ABSTRACT
Mitigation strategies to prevent microbial contamination of crops are lacking. We tested the hypothesis that induction of plant systemic resistance by biological (induced systemic resistance [ISR]) and chemical (systemic acquired resistance [SAR]) elicitors reduces endophytic colonization of leaves by Salmonella enterica serovars Senftenberg and Typhimurium. S. Senftenberg had greater endophytic fitness than S. Typhimurium in basil and lettuce. The apoplastic population sizes of serovars Senftenberg and Typhimurium in basil and lettuce, respectively, were significantly reduced approximately 10- to 100-fold by root treatment with microbial inducers of systemic resistance compared to H2O treatment. Rhodotorula glutinis effected the lowest population increases of S. Typhimurium in lettuce and S. Senftenberg in basil leaves, respectively 120- and 60-fold lower than those seen with the H2O treatment over 10 days postinoculation. Trichoderma harzianum and Pichia guilliermondii did not have any significant effect on S. Senftenberg in the basil apoplast. The chemical elicitors acidobenzolar-S-methyl and dl-β-amino-butyric acid inhibited S. Typhimurium multiplication in the lettuce apoplast 10- and 2-fold, respectively, compared to H2O-treated plants. All ISR and SAR inducers applied to lettuce roots in this study increased leaf expression of the defense gene PR1, as did Salmonella apoplastic colonization in H2O-treated lettuce plants. Remarkably, both acidobenzolar-S-methyl upregulation and R. glutinis upregulation of PR1 were repressed by the presence of Salmonella in the leaves. However, enhanced PR1 expression was sustained longer and at greater levels upon elicitor treatment than by Salmonella induction alone. These results serve as a proof of concept that priming of plant immunity may provide an intrinsic hurdle against the endophytic establishment of enteric pathogens in leafy vegetables.
IMPORTANCE Fruit and vegetables consumed raw have become an important vehicle of foodborne illness despite a continuous effort to improve their microbial safety. Salmonella enterica has caused numerous recalls and outbreaks of infection associated with contaminated leafy vegetables. Evidence is increasing that enteric pathogens can reach the leaf apoplast, where they confront plant innate immunity. Plants may be triggered for induction of their defense signaling pathways by exposure to chemical or microbial elicitors. This priming for recognition of microbes by plant defense pathways has been used to inhibit plant pathogens and limit disease. Given that current mitigation strategies are insufficient in preventing microbial contamination of produce and associated outbreaks, we investigated the effect of plant-induced resistance on S. enterica colonization of the lettuce and basil leaf apoplast in order to gain a proof of concept for the use of such an intrinsic approach to inhibit human pathogens in leafy vegetables.
KEYWORDS: enteric pathogen, foodborne, produce, plant defense, plant immunity, biocontrol, control, herbs, leafy greens, outbreaks
INTRODUCTION
Salmonella enterica is one of the main causal agents of produce-associated outbreaks (1). Among 12 food categories, 17.6% of salmonellosis outbreaks are linked to produce (2). Leafy greens (including lettuce) and herbs (including basil) ranked among the top six produce vehicles of foodborne outbreaks in the United States from 1996 to 2014 (3). In particular, S. Senftenberg has been associated with outbreaks linked to basil in Europe and the United States (4), and large recalls of fresh basil in the United States and of dried basil in Canada and the United States (Canadian Food Inspection Agency, 2012 [http://www.fda.gov/Safety/Recalls/]). Salmonellosis has been linked also to the contamination of lettuce and other leafy greens with S. Typhimurium in Europe and the United States (5–7). S. enterica has the ability to multiply epiphytically on leaves under warm and wet conditions in the phyllosphere (8, 9). The internalization of the pathogen into leaf tissue may occur in the presence of free water through stomata and other natural openings (10, 11). This provides the bacterial cells access to the plant apoplast, where they may interact with plant cells directly, unimpeded by the cuticle barrier of the phylloplane.
Although in most cases plants do not show any disease symptoms after inoculation with S. enterica, leaf chlorosis and wilting in Arabidopsis thaliana (12, 13) and plant biomass reduction in lettuce (14) have been reported. Evidence is increasing that S. enterica may confront the plant immune system upon interaction with plant cells (13, 15–17). The innate immune system of plants depends on a diverse assortment of cell surface and cytoplasmic receptors that respond to invading pathogens (18). These receptors are commonly classified into a group that recognizes conserved microbial signatures and a group that recognizes highly variable effectors; thus, these two systems are referred to as pathogen- or microbe-associated molecular pattern (PAMP or MAMP)-triggered immunity (PTI or MTI) and effector-triggered immunity (ETI), respectively (19, 20).
PAMP activation in A. thaliana by S. enterica flagellin has been reported (21). While current knowledge does not support that S. enterica can translocate effectors into plant cells via its type 3 secretion system (T3SS) (22), mutants in T3SS components of SPI-1 and SPI-2 displayed reduced endophytic colonization of A. thaliana (17, 21) and altered induction of lettuce stomatal response to S. enterica ingression (23). Melotto and coworkers provided evidence that both E. coli O157:H7 and S. enterica Typhimurium modulate stomatal immunity (24) and that the latter pathogen induces stomatal responses depending on plant species and environmental conditions (25). Other mechanisms of plant basal immunity involve the production of reactive oxygen species and nitric oxide, as well as callose deposition, and these also play a role in the leaf apoplast colonization by enteric pathogens (26–28).
Plant innate immune responses may be activated chemically or biologically by inducing systemic signaling cascades that effect resistance in distal plant parts not infected or colonized by a phytopathogen or microbe. This systemic plant resistance relies on phytohormone signaling pathways. Systemic acquired resistance (SAR) is mediated by salicylic acid (SA)-dependent processes and is associated with the production of PR proteins; on the other hand, induced systemic resistance (ISR) is a response to colonization of plant roots by plant growth-promoting bacteria and fungi and is generally mediated by the jasmonic acid (JA) and ethylene pathways (29–31). Because they are rooted in broad immunity to plant disease, SAR and ISR exhibit long-lasting nonspecific resistance effective against a wide range of organisms.
The effect of SAR and ISR on the growth and survival of human enteric pathogens in plants remains unknown. In this study, we tested whether induction of systemic resistance can reduce colonization of leafy vegetables by human pathogens. We investigated this as-yet-unexplored aspect of plant-enteric pathogen interactions by treating lettuce and basil plants with inducers of SAR and ISR and testing their effect on the apoplastic population levels of S. enterica. We also assessed the expression of marker genes belonging to defense signaling pathways to further our understanding of S. enterica-plant interactions upon elicitation.
RESULTS
Salmonella endophytic fitness in lettuce and basil leaves.
Various Salmonella serovars were tested for their ability to colonize lettuce and basil plants endophytically. Serovar Senftenberg strain 127468 colonized leaves of lettuce and basil plants better than serovar Typhimurium strain 14028S, displaying significantly greater endophytic population sizes over a period of 15 to 20 days after inoculation (Student t test; P < 0.05) (Fig. 1A and B). Serovar Senftenberg also colonized the basil leaf apoplast at significantly greater densities at 10 and 20 days postinoculation than did serovars Montevideo strain 125473, Enteritidis strain 124655, Infantis strain 122798, and Dublin strain 135157 (Tukey’s multiple-comparison test; P < 0.05). S. Senftenberg population sizes were 16.0 and 13.0 times greater than those of S. Dublin at 10 and 20 days postinoculation, respectively (Fig. 2).
FIG 1.
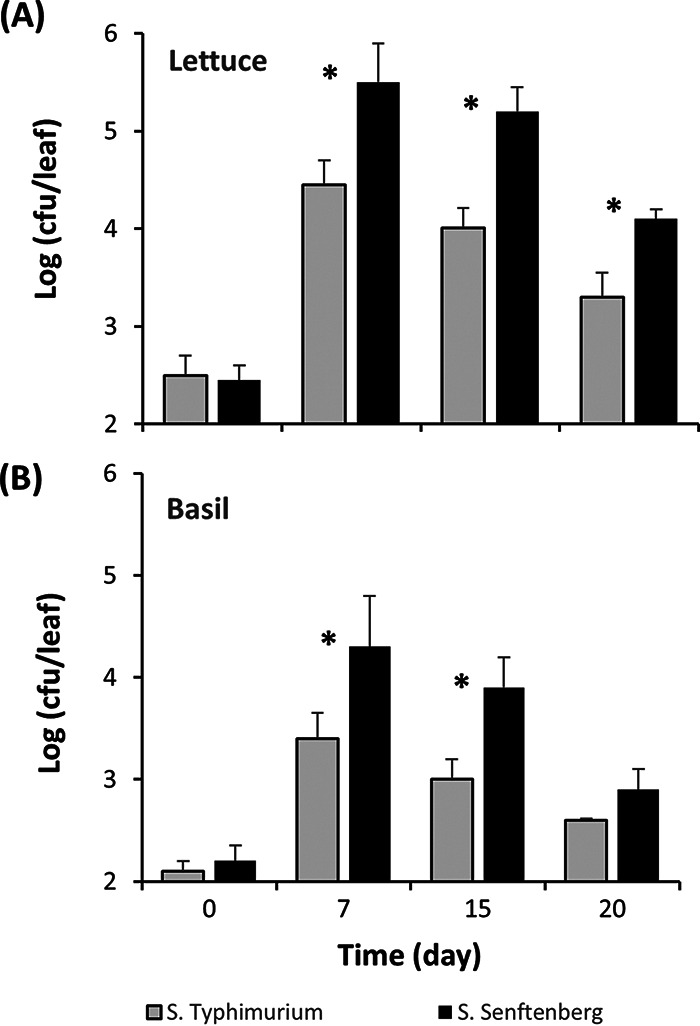
Endophytic population change of S. Typhimurium (gray bars) and S. Senftenberg (black bars) in leaves of lettuce (A) and basil (B) plants over time after passive apoplast inoculation. Bars represent the means of the log10-transformed population sizes for three replicate leaves sampled from three replicate pots, and SEM. *, Significant difference in population sizes between the two serovars (Student t test, P < 0.05).
