Abstract
The highly conserved Saccharomyces cerevisiae Rad51 protein plays a central role in both mitotic and meiotic homologous DNA recombination. Seven members of the Rad51 family have been identified in vertebrate cells, including Rad51, Dmc1, and five Rad51-related proteins referred to as Rad51 paralogs, which share 20 to 30% sequence identity with Rad51. In chicken B lymphocyte DT40 cells, we generated a mutant with RAD51B/RAD51L1, a member of the Rad51 family, knocked out. RAD51B−/− cells are viable, although spontaneous chromosomal aberrations kill about 20% of the cells in each cell cycle. Rad51B deficiency impairs homologous recombinational repair (HRR), as measured by targeted integration, sister chromatid exchange, and intragenic recombination at the immunoglobulin locus. RAD51B−/− cells are quite sensitive to the cross-linking agents cisplatin and mitomycin C and mildly sensitive to γ-rays. The formation of damage-induced Rad51 nuclear foci is much reduced in RAD51B−/− cells, suggesting that Rad51B promotes the assembly of Rad51 nucleoprotein filaments during HRR. These findings show that Rad51B is important for repairing various types of DNA lesions and maintaining chromosome integrity.
Double-strand DNA breaks (DSBs) occur during DNA replication and are produced by ionizing radiation. Since DSBs are so deleterious to the cell, it is not surprising that there are two DSB repair pathways: nonhomologous end joining (NHEJ) and homologous recombination repair (HRR). Repair of DSBs by HRR requires the presence of homologous duplex DNA elsewhere in the genome, i.e., either a homologous chromosome or, more likely, a sister chromatid. NHEJ simply acts to process and ligate broken ends without a requirement for extensive homology. These pathways are conserved from the yeast Saccharomyces cerevisiae to humans (5, 8, 9, 19, 49, 53, 64). While HRR is the primary mechanism of DSB repair in yeast, vertebrate cells use both the NHEJ and HRR pathways extensively (28, 34, 35, 44). The analysis of radiosensitive yeast mutants has revealed a number of genes involved in HRR, which comprise the RAD52 epistasis group (reviewed in references 4, 29, and 51).
Among the members of the RAD52 epistasis group, the structure and function of Rad51 have been conserved to a remarkable degree among all eukaryotes. Rad51 is structurally and functionally related to the Escherichia coli recombination protein RecA (reviewed in reference 32). The functional forms of both RecA and Rad51 are multimeric helical nucleoprotein filaments that form on single-stranded DNA ends produced at DSBs (41). These filaments are involved in the search for homologous sequence, DNA pairing, and strand exchange. Recombination intermediates produced in this way are then processed further in reactions that involve DNA synthesis, branch migration, resolution of Holliday junctions, and ligation (reviewed in reference 4). The conservation of the RAD52 epistasis group genes from yeast to vertebrate cells suggests that the basic mechanism of HRR is maintained during evolution. However, while S. cerevisiae RAD51 mutants are viable, Rad51 deficiency in vertebrate cells causes rapid chromosomal aberrations and cell death (54). One possible explanation for this lethality is that the larger size of the vertebrate genome requires more HRR activity for chromosome stability (22, 54).
RAD51 paralogs (genes related by duplication within a single genome) have been identified in many eukaryotes and constitute the Rad51-related gene family (63, 64). The completion of the S. cerevisiae genomic sequence established that four previously identified proteins, Rad51, Rad55, Rad57, and Dmc1, constitute the complete set of RecA relatives in this organism (1, 3, 7, 37, 50). Thus far, seven members of the Rad51 protein family have been identified in mammals. In human cells, Rad51 (49), Dmc1 (23), XRCC2 (15, 28, 36), XRCC3 (36, 44, 62), Rad51B (also called Rad51L1/hRec2) (2, 14, 47), Rad51C (Rad51L2) (18), and Rad51D (Rad51L3) (14, 30, 45) are highly conserved. While human Dmc1 is ∼50% identical to human Rad51, the other human Rad51 paralogs are 20 to 30% identical to human Rad51. These paralogs are less than 30% identical to each other and to yeast Rad55 and Rad57 (reviewed in reference 63). Overexpression of Rad51 in yeast partially suppresses the DNA repair defect of rad55 and rad57 mutant strains (25, 27), implying that Rad55 and Rad57 may functionally cooperate with Rad51. This idea is supported by physical interactions between Rad51 and Rad55 and between Rad55 and Rad57 (25, 27, 57). Similarly, physical interactions occur between human Rad51 and XRCC3, XRCC3 and Rad51C and between Rad51B and Rad51C (18, 36). These observations argue that Rad51 paralogs may function as Rad51 accessory factors, analogous to yeast Rad55 and Rad57. Just as Rad55 and Rad57 are important for HRR in yeast, in mammalian cells the Rad51 paralogs XRCC2 and XRCC3 have recently been shown to play an important role in radiation resistance through HRR (28, 44). Here we present evidence that the RAD51B gene is also important for HRR.
Very recently, a RAD51B knockout mutation was made in mice but embryonic lethality prevented analysis of the cellular phenotype (52). Transcription of RAD51B was induced following UV or γ irradiation, suggesting that it has a regulated response to DNA damage (42, 47). To investigate the role of Rad51B in vertebrate cells, we generated RAD51B−/− cells from the hyperrecombinogenic chicken DT40 cell line (11, 12). Comparison of the phenotype of this mutant with that of RAD54−/− DT40 cells (5) suggests that Rad51B and Rad54 have distinctly different roles in recombinational repair.
MATERIALS AND METHODS
Construction of targeting and expression vectors, gene targeting, and cell culture.
Partial cDNA clones encoding the chicken RAD51B gene were isolated from a chicken spleen cDNA library (Clontech, Palo Alto, Calif.) by low-stringency hybridization using human RAD51B cDNA as a probe (2). A genomic fragment of the RAD51B locus was PCR amplified from DT40 genomic DNA using a pair of RAD51B-specific primers (5′-CCG TAA GCA TGG GAG GAC TAG ATG GGG C and 5′-CTC GAT ACA GAT GAA TGC TAC GGG TC). The flanking portions of the loci were amplified from a DT40 genomic library constructed in lambda FIX II phage vector (kindly provided by T. Nakayama and Y. Takami, Miyazaki, Japan) using the LA-PCR kit (Takara, Kyoto, Japan) with a combination of either one of the chicken RAD51B-specific primers (5′-GCC CCA TCT AGT CCT CCC ATG CTT ACG G and 5′-GAC CCG TAG CAT TCA TCT GTA TCG AG) and the primer hybridizing with the T7 site of the FIX II phage vector. Amplified fragments were subsequently subcloned into the TOPO-pCRII cloning vector (Invitrogen, Carlsbad, Calif.). The identity of genomic clones was confirmed by sequencing and partial mapping of the clones.
