ABSTRACT
Staphylococcus aureus is an important pathogen that relies on a variety of mechanisms to evade and counteract the immune system. We show that S. aureus uses oleate hydratase (OhyA) to convert host cis-9 unsaturated fatty acids to their 10-hydroxy derivatives in human serum and at the infection site in a mouse neutropenic thigh model. Wild-type and ΔohyA strains were equally infective in the neutropenic thigh model, but recovery of the ΔohyA strain was 2 orders of magnitude lower in the immunocompetent skin infection model. Despite the lower bacterial abundance at the infection site, the levels of interleukin 6 (IL-6), monocyte chemoattractant protein 1 (MCP-1), IL-1β, and tumor necrosis factor alpha (TNF-α) elicited by the ΔohyA strain were as robust as those of either the wild-type or the complemented strain, indicating that the immune system was more highly activated by the ΔohyA strain. Thus, OhyA functions to promote S. aureus virulence.
IMPORTANCE The oleate hydratase protein family was discovered in commensal bacteria that utilize host unsaturated fatty acids as the substrates to produce a spectrum of hydroxylated products. These hydroxy fatty acids are thought to act as signaling molecules that suppress the inflammatory response to create a more tolerant environment for the microbiome. S. aureus is a significant human pathogen, and defining the mechanisms used to evade the immune response is critical to understanding pathogenesis. S. aureus expresses an OhyA that produces at least three 10-hydroxy fatty acids from host unsaturated fatty acids at the infection site, and an S. aureus strain lacking the ohyA gene has compromised virulence in an immunocompetent infection model. These data suggest that OhyA plays a role in immune modulation in S. aureus pathogenesis similar to that in commensal bacteria.
KEYWORDS: Staphylococcus aureus, oleate hydratase, hydroxy fatty acids, unsaturated fatty acids, virulence, soft tissue infection, virulence determinants
OBSERVATION
Commensal organisms of the gut microbiome contain a family of flavin adenine dinucleotide-dependent oleate hydratase genes (ohyA) that produce a spectrum of hydroxylated fatty acids (hFA) from host unsaturated fatty acids (1). Evidence is accumulating that OhyA-derived hFA function to suppress cytokine production and inflammation to create a more tolerant environment for the commensal bacteria (2–5). Staphylococcus aureus is an important pathogen that deploys an array of virulence factors that engage host immune defenses to promote pathogenesis (6–8). S. aureus expresses an OhyA that catalyzes water addition to cis-9 double bonds (9) and protects against palmitoleic acid (16:1) (10), an antimicrobial fatty acid produced by the innate immune system (11–13). The goal of this study was to determine if OhyA has a role in S. aureus pathogenesis and supports the production of hFA at the infection site.
We found that S. aureus OhyA prefers 18:1 over 18:2 as the substrate based on the described in vitro assay (10). Pure OhyA assayed using [14C]oleate (18:1) or [14C]linoleate (18:2) as the substrate yielded specific activities of 8.25 ± 1.45 and 1.19 ± 0.05 nmol/min/mg, respectively. hFA production by S. aureus grown with equal amounts of 18:1 and 18:2 produced 10-hydroxyoctadecanoic acid (h18:0) and 10-hydroxy-cis-12-octadecenoic acid (h18:1) in the 7:1 ratio expected from the enzymology (Fig. S1A). hFA were measured by liquid chromatography-tandem mass spectrometry (LC-MS/MS) as their 3-picolylamide derivatives using an m/z of 109.0 generated from the loss of the common picolylamide moiety (14). The picolylamide approach circumvents the misrepresentation of hFA abundance based on measurements using unique ions generated from breakage at the hydroxyl group position (15). The high efficiency of detecting the m/z of 297.1/185.1 Q1/Q3 ion pair from h18:1 compared to that of detecting the m/z of 271.1/185.1 and m/z of 299.1/185.1 Q1/Q3 ion pairs arising from h16:0 and h18:0, respectively, gives a false view of the relative abundance of the two hFA (Fig. S1B). The conclusion that h18:1 is the most abundant hFA produced by the gut microbiome is based on the latter technique (15).