FIG 2.
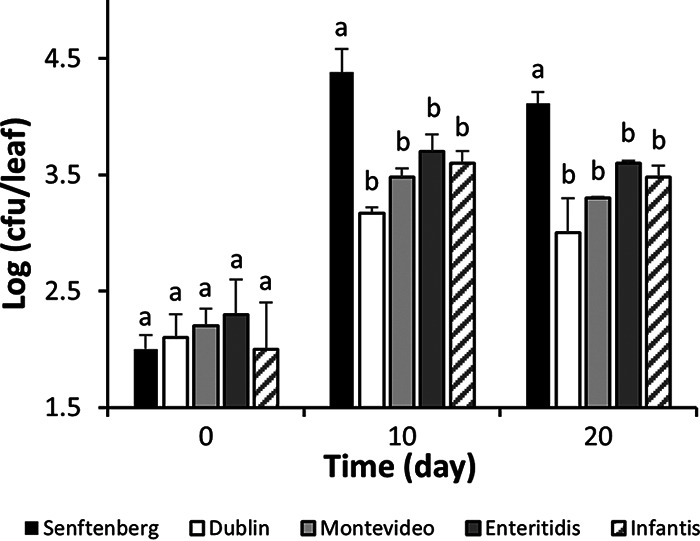
Endophytic population change of S. enterica serovars Senftenberg, Dublin, Montevideo, Enteritidis, and Infantis in the leaves of basil plants immediately and 10 and 20 days after passive apoplast inoculation. Bars represent the means of the log10-transformed population sizes for five replicate leaves sampled from five replicate pots, as well as the SEM. Different letters above the bars indicate significant differences in Salmonella population size within each incubation time by Tukey’s multiple-comparison test (P < 0.05).
Effect of ISR and SAR inducers on endophytic colonization of lettuce and basil leaves.
Plants were treated by soil drench with inducers prior and after Salmonella inoculation and their effect on endophytic populations of the human pathogen in leaves was assessed. In lettuce, all four ISR inducers had a significant effect on S. Typhimurium colonization 10 days postinoculation (Tukey’s multiple-comparison test, P < 0.05) (Fig. 3A). R. glutinis Y13-treatment inhibited S. Typhimurium in lettuce the most effectively, with a 120.2-fold decrease in density over that in control (H2O-treated) plants, compared to 7.1-, 10.7-, and 28.2-fold decreases by treatment with Trichoderma harzianum T39, Pichia guilliermondii S2, and B. subtilis Mel16, respectively. In basil, treatment with R. glutinis Y13 and B. subtilis Mel16 caused the lowest S. Senftenberg population sizes of all four ISR inducers, as revealed by a 63.1- and 10.0-fold differences compared to that in the control plants (Tukey’s multiple-comparison test, P < 0.05) (Fig. 3B).
FIG 3.
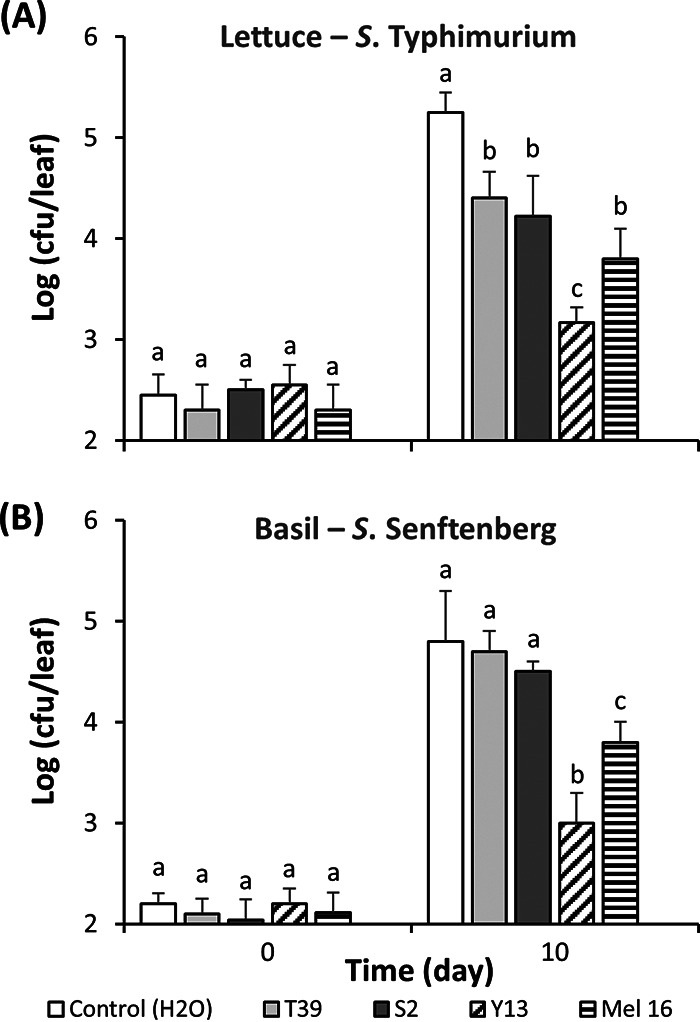
Effect of ISR-inducing microorganisms on endophytic colonization of lettuce leaves by S. Typhimurium (A) and of basil leaves by S. Senftenberg (B). Soil drench treatments were performed with H2O (control) (white bars), T. harzianum T39 (light gray bars), P. guilliermondii S2 (dark gray bars), R. glutinis Y13 (crosshatched bars), and B. subtilis Mel 16 (striped bars). Assessment of endophytic population sizes was performed immediately and 10 days after passive apoplast inoculation with Salmonella. Bars represent the means of the log10-transformed population sizes of three replicate leaves sampled from three replicate pots, as well as the SEM. Different letters above bars illustrate significant difference in population sizes as per Tukey’s multiple-comparison test (P < 0.05).
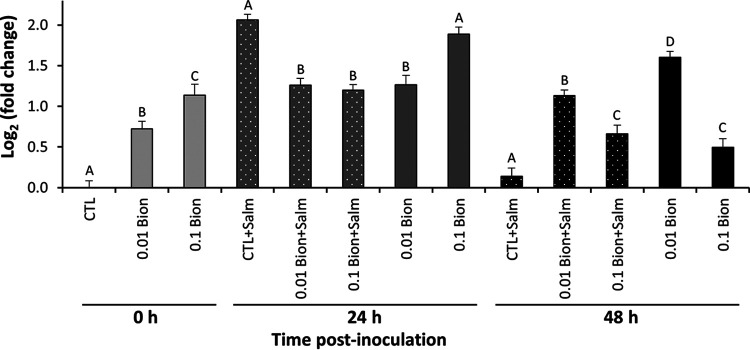
Lettuce plants were also pretreated with the SAR inducers Bion and BABA. The effect of the inducers on S. Typhimurium endophytic populations was measured in a short colonization time course to assess whether they may inhibit Salmonella multiplication. Due to this short colonization period, vacuum infiltration was used rather than passive internalization in order to obtain apoplast inoculum densities that were sufficiently high to prevent stochastic effects, thus reducing leaf-to-leaf variability, but also sufficiently low to enable for amplification of bacterial multiplication. Endophytic multiplication after vacuum infiltration of lettuce plants (without the aid of a surfactant) with low inoculum levels in the mesophyll of approximately 1 × 104 CFU/leaf was evidenced by an increase in S. Typhimurium population sizes in the control plants (H2O treatment) of as much as 53-fold (Fig. 4). Population sizes of this human pathogen showed little or no further increase in the apoplast of the control plants by 48 h. While S. Typhimurium multiplied endophytically in all treatments, the population sizes were significantly lower already at 24 h postinoculation in the leaves of plants treated with low concentrations of SAR inducers (Bion [0.01%] and BABA [0.8 mg/ml]), than in the control plants treated with H2O (Dunnett’s multiple-comparison test; P < 0.05) (Fig. 4). At 48 h postinoculation, S. Typhimurium population sizes in the inducer-treated plants were significantly lower than in control plants, irrespective of the inducer concentration (Dunnett’s multiple-comparison test); at the highest concentrations of Bion and BABA, the population sizes were 0.11- and 0.55-fold, respectively, that observed in control plants. However, no significant effect of inducer dosage was observed (Fig. 4).
FIG 4.
