Significance
Plant cell surface receptors perceive carbohydrate signaling molecules and hereby establish communication with surrounding microbes. Genetic studies have identified two different classes of lysin motif receptor kinases as gatekeepers that together trigger the symbiotic pathway in plants; however, no structural or functional data of the perception mechanisms switching these receptors from resting state into activation is known. In this study, we use structural biology, biochemical, and genetic approaches to demonstrate how the NFP/NFR5 class of lipochitooligosaccharide (LCO) receptors discriminate bacterial symbionts based on a kinetic proofreading mechanism that controls receptor activation and signaling specificity. We show that the LCO binding site can be engineered to support symbiotic functions, which greatly advance future opportunities for receptor engineering in legumes and nonlegumes.
Keywords: LysM receptors, legume symbiosis, lipochitooligosaccharide signaling, receptor–ligand interaction, kinetic proofreading
Abstract
Plants and animals use cell surface receptors to sense and interpret environmental signals. In legume symbiosis with nitrogen-fixing bacteria, the specific recognition of bacterial lipochitooligosaccharide (LCO) signals by single-pass transmembrane receptor kinases determines compatibility. Here, we determine the structural basis for LCO perception from the crystal structures of two lysin motif receptor ectodomains and identify a hydrophobic patch in the binding site essential for LCO recognition and symbiotic function. We show that the receptor monitors the composition of the amphiphilic LCO molecules and uses kinetic proofreading to control receptor activation and signaling specificity. We demonstrate engineering of the LCO binding site to fine-tune ligand selectivity and correct binding kinetics required for activation of symbiotic signaling in plants. Finally, the hydrophobic patch is found to be a conserved structural signature in this class of LCO receptors across legumes that can be used for in silico predictions. Our results provide insights into the mechanism of cell-surface receptor activation by kinetic proofreading of ligands and highlight the potential in receptor engineering to capture benefits in plant–microbe interactions.
Bacteria and fungi surrounding plants synthesize amphiphilic lipochitooligosaccharide (LCO)-signaling molecules consisting of a chitin backbone decorated with a large variety of substitutions at the terminal moieties including hydrophobic acyls at the nonreducing end (1–3). The selectivity and activation mechanisms of single-pass transmembrane receptors involved in deciphering between this myriad of extracellular signals is still unknown. The symbiotic relationship between rhizobia and legumes is an example of selective recognition guided by LCO structural variation, and this plant–bacterial interaction offers a rare opportunity for understanding receptor selectivity and signaling function. Specific legume species recognize and engage in root nodule symbiosis only with a distinct subset of rhizobial strains and vice versa. Among the ∼18,000 legume species, this host/nonhost relationship represents a continuum. At one end of the spectrum, the broad–host-range strain Sinorhizobium fredii NGR234 is capable of infecting and eliciting nodule formation on more than 200 distantly related legume species (4), and at the other end of the spectrum, narrow–host-range rhizobia like Sinorhizobium meliloti primarily nodulate Medicago truncatula (Medicago) and closely related medics such as alfalfa (Medicago sativa). Legume–rhizobia compatibility is dependent on plant recognition of specific bacterial symbiotic LCO-signaling molecules (5), known as Nod factors, with additional contributions by exopolysaccharides, lipopolysaccharides, cyclic β-glucans, and proteinaceous effectors. Nod factor receptors and exopolysaccharide receptors have been identified in many legumes, including the model species Medicago (6–8) and Lotus japonicus (Lotus) (9–15). Purified rhizobial LCOs are directly perceived by two different classes of single-pass transmembrane lysin motif (LysM) receptors (14, 16). One class has an active intracellular kinase (Medicago LYK3/Lotus NFR1), and the other has a pseudokinase domain (Medicago NFP/Lotus NFR5). All plant responses to LCOs are abolished in Medicago nfp and Lotus nfr5 mutants and in the corresponding pea (sym10) (9, 17) and soybean (nfr5α) mutants (18). Genetic and physiological studies examining LCO interactions across four legume species demonstrate that the NFP/NFR5 class of receptors are crucial for initial LCO perception and essential for LCO-mediated root nodule symbiosis (6, 8–10). Consistent with this fundamental role, recent studies show that the emergence of the NFP/NFR5 class of receptors was one of the key innovations in the evolution of nitrogen-fixing symbiosis (19, 20).
Results
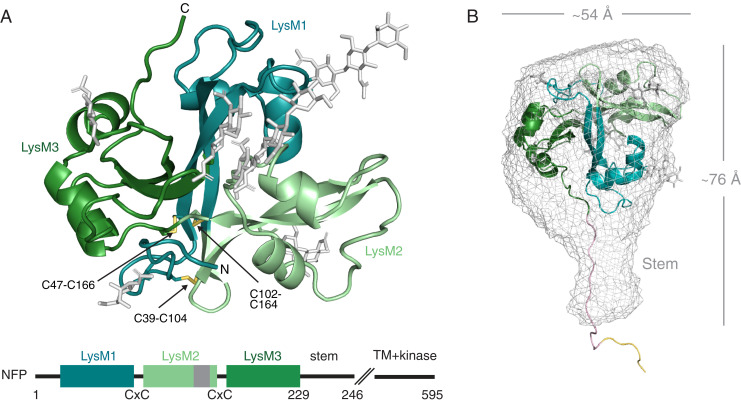
To gain structural insights into LCO perception in symbiosis, we crystallized the ectodomain of Medicago NFP (hereafter referred to as NFP), expressed and purified from insect cells (SI Appendix, Fig. S1). The structure of NFP (residue 31 to 233) was built, including six N-linked glycosylations visible in the 2.55-Å resolution electron density map (SI Appendix, Tables S1 and S2). NFP forms a compact cloverleaf structure where three canonical βααβ LysM domains are tightly interconnected and stabilized by three conserved disulfide bridges (C39-C104, C47-C166, and C102-C164) (Fig. 1A). NFP shares this cysteine bridge connectivity pattern and overall fold with other LysM receptors with active kinases involved in both LCO- and chitin-elicited defense signaling, such as Arabidopsis CERK1, Lotus CERK6, and Medicago LYK3 (SI Appendix, Fig. S2) (15, 16, 21), supporting the hypothesis that these receptors share a common evolutionary origin (22). To examine the structure in solution, we performed small-angle X-ray scattering (SAXS) measurements on NFP (SI Appendix, Table S3 and Fig. S3). The SAXS data and the reconstructed ab initio model fits the crystal structure; however, in addition an elongated stem-like structure is present, similar to that observed for the exopolysaccharide receptor EPR3 (23) (Fig. 1B). This stem region is most likely composed of the C-terminal part of NFP, which is not visible in the crystal structure and might serve to position the ectodomain at the correct distance from the plasma membrane or is involved in transducing the signal to the intracellular side.
Fig. 1.
Structure of the NFP receptor ectodomain. (A) Cartoon representation of the NFP crystal structure with the three LysM domains colored as indicated in the schematic. Glycosylations are shown in gray and disulfide bridges in yellow. On the schematic representation of the protein, the position of the identified hydrophobic patch in LysM2 is indicated in dark gray. (B) Mesh representation of the NFP ab initio SAXS envelope with a rigid body fit of the ectodomain structure. The solution structure reveals a stem-like structure which is not visible in the crystal and a modeled possible configuration of the stem (light pink) and the hexahistidine tag (yellow). The overall dimensions are shown in angstrom (Å).
