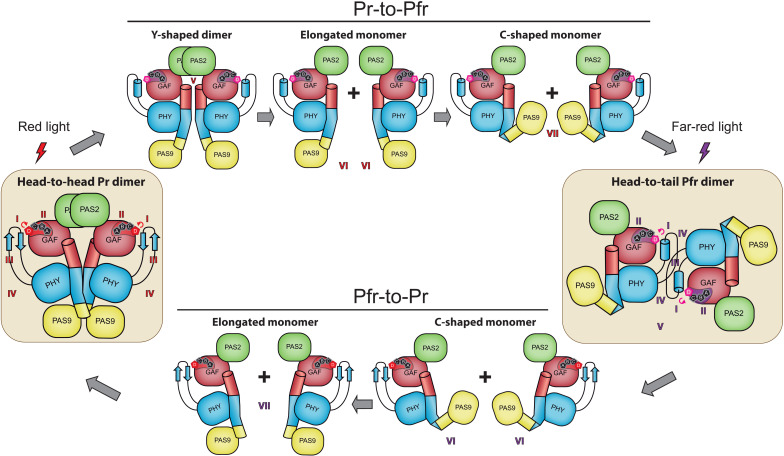
A bacterial phytochrome reversibly interconverts between a parallel and antiparallel dimeric arrangement during photoconversion.
Abstract
Phytochromes constitute a widespread photoreceptor family that typically interconverts between two photostates called Pr (red light–absorbing) and Pfr (far-red light–absorbing). The lack of full-length structures solved at the (near-)atomic level in both pure Pr and Pfr states leaves gaps in the structural mechanisms involved in the signal transmission pathways during the photoconversion. Here, we present the crystallographic structures of three versions from the plant pathogen Xanthomonas campestris virulence regulator XccBphP bacteriophytochrome, including two full-length proteins, in the Pr and Pfr states. The structures show a reorganization of the interaction networks within and around the chromophore-binding pocket, an α-helix/β-sheet tongue transition, and specific domain reorientations, along with interchanging kinks and breaks at the helical spine as a result of the photoswitching, which subsequently affect the quaternary assembly. These structural findings, combined with multidisciplinary studies, allow us to describe the signaling mechanism of a full-length bacterial phytochrome at the atomic level.
INTRODUCTION
Light-sensing mechanisms allow prokaryotes and eukaryotes to gather information about their ever-changing environments. Phytochromes are red light–sensing and far-red light–sensing proteins that constitute a widespread biological photoreceptor family found in plants, algae, fungi, and prokaryotes (1, 2). Bacterial phytochromes [bacteriophytochromes (BphPs)] have gained attention since their discovery (3, 4), with growing numbers of studies addressing their roles in infective and pathogenic organisms.
The prototypical BphP architecture consists of an N-terminal photosensory module (PSM), which autocatalytically binds biliverdin IXα (BV) chromophore to a conserved cysteine residue, and a C-terminal variable output module (OM), responsible for transducing the PSM light-driven conformational changes into a specific physiological signal (5–9). The PSM is typically composed of three modular domains linearly structured sharing topological elements: a Per-Arnt-Sim (PAS) domain, followed by cyclic guanosine monophosphate (GMP)–adenylyl cyclase-FhlA (GAF) and phytochrome-specific (PHY) domains. The OM is often composed of histidine kinase modules (HK) that are assumed to trigger a two-component signaling pathway via a phosphotransfer mechanism to regulate downstream processes such as gene expression. However, other frequent OM architectures include (i) HK–response regulator pairs, (ii) c-di-GMP cyclase/phosphodiesterases, and (iii) PAS domains, among others, evidencing the wide range of signaling outputs in the phytochrome family (8). The PSM-containing phytochromes presenting at least one PAS domain within the OM represent ~26% of the total sequences from the Pfam database (https://pfam.xfam.org/).
BphPs usually show a dimeric quaternary structure, where protomers are docked to each other in a parallel or antiparallel arrangement (10–16) through long helical bundles across the different domains called GAF-PHY and PHY-OM helical linkers, which form the “helical spine” (6, 7). Other outstanding structural features from these photoreceptors are (i) the presence of a figure-of-eight knot that crosses over residues between the PAS and GAF domains, strengthening their association (17), and (ii) a highly conserved hairpin protrusion from the PHY domain, termed as the “tongue,” which encloses the chromophore-binding pocket by interacting electrostatically with the GAF domain (6–9).
The basic principle of BphP photochemistry is their reversible photoconversion, typically between two photostates that exhibit different absorption spectra named Pr (red light–absorbing) and Pfr (far-red light–absorbing) (10, 18). In the dark, canonical-type BphPs exhibit a Pr thermal ground state, while bathy-type BphPs show a Pfr thermal ground state (19). Although the full picture on how the light-induced changes are transduced from the PSM to the OM is still incomplete, there are a series of well-established reversible structural features during the Pr/Pfr photoconversion (20–30).
Upon red (far-red) light absorption in the Pr (Pfr) state, the initial step of the intramolecular signaling mechanism, namely, the BV chromophore photoisomerization, is triggered. This initial step results in the Lumi-R (Lumi-F) intermediate, which involves a Z/E (E/Z) conversion of the C15═C16 double bond between the pyrrole rings C and D along with a series of transient proton translocation (Meta-R or Meta-F intermediates) events at the ring B and C pyrrole nitrogen atoms, as well as protonation dynamic events in the biliverdin-binding pocket. As a result, a ~180° rotation of ring D is produced, defining a ZZZssa or ZZEssa BV configuration in the Pr or Pfr states, respectively. The time scale of the ZZZssa-to-ZZEssa intermediate formation ranges from ultrafast transitions ranging in the picosecond range (Lumi intermediates) toward microseconds (Meta species). The distinct intermediate states have been elucidated in view of their structural and spectroscopic features (16, 31–33).
As the chromophore interacts with residues located at different regions of the PSM, the local physicochemical changes upon photoisomerization perturb the structure of the PSM. Consequently, the tongue reversibly interconverts into two structural conformations: a two-stranded antiparallel β sheet in the Pr state and an α helix in the Pfr state (21, 24–30). Although it is not currently fully understood how the tongue structural changes develop, it is well agreed that the β-sheet/α-helix tongue transition generates a push/pull movement between the GAF and PHY domains, reorienting the GAF-PHY and PHY-OM helical linkers from the helical spine and thus modifying the dimer interface (28–30). This large-scale structural motion is proposed to be critical for modulating the OM signaling activity. Nevertheless, the structural determinants of this photoreception mechanism are still poorly understood, as most of the BphP structures reported to date lack their corresponding OMs and, additionally, no full-length phytochrome structures have been solved at the (near-)atomic level in both pure (nonmixed) Pr and Pfr photostates (9).
Our group studies the BphP from the plant pathogen Xanthomonas campestris pv. campestris (XccBphP), which functions as a virulence regulator by modulating key physiological processes including xanthan production, biofilm formation, and infection capacity (34). XccBphP has been classified as a bathy-like phytochrome as it reaches a mixed Pfr:Pr thermal equilibrium in the dark (~6:1) (35). Moreover, we have solved the crystal structure of the full-length version bearing its PSM (formed by the domain triad PAS2-GAF-PHY) and its complete OM of unknown function (a PAS9 domain) in the Pr state at 3.25-Å resolution (35, 36). However, the structure showed weak electron density at the region corresponding to ring D of the BV molecule, so neither a ZZZssa nor a ZZEssa chromophore configuration could be defined based solely on the x-ray crystallographic data (35). Because of the lack of a XccBphP Pfr structure, we used the Rhodopseudomonas palustris RpBphP1 Pfr structure (sharing a similar domain architecture) (12) to propose a rough model for signal propagation from the PSM to the OM during the XccBphP light-driven conversion mechanism. However, the use of two different BphPs to build a precise mechanistic intramolecular transducing model at the atomic scale is far from optimal.
In this study, we present two previously unidentified crystal structures of XccBphP variants in the Pfr state, including a full-length version. In addition, we have obtained a wild-type full-length Pr structure with an enhanced resolution, with the complete chromophore molecule presenting a ZZZssa configuration. These structures enabled us to identify unprecedented structural rearrangements between both photostates in a complete BphP. Spectroscopic and mutational analyses along with computational modeling and biophysical experiments have allowed us to probe the mechanism at play. Our results enabled us to build a more complete model on the light-driven conformational changes transmitted from the PSM to the OM in the phytochrome photocycle.
RESULTS
To gain insights into the signaling mechanisms at the atomic level during the reversible Pr-Pfr photoswitching in full-length BphPs, we intended to solve the crystal structures of XccBphP in the Pr and Pfr photostates. One of our aims was to obtain a better crystal structure of the previously solved Pr form to define the BV ring D and unambiguously allocate a ZZZssa chromophore configuration. Another aim was to obtain a Pfr crystal structure. The wild-type Pfr crystals failed to grow even under dark conditions, where the highest Pfr proportion (~85%) is reached at equilibrium (35). Under these conditions, there is still a considerable Pr population from which we hypothesize that Pr crystals of XccBphP grow (36). Hence, our strategy to obtain the Pfr crystal structure was to use two Pfr-favored variants, for which the dark conversion rate is increased and/or the equilibrium is shifted. One of these variants is a deletion mutant lacking the C-terminal PAS9 domain corresponding to the 1–511 residue range, here named ΔPAS9(1–511), which generates a photoactive bathy-type PSM with a relatively more stable Pfr state (35). The other variant bears the G454E substitution in the tongue region of the PHY domain, obtained from a saturated mutagenesis library screening for Pfr-favored candidates (37).
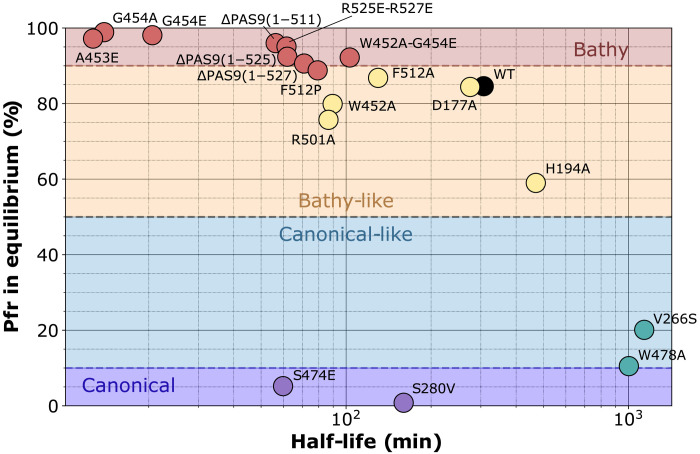
Before the crystallization assays, we performed a brief characterization of protein properties in solution. The XccBphP wild-type and ΔPAS9(1–511) variants have been previously proven to assemble in the Pr form in approximately 5 min (35). In contrast, G454E shows a slower dark assembly also in the Pr form (fig. S1A). ΔPAS9(1–511) and G454E photoconvert upon red and far-red irradiation as the wild-type version (fig. S1B) (35). The dark conversion kinetics of these variants, and others that will be presented during this work, is analyzed in figs. S2 to S4 and summarized in Fig. 1 and Table 1. During dark conversion, both ΔPAS9(1–511) and G454E are enriched in the Pfr form compared to the wild-type version, reaching proportions more than 96% of Pfr at equilibrium and with rapid kinetics, presenting half-life values of ~15 and 50 min for G454E and ΔPAS9(1–511), respectively (the wild-type estimated half-life was ~307 min) (Table 1).
Fig. 1. Kinetic parameters calculated for dark conversion of XccBphP variants.
The photochemical behaviors of all variants studied in this work are summarized in this scatterplot, where the x axis corresponds to the half-life values and the y axis corresponds to the Pfr fractions (%) at equilibrium. These two parameters were estimated by fitting Eq. 1 (monoexponential) and Eq. 2 (biexponential) to the data from fig. S2, respectively (see also fig. S3). Each variant is represented by a different colored dot in the scatterplot. BphP categories were based on Pfr enrichment in equilibrium: bathy (>90%), bathy-like (50 to 90%), canonical-like (10 to 50%), and canonical (0 to 10%). WT, wild type.
Table 1. Kinetic and oligomerization parameters calculated for XccBphP variants.
Half-life was calculated using a monoexponential fit (Eq. 1) for Pr-to-Pfr and Pfr-to-Pr conversions for bathy-like and canonical-like types, respectively. Pr and Pfr relative (%) contributions at equilibrium (Eq.) were estimated using a double exponential fit (Eq. 2). Pr and Pfr pure extrapolated spectra were used to estimate by Gaussian fitting their peak wavelengths (λ max). WT, wild type.
| Variant |
Location
affected |
Type |
Half-life
(min) |
Pr Eq.
(%) |
Pfr Eq.
(%) |
Pr λ max
(nm) |
Pfr λ max
(nm) |
SEC-SLS main peak | SLS secondary peak | ||
| MW (kDa) |
Oligomeric
state |
MW
(kDa) |
Oligomeric
state |
||||||||
| WT | – | Bathy-like | 307 | 15.4 | 84.6 | 684 | 757 | 139 ± 3 | Dimer | – | – |
| D177A | GAF | Bathy-like | 275 | 15.6 | 84.4 | 685 | 756 | – | – | – | – |
| H194A | GAF | Bathy-like | 470 | 41.0 | 59.0 | 684 | 757 | – | – | – | – |
| V266S | GAF | Canonical- like |
1137 | 79.9 | 20.1 | 697 | 753 | – | – | – | – |
| S280V | GAF | Canonical | 160 | 99.2 | 0.8 | 673 | 758 | – | – | – | – |
| W452A | PHY/ tongue |
Bathy-like | 90 | 20.1 | 79.9 | 688 | 751 | 285 ± 63 | Oligomer (n > 2) |
– | – |
| W452A-G454E | PHY/ tongue |
Bathy | 103 | 7.8 | 92.2 | 687 | 749 | 131 ± 15 | Dimer | – | – |
| A453E | PHY/ tongue |
Bathy | 13 | 2.8 | 97.2 | 688 | 748 | 146 | Dimer | – | – |
| G454E | PHY/ tongue |
Bathy | 21 | 1.9 | 98.1 | 688 | 748 | 141 ± 1 | Dimer | – | – |
| G454A | PHY/ tongue |
Bathy | 14 | 1 | 99 | 688 | 751 | 153 ± 3 | Dimer | – | – |
| S474E | PHY/ tongue |
Canonical | 60 | 94.8 | 5.2 | 685 | 739 | – | – | – | – |
| W478A | PHY/ tongue |
Canonical- like |
1003 | 89.5 | 10.5 | 685 | 747 | 139 ± 2 | Dimer | – | – |
| R501A | PHY-OM linker |
Bathy-like | 87 | 24.3 | 75.7 | 685 | 757 | – | – | – | – |
| F512A | PHY-OM linker |
Bathy-like | 130 | 13.2 | 86.8 | 685 | 758 | – | – | – | – |
| F512P | PHY-OM linker |
Bathy-like | 79 | 11.2 | 88.8 | 684 | 757 | 152 ± 5 | Dimer | – | – |
| R525E-R527E | PHY-OM linker |
Bathy | 42 | 3.0 | 97.0 | 684 | 757 | 79 ± 4 | Monomer (~85%) |
142 ± 21 | Dimer (~15%) |
| ΔPAS9(1–511)* | PHY-OM linker |
Bathy | 53 | 3.5 | 96.5 | 684 | 757 | 58 | Monomer (100%) |
– | – |
| ΔPAS9(1–525) | PHY-OM linker |
Bathy | 62 | 7.6 | 92.4 | 684 | 757 | 70 | Monomer (~92%) |
111 | Dimer (~8%) |
| ΔPAS9(1–527) | PHY-OM linker |
Bathy | 71 | 9.4 | 90.6 | 685 | 756 | 75 | Monomer (~89%) |
122 | Dimer (~11%) |
*SEC-SLS MW from Otero et al. (35).
The G454E Pfr-favored variant is a functional photoreceptor in vivo
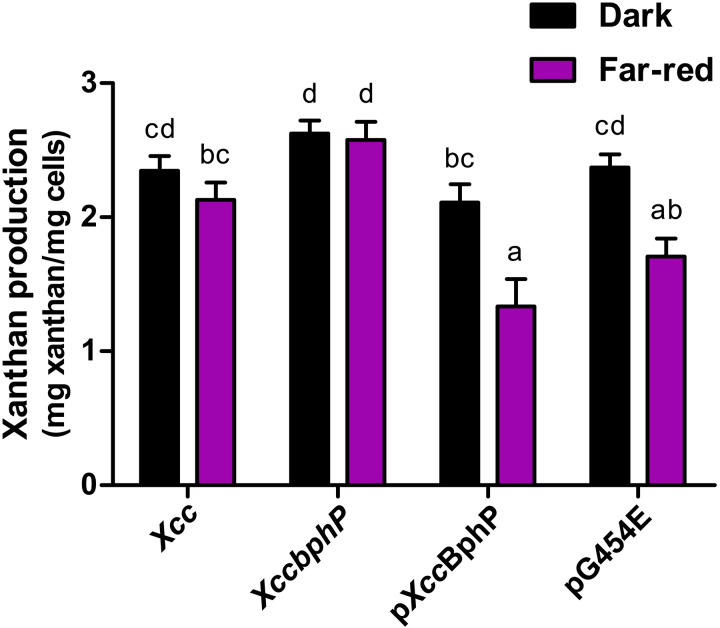
With the aim of evaluating the functionality of the G454E variant in vivo, we studied xanthan production as a proxy for light sensing and signaling. The exopolysaccharide xanthan is a key virulence factor in several Xanthomonas species, including X. campestris pv. campestris (Xcc) (38). In previous works, we have demonstrated that white or far-red light inhibits xanthan production in wild-type Xcc, while in the null knockout mutant (Xccbphp) xanthan production is increased, regardless of the light conditions. These findings show that the Pr state is the active species that inhibits xanthan production (34, 39). The complemented strains either with a wild-type copy of the Xccbphp gene (pXccBphP) or with the genes coding for the G454E variant (pG454E) presented a behavior similar to the wild-type Xcc strain, albeit with a more pronounced response to far-red light (Fig. 2). These pronounced effects can be explained by the differential protein expression that the complemented strains exhibit (fig. S5), leading to a possible amplification of the signaling by the XccBphP pathway (34, 39). Together, the ultraviolet-visible (UV-Vis) spectroscopy and in vivo results indicate that the G454E tongue substitution produces a functional photoreceptor, capable of sensing light and signaling downstream, validating its use as a model to study the full-length Pfr structure of XccBphP.
