Significance
To address a crucial knowledge deficiency concerning the correlation between nanoparticles (NPs) exposure and cardiovascular diseases, here we present a toxicological mechanism for inhaled NPs by delineating their interactions with the biomacromolecules encountered en route. In biological ambience, NPs are known to spontaneously adsorb proteins, thereby acquiring a new biological identity and further entailing pathological prospects. Here, we found that silica nanoparticles (SiNPs) specifically adsorbed apolipoprotein A-I (Apo A-I) in the blood to ameliorate their cytotoxicity, while a rapid clearance of SiNPs from the bloodstream depleted plasma Apo A-I and facilitated SiNPs-induced atherosclerosis. This study is a demonstration of the relationship between plasma protein adsorption and cardiovascular damage induced by engineered NPs.
Keywords: silica nanoparticle, corona, apolipoprotein A-I, cardiovascular damage, atherosclerosis
Abstract
The rapid development of nanotechnology has greatly benefited modern science and engineering and also led to an increased environmental exposure to nanoparticles (NPs). While recent research has established a correlation between the exposure of NPs and cardiovascular diseases, the intrinsic mechanisms of such a connection remain unclear. Inhaled NPs can penetrate the air–blood barrier from the lung to systemic circulation, thereby intruding the cardiovascular system and generating cardiotoxic effects. In this study, on-site cardiovascular damage was observed in mice upon respiratory exposure of silica nanoparticles (SiNPs), and the corresponding mechanism was investigated by focusing on the interaction of SiNPs and their encountered biomacromolecules en route. SiNPs were found to collect a significant amount of apolipoprotein A-I (Apo A-I) from the blood, in particular when the SiNPs were preadsorbed with pulmonary surfactants. While the adsorbed Apo A-I ameliorated the cytotoxic and proinflammatory effects of SiNPs, the protein was eliminated from the blood upon clearance of the NPs. However, supplementation of Apo A-I mimic peptide mitigated the atherosclerotic lesion induced by SiNPs. In addition, we found a further declined plasma Apo A-I level in clinical silicosis patients than coronary heart disease patients, suggesting clearance of SiNPs sequestered Apo A-I to compromise the coronal protein’s regular biological functions. Together, this study has provided evidence that the protein corona of SiNPs acquired in the blood depletes Apo A-I, a biomarker for prediction of cardiovascular diseases, which gives rise to unexpected toxic effects of the nanoparticles.
Silica nanoparticles (SiNPs) have been broadly used in construction, electronics, food packaging, cosmetics, and even the biomedicine field and contribute to extensive exposure pathways of SiNPs in human life (1). There are ∼1.1 billion workers potentially exposed to silica in agricultural or industrial activities (2), and long-term occupational exposure of silica is responsible for various diseases [i.e, silicosis, tuberculosis, airway obstructive pulmonary disease, lung cancer, rheumatoid arthritis, and scleroderma (3–6)]. Occupational diseases not only have a significant impact on individual life expectancy but also lead to serious labor loss to the society (7).
Recent advances have suggested that small, inhaled particles can penetrate the air–blood barrier (ABB) from the lungs to systemic circulation (8–10), and the invasion of foreign particles may have adverse effects on the cardiovascular system (11). Substantial epidemiological studies have revealed the relationship between air pollution and cardiovascular diseases (12–14); however, the interplay of cardiovascular damage and silica exposure is not well understood.
Due to high surface free energy, micro- or nano-size particles tend to adsorb encountered biomacromolecules on their surfaces to form a corona (15, 16). Theoretically, particles primitively coated with pulmonary surfactants (PS) are composed of phospholipids and surfactant proteins in alveoli. During the ABB-crossing process, the composition of the protein corona may undergo drastic exchanges, both quantitatively and qualitatively (17). Thus, inhaled particles may encounter plasma proteins and further acquire a secondary corona, which may potentially remodel the surface properties of particles and influence their subsequent biokinetics and, ultimately, toxicity (18).
Here, we present a toxicological mechanism of SiNPs in cardiovascular diseases and discuss the “Jekyll and Hyde” effect of their protein corona. First, the proinflammatory effect of SiNPs was markedly relieved by corona decoration due to the dysopsonin effect. Intriguingly, the adsorption of apolipoprotein A-I (Apo A-I) on SiNPs drastically depleted Apo A-I in blood, which facilitated SiNPs-induced atherosclerosis. Accordingly, supplementation of an Apo A-I mimic peptide mitigated the atherosclerotic lesion induced by SiNPs. Our results demonstrated the feasibility of in vivo manipulation of the protein corona for mitigating SiNPs-induced cardiovascular damage.
Results
SiNPs Induce Direct Cardiovascular Damage after Respiratory Exposure.
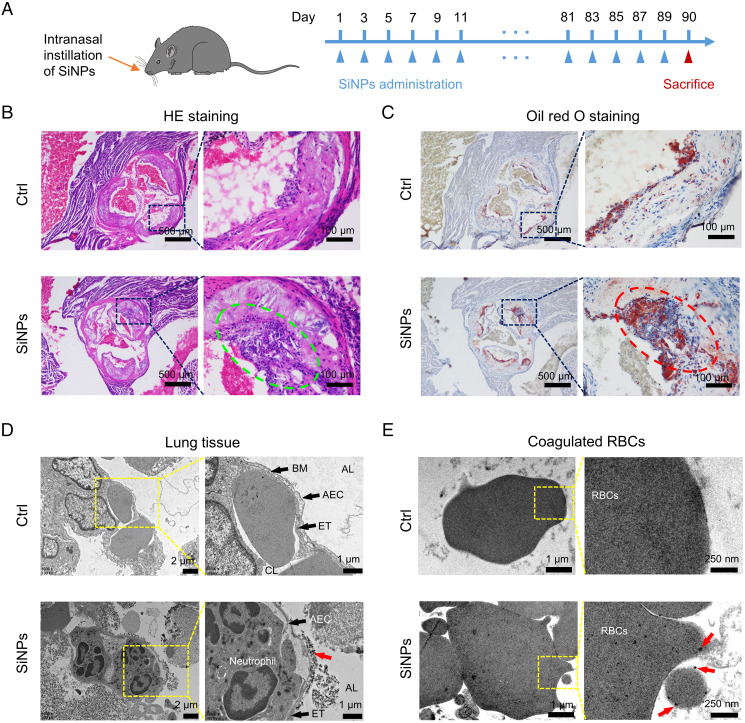
To simulate silica exposure, intranasal instillation of SiNPs (primary size of ∼15 nm; detailed characterizations are shown in SI Appendix, Fig. S1 and Table S1, 10 mg/kg every 2 d) was performed on apolipoprotein E knockout (Apo E−/−) mice with a high-fat diet (Fig. 1A) (19). The dose of SiNPs applied in mice exposure was chosen to ensure SiNPs can surmount the clearance mechanisms of pulmonary system and bring about considerable lung stimulation, as described in previous investigations about silica-induced diseases by intratracheal instillation of 5 to 10 mg silica (20, 21). After 3 mo of SiNPs exposure, significant histopathological lesions were found in mice blood vessels as indicated by hematoxylin-eosin (HE) staining (Fig. 1B) and oil red O staining (Fig. 1C). Aggravated inflammatory cells infiltration along with plaque lesions on blood vessel walls and severe plaque lipidation and lipid storage of aortic wall were observed, indicating cardiovascular injury triggered by SiNPs. Increased inflammatory cell infiltration and plaque lesions (green circle) can be observed in HE staining of mice aortic roots upon SiNPs exposure, and obvious plaque lipidation (red circle) was found in oil red O staining images. Hence, a linkage between SiNPs respiratory exposure and cardiovascular diseases was confirmed. Apart from the lungs, obvious lesions were also observed in the livers and kidneys in SiNPs-exposed mice, while no evident injury was found in the spleen and heart of the animals (SI Appendix, Fig. S2).
Fig. 1.
SiNPs exposure induces mice aortic root injury. (A) Schematic diagram of the SiNPs exposure model. Male C57BL/6 Apo E−/− mice with high-fat diet were randomly divided into two groups. Mice received intranasal instillation of SiNPs (10 mg/kg, every other day), and the control group received equivalent volume of physiological saline. After 3 mo, the mice were euthanized, and their aortic roots were collected for (B) HE and (C) oil red O staining (n = 1). Translocation of SiNPs from the lungs to bloodstream was observed. Male C57BL/6 mice received intranasal instillation of SiNPs (10 mg/kg) or an equivalent volume of saline once. After 24 h SiNPs instillation, the mice were euthanized for the following analysis. (D) Lung tissues were collected for TEM. SiNPs (indicated by red arrow) were observed in alveolar lumen, endosome-like vesicles of alveolar epithelial cells (AECs), and the surface of the AECs as well as endothelial cells at the sites of alveolar walls. BM: basement membrane, ET: endothelial cell. CL: capillary lumen. AL: alveolar lumen. (E) Whole blood was collected and stood for coagulation, and the samples were diced for TEM analysis. SiNPs (black dots pointed by red arrows) were associated with red blood cells.
