Abstract
The E7 protein encoded by human papillomavirus type 16 is one of the few viral genes that can immortalize primary human cells and thereby override cellular senescence. While it is generally assumed that this property of E7 depends on its interaction with regulators of the cell cycle, we show here that E7 targets insulin-like growth factor binding protein 3 (IGFBP-3), the product of a p53-inducible gene that is overexpressed in senescent cells. IGFBP-3 can suppress cell proliferation and induce apoptosis; we show here that IGFBP-3-mediated apoptosis is inhibited by E7, which binds to IGFBP-3 and triggers its proteolytic cleavage. Two transformation-deficient mutants of E7 failed to inactivate IGFBP-3, suggesting that inactivation of IGFBP-3 may contribute to cell transformation.
Human papillomaviruses (HPVs) of the high-risk group (e.g., HPV-16) cause cancers in humans, while papillomaviruses of the low-risk group (e.g., HPV-11) cause benign epithelial hyperproliferation (90). Cell transformation by high-risk HPVs requires expression of the viral genes E6 and E7 (for a review, see reference 1). Coexpression of HPV-16 E6 and E7 is sufficient to immortalize primary human keratinocytes (35, 57), the natural host cells for papillomavirus infection. The E6 protein of HPV-16 interacts with the p53 tumor suppressor, which leads to recruitment of the ubiquitin ligase E6AP (39), resulting in the ubiquitination and subsequent degradation of p53 (72). Consequently, p53-dependent upregulation of growth-inhibitory genes, such as p21WAF-1 (27), is abrogated. A major target for the E7 oncoprotein of HPV-16 appears to be the p16/Rb pathway, as it is known that E7 binds to all three members of the retinoblastoma protein family and abrogates their growth-suppressive function (for a review, see reference 83); consequently, E7-expressing cells are refractory to growth inhibition by the cyclin-dependent kinase (cdk) inhibitor p16INK4 (49, 50). The identification of specific target proteins for E6 and E7 suggests that both viral oncoproteins target nonidentical regulatory pathways and that immortalization depends on the combined action of both gene products. However, it is known that expression of E7 alone is sufficient to immortalize human cells (85), albeit at reduced efficiency, compared to the simultaneous expression of both E6 and E7 (35). This indicates that E7 may also impinge on growth regulatory pathways that are principal targets for E6. In support of this hypothesis, it was shown that E7 binds and inactivates the cdk inhibitor p21WAF-1 (28, 42), which is encoded by a p53-inducible gene (27). This observation provides an explanation for how p53-mediated growth arrest can be undermined by E7 in the absence of E6, i.e., in cells where p53 is present and functional.
Immortalization of mammalian cells is considered the first step in tumorigenesis (88) which abrogates a cellular senescence program that is characterized by irreversible cell cycle exit after extended passaging. There is evidence that mitogenic signal transduction is disturbed in senescent fibroblasts. Thus, expression of early-growth response genes, e.g., the c-fos gene, cannot be induced by serum growth factors in senescent cells (73). Furthermore, insulin-like growth factor binding protein 3 (IGFBP-3), a member of a protein family that regulates the mitogenic activity of IGF-I (for a review, see reference 11), is strongly overexpressed in senescent cells (30, 31, 55). IGFBP-3 can block the proliferation of various cell types in vitro (for a review, see reference 59) by at least two distinct ways. As mentioned above, IGFBP-3 binds IGF-I and thereby regulates IGF-I dependent signaling. Second, there is evidence that mutants of IGFBP-3 that fail to interact with IGF-I are still able to induce apoptosis in PC-3 cells (67). It is assumed that this second IGF-I-independent function of IGFBP-3, which is also effective in cells lacking the IGF receptor (82), involves the uptake of extracellular IGFBP-3 through a cellular IGFBP-3 receptor (45) and subsequent localization in the nucleus (48, 71, 87); however, nuclear targets for IGFBP-3 have not been described so far. The IGFBP-3 gene is transcriptionally activated by p53 through a p53 binding site (15), and it is believed that increased expression of IGFBP-3 contributes to p53-dependent apoptosis (67; for a review, see reference 16).
Genetic evidence suggests that multiple genetic loci, present in four distinct complementation groups, contribute to cellular senescence (64). Expression of the cdk inhibitors p16INK4 (2, 34) and p21WAF-1 (2, 78) is considerably increased in senescent cells, and it is assumed that the p16/pRb and the ARF/p53/p21 pathways play key roles in establishing cellular senescence (for recent review, see references 74 and 76). In addition, a telomere maintenance mechanism, in most cases achieved through reactivation of telomerase (hTERT) gene expression (18), is required for immortalization (for a review, see reference 22). Hence, several growth-suppressing pathways must be independently subverted in all immortalized cells. While it is possible that a high frequency of random genetic alterations triggers the immortalization of human cells that express E7 as the sole viral oncogene, we reasoned that, in addition to pRb and p21, E7 may target additional factors in senescence-inducing pathways that are currently not recognized as E7 targets. Applying a two-hybrid screen with E7 as bait, we identified the human IGFBP-3 as a new interaction partner for E7. We also present evidence here that E7 suppresses a major function of IGFBP-3, namely, its ability to induce apoptosis.
MATERIALS AND METHODS
Interaction analysis in yeast.
The C-terminal part of HPV-16 E7, fused in frame to the DNA-binding domain of LexA, was used as bait in the yeast two-hybrid system (92) to identify cDNAs for E7-binding proteins from a human cDNA expression library (6). Yeast strain EGY48/pSH1834 (MATα his3 ura3 trp1 leu2::lexAo6-pLEU2/LexAo8-GAL1-lacZ::URA3) (92) was used for both the LEU2 and β-galactosidase reporter gene assays. For determination of reporter gene activity, EGY48/pSH1834 was transformed with plasmids expressing the LexA-E7(39-98), LexA-16E7, LexA-11E7, LexA-Myc, or LexA-Bicoid fusion protein, together with the plasmid pB42-IGFBP3::TRP1, and it was selected for leucine prototrophy as previously described (6); alternatively, β-galactosidase activity was determined in cellular extracts as previously described (6). The various LexA fusion proteins were expressed to the same level, as confirmed by direct immunoblotting, using a polyclonal antibody to LexA (92).
In vitro interaction analysis.
Different quantities of recombinant glutathione S-transferase (GST), GST-E7 (93), and IGF-I (a gift from Pharmacia, Uppsala, Sweden) proteins were immobilized on nitrocellulose filters and probed with radiolabeled IGFBP-3, as previously described (37). Subsequently, the membranes were washed and exposed for autoradiography. For the determination of binding constants, the amount of filter-bound IGFBP-3 was calculated by comparison with a control curve.
For GST pull-down assays, GST and GST fusion proteins containing various mutants of E7 (93) were loaded (each at 10 ng/μl) on glutathione-Sepharose 4B beads. IGFBP-3 protein was produced in yeast cells or transiently transfected U-2OS cells; extracts were incubated with GST-E7 fusion proteins, and bound proteins were analyzed by Western blotting (89), using anti-IGFBP-3 antibodies (Diagnostic Systems Laboratory Inc., Webster, Tex.) or anti-HA1 antibodies (94). Input of the various GST-derived fusion proteins was controlled either by Western blot analysis using a monoclonal antibody against GST (clone GST-2; Sigma Chemical Co., St. Louis, Mo.) or by Coomassie staining.
Cell culture and transfection.
The human osteosarcoma cell line U-2OS was cultured in Dulbecco modified Eagle medium plus 10% fetal calf serum (FCS) (93). The human PC-3 cells were originally initiated from a grade IV prostatic adenocarcinoma from a 62-year-old male Caucasian. PC-3 cells were cultured in RPMI or FK-12 medium, both supplemented with 10% FCS (67). Primary human keratinocytes were isolated from human newborn foreskin and were immortalized by infection with the pLXSN retroviral vector expressing HPV-16 E7, as previously described (12). Primary keratinocytes and E7-immortalized keratinocytes were cultured in keratinocyte growth medium (Promocell, Heidelberg, Germany). Transfection efficiency was routinely controlled by cotransfecting a cytomegalovirus-driven expression vector for β-galactosidase. Subsequent to transfection, we determined β-galactosidase activity, which varied less than 10% between individual experiments.
Apoptosis assays.
The human IGFBP-3 cDNA (a gift of A. Groyer, Paris, France) was subcloned in the cytomegalovirus-driven expression vector pX (89) to yield pX-IGFBP3. PC-3 cells were transfected with pX-IGFBP-3, pX-16E7, and green fluorescent protein (Clontech, Heidelberg, Germany) as a marker of cells successfully transfected using the FuGENE6 reagent (Roche Diagnostics, Mannheim, Germany); transfection with the empty pX vector was included as a control. For apoptosis detection, cells were harvested 24 h after transfection, fixed in ethanol, and incubated with a buffer containing 40 μg of RNase A/ml and 50 μg of propidium iodide/ml for 30 min on ice. Fluorescence was measured in a flow cytometer, and green-fluorescent protein-transfected cells with sub-G1 DNA content were considered to be apoptotic. The abundance of transfected proteins was determined by Western blotting, using a polyclonal goat antiserum to IGFBP-3 and a monoclonal antibody to HPV-16 E7 (Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.).
Analysis of IGFBP-3 in the cell culture medium.
For the analysis of IGFBP-3 in the cell culture medium, conditioned serum-free medium was treated with an equal volume of 20% cold trichloroacetic acid and incubated on ice for 10 min. After centrifugation, precipitated proteins were neutralized using 1 M Tris solution and subjected to gel electrophoresis and Western blotting.
Northern blot analysis.
For Northern blot analysis, total RNA was prepared, separated on a 1.2% formaldehyde agarose gel, and probed with a 32P-labeled probe derived from human IGFBP-3 cDNA as previously described (89).
Metabolic labeling experiments.
U-2OS cells were transfected by pX-IGFBP3 and pX-16E7 using the FuGene 6 reagent and were pulse-labeled with [35S]methionine plus [35S]cysteine (Amersham Pharmacia Biotech) for 20 min, followed by a chase period of variable length. IGFBP-3 was immunoprecipitated from cellular extracts by IGFBP-3 antisera and quantitated by autoradiography. To inhibit the proteolytic activity of the proteasome, N-acetyl-Leu-Leu-norleucinal (LLnL; Sigma Chemical Co.) was added to the cells at a concentration of 100 μM.
