Abstract
Background
The molecular pathology underlying posttraumatic stress disorder (PTSD) remains unclear mainly due to a lack of human PTSD postmortem brain tissue. The orexigenic neuropeptides ghrelin, neuropeptide Y, and hypocretin were recently implicated in modulating negative affect. Drawing from the largest functional genomics study of human PTSD postmortem tissue, we investigated whether there were molecular changes of these and other appetitive molecules. Further, we explored the interaction between PTSD and body mass index (BMI) on gene expression.
Methods
We analyzed previously reported transcriptomic data from 4 prefrontal cortex regions from 52 individuals with PTSD and 46 matched neurotypical controls. We employed gene co-expression network analysis across the transcriptomes of these regions to uncover PTSD-specific networks containing orexigenic genes. We utilized Ingenuity Pathway Analysis software for pathway annotation. We identified differentially expressed genes (DEGs) among individuals with and without PTSD, stratified by sex and BMI.
Results
Three PTSD-associated networks (P < .01) contained genes in signaling families of appetitive molecules: 2 in females and 1 in all subjects. We uncovered DEGs (P < .05) between PTSD and control subjects stratified by sex and BMI with especially robust changes in males with PTSD with elevated vs normal BMI. Further, we identified putative upstream regulators (P < .05) driving these changes, many of which were enriched for involvement in inflammation.
Conclusions
PTSD-associated cortical transcriptomic modules contain transcripts of appetitive genes, and BMI further interacts with PTSD to impact expression. DEGs and inferred upstream regulators of these modules could represent targets for future pharmacotherapies for obesity in PTSD.
Keywords: PTSD, inflammation, BMI, transcriptomics
Significance Statement.
Posttraumatic stress disorder (PTSD) is a debilitating psychiatric disease that can afflict individuals who have witnessed or experienced trauma. It is characterized by disturbing feelings and thoughts associated with the event, persisting for months to years after. Although PTSD impacts a substantial proportion of our society, our understanding of underlying molecular mechanisms is limited, and effective treatments are lacking. Recently, the largest transcriptomic study of the human PTSD brain was completed. We employed this dataset to study the involvement of hunger-related molecules in PTSD pathophysiology, as there is evidence that they can impact emotion. We show that unique networks of genes in the brains of individuals with PTSD contain hunger molecules and that body mass index may impact brain gene expression. These findings enhance our understanding of PTSD and may facilitate the development of medications to target obesity in this population.
Introduction
Posttraumatic stress disorder (PTSD) is a devastating psychiatric illness characterized by a fearful response to traumatic events and protracted symptoms such as avoidance, arousal, and re-experiencing persisting for years after the events. The estimated lifetime prevalence of PTSD among Americans is approximately 7% (Kessler et al., 2005), and the rate is even higher among particular groups such as combat veterans (23% of returning soldiers from the wars in Iraq and Afghanistan, as a recent meta-analysis revealed) (Fulton et al., 2015) and individuals residing in inner city neighborhoods (between 43% and 51%) (Schwartz et al., 2005; Alim et al., 2006; Gillespie et al., 2009). Despite these striking numbers, there are few effective medications to treat symptoms or for prophylaxis. PTSD manifests as a result of emotion regulatory dysfunction, likely due to perturbation in fear circuits (comprised of the prefrontal cortex [PFC], amygdala, and hippocampus) (Abdallah et al., 2019). The molecular neurobiology of fear memory in rodents has been significantly studied (Milad and Quirk, 2002; Wilensky et al., 2006; Parsons and Ressler, 2013); however, the molecular pathophysiology of PTSD in humans remains poorly understood. This dearth in the literature has in part been a consequence of the inability to examine the brains of humans with PTSD (Krystal and Duman, 2004; Girgenti and Duman, 2018).
An emerging literature suggests that neuropeptide Y (NPY), hypocretin, and ghrelin, all of which are orexigenic neuropeptides, may represent potentially promising targets for research on the pathophysiology of PTSD (Morgan et al., 2000; Zhou et al., 2008; Sah et al., 2009; Strawn et al., 2010; Meyer et al., 2014; Sah et al., 2014; Cohen et al., 2016; Harmatz et al., 2017; Yousufzai et al., 2018). Multiple lines of evidence suggest a role for these neuropeptides in the context of modulation of negative affective states, specifically in the context of exposure to trauma. For instance, NPY promotes stress resilience in humans, as military soldiers with elevated plasma levels demonstrate enhanced coping following extreme stress (Morgan et al., 2000), and individual variation in expression of NPY mRNA modulates emotion and resiliency (Zhou et al., 2008). Levels of NPY in the cerebrospinal fluid (CSF) are reduced in individuals with combat-associated PTSD (relative to healthy subjects) (Sah et al., 2009) and in veterans exposed to combat who develop PTSD relative to those who do not (Sah et al., 2014). Orexin-A (hypocretin-1) is also decreased among combat veterans with PTSD in both plasma and the CSF, relative to neurotypical comparison subjects, and CSF levels negatively correlate with PTSD symptom severity (Strawn et al., 2010). While little is known about levels of ghrelin in the CSF in humans, rodent research has demonstrated that resistance to ghrelin in the central nervous system promotes fear enhancement following chronic stress in a rodent model of PTSD (in comparison, among unstressed rodents, ghrelin reduces fear) (Meyer et al., 2014; Harmatz et al., 2017). Severe stressor exposure among adolescent humans facilitates an increase in circulating serum ghrelin for at least 4.5 years, and a parallel sustained elevation in ghrelin in rodents promotes central ghrelin resistance via a decrease in available receptors (Harmatz et al., 2017; Yousufzai et al., 2018). Thus, reduced central actions of NPY, hypocretin, and ghrelin may induce vulnerability to PTSD.
The circuit-based interactions between ghrelin, NPY, and hypocretin, as well as their widespread expression in the brain, increase interest in their possible role in the biology of PTSD. Ghrelin and orexin-A stimulate NPY neurons in the arcuate nucleus of the hypothalamus (Kohno et al., 2003), a major hunger center of the brain (Krashes et al., 2014). Further, members of the signaling families of these neuropeptides are expressed in the cerebral cortex, and NPY- and hypocretin-related molecules are specifically expressed in regions of interest for PTSD, such as the anterior cingulate cortex (Talebizadeh et al., 2005; Sommer et al., 2010; Lu et al., 2017). Moreover, the resilience-promoting effects of modafinil in a rodent stress model were associated with upregulation of both hypocretin and NPY systems in the hypothalamus (Cohen et al., 2016).
Here we report the results of an exploratory gene coexpression network analysis examining NPY, hypocretin, and ghrelin signaling pathways associated with PTSD. This analysis was based on a reanalysis of RNA sequencing results from human postmortem PFC tissue obtained from individuals with and without PTSD from the National PTSD Brain Bank of the United States Department of Veterans Affairs (Friedman et al., 2017; Girgenti et al., 2020). Because these neuropeptides are implicated in the regulation of appetite and weight (Sahu and Kalra, 1993; Willie et al., 2001; Muller et al., 2015), we also explored the impact of body mass index (BMI) on the gene expression changes in the PTSD brain.
Methods
Research Subjects and Clinical Assessment
The human postmortem brain tissue samples analyzed in this study were obtained from 52 individuals with PTSD (26 males, 26 females) and 46 matched neurotypical comparison subjects (20 females, 26 males) through the University of Pittsburg Medical Center and the National Center for PTSD Brain Bank. The demographic and tissue characteristics of the sample are presented in Table 1. Tissue samples were group-matched for age, postmortem interval (PMI), and pH. Inclusion criteria were PMI less than 30 hours and age older than 18 but less than 70. Demographic and clinical characteristics were determined by psychological autopsy, that is, diagnoses were determined in accordance with the DSM-IV via SCID-1 interviews for use in psychological autopsy. Between-group differences in these parameters were assessed using Student’s t test, employing P < .05 for statistical significance.
Table 1.
