Significance
The Murine Double Minute 2–Murine Double Minute X (MDM2–MDMX) heterodimer is well known as one of the major negative regulators of the tumor suppressor p53, but the p53-independent functions of MDM2 and MDMX are not as well studied. Our work demonstrates that MDM2 and MDMX, both independently and as a heterocomplex, are required for cell-cycle progression in cancer cells either expressing certain mutant forms of p53 or lacking p53 expression entirely. MDM2 and MDMX further regulate the expression of p73 and certain E2F family activators, both of which are linked to cell division. The p53-independent activity of MDM2 and MDMX may be a potential therapeutic target in cancers lacking wild-type p53.
Keywords: cell cycle, MDM2, E2F1, MDMX, p73
Abstract
The p53 tumor suppressor protein, known to be critically important in several processes including cell-cycle arrest and apoptosis, is highly regulated by multiple mechanisms, most certifiably the Murine Double Minute 2–Murine Double Minute X (MDM2–MDMX) heterodimer. The role of MDM2–MDMX in cell-cycle regulation through inhibition of p53 has been well established. Here we report that in cells either lacking p53 or expressing certain tumor-derived mutant forms of p53, loss of endogenous MDM2 or MDMX, or inhibition of E3 ligase activity of the heterocomplex, causes cell-cycle arrest. This arrest is correlated with a reduction in E2F1, E2F3, and p73 levels. Remarkably, direct ablation of endogenous p73 produces a similar effect on the cell cycle and the expression of certain E2F family members at both protein and messenger RNA levels. These data suggest that MDM2 and MDMX, working at least in part as a heterocomplex, may play a p53-independent role in maintaining cell-cycle progression by promoting the activity of E2F family members as well as p73, making them a potential target of interest in cancers lacking wild-type p53.
It is well established that the gene encoding the p53 tumor suppressor protein is mutated in human cancer with extraordinary frequency (1). Under nonmalignant conditions, p53 is closely regulated by the E3 ubiquitin ligase Murine Double Minute 2 (MDM2), working in concert with Murine Double Minute X (MDMX); both inhibit p53 transactivation activity by binding to its N-terminal transactivation domain (TAD) (reviewed in ref. 2). Although MDM2 and MDMX possess highly similar C-terminal RING domains, only MDM2 has E3 ubiquitin ligase activity through its RING domain; consequently, only MDM2 is capable of ubiquitinating p53 and targeting it for degradation by the proteasome (3–5) (reviewed in ref. 6). MDMX heterodimerizes with MDM2 to promote MDM2 activity and stability (7–9). The MDM2–MDMX complex classically regulates p53 through multiple mechanisms beyond modulation of protein stability, including subcellular localization and inhibition of p53 transactivation (10–12) (reviewed in ref. 13).
Due to seminal discoveries that embryonic lethality in mice caused by deletion of MDM2 or MDMX (or both together) is rescued by codeletion of p53 (14–17) (reviewed in ref. 18), MDM2 and MDMX are most frequently studied in the context of their p53-related activities. However, MDM2 and MDMX have p53-independent functions as well, playing roles in epithelial–mesenchymal transition, response to DNA damage, initiation of chromosome instability, and regulation of the cell cycle, among others (2, 19–23). Of note, MDM2 was reported to bind to well-known regulators of the cell cycle, including pRb and E2F1 (24, 25) (reviewed in ref. 20).
E2F1 is one member of the E2F family of eight transcription factors, all of which play key roles in regulating the cell cycle, along with their binding partners of the DP family (26–28) and the pRb family of pocket proteins (29). The eight E2Fs are classified into three subgroups: activating E2Fs (E2F1, E2F2, and E2F3a), repressive E2Fs (E2F3b, E2F4, E2F5, and E2F6), and atypical E2Fs (E2F7 and E2F8). Only the activating E2Fs are responsible for transitioning between G1 and S phases of the cell cycle. pRb keeps cells in G1 by binding to the activating E2Fs and their DP family binding partners, thus sequestering the E2F–DP complexes from their S phase targets. Cyclin-dependent kinases, CDK4/6 and CDK2, in conjunction with cyclins D and E, phosphorylate pRb, releasing the E2Fs and promoting the transition from G1 to S. CDK inhibitors maintain pRb in its hypophosphorylated state, thus keeping the cell in G1 phase (30, 31).
There are multiple links between the MDM2–MDMX–p53 axis and E2F. p53 and E2F transcription factors have an intricate relationship, sometimes antagonizing and at other times augmenting each other’s activities (32). The p53 target p21 (encoded by the CDKN1a gene) inhibits E2F-mediated transcription through multiple ways—both indirectly through the inhibition of CDKs and directly by interacting with E2F—in response to p53 activation (33, 34) (reviewed in ref. 35). Additionally, MDM2 itself regulates E2F1 both negatively and positively in a p53-independent manner via multiple direct and indirect mechanisms (36, 37). MDMX also binds E2F1 and represses its transcriptional activity (38) by preventing it from binding to DNA (39).
p53 is a member of a small family, also composed of p63 and p73 (40, 41), both of which exist as different isoforms including ones generated by alternate promoters and possessing several different C-terminal regions generated by alternative splicing events (42). p63 and p73 are structurally similar to p53 and regulate many of the same genes, although they each have unique transcriptional targets as well (43). While MDM2 and MDMX bind both p63 and p73, their interactions with p63 are reported to be very weak (44–46). However, binding by MDM2 or MDMX was shown to stabilize p73α and, in doing so, to inhibit its transactivation (47–50). While p73, like p53, functions as a sequence-dependent transcriptional activator which can increase expression of a subset of p53 targets and initiate pathways similar to p53 such as cell-cycle arrest and apoptosis, there is some evidence that p73 may also be linked to cell growth, survival, and wound healing (51, 52). Given the overlap in targets of p53 and p73, it may be difficult to distinguish p73 functions in the presence of p53, and MDM2–MDMX regulation of p73 in the absence of p53 has not been extensively characterized. Here we show that genetic ablation or pharmacological inhibition of MDM2 and MDMX in p53-null H1299 non–small-cell lung carcinoma cells results in cell-cycle arrest and down-regulation of E2F1, E2F3a, and p73. Furthermore, arrest induced by inhibition of MDM2 occurs in cells expressing mutant p53, despite the well-known, strongly prooncogenic activity of mutant p53. Here, for the most part, we have focused on endogenous protein for all of our studies. Our observations suggest unexpected capabilities of MDM2, p73, and E2F family members to promote cell growth in p53-null and mutant p53–expressing cancers.
Results
MDM2 and MDMX Promote Cell-Cycle Progression Independent of p53.
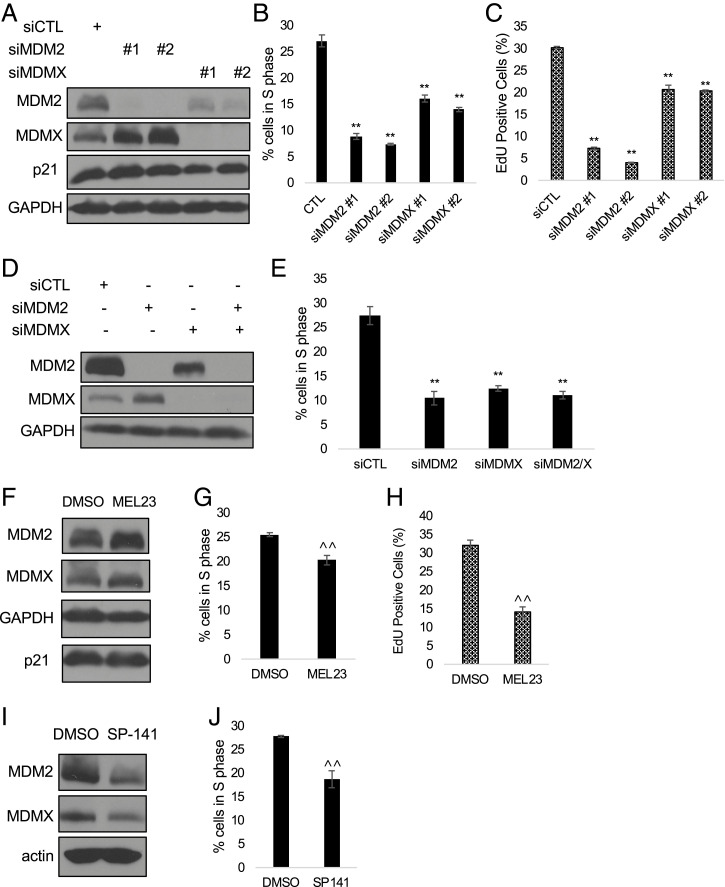
We wanted to explore roles for MDM2 and MDMX, both independently and as a heterocomplex, in regulating cell-cycle progression in a p53-independent fashion. Using RNA interference (RNAi), we knocked down MDM2 or MDMX expression in human H1299 cells, derived from a non–small-cell lung carcinoma, which naturally lack p53. We treated the cells with small interfering RNAs (siRNAs) directed against MDM2, MDMX, or control siRNA for 48 h and assessed the cell cycle by flow cytometry. Inhibiting expression of MDM2 or MDMX protein caused cell-cycle arrest in G1 and a concomitant decrease in the percentage of cells in S phase compared with the control siRNA (Fig. 1 A and B and SI Appendix, Fig. S1A). To control for off-target effects, these results were recapitulated with multiple siRNAs targeting MDM2 (SI Appendix, Fig. S1 B and C). In cells with wild-type p53, inhibition of MDM2 typically increases p53, which, among other targets, induces expression of CDKN1a (p21), leading to arrest in G1. We were curious whether MDM2 still acted through the p21 pathway in the absence of wild-type p53. However, we found that siRNA knockdown of MDM2 or MDMX caused arrest without altering p21 levels in p53-null cells. MDM2 ablation also did not affect pRb, p107, or p130, and direct ablation of pRb family members did not cause arrest (SI Appendix, Fig. S2). Notably, knocking down MDMX had a similar effect, although often somewhat less pronounced, indicating that the observed cell-cycle arrest is specifically mediated by loss of either MDM2 or MDMX and is independent of p53 or induction of p21. Additionally, this effect occurred whether MDM2 and MDMX were ablated individually or together (Fig. 1 D and E). MDMX siRNA also reduced MDM2 levels, in agreement with the reported role of MDMX in stabilizing MDM2 (53, 54).
