ABSTRACT
Phage-based biocontrol of bacteria is considered a natural approach to combat foodborne pathogens. Salmonella spp. are notifiable and highly prevalent pathogens that cause foodborne diseases worldwide. In this study, six bacteriophages were isolated and further characterized that infect food-derived Salmonella isolates from different meat sources. The siphovirus VB_StyS-LmqsSP1, which was isolated from a cow’s nasal swab, was further subjected to in-depth characterization. Phage-host interaction investigations in liquid medium showed that vB_StyS-LmqsSP1 can suppress the growth of Salmonella species isolates at 37°C for 10 h and significantly reduce the bacterial titer at 4°C. A reduction of 1.4 to 3 log units was observed in investigations with two food-derived Salmonella isolates and one reference strain under cooling conditions using multiplicities of infection (MOIs) of 104 and 105. Phage application on chicken skin resulted in a reduction of about 2 log units in the tested Salmonella isolates from the first 3 h throughout a 1-week experiment at cooling temperature and with an MOI of 105. The one-step growth curve analysis using vB_StyS-LmqsSP1 demonstrated a 60-min latent period and a burst size of 50 to 61 PFU/infected cell for all tested hosts. Furthermore, the genome of the phage was determined to be free from genes causing undesired effects. Based on the phenotypic and genotypic properties, LmqsSP1 was assigned as a promising candidate for biocontrol of Salmonella enterica serovar Typhimurium in food.
IMPORTANCE Salmonella enterica is one of the major global causes of foodborne enteritis in humans. The use of chemical sanitizers for reducing bacterial pathogens in the food chain can result in the spread of bacterial resistance. Targeted and clean-label intervention strategies can reduce Salmonella contamination in food. The significance of our research demonstrates the suitability of a bacteriophage (vB_StyS-LmqsSP1) for biocontrol of Salmonella enterica serovar Typhimurium on poultry due to its lytic efficacy under conditions prevalent in food production environments.
KEYWORDS: Salmonella Typhimurium, phage biocontrol, phage therapy, foodborne zoonoses, chicken, biocontrol, phage
INTRODUCTION
Salmonella enterica is one of the major global causes of foodborne enteritis in humans (1). In 2018, it was the leading cause of notified foodborne outbreaks in the European Union, leading to more than 91,000 human cases of salmonellosis. To date, more than 2,500 serovars of S. enterica have been notified (1), but foodborne outbreaks in the United States and Europe are mainly caused by S. enterica serovar Enteritidis and S. enterica serovar Typhimurium (2, 3). The S. Enteritidis serovar is predominantly associated with chicken, whereas S. Typhimurium derives from a wide variety of food animal sources, among them poultry, pigs, and cattle (2). The highest prevalences of Salmonella-positive single samples in the European Union were reported for poultry meat, minced meat, and meat preparations (2).
The use of chemical sanitizers and preservatives for reducing pathogenic bacteria in the food chain can result in the spread of bacterial resistance (4) and in the emergence of undesirable allergenic effects, generation of by-products, and negative impact on the environment (5). Due to these reasons, natural agents for biocontrol of pathogenic bacteria in foodstuff have gained further attention in recent years (6).
Bacteriophage-based biocontrol is considered an alternative and natural intervention strategy for reducing bacterial contamination in food (7–9). While conventional strategies combat both pathogens and the commensal microflora indiscriminately, the use of phages allows a specific reduction in undesirable bacteria (9). Additionally, bacteriophages do not affect organoleptic properties such as the flavor, color, and/or aroma of the treated food products (9, 10).
Phage application in livestock before slaughter (preharvest) and in foods (postharvest) is permitted under FSIS Directive 7120.1 in the United States (9, 11), and commercial phage preparations have been approved as generally recognized as safe (GRAS) for application in raw and ready-to-eat meat and poultry products (9). In Europe, approval of phage products for food applications is still in the preparatory stage due to open regulatory and scientific issues (9, 12, 13). Further phage preparations are needed for reducing foodborne zoonoses, especially multidrug-resistant pathogens. Since they can be produced by relatively easy and economical processes (14), phages are a very suitable measure for this purpose, especially in developing countries (15, 16).
Phage application has been investigated for biocontrol of S. enterica in different food products, as reviewed by Moye et al. (9). Studies on bacteriophage application for control of Salmonella in food products have shown promising results (6, 17–19). Some studies investigated the effect of phages on chicken skin. However, only few studies have determined the phage-based reduction under practical conditions, such as the temperatures of cooled storage, and against Salmonella field isolates from food samples (17, 20, 21). These factors can have a high impact on the reduction of target bacteria in commercial food production settings. Testing phages only against laboratory strains and under laboratory settings that do not resemble conditions during food production might thus lead to biased results and the selection of unsuitable phages (22, 23). For bacteriophage efficacy testing, phages should be tested under realistic conditions, using the strains most similar to those found on the targeted product (17, 21). Testing of phages in commercial production plants is laborious and time consuming, and the results can sometimes be difficult to interpret due to the complexity of influencing factors. Additionally, these factors might lead to scientific results that are hard to reproduce, which thus cannot be extrapolated to other production plants or settings without further experimental analysis (24, 25). In vitro models combining environmental factors of commercial food production settings with controlled experimental conditions should be used to allow and extrapolation of the collected data. Cooling conditions and food-derived Salmonella isolates on food matrices need to be used for testing the lytic phage efficacy of promising candidates in vitro. It has been reported that clinically or environmentally isolated Salmonella can show insensitivity against phage infection that suggests promising results on laboratory Salmonella and higher efficacy in reduction experiments using laboratory strains compared to that using field isolates (17, 21). The former study reported that meat-derived Salmonella isolates were less sensitive than laboratory strains to phage infection (17). Killing of bacteria by phages can be achieved by different mechanisms, e.g., passive inundation or a lytic infection cycle resulting in phage progenies. The first infection step of phage binding to the bacterial cell surface is important for both mechanisms and requires attachment of phages to the bacterial cell by specific recognition between the phage’s receptor binding protein at the tip of the phage tail and a receptor located on the bacterial cell surface. If a multitude of phages bind to the bacterial cell, destabilization can lead to destruction without production of phage progenies (lysis from without), which is often referred to as passive inundation (26). On the other hand, injection of the phage’s nucleic acids, followed by production of structural proteins and phage morphogenesis, may lead to lysis of the bacterial cell, releasing a variable number of progenies. For this complex process, several metabolic prerequisites are necessary, and internal resistance mechanisms of the bacterial cell, which destroy the phage’s nucleic acids or interfere with phage replication, need to be absent. These prerequisites and the general layout of phage defense mechanisms in bacteria may differ between strains from different environments (27). Relatively few studies have investigated the efficacy of Salmonella phages using food-derived strains on food matrices under cooling conditions (17, 20, 28).
The present study aims to isolate bacteriophages suitable for reducing Salmonella on poultry skin and to evaluate their efficiency as potential measures for biocontrol in commercial broiler meat production systems. Six Salmonella-specific phages were investigated regarding their host range on field isolates. Based on its broad host range, one of these phages, vB_StyS-LmqsSP1 (LmqsSP1), was selected for the analysis of morphological and genomic properties as well as burst size and latent period. For potential application purposes in the food chain, the lytic efficacy against different Salmonella isolates was determined at 37°C and 4°C on chicken skin and in culture medium.
RESULTS
Six isolated phages form plaques on Salmonella isolates from meat products.
Overall, 10 bacteriophages were isolated from 52 investigated samples (Table 1). Nine bacteriophages were recovered from fecal samples of chicken flocks located in different parts of northern Germany and one bacteriophage was isolated from a nasal swab of a cow housed at the University of Veterinary Medicine Hannover, Germany, while no phages originated from the cecal contents of various commercial poultry flocks when using the Salmonella host strain LT2 for phage isolation. Based on their higher stability and reliable replication under laboratory conditions, six bacteriophages were selected for further examination, while the others were rejected due to their low concentration increase during propagation (<106 PFU/ml after two propagations) and instability at 4°C (Table 1).
TABLE 1.
Origin and growth characteristics of isolated phages
| Phage isolate | Sample type | Animal origina | Propagation (PFU increase) |
|---|---|---|---|
| LmqsSP1 | Nasal swab | Cow | Fast |
| LmqsSP2 | Feces | Chicken | Fast |
| LmqsSP3 | Feces | Chicken | Fast |
| LmqsSP4 | Feces | Chicken | Fast |
| LmqsSP5 | Feces | Chicken | Lowb |
| LmqsSP6 | Feces | Chicken | Lowb |
| LmqsSP7 | Feces | Chicken | Lowb |
| LmqsSP8 | Feces | Chicken | Lowb |
| LmqsSP9 | Feces | Chicken | Lowb |
| LmqsSP10 | Feces | Chicken | Lowb |
Samples originated from different farms in northern Germany.
A sufficient concentration of >106 PFU/ml was not met after two production cycles.