Wild-type S. aureus strain AH1263 and its derivatives, PDJ68 (ΔohyA) and PDJ68 (ΔohyA)/pOhyA (10), were grown in 50% human serum to stationary phase to assess the capability of S. aureus to produce hFA in the presence of a mixture of mammalian lipids. The total fatty acid composition of the serum lot shows that the OhyA substrates 18:1 and 18:2 were present in equal amounts, whereas palmitoleate (16:1) was an order of magnitude less abundant (Fig. 1A). However, most of these FA are esterified and not available to OhyA unless first released by S. aureus lipases. Geh is a major extracellular lipase that is known to release FA from serum triacylglycerols for incorporation into S. aureus phospholipids (16). An isogenic strain lacking Geh produces significantly less hFA in human serum than either the wild-type or complemented strains (Fig. S2). Geh is only one of many lipases and phospholipases that could contribute to OhyA substrate availability in environments where their specific lipid substrates are abundant. A representative LC-MS/MS analysis of hFA produced by strain PDJ68 (ΔohyA)/pOhyA grown in 50% serum illustrates the raw ion current from the hFA region of the gradient between 11 and 15 min that was absent in experiments with strain PDJ68 (ΔohyA) (Fig. 1B). Experiments using internal standards showed that h18:0 was the major hFA produced, reaching a concentration of 27.56 ± 1.21 μM (Fig. 1C). h18:1 was the next most abundant, and h16:0 was the least abundant. hFA production by strain PDJ68 (ΔohyA) was not detected (Fig. 1C). OhyA expression in strain PDJ68 (ΔohyA)/pOhyA resulted in an increase in the levels of all hFA. These data show the OhyA-dependent production of hFA when grown in human serum.
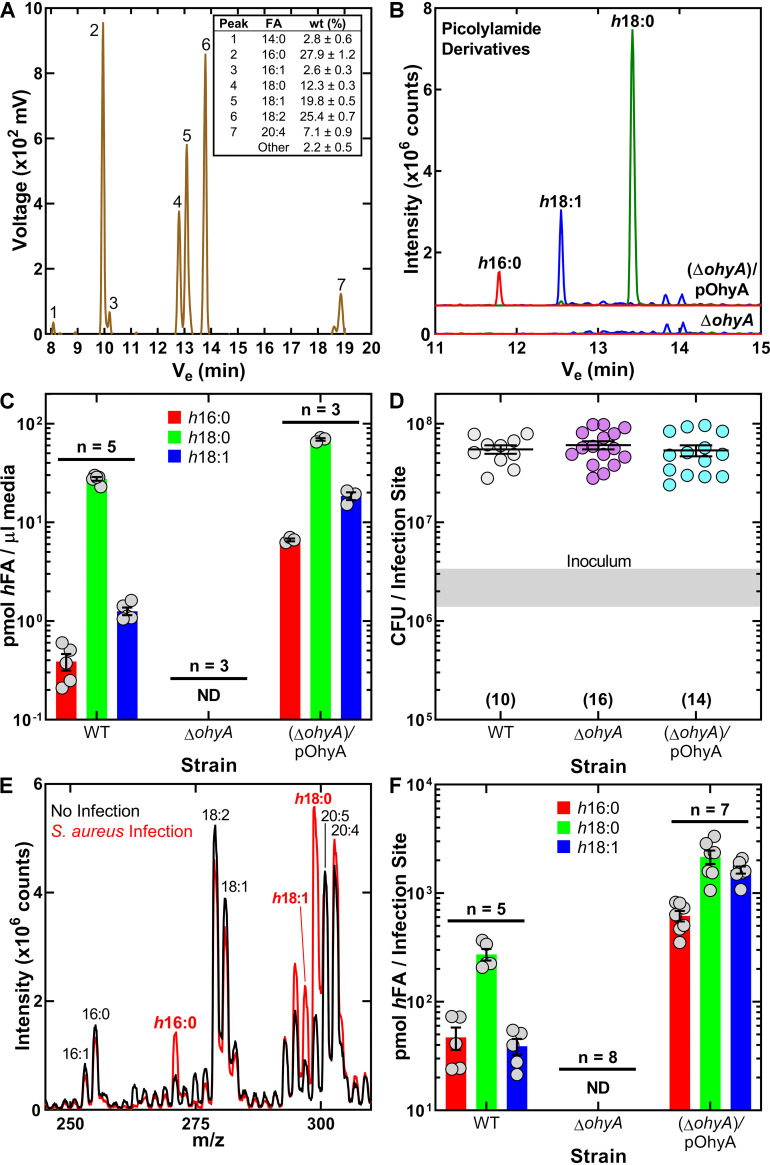
FIG 1.