Fig. 1.
Ablation of either MDM2 or MDMX causes cell-cycle arrest in p53-null lung carcinoma H1299 cells. (A–C) H1299 cells were transfected with siMDM2, siMDMX, or control siRNA for 48 h. (A) Knockdown of MDM2 and MDMX was verified by immunoblot. (B and C) Cell-cycle distribution was analyzed using flow cytometry and the percentage of cells in S phase was plotted, measuring either by PI staining (B) or EdU incorporation assay (C). (D) H1299 cells were transfected with control siRNA, siMDM2, or siMDMX, or cotransfection of siMDM2/siMDMX for 48 h. Knockdown was verified by immunoblot. Coknockdown was performed with siMDM2 no. 2 and siMDMX no. 1. (E) Cell-cycle distribution was analyzed using flow cytometry and the percentage of cells in S phase was plotted. (F–J) H1299 cells were treated with dimethyl sulfoxide (DMSO), MEL23 (F; 15 μM), or SP-141 (I; 250 nM) for 24 h. Drug efficacy was measured by accumulation of MDMX for MEL23 (F) or by decrease of MDM2 (I). Representative graphs of three biological replicates of cell-cycle distribution measured by PI staining (B, E, G, and J) or EdU incorporation (C and H) are shown. **P < 0.01 (one-way ANOVA), ^^P < 0.01 (unpaired t test). All data represent at least three biological replicates (error bars represent SEM).
To extend these observations, we additionally used small-molecule inhibitors of MDM2. Most notably, because ablating MDM2 or MDMX caused the same effect, we considered the possibility that cell-cycle progression relies upon the MDM2–MDMX heterocomplex, rather than functions of either protein that are independent of heterodimerization. To address whether the E3 ubiquitin ligase activity of the heterocomplex is necessary for promoting cell-cycle progression, we treated H1299 cells with MEL23 (MDM2 E3 ubiquitin ligase inhibitor 23), which inhibits catalytic activity specific to the heterocomplex (55). Indeed, blocking the E3 ligase activity of the heterocomplex also caused a cell-cycle arrest prior to S phase (Fig. 1 F and G). To address potential off-target effects of MEL23, we also treated cells with SP-141, an MDM2 inhibitor that functions by inducing autoubiquitination and proteasomal degradation of MDM2 (56). Treatment with SP-141 also caused a cell-cycle arrest (Fig. 1 I and J). Reduced S phase measured by flow cytometry was confirmed by a 5-ethynyl-2′-deoxyuridine (EdU) incorporation assay, indicating that DNA synthesis was inhibited by ablation of MDM2, MDMX, or the heterocomplex (Fig. 1 C and H). Full cell-cycle profiles derived from fluorescence-activated cell sorting (FACS) analysis of H1299 cells treated with MEL23 or SP-141 are shown in SI Appendix, Fig. S1 D and E, respectively. Taken together, these results strongly indicate that MDM2 and MDMX, and most likely the E3 ubiquitin ligase activity of MDM2 in the MDM2–MDMX heterocomplex, are necessary for cell-cycle progression in p53-null H1299 cells.
Mutant p53 Does Not Counteract the Effect of MDM2 Inhibition on the Cell Cycle.
The majority of p53-mutated cancers express p53 with missense mutations, rather than complete loss of p53, and mutant p53 is known for its gain-of-function role in many cancers—including heightened proliferation, resistance to apoptosis, and other promalignant effects (57). Given the prooncogenic role(s) of mutant p53, we asked whether mutant p53 could overcome cell-cycle arrest associated with loss of MDM2.
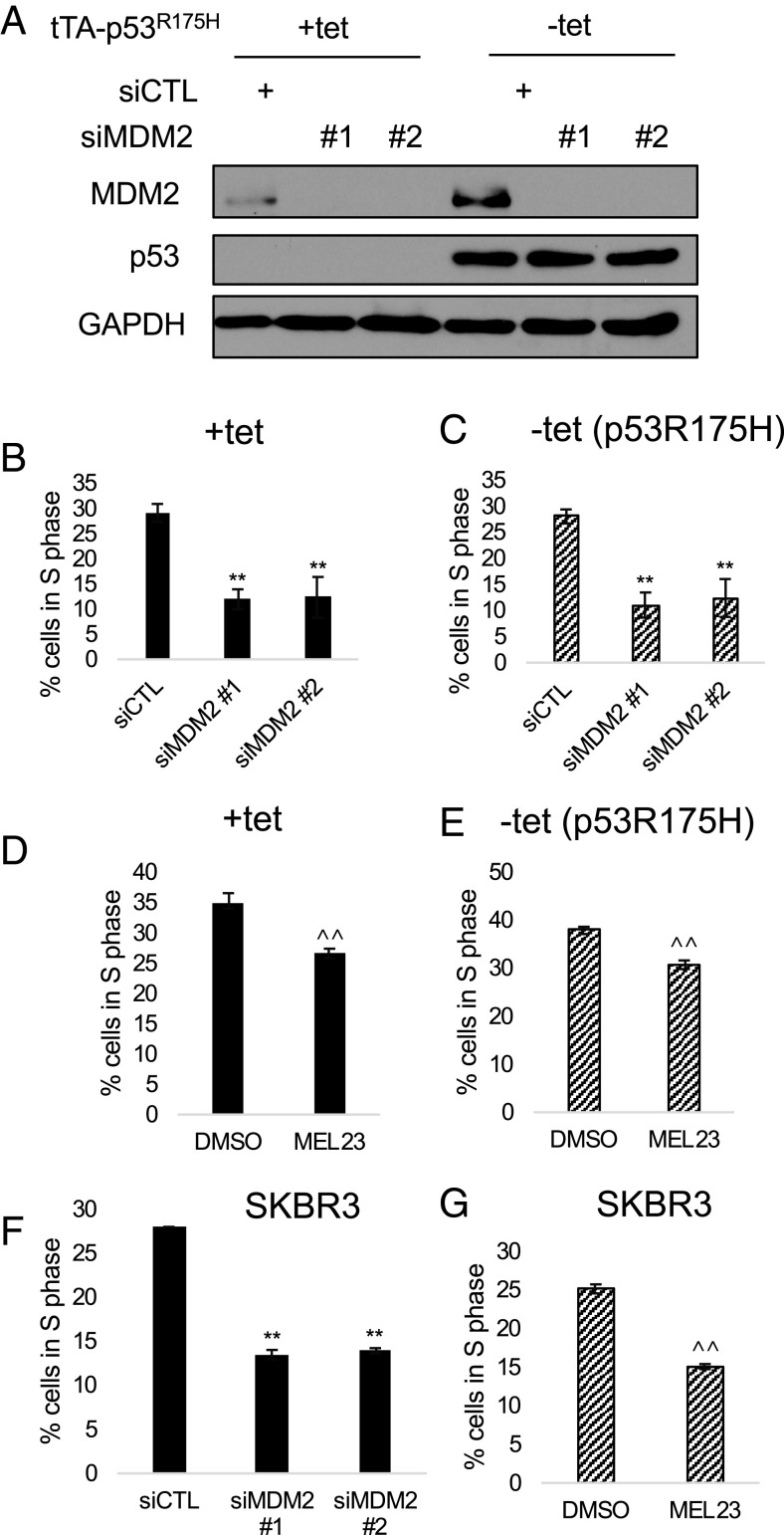
The p53R175H gain-of-function mutation is frequently found in aggressive human cancers; it both exerts a dominant-negative effect over p53 and contributes to significant tumorigenic activity in the absence of wild-type p53 (58). MDM2 expression was knocked down with RNAi in H1299 cells expressing p53R175H controlled by a tetracycline-off promoter system. Expression of p53R175H failed to rescue cell-cycle arrest caused by loss of MDM2 (Fig. 2 A–C) or by pharmacological inhibition of the heterocomplex (Fig. 2 D and E). In SKBR3 cancer cells expressing the p53R175H mutant endogenously, ablation of MDM2 or the heterocomplex by RNAi or MEL23 also caused a cell-cycle arrest, indicating that the effect is not due to the inducible promoter system (Fig. 2 F and G). These results were further recapitulated in cancer cell lines expressing other p53 mutants, both ectopically (H1299 tetracycline-controlled p53R248W) and endogenously (MDA-MB-231) (Table 1 and SI Appendix, Fig. S3). We conclude that MDM2 is needed for cell-cycle progression both in cancers that lack p53 expression entirely and in those that express at least some gain-of-function mutant p53 alleles.