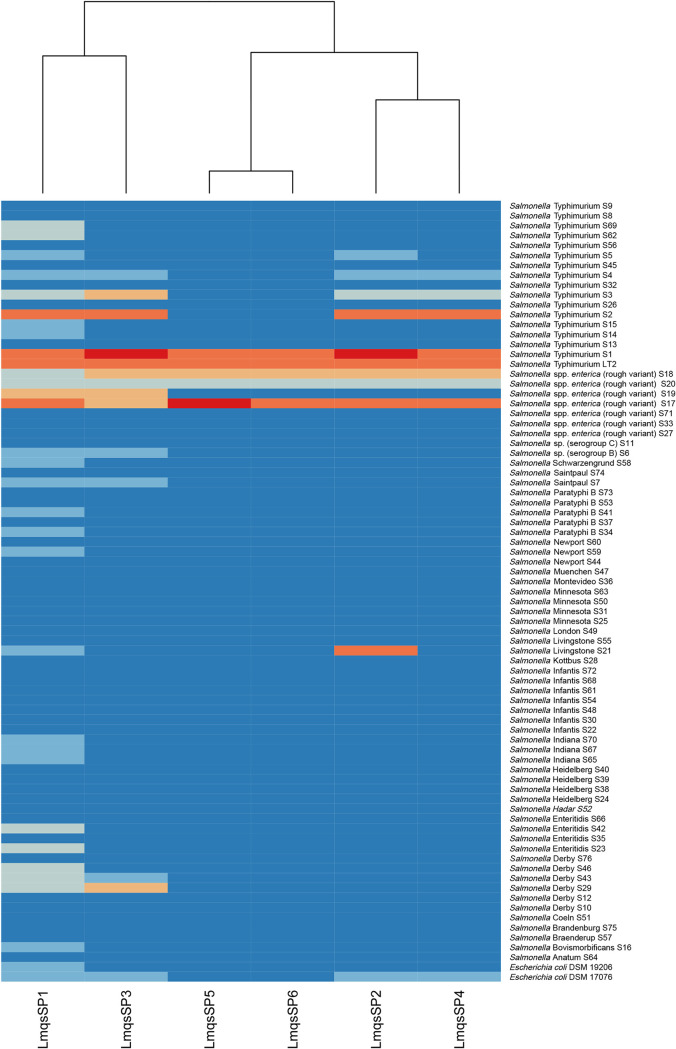
The host ranges of the six phages were tested on 76 food-derived Salmonella isolates (Table 2) representing 26 different serotypes derived from different food sources. Among these isolates, 60 were provided by the national reference laboratory on Salmonella, hosted at the German Federal Institute for Risk Assessment (BfR, Berlin, Germany), 17 isolates originated from the laboratory collection of the University of Veterinary Medicine Hannover, and two Escherichia coli O157:H7 strains were obtained from the German collection of microorganisms and cell cultures (Leibniz Institute DSMZ, Braunschweig, Germany). Salmonella isolates S1 and S2 were characterized as monophasic S. enterica Typhimurium. Isolate S1 was further identified as the most susceptible host, while S2 was very susceptible to four of the phages and exhibited a non-wild-type phenotype against ampicillin (MIC > 64 μg/ml), gentamicin (MIC = 16 μg/ml), sulfamethoxazole (MIC > 1,024 μg/ml) and tetracycline (MIC > 64 μg/ml). The S. enterica subsp. enterica rough variant S17, which was O-antigen-deficient, showed susceptibility toward all examined phages and exhibited phenotypic resistance against ampicillin, chloramphenicol, ciprofloxacin, trimethoprim, sulfamethoxazole, and tetracycline. Efficient plaque production was shown on Salmonella sp. I rough variant S18 from poultry meat. Furthermore, phage LmqsSP2 showed highly efficient plaque formation on the S. Livingstone isolate S21, while LmqsSP1 only showed a weak lytic effect on this isolate. High plaque formation by LmqsSP2 was also observed on S. Typhimurium isolate S3 and S. enterica serovar Derby isolate S29. Both LmqsSP1 and LmqsSP2 showed efficient plaque production on S. enterica subsp. enterica rough variant S19. The efficiency of plating (EOP) was calculated in relation to the plating efficiency on their host strain, S. Typhimurium LT2, as shown in Fig. 1. The six phages showed lytic ability on 30 (LmqsSP1), 13 (LmqsSP3), 10 (LmqsSP2), 8 (LmqsSP4) and 5 (LmqsSP5 and LmqsSP6) of the examined Salmonella isolates, covering 13 of 26 examined serotypes. The phage LmqsSP1 expressed the broadest host range, covering 39% of the investigated Salmonella isolates and both tested E. coli strains. It showed a lytic effect on more than half of the examined S. Typhimurium and S. enterica subsp. enterica rough variants, respectively. Furthermore, lysis by LmqsSP1 was observed on Salmonella serogroup B and the serotypes Schwarzengrund, Saintpaul, Paratyphi B, Newport, Livingstone, Indiana, Enteritidis, Derby, and Bovismorbificans. No lytic ability was observed on 47 of the 77 examined isolates (Fig. 1). Based on its inhibitory effect on the broadest spectrum of Salmonella isolates, we selected phage LmqsSP1 for further characterization and efficacy testing at different temperatures and on food matrices.
TABLE 2.
Origin and serotype of food-derived Salmonella isolates
| Salmonella isolate | Food sample | Animal origin | Serovar or serogroup |
|---|---|---|---|
| S1 | Minced meat | Pig | Typhimurium |
| S2 | Minced meat (mixed) | Cattle, pig | Typhimurium |
| S3 | Heart | Pig | Typhimurium |
| S4 | Meat | Pig | Typhimurium |
| S5 | Liver | Pig | Typhimurium |
| S6 | Minced meat (mixed) | Cattle, pig | Serogroup B |
| S7 | Meat | Deer | Saintpaul |
| S8 | Crust | Pig | Typhimurium |
| S9 | Meat | Poultry | Typhimurium |
| S10 | Minced meat (mixed) | Cattle, pig | Derby |
| S11 | Meat | Poultry | Serogroup C |
| S12 | Minced meat (mixed) | Cattle, pig | Derby |
| S13 | Minced meat (mixed) | Cattle, pig | Typhimurium |
| S14 | Minced meat | Pig | Typhimurium |
| S15 | Minced meat | Pig | Typhimurium |
| S16 | Meat | Turkey | Bovismorbificans |
| S17 | Minced meat | Pig | subsp. enterica rough variant |
| S18 | Meat | Poultry | subsp. enterica rough variant |
| S19 | Minced meat | Unknown | subsp. enterica rough variant |
| S20 | Meat | Poultry | subsp. enterica rough variant |
| S21 | Minced meat | Pig | Livingstone |
| S22 | Minced meat | Unknown | Infantis |
| S23 | Meat | Poultry | Enteritidis |
| S24 | Meat | Poultry | Heidelberg |
| S25 | Meat | Poultry | Minnesota |
| S26 | Minced meat | Pig | Typhimurium |
| S27 | Meat | Poultry | subsp. enterica rough variant |
| S28 | Meat | Poultry | Kottbus |
| S29 | Meat | Pig | Derby |
| S30 | Meat | Poultry | Infantis |
| S31 | Meat | Poultry | Minnesota |
| S32 | Minced meat | Unknown | Typhimurium |
| S33 | Meat | Poultry | subsp. enterica rough variant |
| S34 | Meat | Poultry | Paratyphi B |
| S35 | Meat | Poultry | Enteritidis |
| S36 | Meat | Poultry | Montevideo |
| S37 | Meat | Poultry | Paratyphi B |
| S38 | Meat | Poultry | Heidelberg |
| S39 | Meat | Poultry | Heidelberg |
| S40 | Meat | Poultry | Heidelberg |
| S41 | Meat | Poultry | Paratyphi B |
| S42 | Meat | Poultry | Enteritidis |
| S43 | Meat | Poultry | Derby |
| S44 | Meat | Poultry | Newport |
| S45 | Minced meat | Unknown | Typhimurium monophasic variant |
| S46 | Minced meat | Pig | Derby |
| S47 | Minced meat (mixed) | Cattle, pig | Muenchen |
| S48 | Minced meat | Cattle | Infantis |
| S49 | Minced meat | Pig | London |
| S50 | Meat | Poultry | Minnesota |
| S51 | Minced meat | Cattle | Coeln |
| S52 | Meat | Poultry | Hadar |
| S53 | Meat | Poultry | Paratyphi B |
| S54 | Meat | Poultry | Infantis |
| S55 | Meat | Poultry | Livingstone |
| S56 | Minced meat | Pig | Typhimurium |
| S57 | Meat | Poultry | Braenderup |
| S58 | Meat | Poultry | Schwarzengrund |
| S59 | Meat | Poultry | Newport |
| S60 | Meat | Poultry | Newport |
| S61 | Ground pork | Pig | Infantis |
| S62 | Ground pork | Pig | Typhimurium monophasic variant |
| S63 | Meat | Poultry | Minnesota |
| S64 | Meat | Poultry | Anatum |
| S65 | Meat | Poultry | Indiana |
| S66 | Meat | Poultry | Enteritidis |
| S67 | Meat | Poultry | Indiana |
| S68 | Meat | Poultry | Infantis |
| S69 | Ground pork | Pig | Typhimurium |
| S70 | Meat | Poultry | subsp. indica |
| S71 | Meat | Poultry | subsp. indica |
| S72 | Meat | Poultry | Infantis |
| S73 | Minced meat | Unknown | Paratyphi B |
| S74 | Meat | Poultry | Saintpaul |
| S75 | Minced meat | Pig | Brandenburg |
| S76 | Ground pork | Pig | Derby |
FIG 1.
Heatmap of phage host range. Phages are displayed on the x axis and bacterial isolates and strains on the y axis. Dark blue, no lysis; medium blue, at highest phage titer, efficiency of plating (EOP) < 0.001 with turbid plaques or inhibitory zone; light blue, EOP < 0.1; salmon pink, 0.1 ≤ EOP < 1; orange, 1 ≤ EOP ≤ 10; red, EOP > 10.
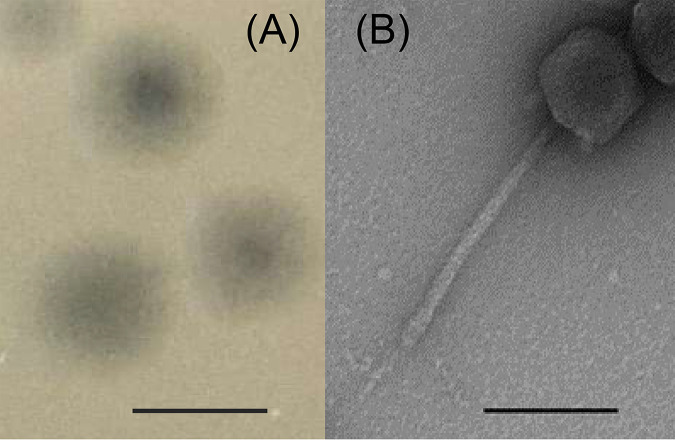
LmqsSP1 exhibited a morphology of the Siphoviridae family and forms plaques on lawns of food-derived Salmonella isolates.
Phage LmqsSP1 was isolated from the nasal swab of a cow and was classified as a siphovirus, based on its morphology with an average head width of 76 nm (standard deviation of 3), an average head length of 80 nm (standard deviation of 4), and an average tail length of 191 nm (standard deviation of 3) (Fig. 2B). LmqsSP1 produced clear plaques (Fig. 2A) on S. Typhimurium LT2, as well as on food-derived S. Typhimurium isolates S1, S2, and S3, and low EOP with turbid plaques on food-derived Salmonella isolates S4, S5, S6, and S7. On S. Typhimurium S1 and S2, LmqsSP1 showed an EOP of ≥1, while on isolate S3, the EOP was determined to be <0.1 (Fig. 1).
FIG 2.
(A) Plaques of examined bacteriophage vB_StyS-LmqsSP1. Bar, 3 mm. (B) Transmission electron micrograph of negatively stained phage vB_StyS-LmqsSP1. Bar, 100 nm.
One-step growth curves show low variance of LmqsSP1 propagation on different susceptible Salmonella isolates.
Growth experiments were performed using S. Typhimurium strain LT2 and the food-derived isolates S1 and S2 as hosts (Fig. 3). The latent period of LmqsSP1 was determined to be ∼60 min in all tested isolates. The burst sizes were determined to be 50 PFU/cell (ranging between 42 and 56 in the three experiments) in Salmonella strain LT2, 55 PFU/cell (between 55 and 58 in the three experiments) in food-derived isolate S1, and 61 PFU/cell (between 52 and 80 in the three experiments) in food-derived isolate S2 (Fig. 3).
FIG 3.