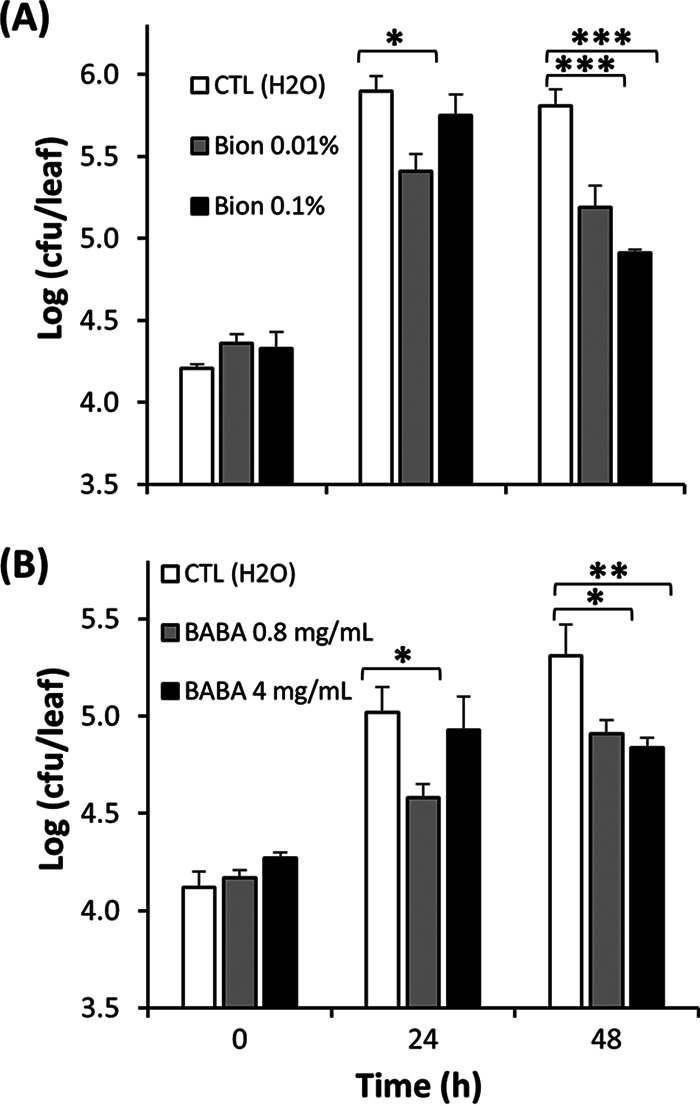
Effect of Bion (A) and BABA (B) at different concentrations as 24-h preinoculation treatments by root drench on the endophytic multiplication of S. Typhimurium in leaves of young potted romaine lettuce plants inoculated by vacuum infiltration. Data represent the means of the log-transformed population sizes of the pathogen for two, three, and four replicate leaves sampled from three replicate pots immediately, and 24 and 48 h after inoculation, respectively, and SEM. *, **, and *** indicate significant differences (P < 0.05, 0.01, and 0.001, respectively) in Salmonella population sizes between elicitor- and H2O-treated plants, as determined by Dunnett’s multiple-comparison test.
Expression of plant defense genes in lettuce treated with SAR and ISR inducers.
Table 1 shows the expression of genes involved in SA and JA signaling pathways in lettuce leaves 24 h after plants were treated with 0.1% Bion or 4 mg/ml BABA compared to that in control plants (H2O treated). Bion induced expression of SAR marker genes PR1, PR2, and EDS1 greater than 3-fold relative to the control, while PAL2 and FLS2 were also upregulated. In contrast, the expression of ERF and DEF, markers for the ethylene and JA signaling pathways, was below the detection limit in Bion-treated plants but was induced 3.73- and 4.99-fold, respectively, upon BABA treatment compared to the control. BABA-treated plants also had 3.89-fold greater expression of PR1 than control plants. The mechanism of BABA-induced plant immunity is broader than that of Bion-mediated SAR and may result from the interplay of several signaling pathways (32, 33).
TABLE 1.
Comparative transcript abundance of defense genes in romaine lettuce leaves following plant treatment with Bion and BABA treatment relative to the control treatment (H2O)
| Gene | Mean comparative expression (SEM)a |
|
|---|---|---|
| Bion | BABA | |
| PR1 | 3.27 (1.05) | 3.89 (1.11) |
| PR2 | 5.27 (1.23) | ND |
| EDS1 | 5.20 (2.00) | ND |
| PAL2 | 3.07 (1.42) | ND |
| FLS2 | 2.28 (1.03) | ND |
| ERF | BD | 3.73 (1.19) |
| DEF | BD | 4.99 (1.06) |
Bion and BABA were applied by soil drench at 0.1% and 4 mg/ml, respectively, and leaves sampled for gene expression 24 h after treatment. Comparative expression was measured by RT-qPCR and is shown as fold change over expression in H2O-treated plants. The data represent the means for three replicate leaves. BD, below detection level; ND, not determined.
Figure 5 illustrates the comparative transcription of PR1 in leaves of lettuce plants treated with 0.01 and 0.1% Bion versus an H2O-treated control at various times after infiltration with S. Typhimurium or with H2O, as measured by RT-qPCR. PR1 increased in expression 24 h after treatment with Bion in a dose-dependent manner since its transcription at that time (0 h) was greater after SAR induction with 0.1% than 0.01% Bion (Tukey’s multiple-comparison test; P < 0.05). After infiltration with H2O (mock-inoculated plants), PR1 expression was the highest 48 h after treatment with 0.1% Bion (24 h after inoculation), with a difference of 3.7-fold compared to control plants prior to infiltration. Inoculation with S. Typhimurium and subsequent endophytic multiplication increased PR1 expression 4.1-fold 24 h postinoculation compared to transcriptional levels in control plants prior to inoculation, thus to a similar extent as treatment with 0.1% Bion at that time. Remarkably, the expression of PR1 was 1.8- and 1.6-fold lower in leaves of plants pretreated with 0.1% Bion and colonized endophytically by S. Typhimurium for 24 h than in leaves of untreated plants inoculated with the pathogen and in leaves of plants treated with 0.1% Bion that were mock inoculated, respectively. Thus, PR1 upregulation by either the pathogen or Bion appeared to be suppressed by the activity of the other. However, the high expression of PR1 triggered by S. Typhimurium colonization (in the control H2O-treated plants) was not sustained and at 48 h postinoculation returned to preinoculation levels (0 h). In contrast, the enhanced transcriptional activity of PR1 persisted in Bion-treated plants for up to 48 h postinoculation (72 h posttreatment; Tukey’s multiple-comparison test; P < 0.05).
FIG 5.
Expression of the SAR marker PR1 in leaves of lettuce plants pretreated with 0.01 or 0.1% Bion, or H2O (control), as a root drench 24 h before vacuum infiltration with S. Typhimurium or with H2O (0 h). PR1 expression at 24 h and 48 h is shown as log2-fold change compared to that in leaves of mock-inoculated control (H2O-treated) plants at 0 h. Expression in inoculated leaves is denoted by +Salm. Solid bars, plants treated with Bion or H2O but noninoculated. Solid bars with dots, plants treated with Bion or H2O and inoculated with S. Typhimurium. Data represent the means for three replicate leaves sampled from three replicate pots, as well as the SEM. Different letters above bars illustrate significant difference within each sampling time as determined by Tukey’s multiple-comparison test (P < 0.05).
The ability of S. Typhimurium to suppress PR1 transcription in lettuce plants primed for basal defense was further tested by elicitation with B. subtilis Mel16 and R. glutinis Y13. While ISR is commonly attributed to the activity of the JA pathway, additional involvement of PR1 has been described in lettuce and other species (34, 35), and SAR is known to be triggered by both biological and chemical inducers (36). Figure 6A illustrates the relative expression of PR1 in lettuce plants at 3 days after root treatment and before apoplast inoculation with S. Typhimurium or mock inoculation with H2O (0 h), as well as 48 h thereafter. Microbial elicitation caused an increase in PR1 expression in lettuce leaves compared to the case in H2O-treated plants by the time of inoculation (0 h), and this induction was sustained 48 h later. Endophytic colonization of the leaves by S. Typhimurium in the absence of treatment with an elicitor of plant resistance increased PR1 expression 4-fold by 48 h postinoculation compared to preinoculation levels (0 h) (Fig. 6A), as we observed also in experiments with Bion (Fig. 5). However, this transcriptional increase was lower than that observed in plants that had been treated with B. subtilis Mel16 and R. glutinis Y13, whether these were inoculated with the human pathogen or mock inoculated (Tukey’s multiple-comparison test; P < 0.05). Furthermore, in R. glutinis-treated plants, colonization by S. Typhimurium significantly decreased PR1 transcription levels by 48 h postinoculation, whereas no effect of the pathogen was detected in B. subtilis-treated plants.
FIG 6.
