Significance
Many bacterial pathogens encode type III protein secretion machines, which have the capacity to inject bacterial proteins into host cells to promote virulence. An important component of these machines is the tip complex, a structure located at the end of the needle filament of the secretion machine. This structure is essential for the delivery of effector proteins into host cells. We have used cryoelectron microscopy to visualize at high resolution the tip complex of a type III secretion machine of the bacterial pathogen Salmonella Typhimurium. This work provides important insight into the assembly and function of type III protein secretion machines and may provide the bases for the development of therapeutic strategies to combat infectious diseases.
Keywords: bacterial pathogenesis, structural biology, cryoelectron microscopy, protein secretion, organelle assembly
Abstract
Type III secretion systems are multiprotein molecular machines required for the virulence of several important bacterial pathogens. The central element of these machines is the injectisome, a ∼5-Md multiprotein structure that mediates the delivery of bacterially encoded proteins into eukaryotic target cells. The injectisome is composed of a cytoplasmic sorting platform, and a membrane-embedded needle complex, which is made up of a multiring base and a needle-like filament that extends several nanometers from the bacterial surface. The needle filament is capped at its distal end by another substructure known as the tip complex, which is crucial for the translocation of effector proteins through the eukaryotic cell plasma membrane. Here we report the cryo-EM structure of the Salmonella Typhimurium needle tip complex docked onto the needle filament tip. Combined with a detailed analysis of structurally guided mutants, this study provides major insight into the assembly and function of this essential component of the type III secretion protein injection machine.
Many pathogenic or symbiotic bacteria for plants or animals have evolved specialized molecular machines known as type III protein secretion systems (T3SSs) (1–3). These machines inject bacterially encoded effector proteins into target eukaryotic cells to modulate cellular processes and ensure the survival and replication of the pathogens or symbionts that encode them (4–6). Although the structural organization of this secretion machine has been largely derived from studies of the T3SSs of the bacterial pathogens Salmonella Typhimurium and Shigella flexneri (7–15), given the conservation of its core components, it is predicted that the T3SSs in other bacteria exhibit a similar architecture. The secretion machine itself is composed of a ∼5-Md multiprotein assembly known as the injectisome (1, 7, 14, 16, 17). This core structure consists of two large substructures, an envelope-associated needle complex (7, 16, 18, 19) and a large cytoplasmic complex known as the “sorting platform” (14, 15, 20).
The needle complex is composed of a multiring hollow base, which anchors the injectisome to the bacterial envelope (18). The base encloses the export apparatus, a helical structure that serves as a conduit for the passage through the inner membrane of the proteins destined to transit this secretion pathway (21). The base is also linked to a needle-like filament, which protrudes several nanometers from the bacterial surface and is formed by a single protein arranged in a helical fashion (17, 22). The needle filament, which is traversed in its entire length by a central channel ∼3 nm in diameter, is linked to the base through a structure known as the inner rod (11) and is capped at its distal end by another substructure known as the “tip complex” (23, 24).
The sorting platform is located in its entirety within the cytoplasm (14, 15, 20). It is made of six pods that form a cage-like enclosure, which is capped on its cytoplasmic side by a wheel-like structure that holds a hexameric ATPase. Also harbored within the sorting platform cage is the large cytoplasmic domain of one of the components of the export apparatus, which is arranged as two concentric rings and forms a conduit for the secreted substrates to reach the entrance of the export channel (25).
A distinctive feature of the T3SSs is that their activation requires contact with the target eukaryotic cell (26, 27). The activation of the T3SS is followed by the deployment of the translocon substructure, which firmly anchors the injectisome to the target cell and serves as the passageway for the effector proteins across the eukaryotic cell membrane. Although little is known about the activation process, it is thought that sensing of the target cell by the tip complex initiates a signaling event that is transduced to the secretion machine by the needle filament itself (26–31). Activation of the secretion machine is then followed by the deployment of the translocon on the target cell membrane, which along with the tip complex and the needle filament, form a continuous passageway through which effector proteins transit from the bacterial cytoplasm to the cytosol of the eukaryotic cell (23, 32, 33). The composition of the tip complex has been the subject of some controversy. While it has been proposed that in the T3SSs of Yersinia spp., Pseudomonas aeruginosa, and Salmonella spp. the tip structure is made up of a single protein, LcrV (24), PcrV (34), and SipD (35), respectively, in the case of Shigella spp., it has been alternatively proposed to be composed of two proteins, IpaB and IpaD (36), or just IpaD (37, 38). The crystal structures of monomeric SipD and close homologs show that these proteins are arranged in three domains: an N-terminal α-helical hairpin, a central coiled-coil, and a mixed α/β carboxyl-terminal domain (39–42). It has been proposed that the N-terminal α-helical hairpin domain functions as a self-chaperone that prevents the self-oligomerization and/or the premature interaction of the tip protein with the needle filament subunit within the bacterial cytoplasm (39). A current hypothesis is that during assembly at the tip of the needle, the N-terminal α-helical hairpin of SipD/IpaD is displaced to allow other domains to interact with the needle. However, there is no structural information of the fully assembled tip complex to support this hypothesis. How the tip protein assembles into the tip complex, and how it is anchored at the distal end of the needle filament, is currently unknown in large part because of the absence of a high-resolution structure of this complex. Understanding of the events that lead to the activation of the secretion machine requires detailed knowledge not only of the structure of the tip complex that caps the needle filament but, importantly, its interface with the needle filament itself.
Advances in cryoelectron microscopy (cryo-EM) have allowed the visualization of most components of the T3SS machine at high resolution, both in isolation as well as in situ (7–15). However, the tip structure has eluded high-resolution visualization, in part because existing needle complex isolation protocols result in the dissociation of the tip complex from the needle filament. Here we report the visualization at high resolution by cryo-EM of the tip structure of the needle complex of the S. Typhimurium T3SS encoded within its pathogenicity island 1. Combined with functional analysis, the structure provides major insight into the potential mechanisms of injectisome assembly and activation and fills one of the remaining gaps in the quest for the high-resolution visualization of the entire T3SS injectisome.
Results
Overall Structure of the Tip Complex.
Because the needle tip substructure is not firmly attached to the needle filament, commonly used protocols to isolate the needle complex result in the loss of the tip structure during purification. We therefore developed a gentler isolation protocol that resulted in the retention of the tip structure in a substantial proportion of the purified needle complexes. Isolated needle complexes were then examined by single particle cryo-EM (SI Appendix, Fig. S1). Reference-free two-dimensional (2D) classification of manually picked needle ends of the needle complexes showed two well-defined groups characterized by the presence or absence of the tip complex (Fig. 1A). Single-particle reconstruction of the two groups gave rise to maps with global resolution of 3.9 Å for the needle filament ends with a tip and 5 Å for those without a tip as determined by the Fourier shell correlation (FSC) at 0.143 cutoff (SI Appendix, Fig. S2). Local resolution estimation of the three-dimensional (3D) map of the needle filament without the tip showed that, although most of the map was resolved to 5-Å resolution, the first turn of the PrgI subunits at the very end of the helical needle filament was resolved at a much lower resolution, with many of the α-helices resolved up to 13 Å (SI Appendix, Fig. S3). One possible explanation for this observation is that, in the absence of the capping tip, stabilizing contacts between the needle protein PrgI at the end of the needle filament are lost, leading to structural flexibility and low resolution.
