Significance
A previous Plasmodium infection enhances the mosquito immune response to subsequent infections. Priming is mediated by a hemocyte differentiation factor (HDF) consisting of lipoxin A4 (LXA4) bound to Evokin, a lipid carrier. Insects produce LXA4 but lack lipoxygenase enzymes. Here we establish that the double peroxidase (DBLOX) enzyme is essential for HDF synthesis and is highly expressed in fat-body oenocytes, a group of specialized cells that increase in number in primed females. The histone acetyltransferase (HAT) AgTip60 is also essential for HDF synthesis, for the persistent increase in oenocyte numbers, and to maintain immune priming. We identified an enzyme essential for lipoxygenase-independent LXA4 synthesis and show that AgTip60 HAT is also critical to maintain the priming response.
Keywords: innate immunity, mosquito priming, double peroxidase, histone acetyltransferase, Anopheles epigenetic
Abstract
Immune priming in Anopheles gambiae is mediated by the systemic release of a hemocyte differentiation factor (HDF), a complex of lipoxin A4 bound to Evokin, a lipid carrier. HDF increases the proportion of circulating granulocytes and enhances mosquito cellular immunity. Here, we show that Evokin is present in hemocytes and fat-body cells, and messenger RNA (mRNA) expression increases significantly after immune priming. The double peroxidase (DBLOX) enzyme, present in insects but not in vertebrates, is essential for HDF synthesis. DBLOX is highly expressed in oenocytes in the fat-body tissue, and these cells increase in number in primed mosquitoes. We provide direct evidence that the histone acetyltransferase AgTip60 (AGAP001539) is also essential for a sustained increase in oenocyte numbers, HDF synthesis, and immune priming. We propose that oenocytes may function as a population of cells that are reprogrammed, and orchestrate and maintain a broad, systemic, and long-lasting state of enhanced immune surveillance in primed mosquitoes.
There is growing evidence that a previous infection can “train” or “prime” the innate immune system, allowing it to respond more effectively to subsequent infections (1–3). This is an ancient response that has been documented in plants (4), insects (5), and humans (6). It is not clear if individual immune cells maintain the training response, or if there are specific cell populations that are persistently reprogrammed and orchestrate a constant retraining of effector immune cells. In Anopheles gambiae, the primary vector of malaria in Africa, Plasmodium infection induces a long-lasting priming response that enhances antiplasmodial immunity (5). Plasmodium midgut invasion allows direct contact between the gut microbiota and midgut epithelial cells, triggering a burst of prostaglandin E2 (PGE2) production by midgut cells. The transient systemic release of PGE2 establishes a long-lasting release of hemocyte differentiation factor (HDF) which, in turn, induces an increase in the proportion of circulating granulocytes, a hallmark of immune priming (7, 8). HDF is a complex of Evokin, a lipid carrier protein of the lipocalin family, and lipoxin A4 (LXA4) (7). Lipoxins are bioactive lipids derived from arachidonic acid that dampen and resolve inflammation in vertebrates (9). At a biochemical level, immune priming can be defined as the enhanced ability of mosquitoes to convert arachidonic acid into LXA4 that results in a permanent functional state of enhanced immune surveillance (7). An effective antiplasmodial response requires the coordinated activation of epithelial, cellular, and complement components of the mosquito immune system (10). PGE2 attracts hemocytes to the basal surface of the midgut (8), and primed hemocytes release more microvesicles, enhancing complement-mediated ookinete lysis (10). Prostaglandins are also lipid mediators derived from arachidonic acid that have multiple biological activities in different organs, and play a key role in the activation of the inflammatory response (9).
In vertebrates, cyclooxygenases (COXs) and lipoxygenases (LOXs) catalyze the synthesis of prostaglandins and lipoxins, respectively (11). Although eicosanoids have been detected in mosquitoes and other insects (8, 12, 13), neither COX nor LOX enzymes are present in insects. We recently showed that two heme peroxidases, HPX7 and HPX8, are necessary for midgut PGE2 synthesis and are essential to establish immune priming in response to Plasmodium infection (8). However, the enzyme(s) mediating LXA4 synthesis in insects remains unknown. In this manuscript, we identify the double peroxidase (DBLOX) enzyme as essential for HDF synthesis and show that DBLOX is highly expressed in oenocytes, a cell population involved in lipid processing. Furthermore, we discovered that the number of oenocytes increases significantly in primed mosquitoes. In vertebrates, monocyte trained immunity is mediated by epigenetic modifications (14, 15). Here, we show that a histone acetyltransferase (HAT) is also essential to maintain the priming response in mosquitoes.
Results
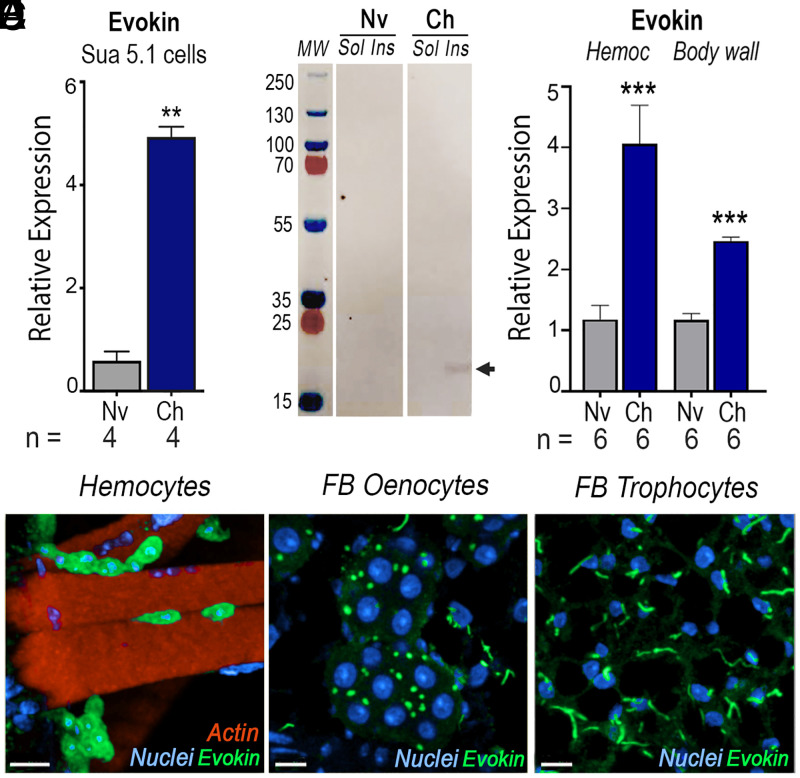
Messenger RNA (mRNA) expression of Evokin, an essential component of HDF (7), was significantly induced in the hemocyte-like immune-responsive A. gambiae Sua 5.1 cell line in response to bacterial challenge (Fig. 1A) (P = 0.0037, Mann–Whitney U test). Additionally, antibodies to recombinant Evokin recognized a single band of the expected size (21 kDa) (SI Appendix, Table S1) in the fraction containing membrane and insoluble proteins but not in the cytoplasm (Fig. 1B and SI Appendix, Fig. S1). Evokin mRNA expression also increased significantly in hemocytes (P = 0.0006, unpaired t test) and the body wall (P = 0.0005, unpaired t test) of primed mosquitoes 7 d postinfection (Fig. 1C). In naïve blood-fed females, Evokin is highly expressed in sessile hemocytes associated with the body-wall surface (Fig. 1D). It is also present in oenocytes, with a dotted vesicle-like pattern (Fig. 1E), while in fat-body trophocytes a string pattern is observed, suggestive of Evokin clustering in specific membrane regions that may represent lipid rafts (Fig. 1F). A similar expression pattern was observed in Plasmodium-challenged females (SI Appendix, Fig. S2).
