Abstract
Cryptosporidium is a life-threating protozoan parasite belonging to the phylum Apicomplexa, which mainly causes gastroenteritis in a variety of vertebrate hosts. Currently, there is a re-emergence of Cryptosporidium infection; however, no fully effective drug or vaccine is available to treat Cryptosporidiosis. In the present study, to better understand the detailed interaction between the host and Cryptosporidium parvum, a large-scale label-free proteomics study was conducted to characterize the changes to the proteome induced by C. parvum infection. Among 4406 proteins identified, 121 proteins were identified as differentially abundant (> 1.5-fold cutoff, P < 0.05) in C. parvum infected HCT-8 cells compared with uninfected cells. Among them, 67 proteins were upregulated, and 54 proteins were downregulated at 36 h post infection. Analysis of the differentially abundant proteins revealed an interferon-centered immune response of the host cells against C. parvum infection and extensive inhibition of metabolism-related enzymes in the host cells caused by infection. Several proteins were further verified using quantitative real-time reverse transcription polymerase chain reaction and western blotting. This systematic analysis of the proteomics of C. parvum-infected HCT-8 cells identified a wide range of functional proteins that participate in host anti-parasite immunity or act as potential targets during infection, providing new insights into the molecular mechanism of C. parvum infection.
Author summary
Cryptosporidium parvum is an emerging zoonotic pathogen transmitted via the fecal–oral route, and is considered a leading cause of moderate-to-severe diarrheal disease in young children in resource limited areas. After infection, C. parvum parasitizes intestinal epithelial cells and evokes an inflammatory immune response, leading to severe damage of the intestinal mucosa. The infection can be lethal to immunosuppressed individuals. However, no fully effective drug or vaccine is available for cryptosporidiosis, and the pathogenesis and immune mechanisms during C. parvum infection are obscure. Thus, an in-depth understanding of host-parasite interaction is needed. Hence, we established a C. parvum-infected HCT-8 cell model and performed comparative quantitative proteomic analyses to profile global host-parasite interactions and determine the molecular mechanisms that are activated during infection, aiming to offer new insights into the treatment of Cryptosporidium.
1. Introduction
Cryptosporidium is an opportunistic pathogen with a worldwide distribution, which infects a variety of vertebrates (including humans, mammals, reptiles, amphibians, and poultry) [1]. Immunocompetent individuals experience a self-limiting illness after C. parvum infection; immunocompromised hosts suffer from more severe and prolonged gastrointestinal disease that can be fatal [2,3]. Nearly 40 genotypes of Cryptosporidium have been established, and it is likely that after further biological and molecular characterization, many of these will eventually be given species status [4].
Cryptosporidium parasitizes mainly the epithelial cells of the gastrointestinal and respiratory tracts, evoking host epithelial defense responses mediated by Toll like receptors (TLRs) [5–8]. In contrast to other apicomplexans, such as Toxoplasma gondii and Plasmodium falciparum, Cryptosporidium has lost the plastid and mitochondrial genomes, and both the asexual and sexual stages are completed within a single host [9–11]. As a result, Cryptosporidium metabolism is almost exclusively based on glycolysis, which is likely to be the reason why it parasitizes mainly gut epithelial cells [9,10]. Interestingly, C. parvum does not fully invade the host cell, but resides intracellularly in the parasitophorous vacuole with an epicellular location.
Cryptosporidium forms an actin-rich disk as a feeder organelle, which is an important strategy for successful nutritional uptake and rapid replication [12–16]. However, the molecular mechanisms in the host and parasite that lead to this epicellular niche remain unknown. Moreover, evidence shows that some host-parasite homologous genes of the host glycolysis/gluconeogenesis pathways are downregulated, while host-exclusive genes are upregulated during invasion and intracellular development. This might suggest parasite-derived competition for metabolic substrates [17]. Although evidence shows that multiple immune cells and effectors participate in resistance to C. parvum infection or parasite clearage, infected cells eventually die because of nutrient deprivation and disruption of the cytoskeleton [18–20]. Our knowledge of the molecular mechanisms of C. parvum-host interactions is limited and no effective anti-cryptosporidial therapies are available to treat cryptosporidiosis in children, patients with HIV/AIDS, and animals [21–25]. Thus, there is an urgent need to gain an in-depth understanding of host-parasite interactions and develop effective drugs and vaccines.
In the present study, we aimed to use a highly sensitive quantitative approach, combining label-free proteomic quantification techniques on a liquid chromatography-dual mass spectrometry (LC-MS/MS) platform with advanced bioinformatic analysis, to analyze the global proteome differences in C. parvum-infected and non-infected HCT-8 cells. Moreover, the authenticity and accuracy of the protein levels identified by the quantitative proteomic examination were further confirmed using western blotting and quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR). This study aimed to identify molecules that potentially play important roles during C. parvum infection.
2. Materials and methods
2.1 Sample preparation and collection
C. parvum oocysts of the Iowa strain were purchased from a commercial source (Waterborne, New Orleans, LA, USA). The HCT-8 human colon adenocarcinoma cell line (ATCC, Manassas, VA, USA) was cultured in Dulbecco’s modified Eagle’s medium (DMEM) nutrient mix F-12 (Gibco, Grand Island, NY, USA) containing 10% fetal bovine serum (FBS, Gibco). For infection, C. parvum oocysts were suspended in phosphate-buffered saline (PBS) with 20% sodium hypochlorite, centrifuged at 3, 667 × g, and washed twice with PBS. Infection was performed in culture medium containing viable C. parvum oocysts (at an oocyst to host cell ratio of 2:1) at 37°C in a humidified 5% CO2 incubator for 36 h. An equal volume of culture medium was added to the control group. After 36 h of incubation, the cells were washed three times with cold PBS and collected using a cell scraper. The cells were centrifuged at 200 × g for 10 min to obtain the cell pellet, which was stored at -80°C until analysis.
2.2 Quantitative real-time reverse transcription PCR (qRT-PCR)
Total RNA from the HCT-8 cells was extracted using the TRIzol reagent according to the manufacturer’s protocol (Takara Biotechnology, Inc., Shiga, Japan). The primers were designed by EnzyArtisan Biotech Co., Ltd. (Shanghai, China, Table 1). The cDNA was prepared using a PrimeScript RT–PCR Kit (Takara Biotechnology, Inc.) and then amplified using a SYBR qPCR Mix (EnzyArtisan Biotech). The ACTB (β-actin) gene was used as an endogenous control for normalization.
Table 1. List of primers used for qRT-PCR analysis.
| Gene | Primer sequence |
|---|---|
| ISG15 | Forward: CTCTGAGCATCCTGGTGAGGAA Reverse: AAGGTCAGCCAGAACAGGTCGT |
| IFITM3 | Forward: GTGCTGATCTTCCAGGCCTATG Reverse: TGGAGTACGTGGGATACAGGTCAT |
| PLSCR1 | Forward: ATTAAGAACAGCTTTGGACAGAGG Reverse: TCCTCAAGGTAAAAGGTCTAGATGG |
| NMI | Forward: GAAACGGAGTTACAAGAGGCTAC Reverse: GACAACTGGCTGTCATTCTCAGG |
| SFPQ | Forward:CTGTGTCATCCGCCATTTTGTG Reverse: GGAACCGATCCCGAGACATG |
| RBMX | Forward: CACCTCGAAGGGAACCGCTG Reverse: TCGTGGTGGTGGTGCATAATCTCTA |
| IFI35 | Forward: CACGATCAACATGGAGGAGTGC Reverse: GGCAGGAAATCCAGTGACCAAC |
| HELZ2 | Forward: GAGGTGCATCTGTGTCGTTTCC Reverse: CAGGATCTCAAAACTGCCGACAG |
| ATP5PO | Forward: GCCTAAATGACATCACAGCAAAAG Reverse: AGGCAGAAACGACTCCTTGGGT |
| ACTB | Forward: CACCATTGGCAATGAGCGGTTC Reverse: AGGTCTTTGCGGATGTCCACGT |
2.3 Protein extraction and trypsin digestion
More than 2 × 107 cells of each group were collected and sonicated three times on ice using a high intensity ultrasonic processor (Scientz, Zhejiang, China) in lysis buffer (8 M urea, 1% Protease Inhibitor Cocktail). The lysate was centrifuged at 12, 000 × g at 4°C for 10 min to separate the insoluble debris. Finally, the soluble supernatant was collected and the protein concentration was determined using a bicinchoninic acid (BCA) kit according to the manufacturer’s instructions (Beyotime, Jiangsu, China). Then, the protein supernatant was thoroughly digested using trypsin for further analysis.
