Abstract
The KRAB domain is a highly conserved transcription repression module commonly found in eukaryotic zinc finger proteins. KRAB-mediated repression requires binding to the KAP-1 corepressor, which in turn recruits members of the heterochromatin protein 1 (HP1) family. The HP1 proteins are nonhistone chromosomal proteins, although it is unclear how they are targeted to unique chromosomal domains or promoters. In this report, we have reconstituted and characterized the HP1–KAP-1 interaction using purified proteins and have compared KAP-1 to three other known HP1 binding proteins: SP100, lamin B receptor (LBR), and the p150 subunit from chromatin assembly factor (CAF-1 p150). We show that the chromoshadow domain (CSD) of HP1 is a potent repression domain that binds directly to all four previously described proteins. For KAP-1, we have mapped the CSD interaction region to a 15-amino-acid segment, termed the HP1BD, which is also present in CAF-1 p150 but not SP100 or LBR. The region of KAP-1 harboring the HP1BD binds as a monomer to a dimer of the CSD, as revealed by gel filtration, analytical ultracentrifugation, and optical biosensor analyses. The use of a spectrum of amino acid substitutions in the human HP1α CSD revealed a strong correlation between CSD-mediated repression and binding to KAP-1, CAF-1 p150, and SP100 but not LBR. Differences among the HP1 binding partners could also be discerned by fusion to a heterologous DNA binding domain and by the potential to act as dominant negative molecules. Together, these results strongly suggest that KAP-1 is a physiologically relevant target for HP1 function.
It is clear that many of the effector domains in eukaryotic transcription factors act as protein-protein interfaces which allow the assembly of macromolecular complexes at various sites in the nucleus. The constellation of transcription factors that are arranged at a gene promoter integrate different complexes via coactivators and corepressors, collectively termed cofactors. Cofactors can act directly upon the process of transcription by regulating components of the RNA polymerase complex or through components of chromatin, often leading to changes in gene expression which are stable through cell division. Core histone modification by histone deacetylases and histone acetyltransferases, which can be targeted, respectively, by corepressors and coactivators, has emerged as a common mechanism for influencing gene expression by altering chromatin (58). Another example is the methylation of CpG islands, which is associated with epigenetically imprinted alleles or repressed genes (8). More recent evidence showing that the methyl-CpG-binding protein is present in a complex with histone deacetylase activity (26, 44) suggests a link between these two chromatin modifications that promote gene silencing. Thus, by acting as links between modifiers of chromatin and site-specific transcription factors, corepressors and coactivators can alter the gene expression profile of a cell in a heritable manner.
To investigate the mechanistic steps through which transcription repressors down-regulate gene expression, we have focused on the KRAB (Kruppel-associated box) domain as a model. The KRAB domain is a potent repression domain present in nearly one-third of the members of a family of zinc finger transcription factors for which there are an estimated 300 to 700 human genes (7, 36). The family is characterized by an amino-terminal KRAB repression domain linked to multiple arrays of Cys2His2-type zinc fingers, which are responsible for DNA binding (29). Like many other repression domains, the KRAB repression domain retains repressor activity when transferred to a heterologous DNA binding domain (35, 62, 65).
KAP-1 is a universal corepressor for the KRAB domain and is the founding member of a small family of cofactors collectively designated the transcriptional intermediary factor 1 (TIF1) family in humans and mice (61). KAP-1 was isolated by affinity chromatography (19) and subsequently by yeast two-hybrid screening (27, 40). It is a 97-kDa nuclear phosphoprotein and contains a number of domains common to the family but also found in other types of transcriptional regulators (see Fig. 1A). At the amino terminus is the RBCC multidomain unit, comprised of a RING finger, two B boxes, and a coiled-coil domain. The RBCC domain is essential for binding to the KRAB domain and participates in multimerization of KAP-1 (19, 48). A plant-like homeodomain (PHD) and a bromodomain are tandemly arranged at the carboxy terminus. The central region of KAP-1 is the least conserved among the family members and is generally rich in prolines, glycines, and serines. KAP-1 itself possesses potent repression activity, and this function is contributed by the carboxy terminus, including the PHD and the bromodomain (19, 40, 61).
FIG. 1.
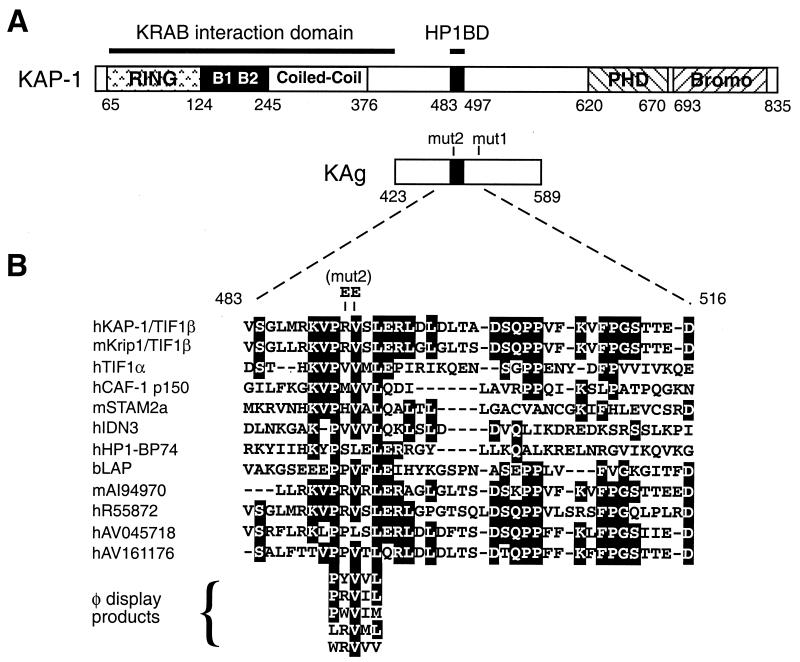
Schematic representations of the KAP-1 protein and the HP1BD. (A) The KAP-1 protein is shown in linear format with the conserved domains noted by shaded boxes and with amino acid positions indicated. A RING finger, two B boxes, and a coiled-coil region comprise the amino-terminal RBCC domain, which is required for binding to the KRAB repressor domain. The PHD, the bromodomain (Bromo), and the HP1BD are also highlighted. KAg is shown below, and the locations of the mut1 (LI519 and 520AA) and mut2 (RV487 and 488EE) double substitutions are indicated. (B) Sequence alignment of the 39-amino-acid segment covering the HP1BD from members of the TIF1 family. Also included in the alignment are other proteins containing a putative HP1BD, as determined by a PHI-BLAST search of nonredundant and EST databases (70). Residues that are identical in at least six members were considered conserved and are highlighted in black. The letters h, m, and b stand for human, murine, and bovine sequences, respectively, and the accession numbers for the ESTs follow these letters. Five different pentamer peptide sequences obtained through phage display analysis of the D. melanogaster HP1 CSD are also included (57).
The basis for KAP-1 function in transcription repression is not yet fully understood, but the ability of KAP-1 to bind heterochromatin protein 1 (HP1) is very likely to have a role. The interaction between these families of proteins was first uncovered in a yeast two-hybrid screen using TIF1α (32). There are three different HP1 proteins in humans and mice: HP1α, HP1β, and HP1γ (mouse HP1β and HP1γ are also referred to as M31 or MOD1 and M32 or MOD2, respectively [25]). All the proteins share a basic structure of an amino-terminal chromodomain (CD) and a carboxy-terminal chromoshadow domain (CSD) linked by a hinge region (see Fig. 2A). The CD is present in numerous proteins, but the CSD has been found only in the HP1 proteins and is thus considered the signature motif for this family (2, 47, 56).
FIG. 2.
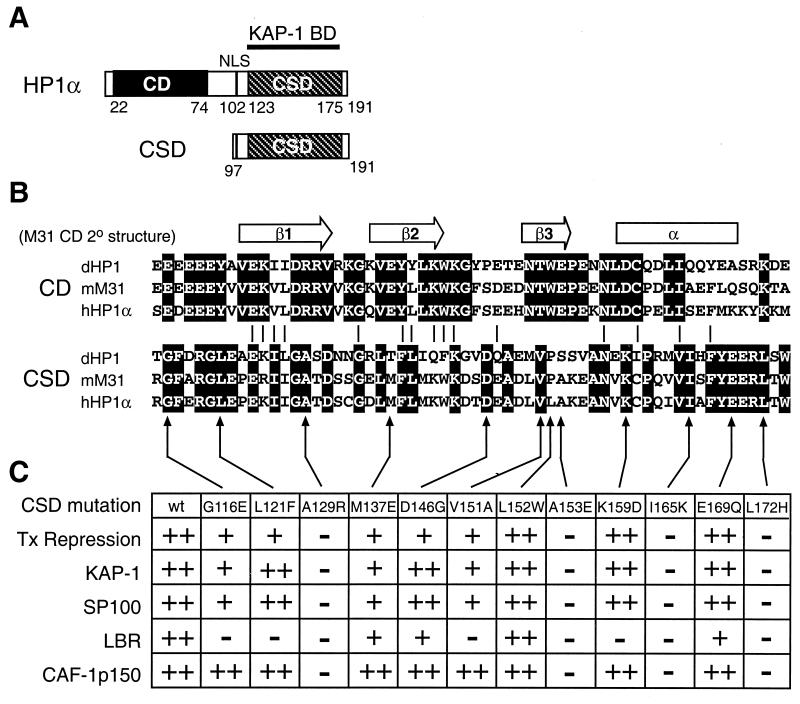
Schematic representations of the HP1 protein, the CD, and the CSD. (A) Diagram of human HP1α depicting the prototypical structure of the HP1 proteins. The relevant domains are noted; NLS, nuclear localization signal. The CSD polypeptide fragment from human HP1α is shown. BD, binding domain. (B) Alignment of the primary amino acid sequences of the CD and the CSD of D. melanogaster HP1 (dHP1), mouse M31 (mM31), and human HP1α (hHP1α) proteins. The secondary structure determined for the CD of M31 is indicated above. Identical residues within either the CD or the CSD are highlighted in black. (C) Point mutations engineered in the human HP1α CSD. The table summarizes the transcription (Tx) repression activity of the mutant CSD polypeptides and their ability to bind to various HP1 partner proteins. The reported values were judged relative to the activity observed for the wild-type (wt) CSD: ++, 100 to 75%; +, 75 to 25%; −, 25 to 0.
Studies of HP1 in Drosophila melanogaster provide one of the best examples of epigenetic mechanisms of gene regulation. Position effect variegation (PEV) in D. melanogaster refers to the process of silencing euchromatic genes that have integrated adjacent to heterochromatin (63). Gene silencing occurs in only a subset of cells, and this state is faithfully inherited by their progeny, leading to variegated or mosaic patterns of expression. The stochastic nature of PEV is hypothesized to be due to the variable spreading of heterochromatin into adjacent regions (55). Genetic screens for modifiers of PEV led to the discovery of the suppressor allele Su(var)2-5, which encoded HP1, a nonhistone chromosomal protein first identified by its localization to heterochromatin in polytene nuclei (14, 23). D. melanogaster HP1 plays a dose-dependent role in PEV and likely contributes to the formation and/or stabilization of heterochromatin (15). More recent studies showing that there is a correlation between overexpression of the CD protein Swi6 and mat locus imprinting in fission yeast and increased expression of an M31 transgene and changes in variegation gene expression in mice indicate that HP1 function has been strongly conserved (17, 43).
Position effects are known to play a role in both human and murine genetic defects (6, 28). In many cases, it appears that positively acting elements are removed and/or that negatively acting (silencing) regions are repositioned near genes. Indeed, boundary elements and locus control regions may function as barriers that prevent inappropriate spreading of heterochromatin or association with heterochromatic regions in nuclei (18, 38, 60). Moreover, enhancer function has been postulated to work by keeping genes out of heterochromatic environs (10). Emerging studies of gene regulation by Ikaros have revealed a colocalization of inactive genes with centromeric heterochromatin and M31 in normal cycling lymphocytes (9). Thus, local compartmentalization of genes in the nucleus is emerging as a common mechanism for repressing transcription in a stable manner.
However, the mechanisms responsible for sequence-specific targeting of this repression are unclear. Heterochromatin, as defined classically by cytological appearance, is interspersed throughout chromosomes but is abundant near centromeres and telomeres and is frequently composed of repetitive sequences (64). The mammalian HP1 proteins have distinct euchromatic or heterochromatic staining patterns in nuclei, suggesting that their roles have become specialized or that targeting to chromatin has been regulated uniquely (39, 52). KAP-1 is dynamically associated with the euchromatic and heterochromatic regions, suggesting that it links heterochromatin-mediated gene regulation to localization in a specific chromosomal territory (52). This model proposes that KRAB-zinc finger proteins recruit the KAP-1 corepressor to DNA; this complex, in turn, binds to HP1, which may then nucleate local heterochromatin formation, resulting in gene silencing.
In order to understand the interaction between KAP-1 and HP1 in more detail, we have reconstituted the complex using recombinant proteins and have comprehensively defined its biochemical properties. We have found that the CSD in HP1 is required for direct binding to KAP-1 and that a stretch of 15 residues in the middle of KAP-1, the HP1BD, is necessary and sufficient for association. We present evidence that the CSD dimerizes and that it binds the KAP-1 HP1BD in a 2:1 stoichiometry, with an apparent Kd of approximately 60 nM. Mutational analysis with the human HP1α CSD and KAP-1 has pinpointed specific residues that are essential for both binding and transcription repression activity in either protein. We also show that interactions with three other HP1 binding proteins are disrupted by the same CSD mutations as those which disrupt KAP-1 binding; however, these domains are not equally able to repress transcription or inhibit HP1-mediated transcription repression.
