Abstract
Drug repositioning has been used extensively since the beginning of the COVID-19 pandemic in an attempt to identify antiviral molecules for use in human therapeutics. Hydroxychloroquine and azithromycin have shown inhibitory activity against SARS-CoV-2 replication in different cell lines. Based on such in vitro data and despite the weakness of preclinical assessment, many clinical trials were set up using these molecules. In the present study, we show that hydroxychloroquine and azithromycin alone or combined does not block SARS-CoV-2 replication in human bronchial airway epithelia. When tested in a Syrian hamster model, hydroxychloroquine and azithromycin administrated alone or combined displayed no significant effect on viral replication, clinical course of the disease and lung impairments, even at high doses. Hydroxychloroquine quantification in lung tissues confirmed strong exposure to the drug, above in vitro inhibitory concentrations. Overall, this study does not support the use of hydroxychloroquine and azithromycin as antiviral drugs for the treatment of SARS-CoV-2 infections.
Keywords: ACE2, Angiotensin-Converting enzyme 2; AZM, Azithromycin; BID, Bis in die; COVID-19, Coronavirus disease 2019; CQ, Chloroquine; EC50, 50% effective concentrations; FCS, Fetal calf serum; HAE, Human airway epithelium; HCQ, Hydroxychloroquine; H&E, Hematoxylin-eosin; IQ, Inhibitory quotient; LDH, Lactate dehydrogenase; MERS-CoV, Middle East Respiratory Syndrome Coronavirus; MOI, Multiplicity of infection; REM, Remdesivir; RT-qPCR, Quantitative real-time RT-PCR; SARS-CoV, Severe Acute Respiratory Syndrome Coronavirus; SARS-CoV-2, Severe Acute Respiratory Syndrome Coronavirus 2; SD, Standard deviation; TCID50, Median tissue culture infectious dose; TMPRSS2, Type II transmembrane serine protease; WHO, World Health Organization
Keywords: SARS-CoV-2, COVID-19, Coronavirus, Antivirals, Hydroxychloroquine, Azithromycin, In vivo, Syrian hamster, Ex vivo, Human airway epithelium
1. Introduction
A novel member of the Betacoronavirus genus, named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), emerged in late 2019 in Wuhan, China. As other emerging betacoronaviruses such as severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV), this new virus causes severe pneumonia and acute respiratory distress, particularly in the elderly population. In March 2020, the World Health Organization (WHO) declared the coronavirus disease 2019 (COVID-19) as pandemic. As of November 2nd, 2021, the numbers of SARS-CoV-2-related infections and deaths listed by the WHO were over 246 million and near 5 million, respectively (WHO, 2021). During the first months of the pandemic, repositioning of drugs has been proposed to offer the possibility of rapid availability of molecules with antiviral activity (Mercorelli et al., 2018). In this regard, significant antiviral activity was reported for chloroquine (CQ) on VeroE6 cells at a clinically relevant concentration (Wang et al., 2020). This article was quickly followed by a press release claiming efficacy in humans (Gao et al., 2020). After these two reports, CQ and derivatives have been viewed by many as serious candidates for therapeutic use.
Chloroquine and its hydroxylated form, hydroxychloroquine (HCQ), have been developed as antimalarial drugs. HCQ is now also used against autoimmune diseases. Both drugs have already been reported to be effective in inhibiting the replication of many viruses in vitro, including emerging coronaviruses (Barnard et al., 2006; Dyall et al., 2014). Their antiviral activity against several RNA viruses has been assessed in rodent models with positive (Li et al., 2017; Yan et al., 2013) and negative results (Falzarano et al., 2015; Vigerust and McCullers, 2007). However, some clinical trials conducted before the current pandemic with several RNA viruses indicated that CQ had no effect on acute virus infection in humans (De Lamballerie et al., 2008; Paton et al., 2011; Tricou et al., 2010). At the beginning of the pandemic, the in vitro antiviral activity of HCQ against SARS-CoV-2 was widely investigated. In VeroE6 cells, 50% effective concentrations (EC50) were ranging from 4.2 to 4.5 μM (Liu et al., 2020; Touret et al., 2020), and in CaCo2 cells EC50 were below 5.0 μM (Touret et al., 2020). However, the inconsistency of the antiviral activity of HCQ and CQ in other models such as in Calu-3 cells, A549 cells and in a reconstituted human airway epithelium (HAE) has raised doubts about their repurposing potential for clinical use (Grosse et al., 2021; Hoffmann et al., 2020; Maisonnasse et al., 2020; Ou et al., 2021). Currently, several hypotheses on the mechanism of antiviral action of HCQ in vitro have been proposed, such as inhibition of virus entry through the ACE2 receptor and GM1 ganglioside pathways (Fantini et al., 2020a, 2020b), and blockage of virus fusion with endosomal membranes by inhibiting endosomal maturation (Liu et al., 2020).
Azithromycin (AZM) is an antibiotic of the macrolide class commonly used to treat respiratory infections and having a strong pulmonary diffusion (Ishida et al., 1994). AZM has a broad-spectrum antiviral activity in cell culture and some studies reported its efficacy against several RNA viruses infections using ex vivo and preclinical models (Madrid et al., 2015; Menzel et al., 2016; Schögler et al., 2015; Wu et al., 2018; Zeng et al., 2019). Notably, AZM demonstrated in vitro antiviral activity against SARS-CoV-2 with an EC50 value of 2.1 μM and less than 5.0 μM in VeroE6 and CaCo2 cell cultures respectively (Touret et al., 2020). Its ability to prevent bacterial superinfection and its immunomodulatory properties have been proposed as an additional argument for therapeutic use in SARS-CoV-2 infection (Beigelman et al., 2010; Menzel et al., 2017; Schögler et al., 2015) and 132 clinical trials assessing HCQ and AZM on COVID-19 patients have been launched (www.clinicaltrials.gov) before the release of first preclinical data on non-human primates (Maisonnasse et al., 2020).
Here, we completed the ex vivo assessment of AZM and the combination of HCQ and AZM by testing their antiviral activity against SARS-CoV-2 in a HAE model and we assessed the antiviral activity of high doses of HCQ and AZM, alone or combined, against SARS-CoV-2 using a Syrian hamster model (Mesocricetus auratus).
2. Material and methods
2.1. Cell line
VeroE6 cells (ATCC CRL-1586) were grown in minimal essential medium (Life Technologies) supplemented with 7 .5% heat-inactivated fetal calf serum (FCS; Life Technologies), incubated at 37 °C and 5% CO2 with 1% penicillin/streptomycin (PS, 5000 U.mL−1 and 5000 μg mL−1 respectively; Life Technologies) and supplemented with 1% non-essential amino acids (Life Technologies).
2.2. Human airway epithelia (HAE)
Mucilair™ HAE recovered from human primary cells of bronchial biopsies were purchased from Epithelix SARSL (Geneva, Switzerland). MucilAir™ HAE is composed of approximately 400,000 cells (Essaidi-Laziosi et al., 2018). Specific Epithelix media had been used to maintain bronchial epithelium in air-liquid interface. Epithelium was derived from a 56-year-old Caucasian female donor with no known pathology. All samples purchased from Epithelix SARL have been obtained with informed consent. These studies received approval from local ethic committees, and were conducted according to the declaration of Helsinki on biomedical research (Hong Kong amendment, 1989).
2.3. Virus
SARS-CoV-2 strain BavPat1 was provided by Christian Drosten (Berlin, Germany) through the European Virus Archive GLOBAL (www.european-virus-archive.com). Virus stock was prepared by inoculating at multiplicity of infection (MOI) of 0.001 a 25 cm2 culture flask of confluent VeroE6 cells with medium supplemented with 2.5% FCS. At the peak of replication, cell supernatant medium was harvested, supplemented with 25 mM HEPES (Sigma) and then stored at −80 °C. Whole-genome sequencing of virus stock was performed as previously described (Driouich et al., 2021) to check the absence of furin-cleavage-site adaptation mutations (Klimstra et al., 2020).
2.4. Antiviral assay using HAE
Before inoculation, epithelia were gently washed with pre-warmed OPTI-MEM medium (Life technologies). Infection with SARS-COV-2 was performed on the apical side at a multiplicity of infection of 0.1 as previously described (Pizzorno et al., 2020; Touret et al., 2021). Epithelial cells were cultivated in basolateral media with different concentrations of azithromycin (Sigma) (from 0 to 80 μM), with hydroxychloroquine (Sigma) (5 and 10 μM) and with remdesivir (BLD pharm) used as a positive compound control (10 μM). For the combination experiment, two concentrations of both hydroxychloroquine and azithromycin were evaluated: 5 and 10 μM. Media were replaced every day during the experiment. At the apical side, samples were collected by washing with 200 μL of pre-warmed OptiMEM medium. A volume of 100 μL of this washing medium was used for RNA extraction (see below) and the remaining volume was used to perform a TCID50 assay. For the cytotoxicity evaluation, LDH (lactate dehydrogenase) release was quantified in the basal medium using the Cytotoxicity Detection KitPLUS (LDH) (Roche) according to the manufacturer instructions at day 4 post infection using a Tecan Infinite 200Pro plate reader (TECAN). Assessing cytotoxicity in HAE model, based on LDH detection has been previously described (Essaidi-Laziosi et al., 2018). The media from each insert was analyzed in duplicate.
