Abstract
The lateral hypothalamic area (LHA) is a highly conserved brain region critical for maintaining physiological homeostasis and goal-directed behavior. LHA neurons that express melanin-concentrating hormone (MCH) are key regulators of arousal, energy balance, and motivated behavior. However, cellular and functional diversity among LHAMCH neurons is not well understood. Previous anatomic and molecular data suggest that LHAMCH neurons may be parsed into at least two distinct subpopulations, one of which is enriched in neurokinin-3 receptor (NK3R), the receptor for neurokinin B (NKB), encoded by the Tac2 gene. This tachykininergic ligand-receptor system has been implicated in reproduction, fear memory, and stress in other brain regions, but NKB interactions with LHAMCH neurons are poorly understood. We first identified how LHAMCH subpopulations may be distinguished anatomically and electrophysiologically. To dissect functional connectivity between NKB-expressing neurons and LHAMCH neurons, we used Cre-dependent retrograde and anterograde viral tracing in male Tac2-Cre mice and identified Tac2/EYFP+ neurons in the bed nucleus of the stria terminalis and central nucleus of the amygdala, the central extended amygdala, as major sources of NKB input onto LHAMCH neurons. In addition to innervating the LHA, these limbic forebrain NKB neurons also project to midbrain and brainstem targets. Finally, using a dual-virus approach, we found that optogenetic activation of these inputs in slices evokes GABA release onto a subset of LHAMCH neurons but lacked specificity for the NK3R+ subpopulation. Overall, these data define parallel tachykininergic/GABAergic limbic forebrain projections that are positioned to modulate multiple nodes of homeostatic and behavioral control.
SIGNIFICANCE STATEMENT The LHA orchestrates fundamental behavioral states in the mammalian hypothalamus, including arousal, energy balance, memory, stress, and motivated behavior. The neuropeptide MCH defines one prominent population of LHA neurons, with multiple roles in the regulation of homeostatic behavior. Outstanding questions remain concerning the upstream inputs that control MCH neurons. We sought to define neurochemically distinct pathways in the mouse brain that may communicate with specific MCH neuron subpopulations using viral-based retrograde and anterograde neural pathway tracing and optogenetics in brain slices. Here, we identify a specific neuropeptide-defined forebrain circuit that makes functional synaptic connections with MCH neuron subpopulations. This work lays the foundation for further manipulating molecularly distinct neural circuits that modulate innate behavioral states.
Keywords: bed nucleus of the stria terminalis, central nucleus of the amygdala, lateral hypothalamic area, melanin-concentrating hormone, neurokinin B, neurokinin-3 receptor
Introduction
The lateral hypothalamic area (LHA) has a critical and conserved role in the adaptive coordination of physiological and behavioral states. Through its position as an integrator of widespread inputs from throughout the brain, neuronal cell types and circuits in the LHA both drive and modulate sleep–wake states, ingestive and motivated behavior, locomotion, autonomic function, and stress responses (Brown et al., 2015; Bonnavion et al., 2016; Stuber and Wise, 2016; Yamashita and Yamanaka, 2017; Qualls-Creekmore and Münzberg, 2018; Arrigoni et al., 2019). Underpinning the diverse functions of the LHA is a heterogeneous population of neurons, one of the best known of which is defined by the expression of the neuropeptide melanin-concentrating hormone (MCH). LHA MCH-expressing neurons (LHAMCH) project widely throughout the brain (Bittencourt et al., 1992), are broadly conserved across mammals, and are implicated in a multitude of physiological functions and behaviors, having important roles to play in the modulation of sleep–wake states, feeding, energy balance, motivated behavior, stress, and memory (Pissios et al., 2006; Barson et al., 2013; Croizier et al., 2013; Monti et al., 2013; Diniz and Bittencourt, 2017; Ferreira et al., 2017; Bandaru et al., 2020; Concetti and Burdakov, 2021).
One explanation for the functional diversity of the LHAMCH system may lie in its cellular and neural circuit-level heterogeneity. Anatomical, developmental, and transcriptomic evidence suggests that LHAMCH neurons may be heterogeneous based on the expression of several neurochemical markers. For example, early anatomic work in rodents demonstrated that a subpopulation of LHAMCH neurons coexpress the neuropeptide cocaine- and amphetamine-regulated transcript (CART; Broberger, 1999; Vrang et al., 1999; Elias et al., 2001). Further, a subpopulation of LHAMCH neurons are enriched in the G-protein-coupled tachykininergic neurokinin-3 receptor (NK3R; Griffond et al., 1997; Brischoux et al., 2002; Cvetkovic et al., 2003, 2004; Croizier et al., 2010). Single-cell transcriptomic analyses suggest that LHAMCH neurons may indeed be diverse (Mickelsen et al., 2017, 2019; Jiang et al., 2020; Kim et al., 2020). For example, our recent work using single-cell RNA sequencing (scRNA-seq) to deconstruct neuronal cell types in the mouse LHA revealed that Pmch+ LHAMCH neurons may be parsed into at least two transcriptionally distinct subpopulations based on large suites of discriminatory genes. Notably, we confirmed that one subpopulation is enriched in both Cartpt (CART) and Tacr3 (NK3R). Furthermore, we found that Tacr3 expression within the LHA is confined to Pmch+ neurons (Mickelsen et al., 2019).
NK3R is a member of the tachykinin receptor family and preferentially binds to the neuropeptide neurokinin B (NKB), encoded by the Tac2 gene in rodents (Shigemoto et al., 1990; Steinhoff et al., 2014). Other members of the tachykinin neuropeptide family, including neurokinin A (NKA) and substance P (SP), are recognized by NK3R with a significantly reduced affinity (Steinhoff et al., 2014). Central NKB/NK3R signaling has been implicated in reproductive physiology and behavior in the hypothalamic arcuate and preoptic regions (Rance et al., 2010, 2013), fear memory consolidation (Andero et al., 2014, 2016; Florido et al., 2021; Marvar et al., 2021; Shaam Al Abed et al., 2021), and appetitive behavior (Kim et al., 2017) in the central nucleus of the amygdala. Most recently, chronic social isolation stress in mice has been shown to induce a striking brainwide upregulation of Tac2 mRNA, along with enhanced aggressive and defensive behaviors (Zelikowsky et al., 2018). Less well understood is the role that this tachykininergic ligand-receptor system plays in the circuit-level function of LHAMCH neurons. Do LHAMCH subpopulations display different functional properties, and are they embedded within distinct brainwide networks? Do NK3R-bearing LHAMCH neurons receive unique NKB+ inputs that may underlie specific operations within the wide functional repertoire ascribed to LHAMCH neurons? Foundational anatomic work in this regard showed that both NKB and SP fibers project onto LHAMCH neurons in the rat brain and, through fluorogold retrograde tracing from the LHA, tachykininergic inputs arise from widespread forebrain and brainstem regions (Cvetkovic et al., 2003). However, the contribution of specific, genetically defined NKB+ inputs to the LHA and evidence for functional synaptic connectivity and neurotransmitter release onto molecularly distinct LHAMCH neurons remain unanswered areas.
To begin to understand differential gene expression and possible functional differences between LHAMCH neuron subpopulations, we conducted an anatomic and in vitro electrophysiological analysis of molecularly distinct LHAMCH neurons. We went on to identify the putative anatomic source(s) of NKB projections onto LHAMCH neuron subpopulations through cell-type-specific retrograde and anterograde viral tracing in a Tac2-Cre mutant mouse. We then examined the possibility of functional synaptic connectivity and fast neurotransmitter release between NKB-expressing neurons and LHAMCH neuron subpopulations using in vitro optogenetics and single-cell quantitative PCR (sc-qPCR) in brain slices. In so doing, we identified two novel limbic forebrain GABAergic pathways, defined by NKB expression, that converge to make prominent descending projections onto LHAMCH neurons in addition to innervating other key midbrain and brainstem targets implicated in homeostatic and behavioral control.
Materials and Methods
Ethics statement
All experiments were performed in accordance with the ethical guidelines described in the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee of the University of Connecticut.
Animals
To identify and visualize MCH neurons, we used Pmch-Cre transgenic mice (Tg(Pmch-cre)1Lowl/J transgenic mice; stock #014099, The Jackson Laboratory; RRID:IMSR_JAX:014099; Kong et al., 2010). These mice were crossed to a Cre recombinase (Cre)-dependent tdTomato (tdT) reporter line (B6;Cg-Rt(ROSA)26Sortm14(CAG-tdTomato)Hze/J or Ai14; stock #007914, The Jackson Laboratory; RRID:IMSR_JAX:007914) or EYFP (enhanced yellow fluorescent protein) reporter line (B6.Cg-Gt(ROSA)26Sortm3(CAG-EYFP)Hze/J or Ai3; stock #007903, The Jackson Laboratory; RRID: IMSR_JAX:007903; Madisen et al., 2010), which selectively expresses tdT or EYFP, respectively, following Cre-dependent recombination. The resulting crosses are referred to here as Pmch-Cre;tdT and Pmch-Cre;EYFP respectively. To identify and visualize Tac2/NKB neurons for fluorescence in situ hybridization (FISH), anterograde labeling, and in vitro electrophysiology experiments, we used Tac2-IRES-Cre-D mutant mice, referred to here as Tac2-Cre (B6;129S-Tac2tm1.1(cre)Hze/J; stock #021878, The Jackson Laboratory; RRID: IMSR_JAX:021878; Harris et al., 2014). For immunolabeling and retrograde labeling experiments, Tac2-Cre mice were crossed to either Ai14 or Ai3 lines (Madisen et al., 2010). The resulting crosses are referred to here as either Tac2-Cre;tdT or Tac2-Cre;EYFP, respectively. For other FISH, viral specificity, and in vitro electrophysiology experiments, wild-type C57BL/6 (stock #000664, The Jackson Laboratory) mice were used. All mice were 1- to 5-month-old males and either group housed or singly housed following stereotactic viral injections. Cages of singly housed mice were placed in the same racks as group housed mice. No other modifications were made to the cages or the vivarium between the two housing conditions. All mice were fed ad libitum and kept on a 12/12 h light/dark cycle.
Single-cell data analysis
Raw expression data were downloaded from the Gene Expression Omnibus (record GSE125065, corresponding to Mickelsen et al., 2019) and processed using Cell Ranger (version 3.0.0, 10× Genomics) counted and aggregated as described in Mickelsen et al. (2019). Finalized cell bar codes, cluster labels, and t-distributed stochastic neighbor embedding (t-SNE) coordinates corresponding to the original analysis were obtained from the authors. Together, these were used to subset and embed the raw expression into the original t-SNE space shown in Mickelsen et al. (2019) to identify a single cluster of Pmch+ neurons. We then performed a separate analysis on just the Pmch+ cells using the Scanpy toolkit (version 1.4.6; Wolf et al., 2018). Raw expression at these cells was subjected to library-size normalization (scaled to the median unique molecular identifiers per cell across Pmch+ cells) and natural-log transformation, then the 500 most highly variable genes were used to compute principal components, build a k = 5 nearest neighbor graph, assign clusters with Leiden community detection (resolution = 0.1), and embed with uniform manifold approximation and projection (UMAP).
FISH
For FISH experiments, 1- to 4-month-old male wild-type and Tac2-Cre mice were used. For tissue preparation, mice were anesthetized with isoflurane, and harvested brains were placed in ice-cold sucrose. Brains were then flash frozen on aluminum foil on dry ice, embedded in optimal cutting temperature (OCT) compound (Fisher Scientific) and cryosectioned coronally at a thickness of 14 µm and mounted on Superfrost Plus microscope slides (Fisher Scientific). Collected sections were fixed in 4% paraformaldehyde (PFA) for 15 min at 4°C and dehydrated with increasing concentrations of ethanol (50, 70, 100%). For all experiments, RNAscope 2.5 Assay [Advanced Cell Diagnostics (ACD); Wang et al., 2012] was used, and RNAscope probes were designed and validated by ACD. The following probes from ACD were used: Cartpt (catalog #432001), Cre (catalog #312281), Pmch (catalog #478721-C2), Tac2 (catalog #446391-C2), and Tacr3 (catalog #481671-C3). Slides were coverslipped with ProLong Gold Antifade Mountant with DAPI (Thermo Fisher Scientific). All anatomic maps were modified from Paxinos and Franklin (2012), and selected in situ hybridization images were acquired from the publically available resource, the Allen Mouse Brain Atlas from the Allen Brain Institute (https://mouse.brain-map.org/; Lein et al., 2007) as indicated.
Transcardial perfusions, fixation, and sectioning
Animals were anesthetized with a two-step process using inhaled isoflurane and ketamine/xylazine intraperitoneally. Animals were then transcardially perfused using 5–10 ml of 0.125 m saline at room temperature, followed by 25–30 ml of cold 4% PFA in 1× PBS. Animals were then decapitated, and brains were dissected and stored in 4% PFA overnight at 4°C for postfixation. Brains were then cryoprotected and transferred into 30% sucrose at 4°C for 24–48 h. After cryoprotection, the sucrose solution was decanted, and brains were placed in isopentane on dry ice for rapid freezing and stored at −80°C. For sectioning, frozen brains were placed on OCT compound and sectioned on a cryostat (Leica 3050 S) at −20°C at a thickness of 20–40 µm in either the coronal or parasagittal orientation. Slices were collected in 1× PBS and mounted on glass microscope slides with hard-set Vectashield mounting medium with DAPI (Vector Laboratories) or prepared for immunohistochemistry (IHC).
Immunohistochemistry
Brain slices containing the LHA and midbrain for immunostaining were washed in PBS, then in 0.2% Tween 20 plus PBS (PBST), followed by blocking in 2% donkey normal serum (DNS) plus PBST for 2 h. LHA and midbrain sections were then incubated in diluted primary antibody on an orbital shaker overnight (∼17 h) at room temperature. The following antibodies and dilutions were used for immunostaining: Rb anti-MCH (1:1000; Phoenix Pharmaceuticals), Rb anti-NK3R (1:5000; Novus Biologicals), and Ck anti-TH (1:1000; Aves Labs). Following primary incubation, sections were washed in PBST, then incubated in one of the following diluted secondary antibodies for 2 h: Dk anti-Rb Alexa Fluor 488 (1:500; Abcam), Dk anti-Rb Alexa Fluor 594 (1:500; Abcam), and Gt anti-Ck Alexa Fluor 594 (1:500; Abcam). All sections were then washed in PBST then PBS and mounted onto glass microscope slides with hard-set Vectashield mounting medium with DAPI (Vector Laboratories). For immunostaining against NKB and NK3R, LHA sections were washed in PBS, then blocked in 10% DNS plus 0.3% Triton X-100 for 1 h. Sections were then incubated in the primary antibody Rb anti-NK3R or anti-NKB (1:1000; Invitrogen) and incubated overnight (∼17 h) at room temperature. Following incubation, sections were washed in PBS and incubated with the secondary antibody Dk anti-Rb Alexa 488 (1:500; Abcam) for 2 h. After washing the sections in PBS, sections were mounted onto glass microscope slides with hard-set Vectashield mounting medium with DAPI (Vector Laboratories).
Stereotaxic viral injections
For retrograde tracing experiments, Tac2-Cre;EYFP mice were bilaterally injected with 200 nl of retrograde adeno-associated virus (retroAAV)-Flex-tdT; Tervo et al., 2016; catalog #28306-AAVrg, Addgene) in the LHA [anteroposterior (AP), −1.55; mediolateral (ML), ±1.1; dorsoventral (DV), −4.75 mm] and were incubated for 4 weeks. To determine the specificity of the MCH virus, and for slice electrophysiology experiments, wild-type C57BL/6 mice were bilaterally injected in the LHA (AP, −1.6; ML, ±1.2; DV, −5.3 and −5.0 mm) with 400 nl of AAV2-rMCHp-mCherry (Vector BioLabs, Kanoski Lab). Mice were incubated for 4 weeks before immunostaining against MCH. For anterograde tracing experiments, Tac2-Cre mice were bilaterally injected in the bed nucleus of the stria terminalis (BNST; AP, 0.05; ML, ±0.9; DV, −4.8 and −4.3 mm) or central nucleus of the amygdala (CeA; AP,−1.45; ML, ±2.4; DV, −4.55 mm) with 300–600 nl of AAV2-EF1α-DIO-hChR2(H134R)-EYFP (University of North Carolina Vector Core, Deisseroth Lab). Mice were perfused after a 6–8 week incubation period. For slice electrophysiology experiments, dual injections were performed with bilateral injections in the BNST or CeA with AAV2-EF1α-DIO-hChR2(H134R)-EYFP, followed by 400 nl of AAV2-rMCHp-mCherry in the LHA (AP, −1.7; ML, ±1.2, DV, −5.3 and −5.0 mm). Mice were housed for at least 6 weeks for long-range anterograde tracing and slice electrophysiology experiments.
Imaging and cell counting
Fluorescence images at 10× were obtained using a fluorescence microscope (AxioZoom.V16 or Keyence BZ-X700). Higher magnification images at 40× were acquired using a laser-scanning confocal microscope (Leica TSC SP8). For quantification of immunostained images, confocal image files (.lif) were used with the Cell Counter plug-in for ImageJ (Kurt De Vos, University of Sheffield). In the retrograde labeling experiments, fluorescent cells in various brain regions were quantified based on tdT, EYFP, and colocalized expression. The specificity of the AAV2-rMCHp-mCherry virus to MCH neurons has previously been confirmed in rats (Noble et al., 2018, 2019). To confirm the specificity in mice, coronal sections were obtained and immunostained with the primary antibody Rb anti-MCH (1:1000) and the secondary antibody Dk anti-Rb Alexa Fluor 488 (1:500; see above, Immunohistochemistry, for procedures). Efficiency of the virus is dependent on the region and the volume of the injection and was therefore not quantified. For the FISH experiments, images were analyzed using CellProfiler 3.1.9 with a custom pipeline. In the Pmch/Tacr3/Cartpt images, Pmch intensity and morphology determined the regions of interest to further analyze Tacr3 and Cartpt mean intensity within each Pmch+ cell. For image analysis of the Cre/Tac2 FISH experiment, a custom code written in MATLAB R2019a (MathWorks) was used to overlay the Cre and Tac2 images and create a composite cell template image. The intensity and morphology in the template image similarly determined the regions to analyze Cre and Tac2 expression within each cell. Each image was screened to ensure that background signals were not detected as cells and that any images with high background were removed from the analysis.
In vitro slice electrophysiology
Male 2- to 5-month-old wild-type C57BL/6, Tac2-Cre and Pmch-Cre;tdT mice were anesthetized with isoflurane, followed by ketamine/xylazine injected intraperitoneally. Mice were transcardially perfused with 20 ml of ice-cold sucrose containing the following (in mm): 87 NaCl, 75 sucrose, 25 glucose, 25 NaHCO3, 7.5 MgCl2, 2.5 KCl, 1.25 NaH2PO4, 0.5 CaCl2, and 5 ascorbic acid. After decapitation, the brain was dissected and immersed in ice-cold sucrose solution. Coronal LHA sections at a thickness of 225 µm were obtained using a vibrating microtome (7000smz-2, Lafayette Instrument). Slices were incubated for 45 min at 34°C in artificial CSF (ACSF) containing the following (in mm): 125 NaCl, 25 NaHCO3, 11 glucose, 2.5 KCl, 1.25 NaH2PO4, 1 MgCl2, and 2 CaCl2. After recovery, slices were then transferred to room temperature for at least 30 min before recordings. All solutions were continuously bubbled with 95%O2/5%CO2.
Slices were placed in a recording chamber and continuously superfused with oxygenated ACSF at physiological temperature (34–35°C). Drug application recordings were performed at room temperature. Cells were identified based on mCherry, tdTomato, or EYFP fluorescence using an upright Olympus microscope (Olympus America). All recordings were performed using a Multiclamp 700B amplifier and pClamp10 acquisition software (Molecular Devices). For recordings using Pmch-Cre;tdT or Pmch-Cre;EYFP mice, whole-cell recordings were performed in current-clamp mode using the following potassium-gluconate-based internal solution containing the following (in mm): 130 K-gluconate, 10 HEPES, 0.1 EGTA, 10 NaCl, 2 MgCl2, 10 phosphocreatine (tris), 4 Mg-ATP, and 0.3 Na-GTP (290 mOsm/L, pH 7.3). Senktide (100 or 500 nm) was puff applied onto identified tdT+ or EYFP+ neurons with a secondary pipette. For recordings using Tac2-Cre mice, whole-cell voltage-clamp recordings were performed from identified Pmch (mCherry+) neurons using the following low-chloride, cesium-based internal solution containing the following (in mm): 135 CsMeSO3, 10 HEPES, 1 EGTA, 1 MgCl2, 3.2 TEA-Cl, 0.1 spermine, 5 Na2-phosphocreatine, 4 Mg-ATP, and 0.3 Na-GTP (288 mOsm/l, pH 7.3). Blue light was used to stimulate ChR2-EYFP-expressing fibers. Cells were held at 0 mV and −70 mV, and ChR2-EYFP-expressing fibers were photostimulated (5 ms pulses, 3 × 5 Hz stimulation pattern). Kynurenic acid (1 mm) and picrotoxin (PTX; 100 μm) were applied with the use of a perfusion pencil (AutoMate Scientific). For current-clamp experiments, in conjunction with optogenetic stimulation, the previously mentioned K-gluconate-based internal was used. For optogenetic experiments combined with cytoplasm harvest, we used previously described procedures (Mickelsen et al., 2017). Recordings were performed using an internal solution made with RNase-free H2O containing the following (in mm): 135 CsMeSO3, 10 HEPES, 1 EGTA, 1 MgCl2, 3.2 TEA-Cl, 0.1 spermine with 20 µg/ml glycogen, and 1 U/µl RNase inhibitor. For recordings using wild-type mice, current-clamp experiments combined with the cytoplasm harvest were performed using the following internal solution made with RNase-free H2O (in mm): 130 K-gluconate, 10 HEPES, 0.1 EGTA, 10 NaCl, 2 MgCl2, and 1U/µl RNase inhibitor. Pipettes were backfilled with 1 µl of the internal for the cytoplasm harvest experiments. At the end of the recording, light suction was applied to harvest the cytoplasm. After collection, the tip of the pipette was cracked, and positive pressure was applied to collect the contents in an RNase free PCR tube containing 2.5 µl lysis buffer [5% NP-40, 1U/µL RNasin+ (Promega), 0.2× pooled TaqMan assays]. Tubes were then snap frozen on dry ice and stored at −80°C for further processing for single-cell qPCR.