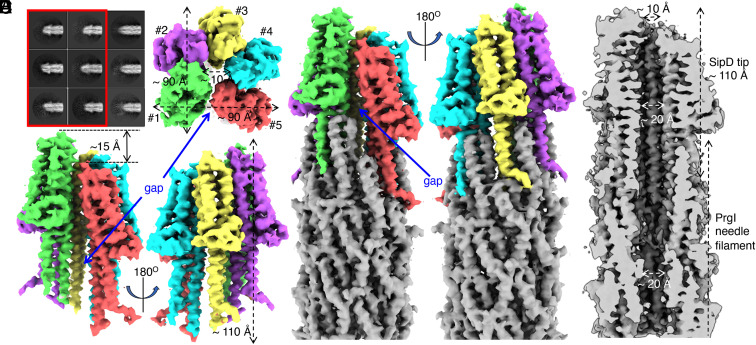
Fig. 1.
Structure of the type III secretion needle filament tip complex. (A) Representative 2D class averages of needle filaments with (red box) and without tip complexes. (B and C) Top (B) and side (C) views of the SipD pentamer. Each subunit is rendered in a different color. The dimensions of the complex and the gap between the first and the fifth SipD subunits are indicated. The five SipD subunits are in helical arrangement and the distance along the filament axis between the first and the fifth subunits is about 15 Å (indicated in C). (D) Side views of the SipD pentamer sitting on top of the needle filament. The five SipD subunits are rendered in different colors. The needle filament is colored in gray. The gap between the first and the fifth SipD subunits is indicated. (E) Cut-through view of the tip complex and needle filament. The diameters of the pore and the central lumen are indicated.
The 3D reconstruction of the needle filament with its tip showed five SipD subunits at the end of the needle filament organized in an open ring structure with an approximately helical configuration (Fig. 1 B–E). The translational distance along the helical axis between the first and the fifth subunits is about 15 Å (Fig. 1C). The dimension of the SipD ring structure is about 90 Å in length, 90 Å in width, and 110 Å in height (Fig. 1 B and C). Each SipD molecule is inserted in between two PrgI subunits (Fig. 1D), which is consistent with a previously proposed model based on NMR data (35). The highest resolution was observed at the center of the structure, while lower resolution was observed at its surface, with the resolution of the loop region at the top of the SipD pentamer dropping to around 7 Å (SI Appendix, Fig. S4). Compared to the SipD pentamer, the resolution of the PrgI filament is higher, ranging from 3.7 Å to 4.5 Å (SI Appendix, Fig. S4). A notable feature of the tip structure is the presence of a gap between the first and fifth subunits of the SipD pentamer, which is 10 Å wide at its widest (Fig. 1 C and D). The gap is closed by a small interface between the first and the fifth SipD subunits at the distal end of the tip structure and broadens toward its needle-proximal end. Two PrgI subunits border the gap at its intersection with the needle filament end, and as a result, the symmetry mismatch between the SipD pentamer and the PrgI filament is solved (Fig. 1D). The SipD pentamer encloses a pore or channel, which is 20 Å in diameter on its needle filament proximal side and ∼10 Å at its opening on the distal side (Fig. 1E). The central channel of the tip complex ring seamlessly connects to the central lumen of the needle filament. Unlike what has been observed in the transition between the needle filament channel on its cytoplasmic side and the export apparatus (12), we observed no narrowing of the diameter of the channel in the transition between the needle filament and the tip complex (Fig. 1E).
Atomic Model of the Tip Complex.
To build an atomic model of the tip complex, we docked the crystal structure of the SipD monomer (PDB ID: 2YM9) (42) into the EM map using Chimera (43) and performed several rounds of model building and refinement with COOT (44) and Phenix (45). The arrangement of SipD subunits in the tip complex agrees with a previously proposed NMR model (35) and a low-resolution EM structure of IpaD tip complex (37), with the central coiled-coil domain lining along the central lumen and the C-terminal domain facing outwards (Fig. 2A and SI Appendix, Fig. S5). Similar to what has been observed in the crystal structure of the SipD monomer (PDB IDs: 2YM9 and 3NZZ) (40, 42), the N-terminal domain (residues 1 to 123) of SipD could not be resolved in our map presumably due to its intrinsic flexibility. In the close homolog IpaD, this domain has been proposed to exert a self-chaperoning function to prevent its self-oligomerization in the bacterial cytoplasm, or its premature interaction with the needle filament protein (39). It has been proposed that this domain would not participate in interactions with the needle filament but rather, it would have to be displaced during tip complex assembly to allow docking of the SipD subunits onto the tip of the needle filament. The inability to see this domain in our structure is consistent with this hypothesis. All the five SipD subunits in the complex are in the same conformation, with a Cα root-mean-square deviation (rmsd) of less than 0.582 Å over 216 residues (Fig. 2B). However, SipD within the complex exhibits significant structural differences with the two available crystal structures of monomeric SipD (PDB IDs: 3NZZ and 2YM9), with a Cα rmsd of 1.2 Å over 207 residues and 1.5 Å over 211 residues, respectively. The major differences are within the two helices (helices 4 and 8) of the central coiled-coil domain, which make contact with the needle filament subunit PrgI (Fig. 2C). In particular, in the complex, helix 8 is bent by the presence of a 310 helix, resulting in a deviation by ∼22° from the path followed by the same helix in the structure of the monomeric form of SipD (Fig. 2C). The trajectory of helix 4 is also bent by ∼20° relative to its trajectory in the monomeric structure (Fig. 2C). There are also differences with the structure of the monomer of the IpaD homolog, which also lacks the 310 helix (46). However, the functional relevance of this difference is uncertain.
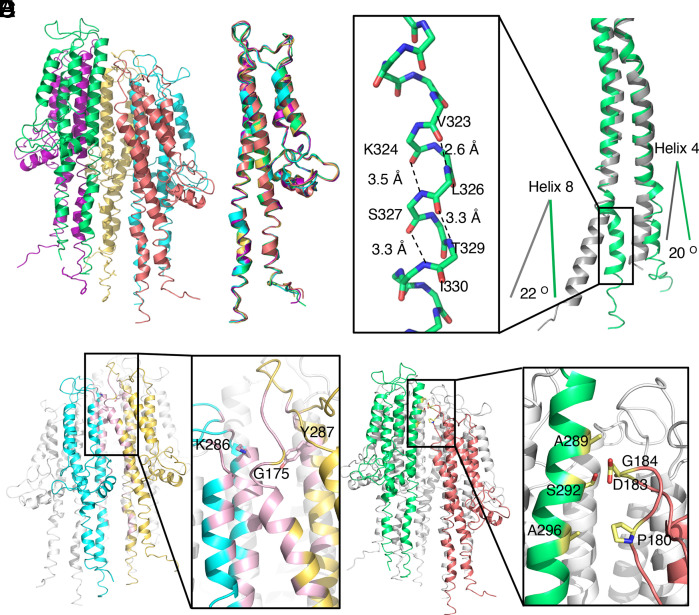
Fig. 2.
Atomic model of the tip complex. (A) Atomic model of the SipD pentamer. Each SipD subunit is colored following the same color scheme used in Fig. 1. (B) Structural alignment of the five SipD subunits. (C) Structural differences between SipD in the tip complex (green) and the crystal structure of monomeric SipD (gray) (PDB ID: 3NZZ). Shown in the Inset is a 310 helix that is present in SipD in the tip complex but absent in the monomeric structure. Hydrogen bonds are denoted as dashed lines. The differences between the orientations of residues 136 to 146 of helix 4 and residues 318 to 340 of helix 8 in the complex and monomeric structures of SipD are shown. (D) Common intersubunit interface in the SipD tip complex. Two of the SipD subunits in the complex are colored in yellow and cyan and the other three are colored in white. The interface between the two neighboring SipD subunits is highlighted in pink. The Inset shows a detail of the interface indicating the relevant residues involved. (E) SipD1–SipD5 interface. SipD1 and SipD5 are colored in green and deep salmon, respectively. The interface, depicted in the Inset, is highlighted in yellow. Side chains of the interacting residues targeted for functional analysis are shown as a stick model.