Coimmunoprecipitation experiments.
U-2OS cells were cotransfected with pX-IGFBP-3 and pJ4ΩHPV16E7 (53), using the Effectene transfection kit (Qiagen, Hilden, Germany). The proteasome activity was blocked by addition of LLnL to a final concentration of 100 μM. Extracts from 106 cells were prepared in lysis buffer (1.5 mM KH2PO4, 8.1 mM Na2HPO4 [pH 7.4], 2.7 mM KCl, 137 mM NaCl, 0.2 mM phenylmethylsulfonyl fluoride [PMSF], 1 mM NaF, 100 μM LLnL, 1 mM dithiothreitol [DTT], 0.1% Triton X-100, protease inhibitor mix no. 1836170 [Roche Diagnostics]) and subjected to immunoprecipitation with antibodies raised against HPV-16 E7 (clone D6, a gift from R. Tindle, Brisbane, Australia), with an antiserum against IGFBP-3 (Diagnostic Systems Laboratory, Inc.), or with a mixture of a monoclonal anti-GST antibody (clone GST-2; Sigma Chemical Co.) and a polyclonal antiserum against cdk4 (Santa Cruz Biotechnology Inc.). Beads were washed three times in wash buffer 1 (20 mM Tris-Cl [pH 7.5], 75 mM NaCl, 0.2 mM PMSF, 1 mM NaF, 1 mM DTT, 0.1% NP-40, 10% glycerol, protease inhibitor mix no. 1836170 [Roche Diagnostics]) and once in wash buffer 2 (10 mM Tris-Cl [pH 7.5], 0.2 mM PMSF, 1 mM NaF, 1 mM DTT, 0.1% NP-40, 10% glycerol, protease inhibitor mix no. 1836170 [Roche Diagnostics]). Precipitated proteins were visualized by Western blotting.
Indirect immunofluorescence analysis.
U-2OS cells were cultured in Dulbecco modified Eagle medium plus 10% FCS. For transient expression of cDNAs, cells were grown to about 80% confluence on glass coverslips coated with 0.04% gelatin. Transfection of the expression vectors pX-IGFBP-3 and pJ4ΩHPV16-E7 (53) was performed using the Effectene transfection kit (Qiagen). At 36 h posttransfection, cells were prepared for indirect immunofluorescence according to standard protocols, including methanol fixation. After incubation with primary antibodies (polyclonal rabbit anti-HPV16-E7 antibodies [W. Zwerschke, unpublished results] and polyclonal goat anti-IGFBP-3 antibodies [Diagnostic Systems Laboratory Inc.], respectively) and secondary antibodies tetramethyl rhodamine isocyanate (TRITC)-conjugated anti-goat immunoglobulin (IgG) and fluorescein isothiocyanate (FITC)-conjugated anti-rabbit IgG (both from Dianova, Hamburg, Germany), cells were washed and embedded in Fluoromount G (Biozol, Eching, Germany). Samples were viewed by indirect immunofluorescence microscopy using the confocal scanning system MicroRadiance (Bio-Rad, Hemel Hempstead, Hertfordshire, Great Britain) in combination with a Zeiss Axiophot microscope. The following filters were used for FITC-derived and TRITC-derived fluorescence: excitation for FITC at 488 nm and for TRITC at 543 nm and emission for FITC at 515 to 530 nm and for TRITC at >570 nm.
RESULTS
Identification of IGFBP-3 as E7 binding protein.
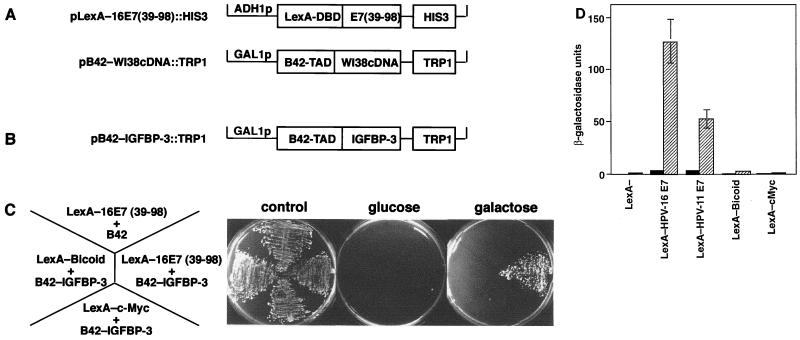
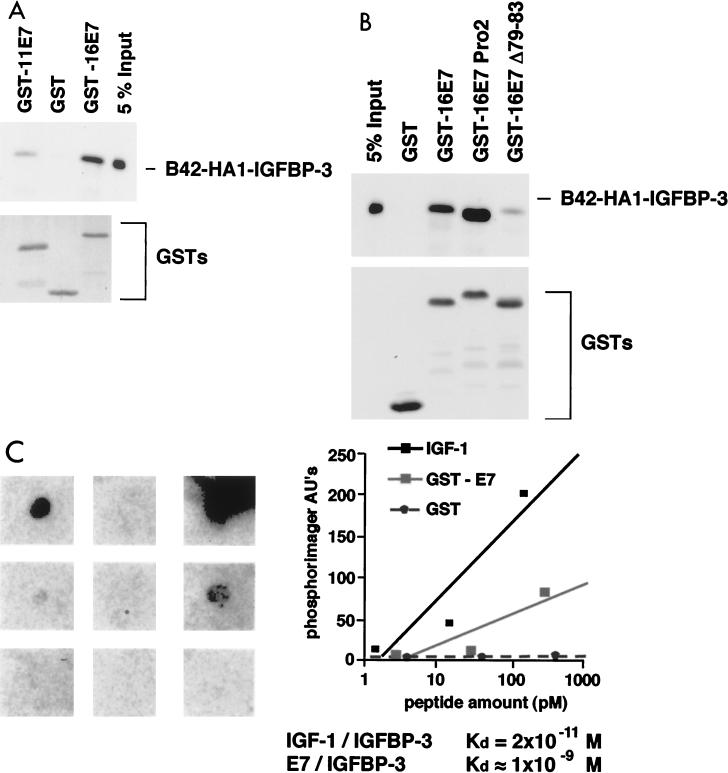
To identify regulatory pathways relevant for immortalization by HPV-16 E7, we screened a human cDNA library (32) for cellular E7-binding proteins. In a yeast two-hybrid screen (Fig. 1A), we repeatedly isolated the cDNA encoding IGFBP-3 (Fig. 1B). Specific interaction of IGFBP-3 with E7, but not with two unrelated fusion proteins, was confirmed by coexpression in yeast by using two distinct reporter gene assays (Fig. 1C and D). The ability of the E7 protein of HPV-11, a low-risk HPV type, to interact with IGFBP-3 was significantly reduced, suggesting that the ability of E7 proteins to bind IGFBP-3 may correlate with the oncogenic potential of the virus (Fig. 1D and 2A). To determine if HPV-16 E7 and IGFBP-3 can also interact in vitro, GST pull-down experiments were performed. As is shown in Fig. 2A, IGFBP-3 fusion protein isolated from yeast cells showed significant interaction with GST-16E7 fusion proteins but not GST alone. The binding affinity of HPV-11 E7 to IGFBP-3 was significantly reduced, in accordance with the results shown in Fig. 1D. Similarly, a mutated version of the HPV-16 E7 protein, Δ79-83 (53), in which four amino acids from the C-terminal domain were deleted, showed strongly reduced binding affinity for IGFBP-3 (Fig. 2B). To further characterize the interaction of IGFBP-3 with HPV-16 E7, a ligand-binding blotting (37) was performed using 125I-labeled IGFBP-3. GST-tagged HPV-16 E7 protein was immobilized on nitrocellulose filters and probed with radiolabeled IGFBP-3. Subsequently, the membranes were washed and exposed for autoradiography. In this experiment, a binding constant of 10−9 M was determined (Fig. 2C), suggesting that both proteins strongly interact in vitro.
FIG. 1.
Identification of IGFBP-3 as E7-binding protein. (A) Plasmid pLexA-16E7(39-98)::HIS3 encodes the DNA binding domain of LexA fused in frame to the C-terminal part of HPV-16 E7 and the HIS3 selectable marker. The structure of the expression library, derived from vector pJG4-5 (32) by insertion of cDNA fragments from a WI-38 cDNA library, is indicated. B42-TAD refers to the B42 transactivation domain. B42 fusion proteins are expressed from the inducible GAL1 promoter, and these plasmids contain the TRP1 gene as a selectable marker. (B) The plasmid designated pB42-IGFBP-3::TRP1 was isolated during the interaction screen; it contains the full-length cDNA for human IGFBP-3 (86). (C) Derivatives of yeast strain EGY48/pSH1834, expressing various LexA fusion proteins as indicated, were transformed with the plasmid pB42-IGFBP-3::TRP1. pJG4-5, expressing the unfused B42 transactivation domain, was used as a negative control. Transformants were selected for uracil, histidine, and tryptophan prototrophy and were grown in glucose minimal medium (Ura−, His−, Trp−). Yeast cells were then streaked out onto each of three plates and incubated for 4 days at 30°C under the following nutrient conditions: control, glucose minimal medium with leucine, all strains grew; glucose, glucose minimal medium without leucine, selection for B42 fusion protein-independent activation of the LexAo6-pLEU2 reporter; galactose, minimal medium without leucine, selection for B42 fusion protein-dependent activation of the LexAo6-pLEU2 gene. (D) Saccharomyces cerevisiae strain EGY48/pSH1834, containing a LexA operator-lacZ gene (LexAo8-GAL1-LacZ::URA3), was transformed with the plasmid pB42-IGFBP-3::TRP1 expressing IGFBP-3 (striped bars) or pJG4-5 expressing the unfused B42 transactivation domain (solid bars). Plasmids encoding LexA fusion proteins were coexpressed, as indicated, and β-galactosidase activity was determined. LexA-Bicoid and LexA-C-Myc (C terminus) were used as negative controls.
FIG. 2.
