Significance
The human ERG (hERG) voltage-gated K+ channel has an important role in heart rhythm, and channel malfunction can result in arrhythmias and sudden death. The Per-Arnt-Sim (PAS) and cyclic nucleotide binding homology (CNBh) domains in hERG influence some of the channel’s unique functional properties. However, the role of these domains is not completely understood. We use a conformation-sensitive antibody to show that the PAS/CNBh assembly is an active participant in the mechanism of channel gating: the assembly is destabilized when the channel gate opens and stabilized when the gate is closed. This finding adds to the understanding of hERG and provides avenues to correct channel function in disease.
Keywords: FRET, PAS domain, CNBh domain, scFv antibodies, LQTS
Abstract
The human ERG (hERG) K+ channel has a crucial function in cardiac repolarization, and mutations or channel block can give rise to long QT syndrome and catastrophic ventricular arrhythmias. The cytosolic assembly formed by the Per-Arnt-Sim (PAS) and cyclic nucleotide binding homology (CNBh) domains is the defining structural feature of hERG and related KCNH channels. However, the molecular role of these two domains in channel gating remains unclear. We have previously shown that single-chain variable fragment (scFv) antibodies can modulate hERG function by binding to the PAS domain. Here, we mapped the scFv2.12 epitope to a site overlapping with the PAS/CNBh domain interface using NMR spectroscopy and mutagenesis and show that scFv binding in vitro and in the cell is incompatible with the PAS interaction with CNBh. By generating a fluorescently labeled scFv2.12, we demonstrate that association with the full-length hERG channel is state dependent. We detect Förster resonance energy transfer (FRET) with scFv2.12 when the channel gate is open but not when it is closed. In addition, state dependence of scFv2.12 FRET signal disappears when the R56Q mutation, known to destabilize the PAS–CNBh interaction, is introduced in the channel. Altogether, these data are consistent with an extensive structural alteration of the PAS/CNBh assembly when the cytosolic gate opens, likely favoring PAS domain dissociation from the CNBh domain.
Members of the KCNH superfamily of voltage-gated K+ channels contribute to neuronal excitability, cardiac repolarization, and cellular proliferation and are linked to human disease (1–12). In particular, the human ERG (hERG) channel has a crucial role in repolarization of the cardiac action potential; channel malfunction, either from genetic alterations or from unwanted pharmacological channel block, results in long QT syndrome (LQTS), a condition associated with cardiac arrhythmias and sudden death (2, 13, 14).
KCNH channels are characterized by a “non-domain swapped” architecture, where the voltage-sensor domain interfaces with the pore domain in the same subunit (15). While the domain architecture is present in other channels (16), what distinguishes KCNH channels is the conserved cytosolic assembly formed by the N-terminal PAS (Per-Arnt-Sim) domain and the C-terminal cyclic nucleotide binding homology (CNBh) domain (17–24). PAS domains are widespread in nature, sensing light, redox potential, or small molecules and mediating protein–protein interactions (25–27). CNBh domains closely resemble CNB domains but lack the ability to bind nucleotides (28–32). Instead, the C-terminal tail of the CNBh domain acts as an intrinsic ligand, occupying the same position as a cyclic nucleotide in a bona fide CNB domain (30, 31).
It is well established that PAS and CNBh domains interact with each other in the channel (15, 22, 23, 33, 34). Mutations that interfere with this interaction, disrupt the CNBh intrinsic ligand, or destabilize the fold of the domains give rise to changes in hERG gating. Many are associated with type 2 LQTS (LQT2) (29–31, 35–41). It is also clear that hERG function is modulated by variation in the number of PAS/CNBh assemblies present in individual channels, resulting from heteromers of two isoforms, one isoform with the PAS domain (hERG1a) and another without (hERG1b) (42–47).
Further clues about the role of PAS and CNBh domains have been provided by ligands that target the cytosolic channel domains. We have demonstrated that allosteric modulation of hERG channel function through the PAS domain is possible by using scFv (single-chain variable fragment) antibodies (scFv2.10 and scFv2.12) that bind to PAS at distinct epitopes (48). scFv2.10 binds to residues R4 and R5 of hERG, in the PAS-cap region that spans the first 25 residues of the channel N terminus, just before the globular region of the PAS domain. In contrast, scFv2.12 binds to a region in the globular domain. In addition, small-molecule screening campaigns have identified ligands of PAS and CNBh domains that affect channel function (49–52). These data suggest that the assembly formed by the PAS and CNBh domains has an important role in the mechanism of gating of KCNH channels.
Comparison of the cryoelectron microscopy (cryo-EM) structures of hERG, with an open cytosolic gate, and the calmodulin-inhibited rat EAG (another KCNH channel), with a closed gate, provides clear insights about the role of the PAS-cap in gating (15, 34). The PAS-cap engages the channel gating machinery when the gate opens, with its N terminus trapped between the C-linker, the S4-S5 linker, and the S2–S3 cytosolic loop. When the channel closes, the C-linker moves away, widening the PAS-cap binding site and releasing the N terminus. In contrast, the cryo-EM structure comparison shows that the PAS/CNBh assembly is not altered, even relative to the crystal structure of the isolated complex (33), undergoing only a simple rigid-body rotation (15, 34, 53). The overall view is that that role of the PAS/CNBh assembly is limited to correctly position the N-terminal PAS-cap for engagement with the gating machinery.
Here, we propose a model in which the PAS/CNBh domain assembly is an active participant in the mechanism of hERG channel gating, undergoing a stabilization/destabilization cycle during hERG gating. This proposal results from the characterization of the molecular basis for the functional effect of scFv2.12 on the hERG channel—defining the antibody’s epitope, determining the impact of antibody binding on the PAS interaction with the CNBh domain in vitro and in the cell, and finally from monitoring the association of a fluorescent scFv2.12 antibody to the PAS domain during hERG gating.
Results
scFv2.12 Interferes with the PAS–CNBh Interaction.
We have previously demonstrated that exposing the cytosolic face of functional hERG channels to the scFv2.12 antibody, perfused into the cell through the patch pipette, alters the channel’s functional properties at 37 °C, accelerating activation and recovery from inactivation (48). The scFv2.12 binds to the natively folded globular region of the PAS domain, and we asked how this interaction could affect channel function, wondering whether it reflected a yet uncharacterized molecular role of PAS in hERG gating.
