ABSTRACT
Florfenicol is an important antibiotic commonly used in poultry production to prevent and treat Salmonella infection. However, oral administration of florfenicol may alter the animals’ natural microbiota and metabolome, thereby reducing intestinal colonization resistance and increasing susceptibility to Salmonella infection. In this study, we determined the effect of florfenicol (30 mg/kg of body weight) on gut colonization of neonatal chickens challenged with Salmonella enterica subsp. enterica serovar Enteritidis. We then analyzed the microbial community structure and metabolic profiles of cecal contents using microbial 16S amplicon sequencing and liquid chromatography-mass spectrometry (LC-MS) untargeted metabolomics, respectively. We also screened the marker metabolites using a multi-omics technique and assessed the effect of these markers on intestinal colonization by S. Enteritidis. Florfenicol administration significantly increased the loads of S. Enteritidis in cecal contents, spleen, and liver and prolonged the residence of S. Enteritidis. Moreover, florfenicol significantly affected cecal colony structures, with reduced abundances of Lactobacillus and Bacteroidetes and increased levels of Clostridia, Clostridium, and Dorea. The metabolome was greatly influenced by florfenicol administration, and perturbation in metabolic pathways related to linoleic acid metabolism (linoleic acid, conjugated linoleic acid [CLA], 12,13-EpOME, and 12,13-diHOME) was most prominently detected. We screened CLA and 12,13-diHOME as marker metabolites, which were highly associated with Lactobacillus, Clostridium, and Dorea. Supplementation with CLA maintained intestinal integrity, reduced intestinal inflammation, and accelerated Salmonella clearance from the gut and remission of enteropathy, whereas treatment with 12,13-diHOME promoted intestinal inflammation and disrupted intestinal barrier function to sustain Salmonella infection. Thus, these results highlight that florfenicol alters the intestinal microbiota and metabolism of neonatal chickens and promotes Salmonella infection mainly by affecting linoleic acid metabolism.
IMPORTANCE Florfenicol is a broad-spectrum fluorine derivative of chloramphenicol frequently used in poultry to prevent/treat Salmonella. However, oral administration of florfenicol may lead to alterations in the microbiota and metabolome in the chicken intestine, thereby reducing colonization resistance to Salmonella infection, and the possible mechanisms linking antibiotics and Salmonella colonization in poultry have not yet been fully elucidated. In the current study, we show that increased colonization by S. Enteritidis in chickens administered florfenicol is associated with large shifts in the gut microbiota and metabolic profiles. The most influential linoleic acid metabolism is highly associated with the abundances of Lactobacillus, Clostridium, and Dorea in the intestine. The screened target metabolites in linoleic acid metabolism affect S. Enteritidis colonization, intestinal inflammation, and intestinal barrier function. Our findings provide a better understanding of the susceptibility of animal species to Salmonella after antibiotic intervention, which may help to elucidate infection mechanisms that are important for both animal and human health.
KEYWORDS: Salmonella, chicken, florfenicol, gut microbiota, metabolome
INTRODUCTION
Foodborne infectious diseases are associated with high morbidity and mortality rates worldwide and pose great threats to human and animal health. Among all the foodborne pathogens, Salmonella is one of the most widespread pathogens for livestock and humans and is a leading cause of foodborne outbreaks and infections around the world (1–3). More than 2,600 Salmonella serovars (2,500+ serotypes classified) are known, and most of them are not host restricted and have the ability to colonize a wide variety of animal species. Salmonella enterica subsp. enterica serovar Enteritidis is a typical representative of non-host-specific Salmonella found in poultry and has a significant impact on the poultry industry. In 2010, the CDC reported that more than 500 million eggs were recalled due to an outbreak of human foodborne salmonellosis caused by S. Enteritidis contamination of retail eggs (https://www.cdc.gov/salmonella/2010/shell-eggs-12-2-10.html). In addition, S. Enteritidis intestinal colonization is mainly through the fecal-oral route and can cause intestinal inflammation and barrier dysfunction in chickens.
Due to the widespread use of antibiotics in large-scale intensive rearing systems in the livestock industry, Salmonella resistance has rapidly increased (4). In 2003, China alone consumed approximately 927,000 tons of antibiotics, and veterinary antibiotics accounted for 84.3% of the total usage of 36 target antibiotics, whereas human antibiotics comprised only 15.6% (5). It is estimated that the global application of antibiotics for chickens, pigs, and cattle will increase by 67%, from 63,151 tons in 2010 to 105,596 tons in 2030 (6). Among them, florfenicol (FFC), lincomycin, enrofloxacin, and amoxicillin are among the most commonly consumed veterinary antibiotics. Florfenicol is a broad-spectrum fluorine derivative of chloramphenicol frequently used in poultry in many countries, and the residual concentrations of florfenicol in chicken meat and liver samples were found to be up to 311.42 and 1,759.71 μg/kg, respectively, which are significantly higher than their respective maximum residual limits (7).
The intestine harbors an ecosystem composed of the commensal microbiota that promotes development, aids digestion, and protects the host from pathogens. The balance of the gut microbiota is important for reducing colonization by pathogens and preventing the proliferation of existing pathogens (8, 9). However, antibiotics shift the composition and number of bacterial species in the gut, thereby altering resistance to pathogen invasion (10). In early clinical studies, it was found that antibiotics change the composition of the gut microbiota, which is more conducive to colonization by conditioned pathogens in the intestine (11, 12). Streptomycin and vancomycin can significantly alter the composition of the gut microbiota, and greater preinfection perturbations in the gut microbiota lead to increased mouse susceptibility to Salmonella enterica serovar Typhimurium gut colonization, greater postinfection alterations in the gut microbiota, and more serious intestinal pathology (13). Vitamin B6 produced by Bacteroides spp. in the colon accelerated S. Typhimurium clearance from the gut and remission of enteropathy, and the antibiotic prolongs Salmonella infection by suppressing Bacteroides and reducing vitamin B6 levels in the gut (14).
Furthermore, antibiotics can also affect intestinal metabolism by altering the composition of the intestinal microbiota. Streptomycin treatment reduced the number of short-chain fatty acid (SCFA)-producing Clostridia from the mouse intestinal lumen, leading to decreased butyrate levels, increased epithelial oxygenation, and aerobic expansion of Salmonella. Also, oral antibiotics inhibit the growth of butyrate-producing Clostridium perfringens in the gut microbiota, switching host cells from oxidative metabolism to lactate fermentation and thus supporting Salmonella infection (15). Also, the use of antibiotics leads to disruption of the resident microbiota and subsequently increases fucose and sialic acid contents in the intestine, thereby affecting Salmonella enterica serovar Typhimurium and Clostridium difficile colonization (16). Similarly, antibiotics also alter the gut microbiota and decrease secondary bile acid production, allowing Clostridium difficile spore germination and outgrowth in the gastrointestinal tract (17). Consequently, the change in the composition of the gut microbiota caused by antibiotics may cause disorder in the ecological balance between the microorganism and the host, increase the sensitivity to exogenous pathogen colonization, and cause serious clinical implications. Thus, it is of great clinical significance to understand the effects of specific antibiotics on the intestinal flora and the colonization mechanisms of exogenous pathogens after antibiotic intervention.
So far, most of the research on the mechanisms of Salmonella intestinal colonization after antibiotic intervention has been conducted only in mouse models. However, the gut microbiota of poultry is different from that of mammals, and the mechanisms linking veterinary antibiotics and Salmonella colonization in poultry have not yet been fully understood. Thus, we are interested in florfenicol since it is the most commonly used veterinary antibiotic in poultry in many countries. In China, florfenicol ranks first among commonly used antibiotics in laying and broiler chicken farms, with utilization rates of 64% (18) and 78% (19), respectively. It is often used as an opening medicine for chicks and an important therapeutic drug for the prevention and treatment of bacterial diseases in poultry farming. The floR gene is widely distributed in Gram-negative antibiotic-resistant strains, which mediates the resistance of Escherichia coli, Klebsiella pneumoniae, and Salmonella to florfenicol. A large number of studies have found that floR is the most common gene in poultry multidrug-resistant Salmonella strains isolated from the chicken intestinal tract, chicken breeding environment, or slaughterhouse, and its carriage rate is even >90% (20–23). Antibiotic-resistant Salmonella strains are a leading cause of foodborne diseases and serious human and animal health concerns worldwide. However, there are few studies on intestinal colonization by antibiotic-resistant Salmonella.
Therefore, in the present study, we investigate the influence of florfenicol on florfenicol-resistant (floR mutant) Salmonella colonization in chickens and the relationship between the gut microbiota and S. Enteritidis colonization. We also evaluated changes in the metabolome in the cecum of florfenicol-treated chickens infected with Salmonella and used multi-omics association analyses to determine the correlations between the microbiome and metabolome and the potential factors that facilitate infection by S. Enteritidis in the cecum.
RESULTS
Shedding levels of Salmonella in response to florfenicol treatment.
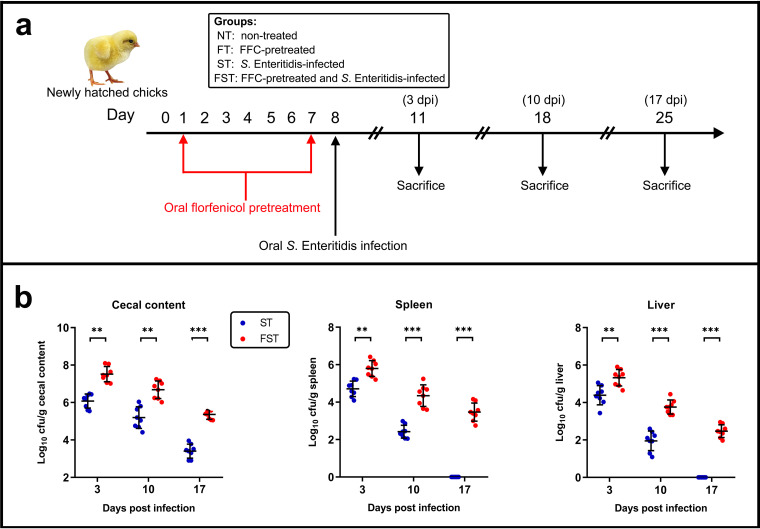
We first evaluated the abundance of S. Enteritidis in the samples. In a specific-pathogen-free (SPF) environment, in which chickens were reared in an individual SPF isolator and fed sterile feed and water, all the chickens were culture negative for Salmonella spp. until experimental infection with S. Enteritidis. During the experiment, we evaluated the shedding levels of Salmonella in the nontreated (NT) and florfenicol (FFC)-treated (FT) groups, and these groups remained culture negative for Salmonella spp. throughout the study. At 3 days postinfection (dpi), the number of S. Enteritidis bacteria in the cecal contents of the S. Enteritidis-infected and florfenicol-treated (FST) group (7.516 log10 CFU/g) was significantly higher than that in the S. Enteritidis-infected (ST) group (6.079 log10 CFU/g [P < 0.01]). The abundances of S. Enteritidis in the spleen (5.795 log10 CFU/g) and liver (5.325 log10 CFU/g) of the FST group were also significantly higher than those in the ST group (4.709 log10 CFU/g spleen [P < 0.01] and 4.389 log10 CFU/g liver [P < 0.001]). On day 18 (10 dpi), the abundances of S. Enteritidis in the cecal contents (6.683 log10 CFU/g), spleen (4.344 log10 CFU/g), and liver (3.759 log10 CFU/g) of the FST group were still significantly higher than those in the ST group (5.195 log10 CFU/g cecal content [P < 0.01], 2.42 log10 CFU/g spleen [P < 0.001], and 1.952 log10 CFU/g liver [P < 0.001]). Moreover, on day 25 (17 dpi), the relative loads of S. Enteritidis were 5.361, 3.467, and 2.473 log10 CFU/g in the FST group (cecal contents, spleen, and liver, respectively), which were significantly higher than those in the ST group. However, no S. Enteritidis bacteria were detected in the spleen and liver samples of the ST group (Fig. 1).
FIG 1.
Effect of florfenicol on S. Enteritidis infection in neonatal chickens. (a) Experimental design. Newly hatched chickens (n = 8 at each sampling point) were randomly divided into four groups (NT, FT, ST, and FST). The chickens were either administered a 7-day treatment (30 mg/kg of body weight) of florfenicol or infected with ∼108 CFU of the S. Enteritidis challenge strain by oral gavage. Sampling points for cecal microbiota analysis during infection are indicated. (b) At 3, 10, or 17 days postinfection (dpi), chicks were sacrificed, and S. Enteritidis loads in cecal contents, spleen, and liver were determined by the plate counting method. Data are expressed as means ± standard deviations. **, P < 0.01; ***, P < 0.001 (by a Mann-Whitney U test).
Florfenicol administration aggravates S. Enteritidis-induced intestinal morphology and barrier injury.
