Ligand-stimulated enzymatic release or cellular degradation of PrP may interfere with neurodegeneration-associated toxicity.
Abstract
The prion protein (PrPC) is a central player in neurodegenerative diseases, such as prion diseases or Alzheimer’s disease. In contrast to disease-promoting cell surface PrPC, extracellular fragments act neuroprotective by blocking neurotoxic disease-associated protein conformers. Fittingly, PrPC release by the metalloprotease ADAM10 represents a protective mechanism. We used biochemical, cell biological, morphological, and structural methods to investigate mechanisms stimulating this proteolytic shedding. Shed PrP negatively correlates with prion conversion and is markedly redistributed in murine brain in the presence of prion deposits or amyloid plaques, indicating a sequestrating activity. PrP-directed ligands cause structural changes in PrPC and increased shedding in cells and organotypic brain slice cultures. As an exception, some PrP-directed antibodies targeting repetitive epitopes do not cause shedding but surface clustering, endocytosis, and degradation of PrPC. Both mechanisms may contribute to beneficial actions described for PrP-directed ligands and pave the way for new therapeutic strategies against currently incurable neurodegenerative diseases.
INTRODUCTION
Neurodegenerative diseases, such as Alzheimer’s disease (AD) and Parkinson’s disease (PD), as well as less frequent prion diseases, not only share mechanisms of protein misfolding, protein aggregation, and progressive spreading of pathology (1, 2) but also involve common molecular players (3, 4). One example is the cellular prion protein (PrPC), a highly conserved cell surface glycoprotein with high (yet not exclusive) expression in the nervous system (5).
Apart from its physiological functions, PrPC plays a key role in prion diseases of humans [e.g., Creutzfeldt-Jakob disease (CJD)] and animals (e.g., chronic wasting disease in elk and deer and bovine spongiform encephalopathy in cattle). In these transmissible diseases, PrPC misfolds into a pathogenic and partially proteinase K (PK)–resistant conformation (PrPSc) (6, 7) due to either (i) a sporadic event, (ii) mutations in the coding Prn-p gene (causing genetic/familial disease forms), or (iii) contact with infectious “prions” (i.e., misfolded PrP species acting as “seeds” to template further PrPC misfolding in acquired forms). More recently, glycosylphosphatidylinositol (GPI)–anchored PrPC has emerged as an important cell surface receptor for neurotoxic oligomers of β sheet–rich peptides/proteins (5, 8–11) such as PrPSc itself, amyloid β (Aβ), tau, and α-synuclein, which are all mediators of neuronal dysfunction found in neurodegenerative diseases such as prion diseases, AD, tauopathies, and PD, respectively (11, 12). The plasma membrane is the primary site for the detrimental interactions of such extracellular toxic conformers with the disordered N-terminal part of signaling-competent PrPC (13–15). This binding causes synapto- and neurotoxic signaling [enabled by certain transmembrane proteins associating with PrPC (16, 17)] and, in the case of PrPSc seeds, subsequent templated misfolding of native PrPC. In prion diseases, the survival time is inversely correlated with PrPC expression levels (18, 19). For these reasons, approaches to lower total or cell surface PrPC levels are considered as promising therapeutic options with potential benefit also in the other abovementioned protein misfolding diseases (20–25).
Notably, surface levels of PrPC are tightly regulated by various cellular mechanisms (26). Among those is the proteolytic cleavage and extracellular release (shedding) by the metalloproteinase ADAM10 (27–29). The latter is yet another example of a protein with relevance in different proteinopathies: Acting as the main “α-secretase,” ADAM10 is responsible for the non-amyloidogenic processing of the Aβ precursor protein (APP), thus competing with the generation of toxic Aβ in the first place. Hence, it has been proposed and investigated as a potential target in AD therapy (30–32). Furthermore, by also lowering surface PrPC levels, ADAM10 stimulation impairs the binding of Aβ to neurons and thus reduces toxicity (25). In experimental prion diseases in mice, ADAM10 similarly confers protection as its expression correlates with survival time (33, 34). Last, once being released from the surface, shed PrP (sPrP), which has all relevant binding sites, may interfere with PrPSc formation (in prion diseases) and block or neutralize various toxic conformers in the extracellular space. Similar effects have already been described for recombinant or anchorless PrP versions, artificial PrP dimers, or the soluble N-terminal fragment (N1), resulting from the constitutive α-cleavage in the middle of PrPC (see references I in fig. S2), which may all be considered as a proxy for physiologically acting bona fide sPrP. In support of this, we here provide data obtained with murine disease models indicating that physiological sPrP acts protective in neurodegenerative diseases by blocking and sequestering toxic oligomers.
When considering new treatment options against currently incurable diseases: Why not using a potentially protective process already provided by nature? While ADAM10 has been suggested as a therapeutic target in AD (30, 32), apparent problems arise from its rather broad expression pattern, the multitude of substrates in different tissues, and its involvement in various important physiological and pathological processes ranging from development and tissue homeostasis to intercellular communication and cancer (35). Therefore, directly manipulating this protease may cause substantial side effects, whereas a substrate-specific approach to stimulate the ADAM10-mediated shedding of PrPC would likely be superior.
On the basis of two earlier yet so far independent data-based concepts of (i) an increased ADAM10-mediated cleavage of some other ADAM10 substrates upon specific antibody binding or dimerization (36–38) and (ii) protective effects of PrPC-directed antibodies in various models of AD and prion disease (see references II in fig. S2), we aimed at investigating how ligands binding to PrPC would affect its supposed protective release by ADAM10. We show that a wide range of full-length immunoglobulin G (fl-IgG) antibodies binding to central epitopes of PrPC and some other PrP-directed ligands increase the ADAM10-mediated PrP shedding. In contrast, an fl-IgG antibody targeting repetitive epitopes within the octarepeat region of PrPC leads to strong PrPC surface clustering and subsequent internalization and lysosomal degradation of the PrPC-antibody complex, whereas an identical derivative in its single-chain form increases shedding similar to abovementioned ligands. Moreover, we provide structural insight suggesting that shedding-stimulating effects of a PrP-directed antibody are enabled by a moderate conformational change in the relative positioning of the N- and C-terminal halves of PrPC.
Collectively, our data suggest that PrPC-to-ligand interactions play key roles in determining the fate of PrPC regarding strong surface clustering followed by internalization and degradation on the one hand or increased ADAM10-mediated shedding on the other hand. Both mechanisms may pave the way for future therapeutic options against a wide range of dementias.
RESULTS
Effects of ADAM10 and sPrP in mouse models of neurodegenerative diseases
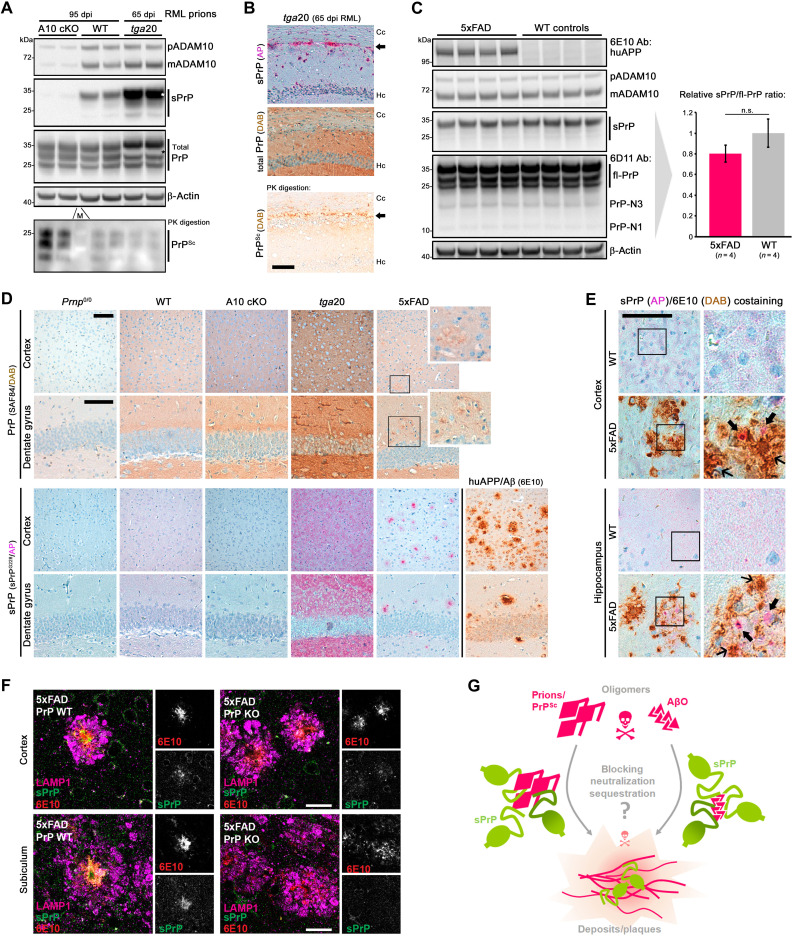
The role of ADAM10 in prion diseases has so far only been addressed by two studies in mice. Both studies, one using ADAM10-overexpressing mice (33) and one from our group using a conditional, neuron-specific knockout model [A10 cKO; (34)], found a notable correlation between expression of the protease and survival times of prion-infected mice, thus pointing toward protective effects. In the latter study, we have also shown that PrPSc production was highest in A10 cKO mice, whereas only little PrPSc was detected in PrP-overexpressing tga20 animals, even at a terminal stage of disease (34). To directly assess sPrP and how this correlates with PrPSc amounts, we performed a biochemical comparison of brain homogenates of prion-infected A10 cKO mice and wild-type (WT) littermate controls [at 95 days post-inoculation (dpi)] and tga20 mice (at 65 dpi, which, in these mice, corresponds to terminal disease). In agreement with our earlier study, highest PrPSc levels (upon sample digestion with PK) were found in A10 cKO, followed by moderate amounts in WT and lowest levels in tga20 mice (Fig. 1A). Analysis of the respective nondigested samples—as expected for the genotypes—revealed that amounts of sPrP [assessed using our recently generated sPrP-specific sPrPG228 antibody (26)] were highest in tga20 but hardly detectable in A10 cKO mice. This suggests an inverse correlation between this released PrP form and PrPSc production and may indicate that sPrP interferes with the conversion process. The seemingly contradictory fact that tga20 mice, despite strongly elevated sPrP levels, still develop a rapidly progressive prion disease is in line with recent studies (19) and supports the concept that the amount of membrane-associated PrPC [which is increased in this model despite efficient shedding (34)] rather than net PrPSc levels determines neurotoxicity. Next, we assessed localization of sPrP in terminally prion-diseased tga20 mice by immunohistochemistry (IHC). Despite exhibiting relatively low total amounts of PrPSc (Fig. 1A) (34), deposits of PK-resistant PrP are mainly found in a layer between the hippocampus and corpus callosum (Fig. 1B, bottom picture). In contrast to the diffuse staining of sPrP in noninfected tga20 brain described earlier (26) and shown for comparison in Fig. 2D, in prion disease, sPrP is redistributed and clusters around those PrPSc aggregates (Fig. 1B), pointing toward close interaction between sPrP and PrPSc in respective deposits.