The distribution of SiNPs in the gut was evaluated since intranasally instilled SiNPs might enter digestive organs apart from the pulmonary system, while no significant difference was found in both intestine and stomach after 15 d of exposure, indicating the extremely low distribution of SiNPs in the gut (SI Appendix, Fig. S3).
To investigate the penetration of SiNPs via the ABB, lung tissues were dissected for transmission electron microscopy (TEM) imaging 24 h after SiNPs exposure. SiNPs are electron-dense spherical particles and can be easily distinguished from cellular structures. SiNPs appearing as individual particles or agglomerations were dispersed in alveolar lumen, endosome-like vesicles of alveolar epithelial cells, and endothelial cells at the site of alveolar walls (Fig. 1D). In addition, blood samples from SiNPs-treated mice were extracted and stood for coagulation. TEM images revealed that substantial black dots were associated with red blood cells, indicating the presence of SiNPs in blood circulation (Fig. 1E).
PS Facilitates Apo A-I Adsorption on SiNPs.
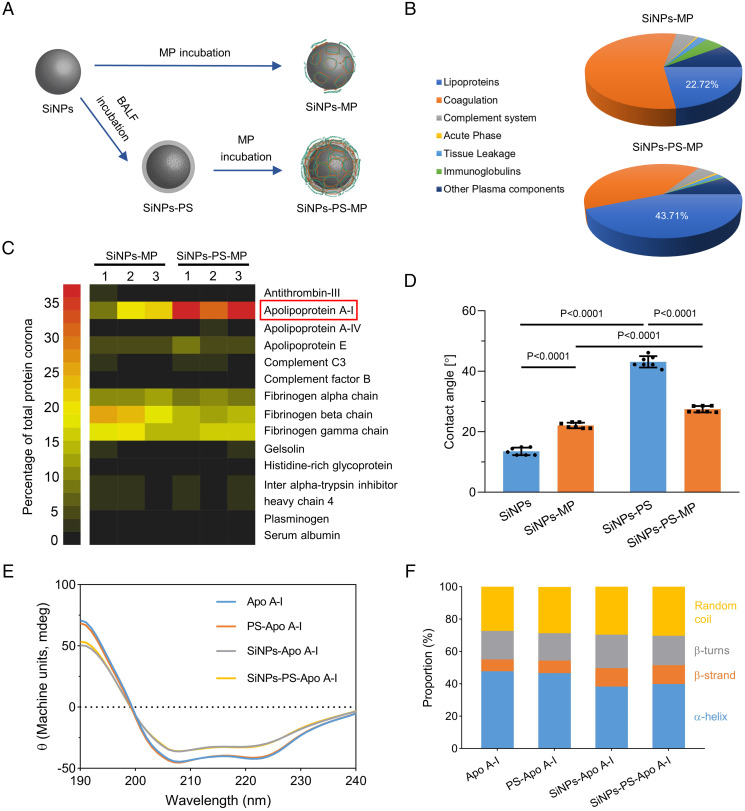
Inhaled SiNPs first reached the pulmonary system and interacted with alveolar fluids, among which PS is the principal reactive component that functions to lower surface tension during respiration (22). It is rational to speculate that the presence of PS alters the protein-binding pattern of NPs in blood (15). Interestingly, silica inhalation is associated with increased phospholipid and surfactant protein productions in the lungs, which may amplify the influence of PS on the secondary corona (23). The plasma protein adsorption behavior of SiNPs in the presence/absence of PS coating was investigated through an ex vivo assay. PS was obtained by bronchoalveolar lavage performed on healthy mice and collected in the bronchoalveolar lavage fluid (BALF). An incubation of SiNPs with BALF was performed to generate PS-covered SiNPs (SiNPs-PS). Then, SiNPs-PS was incubated with 10% mice plasma (MP) to further render a SiNPs-corona complex, termed as SiNPs-PS-MP. As a parallel control, SiNPs-MP represented a complex obtained by directly incubating SiNPs with 10% MP (Fig. 2A). The total amount of proteins adsorbed on the SiNPs was determined. As shown in SI Appendix, Fig. S4A, small amounts of proteins existed for the SiNPs-PS group (due to the presence of proteins in BALF), while SiNPs-PS-MP possessed comparable but slightly higher amounts of proteins than SiNPs-MP since BALF contains protein components. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) analysis showed consistent results, SI Appendix, Fig. S4B. Interestingly, compared to the MP group, the proteins adsorbed on SiNPs-MP and SiNPs-PS-MP showed different patterns. Notably, proteins with molecule weight ∼28 kDa were adsorbed on SiNPs-MP and further enriched on SiNPs-PS-MP.
Fig. 2.
PS facilitates the adsorption of Apo A-I on SiNPs through increasing the surface hydrophobicity of SiNPs. (A) Schematic diagram of SiNPs-PS, SiNPs-MP, and SiNPs-PS-MP preparations. (B) Classification and quantification of the protein corona components of SiNPs-MP and SiNPs-PS-MP using LC-ESI-MS/MS, with the percentages specifically indicated for Apo A-I. The classification of proteins was based on their functions in biological processes, where the proportion of lipoproteins was drastically increased when PS was preincubated with SiNPs. Data were calculated based on three individual samples. (C) Heat map of the most abundant proteins in the protein coronas of SiNPs-MP and SiNPs-PS-MP measured by LC-ESI-MS/MS. Apo A-I was significantly enriched in the protein corona of SiNPs-PS-MP compared to SiNPs-MP. Proteins that constituted more than 1% in the protein corona in at least one sample were shown. (D) Contact angle test for the determination of the hydrophobicity of SiNPs, SiNPs-MP, SiNPs-PS, and SiNPs-PS-MP. Data were represented as the mean ± SD (n = 7). (E) CD spectroscopy of Apo A-I, PS-Apo A-I, SiNPs-Apo A-I, and SiNPs-PS-Apo A-I. PS-Apo A-I was obtained by mixing BALF with Apo A-I, SiNPs-Apo A-I was obtained by adding Apo A-I into SiNPs suspension, while SiNPs-PS-Apo A-I was prepared by adding Apo A-I into SiNPs-PS suspension. (F) Proportion of Apo A-I secondary structure (α-helices, β-strands, β-turns, and random coils) obtained from CD spectroscopy result.
Label-free quantitative liquid chromatography–electron spray ionization-tandem mass spectrometry (LC-ESI-MS/MS) was employed to identify and quantify the protein corona components. As shown in Fig. 2B, coagulation proteins were found to be the most abundant in the protein corona of SiNPs-MP, while lipoproteins were markedly enriched from 22.7 to 42.7% upon the preincubation with PS. Among the top abundant adsorbed proteins, the proportion of Apo A-I drastically increased from 16.7% for SiNPs-MP to 35.1% for SiNPs-PS-MP (Fig. 2C). In addition, SI Appendix, Fig. S5 showed parallel results obtained from Apo A-I blotting. Apolipoproteins are the primary protein components of lipoproteins, which mediate the transfer of lipids and balance the contents of lipids and lipoproteins (24). As the major structural and functional protein in high-density lipoprotein (HDL), Apo A-I has been shown to protect against atherosclerosis (25). Apo A-I has also been reported to adsorb on various NPs with a preference for hydrophobic surfaces (18, 26–28). Therefore, it is of interest to explore a connection between PS and Apo A-I adsorption.
Lipids account for over 90% of the total components of PS, and the formation of a lipid corona has been demonstrated for different types of NPs (22, 29). Dipalmitoyl phosphatidylcholine (DPPC) is the predominant lipid component of PS, which consists of two palmitic acids attached to a phosphatidylcholine head-group (30). The presence of DPPC on SiNPs-PS or SiNPs-PS-MP was confirmed using thin-layer chromatography (TLC). The result indicated that DPPC was indeed adsorbed on the surface of SiNPs after incubation with BALF and remained on SiNPs after MP incubation (SI Appendix, Fig. S6). The amphiphilic DPPC has a propensity to attach its hydrophilic head on the surface of SiNPs to render a lipid corona, leaving its hydrophobic tail outside to increase the hydrophobicity of SiNPs (31). As expected, the contact angle of SiNPs suspension was apparently increased after incubating with PS (Fig. 2D and SI Appendix, Fig. S7), which explained the preferential binding of Apo A-I (32, 33). Structural changes in coronal proteins may occur upon adsorption, potentially altering the function of the proteins (34). On the other hand, the protein corona frequently alters the biological fate of NPs, such as their biodistribution and elimination. Circular dichroism (CD) analysis indicated the secondary structure changes of Apo A-I after its binding with SiNPs (Fig. 2 E and F). Upon surface adsorption, a decreased proportion of α-helices alongside increased proportions of β-strands and random coils were recorded in Apo A-I, indicating the secondary structure changes of Apo A-I resulting from the protein corona (35). PS itself played a nonmajor role in the secondary structure of Apo A-I, as SiNPs-PS entailed an identical CD spectrum compared with bare SiNPs.
