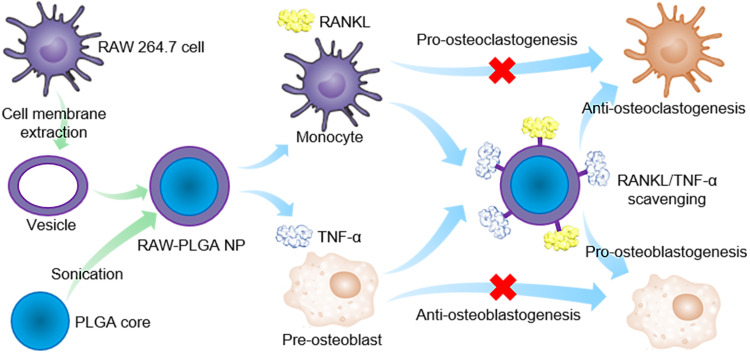
Biomimetic nanodecoys reconstruct the osteoclast/osteoblast balance to treat postmenopausal osteoporosis.
Abstract
Imbalance between osteoblasts and osteoclasts accounts for the incidence and deterioration of postmenopausal osteoporosis. Abnormally elevated RANKL and TNF-α levels after menopause promote osteoclast formation and inhibit osteoblast differentiation, respectively. Here, nanodecoys capable of scavenging RANKL and TNF-α were developed from preosteoclast (RAW 264.7 cell) membrane–coated poly(lactic-co-glycolic acid) (PLGA) nanoparticles, which inhibited osteoporosis and maintained bone integrity. The nanodecoys effectively escaped from macrophage capture and enabled prolonged blood circulation after systemic administration. The abundant RANK and TNF-α receptor (TNF-αR) on the cell membranes effectively neutralized RANKL and TNF-α to prevent osteoclastogenesis and promote osteoblastogenesis, respectively, thus reversing the progression of osteoporosis in the ovariectomized (OVX) mouse model. These biomimetic nanodecoys provide an effective strategy for reconstructing the osteoclast/osteoblast balance and hold great potentials for the clinical management of postmenopausal osteoporosis.
INTRODUCTION
Osteoporosis is a progressive skeletal disorder characterized by the decline of bone mineral density (BMD) and quality, destruction of bone microstructure, and increase of bone fragility (1, 2). For women, postmenopausal osteoporosis resulting from estrogen deficiency is the most common type, and approximately 50% of women experience at least one bone fracture after menopause (3, 4).
Bone mass regulation relies on the dynamic balance between osteoblast-driven bone formation and osteoclast-driven bone resorption (5–7). In menopausal women, estrogen deficiency up-regulates the receptor activator of nuclear factor κB ligand (RANKL) (8–11), which further activates the nuclear factor κB (NF-κB) pathway by binding to RANK on the monocyte membrane to up-regulate c-Fos gene expression (12–15). In addition, RANKL activates the mitogen-activated protein kinase (MAPK) pathway by initiating phosphorylation of a series of signaling molecules such as p38, extracellular signal–regulated kinase (ERK), and c-Jun N-terminal kinase (JNK) (16–18). Activation of both pathways leads to osteoclast differentiation from monocytes and initiates the transcription program of genes such as matrix metalloproteinase (MMP) and alkaline phosphatase (ALP), ultimately promoting bone resorption and inducing bone loss (19). In the meantime, estrogen deficiency will cause immune network imbalance and up-regulate tumor necrosis factor–α (TNF-α) to inhibit bone formation (20, 21). Elevated TNF-α level will induce apoptosis of osteoblasts (22, 23). In addition, TNF-α can down-regulate the transcription of runt-related transcription factor 2 (RUNX2) and osteoblast transcription factor osterix (OSX) (1, 24–28), resulting in the failure of osteoblast differentiation and inhibition of mineralized nodule production.
Given the important roles of RANKL and TNF-α in osteoporosis, therapies that can scavenge RANKL and TNF-α hold great potential for anti-osteoporosis treatment (29, 30). In the clinic, osteoprotegerin (OPG) and antibodies such as denosumab have been widely applied for scavenging RANKL and suppressing osteoporosis (9, 31). Moreover, leucine-rich repeat-containing G protein–coupled receptor 4 (LGR4), which can compete with RANK in binding with RANKL, and TNF-α antagonist etanercept, which can inhibit TNF-α function, are considered beneficial for the treatment of osteoporosis (9, 32). However, these RANKL/TNF-α inhibitors, mostly protein drugs, often suffer from shortcomings such as short blood circulation time, suboptimal biodistribution, complex manufacturing processes, and resistance of antibodies (33–35). In addition, the pathological background of osteoporosis is often related to multiple targets, and thus, a single-target therapeutic modality may be suboptimal (36). Therefore, there is an urgent demand for safer and more effective medications for osteoporosis treatment.
Recently, cell membrane–coated nanodecoys have shown great promise for cytokine/antibody clearance and toxin neutralization through interactions with membrane proteins (36–39). These cell membrane–coated nanodecoys that inherit the antigenic profile of the source cell can absorb and neutralize pathological molecules (37, 40–42). For example, red blood cell (RBC) membrane–coated nanodecoys can bind and scavenge bacterial pore-forming toxins and pathological autoantibodies, preventing their attack on healthy RBCs (41). In addition, cell membrane provides diversified binding sites to allow scavenging of multiple hazardous substances, making it superior to single-target antibodies. For example, cell membranes derived from leukocytes, such as macrophages and neutrophils, can bind and neutralize lipopolysaccharide as well as varieties of proinflammatory cytokines, thereby preventing the initiation of inflammation (40, 43–45). The unique properties of cell-mimicking nanodecoys in biodetoxification suggest their promising applications for anti-osteoporosis treatment, which, according to the best of our knowledge, are still lacking.
Here, preosteoclast (RAW 264.7 cell) membrane–coated poly(lactic-co-glycolic acid) (PLGA) nanodecoys (denoted as RAW-PLGA nanodecoys) were developed as RANKL- and TNF-α–scavenging agents for the management of postmenopausal osteoporosis. The RAW-PLGA nanodecoys were anticipated to mimic the source cells, displaying abundant RANK and TNF-α receptor (TNF-αR) on the nanosurfaces. Thus, the nanodecoys could bind and scavenge RANKL and TNF-α that would otherwise target monocytes and preosteoblasts, respectively, hampering osteoclastogenesis yet promoting osteoblastogenesis to suppress osteoporosis (Fig. 1).
Fig. 1. Schematic illustration of preosteoclast membrane–coated nanodecoys for the anti-osteoporosis management.
PLGA NPs were coated with RAW 264.7 cell membrane to form the RAW-PLGA nanodecoys, and the abundant RANK and TNF-αR on the membrane surfaces allowed efficient scavenging of RANKL and TNF-α, which inhibits osteoclastogenesis and promotes osteoblastogenesis to rebalance osteoclasts and osteoblasts.
RESULTS
Preparation and characterization of RAW-PLGA nanodecoys
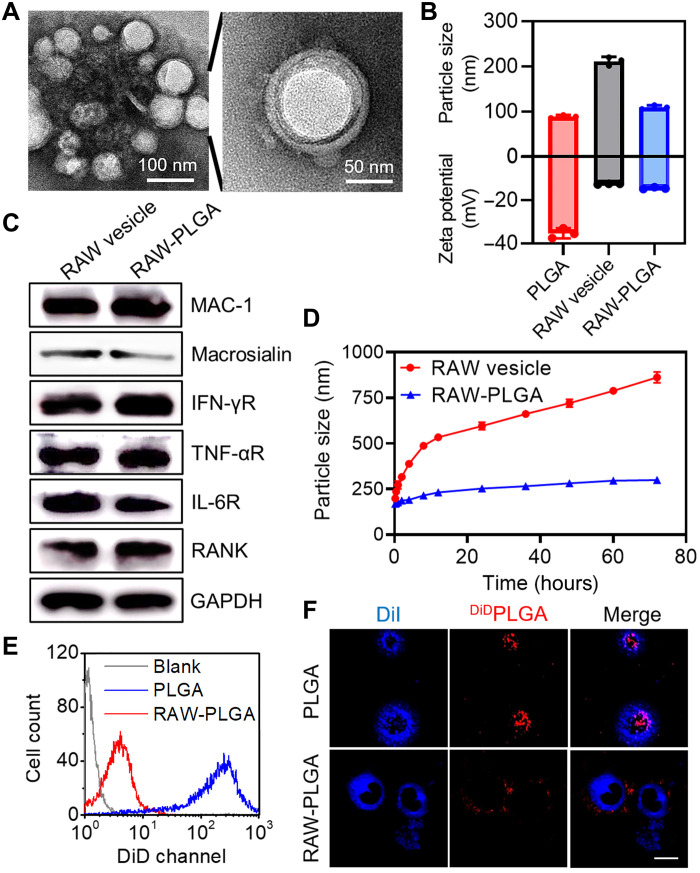
PLGA nanoparticles (NPs) with an average diameter of 88.9 nm and zeta potential of −35.3 mV were synthesized according to reported procedures (40). Then, using the sonication-triggered membrane fusion method (43), cell membranes derived from RAW 264.7 cells were allowed to form membrane vesicles (denoted as RAW vesicles) and were subsequently decorated onto PLGA NPs to obtain RAW-PLGA nanodecoys. When the membrane protein/PLGA weight ratio increased from 1:64 to 1:4, the particle size remained constant, while the negative zeta potential gradually decreased (fig. S1). Further increment of the weight ratio led to appreciable enhancement of the particle size, indicating excessive addition of the cell membranes. At the optimal weight ratio of 1:4, RAW-PLGA nanodecoys with spherical morphology and distinct membrane structure were obtained, as shown in the transmission electron microscopy (TEM) images (Fig. 2A). The hydrodynamic diameter of RAW-PLGA nanodecoys (109.5 nm) was slightly larger than that of PLGA NPs (88.9 nm), while its zeta potential (−14.5 mV) was close to that of RAW vesicles (−12.5 mV), indicating successful surface coating with cell membrane (Fig. 2B). The obtained RAW-PLGA nanodecoys could be stored at room temperature (RT) for longer than 1 month without precipitation or size change. After RAW-DiDPLGA nanodecoys (containing DiD-labeled PLGA) or DiDRAW-PLGA nanodecoys (containing DiD-labeled cell membrane) were centrifuged, the fluorescence intensity in the supernatant markedly decreased, further indicating high efficiency of membrane coating and excellent integrity of the nanodecoys (fig. S2).
