Abstract
The extracellular vesicle (EV) surface proteome (surfaceome) acts as a fundamental signalling gateway by bridging intra‐ and extracellular signalling networks, dictates EVs’ capacity to communicate and interact with their environment, and is a source of potential disease biomarkers and therapeutic targets. However, our understanding of surface protein composition of large EVs (L‐EVs, 100–800 nm, mean 310 nm, ATP5F1A, ATP5F1B, DHX9, GOT2, HSPA5, HSPD1, MDH2, STOML2), a major EV‐subtype that are distinct from small EVs (S‐EVs, 30–150 nm, mean 110 nm, CD44, CD63, CD81, CD82, CD9, PDCD6IP, SDCBP, TSG101) remains limited. Using a membrane impermeant derivative of biotin to capture surface proteins coupled to mass spectrometry analysis, we show that out of 4143 proteins identified in density‐gradient purified L‐EVs (1.07–1.11 g/mL, from multiple cancer cell lines), 961 proteins are surface accessible. The surface molecular diversity of L‐EVs include (i) bona fide plasma membrane anchored proteins (cluster of differentiation, transporters, receptors and GPI anchored proteins implicated in cell‐cell and cell‐ECM interactions); and (ii) membrane surface‐associated proteins (that are released by divalent ion chelator EDTA) implicated in actin cytoskeleton regulation, junction organization, glycolysis and platelet activation. Ligand‐receptor analysis of L‐EV surfaceome (e.g., ITGAV/ITGB1) uncovered interactome spanning 172 experimentally verified cognate binding partners (e.g., ANGPTL3, PLG, and VTN) with highest tissue enrichment for liver. Assessment of biotin inaccessible L‐EV proteome revealed enrichment for proteins belonging to COPI/II‐coated ER/Golgi‐derived vesicles and mitochondria. Additionally, despite common surface proteins identified in L‐EVs and S‐EVs, our data reveals surfaceome heterogeneity between the two EV‐subtype. Collectively, our study provides critical insights into diverse proteins operating at the interactive platform of L‐EVs and molecular leads for future studies seeking to decipher L‐EV heterogeneity and function.
Keywords: extracellular vesicles, mass spectrometry‐based proteomics, surface proteins, surfaceome, vesicle heterogeneity
1. INTRODUCTION
Extracellular vesicles (EVs), membranous vesicles released by cells into extracellular space, are laden with proteins and nucleic acids which they can transfer between cells to elicit functional response and therefore serve as an effective means of intercellular signalling, both in physiological as well as pathological conditions (Xu et al., 2018; Claridge, Lozano et al., 2021). Critical to EV function are the EV surface proteins which regulate their interaction with the extracellular environment (recipient cells, ECM) (Hu et al., 2020), dictate biodistribution (Hoshino et al., 2015), half‐life in circulation (Buzás et al., 2018) and pharmacokinetics (Charoenviriyakul et al., 2018). In cancer, EV surface proteins not only mobilize pro‐metastatic bone marrow cells (through transfer of active surface MET) (Peinado et al., 2012) but also regulate EV homing to specific organs (to lungs via surface integrins ITGA6/B4 and ITGA6/B1, and to liver via integrins ITGAV/B5) (Hoshino et al., 2015) to establish pre‐metastatic niches and enhance metastasis. EV surface proteins (e.g., PD‐L1) also regulate immune escape to promote tumorigenesis in lung cancer (Kim et al., 2019). Moreover, EV surface proteins are not only a source of potential disease biomarkers (Al‐Nedawi et al., 2008; Melo et al., 2015) but also enable capture of EVs as liquid biopsy (Yoshioka et al., 2014) and systemic clearance of pathogenic EVs (Zaborowski et al., 2019) in the clinic. EV surface proteins also have therapeutic potential in bone healing (Furuta et al., 2016), provide cardio protection (Vicencio et al., 2015), and rescue neuronal function impairment (Yang et al., 2017). Recently, functionalization of EV surface proteins is emerging as an effective means to design vehicles that can potentially deliver therapeutic drugs to target sites (Gao et al., 2018; Mentkowski & Lang, 2019; Ohno et al., 2013; Tian et al., 2018) or to develop potential cancer vaccines (Yaddanapudi et al., 2019). Because EVs are released in high number, their collective surface area represents a large interactive platform, engaging proteins, lipids and glycans; understanding players of this dynamic interactome is thus pivotal.
Based on their size, EVs can be broadly categorized into large EVs (L‐EVs, 100–1000 nm) and small EVs (S‐EVs, 30–150 nm) (Kowal et al., 2016; Xu et al., 2015). While several studies have catalogued role of S‐EVs in many pathologies (e.g., cancer [Xu et al., 2018], cardiovascular disease [Bellin et al., 2019], and neurodegenerative diseases [Howitt & Hill, 2016]), L‐EVs—although relatively understudied compared to S‐EVs—are now also starting to gain prominence for their signalling role, either complementing or contrasting S‐EV function (Tkach et al., 2018). We have previously shown that L‐EVs can be further segregated based on their varying densities, namely L‐EVs displaying buoyant density of 1.07–1.11 g/ml which are distinct from L‐EVs of relatively higher buoyant density (1.22–1.30 g/ml) (Rai, Greening, et al., 2021). While heavy density L‐EVs represent midbody remnants of cytokinetic origin, our understanding of low‐density L‐EVs remains limited.
Besides their size‐based categorization, it is important to note that cells release heterogeneous sub‐populations of EVs (Ji et al., 2014; Rai, Poh, et al., 2021; Tauro et al., 2013; Théry et al., 2018) which can be categorised based on their origin (e.g., plasma membrane budding that give rise to shed microvesicles [Al‐Nedawi et al., 2008], EVs of endosomal origin called exosomes [Jeppesen et al., 2019; Xu et al., 2018], shed midbody remnants derived from cytokinetic bridges [Rai, Greening, et al., 2021], migrasomes released by migrating cells [Wiklander et al., 2015]), floatation density (e.g., light vs. heavy density EVs [Kowal et al., 2016; Rai et al., 2019; Rai, Poh, et al., 2021]) and/or biochemical compositions (Jeppesen et al., 2019; Kowal et al., 2016) (e.g., EPCAM/A33+ EVs [Tauro et al., 2013]). Depending on isolation strategies employed, heterogeneous vesicles co‐purify (e.g., S‐EVs can arise from plasma membrane budding or are endosomally‐derived) due to overlapping characteristics (Théry et al., 2018; Tkach et al., 2018). Thus, while these subtype categorizations are useful at an operational level, there is growing awareness of EV heterogeneity and the need to study it. Fundamental questions regarding L‐EV form and function remain unanswered, these include our insight into their surface proteome landscape.
By definition, cellular surfaceome encompasses all plasma membrane proteins with at least one amino acid residue exposed to the extracellular space (Bausch‐Fluck et al., 2018). Several studies have characterized S‐EVs surfaceome (Castillo et al., 2018; Cvjetkovic et al., 2016; Hu et al., 2018; Jeppesen et al., 2014; Jung et al., 2020; Wu et al., 2019; Xu et al., 2019; Zaborowski et al., 2019); we previously characterized surfaceome of S‐EVs by proteolytically “shaving” surface proteins using proteinase K (Xu et al., 2019). However, we noted that proteinase K treatment compromises L‐EVs integrity, calling for an alternative strategy to define surface landscape of L‐EVs. The focus of this paper is directed at defining surface protein landscape of L‐EVs (buoyant density of 1.07–1.11 g/mL) by using membrane impermeant biotin to label and enrich for surface proteins and identify them using mass spectrometry.
2. METHODS
2.1. Cell culture
SW620 (CCL‐227, ATCC) and LIM1863 cells (Whitehead et al., 1987) (Ludwig Institute for Cancer Research, Melbourne) cells were cultured in RPMI‐1640 (Life Technologies). MDA MB 231 (HTB‐26, ATCC) and U87 (HTB‐14, ATCC) cells were cultured in DMEM (Life Technologies). Complete culture media included media supplemented with 5% (v/v) Fetal Bovine Serum (FBS, Life Technologies) and 1% (v/v) Penicillin Streptomycin (Pen/Strep, Life Technologies) at 37°C with 10% CO2. Cells were passaged with trypsin‐EDTA (Gibco).
2.2. Generation of cell conditioned media
Cells (SW620, LIM1863, MDA MB 231, and U87) were cultured in CELLine AD‐1000 Bioreactor classic flasks (Integra Biosciences) as previously described (Rai, Greening, et al., 2021). Cells (3 × 107) in 15 ml of complete culture media were added to the lower cell‐cultivation chamber for 72 h at 37°C with 10% CO2. The upper nutrient supply chamber contained 500 ml RPMI or DMEM (5% FBS, 1% Pen/Strep) that was replaced every four days. The lower chamber was washed three times with serum‐free media. For LIM1863 cells, which grow as floating organoids, cells were recovered and washed in serum free media (150 g, 5 min) and re‐introduced back into the lower chamber. Cells in the lower chamber were then cultured in 15 ml of media supplemented with 0.5% (v/v) insulin transferrin selenium (Invitrogen) and 1% Pen/Strep). Thereafter, conditioned medium (CM) in the cell cultivation chamber was collected every 2 days.
2.3. Isolation of large and small EVs
CM was centrifuged at 500 g (5 min, 4°C) and 2,000 g (10 min, 4°C) and the supernatant either processed for EV isolation or stored at ‐20°C until further use. The supernatant was centrifuged at 10,000 g (30 min, 4°C, SW28 rotor; Optima XPN Ultracentrifuge, Beckman Coulter) to pellet crude L‐EVs and then at 100,000 g (1 h, 4°C, 41 Ti rotor; Optima XPN Ultracentrifuge) to pellet crude S‐EVs. EV‐pellets were resuspended in ∼200 μl PBS and subjected to top‐down isopycnic (iodixanol‐density) ultracentrifugation (Ford et al., 1994; Tauro et al., 2013), whereby EVs were then over (discontinuous gradient of OptiPrep™ , 40% [3 ml], 20% [3 ml], 10% [3 ml], and 5% [2.5 ml] [diluent: 0.25 M sucrose/PBS solution]) and ultracentrifuged at 100,000 g for 18 h (4°C, 41 Ti rotor; Optima XPN Ultracentrifuge). Twelve equal fractions were collected, diluted in PBS (2 ml) and centrifuged at either 10,000 g (30 min, 4°C, Eppendorf 5430R) or 100,000 g (1 h, 4°C, TLA‐55 rotor; Optima MAX‐MP Tabletop Ultracentrifuge) to collect L‐EV and S‐EV containing fractions, respectively. Density of each fraction was determined as previously described (Rai, Fang, et al., 2021; Rai, Poh, et al., 2021). Pellets were further washed in PBS, reconstituted in PBS and stored at ‐80°C until further use.
2.4. Immunoblotting
Protein quantification (microBCA™ Protein Assay Kit (23235, Thermo Fisher Scientific)) and Western blotting (iBlot 2 Dry Blotting System, Thermo Fisher Scientific) were performed as per manufacturer's instructions. Dot blot analysis was performed using 96‐well Bio‐Dot (Bio‐Rad Laboratories) as per manufacturer's instructions with proteins were lysed in 50 mM HEPES (1% SDS). Rabbit antibodies raised against, MET (Santa Cruz Biotechnology), CD63 (Santa Cruz), ANXA1 (Abcam), GAPDH (Cell Signalling), and GFP (Abcam) were used. Mouse antibodies ALIX (BD Biosciences), TSG101 (BD Biosciences) were used. Secondary antibodies used were IRDye 800 goat anti‐mouse IgG or IRDye 700 goat anti‐rabbit IgG (1:15000, LI‐COR Biosciences).
2.5. Biophysical characterization of EVs
Cryo‐electron microscopy (Tecnai G2 F30) on EVs (2 μg) was performed as described (Rai, Fang, et al., 2021; Xu et al., 2015). Vesicle particle size was determined using a NanoSight NS300, Nanoparticle tracking analysis (NTA) (Malvern) system fitted with a NS300 flow‐cell top plate with a 405 nm laser as described (Rai, Greening, et al., 2021). Samples (1 μg μl–1) were diluted in 500 μl PBS (1:10,000) and injected using 1 ml syringes (BD Biosciences) (detection threshold = 10, flowrate = 50, temperature = 25°C). Each analysis consisted of 60 s video captures. Data was analysed using NTA software 3.0 (Malvern).
2.6. Global proteomic sample preparation of EVs
Global mass spectrometry‐based proteomics of EVs (10 μg in 50 μl) was performed as previously described (Poh et al., 2021) using single‐pot solid‐phase‐enhanced sample preparation (SP3) method (Claridge, Lozano et al., 2021; Hughes et al., 2019). Briefly, samples were solubilised in 1% (v/v) sodium dodecyl sulphate (SDS), 50 mM HEPES pH 8.0, incubated at 95°C for 5 min and cooled. Samples were reduced with 10 mM dithiothreitol (DTT) for 45 min at 25°C followed by alkylation with 20 mM iodoacetamide for 30 min at 25°C in the dark. The reaction was quenched to a final concentration of 20 mM DTT. Magnetic beads were prepared by mixing SpeedBeads™ magnetic carboxylate modified particles (65152105050250, 45152105050250, Cytiva) at 1:1 (v:v) ratio and washing twice with 200 μl MS‐water. Magnetic beads were reconstituted to a final concentration of 100 μg/μl. Magnetic beads were added to the samples at 10:1 beads‐to‐protein ratio and 100% ethanol (EA043, ChemSupply) added for a final concentration of 50% ethanol (v/v). Protein‐bound magnetic beads were washed three times with 200 μl of 80% ethanol and reconstituted in 50 mM TEAB and digested with trypsin (Promega, V5111) at a 1:50 enzyme‐to‐substrate ratio for 16 h at 37°C with constant shaking (1000 rpm). The peptide mixture was acidified to a final concentration of 2% formic acid (pH ∼1‐2) and centrifuged at 20,000g for 1 min. The peptide digests were frozen at ‐80°C and dried by vacuum centrifugation (Savant SPD121P, Thermo Fisher Scientific), reconstituted in 0.07% trifluoroacetic acid, and quantified by Fluorometric Peptide Assay (23290, Thermo Fisher Scientific) as per manufacturer's instruction.
2.7. Surface biotin‐labelling of EVs and proteomic sample preparation
EV surface proteins were biotinylated using Pierce™ Cell Surface Biotinylation and Isolation Kit (A44390, Thermo Fisher Scientific) as per manufacturer's recommendation. Briefly, 100 μg EVs (L‐EVs or S‐EVs) were labeled with 0.25 mg/ml EZ‐Link Sulfo‐NHS‐SS‐Biotin for 10 min at room temperature. EVs were then washed twice with ice‐cold TBS (10,000 g for 30 min for L‐EVs or 100,000 g for 1 h for S‐EVs). Labelled EVs were then lysed (30 min on ice) as per manufacturer's instructions. To assess bound and released EV surface proteins, biotinylated EVs in PBS were treated with 5 mM EDTA for 15 min at room temperature (Rogers et al., 2020). Samples were centrifuged at either 10,000 g (30 min, 4°C, Eppendorf 5430R) or 100,000 g (1 h, 4°C, TLA‐55 rotor; Optima MAX‐MP Tabletop Ultracentrifuge) to collect L‐EV and S‐EV with their membrane bound proteins (pellet fraction), respectively. Supernatants (containing the released proteins) were processed for SP3‐based tryptic protein digestion (as above). The pellet fractions were resuspended in 1 ml PBS, re‐centrifuged and pellet fractions reconstituted in 50 μl of PBS.
Biotin‐labelled proteins were then captured onto NeutrAvidin Agarose slurry (30 min at room temperature with end‐over‐end mixing on a rotor). Samples were loaded onto epTIPS (Eppendorf, 200 μl) fitted with 20 μm nylon net (NY2004700, Merck Millipore), washed three times as per manufacturers instruction, and reduced in 10 mM DTT in 100 mM triethylamonium bicarbonate (TEAB) for 45 min at 25°C. Eluted proteins were then alkylated with 20 mM iodoacetamide (IAA) for 30 min at 25°C in the dark. The reaction was quenched to a final concentration of 20 mM DTT and digested with trypsin (Promega, V5111) at a 1:50 enzyme‐to‐substrate ratio for 16 h at 37°C. The resultant peptides were acidified to a final concentration of 2% formic acid (FA), peptides desalted using SDB‐RPS Stage‐Tips (Rai, Poh et al., 2021) followed by elution with 30%–80% acetonitrile (ACN), 0.1% trifluoroacetic acid, and dried by vacuum centrifugation. Peptides were reconstituted in 0.07% trifluoroacetic acid and quantified by Fluorometric Peptide Assay (23290, Thermo Fisher Scientific). SP3‐based tryptic protein digestion for global proteome was performed as previously described (Claridge, Rai et al., 2021).