Apo A-I Decoration Decreases Cellular Internalization and Relieves Cytotoxicity and Proinflammatory Effect of SiNPs.
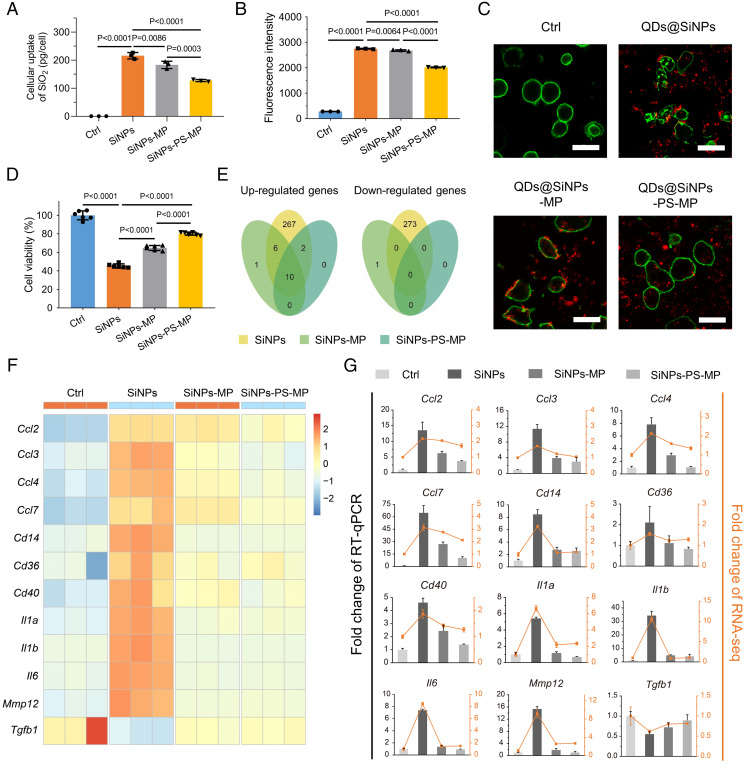
The cellular uptake of NPs is significantly influenced by the presence of coronal proteins, thus impacting the cytotoxicity of pristine NPs (36). Interestingly, the effects of the protein corona on NP internalization are divergent in literature (29, 37, 38). Since apolipoproteins are referred to as dysopsonins that decrease macrophage recognition (39), we speculated the enrichment of coronal Apo A-I could decrease the cellular engulfment of NPs. According to inductively coupled plasma-optical emission spectroscopy (ICP-OES), the cellular uptake of SiNPs by J774A.1 macrophages was decreased in both SiNPs-MP and SiNPs-PS-MP groups. Notably, the most significant difference was also found between the SiNPs-MP and SiNPs-PS-MP groups (P = 0.0003), confirming the role of Apo A-I in blocking SiNPs internalization, Fig. 3A. For a comprehensive evaluation of the cellular uptake of SiNPs, fluorescent SiNPs were manufactured by coating CdSe/ZnS quantum dots (QDs) with a SiO2 shell [characterized in SI Appendix, Fig. S8 and Table S2 (40)]. The flow cytometry result indicated the average fluorescence intensity of macrophages, and the confocal images reflected the engulfment condition of QDs@SiNPs-exposed macrophages, respectively. As shown in Fig. 3 B and C, the average fluorescence intensity and the internalization behavior in macrophages were both significantly declined in the QDs@SiNPs-MP and QDs@SiNPs-PS-MP groups compared to the QDs@SiNPs group. Besides, morphological shrinkage was observed on J774A.1 macrophages exposed to pristine SiNPs. These features were absent in both SiNPs-MP- and SiNPs-PS-MP-treated cells, indicating the protective role of their protein coronas (Fig. 3C). We further investigated the cytotoxicity of SiNPs with different surface decorations. In line with the uptake assay, SiNPs significantly decreased the J774A.1 cell viability, which were partially recovered in the SiNPs-MP and SiNPs-PS-MP groups. Precoating of PS on SiNPs (SiNPs-PS-MP) further elevated cell viability compared with SiNPs-MP (Fig. 3D).
Fig. 3.
Apo A-I decoration decreases cellular internalization and relieves cytotoxicity and proinflammatory effect of SiNPs. (A) J774A.1 macrophages were treated with SiNPs, SiNPs-MP, and SiNPs-PS-MP (50 μg/mL, 6 h). Cellular uptake was determined via the detection of Si element by ICP-OES. Data are represented as the mean ± SD (n = 3). (B) In parallel, QDs@SiNPs were applied for uptake detection via fluorescent signaling. Flow cytometry was employed to determine the average fluorescence intensity of J774A.1 macrophages. Data were represented as the mean ± SD (n = 3). (C) Representative confocal images of cellular uptake of QDs@SiNPs, QDs@SiNPs-MP, and QDs@SiNPs-PS-MP were shown. Green, DiO for cell membranes; red, QDs@SiNPs; n = 3. (Scale bar, 25 μm.) J774A.1 macrophages were treated with SiNPs, SiNPs-MP, and SiNPs-PS-MP (50 μg/mL, 6 h). Morphological changes were evident to cells exposed to QDs@SiNPs. (D) Cell viability. Data were represented as the mean ± SD (n = 6). (E) RNA-seq. Venn diagrams of up-regulated genes and down-regulated genes. Gene expression data from RNA-seq was filtered by |log2FC| > 1 and FDR < 0.05 between two groups and recorded as up-/down-regulated genes. FC: fold change. Data were represented as the mean ± SD (n = 3). (F) Heat map of selected genes expression levels based on RNA-seq data. (G) RT-qPCR. Columns represent the fold change of RT-qPCR results and lines represent the fold change of RNA-seq data. Data were represented as the mean ± SD (n = 3).
Subsequently, an RNA sequencing (RNA-seq) workflow was performed to investigate the specific biological effects of SiNPs, SiNPs-MP, and SiNPs-PS-MP on J774A.1 macrophages. Analysis of different expressed genes revealed that the naked SiNPs group expressed very different sets of genes, but SiNPs-MP and SiNPs-PS-MP induced insignificant modifications (Fig. 3E and SI Appendix, Fig. S9). Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis discovered that pristine SiNPs exposure stimulated the immune system, including stimulus response and inflammatory signaling pathways, while SiNPs-MP and SiNPs-PS-MP exposures rendered ameliorative effects (Fig. 3F and SI Appendix, Figs. S10–S12). We next validated these genes via a RT-qPCR assay (Fig. 3G). During the initiation of atherosclerotic lesion, both pattern recognition receptors like CD14 and proinflammatory mediators including interleukin-1α (IL-1α), IL-1β, and IL-6 made essential contributions, and chemokines such as chemokine (C-C motif) ligand (CCL) family (CCL2, CCL3, CCL4, and CCL7) attracted leukocytes traveled to the inflammation sites (41). Once the recruitment of macrophages within the lesion occurred, CD36 played a crucial role in triggering foam cell formation prior to the emergence of a plaque (42). Afterward, CD40 participated in plaque fibrosis while disruption of elastic lamina could be induced by matrix metallopeptidase 12 (MMP12) (43). The gene expressions of aforementioned atherogenic factors were significantly up-regulated, whereas the up-regulation of these genes were inhibited in the SiNPs-MP and SiNPs-PS-MP groups (Fig. 3 F and G). Instead, anti-inflammatory and atheroprotective cytokines such as transforming growth factor-β were down-regulated in SiNPs-treated group and were restored in the SiNPs-MP and SiNPs-PS-MP groups (44). In conclusion, SiNPs exposure induced intense inflammation and promoted atherogenesis to macrophages, whereas these effects were markedly relieved in the presence of the protein corona on SiNPs.
SiNPs Exposure Induces Apo A-I Depletion in Blood.