Single-cell qPCR procedure and analysis
As described previously (Mickelsen et al., 2017), tubes of the harvested cytoplasm in lysis buffer were thawed and then denatured at 70°C/10 min, 4°C/5 min. Reverse transcription (RT) was performed by adding 2.5 µl of the RT mixture [1 µl 5× RT buffer (Promega), 0.6 µl 25 mm MgCl2, 0.25 µl Moloney murine leukemia virus (Promega), 0.1 µl 25 mm dNTPs, 0.5 µl H2O, 0.05 µl RNasin plus (Promega)] and incubated at 37°C/2 min, 42°C/1 min, 50°C/1 s for 40 cycles, then 85°C/5 min and 4°C hold. cDNA was then preamplified (PreAmp) by adding 2 µl of the cDNA from the RT step to 8 µl of the PreAmp mixture [5 µl of the Taq DNA polymerase (Clontech), 2.5 µl of the 0.2× TaqMan pooled assay, 0.5 µl H2O] and incubated at 95°C/3 min, 55°C/2 min, 72°C/2 min, then 95°C/15 s, 60°C/2 min, 72°C/2 min for 16 cycles and 4°C hold. qPCR was then performed against the selected panel of genes with 2 µl of the 1:50 diluted amplified cDNA added to the qPCR master mixture (5 µl 2× PCR master mixture, 0.5 µl 20× TaqMan assay, 2.5 µl H2O) on the QuantStudio 3 qPCR instrument (Thermo Fisher Scientific). To analyze gene expression, raw cycle threshold (Ct) values were inverted (40-Ct) to generate a log2-based scale. Cells were filtered based on the 95% confidence interval for Gapdh and Pmch expression. Hierarchical cluster analysis was performed using R software (https://www.r-project.org). Euclidean distance was calculated to measure dissimilarity between cells, which is represented by the length of the branch in the dendrogram. Ward's (1963) method minimizing within-group variance was used to cluster cells.
Analysis of slice electrophysiology data
In voltage-clamp experiments, passive properties were determined from a 100 ms hyperpolarizing step of −5 mV, where the cell was initially held at a holding potential, Vh = −85 mV. Input resistance was calculated using the change in current 50 ms before the step and the last 50 ms during the step. After subtracting the holding current off-line, the integrated area under the transient from the same trace was used to determine membrane capacitance. Multiple sweeps were averaged to remove noise.
In current-clamp recordings, single action potential (AP) properties were determined from a 1 s depolarizing step, in which the cell was also held around Vh =−85 mV. The first evoked spike was used to extract AP features. Spike threshold was defined as the voltage at 2% of the maximum time derivative of the voltage (dV/dt). AP and afterhyperpolarization (AHP) amplitudes were measured from the peak and the trough of the AP waveform, relative to the threshold. Rise and decay times were also based on the threshold. AP half-width was defined as the AP width at the midpoint between AP threshold and AP peak. Membrane potential values were corrected for a liquid junction potential of −14.9 mV, calculated using pClamp10 (Molecular Devices). Statistical comparisons between groups was based on the nonparametric Mann–Whitney test. Statistical significance was determined as p < 0.05. All data were analyzed with ClampFit (Molecular Devices), MATLAB R2020b (MathWorks), and R (https://www.r-project.org).
Results
A molecularly distinct subpopulation of MCH neurons expresses Tacr3 mRNA and functional NK3R receptors
Consistent with previous anatomic evidence for neurochemical heterogeneity among MCH neurons (Griffond et al., 1997; Broberger, 1999; Vrang et al., 1999; Elias et al., 2001; Cvetkovic et al., 2003, 2004; Harthoorn et al., 2005; Hanriot et al., 2007; Croizier et al., 2010), our recent scRNA-seq analysis of the murine LHA demonstrated that Pmch+ neurons may be parsed into two transcriptionally distinct subpopulations, with Cartpt and Tacr3 mRNA enriched in one of them (Mickelsen et al., 2019). We first set out to confirm Tacr3/NK3R expression in MCH neurons with transcriptomic, anatomic, and electrophysiological data. Using our previous transcriptomic dataset (Mickelsen et al., 2019), we illustrate that Pmch+ neurons represent one of 30 distinct neuronal clusters in the LHA (Fig. 1A) and that further unbiased clustering of Pmch+ neurons reveal two subclusters defined by a suite of neurochemical signatures, represented as average expression in each subcluster. These include Pmch, common to both subclusters, and Cartpt, Tacr3, Nptx1, and Gfra1, which are all enriched in subcluster 1 (Fig. 1A). In contrast, subcluster 2 is enriched in a set of markers that includes Scg2, Rmst, Nrxn3, and Pcdh11x, among others (Mickelsen et al., 2019). Normalized expression of Tacr3 and Cartpt among Pmch+ neurons reveals a subpopulation of neurons that strongly coexpress both transcripts, which constitutes 58.8% of all Pmch+ neurons sampled. An additional 12.6% of Pmch+ neurons express Tacr3 but lack Cartpt (Fig. 1B). These single-cell transcriptomic data independently demonstrate that a subpopulation of Pmch+ neurons coexpress Cartpt and Tacr3 transcripts, consistent with previous anatomic data.
Figure 1.
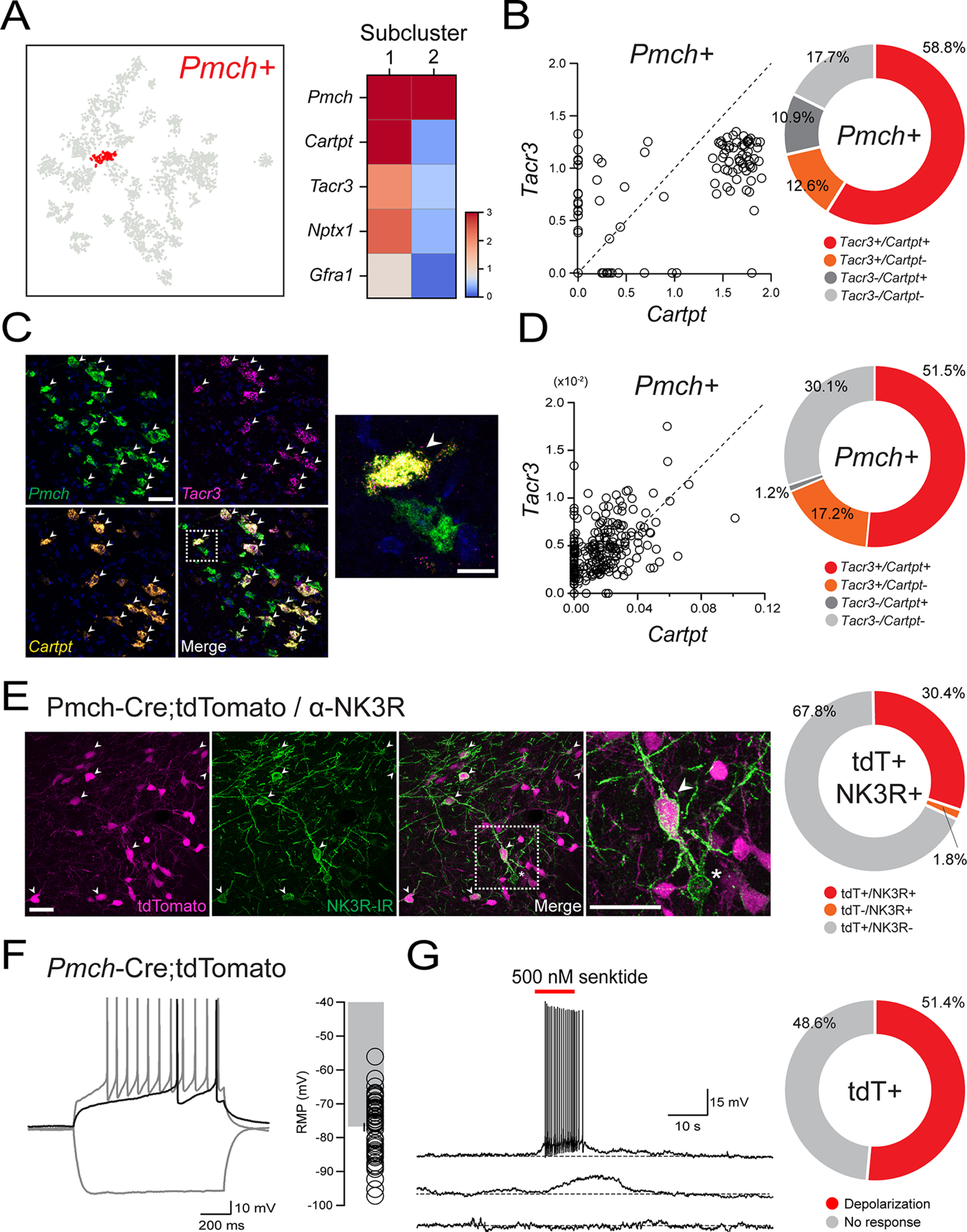
Coexpression of Cartpt and Tacr3 in a subpopulation of Pmch+ neurons. A, Pmch-expressing neuronal cluster (red) visualized on a t-SNE plot after scRNA-seq of the LHA (n = 3589 cells, 5 mice) and corresponding heatmap displaying normalized, natural-log transformed expression of marker genes that distinguish two subpopulations of Pmch+ neurons (n = 119 cells, 5 mice), using published scRNA-seq data (Mickelsen et al., 2019). B, Scatter plot showing normalized, natural-log transformed expression of Tacr3 and Cartpt in 119 Pmch+ neurons (left) using data from Mickelsen et al. (2019). Donut plot showing binarized representation of the scatter plot data with positive (+) expression of either gene corresponds to detection of more than zero transcripts of that gene (right). C, Confocal micrographs (40×) showing the expression of Pmch (green), Cartpt (orange), and Tacr3 (magenta) and merged expression (left). White arrows indicate cells that coexpress Pmch, Cartpt, and Tacr3. Scale bar, 50 µm. Higher magnification confocal micrograph (100×, right) of the boxed region, showing a cell that coexpresses Pmch, Cartpt, and Tacr3 (white arrow), next to a cell that expresses Pmch alone. Scale bar, 15 µm. D, Scatter plot (left) and corresponding donut plot (right) quantifying the mean intensity and proportion of Tacr3 and Cartpt expression in Pmch+ cells (n = 342 cells, 2 mice). E, Representative confocal micrographs (40×) of the LHA showing expression of tdT (magenta), NK3R-IR (green), and merge in Pmch-Cre;tdTomato mice with the boxed region enlarged. Scale bars: 50 µm. Corresponding donut plot (right) depicting the proportion of tdT+ and NK3R-IR cells in Pmch-Cre;tdT mice (n = 729 cells, 3 mice). F, Representative traces of whole-cell recordings from a tdT+ neuron in a Pmch-Cre;tdT mouse held at −85 mV with 1 s of current injection (−120 pA, +40 pA, +120 pA, left). Bar plot displaying RMP values of silent tdT+ and EYFP+ neurons (n = 53/81 cells, 10 mice), with individual values (right). Error bars represent ± SEM. G, Representative current-clamp traces from tdT+ neurons held at −85 mV in Pmch-Cre;tdT mice (left) showing differential responses to local puff application of senktide (red line, 500 nm); depolarization to threshold (top), subthreshold depolarization (middle), and no response (bottom). Corresponding donut plot (right) showing differential responses to either 100 nm or 500 nm senktide (n = 35 cells, 9 mice).
To further confirm the neurochemical phenotype of this LHAMCH neuron subpopulation, we performed multiplex FISH for Pmch, Tacr3, and Cartpt (Fig. 1C). In close correspondence with our scRNA-seq data, 51.5% of Pmch+ cells express both Tacr3 and Cartpt, whereas an additional 17.2% express Tacr3 without Cartpt (Fig. 1D) across the extent of the LHAMCH field. Next, using IHC in Pmch-Cre;tdT mice, we confirmed the detection of NK3R protein (encoded by Tacr3) in a subset of LHAMCH cells. We found that 30.4% of tdT+ neurons exhibited NK3R-immunoreactivity (IR; Fig. 1E), which suggests either a mismatch between Tacr3 mRNA and NK3R protein, or a difference in detection between scRNA-seq/FISH and IHC. Regardless, NK3R-IR was evident as robust perisomatic and dendritic labeling, qualitatively similar to previous reports (Griffond et al., 1997; Brischoux et al., 2002; Cvetkovic et al., 2003; Croizier et al., 2010), in a subpopulation of tdT+ neurons, whereas other tdT+ neurons were devoid of any detectable labeling.
Finally, we examined the functional expression of NK3R in a subset of LHAMCH neurons. Using brain slices that include the LHA obtained from Pmch-Cre;tdT or Pmch-Cre;EYFP mice, we recorded from visually identified tdT+ or EYFP+ cells to survey the characteristic electrophysiological properties of the cells (Fig. 1F). In representative traces from a tdT+ MCH+ neuron, we found that increasing depolarizing steps (1 s) elicited slow firing with a prominent delay to the first spike. Furthermore, a majority of the cells exhibited consistently hyperpolarized resting membrane potentials (RMP; mean ± SEM, −76.94 ± 1.23 mV; Fig. 1F), broadly consistent with the distinctive properties of rodent LHAMCH neurons in hypothalamic slices, as previously described (van den Pol et al., 2004; Burdakov et al., 2005; Linehan and Hirasawa, 2018). In whole-cell current-clamp configuration, we puff applied a selective NK3R agonist senktide (100 or 500 nm) in close proximity to LHAMCH neuron cell bodies in the presence of blockers of fast synaptic transmission, kynurenic acid (1 mm) and picrotoxin (100 μm). Across both concentrations, 51.4% of tdT+ or EYFP+ LHAMCH neurons were depolarized by senktide, resulting in a depolarization from Vrest that was either subthreshold or suprathreshold, accompanied by a burst of action potentials (Fig. 1G). These data suggest that senktide application has a depolarizing effect on approximately half the population of tdT+ or EYFP+ LHAMCH cells, which is broadly congruent with the expression of Tacr3 mRNA/NK3R-IR in Pmch+ cells from our scRNA-seq, FISH, and IHC data.
Electrophysiological differences between molecularly distinct MCH neuron subpopulations
We next asked whether the two transcriptionally distinct MCH+ neuron subpopulations, distinguished by differential gene expression, also differ electrophysiologically. To accomplish this, we recorded the intrinsic membrane properties of visually identified LHAMCH neurons in brain slices. To identify LHAMCH neurons in brain slices, we injected the LHA of wild-type mice with a Pmch promoter-driven AAV expressing mCherry (AAV2-MCH-mCherry; Fig. 2A), previously characterized in rats (Noble et al., 2018, 2019). Before using this virus, we validated its specificity for selectively labeling LHAMCH neurons in mice by immunostaining for anti-MCH in wild-type mice (Fig. 2B). We quantified the percentage of mCherry+ cells that were MCH-IR and found the specificity of the virus to be 92.94% (Fig. 2B), indicating its high specificity and reliable labeling of MCH+ neurons.
Figure 2.
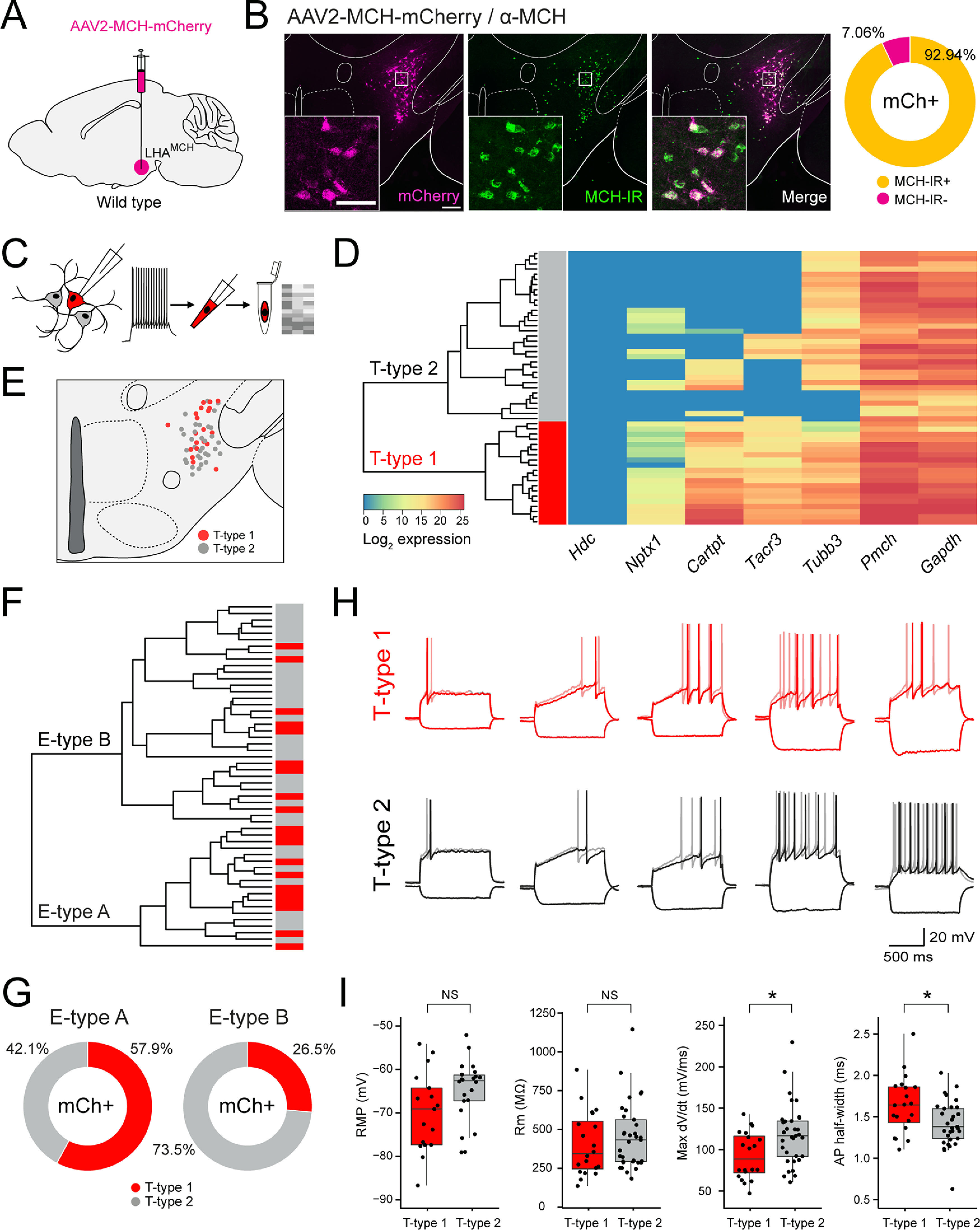
Defining LHAMCH subpopulations through molecular markers and electrophysiological signatures. A, Diagram of the injection site in the LHA of wild-type mice, with AAV2-MCH-mCherry to visually identify LHAMCH neurons in slice recordings. B, Fluorescence micrographs (left) of a representative coronal section of the LHA showing expression of mCherry (magenta), MCH-IR (green), and merged. Scale bar, 200 µm. Insets, Confocal micrographs (40×) of the boxed regions. Scale bar, 80 µm. Donut plot (right) displaying the specificity (% mCherry+ cells that are MCH-IR) of the AAV-MCH-mCherry virus (n = 583 cells, 2 mice). C, Diagram of the electrophysiological recording and sc-qPCR procedure—current-clamp and voltage-clamp recordings from mCherry+ neurons, cytoplasm harvest, and sc-qPCR for key molecular markers. D, Dendrogram of unsupervised cluster analysis and corresponding heatmap of gene expression in recorded mCherry+ neurons (n = 53 cells, 5 mice). The two major clades are classified as transcriptomic T-type 1 (red) and T-type 2 (gray) based on the seven markers. E, Anatomical map of the LHA illustrating the location of the recorded mCherry+ neurons classified based on the transcriptomic type. F, Dendrogram of unsupervised cluster analysis based on the intrinsic membrane properties of recorded mCherry+ neurons. The two major clades are classified as electrophysiological E-type A and E-type B. The corresponding bar chart denotes the T-type (T-type1, red; T-type 2, gray). G, Donut plots depicting the proportion of T-type classified cells in E-type A and E-type B. H, Comparison of representative current-clamp traces among T-type 1 and T-type 2 cells at or near rheobase and a hyperpolarizing current step of −50 pA for 1 s. I, Box and whisker plots, with individual data points, of the RMP, Rm, max dV/dt, and AP half-width across T-type 1 and T-type 2 cells. Asterisks indicate statistical significance, *p < 0.05. NS, Not significant.
