Abstract
Background
Throughout the SARS-CoV-2 pandemic, a rapid identification of the virus was essential to quickly recognize positive cases and limit further spread by applying appropriate infection prevention. Many diagnostic laboratories use a multiplex Real-Time PCR assay, as they are not only highly sensitive but also specific. Currently, there are several assays and platforms in the market available which target different SARS-CoV-2 genes. The aim of this study was to validate and verify the GeneFinder™ COVID-19 PLUS RealAmp kit on the ELITe InGenius® instrument and compare to the national reference method.
Methods
GeneFinder™ COVID-19 PLUS RealAmp kit was evaluated against the routine WHO in- house Real-Time PCR assay, which is also the national reference method in the Netherlands and used in our laboratory. The sensitivity was tested using the analytical panel from Qnostics (Glasgow, United Kingdom) and the specificity was tested with patient material comprising of other seasonal respiratory viruses. In addition, 96 clinical samples initially analyzed by routine Real-Time PCR were tested using the GeneFinder™ COVID-19 PLUS RealAmp kit on the ELITe InGenius® instrument.
Results
The GeneFinder™ COVID-19 PLUS RealAmp kit had a similar performance compared to routine in-house testing, with a limit of detection of 500 dC/mL for the RdRp-gene and E gene. Meanwhile, the N gene showed a limit of detection of 50 dC/mL. The SARS-CoV-2 test was highly specific and detected no other respiratory viruses. The results of the clinical samples were comparable between both assays with similar Ct values observed for the in-house Real-Time-PCR and the GeneFinder™ COVID-19 PLUS RealAmp kit for the N gene.
Conclusion
The GeneFinder™ COVID-19 PLUS RealAmp kit on the ELITe InGenius® instrument had an appropriate sensitivity and specificity that could be used in small scale laboratories or during night shifts where accurate diagnostics are crucial.
Keywords: SARS-CoV-2, GeneFinder™ COVID-19 PLUS RealAmp kit, ELITe InGenius® instrument
1. Introduction
The emergence of a novel beta-coronavirus in the beginning of 2020 that could both infect and spread more rapidly in humans has become a major threat worldwide. Within 6 months, the number of infected cases confirmed by molecular tests significantly increased and led to 404 396 human deaths (WHO, 2020. Coronavirus Disease 2019 (COVID-19)). The novel corona virus can cause a spectrum of symptoms, from a simple cold to a severe acute respiratory syndrome (COVID-19) and even death (Wang et al., 2020). The origin of the SARS-CoV-2 virus remains unknown, however it has been supposedly linked to an animal host in China (Benvenuto et al., 2020; Chen et al., 2020; Guo et al., 2020). Relatively quick molecular tests such as Real-Time PCR (qPCR) were designed and implemented into routine diagnostic laboratories worldwide to rapidly identify the virus in human specimens. This would allow diagnostics to not only apply appropriate measures but also monitor future cases to avoid further spread. Crucially, a short turnaround time is also necessary to reduce the amount of time someone needs to be in quarantine.
There are currently several commercial qPCR available, each with different advantages (e.g. less hands-on time and results within 1 h) and disadvantages (e.g. not fully automated process and time consuming). Most of these assays target one or more of the following genes: the RNA-dependent RNA polymerase- (RdRp), envelope- (E), nucleotidecapsid- (N), spike- (S) or membrane protein (M) (van Kasteren et al., 2020). It typically depends on the laboratory, as well as the population being tested, that determines the most suitable test and system. The worldwide introduction of the SARS-CoV-2 antigenic tests offered even faster results, usually within 15 min. However, these tests were subsequently revealed to be less sensitive compared to nucleic acid amplification tests (NAT’s), as a result they are not currently used for healthcare workers and patient care. For these populations, it is necessary to have tests with high sensitivity and specificity, such as NAT’s (Ferté et al., 2021; Olearo et al., 2021). The aim of this study was to validate and verify the GeneFinder™ COVID-19 PLUS RealAmp kit on the ELITe InGenius® instrument and compare to the national reference qPCR.