To construct RAD51B targeting vectors, ∼1.8-kb and ∼2.4-kb fragments at the RAD51B locus were cloned in the pBS vector (Stratagene, La Jolla, Calif.). A unique BamHI site was generated artificially between the fragments. The bsr or puro drug resistance cassette (56) was inserted into this BamHI site. Gene targeting of these constructs was expected to disrupt two exons and delete the intervening intron, resulting in deletion of the coding sequence corresponding to published human Rad51B amino acids 137 to 173 (2). Cell culture and transfection were done as previously described (12). Before transfection, targeting vectors Rad51B-bsr and Rad51B-puro were linearized by NotI and SacI, respectively. Transfectants were selected in the presence of blastocidin-S (Calbiochem, La Jolla, Calif.) at 25 μg/ml or puromycin (Sigma, St. Louis, Mo.) at 0.5 μg/ml.
To determine gene targeting frequencies, KU70-his (58), XRCC2-his (unpublished), and OVA-his (55) vectors were linearized and transfected into wild-type and mutant cells. Genomic DNA was extracted from each selected clone and analyzed for targeting events by genomic Southern blot analysis. To construct the expression vectors, hRAD51 (49) or hRAD51B (2) cDNA was inserted into the EcoRI site of pAneo (56).
Western and Northern blot analysis.
Cells were harvested and resuspended in lysis buffer (20 mM Tris-HCl [pH 7.5], 140 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 20 μg of aprotinin per ml, 20 μg of leupeptin per ml, 1 mM phenylmethylsulfonyl fluoride, 1% NP-40). After 30 min of incubation on ice, the cell lysates were centrifuged for 15 min at 4°C. The protein concentration of the lysates was determined by the bicinchoninic acid protein assay (Pierce, Rockford, Ill.). The lysates (10 μg/lane) were separated by sodium dodecyl sulfate–12.5% polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane, and probed with anti-Rad51 antibody. The membrane was developed using horseradish peroxidase-conjugated anti-rabbit immunoglobulin G (IgG) antibody and Super Signal chemiluminescent substrate (Pierce). Total RNA was extracted using TRIZOL (Gibco-BRL, Grand Island, N.Y.) following the manufacturer's instructions. Total RNA (20 μg/lane) was separated in a formaldehyde–1.2% agarose gel, transferred to a nylon membrane, and then hybridized with a 32P-labeled RAD51B cDNA probe corresponding to the amino acid sequences shown in Fig. 1A.
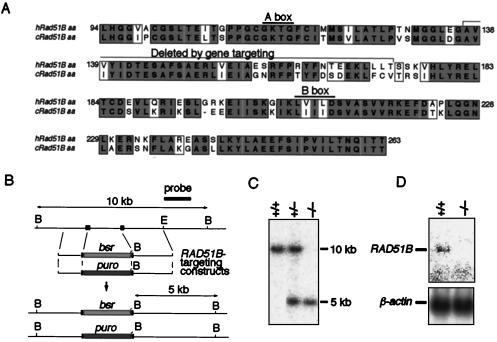
FIG. 1.
Gene targeting of RAD51B loci. (A) Amino acid (aa) sequence comparison between human and chicken Rad51B cDNAs. The Walker A and B motifs for nucleotide binding are overlined, and the sequence deleted by gene targeting is indicated. Letters in gray boxes represent identical amino acids in the two species, whereas those in open boxes represent similar (P, A, G, S, and T; E, D, N, and Q; V, I, L, and M; F, W, and Y; R, K, and H) amino acids. Numbers denote amino acid positions. (B) Schematic representation of part of the RAD51B locus, the gene disruption constructs, and the configuration of the targeted alleles. B, BamHI site; E, EcoRI site. Solid boxes indicate the positions of the exons. Only disrupted exons are indicated. (C) Southern blot analysis of BamHI-digested genomic DNA from cells with the indicated genotypes of the RAD51B gene with the probe shown in panel B. The positions and sizes of the hybridizing fragments of the wild-type and targeted loci are indicated. (D) Northern blot analysis of total RNA with a chicken RAD51B cDNA fragment as a probe. The same membrane was reprobed with the chicken β-actin fragment (12).
Flow cytometric analysis of cell viability, cell counting, and rate of Ig gene conversion.
To determine cell viability, cells were resuspended in phosphate-buffered saline containing propidium iodide at 5 μg/ml and analyzed by FACScaliber (Becton Dickinson, Mountain View, Calif.). Cells were counted in triplicate cultures using a fixed number of plastic microbeads and a FACScaliber as previously described (58). The rate of Ig gene conversion was examined as previously described (11, 59).
Analysis of chromosome aberrations and SCEs.
Chromosome and sister chromatid exchange (SCE) analysis was carried out as previously described (54, 55). To analyze mitomycin C (MMC)-induced SCEs, cells were incubated in medium containing MMC at 0.05 μg/ml for 12 h (the length of a single cell cycle of DT40 cells is ∼8 h). Colcemid at 0.1 μg/ml was added for the last 1.5 h of this incubation before harvest.
Measurement of cells surviving γ irradiation or treatment with cisplatin or MMC.
Clonogenic survival was monitored by colony formation assay as previously described (58). Briefly, following various genotoxic treatments, appropriate numbers of cells were plated into six-well cluster plates containing complete medium supplemented with 1.5% methylcellulose (Aldrich, Milwaukee, Wis.). Colony numbers were counted after 7 to 10 days, and percent survival was determined relative to numbers of colonies of untreated cells. To measure sensitivities to cisplatin (Nihon-Kayaku, Tokyo, Japan) and MMC (Kyowa-Hakkou, Tokyo, Japan), cells were incubated at 39.5°C in complete medium containing these drugs for 1 h and then washed three times with warm medium. 137Cs γ irradiation (Gammacell 40E; Nordion International, Kanata, Ontario, Canada) was done after cells were plated on six-well cluster plates.
Visualization of Rad51 foci.
Cells were harvested at the indicated time points after γ irradiation or MMC treatment. Cytospin slides were prepared using Cytospin 3 (Shandon, Pittsburgh, Pa.). Staining and visualization of Rad51 foci were performed as previously described (65).
RESULTS
Generation of RAD51B−/− mutant cells and their proliferative properties.
To construct RAD51B disruption vectors, we isolated a partial cDNA encoding chicken RAD51B (Fig. 1A). The sequence surrounding the putative ATP binding sites is highly conserved between the chicken and human Rad51B proteins (10) (Fig. 1A). However, the extremely well-conserved GXXXXGKTQ motif in the Walker A box is changed to SXXXXGKTQ in the chicken sequence in both cDNA and genomic isolates. Two RAD51B disruption constructs, RAD51B-puro and RAD51B-bsr (Fig. 1B), were expected to replace the chicken RAD51B coding sequence for amino acids 137 to 173 with the selection markers. The disruption of both RAD51B alleles in two independently isolated clones was verified by Southern and Northern blot analyses (Fig. 1C and D).