Fig. 2. Characterization and decreased macrophage uptake of RAW-PLGA nanodecoys.
(A) TEM images of RAW-PLGA nanodecoys. (B) Size and zeta potential of PLGA NPs, RAW vesicles, and RAW-PLGA nanodecoys (n = 3). (C) Characteristic protein bands of RAW vesicles and RAW-PLGA nanodecoys resolved by Western blot. (D) Size alteration of RAW vesicles and RAW-PLGA nanodecoys after incubation in DMEM containing 10% FBS for different times (n = 3). Flow cytometric diagrams (E) and CLSM images (F) of RAW 264.7 cells following 4-hour incubation with DiDPLGA NPs and RAW-DiDPLGA nanodecoys (100 μg DiDPLGA/ml). The cell membrane was stained with DiI (scale bar, 10 μm).
PLGA NPs formed aggregates in phosphate-buffered saline (PBS) at 1 mg/ml, while clear suspension was noted for RAW-PLGA nanodecoys at the membrane protein/PLGA weight ratio ≥ 1:32, indicating that cell membrane coating would greatly enhance the colloidal stability against salt (fig. S3). In addition, after incubation in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS) for up to 72 hours, size and polydispersity index (PDI) of the RAW vesicles markedly increased, while those of the RAW-PLGA nanodecoys remained almost unaltered (Fig. 2D and fig. S4). It could be explained that the rigid PLGA core could limit the flow of membrane components, thus improving the stability in serum.
To further verify that membrane proteins were retained after the membrane coating process, representative surface markers of RAW 264.7 cells, including integrins (MAC-1 and macrosialin) and cytokine-binding receptors [interferon-γ receptor (IFN-γR), TNF-αR, interleukin-6 receptor (IL-6R), and RANK], were detected via Western blot. As illustrated in Fig. 2C, RAW membranes coated on PLGA NPs showed similar marker expression to the native membranes. In particular, the presence of RANK and TNF-αR would provide the premise for RANKL and TNF-α scavenging.
Decreased macrophage uptake of RAW-PLGA nanodecoys
NPs coated with blood cell membrane can decrease the probability of being cleared by macrophages to evade the immune system, thus prolonging the blood circulation time (45–48). As revealed by both spectrofluorimetric and flow cytometric analyses, the uptake level of RAW-DiDPLGA nanodecoys in RAW 264.7 cells was 20-fold lower than that of DiDPLGA NPs (fig. S5 and Fig. 2E). Confocal laser scanning microscopy (CLSM) observation further revealed that the fluorescence of DiDPLGA NPs was largely spread into the cytoplasm, while that of RAW-DiDPLGA nanodecoys was much weaker and mainly situated on the DiI-stained cell membrane (Fig. 2F). These results collectively demonstrated that coating of NPs with RAW 264.7 cell membrane could efficiently inhibit phagocytosis by macrophages. Furthermore, RAW-PLGA nanodecoys showed negligible cytotoxicity at increased PLGA concentrations up to 160 μg/ml in both preosteoclasts (RAW 264.7 cells) and preosteoblast (MC3T3-E1 cells), indicating their desired cytocompatibility (fig. S6). In addition, RAW-PLGA nanodecoys did not cause the up-regulation of TNF-α or IL-6 in RAW 264.7 cells (fig. S7), indicating their low immunologically stimulatory capability.
RAW-PLGA nanodecoy–mediated RANKL scavenging in vitro
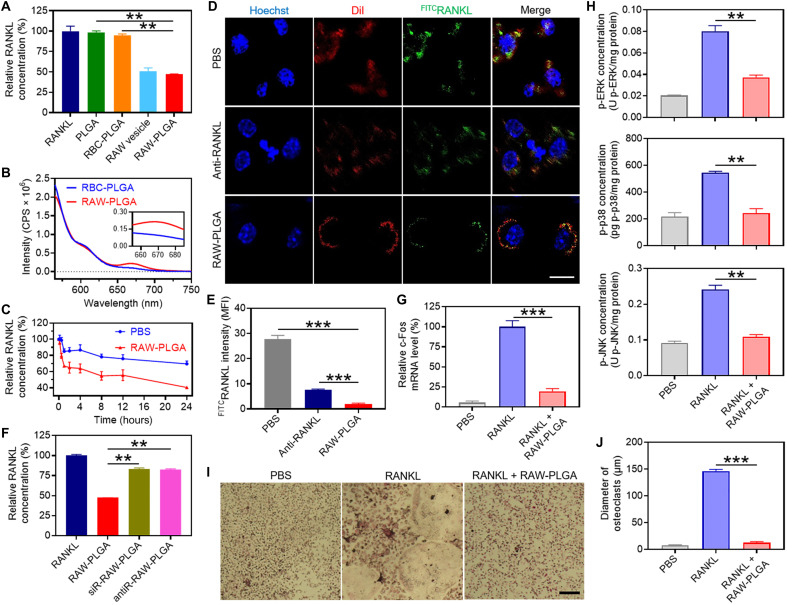
The RAW-PLGA nanodecoy–mediated RANKL scavenging was first probed via determination of the remaining RANKL concentration after coincubation of nanodecoys and RANKL solution. As a negative control, RBC membrane–coated PLGA (RBC-PLGA) NPs were prepared. As illustrated in Fig. 3A and fig. S8, RAW-PLGA nanodecoys mediated remarkable and concentration-dependent scavenging of RANKL, conferring an IC50 (half-maximal inhibitory concentration) value of 2349 μg/ml. In comparison, PLGA NPs and RBC-PLGA NPs showed negligible scavenging effect. The binding between RANKL and RAW-PLGA nanodecoys was strong, wherein minimal amount (~5%) of RANKL was released from the RANKL/RAW-PLGA complexes after incubation in DMEM containing 10% FBS for up to 48 hours (fig. S9A). The fluorescence resonance energy transfer (FRET) assay further confirmed the binding of RAW-PLGA nanodecoys to RANKL. Briefly, Cy5RAW-PLGA nanodecoys or Cy5RBC-PLGA NPs were incubated with Cy3RANKL before being subjected to fluorescence detection. As illustrated in Fig. 3B, the fluorescence intensity of Cy5 in the RAW-PLGA nanodecoys/RANKL solution was notably higher than that in the RBC-PLGA NPs/RANKL solution, indicating that these two dyes, Cy3 and Cy5, were in sufficient proximity in the RAW-PLGA nanodecoys/RANKL solution to induce FRET (49).
Fig. 3. In vitro RANKL-scavenging and anti-osteoclastogenesis efficiencies of RAW-PLGA nanodecoys.
(A) Relative RANKL concentration after incubation with PLGA NPs, RBC-PLGA NPs, RAW vesicles, or RAW-PLGA nanodecoys at the initial RANKL concentration of 200 pg/ml (n = 3). (B) FRET analysis of Cy5RAW-PLGA nanodecoys (or Cy5RBC-PLGA NPs) and Cy3RANKL (λex = 550 nm). (C) Extracellular RANKL concentration of RAW 264.7 cells after incubation with PBS or RAW-PLGA nanodecoys for different times at the initial RANKL concentration of 100 ng/ml (n = 3). (D) CLSM images of RAW 264.7 cells after 4-hour incubation with PBS, RAW-PLGA nanodecoys, or anti-RANKL in the presence of FITCRANKL (scale bar, 10 μm). Cell nuclei and membrane were stained with Hoechst 33258 and DiI, respectively. (E) Quantified mean fluorescence intensity of FITCRANKL in RAW 264.7 cells in (D) (n = 3). (F) RANKL-scavenging efficiencies of RAW-PLGA nanodecoys, antiR-RAW-PLGA nanodecoys, and siR-RAW-PLGA nanodecoys (n = 3). Relative c-Fos mRNA level (G) and p-ERK, p-p38, and p-JNK levels (H) in BMMs (n = 3). (I) TRAP staining images of BMM-derived osteoclasts (scale bar, 100 μm). (J) Size of TRAP-positive multinucleated osteoclasts (≥3 nuclei) (n = 3). In (G) to (J), cells were treated with RAW-PLGA nanodecoys (100 μg PLGA/ml) or PBS for 24 hours in the presence of RANKL (100 ng/ml), and cells treated with PBS served as the control.
Moreover, RAW-PLGA nanodecoys were able to scavenge the extracellular RANKL added to the culture medium of RAW 264.7 cells (Fig. 3C). As a result, binding of extracellular RANKL to RAW 264.7 cells was markedly inhibited by RAW-PLGA nanodecoys, as shown by CLSM images. Particularly, FITCRANKL (green fluorescence) situated on the DiI-stained cell membrane (red fluorescence) was effectively inhibited in the presence of RANKL antibody (anti-RANKL) or RAW-PLGA nanodecoys (Fig. 3D), and quantification of the mean fluorescence intensity of FITCRANKL in RAW 264.7 cells further indicated that RAW-PLGA nanodecoys inhibited RANKL binding by 93%, outperforming anti-RANKL (74%; Fig. 3E).
To demonstrate that the RANKL-scavenging capability of RAW-PLGA nanodecoys was related to the RANK on RAW 264.7 cell membranes, RAW 264.7 cells were transfected with RANK small interfering RNA (siRNA) (siRANK) or treated with anti-RANK to down-regulate the RANK level before the isolation of cell membrane and fabrication of RAW-PLGA nanodecoys. As illustrated in fig. S10, siRANK decreased the RANK mRNA and protein levels in RAW 264.7 cells by 73 and 59%, respectively, and anti-RANK blocked the RANK protein level by 47%. The RAW-PLGA nanodecoys containing siRANK- or anti-RANK–treated membrane were prepared and denoted as siR-RAW-PLGA nanodecoys or antiR-RAW-PLGA nanodecoys, respectively, and their RANKL-scavenging efficiencies were determined as described above. As illustrated in Fig. 3F, siR-RAW-PLGA nanodecoys and antiR-RAW-PLGA nanodecoys could only scavenge RANKL by 17 and 18%, respectively, much weaker than RAW-PLGA nanodecoys, suggesting that RANK on the cell membrane was critical to the RANKL-scavenging capability.