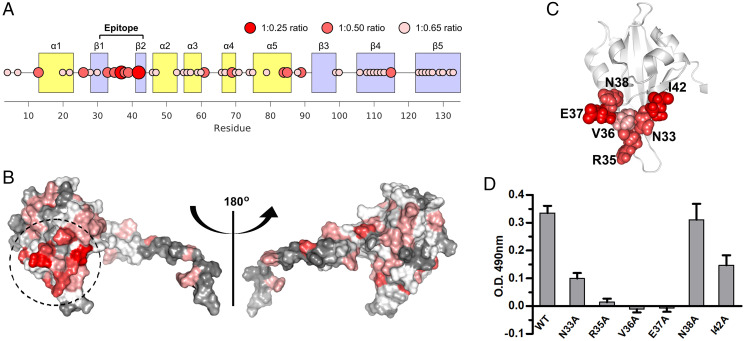
To address this possibility, we started by defining the epitope of the scFv2.12. We produced 15N-labeled PAS domain as a binding reporter and used NMR spectroscopy to monitor the antibody titration. scFv2.12 binding to PAS produced two main physical changes detected by NMR (SI Appendix, Fig. S1): 1) chemical shift perturbations in the heteronuclear single quantum correlation (1H-15N-HSQC) spectrum that are derived from changes in the chemical environment of residues due to antibody binding and 2) signal broadening and loss of peak intensity, resulting from changes in the rotational correlation time with formation of the protein complex. Analyzing both of these properties at the lowest PAS:scFv2.12 titration ratios, at which the PAS domain is not yet fully saturated (ratios 1:0.25 and 1:0.5), reveals the residues that are strongly affected by the scFv2.12 interaction. In particular, at ratios 1:0.25 and 1:0.50 V36 shows the largest chemical shift observed. In addition, E37 and I42 show large changes in peak intensity such that their peaks are no longer detectable at 1:0.25, most likely due to both chemical shift perturbation and signal broadening. Altogether these data pinpoint a group of residues formed by N33, R35,V36, E37, N38, and I42 that are clustered on the surface of PAS and exposed to the solvent (Fig. 1 A and B), suggestive of the binding epitope of scFv2.12. Residues N33 to N38 form the loop linking β-strands Aβ and Bβ (Fig. 1C), and I42 is positioned nearby, on the solvent-exposed hydrophobic patch of the PAS domain (18).
Fig. 1.
Epitope of scFv2.12. (A) Cartoon representing the intensity decays across the PAS residues monitored by NMR spectroscopy upon scFv2.12 titration. Circle width and color define the number of titrant equivalents at which a given residue is titrated out, as indicated. Epitope and secondary structure elements are labeled. (B) Two views of PAS domain structure with the number of equivalents of titrant required to titrate out the residue (as determined in A) color coded on the surface of the domain in a gray–white–red color scale (gray, 1:0.65; red, 1:0.25). Dashed circle shows the cluster of PAS residues that are more sensitive to scFv2.12. Views are related by 180° rotation. (C) Cartoon of PAS domain structure with residues in cluster shown in B as spheres. (D) Residues in cluster were individually mutated to alanine, and mutant proteins evaluated for binding to scFv2.12 by ELISA. Plates were coated with scFv2.12 SNAPHis6 protein and binding of wild-type GST-PAS (WT) or GST-PAS protein with single point mutations in the PAS domain (N33A, R35A, V36A, E37A, N38A, and I42A) evaluated. Error bars denote mean ± SD and n = 12. Reference SI Appendix, Fig. S1.
To confirm that these residues are involved in the interaction between the PAS domain and scFv2.12, we performed an in vitro alanine-scanning mutagenesis experiment. We developed an enzyme linked immunosorbent assay (ELISA) using a fusion protein of scFv2.12 to the SNAP-tag protein, which increased the protein expression level in bacteria. The scFv2.12-SNAP was coated onto the surface of a microplate and incubated with either wild-type GST-PAS, PAS mutant fusions (N33A, R35A, V36A, E37A, N38A, or I42A), or a GST control. Bound GST-PAS was measured using an anti–GST-HRP (horseradish peroxidase) antibody, and detection was reported using a colorimetric assay (Fig. 1D). N38A did not affect binding and functioned as a negative control for this experiment. In contrast, mutating residues N33, R35, V36, and E37 drastically reduced the amount of PAS fusion interacting with scFv2.12, with I42 also having a significant reduction, supporting the conclusion from the NMR experiments that these residues are part of the scFv2.12 epitope on the PAS domain.
Mapping the scFv2.12 epitope residues on the cryo-EM structure of hERG showed that they are positioned close to the interface with the CNBh domain (Fig. 2A). Accessible surface area calculations using a water molecule as a probe (radius 1.4 Å) reveal that in the cryo-EM structure of hERG, 80% of the accessible surface area of N33 is buried in the PAS/CNBh interface. For V36 and I42, these values are 25 and 99%, respectively. In contrast, only 12% of the R35 accessible surface is buried in the interface and E37 is fully accessible. This demonstrates that N33, V36, and I42 are partially or completely occluded in the PAS/CNBh interface, raising the possibility that scFv2.12 binding to the PAS domain disturbs the assembly.
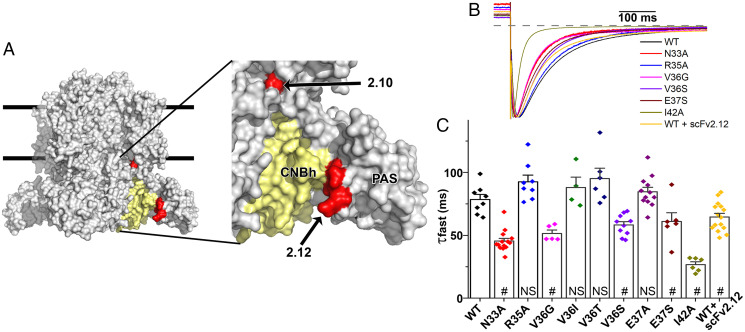
Fig. 2.
Mutations in scFv2.12 epitope destabilize PAS–CNBh functional interactions. (A) Surface representation of hERG channel cryo-EM structure and zoom view (Right) with scFv2.10 and scFv2.12 epitopes mapped (red surfaces) onto a single subunit of the channel. The epitope of scFv2.12 is at the interface between a CNBh domain (in yellow) and its PAS domain partner. Lines represent potential limits of membrane bilayer. (B) Scaled tail currents of wild-type (WT) and N33A, V36G, V36S, E37S, R35A, and I42A mutant channels and of WT channels coexpressed with scFv2.12 antibody. Tail currents were evoked at −120 mV after a step to +60 mV at room temperature. Dotted line indicates zero current level. (C) Fast component of deactivation in control, mutant channels, and WT hERG1a channels coexpressed with the scFv2.12 antibody (#, P < 0.05; NS, not significant). Data are mean ± SEM. Reference SI Appendix, Fig. S2.
To test this hypothesis, we used two-electrode voltage clamp recordings to measure whether mutations within the epitope of scFv2.12 would increase the apparent rate of channel deactivation, a proxy for destabilization of the PAS/CNBh interaction and disengagement of the PAS domain (35, 40). Alanine mutations at N33 and I42 accelerated the fast component of deactivation without large changes in the current–voltage relationship, reminiscent of the LQT2 mutations N33T and I42N (35, 41) (Fig. 2 B and C and SI Appendix, Fig. S2). No changes were observed when residue V36 was mutated to isoleucine or threonine, or residue E37 to alanine, but on mutation to glycine or serine (V36G, V36S, and E37S) deactivation clearly accelerated. Mutant residues on the surface of the PAS domain that destabilize the interaction with the CNBh domain cause, in many cases, an increase in the rate of deactivation (35, 40). Therefore, our mutagenesis results suggest that some residues within the scFv2.12 epitope form important interactions between the PAS and CNBh domains and that association of the scFv2.12 antibody to the epitope will impact the PAS/CNBh assembly. In addition, coexpression of the antibody with wild-type hERG channel accelerates deactivation but to a lesser degree than mutations of the residues forming close contact with the CNBh domain (Fig. 2 B and C), in agreement with the relatively low affinity of the antibody for its epitope (Kd = 4 μM) (48).