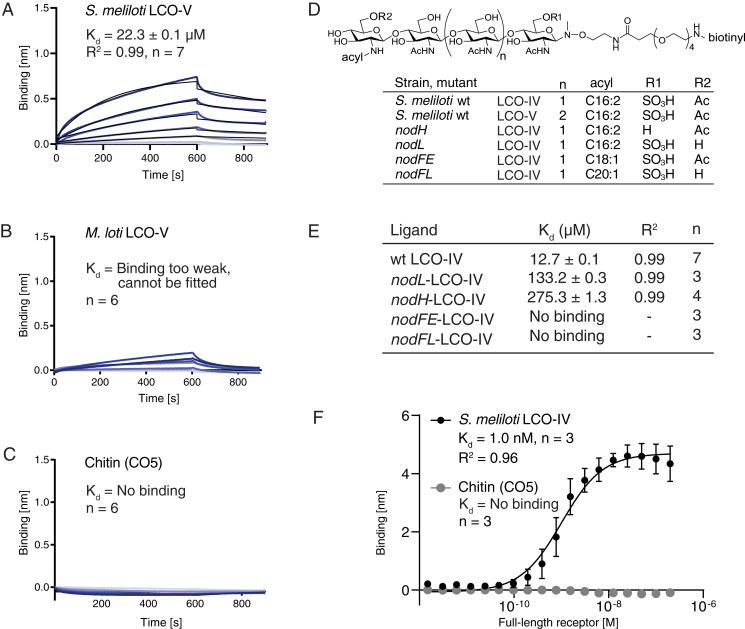
To understand the molecular determinants of symbiont recognition, we investigated the ligand-binding capabilities of NFP. LCO ligands are difficult to handle in solution because of their amphiphilic nature and tendency to form micelles. To overcome this problem, a biotinylated linker was specifically attached to the reducing end of isolated LCO ligands by chemoselective glycosyl-N-(methyl)oxyamine formation, enabling immobilization on streptavidin biosensors (24) (see all ligands in SI Appendix, Figs. S4–S6). This was accomplished without affecting the highly labile but functionally critical sulfate group present on S. meliloti LCOs. The functionality of the biotin-modified Nod factor was tested in an pEnod11-GUS reporter assay, in which the conjugate was able to induce downstream responses in Medicago roots to a similar degree as unconjugated S. meliloti Nod factor or S. meliloti bacteria (SI Appendix, Fig. S7). In biolayer interferometry (BLI) experiments, NFP-bound LCO-V and LCO-IV from its symbiont S. meliloti with Kd = 22.3 ± 0.1 µM and Kd = 12.7 ± 0.1 µM, respectively (Fig. 2 A and E). In parallel experiments, binding of NFP to LCO-V produced by Mesorhizobium loti (11, 14, 25), the symbiont of Lotus, was too weak to be quantified (Fig. 2B), which suggests that Medicago discriminates symbionts based on direct LCO binding by NFP. Additionally, NFP was unable to bind chitin (CO5; Fig. 2C), maltodextrin, or S. meliloti succinoglycan (EPS-I), an exopolysaccharide ligand devoid of N-acetylglucosamine (SI Appendix, Fig. S8), demonstrating that NFP is a selective LCO receptor. The comparison of NFP and the chitin-elicited immunity receptor CERK1 from Arabidopsis illustrates that both ectodomains bind their ligands with low-micromolar affinities (21), while their in vivo signaling is initiated by nanomolar concentrations (26, 27). These observations highlight the fact that, in both immunity and symbiotic LysM receptor–mediated signaling, specific but lower-affinity ligand interactions in the ectodomains are amplified by full-length receptors or receptor complexes in order to initiate and determine the cellular response.
Fig. 2.
NFP has ligand specificity and directly monitors LCO decorations in BLI experiments. (A) NFP binding to S. meliloti LCO-V. (B) NFP binding to M. loti LCO-V is too weak and cannot be fitted. (C) NFP does not bind chitopentaose (CO5) in BLI experiments. A concentration range of analyte (100 to 1.56 µM) was used for each experiment. Experimental binding curves are represented in blue and fitting curves in black. The goodness of fit is described by the global fit R square of the mean value for each point. Numbers of replicates performed using independent protein preparations (n) are indicated. (D) Structure of biotinylated S. meliloti LCO-IV conjugate and overview of S. meliloti mutants associated with variations in LCO structure. S. meliloti LCO-IV has a tetrameric N-acetylglucosamine backbone, is O-sulfated on the reducing end, O-acetylated on the nonreducing terminal residue, and monoN-acylated by a hexadecadienoyl (C16:2) group. (E) BLI data showing NFP binding to S. meliloti LCO-IV variants. (F) Steady-state BLI data of full-length receptor binding at t = 595 s of association to immobilized LCO-IV and CO5, respectively. The binding follows a sigmoidal dose–response model with Kd = 1.0 ± 0.37 nM. Error bars indicate SD. A total of 16 twofold dilution series of analyte (200 to 0.0061 nM) were used for each experiment.
To experimentally address the ligand-binding properties of full-length receptors and compare them to isolated ectodomains, we expressed and purified a full-length NFP receptor construct using tobacco infiltration (SI Appendix, Fig. S9). We were able to obtain sufficient material for these biochemical studies when fusing the NFP ectodomain to the intracellular domain of NFR5. The purified full-length receptors bound S. meliloti LCO-IV with high affinity (Kd =1.0 nM), while no binding to chitin was observed (Fig. 2F and SI Appendix, Fig. S9). The binding stoichiometry is different from the 1:1 stoichiometry we observe for the isolated ectodomains (SI Appendix, Fig. S9 D and E), suggesting that full-length receptors form dimers or higher-order oligomers when stabilized in membrane-mimicking micelles and therefore have more LysM2 binding sites per receptor complex. These data correlate our previously published data on the full-length NFR5 and NFR1 receptors that also bind LCOs with dissociation constants in the nanomolar range, corresponding to physiological Nod factor responses (14). Our data support the hypothesis that full-length receptors form oligomers that are responsible for the absence of 1:1 binding, which can explain the observed higher affinity due to avidity effects.
Studies of mutant rhizobial strains and measurements of calcium spiking in root hairs after LCO application have established that side-chain decorations on N-acetylglucosamine residues present at both reducing and nonreducing ends of Nod factors are functionally important and have a role in symbiotic signaling, although functions including protection against plant chitinases/hydrolases and insertion of the hydrophobic acyl chain in the plant plasma membrane have also been suggested (26, 28–30). To assess the role of LCO decorations directly and to understand how these contribute to selectivity, we measured NFP ectodomain binding to LCOs purified from the S. meliloti mutants nodL, nodH, nodFE, and nodFL (Fig. 2D and SI Appendix, Fig. S10), in which one or more modifications are either missing or altered (31). Elimination of the O-acetyl group on the nonreducing end in nodL–LCO-IV reduced binding to NFP by more than 10-fold (Kd = 133.2 ± 0.3 µM) compared to wild-type S. meliloti LCO-IV (Fig. 2E and SI Appendix, Fig. S10). The lack of the reducing end sulfate in nodH–LCO-IV drastically lowered binding to NFP by more than 21-fold (Kd = 275.3 ± 1.3 µM; Fig. 2E and SI Appendix, Fig. S10), which could explain the reduced calcium spiking observed with this nonsulfated LCO in Medicago (26). Similarly, both nodFE–LCO-IV containing a C18:1 instead of C16:2 acyl chain and nodFL–LCO-IV lacking the acetyl group and having a C20:1 acyl chain showed no significant binding, reflecting the inability to induce both nodule development and infection thread formation after inoculation of the respective S. meliloti mutants (32). These findings highlight the importance of the LCO Δ2 unsaturation for NFP binding, which is absent from both C18:1 and C20:1 fatty acids. These data support the conclusion that NFP monitors the integrity of the ligand by recognizing all individual decorations present on its cognate LCO ligand, making NFP a highly selective receptor.