Fig. 2.
Mutant p53 does not counteract the effect of MDM2 inhibition on the cell cycle. (A) H1299 cells with a tet-off promoter controlling expression of p53R175H were transfected with siMDM2 or control siRNA for 48 h in the presence (p53R175H-suppressed) or absence (p53R175H-induced) of tetracycline. Knockdown of MDM2 and expression of p53R175H upon removal of tetracycline were verified by immunoblot. (B and C) Cell-cycle distribution was analyzed using flow cytometry and the percentage of cells in S phase was plotted for cells lacking p53 expression (B) and cells in which p53R175H was induced by withdrawal of tetracycline (C). Graphs show the mean of four biological replicates (error bars represent SEM). (D and E) H1299 cells with or without expression of p53R175H were treated with DMSO or MEL23 (15 μM) for 24 h. A representative graph of four biological replicates of cell-cycle distribution in cells without p53 (D) and with p53R175H expression (E) is shown. (F and G) SKBR3 cells were transfected with siMDM2 or control siRNA for 48 h (F) or treated with DMSO or MEL23 (15 μM) for 24 h (G). Representative examples of three biological replicates of cell-cycle distribution are shown. **P < 0.01 (one-way ANOVA), ^^P < 0.01 (unpaired t test). All data represent at least three biological replicates (error bars represent SEM).
Table 1.
Multiple cancer cell lines lacking p53 require MDM2 and MDMX to maintain the cell cycle and survival
| Cell line | p53 status | Response to siMDM2 | Response to MEL23 |
| H1299 | Null | Arrest | Arrest and death |
| MDA-MB-231 CRISPR p53KO | Null | Arrest | Arrest and death |
| MCF7 CRISPR p53KO | Null | Arrest | Arrest and death |
| A549 CRISPR p53KO | Null | Arrest | No effect |
| H1299 tet-R175H | R175H | Arrest | Arrest and death |
| H1299 tet-R248W | R248W | Arrest | Arrest and death |
| MDA-MB-231 | R280K | Arrest | Arrest and death |
| SKBR3 | R175H | Arrest | Arrest and death |
Cell-cycle arrest in response to MDM2 knockdown or MEL23 treatment in different cancer cells lacking p53 expression or expressing different p53 point mutants. The cell cycle was measured as in Figs. 1, 2, and 3. “Arrest” indicates reduction in the percentage of cells in S phase. “Death” was measured by the percentage of cells in sub-G1. All data represent at least three biological replicates.
MDM2 and MDMX Maintain the Cell Cycle and Survival in Multiple p53-Null Cell Types.
To demonstrate that the effect seen in H1299 cells was applicable to p53-null cancers beyond non–small-cell lung carcinoma, we knocked out p53 using CRISPR-Cas9 editing in cells derived from other human lung and breast cancers.
Several cancer cell lines were engineered to lose p53 expression through CRISPR-Cas9 editing, generating p53 knockout (KO) versions. Upon RNAi knockdown of MDM2, all of these KO cell lines arrested in S phase (Table 1 and SI Appendix, Fig. S4). MEL23 also caused cell-cycle arrest in MDA-MB-231 KO and MCF7 KO cells, as well as H1299 cells. MEL23 likely did not inhibit the E3 ligase activity of MDM2 in A549 cells—as measured by changes in MDMX protein stabilization—and thus the effect of pharmacological inhibition of MDM2 could not be tested in these cells. Nevertheless, the broad response to loss of MDM2 in various p53-null cancer cell lines indicates that MDM2 and MDMX are required for cell-cycle progression and survival in multiple human cancer types.
MDM2 Maintains E2F1 and E2F3a Levels in H1299 Cells.
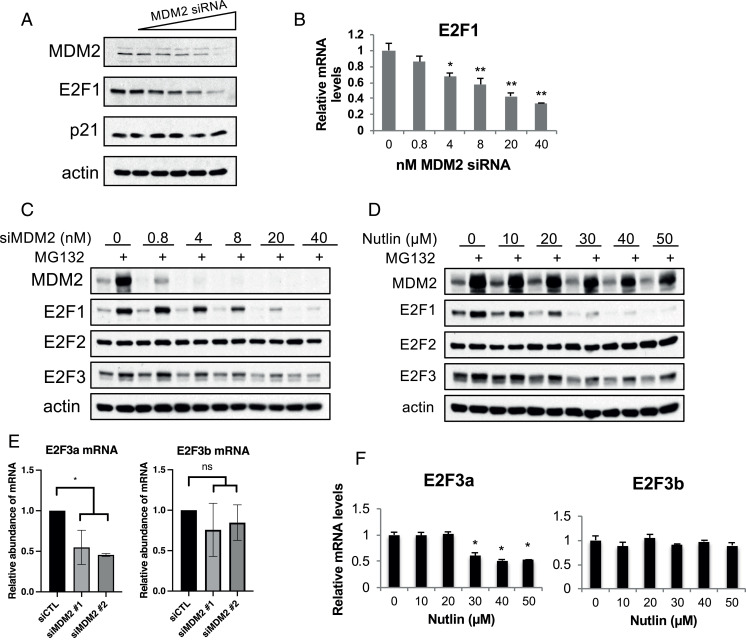
MDM2 was previously reported to regulate E2F1 both positively and negatively through multiple mechanisms (36). As E2F1 is a key regulator of the cell cycle and apoptosis, we examined a dose–response for the effect of MDM2 knockdown on E2F1 protein and messenger RNA (mRNA) levels. E2F1 protein levels were reduced by MDM2 siRNA in a dose-dependent manner (Fig. 3A), suggesting MDM2 maintains E2F1 levels. Reduced MDM2 levels did not affect p21 levels (also shown in Fig. 1). Transcription of the E2F1 gene is regulated by E2F transcription factors, including E2F1, generating a positive feedback loop (59). Accordingly, E2F1 mRNA levels tracked with the protein, and showed a dose-dependent reduction of E2F1 mRNA upon MDM2 siRNA treatment (Fig. 3B). Pharmacological inhibition of the heterocomplex with MEL23 also resulted in lower E2F1 protein (Fig. 4B). Treatment with the MDM2 inhibitor SP-141 also decreased protein levels of E2F1 in a dose-dependent fashion, indicating the effect is not specific to genetic ablation of MDM2 (Fig. 4C).
Fig. 3.
MDM2 is required for maintaining E2F1 and E2F3a levels. H1299 cells were transfected with increasing amounts of MDM2 siRNA as stated in the figure (siRNA concentration was balanced up to 40 nM with siLuc in each sample) (A–C), with increasing concentrations of Nutlin-3 for 20 h (D and F), or with control or MDM2 siRNA (each at 15 nM) for 24 h (E) as indicated. (A) Protein levels of MDM2, E2F1, p21, and actin were measured by immunoblot. (B) E2F1 mRNA levels were quantitated by qRT-PCR analysis. (C and D) Following either MDM2 siRNA (C) or Nutlin-3 (D) treatment, the cells were then either mock treated or treated with MG132 (25 μM) for an additional 4 h. Protein levels were analyzed by immunoblot using antibodies against MDM2, E2F1, E2F2, E2F3, and actin. (E and F) mRNA levels of E2F3a or E2F3b following MDM2 siRNA (15 nM) treatment (E) or Nutlin-3 treatment (F) were quantified by qRT-PCR. *P < 0.05 (one-way ANOVA), **P < 0.01 (one-way ANOVA); ns, not significant (one-way ANOVA). All data represent at least three biological replicates (error bars represent SEM).
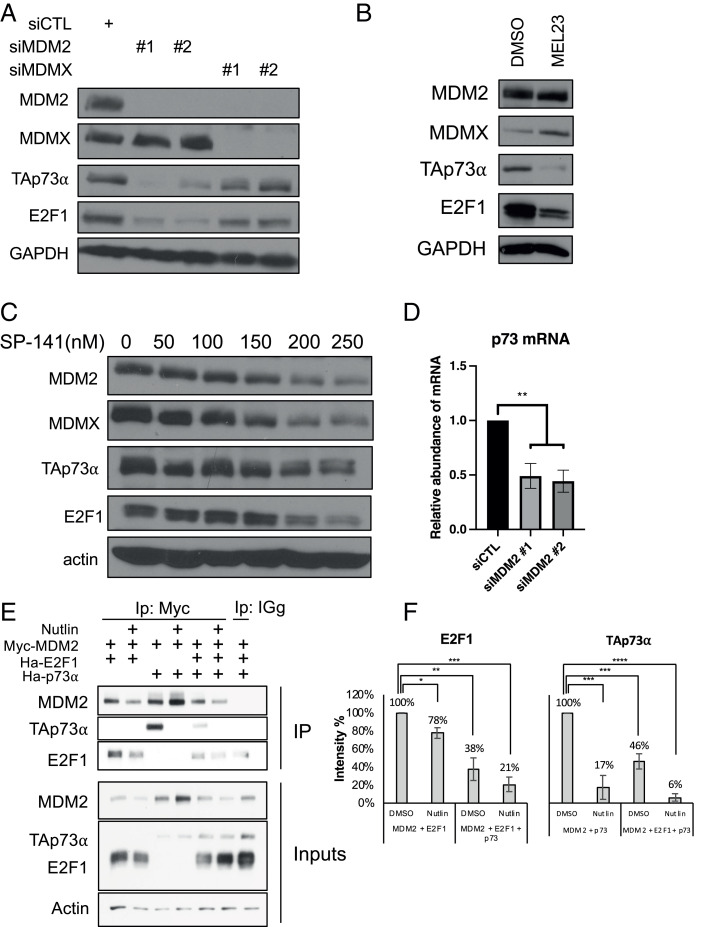
Fig. 4.