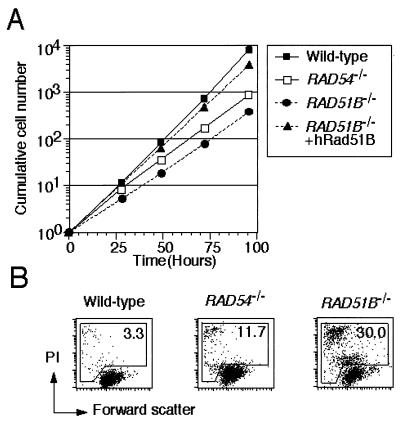
The proliferative properties of RAD51B−/− cells were monitored by growth curves and by cell cycle analysis. The growth rate of RAD51B−/− clones was significantly lower than that of wild-type cells. High expression of human Rad51B in RAD51B−/− clones restored their growth rate to a wild-type level (Fig. 2A). A pulse-chase experiment with bromodeoxyuridine showed that the length of a single cell cycle is comparable between wild-type and RAD51B−/− cells (data not shown). On the other hand, flow cytometric analysis showed that more dead cells were present in RAD51B−/− cultures than in wild-type cultures (Fig. 2B). Thus, the lower proliferation rate of RAD51B−/− cell cultures is likely caused by an elevated rate of cell death. The fraction of RAD51B−/− cells dying spontaneously during a single cell cycle was calculated to be 20% (58), which is not significantly different from the percentage of metaphase cell having chromosomal aberrations (Table 1). The plating efficiencies of cells in methylcellulose plates were 100% for wild-type cells and ∼50% for RAD51B−/− cells.
FIG. 2.
Growth rate and viability of RAD51B−/− cells. (A) Growth curves of cells of the indicated genotypes. Means ± standard deviations of triplicate cultures are shown. (B) The level of spontaneous cell death was assessed by flow cytometric analysis of propidium iodide uptake and forward scatter representing cell size. The values shown are percentages of dead (propidium iodide-bright and propidium iodide-dim) cells.
TABLE 1.
Spontaneous chromosomal aberrations per 100 cellsa
| Cell clone | Chromatid
|
Chromosome
|
Exchange | Total ± SE | ||
|---|---|---|---|---|---|---|
| Breaks | Gaps | Breaks | Gaps | |||
| Wild type | 0 | 1.3 | 0 | 0 | 0 | 1.3 ± 0.9 |
| RAD54−/− | 0 | 1.3 | 0 | 1.3 | 0.67 | 3.3 ± 1.5 |
| RAD51B−/− | 1.3 | 6.7 | 0 | 6.7 | 0 | 15 ± 3.1 |
| RAD51B−/− + hRad51 | 0 | 1.3 | 0 | 0 | 0 | 1.3 ± 0.9 |
One hundred fifty mitotic cells were analyzed for each genotype. The total number of aberrations per cell ± the standard error was calculated as x/N ± √x/N as previously described (58).
Given the possible role of Rad51B in HRR of DSBs, cell death in RAD51B−/− cultures may be caused by a defect in the removal of DSBs that normally arise during DNA replication (reviewed in references 17, 22, and 31). To test this hypothesis, we performed chromosome analysis of metaphase-arrested cells (54). RAD51B−/− cells displayed a significantly increased level of spontaneous chromosome breaks (Table 1), which likely lead to cell death, and the mutant cells exhibited more chromosomal aberrations than did RAD54−/− cells. This difference is consistent with the slower growth rate and higher level of death of RAD51B−/− cells compared to RAD54−/− cells.
Defective homologous recombination in RAD51B−/− cells.
The spontaneous chromosomal aberrations in RAD51B−/− cells may be caused by defective HRR of replication-associated DSBs. To estimate the HR capacity of RAD51B−/− cells, we examined the rate of intragenic homologous recombination at the Ig light-chain (IgL) locus (i.e., Ig gene conversion), the efficiency of targeted integration of transfected genomic DNA constructs, and the frequency of SCE.
In a manner similar to the process of B-cell diversification in the bursa of Fabricius, DT40 cells continue to diversify their Ig loci by gene conversion with pseudogenes serving as donors. To measure gene conversion, RAD51B−/− clones were generated from a surface IgM-negative (sIgM−) variant called clone 18. This clone contains a frameshift in the rearranged V segment of its IgL locus (11, 59). Since overlapping gene conversion events leading to re-expression of sIgM can repair this mutation, we measured the percentage of sIgM+ revertants among 40 subclones of wild-type, RAD51B−/−, and RAD54−/− cells. The calculated Ig gene conversion rate was 8.3 × 10−4 for wild-type cells and 1.70 × 10−4 for HRR-deficient RAD54−/− control cells (5). Like RAD54−/− cells, RAD51B−/− cells also exhibited a significant reduction in intragenic HRR, i.e., to 3.5 × 10−4. We next measured the frequencies of targeted integration at the KU70, OVALBUMIN, and XRCC2 loci and found no targeting events in RAD51B−/− cells (Table 2). The complementation of RAD51B−/− cells with human RAD51B cDNA restored the targeting frequency, showing that Rad51B is indeed involved in HRR.
TABLE 2.
Targeted integration frequenciesa
| Targeted locus | Wild type |
RAD51B−/−
|
RAD51B−/− + hRad51B | RAD51B−/− + hRad51 | |
|---|---|---|---|---|---|
| 1 | 2 | ||||
| KU70 | 3/20 (15) | 0/82 | NDb | 12/58 (21) | ND |
| XRCC2 | 15/24 (63) | 0/37 | 0/14 | 6/21 (29) | 0/35 |
| OVALBUMIN | 28/48 (58) | 0/12 | ND | 5/14 (36) | 0/30 |
The data shown are the number of targeted clones at each locus divided by the number of drug-resistant clones analyzed. The percent frequency is in parentheses.
ND, not determined.
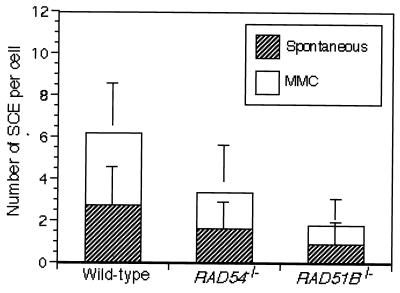
We previously showed that at least a portion of SCEs reflects postreplicational HRR of spontaneous DNA lesions and involves crossing over between sister duplexes (55). RAD51B−/− cells, together with control RAD54−/− cells, exhibited a significant reduction in the spontaneous level of SCEs (Fig. 3). A similar reduction in SCE levels was observed after treatment of the RAD51B−/− cells with MMC, which is known to stimulate SCE.
FIG. 3.