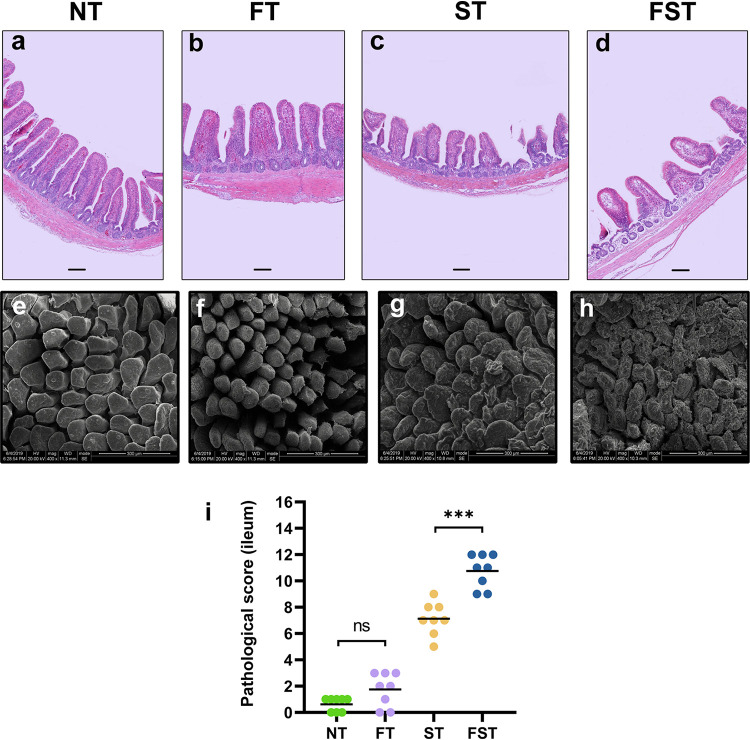
As FFC intervention made the chicks more susceptible to Salmonella infection, it is possible that antibiotics disrupted the immature intestinal barrier homeostasis of the chicks, altered intestinal permeability, and facilitated greater translocation of Salmonella to their internal organs. Therefore, we investigated the effects of FFC administration on S. Enteritidis-induced changes in intestinal morphology. Hematoxylin and eosin (H&E) staining and scanning electron microscopy (SEM) showed that the NT group had complete ileal villi, forming full and closely arranged structures, whereas the FT group also had intact ileal villi, but their arrangement was relatively sparse. The ST group displayed an incomplete structure of the ileal mucosa: villi were shorter and had a sparse distribution, and crypts were shallow. However, the morphology in the FST group included the loss of mucosal structures, atrophic crypts, and lamina propria bowel edema (Fig. 2). Pathological scores showed that the score of the FT group was higher than that of the NT group, but there was no significant difference. The score for the ST group was significantly higher than that for the control group. Relative to the ST group, FFC preadministration significantly increased the ileum injury score (Fig. 2).
FIG 2.
Florfenicol pretreatment exacerbates the development of Salmonella-induced ileal mucosal injury in neonatal chicks at 3 dpi. (a to h) After 3 days of infection, the histopathology of the intestinal tissue was analyzed by H&E staining (a to d) and SEM (e to h). Bars, 100 μm (a to d). (i) Ileal pathology was scored in H&E-stained ileal tissue sections. Bars indicate medians. ns, not significant; ***, P < 0.001 (by a Mann-Whitney U test).
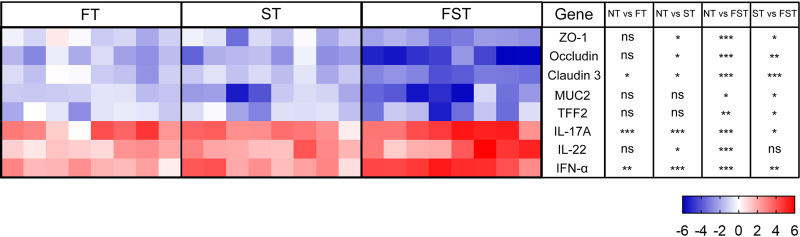
FFC intervention also exacerbates S. Enteritidis-induced intestinal barrier function impairment. FFC pretreatment can exacerbate S. Enteritidis-induced increases in the permeability of the ileum (see Fig. S1a to c in the supplemental material). The serum diamine oxidase (DAO) and lipopolysaccharide (LPS) levels of the FT group were significantly higher (P < 0.001 and P < 0.05, respectively) than those of the NT group. In the case of Salmonella infection, serum d-lactate, DAO, and LPS levels in both the ST and FST groups were significantly increased (P < 0.001) relative to those in the NT group. However, FFC treatment significantly increased (P < 0.001) serum d-lactate, DAO, and LPS levels in chicks exposed to Salmonella. Transcriptional analysis of a range of relevant intestinal barrier genes (Fig. 3) showed that FFC treatment significantly altered the transcription of claudin 3, interleukin-17A (IL-17A), and interferon alpha (IFN-α) in the FT group. However, in the presence of Salmonella, FFC significantly reduced the expression of ZO-1, occludin, claudin 3, MUC2, and TFF2 and significantly increased the expression of IL-17A, IL-22, and IFN-α. Furthermore, FFC also exacerbated the Salmonella-induced inflammatory response. FFC-treated chicks exhibited higher levels of gut inflammation after Salmonella infection (Fig. S1d to i). Salmonella infection after FFC pretreatment significantly increased the levels of the cytokines IL-1β, IL-6, IL-8, tumor necrosis factor alpha (TNF-α), and IFN-γ, whereas the level of IL-10 was significantly decreased. Moreover, the levels of inflammatory cytokines (IL-1β, IL-6, TNF-α, and IFN-γ) were significantly increased in the FT group.
FIG 3.
Gene expression profiles in response to FFC pretreatment or S. Enteritidis infection by qPCR at 3 dpi. Data are represented as log2 fold changes between the treatment and control (NT) groups. Statistical analysis was conducted using one-way ANOVA and Dunnett’s multiple-comparison test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
Effect of florfenicol on microbial composition and structure.
The 16S rRNA of the cecal microbiota was sequenced and analyzed for the species diversity of individual samples and differences in diversity between samples. The α-diversity of microbial communities was measured using Shannon, observed species, and Pielou indices. The Shannon index represents how much the difference is among the abundances of different taxa (diversity). The observed species index reflects how many different taxa are present in the sample (richness), while the Pielou index represents the distribution of the number of individuals of all species in a community (evenness). Figure S2 in the supplemental material shows that the α-diversity was affected by neither florfenicol treatment nor S. Enteritidis infection at 3 dpi (Fig. S2a). However, a significant decrease in α-diversity was observed in the FST group at 10 dpi (Fig. S2b). Figure S2c shows that the Shannon and Pielou indices of the cecal microbial communities were significantly increased in the FST group at 17 dpi. These results indicated that the α-diversity of the gut microbiota from chicks is not significantly affected by a single FFC treatment or Salmonella challenge. However, the combination of both treatments significantly disturbed cecal α-diversity.
The similarity of microbial communities (β-diversity) between groups was visualized using principal-coordinate analysis (PCoA) of Bray-Curtis distances. β-Diversity represents the alterity between individuals in different microbial communities. PCoA plots for 3 dpi showed that the microbial communities from Salmonella- or florfenicol-treated chicks are clearly different from those of untreated chicks. The first axis of the PCoA plot shows 19.0% variation in bacterial diversity, while the second axis shows 13.0% (see Fig. 6c). The first axis roughly distinguishes the antibiotic-pretreated chicks and nonpretreated chicks, and the second axis roughly distinguishes the infected and noninfected chicks. PCoA at 10 dpi shows that the microbiota compositions were very similar between the NT and FT groups, whereas the ST and FST groups were still obviously distinguished from the NT group (Fig. S3a). Intriguingly, at day 25 (17 dpi), the PCoA plot showed that the microbiota compositions of both the ST and FT groups trended toward that of the NT group, whereas the composition of the FST group was still strikingly divergent from that of the NT group (Fig. S3b).
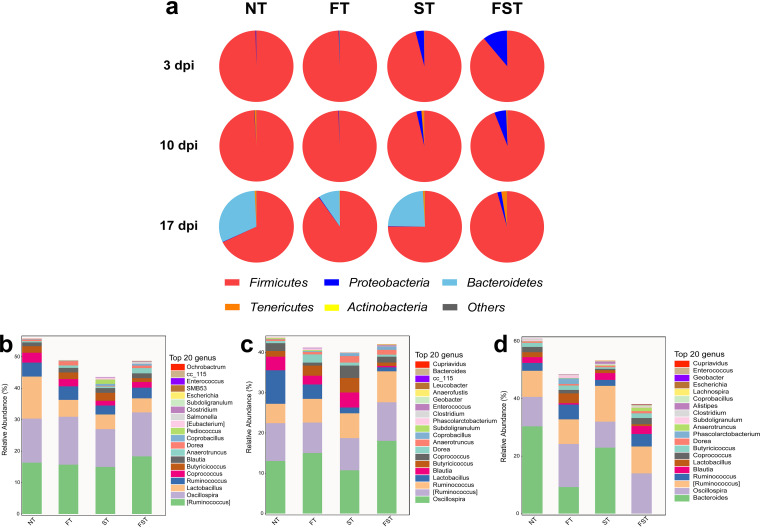
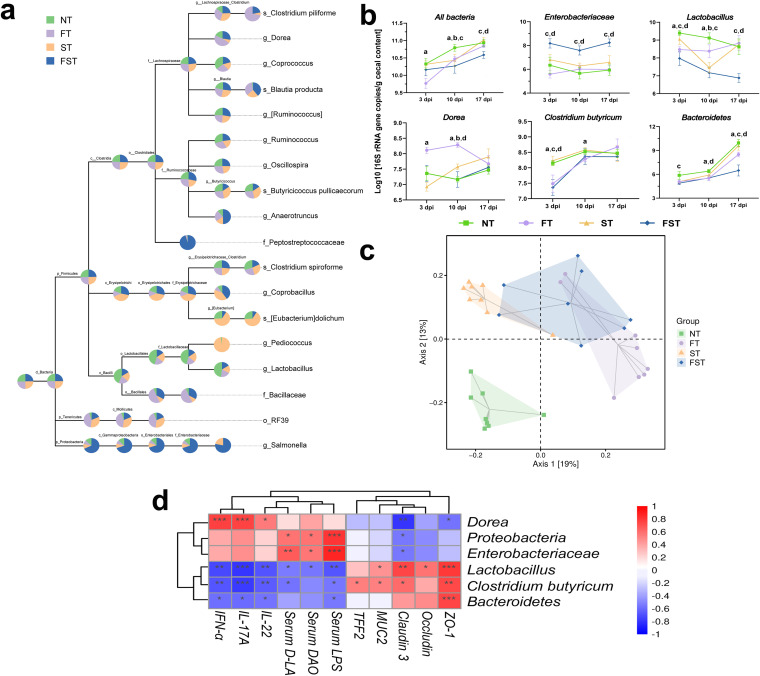
Phylum and genus distributions of microbial compositions are shown in Fig. 4. At the phylum level, Firmicutes, Bacteroidetes, and Proteobacteria were the three dominant members of the microbiota. Among them, Firmicutes (67.85 to 99.62%) dominated the microbiota in all four groups at three different stages of infection (Fig. 4a). At 3, 10, and 17 dpi, the FST chickens had the highest relative abundance of Proteobacteria (10.68%, 5.13%, and 1.75%, respectively) relative to the other three groups (Fig. 4a). At 17 dpi, the FFC-treated (9.23%) chickens had a significantly reduced relative abundance of Bacteroidetes relative to the NT group (31.26%). Salmonella infection (23.87%) had a negligible effect on Bacteroidetes abundance. However, infection with Salmonella after pretreatment with florfenicol almost eliminated the growth of Bacteroidetes (0.01%) (Fig. 4a). At the genus level, Ruminococcus, Oscillospira, and Lactobacillus were the three dominant genera at days 11 and 18, and the genera Ruminococcus, Oscillospira, and Bacteroides were dominant at day 25.
FIG 4.
Cecal bacterial communities. (a) Compositions of the gut microbiota at the phylum level (relative abundance) based on 16S rRNA gene sequencing of DNA isolated from cecal contents at the indicated time points after infection. (b to d) Relative abundances of the top 20 cecal microbiota members at the genus level at 3 dpi (b), 10 dpi (c), and 17 dpi (d).