MDM2 and MDMX are required for maintaining levels of p73 and E2F1 in cells. (A) H1299 cells were transfected with siMDM2, siMDMX, or control siRNA for 48 h and levels of the indicated proteins were determined by immunoblotting. (B) H1299 cells were treated with DMSO or MEL23 (15 μM) for 24 h and levels of the indicated proteins were determined by immunoblotting. (C) H1299 cells were treated with increasing concentrations of SP-141 as indicated. Levels of MDM2, MDMX, TAp73α, and E2F1 were measured by immunoblotting. (D) H1299 cells were treated with siMDM2 or control siRNA and p73 mRNA was quantified by qRT-PCR. **P < 0.01 (one-way ANOVA). (E) H1299 cells were transfected with constructs expressing Myc-MDM2 (2 μg), HA-E2F1 (1.7 μg), and/or HA-TAp73 (0.3 μg), as indicated. Twenty-four hours later, cells were treated with MG132 (20 μM) for 4 h, in the absence or presence of Nutlin-3 (40 μM), as indicated. Cell lysates were prepared and subjected to immunoprecipitation with anti-Myc antibody to pull down MDM2, followed by immunoblotting with anti-HA antibody to detect E2F1 and TAp73. (F) The ratios of MDM2:E2F1 or MDM2:p73 (performed either with Myc-MDM2 or untagged MDM2) were quantified by densitometry and graphed as averages of six independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.00001. All data represent at least three biological replicates (error bars represent SEM).
As mentioned in the Introduction, E2F1, E2F2, and E2F3a are activator E2Fs that share common regulation and functional characteristics, including cell-cycle activation (26, 28, 60). MDM2 has previously been reported to bind and stabilize E2F1 (25). Hence, we examined whether MDM2 influences the levels of E2F2 and E2F3. Additionally, to test whether MDM2 protects the E2Fs from proteasomal degradation, we treated the cells with the proteasome inhibitor MG132. H1299 cells were transfected with increasing amounts of MDM2 siRNA and then treated with vehicle or MG132 for an additional 4 h (Fig. 3C). Protein levels for E2F1 and E2F3, but not E2F2, were reduced in siMDM2-treated cells compared with controls. Treatment with the proteasome inhibitor MG132 stabilized MDM2, E2F1, and E2F3, suggesting an involvement of the proteasome in the down-regulation of E2F1 and E2F3 upon MDM2 ablation. E2F2 and actin, on the other hand, were not affected by the MG132 treatment.
The E2F1 TAD was reported to interact with MDM2 in a manner similar to p53 (61). E2F2 and E2F3 are highly homologous to E2F1 (62) and may also interact with MDM2. To gain further insight into the regulation of E2F family members by MDM2, we utilized the small molecule Nutlin-3, which disrupts MDM2 interaction with E2F1 (63). We hypothesized that if MDM2 stabilizes E2F1 by a direct protein–protein interaction, then Nutlin-3 treatment would mimic the effect of MDM2 knockdown and reduce E2F1 and E2F3 levels. H1299 cells were treated with increasing amounts of Nutlin-3 and then either left untreated or cotreated with MG132 (Fig. 3D). Similar to MDM2 depletion, E2F1 and E2F3, but not E2F2, levels were reduced in response to Nutlin-3 treatment in a dose-dependent manner, and were stabilized by cotreatment with MG132. Since we also found that in p53-null PC-3 prostate cancer cells, Nutlin-3 treatment led to lower E2F1 protein levels (SI Appendix, Fig. S5A) and cell-cycle arrest (SI Appendix, Fig. S5B), this is not a cell line–specific effect.
Following both MDM2 siRNA and Nutlin-3 treatment, E2F3 protein stabilization by MG132 behaved similar to E2F1 stabilization, suggesting MDM2 may regulate E2F3 transcription as well. We used increasing concentrations of Nutlin-3 and examined E2F3 mRNA levels by qRT-PCR. E2F3 has two isoforms, E2F3a and E2F3b, generated by alternate promoter sites (64). While E2F3a is regulated by the cell cycle and has a cell cycle–activating role similar to E2F1, E2F3b levels are constant in cycling and quiescent cells (28). Moreover, E2F1−/− E2F3b−/− mice are viable whereas E2F1−/− E2F3a−/− mice are not, suggesting functional similarity of E2F1 and E2F3a (65). E2F3a mRNA levels were lowered upon knockdown of MDM2 while E2F3b levels were not significantly reduced (Fig. 3E). Similarly, Nutlin-3 treatment resulted in a reduction in E2F3a mRNA levels but not E2F3b mRNA levels (Fig. 3F). Interestingly, ablation of MDMX did not cause decreased E2F activator or E2F3b mRNA levels (SI Appendix, Fig. S6 A–D).
Taken together, our results indicate that E2F1 and E2F3a are subject to at least two modes of regulation by MDM2, one involving protection from proteasomal degradation and the other functioning at the level of RNA accumulation.
p73 Expression Is Maintained by MDM2 and Is Required for Cell-Cycle Progression.
The p53 family member p73 is similar to p53 in both upstream and downstream aspects. Upstream, MDM2 antagonizes both p53 and p73; however, while MDM2 promotes degradation of p53, both MDM2 and MDMX bind and stabilize p73, and inhibit its transcriptional activity (47–50). Both have affinity for p73 that is reported to be comparable to the affinity of MDM2 for p53 (66) and MDM2 interaction with p73 can be disrupted with Nutlin-3 treatment (67). Downstream, p73 canonically activates several transcriptional targets of p53, including factors typically associated with proarrest or proapoptotic pathways, such as p21, 14-3-3σ, BAX, NOXA, and PUMA (68). Note, however, that p73 has also been shown to have targets distinct from those of p53, including antiapoptotic and progrowth genes (51). Given the relationship between MDM2, MDMX, and p73, and the role of MDM2 and MDMX in maintenance of cell-cycle progression, we considered that p73 might have heretofore little explored functions in cell-cycle progression in p53-null cells.
We knocked down MDM2 or MDMX with RNAi and observed a decrease in p73 protein (Fig. 4A). As the commercial antibody we used detects multiple p73 isoforms, we also immunoblotted with an antibody specific to TAp73 isoforms (SI Appendix, Fig. S7 A and B). The effect was notably stronger with siMDM2 than with siMDMX. Inhibiting the E3 ligase activity of the MDM2–MDMX heterocomplex with MEL23 also decreased the levels of p73 and E2F1 protein (Fig. 4B), suggesting that the E3 ligase activity of the MDM2–MDMX heterocomplex is specifically required for maintaining p73 expression. Ablating MDM2 with SP-141 also decreased p73 and E2F1 expression in a dose-dependent fashion (Fig. 4C). A possible explanation for the down-regulation of p73 expression was obtained when we found that ablation of MDM2 was correlated with a significant drop in the levels of p73 mRNA (Fig. 4D). Interestingly, the effects on p73 protein were more pronounced with siMDM2 than with siMDMX; depletion of MDMX did not affect p73 mRNA levels (SI Appendix, Fig. S6E), which suggests that MDMX may not directly regulate p73.
To gain more insight into the relationship between MDM2, E2F1, and p73, we expressed Myc-tagged MDM2, along with either hemagglutinin (HA)–tagged E2F1 or HA-tagged TAp73α, exogenously in H1299 cells, and subjected cell extracts to immunoprecipitation with anti-Myc antibody (Fig. 4E). In this setting, we found that MDM2 and E2F1 could be coimmunoprecipitated as expected, and that the amount of E2F1 associated with MDM2 was reduced upon Nutlin-3 treatment, as was the total amount of ectopic E2F1. (This is consistent with our data examining endogenously expressed proteins; Fig. 3D.) Reciprocally, the level of ectopically expressed MDM2 was reduced when coexpressed with E2F1 (SI Appendix, Fig. S7C). This further supports the likelihood that MDM2 can bind and stabilize E2F1 protein and that disruption of their interaction results in the proteasomal degradation of E2F1, as seen in Fig. 3C. Additionally, the association between MDM2 and E2F1, which can be disrupted by Nutlin-3, occurred whether p73 was present or absent. Nutlin-3 treatment also reduced the amount of p73 in the complex with MDM2 independent of E2F1. The amounts of E2F1 and p73 in the complex were reduced further when they were both coexpressed with MDM2 (Fig. 4 E and F). Although the levels of MDM2 were marginally decreased upon Nutlin-3 administration in the presence of E2F1, the association of both E2F1 and p73 with MDM2 was attenuated to a greater relative extent (quantified in Fig. 4F). Overall, this suggests that E2F1 and p73 likely bind to MDM2 in the vicinity of the Nutlin-3 interaction sites on MDM2. Altogether, these findings indicate that MDM2 may regulate p73, as well as E2F1, at both the protein and mRNA levels.