Fig. 1. sPrP may interfere with toxic oligomers in neurodegenerative diseases.
(A) Immunoblots of premature and mature ADAM10, sPrP, and total PrP in frontal brain of prion-infected ADAM10 knockout (A10 cKO), WT [both 95 days post-infection (dpi)], and tga20 mice (terminal disease; 65 dpi). Loading control: β-actin. Corresponding PrPSc shown in lower blot. Asterisks: overexposed sPrP signals (in tga20) caused white area upon reprobing. M, protein marker. (B) IHC of sPrP [pink, alkaline phosphatase (AP)], total PrP (brownish, DAB), and PrPSc in hippocampus (Hc) and corpus callosum (Cc) of a prion-diseased tga20 mouse. Arrows indicate PrPSc deposition (bottom) and similarly clustered sPrP (top) contrasting with diffuse sPrP in noninfected mice [see (D) for reference]. (C) ADAM10, sPrP, and PrP (including N-terminal fragments resulting from other cleavages) in brains of 6-month-old 5xFAD mice and WT littermates. Actin detected as loading control; human APP [huAPP; 6E10 antibody (Ab)] for genotype confirmation. Quantified sPrP shows no significant (n.s.) differences (mean ± SE; P > 0.05, Student’s t test). (D) PrP (top) and sPrP (bottom) in Prnp0/0, WT, A10 cKO, tga20, and 5xFAD mice. Plaque-like structures are vaguely perceived with pan-PrP staining (magnifications for 5xFAD). No sPrP detected in Prnp0/0 and A10 cKO. Diffuse sPrP (WT and tga20) is converted to a clustered pattern in 5xFAD similar to amyloid plaques detected by 6E10 antibody. (E) Costaining of sPrP and amyloid in WT and 5xFAD. Again, diffuse sPrP changes to clustered signals in 5xFAD. Bold arrows: sPrP in diffuse Aβ deposits. In dense plaques, sPrP is masked by strong brownish signal (thin arrows). Scale bars, 100 μm. (F) Immunofluorescently stained sPrP, Aβ plaques (6E10), and LAMP1 (dystrophic neurites marker) in 5xFAD brains with (PrP-WT) and without PrP expression (PrP-KO). Note that sPrP colocalizes with Aβ plaques, whereas only background is detected in the negative controls. Scale bars, 30 μm. (G) Scheme showing potential neurotoxicity-lowering binding of sPrP to and sequestration of harmful extracellular oligomers.
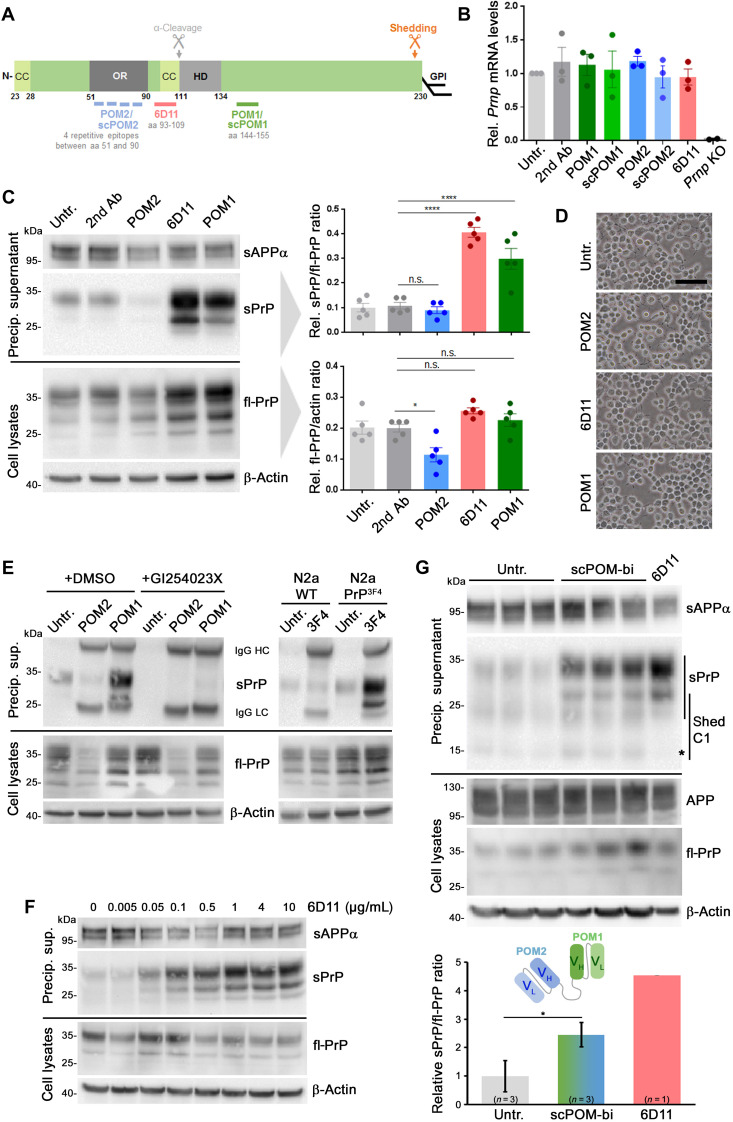
Fig. 2. PrP-directed antibodies cause increased ADAM10-mediated PrP shedding in N2a cells.
(A) PrP scheme showing important domains (CC, charged cluster; OR, octameric repeat region; HD, hydrophobic domain), GPI anchor position, shedding, and α-cleavage sites, plus epitopes for antibodies used here. aa, amino acids. (B) Prnp mRNA levels in cells either untreated or treated for 16 hours with indicated antibodies or single-chain (sc) derivates. Negative controls: PrP-depleted cells (Prnp KO). n = 3 independent experiments (n = 2 for Prnp KO) with three technical replicas each. No significant differences in Prnp mRNA levels were found among different treatments and untreated controls. (C) Representative immunoblot analysis of fl-PrP in lysates (bottom) and sPrP in precipitated medium (top) after 16 hours of incubation with different PrP-directed IgGs. Loading controls: β-actin (lysates) and sAPPα (medium). fl-PrP levels were reduced (P ≤ 0.05) only in POM2-treated cells compared to secondary antibody controls, whereas significantly increased sPrP/fl-PrP ratios were observed for 6D11 and POM1 treatment (P ≤ 0.0001). Data show means ± SEM of n = 5 independent experiments; statistical significance was estimated with analysis of variance (ANOVA) followed by Bonferroni’s multiple comparisons test. (D) Microscopy of untreated and treated cells showing no alterations in density or overall morphology (scale bar, 100 μm). (E) Treatment with POM2 or POM1 in the presence (+GI254023X) or absence [+DMSO (dimethyl sulfoxide), as diluent control] of an ADAM10 inhibitor (left). Right: N2a WT or N2a stably expressing murine PrP with the human 3F4 epitope (N2a PrP3F4) treated or not with 3F4 antibody targeting that motif. Shedding only increased in PrP3F4-expressing cells. (F) Ascending concentrations of 6D11 reveal a dose dependency of the shedding-stimulating effect (reaching saturation at ~1 μg/ml). (G) The bispecific immunotweezer (scPOM-bi; fused complementarity-determining regions VH/VL of POM2 and POM1; see scheme) increases shedding compared to untreated controls. Quantification with controls set to 1 (mean ± SE; *P = 0.024, Student’s t test). Positive control: 6D11 treatment [reduced levels of sPrP-C1 fragment (asterisk) possibly due to 6D11 sterically hindering α-cleavage before shedding]. *P < 0.05 and ****P < 0.0001.