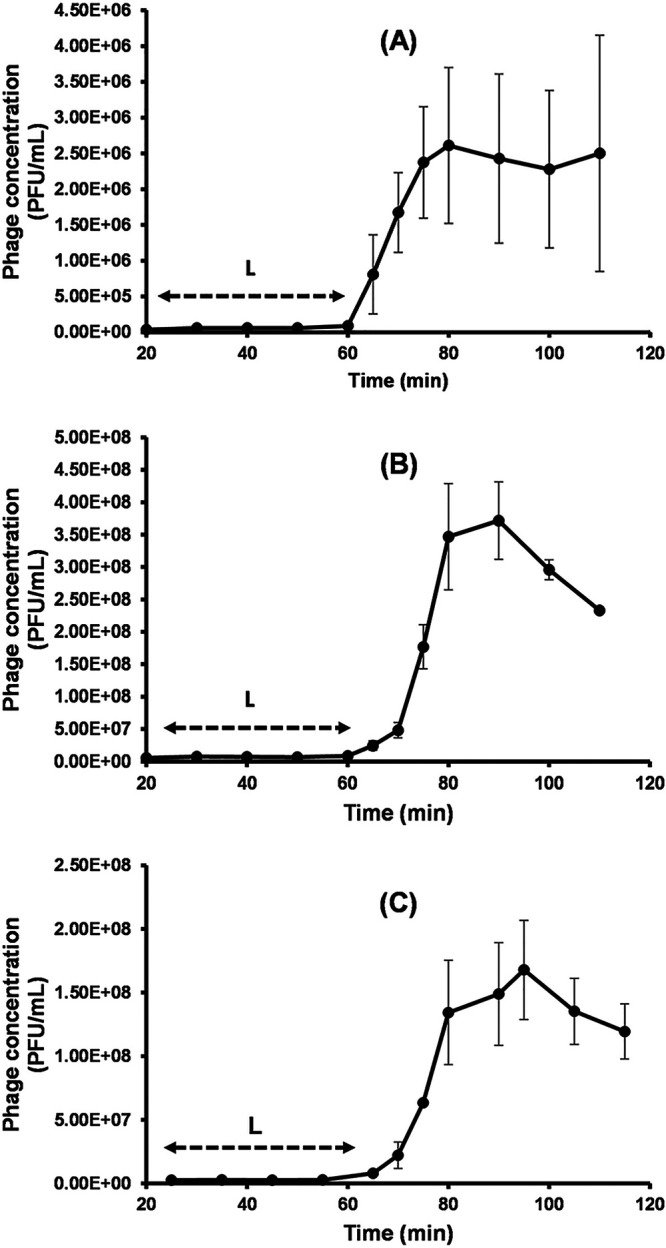
Growth of vB_StyS-LmqsSP1. One-step growth experiments at a multiplicity of infection of 0.1 using Salmonella LT2 (A) and field isolates S1 (B) and S2 (C) as bacterial hosts. Each experiment was performed in triplicate. The highest concentration (PFU/ml) indicates the burst size (S), and double-headed arrows indicate the latent period (L). Error bars represent the standard error of the mean (SEM).
Dose-dependent inhibition of bacterial growth at 37°C.
Results of tests on inhibiting growth of Salmonella populations at 37°C demonstrated a significant time- and dose-dependent effect of the phage (Fig. 4). Even at an MOI of 0.0001, LmqsSP1 significantly inhibited bacterial growth compared to that in the control. Lower optical density compared to that of the control was observed after 8.5 h in cultures with added phages and Salmonella LT2 (P = 0.04), after 6.5 h in cultures with field isolate S1 (P = 0.03), and after 4.5 h in cultures with field isolate S2 (P = 0.03) (Fig. 4). Higher MOIs resulted in faster and more efficient inhibition of bacterial growth compared to that observed at lower MOIs. When LmqsSP1 was applied at an MOI of 10, growth of Salmonella strain LT2 (P = 0.001) and of field isolates S1 (P = 0.05) and S2 (P = 0.009) was completely suppressed (Fig. 4). However, field isolate S2 showed regrowth after 7.5 h and reached an optical density at 600 nm (OD600) of 0.19 after 10 h (Fig. 4C). When LmqsSP1 was applied at MOIs equal to or greater than 0.001, growth inhibition was more effective on the field isolates than on LT2. At MOIs of 0.01 after 10 h, the optical density of S1 and S2 cultures was 0.2 and that of LT2 was 0.3 (Fig. 4).
FIG 4.
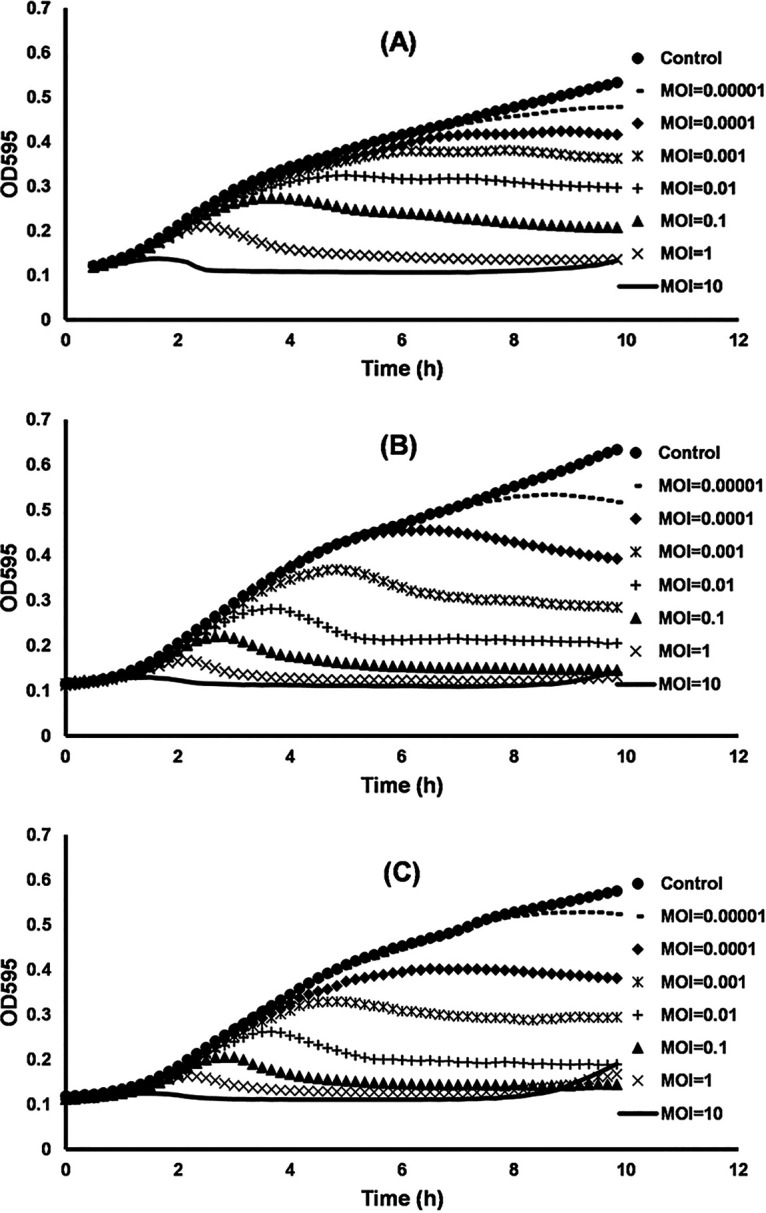
Efficacy of the bacteriophage vB_StyS-LmqsSP1 in inhibiting growth of S. Typhimurium LT2 (A) and field isolates S1 (B) and S2 (C) at different multiplicities of infection (MOIs) in LB. Graphs represent mean values of optical density of Salmonella cultures infected with bacteriophage vB_StyS-LmqsSP1 from three experiments.
LmqsSP1 reduced the Salmonella titer in liquid cultures at 4°C.
When LmqsSP1 was inoculated into lysogeny broth (LB) containing approximately 103 CFU/ml of the respective Salmonella isolate or strain (MOI of 105), significant reductions were observed after 24 h in all experiments. Compared to the control, a reduction of 3 log units was observed in isolate S1, 2.6 log units in isolate S2, and 1.8 log units in Salmonella LT2 (Fig. 5, significance level of P < 0.0001 for all reductions). A higher bacterial density of approximately 104 CFU/ml (MOI of 104) resulted in significant reductions of 3 log units in isolate S2, 2.6 log units in isolate S1, and 1.4 log units in Salmonella LT2 (Fig. 5, significance level of P < 0.0001 for all reductions).
FIG 5.
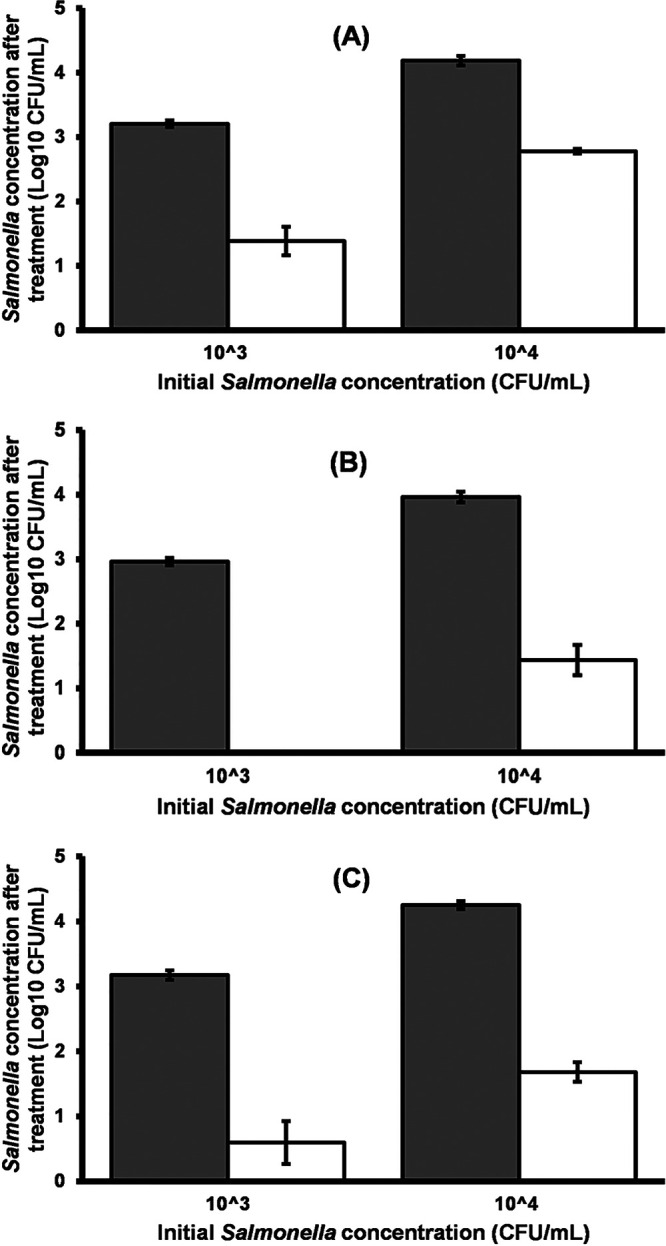
Efficacy of bacteriophage vB_StyS-LmqsSP1 in reducing the quantity of S. Typhimurium LT2 (A) and field isolates S1 (B) and S2 (C) in LB at 4°C. Exponentially growing Salmonella cultures at concentrations of 103 and 104 CFU/ml were inoculated with 109 PFU/ml vB_StyS-LmqsSP1. Gray and white bars indicate the mean concentrations of Salmonella in the control and in the experiment, respectively. Experiments were performed in triplicate. Error bars indicate the standard error of the mean (SEM).