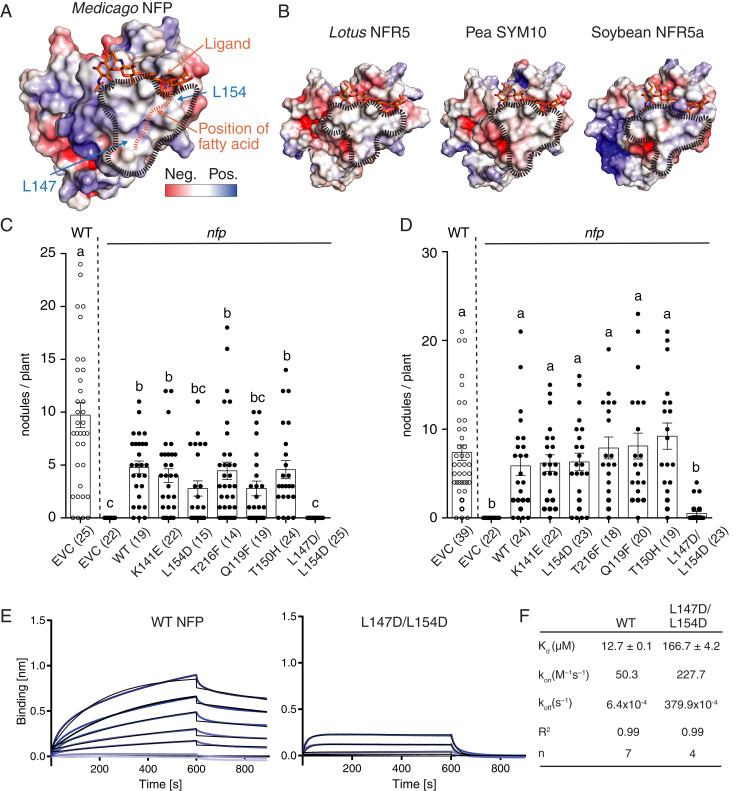
The crystal structure allows accurate calculation of the electrostatic surface potential of NFP, which revealed a hydrophobic patch in the LysM2 domain (Fig. 3A). Structural superposition of NFP with Arabidopsis CERK1 places chitin in the LysM2 region corresponding to the ligand-binding groove of CERK1 without spatial clashes. Interestingly, when applying this chitin backbone orientation on an LCO ligand, the acyl moiety on the nonreducing end of the LCO is positioned into the hydrophobic patch area (Fig. 3A). To investigate if this is a common structural feature, we examined the Lotus NFR5, pea SYM10, and soybean NFR5-α receptors previously demonstrated to be crucial for LCO perception (9, 14, 18, 33). Homology models of these LCO receptors, based on the NFP crystal structure, revealed that the hydrophobic patch is indeed present in the equivalent position in LysM2 (Fig. 3B), supporting the concept of a conserved signature in this class of pseudokinase LCO receptors. The functional importance of the hydrophobic patch for symbiosis was assessed by obtaining structure-guided amino acid substitution variants of NFP that were tested for functional complementation in a Medicago nfp mutant background using hairy root transformation. The complementation with wild-type Nfp resulted in an average of five nodules per plant 49 d after inoculation with S. meliloti 1021. Corresponding experiments with NFP substitution variants replacing residues outside the hydrophobic patch in LysM2 (Q119F, K141E, and T150H) or in LysM3 (T216F) did not affect nodulation (Fig. 3C). Two surface-exposed leucine residues (L147 and L154) give NFP its hydrophobic character in LysM2. To test their contribution, we replaced these with aspartate residues, which preserves the size of the side-chain but adds a negative charge (Fig. 3A). Interestingly, roots transformed with the double mutant construct (L147D/L154D) failed to complement nfp, and no nodules developed after inoculation with S. meliloti 1021 (Fig. 3C). The complementation experiments were repeated using Sinorhizobium medicae, which is reported to nodulate Medicago with higher efficiency (34). These experiments confirmed that the hydrophobic patch is required for functional symbiotic signaling in planta, as the number of nodules were dramatically reduced in the double mutant (L147D/L154D) (Fig. 3D). This conclusion is strengthened by independent results from in planta studies showing that an L154P amino acid substitution in NFP LysM2 failed to complement nfp in Medicago (35). The biochemical analysis of LCO binding to the hydrophobic patch mutant revealed that the purified NFP L147D/L154D ectodomain (SI Appendix, Fig. S11) bound S. meliloti LCO-IV with a 13-fold lower affinity (Kd = 166.7 ± 4.2 µM) than wild-type NFP (Fig. 3 E and F). The association rate (kon) was 4.5-fold faster and the dissociation rate (koff) was increased by 59-fold in the double mutant compared to the wild-type NFP (Fig. 3 E and F), suggesting that the hydrophobic patch has a stabilizing effect on LCO binding mediated by the acyl chain. Together, these data provide evidence that the hydrophobic patch in NFP is a conserved structural signature critical for LCO perception and symbiotic signaling. To assess whether this feature distinguishes the LCO perception mechanism by NFP from chitin perception, the kinetics of Arabidopsis CERK1 binding to a chitin fragment (CO5) were determined (SI Appendix, Fig. S12). These kinetic measurements revealed fast association and dissociation rates, reminiscent of the kinetics we observed for the hydrophobic patch mutant of NFP-binding LCO (Fig. 3F). This emphasizes differences in perception mechanisms between CERK1 and NFP and highlights that the role of the acyl chain is to enhance the dwell time of the NFP–LCO complex.
Fig. 3.
A hydrophobic patch in LysM2 is important for LCO binding and symbiotic signaling. (A) Molecular docking of a chitotetraose molecule (orange sticks) onto the structure of NFP. The surface of NFP is colored according to its electrostatic potential (±5 kT/e), and the hydrophobic patch is highlighted (black dashes). A possible position of the fatty acid chain on the hydrophobic patch is indicated (orange dashes). (B) Homology models of characterized LCO receptor ectodomains: L. japonicus NFR5, Pisum sativum (Pea) SYM10, and Glycine max (Soybean) NFR5α. All have a characteristic hydrophobic patch in LysM2. (C and D) Complementation analysis of NFP variants in an nfp Medicago background underlines that the hydrophobic patch is a prerequisite for functional symbiotic signaling. Columns represent mean nodule numbers after 49 d post infection (S. meliloti) (C) or 28 dpi (Sinorhizobium medicae) (D). Circles indicate individual counts. Empty circles: Medicago Jemalong wild-type background. Filled circles: nfp mutant background. EVC: empty vector control, WT: wild-type NFP. Error bars represent the SEM. Letters indicate statistical significance (ANOVA, Tukey, P < 0.05). Number of plants are indicated in parentheses. (E) BLI experiments of NFP WT and hydrophobic patch mutant (L147D/L154D) binding to S. meliloti LCO-IV. A concentration range of analyte (100 to 1.56 µM) was used for each experiment. Experimental binding curves are represented in blue and fitting curves in black. (F) Table summarizing the kinetic parameters for data in E. The goodness of fit is described by the global fit R square on the mean value of each point. Numbers of replicates performed using independent protein preparations (n) are indicated.
To investigate whether the presence of the hydrophobic patch can serve as a prediction tool for as-yet uncharacterized LCO receptors, we performed molecular modeling of the closest NFP homologs obtained from the chickpea, bean, and peanut genomes. The resulting structures show that the hydrophobic patch in LysM2 is conserved (SI Appendix, Fig. S13A), consistent with the fact that these proteins are members of the NFP/NFR5 class of LCO receptors. Additionally, the entire Medicago and Lotus NFP/NFR5 receptor families were evaluated for presence of the hydrophobic patch. The pseudokinase receptors involved in chitin-elicited defense responses, Medicago LYK4 and Lotus LYS13/14 (15), did not feature the hydrophobic patch, which suggests that this approach represents a valuable tool for the discovery of putative LCO receptors. Other receptors (LYR2/LYS15/LYS16) had a highly negatively charged surface in the same area, which we predict would exclude their function as primary LCO receptors (SI Appendix, Fig. S13 B and C). Studies of membrane fractions suggested that the LysM3 domain of Medicago LYR3 constitutes a high-affinity LCO receptor showing LCO binding independent of the side-chain decorations (36). Modeling does not predict a hydrophobic patch in LysM3 of LYR3 or in the Lotus ortholog LYS12 (SI Appendix, Fig. S13 C and D), and lys12 mutants have no symbiotic phenotype (37). However, our results do not exclude other modes of LCO perception in these receptors as was recently found for the LYK3/NFR1 receptor class, which acts in complex with NFP/NFR5, respectively (16, 38, 39). We found that the hydrophobic patch is preserved in Medicago LYR1 and in its Lotus homolog LYS11 (SI Appendix, Fig. S13 B and C), although the native LYS11 receptor could not be assigned a role in rhizobial symbiosis based on mutant studies and its inability to complement nfr5 mutants (40). Our predictions nevertheless agree with independent results showing that a chimeric receptor consisting of the ectodomain of Lotus LYS11 fused to the kinase domain of NFR5 can complement nfr5 when overexpressed, while the same construct under the Nfr5 promoter cannot complement nfr5 (40).
