Significance
Breast cancer stem cells (BCSCs) have the property of infinite self-renewal. When these cells divide, they give rise to one BCSC and one transient amplifying cell that can rapidly proliferate but only for a limited number of cell divisions. Only BCSCs can give rise to secondary (recurrent and/or metastatic) tumors. It is believed that breast cancer recurrence is due to the survival of a small number of BCSCs. Because the BCSCs represent a very small minority of cells within the primary tumor, their unique characteristics may not be detected by bulk tumor analyses. BCSCs are often located within regions of intratumoral hypoxia, and hypoxia-inducible factors activate the transcription of genes that promote BCSC specification and/or maintenance.
Keywords: breast cancer stem cell, hypoxia-inducible factors, chemosensitivity, EMT
Abstract
Calreticulin (CALR) is a multifunctional protein that participates in various cellular processes, which include calcium homeostasis, cell adhesion, protein folding, and cancer progression. However, the role of CALR in breast cancer (BC) is unclear. Here, we report that CALR is overexpressed in BC compared with normal tissue, and its expression is correlated with patient mortality and stemness indices. CALR expression was increased in mammosphere cultures, CD24−CD44+ cells, and aldehyde dehydrogenase–expressing cells, which are enriched for breast cancer stem cells (BCSCs). Additionally, CALR knockdown led to BCSC depletion, which impaired tumor initiation and metastasis and enhanced chemosensitivity in vivo. Chromatin immunoprecipitation and reporter assays revealed that hypoxia-inducible factor 1 (HIF-1) directly activated CALR transcription in hypoxic BC cells. CALR expression was correlated with Wnt/β-catenin pathway activation, and an activator of Wnt/β-catenin signaling abrogated the inhibitory effect of CALR knockdown on mammosphere formation. Taken together, our results demonstrate that CALR facilitates BC progression by promoting the BCSC phenotype through Wnt/β-catenin signaling in an HIF-1–dependent manner and suggest that CALR may represent a target for BC therapy.
Breast cancer (BC) is the most frequently diagnosed cancer and the leading cause of cancer-related death among women worldwide (1). Although BC therapy has made great progress, there are still large numbers of patients who die due to recurrent and metastatic disease. Breast cancer stem cells (BCSCs), which are defined as cells with tumor-initiating properties and infinite self-renewal/proliferative potential, contribute crucially to tumor metastasis and recurrence (2). Thus, understanding the molecular mechanisms involved in BCSC specification and maintenance is critical for developing more successful therapeutic strategies and improving the overall survival of BC patients.
BC progression is not only the result of somatic mutation but is also related to the tumor microenvironment, and one of the driving forces leading to cancer progression is intratumoral hypoxia. Hypoxia-inducible factor 1 (HIF-1) is a heterodimeric protein, composed of HIF-1α and HIF-1β subunits, which activates the transcription of numerous genes that encode proteins involved in angiogenesis, extracellular matrix remodeling, migration, invasion, and metastasis (3). HIF-1 plays a pivotal role in promoting specification and maintenance of the BCSC phenotype in response to hypoxia, and knockdown of HIF-1α abrogates this effect of hypoxia and significantly impairs the tumor-initiating ability of BC cells (2–4). The complex molecular mechanisms by which hypoxia promotes the BCSC phenotype (2) point to the importance of these cells in BC progression and the key role of intratumoral hypoxia as an microenvironmental signal for BCSC specification.
Calreticulin (CALR), a 46-kDa Ca2+-binding protein and molecular chaperone, is critical for maintaining Ca2+ homeostasis and glycoprotein folding in the endoplasmic reticulum (5, 6). CALR expression is positively correlated with clinical stage and lymph node metastasis in esophageal cancer and nasopharyngeal carcinoma (7, 8). In this study, we demonstrate that CALR expression is activated by HIF-1 when human BC cells are exposed to hypoxia. Knockdown of CALR expression impaired BC cell migration/invasion, decreased the number of BCSCs, and impaired Wnt/β-catenin pathway activation. CALR expression in human BC biopsies was correlated with patient mortality. Our results implicate CALR as a mediator of BC progression and a rational target for BC therapy.
Results
Elevated CALR Expression in BC Is Correlated with Poor Prognosis.
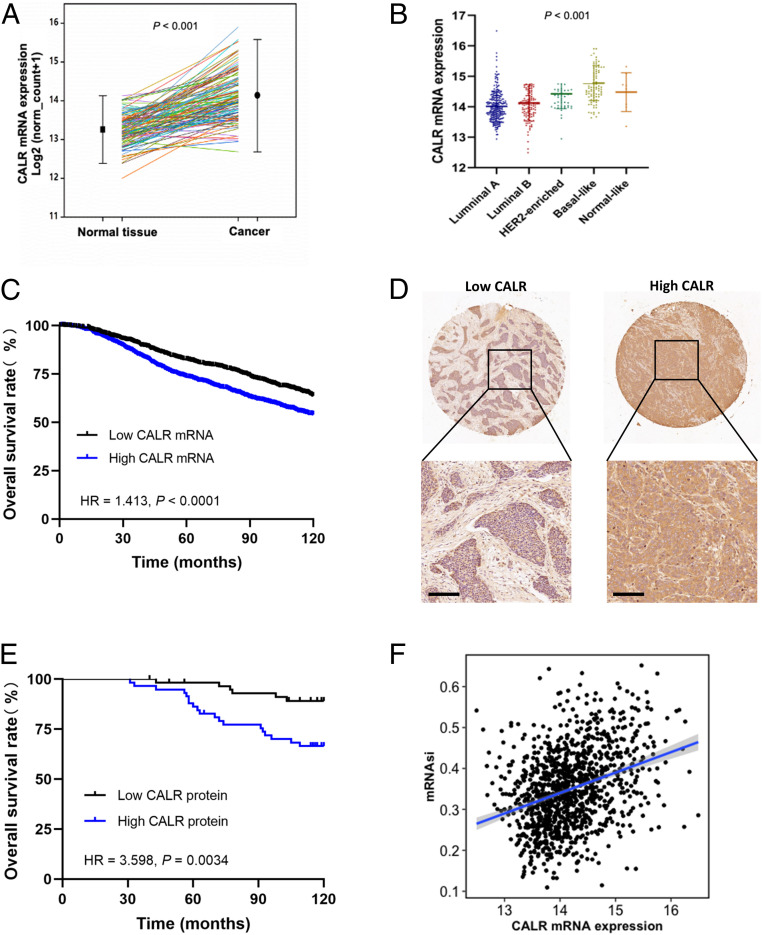
Analysis of CALR messenger RNA (mRNA) levels in 114 paired BCs and adjacent nontumor tissue from the BRCA dataset of The Cancer Genome Atlas (TCGA) revealed increased CALR mRNA levels in BC tissue (Fig. 1A). Stratification of BCs in TCGA database into luminal A, luminal B, HER2-enriched, basal-like, and normal-like molecular subgroups according to the expression of a 50-mRNA signature (PAM50) (9) revealed that the highest expression of CALR was observed in the basal-like subgroup (Fig. 1B), which is enriched for BCSCs (10). Stratification of 1,904 BCs in the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) database based on the median CALR mRNA expression level revealed that high CALR expression was associated with larger tumor size (P = 0.005), lymph node positivity (P = 0.025), grade III tumors (P < 0.001), and invasive ductal carcinoma histological type (P < 0.001) (SI Appendix, Table S1). Kaplan–Meier analysis showed that patients with high CALR mRNA expression exhibited decreased overall survival compared with those with low CALR mRNA expression, with a hazard ratio of 1.41 (P < 0.0001) (Fig. 1C).
