of B-ALL patients treated with CD19-directed therapy, patients with MLL-r have manifested the emergence of AML, often clonally related to the B-ALL, suggesting that the inherent heterogeneity or lineage plasticity of MLL-r B-ALL may predispose patients to a myeloid relapse. Understanding the factors that enable and drive myeloid relapse may be important to devise strategies to improve durability of remissions. In this review, we summarize clinical observations to date with MLL-r B-ALL and generally discuss lineage plasticity as a mechanism of escape from immunotherapy.
Cellular and immunotherapy approaches show promise for MLL-r B-ALL
Relapsed and refractory B-cell acute lymphoblastic leukemia (B-ALL) remains a leading cause of cancer mortality in children despite the successful iterative development of risk-adapted, multi-agent chemotherapeutic regimens [1]. Years of sophisticated molecular characterization of childhood ALL [2,3] make it possible to identify subsets of patients with a high likelihood of relapse at diagnosis. Among the poor prognostic groups are those with rearrangements of the mixed lineage leukemia (MLL1, KMT2A) gene at 11q23 [4,5]. MLL rearrangements occur as the initial or only genetic lesion in >75% of infants with B-ALL [1]. Because of the elevated risk of relapse and resistance, the development of targeted therapies has been a priority to improve outcomes for this population. Although multiple small molecule inhibitors thought to be selective for MLL fusion oncoproteins have reached clinical trials, immunotherapy has also begun to have an impact in this patient group.
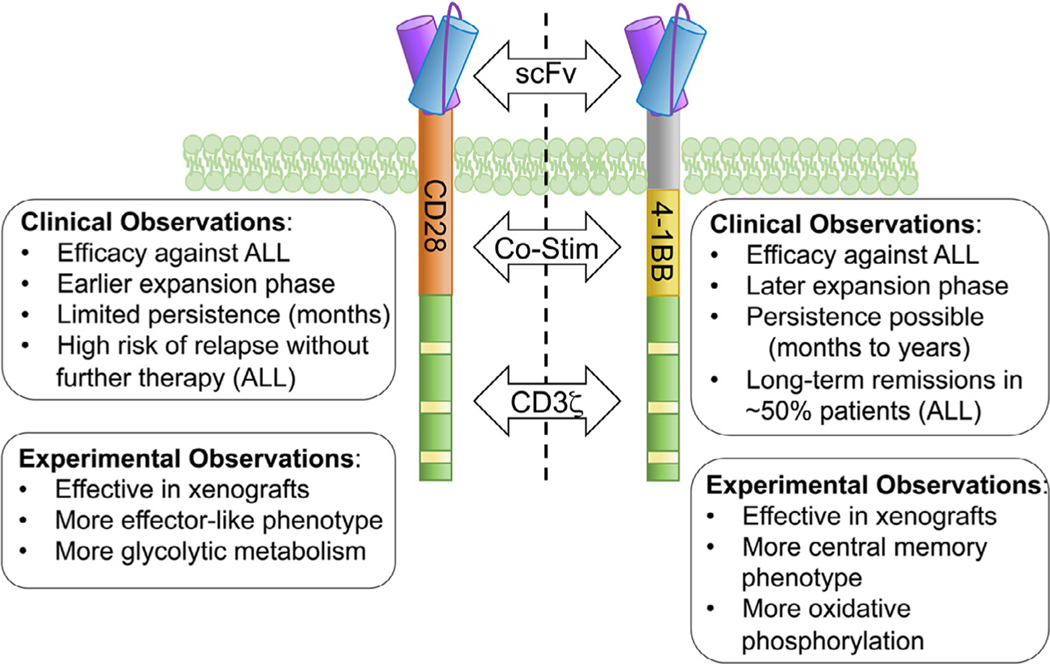
Two relatively new immunotherapies use the patient’s immune system to target the CD19 cell surface protein, which is coupled to B-cell identity and therefore highly expressed on B-ALL. First, blinatumomab is a CD3/CD19 bispecific T-cell engager (BiTE), or bispecific antibody that redirects a patient’s T cells to kill CD19+ cells. Blinatumomab was approved by the U.S. Food and Drug Administration (FDA) for relapsed, refractory, or Philadelphia chromosome−positive B-ALL in 2014, and approval was expanded in 2018 for broader use in B-ALL as a second-line treatment [6,7]. Second, chimeric antigen receptors (CARs) recognizing CD19 can direct the patient’s T cells to kill CD19+ B-ALL. The CAR construct is introduced into patient T cells during ex vivo manufacturing, which endows the T cell with directed specificity using an antibody-derived target binding domain and T-cell receptor signaling domains (Figure 1). Tisagenlecleucel (formerly CTL019) was approved by the FDA in 2017 for the treatment of relapsed or refractory B-ALL in pediatric/young adult patients based on the remarkable success of the phase II trial (NCT02435849) [8]. Multiple clinical trials using a variety of CD19-directed CAR T cell products have indicated complete remission rates of 70%−90% in pediatric patients with multiply relapsed and/or highly refractory B-ALL [8−12]. Longer follow-up in these trials revealed that patients receiving CD28-containing CAR T cells lost functional CAR T activity within 2 months of infusion. These patients had a high risk of post-CAR relapse without further treatment with a consolidative hematopoietic stem cell transplant [11]. For patients receiving CARs containing the 4–1BB co-stimulatory signaling domain (Figure 1), persistence of CAR T cells could be observed for months to years [8−10]. Follow-up studies of patients who received 4–1BB CAR T cells in clinical trials, as well as in postapproval “real world” studies, have reported that, despite the high initial rate of complete remission, only ~50% of patients remain leukemia-free 1 year after treatment because of post-CAR T cell relapses [9,13−15]. For patients receiving either CD28- or 4–1BB-containing CARs, two major patterns of relapse have been observed: antigen-positive (CD19+) relapse occurring in the absence of ongoing CAR T cell activity, and CD19neg relapse in which the loss of the target antigen allows the leukemic cells to survive and expand in the presence of a persistent and functional CAR T cell population [16]. Multiple studies have now found that patients treated with CD19-directed BiTEs or CD19-directed CAR T cells can relapse with CD19neg disease, which can arise via multiple mechanisms [9,16−18]. A poorly understood mechanism of CD19neg relapse is “lineage switching,” in which leukemia undergoes global changes resulting in the loss of multiple lymphoid markers and the acquisition of a myeloid phenotype (Table 1) 18−29,30−33 Lineage switching relapses have been reported after both CD19-directed BiTE and CAR T-cell therapy and tend to be enriched in MLL rearrangements, although cases harboring other translocations have been reported [18,31,33,34]. Because of the relatively recent implementation of immunotherapy and the rarity of MLL-r subsets within all B-ALL, it is too early to know whether CD19neg relapse occurs more or less frequently than in other subtypes. However, MLL rearrangements are associated with increased risk of relapse in pediatric ALL patients and occur in most infant ALL patients [35]; thus, it is increasingly likely that more children and infants harboring MLL rearrangements will receive immune-based therapies, possibly resulting in an increased number of patients experiencing lineage switch relapses. Interestingly, infants with MLL-r B-ALL and detectable residual disease at the end of induction chemotherapy may have better outcomes if myeloid consolidation regimens are used, suggesting that preventing myeloid relapse would result in an overall benefit [36]. Understanding the factors contributing to relapse from this otherwise effective therapy will be critical to improving on the initial success of this approach. Below, we address the clinical observations and potential underlying mechanisms of relapse. Specifically, we focus on the concept of lineage plasticity in generating CD19neg relapse in a poor-prognosis patient group in which this phenomenon has been documented.
Figure 1.