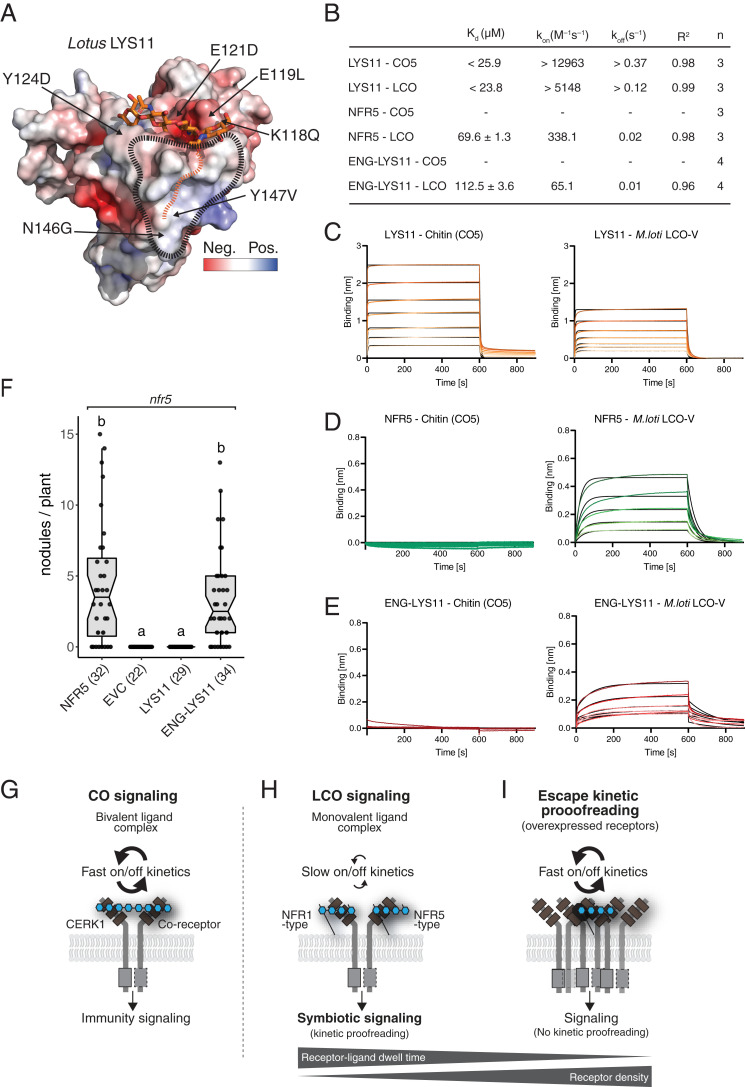
The ability to complement nfr5 when overexpressed confirm that LYS11 has the basic capacity to perceive LCOs and thus provides a test system to examine the function of the hydrophobic patch, the impact of binding kinetics and for validating our in silico prediction tool. We therefore expressed and purified the LYS11 ectodomain (hereafter LYS11) and determined its crystal structure to 2.8-Å resolution (SI Appendix, Fig. S14 and Tables S1 and S2). The calculation of the accurate electrostatic surface potential revealed that LYS11 indeed contains a hydrophobic patch in LysM2 (Fig. 4A), providing direct experimental support for the predictive power of our modeling approach. LYS11 is able to bind M. loti LCO (Kd < 25.9 µM) but, in contrast to NFP, LYS11 is also capable of binding chitin (Kd < 23.8 µM) (Fig. 4 B and C). Both ligands have very fast association and dissociation rates, similar to the chitin kinetics observed for Arabidopsis CERK1 (SI Appendix, Fig. S12). To investigate whether these fast, ligand-binding kinetics are responsible for the inability of Nfr5 promoter–expressed LYS11 to complement nfr5 mutants (40), we expressed and purified the ectodomain of Lotus NFR5 (hereafter NFR5) (SI Appendix, Fig. S14) and characterized its ligand-binding behavior. NFR5 does not bind chitin but recognizes M. loti LCO (Kd = 69.6 ± 1.3 µM) with a slower kinetic profile compared to LYS11 (Fig. 4 B and D). To explore the importance of these differences, we engineered the LYS11 ligand binding site (Fig. 4A) by substituting six surface-exposed residues from NFR5 onto the LYS11 framework. This engineered LYS11 (ENG-LYS11) completely lost the ability to bind chitin but gained a specific and slower kinetic profile to M. loti LCO (Kd = 112.5 ± 3.6 µM) (Fig. 4 B and E and SI Appendix, Fig. S14). These binding experiments show that it is possible to engineer and fine-tune LCO specificity and binding kinetics by modifying the LysM2 binding site. Functionality of the engineered LYS11 was assayed by complementation of Lotus nfr5 mutants. LYS11 under the control of the Nfr5 promoter is unable to complement nfr5, but our engineered LYS11 showed full complementation of nfr5 (Fig. 4F). Interestingly, our data show that LYS11 has a higher affinity for LCO compared to NFR5 but is still unable to functionally complement NFR5 in symbiotic signaling. Furthermore, the engineered LYS11 has an even lower affinity for LCO compared to NFR5 but has both gained specific LCO recognition and slow binding kinetics, which is enough to support symbiotic signaling. We infer that symbiotic signaling requires specific and optimal, LCO-binding kinetics for activation, characterized by slower ligand dissociation and long receptor–ligand dwell time.
Fig. 4.
Engineering ligand specificity and binding kinetics enables LYS11 to support symbiotic signaling. (A) Electrostatic surface potential (±5 kT/e) representation of the crystal structure of LYS11. The chitin ligand of CERK1 is docked into the LysM2 chitin-binding groove. The hydrophobic patch and the proposed acyl chain binding site are indicated. Residues substituted in the engineered LYS11 version are indicated. (B) Table overview of ligand-binding parameters measured by BLI. A concentration range of analyte (100 to 1.56 µM) was used for each experiment. Experimental binding curves are represented in colors and fitting curves in black. The binding events were faster than the instrument sensitivity, so the values for LYS11 are given here as a minimal (kinetic parameters) or maximal (dissociation constant). (C) BLI experiments of LYS11 binding to CO5 and M. loti LCO-V. (D) BLI experiments of NFR5 binding to CO5 and M. loti LCO-V. (E) BLI experiments of engineered LYS11(ENG-LYS11) binding to CO5 and M. loti LCO-V. (F) Complementation analysis in an nfr5 Lotus background shows that the ligand binding site of LYS11 can be engineered to support symbiotic function. Columns represent mean nodule numbers after 35 dpi with M. loti. Circles indicate individual counts. EVC: empty vector control. Error bars represent the SEM. Letters indicate statistical significance (ANOVA, Tukey, P < 0.05). Numbers of plants are indicated in parentheses. (G–I) Models of ligand perception and signaling in immunity and symbiosis. (G) Perception of a bivalent chitin ligand with fast kinetics by Arabidopsis CERK1/coreceptor complex leading to immunity signaling. (H) LCO perception by legume NFR1/NFR5 class receptors on a monovalent LCO ligand with slow binding and kinetic proofreading leading to symbiotic signaling. (I) Overexpression of LYS11 leads to a bypass of the kinetic proofreading mechanism despite fast on/off binding kinetics (40). Altering the hydrophobic patch in NFP leads to a faster binding kinetic and no symbiotic signaling (equal to receptor resting state) and engineering LYS11 with slow and specific LCO binding restores kinetics proofreading and symbiotic signaling even at low-receptor density when expressed from Nfr5 promoter.
Discussion
This study provides structural and kinetic insights into the LCO perception and signaling mechanisms that enable legumes to activate a symbiotic state in root cells. We found that strain-specific LCO decorations are directly monitored and that specific LCO perception is dependent on the presence of a conserved structural signature, the hydrophobic patch in LysM2 of LCO receptors of the NFP/NFR5 class. We recently discovered that the LYK3/NFR1 class of receptors with an active kinase uses LysM1 to recognize ligands (14, 16), and in this study we show that the NFP/NFR5 class of receptors with a pseudokinase uses LysM2 as the ligand binding site. Ligand-induced complex formation between NFR1 and NFR5 receptors is probably part of signal transduction, but the molecular mechanisms and how complex formation between the full-length receptors occur remains to be determined. Our data support that full-length receptors form complexes, resulting in high-ligand affinity, which suggests that multiple ectodomains cooperate to give stringent signaling in plants. We further demonstrate that receptors from the pseudokinase class of LCO receptors differ in their binding properties and that slower dissociation kinetics are crucial for their signaling capacities. Our data show that slow LCO-binding kinetics is even more important for signaling than the affinity in itself. This perception mechanism contrasts with the chain length–mediated mechanism for plant pathogen–related chitin perception shown for Arabidopsis CERK1 that has fast on/off rate kinetics and receptor dimerization on a bivalent chitin ligand (Fig. 4G). We infer that the requirement for slower dissociation kinetics in LCO signaling allows the receptor–ligand complexes to stabilize and reach a threshold dwell time required for transition from resting state to activation state and recruitment of intracellular signaling components (Fig. 4H). Our results support a receptor proofreading mechanism in which ligand specificity and binding kinetics are balanced against receptor abundance to ensure a sensitive-yet-restrained perception system defining the default setting of a dynamic signaling equilibrium (Fig. 4 H and I). Increasing receptor density by overexpression leads to escape of kinetic proofreading as the requirement for slow LCO-binding kinetics are overruled (Fig. 4I) (40). Importantly, when LYS11 receptors are engineered in LysM2 to have a slow-but-specific LCO-binding behavior, they restore symbiotic signaling even at low expression levels. The identified hydrophobic patch in LYR1 and LYS11 suggests that these receptors have a role in recognition of LCOs secreted by arbuscular mycorrhizal fungi (Myc factors) (40) and may suggest that there is a different balance between ligand dwell time and receptor abundance in the plant interaction with arbuscular mycorrhizal fungi. Alternatively, these receptors could be involved in interactions with other LCO-producing fungi. Finally, we demonstrate that the hydrophobic patch enables in silico identification of LCO receptors of the NFP/NFR5 class. The expansion of the LysM receptor kinase family within plants highlights the importance of such a prediction tool for identifying and analyzing LysM receptors whose binding properties and signaling competence can be altered, providing targets and a template for future engineering of plant receptors.