In experimental models of AD, released or recombinant forms of PrP exert protective effects by blocking neurotoxic Aβ oligomers (AβO) (references I in fig. S2). Stimulation of ADAM10 protects cells from AβO toxicity by reducing plasma membrane levels of PrPC as receptor and toxicity transducer (25). We aimed at assessing a potentially beneficial role of sPrP itself by blocking and sequestering toxic conformers in the extracellular space. Comparing levels of sPrP, total PrP and ADAM10 in brains of 6-month-old AD model mice (i.e., 5xFAD mice expressing human mutated APP among other manipulations) and WT littermate controls did not reveal significant differences in overall steady-state levels of these proteins (Fig. 1C). In contrast to a previous report showing up-regulation of the α-cleavage product PrP-N1 in human AD brain (39), we did not observe any similar effect in 5xFAD mice. However, upon histological assessment (Fig. 1D), the expected diffuse staining pattern for sPrP observed in WT and tga20 mice shifted to a clustered distribution in 5xFAD brains, strongly reminiscent of the bona fide amyloid plaques also detected in these mice (6E10-positive signals). As expected, Prnp0/0 and A10 cKO mice showed no sPrP signal. To exclude unspecific plaque binding of our sPrPG228 antibody, we performed additional stainings comparing 5xFAD mice with matched PrP-depleted 5xFAD/Prnp0/0 mice (fig. S1). Moreover, when costaining of sPrP and human APP/Aβ was performed on 5xFAD and WT control sections, the diffuse signal for sPrP in the brain parenchyma observed in WT mice vanished at the cost of a more concentrated and plaque-associated pattern (Fig. 1E, thick arrows). Because the presence of sPrP in dense plaques could not be addressed by IHC due to dominant brownish 6E10-positive 3,3’-diaminobenzidine (DAB) signal (Fig. 1E, slim arrows) and to gain insight at higher resolution, we additionally performed immunofluorescence (IF) stainings on free-floating sections (Fig. 1F). sPrP clustered and colocalized within the cores of many (yet not all) 6E10-positive amyloid plaques surrounded by dystrophic neurites (indicated by the lysosomal marker LAMP1) (Fig. 1F). Again, besides weak unspecific background signal, no specific clustered pattern was observed with the sPrPG228 antibody in 5xFAD/Prnp0/0 mice used as negative controls (Fig. 1F).
Notably, it has repeatedly been described in human AD brain and respective mouse models that PrPC (detected with pan-PrPC antibodies) colocalizes with certain amyloid plaques and may even promote their formation (see references III in fig. S2). Here, we provide first evidence that it is specifically sPrP (generated by ADAM10), which, as a diffusible factor in the extracellular space, is redistributed to the center of Aβ plaques in a murine model for AD-associated amyloid formation.
In sum, published data on protective effects of soluble released or recombinant PrP (recPrP) forms in prion diseases and AD, and published reports in addition to our findings shown here on the role of physiologically sPrP, support the view that sPrP may block formation of (in the case of prion diseases) and could sequester/detoxify (in prion diseases and other proteinopathies) harmful oligomeric protein conformers (scheme in Fig. 1G and fig. S2). This role of released PrP stands in clear contrast to the one of cell surface PrPC acting as a toxicity mediator in these diseases (5, 8, 9, 11, 12).
A substrate-specific approach to stimulate the ADAM10-mediated shedding of PrP
Several earlier studies and our data presented above may point to a protective role of PrP shedding in neurodegenerative diseases. Direct stimulation of ADAM10 is currently pursued in clinical AD trials, albeit with the primary goal to stimulate the non-amyloidogenic processing of the APP (32). However, the multitude of ADAM10 substrates with (patho)physiological relevance throughout the body poses major challenges regarding potentially severe side effects (35).
For some ADAM10 substrates acting as cell surface receptors, binding of antibodies to their extracellular domains leads to their increased proteolytic release (36, 37). Furthermore, as introduced earlier, PrP-directed antibodies show beneficial effects in different models of AD and prion disease and are even used in the framework of a clinical trial (references II in fig. S2). These two seemingly “unrelated” aspects prompted us to assess whether some selected PrP-directed antibodies (scheme in Fig. 2A) and ligands would stimulate its ADAM10-mediated shedding.
Treatment of murine neuroblastoma (N2a) cells with antibodies directed against central parts of PrP [6D11, recognizing amino acids 93 to 109, and POM1, recognizing amino acids 144 to 155 (40)] did not change overall PrPC expression, as assessed by mRNA levels (Fig. 2B). However, levels of sPrP in medium supernatants were significantly increased, whereas cell-associated PrP levels were unchanged (Fig. 2C). In contrast, treatment with an antibody directed against the flexible N-terminal part of PrPC [POM2, recognizing repetitive epitopes between amino acids 51 and 90 (40)] led to a significant reduction in total PrPC (Fig. 2C). As further supported by figs. S3A and S12A, expression or processing of another neuronal ADAM10 substrate, APP, was not affected by treatment with PrP-directed antibodies. Moreover, cell surface PrPC levels, as assessed by a surface biotinylation assay, were increased upon POM1 and decreased after POM2 treatment (fig. S4).
Antibody treatment did not cause any obvious deleterious effects as judged by overall cell morphology (Fig. 2D) and an annexin V apoptosis assay (fig. S5). To exclude cell line–specific effects, we also performed these experiments in another murine neuronal cell line (mHippo) and could essentially reproduce the observed effects regarding levels of sPrP, levels of cell-associated PrP (albeit with higher variation between experiments), and the absence of overt toxicity (fig. S6).
As expected, cotreatment with the ADAM10 inhibitor GI254023X abolished the shedding-stimulating effect of POM1, confirming strict dependence of this process on this protease (Fig. 2E, left) (26). This experiment also confirmed the reduction in total PrPC upon treatment with POM2, which is independent of ADAM10 activity. We next treated cells with an antibody binding to an epitope in human PrPC (3F4), which is absent in murine PrPC (Fig. 2E, right). While no stimulating effect on shedding was observed in murine WT N2a cells, this antibody caused a strong increase of sPrP in N2a cells genetically modified to express 3F4-tagged PrP. Thus, the stimulated shedding observed here is executed by ADAM10 and specifically mediated by the binding of certain antibodies to PrPC.
We then addressed a potential dose dependency of the shedding-stimulating effect using 6D11, which consistently caused the strongest stimulation among the PrPC-directed antibodies used in this study (Fig. 2C and fig. S3A). 6D11-mediated effects on PrPC shedding are dose dependent, with effects seen at concentrations as low as 0.005 μg/ml and reaching saturation at approximately 1 μg/ml (Fig. 2F; a quantified experiment is shown in fig. S3B).
Next, we tested a bispecific immunotweezer (scPOM-bi), which was recently shown to interfere with the formation of toxic prion species (41). This chimeric antibody is composed of the complementarity-determining regions of POM1 and POM2. Being directed against both, the globular C-terminal and the flexible N-terminal half of PrP, this molecule may act intra- and intermolecularly, thus bridging the two dissimilar halves within one or between two PrP molecules, respectively. As shown in Fig. 2G, similar to POM1 and 6D11, treatment with scPOM-bi caused a clear increase in sPrP. Shedding of the N-terminally truncated C1 fragment, which only displays one binding site for scPOM-bi, is only mildly increased when compared to nontruncated sPrP.
In sum, with the exception of POM2 (an anti-PrPC antibody recognizing repetitive N-terminal epitopes further investigated and discussed below), all PrPC-directed antibodies tested here caused a significant increase in the ADAM10-mediated shedding. As expected and demonstrated, this effect is substrate specific, given that no significant alterations in APP processing were observed.
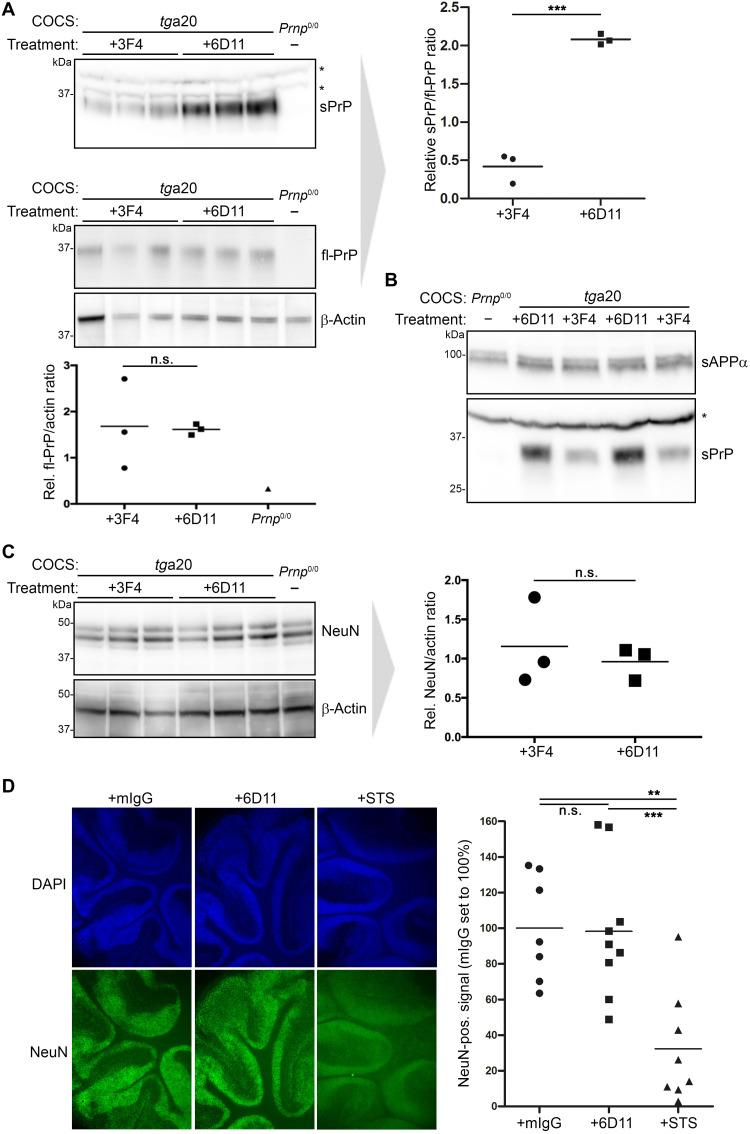
The PrPC-directed antibody 6D11 causes increased shedding in the absence of toxicity in organotypic brain slice cultures
After confirming the shedding-stimulating effect of some antibodies in two neuronal cell lines, we investigated whether this also holds true in a more complex biological system. We therefore tested the effects of the 6D11 antibody (as this IgG led to highest levels of sPrP in cell lines) in murine cerebellar organotypic slice cultures (COCS) (42) derived from tga20 mice. As expected, treatment with 6D11, but not with 3F4 antibody (an anti-PrPC antibody not binding to murine PrPC and therefore used as negative control), significantly increased levels of sPrP in the culture medium surrounding the COCS (Fig. 3A, top). No alterations in levels of fl-PrPC were observed in the corresponding COCS homogenates (Fig. 3A, bottom). Similar to the results obtained with N2a cells (Fig. 2, C, F, and G), the increase in PrP shedding in COCS upon treatment with 6D11 did not affect levels of sAPPα, thus confirming the substrate specificity of this strategy (Fig. 3B).
