ABSTRACT
Lacticaseibacillus rhamnosus GG is a widely marketed probiotic with well-documented probiotic properties. Previously, deletion of the mucus-adhesive spaCBA-srtC1 genes in dairy isolates was reported. In this study, we examined the genome preservation of industrially produced L. rhamnosus GG (DSM 33156) cofermented in yogurts. In total, DNA of 66 samples, including 60 isolates, was sequenced. Population samples and 59 isolates exhibited an intact genome. One isolate exhibited loss of spaCBA-srtC1. In addition, we examined phenotypes related to the probiotic properties of L. rhamnosus GG either from frozen pellets or cofermented in yogurt. L. rhamnosus GG from frozen pellets induced a response in intestinal barrier function in vitro, in contrast to frozen pellets of the starter culture. Yogurt matrix, containing only the starter culture, induced a response, but cofermentation with L. rhamnosus GG induced a higher response. Conversely, only the starter culture stimulated cytokine secretion in dendritic cells, and it was observed that the addition of L. rhamnosus GG to the starter culture reduced the response. We conclude that the L. rhamnosus GG genome is preserved in yogurt and that common in vitro probiotic effects of L. rhamnosus GG are observed when examined in the yogurt matrix.
IMPORTANCE Lacticaseibacillus rhamnosus GG is a well-documented probiotic strain recognized for its high acid and bile tolerance and properties of adhesion to enterocytes and mucus. The strain exhibits SpaCBA pili, which have been demonstrated to play an important role in adhesion and therefore are relevant for persistence in the gastrointestinal tract. Recently we demonstrated that the genome and phenotypes of L. rhamnosus GG are preserved throughout an industrial production pipeline. However, as gene deletions in L. rhamnosus GG were previously reported for isolates from dairy products, a key question on the genomic stability of L. rhamnosus GG in a yogurt matrix remained. The aim of this study was to analyze genome stability and phenotypic characteristics of L. rhamnosus GG in yogurt. We found that the genome of L. rhamnosus GG is well conserved when the organism is cofermented in yogurt. Some phenotypic characteristics are consistent in all product matrixes, while other characteristics are modulated.
KEYWORDS: Lacticaseibacillus rhamnosus GG, industrial fermentation, probiotics, yogurt, genome stability, SpaCBA
INTRODUCTION
The probiotic potential of Lacticaseibacillus rhamnosus GG was initially described in 1985 by Sherwood Gorbach and Barry Goldin, and in 1989, the organism became the first Lactobacillus strain to be patented based on its high acid and bile tolerance and properties of adhesion to enterocytes and mucus (1, 2). Over the past 30 years, health-promoting effects of the strain have been demonstrated in hundreds of clinical trials, including shortening the duration of acute infectious or antibiotic-associated diarrhea (3–5), eliminating gastrointestinal carriage of vancomycin-resistant enterococci (6), and acting as an adjuvant to improve influenza vaccine immunogenicity (7).
Prolonged persistence time and colonization in the gastrointestinal tract are considered of great importance for L. rhamnosus GG to confer its reported health benefits. The ability to adhere has therefore been widely studied in L. rhamnosus GG (8–10). In 1992, Coconnier et al. discovered that the mechanism of adhesion of L. rhamnosus GG was sensitive to protease activity (11), and later, several studies characterized the adhesion properties of the surface protein appendages on L. rhamnosus GG called pili (9, 10, 12). Importantly, pilus-deficient mutants of L. rhamnosus GG, produced using chemical mutagenesis, lose the ability to adhere to porcine and human intestinal mucus (decreased from 37.9% to 0.5 to 1.8% adherence) (13).
The L. rhamnosus GG pili are encoded by the spaCBA-srtC1 gene cluster, which is located in a region rich in insertion sequences (IS) that can mediate larger genomic changes (14–16). Accordingly, the genomic stability of L. rhamnosus GG, in particular the spaCBA-srtC1 gene cluster, has been examined in several studies. One study showed high genomic stability of L. rhamnosus GG in general, but in some isolates continuously exposed to bile or mechanical stress, a mutation that led to the loss of the spaCBA-srtC1 gene cluster was detected after 1,000 generations (14). In a different study, where isolates from three different liquid dairy products were sequenced, two isolates showed large genomic deletions that included the spaCBA-srtC1 gene cluster (17). Subsequently, we demonstrated high genomic stability of L. rhamnosus GG throughout an industrial production process from the original stock in the Chr. Hansen culture collection (CHCC) to the freeze-dried product (18).
Whereas genomic stability is important to ensure that probiotic potential is retained, the composition of the delivery matrix of the strain may influence how the strain behaves phenotypically. In addition to delivery as a freeze-dried product in tablets or capsules, probiotic yogurts are an increasingly common and popular food vehicle consumed for their potential health-promoting effects. L. rhamnosus GG is one of the most prominent probiotic yogurt cultures on the market (19). However, only a few in vivo studies have compared the probiotic effect of L. rhamnosus GG when delivered in different matrices. Two studies have shown similar fecal recovery of L. rhamnosus GG when it was ingested either as a freeze-dried product in tablets or capsules or in fermented milk, indicating no significant matrix effect on survival and colonization (20, 21).
Several in vitro assays have been used to study the mode of action of L. rhamnosus GG, demonstrating various probiotic features, including pathogen inhibition (8), production of mucosa-protecting biofilm (23), superior adhesion properties compared to those of other lactobacilli (24), and modulation of immune responses by reducing several inflammation markers in human intestinal cells (25, 26). Generally, in vitro assays are performed on overnight cultures of probiotic strains, either from a frozen stock or isolated from a product matrix such as yogurt, which enables a high concentration of the investigated strain without the interference of other strains and a potential matrix effect. Only a few studies have examined in vitro characteristics of probiotic strains derived directly from industrially produced products.
In clinical studies, L. rhamnosus GG has been administered either as freeze-dried bacteria in capsules, tablets, or powder or in fermented milk products. While bacteria grown as overnight cultures will reflect laboratory conditions, industrially produced frozen bacteria will remain in a dormant state preserving the physiological profile obtained during production and bacteria in yogurt will adapt to the conditions in the yogurt matrix. Several studies have demonstrated that bacteria can have a dynamic surface architecture and that even minor differences in environmental conditions can affect probiotic properties (27–29). Accordingly, differences in physiological conditions of frozen bacteria and bacteria in a yogurt matrix could result in different phenotypic characteristics.
As mentioned above, high genomic stability was demonstrated in freeze-dried product (18), but the genomic deletions previously identified originated from liquid dairy isolates (17). We hypothesize that the low number of generations of L. rhamnosus GG that occur during yogurt production would not allow a mutated variant to become a noteworthy proportion of the population, even if the mutation provides a niche fitness improvement. In this study, we aimed to assess genome stability of L. rhamnosus GG after cofermentation in a yogurt product in order to validate our hypothesis as a quality assurance of the application of L. rhamnosus GG in yogurt.
In addition, we wanted to determine how specific phenotypic characteristics relevant for probiotic functionality are influenced by matrix.
RESULTS
Genome integrity of L. rhamnosus GG was conserved in all product samples, and a gene deletion was detected in one isolate.
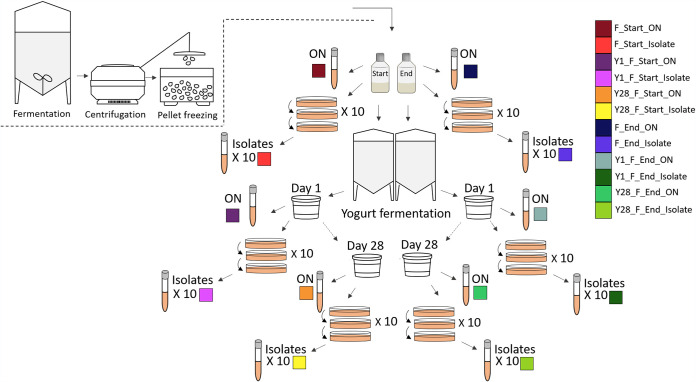
To analyze the genome integrity of L. rhamnosus GG after yogurt fermentation and subsequent cold storage throughout the shelf life of 28 days, the genomes of 66 samples were sequenced. In this industrial production process, L. rhamnosus GG is batch fermented. The strain is inoculated into a closed system filled with a sterilized nutrient solution and fermented under continuously controlled parameters such as pH and temperature. Upon harvesting, the bacterial culture is concentrated and mixed with cryoprotective reagents before pellet freezing, where droplets of the culture are frozen instantaneously in liquid nitrogen. Samples included frozen pellets of L. rhamnosus GG from both the start (F_Start) and the end (F_End) of pellet freezing of an industrial production batch and from yogurts fermented by Lactobacillus delbrueckii subsp. bulgaricus (L. bulgaricus) and Streptococcus thermophilus together with the L. rhamnosus GG samples. Samples of yogurt were prepared for sequencing on the day of yogurt production (Y1_F_start_ON and Y1_F_end_ON) and at the end of shelf life (Y28_F_start_ON and Y28_F_end_ON). In addition, 10 single colony isolates were prepared from each of the six full population samples (isolates 1 to 10) (Fig. 1).
FIG 1.
Overview of samples included in genome sequencing. Samples of frozen pellets of L. rhamnosus GG were collected from start of pelletizing (F_Start) and end of pelletizing (F_end) in a production batch and included in yogurt fermentation. Samples of yogurt were collected on the day of production (Y1) and after 28 days of refrigerated storage (Y28). Full-genome sequencing was performed on an overnight culture (ON) of the frozen pellets and the four samples of yogurt (Y1_F_Start, Y1_F_end, Y28_F_start, and Y28_F_end). Ten isolates were purified from each of the six full population samples. Sample identification and colors are defined in the key.