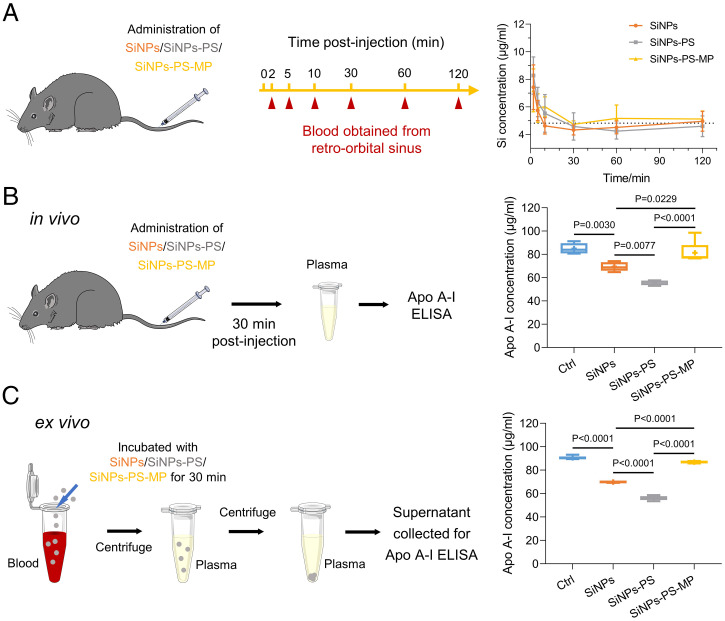
Since reduced cellular internalization was observed for coronal Apo A-I, we further examined whether coronal Apo A-I would affect the elimination of SiNPs from blood circulation in a mouse model. As shown in Fig. 4A, SiNPs, SiNPs-PS, and SiNPs-PS-MP were mostly eliminated from bloodstream in 0.5 h. Due to the enrichment of coronal Apo A-I and the rapid elimination of SiNPs from systemic circulation, a correlation between the intrusion of SiNPs and Apo A-I level in bloodstream ought to be investigated. To this end, the concentrations of Apo A-I in mouse blood were determined 30 min postintravenous injection of SiNPs, SiNPs-PS, and SiNPs-PS-MP. As shown in Fig. 4B, the Apo A-I level was remarkably declined after SiNPs injection, compared to the control group (P = 0.0030). Interestingly, administration of equivalent quantities of SiNPs-PS resulted in a further depletion of Apo A-I in blood compared to naked SiNPs (P = 0.0077), which may be due to the enrichment of Apo A-I by primary PS corona. However, compared with SiNPs and SiNPs-PS, SiNPs-PS-MP significantly prevented Apo A-I depletion, suggesting saturation of Apo A-I in their protein corona by preincubation with the mouse plasma.
Fig. 4.
SiNPs exposure induces Apo A-I depletion in blood. (A) SiNPs, SiNPs-PS, and SiNPs-PS-MP clearance from the circulation. Male C57BL/6 mice were intravenously injected with SiNPs, SiNPs-PS, and SiNPs-PS-MP (250 μg), respectively. The mice blood was collected from retro-orbital sinus at different time points for Si element analysis by ICP-OES. Si concentration of blank mice blood was presented as dashed line. Data were represented as the mean ± SD (n = 5). (B) SiNPs and SiNPs-PS exposure induced in vivo Apo A-I depletion. Male C57BL/6 mice received intravenous injection of saline, SiNPs, SiNPs-PS (500 μg, calculated by the mass of SiNPs), and SiNPs-PS-MP, respectively. The mice were euthanized 30 min postinjection, and the concentration of Apo A-I in MP was determined. Data were represented as the mean ± SD (n = 5). (C) SiNPs-induced ex vivo Apo A-I depletion. The whole blood of mice was collected and pooled. SiNPs, SiNPs-PS, and SiNPs-PS-MP (500 μg SiNPs/mL blood) were added. The mixture was incubated at 37 °C for 30 min. Blood was centrifuged at 800 g for 10 min to obtain plasma, then centrifuged at 30,000 g for 15 min to separate SiNPs. The supernatant was collected for Apo A-I determination (n = 5).
To rule out other factors that might affect the depletion of Apo A-I in vivo, we further verified the effect of SiNPs on the depletion of Apo A-I via a parallel ex vivo experiment. The whole blood of mice was collected and incubated with SiNPs and SiNPs-PS for 30 min. The mixtures were centrifugated to remove NPs and debris, and then corresponding supernatants were collected for Apo A-I measurement. Consistent with the in vivo results, the ex vivo data further demonstrated that both SiNPs and SiNPs-PS apparently eliminated Apo A-I, especially for SiNPs-PS (Fig. 4C and SI Appendix, Fig. S13). To better understand the Apo A-I depletion capabilities of SiNPs and SiNPs-PS in vivo and ex vivo, the depletion quantification was processed based on the calculation of depleted Apo A-I concentration. The Apo A-I depletion capabilities of SiNPs and SiNPs-PS were found to be compatible in the in vivo and ex vivo studies, which increased the feasibility of our experimental design (SI Appendix, Fig. S14). Together, these findings illustrated that SiNPs were eliminated from bloodstream rapidly along with the depletion of coronal Apo A-I, and SiNPs-PS possessed an enhanced capacity in Apo A-I depletion. Similar results were obtained on the Apo A-I depletion effect of copper oxide (CuO) NPs, signifying the potential universality of coronal protein depletion (SI Appendix, Fig. S15).
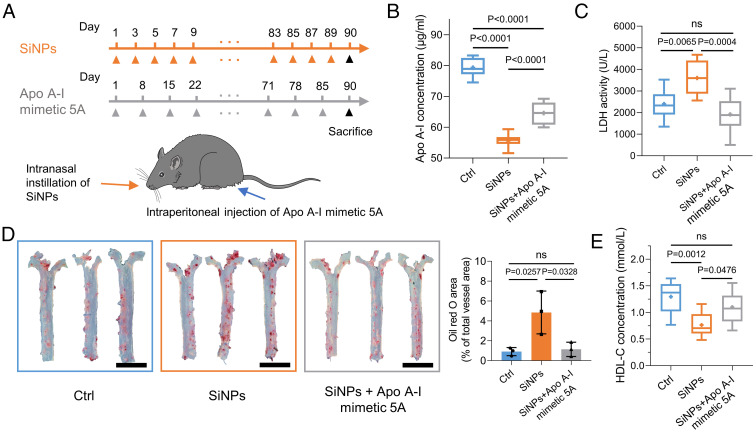
Apo A-I Mimetic Supplementation Alleviates SiNPs-Induced Apo A-I Depletion and Atherosclerosis.
Apo A-I is beneficial for HDL formation, cholesterol efflux elevation, and oxidized lipid elimination (45). With these properties, Apo A-I has become a potential pharmacologic interference against atherosclerosis (46, 47). Interestingly, several reports have indicated the protective role of a synthetic Apo A-I mimetic peptide (45, 48, 49). Compared to Apo A-I, Apo A-I mimetic peptides have several advantages [i.e., better accessibility and higher efficiency of cholesterol offloading (50)]. Therefore, the therapeutic effects of Apo A-I mimetic peptide 5A in preventing the SiNPs-induced atherogenicity were investigated (Fig. 5A). Indeed, Apo A-I concentration in mice with long-term exposure (3 mo) of SiNPs was significantly decreased. However, Apo A-I mimetic 5A supplementation restored Apo A-I level to some extent, which may be related to the anti-inflammatory effect of Apo A-I mimetic 5A that lowering the lactate dehydrogenase (LDH) activity in mouse blood (Fig. 5 B and C). Compared with mice that only received SiNPs, the SiNPs + Apo A-I mimetic group exhibited a lower lipid storage in aorta, indicating that Apo A-I mimetic 5A endorsed the capability of HDL to reduce lipid storage on the aortic wall in SiNPs-induced atherosclerosis (Fig. 5D). Besides, fibrosis of aortic wall was mitigated in Apo A-I mimetic 5A–administrated mice (SI Appendix, Fig. S16). Apo A-I is the predominant composition of HDL cholesterol (HDL-C). We further found SiNPs exposure decreased HDL-C concentration, and Apo A-I mimetic supplementation reversed this effect accordingly (Fig. 5E). Together, these data suggest the supplementation of Apo A-I mimetic peptide counteracts SiNPs-caused Apo A-I depletion and corresponding cardiovascular injury (51, 52).
Fig. 5.