Demographic and Tissue Characteristics
| Neurotypical control (n = 46) | Posttraumatic stress disorder (n = 52) | |
|---|---|---|
| Sex (% male) | 56.5 | 50.0 |
| Body mass index (kg/m2) | 33.3 ± 1.0 | 29.2 ± 1.4 |
| Race (% Caucasian) | 65.2 | 81.0 |
| Tobacco (% use) | 21.7 | 65.0 |
| Alcohol (% use) | 0.0 | 35.0 |
| Opioids (% use) | 8.7 | 15.4 |
| Antidepressant (% use) | 0.0 | 60.0 |
| Average age at death (y) | 48.50 ± 12.40 | 42.80 ± 11.50 |
| Postmortem interval (h) | 21.80 ± 6.60 | 20.50 ± 6.30 |
| Manner of death (% suicide/% natural) | 0.0 | 12.0/88.0 |
| Drug-related death (%) | 0.0 | 52.0 |
| RNA Integrity Number: OFC | 7.70 ± 0.97 | 7.90 ± 1.00 |
| RNA Integrity Number: sgPFC | 8.10 ± 0.85 | 7.90 ± 0.99 |
| RNA Integrity Number: dACC | 7.40 ± 0.98 | 7.30 ± 1.06 |
| RNA Integrity Number: dlPFC | 7.40 ± 1.19 | 7.70 ± 0.92 |
Abbreviations: dACC, dorsal anterior cingulate; dlPFC, dorsolateral PFC; OFC, orbitofrontal cortex; sgPFC, subgenual PFC.
Results for categorical variables are presented as percentages. Results for continuous variables are presented as mean ± SEM. All P values for between-group differences are nonsignificant (P > .05) except for in the case of body mass index (P = .02). Some of these demographic and tissue characteristics have been previously reported (Girgenti et al., 2020); permission has been obtained from the authors.
Fresh frozen tissue (20 mg) from 4 PFC regions—the dorsal anterior cingulate (dACC; BA24), orbitofrontal cortex (OFC; BA11), dorsolateral PFC (dlPFC; BA9/46), and subgenual PFC (sgPFC; BA25)—was obtained from each brain.
RNA Extraction and Sequencing Procedures
These procedures are detailed in previous work (Girgenti et al., 2020) and are summarized here. RNA was obtained with an RNeasy Mini Kit with gDNA elimination according to instructions from the manufacturer (Qiagen). RNA integrity number (RIN) and concentration were identified with a Bioanalyzer (Agilent). Libraries were generated using the SMARTer Stranded RNA-seq Kit (Takara Bio) preceded by rRNA depletion using 1 μg of RNA. Samples were barcoded for multiplexing and sequenced at 75 bp paired-end on an Illumina HiSeq4000 and pooled 8 per land and sequenced at a depth of 50 million reads. For quality control, sequences in FASTQ files were mapped to the human genome with STAR (version 2.5.3a) with reference genome and annotation GTF file downloaded from ENSEMBL (release 79, GRCh38) and counted with featureCounts (version 1.5.3). ENSEMBL IDs were mapped with gene annotation using the biomaRt package in R. Samples with overexpressed mitochondria genes accounting for over one-half of total counts were removed. Gene filtering was unsupervised and nonspecific. Genes with no expression (0 counts) in more than one-half of samples in a given group were dropped.
Differentially Expressed BMI-Related Genes
Differentially expressed genes (DEGs) were determined for males and females and for each brain region with the DESeq2 package in R (Love et al., 2014). This package calculated log2fold-change values per gene and disease, factoring in covariates such as RIN, race, PMI, and age at time of death (supplementary Figure 1 depicts the contribution of covariates to gene expression variance). DEGs were identified by achieving false discovery rate (FDR) < 0.05 with Benjamini-Hochberg multiple comparisons correction. The following comparisons were conducted in all subjects, males only, and females only—neurotypical comparison subjects with normal BMI vs neurotypical comparison subjects with high BMI (CNH), PTSD subjects with normal BMI vs PTSD subjects with high BMI (PNH), neurotypical comparison subjects with normal BMI vs PTSD subjects with normal BMI (NCP), and neurotypical comparison subjects with high BMI vs PTSD subjects with high BMI (HCP).
Gene Coexpression Network Analysis
Following identification of gene expression alterations in PTSD, weighted gene coexpression network analysis (WGCNA) was performed in males and females separately and together to reveal transcript coexpression modules across the 4 PFC regions (Langfelder and Horvath, 2008). Data were quantile normalized for GC content of sequences using R software cqn and corrected for batch effects with ComBat in package sva. Outliers were considered to be samples with standardized sample network connectivity Z scores < −2 and were not included. A soft-threshold power of 6 was employed for studies to achieve approximate scale-free topology (R2 > 0.8). Networks were created with the blockwiseModules function. The network dendrogram was constructed with average linkage hierarchical clustering of the topological overlap dissimilarity matrix. Modules were defined as branches of the dendrogram with the hybrid dynamic tree-cutting method, with a minimum module size of 20, merge threshold of 0.1, and negative pamStage. Each module was given an arbitrary color by the software. Linear regression was employed to determine association between genes in the modules and confounders or covariates. Fisher’s exact test was used to determine if DEGs were enriched in a particular module, and significance values were FDR corrected to adjust for multiple comparisons. Significant modules were probed for any association with NPY, ghrelin, and hypocretin and to determine levels of transcripts with close network associations. We also generated a list of genes with functional annotation related to appetite using the Ingenuity Pathway Analysis (IPA) program (version 01-14; Qiagen) and GeneCards (www.genecards.org).
Algorithm for the Reconstruction of Accurate Cellular Networks (Margolin et al., 2006) was used to create 2-dimensional reconstructions of the networks and reveal key drivers of the modules: highly interconnected DEGs that likely drive transcriptional alterations (Zhang et al., 2013). The organization of significant modules with upstream regulators was generated with igraph in R.
Gene Ontology and Pathway Analysis
IPA was employed for determination of the biological functions/pathways of the genes examined in this study as well as to predict networks that contain them. For gene ontology enrichment, a P value was calculated to assess the degree of overlap between the genes in the study and networks of genes with known biological functions. For determination of likely upstream regulators of transcriptional alterations, IPA assessed the number of known targets of regulators present in the study sample and compared the direction of change with that demonstrated in existing relationships in the literature. The resulting P value indicates the degree of overlap between the genes in the dataset and established targets of regulators to predict regulators. IPA was employed to identify only regulators of DEGs in the comparisons between groups of subjects stratified by sex and BMI. Finally, inferred network analysis was performed using IPA for the sex- and BMI-related genes. For this, the software forms networks surrounding “seed” molecules in the dataset that are highly interconnected based on known biological relationships, and identifies additional molecules known to interact, to generate groups of relationships.
All P values presented are FDR < 0.05 in Fisher’s exact test with Benjamini-Hochberg multiple comparisons correction.
Results
Sex-Specific Organization of the PTSD Transcriptome in the PFC in Association With Appetitive Neuropeptides
In our previous well-powered transcriptome-wide characterization of gene expression alterations in postmortem tissue of individuals with PTSD, there was a significant effect (on principal component [PC] 1) of sex on variance in gene expression with PTSD (P = 2.2 × 10−29) (Girgenti et al., 2020). We identified significant transcriptomic alterations in the dlPFC, OFC, dACC, and sgPFC in males and females with PTSD and characterized those changes by performing WGCNA, revealing 66 gene coexpression modules in the combined-sex comparison, 69 modules in the female-only comparison, and 59 modules in the male-only comparison. WGCNA was performed across all regions. Here we report that 3 of those PTSD-associated modules (P < .01) contained individual genes related to ghrelin and NPY. Two modules were female-specific—darkgreen (containing NPY ligand [NPY] and 135 total genes) (Figure 1; extended network found in supplementary Figure 2) and lightcyan (containing ghrelin ligand [GHRL] and 173 total genes) (Figure 2; extended network found in supplementary Figure 3)—and 1 was identified in the combined sex group: darkgrey (containing NPY receptor type 2 [NPY2R] and 164 total genes) (Figure 3; extended network found in supplementary Figure 4). (The software randomly assigned colors to each module.) We also probed for hypocretin-related genes (hypocretin ligand [HCRT], and hypocretin receptor type 1 [HCRTR1] and type 2 [HCRTR2]), however none were found to be significantly expressed for co-expression analysis. Across the 3 significant modules, 1 DEG was shared: C2 calcium dependent domain containing 4B (C2CD4B) was a member of both the darkgreen (female) and darkgrey (combined sex) modules, and it was upregulated 1.4 fold in the combined-sex comparison.