MATERIALS AND METHODS
Plasmids.
The bacterial expression plasmids were based either on the pGEX vectors (Pharmacia) for glutathione S-transferase (GST) fusion proteins or on the pQE30 series of vectors (Qiagen) for fusion to polyhistidine at the amino terminus. For expression in mammalian cells, fusions to the GAL4 DNA binding domain (G4DBD) were based on the pM1 vectors (53). The pcDNA3.1 vector (Invitrogen) was used for six-His fusion protein expression in mammalian cells. By convention, plasmids are named with a prefix for the vector backbone, followed by the gene name and numbers referring to the amino acids encoded by the cDNA insert. To create pM-HP1α2-191, the full-length human HP1α cDNA was released from the pC3flagHP1α plasmid (52) by digestion with EcoRI and XhoI, and the fragment was ligated into the EcoRI and SalI sites of pM2. An XmaI-XhoI fragment from pC3flagHP1α was used to subclone the full-length HP1α insert into the XmaI and SalI sites of pQE32 (Qiagen) to create pQE-HP1α2-191. The pM-HP1α2-191 and pQE-HP1α2-191 plasmids were digested with HindIII, incubated with the Klenow enzyme and deoxynucleoside triphosphates, digested with EcoRV, and religated to produce the pQE-HP1α2-113 or pM-HP1α2-113 plasmid. To create the pQE-HP1α97-191 and pQE-HP1α73-191 plasmids, human HP1α sequences were amplified by PCR using the following respective forward primers incorporating a BamHI site: HP1α-S97-for. (5′-GCGGATCCAGATGAAGGAGGGTGAAAATAAT-3′) and HP1α-M73-for. (5′-GCGGATCCACAGTGCCGATGACATCAAATC-3′). The HP1α-stop-rev. primer (5′-GGCTCGAGTTAGCTCTTTGCTGTTTCTTTCTC-3′), incorporating an XhoI site after the stop codon, was used for both amplification reactions. The PCR products were digested with BamHI and XhoI and cloned into the BamHI and SalI sites of pQE32 and pM1 or the BamHI and XhoI sites of pGEX-5X-1 (Pharmacia). The D. melanogaster HP1 cDNA sequence encoding amino acids 118 to 206 was amplified by PCR from the plasmid pVβ206-1.1 (which carries a point mutation, V26M, but is wild type in the amplified region [49]). BamHI and HindIII sites were included in the forward (5′-GATCTAATACGACTCACTATAGGGGGATCCGCGCCCTCTGGCAATAAATC-3′) and reverse (5′-AAGCTTCTAATCTTCATTATCAGAGTACC-3′) primers, respectively. The PCR product was cloned into pCR2.1 (Invitrogen), and the DNA was sequenced to confirm that the wild-type sequence was present. The plasmid was then digested with BamHI and XhoI and cloned into the BamHI and SalI sites of pQE32.
The point mutations in human HP1α shown in Fig. 2C were generated by site-directed PCR mutagenesis with the following overlapping internal primers (the codon change is shown in boldface; for., forward; rev., reverse): G116E, for., 5′-GCAATGATATCGCTCGGGAATTTGAGAGAGGACTG-3′, and rev., 5′-CCACTCCTCTCTCAAATTCCCGAGCGATATCATTGC-3′; L121F, for., 5′-GGCTTTGAGAGAGGATTCGAACCAGAAAAGATCATTGG-3′, and rev., 5′-CCAATGATCTTTTCTGGTTCGAATCCTCTCTCAAAGCC-3′; A129R, for., 5′-CCAGAAAAGATCATTGGGCGCACAGATTCCTGTGGTG-3′, and rev., 5′-CACCACAGGAATCTGTGCGCCCAATGATCTTTTCTGG-3′; M137E, for., 5′-GATTCCTGTGGTGATTTAGAGTTCCTAATGAAATGG-3′; and rev., 5′-CCATTTCATTAGGAACTCTAAATCACCACAGGAATC-3′; D146G, for., 5′-CTAATGAAATGGAAAGACACAGGTGAAGCTGACCTGGT-3′, and rev., 5′-GAACCAGGTCAGCTTCACCTGTGTCTTTCCATTTCATT-3′; V151A, for., 5′-GGAGCTGACCTGGCTCTTGCAAAG-3′, and rev., 5′-CTTTTGCAAGAGCCAGGTCAGCTTC-3′; L152W, for., 5′-GATGAAGCTGACCTGGTTTGGGCAAAAGAAGCTAATGT-3′, and rev., 5′-CACATTAGCTTCTTTTGCCCAAACCAGGTCAGCTTCAT-3′; A153E, for., 5′-GAAGCTGACCTGGTTCTTGAAAAAGAAGCTAATGTG-3′, and rev., 5′-CACATTAGCTTCTTTTTCAAGAACCAGGTCAGCTTC-3′; K159D, for., 5′-GCAAAAGAAGCTAATGTGGACTGTCCACAAATTGTG-3′, and rev., 5′-CACAATTAGTGGACAGTCCACATTAGCTTCTTTTGC-3′; I165K, for., 5′-GAAATGTCCACAAATTGTGAAAGCATTTTATGAAGAGA-3′, and rev., 5′-CTCTCTTCATAAAATGCTTTCACAATTTGTGGACATTT-3′; E169Q, for., 5′-TTGTGATAGCATTTTATCAAGAGAGACTGACATGGC-3′, and rev., 5′-GCCATGTCAGTCTCTCTTGATAAAATGCTATCACAA-3′; and L172H, for., 5′-GCATTTTATGAAGAGAGACACACATGGCATGCATATCC-3′, and rev., 5′-GGATATGCATGCCATGTGTGTCTCTCTTCATAAAATGC-3′.
Except for the V151A mutation, each mutational primer was paired with the same outside primers (HP1α-S97-for. and HP1α-stop-rev.). The purified 5′ and 3′ PCR products were used in a second PCR with the outside primers to produce fragments encoding amino acids 97 to 191 of HP1α. These DNAs were digested with BamHI and XhoI and ligated into pQE32, pM1, or pGEX-5X-1. The V151A mutation was first incorporated into the full-length HP1α cDNA in pC3flagHP1α using the plasmid pBTF4HP1α (kindly provided by H. Worman) as a template and outside primers complementary to T7 and T3. Nested primers generating amino acids 97 to 191 were then used to subclone a BamHI-XhoI fragment into pQE32 and pM1. The human HP1α CD point mutation V21M was generated by PCR using the mutational primers HP1αV21Mfor. (5′-GGAGGAGTATGTTATGGAGAAGGTGCTAGAC-3′) and HP1αV21Mrev. (5′-CTAGCACCTTCTCCATAACATACTCCTCCTC-3′). The outside primers flag-HP1α-for. (5′-GCGGATTCGCCACCATGGACTACAAGGACGACGATGACAAAGAT-3′) and EcoRV-HP1α-rev. (5′-GCCCCGAGCGATATCATTGCTCTGCTC-3′) were used for a second PCR, and the fragment was digested with BamHI and EcoRV and cloned into the corresponding sites of pM-HP1α2-191.
Mouse M31 (mMOD1) cDNA fragments were amplified by PCR from the full-length cDNA in pGEX-M31 (kindly provided by H. Worman) and subcloned into pQE32, pM1, and pGEX-5X-1 using the following forward (for.) and reverse (rev.) primers: M31for.A, 5′-GCGGATCCTGGGGAAAAAGCAAAACAAGAAAGTG-3′; M31for.B, 5′-GCGGATCCAAAGCAAACCAAAGAAGAAGAAAG-3′; M31rev.A, 5′-GGCTCGAGCTAATTCTTGTCGTCTTTTTTG-3′; and M31rev.B, 5′-GGCTCGAGCTACTTTTCTGACTCTTCTTTCTTC-3′. The primer pair M31for.A and M31rev.A produced a fragment encoding amino acid residues 2 to 109; M31for.A and M31rev.B produced a fragment encoding amino acid residues 2 to 185 (full length); and M31for.B and M31rev.B produced a fragment encoding amino acid residues 98 to 185.
The pM2-KAP-1(293-835) plasmid (19) and mut1 and mut2 derivatives (52) were used as templates for PCR with the following respective primer pairs, KAP-1/for.408, 5′-CCGGGATCCAGATTGTGGCAGAGCGTCCTG-3′, and KAP-1/rev.584, 5′-CCGCTCGAGTTAACCCTCCGCAAGAGCCATAAGC- 3′; and KAP-1/for.478, 5′-CCGGGATCCAGGTGAGCGGCCTTATGCGC-3′, and KAP-1/rev.516, 5′-CCGCTCGAGCTAGTCCTCAGTGGTACTGCC-3′. The BamHI and XhoI sites incorporated in the primers were used for insertion into pGEX-5X-1. The following primers were used to generate the series of deletions of the KAP-1 shown in Fig. 3C: KAP-1/R483for, 5′-CGCGGATCCGCAAGGTGCCACGAGTGAGCCTTGAACG-3′; KAP-1/L490for, 5′-CGCGGATCCTTGAACGCCTGGACCTGGACCTAACAGCTG-3′; KAP-1/ G511rev, 5′-CCGCTCGAGCTAGCCTGGTAAGACCTTGAAGACG-3′; KAP-1/P504rev, 5′-CCGCTCGAGCTAGGGTGGCTGGCTGTCAGCTGTTAGGTC-3′; and KAP-1/L497rev, 5′-CCGCTCGAGCTATAGGTCCAGGTCCAGGCGTTCAAGGC-3′. The PCR products generated from different pairwise combinations of forward and reverse primers were digested with BamHI and XhoI and ligated into pGEX-5X-1. The pC3-KAP-1(381-618) plasmids (wild type, mut1, and mut2) were constructed by subcloning EcoRI-HindIII fragments from the pQE-KAP-1(381-618) plasmid series (52) into the corresponding sites of pcDNA3.1. The pQE-KAP-1(423-589) and pC3-KAP-1(423-589) plasmids, encoding the KAP-1 antigen (KAg), have been described elsewhere (19, 52).
FIG. 3.
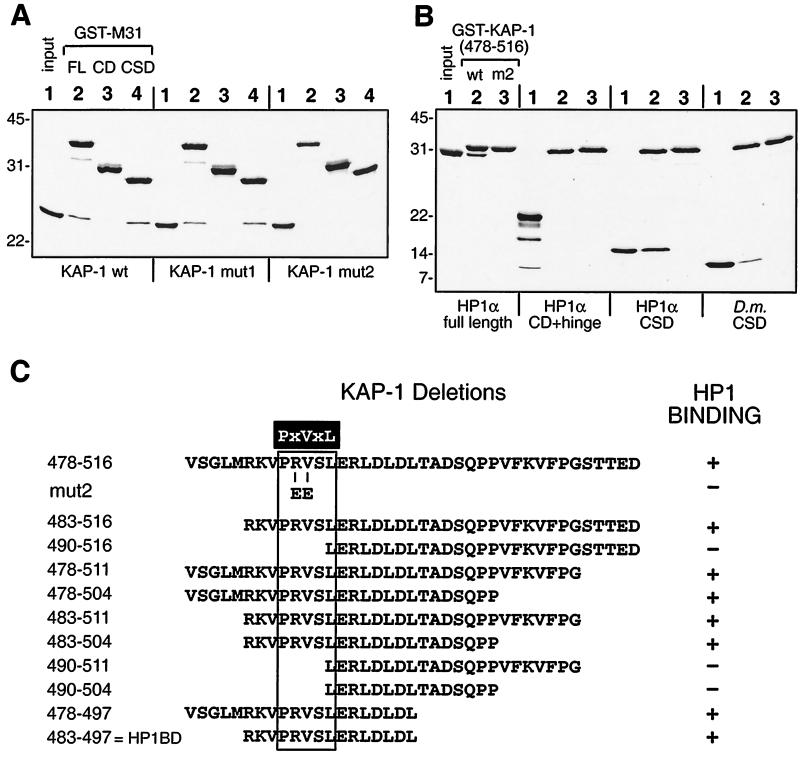
Direct interactions between recombinant KAP-1 and HP1 proteins in vitro and definition of binding domains. (A) GST fusion proteins encompassing different portions of the M31 protein were used in binding reactions with six-His fusion polypeptides containing amino acids 408 to 584 for wild-type (wt), mut1, and mut2 versions of KAP-1. The recombinant proteins were affinity purified to homogeneity, and 2 μg of each GST fusion and 4 μg of each six-His KAP-1 fusion were used in the binding reactions. Complexes retained on glutathione-Sepharose beads were separated by SDS-PAGE and detected by Coomassie blue staining. GST-M31 fusions to the full-length protein (FL, amino acids 2 to 185), the CD (amino acids 2 to 109), and the CSD (amino acids 98 to 185) are indicated above lanes 2, 3, and 4 respectively. Fifty percent of the input KAP-1 polypeptide was loaded into lanes 1. Molecular mass standards (kilodaltons) are indicated to the left. Direct binding is seen only with the full-length protein and a CSD portion of M31, and the KAP-1 mut2 version is never observed to interact. (B) Analysis like that in panel A but with GST–KAP-1 fusions (amino acids 478 to 516) in binding reactions with six-His HP1 fusion polypeptides. Full-length human HP1α (amino acids 2 to 191) or a polypeptide that contains the CSD (amino acids 97 to 191) binds to the wild-type GST–KAP-1 fusion. No binding is seen for the CD-hinge fragment (amino acids 2 to 113) of human HP1α. The CSD fragment of D. melanogaster HP1 (D.m.CSD) also retains specific binding to GST–KAP-1. (C) Deletion analysis of KAP-1 defines a 15-amino-acid HP1BD. GST–KAP-1 fusion proteins were purified from bacteria and used in binding reactions with purified six-His HP1α polypeptides (full-length protein or CSD). The starting 39 residues in KAP-1 are shown at the top, and below are shown the various amino- and carboxy-terminal deletions which were examined. The mut2 double substitution in KAP-1 is also indicated. The minimal CSD binding motif, PXVXL, found in KAP-1/TIF1 and CAF-1 p150 family members, is highlighted in black and boxed. The residue positions in KAP-1 are indicated to the left.