Materials and Methods
Expression and Purification of Full-Length Receptor.
Full-length receptor construct, with NFP ectodomain and NFR5 intracellular domain, was produced using protocols established for full-length NFR1/NFR5 (14). Nicotiana benthamiana plants, 3- to 4-wk-old, were syringe infiltrated in infiltration buffer (10 mM 2-(N-morpholino)ethanesulfonic acid [MES], 10 mM MgCl2, and 0.15 mM acetosyringone) with Agrobacterium tumefaciens strain AGL-1 containing the receptor construct fused to a C-terminal eYFP-decahistidine tag in a modified vector pEarleyGate101 expressed from the 35S promoter. An A. tumefaciens AGL-1 strain expressing the tomato bushy stunt virus post-transcriptional gene silencing suppressor P19 was coinfiltrated at an optical density at 600 nm ratio of 0.2:0.02 receptor construct:P19. Leaves were harvested after 3 d by flash freezing in liquid nitrogen. Membranes were prepared as follows: Plant material was homogenized at 4 °C in a blender in 2 mL/g plant material extraction buffer (50 mM 3-(N-morpholino)propanesulfonic acid (MOPS) ⋅ KOH, pH 7.5, 0.3 M sucrose, 0.6% [weight/volume] PVPP, 2 mM β-mercaptoethanol, 1 mM PMSF, and protease inhibitor tablets [Sigma]). The homogenate was filtered through rayon-polyester fabric (Miracloth, Millipore) and cleared by centrifugation at 10,000 × g for 10 min at 4 °C. The membrane pellet was resuspended in 0.07 mL resuspension buffer per gram plant material used (5 mM potassium phosphate at pH 7.8, 2 mM 2- β-mercaptoethanol, 0.3 M sucrose, 1 mM PMSF, and one Sigma protease inhibitor tablet per 100 mL). Plasma membrane enrichment was done by a modified two-phase aqueous partition protocol, as described previously (41), with 6.5% (weight/weight) Dextran-100 and 6.5% (weight/weight) PEG-3350 in the bulk phase system. The final upper phase containing enriched plasma membrane was precipitated by centrifugation at 100,000 × g for 1 h at 4 °C. Plasma membranes were solubilized in 8× critical micelle concentration (CMC) Fos-choline 10 (Affymetrix) by incubation for 1 h at 4 °C. Insoluble membrane parts were removed by centrifugation at 100,000 × g for 1 h. The supernatant was diluted to 1 to 2× CMC Fos-choline 10 concentration with Wash buffer (50 mM Tris ⋅ HCl pH 8, 500 mM NaCl, 10 mM Imidazole, 2 mM β-mercaptoethanol, 1 mM PMSF, and Sigma protease inhibitor tablets). Ni-immobilized metal affinity chromatography (IMAC) was performed using 1-mL Protino Ni-NTA columns (Machery-Nagel), with an elution buffer containing 50 mM Tris ⋅ HCl pH 8, 500 mM NaCl, 500 mM Imidazole, 2 mM β-mercaptoethanol, 1× CMC Fos-Choline 10, 1 mM PMSF, and Sigma protease inhibitor tablets. Purification was finalized using a Superose 6 10/300 GL column on an ÄKTA Pure system (GE Healthcare) in gel filtration buffer (50 mM Tris ⋅ HCl pH 8, 200 mM NaCl, 2 mM β-mercaptoethanol, 1× CMC Fos-Choline 10, 1 mM PMSF, and Sigma protease inhibitor tablets). Purity and yield were assayed using SDS-polyacrylamide gel electrophoresis (PAGE) by detecting eYFP in-gel fluorescence on a Typhoon Trio Variable Mode Imager (Amersham Biosciences) and a titration series of GFP to estimate protein concentration. Typical yields were 5 to 10 ng receptor construct per 500 g plant material.
Expression and Purification of Receptor Ectodomains.
M. truncatula NFP (residues 28 to 246) was codon-optimized for insect cell expression (Genscript) and cloned into the pOET4 baculovirus transfer vector (Oxford Expression Technologies). The native NFP signal peptide (residues 1 to 27, predicted by SignalP4.1) was substituted by the AcMNPV gp67 signal peptide to facilitate secretion, and a hexa-histidine tag was added to the C terminus. Point mutants of NFP were engineered by site-directed mutagenesis. Recombinant baculoviruses were produced in Sf9 cells using the FlashBac Gold kit (Oxford Expression Technologies) according to the manufacturer’s instructions with Lipofectin (Thermo Fisher Scientific) as transfection reagent. Protein expression was performed as follows. Suspension-cultured Sf9 cells were maintained shaking at 26 °C in serum-free MAX-XP (BD-Biosciences, discontinued) or HyClone SFX (GE Healthcare) medium supplemented with 1% Pen-Strep (10,000 U/mL, Life Technologies) and 1% chemically defined lipid concentrate(Gibco). Protein expression was induced by addition of recombinant passage 3 virus at a cell density of 106 cells/mL. After 5 to 7 d of expression, medium containing NFP was harvested by centrifugation followed by an overnight dialysis step against 50 mM Tris ⋅ HCl pH 8 and 200 mM NaCl at 4 °C. NFP was enriched by two subsequent steps of Ni-IMAC purification (first HisTrap excel then HisTrap HP; GE Healthcare). For crystallography experiments, N-linked glycans were trimmed using the endoglycosidase PNGase F (1:15 [weight/weight] PNGase F:NFP, room temperature, overnight). NFP was then purified by gel filtration on a Superdex 200 increase 10/300, Superose 6 10/300 GL, or HiLoad Superdex 200 16/600 (all GE Healthcare) in phosphate-buffered saline, pH 7.2 supplemented to a total of 500 mM NaCl (for BLI-binding assays) or 50 mM Tris ⋅ HCl, 200 mM NaCl (for crystallography). NFP elutes as a single, homogeneous peak corresponding to a monomer. The NFP-L147D/L154D variant, NFR5, LYS11, engineered LYS11, and CERK1 (11) were expressed and purified using the same protocol. For crystallography experiments, LYS11 was treated with PNGase F at a molar ratio of 1:25 (PNGase F:LYS11) for 3 h at room temperature, followed by 20 h incubation at 4 °C. Deglycosylated LYS11 was then gel filtrated into a buffer containing 50 mM sodium-Hepes pH 7, 150 mM NaCl on a HiLoad Superdex 75 16/600 (GE Healthcare) column.
Crystallization and Structure Determination.
Crystals of PNGaseF-treated NFP were obtained using a vapor diffusion setup at 3 to 5 mg/mL in 0.2 M sodium acetate, 0.1 M sodium-cacodylate pH 6.5, 30% (wild-type/volume) PEG-8000 at 19 °C. Crystals were cryoprotected in their crystallization condition supplemented with 5% (weight/volume) PEG-400 before being flash cooled in liquid nitrogen. Diffraction data to 2.55-Å resolution were obtained at the MaxLab I911-3 beamline in Lund, Sweden. Due to strong anisotropy, data were truncated elliptically with a resolution cutoff of 2.55 Å in the a* and c* directions and 3.2 Å in the b* direction using the University of California, Los Angeles(UCLA) Diffraction Anisotropy Server (42). The phase problem was solved by molecular replacement using Phaser (43) with a homology model, generated with Phyre2 (44) based on the AtCERK1 ectodomain structure (Protein Data Bank[PDB] coordinates 4EBZ), as a search model. Crystals of PNGaseF-treated LYS11 were obtained by vapor diffusion at 6.8 mg/mL in 0.1 M sodium malonate pH 6.0, 12% PEG-3350 at 4 °C. Cryoprotection was achieved by adding reservoir buffer supplemented with 40% glycerol to the crystallization drop prior to crystal mounting and flash cooling in liquid nitrogen. Diffraction data to 2.88-Å resolution were obtained at the Biomax beamline in Lund, Sweden. Because of strong anisotropy, data were truncated elliptically with a resolution cutoff of 3.3 Å in the a* direction, 3.4 Å in the b* direction, and 2.88 Å in the c* direction using the UCLA Diffraction Anisotropy Server. The phase problem was solved by molecular replacement using Phaser (43) with NFP as search model. Model building of both NFP and LYS11 was done in COOT (45) and refined using the PHENIX suite (46). Data collection statistics for both ellipsoidal and spherical datasets and refinement statistics are reported in SI Appendix, Tables S1 and S2. Figures were prepared using PyMOL version 2.4.1. Electrostatic surface potentials were calculated using PDB2PQR and APBS webservers and visualized in PyMOL using APBS tools 2.1.
SAXS.