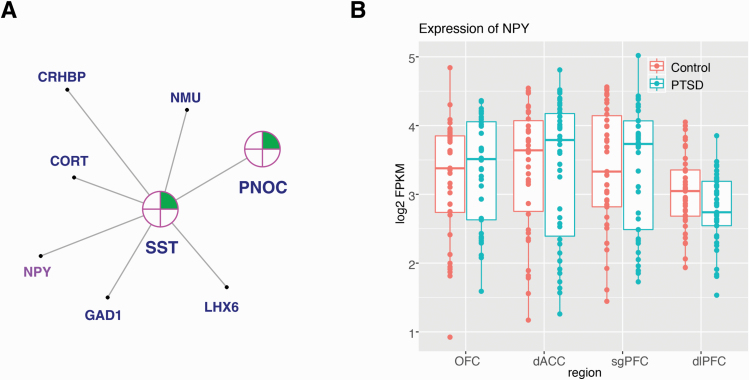
Figure 1.
Network coexpression analysis across prefrontal cortex regions revealed the posttraumatic stress disorder (PTSD)-associated darkgreen module, enriched for differentially expressed genes (female) and containing neuropeptide Y (NPY). (A) A 2-dimensional representation of the local gene network surrounding NPY is depicted (full organization of the module presented in supplementary Figure 2). Nearest key drivers are outlined in pink. Colors inside the circles signify regulation of gene expression (green = downregulation) in brain regions as represented by numbers: in the dorsal anterior cingulate (dACC), dorsolateral PFC (dlPFC), orbitofrontal cortex (OFC), and subgenual PFC (sgPFC). (B) Box plot displays the expression level (fragments per kilobase of exon model per million reads mapped [FPKM]) of NPY in individuals with PTSD vs neurotypical comparison subjects in the dACC, dlPFC, OFC, and sgPFC. Individual sample FPKMs are indicated and error bars indicate ± SEM.
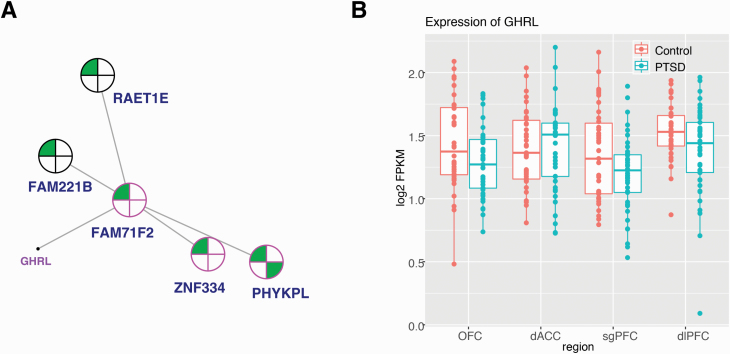
Figure 2.
Network coexpression analysis across prefrontal cortex regions revealed the posttraumatic stress disorder (PTSD)-associated lightcyan module, enriched for differentially expressed genes (female) and containing ghrelin (GHRL). (A) A 2-dimensional representation of the local gene network surrounding GHRL is depicted (full organization of the module presented in supplementary Figure 3). Nearest key drivers are outlined in pink and nodes are outlined in black. Colors inside the circles signify regulation of gene expression (green = downregulation) in brain regions as represented by numbers: in the dorsal anterior cingulate (dACC), dorsolateral PFC (dlPFC), orbitofrontal cortex (OFC), and subgenual PFC (sgPFC). (B) Box plot displays the expression level (fragments per kilobase of exon model per million reads mapped [FPKM]) of GHRL in individuals with PTSD vs neurotypical comparison subjects in the dACC, dlPFC, OFC, and sgPFC. Individual sample FPKMs are indicated and error bars indicate ± SEM.
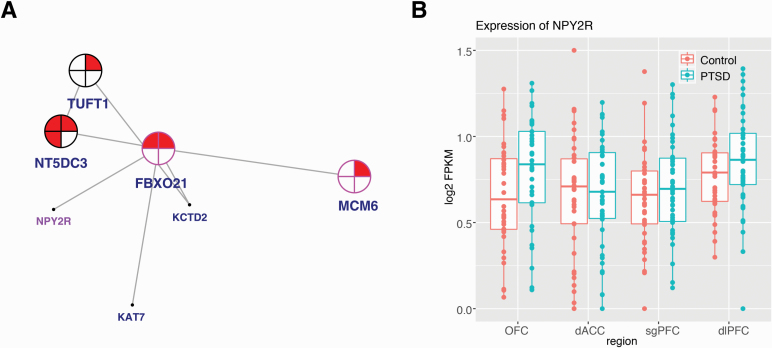
Figure 3.
Network coexpression analysis across prefrontal cortex regions revealed the posttraumatic stress disorder (PTSD)-associated darkgrey module, enriched for differentially expressed genes (combined sex) and containing NPY receptor type 2 (NPY2R). (A) A 2-dimensional representation of the local gene network surrounding NPY2R is depicted (full organization of the module presented in supplementary Figure 4). Nearest key drivers are outlined in pink and nodes are outlined in black. Colors inside the circles signify regulation of gene expression (red = upregulation) in brain regions as represented by numbers: in the dorsal anterior cingulate (dACC), dorsolateral PFC (dlPFC), orbitofrontal cortex (OFC), and subgenual PFC (sgPFC). (B) Box plot displays the expression level (fragments per kilobase of exon model per million reads mapped [FPKM]) of NPY2R in individuals with PTSD vs neurotypical comparison subjects in the dACC, dlPFC, OFC, and sgPFC. Individual sample FPKMs are indicated and error bars indicate ± SEM.
NPY, GHRL, and NPY2R each had moderately high kME values for positive or negative association with PTSD: 0.80 (NPY), 0.59 (GHRL), and −0.51 (NPY2R). We compared the canonical signaling pathways for these 3 genes (using STRING and IPA to generate lists of genes empirically shown to interact) with the members of our modules (Szklarczyk et al., 2019). As expected, there is significant overlap between the canonical pathways of NPY and NPY2R, and each is present in the pathway of the other. Moreover, cortistatin (CORT), hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 (HSD3B2), 5-hydroxytryptamine receptor 1 (HTR1F), LSM5 homolog, U6 small nuclear RNA and mRNA degradation associated (LSM5), nerve growth factor (NGF), neuromedin U (NMU), prepronociceptin (PNOC), prolactin releasing hormone receptor (PRLHR), and somatostatin (SST) are members of the signaling network of NPY and are present in the darkgreen (NPY) module. (CORT, HTR1F, NMU, PNOC, and SST are also members of the signaling network of NPY2R.) Adenylate cyclase 4 (ADCY4), C-X-C motif chemokine ligand 12 (CXCL12), endothelin 3 (EDN3), fms related receptor tyrosine kinase 4 (FLT4), G protein subunit gamma 11 (GNG11), and opioid receptor mu 1 (OPRM1) are members of the signaling network of NPY2R and are present in the darkgrey (NPY2R) module. (ADCY4, CXCL12, GNG11, and OPRM1 are also members of the signaling network of NPY.) There were no other ghrelin-related signaling genes in the lightcyan module.
After identifying PTSD-associated coexpression modules containing neuropeptides of interest, we reasoned that other appetitive molecules might also be present within these modules. We generated a list of genes with functional annotation related to appetite from IPA and GeneCards and compared this with the list of genes within our PTSD modules. The NPY2R module, darkgrey, contained 37 of these (supplementary Figure 4) and the GHRL module, lightcyan, contained 18 (supplementary Figure 3). The NPY module, darkgreen, contained 36 appetitive molecules (supplementary Figure 2); 6 of the 7 genes of the local gene network surrounding NPY in darkgreen were among these (CORT, Corticotropin Releasing Hormone Binding Protein [CRHBP], Glutamate Decarboxylase 1 [GAD1], NMU, PNOC, SST) (Figure 1).
Interestingly, the combined sex module darkgrey was enriched for endothelial cell markers, and the female modules lightcyan and darkgreen were enriched for endothelial cell markers and neuronal markers, respectively (as predicted by cell type-enrichment analysis). Furthermore, IPA pathway analyses revealed that darkgrey contained 6 significant pathways (P < .05) under the category of the inflammatory response and that darkgreen contained 3 in this category (P < .05). This indicates that there may be sex-specific neuro-molecular alterations with respect to cell type and function of genes in the context of appetitive molecules in PTSD. Furthermore, there were 11 key drivers of darkgreen (supplementary Figure 2), 8 of darkgrey (supplementary Figure 4), and 20 of lightcyan (supplementary Figure 3). Key drivers are highly connected hub genes with likely upstream control of the transcriptomic organization of the module.