2. Materials and methods
2.1. In-house SARS-CoV-2 qPCR (in-house qPCR)
Nucleic acids were extracted from 190 μL of sample material, in addition to 10 μL of internal control (phocine distemper virus (PDV)), using the NucliSense EasyMag or eMAG with the Specific A protocol (bioMerieux, Lyon, France), according to the manufacturer’s instructions.
A SARS-CoV-2 qPCR targeting the E-gene was performed as described by Corman et al. (Corman et al., 2020), with minor modifications. Briefly, the multiplex PCR was performed with a total reaction volume of 25 μL using 10 μL RNA and 15 μL PCR mix which contained 1xTaqMan® Fast Virus 1-Step Master (Applied Biosystems, Foster City, CA, USA), DNAse/RNAse free water (Sigma, The Netherlands), 400 nM of SARS-CoV-2 forward and reverse primer, 200 nM of SARS-CoV-2 probe, 300 nM of PDV forward primer (5’-cgggtgccttttacaagaac), 300 nM of PDV reverse primer (5’-ttctttcctcaacctcgtcc) and 100 nM of PDV probe (NED-aag ggc caa ttc t-MGBNFQ). The ABI PRISM 7500 (Life technologies, USA) was used for the amplification and detection by the following profile: 2 min 50 °C, 20 s 95 °C, 45 cycles of 3 s 95 °C and 32 s 60 °C.
Analysis was performed using a middleware software referred to as FlowG (LabHelp Labautomation), which can be used for interpretation of the results and communicating them back to the laboratory information system (LIS). Samples with a cycle time value (Ct) lower than 34 were considered as positive. Meanwhile, Ct of 34–39 was considered inconclusive and the sample was repeated, which is routine clinical practice in our hospital for any new cases. Finally, Ct values above 40 were considered negative.
2.2. GeneFinder™ COVID-19 PLUS RealAmp kit on the ELITe InGenius® instrument (InGenius)
The GeneFinder™ COVID-19 PLUS RealAmp kit was certified by the manufacturer for Alveolar Lavage Fluid, nasopharyngeal swabs and sputum samples. As a result, only nasopharyngeal swabs and sputum samples were used in this study
Sample extraction and qPCR were performed using the ELITe InGenius® instrument (ELITechGroup, Puteaux, France) with the GeneFinder™ COVID-19 PLUS RealAmp kit, according to the manufacturer’s instructions. Briefly, nucleic acid was extracted from 200 μL of sample in universal virus transport medium (UTM) (HIMEDIA, India) using the ELITe InGenius SP200 Extraction Cartridge. After extraction 5 μl of purified RNA from the sample, along with 15 μl of complete PCR Mix which contained primers and probes (10 μl COVID-19 PLUS Reaction Mixture and 5 μl of COVID-19 PLUS Probe Mixture) was used in the reaction. This assay targeted three SARS-CoV-2 specific genes: the virus polymerase gene (RdRp-gene), the virus envelope gene (E gene), and the virus nucleoprotein gene (N gene). Additionally, it contained an Internal Control that targets the human endogenous RNase P gene.
The interpretation of the results was performed according to manufacturer’s instructions. A sample was considered positive for SARS-CoV-2 if all three genes were detected or the following gene combinations were obtained: RdRP with E or RdRP and N. In cases where a single gene was detected (only RdRP-gene or only N-gene) or a combination of E and N, the result was considered not reliable and tests needed to be repeated to confirm the sample was positive for SARS-CoV-2. If only the E gene was detected, the sample was interpreted as SARS-CoV-2 positive.