Fig. 1.
Effect of immune challenge of Sua 5.1 cells and mosquitoes on Evokin expression. (A and B) Effect of bacterial challenge on Sua 5.1 cells on (A) Evokin mRNA levels measured by qRT-PCR 1 d postchallenge and (B) Evokin protein expression (arrow) analyzed by Western blot. A Coomassie blue–stained gel is shown in SI Appendix, Fig. S1. (C) Effect of P. berghei infection on Evokin mRNA levels measured by qRT-PCR in mosquito hemocytes and body wall 7 d postinfection. (D–F) Localization of Evokin in (D) sessile, body wall–associated hemocytes, (E) oenocytes, and (F) trophocytes analyzed by immunofluorescence staining of naïve mosquitoes. (A and C) Means ± SEM are plotted and groups were compared using Student’s t test (**P < 0.01, ***P < 0.001). Each treatment had at least three biological replicates and the results were confirmed in at least two independent experiments. Cells from a single well of a six-well plate (A) or pools of 15 to 20 mosquitoes (C) were used. [Scale bars, 7 µm (D), 10 µm (E), and 10 µm (F).] Ch, challenged; fat body, FB; Hemoc, hemocytes; Ins, insoluble fraction; Nv, naïve; Sol, soluble fraction.
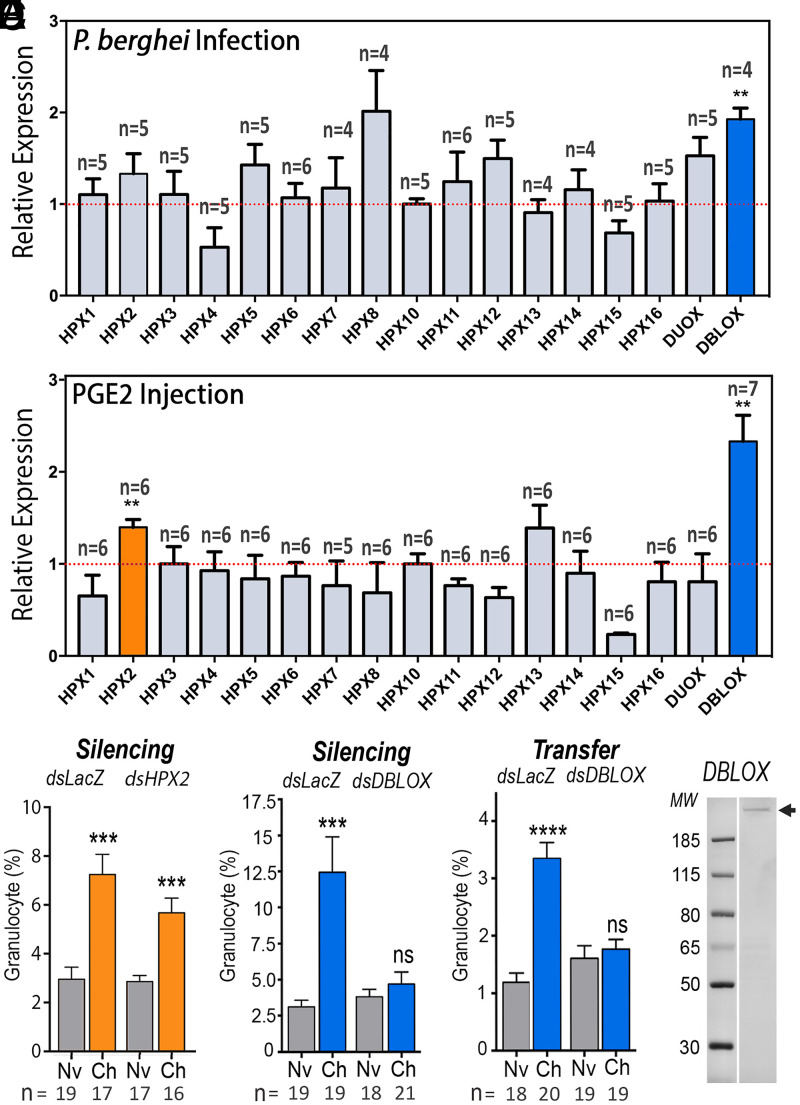
Although COXs and LOXs are unrelated in sequence and three-dimensional structure, they catalyze similar biochemical reactions. They both react polyunsaturated fatty acids, such as arachidonic acid, with oxygen to form peroxide products. Both enzymes employ free radical chemistry reminiscent of hydrocarbon autoxidation but they catalyze a highly effective formation of specific products (16). Given the critical role of HPX7 and HPX8 in PGE2 synthesis, we investigated whether other HPX enzymes could mediate the conversion of arachidonic acid to LXA4. Changes in mRNA expression of 15 HPX enzymes, DBLOX, and dual oxidase in mosquitoes primed by Plasmodium berghei infections (Fig. 2A) or by systemic injection of PGE2 (Fig. 2B) were evaluated. Both treatments resulted in a prolonged and significant increase in DBLOX expression in the body wall (P = 0.0056 and P = 0.0059, respectively, Mann–Whitney U test), whereas HPX2 was only induced following PGE2 injection (P = 0.0058, Mann–Whitney U test) (Fig. 2B). HPX2 silencing did not affect priming, as the characteristic increase in granulocytes was observed in response to Plasmodium infection (P = 0.0001, Mann–Whitney U test) (Fig. 2C). In contrast, DBLOX silencing completely abolished the priming response to infection (Fig. 2D) and to PGE2 injection (SI Appendix, Fig. S3). Furthermore, hemolymph of mosquitoes in which DBLOX was silenced no longer had HDF activity when transferred to naïve mosquitoes (Fig. 2E). Transfer of hemolymph from dsDBLOX donor mosquitoes did not silence DBLOX in the recipients (SI Appendix, Fig. S4). Indeed, a modest increase in DBLOX expression was observed. We also confirmed that DBLOX silencing in the donors had no effect on Evokin expression (SI Appendix, Fig. S5), suggesting that the lack of HDF activity is due to loss of LXA4 synthesis.