Differences in the Sex-Specific PTSD Transcriptome Associated With BMI
We previously observed an effect (PC) of sex on variance in gene expression with PTSD; we also observed a modest effect of BMI on gene expression variance (PC 1, P = .05 and PC 2, P = 2.18 × 10−5), suggesting a role for BMI in the expression changes observed. It is also important to note that individuals with PTSD had a significantly lower mean BMI relative to neurotypical comparison subjects (29.2 vs 33.3 kg/m2 [P = .02]) (Table 1).
Expanding on the observed effects of sex and BMI on variance in gene expression with PTSD, we identified DEGs (P < .05) between subjects when stratified by sex and BMI (supplementary Table 1). Subjects with PTSD with elevated BMI vs normal BMI (PNH) displayed the highest number of DEGs (328 in males and 31 in the combined sex group). Among these DEGs identified in the 2 comparisons, 28 were shared and were all upregulated (Table 2). Of note, few genes reached statistical significance for differential expression among neurotypical comparison subjects with normal BMI vs neurotypical comparison subjects with high BMI (supplementary Table 1); there was also no overlap with genes differentially expressed in any of the comparisons among individuals with PTSD with normal vs high BMI.
Table 2.
Twenty-Eight Genes Were Upregulated in the PNH Comparison in Both Male Subjects and Combined Sex Group
| PNH male | PNH combined sex | ||||
|---|---|---|---|---|---|
| Gene name | ENSEMBL ID | Differential expression level (log2 fold-change) | P adjusted | Differential expression level (log2 fold-change) | P adjusted |
| AOC3 | ENSG00000131471 | 2.20 | 3.31E-04 | 1.31 | 3.88E-02 |
| COL4A1 | ENSG00000187498 | 2.83 | 7.97E-06 | 1.70 | 2.47E-03 |
| COL4A2 | ENSG00000134871 | 1.85 | 3.45E-05 | 1.06 | 8.58E-03 |
| DES | ENSG00000175084 | 3.46 | 2.53E-03 | 2.35 | 2.88E-02 |
| DYNLT1 | ENSG00000146425 | 0.45 | 2.36E-02 | 0.33 | 3.81E-02 |
| ERRFI1 | ENSG00000116285 | 1.03 | 1.80E-05 | 0.55 | 3.81E-02 |
| FAT2 | ENSG00000086570 | 1.09 | 7.48E-04 | 0.74 | 2.82E-03 |
| GPR182 | ENSG00000166856 | 0.59 | 2.83E-02 | 0.63 | 2.47E-03 |
| HILPDA | ENSG00000135245 | 3.09 | 1.04E-05 | 1.67 | 2.91E-02 |
| HK2 | ENSG00000159399 | 2.09 | 2.86E-03 | 1.33 | 2.57E-02 |
| IGFBP4 | ENSG00000141753 | 2.62 | 1.45E-07 | 1.49 | 2.47E-03 |
| IL16 | ENSG00000172349 | 0.96 | 7.48E-04 | 0.66 | 1.78E-02 |
| LAMA4 | ENSG00000112769 | 0.92 | 3.50E-05 | 0.59 | 1.23E-02 |
| LMCD1 | ENSG00000071282 | 3.17 | 6.83E-10 | 1.87 | 1.81E-03 |
| LOX | ENSG00000113083 | 2.95 | 3.64E-11 | 1.66 | 2.47E-03 |
| MEDAG | ENSG00000102802 | 4.13 | 1.18E-05 | 2.77 | 2.47E-03 |
| PHLDB2 | ENSG00000144824 | 1.18 | 4.42E-05 | 0.73 | 2.91E-02 |
| PLCE1 | ENSG00000138193 | 1.15 | 1.39E-05 | 0.67 | 6.74E-03 |
| RASL12 | ENSG00000103710 | 1.54 | 2.86E-03 | 0.91 | 3.81E-02 |
| RHOB | ENSG00000143878 | 1.22 | 4.36E-05 | 0.68 | 1.78E-02 |
| SVIL | ENSG00000197321 | 1.15 | 2.02E-04 | 0.66 | 3.81E-02 |
| TAF1 | ENSG00000147133 | 0.21 | 1.44E-02 | 0.16 | 4.99E-02 |
| TAGLN | ENSG00000149591 | 1.88 | 3.04E-03 | 1.18 | 3.88E-02 |
| TGM2 | ENSG00000198959 | 3.03 | 9.28E-08 | 1.78 | 1.81E-03 |
| TNC | ENSG00000041982 | 2.74 | 7.94E-04 | 1.61 | 3.13E-02 |
| TNNC2 | ENSG00000101470 | 0.95 | 3.18E-02 | 0.71 | 4.99E-02 |
| TTR | ENSG00000118271 | 3.25 | 2.71E-02 | 2.73 | 3.53E-02 |
| ZNF331 | ENSG00000130844 | 1.23 | 5.28E-07 | 0.62 | 2.91E-02 |
Abbreviations: PNH, posttraumatic stress disorder with normal vs high body mass index.
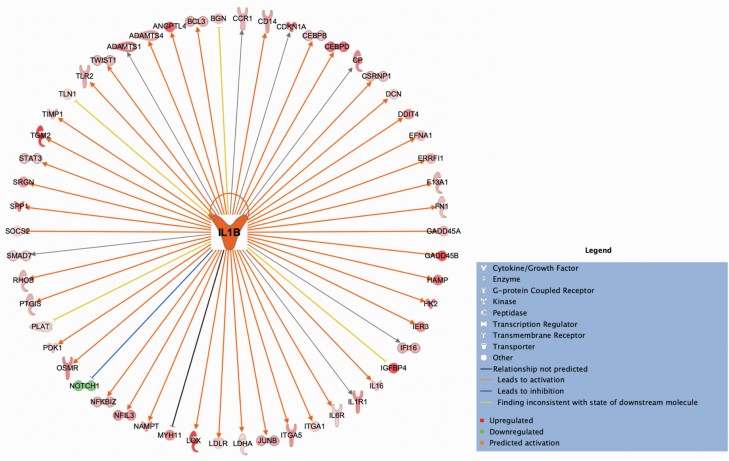
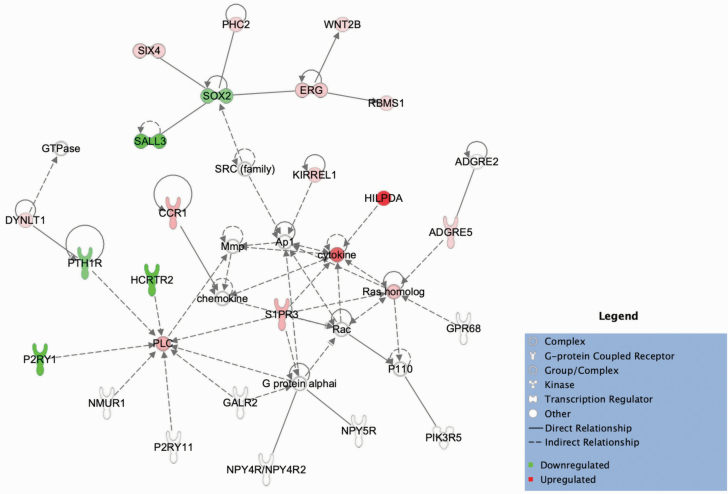
We performed IPA pathway analysis to further characterize the DEGs from the PNH male and PNH combined sex comparisons. The large number of DEGs observed were significantly enriched for inflammatory response pathways (29 in males and 6 in the combined sex group [P < .05]). Furthermore, in the case of the male PNH comparison, we were able to identify the DEG interleukin 1 beta (IL1B) as the most significant upstream regulator (P = 1.9 × 10−15) with 54 downstream targets that were mostly upregulated (53 of 54) (Figure 4). As displayed in Figure 4, 42 of 54 observed relationships were consistent with previous findings on activation of IL1B.
Figure 4.
Interleukin 1 beta (IL1B) was determined to be the most significant inferred upstream regulator of differentially expressed genes in the posttraumatic stress disorder with normal vs high body mass index (PNH) comparison among male subjects. Ingenuity Pathway Analysis software generated a spatial representation of the downstream expression changes observed in 54 target genes in the dataset (P = 1.9 × 10−15) that were mostly upregulated (53 of 54) as a result of IL1B activation; 42 of the relationships associated with activation of IL1B have been empirically observed. Shapes represent the type of molecule. Colors inside the shapes signify regulation of gene expression (red = upregulation; green = downregulation; orange = predicted activation). Line colors signify consistency of the observed relationships with the literature (grey = not predicted; orange = precipitates activation; blue = precipitates inhibition; yellow = observed relationship is not consistent with the state of the downstream molecule).