2.3. AlinityM SARS-CoV-2 assay
Twenty-four nasopharyngeal patient samples (Ct 19–40) were tested with the SARS-CoV-2 assay on the AlinityM platform (Abbott Laboratories, Chicago, IL, USA). The AlinityM SARS-CoV-2 assay is a sample to result platform, where extraction and amplification takes place within the one machine. The assay targets the RdRp- and N-gene and additionally has an internal control. A sequence unrelated to SARS-CoV-2 (RNA from the hydroxypyruvate reductase gene of Cucurbita pepo, a pumpkin plant) was introduced into each specimen at the beginning of sample preparation. Samples were extracted, amplified and interpreted according to the manufacturer’s instructions.
2.4. Specificity
Specificity was determined using two different quality panels, the LEQA 1 panel and the proficiency panel prepared by the National Institute for Public Health and Environment (RIVM). The LEQA 1 panel consisted of different dilutions of inactivated SARS-CoV-2, hCoV-NL63, hCoV- 229E, hCoV-OC43, influenza A virus (H3N2) and SARS-CoV-1. Meanwhile, the proficiency panel consisted of different dilutions of inactivated SARS-CoV-1 virus, hCoV-NL63, hCoV- 229E, hCoV-OC43, influenza A virus (H3N2), Influenza B virus (Victoria) and rhinovirus A16. Both panels were tested with the in-house qPCR and the InGenius SARS-CoV-2 assay. Additionally, the following four different viruses, hCoV-229E, hCoV-OC43, hCoV-NL63 and hCoV-HKU1 were evaluated with three different concentrations (Ct 20, 30 and 35). These viruses were isolated from patient nasopharyngeal samples in UTM medium and stored at −80 °C.
2.5. Sensitivity
The SARS-CoV-2 Analytical Qnostic Panel 01 (SCV2AQP01-A) was used to evaluate sensitivity and contained concentrations ranging from 6 Log10 digital PCR Copies/mL (dC/mL) to 1.7 Log10 dC/mL. The QCMD SARS-CoV-2 EQA 2020 (SCV2_20, QAV204215) was additionally evaluated and had a range of 4.29 Log10 dC/mL to 2.48 Log10 dC/mL. Both panels were tested with the in-house qPCR, as well as the InGenius.
2.6. Precision
The precision was validated by inter- and intra assay variation, which was assessed by testing a nasopharyngeal/oropharyngeal patient sample which was positive for SARS-CoV-2 in triplicate with Ct values of approximately 20 and 31.
2.7. Clinical samples
To test the clinical performance of the InGenius, nasopharyngeal and oropharyngeal swabs (contained in 3 mL of UTM or GLY), along with sputum samples were selected based on the results of the in-house qPCR targeting the E gene (Ct 20–39).
3. Results
3.1. Specificity
The InGenius and the in-house qPCR did not show any cross-reactivity with the following viruses: hCoV-229E, hCoV-OC43, hCoV-NL63 hCoV-HKU1, Influenza A virus (H3N2), Influenza B virus (Victoria) and Rhinovirus A16. As the Proficiency panel contained only SARS-CoV-1, the results using the GeneFinder™ COVID-19 Plus RealAmp kit only showed the presence of the E-gene, as the RdRp-gene and the N gene are specific for SARS-CoV-2.
3.2. Sensitivity
The sensitivity of the InGenius assay for the detection of the RdRp-gene and E-gene was 500 dC/mL (Table 1 ). Meanwhile, the sensitivity was lower for the N-gene, with 50 dC/mL. The in-house qPCR had a higher sensitivity for the detection of the SARS-CoV-2 E-gene, compared to the InGenius assay, with 50 dC/mL and 500 dC/mL respectively. However, the limit of detection for the N-gene was comparable between the in-house qPCR and InGenius.
Table 1.