2.8. Proteomic liquid chromatography–tandem mass spectrometry
Peptides were analysed on a Dionex UltiMate NCS‐3500RS nanoUHPLC coupled to a Q‐Exactive HF‐X hybrid quadrupole‐Orbitrap mass spectrometer equipped with nanospray ion source in positive mode as described (Greening et al., 2019; Kompa et al., 2021). Peptides were loaded (Acclaim PepMap100 C18 3 μm beads with 100 Å pore‐size, Thermo Fisher Scientific) and separated (1.9‐μm particle size C18, 0.075 × 250 mm, Nikkyo Technos Co. Ltd) with a gradient of 2%–28% acetonitrile containing 0.1% formic acid over 95 min at 300 nl min–1 followed by 28%–80% from 95 to 98 min at 300 nl min‐1 at 55°C (butterfly portfolio heater, Phoenix S&T). An MS1 scan was acquired from 350 to 1650 m/z (60,000 resolution, 3 × 106 automatic gain control (AGC), 128 msec injection time) followed by MS/MS data‐dependent acquisition (top 25) with collision‐induced dissociation and detection in the ion trap (30,000 resolution, 1 × 105 AGC, 60 msec injection time, 28% normalized collision energy, 1.3 m/z quadrupole isolation width). Unassigned, 1, 6–8 precursor ions charge states were rejected and peptide match disabled. Selected sequenced ions were dynamically excluded for 30 s. Data was acquired using Xcalibur software v4.0 (Thermo Fisher Scientific). The MS‐based proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository and are available via ProteomeXchange with identifier (PXD026658).
2.9. Data processing
MaxQuant (v1.6.6.0) with its built‐in search engine Andromeda (Cox & Mann, 2008) was used to perform peptide identification and quantification as described (Kompa et al., 2021). Human‐only (UniProt #74,823 entries) sequence database (Jan 2020) with a contaminants database was employed. Cysteine carbamidomethylation was set as a fixed modification and N‐terminal acetylation and methionine oxidations as variable modifications; for biotin surface proteome analysis additional Thioacyl (DSP) (C(3)H(4)OS)) was employed. False discovery rate (FDR) was 0.01 for protein and peptide levels. Enzyme specificity was set as C‐terminal to arginine and lysine using trypsin protease, and a maximum of two missed cleavages allowed. Peptides were identified with an initial precursor mass deviation of up to 7 ppm and a fragment mass deviation of 20 ppm. Protein identification required at least one unique or razor peptide per protein group. Contaminants, and reverse identification were excluded from further data analysis. ‘Match between run algorithm’ in MaxQuant (Nagaraj et al., 2012) and label‐free protein quantitation (maxLFQ) was performed. All proteins and peptides matching to the reversed database were filtered out.
For both L‐EVs and S‐EVs, we investigated three biological replicates for global proteome (three cell lines: LIM1863, SW620, U87), three biological replicates for biotin capture proteome (three cell lines: SW620, MDA MB 231, U87), two biological replicates for EDTA bound biotin capture (two cell lines: MDA MB 231, SW620), two biological replicate for EDTA released biotin capture proteome (two cell lines: MDA MB 231, SW620).
For global surface proteome (Table S1), for each cell line, protein was identified at least once in one biological replicate and detected at least once in EVs from at least two cell lines. For biotin‐captured surfaceome, EVs from three cell lines were analysed, with three biological replicates for each cell line. Proteins selected for downstream bioinformatics analysis include those that were identified in at least 2 biological replicates, irrespective of the donor cells. For biotin‐captured surfaceome that are removed by EDTA, EVs from two cell lines were analysed, with two biological replicates for each cell line. Proteins selected for downstream bioinformatics analysis include those that were identified in at least two biological replicates, irrespective of the donor cells.
Experimental parameters are submitted to EV‐TRACK knowledgebase (EV‐TRACK ID: EV210261) (Van Deun et al., 2017).
2.10. Bioinformatics and statistics
Cellular surfaceome data used include proteins previously experimentally verified as a cell surface protein atlas (CSPA) (Bausch‐Fluck et al., 2015) or predicted as surfaceome proteins based on SURFY (Bausch‐Fluck et al., 2018). Voronoi tree maps (i.e., in silico surfaceome tree map) were generated using http://wlab.ethz.ch/surfaceome. Venn diagrams were created using www.interactivenn.net. Gene Ontologies, KEGG and Reactome pathways were obtained using g:Profiler (Raudvere et al., 2019) or DAVID analysis (Huang et al., 2009). Cytoscape (Shannon, 2003) was used to generate EnrichmentMap (Merico et al., 2010) (plugin v3.7.1) and GeneMania‐based radial interaction map (Warde‐Farley et al., 2010) (plugin v3.5.1). Sankey diagram of ligand‐receptor interactions was generated using http://www.rna‐society.org/cellinker/ (Zhang, Liu, et al., 2021). Volcano plot, principal component analysis plot, Pearson correlation matrix and hierarchical clustering was performed using Perseus (Tyanova et al., 2016). Data were analyzed and bar plots/violin plots generated using GraphPad Prism (v8.0.1) or Microsoft Excel. One‐way ANOVA (multiple comparisons) test was performed using Perseus and statistical significance defined at p < 0.05.
2.11. Fluorescence microscopy
Immunofluorescence was performed, as previously described (Rai, Greening, et al., 2021). Generation of SW620 cells stably‐expressing plasma membrane‐targeting Growth Associated protein 43 or GAP43 (1‐20 a.a.) (Zuber et al., 1989) fused to GFP (SW620‐GAP‐GFP cells) is described previously (Rai, Poh, et al., 2021). Briefly, cells (cultured on Nunc® Lab‐Tek® Chamber Slide™ [Sigma‐Aldrich] to 60%–80% confluency) fixed (4% formaldehyde for 5 min), permeabilized (0.2% [v/v] Triton X‐100 in PBS, 5 min) and blocked (3% [w/v] bovine serum albumin [BSA, Sigma]) in TTBS (0.2% [v/v] Triton X‐100) (blocking solution) for 30 min at room temperature. Cells were then incubated with primary antibodies (1:100) MET (Santa Cruz Biotechnology), ALIX (BD Biosciences), TSG101 (BD Biosciences) or EEA1 (Cell Signalling) antibodies in blocking solution for 1 h at room temperature. Cells were washed and incubated with secondary antibodies (1:200) (Alexa Fluor 488‐conjugated goat anti‐mouse IgG or Alexa Fluor 568‐conjugated goat anti‐rabbit IgG (Invitrogen) in blocking solution for 20 min at room temperature (in the dark). Cells were washed three times in TTBS. Where indicated, nuclei were stained with Hoechst stain (10 μg ml–1) for 1 min. Cells were imaged using a Zeiss AxioObserver Z1 microscope (Zeiss) or Zeiss Confocal LSM 780 PicoQuant FLIM (Zeiss) and images were analysed using Zen 2011 (Blue edition, Zeiss).
3. RESULTS
3.1. Isolation and characterization of large EVs
We isolated large EVs (L‐EVs) and small EVs (S‐EVs) released by four different cell lines using differential centrifugation coupled to density gradient separation (Figure 1a) as previously described (Rai et al., 2019; Suwakulsiri et al., 2019; Tauro et al., 2013) and characterized them (for their buoyant densities, size, morphology, presence of specific proteins) to meet the experimental requirements as set out by the International Society for Extracellular Vesicles (ISEV) guidelines (Théry et al., 2018). While both L‐EVs and S‐EVs displayed buoyant densities of 1.07–1.11 g/ml and were positive for EV marker ANXA1 (Figure S1), S‐EVs were enriched in CD63 consistent with previous reports (Rai, Greening, et al., 2021; Rai et al., 2019; Suwakulsiri et al., 2019). Cryo‐EM revealed that both EV subtypes were spherical in shape and morphologically intact with L‐EVs (mean 310 nm) significantly (p < 0.0001) larger than S‐EVs (mean 110 nm) (Figure 1b,c, Interestingly, there was no striking difference in mode size (i.e., size of the majority of EVs in the samples, e.g., 185.5 nm [L‐EVs] vs. 168.7 nm [S‐EVs] for MDA‐MB‐231, Figures S2 and S3); this could be due to bias of NTA towards certain particle size ranges (especially 50–150 nm [Théry et al., 2018; Van Der Pol et al., 2014]). However, NTA analysis of EVs from three cell lines suggests that percentage of EVs >200 nm was higher in L‐EV versus S‐EVs, similar to cryoEM analysis of EVs from SW620 cells (Figure S4). Although the two EV subtypes from different cell lines clustered based on donor cells (Figure S5), mass‐spectrometry revealed that S‐EVs are enriched in classical markers of small EVs (Kowal et al., 2016; Kugeratski et al., 2021; Rai, Greening, et al., 2021; Xu et al., 2015), (CD44, CD63, CD81, CD82, CD9, PDCD6IP, SDCBP, TSG101) (Figures 1d–f, S6, and Tables S1 and S2) whereas L‐EVs were enriched in previously reported large EV proteins (Kowal et al., 2016; Minciacchi et al., 2015; Rai, Greening, et al., 2021; Xu et al., 2015) (ATP5F1A, ATP5F1B, DHX9, GOT2, HSPA5, HSPD1, MDH2, STOML2), ribosomal (RPS5, RPL9/35), endoplasmic reticulum (CANX, CALR, TMEM33), mitochondrial (TIM44, ALDH2, PUS1), spliceosomal (DDX5, DDX39B, SNRPD1/2/3) and ribonucleoproteins (HNRNPU, HNRNPK) (Figures 1d–f, S6, and Tables S1 and S2), as previously described (Rai, Greening et al., 2021; Rai, Fang et al., 2021; Suwakulsiri et al., 2019).
FIGURE 1.

Isolation and characterization of large and small EVs. (a) Workflow for isolation of L‐EVs and S‐EVs. (b) Cryo‐electron microscopic images of L‐EVs and S‐EVs isolated from SW620 cells. (c) Histogram represents diameter of L‐EVs and S‐EVs based on cryo‐EM images. Data presented as mean ± s.e.m (standard error of mean). (d) Volcano plot of protein abundance between L‐EVs and S‐EVs using mass spectrometry‐based proteomics; comparisons of log2 fold changes versus p‐values (Student's t‐test). (e) Heat map of selected proteins enriched in L‐EVs or S‐EVs from three indicated cell lines (p‐value <0.05). (f) EnrichmentMap of Gene Ontology (cellular components) processes overrepresented in L‐EVs or S‐EVs. Node size represents gene number
3.2. Workflow for defining EV surfaceome
The workflow used in this study to define EV surfaceome using quantitative mass spectrometry‐based proteomics is outlined in Figure 2. We mapped experimentally verified cell‐surface proteins (CSPA) (Bausch‐Fluck et al., 2015) and surfaceome proteins based on cell surfaceome predictor SURFY (Bausch‐Fluck et al., 2018) onto global EV protein profiles. Next, we verified their EV surface localization by enriching surface proteins using amine reactive, membrane‐impermeant and thiol‐cleavable Sulfo‐NHS‐SS‐Biotin and subsequently identifying using mass spectrometry. Because EDTA can successfully chelate metal ions such as calcium to release surface bound proteins (such as ANXA1 [Rogers et al., 2020]) without compromising EV integrity (Rogers et al., 2020; Santucci et al., 2019), we also incubated EVs with EDTA and define the released proteins as the “EV surface‐associated proteome.” This led to the identification of 291 and 456 surface, and 122 and 67 surface‐associated proteins in L‐EVs and S‐EVs, respectively.
FIGURE 2.
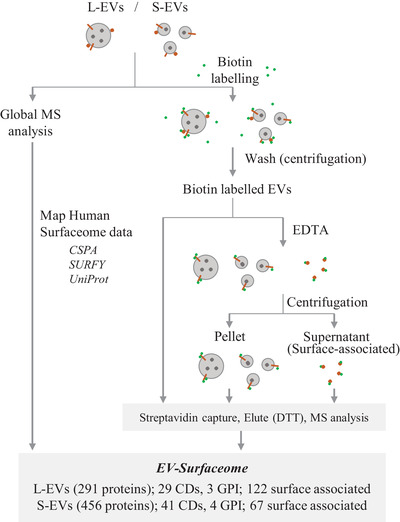
Workflow for capturing surface proteins of large and small EVs. The workflow used in this study to define large and small EV surfaceome is outlined in Figure 3. Experimentally verified cell‐surface proteins (CSPA) (Bausch‐Fluck et al., 2015) and cellular surfaceome predicted by SURFY (Bausch‐Fluck et al., 2018) were mapped onto global EV protein profiles. Surface proteins were captured using membrane‐impermeant Sulfo‐NHS‐SS‐Biotin and subsequently identified by mass spectrometry. We also define a pool of EV‐surface associated proteome by treating EVs with EDTA and assessing released proteins. CD; cluster of differentiation, GPI; Glycosylphosphatidylinositol
3.3. Large EVs contain a sub‐set of cellular surfaceome
We first interrogated distribution of known cellular surface proteins in global proteome of both EV‐subtypes isolated from three different cell lines (SW620, LIM1863, U87) (Figures 3 and S5). In L‐EVs, 2539/4143 proteins were detected in at least 2/3 datasets, of which 284 are cellular surfaceome (CSPA proteins[Bausch‐Fluck et al., 2015]) and 190 predicted as surfaceome (SURFY proteins [Bausch‐Fluck et al., 2018]) (Figure 3a). In S‐EVs, 2449/3086 proteins were detected in at least 2/3 datasets, of which 295 were CSPA and 236 SURFY proteins (Figure 3b). Mapping L‐EVs and S‐EVs data onto the functionally annotated in silico surfaceome tree map revealed striking diversity in protein classes identified, which include transporters, adhesion molecules (cadherins), integrins, ephrins, receptors (including RTKs) and proteases (Figure 3c,d). These proteins include bona fide cell‐surface proteins such as cluster of differentiation (CD) proteins (54 in L‐EVs and 60 in S‐EVs), GPI‐anchored proteins (10 in L‐EVs and 11 in S‐EVs) that do not have transmembrane domains, integrins and membrane transporters (Figure 3c,d and Table S1). Of 638 human surfaceome identified between both EV‐subtypes, 61 and 57 were significantly enriched (q < 0.05) in L‐EVs and S‐EVs, respectively (Figure 3e and Table S1), with their gene ontology (biological process) enrichment analysis presented in Figure S7 Interestingly, we also found that 170/190 SURFY proteins in L‐EVs displayed differential abundance (FDR < 0.05) in L‐EVs from 3 different cell lines (SW620, LIM1863, U87, Figure S8A, Table S3). Moreover, several of these proteins (with higher abundance in L‐EVs from U87 vs. SW620 and LIM1863) were also detected in greater abundance (e.g., APLP2, ERMP1, HM13, LMAN2, RPN1) in U87 L‐EVs as compared to U87 S‐EVs (Figure S8A). Amongst proteins with higher abundance in L‐EVs from CRC cell lines (SW620 or LIM1863) versus U87 L‐EVs, none displayed higher abundance when compared with CRC S‐EVs, however, several of these proteins (MUC13, CDH17, ACE, ACE2, TSPAN8) are reported as intestine‐enriched proteins in Human Protein Atlas (Uhlén et al., 2015) (Figure S8B).
FIGURE 3.

Large EV s contain classical cellular surfaceome. Venn diagram of total proteins identified in (a). L‐EVs or (b). S‐EVs versus surface proteins reported in CSPA (Bausch‐Fluck et al., 2015), SURFY (Bausch‐Fluck et al., 2018) or UniProt. (c) Voronoi tree maps for L‐EV and S‐EV proteomes generated on wlab.ethz.ch/surfaceome. Light colour indicates low expression; dark colour indicates strong expression. White genes are not expressed. (d) Different classes of SURFY (Bausch‐Fluck et al., 2018) predicted cellular surface proteins identified in each EV‐subtype. (e) Volcano plot of differentially abundant surfaceome proteins in L‐EVs and S‐EVs
3.4. Biotin capture of large EVs surface proteome
Next, we employed Sulfo‐NHS‐SS‐Biotin‐based capture coupled with proteomic profiling to ascertain surface localization and identify additional EV surface proteins (not annotated as cellular surfaceome by CSPA/SURFY) (Figure 4 and Table S4). In L‐EVs from different cell lines (SW620, MDA MB 231, U87), 291 proteins were detected in at least two data sets, whereas 456/1365 proteins were identified for S‐EVs (Figures 4a and S9A).
