Abstract
In the nucleus, transcription factors must contend with the presence of chromatin in order to gain access to their cognate regulatory sequences. As most nuclear DNA is assembled into nucleosomes, activators must either invade a stable, preassembled nucleosome or preempt the formation of nucleosomes on newly replicated DNA, which is transiently free of histones. We have investigated the mechanism by which heat shock factor (HSF) binds to target nucleosomal heat shock elements (HSEs), using as our model a dinucleosomal heat shock promoter (hsp82-ΔHSE1). We find that activated HSF cannot bind a stable, sequence-positioned nucleosome in G1-arrested cells. It can do so readily, however, following release from G1 arrest or after the imposition of either an early S- or late G2-phase arrest. Surprisingly, despite the S-phase requirement, HSF nucleosomal binding activity is restored in the absence of hsp82 replication. These results contrast with the prevailing paradigm for activator-nucleosome interactions and implicate a nonreplicative, S-phase-specific event as a prerequisite for HSF binding to nucleosomal sites in vivo.
In the eukaryotic cell, nuclear DNA is packaged into a compact structure known as chromatin, a complex of DNA and histone proteins. The packaging of DNA into chromatin not only serves to confine the genome within the boundaries of the nucleus but also regulates the transcriptional activation of genes. The presence of nucleosomes, the individual subunits of chromatin, can inhibit the binding of sequence-specific activators to upstream elements, as well as impede access of the general transcription machinery to the core promoter (reviewed in reference 35). As a consequence, nucleosomes inhibit transcription, both in vitro (34, 43, 72) and in vivo (27, 29, 61). Thus, an important function of activators is to overcome nucleosomal repression, either by blocking nucleosome formation over promoters, thereby presetting genes for activation, or by remodeling preassembled nucleosomes as the initial step in transcriptional activation.
The mechanisms by which activators recognize and bind their cognate sites within chromatin are varied. Certain factors, like NF-1 and GCN4, can access their sites only within nuclease-hypersensitive, nucleosome-rearranged regions (5, 15, 66, 75). Others, such as PHO4, bind to accessible regions but, once DNA bound, actively remodel neighboring nucleosomes (1, 19). Still others are capable of invading a stable nucleosome and binding to target sites wrapped around the nucleosome core. Glucocorticoid receptor, for example, invades a sequence-positioned nucleosome within the mouse mammary tumor virus promoter following exposure to hormone (5). Receptor binding to its cognate sequence leads to a dramatic reconfiguring of the underlying nucleosome (66) and occurs in the absence of replication (55). The ability of activators to invade a preassembled nucleosome may require the involvement of ATP-dependent chromatin remodeling complexes and/or histone acetyltransferases (reviewed in references 36, 62, 71, and 73). Alternatively, gene-specific activators may access nucleosomal binding sites by preempting the formation of nucleosomes following DNA replication (63). This could occur either by outcompeting histones for binding to newly replicated DNA or by aborting the maturation of a nascent nucleosome. A requirement for DNA replication in gene activation in vitro has been described (7, 33); however, no evidence exists for replication-dependent binding of an activator to a defined nucleosomal site in vivo.
In this study, we used the Saccharomyces cerevisiae HSP82 heat shock gene to investigate transcription factor binding to nucleosomal DNA in vivo. Previous work has shown that the wild-type (WT) promoter is maintained in a DNase I-hypersensitive, nucleosome-disrupted state (24, 64). Within this accessible region, heat shock transcription factor (HSF) constitutively binds to a high-affinity heat shock element, HSE1, and inducibly and cooperatively to two low-affinity sites, HSE2 and HSE3 (18, 23, 25). The TATA element is also constitutively occupied (25, 56). In a promoter mutant lacking HSE1, termed hsp82-ΔHSE1, all detectable sequence-specific interactions are abolished (summarized in Fig. 1) and are replaced by two stable, sequence-positioned nucleosomes. One nucleosome (termed Nuc −2) is centered over the mutated heat shock upstream activation sequence (UAS; comprising HSE2 and HSE3), while the other, Nuc −1, is centered over the TATA initiation site (24). This striking phenotype suggests that the open chromatin structure and transcriptional competence of the WT promoter are maintained by HSF. Consistent with this notion, the protein can act as a high-copy suppressor of hsp82-ΔHSE1. Its ability to activate transcription is obviated by deletion of HSE2 and HSE3, indicating that suppression is mediated through these elements (24). In the present study, we address the mechanism by which HSF binds nucleosomal DNA. In theory, HSF could access its sites by directly invading the preassembled nucleosome (disruption mechanism), by aborting the maturation of a nascent nucleosome (preempt mechanism), or by outcompeting histones at the replication fork (exclusion mechanism). We show here that HSF is incapable of binding nucleosomal HSEs in G1-arrested cells. However, the activated factor can gain access to such sites following release of cells from the G1 block, or after imposition of an early S-phase arrest, and does so in the absence of DNA replication. It also readily binds nucleosomal DNA in cells prearrested in G2/M. This represents, to our knowledge, the first example of cell cycle-specific binding by an activator to a defined nucleosomal site.
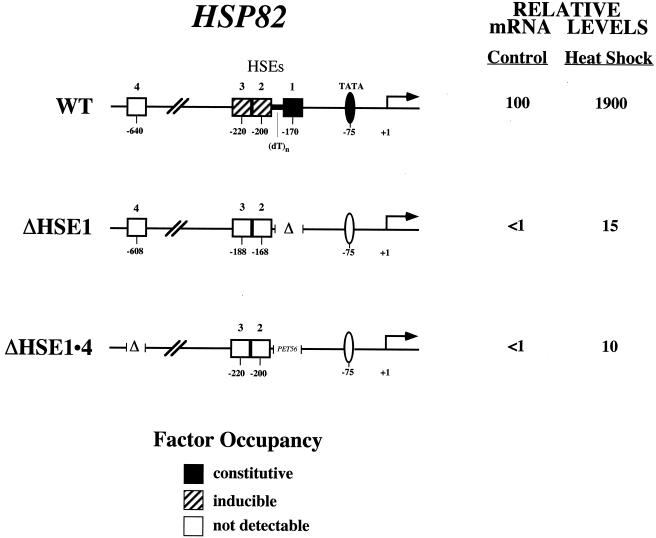
FIG. 1.
Principal HSP82 regulatory elements, their state of occupancy in vivo, and associated transcript levels. The ΔHSE1 mutation consists of a 32-bp deletion of HSE1 and flanking sequence (24). The ΔHSE1 · 4 promoter bears two mutations: a 32-bp substitution of HSE1 for PET56 coding sequence, and a 72-bp deletion of HSE4. +1, principal transcription start site. mRNA expression levels represent the means of at least three independent experiments. Occupancy data are summarized from work described here and elsewhere (18, 23–25, 46, 56).
MATERIALS AND METHODS
Yeast strains and plasmids.
Strain CBY106 was constructed by two-step gene transplacement of the hsp82Δ::CYH2s disruption strain SLY102 (40) (Table 1). The transforming DNA fragment, bearing a 32-bp substitution of HSE1 (−187 to −155) with PET56 coding sequence (24) and a 72-bp deletion of HSE4 (−673 to −612) was constructed by PCR overlap extension and cloned into the URA3 integrating vector, pRS306. Targeting of the hsp82 locus was achieved by linearizing the resultant construct (pCB107) at the unique AvaI site (−914) within HSP82. Gene transplacement was verified by genomic PCR. The BAR1 gene was disrupted by a one-step gene transplacement using the 5.5-kb bar1Δ::LEU2 fragment excised from pZV77 (gift from D. Hagen and G. Sprague). Plasmid pGAL1-HSF (URA3), bearing the PvuII-XhoI fragment of yeast HSF1 under regulation of the GAL1 promoter (gift from P. Sorger), was transformed into strains BSY202 and CBY106, creating CBY101 and CBY107, respectively. Site-directed integration of a TRP1-Kanr reporter (flanked by directly repeated R sites) into chromosome V (6.6 kb upstream of KHS1 and 2.8 kb downstream of ISC10 [45]) was achieved by transforming BSY202 with StuI-linearized p252-KT-DIR (gift from M. K. Raghuraman and W. Fangman). Note that this segment of DNA contains no sequence that can function as an origin of replication in S. cerevisiae (21). An episomal GAL1-regulated R-recombinase gene (YCpG-RURA) from Zygosaccharomyces rouxii was subsequently introduced into this strain, creating CBY120.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| SLY101 | MATα ade−can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | 40 |
| SLY102 | SLY101; cyh2rhsp82Δ::CYH2s | 40 |
| KEY105 | SLY102; hsp82-ΔHSE1 · | 24 |
| BSY202 | SLY102; MATa bar1Δ::LEU2 hsp82-ΔHSE1 | Bruce Stentz |
| CBY100 | BSY202; pRS316 | This study |
| CBY101 | BSY202; pGAL1-HSF | This study |
| CBY106 | SLY102; MATa bar1Δ::LEU2 hsp82-ΔHSE1 · 4 | This study |
| CBY107 | CBY106; pGAL1-HSF | This study |
| CBY108 | CBY106; pRS316 | This study |
| CBY113 | SLY101; MATa bar1Δ::LEU2 | This study |
| CBY114 | CBY113; pGAL1-HSF | This study |
| CBY120 | BSY202; ISC10::TRP1-Kanr; YCpG-RURA | This study |
Cultivation and cell cycle arrest conditions.
Yeast strains were grown at 30°C in synthetic complete medium lacking uracil and containing 2% raffinose to ∼107 cells/ml (determined by cell counting). To effect a G1 arrest, α-factor was added to a final concentration of 500 ng/ml. Strains used in this study were supersensitive to α-factor since they were deleted for BAR1, which encodes a secreted protease that degrades the pheromone. Cultures were incubated in the presence of α-factor for at least 2.5 h at 30°C and then examined by microscopy. When the fraction of shmoos exceeded 95%, an aliquot was removed for RNA analysis (noninduced) directly or following a 15-min, 39°C heat shock (induced). Galactose was then added to a final concentration of 1%, and samples were similarly collected at various time points thereafter. Heat shocks were achieved by adding an equivalent volume of prewarmed (51°C) medium to the culture, resulting in an instantaneous 30→39°C upshift. To effect an arrest in early S phase, hydroxyurea (HU) was added to early log cultures at a final concentration of 200 mM. Cultures were then incubated with shaking for 2.5 to 3 h at 30°C. When 90% of cells contained large buds, cultures were subjected to a galactose shift as described above. To effect an arrest in G2/M, nocodazole (1 mg/ml dissolved in dimethyl sulfoxide [DMSO]) was added to a final concentration of 10 μg/ml, and cells were incubated for 3.5 to 4.0 h prior to addition of galactose, at which time >95% of the cells contained large buds. In all cases, cell density remained constant for the duration of the arrest. Occasionally, asynchronous controls were diluted to maintain a cell density of 107 to 2 × 107 cells/ml. There was no difference in heat shock-induced transcript levels between asynchronous cultures that were diluted and those that were not. To ascertain cellular DNA content, culture aliquots were removed, fixed in 70% ethanol, stained with propidium iodide, and subjected to FACS (fluorescence-activated cell sorting) analysis using a Becton Dickinson (San Jose, Calif.) FACS Calibur. HU arrest was confirmed by a broadening of the 1N peak and a suppression of the 2N peak.