SAXS of NFP was measured in batch at different concentrations (1, 2, 4, and 6 mg/mL for glycosylated NFP in phosphate-buffered saline, pH 7.4, 500 mM NaCl) at the European Molecular Biology Laboratory (EMBL) P12 beamline PETRA III in a temperature-controlled cell at 20 °C at a wavelength of 1.24 Å. Data analysis and modeling were done using BioXTAS RAW, GNOM, and the ATSAS program suite (47–49). The ab initio low-resolution structure was modeled in DAMMIF (slow mode) using 15 individual reconstructions. Envelopes were aligned and averaged with DAMAVER (50). The average was finally refined in DAMMIN (51). NFP models with added N- and C-terminal missing tails and rigid body were fitted into envelopes with Colores (52). Theoretical scattering curves were calculated in CRYSOL (53). Reconstructed NFP was built in COOT and N-glycans visible in the crystal structure expanded to paucimannosidic Man3GlcNAc2 glycans, typical for insect cell expression (54). The molecular weight derived from the forward scattering was determined using an internal fresh bovine serum albumin standard. SAXS data statistics are reported in SI Appendix, Table S3.
BLI.
Binding of NFP, NFP L147D L154D, NFR5, LYS11, engineered LYS11, and CERK1 to ligand conjugates was measured on an Octet RED 96 system (Pall ForteBio) in gel filtration buffer with the addition of 0.01% (volume/volume) Tween-20. Biotinylated ligand conjugates were immobilized on streptavidin biosensors (kinetic quality, Pall ForteBio) at a concentration of 125 to 250 nM for 5 min. Association to the receptor ectodomains was measured for 10 min and dissociation for 5 min. Data analysis was performed in GraphPad Prism 8 (GraphPad software). Kinetic parameters were determined by nonlinear regression (association then dissociation model with interstep correction) on biotin reference subtracted data. The goodness of fit is described by the global fit R square of the mean value for each point. The binding of full-length NFP receptors to biotinylated ligands was performed in the same way. Data were analyzed by plotting the biotin-subtracted response at t = 595 s against receptor concentration. The equilibrium dissociation constant Kd was determined using the sigmoidal dose–response equation in GraphPad Prism.
MST.
NFP was fluorescently labeled (Protein Labeling Kit Blue NHS, NanoTemper Technologies) according to the manufacturer’s instructions. Steady-state binding to a titration series of ligands [0.6 to 20,000 nM for S. meliloti succinoglycan (exopolysaccharide-I [EPS-I]) (55) and 0.6 to 20,000 nM for maltodextrin] was measured using a constant concentration of NFP (12.5 to 50 nM). Data were processed using NT.Analysis (NanoTemper Technologies) and GraphPad Prism 7 (GraphPad Software, Inc.).
Structural Modeling.
Homology modeling was performed with SWISS-MODEL using the crystal structure of NFP as template (56). Electrostatic surfaces were calculated using PDB2PQR and APBS webservers and visualized in PyMOL using APBS tools 2.1.
Complementation.
M. truncatula wild type (Jemalong), M. truncatula nfp-2 mutant, L. japonicus wild type (Gifu), and L. japonicus nfr5-2 mutant containing proNin-Gus (nfr5-2_pNin-gus) lines were used for nodulation assays. For germination, all Medicago and Lotus seeds were scarified with sandpaper and surface-sterilized with diluted bleach solution (0.5% active chlorine). Subsequently, seeds were imbibed overnight in sterile water at room temperature, then germinated on inverted 0.8% water agar plates for 1 d at room temperature in darkness. Seedlings were transferred to slanted agar plates supplied with 1/2× Gamborg’s B5 nutrient solution including vitamins (Duchefa Biochemie). Agrobacterium rhizogenes–mediated hairy root transformations were done on 7- to 10-d-old seedlings using the AR1193 strain (57) carrying the respective construct. After hairy root formation, nontransgenic roots were removed, and plants were transferred to pots with lightweight expanded clay aggregate (2 to 4 mm; Saint-Gobain Weber A/S) and a top layer of vermiculite (size M; Damolin A/S). Pots were watered with nitrogen free 1/4× Broughton and Dilworth nutrient solution. Plants were inoculated with the respective bacterial strain suspended in sterile water to OD600 = 0.02 1 wk after moving to pots. Plants were grown at 21 °C under 16/8-h light/dark conditions.
LCO Production.
M. loti LCO-V and chitopentaose (CO5) conjugates and S. meliloti succinoglycan (EPS-I) were produced as described previously (11, 55). For wild-type LCO extractions, S. meliloti exoY Tn5 mutant containing plasmid pMH682 TcR was used. For extraction of LCO derivatives lacking specific decorations appropriate S. meliloti, 1,021 nod mutant strains containing pMH682 were used (58). Strains were cultured in 10 L glucose-supplemented rhizobium defined medium (G/RDM) broths containing 0.5 μM luteolin for 48 h. The cells were pelleted, and the supernatant was recovered and mixed with 3.5 L (per 10 L supernatant) l-butanol for 12 h. The phases were left to separate at 4 °C for 12 h, the organic phase was recovered, dried by rotary evaporation, resuspended in 15 mL tert-butanol, and freeze dried, yielding ∼2 to 3 g off-white to dark-brown solids. The crude solids containing LCOs were first triturated three times with ethyl acetate, which caused the solids to lose some color. The solids were then dried completely under a nitrogen flow, and resolubilized using H2O–acetonitrile (1:1, volume/volume), producing a light- to dark-brown solution. This solution was then fractionated using semipreparative high-performance liquid chromatography (HPLC) on an UltiMate 3000 instrument fitted with a Waters 996 photodiode detector, using a Phenomenex Luna 5-μm, C18 (2), 100-Å, 250- × 100-mm semipreparative column. A gradient of 5 to 100% acetonitrile in water over 40 min running at 5 mL/min was used. The LCOs eluted from 21 to 23 min (44% acetonitrile) and were collected and lyophilized for a second purification step on semipreparative HPLC. The second program used an isocratic elution of 70% acetonitrile in water (0.1% formic acid). S. meliloti LCO-IV(Ac,C16:2,S) and LCO-V(Ac,C16:2,S) eluted from 13 to 16.5 min and were collected individually and lyophilized. A total of 45 mg LCO-IV(Ac,C16:2,S) and 30 mg LCO-V(Ac,C16:2,S) was obtained from the S. meliloti wild-type strain. Heteronuclear single quantum coherence (HSQC) and total correlation spectroscopy (TOCSY) NMR spectra were recorded on a Bruker Avance 300-MHz spectrometer with a Broadband Observe (BBO) probe on a sample of S. meliloti LCO-IV(Ac,16:2,S) dissolved in D2O-CD3CN, 1:1, at a final concentration of 10 mg/mL. LCOs from S. meliloti nodL, nodH, nodFE, and nodFL mutant strains were cultivated and prepared by butanol extraction in the same way as the S. meliloti wild-type strain. The appearance following this procedure varied between the mutant strains from being light yellow, completely dry granules (S. meliloti nodH and nodFE) to brown and sticky material (S. meliloti nodL and nodFL). The crude material was resolubilized using H2O–acetonitrile (1:1, volume/volume), vortexed, and centrifuged. The supernatant was filtered through a 0.25-μm syringe filter and fractionated using preparative HPLC, using a Phenomenex Jupiter 5-μm, C4, 300-Å, 250- × 21.2-mm column. A gradient of 5 to 100% acetonitrile in water over 30 min running at 10 mL/min was used. The LCOs eluted from 20 to 26 min (40 to 55% acetonitrile) and were collected and lyophilized. A gradient running over a course of 50 min was applied for S. meliloti nodH in order to obtain a better separation. LCOs from S. meliloti nodL (22 mg total) were separated into fractions containing 1) IV(16:2,S), 68%; 2) IV(16:1,S), 20%; and 3) V(16:2,S), 12%. LCOs from S. meliloti nodH (7.5 mg total) were separated into fractions containing 1) IV(Ac, 16:2), 74%; 2) V(Ac, 16:2), 21%; and 3) III(Ac, 16:2), 5%. LCOs from S. meliloti nodFE (40 mg total) were separated into fractions containing 1) IV(Ac,18:1,S), 49%; 2) V(Ac,18:1,S), 17%; 3) IV(Ac,16:2,S), 5%; 4) V(Ac,16:2,S), 5%; 5) IV(Ac,20:0(OH),S), 17%; 6) IV(Ac,18:0(OH),S), 5%; and 7) IV(Ac,16:2) and V(16:2,S), 2%. LCOs from S. meliloti nodFL (8.9 mg total) were separated into fractions containing 1) IV(20:1,S), ≥50%; 2) V(20:1,S) and IV(20:0,S), <30%; 3) IV(22:1,S) and V(22:1,S), <10%; 4) IV(18:1,S), <10%; and 5) IV(22:0(OH),S) and IV(24:0(OH),S), <10%. The main fractions (underlined above) were used for further chemical modification.