RAW-PLGA nanodecoy–mediated anti-osteoclastogenesis in vitro
The RAW-PLGA nanodecoy–mediated anti-osteoclastogenesis efficiency was first evaluated via determination of the c-Fos mRNA level in RANKL-challenged RAW 264.7 cells. As illustrated in fig. S11A, RAW-PLGA nanodecoys markedly down-regulated the c-Fos mRNA level by 73%, close to the level before RANKL challenge. In addition, RAW-PLGA nanodecoys decreased the phosphorylation of ERK, p38, and JNK in RANKL-challenged RAW 264.7 cells by 47, 57, and 57%, respectively (fig. S11, B to D). It therefore demonstrated that RAW-PLGA nanodecoys could efficiently block the RANKL-induced up-regulation of osteoclast differentiation–related genes in the NF-κB pathway and inhibit the phosphorylation of key signaling molecules in the MAPK pathway, which would contribute to the inhibition of osteoclast differentiation.
To further explore the capabilities of RAW-PLGA nanodecoys in inhibiting osteoclast differentiation, mouse bone marrow monocytes/macrophages (BMMs) that represent wider source and more complex composition of preosteoclasts were isolated and allowed to differentiate into osteoclasts in the presence of RANKL and macrophage colony-stimulating factor (M-CSF). Similarly to the results observed in RAW 264.7 cells, RAW-PLGA nanodecoys inhibited the activation of NF-κB and MAPK pathways in RANKL-challenged BMMs, wherein the mRNA level of c-Fos and phosphorylation of ERK, p38, and JNK were obviously down-regulated (Fig. 3, G and H). In addition, tartrate-resistant acid phosphatase (TRAP) staining images showed that BMMs were stimulated to form osteoclasts with large size and multiple nuclei, while this process was notably blocked by RAW-PLGA nanodecoys (Fig. 3, I and J).
RAW-PLGA nanodecoy–mediated TNF-α scavenging in vitro
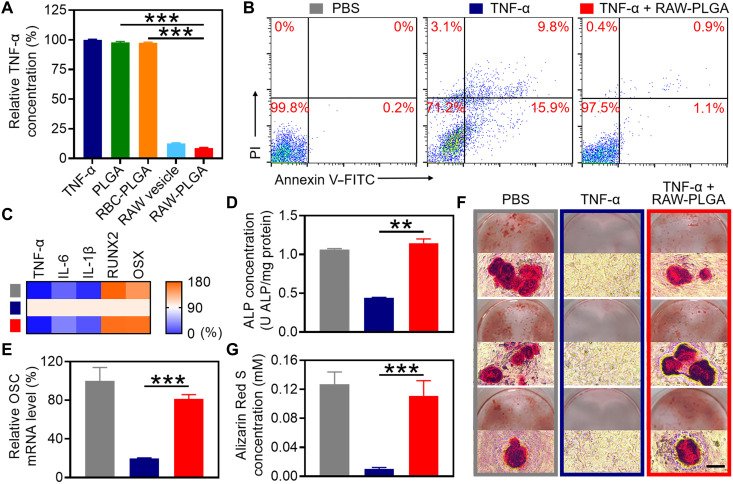
RAW-PLGA nanodecoy–mediated TNF-α scavenging was first explored via determination of the remaining TNF-α concentration after incubation of TNF-α solution with nanodecoys. As illustrated in Fig. 4A and fig. S12, RAW-PLGA nanodecoys showed concentration-dependent capability in scavenging TNF-α, conferring an IC50 value of 396.3 μg/ml. In comparison, PLGA NPs or RBC-PLGA NPs showed negligible scavenging effect. Minimal amount (~5%) of TNF-α was released from the TNF-α/RAW-PLGA complexes after incubation in DMEM containing 10% FBS for up to 48 hours (fig. S9B). Moreover, RAW-PLGA nanodecoys were able to scavenge the extracellular TNF-α added to the culture medium of MC3T3-E1 cells (fig. S13).
Fig. 4. In vitro TNF-α–scavenging and pro-osteoblastogenesis efficiencies of RAW-PLGA nanodecoys.
(A) Relative TNF-α concentration after treatment with PLGA NPs, RBC-PLGA NPs, RAW vesicles, or RAW-PLGA nanodecoys at the initial TNF-α concentration of 500 pg/ml (n = 3). (B) Apoptosis of MC3T3-E1 cells as determined by flow cytometry. (C) Relative mRNA levels of NF-κB–related genes (TNF-α, IL-6, and IL-1β) and osteoblast transcription factor genes (RUNX2 and OSX) in MC3T3-E1 cells (n = 3). (D) ALP protein level in MC3T3-E1 cells (n = 3). (E) Relative OSC mRNA level in MC3T3-E1 cells (n = 3). (F) Alizarin Red S staining images of MC3T3-E1 cells (scale bar, 100 μm). (G) Quantification of Alizarin Red S content in (F) (n = 3). In (C) to (G), cells were treated with RAW-PLGA nanodecoys (100 μg PLGA/ml) or PBS in the presence of TNF-α (10 ng/ml) for 1 day (C), 7 days (D), or 14 days (E to G), and cells treated with PBS served as the control.
To demonstrate that the TNF-α–scavenging capability of RAW-PLGA nanodecoys was related to the TNF-αR on RAW 264.7 cell membranes, RAW 264.7 cells were transfected with TNF-αR siRNA (siTNF-αR) or treated with anti–TNF-αR to down-regulate the TNF-αR level before the isolation of cell membrane and fabrication of RAW-PLGA nanodecoys (denoted as siT-RAW-PLGA nanodecoys or antiT-RAW-PLGA nanodecoys, respectively). As illustrated in fig. S14, siTNF-αR decreased the TNF-αR mRNA and protein levels in RAW 264.7 cells by 68 and 50%, respectively, and anti–TNF-αR blocked the TNF-αR protein level by 58%. In accordance, siT-RAW-PLGA nanodecoys and antiT-RAW-PLGA nanodecoys could only scavenge TNF-α by 46 and 47%, respectively, much weaker than RAW-PLGA nanodecoys (fig. S15), demonstrating that TNF-αR on the cell membrane was critical to the TNF-α–scavenging capability.
RAW-PLGA nanodecoy–mediated pro-osteoblastogenesis in vitro
The RAW-PLGA nanodecoy–mediated anti-apoptosis and NF-κB pathway blocking were first evaluated. RAW-PLGA nanodecoys notably alleviated the TNF-α–induced early and late apoptosis of MC3T3-E1 cells by 93 and 91%, respectively (Fig. 4B). Considering the important role of the NF-κB pathway in TNF-α–induced apoptosis, the mRNA levels of key genes of the NF-κB pathway (TNF-α, IL-6, and IL-1β) were determined in TNF-α–challenged MC3T3-E1 cells. As shown in Fig. 4C, RAW-PLGA nanodecoys markedly down-regulated the TNF-α, IL-6, and IL-1β mRNA levels by 98, 55, and 72%, respectively, indicating comprehensive blockade of the NF-κB pathway to impart anti-apoptosis effect.
The RAW-PLGA nanodecoy–mediated pro-osteoblastogenesis was then explored. As illustrated in Fig. 4C, after treatment with RAW-PLGA nanodecoys, the mRNA levels of osteoblast transcription factors (RUNX2 and OSX) almost recovered to the levels before TNF-α challenge. Similarly, the protein level of osteoblast differentiation-associated early marker (ALP) and mRNA level of late marker [osteocalcin (OSC)] (50, 51) in TNF-α–challenged MC3T3-E1 cells were up-regulated by 2.6- and 4.2-fold, respectively, after treatment with RAW-PLGA nanodecoys, close to the levels of unchallenged control cells (Fig. 4, D and E). It therefore demonstrated that RAW-PLGA nanodecoys could maintain the ability of MC3T3-E1 cells in differentiating into osteoblasts in the presence of TNF-α.
The capability of RAW-PLGA nanodecoys in relieving TNF-α–induced anti-mineralization was further evaluated via Alizarin Red S staining. TNF-α–challenged MC3T3-E1 cells showed minimal red calcium nodules, while, in contrast, nanodecoy-treated, TNF-α–challenged MC3T3-E1 cells developed obvious red calcium nodules, similar to the unchallenged control cells (Fig. 4F). Consistent results were obtained from quantitative analysis, wherein RAW-PLGA nanodecoys restored the Alizarin Red S level to ~87% of that without TNF-α challenge (Fig. 4G). Such findings substantiated that RAW-PLGA nanodecoys could eliminate the inhibitory effect of TNF-α on osteoblast mineralization to mediate pro-osteogenesis.
RAW-PLGA nanodecoy–mediated RANKL down-regulation after TNF-α challenge
In addition to inducing apoptosis of preosteoblasts and inhibiting osteoblastogenesis, TNF-α can also induce RANKL production from cells such as preosteoclasts and preosteoblasts, which, in turn, promotes osteoclastogenesis (19, 52). Here, RAW-PLGA nanodecoys down-regulated the RANKL mRNA level in TNF-α–challenged RAW 264.7 cells and MC3T3-E1 cells by 41 and 65%, respectively, indicating that RAW-PLGA nanodecoys could also inhibit endogenous RANKL production by scavenging TNF-α (fig. S16).
Pharmacokinetics and biodistribution of RAW-PLGA nanodecoys
The pharmacokinetics of DiDRAW vesicles and nanodecoys containing DiDPLGA were probed after intravenous injection. Both RAW-PLGA nanodecoys and RBC-PLGA NPs revealed notably longer blood circulation time than PLGA NPs (fig. S17), conferring the half-time (t1/2) of 13.26, 11.71, and 4.08 hours for RAW-PLGA nanodecoys, RBC-PLGA NPs, and PLGA NPs, respectively. Such enhancement of the blood circulation time could be attributed to the coating of NPs with blood cell membrane that avoided macrophage uptake to enhance the serum stability and evade the clearance by reticuloendothelial tissues such as liver. RAW vesicles showed notably shorter blood circulation time (3.11 hours) than RAW-PLGA nanodecoys, which could be attributed to their lower stability in serum (fig. S17). In support of such finding, live animal fluorescence imaging further revealed that RAW-PLGA nanodecoys had lower accumulation level in the liver than PLGA NPs and RBC-PLGA NPs (fig. S18).
