Severe acute respiratory coronavirus 2 (SARS-CoV-2) pandemic presents new scientific and scale-up challenges for diagnostic capabilities worldwide. The gold standard diagnostic for SARS-CoV-2 infection is an RT-qPCR which targets the SARS-CoV-2 genome, an assay that has now been performed on millions of patient specimens worldwide regardless of symptomatic status. Zhang et al. (1) suggest the possibility that the SARS-CoV-2 N gene could integrate into host cell DNA through the action of the LINE1 retrotransposon, a mobile element potentially active in human somatic cells, thereby calling into question the veracity of N gene−based RT-qPCR for detection of SARS-CoV-2 infection. Multiple studies and a peer-reviewed work (2) have challenged Zhang et al.’s conclusions, attributing the observed viral/human chimeric sequencing reads to artifactual processes happening during library preparation. Moreover, many are the caveats of Zhang et al.’s hypothesis of SARS-COV-2 integration in COVID-19 patients’ genome: 1) rare bona fide viral integrations were only observed using in vitro settings with little evidence in human tissues, 2) no 3′ polyA was observed in the sequenced SARS-COV-2 insertions, and 3) the observed preference of SARS-CoV2 integration in exonic regions does not align with the known unbiased activity of LINE1 retrotransposon. Nevertheless, Zhang et al.’s findings have potentially significant consequences in the interpretation of RT-qPCR measurement of SARS-CoV-2 RNA in patient nasal samples, today’s gold standard procedure for COVID-19 diagnosis. If, indeed, SARS-COV-2 virus can be integrated into the human genome, false positive COVID-19 test results would potentially derive from this event.
To evaluate the impact of possible virus genomic integration on diagnostic testing for COVID-19, we analyzed 768 COVID-positive nasal swab sample remnants, submitted to the Pandemic Response Lab NYC. We performed a two-step RT-qPCR reaction in the presence or absence of reverse transcriptase (RevT). Amplification of target viral sequences will occur in the absence of RevT if integrated into the patient genome. We used primers targeting the 3′ and 5′ SARS-CoV-2 region (Fig. 1) (3) because only the 3′ region was postulated to be retrotransposed (1). Only 2 out of 768 samples (0.26%) reproducibly amplified SARS-COV-2 sequences in the absence of RevT enzyme (Tables 1 and 2). We attributed spurious −RevT signals to amplicon DNA contaminations, a likely source of artifacts in laboratories performing a high volume of RT-qPCR of the same target, that, in diagnostic laboratories, can be controlled with frequent and thorough surveillance and decontamination procedures. Moreover, samples amplified in the absence of RevT enzyme show much higher Ct values than +RevT matched reactions, demonstrating the presence of abundant viral RNA in the analyzed samples. This indicates that, regardless of possible genomic integration of SARS-CoV-2, all patients reported as positive were infected with SARS-CoV-2 virus, demonstrating no impact of possible viral integration on COVID-19 diagnosis.
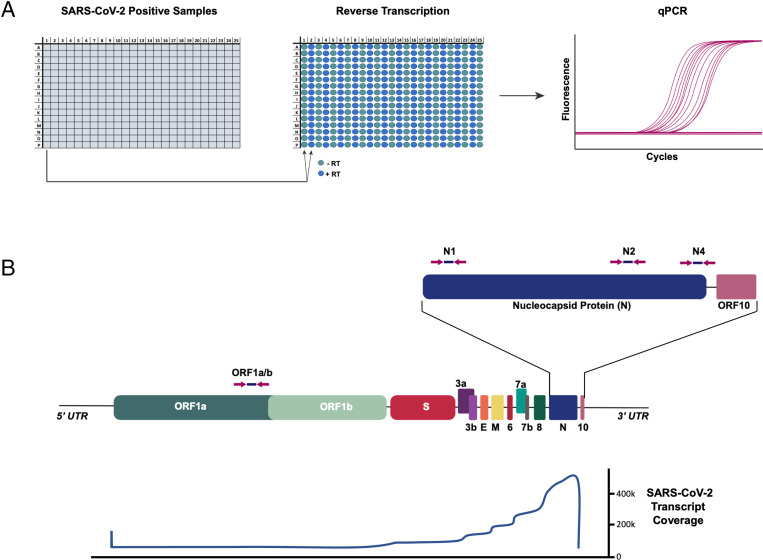
Fig. 1.