E7 binds IGFBP-3 in vitro. (A and B) Purified GST or GST-E7 fusion proteins immobilized on glutathione-Sepharose 4B beads were incubated with whole-cell extracts from yeast cells expressing a B42-HA1-IGFBP-3 fusion protein, as indicated. (Top) After washing, bound proteins were separated by gel electrophoresis and the amount of B42-HA1-IGFBP-3 protein that was retained was determined by direct immunoblotting using a monoclonal antibody against the HA1 epitope (94). For comparison, 5% of the input was also loaded on the gel. (Bottom) Input of the various GST fusion proteins was controlled by either Coomassie staining or Western blotting using anti-GST antibodies. (C) IGF-I and E7 proteins were immobilized on nitrocellulose filters as indicated and were probed with 125I-labeled IGFBP-3. The relative binding constants for both interactions are indicated. AU, arbitrary units.
Abrogation of IGFBP-3 function by E7.
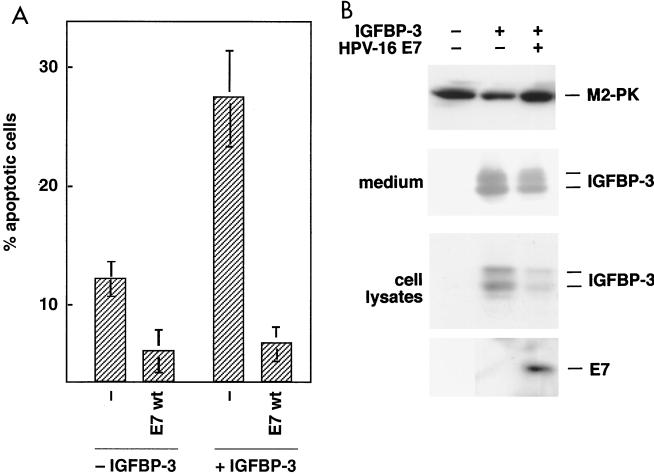
It was shown that IGFBP-3 induces apoptosis in several cell types (29, 67), and our finding that HPV-16 E7 binds to IGFBP-3 raises the possibility that the ability of E7 to modulate apoptotic cell death (80) depends on its interaction with IGFBP-3. To determine if IGFBP-3 is a functional target for E7, a transient-transfection assay which allows the monitoring of the proapoptotic activity of IGFBP-3 was established. After transfection of PC-3 prostate carcinoma cells with an expression vector for IGFBP-3, we found that expression of IGFBP-3 increased the frequency of apoptosis to 28.4% ± 3.6% (Fig. 3A). Coexpression of HPV-16 E7 reduced the amount of apoptotic cells to 6.8% ± 1.2%, indicating that E7 can prevent IGFBP-3-mediated apoptosis.
FIG. 3.
Effects of IGFBP-3 and E7 on apoptosis rate in PC-3 cells. PC-3 cells were transfected with expression vectors for IGFBP-3 and/or E7, as indicated. (A) The rate of apoptosis was determined by flow cytometry. Cells with sub-G1 DNA content were considered to be apoptotic. Values are the means ± standard deviation of three independent experiments (P < 0.005). wt, wild type. (B) The levels of IGFBP-3 in both cellular lysates and conditioned medium and levels of E7 proteins were determined by immunoblotting. M2 pruvate kinase (M2-PK) served as a loading control.
E7 triggers proteolytic degradation of IGFBP-3.
To address the mechanism underlying the functional antagonism between E7 and IGFBP-3, we studied expression of both proteins in transfected PC-3 cells by Western blotting. Since IGFBP-3 can be found both in intracellular compartments and in tissue culture supernatants (30, 48, 71, 87), both cellular extracts and the supernatant were analyzed by Western blotting. We found that IGFBP-3 is well expressed from the respective expression vector in PC-3 cells; however, the abundance of IGFBP-3 protein was considerably reduced when E7 was coexpressed in the same experiment (Fig. 3B). The E7-induced loss of intracellular IGFBP-3 cannot be explained by changes in the secretion of the protein, as the accumulation of extracellular IGFBP-3 was also drastically reduced in the supernatants of E7-expressing cells. The level of M2 pruvate kinase, another E7-binding protein (94) which was analyzed as a control, was not affected by E7 gene expression (Fig. 3B).
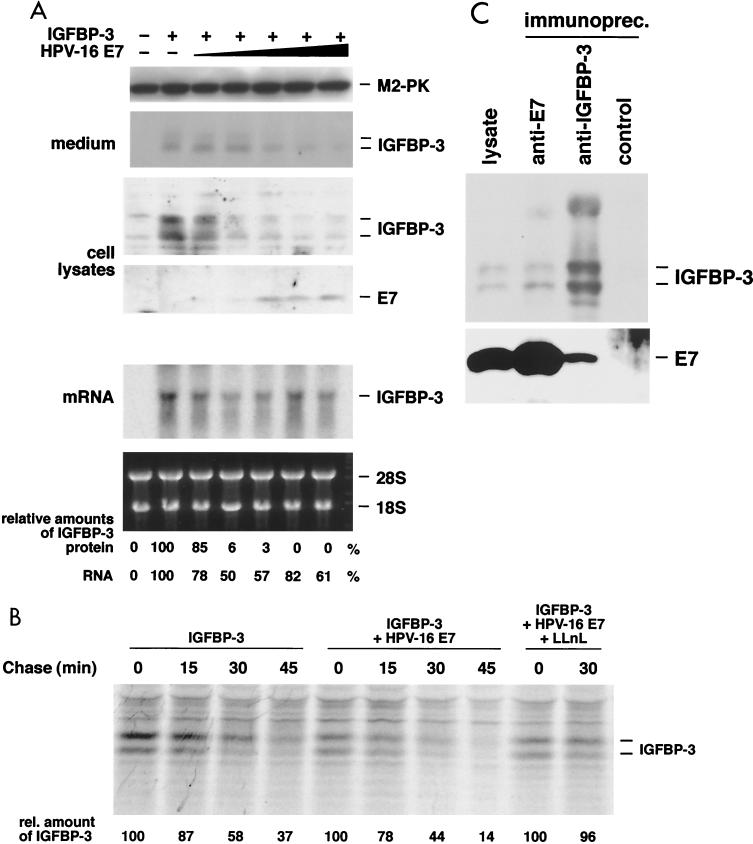
Additional experiments were performed in the human osteosarcoma cell line U-2OS, which displays an intrinsic high transfection efficiency. When U-2OS cells were transfected with expression vectors for E7 and IGFBP-3, both proteins were well expressed. As in the case of the PC-3 cells, coexpression of E7 drastically (more than 100-fold) reduced the expression level of IGFBP-3 in U-2OS cells (Fig. 4A). Again, the E7-induced loss of intracellular IGFBP-3 cannot be explained by changes in the secretion of the protein, as the accumulation of extracellular IGFBP-3 was also drastically reduced in the supernatants of E7-expressing U-2OS cells. We found that E7 only moderately (1.6-fold) reduces the abundance of the IGFBP-3 mRNA in transfected cells (Fig. 4A), suggesting that E7 controls expression of IGFBP-3 at a posttranscriptional level.
FIG. 4.
E7 triggers proteasome-dependent degradation of IGFBP-3. (A) U-2OS cells were transfected with expression vectors for IGFBP-3 and E7 as indicated. The levels of IGFBP-3 in both cellular lysates and conditioned medium and levels of E7 proteins were determined by Western blotting (four top panels), in which M2 pyruvate kinase (M2-PK) served as loading control; the IGFBP-3 mRNA level was determined by Northern blotting (fifth panel). Ethidium bromide staining of the 28S and 18S rRNA served as loading control (bottom panel). Levels of IGFBP-3 mRNA and protein were quantitated by densitometric scanning of the autoradiogram. (B) U-2OS cells were transfected with expression vectors for IGFBP-3 and E7 as indicated and were 35S labeled for 20 min, followed by a chase period, as indicated. IGFBP-3 was immunoprecipitated and detected by autoradiography. The half-life of IGFBP-3 was determined from the average of three independent experiments, as follows: 35 min in the absence of E7, 17 min in the presence of E7, and 44 min in the presence of both E7 and the proteasome inhibitor LLnL. (C) Coimmunoprecipitation experiments. U-2OS cells were cotransfected with expression vectors for IGFBP-3 and E7; proteasome activity was blocked by addition of LLnL. Cell lysates were prepared and subjected to immunoprecipitation with antibodies to E7 and IGFBP-3 and control antibodies, as indicated. Precipitated proteins were detected by Western blotting.
To investigate this further, pulse-chase experiments with 35S-labeled amino acids were performed with U-2OS cells transfected with expression vectors for IGFBP-3 and E7. IGFBP-3 was immunoprecipitated from cellular extracts by an IGFBP-3 antiserum and quantitated by autoradiography (Fig. 4B). This experiment reveals that the rate of synthesis of IGFBP-3 is quite similar in control cells and E7-transfected cells; in contrast, the stability of the protein was reduced in E7-expressing cells. These results suggest that the proteolytic degradation of IGFBP-3 is significantly enhanced by E7. To determine whether E7-induced proteolysis of IGFBP-3 involves the proteasome, the influence of E7 on the stability of IGFBP-3 was determined in the presence of 100 μM LLnL, a potent proteasome inhibitor (68). We found that the stability of the IGFBP-3 protein was significantly increased by the proteasome inhibitor (Fig. 4B). Together, these data suggest that upon expression of E7, degradation of IGFBP-3 by the proteasome is initiated, leading to much lower levels of this growth-inhibiting and apoptosis-inducing protein.
E7 and IGFBP-3 interact in mammalian cells.