Fig. 2.
Effect of Plasmodium infection and PGE2 injection on heme peroxidase (HPX) expression and role of HPXs on immune priming. (A and B) HPX mRNA expression (A) 7 d after P. berghei infection and (B) 6 d after PGE2 injection into mosquito abdominal walls. (C and D) Effect of silencing (C) HPX2 and (D) DBLOX on Plasmodium-induced granulocyte priming 4 to 5 d after blood feeding. (E) Effect of silencing DBLOX on Plasmodium-induced HDF activity 4 to 5 d posthemolymph transfer. (F) DBLOX protein expression (arrow) from hemocyte-like Sua 5.1 cells analyzed by Western blot. A Coomassie blue–stained gel is shown in SI Appendix, Fig. S6. Means ± SEM are plotted and groups were compared using Student’s t test (**P < 0.01, ***P < 0.001, ****P < 0.0001; ns, not significant). (A and B) Each treatment had at least four biological replicates, and the results were confirmed in at least two independent experiments using pools of 15 to 20 mosquitoes. (C–E) Hemocytes were counted in 8 to 11 mosquitoes for each treatment, and the results were confirmed in two independent experiments.
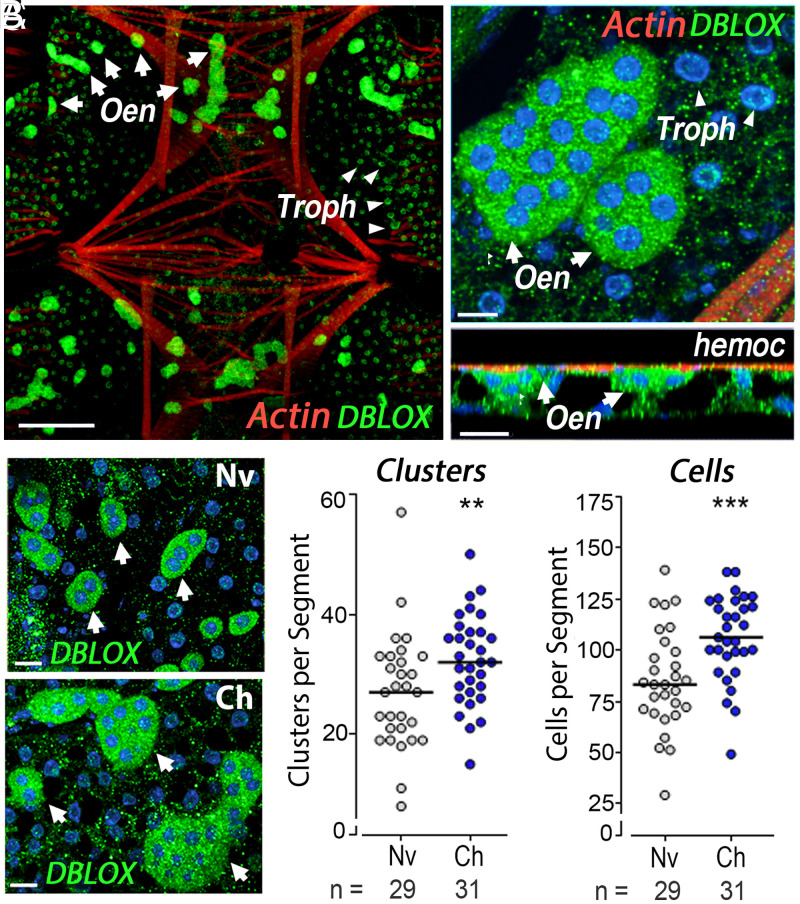
Antibodies against recombinant DBLOX detected a single band under denaturing/nonreducing conditions (Fig. 2F and SI Appendix, Fig. S2), with an estimated size of 110 kDa under denaturing/reducing conditions (SI Appendix, Fig. S6). DBLOX is highly expressed in oenocytes, which form clusters on the fat-body surface facing the hemocoele (Fig. 3 A and B). Oenocytes can be readily distinguished from fat-body trophocytes with an eosin/thiazine stain. Oenocytes have a purple nucleus and a pink-red cytoplasm, indicative of high-protein content, while trophocytes present a purple nucleus and a clear cytoplasm filled with lipid droplets (SI Appendix, Fig. S7).
Fig. 3.
Tissue localization of DBLOX expression and effect of Plasmodium infection on mosquito fat-body oenocytes. (A) DBLOX expression by immunofluorescence staining in the ventral and lateral body wall. Oenocytes (Oen) are indicated by arrows and trophocytes (Troph) are indicated by arrowheads. (B) Oenocyte clusters at higher magnification (Upper) and side view (Lower). The surface of the fat body facing the hemocoele (hemoc) is indicated. (C–E) Effect of Plasmodium infection on DBLOX-expressing oenocytes revealed by immunofluorescence staining (C), the number of oenocyte clusters per body-wall segment (D), and the number of oenocytes per body-wall segment 4 d after feeding (E). (D and E) Lines indicate medians that were compared using the Mann–Whitney U test (**P < 0.01, ***P < 0.001). Cells were counted in 7 to 12 individual mosquitoes in each experiment, and the results were confirmed in three independent experiments (SI Appendix, Table S3). [Scale bars, 100 µm (A), 10 µm (front view) and 20 µm (side view) (B), and 15 µm (C).]
DBLOX staining is cytoplasmic, with a punctate pattern, and is also present in lower abundance in the perinuclear region of fat-body trophocytes (Fig. 3 A and B). The number of oenocytes in the fat body increased significantly in primed mosquitoes 4 d postinfection with a significant increase in both the number of cell clusters (P = 0.0031, Mann–Whitney U test) and number of cells per body-wall segment (P = 0.0001, Mann–Whitney U test) (Fig. 3 C–E). Mosquitoes were fed 5-bromo-2'-deoxyuridine (BrdU) to establish whether oenocytes proliferate and undergo DNA replication in response to priming. Although oenocytes increased in number, BrdU was not incorporated into their nuclei, indicating that DNA replication does not take place. In contrast, BrdU incorporation was often observed in the nuclei of fat-body trophocytes (SI Appendix, Fig. S8). The increase in oenocytes is long-lasting, as significantly higher numbers of clusters and cells per segment were still present 7 d postfeeding (SI Appendix, Fig. S9 and Table S3).