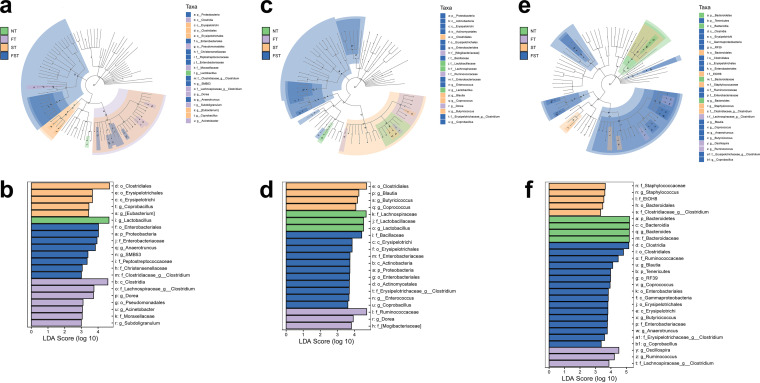
We applied the linear discriminant analysis (LDA) effect size (LEfSe) method to identify abundant bacterial taxa among these groups; only those taxa that obtained a log LDA score of >3 were ultimately considered. A cladogram from phylum- to genus-level abundances is shown in Fig. 5. In total, 21, 21, and 28 differentially abundant taxa were identified at 3, 10, and 17 dpi, respectively (Fig. 5). In the untreated control chickens, the LEfSe method highlighted the greater differential abundances of Lactobacillus at 3 and 10 dpi and Bacteroides at 17 dpi. Notably, the relative abundance of Enterobacteriaceae was significantly higher in the FST group than in the other three groups at all three time points, and the other taxa were altered irregularly at different times in different groups. We also established a taxonomic cladogram at 11 days (3 dpi), with the relative abundance of the taxon node of each group shown as a pie chart; only those taxa with a relative abundance of >0.1% were considered (Fig. 6a). Similarly, the abundance ratio of Lactobacillus was considerably higher in the control group than in the other three groups. Additionally, the abundance ratio of Enterobacteriaceae in the FST group dominated among all four groups. Furthermore, Salmonella was found only in the challenged groups (ST and FST) at the genus level, with the abundance ratio of Salmonella in the FFC pretreatment group being significantly higher than those in the untreated groups (Fig. 6a). We measured the cecal loads of these biomarkers and an intestinal protective bacterium by real-time quantitative PCR (qPCR) (Fig. 6b). At day 11 (3 dpi), florfenicol significantly reduced the densities of total bacteria, Lactobacillus, and Clostridium butyricum and increased the abundance of Dorea. Although Salmonella infection had no effect on cecal bacterial densities, chicks with Salmonella infection after pretreatment with florfenicol harbored much higher densities of Enterobacteriaceae and lower densities of Lactobacillus, Bacteroides, and C. butyricum than the control group. At day 18 (10 dpi), florfenicol still significantly reduced the densities of total bacteria, Lactobacillus, and C. butyricum and increased the abundance of Dorea. Additionally, the FST group still harbored much higher densities of Enterobacteriaceae and lower densities of Lactobacillus and Bacteroides than the control group. At day 25 (17 dpi), C. butyricum was present at equivalent densities in the cecal contents of all four groups. However, significant differences in the bacterial densities of total bacteria, Lactobacillus, Bacteroides, and Enterobacteriaceae were still apparent between the NT and FST groups or between the ST and FST groups (Fig. 6b). Especially, the FST group still harbored much higher densities of Enterobacteriaceae and lower densities of Lactobacillus and Bacteroides than the control group.
FIG 5.
Differences in the gut microbiota of the chicken cecum microbial community determined using the LEfSe analytic method. (a, c, and e) LEfSe plots (P < 0.05; log LDA score of >3) showing microbial strains with significant differences among the four treatment groups at 3 dpi (a), 10 dpi (c), and 17 dpi (e). The different groups are represented by different colors, the microbiota members that play an important role are represented by nodes of corresponding colors, and the organism markers are represented by colored circles. From inside to outside, the circles are ordered by species at the levels of phylum, class, order, family, and genus. (b, d, and f) LDA diagrams at three sampling points. Biomarkers with statistical differences are emphasized, with colors of the histograms representing the respective groups and the lengths representing LDA scores, which represent the magnitude of the effects of significantly different species between groups.
FIG 6.
Impact of florfenicol on the cecal microbial communities in response to S. Enteritidis infection at 3 dpi. (a) Taxonomic cladogram showing the relative abundances of taxon nodes in each group (relative abundance of >0.1%). A larger sector area indicates a higher abundance of taxa in the corresponding group. (b) Quantification of the cecal microbiota at 3, 10, and 17 dpi by qPCR of the 16S or 23S rRNA gene. All bars represent means ± SD. Differences were assessed by ANOVA and denoted as follows: a, P < 0.05, NT vs. FT; b, P < 0.05, NT vs. ST; c, P < 0.05, NT vs. FST; d, P < 0.05, ST vs. FST. (c) PCoA for comparison of the changes in bacterial communities at 3 dpi generated using the Bray-Curtis distance method. (d) Correlation analysis between significantly different strains and intestinal barrier function-related parameters. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (by ANOVA and Dunnett’s multiple-comparison test).
A correlation analysis was then performed to identify associations between the significantly different gut microbiota and intestinal barrier function-related parameters (Fig. 6d). The results of the analysis revealed that Dorea, Proteobacteria, and Enterobacteriaceae, which were significantly increased after florfenicol treatment, were negatively correlated with intestinal tight junction proteins (ZO-1, occludin, claudin 3, MUC2, and TFF2) but positively correlated with intestinal permeability (DAO, d-linoleic acid [d-LA], and LPS) and the inflammatory response (IL-22, IL-17A, and IFN-α), while Lactobacillus, Bacteroidetes, and C. butyricum, which were significantly reduced after florfenicol treated, were positively correlated with tight junction proteins but negatively correlated with intestinal wall permeability and the inflammatory response.
Florfenicol alters metabolic profiles.
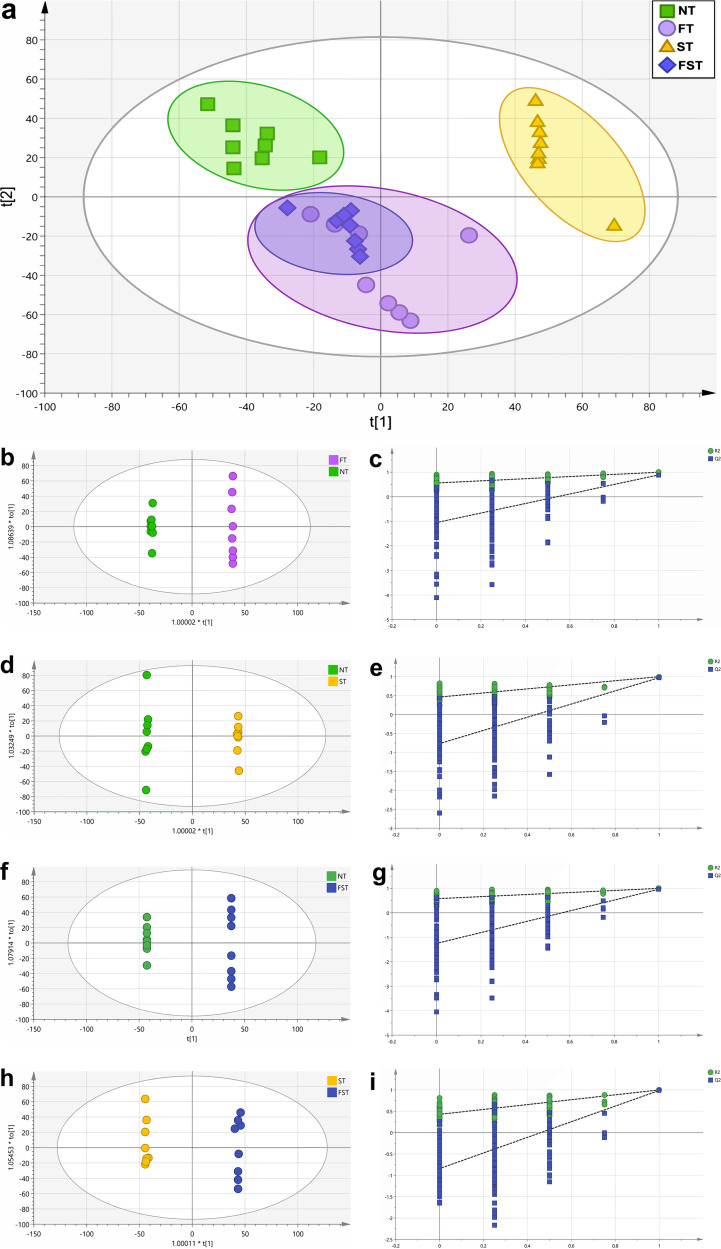
We analyzed metabolomes by liquid chromatography-mass spectrometry (LC-MS) to determine the differential levels of metabolites on day 11 (3 dpi) in cecal contents. The PCoA score plot shows that the FT and ST groups were significantly separated from the NT group, indicating that florfenicol intervention and S. Enteritidis infection can significantly change the intestinal metabolome of chicks. However, there was no clear distinction in cecal metabolites between the FT and FST groups, indicating that after florfenicol intervention, S. Enteritidis infection has little effect on the intestinal metabolome of chicks (Fig. 7a). Orthogonal projections to latent structures discriminant analysis (OPLS-DA) and permutation test plots of OPLS-DA data were performed. As shown in Fig. 7, cecal metabolites of the NT group were clearly distinguished from those of the FT group (Fig. 7b), the ST group (Fig. 7d), and the FST group (Fig. 7f). In addition, there was a clear separation between the FST group and the ST group in cecal metabolites (Fig. 7h).
FIG 7.
PCoA and OPLS-DA plots of cecal metabolomes at 3 dpi. (a) PCoA plot of cecal metabolomes. OPLS-DA score plots of cecal metabolites were based on LC-MS from the different groups. (b, d, f, and h) OPLS-DA plots of four data sets: the FT group versus the NT group (b), the ST group versus the NT group (d), the FST group versus the NT group (f), and the FST group versus the ST group (h). (c, e, g, and i) Validation plots of OPLS-DA data for these four group sets.
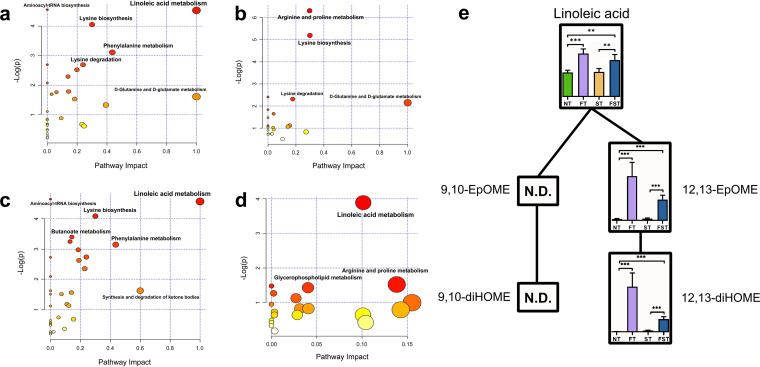
From the OPLS-DA models, we identified 72 differential metabolites between the NT and FT groups, 42 differential metabolites between the NT and ST groups, 69 differential metabolites between the NT and FST groups, and 57 differential metabolites between the FST and ST groups, using thresholds of a variable of importance in projection (VIP) of >1 and a P value of <0.05 (by Welch’s t test). The differential metabolites are listed in Table S2. The effect of florfenicol on metabolic pathway differences according to the Kyoto Encyclopedia of Genes and Genomes (KEGG) is shown in Fig. 8. Linoleic acid metabolism, lysine biosynthesis, lysine degradation, phenylalanine metabolism, d-glutamine and d-glutamate metabolism, and aminoacyl-tRNA biosynthesis were mainly enriched after florfenicol treatment (Fig. 8a). Arginine and proline metabolism, lysine biosynthesis, lysine degradation, and d-glutamine and d-glutamate metabolism were mainly enriched by Salmonella infection (Fig. 8b). Linoleic acid metabolism, aminoacyl-tRNA biosynthesis, lysine biosynthesis, butanoate metabolism, and phenylalanine metabolism were mainly enriched in florfenicol-pretreated, Salmonella-infected chicks (Fig. 8c), and linoleic acid metabolism was mainly enriched between the FST and ST groups (Fig. 8d). These data indicated that linoleic acid metabolism is the most noteworthy metabolic pathway in the FFC-treated groups with or without Salmonella challenge. Next, we mapped the metabolic pathway of linoleic acid based on the identified differential metabolites as well as the relative amounts (means ± standard deviations [SD]) of these metabolites in the four groups (Fig. 8e). The metabolites that affect the metabolic pathways of linoleic acid are primarily linoleic acid, 12,13-epoxyoctadecenoic acid (12,13-EpOME), and 12,13-dihydroxyoctadecenoic acid (12,13-diHOME); the relative levels of these metabolites in the FT and FST groups were significantly higher than those in the NT and ST groups. Notably, the relative levels of 12,13-EpOME and 12,13-diHOME were significantly higher in the FFC-pretreated group but were negligible in the nonpretreated group (Fig. 8e).
FIG 8.
Comparison of differential metabolites and key metabolic pathways at 3 dpi. (a to d) Metabolic pathway analysis of biomarker metabolites. Plots show overrepresented metabolic pathways between the FT and NT groups (a), the ST and NT groups (b), the FST and NT groups (c), and the FST and ST groups (d). The x axis represents pathway impact, and the y axis represents pathway enrichment. Larger sizes and darker colors represent higher pathway enrichment levels and higher pathway impact values, respectively. (e) Linoleic acid metabolomic pathway map. Bars represent relative amounts (means ± SD) of metabolites. N.D., not detected. **, P < 0.01; ***, P < 0.001 (by ANOVA and Dunnett’s multiple-comparison test).
Correlation between the differential gut microbiota and metabolites.