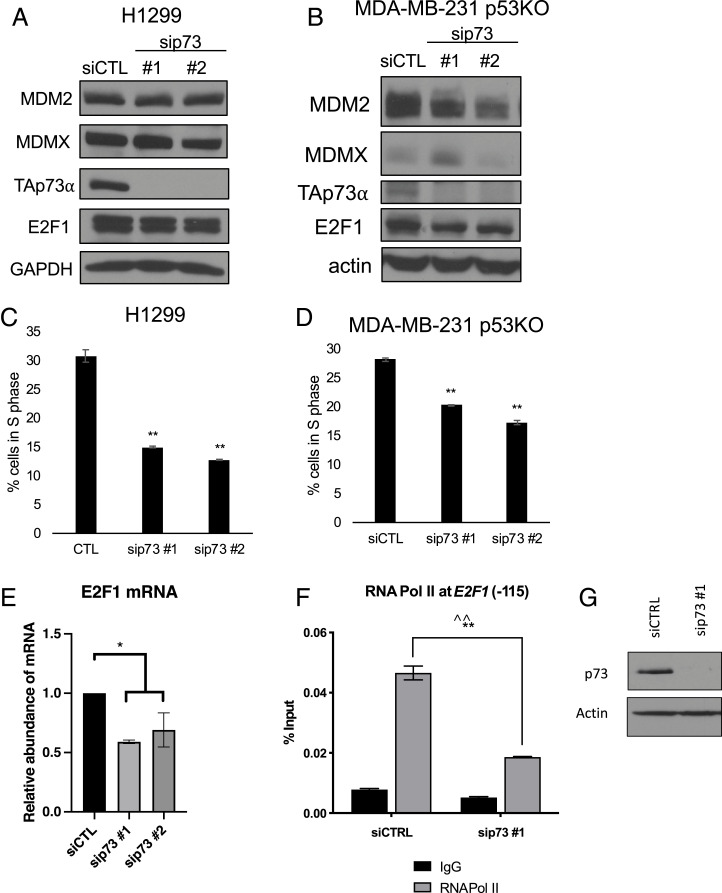
The finding that depletion or inhibition of MDM2 (and in some cases MDMX) led to reduced p73 expression posed the question as to whether p73 itself might have a similar effect on cell-cycle progression. To this end, we used RNAi to knock down p73 directly in H1299 cells and MDA-MB-231 KO cells (Fig. 5 A and B). Remarkably, ablation of p73 caused both a cell-cycle arrest in these cell lines (Fig. 5 C and D)—that appeared to be phenotypically identical to the G1 arrest caused by loss of MDM2 (SI Appendix, Fig. S8A)—as well as down-regulation of E2F1 mRNA (Fig. 5E).
Fig. 5.
Ablation of p73 inhibits cell-cycle progression and reduces the E2F1 message. (A–D) H1299 (A and C) and MDA-MB-231 with p53 knocked out by CRISPR-Cas9 (B and D) cells were transfected with sip73 or control siRNA for 48 h. (A and B) Knockdown was verified by immunoblot. (C and D) Cell-cycle distribution was analyzed with PI staining and the percentage of cells in S phase was plotted as a representative of four replicates. (E) E2F1 mRNA was quantified by qRT-PCR. The graph shows the mean of three biological replicates (error bars represent SEM). (F and G) H1299 cells were transfected with control (siCTRL) siRNA or siRNA against p73 (sip73 no. 1). Chromatin was immunoprecipitated with anti-RNA Pol II or IgG control and subjected to qRT-PCR using a primer −115 from the TSS of E2F1. (F) Binding of RNA Pol II to the −115 region of the E2F1 promoter is graphed as a representative of three biological replicates with two different siRNAs against p73. (G) Knockdown was verified by immunoblot. *P < 0.05 (one-way ANOVA), **P < 0.01 (one-way ANOVA), ^^P < 0.01 (Welch’s t test). All data represent at least three biological replicates (error bars represent SEM).
To determine whether the effect on E2F1 mRNA expression was caused by decreased transcription, we performed chromatin immunoprecipitation (ChIP) of RNA polymerase II (Pol II) at −115 from the transcription start site (TSS) of the E2F1 promoter in the setting of p73 ablation. Indeed, loss of p73 resulted in decreased binding of Pol II to the E2F1 promoter (Fig. 5 F and G). To reduce the chance of off-target effects, we replicated these results with an RNAi pool directed against p73 (SI Appendix, Fig. S8 B and C). E2F1, E2F2, E2F3a, and E2F3b are both regulators and markers of the cell cycle (28). Therefore, we examined other E2F activators following knockdown of p73 and found that E2F2, E2F3a, and E2F3b mRNA levels were not significantly changed (SI Appendix, Fig. S8 D–F), indicating that regulation of the E2F1 message under these conditions is not a consequence of cell-cycle arrest.
Taken together, our findings support a p53-independent pathway regulated by the MDM2–MDMX heterodimer involving E2F1 and a function for p73 in promoting cell-cycle progression in cancer cells.
Discussion
MDM2 and MDMX have been mostly studied in the context of their p53-related activities. Similarly, p73 is classically understood to parallel p53 regulation and function, activating a subset of p53 targets involved in cell-cycle arrest and cell-death pathways. Here we have linked these factors to a p53-independent pathway that controls cell growth and survival. We find that MDM2, MDMX, and the MDM2–MDMX heterocomplex are required for cell-cycle progression in p53-null cancer cells. Our results reveal that the MDM2–MDMX complex maintains expression of p73 and, unexpectedly, depletion of p73 mimics inhibition of MDM2–MDMX by impairing cell-cycle progression in p53-null H1299 cells. MDM2 is also required to maintain expression of E2F1 and E2F3 protein and mRNA, while p73 is required for transcription of E2F1.
While MDM2 has been linked to cell-cycle progression in a p53-independent fashion before, the role of MDMX and the MDM2–MDMX heterocomplex has not been as well explored, despite the fact that heterodimerization is crucial to MDM2 E3 ubiquitin ligase activity and heterodimers are more abundant than homodimers (54, 69). Notably, ablation of MDMX had a less pronounced effect on the cell cycle and on E2F1 and p73 protein or mRNA than ablation of MDM2. MDMX lacks E3 ligase activity but is important for MDM2 stability and E3 activity (9, 54), and thus the loss of MDMX destabilizes the MDM2–MDMX heterodimer and is likely to reduce but not eliminate MDM2 activity. Our work indicates that disruption of the MDM2–MDMX heterocomplex as a distinct entity, as well as specific inhibition of its catalytic activity, has a profound effect on cell-cycle progression.
It is notable that neither ectopic nor endogenous expression of mutant p53 was sufficient to rescue cells from arrest caused by loss of MDM2. Although previous work has indicated that MDM2 may regulate at least some forms of mutant p53 (70), we observed that MDM2 and the MDM2–MDMX heterocomplex regulate the cell cycle independent of both wild-type and mutant p53. The possibility that MDM2 controls a mechanism connected to cell-cycle arrest, independent of both wild-type and mutant p53, is intriguing and merits further exploration. As mutant p53 is responsible for several tumorigenic processes, including unchecked proliferation and survival, targeting MDM2 and the MDM2–MDMX heterocomplex may potentially be an avenue to divert cells from the oncogenic processes enabled by mutant p53.
MDM2 has been proposed to regulate the cell cycle by modulating expression of the CDK inhibitor p21. MDM2 (71–73) and MDMX (74) can target p21 for proteasomal degradation and their ablation in H1299 was previously shown to stabilize p21 protein and arrest cells in G1. However, we did not observe an increase in p21 protein upon knockdown of MDM2 or MDMX, indicating that the cell-cycle arrest seen in our experiments may be independent of p21 as well as p53. Furthermore, the above-mentioned studies described a pathway independent of MDM2-mediated ubiquitylation, whereas we describe a different mechanism, in which MDM2 E3 ubiquitin ligase activity is critical.