Fig. 1.
Elevated CALR expression in BC tissues is correlated with poor prognosis. (A) CALR mRNA expression was analyzed in paired specimens of BC and adjacent nontumor tissue using TCGA data (n = 114, mean ± SEM, P < 0.001). (B) CALR mRNA expression was analyzed using BC data from TCGA, which was stratified according to PAM50 subtype (n = 522, mean ± SEM, P < 0.001). (C) Kaplan–Meier curves were constructed to compare overall survival of BC patients with high (n = 952) and low (n = 952) CALR mRNA expression using METABRIC data (log rank test, n = 1,904, P < 0.0001). (D) CALR expression was determined by immunohistochemical staining of a BC tissue microarray. (Scale bars: 100 μm.) (E) Kaplan–Meier analysis was performed to compare overall survival of BC patients with high (n = 58) and low (n = 59) CALR expression from BC tissue microarray assay (log rank test, n = 117, P < 0.01). (F) Correlation analysis of CALR mRNA expression and mRNAsi using TCGA data (n = 1,083, R = 0.33, P < 2.2e-16). HR, hazard ratio.
Immunohistochemistry was performed to analyze CALR expression in a BC tissue microarray (Fig. 1D). Stratification of 117 BCs from the tissue microarray based on the median CALR protein expression revealed that low CALR expression was correlated with the luminal A molecular subtype and that high CALR expression was associated with the luminal B subtype (SI Appendix, Table S2). Most importantly, high CALR protein expression was correlated with decreased overall survival, with a hazard ratio of 3.60 (Fig. 1E), which is in agreement with the CALR mRNA expression data presented above (Fig. 1C). Because BCSCs are critical for tumor initiation and metastasis, thereby affecting patient survival (4, 11, 12), we analyzed CALR mRNA expression and an mRNA-based stemness index (mRNAsi) using TCGA BRCA data (13), which revealed a highly significant positive correlation between them (R = 0.33; P < 2.2e-16) (Fig. 1F).
CALR Promotes the BCSC Phenotype.
Next, we analyzed CALR expression in immortalized breast epithelial cells (MCF-10A) and six different human BC cell lines, which encompassed the major clinical categories of BC based on expression of the estrogen receptor (ER), progesterone receptor (PR), and HER2. MCF-7 is ER+PR+ HER2− (luminal A molecular subtype); MDA-MB-453 and HCC1954 are ER−PR−HER2+ (HER2-enriched subtype), and MDA-MB-231, SUM159, and BT549 are ER−PR−HER2− (i.e., triple-negative breast cancer [TNBC]; basal-like subtype) (14). Reverse transcription and quantitative real-time PCR (RT-qPCR) and immunoblot assays revealed that CALR mRNA and protein were more highly expressed in all six BC cell lines compared with MCF-10A cells, and highest expression was observed in the TNBC cell lines (SI Appendix, Fig. S1A).
It has been reported that CALR expression was increased in pancreatic cancer stem-like cells compared with the bulk cancer cells (15). CD24−CD44+ populations, aldehyde dehydrogenase–expressing (ALDH+) populations, and mammosphere cultures are regarded as BCSC-enriched subsets (16, 17). Analysis of MDA-MB-231, HCC1954, and SUM159 cells revealed that the expression of CALR was significantly increased in the ALDH+ vs. ALDH− cells, CD24−CD44+ vs. non-CD24−CD44+ cells, and mammosphere vs. adherent (monolayer) cultures (Fig. 2A).
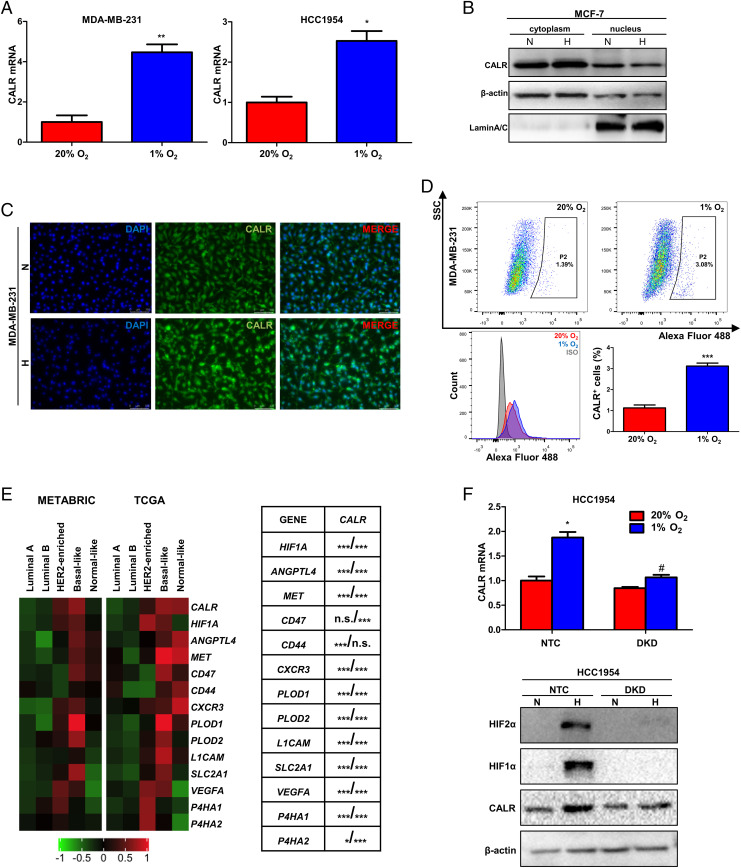
Fig. 2.
CALR promotes the BCSC phenotype. (A) CALR mRNA expression was determined by qRT-PCR in MDA-MB-231, HCC1954, and SUM159 cells, which were sorted by flow cytometry into ALDH+ and ALDH− populations (Left; mean ± SEM, n = 3; **P < 0.01 vs. ALDH−) or CD24−CD44+ and non-CD24−CD44+ populations (Center; mean ± SEM, n = 3; *P < 0.05 vs. non-CD24−CD44+; **P < 0.01 vs. non-CD24−CD44+; ***P < 0.001 vs. non-CD24−CD44+) or were cultured on standard (adherent) vs. ultralow adherence (mammosphere) plates for 7 d (Right; mean ± SEM, n = 3; *P < 0.05 vs. adherent; **P < 0.01 vs. adherent). (B) (Left) Mammosphere formation was analyzed by Leica inverted phase contrast microscopy. (Scale bars in photomicrographs: 100 μm.) (Right) The number of mammospheres per well was counted (bar graphs, mean ± SEM, n = 3; ***P < 0.001 vs. NTC). (C) CD24−CD44+ populations were detected by flow cytometry (scatterplot in Left) and analyzed (bar graphs in Right; mean ± SEM, n = 3; *P < 0.05 vs. NTC). (D) The percentage of ALDH+ cells was determined by flow cytometry (mean ± SEM, n = 3; ***P < 0.001 vs. NTC). (E) Expression of CALR and pluripotency factors (Oct4, NANOG, SOX2) was analyzed in whole-cell lysates subject to immunoblot assay. (F) Tumor-initiating cell frequency (TCF) in MDA-MB-231 orthotopic tumors was calculated.