Detection limits using the SARS-CoV-2 Analytical Qnostic panel 01. The limit of detection for the SARS-CoV-2 in-house qPCR and the N-gene using the GeneFinder was 50 dC/mL. The RdRp-gene and E-gene using the GeneFinder was 500 dC/mL.
| SARS-CoV-2 Qnostic analytical panel |
In-house SARS-CoV-2 | GeneFinderTM COVID-19 Plus RealAmp kit |
||||
|---|---|---|---|---|---|---|
| Panel number | dC/mL | Log10 dC/mL | E- gene (Ct value) | RdRP- gene (Ct value) | N- gene (Ct value) | E-gene (Ct value) |
| SCVA2AQP01-S01 | 1000000 | 6,00 | 21.87 | 24.22 | 24.17 | 22.27 |
| SCVA2AQP01-S02 | 100000 | 5,00 | 24.66 | 27.09 | 27.11 | 25.46 |
| SCVA2AQP01-S03 | 10000 | 4,00 | 27.87 | 30.07 | 29.74 | 28.77 |
| SCVA2AQP01-S04 | 5000 | 3.7 | 28.52 | 31.11 | 30.62 | 29.73 |
| SCVA2AQP01-S05 | 1000 | 3,00 | 30.57 | 34.39 | 32.8 | 32.88 |
| SCVA2AQP01-S06 | 500 | 2.7 | 31.89 | 43.97 | 34.08 | 35.45 |
| SCVA2AQP01-S07 | 100 | 2,00 | 34.26 | Not detected | 36.4 | Not detected |
| SCVA2AQP01-S08 | 50 | 1.7 | 35.01 | Not detected | 40.14 | Not detected |
| SCVA2AQP01-S09 | Negative | – | Not detected | Not detected | Not detected | Not detected |
This finding was similarly observed following the evaluation using the QCMD SARS-CoV-2 EQA 2020 panel. Only the in-house qPCR and the InGenius assay targeting the N-gene could detect SARS-CoV-2 until a viral load of 2.48 dC/mL. The InGenius assay targeting the RdRP-gene and the E-gene were similarly able to detect SARS-CoV-2 until a viral load of 3.16 dC/mL (Table 2 ).
Table 2.
Detection limits using the QCMD SARS-CoV-2 EQA 2020. The SARS-CoV-2 in-house qPCR can detect a viral load of up to log 2.48 dC/mL. The GeneFinder ™ COVID-19 Plus RealAmp kit can detect a viral load of up to log 2.48 dC/mL only with the N-gene.
| QCMD SARS-CoV-2 EQA 2020 |
In-house SARS-CoV-2 | GeneFinderTM COVID-19 Plus RealAmp kit |
||||
|---|---|---|---|---|---|---|
| Panel number | Log10 dC/mL | Sample content | E- gene (Ct value) | RdRP- gene (Ct value) | N- gene (Ct value) | E- gene (Ct value) |
| SCV2_101S-01 | 3.16 | SARS-CoV-2 | 30 | negative | 33,98 | 36,9 |
| SCV2_101S-02 | 3.93 | hCoV-229E | negative | Negative | negative | Negative |
| SCV2_101S-03 | 2.48 | SARS-CoV-2 | 33 | Negative | 35,77 | Negative |
| SCV2_101S-04 | 4.29 | SARS-CoV-2 | 27 | 29,51 | 29,56 | 28,49 |
| SCV2_101S-05 | 3.27 | SARS-CoV-2 | 30 | 42,04 | 32,64 | 33 |
| SCV2_101S-06 | 2.48 | SARS-CoV-2 | 33 | Negative | 36,49 | Negative |
| SCV2_101S-07 | 3.27 | SARS-CoV-2 | 30 | 35,73 | 32,83 | 32,9 |
| SCV2_101S-08 | 3.16 | SARS-CoV-2 | 32 | 42,01 | 33,62 | 34,46 |
| SCV2_101S-09 | Neg | Coronavirus negative | negative | Negative | negative | Negative |
| SCV2_101S-10 | 4.0 | hCoV-OC43 | negative | Negative | negative | Negative |
3.3. Precision
The intra- and inter assay variation was based on the Ct values of high and low viral loads of SARS-CoV-2. The coefficient of variation (CV %) was between 0.25–5.23 % for the intra-assay and 2.59–5.92 % for the inter-assay over 3 different days (Table 3 ).
Table 3.