A segment of the human SP100 cDNA (amino acid residues 270 to 346, comprising the HP1BD [33]) was amplified by PCR with forward and reverse primers having the sequences 5′-CGGAGATCTGTGATGAAGAAAGCCCAGAG-3′ and 5′-GCCTCGAGGTTATGAGATGAAGACTTCTAAGGG-3′, respectively, and with the pCMX-SP100 plasmid (37) as a template. The PCR product was ligated into the pCR2.1 vector and subcloned into pGEX-5X-1 and pQE32 via the BglII and XhoI sites included in the primer sequences.
The human lamin B receptor (LBR) cDNA sequence (amino acid residues 96 to 174 [41, 67]) was amplified using the primers LBR for. (5′-CGCGGATCCGATCTGCTTCTGCTTCCCAC-3′) and LBR rev. (5′-GCGCTCGAGCTATTCTTCTCTTCTTGGACGAAGGC-3′). Likewise, the HP1BD from the human CAF-1 p150 subunit was produced with the following respective primer pairs for residues 187 to 248 and 124 to 315: CAF150 for1., 5′-GCGGATCCGGAGCTGCCCGGAGCTGACG-3′; CAF150 rev1., 5′-CGGCTCGAGCTAGGTCATGTTCTTGCCTTGGGG-3′; CAF150 for2., 5′-GCGGATCCCCTCCAGGGAGGCAATAAATGG-3′; and CAF150 rev2., 5′-GGTCTCGAGCTAGTTCTTCTCTGTAGAGCC-3′. The templates for PCR were generated via reverse transcription from total RNA isolated from human Caco2 cells with a Ready To Go kit (Pharmacia) according to the manufacturer's instructions. The PCR products were digested with BamHI and XhoI and cloned into the pQE32, pGEX-5X-1, and pM1 vectors. EcoRI and HindIII fragments from all of the pQE32-based plasmids were subcloned into the pcDNA3.1 vector to provide expression plasmids controlled by T7 and cytomegalovirus promoters. All plasmids generated by PCR were sequenced to confirm the integrity of the coding sequence and the fusion junctions.
Recombinant proteins.
The GST fusion proteins were expressed in Escherichia coli BL21(DE3) cells, and six-His fusion proteins were expressed in E. coli SG13009 cells (Qiagen). Expression of recombinant proteins was induced in cultures grown at 37°C to an optical density at 600 nm of 0.6 with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). After 1 to 3 h, the bacteria were pelleted by centrifugation at 6,000 × g, washed once in PBS (10 mM Na2HPO4, 1.4 mM KH2PO4, 2.7 mM KCl, 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride [PMSF]), and repelleted. These and subsequent steps were carried out at 4°C or on ice unless indicated otherwise. To purify GST fusion proteins, the cell pellets were resuspended in GST lysis buffer (PBS, 25% sucrose, 1 mg of lysozyme per ml, 1 mM dithiothreitol [DTT]) containing a protease inhibitor cocktail (PIC; 1 mM PMSF, 2 μg of aprotinin per ml, 1 μg of pepstatin per ml, 1 μg of leupeptin per ml). After 10 min, a 1/10 volume of 0.5 M EDTA was added and incubation was continued for another 10 min. The lysate was subjected to a single freeze-thaw cycle and then sonicated in an ice-water bath for 30 s using a Macrotip at setting 4 on the 50% duty cycle (Branson). Sonication was repeated twice, with cooling on ice between applications. Following the addition of a 1/10 volume of PBS containing 10% Triton X-100 and PIC, the lysates were mixed for 5 to 10 min, and the insoluble debris was removed by centrifugation at 10,000 × g for 30 min. GST fusion proteins were collected from extracts by the addition of glutathione-Sepharose beads (Pharmacia) and the beads were washed in batch form first with 10 to 20 volumes of BB500 buffer (20 mM Tris-HCl [pH 8.0], 500 mM NaCl, 0.2 mM EDTA, 0.1% Nonidet P-40, 10% glycerol, 1 mM DTT, PIC) and then with the same buffer containing 100 mM NaCl (BB100 buffer). The purified proteins were either stored on beads as a 50% slurry or eluted with 2.5 volumes of 20 mM reduced glutathione in BB100 buffer and then dialyzed exhaustively against dialysis buffer (see below).
Purification of six-His fusion proteins was performed under native conditions using native lysis buffer (300 mM NaCl, 50 mM NaH2PO4, 10% glycerol, 5 mM imidazole [pH 7.8], PIC). Cell suspensions were sonicated and debris was removed as described above. Ni-nitrilotriacetic acid resin (Qiagen) which was equilibrated in PBS was added to the supernatant and mixed in batch form for 1 to 2 h. The resin was washed first with 20 volumes of native lysis buffer and then with 20 volumes of native lysis buffer adjusted to pH 6.0. Proteins were eluted using increasing imidazole concentrations in native lysis buffer in batch form, after a small aliquot of resin was tested to determine the optimal elution conditions. The wild-type and mutant human HP1α97-191 (hHP1α97-191) polypeptides demonstrated similar expression levels and solubility, except for V151A, which produced little soluble protein. As an alternative, six-His fusion proteins were purified under denaturing conditions using denaturing lysis buffer (8 M urea, 100 mM NaCl, 20 mM HEPES [pH 8.0], 10 mM imidazole, 10% glycerol). Binding to Ni-nitrilotriacetic acid resin, washing, and protein elution were carried out as described for native purification, except that the steps were performed at room temperature and denaturing lysis buffer was substituted. The eluted proteins were renatured via step dialysis against decreasing concentrations of urea in dialysis buffer (100 mM NaCl, 20 mM HEPES [pH 8.0], 0.2 mM EDTA, 10% glycerol, 1 mM DTT, 1 mM PMSF). Dialysis renaturation was completed by three sequential exchanges against dialysis buffer lacking urea. The typical yields of soluble six-His fusion proteins purified under either native or denaturing conditions were 0.5 mg (e.g., hHP1α2-191), 5.0 mg (e.g., hKAP423-589 or hKAg), and 15 mg (e.g., hHP1α97-191 or hCSD) per liter of bacterial culture.
The GST binding experiments were performed using equal amounts of GST fusion proteins, which were estimated from Coomassie blue-stained gels. The binding reactions were carried out with 0.3 ml of BB100 buffer which contained 1 mg of bovine serum albumin per ml to block nonspecific interactions. Purified six-His fusion proteins were incubated for 1 h, and the glutathione-Sepharose beads were washed twice with BB500 buffer and twice with BB100 buffer and then eluted by boiling in 2× protein sample buffer (100 mM Tris-HCl [pH 6.8], 4% sodium dodecyl sulfate [SDS], 20% glycerol, 5% β-mercaptoethanol, 0.002% bromophenol blue). The proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and detected by Coomassie blue staining.
Gel filtration analysis.
Gel filtration analysis of purified hKAP423-589 and hHP1α97-191 proteins was carried out at 4°C on a Superdex 200 HR 10/30 column equilibrated with dialysis buffer. The flow rate (0.25 ml/min) and fraction size (1 ml) were controlled using a Pharmacia fast protein liquid chromatography system. An aliquot of each fraction, including the voided volume, was analyzed by SDS-PAGE and Coomassie blue staining. For individual proteins and complexes, 0.2 to 1.0 mg was applied in a volume of 0.5 to 1.0 ml of dialysis buffer. Protein complexes were prepared by refolding the recombinant KAP-1 and HP1α polypeptides in different molar ratios after separate purifications under denaturing conditions and quantitation. Protein complexes were also obtained from proteins that had been purified under native conditions by incubating the preparations together for 1 h on ice. The behavior of the individual HP1α and KAP-1 polypeptides and the complex in gel filtration analysis was not altered by dialysis buffer containing 500 mM NaCl.
Analytical ultracentrifugation.
Sedimentation equilibrium experiments were performed with an Optima XL-I analytical ultracentrifuge using interference optics to measure the protein concentration gradient. For each experiment, three cells were assembled with double-sector 12-mm centerpieces and sapphire windows. Prior to analytical ultracentrifugation, the polypeptides were purified by gel filtration chromatography as described above. The cells were loaded with dialysis buffer as the reference and three different protein concentrations. A blank scan of distilled water was done before the run to correct for the effects of window distortion on the fringe displacement data (69). The experiments were performed at 4°C and 17,000 to 35,000 rpm. Fringe displacement data were collected every 4 to 6 h until equilibrium was reached.
For all experiments, attainment of the sedimentation equilibrium was verified by comparison of successive scans using the MATCH v.7 program, and the data were edited using the REEDIT v.9 program (both programs were kindly provided by David Yphantis). We used nonlinear regression fitting of the data with various models from the NONLIN program (24). The reduced molecular weight of a protein is defined as [M(1 − υρ)ω2]/RT, where M and υ are the molecular weight and the partial specific volume of the protein, respectively, and ρ is the density of the solvent. The program SEDNTERP (31) was used to calculate the solvent density and the υ of the protein using the known amino acid sequence of the protein. At least three data sets from different loading concentrations and/or rotor speeds were fitted simultaneously. Goodness of fit was determined by examination of the residuals and minimization of variance. The equilibrium constants returned by NONLIN from the association models were converted to the molar scale using the molecular weight of the polypeptide and a specific fringe displacement of 3.26 fringesL/g for interference data (34).
Optical biosensor analysis.
Analysis of the binding kinetics between human HP1α and KAP-1 polypeptides was performed at 25°C on BIAX and BIA3000 instruments using CM5 sensor surface chips (Biacore Inc., Uppsala, Sweden). Anti-GST polyclonal antibody (1 to 5 μg) in 10 mM sodium acetate (pH 5.0) was coupled to the sensor surfaces at a flow rate of 5 μl/min after activation with N-hydroxysuccinimide and N-ethyl-N′-(dimethlyaminopropyl)carbodiimide. The coupling reaction was terminated with 1.0 M ethanolamine, and the amount of anti-GST antibody captured was 500 to 1,000 response units (RU). Sensor surfaces were equilibrated in dialysis buffer, and between 50 and 600 RU of GST–HP1α97-191 or GST–KAP-1(408-584) was captured. The I165K mutant version of GST-HP1α and the mut2 version of GST–KAP-1 (control surfaces) were captured at equivalent RU. Alternatively, direct coupling of six-His human HP1α CSD (hCSD) polypeptide (or the control I165K mutant polypeptide) resulted in 600 RU attached to the sensor surface. The analyte for the sensor surfaces composed of GST-HP1α polypeptides was human KAg (hKAg), while hCSD was used as the analyte for the GST–KAP-1 sensor surfaces. The analyte (100 μl) was injected over the sensor surface at a flow rate of 30 μl/min for a total of 360 s, followed by washing with dialysis buffer for the dissociation phase. Sensor surfaces were regenerated with 1.0 M NaCl (pH 2.5) and then equilibrated in dialysis buffer for subsequent capture of GST proteins and analyte binding steps.
The biosensor data (see Table 1) were collected from three independent experiments and analyzed using BIAevaluation v3.0 software (Biacore). All sensorgrams were corrected for nonspecific binding on the control surfaces, and the data points for binding were fitted globally to different models of binding kinetics. These included the one-step Langmuir 1:1 integrated equation, a bivalent analyte model, and parallel or independent heterogeneous ligand interactions.
TABLE 1.
Biosensor-derived kinetic constants for interaction of the recombinant CSD dimer and KAg
| Ligand (RU) | Analyte (M−6) | kona (M−1 s−1) | koffa (s−1) | Kdb (M) | χ2 |
|---|---|---|---|---|---|
| GST-CSDc (500) | hKAg (0.08–1.0) | (3.31 ± 0.042) × 105 | (1.46 ± 0.007) × 10−2 | 4.43 × 10−8 | 9.0 |
| (9.73 ± 0.099) × 103 | (1.17 ± 0.007) × 10−2 | 1.20 × 10−6 | |||
| GST-CSD (150) | hKAg (0.08–1.0) | (1.34 ± 0.032) × 105 | (3.82 ± 0.083) × 10−2 | 5.89 × 10−8 | 3.12 |
| GST-KAg (<50) | hCSD (1.0–10.0) | (1.44 ± 0.067) × 105 | (1.12 ± 0.011) × 10−2 | 7.82 × 10−8 | 0.38 |
Standard errors (SE) calculated for globally fitted kinetic constants are shown. Values are means ± SE.
Apparent equilibrium dissociation constant calculated from koff/kon.
The data were best described by a model in which two independent interaction sites of lower and higher affinities were present (the two sets of values).
Mammalian cell transfection.