FIGURE 4.

Proteome p rofiling of biotin accessible and inaccessible large EV proteomes. (a) Venn diagram of biotin‐captured surface proteins in L‐EVs versus surface proteins reported in CSPA (Bausch‐Fluck et al., 2015), SURFY (Bausch‐Fluck et al., 2018) or UniProt. (b) Bar plot of relative abundance of number of indicated classes of proteins identified versus total number of proteins identified in global or biotin‐captured surface proteome. (c) Venn diagram of biotin captured L‐EV surface proteins and global L‐EV proteome. Inner circle in L‐EV proteome represents a subset of SURFY/CSPA proteins detected in L‐EV global proteome. (d) Enrichment Map of Gene Ontology (cellular components) terms overrepresented in L‐EV surface proteome versus biotin inaccessible L‐EV proteome. (e) KEGG and Reactome pathways overrepresented in L‐EV surface proteome versus biotin inaccessible L‐EV proteome
An important feature of membrane anchored proteins is the presence of membrane spanning transmembrane domain (TM). Upon cursory inspection, within the biotin‐captured L‐EV proteome, 36/291 proteins were annotated by UniProt to contain at least single TM region and 37/291 as surface PM protein by SURFY (Figure 4a and Table S4); proteins ranged from “containing single” to multiple TM domains (e.g., one TM for EPHA, four for ABCC1, 13 for NPC1L1) (Table S4). In contrast, 238/291 L‐EVs surface biotin proteins not predicted to contain TM domain (by UniProt/SURFY) are cellular surface proteins demonstrated by CSPA (Figure 4a).
Overall, 86/291 proteins are CSPA/SURFY proteins (i.e., plasma membrane proteins) versus 5/291 annotated as non‐surface membrane proteins by SURFY, that is, intracellular origin; this suggests that L‐EVs mainly arise from plasma membrane (Figure 4a and Table S4). We refer to CSPA/SURFY proteins (86/291) as “classical surface proteins” and the rest 205/291 as “non‐classical surface proteins.” Cursory inspection revealed that classical proteins were mainly membrane anchored proteins (e.g., integrins, solute carrier [SLC] transporters, cluster of differentiation [CDs]), whereas non‐classical were proteins that themselves lack TM domain but help in organization of junction proteins (e.g., DSG1, RAC1, RHOA, CTNNA1) and secreted proteins such as annexins, chemokine (S100A8), 14‐3‐3 proteins and enzymes (PGK1) (Table S4).
Differential abundance of biotin surface proteins in S‐EVs and L‐EVs is also provided in Figure S5 and Table S4. We note that compared to L‐EVs, several receptors were enriched in S‐EVs (Figure S9B–E). Moreover, fluorescence microscopy revealed, that consistent with their enrichment in S‐EVs, MET (a receptor tyrosine kinase) co‐localized with endosomal/exosomal markers (EEA1, ALIX and TSG101) (Figure S9F), which supports their active sorting into S‐EVs.
Next, we compared L‐EV surface proteome with global EV proteome; biotin captured proteomes were significantly enriched in CD proteins, receptors and transporters, which further demonstrates successful enrichment of membrane surface proteins (Figure 4b). Importantly, 75 proteins that were annotated as CSPA/SURFY surface proteins in the global L‐EV proteome, we verify as surface accessible proteins (Figure 4c). An additional 168 proteins in global L‐EV proteome not annotated as cell surface proteins were also biotin accessible (Figure 4c). Biotin capture also enabled identification of an additional 48 surface proteins which were not detected in the global EV proteome (Table S5).
3.5. Bioinformatic assessment of biotin accessible/inaccessible proteomes in L‐EVs
We next constructed EnrichmentMap of Cellular Ontologies in 291 biotin captured proteins vs 2052 biotin inaccessible proteins (Figure 4d). Biotin captured proteins were enriched for “plasma membrane” proteins, “membrane raft” proteins, “cell projection,” “leading edge,” and “trailing edge” (Figure 4d and Table 1), which again suggests that these L‐EVs likely arise from budding of plasma membrane. KEGG/Reactome terms enriched include “cell adhesion,” “tight junction,” “glycolysis,” “MAPK signalling,” and “cell‐ECM interactions” (Figure 4e, Tables 1 and S6).
TABLE 1.
Cellular components and functional pathways enriched in L‐EV surface accessible and inaccessible proteome
| Pathways | Number of proteins | G:Profiler terms | ‐Log10 (q‐value) | Gene names | ||
|---|---|---|---|---|---|---|
| Surface biotin captured proteome | Cellular component | Plasma membrane | 177 | GO:0005886 | 25.6 | AFDN, ANXA3, ARPC2, CAP1, CFL1, CTTN, DLG1, EEF2, ENAH, FSCN1, GNAI2, GPI, ITGA2, ITGA6, LMO7, RTN4, SLC16A3, SLC29A1, TPBG, TWF1 |
| Cell surface | 56 | GO:0009986 | 16.6 | ANXA1, ANXA2, HLA‐A, HLA‐B, HSP90AB1, ICAM1, IGHG1, ITGA1, ITGA2, ITGA3, ITGA6, ITGAV, ITGB1, ITGB5, KRT4, LMO7, MSN, PDIA3, RALA, TPBG | ||
| Cell projection | 81 | GO:0120025 | 11.1 | ANXA3, ARPC2, CAPG, CAPZB, CFL1, CTTN, DLG1, ENAH, FSCN1, GNAI2, GNB1, GOT1, GPI, HSP90AB1, ITGA2, ITGA6, LCP1, MSN, TPBG, TWF1 | ||
| Cytoskeleton | 101 | GO:0005856 | 20.7 | ALDOC, ARPC2, CAP1, CAPG, CAPZB, CFL1, CTTN, DLG1, ENAH, FLG, FSCN1, GNAI2, KRT17, LASP1, MYL12A, NPM1, PLS3, POF1B, STOML2, TWF1 | ||
| Membrane raft | 34 | GO:0045121 | 15.3 | ATP1B1, ATP1B3, ATP2B1, BSG, CD177, DLG1, EEF2, GNAI2, GNAI3, ICAM1, ITGA1, LDHB, MYOF, PGK1, S100A10, SLC2A1, SLC9A3R1, STOML2, VCL, VDAC1 | ||
| Cell trailing edge | 5 | GO:0031254 | 3.6 | EZR, FLOT1, FLOT2, MSN, MYH9 | ||
| Functional pathways | Neutrophil degranulation | 67 |
|
31.7 | ADGRE5, CAP1, CD59, DNAJC5, DSC1, DSG1, EEF2, FABP5, FLG2, GSN, IQGAP1, ITGAV, MME, RAC1, RHOA, S100A8, SERPINB6, SPTAN1, TXNDC5, VCL | |
| Glycolysis and gluconeogenesis | 16 | WP:WP534 | 11.6 | ALDOA, ALDOC, ENO1, ENO2, GAPDH, GOT1, GPI, LDHA, LDHB, MDH1, MDH2, PGAM1, PGK1, PKM, SLC2A1, TPI1 | ||
| Regulation of actin cytoskeleton | 29 | KEGG:04810 | 11.4 | ACTB, ACTG1, ACTN1, ACTN4, ARPC2, BAIAP2, CDC42, CFL1, ENAH, EZR, GSN, IQGAP1, ITGA1, ITGA2, ITGA6, ITGB4, MSN, MYH9, MYL12A, PFN1 | ||
| Focal adhesion | 23 | KEGG:04510 | 7.3 | ACTB, ACTG1, ACTN1, ACTN4, CDC42, CTNNB1, FLNA, FLNB, ITGA1, ITGA2, ITGA3, ITGA5, ITGA6, ITGAV, ITGB1, ITGB4, ITGB5, MYL12A, RAC1, RAP1B | ||
| Adherens junction | 14 | KEGG:04520 | 6.9 | ACTB, ACTG1, ACTN1, ACTN4, AFDN, BAIAP2, CDC42, CTNNA1, CTNNB1, IQGAP1, LMO7, RAC1, RHOA, VCL | ||
| Platelet activation & aggregation | 24 |
|
5.6 | ACTN1, ACTN4, ALB, ALDOA, ANXA5, CAP1, CD63, CD9, CDC42, CFL1, FLNA, GNAI3, GNB1, HSPA5, PFN1, PPIA, RAC1, RAP1B, TAGLN2, YWHAZ | ||
| Integrin‐mediated cell adhesion | 15 | WP:WP185 | 4.8 | CAPNS1, CDC42, ITGA1, ITGA2, ITGA3, ITGA5, ITGA6, ITGAV, ITGB1, ITGB4, ITGB5, RAC1, RAP1B, TLN1, VCL | ||
| Biotin inaccessible proteome | Cellular component | Ribosome | 87 | GO:0005840 | 31.1 | EIF2AK2, RPL10, RPL13A, RPL17, RPL18A, RPL19, RPL21, RPL23, RPL24, RPL29, RPL3, RPL31, RPL32, RPL34, RPL37A, RPL6, RPL7, RPL8, RPS26, RPS27L |
| Proteasome complex | 42 | GO:0000502 | 26.7 | ADRM1, ECPAS, PSMB3, PSMC2, PSMC3, PSMC5, PSMC6, PSMD11, PSMD12, PSMD13, PSMD2, PSMD3, PSMD4, PSMD5, PSMD6, PSMD7, PSME1, TXNL1, UCHL5, VCP | ||
| Ribonucleoprotein granule | 55 | GO:0035770 | 8.9 | AGO2, CAPRIN1, CKAP4, DDX3X, DDX6, DHX9, EIF2S1, FMR1, GRB7, HNRNPK, PSMC2, PSMC3, ROCK1, RPL6, RPLP0, RPS4X, RPS6, SERPINB1, TUBB, VCP | ||
| Eukaryotic 43s preinitiation complex | 13 | GO:0016282 | 8.5 | EIF1, EIF3A, EIF3B, EIF3C, EIF3D, EIF3E, EIF3F, EIF3G, EIF3H, EIF3I, EIF3K, EIF3L, EIF3M | ||
| Spliceosomal complex | 45 | GO:0005681 | 7.9 | DDX5, EFTUD2, HNRNPA3, HNRNPC, HNRNPF, HNRNPK, HNRNPM, HNRNPR, HNRNPU, LGALS3, MAGOHB, PABPC1, PRPF8, RALY, RHEB, SNRNP200, SNRPE, SRSF1, U2AF1, UPF1 | ||
| Clathrin‐coated vesicle | 45 | GO:0030136 | 7.7 | AP1B1, AP1G1, AP1S1, AP2A2, AP3B1, BCAP31, CLTA, HIP1R, M6PR, MYO6, PIK3C2A, RAB14, SNX3, SPG21, TMED10, TMED9, VPS11, VPS16, VPS18, VPS33B | ||
| Golgi‐associated vesicle | 28 | GO:0005798 | 6.7 | ACTR1A, ADAM10, AP1G1, AP1S1, ARCN1, CLTA, CLTC, COPA, COPB1, COPB2, COPG1, COPG2, COPZ1, NUCB1, RAB14, SPG21, TMED10, TMED2, TMED7, TMED9 | ||
| COPI‐coated vesicle | 13 | GO:0030137 | 4.6 | ACTR1A, ARCN1, COPA, COPB1, COPB2, COPE, COPG1, COPG2, COPZ1, SEC22B, TMED10, TMED2, TMED7 | ||
| Exocyst | 10 | GO:0000145 | 3.5 | EXOC1, EXOC2, EXOC3, EXOC4, EXOC5, EXOC6, EXOC7, EXOC8, RAB10, SEPTIN2 | ||
| Actin filament | 24 | GO:0005884 | 2.2 | ARPC3, CARMIL1, CORO1B, COTL1, EHBP1L1, FYN, GNG12, MARK2, MYO1B, MYO1C, MYO6, NCKAP1, PDLIM5, SRC, TPM1, TPM2, TWF2, VPS11, VPS18 | ||
| Cytoplasmic side of PM | 30 | GO:0009898 | 2.0 | AKT1, EXOC1, FYN, GNA11, GNA13, GNAI1, GNAS, GNG12, KRAS, LYN, MAP2K2, PKP4, PPP3CA, RAB21, RACGAP1, S100A6, SNX5, SRC, SYK, YES1 | ||
| Chromosome, telomeric region | 29 | GO:0000781 | 1.9 | CBX3, CDK1, FEN1, H3‐3A, HAT1, MACROH2A1, MCM2, MCM4, MCM5, MCM7, PARP1, PCNA, POLR2B, PPP1CA, PPP1CC, PRKDC, PTGES3, RPA2, SSB, XRCC5 | ||
| Rough ER membrane | 10 | GO:0030867 | 1.7 | CCDC47, PLOD1, RPL27, RPS26, SEC61A1, SEC61G, SEC63, SRPRA, SSR4, UBA1 | ||
| Functional pathways | Translation | 140 | REAC:R‐HSA‐72766 | 52.4 | DDOST, RPL10, RPL13A, RPL17, RPL19, RPL21, RPL23, RPL29, RPL3, RPL31, RPL32, RPL34, RPL37A, RPL7, RPL8, RPN1, RPS27L, SEC61A1, SSR1, SSR4 | |
| Membrane trafficking | 174 | REAC:R‐HSA‐199991 | 27.4 | COPE, COPG2, CSNK1D, FNBP1L, LMAN1, LMAN2, RAB1A, SAR1B, SEC13, SEC22B, SEC23A, SEC23IP, SEC24C, SEC31A, TJP1, TMED10, TMED2, TMED7, TMED9, TRIP10 | ||
| Neutrophil degranulation | 135 | REAC:R‐HSA‐6798695 | 21.5 | ACLY, CAPN1, CKAP4, DDOST, DERA, DIAPH1, GGH, GUSB, LTA4H, MLEC, NAPRT, NME2, PDXK, PNP, PRDX4, PTPN6, ROCK1, SLC44A2, UBR4, VCP | ||
| Vesicle‐mediated transport | 176 | REAC:R‐HSA‐5653656 | 20.7 | COPE, COPG2, CSNK1D, FNBP1L, HSPH1, HYOU1, LMAN1, LMAN2, RAB1A, SAR1B, SEC13, SEC22B, SEC23A, SEC24C, SEC31A, TJP1, TMED10, TMED2, TMED7, TRIP10 | ||
| Separation of sister chromatids | 72 | REAC:R‐HSA‐195258 | 18.3 | CKAP5, DYNC1H1, DYNC1I2, DYNC1LI1, DYNLL1, KIF2A, KNTC1, PAFAH1B1, PPP1CC, PPP2CB, PPP2R1A, PSMD5, RCC2, RPS27, SEC13, SMC1A, TUBA1B, TUBB3, TUBB4B, ZW10 | ||
| Ubiquitination & proteasome degradation | 65 | REAC:R‐HSA‐983168 | 3.4 | CUL1, CUL3, DTX3L, GLMN, LNPEP, LRSAM1, NPEPPS, PSMD5, RBX1, RNF114, SKP1, STUB1, THOP1, TPP2, UBA1, UBA5, UBE2L3, UBE2N, UBE2O, UFL1 | ||
| Regulation of actin cytoskeleton | 50 | KEGG:04810 | 3.2 | ARHGAP35, ARPC1B, ARPC5, DIAPH1, GNG12, IQGAP3, ITGB6, KRAS, MAP2K1, MAP2K2, MYL12B, PAK4, PIP4K2A, PIP4K2C, PPP1CA, PPP1CB, PPP1CC, ROCK2, SLC9A1, SSH3 |
We argue that biotin inaccessible proteins represent luminal proteins. In this regard, biotin inaccessible proteins were enriched for “COPI/II‐coated vesicles,” “proteasome,” “ribosomes,” “EIF complex,” “spliceosome,” and “ER/Golgi” (Figure 4d and Table 1). These proteins were implicated in “translation,” “protein processing in ER,” and “MAPK signalling cascade” (Figure 4e). While how these proteins/structures are sorted into EVs is not understood, enrichment of “membrane trafficking,” and “vesicle‐mediated transport” in biotin inaccessible proteome supports for an active sorting mechanism.
3.6. Surface associated proteins of L‐EVs
Previous studies have shown that a pool of proteins can associate with EV surface and are functional (Costa‐Silva et al., 2015; Ringuette Goulet et al., 2018; Rogers et al., 2020; Sung et al., 2015). Presence of CSPA/SURFY proteins (86/291 as classical surface proteins) versus 205/291 as non‐CSPA/SURFY proteins strongly suggests that L‐EVs also contain a subset of proteins that are associated on the surface. Indeed, we found that EDTA‐treatment effectively released 122 proteins (now referred to as EV surface‐associated) from the surface of L‐EVs (Figure 5a, Tables 2 and S4), including annexins and 14‐3‐3 proteins (Figure 5b), but not membrane proteins including CDs, receptors and transporters (Figure 5c).