2.5. In vivo experiments
In vivo experiments were approved by the local ethical committee (C2EA—14) and the French ‘Ministère de l’Enseignement Supérieur, de la Recherche et de l’Innovation’ (APAFIS#23975) and performed in accordance with the French national guidelines and the European legislation covering the use of animals for scientific purposes. All experiments were conducted in biosafety level 3 laboratory.
2.5.1. Animal handling
Three-week-old female golden Syrian hamsters were provided by Janvier Labs. Animals were maintained in ISOcage P - Bioexclusion System (Techniplast) with unlimited access to water/food and 14 h/10 h light/dark cycle. Animals were weighed and monitored daily for the duration of the study to detect the appearance of any clinical signs of illness/suffering.
2.5.2. Infection
Four-week-old anesthetized (isoflurane) animals were intranasally infected with 104 TCID50 of virus diluted in a final volume of 50 μL with 0.9% sodium chloride solution. The mock-infected group was intranasally inoculated with 50 μL of 0.9% sodium chloride solution.
2.5.3. Drug administration
AZM for oral suspension (Zithromax, Pfizer) was dissolved in ultra-pure distilled water to obtain a 20 mg/mL suspension. Two tablets of 200 mg of HCQ (Plaquenil, Sanofi) were crushed and then dissolved in aqueous vehicle solution supplemented with 0.5% of hydroxymethylpropyl cellulose and 0.1% of Tween80 to obtain a 10 mg/mL solution. AZM and HCQ were orally administrated under general anesthesia (isoflurane). Untreated animals received the vehicle of HCQ only. As a positive drug control, a distinct group of hamsters was treated with intraperitoneal favipiravir twice a day (25 mg/mL; solubilized in 0.9% sodium chloride solution; Courtesy of Toyama-Chemical) as previously described (Driouich et al., 2021).
2.5.4. Lung and plasma collection
Lung and blood samples were collected directly after the time of sacrifice. The left lung lobe was washed in 10 mL of 0.9% sodium chloride solution, blotted with filter paper, weighed and transferred into a 2 mL tube that contain 1 mL of 0.9% sodium chloride solution and 3 mm glass beads. It was crushed using a Tissue Lyser machine (Retsch MM400) at 30 cycles/s for 20min and centrifuged at 16,200 g for 10min. The supernatant medium was transferred into a 1.5 mL tube, centrifuged at 16,200 g for 10 min. For plasma collection, 1 mL of blood was harvested into a 2 mL tube containing 100 μL of 0.5 M EDTA (ThermoFischer Scientific), centrifuged at 16,200 g for 10 min and plasma was transferred into a 1.5 mL tube. Lung clarified homogenates and plasma were stored at −80 °C.
2.6. Quantitative real-time RT-PCR (RT-qPCR) assays
To prevent contaminations, all experiments were conducted in a molecular biology laboratory specifically designed for clinical diagnosis which includes separate laboratories to respect walking forward principle. All animal samples were spiked with 10 μL of internal control (bacteriophage MS2) before acid nucleic extraction as previously described (Ninove et al., 2011). Nucleic acid from HAE apical side washes, lung clarified homogenates and plasma were extracted using the Qiacube HT robot and the QIAamp 96 DNA kit (both from Qiagen) following manufacturer's instructions. Before extraction, 100 μL of each sample was transferred in a S-Block (Qiagen) previously loaded with VXL lysis buffer containing RNA carrier and proteinase K.
Real-time RT-qPCR assays (GoTaq 1-step qRT-PCR, Promega) were performed using 3.8 μL of extracted RNA, 6.2 μL of RT-qPCR mix and standard fast cycling parameters (i.e. 10 min at 50 °C, 2 min at 95 °C, and 40 amplification cycles at 95 °C for 3 sec followed by 30 sec at 60 °C). RNA quantities were determined using four serial dilutions of appropriate quantified T7-generated synthetic RNA standards. RT-qPCR reactions were performed on QuantStudio 12 K Flex Real-Time PCR System (Applied Biosystems) and analyzed with the QuantStudio 12 K Flex Applied Biosystems software v1.2.3. For antiviral assay using HAE, primers and probe sequences, which target SARS-CoV-2 N gene, were: Forward: 5′-GGCCGCAAATTGCACAAT-3’; Reverse: 5′-CCAATGCGCGACATTCC-3’; Probe: FAM-CCCCCAGCGCTTCAGCGTTCT-BHQ1. For animal experiments, primers and probe sequences, which target SARS-CoV-2 Rdrp gene, were: Forward: 5′-GTGARATGGTCATGTGTGGCGG-3'; Reverse 5′-CARATGTTAAASACACTATTAGCATA-3'; Probe: 5′-FAM-CAGGTGGAACCTCATCAGGAGATGC-TAMRA-3 (Corman et al., 2020).
2.7. Tissue-culture infectious dose 50 (TCID50) assays
To determine infectious titers, 96-well culture plates of confluent VeroE6 cells were inoculated with serial dilutions of samples (ten-fold or four-fold dilutions for HAE apical side washes and animal samples respectively). Each sample dilution was performed in sextuplicate. Plates were read for presence or absence of cytopathic effect after an incubation of 4–6 days at 37 °C and 5% CO2. Infectious titers were estimated using the method described by Reed & Muench and expressed as TCID50/ml or TCID50/g of lung (Reed, 1938).
2.8. Histology
Animal handling, hamster infections and drug administration were performed as described above in the ‘in vivo experiments’ section. Lungs were collected following intratracheal instillation of 4% (w/v) formaldehyde solution and fixed 72 h at room temperature with a 4% (w/v) formaldehyde solution and then embedded in paraffin. Histological analysis was performed as previously described (Driouich et al., 2021). Briefly, tissue sections of 3.5 μm were stained with hematoxylin-eosin (H&E) and blindly analyzed by a certified veterinary pathologist. Different lung compartments were examined: (1) regarding bronchial and alveolar walls, a score of 0–4 was assigned based on inflammation severity; (2) for alveoli, a score of 0–2 was assigned based on presence and severity of hemorrhagic necrosis; (3) for vessel changes (leukocyte accumulation in sub-endothelial space and tunica media), absence or presence was scored 0 or 1 respectively. A cumulative score (see Supplemental Table 1) was then calculated and assigned to a grade of severity.
2.9. Determination of HCQ concentrations in plasma and lung of hamsters
Quantification of hydroxychloroquine in plasma and lung tissues of infected hamsters was performed by a sensitive and selective validated high-performance liquid chromatography coupled with tandem mass spectrometry method (Quattro Premier XE LC-MS/MS, Waters), with lower limits of quantification of 0.015 μg/mL for plasma and 0.05 μg/mL for lung tissue (Doudka et al., 2020). Plasma samples and lung tissue homogenates were collected at euthanasia, 14 h after last drug intake of 3 days of treatment. Hydroxychloroquine was extracted by a simple protein precipitation method, using methanol and ice-cold acetonitrile for plasma and lung tissue homogenates respectively, as previously described (Maisonnasse et al., 2020). Briefly, 100 μL of sample matrix spiked with 10 μL of internal standard working solution (HCQ-d5, Alsachim) was vortexed for 2 min and then centrifuged at 4 °C for 10 min. Tissue-homogenate supernatants were evaporated. Dry residues or plasma supernatants were then transferred to 96-well plates and 5 μL was injected. To assess the selectivity and the specificity of the method and matrix effect, blank plasma and lung tissue homogenates from untreated hamsters were processed and compared with HCQ and internal standard-spiked samples from the same untreated controls animals.
2.10. Statistical analysis
Statistical analyses and graphical representations were realized using Graphpad Prism 7 (Graphpad software). When relevant, two-sided statistical tests were always used. P-values lower than 0.05 were considered statistically significant.