Level of SCE per cell. Cells were incubated with or without MMC (0.05 μg/ml) for 12 h and treated with colcemid during the last 1.5 h of this incubation to enrich mitotic cells. One hundred fifty cells were analyzed in each preparation. Error bars represent standard errors calculated as previously described (58).
Rad51B promotes the formation of Rad51 foci after genotoxic treatment.
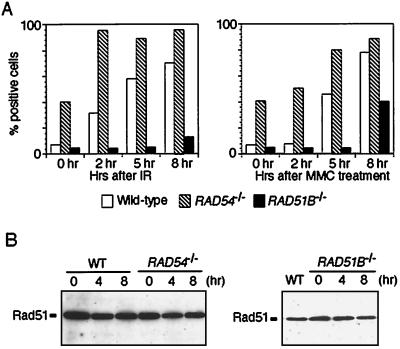
To further assess the role of Rad51B in recombinational repair, we analyzed Rad51 focus formation. Immunostaining of vertebrate cell nuclei has shown that they form visible complexes of Rad51 during meiotic recombination and mitotic DNA repair (6, 16, 21, 33, 38, 61, 66). It is believed that Rad51 foci represent nucleoprotein filaments engaged in recombinational repair (46). We exposed cycling wild-type and RAD51B−/− DT40 clones to γ rays and MMC and then immunostained the cells with anti-Rad51 serum. We found that the formation of Rad51 foci was severely impaired in RAD51B−/− cells following γ irradiation and MMC treatment (Fig. 4 and 5A). Western blot analysis of the Rad51 protein revealed normal steady-state levels of Rad51 in RAD51B−/− DT40 cells after the genotoxic treatments (Fig. 5B). Thus, RAD51B−/− cells are defective in the damage-induced redistribution of Rad51 within the nucleus.
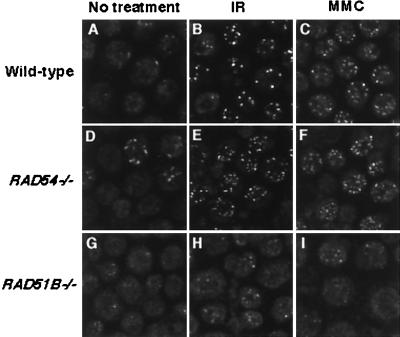
FIG. 4.
Immunofluorescent visualization of Rad51 subnuclear foci. Wild-type (A to C), RAD54−/− (D to F), and RAD51B−/− (G to I) cells were analyzed 5 h after genotoxic treatments. Cells were treated with either 8 Gy of γ radiation (IR) (B, E, and H) or 500 ng of MMC per ml (for 1 h) (C, F, and I).
FIG. 5.
Induction of Rad51 focus-positive cells after genotoxic treatments. (A) Cells with the indicated genotypes were analyzed at the indicated time points after either γ irradiation (IR; 8 Gy) or MMC treatment (500 ng/ml, 1 h). A cell containing more than four distinct foci was scored as positive. Each bar represents the result of scoring at least 100 cells. (B) Rad51 protein expression before and after exposure to 8 Gy of γ irradiation. At the indicated time points, total cell lysates were prepared and the same amount of protein was loaded into each lane. WT, wild type.
As a control, Rad51 focus formation was assayed in RAD54−/− cells. Interestingly, we observed clear differences between RAD51B−/− and RAD54−/− cells with respect to Rad51 focus formation. First, the percentage of RAD54−/− cells containing spontaneous Rad51 foci was about fivefold higher than for wild-type and RAD51B−/− cells (Fig. 4 and 5A). Second, the absence of the Rad54 protein did not abrogate the induction of Rad51 foci; in fact, more Rad51 foci were detected in RAD54−/− cells than in wild-type cells (Fig. 4 and 5A). A simple explanation for these findings is that in the absence of Rad54, HRR is arrested immediately after the formation of nucleoprotein filaments, resulting in their accumulation. This explanation is consistent with a role for Rad54 protein in the synaptic phase of the homologous DNA pairing reaction (43) and is also supported by the presence of Rad51 foci during meiosis in Rad54-deficient yeast (20). In contrast, Rad54 is required for Rad51 focus formation in murine embryonic stem cells (60). Thus, the role of Rad54 in Rad51 focus formation may differ among species and/or among cell lines.
Increased sensitivities of RAD51B−/− cells to various genotoxic treatments.
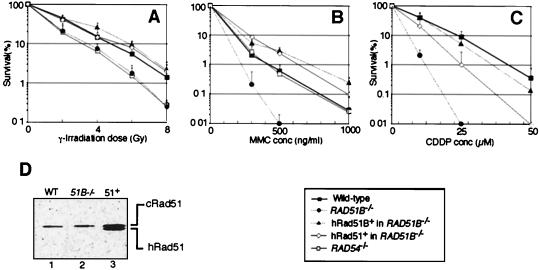
The DNA repair capacity of RAD51B−/− cells was analyzed in colony survival assays. As a control for these experiments, we included HRR-deficient RAD54−/− DT40 cells (5). We examined the cellular sensitivity to ionizing radiation and to the DNA cross-linking agents MMC and cisplatin. RAD51B−/− and RAD54−/− cells were comparable in radiosensitivity (Fig. 6A), whereas RAD51B−/− cells showed higher sensitivity to both MMC and cisplatin compared with RAD54−/− or wild-type cells (Fig. 6B and C). The elevated level of chromatid breaks induced by γ irradiation in RAD51B−/− cells suggests that their defective DSB repair (Table 3) causes their elevated radiosensitivity. Human Rad51B expression in RAD51B−/− cells substantially increased resistance to the three agents (Fig. 6A to C).
FIG. 6.
Sensitivity of clones of the indicated genotypes to DNA-damaging agents. The fractions of the surviving colonies after the indicated treatment of cells compared to nontreated controls of the same genotype are shown on the y axis on a logarithmic scale. Panels: A, γ irradiation; B, MMC; C, cisplatin (CDDP). The dose of radiation treatment and concentrations (conc) of MMC and cisplatin are displayed on the x axis on a linear scale in each graph. The data shown are means ± standard deviations of at least three separate experiments. The cisplatin sensitivity data of RAD54−/− cells are not shown here but were previously described (65). (D) Western blot analysis with rabbit anti-Rad51 serum showing the amounts of endogenous chicken Rad51 and human Rad51 proteins derived from the transgene. Lanes: 1, wild-type (WT) DT40; 2, RAD51B−/− cells; 3, hRad51-expressing RAD51B−/− cells.
TABLE 3.