The results shown in Fig. 1 and 2 demonstrate that E7 and IGFBP-3 strongly interact both in vitro and in yeast cells. In mammalian cells, both IGFBP-3 and E7 are found in both nuclear and nonnuclear sites (44, 48, 71, 75, 87). Since in mammalian cells the binding of E7 to IGFBP-3 results in the proteolytic degradation of the latter (Fig. 3B and 4A and B), a prominent durable physical interaction between both proteins cannot be expected. To address the question of whether E7 can bind to IGFBP-3 in mammalian cells, U-2OS cells were cotransfected with expression vectors for IGFBP-3 and E7. Proteasome function was blocked by the addition of LLnL to transfected cells; subsequently, coimmunoprecipitation experiments were carried out. Under these conditions, we were able to coprecipitate IGFBP-3 with a monoclonal E7 antibody and to coprecipitate E7 with a polyclonal IGFBP-3 antiserum (Fig. 4C). A mixture of (polyclonal rabbit) anti-cdk4 antisera and (monoclonal mouse) anti-GST antibodies did not precipitate any proteins in this experiment (Fig. 4C). Furthermore, coimmunoprecipitation of both E7 and IGFBP-3 was strictly dependent on the coexpression of both proteins (data not shown). These results confirm that an interaction between E7 and IGFBP-3 can indeed be detected within mammalian cells when proteasome function is blocked.
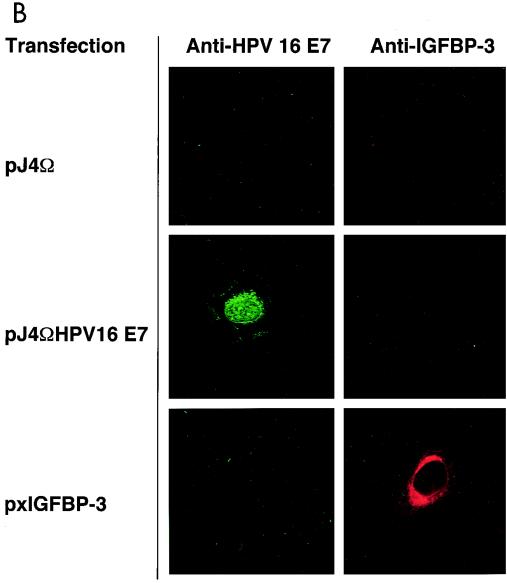
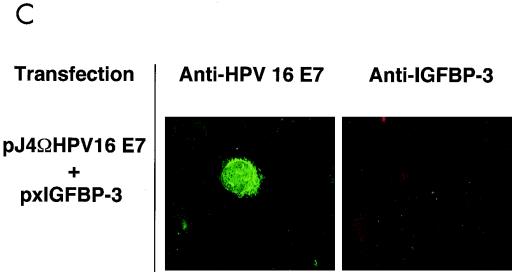
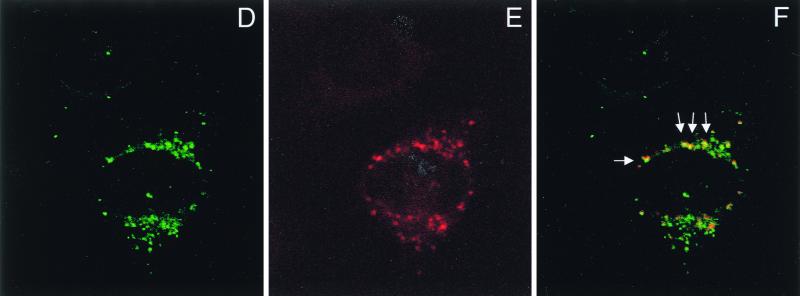
To further study this interaction, we set out to determine the interaction within living cells by indirect immunofluorescence analysis. We used a system in which the level of each of the proteins can be varied individually during the experiment. Furthermore, we used a newly generated polyclonal anti-E7 antiserum for these experiments. As shown by Western blot analysis, this antiserum, which was raised against highly purified recombinant E7 protein (M. Fiedler et al., unpublished results), recognizes a single band in extracts from E7-expressing cells but not control cells, while the preimmune serum is negative (data not shown). U-2OS cells were transiently transfected with expression vectors for E7 and IGFBP-3. Cells were stained with preimmune serum (Fig. 5A) and antibodies to E7 and/or IGFBP-3, respectively. Staining was visualized by either green (E7) or red (IGFBP-3) fluorescence. Under these conditions, we were able to establish specific immunofluorescence detection for both proteins (Fig. 5A and B). Transfection of IGFBP-3 alone yielded cells staining strongly positive for IGFBP-3 (Fig. 5B), while coexpression of E7 abolished IGFBP-3 staining in most cases (Fig. 5C), reflecting the ability of E7 to trigger degradation of IGFBP-3 (Fig. 4A). However, in cells where E7 was expressed at a relatively low level (Fig. 5D), some IGFBP-3 was still detectable (Fig. 5E). Under these conditions, we found clear indications of colocalization of E7 and IGFBP-3 (Fig. 5D and E), as evidenced by yellow spots generated by superimposing the corresponding immunofluorescence signals (Fig. 5F, arrows). Together, these results suggest that binding of E7 to IGFBP-3 may precede IGFBP-3 degradation in situ.
FIG. 5.
Colocalization of HPV-16 E7 and IGFBP-3 after transient expression in human cells. (A and B) U-2OS cells were transiently transfected with expression vectors for HPV-16 E7 and/or IGFBP-3, as indicated. At 36 h posttransfection, cells were processed for indirect immunofluorescence microscopy and viewed by using a confocal scanning system. Cells were stained with preimmune serum (A) and anti-E7 antibodies (B, left) or anti-IGFBP-3 antibodies (B, right), as indicated. (C to F) U-2OS cells were cotransfected with expression vectors for both HPV-16 E7 and IGFBP-3. IGFBP-3 was not detectable in cells that express high levels of E7 (C). In cells where E7 was expressed at a relatively low level, weak IGFBP-3 staining was detectable. The structures stained with anti-E7 (D) and anti-IGFBP-3 (E) antibodies are colocalized, as shown by costaining (F; yellow); arrows indicate areas of colocalization.
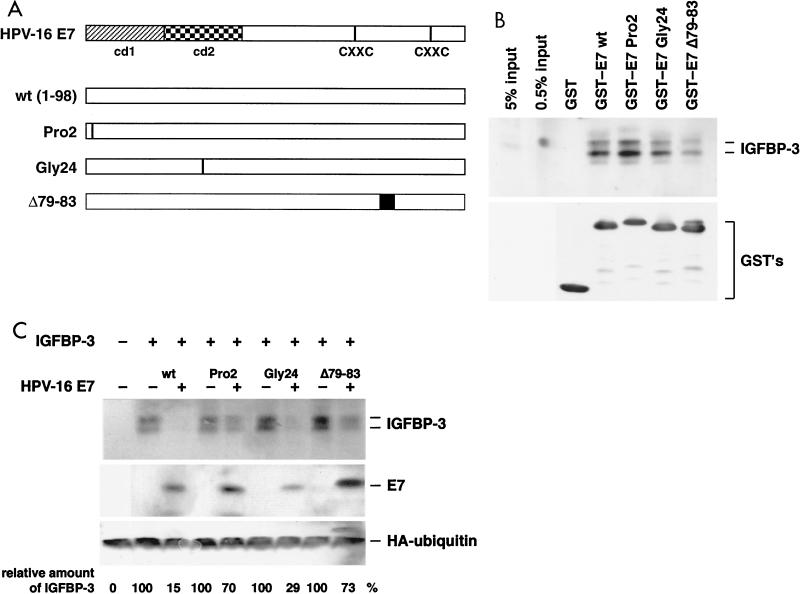
IGFBP-3 degradation by E7 mutants.
To determine whether the ability of E7 to bind and inactivate IGFBP-3 may be relevant for its oncogenic potential, several transformation-deficient E7 mutants were analyzed. Two domains in the E7 oncoprotein have been defined as essential for its transforming potential. The domains are referred to as conserved domain 1 (cd1) and cd2 (Fig. 6A). These domains have been operationally defined by inactivating point mutations in cd1 (PRO2; histidine at position 2 changed to Pro) and cd2 (GLY24; cysteine at position 24 changed to Gly) (for a review, see reference 80). The precise molecular defect of cd1 mutants, such as PRO2, is unknown; in contrast, it was shown that cd2 mutants, such as GLY24, are deficient in binding to the retinoblastoma protein (19, 24). However, there is also evidence that the C terminus of E7 contributes to cell transformation, and recently, it was described that deletion of four amino acids in the C-terminal part of E7, as in mutant E7Δ79-83, strongly reduces the transforming potential of E7 (53). We first analyzed binding of the E7 mutants to IGFBP-3 in a GST pull-down experiment. As shown in Fig. 6B, GST-16E7 was able to bind IGFBP-3 with high affinity, whereas GST alone displayed no binding capacity. The E7 mutants PRO2 and GLY24 both bound IGFBP-3 with an affinity similar to that of the wild type (Fig. 6B), while the binding affinity of mutant E7Δ79-83 was severely reduced, consistent with the results obtained with IGFBP-3–B42 fusion proteins derived from yeast cells (Fig. 2B). Subsequently, the ability of the E7 mutants to trigger IGFBP-3 degradation was analyzed in cotransfection experiments. We found that, while cotransfection of both wild-type E7 and the GLY24 mutant potently induced degradation of IGFBP-3 (reduction to 15.3% ± 4.7% and 28.7% ± 10.7% of the IGFBP-3 levels in control cells, respectively), both the mutants PRO2 (70.3% ± 3.3% of control values) and E7Δ79-83 (73.4% ± 3.8% of control values) were much less active in this respect (Fig. 6C).
FIG. 6.
Analysis of E7 mutants for IGFBP-3 binding and degradation. (A) Structure of the HPV-16 E7 protein. cd1 and cd2 and the putative zinc finger motifs (CXXC) (81) are indicated. The mutants of E7 used in the assays are shown. (B) IGFBP-3 from transiently transfected U-2OS cells was incubated with glutathione- Sepharose 4B beads containing various GST-E7 fusion proteins as indicated. (Top) After elution from the beads, bound proteins were separated by gel electrophoresis and IGFBP-3 was detected by Western blotting; for comparison, samples with 0.5 and 5% of the IGFBP-3 input were also loaded on the gel. (Bottom) Input of the various GST fusion proteins was controlled by anti-GST immunoblotting. (C) U-2OS cells were transfected with expression vectors for IGFBP-3, wild-type E7, and E7 mutants, as indicated. The levels of IGFBP-3 and E7 proteins were determined by Western blotting. Relative IGFBP-3 protein levels are given. Values are the means of three independent experiments. To control transfection efficiency, HA-tagged ubiquitin was cotransfected and detected by Western blotting.