NIH 3T3 cells were cultured in Dulbecco modified Eagle medium (DMEM) plus 10% calf serum and were transfected with plasmids using the Lipofectamine reagent as described elsewhere (52). For luciferase reporter gene assays, 3 × 105 cells were seeded per 60-mm dish and transfected with a total of 6 μg of plasmid DNA, which routinely included 1.0 μg of the luciferase reporter and 0.5 μg of pcDNAlacZ to monitor transfection efficiency. In transfections using increasing doses of effector plasmids, the total amount of plasmid containing the simian virus 40 or cytomegalovirus enhancer was maintained by addition of the empty expression vector. Cells were harvested 24 to 36 h posttransfection, washed with PBS, and then lysed in 250 μl of luciferase reporter buffer (Promega). The lysates were subjected to a freeze-thaw cycle and then clarified by centrifugation at 12,000 × g at 4°C. Twenty microliters of the supernatant was used to measure protein content, β-galactosidase activity, and luciferase activity. To examine protein expression, transiently transfected NIH 3T3 cells were metabolically labeled with 35S-Translabel (ICN) in methionine- and cysteine-free DMEM supplemented with 2% dialyzed calf serum. Cell extracts were prepared in radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 50 mM Tris [pH 8.0], 1.0% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS, 1 mM DTT, PIC) and subjected to immunoprecipitation with G4DBD rabbit antiserum (SC-577; Santa Cruz) or mouse monoclonal antibody RGS-His (Qiagen). The immune complexes were collected with protein G-Sepharose (Pharmacia), washed with RIPA buffer, and eluted by boiling in 2× protein sample buffer. The proteins were resolved by SDS-PAGE, and the gels were subjected to fluorography, dried, and exposed to X-ray film.
RESULTS
Defining the interaction domains of KAP-1 and HP1 proteins.
Our previous work demonstrated that the HP1 family of heterochromatin proteins could interact with the KAP-1 corepressor (52). The KAP-1 molecule is comprised of several conserved motifs at both its amino and its carboxy termini (Fig. 1A). The amino-terminal portion, the RBCC domain, is required for binding to the KRAB domain, while the carboxy-terminal region is involved in repressor function (19, 40, 48, 61). The central region of KAP-1 has been demonstrated to be involved in HP1 interaction, but no well-defined structural motifs have been assigned to this region (32, 41, 52).
We initially assessed the interaction using GST-M31 fusion proteins and a portion of KAP-1 spanning amino acid residues 408 to 584, corresponding approximately to the KAg polypeptide. Two different mutant versions of this KAP-1 polypeptide were also purified to homogeneity and served as specificity controls. The results demonstrated that the wild-type KAP-1 polypeptide bound specifically to the GST-M31 full-length and CSD fusions but not to the fusion with the CD and hinge region (Fig. 3A). The same result was observed with the mut1 version of KAP-1, while the mut2 version was completely unable to bind the GST-M31 fusion, consistent with our previous studies of these mutants (52). To further define the segment of KAP-1 responsible for binding, we prepared GST–KAP-1 fusion proteins carrying nested deletions in the region between the RBCC domain and the PHD and used human HP1α polypeptides in binding reactions. We found that a 39-amino-acid segment of KAP-1 (amino acids 478 to 516) was sufficient for direct binding of human HP1α polypeptides that contained the CSD (Fig. 3B). A polypeptide containing the CD and hinge region of human HP1α was unable to interact with this GST–KAP-1 affinity resin. All forms of the GST–KAP-1 protein with the mut2 double substitution (RV487 and 488EE) completely eliminated the interaction between KAP-1 and human HP1α polypeptides (Fig. 3 and data not shown). Together, the data indicate that the carboxy-terminal CSD is solely responsible for the interaction between mammalian HP1 proteins and KAP-1. We next tested the species specificity of this interaction using the CSD from the D. melanogaster HP1 protein (Fig. 3B). KAP-1 was clearly able to bind a D. melanogaster HP1 CSD polypeptide, although the level of binding was somewhat lower than that observed with the CSD from mammalian HP1 proteins.
Examination of the region required for HP1 binding in the TIF1 family revealed short stretches of similarity (Fig. 1B) within the 39 amino acid residues of KAP-1 used as a GST affinity resin. We made further deletions in this fragment to continue to define the minimal stretch of residues required for HP1 binding. A summary of the data from these experiments is presented in Fig. 3C. The shortest segment of KAP-1 which is sufficient for human HP1α binding spans 15 residues (amino acids 483 to 497). This peptide segment, designated the HP1BD, overlaps the M31 interaction region (MIR) defined in mouse TIF1β and the mouse chromatin assembly factor p150 subunit (CAF-1 p150) (41). In addition, a pentamer motif which was discovered in peptides selected by phage display with the CSD from D. melanogaster HP1 (57) is very similar to the HP1BD or MIR. The PXVXL sequence may represent the minimal motif required for interaction with the CSD, and it is noteworthy that the conserved valine residue is replaced by a glutamate residue in the mut2 double substitution. While other sequences in expressed sequence tag databases contain this putative binding motif (Fig. 1B), several HP1 binding proteins, such as SP100 and LBR, do not contain this motif.
Mutational analysis of the human HP1α CSD.
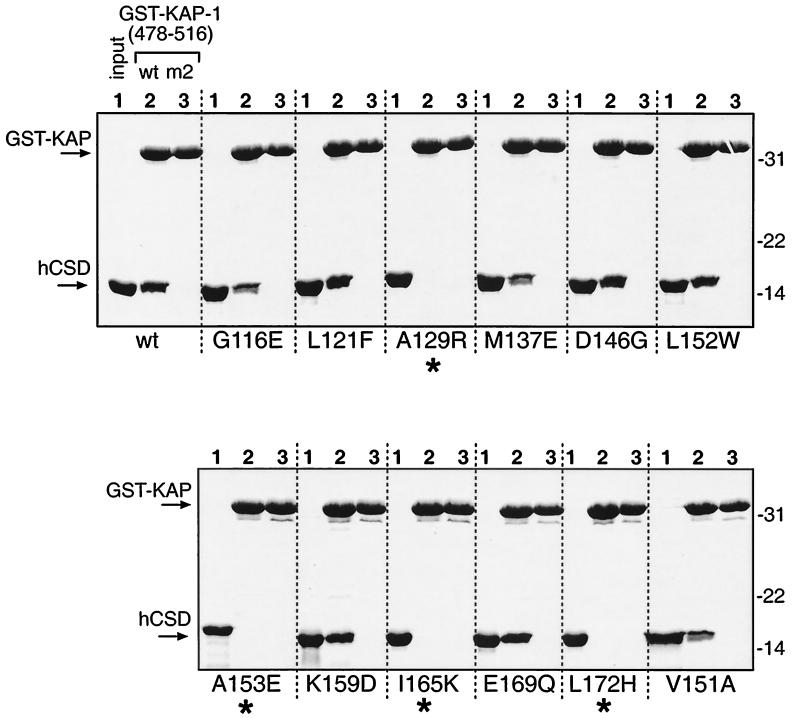
Since our studies indicated that the distantly related D. melanogaster CSD could bind KAP-1, we sought to determine which residues in the CSD were important for this interaction. The CSD at the carboxy terminus of HP1 was identified as a conserved motif related to the CD (2, 56) and is thought to have a similar structure or globular topology (5, 67). However, our results show that the two domains are dissimilar with respect to KAP-1 binding. We created a set of mutations in the human HP1α CSD which, in general, substituted conserved CSD residues with CD-like residues (Fig. 2B). The mutants, in the context of the CSD polypeptide (amino acids 97 to 191), were expressed as recombinant six-His fusion proteins (hereafter designated hCSD), purified to homogeneity, and analyzed for binding to the GST–KAP-1 affinity resin. The 39-amino-acid segment of KAP-1 containing the HP1BD was used, and the mut2 version served as a negative control in these experiments. The results of this analysis show that 4 out of 12 mutants were completely unable to bind KAP-1, while 3 had reduced binding and 5 appeared to bind at the wild-type level (Fig. 4 and 2C). When the mutants were tested as GST fusions, identical results were found, again demonstrating that the configuration of the tags on the recombinant proteins does not affect binding between HP1 and KAP-1 (data not shown). This analysis showed that four residues in the CSD (A129, A153, I165, and L172) are essential for binding to KAP-1 and may be important for distinction between the CSD and the CD.
FIG. 4.
KAP-1 binding by human HP1α CSD mutants. Site-directed mutations in the CSD (Fig. 2C) were produced as recombinant six-His fusion polypeptides in bacteria and purified to homogeneity. Equal amounts of the 12 mutant polypeptides and wild-type (wt) hCSD were used in binding reactions with GST–KAP-1 affinity resins as described in the legend to Fig. 3; the Coomassie blue-stained gels are shown. Molecular mass standards (kilodaltons) are shown to the right. Asterisks indicate four mutants which have completely lost binding to KAP-1.
Interaction between the human HP1α CSD and LBR, SP100, and CAF-1 p150 proteins.
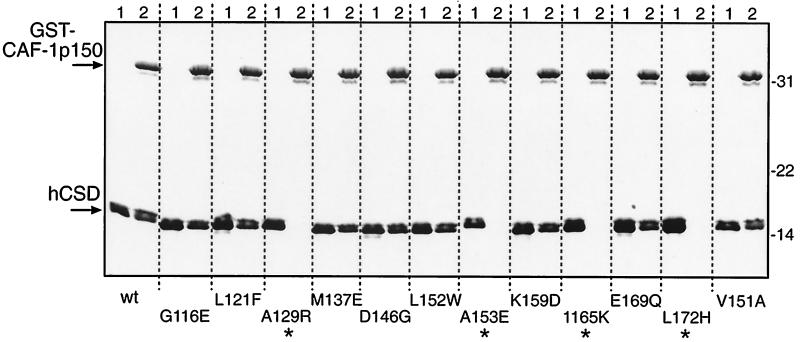
HP1 proteins have been found to interact with a number of other nuclear proteins in addition to the molecules in the TIF1 family. These include the autoantigen SP100 (33, 54), LBR (68), and CAF-1 p150 (41). We produced recombinant proteins carrying the HP1 binding regions from these three proteins and tested them for direct binding to wild-type and mutant hCSD polypeptides. Typical results from one of these experiments using a GST–CAF-1 p150 affinity resin are presented in Fig. 5. Strong binding of wild-type hCSD was observed, and the mutants which failed to bind GST–CAF-1 p150 were the same as those which failed to bind GST–KAP-1, namely, A129R, A153E, I165K, and L172H. Both GST-SP100 and GST-LBR resins were able to bind wild-type hCSD, although the amount bound was reproducibly lower (<50%) than that seen with the GST–KAP-1 and GST–CAF-1 p150 resins (data not shown). The same four hCSD mutants noted above also failed to bind GST-SP100 and GST-LBR. However, GST-LBR failed to bind three other mutants: G116E, K159D, and V151A (see summary in Fig. 2C). Similar results were observed when GST fusions of the CSD mutants were used to bind six-His fusion polypeptides from CAF-1 p150, SP100, and LBR (data not shown). Thus, four different point mutations in the CSD consistently eliminate the interaction between HP1 and four of its potential protein partners (summarized in Fig. 2C).
FIG. 5.
CAF-1 p150 binding by HP1α CSD mutants. A GST fusion protein bearing the HP1 binding region from the p150 subunit of human CAF-1 (amino acids 187 to 248) was purified to homogeneity from bacteria and used in binding reactions with purified hCSD polypeptides as described in the legend to Fig. 4. The Coomassie blue-stained gel is shown, and the input hCSD was loaded into lanes 1. The migration of molecular mass standards (kilodaltons) is indicated to the right. Asterisks mark reactions where no hCSD binding was observed. wt, wild type.
Stoichiometry of the KAP-1–HP1α CSD complex.
To begin to understand the molecular features of KAP-1 and HP1α binding, we examined the biochemical behavior of recombinant polypeptides purified individually or as a reconstituted complex. For these experiments, we chose small domains which were involved in binding and used a refolding strategy which eliminated the formation of aggregates by these recombinant proteins. The polypeptide segment of KAP-1 spanned amino acid residues 423 to 589 (hereafter designated hKAg) (Fig. 1A). The HP1α polypeptide used was hCSD, described above (Fig. 2A). Based on the primary amino acid sequence, the size predicted for hKAg is 18.3 kDa, although its mobility in SDS-PAGE is retarded and it migrates at 28 kDa. The size for hCSD is predicted to be 12.3 kDa, and it migrates near the 14.4-kDa marker in SDS-PAGE.