3. Results
3.1. AZM exhibit no antiviral activity in an ex vivo model of HAE
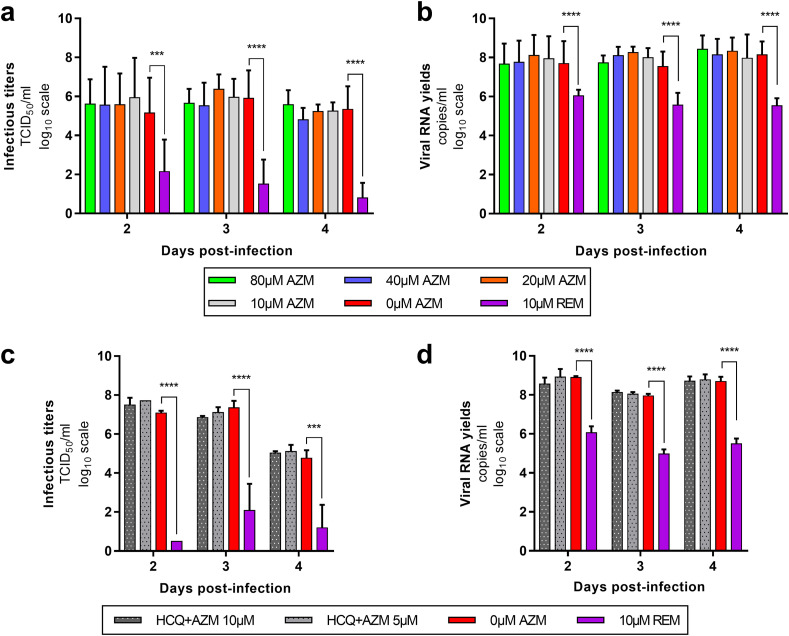
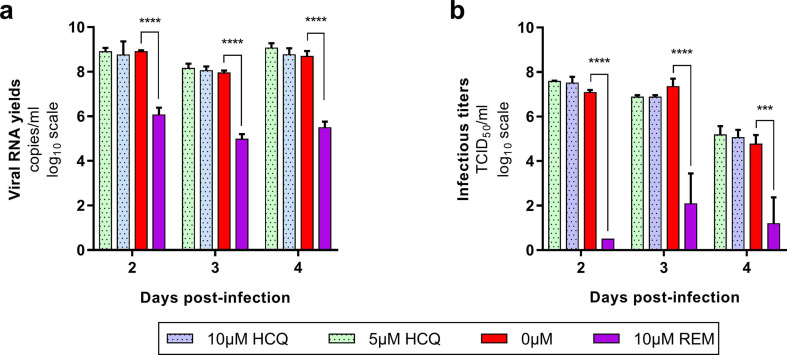
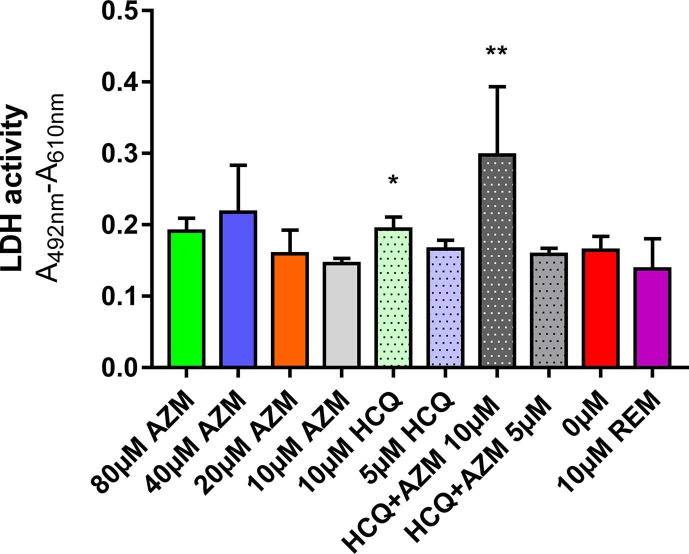
As previously described, the ex vivo model of reconstituted HAE is a relevant tool to assess the antiviral activity of compounds against SARS-CoV-2 (Pizzorno et al., 2020; Touret et al., 2021). In this section, we focused on AZM and HCQ-AZM combination because we previously demonstrated that HCQ alone does not exhibit antiviral activity with that model (Maisonnasse et al., 2020) (Fig. 1 ). Remdesivir was used as antiviral positive control at 10 μM. In a first experiment, several AZM concentrations (0; 10; 20; 40; 80 μM) were tested. This experiment was repeated once, concomitantly with two HCQ-AZM combinations (5 and 10 μM) and HCQ alone (5 and 10 μM) (Fig. 1 and Supplemental Fig. 1). Epithelia were exposed to drugs through their basolateral side from the day of infection. Viral excretion was monitored by performing apical side washes from 2 to 4 days post-infection (dpi) by measuring infectious titres (Fig. 1a,c) and viral RNA yields (Fig. 1b,d) using TCID50 and quantitative real time RT-qPCR assays respectively. The cytotoxic effect of each condition (measure of lactate dehydrogenase activity) was also assessed to strengthen the interpretation of the results as previously described (Essaidi-Laziosi et al., 2018) (Supplemental Fig. 2).
Fig. 1.
Antiviral activity of AZM and AZM-HCQ combination in a bronchial human airway epithelium (HAE).
Kinetics of virus excretion at the apical side of the epithelium in presence of AZM (a,b) and HCQ-AZM combination (c,d). Kinetics were measured using a TCID50 assay (a,c) and a RT-qPCR assay (b,d). For panels a and b, data represent mean ± SD of two independent experiments each performed in duplicate (n = 4). For panels c and d, data represent mean ± SD of on experiment performed in triplicate (details in Supplemental Table 2). **** and *** symbols indicate that infectious titers or RNA yields are significantly smaller than those for untreated (0 μM AZM) epithelia with a p-value inferior to 0.0001 or ranging between 0.0001 and 0.001, respectively (Multiple comparison t-test) (details in Supplemental Table 3).
Taken together, results from all experiments showed that AZM alone or combined with HCQ did not significantly reduce (i) infectious titres and RNA yields in apical washes from 2 to 4 dpi (p ≥ 0.8926 and p ≥ 0.4614 respectively) (Fig. 1). Results also confirmed that HCQ alone was not effective in this model (Maisonnasse et al., 2020) (Supplemental Fig. 1). By contrast, remdesivir significantly reduced infectious titers at 3 and 4 dpi (p ≤ 0.0001) and RNA yields in apical washes from 2 to 4 dpi (p ≤ 0.0482). HCQ at 10 μM when used alone or in combination with AZM induced cytotoxicity (p ≤ 0.0397) (Supplemental Fig. 2) but this did not impact viral replication. The overall results indicate a lack of antiviral activity of AZM, HCQ and combination of both drugs using this ex vivo model of HAE.
3.2. HCQ and AZM alone or combined have no antiviral effect in a hamster model
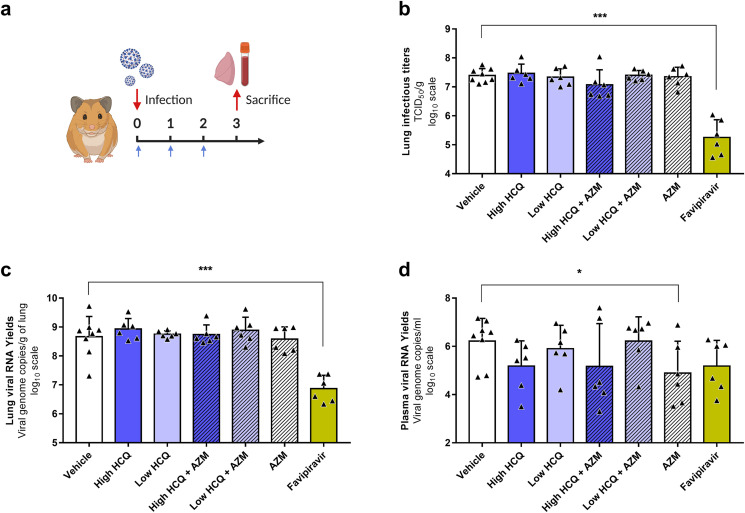
To assess in vivo efficacy of HCQ and AZM alone or combined, groups of Syrian hamsters were intranasally infected with 1 × 104 TCID50 of SARS-CoV-2 and orally treated following a preemptive strategy (infection and first dose of treatment at the same time; Fig. 2 a). Several dosing regimens were tested: high dose of HCQ (176.9 ± 16.3 mg/kg/day BID); low dose of HCQ (135.6 ± 11.6 mg/kg/day BID); AZM (90.6 ± 6.9 mg/kg/day QD); high dose of HCQ combined with AZM; low dose of HCQ combined with AZM.
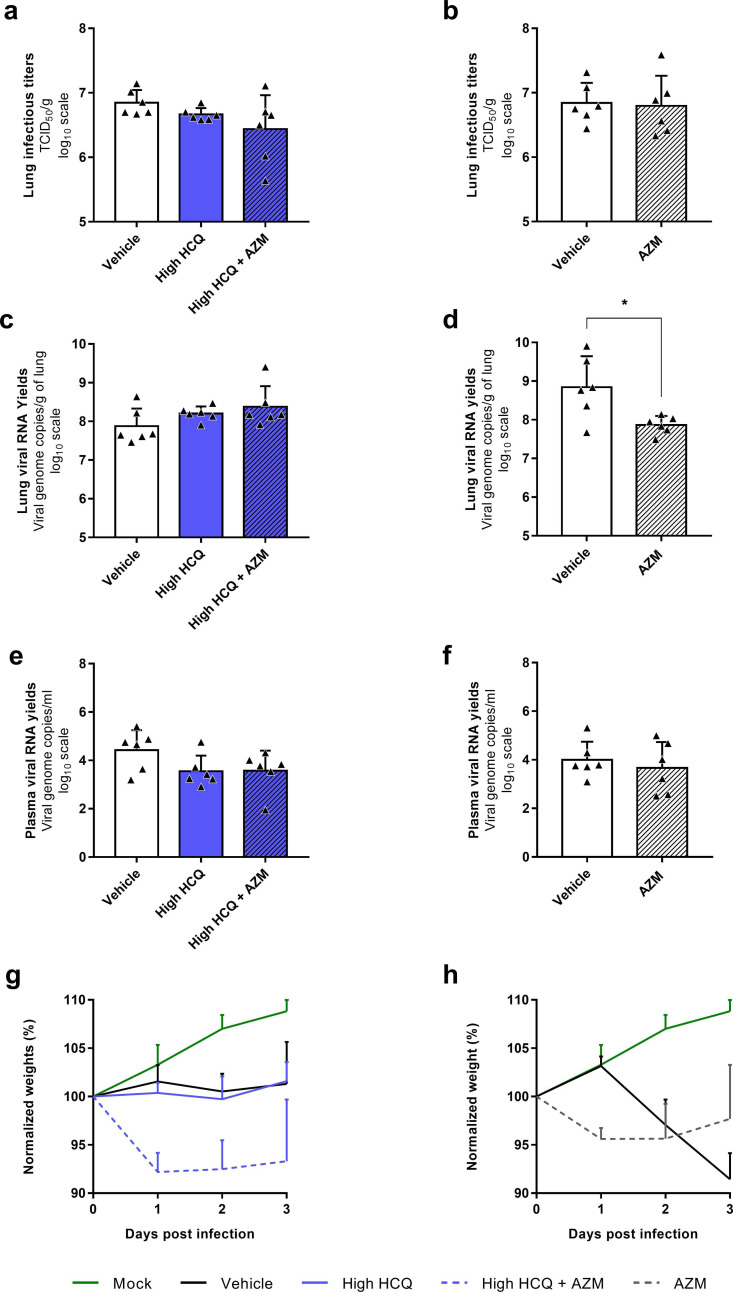
Fig. 2.