The interfaces between neighboring SipD subunits are in all cases identical except the interface between the first and the fifth subunits of the pentameric complex, which is separated by a gap (see below) thus resulting in a much smaller interface (Fig. 2 D and E and SI Appendix, Table S1). The common intersubunit interface involves mostly elements of its central coiled-coil domain (helix 4 and helix 8) in one subunit and helix 8 of the central coiled-coil domain and helix 7 of the C-terminal domain in the neighboring subunit (Fig. 2D). This intersubunit interface, which buries ∼790 Å2, involves mostly hydrophobic interactions mediated by residues D267, Q266, G270, and L271 in helix 7; A285, K286, A289, W290, G293, Q297, and N314 in helix 8 of one subunit; residues of S168, L171, S172, K173, M174, G175, G176, W177, and L178 in helix 4; and residues Y287, S313, and D320 in helix 8 in the neighboring subunit. The interface is further stabilized by several salt bridges, including those formed between E299 in helix 8 of one subunit and K302 in helix 8 of the neighboring subunit, D267 in helix 7 of one subunit and K173 in helix 4 of the neighboring subunit, and N321 in helix 8 of one subunit and D320 and K324 in helix 8 of the neighboring subunit. The interaction between the first and fifth subunits, on the other hand, is much more limited, burying only ∼100 Å2 (Fig. 2E). This interface involves residues P180, D183, and G184 in the loop region between helix 4 and helix 5 of the carboxyl-terminal domain of the first subunit and residues A289, S292, and A296 in helix 8 of the central coiled-coil of the fifth subunit.
Electrostatic Features of the Needle Tip Complex Lumen.
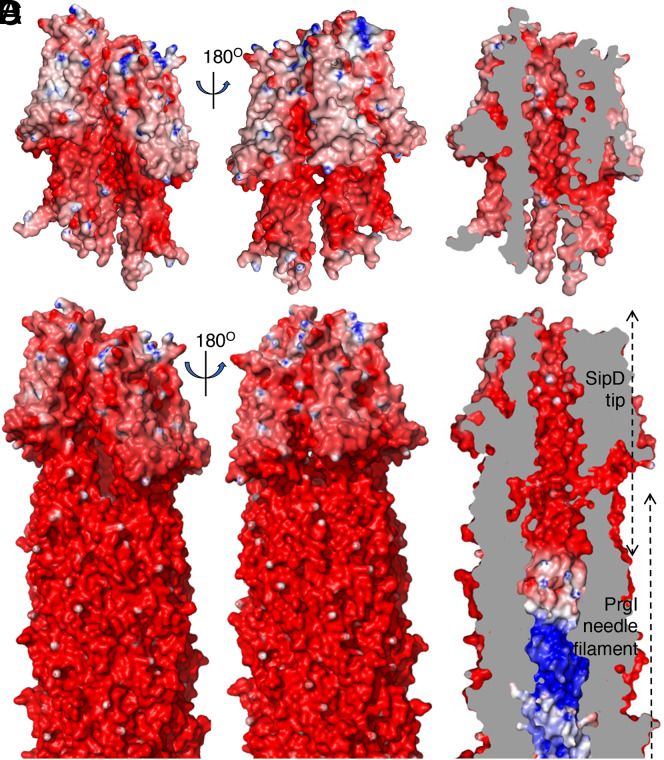
The surface electrostatic potential of the tip complex structure indicates that the outside surface is largely negatively charged (Fig. 3 A and C). The surface of the lumen of the tip complex is also negatively charged (Fig. 3 B and D). The SipD lumen residues include N185 in the loop region between helix 4 and helix 5, and N284, Q288, S292, K295, E299, K302, Q306, T309, Q310, S313, S317, D320, N321, K324, S327, I330, S331, L334, T336, A337, and F340 in helix 8 (SI Appendix, Fig. S6). The negatively charged nature of the lumen surface of the tip complex stands in sharp contrast with the surface of the lumen of the needle filament, which is composed of a spirally arranged hydrophobic groove separated by positively charged residues (17, 47) (Fig. 3D). This arrangement has been proposed to play an important role in the progression of substrates through the central lumen of the needle filament (1, 47). Consistent with this hypothesis, mutations in needle filament lumen residues lead to altered secretion phenotypes (47). The functional significance of the differences in the electrostatic properties of the lumen surfaces of the tip complex and needle filament is unclear. Effector proteins transit the tip complex segment of the continuous channel that transports them from the export apparatus only at the very end of their journey, immediately prior to their translocation through the channel formed by the protein translocases (SipB and SipC in the case of the SPI-1 T3SS) that will move them through the eukaryotic cell membrane. It is therefore reasonable to hypothesize that the electrostatic properties of the lumen of the tip complex may be important to prepare substrates for the translocation step across the eukaryotic cell membrane, perhaps by slowing down their progression through the central lumen. In this context it is noteworthy that residues that make up the lumen of the tip complex are highly conserved across homologs in other species (SI Appendix, Figs. S7 and S8), and that mutagenesis studies have shown that they are essential for effector delivery (35).
Fig. 3.
Electrostatic map of the lumen and outer surface of the SipD tip complex. (A and B) Electrostatic potential map of the SipD pentamer. A cut-through view showing the electrostatic surface potential of the lumen is shown in B. (C and D) Electrostatic surface potential map of the needle filament and tip complex. A cut-through view highlighting the differences in the electrostatic properties of the needle filament and tip complex lumen is shown in D.
Atomic Interface between the Tip Complex and the Needle Filament.
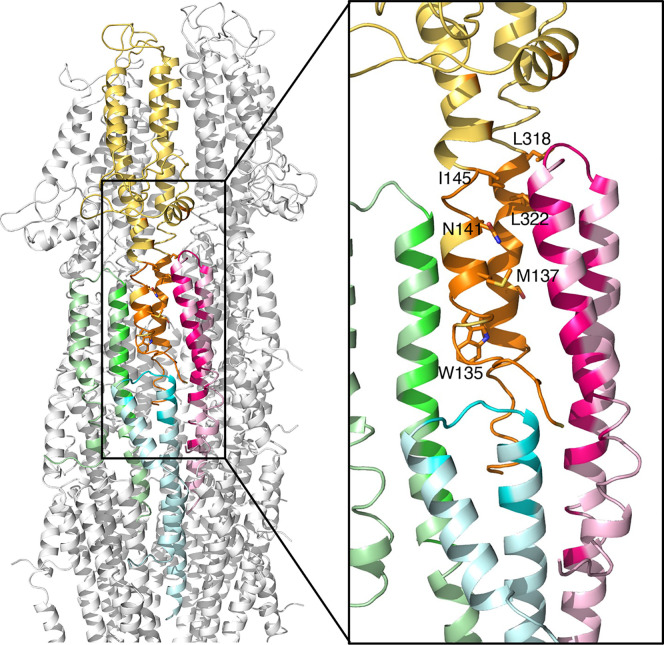
The SipD pentamer is firmly docked onto the needle filament by various hydrophobic interactions and salt bridges between each SipD monomer and the PrgI subunits in the first and second turns of the helical needle filament (Fig. 4 and SI Appendix, Table S2). Each SipD interacts with three PrgI subunits. The interface between SipD (#N) and PrgI (#N) buries about 1,100 Å2, involving helix 4 and helix 8 of the central coiled-coil domain of SipD; helix 6 of its C-terminal domain of SipD; and helix 1, kink region; helix 2, linker region; and helix 3 of PrgI. The interface between SipD (#N) and PrgI (#N+1) buries about 600 Å2 and involves helix 4 and helix 8 of the central coiled-coil domain in SipD and helix 3 of PrgI. Finally, the interface between SipD (#N) and PrgI (#N+6) is also relatively large, burying about 400 Å2, involving helices 4 and 8 of the central coiled-coil domain of the SipD and helix 2, linker region, and helix 3 of PrgI. Notably, most of the interacting residues in PrgI are involved in the PrgI–PrgI interaction in the needle filament, which is consistent with the notion that the tip protein docks onto the tip of the needle filaments through protein–protein interactions similar to those that drive the oligomerization of the needle filament itself.