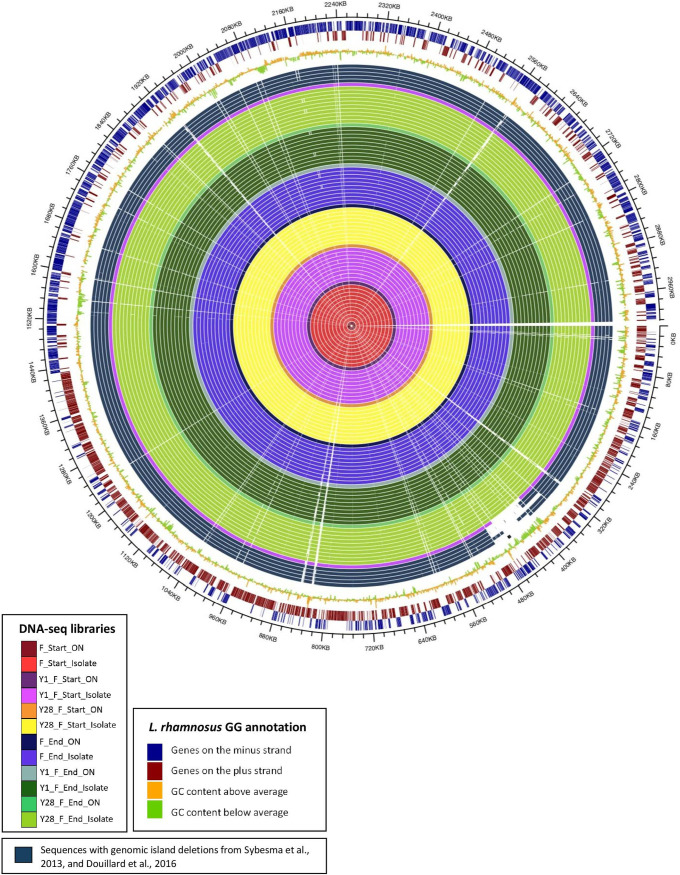
Sequencing data from all samples were assembled de novo, resulting in 66 high-quality draft genomes with a minimum average base coverage of 118×. A hierarchical clustering was performed based on gene frequency for all the samples sequenced in this study, three publicly available reference genomes of L. rhamnosus GG on NCBI RefSeq database and five previously published L. rhamnosus GG genomes of isolates in which the spaCBA-srtC1 gene cluster was lost (Sequence Read Archive [SRA] identifiers SRR645838, SRR3098054, SRR645868, SRR3096056, and SRR3096092). All 66 samples clustered with the reference genomes except one isolate, Y1_F_Start_I9 (isolate 9), which was purified from yogurt and clustered with the sequences of the five isolate samples from literature in which spaCBA-srtC1 was lost (Fig. 2).
FIG 2.
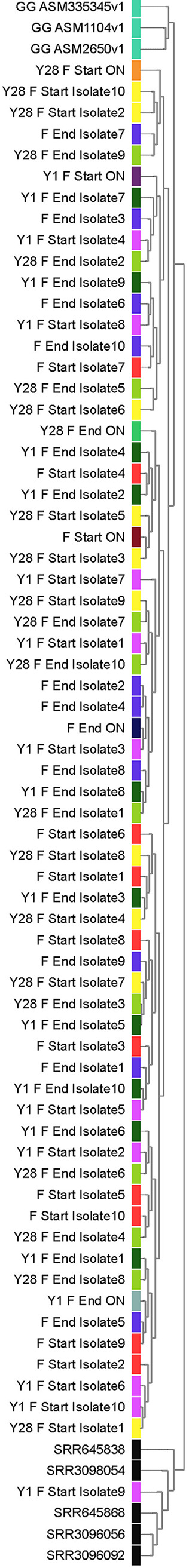
Genome similarity between sequenced samples of L. rhamnosus GG. Three publicly available reference genomes for L. rhamnosus GG on NCBI (ASM2650v1, ASM335345v1, and ASM1104v1) and five previously published genomes of L. rhamnosus GG in which the spaCBA-srtC1 gene cluster was lost (SRR645838, SRR3098054, SRR645868, SRR3096056, and SRR3096092 in NCBI Sequence Read Archive [SRA]) are included. Hierarchical clusters of genomes are based on gene similarities. Sample identifiers and colors are the same as in Fig. 1.
Alignment of the contigs derived from the 66 samples showed consistent coverage across genomes and high identity scores compared to the reference (FM179322 [https://www.ncbi.nlm.nih.gov/assembly/GCF_000026505.1/]). No gene loss was observed in any of the six overnight cultures containing the full population of L. rhamnosus GG prepared from the frozen products and their respective yogurt productions, both on the day of production and at the end of shelf life. In contrast, contig alignment of sequences of the 60 single colony isolate samples showed that one isolate, isolate 9, was missing a region containing the spaCBA-srtC1 cluster (Fig. 3).
FIG 3.
Circular representation of alignments of DNA sequence contigs derived from the 66 samples sequenced. Each circle indicates the sequence from one sample, color-coded as defined in the key. For comparison, contigs from five sequences with gene deletions reported previously by Sybesma et al. (17) and Douillard et al. (14) are included. The three outer circles represent GC content and annotated genes on both strands, as indicated in the key.
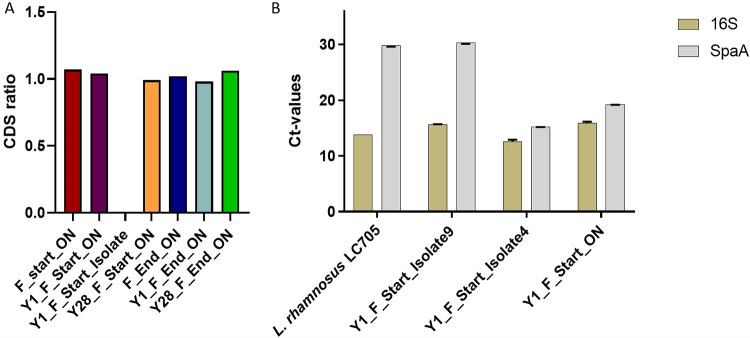
The gene deletion in isolate 9 could have been present in a subpopulation of the L. rhamnosus GG population in the yogurt sample, or it could have arisen during the restreaking steps of the isolation process. To examine if a subpopulation of L. rhamnosus GG in the yogurt from which isolate 9 was purified was missing the spaCBA-srtC1 gene cluster, the ratio of coverage of average coding sequences (CDS) between the spaCBA-srtC1 region and the remaining genes in the genome was calculated for each sample (Fig. 4A). All ratios remained on a level of approximately 1 from the frozen L. rhamnosus GG products to their respective yogurt productions, both on the day of yogurt production and at the end of shelf life. With an average depth of >100-fold coverage, these results show that if a subpopulation was missing the spaCBA-srtC1 cluster, it would be less than 1% of the full population. Isolate 9 was included in the ratio analysis and showed a ratio of 0, consistent with the notion that the spaCBA-srtC1 cluster was missing in the genome sequence of this single colony isolate.
FIG 4.
Validation of the gene deletion. (A) Proportion of genomes containing the spaCBA-srtC1 gene cluster presented as the ratio of coding sequence (CDS) coverages of the genes deleted in isolate 9 and the remaining genes. The samples included all sequences of overnight cultures (_ON) and Y1_F_start_I9. Ratios of CDS are given on the y axis. (B) qPCR on the spaA gene representing the spaCBA-srtC1 gene cluster and 16S rRNA included as a control of DNA. Samples included Y1_F_Start_I4 (positive control), Y1_F_Start_ON (population from which Y1_F_Start_I9 was purified), L. rhamnosus LC705 (strain without the spaCBA-srtC1 gene cluster as a negative control), and Y1_F_Start_I9 where the spaCBA-srtC1 gene cluster was lost. CT values are given on the y axis.
To validate the sequencing results with an independent analysis on separately isolated DNA, loss of the spaCBA-srtC1 gene cluster in isolate 9 was verified by quantitative PCR (qPCR) (Fig. 4B). The presence of spaA was used as a measure of the spaCBA-srtC1 gene cluster, and Lacticaseibacillus rhamnosus 16S rRNA gene was used as a positive control. Lacticaseibacillus rhamnosus LC705 was included as a negative control, as its genome does not contain the spaCBA-srtC1 cluster (10), and a different single colony isolate, Y1_F_start_I4 (isolate 4), which retained the spaCBA-srtC1 genes, was included as a positive control. In addition, the full population sample Y1_F_start_ON, from which isolate 9 was purified, was included. The presence of spaA was confirmed in both the positive control, isolate 4, and the full population sample, with similar threshold cycle (CT) values between 16S and spaA in both isolate 4 and Y1_F_start_ON (Fig. 4B). The negative control, L. rhamnosus LC705, displayed a CT value of 13.8 for 16S rRNA, confirming the presence of DNA in the sample, whereas spaA gave a CT value of 29.8, which was ascribed to nonspecific binding and primer dimers. Based on the results from the negative control, CT values of ≥29.8 were considered to indicate absence of the gene. For isolate 9, a CT value of 15.7 for 16S rRNA was observed, whereas spaA was not present, with a value above 29.8 (CT = 30.3). Therefore, the qPCR results confirm that isolate 9 indeed lacks the spaA gene, consistent with the results from DNA sequencing and ratio analysis.