Although hypocretin-related genes were not contained within PTSD-specific modules, HCRTR2 was significantly downregulated in male subjects with PTSD with elevated BMI vs normal BMI (PNH) (supplementary Table 1). Pathway analysis of the DEGs in this comparison demonstrated that HCRTR2 may contribute to a network incorporating other DEGs involved in inflammation as well as potentially members of the NPY signaling pathway: receptor Y4 (NPY4R/NPY4R2) and receptor Y5 (NPY5R) (Figure 5).
Figure 5.
Hypocretin receptor type 2 (HCRTR2) was a differentially expressed gene in the posttraumatic stress disorder with normal vs high body mass index (PNH) comparison among male subjects and was determined to form a network with other genes differentially expressed in the dataset involved in inflammation as well as with members of the NPY signaling pathway: receptor Y4 (NPY4R/NPY4R2) and receptor Y5 (NPY5R). Ingenuity Pathway Analysis software was used to generate a spatial representation of this inferred network, formed surrounding “seed” molecules in the dataset that are highly interconnected based on known biological relationships and including other molecules known to interact. Shapes represent the type of molecule. Colors inside the shapes signify regulation of gene expression (red = upregulation; green = downregulation). Solid line signifies a direct relationship; dotted line signifies an indirect relationship. dACC, dorsal anterior cingulate; dlPFC, dorsolateral PFC; OFC, orbitofrontal cortex; sgPFC, subgenual PFC.
We hypothesized that our BMI DEGs and/or genes that were members of the modules could also be risk variants for adiposity-related traits or eating disorders such as anorexia nervosa. We used previous genome-wide association studies to identify the following risk variants for adiposity-related traits that are in the modules: ATP/GTP binding protein like 4 (AGBL4), coiled-coil domain containing 39 (CCDC39), cordon-bleu WH2 repeat protein like 1 (COBLL1), EYA transcriptional coactivator and phosphatase 4 (EYA4), PATJ crumbs cell polarity complex component (INADL), nischarin (NISCH), NMU, protein kinase C eta (PRKCH), sideroflexin 2 (SFXN2), and tryptophanyl tRNA synthetase 2, mitochondrial (WARS2); AGBL4 is also a key driver of the darkgreen module (supplementary Table 2). In addition, we found that the following risk variants for adiposity-related traits are among the male PNH BMI DEGs: ADAM metallopeptidase with thrombospondin type 1 motif 9 (ADAMTS9), family with sequence similarity 13 member A (FAM13A), 1,4-alpha-glucan branching enzyme 1 (GBE1), Kruppel like factor 9 (KLF9), and phospholipase C epsilon 1 (PLCE1); PLCE1 also overlaps with the combined sex PNH BMI comparison. Furthermore, cyclin dependent kinase 2 (CDK2) and myosin light chain 6 (MYL6) are risk variants for anorexia nervosa that are among the male PNH BMI DEGs (supplementary Table 2).
Discussion
Here we expand on on our recent sex-specific characterization of the transcriptome of the human PTSD brain (Girgenti et al., 2020). We provide evidence that appetitive molecules may be altered in relation to BMI and PTSD and are members of several PTSD-associated coexpression modules.
We found that the transcriptomic organization of PTSD in the human brain includes 3 coexpression modules that contain members of the NPY and ghrelin signaling families (Figures 1–3; supplementary Figures 2–4). Two of the 3 modules (containing NPY and ghrelin) were observed in females. This result expands on prior NPY and ghrelin research that has primarily included only male rodents and humans and has mainly focused on levels of these peptides in peripheral blood and CSF as opposed to central gene expression (Morgan et al., 2000; Sah et al., 2009, 2014; Meyer et al., 2014; Harmatz et al., 2017; Yousufzai et al., 2018). In the combined sex group, we identified a unique network containing the NPY2R gene. Rodent studies indicate that this receptor inhibits the release of NPY, glutamate, and GABA (Tasan et al., 2010; Stanic et al., 2011). Human studies suggest it may be a candidate gene for type 2 diabetes (in men) (Campbell et al., 2007), and variation in this gene may predispose to obesity (Hunt et al., 2011).
Highlighting the importance of these modules in appetitive signaling, we identified other appetitive molecules within these modules: module darkgrey contained 37 functionally characterized appetitive molecules (supplementary Figure 4) and module lightcyan contained 18 (supplementary Figure 3). Module darkgreen contained 36 (supplementary Figure 2), and 6 of 7 genes of the local gene network surrounding NPY in darkgreen were among these (CORT, CRHBP, GAD1, NMU, PNOC, SST) (Figure 1). These findings suggest that there is systems-level transcriptional control of these appetitive molecules and that PTSD may disrupt this regulation.
Moreover, the 2 modules containing NPY-related genes were enriched with inflammatory response pathways. These 2 modules had 1 shared member, C2CD4B. We show that C2CD4B is significantly upregulated in the PFC among subjects with PTSD. Increased expression has been found to associate with heightened response to inflammatory cytokines such as IL1B; further, in human pancreatic islet cells, C2CD4B has been linked to pathologic metabolic states such as genetic risk of type 2 diabetes (Warton et al., 2004; Kycia et al., 2018).
We further demonstrated that there was an effect of BMI on gene expression (supplementary Table 1). When stratifying subjects by BMI and sex, the majority of DEGs were found in the PNH comparison among males and in the combined sex group. Moreover, the DEGs in the 2 PNH groups were enriched for pro-inflammatory genes. IL1B was a DEG in the male analysis specifically and was identified as a highly significant predicted upstream regulator (Figure 4). (IPA demonstrates that IL1B is also part of the canonical signaling pathways of NPY and GHRL.) Although many studies indicate that PTSD is characterized by a pro-inflammatory phenotype in the periphery (see a meta-analysis) (Passos et al., 2015), gene expression of inflammatory markers in the PTSD brain has only recently begun to be described (Morrison et al., 2019). Thus, a significant implication of our work is the identification of BMI-dependent neuroinflammatory expression changes specific to PTSD.
Appetitive molecule module members AGBL4, CCDC39, COBLL1, EYA4, INADL, NISCH, NMU, PRKCH, SFXN2, and WARS2 have been identified in genome-wide association studies focusing on adiposity-related traits; remarkably, AGBL4 was also a key driver in the darkgreen module (supplementary Table 2). The PNH BMI DEGs ADAMTS9, FAM13A, GBE1, KLF9, and PLCE1 have been identified in genome-wide association studies focusing on adiposity-related traits, and CDK2 and MYL6 have been identified in studies focusing on anorexia nervosa (supplementary Table 2). Collectively, these results suggest that some transcriptomic changes associated with PTSD are occurring at genes with significant risk for metabolic disorders and further highlight the link between PTSD and abnormal BMI.
Contrary to our original hypothesis, only genes associated with the hypocretin family (and not NPY or ghrelin) were regulated in association with BMI change. HCRTR2 was a DEG in the male PNH comparison (supplementary Table 1) and appeared to interact with other genes involved in inflammation in a network predicted to include NPY receptors Y4 and Y5 (Figure 5) (though these NPY-related genes were not differentially expressed in any BMI comparison). Genetic variation and expression changes in Y4 and Y5 have been linked to human obesity (Aerts et al., 2016; Chatree et al., 2018). Preclinical models suggest that enhanced HCRTR2 signaling promotes resistance to diet-induced obesity (Teske et al., 2006; Funato et al., 2009), consistent with our finding of HCRTR2 downregulation in the setting of an elevated BMI.
Rodent neural cell culture experiments demonstrate that the HCRTR2 pathway is suppressed when inflammatory cytokines such as tumor necrosis factor-ɑ (TNF-ɑ) are elevated (Zhan et al., 2011, 2019). Evidence suggests that the HCRTR2 pathway is anti-inflammatory given that in a rodent model of a pro-inflammatory state, treatment with orexin-A (hypocretin-1)—1 of the ligands for the HCRTR2 receptor—decreases neuroinflammation in the brain (specifically, IL1B and TNF-ɑ) (Modi et al., 2017). The pro-inflammatory state evidenced by males with PTSD with elevated vs normal BMI (PNH) in our study may thus in part have been related to downregulated HCRTR2. Intriguingly, these subjects also demonstrated upregulated complement C1q tumor necrosis factor-related protein 1 (C1QTNF1). C1QTNF1 is mainly expressed in adipose tissue (Wong et al., 2008) and is a pro-inflammatory factor positively associated with IL1B and TNF-ɑ secretion in human peripheral samples (Wang et al., 2016; Shen et al., 2019). Thus, the increased expression of C1QTNF1 and IL1B and decreased expression of HCRTR2 in the males with PTSD with elevated vs normal BMI are in general agreement with findings in the periphery and extend our understanding of the role of BMI in brain function, specifically in PTSD.