Antiviral activity of HCQ and AZM alone or combined in a Syrian hamster model at 3 dpi.
(a) Experimental timeline (realized on biorender.com). Groups of 6 or 8 hamsters were intranasally infected with 1 × 104 TCID50 of SARS-CoV-2, treated from 0 to 2 dpi (blue arrows) and sacrificed at 3 dpi. (b) Lung infectious titers measured using a TCID50 assay. (c–d) Lung viral RNA yields (c) and plasma viral loads (d) measured using a RT-qPCR assay. Data represent mean ± SD of individual data of hamsters (n = 6 to 8; details in Supplemental Table 4). *** and * symbols indicate that lung infectious titers, lung viral RNA yields or plasma viral loads are significantly smaller than those for the untreated group (vehicle) with a p-value ranging between 0.0001-0.001 and 0.01–0.05, respectively (Unpaired and Welch's t tests). Clinical follow-up of this experiment is presented in Supplemental Fig. 3.
All treatments were initiated at the time of infection (afternoon of day 0). HCQ treatments began with a single drug administration of 176.9 or 135.6 mg/kg for high and low dose treatments respectively at day 0. Then, similar daily drug amounts were administrated but following a BID scheme from day 1 post-infection (ie. 176.9 or 135.6 mg/kg/day BID for high and low dose treatments respectively). In all animal experiments, an untreated group of hamsters was infected and orally received the HCQ vehicle twice a day (called “vehicle” in graphs and “vehicle group” below). A positive control group was used in some experiments and received intraperitoneal favipiravir (863.9 ± 79.5 mg/kg/day BID) following a preemptive strategy as previously described (Driouich et al., 2021).
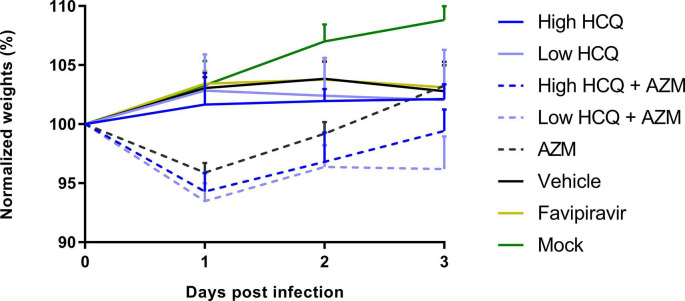
In a first experiment (Fig. 2), groups of 6 animals treated with high dose of HCQ, low dose of HCQ, AZM, high dose of HCQ combined with AZM or low dose of HCQ combined with AZM were sacrificed at 3 dpi (Fig. 2a). Two control groups of 8 and 6 animals received the vehicle only or the favipiravir respectively. Compared to the vehicle group, no treatment regimen induced a significant reduction of infectious titers and viral RNA yields in lungs, while significant reductions were observed in the group treated with favipiravir (p = 0.0002) (Fig. 2b-c). When assessing the viral replication in plasma, a slight significant reduction of viral RNA yields was observed in the group treated with AZM alone (p = 0.0483) (Fig. 2d). To confirm these results, we replicated this experiment in two steps, with only three dosing regimens (groups of 6 animals) (Supplemental Fig. 4). Firstly, with high dose of HCQ, high dose of HCQ combined with AZM and the vehicle only as control (Supplemental Fig. 4a,c,e,g). Secondly, with AZM and the vehicle only as control (Supplemental Fig. 4b,d,f,h). For high dose of HCQ, alone or combined with AZM, no significant reduction was found. For AZM, the results did not confirm the significant reduction observed during the first experiment in plasma, but another non-reproducible slightly significant reduction of viral RNA yields in lungs was detected (p = 0.0290).
During this first set of experiments, a lack of weight gain was observed from 2 dpi with all infected animals as previously described (p ≤ 0.0208) (Driouich et al., 2021). Moreover, immediately after the first administration, all groups treated with AZM alone or combined with HCQ exhibited weight losses (−9.8% to −2.9% of initial weight at 1 dpi), while the others infected groups did not (Supplemental Figs. 3 and 4g-h).
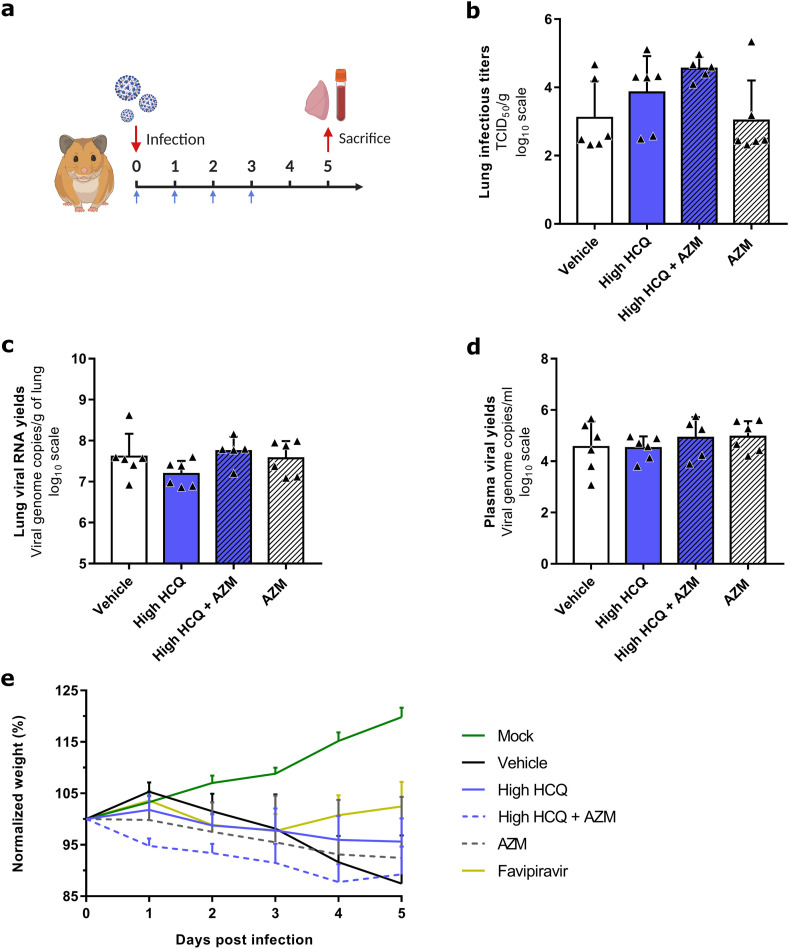
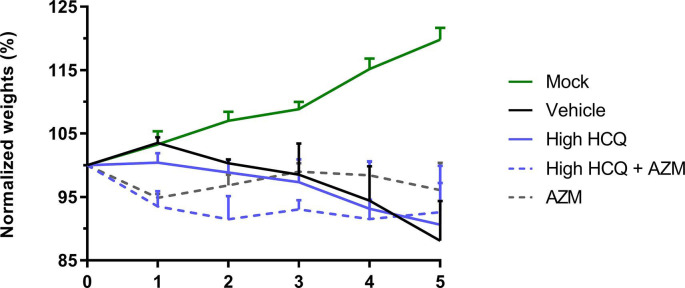
To further investigate the effect of three treatment regimens (high dose of HCQ, AZM alone and high dose of HCQ combined with AZM) on the viral clearance and on the clinical course of the disease, groups of 6 animals were treated from 0 to 3 dpi and sacrificed at 5 dpi (Fig. 3 a). Two control groups of 6 animals received the vehicle only or the favipiravir. The favipiravir group was used only for clinical follow-up because animals were sacrificed at 7 dpi since we combined different experiments. One animal in the group treated with high dose of HCQ combined with AZM and one animal in the favipiravir group died at 1 dpi (unexplained death). Compared to the vehicle group, no treatment regimen induced a significant reduction of lung infectious titers (Fig. 3b) (p ≥ 0.0519), lung viral RNA yields (Fig. 3c) (p ≥ 0.1371) and plasma viral loads (Fig. 3d) (p ≥ 0.4165). Only animals treated with favipiravir gained weight until 4 dpi (p ≤ 0.0143 at 4 and 5 dpi when compared to vehicle group), in accordance with previous experiments with this drug (Driouich et al., 2021) (Fig. 4 e). As described above, animals treated with AZM, alone or combined with HCQ, were subjected to early weight losses (Fig. 4e).