To determine whether CALR was required for BCSC specification/maintenance, we analyzed MCF-7, HCC1954, MDA-MB-231, and SUM159 subclones, which were stably transfected with an expression vector encoding a nontargeting control (NTC) short hairpin RNA (shRNA) or a vector encoding either of two different shRNAs targeting calreticulin (shCALR; shCALR-1 and -2) (SI Appendix, Fig. S1 B–E). Knockdown subclones formed fewer and smaller mammospheres than NTC cells (Fig. 2B and SI Appendix, Fig. S1F). Moreover, the proportions of CD24−CD44+ cells were significantly lower in HCC1954-shCALR-1 (10.3 vs. 3.57%), SUM159-shCALR-1 (28.2 vs. 12.1%), and MDA-MB-231-shCALR-1 (95.6 vs. 85.1%) than in the respective NTC subclones (Fig. 2C and SI Appendix, Fig. S1G). The percentage of ALDH+ cells was decreased from 12.3 to 5.2% in HCC1954, from 5.2 to 0.3% in SUM159, and from 11.5 to 5.0% in MDA-MB-231 after CALR knockdown (Fig. 2D and SI Appendix, Fig. S1 H and I). In addition, expression of the stem cell pluripotency factors Oct4, Nanog, and Sox2 was decreased in shCALR subclones (Fig. 2E), which provides a molecular basis for the observed decrease in BCSCs.
To investigate the role of CALR in regulating tumor initiation, limiting numbers (1 × 103 or 1 × 104) of NTC or shCALR-1 MDA-MB-231 cells were implanted in the mammary fat pad of female NOD-SCID mice, and tumor growth was monitored for 2 mo. The frequency of tumor-initiating cells was decreased by greater than 1,000-fold in the shCALR as compared with the NTC subclone (Fig. 2F). Taken together, the data presented in Fig. 2 and SI Appendix, Fig. S1 demonstrate that CALR is required for BCSC specification.
CALR Knockdown Increases Sensitivity to Cytotoxic Chemotherapy.
BCSCs play an important role in drug resistance, and we found that CALR knockdown significantly enhanced the susceptibility of MDA-MB-231, MCF-7, and SUM159 cells to killing by doxorubicin (DOX), when shCALR-1 cells were compared with NTC cells (Fig. 3 A and B and SI Appendix, Fig. S2). Next, we implanted 5 × 105 NTC or shCALR-1 MDA-MB-231 cells (so that BCSCs were not limiting for tumor initiation) in the mammary fat pad of female NOD-SCID mice, and when each group of tumors became palpable (designated day 0), the mice were treated by intraperitoneal injection of saline (control) or DOX (2.5 mg/kg weekly). There was no significant effect of DOX on the growth of NTC tumors (Fig. 3 C–E). In contrast, in mice implanted with shCALR cells and treated with DOX, a significant decrease in tumor growth was observed (Fig. 3C), and in three of five mice implanted with shCALR cells and treated with DOX, tumors were completely eradicated (Fig. 3 D and E).
Fig. 3.
CALR regulates the chemosensitivity of BC cells. (A) NTC (Left) and shCALR-1 (Right) subclones of MDA-MB-231 cells were cultured for 4 d in the presence of the indicated concentration of DOX, and viable cell numbers were determined by CCK-8 assay. (B) Data are presented according to DOX concentration (mean ± SEM, n = 3; *P < 0.05 vs. NTC; **P < 0.01 vs. NTC; ***P < 0.001 vs. NTC). (C) In total, 5 × 105 cells of each MDA-MB-231 subclone were implanted into the mammary fat pad. When tumors were palpable (day 0), mice were treated with saline or 2.5 mg/kg of DOX every 7 d. Tumor volumes were determined weekly (mean ± SEM, n = 4 in the NTC + saline and shCALR-1 + saline groups, n = 5 in the NTC + DOX and shCALR-1 + DOX groups; *P < 0.05; **P < 0.01). (D and E) On day 35, tumors were excised, (D) photographed, and (E) weighed (mean ± SEM, n = 4 in the NTC + saline and shCALR-1 + saline groups, n = 5 in the NTC + DOX and shCALR-1 + DOX groups; **P < 0.01; n.s., not significant).
CALR Facilitates Cell Motility, Invasion, and Metastasis.
Next, we examined the effect of CALR knockdown on BC cell motility and invasion. CALR knockdown impaired the ability of MDA-MB-231 and SUM159 cells to invade through Matrigel in a modified Boyden chamber assay (Fig. 4A and SI Appendix, Fig. S3A). Cell motility was analyzed by live cell imaging, which revealed that the random motility of shCALR-1 cells was significantly decreased (Fig. 4B and SI Appendix, Fig. S3B). We also studied transendothelial migration by testing the ability of BC cells to invade through a confluent monolayer of human umbilical vein endothelial cells (HUVECs). The NTC cells (red arrows in Fig. 4C) all penetrated through the HUVEC monolayer and disappeared from view by 5 to 6 h, whereas many of the shCALR-1 cells remained above the HUVEC monolayer after 7 h (Fig. 4C, SI Appendix, Fig. S3C, and Movies S1–S6).
Fig. 4.
CALR facilitates BC cell motility, invasion, and metastasis. (A) Invasion of MDA-MB-231 cells was assessed using a modified Boyden chamber assay. Invading cells were stained and photographed under 200× (Left). Five fields were selected randomly for quantification (Right; mean ± SEM, n = 3; ***P < 0.001 vs. NTC). (B) Cell movement was tracked by live cell imaging with time lapse. The velocity and accumulated distance of cell movement were analyzed. Three fields were selected randomly for quantification (mean ± SEM; **P < 0.01 vs. NTC). (Scale bar [thickened line at center top]: 50 μm.) (C) Time-lapse images of cells were captured every 15 min. The elapsed time by which the BC cells invaded through the HUVEC monolayer was recorded. BC cells are marked with red arrows. The rate of cell invasion in three fields selected randomly was determined (mean ± SEM; **P < 0.01 vs. NTC). (Scale bars: 50 μm.) (D) The protein levels of EMT markers were determined by immunoblot assay in MDA-MB-231 and HCC1954 subclones. (E) Hematoxylin and eosin (H&E) staining was performed to identify metastatic nodules (arrows) in the lungs of mice that received mammary fat pad injection of the indicated MDA-MB-231 subclone. (F) The number of metastatic foci per lung section was determined (mean ± SEM, n = 4; **P < 0.01 vs. NTC).
The epithelial–mesenchymal transition (EMT) refers to a reprogramming of cancer cells, which involves altered expression of cell adhesion proteins (switch from E-cadherin to N-cadherin expression) and cytoskeletal proteins (switch from epithelial keratins to vimentin) that occurs in concert with acquisition of BCSC properties (18). As shown in Fig. 4D and SI Appendix, Fig. S3D, CALR knockdown in MDA-MB-231 and HCC1954 cells led to increased expression of the epithelial marker E-cadherin and decreased expression of the mesenchymal markers vimentin and N-cadherin.
Only BCSCs are capable of forming clinically relevant metastases (4). To analyze the effect of CALR knockdown on the ability of BC cells to colonize the lung, we implanted 5 × 105 cells in the mammary fat pad and harvested the lungs 8 wk later. Hematoxylin and eosin staining of lung sections revealed that significantly fewer BC foci were present in lungs of mice implanted with shCALR-1 cells (Fig. 4 E and F). Taken together, the data presented in Fig. 4 and SI Appendix, Fig. S3 demonstrate that CALR is crucial for BC cell motility, invasion, and metastasis.
Hypoxia Induces Increased CALR Expression in an HIF-Dependent Manner.