Clinical and biological differences in CD28- and 4–1BB−containing CAR T cells. Second-generation CARs, consisting of an antigen-binding domain (scFv) connected via an extracellular and transmembrane domain to a co-stimulatory domain (derived from either CD28 or 4–1BB) and the intracellular portion of the CD3z chain. Both CAR formats successfully activate T cells leading to leukemic clearance in preclinical models and in patients; however, each co-stimulatory molecule elicits differences in persistence, T-cell phenotype, and metabolism.
Table 1.
Published cases of CD19neg relapse with myeloid phenotype after CD19-directed immunotherapies, both CAR T and BiTEs
| Immunotherapy | Total relapse cases | CD19-negative relapse cases | No. of myeloid switch cases | Phenotype after CD19-directed immunotherapy | IgH clonal relationship | Time between immunotherapy and lineage switch | Cytogenetics | Age/Sex | Ref. | |
|---|---|---|---|---|---|---|---|---|---|---|
| CD19 CAR-T | 2 | 2 | 2 | Case 1 | CD19−, CD13 (dim)+, CD64+, HLA-DR (dim)+, CD15+, CD33+, CD71, MPO+ | Yes | 22 d | t(4;11)(q21;q23) MLL/AF4 | 52 y/F | 18 |
| Case 2 | CD19−, CD4+, CD56+, CD64+, CD13+, CD33+, CD38+, HLA-DR+, CD34+, CD45+,CD71+ | No | 21 d | ins(11;10)(q23; p12p1?1.2) MLL/MLLT10 | 18 mo/F | |||||
| 4 | 2 | 1 | CD19 negative myeloid phenotype switch | Yes | No data reported | t(4;11)(q21;q23) MLL/AF4 | 52 y/not reported | 33 | ||
| 1 | 1 | 1 | CD13+, CD34+. CD117+, CD123+, CD11b+, CD38(mod), CD7+, CD19−, CD10−, CD22−, TdT−, CD24−, CD20−, MPO− | No data reported | 8 mo | TCF3(Ex11)-ZNF384(Ex2) fusion | 13 mo/M | 31 | ||
| BiTEs | 1 | 1 | 1 | CD19−, CD34−, CD10−, CD3−, CD16−, CD117 −, HLADR−, nTdT−, CD2−, CD7−, CD38−, cCD22−, cCD79a−, cCD3−, CD45+, CD13+, CD15+, CD33+, CD56+, CD36+, CD64 (partial)+, cMPO+ | No data reported | 9 d | t(4;11)(q21;q23) MLL/AF4 | 40 y/F | 20 | |
| 1 | 1 | 1 | sCD19low, CD33+;CD34, CD14++, CD15++, CD11b++, CD64+ | No data reported | 9 d | t(4;11)(q21;q23) MLL/AF4 | 5 mo/F | 21 | ||
| 1 | 1 | 1 | CD19−, PAX5−, CD33+, CD43+, lysozyme+ | No data reported | 8 mo | t(4;11)(q21;q23) MLL/AF4 | 77 y/M | 22 | ||
| 1 | 1 | 1 | CD19−, CD34−, CD10−, CD38+, cMPO+, CD33+, CD13(low)+, CD64 +, CD65+, cCD79+ | Yes | 53 d | t(4;11)(q21;q23) MLL/AF4 | 46 y/F | 23 | ||
| 1 | 1 | 1 | CD19−, CD34−, CD79a−, TdT−,CD33+ CD11b+, CD14 (subset/dim)+, CD64+, MPO+;CD13(dim)+, CD22(dim),+ CD33(dim),+ CD38+, HLA-DR+ | No data reported | 15 d | t(4;11)(q21;q23) MLL/AF4 | 3 mo/not reported | 24 | ||
| 1 | 1 | 1 | CD19−, CD20−, CD22−, CD24+;cyIGM−, cyCD79a−, CD2−, CD3−, CD7−, CD8−, cyCD3−, CD13+, CD33+, CD15+, cyMPO+, CD117−, CD66c+, CD10+, CD34−, CD45+, TdT−, Cd38+, CD52− | No data reported | 3 wk | No | 8 y/F | 25 | ||
| 1 | 1 | 1 | CD19−, cCD79a−, CD22−, CD34−, CD33(low)+, CD65+, CD15+ | Yes | 28 d | t(4;11)(q21;q23) MLL/AF4 | 15 y/M | 26 | ||
| 1 | 1 | 1 | CD19−, PAX5−, CD34−, lysozyme+, CD33+, CD64(dim)+, CD13+, myeloperoxidase+, cytoCD79a- | No data reported | 1 mo | t(4;11)(q21;q23) MLL/AF4 | 40 y/F | 27 | ||
| 1 | 1 | 1 | CD19−,CD33+, CD11b+, CD14+, CD64+, HLADR+, CD38+, CD56+, CD4+, minor clone CD19+, CD22+, CD24+, CD38+ | Yes | 15 d | t(1;11)(p32;q23) MLL/EPS15 | 6 m/F | 32 | ||
F=Female; M=Male; y=year; mo=month; d=day
Lineage identity and plasticity in MLL-r B-ALL
Rearrangements of the MLL1 gene, including internal tandem duplications, occur in adults and children, producing leukemia with mixed myeloid, B-lymphoid, or T-lymphoid characteristics, hence the original designation of “mixed lineage” or “bi-phenotypic” leukemia. Whereas about half of older adults present with acute myelogenous leukemia (AML), the ratio of ALL to AML in infants with MLL-r leukemia is nearly 6 to 1 [37]. In infants, approximately 90% of MLL-r ALL is arrested at a CD19+ pro-B/pre-B stage [37]. Coexpression of myeloid genes and stage of B-cell differentiation (pro-B/pre-B cells) varies as a function of age of the patient [38]. As early as 1986, undifferentiated, mixed lineage features of MLL-r leukemia were appreciated and interpreted to reflect transformation of a multipotent progenitor [39]. The first transcriptional signatures illustrated the distinct identity of pediatric MLL-r B-ALL overall, which occupied a position in principal component space (cell identity) between lymphoid and myeloid leukemia [40]. Both immunophenotypic and genomic features of childhood MLL-r ALL have been interpreted to indicate that the cell of origin is a primitive fetal progenitor rather than a committed B-cell progenitor, given the association of MLL rearrangements with young age [41,42].
It is thus not surprising that MLL-r leukemia is common among cases of relapse-related lineage switching [43]. Reviewing pediatric cases from the 1980s to the 2010s, two groups carefully documented lineage switching after chemotherapy and found frequencies of 1%−6% of any lineage switch posttreatment, which was predominantly from pro-/pre-B-ALL to AML [44,45]. MLL-r leukemia accounted for 78% of the cases that switched lineages posttherapy in the latter study [45]. These clinical observations suggest an underlying heterogeneity or lineage plasticity inherent in cells transformed by MLL-fusion oncoproteins. The current designation of mixed phenotype acute leukemia (MPAL) includes a specific category for MLL-r, as well as BCR-ABL+ leukemia, because of their prevalence in this mixed lineage group [46,47]. Consistent with this concept of high lineage plasticity in MLL-r leukemia, evasion of CD19-targeted immunotherapy through relapse with myeloid markers has been reported under treatment with blinatumimab [20−27,32] or treatment with CD19 CAR T cells (Table 1) [10,18,30,31,33]. Loss of not only the CD19 cell surface protein but complete loss of all B-lineage markers and acquisition of a myeloid phenotype characterizes these cases (Table 1). In addition to these published data, two recent abstracts focusing on infant B-ALL revealed that of the 14 CD19 CAR-treated patients, the majority (79%) were MLL-r, and 4 exhibited a conversion to AML either during the primary CAR response or during relapse, and two of these were MLL-r [48]; and of 14 MLL-r infants treated with CD19 or CD19 × CD22 CARs, three relapsed with B-ALL and one relapsed with AML, overall suggesting a frequency that might be higher than that observed for chemotherapy [30]. However, it is important to emphasize that most patients in these studies remained in remission, and it is unclear how the lineage of the relapse relates to overall outcome.