IL1B was also found to be important in males with PTSD with elevated BMI as an inferred upstream regulator of 54 DEGs that did not include HCRTR2. Previous research did not find IL1B to be differentially regulated in the dlPFC in PTSD (though that study combined sexes, it was relatively underpowered and was not stratified by BMI) (Morrison et al., 2019). IL1B is only known to be elevated in the periphery in PTSD, based on a meta-analysis (Passos et al., 2015). Moreover, in a study of subjects without PTSD of different BMI groupings (underweight, normal, overweight, obese, and morbidly obese), BMI did not relate to gene expression of IL1B in the PFC (Lauridsen et al., 2017). It is attractive to speculate that cortical regulation of IL1B represents a molecular intersection between PTSD and high BMI. This relationship may have functional implications, as genetic variation in IL1B has been linked to risk of PTSD (in males) (Hovhannisyan et al., 2017), and circulating IL1B levels are positively associated with PTSD illness duration (Passos et al., 2015). Among those without PTSD, genetic variation and expression in IL1B in human peripheral samples is linked to obesity (Suzuki et al., 2009; He et al., 2019). Altered IL1B expression and likely associated downstream alterations in the PFC in PTSD that we observed may contribute to medical conditions afflicting the PTSD population, including obesity and the metabolic syndrome (Rasmusson et al., 2010; van den Berk-Clark et al., 2018). Therefore, it is possible that medications such as canakinumab, a human monoclonal antibody that targets IL1B (Ridker et al., 2017), may eventually prove useful in this setting.
Our study has several limitations that are important to acknowledge. We did not obtain peripheral samples from subjects, preventing us from comparing our central findings to the periphery. In addition, the strongest evidence of the interaction between NPY, ghrelin, and hypocretin stems from research on the arcuate nucleus of the hypothalamus (Kohno et al., 2003), which is involved in regulating food intake (Krashes et al., 2014). Future studies should examine gene expression changes in this region.
In conclusion, we demonstrate that transcriptomic change in association with orexigenic molecules, and with alterations in BMI, should be further investigated in the context of the neuropathology underlying PTSD, particularly in terms of potential pathways that may contribute to or result from obesity. Such augmentations of mechanistic understanding may eventually pave the way to identification of novel targets for pharmacological therapies for the treatment of PTSD.
Supplementary Materials
Supplementary data are available at International Journal of Neuropsychopharmacology (IJNPPY) online.
Supplementary Figure S1. Contribution of covariates to gene expression variance. Differentially expressed genes were determined for males and females, and for each brain region. Log2fold-change values per gene and disease were determined, factoring in the covariates displayed.
Abbreviations: agedeath, age at death; ADP, antidepressant use; dx, disease; PMI, postmortem interval; PC, principal component; RIN, RNA integrity number.
Supplementary Figure S2. Network co-expression analysis across prefrontal cortex regions revealed the posttraumatic stress disorder (PTSD)-associated darkgreen module, enriched for differentially expressed genes (female) and containing neuropeptide Y (NPY). Two-dimensional representation of the full organization of the module depicts nodes and key drivers. Pink outline = key drivers; black outline = nodes. Colors inside the circles signify regulation of gene expression (red = upregulation; green = downregulation) in brain regions as represented by numbers.
Abbreviations: dACC, dorsal anterior cingulate; dlPFC, dorsolateral PFC; OFC, orbitofrontal cortex; sgPFC, subgenual PFC.
Supplementary Figure S3. Network co-expression analysis across prefrontal cortex regions revealed the posttraumatic stress disorder (PTSD)-associated lightcyan module, enriched for differentially expressed genes (female) and containing ghrelin (GHRL). Two-dimensional representation of the full organization of the module depicts nodes and key drivers. Pink outline = key drivers; black outline = nodes. Colors inside the circles signify regulation of gene expression (red = upregulation; green = downregulation) in brain regions as represented by numbers.
Abbreviations: dACC, dorsal anterior cingulate; dlPFC, dorsolateral PFC; OFC, orbitofrontal cortex; sgPFC, subgenual PFC.
Supplementary Figure S4. Network co-expression analysis across prefrontal cortex regions revealed the posttraumatic stress disorder (PTSD)-associated darkgrey module, enriched for differentially expressed genes (combined sex) and containing NPY receptor type 2 (NPY2R). Two-dimensional representation of the full organization of the module depicts nodes and key drivers. Pink outline = key drivers; black outline = nodes. Colors inside the circles signify regulation of gene expression (red = upregulation; green = downregulation) in brain regions as represented by numbers.
Acknowledgments
This work was supported with the resources and use of facilities at the Veterans Affairs Connecticut Health Care System, West Haven, CT. The views expressed here are those of the authors and do not necessarily reflect the position or policy of the Department of Veterans Affairs or the United States government.
This work was supported by the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health (grant no. T35DK104689 to L.A.S.); the Department of Veterans Affairs, Veteran Health Administration, VISN1 Career Development Award and a Brain and Behavior Research Foundation Young Investigator award (to M.J.G.); and the National Institute of Mental Health (grant nos. MH093897, MH105910 to R.S.D.). J.W., D.J., H.Z., and J.H.K. do not report any funding sources.
List of authors and affiliations provided in supplementary Material (File S1).
Statement of Interest
J.H.K. has consulting agreements (<$10 000/y) with the following: AstraZeneca Pharmaceuticals, Biogen, Idec, MA, Biomedisyn Corporation, Bionomics, Limited (Australia), Boehringer Ingelheim International, COMPASS Pathways, Limited, United Kingdom, Concert Pharmaceuticals, Inc., Epiodyne, Inc. EpiVario, Inc. Heptares Therapeutics, Limited (UK), Janssen Research & Development, Otsuka America Pharmaceutical, Inc. Perception Neuroscience Holdings, Inc. Spring Care, Inc., Sunovion Pharmaceuticals, Inc., Takeda Industries, and Taisho Pharmaceutical Co., Ltd. J.H.K. serves on the scientific advisory boards of Bioasis Technologies, Inc., Biohaven Pharmaceuticals, BioXcel Therapeutics, Inc. (Clinical Advisory Board), BlackThorn Therapeutics, Inc., Cadent Therapeutics (Clinical Advisory Board), Cerevel Therapeutics, LLC, EpiVario, Inc., Eisai, Inc., Lohocla Research Corporation, Novartis Pharmaceuticals Corporation, and PsychoGenics, Inc. J.H.K. owns stock in Biohaven Pharmaceuticals, Sage Pharmaceuticals, and Spring Care, Inc. He has stock options with Biohaven Pharmaceuticals Medical Sciences, BlackThorn Therapeutics, Inc., EpiVario, Inc., and Terran Life Sciences. J.H.K. is the editor of Biological Psychiatry with an income greater than $10 000. He has the following patents and inventions: (1) Seibyl JP, Krystal JH, Charney DS. Dopamine and noradrenergic reuptake inhibitors in treatment of schizophrenia. US Patent #:5,447,948. September 5, 1995. (2) Vladimir, Coric, Krystal, John H, Sanacora, Gerard – Glutamate Modulating Agents in the Treatment of Mental Disorders US Patent No. 8,778,979 B2 Patent Issue Date: July 15, 2014. US Patent Application No. 15/695,164: Filing Date: September 5, 2017. (3) Charney D, Krystal JH, Manji H, Matthew S, Zarate C. Intranasal Administration of Ketamine to Treat Depression United States Application No. 14/197,767 filed on March 5, 2014; United States application or Patent Cooperation Treaty (PCT) International application No. 14/306,382 filed on June 17, 2014
(4) Zarate, C, Charney, DS, Manji, HK, Mathew, Sanjay J, Krystal, JH, Department of Veterans Affairs “Methods for Treating Suicidal Ideation,” Patent Application No. 14/197.767 filed on March 5, 2014 by Yale University Office of Cooperative Research. (5) Arias A, Petrakis I, Krystal JH. Composition and methods to treat addiction. Provisional Use Patent Application no.61/973/961. April 2, 2014. Filed by Yale University Office of Cooperative Research. (6) Chekroud, A., Gueorguieva, R., & Krystal, JH. “Treatment Selection for Major Depressive Disorder” [filing date 3rd June 2016, USPTO docket number Y0087.70116US00]. Provisional patent submission by Yale University. (7) Gihyun, Yoon, Petrakis I, Krystal JH. Compounds, Compositions and Methods for Treating or Preventing Depression and Other Diseases. U. S. Provisional Patent Application No. 62/444,552, filed on January 10, 2017 by Yale University Office of Cooperative Research OCR 7088 US01. (8) Abdallah, C, Krystal, JH, Duman, R, Sanacora, G. Combination Therapy for Treating or Preventing Depression or Other Mood Diseases. U.S. Provisional Patent Application No. 62/719,935 filed on August 20, 2018 by Yale University Office of Cooperative Research OCR 7451 US01.J.H.K. receives the following NON Federal Research Support: AstraZeneca Pharmaceuticals provides the drug, Saracatinib, for research related to NIAAA grant “Center for Translational Neuroscience of Alcoholism [CTNA-4].” Novartis provides the drug, Mavoglurant, for research related to NIAAA grant “Center for Translational Neuroscience of Alcoholism [CTNA-4].”