The analysis of whole-genome integrity was complemented with an assessment of the presence of single nucleotide polymorphisms (SNPs) in order to identify potential mutational hot spots in the genomes that could result in genomic instability. In total, 21 different SNPs were identified across the 60 isolates sequenced (see Table S3 in the supplemental material); 5 of the SNPs were identified previously and shown to represent a subpopulation in the original stock from the culture collection used for this production (18). The remaining 16 SNPs were distributed across isolates (1 or 2 SNPs per isolate). All SNPs were detected in 1 or 2 isolates only, except for one SNP that was identified in 13 isolate sequences. To examine whether strain phenotype was affected by the identified SNPs, the consequence of a potential amino acid substitution was predicted using the PROVEAN (Protein Variation Effect Analyzer) method based on alignments and chemical properties of amino acids. Of the eight SNPs that were identified in more than one isolate, seven were predicted to have a neutral effect and one was predicted to have a deleterious consequence (genome position 2253036) (Table S3).
L. rhamnosus GG exhibits high acid tolerance and moderate bile tolerance regardless of matrix.
In addition to examining genome stability of L. rhamnosus GG in yogurt, several in vitro assays were performed to investigate the phenotypic characteristics of L. rhamnosus GG as a frozen product and in a yogurt matrix. CFU of L. rhamnosus GG in the yogurt products were counted on the day of production (3.7 × 108 CFU/ml) and at end of shelf life (3.4 × 108 CFU/ml) to ensure that no substantial decrease in viable cells occurred during cold storage.
To examine possible effects on gastrointestinal survival of L. rhamnosus GG due to the yogurt matrix or adaptation of the strain during fermentation and subsequent storage in yogurt, in vitro tolerance to gastric acid and bile was measured. Samples of the frozen L. rhamnosus GG and yogurt cofermented with L. rhamnosus GG from the day of production and at the end of shelf life were tested. To distinguish the possible effect of the yogurt matrix on strain survival from the effect of strain adaptation to the yogurt conditions, a sample of frozen L. rhamnosus GG mixed with a control yogurt (same recipe but without L. rhamnosus GG) immediately before the assay was included.
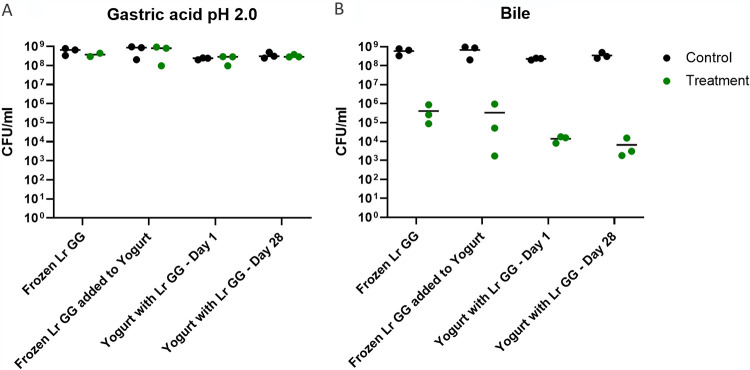
All samples exhibited high tolerance to gastric acid solution at pH 2.0, with no significant decrease (P > 0.05) in CFU after 1 h of incubation compared with their respective controls (Fig. 5A). In contrast, 1 h of incubation in 1% bile solution resulted in decreased survival in all samples with a reduction in CFU of 4 to 5 logs (Fig. 5B). When comparing the mean values in survival from three replicate experiments, a nonsignificant trend toward increased bile tolerance for frozen L. rhamnosus GG compared with L. rhamnosus GG cofermented in yogurt was observed. Therefore, matrix appears to have little to no effect on acid tolerance and a modest effect on bile tolerance.
FIG 5.
Tolerance to gastric acid and bile of L. rhamnosus GG. Samples included L. rhamnosus GG as frozen product (Frozen Lr GG [F_start]) and in yogurt. Yogurt samples included frozen L. rhamnosus GG added to yogurt immediately before treatment (Frozen Lr GG added to Yogurt) and yogurt cofermented with L. rhamnosus GG, both from the day of production (Yogurt with Lr GG - Day 1) and at the end of shelf life (Yogurt with Lr GG - Day 28) (x axis). Results are shown as CFU per milliliter after 1 h of incubation in gastric acid solution at pH 2.0 (A) and 1% porcine bile solution (B). For each treatment a control is included, shown as CFU per milliliter after 1 h of incubation in 10% MRS broth (y axis). Data points represent the means for technical duplicates, and horizontal lines represent the means from three experiments. One-way ANOVA followed by Tukey’s test comparing the different sample sets was performed on treatment normalized to control.
A time course of bile tolerance was performed in order to determine the rate of decrease in survival. In this experiment, survival was measured after 7.5, 15, 30, and 60 min. The results showed that the decrease in survival had already occurred after 7.5 min and after then no further decrease was observed (Fig. S1).
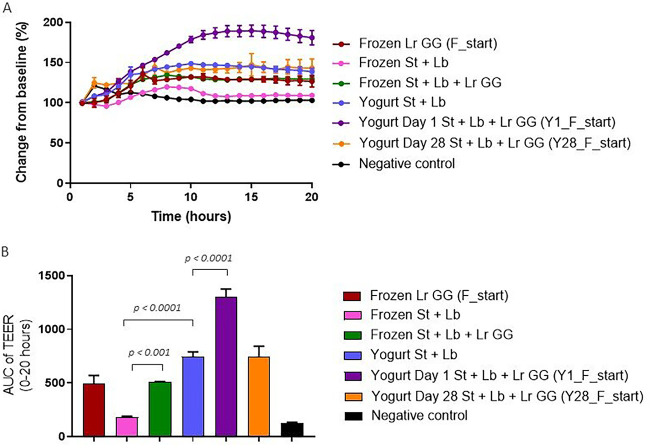
L. rhamnosus GG in yogurt stimulates a greater increase in epithelial barrier function in vitro than L. rhamnosus GG alone.
As a measure of the effect on epithelial cell barrier function, transepithelial electrical resistance (TEER) was assessed across a differentiated monolayer of Caco-2 cells incubated for 20 h with L. rhamnosus GG alone, the starter culture alone (S. thermophilus and L. bulgaricus), and the three strains (L. rhamnosus GG, S. thermophilus, and L. bulgaricus) in combination. The effects of the strains on TEER were measured both for frozen cultures and for yogurt samples. As the concentration of the starter culture was higher than the concentration of L. rhamnosus GG in the yogurt (2 × 109 CFU/ml compared to 4 × 108 CFU/ml), the same proportion was used for the mix of all three strains as frozen cultures. After adjusting for the typical bacterium/cell ratio used in the TEER assay, the final concentrations used were 2 × 108 CFU/ml for the starter culture and 4 × 107 CFU/ml for L. rhamnosus GG.
In contrast to the starter culture alone, L. rhamnosus GG alone induced a substantial response in TEER, with 30% increase relative to baseline after 15 h (Fig. 6A). The induced response measured from a mixture of the three strains was similar to the effect observed for L. rhamnosus GG alone. Although the starter culture strains from the frozen product did not induce a response, yogurt containing only the starter culture induced an increase in TEER of 44% relative to baseline. The sample of freshly produced yogurt cofermented with L. rhamnosus GG induced an additional response, with an increase of 90% relative to baseline (Fig. 6A). However, by end of shelf life, the additional response observed in yogurt cofermented with L. rhamnosus GG had diminished, showing an increase of 47% relative to baseline after 15 h, as seen for the control yogurt.
FIG 6.
Transepithelial electrical resistance (TEER) of L. rhamnosus GG and the starter culture as frozen product and in yogurt. (A) TEER of Caco-2 cell monolayers stimulated with frozen pellets of the starter culture (Frozen St + Lb), frozen pellets of L. rhamnosus GG (Frozen Lr GG [F_start]), or a combination of the two samples (Frozen St + Lb + Lr GG). In addition, yogurt containing only the starter culture (Yogurt St + Lb) and yogurt cofermented with L. rhamnosus GG on the day of production (Yogurt Day 1 St + Lb + Lr GG [Y-1_F_start]) and by end of shelf life (Yogurt Day 28 St + Lb + Lr GG [Y28_F_start]) were included in the experiment. The final concentrations used for all samples were 2 × 108 CFU/ml and 4 × 107 CFU/ml for the starter culture and L. rhamnosus GG, respectively. (A) The y axis shows the percent change in TEER relative to the baseline, which is the TEER reading just prior to adding the bacterial strains (time = 0 h). The x axis shows the time in hours. Each data point represents the mean ± SD of results from technical triplicates. (B) Area under the curve (AUC) of TEER measurements from 0 to 20 h relative to the baseline (100%) (y axis) for the measurement presented in panel A. Each bar represents the mean ± SD for technical triplicates. One-way ANOVA followed by Tukey’s test was performed, and statistically significant differences are shown.
Quantification and statistical analysis of the area under the curve (AUC) indicated that yogurt matrix induced a significantly greater TEER response than the starter culture strains alone (P < 0.0001) and that addition of L. rhamnosus GG to the starter culture, both as frozen products and in yogurt on the day on the day of production, resulted in significantly greater TEER responses (P < 0.001 and P < 0.0001, respectively) (Fig. 6B).