Ionizing radiation-induced chromosomal aberrations per 100 mitotic cellsa
| Strain and colcemid treatment time (h) | Chromatid
|
Chromosome
|
Exchange(s) | Total ± SE | ||
|---|---|---|---|---|---|---|
| Breaks | Gaps | Breaks | Gaps | |||
| Wild type | ||||||
| 0–3 | 17 | 1.3 | 2.7 | 4.7 | 0.7 | 26 ± 4 |
| 3–6 | 4.7 | 0.7 | 1.3 | 4.7 | 0 | 11 ± 3 |
| 6–9 | 1.3 | 0.7 | 7.3 | 2.7 | 1.3 | 13 ± 3 |
| 9–12 | 0 | 1.3 | 3.3 | 4 | 2.7 | 11 ± 3 |
| RAD51B−/− | ||||||
| 0–3 | 70 | 9 | 16 | 8 | 5 | 108 ± 10 |
| 3–6 | 4 | 1 | 11.3 | 9.3 | 1.3 | 27 ± 5 |
| 6–9 | 1.3 | 0 | 6.7 | 10 | 1.3 | 19 ± 4 |
| 9–12 | 2 | 0 | 13.3 | 8.7 | 0 | 24 ± 5 |
One hundred fifty mitotic cells were analyzed for each genotype after treatment with 2 Gy of ionizing radiation. The data shown are the numbers of chromosomal aberrations induced by radiation treatment minus the number of spontaneous chromosomal aberrations. Cells were treated with colcemid for the indicated periods after irradiation. The number of total aberrations per cell ± the standard error was calculated as described in Table 1, footnote a. The data for wild-type cells are from our previous study (58).
In yeast, the overexpression of Rad51 partially suppresses the γ-ray sensitivity of rad55 and rad57 mutant strains (25, 27). Therefore, we overexpressed human Rad51 in RAD51B−/− cells and examined various phenotypes. Remarkably, although the expression level of the human Rad51 transgene was comparable to that of endogenous chicken Rad51 (Fig. 6D), the sensitivity of the RAD51B−/− cells to γ rays and MMC was restored to near wild-type levels (Fig. 6A and B). Similarly, human Rad51 expression partially suppressed cisplatin sensitivity (Fig. 6C). Interestingly, expression of human Rad51 ameliorated the chromosomal aberrations (Table 1) and cell death in the mutant (data not shown) but the defect in gene targeting efficiency was not suppressed (Table 2).
To further investigate the role of Rad51B in recombinational repair, we analyzed chromosomal aberrations following genotoxic treatments (Table 3). RAD51B−/− cells exhibited higher levels of chromosomal aberrations than wild-type cells after ionizing radiation, as did RAD54−/− cells (58). Thus, the HRR pathway(s) involving Rad51B and Rad54 plays an important role in repairing radiation-induced DSBs.
DISCUSSION
Requirement for Rad51B in HRR and Rad51 focus formation.
Our data show that Rad51B is involved in recombinational repair of diverse types of DNA lesions, including spontaneous DSBs that produce chromosome aberrations. Three types of recombinational processes were impaired by Rad51B deficiency: intrachromosomal gene conversion, gene targeting, and SCE. The involvement of two other Rad51 paralogs, XRCC2 and XRCC3, in HRR was suggested by the increased radiosensitivity of hamster xrcc2/3 mutants (36, 62), the absence of Rad51 focus formation in XRCC3-deficient cells (6), and the interaction between XRCC3 and Rad51 (36). More recently, this involvement was demonstrated directly using assays that detect intrachromosomal HRR (28, 44). By analogy, we predicted that the Rad51B, Rad51C, and Rad51D proteins should have similar roles. Our findings constitute the first direct genetic evidence for a role of the Rad51B protein in HRR in vertebrate cells.
The appearance of Rad51 nuclear foci is strongly correlated with active DNA repair and likely reflects Rad51 nucleoprotein filament formation (6, 16, 21, 33, 46). In keeping with RAD51B participation in HRR, we have shown that Rad51B is required for Rad51 focus formation in response to ionizing radiation or MMC treatment. However, it should be noted that the absence of Rad51 foci does not necessarily imply the absence of Rad51 nucleoprotein filaments. A minimum nucleoprotein filament size is necessary for visualization of a focus (46), so presumably sufficient Rad51 might still be assembled to repair some spontaneous DNA lesions without producing obvious foci.
Role of Rad51B in HRR and gene targeting.
Protein-protein interactions between Rad51 and Rad55 and between Rad55 and Rad57 suggest that these molecules act in multiprotein complexes (25, 27, 57). The repair defects of rad55/57 mutants are partially suppressed by the overexpression of RAD51 but not vice versa (25, 27). Also, biochemical analysis suggests that the Rad55/57 heterodimer acts as a cofactor to promote Rad51-single-stranded DNA nucleoprotein filament assembly in the presence of replication protein A (57). While direct physical interaction of Rad51B with human Rad51 has not been detected, Rad51B could still cooperate with human Rad51, through its association with Rad51C and XRCC3 (18), to form nucleoprotein filaments. This idea is consistent with the defective Rad51 focus formation in RAD51B−/− cells. A similar situation applies in yeast, where mutations in RAD55 and RAD57 prevent the appearance of Rad51 foci during meiosis (20). These observations suggest that the Rad51 paralogs either help assemble Rad51 into oligomeric complexes or stabilize the complexes once formed. Likewise, in vertebrates, Rad51B may lead to more extensive nucleoprotein filament formation by Rad51 and, as a result, more efficient homology searching.
Although our RAD51B−/− cells expressing human Rad51 were able to cope well with DNA lesions from genotoxic agents, the modest increase in the level of Rad51 (chicken plus human) expression in this transformant (Fig. 6D) was insufficient to fully restore gene targeting efficiency in these cells (Table 2). Perhaps targeted integration requires longer nucleoprotein filaments than can be formed in this transformant. In this case, the repair of damage may be more efficient than gene targeting because sister chromatids are in close proximity whereas gene targeting requires a search for homology throughout the nucleus.
Role of HRR in removing DNA cross-links.
The cross-linking agents cisplatin and MMC form covalent adducts with many biological molecules, but their principal target is DNA. They form a variety of DNA adducts: intrastrand cross-links, interstrand cross-links, and protein-DNA cross-links (67). Yeast mutants deficient in nucleotide excision repair, HRR, or the postreplication repair pathway show increased sensitivity to cisplatin, indicating that these repair pathways are involved in removal of the DNA lesions it causes (24). The phenotype of sensitivity to cross-linking agents, displayed by our Rad51B-deficient cells, shows that some types of DNA lesions induced by cross-linking agents are repaired by HRR involving Rad51B. Since Rad54-deficient DT40 cells show less MMC sensitivity than Rad51B-deficient cells (Fig. 6) (5), it is possible that Rad54 is required for removal of only a subset of the damage that is removed by Rad51B. Alternatively, a recently identified relative of Rad54, Rad54B (26), might have a role that overlaps that of Rad54.
Chromosome instability and cancer.