In some reports, clonal relationships between the B-ALL and the AML are discussed through detailed characterization of the MLL rearrangement as well as immunoglobulin heavy chain rearrangements (Table 1) [18,23,26,32,33]. Recent single-cell genomics data suggest that pre-existing myeloid-primed B-ALL cells could be the source of such relapses (see Figure 2) [49]. In addition to MLL-r B-ALL, myeloid lineage-switched relapse has been reported for B-ALL harboring a Ph+ or Ph-like phenotype [35], as well as several other B-ALL categories with unique cytogenetics [31,50].
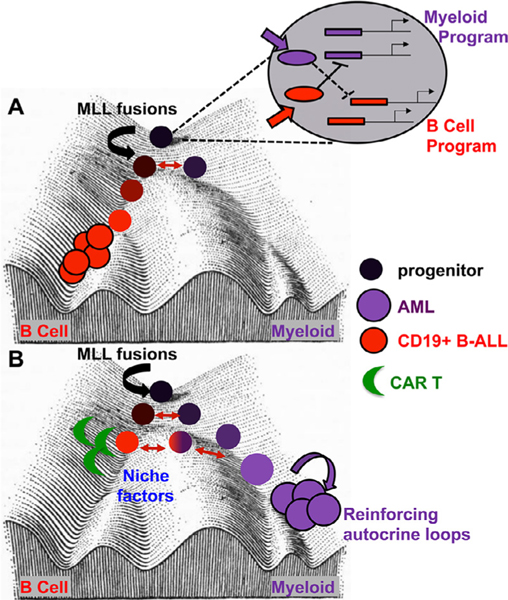
Figure 2.
Waddington landscape depicting the impact of MLL fusion oncoproteins and extrinsic factors on leukemia lineage. (A) The expanded progenitor cell diagram reveals myeloid- or lymphoid-promoting signals (filled arrows) promoting transcription factors (ovals) acting on lineage-directing enhancers (filled rectangles) to maintain exclusive lineage identity. Red double-headed arrows indicate the latent myeloid potential of transformed B-ALL which can overcome the activation energy to lose B-cell characteristics and gain myeloid identity. (B) Lineage switching on CD19-directed therapy (green crescent) as influenced by direct killing of the CD19+ B-ALL and/or impact of the immunotherapy and the niche on lineage decisions within the remaining B-ALL cells.
Model systems for studying MLL-r B-ALL evolution under CAR T cell pressure
Given the incomplete understanding of mechanisms that control lineage plasticity in MLL-r or other acute leukemias, a model system to study mechanisms of escape from CD19-directed immunotherapy would be immensely informative. Although MLL-r B-ALL can be efficiently generated in vitro from human cord blood or human fetal liver progenitors, [51,52] B-ALL, as modeled in xenograft systems, lacks the appropriate niche and endogenous immune components to accurately model in vivo evolution. Furthermore, xenograft models fail to recapitulate many conditions that impact CAR T-cell responses in vivo, such as the lack of CAR stimulation by non-malignant B cells expressing human CD19 antigen, limited cognate interactions between human T cells and the murine innate immune system, requirement for supraphysiological doses of CAR T cells, and the presence of xeno-reactivity between the human TCR and murine MHC complexes, which confounds the study of immune interactions and limits long-term studies because of lethal xenograft-versus-host disease. Ideally, a murine syngeneic system would allow testing of a variety of leukemia-intrinsic, niche, or CAR T cell pathways in the progression and relapse of B-ALL.
Some progress in developing such a system was presented in 2018, with the development of a TCF3-PBX1 cell line based on a model initially developed by Bijl and Sauvageau [53]. This in vitro−adapted CD19+ B-ALL consistently engrafts with as few as 100 cells in a syngeneic murine system. CD19 CAR T cells at doses comparable to those used clinically (2−2.5 × 106/kg) can completely cure animals engrafted with this TCF3-PBX1 line [54,55]. However, CD19neg “late relapses” were observed exhibiting features of a myeloid gene expression program [56]. These CD19neg relapse samples remain CD19 neg on secondary transfer and at the genomic level, reflect an epigenomic reprogramming away from B-cell identity toward myeloid identity [56]. This novel model system provided initial insights into the conditions that promote CD19neg relapse; however, it is unclear whether it reflects the actual processes underlying myeloid relapse in patients. First, TCF3−PBX1 fusions are not common and do not represent a poor prognostic group within pediatric B-ALLs, and patients harboring this fusion oncoprotein are not particularly prone to relapse with AML [57−59] Second, the CD19+ B-ALL line is adapted to culture and is unlikely to exhibit the same genomic plasticity as observed for primary B-ALL. Nonetheless, this model system provided a platform to study the epigenomic changes that occur on CD19neg relapse and the impact of CAR T-cell dose on this process.
In contrast to TCF3-PBX1 translocations, MLL translocations are enriched in myeloid relapses in B-ALL [18,45]. The propensity for lineage switching, particularly toward a myeloid identity, is likely an underlying property of MLL-r B-ALL, based on historical clinical observations [18,20−27,31−33]. It would clearly be beneficial to study processes leading to CD19neg relapse under immunotherapeutic pressure using a model system in which B-ALL is driven by MLL-r. B-ALL initiated by MLL fusion oncoproteins has been surprisingly difficult to produce in mouse models despite nearly 30 years of investigator efforts [60]. The first animal models of MLL-r leukemia in the 1990s used γ retroviruses to introduce MLL-fusion oncoproteins into murine bone marrow progenitors [61]. Retroviral introduction of MLL fusions such as MLL-AF9 and MLL-ENL was sufficient to induce leukemia in 100% of mice, but AML was produced, independent of cell type transduced [60]. In addition, transgenic expression of fusion oncoproteins from the endogenous Mll1 locus generally produced myeloid leukemia, even in conditions expressing fusions predominantly found in human B-ALL, and even when the fusion oncoprotein is directed selectively to lymphoid progenitors [62]. In some cases, B-ALL (defined by immunophenotype) can be produced using γ retroviral models of MLL-r leukemia, best exemplified by So et al. [63] in which an MLL-GAS7 fusion produced mixed lineage/B-ALL in conjunction with added FMS-like tyrosine kinase 3 ligand (FLT3L) and interleukin (IL)-7. Generally, the murine system (together with the gene expression program imposed by MLL fusion oncoproteins) strongly drives AML rather than B-ALL.
Nonetheless, recent publications have reported that using fetal cell types and carefully regulated levels of MLL fusion oncoproteins may improve the lineage fidelity of murine MLL-r leukemia model systems. In one case, inducible Mll-AF4 knock-in models that must be kept on a mixed genetic background exhibit ~30% B-ALL with many still succumbing to AML [64,65] Using knock-in models from the Rabbitts’ laboratory, several groups have found that progenitors exhibit an embryonic period of sensitivity to Mll-AF4 or Mll-ENL transformation [66,67]. For example, Malouf and Ottersbach [66] and Barrett et al. [68] reported that fetal liver lymphoid-primed progenitors exhibit a preleukemic phenotype dependent on Mll-AF4 expression but fall short of producing full-blown B-ALL. Okeyo-Owuor et al. [67] similarly found, using a distinct MLL-ENL knock-in, that a perinatal progenitor exhibited the peak sensitivity to MLL-ENL-mediated transformation; however, AML was the outcome in these mice. In an effort to study the role of the leukemia-extrinsic environment in lineage specification, Rowe et al. [69] found that serially transplanting MLL-AF9- or MLL-ENL−transduced progenitors through newborn mouse recipients (rather than adult recipients) enhanced the frequency of B-ALL−like phenotypes. This effect was attributed to an excess of myeloid-promoting chemokines and cytokines such as Ccl5 in the adult bone marrow niche relative to the newborn niche.