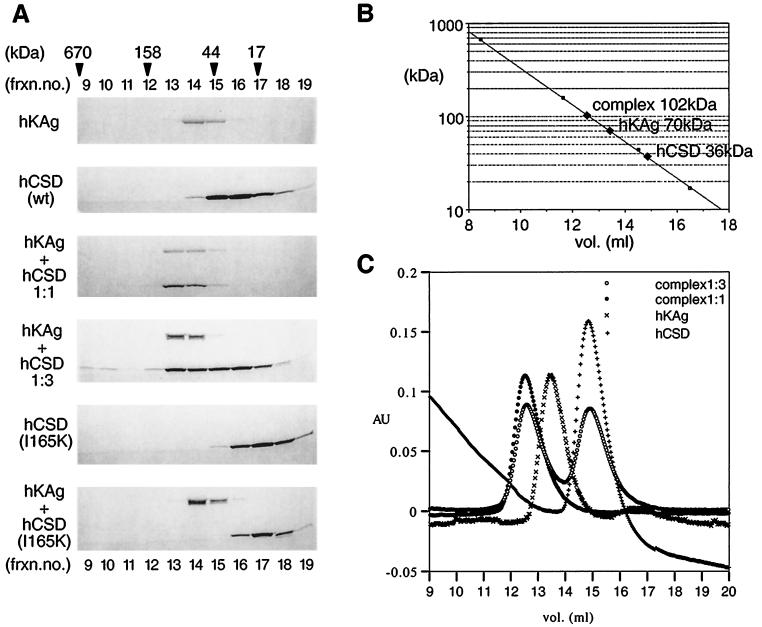
Gel filtration chromatography was performed to characterize the hydrodynamic properties of the purified proteins. Initial gel filtration experiments had shown that hCSD purified under native conditions contained a significant amount of aggregated material in addition to a peak fraction which eluted at an apparent molecular size of 36 kDa. Purification under denaturing conditions followed by step refolding, however, yielded a single species through gel filtration which had the same molecular size, 36 kDa, roughly three times the size of the monomeric polypeptide (Fig. 6). The hKAg polypeptide (native or renatured) behaved as a single species in gel filtration and eluted like a 70-kDa globular protein, much larger than its predicted mass. The unusual mobility of hKAg in this analysis may have been due to a highly asymmetrical shape of this molecule (see Discussion).
FIG. 6.
Gel filtration analyses of the binding domains of the KAP-1–HP1α complex. (A) Six-His fusion polypeptides, hKAg (amino acids 423 to 589), and hCSD (amino acids 97 to 191) were expressed in bacteria, purified under denaturing conditions, and refolded as described in Materials and Methods. The polypeptides were applied to a Superdex 200 gel filtration column, and fractions surrounding the elution peaks were analyzed by SDS-PAGE and Coomassie blue staining. The formation of an hKAg-hCSD complex was obtained by refolding the polypeptides together in 1:1 or 1:3 molar ratios (middle panels). The same method was used for forming an hKAg-hCSD mutant I165K complex (bottom panel). Mixing the separately purified polypeptides resulted in identical elution profiles. Fraction (frxn.) numbers corresponding to elution volume (milliliters) are shown above and below. The elution profile of molecular mass standards is indicated at the top. Note that the complex containing the two wild-type (wt) polypeptides is found in a novel higher-molecular-weight fraction and is not observed with the hCSD I165K mutant. (B) Semilog plot of the molecular masses based upon the protein standards versus elution volume and the apparent molecular masses of the individual polypeptides and the complex. (C) Plot of the protein content (absorbance at 280 nm [AU]) versus elution volume. The traces near the elution peaks are shown for the hKAg and hCSD polypeptides and the complexes. Note that the hKAg-hCSD (1:3) complex contains free hCSD. The trace for hCSD alone has a baseline drift and does not indicate the presence of protein in these fractions.
We then subjected a complex of the hCSD and hKAg polypeptides to gel filtration using material that was either refolded together or mixed (1:1 molar ratio) after purification and refolding of the polypeptides separately. Figure 6 shows that a novel peak containing both polypeptides was obtained at an apparent size of 102 kDa, close to the sum of the apparent sizes of the individual components. The peak fraction containing the complex was rerun over the gel filtration column, resulting in an identical elution profile, without any appreciable dissociation of the complex (data not shown).
These data strongly suggest that the hCSD and hKAg polypeptides form a tight complex that represents the assembly of the individual species. When the complex was formed using a 3:1 molar excess of hCSD-hKAg and subjected to gel filtration, we found two elution peaks corresponding to the complex at an apparent size of 102 kDa and free hCSD polypeptide at an apparent size of 36 kDa (Fig. 6A and C). A 2:1 molar ratio of hCSD-hKAg did not result in a separate peak for the CSD, suggesting that this ratio represents the stoichiometry of the complex. Thus, at a 1:1 molar ratio, an excess of free hKAg polypeptide is predicted to exist; however, this is difficult to ascertain, since there is some overlap between the elution peaks for this species and the complex. To verify the specificity of the complex, we used one of the mutant versions of the hCSD polypeptide (I165K) which cannot bind KAP-1 and prepared the complex in an identical manner. The results of this analysis are shown in the two lower panels of Fig. 6A. The data clearly demonstrate that the mutant hCSD and hKAg polypeptides remained as separate elution peaks and were unable to bind and generate any complex. Interestingly, this analysis also reveals that the I165K mutant hCSD polypeptide had an apparent size of 16 kDa and might possess an aberrant structure. The three other hCSD mutants which were unable to bind KAP-1, A129R, A153E, and L172H, had sizes consistent with globular proteins of 36, 24, and 33 kDa, respectively. Individually, these mutants also failed to generate a complex with the hKAg polypeptide when assayed by gel filtration chromatography (data not shown).
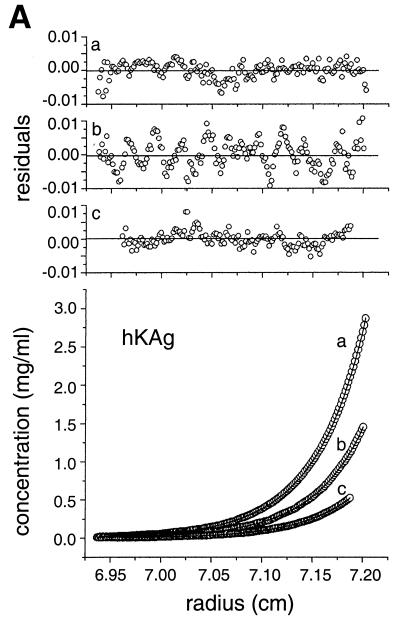
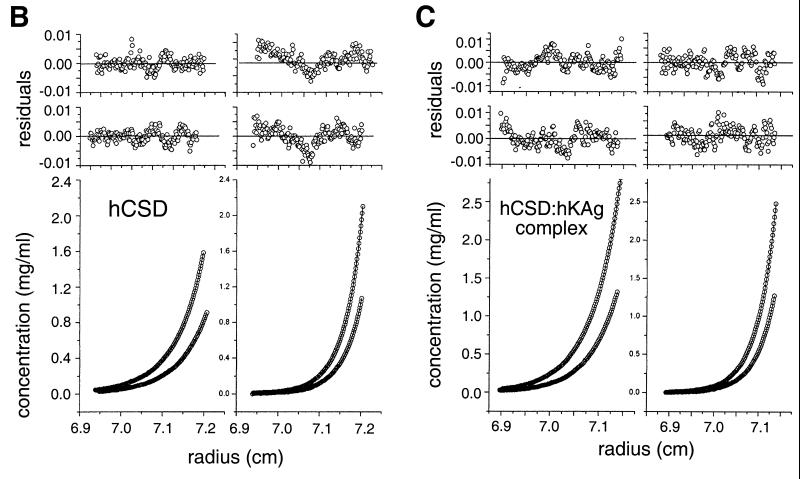
To accurately determine the multimeric status of the hKAg and hCSD polypeptides, analytical ultracentrifugation was performed, and the concentration-versus-radius data at sedimentation equilibrium were fitted with various models using nonlinear regression (24). The data for hKAg fitted well to a model describing a very weak, reversible monomer-dimer association, with an estimated Kd of 3.7 mM (Fig. 7A). The hCSD data were best described by a model containing dimers and tetramers (Fig. 7B). Although the apparent equilibrium constants returned by the fitting program for the dimer-tetramer association were quite similar (0.5 to 0.7 mM for four data sets), the data sets could not be fitted well with a single equilibrium constant. This finding indicates that this weak association is not completely reversible due to a small proportion of the hCSD polypeptide that is incompetent to either dissociate or associate. When a hCSD-hKAg complex was analyzed, the data fitted to a model containing a 2:1 complex and another species corresponding to the hCSD tetramer (Fig. 7C). While some hCSD may exist in a tetrameric state that is not involved in KAP-1 binding, the results are consistent with a dimer of hCSD binding to one molecule of hKAg.
FIG. 7.
Analytical ultracentrifugation analyses of the binding domains from the HP1α–KAP-1 complex reveal 2:1 stoichiometry. (A) Sedimentation equilibrium data for three concentrations of hKAg at 34,800 rpm and 4°C. The three data sets were fitted simultaneously using the nonlinear regression program NONLIN (24). The raw data (circles) and the global fit of an ideal monomer-dimer model (lines) are shown. The upper panels (a to c) show the residuals of the curves fitted to the data points for the three protein concentrations, from highest to lowest (top to bottom). (B) Sedimentation equilibrium data for hCSD at 4°C. The four data sets were fitted simultaneously with a model describing dimers and tetramers using nonlinear regression. The lower panels show the concentration-versus-radius data for two loading concentrations, 1.5 and 0.7 mg/ml, at rotor speeds of 24,000 rpm (left) and 31,000 rpm (right). The calculated fit and raw data are shown as described for panel A. The upper panels show the residuals of the curves fitted to the data points for the high (top) and low (bottom) protein concentrations. (C) Analysis like that in panel B but with an hCSD-hKAg complex isolated by gel filtration at two concentrations, 1.2 and 0.6 mg/ml, and rotor speeds of 21,000 rpm (left) and 25,000 rpm (right). The best fit of the data was to a model describing a 2:1 complex of the hCSD and hKAg polypeptides and a small amount of a tetrameric hCSD species. The upper panels correspond to residuals of the fitted data for the high (top) and low (bottom) protein concentrations.
Biosensor analysis of KAP-1 and HP1 binding kinetics.
To further understand the interaction between the KAP-1 and human HP1α proteins, we examined the real-time kinetics of binding using a surface plasmon resonance optical biosensor assay (11, 42). For the initial biosensor experiments, we attached hCSD from human HP1α directly to the sensor surface. A nonbinding hCSD mutant (I165K) was used as a negative control or reference surface. The analyte for these surfaces was the hKAg polypeptide. hKAg was monomeric on the surface, since its concentration was too low to produce significant dimers, based on the Kd obtained by analytical ultracentrifugation experiments. While specific binding in a dose-dependent manner was observed, we wished to eliminate artifacts caused by direct immobilization of the ligand on the sensor surface and thus oriented the CSD ligand by capturing GST-CSD fusion protein via anti-GST antibodies. We also examined the reverse configuration by capturing GST-KAg (amino acids 408 to 584) on the surface and used hCSD as the analyte. Here, the reference surface was GST-KAg bearing the mut2 double substitution. Table 1 provides a summary of the data and the kinetic parameters obtained. Overall, a model of a preformed CSD dimer interacting with a KAg monomer with single-step 1:1 Langmuir binding kinetics best described the data. However, the binding of hKAg to high levels of the GST-CSD ligand fitted best to a model with a heterogeneous ligand having two different affinities. The association rate constants (kon) derived for this binding showed apparent low- and high-affinity sites with a 100-fold difference, while the dissociation rate constants (koff) did not vary more than 2-fold. Indeed, the koff values for all of the interactions in the optical biosensor assay were high and consistently in the range of 10−2 second−1. This finding was true for hKAg binding to directly coupled hCSD ligand, although the kon value was lower than that obtained with GST-CSD ligand and could have been due to inactivation of ligand binding activity as a result of chemical cross-linking (data not shown). While the association rate constants for binding between hKAg and the CSD dimer could not be fit precisely to a 1:1 interaction model, no other model was able to more accurately describe the association phase. Nevertheless, the koff and apparent Kd values determined by global analysis are similar, with an approximate Kd of 60 nM. This value is also consistent with the tight hCSD-hKAg complex observed in the analytical ultracentrifugation experiments.
Repressor function of HP1 proteins.
Since our data suggest that the KAP-1 corepressor exerts part of its activity through HP1 proteins, we directly tested the effect of human HP1α on transcription by fusing it to a heterologous DNA binding moiety. NIH 3T3 cells were transiently transfected with effector plasmids encoding a G4DBD-HP1 chimera and a luciferase reporter plasmid carrying a minimal promoter linked to five copies of the GAL4 upstream activating sequence (UAS). The results of these assays, shown in Fig. 8, indicate that full-length human HP1α and M31 are potent repressors of transcription. Transcription repression can be achieved in a dose-dependent manner, and the magnitude is similar to that produced by a G4DBD–KAP-1 fusion. The results shown were obtained with a luciferase reporter based on the herpes simplex virus thymidine kinase (TK) promoter, but repression was also observed when the simian virus 40 early promoter or the adenovirus major late promoter was used (data not shown). In each case, the distance between the GAL4 UAS elements and the promoter is less than 200 bp. Deletion of the CSD from HP1α eliminated all repressor activity. The same deletion reduced but did not eliminate repressor activity in M31, demonstrating that the CSD is important for transcription repression by the HP1 proteins. Chimeras expressing the hinge region plus the CSD or the CSD alone were able to repress transcription to the same extent as the full-length protein (Fig. 8). Together, the data strongly suggest that the HP1 proteins can function as transcription repressors when targeted to DNA and that the CSD is necessary and sufficient for this activity.
FIG. 8.