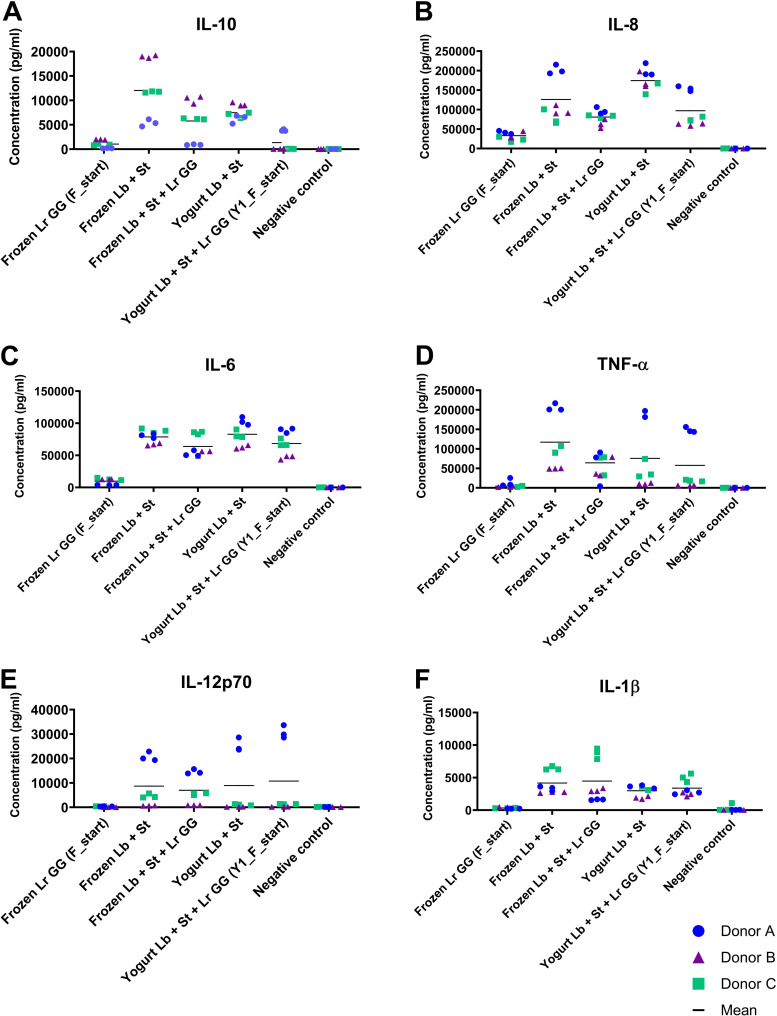
L. rhamnosus GG inhibits cytokine response from the starter culture in dendritic cells.
The ability of the starter culture and L. rhamnosus GG to modulate secretion of cytokines in dendritic cells (DCs) was evaluated in vitro by measuring the regulation of six inflammation-related cytokines in human monocyte-derived DCs from three different donors. As with the barrier function assay, samples of L. rhamnosus GG alone, the starter culture, and the combination of all three strains were examined both as frozen cultures and in the yogurt matrix. DCs were exposed to the starter culture at a ratio of 1:15 and to L. rhamnosus GG at a ratio of 1:50 to obtain the ratio between L. rhamnosus GG and the starter culture present in the yogurt. Donor variation is typical for in vitro DC assays, and therefore, the three donors are presented individually. In contrast to the low response induced by the starter culture in the intestinal barrier assay, the starter culture induced higher secretion of all six cytokines measured in the dendritic cell assay than L. rhamnosus GG, which induced relatively modest responses compared with the negative control (Fig. 7). Compared with the negative control, the starter culture induced significant secretion of all six cytokines, whereas L. rhamnosus GG induced significant secretion of only interleukin 6 (IL-6) and IL-8 (Table S2). Addition of L. rhamnosus GG to the starter culture, both for the frozen product and in the yogurt matrix, resulted in decreased responses of the cytokines IL-10, IL-8, and IL-6 (except for the frozen sample in donor C in IL-8 and IL-6) compared with the starter culture alone. The tendency was most pronounced for IL-10 and IL-8 (Fig. 7A and B and Table S2). In contrast, the IL-12p70 and IL-1β responses stimulated by the starter culture and the combination of starter culture and L. rhamnosus GG were similar, both for frozen samples and in yogurt, with only donor A in IL-12p70 showing a significant difference.
FIG 7.
Cytokine response to L. rhamnosus GG and starter culture as frozen product and in yogurt. Cytokine concentrations (y axis) of IL-10 (A), IL-8 (B), IL-6 (C), TNF-α (D), IL-12p70 (E), and IL-1β (F) secreted by monocyte-derived human dendritic cells derived from three different donors. Dendritic cells were stimulated with the following samples: frozen pellets of L. rhamnosus GG (Frozen Lr GG [F_start]), frozen pellets of the starter culture (Frozen St + Lb), a combination of the frozen samples (Frozen St + Lb + Lr GG), yogurt containing only the starter culture (Yogurt St + Lb), and yogurt cofermented with L. rhamnosus GG [Yogurt St + Lb + Lr GG (F_start)] (x axis). The final concentrations used for all samples were 5 × 107 CFU/ml and 1.5 × 107 CFU/ml for the starter culture and L. rhamnosus GG, respectively. Negative controls were incubated in cell culture medium only. Each data point represents the mean of technical replicate measurements, and biological triplicates were assessed for each donor. Black bars represent the mean concentrations for the three donors.
DISCUSSION
The analysis of the genome integrity of L. rhamnosus GG after yogurt fermentation and subsequent cold storage samples included 66 full genome-sequenced samples, including 6 full population overnight cultures from two frozen culture products and four yogurt products (two from the day of production and two at the end of shelf life) as well as 10 single colony isolates from each product.
All full population samples displayed genomes with no loss of genes detected. The spaCBA-srtC1 gene cluster was of particular interest, as these genes are responsible for the adhesive pili present on the cell surface and gene deletions have been observed in this region previously (10, 14, 17). Therefore, a calculation of the ratio between CDS coverage for the deletion including the spaCBA-srtC1 gene cluster and the remaining genes was performed. The results showed a ratio of approximately 1 for all full population samples, indicating that the number of reads of spaCBA-srtC1 genes approximately corresponds to the number of reads of the remaining coding genes present in the genome. Accordingly, it can be concluded that no larger proportion of the genomes is missing the gene cluster, since a fraction of less than 1% of genomes missing the genes would not be detectable from data analysis on the sequences alone based on the average depth of >100-fold coverage.
From the 40 single colony isolates from the yogurt, loss of the spaCBA-srtC1 gene cluster was identified in one isolate, referred to as isolate 9. Deletion of the gene cluster could have either (i) occurred during the purification of the isolate or (ii) been present already in a nondetectable subpopulation.
As the L. rhamnosus GG genome is rich in IS in the region containing the spaCBA-srtC1 gene cluster, the region is expected to be more susceptible to gene deletions (12). Stressful conditions are generally thought to increase the rate of mutation, and the gene deletions leading to the loss of spaCBA-srtC1 observed by Douillard et al. (14) also arose during prolonged exposure to stress. The purification of single colony isolates can be stressful since nutrients are depleted in the area around colonies during growth in each of the three purification steps, and this has been demonstrated to induce high plasticity of a genome (30, 31). If a minor subpopulation of the full population below the detection level exists, the limited number of generations during the production process and yogurt fermentation will ensure that the subpopulation will remain minor.
Interestingly, it was observed that the deletions identified by Sybesma et al. (17) and Douillard et al. (14) varied in length. This observation suggests that the deletions occurred independently, and the gene deletion cannot be ascribed to a common ancestor. This was consistent with the observation by Sybesma et al. (17) that two isolates that were purified from different dairy products had deletions varying in length. Douillard et al. (14) also identified several independent isolates with three different lengths of deletions that all included the spaCBA-srtC1 gene cluster. While no gene loss was detected in samples continuously incubated in the presence of bile, salt, or shear stress for 100 generations, all the isolates purified from the culture exposed to continuous bile stress for 1,000 generations exhibited gene loss. These observations indicate that the strain is susceptible to gene deletions in this region, particularly after prolonged exposure to stressful conditions. However, whether the mutation occurred during purification or already existed in a nondetectable subpopulation in the frozen culture cannot be determined.
Based on their findings, both studies reported the need for a controlled production process and quality control for the industrial production of L. rhamnosus GG. Previously we demonstrated that L. rhamnosus GG exhibits high genomic stability throughout the production process and that spaCBA-srtC1 is fully conserved from the original stock to the freeze-dried product (18). During production, after pelletizing in liquid nitrogen, the bacteria are freeze-dried or stored at ≤–30°C to stop growth and slow down chemical reactions. When the frozen cultures are added to a milk base and included in a fermentation process, they rehydrate and become physiologically active again. However, growth of L. rhamnosus GG during yogurt fermentation was calculated to be less than three generations (Fig. S2). Therefore, even if a deletion emerges early during fermentation, it would not be able to increase to a noteworthy proportion of the population unless a subpopulation close to the detection level already exists. We previously demonstrated that no deletions in the spaCBA-srtC1 gene cluster could be detected in the populations of seven production batches of freeze-dried L. rhamnosus GG (18). Similarly, no gene deletions could be detected in the frozen product or in the yogurt in this study, as expected, since the number of generations during yogurt production is limited.
As expected, we could identify the five SNPs that we previously demonstrated to constitute a part of the original culture collection used for production (18). However, in 27 out of 60 sequenced isolates, 1 or 2 SNPs were detected which had not been identified previously. Similar to the gene deletion observed in the whole-genome analysis, these may represent minor subpopulations that are stochastically sampled, or the SNPs may have emerged during the purification process. Surprisingly, we also found one SNP (position 2253036) that was detected in 13 isolates, indicating that approximately 22% of the full population had the alternative allele. The SNP was observed in both frozen isolates and yogurt isolates and therefore already existed before inoculation of the milk substrate. The consequence of the amino acid substitution for this SNP was predicted to be deleterious. The gene encodes an ABC transporter, multidrug transporter ATPase. Since a large fraction of a population from an industrial fermentation possesses the SNP, it most likely confers a neutral effect on growth fitness. Even if the mutation conferred a fitness benefit, the number of generations required to accumulate such a fraction exceeds the number of generations in this production process by far. Therefore, it is likely that the SNP represents a subpopulation of the original strain, which was not detected in the previous study.