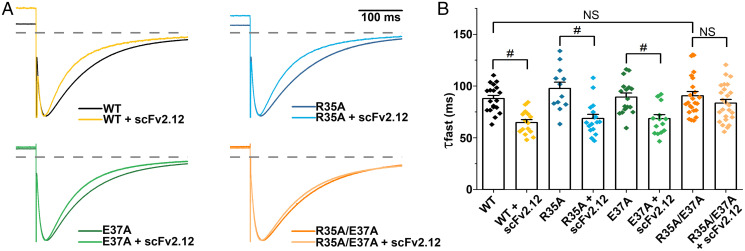
It is possible that the scFv2.12 epitope identified is not the sole binding site of the scFv2.12 antibody and that additional interaction sites present in the full-length hERG channel could contribute to the observed changes in gating. To address this possibility, we targeted two of the more exposed epitope residues, R35 and E37, which when mutated to alanines, caused a reduction in the amount of PAS fusion interacting with scFv2.12 in the ELISA (Fig. 1D) but did not disrupt deactivation (Fig. 2 B and C). These single point mutants maintained wild-type–like sensitivity to scFv2.12 (Fig. 3). Thus, in the context of the cell, the epitope was sufficiently intact in each case to bind antibody. However, when combined in the same channel, these mutations disrupted the ability of the antibody to accelerate deactivation, indicating that the epitope was compromised (Fig. 3B). The mutations per se did not disrupt slow deactivation, even when combined. The failure of the antibody to disrupt slow deactivation indicates that the double mutation effectively disrupted the epitope, and the antibody does not affect deactivation by binding elsewhere. These results attest to the specificity of antibody binding to the epitope defined by NMR and ELISA.
Fig. 3.
Antibody affects deactivation exclusively via interactions with defined epitope. (A) Scaled tail currents of wild-type (WT) and mutant channels R35A, E37A, and R35A/E37A with and without coexpressed scFv2.12 antibody. Tail currents were evoked at −120 mV after a step to +60 mV at room temperature. Dotted line indicates zero current level. (B) Fast component of deactivation in WT and mutant channels with and without coexpressed scFv2.12 antibody (#, P < 0.05; NS, not significant). Data are mean ± SEM.
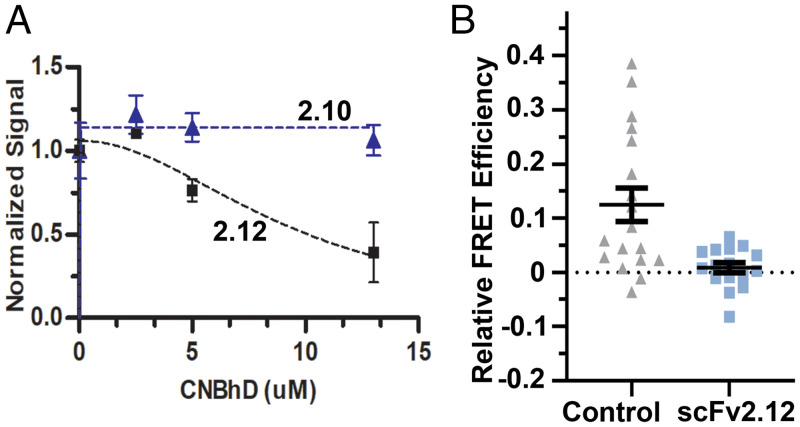
Next, we performed a competition ELISA based upon the one described above but where the amount of wild-type GST-PAS protein bound to scFv2.12 was quantified in the presence of increasing amounts of purified CNBh (residues 712 to 880) domain protein (Fig. 4A). The ELISA signal was reduced with increasing concentration of CNBh domain, indicating a diminution in the amount of GST-PAS bound to scFv2.12 adsorbed on the plate. As a control, we performed the same experiment with another scFv antibody fragment, scFv2.10, which binds to the N-terminal residues (R4 and R5) of PAS-cap (48). The X-ray crystal structure of the PAS/CNBh complex and cryo-EM structures of KCNH channels show no interactions between the residues of the scFv2.10 epitope and the CNBh domain (15, 33, 34). As expected, there was no impact of the CNBh protein on the amount of PAS domain associated with scFv2.10 in our ELISA (Fig. 4A). These in vitro experiments demonstrate that scFv2.12 and CNBh domain compete for binding to the PAS domain.
Fig. 4.
scFv2.12 and CNBh domain compete for binding to the PAS domain. (A) Competition ELISA evaluating wild-type PAS domain (1 μM) binding to scFv2.10 or scFv2.12 (coated on the plate) in the presence of increasing concentration of CNBh domain protein. Error bars denote mean ± SD and n = 8. (B) Plot of relative FRET efficiency in cells coexpressing PAS-CFP (donor) and ΔPAS-hERG Citrine (acceptor) in the absence (control) and presence of scFv2.12, administered through the patch pipette at room temperature. Error bars denote mean ± SEM.
To verify that the scFv2.12 antibody interferes with the interaction between the PAS and CNBh domains in the full-length channel, we performed live-cell, sensitized-emission Förster resonance energy transfer (FRET) experiments using the experimental configuration established by Trudeau and colleagues (54). They have shown that coexpression of PAS-CFP (FRET donor) and ΔPAS-hERG Citrine (FRET acceptor) results in the reconstitution of full-length channel that is reflected in detection of FRET signal and reacquisition of the characteristic slow deactivation of wild-type hERG. We observed a FRET signal corresponding to ∼12% relative FRET efficiency (Fig. 4B), similar to published values (54, 55) and characteristic of the reconstituted channel. Strikingly, when scFv2.12 protein was administered intracellularly via the patch pipette, no FRET signal was observed (Fig. 4B). The absence of FRET suggests that the PAS domain in the presence of scFv2.12 was no longer able to interact with the CNBh domain and reconstitute the hERG channel.
Our results establish that the interactions of scFv2.12 and CNBh with PAS are mutually exclusive and consistent with the proposal that scFv2.12 binding interferes with the PAS/CNBh assembly. Crucially, a consequence of this conclusion is that for scFv2.12 to bind to the PAS domain and affect the function of full-length hERG, the channel must visit a state where the PAS/CNBh assembly is altered or destabilized, allowing access to the residues of the epitope.
PAS/CNBh Assembly Is Altered When the Gate Is Open.