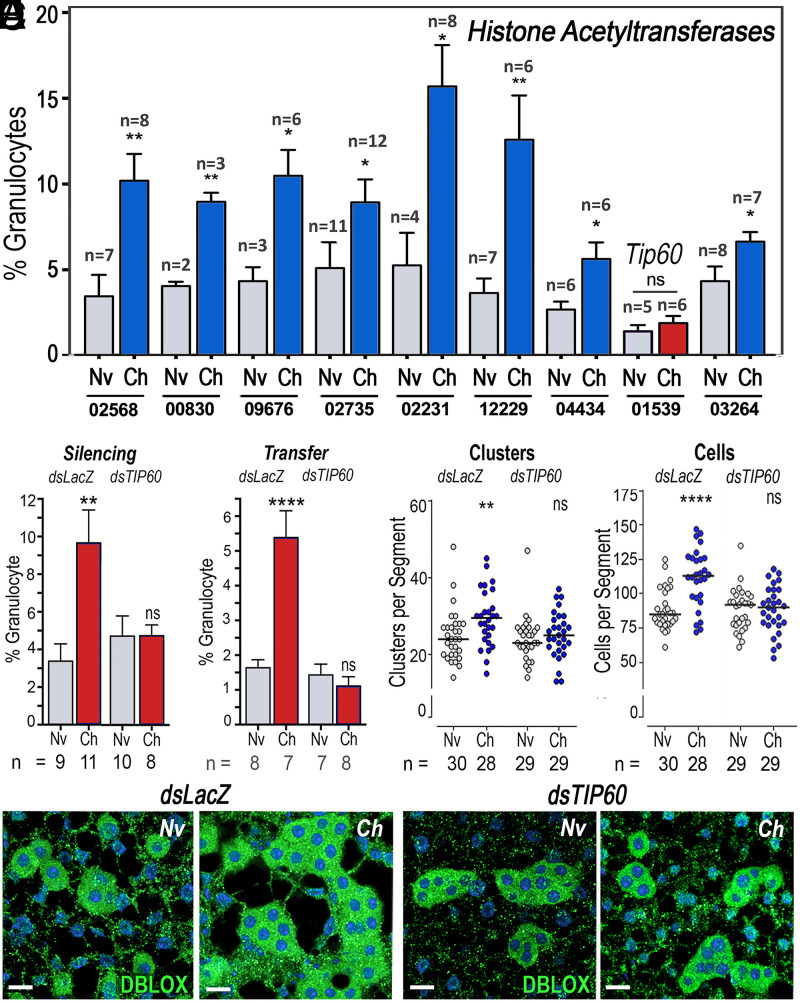
We explored whether the establishment and persistence of immune priming are mediated by HATs, enzymes known to catalyze epigenetic chromatin modifications associated with long-lasting changes in transcriptional regulation (17, 18). We evaluated the effect of silencing each of the 10 HATs present in the A. gambiae genome (AgHATs) on immune priming following P. berghei infection (Fig. 4A). Silencing AGAP000029 resulted in high mortality after blood feeding; thus, it could not be further evaluated. Of the other nine AgHATs, only silencing of the homolog of Drosophila Tip60 (AGAP001539; AgTip60) abolished immune priming (Fig. 4A). We confirmed that dsLacZ injection had no effect on priming, while the proportion of granulocytes no longer increased when AgTip60 was silenced (Fig. 4B). Furthermore, hemolymph of primed AgTip60-silenced females no longer had HDF activity when transferred to naïve mosquitoes (Fig. 4C). The loss of HDF activity following AgTip60 silencing was also associated with a lack of increase in the number of fat-body oenocytes, as both the numbers of clusters and of cells per segment in naïve females were not significantly different from those challenged with P. berghei (Fig. 4 D–F and SI Appendix, Table S4).
Fig. 4.
Effect of HAT silencing on priming and proliferation of DBLOX-expressing oenocytes. (A) Effect of silencing each A. gambiae HAT on granulocyte priming 4 to 5 d after blood feeding. The numbers below the x-axis indicate the last five digits (after “AGAP0”) of the six-digit reference accession number of each silenced gene. (B–E) Effect of silencing AgTip60 (AGAP001539) on Plasmodium-induced granulocyte priming 4 to 5 d after blood feeding (B), HDF activity 4 to 5 d posthemolymph transfer (C), number of oenocyte clusters (D), and number of oenocytes per segment 4 d after blood feeding (E). (F) Effect of silencing AgTip60 (TIP60) on immunofluorescence staining of DBLOX in the body wall of naïve and challenged mosquitoes 4 d after blood feeding. Means ± SEM are plotted and groups were compared using Student’s t test (*P < 0.05, **P < 0.01, ****P < 0.0001). (D and E) Cells and clusters were counted in 13 to 15 individual mosquitoes in each experiment, and the results were confirmed in two independent experiments (SI Appendix, Table S4). (Scale bars, 15 µm.)
Discussion
DBLOX is a unique enzyme with a duplicated heme peroxidase domain that is present in insects but not in vertebrates. The first domain has the predicted substrate binding sites but lacks the functional residues present in catalytically active enzymes and has two integrin-binding motifs, typical of peroxinectins (19). In contrast, the second domain has all the features of a functional heme peroxidase and one integrin-binding motif (19). DBLOX is essential for HDF synthesis, but the biochemical mechanism of LOX-independent lipoxin synthesis in insects and whether DBLOX directly catalyzes LXA4 synthesis remain to be determined.