LCO Conjugation.
LCO-biotin conjugates were synthesized using a two-step procedure from isolated LCOs, as shown in (SI Appendix, Fig. S5). Reagents and solvents were purchased from Sigma-Aldrich and used as received. O-(2-Aminoethyl)-N-methyl hydroxylamine trifluoroacetic acid salt was prepared, as described previously (59). Purified LCOs (1- to 7-mg scale, final concentration of 5 mM) were dissolved in 0.62 M sodium acetate buffer, pH 4.5, containing 50% acetonitrile, and O-(2-aminoethyl)-N-methyl hydroxylamine trifluoroacetic acid salt (150 mM, 30 eq.) was added. The resulting mixture was allowed to react at room temperature for 16 h. The N-glycosyl-N-(methyl)oxyamine intermediate was purified by semipreparative HPLC on an UltiMate 3000 instrument fitted with a Waters 996 photodiode detector, using a Phenomenex Luna 5-μm, C18 (2), 100-Å, 250- × 100-mm semipreparative column. An isocratic elution at 40% acetonitrile in water, 5 mL/min for 30 min was used. The intermediates eluted after ∼15 to 25 min. N-Glycosyl-N-(methyl)oxyamine formation was confirmed by mass spectrometry. The purified intermediate was dissolved in 50 mM sodium tetraborate buffer, pH 8.5, at a concentration of 10 mM. NHS-dPEG4-biotin (15 mM, 1.5 equiv.) was then added. The resulting mixture was allowed to react at room temperature for 16 h. The biotin conjugate product was purified by semipreparative HPLC. A gradient of 5 to 100% acetonitrile in water over 40 min running at 5 mL/min was used. The conjugates eluted after ∼15 to 20 min (50 to 60% acetonitrile). The biotin-LCO conjugates were quantified using the 4'-hydroxyazobenzene-2-carboxylic acid (HABA)/avidin biotin quantification kit from Pierce. Positive-mode high-resolution mass spectra (HR-MS) were obtained using a Bruker Impact HDII Quadrupole-Time of Flight (QTOF) instrument, and negative-mode HR-MS were obtained using a Bruker Solarix XR electrospray ionization Fourier transform ion cyclotron resonance (ESI-FT-ICR) instrument, equipped with a 7-T magnet. The following data were obtained for biotin-LCO conjugates. S. meliloti LCO-IV(Ac,C16:2,S): HR-MS (ES−) calculated for C72H123N9O32S2, [M−H]−: 1,688.7643; found: 1,688.7658. S. meliloti LCO-V(Ac,C16:2,S): HR-MS (ES−) calculated for C80H136N10O37S2, [M−H]−: 1,891.8437; found: 1,891.8427. S. meliloti nodL-LCO-IV(C16:2,S): HR-MS (ES+) calculated for C70H121N9O31S2, [M+H]+: 1,648.7683; found: 1,648.7667, and calculated for [M+2H]2+: 824.8878; found: 824.8876. S. meliloti nodH-LCO-IV(Ac,C16:2): HR-MS (ES+) calculated for C72H123N9O29S2, [M+H]+: 1,610.8220; found: 1,610.8210, and calculated for [M+2H]2+: 805.9147; found: 805.9146. S. meliloti nodFE-LCO-IV(Ac,C18:1,S): HR-MS (ES+) calculated for C74H129N9O32S2, [M+H]+: 1,720.8258; found: 1,720.8227, and calculated for [M+2H]2+: 860.9166; found: 860.9166. S. meliloti nodFL-LCO-IV(C20:1,S): HR-MS (ES+) calculated for C74H131N9O31S2, [M+H]+: 1,706.8465; found: 1,706.8534, and calculated for [M+2H]2+: 853.9269; and found: 853.9273. In-source fragmentation analyses were performed at 75 or 150 eV (m/z ± 0.5 accuracy) using a MSQ Plus electrospray ionization (ESI) mass spectrometer from Thermo Fisher.
Enod11-GUS Reporter Assay.
M. truncatula plants, 5-d-old, containing the pEnod11-GUS reporter (60) were treated for 12 h with either S. meliloti 1021 bacteria (OD600 = 0.02), 10−8 M S.meliloti LCO-IV (Ac,16:2, S), 10−8 M biotin-tagged S. meliloti LCO-IV (Ac,16:2, S), or water. The roots were stained with 0.5 mg/mL 5- bromo-4-chloro-3-indolyl-β-D-glucuronic acid (X-Gluc), 100 mM potassium phosphate buffer (pH 7.0), 10 mM ethylenediaminetetraacetrate (EDTA, pH 8.0), 1 mM potassium ferricyanide, 1 mM potassium ferrocyanide, and 0.1% Triton X-100 and were incubated at 37 °C overnight. Roots were washed with EtOH 70% twice before image acquisition.
Acknowledgments
We thank Theis Brock-Nannestad and Kasper K. Sørensen for HR-MS data, Ola Blixt and Simon B. Hansen for helpful discussions, Angelique Jakobsen and Lene Krusell for the full-length NFP receptor expression construct, and Leila Kathleen Walker for manuscript editing. We thank Marjolein Thunnissen, Thomas Ursby, and Jie Nan for beamline support during synchrotron data collection at beamlines I911-3 (MAX-lab II) and BioMAX at the MAX IV Laboratory and Cy Jeffries for the assistance during synchrotron SAXS data collection at beamline P12 operated by EMBL Hamburg at the PETRA III storage ring (Deutsches Elektronen-Synchrotron). This work was funded by the project Engineering Nitrogen Symbiosis for Africa currently supported through a grant to the University of Cambridge by the Bill and Melinda Gates Foundation and the UK Government’s Department for International Development and the Danish National Research Foundation (DNRF79) and in part by the US Department of Energy grant (Grant No. DE-SC0015662).
Footnotes
Competing interest statement: K.G., Z.B., L.H.M., S.R., J.S., and K.R.A., are inventors on patent application 62/718,186 submitted by Aarhus University that covers LysM receptors: genetically altered LysM receptors with altered agonist specificity and affinity. M.L., D.L., S.R., J.S., and K.R.A., are inventors on patent application 63/027,151 submitted by Aarhus University that covers LysM receptor motifs.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2111031118/-/DCSupplemental.
Data Availability
Coordinates and structure factors for NFP (PDB: 7AU7) (61) and LYS11 (PDB: 7BAX) (62) are deposited in the PDB. All other study data are included in the article and/or SI Appendix.