Effect of yogurt matrix on phenotypic characteristics.
While most in vitro studies measure the phenotypic characteristics of probiotic L. rhamnosus strains prepared from overnight cultures (23, 32, 33), the physiological profile of probiotic products on the market will reflect conditions encountered in the production process and the matrix in which they are delivered. In this study, common phenotypic assays were performed on samples of industrially produced L. rhamnosus GG in frozen pellets and cofermented in yogurt.
High tolerance to acid and bile was one of the criteria that were used initially to select L. rhamnosus GG as a potential probiotic (1). As expected, tolerance to gastric acid solution was high, with no decrease in survival regardless of whether the strain was frozen or cofermented in a yogurt matrix. Our results showed that L. rhamnosus GG is more sensitive to bile exposure, with a loss of 3 to 5 logs of viability that occurred within the first 7.5 min of the 1-h incubation. Bacterial persistence of a fraction of a population is well known from antibiotic treatments. The cells are commonly named persister cells and are characterized by the subpopulation exhibiting a resistance that is not genetically acquired. From the surviving cells, a new population would regrow with the same sensitivity (34, 35). Based on our findings of high tolerance to gastric acid solution, we would expect that consumption of L. rhamnosus GG in yogurt would result in delivery of a large number of viable cells into the upper small intestine.
The samples of L. rhamnosus GG cofermented and stored in yogurt showed a tendency toward decreased tolerance to bile stress compared to the frozen samples. This may be due to a hurdle effect, where a combination of mild factors, such as cold storage, low pH, and competitive flora encountered in yogurt, may increase sensitivity toward other conditions (36). However, the tendency toward decreased bile tolerance in yogurt was nonsignificant, and clinical studies have demonstrated that fecal recovery of L. rhamnosus GG was similar whether it was ingested as a freeze-dried product or in fermented milk (20, 21).
As previously observed (18), epithelial barrier function was increased by the probiotic strain L. rhamnosus GG, whereas the common yogurt starter culture consisting of S. thermophilus and L. bulgaricus did not induce any response. As the response for all three strains combined was similar to that of L. rhamnosus GG alone, we conclude that the detected response is from L. rhamnosus GG alone and that coincubation does not affect the ability of any of the strains to induce TEER. In contrast to the S. thermophilus and L. bulgaricus strains from the frozen product, the control yogurt containing S. thermophilus and L. bulgaricus as starter culture induced a response in TEER. Since the strains in the starter culture do not induce any response, we propose that the observed effect stems mostly from the yogurt matrix or that the physiochemical properties of the yogurt changed the probiotic properties of the starter culture. In yogurt cofermented with L. rhamnosus GG, the increase in TEER was higher than in the control yogurt on the day of production, indicating a potentially additive or synergistic effect, where yogurt might enhance characteristics of L. rhamnosus GG responsible for inducing TEER. Although the number of viable cells of L. rhamnosus GG was preserved, the TEER response had diminished by end of shelf life. This finding correlated with an observation by Deepika et al., that adhesion properties of L. rhamnosus GG in yogurt had decreased after storage under cold conditions for 7 days (37). In this study, adhesion capacity correlated with hydrophobicity, which also had decreased after 7 days and had been correlated previously with adhesion properties (38).
For these experiments, the acidic yogurt samples were neutralized immediately before they were applied to the system to ensure that TEER responses were not simply a result of reduced pH. However, some of the organic acids produced during yogurt fermentation, or their neutral anion counterparts, may be responsible for inducing a response in TEER. The capability to recover intestinal barrier function after 72 h when disrupted with inflammatory stimulus has been demonstrated previously with yogurt containing only a starter culture (39). Our results indicate that an enhanced effect of L. rhamnosus GG can be obtained when the organism is ingested in a yogurt matrix, which could possibly exert a higher protection of intestinal barrier function than the effect obtained from either L. rhamnosus GG or a yogurt matrix alone.
Although all bacterial samples induced secretion of the six cytokines examined, the response stimulated by L. rhamnosus GG was relatively modest compared with the response stimulated by the starter culture. However, modest induction of these cytokines by L. rhamnosus GG has been observed previously in similar in vitro assays (40, 41). For IL-12 and tumor necrosis factor alpha (TNF-α), L. rhamnosus GG was reported as a weakly inducing strain compared with other lactic acid bacteria, in line with what we saw for all cytokines examined in this study (41). Moreover, Zeuthen et al. demonstrated that weakly IL-12- and TNF-α-inducing strains of lactobacilli, including L. rhamnosus GG, inhibited the response from strongly inducing strains (41). In this study, similar inhibitory properties on the response induced by the starter culture were identified for IL-10, IL-6, and IL-8 but not for TNF-α, IL-12p70, or IL-1β. As relatively high responses by the starter culture alone were observed for all cytokines, it is proposed that L. rhamnosus GG may have a role in moderating cytokine responses. A functional role in balancing cytokine expression has been suggested previously for L. rhamnosus GG, where the ability of L. rhamnosus GG to both induce and reduce IL-8 expression in Caco-2 cells depended on which of the surface molecules interacted with host pattern recognition receptors (42).
Conclusion.
In conclusion, our results demonstrate genomic integrity of L. rhamnosus GG, with no deletions in any of the frozen product population samples or the yogurt samples. As one of the isolates had lost the spaCBA-srtC1 gene cluster, it was confirmed that L. rhamnosus GG has the propensity to lose genomic islands in the region containing the spaCBA-srtC1 gene cluster. Therefore, a well-constructed and efficiently regulated production process may be important to ensure preservation of the intact genome. If genome integrity is preserved in L. rhamnosus GG used as inoculation material for probiotic yogurt, the few generations that occur during yogurt production and during shelf life are not enough for a mutation to establish a noteworthy proportion in a yogurt product. In addition, the number of viable L. rhamnosus GG cells was preserved throughout shelf life. In vitro examination of common probiotic features of L. rhamnosus GG indicated that cofermentation with yogurt could improve some of the probiotic features that are characteristic for the strain, such as the increased induction of TEER demonstrated here. In addition, we observed an immunomodulatory capacity for L. rhamnosus GG when it was coincubated with starter culture in an in vitro dendritic cell assay, regardless of whether the strain was examined as frozen product or in a yogurt matrix. Altogether, the results demonstrate that the L. rhamnosus GG genome is largely preserved in yogurt and common in vitro probiotic effects of L. rhamnosus GG are observed when examined in the yogurt matrix.
MATERIALS AND METHODS
Bacterial strains used for the study.
For this study, the effect of L. rhamnosus GG, commercially recognized as the trademark LGG (DSM 33156) from Chr. Hansen, was examined when added to a milk base fermented into a yogurt with the starter culture YF-L901 containing S. thermophilus and L. bulgaricus. Frozen pellets of L. rhamnosus GG were collected both at the beginning and at the end of the process of pellet freezing in liquid nitrogen, providing a sample of L. rhamnosus GG that was frozen directly after addition of cryoprotectants (F_Start) and a sample that was kept in a holding tank for 3 h after addition of cryoprotectants (F_End) (Fig. 1). L. rhamnosus LC705 was used as a negative control for the presence of the spaCBA-srtC1 gene cluster.
Production of yogurt.
Yogurt was produced on a 3-liter scale in an application technology center on a part-skimmed milk base at 1.5%. The milk base was pasteurized at 95°C for 5 min/80°C for 1 min. The milk was homogenized at 200/50 bar at 65°C. YF-L901 was used as starter culture and added according to the instruction (500U/2500L). L. rhamnosus GG was added together with the starter culture and fermented at 43°C until pH reached 4.55. Hereafter the yogurt was mechanically broken with a spoon and cooled to 25°C during transfer to the tap with a pressure of 2 × 105 Pa. Yogurt was subsequently stored refrigerated at 4 to 6°C throughout shelf life (28 days).
Preparation of samples for genome sequencing.
L. rhamnosus GG was continuously grown overnight at 37°C in de Man-Rogosa-Sharpe (MRS) broth or for 3 days on MRS agar. For selective growth of L. rhamnosus GG from yogurt samples, 2% vancomycin (100 mg/ml; Merck) was added to the media, since L. rhamnosus GG has been shown to have intrinsic vancomycin resistance (43).
For genome sequencing, DNA was extracted from an overnight culture of the frozen L. rhamnosus GG samples from start and end of the production batch and from an overnight culture selective to L. rhamnosus GG prepared from yogurt, both on the day of production and by the end of shelf life. From each of the six overnight cultures, 10 single colony isolates were prepared. In this procedure, each sample was inoculated onto an agar plate from which 10 colonies were randomly picked, followed by three successive streaking procedures to ensure purification of isolates. The 60 isolates were subsequently grown in an overnight culture. All overnight cultures were pelletized by centrifugation and dissolved in DNA/RNA Shield (Zymo) for frozen storage until extraction.
DNA extraction and sequencing.
DNA was extracted using the DNeasy blood and tissue minikit (Qiagen) and paired-end (2 × 150 bp) whole-genome sequenced at BaseClear using an Illumina HiSeq 2500. The PhiX control genome was removed and adapter sequences were clipped for de novo assembly. Quality of the remaining reads was assessed with FastQC.