Fig. 2. The G454E variant restores light-dependent xanthan production phenotype in vivo.
Wild-type (Xcc), Xccbphp, pXccBphP, and pG454E strains were grown to stationary phase in 20 ml of PYM media supplemented with 2% glucose under darkness or far-red (733 nm) light conditions. Xanthan was purified through ethanol precipitation of cell-free supernatants. Cell pellets and precipitated extracellular xanthan were dried and weighed. Data are represented by mean values ± SEM from nine independent experiments (three replicates per experiment). Data were analyzed using the Kruskal-Wallis test. Different letters indicate significant differences between groups (P < 0.05).
The XccBphP crystal structures display the classical phytochrome features of the Pr and Pfr states
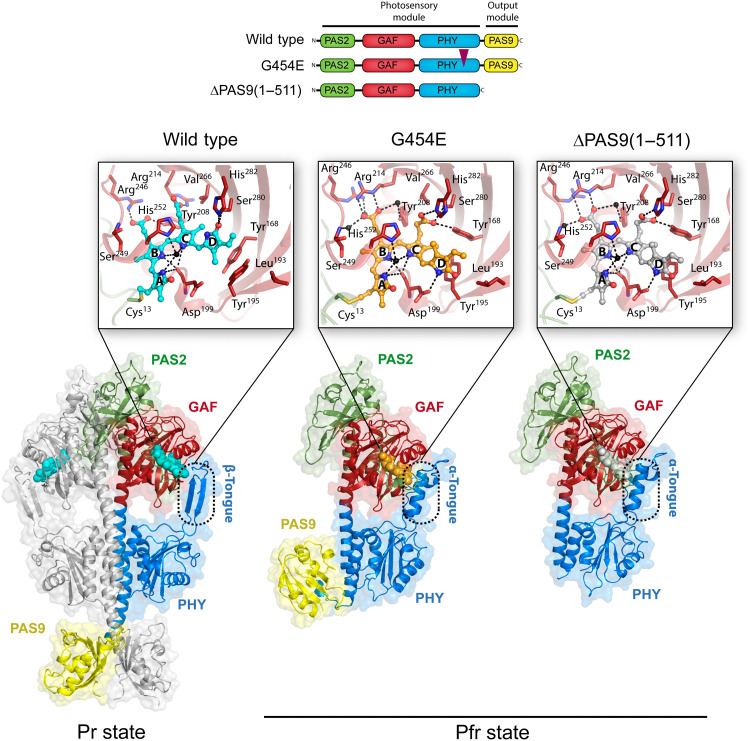
The three crystallographic structures were solved by molecular replacement using the previously reported wild-type Pr structure [Protein Data Bank (PDB) entry 5AKP] as starting model and refined at 2.68 Å (G454E), 2.95 Å [ΔPAS9(1–511)], and 2.96 Å (wild-type protein) with favorable stereochemistry and good refinement statistics (Fig. 3 and table S1). The final refined models show two chains (A and B) in the asymmetric unit in the wild type and only one in the G454E and ΔPAS9(1–511) variants (Fig. 3).
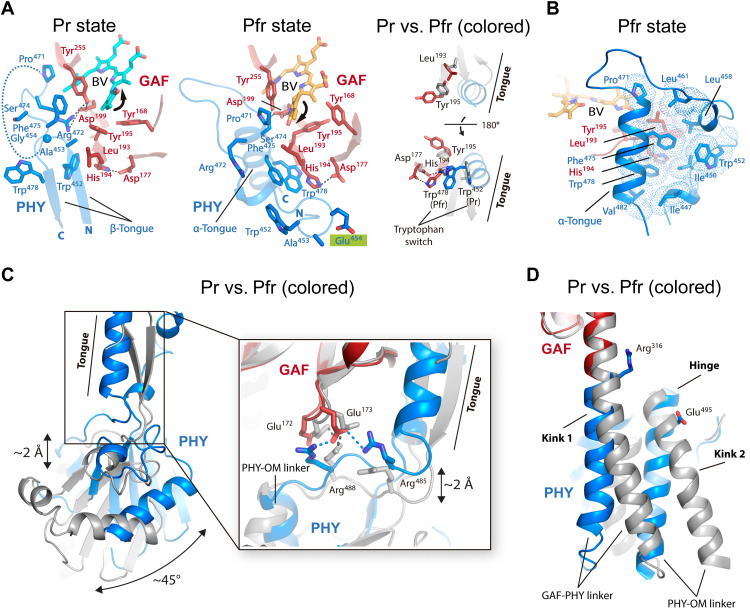
Fig. 3. Crystal structures of XccBphP variants in the Pr and Pfr states.
The domain architecture of the wild-type, G454E, and ΔPAS9(1–511) variants is schematized, showing the PAS2 (green), GAF (red), PHY (blue), and PAS9 (yellow) domains; the same color code is applied to all figures in this work. The domain composition of the photosensory module (PSM) and output module (OM) is indicated. A purple triangle indicates the position of the single amino acid change within the G454E variant. Ribbon representations of the different XccBphP variants: wild type (Pr state), G454E (Pfr state), and ΔPAS9(1–511) (Pfr state) as defined in their respective asymmetric units. The tongue regions are boxed in dashed rounded rectangles, exposing a two-stranded antiparallel β-sheet conformation in the Pr state and an α-helical conformation in the Pfr state. The different domains are labeled, and the solvent-accessible surface calculated by PyMOL is shown in the background. Insets: Detailed views of the chromophore-binding pocket from each structure. The BV chromophore is covalently bound to Cys13 (from the PAS2 domain) and buried into the GAF domain. BV is shown as spheres with carbon atoms in cyan (wild type), orange (G454E), or gray [ΔPAS9(1–511)], oxygen atoms in red, and nitrogen atoms in blue. The four BV pyrrolic rings are indicated as A, B, C, and D. The most relevant residues are depicted as sticks. Structural water molecules (including the pyrrole water molecule coordinated by the NH groups of rings A, B, and C and the Asp199 carbonyl group) are represented as black spheres. Important polar interactions are shown as dashed lines.
The wild-type full-length protein bearing the PSM and the complete C-terminal OM crystallized in a characteristic Pr state, which is evidenced by (i) the ZZZssa chromophore configuration, (ii) the side-chain conformation of the conserved Tyr168-Tyr195 pair that surrounds ring D, and (iii) the two-stranded antiparallel β-sheet tongue conformation (Fig. 3). The Pr crystal structure obtained in this work is essentially equivalent to the former wild-type protein structure (35), revealing a head-to-head parallel dimer with Cα root mean square deviation (RMSD) values of 0.57 and 0.56 Å for chains A (590 aligned residues) and B (599 aligned residues), respectively. Moreover, no substantial structural differences are perceived in the BV interaction pocket. However, the electron density in ring D from the BV chromophore is clearly defined, allowing an unequivocal determination of the chromophore in the ZZZssa configuration and therefore unambiguously validating the Pr state (Fig. 3, inset, and figs. S6 and S7).
The structures of the full-length G454E (PSM and the complete C-terminal OM) and ΔPAS9(1–511) (only PSM) proteins are congruent with other PSMs from BphPs solved in the Pfr state (12, 16, 18, 28, 30, 40) revealed by (i) the ZZEssa chromophore configuration, (ii) the side-chain conformation of the conserved Tyr168-Tyr195 pair surrounding ring D, and (iii) the α-helical tongue conformation, among other Pfr structural hallmark features (Fig. 3 and fig. S8). The OM (PAS9 domain) from G454E is found in a contracted position with respect to the PSM as a result of a conspicuous break in the PHY-OM helical linker (Fig. 3). This key structural feature will be developed in further sections.
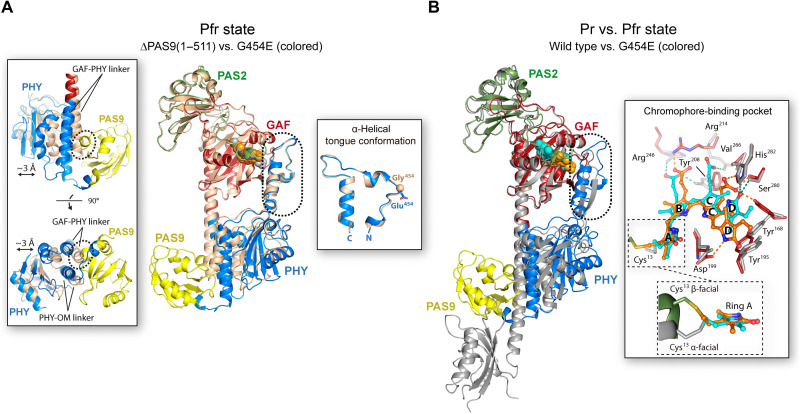
Both proteins show highly similar PAS, GAF, and PHY domain structures, revealed by a pairwise alignment between their main-chain Cα atoms yielding RMSD values of 0.41 Å (113 aligned residues), 0.39 Å (166 aligned residues), and 1.08 Å (164 aligned residues), respectively (fig. S9, top). Furthermore, the conserved figure-of-eight knot that encloses the chromophore pocket remains unaltered between both structures (fig. S10). However, a global comparison shows that the most prominent differences between both Pfr structures lie in the position of their PHY domains, which display a slight translational displacement of ~3 Å between each other, along with a subtle change on the trajectories of the GAF-PHY and PHY-OM helical linkers (Fig. 4A). These structural differences, possibly inflicted by the presence/absence of the PAS9 domain (Fig. 4A, left inset), do not alter the tongue conformation between both proteins (Fig. 4A, right inset). No changes are observed around position 454, which is located in the tongue loop, demonstrating that the G454E substitution does not affect the tongue overall structure. Moreover, both the residue positions and the bilin ligand location on the chromophore-binding pocket are identical in the two structures, showing the expected interactions for the Pfr state around the BV molecule (see details in the next section) (Fig. 3, insets, and figs. S6 to S8).
Fig. 4. Global comparison of the tertiary structure of XccBphP variants in the Pr and Pfr states.
(A) Alignment between Pfr structures: ΔPAS9(1–511) (wheat) and G454E (domains colored according to Fig. 3). The BVs are represented as gray [ΔPAS9(1–511)] and orange (G454E) spheres. Left inset: Most prominent differences are shown in two orientations. The displacement between the PHY domains and the GAF-PHY and PHY-OM helical linkers is indicated. The potential clashes between the GAF-PHY helical linker, as defined in ΔPAS9(1–511), and the PAS9 domain, as defined in G454E, are highlighted by dashed ovals. Right inset: α-Helical tongue conformation from both proteins rotated 90° with respect to the main panel. Gly454 from ΔPAS9(1–511) and Glu454 from G454E are depicted as a sphere and sticks, respectively. N and C termini are indicated. (B) Full-length structural alignment between the wild-type Pr (gray) and G454E Pfr. The BVs are represented as cyan (wild type) and orange (G454E) spheres. Inset: Structural differences on the chromophore-binding pocket from both proteins. The most relevant residues are depicted as sticks; the color code for G454E is according to Fig. 3; wild-type residues are gray-colored. Polar interactions are shown as dashed lines colored in cyan (wild type) or orange (G454E). BV is shown as capped sticks with carbon atoms in cyan (wild type) or orange (G454E), oxygen atoms in red, and nitrogen atoms in blue. The four BV pyrrolic rings are indicated. Two views of the orientation of α-facial (Pr) and β-facial (Pfr) Cys13 binding to BV are enclosed in dashed boxes. (A and B) Structural alignments were performed on the GAF domain; the tongue is boxed in a dashed rounded rectangle.
A structural contrast between the Pr and Pfr structures reveals notable changes in different regions of the protein (Fig. 4B). These include a reorganization of the interaction networks within and around the chromophore-binding pocket promoted by the BV photoisomerization, an α-helix/β-sheet tongue interconversion, and PHY domain reorientations, along with notable perturbations at the helical spine, which define the quaternary assembly in a parallel (Pr) or antiparallel (Pfr) manner.
The BV photoisomerization promotes the flip-and-rotate movement of the conserved tyrosine pair next to ring D and the reconfiguration of the H-bond interaction network in the chromophore-binding pocket
In the Pr and Pfr structures, the BV chromophore is embedded in a cleft inside the GAF domain and covalently bound by a thioether linkage between the C32 atom of ring A and a conserved cysteine residue (Cys13) in the N-terminal extension of the PAS2 domain (Fig. 3, insets, and figs. S7 and S8). The global folding of the PAS2 and GAF domains, including the figure-of-eight knot, remains unaltered between both photostates (Fig. 4B and figs. S9, bottom, and S10).
As noted previously, the Pr state presents a ZZZssa chromophore configuration, whereas the transition to the Pfr state involves an isomerization of the ligand with a ~180° rotation in ring D, showing a ZZEssa configuration (Figs. 3, insets, and 4B, inset). In concordance with other reported BphP structures (8), this configurational motion is accompanied by a synchronized flip-and-rotate movement of two highly conserved aromatic residues, Tyr168 and Tyr195, located next to ring D, along with a slight rearrangement of the Tyr255 side chain (Figs. 4B, inset, and 5A). This local reorganization is associated with a series of structural motions outside the chromophore-binding pocket explained in the next section. Moreover, the hydrogen bonds formed among the ring D carbonyl oxygen and the side chains of Ser280 and His282 in the Pr state are replaced in the Pfr structures by a hydrogen bond between the NH group from ring D and the Asp199 carboxylate, which is part of the highly conserved DIP (Asp-Ile-Pro) motif (residues 199 to 201) (Figs. 3, insets, and 4B, inset, and fig. S7). Accordingly, it has been reported in other BphPs such as Agp1 from Agrobacterium fabrum (also known as Agrobacterium tumefaciens), DrBphP from Deinococcus radiodurans, and PaBphP from Pseudomonas aeruginosa that the abolition of the aspartate residue in this motif typically blocks the protein photocycle in an intermediate state that disrupts the Pr-to-Pfr progression (24, 41, 42).
Fig. 5. Structural rearrangements around the chromophore-binding pocket, the tongue interconversion, and the helical spine reconfiguration between the full-length Pr and Pfr structures.
(A) Left and middle: Main interactions in wild-type Pr and G454E Pfr. Residues and BV are depicted as sticks and colored following Fig. 3. Gly454 and Glu454 (highlighted in green) are depicted as a sphere and sticks, respectively. Polar interactions are shown as dashed lines. The disordered region of the β-hairpin loop in the Pr structure is represented by a curved dashed line. Ring D movements are represented with arrows. The N and C termini are indicated. Right: Structural contrast between both structures (Pr in gray) displayed in two orientations, emphasizing the interactions by Leu193 and His194. (B) Hydrophobic pocket in the tongue-GAF interface of the Pfr. Hydrophobic contacts are represented by a cloud of dots. (C) Tongue-fold interconversion and PHY domain repositioning. The Pr (gray) and Pfr structures (colored according to Fig. 3) are aligned on the GAF domain. The displacement between the GAF and PHY domains and the reorientation of the PHY domain are indicated. Inset: Changes in the salt-bridge clusters from the interfacing GAF/PHY network. Interactions (dashed lines) are shown in gray (wild type) or blue (G454E). (D) Changes on the helical spine. “Kink 1” and “Kink 2” and “Hinge” regions are indicated. The residues Arg316 (GAF domain) and Glu495 (PHY domain) involved in the Pr dimer interface (see Fig. 7A) are shown as references.
A network of interactions among the NH groups of rings A, B, and C; the Asp199 carbonyl group; the conserved pyrrole water molecule; and the Nδ atom of His252 is observed in both the Pr and Pfr states (Fig. 3, insets, and fig. S7). However, the positions of BV rings B and C (but not A) are displaced between both photostates, altering the hydrogen-bond network of their propionate groups (Fig. 4B, inset). In the Pr state, the two BV propionates are within hydrogen-bonding distance from the side chains of Tyr208, Arg214, and Arg246; however, the chromophore displacement observed in the Pfr state is associated with a different interaction network (Figs. 3, insets, and 4B, inset, and fig. S7). In the Pfr state, the propionate group of ring B is directly stabilized by the side chain of Arg214 and indirectly by Tyr208, Arg246, and Ser249 via ordered water molecules. Furthermore, the propionate group of ring C is relocated into a hydrogen-bonding environment constituted by the side chains of Tyr168 (from the tyrosine flip-and-rotate mechanism), Ser280, and His282 (Figs. 3, insets, and 4B, inset, and fig. S7). All these structural changes are accompanied by a transition from an α-facial to a β-facial orientation of the BV-binding Cys13 residue in the Pr and Pfr states, respectively (Fig. 4B, inset), as previously seen in other BphPs (16).