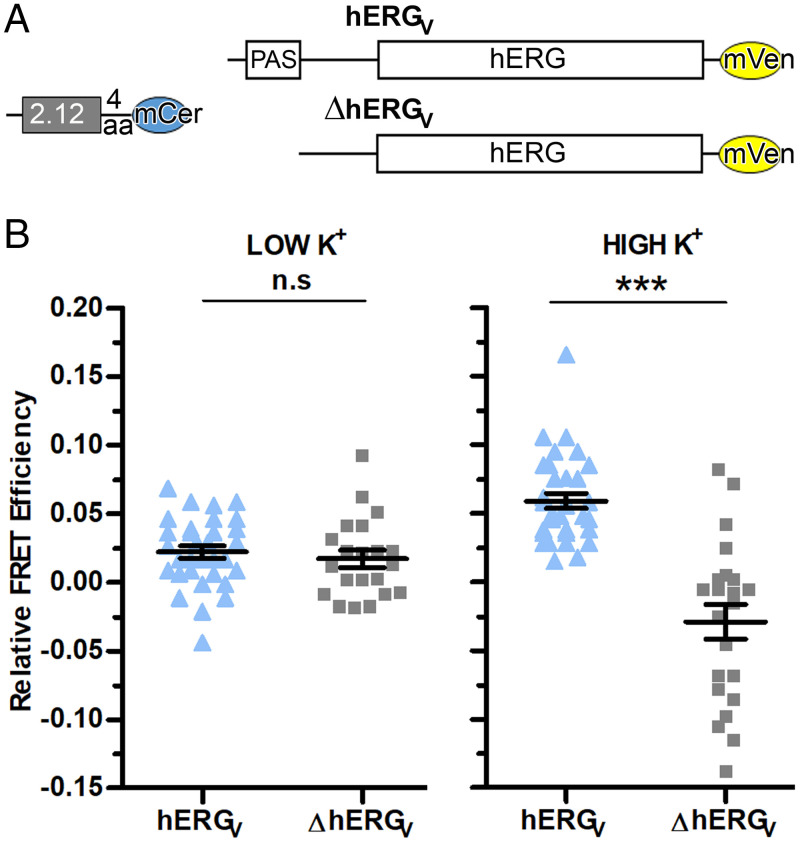
To directly examine the interaction of scFv2.12 with the PAS domain in full-length channels, we took a different live-cell FRET approach. In this case, the FRET donor (2.12(4aa)mCer) was the antibody scFv2.12 fused to mCerulean through a four–amino acid linker. It was paired with the acceptor hERGV, formed by fusion of mVenus to the C terminus of the full-length channel. As a negative control, we used a hERGmVenus channel acceptor lacking the PAS domain (ΔhERGV) that was thus unable to bind the antibody (Fig. 5A).
Fig. 5.
scFv2.12 is a gating sensor. (A) Schematic diagram of donor scFv2.12(4aa)mCerulean and acceptor hERGmVenus (hERGV) or ΔPAS-hERGmVenus (ΔhERGV) constructs used for cotransfection experiments in HEK293 cells. (B) Dot plot of relative FRET efficiency values from individual cells cotransfected with scFv2.12(4aa)mCerulean and hERGV or ΔhERGV channels subjected to 5 mM K+ (LOW K+) or 150 mM K+ (HIGH K+) at 37 °C. Error bars denote mean ± SEM and n ≥ 10. Statistical analysis with unpaired t test and P ≤ 0.001 (***) and P > 0.05 (n.s). Reference SI Appendix, Figs. S3–S5 and Table S1.
We measured sensitized-emission FRET (SI Appendix, Fig. S3) in live cells and evaluated the FRET signal in low K+ for hyperpolarized conditions and in high K+ for a depolarized membrane. Low and high K+ conditions have been used by others to study KCNH channels and, in particular, hERG (56–59). Data were collected at 37 °C for each FRET pair and respective negative control during the same microscope session.
In low K+ conditions, where the membrane is hyperpolarized and the channel therefore has a closed cytosolic gate, there is no difference in FRET signals collected with 2.12(4aa)mCer and hERGV and the negative control, all having relative FRET efficiencies close to 0%. Strikingly, in high K+ conditions, where depolarization will lead to an open cytosolic gate, there is a clear FRET signal compared to the negative control (Fig. 5B and SI Appendix, Table S1). This experiment was repeated using a different FRET donor with a longer linker, yielding similar results (SI Appendix, Fig. S4 and Table S1).
We performed a number of control experiments to rule out nonspecific effects of the different potassium media on the FRET probes. First, we measured the FRET signal from heterotetrameric hERG channels assembled from full-length subunits fused to mCerulean or mVenus at the C termini (SI Appendix, Fig. S5A) and found it was not altered in either low or high K+ conditions (SI Appendix, Fig. S5B and Table S1). In addition, emission spectra of donor 2.12(4aa)mCerulean and acceptor hERGV did not change between low and high K+ conditions (SI Appendix, Fig. S5 C and D). Lastly, FRET signals measured from cells coexpressing 2.12(4aa)mCerulean and hERGV did not vary during the 25-min time period that the cells were exposed to high K+ solution (SI Appendix, Fig. S5E).
Overall, these experiments reveal state-dependent FRET that is related to channel gating and not the result of cellular mechanisms of adaptation to low or high K+ conditions.
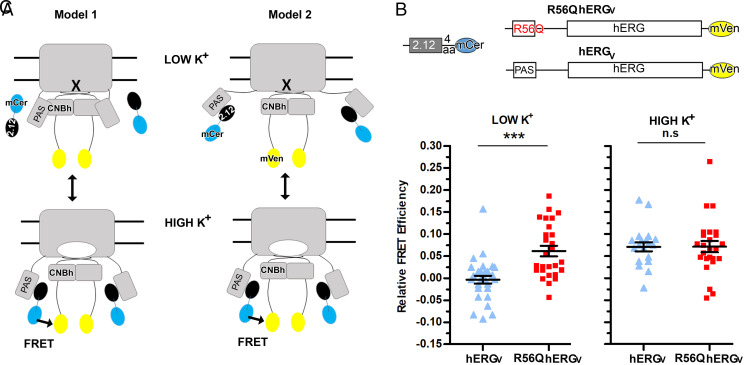
To understand the state-dependent FRET results, we considered two alternative models (Fig. 6A) that take into account our findings that the antibody cannot bind to its epitope unless the interaction between PAS and CNBh is altered. We show the two models in Fig. 6A and for clarity, depict the alteration undergone by the PAS/CNBh assembly, which allows exposure of the epitope for scFv2.12 binding, as a detached PAS domain. In Model 1, the occupation of the epitope on PAS by antibody depends on the state of the gate. Opening the cytoplasmic gate results in alteration and destabilization of the interactions formed by the globular region of PAS and the CNBh domain so that scFv2.12 can bind, allowing FRET signal to be observed. With gate closure, the PAS/CNBh assembly is stabilized as scFv2.12 unbinds and the FRET signal is lost. In Model 2, the epitope of scFv2.12 on PAS is permanently occupied by antibody. In this situation, the PAS/CNBh assembly is altered both when the gate is open and closed, and state-dependent FRET results from relative distance or orientation changes between the fluorophores when the different channel parts move during gating.
Fig. 6.