Apo A-I mimetic supplementation relieves SiNPs-induced Apo A-I depletion and atherosclerosis. (A) Schematic diagram of the experiment design. Male C57BL/6 Apo E−/− mice with high-fat diet were randomly divided into three groups. Control group received equivalent volume of physiological saline compared with SiNPs group. SiNPs group received intranasal instillation of SiNPs (10 mg/kg, every other day). SiNPs + Apo A-I mimetic 5A group received intranasal instillation of SiNPs (10 mg/kg, every other day) plus intraperitoneal injection of Apo A-I mimetic 5A (1 mg/kg, every week). Apo A-I mimetic 5A peptide (DWLKAFYDKVAEKLKEAFPDWAKAAYDKAAEKAKEAA) was synthesized by GL Biochem Ltd., and its purity of >96.3% was confirmed by HPLC. Mice were euthanized after 3 mo. (B) Chronic SiNPs exposure decreased Apo A-I in MP. (C) LDH activity in MP. (D) Mice aortas were collected for oil red O staining, and the percentages of oil red O area to total vessel area were quantified. (Scale bar, 5 mm.) (E) MP was used to determine the concentrations of HDL-C. Data were represented as the mean ± SD (n = 10 in control group, n = 8 in SiNPs group, and n = 9 in SiNPs + Apo A-I mimetic 5A group).
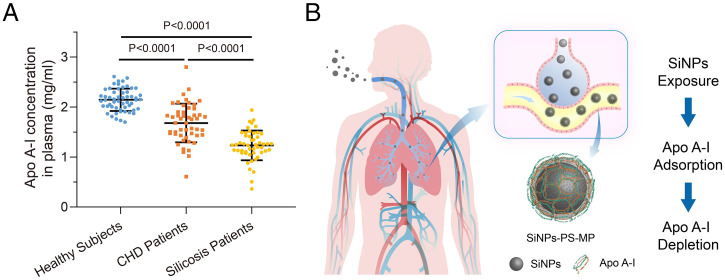
Further Depletion of Apo A-I Was Found in Silicosis Patients over Coronary Heart Disease Patients.
Silicosis patients have been exposed to high concentration of free silica for decades, it is possible that inhaled smaller silica transported via the lungs to blood circulation, we thus hypothesized that the elimination of silica in the blood circulation is accompanied by the depletion of Apo A-I. Coronary heart disease (CHD) patients are usually associated with atherosclerosis and cardiovascular damage (53). Epidemiological studies have suggested an inverse correlation of HDL-C and risk of CHD (54). In addition to the mouse model described in the section SiNPs Exposure Induces Apo A-I Depletion in Blood, the plasma Apo A-I levels from healthy subjects, clinical CHD, and silicosis patients were compared. As shown in Fig. 6A, CHD patients exhibited significantly lower Apo A-I levels relative to healthy subjects (P < 0.0001), signifying the contribution of decreased Apo A-I levels to CHD risk. Next, we evaluated the Apo A-I levels in silicosis patients and found declined Apo A-I levels relative to controls (P < 0.0001). Interestingly, a further depletion of Apo A-I levels in silicosis patients compared to CHD patients was found (P < 0.0001). Therefore, as the major protein component of HDL, Apo A-I appears to be a rational predictor of clinical CHD patients, which has been confirmed by previous prospective cohort studies (55–57). Although this current study did not rule out the diagnosis on artery plaque volume and blood lipids that may affect Apo A-I level, the combination of animal and clinical investigation corroborated that the adsorption of Apo A-I by SiNPs contributed to the loss of plasma Apo A-I, which offered evidence in delineating the potential cardiovascular injury induced by SiNPs exposure (Fig. 6B).
Fig. 6.
Depletion of Apo A-I in CHD and silicosis patients. (A) Human plasma samples from healthy people (n = 55), CHD patients (n = 50), and silicosis patients (n = 50) were collected, and the concentration of Apo A-I in plasma was determined using human Apo A-I ELISA kit (Abcam). (B) Proposed mechanism of SiNPs exposure–induced Apo A-I depletion in silicosis patients.
Discussion
Previous protein corona studies have been largely focused on addressing the biological and toxicological fate of NPs, while much less is known regarding how the NPs may affect the biological functions of their coronal proteins and trigger pathological outcomes. Both latter aspects have been examined in the current study concerning the SiNPs protein corona. Specifically, for pulmonary exposure to SiNPs, the primary lipid-based corona formed in alveoli enhanced the adsorption of Apo A-I in bloodstream, which in turn decreased both cellular internalization and cytotoxicity of SiNPs. Alternatively, elimination of SiNPs automatically led to the depletion of coronal Apo A-I, which were verified in vitro, ex vivo, in vivo, and in clinical silicosis patients. The decreased Apo A-I level in blood potentially caused adverse effects on the cardiovascular system as identified in this study. Accordingly, supplementation of Apo A-I mimetic 5A counteracted the SiNPs-induced cardiovascular injury effectively, representing a successful in vivo strategy at manipulating the NPs exposure–related serum protein depletion for a major health benefit.
Together, the current study has put forth a mechanism concerning SiNPs exposure and cardiovascular damage. By linking and manipulating the formation of an Apo A-I–enriched protein corona and SiNPs elimination from bloodstream, this current study has developed a mechanism for predicting other ABB-associated NP exposure and their pathological implications.
Materials and Methods
Animal Care.
Male C57BL/6 mice (8 wk old) were purchased from Chongqing Academy of Chinese Materia Medica. Male C57BL/6 background Apo E−/− mice (6 wk old) were purchased from the Model Animal Research Center of Nanjing University. All animals were given tap water ad libitum. C57BL/6 mice were fed with a standard chow diet, and Apo E−/− mice were fed with a high-fat diet. Animal study was approved by the Southwest University Animal Care and Use Committee. Experiments were conducted according to the NIH’s guidelines for the care and use of laboratory animals (58).
Preparation of QDs@SiNPs.
QDs@SiNPs were prepared according to our previous publication (59). Briefly, 15 mL cyclohexane, 4 mL Triton X-100, 4 mL n-hexane, 0.5 mL CdSe/ZnS QDs (3 mg/mL), and 200 μL tetraethyl orthosilicate (TEOS) were added and mixed in a round-bottom flask. After gentle stirring for 30 min, 0.5 mL ammonium hydroxide (28%) was added, and then the reaction system was stirred at 40 °C for 24 h in the dark. QDs@SiNPs were obtained after precipitation with acetone and centrifugation at 9,000 g. Finally, the collected QDs@SiNPs were washed with methanol, ethanol, and water, successively, and the final product was dispersed in ultra-pure water for subsequent characterization.
Characterization of SiNPs and QDs@SiNPs.
SiNPs were obtained from Sigma-Aldrich. SiNPs and QDs@SiNPs solutions were deposited onto holey carbon film on copper grids (TED PELLA Inc. Ultrathin Carbon Film on Lacey Carbon Support Film, 400 mesh, Copper) and then visualized by JEM-1200EX transmission electron microscope (JEOL) at an acceleration voltage of 120 kV. After resuspension in phosphate buffered saline (PBS) or Dulbecco’s modified Eagle medium (DMEM) at a concentration of 100 µg/mL, the zeta potential and hydrodynamic diameters of SiNPs and QDs@SiNPs were measured on a Mastersizer Micro. SiNPs and QDs@SiNPs suspension was sonicated at 25 °C for 15 min before detection.
Intranasal Instillation.
Intranasal instillation of SiNPs was carried out to simulate the pulmonary exposure of SiNPs. Male Apo E−/− mice were fed with a high-fat diet and randomly divided into two groups: control and SiNPs group. SiNPs group were intranasally instilled with SiNPs saline suspension (10 mg/kg body weight, every other day for three consecutive months), and control group mice received saline accordingly. The intranasal instillation procedure was carried out after anesthetizing animals using diethyl ether.
Histopathological Examination.
After indicated treatment, mice were euthanized, and blood was extracted through orbit. Next, the aorta, heart, liver, spleen, lung, and kidney tissues were harvested and immediately put into 4% paraformaldehyde solution. The histopathological tests were conducted using standard laboratory procedure. Briefly, tissues were embedded in paraffin blocks, sectioned into 3- to 5-μm slices, and then mounted onto glass slides. After HE staining, oil red O staining, or Masson’s trichrome staining, sections were evaluated, and photos were taken using an optical microscope.
Oil Red O Staining.
Blood vessels were harvested and immersed into 4% paraformaldehyde for 24 h. Then blood vessels were washed twice with PBS, dissected longitudinally along the vessel wall. The blood vessel was immersed in 60% isopropanol for 3 s and stained in oil red O staining solution (0.5 g oil red O in 100 mL 98% isopropanol) in the dark at 37 °C for 1 h. Then, the blood vessel was immersed in 60% isopropanol for differentiation. The plaque formation was observed using a microscope (OLYMPUS IX71). The percentages of oil red O area to total vessel area were quantified using Adobe Photoshop CC 2019.