To assess the contribution of relevant residues for Pr and Pfr stability, as mentioned above and in the following sections, we designed a set of mutations and evaluated their photochemical behavior, using the dark conversion rate (half-life) and the Pr:Pfr proportions at thermal equilibrium as proxies for stability (Fig. 1, Table 1, and figs. S2 to S4). For the sake of simplicity in the photochemical classification of the variants, we define the types as “bathy,” “bathy-like,” “canonical-like,” and “canonical” when they exhibit a Pfr enrichment in thermal equilibrium above 90%, between 50 and 90%, between 10 and 50%, and below 10%, respectively (Fig. 1).
To validate the displacement of the propionate group of ring C observed between both photostates, we designed two substitutions that exchange the polarity and hydrophobicity in two close residues, Val266 by serine (V266S) and Ser280 by valine (S280V) (Figs. 3, insets, and 4B, inset). Both substitutions make a considerable change in the preferred state in equilibrium, turning the XccBphP into canonical-like and canonical types, respectively (Fig. 1 and Table 1). As observed in the structures, the Val266 side chain is near the ring C propionate in either the Pr or Pfr conformations. However, the extra hydrogen bond introduced by the serine hydroxyl group in V266S seems to stabilize this propionate group from BV in the ZZZssa configuration. In contrast, the elimination of the serine hydroxyl group in S280V may destabilize this group from BV in the ZZEssa configuration, impairing the Pfr state.
Key residues on a second shell assist the conserved tryptophan switch, functionally connecting the reconfiguration of the chromophore-binding pocket and the α-helix/β-sheet tongue interconversion
In XccBphP, the tongue is constituted by residues 446 to 483, which are organized in the highly conserved motifs W(G/A)G (452–454), PRxSF (471–475), and (W/Y/F)x(E/Q) (478–480) (35). In the Pr state, the tongue adopts a β-hairpin fold, and the Trp452 residue is located in the entrance β strand within the tongue-GAF interface, while Trp478 is situated in the exit β strand and exposed to the protein surface (Fig. 5A, left). Furthermore, Ala453, Gly454, Pro471, Arg472, and Phe475 are positioned in the β-hairpin loop adjacent to the chromophore-binding pocket and stabilized mainly by hydrogen bonds among the side chains of Arg472, Asp199, and Tyr255 (Fig. 5A, left). Residues 456 to 470 from the β-hairpin loop were not visible in the electron density map, indicating a high flexibility for this region. On the contrary, in the Pfr state, the tongue undergoes a conformational change interconverting the entrance and exit β strands in a loop and an α helix, respectively (Figs. 4B and 5A, middle). As a result, an exchange of the conserved residues Trp452/Trp478 in the tongue-GAF interface takes place in concordance with the Trp switch model previously proposed (13, 29). Concordantly, the Trp478 residue located in the tongue α helix is now placed within the tongue-GAF interface, and Trp452 is exposed in the tongue loop facing the solvent along with Ala453 and the variant residue Glu454 (Fig. 5A, middle). In addition, Pro471, Ser474, and Phe475, which are settled also in the α helix of the tongue, close the tongue-chromophore pocket interface more tightly by setting the following interactions: (i) Pro471 contacts the vinyl side group of BV ring D and Tyr255, (ii) Ser474 is located at van der Waals distance from the Asp199 and Tyr255 side chains (Fig. 5A, middle), and (iii) Phe475 is associated to a predominantly hydrophobic pocket constituted by Leu193 and the tongue residues Ile447, Ile450, Trp452, Leu458, Leu461, Pro471, Trp478, and Val482 (Fig. 5B).
As already stated, the structural connection between the bilin photoisomerization and the tongue interconversion is not completely understood. The crystal structures reported here indicate that upon BV photoisomerization (ring D rotation), the phenolic side chains of the tyrosine triad 168-195-255 in the immediate surroundings of ring D change their positions. The Tyr195 residue lies at a β strand along with its preceding residues Leu193 and His194, which participate in the tongue-GAF interface (Fig. 5A). Because of the alternating side-chain directions in the β-strand structure, the side chains of Leu193 and Tyr195 are inside the chromophore-binding pocket, whereas the side chain of His194 is located outside. This structural organization places the side chains of Leu193 and Tyr195 in close distance. Accordingly, in the Pr state, the OH group of Tyr195 seems to exclude the Leu193 aliphatic side chain from the chromophore-binding domain by steric effects (Fig. 5A, left and right), while in the Pfr state its location allows Leu193 to position its side chain in that area next to ring D (Fig. 5A, middle and right). As a result, the distance between the Leu193 side chain and the BV ring D decreases considerably from ~7 Å in Pr to ~3 Å in Pfr. This movement in the Leu193 side chain seems to affect the hydrophobic interactions observed in the tongue-GAF interface of the Pfr state (Fig. 5B). Consequently, the side chain of His194 flips in the opposite direction (probably by a subtle main-chain torsion due to the movement of the adjacent residues Leu193 and Tyr195) and pulls the Asp177 residue by a hydrogen bond (Fig. 5A). Notably, the His194 motion is accompanied by the Trp switch model described above, through a cation-π or π-π stacking interaction between its imidazole ring and the indole ring of Trp452 in the Pr state or Trp478 in the Pfr state. Moreover, the Tyr255 movement allows or hinders the Phe475 side-chain positioning near the BV molecule in the Pr and Pfr states, respectively.
To challenge these observations and to gain insight into their roles in the Pr-Pfr photocycle, we designed another set of mutations affecting some key residues. The replacement of His194 by Ala (H194A) presents a ~2.7-fold increase of the Pr proportion at equilibrium, indicating that abolition of the cationic imidazole ring affects the Pfr state stabilization more (Fig. 1 and Table 1). In this line, the contacting residue Asp177 does not seem to play a major role in the His194 flipping, as revealed in the D177A variant, whose photochemical behavior is very similar to the wild-type protein. In addition, W478A markedly shifts the Pr:Pfr ratio at equilibrium to 9:1, turning it into a canonical-like type. This evidence supports the notion that there is a functional connection between His194 and Trp478 (linking the chromophore-binding pocket and the tongue), which is crucial for the Pfr state. Similarly, the Ser474 by Glu substitution (S474E) converts the phytochrome into a canonical type, driving the dark equilibrium toward an almost full conversion to the Pr state and with a surprisingly low half-life (60 min; Fig. 1 and Table 1). In concordance with the Pfr structure, the insertion of a carboxylate group in this position might provoke a repulsive electrostatic effect with the Asp199 side chain, validating the Ser474 role in the tongue-GAF interface in this photostate (Fig. 5A, middle).
The Trp452 by Ala substitution (W452A) exhibits a slight change in the Pr:Pfr ratio at equilibrium, indicating that Trp452 does not play a critical role in the stabilization of the Pr state. Nevertheless, the increase of the Pr-to-Pfr dark conversion rate (3.4-fold) relative to the wild type suggests that it may be participating in the stabilization of a tongue intermediate state. It is noteworthy that substitutions on nearby residues (i.e., A453E and G454E) transform the phytochrome into a typical bathy type, where an almost complete Pr-to-Pfr dark conversion occurs and at accelerated rates compared to the wild type with ~24- and ~15-fold increases, respectively (Fig. 1 and Table 1). These results are in total agreement with the structural data, where both residues are involved in the GAF-PHY interface in the Pr state but not in the Pfr (Fig. 5A, left and middle). Therefore, the incorporation of a negative charge in both positions may dislocate the GAF-PHY interface observed in the Pr state, thus favoring the Pfr state. Regarding Gly454, it is likely that the high conformational flexibility of this residue due to its side-chain–less backbone is necessary for Pr stabilization. This scenario is supported by a recent work from our group in which we showed that substituting Gly454 by almost any other amino acid resulted in Pfr-favored variants (37).
The α-helix/β-sheet tongue interconversion promotes reorientations of the PHY domain and large-scale conformational changes in the helical spine
As previously reported, the tongue-fold interconversion between the β-sheet and α-helix conformations implies an elongation or a shortening of this peptide segment, respectively (28). As a consequence, in XccBphP, a push (Pr)/pull (Pfr) displacement of around 2 Å between the GAF and PHY domains takes place, along with a prominent reorientation of the PHY domain of ~45°, altering the GAF/PHY interface between both photostates (Figs. 4B and 5C). In the Pr state, a network of salt bridges involving residues Glu172 and Glu173 (from the GAF domain) and Arg488 (from PHY) is observed, while in the Pfr state the interfacing GAF/PHY network includes the residues Glu173 (from GAF) and Arg485 and Arg488 (from PHY) (Fig. 5C, inset). The PHY domain repositioning does not affect the topology of the secondary structure elements from its globular part, although local changes can be observed on their three-dimensional arrangement (fig. S9, bottom), as detailed below.
In line with the PHY domain rearrangement, changes on the helical spine are noted. In the full-length Pfr structure, the GAF-PHY helical linker is fairly straight, while in the Pr structure it is slightly kinked (kink 1) (Figs. 4B and 5D). Moreover, a pivot point at the beginning of the PHY-OM helical linker (hinge) is observed, which affects the helix trajectory (Figs. 4B and 5D). As the Pr head-to-head dimer interface mainly lies on the GAF-PHY and PHY-OM helical linkers (see the section, “The XccBphP Pr photostate assembles as a parallel head-to-head dimer,” below), their structural rearrangements affect the positions of the interfacing residues and, consequently, the dimeric interactions. Among them, the Glu328/Arg501′ and Glu328′/Arg501 salt bridges, which are found anchoring both helical linkers in the Pr dimer, are worth noting. On this photostate, Arg501 is arranged in the PHY-OM helical linker where another kink is noted (kink 2; Figs. 5D and 6A). Conversely, in the Pfr state, Arg501 is settled on a straight α helix forming a salt bridge with Glu505 from the same protomer.
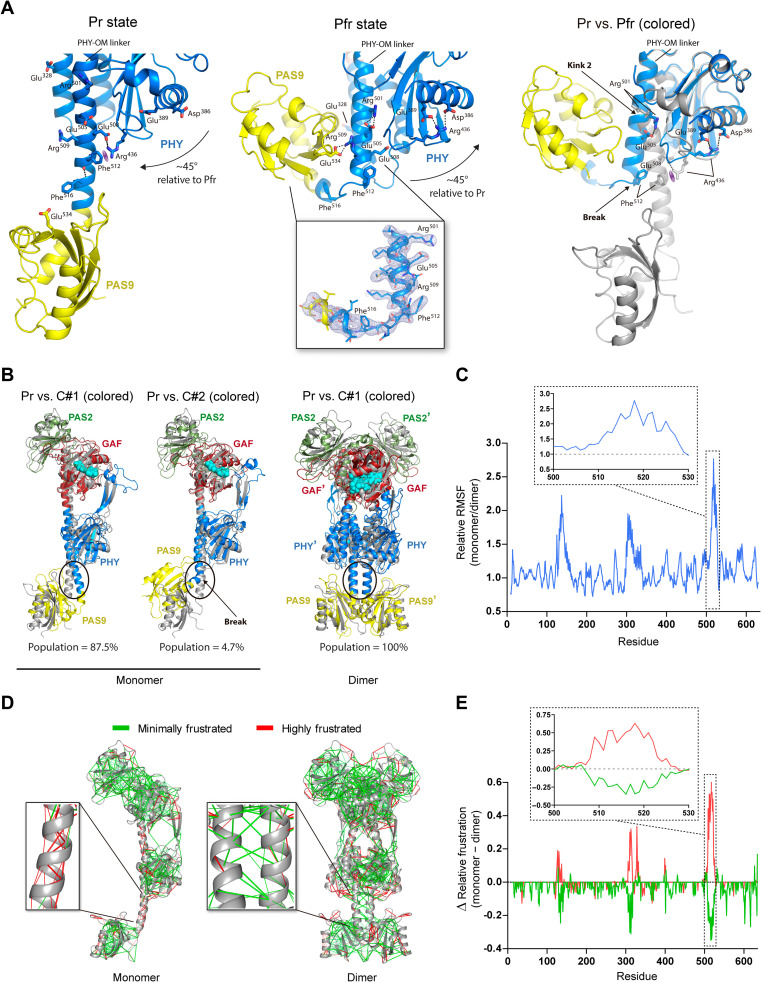
Fig. 6. Large-scale conformational changes in the PHY-OM helical linker and the OMs between the full-length Pr and Pfr structures.
(A) Left and middle: Main interactions from wild-type Pr and G454E Pfr, respectively. The PHY domain repositioning is indicated with arrows. Polar interactions are shown (dashed lines). The cation-π interaction between Arg436 and Phe512 is represented (purple meshed oval). Middle inset: The 2mFo-DFc electron density map (light blue mesh) contoured at the 1.0 σ level defines the break at the PHY-OM helical linker in the Pfr. Right: Contrast between the Pr (gray) and Pfr (colored) structures, aligned on the PHY domain. The bending (Pr) and the disruption (Pfr) in the PHY-OM helical linker are denoted as Kink 2 and Break, respectively. (B) Alignment between the Pr crystal structure (gray) and MD averaged models (colored). Left: MD clusters C#1 and C#2 from a monomer of the Pr structure. Right: MD cluster C#1 from the dimeric Pr crystal structure with chains A and B (prime). The region around the break in the Pfr structure is enclosed in an oval. (C) Relative RMSF per residue from the Pr monomer and Pr dimer (chain A) derived from the MD clusters obtained in (B). Inset: Region around the break. (D) Local frustration patterns on the monomeric (left) and dimeric (right) Pr scaffolds using the Frustratometer server (43). The minimally (green) and highly (red) frustrated interactions are represented. Insets: Expanded view around the break. (E) Difference (Δ) between monomer and dimer densities from minimally (green) and highly (red) frustrated contacts per residue expressed in relative terms (specific contacts/total contacts). Inset: Region around the break.
To probe the relevance of the Glu328/Arg501 interactions in the photochemical mechanism, we eliminated the salt bridges by replacing Arg501 by Ala (R501A) and evaluated the photochemical properties of this variant. Although slight changes were observed regarding the Pr:Pfr relative abundances at dark equilibrium, we observed a change of 3.5-fold reduction in the Pr-to-Pfr conversion rate compared to the wild type (Fig. 1 and Table 1). Hence, the abolition of the Glu328/Arg501 contacts could be leading to a faster Pr-to-Pfr transition by generating a less stabilized PHY-OM helical linker kink in the Pr state.
A break at the PHY-OM helical linker in the Pfr state disrupts the helical spine from the Pr state
A ~90° break at the central region of the PHY-OM helical linker is noted in the Pfr state (hereafter referred to as “break”), which places the PAS9 domain into a contracted position in comparison with the Pr state but preserving its global folding (Figs. 3, 4B, and 6A). This unprecedented helical disruption, unambiguously defined in the electron density map (Fig. 6A and fig. S11), turns the PHY-OM helical linker and the PAS9 domain closer in space, resembling a C-shaped protein global folding (Fig. 4B). The electron density for the PAS9 domain in this position is slightly less defined with relatively higher B-factor values (fig. S12), which denotes higher conformational flexibility favored by lacking intermolecular contacts within the crystal packing (fig. S13). To assess the stability of the experimentally determined helical break, we performed molecular dynamics (MD) simulations (500 ns, four replicas) on the G454E crystal structure. The analysis revealed four major clusters that encompass minor structural changes with respect to the crystallographic structure (Cα-RMSD values ranging from 1.78 to 2.10 Å; fig. S14), suggesting that the PAS9 domain position defined in the Pfr state is conserved under no confinement conditions as those present in the crystal.
A salt bridge between Arg509 and Glu534 is found stabilizing the contracted PAS9 domain with the PHY-OM helical linker in the Pfr state. Arg509 is part of the Pr dimer interface by interacting with Glu505, the counterpart of Arg501 in Pfr, as described above (Fig. 7). Therefore, there is a salt-bridge switch between Glu505 with Arg501 and Arg509 (from the neighboring protomer) in the Pfr and Pr states, respectively. Consequently, when the Pr dimer is assembled, Arg501 and Arg509 are trapped in the dimer interface, but in the Pfr state, both are free to be contacted by Glu505 and Glu534, respectively.