We showed that endogenous p73 is down-regulated by loss of MDM2, MDMX, or MDM2–MDMX. Furthermore, direct ablation of endogenous p73 phenocopies the cell-cycle arrest caused by knockdown of MDM2 or MDMX, or direct inhibition of the MDM2–MDMX heterocomplex. Our finding that ablation of endogenous p73 causes a cell-cycle arrest was an unexpected result, given the broad body of work showing that p73 activates various arrest and death pathways, as well as its frequent classification as a tumor suppressor (75, 76). For example, p73 has recently been proposed to mediate the proapoptotic activity of protoporphyrin IX (77). Additionally, Feeley et al. (78) have also reported that loss of MDM2 causes arrest in p53-null mouse lymphoma, sarcoma, and fibroblast cells, which is accompanied by up-regulation of p73 protein. As one possible explanation for the difference between their findings and ours is that this group examined nonepithelial mouse cell types (78), whereas our results were obtained in epithelial-derived human carcinomas. Alternately, since multiple N-terminal isoforms of p73 are generated from alternate promoter sites [TAp73 and ΔNp73, each with distinct TADs (79)], as well as C-terminal isoforms generated from different splicing sites [e.g., p73α, p73β, and p73γ (68)], and these isoforms have different transcriptional activities (42), it is possible that the cell lines we have tested express p73 isoforms with functions that promote the cell cycle. We used a TAp73-specific antibody (Fig. 4B) to identify and follow TAp73α, but it is possible that other TAp73 C-terminal isoforms are responsible for different effects on the cell cycle.
p73 levels may also respond to E2F ablation. p73 is a transcriptional target of E2F, required for the activation of E2F1-induced apoptosis (80–82). Moreover, MDM2 cooperates with E2F1 in the activation of TAp73 (83). We demonstrated that loss of MDM2, MDMX, or MDM2–MDMX activity down-regulates E2F1 and E2F3a, but not E2F2, both transcriptionally and at the proteasomal level. These results agree with previous reports where an MDM2 antisense oligonucleotide reduced E2F1 protein levels in PC-3 cells (25). However, direct ablation of p73 also down-regulated E2F1 mRNA and inhibited transcription initiation by RNA Pol II at the −115 TSS in the E2F1 promoter. While this is the reverse of the classically described E2F1 transactivation of p73, it may be that, under unstressed conditions, p73 requires E2F1 activity to promote cell-cycle progression or that the two factors cooperate in advancing the cell cycle. Furthermore, given that MDM2 is also a transcriptional target of p73 (84), it is also possible that MDM2, p73, and E2F1 cooperate in a feedback loop, in which MDM2 promotes p73 activity, which in turn promotes E2F1 and MDM2 expression—ultimately stimulating cell-cycle progression. Further exploration of the relationship between MDM2, E2F1, and p73 may elucidate targetable mechanisms that can ameliorate unchecked cell growth in cancer.
The relationship between MDM2, MDMX, and their putative targets E2F1, E2F3a, and p73 suggests a mechanism for regulation of the cell cycle in the absence of wild-type p53. Although further exploration is necessary for defining these interactions, our findings provide an intriguing glimpse at potential therapeutic targets for p53-null and some mutant p53-expressing cancers.
Materials and Methods
Cell Culture.
H1299, MDA-MB-231, A549, MCF7, and PC-3 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS). SKBR3 cells were maintained in McCoy’s 5a medium modified supplemented with 10% FBS. H1299 cells with tetracycline-off promoters were maintained in DMEM supplemented with 10% FBS with 1 μg/mL puromycin and 100 μg/mL G418, with 2.25 μg/mL tetracycline to maintain no expression of mutant p53, or no tetracycline when mutant p53 expression was induced. Drug treatments were as follows: Nutlin-3 (10 to 50 μM as indicated; Sigma-Aldrich) was administered for 20 h. Treatment with MG132 (20 μM; Calbiochem) was carried out for 4 h. MEL23 (15 μM; Millipore Sigma; 373227) was administered for 24 h. SP-141 (250 nM; Tocris; 5332) was administered for 24 h.
Transfection.
siRNA duplexes (QIAGEN and Invitrogen) were transfected into cells with Lipofectamine RNAiMAX transfection reagent (Thermo Fisher Scientific) or DharmaFECT 1 reagent (Dharmacon; Thermo Fisher Scientific) for 48 h. siRNA was transfected at a concentration of 15 nM for single transfections and 30 nM for cotransfections, except where noted in dose curves. siLuc (80), Silencer Select negative control (Thermo Fisher Scientific), or ON-TARGETplus Nontargeting Pool (Dharmacon): 5′-UGGUUUACAUGUCGACUAA-3′, 5′-UGGUUUACAUGUUGUGUGA-3′, 5′-UGGUUUACAUGUUUUCUGA-3′, and 5′-UGGUUUACAUGUUUUCCUA-3′, were used for negative control. siMDMX no. 1: Hs_MDM4_4 FlexiTube siRNA (QIAGEN); siMDMX no. 2: 5′-AGAUUCAGCUGGUUAUUAA-3′ (85); siMDM2 no. 1: Hs_MDM2_5 FlexiTube siRNA; siMDM2 no. 2: 5′-AAGCCAUUGCUUUUGAAGUUA-3′ (71); siMDM2 no. 3: Hs_MDM2_9 FlexiTube siRNA; sipRb: 5′-AAGATACCAGATCATGTCAGA-3′ (86); sip107: 5′-CAAGAGAAGUUGUGGCAUAUU-3′ (87); sip130: 5′-GAGCAGAGCUUAAUCGAAUUU-3′ (87); sip73 no. 1: Thermo Fisher Silencer Select s14319; sip73 no. 2: Thermo Fisher Silencer Select s14320; sip73 pool: ON-TARGETplus Human TP73 (7161) siRNA-SMARTpool (Dharmacon): 5′-GAGACGAGGACACGUACUA-3′, 5′-GCAAUAAUCUCUCGCAGUA-3′, 5′-GAACUUUGAGAUCCUGAUG-3′, 5′-CCACCAUCCUGUACAACUU-3′.
qRT-PCR.
The QIAGEN RNeasy Mini Kit was used for RNA isolation and QuantiTect Reverse Transcription Kit (QIAGEN) was used for complementary DNA synthesis. PCR was performed with the StepOnePlus Real-Time PCR Machine using Power SYBR Green PCR Master Mix (Applied Biosystems; Life Technologies). Relative mRNA levels were calculated using the ΔΔCt method, and the data were normalized first to the levels of control RPL32 mRNA and then to the levels of the respective mRNA in untreated siLuc or Silencer Select negative control samples. Graphs are representative of multiple independent experiments, with error bars representing technical PCR replicates. Primer sequences: L32 forward (F): 5′-TTCCTGGTCCACAACGTCAAG-3′; L32 reverse (R): 5′-TGTGAGCGATCTCGGCAC-3′; E2F1 (F): 5′-AGATGGTTATGGTGATCAAAGCC-3′; E2F1 (R): 5′-ATCTGAAAGTTCTCCGAAGAGTCC-3′; E2F3a (F): 5′-TTTAAACCATCTGAGAGGTACTGATGA-3′; E2F3a (R): 5′-CGGCCCTCCGGCAA-3′; E2F3b (F): 5′-TTTAAACCATCTGAGAGGTACTGATGA-3′; E2F3b (R): 5′-CCCTTACAGCAGCAGGCAA-3′.
Immunoblot Analysis: Western Blot.
Whole-cell lysates were analyzed by a standard immunoblotting procedure as described (80, 88), with lysis buffer described separately (89). Commercially obtained antibodies used in this study were as follows: pRB (IF-8) SC-102, p107 (C-18) SC-318, p130 (C-20) SC-317, E2F1 (KH95) SC-251, E2F2 (C-20) SC-318, E2F3 (C-18) SC-878, p21 (C-19) SC-397 (all SC are antibodies from Santa Cruz Biotechnology), p21 (12D1) (Cell Signaling Technology; 2947), p73 EP436Y (Abcam; ab40658), GAPDH 14C10 (Abcam; 2118), mouse HA-Tag (Cell Signaling Technology; C29F4), rabbit HA-Tag (BioLegend; 905102), actin (Sigma-Aldrich; A2066), and MDM2 DIV2Z (Cell Signaling Technology; 86934). The monoclonal MDMX antibody (8C6) was a kind gift from Jiandong Chen, H. Lee Moffitt Comprehensive Cancer Center, Tampa, FL. The following mouse monoclonal antibodies were used as hybridoma supernatants: p53 (DO-1, 1801) and MDM2 (3G5, 5B10, 4B11). The affinity-purified rabbit polyclonal TAp73-specific antibody was described previously (80).
Immunoprecipitation.
H1299 cells were transfected with myc-MDM2 (2 μg), untagged MDM2 (2 μg), HA-E2F1 (1.7 μg), or HA-p73α (0.3 μg) (Addgene; 22102) constructs, as indicated. The cells were harvested in lysis buffer A (10 mM Tris·HCl, pH 7.5, 137 mM NaCl, 10% glycerol, and 1% Nonidet P-40 with 50 nM phenylmethanesulfonylfluoride and inhibitor mixture containing 100 μM benzamidine, 300 μg/μL leupeptin, 100 mg/mL bacitracin, and 1 mg/mL a2-macroglobulin), and cleared by centrifugation (13,000 rpm for 10 min at 4 °C). The total protein concentration was measured using the Bio-Rad protein assay (Life Science Research). All the following steps were performed at 4 °C. Equivalent amounts (200 to 400 μg) of each clarified cell lysate was subjected to immunoprecipitation with 1 μg of purified MDM2 antibody as indicated for 2 h (DIV2Z; Cell Signaling Technology; 86934 for untagged MDM2; or Myc-Tag; Cell Signaling Technology; 9B11 for Myc-MDM2). Protein G beads (40 μL) (GE Healthcare) preblocked with bovine serum albumin (BSA) (New England BioLabs) were added for an additional hour. Following three washes with 1 mL of lysis buffer A (without protease inhibitors), proteins were eluted by adding protein sample buffer and incubated at 95 °C for 10 min. Immunoblotting analysis was performed as described previously. Quantification of immunoblotting data was carried out using ImageJ software. In each case, the protein intensity value of E2F1 or p73 was compared with the value of MDM2 in the corresponding condition. P values were calculated by comparing the ratio of MDM2:E2F1 or MDM2:p73 with the initial levels without Nutlin-3.