Blot hybridization.
For Northern blots total RNA was isolated, electrophoresed, blotted, and hybridized as described elsewhere (18). HSP82 and HSP26 mRNA levels, internally normalized with respect to ACT1, were quantitated using a PhosphorImager. For Southern blots, genomic DNA was isolated by glass bead lysis (30), digested with EcoO109I, and deproteinized; then 1.5 μg of DNA was applied to a 1% agarose–1× Tris-borate-EDTA gel. Following alkaline transfer and UV cross-linking to GeneScreen, hybridization was conducted at 65°C with the following gene-specific probes: a 250-nucleotide (nt) riboprobe (p5L) spanning −1300 to −1049 of HSP82 (24); a 1.4-kb PCR fragment spanning Kanr; and a 0.4-kb PCR fragment that hybridizes to a region immediately downstream of KEX2.
MNase genomic footprinting.
Cells were cultivated at 30°C to mid-log phase in 500 ml of YPD, metabolically poisoned through addition of sodium azide to 20 mM, harvested, and washed in 50 ml of ice-cold TA buffer (20 mM Tris-HCl [pH 8.0], 20 mM sodium azide). They were then resuspended in 5 ml of spheroplast buffer (1.4 M sorbitol, 40 mM HEPES [pH 7.5], 0.5 mM MgCl2, 0.5% 2-mercaptoethanol, 20 mM NaN3, 1 mM phenylmethylsulfonyl [PMSF], 2 mM N-ethylmaleimide, 2 mM benzamidine). Lyticase (2.5 mg/g of cells) was added, and spheroplasting proceeded for 30 to 60 min at 30°C with mild agitation. Spheroplasts were harvested and resuspended in 10 ml of ice-cold 18% Ficoll buffer (18% Ficoll, 20 mM PIPES [pH 6.5], 0.5 mM MgCl2, 1 mM PMSF, 2 mM N-ethylmaleimide, 2 mM benzamidine). Nuclei were isolated by layering the spheroplast suspension on top of 15 ml of ice-cold glycerol-Ficoll buffer (7% Ficoll, 20% glycerol, 20 mM PIPES [pH 6.5], 0.5 mM MgCl2, 1 mM PMSF, 2 mM N-ethylmaleimide, 2 mM benzamidine) and centrifuged at 12,500 rpm in a Sorvall HB-4 swinging bucket rotor for 30 min. Nuclear pellets were resuspended in 1 ml of micrococcal nuclease (MNase) digestion buffer (40 mM HEPES [pH 7.5], 1 mM MgCl2, 1 mM CaCl2, 1 mM PMSF, 2 mM N-ethylmaleimide, 2 mM benzamidine) and divided into five equal aliquots. To each aliquot was added 15, 30, 60, 120, or 240 U of MNase (Pharmacia Biotech product no. 27-0584-01). Nuclei were digested for 10 min at room temperature; digestions were terminated by the addition of an equal volume of stop buffer (2% sodium dodecyl sulfate [SDS], 1 M NaCl, 50 mM EDTA, 50 mM Tris [pH 7.5]). Digestion of naked DNA was accomplished by reacting 2 μg of DNA with 6 × 10−4 U of MNase in 500 μl of 1 mM CaCl2–1 mM MgCl2–40 mM HEPES (pH 7.5). This digestion was conducted at room temperature for 5 min and terminated by the addition of EDTA to 10 mM. DNA samples were purified as described elsewhere (17). HSP82-specific cleavages were detected by linear PCR using both upper- and lower-strand complementary primers (+26→−11 and −342→−315, respectively). Bands were detected on a PhosphorImager, and their intensities were quantitated (in terms of peak amplitudes) on a Macintosh G3 using ImageQuant 1.1.
DMS in vivo footprinting.
Cells were grown in raffinose Ura-deficient medium to a density of ∼107 cells/ml and subjected to cell cycle arrest and heat shock as described above. Samples were treated with a final concentration of either 0.4 or 0.8% dimethyl sulfate (DMS) for 2 min, with rapid agitation at 39°C. Reactions were terminated, DNA was isolated, and linear PCR was conducted using the upper-strand complementary primer +26→−11 as previously described (56).
Restriction enzyme accessibility assay.
Cells were grown in raffinose complete medium to a density of 107 cells/ml and subjected to cell cycle arrest as described above. Nuclei were isolated and digested at 30°C for 1 h using 12, 40, or 100 U of HindIII per μg of DNA. The amount of DNA in each sample was measured spectrophotometrically prior to addition of enzyme. A 5-μl aliquot of nuclear suspension was added to 495 μl of 1 N NaOH, and DNA concentration was estimated by using the formula (A260 − 1.6 × A320)/27 (17). Purified genomic DNA was digested to completion with MspI and subjected to linear PCR using the upper-strand complementary primer +26→−11.
Protein isolation and Western analysis.
Immunoblot analysis was performed using total protein from whole-cell extracts, obtained by breaking cells with glass beads in the presence of 5% trichloroacetic acid. Proteins were pelleted and resuspended in 6 M urea buffer (14), then electrophoresed on an SDS–10% polyacrylamide gel, blotted to nitrocellulose, and sequentially probed with polyclonal rabbit antisera raised against either glutathione S-transferase–HSF (17) or Leu4p (gift from Gunter Kohlhaw, Purdue University). Horseradish peroxidase-conjugated goat anti-rabbit antibody (Amersham) was then added, and the secondary antibody was detected either by autoradiography using enhanced chemiluminescence (ECL; Amersham) or by PhosphorImager using ECL Plus.
RESULTS
The hsp82-ΔHSE1 promoter assembles into a dinucleosome in vivo.
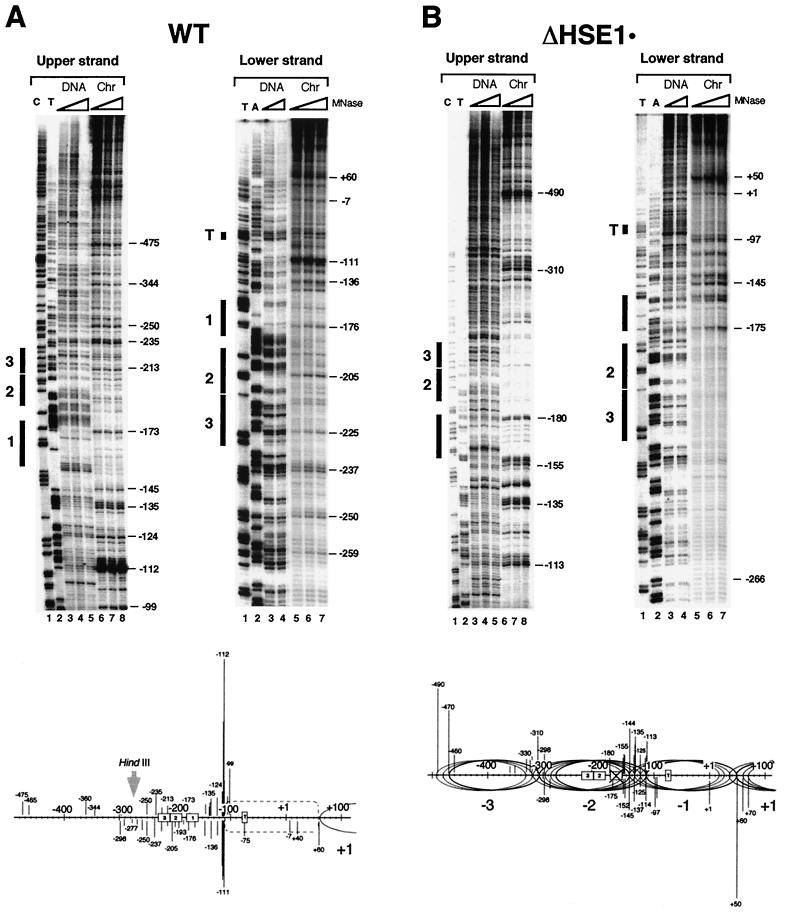
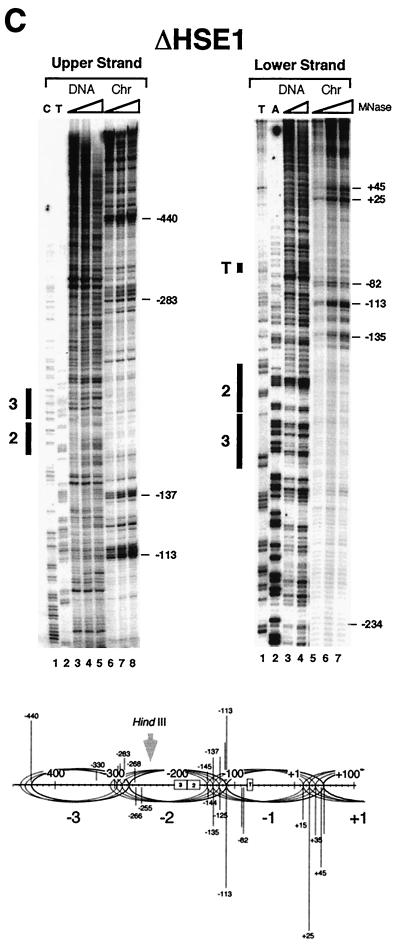
Our previous work has shown that the nucleosome centered over the core promoter of hsp82-ΔHSE1, termed Nuc −1, is rotationally phased (24). However, its translational position and that of the adjacent, UAS-containing nucleosome (Nuc −2) are less well characterized. Such information is important, since accessibility of a binding site decreases as it is moved from the edge to the center (dyad) of a nucleosome core (58). Therefore, the most rigorous test of HSF's nucleosome binding activity will be presented if the target HSEs are proximate to the dyad. To determine the translational setting of these nucleosomes, we digested purified nuclei with MNase and mapped cleavage sites at nucleotide resolution using linear PCR (18). As illustrated in the genomic footprints of Fig. 2A, the heat shock UAS of the WT allele, defined as sequence lying between −235 and −150 (18), is accessible to MNase. This accessibility is evident on both strands and culminates in a series of intense cleavages centered at positions −112 and −111. The cutting pattern is specific to chromatin and argues against the presence of a canonical nucleosome over the UAS (see also Fig. 10A and reference 24). Moreover, the core promoter is assembled into an MNase-resistant structure spanning ∼170 bp (−111 to +60; see schematic), whose precise nature, given its hypersensitivity to DNase I (25, 64), is unclear. A dramatically different cleavage profile is seen with hsp82-ΔHSE1 · , an allele of HSP82 bearing a 32-bp substitution of the UAS (24). MNase cleavage is suppressed over much of the upstream promoter, restricted to two foci centered at −310 and −135 (Fig. 2B). These chromatin-specific cleavages are consistent with the presence of linkers flanking a novel, translationally positioned nucleosome (Nuc −2; see schematic). Notably, the cut sites are spaced at intervals of ∼10 nt, suggesting that Nuc −2 adopts several, equally preferred phasing frames. Alternatively, the 10-nt periodicity of cleavage may arise from the exonuclease activity of MNase (12). Assuming multiple phasing frames, the inferred dyad axes (−238, −227, −217, and −207) of Nuc −2 overlap HSE2 and HSE3 (which span −229 to −188 [18]). The core promoter-associated nucleosome (Nuc −1) may also adopt more than one translational setting, as indicated. Analysis of the 32-bp deletion allele (ΔHSE1 [Fig. 2C]) suggests the presence of two similarly positioned nucleosomes, each occupying multiple phasing frames. As above, the dyad axes of the four principal phasing frames of Nuc −2 overlap HSE2 and HSE3, which in this allele span −197 to −156. Thus, in both substitution and deletion mutants, the dyad axis of Nuc −2 overlaps or is in close proximity to HSE2 and HSE3.