2.4 LC-MS analysis and database searching
The tryptic peptides were loaded onto a laboratory-prepared reversed-phase analytical column (C18), subjected to a nanospray ionization (NSI) source followed by tandem mass spectrometry (MS/MS) in a Q Exactive Plus apparatus (ThermoScientific, Waltham, MA, USA) coupled online to an ultra-performance liquid chromatography (UPLC) column, and then selected for MS/MS using a normalized collision energy (NCE) setting of 28. The fragments were detected in the Orbitrap at a resolution of 17, 500. Automatic gain control (AGC) was set at 5e4 and the fixed first mass was set as 100 m/z. Then, the Maxquant search engine (v.1.5.2.8, Max Planck Institute of Biochemistry, Munich, German) was used for secondary mass spectral data process. All tandem mass (MS/MS) spectra were searched against human and C. parvum data in the Uniprot database, which was concatenated with a reverse decoy database to calculate the false discovery rate (FDR) caused by random matching. Trypsin/P was specified as the cleavage enzyme allowing up to two missing cleavages and the minimum length of the peptide segment was set as seven amino acid residues. 20 ppm and 5 ppm were set as the mass error tolerance of primary parent ions for the first and main search, respectively, and the mass error tolerance was set as 0.02 Da for secondary fragment ions.
2.5 Bioinformatic analysis
The UniProt-GOA database (http://www.ebi.ac.uk/GOA/), InterPro domain database (http://www.ebi.ac.uk/interpro/), and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/kaas-bin/kaas_main; http://www.kegg.jp/kegg/mapper.html) were used separately for Gene Ontology (GO) annotation, characterization of the proteins basic functions, domain functional descriptions, and annotation of biological processes or pathways. Then, a two-tailed Fisher’s exact test was employed to test the enrichment of the differentially expressed proteins (DEPs) against all identified proteins. A P-value < 0.05 was considered significant. All the categories obtained after enrichment were collected and then filtered for those categories that were at least enriched in one of the clusters with a P-value < 0.05. This filtered P-value matrix was transformed and then clustered using one-way hierarchical clustering in Genesis. Clusters were visualized using the R Package, pheatmap (https://cran.r-project.org/web/packages/cluster/).
We used Wolfpsort (http://www.genscript.com/psort/wolf_psort.html) to predict the subcellular locations of the proteins. COG (Clusters of Orthologous Groups of proteins)/KOG (euKaryotic Ortholog Groups) functional classification of the identified proteins was conducted through database comparison and analysis.
2.6 Protein-protein interaction network
All DEPs’ database accession numbers or sequences were searched against STRING database version 11[26] for protein-protein interaction (PPI) analysis. Only interactions between the proteins belonging to the searched data set were selected, thereby excluding external candidates. The interaction network from STRING was visualized in Cytoscape [27].
2.7 Western blotting
C. parvum infected and non-infected HCT-8 cells were incubated for 36 h before harvesting. Equal amounts of protein from total-cell lysates were separated using 12% SDS-PAGE (Beyotime) and transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). The membranes were then blocked at room temperature for 2.5 h in 5% nonfat dry milk in Tris-buffered saline-Tween 20 (TBS-T) buffer. Membranes were incubated with gentle rocking 1.5 h at room temperature with primary antibodies recognizing interferon-induced 15 KDa protein (ISG15) (1/5000, Abcam, Cambridge, MA, USA), interferon induced transmembrane protein 3 (IFITM3) (1/5000, Abcam), phospholipid scramblase 1 (PLSCR1) (1/5000, Abcam), N-Myc and STAT interactor (NMI) (1/1000, Proteintech, Wuhan, China), signal transducer and activator of transcription (STAT1) (1/2000, Proteintech), STAT2 (1/2000, Abcam), splicing factor proline and glutamine rich (SFPQ) (1/500, Abcam), RNA binding motif protein X-linked (RBMX) (1/5000, Abcam), S100 calcium binding protein A10 (S100A10) (1/2000, Abcam), and β-actin (1/20000, Proteintech) as a loading control. Membranes were washed with TBS-T five times for 10 min each, and then incubated with the appropriate secondary antibody for 1 h at room temperature. The immune complexes were then visualized using an enhanced chemiluminescence (ECL) detection kit (Tanon, Shanghai, China). The densities of the immunoreactive protein bands were determined using Image J software (NIH, Bethesda, MD, USA).
2.8 Statistical analysis
Data are presented as means ± SD. Statistical tests were performed using the GraphPad Prism 8 Statistics software package (GraphPad Inc., La Jolla, CA, USA). A value of P < 0.05 was considered statistically significant.
3. Results
3.1 Proteomic analysis of DEPs in non-infected and infected samples
A total of 4406 human proteins were identified, of which 3307 contained quantitative information, ranging in molecular weight from 1.9 kDa to 964.8 kDa (S1 Fig). Among them, 33, 812 acquired unique peptides belonged to 3307 proteins, which served as reliable evidence for these predicted proteins (S1 Fig). Most of the identified proteins contained no less than two unique peptides as multiple peptide-level evidence for protein identification (S1 Fig). By searching the database of C. parvum, we obtained 231 proteins and 563 unique peptides, indicating that a considerable number of proteins are single peptide chains (S2 Fig).
3.2 Bioinformatic analysis of host proteomic data
3.2.1 Subcellular localization of DEPs
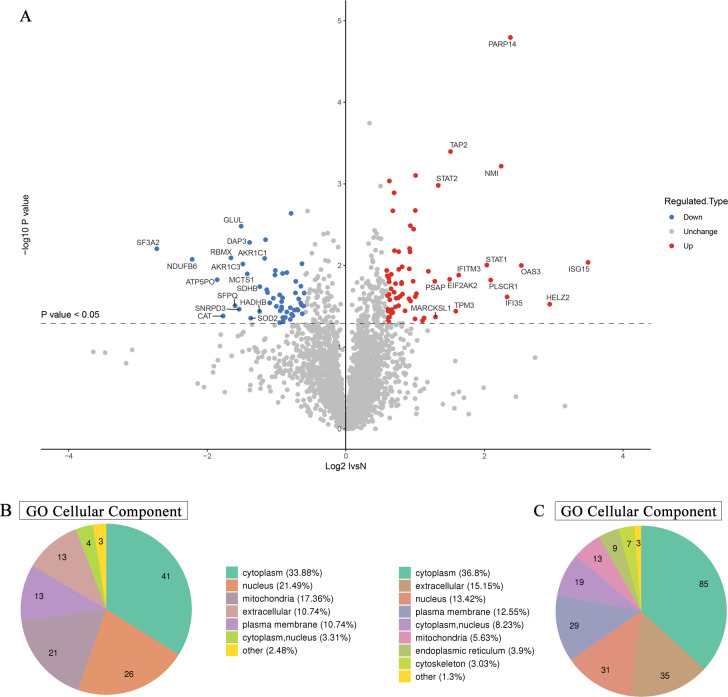
A mass spectrometry comparison between infected and non-infected HCT-8 cells identified 121 DEPs (1.5-fold difference in abundance), of which 64 were upregulated and 57 were downregulated after infection (Fig 1A and 1B). Interestingly, among the DEPs, those located on the plasma membrane were all upregulated after infection (S1 Table). Besides, the upregulated and downregulated proteins had various locations in the cellular compartments, including the cytoplasm, nucleus, mitochondria, and extracellular region (Fig 1B). In-depth analysis further demonstrated the different locations of DEPs caused by infection. Functional enrichment analysis indicated that the upregulated proteins were concentrated in the transporters associated with antigen processing (TAP) complex, extracellular space, intrinsic component of plasma membrane, and MHC class I peptide loading complex. The downregulated proteins were found to be mostly located in the organelle lumen, membrane-enclosed lumen, intracellular organelle lumen, microbody lumen, and peroxisomal matrix (S3 Fig). The obvious local distinction between upregulated and downregulated proteins suggested the special effect of C. parvum infection on various components of the cell.
Fig 1. C. parvum infection altered the levels of multiple proteins in host cells.
(A) Volcano plot of the most significantly DEPs between C. parvum infected and uninfected HCT-8 cells. The horizontal axis shows the relative quantitative value of protein levels after Log2 logarithmic conversion, the vertical axis show the P values of significant difference test after Log10 logarithmic conversion. In this figure, red dots represent proteins with significantly upregulated levels, while blue dots represent downregulated proteins. (B) Subcellular localization and distribution of DEPs in HCT-8 cells. (C) Location and classification of subcellular structures analysis of identified C. parvum proteins.
In addition, we conducted bioinformatic analysis on the intracellular parasite components, and 231 C. parvum-derived proteins were identified at 36 h post infection (Figs 1C and S4). These parasitic proteins were enriched significantly in the cytoplasm, extracellular space, nucleus, plasma membrane, nucleus, mitochondria, and endoplasmic reticulum (Fig 1C), which constitute the core components of C. parvum.