Hypoxia is one of the most prominent microenvironmental factors driving cancer progression (11, 19). HIFs mediate the specification and maintenance of the BCSC phenotype in response to hypoxia (2, 20). After exposure to hypoxic conditions (1% O2) for 24 h, CALR mRNA expression was induced in MDA-MB-231 and HCC1954 cells (Fig. 5A). Cell fractionation and immunofluorescence experiments indicated that when MCF-7 and MDA-MB-231 cells were exposed to 1% O2 for 24 h, increased CALR protein was observed primarily in the cytoplasm (Fig. 5 B and C and SI Appendix, Fig. S4A). Hypoxia exposure also increased CALR cell-surface expression in MCF-7 and MDA-MB-231 cells as determined by flow cytometry (Fig. 5D and SI Appendix, Fig. S4B). We performed Pearson's correlation test using METABRIC and TCGA BC data to compare expression of CALR mRNA with HIF1A and 12 HIF target genes (ANGPTL4, MET, CD47, CD44, CXCR3, PLOD1, PLOD2, L1CAM, SLC2A1, VEGFA, P4HA1, and P4HA2). CALR mRNA levels were significantly correlated with HIF-1α mRNA and 11 of 12 mRNAs encoded by HIF target genes in each human BC dataset (Fig. 5E).
Fig. 5.
Hypoxia induces CALR expression in an HIF-dependent manner. (A) MDA-MB-231 and HCC1954 cells were exposed to 20 or 1% O2 for 24 h, and qRT-PCR was performed to determine CALR mRNA expression (mean ± SEM, n = 3; *P < 0.05 vs. 20% O2; **P < 0.01 vs. 20% O2). (B) CALR expression was analyzed in MCF-7 cells exposed to 20 or 1% O2 for 24 h by immunoblot assay; β-actin and LaminA/C were used as controls for purity of cytoplasmic and nuclear fractions, respectively. (C) CALR expression was analyzed by immunofluorescence in MDA-MB-231 cells exposed to 20 or 1% O2 for 24 h. (Scale bars: 100 μm.) (D) MDA-MB-231 cells were exposed to 20 or 1% O2 for 24 h, and CALR expression on the cell surface was determined by flow cytometry using the anti-CALR antibody or immunoglobulin G isotype control (IgG). The percentage of CALR+ cells is shown (bar graph, mean ± SEM, n = 3; ***P < 0.001 vs. 20% O2). SSC, side scatter. (E) Heat maps (Left) show mRNA expression of CALR, HIF1A, and 12 HIF target genes in the METABRIC (n = 1,898) and TCGA BRCA (n = 522) datasets stratified according to molecular subtype (red, increased expression; green, decreased expression). CALR mRNA expression in each primary BC specimen was compared with the expression of HIF1A mRNA and 12 HIF-regulated mRNAs (table in Right) using Pearson’s correlation test (shown as P value from METABRIC analysis/P value from TCGA analysis; *P < 0.05; ***P < 0.001). n.s., not significant. (F) qRT-PCR (Upper) and immunoblot (Lower) assays were performed to analyze CALR expression in HCC1954 NTC and DKD subclones, which were exposed to 20 or 1% O2 for 24 h (mean ± SEM, n = 3; *P < 0.05 vs. NTC at 20% O2; #P < 0.05 vs. NTC at 1% O2). N, 20% O2; H, 1% O2.
To determine whether hypoxia-induced CALR expression is HIF dependent, HCC1954 cells were stably transfected with expression vectors encoding shRNA targeting HIF-1α and HIF-2α (double-knockdown [DKD]) or NTC shRNA. Hypoxia-induced CALR mRNA and protein expression were lost in the DKD subclone (Fig. 5F). Taken together, the data presented in Fig. 5 and SI Appendix, Fig. S4 demonstrate that CALR mRNA and protein expression are induced by hypoxia in BC cells in an HIF-dependent manner.
CALR Is a Direct HIF-1 Target Gene.
We searched the human genome sequence for matches to the consensus HIF binding site sequence 5′-(A/G)CGTG-3′ (21) that were located within DNase I–hypersensitive domains at the CALR locus to investigate whether HIF-1 directly regulates CALR gene transcription. Chromatin immunoprecipitation (ChIP) assays revealed hypoxia-induced binding of HIF-1 to a DNA sequence, which encompassed the palindrome 5′-CACGCGTG-3′ located at −227 bp relative to the CALR transcription start site (Fig. 6A), when chromatin from HCC1954 or SUM159 cells was immunoprecipitated with HIF-1α or HIF-1β antibodies (Fig. 6B and SI Appendix, Fig. S4C), demonstrating that hypoxia induces direct binding of HIF-1 to the CALR promoter. In contrast, hypoxia did not induce the binding of HIF-2α to the CALR promoter (Fig. 6C and SI Appendix, Fig. S4D).
Fig. 6.
CALR is a direct HIF-1 target gene. (A) A candidate palindromic HIF-1 binding site was identified 227 bp 5′ to the transcription start site of the human CALR gene. (B) HCC1954 cells were exposed to 20 or 1% O2 for 24 h, and ChIP assays were performed using IgG or antibody against HIF-1α or HIF-1β (mean ± SEM, n = 3; *P < 0.05 vs. 20% O2; **P < 0.01 vs. 20% O2). (C) HCC1954 cells were exposed to 20 or 1% O2 for 24 h, and ChIP assays were performed using IgG or antibody against HIF-2α or HIF-1β (mean ± SEM, n = 3; ***P < 0.001 vs. 20% O2). n.s., not significant. (D) HCC1954 and SUM159 cells were transfected with reporter plasmid pEZX-HRE, which contains a 55-bp oligonucleotide encompassing the putative hypoxia response element (HRE), and exposed to 20 or 1% O2 for 24 h, and luciferase activity in cell lysates was determined (mean ± SEM, n = 3; *P < 0.05 vs. 20% O2). (E) NTC and DKD subclones of HCC1954 were transfected with pEZX-HRE, exposed to 20 or 1% O2 for 24 h, and luciferase activity was determined (mean ± SEM, n = 3, **P < 0.01 vs. NTC at 20% O2; ##P < 0.01 vs. NTC at 1% O2).
To determine whether DNA sequences encompassing the site functioned as a hypoxia response element (HRE), a 55-bp oligonucleotide spanning the HIF binding site was inserted into pEZX-FR03 upstream of the human cytomegalovirus immediate-early promoter and firefly luciferase coding sequences to generate the reporter pEZX-HRE. Hypoxia induced increased luciferase activity in HCC1954 and SUM159 cells transfected with pEZX-HRE (Fig. 6D), demonstrating that the 55-bp sequence site functions as an HRE. Furthermore, hypoxia induced increased luciferase activity in HCC1954-NTC cells but not in HCC1954-DKD cells, which were transfected with pEZX-HRE (Fig. 6E). Taken together, the data in Fig. 6 demonstrate that HIF-1 binds directly to the CALR promoter to activate gene transcription in hypoxic BC cells.
CALR Promotes the BCSC Phenotype by Activating the Wnt/β-Catenin Pathway.