In contrast to these murine systems, both viral transduction of human umbilical cord blood progenitors [51,52,70,71] and CRISPR/Cas9 editing of human fetal cells [72] easily produce a B-ALL depending on the cytokines supplied during in vitro culture and on ontogeny [71] These observations collectively argue that the combination of fusion oncoprotein and the murine micro-environment produce a myeloid bias that does not accurately reflect the conditions found during early human development (Figure 2). Altering the collective conditions such that pediatric-relevant B-ALLs can be reliably and reproducibly generated in a mouse model would contribute significantly to discovering better cellular or immunotherapeutics for the most common MLL-r pediatric leukemia and, importantly, enable discovery of more effective methods for inducing long-term remission following targeted immunotherapy.
Can we anticipate and prevent lineage switching-related relapse?
To address this critical clinical question, it is important to understand key features of early B-cell fate commitment and resolution of B-cell versus myeloid identity. Hematopoietic differentiation occurs through a continuum of fate restriction events, and some of the earliest-defined B-cell progenitor−enriched populations including common lymphocyte progenitors (CLPs) and lymphoid primed multipotent progenitors (LPMPs) retain myeloid potential [73−76]. Interestingly, murine fetal CLPs and LPMPs exhibit more robust myeloid priming or myeloid potential than their adult counterparts [77,78], a phenomenon shared with human fetal progenitors [79]. The molecular basis for B-cell commitment is understood best at the level of transcriptional antagonistic and feed-forward hierarchies featuring well-studied transcription factors such as E2A, EBF, and PAX5 [80]. Latent myeloid potential of committed or even transformed pre-B cells was revealed in classic experiments manipulating PAX5 and/or the transcription factors of the C/EBPα family [81−85] through a mechanism that involves coordinated regulation of enhancers co-bound by EBF and C/EBPα. From these studies, it is clear that stochastic fluctuations in key fate-determining transcription factors could have a dramatic impact on the propensity of B-ALL to escape CD19-directed therapy through myeloid differentiation. In addition, the MLL fusion−dependent transcriptional network may perturb the B-cell fate network to tip the balance toward myelopoiesis, or the transformation of a particular progenitor stage may preserve an active enhancer network that retains the ability to respond to myeloid transcription factor networks (Figure 2).
Findings from the murine TCF3−PBX1 model system described earlier [54,86] suggested that late CD19neg relapses may have arisen from cellular reprogramming in the leukemia niche in which the CAR T cells are just one component. Extracellular signals elaborated in the CAR T cell/leukemia/niche such as inflammatory cytokines could promote activation of the Cebpa enhancer in B-ALL, influencing the cell’s propensity to adopt a myeloid fate. How could one apply these fundamental observations to the treatment of B-ALL? One approach could be to identify and target the signaling pathways leading to Cebpa enhancer activation, specifically signaling to activate myeloid-specific Cebpa enhancer elements [87]. Targeting such signals may block feed-forward transcriptional networks that promote myelopoiesis.
Another strategy for patients considered to be at risk for relapse of AML would be to anticipate this problem by applying immunotherapies that simultaneously target CD19 and myeloid-lineage antigens such as FLT3, CD33, or CD123 [88−90]. Given the ongoing development of myeloid-directed CAR T cells for AML, this approach may be closest to clinical use, but combinatorial toxicity would have to be carefully considered. FLT3 CARs have been effective in preclinical studies [91] and are particularly relevant because of the high expression levels on pediatric MLL-r B-ALL [91−96] as well as myeloid leukemias [98−100]. The optimal strategy for simultaneously targeting FLT3 and CD19 has not yet been determined, because approaches using a mixture of antigen-specific CAR T cells, CAR molecules with multiple single-chain variable fragments (ScFvs: antigen-binding domains), and T cells expressing multicistronic CAR molecules have shown efficacy in preclinical models as well as in patients [86,100−106]. Although these approaches would be expected to broaden the CAR response, allowing for the targeting of leukemia cells with either a B-ALL or myeloid phenotype, this would also broaden the “on-target, off-tumor” toxicity beyond B-cell aplasia to likely include myelosuppression. To test any of these strategies, there is a need for suitable preclinical model systems in which to test combinatorial B- and myeloid-directed CAR T cell strategies in a syngeneic, immunocompetent model to carefully test efficacy and toxic effects, because such studies are not possible in existing human−murine xenografts.
Outlook
Immunotherapies including CAR T cell therapies have provided new hope for treating many refractory malignancies. Despite remarkable successes in B-ALL, relapses remain a significant challenge. The mechanisms of resistance to immunotherapy are beginning to be elucidated [16,107,108], and it appears that lineage plasticity plays a role not only in hematologic cancers as described here but also in multiple tumor types; therefore, understanding the factors that influence lineage plasticity should lead to better anticipatory treatment strategies to combine with immunotherapies [109−112].
In the case of MLL-r B-ALL, there remain several unanswered questions relating to the impact of lineage switching and relapse with AML. First, the actual frequency of this event (as opposed to CD19neg relapse with B-cell phenotype) is unknown because of the lack of consistent documentation of this phenomenon in clinical trials and the relatively short time that CAR T cells have been in use. Because of the sporadic nature of case reports, it is unlikely that the true frequency of relapse as AML on CD19-directed therapies is well represented in the literature, particularly because MLL rearrangements are relatively rare. We would advocate for a more uniform, international system for documenting AML relapses using CD19-directed therapies such that frequencies and outcomes can be quantitatively determined. Cases of incomplete lineage conversion during relapse may go undetected because of the limited phenotypic analysis typically performed by standard flow cytometry, which may result in an underappreciation of the true frequency of lineage switch post-CAR treatment, a feature that genomic approaches may better capture [49]. Furthermore, whether lineage-switching relapse is worse than other forms of relapse remains an open question. Relevant to this question, a retrospective study of more than 200 infants diagnosed with MLL-r B-ALL treated on the Interfant-06 protocol provides clues that myeloid fate postinduction requires more aggressive therapy. Eighty percent of patients with high minimal residual disease (MRD) at the end of induction chemotherapy had B-ALL expressing at least one myeloid marker and shorter survival times as compared with patients with low MRD, who had lower frequencies of myeloid markers and survived longer. Indeed, use of myeloid-type chemotherapy was subsequently reported to improve outcomes in these patients [113]. These observations suggest that an underlying capacity for myeloid fate conversion in MLL-r B-ALL corresponds to poor survival and that this would pose a challenge for CD19-targeted immunotherapy as well. Understanding these issues for immunotherapy for B-ALL may have a broad impact as targeted immunotherapies are extended to other forms of cancer with underlying lineage plasticity.
An additional unanswered question is whether lineage switching relapse frequency differs between adult and infant B-ALL. One might expect that the developmental origin or history of the transformed cell could influence lineage fidelity, even under the influence of MLL fusion oncoproteins, based on the human and murine studies discussed earlier [67−69,71,73]. Again, the small numbers of such patients treated with immunotherapy/CAR T cell therapy lineage-switch relapses preclude such analysis at this time. An interesting approach to deducing the developmental history of the cell of origin in B-ALL was to derive a transcriptional signature distinguishing murine B1 (more prevalent during fetal development) versus B2 B-cell subtypes and ask whether pediatric B-ALL subtypes were enriched in this signature. Surprisingly, such an analysis revealed that MLL-r B-ALL exhibits a more B2-like transcriptional signature, despite its fetal origins [114]. Whether such a signature reflective of distinct fetal origins can be used to infer lineage fidelity or not must be experimentally determined.