The occurrence of HRR during the vertebrate mitotic cell cycle is suggested by the appearance of Rad51 foci in S phase (61) and by spontaneous SCE. SCEs are mediated at least partially by HRR (55) and occur at a frequency of about three exchanges per cell cycle in mammalian cells. Additionally, the presence of excessive chromosome breaks in RAD51−/− (54) and RAD54−/− (5) chicken cells indicates that HRR plays an essential role in the repair of potentially lethal chromosomal breaks that likely occur during DNA replication (54). Here we show that Rad51B deficiency causes elevated frequencies of spontaneous chromosomal aberrations, indicating that Rad51B also plays a major role in the maintenance of genomic stability. Our cell survival data (Fig. 6) suggest that Rad51B acts by promoting Rad51's function. The properties of the xrcc2 and xrcc3 hamster mutants (36, 42) suggest that the XRCC2 and XRCC3 proteins have roles similar to that of Rad51B. These mutants show most of the phenotypes of rad51b, including chromosome instability. RAD51B was recently knocked out in mice, but no cellular phenotype has yet been described since embryogenesis was arrested at about day 5 and embryonic cells in culture did not proliferate (52).
Every human genetic disorder that features chromosomal breakage is associated with an increased incidence of cancer (40). Rapidly accumulating evidence suggests that defects in HRR play a significant role in promoting tumorigenesis through genomic instability. For example, chromosomal translocations involving RAD51B are frequently observed in uterine leiomyoma (48). Mutations of RAD54, or its homolog RAD54B, are observed in lymphoma, colon cancer, and breast cancer (26, 39). The breast cancer-linked Brca2 protein interacts with Rad51 in vivo (16) and is required for the formation of visible Rad51 focus formation (66). Since RAD51B-deficient cells exhibit elevated chromosomal breakage, it will be of interest to screen tumors for mutations in the RAD51B locus.
ACKNOWLEDGMENTS
We thank M. Hashishin, Y. Sato, O. Koga, and M. Hirao for excellent technical assistance and H. Kurumizaka and T. Shibata (Riken, Wako, Japan), S. C. West (Imperial Cancer Research Fund, South Mimms, United Kingdom), and D. Schild (Lawrence Berkeley National Laboratory, Berkeley, Calif.) for discussion and critical reading of the manuscript.
C. M. is the recipient of a JSPS postdoctoral fellowship. The Bayer-Chair Department of Molecular Immunology and Allergy is supported by Bayer Yakuhin, Kyoto, Japan. This work was supported in part by CREST, JST (Saitama, Japan); a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science and Culture of Japan; and grants from The Mochida Memorial Foundation for Medical and Pharmaceutical Research and from The Uehara Memorial Foundation. A portion of this work was prepared under the auspices of the U.S. Department of Energy under contract W-7405-ENG-48 (L.H.T.).
REFERENCES
- 1.Aboussekhra A, Chanet R, Adjiri A, Fabre F. Semidominant suppressors of Srs2 helicase mutations of Saccharomyces cerevisiae map in the RAD51 gene, whose sequence predicts a protein with similarities to procaryotic RecA proteins. Mol Cell Biol. 1992;12:3224–3234. doi: 10.1128/mcb.12.7.3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albala J S, Thelen M P, Prange C, Fan W, Christensen M, Thompson L H, Lennon G G. Identification of a novel human RAD51 homolog, RAD51B. Genomics. 1997;46:476–479. doi: 10.1006/geno.1997.5062. [DOI] [PubMed] [Google Scholar]
- 3.Basile G, Aker M, Mortimer R K. Nucleotide sequence and transcriptional regulation of the yeast recombinational repair gene RAD51. Mol Cell Biol. 1992;12:3235–3246. doi: 10.1128/mcb.12.7.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumann P, West S C. Role of the human RAD51 protein in homologous recombination and double-stranded-break repair. Trends Biochem Sci. 1998;23:247–251. doi: 10.1016/s0968-0004(98)01232-8. [DOI] [PubMed] [Google Scholar]
- 5.Bezzubova O Y, Silbergleit A, Yamaguchi-Iwai Y, Takeda S, Buerstedde J M. Reduced X-ray resistance and homologous recombination frequencies in a RAD54−/− mutant of the chicken DT40 cell line. Cell. 1997;89:185–193. doi: 10.1016/s0092-8674(00)80198-1. [DOI] [PubMed] [Google Scholar]
- 6.Bishop D K, Ear U, Bhattacharyya A, Calderone C, Beckett M, Weichselbaum R R, Shinohara A. Xrcc3 is required for assembly of Rad51 complexes in vivo. J Biol Chem. 1998;273:21482–21488. doi: 10.1074/jbc.273.34.21482. [DOI] [PubMed] [Google Scholar]
- 7.Bishop D K, Park D, Xu L, Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- 8.Boulton S J, Jackson S P. Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res. 1996;24:4639–4648. doi: 10.1093/nar/24.23.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boulton S J, Jackson S P. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 1996;15:5093–5103. [PMC free article] [PubMed] [Google Scholar]
- 10.Brendel V, Brocchieri L, Sandler S J, Clark A J, Karlin S. Evolutionary comparisons of RecA-like proteins across all major kingdoms of living organisms. J Mol Evol. 1997;44:528–541. doi: 10.1007/pl00006177. [DOI] [PubMed] [Google Scholar]
- 11.Buerstedde J M, Reynaud C A, Humphries E H, Olson W, Ewert D L, Weill J C. Light chain gene conversion continues at high rate in an ALV-induced cell line. EMBO J. 1990;9:921–927. doi: 10.1002/j.1460-2075.1990.tb08190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buerstedde J M, Takeda S. Increased ratio of targeted to random integration after transfection of chicken B cell lines. Cell. 1991;67:179–188. doi: 10.1016/0092-8674(91)90581-i. [DOI] [PubMed] [Google Scholar]
- 13.Caldecott K, Jeggo P. Cross-sensitivity of γ-ray-sensitive hamster mutants to cross-linking agents. Mutat Res. 1991;255:111–121. doi: 10.1016/0921-8777(91)90046-r. [DOI] [PubMed] [Google Scholar]