Fig. 4.
Atomic interfaces between the SipD pentamer and the PrgI needle filament. Ribbon representation of the SipD–PrgI interface. One SipD subunit (yellow) interacts with three PrgI subunits (light green, light cyan, and light pink). The interfaces are highlighted in orange in SipD, and in green, cyan, and pink in PrgI, respectively. The Inset shows the interacting residues (shown as a stick model) targeted for functional analysis.
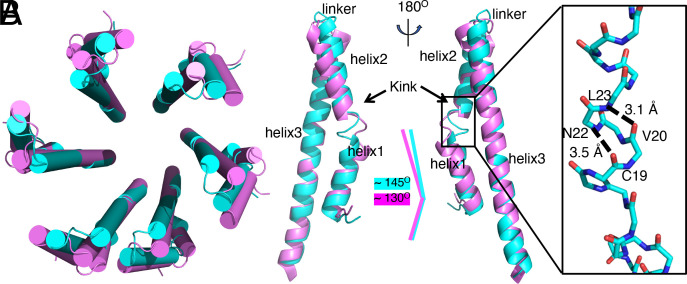
Conformational Changes in the Needle Filament Subunit upon Tip Complex Assembly.
The structures of the PrgI subunits extracted from the second and third turns of the needle filament are identical to the previously reported structures of PrgI in the main body of the needle filament (PDB ID: 6OFH) (47) (SI Appendix, Fig. S9). However, the structure of PrgI in the first turn is in a conformation that is different from all available structures (Fig. 5 A and B). Although the first turn and second turn of PrgI subunits are poorly aligned, the helical parameters stay the same (Fig. 5A). A major difference resides in the kinked region located within the N-terminal helix (residues F16-N22) of PrgI, which instead of a flexible loop, is organized into a helix (Fig. 5B). As a result, the angle between helix 1 and helix 2 is about 15° larger than that of other subunits in the needle complex (Fig. 5B). This reorganization of the helices allows PrgI to engage in critical interactions with SipD, which would be incompatible with the alternative conformation observed in the main body of the needle filament. This observation has biological significance, since it has long been hypothesized that the kink region in the N-terminal helix of PrgI plays a role in the signal transduction process that leads to the activation of the secretion machine upon contact with eukaryotic cells (1, 47). Our structure suggests a different function for the kink region, which is to provide the structural flexibility in PrgI necessary to accommodate interactions with SipD at the end of the needle filament. It remains to be determined whether this region also plays a role in signal transduction. Besides the kink region, there are also differences in the structure of the loop region between helices 2 and 3 (Fig. 5B). This region is at the interface with the SipD complex and therefore undergoes conformational changes upon tip-complex docking and assembly. Finally, helix 3 in the PrgI subunits of the first turn of the needle filament is “bent” relative to its structure in the main body of the filament, a conformational change that is also due to its interaction with SipD (Fig. 5B).
Fig. 5.
Conformational changes in the needle filament protein PrgI upon assembly and docking of the needle tip complex. (A) Cartoon representation (alpha-helices shown as cylinders) of the top view of superimposed structures of the first (cyan) and second (violet) turns of the PrgI subunits in the needle filament structure. (B) Superimposition of the structures of a PrgI subunit in the first (cyan) and second (violet) turns of the needle filament structure. Conformation changes in the kink and linker regions as well as helix 3 are denoted. The Inset shows a 310 helix present in the kink region of PrgI subunits in the first turn, with relevant residues denoted.
Capped and Uncapped Needle Filaments Exhibit a Similar Structure.
It is broadly accepted that the needle filament serves as a signal transduction device (29), a notion that is largely supported by studies that have identified needle filament mutants with altered secretion phenotypes (47–50). The absence of the tip complex results in a phenotype similar to that of an “activated” T3SS, characterized by enhanced secretion of effector proteins. One hypothesis that has been proposed to explain this phenotype is that the tip complex acts as a “plug” that prevents the export of all other proteins. However, our structure showing a continuous open conduit involving the central lumen of the needle filament and the lumen of the pentameric ring of the tip complex (Fig. 1E) rules out the plug hypothesis. An alternative hypothesis proposes that the absence of the tip protein results in conformational changes in the needle filament that trigger signaling events akin to those triggered by physiological activating signals such as contact with mammalian cells. If this is the case, needle filaments from capped and uncapped filaments may show a different conformation. To gain insight in this issue we compared the structures of the central portion (i.e., away from the tip) of the PrgI filaments with or without the tip complex. Comparison of the 2D and 3D maps of capped and uncapped filaments did not show any detectable differences (SI Appendix, Fig. S9), closely matching the published structures of needle filaments attached to the needle complex base (47). This observation indicates that the presence or absence of the tip complex does not result in significant conformational changes in the needle filament itself and, therefore, aberrant signal transduction due to structural changes in the needle filament may not explain the altered secretion phenotype observed in the absence of the needle tip complex.
Bile Salts and the Needle Tip Complex.
Bile salts (i.e., sodium deoxycholate) have been shown to have an activating effect on the T3SS of S. flexneri (51). Although the mechanisms of this activation and its potential physiological significance are unknown, it is believed that sodium deoxycholate acts directly on the tip protein triggering conformational changes and signal transduction events that lead to the stimulation of secretion (40, 52, 53). Studies conducted with the tip complex proteins of Salmonella spp. (SipD) and Shigella spp. (IpaD) have proposed their specific binding sites to deoxycholate (40, 53). However, the proposed binding sites, which were determined in studies with the monomeric forms of these proteins, are incompatible with our structure, since the proposed surfaces would not be available for binding on the assembled tip complex and would result in steric clashes (SI Appendix, Fig. S10). In the case of SipD, the site was derived from the structure of a SipD dimer in complex with deoxycholate (40). However, the dimer in the crystal is incompatible with the assembled SipD pentameric complex and is therefore likely a crystallization artifact (SI Appendix, Fig. S10). It should be stated that, unlike the case of Shigella spp., addition of sodium deoxycholate does not lead to the activation of the S. Typhimurium T3SS. Therefore, more studies will be required to ascertain whether sodium deoxycholate binds to the assembled tip complex in Salmonella, and if it does, to define its binding site.
Structure and Function Analyses of the Tip Structure and Its Interface with the Needle Filament.