Our experimental procedure was to record from mCherry+ neurons in brain slices using a battery of voltage-clamp and current-clamp protocols to identify basic intrinsic membrane properties, followed by cytoplasm harvest for sc-qPCR analysis to identify the molecular signatures (Fig. 2C). We recorded from a total of 53 mCherry+ cells (n = 5 mice) and harvested the cytoplasm of the recorded cells for sc-qPCR analysis. After recording, the samples were snap frozen in lysis buffer and subjected to sc-qPCR with a selected panel of genes with prevalidated primers, including Pmch, Tacr3, Cartpt, and Nptx1. We also included positive controls, such as the neuronal marker Tubb3 and the housekeeping gene Gapdh, in addition to the negative control histidine decarboxylase, Hdc, which should be absent in Pmch+ cells. As illustrated in the heatmap showing the expression of these transcripts in recorded mCherry+ LHAMCH cells (Fig. 2D), all the cells express Pmch, whereas the negative control, Hdc, is undetectable. Using the gene-expression profiles of these cells, we performed unsupervised hierarchical clustering to determine heterogeneity among the recorded cells. At the first branch point of the dendrogram, the clustering reveals two major clades of Pmch+ neurons, one enriched in both Tacr3 and Cartpt, which we refer to as transcriptomic or T-type 1. In the other, Tacr3 and Cartpt exhibit much lower expression levels and are never coexpressed, referred to as T-type 2. This classification, based on differential marker expression, is consistent with our FISH and scRNA-seq results (Fig. 1A–D). The anatomic distribution of recorded T-type 1 and 2 cells is shown in Figure 2E
In each mCherry+ neuron, we evaluated 14 features of the basic electrophysiological properties of the neurons including both passive and active membrane properties in the presence of blockers of synaptic transmission, as show in Tables 1 and 2. All recorded neurons were systematically subjected to the same protocols and analysis routines. On the basis of these properties, we conducted unsupervised hierarchical cluster analysis and found that the data parsed into two major clades, which we refer to as electrophysiological or E-type A and E-type B (Fig. 2F). E-type A (n = 19) and E-type B neurons (n = 34) exhibited significant differences in both aspects of passive and active membrane properties including capacitance, max dV/dt, action potential threshold, trough, and half-width (Table 1). We next asked how the transcriptomic (T) types aligned with the electrophysiological (E) types by color coding each electrophysiologically classified cell with its corresponding T-type (Fig. 2F). Although we found that the classifications were mixed, E-type A was enriched with LHAMCH neurons classified as T-type 1 (11/19 or 57.9%), whereas E-type B was populated by proportionately fewer T-type 1 neurons (9/34 or 26.5%; Fig. 2G).
Table 1.
Passive and active membrane properties of MCH electrophysiological (E)-type A and B neurons
| E-type A (n = 19) |
E-type B (n = 34) |
p value | |||
|---|---|---|---|---|---|
| Mean | SEM | Mean | SEM | ||
| Passive membrane properties | |||||
| Capacitance (pF) | 69.61 | 3.50 | 57.69 | 3.48 | 0.0180* |
| Time constant (ms) | 29.95 | 2.19 | 31.69 | 2.05 | 0.5019 |
| Input resistance (MΩ) | 424.95 | 49.92 | 440.80 | 35.50 | 0.5749 |
| Resting membrane potential (mV) | −67.31 | 1.77 (n = 18) | −67.15 | 1.54 (n = 21) | 1 |
| Active membrane properties | |||||
| Max dV/dt (mV/ms) | 72.82 | 2.93 | 128.50 | 4.82 | < 0.0001* |
| AP threshold (mV) | −39.21 | 0.92 | −45.48 | 1.08 | 0.0004* |
| AP amplitude (mV) | 54.32 | 0.71 | 63.85 | 0.97 | < 0.0001* |
| AP peak (mV) | 15.11 | 1.18 | 18.38 | 1.28 | 0.1011 |
| AHP amplitude (mV) | 17.26 | 0.64 | 18.66 | 0.51 | 0.2196 |
| AP trough (mV) | −56.46 | 0.75 | −64.16 | 0.97 | < 0.0001* |
| AP half-width (ms) | 1.81 | 0.06 | 1.33 | 0.04 | < 0.0001* |
| 10–90% rise time (ms) | 0.91 | 0.10 | 0.58 | 0.02 | < 0.0001* |
| 90–10% decay time (ms) | 1.58 | 0.05 | 1.27 | 0.04 | < 0.0001* |
| Sag ratio (at −50 pA) | 0.98 | <0.01 | 0.98 | <0.01 | 0.8327 |
*p < 0.05 across groups based on the Mann–Whitney test. pF, Picofarad.
Table 2.
Passive and active membrane properties of MCH transcriptomic (T)-type 1 and 2 neurons
| T-type 1 (n = 20) |
T-type 2 (n = 33) |
p value | |||
|---|---|---|---|---|---|
| Mean | SEM | Mean | SEM | ||
| Passive membrane properties | |||||
| Capacitance (pF) | 66.46 | 4.71 | 59.23 | 3.14 | 0.2246 |
| Time constant (ms) | 26.58 | 1.69 | 33.78 | 2.10 | 0.0157* |
| Input resistance (MΩ) | 404.10 | 45.64 | 453.91 | 37.02 | 0.3293 |
| Resting membrane potential (mV) | −70.04 | 2.04 (n = 17) | −65.05 | 1.23(n = 22) | 0.0765 |
| Active membrane properties | |||||
| Max dV/dt (mV/ms) | 92.85 | 6.35 | 118.04 | 6.43 | 0.0165* |
| AP threshold (mV) | −42.45 | 1.22 | −43.70 | 1.19 | 0.9927 |
| AP amplitude (mV) | 58.07 | 1.34 | 61.87 | 1.18 | 0.0676 |
| AP peak (mV) | 15.62 | 1.27 | 18.17 | 1.29 | 0.2246 |
| AHP amplitude (mV) | 18.53 | 0.69 | 17.93 | 0.51 | 0.8344 |
| AP trough (mV) | −60.98 | 1.38 | −61.65 | 1.08 | 0.8472 |
| AP half-width (ms) | 1.65 | 0.08 | 1.42 | 0.05 | 0.0165* |
| 10–90% rise time (ms) | 0.82 | 0.10 | 0.63 | 0.03 | 0.0086* |
| 90–10% decay time (ms) | 1.47 | 0.06 | 1.33 | 0.05 | 0.0863 |
| Sag ratio (at −50 pA) | 0.99 | <0.01 | 0.98 | <0.01 | 0.0003* |
*p < 0.05 across groups based on the Mann–Whitney test. pF, Picofarad.
Next, we systematically compared how T-type 1 and 2 LHAMCH neurons differ from one another electrophysiologically. With a side-by-side comparison of a selection of qualitatively similar current-clamp traces at or near rheobase (Fig. 2H), we observed a broad range of electrophysiological phenotypes. As shown in Table 2, a comparison of T-type 1 and 2 neurons shows no significant difference in properties such as RMP and input resistance (Rm; Fig. 2I), whereas other features such as max dV/dt and AP half-width do show a significant difference (Fig. 2I). Overall, these data suggest that although the passive and active membrane properties of LHAMCH neurons are diverse and likely represent a continuum of electrophysiological phenotypes, there are quantifiable differences in the electrical behavior of molecularly distinct subpopulations.
The LHA is broadly innervated by NKB+ fibers, which are closely apposed to MCH+ neurons
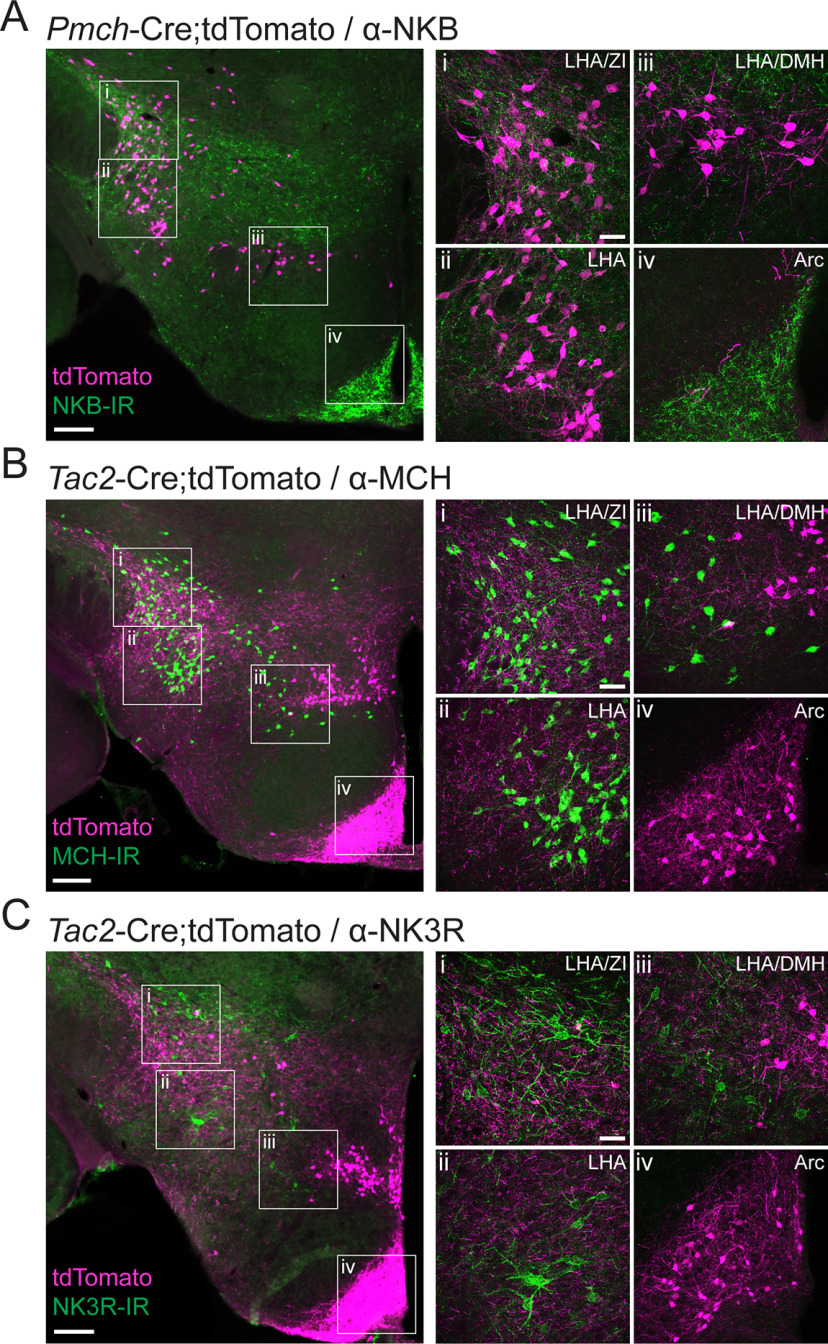
NKB is the preferred endogenous ligand for the receptor NK3R (Shigemoto et al., 1990; Steinhoff et al., 2014). We therefore set out to confirm the presence of NKB+ fibers in the LHA using both immunohistochemical and genetic tools. We first investigated NKB-IR relative to tdT+ LHAMCH neurons in the LHA of Pmch-Cre;tdT mice (Fig. 3A). We observed widespread NKB-IR throughout the LHA, with particularly dense labeling in dorsolateral regions of the LHA, including the parasubthalamic nucleus near the cerebral peduncle and neighboring zona incerta (ZI), in close proximity to tdT+ LHAMCH perikarya (Fig. 3A, i, ii). We also found NKB+ fibers extending into the medial LHA and dorsomedial hypothalamus (DMH), which corresponds less closely with tdT+ LHAMCH neurons (Fig. 3A, iii). This pattern of NKB-IR is in agreement with previous anatomic work in the rat, which showed dense NKB fibers in the LHA apposing MCH perikarya (Cvetkovic et al., 2003). Strong NKB-IR was also identified in the arcuate nucleus (Arc; Fig. 3A, iv), where kisspeptin/NKB/dynorphin (KNDy) neurons are found and are implicated in regulating reproductive behavior (Rance et al., 2010, 2013). NKB-IR found in the Arc would be expected to be evident as both labeled perikarya and dense fibers.
Figure 3.
A subpopulation of LHAMCH neurons are innervated by NKB fibers. A, Fluorescence micrograph (10×) of a coronal section of the LHA immunostained with anti-NKB (green) in a Pmch-Cre;tdT (magenta) mouse with higher magnification confocal micrographs (40×) of the boxed regions (i–iv). B, Fluorescence micrograph (10×) of a coronal section of the LHA immunostained with anti-MCH (green) in a Tac2-Cre;tdT (magenta) mouse with higher magnification confocal micrographs (40×) of the boxed regions (i–iv). C, Fluorescence micrograph (10×) of a coronal section of the LHA immunostained with anti-NK3R (green) in a Tac2-Cre;tdT (magenta) mouse with higher magnification confocal micrographs (40×) of the boxed regions (i–iv). Scale bars: 200 µm, low-magnification images; 50 µm, high-magnification images (i–iv).
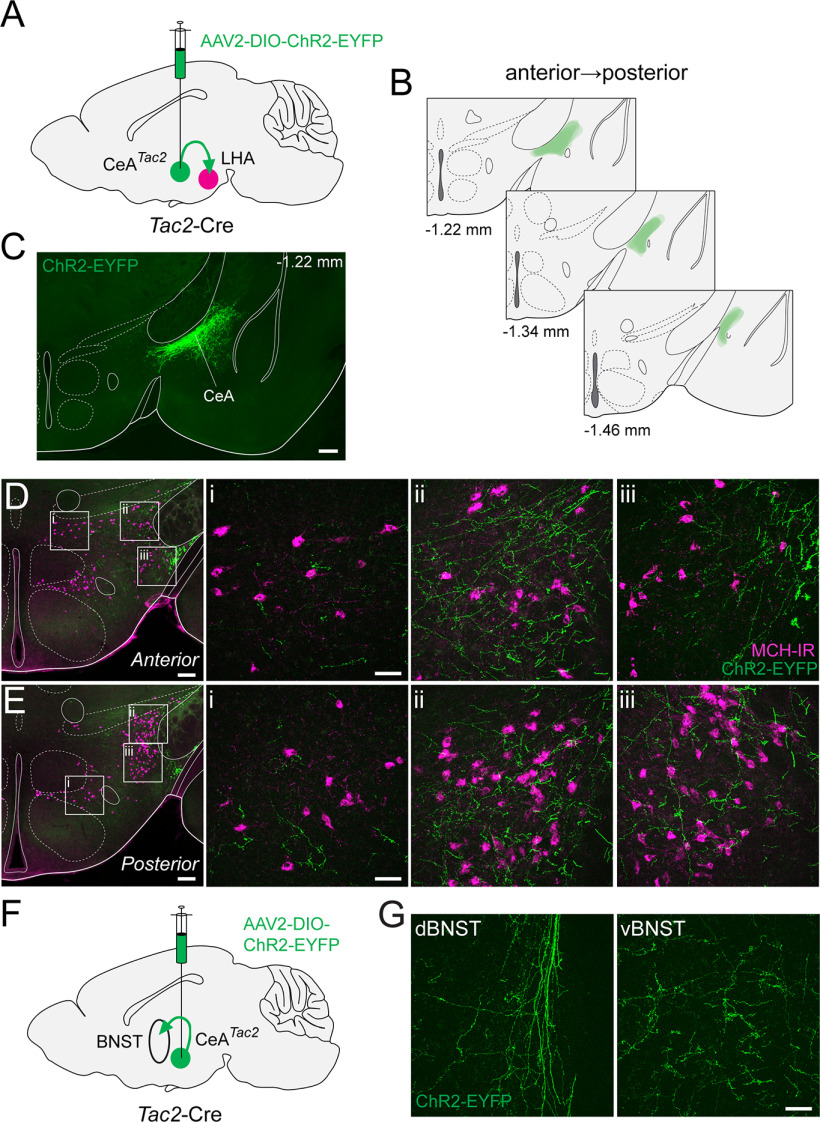
Next, we set out to determine whether the pattern of NKB-IR fibers observed in the LHA may be recapitulated by genetically defined targeting of Tac2. Using a Tac2-Cre mouse line (Harris et al., 2014) crossed to a Cre-dependent reporter line expressing tdT (Ai14; with the resulting cross referred to as Tac2-Cre;tdT), we examined tdT+ fibers relative to MCH-IR perikarya in the LHA (Fig. 3B). Similar to the immunostaining of NKB, tdT+ fibers in Tac2-Cre;tdT mice are present throughout the dorsolateral LHA and intermingled with MCH-IR perikarya (Fig. 3B, i, ii). In contrast to our NKB-IR results, clusters of tdT+ perikarya are apparent in the DMH (Fig. 3B, iii), and more clearly resolved in the Arc (Fig. 3B, iv) of Tac2-Cre;tdT sections. Finally, to better understand the relationship between tdT+ fibers and expression patterns of its cognate receptor NK3R, we immunostained for NK3R in Tac2-Cre;tdT mice (Fig. 3C). Based on the overlaid expression of tdT and NK3R-IR, we observed that NK3R-IR neurons are found in the vicinity of tdT+ fibers without any clear juxtapositions between the two (Fig. 3C, i–iii). Although NK3R-IR is present throughout the LHA and the DMH, we surprisingly failed to observe detectable NK3R-IR among cells in the Arc (Fig. 3C, iv). These data demonstrate that the pattern of NKB+ fibers innervating the LHA shown through immunohistochemistry is largely recapitulated in the Tac2-Cre mouse crossed to a Cre-dependent fluorescent reporter. Together, these data suggest that the dense innervation of the LHA, with close appositions to LHAMCH neurons, originates in one or more populations of NKB+ neurons in the brain and may be recapitulated using a Tac2-Cre mutant mouse.