3.2.2 GO enrichment and KEGG pathway enrichment of DEPs
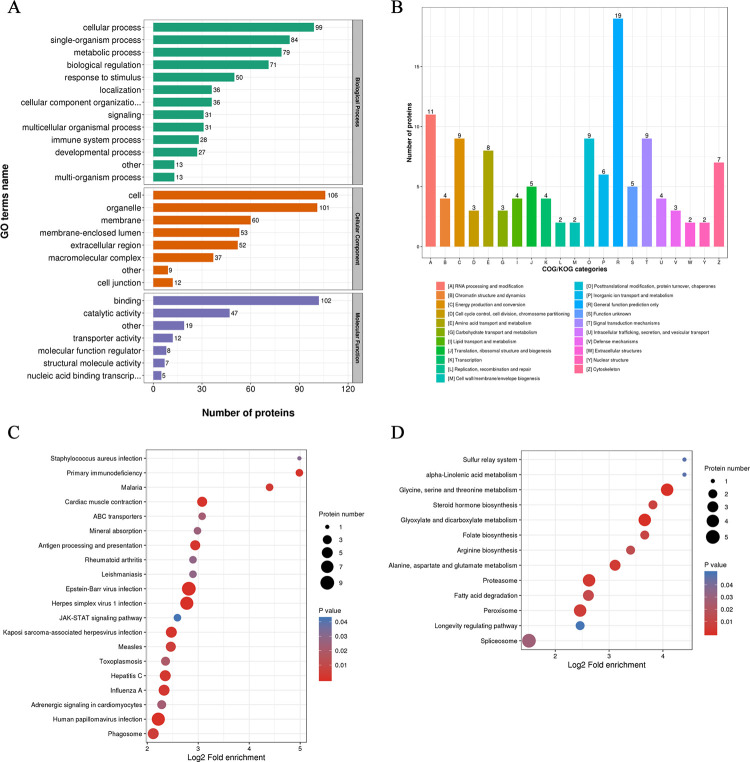
We further annotated and investigated the functions of the identified DEPs, with the aim of discovering important molecules or signaling pathways that play key roles during C. parvum parasitism. According to biological process analysis from the GO classification, the DEPs were closely involved in cellular process, single-organism process, metabolic process, response to stimulus, and biological regulation (Fig 2A). Among the 121 DEPs, 102 were identified as having binding functions and 47 had catalytic activity (Fig 2A). COG/KOG categories provided detailed functional classification of DEPs, suggesting that the C. parvum infection has extensive effects on various physiological activities of host cells, such as RNA processing and modification, signal transduction mechanisms, posttranslational modification, protein turnover, chaperones, energy production and conversion, amino acid transport, and metabolism (Fig 2B).
Fig 2. Bioinformatic characterization of the DEPs in host cells.
(A) GO classification of DEPs according to their associated biological process, cellular component, and molecular function terms. (B) COG/KOG functional classification of DEPs. (C) KEGG pathways enrichment analysis of the upregulated proteins. (D) KEGG pathways enrichment analysis of the downregulated proteins. The color represents the significance P-value of enrichment. The size of the circle represents the number of proteins involved.
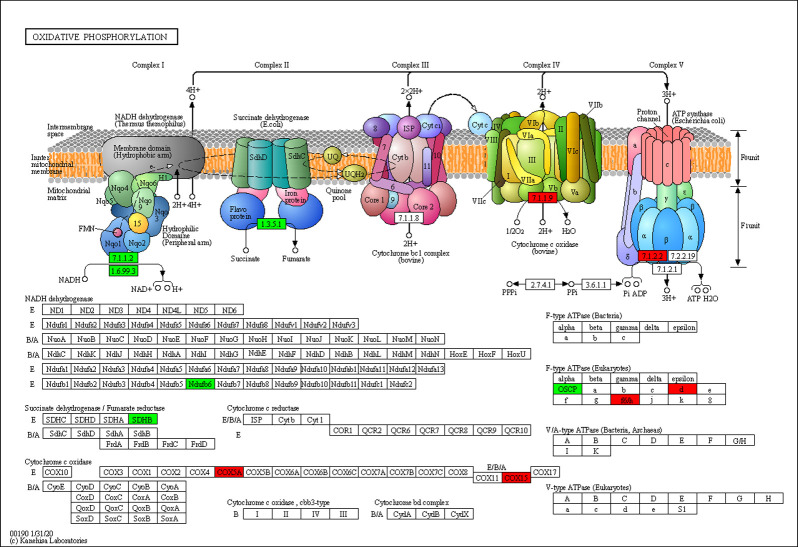
Based on further GO enrichment analysis, C. parvum infection seemed to evoke a series of cell defense reactions, in that the upregulated proteins were related to cellular response to type I interferon, and the defense response to virus and other organisms. By contrast, the downregulated proteins were functionally enriched for antibiotic metabolic processes (including aminoglycoside antibiotic and doxorubicin metabolic process), positive regulation of inclusion body assembly, protein oligomerization, and hypoxia response due to infection. Moreover, several signaling pathways were affected by C. parvum infection. KEGG pathway enrichment analysis of upregulated and downregulated proteins was conducted separately, which indicated an obvious distinction in functional clusters between the DEPs (Fig 2C and 2D). Remarkably, the upregulated proteins exhibited significant enrichment for inflammatory signaling pathways, and were involved in many pathogenic pathways of virus and bacterial infections, Malaria, Leishmaniasis, and Toxoplasmosis (Fig 2C). These findings suggested that in addition to some specific pathways, different parasites or viruses might target common pathways and machineries, which forms the basis for the discovery of broad range vaccines or drugs. By contrast, the downregulated proteins caused by C. parvum infection were found to be markedly enriched in multiple metabolic pathways, including metabolism of many amino acids and propanoate, biosynthesis of folate, unsaturated fatty acids and steroid hormone, and the peroxisome pathway (Fig 2D). Besides, C. parvum had a significant effect on the host oxidative phosphorylation process, with up to 12 related proteins being differentially abundant after infection (Fig 3). The extensive influence of C. parvum on host metabolic process revealed C. parvum’s need for a special cellular microenvironment, and also reflected its dependence on certain nutrients, which provided ideas for identifying effective drugs or inhibitors.
Fig 3. C. parvum affected the oxidative phosphorylation process of host cells.
The protein names in red showed upregulated levels and those in green showed downregulated levels in C. parvum-infected HCT-8 cells compared with uninfected cells.
3.3 Bioinformatic analysis of C. parvum proteomic data
To gain a deeper understanding of the biological characteristics of C. parvum, we also conducted a bioinformatic analysis on the identified C. parvum-derived proteins. The COG/KOG annotation revealed the predominant involvement of identified C. parvum-derived proteins in translation, ribosomal structure and biogenesis, posttranslational modification, protein turnover, and signal transduction processes (S4 Fig). During parasitism by C. parvum, identified proteins were enriched in the myelin sheath, lipid particle, and ATPase complex and were functionally specialized in the metabolic process of ATP, nicotinamide nucleotide, and ribonucleoside triphosphate (S4 Fig). KEGG pathway enrichment further illustrated a wide distribution of identified C. parvum-derived proteins in the ribosome after infection, which could be important for successful parasitism and replication (S4 Fig).
3.4 The DEPs exhibited close interactions in protein-protein interaction network
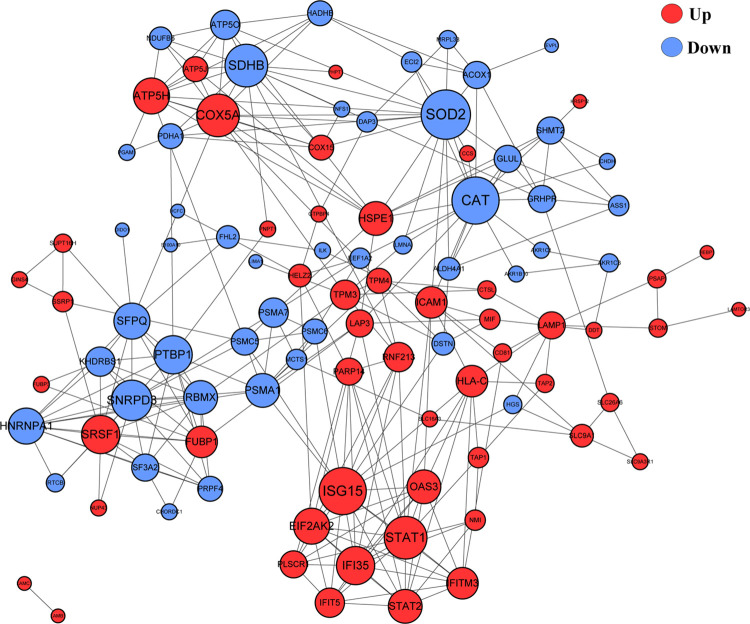
The variety and diverse functions of the identified proteins prompted us to construct a PPI network to better characterize the potential interaction of the host and C. parvum proteins. In total, 100 DEPs were mapped to a PPI network, comprising a dense interaction network of host proteins. Noticeably, several sub-networks with strong interactions were identified (Fig 4). The significant hub proteins included interferon stimulated gene 15 (ISG15), signal transducer and activator of transcription 1 (STAT1), cytochrome C oxidase subunit 5A (COX5A), succinate dehydrogenase (SDHB), superoxide dismutase 2 (SOD2), catalase (CAT), small nuclear ribonucleoprotein D3 polypeptide (SNRPD3), heat shock protein family E member 1 (HSPE1), proteasome subunit alpha type 1 (PMSA1), and polypyrimidine tract-binding protein 1 (PTBP1). All the hub proteins have potential roles in host-C. parvum interplay; however, the detailed mechanisms require further study. Besides, 109 C. parvum-derived proteins were cross referenced with the STRING and Cytoscape databases for protein interaction analysis (S5 Fig). Thirty-two proteins were determined and clustered with a high degree of network interaction, and the hub proteins were identified, which included cgd7_2280, elongation factor 2, and cgd3_2090, identifying a series of parasitic proteins of vital importance, which might represent the core functional proteins of C. parvum.