Phage treatment significantly reduced the concentration of Salmonella isolates on chicken skin at 4°C.
Salmonella-contaminated chicken skin was treated with LmqsSP1 at an MOI of 105 to examine the efficiency and stability of the phage on food samples at cooling temperature for 1 week. Results of this experiment are shown in Fig. 6. Concentrations of Salmonella LT2 and the food-derived isolates S1 and S2 on treated chicken skin samples were reduced by more than 1.8 log units compared to the control after 3 h. After 24 h, bacterial counts on phage-treated samples were reduced by 2.2 log units when containing the food-derived isolates S1 (P = 0.021) or S2 (P = 0.032) and by 1.9 log units on samples containing Salmonella LT2 (P = 0.0007) (Fig. 6). In subsequent experiments on phage susceptibility of recovered Salmonella colonies from the tested skin pieces, no phage-resistant colonies were observed. The numbers of plaques on the lawns from those colonies did closely resemble those of the original strains or isolates (data not shown). After 24 h, a mean phage concentration of log10 9.6 ± 0.08 PFU/ml (mean ± standard deviation [SD]) was measured on chicken skin contaminated with the food-derived Salmonella S1 and a concentration of log10 9.5 ± 0.18 PFU/ml was measured on chicken skin contaminated with S2. A mean concentration of log10 9.3 ± 0.56 PFU/ml was detected on the controls containing phages only. In the samples containing Salmonella LT2 and its control, mean phage concentrations of log10 9.2 ± 0.11 PFU/ml and log10 9.4 ± 0.07 PFU/ml were detected, respectively. The measured concentrations represented increases in phage concentration of 51% and 44% during the experiments with S1 and S2, respectively, and a decrease of 51% in the experiment with Salmonella LT2.
FIG 6.
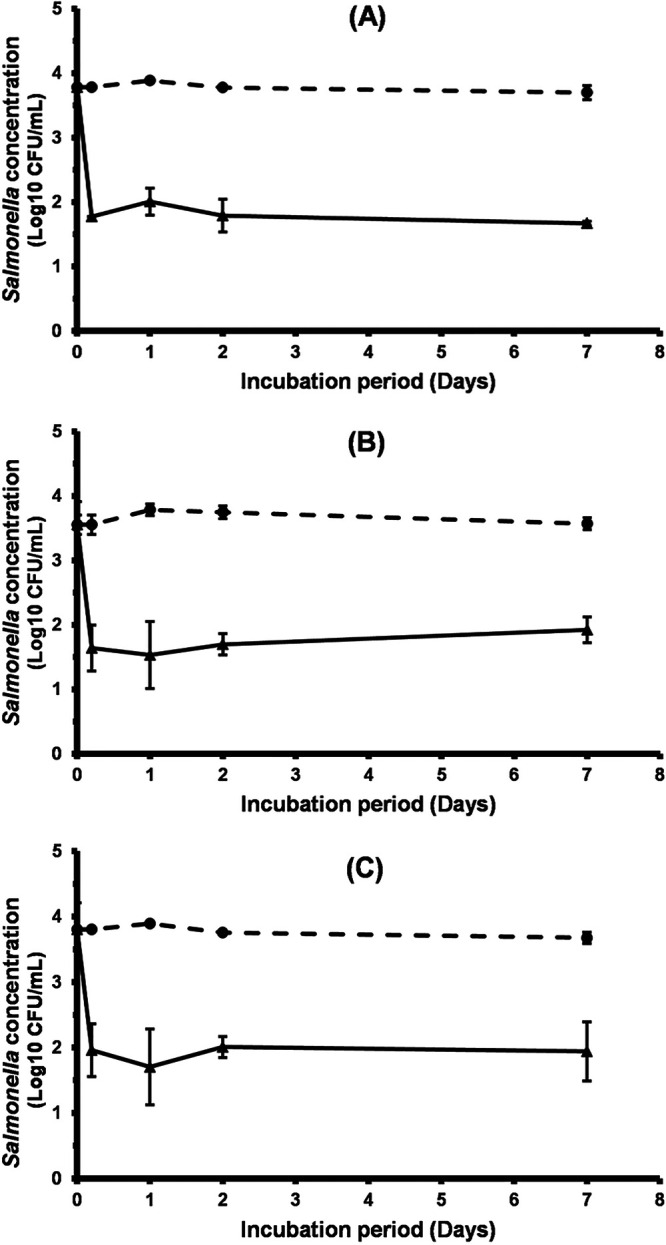
Efficacy of vB_StyS-LmqsSP1 in reducing the prevalence of S. Typhimurium LT2 (A) and field isolates S1 (B) and S2 (C) on chicken skin. Skin pieces were treated with phages at an MOI of 105 and stored for 1 week at 4°C. Mean concentrations of Salmonella on treated skin pieces (triangles, solid lines) and controls (dots, dashed lines) are presented as mean log10 CFU/ml ± standard error of the mean (SEM) of 3 (A) or 4 (B and C) experiments.
In an additional experiment, a lower MOI of 10 was used for treatment of contaminated skin pieces (2.5 × 104 PFU/cm2). No significant reduction in Salmonella counts was observed in this experiment (data not shown).
LmqsSP1 is a new member of the T5-like phages (Tequintavirus genus).
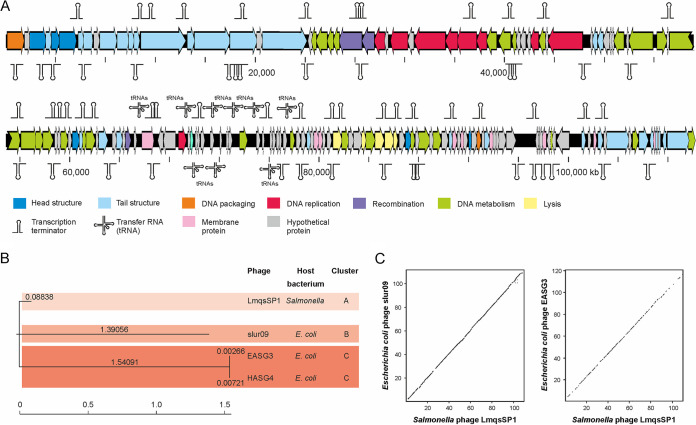
Whole-genome sequencing (WGS) and de novo assembly of the extracted DNA from purified virions resulted in a single contig of ∼110 kb with an average coverage of 200 per consensus base. Manual curation of the assembled consensus sequence resulted in a determination of a final genome size of 109,938 bp. The LmqsSP1 genome exhibited a GC content of 38.8%, which was significantly lower than the GC content of its host bacterium (∼52%). Further DNA alignments against available virus genomes in the GenBank database, conducted using the BLASTn tool of the National Center for Biotechnology Information (NCBI), showed that the Escherichia coli phages slur09 (total score, 1.478 × 105; query coverage, 87%; E value, 0.0; identity: 95.34%; accession no. LN887948.1), vB_EcoS_EASG3 (total score, 1.631 × 105; query coverage, 89%; E value, 0.0; identity, 95.13%; accession no. MK373799.1) and vB_EcoS_HASG4 (total score, 1.620 × 105; query coverage, 89%; E value, 0.0; identity, 95.30%; accession no. MK373797.1), exhibited a close relationship to LmqsSP1. However, the phylogenetic analysis of the phages showed that despite their close relationship, LmqsSP1 clustered independently from slur09 and the almost identical phages vB_EcoS_EASG3 and vB_EcoS_HASG4 (Fig. 7B and C). Nevertheless, due to the fact that all related phages belong to the same taxonomical lineage (viruses; Caudovirales; Siphoviridae; Tequintavirus [synonym T5-like viruses]), we suggest allocating LmqsSP1 to the same viral genus.
FIG 7.
vB_StyS-LmqsSP1 genome analysis and similarities to other phages. (A) Genetic map of examined bacteriophage LmqsSP1. Putative genes are colored according to the predicted functions of their products. (B) Phylogenetic analysis of LmqsSP1 and the related Escherichia coli phages slur09, EASG3, and HASG4. (C) Agreement of the genome organization of LmqsSP1 and the T5-like phages slur09 and EASG3.
Further bioinformatic analysis revealed that the LmqsSP1 genome exhibited 161 putative open reading frames (ORFs), 15 transfer RNAs (tRNAs), and 64 rho-independent transcription terminators (TTs) (see Data Sets S1 and S2 in the supplemental material). Overall, many of the predicted LmqsSP1 gene products showed a close relationship to gene products of phage T5, which represents the prototype and the most prominent member of the tequintaviruses, or other members of T5-like phages (Data Set S1). In general, LmqsSP1 possesses the typical components of T5-like phages. An overview of the functional prediction of the phage-encoded gene products is provided in Fig. 7A. Overall, the majority of the gene products are organized in complex units (i.e., units for DNA metabolism and genome replication lysis of the host cells and virion assembly), but some genes are localized far from their functional units. However, as this organization is in good agreement with other members of T5-like phages (i.e., E. coli phages slur09 and EASG3; Fig. 7C), the delocalization of the genes might not affect their intended function or their concerted activity (i.e., for the generation of virion particles). To assess the suitability of LmqsSP1 for biocontrol or therapeutical applications, in-depth functional predictions of the individual gene products were made. However, we found no gene product involved in a lysogenic behavior (i.e., prophage repressor) and/or chromosomal integration (i.e., phage integrase), supporting the assumption that LmqsSP1 only performs a lytic lifestyle. Furthermore, there was no indication that any LmqsSP1 gene product might be associated with virulence or resistance development. Nevertheless, some gene products showed a structural and functional relationship to homing endonucleases (Data Set S1). As the genes of these nucleases are described as being mobile, copies of them can be scattered on a phage genome and/or within its bacterial host. Genome analysis of DNA from the phage LmqsSP1 after 90 consecutive inoculation cycles with Salmonella host strain LT2 revealed no changes in its genomic traits and thus demonstrated distinct genome stability of the phage.
DISCUSSION
According to the World Health Organization [https://www.who.int/news-room/fact-sheets/detail/salmonella-(non-typhoidal)], Salmonella is one of four key global causes of diarrheal diseases, and approximately one million human cases caused by Salmonella spp. in the United States are food derived (29).