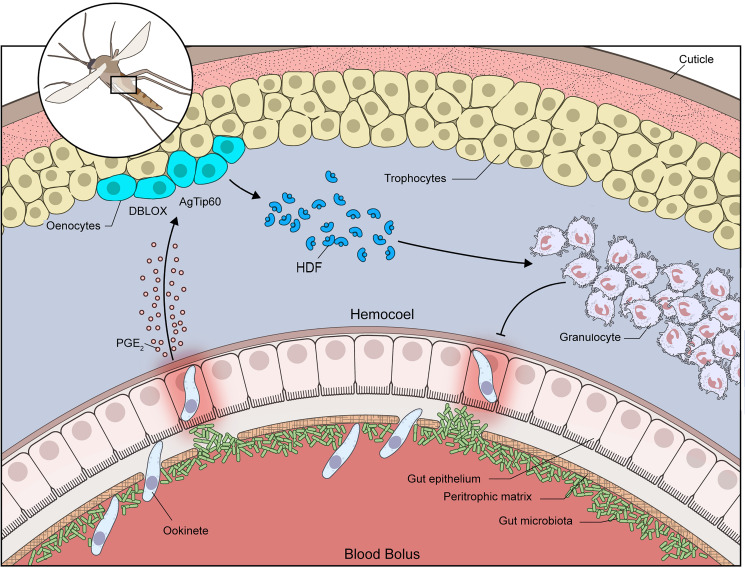
Our findings could be explained by a working model in which the systemic burst of PGE2 released into the hemolymph when ookinete invasion allows direct contact between the microbiota and gut epithelial cells (Fig. 5) triggers epigenetic modifications mediated by AgTip60. Given the essential role of DBLOX and AgTip60 in HDF synthesis and immune priming, the high expression of DBLOX in oenocytes, and the sustained increase in oenocyte numbers mediated by AgTip60, we propose that the documented enhanced ability of primed mosquitoes to synthesize HDF is achieved, at least in part, through functional changes in oenocytes. The function of these cells is poorly understood, but they are known to be involved in biosynthesis of cuticular hydrocarbon and pheromones (20).
Fig. 5.
Working model of innate immune memory in primed A. gambiae mosquitoes. Ookinete invasion allows direct contact between the gut microbiota and midgut epithelial cells by disrupting the peritrophic matrix, and triggers the systemic release of PGE2 by the midgut. High levels of PGE2 result in higher expression of DBLOX in the abdominal wall. DBLOX is highly expressed in oenocytes, and the numbers of oenocyte clusters and cells per segment increase constitutively in primed mosquitoes. This increase in oenocytes requires the activity of the HAT AgTip60. Silencing either DBLOX or AgTip60 disrupts the synthesis of the HDF and immune priming. Constitutively high levels of HDF promote hemocyte differentiation to granulocytes and enhance the immune response to subsequent infections.
Polyploidy has been documented in several insect tissues such as the fat body, midgut, muscle, Malpighian tubules, and nurse cells, and has been reported in several insect species, including Drosophila melanogaster, Locusta migratoria, A. gambiae, and Aedes aegypti (21). This is the result of an endoreplicative cycle, also referred to as an “endocycle,” in which cells undergo successive rounds of DNA replication without an intervening mitosis. Often the endocycle is used to expand the genome of a group of specialized cells that are highly biosynthetically active (22). In lepidoptera, oenocytes undergo terminal differentiation in larval stages. Their nuclear volume increases by 100-fold in early larval stages, as a result of multiple rounds of genome endoreplication without cell division, and DNA synthesis is no longer observed after the fifth larval instar (23). Mosquito larval midgut cells can undergo somatic reduction by cell division of polyploid cells. It is documented that polyploid ileum cells in larvae divide and give rise to diploid cells without going into S phase, the mitotic phase where DNA replication normally takes place (24–27). Thus, the observed increase in the number of oenocytes in primed adult mosquitoes appears to also be the product of somatic reduction due to cell division without DNA replication. The functional changes of oenocytes in response to priming, besides the increase in the number of cells, remain to be defined.
Immune training of human monocytes involves transcriptional and metabolic reprogramming mediated by epigenetic modifications that allow challenged monocytes to maintain a prolonged state of enhanced immune function (14, 28). In mosquitoes, the persistent release of HDF appears to constantly retrain circulating mosquito hemocytes. Taken together, our findings suggest that oenocytes may be functioning as a population of cells that are reprogrammed, and orchestrate a long-lasting state of enhanced immune surveillance in primed mosquitoes, probably as an important site of LXA4 synthesis. The persistent enhancement of the innate immune response of primed mosquitoes involves changes in histone acetylation mediated by AgTip60. Careful characterization of the functional changes in fat-body oenocytes of primed mosquitoes is warranted, as it could provide new insights into the mechanisms that enhance HDF synthesis and allow mosquitoes to “remember” a previous Plasmodium infection. This is a working model to be refined, challenged, and modified as we learn more about innate immune memory in insects. Recently, trained immunity in humans induced by vaccination with the antituberculosis bacillus Calmette–Guérin vaccine has received much attention (28), but little is known about the mechanisms that establish and maintain this type of broad enhancement of innate immunity . In A. gambiae, prostaglandins are key to establish the priming response, while lipoxins maintain a broad general state of enhanced immune surveillance that is not pathogen-specific. This begs the question of whether eicosanoids may also be important mediators of trained immunity in humans.
Methods
Animals, Priming, and Granulocyte Counting.
A. gambiae (G3 strain) mosquitoes were reared at 28 °C, 80% humidity, 12-h light/dark cycle. Mosquitoes were fed 10% Karo syrup solution ad libitum. For Plasmodium infection–induced priming, the P. berghei Anka GFP strain was used. Mouse infections were performed through serial passage in 4- to 5-wk-old female BALB/c mice. A drop of blood was taken from mouse tails 48 to 72 h after passage, and parasitemia was evaluated on Giemsa-stained, methanol-fixed blood smear slides. For mosquito infections, mice carrying 3 to 5% parasitemia infections were chosen. After infection, mosquitoes were kept at 19 °C, 80% humidity, 12-h light/dark cycle until the day of sample processing. Animal studies were done according to the NIH animal study protocol (ASP) approved by the NIH Animal Care and Use Committee, with approval ID ASP-LMVR5. Public Health Service Animal Welfare Assurance A4149-01 guidelines were followed according to the NIH Office of Animal Care and Use.
Alternatively, mosquitoes were primed by injection of cell-free hemolymph transfer from challenged mosquitoes as previously described. Hemolymph was collected from pools of 10 females perfused with 10 µL of a modified anticoagulant buffer (95% Schneider’s medium, 5% citrate buffer). The pool of hemolymph was centrifuged at 9,300 × g for 10 min at 4 °C and the cell-free supernatant was transferred and stored at −80 °C. Cell-free hemolymph challenge was done by injecting 138 nL cell-free hemolymph into a 3-d-old sugar-fed naïve mosquito. For PGE2 priming, an aliquot of ethanol-diluted PGE2 was dried in a nitrogen gas stream and resuspended in anticoagulant buffer, and PGE2 was injected into 3-d-old sugar-fed naïve mosquitoes.