Fig. 3. Stimulated shedding and lack of toxicity in antibody-treated murine organotypic brain slice cultures.
(A) Cerebellar COCS prepared from tga20 mice (or a Prnp0/0 mouse as negative control) and exposed to either 3F4 IgG (as negative control treatment) or 6D11 antibody. Western blot analysis showing levels of PrP in COCS homogenates (bottom panel including quantification; actin was used as loading control) and sPrP in the culture medium (top panel with quantification on the right). (B) Biochemical assessment of sPrP and sAPPα in culture medium after treatment as above. Asterisks in (A) and (B) indicate the presence of unspecific bands (note the presence in Prnp0/0 samples) detected with the sPrPG228 antibody in COCS medium. (C) Levels of the neuronal marker NeuN in abovementioned COCS homogenates and densitometric quantification (actin used as reference). (D) Morphological analysis of antibody-treated COCS sections prepared from WT mice. Non–PrP-directed mouse antibodies (mIgG) were used as negative control, whereas STS was used to induce toxicity and neuronal loss. DAPI (4′,6-diamidino-2-phenylindol) staining (blue) reveals nuclei of all cells, while NeuN staining indicates the presence of neuronal nuclei. Representative sections are shown. Quantifications of the NeuN-positive signal are presented on the right (mIgG, n = 7; 6D11, n = 9; STS, n = 8). Significance was assessed using unpaired two-tailed Student’s t test (A and C) and one-way ANOVA with Dunnett’s post hoc test (D).
We also addressed potential toxic side effects caused by 6D11 antibody treatment. However, neither biochemical (Fig. 3C) nor morphological assessment (Fig. 3D) of the neuronal marker NeuN gave evidence for enhanced neuronal cell death in 6D11-treated COCS when compared to controls incubated with 3F4 antibody or nonspecific murine IgGs, whereas this could readily be demonstrated in COCS treated with staurosporine (STS) as a positive control (Fig. 3D). In summary, substrate-specific and nontoxic stimulation of PrP shedding can be achieved by treatment with anti-PrP antibodies, not only in neuronal cell lines but also in more complex biological settings such as COCS.
Structural rearrangements in PrP caused by 6D11 binding
Antibody-mediated cross-linking induces proteolytic release of other ADAM10 substrates (36, 37). Fittingly, artificial homodimerization of PrPC was shown to result in increased proteolytic processing (43) and to protect cells from prion propagation (44). Thus, given the elevated PrPC surface levels found upon treatment with POM1 (fig. S4), we cannot exclude that dimerization and prolonged surface retention of PrPC also play a role in induced shedding caused by POM1 and 6D11 IgGs. However, as demonstrated by the use of single-chain antibodies in the next paragraph, cross-linking is clearly not a prerequisite for stimulated shedding. Therefore, we aimed to gain further insight into shedding-stimulating mechanisms upon antibody binding at the structural level.
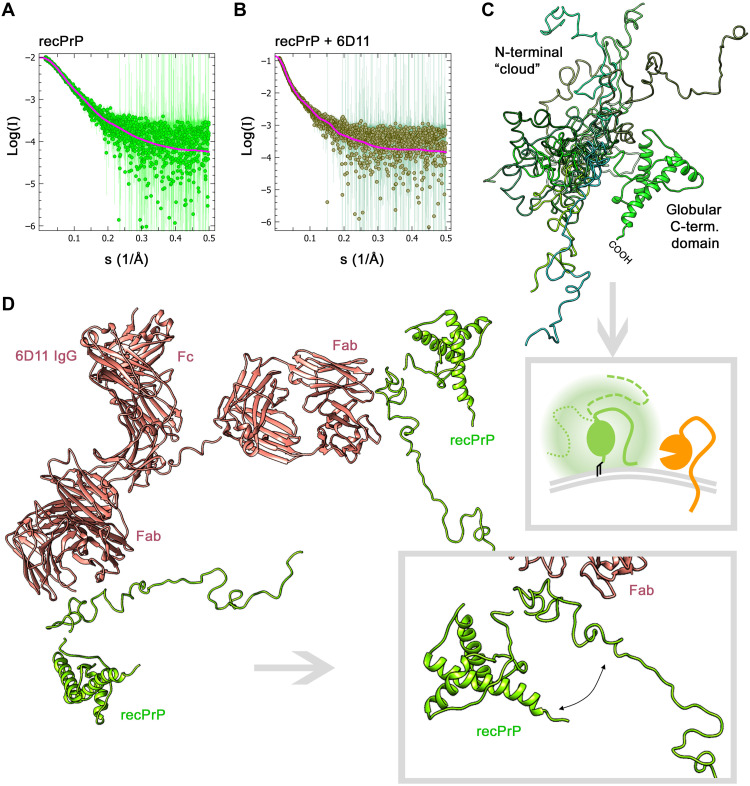
The 6D11 antibody has shown neuroprotective effects in various studies and turned out to be the most efficient shedding stimulator among the PrPC ligands tested here. To assess the structure of the PrP-6D11 complex (Fig. 4B) and compare it to PrP alone (Fig. 4A), we used state-of-the-art small-angle x-ray scattering (SAXS). This method allows structural analysis in solution and to obtain low-resolution three-dimensional (3D) models. In our measurements, recPrP scattering was well fitted using a structured, mostly α-helical C-terminal domain and a flexible N-terminal tail partially flanking the globular domain in relatively close proximity (Fig. 4, A and C; for more information, see fig. S7 and table S1). In line with earlier SAXS data (45), this suggests the presence of a shielding “cloud” formed by the N-terminal part surrounding the globular domain. Depending on the movement and position of the flexible tail (FT) at a given moment, this may regulate access of ADAM10’s catalytic domain to the C-terminal cleavage site in PrPC (framed scheme in Fig. 4C). In addition, the N terminus was shown to interact with the plasma membrane (46–49), which may further block the membrane-proximate shedding event. Notably, for recPrP in complex with 6D11 IgG (here modeled bound to two recPrP molecules), we observed a much more extended N-terminal conformation of recPrP at a wider angle from the globular C-terminal domain (Fig. 4D). Thus, despite their own size, it appears possible that PrP-directed antibodies allow better accessibility of ADAM10 by decreasing the sterical hindrance posed by the FT of PrP.
Fig. 4. SAXS curves and modeling for recPrP (23–230) and the recPrP/6D11 antibody complex.
Experimental SAXS profiles (dots) and fits (solid lines) for the best-fitting model of recPrP; data source: SASBDB accession code: SASDHV9 (χ2 = 0.85) (A) and the complex of two recPrP bound to 6D11 IgG (χ2 = 0.83) (B). (C) Overlay of recPrP models resulting from SAXS measurements and showing multiple possible conformations of the flexible N-terminal tail (different shades of green) flanking the structured C-terminal domain. Framed scheme below outlines that movement of the flexible tail may create a cloud (shadowy corona) surrounding the globular domain and partially shielding PrP (green) from being shed by ADAM10 (orange), depending on the actual positioning and potential membrane interactions of the flexible tail (solid versus intermitted lines). (D) Model of the recPrP (23–230)/6D11 IgG complex. The magnified view (framed box) highlights an extended conformation of the flexible N-terminal region and an increased angle and distance to the C-terminal domain (as required to form a complex consistent with the SAXS data). Note that posttranslational modifications such as N-glycans and the GPI anchor are lacking in these analyses using recPrP.
Stimulated shedding is also achieved by treatment with single-chain antibodies
For some ADAM10 substrates, such as CD44, antibody-mediated cross-linking causes increased cleavage (38). Because, apart from POM2, all PrPC-directed IgGs tested here (POM1 and 6D11; as well as 3F4 in respective cells expressing 3F4-tagged PrPC) and the bispecific immunotweezer (scPOM-bi) stimulated shedding, it was obvious to consider cross-linking of two PrPC molecules as one underlying principle. In this scenario (and supported by the finding of increased PrPC membrane levels caused by POM1 treatment mentioned above; fig. S4), dimerization would cause PrPC to escape its usually high endocytosis rate (50) by stabilizing PrPC and prolonging its presence at the cell surface, where ADAM10-mediated shedding is thought to occur. Accordingly, treatment with single-chain antibodies, which are unable to cross-link two molecules, should not yield elevated sPrP levels.