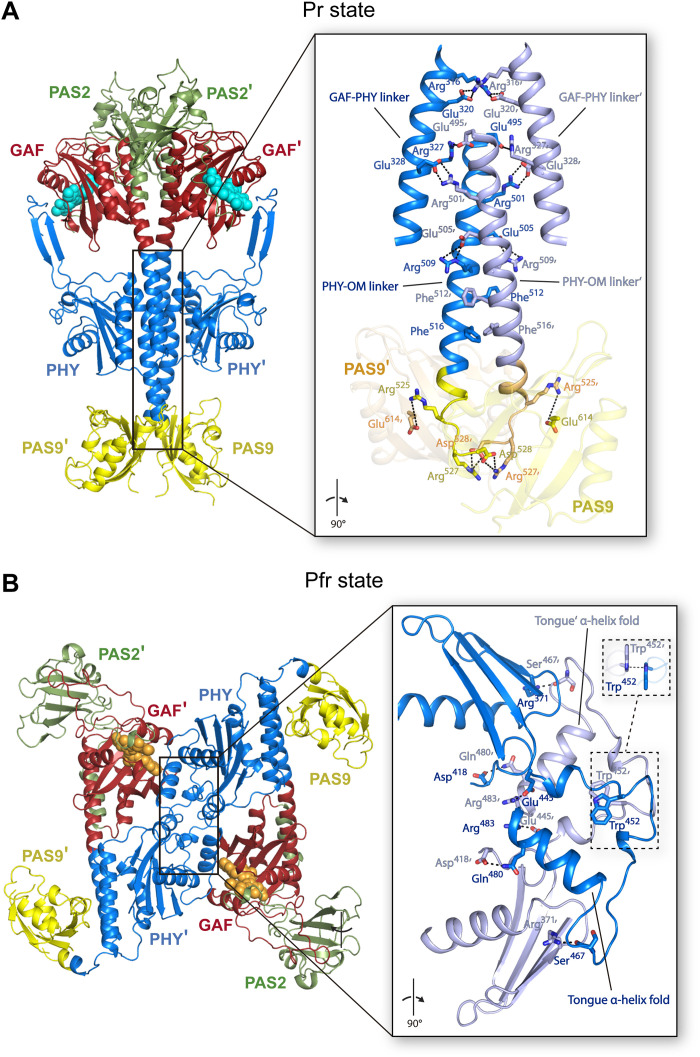
Fig. 7. The XccBphP full-length Pr and Pfr structures reveal different quaternary arrangements.
(A) Head-to-head dimer assembly of the wild-type Pr crystal structure as found in its asymmetric unit. (B) Head-to-tail crystallographic dimer assembly of the G454E Pfr crystal structure constructed by means of the -x, -y, z symmetry operator of the I41 space group. (A and B) Structures are shown in ribbon representation and colored following Fig. 3 domain color code with the two protomers depicted in different shades. Insets: Detailed view (rotated 90° with respect to main panels) of the respective dimerization interfaces encompassing the GAF-PHY and PHY-OM helical linkers from the helical spine in the Pr structure and the tongue region in the Pfr structure. Interfacing residues are depicted as sticks and colored according to their corresponding chain. Important polar interactions are shown as black dashed lines. Two views of the π-π stacking interaction between Trp452/Trp452′ (from the Trp switch) in the Pfr structure are enclosed in dashed boxes.
The breaking point for the PHY-OM helical linker is located at the Phe512 residue (Fig. 6A, middle, and fig. S11, inset). In the Pr state, the phenyl group of Phe512 is within cation-π interaction distance with the cationic guanidine moiety of Arg436 from the PHY domain, which is, in turn, stabilized by a salt-bridge contact with Glu508 (Fig. 6A, left). Phe512 is found buried at the dimer interface close to Phe516 from the neighboring chain (Fig. 7). Following the mutational strategy, we aimed to simulate the PHY-OM helical linker break by introducing a proline residue at position 512 (F512P), altering the native hydrogen-bond network of the helix. The F512P variant photochemistry did not show a marked change of the Pr and Pfr relative stabilities at equilibrium compared to the wild type (Fig. 1 and Table 1), although the kinetics of its Pr-to-Pfr conversion did show a ~4-fold decrease, which is indicative of an alteration of one or more transition states. In addition, the molecular weight (MW) determination of the F512P apoprotein by static light scattering coupled to size exclusion chromatography (SLS-SEC) is consistent with a dimer configuration (Table 1 and fig. S15). Therefore, a likely helical destabilization at this position still allows for a photoswitchable XccBphP phytochrome, with an unaltered dimeric arrangement.
In the Pfr state, the pulled PHY domain moves Arg436 ~18 Å away from Phe512, relocating it at salt-bridge distances with the intradomain residues Asp386 and Glu389 (Fig. 6A, middle). Note that in the Pr state, the Phe512 carbonyl group interacts with the Phe516 (i + 4) amide group in a turn from the PHY-OM helical linker, while in Pfr this interaction is absent as a result of the breaking (Fig. 6A). According to the mutational analysis, the abolition of the Phe512 phenyl group (F512A) accelerates the Pr-to-Pfr dark conversion (~2.4-fold) without changing drastically the Pr:Pfr equilibrium (Fig. 1 and Table 1), in a similar fashion to the F512P variant, also suggesting an alteration of one or more transition states. Thus, the cation-π interaction between Arg436 and Phe512 might not play a major role in Pr stability, leaving this role potentially to the nearby salt-bridged contact Arg436-Glu508 (Fig. 6A), but it would be involved in the photoconversion pathway.
In the same line, MD simulations on the monomeric Pr structure resulted in two clusters: one major cluster showing a bend in the PHY-OM helical linker, and another exhibiting a break in a similar region to that observed in the Pfr full-length structure (Fig. 6B, left, and fig. S16, top). The same computational studies performed on the dimeric Pr structure exposed no changes in this region (Fig. 6B, right, and fig. S16, top). Root mean square fluctuation (RMSF) calculations obtained from the monomer and dimer (chain A) MD simulations highlight the higher relative stability in the dimeric conformation with respect to the monomer around the helical break (Fig. 6C and fig. S16, bottom), suggesting that dimer dissociation is needed for the breaking to occur.
In addition, we calculated the local frustration patterns (i.e., total, minimally, neutrally, and highly frustrated interactions per residue) on the monomeric and dimeric Pr scaffolds using the Frustratometer server (43) to analyze how the frustration distribution in this helical region might be affected by the oligomeric state. The local protein frustration concept is used to learn about the biological behavior of proteins by analyzing how energy is distributed within their structures. Sites of high local frustration often indicate functionally important regions, such as binding or allosteric sites, while minimally frustrated bonds create stable protein cores within the molecule. Moreover, it has been shown that high local frustration often correlates with regions that undergo structural transitions that are prone to locally unfold or crack (44). In the Pr monomer, the region corresponding to the Pfr helical break is consistently (i) enriched in highly frustrated interactions and (ii) reduced in minimally frustrated interactions, with respect to the Pr dimer (Fig. 6, D and E, and fig. S17). The different frustration patterns in the network of local interactions in this particular region between both structural scenarios are consistent with the finding that highly frustrated patches correspond with binding sites. Therefore, the local frustration would be relieved upon binding, guiding the protein dimerization (45). In other words, the breaking area would switch from less to more frustrated if its enclosed protomer-protomer interactions are abolished. Together, the computational studies suggest that the PHY-OM helical linker in this region is flexible, highly frustrated, and prone to be kinked when it is not part of the helical dimeric interface as in the Pr state.
The XccBphP Pr photostate assembles as a parallel head-to-head dimer
The full-length XccBphP Pr state assembles as a parallel head-to-head dimer (35). The dimeric interface (3263 Å2) involves 89 residues per protomer (15% of the total residues) mainly settled along the helical spine that interact by a series of salt bridges, hydrogen bonds, and nonbonded contacts from side to side at the dimer interface. Approximately 65% of the interfacing residues are located between neighboring GAF-PHY linkers, PHY-OM linkers, and PAS9 domains that include 12 potential salt-bridged pair residues (Fig. 7A).
To study this dimer interface, we performed a series of residue substitutions on XccBphP and evaluated their oligomeric state by SLS-SEC in the apoprotein. The MW of the holoproteins could not be calculated because of the interference produced by BV in the light scattering measurements as it absorbs in the range of the laser wavelength (685 nm). This clarification is especially important because of the key role of the BV and the photoconversion in the interchange between the different oligomeric assemblies, as addressed below. Nevertheless, the study of the role of specific regions and residues of the apoprotein on the oligomeric state is still informative and allows us to validate the dimer-dimer interface observed by x-ray crystallography.
We focused on the Arg525/Glu614′, Arg525′/Glu614, Arg527/Asp528′, and Arg527′/Asp528 salt bridges, which stabilize the neighboring PHY-OM linkers and PAS9 domains. To assess their relevance in the Pr dimer formation, we generated a full-length variant replacing the Arg525 and Arg527 residues by Glu (opposite charge and similar side-chain size), named R525E-R527E, as well as two truncated versions lacking the OM, named ΔPAS9(1–525) and ΔPAS9(1–527). SLS-SEC experiments revealed that the three protein constructs predominantly exhibit monomeric species in solution with a minor dimeric population (Table 1 and fig. S15), in line with ΔPAS9(1–511), which is a monomer exclusively (35). This is indicative that the salt bridges together with the hydrogen bonds and nonbonded contacts along with the neighboring PHY-OM linkers and PAS9 domains are individually necessary but not sufficient to sustain the head-to-head dimer assembly. Analogously to the SLS-SEC data, ΔPAS9(1–525) and ΔPAS9(1–527) show lower Pr-to-Pfr dark conversion half-lives and a higher Pr:Pfr at equilibrium compared to the wild-type version, although these changes are slightly less pronounced than in ΔPAS9(1–511) (Fig. 1 and Table 1). R525E-R527E exhibited a considerably faster Pr-to-Pfr dark conversion rate (7.3-fold) compared to the wild-type version and an almost complete Pfr enrichment at dark equilibrium (97%) (Fig. 1 and Table 1). Together, these results indicate that the Pfr state is favored with respect to the Pr state when the integrity of the Pr dimeric arrangement is hindered.
The XccBphP Pfr photostate presents an antiparallel head-to-tail dimeric arrangement
In contrast to the noncrystallographic parallel dimer assembled in the Pr state, the G454E Pfr structure shows only one polypeptide chain in the crystal asymmetric unit (Fig. 3). However, the apoprotein G454E variant undoubtedly behaves as a dimer in solution according to SLS-SEC experiments (Table 1 and fig. S15). An in-depth search for symmetry-related crystallographic partners in the G454E crystal packing revealed an unusual head-to-tail quaternary assembly where both protomers are aligned nearly antiparallel and related by a twofold symmetry axis that can be constructed by means of the -x, -y, z symmetry operator of the I41 space group (Fig. 7B and fig. S13). This potential quaternary structure is exceptionally settled on the PHY domain from both protomers predominantly at the tongue regions sharing an interface area of 1080 Å2 (7.0% total residues of each chain). Outstandingly, the dimer interface displays an antiparallel-displaced (or offset stacked) π-π stacking interaction between Trp452/Trp452′ (from the Trp switch) along with the salt-bridge contacts Glu445/Arg483′ and Glu445′/Arg483 and a series of hydrogen bonds constituted by Arg371/Ser467′, Arg371′/Ser467, Asp418/Gln480′, and Asp418′/Gln480 (Fig. 7B). The substitution W452A generates a formation of higher-order oligomeric arrangements by SLS-SEC measurements (Table 1 and fig. S15), demonstrating that Trp452 plays an important role in the XccBphP oligomerization. Note that the W478A substitution affecting the other residue involved in the Trp switch does not alter the dimeric arrangement of the apoprotein (Table 1 and fig. S15), in agreement with the observation that this residue is not involved in the dimer interface in either photostate.
The substituted Glu454 residue is also located at the tongue region in the dimer interface likely interacting with the protomer partner Lys460. However, no clearly defined electron density was observed around the latter residue side chain to unambiguously determine this interaction. Moreover, the A453E and G454A variants, which exhibit a highly similar photochemical behavior to G454E, also show a dimeric assembly in solution (Table 1 and fig. S15), suggesting that the glutamate substitution does not drive the crystallographic head-to-tail dimer but might contribute to its stabilization. Noticeably, the double substitution W452A-G454E restores the dimeric state (Table 1 and fig. S15), which indicates that G454E governs the dimeric arrangement, in agreement with the fact that the G454E variant favors more the Pfr photostate than the W452A variant (Fig. 1 and Table 1). Collectively, these findings suggest that the effect of the G454E substitution is mainly on the photochemical behavior, which is ultimately driving the dimeric head-to-tail arrangement.
The crystallographic dimer observed in G454E is absent in the ΔPAS9(1–511) crystal packing (fig. S13), which is in agreement with the monomeric behavior observed in solution for this construct (35). As shown before, the PHY domain in G454E is more exposed than in ΔPAS9(1–511), possibly pushed by the OM (Fig. 4A, left inset). As the protomer-protomer antiparallel interactions in G454E are between the neighboring PHY domains, as mentioned above, we hypothesize that the difference in the oligomeric state between both constructs is a consequence of the PHY domain displacement.
The Pr head-to-head dimeric arrangement is not possible in the Pfr tertiary structure reported here because of steric clashes between the PAS9 domains in the contracted position and the changes in the helical spine trajectory (GAF-PHY and PHY-OM helical linkers). Nevertheless, to evaluate the feasibility of the crystallographic tongue-settled antiparallel assembly, Gaussian accelerated MD (GaMD) calculations combined with adaptive Poisson-Boltzmann solver (APBS) electrostatic calculations were performed and compared to a projected parallel head-to-head model without the PHY-OM helical linker break (see fig. S18 and details in Materials and Methods). The results show that the configurational and binding energies are similar both in quaternary arrangements and within the range of a stable dimer (table S2). Hence, we cannot completely discard the existence of Pfr parallel dimer arrangements, taking into account the calculated stability of the one projected here.
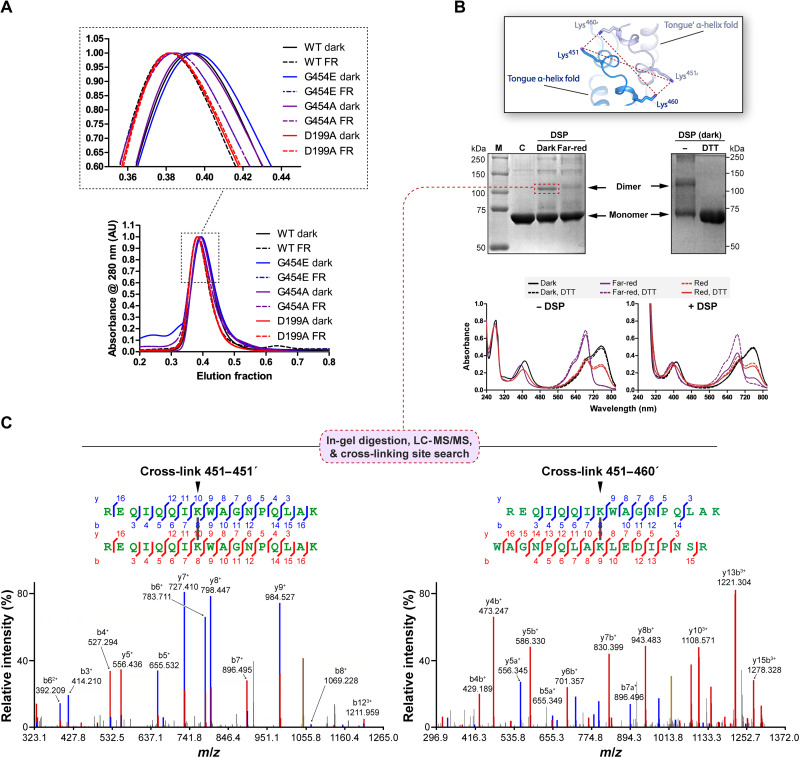
To probe the existence of different dimeric arrangements in solution for the Pr and Pfr states, we performed SEC experiments with wild-type, G454E, and G454A holoproteins under far-red light or dark conditions, maximizing the relative abundance of these photostates, respectively. All variants showed a single and symmetric peak and present almost identical elution fractions that vary with the light conditions. The three samples illuminated with far-red light (before and during the SEC run) showed a clear shift toward lower elution fractions compared to the dark-adapted samples (Fig. 8A). In addition, the peak of the control variant D199A, which does not photoconvert and is locked in a Pr-like state (39), overlaps with the peaks of the far-red–irradiated bathy samples, regardless of the light treatment (Fig. 8A). Together, these results are in agreement with XccBphP transitioning between two dimeric assemblies in response to light.
Fig. 8. Experiments in solution validate the head-to-tail arrangement in the wild-type full-length XccBphP.
(A) SEC of dark-adapted (solid lines) or far-red–irradiated (FR; dashed lines) of wild-type XccBphP, G454E, G454A, and D199A holoprotein variants. Inset: Peak region detailed. (B) Top: Representation of the potential interactions (red dashed lines) between the only pair of primary amines (here Lys residues) located at cross-linking distance in the dimeric interface of the Pfr G454E crystal structure colored following Fig. 7B (inset). Middle: Dimer cross-linking of wild-type XccBphP. The dark-adapted or far-red–illuminated holoproteins were incubated with the cross-linker DSP in the absence or presence of DTT and subjected to SDS-PAGE (M, markers; C, control of dark-adapted wild-type protein without DSP). Bottom: UV-Vis spectra of XccBphP samples previously incubated with DSP and/or DTT were sequentially recorded in the dark, after far-red light and red light treatments. (C) MS/MS spectra corresponding to the top-scoring cross-links of the dimeric band [indicated in (B) with red dashed box] after in-gel trypsin digestion. Left: Spectrum of a 6H+ ion (precursor mass, 4189.1840) matched to peptide 444–460 linked to the same peptide in the other subunit, with a DSP cross-link between Lys451-Lys451′ (top), with a SIM-XL score of 7.59. Right: Spectrum of a 4H+ ion (precursor mass, 4090.0669) matched to peptide 444–460 in one subunit linked to peptide 452–468 in the other subunit, with a DSP cross-link between Lys451-Lys460′ (top), with a SIM-XL score of 4.53. In the spectra, ions are colored red or blue according to their peptide of origin (α or β). Peaks in yellow correspond to the precursor ion. AU, arbitrary units; LC-MS/MS, liquid chromatography–MS/MS.