FIG. 2.
MNase genomic footprinting reveals that the promoter region of hsp82 alleles lacking HSE1 is assembled into a novel dinucleosome. Illustrated are strand-specific genomic footprints for HSP82+, hsp82-ΔHSE1 · (a 32-bp substitution mutant), and hsp82-ΔHSE1 (a 32-bp deletion mutant) (A to C, Chr lanes). Cells were cultivated at 30°C, nuclei were purified and digested with MNase, and genomic DNA was isolated. For the naked DNA controls (DNA), genomic DNA was isolated from each strain and digested with MNase. Each digestion series represents a set of twofold serial dilutions. Cut sites were mapped by linear PCR using both upper-strand- and lower-strand-specific primers (+26→−11 and −342→−315, respectively). Cleavage profiles were visualized using a PhosphorImager, and intensities of all major cut sites were quantitated. They are schematically illustrated below each set of footprints, with line lengths proportionate to cleavage intensities. Not all data summarized in the schematics are from the portions of the gels shown. Ovals, inferred translational positions of nucleosomes; dashed rectangle, an MNase-resistant, DNase I-hypersensitive structure. HindIII designations pertain to the nuclear accessibility assay of Fig. 10. The HindIII site is at −274 in the WT allele and at −242 in the deletion allele.
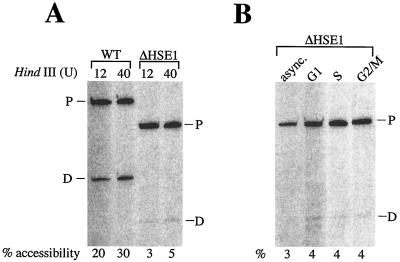
FIG. 10.
The HindIII site is virtually inaccessible within the hsp82-ΔHSE1 promoter in both asynchronous and cell cycle-arrested cells. (A) Asynchronous SLY101 (WT) and BSY202 (ΔHSE1) cells were grown at 30°C in complete synthetic medium supplemented with 2% raffinose. Nuclei, isolated from spheroplasts and quantitated as previously described (17), were digested with either 12 or 40 U of HindIII per μg of DNA at 30°C for 60 min. Genomic DNA was purified, digested to completion with MspI, and subjected to linear PCR using the +26→−11 primer, and the amplified products were electrophoresed on an 8% sequencing gel. Bands were detected and quantitated using a PhosphorImager. Percent accessibility was calculated by dividing the total signal present in the daughter (D) fragment (resulting from HindIII cleavage) by that present in the sum of the parental (P) fragment plus daughter fragment. (B) As in panel A except that BSY202 cells were either maintained in log growth (async.) or arrested at the indicated phases in the cell cycle prior to isolation of nuclei. Samples were digested with 40 U of HindIII per μg of DNA and processed as above. See Fig. 2 for location of HindIII site with respect to mapped chromatin structure.
The null transcription phenotype of ΔHSE1 can be suppressed by overexpression of HSF.
To investigate whether overexpressed HSF could rescue the null transcription phenotype of hsp82-ΔHSE1, we transformed a strain bearing this allele with a galactose-inducible HSF1 gene (creating strain CBY101 [Table 1]). Consistent with previous observations (24), addition of galactose results in a striking increase (25- to 30-fold) in heat-shocked HSP82 transcript levels, paralleling a similar increase in intracellular HSF levels (data not shown; see below). Only cells harboring the GAL1-HSF1 plasmid experienced this activation, indicating that HSF—and not the product of some other galactose-induced gene—was responsible. These results are consistent with the possibility that HSF binds the nucleosomal HSEs. Nonetheless, an extended, degenerate HSE centered at −640 (HSE4 [Fig. 1]) might facilitate HSF-mediated suppression of the ΔHSE1 null phenotype since (i) overproduction of HSF results in the occupancy of HSE4, which resides within a DNase I-hypersensitive region (24); and (ii) HSF binds its target HSEs cooperatively, both in vitro and in vivo (2, 9, 18). Thus, HSF binding at HSE2 and HSE3 might be cooperative with its occupancy at HSE4. To address this possibility, we created an allele bearing both a substitution of HSE1 and a deletion of HSE4 (hsp82-ΔHSE1 · 4 [Fig. 1]). As indicated by MNase nucleotide resolution mapping (data not shown), the promoter of this allele is packaged into a chromatin structure very similar to that inferred for hsp82-ΔHSE1 · (Fig. 2B). A strain containing this allele was transformed with GAL1-HSF1, creating CBY107. This strain was then subjected to a galactose shift. The heat-shocked hsp82-ΔHSE1 · 4 gene, similar to hsp82-ΔHSE1, is effectively activated by HSF, with transcript levels significantly increasing (5- to 10-fold) over a 5-h time course (data not shown; see below). We conclude that overexpressed HSF can restore function exclusively through its interaction with the nucleosomal HSEs.
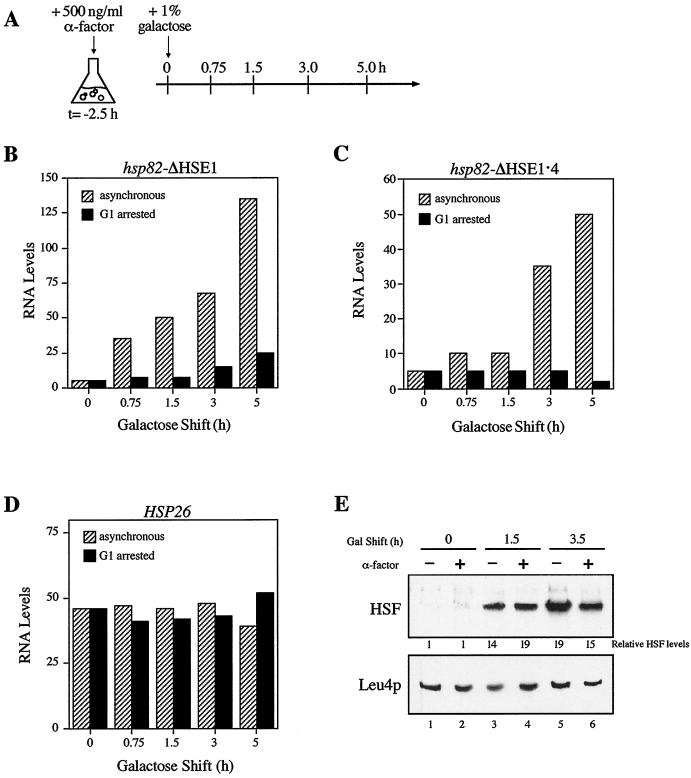
Prior arrest in G1 blocks HSF-mediated suppression of the ΔHSE1 null phenotype.
The ability of HSF to bind HSE2 and HSE3 allowed us to investigate the mechanism by which the protein accesses nucleosomal sites. If HSF invades Nuc −2 and directly binds its target sequences, then its binding to nucleosomal DNA would be cell cycle independent. Alternatively, HSF might preempt the assembly of Nuc −2, either by binding DNA immediately following replication, when histone octamers are transiently displaced from the newly synthesized DNA (13, 32, 63; but see reference 8), or by binding to the nascent nucleosome shortly thereafter, aborting its maturation. Either way, such binding would be cell cycle dependent. More complex mechanisms are also possible. To gain insight into the HSF–Nuc −2 interaction, CBY101 cells were arrested in G1 with the mating pheromone, α-factor. Following a 2.5-h incubation in the presence of α-factor, >95% of CBY101 cells were arrested in G1, as assayed by microscopy. At this point, galactose was added directly to the medium to a final concentration of 1%, and aliquots were removed at various times and heat shocked (protocol summarized in Fig. 3A). As shown in Fig. 3B, HSF-mediated activation is virtually eliminated in G1-arrested cells, even though HSF overexpression is unaffected by a prior G1 arrest (Fig. 3E). Nonetheless, a modest increase in hsp82 transcription is detected. Such activation could stem from HSF interacting with HSE2 and -3 at reduced efficiency in G1-arrested cells, perhaps by binding to Nuc −2 in certain phasing frames but not in others. Alternatively, it could be a consequence of HSF binding to the accessible, far-upstream HSE4. To help distinguish between these possibilities, we subjected the HSE4-deleted strain CBY107 to a similar protocol. As can be seen in Fig. 3C, induced HSP82 transcript levels do not increase in G1-arrested hsp82-ΔHSE1 · 4 cells following addition of galactose, whereas substantial activation of the gene takes place in the parallel, asynchronous culture. Therefore, the slight increase in hsp82-ΔHSE1 transcription following long periods of galactose induction in G1-arrested cells is likely due to HSF binding to HSE4.
FIG. 3.