PAS–CNBh interaction is state dependent. (A) Cartoon representation of two models proposed for interpretation of the state-dependent FRET data collected with scFv2.12. The altered PAS–CNBh assembly is depicted as a dissociated PAS domain for clarity of representation. scFv2.12 fused to mCerulean is represented by black and cyan ovals; hERG channel fused to mVenus is shown in gray and yellow. Closed gate is indicated by cross, open gate by white oval. hERG cytoplasmic domains are indicated in a single subunit. Arrow and FRET label indicate spatial conditions where FRET occurs. Double arrow indicates that the conformation of hERG changes with K+ conditions. (B) Schematic diagram of donor scFv2.12(4aa)mCerulean and acceptor R56Q hERGmVenus (R56Q hERGV) or wild-type hERGmVenus (hERGV) constructs used for cotransfection experiments in HEK293 cells. (C) Dot plot of relative FRET efficiency values from individual cells cotransfected with scFv2.12(4aa)mCerulean and hERGmVenus (hERGV) or R56Q hERG mVenus (R56Q hERGV) channels subjected to 5 mM K+ (LOW K+) or 150 mM K+ (HIGH K+) at 37 °C. Error bars denote mean ± SEM and n ≥ 10. Statistical analysis with unpaired t test and P ≤ 0.001 (***) and P > 0.05 (n.s). Reference SI Appendix, Table S1.
To distinguish these two models, we repeated the live-cell sensitized-emission FRET experiments with 2.12(4aa)mCer as donor and the R56Q mutant of the hERGmVenus channel (R56Q hERGV) as the acceptor (Fig. 6B). The R56Q mutation in the PAS domain has been shown to weaken the PAS–CNBh interaction: 1) the isolated R56Q mutant PAS domain no longer reconstitutes with ΔPAS-hERG channel (54), and 2) isolated wild-type PAS can replace the mutant PAS domain in the R56Q-hERG full-length channel (40, 54). Crucially, this mutation does not alter PAS domain protein stability (35, 39) or affect channel trafficking (35, 39, 40) and is at a position that is distant from the epitope of scFv2.12, with Cα-to-Cα distances between R56 and either N33 or V36 of 16 and 19 Å, respectively.
If Model 1 is correct, R56Q will destabilize the PAS/CNBh assembly when the gate is closed, allowing interaction between scFv2.12 and PAS and giving rise to FRET signal in low K+ conditions. In high K+, with the gate open, FRET efficiency will not be altered if PAS/CNBh destabilization is already extensive. However, if Model 2 is correct, the state-dependent FRET observed for the wild-type channel will be preserved in the R56Q mutation since the PAS/CNBh assembly is already altered in both states.
We performed parallel live-cell FRET experiments with 2.12(4aa)mCer as donor paired with either R56Q mutant hERGmVenus (R56Q hERGV) channel or wild-type hERGmVenus (hERGV) channel as acceptors, in low K+ and high K+ conditions (Fig. 6C and SI Appendix, Table S1). Strikingly, in contrast to the wild-type channel, the R56Q mutant hERG channel now produced a FRET signal (6%) in low K+. In contrast, in high K+ when the gate is open, we observed a similar FRET signal for both the mutant (7%) and wild-type channel (7%), showing that the detected FRET signal between the wild-type form of the channel and scFv2.12 is at maximum.
These results support Model 1, demonstrating that the scFv binds to the channel in a state-dependent fashion. Importantly, this reflects a state-dependent alteration in the PAS/CNBh assembly, where the epitope on the PAS domain surface becomes accessible or occluded during hERG gating. The data also suggest that the alteration in the PAS/CNBh assembly is extensive since the addition of a destabilizing mutation does not further promote antibody binding. Therefore, the PAS/CNBh assembly is dynamic, forming when the gate is closed and becoming extensively altered when the gate opens.
Discussion
Our previous electrophysiology studies at 37 °C indicated that the antibody scFv2.12 accelerates activation and recovery from inactivation (48), abolishing known effects of the PAS domain on hERG gating that limit open probability (44). We have now demonstrated that the binding sites of scFv2.12 and CNBh overlap on the surface of the PAS domain and that their interaction with PAS is mutually exclusive, both in vitro and in the cell. Therefore, our data show that for scFv2.12 to exert its functional effect the PAS/CNBh assembly must undergo an alteration in the fully folded functional channel to make the epitope accessible to the antibody. In addition, we showed using a fluorescently tagged scFv2.12 that the FRET signal between antibody and the channel is state dependent, detected only when the cytosolic gate is open. Finally, a mutation (R56Q) in the PAS domain that is known to destabilize the interaction with the CNBh domain abolishes state-dependent FRET by allowing binding of scFv2.12 when the gate is closed without altering the FRET signal when the gate is open. Altogether, these data demonstrate that the cytosolic PAS/CNBh assembly undergoes a structural change that decreases access to the antibody epitope when the cytoplasmic gate is closed and increases accessibility when the gate opens.
We do not know the exact nature of the structural alteration undergone by the PAS/CNBh assembly. However, our data, together with other published evidence, suggest that it is consistent with a dissociation of PAS from the CNBh domain. The arguments supporting this conclusion are twofold and are presented below.
Structural analysis shows that 1) a large portion (30% of the surface area) of the scFv2.12 epitope on PAS is directly involved in the interface with the CNBh domain and occluded in the hERG structure. In particular, more than 80% of the surface area of residues N33 and I42, which we have shown to be important for antibody binding, is occluded by the interface with the CNBh domain. Moreover, the buried surface area of the epitope residues corresponds to a substantial proportion (17%) of the PAS surface involved in the interaction with CNBh; 2) the scFv2.12 protein is as large as the PAS domain. A model of scFv2.12 (SI Appendix, Fig. S6) shows that the cross-section of the face containing the paratope spans 38 Å by 25 Å; and 3) in antibodies, the surface that includes the antigen recognition region (the paratope) tends to be relatively flat or even concave (60), and therefore, an antibody has a considerable steric “footprint” on an antigen. Altogether, these stereochemical arguments support dissociation of PAS, since exposure of the epitope to allow interaction with the scFv2.12 will require a significant displacement of the PAS domain relative to the CNBh domain, severely affecting the stability of their interaction.
Mutagenesis analysis results reveal that N33 and I42 are important residues both for the interaction of PAS with the scFv2.12 (Figs. 1 and 2) and for the interaction of PAS with CNBh. In particular, N33 and I42 mutations cause acceleration of deactivation (Fig. 2), a proxy for detachment of PAS from CNBh. Moreover, it has been shown that coexpression of the N33T mutant full-length channel with a separate PAS polypetide results in recovery of slow deactivation (40), indicating that the PAS/CNBh interaction is destabilized to such an extent that the channel-tethered mutant PAS domain dissociates from the CNBh domain and permits replacement by a separate wild-type PAS domain. If replacement of N33 by a threonine has such a disruptive impact on the PAS/CNBh assembly, it is very likely that binding of a large protein molecule (the antibody) to a surface region on PAS that includes N33 will also involve dissociation of PAS from CNBh. Finally, our FRET experiments showed that the R56Q mutation did not increase FRET efficiency in the open channel (Fig. 6), consistent with extensive destabilization of the PAS/CNBh interface in the wild-type channel when the gate opens.