L. rhamnosus GG reference genomes.
L. rhamnosus GG published by Kankainen et al. (ASM2650v1 [10]), obtained from the Valio culture collection (Valio Ltd.), was used as a reference genome for the alignment. For the hierarchical clustering based on the frequency of gene calls, all L. rhamnosus GG genomes on the NCBI database (ASM2650v1 [https://www.ncbi.nlm.nih.gov/assembly/GCF_000026505.1/], ASM335345v1 [https://www.ncbi.nlm.nih.gov/assembly/GCF_003353455.1/], and ASM1104v1 [https://www.ncbi.nlm.nih.gov/assembly/GCF_000011045.1/]) were included as references.
Assembly and circle visualization.
For analysis of L. rhamnosus GG population samples from yogurt, genomic contamination from the cofermented strains L. bulgaricus and S. thermophilus was removed using BMTagger (44). Clean reads were de novo assembled using Spades (45) under the “—isolate” mode. Coverage of assembled contigs was calculated with BWA (46) and SAMtools (47), and contigs with lower coverage were considered low quality or DNA contamination and discarded. Cutoff differed based on the depth of sequencing: 60× coverage was used for all the samples from this study, while 5× coverage was used for samples described by Sybesma et al. (17) and Douillard et al. (14) (SRR645838, SRR3098054, SRR645868, SRR3096056, and SRR3096092 in the NCBI Sequence Read Archive [SRA]). The majority of the filtered contigs were short contigs. Quast (48) was used to check the quality of alignments to the reference genome (ASM2650v1) before and after the filtering. The remaining contigs from all samples in this study and the samples from Sybesma et al. (17) and Douillard et al. (14) were aligned to the L. rhamnosus GG reference genome. The Circlize package was used to draw the circular plot of genomic alignments (49).
Gene calling and genome similarity hierarchical clustering.
Prokka (v1.14.5) was used for complete gene calling of all the assembled genomes and the L. rhamnosus GG references under the species L. rhamnosus. Anvio (v6.1) was used to visualize the genomic similarity clustering based on the frequency of the aligned genes across the genomes.
Coverage comparison.
For the calculation of coverage of L. rhamnosus GG genes within each sample, a strategy similar to gene catalogue in metagenomic studies was adopted. Gene calls from the complete reference genome (ASM2650v1) were concatenated together to construct the representative L. rhamnosus GG gene catalogue. Clean reads were used instead of contigs in the case of contigs that were lost in the filtering. Reads from each sample were directly mapped to the gene catalogue using BWA (46). Gene coverage was calculated with BEDTools (50) and normalized by the length of genes. Gene calls were divided into two groups, the genes present in isolate 9 and the genes absent in isolate 9, according to the alignment statistics of isolate 9 with the deletion including the spaCBA-srtC1 gene cluster. For each sample, the ratio of average coverage of CDS for the two groups was calculated.
Mapping and variant calling.
To identify the presence of SNPs, sequences were mapped to the reference genome followed by variant calling. Initially, Smalt (v0.7.6) (51) and Bowtie 2 (v2.4.2) (52) were applied to map all sequence libraries to the reference genome (ASM2650v1). Mutation analysis was done by using three different tools: gatk (v4.2.0.0) (53), bcftools (v1.12) (54), and freebayes (v1.3.5) (55) in haploid mode. Coupled with the two above-described mapping tools, this resulted in six ways of variant calling. In addition, breseq (v0.35.6) (56) was used to validate the outputs from the above-described methods. The intersects from all seven approaches were used as final results. Bcftools (v0.35.6) was used for quantifying read support for the reference and alternative alleles (53–56). The impact of amino acid substitutions was predicted using the PROVEAN web server (v1.1.3 [http://provean.jcvi.org/seq_submit.php]) (57).
qPCR validation.
DNA was extracted from a sample of L. rhamnosus LC705, Y1_F_start_I9, Y1_F_start_I4, and Y1_F_start_ON (Fig. 1) using the DNeasy blood and tissue minikit (Qiagen) according to the manufacturer’s protocol.
Specific primers for spaA (forward, TCGCTAGACGAATTATCAGCTTTA; reverse, AAACGTGCCGGATGGTTAT) and 16S rRNA (right, GAAAGCCACGGCTAACTACG; left, GATAACGCTTGCCACCTACG) were identified using Universal ProbeLibrary Assay Design Center (Roche). Probes were purchased from Roche and primers produced at TAG Copenhagen.
The PCR mix was prepared using FastStart Essential Probes Master (Roche) according to the manufacturer’s protocol. A total of 250 ng of DNA was added for each sample, loaded into the LightCycler 96 instrument, and preheated for 600 s at 95°C. Subsequently 45 cycles were run with 95°C for 10 s, 60°C for 10 s, and 72°C for 1 s. qPCR was performed using technical and biological duplicates.
CFU cell counts for yogurt.
Serial dilutions were prepared and plated on MRS agar (pH 6.5) with 2% vancomycin for selective growth of L. rhamnosus GG for cell counts. Plates were grown anaerobically for 3 days at 37°C before colonies were counted.
Acid and bile tolerance assay.
The assay included samples of frozen L. rhamnosus GG pellets, yogurt cofermented with L. rhamnosus GG and samples where L. rhamnosus GG was added to a yogurt matrix immediately before the assay to determine whether a potential effect could be ascribed to a matrix effect alone or a more long-term adaptation from fermentation or storage. Frozen samples of L. rhamnosus GG were dissolved at a concentration of 4 × 108 CFU/ml in the control media consisting of MRS broth diluted to 10% (vol/vol) with demineralized water. For the yogurt samples with bacteria added immediately before the assay, frozen L. rhamnosus GG was mixed with control yogurt for 2 min in a stomacher at 230 rpm. The yogurt samples fermented with L. rhamnosus GG were also mixed before the assay to ensure an even distribution of the bacterial cells. A 1-ml volume of the frozen samples and 1.00 g of the yogurt samples were transferred to four separate tubes to which 9 ml of three different media were added: (i) control medium (10% MRS broth), (ii) gastric acid solution (control medium containing 3.2 mg/ml of pepsin and 34 mM sodium chloride) adjusted to pH 2 with hydrochloric acid, and (iii) bile solution (control medium with 1% [wt/vol] porcine bile, adjusted to pH 6.5). The cultures were incubated for 1 h at 37°C, and subsequently, serial dilutions were plated on MRS agar (pH 6.5) with 2% vancomycin for selective growth of L. rhamnosus GG for cell counts. An additional bile experiment using the same setup was performed, where samples were also collected during incubation (after 7.5, 15, and 30 min) for cell counts as described above.
Transepithelial electrical resistance assay.
The human colon adenocarcinoma cell line Caco-2 (ACC 169; DSMZ GmbH, Germany) was maintained at 37°C and 5% CO2 in Dulbecco’s modified Eagle medium (DMEM) plus GlutaMAX (Gibco by Life Technologies, UK) supplemented with 20% (vol/vol) fetal bovine serum (Gibco by Life Technologies; South American origin), 1% minimal essential medium (MEM) nonessential amino acid solution (Sigma-Aldrich), 100 U/ml of penicillin G, 100 μg/ml of streptomycin, and 0.25 μg/ml of amphotericin B (PenStrep) (Biological Industries BI, Israel). Caco-2 cells (0.5 ml of 1 × 105 cells/ml) were cultured on Transwell membrane inserts in 12-well cell culture plates (1.12 cm2, polyethylene terephthalate membranes with 0.4-μm pores; Corning) for 21 days before they were transferred to a CellZscope instrument (NanoAnalytics GmbH, Germany) for transepithelial electrical resistance (TEER) measurements.
Upon transfer to the CellZscope, the cells were replenished with cell culture medium containing 10 μg/ml of tetracycline (Sigma-Aldrich, Germany) instead of PenStrep in both the apical and basolateral compartments. During an overnight adaptation period, TEER was measured every hour to establish baseline TEER prior to addition of bacterial samples. For bacterial stimulation, 300 μl of cell culture medium was removed from apical compartments and replaced with 300 μl of either fresh cell culture medium with tetracycline (unstimulated controls) or 300 μl of fresh cell culture medium with tetracycline containing either frozen bacterial samples (of L. rhamnosus GG, the starter culture, or a combination of all three strains) or mixed with a volume of the yogurt samples (containing the starter culture alone or the starter culture cofermented with L. rhamnosus GG). Bacterial cell concentrations were adjusted so the numbers of bacterial cells in the frozen samples and in the yogurt samples corresponded. To reduce viscosity of the yogurt samples, they were diluted in cell culture media with tetracycline, resulting in 4 × 107 or 2 × 108 bacterial cells/sample for L. rhamnosus GG and the starter culture, respectively. Amounts and proportions of the bacterial strains added to the TEER wells were based on the final concentrations and proportions of the strains in 100 μl of yogurt, which contained a smaller amount of the probiotic L. rhamnosus GG strain (4 × 108 CFU/ml) and larger amounts of the yogurt-fermenting strains S. thermophilus and L. bulgaricus (1 × 108 CFU/ml of each strain). The pH of the yogurt samples was neutralized using 0.1 M NaOH to a pH of 7.1, corresponding to the pH of the bacterial samples and the control media. For each condition, triplicate wells were analyzed. Cells were stimulated for 20 h at 37°C and 5% CO2, with TEER measured every hour.
Cytokine secretion from human dendritic cells.