Fig. 3.
Antiviral activity and clinical impact of HCQ and AZM alone or combined in a Syrian hamster model at 5 dpi.
(a) Experimental timeline (realized on biorender.com). Groups of 6 hamsters were intranasally infected with 1 × 104 TCID50 of SARS-CoV-2, treated from 0 to 3 dpi (blue arrows) and sacrificed at 5 dpi. (b) Lung infectious titers measured using a TCID50 assay. (c–d) Lung viral RNA yields (c) and plasma viral loads (d) measured using a RT-qPCR assay. (e) Clinical follow-up. Animal weights are expressed as normalized weights (i.e. % of initial weight). Data represent mean ± SD of individual data of hamsters (n = 5 to 6; details in Supplemental Table 4)
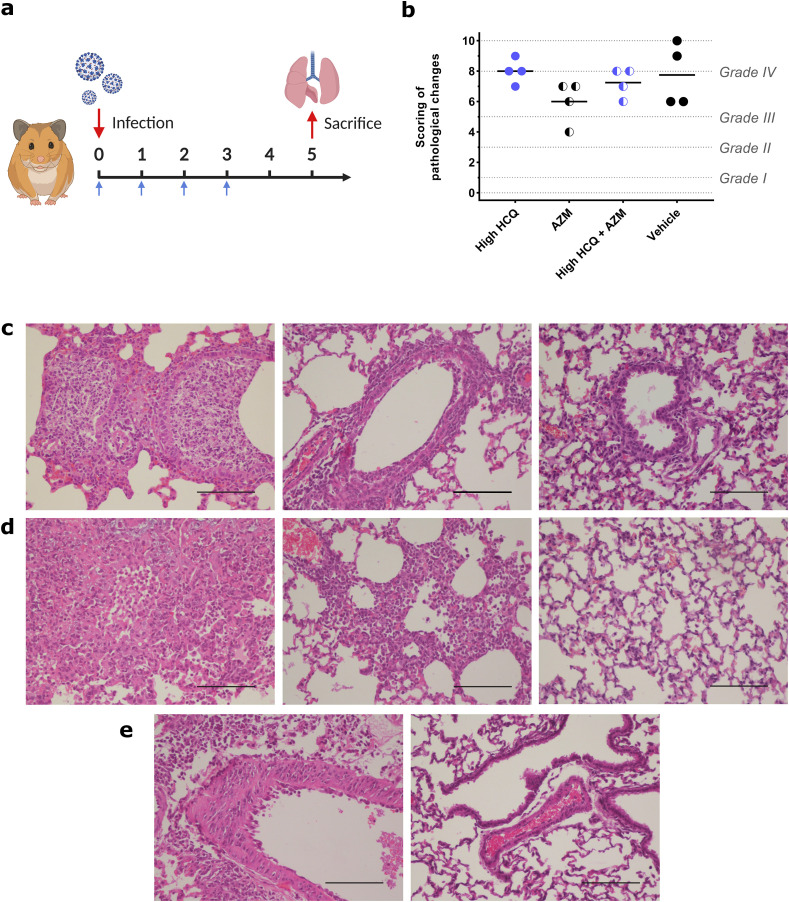
Fig. 4.
Impact of HCQ and AZM alone or combined on lung histological impairments in a Syrian hamster model at 5 dpi.
(a) Experimental timeline (realized on biorender.com). Groups of 4 hamsters were intranasally infected with 1 × 104 TCID50 of SARS-CoV-2, treated from 0 to 3 dpi and sacrificed at 5 dpi (blue arrows). (b) Scores of histopathological changes measured following criteria presented in Supplemental Table 1, as previously described (Driouich et al., 2021). Dashes represent mean scores for each group (n = 4; details in Supplemental Table 5). (c) Representative images of bronchial inflammation (scale bar: 100 μ). From left to right, pictures represent marked neutrophilic bronchitis (high HCQ + AZM animal), mild peribronchial leucocytic infiltration (high HCQ + AZM animal) and no inflammation (mock-infected animal). (d) Representative images of alveolar inflammation (scale bar: 100 μ). From left to right, pictures represent severe interstitial pneumonia (high HCQ animal), marked interstitial changes (high HCQ + AZM animal) and no inflammation (mock-infected animal). (e) Representative images of vessel changes (leukocyte accumulation within vascular walls). From left to right, pictures represent presence (high HCQ animal) and absence of lesions (mock-infected animal) (scale bars: 100 μ). Clinical follow-up is represented in Supplemental Fig. 5.
In a last experiment (Fig. 4), we investigated the impact of high dose of HCQ, AZM alone and high dose of HCQ combined with AZM on lung pathological changes induced by SARS-CoV-2 infection. Groups of 4 animals were infected and treated from 0 to 3 dpi and sacrificed at 5 dpi (Fig. 4a). As a control, a group of 4 animals received the vehicle only. For each animal, a cumulative score from 0 to 10 was calculated and assigned to a grade of severity (normal; I = mild; II = moderate; III = marked and IV = severe; details in Supplemental Table 5), based on the severity of broncho-interstitial inflammation, alveolar haemorrhagic necrosis and vessel lesions as previously described (Driouich et al., 2021). Almost all animals exhibited marked or severe broncho-interstitial pneumonia, except one in the group treated with AZM which displayed moderate pulmonary pathological changes (Fig. 4b). Compared to the vehicle group, no treatment regimen induced a significant reduction of cumulative scores (p ≥ 0.2110).
Finally, the exposure to HCQ in plasma and lungs was assessed from animals sacrificed during the first experiment (i.e. determination of trough concentrations after multiple administration; Table 1 ). With both high and low doses of HCQ, alone or combined with AZM, a high accumulation of HCQ was observed with lung/plasma ratios ranging from 57.5 to 362. At both dosing regimens, lung concentrations of HCQ were above the EC50. The inhibitory quotient (IQ: ratio between the concentration and EC50 of 5 μM) in plasma and lungs was 0.07 and 9.14 for treatment with high dose of HCQ, 0.04 and 8.40 for treatment with low dose of HCQ, 0.13 and 10.3 for treatment with high dose of HCQ combined with AZM and 0.09 and 11.01 for treatment with low dose of HCQ combined with AZM, respectively.
Table 1.
Plasma and lung concentrations of HCQ after multiple drug administrations in infected Syrian hamsters. Results are expressed by mean ± SD (Details in Supplemental Table 6).
| Hydroxychloroquine groups | Hydroxychloroquine + Azithromycin groups | ||||
|---|---|---|---|---|---|
| Plasma (μg/mL) | Lung (μg/g) | Lung/plasma ratio | Plasma (μg/mL) | Lung (μg/g) | Lung/plasma ratio |
| High dose of HCQ | High dose of HCQ + AZM | ||||
| 0.12 ± 0.02 | 15.4 ± 6.07 | 132 ± 63.8 | 0.21 ± 0.08 | 17.3 ± 3.51 | 97.4 ± 47.2 |
| Low dose of HCQ | Low dose of HCQ + AZM | ||||
| 0.07 ± 0.02 | 14.1 ± 3.03 | 242 ± 106 | 0.15 ± 0.07 | 18.5 ± 4.32 | 128 ± 32.4 |
4. Discussion
Early in the pandemic, HCQ and AZM were found to inhibit SARS-CoV-2 replication in vitro (Touret et al., 2020; Wang et al., 2020), leading to numerous clinical trials assessing the efficacy of HCQ alone or in combination with AZM to treat COVID-19. In the present study, we investigate the efficacy of HCQ, AZM and combination of both drugs ex vivo and in vivo, in a hamster model. The doses used in the present study were chosen in order to induce high lung exposition with both molecules and consequently avoid ambiguity regarding their antiviral activity in vivo.
Regarding azithromycin, we completed previous in vitro investigations by using an ex vivo model of human airway epithelium with drug concentrations up to 16 times the EC50 identified in VeroE6 and CaCo2 cells (i.e. 2.1 μM and 5.0 μM, respectively) (Touret et al., 2020). We found that AZM does not inhibit SARS-CoV-2 replication in this model which is relevant for the evaluation of antiviral molecules in SARS-CoV-2 infection (Pizzorno et al., 2020; Touret et al., 2021). In vivo, AZM has previously been evaluated only in combination with HCQ, with doses of 10 mg/kg/day in Syrian hamsters (Kaptein et al., 2020) and 18 mg/kg/day after a loading dose of 36 mg/kg in cynomolgus macaques (Maisonnasse et al., 2020; Rosenke et al., 2020). In the current study, we used a higher dose monotherapy of AZM (91 mg/kg/day) and found no reproducible antiviral effect, with unmodified lung infectious titres and viral RNA yields, and no improvement of pulmonary histopathological impairments. The good pulmonary diffusion of the molecule has been well documented in several animal models (Davila et al., 1991; Resendiz et al., 2020; Rivulgo et al., 2013). In particular, a study assessed the lung diffusion of the drug after a single oral intake of 50 mg/kg in Syrian hamsters. Lung maximal concentrations reached 28.5 μg/g 1 h after the administration of the drug (corresponding to 38.1 μM) and a long half-life of AZM in lungs was reported (22.5 h). These data suggest that the AZM dosing regimen used in the present study, around 2 times higher than the dose used in the work previously cited (Ishida et al., 1994), can induce a lung concentration at least 3.8 times higher than EC50 values between the first drug intake and the sacrifice of the animals used in our investigation.