Importantly, our proposal of a dissociating PAS domain provides a molecular model that is consistent with hERG information in the literature that has remained unexplained. In particular, dissociation of the PAS domain from CNBh provides an explanation for fluorescence measurements indicating that PAS and neighboring sequences undergo a movement toward the membrane plane when the hERG gate opens (61). Our model is also consistent with studies demonstrating that the PAS–CNBh interaction in hERG is easily perturbed by mutations far away from the interface. More specifically, using fluorescent PAS and ΔPAS-hERG channel it was shown that mutation of residues in the N terminus of the PAS-cap (V3, G6, and V8) or around its binding pocket (Y542 in the S4-S5 linker) (62), which in the hERG cryo-EM structure are more than 30 Å from R56 (Cα-to-Cα) on the PAS–CNBh interface, inhibited the assembly of fluorescent PAS with ΔPAS-hERG channel and recovery of slow deactivation. With a different experimental setup, Kubo and colleagues (63) showed that mutation of R752 (on the A helix of the CNBh domain) and D727 (on the C-linker), which are thought to form an electrostatic-interaction pair that is 28 Å away from R56, accelerate deactivation and decrease FRET signal, also suggesting a weakening of the PAS–CNBh interaction. These results reinforce the idea that the PAS/CNBh assembly is not tightly formed and is responsive to molecular events in its surrounding neighborhood.
This model is also consistent with measurements performed in other KCNH channels. In vitro measurement of the association between PAS and CNBh domains from mouse (m)EAG channel revealed a weak interaction, with a KD in the micromolar range (33, 64). Transition-metal FRET measurements in full-length zebrafish ELK (zELK) reported conformational rearrangements between the PAS and CNBh domains during gating (65). In zELK, the relative movement between the PAS and CNBh domains appears to be limited to a couple of angstroms. Nevertheless, the parallels in the rearrangement of the PAS/CNBh assembly in hERG and in zELK suggest that this is a general property of KCNH channel gating.
Why is it that the cryo-EM structure of hERG shows no evidence of an altered PAS/CNBh assembly or of a detached PAS domain? We speculate with two scenarios. Multiple open states have been postulated for hERG (44, 66); the cryo-EM structure may correspond to one in which the PAS/CNBh assembly remains intact, while our functional and fluorescence readout reflects the dynamic dissociation. Alternatively, PAS dissociation may be disabled by the truncation of 200 N-linker residues in the construct used for cryo-EM studies and/or from the conformational relaxation that is likely to occur during cooling of cryo-EM samples to 100 Kelvin. In contrast, we have performed live-cell FRET experiments at 37 °C with the full-length hERG channel.
In conclusion, using scFv2.12, which has low affinity for the PAS domain, we have generated a fluorescent sensor that reports molecular events in hERG gating and demonstrated that low affinity antibodies have potential applications as molecular sensors. We also showed that formation of the PAS/CNBh assembly is dynamic and coupled to channel gating. Thus, native molecules may be found that regulate gating in KCNH channels by modulating the association equilibrium of the PAS/CNBh assembly, as we have demonstrated with scFv2.12. By extension, this interface represents a potential target for therapeutic development of modulators in the treatment of ion channelopathies.
Materials and Methods
A full description of the materials and methods is presented in SI Appendix.
Protein Expression.
For expression in HEK293 cells, hERG1a channel and scFv2.12 fluorescent fusions were cloned in pcDNA3.1. Bacterial expression was done in Escherichia coli strain BL21(DE3) transformed with plasmids enconding scFv2.12 and 2.10 SNAP fusions, GST-PAS, CNBh domain, or scFv2.12 alone.
FRET Experiments.
FRET measurements used the sensitized-emission method. For low and high K+ experiments, HEK293 cells transiently transfected with plasmid DNA at a donor to acceptor ratio of 5:1 or 3:1 (depending on the construct) were analyzed within 48 h of transfection using an inverted Leica Laser Scanning Confocal SP5 II (or SP8) with control of temperature (37 °C) and CO2 (5%). Measurements were performed using sequentially the Argon 458-nm and 488-nm laser lines to excite the fluorophores. The culture media was replaced immediately before imaging with either a low K+ solution (150 mM NaCl, 5.4 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, 15 mM Glucose, 15 mM Hepes, 1 mM sodium pyruvate, pH 7.4) or a high K+ solution (5.4 mM NaCl, 150 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, 15 mM Glucose, 15 mM Hepes, 1 mM sodium pyruvate, pH 7.4) at 37 °C and 5% CO2.
PAS/ΔPAS-hERG assembling FRET experiments were performed at room temperature with HEK293 cells transiently transfected with PAS domain fused at the C terminus with a cyan fluorescent protein (donor) and with S620T-hERG channel with the PAS domain deleted, fused to a citrine fluorescent protein at the C terminus (acceptor), patched, and held at −80 mV. scFv2.12 antibody protein was perfused through the patch pipette. Experiments were performed on an inverted microscope (Nikon TE-2000). Cells were illuminated with a 120-W X-Cite lamp (Lumen Dynamics). Spectra for FRET analysis were gathered using two filter cubes: 1) a custom FRET cube (436/20, 455dclp, D460lp), and 2) a YFP cube (HQ500/20, Q515lp, HQ520lp), both from Chroma Technology.
ELISA.
ELISA plates were coated with scFv2.12-SNAPHis6 or scFv2.10-SNAPHis6 protein overnight at 4 °C. The plate was incubated for 2 h at room temperature with the following proteins: GST-PAS wild-type, N33A, R35A, V36A, E37A, N38A, I42A, or GST alone (for background subtraction) in the absence or presence of CNBh domain protein followed by incubation with Rabbit anti–GST-HRP antibody before colorimetric detection.
NMR Experiments.
NMR experiments were performed with a Bruker Avance III 800 MHz spectrometer equipped with a triple resonance 1H, 13C, 15N-cryoprobe. Two-dimensional selective optimized flip angle short transient technique coupled to heteronuclear multiple quantum correlation (1H-15N SOFAST-HMQC) spectra (1-h duration) were recorded at 298 K (∼25 °C) using 15N-labeled PAS protein at 75 μM and varying the unlabeled scFv2.12 antibody quantity so that the molar ratio was 1:0, 1:0.25, 1:0.5, 1:0.65, 1:0.75, 1:1, and 1:3. Assignment was done using CCPNMR software using reference chemical shift values obtained from the Biological Magnetic Resonance Data Bank with code BMRB: 17066 (Protein Data Bank code: 2L0W). Chemical shift and peak intensity values were extracted for each titration point and analyzed with in-house built MatLab scripts.
Two-Electrode Voltage Clamp.
RNA for wild-type and mutant hERG1a in pGH19 and scFv2.12 in pcDNA3.1 were injected into Stage IV and V oocytes. For hERG1a/scFv2.12 coexpression experiments, cRNAs were injected at 1:1 ratio. All recordings were performed at room temperature (21 °C ± 2 °C) and held at −80 mV between recordings. Deactivation tail currents were evoked at −120 mV after a depolarizing step to +60 mV. The resistance of the electrodes ranged from 0.5 to 2.0 MΩ in 2 M KCl. Recording solution (5 mM KCl, 93 mM N-methyl-D-glucamine, 1 mM MgCl2, 0.3 mM CaCl2, and 5 mM Hepes, with pH adjusted to 7.4 with MES) was perfused through the bath chamber during the recording. Data were analyzed using pCLAMP 10.4, Origin 2021b, and SigmaPlot 13.0. Results were considered statistically significant when P < 0.05. No blinding or randomization was conducted for the experiments. No data were excluded as outliers.