One approach to preventing relapse from an inherently lineage-plastic B-ALL is to employ bispecific CARs to achieve higher selectivity and deeper killing of variants. In fact, CARs including CD133 have been suggested for MLL-r leukemia [115] because CD133 is a direct target of MLL-fusion oncoproteins [116] therefore, loss of both CD19 and CD133 would be very unlikely even if leukemia evolves. Similarly, FLT3 is highly expressed on most MLL-r B-ALLs, and FLT3 CARs are already well developed for use in AML [90,91]. Alternative cellular strategies such as CAR-NK and CAR-iNKT cells [117] may differentially affect the immune/niche/leukemia microenvironment and therefore have a distinct impact on AML relapse, but these strategies should be methodically tested in an appropriate model system.
There are two main hurdles to using the MLL-r leukemia paradigm to understand how to predict risk for and prevent relapse through lineage switch evasion of CD19 CAR T killing. First, an animal model system is desperately needed in which syngeneic or spontaneously arising B-ALL can be produced through induction of MLL fusion oncoproteins. Current model systems favor myelopoiesis and therefore do not accurately model the cellular or genomic features of human pediatric B-ALL driven by MLL fusion oncoproteins. Exciting new data are emerging that describe the B-ALL niche and how it changes postrelapse [49,118], but these systems are not as experimentally tractable as a syngeneic murine system, which would enable dynamic and robust assessments of the complete leukemia niche, as well as genetic manipulation of individual niche components.
Second, although there is a wealth of data surrounding transcriptional and epigenetic networks that control B-cell differentiation and latent myeloid potential, how to realistically manipulate those pathways in a clinical setting is unclear. Further characterization of the signals that exist in the leukemia niche and how they impact myeloid versus B-cell enhancer activity, for example, will be important to identify strategies to activate or suppress particular transcriptional networks. In this sense, reprogramming cancer cells could take a page from the pluripotency field, where cellular reprogramming studies have moved toward identifying small molecules that can replace some of the initial transcription factor manipulations [119−123]. In addition to preventing or treating antigen-negative relapse, such fate-regulating small molecules may also be helpful in combination with other targeted therapeutics to control or reverse tumor plasticity.
Acknowledgments
We thank Michael Verneris, Kathrin Bernt, and Matt Witkowski for their critical review and the anonymous reviewers for very constructive suggestions. We also thank Amy Moskop and Rebecca Gardner for communicating results before publication. This work was supported by funds from the Kate Amato and Morgan Adams Foundations (to Dr P Ernst), the Lady Tata Memorial Trust (to Dr W Liao) Hyundai Hope on Wheels, and K12 CA086913–18 (to Dr ME Kohler).
Footnotes
Conflict of interest disclosure
PE owns Amgen stock. TF is employed by Sana Biotechnology and is an inventor on immunotherapy patents owned by the National Institutes of Health.
References
- 1.Hunger SP, Mullighan CG. Acute lymphoblastic leukemia in children. N Engl J Med. 2015;373:1541–1552. [DOI] [PubMed] [Google Scholar]
- 2.Mullighan CG. Genomic characterization of childhood acute lymphoblastic leukemia. Semin Hematol. 2013;50:314–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hunger SP, Mullighan CG. Redefining ALL classification: toward detecting high-risk ALL and implementing precision medicine. Blood. 2015;125:3977–3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersson AK, Ma J, Wang J, et al. The landscape of somatic mutations in infant MLL-rearranged acute lymphoblastic leukemias. Nat Genet. 2015;47:330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gu Z, Churchman ML, Roberts KG, et al. PAX5-driven subtypes of B-progenitor acute lymphoblastic leukemia. Nat Genet. 2019;51:296–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Topp MS, Gökbuget N, Stein AS, et al. Safety and activity of blinatumomab for adult patients with relapsed or refractory B-precursor acute lymphoblastic leukaemia: a multicentre, single-arm, phase 2 study. Lancet Oncol. 2015;16:57–66. [DOI] [PubMed] [Google Scholar]
- 7.Pulte ED, Vallejo J, Przepiorka D, et al. FDA Supplemental Approval: blinatumomab for treatment of relapsed and refractory precursor B-cell acute lymphoblastic leukemia. Oncologist. 2018;23:1366–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maude SL, Laetsch TW, Buechner J, et al. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N Engl J Med. 2018;378:439–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gardner RA, Finney O, Annesley C, Brakke H. Intent-to-treat leukemia remission by CD19 CAR T cells of defined formulation and dose in children and young adults. Blood. 2017;129:3322–3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maude SL, Frey N, Shaw PA, et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N Engl J Med. 2014;371:1507–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee DW, Kochenderfer JN, Stetler-Stevenson M, et al. T cells expressing CD19 chimeric antigen receptors for acute lymphoblastic leukaemia in children and young adults: a phase 1 dose-escalation trial. Lancet. 2015;385:517–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mackall CL, Merchant MS, Fry TJ. Immune-based therapies for childhood cancer. Nat Rev Clin Oncol. 2014;11:693–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maude SL, Teachey DT, Rheingold SR, et al. Sustained remissions with CD19-specific chimeric antigen receptor (CAR)-modified T cells in children with relapsed/refractory ALL. J Clin Oncol. 2016;34. 3011–3011. [Google Scholar]
- 14.Lee DW III, Stetler-Stevenson M, Yuan CM, et al. Long-term outcomes following CD19 CAR T cell therapy for B-ALL are superior in patients receiving a fludarabine/cyclophosphamide preparative regimen and post-CAR hematopoietic stem cell transplantation. Blood. 2016;128. 218–218. [Google Scholar]
- 15.Pasquini MC, Hu ZH, Curran K, et al. Real-world evidence of tisagenlecleucel for pediatric acute lymphoblastic leukemia and non-Hodgkin lymphoma. Blood Adv. 2020;4:5414–5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shah NN, Fry TJ. Mechanisms of resistance to CAR T cell therapy. Nat Rev Clin Oncol. 2019;16:372–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sotillo E, Barrett DM, Black KL, et al. Convergence of acquired mutations and alternative splicing of CD19 enables resistance to CART-19 immunotherapy. Cancer Discov. 2015;5:1282–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gardner R, Wu D, Cherian S, et al. Acquisition of a CD19-negative myeloid phenotype allows immune escape of MLL-rearranged B-ALL from CD19 CAR-T-cell therapy. Blood. 2016;127:2406–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jabbour E, Düll J, Yilmaz M, et al. Outcome of patients with relapsed/refractory acute lymphoblastic leukemia after blinatumomab failure: no change in the level of CD19 expression. Am J Hematol. 2018;93:371–374. [DOI] [PubMed] [Google Scholar]