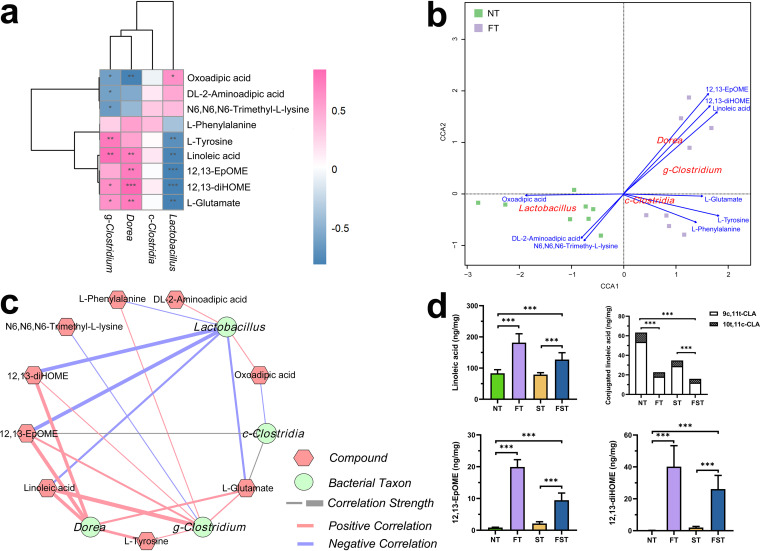
After observing marked differences in metabolite contents as well as microbial compositions after florfenicol treatment, we tested for specific correlations between the microbial taxa and key metabolites using multi-omics. This mainly includes 9 metabolites in 4 significantly differential metabolic pathways, linoleic acid metabolism (linoleic acid, 12,13-EpOME, and 12,13-diHOME), lysine metabolism (oxoadipic acid, N6,N6,N6-trimethyl-l-lysine, and dl-2-aminoadipic acid), phenylalanine metabolism (l-phenylalanine and l-tyrosine), and d-glutamine and d-glutamate metabolism (l-glutamate), and four significantly differential bacterial taxa, Clostridia, Clostridium, Dorea, and Lactobacillus. Spearman correlation analysis revealed an association between bacterial genera and metabolites in FFC-pretreated chicks (Fig. 9a). The results showed that Dorea and Clostridium are strongly positively correlated with linoleic acid metabolism (linoleic acid, 12,13-EpOME, and 12,13-diHOME) and d-glutamine and d-glutamate metabolism (l-glutamate), whereas lysine metabolism (oxoadipic acid, N6,N6,N6-trimethyl-l-lysine, and dl-2-aminoadipic acid) is negatively correlated. Furthermore, the genus Lactobacillus was negatively correlated with linoleic acid metabolism (linoleic acid, 12,13-EpOME, and 12,13-diHOME), phenylalanine metabolism (l-phenylalanine and l-tyrosine), and d-glutamine and d-glutamate metabolism (l-glutamate), whereas it was positively correlated with lysine metabolism (oxoadipic acid, N6,N6,N6-trimethyl-l-lysine, and dl-2-aminoadipic acid). Finally, a weaker positive or no correlation was detected between Clostridia and these 9 metabolites. Canonical correspondence analysis (CCA) showed that Dorea was the most important bacterial taxon influencing linoleic acid metabolism (including linoleic acid, 12,13-EpOME, and 12,13-diHOME) after florfenicol treatment (Fig. 9b). The correlation network between differential bacterial taxa and metabolites consisted of 13 nodes and 22 edges. We found that the metabolic pathway of linoleic acid has a strong positive correlation with Dorea, whereas Lactobacillus has a negative correlation with it (Fig. 9c).
FIG 9.
Correlation between key bacterial taxa and differential metabolites and their absolute abundances at 3 dpi. (a) Spearman correlations between differential metabolites and bacterial taxa calculated for the FT and NT groups (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Positive correlations are labeled in pink, and negative correlations are labeled in blue. (b) CCA plot of differential metabolites and bacterial taxa for the FT and NT groups. The CCA ordination plot shows correlations between bacterial community structures and metabolite factors. Correlations between metabolite factors and key bacterial taxa are represented by the lengths and angles of arrows. (c) Correlation network analysis of the key bacterial taxa and differential metabolites for the FT and NT groups. The hexagons indicate metabolites, and the circles indicate bacterial taxa. Lines connecting each node represent Spearman correlation coefficient values. Red lines represent positive correlations, blue lines represent negative correlations, and the thickness of the edge represents the strength of the correlations. (d) Concentrations of key metabolites in cecal contents. Bars indicate means ± SD. ***, P < 0.001 (by ANOVA and Dunnett’s multiple-comparison test).
As linoleic acid can be converted into conjugated linoleic acid (CLA) by Lactobacillus (24), and a significantly negative correlation between linoleic acid and Lactobacillus was observed in our study, we hypothesized that the non-FFC-pretreated chicks (higher abundance of Lactobacillus) may have higher CLA levels. However, CLA is an isomer of linoleic acid, so the use of untargeted metabolomics cannot distinguish between these compounds. Therefore, we employed targeted LC-MS to detect compounds, including linoleic acid, 9c,11t-CLA, 10t,11c-CLA, 12,13-EpOME, and 12,13-diHOME. In line with the results of metabolic profiling, linoleic acid, 12,13-EpOME, and 12,13-diHOME levels were higher in the FFC-pretreated groups. Moreover, we observed higher CLA concentrations in the cecal contents of non-FFC-pretreated chicks, with the levels of 9c,11t-CLA being significantly higher than those of 10t,11c-CLA (Fig. 9d). Spearman correlation analysis showed a strong association between the abundances of Lactobacillus and CLA concentrations (Fig. S4).
Contrasting effects of conjugated linoleic acid and 12,13-diHOME on S. Enteritidis infection.
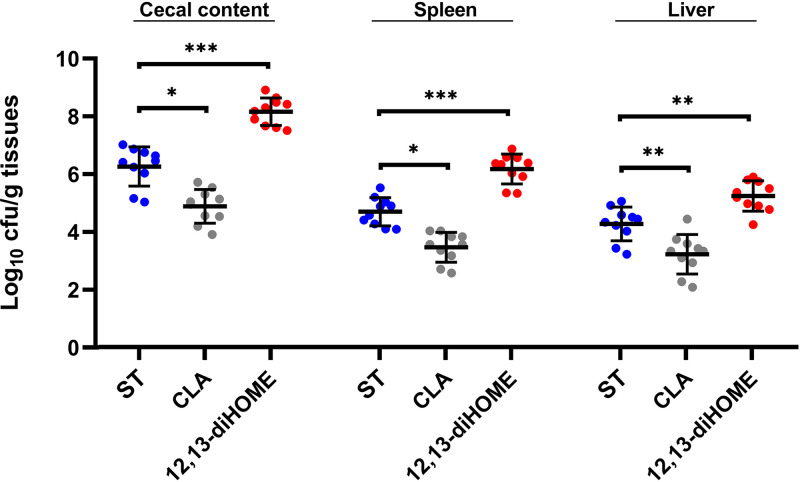
Next, we investigated whether CLA and 12,13-diHOME, the end product of linoleic acid, have an effect on Salmonella infection in newly hatched chicks. Thus, neonatal chickens were pretreated with CLA and 12,13-diHOME for 7 days and infected with S. Enteritidis on day 8. By 3 dpi, Salmonella loads in the cecum (6.264 log10 CFU/g), spleen (4.703 log10 CFU/g), and liver (4.278 log10 CFU/g) were significantly reduced in the chicks pretreated with CLA (4.887 log10 CFU/g cecal content [P < 0.05], 3.473 log10 CFU/g spleen [P < 0.05], and 3.229 log10 CFU/g liver [P < 0.01]), whereas they were significantly increased by pretreatment with 12,13-diHOME (8.165 log10 CFU/g cecal content [P < 0.001], 6.177 log10 CFU/g spleen [P < 0.001], and 5.245 log10 CFU/g liver [P < 0.01]) (Fig. 10).
FIG 10.
Effects of CLA and 12,13-diHOME on S. Enteritidis infection in neonatal chicks. The S. Enteritidis loads in the cecal contents, spleen, and liver at 3 dpi are shown. Bars indicate medians. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (by a Mann-Whitney U test).
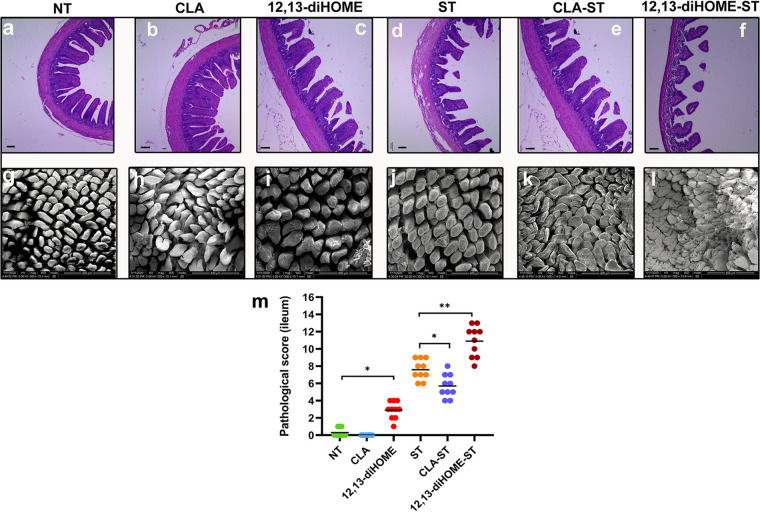
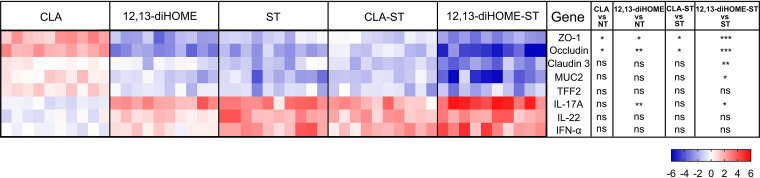
Consistent with the fecal Salmonella loads, pretreatment with CLA significantly reduced Salmonella-induced intestinal injury and enteropathy at 3 dpi, whereas 12,13-diHOME significantly increased them. In addition, in the absence of Salmonella infection, pretreatment with 12,13-diHOME can also cause minor intestinal injury (Fig. 11). Moreover, in the case of Salmonella infection, CLA-pretreated chicks exhibited decreases in intestinal permeability (serum d-lactate, DAO, and LPS levels) and proinflammatory factors (IL-1β, IL-6, IL-8, TNF-α, and IFN-γ) as well as a significant increase in IL-10 levels, and in the absence of Salmonella infection, pretreatment with CLA also decreased serum d-lactate and LPS levels and the expression of IL-1β and IL-8 and increased IL-10 levels. Notably, 12,13-diHOME-pretreated chicks exhibited contrasting results (Fig. S5). We also compared the effects of these two metabolites on the expression of genes related to intestinal barrier function after Salmonella infection (Fig. 12). We found that CLA significantly increased the expression of ZO-1 and occludin, whereas 12,13-diHOME significantly reduced the expression of ZO-1, occludin, claudin 1, and MUC2, and 12,13-diHOME also significantly increased the expression of IL-17A. Furthermore, in the absence of Salmonella infection, pretreatment with CLA also significantly increased the ZO-1 and occludin levels, whereas 12,13-diHOME significantly reduced the expression of ZO-1 and occludin and also significantly increased the expression of IL-17A (Fig. 12).
FIG 11.
CLA pretreatment attenuates yet 12,13-diHOME exacerbates the development of Salmonella-induced ileal mucosal injury in neonatal chicks at 3 dpi. (a to l) After 3 days of infection, the histopathology of the intestinal tissue was analyzed by H&E staining (a to f) and SEM (g to l). Bars, 100 μm (a to f). (m) Ileal pathology was scored in H&E-stained ileal tissue sections. Data are expressed as means ± standard deviations. *, P < 0.05; **, P < 0.01 (by a Mann-Whitney U test).
FIG 12.