Fig 4. Association networks of DEPs using Cytoscape.
PPI network profiles the interplay of the identified host proteins. The color of the circle represents upregulated proteins (red) and downregulated proteins (blue) respectively. The size of the circle represents the significance of the protein.
3.5 Validation of the proteomic results
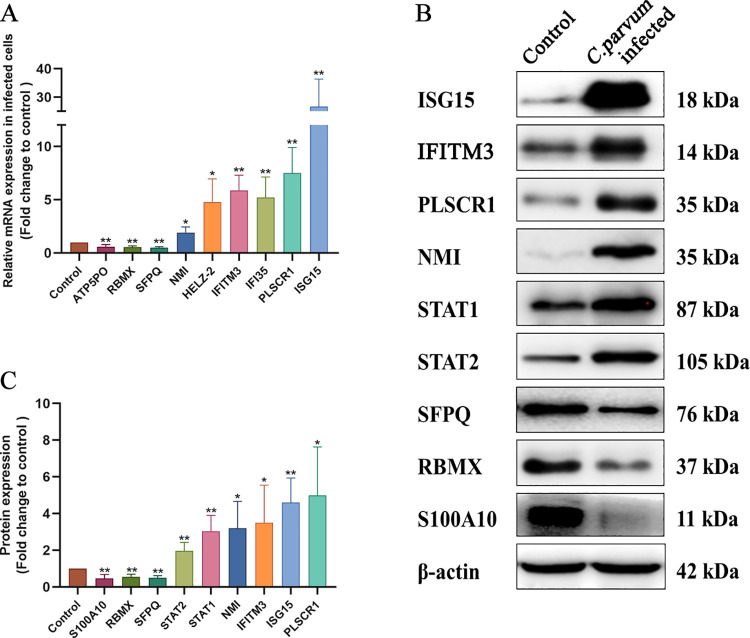
To assess the proteins that were identified as being closely related to the host anti-parasite mechanism, or associated with parasite development and pathogenicity during C. parvum infection, we verified their expression using qRT-PCR and western blotting. Firstly, the mRNA expression level of nine genes, including ISG15, IFITM3 (interferon-induced transmembrane protein 3), PLSCR1 (phospholipid scramblase 1), NMI (N-Myc and STAT interactor), IFI35 (interferon-induced protein 35), HELZ2 (helicase with zinc finger 2), SFPQ (splicing factor, proline- and glutamine-rich), RBMX (RNA-binding motif protein), and ATP5PO (ATP synthase peripheral stalk subunit OSCP) were compared in normal and infected HCT-8 cells, and the results were in accordance with the proteomic analysis (Fig 5A). Then, several proteins were chosen and further confirmed using western blotting, which showed that the levels of ISG15, IFITM3, PLSCR1, NMI, STAT1, signal transducer and activator of transcription 2 (STAT2), SFPQ, RBMX, and protein S100-A10 (S100A10) accorded with those in the proteomic analysis (Fig 5B and 5C).
Fig 5. Verification of the DEPs in normal and C. parvum-infected host cells.
Each experiment was repeated three times. (A) Relative mRNA expression levels of ATP5PO, RBMX, SFPQ, NMI, HELZ2, IFITM3, IFI35, PLSCR1, and ISG15 in normal and C. parvum-infected HCT-8 cells were verified using quantitative real-time reverse transcription PCR. The error bars represent the mean ± SD, from one experiment run in triplicate. Statistical significance was analyzed using an unpaired Student’s t-test (*P < 0.05, **P < 0.01). (B) Western blotting analysis was used to reconfirm the levels of the identified proteins with biological significance in infected and uninfected HCT-8 cells. (C) Histogram analysis of the western blotting results in infected and uninfected HCT-8 cells. Statistical significance was analyzed using an unpaired Student’s t-test (*P < 0.05, ** P < 0.01).
4. Discussion
As an enteric pathogen, C. parvum primarily infects the apical end of small intestinal enterocytes, where they are enveloped by host membranes but remain extracytoplasmic [28,29]. Intestinal epithelial cells (IECs) can sense and participate in the coordination of appropriate immune responses, ranging from tolerance to anti-pathogen immunity [30,31]. Hence, IECs are considered as the frontline sensors for pathogenic microorganism encounters [32,33]. The HCT-8 cell line possesses an excellent ability to support C. parvum growth after infection [34–36]. In this study, we established a C. parvum infection model using HCT-8 cells to mimic the pathogenesis and immune reaction during C. parvum invasion and development, aiming to provide key insights into how the host reacts effectively to infection. Our data revealed that the levels of 121 host proteins were altered during C. parvum infection, which possessed diverse functions, and profiled various biological process changes in infected host cells globally.
Based on bioinformatic analysis and recent research evidence, we focused on several significant signaling pathways and molecules that might serve as important targets during the anti- C. parvum immune response. GO analysis revealed that the interferon (IFN) signaling pathway was the most enriched biological process, in that seven (10%) of the 67 significantly upregulated proteins were identified as targets of IFN signaling; specifically, ISG15, 2’-5’-oligoadenylate synthetase 3 (OAS3), NMI, IFITM3, STAT1, STAT2, and IFI35. Interferon-induced protein ISG15 has been implicated as a core player during the host antiviral response by directly disrupting viral replication, budding, and release, as well as by limiting tissue damage and modulating human type I interferon signaling [37–43]. The biological relevance of ISG15 in parasitic infection has been demonstrated. Novel findings suggest that ISG15 expression was induced during Theileria annulata and Leishmania brazilensis infections, but not in L. amazonensis infection [44,45]. Recently, ISG15 was found to participate in autophagy-mediated ubiquitinoylation of the vacuole during T. gondii infection, as a bridge that links the ATG pathway with IFN-γ-dependent restriction of T. gondii in host cells [46]. In the present study, significantly increased ISG15 expression was confirmed, indicating a potential anti-parasitic function of ISG15 during C. parvum infection. The PPI network further exhibited strong interaction among ISG15 and other upregulated proteins including OAS3, IFITM3, IFI35, intercellular adhesion molecule-1 (ICAM-1), STAT1, STAT2, PLSCR1, eukaryotic translation initiation factor 2 alpha kinase 2 (EIF2AK2), and NMI, most of which are interferon-induced proteins. The upregulation of IFITM3, STAT1, STAT2, PLSCR1, and NMI after infection was validated using western blotting and qRT-PCR. Based on these results, we identified that the prevailing immune response of HCT-8 cells to C. parvum is centered on IFN signaling, revealing the involvement of unique proteins and a distinct immune process in C. parvum infection, which might be key regulators in the anti-C. parvum immune reaction.
ICAM-1 is a cell surface glycoprotein and an adhesion receptor that is expressed widely by endothelial, epithelial, and immune cells, and is responsible for leukocyte recruitment and signal transduction [47]. Under inflammatory conditions or stimulation by injury, the expression of ICAM-1 markedly increased in immune cells and epithelial cells [48–50]. Recently, ICAM-1 has also been proven to be an important cytoadherence receptor for Plasmodium chabaudi infection, and binds to Plasmodium falciparum erythrocyte membrane protein 1, suggesting its important role in parasitic protozoa infection [51,52]. Furthermore, ICAM-1 expression was induced in the rat jejunum during acute T. gondii infection [53]. Moreover, Chen et al. demonstrated that the upregulation of ICAM-1 in epithelial cells was modulated by microRNA-221 following C. parvum infection. ICAM-1 also enhanced the attachment of infected host cells [54]. In the present study, the robustly enhanced expression of ICAM-1 in C. parvum infected cells suggested that ICAM-1 might serve as a biosensor in the response to C. parvum infection and participate in the recruitment of immune cells. However, the detailed mechanism requires in-depth investigation.
In the present study, we observed that IFITM3 was upregulated at both the mRNA and protein levels after C. parvum infection. IFITM3 is an innate immune response protein generally known to inhibit the entry and replication of many viruses [55–57]. Individuals lacking IFITM3 are highly susceptible to infection, even when challenged with influenza virus of low pathogenicity [58]. Therefore, IFITM3 is considered as a first line of cell defense in response to viruses. Besides, the protective effect of IFITM3 is also reflected in the long survival of lung resident memory T cells, in which sustained IFITM3 expression facilitated their survival and protection from viral infection during subsequent exposure [59]. In addition, IFITM3 could mediate the autophagic degradation of interferon regulatory factor 3 (IRF3) and negatively regulates tissue-damaging inflammatory production of type I IFNs induced by the virus [60]. Accordingly, we hypothesized that the enhancement of IFITM3 in response to C. parvum infection is the result of host immune defense and a self-regulating mechanism to prevent fatal inflammation after C. parvum infection; however, the mechanism awaits future exploration.