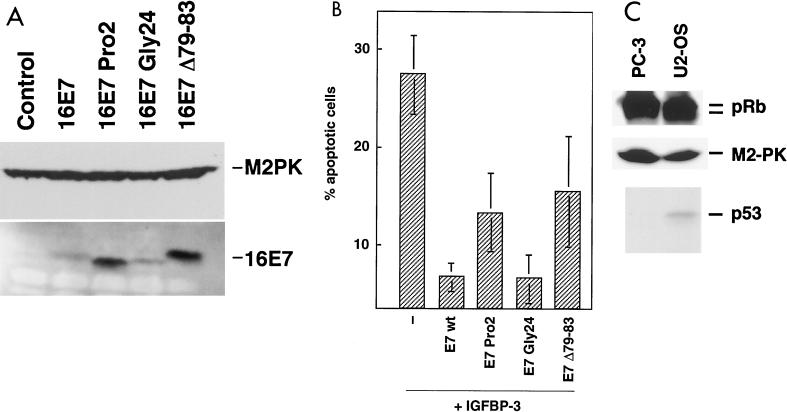
To further study the relationship between inactivation of IGFBP-3 and the transforming potential of E7, we tested the ability of the E7 mutants to counteract apoptosis induction by IGFBP-3 in PC-3 cells. All E7 proteins were well expressed in PC-3 cells; as in U-2OS cells, the mutants Pro2 and Δ79-83 reached even higher levels than wild-type E7 (Fig. 7A). We found that GLY24, the Rb-binding deficient mutant of E7, which interacts with IGFBP-3 and induces its degradation (Fig. 6C) but fails to inactivate pRb (43), suppressed IGFBP-3-induced apoptosis to a similar extent as was observed with wild-type E7. In contrast, both of the PRO2 and Δ79-83 mutants, which failed to induce IGFBP-3 degradation (Fig. 6C), are strongly impaired in their antiapoptotic activity (Fig. 7B). These experiments indicate that the ability of E7 to induce IGFBP-3 degradation is involved in the inhibition of IGFBP-3-driven apoptosis. Since both p53 and pRb were implicated in apoptotic signaling, the status of both proteins was determined in our experimental cells. We found that pRb is present in both the hyper- and hypophosphorylated forms in both cell lines, suggesting that the cells contain wild-type retinoblastoma protein (for a review, see reference 10). The p53 Western blotting reveals that, as was described in the literature (21, 47, 70), U-2OS cells are p53 positive, whereas PC-3 cells lack p53 (Fig. 7C).
FIG. 7.
Functional inactivation of IGFBP-3 by E7 mutants. (A) Expression of E7 mutants. PC-3 cells were transfected with expression vectors for wild type-E7 (16E7) and E7 mutants as indicated. Expression of E7 mutants was analyzed by immunoblotting. Expression of M2 pyruvate kinase (M2PK) was analyzed to control for equal loading. (B) Apoptosis suppression by E7 mutants. PC-3 cells were transfected with expression vectors for IGFBP-3 (−), wild-type E7 (E7 wt), and E7 mutants. Green fluorescent protein served as a transfection marker. Values are means ± standard deviation of three independent experiments. (C) Expression analysis of pRB and p53 in U-2OS and PC-3 cells. Expression of the pRB and p53 proteins was analyzed in extracts from U-2OS and PC-3 cells by immunoblotting. Expression of M2 pyruvate kinase (M2-PK) was analyzed to control for equal loading.
Expression and stability of IGFBP-3 is reduced in E7-immortalized human keratinocytes.
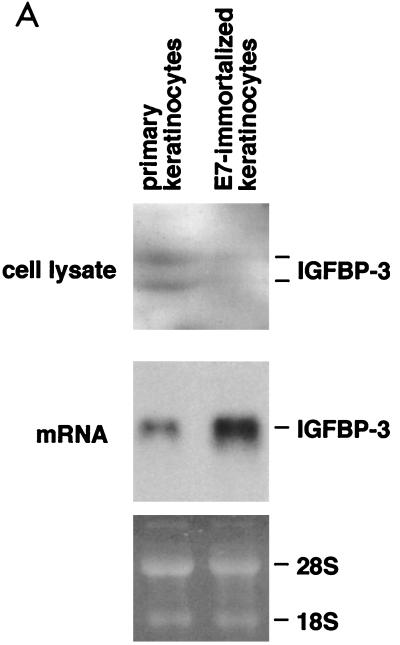
To address the potential significance of the E7–IGFBP-3 interaction for the E7-induced immortalization of human cells, the expression of IGFBP-3 was also studied for normal human keratinocytes by Northern blot and Western blot analyses. We found that primary keratinocytes express detectable levels of IGFBP-3 mRNA and protein (Fig. 8A). Similar experiments were carried out with human keratinocytes which had been immortalized by retroviral expression of the HPV-16 E7 oncogene (12). This experiment revealed that, compared to normal keratinocytes, E7-expressing keratinocytes display a higher abundance of IGFBP-3 mRNA but express drastically reduced levels of the IGFBP-3 (Fig. 8A). These data suggest that IGFBP-3 is present in normal keratinocytes but eliminated from E7-expressing keratinocytes at the posttranscriptional level.
FIG. 8.
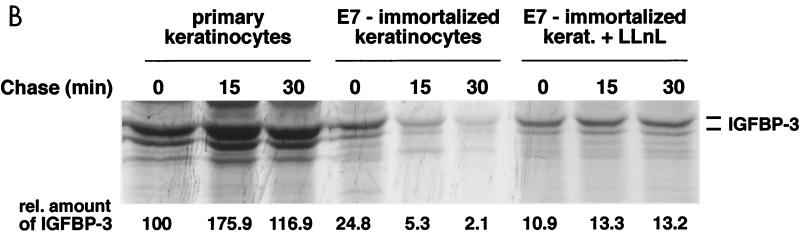
Expression and stability of IGFBP-3 in human keratinocytes. (A) IGFBP-3 level is reduced in E7-immortalized keratinocytes. Primary keratinocytes and E7-immortalized keratinocytes were cultured in keratinocyte growth medium (Promocell) for six passages. In each case, 70,000 cells were lysed and the levels of IGFBP-3 were determined by Western blotting (top two panels). The levels of IGFBP-3 mRNA were determined by Northern blotting (bottom). (B) Metabolic stability of IGFBP-3 is reduced in E7-immortalized keratinocytes. Primary keratinocytes and E7-immortalized keratinocytes were 35S labeled for 20 min, followed by a 30-min chase period, as indicated. Cells were lysed, and IGFBP-3 was immunoprecipitated and detected by autoradiography. Relative protein levels are indicated.
To further address the mechanism by which IGFBP-3 is eliminated, we have analyzed the metabolic stability of IGFBP-3 in human keratinocytes as a function of E7 expression. As is demonstrated in Fig. 8B, the metabolic stability of IGFBP-3 is drastically reduced in E7-expressing keratinocytes; moreover, degradation of IGFBP-3 in E7-expressing keratinocytes is efficiently blocked by the proteasome inhibitor LLnL. These results clearly establish that immortalization of human keratinocytes by HPV-16 E7 involves a significant destabilization of IGFBP-3. The biological significance of the E7–IGFBP-3 interaction for immortalization is clearly established by these experiments. The data raise the possibility that E7-dependent degradation of IGFBP-3 may play a role during E7-dependent immortalization of human keratinocytes.
DISCUSSION
It is shown here that the E7 protein encoded by HPV-16 targets IGFBP-3, a well-known antiproliferative and proapoptotic factor. E7 interacts with IGFBP-3 in vitro as well as in mammalian cells. Expression of E7 in vivo suppresses the ability of IGFBP-3 to induce apoptosis, and this correlates with the ability of E7 to trigger the proteolytic degradation of IGFBP-3. These results identify IGFBP-3 as a new functional target for HPV-16 E7.
The ability of E7 to trigger IGFBP-3 degradation and to block IGFBP-3-dependent apoptosis is severely impaired by point mutations in either cd1 (mutant PRO2) or the C terminus of E7 (mutant Δ79-83) that are both known to severely reduce the oncogenic potential of E7 (25, 53). This finding raises the possibility that the ability of E7 to abrogate the function of IGFBP-3 may contribute to its transforming capacity. Another transformation-deficient mutant, GLY24, which is impaired for binding the retinoblastoma protein family (23), is able to bind IGFBP-3 and trigger its degradation. This finding suggests that the interaction of E7 with the retinoblastoma family of proteins is not required for inactivation of IGFBP-3 and strengthens the notion that several distinct regulatory pathways must be subverted by E7 in order to transform mammalian cells, in keeping with the results from numerous genetic (7, 8, 17, 20, 25, 41, 84) and biochemical (4, 5, 14, 19, 24, 28, 42, 54, 65, 79, 89, 93, 94) studies (for a recent review, see reference 91).
There is precedent for an involvement of IGF signaling in cell transformation. It was shown that disruption of the IGF-I receptor renders cells resistant to transformation by several viral oncoproteins, including HPV-16 E7 (for a review, see reference 9). Reduced expression of IGFBP-3 is an early event in prostatic carcinogenesis (33), raising the possibility that IGFBP-3 may act as an early-phase tumor suppressor in prostate cancer and probably other epithelial cancers. In keeping with a role for IGFBP-3 in tumor suppression, it was shown that expression of the IGFBP-3 gene is induced by the p53 tumor suppressor protein (15) and growth-inhibitory or apoptosis-inducing factors, such as tumor necrosis factor alpha and transforming growth factor beta (60, 67, 69), as well as by anticancer drugs, such as Taxol (Cohen et al., unpublished data). There is precedent for the functional inactivation of IGFBP-3 by oncogenes. Thus, it was shown that malignant transformation of MCF-10A cells by an activated Ha-ras oncogene involves increased resistance to IGFBP-3; however, unlike in the case of E7-mediated resistance to IGFBP-3 (this report), ras-mediated resistance to IGFBP-3 is not due to increased degradation of IGFBP-3 but is associated with increased secretion of IGFBP-3 (52).