Intra- and inter- assay variation of the GeneFinder ™ COVID-19 Plus RealAmp kit using the ELITe InGenius platform. The intra- assay variation of a sample with a high viral load (Ct around 20) was lower compared to a sample with a low viral load (Ct around 31). In contrast, the inter assay variation was comparable for the high and low viral load samples.
| Gene | Sample type | Intra-assay | Inter-assay |
|---|---|---|---|
| Average Ct ± SD (%CV) | Average Ct ± SD (%CV) | ||
| E-gene | high positive | 19.6 ± 0.48 (2.45) | 18.97 ± 1.12 (5.92) |
| low positive | 30.93 ± 1.44 (4.66) | 31.32 ± 0.98 (3.11) | |
| RdRp-gene | high positive | 22.17 ± 0.29 (1.31) | 22.44 ± 0.89 (3.95) |
| low positive | 34.55 ± 1.22 (3.54) | 34.15 ± 1.57 (4.58) | |
| N-gene | high positive | 20.25 ± 0.15 (0.72) | 20.20 ± 1.02 (5.07) |
| low positive | 31.09 ± 1.63 (5.23) | 31.87 ± 0.82 (2.59) |
3.4. Clinical samples
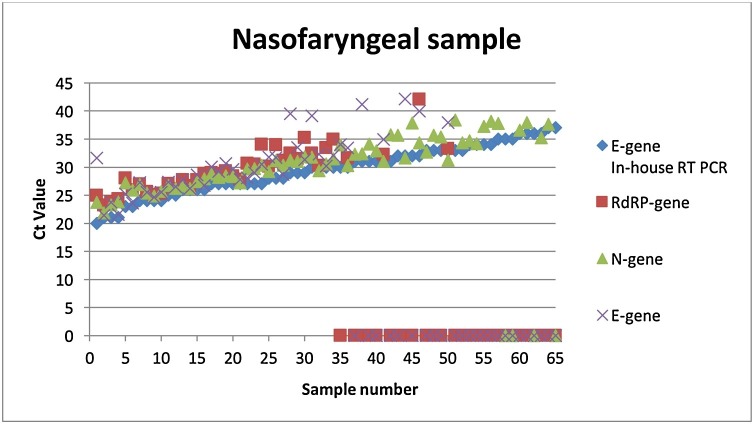
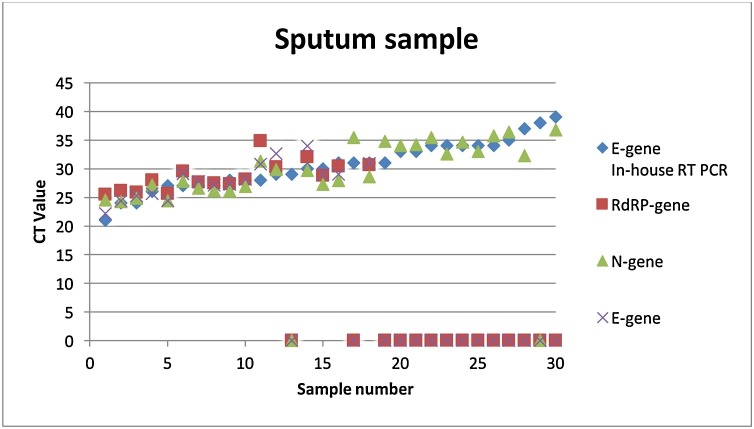
A total of 96 patient samples (66 nasopharyngeal and 30 sputum samples) were randomly selected and tested with the InGenius platform and compared with the in-house qPCR (Fig. 1, Fig. 2 ). Overall, the in-house qPCR had a lower Ct value. Samples with a Ct < 30 were detected with all three genes (RdRp-, N- and E-gene). Most of the samples with a Ct >30 were only detected with the N-gene. Additionally, 24 nasopharyngeal patient samples (Ct 19–40) were tested for SARS-CoV-2 and compared between the following three platforms: the in-house qPCR, the InGenius and the AlinityM. Similarly, the in-house qPCR also yielded lower Ct values compared to the other assays. Twelve samples with a Ct range of 19–31 were detected by all three assays for all three genes. Meanwhile, five samples with a Ct range of 34–37 were only detected with the in-house SARS-CoV-2 assay, the SARS-CoV-2 AlinityM assay (Abbott) and the N-gene from the GeneFinder ™ COVID-19 Plus RealAmp kit assay. Furthermore, samples with a Ct value higher than 37 were found to be above the detection limit of all three assays. Finally, samples with a Ct value higher than 34 were considered inconclusive and a new sample was required.