For granulocyte counting, hemolymph was collected from females perfused with 10 µL of hemocyte counting buffer (10% fetal bovine serum, 30% citrate buffer in Schneider’s medium). Collected samples were applied to sterile hemocytometer slides and granulocytes were counted under a light microscope as the relative number of cells against total hemocyte numbers.
Sua Cell Culture and Priming.
Sua cells were grown on Sua cell media (10% fetal bovine serum, 1,000 IU/mL penicillin, 1 mg/mL streptomycin on Schneider’s media) at 28 °C and passed at 80 to 90% confluence. For bacteria challenge, 10 mg/mL Escherichia coli acetone powder was added to the cell media and cells were incubated for 20 h. Then, cells were extensively washed with cell media and incubated overnight. The next day, media were removed and cells were processed.
Gene Expression Analysis.
For gene expression analysis, midgut and abdominal fat-body specimens from 15 to 20 females per group were dissected. RNA samples from dissected midguts were obtained using the RNeasy RNA Extraction Kit (Qiagen) following a standard protocol. Hemolymph samples were prepared by perfusing mosquitoes with 5 µL of a modified anticoagulant buffer (95% Schneider’s medium, 5% citrate buffer) into TRIzol (Invitrogen). Fat body RNA and Sua cell samples were prepared by a standard TRIzol (Invitrogen) protocol. The RNA concentration was measured using a NanoDrop (Thermo Fisher Scientific) and complementary DNA (cDNA) samples were prepared with the QuantiTect Reverse Transcription Kit with DNA wipeout (Qiagen) following a standard protocol. Relative gene expression was analyzed by qRT-PCR using the DyNamo SYBR Green qPCR Kit (Thermo Fisher Scientific) and a CFX96 Real-Time PCR Detection System (Bio-Rad). Data analysis was done using the ΔΔCt method. The list of primers used is available in SI Appendix, Table S5.
Sua Cell Protein Fractionation.
For Sua cell protein fractionation, the cell media were removed and cold 0.2 mM phenylmethanesulfonylfluoride (PMSF) in phosphate-buffered saline (PBS) was added. Cells were scraped and centrifuged (5 min, 5,000 rpm, 4 °C). The pellet was resuspended in protease inhibitor–supplemented cold buffer A (1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol [DTT], 0.2 mM PMSF, 10 mM Hepes, pH 7.9) and incubated on ice for 10 min. Then, cells were vortexed and centrifuged (5 min, 5,000 rpm, 4 °C). The supernatant (soluble; cytoplasmic fraction) was stored. The pellet was resuspended in protease inhibitor–supplemented cold buffer C (25% glycerol, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM ethylenediaminetetraacetate [EDTA], 0.5 mM DTT, 0.2 mM PMSF, 20 mM Hepes, pH 7.9), incubated on ice for 20 min, and centrifuged (10 min, 20,000 × g, 4 °C). The supernatant was stored and the pellet (insoluble; membrane fraction) was resuspended in solubilization buffer (150 mM NaCl, 5 mM EDTA, 2% sodium dodecyl sulfate [SDS], 1 mM Tris·HCl, pH 7.5) and stored at −70 °C.
Recombinant Evokin Protein Expression and Purification.
The mature full-length AGAP009281-PA coding sequence was codon-optimized for E. coli cell expression and synthesized by Bio Basic, Amherst, NY, a gift from Eric Calvo (LMVR, National Institute of Allergy and Infectious Diseases [NIAID], NIH). A histidine tag was added to the AGAP009281-PA construct by PCR amplification (SI Appendix, Table S1) and subcloned by In-Fusion (Clontech) into a pET17 vector (EMD Millipore) between NdeI and XhoI sites, with primers INFEVO17 (SI Appendix, Table S1). The construct was expressed in BL21 E. coli (Invitrogen). A Hepes-buffered saline (20 mM Hepes, pH 7.5, 150 mM NaCl) 8 M urea solution containing inclusion bodies of cultures induced for 3 h at 37 °C with 1 mM isopropyl β-d-thiogalactopyranoside (IPTG) (Sigma) was purified by nickel affinity chromatography and stored after being dialyzed in Hepes-buffered saline.
Recombinant DBLOX Protein Expression and Purification.
A region of the second domain of the A. gambiae DBLOX (AGAP008350/DBLOX) coding sequence was amplified (N693 to T1319) and subcloned into a pET17B plasmid with primers containing a hexahistidine tag at the C terminus (SI Appendix, Table S2). The DBLOX domain 2 construct was expressed in One Shot BL21(DE3)pLysE T7 Competent E. coli (Thermo Fisher Scientific) (SI Appendix, Table S2). E. coli transformed with the DBLOX domain 2 construct were grown at 37 °C with vigorous shaking until reaching an optical density of 0.4 to 0.6, at which point 1 mM IPTG was added to induce expression for 4 h. Pelleted E. coli were sonicated in a solution containing 8 M urea in Hepes-buffered saline (20 mM Hepes, pH 7.5, 150 mM NaCl) to solubilize inclusion bodies, and then purified in a column using HisPur Ni-NTA resin (Thermo Fisher Scientific) for affinity chromatography.
Mouse and Rabbit Immunizations.
All animal procedures were performed according to protocols approved by the NIAID and NIH Animal Care and Use Committee. Female BALB/c mice, 5- to 8-wk-old naïve, were purchased from Charles River and maintained at a facility at the NIH. Groups (n = 5) of female BALB/c mice were immunized subcutaneously in their ears with a 20-µL solution that contained 5 µg of Hepes-buffered saline-dialyzed recombinant AGAP009281-PA protein emulsified in Magic mouse adjuvant (Creative Diagnostics; CDN-A001). The immunized mice were boosted twice at 2-wk intervals with the same quantity of protein. Blood was collected on day 0 (preimmune sera) and 2 wk after each subsequent immunization for analysis of antibody titer or terminal bleeding. Purified recombinant DBLOX protein was run by SDS–polyacrylamide-gel electrophoresis, excised from the gel, homogenized, and used to generate rabbit antiserum (Noble Sciences). Briefly, rabbits were immunized four times at 3-wk intervals. The initial injection was performed with Freund’s complete adjuvant and subsequent injections were with Freund’s incomplete adjuvant. Blood was collected on day 0 (preimmune sera) and 2 wk after each subsequent immunization. The antiserum from the final blood collection was used for immunostaining and Western blotting experiments.
Western Blot.