OhyA-dependent hFA formation in human serum and the neutropenic thigh infection model. (A) A representative gas chromatogram illustrating the fatty acid composition of the human serum lot with the average fatty acid composition from three replicates (inset). (B) Representative LC-MS/MS chromatograms of picolylamide derivatized hFA recovered from the medium following growth of S. aureus strains PDJ68 (ΔohyA) and PDJ68 (ΔohyA)/pOhyA in 50% human serum. (C) Quantification of hFA recovered from the medium following growth of S. aureus strains AH1263 (WT), PDJ68 (ΔohyA), and PDJ68 (ΔohyA)/pOhyA in 50% human serum. (D) Enumeration of the bacteria recovered from infected neutropenic thighs. The gray shaded bar represents the range of initial inoculum. Numbers of animals are in parentheses. (E) Representative total ion chromatograms of the fatty acid fraction in the LC-MS/MS analysis of mock-infected (black) and infected (red) neutropenic thighs. (F) Quantification of hFA recovered from neutropenic thighs infected with the strain set. ND means <5 pmol. Mean ± standard error of the mean (SEM).
A neutropenic thigh infection model was used to address the formation of hFA in vivo. This model was selected to assess hFA formation in vivo in the presence of equal numbers of bacteria at the infection site. There was no significant difference in the bacterial titers among the strains 24 h after infection (Fig. 1D). We first used shotgun lipidomics profiling (17) to determine if there are changes to the composition of the free fatty acid fraction in the thigh following infection. A comparison of the mock-infected to infected thigh samples showed the appearance of three new peaks that correspond in molecular weight to h16:0, h18:0, and h18:1 (Fig. 1E). Quantitation of the hFA composition using picolylamide derivatization showed that the wild-type strain produced predominantly h18:0, 7-fold less h18:1, and 6-fold less h16:0 (Fig. 1F). hFA were not detected in thighs inoculated with the ΔohyA strain. hFA abundance in thighs infected with the ΔohyA/pOhyA strain was higher than that in thighs infected with the wild type. These data show that hFA formation at the infection site is OhyA-dependent.
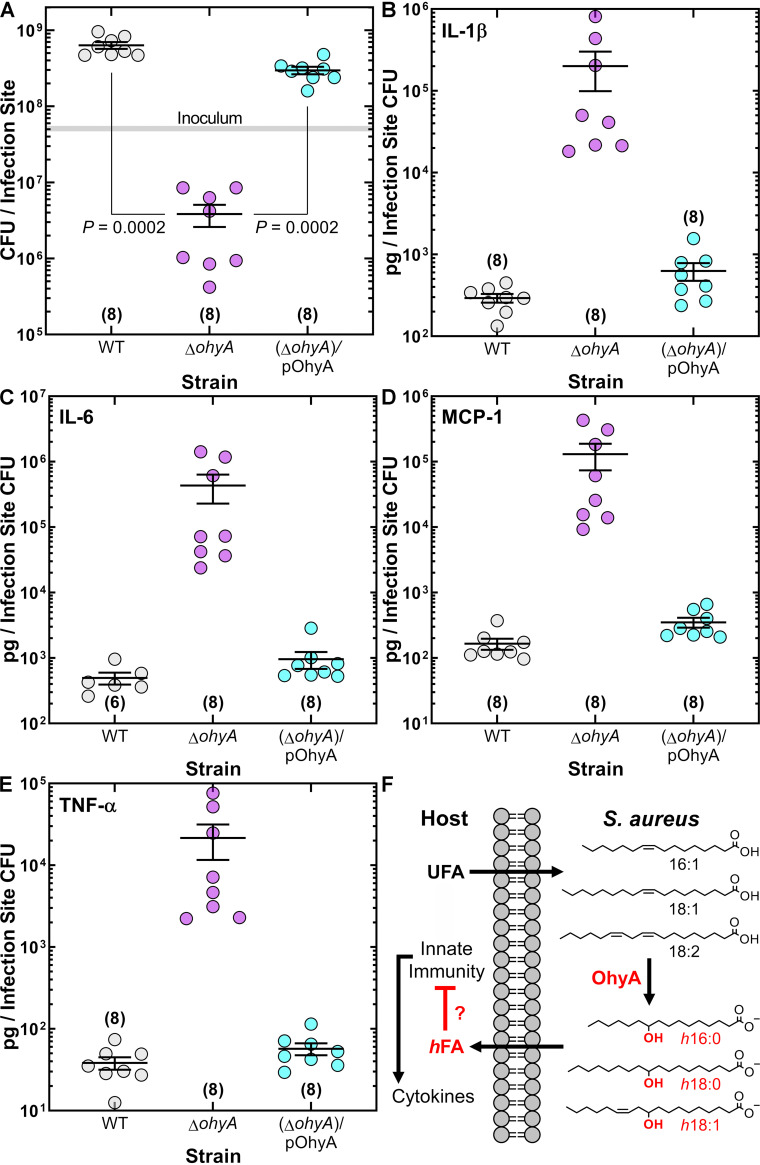
The impact of ohyA deletion on virulence was assessed in an immunocompetent skin/soft tissue (SSTI) infection model that showed that the wild-type and complemented strains established an infection but the bacterial burden from ΔohyA knockout was 2 orders of magnitude lower (Fig. 2A). The impact of OhyA expression on the formation of selected cytokines was assessed using a mouse-specific Milliplex cytokine assay platform to measure levels of proinflammatory cytokines that are produced in response to infection (18–20). Although all three strains elicited large, comparable elevations in cytokines (Fig. S3), the immune response to the ΔohyA strain was 2 orders of magnitude higher than that to the others when the data were normalized to the number of cells present (Fig. 2B to E). These data suggest that OhyA suppresses cytokine production.