FIGURE 5.

Surface‐associated proteins in large EVs. (a) Venn diagram of proteins identified in L‐EV surface proteome, and proteins that remain either bound or are released from L‐EV surface following EDTA treatment. Bar plot of protein numbers of indicated classes that are either released (b) or remain bound (c) with EVs following EDTA treatment. (d) L‐EV biotin surface proteome was divided into CSPA/SURFY proteins (i.e., classical cellular surfaceome) or non‐CSPA/SURFY proteins (i.e., non‐classical surface proteins); Venn diagram reveals pool of these proteins that remain either bound or are released from L‐EV surface following EDTA treatment. (e) EnrichmentMap of KEGG and Reactome pathways overrepresented in L‐EV proteins that remain either bound or are released from L‐EV surface following EDTA treatment. (f) GeneMania‐based radial interaction map of membrane‐bound and membrane‐associated (i.e., released) proteins in L‐EVs. Nodes represent the proteins, and the edges represent evidence‐based direct physical interactions. Pathway involvement for indicated proteins are asterisk‐colour coded
TABLE 2.
L‐EV and S‐EV surface‐bound and surface‐associated proteome
| L‐EVs | S‐EVs | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene_Name | Protein_Name | Uniprot ID | Biotin (n = 9) | Bound (n = 4) | Released (n = 4) | Biotin (n = 9) | Bound (n = 4) | Released (n = 4) | CSPA | CDs | Membranome Almen main‐class |
| ACTB | Actin, cytoplasmic 1 | ACTB_HUMAN | 5 | 2 | 6 | 1 | 1 | Y | |||
| ATP1B1 | Sodium/potassium‐transporting ATPase subunit beta‐1 | AT1B1_HUMAN | 5 | 3 | 8 | 1 | Y | Transporters | |||
| ATP1B3 | Sodium/potassium‐transporting ATPase subunit beta‐3 | AT1B3_HUMAN | 6 | 4 | 8 | 1 | Y | CD298 | Transporters | ||
| BSG | Basigin | BASI_HUMAN | 8 | 4 | 9 | 3 | 1 | Y | CD147 | Unclassified | |
| CALR | Calreticulin | CALR_HUMAN | 4 | 2 | 2 | Y | |||||
| CAPZA1 | F‐actin‐capping protein subunit alpha‐1 | CAZA1_HUMAN | 3 | 2 | 6 | 2 | 1 | ||||
| CAPZA2 | F‐actin‐capping protein subunit alpha‐2 | CAZA2_HUMAN | 3 | 2 | 5 | 1 | |||||
| CD44 | CD44 antigen | CD44_HUMAN | 3 | 2 | 7 | Y | CD44 | Unclassified | |||
| CD59 | CD59 glycoprotein | CD59_HUMAN | 3 | 2 | 8 | 1 | Y | CD59 | |||
| EPCAM | Epithelial cell adhesion molecule | EPCAM_HUMAN | 3 | 3 | 6 | 2 | Y | CD326 | Unclassified / GPI | ||
| F11R | Junctional adhesion molecule A | JAM1_HUMAN | 7 | 3 | 7 | 2 | Y | CD321 | Miscellaneous | ||
| FLOT1 | Flotillin‐1 | FLOT1_HUMAN | 8 | 4 | 7 | 1 | |||||
| FLOT2 | Flotillin‐2 | FLOT2_HUMAN | 5 | 3 | 6 | 2 | |||||
| ICAM1 | Intercellular adhesion molecule 1 | ICAM1_HUMAN | 4 | 2 | 7 | 1 | Y | CD54 | Unclassified | ||
| ITGA2 | Integrin alpha‐2 | ITA2_HUMAN | 5 | 4 | 9 | 2 | 1 | Y | CD49b | Receptors | |
| ITGA3 | Integrin alpha‐3 | ITA3_HUMAN | 4 | 2 | 8 | Y | CD49c | Receptors | |||
| ITGA5 | Integrin alpha‐5 | ITA5_HUMAN | 3 | 2 | 7 | 2 | Y | CD49e | Receptors | ||
| ITGAV | Integrin alpha‐V | ITAV_HUMAN | 4 | 2 | 8 | 2 | Y | CD51 | Receptors | ||
| ITGB1 | Integrin beta‐1 | ITB1_HUMAN | 8 | 4 | 9 | 3 | Y | CD29 | Receptors | ||
| LAMP1 | Lysosome‐associated membrane glycoprotein 1 | LAMP1_HUMAN | 3 | 2 | 6 | 1 | Y | CD107a | Receptors | ||
| MELTF | Melanotransferrin | TRFM_HUMAN | 4 | 3 | 7 | 1 | Y | CD228 | |||
| MFGE8 | Lactadherin | MFGM_HUMAN | 3 | 2 | 7 | 1 | Y | GPI anchored | |||
| MVP | Major vault protein | MVP_HUMAN | 5 | 2 | 6 | ||||||
| P4HB | Protein disulfide‐isomerase | PDIA1_HUMAN | 5 | 2 | 4 | 1 | |||||
| PDIA6 | Protein disulfide‐isomerase A6 | PDIA6_HUMAN | 4 | 3 | 5 | ||||||
| PLEC | Plectin | PLEC_HUMAN | 3 | 2 | 5 | 1 | |||||
| SET | Protein SET | SET_HUMAN | 3 | 2 | 1 | ||||||
| SLC3A2 | 4F2 cell‐surface antigen heavy chain | 4F2_HUMAN | 8 | 4 | 9 | 3 | 1 | Y | CD98 | Transporters | |
| SLC7A5 | Large neutral amino acids transporter small subunit 1 | LAT1_HUMAN | 6 | 3 | 7 | 2 | 1 | Y | Transporters | ||
| TFRC | Transferrin receptor protein 1 | TFR1_HUMAN | 4 | 2 | 6 | 1 | Y | ||||
| TXN | Thioredoxin | THIO_HUMAN | 5 | 2 | 6 | 2 | |||||
| ACTC1 | Actin, alpha cardiac muscle 1 | ACTC_HUMAN | 6 | 3 | 3 | 4 | 1 | 1 | |||
| ACTG1 | Actin, cytoplasmic 2 | ACTG_HUMAN | 9 | 4 | 4 | 9 | 4 | 4 | |||
| ACTN4 | Alpha‐actinin‐4 | ACTN4_HUMAN | 9 | 4 | 3 | 9 | 3 | 1 | Y | ||
| ALB | Albumin | ALBU_HUMAN | 9 | 4 | 4 | 8 | 3 | 3 | |||
| ALDOA | Fructose‐bisphosphate aldolase A | ALDOA_HUMAN | 8 | 4 | 3 | 9 | 3 | 2 | |||
| ANXA1 | Annexin A1 | ANXA1_HUMAN | 8 | 4 | 3 | 9 | 3 | 1 | |||
| ANXA2 | Annexin A2 | ANXA2_HUMAN | 9 | 4 | 4 | 9 | 4 | 4 | |||
| ANXA5 | Annexin A5 | ANXA5_HUMAN | 7 | 4 | 3 | 9 | 2 | 1 | |||
| AZGP1 | Zinc‐alpha‐2‐glycoprotein | ZA2G_HUMAN | 3 | 3 | 3 | 6 | 1 | ||||
| CALML5 | Calmodulin‐like protein 5 | CALL5_HUMAN | 7 | 4 | 3 | 5 | 2 | 3 | |||
| CASP14 | Caspase‐14 | CASPE_HUMAN | 7 | 4 | 3 | 7 | 2 | 3 | |||
| CAT | Catalase | CATA_HUMAN | 5 | 3 | 3 | 6 | 1 | 2 | |||
| DCD | Dermcidin | DCD_HUMAN | 6 | 3 | 3 | 7 | 2 | 3 | |||
| DSC1 | Desmocollin‐1 | DSC1_HUMAN | 6 | 4 | 3 | 7 | 1 | 3 | Miscellaneous | ||
| DSG1 | Desmoglein‐1 | DSG1_HUMAN | 8 | 4 | 4 | 9 | 2 | 4 | Miscellaneous | ||
| DSP | Desmoplakin | DESP_HUMAN | 9 | 4 | 3 | 9 | 3 | 4 | |||
| ENO1 | Alpha‐enolase | ENOA_HUMAN | 7 | 4 | 3 | 9 | 3 | 2 | Y | ||
| GSTP1 | Glutathione S‐transferase P | GSTP1_HUMAN | 4 | 4 | 3 | 7 | 1 | 1 | |||
| H4C1 | Histone H4 | H4_HUMAN | 3 | 4 | 3 | 5 | 1 | ||||
| HSPA8 | Heat shock cognate 71 kDa protein | HSP7C_HUMAN | 7 | 4 | 3 | 9 | 4 | 2 | Y | ||
| JUP | Junction plakoglobin | PLAK_HUMAN | 9 | 4 | 3 | 9 | 2 | 3 | |||
| MSN | Moesin | MOES_HUMAN | 9 | 4 | 3 | 9 | 3 | 3 | |||
| MYH6 | Myosin‐6 | MYH6_HUMAN | 7 | 3 | 3 | 5 | 1 | ||||
| MYH9 | Myosin‐9 | MYH9_HUMAN | 8 | 4 | 3 | 8 | 2 | 1 | Y | ||
| PDIA3 | Protein disulfide‐isomerase A3 | PDIA3_HUMAN | 7 | 3 | 4 | 7 | 1 | ||||
| PGK1 | Phosphoglycerate kinase 1 | PGK1_HUMAN | 7 | 4 | 3 | 9 | 2 | 2 | |||
| PKM | Pyruvate kinase PKM | KPYM_HUMAN | 5 | 4 | 3 | 9 | 2 | 2 | Y | ||
| PKP1 | Plakophilin‐1 | PKP1_HUMAN | 7 | 3 | 3 | 6 | 1 | 2 | |||
| SERPINB12 | Serpin B12 | SPB12_HUMAN | 6 | 3 | 4 | 6 | 1 | 3 | |||
| SFN | 14‐3‐3 protein sigma | 1433S_HUMAN | 5 | 3 | 3 | 7 | 3 | 2 | |||
| TGM1 | Protein‐glutamine gamma‐glutamyltransferase K | TGM1_HUMAN | 6 | 3 | 3 | 7 | 2 | 2 | |||
| TGM3 | Protein‐glutamine gamma‐glutamyltransferase E | TGM3_HUMAN | 7 | 3 | 3 | 7 | 1 | 3 | |||
| TPI1 | Triosephosphate isomerase | TPIS_HUMAN | 8 | 4 | 3 | 9 | 3 | 1 | Y | ||
| VCL | Vinculin | VINC_HUMAN | 6 | 3 | 3 | 8 | 2 | 1 | |||
| YWHAE | 14‐3‐3 protein epsilon | 1433E_HUMAN | 7 | 4 | 3 | 9 | 3 | 2 | |||
| YWHAZ | 14‐3‐3 protein zeta/delta | 1433Z_HUMAN | 9 | 4 | 3 | 9 | 3 | 2 | |||
EV surface‐associated proteome included both classical and non‐classical surface proteins (Figure 5d); we noted that a subset of both classical and non‐classical proteins could be partly released by EDTA treatment. Bound classical L‐EV surface proteins that are prone to removal by EDTA (24, set 1) are predominantly non‐membrane anchored and include actinins (ACTN1/4) and enzymes (GAPDH, PKM, ENO1, PGK), whereas bound non‐classical proteins (non‐CSPA/SURFY proteins) that are removed by EDTA (83, set 2) include annexins, enzymes (CALM3/5, CAT, LDHA/B, TGM1/3), junction assembly proteins (DSG1), cytoskeleton proteins (EZR, ACTC1/G1, MSN), chemokine (S100A9), 14‐3‐3 proteins; these proteins are implicated in “glycolysis,” “regulation of actin cytoskeleton,” “junction organization,” and “platelet activation” (Figure 5d,e and Table S7). In contrast, bound classical cellular surface proteins on L‐EVs resistant to removal by EDTA (43, Figure 5d set 3) were membrane proteins (CDs, receptors such as integrins, LAPM1, SLC transporters, GPI anchored proteins), which are implicated in “adhesion” and “ECM organization” (Figure 5e and Table S7). While we do not understand how these surface associated proteins bind to L‐EV surface, they have been previously reported to physically interact with several proteins of classical surface proteome (Figure 5f and Table 2). Furthermore, we also noted that 93/291 biotin‐capture L‐EV proteome displayed differential abundance in L‐EVs from different cell lines (SW620, U87, MDA MB 231, Table S8) which also includes several proteins (CTNNB1, EZR, SLCA5, SLC9A3R1, VIL1) that were readily removed by EDTA (Figure S10).
3.7. Bioinformatics assessment of large EV surface proteome interactome
Interaction of surface proteins in cancer S‐EVs to their cognate binding partners at distal sites have been shown to dictate EV‐homing to specific organs (Hoshino et al., 2015) to develop pre‐metastatic niches and enhance metastasis (Costa‐Silva et al., 2015; Grange et al., 2011; Hood et al., 2011; Hoshino et al., 2015; Jung et al., 2009; Peinado et al., 2012). To gain insight into potential interactome of the surfaceome of cancer L‐EVs, we next employed manually curated resource Cellinker that catalogues literature‐supported ligand‐receptor interactions (Zhang, Wang, et al., 2021) to retrieve potential surface engagements for 52 classical surfaceome (detected in >3 biotin capture experiments and annotated as SURFY/CSPA proteins). This resulted in 172 experimentally verified binding partners, encompassing 108 cell‐adhesion interactions, 65 ECM‐receptor interactions, 39 cytokine‐cytokine receptor interactions and 36 secreted protein‐receptor interactions (Figure 6a–c and Table S9). Protein receptor interactions for L‐EV surface‐associated proteins are listed in Table S10. DAVID‐based analysis of the cognate binding partners revealed tissue enrichment for liver (Figure 6d). These interactions involved L‐EVs ITGAV/ITGB1 with ANGPTL3, PLG and VTN, which display tissue enrichment in the liver (Figure 6e,f), a primary metastatic site for colorectal and breast cancers (Chen et al., 2017; Ku et al., 2019; Liu et al., 2018). Because exosomal integrin ITGAV/B5 was shown to be linked to liver metastasis (Hoshino et al., 2015), whether both EV‐subtypes may share similar organ‐specific homing and enhance metastasis warrants future investigation.
FIGURE 6.

Receptor‐ligand interactome of large EV surface proteome. (a) Venn diagram of L‐EV biotin surface proteins (classical) and their experimentally‐verified cognate binding partners based on Cellinker that catalogues literature‐supported ligand‐receptor interactions (C. Zhang, Wang, et al., 2021). (b) Distribution of interaction‐type identified in L‐EVs. (c) Sankey diagram of ligand‐receptor interactions identified in L‐EVs; complete list provided in Table S7. (d) Bar plot represents tissue enrichment (based on DAVID analysis) of 172 cognate interacting partners of L‐EV surface proteins. (e) Cellinker‐based interacting ligands identified for L‐EV surface ITGB1/ITGAV proteins. (f) RNA expression and tissue specificity for ANGPTL3, PLG and VTN in different human tissues was obtained from The Human Protein Atlas (http://www.proteinatlas.org, image credit: Human Protein Atlas [Uhlén et al., 2015])
4. DISCUSSION
In this study, we found that the protein diversity on surface of L‐EVs (buoyant density of 1.07–1.11 g/ml) includes bona fide plasma membrane‐anchored proteins (CDs, transporters, receptors) implicated in cell‐cell and cell‐ECM interactions, as well as membrane surface‐associated proteins (actin cytoskeleton components, enzymes, chemokines) implicated in actin filament organization, platelet activation and glycolysis. A pre‐requisite for successful identification of surface proteins by membrane impermeant Sulfo‐NHS‐SS‐biotin, a well‐established agent which has been extensively used to identify cell surface proteins (Li et al., 2019) as well as small EVs without compromising their integrity (Cvjetkovic et al., 2016; Diaz et al., 2016), is purified EV preparations that are morphologically intact as demonstrated in Figure 1. Significant enrichment of bona fide membrane proteins in affinity‐purification using avidin‐beads, as compared to global L‐EV proteome, indicates high surface labeling specificity (Figures 3 and 4). Based on these results, we are confident that our experimental data reliably provides a comprehensive map of L‐EV surfaceome.