Acknowledgments
Access to the Advance Light Microscopy platform at i3S (member of the national infrastructure PPBI–Portuguese Platform of BioImaging) was supported by POCI-01-0145-FEDER-022122. We thank Jin Li for technical support. Work was supported by NIH Grants 1R01NS081320 (G.A.R., J.H.M.-C.), 1R01HL131403 (G.A.R.), National Institute of Neurological Disorders and Stroke F99NS125824 (W.A.S.-S.), R00HL133482 (D.K.J.), and R01 GM130701 (M.C.T.), and Agencia Estatal de Investigación Grant RTI2018‐101269‐B‐I00 (O.M.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2108796118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Sanguinetti M. C., Jurkiewicz N. K., Two components of cardiac delayed rectifier K+ current. Differential sensitivity to block by class III antiarrhythmic agents. J. Gen. Physiol. 96, 195–215 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanguinetti M. C., Jiang C., Curran M. E., Keating M. T., A mechanistic link between an inherited and an acquired cardiac arrhythmia: HERG encodes the IKr potassium channel. Cell 81, 299–307 (1995). [DOI] [PubMed] [Google Scholar]
- 3.Sacco T., Bruno A., Wanke E., Tempia F., Functional roles of an ERG current isolated in cerebellar Purkinje neurons. J. Neurophysiol. 90, 1817–1828 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Crociani O., et al., Cell cycle-dependent expression of HERG1 and HERG1B isoforms in tumor cells. J. Biol. Chem. 278, 2947–2955 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Furlan F., et al., Interneurons transiently express the ERG K+ channels during development of mouse spinal networks in vitro. Neuroscience 135, 1179–1192 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Huang X., et al., Voltage-gated potassium channel EAG2 controls mitotic entry and tumor growth in medulloblastoma via regulating cell volume dynamics. Genes Dev. 26, 1780–1796 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simons C., et al., Mutations in the voltage-gated potassium channel gene KCNH1 cause Temple-Baraitser syndrome and epilepsy. Nat. Genet. 47, 73–77 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Kortüm F., et al., Mutations in KCNH1 and ATP6V1B2 cause Zimmermann-Laband syndrome. Nat. Genet. 47, 661–667 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Huang X., et al., EAG2 potassium channel with evolutionarily conserved function as a brain tumor target. Nat. Neurosci. 18, 1236–1246 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pointer K. B., et al., Administration of non-torsadogenic human ether-à-go-go-related gene inhibitors is associated with better survival for high hERG-expressing glioblastoma patients. Clin. Cancer Res. 23, 73–80 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Francisco M. A., et al., Chloride intracellular channel 1 cooperates with potassium channel EAG2 to promote medulloblastoma growth. J. Exp. Med. 217, 217 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pardo L. A., et al., Oncogenic potential of EAG K(+) channels. EMBO J. 18, 5540–5547 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trudeau M. C., Warmke J. W., Ganetzky B., Robertson G. A., HERG, a human inward rectifier in the voltage-gated potassium channel family. Science 269, 92–95 (1995). [DOI] [PubMed] [Google Scholar]
- 14.Curran M. E., et al., A molecular basis for cardiac arrhythmia: HERG mutations cause long QT syndrome. Cell 80, 795–803 (1995). [DOI] [PubMed] [Google Scholar]
- 15.Whicher J. R., MacKinnon R., Structure of the voltage-gated K+ channel Eag1 reveals an alternative voltage sensing mechanism. Science 353, 664–669 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee C. H., MacKinnon R., Activation mechanism of a human SK-calmodulin channel complex elucidated by cryo-EM structures. Science 360, 508–513 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schönherr R., Heinemann S. H., Molecular determinants for activation and inactivation of HERG, a human inward rectifier potassium channel. J. Physiol. 493, 635–642 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morais Cabral J. H., et al., Crystal structure and functional analysis of the HERG potassium channel N terminus: A eukaryotic PAS domain. Cell 95, 649–655 (1998). [DOI] [PubMed] [Google Scholar]
- 19.Spector P. S., Curran M. E., Zou A., Keating M. T., Sanguinetti M. C., Fast inactivation causes rectification of the IKr channel. J. Gen. Physiol. 107, 611–619 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dai G., Zagotta W. N., Molecular mechanism of voltage-dependent potentiation of KCNH potassium channels. eLife 6, 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Terlau H., Heinemann S. H., Stühmer W., Pongs O., Ludwig J., Amino terminal-dependent gating of the potassium channel rat eag is compensated by a mutation in the S4 segment. J. Physiol. 502, 537–543 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gianulis E. C., Liu Q., Trudeau M. C., Direct interaction of eag domains and cyclic nucleotide-binding homology domains regulate deactivation gating in hERG channels. J. Gen. Physiol. 142, 351–366 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gustina A. S., Trudeau M. C., hERG potassium channel gating is mediated by N- and C-terminal region interactions. J. Gen. Physiol. 137, 315–325 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao Y., Goldschen-Ohm M. P., Morais-Cabral J. H., Chanda B., Robertson G. A., The intrinsically liganded cyclic nucleotide-binding homology domain promotes KCNH channel activation. J. Gen. Physiol. 149, 249–260 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henry J. T., Crosson S., Ligand-binding PAS domains in a genomic, cellular, and structural context. Annu. Rev. Microbiol. 65, 261–286 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Möglich A., Ayers R. A., Moffat K., Structure and signaling mechanism of Per-ARNT-Sim domains. Structure 17, 1282–1294 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McIntosh B. E., Hogenesch J. B., Bradfield C. A., Mammalian Per-Arnt-Sim proteins in environmental adaptation. Annu. Rev. Physiol. 72, 625–645 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Robertson G. A., Warmke J. M., Ganetzky B., Potassium currents expressed from Drosophila and mouse eag cDNAs in Xenopus oocytes. Neuropharmacology 35, 841–850 (1996). [DOI] [PubMed] [Google Scholar]
- 29.Brelidze T. I., Carlson A. E., Zagotta W. N., Absence of direct cyclic nucleotide modulation of mEAG1 and hERG1 channels revealed with fluorescence and electrophysiological methods. J. Biol. Chem. 284, 27989–27997 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brelidze T. I., Carlson A. E., Sankaran B., Zagotta W. N., Structure of the carboxy-terminal region of a KCNH channel. Nature 481, 530–533 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marques-Carvalho M. J., et al., Structural, biochemical, and functional characterization of the cyclic nucleotide binding homology domain from the mouse EAG1 potassium channel. J. Mol. Biol. 423, 34–46 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Brelidze T. I., Gianulis E. C., DiMaio F., Trudeau M. C., Zagotta W. N., Structure of the C-terminal region of an ERG channel and functional implications. Proc. Natl. Acad. Sci. U.S.A. 110, 11648–11653 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haitin Y., Carlson A. E., Zagotta W. N., The structural mechanism of KCNH-channel regulation by the eag domain. Nature 501, 444–448 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang W., MacKinnon R., Cryo-EM structure of the open human ether-à-go-go-related K+ channel hERG. Cell 169, 422–430.e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ke Y., et al., Trafficking defects in PAS domain mutant Kv11.1 channels: Roles of reduced domain stability and altered domain-domain interactions. Biochem. J. 454, 69–77 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Codding S. J., Trudeau M. C., The hERG potassium channel intrinsic ligand regulates N- and C-terminal interactions and channel closure. J. Gen. Physiol. 