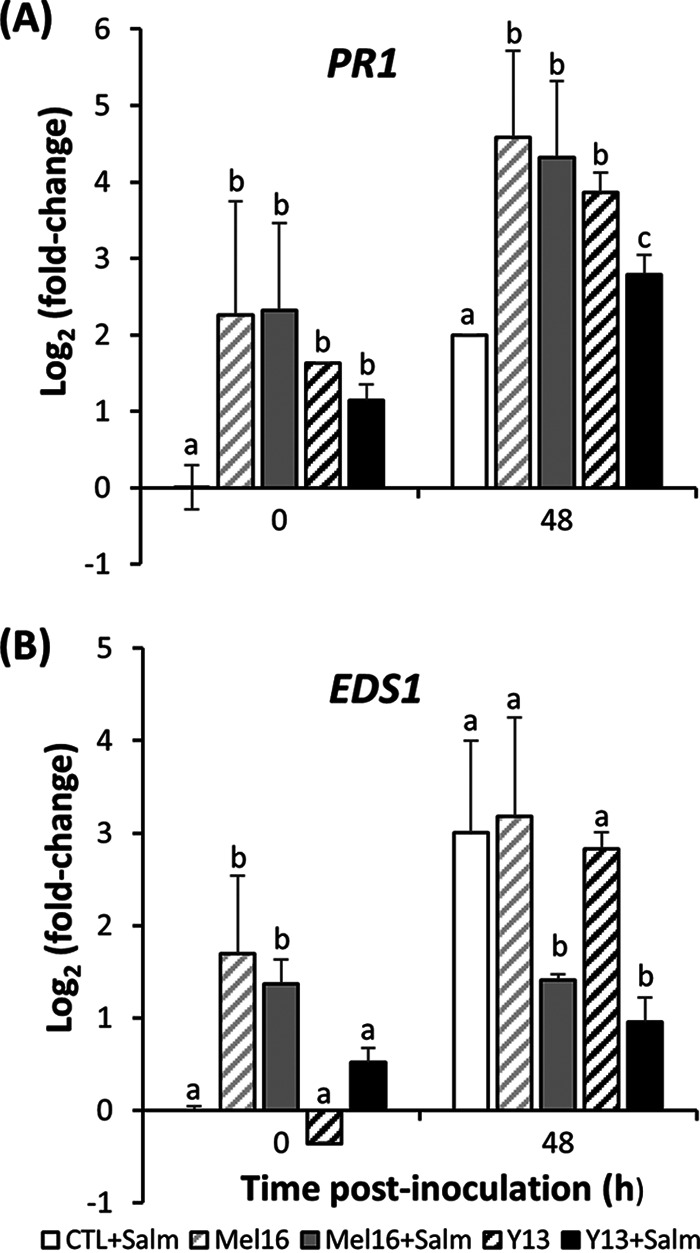
Relative expression of SA-pathway genes in lettuce leaves in response to root treatment with H2O (control) (white bar), B. subtilis Mel16 (gray bars, crosshatched or solid), or R. glutinis Y13 (black bars, crosshatched or solid), followed by leaf apoplast mock inoculation with H2O (crosshatched bars) or inoculation with S. Typhimurium (solid bars). Gene expression in inoculated leaves is denoted by +Salm. PR1 (A) and EDS1 (B) transcript quantification by RT-qPCR was carried out in five replicate leaves sampled from five replicate pots 3 days after root treatment at 0 h and 48 h after mock inoculation or inoculation with S. Typhimurium. Expression is shown relative to the transcriptional level in mock-inoculated control (H2O treated) plants at 0 h. Different letters above bars illustrate significant difference in relative gene expression within each time point as per Tukey’s multiple-comparison test (P < 0.05).
As observed for PR1, the expression of EDS1 was increased significantly by the presence of S. Typhimurium in the leaf apoplasts of unelicited plants (control) (Fig. 6B). While the leaves of plants elicited with B. subtilis Mel16 and R. glutinis Y13 showed enhanced EDS1 transcription at 48 h after mock inoculation and inoculation with S. Typhimurium, this transcriptional activity was lower in leaves colonized by the pathogen compared to that observed in mock-inoculated leaves (Tukey’s multiple-comparison test; P < 0.05).
DISCUSSION
While good agricultural practices and postharvest sanitization contribute to improving produce safety, mitigation strategies to inhibit foodborne pathogens on produce are greatly lacking. Approaches based on the enhanced capability of plants to respond to the presence of enteric pathogens and inhibit their colonization may provide an inherent in planta first step to minimize preharvest contamination of crops. The use of plant breeding to enhance microbial produce safety has been proposed (37). This approach is based on the premise that plant traits related to the physicochemical nature of plants as a bacterial habitat or to plant innate immune responses that inhibit enteric pathogens may be selected for in breeding programs to lower microbial contaminant loads in produce and reduce the risk of human infection.
For their presence to be sensed by the plant immune system, microbes must breach or bypass the cuticle barrier of the leaf to come into direct contact with plant cells in the apoplast. We show here that the ability of Salmonella to get established and survive in the plant apoplast varies among strains of different serovars. In both lettuce and basil plants, S. Senftenberg strain 127468 colonized leaves endophytically at greater population densities than S. Typhimurium 14028s. S. Senftenberg also displayed greater apoplastic population sizes than strains of serovars Montevideo, Enteritidis, Infantis, and Dublin in basil leaves 10 and 20 days after inoculation. Infiltration of Arabidopsis thaliana leaves with S. Senftenberg and S. Typhimurium strain SL1344 failed to reveal any difference in colonization between the two serovars since their population sizes remained stable after inoculation, although the former strain caused leaf chlorosis and wilting, while the latter did not (12). Plant disease symptoms were not observed in basil or lettuce in our study. On the other hand, we observed in this study, increases in endophytic population sizes of approximately 10- to 50-fold within 24 to 48 h after infiltration into lettuce leaves for S. Typhimurium and also for S. Senftenberg (data not shown), indicating that active Salmonella population growth can occur in the leaf apoplast when inoculum levels are low. Such active bacterial multiplication may allow for differences in the interaction of various Salmonella strains with plants during colonization. Delaquis and coworkers demonstrated differences in the colonization of two lettuce genotypes by 43 S. enterica strains from 29 serovars (38), indicating that bacterial and plant factors may affect the outcome of Salmonella-plant interactions.
Mechanisms of plant immunity rely broadly on phytohormone signaling pathways. These signals can effect long-distance resistance away from the infection site and be triggered by chemical elicitors (SAR) or biological inducers (ISR) (39, 40). Although enteric pathogens by themselves have the ability to elicit plant defense responses, induced plant resistance via SAR and ISR may potentiate inhibition against these microbial contaminants and hence, crop safety. Of the four microbial strains inoculated onto roots prior to inoculation of the leaves with Salmonella, the yeast R. glutinis Y13 caused the greatest inhibition of plant colonization in both serovars, with reductions of 63- and 120-fold in Salmonella endophytic population sizes in basil and lettuce apoplasts, respectively, compared to H2O-treated plants. R. glutinis suppresses fungal diseases on a variety of crops (41, 42). Increased activity of the PAL, PPO, and POD enzymes, which are involved in the production of plant secondary metabolites that have antimicrobial activity, was reported in tomato wounds upon treatment with R. glutinis (43). Increased PPO expression, as well as the production of phenolic compounds, was observed after exposure of basil plants to methyl-JA, an analog of the JA signal that is involved in ISR (44). Furthermore, treatment with B. subtilis, an effective ISR-inducing microorganism, similarly enhanced PAL expression in rice cell cultures (45), and significantly reduced Salmonella colonization on basil and lettuce in this study. One could hypothesize that enhanced production of phenolic compounds via increased PAL and related enzymatic activity (PPO and POD) as a result of root colonization by R. glutinis and B. subtilis inhibited endophytic Salmonella cells in the aerial parts of basil and lettuce. In addition, the production of cyclic lipopeptides by B. subtilis and other species is involved in the establishment and maintenance of ISR, thereby protecting many plant species from a wide variety of pathogens, including leaf-spot bacterial pathogens (34, 45, 46).
The ISR-inducing microorganisms T. harzianum T39 and P. guilliermondii S2 had a smaller, but significantly inhibitory effect on S. Typhimurium apoplastic colonization of lettuce. Various species of Trichoderma have been shown to protect crops against fungal diseases and this endophytic fungus has been implicated in induced resistance through a spectrum of strategies (47, 48). In lettuce, the biocontrol of leaf spot diseases by T. asperellum T1 was mediated by the enhanced activity of defense-related enzymes such as PPO and POD (49). The induced resistance by Pichia spp. has been poorly described but may proceed via several signals, including ethylene, JA, and SA signal transduction pathways (50). Although ISR has been investigated in great part for the control of fungal diseases, its commonly observed enhanced synthesis of phenolic and oxidative species in plants suggests a mode of action in the inhibition of endophytic leaf colonization by Salmonella observed here.
Bion and BABA, two common chemical inducers of SAR that are effective against a wide range of plant diseases (36, 51), were also tested as soil drenches in our system, but their effect was investigated over a shorter time after S. Typhimurium vacuum infiltration of lettuce leaves. Both elicitors significantly inhibited multiplication of the pathogen after infiltration compared to the control treatment (soil drench with H2O), although only the lower concentrations exerted a more immediate inhibition at 24 h postinoculation. Bion has been shown to protect lettuce from bacterial infection by the leaf spot pathogen, Xanthomonas campestris pv. vitians (52) and basil from downy mildew disease (53). In our preliminary experiments, 0.03% Bion as soil drench lowered the population sizes of S. Senftenberg in the basil apoplast by 6.2-fold compared to H2O (control) treatment by 10 days postinoculation (data not shown). While BABA provided good control of the oomycete pathogen causing downy mildew in lettuce (54), it also reduced disease symptoms by the bacterial pathogens Pseudomonas syringae pv. tomato (55) and Pectobacterium carotovorum in A. thaliana leaves (56). In our system, S. Typhimurium inhibition by BABA and Bion was approximately in the same range as BABA inhibition of bacterial pathogens in A. thaliana leaves (55, 56). It is unclear if their effect would have been greater, perhaps in the same range as the effect caused by some ISR strains that we report herein, had longer periods of S. Typhimurium colonization been tested.