Transactivation of hsp82-ΔHSE1 is blocked during G1 arrest. (A) Experimental strategy. Cells pregrown to early log phase in the presence of 2% raffinose were arrested in G1 by addition of α-factor. Following a 2.5-h incubation at 30°C, >95% of the cells were unbudded shmoos, indicative of G1 arrest. At that point (t = 0 h), galactose was added, and at the indicated times aliquots were heat-shocked and RNA was isolated. (B) HSP82 transcript levels in asynchronous and G1-arrested CBY101 cells subjected to galactose shift for the times indicated. (C) Same as panel B except that strain CBY107 was used. (D) HSP26 transcript levels in CBY101 cells, assayed from the RNA samples of panel B. (E) Immunoblot analysis. Total protein was isolated from asynchronous (−) and G1-arrested (+) CBY101 cells, electrophoretically separated on an SDS–10% polyacrylamide gel, blotted to nitrocellulose, and sequentially probed with anti-HSF and anti-Leu4p antibodies. HSF levels, quantitated on a Storm PhosphorImager, are normalized with respect to Leu4p. Data depicted in panels B to D are from representative experiments; the trends shown were found to be highly reproducible (five independent experiments for panel B, three for panel C, and two for panel D).
One explanation for the impaired activation of hsp82 is that HSF is functionally compromised during G1. However, as revealed by rehybridization of the Northern blot in Fig. 3B, transcriptional induction of a second HSF-regulated gene, HSP26, is unaffected by pretreatment with α-factor: its transcript levels increase 40- to 50-fold during a 15-min heat shock irrespective of the extent of HSF overexpression, indistinguishable from the parallel, asynchronous culture (Fig. 3D). A potentially significant difference between hsp82-ΔHSE1 and HSP26 is that the HSEs of the latter reside within accessible, DNase I-hypersensitive sites (10; D. Pederson, personal communication). Thus it is possible that in G1-arrested cells, HSF is incapable of activating a nucleosomal promoter while exhibiting full activity at a second, accessible one. Indeed, heat shock induction of HSP82+, like that of HSP26, is unaffected by a prior arrest in G1 (data not shown).
Prior G1 arrest blocks HSF binding to Nuc −2.
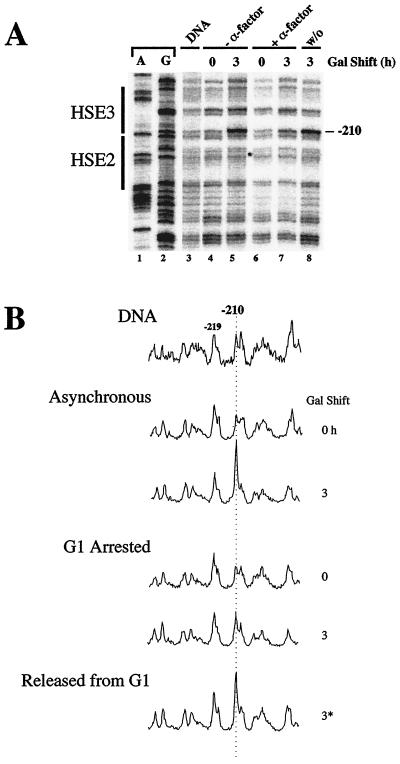
While HSF is unable to activate transcription of the hsp82-ΔHSE1 allele during G1, it is not clear whether this block is at the level of HSF binding or at a subsequent step in the transcription pathway. To help clarify this, we used DMS in vivo footprinting. Previous work has demonstrated that HSF inducibly binds HSE2 and HSE3 within the WT promoter, with the signature of its interaction being hyperreactivity of guanine −210 (18, 23). We therefore subjected asynchronous and G1-arrested CBY101 cultures to a 3-h galactose shift and then heat shocked cells for 15 min at 39°C. During the final 2 min, protein-DNA interactions were probed with DMS; genomic DNA was then isolated to permit mapping of methylated sites. Following incubation in the presence of galactose, HSF binding to the nucleosomal HSEs is readily detectable in asynchronous cells (Fig. 4). Similar but less robust binding is evident at the activated hsp82-ΔHSE1 · 4 promoter (see Fig. 8), paralleling its lower expression level. There is no detectable −210 G hyperreactivity in non-heat-shocked cells (data not shown; see also Fig. 8B), indicating that virtually all binding to HSE2 and HSE3 occurs upon heat shock, as is the case for the WT promoter. In contrast to the asynchronous cultures, the G1-arrested culture is largely refractory to activated HSF binding (Fig. 4A, lane 7 versus lane 5; Fig. 4B, 3-h scans, asynchronous versus G1 arrested), despite attaining comparable HSF levels (Fig. 3E). The slight increase in modification of −210 G during the 3-h galactose shift is most likely due to cells escaping from G1 arrest and/or a low level of nucleosomal binding facilitated by cooperative interactions with HSF bound to the accessible HSE4 site. We conclude that the G1-induced block in HSF activity is at the level of HSF binding to nucleosomal DNA.
FIG. 4.
In vivo DMS footprinting analysis of the heat shock UAS of hsp82-ΔHSE1 in asynchronous and G1-arrested CBY101 cells. (A) Early-log-phase cultures grown in 2% raffinose were arrested in G1 (+α-factor) or left untreated (−α-factor). Galactose was then added to a final concentration of 1% to induce HSF overexpression. Following a 2-h incubation, the arrested culture was washed; one half was resuspended into medium containing pheromone (+α-factor), the remaining half was resuspended in medium alone (washout [w/o]), and the incubation continued for an additional hour. Aliquots were removed, heat shocked, and reacted with DMS. Genomic DNA was isolated and subjected to linear PCR using the +26→−11 primer, detecting the upper-strand methylation pattern. The −210 G, whose reactivity is diagnostic of bound HSF (18, 23), is indicated. DNA, deproteinized genomic DNA methylated in vitro and analyzed as above; A and G, dideoxy sequencing ladders. (B) Densitometric analysis of the data presented in panel A. Amplitudes are normalized to that of −219 G. 3*, 3-h galactose-shifted sample washed free of α-factor during the final hour.
FIG. 8.
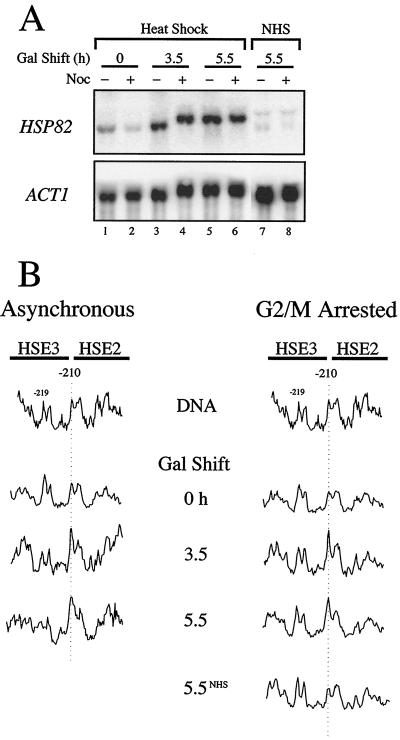
HSF activation of hsp82-ΔHSE1 · 4 is not affected by prior arrest in G2/M. (A) Northern analysis of asynchronous (−) and G2/M-arrested (+) CBY107 cells subjected to a 15-min heat shock (lanes 1 to 6) or maintained under non-heat shock conditions (NHS; lanes 7 and 8) following a 0-, 3.5-, or 5.5-h galactose shift. (B) DMS in vivo footprinting analysis. Cells from the heat-shocked cultures analyzed in panel A were reacted with DMS, and the DNA was purified. The presence of methylated guanines was detected as for Fig. 4; scans of the pertinent region of the sequencing gel are shown. 5.5NHS, nocodazole arrested, 5.5-h galactose shifted, non-heat-shocked culture. DNA, genomic DNA isolated from CBY107 and reacted with DMS and analyzed as above.
Washout of α-factor relieves the transcriptional block.
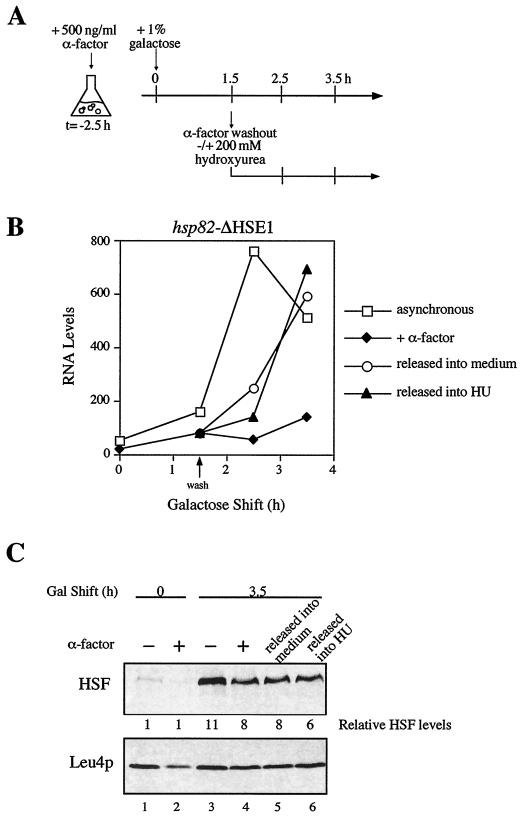
If replication (or some other S-phase-associated event) were required for HSF to access HSE2 and HSE3, then release from α-factor-induced-arrest should restore HSF's ability to transactivate the mutant alleles. To test this idea, cells bearing the HSE1 deletion were arrested in G1 with α-factor as described above. Following a 1.5-h incubation in the presence of galactose, the arrested culture was split into two aliquots. One was washed and resuspended in galactose-containing medium lacking α-factor, while the other was washed and resuspended in medium containing α-factor (experimental strategy summarized in Fig. 5A). Consistent with the above prediction, within 60 to 120 min of washout (i.e., as soon as budded cells were visible), induced transcript levels in the released culture were substantially increased (Fig. 5B). Paralleling this increase is significant binding of HSF to HSE2 and HSE3, as monitored by −210 hyperreactivity (Fig. 4A, lane 8; Fig. 4B, scan 3*). Importantly, the potentiating effect of releasing cells from G1 arrest cannot be explained by an alteration in intracellular HSF levels (Fig. 5C). These results thus indicate that HSF binding to and transactivation of hsp82-ΔHSE1 requires relief from the G1 block.
FIG. 5.
Release from G1 arrest alleviates the transcriptional block at hsp82-ΔHSE1. (A) Experimental strategy. CBY101 cells were arrested in G1 as for Fig. 3; an asynchronous culture (not indicated) was grown in parallel. At t = 0 h, galactose was added to each culture to induce HSF overexpression. At t = 1.5 h, the α-factor-arrested culture was split into three aliquots; one was washed and resuspended into medium lacking α-factor, one was washed and resuspended into medium containing α-factor, and the third was washed and resuspended into medium containing HU. The asynchronous culture was also harvested and resuspended into medium alone. Samples were removed at the indicated times and subjected to heat shock. (B) Plot of Northern data from the experiment depicted in panel A. (C) Immunoblot analysis. Total protein was isolated from aliquots of CBY101 cells subjected to the indicated treatments and harvested at the indicated times and then assayed for relative HSF concentration.
hsp82-ΔHSE1 is reactivated during early S phase.