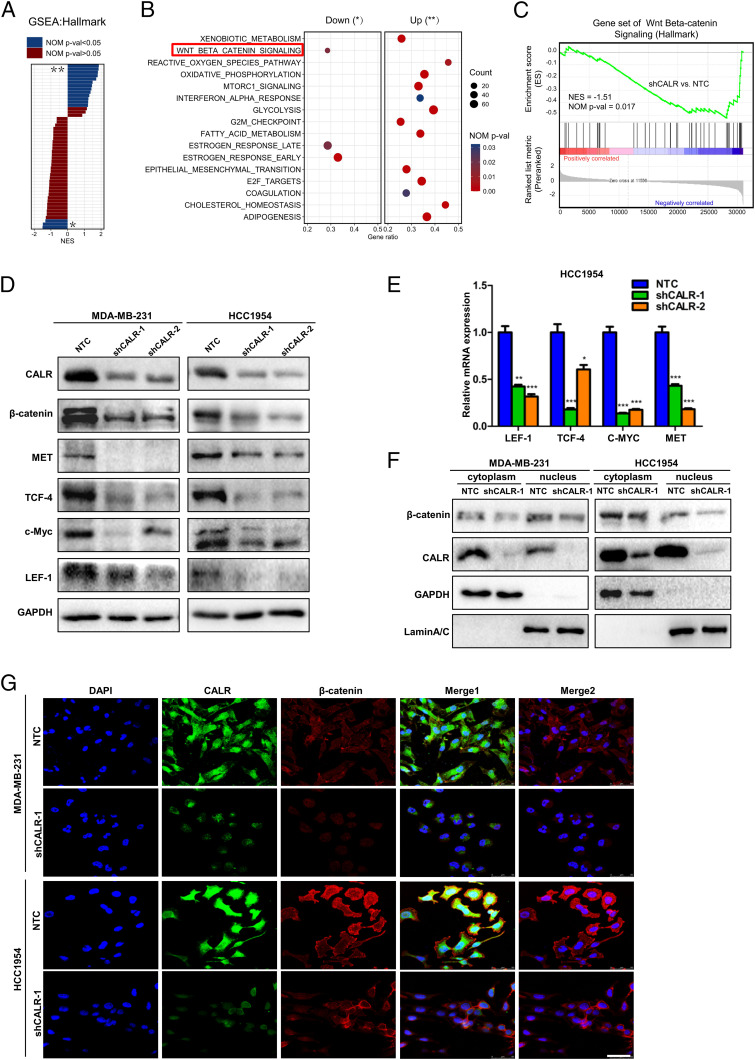
To unveil the potential molecular mechanisms underlying the promotion of the BCSC phenotype by CALR, global gene expression in NTC and shCALR subclones was examined by RNA sequencing (RNA-seq). Gene set enrichment analysis (GSEA) of the RNA-seq data from NTC and shCALR subclones was performed. A total of 16 hallmark pathways were significantly dysregulated (nominal [NOM] P value < 0.05; 13 pathways up-regulated and 3 down-regulated) in the shCALR subclone relative to the NTC cells (Fig. 7 A and B). Compared with NTC, the top down-regulated hallmark in shCALR cells was the Wnt/β-catenin pathway (Fig. 7C and Dataset S1). Among genes implicated in EMT (22), the expression of EGFR, PDGFB, ZEB2, and CXCR4 was decreased in shCALR cells (SI Appendix, Fig. S5 A, Upper). Expression of API5 and BACH1, which have been implicated in DOX resistance (23), was also decreased in shCALR cells (SI Appendix, Fig. S5 A, Lower).
Fig. 7.
CALR knockdown inhibits Wnt/β-catenin pathway activation in BC cells. (A) GSEA shows differentially expressed mRNA hallmark pathways in shCALR as compared with the NTC subclone of MDA-MB-231. Normalized enrichment scores (NESs) of each GSEA hallmark up-regulated (**) or down-regulated (*) category are plotted, with significantly enriched terms (NOM P value < 0.05) shown in blue and nonenriched terms (NOM P value > 0.05) shown in crimson. (B) The significantly dysregulated hallmark pathways in shCALR cells are labeled as down (*) or up (**), with the gene ratio plotted on the x axis. (C) A representative GSEA plot showing the significantly down-regulated Wnt/β-catenin signaling pathway in shCALR cells vs. paired NTC samples. (D) Protein levels of β-catenin and Wnt/β-catenin downstream targets were determined by immunoblot assay. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (E) The mRNA levels of Wnt/β-catenin downstream targets were analyzed by qRT-PCR (mean ± SEM, n = 3; *P < 0.05 vs. NTC; **P < 0.01 vs. NTC; ***P < 0.001 vs. NTC). (F) The expression of β-catenin was assessed in MDA-MB-231 and HCC1954 cells by immunoblot assay. GAPDH and LaminA/C were used as controls for cytoplasmic and nucleus fractional purity, respectively. (G) The expression and subcellular localization of CALR (green) and β-catenin (red) were determined by confocal laser-scanning microscopy. DAPI was used for nuclear staining (blue). Merge 1 consists of DAPI, CALR, and β-catenin images. Merge 2 consists of DAPI and β-catenin images. (Scale bar: 50 μm.)
Based on the strong correlation between CALR expression and Wnt/β-catenin signaling indicated by RNA-seq, we analyzed the expression of β-catenin and Wnt downstream targets (Met, c-Myc, T cell factor 4 [TCF-4], and lymphoid enhancer factor 1 [LEF-1]) in NTC vs. shCALR subclones by immunoblot assays. Knockdown of CALR led to decreased expression of Wnt/β-catenin pathway proteins and downstream targets in MDA-MB-231-shCALR and HCC1954-shCALR cells (Fig. 7D). The mRNA expression of Wnt downstream targets was also significantly decreased in HCC1954-shCALR cells (Fig. 7E).
Cell fractionation studies revealed that β-catenin levels were decreased in both the cytoplasm and the nucleus by CALR knockdown in MDA-MB-231 and HCC1954 cells (Fig. 7F). Immunofluorescence staining showed the colocalization of CALR with β-catenin in NTC cells, and CALR knockdown was associated with decreased nuclear β-catenin expression (Fig. 7G). Immunofluorescent staining of frozen sections of tumor xenografts with antibody against β-catenin and confocal laser microscope scanning revealed that CALR knockdown led to decreased β-catenin expression (SI Appendix, Fig. S5 B and C). These results demonstrate that CALR knockdown inhibits Wnt/β-catenin pathway activation by loss of β-catenin expression.
To investigate whether Wnt/β-catenin signaling plays a role in CALR-induced promotion of the BCSC phenotype, we utilized pharmacologic agents to modulate Wnt/β-catenin pathway activation. XAV939 inhibits tankyrase 1/2, leading to β-catenin phosphorylation and subsequent degradation (24), whereas SKL2001 activates the Wnt/β-catenin pathway by blocking the inhibitory interaction between Axin and β-catenin. MDA-MB-231 and HCC1954 cells were treated with XAV939 (20 μM) or SKL2001 (20 μM) for 24 h. NTC cells treated with XAV939 formed significantly fewer and smaller spheres, while SKL2001 treatment abrogated the inhibitory effect of CALR knockdown on mammosphere formation (Fig. 8 A and B). Immunoblot assays further confirmed that the decreased β-catenin expression in shCALR-1 cells was rescued by treatment with SKL2001 (Fig. 8C), which was consistent with its ability to rescue mammosphere formation by shCALR-1 cells (Fig. 8 A and B). Taken together, these results indicate that CALR induces the BCSC phenotype by activating Wnt/β-catenin signaling (Fig. 8D).
Fig. 8.
CALR promotes the BCSC phenotype by activating Wnt/β-catenin signaling. (A) MDA-MB-231 and HCC1954 cells were pretreated with 20 μM XAV939 or 20 μM SKL2001 for 24 h. Mammosphere formation was quantified by phase contrast microscopy at 40× magnification. (B) The number of mammospheres per well was counted (mean ± SEM, n = 3; **P < 0.01 vs. NTC; ***P < 0.001 vs. NTC; ##P < 0.01 vs. shCALR-1; ###P < 0.001 vs. shCALR-1). (C) The cells were treated with 20 μM XAV939 or 20 μM SKL2001 for 24 h; β-catenin levels in MDA-MB-231 and HCC1954 cells were determined by immunoblot assay. (D) Hypoxia induces HIF-1–dependent expression of CALR in BC cells, and CALR activates Wnt/β-catenin signaling, which promotes the BCSC phenotype, metastasis, and chemoresistance. TCF, T cell factor; LEF, lymphoid enhancer factor.