Cell-Cycle Analysis by Flow Cytometry.
Cell pellets were washed with phosphate-buffered saline (PBS) and fixed/permeabilized with 50% ice-cold ethanol. Pellets were washed with PBS and resuspended in PBS containing 50 μg/mL ribonuclease A and 62.5 μg/mL propidium iodide (PI; Sigma-Aldrich). Samples were analyzed using FACSCalibur (Becton Dickinson). ModFit LT version 3.0 program (Verity Software House) and FlowJo (Becton Dickinson) were used to determine the percentage of cells in each stage of the cell cycle. EdU labeling was performed with the Click-iT Plus EdU Alexa Fluor 647 Flow Cytometry Assay Kit (Thermo Fisher Scientific; C10634) according to the manufacturer’s instructions.
Chromatin Immunoprecipitation.
ChIP experiments were carried out as previously described (90). Briefly, H1299 cells were lysed by incubating on ice for 20 min in RIPA buffer (150 mM NaCl, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate, 50 mM Tris·HCl, pH 8.0, 5 mM ethylenediaminetetraacetate, and protease inhibitors). Lysed cells were sonicated to generate DNA fragments with an average length of ∼500 to 1,000 bp. Sonicated samples were then incubated at 4 °C overnight with the indicated antibodies and protein A/G beads were preblocked with 1 mg/mL BSA and 0.3 mg/mL salmon sperm DNA. Antibodies used for ChIP were as follows: anti-RNA Pol II C-terminal domain (phospho Ser5) antibody (Active Motif; 39749) and rabbit immunoglobulin G (IgG) (Diagenode; C15410206) for a negative control. DNA was purified using the QIAquick PCR Purification Kit (QIAGEN; 28106). Quantitative ChIP was performed on a StepOnePlus Real-Time PCR System (Thermo Fisher Scientific) using Power SYBR Green Master Mix (Thermo Fisher Scientific; 4367659) versus genomic standard DNA and input DNA. ChIP primers designed with Primer Express (Applied Biosystems) were derived from the University of California, Santa Cruz (UCSC) Human Genome Browser hg19 assembly and sequence specificity was confirmed with the UCSC Human Genome Browser in silico PCR tool. ChIP-qPCR primer sequences were as follows: E2F1 TSS (−115) (F): 5′-AAAGTCCCGGCCACTTTTAC-3′; (R): 5′-GCCAATTGTGGCGGCG-3′.
Acknowledgments
We thank Ella Freulich for technical assistance and many Prives laboratory members for thought-provoking discussions. We are particularly indebted to Dr. Jiandong Chen for supplying an antibody directed against MDMX. This work was supported by Grant CA220526 from the NIH to C.P.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2102420118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Olivier M., Hollstein M., Hainaut P., TP53 mutations in human cancers: Origins, consequences, and clinical use. Cold Spring Harb. Perspect. Biol. 2, a001008 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karni-Schmidt O., Lokshin M., Prives C., The roles of MDM2 and MDMX in cancer. Annu. Rev. Pathol. 11, 617–644 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haupt Y., Maya R., Kazaz A., Oren M., Mdm2 promotes the rapid degradation of p53. Nature 387, 296–299 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Honda R., Tanaka H., Yasuda H., Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420, 25–27 (1997). [DOI] [PubMed] [Google Scholar]
- 5.Stad R., et al., Hdmx stabilizes Mdm2 and p53. J. Biol. Chem. 275, 28039–28044 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Leslie P. L., Zhang Y., MDM2 oligomers: Antagonizers of the guardian of the genome. Oncogene 35, 6157–6165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharp D. A., Kratowicz S. A., Sank M. J., George D. L., Stabilization of the MDM2 oncoprotein by interaction with the structurally related MDMX protein. J. Biol. Chem. 274, 38189–38196 (1999). [DOI] [PubMed] [Google Scholar]
- 8.Li J., Kurokawa M., Regulation of MDM2 stability after DNA damage. J. Cell. Physiol. 230, 2318–2327 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wade M., et al., Functional analysis and consequences of Mdm2 E3 ligase inhibition in human tumor cells. Oncogene 31, 4789–4797 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyd S. D., Tsai K. Y., Jacks T., An intact HDM2 RING-finger domain is required for nuclear exclusion of p53. Nat. Cell Biol. 2, 563–568 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Geyer R. K., Yu Z. K., Maki C. G., The MDM2 RING-finger domain is required to promote p53 nuclear export. Nat. Cell Biol. 2, 569–573 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Momand J., Zambetti G. P., Olson D. C., George D., Levine A. J., The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 69, 1237–1245 (1992). [DOI] [PubMed] [Google Scholar]
- 13.Wade M., Li Y. C., Wahl G. M., MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat. Rev. Cancer 13, 83–96 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Francoz S., et al., Mdm4 and Mdm2 cooperate to inhibit p53 activity in proliferating and quiescent cells in vivo. Proc. Natl. Acad. Sci. U.S.A. 103, 3232–3237 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones S. N., Roe A. E., Donehower L. A., Bradley A., Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 378, 206–208 (1995). [DOI] [PubMed] [Google Scholar]
- 16.Montes de Oca Luna R., Wagner D. S., Lozano G., Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 378, 203–206 (1995). [DOI] [PubMed] [Google Scholar]
- 17.Parant J., et al., Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat. Genet. 29, 92–95 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Moyer S. M., Larsson C. A., Lozano G., Mdm proteins: Critical regulators of embryogenesis and homoeostasis. J. Mol. Cell Biol. 9, 16–25 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wienken M., Moll U. M., Dobbelstein M., Mdm2 as a chromatin modifier. J. Mol. Cell Biol. 9, 74–80 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bohlman S., Manfredi J. J., p53-independent effects of Mdm2. Subcell. Biochem. 85, 235–246 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ganguli G., Wasylyk B., p53-independent functions of MDM2. Mol. Cancer Res. 1, 1027–1035 (2003). [PubMed] [Google Scholar]
- 22.Miranda P. J., et al., MDM4 is a rational target for treating breast cancers with mutant p53. J. Pathol. 241, 661–670 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Klein A. M., de Queiroz R. M., Venkatesh D., Prives C., The roles and regulation of MDM2 and MDMX: It is not just about p53. Genes Dev. 35, 575–601 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao Z. X., et al., Interaction between the retinoblastoma protein and the oncoprotein MDM2. Nature 375, 694–698 (1995). [DOI] [PubMed] [Google Scholar]
- 25.Zhang Z., et al., Stabilization of E2F1 protein by MDM2 through the E2F1 ubiquitination pathway. Oncogene 24, 7238–7247 (2005). [DOI] [PubMed] [Google Scholar]
- 26.DeGregori J., The genetics of the E2F family of transcription factors: Shared functions and unique roles. Biochim. Biophys. Acta 1602, 131–150 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Rowland B. D., Bernards R., Re-evaluating cell-cycle regulation by E2Fs. Cell 127, 871–874 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Kent L. N., Leone G., The broken cycle: E2F dysfunction in cancer. Nat. Rev. Cancer 19, 326–338 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Frolov M. V., Dyson N. J., Molecular mechanisms of E2F-dependent activation and pRB-mediated repression. J. Cell Sci. 117, 2173–2181 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Iaquinta P. J., Lees J. A., Life and death decisions by the E2F transcription factors. Curr. Opin. Cell Biol. 19, 649–657 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen H. Z., Tsai S. Y., Leone G., Emerging roles of E2Fs in cancer: An exit from cell cycle control. Nat. Rev. Cancer 9, 785–797 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Polager S., Ginsberg D., p53 and E2f: Partners in life and death. Nat. Rev. Cancer 9, 738–748 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Dotto G. P., p21(WAF1/Cip1): More than a break to the cell cycle? Biochim. Biophys. Acta 1471, M43–M56 (2000). [DOI] [PubMed] [Google Scholar]
- 34.Löhr K., Möritz C., Contente A., Dobbelstein M., p21/CDKN1A mediates negative regulation of transcription by p53. J. Biol. Chem. 278, 32507–32516 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Abbas T., Dutta A., p21 in cancer: Intricate networks and multiple activities. Nat. Rev. Cancer 9, 400–414 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Biderman L., Manley J. L., Prives C., Mdm2 and MdmX as regulators of gene expression. Genes Cancer 3, 264–273 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Senturk E., Manfredi J. J., Mdm2 and tumorigenesis: Evolving theories and unsolved mysteries. Genes Cancer 3, 192–198 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wunderlich M., Ghosh M., Weghorst K., Berberich S. J., MdmX represses E2F1 transactivation. Cell Cycle 3, 472–478 (2004). [PubMed] [Google Scholar]
- 39.Strachan G. D., Jordan-Sciutto K. L., Rallapalli R., Tuan R. S., Hall D. J., The E2F-1 transcription factor is negatively regulated by its interaction with the MDMX protein. J. Cell. Biochem. 88, 557–568 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Kaghad M., et al., Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell 90, 809–819 (1997). [DOI] [PubMed] [Google Scholar]