The time course of PR1 expression in Bion-treated lettuce plants revealed that this SAR marker gene was induced in lettuce leaves in a dose-dependent manner 24 h after soil drench (as assessed at the time of inoculation), thus confirming that our system was functional. Expression continued to increase in mock-inoculated plants compared to that before inoculation. However, in elicited plants that were also inoculated with S. Typhimurium, the change in PR1 expression was smaller at 24 h postinoculation than the change in S. Typhimurium-inoculated plants that were not elicited (H2O treatment), as well as compared to the change in leaves induced with 0.1% Bion in the absence of S. Typhimurium (mock inoculation). This apparent antagonism in PR1 expression suggests that the occurrence of cross talk between defense signaling activities triggered by S. Typhimurium colonization of the lettuce apoplast and by Bion elicitation. Notably, this suppressive effect by S. Typhimurium colonization on SA signaling was evidenced also by a decrease in PR1 expression in leaves of lettuce roots inoculated with the ISR elicitor R. glutinis, the most effective ISR inducer among the four agents that we tested. Likewise, EDS1 transcription in the leaves of lettuce plants elicited with B. subtilis and R. glutinis was suppressed when leaves were colonized by this enteric pathogen. These common observations with different induced plant resistance approaches and different lettuce cultivars in our study strongly suggest the occurrence of interference in plant signaling by different concomitant defense pathways in the presence of S. Typhimurium.
The upregulation of PR1 in lettuce leaves during apoplastic colonization by S. Typhimurium is in line with previous reports that plant basal defenses, as reflected partly by enhanced transcription of PR1 and other components of the SA signaling pathway, are elicited by this human pathogen, e.g., in A. thaliana (13, 24, 27). Of note is our observation in this study that PR1 upregulation triggered solely by S. Typhimurium did not persist at levels comparable to those triggered by chemical elicitors and microbial inducers. Such a transient induction of PR1 by S. Typhimurium was observed similarly in A. thaliana (13). This sustained defense response upon chemical or microbial elicitation of systemic resistance may underlie its inhibitory effect on Salmonella colonization in lettuce and basil leaves in our study. However, given its pattern of expression in Bion- and Y13-treated plants and the potential occurrence of defense signaling cross talk in the presence of S. Typhimurium, the production of PR1 cannot fully explain the inhibition of Salmonella colonization observed in this study, so additional defense-related mechanisms or pathways are very likely at play. Salmonella flagellin is recognized via FLS2 in plants, and its flagellin peptides, as well as Salmonella colonization itself, induce ROS production in A. thaliana and tomato (21, 28, 57). Furthermore, activation of the SA signaling pathway by elicitor treatment increased flg22-triggered oxidative burst and callose deposition in A. thaliana (58). Bion treatment also enhanced FLS2 transcription in lettuce in our study, while BABA and ISR-inducing microorganisms augmented callose deposition to control plant pathogens infection in lettuce and other species in previous studies (54, 59, 60). Since differential colonization of S. Typhimurium in lettuce cultivars and A. thaliana is partly a function of ROS production and callose deposition upon apoplast inoculation (26, 27), these plant defense responses are worthy of investigating further to enhance our understanding of the inhibitory effect of SAR and ISR on endophytic Salmonella.
Plant defense priming has been described by Mauch-Mani and collaborators as the induction of a plant physiological readiness to mount a faster, stronger or more sustained attack on a target by exposure to low doses of stimulus (61). In the primed state, the plant is conditioned to respond to an invader with minimal activation of defenses, which are then potentiated upon the presence of the invader, thus minimizing allocation costs to the plant (62), which would otherwise be incurred by a persistent heightened state of defense (63). The potential for plant defense priming to minimize microbial contamination of crops could be of significant interest, particularly since it may concomitantly reduce the incidence of certain plant diseases. Future plant technologies may provide the means to engineer or exploit intrinsic physiological traits for an enhanced and timed defensive capability against microbial contaminants on plants. The results we described here serve as a proof of concept that enhanced plant immunity through elicitation may contribute to a spectrum of plant-based tools to weaken the probability of enteric pathogen survival and multiplication on crops.
MATERIALS AND METHODS
Microbial strains and growth conditions.
Salmonella enterica serovar Typhimurium strain 14028s was obtained from M. McClelland (University of California, Irvine, CA). S. enterica serovars Senftenberg strain 127468 (clinical isolate, 2010, Israel), Montevideo strain 125473 (clinical isolate, 2009, Israel), Enteritidis strain 124655 (clinical isolate, 2009, Israel), Infantis strain 122798 (chicken isolate, 2009, Israel), and Dublin strain 135157 (milk isolate, 2010, Israel) were obtained from L. Valinsky (Central Laboratories, Ministry of Health, Israel). These Salmonella strains are here referred to as S. Typhimurium, S. Senftenberg, S. Montevideo, S. Enteritidis, S. infantis, and S. Dublin. S. Typhimurium 14028s was marked with rifampin resistance by selection of a spontaneous rifampin-resistant mutant on growth medium containing rifampin at 100 μg/ml and used in SAR experiments. All ISR and Salmonella serovar comparison experiments were carried out with wild-type strains. All strains were cultured in Luria-Bertani half-salt broth (5 g of NaCl/liter; LBHS) by agitation at 28°C overnight to early stationary phase. Inoculum suspensions were prepared by washing the cells first in 10 mM potassium phosphate buffer (pH 7; KPB) and again in 1 mM KPB using centrifugation (17,000 × g) for 3 min at 24°C and then resuspending them in distilled deionized (DDI) H2O for inoculation at concentrations detailed below.
The biological inducers of ISR used in this study were isolated in the laboratory of Y. Elad and as described previously (41, 42, 48, 64). Trichoderma harzianum strain T39 was grown on potato dextrose agar (PDA) for 10 days, while inducers Pichia guilliermondii strain S2 and Rhodotorula glutinis strain Y13 were grown on PDA for 4 days. Bacillus subtilis strain Mel16 was cultured on nutrient agar for 2 days. All biological inducers were cultured at 28°C, washed, and resuspended in DDI H2O to reach the inoculum cell concentration described below in “Plant Treatment and Leaf Apoplast Inoculation.” The inoculum concentration was adjusted based on T. harzianum conidium counts using a hemocytometer and based on the optical density at 600 nm (OD600) for bacterial and yeast strains.
Plant material and growth conditions.
Lettuce plants (Lactuca sativa, romaine cv. Parris island [USA] and cv. Noga 936 [Israel]) were grown for 2 months in 500-ml pots to the four- to six-leaf stage in a plant growth chamber at 22°C (USA) and in a greenhouse at 20 to 25°C (Israel), with 14-h daylight period. The mean leaf weight and the standard errors of the mean (SEM) were 1,197 ± 20 and 578 ± 4 mg for lettuce and basil, respectively. Basil plants (Ocimum basilicum cv. Peri) were obtained from a commercial nursery (Hishtil, Israel), cultivated in a greenhouse at 20 to 25°C, and grown in 750-ml pots for 1 month to the four- to six-leaf-stage. All plants were cultivated in greenhouse soil composed of peat moss and perlite (approximate ratio, 80:20).
Plant treatment and leaf apoplast inoculation. (i) Passive apoplast inoculation.
The effect of ISR strains on endophytic colonization of basil by S. Senftenberg and of lettuce cv. Noga 936 by S. Typhimurium was tested as follows: basil or lettuce plants were treated by soil drench with 20 ml/plant of cell suspension of the biological inducers T. harzianum T39 (106 conidia/ml), B. subtilis strain Mel16 (108 CFU/ml), R. glutinis strain Y13 (108 CFU/ml), and P. guilliermondii strain S2 (108 CFU/ml) in DDI H2O. The control treatment consisted of a soil drench with sterile DDI H2O. Plants were then kept at 20 to 25°C in a greenhouse. After 3 days, all plants, including the control plants, were inoculated by submerging the upper part of the plants in a suspension (108 CFU/ml DDI H2O) of S. Senftenberg or S. Typhimurium for 30 min. The plants were then rinsed thoroughly by sequential immersions in sterile DDI H2O to enrich for inoculant in the apoplast. Application of the inducer agents was repeated as soil drench immediately after inoculation before the plants were returned to the greenhouse, as previously described by Elad and Rav-David (64). Inoculation for comparative lettuce and basil apoplast colonization by various Salmonella serovars was performed using the approach described above, except that the plants did not receive any soil drench treatment.
(ii) Apoplast inoculation by vacuum infiltration.