Dendritic cells (DCs) were generated in vitro from immature monocytes using a protocol modified from the method described by Zeuthen et al. (58). Human peripheral blood mononuclear cells (PBMCs) were separated from buffy coats (Rigshospitalet, Denmark) of three healthy donors by a density gradient centrifugation using Ficoll-Paque PLUS (GE Healthcare, Freiburg, Germany). Monocytes were isolated by positive selection for CD14 using magnetically activated cell sorting with CD14 microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany) and cultured at a density of 2 × 106 cells/ml in complete DC medium (RPMI 1640 supplemented with 10 mM HEPES [Sigma-Aldrich, Schnelldorf, Germany], 50 μM 2-mercaptothanol [2-ME; Sigma-Aldrich, Schnelldorf, Germany], 2 mM l-glutamine [Life Technologies Ltd., Paisley, UK], 10% heat-inactivated fetal bovine serum [Invitrogen, Paisley, UK], 100 U/ml of penicillin [Biological Industries, Kibbutz Beit-Haemek, Israel], and 100 μg/ml of streptomycin [Biological Industries]). The DC medium also contained 30 ng/ml of human recombinant IL-4 and 20 ng/ml of human recombinant granulocyte-macrophage colony-stimulating factor (GM-CSF) (both from Sigma-Aldrich, St. Louis, MO). Cells were kept at 37°C with 5% CO2. After 3 days, fresh complete DC medium containing full doses of IL-4 and GM-CSF was added. At day 6, differentiation to immature DCs was verified by surface marker expression analysis (CD14 < 3%, CD11c > 97%, and CD1a > 95%).
Immature DCs were replenished with antibiotic-free culture medium and seeded in 48-well plates at 1 × 106 cells/well. Before bacterial stimulation, the DCs were allowed to acclimate at 37°C and 5% CO2 for at least 1 h. DCs were stimulated for 20 h with frozen bacterial samples (of L. rhamnosus GG, the starter culture or a combination of all three strains) or yogurt samples (containing the starter culture alone or the starter culture and L. rhamnosus GG) adjusted to approximately 1.5 × 107 or 5 × 107 bacterial cells/well for L. rhamnosus GG and the starter culture, respectively. The pH of the yogurt samples was neutralized using 0.1 M NaOH to a pH of 7.1, corresponding to the pH of the bacterial samples and the control. For the control, DCs were left unstimulated. Cells were kept at 37°C and 5% CO2. After stimulation, DC supernatants were sterile filtered through a 0.2-μm Acro-Prep Advance 96-well filter plate (Pall Corporation, Ann Arbor, MI) and stored at −80°C until cytokine quantification.
Secreted levels of IL-10, IL-12p70, IL-1β, IL-6, IL-8, and TNF-α were quantified using a customized U-plex human proinflammatory panel 1 from Meso Scale Discovery (catalogue number K15049K; MSD, Rockville, MD), according to the manufacturer’s instructions. The means of lower detection limits were 0.07, 0.26, 0.12, 0.14, 0.06, and 0.30 pg/ml, respectively.
Statistical analysis.
All data from the phenotypic experiments were analyzed for statistically significant differences in GraphPad Prism 8 software (GraphPad Software, La Jolla, CA) using one-way analysis of variance (ANOVA) followed by Tukey’s test for multiple-comparison testing.
In the acid and bile experiments, mean values of cell counts after treatment and cell counts from the control were compared among all samples within each treatment group. In the intestinal barrier experiment, the TEER measure of each well was normalized to the baseline TEER of that well before stimulation and expressed as percent increase relative to baseline, where baseline is equal to 100%. Mean and standard deviation of the triplicates are displayed for each time point and each condition. Values for area under the curve (AUC) were calculated for each well using GraphPad Prism 8 software, and means and standard deviations of the triplicates are shown. For the cytokine experiment, statistical analysis was performed separately for each donor, as significant donor variation typically exists. For each donor, cytokine secretion levels (in picograms per milliliter) stimulated by the different samples were compared (Table S2). Values below the detection limit or below fitting curve were replaced by half-limit of detection.
Data availability.
All DNA sequence data are stored at the National Center for Biotechnology Information (NCBI) BioProject database under accession number PRJNA762716 (Table S1).
ACKNOWLEDGMENTS
The present study was supported by the Innovation Foundation Denmark (grant 5139-00024B).
We thank Eric Johansen for valuable comments on the manuscript.
M.S.S., D.S.N., and A.W. designed the experiments. M.S.S., J.O., and N.I.V.-J. performed the experiments. Y.H. and M.S.S. performed the data analysis. M.S.S. wrote the paper. D.S.N., A.S., and A.W. revised the paper. N.I.V.-J. and J.O. are employed by Chr. Hansen A/S, which produces and markets the LGG strain. A.W. and M.S.S. were employed by Chr. Hansen A/S until April 2021 and July 2021, respectively.
Footnotes
Supplemental material is available online only.
Contributor Information
Anita Wichmann, Email: anitawichmann@yahoo.com.
Danilo Ercolini, University of Naples Federico II.
REFERENCES
- 1.Goldin BR, Gorbach SL, Saxelin M, Barakat S, Gualtieri L, Salminen S. 1992. Survival of Lactobacillus species (strain GG) in human gastrointestinal tract. Dig Dis Sci 37:121–128. 10.1007/BF01308354. [DOI] [PubMed] [Google Scholar]
- 2.Silva M, Jacobus NV, Deneke C, Gorbach SL. 1987. Antimicrobial substance from a human Lactobacillus strain. Antimicrob Agents Chemother 31:1231–1233. 10.1128/AAC.31.8.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arvola T, Laiho K, Torkkeli S, Mykkänen H, Salminen S, Maunula L, Isolauri E. 1999. Children with respiratory infections: a randomized study. Pediatrics 104:e64. 10.1542/peds.104.5.e64. [DOI] [PubMed] [Google Scholar]
- 4.Isolauri E, Juntunen M, Rautanen T, Sillanaukee P, Koivula T. 1991. A human Lactobacillus strain (Lactobacillus casei sp strain GG) promotes recovery from acute diarrhea in children. Pediatrics 88:90–97. [PubMed] [Google Scholar]
- 5.Fox MJ, Ahuja KDK, Robertson IK, Ball MJ, Eri RD. 2015. Can probiotic yogurt prevent diarrhoea in children on antibiotics? A double-blind, randomised, placebo-controlled study. BMJ Open 5:e006474. 10.1136/bmjopen-2014-006474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manley KJ, Fraenkel MB, Mayall BC, Power DA. 2007. Probiotic treatment of vancomycin-resistant enterococci: a randomised controlled trial. Med J Aust 186:454–457. 10.5694/j.1326-5377.2007.tb00995.x. [DOI] [PubMed] [Google Scholar]
- 7.Davidson LE, Fiorino AM, Snydman DR, Hibberd PL. 2011. Lactobacillus GG as an immune adjuvant for live-attenuated influenza vaccine in healthy adults: a randomized double-blind placebo-controlled trial. Eur J Clin Nutr 65:501–507. 10.1038/ejcn.2010.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tytgat HLP, Douillard FP, Reunanen J, Rasinkangas P, Hendrickx APA, Laine PK, Paulin L, Satokari R, de Vos WM. 2016. Lactobacillus rhamnosus GG outcompetes Enterococcus faecium via mucus-binding pili: evidence for a novel and heterospecific probiotic mechanism. Appl Environ Microbiol 82:5756–5762. 10.1128/AEM.01243-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vargas G, Cynthia E, Petrova M, Claes IJJ, De Boeck I, Verhoeven TLA, Dilissen E, von Ossowski I, Palva A, Bullens DM, Vanderleyden J, Lebeer S. 2015. Piliation of Lactobacillus rhamnosus GG promotes adhesion, phagocytosis, and cytokine modulation in macrophages. Appl Environ Microbiol 81:2050–2062. 10.1128/AEM.03949-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kankainen M, Paulin L, Tynkkynen S, Von Ossowski I, Reunanen J, Partanen P, Satokari R, Vesterlund S, Hendrickx APA, Lebeer S, De Keersmaecker SCJ, Vanderleyden J, Hämäläinen T, Laukkanen S, Salovuori N, Ritari J, Alatalo E, Korpela R, Mattila-Sandholm T, Lassig A, Hatakka K, Kinnunen KT, Karjalainen H, Saxelin M, Laakso K, Surakka A, Palva A, Salusjärvi T, Auvinen P, De Vos WM. 2009. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human-mucus binding protein. Proc Natl Acad Sci USA 106:17193–17198. 10.1073/pnas.0908876106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coconnier MH, Klaenhammer TR, Kerneis S, Bernet MF, Servin AL. 1992. Protein-mediated adhesion of Lactobacillus acidophilus BG2FO4 on human enterocyte and mucus-secreting cell lines in culture. Appl Environ Microbiol 58:2034–2039. 10.1128/aem.58.6.2034-2039.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Segers ME, Lebeer S. 2014. Towards a better understanding of Lactobacillus rhamnosus GG-host interactions. Microb Cell Fact 13:S7. 10.1186/1475-2859-13-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rasinkangas P, Reunanen J, Douillard FP, Ritari J, Uotinen V, Palva A, de Vos WM. 2014. Genomic characterization of non-mucus-adherent derivatives of Lactobacillus rhamnosus GG reveals genes affecting pilus biogenesis. Appl Environ Microbiol 80:7001–7009. 10.1128/AEM.02006-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Douillard F, Ribbera A, Xiao K, Ritari J, Rasinkangas P, Paulin L, Palva A, Hao Y, De Vos Willem M. 2016. Polymorphisms, chromosomal rearrangements, and mutator phenotype development during experimental evolution of Lactobacillus rhamnosus GG. Appl Environ Microbiol 82:3783–3792. 10.1128/AEM.00255-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schneider D, Lenski RE. 2004. Dynamics of insertion sequence elements during experimental evolution of bacteria. Res Microbiol 155:319–327. 10.1016/j.resmic.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 16.Thomas CM, Nielsen KM. 2005. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol 3:711–721. 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- 17.Sybesma W, Molenaar D, van IJcken W, Venema K, Kort R. 2013. Genome instability in Lactobacillus rhamnosus GG. Appl Environ Microbiol 79:2233–2239. 10.1128/AEM.03566-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stage M, Wichmann A, Jørgensen M, Vera-Jimenéz NI, Wielje M, Nielsen DS, Sandelin A, Chen Y, Baker A. 2020. Lactobacillus rhamnosus GG genomic and phenotypic stability in an industrial production process. Appl Environ Microbiol 86:e02780-19. 10.1128/AEM.02780-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nyanzi R, Jooste PJ, Buys EM. 2021. Invited review: probiotic yogurt quality criteria, regulatory framework, clinical evidence, and analytical aspects. J Dairy Sci 104:1–19. 10.3168/jds.2020-19116. [DOI] [PubMed] [Google Scholar]