STAT1 and STAT2 proteins are key mediators of IFN signaling, and are essential components in the cellular antiviral response and adaptive immunity [61]. In various parasite infections, such as those by T. gondii and Schistosoma japonicum, STAT1 and STAT2 are evoked and mediate the immune response [62,63]. NMI and IFI35 are both IFN-induced proteins: NMI enhances STAT-mediated transcription of downstream genes in the Janus kinase (JAK)-STAT pathway [64]. IFI35 is known to interact with N-Myc and NMI [65]. However, IFI35 has been proven to negatively regulate retinoic acid-inducible gene I protein (RIG-I) antiviral signaling and promote the replication of vesicular stomatitis virus. Therefore, IFI35 might serve as a flexible immunological regulator during pathogen defense [66,67]. In the present study, the upregulation of IFN-induced proteins STAT1, STAT2, NMI, and IFI35 was observed and validated, and the strong interactions in the PPI network further demonstrated the IFN-centered immune reaction against C. parvum. The inflammatory molecules mentioned above might be key targets in the anti-C. parvum response.
Apart from IFN-centered signaling pathways, the remarkable downregulation of proteins involved in multiple metabolic pathways also reflects C. parvum’s dependency on certain nutrients, which also provides candidates for effective drugs or inhibitors. Possessing highly streamlined metabolic pathways, but an inability to de novo synthesize nucleosides, fatty acids, and any amino acids, C. parvum hijacks and remodels existing host metabolic pathways for its own benefit [10,68]. In the present study, we found that the levels of multiple host metabolic enzymes were downregulated in response to C. parvum infection, and then an in-depth exploration was conducted to characterize the proteins’ functions. The data revealed widely decreased levels of host metabolic enzymes such as argininosuccinate synthase (ASS1), glutamine synthetase (GLUL), phosphoglycerate mutase 1 (PGAM1), glyoxylate and hydroxypyruvate reductase (GRHPR), NADH: ubiquinone oxidoreductase subunit B6 (NDUFB6), NFS1 cysteine desulfurase (NFS1), pyruvate dehydrogenase E1 component subunit alpha (PDHA1), SDHB, and serine hydroxymethyltransferase 2 (SHMT2) after infection. Folate and thymidylate biosynthesis are of vital importance during DNA replication, and dihydrofolate reductase (DHFR) is a key enzyme in folate metabolism, catalyzing the oxidation of NADPH and reduction of dihydrofolate to NADP and tetrahydrofolate [69,70]. The important roles of folate metabolism and nucleic acid synthesis in cryptosporidiosis, mean that currently, inosine monophosphate dehydrogenase (IMPDH) and dihydrofolate reductase-thymidylate synthase (DHFR-TS) are common targets for the treatment of cryptosporidiosis [71,72]. However, the parasite has been found to tolerate the loss of these classical targets, possibly via as-yet-undiscovered purine transporters and salvage enzymes. In the present study, the data revealed downregulation of SHMT2 in host cells, which is a pyridoxal phosphate (PLP) binding protein catalyzing the cleavage of serine to glycine, accompanied with the production of 5, 10-methylenetetrahydrofolate (5, 10-CH2-THF) [73,74]. The downregulation of SHMT2 by C. parvum revealed not only obstruction of the host metabolism, but also a potential parasitism strategy of C. parvum to sustain an essential nutrient supply. An early study reported the promotion of exogenous purine nucleosides during C. parvum infection [75]. However, genomic analysis indicated the loss of pyrimidine de novo synthesis in C. parvum; therefore, this parasite is almost entirely dependent on import from the host for its purine and pyrimidine requirements [76,77]. SHMT2 is also an essential intermediate for purine biosynthesis; therefore, we concluded that inhibition of SHMT2 caused by C. parvum serves as one of the strategies to secure a purine supply from the host. Similarly, we found that ASS1, a urea cycle enzyme that converts nitrogen from ammonia and aspartate to urea, was downregulated after C. parvum infection. A recent publication on hepatocellular carcinoma showed that downregulation of ASS1 is associated with a more malignant cancerous phenotype and poor prognosis [78]. In-depth investigation further revealed that ASS1 facilitates pyrimidine synthesis during cancerous proliferation by activating CAD (carbamoyl-phosphate synthase 2, aspartate transcarbamylase, and dihydroorotase complex), through the regulation of aspartate levels [79]. Based on these processes, C. parvum would obtain sufficient purines and pyrimidines, which would ensure successful parasitism and replication, possibly by interfering with the host’s purine and pyrimidine metabolism.
Besides, we observed significant reductions in the abundances of enzymes involved in the cell oxidative respiratory chain, such as NFS1, PDHA1, and SDHB. Mounting evidence demonstrates the C. parvum is an energy-requiring parasite. It is generally accepted that Cryptosporidium spp. has lost mitochondria-like organelle-derived energy metabolic capabilities via reductive evolution. Lacking constitutive oxidative phosphorylation, C. parvum is unable to complete the tricarboxylic acid (TCA) cycle and cytochrome-based electron transport processes [10,68,80,81]. Thus, gluconeogenesis and glycolysis are the most important energy producing processes in parasites [15,80]. Genomic and biochemical evidence further confirmed the dependence of Cryptosporidium on glycolysis as the main energy source and an overall reliance on the host for basic nutrients [10,82–85]. PGAM1 was among the proteins identified in our study, which is an important glycolytic enzyme coordinating glycolysis and biosynthesis, including the pentose phosphate pathway and serine synthesis pathway [86,87]. Evidence showed that some host-parasite homologous genes of the host glycolysis/gluconeogenesis pathways were downregulated, while host-exclusive genes were upregulated during invasion and intracellular development, suggesting parasite-derived competition for metabolic substrates, which might explain the reduction of host enzymes in our study [17]. Another study suggested that sugar transportation occurs between Cryptosporidium and host intestine epithelial cells [88]. Parasites further deprive the host cells of the substrates of glycolysis by transportation, and might correspondingly affect the expression of the host genes in the glycolysis pathway. Recent research showed that C. parvum could interfere with host glucose transporters (GLUT) 1/2 and Na+-coupled glucose transporter (SGLT) 1 expression, and a significantly higher intracellular glucose level was observed in infected cells, which would point to an adaptation of the host cells’ glucose uptake after infection [89]. Based on this, PGAM1 might be a new target for C. parvum to inhibit glycolysis and biosynthesis in host cells.
Research has also proven that glutaminolysis and lactate are necessary for parasite replication [15]. In this study, the downregulation of GLUL after C. parvum infection would hinder host glutamine synthesis from glutamate and ammonia, and might even promote parasite replication, which is dependent on glutamine catabolism. All these identified enzymes indicate modification of host energy production and conversion by C. parvum to sustain its own growth, which might also provide new insights into anti-Cryptosporidium targets.
In summary, we observed significant enrichment of interferon-centered signaling pathways and extensive inhibition of metabolism-related enzymes in host cells caused by C. parvum infection, providing a deeper understanding of the molecules and their functions involved in the host-C. parvum interaction. The identified DEPs and signaling pathways narrowed the range of important functional molecules during anti-parasite immune responses and also confirmed C. parvum’s deprivation of certain host nutrients, which might be potential targets for Cryptosporidium treatment. Moreover, the mechanism by which the identified factors participate in C. parvum parasitism and the complex interplay between the identified factors require further investigation.
Supporting information
(XLSX)
(A) Basic statistics of the mass spectrometry data. (B) Length distribution of identified host peptides. (C) Mass and sequences coverage ratio of host proteins. (D) Mass distribution of identified proteins in host cells.
(TIF)
(A) Basic statistics of the mass spectrometry data of identified C. parvum proteins. (B) Length distribution of identified C. parvum derived-peptides from host cells. (C) Mass distribution of identified C. parvum proteins from host cells. (D) Distribution of the number of peptides per protein.
(TIF)
GO enrichment analysis of upregulated (A) and downregulated proteins (B) in the Cellular Component category. The size of the circle area represents the number of DEPs, and the color represents the P value of the enrichment significance of the DEPs under the GO classification. The redder and more distributed to the right the circles, the more important their classification.
(TIF)
(A) COG/KOG analysis of identified C. parvum proteins. (B) GO enrichment analysis of parasite-derived proteins. The size of the circle represents the number of DEPs in that functional class or pathway, and the color represents the significance of the enrichment (P value). (C) GO enrichment analysis of identified C. parvum proteins in the Biological Process category. (D) C. parvum-derived proteins that were enriched significantly in the ribosome pathway from the KEGG pathway analysis. Protein names in red are parasite-derived proteins identified from infected HCT-8 cells.
(TIF)
The degree represents the strength of the protein interaction.