FIG 2.
OhyA is a virulence determinant in an SSTI infection model. (A) Enumeration of the bacteria recovered from the infection site. S. aureus strains AH1263 (WT), PDJ68 (ΔohyA), and PDJ68 (ΔohyA)/pOhyA were used to infect mice by subcutaneous infection. Mock-infected mice were given an injection of sterile phosphate-buffered saline (PBS). Kruskal-Wallis test determined whether overall differences between groups have statistical significance, and P values were calculated using Mann-Whitney test. The gray shaded bar represents the range of initial inoculum determined by serial dilution. (B to E) Measurements of cytokine analytes that were recovered from the infection sites in the SSTI model. Data are normalized to the number of bacteria recovered. The cytokine levels per infection site are shown in Figure S3. (F) Model for OhyA-dependent hFA production at the infection site. Unsaturated cis-9 fatty acids (UFA) (16:1, 18:1, 18:2) are converted to hydroxy fatty acids (hFA) (h16:0, h18:0, h18:1) by OhyA. hFA are released into the extracellular environment. This process inactivates the antimicrobial fatty acids (16:1 and 18:2) and generates mediators that inhibit cytokine production by mechanisms that remain to be delineated. Numbers of animals are in parentheses.
Conclusions. This work establishes OhyA as a determinant of virulence in S. aureus. The importance of OhyA to S. aureus pathogenesis is corroborated by Malachowa et al. (21), who identified a gene they called sok that is required for virulence in a rabbit endocarditis infection model. At the time, sok was a gene of unknown function, but we now know that it corresponds to ohyA. A model for the OhyA-dependent metabolism of host unsaturated fatty acids is diagrammed in Figure 2F. Host unsaturated fatty acids are hydroxylated by OhyA. The hFA are not utilized by the pathogen; rather, they are released into the environment. Purified OhyA is a soluble protein, but imaging experiments suggest that it is membrane associated in vivo (21) and OhyA is enriched in S. aureus exosomes (22), showing that OhyA is also exported from the cells where it can act on host unsaturated fatty acids. The challenge ahead is to define the mechanism(s) by which the hFA interact with the immune system. The inactivation of antimicrobial fatty acids is one clearly identified mechanism (10), but the specifics of how hFA interfere with TLR signaling and other arms of the innate immune response remain to be elucidated. PPARγ activation has an established role in regulating host lipid metabolism and inflammation (23), and hFA are known agonists of this transcriptional regulator (24). GPR40 and GRP120 are engaged by h18:1 (3, 25), but it is not obvious how these nutrient sensors (26) would affect S. aureus virulence. Most intriguing are the published reports of hFA suppression of cytokine formation in response to lipopolysaccharide (LPS) and Toll-like receptor (TLR) activation (2–5). More work is needed to define the step(s) in the TLR signaling pathway modulated by hFA.
ACKNOWLEDGMENTS
We thank Amy Iverson and Aaron Poole for assistance with murine infection models and Pam Jackson for strain construction. This work was supported by National Institutes of Health grant GM034496 (C.O.R.), Cancer Center Support Grant CA21765, and the American Lebanese Syrian Associated Charities. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We declare no conflicts of interest and no competing financial interests.
Footnotes
Supplemental material is available online only.