Analysis of SARS-CoV-2−positive samples for genomic SARS-CoV-2 DNA. (A) SARS-CoV-2−positive samples were consolidated in a 384-well plate. Each sample was run in duplicate RT reactions with or without RevT. Resulting reactions were assessed for SARS-CoV-2 complementary DNA by qPCR. (B) (Top) Schematic of SARS-CoV-2 genome. The qPCR primer/probe locations for N1, N2, N4 (3′ end), and ORF1ab (5′ end) are shown with arrows. (Bottom) Profile of RNA read depth across the viral genome as reported by refs. 4 and 5, showing strong bias of expression toward the 3′ end of the viral genome.
Table 1.
Initial RT-qPCR evaluation for SARS-CoV-2 DNA in COVID-19−positive samples
| Number of samples (percent of +RevT) |
Average Ct (−RevT amplified samples) |
|
| N1 +RevT | 523 | 24.56 |
| N1 −RevT | 11 (2.10) | 33.76 |
| N2 +RevT | 475 | 27.47 |
| N2 −RevT | 13 (2.74) | 33.02 |
| RP +RevT | 756 | 27.40 |
| RP −RevT | 759 | 30.44 |
| Total samples | 768 |
RT-qPCR was performed on SARS-CoV-2−positive samples either with (+RevT) or without (−RevT) reverse transcriptase enzyme (as described in Fig. 1). The qPCRs using N1 and N2 primers/probes sets were initially performed, and numbers of positive samples and average Ct values (for −RevT−positive reactions) are shown.
Table 2.
Confirmatory RT-qPCR analysis of SARS-CoV-2 DNA in COVID-19−positive samples
| Sample number | N1 | N2 | N4 | ORF1ab | −RevT/qPCR repeat | −RevT/qPCR repeat +DNase |
| 1 | X | X | n/a | n/a | — | — |
| 2 | X | n/a | n/a | — | — | |
| 3 | X | X | n/a | n/a | — | — |
| 4 | X | n/a | n/a | — | — | |
| 5 | X | — | — | |||
| 6 | X | — | — | |||
| 7 | X (1/2) | — | — | |||
| 8 | X (1/2) | — | — | |||
| 9 | X (1/2) | — | — | |||
| 10 | X (2/2) | — | — | |||
| 11 | X | X (2/2) | X | X (2/2) | — | — |
| 12 | X | — | — | |||
| 13 | X | X (1/2) | — | — | ||
| 14 | X (1/2) | X | X (2/2) | N2, N4 | — | |
| 15 | X | N4 | — | |||
| 16 | X (1/2) | X (1/2) | — | — | ||
| 17 | X (1/2) | — | — | |||
| 18 | X (1/2) | — | — | |||
| 19 | X | — | — | |||
| 20 | X | X (2/2) | X | X (2/2) | — | — |
| 21 | X | X (2/2) | X | X (2/2) | — | — |
| 22 | X (1/2) | — | — |
The qPCR for N1, N2, N4, and ORF1ab was repeated for all the samples with amplifications in the −RevT condition. Parentheses are for N2 and ORF1ab display repetition in technical qPCR replicates. An aliquot of −RevT samples was also used for DNase treatment prior to qPCR (last column) to show the DNA nature of possible −RevT signals (X indicates a detected amplification; — indicates no amplification detected; bolded results indicate possible virus genomic insertions; n/a indicates analysis was not performed).
These data suggest that LINE1-mediated retrotransposition of the SARS-CoV-2 genome into the host DNA is a rare event with no practical impact on RT-PCR−based diagnostic capability. Our data suggest that purported SARS-CoV-2 integrations are likely artifactual, stemming from amplicon DNA contaminations and/or other unintended processes.
Footnotes
Competing interest statement: The authors of this study are employees of the Pandemic Response Lab/ReOpen Diagnostics, a private company performing SARS-CoV-2 RT-qPCR based testing, an area of interest of this study.
References
- 1.Zhang L., et al., Reverse-transcribed SARS-CoV-2 RNA can integrate into the genome of cultured human cells and can be expressed in patient-derived tissues. Proc. Natl. Acad. Sci. U.S.A. 118, e2105968118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yan B., et al., Host-virus chimeric events in SARS-CoV2 infected cells are infrequent and artifactual. J. Virol. 95, e0029421 (2021). [DOI] [PMC free article] [PubMed]
- 3.Arena F., Pollini S., Rossolini G. M., Margaglione M., Summary of the available molecular methods for detection of SARS-CoV-2 during the ongoing pandemic. Int. J. Mol. Sci. 22, 1298 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim D., et al., The architecture of SARS-CoV-2 transcriptome. Cell 181, 914–921.e10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davidson A. D., et al., Characterisation of the transcriptome and proteome of SARS-CoV-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein. Genome Med. 12, 68 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]