Validation of a Tac2-Cre mouse
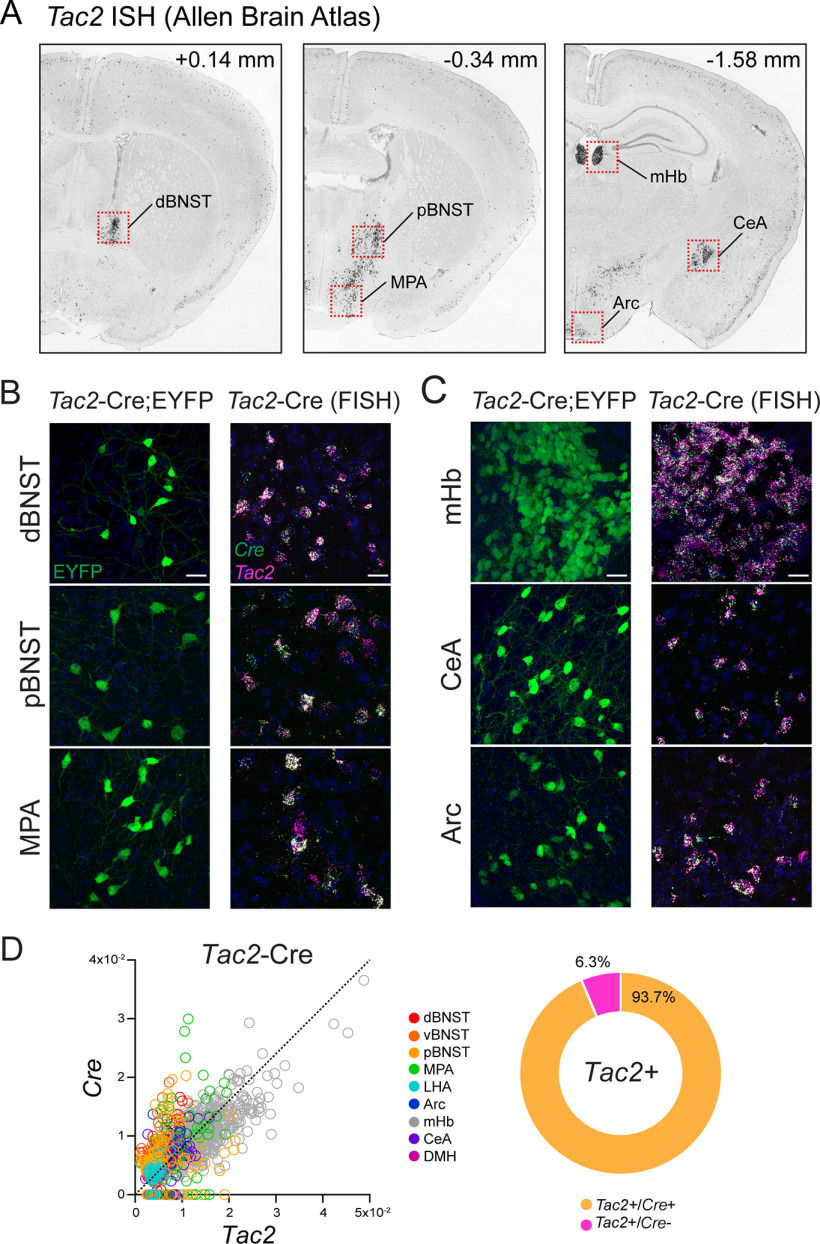
To identify the source of Tac2 (NKB) fibers innervating the LHA, we first asked where Tac2 mRNA is natively expressed in the adult mouse brain. Consistent with the localization of Tac2 mRNA and NKB protein in discrete regions of the rat brain (Warden and Young, 1988; Marksteiner et al., 1992), Tac2 ISH images, obtained from the Allen Mouse Brain Atlas (Lein et al., 2007), show that Tac2 is robustly expressed in select regions of the mouse forebrain. These include subregions of the BNST, medial preoptic area (MPA), medial habenula (mHb), CeA, LHA, DMH, and Arc (Fig. 4A). To both visualize Tac2+ neurons and assess how well the expression of Cre recombinase in Tac2-Cre mice recapitulates native expression of Tac2 mRNA in the mouse brain, we crossed it to a Cre-dependent reporter line expressing EYFP (Ai3, referred to as Tac2-Cre;EYFP; Fig. 4B,C). In the Tac2-Cre;EYFP mouse, EYFP expression largely corresponds to the anatomic regions that express Tac2 mRNA, based on the Allen Mouse Brain Atlas (Lein et al., 2007), with enrichment of EYFP+ neurons in the dorsal BNST (dBNST), posterior BNST (pBNST), MPA, mHb, CeA, Arc, and other regions (Fig. 4B,C). One diencephalic region that expresses Tac2 mRNA but is lacking in reporter expression is the lateral mammillary nucleus (not shown). Finally, to evaluate the specificity of this mouse line, we performed two-plex FISH in Tac2-Cre mice and probed for both Tac2 and Cre to quantify the penetrance (expression of Cre in Tac2+ cells) and the specificity (expression of Tac2 in Cre+ cells) of this line (Fig. 4B,C). Using image analysis of the FISH data across nine selected regions of the forebrain in which we observed high expression of Tac2, we found that the vast majority of Tac2+ cells are Cre+ (93.7%), whereas a small proportion of Tac2+ cells are Cre− (6.3%). Finally, we found no detectable Tac2−/Cre+ cells. Overall, these data show that the Tac2-Cre mouse exhibits high specificity and penetrance (Fig. 4D).
Figure 4.
Validation of a Tac2-Cre mouse. A, Coronal sections showing Tac2 ISH expression in select brain regions from the Allen Mouse Brain Atlas (Lein et al., 2007). B, Confocal micrographs (40×) of the dBNST, pBNST, and MPA showing EYFP (green) expression in Tac2-Cre;EYFP mice and FISH expression for Tac2 (magenta) and Cre (green) in Tac2-Cre mice. Scale bars: 50 µm. C, Confocal micrographs (40×) of the mHb, CeA, and Arc showing EYFP (green) expression in Tac2-Cre;EYFP mice and FISH expression for Tac2 (magenta) and Cre (green) in Tac2-Cre mice. Scale bars: 50 µm. D, Scatter plot (left) showing mean intensity of Cre and Tac2 coexpression in single cells from Tac2-Cre mice, color coded for anatomic region (n = 1157 cells, 2 mice). Donut plot (right) showing proportion of Tac2+ cells that were Cre+ or Cre−. Note that there are no detectable Tac2−/Cre+ cells.
Retrograde tracing to determine the source(s) of Tac2/NKB innervation of the LHA
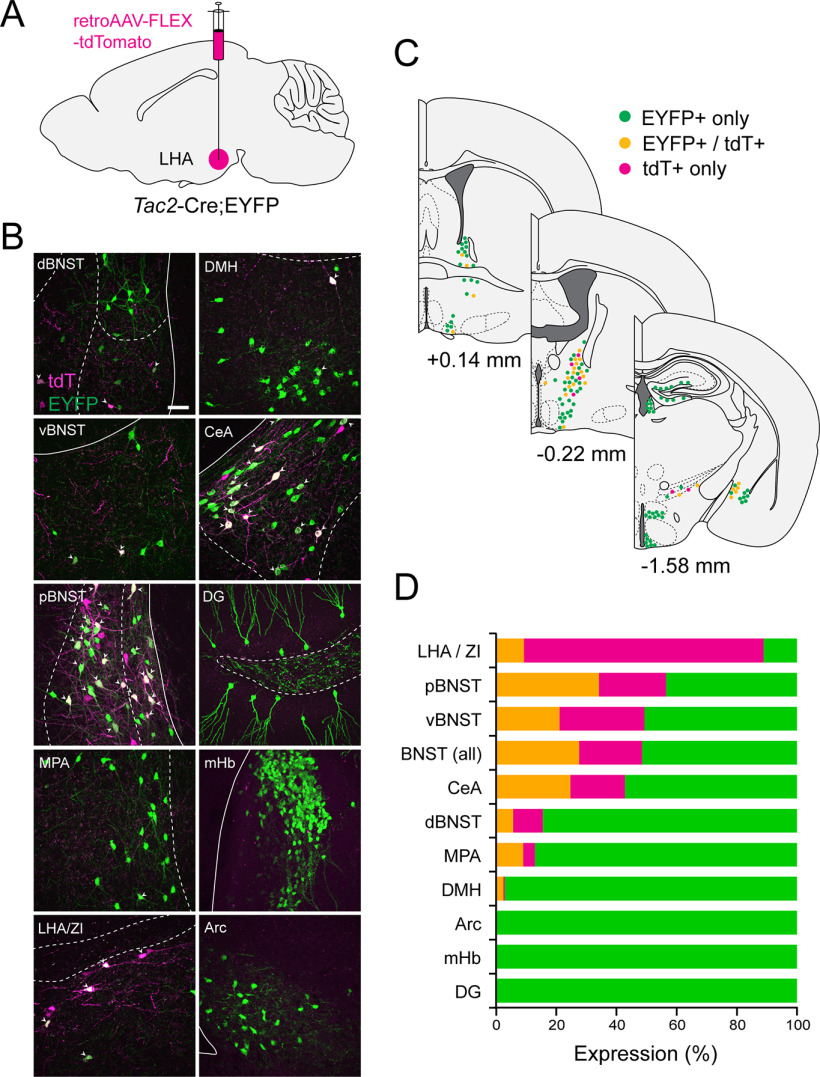
To identify the specific source(s) of NKB+ input onto MCH+ neurons in the LHA, we performed injections of a Cre-dependent retrograde virus expressing tdT (retroAAV-Flex-tdT; Tervo et al., 2016) in the LHA of Tac2-Cre;EYFP mice (Fig. 5A, diagram). After collecting serial sections throughout the brain, we quantified tdT+, EYFP+, and tdT+/EYFP+ expression (Fig. 5B–D). In our anatomic analysis of retrogradely labeled tdT+ cells, we identified several Tac2/NKB-expressing brain regions with strong retrograde labeling (BNST and CeA in particular), with other regions (e.g., mHb) in which retrograde labeling was undetectable, as described in detail below. To address the possibility that housing conditions and social isolation stress (Zelikowsky et al., 2018) could result in differences in retrograde labeling, we compared group housed and singly housed mice. We failed to observe any qualitative difference in the extent of retrograde labeling between group housed and singly housed mice. We also noted that in regions where we observed colocalization of green and red fluorescence, we found cells that were solely tdT+, which were most prevalent at the site of injection in the LHA (Fig. 5D). One possible explanation for this, given the tight correlation between Cre and Tac2 expression in Tac2-Cre mice (Fig. 4B,C), is that acute Cre-dependent viral expression of tdT, introduced in adulthood, may be more robust than constitutive EYFP expression and thus overwhelm it. Notably, we did not observe tdT+ cells in brain regions where native Tac2 is not typically expressed, making ectopic expression a less likely explanation.
Figure 5.
Retrograde tracing to determine sources of NKB innervation of the LHA. A, Diagram of the injection site with retroAAV-FLEX-tdTomato in the LHA of Tac2-Cre;EYFP mice. B, Confocal micrographs (40×) of select regions showing coexpression of EYFP (green) and tdT (magenta). Arrowheads indicate colocalization. Scale bar, 50 µm. C, Diagram illustrating the distribution of EYFP+, tdT+, and colocalized neurons at the following distances from bregma (in mm): +0.14, −0.22, −1.58. D, Bar plot showing quantified percentages of EYFP+ (green), tdT+ (magenta), and colocalized (yellow) expression across different brain regions (n = 4 mice).
We observed significant retrograde labeling in both the BNST and CeA. In our anatomic identification of the BNST, we examined labeling in the dorsal, ventral (vBNST), and posterior regions of the BNST, in which the dorsal and ventral divisions (dBNST and vBNST, respectively) were relative to the anterior commissure, followed by the more caudal pBNST (approximate distance from bregma, 0.14 to −0.34 mm; Paxinos and Franklin, 2012). EYFP+ Tac2/NKB+ neurons were especially abundant in the pBNST, and this region showed the highest density of retrogradely labeled neurons (34.21 ± 2.93%). In more anterior sections, we also observed colocalization of EYFP and tdT in the vBNST (21.07 ± 2.80%) with a few colocalized cells in the dBNST (5.66 ± 1.72%). The boundaries of the pBNST eventually merged ventrally with those in the MPA, where some tdT+/EYFP+ cells are also found. Despite observing a high degree of retrograde labeling in the pBNST, we only identified a few retrogradely labeled cells in the MPA (9.06 ± 0.73%; Fig. 5B–D).
More caudally, we also observed a high degree of colocalization in the CeA (24.73 ± 2.02%), suggesting that the CeA may be another significant source of Tac2/NKB+ innervation of the LHA. Tac2 has been identified in both the medial (CeM) and lateral compartments (CeL) of the CeA (Andero et al., 2014; Kim et al., 2017; McCullough et al., 2018), and interestingly, we found dense retrograde labeling of Tac2 cells specifically in the CeM as opposed to the CeL. More locally, the LHA and the ZI showed scattered EYFP expression with a few colocalized cells (9.28 ± 2.74%). In the same region, our quantification revealed a high percentage of cells that only expressed tdT (79.64 ± 6.40%). In the nearby DMH, we observed only a few retrogradely labeled cells (2.44 ± 0.69%; Fig. 5B–D).
In several other brain regions known to express Tac2/NKB, we observed only sparse to undetectable retrograde labeling. For example, the mHb exhibits an especially dense concentration of EYFP+ Tac2/NKB neurons but no detectable retrograde labeling (0.00%). Similarly, we observed little to no retrograde labeling in the hippocampal dentate gyrus (0.00%) or Arc (0.10 ± 0.03%; Fig. 5B–D). These data indicate that despite the abundance of Tac2/NKB in these regions, they are unlikely to contribute to Tac2/NKB projections to the LHA. Together, we found that Tac2+ neurons in the BNST and CeA, collectively known as the central extended amygdala, are the likeliest long-range sources of NKB input to the LHA. Local signaling from Tac2+ neurons in the LHA/ZI may be yet another source of NKB within the LHA.
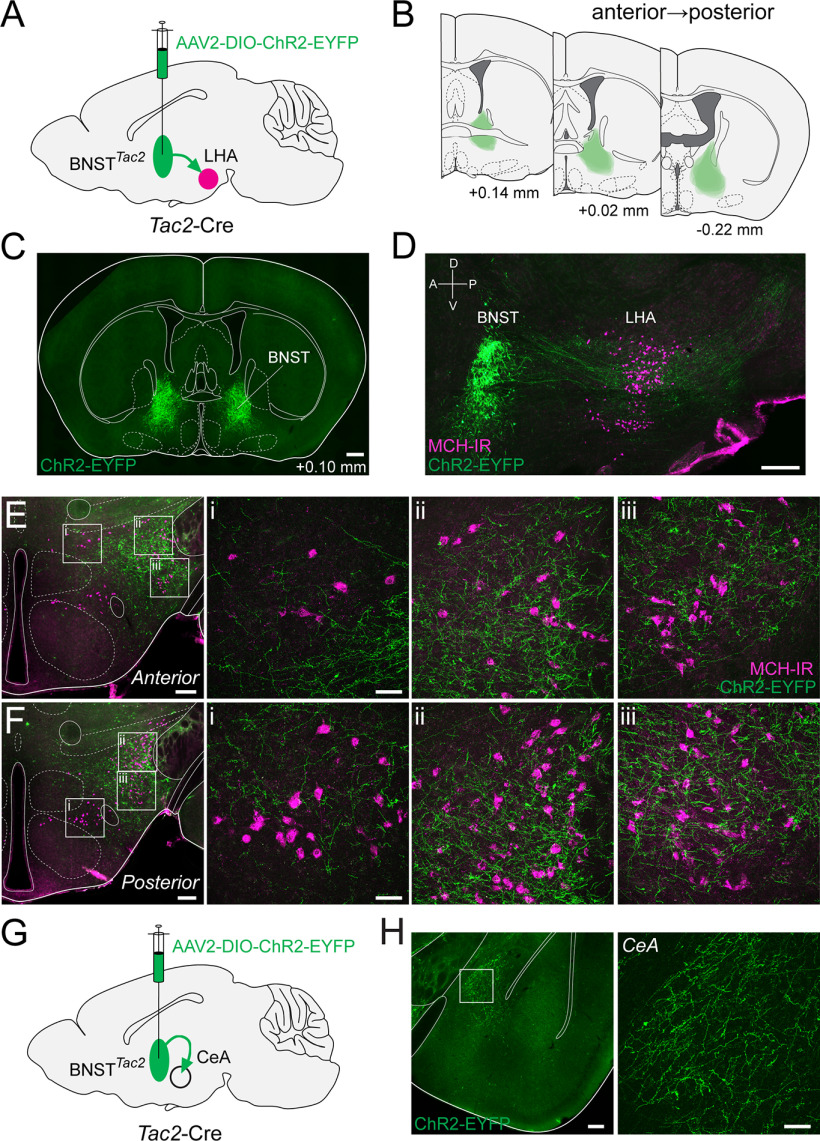
Anterograde tracing from BNSTTac2 → LHAMCH
After identifying retrogradely labeled BNSTTac2 neurons, we proceeded to validate putative projections to the LHA and relationship with LHAMCH neurons through genetically defined anterograde tracing from the BNST. To this end, we bilaterally injected an anterograde Cre-dependent ChR2-EYFP virus (AAV-EF1α-DIO-ChR2-EYFP) in the BNST of Tac2-Cre mice (Fig. 6A). Following an incubation period, we verified and mapped the injection site targeting a large proportion of the BNST (Fig. 6B,C). We then obtained coronal and sagittal sections of the LHA and examined the relationship between ChR2-positive fibers and MCH-IR perikarya (Fig. 6D–F). The BNSTTac2 pathway is visible in parasagittal sections in which we can observe fibers from the BNST projecting rostrocaudally, sweeping through the MCH field, and traversing toward the midbrain and brainstem (Fig. 6D). In coronal sections, a high density of fibers is observed throughout the dorsolateral LHA surrounding the cerebral peduncle and overlapping with the distribution of MCH-IR perikarya in two anteroposterior levels of the LHA (Fig. 6E,F). Higher magnification images of the white boxed regions from Figure 6, E and F, show a concentration of fibers throughout the dorsolateral and ventrolateral regions of the LHA (Fig. 6E, ii, iii, F, ii, iii). In medial regions of the LHA, fibers were sparse and were not intermingled with MCH-IR perikarya (Fig. 6E, i, F, i). Neighboring the LHA, we also identify ChR2-positive terminals throughout the CeA (Fig. 6G,H) suggestive of BNSTTac2 → CeA projections. This observation is in agreement with previous studies showing subdivisions of the BNST projecting to the CeA and regulating emotional behaviors (Dong et al., 2000; Dong and Swanson, 2003, 2004a; Yamauchi et al., 2018; Ye and Veinante, 2019). Finally, we found no qualitative difference in fiber distribution between housing conditions. Together, these data show that BNSTTac2 neurons extend fibers into the LHA, in close proximity to MCH-IR neurons, as well as projecting to the CeA.
Figure 6.
Anterograde tracing from BNSTTac2 → LHAMCH. A, Diagram of the BNST injection site with AAV-DIO-ChR2-EYFP in Tac2-Cre mice. B, Overlay of the injection site (green) throughout the BNST from anterior to posterior at the following distances from bregma (in mm): +0.14, +0.02, −0.22 (n = 6 mice). C, Fluorescence micrograph (10×) of a representative coronal section showing ChR2-EYFP expression in the injection site in the BNST. Scale bar, 500 µm. D, Fluorescence micrograph (10×) of a representative parasagittal section showing the BNST injection site and descending projections (green) overlaid with immunostaining for anti-MCH (magenta) in the LHA. Scale bar, 100 µm. E, Fluorescence micrograph (10×) of ChR2-EYFP fibers and MCH immunostaining in a representative coronal section of the anterior LHA with confocal micrographs (40×) of the boxed regions (i–iii). Scale bar, 200 µm, low-magnification images; 50 µm, high-magnification images. F, Fluorescence micrograph (10×) of ChR2-EYFP fibers and MCH immunostaining in a representative coronal section of the posterior LHA with confocal micrographs (40×) of the boxed regions (i–iii). Scale bar, 200 µm, low-magnification images; 50 µm, high-magnification images. G, Diagram illustrating the BNST injection site and its projections to the CeA. H, Fluorescence micrograph (10×) of EYFP-expressing BNSTTac2 terminals in the CeA (left) with a confocal micrograph (40×) of the boxed region (right). Scale bar, 200 µm, low-magnification images; 50 µm, high-magnification images.
Anterograde tracing from CeATac2 → LHAMCH
In addition to BNSTTac2 neurons, our retrograde tracing revealed a subset of CeATac2 neurons that putatively project to the LHA. To examine the projection patterns of CeATac2 neurons, we injected AAV-EF1α-DIO-ChR2-EYFP bilaterally into the CeA of Tac2-Cre mice (Fig. 7A). In our injection site, we attempted to capture the medial CeA adjacent to the cerebral peduncle (Fig, 7B,C), where we previously identified retrogradely labeled cells from the LHA. After an incubation period, we observed ChR2-positive terminals in the LHA relative to MCH-IR perikarya in two anteroposterior levels of the LHA (Fig. 7D,E). Although not as prominent compared with BNSTTac2 fibers, we observed a moderate density of fibers either appearing to directly project ventromedially from the injection site or dorsally through the stria terminalis, along the length of the cerebral peduncle. Higher magnification images display fibers preferentially in the dorsolateral region of the LHA intermingled with LHAMCH perikarya, in addition to sparser fibers closer to the DMH (Fig. 7D, i–iii, 7E, i–iii). We also examined the presence of ChR2-positive CeATac2 terminals in the BNST. We identified fibers in both the dorsal and ventral BNST specifically along the stria terminalis, adjacent to the internal capsule, and between the anterior commissure (Fig. 7F,G). Again, we found no qualitative difference in fiber distribution between housing conditions. Together, these anterograde tracing experiments further confirm the results of our retrograde viral tracing and show that CeATac2 neurons project into the LHA in close proximity to MCH-IR neurons, as well as projecting to the BNST.
Figure 7.
Anterograde tracing from CeATac2→ LHAMCH. A, Diagram of the CeA injection site with AAV-DIO-ChR2-EYFP in Tac2-Cre mice. B, Overlay of the injection site (green) throughout the CeA from anterior to posterior at the following distances from bregma (in mm): −1.22, −1.34, −1.46 (n = 4 mice). C, Fluorescence micrograph (10×) of a representative coronal section showing ChR2-EYFP expression of the injection site (green) in the CeA. Scale bar, 300 µm. D, Fluorescence micrograph (10×) of ChR2-EYFP fibers (green) and MCH immunostaining (magenta) in a representative coronal section of the anterior LHA with confocal micrographs (40×) of the boxed regions (i–iii). Scale bars: 200 µm, low-magnification images; 50 µm, high-magnification images. E, Fluorescence micrograph (10×) of ChR2-EYFP fibers and MCH immunostaining in a representative coronal section of the posterior LHA with confocal micrographs (40×) of the boxed regions (i–iii). Scale bars: 200 µm, low-magnification images; 50 µm, high-magnification images. F, Diagram illustrating the CeA injection site and its projections to the BNST in Tac2-Cre mice. G, Confocal micrograph (40×) of the EYFP-expressing CeATac2 terminals in the dBNST (left) and vBNST (right). Scale bar, 50 µm.