- 20.Saxelin M, Ahokas M, Salminen S. 1993. Dose response on the faecal colonisation of Lactobacillus strain GG administered in two different formulations. Microb Ecol Health Dis 6:119–122. [Google Scholar]
- 21.Saxelin M, Lassig A, Karjalainen H, Tynkkynen S, Surakka A, Vapaatalo H, Järvenpää S, Korpela R, Mutanen M, Hatakka K. 2010. Persistence of probiotic strains in the gastrointestinal tract when administered as capsules, yoghurt, or cheese. Int J Food Microbiol 144:293–300. 10.1016/j.ijfoodmicro.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 22.Reference deleted. [Google Scholar]
- 23.Lebeer S, Claes IJJ, Verhoeven TLA, Vanderleyden J, De Keersmaecker SCJ. 2011. Exopolysaccharides of Lactobacillus rhamnosus GG form a protective shield against innate immune factors in the intestine. Microb Biotechnol 4:368–374. 10.1111/j.1751-7915.2010.00199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tuomola E, Crittenden R, Playne M, Isolauri E, Salminen S. 2001. Quality assurance criteria for probiotic bacteria. Am J Clin Nutr 73:393–398. [DOI] [PubMed] [Google Scholar]
- 25.Zhang L, Li N, Caicedo R, Neu J. 2005. Alive and dead Lactobacillus rhamnosus GG decrease tumor necrosis factor-α–induced interleukin-8 production in Caco-2 cells. J Nutr 135:1752–1756. 10.1093/jn/135.7.1752. [DOI] [PubMed] [Google Scholar]
- 26.Capurso L. 2019. Thirty years of Lactobacillus rhamnosus GG. A review. J Clin Gastroenterol 53:S1–S41. 10.1097/MCG.0000000000001170. [DOI] [PubMed] [Google Scholar]
- 27.Chavant P, Martinie B, Meylheuc T, Bellon-Fontaine M-N, Hebraud M. 2002. Listeria monocytogenes LO28: surface physicochemical properties and ability to form biofilms at different temperatures and growth phases. Appl Environ Microbiol 68:728–737. 10.1128/AEM.68.2.728-737.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sengupta R, Altermann E, Anderson RC, McNabb WC, Moughan PJ, Roy NC. 2013. The role of cell surface architecture of lactobacilli in host-microbe interactions in the gastrointestinal tract. Mediators Inflamm 2013:237921. 10.1155/2013/237921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maassen CBM, Boersma WJA, van Holten-Neelen C, Claassen E, Laman JD. 2003. Growth phase of orally administered Lactobacillus strains differentially affects IgG1/IgG2a ratio for soluble antigens: implications for vaccine development. Vaccine 21:2751–2757. 10.1016/S0264-410X(03)00220-2. [DOI] [PubMed] [Google Scholar]
- 30.Finkel SE, Kolter R. 1999. Evolution of microbial diversity during prolonged starvation. Proc Natl Acad Sci USA 96:4023–4027. 10.1073/pnas.96.7.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosenberg SM, Shee C, Frisch RL, Hastings PJ. 2012. Stress-induced mutation via DNA breaks in Escherichia coli: a molecular mechanism with implications for evolution and medicine. Bioessays 34:885–892. 10.1002/bies.201200050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gopal PK, Prasad J, Smart J, Gill HS. 2001. In vitro adherence properties of Lactobacillus rhamnosus DR20 and Bifidobacterium lactis DR10 strains and their antagonistic activity against an enterotoxigenic Escherichia coli. Int J Food Microbiol 67:207–216. 10.1016/S0168-1605(01)00440-8. [DOI] [PubMed] [Google Scholar]
- 33.Evrard B, Coudeyras S, Dosgilbert A, Charbonnel N, Alamé J, Tridon A, Forestier C. 2011. Dose-dependent immunomodulation of human dendritic cells by the probiotic Lactobacillus rhamnosus lcr35. PLoS One 6:e18735. 10.1371/journal.pone.0018735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kussell E, Kishony R, Balaban NQ, Leibler S. 2005. Bacterial persistence: a model of survival in changing environments. Genetics 169:1807–1814. 10.1534/genetics.104.035352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622–1625. 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 36.Leistner L. 2000. Basic aspects of food preservation by hurdle technology. Int J Food Microbiol 55:181–186. 10.1016/s0168-1605(00)00161-6. [DOI] [PubMed] [Google Scholar]
- 37.Deepika G, Rastall RA, Charalampopoulos D. 2011. Effect of food models and low-temperature storage on the adhesion of Lactobacillus rhamnosus GG to Caco-2 cells. J Agric Food Chem 59:8661–8666. 10.1021/jf2018287. [DOI] [PubMed] [Google Scholar]
- 38.Deepika G, Green RJ, Frazier RA, Charalampopoulos D. 2009. Effect of growth time on the surface and adhesion properties of Lactobacillus rhamnosus GG. J Appl Microbiol 107:1230–1240. 10.1111/j.1365-2672.2009.04306.x. [DOI] [PubMed] [Google Scholar]
- 39.Putt KK, Pei R, White HM, Bolling BW. 2017. Yogurt inhibits intestinal barrier dysfunction in Caco-2 cells by increasing tight junctions. Food Funct 8:406–414. 10.1039/C6FO01592A. [DOI] [PubMed] [Google Scholar]
- 40.Cai S, Kandasamy M, Rahmat JN, Tham SM, Bay BH, Lee YK, Mahendran R. 2016. Lactobacillus rhamnosus GG activation of dendritic cells and neutrophils depends on the dose and time of exposure. J Immunol Res 2016:7402760. 10.1155/2016/7402760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zeuthen LH, Christensen HR, Frøkiaer H. 2006. Lactic acid bacteria inducing a weak interleukin-12 and tumor necrosis factor alpha response in human dendritic cells inhibit strongly stimulating lactic acid bacteria but act synergistically with gram-negative bacteria. Clin Vaccine Immunol 13:365–375. 10.1128/CVI.13.3.365-375.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lebeer S, Claes I, Tytgat HLP, Verhoeven TLA, Marien E, von Ossowski I, Reunanen J, Palva A, de Vos WM, De Keersmaecker SCJ, Vanderleyden J. 2012. Functional analysis of Lactobacillus rhamnosus GG pili in relation to adhesion and immunomodulatory interactions with intestinal epithelial cells. Appl Environ Microbiol 78:185–193. 10.1128/AEM.06192-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tynkkynen S, Singh KV, Varmanen P. 1998. Vancomycin resistance factor of Lactobacillus rhamnosus GG in relation to enterococcal vancomycin resistance (van) genes. Int J Food Microbiol 41:195–204. 10.1016/S0168-1605(98)00051-8. [DOI] [PubMed] [Google Scholar]
- 44.Rotmistrovsky K, Agarwala R. 2011. BMTagger: Best Match Tagger for removing human reads from metagenomics datasets.
- 45.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997 [q-bio.GN].
- 47.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. Bioinformatics application note. Genome analysis QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gu Z, Gu L, Eils R, Schlesner M, Brors B. 2014. Circlize implements and enhances circular visualization in R. Bioinformatics 30:2811–2812. 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- 50.Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ponsting H, Ning H. 2010. SMALT—a new mapper for DNA sequencing reads. F1000 poster L313.
- 52.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van der Auwera G, O’Connor BD. 2020. Genomics in the Cloud: using Docker, Gatk, and Wdl in Terra. O’Reilly Media, Sebastopol, CA. [Google Scholar]
- 54.Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27:2987–2993. 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garrison E, Marth G. 2012. Haplotype-based variant detection from short-read sequencing. arXiv:1207.3907 [q-bio.GN].
- 56.Barrick JE, Colburn G, Deatherage DE, Traverse CC, Strand MD, Borges JJ, Knoester DB, Reba A, Meyer AG. 2014. Identifying structural variation in haploid microbial genomes from short-read resequencing data using breseq. BMC Genomics 15:1039–1017. 10.1186/1471-2164-15-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Choi Y, Chan AP, Craig TJ. 2015. Sequence analysis PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 31:2745–2747. 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zeuthen LH, Fink LN, Frøkiaer H. 2008. Toll-like receptor 2 and nucleotide-binding oligomerization domain-2 play divergent roles in the recognition of gut-derived lactobacilli and bifidobacteria in dendritic cells. Immunology 124:489–502. 10.1111/j.1365-2567.2007.02800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 and S2, Tables S1 to S3. Download aem.01575-21-s0001.pdf, PDF file, 0.4 MB (362.5KB, pdf)
Data Availability Statement
All DNA sequence data are stored at the National Center for Biotechnology Information (NCBI) BioProject database under accession number PRJNA762716 (Table S1).