Gene expression profiles in response to CLA and 12,13-diHOME pretreatment following S. Enteritidis infection at 3 dpi. Data are represented as log2 fold changes between the treatment group and the control group. Statistical analysis was conducted using one-way ANOVA and Dunnett’s multiple-comparison test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
DISCUSSION
The gut microbiome plays vital roles in resistance to exogenous pathogens via phage deployment, the secretion of antibacterial substances, competing nutrients, and intestinal barrier function. Antibiotic administration perturbs the gut bacterial community and metabolome, resulting in weakened resistance to gut colonization by pathogens. However, their effects on the gut microbiota and metabolome correlations and how they promote pathogen infection remain unclear. In this study, we investigated the effect of florfenicol on intestinal Salmonella colonization of chicks and how it affects Salmonella colonization by microbiome and metabolomics analyses. In our previous study, we used enrofloxacin to feed 1-day-old chickens followed by oral challenge with a non-antibiotic-resistant Salmonella strain to study the effect of enrofloxacin on Salmonella colonization in the chicks. The results showed that although enrofloxacin reduced Salmonella colonization at 1 dpi, the abundance of Salmonella was significantly higher in the enrofloxacin-treated group than in the untreated group at 7 and 14 dpi. We also found that enrofloxacin significantly altered the gut microbiota and metabolome, but we did not explore the underlying mechanism (25). In this study, we also used a non-florfenicol-resistant Salmonella strain (ATCC 13076) to infect chicks, and the same result was obtained (data not shown). The purpose of our research is to explore how florfenicol changes the structure of the gut microbiota and its metabolism, thereby affecting Salmonella colonization. Here, we used florfenicol-resistant Salmonella to exclude a direct effect of florfenicol on Salmonella, to more directly explore how florfenicol disturbs the host microbiome and metabolome to affect intestinal Salmonella colonization. Our results showed that the abundance of Salmonella was significantly higher in the FFC-treated group than in the untreated group, and Salmonella persisted longer. Thus, our findings indicated that florfenicol altered the ability of S. Enteritidis to colonize the gut in chickens, and florfenicol can cause severe Salmonella infection and prolonged gut colonization. Furthermore, we found that FFC pretreatment exacerbated Salmonella-induced defects in morphology, decreased intestinal barrier function, and increased intestinal barrier permeability. Previous studies showed that intestinal inflammation provides a growth advantage for Salmonella (26–28), and the reason for this inflammation may be related to the significantly increased LPS level after florfenicol pretreatment (29). Taken together, these findings imply that FFC pretreatment increased intestinal permeability and inflammation and aggravated Salmonella-induced intestinal barrier damage. These changes collectively promoted Salmonella colonization in neonatal chicks.
As the gut microbiota plays an important role in combating Salmonella invasion and maintaining intestinal immunity (26, 30), we characterized the intestinal flora of neonatal chicks in the treated groups. Our results showed that florfenicol intervention did not clearly change α-diversity but significantly altered β-diversity. This finding is consistent with previous studies. Saenz et al. reported that the input of florfenicol did not clearly change metagenomic diversity but altered the abundance of bacterial families in the gut of fish (31). Yang et al. found that α-diversity was not significantly affected by florfenicol intervention, while the percentage of Flavobacteriia in sea cucumber intestines was significantly decreased (32). Our results show that there is a similar phenomenon in the effect of florfenicol on the diversity of the gut microbiota in chickens, where florfenicol has no significant effect on α-diversity, while it alters the abundances of specific bacterial species. The β-diversity results indicated that the gut microflora structure of chicks was significantly changed by a single florfenicol treatment or Salmonella infection. The microbial community of chickens is complex and relatively stable, and the restoration of the microbiota after antibiotic withdrawal can be expected (33, 34). However, florfenicol administration at an early age in chickens may have a profound effect on microbial composition that would hinder its restoration (see Fig. S3 in the supplemental material). Gao et al. found that feeding antibiotics in the chick stage significantly delayed the maturation of the gut flora and formed less and more fragile correlation networks of bacteria, which tended to be less resistant to pathogen colonization (35). Our previous study using enrofloxacin on chicks found the same phenomenon (25), and in this study, we also demonstrated that the maturation of the intestinal microbiota is significantly retarded and eventually delayed by florfenicol intervention in chicks at early ages.
FFC administration significantly decreased the abundance of Lactobacillus at days 11 and 18 (3 and 10 days posttreatment) and that of Bacteroides at day 25 (17 days posttreatment). Lactobacillus spp. are considered probiotic in nature and have been used in livestock feed processing for decades because of their beneficial effects on immunity, growth, and intestinal colonization resistance (36–38). For example, Lactobacillus rhamnosus reduces colonization by pathogenic Salmonella, Clostridium, and E. coli strains in porcine intestinal mucus (39). Lactobacillus acidophilus binds to cultured human intestinal cell lines and inhibits cell invasion by enterovirulent bacteria, including Salmonella Typhimurium (40). Another study showed that Lactobacillus plantarum exerts an antagonistic effect on pathogenic bacteria by increasing the content of secretory IgA (SIgA) (41). In our study, FFC treatment significantly decreased the abundance of Lactobacillus in chicks, suggesting that this genus may be the main target bacteria of FFC, and this reduction may be responsible for the promotion of Salmonella colonization after FFC pretreatment. The phylum Bacteroidetes is the dominant phylum in the mature microbiota of chickens (42) and may have some inhibitory effects on gut colonization by Salmonella. Miki et al. reported that Bacteroides spp. accelerate the elimination of S. Typhimurium from the intestinal lumen of mice by producing vitamin B6 (14). Another study demonstrated that Bacteroides species confer colonization resistance to S. Typhimurium infection by producing propionate, which directly limits Salmonella growth by disrupting intracellular pH homeostasis (43). Thus, the use of florfenicol may delay the maturation of the chicken intestinal flora and hinder the clearance of Salmonella by reducing the abundance of Bacteroidetes. Furthermore, chicks with Salmonella infection after FFC pretreatment had the highest relative abundance of Proteobacteria, which are known to be potential pathogens of poultry and humans. A recent study showed that preventive treatment of calves with florfenicol resulted in a 10-fold increase in facultative anaerobic Escherichia spp., which is a signature of an imbalanced microbiota (44). Saenz et al. reported that oral administration of florfenicol to fish resulted in a shift in the gut microbiome toward well-known putative pathogens such as Salmonella, Plesiomonas, and Citrobacter (31). Combined with our results, we conclude that florfenicol administration changed the overall structure of the gut microbiota, and these changes make it easier for Proteobacteria to gain a competitive advantage in cases of Salmonella infection. Furthermore, correlation analysis found that these intestinal microbes that were significantly altered after florfenicol treatment were associated with changes in intestinal barrier function. Of these, Dorea was found to be significantly enriched in patients diagnosed with irritable bowel syndrome (45), multiple sclerosis (46), and nonalcoholic fatty liver disease (47), suggesting that the overgrowth of this genus has adverse health effects, and in our study, Dorea showed a significant positive correlation with the inflammatory response, while C. butyricum effectively attenuated inflammation and intestinal barrier damage in the intestine of Salmonella-infected chickens (48). The significantly altered intestinal microbiota after florfenicol administration may be a potential mechanism for disrupting intestinal barrier function and promoting intestinal colonization by Salmonella.
Next, we used metabolomics to determine how florfenicol affects Salmonella gut colonization. Our data indicated that arginine and proline metabolism is the most significant pathway affected by Salmonella infection, and linoleic acid metabolism is the most notable pathway affected by florfenicol intervention. A recent study also showed that arginine and proline metabolism is the most prominent pathway in chickens infected with S. Enteritidis (49). Arginine is a common amino acid substrate, and arginase can be used for the synthesis of ornithine and proline and also for inducible nitric oxide (NO) synthase (iNOS) for NO production (50). NO produced by iNOS during Salmonella infection is an innate immune response that is part of the host defense mechanism, and inhibition or downregulation of iNOS can enhance the proliferation and survival of Salmonella (51, 52). Arginase can negatively regulate the production of NO by competing with the common substrate arginine. Therefore, those authors think that the upregulation of the arginine and proline metabolism pathway after Salmonella infection may be part of the host metabolic regulation strategy to inhibit intestinal inflammation during Salmonella infection (49). Linoleic acid metabolism is the most notable pathway affected by florfenicol pretreatment. Linoleic acid is first metabolized to 12,13-EpOME by cytochrome P450 (CYP) epoxygenase, followed by hydrolysis catalyzed by soluble epoxide hydrolases (sEHs) to form the diol 12,13-diHOME (53). diHOME compounds have multiple pathological features, such as decreasing postischemic cardiac recovery, participating in vascular cognitive impairment, increasing skeletal muscle fatty acid uptake, and impeding immune tolerance in asthmatic children (54–57). The 12,13-diHOME produced by sEH hydrolysis of 12,13-EpOME showed stronger cytotoxicity (53, 58). Besides liver, a variety of gut bacteria also produce sEHs (57). Correlation analysis results showed that the concentration of 12,13-diHOME was positively correlated with Dorea and Clostridium, so we suspected that sEH may be produced by these bacteria in the gut. A recent study showed that the sEH and sEH-derived lipid metabolites induce intestinal barrier dysfunction, bacterial translocation, and colonic inflammation in mice (59). In this study, we pretreated neonatal chicks with 12,13-diHOME and observed significantly higher Salmonella colonization in the gut. Intestinal inflammation, particularly that due to proinflammatory cytokines, disrupts barrier function, leads to intestinal permeability, and promotes colonization by pathogens (27, 60, 61). Previous studies showed that diHOME compounds exhibit proinflammatory effects on vascular endothelial cells (62), lung (57), and peripheral nervous tissue (63). Our study indicated that 12,13-diHOME also exhibits a proinflammatory effect on intestinal epithelial cells. Moreover, the diHOME compounds also disrupt mitochondrial function by altering mitochondrial permeability and inducing cellular apoptosis (64, 65), and this may be why 12,13-diHOME exacerbates intestinal barrier damage. Therefore, we suggest that 12,13-diHOME contributes to Salmonella colonization of chick intestines by promoting intestinal inflammation and disrupting intestinal barrier function.
CLA is the second factor affecting Salmonella gut colonization in chicks preadministered FFC. Our correlation analysis combined with targeted metabolomics revealed that Lactobacillus and CLA showed a significant positive correlation, and FFC pretreatment reduced the abundances of both Lactobacillus and CLA in the gut lumen. CLA is formed from linoleic acid by Lactobacillus and can inhibit the growth of pathogenic bacteria (66). We pretreated neonatal chicks with CLA and observed that it effectively reduced Salmonella colonization. Recently, de Barros et al. also found that CLA pretreatment can reduce intestinal permeability, bacterial translocation, and the inflammatory response to prevent damage caused by intestinal mucositis induced by 5-fluorouracil in a mouse model (67). We suggest that CLA reduces intestinal colonization by Salmonella by influencing several processes. First, CLA treatment considerably upregulated the concentrations of tight junction proteins (ZO-1, occludin, E-cadherin 1, and claudin 3) and ameliorated epithelial apoptosis, which protects intestinal cells from impairment caused by Salmonella infection. Second, CLA modulates gut inflammation by attenuating the expression of proinflammatory cytokines. Finally, studies also show that CLA directly inhibits the growth of pathogenic bacteria, including Salmonella. Byeon et al. showed that CLA possesses antimicrobial activity against the growth of a variety of foodborne pathogens; 1.8 mM CLA completely inhibits the growth of S. Typhimurium (66). Peng et al. indicated that CLA produced by Lactobacillus competitively excludes Salmonella under mixed-culture conditions (68). Additionally, Tabashsum et al. also showed that CLA produced by Lactobacillus inhibits the growth and survival of Salmonella by altering the relative expression of genes related to Salmonella virulence (69). Thus, our results suggest that CLA maintains intestinal integrity, reduces intestinal inflammation, and inhibits Salmonella growth to effectively reduce gut colonization by Salmonella in chicks. Therefore, FFC may reduce the production of CLA by inhibiting Lactobacillus growth, thereby reducing the colonization resistance of neonatal chicks to Salmonella infection.
In conclusion, our results indicated that florfenicol administration affected the colony structure and metabolite composition in the neonatal chicken cecum, and the abundances of Lactobacillus, Clostridium, and Dorea and linoleic acid metabolism were significantly affected. Florfenicol reduces the production of CLA by inhibiting Lactobacillus growth and increases 12,13-diHOME levels in the intestine, thereby reducing the colonization resistance of neonatal chicks to Salmonella infection. It is suggested that CLA or Lactobacillus may replace florfenicol to reduce intestinal colonization by Salmonella, which is of great significance to ensure the healthy growth of chicks and the prevention and control of Salmonella. We provide a better understanding of the susceptibility of animal species to Salmonella after antibiotic intervention that may help to elucidate infection mechanisms that are important in both animal and human health.
MATERIALS AND METHODS
Bacterial strains.
An S. Enteritidis (ATCC 13076) floR mutant (florfenicol-resistant strain) was used as the challenge strain and stored in our laboratory. The S. Enteritidis florfenicol-resistant strain was constructed by inserting the floR gene (GenBank accession no. NG_047860.1) using a plasmid-based homologous-recombination integration method as previously described (70). Prior to inoculation, S. Enteritidis was grown overnight in Luria-Bertani broth at 37°C.
Florfenicol intervention and S. Enteritidis infection.
The experimental protocols used in this experiment, including animal care and use, were reviewed and approved by the Animal Care and Use Ethics Committee of Sichuan University. Leghorn layer chicks (1 day old) were hatched from the same batch of eggs of SPF birds (Beijing Boehringer Ingelheim Vital Biotechnology Co. Ltd., China), and each assigned group was reared in an individual GJ-1 SPF isolator (Suzhou Fengshi Laboratory Animal Equipment Co. Ltd., China). The Leghorn chicken is a famous layer breed that is widely distributed throughout the world; these chickens grow to about 150 days to reach the stage for laying eggs, and the annual egg output is 220 to 300 eggs. Animals received the same nonmedicated chick feed and water ad libitum; they were raised under controlled environmental conditions with a 16-h lighting cycle and a temperature of 32°C at day 1, which was gradually reduced and maintained at 24°C from day 10.
(i) Animal protocol 1: effect of florfenicol exposure on Salmonella infection.