BNSTTac2 and CeATac2 projections → midbrain and the brainstem
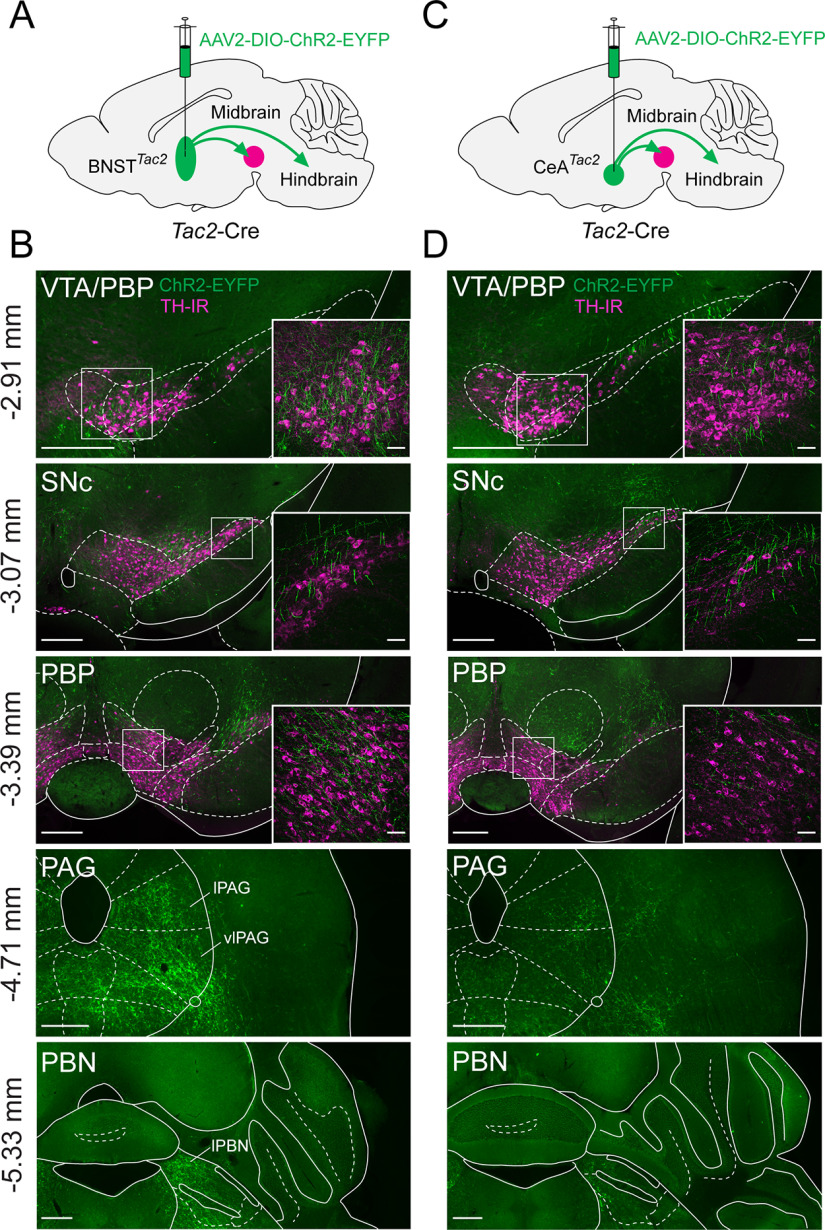
In our anterograde mapping of BNSTTac2 and CeATac2 to the LHA, we found that the fibers not only project to the LHA but appear to project more caudally as well. We collected coronal sections from both the midbrain and the brainstem to further examine these projection patterns. To further delineate the anatomic borders that make up the midbrain, including the ventral tegmental area (VTA), the substantia nigra pars compacta (SNc) and the parabrachial pigmented nucleus (PBP), we immunostained these brain sections with anti-tyrosine hydroxylase (TH) to label dopaminergic (DA) neurons. We used the same Tac2-Cre mice that were injected in the BNST, or the CeA, with the virus AAV-EF1α-DIO-ChR2-EYFP (Fig. 8A,C) and identified fibers in the midbrain that were in the field of TH-IR perikarya (Fig. 8B,D). Higher magnification images show a dense plexus of BNSTTac2 fibers intermingled with TH-IR perikarya in the VTA and the PBP, with sparse fibers in the SNc that are roughly orthogonal to the plane of the brain section (Fig. 8B). Dorsal to the SNc, we found a high concentration of BNSTTac2 fibers in the midbrain reticular nucleus (MRN). More caudally, we observed robust BNSTTac2 fibers throughout the periaqueductal gray (PAG) that span the lateral portion and spread in higher density to the ventrolateral regions of the PAG. BNSTTac2 fibers were also identified in the medial and lateral divisions of the parabrachial nucleus (PBN), surrounding the superior cerebral peduncles and adjacent to the locus ceruleus. Similarly, we observed CeATac2 fibers in the midbrain and the brainstem with varying levels of density (Fig. 8D). In the VTA and the PBP, we found sparse and scattered CeATac2 fibers throughout the regions. However, a higher density of CeATac2 fibers was identified in the MRN and the SNc, but the SNc fibers appear truncated because of the orthogonal alignment of the fibers. Caudally, within the regions of the brainstem, CeATac2 fibers are substantially lower in density and extend outside the PAG into the neighboring regions of the MRN. Collectively, these data demonstrate that the BNSTTac2 and CeATac2 neurons that project to the LHA also project to other key nodes of behavioral state regulation in the midbrain and brainstem, including close appositions to midbrain DA neurons.
Figure 8.
BNSTTac2 and CeATac2 projections to the midbrain and brainstem. A, Diagram of the BNST injection site and projections with AAV-DIO-ChR2-EYFP in Tac2-Cre mice. B, Fluorescence micrographs (10×) of ChR2-EYFP fibers (green) in the VTA/PBP, SNc, PBP, PAG, and PBN with the top three regions immunostained for anti-TH (magenta). Insets, Confocal micrographs (40×) of the boxed regions. C, Diagram of the CeA injection site and projections with AAV-DIO-ChR2-EYFP in Tac2-Cre mice. D, Fluorescence micrographs (10×) of ChR2-EYFP fibers in the VTA/PBP, SNc, PBP, PAG, and PBN with the top three regions immunostained for anti-TH. Insets, Confocal micrographs (40×) of the boxed regions. Scale bars: 400 µm, low-magnification images; 50 µm, high-magnification images. lPBN, lateral parabrachial nucleus.
Functional synaptic connectivity between BNSTTac2/CeATac2 → LHAMCH neurons
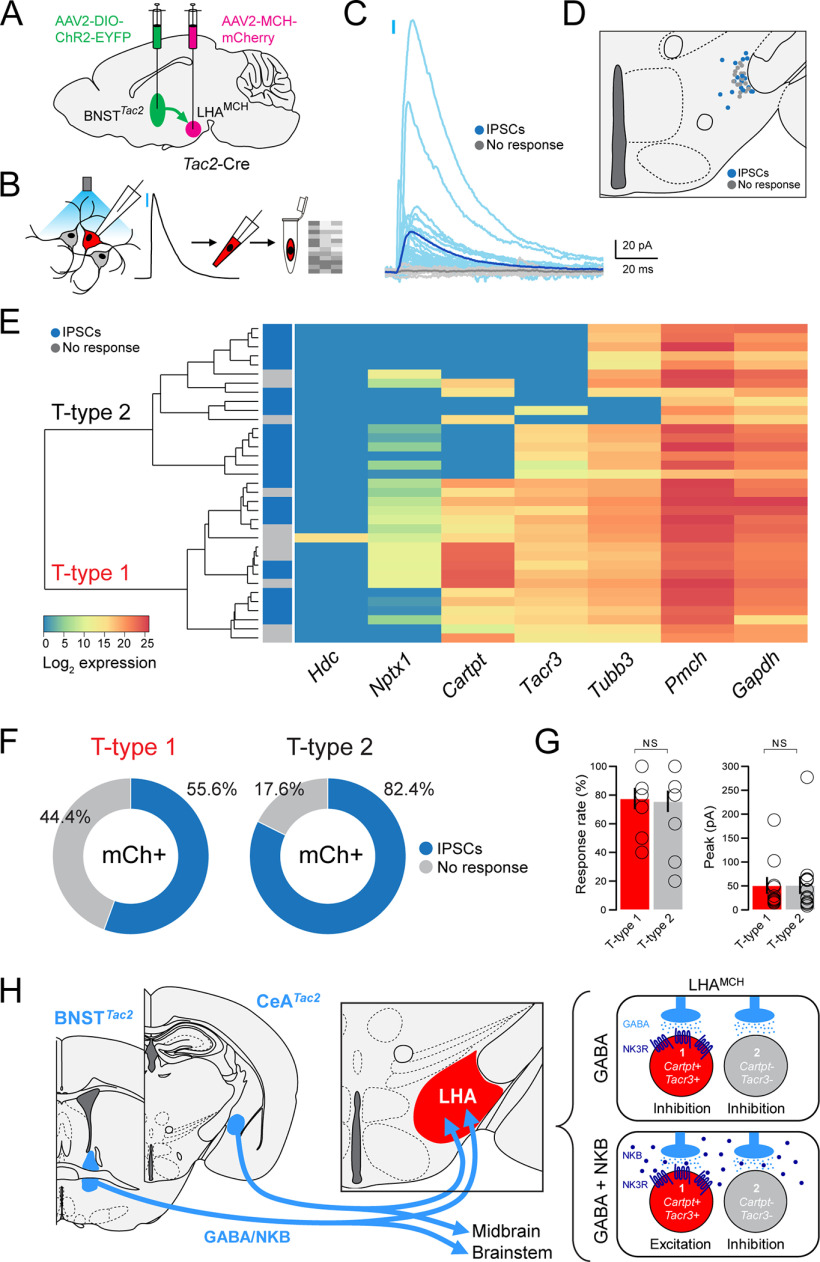
We next investigated whether BNSTTac2 and CeATac2 projections to the LHA exhibit functional synaptic connectivity with LHAMCH neurons. Our goal was to record from visually identified, genetically defined LHAMCH neurons in brain slices while optogenetically stimulating BNSTTac2 or CeATac2 fibers in the LHA. To accomplish this, we took a dual-virus approach; in Tac2-Cre mice, we injected the Cre-dependent ChR2-EYFP virus in either the BNST or CeA, as in the previous anterograde tracing experiments, in conjunction with AAV-MCH-mCherry injected into the LHA, as shown in Figure 2A.
We proceeded to probe the functional synaptic connectivity of BNSTTac2 → LHAMCH projections using the dual-virus approach. We investigated fast neurotransmitter release by BNSTTac2 terminals in the LHA with blue-light photostimulation while recording from mCherry+ MCH+ neurons, the locations of which were mapped anatomically (Fig. 9A). In whole-cell voltage-clamp recordings, we examined possible photostimulation-evoked EPSCs at −70 mV and IPSCs at 0 mV in each visually identified LHAMCH neuron. Holding at −70 mV, we failed to observe any evoked current indicative of glutamatergic EPSCs (Fig. 9B). However, at 0 mv, photostimulation of BNSTTac2 terminals evoked time-locked IPSCs in 54.5% of mCherry+ LHAMCH neurons (Fig. 9B). We performed the same analysis in CeATac2 → LHAMCH projections (Fig. 9C). Similarly, at −70 mV, photostimulation of CeATac2 terminals failed to evoke EPSCs, however, at 0 mV, we observed IPSCs in 43.1% of mCherry+ LHAMCH neurons (Fig. 9D). These results suggest functional inhibitory synaptic transmission from both BNSTTac2 and CeATac2 neurons onto subpopulations of LHAMCH neurons.
Figure 9.
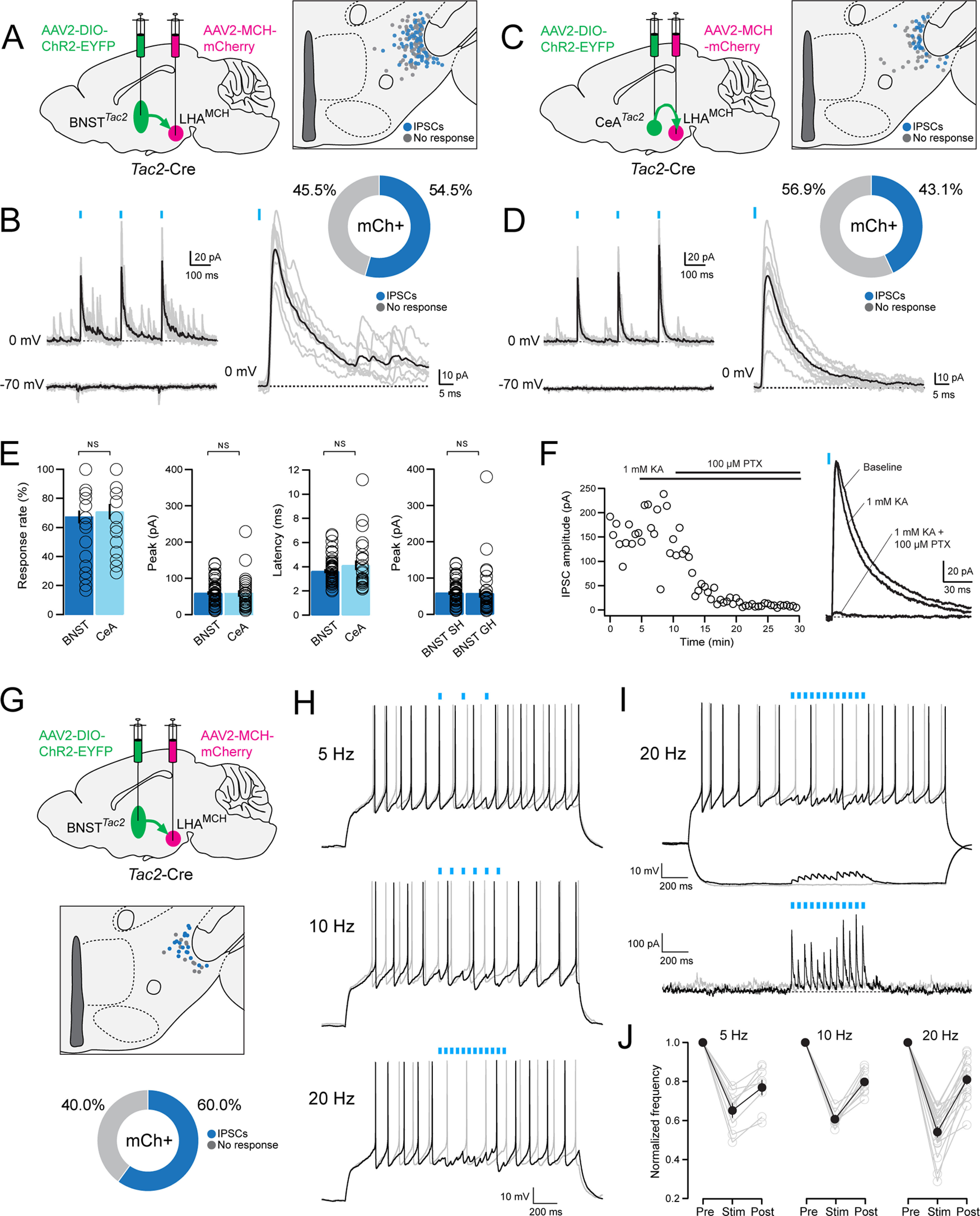
Functional connectivity between BNSTTac2/CeATac2 neurons and LHAMCH neurons. A, Diagram illustrating dual, bilateral stereotactic injections in a Tac2-Cre mouse with AAV-DIO-ChR2-EYFP in the BNST and AAV-MCH-mCherry in the LHA (left). Anatomical map of the LHA illustrating the location of the recorded mCherry+ neurons classified based on the response to the photostimulation (gray, no response; blue, evoked IPSCs; n = 145 cells, 10 mice; right). B, Overlay of average (black) and individual (gray) filtered traces of a representative photostimulated (blue lines) mCherry+ neuron held at 0 mV (top) and −70 mV (bottom, left). Magnified view of the evoked IPSC at the first pulse (blue line) held at 0 mV from the same mCherry+ neuron (bottom) and a corresponding donut plot illustrating the responses of all recorded mCherry+ neurons (top). C, Diagram illustrating dual, bilateral stereotactic injections in a Tac2-Cre mouse with AAV-DIO-ChR2-EYFP in the CeA and AAV-MCH-mCherry in the LHA (left). Anatomical map of the LHA illustrating the location of the recorded mCherry+ neurons classified based on the response to the photostimulation (gray, no response; blue, evoked IPSCs; n = 65 cells, 4 mice; right). D, Overlay of average (black) and individual (gray) filtered traces of a representative photostimulated (blue lines) mCherry+ neuron held at 0 mV (top) and −70 mV (bottom; left). Magnified view of the evoked IPSC at the first pulse (blue line) held at 0 mV from the same mCherry+ neuron (bottom) and a corresponding donut plot illustrating the responses of all recorded mCherry+ neurons (top). E, Bar plots quantifying (left to right) the response rate (%), peak amplitude (pA), and latency (ms) of recorded mCherry+ neurons with photostimulation of BNSTTac2 (dark blue; n = 95 cells, 6 mice) or CeATac2 terminals (light blue; n = 65 cells, 4 mice; Mann–Whitney test; response rate, p = 0.536; peak amplitude, p = 0.461; latency, p = 0.721). Bar plot quantifying the peak amplitude of recorded mCherry+ neurons with photostimulation of BNSTTac2 terminals between group housed (GH; n = 50 cells, 4 mice) and singly housed mice (SH; n = 95 cells, 6 mice; Mann–Whitney test; p = 0.0653; far right). F, Plot displaying the IPSC amplitude (pA) of a representative mCherry+ neuron during baseline, followed by application of KA (1 mm) and PTX (100 μm; left) and the corresponding overlay of the average traces from each condition (right). G, Diagram illustrating dual, bilateral stereotactic injections in a Tac2-Cre mouse with AAV-DIO-ChR2-EYFP in the BNST and AAV-MCH-mCherry in the LHA (top). Anatomical map of the LHA illustrating the location of the recorded mCherry+ neurons classified based on the response to the photostimulation (gray, no response; blue, evoked IPSCs; n = 30 cells, 3 mice; middle). Donut plot depicting the responses of all recorded mCherry+ neurons (bottom). H, Representative current-clamp traces of a recorded mCherry+ neuron without (light gray) and with (black) 5, 10, and 20 Hz photostimulation (blue lines) in conjunction with a depolarizing step. I, Representative current-clamp traces of a recorded mCherry+ neuron without (light gray) and with (black) 20 Hz photostimulation (blue lines) during a depolarizing and hyperpolarizing step (top). Corresponding voltage-clamp trace in the same cell without (light gray) and with (black) 20 Hz photostimulation (blue lines). J, Plot of the averaged normalized frequency before (pre), during (stim), and after (post) photostimulation at varying frequencies of every recorded mCherry+ neuron (light gray) and the average of each frequency group overlaid (black).
We further analyzed the parameters of IPSCs evoked by the first blue-light pulse on BNSTTac2 and CeATac2 fibers by quantifying the response rate, peak amplitude, and latency. In comparing BNSTTac2 and CeATac2 with LHAMCH photostimulation-evoked IPSC parameters, we found no significant differences (Fig. 9E). Finally, to control for the possibility that housing conditions may affect inhibitory synaptic strength, we compared BNSTTac2 → LHAMCH IPSC amplitudes between group housed and singly housed mice and found no significant difference (Fig. 9E). Together, our functional connectivity data show that across projections from two separate regions, the connectivity strength of the tachykininergic pathway in the BNSTTac2 → LHAMCH and CeATac2 → LHAMCH circuits show remarkable consistency between these two distinct sources of synaptic input.
To further confirm that the light-evoked IPSCs are attributable to monosynaptic release of GABA and not disynaptic feedforward inhibition, we bath applied kynurenic acid (KA), a broad-spectrum ionotropic glutamate receptor antagonist, in a subset of cells (Fig. 9F). The addition of KA (1 mm) did not block the outward currents in a representative example, suggesting that this functional connection is unlikely to be the result of feedforward inhibition. The GABAA receptor antagonist PTX (100 μm) was subsequently washed in and blocked photostimulation-evoked IPSCs, confirming that this response is because of the activation of GABAA receptors. These findings are in agreement with existing evidence suggesting that BNST and CeA neurons are predominantly GABAergic (Sun and Cassell, 1993; Swanson and Petrovich, 1998; Kudo et al., 2012; Duvarci and Pare, 2014; Lebow and Chen, 2016; Welch et al., 2019).
Finally, to examine the impact of limbic Tac2+ GABAergic inputs on the excitability of LHAMCH neurons, we recorded from LHAMCH neurons in current-clamp mode while stimulating BNSTTac2 inputs at various frequencies. We used the dual-virus approach as previously described, marked the anatomic location of recorded cells, and found that 60.0% of mCherry+ cells displayed photostimulation-evoked IPSCs (Fig. 9G). As we found that LHAMCH neurons were often electrically silent in slices, we applied a 2 s depolarizing pulse to evoke regular firing while carrying out photostimulation at 5, 10, and 20 Hz (Fig. 9H). We found that photostimulation enhances GABAergic inhibition of firing in a frequency-dependent manner, whereas in the examples shown, 20 Hz stimulation is especially effective at reversibly diminishing or silencing evoked firing (Fig. 9H–J). We further observed that the average poststimulation firing rate was ∼20% lower than prestimulation at all frequencies, suggesting a modest although longer-lasting hyperpolarization following the train of IPSCs. Overall, these data demonstrate that BNSTTac2 GABAergic inputs are capable of suppressing evoked firing in a large subset of LHAMCH neurons in a frequency-dependent manner.