Last, with the aim of validating that the antiparallel quaternary arrangement observed in the G454E crystal exists in solution (discarding a crystallographic artifact) and whether it is also detected in the wild-type protein, we designed a cross-linking strategy based on the differential disposition of the residues at the protomer-protomer interface between the two dimer configurations (Fig. 8B). We used dithiobis(succinimidyl propionate) (DSP), a cross-linker that mainly reacts with primary amines (i.e., side chains of Lys residues and N termini). Taking into account that in the crystallographic quaternary assembly of the full-length Pfr structure only two pairs of primary amines are within the cross-linking distance range between subunits, namely, Lys451/Lys460′, Lys451′/Lys460, Lys451/Lys451′, and Lys460/Lys460′ (Fig. 8B, top, and fig. S19, bottom), and that in the Pr crystallographic structure all primary amines between subunits are distant from each other (fig. S19, top), the presence of a cross-linked dimer would be indicative of the existence of the antiparallel Pfr dimer (fig. S19, bottom). Wild-type dark-adapted or far-red–irradiated holoprotein samples were incubated in the presence of DSP, and then the samples were resolved in a denaturing nonreducing SDS–polyacrylamide gel electrophoresis (SDS-PAGE), breaking noncovalently linked dimers. A band corresponding to the cross-linked dimer was observed predominantly in the dark-adapted samples, enriched in the Pfr form, whereas the samples irradiated with far-red light, enriched in the Pr form, showed a clear curtail in the dimeric population (Fig. 8B, middle left). As expected, when the cross-linked samples incubated in the dark were treated with dithiothreitol (DTT), the dimeric band was abolished as a result of the reduction of the DSP disulfide bond (Fig. 8B, middle right, and fig. S20A).
To corroborate that the cross-linked sites in the Pfr state are among the Lys residues mentioned above, we conducted a mass spectrometry (MS) determination of the band corresponding to the dark-adapted DSP-treated dimer (Fig. 8B, middle left). The SDS-PAGE band was subjected to in-gel trypsin digestion, followed by MS/MS analysis of the peptides and cross-linked site identification (fig. S20A). The unbiased search using a specialized software for all possible cross-linked peptides revealed that the region from Arg443/Arg444 to Arg468 (containing Lys451 and Lys460) is a cross-linking “hotspot” between subunits. In particular, the top-scoring cross-links Lys451-Lys451′ and Lys451-Lys460′ (or Lys451′-Lys460) are the most prominent, supported by a large number of experimental observations (Fig. 8C). Note that in a homodimer, the cross-links between peptides that overlap in sequence cannot be from the same molecule, and thus, they must originate from different subunits (intersubunit interaction sites) exclusively. When data from the monomer fraction were subjected to the same analysis, none of these intersubunit cross-links could be found (fig. S20B). The cross-linking site 451–460, within a single tryptic peptide (loop-link), was found only in the monomeric band (fig. S20B).
Then, we sought to determine whether the cross-linking between protomers affects the dimer photochemistry. To that aim, we performed three sequential UV-Vis measurements of (i) dark-adapted samples incubated with DSP, (ii) after far-red irradiation, and (iii) after red light treatment, in the presence or absence of DTT (reducing agent) (Fig. 8B, bottom). All the dark-adapted samples show typical Pfr UV-Vis spectra with a peak at ~752 nm. In the control samples without DSP, the 752-nm peak completely disappears after far-red light treatment, indicating a full transition to Pr. In contrast, the DSP-treated XccBphP sample that was irradiated with far-red light presented an incomplete Pr photoconversion, presumably corresponding to the cross-linked fraction of the XccBphP sample (Fig. 8B, bottom right). The remaining 752-nm peak disappears with the addition of DTT by reduction of the cross-linker arm. On the other hand, upon red light irradiation, the DSP- or DSP/DTT-treated XccBphP samples did not differ from the control (−DSP), which presents both bands at ~685 and ~752 nm (Fig. 8B, bottom), suggesting that the Pr form was not cross-linked. Overall, the results of the cross-link XccBphP UV-Vis spectroscopic analysis indicate that the cross-linked head-to-tail dimer corresponds to the Pfr form. In addition, this experiment also supports that the quaternary arrangement governs the photochemical behavior of this phytochrome (that is, blocking the XccBphP head-to-tail configuration also blocks the Pfr-to-Pr photoconversion), an aspect of the allostery that may have important functional implications. In conclusion, the cross-linking/MS data, together with the spectroscopic and SEC experiments, validate the head-to-tail arrangement of the Pfr G454E crystal structure in the wild-type version, as well as the light-gated transition between the two dimeric assemblies.
DISCUSSION
Despite the vast structural information reported on photoreceptors, their dynamic protein structures pose a challenging matter in structural photobiology. To date, some valuable studies have reported structural data of a full-length phytochrome in both its Pr and Pfr photostates. From those, Vierstra’s and Westenhoff’s groups have revealed models (at >10-Å resolution) of the two forms of DrBphP by electron microscopy (25, 46) and small-angle x-ray scattering (47), respectively, but with different proposed mechanisms for the OM activation (“opening model” and “rotational model,” respectively). On the other hand, Winkler’s group has recently solved the crystal structures of the Idiomarina sp. A28L IsPadC in both photostates, from which another mechanism for the OM activation has been proposed (“register model”); however, although the Pr state was crystallized as a homodimer (48), the Pfr state was defined in a heterodimeric assembly (one protomer in Pfr and the other one in Pr) derived from a constitutively active mutant variant (40).
The lack of full-length structures solved at the (near-)atomic resolution in both pure (nonmixed) Pr and Pfr states leaves gaps in the structural mechanisms involved in the signal transmission pathways during phytochrome photoconversion. The XccBphP crystal structures reported here in both photostates, combined with experimental and computational studies, allowed us to describe a more complete and precise landscape on the light-driven conformational changes from the chromophore to the OM during the reversible photoswitching in a full-length phytochrome.
The structural changes in the chromophore-binding pocket and the tongue region upon the BV isomerization from the Pr ZZZssa to the Pfr ZZEssa configuration agreed well with those reported in other BphP structures solved in each photostate (8, 9). From those, definitely the most notable ones are the orientation of the chromophore-binding cysteine residue (Cys13 in XccBphP) from α-facial to β-facial, the flip-and-rotate movements of two highly conserved tyrosine residues surrounding the chromophore ring D (Tyr168 and Tyr195 in XccBphP), and the tongue adopting an antiparallel β-sheet to an α-helix conformation. This tongue-fold interconversion, elegantly confirmed in the same BphP using dark and illuminated crystal structures of the PSM from DrBphP (28), is a hallmark contrasting both structural photostates.
The extended protein elements associating the photo-induced changes in the chromophore-binding pocket with the tongue conformation have not been fully characterized yet. Despite this, a series of structural rearrangements that associate both regions in the different photostates have been exposed in most of the BphP structures solved to date (8, 9). Our structures are in complete concordance with these structural features, confirming that the contrasts between the Pr and Pfr structures previously reported are preserved in a full-length BphP. Among these, the most relevant ones involve the aspartate residue from the DIP motif (Asp199 in XccBphP) located in the chromophore-binding pocket, which is found to be interacting with the arginine and serine residue from the highly conserved PRxSF motif at the PHY tongue (Arg472 and Ser474 in XccBphP), in the Pr and Pfr states, respectively. This interdomain contact swapping is accompanied by an interchange in the position of two conserved tryptophan residues from the motifs W-G/A-G (Trp452 in XccBphP) and WxQ (Trp478 in XccBphP) from the Trp switch (13).
According to our results, the residues Leu193 and His194, which dwell outside the chromophore-binding pocket but close to Tyr195 (from the tyrosine flip-and-rotate mechanism), are also associative structural elements between these regions. The Leu193 side-chain movement, driven by the flipping of Tyr195, seems to affect the hydrophobic pocket settled in the tongue-GAF interface observed in the Pfr state. We postulate that this pocket, not previously described for other BphPs, is interconnected with the hydrophobic area fashioned around ring D by the conserved tyrosine pair. XccBphP polar variants at position 193 (i.e., L193H, L193T, L193R, L193E, and L193C) produced Pr-favored proteins, while hydrophobic substitutions (i.e., L193F and L193M) generated Pfr-favored variants (37). This supports the observation that the substitution of this residue by polar ones may cause a destabilization of the interdomain hydrophobic pocket, arranged in the Pfr state. Leucine is the most frequent residue present at the homologous position from Leu193 in other phytochromes (fig. S21), indicating that the above mechanism might be shared among them. This position has been previously described to be key in the Pr-to-Pfr photoconversion by other groups. Yang et al. (18, 24) have shown in the bathy-type PaBphP that a substitution in the homologous position (Gln188) by a leucine residue affected its photocycle, producing a mixed Pr-Pfr state. In the same line, Burgie et al. (30) have confirmed that substitutions of the homologous residue in DrBphP (His201) markedly compromise its full Pr-to-Pfr photoconversion. More recently, a study on the PSM from Agp2-PAiRFP2, a fluorescence-optimized BphP derived from A. fabrum Agp2, has revealed that Phe192 flipping (Tyr195 in XccBphP) upon BV isomerization produces a positional shift of Gln190 (Leu193 in XccBphP), which would induce a tongue refolding to the Pr state by steric hindrance with Trp440 (Leu458 in XccBphP) (16). This mechanism might share some analogous features to the one proposed here for XccBphP, because Leu458 is structured inside in the hydrophobic pocket in the Pfr state and disordered in the Pr state (Fig. 5B).
On the other hand, during the Trp switch, His194 is found to be interacting with Trp452 in Pr or Trp478 in Pfr. This interaction seems to be more crucial in the Pfr state, as revealed by the photochemical behavior of the H194A variant (Fig. 1 and Table 1). Because His194 can act as a cation or as a π-system depending on its protonation state, the nature of its interaction cannot be assigned solely on the basis of the structural data. Nevertheless, positive residues or residues carrying a partial positive charge are mostly found at the homologous position of His194 in phytochromes (fig. S21). Thus, it is reasonable to propose that the contact between this position and the aromatic residues from the tongue motifs W(G/A)G and (W/Y/F)x(E/Q) may be a conserved cation-π interaction. However, further research is needed to evaluate the nature of this GAF-tongue contact and its influence on downstream signal transduction. Together, our structures suggest that residues −2 and −1 from the second (from N to C terminus) flipping tyrosine position may be important linkers between the tongue and the chromophore-binding pocket during the Pr-to-Pfr photoconversion.
Undoubtedly, one of the most relevant concerns is how the long-range structural changes are propagated from the PSM to the OM during the Pr-Pfr photoconversion. There is a broad consensus that the tongue-fold interconversion and the associated structural motions affect the PHY domain position by push/pull movements and thus the main helical spine trajectory (9, 28). This large-scale structural rearrangement in the PSM, also termed “toggle model” (7, 29), is proposed to be essential for modulating the OM position and its activation/deactivation after the photoactivation cascade. However, this notion mostly arises from structures derived from truncated BphP versions (without a complete OM). A recent characterization of the PSM from DrBphP by nuclear magnetic resonance experiments in solution proposed a pathway for the signal transduction from the chromophore-binding pocket to the helical spine via the figure-of-eight knot along with the PAS and GAF domains (49). These regions remain invariable in the Pr and Pfr structures reported here. Thus, this mechanism is absent or is consigned to intermediate states during the XccBphP photoconversion.
Our structures reveal a prominent reorientation in the PHY position with alterations in the GAF-PHY and PHY-OM helical linkers along with a likely inversion of the quaternary assembly when contrasting both states. Helical deviations have been frequently reported in sensory histidine kinases, comprising photoreceptors, which are crucial for protein signaling (50, 51), and helical hinges are common in transmembrane proteins, being associated with their functional dynamics (52).
The GAF-PHY helical linker has been proposed to be correlated with the photostate, being straight in Pfr and curved in Pr (9, 15). We observed the same structural behavior in both full-length photostates, where the Pr structure shows a helix bending (kink 1). The comparison between the Pfr structures from G454E and ΔPAS9(1–511) also displays a torsion (albeit less pronounced), which indicates that this region has a particular flexibility and may be affected by the OM omission. Thus, changes in the GAF-PHY helical linker from PSM fragments should be cautiously analyzed when comparing different photostates.
The PHY-OM helical linker connects the PSM with the OM, and thus, it is a key structural element for coupling the light perception and the allosteric response. In this region, we noted three variations between both photostates: a change in the trajectory (hinge), a helical bending in Pr (kink 2), and an unprecedented helical disruption in Pfr (break). The analysis of the XccBphP structures revealed that the PHY-OM helical linker trajectory is altered by the push/pull effect on the PHY domain generated by the tongue interconversion, validating the toggle model in a full-length BphP (please refer to Fig. 5D for the structures aligned at the GAF domains and to Fig. 6A, right, for the structures aligned at the PHY domains).
As the Pr dimer interface is largely arranged on the helical spine, these helical rearrangements on the GAF-PHY and PHY-OM helical linkers should partially or completely destabilize the head-to-head dimeric assembly and, consequently, open it. Strong and crucial Pr dimer interactions such as Glu328/Arg501 and Glu505/Arg509 stabilizing the GAF-PHY/PHY-OM and PHY-OM/PHY-OM helical linkers, respectively, are switched by intramolecular interactions in the Pfr state. This scenario is also supported by the Pfr structures of the PSM from DrBphP and the (near) full-length RpBphP1, which showed an open dimer conformation (12, 28). Moreover, previous works on DrBphP revealed that some of the dimers were opened at the histidine kinase region (25) and that the dimerization interface between the OMs could be broken by light absorption (53). Therefore, the dissociation of the dimer interface is a possible way of action during photoconversion. In this regard, the contribution of the OMs to the dimer interface in the Pr head-to-head quaternary assembly would respond to why the constructs ΔPAS9(1–511), ΔPAS9(1–525), and ΔPAS9(1–527) mainly behave as monomers with preference to the Pfr. In addition, the two point substitutions in the R525E-R527E variant, which were designed to disturb the Pr dimer interface involving the OMs, generated a monomeric full-length Pfr-favored protein. These results indicate that the Pfr state is favored with respect to the Pr state when the dimeric interface interactions from the OMs are absent, which is in total agreement with the crystallographic structures. Accordingly, our computational analysis showed lower stability on the PHY-OM helical linker in a hypothetical monomeric scenario, compared to the dimer, which would facilitate the unprecedented disruption (break) in the helical spine and subsequently generate an abrupt change in the OM position.
The breaking point of the PHY-OM helical linker is located at Phe512. This residue has been previously identified by our group as a potential key structural element in the light-driven conformational changes of XccBphP (35). Aromatic residues have high propensity to be on helix kinks, and the (F/Y/W)xxx(F/Y/W) motif (512FQQDF516 in XccBphP) has been frequently observed in kinked or broken helices (54). Moreover, the probability of helical disruption for phenylalanine is larger than for other aromatic residues (55). It is unclear how the helical breaking is established. A possible scenario is that the abolition of the Arg436-Phe512 cation-π interaction, triggered by the PHY domain relocation in the Pfr state, provokes an exposition of the Phe512 hydrophobic side chain to the solvent. This change in the hydration environment may generate a conformational rearrangement in Phe512 that breaks the hydrogen bond between its carbonyl group and the Phe516 (i + 4) amide group of the helical turn and thus introducing a destabilizing kink in the α helix.
The combined structural motions described here might be followed by a dimer arrangement switch from head to head settled on the helical spine in Pr to head to tail settled on the tongue in Pfr. The functional dimerization assembly in this kind of photoreceptors is still controversial but could be crucial in the regulation of the OM activation. Although parallel assemblies are the most observed in BphPs, antiparallel arrangements have also been reported (10–16). However, in both cases, the dimer interface is typically arranged on the helical spine from both protomers. The parallel/antiparallel quaternary remodeling between the Pr and Pfr states reported here and supported by spectroscopic, biophysical, biochemical, and computational approaches associates three major structural players of BphP upon photoconversion: the tongue, the helical spine, and the OMs. In addition, the remodeling is in line with a proposed signal output (de-)activation in which both OMs might be needed to work in concert or separated in the different photostates (7). The in vivo experiments from this and previous studies demonstrating that the Pr state acts as a negative regulator of virulence-associated mechanisms (34, 39), combined with the structural information about the Pr and Pfr states, indicate that the PAS9 domains from the OM are active in the parallel configuration within the XccBphP dimer. The antiparallel Pfr configuration dissociates the PAS9 domains, which would disrupt their inhibitory activity.
PAS domains play different roles in bacterial proteins, such as signal sensors, modulators and transducers, dimerization motifs, protein interaction motifs, and cellular localization determinants (56). However, its function as OM in XccBphP still remains to be elucidated. Taking into account that XccBphP PAS9 domain does not present a recognizable biochemical activity, it may interact via protein-protein interactions with downstream signaling partners. It has been reported that c-di-GMP intracellular levels in Xcc regulate processes including xanthan production and biofilm formation (57, 58), which are involved in the light-gated signaling pathway through XccBphP (34, 39). Therefore, we hypothesize that the XccBphP partner may bear phosphodiesterase or diguanylate cyclase activities. The identification of the XccBphP partner system will not only shed light on the signaling pathway but also help to understand the functional implications of the photoconversion model proposed here.