Fig. 1.
Overview of the nasopharyngeal samples tested with the in-house SARS-CoV-2 PCR and the GeneFinder ™ COVID-19 Plus RealAmp kit on the ELITe InGenius platform. Overall, twelve samples in the Ct range of 20-25 (number 1-12), twenty-four samples in the Ct range of 26–30 (number 13–36), twenty samples in the Ct range of 31-34 (number 37-56) and ten samples in the Ct range of 35–39 (57–66) were included.
Fig. 2.
Overview of the sputum samples tested with the in-house SARS-CoV-2 qPCR and the GeneFinder ™ COVID-19 Plus RealAmp kit on the ELITe InGenius platform. Overall, three samples in the Ct range of 20-25 (number 1–3), twelve samples in the Ct range of 26–30 (number 4–15), eleven samples in the Ct range of 31-34 (number 16–25) and four samples in the Ct range of 35-39 (26–30) were included.
4. Discussion
In this study, we demonstrated that the InGenius platform performs well in the detection of SARS-CoV-2. Only nasopharyngeal swabs and sputum samples were used for this validation and verification. The InGenius platform was shown to be SARS-CoV-2 specific and did not detect other coronaviruses.
The sensitivity of the RdRp-gene and the E gene were comparable and detected samples with up to 500 dC/mL of the virus. The InGenius had a higher sensitivity for the N gene, detecting up to 50 dC/mL of the virus, which was comparable to the level of sensitivity for the E gene in the national reference method. The difference in sensitivity between these three genes may be related to a higher presence of subgenomic N gene messenger RNA, which is related to the replication and transcription process, compared to the E gene and RdRp-gene (Ogando et al., 2020). As far as we know, only D.S.Y. Ong et al. (Ong et al., 2020) looked at the added value for the detection of the N gene to determine a SARS-CoV-2 infection. Furthermore, it was subsequently shown that solitary N gene positives, previously tested negative by the reference method, could be considered positive given the radiological findings or other respiratory materials positive for SARS-CoV-2 (Ong et al., 2020). The question remains regarding the clinical relevance. If only the N gene is detected, it can indicate a lower viral load. When samples have a higher viral load, the other genes in the assay are more likely to be detected. Additionally, it is still unclear to what extent a low viral load can contribute to the spread of SARS-CoV-2. It is believed that a viral load of Ct 30 or higher may have a lower level of transmission compared to higher viral loads (Kampen et al., 2021; Rhee et al., 2021).
In our hospital, routine clinical practice dictates in case of low positives, to take a new sample from the individual on the day or the day after to confirm the result. Such confirmation is important, especially if someone is at the beginning or at the end of an infection.
The difference in sensitivity between the E-gene of the InGenius platform and the national reference method could be due to the differences in the extraction method and/or the PCR master mix. Nevertheless, the sensitivity of the InGenius platform is good enough to be used for the diagnosis of SARS-CoV-2.
We also demonstrated a difference in sensitivity between the in-house qPCR and the InGenius platform in the detection of SARS-CoV-2 in 96 clinical samples. All three genes were detected in the majority of samples with a Ct value of up to 33 from the in-house qPCR.
The InGenius is a fully automated device. As a result, the InGenius has the advantage that the extraction, PCR and interpretation can take place within one platform. The software is easy to operate and the curves are clearly displayed, rendering them easy to interpret.