Homogenate samples were boiled in NuPAGE LDS sample buffer (Thermo Fisher Scientific) and separated using a 12% Bis-Tris gel. The proteins were transferred to a nitrocellulose (Evokin) or polyvinylidene fluoride membrane (DBLOX), and blocked for 4 h in blocking buffer (5% nonfat dry milk in 0.1% Tween 20, Tris-buffered saline; TBST). Proteins were incubated with primary antibodies diluted in blocking buffer at the following concentrations: anti-Evokin: 1:1,000; anti-DBLOX: 1:2,000. The membranes were subsequently washed three times in TBST and incubated with goat anti-mouse or anti-rabbit secondary antibodies conjugated to alkaline phosphatase at a 1:5,000 dilution. The membranes were developed using Western Blue Stabilized Substrate for Alkaline Phosphatase (Promega).
To remove unspecific antibodies that could target E. coli proteins, DBLOX serum was diluted and preabsorbed twice for 30 min with E. coli acetone powder immediately before it was used. Briefly, One Shot BL21(DE3)pLysE T7 Competent E. coli (Thermo Fisher Scientific) were grown overnight at 37 °C, pelleted, and resuspended on 0.9% NaCl. The E. coli were lysed and dehydrated using acetone, pelleted, and crushed into a fine powder and washed three times with PBS. For preabsorption, 1% (weight/volume) of the powder pellet was used.
Immunohistochemistry.
Mosquitoes were individually injected with 207 nL of 16% paraformaldehyde and left for 20 s. Then, in 1× PBS, the mosquito abdomen was separated from the head and thorax, and the midgut, ovaries, and Malpighian tubes were removed. The abdomens were cut longitudinally through the dorsal region resulting in a flat, open tissue with an exposed fat body. Opened abdomens were immediately placed into 4% paraformaldehyde and left to fix for 30 min. The tissues were washed twice with 1× PBS and then delipidated in ice-cold ethanol in a stepwise manner starting with 20% up to 80% in 20% intervals, and then back down. Tissues were washed twice with 1× PBS and then placed into 4% paraformaldehyde and left for 1 h with gentle shaking. Fixed tissues were washed twice with 1× PBS, and washed three times with PBST (PBS, 0.1% Triton) for 5 min each. Tissues were blocked in blocking buffer (2% bovine serum albumin [BSA], 0.1% gelatin, in PBST) for 2 h at room temperature and incubated overnight at 4 °C with the rabbit DBLOX primary antibody diluted 1:1,500 in blocking buffer and preabsorbed with E. coli acetone powder twice, as previously described. Tissues were washed with blocking buffer and then blocked in blocking buffer with 20% goat serum for 1 h and incubated for 2 h at room temperature with a goat anti-rabbit Alexa Fluor 594 secondary antibody (Thermo Fisher Scientific) diluted 1:1,000 in blocking buffer with 20% goat serum. Tissues were washed with blocking buffer and then PBST and labeled with phalloidin 488 (1:40) and Hoechst (1:10,000) for 30 min and then incubated in PBS, 10% glycerol for 20 min before mounting on slides in ProLong Gold mounting media (Molecular Probes).
Double-Stranded RNA Synthesis and Gene Silencing.
To prepare double-stranded RNA (dsRNA) for gene silencing, a cDNA template of challenged mosquitoes was first amplified by PCR using primers with flanking T7 promoter recognition sequences (SI Appendix, Table S5). Following amplification, PCR products were purified using the QIAquick PCR Purification Kit (Qiagen) and dsRNA was synthesized using the MEGAscript RNAi Kit (Ambion) according to the manufacturer’s instructions. For gene silencing, 69 to 128 nL of 3 µg/µL dsRNA was injected into 2- to 3-d-old naïve females.
BrdU Proliferation Assay for Body Walls.
Three- to 4-d-old females were treated with 1 mg/mL BrdU (Sigma-Aldrich) in sugar for 3 d before feeding on a naïve or P. berghei–infected mouse. Mouse feeding and infection were performed as described above. Mosquitoes were placed at 21 °C for 48 h to allow ookinete invasion and priming, and then cages were transferred to 28 °C until dissection, 4 d postinfection. Body walls were dissected in PBS and immediately placed in 4% paraformaldehyde for 30 min at room temperature, briefly transferred to 80% ethanol for 3 min, and fixed again in 4% paraformaldehyde for 1 h at room temperature. The tissues were treated with sodium citrate to open the chromatin and allow BrdU detection. Tissues were placed in tubes with 1 mM sodium citrate (pH 6) and steamed at 99 °C for 15 min. After this treatment, tissues were rinsed twice and washed three times with PBS, 0.1% Tween for 5 min each, and blocked for 1 h at room temperature with PBS, 0.1% Tween, 2% BSA. They were then placed in blocking solution containing rabbit anti-DBLOX antibody (1:1,000) and mouse anti-BrdU antibody (1:100) (Invitrogen; B35128; MoBU-1) overnight at 4 °C. The next day, body walls were washed three times with blocking buffer (10 min each), and then incubated with goat anti-rabbit immunoglobulin G (IgG)–Alexa Fluor 594 (Invitrogen) (1:1,000) and goat anti-mouse IgG–Alexa Fluor 488 (1:2,000) (Invitrogen) diluted in blocking buffer at room temperature for 2 h. Tissues were washed twice in blocking buffer and then twice more in PBS, 0.1% Tween before being placed in ProLong Gold mounting media (Invitrogen). Counterstaining was performed with phalloidin 647 (1:40) and Hoechst 405 (1:10,000).
Whole-Mosquito Sample Preparation, Pretreatment, and Bright-Field Microscopy.
Whole mosquitoes were fixed in 4% paraformaldehyde, processed on the Leica ASP 6025, and then embedded in histology-grade paraffin. Samples were sectioned at 5 µm. Tissue section pretreatment was performed on the Leica Bond RX. Sections were baked for 30 min at 60 °C and then dewaxed for 30 s in Bond Dewax solution (Leica) heated to 72 °C and rehydrated with absolute ethanol washes and 1× ImmunoWash (StatLab). Sections subsequently underwent a 20-min treatment in Epitope Retrieval solution 1 (Leica) heated to 100 °C. Slides were placed in 1× PBS (Roche) following the pretreatment steps. Then, sections were stained with Hemacolor Rapid staining solutions II and III (Sigma-Aldrich). Briefly, after sample dewaxing and rehydration, sections were removed from the wash buffer and incubated with Hemacolor solution II (eosin Y) for 2 min, followed by incubation with Hemacolor solution III (thiazine) for 3 min. Slides were rinsed in distilled water and dipped twice in differentiation solution (0.25% acetic acid in water), rinsed in water, dipped twice in 70% ethanol, then twice in 100% ethanol, and finally twice in a xylene:ethanol (1:1) solution. Slides were air-dried and mounted with a coverslip using Entellan (Sigma-Aldrich). Mosquito sections were observed and imaged using bright-field microscopy on a Leica DMI6000 (Leica Microsystems).