Discussion
BCSCs represent a small subgroup of BC cells that can both self-renew and generate the diverse cell types that comprise the bulk tumor. BCSCs are required for recurrent and metastatic disease (25, 26), and the inability to target BCSCs has been suggested as a major cause of treatment failure (4, 12, 27). In the present study, we have demonstrated by analysis of public databases that CALR is overexpressed in BCs and correlates with BC patient mortality. At the molecular level, CALR, an HIF target gene, promotes the BCSC phenotype, resulting in increased tumor initiation, chemoresistance, cellular motility, invasion, and metastasis. Further, we have elucidated that CALR loss of function inhibits WNT/β-catenin pathway activation. Thus, CALR provides a missing link between HIF-dependent and hypoxia-induced Wnt/β-catenin pathway activation and BCSC enrichment (20).
CALR was reported to be an endoplasmic reticulum–associated chaperone and Ca2+-binding lectin-like protein involved in quality control during the synthesis, maturation, and transport of secretory and membrane proteins to regulate Ca2+ homeostasis, mRNA stability, cell adhesion/migration, and phagocytosis (28, 29). In this study, we have demonstrated that CALR is also required for expression of pluripotency factors that specify the BCSC phenotype. Although previous studies indicated that CALR is correlated with the cancer stem cell phenotype, the underlying mechanism remained unknown. Here, we report that CALR promotes the BCSC phenotype by supporting Wnt/β-catenin activity, leading to β-catenin/TCF-dependent transcription. Previous studies have reported that hypoxia induces β-catenin activity in an HIF-dependent manner (20) and that Wnt/β-catenin signaling plays a critical role in BCSC self-renewal, tumor initiation, metastasis, and chemotherapy resistance (30–38), indicating that targeting the Wnt/β-catenin signaling pathway might be a promising therapeutic approach for BCSC inhibition.
β-catenin colocalized with CALR in hearts affected by right ventricular cardiomyopathy (39). In CALR-deficient embryonic stem (ES) cells, β-catenin is predominately junctional and absent from nuclei (40). CALR overexpression rescued β-catenin expression induced by epidermal growth factor in CALR-silenced pancreatic cancer cells (41). These prior studies established the ability of CALR to regulate β-catenin in different cell types. However, the mechanism by which CALR modulated β-catenin expression in these cells was not determined. A previous study found that CALR inhibited translation of CCAAT/enhancer-binding protein α (C/EBPα) and C/EBPβ mRNA (42). C/EBPβ induces the expression of C/EBPα and peroxisome proliferator-activated receptor γ (PPARγ), which in turn, regulate the expression of Axin2, a Wnt signaling inhibitor (43). Thus, CALR might activate Wnt/β-catenin signaling by repressing C/EBP expression. WNT3a ligand was retained in the endoplasmic reticulum in CALR-deficient ES cells, leading to impaired WNT/β-catenin signaling (44). CALR also interacts with the chromosome region maintenance 1 (CRM1) protein (45, 46). CRM1-dependent nuclear export of β-catenin has been reported (47). Nuclear accumulation of p21-activated kinase 4 (PAK4) up-regulated β-catenin nuclear translocation, and PAK4 nuclear export was CRM1 dependent (48). Given these findings, further studies are required to investigate the mechanism by which CALR activates Wnt/β-catenin signaling.
Metabolic alterations appear to represent another major consequence of CALR knockdown based on GSEA, which revealed increased expression of genes involved in adipogenesis, cholesterol homeostasis, fatty acid metabolism, glycolysis, oxidative phosphorylation, and mechanistic target of rapamycin complex 1 (mTORC1) signaling. Thus, further investigation into the role of CALR in BC metabolism is warranted.
Finally, prior studies have revealed that CALR expressed on the surface of cancer cells interacts with low-density lipoprotein receptor–related protein (LRP) on the surface of macrophages to trigger phagocytosis, unless the cancer cells also express CD47, which interacts with signal regulatory protein α (SIRPα) on macrophages to counteract CALR–LRP-dependent signaling (49). We previously reported that hypoxia induces CD47 expression in BC cells in an HIF-dependent manner (50). Coordinate HIF-dependent regulation of CALR and CD47 may provide a mechanism to promote the BCSC phenotype while evading innate immunity.
Materials and Methods
Tissue Culture and Reagents.
MCF-7 and MDA-MB-231 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS) (HyClone). HCC1954 cells were cultured in 1640 medium with 10% FBS. SUM159 cells were cultured in F-12 medium with 10% FBS. Cells were maintained under standard culture conditions (5% CO2, 95% air, and 37 °C). For hypoxia exposure, cells were incubated in a workstation maintained at 1% O2, 5% CO2, 94% N2, and 37 °C. Cell viability was determined by CCK-8 assay (Dojindo Molecular Technologies). XAV939 and SKL2001 were purchased from TargetMol.
Lentiviral Transduction, RT-qPCR, and Immunoblot Assays.
These procedures were performed using standard techniques as described in the SI Appendix.
ChIP.
Cells were exposed to 20 or 1% O2 for 24 h; then, ChIP assays were performed following the protocol of the EZ-Magna-ChIP A/G kit (Millipore; 17-10086). HIF-1α antibody (Abcam; ab1), HIF-2α antibody (Abcam; ab243861), HIF-1β antibody (Cell Signaling Technology; 5537S), or control rabbit IgG was incubated with sheared chromatin. Precipitated DNAs containing fragments of the CALR promoter were analyzed by qPCR using flanking primers with the following nucleotide sequences: 5′-GGG TTC AGG TCT GGT CAC AT-3′ and 5′-CCG ATT TCT ATT GGC CTC AC-3′.
Luciferase Reporter Assay.
The following double-stranded DNA sequence encompassing the HIF-1 binding site in the CALR gene was inserted into pEZX-FR03: 5′-ACC CCA CCA GTG GGC GTC CCC CCC ACG CGT GGT CGA CCA TCA TTG GTC GGT GGT G-3′. The recombinant plasmid was transfected into HCC1954 cells; 24 h after transfection, cells were cultured in 20 or 1% O2 for 24 h, and luciferase activity was quantified using the Dual-Luciferase Reporter Assay System (Promega).
Flow Cytometry.
After exposure to 20 or 1% O2 for 24 h, cells were harvested to make a single-cell suspension, incubated with CALR antibody (Abcam; ab2907, 1:200) or isotype control IgG antibody (BIOSS; bs-0295P) for 20 min, incubated with Alexa Fluor 488–conjugated secondary antibody (Proteintech; SA00006-2, 1:200), and subjected to flow cytometry (FACScalibur; BD Biosciences). Dead cells were gated out by side-scatter and forward-scatter analyses.
Mammosphere Assay.
As previously described (51), 1 × 104 cells were seeded per well on ultralow attachment (Corning) six-well plates in complete Mammocult medium (Stem Cell Technologies). Mammospheres (diameter ≥70 µM) were counted after 7 d using a DM18 inverted phase microscope and LAS AF Lite software (Leica).
ALDH Assay.
The ALDEFLUOR kit (Stem Cell Technologies) was used to determine intracellular ALDH enzyme activity. As previously described (51), cells were harvested to make a single-cell suspension in assay buffer containing the ALDH substrate and incubated for 45 min at 37 °C. As a negative control, for each sample an aliquot of cells was treated with the ALDH inhibitor diethylaminobenzaldehyde. Samples were subjected to flow cytometry analysis (FACScalibur; BD Biosciences).
Invasion Assays.