- 20.Haddox CL, Mangaonkar AA, Chen D, et al. Blinatumomab-induced lineage switch of B-ALL with t(4:11)(q21;q23) KMT2A/AFF1 into an aggressive AML: pre- and post-switch phenotypic, cytogenetic and molecular analysis. Blood Cancer J. 2017;7:e607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wölfl M, Rasche M, Eyrich M, Schmid R, Reinhardt D, Schlegel PG. Spontaneous reversion of a lineage switch following an initial blinatumomab-induced ALL-to-AML switch in MLL-rearranged infant ALL. Blood Adv. 2018;2:1382–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aldoss I, Song JY. Extramedullary relapse of KMT2A(MLL)-rearranged acute lymphoblastic leukemia with lineage switch following blinatumomab. Blood. 2018;131:2507. [DOI] [PubMed] [Google Scholar]
- 23.Fournier E, Inchiappa L, Delattre C, et al. Increased risk of adverse acute myeloid leukemia after anti-CD19-targeted immunotherapies in KMT2A-rearranged acute lymphoblastic leukemia: a case report and review of the literature. Leuk Lymphoma. 2019;60:1827–1830. [DOI] [PubMed] [Google Scholar]
- 24.Rayes A, McMasters RL, O’Brien MM. Lineage switch in MLL-rearranged infant leukemia following CD19-directed therapy. Pediatr Blood Cancer. 2016;63:1113–1115. [DOI] [PubMed] [Google Scholar]
- 25.Zoghbi A, Zur Stadt U, Winkler B, Muller I, Escherich G. Lineage switch under blinatumomab treatment of relapsed common acute lymphoblastic leukemia without MLL rearrangement. Pediatr Blood Cancer. 2017;64. 10.1002/pbc.26594. [DOI] [PubMed] [Google Scholar]
- 26.Balducci E, Nivaggioni V, Boudjarane J, et al. Lineage switch from B acute lymphoblastic leukemia to acute monocytic leukemia with persistent t(4;11)(q21;q23) and cytogenetic evolution under CD19-targeted therapy. Ann Hematol. 2017;96:1579–1581. [DOI] [PubMed] [Google Scholar]
- 27.He RR, Nayer Z, Hogan M, et al. Immunotherapy- (blinatumomab-) related lineage switch of KMT2A/AFF1 rearranged B-lymphoblastic leukemia into acute myeloid leukemia/myeloid sarcoma and subsequently into B/myeloid mixed phenotype acute leukemia. Case Rep Hematol. 2019: 7394619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park JH, Riviere I, Gonen M, et al. Long-term follow-up of CD19 CAR therapy in acute lymphoblastic leukemia. N Engl J Med. 2018;378:449–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Braig F, Brandt A, Goebeler M, et al. Resistance to anti-CD19/ CD3 BiTE in acute lymphoblastic leukemia may be mediated by disrupted CD19 membrane trafficking. Blood. 2017; 129:100–104. [DOI] [PubMed] [Google Scholar]
- 30.Annesley C, Summers C, Pulsipher MA, et al. Clinical experience of CAR T cell immunotherapy for relapsed and refractory infant ALL demonstrates feasibility and favorable responses. Blood. 2019;134:3869. [Google Scholar]
- 31.Oberley MJ, Gaynon PS, Bhojwani D, et al. Myeloid lineage switch following chimeric antigen receptor T-cell therapy in a patient with TCF3-ZNF384 fusion-positive B-lymphoblastic leukemia. Pediatr Blood Cancer. 2018;65:e27265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du J, Chisholm KM, Tsuchiya K, et al. Lineage switch in an infant B-lymphoblastic leukemia with t(1;11)(p32;q23); KMT2A/EPS15, following blinatumomab therapy. Pediatr Dev Pathol. 2021;24:378–382. [DOI] [PubMed] [Google Scholar]
- 33.Turtle CJ, Hanafi LA, Berger C. CD19 CAR-T cells of defined CD4+:CD8+ composition in adult B cell ALL patients. J Clin Invest. 2016;126:2123–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagel I, Bartels M, Duell J, et al. Hematopoietic stem cell involvement in BCR-ABL1-positive ALL as a potential mechanism of resistance to blinatumomab therapy. Blood. 2017;130:2027–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pieters R, De Lorenzo P, Ancliffe P, et al. Outcome of infants younger than 1 year with acute lymphoblastic leukemia treated with the Interfant-06 Protocol: results from an international phase III randomized study. J Clin Oncol. 2019;37:2246–2256. [DOI] [PubMed] [Google Scholar]
- 36.Kanemitsu Y, Shitara K, Mizusawa J, et al. Primary tumor resection plus chemotherapy versus chemotherapy alone for colorectal cancer patients with asymptomatic, synchronous unresectable metastases (JCOG1007; iPACS): a randomized clinical trial. J Clin Oncol. 2021;39:1098–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marschalek R. Systematic classification of mixed-lineage leukemia fusion partners predicts additional cancer pathways. Ann Lab Med. 2016;36:85–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jansen MWJC, Corral L, van der Velden VHJ, et al. Immunobiological diversity in infant acute lymphoblastic leukemia is related to the occurrence and type of MLL gene rearrangement. Leukemia. 2007;21:633–641. [DOI] [PubMed] [Google Scholar]
- 39.Mirro J, Kitchingman G, Williams D, et al. Clinical and laboratory characteristics of acute leukemia with the 4;11 translocation. Blood. 1986;67:689–697. [PubMed] [Google Scholar]
- 40.Armstrong SA, Staunton JE, Silverman LB, et al. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30:41–47. [DOI] [PubMed] [Google Scholar]
- 41.Ford AM, Ridge SA, Cabrera ME, et al. In utero rearrangements in the trithorax-related oncogene in infant leukaemias. Nature. 1993;363:358–360. [DOI] [PubMed] [Google Scholar]
- 42.Greaves MF, Maia AT, Wiemels JL, Ford AM. Leukemia in twins: lessons in natural history. Blood. 2003;102:2321–2333. [DOI] [PubMed] [Google Scholar]
- 43.Dorantes-Acosta E, Pelayo R. Lineage switching in acute leukemias: a consequence of stem cell plasticity? Bone Marrow Res. 2012;2012:406796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stass S, Mirro J, Melvin S, Pui CH, Murphy SB, Williams D. Lineage switch in acute leukemia. Blood. 1984;64:701–706. [PubMed] [Google Scholar]
- 45.Rossi JG, Bernasconi AR, Alonso CN, et al. Lineage switch in childhood acute leukemia: an unusual event with poor outcome. Am J Hematol. 2012;87:890–897. [DOI] [PubMed] [Google Scholar]
- 46.Weinberg OK, Arber DA. Mixed-phenotype acute leukemia: historical overview and a new definition. Leukemia. 2010;24:1844–1851. [DOI] [PubMed] [Google Scholar]
- 47.Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–2405. [DOI] [PubMed] [Google Scholar]
- 48.Moskop A, Breese E, Guest E, et al. Real-world use of tisagenlecleucel in infant acute lymphoblastic leukemia. Transplant Cell Ther. 2021;27:S73–S74. [Google Scholar]
- 49.Chen C, Yu W, Alikarami F, et al. Single-cell multi-omics reveals elevated plasticity and stem-cell-like blasts relevant to the poor prognosis of KMT2A-rearranged leukemia. bioRxiv. 2020. 10.1101/2020.12.06.413930. Online ahead of print. [DOI] [Google Scholar]
- 50.Novákova M, Žaliova M, Suková M, et al. Loss of B cells and their precursors is the most constant feature of GATA-2 deficiency in childhood myelodysplastic syndrome. Haematologica. 2016;101:707–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin S, Luo RT, Ptasinska A, et al. Instructive role of MLL-fusion proteins revealed by a model of t(4;11) pro-B acute lymphoblastic leukemia. Cancer Cell. 2016;30:737–749. [DOI] [PubMed] [Google Scholar]
- 52.Barabe F, Kennedy JA, Hope KJ, Dick JE. Modeling the initiation and progression of human acute leukemia in mice. Science. 2007;316:600–604. [DOI] [PubMed] [Google Scholar]
- 53.Bijl J, Sauvageau M, Thompson A, Sauvageau G. High incidence of proviral integrations in the Hoxa locus in a new model of E2a-PBX1-induced B-cell leukemia. Genes Dev. 2005;19:224–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kohler ME, Yang Y, Sepulveda G, Fry TJ. A direct comparison of the in vivo efficacy and in vitro signaling of chimeric antigen receptors and endogenous T cell receptors. Blood. 2017;130:4451. [Google Scholar]