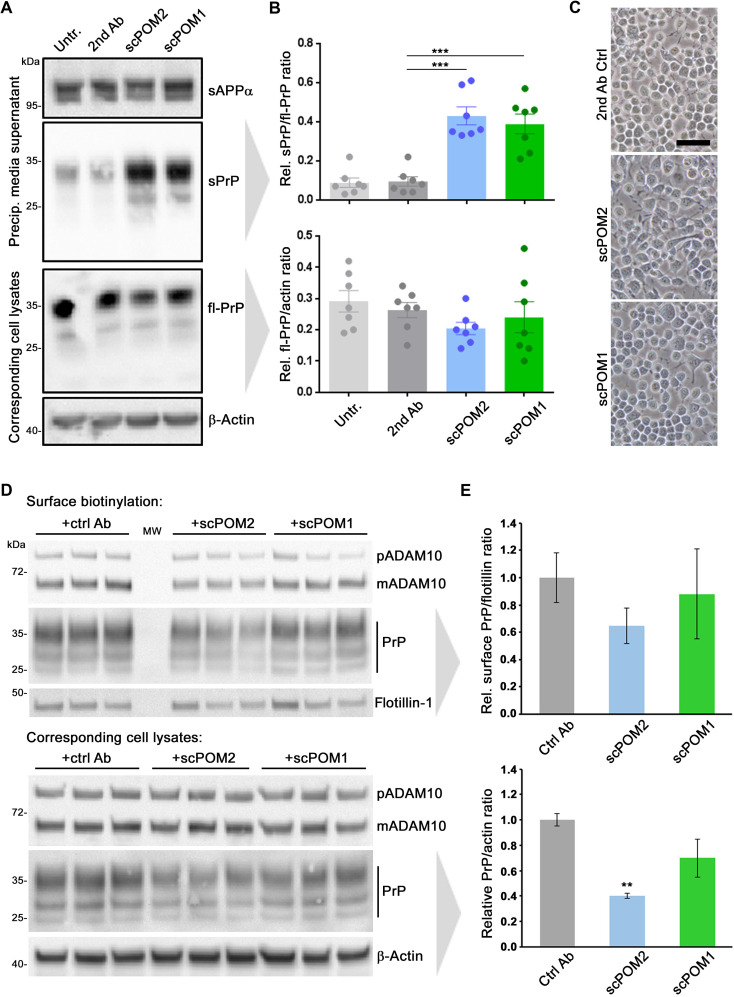
To test for this, we treated N2a cells with single-chain versions of the two contrarily acting “ancestor” IgGs POM1 (scPOM1) and POM2 (scPOM2). Both single-chain antibodies resulted in increased shedding compared to controls (Fig. 5, A and B). Again, no toxic effects were observed on the basis of overall cell morphology (Fig. 5C) and an annexin V toxicity assay (fig. S5). To exclude stabilizing effects of these treatments on surface PrP levels, we again performed a surface biotinylation assay yet did not detect significant differences compared to the control treatment (Fig. 5, D and E).
Fig. 5. Single-chain antibodies induce shedding without causing PrP surface retention.
(A) Representative Western blot analysis showing levels of sPrP (and sAPPα as loading control) in precipitated medium and fl-PrP (and actin as loading control) in respective lysates of N2a cells treated with single-chain variable fragments of POM2 (scPOM2) and POM1 (scPOM1) antibodies. Untreated (Untr.) and anti-mouse secondary antibody–treated (2nd Ab) cells served as controls. (B) Densitometric quantification of sPrP (top diagram) and cell-associated PrP levels (bottom diagram). Plotted data show means ± SEM for n = 7 independent repetitions. One-way ANOVA and Bonferroni’s multiple comparisons test were used to calculate significances. Relative sPrP/fl-PrP level was found to be increased in both scPOM-treated cells in comparison to that of secondary antibody controls (P ≤ 0.001). (C) Microscopic assessment of cell density and morphology (scale bar, 50 μm). (D) Cell surface biotinylation assay (top) revealing membrane levels of ADAM10, PrP, and flotillin. Total levels of ADAM10 and PrP in respective cell lysates are shown below. Actin served as loading control in lysates. Note the relative shift toward diglycosylated PrP and mature ADAM10 in biotinylated samples (compared to lysates) as these forms are thought to primarily locate at the cell surface. MW, lane used for molecular weight ladder. (E) Densitometric quantification of PrP levels presented in (D). Plotted data show means ± SEM for n = 3 technical replicas [shown in (D)]. Significance was assessed using Student’s t test (**P < 0.005).
These data strongly suggest that dimerization and an associated stabilization of PrPC at the cell surface (as detected for POM1 IgG; fig. S4) are at least not a prerequisite for the shedding-stimulating effect observed in this study. Our results also speak against an epitope specificity of this effect. Instead, the combined data rather point toward a more general role of ligand binding to PrPC in stimulating its shedding. Moreover, it is intriguing that a single-chain version of POM2, despite targeting the very same epitopes, acts completely opposite to its IgG ancestor with regard to the ADAM10-mediated shedding. This aspect will be further outlined below.
Shedding is not stimulated by treatment with four chemical compounds known to bind PrPC
The finding of stimulated shedding caused even by single-chain antibodies highlighted the possibility that various—and maybe even much smaller—ligands of PrPC could likewise cause this effect. This would be particularly tempting considering potential therapeutic approaches and known difficulties associated with the use of antibodies in that regard (such as routes and doses of administration, costs, biostability, and passage through the blood-brain barrier). In an initial attempt, we therefore investigated four small chemical compounds shown to bind to different regions within PrPC (highlighted in fig. S8A) and described to exert anti-prion activity, at least in vitro [reviewed in (51)]: (i) GJP49, an anti-prion molecule identified through in silico analyses aimed at directly identifying pharmacological chaperones for PrPC (52); (ii) chlorpromazine, an antipsychotic drug originally claimed to inhibit prion propagation by directly binding to PrPC (53) but more recently shown to rather promote relocalization of PrPC from the cell surface (54); (iii) the porphyrin Fe(III)-TMPyP, perhaps the only extensively validated PrPC ligand, reported to inhibit prion propagation in a strain-independent fashion (55, 56); and, last, (ii) quinacrine, another tricyclic acridine derivative traditionally used as an anti-malaria drug and then identified as an anti-prion compound capable of directly binding to PrPC (57, 58).
These candidates were tested using ascending concentrations (0.1, 0.3, 1, and 3 μM) for treatment (24 hours) in an established screening system using human embryonic kidney (HEK) 293 cells stably expressing murine PrPC (fig. S8B) (59). None of these compounds significantly altered levels of cell-associated PrPC. Likewise, no relevant changes for sPrP were detected with the unexpected exception of quinacrine, which rather caused a reduction in shedding. This could be due to its expected binding region at the C terminus of PrPC (fig. S8A) overlapping with the cleavage site and, thus, potential hindrance of the protease. However, because we were rather interested in ligands increasing the shedding, we did not further investigate this aspect. Although we are continuing to screen for small compounds that stimulate shedding, the findings presented here may indicate that ligands of PrPC are required to exceed a critical size or fit certain sterical characteristics to stimulate its proteolytic shedding.
The PrPC-reducing effect of POM2 IgG is linked to its special binding characteristics
Among the antibodies tested here, POM2 IgG (yet not its single-chain variant) represents an exception, as it did not cause increased shedding but rather reduced levels of cell surface and total PrPC. This is interesting given that lowering (cell-associated) PrPC amounts is considered one of the most promising strategies for prion disease therapy (19, 24, 60, 61). Moreover, POM2 was shown to be neuroprotective, as it impairs detrimental interactions of the flexible N terminus with the plasma membrane caused by prions (46, 48, 49).