To define more precisely at which point in the cell cycle HSF can activate hsp82-ΔHSE1, HU, an inhibitor of ribonucleotide reductase (16), was used to block cells in early S phase following their release from G1 arrest. The experimental strategy used is similar to that described above (Fig. 5A). Northern analysis shows that cells released into HU are induced to levels similar to those of either the asynchronous culture or the α-factor-arrested culture released into medium alone (Fig. 5B). Moreover, as above, HSF levels in such cells are virtually identical to those seen in the parallel, α-factor-arrested cells (Fig. 5C). Taken together, these results indicate that an early S-phase event is both necessary and sufficient for HSF to transactivate the nucleosomal promoter.
Prior arrest in early S phase potentiates the nucleosomal heat shock promoter for transactivation.
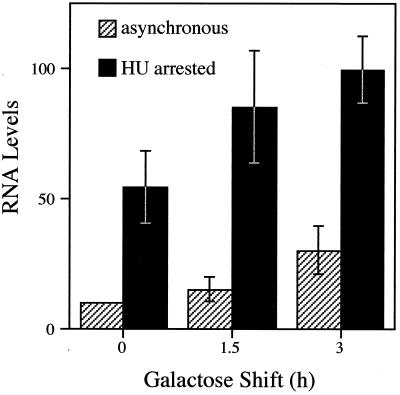
An implication of the above is that cells which transit from late G1 (Start) to a point in early S phase are permissive to HSF transactivation. To determine whether asynchronous cells treated with HU are similarly permissive, and to extend the analysis to the hsp82-ΔHSE1 · 4 allele, we arrested CBY107 cells with HU prior to inducing HSF overexpression. Inhibition of DNA replication was confirmed by FACS analysis of propidium iodide-stained cells (see Materials and Methods). Galactose was then added to induce HSF overexpression, and aliquots were removed and heat shocked at various times thereafter. Quite strikingly, hsp82 transcription is strongly activated in cells synchronized in S phase, even in the absence of galactose induction (Fig. 6). Following galactose shift, a further increase in HSP82 transcription is seen. These data suggest that activated HSF efficiently binds nucleosomal HSEs during early S phase. The possibility that the enhanced transcription seen in the treated cultures is due to HU induction of hsp82 beyond that elicited by heat shock is unlikely, since addition of HU to noninduced CBY107 cells does not discernibly increase hsp82 transcript levels (data not shown). Moreover, HSF protein levels are induced to similar levels in asynchronous and HU-arrested cells (data not shown; Fig. 5C). Taken together, these data implicate an event in early S phase as potentiating HSF activation of the nucleosomal promoter.
FIG. 6.
Prior arrest in S phase potentiates transcriptional activation of the hsp82-ΔHSE1 · 4 promoter. CBY107 cells pregrown in 2% raffinose-containing medium were arrested in S phase by incubation in the presence of 200 mM HU for 2.5 h. Galactose was added to the arrested culture and to a parallel asynchronous culture, and aliquots were removed at the indicated times for heat shock and subsequent RNA analysis. Depicted are mean values of three independent Northern analyses (normalized to the 0-h asynchronous sample; bars indicate standard error of the mean).
HSF efficiently activates hsp82-ΔHSE1 in G2/M-arrested cells.
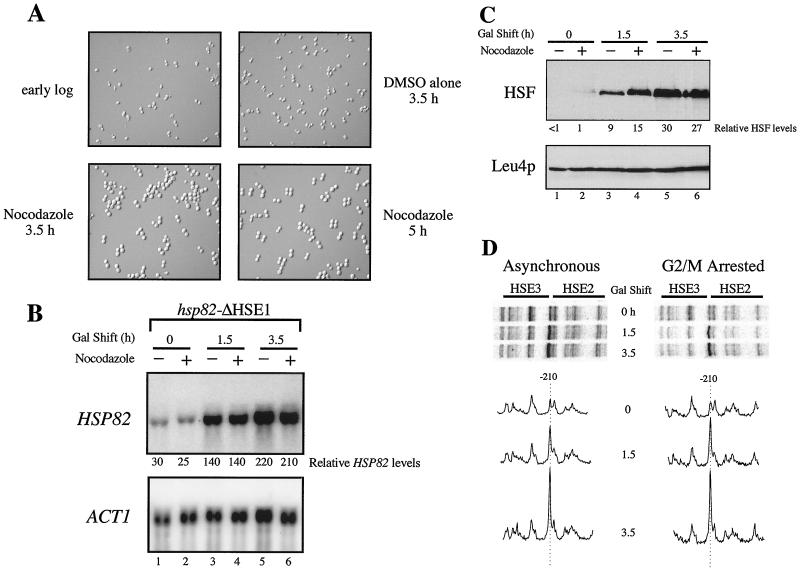
To extend the above findings, we tested whether HSF could transactivate hsp82-ΔHSE1 at a later point in the cell cycle. We used nocodazole, an inhibitor of microtubule assembly (53), which imposes a synchronous arrest in late G2/early M. Given the short G2 phase in yeast (41), activation of the nucleosomal promoter might be anticipated if nucleosome maturation is regulated and, as previously observed in metazoans (59, 70), proceeds over an extended period of time. On the other hand, as chromatin condensation is maximal during mitosis (26), HSF binding to a stable nucleosome might be severely impaired. Indeed, in vivo DMS footprints of sequence-specific activators are erased at a variety of mammalian promoters during metaphase (28, 44, 47).
To effect a G2/M arrest, nocodazole was added to logarithmically growing CBY101 cells, which were allowed to incubate in its presence for 3.5 h. Addition of drug, but not of vehicle alone, resulted in the arrest of >95% of cells, as assayed by microscopic examination of cell morphology (Fig. 7A). Following either a 1.5- or 3.5-h galactose shift, hsp82-ΔHSE1 activation in G2/M-arrested cells occurs at levels indistinguishable from the asynchronous control (Fig. 7B), correlating with the respective levels of HSF overexpression (Fig. 7C). Also unaffected is HSF binding: reactivity of −210 G in response to heat shock is equally robust in asynchronous cultures and those prearrested in G2/M (Fig. 7D). Identical results were seen with the HSE double mutant, hsp82-ΔHSE1 · 4, since neither transcriptional activation nor HSF binding was affected by prearresting cells in G2/M phase (Fig. 8). The latter data indicate that HSF is capable of binding the nucleosomal elements independently of HSE4. They also show that even when overexpressed, HSF binding to HSE2 and HSE3 and transactivation of hsp82 is detectable only upon heat shock (Fig. 8A, compare lanes 7 and 8 with lanes 3 to 6; Fig. 8B, compare scan 5.5NHS with scan 5.5). We conclude that the ability of HSF to bind nucleosomal DNA and transactivate either hsp82-ΔHSE1 or hsp82-ΔHSE1 · 4 is not restricted to early S phase but rather extends to at least late G2/early M.
FIG. 7.
HSF activation of hsp82-ΔHSE1 is not affected by prior arrest in G2/M. (A) Cell morphology of asynchronous and G2/M-arrested CBY101 cells. Cells grown to early log phase in 2% raffinose (upper left) were split into two aliquots. Nocodazole was added to one; vehicle alone (DMSO) was added to the other. Following incubation for 3.5 h at 30°C, >95% of cells in the nocodazole-treated culture had arrested with large buds (lower left), while the mock-treated culture contained ∼25% large-budded cells (upper right). Galactose was then added to a final concentration of 1%, and cells were allowed to incubate for a further 1.5 h (lower right). (B) Northern analysis of asynchronous (−) and G2/M-arrested (+) CBY101 cells subjected to heat shock following a 0-, 1.5-, or 3.5-h galactose shift. Transcript levels of HSP82 and the internal loading control, ACT1, were detected by blot hybridization. (C) Immunoblot analysis. Total protein was isolated from samples depicted in panel B, and sequential detection of HSF and Leu4p was performed as before. (D) DMS in vivo footprinting analysis. Cells from the cultures analyzed in panel B were reacted with DMS, DNA was purified, and the presence of methylated guanines was detected as described for Fig. 4.
Activation of hsp82-ΔHSE1 is uncoupled from its replication.
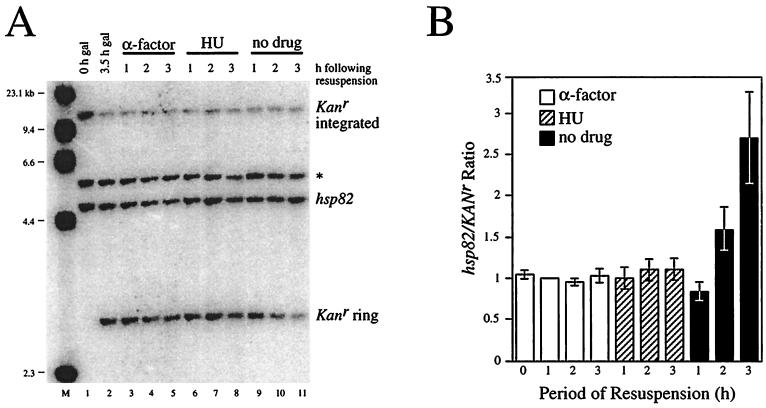
The foregoing experiments demonstrate that cells which transit from late G1 to a point in early S phase are permissive to HSF binding and transactivation. One mechanism that could account for this is DNA replication. In eukaryotes, the temporal order in which genes are replicated during each S phase is preset (20, 21); therefore, it is possible that hsp82 is replicated early, prior to the HU block. To investigate this possibility, we used quantitative Southern blotting. The DNA content of hsp82-ΔHSE1 was measured with respect to that of a chromosomal reporter gene (Kanr) excised during G1 arrest as a nonreplicating DNA ring (see Materials and Methods). A strain bearing both the hsp82-ΔHSE1 and Kanr alleles was arrested in G1 using α-factor as above, then a GAL1–R-recombinase gene fusion was induced through addition of galactose. Following a 3.5-h galactose shift, virtually complete excision of Kanr is achieved (Fig. 9A, lane 2 versus 1). Cells released from G1 arrest and resuspended into medium alone replicate hsp82, but not Kanr, as expected, with the hsp82/Kanr ratio nearly doubling within 120 min and tripling within 180 min (Fig. 9A, lanes 10 and 11; mean values from four independent experiments are graphically illustrated in Fig. 9B). In contrast, there is no increase in the hsp82/Kanr ratio in cells maintained in the presence of α-factor (Fig. 9A, lanes 3 to 5; Fig. 9B), as expected. Importantly, cells subjected to a subsequent HU arrest also show no increase in relative hsp82 DNA content (Fig. 9A, lanes 6 to 8) since hsp82/Kanr ratios are statistically indistinguishable from those obtained for the G1-arrested samples (Fig. 9B). This observation argues against DNA replication being the S-phase requirement for HSF binding and transactivation of hsp82-ΔHSE1.