- 14.Cartwright R, Dunn A M, Simpson P J, Tambini C E, Thacker J. Isolation of novel human and mouse genes of the recA/RAD51 recombination-repair gene family. Nucleic Acids Res. 1998;26:1653–1659. doi: 10.1093/nar/26.7.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cartwright R, Tambini C E, Simpson P J, Thacker J. The XRCC2 DNA repair gene from human and mouse encodes a novel member of the recA/RAD51 family. Nucleic Acids Res. 1998;26:3084–3089. doi: 10.1093/nar/26.13.3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J, Silver D P, Walpita D, Cantor S B, Gazdar A F, Tomlinson G, Couch F J, Weber B L, Ashley T, Livingston D M, Scully R. Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol Cell. 1998;2:317–328. doi: 10.1016/s1097-2765(00)80276-2. [DOI] [PubMed] [Google Scholar]
- 17.Cox M M. A broadening view of recombinational DNA repair in bacteria. Genes Cells. 1998;3:65–78. doi: 10.1046/j.1365-2443.1998.00175.x. [DOI] [PubMed] [Google Scholar]
- 18.Dosanjh M K, Collins D W, Fan W, Lennon G G, Albala J S, Shen Z, Schild D. Isolation and characterization of RAD51C, a new human member of the RAD51 family of related genes. Nucleic Acids Res. 1998;26:1179–1184. doi: 10.1093/nar/26.5.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Essers J, Hendriks R W, Swagemakers S M A, Troelstra C, de Wit J, Bootsma D, Hoeijmakers J H J, Kanaar R. Disruption of mouse RAD54 reduces ionizing radiation resistance and homologous recombination. Cell. 1997;89:195–204. doi: 10.1016/s0092-8674(00)80199-3. [DOI] [PubMed] [Google Scholar]
- 20.Gasior S L, Wong A K, Kora Y, Shinohara A, Bishop D K. Rad52 associates with RPA and functions with rad55 and rad57 to assemble meiotic recombination complexes. Genes Dev. 1998;12:2208–2221. doi: 10.1101/gad.12.14.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haaf T, Golub E I, Reddy G, Radding C M, Ward D C. Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc Natl Acad Sci USA. 1995;92:2298–2302. doi: 10.1073/pnas.92.6.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haber J E. DNA recombination: the replication connection. Trends Biochem Sci. 1999;24:271–275. doi: 10.1016/s0968-0004(99)01413-9. [DOI] [PubMed] [Google Scholar]
- 23.Habu T, Taki T, West A, Nishimune Y, Morita T. The mouse and human homologs of DMC1, the yeast meiosis-specific homologous recombination gene, have a common unique form of exon-skipped transcript in meiosis. Nucleic Acids Res. 1996;24:470–477. doi: 10.1093/nar/24.3.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hartwell L H, Szankasi P, Roberts C J, Murray A W, Friend S H. Integrating genetic approaches into the discovery of anticancer drugs. Science. 1997;278:1064–1068. doi: 10.1126/science.278.5340.1064. [DOI] [PubMed] [Google Scholar]
- 25.Hays S L, Firmenich A A, Berg P. Complex formation in yeast double-strand break repair: participation of Rad51, Rad52, Rad55, and Rad57 proteins. Proc Natl Acad Sci USA. 1995;92:6925–6929. doi: 10.1073/pnas.92.15.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hiramoto T, Nakanishi T, Sumiyoshi T, Fukuda T, Matsuura S, Tauchi H, Komatsu K, Shibasaki Y, Inui H, Watatani M, Yasutomi M, Sumii K, Kajiyama G, Kamada N, Miyagawa K, Kamiya K. Mutations of a novel human RAD54 homologue, RAD54B, in primary cancer. Oncogene. 1999;18:3422–3426. doi: 10.1038/sj.onc.1202691. [DOI] [PubMed] [Google Scholar]
- 27.Johnson R D, Symington L S. Functional differences and interactions among the putative RecA homologs Rad51, Rad55, and Rad57. Mol Cell Biol. 1995;15:4843–4850. doi: 10.1128/mcb.15.9.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson R D, Liu N, Jasin M. Mammalian XRCC2 promotes the repair of DNA double-strand breaks by homologous recombination. Nature. 1999;401:397–399. doi: 10.1038/43932. [DOI] [PubMed] [Google Scholar]
- 29.Kanaar R, Hoeijmakers J H, van Gent D C. Molecular mechanisms of DNA double strand break repair. Trends Cell Biol. 1998;8:483–489. doi: 10.1016/s0962-8924(98)01383-x. [DOI] [PubMed] [Google Scholar]
- 30.Kawabata M, Saeki K. Sequence analysis and expression of a novel mouse homolog of Escherichia coli recA gene. Biochim Biophys Acta. 1998;1398:353–358. doi: 10.1016/s0167-4781(98)00061-x. [DOI] [PubMed] [Google Scholar]
- 31.Kogoma T. Stable DNA replication: interplay between DNA replication, homologous recombination, and transcription. Microbiol Mol Biol Rev. 1997;61:212–238. doi: 10.1128/mmbr.61.2.212-238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kowalczykowski S C. In vitro reconstitution of homologous recombination reactions. Experientia. 1994;50:204–215. doi: 10.1007/BF01924003. [DOI] [PubMed] [Google Scholar]
- 33.Li M J, Maizels N. Nuclear Rad51 foci induced by DNA damage are distinct from Rad51 foci associated with B cell activation and recombination. Exp Cell Res. 1997;237:93–100. doi: 10.1006/excr.1997.3761. [DOI] [PubMed] [Google Scholar]
- 34.Liang F, Han M, Romanienko P J, Jasin M. Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc Natl Acad Sci USA. 1998;95:5172–5177. doi: 10.1073/pnas.95.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin Y, Lukacsovich T, Waldman A. Multiple pathways for repair of DNA double-strand breaks in mammalian chromosomes. Mol Cell Biol. 1999;19:8353–8360. doi: 10.1128/mcb.19.12.8353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu N, Lamerdin J E, Tebbs R S, Schild D, Tucker J D, Shen M R, Brookman K W, Siciliano M J, Walter C A, Fan W, Narayama L S, Zhou Z-Q, Adamson A W, Sorensen K J, Chen D J, Jones N J, Thompson L H. XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA crosslinks and other damages. Mol Cell. 1998;1:783–793. doi: 10.1016/s1097-2765(00)80078-7. [DOI] [PubMed] [Google Scholar]
- 37.Lovett S T. Sequence of the RAD55 gene of Saccharomyces cerevisiae: similarity of RAD55 to prokaryotic RecA and other RecA-like proteins. Gene. 1994;142:103–106. doi: 10.1016/0378-1119(94)90362-x. [DOI] [PubMed] [Google Scholar]
- 38.Maser R S, Monsen K J, Nelms B E, Petrini J H. hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Mol Cell Biol. 1997;17:6087–6096. doi: 10.1128/mcb.17.10.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsuda M, Miyagawa K, Takahashi M, Fukuda T, Kataoka T, Asahara T, Inui H, Watatani M, Yasutomi M, Kamada N, Dohi K, Kamiya K. Mutations in the RAD54 recombination gene in primary cancers. Oncogene. 1999;18:3427–3430. doi: 10.1038/sj.onc.1202692. [DOI] [PubMed] [Google Scholar]
- 40.Meyn M S. Chromosome instability syndromes: lessons for carcinogenesis. Curr Top Microbiol Immunol. 1997;221:71–148. doi: 10.1007/978-3-642-60505-5_6. [DOI] [PubMed] [Google Scholar]