Ninety-six newly hatched chicks were assigned at random to four groups, and each group included 24 chicks for three time points (n = 8 chicks at each time point). Each chicken was weighed, and the average body weight of the chickens was approximately 50 g. From 1 to 7 days of age, chicks were pretreated with 30 mg/kg of body weight of florfenicol (FFC) by oral gavage. At 8 days of age, chicks were infected by the oral administration of 108 CFU of the challenge strain of S. Enteritidis. The following groups were included: (i) the NT group (control group that was neither FFC treated nor S. Enteritidis infected), (ii) the FT group (FFC-treated group), (iii) the ST group (S. Enteritidis-infected group), and (iv) the FST group (FFC-pretreated and S. Enteritidis-infected group). Chickens were euthanized on days 11, 18, and 25, and cecal contents, spleen, liver, ileum, and serum were collected (Fig. 1a; see also Fig. S6 in the supplemental material).
(ii) Animal protocol 2: effect of conjugated linoleic acid and 12,13-diHOME on Salmonella infection.
Sixty newly hatched chicks were assigned at random to six groups (n = 10 chicks per group) and treated with conjugated linoleic acid (CLA) or 12,13-diHOME by gastric gavage. CLA (purity of >99%; Nu-Chek Prep, Elysian, MN, USA) is a mixture of 65.5% c9,t11-CLA and 34.5% t10- and c12-CLA isomers by LC-MS detection (data not shown). The 12,13-diHOME (purity of ≥98%; Cayman Chemical, Ann Arbor, MI, USA) solution was prepared as described in a previous report (57). The doses of CLA and 12,13-diHOME corresponded to the amount of consumed diet supplemented with 1% CLA and 12,13-diHOME (1% [10 mg/g diet] × 3 − 9 [grams of diet consumed on average by chicks at days 1 to 7]). From 1 to 7 days of age, chicks were treated with CLA or 12,13-diHOME. At 8 days of age, chickens were infected by the oral administration of 108 CFU of S. Enteritidis. Chicks were divided into six groups: (i) the NT group (control group), (ii) the CLA group (CLA-pretreated group), (iii) the 12,13-diHOME group (12,13-diHOME-pretreated group), (iv) the ST group (S. Enteritidis-infected group), (v) the CLA-ST group (CLA-pretreated and S. Enteritidis-infected group), and (vi) the 12,13-diHOME-ST group (12,13-diHOME-pretreated and S. Enteritidis-infected group). On day 11, the chicks were euthanized for analysis (Fig. S6).
Salmonella identification and enumeration.
After chicks were euthanized, the cecal contents and internal organs were aseptically collected and homogenized in phosphate-buffered saline (PBS). For enumerating Salmonella loads, an aliquot (100 μl) of the appropriate dilution was spread onto XLT4 agar plates (50 μg/ml florfenicol); Salmonella appeared as typical black colonies after incubation at 37°C for 24 h. S. Enteritidis colonies were counted, the CFU per gram of tissue were calculated, and the results are expressed as log10 CFU per gram.
DNA extraction, 16S rRNA gene sequencing, and data analysis.
Seven or eight chicks per treatment were randomly chosen at three time points, 11, 18, and 25 days of age, and euthanized by carotid artery bleeding. The cecal contents were collected within 5 min of euthanasia, immediately placed into precooling cryogenic vials, and stored at −80°C until DNA extraction. Total genomic DNA was extracted from cecal contents using the QIAamp DNA stool minikit (Qiagen, Germany) according to the manufacturer’s protocols and stored at −20°C until analysis. The concentration and quality of extracted DNA samples were measured by using a Nanodrop 2000 system (Thermo Fisher Scientific, Waltham, MA, USA) and agarose gel electrophoresis, respectively.
Using the isolated genomic DNA as the template, the V3-V4 hypervariable regions of the bacterial 16S rRNA genes were PCR amplified with primers 338F (5′-ACTCCTACGGGAGGCAGCA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) according to a previously described method (71). Amplicons were then sequenced on the Illumina MiSeq platform (Illumina Inc., USA) using 2- by 250-bp cycles. QIIME was employed to process the sequencing data. Briefly, raw sequencing reads with exact matches to the barcodes were assigned to the respective samples and identified as valid sequences. The low-quality sequences were filtered according to the following criteria (72, 73): sequences that had a length of <150 bp, sequences with average Phred scores of <20, sequences that contained ambiguous bases, and sequences that contained mononucleotide repeats of >8 bp. Paired-end reads were assembled using FLASH (74). After chimera detection, the remaining high-quality sequences were clustered into operational taxonomic units (OTUs) at 97% sequence identity by UCLUST (75). A representative sequence was selected from each OTU using default parameters. OTU taxonomic classification was conducted by BLAST searches of the representative sequence set against the Greengenes database (76), and the best hit was used for further analysis (77). An OTU table was generated to record the abundance of each OTU in each sample and the taxonomy of these OTUs. OTUs containing <0.001% of total sequences across all samples were discarded. To minimize differences in sequencing depth across samples, an averaged, rounded, rarefied OTU table was generated by averaging 100 evenly resampled OTU subsets under 90% of the minimum sequencing depth for further analysis.
Sequence data were analyzed using the QIIME and R software packages (v3.2.0) (78). OTU-level α-diversity indices, including Shannon indices, species abundance, and Pielou indices, were calculated using the OTU table in QIIME. β-Diversity analysis was performed to investigate the structural variation of microbial communities across samples using Bray-Curtis distance metrics and visualized via principal-coordinate analysis (PCoA). Taxonomic compositions and relative abundances were visualized using MEGAN (79) and GraPhlAn (80). The LEfSe method was performed to detect differentially abundant taxa across groups using default parameters (81).
Quantitative PCR for microbiota analysis.
The bacterial composition of the microbiota was validated by qPCR as previously described (14, 82–84). All qPCRs were performed using the Bio-Rad real-time PCR detection system (CFX Maestro 1.1, 3.0; Bio-Rad Inc., USA) and SsoFast EvaGreen supermix (Bio-Rad Inc., USA) according to the manufacturer’s instructions. Genomic DNA from cecal samples was used as a template for qPCR using the main group-specific primers (Table S1) for all Eubacteria, Enterobacteriaceae, Lactobacillus, Dorea, Clostridium butyricum, and Bacteroidetes. Serial dilutions of plasmids containing the target gene cloned into the pMD-19 T cloning vector (TaKaRa, Dalian, China) were analyzed to generate standard curves and calculate absolute counts of target genes.
Metabolomics for chicken cecal contents. (i) Untargeted metabolomics.
Chickens were sacrificed, and the cecum was resected. The cecal contents were obtained and stored at −80°C until analysis. Sample preparation for LC-MS was performed as previously described (85). Briefly, 50 mg freeze-dried sample, 800 μl methanol, and 5 μl dl-o-chlorophenylalanine (internal standard) were added to a 1.5-ml Eppendorf tube. All samples were ground to a fine powder using a grinding mill at 65 Hz for 90 s, vortexed for 30 s, and centrifuged at 12,000 rpm at 4°C for 15 min. Next, 200 μl of the supernatant was transferred to a new vial for LC-MS. A total of 10 μl of the sample solution at 4°C was injected into the LC-MS system (Ultimate 3000LC, Exactive Orbitrap; Thermo) with an Agilent C18 column (Hypergod C18, 100 by 2.1 mm by 1.9 μm), with the column temperature maintained at 40°C. The mobile phase consisted of solutions A and B: solution A was 0.1% formic acid–5% acetonitrile–water (vol/vol/vol), and solution B was 0.1% formic acid–acetonitrile (vol/vol). The flow rate was 350 μl/min. The gradient was set as 0% solution B at 0 min, 20% solution B at 1.5 min, 100% solution B at 9.5 min to 14.5 min, and 0% solution B at 14.6 min to 18 min. Samples were analyzed in positive- and negative-ion modes using a 300°C heater temperature, a 350°C capillary temperature, and a 3.0-kV spray voltage. The flow rates of sheath gas, auxiliary gas, and sweep gas were 45, 15, and 1 arb, respectively. Peaks were aligned according to m/z values and the normalized migration time. Peak areas were calculated by normalization against the internal standards. Metabolites were identified by searches against the database based on m/z values and the normalized migration time. Compound Discoverer software (Thermo) was used to process the Thermo raw files. The data after editing were subjected to multivariate analysis using SIMCA-P 14.0 software (Umetrics AB, Umea, Sweden). Metabolites selected as biomarker candidates were identified on the basis of a VIP threshold of 1 from the 7-fold-cross-validated OPLS-DA model, which was validated at a univariate level with an adjusted P value of <0.05. MetaboAnalyst (version 3.0) was used for the identification of metabolic pathways (86).
(ii) Targeted metabolite detection.
A 50-mg sample of dried cecal contents and 800 μl methanol were added to a 1.5-ml Eppendorf tube. The sample was ground to a fine powder using a grinding mill at 65 Hz for 90 s followed by being vortexed for 30 s and centrifuged at 12,000 rpm at 4°C for 15 min. Next, 200 μl of the supernatant was used for detection.
For quantitative detection of linoleic acid and CLA, 1 μl of each sample was injected into a DB-5 column (60 mm by 0.25 mm by 0.25 μm) using a Thermo Trace 1300 gas chromatography (GC) system (Thermo Fisher Scientific, USA) online with a mass spectrometer (ISQ7000; Thermo Fisher Scientific, USA) (GC-MS). The temperature program was as follows: an initial oven temperature of 140°C held for 5 min, increased at 10°C/min to 180°C and at 4°C/min to 210°C, finally reaching 260°C at a rate of 10°C/min, and then held for 20 min. Helium (99.999% purity) was used as the carrier gas, with a flow rate of 1.5 ml/min. The MS inlet line and the ion source temperatures were maintained at 260°C and 230°C, respectively, and the MS ionization energy was 70 eV. A full-scan mode set from 5 min to 20 min, monitoring the m/z range from 33 to 550 Da, was used for the identification of possible interferences from the matrix extract.
For the quantitative detection of 12,13-EpOME and 12,13-diHOME, 4 μl of each sample was injected into an Acquity ultraperformance liquid chromatography (UPLC) BEH C18 column (100 mm by 2.1 mm by 1.7 μm) using an Acquity UPLC system (Waters Corporation, USA) coupled with a triple-quadrupole mass spectrometry system (API5500; AB Sciex LLC, USA) (UPLC-QqQ-MS). The mobile phase consisted of solution A (water) and solution B (acetonitrile), and the flow rate was 300 μl/min. The gradient was set as follows: 10% solution B at 0 min, 10% solution B at 1.0 min, 90% solution B at 1.5 min, 90% solution B at 5.0 min, 10% solution B at 6.0 min, and 10% solution B at 7.0 min. Samples were analyzed in negative-ion modes using a 550°C atomizing temperature, a −4.5-kV spray voltage, and multiple-reaction monitoring (MRM) of the scanning method. The flow rates of curtain gas, collision gas, GS1 (atomizing gas), and GS2 (auxiliary gas) were 35, 9, 55, and 55 arb, respectively.
Histopathology and microscopic analysis of the intestine.
Parts of the ileal tissue were perfusion fixed with formalin for 24 h. After gradient dehydration with ethanol, specimens were embedded in paraffin. Subsequently, 5-μm sections were rehydrated and stained with alcian blue. Representative images were obtained with a BA400 digital microscope (Motic Group Co. Ltd., China). To determine the degree of lesions, the pathological score was monitored as previously described (87). SEM (Inspect; FEI Ltd., USA) of the intestinal villi was performed as previously described (88). Statistical significance was determined by one-way analysis of variance (ANOVA). A P value of less than 0.05 was considered statistically significant.
RNA isolation and RT-qPCR.
Total RNA from the ileum tissue was extracted using TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA) according to the manufacturer’s instructions. The quality and concentration of RNA were measured using a Nanodrop 2000 spectrophotometer. One microgram of total RNA from each sample was reverse transcribed into cDNA using a SuperScript II kit (Invitrogen Life Technologies, Carlsbad, CA, USA) using oligo(dT) primers and random hexamer primers. qPCR was performed with SsoFast EvaGreen supermix using a Bio-Rad CFX real-time PCR detection system according to the manufacturer’s protocols. Primers are listed in Table S1 (89). Relative mRNA expression levels of each target gene were calculated using the log2 fold change method. Triplicate parallel reactions were run for all samples.
ELISA detection.
Concentrations of chicken IL-1β, IL-6, IL-8, IL-10, IFN-γ, and TNF-α in the ileum tissue and serum LPS, DAO, and d-lactate were determined using the mlbio enzyme-linked immunosorbent assay (ELISA) kit (Shanghai Enzyme-Linked Biotechnology Co. Ltd., China) according to the manufacturer’s instructions. Concentrations were calculated from the standard curves.
Data and statistical analysis.