References
- 1.D’Haeze W., Holsters M., Nod factor structures, responses, and perception during initiation of nodule development. Glycobiology 12, 79R–105R (2002). [DOI] [PubMed] [Google Scholar]
- 2.Cope K. R., et al., The ectomycorrhizal fungus Laccaria bicolor produces lipochitooligosaccharides and uses the common symbiosis pathway to colonize Populus roots. Plant Cell 31, 2386–2410 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rush T. A., et al., Lipo-chitooligosaccharides as regulatory signals of fungal growth and development. Nat. Commun. 11, 3897 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pueppke S. G., Broughton W. J., Rhizobium sp. strain NGR234 and R. fredii USDA257 share exceptionally broad, nested host ranges. Mol. Plant Microbe Interact. 12, 293–318 (1999). [DOI] [PubMed] [Google Scholar]
- 5.Kelly S., Radutoiu S., Stougaard J., Legume LysM receptors mediate symbiotic and pathogenic signalling. Curr. Opin. Plant Biol. 39, 152–158 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Arrighi J.-F., et al., The Medicago truncatula lysin [corrected] motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 142, 265–279 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Limpens E., et al., LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302, 630–633 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Amor B. B., et al., The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J. 34, 495–506 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Madsen E. B., et al., A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425, 637–640 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Radutoiu S., et al., Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425, 585–592 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Murakami E., et al., Epidermal LysM receptor ensures robust symbiotic signalling in Lotus japonicus. eLife 7, e33506 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawaharada Y., et al., Receptor-mediated exopolysaccharide perception controls bacterial infection. Nature 523, 308–312 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Lohmann G. V., et al., Evolution and regulation of the Lotus japonicus LysM receptor gene family. Mol. Plant Microbe Interact. 23, 510–521 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Broghammer A., et al., Legume receptors perceive the rhizobial lipochitin oligosaccharide signal molecules by direct binding. Proc. Natl. Acad. Sci. U.S.A. 109, 13859–13864 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bozsoki Z., et al., Receptor-mediated chitin perception in legume roots is functionally separable from Nod factor perception. Proc. Natl. Acad. Sci. U.S.A. 114, E8118–E8127 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bozsoki Z., et al., Ligand-recognizing motifs in plant LysM receptors are major determinants of specificity. Science 369, 663–670 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Zhukov V., et al., The pea Sym37 receptor kinase gene controls infection-thread initiation and nodule development. Mol. Plant Microbe Interact. 21, 1600–1608 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Indrasumunar A., et al., Inactivation of duplicated nod factor receptor 5 (NFR5) genes in recessive loss-of-function non-nodulation mutants of allotetraploid soybean (Glycine max L. Merr.). Plant Cell Physiol. 51, 201–214 (2010). [DOI] [PubMed] [Google Scholar]
- 19.van Velzen R., et al., Comparative genomics of the nonlegume Parasponia reveals insights into evolution of nitrogen-fixing rhizobium symbioses. Proc. Natl. Acad. Sci. U.S.A. 115, E4700–E4709 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griesmann M., et al., Phylogenomics reveals multiple losses of nitrogen-fixing root nodule symbiosis. Science 361, eaat1743 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Liu T., et al., Chitin-induced dimerization activates a plant immune receptor. Science 336, 1160–1164 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Zhang X.-C., et al., Molecular evolution of lysin motif-type receptor-like kinases in plants. Plant Physiol. 144, 623–636 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wong J. E. M. M., et al., Structural signatures in EPR3 define a unique class of plant carbohydrate receptors. Nat. Commun. 11, 3797 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hjuler C. T., et al., Preparation of glycoconjugates from unprotected carbohydrates for protein-binding studies. Nat. Protoc. 12, 2411–2422 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Bek A. S., et al., Improved characterization of nod factors and genetically based variation in LysM receptor domains identify amino acids expendable for nod factor recognition in Lotus spp. Mol. Plant Microbe Interact. 23, 58–66 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Oldroyd G. E. D., Mitra R. M., Wais R. J., Long S. R., Evidence for structurally specific negative feedback in the Nod factor signal transduction pathway. Plant J. 28, 191–199 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Felix G., Regenass M., Boller T., Specific perception of subnanomolar concentrations of chitin fragments by tomato cells: Induction of extracellular alkalinization, changes in protein phosphorylation, and establishment of a refractory state. Plant J. 4, 307–316 (1993). [Google Scholar]
- 28.Wais R. J., Keating D. H., Long S. R., Structure-function analysis of nod factor-induced root hair calcium spiking in Rhizobium-legume symbiosis. Plant Physiol. 129, 211–224 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ovtsyna A. O., et al., Nod factors of Rhizobium leguminosarum bv. viciae and their fucosylated derivatives stimulate a nod factor cleaving activity in pea roots and are hydrolyzed in vitro by plant chitinases at different rates. Mol. Plant Microbe Interact. 13, 799–807 (2000). [DOI] [PubMed] [Google Scholar]
- 30.Demont-Caulet N., et al., Nodule-inducing activity of synthetic Sinorhizobium meliloti nodulation factors and related lipo-chitooligosaccharides on alfalfa. Importance of the acyl chain structure. Plant Physiol. 120, 83–92 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dénarié J., Cullimore J., Lipo-oligosaccharide nodulation factors: A minireview new class of signaling molecules mediating recognition and morphogenesis. Cell 74, 951–954 (1993). [DOI] [PubMed] [Google Scholar]
- 32.Ardourel M., et al., Rhizobium meliloti lipooligosaccharide nodulation factors: Different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses. Plant Cell 6, 1357–1374 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walker S. A., Viprey V., Downie J. A., Dissection of nodulation signaling using pea mutants defective for calcium spiking induced by nod factors and chitin oligomers. Proc. Natl. Acad. Sci. U.S.A. 97, 13413–13418 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Terpolilli J. J., O’Hara G. W., Tiwari R. P., Dilworth M. J., Howieson J. G., The model legume Medicago truncatula A17 is poorly matched for N2 fixation with the sequenced microsymbiont Sinorhizobium meliloti 1021. New Phytol. 179, 62–66 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Bensmihen S., de Billy F., Gough C., Contribution of NFP LysM domains to the recognition of Nod factors during the Medicago truncatula/Sinorhizobium meliloti symbiosis. PLoS One 6, e26114 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gough C., Cottret L., Lefebvre B., Bono J.-J., Evolutionary history of plant LysM receptor proteins related to root endosymbiosis. Front. Plant Sci. 9, 923 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fuechtbauer W., et al., LYS12 LysM receptor decelerates Phytophthora palmivora disease progression in Lotus japonicus. Plant J. 93, 297–310 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Madsen E. B., et al., Autophosphorylation is essential for the in vivo function of the Lotus japonicus Nod factor receptor 1 and receptor-mediated signalling in cooperation with Nod factor receptor 5. Plant J. 65, 404–417 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Pietraszewska-Bogiel A., et al., Interaction of Medicago truncatula lysin motif receptor-like kinases, NFP and LYK3, produced in Nicotiana benthamiana induces defence-like responses. PLoS One 8, e65055 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rasmussen S. R., et al., Intraradical colonization by arbuscular mycorrhizal fungi triggers induction of a lipochitooligosaccharide receptor. Sci. Rep. 6, 29733 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alexandersson E., et al., Purification and proteomic analysis of plant plasma membranes. Methods Mol. Biol. 432, 161–173 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Strong M., et al., Toward the structural genomics of complexes: Crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 103, 8060–8065 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCoy A. J., et al., Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kelley L. A., Mezulis S., Yates C. M., Wass M. N., Sternberg M. J. E., The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adams P. D., et al., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hopkins J. B., Gillilan R. E., Skou S., BioXTAS RAW: Improvements to a free open-source program for small-angle X-ray scattering data reduction and analysis. J. Appl. Cryst. 50, 1545–1553 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Svergun D. I., Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Cryst. 25, 495–503 (1992). [Google Scholar]
- 49.Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H. J., Svergun D. I., PRIMUS: A Windows PC-based system for small-angle scattering data analysis. J. Appl. Cryst. 36, 1277–1282 (2003). [Google Scholar]
- 50.Franke D., Svergun D. I., DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Cryst. 42, 342–346 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Svergun D. I., Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 76, 2879–2886 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wriggers W., Chacón P., Using Situs for the registration of protein structures with low-resolution bead models from x-ray solution scattering. J. Appl. Cryst. 34, 773–776 (2001). [Google Scholar]
- 53.Svergun D., Barberato C., Koch M. H. J., CRYSOL – A program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Cryst. 28, 768–773 (1995). [Google Scholar]
- 54.Shi X., Jarvis D. L., Protein N-glycosylation in the baculovirus-insect cell system. Curr. Drug Targets 8, 1116–1125 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Muszyński A., et al., Structures of exopolysaccharides involved in receptor-mediated perception of Mesorhizobium loti by Lotus japonicus. J. Biol. Chem. 291, 20946–20961 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Biasini M., et al., SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stougaard J., Abildsten D., Marcker K. A., The Agrobacterium rhizogenes pRi TL-DNA segment as a gene vector system for transformation of plants. Mol. Gen. Genet. 207, 251–255 (1987). [Google Scholar]
- 58.Honma M. A., Asomaning M., Ausubel F. M., Rhizobium meliloti nodD genes mediate host-specific activation of nodABC. J. Bacteriol. 172, 901–911 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bohorov O., Andersson-Sand H., Hoffmann J., Blixt O., Arraying glycomics: A novel bi-functional spacer for one-step microscale derivatization of free reducing glycans. Glycobiology 16, 21C–27C (2006). [DOI] [PubMed] [Google Scholar]
- 60.Journet E. P., et al., Medicago truncatula ENOD11: A novel RPRP-encoding early nodulin gene expressed during mycorrhization in arbuscule-containing cells. Mol. Plant Microbe Interact. 14, 737–748 (2001). [DOI] [PubMed] [Google Scholar]
- 61.K. Gysel, M. Blaise, K. R. Andersen, Crystal structure of Nod Factor Perception ectodomain. RCSB. http://www.rcsb.org/pdb/explore/explore.do?structureId=7AU7. Deposited 2 November 2020. [Google Scholar]
- 62.M. Laursen, J. Cheng, K. Gysel, M. Blaise, K. R. Andersen, Crystal structure of LYS11 ectodomain. RCSB. http://www.rcsb.org/pdb/explore/explore.do?structureId=7BAX. Deposited 16 December 2020. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Coordinates and structure factors for NFP (PDB: 7AU7) (61) and LYS11 (PDB: 7BAX) (62) are deposited in the PDB. All other study data are included in the article and/or SI Appendix.