Western Blot Assay.
The concentrations of lung tissue and cellular proteins were detected by the BCA Protein Assay Kit (Sangon Biotech) according to the manufacturer’s instruction. Molecular weight marker (Sangon Biotech) and samples were loaded and proceeded with gel electrophoresis. Cellular protein (30 μg) from each group were blotted onto nitrocellulose membrane after separation with 10 to 12.5% SDS-PAGE. The immunoblot was incubated with the blocking solution (5 to 10% skim milk) at 25 °C for 2 h, followed by incubation with Apo A-I antibody (Bioss) (1:1,000) at 25 °C for 3 h. After washing with Tween 20/Tris-buffered saline three times, the immune-blot was incubated with secondary antibody (Sangon Biotech) (1:5,000) at 25 °C for 2 h. Followed by visualization using the electrochemiluminescence (ECL) system.
TEM Analysis of Trachea/Alveoli and Blood.
Male C57BL/6 mice were intranasally instilled with 10 mg/kg SiNPs and euthanized 24 h after instillation. Lung tissues from the trachea/alveoli were fixed in 2.5% glutaraldehyde solution overnight. Blood sample was extracted and stood for coagulation. Cubes ranging from 1 to 2 mm3 were taken from trachea/alveoli or blood clot. After rinsing in 0.1 M PBS (pH = 7.0), cubes were postfixed in 1% osmium tetroxide for 1 h, dehydrated in alcohol, and embedded in epoxy resin. Ultrathin sections (70 to 90 nm) were obtained using ultramicrotome (Leica EM UC7) and stained with lead citrate and uranyl acetate for 5 min. Samples were examined by TEM (Hitachi-7500, Hitachi).
LDH Release Assay.
Briefly, the cell supernatant or mouse serum (5× dilution) were collected for LDH activity analysis according to the manufacturer’s protocol (Nanjing Jiancheng Bioengineering Institute). The absorbance at 450-nm wavelength was measured by microplate reader (Bio Tek ELX800). The amount of LDH released was expressed as LDH activity (U/L) in mice serum.
BALF and MP Preparation.
Briefly, mice trachea was exposed, and a self-made plastic canula was inserted to lungs through trachea, then 1 mL sterile physiological saline was used to flush the lung three times. The extracted BALF containing PS was lyophilized and stored at −80 °C. Mice blood was collected through orbit for MP preparation (3,000 g, 10 min, 1% heparin was used for anticoagulation).
Preparation of SiNPs-PS, SiNPs-MP, and SiNPs-PS-MP.
BALF powder extracted from mice lung was dissolved in ultra-pure water and filtered by 0.22-μm filter membrane to remove aggregated particles. SiNPs were added into BALF solution and incubated with gentle shake at 37 °C for 1 h. For SiNPs-MP preparation, SiNPs were incubated with 10% MP. SiNPs-PS-MP were obtained by incubating SiNPs-PS with BALF, then 10% MP. The mixtures were centrifuged at 16,000 g for 20 min, supernatant was discarded, and precipitation was washed by PBS three times and resuspended in PBS.
Coomassie Brilliant Blue Staining on SDS-PAGE.
The relative semiquantification of adsorbed proteins on SiNPs or SiNPs-PS was performed by SDS-PAGE and Coomassie brilliant blue staining. Molecular weight marker (Sangon Biotech) and samples treated with loading buffer were loaded into 12.5% SDS-PAGE. After separation, SDS-PAGE was stained by Coomassie brilliant blue R 250 (Solarbio) for 1 h and then visualized using a gel imaging system (Vilber Lourmat).
LC-ESI-MS/MS Analysis of Coronal Protein.
SiNPs-MP and SiNPs-PS-MP samples were incubated with loading buffer, heated at 97 °C for 10 min to fully denature. Proteins were reduced by dithiothreitol. Cysteine residues were alkylated by iodoacetamide and cleaned by desalting columns or ethanol precipitation. The sample was then digested with sequencing-grade modified trypsin (Promega) in the digestion buffer (ammonium bicarbonate 100 mM, pH = 8.5). A dissolved peptide sample was then analyzed by a Nano LC-ESI-MS/MS system. Nano LC-ESI-MS/MS analysis of a digested protein sample was carried out by a high-performance liquid chromatography (HPLC) system (Agilent) with a 75-μm diameter and 8-cm length in-house–packed reverse-phase C18 capillary column. The particle size of the C18 was 3 μm, and the pore size was 300 Å. The sample injection time was 20 min. The HPLC Solvent A contained 97.5% water, 2% acetonitrile, and 0.5% formic acid. HPLC Solvent B contained 9.5% water, 90% acetonitrile, and 0.5% formic acid. The gradation time was 60 min from 2% Solvent B to 90% solvent B, plus 20 min for sample loading, and 20 min for column washing. The column flow rate was around 800 nanoliter per minute after splitting. Typical sample injection volume was 3 μL.
The HPLC system was on-line coupled with a linear ion trap mass spectrometer (LTQ, Thermo Fisher) in a way that a sample eluted from HPLC column was directly ionized by an electrospray ionization (ESI) process and enter the mass spectrometer. The ionization voltage was often optimized in the instrument tuning process and normally in a range of 1.5 to 1.8 kV. The capillary temperature was set at 100 °C. The mass spectrometer was set at the data-dependent mode to acquire MS/MS data via a low-energy collision–induced dissociation process. The default collision energy was 33%, and the default charge state was 3. One full scan with one microscan with a mass range of 350 to 1,650 amu was acquired, followed by nine MS/MS scans of the nine most intense ions with a full mass range and three microscans. The dynamic exclusion feature was set as follows: repeat count of 1 and exclusion duration of 1 min. The exclusion width was 4 Da. The mass spectrometric data were used to search against the UniProt protein database with ProtTech’s ProQuest software suite, and relative protein abundance was calculated based on a published method (60).
TLC.
TLC was applied to separate the surfactant lipids that bound to SiNPs. Pristine SiNPs and DPPC were dispersed in resolving agent composed of CHCl3 and CH3OH (2:1), and 10-μL dispersion were loaded onto silica gel plate. Two developing agents (CHCl3/CH3OH/CH3COOH/H2O, 56:33:9:2 and hexane/Ether/CH3COOH, 80:20:1) were applied for a tandem chromatographical separation. Finally, the plate was placed in color developing reagent containing 8% H3PO4 and 10% CuSO4, developed at 180 °C for 5 min.
CD Spectroscopy.
Equal volumes of 200 μg/mL Apo A-I protein solution were mixed with saline, PS, SiNPs, and SiNPs-PS (100 μg/mL), respectively. After incubation for 1 h, the mixed solution was immediately determined by CD spectrometer (MOS-450, Bio-Logic) with 1-mm path length quartz cuvette at 20 °C. The spectra from 190 to 260 nm were scanned, and the recorded data were analyzed for protein secondary structure. The proportion of secondary structure of each sample (α-helix, β-sheet, and β-turn and random coil) was obtained using online CD analysis (DichroWeb).
Cell Culture.
J774A.1 murine macrophage was obtained from the American Type Culture Collection, cultured in DMEM with 10% fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin. Cells were incubated at 37 °C in a humidified atmosphere containing 5% CO2.
Cell Viability Assay.
J774A.1 cells were seeded in a 96-well plate with proper density and incubated at 37 °C overnight. After adhesion, cells were treated with SiNPs, SiNPs-MP, and SiNPs-PS-MP (50 μg/mL) for 6 h. Cell viability was determined using the cell counting kit-8 (TargetMol) according to the manufacturer’s instruction.
Cellular Uptake Assays.
ICP-OES.
J774A.1 cell was exposed to SiNPs, SiNPs-MP, and SiNPs-PS-MP (50 μg/mL) for 6 h. After discarding the supernatant, cells were collected and washed with PBS three times. Decomposition reagent containing HNO3, H2O2, and HF (3:1:1) was added and the mixture was heated at 70 °C for 6 h. The mixture was diluted, and the concentration of Si element was determined by ICP-OES (Agilent ICP-OES 730).
Confocal laser scanning microscope and flow cytometry analysis.