Contributor Information
Charles O. Rock, Email: charles.rock@stjude.org.
Christopher N. LaRock, Emory University School of Medicine
REFERENCES
- 1.Ogawa J, Kishino S, Ando A, Sugimoto S, Mihara K, Shimizu S. 2005. Production of conjugated fatty acids by lactic acid bacteria. J Biosci Bioeng 100:355–364. doi: 10.1263/jbb.100.355. [DOI] [PubMed] [Google Scholar]
- 2.Ikeguchi S, Izumi Y, Kitamura N, Kishino S, Ogawa J, Akaike A, Kume T. 2018. Inhibitory effect of the gut microbial linoleic acid metabolites, 10-oxo-trans-11-octadecenoic acid and 10-hydroxy-cis-12-octadecenoic acid, on BV-2 microglial cell activation. J Pharmacol Sci 138:9–15. doi: 10.1016/j.jphs.2018.06.015. [DOI] [PubMed] [Google Scholar]
- 3.Miyamoto J, Mizukure T, Park SB, Kishino S, Kimura I, Hirano K, Bergamo P, Rossi M, Suzuki T, Arita M, Ogawa J, Tanabe S. 2015. A gut microbial metabolite of linoleic acid, 10-hydroxy-cis-12-octadecenoic acid, ameliorates intestinal epithelial barrier impairment partially via GPR40-MEK-ERK pathway. J Biol Chem 290:2902–2918. doi: 10.1074/jbc.M114.610733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaikiri H, Miyamoto J, Kawakami T, Park SB, Kitamura N, Kishino S, Yonejima Y, Hisa K, Watanabe J, Ogita T, Ogawa J, Tanabe S, Suzuki T. 2017. Supplemental feeding of a gut microbial metabolite of linoleic acid, 10-hydroxy-cis-12-octadecenoic acid, alleviates spontaneous atopic dermatitis and modulates intestinal microbiota in NC/nga mice. Int J Food Sci Nutr 68:941–951. doi: 10.1080/09637486.2017.1318116. [DOI] [PubMed] [Google Scholar]
- 5.Yang HE, Li Y, Nishimura A, Jheng HF, Yuliana A, Kitano-Ohue R, Nomura W, Takahashi N, Kim CS, Yu R, Kitamura N, Park SB, Kishino S, Ogawa J, Kawada T, Goto T. 2017. Synthesized enone fatty acids resembling metabolites from gut microbiota suppress macrophage-mediated inflammation in adipocytes. Mol Nutr Food Res 61:e17000641. doi: 10.1002/mnfr.201700064. [DOI] [PubMed] [Google Scholar]
- 6.Thammavongsa V, Kim HK, Missiakas D, Schneewind O. 2015. Staphylococcal manipulation of host immune responses. Nat Rev Microbiol 13:529–543. doi: 10.1038/nrmicro3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung GYC, Bae JS, Otto M. 2021. Pathogenicity and virulence of Staphylococcus aureus. Virulence 12:547–569. doi: 10.1080/21505594.2021.1878688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ford CA, Hurford IM, Cassat JE. 2020. Antivirulence strategies for the treatment of Staphylococcus aureus infections: a mini review. Front Microbiol 11:e632706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Radka CD, Batte JL, Frank MW, Young BM, Rock CO. 2021. Structure and mechanism of Staphylococcus aureus oleate hydratase (OhyA). J Biol Chem 296:e100252. doi: 10.1074/jbc.RA120.016818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Subramanian C, Frank MW, Batte JL, Whaley SG, Rock CO. 2019. Oleate hydratase from Staphylococcus aureus protects against palmitoleic acid, the major antimicrobial fatty acid produced by mammalian skin. J Biol Chem 294:9285–9294. doi: 10.1074/jbc.RA119.008439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drake DR, Brogden KA, Dawson DV, Wertz PW. 2008. Skin lipids. Antimicrobial lipids at the skin surface. J Lipid Res 49:4–11. doi: 10.1194/jlr.R700016-JLR200. [DOI] [PubMed] [Google Scholar]