As shown in Fig. 1, the tested bacteriophages LmqsSP1, SP2, SP3, SP4, SP5, and SP6 were able to form plaques on 4 out of 76 food-derived Salmonella isolates with similar or higher efficacy compared to that of the laboratory host strain LT2. LmqsSP1 showed a 10- to 100-fold reduced plaque formation on 11 Salmonella isolates (Fig. 1). On the tested food-derived isolates, the Salmonella phages in this study showed a moderate range of lysed host bacteria, which is comparable to the host range reported for other phages of the Siphoviridae family (30, 31). Highly efficient plaque formation on a broad range of bacterial hosts is known to be an uncommon feature among most phages, especially when bacterial field isolates are tested (32). Thus, phages with different host ranges can be applied simultaneously to increase the range of affected bacterial strains. This application of phage cocktails by adding phages with different host ranges and receptors is especially recommended in commercial settings when the number and susceptibility of bacterial pathogens cannot be determined prior to treatment. Synergistic effects of phage cocktails containing different phages compared to single phages were reported in previous studies (18, 33–35). Phages showing a narrow host range might be especially suitable for targeted reduction of previously identified bacterial strains (36).
The mean burst sizes of LmqsSP1 on the three tested Salmonella cultures were measured to be 50 to 61 PFU/cell. This is in accordance with results from previous studies (20, 37–39), while the measured latent periods of approximately 60 min were longer than those previously reported for Salmonella-specific phages, which showed latent periods of 15 to 25 min (20, 37–39). However, results were consistent with those from studies on the Salmonella-specific siphoviruses STm101, STm118, and PHB06 (19, 38). Comparable with LmqsSP1, STm118 showed a burst size of 48 PFU/cell (38). A phage infecting S. Typhimurium that showed a latent period of 50 ± 5 min was reported previously by Carey-Smith et al. However, this phage failed to lyse its host strain at 5°C even when the MOI exceeded 104 (30).
A short latent period and a large burst size were suggested as indicators for highly efficient phages (22). Nonetheless, the results showed that LmqsSP1 reduces the Salmonella concentration at 4°C on chicken skin samples and in broth if a sufficient concentration of phage was applied.
LmqsSP1 demonstrated a dose-dependent effect in broth, as shown in Fig. 4. This is consistent with the finding that bacterial reduction by LmqsSP1 on chicken skin was dependent on its concentration. For a significant reduction of more than 1 log unit, other authors suggested that a minimum MOI of 103 or 104 is required (40, 41).
After 3 h at 4°C, LmqsSP1 reduced the prevalence of food-derived salmonellae on chicken skin by 1.9 log units. The reduction lasted for the entire storage time. To our knowledge, a limited number of studies with conditions similar to those of our experiments have been published to date (39, 42–44). A study using a siphovirus at a concentration of 107 PFU/ml at 4°C on S. enterica-contaminated chicken skin found that it reduced Salmonella contamination by 2 log units from day 1 to day 7 (42). However, another study using a myovirus at a concentration of 1010 PFU/ml reduced S. Typhimurium contamination on chicken skin by only 1.2 log units from 6 h to 24 h after treatment (39). Other studies applying phages at cooling temperatures on chicken skin detected reductions equal to or less than one log unit (43, 44). At a higher temperature of 8°C, which allows for the growth of Salmonella, application of a siphovirus at an MOI of 104 on chicken skin resulted in a decrease in Salmonella prevalence by 3 log units after 24 h, but after 7 days, the reduction accounted for only 2.43 log units (20). Other studies applied phages on chicken meat instead of skin to test their efficacy at the retail level. Two phages from the Myoviridae family, individually and as a cocktail, decreased S. Enteritidis levels on chicken breast by 1.5 and 2.5 log units after 1 h (33). Another phage at an MOI of 104 caused a reduction of about 1.7 log10 CFU/g in S. Typhimurium LT2 counts following 24 h of treatment (28). A cocktail of a myovirus and a siphovirus eradicated S. Typhimurium and S. Enteritidis within 15 min when chicken breast samples were wrapped in plastic coated with ∼1012 PFU/ml (108 PFU/cm2), and this effectiveness remained stable for 1 week (45). Complete elimination of S. Typhimurium and S. Enteritidis on chicken breast meat was also observed at lower MOIs of 104 to 102 from 3 h to 16 h after phage treatment when phage cocktails were used (46, 47). However, other studies reported that S. Typhimurium and S. Enteritidis populations were reduced by 0.53 and 1.39 log units only, after using a phage cocktail on chicken breast meat in other studies (48). These results show that the food matrix and the temperature at which phages are applied can have a major impact on the observed reduction level.
Phage concentrations on chicken skin after treatment in our study indicated that phage numbers increased by log10 0.3 and 0.2 PFU/ml (51% and 44%), respectively, during the experiments with S1 and S2 and decreased by log10 0.2 PFU/ml (minus 51%) with Salmonella strain LT2 compared to the controls. Considering the high MOI applied, this might indicate that reduction of the strain LT2 may have occurred by inundation without phage replication. In the experiments with field isolates S1 and S2, replication occurred. The resulting increase in total phage numbers exceeded the expected increase after one replication cycle in all available bacterial cells (104 CFU/chicken skin or 10,000-fold, based on the determined burst size of the phages). However, considering the large number of spatial segments on the chicken skin and the likely nonuniform distribution of phages and bacteria in these segments (49), killing by phage inundation without phage replication and killing by a phage infection that resulted in final phage progeny release might have occurred simultaneously in different segments of the matrix. The outcome of the experiments might thus be a result of the predominating process rather than of killing by either inundation or productive phage infection (25).
In poultry meat production, processes are tightly connected, and time intervals for the treatment of meat using phages are limited. However, some studies have reported that reduction in Salmonella concentrations was dependent on extended times of incubation (35, 50, 51). The characteristics of chicken skin include feather follicles, high fat content, and large folded surfaces, which provide protection for Salmonella from eradication by antimicrobial treatments (42, 49, 52). The recovered colonies from treated skins were susceptible to LmqsSP1, and thus full reduction by the phages would have been expected if no protection by the matrix occurred. In addition, the reducing effect on Salmonella load on chicken skin was significantly smaller than that in broth medium at the same temperature, indicating that the efficiency of phages was negatively affected on chicken skin. Short periods of phage activity may limit the efficiency of bacterial reduction under commercial conditions when lytic phage infection is required for reduction (17). This practical aspect suggests that using a high titer of bacteriophage is crucial for achieving efficient results by passive inundation or a combination of lytic infection and passive inundation under commercial conditions of poultry meat production. It has also been emphasized that a homogeneous application of phages is necessary for a good coverage of all host bacteria present (20, 44, 53). To achieve this, an appropriate spray equipment (44) and high volumes of liquid (53) have been recommended. However, it has been proposed that phage application in chilling water at chicken slaughtering plants could be more efficient than spraying (20). While extended use of current chemical treatments, such as chlorine, can lead to a decline in the organoleptic quality of the meat (43), application of phages does not affect the organoleptic properties of chicken carcasses (9). The latter could, additionally, increase the possibility of bacterial reduction through the mechanism of passive inundation, particularly for low bacterial concentrations on chicken skin and, when followed by refrigeration, could consequently restrict the emergence of bacteriophage-insensitive mutants throughout the process until use (54).
Results suggest that bacteriophage-insensitive mutants did not occur after application of LmqsSP1 under the applied conditions. However, while none of the investigated Salmonella isolates recovered from phage-treated skin pieces after phage treatment showed reduced susceptibility to LmqsSP1, all tested reisolates from liquid culture were insensitive after an extended 1-week incubation of LmqsSP1 with S. Typhimurium LT2 at 37°C. Bacterial growth and long storage, optimal nutrient supply, and growth temperatures do not reflect conditions of practical application settings. These conditions might enhance the spread of phage-resistant isolates due to an increased number of replication cycles compared to that in practical settings or to a short incubation period (27, 37). However, further research on conditions that favor the occurrence of reduced phage susceptibility and influence the fitness of resistant isolates is urgently needed (27). After application on chicken skin, the few sensitive bacteria remaining after treatment with phages might result from the lacking adsorption of phages to bacteria hidden in holes and pores in the chicken skin. This is in accordance with other studies (9, 42, 53). In agreement with these results, growth of the tested Salmonella isolates in liquid medium at 37°C was inhibited for a minimum of 7.5 h without regrowth, as shown in Fig. 4. No isolates were recovered for susceptibility testing after this experiment, but results indicate that resistance might only occur after prolonged incubation in liquid culture. In contrast to these findings, regrowth was reported 5 to 8 h after significant reductions or shortly after phage application in other studies (37, 55), and O’Flynn et al. detected bacteriophage-insensitive mutants after 99% reduction of an S. Typhimurium strain by the commercial phage Felix 01 (55). Bai et al. concluded that the rapid occurrence of bacteriophage resistance in bacterial populations is a significant limitation for the efficacy of phage application (6). However, the findings of other studies were in accordance with our results (7, 42).
In conclusion, the rapid and significant reduction in the number of foodborne S. Typhimurium isolates in liquid medium for 10 h without considerable bacterial regrowth and under conditions resembling the situation in the food production chain on chicken skin demonstrates that LmqsSP1 is a potential candidate for biocontrol of Salmonella in food. LmqsSP1 could be used for reducing the prevalence of known Salmonella strains in bacteriophage cocktails or in combination with other techniques as a multihurdle approach. Based on the genome sequence, the phage LmqsSP1 was assessed to be well suited for phage application, as no genes with undesired effects were identified. Some gene products showed a structural and functional relationship to homing endonucleases. These gene copies might represent target sites for homologous recombination, potentially leading to genomic adaptions of the bacterial host and/or the phage. In our study, we found no indication of a genetic instability of the phage genome that might be associated with potential changes of either its lytic activity or the spectrum of the infected bacterial hosts. However, further long-term analyses are necessary to assess the genetic stability of the phage in detail. Based on the prevailing results from the bioinformatics analysis, LmqsSP1 seemed to be suited for application uses.
Nonetheless, the presence of some mobile genes might influence the genetic stability of the phage during successive or long-term usage. To date, there has been no indication that the phage sequence and/or the organization of the genes is significantly changed during in vitro reduction tests. Further experiments are necessary to determine genetic stability in vivo. In general, members of the T5-like phages are broadly assigned as promising tools for biocontrol or therapeutic applications against E. coli, Salmonella, and other bacteria (56–58).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The S. enterica subsp. enterica serovar Typhimurium strain LT2 was used for bacteriophage detection and isolation. Host range analysis was conducted using 76 Salmonella isolates originating from current food samples of different animal species (Table 2), S. Typhimurium host strain LT2, and two E. coli O157:H7 strains (DSM 19206 and DSM 17076). Host bacteria were cultivated on blood agar (Columbia agar with 5% sheep blood; Thermo Scientific, Inc., Waltham, MA) at 37°C overnight for all experiments. Thereafter, one colony was used for a subsequent culturing in lysogeny broth (LB; Carl Roth GmbH, Karlsruhe, Germany) supplemented with CaCl2 (at a final concentration of 1 mM/liter) at 37°C for 3 h for an approximate optical density at 600 nm (OD600) of 0.5. This early-logarithmic-phase culture was then used for phage infection assays.
Bacteriophage isolation and propagation.