FIG. 9.
The hsp82-ΔHSE1 locus is not detectably replicated between late G1 (Start) and early S phase, as revealed by Southern blot hybridization. (A) CBY120 cells (Table 1) growing in the presence of 2% raffinose were arrested in G1 with α-factor (lane 1). Galactose was then added for 3.5 h, and cells were harvested (lane 2). One third of the cell pellet was resuspended in 2% glucose medium containing α-factor, one third was resuspended in glucose medium containing HU, and one third was resuspended in glucose medium alone (no drug). At the indicated times, cells were harvested. DNA was then purified and digested to completion with EcoO109I, and 1.5 μg was separated on a 1% agarose gel, blotted to nylon, and simultaneously hybridized with probes specific for the hsp82 and Kanr loci. Radioactivity was detected using a Storm PhosphorImager; bands corresponding to the EcoO109I fragments of hsp82 (4.5 kb) and the Kanr excised ring (2.9 kb) were quantitated using ImageQuant 3.3. *, band which cross-hybridizes to the HSP82 riboprobe. M, molecular weight standard (HindIII-cut λ DNA). Note that equivalent amounts of DNA were loaded in all lanes; thus, the nonreplicating Kanr sequence is diluted in growing cells (lanes 10 and 11). (B) Bar graph summary of hsp82/Kanr quotients from four independent experiments. Presented are means ± standard error of the means, with values normalized to α-factor 1-h samples.
Integrity of Nuc −2 persists in G1-, early S-, and G2/M-arrested cells.
An alternative explanation for the cell cycle-specific binding of HSF is that alterations in chromatin structure occur at Nuc −2 which impair HSF binding to HSE2 and HSE3 at one point in the cell cycle (late G1) while enhancing it at another (early S phase). To test this idea, we measured accessibility of Nuc −2 in nuclei isolated from asynchronous hsp82-ΔHSE1 cells or cells arrested in late G1, early S, or G2/M. Following purification from spheroplasts, nuclei were digested with HindIII, whose recognition site (position −242 at hsp82-ΔHSE1 [Fig. 2C]) lies within the mapped location of Nuc −2. As expected, the HindIII site is largely inaccessible in nuclei isolated from noninduced, nonoverexpressing BSY202 cells (3 to 5% cleavage versus 20 to 30% cleavage in WT nuclei [Fig. 10A]). A similar, low level of accessibility is seen in ΔHSE1 nuclei isolated from noninduced cells arrested in late G1, early S, or G2/M (Fig. 10B). Therefore, the basic integrity of Nuc −2 is retained during these phases of the cell cycle and cannot, by itself, account for the cell cycle-specific phenomena described above. These results are consistent with those of the quantitative Southern analysis; both argue that enhanced HSF binding and transactivation of hsp82-ΔHSE1 occur independently of its replication and attendant chromatin disassembly.
DISCUSSION
HSF binds nucleosomal HSEs in a cell cycle-dependent fashion.
Using a combination of in vivo footprinting and RNA expression analysis, we have obtained compelling evidence that S. cerevisiae HSF binds nucleosomal HSEs, and transactivates the linked gene, in a cell cycle-dependent fashion. To demonstrate this, we have used the hsp82-ΔHSE1 gene, whose promoter is assembled into a stable dinucleosome, as our model system. The ability of HSF to access HSE2 and HSE3 is dependent on the protein being (i) overexpressed and (ii) activated by heat shock, which enhances its intrinsic DNA binding activity 10- to 25-fold (18, 23). These two requirements are consistent with previous in vitro binding competition assays which indicate that the overall affinity of the hsp82-ΔHSE1 promoter for HSF is <4% that of the WT promoter (18). They have also provided us with a means to assay HSF binding at precisely defined points during the cell cycle.
Using this approach, we have found that HSF binding to nucleosomal HSEs and concomitant transcriptional activation of hsp82-ΔHSE1 is greatly impaired in cells prearrested in late G1. However, factor binding and activation of hsp82-ΔHSE1 are restored following release of cells from the G1 block, even when such cells are subsequently arrested in early S phase. Indeed, prearresting cells in early S actually potentiates hsp82-ΔHSE1 for transcriptional activation, implying that HSF binds nucleosomal HSEs with increased efficiency in early S (see below). Cells prearrested in late G2/early M are neither suppressed nor enhanced for HSF binding and transactivation.
It is possible that the G1 block in HSF activity stems from an α-factor-specific effect. However, we have found that addition of α-factor to cells prearrested in G2/M has no effect on the capacity of subsequently overexpressed HSF to suppress the ΔHSE1 null phenotype (data not shown). In light of previous work indicating that α-factor is capable of eliciting a normal signal transduction cascade in nocodazole-arrested cells (49), an α-factor-specific effect is unlikely to explain the G1 block in HSF binding.
Cell cycle-dependent binding by HSF to a defined nucleosomal site represents, to the best of our knowledge, the first demonstration of such behavior by a transcriptional activator. In particular, HSF provides a striking contrast to GAL4 and Bicoid, which are able to bind to well-defined nucleosomal sites in G1-arrested S. cerevisiae cells (6) and to nuclear hormone receptors and NF-κB, which appear capable of binding target nucleosomal sites in vertebrate cells irrespective of the phase of the cell cycle (55, 67, 69). Similar to the case for HSF, cell cycle-dependent binding by yeast PPR1 has been inferred from genetic studies of a telomere-linked URA3 allele. These showed that conditionally overexpressed PPR1 could transactivate URA3-TEL in cells prearrested in G2/M but not in cells prearrested in G1, early S, or G0 (4). The chromatin structure of the URA3-TEL promoter is unknown; nevertheless, it is probable that this structure undergoes disassembly during DNA replication, which may account for its accessibility in cells blocked in G2/M.
Is DNA replication required for HSF binding to nucleosomal sites?
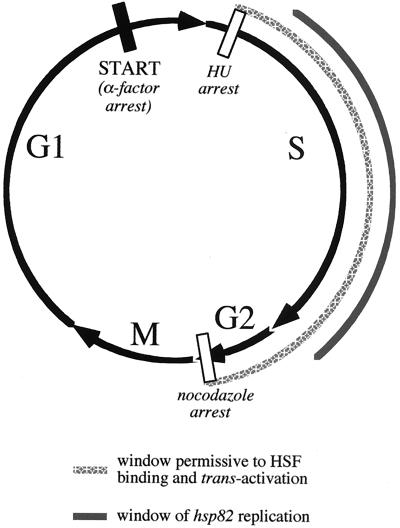
Using quantitative Southern blotting, we have tested whether HSP82 is replicated between its release from α-factor-induced arrest in G1 and its subsequent HU-induced arrest in S phase. We have found that HSP82 DNA content remains constant with respect to both a very late replicating locus, KEX2 (20) (data not shown) and a reporter gene excised during G1 arrest as a nonreplicating DNA ring using an inducible site-specific recombinase system (Fig. 9). While we cannot rule out a small (<20%) increase in hsp82 DNA content between Start and HU arrest, we can rule out a doubling of gene copy number. Consistent with the idea that replication is unnecessary for restoring HSF binding activity, enhanced DMS reactivity of the −210 G residue, diagnostic of the HSF-HSE2/3 interaction, is seen within 1 h of release from α-factor arrest, a time when replication has yet to be detected in cells resuspended in medium alone. Also arguing against replication is that the integrity of Nuc −2, as assayed by its accessibility to HindIII, persists through the HU block. Thus, HSF binding to Nuc −2 is most likely replication independent. The temporal uncoupling of DNA replication from HSF binding argues for a more complex mechanism than the simple disruption, preempt, or exclusion models discussed earlier. HSF might use aspects of all three, with the operative mechanism dependent on the phase of the cell cycle. A summary of our findings is presented in Fig. 11.
FIG. 11.
HSF binding and transactivation of a nucleosomal regulatory site as a function of the cell cycle: summary of Northern, DNA footprinting, and Southern data. Restriction to HSF binding/transactivation is indicated by a filled rectangle; points which are permissive are indicated by open rectangles. The window of opportunity for HSF binding and transactivation could extend beyond the G2/M transition but has not yet been examined. As indicated, replication of hsp82 occurs subsequent to the HU block (see text). The relative lengths of each cell cycle phase have not been determined for the strains used in this study and represent an approximation only.
If replication is unlikely to account for the cell cycle-dependent binding of HSF to HSE2 and -3, what can? One possibility is that HSF itself is cell cycle regulated. However, since HSF's ability to bind accessible promoters during late G1 is not impaired—as implied by its efficient induction of either HSP26 (Fig. 3) or HSP82+ (data not shown)—such a G1-specific defect would be restricted to nucleosomal target sequences. Alternatively, a coactivator of HSF might be cell cycle regulated, inactive during G1, and reactivated following entry into S phase. Indeed, activity of the human SWI/SNF remodeling complex is cell cycle regulated (57); however, in this case, transitional inactivation and reactivation occur in G2/M and G1, respectively. Relevant to this, in experiments to be reported elsewhere, we find that nuclei isolated from early S-phase-arrested, heat-shocked BSY202 cells show a pronounced (three- to fivefold) increase in HindIII accessibility over that seen in non-heat-shocked cells. No significant increase in accessibility is seen in nuclei isolated from heat-shocked, asynchronous cells or from those arrested in other points of the cell cycle (data not shown). These observations parallel the expression data; together they indicate that HSF binds, remodels, and transactivates the nucleosomal promoter most efficiently during early S phase. Thus a coactivator with which HSF associates could be regulated such that it is maximally active or abundant during this point in the cell cycle. Alternatively, an S-phase-specific event might globally prepare chromatin for replication (37); activated HSF might exploit this remodeled state to bind and perturb Nuc −2. Current experiments are addressing these and other possibilities.