(TIF)
Acknowledgments
We thank Jingjie PTM Bio (Hangzhou, China) CO., LTD for technical support.
Data Availability
The mass spectrometry proteomics data have been deposited at iProX with the data set identifier IPX0003333000, subproject IPX0003333001. https://www.iprox.cn/page/project.html?id=IPX0003333000.
Funding Statement
This work was supported by the National Key R&D Program of China (No. 2017YFD0500400 to HL), the National Natural Science Foundation of China (Nos. 81772225 and 81971969 to JC), the National Science and Technology Major Project (No. 2018ZX10713001-004 to YS) and the Fifth Round of Three-Year Public Health Action Plan of Shanghai, China (No. GWV-10.1-XK13 to JC). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Plutzer J, Karanis P. Genetic polymorphism in Cryptosporidium species: an update. Vet parasitol. 2009; 165(3–4):187–99. doi: 10.1016/j.vetpar.2009.07.003 [DOI] [PubMed] [Google Scholar]
- 2.Hunter PR, Nichols G. Epidemiology and clinical features of Cryptosporidium infection in immunocompromised patients. Clin Microbiol Rev. 2002; 15(1):145–54. doi: 10.1128/CMR.15.1.145-154.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manabe YC, Clark DP, Moore RD, Lumadue JA, Dahlman HR, Belitsos PC, et al. Cryptosporidiosis in patients with AIDS: correlates of disease and survival. Clin Infect Dis. 1998; 27(3):536–42. doi: 10.1086/514701 [DOI] [PubMed] [Google Scholar]
- 4.Feng Y, Ryan UM, Xiao L. Genetic diversity and population structure of Cryptosporidium. Trends parasitol. 2018; 34(11):997–1011. doi: 10.1016/j.pt.2018.07.009 [DOI] [PubMed] [Google Scholar]
- 5.Shrikhande SN, Chande CA, Shegokar VR, Powar RM. Pulmonary cryptosporidiosis in HIV negative, immunocompromised host. Indian J Pathol Microbiol. 2009; 52(2):267–8. doi: 10.4103/0377-4929.48942 [DOI] [PubMed] [Google Scholar]
- 6.Tzipori S, Ward H. Cryptosporidiosis: biology, pathogenesis and disease. Microbes Infect. 2002; 4(10):1047–58. doi: 10.1016/s1286-4579(02)01629-5 [DOI] [PubMed] [Google Scholar]
- 7.Yang Z, Fu Y, Gong P, Zheng J, Liu L, Yu Y, et al. Bovine TLR2 and TLR4 mediate Cryptosporidium parvum recognition in bovine intestinal epithelial cells. Microb Pathog. 2015; 85:29–34. doi: 10.1016/j.micpath.2015.05.009 [DOI] [PubMed] [Google Scholar]
- 8.Chen XM, O’Hara SP, Nelson JB, Splinter PL, Small AJ, Tietz PS, et al. Multiple TLRs are expressed in human cholangiocytes and mediate host epithelial defense responses to Cryptosporidium parvum via activation of NF-kappaB. J Immunol. 2005; 175(11):7447–56. doi: 10.4049/jimmunol.175.11.7447 [DOI] [PubMed] [Google Scholar]
- 9.Zhu G, Marchewka MJ, Keithly JS. Cryptosporidium parvum appears to lack a plastid genome. Microbiology (Reading). 2000; 146 (Pt 2):315–21. doi: 10.1099/00221287-146-2-315 [DOI] [PubMed] [Google Scholar]
- 10.Abrahamsen MS, Templeton TJ, Enomoto S, Abrahante JE, Zhu G, Lancto CA, et al. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science. 2004; 304(5669):441–5. doi: 10.1126/science.1094786 [DOI] [PubMed] [Google Scholar]
- 11.Bouzid M, Hunter PR, Chalmers RM, Tyler KM. Cryptosporidium pathogenicity and virulence. Clin Microbiol Rev. 2013; 26(1):115–34. doi: 10.1128/CMR.00076-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leander BS, Lloyd SA, Marshall W, Landers SC. Phylogeny of marine gregarines (Apicomplexa)-Pterospora, Lithocystis and Lankesteria-and the origin(s) of coelomic parasitism. Protist. 2006; 157(1):45–60. doi: 10.1016/j.protis.2005.10.002 [DOI] [PubMed] [Google Scholar]
- 13.Barta JR, Thompson RC. What is Cryptosporidium? Reappraising its biology and phylogenetic affinities. Trends Parasitol. 2006; 22(10):463–8. doi: 10.1016/j.pt.2006.08.001 [DOI] [PubMed] [Google Scholar]
- 14.Borowski H, Clode PL, Thompson RC. Active invasion and/or encapsulation? A reappraisal of host-cell parasitism by Cryptosporidium. Trends Parasitol. 2008; 24(11):509–16. doi: 10.1016/j.pt.2008.08.002 [DOI] [PubMed] [Google Scholar]
- 15.Vélez J, Velasquez Z, Silva LMR, Gärtner U, Failing K, Daugschies A, et al. Metabolic signatures of Cryptosporidium parvum-infected HCT-8 cells and impact of selected metabolic inhibitors on C. parvum infection under physioxia and hyperoxia. Biology. 2021; 10(1). doi: 10.3390/biology10010060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson RC, Olson ME, Zhu G, Enomoto S, Abrahamsen MS, Hijjawi NS. Cryptosporidium and cryptosporidiosis. Adv Parasitol. 2005; 59:77–158. doi: 10.1016/S0065-308X(05)59002-X [DOI] [PubMed] [Google Scholar]
- 17.Xu T, Ping J, Yu Y, Yu F, Yu Y, Hao P, et al. Revealing parasite influence in metabolic pathways in Apicomplexa infected patients. BMC Bioinformatics. 2010; 11 Suppl 11(Suppl 11):S13. doi: 10.1186/1471-2105-11-S11-S13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pollok RC, Farthing MJ, Bajaj-Elliott M, Sanderson IR, McDonald V. Interferon gamma induces enterocyte resistance against infection by the intracellular pathogen Cryptosporidium parvum. Gastroenterology. 2001; 120(1):99–107. doi: 10.1053/gast.2001.20907 [DOI] [PubMed] [Google Scholar]
- 19.Dann SM, Wang HC, Gambarin KJ, Actor JK, Robinson P, Lewis DE, et al. Interleukin-15 activates human natural killer cells to clear the intestinal protozoan Cryptosporidium. J Infect Dis. 2005; 192(7):1294–302. doi: 10.1086/444393 [DOI] [PubMed] [Google Scholar]
- 20.Lantier L, Lacroix-Lamandé S, Potiron L, Metton C, Drouet F, Guesdon W, et al. Intestinal CD103+ dendritic cells are key players in the innate immune control of Cryptosporidium parvum infection in neonatal mice. PLoS pathog. 2013; 9(12):e1003801. doi: 10.1371/journal.ppat.1003801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amadi B, Mwiya M, Musuku J, Watuka A, Sianongo S, Ayoub A, et al. Effect of nitazoxanide on morbidity and mortality in Zambian children with cryptosporidiosis: a randomised controlled trial. Lancet. 2002; 360(9343):1375–80. doi: 10.1016/S0140-6736(02)11401-2 [DOI] [PubMed] [Google Scholar]
- 22.Amadi B, Mwiya M, Sianongo S, Payne L, Watuka A, Katubulushi M, et al. High dose prolonged treatment with nitazoxanide is not effective for cryptosporidiosis in HIV positive Zambian children: a randomised controlled trial. BMC Infect Dis. 2009; 9:195. doi: 10.1186/1471-2334-9-195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shahiduzzaman M, Daugschies A. Therapy and prevention of cryptosporidiosis in animals. Vet parasitol. 2012; 188(3–4):203–14. doi: 10.1016/j.vetpar.2012.03.052 [DOI] [PubMed] [Google Scholar]
- 24.Checkley W, White AC Jr., Jaganath D, Arrowood MJ, Chalmers RM, Chen XM, et al. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for Cryptosporidium. Lancet Infect Dis. 2015; 15(1):85–94. doi: 10.1016/S1473-3099(14)70772-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Innes EA, Chalmers RM, Wells B, Pawlowic MC. A One Health approach to tackle cryptosporidiosis. Trends Parasitol. 2020; 36(3):290–303. doi: 10.1016/j.pt.2019.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019; 47(D1):D607–d13. doi: 10.1093/nar/gky1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003; 13(11):2498–504. doi: 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Umemiya R, Fukuda M, Fujisaki K, Matsui T. Electron microscopic observation of the invasion process of Cryptosporidium parvum in severe combined immunodeficiency mice. J Parasitol. 2005; 91(5):1034–9. doi: 10.1645/GE-508R.1 [DOI] [PubMed] [Google Scholar]
- 29.Current WL, Reese NC. A comparison of endogenous development of three isolates of Cryptosporidium in suckling mice. J Protozool. 1986; 33(1):98–108. doi: 10.1111/j.1550-7408.1986.tb05567.x [DOI] [PubMed] [Google Scholar]