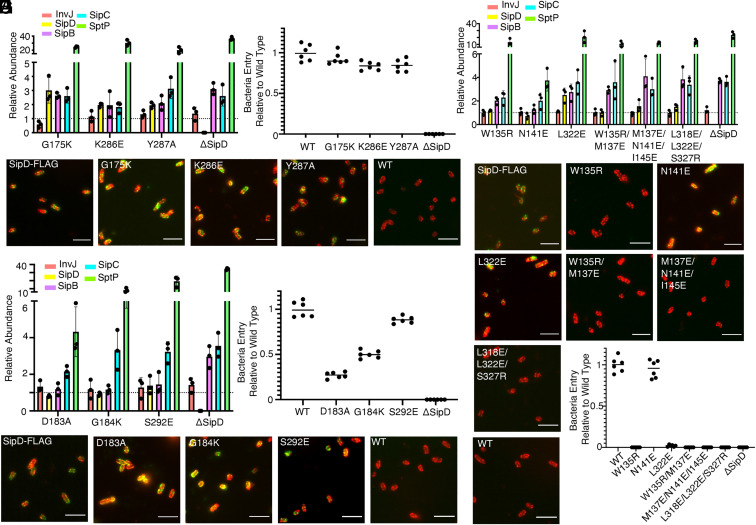
To validate our cryo-EM structure, we carried out structure-guided mutagenesis targeting key amino acids in the different interacting surfaces mapped by our structural analysis. We first targeted the common interface between SipD subunits. As described above, this large interface is stabilized by several hydrophobic interactions between amino acids on helix 4 and 8 of the central coiled-coil domain (Fig. 2D). Consistent with the redundant nature of the interactions that establish this interface, introduction of single mutations in several of the key amino acids involved did not result in the loss of the needle tip complex, or in the loss of type III secretion function as measured by the ability of S. Typhimurium to invade cultured epithelial cells (Fig. 6 B and C). However, these mutations resulted in dysregulation of type III secretion reflected in enhanced secretion of effectors and protein translocases, although none of the mutations affected the stability of SipD (Fig. 6A and SI Appendix, Fig. S11 and Dataset S1). This phenotype is of interest since it is equivalent to the secretion phenotype observed in the absence of the tip complex protein SipD, or as a consequence of mutations that disrupt its association with the needle filament (47). However, in the case of the mutations in residues involved in the SipD–SipD interface, the enhanced secretion was observed despite the presence of the tip complex (Fig. 6 A and B and SI Appendix, Fig. S11 and Dataset S1), suggesting that these mutations may affect signal transduction relay points in the needle tip complex that influence type III secretion.
Fig. 6.
Structure and function analyses of the tip structure and its interface with the needle filament. (A–C) Functional analysis of S. Typhimurium strains expressing SipD variants with the indicated mutations in residues involved in the formation of the common SipD intersubunit interface. (A) Bar graph detailing the relative abundance of the indicated secreted substrates in culture supernatants standardized relative to the values of the WT strains (given a value of 1 and demarcated by a gray dashed line). All values represent the mean ± the SD of three independent experiments. The data were compiled from the data presented in Dataset S1. (B) Fluorescence microscopy images of intact, nonpermeabilized S. Typhimurium expressing FLAG-tagged wild-type SipD (SipD-FLAG), or the indicated mutant forms. Bacterial cells were probed with antibodies to the FLAG tag (green) and to lipopolysaccharide (red). The wild-type strain (with no tag) (WT) was included as a negative control. (Scale bars, 5 μm.) (C) Cultured epithelial cell invasion ability of S. Typhimurium strains expressing the indicated SipD mutants. Numbers represent the percentage of the inoculum that survived antibiotic treatment due to internalization and are the mean ± SD of six independent experiments normalized to wild type, which was set to 1. T3SS, type III secretion system; WT, wild type. (D) Secretion profile of different S. Typhimurium SipD1–SipD5 interface mutant strains shown by bar graph. The relative abundance of the secreted substrates has been standardized relative to the values of the WT strains, which were given a value of 1 and are demarcated by a gray dashed line. All values represent the mean ± the SD of three independent experiments. (D and E) Functional analysis of S. Typhimurium strains expressing SipD variants with the indicated mutations in residues involved in the formation of the SipD1–SipD5 interface. Secretion profiles (D), culture epithelial cell invasion (E), and fluorescence microscopy images (F) are shown. Experiments were conducted as indicated in A–C. (F–H) Functional analysis of S. Typhimurium strains expressing SipD variants with the indicated mutations in residues involved in the formation of the SipD–PrgI interface. Secretion profiles (G), fluorescence microscopy images (H), and culture epithelial cell invasion (I) data are shown. Experiments were conducted as indicated in A–C.
Because of the presence of a gap, the interface between subunits 1 and 5 of the needle tip complex is much more limited, involving only a small number of residues (Fig. 2E). To assess the functional importance of this interface, we introduced mutations in the key residues involved in this interaction. We found that none of the mutations affected the assembly of the tip complex as demonstrated by the detection of SipD on the bacterial surface (Fig. 6F), nor did they significantly affect the pattern of protein secretion (Fig. 6D) or the stability of SipD (SI Appendix, Fig. S12 and Dataset S1). However, two of the mutants drastically increased the secretion of the effector protein SptP, (Fig. 6D and SI Appendix, Fig. S12 and Dataset S1). In addition, two of the mutations measurably affected type III secretion function (i.e., protein injection) as assayed by measuring the ability of S. Typhimurium mutants to invade cultured epithelial cells (Fig. 6E). The reasons for the specific defect in effector protein injection of these mutants are unclear. These mutants are located at the boundaries of the gap between two SipD subunits. Therefore, it is possible that this region of SipD may serve as a platform to accommodate interactions with other translocases (i.e., SipB or SipC), which are expected to be crucial for effector protein translocation.
The tip complex is firmly anchored at the distal end of the needle filament by various interactions between the central coiled-coil domain of SipD and PrgI subunits in the first and second turns of the helical filament. We found that introduction of mutations in most of the residues involved in the formation of this large intermolecular interface resulted in the loss of the tip complex from the bacterial surface and a complete loss of function, characterized by hypersecretion of effectors and inability to invade cultured epithelial cells, although none of the mutations affected the stability of SipD (Fig. 6 G, H, and I and SI Appendix, Fig. S13 and Dataset S1). The strong phenotypes observed in most of the mutants is surprising, given the apparent redundancy in the interactions between SipD and PrgI. These observations indicate that even small structural changes in SipD due to the introduced mutations may lead to perhaps larger conformational changes that prevent the engagement and proper docking of SipD onto the needle filament. These results in turn suggest a cooperative process driving the assembly and docking of the tip complex onto the needle filament.
Discussion
We have obtained the cryo-EM structure of the tip complex of the S. Typhimurium T3SS encoded within its pathogenicity island 1. The structure is composed of five subunits of SipD arranged in a helical ring configuration with its central lumen seamlessly connected to the lumen of the needle filament, thus configuring a continuous open conduit. This observation has functional implications for the understanding of the mechanisms that establish a hierarchy in the type III secretion process. It is well established that type III protein secretion occurs in a hierarchical fashion in that the secretion machine can sequentially recognize a distinct set of secreted proteins (1, 20, 54). Thus, the first proteins recognized by the secretion machine (i.e., “early substrates”) are those involved in the building of the needle filament. This is then followed by the secretion of the tip protein and the protein translocases (“middle substrates”), which will form the tip complex at the end of the needle filament and the translocon embedded within the target eukaryotic cell membrane. Finally, the secretion machine engages the effector proteins that are destined to be delivered directly into the target eukaryotic cells. The mechanisms by which the secretion machine establishes the secretion hierarchy are poorly understood and likely multifactorial. However, it has been shown that the absence of the tip complex results in a breakdown in the secretion hierarchy leading to constitutive secretion of effector protein (26). Consequently, it has been proposed that the tip complex may serve as a plug that prevents secretion of effectors until bacterial contact with a target eukaryotic cell. However, our structure showing a continuously open channel spanning the needle filament and the tip complex is not consistent with this hypothesis. Rather, the structure is more consistent with an alternative model in which conformational changes in the tip complex that may occur upon bacterial contact with the target cell may serve as a signal that is transduced through the needle filament to the secretion machine to trigger the secretion of effectors.