However, the InGenius platform also has some disadvantages, compared to other platforms currently on the market. Firstly, no internal control is included in the process, from extraction to amplification. This indicates that there was no monitoring of the effectiveness of the process. However, there is an endogenous internal control included (RNaseP gene). This control will indicate if there are sufficient human cells present. It is typically included to determine whether the sampling has been carried out correctly, which is essential for proper diagnostics.
Secondly, as only 12 samples can be tested per run, the InGeniys may not be suitable for large scale screening. Currently there are several platforms available where more samples could be tested simultaneously with the results known within 1 h. For example, the FilmArray (bioMerieux) and the Gene-Xpert (Cepheid) platforms (Eckbo et al., 2021; Wolters et al., 2020). These platforms are easy to operate and generate the results within 1 h. Additionally, these platforms include separate modules, which indicates that they can be easily expanded, according to the needs of the laboratory and the population being screened. However, similarly to the InGenius, these platforms are also unsuitable for large population screening.
The BDmax (BD) platform (Chung et al., 2021) has a similar workflow to the InGenius. Twenty-four samples can be tested simultaneously, with new samples being able to be loaded after extraction, while the qPCR is still running. As a result, there is more of a continuous flow, compared to the InGenius.
Our in-house qPCR uses the eMag (bioMerieux), which allows for 46 samples to be extracted at once. Compared to the InGenius, additional manual actions are required prior to qPCR. The time from sample to result is approximately 170 min for both platforms.
The use of the InGenius platform for SARS-CoV-2 in routine diagnostics has another drawback in our setting. It is possible to connect the in-house qPCR with FlowG (a middleware software) to remotely view the curves, interpret results and communicate the results to the LIS, including Ct values. Currently, ELITech-group contains a middleware software which connects multiple devices, however it is not possible to view the curves remotely. In addition, Ct values are not sent to the LIS, which is desirable to monitor viral loads in patients. The selection of tests and platforms will typically depend on the population being tested, the size of the laboratory and the expertise of the operational staff.
An advantage of InGenius platform is that multiple genes could be targeted simultaneously for the detection of SARS-CoV-2. In the current situation, where additional variants are being observed and mutation frequency in the targeted genes are increasing, it is beneficial to use a single assay targeting multiple genes to avoid false-negative results. In conclusion, the GeneFinder™ COVID-19 PLUS RealAmp kit on the ELITe InGenius® instrument is a sensitive test and suitable for diagnosing SARS-CoV-2 infections. The platform could be used in smaller laboratories that do not perform 'bulk' SARS-CoV-2 screening and are not dependent on a diagnosis within 1 h.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Benvenuto D., Giovanetti M., Salemi M., Prosperi M., De Flora C., Junior Alcantara L.C., Angeletti S., Ciccozzi M. The global spread of 2019-nCoV: a molecular evolutionary analysis. Pathog. Glob. Health. 2020;114:64. doi: 10.1080/20477724.2020.1725339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H., Jian M., Chang C., Lin J., Yeh K., Chen C., Chiu S., Wang Y., Liao S., Li S., Hsieh S., Tsai S., Perng C., Yang J., Liu M., Chang F., Shang H. Novel dual multiplex real-time RT-PCR assays for the rapid detection of SARS-CoV-2, influenza A/B, and respiratory syncytial virus using the BD MAX open system. Emerg. Microbes Infect. 2021;10:161–166. doi: 10.1080/22221751.2021.1873073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brunink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B., Veer B., van den Brink S., Wijsman L., Goderski G., Romette J., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M., sten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveillance: bulletin européen sur les maladies transmissibles. 