Specificity of BNSTTac2 GABAergic input → LHAMCH neuron subpopulations
Thus far, we found in our scRNA-seq and FISH experiments that more than half of LHAMCH neurons coexpress Tacr3 and Cartpt. Furthermore, we found that photostimulation of BNSTTac2 and CeATac2 terminals induced IPSCs in roughly half of the LHAMCH neurons recorded. One remaining question is whether the functional inhibitory connections between limbic forebrain Tac2/NKB+ neurons and LHAMCH neurons are specific to the subpopulation of LHAMCH neurons that coexpress Cartpt/CART and Tacr3/NK3R. To address this question, we focused on BNSTTac2 to LHAMCH inhibitory connectivity using the same dual-virus approach (Fig. 10A) shown previously (Fig. 9A,G). However, immediately following photostimulation, we harvested the cytoplasm of the recorded LHAMCH neurons for sc-qPCR analysis (Fig. 10B) as previously described (Fig. 2). Among 35 visually identified mCherry+ cells in the LHA, 24 exhibited photostimulation-evoked IPSCs. Individual traces and the average trace are shown in Figure 10C, whereas the anatomic location of each recorded cell is displayed in Figure 10D. The resulting sc-qPCR data are highly consistent with those shown in Figure 2D. Using a panel of seven markers (Hdc, Nptx1, Cartpt, Tacr3, Tubb3, Pmch, and Gapdh), unsupervised hierarchical clustering showed two major clades representing T-type 1 (Cartpt/Tacr3 coexpressing) and T-type 2 (Cartpt/Tacr3 noncoexpressing; Fig. 10E). However, in comparison with the vertical plot denoting the presence or absence of photostimulation-evoked IPSCs (Fig. 10E), transcriptomic classification did not appear to track with cells that exhibited IPSCs. Among T-type 1 neurons, 55.6% exhibited IPSCs (Fig. 10F). In contrast, among T-type 2 neurons, 82.4% exhibited IPSCs (Fig. 10F). Furthermore, in comparing the response rate and peak amplitude of photostimulation-evoked IPSCs in T-type 1 and 2 neurons, there was no significant difference in synaptic input (Fig. 10G). These data suggest that BNSTTac2 inhibitory projections onto LHAMCH neurons may be rather indiscriminate, making functional GABAergic synapses onto both transcriptomically defined LHAMCH neuron subpopulations (Fig. 10H, diagram). However, in the hypothetical case that GABA and NKB are coreleased, NK3R-expressing T-type 1 neurons would likely be preferentially depolarized (Fig. 10H), leading to selective activation of a molecularly distinct LHAMCH subcircuit.
Figure 10.
Assessment of the specificity of BNSTTac2 GABAergic inputs onto transcriptionally distinct LHAMCH neuron subpopulations. A, Diagram illustrating dual, bilateral stereotactic injections in a Tac2-Cre mouse with AAV-DIO-ChR2-EYFP in the BNST and AAV-MCH-mCherry in the LHA (top). B, Diagram of optogenetic stimulation and sc-qPCR procedure—voltage-clamp recording from mCherry+ cells, photostimulation to evoke IPSCs, cytoplasm harvest, and sc-qPCR for key molecular markers. C, Overlay of the averaged trace of every photostimulated mCherry+ neuron classified based on the response (light gray, no response; light blue, evoked IPSCs) with the average of each response group overlaid (gray, no response; dark blue, evoked IPSCs). D, Anatomical map of the LHA illustrating the location of the recorded mCherry+ neurons classified based on the response to the photostimulation (gray, no response; blue, evoked IPSCs; n = 35 cells, 5 mice). E, Dendrogram of unsupervised cluster analysis and corresponding heatmap of gene expression in recorded mCherry+ neurons following photostimulation. The two major clades are transcriptomic T-type 1 and T-type 2 based on the seven markers. Photostimulation-evoked IPSCs are indicated for each cell (gray, no response; blue, evoked IPSC). F, Donut plots depicting the proportion of responses to the photostimulation in mCherry+ neurons that are either T-type 1 (left, n = 18 cells) or T-type 2 (right, n = 17 cells). G, Bar plots quantifying (left to right) the response rate (%) and peak amplitude (pA) of IPSCs in recorded mCherry+ neurons with photostimulation of BNSTTac2 terminals on T-type 1 (red; 10 cells) or T-type 2 (gray; 14 cells) LHAMCH neurons. (Mann–Whitney test; response rate, p = 0.9519; peak amplitude, p = 0.7961). H, Diagram illustrating key findings. BNSTTac2 and CeATac2 neuron projections converge on the LHA, as well as projecting to midbrain and brainstem targets (left). GABAergic innervation of both LHAMCH subpopulations results in fast synaptic inhibition (top), whereas hypothetical cotransmission of GABA and NKB (bottom) would be predicted to result in excitation of the NK3R+ LHAMCH subpopulation alone (right).
Discussion
Here, we identify parallel limbic forebrain-hypothalamic pathways, which are putative sources of tachykininergic NKB input to the LHA, and define the synaptic connectivity with molecularly distinct NK3R-expressing LHAMCH subpopulations. Consistent with our anatomic data, confirming that a subpopulation of LHAMCH neurons express both Tacr3 mRNA and NK3R protein, application of an NK3R-selective agonist depolarized roughly half of the recorded LHAMCH neurons, thereby demonstrating the expression and functional activity of NK3R receptors in LHAMCH neurons. Through electrophysiological phenotyping of LHAMCH neurons, we found that molecularly defined subpopulations, identified through sc-qPCR, exhibit differences in intrinsic membrane properties. Furthermore, in both Pmch-Cre and Tac2-Cre mice, we observed dense Tac2/NKB fibers apposing MCH- and NK3R-expressing perikarya in the LHA. To identify putative sources of Tac2/NKB fibers onto LHAMCH neurons, we used Cre-dependent retrograde labeling and identified prominent populations of retrogradely labeled Tac2/NKB neurons in the BNST and the CeA, collectively known as the central extended amygdala. We further validated these results through a comparison of cell-type-specific anterograde viral tracing from both BNSTTac2 and CeATac2 fibers, both of which we found to innervate the LHA. Both BNSTTac2 and CeATac2 neuron populations not only project to the LHA but also to targets in the midbrain and brainstem. Optogenetic activation of these Tac2/NKB fibers in the LHA induced IPSCs in roughly half of the recorded LHAMCH neurons. Finally, gene-expression profiles of recorded LHAMCH neurons using sc-qPCR revealed that BNSTTac2 fibers form functional GABAergic synapses onto both LHAMCH neuron subpopulations, suggesting rather indiscriminate GABAergic input but selectivity in terms of putative NKB/NK3R signaling in a subpopulation of LHAMCH neurons. These parallel descending central extended amygdala circuits, projecting to the hypothalamus, midbrain, and brainstem, are well positioned to conjointly modulate multiple nodes of homeostatic and behavioral control, likely through both NKB/NK3R signaling and GABAergic mechanisms.
Cellular and functional diversity of MCH neurons
LHAMCH neurons play fundamental and conserved, yet diverse, roles in physiology and behavior including feeding, arousal, stress/anxiety, and memory (Pissios et al., 2006; Barson et al., 2013; Croizier et al., 2013; Monti et al., 2013; Diniz and Bittencourt, 2017; Ferreira et al., 2017; Bandaru et al., 2020; Concetti and Burdakov, 2021). In terms of feeding and metabolism, central administration of MCH in mice increases food intake (Qu et al., 1996; Rossi et al., 1997), MCH KO mice display a hypophagic, lean phenotype (Shimada et al., 1998), and LHAMCH neurons are key regulators of sucrose preference (Domingos et al., 2013). In the context of sleep and arousal, LHAMCH neurons discharge maximally during rapid eye movement (REM) sleep (Verret et al., 2003; Hanriot et al, 2007; Hassani et al., 2009), whereas optogenetic activation of LHAMCH neurons and projections increase REM sleep duration (Jego et al., 2013; Konadhode et al., 2013; Tsunematsu et al., 2014; Blanco-Centurion et al., 2016; Vetrivelan et al., 2016). Moreover, LHAMCH neurons have been implicated in exploratory behavior related to novel object recognition (González et al., 2016; Blanco-Centurion et al., 2019; Kosse and Burdakov, 2019) and reward behavior (Dilsiz et al., 2020) and have anxiogenic effects attributable to activation of the hypothalamic-pituitary-adrenal axis (Kennedy et al., 2003; Smith et al., 2006). Most recently, LHAMCH projections to the hippocampus have been implicated in both modulating memory during REM sleep (Izawa et al., 2019) and impulsivity (Noble et al., 2019).
One explanation for the extraordinary diversity of LHAMCH function may be that the MCH system operates through parallel subcircuits; that is, LHAMCH neurons may consist of distinct subpopulations, with unique inputs, outputs, signaling mechanisms, and functional repertoires. Anatomical evidence in rodents supports the notion of distinct LHAMCH subpopulations based on neurochemical markers including CART and NK3R (Griffond et al., 1997; Broberger, 1999; Vrang et al., 1999; Elias et al., 2001; Brischoux et al., 2002; Cvetkovic et al., 2003, 2004; Harthoorn et al., 2005; Hanriot et al., 2007; Croizier et al., 2010). Subsequent bulk molecular profiling approaches confirmed the enrichment of Cartpt and Tacr3 in LHAMCH neurons (Knight et al., 2012; Ekstrand et al., 2014; Nectow et al., 2017), whereas our previous scRNA-seq analysis confirmed and extended the transcriptomic basis for at least two LHAMCH subpopulations (Mickelsen et al., 2019). Further single-cell and single-nuclei analyses of the hypothalamus suggest multiple LHAMCH subpopulations (Jiang et al., 2020; Kim et al., 2020). Key insights emerged from developmental fate-mapping experiments, which revealed that LHAMCH neurons are generated from E10 onwards, but subsets begin to express CART at E18, and NK3R expression is eventually observed at P5 (Brischoux et al., 2001). In terms of unique projections, retrograde tract-tracing experiments demonstrated that MCH+/NK3R− neurons project to the spinal cord with some labeled axons in the subthalamic and parabrachial nuclei, whereas MCH+/NK3R+ fibers innervate the telencephalon, including the cerebral cortex, hippocampus, and the medial septum (Brischoux et al., 2002; Cvetkovic et al., 2004). Furthermore, functional evidence suggests that a distinct subpopulation of LHAMCH neurons innervates the cerebral ventricles and operates through volume transmission to modulate feeding behavior (Noble et al., 2018). Our gene-expression, anatomic, and electrophysiological data complement and extend the existing anatomic and developmental work, suggesting that NKB/NK3R signaling in a subpopulation of LHAMCH neurons likely defines a functionally distinct subcircuit within the LHAMCH system.
In terms of functional diversity of LHAMCH neurons, we examined the possibility that transcriptionally distinct subpopulations exhibit divergent electrical phenotypes. Although we found that the overall electrophysiological properties of LHAMCH neurons align with previous descriptions of rodent LHAMCH neurons in hypothalamic slices (van den Pol et al., 2004; Burdakov et al., 2005; Linehan and Hirasawa, 2018), they are also quite heterogeneous. Cluster analysis, based on passive and active intrinsic membrane properties, revealed two broad categories, referred to as E-types A and B. Although E-type A exhibits a moderate enrichment in neurons that coexpress Cartpt and Tacr3 (T-type 1), electrical phenotypes were found to be diverse. Nevertheless, we observed significant differences in the electrophysiological features of transcriptionally distinct LHAMCH subpopulations, perhaps reflecting differences in developmental origin (Brischoux et al., 2001). One limitation of our analysis is that we mostly sampled intermingled LHAMCH subpopulations in the more lateral part of the LHA, whereas a more comprehensive sampling of both medial and lateral LHAMCH neurons may yield more distinct electrophysiological phenotypes. Finally, other neurochemical features of LHAMCH neurons may further functionally distinguish subpopulations. Investigations into the fast neurotransmitter phenotype of LHAMCH neurons, as determined by optogenetic activation of neuron terminals in different target regions, has yielded divergent effects (Jego et al., 2013; Apergis-Schoute et al., 2015; Chee et al., 2015), perhaps suggestive of functional subpopulations. However, our single-cell gene-expression data showing that Pmch+ neurons uniformly express Slc17a6 and Gad1, but not Slc32a1 (Mickelsen et al., 2017, 2019), would argue against GABAergic versus glutamatergic synaptic phenotype being the basis for distinguishing LHAMCH subpopulations. Other functional signatures, such as differential responses to neurotransmitters and neuropeptides (van den Pol et al., 2004; Diniz and Bittencourt, 2017) and perhaps other neuromodulatory factors such as glucose (Burdakov et al., 2005; Kong et al., 2010) may be important in this regard. Future work is required to align the transcriptomic profiles of LHAMCH subpopulations with functional validation of their unique signaling mechanisms.
Identification of the BNST and CeA as key sources of Tac2/NKB input to the LHA
The BNST and the CeA, collectively known as the central extended amygdala, are heterogeneous and functionally intertwined structures implicated in stress, anxiety, fear, arousal, pain, and motivated behavior (Davis et al., 2010; Janak and Tye, 2015; Kash et al., 2015; Tovote et al., 2015; Gafford and Ressler, 2016; Lebow and Chen, 2016; Shackman and Fox, 2016; Ch'ng et al., 2018; Fox and Shackman, 2019; Giardino and Pomrenze, 2021). Detailed anatomic and functional work has demonstrated that both the BNST and CeA broadly innervate the LHA (Hosoya and Matsushita, 1980; Barone et al., 1981; Dong et al., 2000; Dong and Swanson, 2004a,b, 2006; Barbier et al., 2018a,b, 2021; Weera et al., 2021) and terminate onto defined populations of LHA neurons (Nakamura et al., 2009; Jennings et al., 2013a; Giardino et al., 2018; Yoshida et al., 2006; Barbier et al., 2018a, 2021). Cell-type-specific monosynaptic retrograde tracing and optogenetic experiments further identified the BNST and the CeA as two sources of monosynaptic GABAergic input onto LHAMCH neurons (González et al., 2016). Both of these neurochemically complex regions are predominantly composed of GABAergic neurons (Sun and Cassell, 1993; Swanson and Petrovich, 1998; Kudo et al., 2012; Duvarci and Pare, 2014; Lebow and Chen, 2016; Welch et al., 2019), but further deconstruction of GABAergic neuron cell-type identity is necessary to determine specific projection patterns and diverse, often divergent, roles in physiology and behavior. Anatomical and functional work examining cell-type diversity in the CeA suggests that Tac2 is indeed a discriminatory marker that defines a distinct CeA subpopulation (Andero et al., 2014; Kim et al., 2017; McCullough et al., 2018), whereas single-nuclei RNA-seq has shown that Tac2 defines 1 of 41 neuronal populations in the mouse BNST (Welch et al., 2019). Furthermore, translational profiling of retrogradely labeled BNST projections to the brainstem parabrachial nucleus showed enrichment in Tac2 (Luskin et al., 2021), consistent with our anterograde tracing showing BNSTTac2 → lPBN projections. Importantly, recent work has shown that chronic social isolation stress (SIS) induces a widespread enhancement in Tac2 expression in the brain, prominently in the BNST, CeA, and other regions, in conjunction with increased aggressive and defensive behaviors such as freezing (Zelikowsky et al., 2018), highlighting the importance of BNST/CeATac2 neurons in the regulation of affective state. In our work, we found that BNSTTac2 and CeATac2 neurons not only converge on the LHA, but also directly innervate LHAMCH neurons regardless of Tacr3 expression. These findings align with detailed anatomic work in rats demonstrating NKB input to the LHA that may arise from distributed forebrain and brainstem sites, including the BNST, basal forebrain, CeA, and PAG (Cvetkovic et al., 2003).
The BNST serves as a hub for diverse intrinsic and extrinsic cues and is implicated in processing affective state, arousal, motivated behavior, and homeostatic functions. An extensive and evolving literature has described the circuit-level underpinnings of different BNST subdivisions and subpopulations, classified by anatomic location, neurochemical profile, projection patterns, and target-dependent effects on diverse behaviors (Kash et al., 2015; Daniel and Rainnie, 2016; Gafford and Ressler, 2016; Lebow and Chen, 2016; Vranjkovic et al., 2017; Ch'ng et al., 2018; Giardino and Pomrenze, 2021). For example, chemogenetic or optogenetic activation of GABAergic BNST neurons is anxiogenic (Mazzone et al., 2018) and induces wakefulness from non-REM sleep (Kodani et al., 2017), whereas optogenetic activation of BNST GABAergic fibers in the LHA and VTA induces target-region-dependent effects on feeding, reward-related behaviors, and anxiety (Kim et al., 2013; Jennings et al., 2013a,b). These data demonstrate that inhibitory BNST neurons have multimodal roles to play in appetitive and consummatory behaviors, threat responses, and arousal and are both cell-type specific and target dependent. Work that is illustrative of the functional diversity of inhibitory BNST neurons shows that two GABAergic BNST populations, differentiated by cell-type-specific neuropeptide markers, have divergent responses to appetitive and aversive stimuli (Giardino et al., 2018). With regard to BNSTTac2 neurons, Zelikowsky et al. (2018) showed that chemogenetic silencing of BNSTTac2 neurons or selective Tac2 knockdown in the BNST diminished the effects of SIS, whereas a combination of chemogenetic activation and Tac2 overexpression mimicked aspects of SIS. Additional work is required to unravel the neurochemistry of Tac2 neurons in the BNST and identify coexpression with known BNST subpopulations and target-dependent effects of these subpopulations on physiology and behavior. Consistent with the broad projection patterns of the BNST as a whole, we identified similarly widespread BNSTTac2 terminals in regions regulating affective, motivated, and defensive behavior, including the hypothalamus, amygdala, midbrain, and brainstem.
The CeA is critical for processing emotion, primarily in the context of fear/threat responses (Ciocchi et al., 2010; Janak and Tye, 2015; Gafford and Ressler, 2016; Fadok et al., 2018). Tac2 neurons have been identified in the CeM and CeL subdivisions of the CeA (Andero et al., 2014), where CeL neurons also coexpress Crh, Nts, and Sst (Kim et al., 2017; McCullough et al., 2018). Activation of Tac2 neurons in the CeL (using a Crh-Cre line) and the CeM both promote appetitive behaviors (Kim et al., 2017). However, differential neurochemical identity among these subdivisions could also suggest distinct inputs, outputs, and functional roles. For example, CeM Tac2 neurons promote fear memory consolidation without affecting anxiety levels or pain sensitivity (Andero et al., 2014, 2016), whereas CeL Crh neurons are anxiogenic (Pomrenze et al., 2019). In our retrograde viral tracing experiments, we found a higher density of labeled Tac2 neurons in the CeM as opposed to the CeL that may be innervating the LHA. Moreover, we found that CeATac2 neurons send projections to the BNST, midbrain, and brainstem, consistent with observations of CeATac2 projections to the PAG (Kim et al., 2017). Although understanding the functional role of CeATac2 outputs on specific target cell populations will require further investigation, work from Zelikowsky et al. (2018) showed that chemogenetic manipulations of CeATac2 neurons, along with Tac2 knockdown and overexpression experiments in the CeA, largely mirrored the effects seen in the BNST. Interestingly, another study showed that stimulation of GABAergic CeA fibers in the brainstem can result in predatory behavior, where features can be manipulated based on the stimulated region (Han et al., 2017). Therefore, CeATac2 may have distinct functional roles that are cell-type- and target-region-specific, as well as behavioral state dependent, which can modulate the valence of specific affective and motivated behaviors, including the expression of fear behaviors and threat responses.
With regard to BNST/CeATac2 projections to the midbrain, we found dense innervation of the VTA and SNc with fibers closely apposed to DA neurons. This is especially interesting as a subset of DA neurons in the VTA and SNc also express Tacr3 (Bannon and Whitty, 1995; Whitty et al., 1995; Viereckel et al., 2016), and NKB/senktide has been shown to depolarize VTA neurons (Seabrook et al., 1995; Nalivaiko et al., 1997; Werkman et al., 2011). More recently, genetically defined Tacr3-expressing VTADA neurons have been shown to densely project to the shell of the nucleus accumbens and mediate reinforcement (Heymann et al., 2020). Therefore, an intriguing possibility is that the same populations of BNST/CeATac2 neurons that project to the LHA, and make functional synaptic contacts with NK3R+ LHAMCH neurons, simultaneously innervate multiple downstream targets in series, likely including NK3R+ VTADA neurons.