The data reported in Fig. 4B demonstrate that the metabolic stability of IGFBP-3 is reduced upon expression of HPV-16 E7. Addition of LLnL to E7-expressing cells prevented degradation of IGFBP-3. In vitro, LLnL inhibits the proteolytic activity of purified proteasome preparations and has some inhibitory activity towards cathepsin B and calpain; however, when added to living cells, LLnL exclusively blocks the protease activity associated with the 26S proteasome (68). Hence, the inability of E7 to trigger IGFBP-3 degradation in LLnL-treated cells indicates that a functional 26S proteasome is required. Several experimental approaches to visualize IGFBP-3 ubiquitination did not reveal any indication for the occurrence of polyubiquitinated forms of the protein. Thus, an in vitro ubiquitination assay that was used by others to demonstrate polyubiquitination of p27KIP1 (56, 58) did not yield any evidence for polyubiquitinated IGFBP-3; similarly, when we used the very sensitive hemagglutinin (HA) ubiquitin cotransfection assay (61), we found no indication for polyubiquitination of IGFBP-3 in vivo (W. Zwerschke, M. Pagano, et al., unpublished data). At present, it is unclear if the observed proteasome-dependent degradation of IGFBP-3 requires ubiquitination of IGFBP-3 or, alternatively, if the decreased metabolic stability of IGFBP-3 shown in Fig. 4B rather reflects the LLnL-mediated stabilization of some upstream regulators of IGFBP-3. Recently, it was reported that E7 triggers degradation of the retinoblastoma protein (pRb) (13). Similar to our findings with IGFBP-3, E7-induced degradation of pRb is sensitive to proteasome inhibitors, while no ubiquitination of pRb was detectable in that study (13).
Consistent with our finding that the IGFBP-3 protein levels and metabolic stability are strongly downregulated in E7-immortalized keratinocytes (Fig. 8) and in HPV-16-positive tumorigenic cell lines (3, 36), the present data suggest that immortalization of human cells by HPV-16 may involve the functional inactivation of IGFBP-3. What could be the reason that HPV-16 evolved a strategy to get rid of IGFBP-3? It is clear that IGFBP-3 gene expression is induced through p53-dependent (15) as well as p53-independent (26) pathways; furthermore, it was reported that IGFBP-3 can induce apoptosis in cells that contain either wild-type p53 or mutated p53 (67). Thus, while p53-dependent apoptotic pathways are inactivated through E6 in HPV-16-infected cells, proapoptotic signals that cannot be neutralized by E6 may affect the viability of HPV-16-infected cells and thereby also affect the production of viral progeny. In this scenario, E7-driven loss of both intracellular and secreted IGFBP-3 (Fig. 3 and 4) may help to downmodulate the apoptotic response of the infected host cells; this could at least in part explain the evolutionary conservation of this function of the E7 protein.
This hypothesis appears to be in contrast with several earlier reports demonstrating that expression of HPV-16 E7 leads to a strong apoptotic response in several experimental systems (38, 40, 62, 63, 66, 77). However, it was also shown that, in human keratinocytes containing mutated p53 (51), as well as in primary murine astrocytes (46), E7 suppresses apoptosis; this finding indicates that regulation of apoptosis by E7 is complex and may involve more than one regulatory pathway. The data reported here support this notion and suggest that suppression of apoptosis by E7 may involve functional inactivation of IGFBP-3.
ACKNOWLEDGMENTS
We are grateful to Gerald Pfister for expert assistance with confocal microscopy and to A. Groyer for the IGFBP-3 cDNA.
This work has been supported by grants from the Fonds zur Förderung der Wissenschaftlichen Forschung (FWF) and the European Union (Biomed 2 program) to P.J.-D.
REFERENCES
- 1.Alani R M, Munger K. Human papillomaviruses and associated malignancies. J Clin Oncol. 1998;16:330–337. doi: 10.1200/JCO.1998.16.1.330. [DOI] [PubMed] [Google Scholar]
- 2.Alcorta D A, Xiong Y, Phelps D, Hannon G, Beach D, Barrett J C. Involvement of the cyclin-dependent kinase inhibitor p16 (INK4a) in replicative senescence of normal human fibroblasts. Proc Natl Acad Sci USA. 1996;93:13742–13747. doi: 10.1073/pnas.93.24.13742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andreatta, Van L S, Hembree J R, Eckert R L. Regulation of insulin-like growth factor 1 binding protein 3 levels by epidermal growth factor and retinoic acid in cervical epithelial cells. J Cell Physiol. 1994;160:265–274. doi: 10.1002/jcp.1041600208. [DOI] [PubMed] [Google Scholar]
- 4.Antinore M J, Birrer M J, Patel D, Nader L, Mccance D J. The human papillomavirus type 16 E7 gene product interacts with and trans-activates the AP1 family of transcription factors. EMBO J. 1996;15:1950–1960. [PMC free article] [PubMed] [Google Scholar]
- 5.Arroyo M, Bagchi S, Raychaudhuri P. Association of the human papillomavirus type 16 E7 protein with the S-phase-specific E2F-cyclin A complex. Mol Cell Biol. 1993;13:6537–6546. doi: 10.1128/mcb.13.10.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 7.Banks L, Edmonds C, Vousden K H. Ability of the HPV16 E7 protein to bind RB and induce DNA synthesis is not sufficient for efficient transforming activity in NIH3T3 cells. Oncogene. 1990;5:1383–1389. [PubMed] [Google Scholar]
- 8.Barbosa M, Edmonds C, Fisher C, Schiller J T, Lowy D, Vousden K. The region of the HPV E7 oncoprotein homologous to adenovirus E1A and SV40 large T antigen contains separate domains for RB binding and casein kinase 2 phosphorylation. EMBO J. 1990;9:153–160. doi: 10.1002/j.1460-2075.1990.tb08091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baserga R, Resnicoff M, Dews M. The IGF-I receptor and cancer. Endocrine. 1997;7:99–102. doi: 10.1007/BF02778073. [DOI] [PubMed] [Google Scholar]
- 10.Beijersbergen R L, Bernards R. Cell cycle regulation by the retinoblastoma family of growth inhibitory proteins. Biochim Biophys Acta. 1996;1287:103–120. doi: 10.1016/0304-419x(96)00002-9. [DOI] [PubMed] [Google Scholar]
- 11.Binoux M. Insulin-like growth factor binding proteins (IGFBPs): physiological and clinical implications. J Pediatr Endocrinol Metab. 1996;3:285–288. [PubMed] [Google Scholar]
- 12.Blasko I, Wagner M, Whitaker N, Grubeck-Loebenstein B, Jansen-Durr P. The amyloid beta peptide abeta (25-35) induces apoptosis independent of p53. FEBS Lett. 2000;470:221–225. doi: 10.1016/s0014-5793(00)01323-5. [DOI] [PubMed] [Google Scholar]
- 13.Boyer S N, Wazer D E, Band V. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 1996;56:4620–4624. [PubMed] [Google Scholar]
- 14.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 15.Buckbinder L, Talbott R, Velascomiguel S, Takenaka I, Faha B, Seizinger B R, Kley N. Induction of the growth inhibitor IGF-binding protein 3 by p53. Nature. 1995;377:646–649. doi: 10.1038/377646a0. [DOI] [PubMed] [Google Scholar]
- 16.Butt A J, Firth S M, Baxter R C. The IGF axis and programmed cell death. Immunol Cell Biol. 1999;77:256–262. doi: 10.1046/j.1440-1711.1999.00822.x. [DOI] [PubMed] [Google Scholar]
- 17.Chellappan S, Kraus V, Kroger B, Münger K, Howley P M, Phelps W, Nevins J R. Adenovirus E1A, simian virus 40 tumor antigen, and human papillomavirus E7 protein share the capacity to disrupt the interaction between transcription factor E2F and the retinoblastoma gene product. Proc Natl Acad Sci USA. 1992;89:4549–4553. doi: 10.1073/pnas.89.10.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Counter C M, Hahn W C, Wei W, Caddle S D, Beijersbergen R L, Lansdorp P M, Sedivy J M, Weinberg R A. Dissociation among in vitro telomerase activity, telomere maintenance, and cellular immortalization. Proc Natl Acad Sci USA. 1998;95:14723–14728. doi: 10.1073/pnas.95.25.14723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davies R, Hicks R, Crook T, Morris J, Vousden K. Human papillomavirus type 16 E7 associates with a histone H1 kinase and with p107 through sequences necessary for transformation. J Virol. 1993;67:2521–2528. doi: 10.1128/jvi.67.5.2521-2528.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Defeo-Jones D, Vuocolo G, Haskell K, Hanobik M, Kiefer D, McAvoy E, Ivey-Hoyle M, Brandsma J, Oliff A, Jones R. Papillomavirus E7 protein binding to the retinoblastoma protein is not required for viral induction of warts. J Virol. 1993;67:716–725. doi: 10.1128/jvi.67.2.716-725.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diller L, Kassel J, Nelson C E, Gryka M A, Litwak G, Gebhardt M, Bressac B, Ozturk M, Baker S J, Vogelstein B, Friend S H. p53 functions as a cell cycle control protein in osteosarcomas. Mol Cell Biol. 1990;10:5772–5781. doi: 10.1128/mcb.10.11.5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duncan E L, Reddel R R. Genetic changes associated with immortalization. A review. Biochemistry. 1997;62:1263–1274. [PubMed] [Google Scholar]