Anti-osteoporosis efficacy of RAW-PLGA nanodecoys in ovariectomized mice
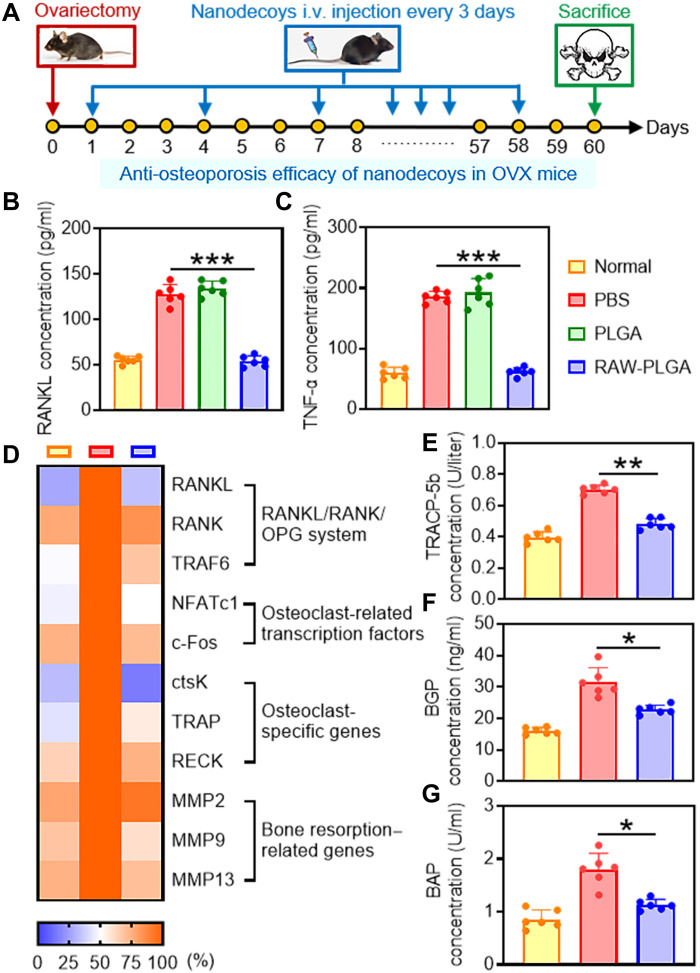
The anti-osteoporosis effect of nanodecoys was evaluated in ovariectomized (OVX) mice after they were intravenously injected every 3 days for a total of 20 injections (Fig. 5A). Compared to PBS treatment, the serum RANKL and TNF-α concentrations in RAW-PLGA nanodecoy–treated OVX mice were decreased by 57 and 69%, respectively, almost back to the normal levels (Fig. 5, B and C). In addition, the mRNA levels of key factors associated with RANKL (RANKL/RANK/OPG system gene), osteoclastogenesis (osteoclast-related transcription factors and osteoclast-specific genes), and bone resorption (MMPs) in the femoral tissues were normalized by RAW-PLGA nanodecoys (Fig. 5D). In particular, the key factors of the RANKL/RANK/OPG system [RANKL, RANK, and TNF receptor–associated factor 6 (TRAF6)] decreased to normal levels, demonstrating that the entire system was rebalanced. In the meantime, RAW-PLGA nanodecoys not only restored the abnormally up-regulated osteoclast-related transcription factors (NFATc1 and c-Fos) to normal levels but also inhibited the expression levels of osteoclast-specific genes (ctsK, TRAP, and RECK), suggesting their potent capabilities in inhibiting osteoclast differentiation. As a consequence of nanodecoy-mediated anti-osteoclastogenesis, the bone resorption–related genes (MMP2, MMP9, and MMP13) were substantially restored to normal levels, which would further benefit anti-osteoporosis as well as bone formation.
Fig. 5. Anti-osteoporosis efficacy of RAW-PLGA nanodecoys in OVX mice.
(A) Time line of the in vivo study. Female C57/BL6 mice were subjected to bilateral ovariectomy. In the next 60 days, PBS, PLGA NPs, or RAW-PLGA nanodecoys were intravenously (i.v.) injected at 10 mg PLGA/kg every 3 days, and mice were sacrificed on day 60 for analysis. Mice without ovariectomy or NP/nanodecoy treatment served as the normal control. RANKL (B) and TNF-α (C) levels in the serum (n = 6). (D) Relative mRNA levels of RANKL/RANK/OPG system genes (RANKL, RANK, and TRAF6), osteoclast-related transcription factors (NFATc1 and c-Fos), osteoclast-specific genes (ctsK, TRAP, and RECK), and bone resorption–related genes (MMP2, MMP9, and MMP13) in the femoral tissues (n = 6). TRACP-5b (E), BGP (F), and BAP (G) levels in the serum (n = 6).
Biochemical markers of bone turnover such as tartrate-resistant acid phosphatase 5b (TRACP-5b), bone Gla proteins (BGP), and bone-specific alkaline phosphatase (BAP) are closely associated with osteoclast activity, bone turnover rate, and hyperfunction of bone cells, respectively, and thus are important indicators for evaluating bone resorption and bone formation during postmenopausal osteoporosis (53–55). As illustrated in Fig. 5 (E to G), RAW-PLGA nanodecoys significantly reduced the serum concentrations of TRACP-5b, BGP, and BAP, which recovered to the levels close to those in normal mice.
Histological and radiographic evaluations
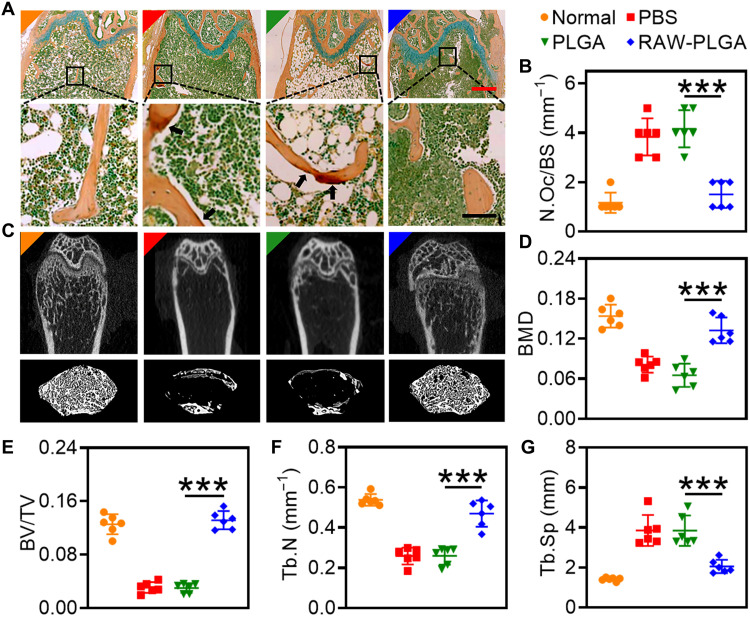
The persistence of osteoporosis may cause irreversible damage to the bone integrity and ultimately lead to an increase in the fracture risk (1, 3). Here, the protective effect of nanodecoys on trabecular bones was first explored by TRAP staining. As illustrated in Fig. 6A, RAW-PLGA nanodecoys efficiently decreased the degree of trabecular bone erosion by osteoclasts. Meanwhile, RAW-PLGA nanodecoys decreased the number of osteoclasts by 61%, almost back to the normal level (Fig. 6B).
Fig. 6. RAW-PLGA nanodecoy–mediated bone protection in OVX mice.
(A) Representative photomicrographs of TRAP-stained femoral sections (scale bars, 500 and 100 μm for top and bottom images, respectively). Arrows point to multiple nuclear osteoclasts. (B) Calculated osteoclast number from (A) (n = 6). (C) Representative micro-CT images of femoral and trabecular bones. BMD (D), BV/TV (E), Tb.N (F), and Tb.Sp (G) of mouse femurs calculated from micro-CT images (n = 6).
The recovery of femur integrity was further analyzed by micro–computed tomography (micro-CT). As illustrated in Fig. 6C, the ovariectomy-induced bone loss was inhibited and the osteopenic phenotypes in trabecular bones were alleviated after administration of RAW-PLGA nanodecoys. In addition, BMD, bone volume density (BV/TV), trabecular number (Tb.N), and trabecular separation (Tb.Sp) largely recovered to the normal levels after treatment with RAW-PLGA nanodecoys (Fig. 6, D to G).
Biocompatibility of nanodecoys
The in vivo biocompatibility of RAW-PLGA nanodecoys was examined after intravenous injection (10 mg PLGA/kg) to female C57/BL6 mice as described above for the efficacy study. Compared to PBS-treated mice, RAW-PLGA nanodecoy–treated mice showed negligible abnormalities in the context of representative hematological parameters and biochemical parameters (fig. S19). In addition, in the hematoxylin and eosin (H&E)–stained cross sections of major organs (heart, liver, spleen, lung, and kidney), no necrosis, inflammation, edema, or other pathological symptoms were detected (fig. S20). These results suggested the desired biocompatibility of RAW-PLGA nanodecoys after systemic administration.
DISCUSSION
Here, we developed preosteoclast membrane–coated nanodecoys that were able to rebalance osteoclasts and osteoblasts. In the in vitro studies, RAW-PLGA nanodecoys exhibited potent anti-osteoclastogenesis and pro-osteoblastogenesis efficacies by scavenging RANKL and TNF-α, respectively. Thus, the reconstructed balance between osteoclasts and osteoblasts contributed to the pronounced anti-osteoporosis effect in OVX mice. Besides RANKL and TNF-α, other cytokines such as IL-6, IL-1β, and IFN-γ also participate in the formation of osteoporosis, while they may not be as important as RANKL or TNF-α. With IL-6 as an example, previous studies have shown that inhibition of IL-6 is less effective for the treatment of osteoporosis than inhibition of TNF-α or RANKL (56, 57). Besides, it has been well documented that TNF-α can synergize with RANKL to induce osteoporosis, while such synergism is not observed for IL-6 (52, 58). In addition, IL-6 induces T helper 17 (TH17) cell differentiation, which is mostly associated with inflammatory osteoporosis and is beyond the scope of this study (58). To this end, the current study is mainly focused on evaluating the scavenging efficiencies of nanodecoys against RANKL and TNF-α and elucidating their mechanisms during anti-osteoclastogenesis and pro-osteoblastogenesis. Further studies on the scavenging of more cytokines toward the treatment of different types of osteoporosis are definitely valuable and will be explained in future studies.
In conclusion, this study provides an effective example for using cell membrane–based nanodecoys to specifically scavenge the molecules involved in osteoporosis induction, and it also renders promising implications into the treatment of postmenopausal osteoporosis by orchestrating the osteomicroenvironment. The simple preparation process and wide source of raw materials will benefit the translation of RAW-PLGA nanodecoys. Composed of the U.S. Food and Drug Administration–approved PLGA and bio-originated cell membrane, the nanodecoys would have desired biocompatibility and safety. The broad-spectrum cytokine neutralization capabilities of RAW-PLGA nanodecoys render obvious advantages over existing drugs that act only on a single target. Owing to the long circulation time of nanodecoys, the dosing frequency could be largely reduced, which would lead to better patient compliance. These unique attributes of the RAW-PLGA nanodecoys thus suggest the profound potentials as safe and effective anti-osteoporosis therapeutics. While further research is needed to detect the long-term effect of nanodecoy-bound cytokines and to expand the applicability to different administration routes, the vast therapeutic potential of cell membrane–based platforms is poised to shift the paradigm in postmenopausal osteoporosis management.