Altogether, our results suggest that despite favourable pharmacokinetics and lung exposure, AZM has no antiviral activity against SARS-CoV-2 in the hamster model, even at concentrations far above in vitro inhibitory concentrations. Of note, AZM treatment was associated with weight losses, especially at 1 dpi, in accordance with previous studies that showed the toxicity of this class of antibiotic in small rodents when administrated orally (DeSalva et al., 1969; Faine and Kaipainen, 1955).
Regarding the HCQ, previous studies have reported antiviral activity in different cell lines (Liu et al., 2020; Touret et al., 2020; Weston et al., 2020) but also reduced or lacking antiviral efficacy of the molecule in cells expressing ACE2 and TMPRSS2 (Dittmar et al., 2021; Grosse et al., 2021; Hoffmann et al., 2020; Ou et al., 2021) and the absence of antiviral activity in the HAE model (Maisonnasse et al., 2020). Here we confirmed that HCQ does not inhibit SARS-CoV-2 replication in the HAE model with concentrations near or at least 2 times above the EC50 values identified in VeroE6 and CaCo2 cells (i.e. from 4.2 to 4.5 μM and below 5.0 μM, respectively). Furthermore, a few preclinical studies investigated the efficacy of HCQ against SARS-CoV-2 in hamster and macaque models (Kaptein et al., 2020; Maisonnasse et al., 2020; Rosenke et al., 2020). Syrian hamsters were treated with doses ranging between 6.5 and 50 mg/kg/day (Kaptein et al., 2020; Rosenke et al., 2020). Rhesus and cynomolgus macaques were treated with maintenance doses ranging between 6.5 and 45 mg/kg/day (Maisonnasse et al., 2020; Rosenke et al., 2020). In these models, preemptive and preventive treatments were ineffective in reducing respiratory viral loads and lung impairments. To ensure a good lung exposure of the drug, we used higher dosing regimens than previously investigated: the high dose of HCQ in our work was 3.5 times higher than the highest dose administrated in the hamster model of SARS-CoV-2 infection (Kaptein et al., 2020; Rosenke et al., 2020).
As expected, the mean lung trough concentration for all treatment was 16.3 μg/g (48.6 μM), well above the EC50 values determined in vitro with a mean IQ of 9.72. In fact, 4-aminoquinolines such as HCQ have a large volume of distribution, leading to a high lung accumulation thanks to their physicochemical properties (Browning, 2014; Mackenzie, 1983; Rowland Yeo et al., 2020). Recent results of bronchoalveolar lavages on COVID-19 patients have shown that HCQ is found in the lung epithelial lining fluid at concentrations above EC50 values, once again suggesting a good exposition of the tissue of concern (Ruiz et al., 2021).
Despite such high lung concentrations, our results were in accordance with previous studies: HCQ treatment had no impact on (i) viral load at peak of replication, (ii) viral clearance at 5 dpi, (iii) clinical disease and (iv) lung pathological changes. However, the ability of HCQ and AZM to prevent transmission between animals has not been studied since no nasal washes were perfomed during in vivo experiments. This could be a limit of the current study.
After providing experimental evidence that both HCQ and AZM have no antiviral activity ex vivo and in vivo against SARS-CoV-2 when used in monotherapy, we observed no antiviral activity with the combination of both drugs in the same models, despite a synergistic effect predicted in silico (Fantini et al., 2020a). Of note, the only report of synergistic effect of these two drugs in vitro failed to demonstrate it in a robust way (Andreani et al., 2020). Indeed, no combination tested with concentrations below the EC50 values showed a significant antiviral effect and no robust synergy evaluation experiments were performed (Meyer et al., 2019). Finally, other preclinical studies have also shown no synergistic effect with these two drugs (Kaptein et al., 2020; Maisonnasse et al., 2020).
Interestingly, recent investigations shed a new light on the link between phospholipidosis and the in vitro activity of antiviral candidates against SARS-CoV-2 (Tummino et al., 2021). A correlation between the induction of phospholipidosis and antiviral activity has been described for several drugs and demonstrated an in vitro antiviral effect solely due to physicochemical properties and not to a specific targeting of SARS-CoV-2. Hence, the in vitro antiviral activity of HCQ and AZM must be rediscussed since these molecules are known or strongly suspected to induce this biological disorder in various models (Baronas et al., 2007; Tsukimura et al., 2021; Tummino et al., 2021; Van Bambeke et al., 1996) and clinical cases (Costa et al., 2013; Khubchandani and Bichle, 2013; Manabe et al., 2021; Obeidat et al., 2020).
In conclusion, our results showed a lack of antiviral activity of HCQ, AZM and combination of both drugs against SARS-CoV-2 ex vivo using a reconstituted HAE model. Moreover, the assessment of efficacy of high doses of HCQ and AZM, used alone or combined, in the hamster model showed no effect on viral replication, viral clearance, clinical disease and pulmonary histopathological impairments. This lack of efficacy in vivo cannot be correlated with an unfavourable pharmacokinetics, since both molecules showed good lung diffusion and lung concentrations above EC50 values determined in vitro. Altogether, these results do not support the use of HCQ and AZM for the treatment of COVID-19.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Laurence Thirion (UVE; Marseille) for providing RT-qPCR systems, Camille Placidi (UVE; Marseille) for her help to manage cell culture experiments and Karine Barthélémy (UVE; Marseille) for pharmacology experiments. We thank Pr. Drosten and Pr. Drexler for providing the SARS-CoV-2 strain through the European Research infrastructure EVA GLOBAL. This work was supported by the Fondation de France “call FLASH COVID-19”, project TAMAC, by “Institut national de la santé et de la recherche médicale” through the REACTing (REsearch and ACTion targeting emerging infectious diseases) initiative (“Preuve de concept pour la production rapide de virus recombinant SARS-CoV-2”), and by European Virus Archive Global (EVA 213 GLOBAL) funded by the European Union's Horizon 2020 research and innovation program under grant agreement No. 871029. This work was also funded by REACTING/ANRS MIE under the agreement No. 21180 (‘Activité des molécules antivirales dans le modèle hamster’). A part of the work was done on the Aix Marseille University antivirals platform “AD2P”.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.antiviral.2021.105212.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Andreani J., Le Bideau M., Duflot I., Jardot P., Rolland C., Boxberger M., Wurtz N., Rolain J.M., Colson P., La Scola B., Raoult D. In vitro testing of combined hydroxychloroquine and azithromycin on SARS-CoV-2 shows synergistic effect. Microb. Pathog. 2020;145:104228. doi: 10.1016/j.micpath.2020.104228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard D.L., Day C.W., Bailey K., Heiner M., Montgomery R., Lauridsen L., Chan P.K., Sidwell R.W. Evaluation of immunomodulators, interferons and known in vitro SARS-coV inhibitors for inhibition of SARS-coV replication in BALB/c mice. Antivir. Chem. Chemother. 2006;17:275–284. doi: 10.1177/095632020601700505. [DOI] [PubMed] [Google Scholar]
- Baronas E.T., Lee J.-W., Alden C., Hsieh F.Y. Biomarkers to monitor drug-induced phospholipidosis. Toxicol. Appl. Pharmacol. 2007;218:72–78. doi: 10.1016/j.taap.2006.10.015. [DOI] [PubMed] [Google Scholar]
- Beigelman A., Mikols C.L., Gunsten S.P., Cannon C.L., Brody S.L., Walter M.J. Azithromycin attenuates airway inflammation in a mouse model of viral bronchiolitis. Respir. Res. 2010;11:90. doi: 10.1186/1465-9921-11-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning D.J. Springer New York; New York, NY: 2014. Pharmacology of Chloroquine and Hydroxychloroquine, Hydroxychloroquine and Chloroquine Retinopathy; pp. 35–63. [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brunink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa R.M., Martul E.V., Reboredo J.M., Cigarrán S. Curvilinear bodies in hydroxychloroquine-induced renal phospholipidosis resembling Fabry disease. Clin. Kidney J. 2013;6:533–536. doi: 10.1093/ckj/sft089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davila D., Kolacny-Babic L., Plavsic F. Pharmacokinetics of azithromycin after single oral dosing of experimental animals. Biopharm Drug Dispos. 1991;12:505–514. doi: 10.1002/bdd.2510120704. [DOI] [PubMed] [Google Scholar]