- 23.Dyson N, Guida P, Münger K, Harlow E. Homologous sequences in adenovirus E1A and human papillomavirus E7 proteins mediate interactions with the same set of cellular proteins. J Virol. 1992;66:6893–6902. doi: 10.1128/jvi.66.12.6893-6902.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dyson N, Howley P M, Munger K, Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 25.Edmonds C, Vousden K. A point mutational analysis of human papillomavirus type 16 E7 protein. J Virol. 1989;63:2650–2656. doi: 10.1128/jvi.63.6.2650-2656.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edmondson S R, Murashita M M, Russo V C, Wraight C J, Werther G A. Expression of insulin-like growth factor binding protein-3 (IGFBP-3) in human keratinocytes is regulated by EGF and TGFbeta1. J Cell Physiol. 1999;179:201–207. doi: 10.1002/(SICI)1097-4652(199905)179:2<201::AID-JCP10>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 27.El-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 28.Funk J, Waga S, Harry J, Espling E, Stillman B, Galloway D. Inhibition of cdk activity and PCNA-dependent DNA replication by p21 is blocked by interaction with the HPV-16 E7 oncoprotein. Genes Dev. 1997;11:2090–2100. doi: 10.1101/gad.11.16.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gill Z P, Perks C M, Newcomb P V, Holly J M. Insulin-like growth factor-binding protein (IGFBP-3) predisposes breast cancer cells to programmed cell death in a non-IGF-dependent manner. J Biol Chem. 1997;272:25602–25607. doi: 10.1074/jbc.272.41.25602. [DOI] [PubMed] [Google Scholar]
- 30.Goldstein S, Moerman E J, Jones R A, Baxter R C. Insulin-like growth factor binding protein 3 accumulates to high levels in culture medium of senescent and quiescent human fibroblasts. Proc Natl Acad Sci USA. 1991;88:9680–9684. doi: 10.1073/pnas.88.21.9680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grigoriev V G, Moerman E J, Goldstein S. Overexpression of insulin-like growth factor binding protein-3 by senescent human fibroblasts: attenuation of the mitogenic response to IGF-I. Exp Cell Res. 1995;219:315–321. doi: 10.1006/excr.1995.1234. [DOI] [PubMed] [Google Scholar]
- 32.Gyuris J, Golemis E, Chertkov H, Brent R. Cdi1, a human G1-phase and S-phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 33.Hampel O Z, Kattan M W, Yang G, Haidacher S J, Saleh G Y, Thompson T C, Wheeler T M, Marcelli M. Quantitative immunohistochemical analysis of insulin-like growth factor binding protein-3 in human prostatic adenocarcinoma: a prognostic study. J Urol. 1998;159:2220–2225. doi: 10.1016/S0022-5347(01)63309-3. [DOI] [PubMed] [Google Scholar]
- 34.Hara E, Smith R, Parry D, Tahara H, Stone S, Peters G. Regulation of p16CDKN2 expression and its implications for cell immortalization and senescence. Mol Cell Biol. 1996;16:859–867. doi: 10.1128/mcb.16.3.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hawley-Nelson P, Vousden K H, Hubbert N L, Lowy D R, Schiller J T. HPV16 E6 and E7 proteins cooperate to immortalize human foreskin keratinocytes. EMBO J. 1989;8:3905–3910. doi: 10.1002/j.1460-2075.1989.tb08570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hembree J R, Agarwal C, Eckert R L. Epidermal growth factor suppresses insulin-like growth factor binding protein 3 levels in human papillomavirus type 16-immortalized cervical epithelial cells and thereby potentiates the effects of insulin-like growth factor 1. Cancer Res. 1994;54:3160–3166. [PubMed] [Google Scholar]
- 37.Hossenlopp P, Seurin D, Segovia Q B, Hardouin S, Binoux M. Analysis of serum insulin-like growth factor binding proteins using Western blotting: use of the method for titration of the binding proteins and competitive binding studies. Anal Biochem. 1986;154:138–143. doi: 10.1016/0003-2697(86)90507-5. [DOI] [PubMed] [Google Scholar]
- 38.Howes K A, Ransom N, Papermaster D S, Lasudry J G, Albert D M, Windle J J. Apoptosis or retinoblastoma: alternative fates of photoreceptors expressing the HPV-16 E7 gene in the presence or absence of p53. Genes Dev. 1994;8:1300–1310. doi: 10.1101/gad.8.11.1300. [DOI] [PubMed] [Google Scholar]
- 39.Huibregtse J M, Scheffner M, Howley P M. Cloning and expression of the cDNA for E6-AP, a protein that mediates the interaction of the human papillomavirus E6 oncoprotein with p53. Mol Cell Biol. 1993;13:775–784. doi: 10.1128/mcb.13.2.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iglesias M, Yen K, Gaiotti D, Hildesheim A, Stoler M H, Woodworth C D. Human papillomavirus type 16 E7 protein sensitizes cervical keratinocytes to apoptosis and release of interleukin-1alpha. Oncogene. 1998;17:1195–1205. doi: 10.1038/sj.onc.1202054. [DOI] [PubMed] [Google Scholar]
- 41.Jewers R J, Hildebrandt P, Ludlow J W, Kell B, McCance D J. Regions of human papillomavirus type 16 E7 oncoprotein required for immortalization of human keratinocytes. J Virol. 1992;66:1329–1335. doi: 10.1128/jvi.66.3.1329-1335.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones D L, Alani R M, Münger K. The human papillomavirus E7 oncoprotein can uncouple cellular differentiation and proliferation in human keratinocytes by abrogating p21CIP1-mediated inhibition of cdk2. Genes Dev. 1997;11:2101–2111. doi: 10.1101/gad.11.16.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones D L, Münger K. Analysis of the p53-mediated G1 growth arrest pathway in cells expressing the human papillomavirus type 16 E7 oncoprotein. J Virol. 1997;71:2905–2912. doi: 10.1128/jvi.71.4.2905-2912.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanda T, Zanma S, Watanabe S, Furuno A, Yoshiike K. Two immunodominant regions of the human papillomavirus type 16 E7 protein are masked in the nuclei of monkey COS-1 cells. Virology. 1991;182:723–731. doi: 10.1016/0042-6822(91)90613-g. [DOI] [PubMed] [Google Scholar]
- 45.Leal S M, Liu Q, Huang S S, Huang J S. The type V transforming growth factor beta receptor is the putative insulin-like growth factor-binding protein 3 receptor. J Biol Chem. 1997;272:20572–20576. doi: 10.1074/jbc.272.33.20572. [DOI] [PubMed] [Google Scholar]
- 46.Lee J E, Kim C Y, Giaccia A J, Giffard R G. The E6 and E7 genes of human papilloma virus-type 16 protect primary astrocyte cultures from injury. Brain Res. 1998;795:10–16. doi: 10.1016/s0006-8993(98)00172-3. [DOI] [PubMed] [Google Scholar]
- 47.Lee W H, Shew J W, Hong F D, Sery T W, Donoso L A, Young L J, Bookstein R, Lee E Y. The retinoblastoma susceptibility gene encodes a nuclear phosphoprotein associated with DNA binding activity. Nature. 1987;329:642. doi: 10.1038/329642a0. [DOI] [PubMed] [Google Scholar]
- 48.Li W L, Fawcett J, Widmer H R, Fielder P J, Rabkin R, Keller G A. Nuclear transport of insulin-like growth factor-I and insulin-like growth factor binding protein-3 in opossum kidney cells. Endocrinology. 1997;138:1763–1766. doi: 10.1210/endo.138.4.5176. [DOI] [PubMed] [Google Scholar]
- 49.Lukas J, Müller H, Bartkova J, Spitkovsky D, Kjerulff A, Jansen-Dürr P, Strauss M, Bartek J. DNA tumour viruses oncoproteins and retinoblastoma gene mutations share the ability to relieve the cell's requirement for cyclin D1 function in G1. J Cell Biol. 1994;125:625–638. doi: 10.1083/jcb.125.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lukas J, Parry D, Aagaard L, Mann D J, Bartkova J, Strauss M, Peters G, Bartek J. Retinoblastoma-protein-dependent cell-cycle inhibition by the tumour suppressor p16. Nature. 1995;375:503–506. doi: 10.1038/375503a0. [DOI] [PubMed] [Google Scholar]
- 51.Magal S S, Jackman A, Pei X F, Schlegel R, Sherman L. Induction of apoptosis in human keratinocytes containing mutated p53 alleles and its inhibition by both the E6 and E7 oncoproteins. Int J Cancer. 1998;75:96–104. doi: 10.1002/(sici)1097-0215(19980105)75:1<96::aid-ijc15>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 52.Martin J L, Baxter R C. Oncogenic ras causes resistance to the growth inhibitor insulin-like growth factor binding protein-3 (IGFBP-3) in breast cancer cells. J Biol Chem. 1999;274:16407–16411. doi: 10.1074/jbc.274.23.16407. [DOI] [PubMed] [Google Scholar]
- 53.Massimi P, Pim D, Banks L. Human papillomavirus type 16 E7 binds to the conserved carboxy-terminal region of the TATA box binding protein and this contributes to E7 transforming activity. J Gen Virol. 1997;78:2607–2613. doi: 10.1099/0022-1317-78-10-2607. [DOI] [PubMed] [Google Scholar]
- 54.Mcintyre M C, Ruesch M N, Laimins L A. Human papillomavirus E7 oncoproteins bind a single form of cyclin E in a complex with cdk2 and p107. Virology. 1996;215:73–82. doi: 10.1006/viro.1996.0008. [DOI] [PubMed] [Google Scholar]
- 55.Moerman E J, Thweatt R, Moerman A M, Jones R A, Goldstein S. Insulin-like growth factor binding protein-3 is overexpressed in senescent and quiescent human fibroblasts. Exp Gerontol. 1993;28:361–370. doi: 10.1016/0531-5565(93)90063-j. [DOI] [PubMed] [Google Scholar]