QDs@SiNPs were applied to evaluate the cellular uptake of SiNPs through fluorescence detection. QDs@SiNPs were incubated with PS and MP using parallel procedure as described in the section Preparation of SiNPs-PS, SiNPs-MP, and SiNPs-PS-MP. For confocal imaging analysis, J774A.1 macrophages were plated in confocal dishes with 70% cell confluency and then incubated with QDs@SiNPs, QDs@SiNPs-MP, or QDs@SiNPs-PS-MP (50 μg/mL) in a serum-free medium at 37 °C for 6 h. After washing with PBS three times, cells were fixed by 4% paraformaldehyde for 20 min and washed with PBS three times. Cells were than stained by 3,3′-dioctadecyloxacarbocyanine perchlorate (DiO, 20 μg/mL) (Solarbio) at 37 °C for 20 min and washed with PBS three times. Cells were observed under confocal laser scanning microscope (Nikon N-SIM). For flow cytometry analysis, J774A.1 cell was collected and detected by flow cytometry (BD FACSMelody). The relative quantification of NPs uptake was evaluated by quantifying the fluorescence intensity using FlowJo version 10.0.7.
Blood Circulation.
The blood circulation of SiNPs and SiNPs-PS were conducted through detecting the concentrations of Si element in blood at specific time points after intravenous injection. Briefly, male C57BL/6 mice were randomly divided into two groups (SiNPs and SiNPs-PS) and intravenously injected with SiNPs and SiNPs-PS (1 μg/μL × 50 μL) through tail, respectively. Blood was collected at specific time points (5, 30, 60, 120, 240, and 480 min). Then, decomposition reagent containing 3 mL HNO3, 1 mL H2O2, and 1 mL HF was added into 0.5-mL blood sample, heated at 120 °C for 30 min. Finally, samples were volumed with ultra-pure water to 10 mL, and the concentration of Si element was determined by ICP-OES (Avio 200, PerkinElmer).
RNA-Seq.
J774A.1 cell was treated with SiNPs, SiNPs-MP, and SiNPs-PS-MP (50 μg/mL) for 6 h in a serum-free medium. TRIzol reagent kit was applied for total RNA extraction according to the manufacturer’s protocol (Invitrogen). Total RNA quality was evaluated by Agilent 2100 Bioanalyzer (Agilent Technologies) and RNase free agarose gel electrophoresis. After RNA extraction, messenger RNA was enriched by Oligo (dT) beads and fragmented into short fragments, then reverse transcribed into complementary DNA (cDNA) with random primers. Then, the cDNA fragments were purified with QiaQuick PCR extraction kit (Qiagen), end repaired, poly (A) added, and ligated to Illumina sequencing adapters. The size-selected products were sequenced by Illumina HiSEq. 2500 platform by Gene Denovo Biotechnology Co.
Differential Expressed Genes Analysis.
The differential expressed genes (DEGs) analysis of RNA between different groups was performed by DESeq2 software. Genes with absolute fold change ≥ 2 and false discovery rate (FDR) < 0.05 were considered as DEGs.
GO and KEGG Analysis.
All DEGs were mapped for GO enrichment analysis (http://www.geneontology.org/). Pathway enrichment analysis was performed by applying KEGG database. Significantly enriched GO terms and pathways in DEGs were filtered by FDR at the threshold below 0.05.
RT-qPCR.
A total RNA extraction kit was used for total RNA extraction. cDNA was obtained from extracted RNA through reverse transcription. Next, the obtained cDNA was applied to run RT-qPCR analysis programs following the LightCycler 96 instrument instruction. After that, the denaturation, annealing, extension, and incubation times as well as corresponding temperatures were set to appropriate parameters. Each experiment was performed three times, and data were analyzed based on three independent experiments. The relative gene expression was summarized via the method of 2-ΔΔCt, which was normalized by β-actin expression. Primers used in this study that provided by Sangon Biotech were listed in SI Appendix, Table S3.
Human Blood Samples.
Human plasma samples of silicosis patients were obtained from Zhejiang Academy of Medical Sciences, and procedures were performed in accordance with the protocol approved by the ethics committee of Zhejiang Academy of Medical Sciences (2019-012). Basic information of all silicosis patients (n = 50) was listed in SI Appendix, Table S4. Plasma samples of CHD patients with an average age of 63.6 were collected from Beijing Friendship Hospital, Capital Medical University, and procedures were performed in accordance with the protocol approved by the ethics committee of Beijing Friendship Hospital, Capital Medical University (2020-P2-165-01). CHD patients (n = 50) underwent initial diagnostic coronary angiography and demonstrated at least 70% stenosis in a major epicardial artery or 50% stenosis in the left main coronary artery. Healthy plasma samples were collected from volunteer donors (n = 55) with an average age of 51.7. All participants provided written informed consent after complete description of the study. All plasma samples were obtained after centrifuging at 800 g for 15 min and stored at −80 °C.
Statistical Analysis.
Statistical analysis was performed using GraphPad Prism 8.0 and Microsoft Excel 2016. Shapiro–Wilk test was applied for normality test. Two-tailed Student’s t test was used for parametric data, and Mann–Whitney U test was used for nonparametric data in comparison of two groups. One-way ANOVA followed by Tukey post hoc test was utilized in comparisons of more than two groups. Experiments were repeated for at least three times. P < 0.05 was considered to show statistical significance.
Acknowledgments
This work is supported by the National Natural Science Foundation of China (Grants 21976145 and 22176206) and Fundamental Research Funds for the Central Universities (Grant XDJK2019TJ001).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2108131118/-/DCSupplemental.
Data Availability
The transcriptomic data have been deposited in National Center for Biotechnology Information (NCBI)’s Sequence Read Archive and are accessible through BioProject accession no. PRJNA682685. All other study data are included in the article and SI Appendix.
References
- 1.Li Z., Barnes J. C., Bosoy A., Stoddart J. F., Zink J. I., Mesoporous silica nanoparticles in biomedical applications. Chem. Soc. Rev. 41, 2590–2605 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Liu Y., et al., Long-term exposure to crystalline silica and risk of heart disease mortality. Epidemiology 25, 689–696 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Leung C. C., Yu I. T. S., Chen W., Silicosis. Lancet 379, 2008–2018 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Riley L., Urbine D., Chronic silicosis with progressive massive fibrosis. N. Engl. J. Med. 380, 2256–2256 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Calvert G. M., Rice F. L., Boiano J. M., Sheehy J. W., Sanderson W. T., Occupational silica exposure and risk of various diseases: An analysis using death certificates from 27 states of the United States. Occup. Environ. Med. 60, 122–129 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding M., et al., Diseases caused by silica: Mechanisms of injury and disease development. Int. Immunopharmacol. 2, 173–182 (2002). [DOI] [PubMed] [Google Scholar]
- 7.GBD 2017 DALYs and HALE Collaborators, Global, regional, and national disability-adjusted life-years (DALYs) for 359 diseases and injuries and healthy life expectancy (HALE) for 195 countries and territories, 1990-2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 392, 1859–1922 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsuda A., et al., Age-dependent translocation of gold nanoparticles across the air-blood barrier. ACS Nano 13, 10095–10102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi H. S., et al., Rapid translocation of nanoparticles from the lung airspaces to the body. Nat. Biotechnol. 28, 1300–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kreyling W. G., et al., Air-blood barrier translocation of tracheally instilled gold nanoparticles inversely depends on particle size. ACS Nano 8, 222–233 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang G. S., et al., Long-term inhalation exposure to nickel nanoparticles exacerbated atherosclerosis in a susceptible mouse model. Environ. Health Perspect. 119, 176–181 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brook R. D., et al.; American Heart Association Council on Epidemiology and Prevention, Council on the Kidney in Cardiovascular Disease, and Council on Nutrition, Physical Activity and Metabolism, Particulate matter air pollution and cardiovascular disease: An update to the scientific statement from the American Heart Association. Circulation 121, 2331–2378 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Dominici F., et al., Fine particulate air pollution and hospital admission for cardiovascular and respiratory diseases. JAMA 295, 1127–1134 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brunekreef B., Holgate S. T., Air pollution and health. Lancet 360, 1233–1242 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Monopoli M. P., Aberg C., Salvati A., Dawson K. A., Biomolecular coronas provide the biological identity of nanosized materials. Nat. Nanotechnol. 7, 779–786 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Lynch I., Salvati A., Dawson K. A., Protein-nanoparticle interactions: What does the cell see? Nat. Nanotechnol. 4, 546–547 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Pinals R. L., Yang D., Lui A., Cao W., Landry M. P., Corona exchange dynamics on carbon nanotubes by multiplexed fluorescence monitoring. J. Am. Chem. Soc. 142, 1254–1264 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tenzer S., et al., Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology. Nat. Nanotechnol. 