- 41.Ogawa T, Shinohara A, Nabetani A, Ikeya T, Yu X, Egelman E H, Ogawa H. RecA-like recombination proteins in eukaryotes: functions and structures of RAD51 genes. Cold Spring Harb Symp Quant Biol. 1993;58:567–576. doi: 10.1101/sqb.1993.058.01.063. [DOI] [PubMed] [Google Scholar]
- 42.Peng L, Rice M C, Kmiec E B. Analysis of the human RAD51L1 promoter region and its activation by UV light. Genomics. 1998;54:529–541. doi: 10.1006/geno.1998.5536. [DOI] [PubMed] [Google Scholar]
- 43.Petukhova G, Stratton S, Sung P. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature. 1998;393:91–94. doi: 10.1038/30037. [DOI] [PubMed] [Google Scholar]
- 44.Pierce A J, Johnson R D, Thompson L H, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pittman D L, Weinberg L R, Schimenti J C. Identification, characterization, and genetic mapping of Rad51d, a new mouse and human RAD51/RecA-related gene. Genomics. 1998;49:103–111. doi: 10.1006/geno.1998.5226. [DOI] [PubMed] [Google Scholar]
- 46.Raderschall E, Golub E I, Haaf T. Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage. Proc Natl Acad Sci USA. 1999;96:1921–1926. doi: 10.1073/pnas.96.5.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rice M C, Smith S T, Bullrich F, Havre P, Kmiec E B. Isolation of human and mouse genes based on homology to REC2, a recombinational repair gene from the fungus Ustilago maydis. Proc Natl Acad Sci USA. 1997;94:7417–7422. doi: 10.1073/pnas.94.14.7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schoenmakers E F, Huysmans C, Van de Ven W J. Allelic knockout of novel splice variants of human recombination repair gene RAD51B in t(12;14) uterine leiomyomas. Cancer Res. 1999;59:19–23. [PubMed] [Google Scholar]
- 49.Shinohara A, Ogawa H, Matsuda Y, Ushio N, Ikeo K, Ogawa T. Cloning of human, mouse and fission yeast recombination genes homologous to RAD51 and recA. Nat Genet. 1993;4:239–243. doi: 10.1038/ng0793-239. [DOI] [PubMed] [Google Scholar]
- 50.Shinohara A, Ogawa H, Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 51.Shinohara A, Ogawa T. Homologous recombination and the roles of double-strand breaks. Trends Biochem Sci. 1995;20:387–391. doi: 10.1016/s0968-0004(00)89085-4. [DOI] [PubMed] [Google Scholar]
- 52.Shu Z, Smith S, Wang L, Rice M C, Kmiec E B. Disruption of muREC2/RAD51L1 in mice results in early embryonic lethality which can be partially rescued in a p53−/− background. Mol Cell Biol. 1999;19:8686–8693. doi: 10.1128/mcb.19.12.8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Siede W, Friedl A A, Dianova I, Eckardt-Schupp F, Friedberg E C. The Saccharomyces cerevisiae Ku autoantigen homologue affects radiosensitivity only in the absence of homologous recombination. Genetics. 1996;142:91–102. doi: 10.1093/genetics/142.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sonoda E, Sasaki M S, Buerstedde J-M, Bezzubova O, Shinohara A, Ogawa H, Takata M, Yamaguchi-Iwai Y, Takeda S. Rad51 deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 1998;17:598–608. doi: 10.1093/emboj/17.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sonoda E, Sasaki M S, Morrison C, Yamaguchi-Iwai Y, Takata M, Takeda S. Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol Cell Biol. 1999;19:5166–5169. doi: 10.1128/mcb.19.7.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sugawara H, Kurosaki M, Takata M, Kurosaki T. Genetic evidence for involvement of type 1, type 2 and type 3 inositol 1,4,5-trisphosphate receptors in signal transduction through the B-cell antigen receptor. EMBO J. 1997;16:3078–3088. doi: 10.1093/emboj/16.11.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sung P. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 1997;11:1111–1121. doi: 10.1101/gad.11.9.1111. [DOI] [PubMed] [Google Scholar]
- 58.Takata M, Sasaki M S, Sonoda E, Morrison C, Hashimoto M, Utsumi H, Yamaguchi-Iwai Y, Shinohara A, Takeda S. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 1998;17:5497–5508. doi: 10.1093/emboj/17.18.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takeda S, Masteller E L, Thompson C B, Buerstedde J M. RAG-2 expression is not essential for chicken immunoglobulin gene conversion. Proc Natl Acad Sci USA. 1992;89:4023–4027. doi: 10.1073/pnas.89.9.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tan T L R, Essers J, Citterio E, Swagemakers S M A, De Wit J, Benson F E, Hoeijmakers J H J, Kanaar R. Mouse Rad54 affects DNA conformation and DNA-damage-induced Rad51 foci formation. Curr Biol. 1999;9:325–328. doi: 10.1016/s0960-9822(99)80142-0. [DOI] [PubMed] [Google Scholar]
- 61.Tashiro S, Kotomura N, Shinohara A, Tanaka K, Ueda K, Kamada N. S phase specific formation of the human Rad51 protein nuclear foci in lymphocytes. Oncogene. 1996;12:2165–2170. [PubMed] [Google Scholar]
- 62.Tebbs R S, Zhao Y, Tucker J D, Scheerer J B, Siciliano M J, Hwang M, Liu N, Legerski R J, Thompson L H. Correction of chromosomal instability and sensitivity to diverse mutagens by a cloned cDNA of the XRCC3 DNA repair gene. Proc Natl Acad Sci USA. 1995;92:6354–6358. doi: 10.1073/pnas.92.14.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thacker J. A surfeit of RAD51-like genes? Trends Genet. 1999;15:166–168. doi: 10.1016/s0168-9525(99)01733-3. [DOI] [PubMed] [Google Scholar]
- 64.Thompson L H, Schild D. The contribution of homologous recombination in preserving genome integrity in mammalian cells. Biochimie. 1999;81:87–105. doi: 10.1016/s0300-9084(99)80042-x. [DOI] [PubMed] [Google Scholar]
- 65.Yamaguchi-Iwai Y, Sonoda E, Buerstedde J-M, Bezzubova O, Morrison C, Takata M, Shinohara A, Takeda S. Homologous recombination, but not DNA repair, is reduced in vertebrate cells deficient in RAD52. Mol Cell Biol. 1998;18:6430–6435. doi: 10.1128/mcb.18.11.6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yuan S S, Lee S Y, Chen G, Song M, Tomlinson G E, Lee E Y. BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res. 1999;59:3547–3551. [PubMed] [Google Scholar]
- 67.Zamble D B, Lippard S J. Cisplatin and DNA repair in cancer chemotherapy. Trends Biochem Sci. 1995;20:435–439. doi: 10.1016/s0968-0004(00)89095-7. [DOI] [PubMed] [Google Scholar]