Expectedly, L‐EVs surface proteome comprised proteins of TM domain containing surface receptors, CDs, transporters and GPI anchored proteins. Several of these surface proteins are drivers of cancer progression from primary tumour growth, remodelling of the tumour microenvironment, invasive outgrowth (MET [Peinado et al., 2012], EGFR [Liu et al., 2009]), immune regulation and metastatic spread (integrins [Sökeland & Schumacher, 2019]). The diversity of transporters encompassing ABC transporters, metal transporters (CNNM3/4), lactate transporters (SLC16A3), carboxylic acids transporters (SLC16A1/MTC) and SLC transporters (amino acid). S‐EV‐mediated horizontal transfer of membrane‐embedded drug efflux pumps to sensitive cancer cells is well‐known, leading to acquired drug resistance in vitro and in vivo (Bebawy et al., 2009; Levchenko et al., 2005; Lv et al., 2014; Sousa et al., 2015; Zhao et al., 2021). Besides their role in chemoresistance, membrane transporters also dictate organ tropism of circulating EVs (Wu et al., 2020).
Resonating S‐EVs capturing tissue‐specific signatures of organs they originate from Hoshino et al. (2020), we also noted that L‐EVs from different cell lines clustered based on donor cells (Figure S5). Moreover, we identified 170/190 SURFY proteins in L‐EVs displaying differential abundance (FDR < 0.05) in L‐EVs based on donor cells (SW620, LIM1863, U87). Several of these proteins, for example, ICAM1 (Piao et al., 2017), SLC9A1 (Guan et al., 2018), and TPBG (Zhang, Liu, et al., 2021) enriched in U87 L‐EVs (vs. SW620 and LIM186 L‐EVs) have been implicated in glioblastoma disease progression. Interestingly, similar abundance was observed in U87‐derived S‐EVs and L‐EVs for these proteins (ICAM1, SLC9A1, TPBG), highlighting potential of both EV‐subtypes to recapitulate parental cell proteome. On the other hand, several proteins (APLP2, HM13, LMAN2 and RPN1) displayed higher abundance in U87 L‐EVs compared to U87 S‐EVs. HM13 was recently shown to enhance tumour progression by regulating secretion profile in glioblastoma (Wei et al., 2017). APLP2 (Amyloid precursor‐like protein 2, a type I transmembrane protein and a member of the amyloid precursor protein family) have been shown to exert oncogenic function in diverse types of cancer (Pandey et al., 2016). Although APLP2 expression is similar between glioblastoma and normal brain, however, a high level of APLP2 is associated with poor prognosis in the glioblastoma patient group (Chen et al., 2018). On the other hand, amongst proteins with higher abundance in L‐EVs from CRC cell lines (SW620 or LIM1863) versus U‐87 L‐EVs, none displayed greater abundance when compared with CRC cell line S‐EVs, however, several of these proteins (MUC13, CDH17, ACE, ACE2, TSPAN8) are reported as intestine‐enriched proteins in Human Protein Atlas (Uhlén et al., 2015). Moreover, in our study, upon manual inspection, we noted that colon cancer cell‐derived EVs (compared to EVs derived from U87 glioma or MDA MB 231 breast cancer cells) carried 17 intestinal specific proteins, including ACE/CD143, CDHR2 and MUC13. CDHR2 is a cell adhesion molecule expressed in intestinal epithelium in a tissue‐specific way (https://www.proteinatlas.org/ENSG00000074276‐CDHR2/tissue) (Uhlén et al., 2015) and has the potential to diagnose gastrointestinal adenocarcinomas (Panarelli et al., 2012; Su et al., 2008). Thus, L‐EV surface proteins such as CDHR2 could potentially enable capture/detection of intestinal EV sub‐population (amongst the bulk circulating EVs). Combined with our previous report of L‐EVs carrying oncogenic KRAS (G12V mutant variant) (Rai, Greening, et al., 2021), L‐EVs surfaceome could potentially provide immunocapture‐based avenue for cancer biomarker diagnostics.
Besides transmembrane anchored proteins, we also identified proteins that were associated with the surface of L‐EVs. Several surface‐associated proteins have been documented to dictate EV function; whereas EV surface ANXA1 promotes microcalcification (Rogers et al., 2020), FN1 facilitates cellular migration through tissues (Sung et al., 2015), TGFB1 triggers fibroblast differentiation (Ringuette Goulet et al., 2018) and MIF establishes pre‐metastatic niche in the liver (Costa‐Silva et al., 2015). We show that major components of glycolytic pathways were also present on L‐EV surface (ALDH7A1, ALDOA, ENO1, GAPDH, LDHA, LDHB, PGAM1, PGK1, PKM, TPI1). Secretion of glycolytic enzymes (GAPDH, PKM, TPI1) in sera of colon cancer patients has been associated with 5‐fluorouracil resistance (Shin et al., 2009). The mechanism of glycolytic protein secretion is only beginning to emerge, which includes export via EVs (exosomes via tetraspanins (Perez‐Hernandez et al., 2013) and shed microvesicles via caveolin‐1 (Raikar et al., 2006)) or via SNARE‐driven unconventional secretion (Miura et al., 2012). These proteins then localize to surface of cells and/or potentially EVs (Gómez‐Arreaza et al., 2014). Besides their well‐known role in glycolysis, these proteins (GAPDH, TPI, PKM) also carry out multiple moonlighting functions in cancer such as attachment (Gründel et al., 2016), proliferation (Hsu et al., 2016) and migration via surface localization (Gómez‐Arreaza et al., 2014). Currently, how glycolytic proteins associate with cell or EV outer surface remains unknown, but are potentially mediated by direct interactions with surface proteins (Figure 6); indeed, extracellular PKM is a ligand for cell surface EGFR (Hsu et al., 2016).
We also found that non‐classical surfaceome of L‐EVs include proteins that assist in the assembly of junction complex (DLG1/3, CTNNA1/B1/D1, RHOA, NECTIN2, VCL) and components of actomyosin bundle (ACTN1, ACTN4, MYH6, MYH9) (Table 1). While these are classically annotated in the cytoplasmic side of plasma membrane, they were also removed from EV surface following EDTA treatment (Table 1); likely due to a subpopulation of EVs displaying inside‐out topology (Cvjetkovic et al., 2016). Along this line, L‐EV surface associated proteins also include actin and actin binding proteins that are typically intracellular proteins. While cytosolic proteins likely originate from damaged cells, attach to the surface of EVs and are hence co‐purified, it is not uncommon for cytosolic proteins such as functional histones to be released extracellularly and bind to extracellular structure such as neutrophil extracellular traps or protein granules (Nakazawa et al., 2017; Yousefi et al., 2020). Alternatively, actins are also found extracellularly, co‐localise with and is a ligand for TREM‐1 (a potent amplifier of pro‐inflammatory innate immune response) on the surface of activated mouse macrophage (Fu et al., 2017). Our current data also concurs our previous finding that actins are peripheral surface exposed proteins on S‐EVs removed by proteinase K treatment (Xu et al., 2019), a finding that is also corroborated by other groups employing biotin labelling of surface proteins (Castillo et al., 2018; Cvjetkovic et al., 2016). In contrast, classical CD63+, CD81+, and CD9+ S‐EVs were shown to lack cytoskeletal constituents that make up actin filaments, microtubules, or intermediate filaments (Jeppesen et al., 2019), however we have previously shown that cytoskeleton constituents including actins are found in EPCAM+/A33+ S‐EVs (Tauro et al., 2013).
EVs can arise from different parts of cells (e.g., exosomes are endosomally derived, microvesicles are plasma membrane derived, midbody remnants are derived from cytokinetic bridges), with several extracellular EV‐subtypes being discovered whose route of biogenesis is yet to be unravelled. Based on our data, we propose that the majority of L‐EVs arise from plasma membrane as we see striking enrichment of plasma membrane proteins in L‐EV surfaceome. Their involvement in “cell projection,” “leading edge,” and “trailing edge” (Figure 4d and Table 1) also suggests that a subset of L‐EVs could also represent EVs released by migrating cells also known as migrasomes (Jiang et al., 2019). It should be noted that although midbody remnants are also L‐EVs by definition (200–600 nm) (Rai, Greening, et al., 2021), midbody remnants display higher buoyant density (1.22–1.30 g/ml) compared to L‐EVs purified in this study (1.07–1.11 g/ml). Whether these L‐EV subtypes have distinct surfaceome warrants future investigation.
Phenotypic diversity in EVs is gaining vested interest with S‐EVs carrying multiple smaller EVs within its lumen.: such morphological diversity is observed in EVs released by single cell‐type (Théry et al., 2018) as well isolated from body fluids (plasma [Gallart‐Palau et al., 2015], serum [Chen et al., 2013], cerebrospinal fluid [Chen et al., 2013; Emelyanov et al., 2020] and semen [Poliakov et al., 2009]). Some studies suggest that such EV phenotypic diversity could be an artefact due to high speed centrifugation (Issman et al., 2013), however, cells have been shown to release EVs encapsulating smaller EVs (Wiklander et al., 2015; Zabeo et al., 2017), with such EV diversity also evident in unprocessed EVs from ejaculates (Höög & Lötvall, 2015)., In our data, assessment of biotin inaccessible proteins (enriched for “COPI/II‐coated vesicles,” “ribosomes” that are associated with “ER/Golgi”) strongly suggest that sub‐populations of L‐EVs could potentially contain smaller EVs within its lumen. Indeed, mitochondria can be actively sorted into plasma membrane blebs that are subsequently released in EVs (Islam et al., 2012; Nicolás‐Ávila et al., 2020). Alternatively, L‐EVs not only encapsulate mitochondria but also co‐purify with free mitochondria themselves (Puhm et al., 2019). While enrichment of mitochondrial proteins in L‐EVs is in accord with our previous findings (Xu et al., 2015), we did not find discernible cristae‐containing mitochondria from our cryo EM imaging. In our data, we do note that VDAC1/2 were also labelled with biotin, thus L‐EV subtype, in the absence of cristae phenotypes, could potentially include mitochondria‐derived vesicles (VDAC enriched, ∼200 nm, lack cristae) (Soubannier et al., 2012) that package mitochondrial matrix proteins (hence explaining their biotin inaccessibility). Mito‐vesicles are known to selectively package mitochondrial proteins (exclude complex proteins and TOM20 which were also not detected in L‐EV biotin surface proteome) (Soubannier et al., 2012) and are released extracellularly (D'Acunzo et al., 2021). Although, we do not know the mechanisms underlying their biogenesis, intracellular mitovesicles are targeted either to lysosomes or MVB (Matheoud et al., 2016; Sugiura et al., 2014) which could facilitate their release to extracellular space.
In summary, our study provides a first comprehensive map of L‐EV surface proteins. While density gradient‐based purifications of EVs are regarded as a gold‐standard by ISEV (Théry et al., 2018) and can separate EVs from soluble secreted proteins and heavy density EVs/protein aggregates, it is now apparent that other cellular organelles could potentially share biophysical properties which results in co‐enrichment and preclude L‐EV assessment for their form and function. Moreover, while size‐based (i.e., large versus small) broad categorization of EVs is useful at an operational level, future dissection calls for immunocapture of L‐EV sub‐populations if we are to resolve their origins, cargo and function ((an approach successfully used to classify S‐EV subpopulations) successfully used to classify S‐EV subpopulations).
AUTHOR CONTRIBUTION
A.R. and D.W.G conceptualised the idea. A.R. designed the experiments and wrote the manuscript. A.R., H.F., B.C. and D.W.G. performed experiments. A.R. performed bioinformatics analysis A.R., H.F., B.C., R.J.S. and D.W.G. authors reviewed manuscript for submission.
COMPETING INTERESTS
The authors have no conflicts to declare.
DATA AND SOFTWARE AVAILABILITY
The MS‐based proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (Jarnuczak & Vizcaino, 2017) and are available via ProteomeXchange with identifier PXD024733. Experimental parameters submitted to EV‐TRACK knowledgebase (EV‐TRACK ID: EV210261) (Van Deun et al., 2017).
Supporting information
Supplementary Figure 1. A. Immunoblot detection of CD63 and ANXA1 in large and small EVs purified using density‐gradient separation from conditioned media of SW620 cells. B. Immunoblot detection of CD63 and ANXA1 in MDA MB 231 whole cell lysate and derived large and small EVs.
Supplementary Figure 2. Nanoparticle tracking analysis of purified large and small EVs from indicated cell lines.
Supplementary Figure 3. Nanoparticle tracking analysis data of purified large and small EVs from indicated cell lines represented as log10 transformed data (concentration (particle/mL) and diameter (size, nm).
Supplementary Figure 4. Bar plot depicting percentage of EVs of indicated size (>200 nm, >300 nm, >400 nm) within each EV‐subtype from indicated cell lines based on A. cryo‐EM analysis or B. single particle tracking (NTA) data.
Supplementary Figure 5. A. Venn diagram of proteins identified in L‐EVs and S‐EVs. B. Violin plot of LFQ intensities of proteins identified in L‐EVs and S‐EVs. C. Pearson correlation matrix of L‐EV and S‐EV proteomes for indicated cell lines.
Supplementary Figure 6. Bar plot depicting normalised LFQ intensities of indicated proteins in L‐EVs or S‐EVs from three cell lines MDA MB 231, SW620 and U87. p‐value < 0.05 (*), 0.005 (**), 0.001 (***), 0.0001 (****), ns representing non‐significant.
Supplementary Figure 7. Bar plot of Gene ontology‐based biological processes overrepresented in surface proteins enriched in L‐EVs or S‐EVs. Middle panel represents biological processes enriched in surface proteins commonly identified in both L‐EVs and S‐EVs.
Supplementary Figure 8. A. Hierarchical clustering analysis of differentially abundant SURFY proteins (FDR < 0.05) in global proteome of L‐EVs from U87, SW620 and LIM1863 cell lines. Corresponding boxes list proteins that are found in associated clusters. *Proteins that are found in higher abundance in U87 L‐EVs vs U87 S‐EVs. B. Venn diagram of Human Protein Atlas‐based “brain enriched” or “intestine enriched” proteins and Cluster 2 (U87 enriched) and CRC cell lines enriched (Cluster 1/3/4) proteins. Bar plots depicts LFQ intensities of indicated proteins in L‐EVs and S‐EVs from LIM1863.
Supplementary Figure 9. A. Venn diagram of biotin‐captured surface proteins in S‐EVs versus surface proteins reported in CSPA (Bausch‐Fluck et al., 2015), SURFY (Bausch‐Fluck et al., 2018) or UniProt. B. Relative abundance of transporters, CDs and receptors in L‐EVs and S‐EVs biotin‐captured proteome. C. Volcano plot of differentially abundant biotin‐captured receptors in L‐EVs and S‐EVs. D. Relative abundance of LFQ intensities of indicated proteins in S‐EVs versus L‐EVs. E. Western blot analysis of L‐EVs and S‐EVs released by SW620 cells using indicated antibodies. F. Fluorescence microscopic analysis of SW620 cells expressing plasma membrane targeting GAP‐GFP proteins using indicated antibodies.
Supplementary Figure 10. Heat map of differentially abundant biotin captured proteins (FDR < 0.05) in L‐EVs from U87, SW620 and MDA MB 231 cell lines. Corresponding boxes list proteins that are found in associated clusters. *Proteins that are released from MDA MB 231 L‐EVs following EDTA treatment.
Supplementary Table 1. Global proteomic analysis of L‐EVs and S‐EVs
Supplementary Table 2. Gene ontology (cellular component) enriched in L‐EVs and S‐EVs
Supplementary Table 3. Differentially abundant SURFY proteins in L‐EVs
Supplementary Table 4. Proteomic analysis of EV‐surface proteins captured using membrane impermeant biotin
Supplementary Table 5. List of surface proteins identified in L‐EVs
Supplementary Table 6. Pathways enriched in L‐EVs and S‐EVs
Supplementary Table 7. List of proteins from Figure 5D
Supplementary Table 8. Differentially abundant biotin captured proteins in L‐EVs
Supplementary Table 9. Receptor‐ligand interactome of L‐EV surface proteins
Supplementary Table 10. Receptor‐ligand interactome of L‐EV surface‐associated proteins
ACKNOWLEDGMENTS
We thank Bio21 Molecular Science and Biotechnology Institute for assisting cryo‐electron microscopy (University of Melbourne). This work was supported by National Health and Medical Research Council Project grants 1057741 and 1139489 (to D.W.G), and Medical Research Future Fund grant 1201805 (to D.W.G). We note further support by Helen Amelia Hains Fellowship (to D.W.G), and by the Victorian State Government Operational Infrastructure funding to the Baker Institute. BC is supported by an Australian Government Training Program (RTP) scholarship and Baker Institute Bright Sparks Scholarship Top Up.