Our structure also clarifies a hitherto unsettled issue related to the composition of the needle tip complex. Previous functional and structural modeling studies have proposed that the tip complex of a highly related T3SS in Shigella spp. is a heteromeric structure composed of four molecules of IpaD, a close homolog of SipD, and one molecule of IpaB, a close homolog of SPI-1 T3SS translocase SipB (36). In support of this model, previous studies have detected IpaB on the bacterial surface in a proportion of bacteria prior to contact with eukaryotic cells after examination by immunofluorescence or crude biochemical isolation (36, 55). However, our structure is incompatible with this model and indicates that, in its resting state, the tip complex is a homopentameric structure exclusively composed of SipD. This configuration is also consistent with our inability to detect SipB on the bacterial surface prior to bacterial contact with host cells (56). It is possible that the Shigella T3SS may be more prone to stochastic activation prior to contact with eukaryotic cells leading to the secretion and deposition of IpaB on the bacterial surface. A model has been proposed in which upon activation of the T3SS in Shigella, a molecule of SipB would be exchanged with a molecule of SipD on the pentameric structure as an intermediate step in the insertion of IpaB on the eukaryotic target membrane (36). However, our structure indicates that such an exchange is unlikely to occur as each SipD subunit is stabilized on the tip complex by multiple SipD–SipD as well as SipD–PrgI interactions resulting in over 3,000 Å2 of surface buried between these proteins. The stability conferred by these interactions makes the displacement model highly unlikely. Rather, we favor a model in which the interaction of the tip complex with the eukaryotic cell membrane generates a signal that leads to the secretion of SipB (and SipC) and its subsequent insertion on the plasma membrane. This process would not require an exchange with SipD on the tip complex. Instead, we propose that SipB would remain tethered to the pentameric complex by interactions with SipD. More specifically, the structure of the tip complex shows the presence of a gap between the SipD subunits #1 and #5. Although this gap could not accommodate an entire molecule of SipB, it could potentially accommodate a small domain of this translocase, thus allowing the firm docking of the tip complex to the inserted translocases. What the domain of SipB might be is unclear but the elongated coiled coil of SipB observed in the crystal structure of a portion of SipB may be a candidate to fulfill this role (57). It is known that once the translocases are deployed, bacteria make very intimate contact with the mammalian cell through its T3SS and that this intimate interaction is entirely dependent on the protein translocases and the tip complex (56). The structural organization of the tip complex shown here and the model we propose would allow the intimate interaction between the tip complex and the translocases. Furthermore, the T3SS-mediated Salmonella–host cell interface observed by cryoelectron tomography is entirely consistent with this model (58).
Our structure shows that each SipD subunit in the tip pentameric complex is firmly attached to the needle filament through various interactions that buried over 2,000 Å2. The way in which SipD engages the needle filament is reminiscent of the way in which the PrgI monomers interact with one another to form the needle filament. The insertion of the tip complex on the distal end of the filament results in significant conformational changes in PrgI. Within the main body of the needle filament, this protein adopts a hairpin conformation, with amino terminal and carboxy terminal helices bridged by a flexible loop. Of note, the amino terminal helix is interrupted by a pronounced kink that bents the helix at a 130° angle. This feature has garnered some attention and has been hypothesized to play an important role in signal transduction (1). However, no evidence has so far been presented in support of this hypothesis. The structure of the tip complex, however, presents an alternative explanation for the presence of the kink in the amino terminal helix of PrgI. The structure shows that insertion of the SipD subunits at the tip of the needle filament results in the straightening of the amino terminal helix of PrgI, which leads to a significant change in the angle between helix 1 and helix 2. These changes are essential for the insertion of SipD at the terminal end of the filament. We therefore propose that the kink in the amino terminal helix of PrgI is there to provide the structural flexibility necessary to accommodate the tip complex at the end of the needle filament.
The cryo-EM structure of the tip complex that caps the needle filament presented here will be instrumental in better understanding the signal transduction events that lead to T3SS activation as well as the deployment of the protein translocases on the target eukaryotic cell membrane. In addition, this information could help the development of antimicrobial strategies aimed at targeting this surface-exposed structure that is essential for the function of T3SSs.
Methods
Bacterial Strains and Plasmids.
All S. Typhimurium mutant strains were derived from the wild-type strain SL1344 (59), constructed by allelic exchange as previously described (60), and are listed in SI Appendix, Table S3. All plasmids were constructed by standard cloning methodologies using the low copy vector pWSK29 (61) as the backbone and are listed in SI Appendix, Table S4.
Needle Complex Isolation.
Needle complex isolation using affinity purification has been described elsewhere (62). Briefly, an S. Typhimurium strain expressing MBP-tagged PrgH and FLAG-epitope tagged SipD was grown in Luria-Bertani (LB) broth overnight and cells were recovered by centrifugation and resuspended in lysis buffer (200 mM Tris pH 7.5, 20% sucrose, 1 mM ethylenediaminetetraacetic acid (EDTA), 0.25 mg/mL of lysozyme, and complete EDTA-free protease inhibitor mixture [Sigma 4693159001]). After a 1-h incubation on ice, cells were lysed by the addition of 0.5% N-dodecyl-β-D-maltoside (DDM) (Anatrace D310S), and the lysates were clarified by centrifugation at 10,000 rpm for 1 h. Needle complexes were then recovered from the cell lysates by affinity purification on amylose resin as previously described (62).
Cryo-EM Sample Preparation, Data Acquisition, and Image Processing.
Sample preparation was carried out as previously described (47). Cryo-EM data were acquired on a Titan Krios electron microscope (Thermo Fisher) operating at 300 kV, at a calibrated magnification of 47,000 × g using a K2 Summit direct electron detector (Gatan, Inc.) in superresolution mode and a pixel size of 1.715 Å. Three datasets were collected. The first dataset was used previously for the structure calculation of needle filament (47). For the second and third datasets, a total of 3,847 movies were recorded with defocus values in the range of −1.0 to −3.5 μm and a dose rate of eight electrons per pixel2 per second. Each movie had 48 frames and each frame time was 0.375 s. Image processing was carried out also as previously described (47). Briefly, dose-fractionated image stacks from all three datasets were motion corrected using MotionCor2 (63) and the contrast transfer function (CTF) parameters for each micrograph were determined by Gctf (64). Particles were manually picked using sxhelixboxer.py in SPARX (65). A total of 72,768 particles were exported to Relion3 (66) and subjected to reference-free 2D classification. Among the particles categorized in well-defined classes, 27,737 had the tip complex and 23,121 did not. The 3D calculations were performed using the map of needle filaments (EMD-20046) as initial reference model for both structures. In the case of needle filaments with the tip complex, all the particles were subjected to 3D refinement and postprocessing yielding a final map with a global nominal resolution of 3.9 Å. For needle filaments without the tip structure, all the particles were subjected to 3D refinement and postprocessing and led to one map with a global nominal resolution of 5.0 Å. After 2D classification, all the particles were sorted into two groups, with or without the tip complex. The PrgI filament segment between the needle tip and the base was extracted for helical reconstructions using Relion3 (67). In total 44,958 and 34,305 segments in the groups of needles with and without the tip complex, respectively, were subjected to further 3D calculations. The map of needle filaments (EMD-20046) was used as an initial reference model. The known helical parameters of the needle filament were imposed for 3D calculations. The final resolutions of the structures of the needle with and without the tip complex were 3.7 Å and 3.8 Å, respectively. The statistics for data collection, map reconstruction, and model refinement are listed in SI Appendix, Table S5.
Model Building and Fitting.
For the tip protein complex, the initial template was derived from the SipD structure solved by X-ray crystallography (PDB ID: 2YM9) (42). For the PrgI needle, the initial template was derived from the PrgI structure solved by cryo-EM (PDB ID: 6OFH) (47). Models were docked into the EM maps using Chimera (43), adjusted manually in Coot (44), and further refined with Phenix (45). The parameters of the final model are described in SI Appendix, Table S5. Figures of the different structures were generated with PyMOL (68) and Chimera. Interface areas and interacting residues were determined with PISA (69).
Type III Protein Secretion Assays and Quantitative Immunoblotting.