R.S.D. has received consulting fees from Taisho, Johnson & Johnson, and Naurex, and grant support from Taisho, Johnson & Johnson, Naurex, Navitor, Allergan, Lundbeck, and Lilly. None of the above listed companies or funding agencies had any influence on the content of this article. L.A.S., M.J.G., J.W., D.J., and H.Z. have nothing to disclose.
References
- Abdallah CG, Averill LA, Akiki TJ, Raza M, Averill CL, Gomaa H, Adikey A, Krystal JH (2019) The neurobiology and pharmacotherapy of posttraumatic stress disorder. Annu Rev Pharmacol Toxicol 59:171–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aerts E, Beckers S, Zegers D, Van Hoorenbeeck K, Massa G, Verrijken A, Verhulst SL, Van Gaal LF, Van Hul W (2016) CNV analysis and mutation screening indicate an important role for the NPY4R gene in human obesity. Obesity (Silver Spring) 24:970–976. [DOI] [PubMed] [Google Scholar]
- Alim TN, Graves E, Mellman TA, Aigbogun N, Gray E, Lawson W, Charney DS (2006) Trauma exposure, posttraumatic stress disorder and depression in an African-American primary care population. J Natl Med Assoc 98:1630–1636. [PMC free article] [PubMed] [Google Scholar]
- Campbell CD, Lyon HN, Nemesh J, Drake JA, Tuomi T, Gaudet D, Zhu X, Cooper RS, Ardlie KG, Groop LC, Hirschhorn JN (2007) Association studies of BMI and type 2 diabetes in the neuropeptide Y pathway: a possible role for NPY2R as a candidate gene for type 2 diabetes in men. Diabetes 56:1460–1467. [DOI] [PubMed] [Google Scholar]
- Chatree S, Sitticharoon C, Maikaew P, Uawithya P, Chearskul S (2018) Adipose Y5R mRNA is higher in obese than non-obese humans and is correlated with obesity parameters. Exp Biol Med (Maywood) 243:786–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Ifergane G, Vainer E, Matar MA, Kaplan Z, Zohar J, Mathé AA, Cohen H (2016) The wake-promoting drug modafinil stimulates specific hypothalamic circuits to promote adaptive stress responses in an animal model of PTSD. Transl Psychiatry 6:e917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman MJ, Huber BR, Brady CB, Ursano RJ, Benedek DM, Kowall NW, McKee AC; Traumatic Stress Brain Research Group (2017) VA’s National PTSD Brain Bank: a national resource for research. Curr Psychiatry Rep 19:73. [DOI] [PubMed] [Google Scholar]
- Fulton JJ, Calhoun PS, Wagner HR, Schry AR, Hair LP, Feeling N, Elbogen E, Beckham JC (2015) The prevalence of posttraumatic stress disorder in Operation Enduring Freedom/Operation Iraqi Freedom (OEF/OIF) Veterans: a meta-analysis. J Anxiety Disord 31:98–107. [DOI] [PubMed] [Google Scholar]
- Funato H, Tsai AL, Willie JT, Kisanuki Y, Williams SC, Sakurai T, Yanagisawa M (2009) Enhanced orexin receptor-2 signaling prevents diet-induced obesity and improves leptin sensitivity. Cell Metab 9:64–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie CF, Bradley B, Mercer K, Smith AK, Conneely K, Gapen M, Weiss T, Schwartz AC, Cubells JF, Ressler KJ (2009) Trauma exposure and stress-related disorders in inner city primary care patients. Gen Hosp Psychiatry 31:505–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girgenti MJ, Duman RS (2018) Transcriptome alterations in posttraumatic stress disorder. Biol Psychiatry 83: 840–848. [DOI] [PubMed] [Google Scholar]
- Girgenti MJ, Wang J, Ji D, Cruz D, Stein MB, Gelernter J, Young K, Huber BR, Williamson DE, Friedman MJ, Krystal JH, Zhao H, Duman RS (2020) Transcriptomic organization of human posttraumatic stress disorder. bioRxiv. doi: 10.1101/2020.01.27.921403. [DOI] [Google Scholar]
- Harmatz ES, Stone L, Lim SH, Lee G, McGrath A, Gisabella B, Peng X, Kosoy E, Yao J, Liu E, Machado NJ, Weiner VS, Slocum W, Cunha RA, Goosens KA (2017) Central ghrelin resistance permits the overconsolidation of fear memory. Biol Psychiatry 81:1003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Yuan T, Maedler K (2019) Macrophage-associated pro-inflammatory state in human islets from obese individuals. Nutr Diabetes 9:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovhannisyan L, Stepanyan A, Arakelyan A (2017) Genetic variability of interleukin-1 beta as prospective factor from developing post-traumatic stress disorder. Immunogenetics 69:703–708. [DOI] [PubMed] [Google Scholar]
- Hunt SC, Hasstedt SJ, Xin Y, Dalley BK, Milash BA, Yakobson E, Gress RE, Davidson LE, Adams TD (2011) Polymorphisms in the NPY2R gene show significant associations with BMI that are additive to FTO, MC4R, and NPFFR2 gene effects. Obesity (Silver Spring) 19:2241–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler RC, Berglund P, Demler O, Jin R, Merikangas KR, Walters EE (2005) Lifetime prevalence and age-of-onset distributions of DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiatry 62:593–602. [DOI] [PubMed] [Google Scholar]
- Kohno D, Gao HZ, Muroya S, Kikuyama S, Yada T (2003) Ghrelin directly interacts with neuropeptide-Y-containing neurons in the rat arcuate nucleus: Ca2+ signaling via protein kinase A and N-type channel-dependent mechanisms and cross-talk with leptin and orexin. Diabetes 52:948–956. [DOI] [PubMed] [Google Scholar]
- Krashes MJ, Shah BP, Madara JC, Olson DP, Strochlic DE, Garfield AS, Vong L, Pei H, Watabe-Uchida M, Uchida N, Liberles SD, Lowell BB (2014) An excitatory paraventricular nucleus to AgRP neuron circuit that drives hunger. Nature 507:238–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krystal JH, Duman R (2004) What’s missing in posttraumatic stress disorder research? Studies of human postmortem tissue. Psychiatry 67:398–403. [DOI] [PubMed] [Google Scholar]
- Kycia I, et al. (2018) A common type 2 diabetes risk variant potentiates activity of an evolutionarily conserved Islet stretch enhancer and increases C2CD4A and C2CD4B expression. Am J Hum Genet 102:620–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauridsen JK, Olesen RH, Vendelbo J, Hyde TM, Kleinman JE, Bibby BM, Brock B, Rungby J, Larsen A (2017) High BMI levels associate with reduced mRNA expression of IL10 and increased mRNA expression of iNOS (NOS2) in human frontal cortex. Transl Psychiatry 7:e1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Zhao J, Balesar R, Fronczek R, Zhu QB, Wu XY, Hu SH, Bao AM, Swaab DF (2017) Sexually dimorphic changes of hypocretin (orexin) in depression. Ebiomedicine 18:311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin AA, Nemenman I, Basso K, Wiggins C, Stolovitzky G, Dalla Favera R, Califano A (2006) ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics 7(Suppl 1):S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer RM, Burgos-Robles A, Liu E, Correia SS, Goosens KA (2014) A ghrelin-growth hormone axis drives stress-induced vulnerability to enhanced fear. Mol Psychiatry 19:1284–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milad MR, Quirk GJ (2002) Neurons in medial prefrontal cortex signal memory for fear extinction. Nature 420:70–74. [DOI] [PubMed] [Google Scholar]
- Modi HR, Wang Q, Gd S, Sherman D, Greenwald E, Savonenko AV, Geocadin RG, Thakor NV (2017) Intranasal post-cardiac arrest treatment with orexin-A facilitates arousal from coma and ameliorates neuroinflammation. Plos One 12:e0182707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan CA 3rd, Wang S, Southwick SM, Rasmusson A, Hazlett G, Hauger RL, Charney DS (2000) Plasma neuropeptide-Y concentrations in humans exposed to military survival training. Biol Psychiatry 47:902–909. [DOI] [PubMed] [Google Scholar]