Rai, A. , Fang, H. , Claridge, B. , Simpson, R. J. , & Greening, D. W. (2021). Proteomic dissection of large extracellular vesicle surfaceome unravels interactive surface platform. Journal of Extracellular Vesicles, 11, e12164. 10.1002/jev2.12164
Contributor Information
Alin Rai, Email: alin.rai@baker.edu.au.
David W Greening, Email: david.greening@baker.edu.au.
REFERENCES
- Al‐Nedawi, K. , Meehan, B. , Micallef, J. , Lhotak, V. , May, L. , Guha, A. , & Rak, J. (2008). Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nature Cell Biology, 10, 619–624. [DOI] [PubMed] [Google Scholar]
- Bausch‐Fluck, D. , Goldmann, U. , Müller, S. , Van Oostrum, M. , Müller, M. , Schubert, O. T. , & Wollscheid, B. (2018). The in silico human surfaceome. PNAS, 115, E10988–E10997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bausch‐Fluck, D. , Hofmann, A. , Bock, T. , Frei, A. P. , Cerciello, F. , Jacobs, A. , Moest, H. , Omasits, U. , Gundry, R. L. , Yoon, C. , Schiess, R. , Schmidt, A. , Mirkowska, P. , Härtlová, A. , Van Eyk, J. E. , Bourquin, J.‐P. , Aebersold, R. , Boheler, K. R. , Zandstra, P. , & Wollscheid, B. (2015). A mass spectrometric‐derived cell surface protein atlas. Plos One, 10, e0121314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebawy, M. , Combes, V. , Lee, E. , Jaiswal, R. , Gong, J. , Bonhoure, A. , Grau, G. E. R. (2009). Membrane microparticles mediate transfer of P‐glycoprotein to drug sensitive cancer cells. Leukemia, 23, 1643–1649. [DOI] [PubMed] [Google Scholar]
- Bellin, G. , Gardin, C. , Ferroni, L. , Chachques, J. , Rogante, M. , Mitrečić, D. , Ferrari, R. , & Zavan, B. (2019). Exosome in cardiovascular diseases: A complex world full of hope. Cells, 8, 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzás, E. I. , Tóth, E. Á. , Sódar, B. W. , & Szabó‐Taylor, K. É. (2018). Molecular interactions at the surface of extracellular vesicles. Seminars in iImmunopathology, 40, 453–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo, J. , Bernard, V. , San Lucas, F. A. , Allenson, K. , Capello, M. , Kim, D. U. , Gascoyne, P. , Mulu, F. C. , Stephens, B. M. , Huang, J. , Wang, H. , Momin, A. A. , Jacamo, R. O. , Katz, M. , Wolff, R. , Javle, M. , Varadhachary, G. , Wistuba, I. I. , Hanash, S. , … Alvarez, H. (2018). Surfaceome profiling enables isolation of cancer‐specific exosomal cargo in liquid biopsies from pancreatic cancer patients. Annals of Oncology, 29, 223–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charoenviriyakul, C. , Takahashi, Y. , Morishita, M. , Nishikawa, M. , & Takakura, Y. (2018). Role of extracellular vesicle surface proteins in the pharmacokinetics of extracellular vesicles. Molecular Pharmaceutics, 15, 1073–1080. [DOI] [PubMed] [Google Scholar]
- Chen, M.‐T. , Sun, He‐F. , Zhao, Y. , Fu, W.‐Y. , Yang, Li‐P. , Gao, S.‐P. , Li, L.‐D. , Jiang, H.‐L. , & Jin, W. (2017). Comparison of patterns and prognosis among distant metastatic breast cancer patients by age groups: A SEER population‐based analysis. Scientific Reports, 7, 9254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W. W. , Balaj, L. , Liau, L. M. , Samuels, M. L. , Kotsopoulos, S. K. , Maguire, C. A. , Loguidice, L. , Soto, H. , Garrett, M. , Zhu, L. D. , Sivaraman, S. , Chen, C. , Wong, E. T. , Carter, B. S. , Hochberg, F. H. , Breakefield, X. O. , & Skog, J. (2013). BEAMing and droplet digital PCR analysis of mutant IDH1 mRNA in glioma patient serum and cerebrospinal fluid extracellular vesicles. Molecular Therapy ‐ Nucleic Acids , 2, e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. , Wang, H. , Tan, C. , Yan, Y. , Shen, J. , Huang, Q. , Xu, T. , Lin, J. , & Chen, J. (2018). Expression of amyloid precursor‐like protein 2 (APLP2) in glioblastoma is associated with patient prognosis. Folia Neuropathologica, 56, 30–38. [DOI] [PubMed] [Google Scholar]
- Claridge, B. , Rai, A. , Fang, H. , Matsumoto, A. , Luo, J. , Mcmullen, J. R. , & Greening, D. W. (2021). Proteome characterisation of extracellular vesicles isolated from heart. Proteomics, 21, 2100026. [DOI] [PubMed] [Google Scholar]
- Claridge, B. , Lozano, J. , Poh, Q. H. , Greening, D. W. (2021). Development of Extracellular Vesicle Therapeutics: Challenges, Considerations, and Opportunities. Front Cell Dev Biol. 9, 734720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa‐Silva, B. , Aiello, N. M. , Ocean, A. J. , Singh, S. , Zhang, H. , Thakur, B. . K. , Becker, A. , Hoshino, A. , Mark, M. T. , Molina, H. , Xiang, J. , Zhang, T. , Theilen, T.‐M. , García‐Santos, G. , Williams, C. , Ararso, Y. , Huang, Y. , Rodrigues, G. , Shen, T.‐L. , … Lyden, D. (2015). Pancreatic cancer exosomes initiate pre‐metastatic niche formation in the liver. Nature Cell Biology, 17, 816–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, J. , & Mann, M. (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.‐range mass accuracies and proteome‐wide protein quantification. Nature Biotechnology, 26, 1367–1372. [DOI] [PubMed] [Google Scholar]
- Cvjetkovic, A. , Jang, Su C. , Konečná, B. , Höög, J. L. , Sihlbom, C. , Lässer, C. , & Lötvall, J. (2016). Detailed analysis of protein topology of extracellular vesicles‐evidence of unconventional membrane protein orientation. Scientific Reports, 6, 36338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Acunzo, P. , Pérez‐González, R. , Kim, Y. , Hargash, T. , Miller, C. , Alldred, M. J. , Erdjument‐Bromage, H. , Penikalapati, S. C. , Pawlik, M. , Saito, M. , Saito, M. , Ginsberg, S. D. , Neubert, T. A. , Goulbourne, C. N. , & Levy, E. (2021). Mitovesicles are a novel population of extracellular vesicles of mitochondrial origin altered in Down syndrome. Science Advances, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz, G. , Wolfe, L. M. , Kruh‐Garcia, N. A. , & Dobos, K. M. (2016). Changes in the membrane‐associated proteins of exosomes released from human macrophages after mycobacterium tuberculosis infection. Scientific Reports, 6, 37975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emelyanov, A. , Shtam, T. , Kamyshinsky, R. , Garaeva, L. , Verlov, N. , Miliukhina, I. , Kudrevatykh, A. , Gavrilov, G. , Zabrodskaya, Y. , Pchelina, S. , & Konevega, A. (2020). Cryo‐electron microscopy of extracellular vesicles from cerebrospinal fluid. Plos One, 15, e0227949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford, T. , Graham, J. , & Rickwood, D. (1994). Iodixanol: A nonionic iso‐osmotic centrifugation medium for the formation of self‐generated gradients. Analytical Biochemistry, 220, 360–366. [DOI] [PubMed] [Google Scholar]
- Fu, L. , Han, Li , Xie, C. , Li, W. , Lin, L. , Pan, S. , Zhou, Y. , Li, Z. , Jin, M. , & Zhang, A. (2017). Identification of extracellular actin as a ligand for triggering receptor expressed on myeloid cells‐1 signaling. Frontiers in immunology, 8, 917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta, T. , Miyaki, S. , Ishitobi, H. , Ogura, T. , Kato, Y. , Kamei, N. , Miyado, K. , Higashi, Y. , & Ochi, M. (2016). Mesenchymal stem cell‐derived exosomes promote fracture healing in a mouse model. Stem Cells Translational Medicine , 5, 1620–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallart‐Palau, X. , Serra, A. , Wong, A. S. W. , Sandin, S. , Lai, M. K. P. , Chen, C. P. , Kon, Oi L. , & Sze, S. K. (2015). Extracellular vesicles are rapidly purified from human plasma by PRotein Organic Solvent PRecipitation (PROSPR). Scientific Reports, 5, 14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, X. , Ran, N. , Dong, X. , Zuo, B. , Yang, R. , Zhou, Q. , Moulton, H. M. , Seow, Y. , & Yin, H. (2018). Anchor peptide captures, targets, and loads exosomes of diverse origins for diagnostics and therapy. Science Translational Medicine, 10(444), eaat0195. [DOI] [PubMed] [Google Scholar]
- Gómez‐Arreaza, A. , Acosta, H. , Quiñones, W. , Concepción, J. L. , Michels, P. A.M. , & Avilán, L. (2014). Extracellular functions of glycolytic enzymes of parasites: Unpredicted use of ancient proteins. Molecular and Biochemical Parasitology, 193, 75–81. [DOI] [PubMed] [Google Scholar]
- Grange, C. , Tapparo, M. , Collino, F. , Vitillo, L. , Damasco, C. , Deregibus, M. C. , Tetta, C. , Bussolati, B. , & Camussi, G. (2011). Microvesicles released from human renal cancer stem cells stimulate angiogenesis and formation of lung premetastatic niche. Cancer Research, 71, 5346–5356. [DOI] [PubMed] [Google Scholar]
- Greening, D. W. , Notaras, M. , Chen, M. , Xu, R. , Smith, J. D. , Cheng, L. , Simpson, R. J. , Hill, A. F. , & Van Den Buuse, M. (2019). Chronic methamphetamine interacts with BDNF Val66Met to remodel psychosis pathways in the mesocorticolimbic proteome. Molecular Psychiatry, In Press, 1–17. [DOI] [PubMed] [Google Scholar]
- Gründel, A. , Jacobs, E. , Dumke, R. (2016). Interactions of surface‐displayed glycolytic enzymes of Mycoplasma pneumoniae with components of the human extracellular matrix. International Journal of Medical Microbiology, 306, 675–685. [DOI] [PubMed] [Google Scholar]
- Guan, X. , Luo, L. , Begum, G. , Kohanbash, G. , Song, Q. , Rao, A. , Amankulor, N. , Sun, B. , Sun, D. , & Jia, W. (2018). Elevated Na/H exchanger 1 (SLC9A1) emerges as a marker for tumorigenesis and prognosis in gliomas. Journal of Experimental & Clinical Cancer Research, 37(255), 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood, J. L. , San, R. S. , & Wickline, S. A. (2011). Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Research, 71, 3792–3801. [DOI] [PubMed] [Google Scholar]
- Höög, J. L. , & Lötvall, J. (2015). Diversity of extracellular vesicles in human ejaculates revealed by cryo‐electron microscopy. Journal of Extracellular Vesicles , 4, 28680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino, A. , Costa‐Silva, B. , Shen, T.‐L. , Rodrigues, G. , Hashimoto, A. , Tesic Mark, M. , Molina, H. , Kohsaka, S. , Di Giannatale, A. , Ceder, S. , Singh, S. , Williams, C. , Soplop, N. , Uryu, K. , Pharmer, L. , King, T. , Bojmar, L. , Davies, A. E. , Ararso, Y. , … Lyden, D. (2015). Tumour exosome integrins determine organotropic metastasis. Nature, 527, 329–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino, A. , Kim, H. S. , Bojmar, L. , Gyan, K. E. , Cioffi, M. , Hernandez, J. , Zambirinis, C. P. , Rodrigues, G. , Molina, H. , Heissel, S. , Mark, M. T. , Steiner, L. , Benito‐Martin, A. , Lucotti, S. , Di Giannatale, A. , Offer, K. , Nakajima, M. , Williams, C. , Nogués, L. , … Lyden, D. (2020). Extracellular Vesicle and Particle Biomarkers Define Multiple Human Cancers. Cell, 182, 1044–1061.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howitt, J. , & Hill, A F. (2016). Exosomes in the pathology of neurodegenerative diseases. Journal of Biological Chemistry, 291, 26589–26597. https://pubmed.ncbi.nlm.nih.gov/27852825/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, M. C. , Hung, W.‐C. , Yamaguchi, H. , Lim, S‐Oe , Liao, H.‐W. , Tsai, C.‐H. , & Hung, M.‐C. (2016). Extracellular PKM2 induces cancer proliferation by activating the EGFR signaling pathway. American Journal of Cancer Research , 6, 628–638. [PMC free article] [PubMed] [Google Scholar]
- Hu, Q. , Su, H. , Li, J. , Lyon, C. , Tang, W. , Wan, M. , & Hu, T. Y.e (2020). Clinical applications of exosome membrane proteins. Precision Clinical Medicine , 3, 54–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, S. , Musante, L. , Tataruch, D. , Xu, X. , Kretz, O. , Henry, M. , Meleady, P. , Luo, H. , Zou, H. , Jiang, Y. , & Holthofer, H. (2018). Purification and Identification of Membrane Proteins from Urinary Extracellular Vesicles using Triton X‐114 Phase Partitioning. Journal of Proteome Research, 17, 86–96. [DOI] [PubMed] [Google Scholar]
- Huang, Da W. , Sherman, B. T. , & Lempicki, R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols, 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Hughes, C. S. , Moggridge, S. , Müller, T. , Sorensen, P. H. , Morin, G. B. , & Krijgsveld, J. (2019). Single‐pot, solid‐phase‐enhanced sample preparation for proteomics experiments. Nature Protocols, 14, 68–85. [DOI] [PubMed] [Google Scholar]
- Islam, M. N. , Das, S. R. , Emin, M. T. , Wei, M. , Sun, Li , Westphalen, K. , Rowlands, D. J. , Quadri, S. K. , Bhattacharya, S. , & Bhattacharya, J. (2012). Mitochondrial transfer from bone‐marrow‐derived stromal cells to pulmonary alveoli protects against acute lung injury. Nature Medicine, 18, 759–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issman, L. , Brenner, B. , Talmon, Y. , & Aharon, A. (2013). Cryogenic transmission electron microscopy nanostructural study of shed microparticles. Plos One, 8, e83680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarnuczak, A. F. , & Vizcaino, J. A. (2017). Using the PRIDE database and pProteomeXchange for submitting and accessing public proteomics datasets. Current Protocols in Bioinformatics , 59, 13 31 11–13 31 12. https://pubmed.ncbi.nlm.nih.gov/28902400/ [DOI] [PubMed] [Google Scholar]
- Jeppesen, D. K. , Fenix, A. M. , Franklin, J. L. , Higginbotham, J. N. , Zhang, Q. , Zimmerman, L. J. , Liebler, D. C. , Ping, J. , Liu, Qi , Evans, R. , Fissell, W. H. , Patton, J. G. , Rome, L. H. , Burnette, D. T. , & Coffey, R. J. (2019). Reassessment of exosome composition. Cell, 177, 428–445.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen, D. K. , Nawrocki, A. , Jensen, S. G. , Thorsen, K. , Whitehead, B. , Howard, K A. , Dyrskjøt, L. , Ørntoft, T. F. , Larsen, M R. , & Ostenfeld, M. S. (2014). Quantitative proteomics of fractionated membrane and lumen exosome proteins from isogenic metastatic and nonmetastatic bladder cancer cells reveal differential expression of EMT factors. Proteomics, 14, 699–712. [DOI] [PubMed] [Google Scholar]
- Ji, H. , Chen, M. , Greening, D. W. , He, W. , Rai, A. , Zhang, W. , & Simpson, R. J. (2014). Deep sequencing of RNA from three different extracellular vesicle (EV) subtypes released from the human LIM1863 colon cancer cell line uncovers distinct miRNA‐enrichment signatures. Plos One, 9, e110314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, D. , Jiang, Z. , Lu, Di , Wang, X. , Liang, H. , Zhang, J. , Meng, Y. , Li, Y. , Wu, D. , Huang, Y. , Chen, Y. , Deng, H. , Wu, Q. , Xiong, J. , Meng, A. , & Yu, Li (2019). Migrasomes provide regional cues for organ morphogenesis during zebrafish gastrulation. Nature Cell Biology, 21, 966–977. [DOI] [PubMed] [Google Scholar]