To assess protein secretion through the T3SS, the different S. Typhimurium strains were grown in LB supplemented with 0.3 M NaCl to an optical density at 600 nm of 0.9. Culture supernatants were obtained by centrifugation, filtered through a 0.45-mm-pore-size filter, and proteins were precipitated with trichloroacetic acid, washed by acetone, dried, and resuspended in Laemmli buffer. Culture supernatant proteins were then analyzed by Western blotting with antibodies to detect early (InvJ), middle (SipD, SipB, and SipC), and late (SptP) substrates. The LI-COR Odyssey imaging system was utilized to detect the infrared-fluorescent DyLight-conjugated secondary antibodies (emission 800 nm [Thermo Fisher Scientific]). Quantitation of specific bands was performed with Odyssey v3.0 software (LI-COR). The abundance of each substrate was normalized relative to the wild-type control.
SipD Surface Display Assay.
Immunofluorescence assay to detect SipD at the tip of T3SSs in Salmonella was performed as previously described (47). Briefly, different Salmonella SipD mutant strains were grown to an optical density at 600 nm of 0.9, harvested by centrifugation, washed with PBS , and fixed in 4% paraformaldehyde for 20 min. The fixed bacteria were then washed and blocked with 250 μL of 3% bovine serum albumine (BSA) in 1× phosphate buffered saline (PBS) for 30 min, followed by overnight incubation with an antibody against the FLAG epitope to detect SipD (Sigma-Aldrich, dilution of 1:1,000) and an antibody against Salmonella lipopolysccharide (LPS) (SIFIN, catalog no.: TS 1624). The samples were washed and incubated with conjugated antibodies Alexa Fluor 594 anti-rabbit and Alexa Fluor 488 anti-mouse to detect LPS and SipD, respectively (Invitrogen) for 2 h at room temperature. Following one more wash, the bacteria were added (1:5 dilution) to a polylysine-coated glass coverslip submerged in 500 μL PBS in a 12-well culture plate overnight at 4 °C. The following morning, coverslips were mounted onto slides and examined by fluorescence microscopy on a Nikon TE2000 microscope. Images were acquired using an Andor Zyla 5.5 sCMOS camera run by the software Micromanager (https://www.micro-manager.org).
Cell Invasion Assay.
The ability of the different S. Typhimurium strains to invade cultured epithelial cells, a measure of the functionality of the SPI-1 T3SS, was carried out using the gentamicin protection assay as previously described (47) with some modifications. The different S. Typhimurium strains were grown in LB supplemented with 0.3 M NaCl to an optical density at 600 nm of 0.9. Bacteria were then applied to Henle-407 cells grown on 24-well plates with a multiplicity of infection (MOI) of 10 and incubated for 1 h. The cells were then washed with prewarmed PBS and incubated for 2 h with Dulbecco's Modified Eagel Medium (DMEM) supplemented with 10% fetal bovine serum and 50 μg/mL of gentamicin to kill any extracellular bacteria. After the incubation, the cells were washed again and lysed with 0.1% sodium deoxycholate (DOC) to release intracellular bacteria, followed by plating dilutions onto LB agar plates. The numbers of Salmonella recovered were counted and the percentage of the inoculum that survived the gentamicin treatement as a consequnece of cell invasion was calculated. Values for each mutant were normalized relative to those determined for wild type.
Acknowledgments
We thank Dr. Shenping Wu at the Yale West Campus Advanced Biosciences Center for assistance with the data collection. This work was supported by NIH Grant AI030492 (to J.E.G.).
Footnotes
Reviewers: M.B., Biozentrum, Basel; and W.D.P., The University of Kansas.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2114552118/-/DCSupplemental.
Data Availability
Protein structure coordinates and maps data have been deposited in the Protein Data Bank (https://www.rcsb.org/) (accession no. 7RYE), and the Electron Microscopy Data Bank (https://www.ebi.ac.uk/emdb/) (accession nos. EMD-24734, EMD-24735, EMD-24736, and EMD-24737). All other data are included in the article and/or in SI Appendix. Cryo-EM structure data have been deposited in PDB Protein Data Bank Deposition ID: D_1000258230; D_1000258231; D_1000258234; and D_1000258235.
References
- 1.Galán J. E., Lara-Tejero M., Marlovits T. C., Wagner S., Bacterial type III secretion systems: Specialized nanomachines for protein delivery into target cells. Annu. Rev. Microbiol. 68, 415–438 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deng W., et al., Assembly, structure, function and regulation of type III secretion systems. Nat. Rev. Microbiol. 15, 323–337 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Notti R. Q., Stebbins C. E., The structure and function of type III secretion systems. Microbiol. Spectr. 4, 1–48 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schreiber K. J., Chau-Ly I. J., Lewis J. D., What the wild things do: Mechanisms of plant host manipulation by bacterial type III-secreted effector proteins. Microorganisms 9, 1029 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galán J. E., Common themes in the design and function of bacterial effectors. Cell Host Microbe 5, 571–579 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pinaud L., Sansonetti P. J., Phalipon A., Host cell targeting by enteropathogenic bacteria T3SS effectors. Trends Microbiol. 26, 266–283 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Worrall L. J., et al., Near-atomic-resolution cryo-EM analysis of the Salmonella T3S injectisome basal body. Nature 540, 597–601 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Majewski D. D., et al., Cryo-EM structure of the homohexameric T3SS ATPase-central stalk complex reveals rotary ATPase-like asymmetry. Nat. Commun. 10, 626 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu J., et al., Cryo-EM analysis of the T3S injectisome reveals the structure of the needle and open secretin. Nat. Commun. 9, 3840 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marlovits T. C., et al., Structural insights into the assembly of the type III secretion needle complex. Science 306, 1040–1042 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marlovits T. C., et al., Assembly of the inner rod determines needle length in the type III secretion injectisome. Nature 441, 637–640 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Miletic S., et al., Substrate-engaged type III secretion system structures reveal gating mechanism for unfolded protein translocation. Nat. Commun. 12, 1546 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson S., Kuhlen L., Deme J. C., Abrusci P., Lea S. M., The structure of an injectisome export gate demonstrates conservation of architecture in the core export gate between flagellar and virulence type III secretion systems. MBio 10, e00818–e00819 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu B., Lara-Tejero M., Kong Q., Galán J. E., Liu J., In situ molecular architecture of the Salmonella type III secretion machine. Cell 168, 1065–1074.e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu B., et al., Visualization of the type III secretion sorting platform of Shigella flexneri. Proc. Natl. Acad. Sci. U.S.A. 112, 1047–1052 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schraidt O., Marlovits T. C., Three-dimensional model of Salmonella’s needle complex at subnanometer resolution. Science 331, 1192–1195 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Loquet A., et al., Atomic model of the type III secretion system needle. Nature 486, 276–279 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kubori T., et al., Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280, 602–605 (1998). [DOI] [PubMed] [Google Scholar]