- 30.Kagnoff MF. The intestinal epithelium is an integral component of a communications network. J Clin Invest. 2014; 124(7):2841–3. doi: 10.1172/JCI75225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Price AE, Shamardani K, Lugo KA, Deguine J, Roberts AW, Lee BL, et al. A Map of toll-like receptor expression in the intestinal epithelium reveals distinct spatial, cell type-specific, and temporal patterns. Immunity. 2018; 49(3):560–75.e6. doi: 10.1016/j.immuni.2018.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gallo RL, Hooper LV. Epithelial antimicrobial defence of the skin and intestine. Nat Rev Immunol. 2012; 12(7):503–16. doi: 10.1038/nri3228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin-Gallausiaux C, Larraufie P, Jarry A, Béguet-Crespel F, Marinelli L, Ledue F, et al. Butyrate produced by commensal bacteria down-regulates indolamine 2,3-dioxygenase 1 (IDO-1) expression via a dual mechanism in human intestinal epithelial cells. Front Immunol. 2018; 9:2838. doi: 10.3389/fimmu.2018.02838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Upton SJ, Tilley M, Brillhart DB. Comparative development of Cryptosporidium parvum (Apicomplexa) in 11 continuous host cell lines. FEMS Microbiol Lett. 1994; 118(3):233–6. doi: 10.1111/j.1574-6968.1994.tb06833.x [DOI] [PubMed] [Google Scholar]
- 35.Sifuentes LY, Di Giovanni GD. Aged HCT-8 cell monolayers support Cryptosporidium parvum infection. Appl Environ Microbiol. 2007; 73(23):7548–51. doi: 10.1128/AEM.01579-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morada M, Lee S, Gunther-Cummins L, Weiss LM, Widmer G, Tzipori S, et al. Continuous culture of Cryptosporidium parvum using hollow fiber technology. Int J Parasitol. 2016; 46(1):21–9. doi: 10.1016/j.ijpara.2015.07.006 [DOI] [PubMed] [Google Scholar]
- 37.Kuang Z, Seo EJ, Leis J. Mechanism of inhibition of retrovirus release from cells by interferon-induced gene ISG15. J Virol. 2011; 85(14):7153–61. doi: 10.1128/JVI.02610-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pincetic A, Kuang Z, Seo EJ, Leis J. The interferon-induced gene ISG15 blocks retrovirus release from cells late in the budding process. J Virol. 2010; 84(9):4725–36. doi: 10.1128/JVI.02478-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okumura A, Lu G, Pitha-Rowe I, Pitha PM. Innate antiviral response targets HIV-1 release by the induction of ubiquitin-like protein ISG15. Proc Natl Acad Sci U S A. 2006; 103(5):1440–5. doi: 10.1073/pnas.0510518103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okumura A, Pitha PM, Harty RN. ISG15 inhibits Ebola VP40 VLP budding in an L-domain-dependent manner by blocking Nedd4 ligase activity. Proc Natl Acad Sci U S A. 2008; 105(10):3974–9. doi: 10.1073/pnas.0710629105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malakhova OA, Zhang DE. ISG15 inhibits Nedd4 ubiquitin E3 activity and enhances the innate antiviral response. J Biol Chem. 2008; 283(14):8783–7. doi: 10.1074/jbc.C800030200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu D, Zhang T, Xiao J, Zhu K, Wei R, Wu Z, et al. Modification of BECN1 by ISG15 plays a crucial role in autophagy regulation by type I IFN/interferon. Autophagy. 2015; 11(4):617–28. doi: 10.1080/15548627.2015.1023982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katzenell S, Leib DA. Herpes simplex virus and interferon signaling induce novel autophagic clusters in sensory neurons. J Virol. 2016; 90(9):4706–19. doi: 10.1128/JVI.02908-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oura CA, McKellar S, Swan DG, Okan E, Shiels BR. Infection of bovine cells by the protozoan parasite Theileria annulata modulates expression of the ISGylation system. Cell Microbiol. 2006; 8(2):276–88. doi: 10.1111/j.1462-5822.2005.00620.x [DOI] [PubMed] [Google Scholar]
- 45.Vargas-Inchaustegui DA, Xin L, Soong L. Leishmania braziliensis infection induces dendritic cell activation, ISG15 transcription, and the generation of protective immune responses. J Immunol. 2008; 180(11):7537–45. doi: 10.4049/jimmunol.180.11.7537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bhushan J, Radke JB, Perng YC, McAllaster M, Lenschow DJ, Virgin HW, et al. ISG15 connects autophagy and IFN-γ-dependent control of Toxoplasma gondii infection in human cells. mBio. 2020; 11(5). doi: 10.1128/mBio.00852-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hubbard AK, Rothlein R. Intercellular adhesion molecule-1 (ICAM-1) expression and cell signaling cascades. Free Radic Biol Med. 2000; 28(9):1379–86. doi: 10.1016/s0891-5849(00)00223-9 [DOI] [PubMed] [Google Scholar]
- 48.Gorina R, Lyck R, Vestweber D, Engelhardt B. β2 integrin-mediated crawling on endothelial ICAM-1 and ICAM-2 is a prerequisite for transcellular neutrophil diapedesis across the inflamed blood-brain barrier. J Immunol. 2014; 192(1):324–37. doi: 10.4049/jimmunol.1300858 [DOI] [PubMed] [Google Scholar]
- 49.Liu RR, Li J, Gong JY, Kuang F, Liu JY, Zhang YS, et al. MicroRNA-141 regulates the expression level of ICAM-1 on endothelium to decrease myocardial ischemia-reperfusion injury. Am J Physiol Heart Circ Physiol. 2015; 309(8):H1303–13. doi: 10.1152/ajpheart.00290.2015 [DOI] [PubMed] [Google Scholar]
- 50.Sumagin R, Robin AZ, Nusrat A, Parkos CA. Transmigrated neutrophils in the intestinal lumen engage ICAM-1 to regulate the epithelial barrier and neutrophil recruitment. Mucosal Immunol. 2014; 7(4):905–15. doi: 10.1038/mi.2013.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lennartz F, Smith C, Craig AG, Higgins MK. Structural insights into diverse modes of ICAM-1 binding by Plasmodium falciparum-infected erythrocytes. Proc Natl Acad Sci U S A. 2019; 116(40):20124–34. doi: 10.1073/pnas.1911900116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cunningham DA, Lin JW, Brugat T, Jarra W, Tumwine I, Kushinga G, et al. ICAM-1 is a key receptor mediating cytoadherence and pathology in the Plasmodium chabaudi malaria model. Malar J. 2017; 16(1):185. doi: 10.1186/s12936-017-1834-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pastre MJ, Casagrande L, Gois MB, Pereira-Severi LS, Miqueloto CA, Garcia JL, et al. Toxoplasma gondii causes increased ICAM-1 and serotonin expression in the jejunum of rats 12 h after infection. Biomed Pharmacother. 2019; 114:108797. doi: 10.1016/j.biopha.2019.108797 [DOI] [PubMed] [Google Scholar]
- 54.Gong AY, Hu G, Zhou R, Liu J, Feng Y, Soukup GA, et al. MicroRNA-221 controls expression of intercellular adhesion molecule-1 in epithelial cells in response to Cryptosporidium parvum infection. Int J Parasitol. 2011; 41(3–4):397–403. doi: 10.1016/j.ijpara.2010.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feeley EM, Sims JS, John SP, Chin CR, Pertel T, Chen LM, et al. IFITM3 inhibits influenza A virus infection by preventing cytosolic entry. PLoS pathog. 2011; 7(10):e1002337. doi: 10.1371/journal.ppat.1002337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bailey CC, Zhong G, Huang IC, Farzan M. IFITM-family proteins: The cell’s first line of antiviral defense. Annu Rev Virol. 2014; 1:261–83. doi: 10.1146/annurev-virology-031413-085537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Desai TM, Marin M, Chin CR, Savidis G, Brass AL, Melikyan GB. IFITM3 restricts influenza A virus entry by blocking the formation of fusion pores following virus-endosome hemifusion. PLoS pathog. 2014; 10(4):e1004048. doi: 10.1371/journal.ppat.1004048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Everitt AR, Clare S, Pertel T, John SP, Wash RS, Smith SE, et al. IFITM3 restricts the morbidity and mortality associated with influenza. Nature. 2012; 484(7395):519–23. doi: 10.1038/nature10921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wakim LM, Gupta N, Mintern JD, Villadangos JA. Enhanced survival of lung tissue-resident memory CD8+ T cells during infection with influenza virus due to selective expression of IFITM3. Nat Immunol. 2013; 14(3):238–45. doi: 10.1038/ni.2525 [DOI] [PubMed] [Google Scholar]