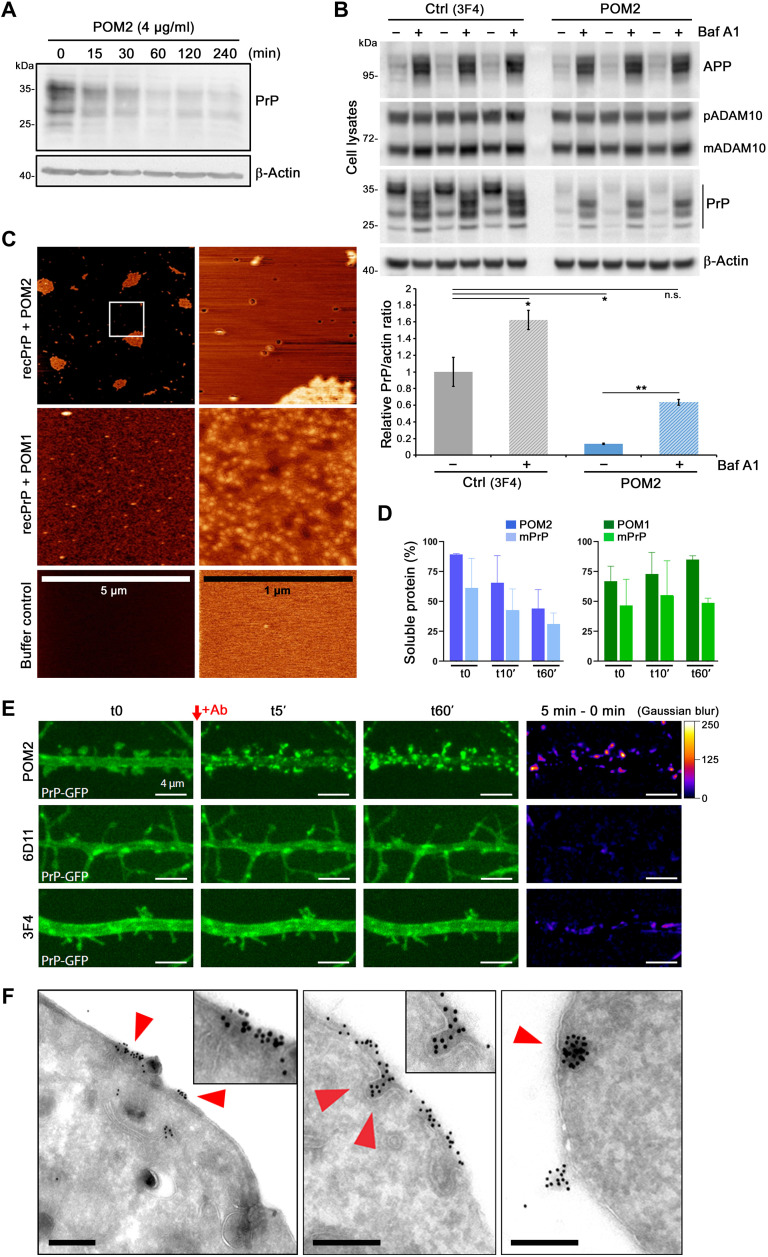
To further investigate the decrease in PrPC caused by treatment with this antibody, we treated N2a cells with POM2 and assayed total PrPC levels by Western blot over time. PrPC amounts were reduced quickly following POM2 treatment and subsequently reached a stable plateau at low levels (Fig. 6A; a quantified experiment shown in fig. S9). Because a reduction in cell-associated PrPC with no parallel increase in medium supernatants (Fig. 2, C and E) indicated enhanced cellular degradation, we performed antibody treatments in the presence or absence of bafilomycin (Baf A1), an antibiotic targeting the vacuolar H+-ATPase (H+-dependent adenosine triphosphatase), thus impairing lysosomal acidification and functioning (Fig. 6B). Cotreatment with Baf A1 caused increased cellular levels of PrP and APP in cells treated with a control antibody (3F4) or POM2 (and an altered banding pattern of particularly diglycosylated PrP, suggesting effects of bafilomycin treatment on PrP glycosylation or N-glycan degradation). Notably, in POM2-treated cells, Baf A1 partially restored physiological PrP levels. This indicated that POM2 treatment leads to uptake and lysosomal degradation of PrPC rather than induced shedding.
Fig. 6. POM2 IgG treatment results in strong (surface) clustering, uptake, and degradation of PrP.
(A) Time course experiment showing cell-associated PrP levels in N2a cells lysed at different time points after treatment with POM2. (B) Western blot showing cellular levels of PrP, APP, and ADAM10 upon treatment with POM2 or control antibody (3F4) in the presence (+) or absence (−) of the lysosomal inhibitor bafilomycin (Baf A1). Densitometric quantification (below) shows mean (untreated controls set to 1) ± SE; Student’s t test results are considered significant at *P < 0.05 and **P < 0.005. Actin served as loading control in (A) and (B). (C) AFM of recPrP incubated with POM2 or POM1 antibody in overview (left column; scale is indicated) and fivefold further magnification (right column). Bottom: Mica surface treated with protein-free buffer only. (D) Quantification of a solubility assay of a mixture of mouse recPrP (mPrP) with either POM2 (blue graphs) or POM1 (green graphs). Recovery of respective antibodies or PrP alone in solution immediately (t0), 10 min (t10′), or 60 min (t60′) after mixture was set to 100%. (E) Individual frames of confocal time-lapse imaging showing dendrites of rat hippocampal neurons expressing GFP-tagged PrP (green) taken directly before (t0) and at several indicated time points after treatment with POM2, 6D11, or 3F4 antibody (for a complete overview, refer to fig. S10A and movie S1). Right lane: Subtraction image of time point 5 min minus “before treatment” in pseudocolor scale (right) to visualize changes in PrP-GFP localization. Scale bars, 4 μm. (F) Representative electron microscopy pictures of N2a cells after 5 min (left) or 30 min (middle and right) of treatment with POM2 antibody showing clustering and uptake of PrP-directed immunogold particles. Scale bars, 250 nm.
We reasoned that the exceptional behavior of POM2 must result from its specific binding characteristics. In contrast to all other antibodies assessed here, POM2 can bind to four repetitive epitopes all located in the structurally disordered N-terminal part. Hence, POM2 IgGs (but not scPOM2) with their double valance and four epitopes, supported by the extreme structural flexibility in this region of PrPC, might be able to cluster large complexes of several PrPC (and antibody) molecules. To test for this in vitro, we performed atomic force microscopy (AFM) of recPrP incubated with either POM2 or POM1 antibodies (Fig. 6C). In contrast to the latter, which—at best—is able to cross-link two PrP molecules and causes a dotty staining on the mica surface, POM2 treatment resulted in the formation of large clusters (on cost of the dotty pattern observed for POM1/recPrP).
We next confirmed these findings by a solubility assay (Fig. 6D), where recovery of both components in the soluble fraction of a mixture of either POM2 or POM1 with recombinant murine PrP was assessed over time (normalized against respective POM antibody or mPrP alone set to 100%). A clear trend of a progressive decrease of both components was observed for POM2, yet not for POM1, thus further supporting the abovementioned AFM findings of strong aggregation of POM2/recPrP complexes.
To investigate whether comparable clusters are also formed at the surface of cells upon POM2 treatment, we performed live microscopy of primary rat neurons transfected with green fluorescent protein (GFP)–tagged PrPC. As shown in movie S1, the representative snapshots in Fig. 6E, and the respective overview presented in fig. S10A, POM2 treatment caused a fast and strong surface clustering of PrPC in all examined neurons, which was not observed in any neurons incubated with 6D11 or 3F4 antibodies as controls. To exclude that this clustering by POM2 at the neuronal surface was only due to transgenic overexpression (PrP-GFP), we also performed staining of endogenous PrPC after treatment with POM2 and found a comparable pattern of clusters (fig. S10B). To address consequences of this clustering at a subcellular resolution, we also used electron microscopy analysis upon immunogold labeling. As early as 5 min after incubation of N2a cells with POM2, we found large immunogold-positive clusters of PrPC at the cell surface and many instances revealing subsequent endocytosis of these complexes (as represented in Fig. 6F), thus supporting POM2-stimulated uptake and degradation of PrPC (Fig. 6, A and B). In another set of experiments, this time directly comparing POM2- with POM1-treated cells, we confirmed cluster formation for POM2 yet could not find evidence for this in cells incubated with POM1 antibody (fig. S11).
During revision, we also performed a direct comparison between POM2 and its single-chain form (fig. S12A). This confirmed the contrary effects of total PrP reduction (POM2) and stimulated shedding (scFvPOM2). POM2-dependent reduction of total PrP in lysates was rescued upon bafilomycin treatment, whereas scFvPOM2 caused a pattern similar to the ones observed in cells treated with no antibody or 6D11 (fig. S12B). In addition, large multimolecular clusters were only found in the case of POM2 treatment yet not for its single-chain form (fig. S12C). Although the very same epitopes in the N-terminal tail of PrP are targeted by POM2 and scFvPOM2, valence of the ligand (and cross-linking potential) seems critical for the exceptional effects of POM2.
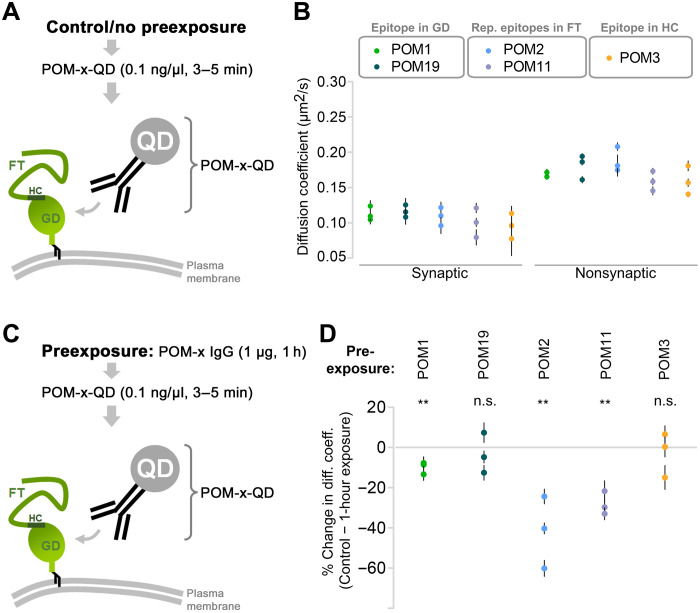
Membrane proteins are highly dynamic because of lateral diffusion on the plasma membrane (62). Clustering on the plasma membrane is either a 2D (complex formation between laterally diffusing membrane proteins) or 3D event (complex formation between membrane protein and scaffolds). Using SPT-QD (single-particle tracking using quantum dots) in primary neurons, we monitored the events leading to PrPC clustering by POM2 IgG. SPT-QD is a powerful method that allows the measurement of lateral diffusion and protein-protein interaction at a single-molecule resolution. Besides other POM-IgGs used as controls, we here also included POM11, which shares two important features with POM2: (i) having more than just one epitope, which are (ii) located within PrP’s FT. First, we measured the diffusion coefficient of endogenous PrPC using five different POM antibodies [POM-x: POM1/19 (single epitopes within the C-terminal half/globular domain), POM2/POM11 (FT-binders with ≥2 epitopes as described above), or POM3: epitope in the central part/hydrophobic core (HC)] without prior exposure to the same POM-x antibodies (Fig. 7A). Synapses were labeled using FM4-64 dye, and single-molecule trajectories were obtained and analyzed both in and out of synapses. Compared to the extrasynaptic sites, PrPC diffusion was generally slower at the synapses, which is likely due to the crowded environment. Similar diffusion coefficient values of PrPC were obtained with all POM antibodies (Fig. 7B; each data point represents an average value measured for hundreds of QDs; also refer to table S2). Thus, without prior exposure to high POM-x concentrations, PrPC mobility remains largely unaltered. Next, neurons were exposed to POM-x antibodies (1 μg for 1 hour) before PrPC diffusion measurement (Fig. 7C). In neurons preexposed to FT-directed IgGs with two (POM11) or four (POM2) repetitive epitopes, diffusion of PrPC showed a 20 to 60% slowdown (most pronounced for POM2). On the contrary, preexposure to the other IgGs had no (POM3/POM19) or only moderate effects (<10% for POM1) on PrPC diffusion. Each data point represents the difference (mean ± SEM) between unexposed control and POM-x–exposed condition for a given experiment. The absolute diffusion coefficient values for all QDs are provided in table S2. In sum, the strong impact of POM2 IgG (and—albeit to lesser extent—the closely related POM11 binding to overlapping epitopes) on PrP mobility supports the idea that these IgGs drive freely diffusing PrPC to form larger complexes, resulting in cluster formation.