Recently, Takala and colleagues (9) summarized the three probable structural mechanisms for OM activation: (i) opening model (25, 46), where a partial opening of the dimeric arrangement might take place; (ii) rotation model (47), which may involve a rotation of the dimer around the helical spine; and (iii) register model (40, 48), which might affect the helical spine register. According to our results, the XccBphP photoactivation may contain elements of the opening model in the aperture of the Pr dimer (“Y-shaped dimer”), followed by a previously unknown defined “joint-inversion model” to explain the events involved in the PHY-OM helical linker and the quaternary reassembly. As a whole, we propose a general mechanistic model for the photoconversion in XccBphP, starting in the photoreaction center and culminating with the long-range allosteric reorientation of the OM (Fig. 9). The main series of events comprises (i) the ring D isomerization of the BV molecule upon the absorption of a photon or dark conversion, (ii) the reorganization of the networks within and around the chromophore-binding pocket, (iii) the β-sheet/α-helix tongue transition, (iv) the PHY reorientation, with the concomitant GAF-PHY and PHY-OM helical linker changes, (v) the dimer dissociation, (vi) the PHY-OM helical linker rectify/breaking with the reorientation of the PAS9 domain, and (vii) the parallel/antiparallel dimer inversion assembly.
Fig. 9. Proposed XccBphP photoconversion model.
The Pr and Pfr structures are schematically represented on the left and right rounded panels, and the hypothetical steps involved in the photoconversion are depicted in between. Domains are colored according to Fig. 3. The main series of events comprises (i) BV ring D isomerization, (ii) reorganization of the networks within and around the chromophore-binding pocket, (iii) β-sheet/α-helix tongue transition, (iv) PHY reorientation, with the concomitant changes in the GAF-PHY and PHY-OM helical linkers, (v) dimer dissociation, (vi) PHY-OM helical linker rectification/break with the reorientation of the PAS9 domain, and (vii) parallel/antiparallel dimer inversion assembly. In the Pr-to-Pfr photoconversion, ring D isomerizes to the ZZEssa configuration followed by an α-helix conformational transition of the tongue. As a result, a pull movement of the PHY domain along with a straightening of the GAF-PHY and PHY-OM helical linkers triggers the detachment of the intertwined OMs (PAS9), generating a Y-shaped dimer. The loss of dimeric interactions dissociates the dimer, and the PHY-OM helical linker suffers a break, generating C-shaped monomers. These interact through a dimerization surface generated mainly by the tongue in the α-helix conformation, forming an antiparallel dimer. The Pfr-to-Pr photoconversion starts by ring D isomerization to the ZZZssa configuration, followed by a β-sheet transition of the tongue, disrupting the dimeric interface and pushing the PHY domain. As a result, a bending of the helical linkers occurs (Kinks 1 and 2) and the PHY-OM helical linker suffers a rectification, reconstituting the helical spine dimeric interface and the Pr dimer.
As the PSM overall architecture and the helical spine are conserved in several phytochromes, the mechanistic features exposed in this work can help to elucidate the downstream elements triggered by light in other members of this photoreceptor family, which are required to transduce the signal from photoreception to the biological response. However, as the nature of the OMs is variable and, consequently, their extended dimerization interfaces might have strong regulatory differences for the long-range signal transduction, diverse OM photoactivation mechanisms can occur depending on their specific function, thus preventing a universal common principle.
MATERIALS AND METHODS
Generation of the bacterial strains and culture conditions
The X. campestris pv. campestris 8004 (Xcc), Xccbphp knockout mutant, and Xccbphp complemented strains derive from a previous work (34). Briefly, the Xccbphp gene (XC_4241) was partially deleted and replaced by a 2-kb Smr/Spcr cassette (Ω) using two fragments from the flanking regions of Xccbphp. Complementation of Xccbphp was achieved by cloning a 2.7-kb fragment containing the complete XC_4242-XC_4241 operon and its regulatory sequences into the pBBR1MCS2 vector (yielding the pBBR-XccBphP vector), which was used to transform Xccbphp to generate the pXccBphP strain. The site-directed mutant G454E complemented strain was generated first by constructing the mutant vector pBBR-XccBphP-G454E (pG454E) by whole-plasmid amplification using Q5 High-Fidelity DNA Polymerase (New England Biolabs, Ipswich, MA, USA), the corresponding set specific primers (table S3), and the pBBR-XccBphP vector as template. Last, the template was digested from the reaction mix with the Dpn I restriction enzyme (New England Biolabs). Clones were isolated, and mutations were corroborated by Sanger sequencing. The resulting mutant vector was used to transform Xccbphp.
The Xcc and Xcc-derived strains were cultured in peptone-yeast extract-malt extract (PYM) medium (59) at 28°C with agitation (250 rpm). When required, antibiotics were used [rifampicin (50 μg/ml), kanamycin (50 μg/ml), and spectinomycin (100 μg/ml)]. Bacteria were grown under far-red (733 nm) continuous light at approximately 0.2 and 0.6 μmol m−2 s−1, respectively, from light-emitting diode (LED) sources or in darkness by covering the plates or flasks with double aluminum foil. All strains presented similar growth curves under the different light treatments (fig. S22).
Xanthan production
Xanthan quantification in liquid culture was performed as previously described (34). Briefly, bacterial strains were cultured in the dark or under red or far-red illumination for 48 hours at 28°C in 15 ml of PYM liquid medium with the addition of 2% glucose in 50-ml flasks, using an orbital shaker rotating at 130 rpm. After recording their optical density at 600 nm (OD600) values, cultures were centrifuged at 25,000g for 40 min and KCl was added to the supernatants to a final concentration of 1%. The crude xanthan was precipitated by adding 30 ml of 96% ethanol. All crude xanthan extracts and their corresponding cell pellets were collected, dried, and weighed. Results are expressed as milligrams of xanthan per milligram of cell pellet.
Western blot
Bacterial extracts from pG454E, pXccBphP, Xccbphp, and wild-type (Xcc) strains normalized by OD600 were loaded and separated by SDS-PAGE (12.5% gel). Proteins were transferred to Immobilon-P polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA). Membranes were blocked with nonfat milk in tris-buffered saline and 0.05% Tween 20 and incubated with an anti-XccBphP polyclonal antibody (1:500) and then with anti-mouse immunoglobulin G (Fc specific)–peroxidase antibody produced in goat (1:5000) (Sigma-Aldrich). Detection was achieved using the SuperSignal West Pico chemiluminescent substrate (Life Technologies Corporation, Carlsbad, CA, USA) on a Syngene G:BOX Chemi XRQ apparatus (Syngene, Cambridge, UK) (fig. S5).
Generation and purification of recombinant XccBphP variants
The pET24a-XccBphP expression vector coding for the wild-type version of the XccBphP protein [open reading frame (ORF) XC_4241] from X. campestris pv. campestris 8004 is from a previous work (36). The cloned ORF includes an N-terminal methionine and His-tag followed by the complete coding sequence (with the exception of its starting residue: 2 to 634), totaling 640 residues. Some mutant constructs were generated using pET24a-6xHis-XccBphP (36) as a template, through a Gibson assembly approach (namely, D177A, H194A, S280V, W452A, W452A-G454E, A453E, W478A, R501A, F512A, and F512P). Briefly, two independent amplicons, with 3′ and 5′ complementary regions, were generated from the template using specific primers (one set of primers bearing the specific mutation, while the other corresponds to the kanamycin cassette on the vector backbone; table S3). Then, both polymerase chain reaction (PCR) fragments were assembled following the Gibson assembly protocol, where only whole reconstituted vectors were able to transform bacteria. The R525E-R527E mutant plasmid was generated using pET-24a-XccBphP as a template for mutagenesis using whole-plasmid amplification followed by Dpn I template digestion (New England Biolabs). This was achieved using Q5 High-Fidelity DNA Polymerase (New England Biolabs), the corresponding set of specific primers (table S3), and digestion of the template from the reaction mix with Dpn I (New England Biolabs), following the manufacturer’s instructions. The mix was later used to transform bacteria. Last, the vectors coding for the truncated versions ΔPAS9(1–525) and ΔPAS9(1–527) were generated first by amplifying the pET-24a-XccBphP template by PCR using specific primers (table S3). The resulting amplicons were digested with Nde I and Bam HI and ligated with T4 ligase (New England Biolabs) into a pET-24a empty vector digested with the same restriction enzymes. The ligation was used for bacterial transformation. All cloning was performed in Escherichia coli DH10B, and the resulting mutations were confirmed by Sanger’s DNA sequencing. The constructs corresponding to pET-24a-XccBphP D199A, V266S, G454E, G454A, S474E, and ΔPAS9(1–511) were obtained in previous works (35, 37). All resulting vectors were used to transform E. coli BL21(DE3)pLysE for recombinant protein production.
Bacterial strains harboring each expression vector were cultured at 37°C in LB medium. When required, the antibiotics kanamycin (Km) and chloramphenicol (Cm) were added in final concentrations of 35 and 25 μg ml−1, respectively, and induced with a final concentration of 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) overnight at 20°C with agitation (250 rpm). Cells were harvested and ruptured by sonication, and apoproteins and holoproteins were purified by means of nickel-NTA affinity and SEC steps as described previously (36). For the case of holoproteins, a 1-hour incubation with BV at room temperature in the dark (Sigma-Aldrich) was performed before SEC. Protein concentration was estimated using the calculated molar extinction coefficients at 280 nm provided by the ExPASy ProtParam tool (https://web.expasy.org/protparam/) based on the polypeptide sequences. In the UV-Vis spectroscopy experiments, the protein buffer composition was 50 mM tris-HCl and 250 mM NaCl (pH 7.5). For crystallization, the final protein buffer composition was 10 mM tris-HCl and 25 mM NaCl (pH 7.6).
UV-Vis spectroscopy and data analyses
Dark assembly of wild-type XccBphP and G454E was performed by incubating the apoproteins with BV in a 2:1 molar ratio inside quartz cuvettes, in 50 mM tris-HCl and 250 mM NaCl (pH 7.5) at 25°C. UV-Vis spectra were recorded in the dark since the addition of BV, and the spectra were collected every 12 s for 0 to 15 min, every 30 s for 15 to 30 min, every 1 min for 30 to 45 min, every 2.5 min for 45 to 60 min, and every 10 min for 60 to 180 min. All UV-Vis measurements in this work were performed in a spectrophotometer (model Cary 60, Agilent Technologies) with an installed temperature control Peltier module (fig. S1).
Dark conversion experiments were initiated in quartz cuvettes containing holoprotein solutions of XccBphP or its variants at ~1 mg ml−1 in 50 mM tris-HCl and 250 mM NaCl (pH 7.5) at 25°C. Samples were irradiated for 20 min with red LED light (630 nm; fluence, 0.2 μmol m−2 s−1) or for 7 min with far-red LED light (733 nm; fluence, 0.6 μmol m−2 s−1). Sequential absorption spectra were recorded in the dark (fig. S2).
The pure-Pr and pure-Pfr spectra were calculated using data derived from the dark conversion datasets initially illuminated with far-red light and/or red light. The spectra most enriched in Pr (highest Abs684nm:Abs754nm ratio) or Pfr (lowest Abs684nm:Abs754nm ratio) in the variant datasets were selected (Pr-enriched and Pfr-enriched, respectively). The pure Pfr was calculated similarly as previously described by Assafa et al. (60). Briefly, a series of spectra was generated by subtracting the Pfr-enriched spectrum with increment fractions (0 to 1 with increment steps of 0.001) of the Pr-enriched spectrum. The derivative spectra of the subtraction series were subsequently calculated, and the minima and maxima around the wavelength corresponding to the Pfr maximum were determined. Last, the pure Pfr was estimated as the subtraction that minimized the difference of the derivatives corresponding to the abovementioned minima and maxima wavelengths (fig. S3A).
The pure Pr was estimated, first, by calculating a series of spectra subtracting the Pr-enriched spectrum with increment fractions (0 to 1 with increment steps of 0.001) of the pure-Pfr spectrum. Then, the derivative spectra of the subtractions were subsequently calculated, and for each one of them, a mean and SD were calculated in the range between the wavelength corresponding to the Pfr maxima and the highest wavelength in the dataset (820 nm). Last, the pure-Pr spectrum was calculated by the subtraction of the pure-Pfr fraction corresponding to the local maxima produced by the multiplication of the mean and SD values of the derivative spectra in the selected range (fig. S3B).
Then, for each spectrum, assuming to be composed of two pure components, we calculated the linear combinations of their pure-Pfr and pure-Pfr spectra to best fit the dataset using data corresponding to wavelengths above 510 nm. This allowed us to obtain the individual Pr and Pfr spectra at each time point that, added together, reconstituted the experimental dataset (fig. S4). The Pr and Pfr abundances (%) at each time were estimated as the relative spectral area corresponding to the pure Pr and the pure Pfr with respect to the sum of the areas (using the data corresponding to wavelengths above 510 nm).
The kinetic analysis was carried out by optimizing the parameters of a double exponential model (Eq. 1) and a monoexponential model (Eq. 2) to fit the Pr/Pfr abundance data. The Pr or Pfr equilibrium values were derived from parameter A from the double exponential fitting, while the monoexponential model was used to estimate half-life values using parameter H (Table 1)
| (1) |
| (2) |
As an example, the processed datasets for the wild-type version are available in GIF format as Supplementary Materials (GIFs S1 and S2), overlaying individual pure-Pr and Pfr spectral components, and their relative abundances at each time point. All steps of the analysis were performed using custom-made scripts developed in MATLAB software (version 2015b).
SLS-SEC measurements
The average MWs of XccBphP and its variants were determined using a PD2010 90° light scattering instrument (Precision Detectors) connected in tandem to a high-performance liquid chromatography system and an LKB 2142 differential refractometer detector (Pharmacia). Only MWs for apoproteins could be calculated due to the interference produced by BV in the 90° light scattering measurements because of its absorption in the laser wavelength (685 nm, AlGaInP). A Superdex 200 HR-10/30 column (24 ml; GE Healthcare Life Sciences) was used with isocratic elution in 50 mM tris-HCl and 250 mM NaCl (pH 7.7) at a flow rate of 0.4 ml min−1 at 20°C, with 0.08 to 0.22 mg of injected protein sample. The MW was calculated relating its 90° to the infrared signals using the software Discovery32 supplied by Precision Detectors. Bovine serum albumin (MW, 66.5 kDa) was used as a standard. The MW values were averaged as follows: (i) If the peak was clean (no major overlapping peak present), all the values corresponding to the upper half of the peak were included; (ii) if the peak was partially overlapped with another one, the MW values corresponding to the overlapping peak projection were discarded. For the sake of homogeneity, we chose the range of MW values for which the slope is null or almost null and the dispersion is minimal, in an MW versus elution fraction plot.
SEC elution fraction determination
Holoprotein samples of wild-type XccBphP and its variants G545E, G454A, and D199A were incubated for 24 hours in the dark and then either kept in the dark or irradiated for 7 min with a far-red LED source (733 nm; fluence, 0.6 μmol m−2 s−1) before the experiment. The dark-adapted samples were subjected to fast protein liquid chromatography (FPLC)–SEC at room temperature in a Superdex 200 HR-10/30 column. The total amount of the protein samples injected in the column was 0.1 mg. The column was covered in aluminum foil for the dark experiments, while the irradiated samples were kept under far-red light during the SEC. The SEC buffer was 50 mM tris-HCl and 250 mM NaCl (pH 7.5) at a flow rate of 0.6 ml min−1, and the recorded signal was absorbance at 280 nm. Data were normalized and plotted using GraphPad Prism software. The manipulation of the dark samples was performed under dim green-filtered light using a green acetate band-pass filter (LEE Lighting, catalog no. 090) with a transmission maximum at 525 nm. Figure 8A shows a representative experiment of three independent replicates.
Dimer cross-linking, electrophoresis, and UV-Vis spectroscopy
Dark-adapted or far-red–irradiated (pre-irradiation for 7 min and irradiation during the experiment with a far-red LED source of 733 nm; fluence, 0.6 μmol m−2 s−1) XccBphP wild-type samples at 1 mg ml−1 (14 μM) were incubated with 5- to 25-fold molar excess of DSP (Thermo Fisher Scientific) for 10 to 60 min at 22°C in a buffer containing 50 mM phosphate and 250 mM NaCl (pH 7.5) and in the absence or presence of 100 mM DTT. The cross-linking reactions were quenched with 100 mM tris-HCl (pH 7.5) (final concentration). Then, the loading buffer was added to the samples and resolved by SDS-PAGE (11% gel). The manipulation of the dark samples was performed under dim green-filtered light using a green acetate band-pass filter (LEE Lighting, catalog no. 090) with a measured maximum at 525 nm. The DSP cross-linking arm length is approximately 12 Å according to the manufacturer.
The UV-Vis experiments were performed in a way similar to the one described above (in the “UV-Vis spectroscopy and data analyses” section) and consisted of single scans. Samples of wild-type XccBphP (1 mg ml−1) in a buffer containing 50 mM tris-HCl and 250 mM NaCl (pH 7.5) at 25°C were first measured in the dark, then irradiated with far-red LED light (733 nm; fluence, 0.6 μmol m−2 s−1) for 7 min, and finally irradiated with red LED light (630 nm; fluence, 0.2 μmol m−2 s−1) for 20 min. Dark-adapted samples were previously incubated at 22°C with 30-fold molar excess DSP and/or 10 mM DTT for 3 hours in the dark.