Filters with 8-µm-diameter pores were coated with Matrigel (Corning), which was diluted 1:10 with FBS-free DMEM and inserted into a 24-well companion plate. BC cells were suspended at 1 × 105/mL in FBS-free DMEM, and a 200-µL aliquot was added to the upper chamber, whereas 800 µL of DMEM with 10% FBS was added to the lower chamber. The plates were incubated in 5% CO2 at 37 °C and observed every 4 h. When the appearance of cells was observed in the lower chamber, the cells were fixed in methanol. After 15 min, cells were stained for 30 min with 0.4% crystal violet. After gently removing the cells from the upper chamber, the invaded cells were imaged using an inverted phase contrast microscope and counted in five randomly selected fields.
Live Cell Imaging.
For the motility assay, BC cells were plated on 6-cm dishes and placed on the stage of a DM16000 B microscope (Leica), which was maintained at 5% CO2, 95% air, and 37 °C. For the transendothelial migration assay (52), BC cells were plated onto a confluent HUVEC monolayer and placed immediately on the microscope stage, and three fields were monitored every 15 min at 200× magnification. These images were compiled into time-lapse movies using the LAS X Hardware Configurator (Leica).
In Vivo Assays.
NOD-SCID female mice aged 8 wk were purchased from the Experimental Animal Center of the Medical College of Xi’an Jiaotong University. For the tumor initiation assay, 1 × 104 or 1 × 103 cells were injected into the fourth mammary fat pad. Tumor-initiating cell frequency was calculated by the limited dilution assay according to extreme limiting dilution analysis (http://bioinf.wehi.edu.au/software/elda/). For xenograft assays, 5 × 105 cells diluted in 50 μL of FBS-free DMEM and 50 μL of Matrigel were injected into the fourth mammary fat pad. Tumor length (L) and width (W) were measured (in millimeters) with calipers weekly, and volume (V) was determined using the formula V = 1/2 × L × W2.
RNA Sequencing.
NTC and shCALR-1 subclones of MDA-MB-231 cells were analyzed by RNA sequencing using the Illumina HiSeq4000 system to generate 151-bp paired-end reads. The data were uploaded to the Gene Expression Omnibus (accession no. GSE181431).
RNA-seq Data Analysis.
Adaptors and low-quality bases were trimmed from raw sequencing reads using the FASTX-Toolkit (Version 0.0.13), reads <16 nt were discarded, and the remaining reads were aligned to the GRCh38 genome by TopHat2 (53) with a maximum of four mismatches. Uniquely mapped reads were used to calculate read number and paired-end fragments per kilobase of exon per million fragments mapped value for each gene.
GSEA.
To identify functional enrichment profiles in the RNA-seq data (54), we used annotated gene collections downloaded from the Molecular Signature Database (version 7.2) for the hallmark collection and GSEA software (version 4.0.3). We determined normalized enrichment scores with the total gene list as background, with n = 1,000 permutations, where NOM P value < 0.05 was considered significant.
Tissue Microarray Immunohistochemistry.
BC microarray slides (Shanghai Outdo Biotech Company; HBreD140Su05) were stained with anti-CALR primary antibody (Abcam; ab2907), followed by biotin-labeled anti-rabbit IgG and then peroxidase-conjugated streptavidin. The stained samples were visualized using a DAB Detection Kit. The slides were scanned using Panoramic MIDI (3D HISTECH). Immunohistochemistry results were quantified using integrated optical density.
Immunofluorescent Staining.
Cells were fixed in 4% paraformaldehyde for 10 min, then permeabilized with 0.2% Triton X-100 for 10 min on ice, and blocked in 5% bovine serum albumin for 1 h. Cells were incubated with CALR antibody (Abcam; ab2907, 1:200) or β-catenin antibody (CST; sc7963, 1:100), followed by Alexa Fluor 488–conjugated (Proteintech; SA00006-2, 1:200) or Alexa Fluor 649–conjugated (Abbkin; A23610, 1:200) secondary antibody. DNA staining was performed using 4’,6-diamidino-2-phenylindole (DAPI). Microscopic analyses were conducted with a confocal laser-scanning microscope (Leica TCS SP5).
Databases and Statistical Analysis.
The human BC gene expression data analyzed in the current study are available from the METABRIC (downloaded from cBioPortal; http://www.cbioportal.org/) and TCGA (downloaded from UCSC XENA; https://xena.ucsc.edu/). Data are presented as mean ± SEM. The Student's t test was used to analyze the significance of differences between two groups, whereas ANOVA was used for multiple groups. Kaplan–Meier curves were generated using the METABRIC dataset, and the log rank test was performed to determine whether observed differences between two groups were statistically significant. P < 0.05 was considered to be statistically significant.
Ethics Approval and Consent to Participate.
This research was carried out at the Center for Translational Medicine, The First Affiliated Hospital of Xi’an Jiaotong University, Shaanxi, China. All experimental schemes were assessed and approved by the Joint Ethics Committee of the Xi’an Jiaotong University and implemented according to national guidelines. Mice were provided food and water ad libitum, maintained on a regular 12-h day/night cycle, and housed in cages with various enrichment materials added, including nesting materials and plastic shelters. All authors have read and approved this paper, which has not been submitted for publication and is not under consideration for publication elsewhere.
Acknowledgments
This study was supported by National Natural Science Foundation of China Grant 81702632 and Natural Science Foundation of Shaanxi Province, China Grant 2018JQ8004. We appreciate the technicians from the Center for Translational Medicine and Key Laboratory for Tumor Precision Medicine of Shaanxi Province for their technical support. G.L.S. is the C. Michael Armstrong Professor of Genetic Medicine at the Johns Hopkins University School of Medicine and an American Cancer Society Research Professor. Research in his laboratory is supported by the American Cancer Society and the Armstrong Family Foundation.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2109144118/-/DCSupplemental.
Data Availability
RNA sequencing data are available in the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/; accession no. GSE181431). All other study data are included in the article and/or supporting information.