The soft agar overlay technique was used to detect Salmonella-specific phages from different sample matrices. Out of 52 samples originating from different farms in northern Germany, the majority were chicken feces (n = 34) or cecal contents of poultry (chickens [n = 6], turkeys [n = 2], and Peking ducks [n = 1]), while only nine samples were nasal swabs (n = 5) or manure (n = 4) from cattle.
For sample preparation, ∼6 g of each matrix was suspended for 1 min in 30 ml of sodium chloride-magnesium sulfate (SM) buffer (50 mM Tris-HCl, 8 mM magnesium sulfate, 100 mM sodium chloride, and 0.01% gelatin [pH 7.5]) using a Turrax homogenizer (IKA-Werke GmbH & Co. KG, Staufen, Germany). After overnight shaking in the refrigerator, the suspensions were centrifuged twice at 4°C (4,000 × g for 20 min and 13,000 × g for 10 min). The remaining supernatant was filtered through a 0.22-μm membrane filter (VWR International GmbH, Darmstadt, Germany). For phage isolation and purification, S. Typhimurium strain LT2 was cultured with the sample using the soft agar overlay technique as described previously (59). Briefly, 100 μl of sample and 100 μl of exponentially growing S. Typhimurium strain LT2 were added to 5 ml of LB soft agar (0.4% [wt/vol] agar bacteriological and 2 mM CaCl2), poured onto petri dishes containing LB agar (1.5% [wt/vol] agar bacteriological and 2 mM CaCl2), and incubated at 37°C overnight. Phages were isolated and purified by a successive 3-fold picking and plating procedure of single plaques. Subsequently, phages were propagated to obtain concentrations of 109 to 1010 PFU/ml and stored at 4°C for further use. The phage titer was determined by plating 100 μl of a 10-fold serial dilution series of the phage suspension on S. Typhimurium LT2 using the soft agar overlay technique. For testing phage genome stability, 100 μl of filtrated phage culture was serially incubated with 100 μl of S. Typhimurium strain LT2 overnight culture for 24 to 72 h at 37°C in 5 ml Mueller-Hinton broth. The phage filtrate of one incubation was used for the subsequent 90 consecutive incubation cycles. The final phage culture was used for phage propagation and DNA extraction, sequencing, and bioinformatics analysis, as described below.
Host range and efficiency of plating.
Serial dilutions were spotted on soft agar overlays to determine the relative efficiency of plating (EOP) as previously described by Kutter et al. (60). Briefly, the bacterial overlay agar was prepared as described above using the food-derived Salmonella isolates, S. Typhimurium strain LT2 and the E. coli strains without the addition of phages. Subsequently, 10 μl of 10-fold serial dilutions of the purified phages were applied on these lawns. The plates were incubated at 37°C for 24 h after the phage suspensions had been absorbed by the medium. Each test was performed in triplicate. The sensitivity of the tested bacteria to the phages was determined by counting the number of plaques in the spots. The relative efficiency of plating (EOP) was defined as the phage titer on a given bacterial lawn divided by the maximum titer observed on the original host S. Typhimurium strain LT2.
Negative staining of phages.
Thin carbon support films were prepared by evaporating a carbon thread onto a freshly cleaved mica surface (SCD 500; Bal-Tec). After cutting small pieces of mica (approximately 3 mm in length), the phages were negatively stained with 4% (wt/vol) aqueous uranyl acetate, pH 5.0, in accordance with the method of Valentine et al. (61). In brief, phages were adsorbed for 15 to 30 sec onto the carbon film, washed in TE buffer (10 mM Tris and 1 mM EDTA [pH 6.9]) and picked up with a 300-mesh nickel grid, blotted dry on a filter paper, and subsequently air-dried. Dried samples were examined in a transmission electron microscope (TEM 910; Carl Zeiss Industrielle Messtechnik GmbH, Oberkochen, Germany) at an acceleration voltage of 80 kV. Images were taken at calibrated magnifications using a line replica. Images were recorded digitally with a slow-scan closed circuit device (CCD) camera (1024 × 1024; Proscan Elektronische Systeme GmbH, Scheuring, Germany) with iTEM software (Olympus Soft Imaging Solutions GmbH, Münster, Germany).
The head diameter and tail length were determined using ImageJ software version 1.51q and calculating the average size from a minimum of 10 measurements.
One-step growth experiments.
One-step growth experiments were performed as previously described by Hyman and Abedon (62), with some modifications. For this purpose, LmqsSP1 was incubated at 37°C with Salmonella LT2 and the food-derived isolates S1 and S2. Briefly, the cultures of the host bacteria were grown to an OD600 of 0.5 and mixed with LmqsSP1 at a multiplicity of infection (MOI) of 0.1. After allowing the phage to adsorb to the target bacteria for 10 min at room temperature, samples were centrifuged at 1,300 × g for 4 min at 4°C and the excess free phages in the supernatant were discarded. The pelleted cells were suspended in fresh LB and placed in a heating block at 37°C for the duration of the experiment. Samples were withdrawn, and concentrations of phages in the samples were measured immediately at 5- or 10-min intervals for up to 2 h. Experiments were performed in duplicate and were repeated three times. The latent period and burst size were calculated as described previously (62).
Bacterial challenge tests.
(i) In vitro growth inhibition experiments at 37°C. Growth inhibition of S. Typhimurium LT2 and of the food-derived isolates S1 and S2 was tested using the method described by O’Flynn et al. (55), with some modifications. Briefly, bacterial cells were grown to a McFarland standard of 2.4 and diluted to McFarland 0.5 in fresh LB (approximately 108 CFU/ml). Three hundred microliters of each aforementioned culture and of LmqsSP1 at concentrations of 103 to 109 PFU/ml were mixed in a 48-well microplate, and after 10 min at room temperature, incubated in a Spark microplate reader (Tecan Group AG, Männedorf, Switzerland) at 37°C. Bacterial cultures inoculated with LB instead of phages served as negative controls. The absorbance at 595 nm was measured every 10 min during a 10-h period.
(ii) Reduction experiments in vitro at 4°C.
To determine the efficacy of LmqsSP1 for the reduction of salmonella prevalence at 4°C, phages were added to bacterial cultures in LB at MOIs of 104 and 105 CFU/ml. Briefly, exponentially growing salmonellae (strain LT2 and the food-derived isolates S1 and S2) were diluted in LB to final concentrations of 104 and 103 CFU/ml, and 100 μl of LmqsSP1 at a titer of 1010 PFU/ml was added. The same amount of SM buffer (50 mM Tris-HCl, 8 mM magnesium sulfate, 100 mM sodium chloride, and 0.01% gelatin [pH 7.5]) was added instead of phage for negative controls. The number of viable Salmonella cells was determined after incubation at 4°C for 24 h by using the pour plate method in accordance with ISO 4833-1:2013 (63) with slight modifications. In brief, LB agar (1.5% agar; Carl Roth GmbH & Co. KG) was poured into petri dishes containing 1-ml serial dilutions of the samples. The agar plates were prepared in duplicate and incubated at 37°C, and colonies were counted after 24 h. Each test was performed three times.
(iii) Reduction experiments on chicken skin at 4°C.
To determine the efficacy of LmqsSP1 for reduction of Salmonella prevalence on chicken skin, the experiments were carried out following the method described by Guenther et al. (53), with some modifications. Briefly, irradiated chicken skin pieces of 25 cm2 (radiation in a Cobalt-60 Gamma irradiation facility 3000 at a dose of 13.77 kGy ± 1 kGy) were inoculated with 100 μl of the respective Salmonella isolate or strain at a concentration of 105 CFU/ml on both sides, aiming at an initial concentration of approximately 2 × 103 CFU/cm2. Skin pieces were incubated at 4°C for 1 h, allowing the bacteria to adapt to the low temperature. Subsequently, 100 μl of LmqsSP1 were applied to the skin pieces, aiming for a concentration of about 2.5 × 108 PFU/cm2. On the negative controls, 100 μl SM buffer (50 mM Tris-HCl, 8 mM magnesium sulfate, 100 mM sodium chloride, and 0.01% gelatin [pH 7.5]) was applied instead of phages, and positive controls were treated with phages without Salmonella contamination. One skin piece was used for confirming the sterility after incubation. Samples were incubated for 1 week at 4°C in a refrigerator. An additional experiment with a phage concentration of 2.5 × 104 PFU/cm2 was carried out for 48 h. The experiments were conducted in three replicates for S. Typhimurium LT2 and repeated four times for field isolates S1 and S2.
Concentrations of viable bacteria on the skin pieces were enumerated after 3 h, 24 h, and 48 h and after 1 week of storage, respectively. For this purpose, 20 ml SM buffer was added to each skin sample in a sterile plastic bag, and samples were shaken and squeezed for 2 min. A 1-ml aliquot of the suspension was serially diluted 1:10 and plated in duplicate on LB agar using the pour plate method. Plates were incubated for 48 h at 37°C, and Salmonella counts were enumerated after 24 h and 48 h.
Phage concentration on the skin pieces was determined after 24 h by using the soft agar overlay technique as described above. Briefly, samples derived from enumeration of bacteria were filtrated through 0.22-μm-pore-size filters (VWR International GmbH, Hannover, Germany), serially diluted (1:10) in SM buffer, and plated in duplicate on LB agar.
Bacterial resistance testing.
To assess the phage susceptibility among bacteria that had survived treatment with LmqsSP1 in the experiments on chicken skin, the soft agar overlay technique was used to verify plaque formation. Salmonella colonies were picked from LB agar and were plated with 100 μl of serially diluted phage suspension containing up to 109 PFU/ml. Isolates were considered to show reduced susceptibility when plaque formation was reduced by more than one logarithmic unit compared to plaque formation on the original Salmonella isolate. Bacterial resistance was tested after challenge tests and in a separate experiment after an extended incubation period of 7 days in Mueller-Hinton broth with Salmonella LT2. After incubation, dilutions of the culture were plated on LB agar, and 50 Salmonella isolates were picked and examined for their susceptibility to LmqsSP1 as described above for host range testing but with only one replicate per isolate.
Extraction of phage DNA, whole-genome sequencing, and bioinformatics analysis.