- 56.Montagnoli A, Fiore F, Eytan E, Carrano A C, Draetta G F, Hershko A, Pagano M. Ubiquitination of p27 is regulated by Cdk-dependent phosphorylation and trimeric complex formation. Genes Dev. 1999;13:1181–1189. doi: 10.1101/gad.13.9.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Münger K, Phelps W C, Bubb V, Howley P M, Schlegel R. The E6 and E7 genes of human papillomavirus type 16 together are necessary and sufficient for transformation of primary human keratinocytes. J Virol. 1989;63:4417–4421. doi: 10.1128/jvi.63.10.4417-4421.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nguyen H, Gitig D M, Koff A. Cell-free degradation of p27kip1, a G1 cyclin-dependent kinase inhibitor, is dependent on CDK2 activity and the proteasome. Mol Cell Biol. 1999;19:1190–1201. doi: 10.1128/mcb.19.2.1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oh Y, Gucev Z, Ng L, Muller H L, Rosenfeld R G. Antiproliferative actions of insulin-like growth factor binding protein (IGFBP)-3 in human breast cancer cells. Prog Growth Factor Res. 1995;6:503–512. doi: 10.1016/0955-2235(95)00025-9. [DOI] [PubMed] [Google Scholar]
- 60.Oh Y, Muller H, Ng L, Rosenfeld R. TGF-β-induced cell growth inhibition in human breast cancer cells is mediated through insulin-like growth factor-binding protein-3 action. J Biol Chem. 1995;270:13589–13592. doi: 10.1074/jbc.270.23.13589. [DOI] [PubMed] [Google Scholar]
- 61.Pagano M. Cell cycle regulation by the ubiquitin pathway. FASEB J. 1997;11:1067–1075. doi: 10.1096/fasebj.11.13.9367342. [DOI] [PubMed] [Google Scholar]
- 62.Pan H, Griep A E. Altered cell cycle regulation in the lens of HPV-16 E6 or E7 transgenic mice: implications for tumor suppressor gene function in development. Genes Dev. 1994;8:1285–1299. doi: 10.1101/gad.8.11.1285. [DOI] [PubMed] [Google Scholar]
- 63.Pan H C, Griep A E. Temporally distinct patterns of p53-dependent and p53-independent apoptosis during mouse lens development. Genes Dev. 1995;9:2157–2169. doi: 10.1101/gad.9.17.2157. [DOI] [PubMed] [Google Scholar]
- 64.Pereira S O, Smith J R. Genetic analysis of indefinite division in human cells: identification of four complementation groups. Proc Natl Acad Sci USA. 1988;85:6042–6046. doi: 10.1073/pnas.85.16.6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Phillips A C, Vousden K H. Analysis of the interaction between human papillomavirus type 16 E7 and the TATA binding protein, TBP. J Gen Virol. 1997;78:905–909. doi: 10.1099/0022-1317-78-4-905. [DOI] [PubMed] [Google Scholar]
- 66.Puthenveettil J A, Frederickson S M, Reznikoff C A. Apoptosis in human papillomavirus16 E7-, but not E6-immortalized human uroepithelial cells. Oncogene. 1996;13:1123–1131. [PubMed] [Google Scholar]
- 67.Rajah R, Valentinis B, Cohen P. Insulin like growth factor (IGF)-binding protein-3 induces apoptosis and mediates the effects of transforming growth factor-beta 1 on programmed cell death through a p53- and IGF-independent mechanism. J Biol Chem. 1997;272:12181–12188. doi: 10.1074/jbc.272.18.12181. [DOI] [PubMed] [Google Scholar]
- 68.Rock K L, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg A L. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 69.Rozen F, Zhang J, Pollak M. Antiproliferative action of tumor necrosis factor-a on MCF-7 breast cancer cells is associated with increased insulin-like growth factor binding protein-3 accumulation. Int J Oncol. 1998;13:865–869. doi: 10.3892/ijo.13.4.865. [DOI] [PubMed] [Google Scholar]
- 70.Rubin S J, Hallahan D E, Ashman C R, Brachman D G, Beckett M A, Virudachalam S, Yandell D W, Weichselbaum R R. Two prostate carcinoma cell lines demonstrate abnormalities in tumor suppressor genes. J Surg Oncol. 1991;46:31–36. doi: 10.1002/jso.2930460108. [DOI] [PubMed] [Google Scholar]
- 71.Schedlich L J, Young T F, Firth S M, Baxter R C. Insulin-like growth factor-binding protein (IGFBP)-3 and IGFBP-5 share a common nuclear transport pathway in T47D human breast carcinoma cells. J Biol Chem. 1998;273:18347–18352. doi: 10.1074/jbc.273.29.18347. [DOI] [PubMed] [Google Scholar]
- 72.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 73.Seshadri T, Campisi J. Repression of c-fos transcription and an altered genetic program in senescent human fibroblasts. Science. 1990;247:205–209. doi: 10.1126/science.2104680. [DOI] [PubMed] [Google Scholar]
- 74.Sharpless N E, DePinho R A. The INK4A/ARF locus and its two gene products. Curr Opin Genet Dev. 1999;9:22–30. doi: 10.1016/s0959-437x(99)80004-5. [DOI] [PubMed] [Google Scholar]
- 75.Smotkin D, Wettstein F O. The major human papillomavirus protein in cervical cancers is a cytoplasmic phosphoprotein. J Virol. 1987;61:1686–1689. doi: 10.1128/jvi.61.5.1686-1689.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stein G H, Dulic V. Origins of G(1) arrest in senescent human fibroblasts. Bioessays. 1995;17:537–543. doi: 10.1002/bies.950170610. [DOI] [PubMed] [Google Scholar]
- 77.Stoppler H, Stoppler M C, Johnson E, Simbulan R C, Smulson M E, Iyer S, Rosenthal D S, Schlegel R. The E7 protein of human papillomavirus type 16 sensitizes primary human keratinocytes to apoptosis. Oncogene. 1998;17:1207–1214. doi: 10.1038/sj.onc.1202053. [DOI] [PubMed] [Google Scholar]
- 78.Tahara H, Sato E, Noda A, Ide T. Increase in expression level of p21(sdi1/cip1/waf1) with increasing division age in both normal and SV40-transformed human fibroblasts. Oncogene. 1995;10:835–840. [PubMed] [Google Scholar]
- 79.Tommasino M, Adamczewski J P, Carlotti F, Barth C F, Manetti R, Contorni M, Cavalieri F, Hunt T, Crawford L. HPV16 E7 protein associates with the protein kinase p33CDK2 and cyclin A. Oncogene. 1993;8:195–202. [PubMed] [Google Scholar]
- 80.Tommasino M, Crawford L. Human papillomavirus E6 and E7: proteins which deregulate the cell cycle. Bioessays. 1995;17:509–518. doi: 10.1002/bies.950170607. [DOI] [PubMed] [Google Scholar]
- 81.Ullman C G, Haris P I, Galloway D A, Emery V C, Perkins S J. Predicted alpha-helix/beta-sheet secondary structures for the zinc-binding motifs of human papillomavirus E7 and E6 proteins by consensus prediction averaging and spectroscopic studies of E7. Biochem J. 1996;319:229–239. doi: 10.1042/bj3190229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Valentinis B, Bhala A, DeAngelis T, Baserga R, Cohen P. The human insulin-like growth factor (IGF) binding protein-3 inhibits the growth of fibroblasts with a targeted disruption of the IGF-I receptor gene. Mol Endocrinol. 1995;9:361–367. doi: 10.1210/mend.9.3.7539889. [DOI] [PubMed] [Google Scholar]
- 83.Vousden K H. Regulation of the cell cycle by viral oncoproteins. Semin Cancer Biol. 1995;6:109–116. doi: 10.1006/scbi.1995.0014. [DOI] [PubMed] [Google Scholar]
- 84.Watanabe S, Kanda T, Sato H, Furuno A, Yoshiike K. Mutational analysis of human papillomavirus type 16 E7 functions. J Virol. 1990;64:207–214. doi: 10.1128/jvi.64.1.207-214.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wazer D E, Liu X L, Chu Q, Gao Q, Band V. Immortalization of distinct human mammary epithelial cell types by human papilloma virus 16 E6 or E7. Proc Natl Acad Sci USA. 1995;92:3687–3691. doi: 10.1073/pnas.92.9.3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wood W I, Cachianes G, Henzel W J, Winslow G A, Spencer S A, Hellmiss R, Martin J L, Baxter R C. Cloning and expression of the growth hormone-dependent insulin-like growth factor-binding protein. Mol Endocrinol. 1988;2:1176–1185. doi: 10.1210/mend-2-12-1176. [DOI] [PubMed] [Google Scholar]
- 87.Wraight C J, Liepe I J, White P J, Hibbs A R, Werther G A. Intranuclear localization of insulin-like growth factor binding protein-3 (IGFBP-3) during cell division in human keratinocytes. J Investig Dermatol. 1998;111:239–242. doi: 10.1046/j.1523-1747.1998.00258.x. [DOI] [PubMed] [Google Scholar]
- 88.Yeager T R, DeVries S, Jarrard D F, Kao C, Nakada S Y, Moon T D, Bruskewitz R, Stadler W M, Meisner L F, Gilchrist K W, Newton M A, Waldman F M, Reznikoff C A. Overcoming cellular senescence in human cancer pathogenesis. Genes Dev. 1998;12:163–174. doi: 10.1101/gad.12.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zerfass-Thome K, Zwerschke W, Mannhardt B, Tindle R, Botz J, Jansen-Dürr P. Inactivation of the cdk inhibitor p27KIP1 by the human papillomavirus type 16 E7 oncoprotein. Oncogene. 1996;13:2323–2330. [PubMed] [Google Scholar]
- 90.zur Hausen H. Human papillomaviruses in the pathogenesis of anogenital cancer. Virology. 1991;184:9–13. doi: 10.1016/0042-6822(91)90816-t. [DOI] [PubMed] [Google Scholar]
- 91.Zwerschke W, Jansen-Dürr P. Cell transformation by the E7 oncoprotein of HPV type 16: interactions with nuclear and cytoplasmic target proteins. Adv Cancer Res. 2000;78:1–29. doi: 10.1016/s0065-230x(08)61022-2. [DOI] [PubMed] [Google Scholar]
- 92.Zwerschke W, Joswig S, Jansen-Dürr P. Identification of domains required for transcriptional activation and protein dimerization in the human papillomavirus type-16 E7 protein. Oncogene. 1996;12:213–220. [PubMed] [Google Scholar]
- 93.Zwerschke W, Mannhardt B, Massimi P, Nauenburg S, Pim D, Nickel W, Banks L, Reuser A J, Jansen-Dürr P. Allosteric activation of acid alpha-glucosidase by the human papillomavirus E7 protein. J Biol Chem. 2000;275:9534–9541. doi: 10.1074/jbc.275.13.9534. [DOI] [PubMed] [Google Scholar]
- 94.Zwerschke W, Mazurek S, Massimi P, Banks L, Eigenbrodt E, Jansen-Dürr P. Modulation of type M2 pyruvate kinase activity by the human papillomavirus type 16 E7 oncoprotein. Proc Natl Acad Sci USA. 1999;96:1291–1296. doi: 10.1073/pnas.96.4.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]