- 55.Qin H, Ishii K, Nguyen S, et al. Murine pre−B-cell ALL induces T-cell dysfunction not fully reversed by introduction of a chimeric antigen receptor. Blood. 2018;132:1899–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jacoby E, Nguyen SM, Fountaine TJ, et al. CD19 CAR immune pressure induces B-precursor acute lymphoblastic leukaemia lineage switch exposing inherent leukaemic plasticity. Nat Commun. 2016;7:12320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burmeister T, Gökbuget N, Schwartz S, et al. Clinical features and prognostic implications of TCF3-PBX1 and ETV6-RUNX1 in adult acute lymphoblastic leukemia. Haematologica. 2010;95:241–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kager L, Lion T, Attarbaschi A, et al. Incidence and outcome of TCF3-PBX1-positive acute lymphoblastic leukemia in Austrian children. Haematologica. 2007;92:1561–1564. [DOI] [PubMed] [Google Scholar]
- 59.Asai D, Imamura T, Yamashita Y, et al. Outcome of TCF3-PBX1 positive pediatric acute lymphoblastic leukemia patients in Japan: a collaborative study of Japan Association of Childhood Leukemia Study (JACLS) and Children’s Cancer and Leukemia Study Group (CCLSG). Cancer Med. 2014;3:623–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li BE, Ernst P. Two decades of leukemia oncoprotein epistasis: the MLL1 paradigm for epigenetic deregulation in leukemia. Exp Hematol. 2014;42:995–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lavau C, Szilvassy SJ, Slany R, Cleary ML. Immortalization and leukemic transformation of a myelomonocytic precursor by retrovirally transduced HRX-ENL. EMBO J. 1997;16:4226–4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cano F, Drynan LF, Pannell R, Rabbitts TH. Leukaemia lineage specification caused by cell-specific Mll−Enl translocations. Oncogene. 2008;27:1945–1950. [DOI] [PubMed] [Google Scholar]
- 63.So CW, Karsunky H, Passegue E, Cozzio A, Weissman IL, Cleary ML. MLL-GAS7 transforms multipotent hematopoietic progenitors and induces mixed lineage leukemias in mice. Cancer Cell. 2003;3:161–171. [DOI] [PubMed] [Google Scholar]
- 64.Krivtsov AV, Feng Z, Lemieux ME, et al. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell. 2008;14:355–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chu SH, Song EJ, Chabon JR, et al. Inhibition of MEK and ATR is effective in a B-cell acute lymphoblastic leukemia model driven by Mll-Af4 and activated Ras. Blood Adv. 2018;2:2478–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Malouf C, Ottersbach K. Fetal liver Mll-AF4+ hematopoietic stem and progenitor cells respond directly to poly(I:C), but not to a single maternal immune activation. Exp Hematol. 2019;76:49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Okeyo-Owuor T, Li Y, Patel RM, et al. The efficiency of murine MLL-ENL-driven leukemia initiation changes with age and peaks during neonatal development. Blood Adv. 2019;3:2388–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Barrett NA, Malouf C, Kapeni C, et al. Mll-AF4 confers enhanced self-renewal and lymphoid potential during a restricted window in development. Cell Rep. 2016;16:1039–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rowe RG, Lummertz da Rocha E, Sousa P, et al. The developmental stage of the hematopoietic niche regulates lineage in MLL-rearranged leukemia. J Exp Med. 2019;216:527–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wei J, Wunderlich M, Fox C, et al. Microenvironment determines lineage fate in a human model of MLL-AF9 leukemia. Cancer Cell. 2008;13:483–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Horton SJ, Jaques J, Woolthuis C, et al. MLL-AF9-mediated immortalization of human hematopoietic cells along different lineages changes during ontogeny. Leukemia. 2013;27:1116–1126. [DOI] [PubMed] [Google Scholar]
- 72.Rice S, Jackson T, Crump NT, et al. A novel human fetal liver-derived model reveals that MLL-AF4 drives a distinct fetal gene expression program in infant ALL. bioRxiv. 2020. 10.1101/2020.11.15.379990. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rumfelt LL, Zhou Y, Rowley BM, Shinton SA, Hardy RR. Lineage specification and plasticity in CD19- early B cell precursors. J Exp Med. 2006;203:675–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fischer U, Yang JJ, Ikawa T, et al. Cell fate decisions: the role of transcription factors in early B-cell development and leukemia. Blood Cancer Discov. 2020;1:224–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Adolfsson J, Ma nsson R, Buza-Vidas N, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential: a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. [DOI] [PubMed] [Google Scholar]
- 76.Iwasaki H, Akashi K. Myeloid lineage commitment from the hematopoietic stem cell. Immunity. 2007;26:726–740. [DOI] [PubMed] [Google Scholar]
- 77.Mansson R, Hultquist A, Luc S, et al. Molecular evidence for hierarchical transcriptional lineage priming in fetal and adult stem cells and multipotent progenitors. Immunity. 2007;26:407–419. [DOI] [PubMed] [Google Scholar]
- 78.Böiers C, Carrelha J, Lutteropp M, et al. Lymphomyeloid contribution of an immune-restricted progenitor emerging prior to definitive hematopoietic stem cells. Cell Stem Cell. 2013;13:535–548. [DOI] [PubMed] [Google Scholar]
- 79.Jackson TR, Ling RE, Roy A. The origin of B-cells: human fetal B cell development and implications for the pathogenesis of childhood acute lymphoblastic leukemia. Front Immunol. 2021;12:637975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Boller S, Grosschedl R. The regulatory network of B-cell differentiation: a focused view of early B-cell factor 1 function. Immunol Rev. 2014;261:102–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rolink AG, Schaniel C, Bruno L, Melchers F. In vitro and in vivo plasticity of Pax5-deficient pre-B I cells. Immunol Lett. 2002;82:35–40. [DOI] [PubMed] [Google Scholar]
- 82.Rolink AG, Nutt SL, Melchers F, Busslinger M. Long-term in vivo reconstitution of T-cell development by Pax5-deficient B-cell progenitors. Nature. 1999;401:603–606. [DOI] [PubMed] [Google Scholar]
- 83.Cirovic B, Schönheit J, Kowenz-Leutz E, et al. C/EBP-Induced transdifferentiation reveals granulocyte−macrophage precursor-like plasticity of B cells. Stem Cell Rep. 2017;8:346–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bussmann LH, Schubert A, Manh TPV, et al. A robust and highly efficient immune cell reprogramming system. Cell Stem Cell. 2009;5:554–566. [DOI] [PubMed] [Google Scholar]
- 85.Di Stefano B, Sardina JL, van Oevelen C, et al. C/EBPalpha poises B cells for rapid reprogramming into induced pluripotent stem cells. Nature. 2014;506:235–239. [DOI] [PubMed] [Google Scholar]
- 86.Qin H, Nguyen SM, Ramakrishna S, et al. Novel CD19/CD22 bicistronic chimeric antigen receptors outperform single or bivalent cars in eradicating CD19+CD22+, CD19−, and CD22 − pre-B leukemia. Blood. 2017;130. 810–810. [Google Scholar]
- 87.Avellino R, Delwel R. Expression and regulation of C/EBPalpha in normal myelopoiesis and in malignant transformation. Blood. 2017;129:2083–2091. [DOI] [PubMed] [Google Scholar]
- 88.Yan LE, Zhang H, Wada M, et al. Targeting two antigens associated with B-ALL with CD19-CD123 compound Car T cell therapy. Stem Cell Rev Rep. 2020;16:385–396. [DOI] [PubMed] [Google Scholar]
- 89.Krupka C, Kufer P, Kischel R, et al. CD33 target validation and sustained depletion of AML blasts in long-term cultures by the bispecific T-cell-engaging antibody AMG 330. Blood. 2014;123:356–365. [DOI] [PubMed] [Google Scholar]