MATERIALS AND METHODS
Materials, cell lines, and animals
PLGA, murine RANKL, M-CSF, trypan blue, L-ascorbic acid, β-glycerophosphate, and cetylpyridinium chloride were purchased from Sigma-Aldrich (St. Louis, MO, USA). Lipofectamine (LPF2000) and bicinchoninic acid (BCA) protein assay kit were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Murine TNF-α was purchased from PeproTech China (Suzhou, China). All primers, subcellular structure protein extraction kit, siRANK (sense, 5′-GCGCAGACUUCACUCCAUAUU-3′; antisense, 5′-UAUGGAGUGAAGUCUGCGCUU-3′), siTNF-αR (sense, 5′-GAAGGAGUUCAUGCGUUUCAUUU-3′; antisense, 5′-AUGAAACGCAUGAACUCCUUCUU-3′), and negative control siRNA with scrambled sequence (sense, 5′-UUCUCCGAACGUGUCACGUTT-3′; antisense, 5′-ACGUGACACGUUCGGAGAATT-3′) were purchased from Sangon Biotech (Shanghai, China). All enzyme-linked immunosorbent assay (ELISA) kits and Alizarin Red S were purchased from Sanjia Biochemical Supplies (Jiangsu, China). TRAP staining kit was purchased from G-CLONE (Beijing, China). An annexin V–fluorescein isothiocyanate (FITC)/propidium iodide (PI) apoptosis detection kit was purchased from Beyotime (Shanghai, China). All antibodies were purchased from Abcam (Cambridge, Britain).
RAW 264.7 cells (mouse preosteoclast) and MC3T3-E1 cells (mouse preosteoclast) were purchased from the American Type Culture Collection (Rockville, MD, USA) and were in DMEM containing 10% FBS.
Female C57/BL6 mice (6 to 8 weeks, 18 to 20 g) were purchased from Shanghai Slaccas Experimental Animal Co. Ltd. (Shanghai, China) and housed in a clean room, four to a cage, with access to water ad libitum, a 12-hour light:12-hour dark cycle (7:00 a.m. to 7:00 p.m.), and a temperature of 25 ± 1°C. The animal experimental protocols were reviewed and approved by the Institutional Animal Care and Use Committee, Soochow University.
Isolation of RAW 264.7 cell membrane
RAW 264.7 cell membrane was isolated according to reported procedures (43). Briefly, RAW 264.7 cells were suspended in the homogenization buffer containing 20 mM tris-HCl (pH 7.5), 10 mM KCl, 75 mM sucrose, 2 mM MgCl2, and one tablet of protease/phosphatase inhibitors. The suspension was disrupted with a JY 92-IIN homogenizer (75 W) and then centrifuged at 20,000g for 25 min. The cell membrane was collected by centrifuging the supernatant (100,000g, 35 min). The protein content of collected cell membrane was determined using a BCA kit. Membranes containing about 5 mg of membrane protein could be extracted from 3 × 107 RAW 264.7 cells. For fluorescence microscopy imaging and FRET analyses, DiD-stained RAW 264.7 cell membrane (DiDRAW) and Cy5-stained RAW 264.7 cell membrane (Cy5RAW) were prepared by mixing the cell membrane with DiD or Cy5-N-hydroxysuccinimide ester (Cy5-NHS) at a membrane protein-to-dye weight ratio of 1000:1 or 20:1, respectively (40, 43).
Isolation of RBC membrane
RBC membrane was isolated according to reported procedures (41). Briefly, RBCs collected from whole blood of female C57/BL6 mice were resuspended in 0.25 × PBS and centrifuged at 800g for 5 min. The RBC membrane was collected by repeating this process until the hemoglobin was completely removed, and the protein concentration was quantified using a BCA kit. For FRET analysis, Cy5-stained RBC membrane (Cy5RBC) was similarly prepared as described above.
Preparation and characterization of nanodecoys
RAW-PLGA nanodecoys were prepared using the sonication method (40, 43). Briefly, to synthesize the nanocore, PLGA in acetone (10 mg/ml, 1 ml) was added dropwise into deionized (DI) water (2 ml). The mixture was then stirred in the open air until the acetone had evaporated completely. DiD-labeled PLGA (DiDPLGA) NPs were prepared by adding DiD to the PLGA acetone solution at a polymer/dye weight ratio of 1000:1 followed by preparation of NPs using the same method. Excessive DiD was removed by centrifugation (20,000g, 15 min). To prepare RAW vesicles, RAW 264.7 cell membrane was sonicated with a bath sonicator (Fisher Scientific, FS30D, 100 W) for 2 min. To prepare the RAW-PLGA nanodecoys, RAW vesicles were mixed with PLGA NPs at the membrane protein/PLGA weight ratio of 1:4. The mixture was then sonicated with a bath sonicator for 2 min (100 W). RBC-PLGA NPs were prepared following the same procedure.
The hydrodynamic size and zeta potential of nanodecoys were determined using Zetasizer Nano ZS90 (Malvern Instruments Ltd., UK). The morphology of nanodecoys was visualized using TEM (TECNAI G2, FEI, USA) after staining with uranyl acetate (0.2 weight %). Macrophage-specific surface markers (MAC-1, macrosialin, IFN-γR, TNF-αR, IL-6R, and RANK) on RAW vesicles and RAW-PLGA nanodecoys were examined by Western blotting. The concentrations of MAC-1, macrosialin, IFN-γR, TNF-αR, IL-6R, RANK, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) primary antibodies were 1:1000, and the concentration of horseradish peroxidase (HRP)–labeled secondary antibody was 1:500. The coating efficiency of RAW membranes on PLGA NPs was determined by monitoring the fluorescence intensity of RAW-DiDPLGA nanodecoys or DiDRAW-PLGA nanodecoys (1 mg PLGA/ml) before and after centrifugation (10,000g, 10 min). The serum stability of nanodecoys was evaluated by measuring the particle size of nanodecoys in DI water or DMEM containing 10% FBS after incubation at RT for different times.
Cell uptake of nanodecoys
The cellular uptake of nanodecoys was monitored by flow cytometry. Briefly, RAW 264.7 cells were seeded on 24-well plates at 5 × 104 cells/well and cultured for 24 hours. DiDPLGA NPs or RAW-DiDPLGA nanodecoys were added at 100 μg DiDPLGA/ml (500 μl per well) and incubated with cells for 4 hours. Cells were then treated with trypan blue solution (0.4%) for 10 min, washed with PBS, and analyzed by flow cytometry.
In a parallel study, RAW 264.7 cells were treated with DiDPLGA NPs or RAW-DiDPLGA nanodecoys similarly before being lysed with the passive lysis buffer. DiDPLGA concentration and protein concentration in the lysate were monitored by spectrofluorimetry (λex = 644 nm, λem = 663 nm) and using the BCA kit, respectively. The uptake level was denoted as ng DiDPLGA per mg protein.
CLSM was further used to observe the cellular uptake of nanodecoys. RAW 264.7 cells were seeded on a cell culture dish (Ф = 20 mm) at 4 × 104 cells per dish and cultured for 24 hours. After staining of the cell membrane with DiI (5 μg/ml), cells were incubated with DiDPLGA NPs or RAW-DiDPLGA nanodecoys (100 μg DiDPLGA/ml) at 37°C for 4 hours. After washing with cold PBS for three times, cells were observed by CLSM (Leica, TCS SP5, Germany).
Cytotoxicity of nanodecoys
RAW 264.7 cells or MC3T3-E1 cells on 96-well plates were similarly incubated with RAW-PLGA nanodecoys (5, 10, 20, 40, 80, or 160 μg PLGA/ml) at 37°C for 24 hours. Cell viability was determined by 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay (49).
Immunostimulatory properties of nanodecoys
RAW 264.7 cells on 96-well plates were similarly incubated with RAW-PLGA nanodecoys (5, 10, 20, 40, 80, or 160 μg PLGA/ml) at 37°C for 24 hours. The TNF-α and IL-6 concentrations in the cell culture medium were determined by ELISA kits.
Quantification of RANKL scavenging
RANKL (200 pg/ml) was mixed with PLGA NPs, RBC-PLGA NPs, RAW vesicles, or RAW-PLGA nanodecoys at 1 mg PLGA/ml or 0.25 mg membrane protein/ml. The mixtures were incubated for 2 hours at 37°C and then centrifuged at 16,100g for 10 min. RANKL concentration in the supernatant was quantified using an ELISA kit. The IC50 value was derived from the variable slope model using GraphPad Prism 8. In a parallel study, the precipitate after centrifugation was redissolved in DMEM containing 10% FBS and incubated at 37°C. At predetermined time intervals, RANKL release from the RANKL/RAW-PLGA complexes was evaluated by measuring the RANKL concentration in the supernatant after centrifugation. The RANKL binding was further monitored by FRET. Briefly, Cy3RANKL (200 pg/ml) and Cy5RBC-PLGA or Cy5RAW-PLGA (1 mg PLGA/ml) as the FRET pair were mixed and incubated as mentioned above. The fluorescence emission spectrum was collected within the range of 565 to 750 nm (λex = 550 nm).
To explore the real-time RANKL-scavenging effect of nanodecoys in the presence of cells, RAW 264.7 cells were seeded on 96-well plates at 1 × 104 cells per well and cultured for 24 hours. Cells were then incubated with RANKL (100 ng/ml) and RAW-PLGA nanodecoys (100 μg PLGA/ml) at 37°C. The RANKL-scavenging efficiency was evaluated by measuring the RANKL concentration in the cell culture medium at predetermined time intervals.