8, 772–781 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Trentin P. G., et al., Annexin A1 mimetic peptide controls the inflammatory and fibrotic effects of silica particles in mice. Br. J. Pharmacol. 172, 3058–3071 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barbarin V., et al., The role of pro- and anti-inflammatory responses in silica-induced lung fibrosis. Respir. Res. 6, 112 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayeux J. M., et al., Silicosis and silica-induced autoimmunity in the diversity outbred mouse. Front. Immunol. 9, 874 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gasser M., et al., The adsorption of biomolecules to multi-walled carbon nanotubes is influenced by both pulmonary surfactant lipids and surface chemistry. J. Nanobiotechnology 8, 31 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamilton R. F. Jr., Thakur S. A., Holian A., Silica binding and toxicity in alveolar macrophages. Free Radic. Biol. Med. 44, 1246–1258 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ashavaid T. F., et al., Lipid, lipoprotein, apolipoprotein and lipoprotein(a) levels: Reference intervals in a healthy Indian population. J. Atheroscler. Thromb. 12, 251–259 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Rosenson R. S., et al., Translation of high-density lipoprotein function into clinical practice: Current prospects and future challenges. Circulation 128, 1256–1267 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Tenzer S., et al., Nanoparticle size is a critical physicochemical determinant of the human blood plasma corona: A comprehensive quantitative proteomic analysis. ACS Nano 5, 7155–7167 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Lundqvist M., et al., Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts. Proc. Natl. Acad. Sci. U.S.A. 105, 14265–14270 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cedervall T., et al., Detailed identification of plasma proteins adsorbed on copolymer nanoparticles. Angew. Chem. Int. Ed. Engl. 46, 5754–5756 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Kapralov A. A., et al., Adsorption of surfactant lipids by single-walled carbon nanotubes in mouse lung upon pharyngeal aspiration. ACS Nano 6, 4147–4156 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leo B. F., et al., The stability of silver nanoparticles in a model of pulmonary surfactant. Environ. Sci. Technol. 47, 11232–11240 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ye X., Hao C., Yang J., Sun R., Influence of modified silica nanoparticles on phase behavior and structure properties of DPPC monolayers. Colloids Surf. B Biointerfaces 172, 480–486 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Hellstrand E., et al., Complete high-density lipoproteins in nanoparticle corona. FEBS J. 276, 3372–3381 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Gessner A., et al., Nanoparticles with decreasing surface hydrophobicities: Influence on plasma protein adsorption. Int. J. Pharm. 196, 245–249 (2000). [DOI] [PubMed] [Google Scholar]
- 34.Park S. J., Protein-nanoparticle interaction: Corona formation and conformational changes in proteins on nanoparticles. Int. J. Nanomedicine 15, 5783–5802 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silva R. A., et al., Structure of apolipoprotein A-I in spherical high density lipoproteins of different sizes. Proc. Natl. Acad. Sci. U.S.A. 105, 12176–12181 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lesniak A., et al., Effects of the presence or absence of a protein corona on silica nanoparticle uptake and impact on cells. ACS Nano 6, 5845–5857 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Cai R., Chen C., The crown and the scepter: Roles of the protein corona in nanomedicine. Adv. Mater. 31, e1805740 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Schöttler S., et al., Protein adsorption is required for stealth effect of poly(ethylene glycol)- and poly(phosphoester)-coated nanocarriers. Nat. Nanotechnol. 11, 372–377 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Ritz S., et al., Protein corona of nanoparticles: Distinct proteins regulate the cellular uptake. Biomacromolecules 16, 1311–1321 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Pan D., Wang Q., Jiang S., Ji X., An L., Synthesis of extremely small CdSe and highly luminescent CdSe/CdS core-shell nanocrystals via a novel two-phase thermal approach. Adv. Mater. 17, 176–179 (2005). [Google Scholar]
- 41.Charo I. F., Taubman M. B., Chemokines in the pathogenesis of vascular disease. Circ. Res. 95, 858–866 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Raghavan S., Singh N. K., Gali S., Mani A. M., Rao G. N., Protein kinase Cθ via activating transcription factor 2-mediated CD36 expression and foam cell formation of Ly6Chi cells contributes to atherosclerosis. Circulation 138, 2395–2412 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liang J., et al., Macrophage metalloelastase accelerates the progression of atherosclerosis in transgenic rabbits. Circulation 113, 1993–2001 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Mallat Z., et al., Inhibition of transforming growth factor-beta signaling accelerates atherosclerosis and induces an unstable plaque phenotype in mice. Circ. Res. 89, 930–934 (2001). [DOI] [PubMed] [Google Scholar]
- 45.Li X., et al., Differential effects of apolipoprotein A-I-mimetic peptide on evolving and established atherosclerosis in apolipoprotein E-null mice. Circulation 110, 1701–1705 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Sherman C. B., Peterson S. J., Frishman W. H., Apolipoprotein A-I mimetic peptides: A potential new therapy for the prevention of atherosclerosis. Cardiol. Rev. 18, 141–147 (2010). [DOI] [PubMed] [Google Scholar]
- 47.Benoit P., et al., Somatic gene transfer of human ApoA-I inhibits atherosclerosis progression in mouse models. Circulation 99, 105–110 (1999). [DOI] [PubMed] [Google Scholar]
- 48.Sviridov D. O., et al., Hydrophobic amino acids in the hinge region of the 5A apolipoprotein mimetic peptide are essential for promoting cholesterol efflux by the ABCA1 transporter. J. Pharmacol. Exp. Ther. 344, 50–58 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Navab M., et al., Oral administration of an Apo A-I mimetic Peptide synthesized from D-amino acids dramatically reduces atherosclerosis in mice independent of plasma cholesterol. Circulation 105, 290–292 (2002). [DOI] [PubMed] [Google Scholar]
- 50.Song X., Fischer P., Chen X., Burton C., Wang J., An apoA-I mimetic peptide facilitates off-loading cholesterol from HDL to liver cells through scavenger receptor BI. Int. J. Biol. Sci. 5, 637–646 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Navab M., et al., Oral D-4F causes formation of pre-beta high-density lipoprotein and improves high-density lipoprotein-mediated cholesterol efflux and reverse cholesterol transport from macrophages in apolipoprotein E-null mice. Circulation 109, 3215–3220 (2004). [DOI] [PubMed] [Google Scholar]
- 52.Navab M., Hama S., Hough G., Fogelman A. M., Oral synthetic phospholipid (DMPC) raises high-density lipoprotein cholesterol levels, improves high-density lipoprotein function, and markedly reduces atherosclerosis in apolipoprotein E-null mice. Circulation 108, 1735–1739 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Ball M. J., The aetiology of atherosclerosis and coronary heart disease. Br. J. Hosp. Med. 38, 404–410 (1987). [PubMed] [Google Scholar]
- 54.Boden W. E., High-density lipoprotein cholesterol as an independent risk factor in cardiovascular disease: Assessing the data from Framingham to the Veterans Affairs High--Density Lipoprotein Intervention Trial. Am. J. Cardiol. 86, 19L–22L (2000). [DOI] [PubMed] [Google Scholar]
- 55.Suematsu Y., et al., Anti-atherosclerotic effects of an improved apolipoprotein A-I mimetic peptide. Int. J. Cardiol. 297, 111–117 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Luc G., et al.; Prospective Epidemiological Study of Myocardial Infarction, Value of HDL cholesterol, apolipoprotein A-I, lipoprotein A-I, and lipoprotein A-I/A-II in prediction of coronary heart disease: The PRIME Study. Arterioscler. Thromb. Vasc. Biol. 22, 1155–1161 (2002). [DOI] [PubMed] [Google Scholar]
- 57.Ishikawa T., Fidge N., Thelle D. S., Førde O. H., Miller N. E., The Tromsø Heart Study: Serum apolipoprotein AI concentration in relation to future coronary heart disease. Eur. J. Clin. Invest. 8, 179–182 (1978). [DOI] [PubMed] [Google Scholar]
- 58.National Research Council, Guide for the Care and Use of Laboratory Animals (National Academies Press, Washington, DC, ed. 8, 2011). [Google Scholar]
- 59.Zhang L., et al., Dual effects of fibrinogen decoration on the tuning of silica nanoparticles-induced autophagic response: The implication of sedimentation and internalization. J. Hazard. Mater. 408, 124467 (2021). [DOI] [PubMed] [Google Scholar]
- 60.Griffin N. M., et al., Label-free, normalized quantification of complex mass spectrometry data for proteomic analysis. Nat. Biotechnol. 28, 83–89 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The transcriptomic data have been deposited in National Center for Biotechnology Information (NCBI)’s Sequence Read Archive and are accessible through BioProject accession no. PRJNA682685. All other study data are included in the article and SI Appendix.