Therefore, an appealing scenario is that the hsp82 dinucleosome is remodeled by a histone acetyltransferase or ATP-dependent remodeling complex following release of cells from α-factor arrest, facilitating HSF binding and transactivation of hsp82-ΔHSE1. Consistent with this, Drosophila HSF is greatly benefited by histone acetylation in binding to an in vitro-reconstituted chromatin template, much more so than other activators (48). This might be a consequence of the fact that HSF binds DNA as a homotrimer (52, 60), occupying both faces of the DNA helix (42). Thus, for the protein to bind a nucleosomal site, particularly one proximate to the dyad, a substantial change in free energy is likely necessary to disengage the double helix from its contacts with the histone octamer (54). This process could be facilitated by acetylation (39, 68) or ATP-dependent nucleosomal remodeling (11, 38). While such a requirement may not be as stringent in situations where the target HSEs are positioned near the edge of a nucleosome, it is noteworthy that only specific rotational variants are permissive to HSF binding in vivo (22, 51).
The HSF-nucleosome interaction: paradigm for cell differentiation-specific regulators.
DNA binding by transcriptional activators is normally an essential, early step in the activation of gene expression. The current paradigm for activator-chromatin interactions posits that either transcription factors possess an inherent ability to invade a preassembled nucleosome and directly bind their cognate sites (at least in a milieu rich in chromatin remodeling activity such as the eukaryotic nucleus) or their binding sites reside within nucleosome-free regions. In either case, when an activator is present, its binding occurs irrespective of the stage of the cell cycle. Supporting this notion, a variety of transcription factors are able to bind nucleosomal sites in a cell cycle-independent fashion (6, 55, 67, 69, 74). Here we have provided evidence for a distinctly different mechanism, one in which the nucleosome per se dictates cell cycle-dependent binding of a gene-specific activator. The HSFNuc −2 interaction may therefore serve as an alternative paradigm for those transcriptional activators which are unable to access their target DNA binding sites when such sites are assembled into stable nucleosomes. This property would prevent genes bearing regulatory sites from being inappropriately activated in G1-phase cells expressing the cognate activator. Cell cycle-dependent gene activation could be integral to such processes as cell differentiation, where a gene-specific activator could theoretically gain access to its target regulatory sites during a “quantal” cell division (31).
It is possible that this cell cycle-dependent mechanism is in fact never used by HSF in the regulation of HSP82+ or other stress-responsive genes in S. cerevisiae. However, three considerations suggest otherwise. First, the intrinsic state of the HSP82 promoter is dinucleosomal. This structure not only forms in vivo at hsp82 alleles lacking HSE1 but is readily reconstituted over the WT promoter in vitro (A. M. Erkine and D. S. Gross, unpublished results). Second, yeast HSF, similar to human HSF (65), is incapable of binding even high-affinity sites assembled within stable nucleosomes in vitro (Erkine and Gross, unpublished). Thus, while it is possible that HSF can invade a G1-phase nucleosome bearing a high-affinity site, the inability of HSF to exhibit such activity in vitro, coupled with the inability of overexpressed HSF to bind low-affinity nucleosomal sites in vivo, argues otherwise. Finally, unlike its metazoan counterpart, yeast HSF constitutively binds high-affinity HSEs. This constitutive binding is responsible not only for maintaining target promoters in an open chromatin configuration (17, 24) but also for driving basal transcription (46, 50). It is thus probable that HSF binds to target promoters soon after they have replicated, well within the S/G2 window observed here. The breadth of the window could reflect the fact that immature nucleosomes (comprised of acetylated H3-H4 tetramers or octamers [3]) are more accessible to transactivators than mature nucleosomes comprised of unacetylated octamers.
ACKNOWLEDGMENTS
We thank Mike Hampsey, Ned Sekinger, and Harpreet Singh for comments on the manuscript and acknowledge Walton Fangman, David Hagen, Gunter Kohlhaw, M. K. Raghuraman, Peter Sorger, George Sprague, and Bruce Stentz for gifts of antibodies, strains, and plasmids.
This work was supported by grants to D.S.G. from the National Institute of General Medical Sciences (GM45842), the American Cancer Society, Inc. (NP-945), and the Center for Excellence in Cancer Research at LSUMC-Shreveport.
REFERENCES
- 1.Almer A, Rudolph H, Hinnen A, Horz W. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 1986;5:2689–2696. doi: 10.1002/j.1460-2075.1986.tb04552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amin J, Fernandez M, Ananthan J, Lis J T, Voellmy R. Cooperative binding of heat shock transcription factor to the Hsp70 promoter in vivo and in vitro. J Biol Chem. 1994;269:4804–4811. [PubMed] [Google Scholar]
- 3.Annunziato A T, Seale R L. Histone deacetylation is required for the maturation of newly replicated chromatin. J Biol Chem. 1983;258:12675–12684. [PubMed] [Google Scholar]
- 4.Aparicio O M, Gottschling D E. Overcoming telomeric silencing: a trans-activator competes to establish gene expression in a cell cycle-dependent way. Genes Dev. 1994;8:1133–1146. doi: 10.1101/gad.8.10.1133. [DOI] [PubMed] [Google Scholar]
- 5.Archer T K, Cordingley M G, Wolford R G, Hager G L. Transcription factor access is mediated by accurately positioned nucleosomes on the mouse mammary tumor virus promoter. Mol Cell Biol. 1991;11:688–698. doi: 10.1128/mcb.11.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balasubramanian B, Morse R H. Binding of Gal4p and Bicoid to nucleosomal sites in yeast in the absence of replication. Mol Cell Biol. 1999;19:2977–2985. doi: 10.1128/mcb.19.4.2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barton M C, Emerson B M. Regulated expression of the β-globin gene locus in synthetic nuclei. Genes Dev. 1994;8:2453–2465. doi: 10.1101/gad.8.20.2453. [DOI] [PubMed] [Google Scholar]
- 8.Bonne-Andrea C, Wong M L, Alberts B M. In vitro replication through nucleosomes without histone displacement. Nature. 1990;343:719–726. doi: 10.1038/343719a0. [DOI] [PubMed] [Google Scholar]
- 9.Bonner J, Ballou C, Fackenthal D. Interactions between DNA-bound trimers of the yeast heat shock factor. Mol Cell Biol. 1994;14:501–508. doi: 10.1128/mcb.14.1.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J, Pederson D S. A distal heat shock element promotes the rapid response to heat shock of the HSP26 gene in the yeast Saccharomyces cerevisiae. J Biol Chem. 1993;268:7442–7448. [PubMed] [Google Scholar]
- 11.Cote J, Quinn J, Workman J L, Peterson C L. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 12.Cuatrecasas P, Fuchs S, Anfinsen C B. Catalytic properties and specificity of the extracellular nuclease of Staphylococcus aureus. J Biol Chem. 1967;242:1541–1547. [PubMed] [Google Scholar]
- 13.Cusick M E, Herman T H, DePamphilis M L, Wassarman P M. Structure of chromatin at deoxyribonucleic acid replication forks: prenucleosomal deoxyribonucleic acid is rapidly excised from replicating simian virus 40 chromosomes by micrococcal nuclease. Biochemistry. 1981;20:6648–6658. doi: 10.1021/bi00526a020. [DOI] [PubMed] [Google Scholar]
- 14.Davis N G, Horecka J L, Sprague G F. Cis- and trans-acting functions required for endocytosis of the yeast pheromone receptors. J Cell Biol. 1993;122:53–65. doi: 10.1083/jcb.122.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Devlin C, Tice-Baldwin K, Shore D, Arndt K T. RAP1 is required for BAS1/BAS2- and GCN4-dependent transcription of the yeast HIS4 gene. Mol Cell Biol. 1991;11:3642–3651. doi: 10.1128/mcb.11.7.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elford H L. Effect of hydroxyurea on ribonucleotide reductase. Biochem Biophys Res Commun. 1968;33:129–135. doi: 10.1016/0006-291x(68)90266-0. [DOI] [PubMed] [Google Scholar]
- 17.Erkine A M, Adams C C, Diken T, Gross D S. Heat shock factor gains access to the yeast HSC82 promoter independently of other sequence-specific factors and antagonizes nucleosomal repression of basal and induced transcription. Mol Cell Biol. 1996;16:7004–7017. doi: 10.1128/mcb.16.12.7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erkine A M, Magrogan S F, Sekinger E A, Gross D S. Cooperative binding of heat shock factor to the yeast HSP82 promoter in vivo and in vitro. Mol Cell Biol. 1999;19:1627–1639. doi: 10.1128/mcb.19.3.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fascher K-D, Schmitz J, Horz W. Role of trans-activating proteins in the generation of active chromatin at the PHO5 promoter in S. cerevisiae. EMBO J. 1990;9:2523–2528. doi: 10.1002/j.1460-2075.1990.tb07432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedman K L, Diller J D, Ferguson B M, Nyland S V M, Brewer B J, Fangman W L. Multiple determinants controlling activation of yeast replication origins late in S phase. Genes Dev. 1996;10:1595–1607. doi: 10.1101/gad.10.13.1595. [DOI] [PubMed] [Google Scholar]
- 21.Friedman K L, Raghuraman M K, Fangman W L, Brewer B J. Analysis of the temporal program of replication initiation in yeast chromosomes. J Cell Sci Suppl. 1995;19:51–58. doi: 10.1242/jcs.1995.supplement_19.7. [DOI] [PubMed] [Google Scholar]
- 22.Geraghty D S, Sucic H B, Chen J, Pederson D S. Evidence that partial unwrapping of DNA from nucleosomes facilitates the binding of heat shock factor following DNA replication in yeast. J Biol Chem. 1998;273:20463–20472. doi: 10.1074/jbc.273.32.20463. [DOI] [PubMed] [Google Scholar]
- 23.Giardina C, Lis J T. Dynamic protein-DNA architecture of a yeast heat shock promoter. Mol Cell Biol. 1995;15:2737–2744. doi: 10.1128/mcb.15.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gross D S, Adams C C, Lee S, Stentz B. A critical role for heat shock transcription factor in establishing a nucleosome-free region over the TATA-initiation site of the yeast HSP82 heat shock gene. EMBO J. 1993;12:3931–3945. doi: 10.1002/j.1460-2075.1993.tb06071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gross D S, English K E, Collins K W, Lee S. Genomic footprinting of the yeast HSP82 promoter reveals marked distortion of the DNA helix and constitutive occupancy of heat shock and TATA elements. J Mol Biol. 1990;216:611–631. doi: 10.1016/0022-2836(90)90387-2. [DOI] [PubMed] [Google Scholar]