Fig. 7. Slowdown in the lateral diffusion of endogenous PrPC by IgGs directed against repetitive epitopes within the flexible tail.
(A) SPT-QD to quantify the diffusion coefficient of endogenous PrPC and using QD precoupled to anti-PrPC antibodies (POM-x-QD). (B) Under control conditions (no preexposure to high concentration of antibodies), similar diffusion coefficient values of PrPC were obtained using various antibodies [globular domain (GD)–directed IgGs: POM1 and POM19; flexible tail (FT)–directed IgGs: POM2 and POM11; or hydrophobic core (HC)–directed: POM3]. Synapses were identified using FM4-64 labeling. Plotted data show means ± SEM values for three independent experiments, and one-way ANOVA test was performed with no significant difference. (C) SPT-QD of PrPC using POM-x-QD antibody following preexposure (1 hour) to a high concentration (1 μg) of POM-x antibodies. (D) Only preexposure to FT-directed IgGs, but not to the others, greatly (>20%) reduced the diffusion coefficient of PrPC. Plotted data show means ± SEM for three independent experiments. Paired t test was performed to compare the difference from control condition [no preexposure, (B)]. Data for all QDs analyzed are shown in table S2.
DISCUSSION
A soluble form of PrP, most likely representing sPrP, has been described decades ago (27). However, convenient and reliable discrimination between sPrP and the usually much higher (and thus masking) amounts of membrane-associated fl-PrPC in biological samples has only been achieved recently, fueled by the generation of antibodies specifically recognizing sPrP (26, 63). Thus, knowledge on potential functions of sPrP is mainly based on data obtained using recPrP. Although recPrP lacks glycosylation and may thus differ structurally from both PrPC and physiologically released sPrP, data obtained using recPrP imply physiological functions in synapse formation (64), neurite outgrowth guidance (65), differentiation, immune signaling, and intercellular communication (66–68). One of the few molecularly well-defined physiological roles of PrPC (i.e., in regulating myelin maintenance in the peripheral nervous system) might be executed by sPrP (and/or PrP’s released N1 fragment) (69).
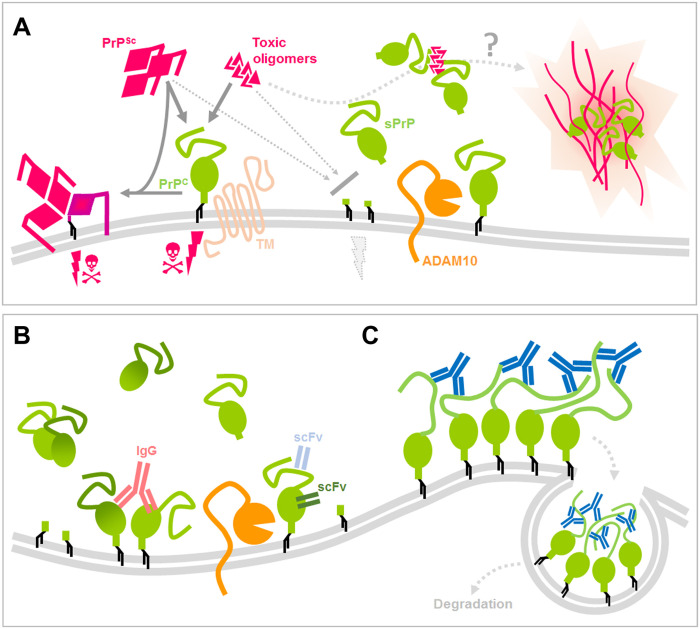
Regarding a role of sPrP in prion diseases, data show that not only released forms of PrP, secreted PrP dimers, but also recPrP confer neuroprotection mainly by blocking the buildup of neurotoxic protein conformers (reference group I in fig. S2). We and others have already shown that levels of the PrP sheddase ADAM10 correlate with incubation times in prion-infected mice and provided indirect evidence for an inverse correlation between ADAM10 expression (and, thus, supposed levels of sPrP) on the one hand and neurotoxic PrPSc formation on the other hand (33, 34). Using our sPrP-specific antibody, we here provide more direct evidence for this. We found sPrP to colocalize with PrPSc in respective deposits in prion-diseased mice. This may indicate that sPrP binds to neurotoxic PrPSc in the extracellular space and helps to sequester those conformers into deposits (Fig. 8A) and could also explain the fact that even mild transgenic overexpression of ADAM10 leads to a relevant decrease in PrPSc production in the brain (33).
Fig. 8. Scheme summarizing potential protective roles of sPrP and effects of PrP-directed ligands.
(A) PrP (green) expressed at the cell surface is a central player in neurodegenerative diseases, as it serves as a substrate for prion conversion and PrPSc (pink) production in prion diseases and (in complex with certain transmembrane proteins) acts as a receptor for toxic protein conformers (pink), such as Aβ in AD, initiating toxic signaling (pink thunderbolts and skulls). As supported by several published reports, mechanisms that lower PrP levels at the plasma membrane, such as the endogenous shedding mediated by ADAM10 (orange), are considered neuroprotective. In addition, several studies have shown that released forms or fragments of PrP interfere with toxic proteins in the extracellular space. Our data suggest that sPrP inversely correlates with PrPSc formation and colocalizes with deposits of PrPSc and Aβ, indicating a blocking and possible sequestrating activity of sPrP toward harmful conformers. As a consequence, stimulated shedding may represent a promising therapeutic option. (B) Regarding the latter, we here provide evidence that several PrP-directed ligands binding to different epitopes cause an increased ADAM10-mediated shedding. While cross-linking may be involved (as in the case of IgGs), it at least is not a prerequisite for this action (scFv). (C) One exception to this shedding-stimulating effect was found for IgGs directed against several (repetitive) epitopes within the disordered and flexible N-terminal half of PrP (e.g., POM2; blue). These ligands rather cause a strong surface clustering [possibly by multimeric cross-linking (as indicated in this scheme) or other structural alterations facilitating tight molecular interaction] followed by fast endocytosis and (lysosomal) degradation of PrP (which may likewise be beneficial against neurodegenerative processes).
With regard to AD, released forms or proteolytic fragments of PrP (such as N1), but likewise recPrP or PrPC on extracellular vesicles, confer neuroprotection in models of AD (references I in fig. S2). How exactly this is achieved on the molecular level is less clear to date. In cell-free systems, recPrP limits the growth of Aβ fibrils, while vesicle-bound PrPC accelerates the formation of these supposedly less toxic Aβ species. In analogy to our finding for prion diseases, data obtained by using a murine AD model and our sPrPG228 antibody speak in favor of a (likely beneficial) role of sPrP in promoting Aβ fibrillization and plaque formation, possibly at the expense of mobile neurotoxic AβO in the extracellular space. We show that, in analogy to what we observed in prion-infected mouse brain, sPrP is completely redistributed to particular deposits in brains of 5xFAD mice, where it is likely bound to Aβ and seen in the center of many amyloid plaques. Similar effects were also observed in AD patients, where PrP localizes to dense cores within Aβ plaques and it was suggested (though not proven) that these may represent “released” forms of PrPC (references III in fig. S2). This reconciles data showing that PrPC in its plasma membrane–bound state acts as a mediator of AβO neurotoxicity (16, 70–72) yet, when overexpressed as a GPI-anchorless version, rather blocks neurotoxicity (73). Our data also suggest that it is sPrP, which is responsible for the plaque-promoting effect initially ascribed to PrPC “in general” in earlier studies (references III in fig. S2). Thus, physiologically, sPrP may act protective in prion diseases and AD by blocking toxic oligomers and/or by precipitating these into less toxic deposits (Fig. 8A). Although not assessed here, it appears likely that sPrP may act similarly against other proteinopathies, given the central role of PrPC as neuronal toxicity receptor (8, 11, 15). It would also be interesting to assess whether sPrP bound to these oligomers and deposits may serve as an “eat-me” signal for internalization by phagocytic cells (74). In sum, stimulation of PrPC shedding may be a promising therapeutic target in neurodegenerative diseases, yet further studies on this are clearly required.