- De Lamballerie X., Boisson V., Reynier J.C., Enault S., Charrel R.N., Flahault A., Roques P., Le Grand R. On chikungunya acute infection and chloroquine treatment. Vector Borne Zoonotic Dis. 2008;8:837–839. doi: 10.1089/vbz.2008.0049. [DOI] [PubMed] [Google Scholar]
- DeSalva S.J., Evans R.A., Marcussen H.W. Lethal effects of antibiotics in hamsters. Toxicol. Appl. Pharmacol. 1969;14:510–514. doi: 10.1016/0041-008x(69)90012-x. [DOI] [PubMed] [Google Scholar]
- Dittmar M., Lee J.S., Whig K., Segrist E., Li M., Kamalia B., Castellana L., Ayyanathan K., Cardenas-Diaz F.L., Morrisey E.E., Truitt R., Yang W., Jurado K., Samby K., Ramage H., Schultz D.C., Cherry S. Drug repurposing screens reveal cell-type-specific entry pathways and FDA-approved drugs active against SARS-Cov-2. Cell Rep. 2021;35:108959. doi: 10.1016/j.celrep.2021.108959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudka N., Giocanti M., Basso M., Ugdonne R., Barthelemy K., Lacarelle B., Blin O., Solas C., Guilhaumou R. Development and validation of a simple and rapid UHPLC-MS/MS method for the quantification of hydroxychloroquine in plasma and blood samples in the emergency context of SARS-CoV-2 pandemic. Ther. Drug Monit. 2020;43(4):570–576. doi: 10.1097/FTD.0000000000000836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driouich J.S., Cochin M., Lingas G., Moureau G., Touret F., Petit P.R., Piorkowski G., Barthelemy K., Laprie C., Coutard B., Guedj J., de Lamballerie X., Solas C., Nougairede A. Favipiravir antiviral efficacy against SARS-CoV-2 in a hamster model. Nat. Commun. 2021;12:1735. doi: 10.1038/s41467-021-21992-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyall J., Coleman C.M., Hart B.J., Venkataraman T., Holbrook M.R., Kindrachuk J., Johnson R.F., Olinger G.G., Jr., Jahrling P.B., Laidlaw M., Johansen L.M., Lear-Rooney C.M., Glass P.J., Hensley L.E., Frieman M.B. Repurposing of clinically developed drugs for treatment of Middle East respiratory syndrome coronavirus infection. Antimicrob. Agents Chemother. 2014;58:4885–4893. doi: 10.1128/AAC.03036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essaidi-Laziosi M., Brito F., Benaoudia S., Royston L., Cagno V., Fernandes-Rocha M., Piuz I., Zdobnov E., Huang S., Constant S., Boldi M.O., Kaiser L., Tapparel C. Propagation of respiratory viruses in human airway epithelia reveals persistent virus-specific signatures. J. Allergy Clin. Immunol. 2018;141:2074–2084. doi: 10.1016/j.jaci.2017.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faine S., Kaipainen W.J. Erythromycin in experimental leptospirosis. J. Infect. Dis. 1955;97:146–151. doi: 10.1093/infdis/97.2.146. [DOI] [PubMed] [Google Scholar]
- Falzarano D., Safronetz D., Prescott J., Marzi A., Feldmann F., Feldmann H. Lack of protection against ebola virus from chloroquine in mice and hamsters. Emerg. Infect. Dis. 2015;21:1065–1067. doi: 10.3201/eid2106.150176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantini J., Chahinian H., Yahi N. Synergistic antiviral effect of hydroxychloroquine and azithromycin in combination against SARS-CoV-2: what molecular dynamics studies of virus-host interactions reveal. Int. J. Antimicrob. Agents. 2020;56:106020. doi: 10.1016/j.ijantimicag.2020.106020. [DOI] [PubMed] [Google Scholar]
- Fantini J., Di Scala C., Chahinian H., Yahi N. Structural and molecular modelling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection. Int. J. Antimicrob. Agents. 2020;55:105960. doi: 10.1016/j.ijantimicag.2020.105960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Tian Z., Yang X. Breakthrough: chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci Trends. 2020;14:72–73. doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- Grosse M., Ruetalo N., Layer M., Hu D., Businger R., Rheber S., Setz C., Rauch P., Auth J., Froba M., Brysch E., Schindler M., Schubert U. Quinine inhibits infection of human cell lines with SARS-CoV-2. Viruses. 2021;13 doi: 10.3390/v13040647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Mösbauer K., Hofmann-Winkler H., Kaul A., Kleine-Weber H., Krüger N., Gassen N.C., Müller M.A., Drosten C., Pöhlmann S. Chloroquine does not inhibit infection of human lung cells with SARS-CoV-2. Nature. 2020;585:588–590. doi: 10.1038/s41586-020-2575-3. [DOI] [PubMed] [Google Scholar]
- Ishida K., Kaku M., Irifune K., Mizukane R., Takemura H., Yoshida R., Tanaka H., Usui T., Suyama N., Tomono K., et al. In vitro and in vivo activities of macrolides against Mycoplasma pneumoniae. Antimicrob. Agents Chemother. 1994;38:790–798. doi: 10.1128/aac.38.4.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaptein S.J.F., Jacobs S., Langendries L., Seldeslachts L., Ter Horst S., Liesenborghs L., Hens B., Vergote V., Heylen E., Barthelemy K., Maas E., De Keyzer C., Bervoets L., Rymenants J., Van Buyten T., Zhang X., Abdelnabi R., Pang J., Williams R., Thibaut H.J., Dallmeier K., Boudewijns R., Wouters J., Augustijns P., Verougstraete N., Cawthorne C., Breuer J., Solas C., Weynand B., Annaert P., Spriet I., Vande Velde G., Neyts J., Rocha-Pereira J., Delang L. Favipiravir at high doses has potent antiviral activity in SARS-CoV-2-infected hamsters, whereas hydroxychloroquine lacks activity. Proc. Natl. Acad. Sci. U. S. A. 2020;117:26955–26965. doi: 10.1073/pnas.2014441117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khubchandani S.R., Bichle L.S. Hydroxychloroquine-induced phospholipidosis in a case of SLE: the wolf in zebra clothing. Ultrastruct. Pathol. 2013;37:146–150. doi: 10.3109/01913123.2012.751950. [DOI] [PubMed] [Google Scholar]
- Klimstra W.B., Tilston-Lunel N.L., Nambulli S., Boslett J., McMillen C.M., Gilliland T., Dunn M.D., Sun C., Wheeler S.E., Wells A., Hartman A.L., McElroy A.K., Reed D.S., Rennick L.J., Duprex W.P. SARS-CoV-2 growth, furin-cleavage-site adaptation and neutralization using serum from acutely infected hospitalized COVID-19 patients. J. Gen. Virol. 2020;101:1156–1169. doi: 10.1099/jgv.0.001481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Zhu X., Ji X., Quanquin N., Deng Y.Q., Tian M., Aliyari R., Zuo X., Yuan L., Afridi S.K., Li X.F., Jung J.U., Nielsen-Saines K., Qin F.X., Qin C.F., Xu Z., Cheng G. Chloroquine, a FDA-approved drug, prevents Zika virus infection and its associated congenital microcephaly in mice. EBioMedicine. 2017;24:189–194. doi: 10.1016/j.ebiom.2017.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Cao R., Xu M., Wang X., Zhang H., Hu H., Li Y., Hu Z., Zhong W., Wang M. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020;6:16. doi: 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie A.H. Pharmacologic actions of 4-aminoquinoline compounds. Am. J. Med. 1983;75:5–10. doi: 10.1016/0002-9343(83)91264-0. [DOI] [PubMed] [Google Scholar]
- Madrid P.B., Panchal R.G., Warren T.K., Shurtleff A.C., Endsley A.N., Green C.E., Kolokoltsov A., Davey R., Manger I.D., Gilfillan L., Bavari S., Tanga M.J. Evaluation of ebola virus inhibitors for drug repurposing. ACS Infect. Dis. 2015;1:317–326. doi: 10.1021/acsinfecdis.5b00030. [DOI] [PubMed] [Google Scholar]
- Maisonnasse P., Guedj J., Contreras V., Behillil S., Solas C., Marlin R., Naninck T., Pizzorno A., Lemaitre J., Gonçalves A., Kahlaoui N., Terrier O., Fang R.H.T., Enouf V., Dereuddre-Bosquet N., Brisebarre A., Touret F., Chapon C., Hoen B., Lina B., Calatrava M.R., van der Werf S., de Lamballerie X., Le Grand R. Hydroxychloroquine use against SARS-CoV-2 infection in non-human primates. Nature. 2020;585:584–587. doi: 10.1038/s41586-020-2558-4. [DOI] [PubMed] [Google Scholar]