2020:25. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckbo E.J., Locher K., Caza M., Li L., Lavergne V., Charles M. Evaluation of the BioFire® COVID-19 test and Respiratory Panel 2.1 for rapid identification of SARS-CoV-2 in nasopharyngeal swab samples. Diagn. Microbiol. Infect. Dis. 2021;99 doi: 10.1016/j.diagmicrobio.2020.115260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferté T., Ramel V., Cazanave C., Lafon M., Bébéar C., Malvy D., Georges-Walryck A., Dehail P. Accuracy of COVID-19 rapid antigenic tests compared to RT-PCR in a student population: the StudyCov study. J. Clin. Virol. 2021;141 doi: 10.1016/j.jcv.2021.104878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Cao Q., Hong Z., Tan Y., Chen S., Jin H., Tan K., Wang D., Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak – an update on the status. Military Med. Res. 2020:7. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampen J., Vijver D., Fraaij P., Haagmans B., Lamers M.M., Okba N., Van Den Akker J.P.C., Endeman H., Gommers D., Cornelissen J., Hoek R., Eerden M., Hesselink D., Metselaar H., Verbon A., Steenwinkel J., Aron G., Gorp E., Boheemen S., Voermans J., Boucher C., Molenkamp R., Koopmans M., Geurts van Kessel C., Eijck A. Duration and key determinants of infectious virus shedding in hospitalized patients with coronavirus disease-2019 (COVID-19) Nat. Commun. 2021;12:267. doi: 10.1038/s41467-020-20568-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogando N.S., Dalebout T.J., Zevenhoven-Dobbe J.C., Limpens, Ronald W.A.L., van der Meer Y., Caly L., Druce J., de Vries, Jutte J.C., Kikkert M., Bárcena M., Sidorov I., Snijder E.J. SARS-coronavirus-2 replication in Vero E6 cells: replication kinetics, rapid adaptation and cytopathology. J. Gen. Virol. 2020;101:925–940. doi: 10.1099/jgv.0.001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olearo F., Nörz D., Heinrich F., Sutter J.P., Roedl K., Schultze A., Schulze zur Wiesch J., Braun P., Oestereich L., Kreuels B., Wichmann D., Aepfelbacher M., Pfefferle S., Lütgehetmann M. Handling and accuracy of four rapid antigen tests for the diagnosis of SARS-CoV-2 compared to RT-qPCR. J. Clin. Virol. 2021;137 doi: 10.1016/j.jcv.2021.104782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong D.S.Y., Claas E.C.J., Breijer S., Vaessen N. Comparison of the GeneFinderTM COVID-19 Plus RealAmp Kit on the sample-to-result Platform ELITe InGenius to the national reference method: An added value of N gene target detection? J. Clin. Virol. 2020;132 doi: 10.1016/j.jcv.2020.104632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee C., Kanjilal S., Baker M., Klompas M., Rhee C. 2021. Duration of SARS-CoV-2 Infectivity: when is it Safe to Discontinue Isolation? [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kasteren P.B., van der Veer B., van den Brink S., Wijsman L., de Jonge J., van den Brandt A., Molenkamp R., Reusken C.B.E.M., Meijer A. Comparison of seven commercial RT-PCR diagnostic kits for COVID-19. J. Clin. Virol. 2020;128 doi: 10.1016/j.jcv.2020.104412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wang Y., Chen Y., Qin Q.Q. Unique epidemiological and clinical features of the emerging 2019 novel coronavirus pneumonia (COVID‐19) implicate special control measures. J. Med. Virol. 2020;92:568. doi: 10.1002/jmv.25748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2020. Coronavirus Disease 2019 (COVID-19). W.H.O. 2020. Coronavirus Disease 2019 (COVID-19) - Situation Report 141, on Wold Health Organization 09.06.2020. [Google Scholar]
- Wolters F., van de Bovenkamp J., van den Bosch B., van den Brink S., Broeders M., Chung N.H., Favié B., Goderski G., Kuijpers J., Overdevest I., Rahamat-Langedoen J., Wijsman L., Melchers W.J., Meijer A. Multi-center evaluation of cepheid xpert® xpress SARS-CoV-2 point-of-care test during the SARS-CoV-2 pandemic. J. Clin. Virol. 2020;128 doi: 10.1016/j.jcv.2020.104426. [DOI] [PMC free article] [PubMed] [Google Scholar]