To demonstrate the critical role of membrane-bound RANK in scavenging RANKL, RAW 264.7 cells on cell culture dish (Ф = 60 mm, 80% cell confluency) were treated with LPF2000/siRANK nanocomplexes (LPF2000/siRANK = 1:1, w/w) or anti-RANK (0.05 mg/ml) in serum-free medium for 4 hours, followed by further incubation in fresh serum-containing medium for 20 hours to deplete the RANK expression on cell membranes. RANK mRNA level in RAW 264.7 cells was then quantified by real-time polymerase chain reaction (PCR) using RANK primers (table S1). The RANK expression level on cell membranes was further determined by ELISA after the membrane proteins were isolated using the subcellular structure protein extraction kit. Then, RAW 264.7 cells treated with LPF2000/siRANK nanocomplexes or anti-RANK were used to isolate cell membrane and prepare the membrane-coated nanodecoys as mentioned above, obtaining the siR-RAW-PLGA nanodecoys and antiR-RAW-PLGA nanodecoys, respectively. RANKL (200 pg/ml) was mixed with RAW-PLGA nanodecoys, siR-RAW-PLGA nanodecoys, or antiR-RAW-PLGA nanodecoys at 1 mg PLGA/ml. The mixtures were incubated for 2 hours at 37°C and then centrifuged at 16,100g for 10 min before determination of the RANKL concentration in the supernatant using an ELISA kit.
In vitro anti-osteoclastogenesis efficiency
RAW 264.7 cells were seeded on six-well plates at 2 × 105 cells per well and cultured for 24 hours. Cells were then incubated with RANKL (100 ng/ml) and RAW-PLGA nanodecoys (100 μg PLGA/ml) for 24 hours. c-Fos mRNA level was then quantified by real-time PCR. In addition, cells were lysed using the lysis buffer [200 μl per well, 50 mM tris-HCl, 0.15 mM NaCl, 1% Triton X-100, and phenylmethylsulfonyl fluoride (100 μg/ml)], and p-ERK, p-p38, and p-JNK protein concentrations were quantified using ELISA kits.
To visualize the nanodecoy-mediated blocking of binding between RANKL and RAW 264.7 cell membrane, RAW 264.7 cells were seeded on a cell culture dish (Ф = 20 mm) at 4 × 104 cells per dish and cultured for 24 hours. Cells were stained with DiI (5 μg/ml, for cell membrane) and then cultured in fresh medium containing FITCRANKL (100 ng/ml). After incubation with anti-RANKL (0.05 mg/ml) or RAW-PLGA nanodecoys (100 μg PLGA/ml) for 4 hours, cells were washed with cold PBS for three times, stained with Hoechst 33258 (5 μg/ml, for nuclei), and observed by CLSM.
To explore the nanodecoy-mediated inhibition of RANKL-induced osteoclast formation, mouse BMMs were isolated from mouse bone marrow, seeded on 96-well plates at 1 × 104 cells per well, and cultured for 24 hours. Cells were then cultured in fresh medium containing M-CSF (30 ng/ml) and RANKL (100 ng/ml) and incubated with RAW-PLGA nanodecoys (100 μg PLGA/ml) for 24 hours. c-Fos mRNA level and p-ERK, p-p38, and p-JNK protein levels were quantified as mentioned above. In a parallel study, BMMs were cultured in fresh medium containing M-CSF (30 ng/ml) and RANKL (100 ng/ml) and incubated with RAW-PLGA nanodecoys (100 μg PLGA/ml) for 96 hours. Cells were then fixed with 4% buffered formalin for 10 min, washed three times with PBS, stained using the TRAP staining kit, and imaged under an inverted microscope (Leica DM4000, Solms, Germany).
Quantification of TNF-α scavenging
TNF-α (500 pg/ml) was mixed with PLGA NPs, RBC-PLGA NPs, RAW vesicles, or RAW-PLGA nanodecoys at 1 mg PLGA/ml or 0.25 mg membrane protein/ml. The mixtures were incubated for 2 hours at 37°C and then centrifuged at 16,100g for 10 min before determination of the TNF-α concentration in the supernatant using an ELISA kit. The IC50 value for TNF-α scavenging and the release of TNF-α from the TNF-α/RAW-PLGA complexes were evaluated using the same method as described above for RANKL.
To explore the real-time TNF-α–scavenging effect of nanodecoys in the presence of cells, MC3T3-E1 cells were seeded on 96-well plates at 1 × 104 cells per well and cultured for 24 hours. Cells were then incubated with TNF-α (100 ng/ml) and RAW-PLGA nanodecoys (100 μg PLGA/ml) at 37°C. The TNF-α–scavenging efficiency was evaluated by measuring the TNF-α concentration in the cell culture medium at predetermined time intervals.
To demonstrate the critical role of membrane-bound TNF-αR in scavenging TNF-α, RAW 264.7 cells were treated with LPF2000/siTNF-αR nanocomplexes (LPF2000/siTNF-αR = 1:1, w/w) or anti-TNF-αR (0.1 mg/ml) before isolation of cell membranes and construction of siT-RAW-PLGA nanodecoys and antiT-RAW-PLGA nanodecoys using the same method as described for siR-RAW-PLGA nanodecoys and antiR-RAW-PLGA nanodecoys. The obtained nanodecoys were subjected to the determination of TNF-α–scavenging efficiencies using an ELISA kit.
In vitro pro-osteoblastogenesis efficiency
MC3T3-E1 cells were seeded on 96-well plates at 1 × 104 cells per well and cultured for 24 hours. Cells were then incubated with TNF-α (50 ng/ml) and RAW-PLGA nanodecoys (100 μg PLGA/ml) at 37°C for 24 hours. Cells were collected, stained with an annexin V–FITC/PI apoptosis detection kit, and subjected to flow cytometry analysis.
To explore the nanodecoy-mediated blocking of TNF-α–induced NF-κB activation, MC3T3-E1 cells were seeded on six-well plates at 2 × 105 cells per well, cultured for 24 hours in the normal medium, and further cultured in the differentiation medium [αMEM (minimum essential medium) containing 10% FBS, L-ascorbate (50 μg/ml), and 10 mM β-glycerophosphate] for 24 hours. Cells were then incubated with TNF-α (10 ng/ml) and RAW-PLGA nanodecoys (100 μg PLGA/ml) for 24 hours. TNF-α, IL-6, and IL-1β mRNA levels were quantified by real-time PCR using specific primers (table S1).
To explore the nanodecoy-mediated blocking of TNF-α–induced anti-osteoblastogenesis, MC3T3-E1 cells were seeded on six-well plates at 2 × 105 cells per well [for 1- and 7-day incubation hereafter] or 1 × 104 cells per well (for 14-day incubation hereafter), cultured for 24 hours in the normal medium, and further cultured in the differentiation medium for 24 hours. Cells were then incubated with TNF-α (10 ng/ml) and RAW-PLGA nanodecoys (100 μg PLGA/ml) for 1, 7, or 14 days, wherein the differentiation medium containing TNF-α and RAW-PLGA nanodecoys was daily refreshed. On day 1, RUNX2 and OSX mRNA levels were quantified by real-time PCR using specific primers (table S1). On day 7, intracellular ALP protein was extracted using the E1A lysis buffer [250 mM NaCl, 0.1% NP-40, and 50 mM Hepes (pH 7.5)], and its concentration was quantified using an ELISA kit. On day 14, OSC mRNA level was quantified by real-time PCR.
In vitro inhibition of TNF-α–induced RANKL up-regulation
RAW 264.7 cells or MC3T3-E1 cells were seeded on 96-well plates at 1 × 104 cells/well and cultured for 24 hours. Cells were then incubated with TNF-α (50 ng/ml) and RAW-PLGA nanodecoys (100 μg PLGA/ml) at 37°C for 24 hours. RANKL mRNA level was then quantified by real-time PCR using RANKL primers (table S1).
Determination of mineralization
MC3T3-E1 cells were incubated with TNF-α (10 ng/ml) and RAW-PLGA nanodecoys (100 μg PLGA/ml) for 14 days, and the differentiation medium containing TNF-α and RAW-PLGA nanodecoys was daily refreshed as described above. Cells were washed with PBS for three times and fixed with 4% buffered formalin for 30 min. After washing with DI water for three times, cells were incubated with Alizarin Red S (40 mM) for 5 min and then washed with PBS for 15 min. Cells were observed with an inverted microscope and imaged. To quantify the intracellular concentration of Alizarin Red S, 10% cetylpyridinium chloride in disodium hydrogen phosphate solution (10 mM, pH 7) was added to cells at 1 ml per well. After 1 hour, the absorbance of the solution at 562 nm was measured.
Pharmacokinetics and biodistribution
Female C57/BL6 mice were intravenously injected with DiDPLGA NPs, DiDRAW vesicles, RBC-DiDPLGA NPs, or RAW-DiDPLGA nanodecoys at 10 mg DiDPLGA/kg or 2.5 mg DiDRAW vesicles/kg. At predetermined time points, blood was collected and the DiDPLGA content or DiDRAW vesicles in the plasma were determined by spectrofluorimetry (λex = 644 nm, λem = 663 nm) as described previously (49). The circulation t1/2 of NPs was calculated according to the previously described method (59). In a parallel study, mice were sacrificed at 24 hours after injection. The major organs were harvested and imaged (λex = 644 nm, λem = 663 nm) using a Maestro in vivo imaging system to evaluate the biodistribution profile.
Establishment of OVX mouse model and drug administration
Female C57/BL6 mice were subjected to bilateral ovariectomy. In the next 60 days, PBS, PLGA NPs, or RAW-PLGA nanodecoys were intravenously injected at 10 mg PLGA/kg every 3 days (six mice per group). Mice without bilateral ovariectomy or NP treatment served as the normal control.
In vivo anti-osteoporosis efficiency
On day 60 after bilateral ovariectomy, mice were sacrificed, and peripheral blood was collected and centrifuged at 500g at 4°C for 10 min to extract the serum. RANKL, TNF-α, TRACP-5b, BGP, and BAP levels in the serum were quantified using ELISA kits.
In addition, mice were sacrificed, and the femoral tissues were frozen in liquid nitrogen and homogenized with the TRIzol reagent to isolate total RNA. The mRNA levels of the RANKL/RANK/OPG system (RANK, RANKL, and TRAF6), osteoclast-related transcription factors (NFATc1 and c-Fos), osteoclast-specific genes (ctsK, TRAP, and RECK), and bone resorption–related factors (MMP2, MMP9, and MMP13) in the femur tissues were determined by real-time PCR.
Histological analysis
On day 60 after bilateral ovariectomy, mice were sacrificed, and femoral tissues were harvested, fixed in 10% buffered formalin, and then incubated in decalcifying solution (14% EDTA) at RT for 1 month for decalcification. Then, the femurs were embedded in paraffin, cross-sectioned at 8 μm in thickness, and stained using a TRAP staining kit. The number of osteoclasts per bone surface (N.Oc/BS) was calculated (35).