The effect of the SAR on S. Typhimurium multiplication in the leaf apoplastic space was investigated using the compounds acibenzolar-S-methyl (ASM) and dl-β-amino-butyric acid (BABA; Sigma) in lettuce cv. Parris Island. Acibenzolar-S-methyl, a functional SA analog that is commercially available as Bion 50 WG (or Actigard 50 WG), was kindly provided by Syngenta Crop Protection, Inc. (Greensboro, NC). Plants were treated by soil drench with 25 ml of 0.01 and 0.1% Bion, as well as 0.8 and 4 mg/ml BABA, 24 h before inoculation. Control treatment consisted of soil drench with DDI H2O. The aerial part of the plants, including that of H2O-treated plants, was then inoculated upside down in the inoculum suspension by vacuum infiltration. Briefly, cotton was placed tightly on the soil surface around the potted plant to prevent soil from falling into the suspension. The lettuce leaves were immersed in 1.8-liter suspension of S. Typhimurium at 104 CFU/ml DDI H2O that did not contain any surfactant, such as Silwet L-77; for infiltration of the suspension into the leaves, the beaker with the suspension and the plant were placed in a vacuum bell jar (Abbess Instruments, Holliston, MA), and a vacuum was applied for 1 min to remove air from the leaf mesophyll. The vacuum was then quickly released to replace the air in the apoplast with the cell suspension (65). After inoculation, the aerial part of the plant was rinsed three times by successive immersion in 1 liter of sterile DDI H2O to remove the inoculum from the leaf surfaces to the greatest extent possible. The plants were placed in a chamber at 28°C and 65% relative humidity, thus ensuring that the surfaces of the leaves were macroscopically dry and that S. Typhimurium multiplication during incubation was limited to the leaf apoplast.
Bacterial cell recovery and quantification of apoplastic population sizes.
Basil leaves (the third or fourth leaf) and lettuce leaves (the fourth or fifth leaf) were sampled at random from different plants and pots immediately after inoculation (0 h) and at various times of incubation after inoculation. The number of replicate leaf samples and replicate pots is provided in each figure legend and varied slightly depending on the number of available plants of very similar age and size. The leaves were surface sterilized by immersion in 2% sodium hypochlorite containing 0.1% Tween 20 for 30 s. Each leaf was rinsed five times in five different beakers containing 250 ml of DDI H2O. The leaves were homogenized with a mortar and pestle in 2 ml of DDI H2O to release the Salmonella cells from the apoplast, and aliquots of the homogenate were dilution plated for colony plate counts. For SAR experiments, which were carried out with the rifampin-resistant S. Typhimurium 14028s, the homogenate was plated onto LBHS agar containing rifampin at 100 μg/ml (LBHS-rif agar). For ISR experiments and comparative Salmonella serovar colonization experiments, the homogenates were plated onto XLD agar. Plate counts were performed after incubation of the plates at 37°C for at least 20 h. No colonies were detected on LBHS-rif agar or XLD agar from homogenates obtained from leaves infiltrated with sterile DDI H2O only, indicating that the plate counts were specific for Salmonella.
In addition, homogenates from leaves that were surface inoculated by rapid immersion of the plant aerial part, rinsed in sterile DDI H2O five times, and surface sterilized after plant incubation and then, prior to bacterial recovery, were plated on LBHS-rif agar. The plates were incubated at least 24 h at 28°C and did not yield any bacterial colonies. This demonstrated (i) the efficacy of our surface sterilization treatment and (ii) the efficacy of our approach in enriching for Salmonella populations in the apoplast; that is, by thoroughly rinsing leaf surfaces after inoculation and then incubating the plants under conditions that do not promote free water on the leaves, the number of cells on the leaf surface immediately after inoculation was restricted, and Salmonella multiplication in the phyllosphere was minimized, respectively.
Measurement of relative plant gene expression.
Expression of select basal plant defense genes that are part of the pathways involved in SAR and ISR was quantified in lettuce leaves at the indicated times after inoculation with Salmonella or mock inoculation (H2O only). Leaves were harvested, frozen in liquid nitrogen, and stored in a −80°C freezer until used for RT-qPCR. Portions (50 mg) of leaf tissue from three or five replicate leaves were ground using a mortar and pestle in liquid nitrogen before RNA was extracted using Ambion TRIzol reagent and a PureLink RNA kit (Life Technologies). The RNA was DNase treated a second time with Turbo DNase I (Ambion), and the absence of DNA was confirmed by RT-qPCR in the absence of reverse transcriptase using primers for ACT7. Gene expression in lettuce treated with Bion and BABA was analyzed by one-step RT-qPCR with the Stratagene Brilliant II SYBR green 1-Step kit (Stratagene) on a MxPro 3000P cycler (Stratagene). Gene expression in lettuce treated with ISR microbial inducers was analyzed by two-step RT-qPCR using a qScript cDNA synthesis kit (Quanta Biosciences, Gaithersburg, MD) in which 1 μg of purified RNA was reverse transcribed to cDNA. The obtained cDNA served as the template for PCR amplification using the Fast SYBR green Master Mix (Applied Biosystems, Foster City, CA). Real-time detection was performed in triplicates with a 7300 Real-Time PCR System (Applied Biosystems). Expression of ACT7 was used to normalize the data between samples. The sequences of the primers were as follows: EDS1-F, 5′-ACCTTGAGGAATACACGCGATCCA; EDS1-R, 5′-AAGGCGTGTGTGAATATCCGGTCA (66); ERF-F, 5′-CCGTTTGATTGTTCCGATTT; ERF-R, 5′-TTCGGCTTCTTCACTGGATT (34); DEF1-F, 5′-GCCATCTTCTCTGCTTTTGAA; DEF1-R, 5′-ACACAAGACACTGCGACGAC (34); FLS2a-F, 5′-ATTCCGGCGTCTATTTTCTGTA; FLS2a-R, 5′-ATTAGTCAGCCACAAAGGGAAA; PAL2-F, 5′-CACCCTCCTTCAAGGTTACTCCGG; PAL2-R, 5′-GGGACGAGATCGCCGGAGGCGG; PR1-F, 5′-TCGCCACAAGACTTTGTTAATG; PR1-R, 5′-GAGGCAAGATTTTCACCATAGG (67); PR2-F, 5′-TTGAGTGGATCCAACATTGAAG; PR2-R, 5′-TCATGGATATTGGTCAAAGCAG; ACT7-F, 5′-GCAATTCAAGCCGTTCTTTC; and ACT7-R, 5′-GATCCAAACGGAGGATAGCA (67). Primers targeting the defense genes FLS2a, PAL2, and PR2 were designed with the Primer3 software. The efficiencies of all primers used in our systems were tested by PCR.
Statistical analysis.
Statistical analysis of the data was performed with Prism version 7.05 (GraphPad Software). The bacterial population sizes were log transformed before analysis. Differences among population sizes and gene expressions were tested with Student t test or one-way analysis of variance, followed by Dunnett’s or Tukey’s multiple-comparison test (P < 0.05). All experiments were replicated at least twice.
ACKNOWLEDGMENTS
We are grateful to Julio Martinez and Li Hui at the USDA, ARS, for technical assistance, and to Lea Valinsky at the Central Laboratories, Ministry of Health, Jerusalem, Israel, for providing Salmonella strains.
This research was supported by grant IS-4862-15 R from the United States-Israel Binational Agricultural Research and Development Fund and by USDA Agricultural Research Service CRIS project 2030-42000-052-00D.
Contributor Information
M. T. Brandl, Email: maria.brandl@usda.gov.
Johanna Björkroth, University of Helsinki.