151, 478–488 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ng C. A., Phan K., Hill A. P., Vandenberg J. I., Perry M. D., Multiple interactions between cytoplasmic domains regulate slow deactivation of Kv11.1 channels. J. Biol. Chem. 289, 25822–25832 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ng C. A., et al., The N-terminal tail of hERG contains an amphipathic α-helix that regulates channel deactivation. PLoS One 6, e16191 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harley C. A., Jesus C. S., Carvalho R., Brito R. M., Morais-Cabral J. H., Changes in channel trafficking and protein stability caused by LQT2 mutations in the PAS domain of the HERG channel. PLoS One 7, e32654 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gianulis E. C., Trudeau M. C., Rescue of aberrant gating by a genetically encoded PAS (Per-Arnt-Sim) domain in several long QT syndrome mutant human ether-á-go-go-related gene potassium channels. J. Biol. Chem. 286, 22160–22169 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen J., Zou A., Splawski I., Keating M. T., Sanguinetti M. C., Long QT syndrome-associated mutations in the Per-Arnt-Sim (PAS) domain of HERG potassium channels accelerate channel deactivation. J. Biol. Chem. 274, 10113–10118 (1999). [DOI] [PubMed] [Google Scholar]
- 42.London B., et al., Two isoforms of the mouse ether-a-go-go-related gene coassemble to form channels with properties similar to the rapidly activating component of the cardiac delayed rectifier K+ current. Circ. Res. 81, 870–878 (1997). [DOI] [PubMed] [Google Scholar]
- 43.Lees-Miller J. P., Kondo C., Wang L., Duff H. J., Electrophysiological characterization of an alternatively processed ERG K+ channel in mouse and human hearts. Circ. Res. 81, 719–726 (1997). [DOI] [PubMed] [Google Scholar]
- 44.Sale H., et al., Physiological properties of hERG 1a/1b heteromeric currents and a hERG 1b-specific mutation associated with Long-QT syndrome. Circ. Res. 103, e81–e95 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McPate M. J., et al., hERG1a/1b heteromeric currents exhibit amplified attenuation of inactivation in variant 1 short QT syndrome. Biochem. Biophys. Res. Commun. 386, 111–117 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Larsen A. P., Olesen S. P., Differential expression of hERG1 channel isoforms reproduces properties of native I(Kr) and modulates cardiac action potential characteristics. PLoS One 5, e9021 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones D. K., et al., hERG 1b is critical for human cardiac repolarization. Proc. Natl. Acad. Sci. U.S.A. 111, 18073–18077 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harley C. A., et al., Enhancement of hERG channel activity by scFv antibody fragments targeted to the PAS domain. Proc. Natl. Acad. Sci. U.S.A. 113, 9916–9921 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brelidze T. I., Carlson A. E., Davies D. R., Stewart L. J., Zagotta W. N., Identifying regulators for EAG1 channels with a novel electrophysiology and tryptophan fluorescence based screen. PLoS One 5, 5 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carlson A. E., Brelidze T. I., Zagotta W. N., Flavonoid regulation of EAG1 channels. J. Gen. Physiol. 141, 347–358 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Z. J., Soohoo S. M., Tiwari P. B., Piszczek G., Brelidze T. I., Chlorpromazine binding to the PAS domains uncovers the effect of ligand modulation on EAG channel activity. J. Biol. Chem. 295, 4114–4123 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Burton M. J., et al., Discovery of a heme-binding domain in a neuronal voltage-gated potassium channel. J. Biol. Chem. 295, 13277–13286 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robertson G. A., Morais-Cabral J. H., hERG function in light of structure. Biophys. J. 118, 790–797 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gustina A. S., Trudeau M. C., A recombinant N-terminal domain fully restores deactivation gating in N-truncated and long QT syndrome mutant hERG potassium channels. Proc. Natl. Acad. Sci. U.S.A. 106, 13082–13087 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu Q. N., Trudeau M. C., Eag domains regulate LQT mutant hERG channels in human induced pluripotent stem cell-derived cardiomyocytes. PLoS One 10, e0123951 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sahoo N., Schönherr R., Hoshi T., Heinemann S. H., Cysteines control the N- and C-linker-dependent gating of KCNH1 potassium channels. Biochim. Biophys. Acta 1818, 1187–1195 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zou A., Xu Q. P., Sanguinetti M. C., A mutation in the pore region of HERG K+ channels expressed in Xenopus oocytes reduces rectification by shifting the voltage dependence of inactivation. J. Physiol. 509, 129–137 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weinsberg F., Bauer C. K., Schwarz J. R., The class III antiarrhythmic agent E-4031 selectively blocks the inactivating inward-rectifying potassium current in rat anterior pituitary tumor cells (GH3/B6 cells). Pflugers Arch. 434, 1–10 (1997). [DOI] [PubMed] [Google Scholar]
- 59.Miranda P., et al., Specificity of TRH receptor coupling to G-proteins for regulation of ERG K+ channels in GH3 rat anterior pituitary cells. J. Physiol. 566, 717–736 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.MacCallum R. M., Martin A. C., Thornton J. M., Antibody-antigen interactions: Contact analysis and binding site topography. J. Mol. Biol. 262, 732–745 (1996). [DOI] [PubMed] [Google Scholar]
- 61.Barros F., Dominguez P., de la Pena P., Relative positioning of Kv11.1 (hERG) K+ channel cytoplasmic domain-located fluorescent tags toward the plasma membrane. Sci. Rep. 8, 15494 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fernández-Trillo J., et al., Molecular determinants of interactions between the N-terminal domain and the transmembrane core that modulate hERG K+ channel gating. PLoS One 6, e24674 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kume S., Shimomura T., Tateyama M., Kubo Y., Two mutations at different positions in the CNBH domain of the hERG channel accelerate deactivation and impair the interaction with the EAG domain. J. Physiol. 596, 4629–4650 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fernandes A. S., Morais-Cabral J. H., Harley C. A., Screening for non-pore-binding modulators of EAG K+ channels. J. Biomol. Screen. 21, 758–765 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dai G., James Z. M., Zagotta W. N., Dynamic rearrangement of the intrinsic ligand regulates KCNH potassium channels. J. Gen. Physiol. 150, 625–635 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kiehn J., Lacerda A. E., Brown A. M., Pathways of HERG inactivation. Am. J. Physiol. 277, H199–H210 (1999). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All study data are included in the article and/or SI Appendix.