For DNA extraction, phage solution was prepared using a cesium chloride density gradient as described previously (64), with some modifications. Subsequently, DNA was extracted by using the Wizard kit in accordance with the manufacturer’s instructions (Promega GmbH, Walldorf, Germany). Briefly, 300 ml of phage lysate from a liquid culture in LB containing about 109 PFU/ml was centrifuged for sedimentation of phages at 24,000 × g for 2 h at 10°C (Avanti J-26S XP; Beckmann Coulter Inc., Brea, CA). After removing the supernatant, phage sediment was suspended overnight in 1.6 ml of SM buffer (50 mM Tris-HCl, 8 mM magnesium sulfate, 100 mM sodium chloride, and 0.01% gelatin [pH 7.5]) on an orbital shaker at 120 rpm and filtered through 0.22-μm-pore-size filters (Rotilabo syringe filter; Carl Roth GmbH & Co. KG). Subsequently, the gradient was prepared by pipetting 500 μl of four cesium chloride solutions at a ρ of 1.6, 1.5, 1.4, or 1.3 g/cm3 sequentially into the bottom of a thin-walled tube. An aliquot (2 ml) of the phage suspension was layered on the top of the CsCl gradient, and tubes were subjected to ultracentrifugation in an Optima XPN-100 with an SW 60 Ti rotor (Beckmann Coulter, Inc.) at 165,100 × g and 4°C for 2 h. Phage bands were collected and dialyzed against SM buffer overnight. The phage suspensions were treated with 10-fold reaction buffer (100 mM Tris-HCl [pH 7.5], 25 mM MgCl2, and 1 mM CaCl2; Thermo Fisher Scientific, Inc., Waltham, MA), 0.2 mg/ml RNase A, and 0.002 U/μl DNase I (Thermo Fisher Scientific, Inc.) at 37°C overnight, and DNA was extracted using the Promega Wizard kit in accordance with the manufacturer’s instructions. Whole-genome sequencing of phage DNA was performed in-house at the German Federal Institute for Risk Assessment (BfR), Berlin, Germany. DNA sequencing libraries were generated using the Nextera XT DNA Library Flex preparation kit (Illumina Inc., San Diego, CA) according to the recommendations of the manufacturer. Short-read, paired-end sequencing was conducted on an Illumina MiSeq benchtop device using the MiSeq Reagent v3 600-cycle kit (Illumina). Long-read WGS was conducted using phage DNA on a MinIon device (Oxford Nanopore, Oxford, UK). The raw reads from both short-read and long-read sequencing platforms were subjected to a hybrid assembly (Unicycler v.0.44). Initial annotation was performed using the Pathosystems Resource Integration Center (www.patricbrc.org). For final prediction of specific gene product functions, BLASTp (NCBI) was used. Further basic sequence analyses and DNA alignments were carried out using DS Gene (Accelrys, Inc., San Diego, CA). Prediction of genetic elements (i.e., open reading frames [ORFs], transcription terminators, and tRNAs) on the phage genome was conducted as described previously (65, 66).
Statistical analysis.
Bacterial concentrations of the groups were compared using a paired t test with Bonferroni correction for detecting significant differences. Comparison of bacterial numbers in treated groups and the control group was performed by using Dunnett’s test. Analysis of trends over time was carried out using mixed models, considering time as the random effect. All statistical analyses of bacterial challenge tests were performed using the R software package version 3.5.3, and clustering (20) of bacteriophage host range was performed using version 3.5.2.
Data availability.
The complete nucleotide sequence of Salmonella phage vB_StyS-LmqsSP1 was deposited in GenBank under accession number MT577844.
ACKNOWLEDGMENTS
Whole-genome sequencing (WGS) of the phage genome was financially supported by grant 43-001 from the German Federal Institute for Risk Assessment. Golshan Shakeri was awarded a scholarship by Iran Ministry of Science, Research and Technology as a visiting scholar at the University of Veterinary Medicine Hannover, Germany.
Footnotes
Supplemental material is available online only.
Contributor Information
Sophie Kittler, Email: sophie.kittler@tiho-hannover.de.
Gladys Alexandre, University of Tennessee at Knoxville.
REFERENCES
- 1.Ferrari RG, Rosario DKA, Cunha-Neto A, Mano SB, Figueiredo EES, Conte-Junior CA. 2019. Worldwide epidemiology of Salmonella serovars in animal-based foods: a meta-analysis. Appl Environ Microbiol 85:e00591-19. 10.1128/AEM.00591-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.EFSA, ECDC. 2019. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFSA J 17:5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marder Mph EP, Griffin PM, Cieslak PR, Dunn J, Hurd S, Jervis R, Lathrop S, Muse A, Ryan P, Smith K, Tobin-D’Angelo M, Vugia DJ, Holt KG, Wolpert BJ, Tauxe R, Geissler AL. 2018. Preliminary incidence and trends of infections with pathogens transmitted commonly through food—Foodborne Diseases Active Surveillance Network, 10 U.S. sites, 2006–2017. MMWR Morb Mortal Wkly Rep 67:324–328. 10.15585/mmwr.mm6711a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leverentz B, Conway WS, Alavidze Z, Janisiewicz WJ, Fuchs Y, Camp MJ, Chighladze E, Sulakvelidze A. 2001. Examination of bacteriophage as a biocontrol method for Salmonella on fresh-cut fruit: a model study. J Food Prot 64:1116–1121. 10.4315/0362-028x-64.8.1116. [DOI] [PubMed] [Google Scholar]
- 5.Pawlowska AM, Zannini E, Coffey A, Arendt EK. 2012. “Green preservatives”: combating fungi in the food and feed industry by applying antifungal lactic acid bacteria. Adv Food Nutr Res 66:217–238. 10.1016/B978-0-12-394597-6.00005-7. [DOI] [PubMed] [Google Scholar]
- 6.Bai J, Jeon B, Ryu S. 2019. Effective inhibition of Salmonella Typhimurium in fresh produce by a phage cocktail targeting multiple host receptors. Food Microbiol 77:52–60. 10.1016/j.fm.2018.08.011. [DOI] [PubMed] [Google Scholar]
- 7.Carlton RM, Noordman WH, Biswas B, de Meester ED, Loessner MJ. 2005. Bacteriophage P100 for control of Listeria monocytogenes in foods: genome sequence, bioinformatic analyses, oral toxicity study, and application. Regul Toxicol Pharmacol 43:301–312. 10.1016/j.yrtph.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 8.Lewis R, Hill C. 2020. Overcoming barriers to phage application in food and feed. Curr Opin Biotechnol 61:38–44. 10.1016/j.copbio.2019.09.018. [DOI] [PubMed] [Google Scholar]
- 9.Moye ZD, Woolston J, Sulakvelidze A. 2018. Bacteriophage applications for food production and processing. Viruses 10:205. 10.3390/v10040205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pietracha D, Misiewicz A. 2016. Use of products containing a phage in food industry as a new method for Listeria monocytogenes elimination from food (Listeria monocytogenes phages in food industry)—a review. Czech J Food Sci 34:1–8. 10.17221/217/2015-CJFS. [DOI] [Google Scholar]
- 11.USDA. 2019. Safe and suitable ingredients used in the production of meat, poultry, and egg products. Related documents for FSIS Directive 71201.
- 12.Debarbieux L, Pirnay JP, Verbeken G, De Vos D, Merabishvili M, Huys I, Patey O, Schoonjans D, Vaneechoutte M, Zizi M, Rohde C. 2016. A bacteriophage journey at the European Medicines Agency. FEMS Microbiol Lett 363:fnv225. 10.1093/femsle/fnv225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.EFSA. 2009. Scientific opinion on “The use and mode of action of bacteriophages in food production.” EFSA J 7:1076. [Google Scholar]
- 14.Nagel TE, Chan BK, De Vos D, El-Shibiny A, Kang’ethe EK, Makumi A, Pirnay JP. 2016. The developing world urgently needs phages to combat pathogenic bacteria. Front Microbiol 7:882. 10.3389/fmicb.2016.00882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kassa T. 2021. Bacteriophages against pathogenic bacteria and possibilities for future application in Africa. Infect Drug Resist 14:17–31. 10.2147/IDR.S284331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khalid A, Lin RCY, Iredell JR. 2020. A phage therapy guide for clinicians and basic scientists: background and highlighting applications for developing countries. Front Microbiol 11:599906. 10.3389/fmicb.2020.599906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grant A, Parveen S, Schwarz J, Hashem F, Vimini B. 2017. Reduction of Salmonella in ground chicken using a bacteriophage. Poult Sci 96:2845–2852. 10.3382/ps/pex062. [DOI] [PubMed] [Google Scholar]
- 18.Augustine J, Bhat SG. 2015. Biocontrol of Salmonella Enteritidis in spiked chicken cuts by lytic bacteriophages PhiSP-1 and PhiSP-3. J Basic Microbiol 55:500–503. 10.1002/jobm.201400257. [DOI] [PubMed] [Google Scholar]
- 19.Chen Y, Sun E, Song J, Tong Y, Wu B. 2018. Three Salmonella enterica serovar Enteritidis bacteriophages from the Siphoviridae family are promising candidates for phage therapy. Can J Microbiol 64:865–875. 10.1139/cjm-2017-0740. [DOI] [PubMed] [Google Scholar]
- 20.Kang HW, Kim JW, Jung TS, Woo GJ. 2013. wksl3, a new biocontrol agent for Salmonella enterica serovars Enteritidis and Typhimurium in foods: characterization, application, sequence analysis, and oral acute toxicity study. Appl Environ Microbiol 79:1956–1968. 10.1128/AEM.02793-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ross A, Ward S, Hyman P. 2016. More is better: selecting for broad host range bacteriophages. Front Microbiol 7:1352. 10.3389/fmicb.2016.01352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gill JJ, Hyman P. 2010. Phage choice, isolation, and preparation for phage therapy. Curr Pharm Biotechnol 11:2–14. 10.2174/138920110790725311. [DOI] [PubMed] [Google Scholar]
- 23.Zampara A, Sørensen MCH, Elsser-Gravesen A, Brøndsted L. 2017. Significance of phage-host interactions for biocontrol of Campylobacter jejuni in food. Food Control 73:1169–1175. 10.1016/j.foodcont.2016.10.033. [DOI] [Google Scholar]
- 24.Chinivasagam HN, Estella W, Maddock L, Mayer DG, Weyand C, Connerton PL, Connerton IF. 2020. Bacteriophages to control Campylobacter in commercially farmed broiler chickens, in Australia. Front Microbiol 11:632. 10.3389/fmicb.2020.00632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weld RJ, Butts C, Heinemann JA. 2004. Models of phage growth and their applicability to phage therapy. J Theor Biol 227:1–11. 10.1016/S0022-5193(03)00262-5. [DOI] [PubMed] [Google Scholar]
- 26.Cairns BJ, Timms AR, Jansen VA, Connerton IF, Payne RJ. 2009. Quantitative models of in vitro bacteriophage-host dynamics and their application to phage therapy. PLoS Pathog 5:e1000253. 10.1371/journal.ppat.1000253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oechslin F. 2018. Resistance development to bacteriophages occurring during bacteriophage therapy. Viruses 10:351. 10.3390/v10070351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zinno P, Devirgiliis C, Ercolini D, Ongeng D, Mauriello G. 2014. Bacteriophage P22 to challenge Salmonella in foods. Int J Food Microbiol 191:69–74. 10.1016/j.ijfoodmicro.2014.08.037. [DOI] [PubMed] [Google Scholar]
- 29.Scallan E, Griffin PM, Angulo FJ, Tauxe RV, Hoekstra RM. 2011. Foodborne illness acquired in the United States—unspecified agents. Emerg Infect Dis 17:16–22. 10.3201/eid1701.091101p2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carey-Smith GV, Billington C, Cornelius AJ, Hudson JA, Heinemann JA. 2006. Isolation and characterization of bacteriophages infecting Salmonella spp. FEMS Microbiol Lett 258:182–186. 10.1111/j.1574-6968.2006.00217.x. [DOI] [PubMed] [Google Scholar]
- 31.Demczuk W, Ahmed R, Ackermann HW. 2004. Morphology of Salmonella enterica serovar Heidelberg typing phages. Can J Microbiol 50:873–875. 10.1139/w04-075. [DOI] [PubMed] [Google Scholar]
- 32.Hyman P, Abedon ST. 2010. Bacteriophage host range and bacterial resistance. Adv Appl Microbiol 70:217–248. 10.1016/S0065-2164(10)70007-1. [DOI] [PubMed] [Google Scholar]
- 33.Bao H, Zhang P, Zhang H, Zhou Y, Zhang L, Wang R. 2015. Bio-control of Salmonella Enteritidis in foods using bacteriophages. Viruses 7:4836–4853. 10.3390/v7082847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan BK, Abedon ST, Loc-Carrillo C. 2013. Phage cocktails and the future of phage therapy. Future Microbiol 8:769–783. 10.2217/fmb.13.47. [DOI] [PubMed] [Google Scholar]
- 35.Hooton SP, Atterbury RJ, Connerton IF. 2011. Application of a bacteriophage cocktail to reduce Salmonella Typhimurium U288 contamination on pig skin. Int J Food Microbiol 151:157–163. 10.1016/j.ijfoodmicro.2011.08.015. [DOI] [PubMed] [Google Scholar]
- 36.Kittler S, Mengden R, Korf IHE, Bierbrodt A, Wittmann J, Plotz M, Jung A, Lehnherr T, Rohde C, Lehnherr H, Klein G, Kehrenberg C. 2020. Impact of bacteriophage-supplemented drinking water on the E. coli population in the chicken gut. Pathogens 9:293. 10.3390/pathogens9040293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park M, Lee JH, Shin H, Kim M, Choi J, Kang DH, Heu S, Ryu S. 2012. Characterization and comparative genomic analysis of a novel bacteriophage, SFP10, simultaneously inhibiting both Salmonella enterica and Escherichia coli O157:H7. Appl Environ Microbiol 78:58–69. 10.1128/AEM.06231-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phothaworn P, Dunne M, Supokaivanich R, Ong C, Lim J, Taharnklaew R, Vesaratchavest M, Khumthong R, Pringsulaka O, Ajawatanawong P, Klumpp J, Brown N, Imam M, Clokie MRJ, Galyov EE, Korbsrisate S. 2019. Characterization of flagellotropic, chi-like Salmonella phages isolated from Thai poultry farms. Viruses 11:520. 10.3390/v11060520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duc HM, Son HM, Yi HPS, Sato J, Ngan PH, Masuda Y, Honjoh KI, Miyamoto T. 2020. Isolation, characterization and application of a polyvalent phage capable of controlling Salmonella and Escherichia coli O157:H7 in different food matrices. Food Res Int 131:108977. 10.1016/j.foodres.2020.108977. [DOI] [PubMed] [Google Scholar]
- 40.Campden BRI. 2016. Systematic and critical review on the potential use of bacteriophage on foods. Project code FS102079. https://www.food.gov.uk/research/research-projects/systematic-and-critical-review-on-the-potential-use-of-bacteriophages-on-foods.