REFERENCES
- 1.Painter JA, Hoekstra RM, Ayers T, Tauxe RV, Braden CR, Angulo FJ, Griffin PM. 2013. Attribution of foodborne illnesses, hospitalizations, and deaths to food commodities by using outbreak data, United States, 1998–2008. Emerg Infect Dis 19:407–415. doi: 10.3201/eid1903.111866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batz MB, Hoffmann S, Morris JG. 2012. Ranking the disease burden of 14 pathogens in food sources in the United States using attribution data from outbreak investigations and expert elicitation. J Food Prot 75:1278–1291. doi: 10.4315/0362-028X.JFP-11-418. [DOI] [PubMed] [Google Scholar]
- 3.FDA. 2015. Final qualitative assessment of risk to public health from on-farm contamination of produce. Food and Drug Administration, Bethesda, MD. [Google Scholar]
- 4.Pezzoli L, Elson R, Little CL, Yip H, Fisher I, Yishai R, Anis E, Valinsky L, Biggerstaff M, Patel N, Mather H, Brown DJ, Coia JE, van Pelt W, Nielsen EM, Ethelberg S, de Pinna E, Hampton MD, Peters T, Threlfall J. 2008. Packed with Salmonella: investigation of an international outbreak of Salmonella Senftenberg infection linked to contamination of prepacked basil in 2007. Foodborne Pathog Dis 5:661–668. doi: 10.1089/fpd.2008.0103. [DOI] [PubMed] [Google Scholar]
- 5.Jackson B, Griffin P, Cole D, Walsh K, Chai S. 2013. Outbreak-associated Salmonella enterica serotypes and food commodities, United States, 199X 2008. Emerg Infect Dis 19:1239–1244. doi: 10.3201/eid1908.121511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horby PW, O’Brien SJ, Adak GK, Graham C, Hawker JI, Hunter P, Lane C, Lawson AJ, Mitchell RT, Reacher MH, Threlfall EJ, Ward LR, PHLS Outbreak Investigation Team. 2003. A national outbreak of multi-resistant Salmonella enterica serovar Typhimurium definitive phage type (DT) 104 associated with consumption of lettuce. Epidemiol Infect 130:169–178. doi: 10.1017/s0950268802008063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takkinen J, Nakari U-M, Johansson T, Niskanen T, Siitonen A, Kuusi M. 2005. A nationwide outbreak of multiresistant Salmonella Typhimurium in Finland due to contaminated lettuce from Spain, May 2005. Euro Surveill 10:E050630.1. doi: 10.2807/esw.10.26.02734-en. [DOI] [PubMed] [Google Scholar]
- 8.Brandl MT, Amundson R. 2008. Leaf age as a risk factor in contamination of lettuce with Escherichia coli O157:H7 and Salmonella enterica. Appl Environ Microbiol 74:2298–2306. doi: 10.1128/AEM.02459-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brandl MT, Mandrell RE. 2002. Fitness of Salmonella enterica serovar Thompson in the cilantro phyllosphere. Appl Environ Microbiol 68:3614–3621. doi: 10.1128/AEM.68.7.3614-3621.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erickson MC. 2012. Internalization of fresh produce by foodborne pathogens. Annu Rev Food Sci Technol 3:283–310. doi: 10.1146/annurev-food-022811-101211. [DOI] [PubMed] [Google Scholar]
- 11.Kroupitski Y, Golberg D, Belausov E, Pinto R, Swartzberg D, Granot D, Sela S. 2009. Internalization of Salmonella enterica in leaves is induced by light and involves chemotaxis and penetration through open stomata. Appl Environ Microbiol 75:6076–6086. doi: 10.1128/AEM.01084-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berger CN, Brown DJ, Shaw RK, Minuzzi F, Feys B, Frankel G. 2011. Salmonella enterica strains belonging to O serogroup 1,3,19 induce chlorosis and wilting of Arabidopsis thaliana leaves. Environ Microbiol 13:1299–1308. doi: 10.1111/j.1462-2920.2011.02429.x. [DOI] [PubMed] [Google Scholar]
- 13.Schikora A, Carreri A, Charpentier E, Hirt H. 2008. The dark side of the salad: Salmonella Typhimurium overcomes the innate immune response of Arabidopsis thaliana and shows an endopathogenic lifestyle. PLoS One 3:e2279. doi: 10.1371/journal.pone.0002279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klerks M, van Gent-Pelzer M, Franz E, Zijlstra C, Van Bruggen A. 2007. Physiological and molecular responses of Lactuca sativa to colonization by Salmonella enterica serovar Dublin. Appl Environ Microbiol 73:4905–4914. doi: 10.1128/AEM.02522-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shirron N, Yaron S. 2011. Active suppression of early immune response in tobacco by the human pathogen Salmonella Typhimurium. PLoS One 6:e18855. doi: 10.1371/journal.pone.0018855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Melotto M, Panchal S, Roy D. 2014. Plant innate immunity against human bacterial pathogens. Front Microbiol 5:411. doi: 10.3389/fmicb.2014.00411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schikora A, Virlogeux-Payan I, Bueso E, Garcia AV, Nilau T, Charrier A, Pelletier S, Menanteau P, Baccarini M, Velge P, Hirt H. 2011. Conservation of Salmonella infection mechanisms in plants and animals. PLoS One 6:e24112. doi: 10.1371/journal.pone.0024112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ronald P, Shirasu K. 2012. Front-runners in plant-microbe interactions. Curr Opin Plant Biol 15:345–348. doi: 10.1016/j.pbi.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 19.Jones JDG, Dangl JL. 2006. Plant immune system. Nature 444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 20.Nicaise V, Roux M, Zipfel C. 2009. Recent advances in PAMP-triggered immunity against bacteria: pattern recognition receptors watch over and raise the alarm. Plant Physiol 150:1638–1647. doi: 10.1104/pp.109.139709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia AV, Charrier A, Schikora A, Bigeard J, Pateyron S, de Tauzia-Moreau M-L, Evrard A, Mithöfer A, Martin-Magniette ML, Virlogeux-Payant I, Hirt H. 2014. Salmonella enterica flagellin is recognized via FLS2 and activates PAMP-triggered immunity in Arabidopsis thaliana. Mol Plant 7:657–674. doi: 10.1093/mp/sst145. [DOI] [PubMed] [Google Scholar]
- 22.Chalupowicz L, Nissan G, Brandl MT, McClelland M, Sessa G, Popov G, Barash I, Manulis-Sasson S. 2018. Assessing the ability of Salmonella enterica to translocate type III effectors into plant cells. Mol Plant Microbe Interact 31:233–239. doi: 10.1094/MPMI-07-17-0166-R. [DOI] [PubMed] [Google Scholar]
- 23.Johnson N, Litt PK, Kniel KE, Bais H. 2020. Evasion of plant innate defense response by Salmonella on lettuce. Front Microbiol 11:500. doi: 10.3389/fmicb.2020.00500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roy D, Panchal S, Rosa BA, Melotto M. 2013. Escherichia coli O157: H7 induces stronger plant immunity than Salmonella enterica Typhimurium SL1344. Phytopathology 103:326–332. doi: 10.1094/PHYTO-09-12-0230-FI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roy D, Melotto M. 2019. Stomatal response and human pathogen persistence in leafy greens under preharvest and postharvest environmental conditions. Postharvest Biol Technol 148:76–82. doi: 10.1016/j.postharvbio.2018.10.013. [DOI] [Google Scholar]
- 26.Jacob C, Melotto M. 2019. Human pathogen colonization of lettuce dependent upon plant genotype and defense response activation. Front Plant Sci 10:1769. doi: 10.3389/fpls.2019.01769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oblessuc PR, Matiolli CC, Melotto M. 2020. Novel molecular components involved in callose-mediated Arabidopsis defense against Salmonella enterica and Escherichia coli O157: H7. BMC Plant Biol 20:1–13. doi: 10.1186/s12870-019-2232-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferelli AMC, Bolten S, Szczesny B, Micallef SA. 2020. Salmonella enterica elicits and is restricted by nitric oxide and reactive oxygen species on tomato. Front Microbiol 11:391. doi: 10.3389/fmicb.2020.00391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Loon LC, Bakker PA, Pieterse CM. 1998. Systemic resistance induced by rhizosphere bacteria. Annu Rev Phytopathol 36:453–483. doi: 10.1146/annurev.phyto.36.1.453. [DOI] [PubMed] [Google Scholar]
- 30.Verhagen BWM, Glazebrook J, Tong Z, Chang H-S, van Loon LC, Pieterse CMJ. 2004. The transcriptome of rhizobacterium-induced systemic resistance in Arabidopsis. Mol Plant Microbe Interact 17:895–908. doi: 10.1094/MPMI.2004.17.8.895. [DOI] [PubMed] [Google Scholar]
- 31.Vlot AC, Dempsey DMA, Klessig DF. 2009. Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol 47:177–206. doi: 10.1146/annurev.phyto.050908.135202. [DOI] [PubMed] [Google Scholar]
- 32.Baccelli I, Mauch-Mani B. 2016. Beta-aminobutyric acid priming of plant defense: the role of ABA and other hormones. Plant Mol Biol 91:703–711. doi: 10.1007/s11103-015-0406-y. [DOI] [PubMed] [Google Scholar]
- 33.Piękna-Grochala J, Kępczyńska E. 2013. Induction of resistance against pathogens by β-aminobutyric acid. Acta Physiol Plant 35:1735–1748. [Google Scholar]