- 41.Zhu J., Jiang J., Zhou W., Chen X., The potential tumor suppressor p73 differentially regulates cellular p53 target genes. Cancer Res. 58, 5061–5065 (1998). [PubMed] [Google Scholar]
- 42.Moll U. M., Slade N., p63 and p73: Roles in development and tumor formation. Mol. Cancer Res. 2, 371–386 (2004). [PubMed] [Google Scholar]
- 43.Harms K., Nozell S., Chen X., The common and distinct target genes of the p53 family transcription factors. Cell. Mol. Life Sci. 61, 822–842 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stindt M. H., et al., Functional interplay between MDM2, p63/p73 and mutant p53. Oncogene 34, 4300–4310 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Calabrò V., et al., The human MDM2 oncoprotein increases the transcriptional activity and the protein level of the p53 homolog p63. J. Biol. Chem. 277, 2674–2681 (2002). [DOI] [PubMed] [Google Scholar]
- 46.Watson I. R., Blanch A., Lin D. C., Ohh M., Irwin M. S., Mdm2-mediated NEDD8 modification of TAp73 regulates its transactivation function. J. Biol. Chem. 281, 34096–34103 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Bálint E., Bates S., Vousden K. H., Mdm2 binds p73 alpha without targeting degradation. Oncogene 18, 3923–3929 (1999). [DOI] [PubMed] [Google Scholar]
- 48.Dobbelstein M., Wienzek S., König C., Roth J., Inactivation of the p53-homologue p73 by the mdm2-oncoprotein. Oncogene 18, 2101–2106 (1999). [DOI] [PubMed] [Google Scholar]
- 49.Ongkeko W. M., et al., MDM2 and MDMX bind and stabilize the p53-related protein p73. Curr. Biol. 9, 829–832 (1999). [DOI] [PubMed] [Google Scholar]
- 50.Zeng X., et al., MDM2 suppresses p73 function without promoting p73 degradation. Mol. Cell. Biol. 19, 3257–3266 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang C., Teo C. R., Sabapathy K., p53-related transcription targets of TAp73 in cancer cells—Bona fide or distorted reality? Int. J. Mol. Sci. 21, 1346 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beeler J. S., et al., p73 regulates epidermal wound healing and induced keratinocyte programming. PLoS One 14, e0218458 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tanimura S., et al., MDM2 interacts with MDMX through their RING finger domains. FEBS Lett. 447, 5–9 (1999). [DOI] [PubMed] [Google Scholar]
- 54.Gu J., et al., Mutual dependence of MDM2 and MDMX in their functional inactivation of p53. J. Biol. Chem. 277, 19251–19254 (2002). [DOI] [PubMed] [Google Scholar]
- 55.Herman A. G., et al., Discovery of Mdm2-MdmX E3 ligase inhibitors using a cell-based ubiquitination assay. Cancer Discov. 1, 312–325 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang W., et al., The pyrido[b]indole MDM2 inhibitor SP-141 exerts potent therapeutic effects in breast cancer models. Nat. Commun. 5, 5086 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Muller P. A., Vousden K. H., Mutant p53 in cancer: New functions and therapeutic opportunities. Cancer Cell 25, 304–317 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oren M., Rotter V., Mutant p53 gain-of-function in cancer. Cold Spring Harb. Perspect. Biol. 2, a001107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Johnson D. G., Ohtani K., Nevins J. R., Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes Dev. 8, 1514–1525 (1994). [DOI] [PubMed] [Google Scholar]
- 60.Attwooll C., Lazzerini Denchi E., Helin K., The E2F family: Specific functions and overlapping interests. EMBO J. 23, 4709–4716 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martin K., et al., Stimulation of E2F1/DP1 transcriptional activity by MDM2 oncoprotein. Nature 375, 691–694 (1995). [DOI] [PubMed] [Google Scholar]
- 62.Trimarchi J. M., Lees J. A., Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol. 3, 11–20 (2002). [DOI] [PubMed] [Google Scholar]
- 63.Ambrosini G., et al., Mouse double minute antagonist Nutlin-3a enhances chemotherapy-induced apoptosis in cancer cells with mutant p53 by activating E2F1. Oncogene 26, 3473–3481 (2007). [DOI] [PubMed] [Google Scholar]
- 64.Leone G., et al., Identification of a novel E2F3 product suggests a mechanism for determining specificity of repression by Rb proteins. Mol. Cell. Biol. 20, 3626–3632 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Danielian P. S., et al., E2f3a and E2f3b make overlapping but different contributions to total E2f3 activity. Oncogene 27, 6561–6570 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zdzalik M., et al., Interaction of regulators Mdm2 and Mdmx with transcription factors p53, p63 and p73. Cell Cycle 9, 4584–4591 (2010). [DOI] [PubMed] [Google Scholar]
- 67.Lau L. M., Nugent J. K., Zhao X., Irwin M. S., HDM2 antagonist Nutlin-3 disrupts p73-HDM2 binding and enhances p73 function. Oncogene 27, 997–1003 (2008). [DOI] [PubMed] [Google Scholar]
- 68.Melino G., De Laurenzi V., Vousden K. H., p73: Friend or foe in tumorigenesis. Nat. Rev. Cancer 2, 605–615 (2002). [DOI] [PubMed] [Google Scholar]
- 69.Kawai H., Lopez-Pajares V., Kim M. M., Wiederschain D., Yuan Z. M., RING domain-mediated interaction is a requirement for MDM2’s E3 ligase activity. Cancer Res. 67, 6026–6030 (2007). [DOI] [PubMed] [Google Scholar]
- 70.Terzian T., et al., The inherent instability of mutant p53 is alleviated by Mdm2 or p16INK4a loss. Genes Dev. 22, 1337–1344 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jin Y., Lee H., Zeng S. X., Dai M. S., Lu H., MDM2 promotes p21waf1/cip1 proteasomal turnover independently of ubiquitylation. EMBO J. 22, 6365–6377 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu H., Zhang Z., Li M., Zhang R., MDM2 promotes proteasomal degradation of p21Waf1 via a conformation change. J. Biol. Chem. 285, 18407–18414 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang Z., et al., MDM2 is a negative regulator of p21WAF1/CIP1, independent of p53. J. Biol. Chem. 279, 16000–16006 (2004). [DOI] [PubMed] [Google Scholar]
- 74.Jin Y., et al., MDMX promotes proteasomal turnover of p21 at G1 and early S phases independently of, but in cooperation with, MDM2. Mol. Cell. Biol. 28, 1218–1229 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tomasini R., et al., TAp73 knockout shows genomic instability with infertility and tumor suppressor functions. Genes Dev. 22, 2677–2691 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rosenbluth J. M., Pietenpol J. A., The jury is in: p73 is a tumor suppressor after all. Genes Dev. 22, 2591–2595 (2008). [DOI] [PubMed] [Google Scholar]
- 77.Sznarkowska A., et al., Reactivation of TAp73 tumor suppressor by protoporphyrin IX, a metabolite of aminolevulinic acid, induces apoptosis in TP53-deficient cancer cells. Cell Div. 13, 10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Feeley K. P., Adams C. M., Mitra R., Eischen C. M., Mdm2 is required for survival and growth of p53-deficient cancer cells. Cancer Res. 77, 3823–3833 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu G., Nozell S., Xiao H., Chen X., DeltaNp73beta is active in transactivation and growth suppression. Mol. Cell. Biol. 24, 487–501 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Urist M., Tanaka T., Poyurovsky M. V., Prives C., p73 induction after DNA damage is regulated by checkpoint kinases Chk1 and Chk2. Genes Dev. 18, 3041–3054 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Irwin M., et al., Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature 407, 645–648 (2000). [DOI] [PubMed] [Google Scholar]
- 82.Stiewe T., Pützer B. M., Role of the p53-homologue p73 in E2F1-induced apoptosis. Nat. Genet. 26, 464–469 (2000). [DOI] [PubMed] [Google Scholar]
- 83.Kasim V., et al., Synergistic cooperation of MDM2 and E2F1 contributes to TAp73 transcriptional activity. Biochem. Biophys. Res. Commun. 449, 319–326 (2014). [DOI] [PubMed] [Google Scholar]
- 84.Wang X. Q., Ongkeko W. M., Lau A. W., Leung K. M., Poon R. Y., A possible role of p73 on the modulation of p53 level through MDM2. Cancer Res. 61, 1598–1603 (2001). [PubMed] [Google Scholar]
- 85.Chen L., Gilkes D. M., Pan Y., Lane W. S., Chen J., ATM and Chk2-dependent phosphorylation of MDMX contribute to p53 activation after DNA damage. EMBO J. 24, 3411–3422 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Barsotti A. M., Prives C., Pro-proliferative FoxM1 is a target of p53-mediated repression. Oncogene 28, 4295–4305 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jackson J. G., Pereira-Smith O. M., Primary and compensatory roles for RB family members at cell cycle gene promoters that are deacetylated and downregulated in doxorubicin-induced senescence of breast cancer cells. Mol. Cell. Biol. 26, 2501–2510 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Biderman L., Poyurovsky M. V., Assia Y., Manley J. L., Prives C., MdmX is required for p53 interaction with and full induction of the Mdm2 promoter after cellular stress. Mol. Cell. Biol. 32, 1214–1225 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Eischen C. M., Weber J. D., Roussel M. F., Sherr C. J., Cleveland J. L., Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 13, 2658–2669 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Moon S. H., et al., p53 represses the mevalonate pathway to mediate tumor suppression. Cell 176, 564–580.e19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All study data are included in the article and/or SI Appendix.