Identification of Oenocytes in the Fat-Body Tissue.
Mosquito oenocytes were identified based on their distinctive morphology, as they form characteristic cell clusters, predominantly ventral, clumps on the hemolymph surface of the fat body (29). Eosin and thiazine staining of the fat body from adult mosquito females revealed trophocytes with a clear cytoplasm, characteristic of cells with large lipid droplets, and a dark purple nucleus. Oenocytes are located facing the surface of the fat body facing the hemocoele, and form cell clusters that have a cytoplasm denser than that of trophocytes and which stains with a red hue, indicative of high-protein content (SI Appendix, Fig. S7).
Acknowledgments
This work was supported by the Intramural Research Program of the Division of Intramural Research (Z01AI000947), NIAID, NIH. We thank Brian Brown, the NIH Library Writing Center, and Asher Kantor for editorial assistance; Ryan Kissinger for the illustration of the working model; Kevin Lee, Yonas Gebremicale, and André Laughinghouse for insectary support; and Dr. Ian Moore and Bianca Nagata from the NIAID/Division of Intramural Research, Comparative Medicine Branch Infectious Disease Pathology Section for preparing the histological sections.
Footnotes
Reviewers: M.J.-L., The Johns Hopkins University Bloomberg School of Public Health; and M.R.K., Kansas State University.
The authors declare no competing interest.
1Present address: Laboratório de Ultraestrutura Celular Hertha Meyer, Instituto de Biofísica Carlos Chagas Filho, Universidade Federal do Rio de Janeiro, Rio de Janeiro, RJ, 21941-902, Brazil.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2114242118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Melillo D., Marino R., Italiani P., Boraschi D., Innate immune memory in invertebrate metazoans: A critical appraisal. Front. Immunol. 9, 1915 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Milutinović B., Kurtz J., Immune memory in invertebrates. Semin. Immunol. 28, 328–342 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Cooper D., Eleftherianos I., Memory and specificity in the insect immune system: Current perspectives and future challenges. Front. Immunol. 8, 539 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muthamilarasan M., Prasad M., Plant innate immunity: An updated insight into defense mechanism. J. Biosci. 38, 433–449 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Rodrigues J., Brayner F. A., Alves L. C., Dixit R., Barillas-Mury C., Hemocyte differentiation mediates innate immune memory in Anopheles gambiae mosquitoes. Science 329, 1353–1355 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mulder W. J. M., Ochando J., Joosten L. A. B., Fayad Z. A., Netea M. G., Therapeutic targeting of trained immunity. Nat. Rev. Drug Discov. 18, 553–566 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramirez J. L., et al., A mosquito lipoxin/lipocalin complex mediates innate immune priming in Anopheles gambiae. Nat. Commun. 6, 7403 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barletta A. B. F., Trisnadi N., Ramirez J. L., Barillas-Mury C., Mosquito midgut prostaglandin release establishes systemic immune priming. iScience 19, 54–62 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandrasekharan J. A., Sharma-Walia N., Lipoxins: Nature’s way to resolve inflammation. J. Inflamm. Res. 8, 181–192 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castillo J. C., Ferreira A. B. B., Trisnadi N., Barillas-Mury C., Activation of mosquito complement antiplasmodial response requires cellular immunity. Sci. Immunol. 2, 2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanna V. S., Hafez E. A. A., Synopsis of arachidonic acid metabolism: A review. J. Adv. Res. 11, 23–32 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim Y., Ahmed S., Stanley D., An C., Eicosanoid-mediated immunity in insects. Dev. Comp. Immunol. 83, 130–143 (2018). [DOI] [PubMed] [Google Scholar]
- 13.García Gil de Muñoz F. L., Martínez-Barnetche J., Lanz-Mendoza H., Rodríguez M. H., Hernández-Hernández F. C., Prostaglandin E2 modulates the expression of antimicrobial peptides in the fat body and midgut of Anopheles albimanus. Arch. Insect Biochem. Physiol. 68, 14–25 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Saeed S., et al., Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science 345, 1251086 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Netea M. G., et al., Trained immunity: A program of innate immune memory in health and disease. Science 352, aaf1098 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneider C., Pratt D. A., Porter N. A., Brash A. R., Control of oxygenation in lipoxygenase and cyclooxygenase catalysis. Chem. Biol. 14, 473–488 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eberharter A., Becker P. B., Histone acetylation: A switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep. 3, 224–229 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imhof A., Wolffe A. P., Transcription: Gene control by targeted histone acetylation. Curr. Biol. 8, R422–R424 (1998). [DOI] [PubMed] [Google Scholar]
- 19.Choudhury T. P., Gupta L., Kumar S., Identification, characterization and expression analysis of Anopheles stephensi double peroxidase. Acta Trop. 190, 210–219 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Makki R., Cinnamon E., Gould A. P., The development and functions of oenocytes. Annu. Rev. Entomol. 59, 405–425 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren D., Song J., Ni M., Kang L., Guo W., Regulatory mechanisms of cell polyploidy in insects. Front. Cell Dev. Biol. 8, 361 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lilly M. A., Duronio R. J., New insights into cell cycle control from the Drosophila endocycle. Oncogene 24, 2765–2775 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Locke M., The ultrastructure of the oenocytes in the molt/intermolt cycle of an insect. Tissue Cell 1, 103–154 (1969). [DOI] [PubMed] [Google Scholar]
- 24.Berger C. A., Additional evidence of repeated chromosome division without mitotic activity. Am. Nat. 71, 187–190 (1937). [Google Scholar]
- 25.Berger C. A., Multiplication and reduction of somatic chromosome groups as a regular developmental process in the mosquito, Culex pipiens. Contrib. Embryol. 27, 200–232 (1938). [Google Scholar]
- 26.Grell M., Cytological studies in Culex I. Somatic reduction divisions. Genetics 31, 60 (1946). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Forer A., Ploidy and division in cancer and mosquito hind-gut cells. Cell Biol. Int. 33, 253 (2009). [DOI] [PubMed] [Google Scholar]
- 28.van der Meer J. W., Joosten L. A., Riksen N., Netea M. G., Trained immunity: A smart way to enhance innate immune defence. Mol. Immunol. 68, 40–44 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Lycett G. J., et al., Anopheles gambiae P450 reductase is highly expressed in oenocytes and in vivo knockdown increases permethrin susceptibility. Insect Mol. Biol. 15, 321–327 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All study data are included in the article and/or SI Appendix.