The Spearman correlation analysis was used to evaluate the correlation between significantly different strains and parameters related to intestinal barrier function, and data are shown as a heat map. The heat map of the interrelationship between the differential flora and the metabolites was generated using the R (3.6.1) pheatmap package. The calculated correlation coefficient (R < 0.5) was used to exclude metabolites and flora with weak correlations and no correlations. Cytoscape (3.7.1) software was used to draw the correlation network diagram; the flora and metabolites were used to form points, and line segments represent the correlation size. The distributions of bacterial communities and their potential correlations with differential metabolites were determined by CCA using the R (3.6.1) vegan package. Statistical analyses were conducted with SPSS 20.0 (SPSS Inc., USA). Data collected are presented as geometric medians or means ± standard deviations. Statistical significance was determined by Mann-Whitney tests or one-way ANOVA. The Mann-Whitney test was used for comparing two groups. One-way ANOVA with Dunnett’s multiple-comparison test was used for pairwise comparison of means from more than two groups in relation to the control group. P values of less than 0.05 were considered statistically significant (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
ACKNOWLEDGMENTS
This work was financially supported by the General Program of the National Natural Science Foundation of China (grant no. 31830098 and 3177131163), the China Agriculture Research System National System for Layer Production Technology (grant no. CARS-40-K14), the National Key R&D Program of China (grant no. 2016YFD0501608), and the Sichuan Science and Technology Program (2020NZZJ001).
Footnotes
Supplemental material is available online only.
Contributor Information
Hongning Wang, Email: whongning@163.com.
Johanna Björkroth, University of Helsinki.
REFERENCES
- 1.Kirk MD, Pires SM, Black RE, Caipo M, Crump JA, Devleesschauwer B, Dopfer D, Fazil A, Fischer-Walker CL, Hald T, Hall AJ, Keddy KH, Lake RJ, Lanata CF, Torgerson PR, Havelaar AH, Angulo FJ. 2015. World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: a data synthesis. PLoS Med 12:e1001921. 10.1371/journal.pmed.1001921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.European Food Safety Authority and European Centre for Disease Prevention and Control. 2019. The European Union One Health 2018 zoonoses report. EFSA J 17:e05926. 10.2903/j.efsa.2019.5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dewey-Mattia D, Manikonda K, Hall AJ, Wise ME, Crowe SJ. 2018. Surveillance for foodborne disease outbreaks—United States, 2009-2015. MMWR Surveill Summ 67:1–11. 10.15585/mmwr.ss6710a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiong W, Wang Y, Sun Y, Ma L, Zeng Q, Jiang X, Li A, Zeng Z, Zhang T. 2018. Antibiotic-mediated changes in the fecal microbiome of broiler chickens define the incidence of antibiotic resistance genes. Microbiome 6:34. 10.1186/s40168-018-0419-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang QQ, Ying GG, Pan CG, Liu YS, Zhao JL. 2015. Comprehensive evaluation of antibiotics emission and fate in the river basins of China: source analysis, multimedia modeling, and linkage to bacterial resistance. Environ Sci Technol 49:6772–6782. 10.1021/acs.est.5b00729. [DOI] [PubMed] [Google Scholar]
- 6.Van Boeckel TP, Brower C, Gilbert M, Grenfell BT, Levin SA, Robinson TP, Teillant A, Laxminarayan R. 2015. Global trends in antimicrobial use in food animals. Proc Natl Acad Sci USA 112:5649–5654. 10.1073/pnas.1503141112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nasim A, Aslam B, Javed I, Ali A, Muhammad F, Raza A, Sindhu ZU. 2016. Determination of florfenicol residues in broiler meat and liver samples using RP-HPLC with UV-visible detection. J Sci Food Agric 96:1284–1288. 10.1002/jsfa.7220. [DOI] [PubMed] [Google Scholar]
- 8.Sullivan A, Edlund C, Nord CE. 2001. Effect of antimicrobial agents on the ecological balance of human microflora. Lancet Infect Dis 1:101–114. 10.1016/S1473-3099(01)00066-4. [DOI] [PubMed] [Google Scholar]
- 9.Looft T, Johnson TA, Allen HK, Bayles DO, Alt DP, Stedtfeld RD, Sul WJ, Stedtfeld TM, Chai B, Cole JR, Hashsham SA, Tiedje JM, Stanton TB. 2012. In-feed antibiotic effects on the swine intestinal microbiome. Proc Natl Acad Sci USA 109:1691–1696. 10.1073/pnas.1120238109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Modi SR, Collins JJ, Relman DA. 2014. Antibiotics and the gut microbiota. J Clin Invest 124:4212–4218. 10.1172/JCI72333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Linsell WD, Fletcher AP. 1950. Laboratory and clinical experience with terramycin hydrochloride. Br Med J ii:1190–1195. 10.1136/bmj.2.4690.1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Most H, Miller JW, Grossman EJ. 1950. Treatment of amebiasis with bacitracin and other antibiotics. Am J Trop Med Hyg 30:491–497. 10.4269/ajtmh.1950.s1-30.491. [DOI] [PubMed] [Google Scholar]
- 13.Sekirov I, Tam NM, Jogova M, Robertson ML, Li YL, Lupp C, Finlay BB. 2008. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect Immun 76:4726–4736. 10.1128/IAI.00319-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miki T, Goto R, Fujimoto M, Okada N, Hardt WD. 2017. The bactericidal lectin RegIIIbeta prolongs gut colonization and enteropathy in the streptomycin mouse model for Salmonella diarrhea. Cell Host Microbe 21:195–207. 10.1016/j.chom.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 15.Gillis CC, Hughes ER, Spiga L, Winter MG, Zhu W, Furtado de Carvalho T, Chanin RB, Behrendt CL, Hooper LV, Santos RL, Winter SE. 2018. Dysbiosis-associated change in host metabolism generates lactate to support Salmonella growth. Cell Host Microbe 23:54–64.e6. 10.1016/j.chom.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM, Sonnenburg JL. 2013. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 502:96–99. 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Theriot CM, Bowman AA, Young VB. 2016. Antibiotic-induced alterations of the gut microbiota alter secondary bile acid production and allow for Clostridium difficile spore germination and outgrowth in the large intestine. mSphere 1:e00045-15. 10.1128/mSphere.00045-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su A, Yu X. 2013. Analysis of livestock and poultry market and the use of antibiotics in egg poultry in China. China Anim Health 15:16–18. [Google Scholar]
- 19.Su A, Yu X. 2013. Analysis of livestock and poultry market and antibiotic use in broiler breeding in China. China Anim Health 15:20–22. [Google Scholar]
- 20.Ćwiek K, Korzekwa K, Tabiś A, Bania J, Bugla-Płoskońska G, Wieliczko A. 2020. Antimicrobial resistance and biofilm formation capacity of Salmonella enterica serovar Enteritidis strains isolated from poultry and humans in Poland. Pathogens 9:643. 10.3390/pathogens9080643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu Y, Lai H, Zou L, Yin S, Wang C, Han X, Xia X, Hu K, He L, Zhou K, Chen S, Ao X, Liu S. 2017. Antimicrobial resistance and resistance genes in Salmonella strains isolated from broiler chickens along the slaughtering process in China. Int J Food Microbiol 259:43–51. 10.1016/j.ijfoodmicro.2017.07.023. [DOI] [PubMed] [Google Scholar]
- 22.Alam SB, Mahmud M, Akter R, Hasan M, Sobur A, Nazir KNH, Noreddin A, Rahman T, El Zowalaty ME, Rahman M. 2020. Molecular detection of multidrug resistant Salmonella species isolated from broiler farm in Bangladesh. Pathogens 9:201. 10.3390/pathogens9030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gu D, Wang Z, Tian Y, Kang X, Meng C, Chen X, Pan Z, Jiao X. 2020. Prevalence of Salmonella isolates and their distribution based on whole-genome sequence in a chicken slaughterhouse in Jiangsu, China. Front Vet Sci 7:29. 10.3389/fvets.2020.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogawa J, Kishino S, Ando A, Sugimoto S, Mihara K, Shimizu S. 2005. Production of conjugated fatty acids by lactic acid bacteria. J Biosci Bioeng 100:355–364. 10.1263/jbb.100.355. [DOI] [PubMed] [Google Scholar]
- 25.Ma B, Mei X, Lei C, Li C, Gao Y, Kong L, Zhai X, Wang H. 2020. Enrofloxacin shifts intestinal microbiota and metabolic profiling and hinders recovery from Salmonella enterica subsp. enterica serovar Typhimurium infection in neonatal chickens. mSphere 5:e00725-20. 10.1128/mSphere.00725-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, Chaffron S, Macpherson AJ, Buer J, Parkhill J, Dougan G, von Mering C, Hardt WD. 2007. Salmonella enterica serovar Typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol 5:2177–2189. 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, Sterzenbach T, Tsolis RM, Roth JR, Baumler AJ. 2011. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci USA 108:17480–17485. 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, Roth JR, Baumler AJ. 2010. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467:426–429. 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, Neyrinck AM, Fava F, Tuohy KM, Chabo C, Waget A, Delmee E, Cousin B, Sulpice T, Chamontin B, Ferrieres J, Tanti JF, Gibson GR, Casteilla L, Delzenne NM, Alessi MC, Burcelin R. 2007. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 56:1761–1772. 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- 30.Hooper LV, Gordon JI. 2001. Commensal host-bacterial relationships in the gut. Science 292:1115–1118. 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 31.Saenz JS, Marques TV, Barone RSC, Cyrino JEP, Kublik S, Nesme J, Schloter M, Rath S, Vestergaard G. 2019. Oral administration of antibiotics increased the potential mobility of bacterial resistance genes in the gut of the fish Piaractus mesopotamicus. Microbiome 7:24. 10.1186/s40168-019-0632-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang G, Peng M, Tian X, Dong S. 2017. Molecular ecological network analysis reveals the effects of probiotics and florfenicol on intestinal microbiota homeostasis: an example of sea cucumber. Sci Rep 7:4778. 10.1038/s41598-017-05312-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson CJ, Bohannan BJ, Young VB. 2010. From structure to function: the ecology of host-associated microbial communities. Microbiol Mol Biol Rev 74:453–476. 10.1128/MMBR.00014-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Macfarlane S. 2014. Antibiotic treatments and microbes in the gut. Environ Microbiol 16:919–924. 10.1111/1462-2920.12399. [DOI] [PubMed] [Google Scholar]
- 35.Gao P, Ma C, Sun Z, Wang L, Huang S, Su X, Xu J, Zhang H. 2017. Feed-additive probiotics accelerate yet antibiotics delay intestinal microbiota maturation in broiler chicken. Microbiome 5:91. 10.1186/s40168-017-0315-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patterson JA, Burkholder KM. 2003. Application of prebiotics and probiotics in poultry production. Poult Sci 82:627–631. 10.1093/ps/82.4.627. [DOI] [PubMed] [Google Scholar]
- 37.Abudabos AM, Al-Batshan HA, Murshed MA. 2015. Effects of prebiotics and probiotics on the performance and bacterial colonization of broiler chickens. S Afr J Anim Sci 45:419–428. 10.4314/sajas.v45i4.8. [DOI] [Google Scholar]
- 38.Surendran Nair M, Amalaradjou MA, Venkitanarayanan K. 2017. Antivirulence properties of probiotics in combating microbial pathogenesis. Adv Appl Microbiol 98:1–29. 10.1016/bs.aambs.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 39.Collado MC, Grześkowiak Ł, Salminen S. 2007. Probiotic strains and their combination inhibit in vitro adhesion of pathogens to pig intestinal mucosa. Curr Microbiol 55:260–265. 10.1007/s00284-007-0144-8. [DOI] [PubMed] [Google Scholar]
- 40.Bernet MF, Brassart D, Neeser JR, Servin AL. 1994. Lactobacillus acidophilus LA 1 binds to cultured human intestinal cell lines and inhibits cell attachment and cell invasion by enterovirulent bacteria. Gut 35:483–489. 10.1136/gut.35.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang L, Zhang J, Guo Z, Kwok L, Ma C, Zhang W, Lv Q, Huang W, Zhang H. 2014. Effect of oral consumption of probiotic Lactobacillus planatarum [sic] P-8 on fecal microbiota, SIgA, SCFAs, and TBAs of adults of different ages. Nutrition 30:776–783.e1. 10.1016/j.nut.2013.11.018. [DOI] [PubMed] [Google Scholar]
- 42.Dong XY, Azzam MMM, Zou XT. 2017. Effects of dietary threonine supplementation on intestinal barrier function and gut microbiota of laying hens. Poult Sci 96:3654–3663. 10.3382/ps/pex185. [DOI] [PubMed] [Google Scholar]