- Jung, A. L. , Jørgensen, M. M. , Bæk, R. , Griss, K. , Han, M. , Brinke, K. A. D. , Timmesfeld, N. , Bertrams, W. , Greulich, T. , Koczulla, R. , Hippenstiel, S. , Suttorp, N. , & Schmeck, B. (2020). Surface proteome of plasma extracellular vesicles as biomarkers for pneumonia and acute exacerbation of chronic obstructive pulmonary disease. Journal of Infectious Diseases, 221, 325–335. [DOI] [PubMed] [Google Scholar]
- Jung, T. , Castellana, D. , Klingbeil, P. , Hernández, I. C. , Vitacolonna, M. , Orlicky, D. J. , Roffler, S. R. , Brodt, P. , & Zöller, M. (2009). CD44v6 dependence of premetastatic niche preparation by exosomes. Neoplasia, 11, 1093–IN17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D. H. , Kim, H. R. , Choi, Y. J. , Kim, S. Y.e , Lee, J.‐E. , Sung, Ki J. , Sung, Y. H. , Pack, C‐Gi , Jung, M. ‐. , Han, B. , Kim, K. , Kim, W. S. , Nam, S. J. , Choi, C.‐M. , Yun, M. , Lee, J. C. , & Rho, J. K. (2019). Exosomal PD‐L1 promotes tumor growth through immune escape in non‐small cell lung cancer. Experimental & Molecular Medicine, 51, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kompa, A. R. , Greening, D. W. , Kong, A. M. , Mcmillan, P. J. , Fang, H. , Saxena, R. , Wong, R. C. B. , Lees, J. G. , Sivakumaran, P. , Newcomb, A. E. , Tannous, B. A. , Kos, C. , Mariana, L. , Loudovaris, T. , Hausenloy, D. J. , & Lim, S. Y. (2021). Sustained subcutaneous delivery of secretome of human cardiac stem cells promotes cardiac repair following myocardial infarction. Cardiovascular Research, 117, 918–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowal, J. , Arras, G. , Colombo, M. , Jouve, M. , Morath, J. P. , Primdal‐Bengtson, B. , Dingli, F. , Loew, D. , Tkach, M. , & Théry, C. (2016). Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. PNAS, 113, E968–E977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku, X. , Xu, Y. , Cai, C. , Yang, Y. , Cui, L. , & Yan, W. (2019). In‐depth characterization of mass spectrometry‐based proteomic profiles revealed novel signature proteins associated with liver metastatic colorectal cancers. Analytical Cellular Pathology (Amsterdam) , 2019, 7653230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugeratski, F. G. , Hodge, K. , Lilla, S. , Mcandrews, K. M. , Zhou, X. , Hwang, R. F. , Zanivan, S. , & Kalluri, R. (2021). Quantitative proteomics identifies the core proteome of exosomes with syntenin‐1 as the highest abundant protein and a putative universal biomarker. Nature Cell Biology, 23, 631–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levchenko, A. , Mehta, B. M. , Niu, X. , Kang, G. , Villafania, L. , Way, D. , Polycarpe, D. , Sadelain, M. , & Larson, S. M. (2005). Intercellular transfer of P‐glycoprotein mediates acquired multidrug resistance in tumor cells. PNAS, 102, 1933–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Wang, Y. , Mao, J. , Yao, Y. , Wang, K. , Qiao, Q. , Fang, Z. , & Ye, M. (2019). Sensitive profiling of cell surface proteome by using an optimized biotinylation method. Journal of Proteomics, 196, 33–41. [DOI] [PubMed] [Google Scholar]
- Liu, B. , Fan, Z. , Edgerton, S. M. , Deng, X.‐S. , Alimova, I. N. , Lind, S. E. , & Thor, A. D. (2009). Metformin induces unique biological and molecular responses in triple negative breast cancer cells. Cell Cycle, 8, 2031–2040. [DOI] [PubMed] [Google Scholar]
- Liu, J. , Wang, D. , Zhang, C. , Zhang, Z. , Chen, X. , Lian, J. , Liu, J. , Wang, G. , Yuan, W. , Sun, Z. , Wang, W. , Song, M. , Wang, Y. , Wu, Q. , Cao, L. , Wang, D. , & Zhang, Y. (2018). Identification of liver metastasis‐associated genes in human colon carcinoma by mRNA profiling. Chinese journal of cancer research = Chung‐kuo yen cheng yen chiu, 30, 633–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv, M.‐M. , Zhu, X‐Ya , Chen, W.‐X. , Zhong, S.‐L. , Hu, Q. , Ma, T.‐F. , Zhang, J. , Chen, L. , Tang, J.‐H. , & Zhao, J.‐H. (2014). Exosomes mediate drug resistance transfer in MCF‐7 breast cancer cells and a probable mechanism is delivery of P‐glycoprotein. Tumour Biology: The Journal of the International Society for Oncodevelopmental Biology and Medicine, 35, 10773–10779. [DOI] [PubMed] [Google Scholar]
- Matheoud, D. , Sugiura, A. , Bellemare‐Pelletier, A. , Laplante, A. , Rondeau, C. , Chemali, M. , Fazel, A. , Bergeron, J. J. , Trudeau, L.‐E. , Burelle, Y. , Gagnon, E. , Mcbride, H. M. , & Desjardins, M. (2016). Parkinson's disease‐related proteins PINK1 and parkin repress mitochondrial antigen presentation. Cell, 166, 314–327. [DOI] [PubMed] [Google Scholar]
- Melo, S. A. , Luecke, L. B. , Kahlert, C. , Fernandez, A. F. , Gammon, S. T. , Kaye, J. , Lebleu, V. S. , Mittendorf, E. A. , Weitz, J. , Rahbari, N. , Reissfelder, C. , Pilarsky, C. , Fraga, M. F. , Piwnica‐Worms, D. , & Kalluri, R. (2015). Glypican‐1 identifies cancer exosomes and detects early pancreatic cancer. Nature, 523, 177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentkowski, K I. , Lang, J K. (2019). Exosomes engineered to express a cardiomyocyte binding peptide demonstrate improved cardiac retention in vivo. Scientific Reports, 9, 10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merico, D. , Isserlin, R. , Stueker, O. , Emili, A. , & Bader, G. D. (2010). Enrichment map: A network‐based method for gene‐set enrichment visualization and interpretation. Plos One, 5, e13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minciacchi, V. R. , You, S. , Spinelli, C. , Morley, S. , Zandian, M. , Aspuria, P.‐J. , Cavallini, L. , Ciardiello, C. , Sobreiro, M. R. , Morello, M. , Kharmate, G. , Jang, Su C. , Kim, D.‐K. , Hosseini‐Beheshti, E. , Guns, E. T. , Gleave, M. , Gho, Y. S. , Mathivanan, S. , Yang, W. , … Di Vizio, D. (2015). Large oncosomes contain distinct protein cargo and represent a separate functional class of tumor‐derived extracellular vesicles. Oncotarget, 6, 11327–11341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura, N. , Kirino, A. , Endo, S. , Morisaka, H. , Kuroda, K. , Takagi, M. , & Ueda, M. (2012). Tracing putative trafficking of the glycolytic enzyme enolase via SNARE‐driven unconventional secretion. Eukaryot Cell, 11, 1075–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraj, N. , Kulak, N. A. , Cox, J. , Neuhauser, N. , Mayr, K. , Hoerning, O. , Vorm, O. , & Mann, M. (2012)System‐wide perturbation analysis with nearly complete coverage of the yeast proteome by single‐shot ultra HPLC runs on a bench top Orbitrap. Molecular & Cellular Proteomics, 11, M111.013722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa, D. , Kumar, S. V. , Marschner, J. , Desai, J. , Holderied, A. , Rath, L. , Kraft, F. , Lei, Y. , Fukasawa, Y. , Moeckel, G. W. , Angelotti, M. L. , Liapis, H. , & Anders, H.‐J. (2017). Histones and neutrophil extracellular traps enhance tubular necrosis and remote organ injury in ischemic AKI. Journal of the American Society of Nephrology, 28, 1753–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolás‐Ávila, J. A. , Lechuga‐Vieco, A. V. , Esteban‐Martínez, L. , Sánchez‐Díaz, M. , Díaz‐García, E. , Santiago, D. J. , Rubio‐Ponce, A. , Li, J. L. , Balachander, A. , Quintana, J. A. , Martínez‐De‐Mena, R. , Castejón‐Vega, B. , Pun‐García, A. , Través, P. G. , Bonzón‐Kulichenko, E. , García‐Marqués, F. , Cussó, L. , A‐González, N. , González‐Guerra, A. … Hidalgo, A. (2020). A network of macrophages supports mitochondrial homeostasis in the heart. Cell, 183, 94–109.e23. [DOI] [PubMed] [Google Scholar]
- Ohno, S.‐I. , Takanashi, M. , Sudo, K. , Ueda, S. , Ishikawa, A. , Matsuyama, N. , Fujita, K. , Mizutani, T. , Ohgi, T. , Ochiya, T. , Gotoh, N. , & Kuroda, M. (2013). Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Molecular Therapy, 21, 185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panarelli, N. C. , Yantiss, R. K. , Yeh, M. M. , Liu, Y. , & Chen, Y.‐T. (2012). Tissue‐specific cadherin CDH17 is a useful marker of gastrointestinal adenocarcinomas with higher sensitivity than CDX2. American Journal of Clinical Pathology, 138, 211–222. [DOI] [PubMed] [Google Scholar]
- Pandey, P. , Sliker, B. , Peters, H. L. , Tuli, A. , Herskovitz, J. , Smits, K. , Purohit, A. , Singh, R. K. , Dong, J. , Batra, S. K. , Coulter, D. W. , & Solheim, J. C. (2016). Amyloid precursor protein and amyloid precursor‐like protein 2 in cancer. Oncotarget, 7, 19430–19444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado, H. , Alečković, M. , Lavotshkin, S. , Matei, I. , Costa‐Silva, B. , Moreno‐Bueno, G. , Hergueta‐Redondo, M. , Williams, C. , García‐Santos, G. , Ghajar, C. M. , Nitadori‐Hoshino, A. , Hoffman, C. , Badal, K. , Garcia, B. A. , Callahan, M. K. , Yuan, J. , Martins, V. R. , Skog, J. , Kaplan, R. N. , … Lyden, D. (2012). Melanoma exosomes educate bone marrow progenitor cells toward a pro‐metastatic phenotype through MET. Nature Medicine, 18, 883–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez‐Hernandez, D. , Gutiérrez‐Vázquez, C. , Jorge, I. , López‐Martín, S. , Ursa, A. , Sánchez‐Madrid, F. , Vázquez, J. , & Yáñez‐Mó, M. (2013). The intracellular interactome of tetraspanin‐enriched microdomains reveals their function as sorting machineries toward exosomes. Journal of Biological Chemistry, 288, 11649–11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao, Y. , Henry, V. , Tiao, N. , Park, S. Y. , Martinez‐Ledesma, J. , Dong, J. W. , Balasubramaniyan, V. , & De Groot, J. F. (2017). Targeting intercellular adhesion molecule‐1 prolongs survival in mice bearing bevacizumab‐resistant glioblastoma. Oncotarget, 8, 96970–96983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poh, Qi H. , Rai, A. , Carmichael, I. I. , Salamonsen, L. A. , & Greening, D. W. (2021). Proteome reprogramming of endometrial epithelial cells by human trophectodermal small extracellular vesicles reveals key insights into embryo implantation. Proteomics, 21, 2000210. [DOI] [PubMed] [Google Scholar]
- Poliakov, A. , Spilman, M. , Dokland, T. , Amling, C. L. , & Mobley, J. A. (2009). Structural heterogeneity and protein composition of exosome‐like vesicles (prostasomes) in human semen. The Prostate, 69, 159–167. [DOI] [PubMed] [Google Scholar]
- Puhm, F. , Afonyushkin, T. , Resch, U. , Obermayer, G. , Rohde, M. , Penz, T. , Schuster, M. , Wagner, G. , Rendeiro, A. F. , Melki, I. , Kaun, C. , Wojta, J. , Bock, C. , Jilma, B. , Mackman, N. , Boilard, E. , & Binder, C. J. (2019). Mitochondria are a subset of extracellular vesicles released by activated monocytes and induce type I IFN and TNF responses in endothelial cells. Circulation Research, 125, 43–52. [DOI] [PubMed] [Google Scholar]
- Rai, A. , Greening, D. W. , Chen, M. , Xu, R. , Ji, H. , & Simpson, R. J. (2019). Exosomes derived from human primary and metastatic colorectal cancer cells contribute to functional heterogeneity of activated fibroblasts by reprogramming their proteome. Proteomics, 19, 1800148. [DOI] [PubMed] [Google Scholar]
- Rai, A. , Greening, D. W. , Xu, R. , Chen, M. , Suwakulsiri, W. , & Simpson, R. J. (2021). Secreted midbody remnants are a class of extracellular vesicles molecularly distinct from exosomes and microparticles. Communications Biology, 4, 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai, A. , Fang, H. , Fatmous, M. , Claridge, B. , Poh, Q. H. , Simpson, R. J. , & Greening, D. W. (2021). A protocol for isolation, purification, characterization, and functional dissection of exosomes. Methods Mol Biol, 2261, 105–149. [DOI] [PubMed] [Google Scholar]
- Rai, A. , Poh, Qi H. , Fatmous, M. , Fang, H. , Gurung, S. , Vollenhoven, B. , Salamonsen, L. A. , & Greening, D. W. (2021). Proteomic profiling of human uterine extracellular vesicles reveal dynamic regulation of key players of embryo implantation and fertility during menstrual cycle. Proteomics, 21, 2000211. [DOI] [PubMed] [Google Scholar]
- Raikar, L. S. , Vallejo, J. , Lloyd, P. G. , & Hardin, C. D. (2006). Overexpression of caveolin‐1 results in increased plasma membrane targeting of glycolytic enzymes: The structural basis for a membrane associated metabolic compartment. Journal of Cellular Biochemistry, 98, 861–871. [DOI] [PubMed] [Google Scholar]
- Raudvere, U. , Kolberg, L. , Kuzmin, I. , Arak, T. , Adler, P. , Peterson, H. , & Vilo, J. (2019). g:PRofiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res., 47, W191–W198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringuette Goulet, C. , Bernard, G. , Tremblay, S. , Chabaud, S. , Bolduc, S. , & Pouliot, F. (2018). Exosomes induce fibroblast differentiation into cancer‐associated fibroblasts through TGFbeta signaling. Molecular Cancer Research, 16, 1196–1204. [DOI] [PubMed] [Google Scholar]
- Rogers, M. A. , Buffolo, F. , Schlotter, F. , Atkins, S. K. , Lee, L. H. , Halu, A. , Blaser, M. C. , Tsolaki, E. , Higashi, H. , Luther, K. , Daaboul, G. , Bouten, C. V.C. , Body, S. C. , Singh, S. A. , Bertazzo, S. , Libby, P. , Aikawa, M. , & Aikawa, E. (2020). Annexin A1‐dependent tethering promotes extracellular vesicle aggregation revealed with single‐extracellular vesicle analysis. Science Advances, 6, eabb1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santucci, L. , Bruschi, M. , Del Zotto, G. , Antonini, F. , Ghiggeri, G. M. , Panfoli, I. , & Candiano, G. (2019). Biological surface properties in extracellular vesicles and their effect on cargo proteins. Scientific Reports, 9, 13048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon, P. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Research, 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, Y.‐K. , Yoo, B. C. , Hong, Y. S. , Chang, H. J. , Jung, K. H. , Jeong, S.‐Y. , & Park, J.‐G. (2009). Upregulation of glycolytic enzymes in proteins secreted from human colon cancer cells with 5‐fluorouracil resistance. Electrophoresis, 30, 2182–2192. [DOI] [PubMed] [Google Scholar]
- Sökeland, G. , & Schumacher, U. (2019). The functional role of integrins during intra‐ and extravasation within the metastatic cascade. Molecular Cancer, 18(1), 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soubannier, V. , Rippstein, P. , Kaufman, B. A. , Shoubridge, E. A. , & Mcbride, H. M. (2012). Reconstitution of mitochondria derived vesicle formation demonstrates selective enrichment of oxidized cargo. Plos One, 7, e52830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa, D. , Lima, R. T. , & Vasconcelos, M. H. (2015). Intercellular transfer of cancer drug resistance traits by extracellular vesicles. Trends in Molecular Medicine, 21, 595–608. [DOI] [PubMed] [Google Scholar]
- Su, M.‐C. , Yuan, R.‐H. , Lin, C.‐Y. , & Jeng, Y.‐M. (2008). Cadherin‐17 is a useful diagnostic marker for adenocarcinomas of the digestive system. Modern Pathology, 21, 1379–1386. [DOI] [PubMed] [Google Scholar]