Micro-CT imaging
The right femur specimens of OVX mice were scanned using micro-CT (Skyscan 1176, Belgium) on day 60 after bilateral ovariectomy. High-resolution scanograms (9 to 20 mm) were obtained (resolution: 8.8 mm, source voltage: 50 kV, source current: 500 mA, rotation step: 0.7°). The dataset was reconstructed using a CT analyzer software (Skyscan) to obtain the three-dimensional images of femoral tissues and to measure morphometric parameters. Bone erosion on micro-CT scans was calculated based on reconstructed data with an in-house written Fiji script. The program determines the bone surface and bone inner space and fills the pores in the bone surface. The region of interest (ROI) in calcaneus was chosen for analysis of the following morphometric parameters including (i) BMD, (ii) BV/TV, (iii) Tb.N, and (iv) Tb.Sp.
Biocompatibility evaluation
PBS (200 μl) or RAW-PLGA nanodecoys (10 mg PLGA/kg, 200 μl) were intravenously injected to female C57/BL6 mice as described above. Blood and major organs (heart, liver, spleen, lung, and kidney) were then collected. The major organs were fixed in 10% buffered formalin, embedded in paraffin, cross-sectioned at 8 μm in thickness, stained with H&E, and observed by overall perspective optical microscopy.
Hematological assessment was performed on a Cobas501 automatic hematology analyzer (Roche, USA). Serum levels of biochemical parameters were determined using a BC-5380 automatic chemistry analyzer (Mindray, China). The abbreviations of various hematological and biochemical parameters were listed in tables S2 and S3.
Statistical analysis
All data were presented as mean ± SD, and statistical analysis was performed using Student’s t test. Differences between the two groups were judged to be significant at *P < 0.05 and very significant at **P < 0.01 and ***P < 0.001.
Acknowledgments
Funding: This study was supported by the National Natural Science Foundation of China (51873142 and 52033006), the Collaborative Innovation Center of Suzhou Nano Science and Technology, The 111 Project, Suzhou Key Laboratory of Nanotechnology and Biomedicine, and the Joint International Research Laboratory of Carbon-Based Functional Materials and Devices.
Author contributions: Methodology: L.Y., Y.Z., Y.D., and M.H. Experiment: Y.Z., Y.D., Z.L., M.Y., and Z.Z. Supervision: L.Y. Writing—original draft: Y.Z. and L.Y. Writing—draft revision and proofreading: Y.Z., X.Z., and L.Y.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S20
Tables S1 to S3
REFERENCES AND NOTES
- 1.Zhang Y., Su J., Wu S., Teng Y., Yin Z., Guo Y., Li J., Li K., Yao L., Li X., DDR2 (discoidin domain receptor 2) suppresses osteoclastogenesis and is a potential therapeutic target in osteoporosis. Sci. Signal. 8, ra31 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Wu M., Chen W., Lu Y., Zhu G., Hao L., Li Y.-P., Gα13 negatively controls osteoclastogenesis through inhibition of the Akt-GSK3β-NFATc1 signalling pathway. Nat. Commun. 8, 13700 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reid I. R., A broader strategy for osteoporosis interventions. Nat. Rev. Endocrinol. 16, 333–339 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Liu J.-H., Chen C.-Y., Liu Z.-Z., Luo Z.-W., Rao S.-S., Jin L., Wan T.-F., Yue T., Tan Y.-J., Yin H., Yang F., Huang F.-Y., Guo J., Wang Y.-Y., Xia K., Cao J., Wang Z.-X., Hong C.-G., Luo M.-J., Hu X.-K., Liu Y.-W., Du W., Luo J., Hu Y., Zhang Y., Huang J., Li H.-M., Wu B., Liu H.-M., Chen T.-H., Qian Y.-X., Li Y.-Y., Feng S.-K., Chen Y., Qi L.-Y., Xu R., Tang S.-Y., Xie H., Extracellular vesicles from child gut microbiota enter into bone to preserve bone mass and strength. Adv. Sci. 8, 2004831 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu Z., Greenblatt M. B., Yan G., Feng H., Sun J., Lotinun S., Brady N., Baron R., Glimcher L. H., Zou W. G., SMURF2 regulates bone homeostasis by disrupting SMAD3 interaction with vitamin D receptor in osteoblasts. Nat. Commun. 8, 14570 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maeda K., Kobayashi Y., Udagawa N., Uehara S., Ishihara A., Mizoguchi T., Kikuchi Y., Takada I., Kato S., Kani S., Nishita M., Marumo K., Martin T. J., Minami Y., Takahashi N., Wnt5a-Ror2 signaling between osteoblast-lineage cells and osteoclast precursors enhances osteoclastogenesis. Nat. Med. 18, 405–412 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Weitzmann M. N., Ofotokun I., Physiological and pathophysiological bone turnover–Role of the immune system. Nat. Rev. Endocrinol. 12, 518–532 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hofbauer L. C., Schoppet M., Clinical implications of the osteoprotegerin/RANKL/RANK system for bone and vascular diseases. JAMA 292, 490–495 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Luo J., Yang Z., Ma Y., Yue Z., Lin H., Qui G., Huang J., Dai W., Li C., Zheng C., Xu L., Chen H., Wang J., Li D., Siwko S., Penninger J. M., Ning G., Xiao J., Liu M., LGR4 is a receptor for RANKL and negatively regulates osteoclast differentiation and bone resorption. Nat. Med. 22, 539–546 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Lee J. W., Hoshino A., Inoue K., Saitou T., Uehara S., Kobayashi Y., Ueha S., Matsushima K., Yamaguchi A., Imai Y., Iimura T., The HIV co-receptor CCR5 regulates osteoclast function. Nat. Commun. 8, 2226 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishikawa K., Iwamoto Y., Kobayashi Y., Katsuoka F., Kawaguchi S., Tsujita T., Nakamura T., Kato S., Yamamoto M., Takayanagi H., Ishii M., DNA methyltransferase 3a regulates osteoclast differentiation by coupling to an S-adenosylmethionine-producing metabolic pathway. Nat. Med. 21, 281–287 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Saito H., Gassers A., Bolamperti S., Maeda M., Matthies L., Jähn K., Long C. L., Schlüter H., Kwiatkowski M., Saini V., Pajevic P. D., Bellido T., van Wijnen A. J., Mohammad K. S., Guise T. A., Taipaleenmäki H., Hesse E., TG-interacting factor 1 (Tgif1)-deficiency attenuates bone remodeling and blunts the anabolic response to parathyroid hormone. Nat. Commun. 10, 1354 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu W., Zhou L., Zhou C., Zhang S., Jing J., Xie L., Sun N., Duan X., Jing W., Liang X., Zhao H., Ye L., Chen Q., Yuan Q., GDF11 decreases bone mass by stimulating osteoclastogenesis and inhibiting osteoblast differentiation. Nat. Commun. 7, 12794 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takayanagi H., Kim S., Koga T., Nishina H., Isshiki M., Yoshida H., Saiura A., Isobe M., Yokochi T., Inoue J., Wagner E. F., Mak T. W., Kodama T., Taniguchi T., Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev. Cell 3, 889–901 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Ping Z., Wang Z., Shi J., Wang L., Guo X., Zhou W., Hu X., Wu X., Liu Y., Zhang W., Yang H., Xu Y., Gu Y., Geng D., Inhibitory effects of melatonin on titanium particle-induced inflammatory bone resorption and osteoclastogenesis via suppression of NF-κB signaling. Acta Biomater. 62, 362–371 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Tian B., Jiang T., Shao Z., Zhai Z., Li H., Fan Q., Liu X., Ouyang Z., Tang T., Jiang Q., Zheng M., Dai K., Qin A., Yu Y., Zhu Z., The prevention of titanium-particle-induced osteolysis by OA-14 through the suppression of the p38 signaling pathway and inhibition of osteoclastogenesis. Biomaterials 35, 8937–8950 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Heo D. N., Ko W. K., Moon H. J., Kim H. J., Lee S. J., Lee J. B., Bae M. S., Yi J. K., Hwang Y. S., Bang J. B., Kim E. C., Do S. H., Kwon I. K., Inhibition of osteoclast differentiation by gold nanoparticles functionalized with cyclodextrin curcumin complexes. ACS Nano 8, 12049–12062 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Chen K., Qiu P., Yuan Y., Zheng L., He J., Wang C., Guo Q., Kenny J., Liu Q., Zhao J., Chen J., Tickner J., Fan S., Lin X., Xu J., Pseurotin A inhibits osteoclastogenesis and prevents ovariectomized-induced bone loss by suppressing reactive oxygen species. Theranostics 9, 1634–1650 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J., Ayoub A., Xiu Y., Yin X., Sanders J. O., Mesfin A., Xing L., Yao Z., Boyce B. F., TGFβ -induced degradation of TRAF3 in mesenchymal progenitor cells causes age-related osteoporosis. Nat. Commun. 10, 2795 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Redlich K., Smolen J. S., Inflammatory bone loss: Pathogenesis and therapeutic intervention. Nat. Rev. Drug Discov. 11, 234–250 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Walsh M. C., Takegahara N., Kim H., Choi Y., Updating osteoimmunology: Regulation of bone cells by innate and adaptive immunity. Nat. Rev. Rheumatol. 14, 146–156 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jilka R. L., Weinstein R. S., Bellido T., Parfitt A. M., Manolagas S. C., Osteoblast programmed cell death (apoptosis): Modulation by growth factors and cytokines. J. Bone Miner. Res. 13, 793–802 (1998). [DOI] [PubMed] [Google Scholar]