- Morrison FG, Miller MW, Wolf EJ, Logue MW, Maniates H, Kwasnik D, Cherry JD, Svirsky S, Restaino A, Hildebrandt A, Aytan N, Stein TD, Alvarez VE, McKee AC, Huber BR; Traumatic Stress Brain Study Group (2019) Reduced interleukin 1A gene expression in the dorsolateral prefrontal cortex of individuals with PTSD and depression. Neurosci Lett 692:204–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller TD, et al. (2015) Ghrelin. Mol Metab 4:437–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons RG, Ressler KJ (2013) Implications of memory modulation for post-traumatic stress and fear disorders. Nat Neurosci 16:146–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passos IC, Vasconcelos-Moreno MP, Costa LG, Kunz M, Brietzke E, Quevedo J, Salum G, Magalhães PV, Kapczinski F, Kauer-Sant’Anna M (2015) Inflammatory markers in post-traumatic stress disorder: a systematic review, meta-analysis, and meta-regression. Lancet Psychiatry 2:1002–1012. [DOI] [PubMed] [Google Scholar]
- Rasmusson AM, Schnurr PP, Zukowska Z, Scioli E, Forman DE (2010) Adaptation to extreme stress: post-traumatic stress disorder, neuropeptide Y and metabolic syndrome. Exp Biol Med (Maywood) 235:1150–1162. [DOI] [PubMed] [Google Scholar]
- Ridker PM, et al. ; CANTOS Trial Group (2017) Antiinflammatory therapy with canakinumab for atherosclerotic disease. N Engl J Med 377:1119–1131. [DOI] [PubMed] [Google Scholar]
- Sah R, Ekhator NN, Strawn JR, Sallee FR, Baker DG, Horn PS, Geracioti TD Jr (2009) Low cerebrospinal fluid neuropeptide Y concentrations in posttraumatic stress disorder. Biol Psychiatry 66:705–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sah R, Ekhator NN, Jefferson-Wilson L, Horn PS, Geracioti TD Jr (2014) Cerebrospinal fluid neuropeptide Y in combat veterans with and without posttraumatic stress disorder. Psychoneuroendocrinology 40:277–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu A, Kalra SP (1993) Neuropeptidergic regulation of feeding behavior Neuropeptide Y. Trends Endocrinol Metab 4:217–224. [DOI] [PubMed] [Google Scholar]
- Schwartz AC, Bradley RL, Sexton M, Sherry A, Ressler KJ (2005) Posttraumatic stress disorder among African Americans in an inner city mental health clinic. Psychiatr Serv 56:212–215. [DOI] [PubMed] [Google Scholar]
- Shen L, Wang S, Ling Y, Liang W (2019) Association of C1q/TNF-related protein-1 (CTRP1) serum levels with coronary artery disease. J Int Med Res 47:2571–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer WH, Lidström J, Sun H, Passer D, Eskay R, Parker SC, Witt SH, Zimmermann US, Nieratschker V, Rietschel M, Margulies EH, Palkovits M, Laucht M, Heilig M (2010) Human NPY promoter variation rs16147:T>C as a moderator of prefrontal NPY gene expression and negative affect. Hum Mutat 31:E1594–E1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanić D, Mulder J, Watanabe M, Hökfelt T (2011) Characterization of NPY Y2 receptor protein expression in the mouse brain. II. Coexistence with NPY, the Y1 receptor, and other neurotransmitter-related molecules. J Comp Neurol 519:1219–1257. [DOI] [PubMed] [Google Scholar]
- Strawn JR, Pyne-Geithman GJ, Ekhator NN, Horn PS, Uhde TW, Shutter LA, Baker DG, Geracioti TD Jr (2010) Low cerebrospinal fluid and plasma orexin-A (hypocretin-1) concentrations in combat-related posttraumatic stress disorder. Psychoneuroendocrinology 35:1001–1007. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Inoue T, Yanagisawa A, Kimura A, Ito Y, Hamajima N (2009) Association between Interleukin-1B C-31T polymorphism and obesity in Japanese. J Epidemiol 19:131–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV (2019) STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47:D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talebizadeh Z, Kibiryeva N, Bittel DC, Butler MG (2005) Ghrelin, peptide YY and their receptors: gene expression in brain from subjects with and without Prader-Willi syndrome. Int J Mol Med 15:707–711. [PMC free article] [PubMed] [Google Scholar]
- Tasan RO, Nguyen NK, Weger S, Sartori SB, Singewald N, Heilbronn R, Herzog H, Sperk G (2010) The central and basolateral amygdala are critical sites of neuropeptide Y/Y2 receptor-mediated regulation of anxiety and depression. J Neurosci 30:6282–6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teske JA, Levine AS, Kuskowski M, Levine JA, Kotz CM (2006) Elevated hypothalamic orexin signaling, sensitivity to orexin A, and spontaneous physical activity in obesity-resistant rats. Am J Physiol Regul Integr Comp Physiol 291:R889–R899. [DOI] [PubMed] [Google Scholar]
- van den Berk-Clark C, Secrest S, Walls J, Hallberg E, Lustman PJ, Schneider FD, Scherrer JF (2018) Association between posttraumatic stress disorder and lack of exercise, poor diet, obesity, and co-occuring smoking: a systematic review and meta-analysis. Health Psychol 37:407–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XQ, Liu ZH, Xue L, Lu L, Gao J, Shen Y, Yang K, Chen QJ, Zhang RY, Shen WF (2016) C1q/TNF-related protein 1 links macrophage lipid metabolism to inflammation and atherosclerosis. Atherosclerosis 250:38–45. [DOI] [PubMed] [Google Scholar]
- Warton K, Foster NC, Gold WA, Stanley KK (2004) A novel gene family induced by acute inflammation in endothelial cells. Gene 342:85–95. [DOI] [PubMed] [Google Scholar]
- Wilensky AE, Schafe GE, Kristensen MP, LeDoux JE (2006) Rethinking the fear circuit: the central nucleus of the amygdala is required for the acquisition, consolidation, and expression of Pavlovian fear conditioning. J Neurosci 26:12387–12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willie JT, Chemelli RM, Sinton CM, Yanagisawa M (2001) To eat or to sleep? Orexin in the regulation of feeding and wakefulness. Annu Rev Neurosci 24:429–458. [DOI] [PubMed] [Google Scholar]
- Wong GW, Krawczyk SA, Kitidis-Mitrokostas C, Revett T, Gimeno R, Lodish HF (2008) Molecular, biochemical and functional characterizations of C1q/TNF family members: adipose-tissue-selective expression patterns, regulation by PPAR-gamma agonist, cysteine-mediated oligomerizations, combinatorial associations and metabolic functions. Biochem J 416:161–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousufzai MIUA, Harmatz ES, Shah M, Malik MO, Goosens KA (2018) Ghrelin is a persistent biomarker for chronic stress exposure in adolescent rats and humans. Transl Psychiatry 8:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan S, Cai GQ, Zheng A, Wang Y, Jia J, Fang H, Yang Y, Hu M, Ding Q (2011) Tumor necrosis factor-alpha regulates the hypocretin system via mRNA degradation and ubiquitination. Biochim Biophys Acta 1812:565–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan S, Che P, Zhao XK, Li N, Ding Y, Liu J, Li S, Ding K, Han L, Huang Z, Wu L, Wang Y, Hu M, Han X, Ding Q (2019) Molecular mechanism of tumour necrosis factor alpha regulates hypocretin (orexin) expression, sleep and behaviour. J Cell Mol Med 23:6822–6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, et al. (2013) Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell 153:707–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, et al. (2008) Genetic variation in human NPY expression affects stress response and emotion. Nature 452:997–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.