- 26.Guacci V, Hogan E, Koshland D. Chromosome condensation and sister chromatid pairing in budding yeast. J Cell Biol. 1994;125:517–530. doi: 10.1083/jcb.125.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han M, Grunstein M. Nucleosome loss activates yeast downstream promoters in vivo. Cell. 1988;55:1137–1145. doi: 10.1016/0092-8674(88)90258-9. [DOI] [PubMed] [Google Scholar]
- 28.Hershkovitz M, Riggs A D. Metaphase chromosome analysis by ligation-mediated PCR: heritable chromatin structure and a comparison of active and inactive X chromosomes. Proc Natl Acad Sci USA. 1995;92:2379–2383. doi: 10.1073/pnas.92.6.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirschhorn J N, Brown S A, Clark C D, Winston F. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 1992;6:2288–2298. doi: 10.1101/gad.6.12a.2288. [DOI] [PubMed] [Google Scholar]
- 30.Hoffman C S, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 31.Holtzer H, Weintraub H, Mayne R, Mochan B. The cell cycle, cell lineages, and cell differentiation. Curr Top Dev Biol. 1972;7:229–256. doi: 10.1016/s0070-2153(08)60073-3. [DOI] [PubMed] [Google Scholar]
- 32.Jackson V. In vivo studies on the dynamics of histone-DNA interaction: evidence for nucleosome dissolution during replication and transcription and a low level of dissolution independent of both. Biochemistry. 1990;29:719–731. doi: 10.1021/bi00455a019. [DOI] [PubMed] [Google Scholar]
- 33.Kamakaka R T, Bulger M, Kadonaga J T. Potentiation of RNA polymerase II transcription by Gal4-VP16 during but not after DNA replication and chromatin assembly. Genes Dev. 1993;7:1779–1795. doi: 10.1101/gad.7.9.1779. [DOI] [PubMed] [Google Scholar]
- 34.Knezetic J A, Luse D S. The presence of nucleosomes on a DNA template prevents initiation by RNA polymerase II in vitro. Cell. 1986;45:95–104. doi: 10.1016/0092-8674(86)90541-6. [DOI] [PubMed] [Google Scholar]
- 35.Kornberg R D, Lorch Y. Irresistible force meets immovable object: transcription and the nucleosome. Cell. 1991;67:833–836. doi: 10.1016/0092-8674(91)90354-2. [DOI] [PubMed] [Google Scholar]
- 36.Kornberg R D, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 37.Krebs J E, Kuo M-H, Allis C D, Peterson C L. Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes Dev. 1999;13:1412–1421. doi: 10.1101/gad.13.11.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kwon H, Imbalzano A N, Khavarl P A, Kingston R E, Green M R. Nucleosome disruption and enhancement of activator binding by a human SWI/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 39.Lee D Y, Hayes J J, Pruss D, Wolffe A P. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993;72:73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- 40.Lee S, Gross D S. Conditional silencing: The HMRE mating-type silencer exerts a rapidly reversible position effect on the yeast HSP82 heat shock gene. Mol Cell Biol. 1993;13:727–738. doi: 10.1128/mcb.13.2.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lew D J, Weinert T, Pringle J R. Cell cycle control in Saccharomyces cerevisiae. In: Pringle J R, Broach J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces. Vol. 3. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 607–695. [Google Scholar]
- 42.Littlefield O, Nelson H C M. A new use for the ‘wing’ of the ‘winged’ helix-turn-helix motif in the HSF-DNA cocrystal. Nat Struct Biol. 1999;6:464–470. doi: 10.1038/8269. [DOI] [PubMed] [Google Scholar]
- 43.Lorch Y, LaPointe J W, Kornberg R D. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell. 1987;49:203–210. doi: 10.1016/0092-8674(87)90561-7. [DOI] [PubMed] [Google Scholar]
- 44.Martinez-Balbas M A, Dey A, Rabindran S K, Ozato K, Wu C. Displacement of sequence-specific transcription factors from mitotic chromatin. Cell. 1995;83:29–38. doi: 10.1016/0092-8674(95)90231-7. [DOI] [PubMed] [Google Scholar]
- 45.Matsuzaki H, Nakajima R, Nishiyama J, Araki H, Oshima Y. Chromosome engineering in Saccharomyces cerevisiae by using a site-specific recombination system of a yeast plasmid. J Bacteriol. 1990;172:610–618. doi: 10.1128/jb.172.2.610-618.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McDaniel D, Caplan A J, Lee M S, Adams C C, Fishel B R, Gross D S, Garrard W T. Basal-level expression of the yeast HSP82 gene requires a heat shock regulatory element. Mol Cell Biol. 1989;9:4789–4798. doi: 10.1128/mcb.9.11.4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Michelotti E F, Sanford S, Levens D. Marking of active genes on mitotic chromosomes. Nature. 1997;388:895–899. doi: 10.1038/42282. [DOI] [PubMed] [Google Scholar]
- 48.Nightingale K P, Wellinger R E, Sogo J M, Becker P B. Histone acetylation facilitates RNA polymerase II transcription of the Drosophila hsp26 gene in chromatin. EMBO J. 1998;17:2865–2876. doi: 10.1093/emboj/17.10.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oehlen L J W M, Cross F R. G1 cyclins CLN1 and CLN2 repress the mating factor response pathway at Start in the yeast cell cycle. Genes Dev. 1994;8:1058–1070. doi: 10.1101/gad.8.9.1058. [DOI] [PubMed] [Google Scholar]
- 50.Park H-O, Craig E A. Positive and negative regulation of basal expression of a yeast HSP70 gene. Mol Cell Biol. 1989;9:2025–2033. doi: 10.1128/mcb.9.5.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pederson D S, Fidrych T. Heat shock factor can activate transcription while bound to nucleosomal DNA in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:189–199. doi: 10.1128/mcb.14.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perisic O, Xiao H, Lis J T. Stable binding of Drosophila heat shock factor to head-to-head and tail-to-tail repeats of a conserved 5 bp recognition unit. Cell. 1989;59:797–806. doi: 10.1016/0092-8674(89)90603-x. [DOI] [PubMed] [Google Scholar]
- 53.Pillus L, Solomon F. Components of microtubular structures in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1986;83:2468–2472. doi: 10.1073/pnas.83.8.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Polach K J, Widom J. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J Mol Biol. 1995;254:130–149. doi: 10.1006/jmbi.1995.0606. [DOI] [PubMed] [Google Scholar]
- 55.Reik A, Schutz G, Stewart A F. Glucocorticoids are required for establishment and maintenance of an alteration in chromatin structure: induction leads to reversible disruption of nucleosomes over an enhancer. EMBO J. 1991;10:2569–2576. doi: 10.1002/j.1460-2075.1991.tb07797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sekinger E A, Gross D S. SIR repression of a yeast heat shock gene: UAS and TATA footprints persist within heterochromatin. EMBO J. 1999;18:7041–7055. doi: 10.1093/emboj/18.24.7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sif S, Stukenberg P T, Kirschner M W, Kingston R E. Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes Dev. 1998;12:2842–2851. doi: 10.1101/gad.12.18.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simpson R T. Nucleosome positioning can affect the function of a cis-acting DNA element in vivo. Nature. 1990;343:387–389. doi: 10.1038/343387a0. [DOI] [PubMed] [Google Scholar]
- 59.Smith P A, Jackson V, Chalkley R. Two-stage maturation process for newly replicated chromatin. Biochemistry. 1984;23:1576–1581. doi: 10.1021/bi00302a036. [DOI] [PubMed] [Google Scholar]
- 60.Sorger P K, Nelson H C M. Trimerization of a yeast transcriptional activator via a coiled-coil motif. Cell. 1989;59:807–813. doi: 10.1016/0092-8674(89)90604-1. [DOI] [PubMed] [Google Scholar]
- 61.Straka C, Horz W. A functional role for nucleosomes in the repression of a yeast promoter. EMBO J. 1991;10:361–368. doi: 10.1002/j.1460-2075.1991.tb07957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Struhl K. Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell. 1999;98:1–4. doi: 10.1016/S0092-8674(00)80599-1. [DOI] [PubMed] [Google Scholar]
- 63.Svaren J, Chalkley R. The structure and assembly of active chromatin. Trends Genet. 1990;6:52–56. doi: 10.1016/0168-9525(90)90074-g. [DOI] [PubMed] [Google Scholar]
- 64.Szent-Gyorgyi C, Finkelstein D B, Garrard W T. Sharp boundaries demarcate the chromatin structure of a yeast heat-shock gene. J Mol Biol. 1987;193:71–80. doi: 10.1016/0022-2836(87)90628-0. [DOI] [PubMed] [Google Scholar]
- 65.Taylor I C, Workman J L, Schuetz T J, Kingston R E. Facilitated binding of GAL4 and heat shock factor to nucleosomal templates: differential function of DNA-binding domains. Genes Dev. 1991;5:1285–1298. doi: 10.1101/gad.5.7.1285. [DOI] [PubMed] [Google Scholar]
- 66.Truss M, Bartsch J, Schelbert A, Hache R J G, Beato M. Hormone induces binding of receptors and transcription factors to a rearranged nucleosome on the MMTV promoter in vivo. EMBO J. 1995;14:1737–1751. doi: 10.1002/j.1460-2075.1995.tb07163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Verdin E, Paras P, Lint C V. Chromatin disruption in the promoter of human immunodeficiency virus type 1 during transcriptional activation. EMBO J. 1993;12:3249–3259. doi: 10.1002/j.1460-2075.1993.tb05994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vettese-Dadey M, Grant P A, Hebbes T R, Crane-Robinson C, Allis C D, Workman J L. Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. EMBO J. 1996;15:2508–2518. [PMC free article] [PubMed] [Google Scholar]
- 69.Wong J, Shi Y-B, Wolffe A P. Determinants of chromatin disruption and transcriptional regulation instigated by the thyroid hormone receptor: hormone-regulated chromatin disruption is not sufficient for transcriptional activation. EMBO J. 1997;16:3158–3171. doi: 10.1093/emboj/16.11.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Worcel A, Han S, Wong M L. Assembly of newly replicated chromatin. Cell. 1978;15:969–977. doi: 10.1016/0092-8674(78)90280-5. [DOI] [PubMed] [Google Scholar]
- 71.Workman J L, Kingston R E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 72.Workman J L, Roeder R G. Binding of transcription factor TFIID to the major late promoter during in vitro nucleosome assembly potentiates subsequent initiation by RNA polymerase II. Cell. 1987;51:613–622. doi: 10.1016/0092-8674(87)90130-9. [DOI] [PubMed] [Google Scholar]
- 73.Wu C. Chromatin remodeling and the control of gene expression. J Biol Chem. 1997;272:28171–28174. doi: 10.1074/jbc.272.45.28171. [DOI] [PubMed] [Google Scholar]
- 74.Xu M, Simpson R T, Kladde M P. Gal4p-mediated chromatin remodeling depends on binding site position in nucleosomes but does not require DNA replication. Mol Cell Biol. 1998;18:1201–1212. doi: 10.1128/mcb.18.3.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu L, Morse R H. Chromatin opening and transactivator potentiation by RAP1 in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:5279–5288. doi: 10.1128/mcb.19.8.5279. [DOI] [PMC free article] [PubMed] [Google Scholar]