Transcriptional repression by HP1 proteins is mediated by the CSD. Plasmids expressing G4DBD fusions were cotransfected into NIH 3T3 cells with a fixed amount of a luciferase reporter carrying five copies of a synthetic GAL4 UAS in front of a minimal herpes simplex virus TK promoter. The amounts of transfected plasmids (0.5, 1.0, and 5.0 μg) expressing the G4DBD fusion proteins are shown as triangles on the y axis. The pcDNAlacZ plasmid was included to normalize transfection efficiencies. The portions of human HP1α, mouse M31, or KAP-1 fused to G4DBD are schematically drawn as in Fig. 2 and indicated by amino acid numbers in the construct names. In KAP-1, H denotes the HP1BD and Br denotes the bromodomain. Three independent transfections were carried out, and the average value for fold repression, along with the standard error, is given. Fold repression was calculated from the basal luciferase activity obtained from cells transfected without a G4DBD plasmid. A parallel series of transient transfections was carried out with a luciferase reporter lacking the GAL4 UAS sites, and this nonspecific fold repression was subtracted from the values obtained for transfections with the GAL4 UAS site-containing luciferase reporter. NLS, nuclear localization signal.
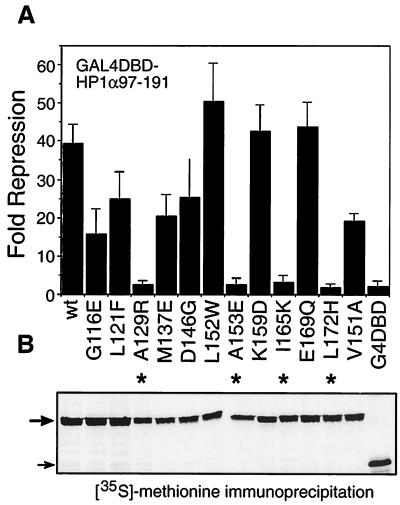
We next tested the effect of the 12 targeted mutations within the CSD on transcription repression in transiently transfected cells. Interestingly, all mutants which were unable to bind KAP-1 were also unable to repress transcription (Fig. 9A). This correlation was also largely true for CSD mutants that retained KAP-1 binding activity; i.e., they showed transcription repression activity approximating the level of KAP-1 binding. For example, the K159D mutant binds KAP-1 strongly and represses transcription at the level seen for the wild-type CSD, while the G116E mutant binds less efficiently to KAP-1 and has repressor activity less than half that of the wild-type CSD. Note that the same correlation exists for CAF-1 p150 and SP100 but not for LBR, since the K159D mutant retains repressor activity but fails to bind LBR (see summary in Fig. 2C). We confirmed that these fusion proteins were expressed at similar levels (Fig. 9B) and retained DNA binding activity (data not shown). Therefore, the differences in the transcription repression activities of these mutants could not be attributed to protein instability or their inability to bind DNA. Taken together, the data suggest that the transcription repression activity of the CSD is related to its ability to bind KAP-1 and underscore the functional significance of proper formation of a KAP-1–HP1 complex.
FIG. 9.
Transcriptional repression by the HP1α CSD is correlated with its ability to bind KAP-1. Three independent transfections were conducted with plasmids (5 μg) expressing G4DBD fusions to human HP1α CSD mutants, and fold repression was calculated as described in the legend to Fig. 8. Asterisks indicate mutants that displayed little or no repression activity, as evidenced by analysis with G4DBD alone. The bottom panel shows the results of an anti-G4DBD immunoprecipitation from a duplicate transfection series in which cells were metabolically labeled with [35S]methionine. The large arrow marks the CSD fusion protein, and the small arrow marks G4DBD alone. wt, wild type.
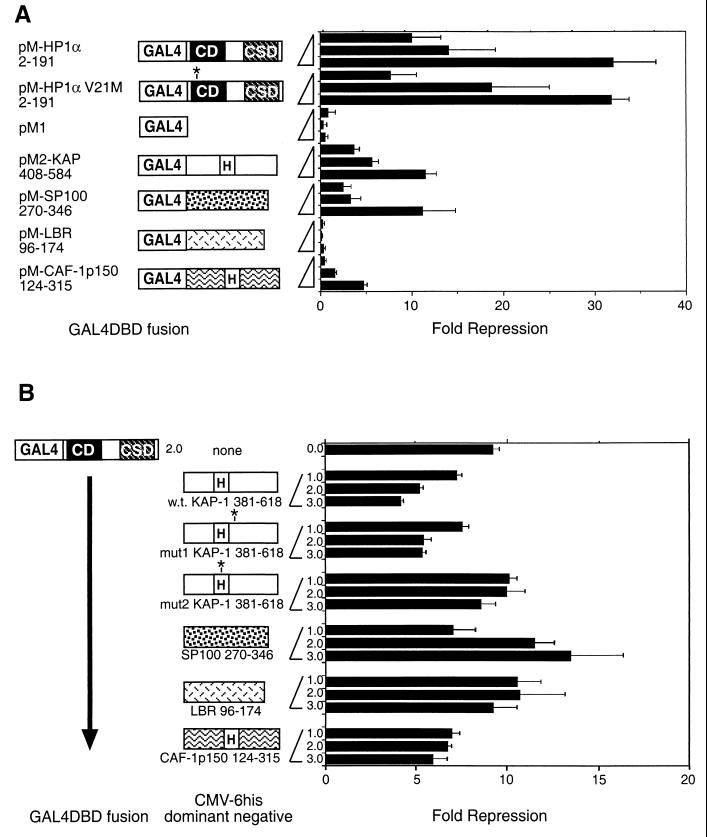
In addition, we tested the transcription repression potential of other HP1 partners by fusing their HP1 binding regions to G4DBD. Fusion proteins were expressed at similar levels in transfected cells, as determined by immunoblotting or immunoprecipitation with G4DBD antisera (data not shown). The data from these transient transfections with the TK-based luciferase reporter are shown in Fig. 10A. We also examined a point mutation in the CD corresponding to the D. melanogaster mutant allele Su(var)2-0502 (14) and found that it did not affect the transcription repression function of human HP1α, supporting the notion that when tethered to DNA, the CSD is largely responsible for repressor function. Both the KAP-1 and the SP100 HP1 binding regions show significant degrees of transcription repression when tethered to DNA, is consistent with our analysis of KAP-1 (52) and earlier work on the SP100 protein (33). A modest degree of transcription repression was observed for the CAF-1 p150 chimera, while no repression activity was observed for the LBR chimera. The reasons for these results are at present unclear, but our results may indicate that these domains cannot or do not work simply as HP1-recruiting moieties in this transcription assay.
FIG. 10.
Modulation of transcription by the HP1 binding regions from KAP-1, SP100, LBR, and CAF-1 p150. (A) Direct assessment of the effect on basal transcription of the HP1 binding regions from KAP-1, SP100, LBR, and CAF-1 p150. Plasmids expressing G4DBD fusions were cotransfected as outlined in the legend to Fig. 8, and fold repression was plotted. The amino acids encoded are indicated below the constructs. For comparison, full-length human HP1α was also tested. Also shown are the results for a point mutation engineered in the human HP1α CD (V21M), corresponding to the D. melanogaster mutant allele Su(var)2-0502 (14). H, HP1BD. (B) Squelching of human HP1α-mediated transcription repression. A series of cotransfections were conducted with a constant amount of pM-HP1α2-191 (2 μg) and increasing amounts of plasmids expressing six-His fusions with the HP1 binding regions from KAP-1, SP100, LBR, and CAF-1 p150 (amounts, in micrograms, are indicated on the y axis). Asterisks indicate the position of mutations in HP1α (V21M) and in KAP-1 (mut1 and mut2).
This point was further addressed by examining the potential for each of the HP1 binding regions from KAP-1, SP100, LBR, and CAF-1 p150 to interfere with or act as a squelcher of HP1-mediated transcription repression. Increasing amounts of plasmids expressing the HP1 binding regions from the aforementioned proteins were cotransfected with constant amounts of a G4DBD-HP1 expression plasmid and a luciferase reporter plasmid. We first assessed the ability of the KAP-1 fragment (amino acids 381 to 618) and found a dose-dependent relief of HP1-mediated repression (Fig. 10B). Significant squelching was also observed with the mut1 version of KAP-1 but not with the mut2 version, which is incapable of binding HP1. A fragment from CAF-1 p150 harboring the MIR was also capable of squelching or relieving HP1-mediated repression. The HP1 binding domains of SP100 and LBR were ineffective in relieving HP1-mediated transcription repression (Fig. 10B). Interestingly, the level of HP1-mediated repression increased moderately with increasing doses of the HP1 binding region from SP100. Thus, these HP1 binding regions appear to act differently in modulating HP1 effects on transcription, and this behavior may relate to the manner in which they bind the CSD.
DISCUSSION
HP1 binding is mediated by a short motif, HP1BD, in KAP-1.
In this report, we examined in detail the interaction between KAP-1 and members of the HP1 family. Using recombinant proteins, we were able to pinpoint a stretch of 15 residues in KAP-1 which is sufficient for direct binding of HP1 proteins (HP1BD). The HP1BD is located in the middle of the KAP-1 molecule, a region that contains a high proportion of prolines and glycines and that does not fit well with secondary structure predictions. Analytical ultracentrifugation shows that hKAg is predominantly a monomer and that dimer formation occurs only well above physiological concentrations. In addition, protease treatment shows that virtually the entire central portion of KAP-1 is susceptible to cleavage, and analysis by circular dichroism has shown that the hKAg polypeptide contains little α-helix or β-sheet structure (X. Li, M. S. Lechner, and F. J. Rauscher III, unpublished data). Thus, it is very likely that this region has a highly extended and perhaps flexible conformation.
This central region of KAP-1 is situated between the RBCC domain and the tandem PHD-bromodomain and is the least conserved among the members of the TIF1 family (61). However, the HP1BD does contain well-conserved residues (Fig. 1B). Mutation of a pair of residues within the minimal consensus sequence (the PXVXL motif) in either KAP-1 or TIF1α completely abolishes HP1 binding (32, 52). Likewise, mutation of conserved residues in the PXVXL motif in the MIR of CAF-1 p150 completely disrupts M31 interactions (41). During the preparation of this article, results reported from phage display experiments revealed a peptide motif which was capable of binding the D. melanogaster HP1 CSD and which was extremely similar to the CAF-1 p150 MIR and the KAP-1 HP1BD (57). The authors also speculated that a sequence similar to the PXVXL motif might also be present in the CSD itself and might provide a basis for the multimerization of HP1 proteins (see below). Database searches reveal that a few expressed sequence tags from previously uncharacterized gene products also have such a motif. We are currently testing whether these polypeptides have similar affinities for HP1 proteins or if certain sequences show tighter binding. This information might indicate that the context of the HP1BD or PXVXL motif within a polypeptide would have an impact on association and that there are preferred partners for HP1.
Different binding motifs and potential transcriptional regulatory mechanisms for HP1 partners.
The CSD of HP1 proteins is responsible for KAP-1 binding. The CSD has been shown to specifically bind several other proteins: CAF-1 p150 (41), LBR (67), and SP100 (33, 54). Interestingly, among these, only CAF-1 p150 appears to contain the HP1BD described above. SP100 is an integral component of a set of discrete nuclear bodies, also referred to as ND10 (37). LBR is found on the inner surface of the nuclear envelope and may participate in the attachment of nuclear lamina and heterochromatin to this membrane (21). For both of these proteins, it is tempting to speculate that they affect the sublocalization of HP1 proteins within the nucleus. Indeed, there are reports that directed localization of genes to the nuclear periphery results in silencing (4). We have analyzed the transcription repression potential of each of the HP1 binding regions from KAP-1, CAF-1 p150, SP100, and LBR by fusing them directly to a heterologous DNA binding domain. We have confirmed that the SP100 chimera has moderate repression activity (33), as does the KAP-1 chimera, noting that when it contains the tandem PHD-bromodomain, more repression activity is observed (Fig. 8 and 10B). The regions from CAF-1 p150 and LBR which were tested are relatively much weaker in this transcription repression assay. While several factors, such as correct protein folding, could affect the output of such an assay, it is possible that the different HP1 binding regions do not recruit HP1 proteins to a promoter in the same manner. That is, when it binds to a particular partner, HP1 may be uniquely regulated. Given that KAP-1 and CAF-1 p150 have essentially identical CSD binding motifs, it is probable that flanking sequences affect HP1 activity upon binding. This idea will be especially interesting to investigate.
Common residues in the HP1 CSD are essential for the repression function and protein interactions.
The CSD was first identified by its resemblance to the CD, which can be found in proteins outside the HP1 family, such as Polycomb, SUVAR39H1, and CHD-1 (2, 47, 56). In contrast, the CSD has been found only in the HP1 family and is always located at the carboxy terminus, suggesting that its function has evolved in a close relationship to that of the CD at the amino terminus. The structure of the CD from the mouse M31 protein has been elucidated by nuclear magnetic resonance spectroscopy (5). The overall folding is similar to that of two archaebacterial proteins, Sac7d and Sso7d, which have DNA binding activity (20, 51), and Ball et al. (5) speculate that the CSD may adopt the same folding pattern. Interestingly, one study has found that human HP1α can bind DNA, although the activity maps only partially to the CD and is without any apparent sequence specificity (59). In vivo studies indicate that the CD from D. melanogaster HP1 or Pc can direct proteins to different sites on chromosomes (49). Other work has shown that the CD is not required for heterochromatin binding and that the CSD is necessary for this activity (50). The matter of the association with heterochromatin or DNA itself is not completely resolved and may be complicated by the fact that the CD and the CSD have some similar attributes, e.g., self-association (see below). Our studies nevertheless point to a clear difference in the two domains; the CSD and not the CD is responsible for both KAP-1 binding activity and transcription repression function.