- 90.Oelsner S, Waldmann A, Billmeier A, et al. Genetically engineered CAR NK cells display selective cytotoxicity against FLT3-positive B-ALL and inhibit in vivo leukemia growth. Int J Cancer. 2019;145:1935–1945. [DOI] [PubMed] [Google Scholar]
- 91.Sommer C, Cheng HY, Nguyen D, et al. Allogeneic FLT3 CAR T cells with an off-switch exhibit potent activity against AML and can be depleted to expedite bone marrow recovery. Mol Ther. 2020;28:2237–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chillón MC, Gomez-Casares MT, López-Jorge CE, et al. Prognostic significance of FLT3 mutational status and expression levels in MLL−AF4+ and MLL− germline acute lymphoblastic leukemia. Leukemia. 2012;26. 2360–2306. [DOI] [PubMed] [Google Scholar]
- 93.Annesley CE, Brown P. The biology and targeting of FLT3 in pediatric leukemia. Front Oncol. 2014;4:263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Brown P, Levis M, Shurtleff S, Campana D, Downing J, Small D. FLT3 inhibition selectively kills childhood acute lymphoblastic leukemia cells with high levels of FLT3 expression. Blood. 2005;105:812–820. [DOI] [PubMed] [Google Scholar]
- 95.Stam RW, den Boer ML, Schneider P, et al. Targeting FLT3 in primary MLL-gene-rearranged infant acute lymphoblastic leukemia. Blood. 2005;106:2484–2490. [DOI] [PubMed] [Google Scholar]
- 96.Brown P, Levis M, McIntyre E, Griesemer M, Small D. Combinations of the FLT3 inhibitor CEP-701 and chemotherapy synergistically kill infant and childhood MLL-rearranged ALL cells in a sequence-dependent manner. Leukemia. 2006;20:1368–1376. [DOI] [PubMed] [Google Scholar]
- 97.Perl AE, Altman JK, Cortes J, et al. Selective inhibition of FLT3 by gilteritinib in relapsed or refractory acute myeloid leukaemia: a multicentre, first-in-human, open-label, phase 1–2 study. Lancet Oncol. 2017;18:1061–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Perl AE, Martinelli G, Cortes JE, et al. Gilteritinib or chemotherapy for relapsed or refractory FLT3-mutated AML. N Engl J Med. 2019;381:1728–1740. [DOI] [PubMed] [Google Scholar]
- 99.Levis M, Ravandi F, Wang ES, et al. Results from a randomized trial of salvage chemotherapy followed by lestaurtinib for patients with FLT3 mutant AML in first relapse. Blood. 2011;117:3294–3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang N, Hu X, Cao W, et al. Efficacy and safety of CAR19/22 T-cell cocktail therapy in patients with refractory/relapsed B-cell malignancies. Blood. 2020;135:17–27. [DOI] [PubMed] [Google Scholar]
- 101.Fry TJ, Shah NN, Orentas RJ, et al. CD22-targeted CAR T cells induce remission in B-ALL that is naive or resistant to CD19-targeted CAR immunotherapy. Nat Med. 2018;24: 20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zah E, Lin MY, Silva-Benedict A, Jensen MC, Chen YY. T cells expressing CD19/CD20 bispecific chimeric antigen receptors prevent antigen escape by malignant B cells. Cancer Immunol Res. 2016;4:498–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schultz LM, Davis KL, Baggott C, et al. Phase 1 study of CD19/CD22 bispecific chimeric antigen receptor (CAR) therapy in children and young adults with B cell acute lymphoblastic leukemia (ALL). Blood. 2018;132. 898–898. [Google Scholar]
- 104.de Larrea CF, Staehr M, Lopez AV, et al. Defining an optimal dual-targeted CAR T-cell therapy approach simultaneously targeting BCMA and GPRC5D to prevent BCMA escape-driven relapse in multiple myeloma. Blood Cancer Discov. 2020;1:146–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shah NN, Johnson BD, Schneider D, et al. Bispecific anti-CD20, anti-CD19 CAR T cells for relapsed B cell malignancies: a phase 1 dose escalation and expansion trial. Nat Med. 2020;26:1569–1575. [DOI] [PubMed] [Google Scholar]
- 106.Bielamowicz K, Fousek K, Byrd TT, et al. Trivalent CAR T cells overcome interpatient antigenic variability in glioblastoma. Neuro Oncol. 2018;20:506–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cheng J, Zhao L, Zhang Y, et al. Understanding the mechanisms of resistance to CAR T-cell therapy in malignancies. Front Oncol. 2019;9:1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Song MK, Park BB, Uhm JE. Resistance mechanisms to CAR T-cell therapy and overcoming strategy in B-cell hematologic malignancies. Int J Mol Sci. 2019;20:5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Breindel JL, Skibinski A, Sedic M, et al. Epigenetic reprogramming of lineage-committed human mammary epithelial cells requires DNMT3A and loss of DOT1L. Stem Cell Rep. 2017;9:943–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Horn LA, Fousek K, Palena C. Tumor plasticity and resistance to immunotherapy. Trends Cancer. 2020;6:432–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Landsberg J, Kohlmeyer J, Renn M, et al. Melanomas resist T-cell therapy through inflammation-induced reversible dedifferentiation. Nature. 2012;490:412–416. [DOI] [PubMed] [Google Scholar]
- 112.Mehta A, Kim YJ, Robert L, et al. Immunotherapy resistance by inflammation-induced dedifferentiation. Cancer Discov. 2018;8:935–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Stutterheim J, van der Sluis IM, de Lorenzo P, et al. Clinical implications of minimal residual disease detection in infants with KMT2A-rearranged acute lymphoblastic leukemia treated on the interfant-06 protocol. J Clin Oncol. 2021;39:652–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fitch B, Roy R, Geng H, et al. Human pediatric B-cell acute lymphoblastic leukemias can be classified as B-1 or B-2-like based on a minimal transcriptional signature. Exp Hematol. 2020;90. 65−71.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li D, Hu Y, Jin Z, et al. TanCAR T cells targeting CD19 and CD133 efficiently eliminate MLL leukemic cells. Leukemia. 2018;32:2012–2016. [DOI] [PubMed] [Google Scholar]
- 116.Godfrey L, Crump NT, O’Byrne S, et al. H3K79me2/3 controls enhancer−promoter interactions and activation of the pan-cancer stem cell marker PROM1/CD133 in MLL-AF4 leukemia cells. Leukemia. 2021;35:90–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rotolo A, Caputo VS, Holubova M, et al. Enhanced anti-lymphoma activity of CAR19-iNKT cells underpinned by dual CD19 and CD1d targeting. Cancer Cell. 2018;34. 596−610. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Witkowski MT, Dolgalev I, Evensen NA, et al. Extensive remodeling of the immune microenvironment in B cell acute lymphoblastic leukemia. Cancer Cell. 2020;37. 867−882.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhang Y, Li W, Laurent T, Ding S. Small molecules, big roles —The chemical manipulation of stem cell fate and somatic cell reprogramming. J Cell Sci. 2012;125:5609–5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kim Y, Jeong J, Choi D. Small-molecule-mediated reprogramming: a silver lining for regenerative medicine. Exp Mol Med. 2020;52:213–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Xie H, Ye M, Feng R, Graf T. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117:663–676. [DOI] [PubMed] [Google Scholar]
- 122.Mikkola I, Heavey B, Horcher M, Busslinger M. Reversion of B cell commitment upon loss of Pax5 expression. Science. 2002;297:110–113. [DOI] [PubMed] [Google Scholar]
- 123.Onder TT, Kara N, Cherry A, et al. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012;483:598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]