- 23.Schett G., David J.-P., The multiple faces of autoimmune-mediated bone loss. Nat. Rev. Endocrinol. 6, 698–706 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Gilbert L., He X. F., Farmer P., Rubin J., Drissi H., van Wijnen A. J., Lian J. B., Stein G. S., Nanes M. S., Expression of the osteoblast differentiation factor RUNX2 (Cbfa1/AML3/Pebp2αA) is inhibited by tumor necrosis factor-α. J. Biol. Chem. 277, 2695–2701 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Krum S. A., Chang J., Miranda-Carboni G., Wang C. Y., Novel functions for NFκB: Inhibition of bone formation. Nat. Rev. Rheumatol. 6, 607–611 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zuo C., Zhao X., Shi Y., Wu W., Zhang N., Xu J., Wang C., Hu G., Zhang X., TNF-α inhibits SATB2 expression and osteoblast differentiation through NF-κB and MAPK pathways. Oncotarget 9, 4833–4850 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu X., Gilbert L., He X., Rubin J., Nanes M. S., Transcriptional regulation of the osterix (Osx, Sp7) promoter by tumor necrosis factor identifies disparate effects of mitogen-activated protein kinase and NF kappa B pathways. J. Biol. Chem. 281, 6297–6306 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Wang Y. L., He H., Liu Z. J., Cao Z. G., Wang X. Y., Yang K., Fang Y., Han M., Zhang C., Huo F. Y., Effects of TNF-α on cementoblast differentiation, mineralization, and apoptosis. J. Dent. Res. 94, 1225–1232 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Warren J. T., Nelson C. A., Decker C. E., Zou W., Fremont D. H., Teitelbaum S. L., Manipulation of receptor oligomerization as a strategy to inhibit signaling by TNF superfamily members. Sci. Signal. 7, ra80 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rao S.-S., Hu Y., Xie P.-L., Cao J., Wang Z.-X., Liu J.-H., Yin H., Huang J., Tan Y.-J., Luo J., Luo M.-J., Tang S.-Y., Chen T.-H., Yuan L.-Q., Liao E.-Y., Xu R., Liu Z.-Z., Chen C.-Y., Xie H., Omentin-1 prevents inflammation-induced osteoporosis by downregulating the pro-inflammatory cytokines. Bone Res. 6, 9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Y., Di Grappa M. A., Molyneux S. D., McKee T. D., Waterhouse P., Penninger J. M., Khokha R., RANKL blockade prevents and treats aggressive osteosarcomas. Sci. Transl. Med. 7, 317ra197 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Ofotokun I., Titanji K., Vikulina T., Roser-Page S., Yamaguchi M., Zayzafoon M., Williams I. R., Weitzmann M. N., Role of T-cell reconstitution in HIV-1 antiretroviral therapy-induced bone loss. Nat. Commun. 6, 8282 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rochette L., Meloux A., Rigal E., Zeller M., Cottin Y., Vergely C., The role of osteoprotegerin in the crosstalk between vessels and bone: Its potential utility as a marker of cardiometabolic diseases. Pharmacol. Ther. 182, 115–132 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Park-Min K.-H., Lim E., Lee M. J., Park S. H., Giannopoulou E., Yarilina A., van der Meulen M., Zhao B., Smithers N., Witherington J., Lee K., Tak P. P., Prinjha R. K., Ivashkiv L. B., Inhibition of osteoclastogenesis and inflammatory bone resorption by targeting BET proteins and epigenetic regulation. Nat. Commun. 5, 5418 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim H., Lee K., Kim J. M., Kim M. Y., Kim J.-R., Lee H.-W., Chung Y. W., Shin H.-I., Kim T., Park E.-S., Rho J., Lee S. H., Kim N., Lee S. Y., Choi Y., Jeong D., Selenoprotein W ensures physiological bone remodeling by preventing hyperactivity of osteoclasts. Nat. Commun. 12, –2258 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu Z., Li T., Gao W., Xu T., Jurado-Sanchez B., Li J., Gao W., He Q., Zhang L., Wang J., Cell-membrane-coated synthetic nanomotors for effective biodetoxification. Adv. Funct. Mater. 25, 3881–3887 (2015). [Google Scholar]
- 37.Hu C. M. J., Fang R. H., Wang K. C., Luk B. T., Thamphiwatana S., Dehaini D., Nguyen P., Angsantikul P., Wen C. H., Kroll A. V., Carpenter C., Ramesh M., Qu V., Patel S. H., Zhu J., Shi W., Hofman F. M., Chen T. C., Gao W. W., Zhang K., Chien S., Zhang L. F., Nanoparticle biointerfacing by platelet membrane cloaking. Nature 526, 118–121 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pang Z. Q., Hu C. M. J., Fang R. H., Luk B. T., Gao W. W., Wang F., Chuluun E., Angsantikul P., Thamphiwatana S., Lu W. Y., Jiang X. G., Zhang L. F., Detoxification of organophosphate poisoning using nanoparticle bioscavengers. ACS Nano 9, 6450–6458 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Z., Wang Z., Dinh P.-U. C., Zhu D., Popowski K. D., Lutz H., Hu S., Lewis M. G., Cook A., Andersen H., Greenhouse J., Pessaint L., Lobo L. J., Cheng K., Cell-mimicking nanodecoys neutralize SARS-CoV-2 and mitigate lung injury in a non-human primate model of COVID-19. Nat. Nanotechnol. 16, 942–951 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Q. Z., Dehaini D., Zhang Y., Zhou J. L., Chen X. Y., Zhang L. F., Fang R. H., Gao W. W., Zhang L. F., Neutrophil membrane-coated nanoparticles inhibit synovial inflammation and alleviate joint damage in inflammatory arthritis. Nat. Nanotechnol. 13, 1182–1190 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Hu C. M. J., Fang R. H., Copp J., Luk B. T., Zhang L. F., A biomimetic nanosponge that absorbs pore-forming toxins. Nat. Nanotechnol. 8, 336–340 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang H. Y., Peng B., Dong C., Liu L. X., Mao J. J., Wei S., Wang X. L., Xu H. S., Shen J., Mao H. Q., Gao X. H., Leong K. W., Chen Y. M., Cationic nanoparticle as an inhibitor of cell-free DNA-induced inflammation. Nat. Commun. 9, 4291 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thamphiwatana S., Angsantikul P., Escajadillo T., Zhang Q. Z., Olson J., Luk B. T., Zhang S., Fang R. H., Gao W. W., Nizet V., Zhang L. F., Macrophage-like nanoparticles concurrently absorbing endotoxins and proinflammatory cytokines for sepsis management. Proc. Natl. Acad. Sci. U.S.A. 114, 11488–11493 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rao L., Xia S., Xu W., Tian R., Yu G. C., Gu C. J., Pan P., Meng Q. F., Cai X., Qu D., Lu L., Xie Y. H., Jiang S. B., Chen X. Y., Decoy nanoparticles protect against COVID-19 by concurrently adsorbing viruses and inflammatory cytokines. Proc. Natl. Acad. Sci. U.S.A. 117, 27141–27147 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rao L., Tian R., Chen X. Y., Cell-membrane-mimicking nanodecoys against infectious diseases. ACS Nano 14, 2569–2574 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dehaini D., Wei X. L., Fang R. H., Masson S., Angsantikul P., Luk B. T., Zhang Y., Ying M., Jiang Y., Kroll A. V., Gao W. W., Zhang L. F., Erythrocyte-platelet hybrid membrane coating for enhanced nanoparticle functionalization. Adv. Mater. 29, 201606209 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fang R. H., Kroll A. V., Gao W. W., Zhang L. F., Cell membrane coating nanotechnology. Adv. Mater. 30, e1706759 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bu L. L., Rao L., Yu G. T., Chen L., Deng W. W., Liu J. F., Wu H., Meng Q. F., Guo S. S., Zhao X. Z., Zhang W. F., Chen G. J., Gu Z., Liu W., Sun Z. J., Cancer stem cell-platelet hybrid membrane-coated magnetic nanoparticles for enhanced photothermal therapy of head and neck squamous cell carcinoma. Adv. Funct. Mater. 29, 1807733 (2019). [Google Scholar]
- 49.Deng Y. K., Zhou Y., Liang Q. J., Ge C. L., Yang J. D., Shan B. C., Liu Y., Zhou X. Z., Yin L. C., Inflammation-instructed hierarchical delivery of IL-4/miR-21 orchestrates osteoimmune microenvironment toward the treatment of rheumatoid arthritis. Adv. Funct. Mater. 31, 2101033 (2021). [Google Scholar]
- 50.Han P., Cheng P., Zhang S., Zhao C., Ni J., Zhang Y., Zhong W., Hou P., Zhang X., Zheng Y., Chai Y., In vitro and in vivo studies on the degradation of high-purity Mg (99.99wt.%) screw with femoral intracondylar fractured rabbit model. Biomaterials 64, 57–69 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Burghardt I., Luthen F., Prinz C., Kreikemeyer B., Zietz C., Neumann H. G., Rychly J., A dual function of copper in designing regenerative implants. Biomaterials 44, 36–44 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Loi F., Cordova L. A., Pajarinen J., Lin T. H., Yao Z. Y., Goodman S. B., Inflammation, fracture and bone repair. Bone 86, 119–130 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Greenblatt M. B., Tsai J. N., Wein M. N., Bone turnover markers in the diagnosis and monitoring of metabolic bone disease. Clin. Chem. 63, 464–474 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakamura Y., Suzuki T., Kamimura M., Ikegami S., Murakami K., Uchiyama S., Taguchi A., Kato H., Two-year clinical outcome of denosumab treatment alone and in combination with teriparatide in Japanese treatment-naive postmenopausal osteoporotic women. Bone Res. 5, 16055 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eastell R., Christiansen C., Grauer A., Kutilek S., Libanati C., McClung M. R., Reid I. R., Resch H., Siris E., Uebelhart D., Wang A., Weryha G., Cummings S. R., Effects of denosumab on bone turnover markers in postmenopausal osteoporosis. J. Bone Miner. Res. 26, 530–537 (2011). [DOI] [PubMed] [Google Scholar]
- 56.Zhu S. Y., He H. C., Gao C. F., Luo G. J., Xie Y., Wang H. M., Tian L., Chen X., Yu X. J., He C. Q., Ovariectomy-induced bone loss in TNFα and IL6 gene knockout mice is regulated by different mechanisms. J. Mol. Endocrinol. 60, 185–198 (2018). [DOI] [PubMed] [Google Scholar]
- 57.Lazzaro L., Tonkin B. A., Poulton I. J., McGregor N. E., Ferlin W., Sims N. A., IL-6 trans-signalling mediates trabecular, but not cortical, bone loss after ovariectomy. Bone 112, 120–127 (2018). [DOI] [PubMed] [Google Scholar]
- 58.Takayanagi H., Osteoimmunology: Shared mechanisms and crosstalk between the immune and bone systems. Nat. Rev. Immunol. 7, 292–304 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Feng L., Gao M., Tao D., Chen Q., Wang H., Dong Z., Chen M., Liu Z., Cisplatin-prodrug-constructed liposomes as a versatile theranostic nanoplatform for bimodal imaging guided combination cancer therapy. Adv. Funct. Mater. 26, 2207–2217 (2016). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S20
Tables S1 to S3