Functional significance of limbic forebrain Tac2/NKB projections to the LHA, midbrain, and brainstem
Collectively, we show that BNST/CeATac2 neurons send parallel, descending projections to the LHA, as well as other key targets in the midbrain and brainstem. Our results indicate that BNST/CeATac2 neurons likely inhibit a proportion of LHAMCH neurons through fast GABA-mediated neurotransmission, but the possible role of NKB cotransmission remains unresolved. Additional studies will be required to determine the conditions under which NKB may be released in vitro and in vivo and, further, how cotransmission of an inhibitory fast neurotransmitter and an excitatory neuropeptide may sculpt postsynaptic excitability in an activity-dependent and/or behavioral state-dependent manner. Experimentally inducing neuropeptide release through optogenetic activation generally requires high-frequency stimulation but can often be technically challenging in vitro (van den Pol, 2012; Arrigoni and Saper, 2014). However, in the arcuate nucleus, high-frequency optogenetic stimulation of KNDy neurons induces a slow depolarization in postsynaptic neurons in vitro that is blocked by a Tacr3 antagonist (Qiu et al., 2016). These findings suggest that changes in firing patterns may contribute to the differential release of transmitters by BNST/CeATac2 neurons that uniquely influence behavioral output. For example, CeACRF neurons have opposing roles in anxiety, where GABA release is anxiolytic and corticotropin-releasing factor and/or dynorphin release is anxiogenic (Pomrenze et al., 2019). An interesting possibility is that cotransmission of GABA and NKB from BNST/CeATac2 neurons may mediate different behavioral states related to emotional valence. Therefore, under baseline conditions, point-to-point synaptic transmission at BSNT/CeATac2 → LHAMCH synapses is likely GABAergic, inhibitory, and seemingly nonselective among LHAMCH subpopulations. However, under conditions of elevated excitability in the central extended amygdala, perhaps associated with particular appetitive or affective states, BSNT/CeATac2 → LHAMCH synapses may be capable of releasing NKB, resulting in a countervailing slow and long-lasting depolarization specific for the Tacr3-expressing LHAMCH subpopulation (Fig. 10H). If indeed Cartpt/Tacr3-expressing LHAMCH neurons make unique projections in the brain, such as the hippocampus and cortex, as suggested by previous anatomic work (Brischoux et al., 2002; Cvetkovic et al., 2004), NKB/NK3R signaling may underlie a specific, context-dependent activation of an LHAMCH subcircuit. Moreover, NK3R activation in LHAMCH neurons may promote the release of MCH and modulate postsynaptic MCH1R neurons, which have also been implicated in stress- and anxiety-related behaviors (Chaki et al., 2005, 2015).
In summary, we have identified genetically defined cell populations in the CeA and BNST as prominent, putative sources of NKB to the LHA and specifically provide monosynaptic GABAergic inputs that converge onto LHAMCH neurons. These results are consistent with the close anatomic and functional interrelationship between the CeA and BNST (Shackman and Fox, 2016) and furthers our understanding of how a limbic forebrain tachykininergic projection system may coordinate and engage multiple behaviorally important downstream targets. Furthermore, these results provide a foundation for deciphering how tachykininergic/GABAergic inputs may differentially modulate LHAMCH subcircuits, perhaps accounting for the diversity of physiological functions and behaviors associated with the LHAMCH system. Additional work will be required to dissect the cellular mechanisms underlying NKB/GABA cotransmission, possible sexual dimorphism in the anatomy and function of these circuits, and their roles in modulating arousal and affective and motivated behavior during different behavioral states.
Footnotes
This work was supported by the National Institutes of Health Grant R01MH112739 (to A.C.J.) and R01DK118402 (to S.E.K.), National Institutes of Health Shared Instrumentation Grant S10OD016435 (to Akiko Nishiyama) for imaging support, and the Connecticut Institute for the Brain and Cognitive Sciences graduate and undergraduate fellowships (to A.F. and L.Z., respectively). We thank all members of the Jackson lab for support, assistance, and discussions; Adriane Mosley for genotyping support; Chris O'Connell for imaging support; and Bruce Bean and Melissa Chee for discussions.
The authors declare no competing financial interests.
References
- Andero R, Dias BG, Ressler KJ (2014) A role for Tac2, NkB, and Nk3 receptor in normal and dysregulated fear memory consolidation. Neuron 83:444–454. 10.1016/j.neuron.2014.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andero R, Daniel S, Guo JD, Bruner RC, Seth S, Marvar PJ, Rainnie D, Ressler KJ (2016) Amygdala-dependent molecular mechanisms of the Tac2 pathway in fear learning. Neuropsychopharmacology 41:2714–2722. 10.1038/npp.2016.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apergis-Schoute J, Iordanidou P, Faure C, Jego S, Schone C, Aitta-Aho T, Adamantidis A, Burdakov D (2015) Optogenetic evidence for inhibitory signaling from orexin to MCH neurons via local microcircuits. J Neurosci 35:5435–5441. 10.1523/JNEUROSCI.5269-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrigoni E, Saper CB (2014) What optogenetic stimulation is telling us (and failing to tell us) about fast neurotransmitters and neuromodulators in brain circuits for wake-sleep regulation. Curr Opin Neurobiol 29:165–171. 10.1016/j.conb.2014.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrigoni E, Chee MJS, Fuller PM (2019) To eat or to sleep: that is a lateral hypothalamic question. Neuropharmacology 154:34–49. 10.1016/j.neuropharm.2018.11.017 [DOI] [PubMed] [Google Scholar]
- Bandaru SS, Khanday MA, Ibrahim N, Naganuma F, Vetrivelan R (2020) Sleep-wake control by melanin-concentrating hormone (MCH) neurons: a review of recent findings. Curr Neurol Neurosci Rep 20:55. 10.1007/s11910-020-01075-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannon MJ, Whitty CJ (1995) Neurokinin receptor gene expression in substantia nigra: localization, regulation, and potential physiological significance. Can J Physiol Pharmacol 73:866–870. 10.1139/y95-119 [DOI] [PubMed] [Google Scholar]
- Barbier M, Fellmann D, Risold PY (2018a) Morphofunctional organization of the connections from the medial and intermediate parts of the central nucleus of the amygdala into distinct divisions of the lateral hypothalamic area in the rat. Front Neurol 9:688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbier M, Fellmann D, Risold PY (2018b) Characterization of McDonald's intermediate part of the central nucleus of the amygdala in the rat. J Comp Neurol 526:2165–2186. 10.1002/cne.24470 [DOI] [PubMed] [Google Scholar]
- Barbier M, González JA, Houdayer C, Burdakov D, Risold PY, Croizier S (2021) Projections from the dorsomedial division of the bed nucleus of the stria terminalis to hypothalamic nuclei in the mouse. J Comp Neurol 529:929–956. 10.1002/cne.24988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barone FC, Wayner MJ, Scharoun SL, Guevara-Aguilar R, Aguilar-Baturoni HU (1981) Afferent connections to the lateral hypothalamus: a horseradish peroxidase study in the rat. Brain Res Bull 7:75–88. 10.1016/0361-9230(81)90101-5 [DOI] [PubMed] [Google Scholar]
- Barson JR, Morganstern I, Leibowitz SF (2013) Complementary roles of orexin and melanin-concentrating hormone in feeding behavior. Int J Endocrinol 2013:983964. 10.1155/2013/983964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittencourt JC, Presse F, Arias C, Peto C, Vaughan J, Nahon JL, Vale W, Sawchenko PE (1992) The melanin-concentrating hormone system of the rat brain: an immuno- and hybridization histochemical characterization. J Comp Neurol 319:218–245. 10.1002/cne.903190204 [DOI] [PubMed] [Google Scholar]
- Blanco-Centurion C, Liu M, Konadhode RP, Zhang X, Pelluru D, van den Pol AN, Shiromani PJ (2016) Optogenetic activation of melanin-concentrating hormone neurons increases non-rapid eye movement and rapid eye movement sleep during the night in rats. Eur J Neurosci 44:2846–2857. 10.1111/ejn.13410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Centurion C, Luo S, Spergel DJ, Vidal-Ortiz A, Oprisan SA, Van Den Pol AN, Liu M, Shiromani PJ (2019) Dynamic network activation of hypothalamic MCH neurons in rem sleep and exploratory behavior. J Neurosci 39:4986–4998. 10.1523/JNEUROSCI.0305-19.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnavion P, Mickelsen LE, Fujita A, de Lecea L, Jackson AC (2016) Hubs and spokes of the lateral hypothalamus: cell types, circuits and behaviour. J Physiol 594:6443–7257. 10.1113/JP271946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brischoux F, Fellmann D, Risold PY (2001) Ontogenetic development of the diencephalic MCH neurons: a hypothalamic “MCH area” hypothesis. Eur J Neurosci 13:1733–1744. 10.1046/j.0953-816x.2001.01552.x [DOI] [PubMed] [Google Scholar]
- Brischoux F, Cvetkovic V, Griffond B, Fellmann D, Risold PY (2002) Time of genesis determines projection and neurokinin-3 expression patterns of diencephalic neurons containing melanin-concentrating hormone. Eur J Neurosci 16:1672–1680. 10.1046/j.1460-9568.2002.02229.x [DOI] [PubMed] [Google Scholar]
- Broberger C (1999) Hypothalamic cocaine- and amphetamine-regulated transcript (CART) neurons: histochemical relationship to thyrotropin-releasing hormone, melanin-concentrating hormone, orexin/hypocretin and neuropeptide Y. Brain Res 848:101–113. 10.1016/s0006-8993(99)01977-0 [DOI] [PubMed] [Google Scholar]
- Brown JA, Woodworth HL, Leinninger GM (2015) To ingest or rest? Specialized roles of lateral hypothalamic area neurons in coordinating energy balance. Front Syst Neurosci 9:9–25. 10.3389/fnsys.2015.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdakov D, Gerasimenko O, Verkhratsky A (2005) Physiological changes in glucose differentially modulate the excitability of hypothalamic melanin-concentrating hormone and orexin neurons in situ. J Neurosci 25:2429–2433. 10.1523/JNEUROSCI.4925-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaki S, Funakoshi T, Hirota-Okuno S, Nishiguchi M, Shimazaki T, Iijima M, Grottick AJ, Kanuma K, Omodera K, Sekiguchi Y, Okuyama S, Tran TA, Semple G, Thomsen W (2005) Anxiolytic- and antidepressant-like profile of ATC0065 and ATC0175: nonpeptidic and orally active melanin-concentrating hormone receptor 1 antagonists. J Pharmacol Exp Ther 313:831–839. 10.1124/jpet.104.081711 [DOI] [PubMed] [Google Scholar]
- Chaki S, Shimazaki T, Nishiguchi M, Funakoshi T, Iijima M, Ito A, Kanuma K, Sekiguchi Y (2015) Antidepressant/anxiolytic potential and adverse effect liabilities of melanin-concentrating hormone receptor 1 antagonists in animal models. Pharmacol Biochem Behav 135:154–168. 10.1016/j.pbb.2015.05.018 [DOI] [PubMed] [Google Scholar]
- Chee MJS, Arrigoni E, Maratos-Flier E (2015) Melanin-concentrating hormone neurons release glutamate for feedforward inhibition of the lateral septum. J Neurosci 35:3644–3651. 10.1523/JNEUROSCI.4187-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ch'ng S, Fu J, Brown RM, McDougall SJ, Lawrence AJ (2018) The intersection of stress and reward: BNST modulation of aversive and appetitive states. Prog Neuropsychopharmacol Biol Psychiatry 87:108–125. 10.1016/j.pnpbp.2018.01.005 [DOI] [PubMed] [Google Scholar]
- Ciocchi S, Herry C, Grenier F, Wolff SBE, Letzkus JJ, Vlachos I, Ehrlich I, Sprengel R, Deisseroth K, Stadler MB, Müller C, Lüthi A (2010) Encoding of conditioned fear in central amygdala inhibitory circuits. Nature 468:277–282. 10.1038/nature09559 [DOI] [PubMed] [Google Scholar]
- Concetti C, Burdakov D (2021) Orexin/hypocretin and MCH neurons: cognitive and motor roles beyond arousal. Front Neurosci 15:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croizier S, Franchi-Bernard G, Colard C, Poncet F, la Roche A, Risold PY (2010) A comparative analysis shows morphofunctional differences between the rat and mouse melanin-concentrating hormone systems. PLoS One 5:e15471. 10.1371/journal.pone.0015471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croizier S, Cardot J, Brischoux F, Fellmann D, Griffond B, Risold PY (2013) The vertebrate diencephalic MCH system: a versatile neuronal population in an evolving brain. Front Neuroendocrinol 34:65–87. 10.1016/j.yfrne.2012.10.001 [DOI] [PubMed] [Google Scholar]
- Cvetkovic V, Poncet F, Fellmann D, Griffond B, Risold PY (2003) Diencephalic neurons producing melanin-concentrating hormone are influenced by local and multiple extra-hypothalamic tachykininergic projections through the neurokinin 3 receptor. Neuroscience 119:1113–1145. [DOI] [PubMed] [Google Scholar]
- Cvetkovic V, Brischoux F, Jacquemard C, Fellmann D, Griffond B, Risold PY (2004) Characterization of subpopulations of neurons producing melanin-concentrating hormone in the rat ventral diencephalon. J Neurochem 91:911–919. 10.1111/j.1471-4159.2004.02776.x [DOI] [PubMed] [Google Scholar]
- Daniel SE, Rainnie DG (2016) Stress modulation of opposing circuits in the bed nucleus of the stria terminalis. Neuropsychopharmacology 41:103–125. 10.1038/npp.2015.178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M, Walker DL, Miles L, Grillon C (2010) Phasic vs sustained fear in rats and humans: role of the extended amygdala in fear vs anxiety. Neuropsychopharmacology 35:105–135. 10.1038/npp.2009.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilsiz P, Aklan I, Sayar Atasoy N, Yavuz Y, Filiz G, Koksalar F, Ates T, Oncul M, Coban I, Ates Oz E, Cebecioglu U, Alp MI, Yilmaz B, Atasoy D (2020) MCH neuron activity is sufficient for reward and reinforces feeding. Neuroendocrinology 110:258–270. 10.1159/000501234 [DOI] [PubMed] [Google Scholar]
- Diniz GB, Bittencourt JC (2017) The melanin-concentrating hormone as an integrative peptide driving motivated behaviors. Front Syst Neurosci 11:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingos AI, Sordillo A, Dietrich MO, Liu ZW, Tellez L. a, Vaynshteyn J, Ferreira JG, Ekstrand MI, Horvath TL, de Araujo IE, Friedman JM (2013) Hypothalamic melanin concentrating hormone neurons communicate the nutrient value of sugar. Elife 2:e01462. 10.7554/eLife.01462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong HW, Swanson LW (2003) Projections from the rhomboid nucleus of the bed nuclei of the stria terminalis: implications for cerebral hemisphere regulation of ingestive behaviors. J Comp Neurol 463:434–472. 10.1002/cne.10758 [DOI] [PubMed] [Google Scholar]
- Dong HW, Swanson LW (2004a) Projections from bed nuclei of the stria terminalis, posterior division: implications for cerebral hemisphere regulation of defensive and reproductive behaviors. J Comp Neurol 471:396–433. 10.1002/cne.20002 [DOI] [PubMed] [Google Scholar]
- Dong HW, Swanson LW (2004b) Organization of axonal projections from the anterolateral area of the bed nuclei of the stria terminalis. J Comp Neurol 468:277–298. 10.1002/cne.10949 [DOI] [PubMed] [Google Scholar]
- Dong H-W, Swanson LW (2006) Projections from bed nuclei of the stria terminalis, dorsomedial nucleus: implications for cerebral hemisphere integration of neuroendocrine, autonomic, and drinking responses. J Comp Neurol 494:75–107. 10.1002/cne.20790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong HW, Petrovich GD, Swanson LW (2000) Organization of projections from the juxtacapsular nucleus of the BST: a PHAL study in the rat. Brain Res 859:1–14. 10.1016/S0006-8993(99)02246-5 [DOI] [PubMed] [Google Scholar]
- Duvarci S, Pare D (2014) Amygdala microcircuits controlling learned fear. Neuron 82:966–980. 10.1016/j.neuron.2014.04.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekstrand MI, Nectow AR, Knight ZA, Latcha KN, Pomeranz LE, Friedman JM (2014) Molecular profiling of neurons based on connectivity. Cell 157:1230–1242. 10.1016/j.cell.2014.03.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias CF, Lee CE, Kelly JF, Ahima RS, Kuhar M, Saper CB, Elmquist JK (2001) Characterization of CART neurons in the rat and human hypothalamus. J Comp Neurol 432:1–19. 10.1002/cne.1085 [DOI] [PubMed] [Google Scholar]
- Fadok JP, Markovic M, Tovote P, Lüthi A (2018) New perspectives on central amygdala function. Curr Opin Neurobiol 49:141–147. 10.1016/j.conb.2018.02.009 [DOI] [PubMed] [Google Scholar]
- Ferreira JGP, Bittencourt JC, Adamantidis A (2017) Melanin-concentrating hormone and sleep. Curr Opin Neurobiol 44:152–158. 10.1016/j.conb.2017.04.008 [DOI] [PubMed] [Google Scholar]
- Florido A, Velasco ER, Soto-Faguás CM, Gomez-Gomez A, Perez-Caballero L, Molina P, Nadal R, Pozo OJ, Saura CA, Andero R (2021) Sex differences in fear memory consolidation via Tac2 signaling in mice. Nat Commun 12: 10.1038/s41467-021-22911-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AS, Shackman AJ (2019) The central extended amygdala in fear and anxiety: closing the gap between mechanistic and neuroimaging research. Neurosci Lett 693:58–67. 10.1016/j.neulet.2017.11.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafford GM, Ressler KJ (2016) Mouse models of fear-related disorders: cell-type-specific manipulations in amygdala. Neuroscience 321:108–120. 10.1016/j.neuroscience.2015.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardino WJ, Eban-Rothschild A, Christoffel DJ, Li SB, Malenka RC, de Lecea L (2018) Parallel circuits from the bed nuclei of stria terminalis to the lateral hypothalamus drive opposing emotional states. Nat Neurosci 21:1084–1095. 10.1038/s41593-018-0198-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardino WJ, Pomrenze MB (2021) Extended amygdala neuropeptide circuitry of emotional arousal: waking up on the wrong side of the bed nuclei of stria terminalis. Front Behav Neurosci 15:613025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González JA, Iordanidou P, Strom M, Adamantidis A, Burdakov D (2016) Awake dynamics and brain-wide direct inputs of hypothalamic MCH and orexin networks. Nat Commun 7:11395. 10.1038/ncomms11395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffond B, Ciofi P, Bayer L, Jacquemard C, Fellmann D (1997) Immunocytochemical detection of the neurokinin B receptor (NK3) on melanin-concentrating hormone (MCH) neurons in rat brain. J Chem Neuroanat 12:183–189. 10.1016/S0891-0618(96)00200-1 [DOI] [PubMed] [Google Scholar]
- Han W, Tellez LA, Rangel MJ, Motta SC, Zhang X, Perez IO, Canteras NS, Shammah-Lagnado SJ, van den Pol AN, De Araujo IE (2017) Integrated control of predatory hunting by the central nucleus of the amygdala. Cell 168:311–324.e18. 10.1016/j.cell.2016.12.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanriot L, Camargo N, Courau AC, Leger L, Luppi PH, Peyron C (2007) Characterization of the melanin-concentrating hormone neurons activated during paradoxical sleep hypersomnia in rats. J Comp Neurol 505:147–157. [DOI] [PubMed] [Google Scholar]
- Harris JA, Hirokawa KE, Sorensen SA, Gu H, Mills M, Ng LL, Bohn P, Mortrud M, Ouellette B, Kidney J, Smith KA, Dang C, Sunkin S, Bernard A, Oh SW, Madisen L, Zeng H (2014) Anatomical characterization of Cre driver mice for neural circuit mapping and manipulation. Front Neural Circuits 8:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harthoorn LF, Sañé A, Nethe M, Van Heerikhuize JJ (2005) Multi-transcriptional profiling of melanin-concentrating hormone and orexin-containing neurons. Cell Mol Neurobiol 25:1209–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassani OK, Lee MG, Jones BE (2009) Melanin-concentrating hormone neurons discharge in a reciprocal manner to orexin neurons across the sleep-wake cycle. Proc Natl Acad Sci U S A 106:2418–2422. 10.1073/pnas.0811400106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymann G, Jo YS, Reichard KL, McFarland N, Chavkin C, Palmiter RD, Soden ME, Zweifel LS (2020) Synergy of distinct dopamine projection populations in behavioral reinforcement. Neuron 105:909–920.e5. 10.1016/j.neuron.2019.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoya Y, Matsushita M (1980) Cells of origin of the descending afferents to the lateral hypothalamic area in the rat, studied with the horseradish peroxidase method. Neurosci Lett 18:231–236. 10.1016/0304-3940(80)90290-6 [DOI] [PubMed] [Google Scholar]