- 60.Jiang LQ, Xia T, Hu YH, Sun MS, Yan S, Lei CQ, et al. IFITM3 inhibits virus-triggered induction of type I interferon by mediating autophagosome-dependent degradation of IRF3. Cell Mol Immunol. 2018; 15(9):858–67. doi: 10.1038/cmi.2017.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stark GR, Darnell JE Jr., The JAK-STAT pathway at twenty. Immunity. 2012; 36(4):503–14. doi: 10.1016/j.immuni.2012.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang Y, He JJ, Hu S, Chang H, Xiang X, Yang JF, et al. Comparative study of transcriptome profiles of mouse livers and skins infected by fork-tailed or non-fork-tailed Schistosoma japonicum. Front Microbiol. 2017; 8:1648. doi: 10.3389/fmicb.2017.01648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Matta SK, Olias P, Huang Z, Wang Q, Park E, Yokoyama WM, et al. Toxoplasma gondii effector TgIST blocks type I interferon signaling to promote infection. Proc Natl Acad Sci U S A. 2019; 116(35):17480–91. doi: 10.1073/pnas.1904637116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhu M, John S, Berg M, Leonard WJ. Functional association of NMI with STAT5 and STAT1 in IL-2- and IFN gamma-mediated signaling. Cell. 1999; 96(1):121–30. doi: 10.1016/s0092-8674(00)80965-4 [DOI] [PubMed] [Google Scholar]
- 65.Zhou X, Liao J, Meyerdierks A, Feng L, Naumovski L, Bottger EC, et al. Interferon-alpha induces NMI-IFP35 heterodimeric complex formation that is affected by the phosphorylation of IFP35. J Biol Chem. 2000; 275(28):21364–71. doi: 10.1074/jbc.M003177200 [DOI] [PubMed] [Google Scholar]
- 66.Shirai K, Shimada T, Yoshida H, Hayakari R, Matsumiya T, Tanji K, et al. Interferon (IFN)-induced protein 35 (IFI35) negatively regulates IFN-β-phosphorylated STAT1-RIG-I-CXCL10/CCL5 axis in U373MG astrocytoma cells treated with polyinosinic-polycytidylic acid. Brain Res. 2017; 1658:60–7. doi: 10.1016/j.brainres.2017.01.018 [DOI] [PubMed] [Google Scholar]
- 67.Das A, Dinh PX, Pattnaik AK. Trim21 regulates NMI-IFI35 complex-mediated inhibition of innate antiviral response. Virology. 2015; 485:383–92. doi: 10.1016/j.virol.2015.08.013 [DOI] [PubMed] [Google Scholar]
- 68.Xu P, Widmer G, Wang Y, Ozaki LS, Alves JM, Serrano MG, et al. The genome of Cryptosporidium hominis. Nature. 2004; 431(7012):1107–12. doi: 10.1038/nature02977 [DOI] [PubMed] [Google Scholar]
- 69.Blakley RL. Eukaryotic dihydrofolate reductase. Adv Enzymol Relat Areas Mol Biol. 1995; 70:23–102. doi: 10.1002/9780470123164.ch2 [DOI] [PubMed] [Google Scholar]
- 70.Anderson DD, Quintero CM, Stover PJ. Identification of a de novo thymidylate biosynthesis pathway in mammalian mitochondria. Proc Natl Acad Sci U S A. 2011; 108(37):15163–8. doi: 10.1073/pnas.1103623108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sharling L, Liu X, Gollapalli DR, Maurya SK, Hedstrom L, Striepen B. A screening pipeline for antiparasitic agents targeting cryptosporidium inosine monophosphate dehydrogenase. PLoS Negl Trop Dis. 2010; 4(8):e794. doi: 10.1371/journal.pntd.0000794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mukerjee A, Iyidogan P, Castellanos-Gonzalez A, Cisneros JA, Czyzyk D, Ranjan AP, et al. A nanotherapy strategy significantly enhances anticryptosporidial activity of an inhibitor of bifunctional thymidylate synthase-dihydrofolate reductase from Cryptosporidium. Bioorg Med Chem Lett. 2015; 25(10):2065–7. doi: 10.1016/j.bmcl.2015.03.091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Giardina G, Brunotti P, Fiascarelli A, Cicalini A, Costa MG, Buckle AM, et al. How pyridoxal 5’-phosphate differentially regulates human cytosolic and mitochondrial serine hydroxymethyltransferase oligomeric state. FEBS J. 2015; 282(7):1225–41. doi: 10.1111/febs.13211 [DOI] [PubMed] [Google Scholar]
- 74.Kim PB, Nelson JW, Breaker RR. An ancient riboswitch class in bacteria regulates purine biosynthesis and one-carbon metabolism. Mol Cell. 2015; 57(2):317–28. doi: 10.1016/j.molcel.2015.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lawton P, Hejl C, Mancassola R, Naciri M, Petavy AF. Effects of purine nucleosides on the in vitro growth of Cryptosporidium parvum. FEMS Microbiol Lett. 2003; 226(1):39–43. doi: 10.1016/S0378-1097(03)00555-X [DOI] [PubMed] [Google Scholar]
- 76.Striepen B, Pruijssers AJ, Huang J, Li C, Gubbels MJ, Umejiego NN, et al. Gene transfer in the evolution of parasite nucleotide biosynthesis. Proc Natl Acad Sci U S A. 2004; 101(9):3154–9. doi: 10.1073/pnas.0304686101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pawlowic MC, Somepalli M, Sateriale A, Herbert GT, Gibson AR, Cuny GD, et al. Genetic ablation of purine salvage in Cryptosporidium parvum reveals nucleotide uptake from the host cell. Proc Natl Acad Sci U S A. 2019; 116(42):21160–5. doi: 10.1073/pnas.1908239116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tan GS, Lim KH, Tan HT, Khoo ML, Tan SH, Toh HC, et al. Novel proteomic biomarker panel for prediction of aggressive metastatic hepatocellular carcinoma relapse in surgically resectable patients. J Proteome Res. 2014; 13(11):4833–46. doi: 10.1021/pr500229n [DOI] [PubMed] [Google Scholar]
- 79.Rabinovich S, Adler L, Yizhak K, Sarver A, Silberman A, Agron S, et al. Diversion of aspartate in ASS1-deficient tumours fosters de novo pyrimidine synthesis. Nature. 2015; 527(7578):379–83. doi: 10.1038/nature15529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Seeber F, Soldati-Favre D. Metabolic pathways in the apicoplast of apicomplexa. Int Rev Cell Mol Biol. 2010; 281:161–228. doi: 10.1016/S1937-6448(10)81005-6 [DOI] [PubMed] [Google Scholar]
- 81.Seeber F, Limenitakis J, Soldati-Favre D. Apicomplexan mitochondrial metabolism: a story of gains, losses and retentions. Trends Parasitol. 2008; 24(10):468–78. doi: 10.1016/j.pt.2008.07.004 [DOI] [PubMed] [Google Scholar]
- 82.Manjunatha UH, Vinayak S, Zambriski JA, Chao AT, Sy T, Noble CG, et al. A Cryptosporidium PI(4)K inhibitor is a drug candidate for cryptosporidiosis. Nature. 2017; 546(7658):376–80. doi: 10.1038/nature22337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mauzy MJ, Enomoto S, Lancto CA, Abrahamsen MS, Rutherford MS. The Cryptosporidium parvum transcriptome during in vitro development. PLoS one. 2012; 7(3):e31715. doi: 10.1371/journal.pone.0031715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang H, Guo F, Zhu G. Cryptosporidium lactate dehydrogenase is associated with the parasitophorous vacuole membrane and is a potential target for developing therapeutics. PLoS pathog. 2015; 11(11):e1005250. doi: 10.1371/journal.ppat.1005250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu S, Roellig DM, Guo Y, Li N, Frace MA, Tang K, et al. Evolution of mitosome metabolism and invasion-related proteins in Cryptosporidium. BMC Genomics. 2016; 17(1):1006. doi: 10.1186/s12864-016-3343-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jacobowitz DM, Jozwik C, Fukuda T, Pollard HB. Immunohistochemical localization of phosphoglycerate mutase in capillary endothelium of the brain and periphery. Microvasc Res. 2008; 76(2):89–93. doi: 10.1016/j.mvr.2008.04.001 [DOI] [PubMed] [Google Scholar]
- 87.Jiang X, Sun Q, Li H, Li K, Ren X. The role of phosphoglycerate mutase 1 in tumor aerobic glycolysis and its potential therapeutic implications. Int J Cancer. 2014; 135(9):1991–6. doi: 10.1002/ijc.28637 [DOI] [PubMed] [Google Scholar]
- 88.Zapata F, Perkins ME, Riojas YA, Wu TW, Le Blancq SM. The Cryptosporidium parvum ABC protein family. Mol Biochem Parasitol. 2002; 120(1):157–61. doi: 10.1016/s0166-6851(01)00445-5 [DOI] [PubMed] [Google Scholar]
- 89.Delling C, Daugschies A, Bangoura B, Dengler F. Cryptosporidium parvum alters glucose transport mechanisms in infected enterocytes. Parasitol Res. 2019; 118(12):3429–41. doi: 10.1007/s00436-019-06471-y [DOI] [PubMed] [Google Scholar]