Cross-linking mass spectrometry
Bands corresponding to the dimer and monomer fractions of dark-adapted DSP-treated wild-type XccBphP were excised from SDS-PAGE gels. Gel slices were destained with a 100 mM ammonium bicarbonate/acetonitrile (1:1, v/v) solution and incubated with MS-grade trypsin for 16 hours at 37°C with an enzyme/protein ratio of 1:20 (w/w). Peptides were extracted, evaporated to near dryness by vacuum centrifugation, and resuspended in water containing 1% (v/v) formic acid. Samples were desalted using C18 ZipTips following the recommendations of the manufacturer (Merck Millipore). The elution volume was 15 μl.
Peptides were separated on reversed-phase C18 columns (precolumn: Acclaim PepMap 100, 300 μm by 5 mm, 5 μm particle size; separation column: 50 cm by 75 μm internal diameter, PepMap RSLC C18, 2 μm particle size). Peptides were washed, eluted, and separated using the following program: 15 min 4% B [solvent A: 0.1% (v/v) formic acid in water; solvent B: 0.1% (v/v) formic acid in acetonitrile], 4 to 35% B linearly in 90 min, 35 to 90% B linearly in 20 min, 5 min 90% B, and 90 to 4% B in 5 min. All steps were carried out at a constant flow of 300 nl min−1. Peptides were analyzed in a Q-Exactive HF mass spectrometer (Thermo Fisher Scientific) equipped with an EASY-Spray source and coupled to an UltiMate 3000 RSLCnano system (Dionex). The mass spectrometer was operated in data-dependent MS/MS mode. Full MS1 scans were acquired at a 120,000 resolution at 200 mass/charge ratio (m/z) with a mass range of 375 to 2000 m/z and automatic gain control set to 3 × 106. Full MS1 scans were followed by MS2 scans of the 15 most intense precursor spectra. MS2 scans were recorded at a 30,000 resolution with automatic gain control set at 1 × 105 and an isolation window of 1.4 Th. Fragmentation was achieved using stepped higher-energy collision-induced dissociation and normalized collision energies of 27 and 30%. Precursor ions with charges of +1 and >8+ were excluded. Dynamic exclusion was set to 15 s.
Data were analyzed using SIM-XL (version 1.5.5), a software specialized in cross-linking analysis (61). The program was set as follows: Three trypsin missed cleavages were allowed, linear peptide minimum length was five, methionine oxidation was set as a variable modification, cross-linker was DSP (monoisotopic mass of modification 173.98092 Da and specificity of lysine residues and N terminus), and MS1 and MS2 mass accuracies were set to 5 and 20 parts per million, respectively. The database consisted of the protein sequence of XccBphP and 10 other nonrelated proteins. SIM-XL uses a scoring scheme to rank cross-linked peptides: Scores above 3 for interpeptide links and 2 for intrapeptide links suggest high-quality identifications. Results from SIM-XL were further analyzed with RawVegetable (62) using its in-built XL-Artifact analysis module to exclude that the identified interpeptide links were not the result of nonspecific interactions (63). A score of 0 indicates that the alleged cross-linked peptide is not an artifact, which was the case for all candidates identified in this work.
Crystallization
XccBphP was crystallized as reported previously (36). However, a slower adaptation of the crystals to the cryoprotectant before the flash-cooling into liquid nitrogen was crucial to reach a better resolution of the diffraction data in comparison with the existing structure (PDB entry 5AKP). For the ΔPAS9(1–511) construct and the G454E mutant, initial crystallization conditions were screened on sitting-drop Greiner 609120 96-well plates using a Honeybee963 robot (Digilab) and commercial kits from Jena Bioscience (Jena) and Hampton Research (Aliso Viejo) at 20 and 13 mg ml−1, respectively. In both cases, thin green bars appeared after a few weeks of equilibration at 21°C in several solutions of the kits. Some of these initial conditions were then optimized in 24-well hanging-drop Hampton Research VDX plates. In this sense, diffraction-quality ΔPAS9(1–511) crystals were grown with a precipitation solution consisting of 6% (w/v) polyethylene glycol (PEG) 4000, 0.1 M Hepes, and 10% (v/v) isopropanol (pH 6.7), whereas G454E crystals appeared with 30% (w/v) pentaerythritol propoxylate (5/4 PO/OH) and 0.1 M MES (pH 6.8). Samples were cryoprotected in their respective mother liquors added with 29% (w/v) PEG 400 for ΔPAS9(1–511) and 5% (w/v) PEG 400 for G454E and then flash-cooled in liquid nitrogen using synthetic mounted cryoloops (Hampton Research). In all cases, crystallization trials were performed in the dark, while crystal handling and vitrifying procedures were carried out under dim green-filtered light using a green acetate band-pass filter (LEE Lighting, catalog no. 090) with a measured maximum at 525 nm. Crystals were observed under the microscope using a 3-W power, 520-nm green LED light filtered with the same green acetate band-pass filter.
Data collection, structure resolution, and refinement
X-ray diffraction datasets were collected at the PROXIMA-1 and PROXIMA-2A beamlines at Synchrotron SOLEIL (France) on several crystals with the help of the MXCuBE application (64) using the classical and helical modes. Datasets were indexed, integrated, and scaled with XDS (65) and AIMLESS (66), leaving 5% of the reflections apart for cross-validation purposes. All structures were solved by the molecular replacement method with PHASER (67) using the coordinates of the wild-type protein as a search model (PDB entry 5AKP) (35) and checked for proper packing. Refinement and manual model building were then performed with BUSTER (68) and COOT (69), respectively. The 2mFo-DFc electron density maps allowed for a fairly complete trace of the protein backbones with the exception for the initial 8 to 10 N-terminal residues and the regions comprising the residues 457 to 469 for the wild-type protein; 393 to 398, 527 to 528, 601 to 606, and 616 to 623 for G454E; and 333 to 340 and 389 to 401 for ΔPAS9(1–511), which correspond to exposed loops. Two alternative chromophore structures were tested: (i) phytochromobilin-like configuration (i.e., tetrahedral C2 carbon at ring A and an exocyclic C3═C31 double bond) and (ii) BV-like configuration (i.e., endocyclic C2═C3 double bond at ring A and an exocyclic C3─C31 single bond). As the chromophore fitting in the electron density maps was better with the BV-like configuration, all the structures were refined with the BLA ligand (www.rcsb.org/ligand/BLA). The final models were validated with MolProbity (70). Table S1 shows the most relevant statistics on the data collection and processing steps, as well as the PDB deposition information.
MD simulations and electrostatic energy calculations
For the classical all-atom MD simulations, starting structures (wild-type Pr state, PDB entry 6PL0, and G454E Pfr state, PDB entry 7L59) were completed for missing loops (124 to 126 and 457 to 469 for 6PL0; 384 to 391, 519 to 520, 593 to 598, and 608 to 615 for 7L59) with the ab initio loop modeling tool of MODELLER (71). Both structures were initially simulated in monomeric states. All structures were solvated with a 10-Å octahedral TIP3P water box. Standard protonation states were assigned to titratable residues (Asp and Glu negatively charged, Lys and Arg positively charged). Histidine protonation was assigned to favor hydrogen-bond formation in the crystal structure. The protein system was modeled using Amber ff14SB as a force field (72), while for the parametrization of the BV chromophore, the electrostatic potential for the ligand was computed using Hartree-Fock and 6-31G* basis set as implemented in Gaussian 09 (73). Antechamber (74) with a RESP scheme was used to compute partial charges for the molecule. All systems were subjected to a mild minimization protocol of 1000 steps to remove bad contacts between atoms. The systems were slowly heated up from 10 to 300 K in 100 ps with a linear ramp in an NVT ensemble. Afterward, density equilibration was performed for 1 ns in an NPT ensemble. Simulations were run with Langevin thermostat with a collision frequency of 5 ps−1, while the pressure was kept constant with the Monte Carlo barostat. During both the heating and the equilibration process, a harmonic restraint (1 kcal mol−1 Å−2) on all Cα atoms was placed. All simulations were performed with Particle Mesh Ewald to treat long-range electrostatic interactions with a cutoff of 9 Å. All MD runs were performed with a 2-fs time step using the SHAKE algorithm to keep heavy atom–hydrogen bonds at equilibrium distances. The graphics processing unit (GPU)-accelerated version of pmemd (v18) was used to run the simulations (75). For each system, four independent 500-ns simulations were performed. To analyze these MD runs, dihedral angle computations and clustering were performed with CPPTRAJ (76), while statistical analysis and plotting were performed with R and ggplot2. For clustering, the dbscan algorithm was used with an epsilon of 0.9 Å.
To assess the dimer stability of the Pfr state, we additionally performed GaMD (77) simulations on the head-to-tail structural model and a projected head-to-head in silico model. While the head-to-tail dimer model was constructed using the G454E full-length crystallographic structure, the head-to-head model was designed via homology modeling, taking as a template the full-length dimeric assembly in the Pr state. Here, the PSM tertiary fold corresponds to that reported for the G454E monomer, and PAS9 was rearranged in the same conformation as found in the Pr state (without the PHY-OM helical linker break), thereby obtaining a full-length Pfr head-to-head dimer similar to the Pr assembly. The trajectories resulting from these simulations were further used for estimating average conformational energies and interaction energies between monomers (listed in table S2).
Simulations of XccBphP in aqueous solution were performed with Amber ff14SB, including BV parameters, as derived in a previous work (78). Minimization, heating, and thermal equilibration were performed following the same protocols described above for the XccBphP monomer models. These conventional MD simulations were extended by 500 ns of unconstrained GaMD simulations, with acceleration threshold σ0 = 5 (77), thereby enhancing conformational sampling. Electrostatic energy calculations on the two dimers were performed with APBS (79), with 545 grid points in the x, y, and z dimensions and a resolution of 0.3 Å on 500 snapshot structures derived from a 500-ns GaMD simulation.
GAF sequence analysis
To generate the GAF domain sequence logo (fig. S21), the Seq2Logo web server (www.cbs.dtu.dk/biotools/Seq2Logo/) was used with default parameters (Kullback-Leibler logo, 0.63 for Hobohm1 clustering, and 200 for weight on prior/pseudo counts) (80). The input was a multiple sequence alignment (MSA) of a diverse and nonredundant dataset of GAF domain sequences from phytochromes, derived from a previous work (37). Briefly, an initial phytochrome dataset was constructed with all sequences extracted from the UniProtKB and mapped UniRef50 database (for avoiding redundancy) and including the XccBphP sequence. Only sequences bearing a PAS2-GAF-PHY domain triad were selected. Subsequently, their corresponding GAF sequences were identified locally using HAMMER 3.2 software (http://hmmer.org/) and extracted using custom MATLAB scripts. This last dataset of 751 entries was used to construct the MSA using the Clustal-O program in the EBI server (www.ebi.ac.uk/Tools/msa/clustalo/) with default parameters. In addition, absolute and relative frequency counts were performed on the MSA for positions corresponding to Leu193 and His194 of XccBphP.
Structure representation
Molecular structures and their electron densities were represented using PyMOL Molecular Graphics System 1.8 (Schrödinger, USA). The electron density maps were generated in PHENIX (81).
Accession numbers
Coordinates and structure factors have been deposited in the PDB (http://wwpdb.org/) with accession numbers 6PL0 (wild-type XccBphP, Pr state), 7L59 (G454E, Pfr state), and 7L5A [ΔPAS9(1–511), Pfr state]. The MS data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (www.ebi.ac.uk/pride/) with the dataset identifier PXD026829.
Acknowledgments
We thank F. Velazquez-Escobar and D. Ferreiro for helpful advice in UV-Vis spectroscopy and frustration analyses, respectively. We also thank E. A. Ceccarelli for support in the MS analysis.
Funding: This work was supported by the Argentinian Ministry of Science (MINCyT), the Argentinian Research Council (CONICET), National Agency for the Promotion of Science and Technology of Argentina (ANPCyT) grants PICT 2015-0621 and PICT 2016-1425, and German Research Foundation (DFG) grant SFB1078. L.H.O., G.L.R., S.K., A.A.V., F.A.G., J.R., and H.R.B. are researchers from CONICET. M.S.-L. is supported by an ANPCyT fellowship. L.H.O. and S.K. acknowledge MINCyT for travel support. L.A.D. is supported by the EMBL Interdisciplinary Postdoc Programme under Marie Curie Actions COFUND 664726. We are grateful for the access to the PROXIMA-1 and PROXIMA-2A beamlines at Synchrotron SOLEIL, France (proposal nos. 20140791 and 20160817).