- Izawa S, Chowdhury S, Miyazaki T, Mukai Y, Ono D, Inoue R, Ohmura Y, Mizoguchi H, Kimura K, Yoshioka M, Terao A, Kilduff TS, Yamanaka A (2019) REM sleep-active MCH neurons are involved in forgetting hippocampusdependent memories. Science 365:1308–1313. 10.1126/science.aax9238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janak PH, Tye KM (2015) From circuits to behaviour in the amygdala. Nature 517:284–292. 10.1038/nature14188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jego S, Glasgow SD, Herrera CG, Ekstrand M, Reed SJ, Boyce R, Friedman J, Burdakov D, Adamantidis AR (2013) Optogenetic identification of a rapid eye movement sleep modulatory circuit in the hypothalamus. Nat Neurosci 16:1637–1643. 10.1038/nn.3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings JH, Rizzi G, Stamatakis AM, Ung RL, Stuber GD, Stmatakis AM, Ung RL, Stuber GD, Stamatakis AM, Ung RL, Stuber GD (2013a) The inhibitory circuit architecture of the lateral hypothalamus orchestrates feeding. Science 341:1517–1521. 10.1126/science.1241812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings JHJ, Sparta DDR, Stamatakis AAM, Ung RL, Pleil KE, Kash TL, Stuber GD (2013b) Distinct extended amygdala circuits for divergent motivational states. Nature 496:224–228. 10.1038/nature12041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Gallet S, Klemm P, Scholl P, Folz-Donahue K, Altmüller J, Alber J, Heilinger C, Kukat C, Loyens A, Müller-Fielitz H, Sundaram S, Schwaninger M, Prevot V, Brüning JC (2020) MCH neurons regulate permeability of the median eminence barrier. Neuron 107:306–319.e9. 10.1016/j.neuron.2020.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kash TL, Pleil KE, Marcinkiewcz CA, Lowery-Gionta EG, Crowley N, Mazzone C, Sugam JA, Hardaway J, McElligott ZA (2015) Neuropeptide regulation of signaling and behavior in the BNST. Mol Cells 38:1–13. 10.14348/molcells.2015.2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy AR, Todd JF, Dhillo WS, Seal LJ, Ghatei MA, O'Toole CP, Jones M, Witty D, Winborne K, Riley G, Hervieu G, Wilson S, Bloom SR (2003) Effect of direct injection of melanin-concentrating hormone into the paraventricular nucleus: further evidence for a stimulatory role in the adrenal axis via SLC-1. J Neuroendocrinol 15:268–272. 10.1046/j.1365-2826.2003.00997.x [DOI] [PubMed] [Google Scholar]
- Kim DW, Washington PW, Wang ZQ, Lin SH, Sun C, Ismail BT, Wang H, Jiang L, Blackshaw S (2020) The cellular and molecular landscape of hypothalamic patterning and differentiation from embryonic to late postnatal development. Nat Commun : 11: 10.1038/s41467-020-18231-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Zhang X, Muralidhar S, LeBlanc SA, Tonegawa S (2017) Basolateral to central amygdala neural circuits for appetitive behaviors. Neuron 93:1464–1479.e5. 10.1016/j.neuron.2017.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S-YY, Adhikari A, Lee SY, Marshel JH, Kim CK, Mallory CS, Lo M, Pak S, Mattis J, Lim BK, Malenka RC, Warden MR, Neve R, Tye KM, Deisseroth K (2013) Diverging neural pathways assemble a behavioural state from separable features in anxiety. Nature 496:219–223. 10.1038/nature12018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight ZAA, Tan K, Birsoy K, Schmidt S, Garrison JLL, Wysocki RWW, Emiliano A, Ekstrand MII, Friedman JMM (2012) Molecular profiling of activated neurons by phosphorylated ribosome capture. Cell 151:1126–1137. 10.1016/j.cell.2012.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodani S, Soya S, Sakurai T (2017) Excitation of GABAergic Neurons in the Bed Nucleus of the Stria Terminalis Triggers Immediate Transition from Non-Rapid Eye Movement Sleep to Wakefulness in Mice. J Neurosci 37:7164–7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konadhode RR, Pelluru D, Blanco-Centurion C, Zayachkivsky A, Liu M, Uhde T, Glen WB, van den Pol AN, Mulholland PJ, Shiromani PJ (2013) Optogenetic stimulation of MCH neurons increases sleep. J Neurosci 33:10257–10263. 10.1523/JNEUROSCI.1225-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong D, Vong L, Parton LE, Ye C, Tong Q, Hu X, Choi B, Brüning JC, Lowell BB (2010) Glucose stimulation of hypothalamic MCH neurons involves KATP channels, is modulated by UCP2, and regulates peripheral glucose homeostasis. Cell Metab 12:545–552. 10.1016/j.cmet.2010.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosse C, Burdakov D (2019) Natural hypothalamic circuit dynamics underlying object memorization. Nat Commun 10:8. 10.1038/s41467-019-10484-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo T, Uchigashima M, Miyazaki T, Konno K, Yamasaki M, Yanagawa Y, Minami M, Watanabe M (2012) Three types of neurochemical projection from the bed nucleus of the stria terminalis to the ventral tegmental area in adult mice. J Neurosci 32:18035–18046. 10.1523/JNEUROSCI.4057-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebow MA, Chen A (2016) Overshadowed by the amygdala: the bed nucleus of the stria terminalis emerges as key to psychiatric disorders. Mol Psychiatry 21:450–463. 10.1038/mp.2016.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen L, Chen T-M, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, et al. (2007) Genome-wide atlas of gene expression in the adult mouse brain. Nature 445:168–176. 10.1038/nature05453 [DOI] [PubMed] [Google Scholar]
- Linehan V, Hirasawa M (2018) Electrophysiological properties of melanin-concentrating hormone and orexin neurons in adolescent rats. Front Cell Neurosci 12:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luskin AT, Bhatti DL, Mulvey B, Pedersen CE, Girven KS, Oden-Brunson H, Kimbell K, Blackburn T, Sawyer A, Gereau RW, Dougherty JD, Bruchas MR (2021) Extended amygdala-parabrachial circuits alter threat assessment and regulate feeding. Sci Adv 7:eabd3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H (2010) A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13:133–140. 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marksteiner J, Sperk G, Krause JE (1992) Distribution of neurons expressing neurokinin B in the rat brain: immunohistochemistry and in situ hybridization. J Comp Neurol 317:341–356. 10.1002/cne.903170403 [DOI] [PubMed] [Google Scholar]
- Marvar PJ, Andero R, Hurlemann R, Lago TR, Zelikowsky M, Dabrowska J (2021) Limbic neuropeptidergic modulators of emotion and their therapeutic potential for anxiety and post-traumatic stress disorder. J Neurosci 41:901–910. 10.1523/JNEUROSCI.1647-20.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzone CM, Pati D, Michaelides M, DiBerto J, Fox JH, Tipton G, Anderson C, Duffy K, McKlveen JM, Hardaway JA, Magness ST, Falls WA, Hammack SE, McElligott ZA, Hurd YL, Kash TL (2018) Acute engagement of Gq-mediated signaling in the bed nucleus of the stria terminalis induces anxiety-like behavior. Mol Psychiatry 23:143–153. 10.1038/mp.2016.218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough KM, Morrison FG, Hartmann J, Carlezon WA, Ressler KJ (2018) Quantified coexpression analysis of central amygdala subpopulations. eNeuro 5:ENEURO.0010-18.2018. 10.1523/ENEURO.0010-18.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickelsen LE, Kolling FW, Chimileski BR, Fujita A, Norris C, Chen K, Nelson CE, Jackson AC (2017) Neurochemical heterogeneity among lateral hypothalamic hypocretin/orexin and melanin-concentrating hormone neurons identified through single cell gene expression analysis. eNeuro 4:ENEURO.0013-17.2017. 10.1523/ENEURO.0013-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickelsen LE, Bolisetty M, Chimileski BR, Fujita A, Beltrami EJ, Costanzo JT, Naparstek JR, Robson P, Jackson AC (2019) Single-cell transcriptomic analysis of the lateral hypothalamic area reveals molecularly distinct populations of inhibitory and excitatory neurons. Nat Neurosci 22:642–656. 10.1038/s41593-019-0349-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monti JM, Torterolo P, Lagos P (2013) Melanin-concentrating hormone control of sleep-wake behavior. Sleep Med Rev 17:293–298. 10.1016/j.smrv.2012.10.002 [DOI] [PubMed] [Google Scholar]
- Nakamura S, Tsumori T, Yokota S, Oka T, Yasui Y (2009) Amygdaloid axons innervate melanin-concentrating hormone- and orexin-containing neurons in the mouse lateral hypothalamus. Brain Res 1278:66–74. 10.1016/j.brainres.2009.04.049 [DOI] [PubMed] [Google Scholar]
- Nalivaiko E, Michaud JC, Soubrié P, Le Fur G, Feltz P (1997) Tachykinin neurokinin-1 and neurokinin-3 receptor-mediated responses in guinea-pig substantia nigra: an in vitro electrophysiological study. Neuroscience 78:745–757. 10.1016/S0306-4522(96)00625-2 [DOI] [PubMed] [Google Scholar]
- Nectow AR, Moya MV, Ekstrand MI, Mousa A, McGuire KL, Sferrazza CE, Field BC, Rabinowitz GS, Sawicka K, Liang Y, Friedman JM, Heintz N, Schmidt EF (2017) Rapid molecular profiling of defined cell types using viral TRAP. Cell Rep 19:655–667. 10.1016/j.celrep.2017.03.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble EE, Hahn JD, Konanur VR, Hsu TM, Page SJ, Cortella AM, Liu CM, Song MY, Suarez AN, Szujewski CC, Rider D, Clarke JE, Darvas M, Appleyard SM, Kanoski SE (2018) Control of feeding behavior by cerebral ventricular volume transmission of melanin-concentrating hormone. Cell Metab 28:55–68.e7. 10.1016/j.cmet.2018.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble EE, Wang Z, Liu CM, Davis EA, Suarez AN, Stein LM, Tsan L, Terrill SJ, Hsu TM, Jung AH, Raycraft LM, Hahn JD, Darvas M, Cortella AM, Schier LA, Johnson AW, Hayes MR, Holschneider DP, Kanoski SE (2019) Hypothalamus-hippocampus circuitry regulates impulsivity via melanin-concentrating hormone. Nat Commun 10:16. 10.1038/s41467-019-12895-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Franklin K, (2012) Paxinos and Franklin's the mouse brain in stereotaxic coordinates. Amsterdam: Elsevier. [Google Scholar]
- Pissios P, Bradley RL, Maratos-Flier E (2006) Expanding the scales: the multiple roles of MCH in regulating energy balance and other biological functions. Endocr Rev 27:606–620. 10.1210/er.2006-0021 [DOI] [PubMed] [Google Scholar]
- Pomrenze MB, Giovanetti SM, Maiya R, Gordon AG, Kreeger LJ, Messing RO (2019) Dissecting the roles of GABA and neuropeptides from rat central amygdala CRF neurons in anxiety and fear learning. Cell Rep 29:13–21.e4. 10.1016/j.celrep.2019.08.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Nestor CC, Zhang C, Padilla SL, Palmiter RD, Kelly MJ, Rønnekleiv OK (2016) High-frequency stimulation-induced peptide release synchronizes arcuate kisspeptin neurons and excites GnRH neurons. Elife 5:e16246. 10.7554/eLife.16246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu D, Ludwig DS, Gammeltoft S, Piper M, Pelleymounter MA, Cullen MJ, Mathes WF, Przypek J, Kanarek R, Maratos-Flier E (1996) A role for melanin-concentrating hormone in the central regulation of feeding behaviour. Nature 380:243–247. 10.1038/380243a0 [DOI] [PubMed] [Google Scholar]
- Qualls-Creekmore E, Münzberg H (2018) Modulation of feeding and associated behaviors by lateral hypothalamic circuits. Endocrinology 159:3631–3642. 10.1210/en.2018-00449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rance NE, Dacks PA, Mittelman-Smith MA, Romanovsky AA, Krajewski-Hall SJ (2013) Modulation of body temperature and LH secretion by hypothalamic KNDy (kisspeptin, neurokinin B and dynorphin) neurons: A novel hypothesis on the mechanism of hot flushes. Front Neuroendocrinol 34:211–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rance NE, Krajewski SJ, Smith MA, Cholanian M, Dacks PA (2010) Neurokinin B and the hypothalamic regulation of reproduction. Brain Res 1364:116–128. 10.1016/j.brainres.2010.08.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi M, Choi SJ, O'Shea D, Miyoshi T, Ghatei MA, Bloom SR (1997) Melanin-concentrating hormone acutely stimulates feeding, but chronic administration has no effect on body weight. Endocrinology 138:351–355. 10.1210/endo.138.1.4887 [DOI] [PubMed] [Google Scholar]
- Seabrook GR, Bowery BJ, Hill RG (1995) Pharmacology of tachykinin receptors on neurones in the ventral tegmental area of rat brain slices. Eur J Pharmacol 273:113–119. 10.1016/0014-2999(94)00681-v [DOI] [PubMed] [Google Scholar]
- Shaam Al Abed A, Reynolds NJ, Dehorter N (2021) A second wave for the neurokinin Tac2 pathway in brain research. Biol Psychiatry 90:156–164. [DOI] [PubMed] [Google Scholar]
- Shackman AJ, Fox AS (2016) Contributions of the central extended amygdala to fear and anxiety. J Neurosci 36:8050–8063. 10.1523/JNEUROSCI.0982-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigemoto R, Yokota Y, Tsuchida K, Nakanishi S (1990) Cloning and expression of a rat neuromedin K receptor cDNA. J Biol Chem 265:623–628. [PubMed] [Google Scholar]
- Shimada M, Tritos NA, Lowell BB, Flier JS, Maratos-Flier E (1998) Mice lacking melanin-concentrating hormone are hypophagic and lean. Nature 396:670–679. 10.1038/25341 [DOI] [PubMed] [Google Scholar]
- Smith DG, Davis RJ, Rorick-Kehn L, Morin M, Witkin JM, McKinzie DL, Nomikos GG, Gehlert DR (2006) Melanin-concentrating hormone-1 receptor modulates neuroendocrine, behavioral, and corticolimbic neurochemical stress responses in mice. Neuropsychopharmacology 31:1135–1145. 10.1038/sj.npp.1300913 [DOI] [PubMed] [Google Scholar]
- Steinhoff MS, von Mentzer B, Geppetti P, Pothoulakis C, Bunnett NW (2014) Tachykinins and their receptors: contributions to physiological control and the mechanisms of disease. Physiol Rev 94:265–301. 10.1152/physrev.00031.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuber GD, Wise RA (2016) Lateral hypothalamic circuits for feeding and reward. Nat Neurosci 19:198–205. 10.1038/nn.4220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N, Cassell MD (1993) Intrinsic GABAergic neurons in the rat central extended amygdala. J Comp Neurol 330:381–404. 10.1002/cne.903300308 [DOI] [PubMed] [Google Scholar]
- Swanson LW, Petrovich GD (1998) What is the amygdala? Trends Neurosci 21:323–331. 10.1016/s0166-2236(98)01265-x [DOI] [PubMed] [Google Scholar]
- Tervo DGR, Hwang BY, Viswanathan S, Gaj T, Lavzin M, Ritola KD, Lindo S, Michael S, Kuleshova E, Ojala D, Huang CC, Gerfen CR, Schiller J, Dudman JT, Hantman AW, Looger LL, Schaffer DV, Karpova AY (2016) A designer AAV variant permits efficient retrograde access to projection neurons. Neuron 92:372–382. 10.1016/j.neuron.2016.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovote P, Fadok JP, Lüthi A (2015) Neuronal circuits for fear and anxiety. Nat Rev Neurosci 16:317–331. 10.1038/nrn3945 [DOI] [PubMed] [Google Scholar]
- Tsunematsu T, Ueno T, Tabuchi S, Inutsuka A, Tanaka KF, Hasuwa H, Kilduff TS, Terao A, Yamanaka A (2014) Optogenetic manipulation of activity and temporally controlled cell-specific ablation reveal a role for MCH neurons in sleep/wake regulation. J Neurosci 34:6896–6909. 10.1523/JNEUROSCI.5344-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Pol AN (2012) Neuropeptide transmission in brain circuits. Neuron 76:98–115. 10.1016/j.neuron.2012.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Pol AN, Acuna-Goycolea C, Clark KR, Ghosh PK (2004) Physiological properties of hypothalamic MCH neurons identified with selective expression of reporter gene after recombinant virus infection. Neuron 42:635–652. 10.1016/s0896-6273(04)00251-x [DOI] [PubMed] [Google Scholar]
- Verret L, Goutagny R, Fort P, Cagnon L, Salvert D, Léger L, Boissard R, Salin P, Peyron C, Luppi PH (2003) A role of melanin-concentrating hormone producing neurons in the central regulation of paradoxical sleep. BMC Neurosci 4:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetrivelan R, Kong D, Ferrari LL, Arrigoni E, Madara JC, Bandaru SS, Lowell BB, Lu J, Saper CB (2016) Melanin-concentrating hormone neurons specifically promote rapid eye movement sleep in mice. Neuroscience 336:102–113. 10.1016/j.neuroscience.2016.08.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viereckel T, Dumas S, Smith-Anttila CJA, Vlcek B, Bimpisidis Z, Lagerström MC, Konradsson-Geuken A, Wallén-Mackenzie A (2016) Midbrain gene screening identifies a new mesoaccumbal glutamatergic pathway and a marker for dopamine cells neuroprotected in Parkinson's disease. Sci Rep 6: 10.1038/srep35203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrang N, Larsen PJ, Clausen JT, Kristensen P (1999) Neurochemical characterization of hypothalamic cocaine- amphetamine-regulated transcript neurons. J Neurosci 19:RC5–RC5. 10.1523/JNEUROSCI.19-10-j0006.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vranjkovic O, Pina M, Kash TL, Winder DG (2017) The bed nucleus of the stria terminalis in drug-associated behavior and affect: a circuit-based perspective. Neuropharmacology 122:100–106. 10.1016/j.neuropharm.2017.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, Wu X, Vo HT, Ma XJ, Luo Y (2012) RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn 14:22–29. 10.1016/j.jmoldx.2011.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JH (1963) Hierarchical grouping to optimize an objective function. J Am Stat Assoc 58:236–244. 10.1080/01621459.1963.10500845 [DOI] [Google Scholar]
- Warden MK, Young WS (1988) Distribution of cells containing mRNAs encoding substance P and neurokinin B in the rat central nervous system. J Comp Neurol 272:90–113. 10.1002/cne.902720107 [DOI] [PubMed] [Google Scholar]
- Weera MM, Shackett RS, Kramer HM, Middleton JW, Gilpin NW (2021) Central amygdala projections to lateral hypothalamus mediate avoidance behavior in rats. J Neurosci 41:61–72. 10.1523/JNEUROSCI.0236-20.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JD, Kozareva V, Ferreira A, Vanderburg C, Martin C, Macosko EZ, Welch JD, Kozareva V, Ferreira A, Vanderburg C, Martin C, Macosko EZ (2019) Single-cell multi-omic integration compares and contrasts features of brain cell identity. Brain Cell Identity 177:1873–1887.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werkman TR, Mccreary AC, Kruse CG, Wadman WJ (2011) NK3 receptors mediate an increase in firing rate of midbrain dopamine neurons of the rat and the guinea pig. Synapse 65:814–826. 10.1002/syn.20908 [DOI] [PubMed] [Google Scholar]
- Whitty CJ, Walker PD, Goebel DJ, Poosch MS, Bannon MJ (1995) Quantitation, cellular localization and regulation of neurokinin receptor gene expression within the rat substantia nigra. Neuroscience 64:419–425. 10.1016/0306-4522(94)00373-d [DOI] [PubMed] [Google Scholar]
- Wolf FA, Angerer P, Theis FJ (2018) SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 19: 10.1186/s13059-017-1382-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita T, Yamanaka A (2017) Lateral hypothalamic circuits for sleep–wake control. Curr Opin Neurobiol 44:94–100. 10.1016/j.conb.2017.03.020 [DOI] [PubMed] [Google Scholar]
- Yamauchi N, Takahashi D, Sugimura YK, Kato F, Amano T, Minami M (2018) Activation of the neural pathway from the dorsolateral bed nucleus of the stria terminalis to the central amygdala induces anxiety-like behaviors. Eur J Neurosci 48:3052–3061. 10.1111/ejn.14165 [DOI] [PubMed] [Google Scholar]
- Ye J, Veinante P (2019) Cell-type specific parallel circuits in the bed nucleus of the stria terminalis and the central nucleus of the amygdala of the mouse. Brain Struct Funct 224:1067–1095. 10.1007/s00429-018-01825-1 [DOI] [PubMed] [Google Scholar]
- Yoshida K, McCormack S, España RA, Crocker A, Scammell TE (2006) Afferents to the orexin neurons of the rat brain. J Comp Neurol 494:845–861. 10.1002/cne.20859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelikowsky M, Hui M, Karigo T, Choe A, Yang B, Blanco MR, Beadle K, Gradinaru V, Deverman BE, Anderson DJ (2018) The neuropeptide Tac2 controls a distributed brain state induced by chronic social isolation stress. Cell 173:1265–1279.e19. 10.1016/j.cell.2018.03.037 [DOI] [PMC free article] [PubMed] [Google Scholar]