- Sugiura, A. , Mclelland, G. ‐. L. , Fon, E. A. , & Mcbride, H. M. (2014). A new pathway for mitochondrial quality control: Mitochondrial‐derived vesicles. Embo Journal, 33, 2142–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung, B. H. , Ketova, T. , Hoshino, D. , Zijlstra, A. , & Weaver, A. M. (2015). Directional cell movement through tissues is controlled by exosome secretion. Nature communications, 6, 7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwakulsiri, W. , Rai, A. , Xu, R. , Chen, M. , Greening, D. W. , & Simpson, R. J. (2019). Proteomic profiling reveals key cancer progression modulators in shed microvesicles released from isogenic human primary and metastatic colorectal cancer cell lines. Biochimica et Biophysica Acta ‐ Proteins and Proteomics, 1867(12), 140171. [DOI] [PubMed] [Google Scholar]
- Tauro, B. J. , Greening, D. W. , Mathias, R. A. , Mathivanan, S. , Ji, H. , & Simpson, R. J. (2013). Two distinct populations of exosomes are released from LIM1863 colon carcinoma cell‐derived organoids. Molecular and Cellular Proteomics, 12, 587–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry, C. , Witwer, K. W. , Aikawa, E. , Alcaraz, M. J. , Anderson, J. D. , Andriantsitohaina, R. , Antoniou, A. , Arab, T. , Archer, F. , Atkin‐Smith, G. K. , Ayre, D. C. , Bach, J.‐M. , Bachurski, D. , Baharvand, H. , Balaj, L. , Baldacchino, S. , Bauer, N. N. , Baxter, A. A. , Bebawy, M. , … Zuba‐Surma, E. K. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of Extracellular Vesicles , 7, 1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, T. , Zhang, H.‐X. , He, C.‐P. , Fan, S. , Zhu, Y.‐L. , Qi, C. , Huang, N.‐P. , Xiao, Z.‐D. , Lu, Zu‐H. , Tannous, B. A. , & Gao, J. (2018). Surface functionalized exosomes as targeted drug delivery vehicles for cerebral ischemia therapy. Biomaterials, 150, 137–149. [DOI] [PubMed] [Google Scholar]
- Tkach, M. , Kowal, J. , & Théry, C. (2018). Why the need and how to approach the functional diversity of extracellular vesicles. Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences, 373(1737), 20160479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova, S. , Temu, T. , Sinitcyn, P. , Carlson, A. , Hein, M. Y. , Geiger, T. , Mann, M. , & Cox, J. (2016). The Perseus computational platform for comprehensive analysis of (prote)omics data. Nature Methods, 13, 731–740. [DOI] [PubMed] [Google Scholar]
- Uhlén, M. , Fagerberg, L. , Hallström, B. M. , Lindskog, C. , Oksvold, P. , Mardinoglu, A. , Sivertsson, Å. , Kampf, C. , Sjöstedt, E. , Asplund, A. , Olsson, I. , Edlund, K. , Lundberg, E. , Navani, S. , Szigyarto, C. A.l‐K. , Odeberg, J. , Djureinovic, D. , Takanen, J. O. , Hober, S. , … Pontén, F. (2015). Tissue‐based map of the human proteome. Science, 347(6220), 12604191‐9. [DOI] [PubMed] [Google Scholar]
- Van Der Pol, E. , Coumans, F. A. W. , Grootemaat, A. E. , Gardiner, C. , Sargent, I. L. , Harrison, P. , Sturk, A. , Van Leeuwen, T. G. , & Nieuwland, R. (2014). Particle size distribution of exosomes and microvesicles determined by transmission electron microscopy, flow cytometry, nanoparticle tracking analysis, and resistive pulse sensing. Journal of Thrombosis and Haemostasis, 12, 1182–1192. [DOI] [PubMed] [Google Scholar]
- Van Deun, J. , Mestdagh, P. , Agostinis, P. , Akay, Ö. , Anand, S. , Anckaert, J. , Martinez, Z. A. , Baetens, T. , Beghein, E. , Bertier, L. , Berx, G. , Boere, J. , Boukouris, S. , Bremer, M. , Buschmann, D. , Byrd, J. B. , Casert, C. , Cheng, L. , Cmoch, A. , … Hendrix, An (2017). EV‐TRACK: Transparent reporting and centralizing knowledge in extracellular vesicle research. Nature Methods, 14, 228–232. [DOI] [PubMed] [Google Scholar]
- Vicencio, J. M. , Yellon, D. M. , Sivaraman, V. , Das, D. , Boi‐Doku, C. , Arjun, S. , Zheng, Y. , Riquelme, J. A. , Kearney, J. , Sharma, V. , Multhoff, G. , Hall, A. R. , & Davidson, S. M. (2015). Plasma exosomes protect the myocardium from ischemia‐reperfusion injury. Journal of the American College of Cardiology, 65, 1525–1536. [DOI] [PubMed] [Google Scholar]
- Warde‐Farley, D. , Donaldson, S L. , Comes, O. , Zuberi, K. , Badrawi, R. , Chao, P. , Franz, M. , Grouios, C. , Kazi, F. , Lopes, C. T. , Maitland, A. , Mostafavi, S. , Montojo, J. , Shao, Q. , Wright, G. , Bader, G D. , Morris, Q. (2010). The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res., 38, W214–W220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, J.‐W. , Cai, J.‐Q. , Fang, C. , Tan, Y‐Li , Huang, K. , Yang, C. , Chen, Q. , Jiang, C‐Lu , & Kang, C.‐S. (2017). Signal peptide peptidase, encoded by HM13, contributes to tumor progression by affecting EGFRvIII secretion profiles in glioblastoma. CNS Neuroscience & Therapeutics, 23, 257–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead, R. H. , Jones, J. K. , Gabriel, A. , & Lukies, R. E. (1987). A new colon carcinoma cell line (LIM1863) that grows as organoids with spontaneous differentiation into crypt‐like structures in vitro. Cancer Research, 47, 2683–2689. [PubMed] [Google Scholar]
- Wiklander, O. P. B. , Nordin, J. Z. , O'loughlin, A. , Gustafsson, Y. , Corso, G. , Mäger, I. , Vader, P. , Lee, Yi , Sork, H. , Seow, Y. , Heldring, N. , Alvarez‐Erviti, L. , Smith, Ci E. , Le Blanc, K. , Macchiarini, P. , Jungebluth, P. , Wood, M. J. A. , & Andaloussi, S. E.l (2015). Extracellular vesicle in vivo biodistribution is determined by cell source, route of administration and targeting. Journal of Extracellular Vesicles, 4, 26316‐1‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, A. Y.‐.T. , Sung, Y. ‐. C. , Chen, Y‐Ju , Chou, S. T.‐Yu , Guo, V. , Chien, J. C.‐.Y. , Ko, J. J.‐.S. , Yang, A. L. , Huang, H. ‐. C. , Chuang, Ju‐C. , Wu, S. , Ho, M‐Ru , Ericsson, M. , Lin, W. ‐. W. , Cheung, C. H. Y. , Juan, H. ‐. F. , Ueda, K. , Chen, Y. , & Lai, C. P.‐.K. (2020). Multiresolution imaging using bioluminescence resonance energy transfer identifies distinct biodistribution profiles of extracellular vesicles and exomeres with redirected tropism. Advanced Science, 7, 2001467‐1‐17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Di , Yan, J. , Shen, X. , Sun, Yu , Thulin, M. , Cai, Y. , Wik, L. , Shen, Q. , Oelrich, J. , Qian, X. , Dubois, K. L. , Ronquist, K. G. , Nilsson, M. , Landegren, U. , & Kamali‐Moghaddam, M. (2019). Profiling surface proteins on individual exosomes using a proximity barcoding assay. Nature communications, 10, 3854 https://pubmed.ncbi.nlm.nih.gov/31451692/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, R. , Greening, D. W. , Chen, M. , Rai, A. , Ji, H. , Takahashi, N. , & Simpson, R. J. (2019). Surfaceome of exosomes secreted from the colorectal cancer cell line SW480: Peripheral and integral membrane proteins analyzed by proteolysis and TX114. Proteomics, 19, 1700453. [DOI] [PubMed] [Google Scholar]
- Xu, R. , Greening, D. W. , Rai, A. , Ji, H. , & Simpson, R. J. (2015). Highly‐purified exosomes and shed microvesicles isolated from the human colon cancer cell line LIM1863 by sequential centrifugal ultrafiltration are biochemically and functionally distinct. Methods (San Diego, Calif.), 87, 11–25. [DOI] [PubMed] [Google Scholar]
- Xu, R. , Rai, A. , Chen, M. , Suwakulsiri, W. , Greening, D. W. , & Simpson, R. J. (2018). Extracellular vesicles in cancer‐implications for future improvements in cancer care. Nature reviews Clinical oncology, 15, 617–638. [DOI] [PubMed] [Google Scholar]
- Yaddanapudi, K. , Meng, S. , Whitt, A. G. , Al Rayyan, N. , Richie, J. , Tu, A. , Eaton, J. W. , & Li, C. (2019). Exosomes from GM‐CSF expressing embryonic stem cells are an effective prophylactic vaccine for cancer prevention. Oncoimmunology, 8, 1561119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Yi , Qin, M. , Bao, P. , Xu, W. , & Xu, J. (2017). Secretory carrier membrane protein 5 is an autophagy inhibitor that promotes the secretion of alpha‐synuclein via exosome. Plos One, 12, e0180892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka, Y. , Kosaka, N. , Konishi, Y. , Ohta, H. , Okamoto, H. , Sonoda, H. , Nonaka, R. , Yamamoto, H. , Ishii, H. , Mori, M. , Furuta, K. , Nakajima, T. , Hayashi, H. , Sugisaki, H. , Higashimoto, H. , Kato, T. , Takeshita, F. , & Ochiya, T. (2014). Ultra‐sensitive liquid biopsy of circulating extracellular vesicles using ExoScreen. Nature communications, 5, 3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefi, S. , Simon, D. , Stojkov, D. , Karsonova, A. , Karaulov, A. , & Simon, H.‐U. (2020). In vivo evidence for extracellular DNA trap formation. Cell death & disease, 11, 300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabeo, D. , Cvjetkovic, A. , Lässer, C. , Schorb, M. , Lötvall, J. , & Höög, J. L. (2017). Exosomes purified from a single cell type have diverse morphology. Journal of Extracellular Vesicles , 6, 1329476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaborowski, M. P. , Lee, K. , Na, Y. J. , Sammarco, A. , Zhang, X. , Iwanicki, M. , Cheah, P. S. , Lin, H.‐Y. , Zinter, M. , Chou, C‐Yu , Fulci, G. , Tannous, B. A. , Lai, C. P.‐.K. , Birrer, M. J. , Weissleder, R. , Lee, H. , & Breakefield, X. O. (2019). Methods for systematic identification of membrane proteins for specific capture of cancer‐derived extracellular vesicles. Cell reports, 27, 255–268.e6 e256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C. , Wang, M. , Ji, F. , Peng, Y. , Wang, Bo , Zhao, J. , Wu, J. , & Zhao, H. (2021). A novel glucose metabolism‐related gene signature for overall survival prediction in patients with glioblastoma. BioMed research international, 2021, 8872977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Liu, T. , Wang, J. , Zou, B. , Li, Le , Yao, L. , Chen, K. , Ning, L. , Wu, B. , Zhao, X. , & Wang, D. (2021). Cellinker: A platform of ligand‐receptor interactions for intercellular communication analysis. Bioinformatics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , Chen, Y. , Wang, J. , & Liu, L. (2021). Effects of ATP‐binding cassette transporter G2 in extracellular vesicles on drug resistance of laryngeal cancer cells in in vivo and in vitro. Oncology letters, 21, 364 10.3892/ol.2021.12625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber, M. X. , Strittmatter, S. M. , & Fishman, M. C. (1989). A membrane‐targeting signal in the amino terminus of the neuronal protein GAP‐43. Nature, 341, 345–348. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. A. Immunoblot detection of CD63 and ANXA1 in large and small EVs purified using density‐gradient separation from conditioned media of SW620 cells. B. Immunoblot detection of CD63 and ANXA1 in MDA MB 231 whole cell lysate and derived large and small EVs.
Supplementary Figure 2. Nanoparticle tracking analysis of purified large and small EVs from indicated cell lines.
Supplementary Figure 3. Nanoparticle tracking analysis data of purified large and small EVs from indicated cell lines represented as log10 transformed data (concentration (particle/mL) and diameter (size, nm).
Supplementary Figure 4. Bar plot depicting percentage of EVs of indicated size (>200 nm, >300 nm, >400 nm) within each EV‐subtype from indicated cell lines based on A. cryo‐EM analysis or B. single particle tracking (NTA) data.
Supplementary Figure 5. A. Venn diagram of proteins identified in L‐EVs and S‐EVs. B. Violin plot of LFQ intensities of proteins identified in L‐EVs and S‐EVs. C. Pearson correlation matrix of L‐EV and S‐EV proteomes for indicated cell lines.
Supplementary Figure 6. Bar plot depicting normalised LFQ intensities of indicated proteins in L‐EVs or S‐EVs from three cell lines MDA MB 231, SW620 and U87. p‐value < 0.05 (*), 0.005 (**), 0.001 (***), 0.0001 (****), ns representing non‐significant.
Supplementary Figure 7. Bar plot of Gene ontology‐based biological processes overrepresented in surface proteins enriched in L‐EVs or S‐EVs. Middle panel represents biological processes enriched in surface proteins commonly identified in both L‐EVs and S‐EVs.
Supplementary Figure 8. A. Hierarchical clustering analysis of differentially abundant SURFY proteins (FDR < 0.05) in global proteome of L‐EVs from U87, SW620 and LIM1863 cell lines. Corresponding boxes list proteins that are found in associated clusters. *Proteins that are found in higher abundance in U87 L‐EVs vs U87 S‐EVs. B. Venn diagram of Human Protein Atlas‐based “brain enriched” or “intestine enriched” proteins and Cluster 2 (U87 enriched) and CRC cell lines enriched (Cluster 1/3/4) proteins. Bar plots depicts LFQ intensities of indicated proteins in L‐EVs and S‐EVs from LIM1863.
Supplementary Figure 9. A. Venn diagram of biotin‐captured surface proteins in S‐EVs versus surface proteins reported in CSPA (Bausch‐Fluck et al., 2015), SURFY (Bausch‐Fluck et al., 2018) or UniProt. B. Relative abundance of transporters, CDs and receptors in L‐EVs and S‐EVs biotin‐captured proteome. C. Volcano plot of differentially abundant biotin‐captured receptors in L‐EVs and S‐EVs. D. Relative abundance of LFQ intensities of indicated proteins in S‐EVs versus L‐EVs. E. Western blot analysis of L‐EVs and S‐EVs released by SW620 cells using indicated antibodies. F. Fluorescence microscopic analysis of SW620 cells expressing plasma membrane targeting GAP‐GFP proteins using indicated antibodies.
Supplementary Figure 10. Heat map of differentially abundant biotin captured proteins (FDR < 0.05) in L‐EVs from U87, SW620 and MDA MB 231 cell lines. Corresponding boxes list proteins that are found in associated clusters. *Proteins that are released from MDA MB 231 L‐EVs following EDTA treatment.
Supplementary Table 1. Global proteomic analysis of L‐EVs and S‐EVs
Supplementary Table 2. Gene ontology (cellular component) enriched in L‐EVs and S‐EVs
Supplementary Table 3. Differentially abundant SURFY proteins in L‐EVs
Supplementary Table 4. Proteomic analysis of EV‐surface proteins captured using membrane impermeant biotin
Supplementary Table 5. List of surface proteins identified in L‐EVs
Supplementary Table 6. Pathways enriched in L‐EVs and S‐EVs
Supplementary Table 7. List of proteins from Figure 5D
Supplementary Table 8. Differentially abundant biotin captured proteins in L‐EVs
Supplementary Table 9. Receptor‐ligand interactome of L‐EV surface proteins
Supplementary Table 10. Receptor‐ligand interactome of L‐EV surface‐associated proteins
Data Availability Statement
The MS‐based proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (Jarnuczak & Vizcaino, 2017) and are available via ProteomeXchange with identifier PXD024733. Experimental parameters submitted to EV‐TRACK knowledgebase (EV‐TRACK ID: EV210261) (Van Deun et al., 2017).