References
- 1.DeSantis C. E., et al., International variation in female breast cancer incidence and mortality rates. Cancer Epidemiol. Biomarkers Prev. 24, 1495–1506 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Xiang L., Semenza G. L., Hypoxia-inducible factors promote breast cancer stem cell specification and maintenance in response to hypoxia or cytotoxic chemotherapy. Adv. Cancer Res. 141, 175–212 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Schito L., Semenza G. L., Hypoxia-inducible factors: Master regulators of cancer progression. Trends Cancer 2, 758–770 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Oskarsson T., Batlle E., Massagué J., Metastatic stem cells: Sources, niches, and vital pathways. Cell Stem Cell 14, 306–321 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu Y. C., Weng W. C., Lee H., Functional roles of calreticulin in cancer biology. BioMed Res. Int. 2015, 526524 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michalak M., Groenendyk J., Szabo E., Gold L. I., Opas M., Calreticulin, a multi-process calcium-buffering chaperone of the endoplasmic reticulum. Biochem. J. 417, 651–666 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Shi F., et al., Calreticulin promotes migration and invasion of esophageal cancer cells by upregulating neuropilin-1 expression via STAT5A. Clin. Cancer Res. 20, 6153–6162 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Ye D., et al., LMP1 Up-regulates calreticulin to induce epithelial-mesenchymal transition via TGF-β/Smad3/NRP1 pathway in nasopharyngeal carcinoma cells. J. Cancer 11, 1257–1269 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parker J. S., et al., Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 27, 1160–1167 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Geldermalsen M., et al., ASCT2/SLC1A5 controls glutamine uptake and tumour growth in triple-negative basal-like breast cancer. Oncogene 35, 3201–3208 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Semenza G. L., The hypoxic tumor microenvironment: A driving force for breast cancer progression. Biochim. Biophys. Acta 1863, 382–391 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brooks M. D., Burness M. L., Wicha M. S., Therapeutic implications of cellular heterogeneity and plasticity in breast cancer. Cell Stem Cell 17, 260–271 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malta T. M.et al.; Cancer Genome Atlas Research Network , Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell 173, 338–354.e15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dai X., Cheng H., Bai Z., Li J., Breast cancer cell line classification and its relevance with breast tumor subtyping. J. Cancer 8, 3131–3141 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsukuma S., et al., Calreticulin is highly expressed in pancreatic cancer stem-like cells. Cancer Sci. 107, 1599–1609 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dontu G., et al., In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 17, 1253–1270 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ginestier C., et al., ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 1, 555–567 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ye X., Weinberg R. A., Epithelial-mesenchymal plasticity: A central regulator of cancer progression. Trends Cell Biol. 25, 675–686 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vaupel P., Höckel M., Mayer A., Detection and characterization of tumor hypoxia using pO2 histography. Antioxid. Redox Signal. 9, 1221–1235 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Conley S. J., et al., Antiangiogenic agents increase breast cancer stem cells via the generation of tumor hypoxia. Proc. Natl. Acad. Sci. U.S.A. 109, 2784–2789 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Semenza G. L., et al., Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J. Biol. Chem. 271, 32529–32537 (1996). [DOI] [PubMed] [Google Scholar]
- 22.Tao C., et al., Genomics and prognosis analysis of epithelial-mesenchymal transition in glioma. Front. Oncol. 10, 183 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Villeneuve D. J., et al., cDNA microarray analysis of isogenic paclitaxel- and doxorubicin-resistant breast tumor cell lines reveals distinct drug-specific genetic signatures of resistance. Breast Cancer Res. Treat. 96, 17–39 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Huang S. M., et al., Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 461, 614–620 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Pan Q., et al., Concise review: Targeting cancer stem cells using immunologic approaches. Stem Cells 33, 2085–2092 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kabakov A., Yakimova A., Matchuk O., Molecular chaperones in cancer stem cells: Determinants of stemness and potential targets for antitumor therapy. Cells 9, 892 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Al-Hajj M., Becker M. W., Wicha M., Weissman I., Clarke M. F., Therapeutic implications of cancer stem cells. Curr. Opin. Genet. Dev. 14, 43–47 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Eggleton P., Bremer E., Dudek E., Michalak M., Calreticulin, a therapeutic target? Expert Opin. Ther. Targets 20, 1137–1147 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Totary-Jain H., et al., Calreticulin destabilizes glucose transporter-1 mRNA in vascular endothelial and smooth muscle cells under high-glucose conditions. Circ. Res. 97, 1001–1008 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Dittmer J., Breast cancer stem cells: Features, key drivers and treatment options. Semin. Cancer Biol. 53, 59–74 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Xu B. S., et al., ALKATI interacts with c-Myc and promotes cancer stem cell-like properties in sarcoma. Oncogene 39, 151–163 (2020). [DOI] [PubMed] [Google Scholar]
- 32.Lee K. M., et al., MYC and MCL1 cooperatively promote chemotherapy-resistant breast cancer stem cells via regulation of mitochondrial oxidative phosphorylation. Cell Metab. 26, 633–647.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang T., et al., LncCCAT1 promotes breast cancer stem cell function through activating WNT/β-catenin signaling. Theranostics 9, 7384–7402 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhuvanalakshmi G., et al., Breast cancer stem-like cells are inhibited by diosgenin, a steroidal saponin, by the attenuation of the Wnt β-catenin signaling via the Wnt antagonist secreted frizzled related protein-4. Front. Pharmacol. 8, 124 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lv C., et al., MiR-31 promotes mammary stem cell expansion and breast tumorigenesis by suppressing Wnt signaling antagonists. Nat. Commun. 8, 1036 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakunrangsit N., Ketchart W., Plumbagin inhibits cancer stem-like cells, angiogenesis and suppresses cell proliferation and invasion by targeting Wnt/β-catenin pathway in endocrine resistant breast cancer. Pharmacol. Res. 150, 104517 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Lopez Almeida L., et al., The SCRIB paralog LANO/LRRC1 regulates breast cancer stem cell fate through WNT/β-catenin signaling. Stem Cell Reports 11, 1040–1050 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma F., et al., MiR-23a promotes TGF-β1-induced EMT and tumor metastasis in breast cancer cells by directly targeting CDH1 and activating Wnt/β-catenin signaling. Oncotarget 8, 69538–69550 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oxford E. M., Danko C. G., Fox P. R., Kornreich B. G., Moïse N. S., Change in β-catenin localization suggests involvement of the canonical Wnt pathway in Boxer dogs with arrhythmogenic right ventricular cardiomyopathy. J. Vet. Intern. Med. 28, 92–101 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pilquil C., Alvandi Z., Opas M., Calreticulin regulates a switch between osteoblast and chondrocyte lineages derived from murine embryonic stem cells. J. Biol. Chem. 295, 6861–6875 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheng W., et al., Calreticulin promotes EGF-induced EMT in pancreatic cancer cells via Integrin/EGFR-ERK/MAPK signaling pathway. Cell Death Dis. 8, e3147 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Timchenko L. T., Iakova P., Welm A. L., Cai Z. J., Timchenko N. A., Calreticulin interacts with C/EBPalpha and C/EBPbeta mRNAs and represses translation of C/EBP proteins. Mol. Cell. Biol. 22, 7242–7257 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Winter T. J. J., Nusse R., Running against the Wnt: How Wnt/β-catenin suppresses adipogenesis. Front. Cell Dev. Biol. 9, 627429 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Groenendyk J., Michalak M., Disrupted WNT signaling in mouse embryonic stem cells in the absence of calreticulin. Stem Cell Rev. Rep. 10, 191–206 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Cruz-Ramos E., Sandoval-Hernández A., Tecalco-Cruz A. C., Differential expression and molecular interactions of chromosome region maintenance 1 and calreticulin exportins in breast cancer cells. J. Steroid Biochem. Mol. Biol. 185, 7–16 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Grespin M. E., et al., Thyroid hormone receptor α1 follows a cooperative CRM1/calreticulin-mediated nuclear export pathway. J. Biol. Chem. 283, 25576–25588 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meng X., et al., Suppressor of fused negatively regulates β-catenin signaling. J. Biol. Chem. 276, 40113–40119 (2001). [DOI] [PubMed] [Google Scholar]
- 48.Li Y., et al., Nucleo-cytoplasmic shuttling of PAK4 modulates β-catenin intracellular translocation and signaling. Biochim. Biophys. Acta 1823, 465–475 (2012). [DOI] [PubMed] [Google Scholar]
- 49.Chao M. P., et al., Calreticulin is the dominant pro-phagocytic signal on multiple human cancers and is counterbalanced by CD47. Sci. Transl. Med. 2, 63ra94 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang H., et al., HIF-1 regulates CD47 expression in breast cancer cells to promote evasion of phagocytosis and maintenance of cancer stem cells. Proc. Natl. Acad. Sci. U.S.A. 112, E6215–E6223 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Samanta D., Semenza G. L., In vitro assays of breast cancer stem cells. Methods Mol. Biol. 1742, 237–246 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Itoh K., et al., An essential part for Rho-associated kinase in the transcellular invasion of tumor cells. Nat. Med. 5, 221–225 (1999). [DOI] [PubMed] [Google Scholar]
- 53.Kim D., et al., TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Subramanian A., et al., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
RNA sequencing data are available in the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/; accession no. GSE181431). All other study data are included in the article and/or supporting information.