- 19.Schraidt O., et al., Topology and organization of the Salmonella typhimurium type III secretion needle complex components. PLoS Pathog. 6, e1000824 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lara-Tejero M., Kato J., Wagner S., Liu X., Galán J. E., A sorting platform determines the order of protein secretion in bacterial type III systems. Science 331, 1188–1191 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhlen L., et al., Structure of the core of the type III secretion system export apparatus. Nat. Struct. Mol. Biol. 25, 583–590 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kubori T., Sukhan A., Aizawa S. I., Galán J. E., Molecular characterization and assembly of the needle complex of the Salmonella typhimurium type III protein secretion system. Proc. Natl. Acad. Sci. U.S.A. 97, 10225–10230 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mueller C. A., Broz P., Cornelis G. R., The type III secretion system tip complex and translocon. Mol. Microbiol. 68, 1085–1095 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Mueller C. A., et al., The V-antigen of Yersinia forms a distinct structure at the tip of injectisome needles. Science 310, 674–676 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Butan C., Lara-Tejero M., Li W., Liu J., Galán J. E., High-resolution view of the type III secretion export apparatus in situ reveals membrane remodeling and a secretion pathway. Proc. Natl. Acad. Sci. U.S.A. 116, 24786–24795 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ménard R., Sansonetti P., Parsot C., The secretion of the Shigella flexneri Ipa invasins is activated by epithelial cells and controlled by IpaB and IpaD. EMBO J. 13, 5293–5302 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zierler M. K., Galán J. E., Contact with cultured epithelial cells stimulates secretion of Salmonella typhimurium invasion protein InvJ. Infect. Immun. 63, 4024–4028 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deane J. E., et al., Molecular model of a type III secretion system needle: Implications for host-cell sensing. Proc. Natl. Acad. Sci. U.S.A. 103, 12529–12533 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blocker A. J., et al., What’s the point of the type III secretion system needle? Proc. Natl. Acad. Sci. U.S.A. 105, 6507–6513 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barta M. L., et al., Identification of the bile salt binding site on IpaD from Shigella flexneri and the influence of ligand binding on IpaD structure. Proteins 80, 935–945 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Veenendaal A. K., et al., The type III secretion system needle tip complex mediates host cell sensing and translocon insertion. Mol. Microbiol. 63, 1719–1730 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Montagner C., Arquint C., Cornelis G. R., Translocators YopB and YopD from Yersinia enterocolitica form a multimeric integral membrane complex in eukaryotic cell membranes. J. Bacteriol. 193, 6923–6928 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dey S., Chakravarty A., Guha Biswas P., De Guzman R. N., The type III secretion system needle, tip, and translocon. Protein Sci. 28, 1582–1593 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sato H., Frank D. W., Multi-functional characteristics of the Pseudomonas aeruginosa type III needle-tip protein, PcrV; comparison to orthologs in other gram-negative bacteria. Front. Microbiol. 2, 142 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rathinavelan T., et al., NMR model of PrgI-SipD interaction and its implications in the needle-tip assembly of the Salmonella type III secretion system. J. Mol. Biol. 426, 2958–2969 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheung M., et al., Three-dimensional electron microscopy reconstruction and cysteine-mediated crosslinking provide a model of the type III secretion system needle tip complex. Mol. Microbiol. 95, 31–50 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Epler C. R., Dickenson N. E., Bullitt E., Picking W. L., Ultrastructural analysis of IpaD at the tip of the nascent MxiH type III secretion apparatus of Shigella flexneri. J. Mol. Biol. 420, 29–39 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Picking W. D., Barta M. L., The tip complex: From host cell sensing to translocon formation. Curr. Top. Microbiol. Immunol. 427, 173–199 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Johnson S., et al., Self-chaperoning of the type III secretion system needle tip proteins IpaD and BipD. J. Biol. Chem. 282, 4035–4044 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chatterjee S., et al., The crystal structures of the Salmonella type III secretion system tip protein SipD in complex with deoxycholate and chenodeoxycholate. Protein Sci. 20, 75–86 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Erskine P. T., et al., High resolution structure of BipD: An invasion protein associated with the type III secretion system of Burkholderia pseudomallei. J. Mol. Biol. 363, 125–136 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Lunelli M., Hurwitz R., Lambers J., Kolbe M., Crystal structure of PrgI-SipD: Insight into a secretion competent state of the type three secretion system needle tip and its interaction with host ligands. PLoS Pathog. 7, e1002163 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pettersen E. F., et al., UCSF Chimera--A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Adams P. D., et al., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bernard A. R., Jessop T. C., Kumar P., Dickenson N. E., Deoxycholate-enhanced Shigella virulence is regulated by a rare π-helix in the type three secretion system tip protein IpaD. Biochemistry 56, 6503–6514 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo E. Z., et al., A polymorphic helix of a Salmonella needle protein relays signals defining distinct steps in type III secretion. PLoS Biol. 17, e3000351 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kenjale R., et al., The needle component of the type III secreton of Shigella regulates the activity of the secretion apparatus. J. Biol. Chem. 280, 42929–42937 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Torruellas J., Jackson M. W., Pennock J. W., Plano G. V., The Yersinia pestis type III secretion needle plays a role in the regulation of Yop secretion. Mol. Microbiol. 57, 1719–1733 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Davis A. J., Mecsas J., Mutations in the Yersinia pseudotuberculosis type III secretion system needle protein, YscF, that specifically abrogate effector translocation into host cells. J. Bacteriol. 189, 83–97 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pope L. M., Reed K. E., Payne S. M., Increased protein secretion and adherence to HeLa cells by Shigella spp. following growth in the presence of bile salts. Infect. Immun. 63, 3642–3648 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Olive A. J., et al., Bile salts stimulate recruitment of IpaB to the Shigella flexneri surface, where it colocalizes with IpaD at the tip of the type III secretion needle. Infect. Immun. 75, 2626–2629 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Y., Nordhues B. A., Zhong D., De Guzman R. N., NMR characterization of the interaction of the Salmonella type III secretion system protein SipD and bile salts. Biochemistry 49, 4220–4226 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Collazo C. M., Galán J. E., Requirement for exported proteins in secretion through the invasion-associated type III system of Salmonella typhimurium. Infect. Immun. 64, 3524–3531 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen D. K., Saurya S., Wagner C., Nishioka H., Blocker A. J., Domains of the Shigella flexneri type III secretion system IpaB protein involved in secretion regulation. Infect. Immun. 78, 4999–5010 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lara-Tejero M., Galan J. E., The Salmonella Typhimurium SPI-1 type III secretion translocases mediate intimate attachment to non-phagocytic cells. Infect. Immun. 77, 2635–2642 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barta M. L., et al., The structures of coiled-coil domains from type III secretion system translocators reveal homology to pore-forming toxins. J. Mol. Biol. 417, 395–405 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Park D., et al., Visualization of the type III secretion mediated Salmonella-host cell interface using cryo-electron tomography. Elife 7, e39514 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hoiseth S. K., Stocker B. A., Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291, 238–239 (1981). [DOI] [PubMed] [Google Scholar]
- 60.Kaniga K., Bossio J. C., Galán J. E., The Salmonella typhimurium invasion genes invF and invG encode homologues of the AraC and PulD family of proteins. Mol. Microbiol. 13, 555–568 (1994). [DOI] [PubMed] [Google Scholar]
- 61.Wang R. F., Kushner S. R., Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100, 195–199 (1991). [PubMed] [Google Scholar]
- 62.Lara-Tejero M., et al., Role of SpaO in the assembly of the sorting platform of a Salmonella type III secretion system. PLoS Pathog. 15, e1007565 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zheng S. Q., et al., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang K., Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hohn M., et al., SPARX, a new environment for Cryo-EM image processing. J. Struct. Biol. 157, 47–55 (2007). [DOI] [PubMed] [Google Scholar]
- 66.Zivanov J., et al., New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.He S., Scheres S. H. W., Helical reconstruction in RELION. J. Struct. Biol. 198, 163–176 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.DeLano W. L., The PyMOL molecular graphics system. www.pymol.org (2002).
- 69.Krissinel E., Henrick K., Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Protein structure coordinates and maps data have been deposited in the Protein Data Bank (https://www.rcsb.org/) (accession no. 7RYE), and the Electron Microscopy Data Bank (https://www.ebi.ac.uk/emdb/) (accession nos. EMD-24734, EMD-24735, EMD-24736, and EMD-24737). All other data are included in the article and/or in SI Appendix. Cryo-EM structure data have been deposited in PDB Protein Data Bank Deposition ID: D_1000258230; D_1000258231; D_1000258234; and D_1000258235.