In our analysis of the human HP1α CSD, we selected several residues for mutation based on the differences which discriminate the CSD from the CD. Although the mutations that we designed were in most cases nonconservative changes, their ability to disrupt KAP-1 binding did not appear to fall into an easily recognizable pattern. The carboxy-terminal half of the CSD shows the most divergence from the CD, and of the 12 mutants that we generated, 3 out of 4 that fail to bind KAP-1 fall within this area. It is possible that the carboxy-terminal region has evolved a specialized function for binding to the KAP-1 corepressor, CAF-1 p150, and other nuclear proteins, such as SP100 and LBR. In the M31 CD structure, an α helix at the carboxy terminus sits in a hydrophobic pocket created by antiparallel β sheets in the amino terminus (5). While many of the mutations that we designed lie within these predicted secondary structure motifs, not all were found to be disruptive, i.e., to eliminate partner protein interactions and the repression activity of the human HP1α CSD. A more detailed study at the atomic level will be necessary to determine the exact folding of the CSD and to show which residues are critical for protein-protein interactions.
We have found that the human HP1α CSD directly binds to recombinant CAF-1 p150, SP100, and LBR proteins and that the mutants that fail to bind KAP-1 also fail to bind these three partners. It is possible that these mutations cause gross changes in the structure of the CSD so that it is no longer able to bind any of its partners. Cross-linking studies show that each of the mutant hCSDs can multimerize essentially like the wild-type polypeptide, suggesting that the overall structure has not been disrupted (M. S. Lechner and F. J. Rauscher III, unpublished results). However, we cannot rule out the possibility that there are still subtle effects on dimerization which cannot be revealed by such assays. The I165K CSD mutation lies within a putative PXVXL-like motif described by Smothers and Henikoff (57). This mutation could affect dimerization, and we did observe a significantly altered elution profile in gel filtration compared to that of wild-type hCSD. It is also possible that the CSD mutations reducing binding coincide with contact residues shared by each of the partner proteins and that they use the same surface for interaction. This situation would be expected for KAP-1 and CAF-1 p150, which appear to use the same HP1 binding motif. In support of this view, we have not been able to isolate hCSD in a ternary complex with a combination of the KAP-1, CAF-1 p150, SP100, and LBR polypeptides (Lechner and Rauscher, unpublished).
The reduced repression activity observed for the V151A CSD mutant is consistent with the results of a previous study which also found that the V151A mutant interacted with SP100 as well as wild-type human HP1α in a yeast two-hybrid assay (33). Our studies using recombinant proteins indicate that the V151A mutant does not bind to SP100 (or to KAP-1 or LBR) as strongly as does wild-type CSD. Thus, the V151A mutation in human HP1α compromises interactions with some partners and may be related to reduced transcription repression activity.
It should be kept in mind that the transcription repression defect in our CSD mutants may be a result of a failure to interact with any of these partners or other factors known to bind HP1 proteins. Candidates include INCENP (3), SU(VAR)3-7 (12), SUV39H1 (1), and origin recognition complex subunits (46). However, the molecular details of the interactions of HP1 proteins and these proteins have yet to be determined.
Oligomerization and function of the CSD.
The ability of the HP1 proteins to self-associate is likely to be critical for their function in heterochromatin regulation. The dose-dependent phenotypes seen in D. melanogaster would support the notion that multimerization of the HP1 proteins is important for the formation and spreading of heterochromatin. A more recent study has shown that the degree of suppression or enhancement of variegating transgenes in mice is correlated with the level of expression of the M31 protein (17). We have shown that the CSD of human HP1α forms oligomers in solution and preferentially exists as dimers. These data are consistent with the results of a previous study demonstrating that the CSD can be cross-linked into species representing dimers, trimers, and tetramers, while the CD exists mainly as a dimer after cross-linking (66). The CD from the murine Polycomb homolog, mPc1, has also been shown to multimerize in solution (13). Other studies have further demonstrated an interaction between CSDs from different HP1 proteins (32, 67). Multimerization (homo- or heterotypic) may be a common feature of these two domains, and it might be feasible to generate dominant negative molecules to interfere with HP1 function.
HP1 isolated from D. melanogaster embryos exists as three distinct oligomeric species (22). M31 and mouse HP1α isolated from stable cell lines can also be found in high-molecular-weight complexes, while M32, which is predominantly located in euchromatin, is restricted to low-molecular-weight fractions (1). The presence of three HP1 species (in mammals or flies) which have distinct subnuclear localization and biochemical properties suggests that they are important for demarcating chromatin territories. Such territories might be characterized not only by transcription output but also by replication timing, recombination potential, and DNA damage susceptibility. Furthermore, HP1 proteins have been implicated in maintaining chromosomal integrity by preventing telomere fusion (16), and it remains to be tested whether any of the HP1 binding partners, such as KAP-1, also play a role in this process.
Stoichiometry of the HP1–KAP-1 complex and macromolecular assembly.
The region of KAP-1 which interacts with HP1 proteins behaves as a monomer in solution and is found in a 1:2 complex with the CSD. Additional biochemical studies of the KAP-1 molecule show that it likely exists as a trimer in solution and that this property is mediated by the RBCC domain (48). While the Kd for dimerization of the KAg fragment is not near physiological levels, trimer formation in the full-length KAP-1 molecule might enhance this weak dimerization function. Preliminary experiments with a trimeric form of KAP-1 indicate that it binds to the CSD with different kinetics than KAg and lower koff values (M. S. Lechner, G. Canziani, and F. J. Rauscher III, unpublished observations). Hence, a functional KAP-1 corepressor might bring together six or more HP1 molecules and further nucleate HP1 multimerization. We have also shown in a previous study that KAP-1 from cell extracts can interact with the KRAB domain bound to DNA and still bind HP1 proteins (52). Hence, there is evidence that KAP-1 is capable of acting as a bridging molecule between DNA-bound factors and HP1 proteins and represents a possible mechanism for gene-specific targeting of HP1 proteins and initiation of heterochromatin formation. Additional experiments indicate that a KRAB–KAP-1–HP1 ternary complex can be formed in vitro with recombinant polypeptides (M. S. Lechner, H. Peng, and F. J. Rauscher III, unpublished observations). The formation of this complex by bacterial proteins indicates that posttranslational modifications are not required for binding. Nonetheless, they may play a role in regulating interactions in vivo, where it is known that the KAP-1 and HP1 proteins exist as phosphoproteins (22, 39). A few candidate protein kinases have recently been implicated, including, perhaps, TIF1 family members themselves (30, 45, 71). Moreover, the PHD and the bromodomain at the carboxy terminus of KAP-1 could integrate additional factors that play a role in HP1 recruitment and regulation. It will be important for future studies to examine how HP1 binding to different partners is regulated in the nucleus and to determine the consequences of these different interactions.
ACKNOWLEDGMENTS
We thank Gabriela Canziani of the Biosensor/Interaction Analysis Structural Biology Cores Group (University of Pennsylvania; Irwin Chaiken, Director, and Jerry Salem, Manager) for the basic surface plasmon resonance training, experiment optimization, and data analysis. We are also grateful to P. B. Singh, H. J. Worman, J. C. Eissenberg, and G. Maul for supplying plasmids. We acknowledge the many helpful discussions from members of the F.J.R. laboratory and W. J. Fredericks, H. Peng, and D. C. Schultz for critical reading of the manuscript. We also thank Sandra L. Harper for help in creating the analytical ultracentrifugation figures.
M.S.L. was supported by Wistar basic cancer research training grant CA 09171. D.W.S. was supported by grants CA 74294 and CA 66671. F.J.R. is supported in part by National Institutes of Health grants CA 52009, CA 10815 (core grant), DK 49210, and GM 54220; ACS grant NP-954; the Irving A. Hansen Memorial Foundation; the Mary A. Rumsey Memorial Foundation; and the Pew Scholars Program in the Biomedical Sciences.
REFERENCES
- 1.Aagaard L, Laible G, Selenko P, Schmid M, Dorn R, Schotta G, Kuhfittig S, Wolf A, Lebersorger A, Singh P B, Reuter G, Jenuwein T. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J. 1999;18:1923–1938. doi: 10.1093/emboj/18.7.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aasland R, Stewart A F. The chromo shadow domain, a second chromo domain in heterochromatin-binding protein 1, HP1. Nucleic Acids Res. 1995;23:3163–3173. doi: 10.1093/nar/23.16.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ainsztein A M, Kandels-Lewis S E, Mackay A M, Earnshaw W C. INCENP centromere and spindle targeting: identification of essential conserved motifs and involvement of heterochromatin protein HP1. J Cell Biol. 1998;143:1763–1774. doi: 10.1083/jcb.143.7.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrulis E D, Neiman A M, Zappulla D C, Sternglanz R. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature. 1998;394:592–595. doi: 10.1038/29100. [DOI] [PubMed] [Google Scholar]
- 5.Ball L J, Murzina N V, Broadhurst R W, Raine A R, Archer S J, Stott F J, Murzin A G, Singh P B, Domaille P J, Laue E D. Structure of the chromatin binding (chromo) domain from mouse modifier protein 1. EMBO J. 1997;16:2473–2481. doi: 10.1093/emboj/16.9.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bedell M A, Jenkins N A, Copeland N G. Good genes in bad neighbourhoods. Nat Genet. 1996;12:229–232. doi: 10.1038/ng0396-229. [DOI] [PubMed] [Google Scholar]
- 7.Bellefroid E J, Poncelet D A, Lecocq P J, Revelant O, Martial J A. The evolutionarily conserved Kruppel-associated box domain defines a subfamily of eukaryotic multifingered proteins. Proc Natl Acad Sci USA. 1991;88:3608–3612. doi: 10.1073/pnas.88.9.3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bird A P, Wolffe A P. Methylation-induced repression—belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 9.Brown K E, Guest S S, Smale S T, Hahm K, Merkenschlager M, Fisher A G. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91:845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- 10.Bulger M, Groudine M. Looping versus linking: toward a model for long-distance gene activation. Genes Dev. 1999;13:2465–2477. doi: 10.1101/gad.13.19.2465. [DOI] [PubMed] [Google Scholar]
- 11.Canziani G, Zhang W, Cines D, Rux A, Willis S, Cohen G, Eisenberg R, Chaiken I. Exploring biomolecular recognition using optical biosensors. Methods. 1999;19:253–269. doi: 10.1006/meth.1999.0855. [DOI] [PubMed] [Google Scholar]
- 12.Cleard F, Delattre M, Spierer P. SU(VAR)3-7, a Drosophila heterochromatin-associated protein and companion of HP1 in the genomic silencing of position-effect variegation. EMBO J. 1997;16:5280–5288. doi: 10.1093/emboj/16.17.5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cowell I G, Austin C A. Self-association of chromo domain peptides. Biochim Biophys Acta. 1997;1337:198–206. doi: 10.1016/s0167-4838(96)00165-3. [DOI] [PubMed] [Google Scholar]
- 14.Eissenberg J C, James T C, Foster-Hartnett D M, Hartnett T, Ngan V, Elgin S C. Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophila melanogaster. Proc Natl Acad Sci USA. 1990;87:9923–9927. doi: 10.1073/pnas.87.24.9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eissenberg J C, Morris G D, Reuter G, Hartnett T. The heterochromatin-associated protein HP-1 is an essential protein in Drosophila with dosage-dependent effects on position-effect variegation. Genetics. 1992;131:345–352. doi: 10.1093/genetics/131.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fanti L, Giovinazzo G, Berloco M, Pimpinelli S. The heterochromatin protein 1 prevents telomere fusions in Drosophila. Mol Cell. 1998;2:527–538. doi: 10.1016/s1097-2765(00)80152-5. [DOI] [PubMed] [Google Scholar]
- 17.Festenstein R, Sharghi-Namini S, Fox M, Roderick K, Tolaini M, Norton T, Saveliev A, Kioussis D, Singh P. Heterochromatin protein 1 modifies mammalian PEV in a dose- and chromosomal-context-dependent manner. Nat Genet. 1999;23:457–461. doi: 10.1038/70579. [DOI] [PubMed] [Google Scholar]
- 18.Festenstein R, Tolaini M, Corbella P, Mamalaki C, Parrington J, Fox M, Miliou A, Jones M, Kioussis D. Locus control region function and heterochromatin-induced position effect variegation. Science. 1996;271:1123–1125. doi: 10.1126/science.271.5252.1123. [DOI] [PubMed] [Google Scholar]