- Manabe S., Mochizuki T., Sato M., Kataoka H., Taneda S., Honda K., Uchida K., Nitta K. Lupus nephritis and hydroxychloroquine-associated zebra bodies: not just in Fabry disease. Kidney Med. 2021;3:442–446. doi: 10.1016/j.xkme.2021.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel M., Akbarshahi H., Bjermer L., Uller L. Azithromycin induces anti-viral effects in cultured bronchial epithelial cells from COPD patients. Sci. Rep. 2016;6:28698. doi: 10.1038/srep28698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel M., Akbarshahi H., Tufvesson E., Persson C., Bjermer L., Uller L. Azithromycin augments rhinovirus-induced IFNβ via cytosolic MDA5 in experimental models of asthma exacerbation. Oncotarget. 2017;8:31601–31611. doi: 10.18632/oncotarget.16364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercorelli B., Palu G., Loregian A. Drug repurposing for viral infectious diseases: how far are we? Trends Microbiol. 2018;26:865–876. doi: 10.1016/j.tim.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer C.T., Wooten D.J., Paudel B.B., Bauer J., Hardeman K.N., Westover D., Lovly C.M., Harris L.A., Tyson D.R., Quaranta V. Quantifying drug combination synergy along potency and efficacy axes. Cell Syst. 2019;8:97–108 e116. doi: 10.1016/j.cels.2019.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninove L., Nougairede A., Gazin C., Thirion L., Delogu I., Zandotti C., Charrel R.N., De Lamballerie X. RNA and DNA bacteriophages as molecular diagnosis controls in clinical virology: a comprehensive study of more than 45,000 routine PCR tests. PLoS One. 2011;6 doi: 10.1371/journal.pone.0016142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obeidat M., Isaacson A.L., Chen S.J., Ivanovic M., Holanda D. Zebra-like bodies in COVID-19: is phospholipidosis evidence of hydroxychloroquine induced acute kidney injury? Ultrastruct. Pathol. 2020;44:519–523. doi: 10.1080/01913123.2020.1850966. [DOI] [PubMed] [Google Scholar]
- Ou T., Mou H., Zhang L., Ojha A., Choe H., Farzan M. Hydroxychloroquine-mediated inhibition of SARS-CoV-2 entry is attenuated by TMPRSS2. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paton N.I., Lee L., Xu Y., Ooi E.E., Cheung Y.B., Archuleta S., Wong G., Wilder-Smith A. Chloroquine for influenza prevention: a randomised, double-blind, placebo controlled trial. Lancet Infect. Dis. 2011;11:677–683. doi: 10.1016/S1473-3099(11)70065-2. [DOI] [PubMed] [Google Scholar]
- Pizzorno A., Padey B., Julien T., Trouillet-Assant S., Traversier A., Errazuriz-Cerda E., Fouret J., Dubois J., Gaymard A., Lescure F.X., Duliere V., Brun P., Constant S., Poissy J., Lina B., Yazdanpanah Y., Terrier O., Rosa-Calatrava M. Characterization and treatment of SARS-CoV-2 in nasal and bronchial human airway epithelia. Cell Rep. Med. 2020;1:100059. doi: 10.1016/j.xcrm.2020.100059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L.J.M.H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938:493–497. [Google Scholar]
- Resendiz A.S., Bernad M.J., Sanchez Lemus J.C., Rodriguez I.J., Carlin Valderrabano S.C., Estrada D.V. Disposition and pharmacokinetics of azithromycin in serum and a lung tissue of two modified-release formulations compared with an immediate-release product on the market. Pak. J. Pharm. Sci. 2020;33:1079–1085. [PubMed] [Google Scholar]
- Rivulgo V., Sparo M., Ceci M., Fumuso E., Confalonieri A., Delpech G., Bruni S.F. Comparative plasma exposure and lung distribution of two human use commercial azithromycin formulations assessed in murine model: a preclinical study. BioMed Res. Int. 2013;2013:392010. doi: 10.1155/2013/392010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenke K., Jarvis M.A., Feldmann F., Schwarz B., Okumura A., Lovaglio J., Saturday G., Hanley P.W., Meade-White K., Williamson B.N., Hansen F., Perez-Perez L., Leventhal S., Tang-Huau T.L., Callison J., Haddock E., Stromberg K.A., Scott D., Sewell G., Bosio C.M., Hawman D., de Wit E., Feldmann H. Hydroxychloroquine prophylaxis and treatment is ineffective in macaque and hamster SARS-CoV-2 disease models. JCI Insight. 2020;5 doi: 10.1172/jci.insight.143174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland Yeo K., Zhang M., Pan X., Ban Ke A., Jones H.M., Wesche D., Almond L.M. Impact of disease on plasma and lung exposure of chloroquine, hydroxychloroquine and azithromycin: application of PBPK modeling. Clin. Pharmacol. Ther. 2020;108:976–984. doi: 10.1002/cpt.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz S., Concordet D., Lanot T., Georges B., Goudy P., Baklouti S., Mané C., Losha E., Vinour H., Rousset D., Lavit M., Minville V., Conil J.M., Gandia P. Hydroxychloroquine lung pharmacokinetics in critically ill patients with COVID-19. Int. J. Antimicrob. Agents. 2021;57:106247. doi: 10.1016/j.ijantimicag.2020.106247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schögler A., Kopf B.S., Edwards M.R., Johnston S.L., Casaulta C., Kieninger E., Jung A., Moeller A., Geiser T., Regamey N., Alves M.P. Novel antiviral properties of azithromycin in cystic fibrosis airway epithelial cells. Eur. Respir. J. 2015;45:428–439. doi: 10.1183/09031936.00102014. [DOI] [PubMed] [Google Scholar]
- Touret F., Driouich J.S., Cochin M., Petit P.R., Gilles M., Barthelemy K., Moureau G., Mahon F.X., Malvy D., Solas C., de Lamballerie X., Nougairede A. Preclinical evaluation of Imatinib does not support its use as an antiviral drug against SARS-CoV-2. Antivir. Res. 2021;193:105137. doi: 10.1016/j.antiviral.2021.105137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touret F., Gilles M., Barral K., Nougairede A., van Helden J., Decroly E., de Lamballerie X., Coutard B. In vitro screening of a FDA approved chemical library reveals potential inhibitors of SARS-CoV-2 replication. Sci. Rep. 2020;10:13093. doi: 10.1038/s41598-020-70143-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tricou V., Minh N.N., Van T.P., Lee S.J., Farrar J., Wills B., Tran H.T., Simmons C.P. A randomized controlled trial of chloroquine for the treatment of dengue in Vietnamese adults. PLoS Neglected Trop. Dis. 2010;4:e785. doi: 10.1371/journal.pntd.0000785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukimura T., Shiga T., Saito K., Ogawa Y., Sakuraba H., Togawa T. Does administration of hydroxychloroquine/amiodarone accelerate accumulation of globotriaosylceramide and globotriaosylsphingosine in Fabry mice? Mol. Genet. Metabol. Rep. 2021;28:100773. doi: 10.1016/j.ymgmr.2021.100773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tummino T.A., Rezelj V.V., Fischer B., Fischer A., O'Meara M.J., Monel B., Vallet T., White K.M., Zhang Z., Alon A., Schadt H., O'Donnell H.R., Lyu J., Rosales R., McGovern B.L., Rathnasinghe R., Jangra S., Schotsaert M., Galarneau J.-R., Krogan N.J., Urban L., Shokat K.M., Kruse A.C., García-Sastre A., Schwartz O., Moretti F., Vignuzzi M., Pognan F., Shoichet B.K. Drug-induced phospholipidosis confounds drug repurposing for SARS-CoV-2. Science. 2021 doi: 10.1126/science.abi4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bambeke F., Montenez J.P., Piret J., Tulkens P.M., Courtoy P.J., Mingeot-Leclercq M.P. Interaction of the macrolide azithromycin with phospholipids. I. Inhibition of lysosomal phospholipase A1 activity. Eur. J. Pharmacol. 1996;314:203–214. doi: 10.1016/s0014-2999(96)00552-3. [DOI] [PubMed] [Google Scholar]
- Vigerust D.J., McCullers J.A. Chloroquine is effective against influenza A virus in vitro but not in vivo. Influenza Other Respir Viruses. 2007;1:189–192. doi: 10.1111/j.1750-2659.2007.00027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston S., Coleman C.M., Haupt R., Logue J., Matthews K., Li Y., Reyes H.M., Weiss S.R., Frieman M.B. Broad anti-coronavirus activity of food and drug administration-approved drugs against SARS-CoV-2 in vitro and SARS-CoV in vivo. J. Virol. 2020;94 doi: 10.1128/JVI.01218-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2021. COVID-19 Weekly Epidemiological Update. [Google Scholar]
- Wu Y.H., Tseng C.K., Lin C.K., Wei C.K., Lee J.C., Young K.C. ICR suckling mouse model of Zika virus infection for disease modeling and drug validation. PLoS Neglected Trop. Dis. 2018;12 doi: 10.1371/journal.pntd.0006848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Zou Z., Sun Y., Li X., Xu K.F., Wei Y., Jin N., Jiang C. Anti-malaria drug chloroquine is highly effective in treating avian influenza A H5N1 virus infection in an animal model. Cell Res. 2013;23:300–302. doi: 10.1038/cr.2012.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng S., Meng X., Huang Q., Lei N., Zeng L., Jiang X., Guo X. Spiramycin and azithromycin, safe for administration to children, exert antiviral activity against enterovirus A71 in vitro and in vivo. Int. J. Antimicrob. Agents. 2019;53:362–369. doi: 10.1016/j.ijantimicag.2018.12.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.