- 43.Jacobson A, Lam L, Rajendram M, Tamburini F, Honeycutt J, Pham T, Van Treuren W, Pruss K, Stabler SR, Lugo K, Bouley DM, Vilches-Moure JG, Smith M, Sonnenburg JL, Bhatt AS, Huang KC, Monack D. 2018. A gut commensal-produced metabolite mediates colonization resistance to Salmonella infection. Cell Host Microbe 24:296–307.e7. 10.1016/j.chom.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dobrzanska DA, Lamaudiere MTF, Rollason J, Acton L, Duncan M, Compton S, Simms J, Weedall GD, Morozov IY. 2020. Preventive antibiotic treatment of calves: emergence of dysbiosis causing propagation of obese state-associated and mobile multidrug resistance-carrying bacteria. Microb Biotechnol 13:669–682. 10.1111/1751-7915.13496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saulnier DM, Riehle K, Mistretta TA, Diaz MA, Mandal D, Raza S, Weidler EM, Qin X, Coarfa C, Milosavljevic A, Petrosino JF, Highlander S, Gibbs R, Lynch SV, Shulman RJ, Versalovic J. 2011. Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology 141:1782–1791. 10.1053/j.gastro.2011.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen J, Chia N, Kalari KR, Yao JZ, Novotna M, Soldan MMP, Luckey DH, Marietta EV, Jeraldo PR, Chen XF, Weinshenker BG, Rodriguez M, Kantarci OH, Nelson H, Murray JA, Mangalam AK. 2016. Multiple sclerosis patients have a distinct gut microbiota compared to healthy controls. Sci Rep 6:28484. 10.1038/srep28484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Del Chierico F, Nobili V, Vernocchi P, Russo A, De Stefanis C, Gnani D, Furlanello C, Zandona A, Paci P, Capuani G, Dallapiccola B, Miccheli A, Alisi A, Putignani L. 2017. Gut microbiota profiling of pediatric nonalcoholic fatty liver disease and obese patients unveiled by an integrated meta-omics-based approach. Hepatology 65:451–464. 10.1002/hep.28572. [DOI] [PubMed] [Google Scholar]
- 48.Zhao X, Yang J, Ju Z, Wu J, Wang L, Lin H, Sun S. 2020. Clostridium butyricum ameliorates Salmonella enteritis induced inflammation by enhancing and improving immunity of the intestinal epithelial barrier at the intestinal mucosal level. Front Microbiol 11:299. 10.3389/fmicb.2020.00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mon KKZ, Zhu Y, Chanthavixay G, Kern C, Zhou H. 2020. Integrative analysis of gut microbiome and metabolites revealed novel mechanisms of intestinal Salmonella carriage in chicken. Sci Rep 10:4809. 10.1038/s41598-020-60892-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wijnands KA, Castermans TM, Hommen MP, Meesters DM, Poeze M. 2015. Arginine and citrulline and the immune response in sepsis. Nutrients 7:1426–1463. 10.3390/nu7031426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ghosh J. 2012. Role of nitric oxide in Salmonella infection. Ind J Clin Biochem 27:306–308. 10.1007/s12291-012-0187-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vazquez-Torres A, Jones-Carson J, Mastroeni P, Ischiropoulos H, Fang FC. 2000. Antimicrobial actions of the NADPH phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. I. Effects on microbial killing by activated peritoneal macrophages in vitro. J Exp Med 192:227–236. 10.1084/jem.192.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moghaddam MF, Grant DF, Cheek JM, Greene JF, Williamson KC, Hammock BD. 1997. Bioactivation of leukotoxins to their toxic diols by epoxide hydrolase. Nat Med 3:562–566. 10.1038/nm0597-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bannehr M, Lohr L, Gelep J, Haverkamp W, Schunck WH, Gollasch M, Wutzler A. 2019. Linoleic acid metabolite diHOME decreases post-ischemic cardiac recovery in murine hearts. Cardiovasc Toxicol 19:365–371. 10.1007/s12012-019-09508-x. [DOI] [PubMed] [Google Scholar]
- 55.Yu D, Hennebelle M, Sahlas DJ, Ramirez J, Gao F, Masellis M, Cogo-Moreira H, Swartz RH, Herrmann N, Chan PC, Pettersen JA, Stuss DT, Black SE, Taha AY, Swardfager W. 2019. Soluble epoxide hydrolase-derived linoleic acid oxylipins in serum are associated with periventricular white matter hyperintensities and vascular cognitive impairment. Transl Stroke Res 10:522–533. 10.1007/s12975-018-0672-5. [DOI] [PubMed] [Google Scholar]
- 56.Stanford KI, Lynes MD, Takahashi H, Baer LA, Arts PJ, May FJ, Lehnig AC, Middelbeek RJW, Richard JJ, So K, Chen EY, Gao F, Narain NR, Distefano G, Shettigar VK, Hirshman MF, Ziolo MT, Kiebish MA, Tseng Y-H, Coen PM, Goodyear LJ. 2018. 12,13-diHOME: an exercise-induced lipokine that increases skeletal muscle fatty acid uptake. Cell Metab 27:1111–1120.e3. 10.1016/j.cmet.2018.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Levan SR, Stamnes KA, Lin DL, Panzer AR, Fukui E, McCauley K, Fujimura KE, McKean M, Ownby DR, Zoratti EM, Boushey HA, Cabana MD, Johnson CC, Lynch SV. 2019. Elevated faecal 12,13-diHOME concentration in neonates at high risk for asthma is produced by gut bacteria and impedes immune tolerance. Nat Microbiol 4:1851–1861. 10.1038/s41564-019-0498-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Newman JW, Morisseau C, Hammock BD. 2005. Epoxide hydrolases: their roles and interactions with lipid metabolism. Prog Lipid Res 44:1–51. 10.1016/j.plipres.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 59.Wang Y, Yang J, Wang W, Sanidad KZ, Cinelli MA, Wan D, Hwang SH, Kim D, Lee KSS, Xiao H, Hammock BD, Zhang G. 2020. Soluble epoxide hydrolase is an endogenous regulator of obesity-induced intestinal barrier dysfunction and bacterial translocation. Proc Natl Acad Sci USA 117:8431–8436. 10.1073/pnas.1916189117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bruewer M, Luegering A, Kucharzik T, Parkos CA, Madara JL, Hopkins AM, Nusrat A. 2003. Proinflammatory cytokines disrupt epithelial barrier function by apoptosis-independent mechanisms. J Immunol 171:6164–6172. 10.4049/jimmunol.171.11.6164. [DOI] [PubMed] [Google Scholar]
- 61.Turner JR. 2009. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol 9:799–809. 10.1038/nri2653. [DOI] [PubMed] [Google Scholar]
- 62.Slim R, Hammock BD, Toborek M, Robertson LW, Newman JW, Morisseau CH, Watkins BA, Saraswathi V, Hennig B. 2001. The role of methyl-linoleic acid epoxide and diol metabolites in the amplified toxicity of linoleic acid and polychlorinated biphenyls to vascular endothelial cells. Toxicol Appl Pharmacol 171:184–193. 10.1006/taap.2001.9131. [DOI] [PubMed] [Google Scholar]
- 63.Zimmer B, Angioni C, Osthues T, Toewe A, Thomas D, Pierre SC, Geisslinger G, Scholich K, Sisignano M. 2018. The oxidized linoleic acid metabolite 12,13-diHOME mediates thermal hyperalgesia during inflammatory pain. Biochim Biophys Acta 1863:669–678. 10.1016/j.bbalip.2018.03.012. [DOI] [PubMed] [Google Scholar]
- 64.Moran JH, Mon T, Hendrickson TL, Mitchell LA, Grant DF. 2001. Defining mechanisms of toxicity for linoleic acid monoepoxides and diols in Sf-21 cells. Chem Res Toxicol 14:431–437. 10.1021/tx000200o. [DOI] [PubMed] [Google Scholar]
- 65.Sisemore MF, Zheng J, Yang JC, Thompson DA, Plopper CG, Cortopassi GA, Hammock BD. 2001. Cellular characterization of leukotoxin diol-induced mitochondrial dysfunction. Arch Biochem Biophys 392:32–37. 10.1006/abbi.2001.2434. [DOI] [PubMed] [Google Scholar]
- 66.Byeon JI, Song HS, Oh TW, Kim YS, Choi BD, Kim HC, Kim JO, Shim KH, Ha YL. 2009. Growth inhibition of foodborne and pathogenic bacteria by conjugated linoleic acid. J Agric Food Chem 57:3164–3172. 10.1021/jf8031167. [DOI] [PubMed] [Google Scholar]
- 67.de Barros PAV, Rabelo Andrade ME, de Vasconcelos Generoso S, Mendes Miranda SE, Dos Reis DC, Lacerda Leocadio PC, de Sales ESEL, Dos Santos Martins F, da Gama MAS, Cassali GD, Alvarez Leite JI, Antunes Fernandes SO, Cardoso VN. 2018. Conjugated linoleic acid prevents damage caused by intestinal mucositis induced by 5-fluorouracil in an experimental model. Biomed Pharmacother 103:1567–1576. 10.1016/j.biopha.2018.04.133. [DOI] [PubMed] [Google Scholar]
- 68.Peng M, Tabashsum Z, Patel P, Bernhardt C, Biswas D. 2018. Linoleic acids overproducing Lactobacillus casei limits growth, survival, and virulence of Salmonella Typhimurium and enterohaemorrhagic Escherichia coli. Front Microbiol 9:2663. 10.3389/fmicb.2018.02663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tabashsum Z, Peng M, Bernhardt C, Patel P, Carrion M, Rahaman SO, Biswas D. 2020. Limiting the pathogenesis of Salmonella Typhimurium with berry phenolic extracts and linoleic acid overproducing Lactobacillus casei. J Microbiol 58:489–498. 10.1007/s12275-020-9545-1. [DOI] [PubMed] [Google Scholar]
- 70.Philippe N, Alcaraz JP, Coursange E, Geiselmann J, Schneider D. 2004. Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid 51:246–255. 10.1016/j.plasmid.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 71.Evans CC, LePard KJ, Kwak JW, Stancukas MC, Laskowski S, Dougherty J, Moulton L, Glawe A, Wang Y, Leone V, Antonopoulos DA, Smith D, Chang EB, Ciancio MJ. 2014. Exercise prevents weight gain and alters the gut microbiota in a mouse model of high fat diet-induced obesity. PLoS One 9:e92193. 10.1371/journal.pone.0092193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. 2006. Metagenomic analysis of the human distal gut microbiome. Science 312:1355–1359. 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen H, Jiang W. 2014. Application of high-throughput sequencing in understanding human oral microbiome related with health and disease. Front Microbiol 5:508. 10.3389/fmicb.2014.00508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Magoc T, Salzberg SL. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 76.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huson DH, Mitra S, Ruscheweyh HJ, Weber N, Schuster SC. 2011. Integrative analysis of environmental sequences using MEGAN4. Genome Res 21:1552–1560. 10.1101/gr.120618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Asnicar F, Weingart G, Tickle TL, Huttenhower C, Segata N. 2015. Compact graphical representation of phylogenetic data and metadata with GraPhlAn. PeerJ 3:e1029. 10.7717/peerj.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. 2011. Metagenomic biomarker discovery and explanation. Genome Biol 12:R60. 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bartosch S, Fite A, Macfarlane GT, McMurdo MET. 2004. Characterization of bacterial communities in feces from healthy elderly volunteers and hospitalized elderly patients by using real-time PCR and effects of antibiotic treatment on the fecal microbiota. Appl Environ Microbiol 70:3575–3581. 10.1128/AEM.70.6.3575-3581.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Matsuda K, Tsuji H, Asahara T, Kado Y, Nomoto K. 2007. Sensitive quantitative detection of commensal bacteria by rRNA-targeted reverse transcription-PCR. Appl Environ Microbiol 73:32–39. 10.1128/AEM.01224-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bergstrom A, Licht TR, Wilcks A, Andersen JB, Schmidt LR, Gronlund HA, Vigsnaes LK, Michaelsen KF, Bahl MI. 2012. Introducing GUt low-density array (GULDA): a validated approach for qPCR-based intestinal microbial community analysis. FEMS Microbiol Lett 337:38–47. 10.1111/1574-6968.12004. [DOI] [PubMed] [Google Scholar]
- 85.Wang P, Xu J, Wang Y, Cao X. 2017. An interferon-independent lncRNA promotes viral replication by modulating cellular metabolism. Science 358:1051–1055. 10.1126/science.aao0409. [DOI] [PubMed] [Google Scholar]
- 86.Xia J, Sinelnikov IV, Han B, Wishart DS. 2015. MetaboAnalyst 3.0—making metabolomics more meaningful. Nucleic Acids Res 43:W251–W257. 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.de Souza M, Baptista AAS, Valdiviezo MJJ, Justino L, Menck-Costa MF, Ferraz CR, da Gloria EM, Verri WA, Jr, Bracarense APFRL. 2020. Lactobacillus spp. reduces morphological changes and oxidative stress induced by deoxynivalenol on the intestine and liver of broilers. Toxicon 185:203–212. 10.1016/j.toxicon.2020.07.002. [DOI] [PubMed] [Google Scholar]
- 88.Xu S, Wang D, Zhang P, Lin Y, Fang Z, Che L, Wu D. 2015. Oral administration of Lactococcus lactis-expressed recombinant porcine epidermal growth factor stimulates the development and promotes the health of small intestines in early-weaned piglets. J Appl Microbiol 119:225–235. 10.1111/jam.12833. [DOI] [PubMed] [Google Scholar]
- 89.Li HX, Liu XL, Chen FY, Zuo KJ, Wu C, Yan YM, Chen WG, Lin WC, Xie QM. 2018. Avian influenza virus subtype H9N2 affects intestinal microbiota, barrier structure injury, and inflammatory intestinal disease in the chicken ileum. Viruses (Basel) 10:270. 10.3390/v10050270. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S6, Tables S1 and S2. Download aem.01681-21-s0001.pdf, PDF file, 1.0 MB (1,006.3KB, pdf)