- 41.Sharma CS, Dhakal J, Nannapaneni R. 2015. Efficacy of lytic bacteriophage preparation in reducing Salmonella in vitro, on turkey breast cutlets, and on ground turkey. J Food Prot 78:1357–1362. 10.4315/0362-028X.JFP-14-585. [DOI] [PubMed] [Google Scholar]
- 42.El-Shibiny A, El-Sahhar S, Adel M. 2017. Phage applications for improving food safety and infection control in Egypt. J Appl Microbiol 123:556–567. 10.1111/jam.13500. [DOI] [PubMed] [Google Scholar]
- 43.Hungaro HM, Mendonça RCS, Gouvêa DM, Vanetti MCD, Pinto CLDO. 2013. Use of bacteriophages to reduce Salmonella in chicken skin in comparison with a chemical agents. Food Res Int 52:75–81. 10.1016/j.foodres.2013.02.032. [DOI] [Google Scholar]
- 44.Goode D, Allen VM, Barrow PA. 2003. Reduction of experimental Salmonella and Campylobacter contamination of chicken skin by application of lytic bacteriophages. Appl Environ Microbiol 69:5032–5036. 10.1128/AEM.69.8.5032-5036.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Phothaworn P, Supokaivanich R, Lim J, Klumpp J, Imam M, Kutter E, Galyov EE, Dunne M, Korbsrisate S. 2020. Development of a broad-spectrum Salmonella phage cocktail containing viunalike and jerseylike viruses isolated from Thailand. Food Microbiol 92:103586. 10.1016/j.fm.2020.103586. [DOI] [PubMed] [Google Scholar]
- 46.Islam MS, Zhou Y, Liang L, Nime I, Liu K, Yan T, Wang X, Li J. 2019. Application of a phage cocktail for control of Salmonella in foods and reducing biofilms. Viruses 11:841. 10.3390/v11090841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Esmael A, Azab E, Gobouri AA, Nasr-Eldin MA, Moustafa MMA, Mohamed SA, Badr OAM, Abdelatty AM. 2021. Isolation and characterization of two lytic bacteriophages infecting a multi-drug resistant Salmonella Typhimurium and their efficacy to combat salmonellosis in ready-to-use foods. Microorganisms 9:423. 10.3390/microorganisms9020423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petsong K, Benjakul S, Chaturongakul S, Switt AIM, Vongkamjan K. 2019. Lysis Profiles of Salmonella phages on Salmonella isolates from various sources and efficiency of a phage cocktail against S. Enteritidis and S. Typhimurium. Microorganisms 7:100. 10.3390/microorganisms7040100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang X, Peng Z, Li P, Mao Y, Shen R, Tao R, Diao X, Liu L, Zhao Y, Luo X. 2020. Complex internal microstructure of feather follicles on chicken skin promotes the bacterial cross-contamination of carcasses during the slaughtering process. Front Microbiol 11:571913. 10.3389/fmicb.2020.571913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sukumaran AT, Nannapaneni R, Kiess A, Sharma CS. 2015. Reduction of Salmonella on chicken meat and chicken skin by combined or sequential application of lytic bacteriophage with chemical antimicrobials. Int J Food Microbiol 207:8–15. 10.1016/j.ijfoodmicro.2015.04.025. [DOI] [PubMed] [Google Scholar]
- 51.Yeh Y, Purushothaman P, Gupta N, Ragnone M, Verma SC, de Mello AS. 2017. Bacteriophage application on red meats and poultry: effects on Salmonella population in final ground products. Meat Sci 127:30–34. 10.1016/j.meatsci.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 52.Atterbury RJ, Connerton PL, Dodd CE, Rees CE, Connerton IF. 2003. Application of host-specific bacteriophages to the surface of chicken skin leads to a reduction in recovery of Campylobacter jejuni. Appl Environ Microbiol 69:6302–6306. 10.1128/AEM.69.10.6302-6306.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guenther S, Huwyler D, Richard S, Loessner MJ. 2009. Virulent bacteriophage for efficient biocontrol of Listeria monocytogenes in ready-to-eat foods. Appl Environ Microbiol 75:93–100. 10.1128/AEM.01711-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Atterbury RJ, Gigante AM, Rubio Lozano MS, Mendez Medina RD, Robinson G, Alloush H, Barrow PA, Allen VM. 2020. Reduction of Salmonella contamination on the surface of chicken skin using bacteriophage. Virology J 17:98. 10.1186/s12985-020-01368-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O’Flynn G, Coffey A, Fitzgerald GF, Ross RP. 2006. The newly isolated lytic bacteriophages st104a and st104b are highly virulent against Salmonella enterica. J Appl Microbiol 101:251–259. 10.1111/j.1365-2672.2005.02792.x. [DOI] [PubMed] [Google Scholar]
- 56.Amarillas L, Rubi-Rangel L, Chaidez C, Gonzalez-Robles A, Lightbourn-Rojas L, Leon-Felix J. 2017. Isolation and characterization of phiLLS, a novel phage with potential biocontrol agent against multidrug-resistant Escherichia coli. Front Microbiol 8:1355. 10.3389/fmicb.2017.01355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Niu YD, Stanford K, Kropinski AM, Ackermann HW, Johnson RP, She YM, Ahmed R, Villegas A, McAllister TA. 2012. Genomic, proteomic and physiological characterization of a T5-like bacteriophage for control of Shiga toxin-producing Escherichia coli O157:H7. PLoS One 7:e34585. 10.1371/journal.pone.0034585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hartman S, Zeng C, O’Leary C, Newkirk H, Kongari R, Gill J, Liu M. 2019. Complete genome sequence of Salmonella enterica serovar enteritidis siphophage seafire. Microbiol Resour Announc 8:e01167-19. 10.1128/MRA.01167-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Adams MH. 1959. Bacteriophages, vol 1. Interscience Publishers, New York, NY. [Google Scholar]
- 60.Kutter E. 2009. Phage host range and efficiency of plating. Methods Mol Biol 501:141–149. 10.1007/978-1-60327-164-6_14. [DOI] [PubMed] [Google Scholar]
- 61.Valentine RC, Shapiro BM, Stadtman ER. 1968. Regulation of glutamine synthetase. XII. Electron microscopy of the enzyme from Escherichia coli. Biochemistry (Mosc) 7:2143–2152. 10.1021/bi00846a017. [DOI] [PubMed] [Google Scholar]
- 62.Hyman P, Abedon ST. 2009. Practical methods for determining phage growth parameters. Methods Mol Biol 501:175–202. 10.1007/978-1-60327-164-6_18. [DOI] [PubMed] [Google Scholar]
- 63.International Organization for Standardization. 2013. ISO 4833-1:2013 Microbiology of the food chain—Horizontal method for the enumeration of microorganisms—Part 1: Colony count at 30 degrees C by the pour plate technique. International Organization for Standardization, Geneva, Switzerland. [Google Scholar]
- 64.Bachrach U, Friedmann A. 1971. Practical procedures for the purification of bacterial viruses. Appl Microbiol 22:706–715. 10.1128/am.22.4.706-715.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jackel C, Hertwig S, Scholz HC, Nockler K, Reetz J, Hammerl JA. 2017. Prevalence, host range, and comparative genomic analysis of temperate Ochrobactrum phages. Front Microbiol 8:1207. 10.3389/fmicb.2017.01207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lesnik EA, Sampath R, Levene HB, Henderson TJ, McNeil JA, Ecker DJ. 2001. Prediction of rho-independent transcriptional terminators in Escherichia coli. Nucleic Acids Res 29:3583–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Sets S1 and S2. Download aem.01424-21-s0001.xls, XLS file, 0.2 MB (214.5KB, xls)
Data Availability Statement
The complete nucleotide sequence of Salmonella phage vB_StyS-LmqsSP1 was deposited in GenBank under accession number MT577844.