Author contributions: L.H.O., J.R., and H.R.B. designed and supervised the project. L.H.O., M.S.-L., S.F., J.R., and H.R.B. designed and performed the mutant constructs. S.F., G.T.A., J.R., and H.R.B. purified the proteins and performed UV-Vis spectroscopy experiments. H.R.B. performed data analysis of UV-Vis data and bioinformatics. G.T.A., J.R., and H.R.B. performed SEC-SLS experiments. J.R. analyzed SLS-SEC data. J.R. and S.F. performed the cross-linking assays. G.T.A., S.S., J.R., and S.K. crystallized the proteins. L.H.O., S.S., S.K., and L.M.G.C. performed x-ray diffraction data collection. L.H.O., S.S., and L.M.G.C. processed crystallographic data. L.H.O. solved and refined the crystallographic structures. L.A.D., G.B., and M.-A.M. performed MD simulations. G.T.A. and V.C. performed the in vivo experiments in Xcc. G.L.R. performed the cross-linking MS analysis and interpreted the data. L.H.O., J.R., and H.R.B. analyzed, interpreted, and discussed all results. L.H.O., A.A.V., F.A.G., L.M.G.C., and H.R.B. financed the project. L.H.O., J.R., and H.R.B. wrote and illustrated the paper with contributions from all authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S22
Tables S1 to S3
Legends for movies S1 and S2
Legends for data files S1 and S2
References
Other Supplementary Material for this manuscript includes the following:
Movies S1 and S2
Data files S1 and S2
REFERENCES AND NOTES
- 1.Rockwell N. C., Su Y.-S., Lagarias J. C., Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 57, 837–858 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Möglich A., Yang X., Ayers R. A., Moffat K., Structure and function of plant photoreceptors. Annu. Rev. Plant Biol. 61, 21–47 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Hughes J., Lamparter T., Mittmann F., Hartmann E., Gärtner W., Wilde A., Börner T., A prokaryotic phytochrome. Nature 386, 663 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Yeh K. C., Wu S. H., Murphy J. T., Lagarias J. C., A cyanobacterial phytochrome two-component light sensory system. Science 277, 1505–1508 (1997). [DOI] [PubMed] [Google Scholar]
- 5.Auldridge M. E., Forest K. T., Bacterial phytochromes: More than meets the light. Crit. Rev. Biochem. Mol. Biol. 46, 67–88 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Ulijasz A. T., Vierstra R. D., Phytochrome structure and photochemistry: Recent advances toward a complete molecular picture. Curr. Opin. Plant Biol. 14, 498–506 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Burgie E. S., Vierstra R. D., Phytochromes: An atomic perspective on photoactivation and signaling. Plant Cell 26, 4568–4583 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gourinchas G., Etzl S., Winkler A., Bacteriophytochromes—From informative model systems of phytochrome function to powerful tools in cell biology. Curr. Opin. Struct. Biol. 57, 72–83 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takala H., Edlund P., Ihalainen J. A., Westenhoff S., Tips and turns of bacteriophytochrome photoactivation. Photochem. Photobiol. Sci. 19, 1488–1510 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Essen L.-O., Mailliet J., Hughes J., The structure of a complete phytochrome sensory module in the Pr ground state. Proc. Natl. Acad. Sci. U.S.A. 105, 14709–14714 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mailliet J., Psakis G., Feilke K., Sineshchekov V., Essen L.-O., Hughes J., Spectroscopy and a high-resolution crystal structure of Tyr263 mutants of cyanobacterial phytochrome Cph1. J. Mol. Biol. 413, 115–127 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Bellini D., Papiz M. Z., Structure of a bacteriophytochrome and light-stimulated protomer swapping with a gene repressor. Structure 20, 1436–1446 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Anders K., Daminelli-Widany G., Mroginski M. A., von Stetten D., Essen L.-O., Structure of the cyanobacterial phytochrome 2 photosensor implies a tryptophan switch for phytochrome signaling. J. Biol. Chem. 288, 35714–35725 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang X., Stojković E. A., Ozarowski W. B., Kuk J., Davydova E., Moffat K., Light signaling mechanism of two tandem bacteriophytochromes. Structure 23, 1179–1189 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagano S., Scheerer P., Zubow K., Michael N., Inomata K., Lamparter T., Krauß N., The crystal structures of the N-terminal photosensory core module of agrobacterium phytochrome Agp1 as parallel and anti-parallel dimers. J. Biol. Chem. 291, 20674–20691 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmidt A., Sauthof L., Szczepek M., Lopez M. F., Escobar F. V., Qureshi B. M., Michael N., Buhrke D., Stevens T., Kwiatkowski D., von Stetten D., Mroginski M. A., Krauß N., Lamparter T., Hildebrandt P., Scheerer P., Structural snapshot of a bacterial phytochrome in its functional intermediate state. Nat. Commun. 9, 4912 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wagner J. R., Brunzelle J. S., Forest K. T., Vierstra R. D., A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome. Nature 438, 325–331 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Yang X., Kuk J., Moffat K., Crystal structure of Pseudomonas aeruginosa bacteriophytochrome: Photoconversion and signal transduction. Proc. Natl. Acad. Sci. U.S.A. 105, 14715–14720 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karniol B., Vierstra R. D., The pair of bacteriophytochromes from Agrobacterium tumefaciens are histidine kinases with opposing photobiological properties. Proc. Natl. Acad. Sci. U.S.A. 100, 2807–2812 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hahn J., Strauss H. M., Schmieder P., Heteronuclear NMR investigation on the structure and dynamics of the chromophore binding pocket of the cyanobacterial phytochrome Cph1. J. Am. Chem. Soc. 130, 11170–11178 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Rohmer T., Lang C., Hughes J., Essen L.-O., Gartner W., Matysik J., Light-induced chromophore activity and signal transduction in phytochromes observed by 13C and 15N magic-angle spinning NMR. Proc. Natl. Acad. Sci. U.S.A. 105, 15229–15234 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dasgupta J., Frontiera R. R., Taylor K. C., Lagarias J. C., Mathies R. A., Ultrafast excited-state isomerization in phytochrome revealed by femtosecond stimulated Raman spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 106, 1784–1789 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rockwell N. C., Shang L., Martin S. S., Lagarias J. C., Distinct classes of red/far-red photochemistry within the phytochrome superfamily. Proc. Natl. Acad. Sci. U.S.A. 106, 6123–6127 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang X., Kuk J., Moffat K., Conformational differences between the Pfr and Pr states in Pseudomonas aeruginosa bacteriophytochrome. Proc. Natl. Acad. Sci. U.S.A. 106, 15639–15644 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H., Zhang J., Vierstra R. D., Li H., Quaternary organization of a phytochrome dimer as revealed by cryoelectron microscopy. Proc. Natl. Acad. Sci. U.S.A. 107, 10872–10877 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song C., Psakis G., Lang C., Mailliet J., Gärtner W., Hughes J., Matysik J., Two ground state isoforms and a chromophore D-ring photoflip triggering extensive intramolecular changes in a canonical phytochrome. Proc. Natl. Acad. Sci. U.S.A. 108, 3842–3847 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang X., Ren Z., Kuk J., Moffat K., Temperature-scan cryocrystallography reveals reaction intermediates in bacteriophytochrome. Nature 479, 428–432 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takala H., Björling A., Berntsson O., Lehtivuori H., Niebling S., Hoernke M., Kosheleva I., Henning R., Menzel A., Ihalainen J. A., Westenhoff S., Signal amplification and transduction in phytochrome photosensors. Nature 509, 245–248 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burgie E. S., Bussell A. N., Walker J. M., Dubiel K., Vierstra R. D., Crystal structure of the photosensing module from a red/far-red light-absorbing plant phytochrome. Proc. Natl. Acad. Sci. U.S.A. 111, 10179–10184 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burgie E. S., Zhang J., Vierstra R. D., Crystal structure of deinococcus phytochrome in the photoactivated state reveals a cascade of structural rearrangements during photoconversion. Structure 24, 448–457 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Toh K. C., Stojkovic E. A., van Stokkum I. H. M., Moffat K., Kennis J. T. M., Proton-transfer and hydrogen-bond interactions determine fluorescence quantum yield and photochemical efficiency of bacteriophytochrome. Proc. Natl. Acad. Sci. U.S.A. 107, 9170–9175 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Escobar F. V., Piwowarski P., Salewski J., Michael N., Lopez M. F., Rupp A., Qureshi B. M., Scheerer P., Bartl F., Frankenberg-Dinkel N., Siebert F., Mroginski M. A., Hildebrandt P., A protonation-coupled feedback mechanism controls the signalling process in bathy phytochromes. Nat. Chem. 7, 423–430 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Velázquez Escobar F., Buhrke D., Michael N., Sauthof L., Wilkening S., Tavraz N. N., Salewski J., Frankenberg-Dinkel N., Mroginski M. A., Scheerer P., Friedrich T., Siebert F., Hildebrandt P., Common structural elements in the chromophore binding pocket of the Pfr state of bathy phytochromes. Photochem. Photobiol. 93, 724–732 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Bonomi H. R., Toum L., Sycz G., Sieira R., Toscani A. M., Gudesblat G. E., Leskow F. C., Goldbaum F. A., Vojnov A. A., Malamud F., Xanthomonas campestris attenuates virulence by sensing light through a bacteriophytochrome photoreceptor. EMBO Rep. 17, 1565–1577 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Otero L. H., Klinke S., Rinaldi J., Velázquez-Escobar F., Mroginski M. A., Fernández López M., Malamud F., Vojnov A. A., Hildebrandt P., Goldbaum F. A., Bonomi H. R., Structure of the full-length bacteriophytochrome from the plant pathogen xanthomonas campestris provides clues to its long-range signaling mechanism. J. Mol. Biol. 428, 3702–3720 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Klinke S., Otero L. H., Rinaldi J., Sosa S., Guimarães B. G., Shepard W. E., Goldbaum F. A., Bonomi H. R., Crystallization and preliminary x-ray characterization of the full-length bacteriophytochrome from the plant pathogen Xanthomonas campestris pv. campestris. Acta Crystallogr. F Struct. Biol. Commun. 70, 1636–1639 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Antelo G. T., Sánchez-Lamas M., Goldbaum F. A., Otero L. H., Bonomi H. R., Rinaldi J., A spectroscopy-based methodology for rapid screening and characterization of phytochrome photochemistry in search of Pfr-favored variants. Photochem. Photobiol. 96, 1221–1232 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Bianco M. I., Toum L., Yaryura P. M., Mielnichuk N., Gudesblat G. E., Roeschlin R., Marano M. R., Ielpi L., Vojnov A. A., Xanthan pyruvilation is essential for the virulence of Xanthomonas campestris pv. campestris. Mol. Plant Microbe Interact. 29, 688–699 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Conforte V., Otero L. H., Toum L., Sirigu S., Antelo G. T., Rinaldi J., Foscaldi S., Klinke S., Chavas L. M. G., Vojnov A. A., Goldbaum F. A., Malamud F., Bonomi H. R., Pr-favoured variants of the bacteriophytochrome from the plant pathogen Xanthomonas campestris hint on light regulation of virulence-associated mechanisms. FEBS J. 288, 5986–6002 (2021). [DOI] [PubMed] [Google Scholar]
- 40.Gourinchas G., Heintz U., Winkler A., Asymmetric activation mechanism of a homodimeric red light-regulated photoreceptor. eLife 7, e34815 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.von Stetten D., Seibeck S., Michael N., Scheerer P., Mroginski M. A., Murgida D. H., Krauss N., Heyn M. P., Hildebrandt P., Borucki B., Lamparter T., Highly conserved residues Asp-197 and His-250 in Agp1 phytochrome control the proton affinity of the chromophore and Pfr formation. J. Biol. Chem. 282, 2116–2123 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Wagner J. R., Zhang J., von Stetten D., Günther M., Murgida D. H., Mroginski M. A., Walker J. M., Forest K. T., Hildebrandt P., Vierstra R. D., . analysis of Deinococcus radiodurans bacteriophytochrome reveals key amino acids necessary for the photochromicity and proton exchange cycle of phytochromes. J. Biol. Chem. 283, 12212–12226 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parra R. G., Schafer N. P., Radusky L. G., Tsai M.-Y., Guzovsky A. B., Wolynes P. G., Ferreiro D. U., Protein Frustratometer 2: A tool to localize energetic frustration in protein molecules, now with electrostatics. Nucleic Acids Res. 44, W356–W360 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ferreiro D. U., Hegler J. A., Komives E. A., Wolynes P. G., On the role of frustration in the energy landscapes of allosteric proteins. Proc. Natl. Acad. Sci. U.S.A. 108, 3499–3503 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferreiro D. U., Hegler J. A., Komives E. A., Wolynes P. G., Localizing frustration in native proteins and protein assemblies. Proc. Natl. Acad. Sci. U.S.A. 104, 19819–19824 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burgie E. S., Wang T., Bussell A. N., Walker J. M., Li H., Vierstra R. D., Crystallographic and electron microscopic analyses of a bacterial phytochrome reveal local and global rearrangements during photoconversion. J. Biol. Chem. 289, 24573–24587 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Björling A., Berntsson O., Lehtivuori H., Takala H., Hughes A. J., Panman M., Hoernke M., Niebling S., Henry L., Henning R., Kosheleva I., Chukharev V., Tkachenko N. V., Menzel A., Newby G., Khakhulin D., Wulff M., Ihalainen J. A., Westenhoff S., Structural photoactivation of a full-length bacterial phytochrome. Sci. Adv. 2, e1600920 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gourinchas G., Etzl S., Göbl C., Vide U., Madl T., Winkler A., Long-range allosteric signaling in red light-regulated diguanylyl cyclases. Sci. Adv. 3, e1602498 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Isaksson L., Gustavsson E., Persson C., Brath U., Vrhovac L., Karlsson G., Orekhov V., Westenhoff S., Signaling mechanism of phytochromes in solution. Structure 29, 151–160.e3 (2021). [DOI] [PubMed] [Google Scholar]
- 50.Möglich A., Signal transduction in photoreceptor histidine kinases. Protein Sci. 28, 1923–1946 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buschiazzo A., Trajtenberg F., Two-component sensing and regulation: How do histidine kinases talk with response regulators at the molecular level? Annu. Rev. Microbiol. 73, 507–528 (2019). [DOI] [PubMed] [Google Scholar]
- 52.Bowie J. U., Membrane protein twists and turns. Science 339, 398–399 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Takala H., Björling A., Linna M., Westenhoff S., Ihalainen J. A., Light-induced changes in the dimerization interface of bacteriophytochromes. J. Biol. Chem. 290, 16383–16392 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilman H. R., Shi J., Deane C. M., Helix kinks are equally prevalent in soluble and membrane proteins. Proteins 82, 1960–1970 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang Y.-H., Chen C.-M., Statistical analyses and computational prediction of helical kinks in membrane proteins. J. Comput. Aided Mol. Des. 26, 1171–1185 (2012). [DOI] [PubMed] [Google Scholar]
- 56.Stuffle E. C., Johnson M. S., Watts K. J., PAS domains in bacterial signal transduction. Curr. Opin. Microbiol. 61, 8–15 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dow J. M., Maxwell Dow J., Fouhy Y., Lucey J. F., Ryan R. P., The HD-GYP domain, cyclic di-GMP signaling, and bacterial virulence to plants. Mol. Plant Microbe Interact. 19, 1378–1384 (2006). [DOI] [PubMed] [Google Scholar]
- 58.An S.-Q., Potnis N., Dow M., Vorhölter F.-J., He Y.-Q., Becker A., Teper D., Li Y., Wang N., Bleris L., Tang J.-L., Mechanistic insights into host adaptation, virulence and epidemiology of the phytopathogen Xanthomonas. FEMS Microbiol. Rev. 44, 1–32 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cadmus M. C., Rogovin S. P., Burton K. A., Pittsley J. E., Knutson C. A., Jeanes A., Colonial variation in Xanthomonas campestris NRRL B-1459 and characterization of the polysaccharide from a variant strain. Can. J. Microbiol. 22, 942–948 (1976). [DOI] [PubMed] [Google Scholar]
- 60.Assafa T. E., Anders K., Linne U., Essen L.-O., Bordignon E., Light-driven domain mechanics of a minimal phytochrome photosensory module studied by EPR. Structure 26, 1534–1545.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 61.Lima D. B., de Lima T. B., Balbuena T. S., Neves-Ferreira A. G. C., Barbosa V. C., Gozzo F. C., Carvalho P. C., SIM-XL: A powerful and user-friendly tool for peptide cross-linking analysis. J. Proteomics 129, 51–55 (2015). [DOI] [PubMed] [Google Scholar]
- 62.Kurt L. U., Clasen M. A., Santos M. D. M., Souza T. A. C. B., Andreassa E. C., Lyra E. B., Lima D. B., Gozzo F. C., Carvalho P. C., RawVegetable—A data assessment tool for proteomics and cross-linking mass spectrometry experiments. J. Proteomics 225, 103864 (2020). [DOI] [PubMed] [Google Scholar]
- 63.Giese S. H., Belsom A., Sinn L., Fischer L., Rappsilber J., Noncovalently associated peptides observed during liquid chromatography-mass spectrometry and their effect on cross-link analyses. Anal. Chem. 91, 2678–2685 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gabadinho J., Beteva A., Guijarro M., Rey-Bakaikoa V., Spruce D., Bowler M. W., Brockhauser S., Flot D., Gordon E. J., Hall D. R., Lavault B., McCarthy A. A., McCarthy J., Mitchell E., Monaco S., Mueller-Dieckmann C., Nurizzo D., Ravelli R. B. G., Thibault X., Walsh M. A., Leonard G. A., McSweeney S. M., MxCuBE: A synchrotron beamline control environment customized for macromolecular crystallography experiments. J. Synchrotron Radiat. 17, 700–707 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kabsch W., XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Evans P. R., Murshudov G. N., How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J., Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.G. Bricogne, E. Blanc, M. Brandl, C. Flensburg, P. Keller, W. Paciorek, P. Roversi, A. Sharff, O. S. Smart, C. Vonrhein, T. Womack , BUSTER Version XYZ (Global Phasing Ltd., 2011).
- 69.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen V. B., Arendall W. B. III, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C., MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Eswar N., Eramian D., Webb B., Shen M.-Y., Sali A., Protein structure modeling with MODELLER. Methods Mol. Biol. 426, 145–159 (2008). [DOI] [PubMed] [Google Scholar]
- 72.Maier J. A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K. E., Simmerling C., ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.M. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, B. Mennucci, G. Petersson, H. Nakatsuji, M. Caricato, X. Li, H. P. Hratchian, A. F. Izmaylov, J. Bloino, G. Zheng, J. L. Sonnenberg, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, J. A. Montgomery Jr., J. E. Peralta, F. Ogliaro, M. Bearpark, J. J. Heyd, E. Brothers, K. N. Kudin, V. N. Staroverov, R. Kobayashi, J. Normand, K. Raghavachari, A. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, N. Rega, J. M. Millam, M. Klene, J. E. Knox, J. B. Cross, V. Bakken, C. Adamo, J. Jaramillo, R. Gomperts, R. E. Stratmann, O. Yazyev,A. J. Austin, R. Cammi, C. Pomelli, J. W. Ochterski, R. L. Martin, K. Morokuma, V. G. Zakrzewski, G. A. Voth, P. Salvador, J. J. Dannenberg, S. Dapprich, A. D. Daniels, Ö. Farkas, J. B. Foresman, J. V. Ortiz, J. Cioslowski, D. J. Fox, Gaussian 09, Revision D.01 (Gaussian Inc., 2009).
- 74.Wang J., Wang W., Kollman P. A., Case D. A., Antechamber: An accessory software package for molecular mechanical calculations. J. Am. Chem. Soc. 222, U403 (2001). [Google Scholar]
- 75.Salomon-Ferrer R., Götz A. W., Poole D., Le Grand S., Walker R. C., Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh Ewald. J. Chem. Theory Comput. 9, 3878–3888 (2013). [DOI] [PubMed] [Google Scholar]
- 76.Roe D. R., Cheatham T. E. 3rd, PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 9, 3084–3095 (2013). [DOI] [PubMed] [Google Scholar]
- 77.Miao Y., Feher V. A., McCammon J. A., Gaussian accelerated molecular dynamics: Unconstrained enhanced sampling and free energy calculation. J. Chem. Theory Comput. 11, 3584–3595 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Battocchio G., González R., Rao A. G., Schapiro I., Mroginski M. A., Dynamic properties of the photosensory domain of Deinococcus radiodurans bacteriophytochrome. J. Phys. Chem. B 124, 1740–1750 (2020). [DOI] [PubMed] [Google Scholar]
- 79.Jurrus E., Engel D., Star K., Monson K., Brandi J., Felberg L. E., Brookes D. H., Wilson L., Chen J., Liles K., Chun M., Li P., Gohara D. W., Dolinsky T., Konecny R., Koes D. R., Nielsen J. E., Head-Gordon T., Geng W., Krasny R., Wei G.-W., Holst M. J., McCammon J. A., Baker N. A., Improvements to the APBS biomolecular solvation software suite. Protein Sci. 27, 112–128 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thomsen M. C. F., Nielsen M., Seq2Logo: A method for construction and visualization of amino acid binding motifs and sequence profiles including sequence weighting, pseudo counts and two-sided representation of amino acid enrichment and depletion. Nucleic Acids Res. 40, W281–W287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liebschner D., Afonine P. V., Moriarty N. W., Poon B. K., Sobolev O. V., Terwilliger T. C., Adams P. D., Polder maps: Improving OMIT maps by excluding bulk solvent. Acta Crystallogr. D Struct. Biol. 73, 148–157 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Engh R. A., Huber R., Accurate bond and angle parameters for x-ray protein structure refinement. Acta Crystallogr. A 47, 392–400 (1991). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S22
Tables S1 to S3
Legends for movies S1 and S2
Legends for data files S1 and S2
References
Movies S1 and S2
Data files S1 and S2