- 34.Chowdhury SP, Uhl J, Grosch R, Alquéres S, Pittroff S, Dietel K, Schmitt-Kopplin P, Borriss R, Hartmann A. 2015. Cyclic lipopeptides of Bacillus amyloliquefaciens subsp. plantarum colonizing the lettuce rhizosphere enhance plant defense responses toward the bottom rot pathogen Rhizoctonia solani. Mol Plant Microbe Interact 28:984–995. doi: 10.1094/MPMI-03-15-0066-R. [DOI] [PubMed] [Google Scholar]
- 35.García-Gutiérrez L, Zeriouh H, Romero D, Cubero J, de Vicente A, Pérez-García A. 2013. The antagonistic strain Bacillus subtilis UMAF6639 also confers protection to melon plants against cucurbit powdery mildew by activation of jasmonate- and salicylic acid-dependent defence responses. Microb Biotechnol 6:264–274. doi: 10.1111/1751-7915.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vallad GE, Goodman RM. 2004. Systemic acquired resistance and induced systemic resistance in conventional agriculture. Crop Sci 44:1920–1934. doi: 10.2135/cropsci2004.1920. [DOI] [Google Scholar]
- 37.Melotto M, Brandl MT, Jacob C, Jay-Russell MT, Micallef SA, Warburton ML, Van Deynze A. 2020. Breeding crops for enhanced food safety. Front Plant Sci 11:428. doi: 10.3389/fpls.2020.00428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wong CWY, Wang S, Lévesque RC, Goodridge L, Delaquis P. 2019. Fate of 43 Salmonella strains on lettuce and tomato seedlings. J Food Prot 82:1045–1051. doi: 10.4315/0362-028X.JFP-18-435. [DOI] [PubMed] [Google Scholar]
- 39.Durrant WE, Dong X. 2004. Systemic acquired resistance. Annu Rev Phytopathol 42:185–209. doi: 10.1146/annurev.phyto.42.040803.140421. [DOI] [PubMed] [Google Scholar]
- 40.Pieterse CM, Zamioudis C, Berendsen RL, Weller DM, Van Wees SC, Bakker PA. 2014. Induced systemic resistance by beneficial microbes. Annu Rev Phytopathol 52:347–375. doi: 10.1146/annurev-phyto-082712-102340. [DOI] [PubMed] [Google Scholar]
- 41.Kalifa HB, Rav-David D, Borenshtein M, Shulchany R, Elad Y. 2012. Climate change effect on plant–pathogen–beneficial microorganisms interaction in high humidity-promoted tomato diseases. IOBC/WPRS Bull 78:15–18. [Google Scholar]
- 42.Agra O, Rav-David D, Borenshtein M, Shulchany R, Elad Y. 2012. Climate effect on pathogen–biocontrol agents interaction in the tomato–powdery mildew (Oidium neolycopersici) pathosystem. IOBC/WPRS Bull 78:233–237. [Google Scholar]
- 43.Yan F, Xu S, Chen Y, Zheng X. 2014. Effect of rhamnolipids on Rhodotorula glutinis biocontrol of Alternaria alternata infection in cherry tomato fruit. Postharvest Biol Technol 97:32–35. doi: 10.1016/j.postharvbio.2014.05.017. [DOI] [Google Scholar]
- 44.Li Z, Wang X, Chen F, Kim H-J. 2007. Chemical changes and overexpressed genes in sweet basil (Ocimum basilicum L.) upon methyl jasmonate treatment. J Agric Food Chem 55:706–713. doi: 10.1021/jf062481x. [DOI] [PubMed] [Google Scholar]
- 45.Chandler S, Van Hese N, Coutte F, Jacques P, Höfte M, De Vleesschauwer D. 2015. Role of cyclic lipopeptides produced by Bacillus subtilis in mounting induced immunity in rice (Oryza sativa L.). Physiol Mol Plant Pathol 91:20–30. doi: 10.1016/j.pmpp.2015.05.010. [DOI] [Google Scholar]
- 46.Choudhary DK, Johri BN. 2009. Interactions of Bacillus spp. and plants – With special reference to induced systemic resistance (ISR). Microbiol Res 164:493–513. doi: 10.1016/j.micres.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 47.Ramírez-Valdespino CA, Casas-Flores S, Olmedo-Monfil V. 2019. Trichoderma as a model to study effector-like molecules. Front Microbiol 10:1030. doi: 10.3389/fmicb.2019.01030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meller Harel Y, Mehari Z, Rav-David D, Elad Y. 2014. Systemic resistance to gray mold induced in tomato by benzothiadiazole and Trichoderma harzianum T39. Phytopathology 104:150–157. doi: 10.1094/PHYTO-02-13-0043-R. [DOI] [PubMed] [Google Scholar]
- 49.Baiyee B, Ito S-i, Sunpapao A. 2019. Trichoderma asperellum T1 mediated antifungal activity and induced defense response against leaf spot fungi in lettuce (Lactuca sativa L.). Physiol Mol Plant Pathol 106:96–101. doi: 10.1016/j.pmpp.2018.12.009. [DOI] [Google Scholar]
- 50.Zhang X, Wu F, Gu N, Yan X, Wang K, Dhanasekaran S, Gu X, Zhao L, Zhang H. 2020. Postharvest biological control of Rhizopus rot and the mechanisms involved in induced disease resistance of peaches by Pichia membranefaciens. Postharvest Biol Technol 163:111146. doi: 10.1016/j.postharvbio.2020.111146. [DOI] [Google Scholar]
- 51.Cohen YR. 2002. β-Aminobutyric acid-induced resistance against plant pathogens. Plant Dis 86:448–457. doi: 10.1094/PDIS.2002.86.5.448. [DOI] [PubMed] [Google Scholar]
- 52.Yigit F. 2011. Acibenzolar-S-methyl induces lettuce resistance against Xanthomonas campestris pv. vitians. Afr J Biotechnol 10:9606–9612. [Google Scholar]
- 53.Patel JS, Zhang S, de Novaes MIC. 2014. Effect of plant age and acibenzolar-S-methyl on development of downy mildew of basil. Horts 49:1392–1396. doi: 10.21273/HORTSCI.49.11.1392. [DOI] [Google Scholar]
- 54.Cohen Y, Rubin AE, Kilfin G. 2010. Mechanisms of induced resistance in lettuce against Bremia lactucae by dl-β-amino-butyric acid (BABA). Eur J Plant Pathol 126:553–573. doi: 10.1007/s10658-009-9564-6. [DOI] [Google Scholar]
- 55.Zimmerli L, Jakab G, Métraux J-P, Mauch-Mani B. 2000. Potentiation of pathogen-specific defense mechanisms in Arabidopsis by β-aminobutyric acid. Proc Natl Acad Sci USA 97:12920–12925. doi: 10.1073/pnas.230416897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Po‐Wen C, Singh P, Zimmerli L. 2013. Priming of the Arabidopsis pattern-triggered immunity response upon infection by necrotrophic Pectobacterium carotovorum bacteria. Mol Plant Pathol 14:58–70. doi: 10.1111/j.1364-3703.2012.00827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meng F, Altier C, Martin GB. 2013. Salmonella colonization activates the plant immune system and benefits from association with plant pathogenic bacteria. Environ Microbiol 15:2418–2430. doi: 10.1111/1462-2920.12113. [DOI] [PubMed] [Google Scholar]
- 58.Yi SY, Shirasu K, Moon JS, Lee S-G, Kwon S-Y. 2014. The activated SA and JA signaling pathways have an influence on flg22-triggered oxidative burst and callose deposition. PLoS One 9:e88951. doi: 10.1371/journal.pone.0088951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van der Ent S, Van Hulten M, Pozo MJ, Czechowski T, Udvardi MK, Pieterse CM, Ton J. 2009. Priming of plant innate immunity by rhizobacteria and β-aminobutyric acid: differences and similarities in regulation. New Phytol 183:419–431. doi: 10.1111/j.1469-8137.2009.02851.x. [DOI] [PubMed] [Google Scholar]
- 60.Flors V, Ton J, Van Doorn R, Jakab G, García-Agustín P, Mauch‐Mani B. 2008. Interplay between JA, SA, and ABA signaling during basal and induced resistance against Pseudomonas syringae and Alternaria brassicicola. Plant J 54:81–92. doi: 10.1111/j.1365-313X.2007.03397.x. [DOI] [PubMed] [Google Scholar]
- 61.Conrath U, Beckers GJ, Flors V, García-Agustín P, Jakab G, Mauch F, Newman M-A, Pieterse CM, Poinssot B, Pozo MJ, Pugin A, Schaffrath U, Ton J, Wendehenne D, Zimmerli L, Mauch-Mani B, Prime-A-Plant Group. 2006. Priming: getting ready for battle. Mol Plant Microbe Interact 19:1062–1071. doi: 10.1094/MPMI-19-1062. [DOI] [PubMed] [Google Scholar]
- 62.Martinez-Medina A, Flors V, Heil M, Mauch-Mani B, Pieterse CM, Pozo MJ, Ton J, Van Dam NM, Conrath U. 2016. Recognizing plant defense priming. Trends Plant Sci 21:818–822. doi: 10.1016/j.tplants.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 63.Bostock RM. 2005. Signal crosstalk and induced resistance: straddling the line between cost and benefit. Annu Rev Phytopathol 43:545–580. doi: 10.1146/annurev.phyto.41.052002.095505. [DOI] [PubMed] [Google Scholar]
- 64.Elad Y, Rav-David D. 2013. Induction of resistance under different water regimes. IOBC-WPRS Bull 89:155–159. [Google Scholar]
- 65.Simmons CW, VanderGheynst JS, Upadhyaya SK. 2009. A model of Agrobacterium tumefaciens vacuum infiltration into harvested leaf tissue and subsequent in planta transgene transient expression. Biotechnol Bioeng 102:965–970. doi: 10.1002/bit.22118. [DOI] [PubMed] [Google Scholar]
- 66.De Cremer K, Mathys J, Vos C, Froenicke L, Michelmore RW, Cammue BPA, De Coninck B. 2013. RNA seq-based transcriptome analysis of Lactuca sativa infected by the fungal necrotroph Botrytis cinerea. Plant Cell Environ 36:1992–2007. doi: 10.1111/pce.12106. [DOI] [PubMed] [Google Scholar]
- 67.Simko I, Zhou Y, Brandl MT. 2015. Downy mildew disease promotes the colonization of romaine lettuce by Escherichia coli O157:H7 and Salmonella enterica. BMC Microbiol 15:1–9. doi: 10.1186/s12866-015-0360-5. [DOI] [PMC free article] [PubMed] [Google Scholar]