- 12.Parsons JB, Yao J, Frank MW, Jackson P, Rock CO. 2012. Membrane disruption by antimicrobial fatty acids releases low molecular weight proteins from Staphylococcus aureus. J Bacteriol 194:5294–5304. doi: 10.1128/JB.00743-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Georgel P, Crozat K, Lauth X, Makrantonaki E, Seltmann H, Sovath S, Hoebe K, Du X, Rutschmann S, Jiang Z, Bigby T, Nizet V, Zouboulis CC, Beutler B. 2005. A toll-like receptor 2-responsive lipid effector pathway protects mammals against skin infections with gram-positive bacteria. Infect Immun 73:4512–4521. doi: 10.1128/IAI.73.8.4512-4521.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X, Franke AA. 2011. Improved LC-MS method for the determination of fatty acids in red blood cells by LC-orbitrap MS. Anal Chem 83:3192–3198. doi: 10.1021/ac103093w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kishino S, Takeuchi M, Park SB, Hirata A, Kitamura N, Kunisawa J, Kiyono H, Iwamoto R, Isobe Y, Arita M, Arai H, Ueda K, Shima J, Takahashi S, Yokozeki K, Shimizu S, Ogawa J. 2013. Polyunsaturated fatty acid saturation by gut lactic acid bacteria affecting host lipid composition. Proc Natl Acad Sci USA 110:17808–17813. doi: 10.1073/pnas.1312937110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delekta PC, Shook JC, Lydic TA, Mulks MH, Hammer ND. 2018. Staphylococcus aureus utilizes host-derived lipoprotein particles as sources of exogenous fatty acids. J Bacteriol 200:e00728-17. doi: 10.1128/JB.00728-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frank MW, Yao J, Batte JL, Gullett JM, Subramanian C, Rosch JW, Rock CO. 2020. Host fatty acid utilization by Staphylococcus aureus at the infection site. mBio 11:e00920. doi: 10.1128/mBio.00920-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fournier B, Philpott DJ. 2005. Recognition of Staphylococcus aureus by the innate immune system. Clin Microbiol Rev 18:521–540. doi: 10.1128/CMR.18.3.521-540.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brandt SL, Putnam NE, Cassat JE, Serezani CH. 2018. Innate immunity to Staphylococcus aureus: evolving paradigms in soft tissue and invasive infections. J Immunol 200:3871–3880. doi: 10.4049/jimmunol.1701574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knuefermann P, Sakata Y, Baker JS, Huang CH, Sekiguchi K, Hardarson HS, Takeuchi O, Akira S, Vallejo JG. 2004. Toll-like receptor 2 mediates Staphylococcus aureus-induced myocardial dysfunction and cytokine production in the heart. Circulation 110:3693–3698. doi: 10.1161/01.CIR.0000143081.13042.04. [DOI] [PubMed] [Google Scholar]
- 21.Malachowa N, Kohler PL, Schlievert PM, Chuang ON, Dunny GM, Kobayashi SD, Miedzobrodzki J, Bohach GA, Seo KS. 2011. Characterization of a Staphylococcus aureus surface virulence factor that promotes resistance to oxidative killing and infectious endocarditis. Infect Immun 79:342–352. doi: 10.1128/IAI.00736-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kengmo Tchoupa A, Peschel A. 2020. Staphylococcus aureus releases proinflammatory membrane vesicles to resist antimicrobial fatty acids. mSphere 5. doi: 10.1128/mSphere.00804-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varga T, Czimmerer Z, Nagy L. 2011. PPARs are a unique set of fatty acid regulated transcription factors controlling both lipid metabolism and inflammation. Biochim Biophys Acta 1812:1007–1022. doi: 10.1016/j.bbadis.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Itoh T, Fairall L, Amin K, Inaba Y, Szanto A, Balint BL, Nagy L, Yamamoto K, Schwabe JW. 2008. Structural basis for the activation of PPARg by oxidized fatty acids. Nat Struct Mol Biol 15:924–931. doi: 10.1038/nsmb.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyamoto J, Igarashi M, Watanabe K, Karaki SI, Mukouyama H, Kishino S, Li X, Ichimura A, Irie J, Sugimoto Y, Mizutani T, Sugawara T, Miki T, Ogawa J, Drucker DJ, Arita M, Itoh H, Kimura I. 2019. Gut microbiota confers host resistance to obesity by metabolizing dietary polyunsaturated fatty acids. Nat Commun 10:4007. doi: 10.1038/s41467-019-11978-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ichimura A, Hirasawa A, Hara T, Tsujimoto G. 2009. Free fatty acid receptors act as nutrient sensors to regulate energy homeostasis. Prostaglandins Other Lipid Mediat 89:82–88. doi: 10.1016/j.prostaglandins.2009.05.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download SPECTRUM01546-21_Supp_1_seq4.pdf, PDF file, 0.4 MB (403.1KB, pdf)