- 19.Friedman J R, Fredericks W J, Jensen D E, Speicher D W, Huang X P, Neilson E G, Rauscher F J. KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 1996;10:2067–2078. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- 20.Gao Y G, Su S Y, Robinson H, Padmanabhan S, Lim L, McCrary B S, Edmondson S P, Shriver J W, Wang A H. The crystal structure of the hyperthermophile chromosomal protein Sso7d bound to DNA. Nat Struct Biol. 1998;5:782–786. doi: 10.1038/1822. [DOI] [PubMed] [Google Scholar]
- 21.Goldberg M, Harel A, Gruenbaum Y. The nuclear lamina: molecular organization and interaction with chromatin. Crit Rev Eukaryot Gene Expr. 1999;9:285–293. doi: 10.1615/critreveukargeneexpr.v9.i3-4.130. [DOI] [PubMed] [Google Scholar]
- 22.Huang D W, Fanti L, Pak D T, Botchan M R, Pimpinelli S, Kellum R. Distinct cytoplasmic and nuclear fractions of Drosophila heterochromatin protein 1: their phosphorylation levels and associations with origin recognition complex proteins. J Cell Biol. 1998;142:307–318. doi: 10.1083/jcb.142.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.James T C, Elgin S C. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol Cell Biol. 1986;6:3862–3872. doi: 10.1128/mcb.6.11.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson M L, Correia J J, Yphantis D A, Halvorson H R. Analysis of data from the analytical ultracentrifuge by nonlinear least-squares techniques. Biophys J. 1981;36:575–588. doi: 10.1016/S0006-3495(81)84753-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones D O, Cowell I G, Singh P B. Mammalian chromodomain proteins: their role in genome organisation and expression. Bioessays. 2000;22:124–137. doi: 10.1002/(SICI)1521-1878(200002)22:2<124::AID-BIES4>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 26.Jones P L, Veenstra G J, Wade P A, Vermaak D, Kass S U, Landsberger N, Strouboulis J, Wolffe A P. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 27.Kim S S, Chen Y M, O'Leary E, Witzgall R, Vidal M, Bonventre J V. A novel member of the RING finger family, KRIP-1, associates with the KRAB-A transcriptional repressor domain of zinc finger proteins. Proc Natl Acad Sci USA. 1996;93:15299–15304. doi: 10.1073/pnas.93.26.15299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kleinjan D J, van Heyningen V. Position effect in human genetic disease. Hum Mol Genet. 1998;7:1611–1618. doi: 10.1093/hmg/7.10.1611. [DOI] [PubMed] [Google Scholar]
- 29.Klug A, Schwabe J W. Protein motifs 5. Zinc fingers. FASEB J. 1995;9:597–604. [PubMed] [Google Scholar]
- 30.Koike N, Maita H, Taira T, Ariga H, Iguchi-Ariga S M. Identification of heterochromatin protein 1 (HP1) as a phosphorylation target by Pim-1 kinase and the effect of phosphorylation on the transcriptional repression function of HP1. FEBS Lett. 2000;467:17–21. doi: 10.1016/s0014-5793(00)01105-4. [DOI] [PubMed] [Google Scholar]
- 31.Laue T M, Shah B, Ridgeway T M, Pelletier S L. Computer-aided interpretation of analytical sedimentation data for proteins. Cambridge, England: Royal Society of Chemistry; 1992. [Google Scholar]
- 32.Le Douarin B, Nielsen A L, Garnier J M, Ichinose H, Jeanmougin F, Losson R, Chambon P. A possible involvement of TIF1 alpha and TIF1 beta in the epigenetic control of transcription by nuclear receptors. EMBO J. 1996;15:6701–6715. [PMC free article] [PubMed] [Google Scholar]
- 33.Lehming N, Le Saux A, Schuller J, Ptashne M. Chromatin components as part of a putative transcriptional repressing complex. Proc Natl Acad Sci USA. 1998;95:7322–7326. doi: 10.1073/pnas.95.13.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luckow E A, Lyons D A, Ridgeway T M, Esmon C T, Laue T M. Interaction of clotting factor V heavy chain with prothrombin and prethrombin 1 and role of activated protein C in regulating this interaction: analysis by analytical ultracentrifugation. Biochemistry. 1989;28:2348–2354. doi: 10.1021/bi00431a055. [DOI] [PubMed] [Google Scholar]
- 35.Margolin J F, Friedman J R, Meyer W K, Vissing H, Thiesen H J, Rauscher F J. Kruppel-associated boxes are potent transcriptional repression domains. Proc Natl Acad Sci USA. 1994;91:4509–4513. doi: 10.1073/pnas.91.10.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mark C, Abrink M, Hellman L. Comparative analysis of KRAB zinc finger proteins in rodents and man: evidence for several evolutionarily distinct subfamilies of KRAB zinc finger genes. DNA Cell Biol. 1999;18:381–396. doi: 10.1089/104454999315277. [DOI] [PubMed] [Google Scholar]
- 37.Maul G G, Yu E, Ishov A M, Epstein A L. Nuclear domain 10 (ND10) associated proteins are also present in nuclear bodies and redistribute to hundreds of nuclear sites after stress. J Cell Biochem. 1995;59:498–513. doi: 10.1002/jcb.240590410. [DOI] [PubMed] [Google Scholar]
- 38.Milot E, Strouboulis J, Trimborn T, Wijgerde M, de Boer E, Langeveld A, Tan-Un K, Vergeer W, Yannoutsos N, Grosveld F, Fraser P. Heterochromatin effects on the frequency and duration of LCR-mediated gene transcription. Cell. 1996;87:105–114. doi: 10.1016/s0092-8674(00)81327-6. [DOI] [PubMed] [Google Scholar]
- 39.Minc E, Allory Y, Worman H J, Courvalin J C, Buendia B. Localization and phosphorylation of HP1 proteins during the cell cycle in mammalian cells. Chromosoma. 1999;108:220–234. doi: 10.1007/s004120050372. [DOI] [PubMed] [Google Scholar]
- 40.Moosmann P, Georgiev O, Le Douarin B, Bourquin J P, Schaffner W. Transcriptional repression by RING finger protein TIF1 beta that interacts with the KRAB repressor domain of KOX1. Nucleic Acids Res. 1996;24:4859–4867. doi: 10.1093/nar/24.24.4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murzina N, Verreault A, Laue E, Stillman B. Heterochromatin dynamics in mouse cells: interaction between chromatin assembly factor 1 and HP1 proteins. Mol Cell. 1999;4:529–540. doi: 10.1016/s1097-2765(00)80204-x. [DOI] [PubMed] [Google Scholar]
- 42.Myszka D G. Improving biosensor analysis. J Mol Recognit. 1999;12:279–284. doi: 10.1002/(SICI)1099-1352(199909/10)12:5<279::AID-JMR473>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 43.Nakayama J-I, Klar A J S, Grewal S I S. A chromodomain protein, Swi6, performs imprinting functions in fission yeast during mitosis and meiosis. Cell. 2000;101:307–317. doi: 10.1016/s0092-8674(00)80840-5. [DOI] [PubMed] [Google Scholar]
- 44.Nan X, Ng H H, Johnson C A, Laherty C D, Turner B M, Eisenman R N, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 45.Nielsen A L, Ortiz J A, You J, Oulad-Abdelghani M, Khechumian R, Gansmuller A, Chambon P, Losson R. Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J. 1999;18:6385–6395. doi: 10.1093/emboj/18.22.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pak D T, Pflumm M, Chesnokov I, Huang D W, Kellum R, Marr J, Romanowski P, Botchan M R. Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell. 1997;91:311–323. doi: 10.1016/s0092-8674(00)80415-8. [DOI] [PubMed] [Google Scholar]
- 47.Paro R, Hogness D S. The Polycomb protein shares a homologous domain with a heterochromatin-associated protein of Drosophila. Proc Natl Acad Sci USA. 1991;88:263–267. doi: 10.1073/pnas.88.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peng H, Begg G E, Schultz D C, Friedman J R, Jensen D E, Speicher D W, Rauscher F J., III Reconstitution of the KRAB-KAP-1 repressor complex: a model system for defining the molecular anatomy of RING-B box-coiled-coil domain-mediated protein-protein interactions. J Mol Biol. 2000;295:1139–1162. doi: 10.1006/jmbi.1999.3402. [DOI] [PubMed] [Google Scholar]
- 49.Platero J S, Hartnett T, Eissenberg J C. Functional analysis of the chromo domain of HP1. EMBO J. 1995;14:3977–3986. doi: 10.1002/j.1460-2075.1995.tb00069.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Powers J A, Eissenberg J C. Overlapping domains of the heterochromatin-associated protein HP1 mediate nuclear localization and heterochromatin binding. J Cell Biol. 1993;120:291–299. doi: 10.1083/jcb.120.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robinson H, Gao Y G, McCrary B S, Edmondson S P, Shriver J W, Wang A H. The hyperthermophile chromosomal protein Sac7d sharply kinks DNA. Nature. 1998;392:202–205. doi: 10.1038/32455. [DOI] [PubMed] [Google Scholar]
- 52.Ryan R F, Schultz D C, Ayyanathan K, Singh P B, Friedman J R, Fredericks W J, Rauscher F J., III KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Kruppel-associated box–zinc finger proteins in heterochromatin-mediated gene silencing. Mol Cell Biol. 1999;19:4366–4378. doi: 10.1128/mcb.19.6.4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sadowski I, Bell B, Broad P, Hollis M. GAL4 fusion vectors for expression in yeast or mammalian cells. Gene. 1992;118:137–141. doi: 10.1016/0378-1119(92)90261-m. [DOI] [PubMed] [Google Scholar]
- 54.Seeler J S, Marchio A, Sitterlin D, Transy C, Dejean A. Interaction of SP100 with HP1 proteins: a link between the promyelocytic leukemia-associated nuclear bodies and the chromatin compartment. Proc Natl Acad Sci USA. 1998;95:7316–7321. doi: 10.1073/pnas.95.13.7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Singh P B, Huskisson N S. Chromatin complexes as aperiodic microcrystalline arrays that regulate genome organisation and expression. Dev Genet. 1998;22:85–99. doi: 10.1002/(SICI)1520-6408(1998)22:1<85::AID-DVG9>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 56.Singh P B, Miller J R, Pearce J, Kothary R, Burton R D, Paro R, James T C, Gaunt S J. A sequence motif found in a Drosophila heterochromatin protein is conserved in animals and plants. Nucleic Acids Res. 1991;19:789–794. doi: 10.1093/nar/19.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smothers J F, Henikoff S. The HP1 chromo shadow domain binds a consensus peptide pentamer. Curr Biol. 2000;10:27–30. doi: 10.1016/s0960-9822(99)00260-2. [DOI] [PubMed] [Google Scholar]
- 58.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 59.Sugimoto K, Yamada T, Muro Y, Himeno M. Human homolog of Drosophila heterochromatin-associated protein 1 (HP1) is a DNA-binding protein which possesses a DNA-binding motif with weak similarity to that of human centromere protein C (CENP-C) J Biochem. 1996;120:153–159. doi: 10.1093/oxfordjournals.jbchem.a021378. [DOI] [PubMed] [Google Scholar]
- 60.van Der Vlag J, den Blaauwen J L, Sewalt R G, van Driel R, Otte A P. Transcriptional repression mediated by polycomb group proteins and other chromatin-associated repressors is selectively blocked by insulators. J Biol Chem. 2000;275:697–704. doi: 10.1074/jbc.275.1.697. [DOI] [PubMed] [Google Scholar]
- 61.Venturini L, You J, Stadler M, Galien R, Lallemand V, Koken M H, Mattei M G, Ganser A, Chambon P, Losson R, de The H. TIF1gamma, a novel member of the transcriptional intermediary factor 1 family. Oncogene. 1999;18:1209–1217. doi: 10.1038/sj.onc.1202655. [DOI] [PubMed] [Google Scholar]
- 62.Vissing H, Meyer W K, Aagaard L, Tommerup N, Thiesen H J. Repression of transcriptional activity by heterologous KRAB domains present in zinc finger proteins. FEBS Lett. 1995;369:153–157. doi: 10.1016/0014-5793(95)00728-r. [DOI] [PubMed] [Google Scholar]
- 63.Wakimoto B T. Beyond the nucleosome: epigenetic aspects of position-effect variegation in Drosophila. Cell. 1998;93:321–324. doi: 10.1016/s0092-8674(00)81159-9. [DOI] [PubMed] [Google Scholar]
- 64.Wallrath L L. Unfolding the mysteries of heterochromatin. Curr Opin Genet Dev. 1998;8:147–153. doi: 10.1016/s0959-437x(98)80135-4. [DOI] [PubMed] [Google Scholar]
- 65.Witzgall R, O'Leary E, Leaf A, Onaldi D, Bonventre J V. The Kruppel-associated box-A (KRAB-A) domain of zinc finger proteins mediates transcriptional repression. Proc Natl Acad Sci USA. 1994;91:4514–4518. doi: 10.1073/pnas.91.10.4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yamada T, Fukuda R, Himeno M, Sugimoto K. Functional domain structure of human heterochromatin protein HP1(Hsalpha): involvement of internal DNA-binding and C-terminal self-association domains in the formation of discrete dots in interphase nuclei. J Biochem. 1999;125:832–837. doi: 10.1093/oxfordjournals.jbchem.a022356. [DOI] [PubMed] [Google Scholar]
- 67.Ye Q, Callebaut I, Pezhman A, Courvalin J C, Worman H J. Domain-specific interactions of human HP1-type chromodomain proteins and inner nuclear membrane protein LBR. J Biol Chem. 1997;272:14983–14989. doi: 10.1074/jbc.272.23.14983. [DOI] [PubMed] [Google Scholar]
- 68.Ye Q, Worman H J. Interaction between an integral protein of the nuclear envelope inner membrane and human chromodomain proteins homologous to Drosophila HP1. J Biol Chem. 1996;271:14653–14656. doi: 10.1074/jbc.271.25.14653. [DOI] [PubMed] [Google Scholar]
- 69.Yphantis D A. Equilibrium ultracentrifugation of dilute solutions. Biochemistry. 1964;3:297–317. doi: 10.1021/bi00891a003. [DOI] [PubMed] [Google Scholar]
- 70.Zhang Z, Schaffer A A, Miller W, Madden T L, Lipman D J, Koonin E V, Altschul S F. Protein sequence similarity searches using patterns as seeds. Nucleic Acids Res. 1998;26:3986–3990. doi: 10.1093/nar/26.17.3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao T, Eissenberg J C. Phosphorylation of heterochromatin protein 1 by casein kinase II is required for efficient heterochromatin binding in Drosophila. J Biol Chem. 1999;274:15095–15100. doi: 10.1074/jbc.274.21.15095. [DOI] [PubMed] [Google Scholar]