In an effort to identify inducers of the ADAM10-mediated PrP shedding, we used a candidate approach and took advantage of known principles of an increased shedding caused by antibodies directed against some other ADAM10 substrates (36, 37). This has, for instance, been shown for CD44, where antibody binding to this protein leads to increased release (38). We found that several PrPC-directed antibodies and derivatives significantly increase its ADAM10-mediated shedding. To investigate the underlying mechanism of this, we assessed whether antibody-induced dimerization (and hence longer retention) of PrPC at the cell surface is a prerequisite. For the ADAM10 substrate CD44, dimerization is required for increased shedding (38). Moreover, some PrP-directed antibodies cause PrPC cross-linking (75), and forced dimerization of PrPC leads to increased proteolytic processing, mostly by elevated α-cleavage of PrPC (43). Although we cannot rule out that dimerization contributes to increased shedding upon binding of fl-IgGs to PrPC, the fact that we also found increased sPrP levels upon treatment with PrP-directed single-chain antibodies rather demonstrates that cross-linking is at least not a general prerequisite for the shedding-stimulating effect of these ligands. A related potential explanation for increased shedding could be antibody-mediated changes in steady-state levels of PrPC at the plasma membrane, the place where ADAM10 and PrPC meet and where shedding occurs. PrPC-binding IgGs (exemplified here for POM1) lead to increased cell surface levels of PrPC, yet again, single-chain antibodies, which basically seem as effective as their full-length counterparts in their ability to stimulate shedding, do not do this. Thus, as above, we conclude that altered PrPC plasma membrane levels may contribute to increased shedding yet are not a prerequisite for this. Regulation of ADAM10 substrate specificity and sheddase activity is complex and poorly understood, and may involve conformational changes of the substrate (76). To investigate whether a ligand-induced conformational change within PrPC occurs upon binding of an antibody, we took advantage of SAXS and focused on antibody-induced structural transitions of PrPC that could modify its interaction with and support subsequent cleavage by ADAM10. Our data using recPrP suggest that the highly flexible N-terminal tail of PrP is relatively moved away from the C-terminal half (and likely the GPI anchor of PrPC) upon antibody binding. This might support increased exposure of the very C-terminal part (containing the cleavage site), previously “shielded” by movements (45) or intramolecular and possibly even described plasma-membrane interactions of the N-terminal tail (46–49), and favor access of the membrane-proximate catalytic domain of active ADAM10. This is in agreement with publications showing that Fab fragments binding to similar PrP domains influence its conformation by preventing intramolecular interactions (45) and data proposing these interactions of N- with C-terminal domains of PrPC in the absence of antibody binding (49).
Lowering PrPC amounts is among the most promising strategies for causal treatment of prion diseases (24, 60, 61). There is a linear inverse correlation between PrPC expression levels and susceptibility toward prion diseases (19) with a complete lack of susceptibility when PrPC is absent (18). Reduction of PrPC levels even has therapeutic potential once the disease becomes clinically apparent (21). However, as with many complex diseases, combination therapy targeting different pathomechanistic aspects will likely be superior over a therapeutic strategy focusing on one mechanism only. Even if current strategies to lower total PrP expression succeed, additional stimulation of PrP shedding may add further protection (by generating sPrP as a potential anti-prion agent) while preserving certain physiological functions potentially carried out by released PrP fragments [such as myelin maintenance in the periphery (69)]. The POM2 antibody was shown to be neuroprotective, and this was linked to impairing harmful interactions of the flexible N-terminal tail of PrP with the plasma membrane occurring in prion disease (46, 48, 49). Here, we show that POM2, because of its unique binding characteristics (partially shared with POM11) and in contrast to other ligands tested here (including its own single-chain version), leads to strong multimeric clustering of PrPC at the plasma membrane and subsequent cellular uptake of these clusters for lysosomal degradation (Fig. 8C). This reconciles abovementioned concepts of lowering PrP levels and implies that, in addition to POM2’s role in preventing toxic PrP N terminus–to–membrane interaction, it efficiently causes removal of PrPC from the cell surface and a reduction in total PrPC levels. Because administration of antibody would allow to do this in a reversible and potentially controllable fashion, therapeutic applications are also conceivable here.
As introduced in detail above, anti-PrP antibodies have been proposed as potential prion and AD therapeutics, and in a number of studies, anti-PrP antibodies led to delay of disease onset and reduced signs of neurodegeneration (references II in fig. S2). Although details are not completely understood, for prion diseases, this beneficial effect is generally attributed to reduced PrPSc replication thought to be caused by either direct sterical hindrance of the critical PrPSc-to-PrPC interaction, stabilization of the native conformation, and/or altered cellular trafficking of PrPC. For AD, binding of antibodies to membrane-bound PrPC is likewise thought to block the interaction with toxic AβO species and, thus, to restrict respective surface PrPC–dependent synaptotoxic signaling cascades. Our finding that the same anti-PrP antibodies, which have been used in some of the studies mentioned above, also induce the ADAM10-mediated shedding of PrPC (Fig. 8B) adds another level of complexity and potential explanations to these data and may indicate that stimulated shedding raising extracellular amounts of seemingly neutralizing sPrP, at least in part, accounts for the protective effects.
For prion diseases, it is conceivable that sPrP, similar to what was shown for a secreted PrP dimer (77), contributes to stopping the buildup of PrPSc by binding to critical seeds thereof, thus acting dominant negative against the misfolding of cell-associated PrPC. This is only seemingly in conflict with data from transgenic mice showing that anchorless PrP readily misfolds into PrPSc, which is then preferentially deposited as plaques (78). So, how does physiologically produced sPrP differ from the highly misfolding-prone anchorless PrP? Although this certainly deserves further investigation, a likely explanation may lie in the apparently different glycosylation state. While physiological sPrP is preferentially fully (i.e., di-) glycosylated (26), transgenically expressed secreted PrP is mainly un- and monoglycosylated. Fittingly, for many prion strains [such as the Rocky Mountain Laboratories (RML) isolate used here], diglycosylated PrP is a relatively poor substrate (79–82), which is also reflected by relative differences in the glycopattern between PrPC and PK-digested PrPSc in brain samples. Moreover, earlier cell culture experiments, where a broad range of anti-PrP antibodies inhibited PrPSc replication, revealed that “PrP levels” in the medium were increased (83). According to the data presented here, this most likely represents sPrP. In the case of AD, it may well be that anti-PrP antibodies contribute to mitigating neuroprotection threefold, namely, (i) by directly blocking interaction with toxic AβO species, (ii) by removing PrP, the receptor for toxic AβO, from the plasma membrane, and (iii) by hence increasing levels of sPrP, which, in turn, precipitates toxic AβO into possibly less-toxic fibrillary species as described earlier for recPrP or the PrP-N1 fragment. In the latter regard and to eventually develop therapeutic approaches, it will be important to further explore ligands increasing the shedding without occupying critical PrP binding sites for toxic conformers.
We found that single-chain anti-PrP antibodies efficiently induce the ADAM10-mediated shedding of PrP. Thus, not only large antibodies but also smaller molecules resembling PrP ligands are able to induce shedding (Fig. 8B). On the other hand, the four small chemical compounds described to bind PrP and tested here did not induce increased release. The latter may fit a recent report suggesting that PrP is a difficult-to-target protein, at least with small ligands, and implying that, for PrP ligands to be therapeutically effective, they have to have a certain size and binding characteristics (84). This, in turn, reinforces the use of non–small molecules, such as antibodies or their derived smaller fragments. Our finding of shedding-stimulating ligands is also of interest in two different perspectives. First, it may give hints to further physiological functions of PrP, which, upon ligand binding and release, may act as a ligand itself serving intercellular communication, possibly in complex with the bound partner protein. It will thus be up to future studies to look for endogenous PrP ligands or interacting partners promoting its shedding. Using our sPrP-specific antibody in appropriate paradigms, this should be a feasible task. Secondly, it opens up the possibility to screen for or custom-design anti-PrP ligands promoting the shedding, which may then be used in therapeutic settings. Given the fact that these ligands could potentially be smaller and pharmacokinetically more favorable than antibodies, this could be an attractive aim for future studies on new therapeutic options against prion diseases and other neurodegenerative proteinopathies alike.
MATERIALS AND METHODS
Animals and ethics statement
Mice and rats were housed at standard laboratory conditions (12-hour light/12-hour dark cycle, constant room temperature and humidity, with food and water access ad libitum) in authorized animal facilities at Christian Albrechts University Kiel, Georg August University Göttingen, University of Zurich, and University Medical Center Hamburg-Eppendorf (UKE) (Table 1). Breeding, sacrification for the sake of organ removal (slice cultures and primary neurons), and all experimental procedures were approved by the respective ethical research committees of local German, French, or Swiss authorities [Freie und Hansestadt Hamburg–Behörde für Gesundheit und Verbraucherschutz (permit numbers 48/09, 84/13, ORG587, and ORG739); Schleswig-Holsteinisches Ministerium für Energiewende, Landwirtschaft, Umwelt und ländliche Räume, Kiel (V241-25481/2018(30-3/16); federal state of Niedersachsen (permit 13/1232); Comité d’éthique pour l’expérimentation animale, Paris (permits Ce5/2012/018 and #1339-2015073113467359 v4); and the veterinary office of the Canton of Zurich (permit ZH236/19)]. Procedures were in agreement with the principles of good laboratory animal care (National Institutes of Health publication no. 86-23, revised 1985) and the respective rules in the Guide for the Care and Use of Laboratory Animals of the German Animal Welfare Act on protection of animals. 5xFAD/Prnp0/0 mice were obtained by crossing Prnp0/0 mice with 5xFAD mice and crossed back with Prnp0/0 for at least six generations (mixed 129/Sv and C57BL/6J background). As control mice, we used 5xFAD mice crossed back with WT mice for at least six generations (mixed 129/Sv and C57BL/6J background). Genotyping was performed by polymerase chain reaction (PCR) from the tail biopsy. Intracerebral inoculations of mice with the mouse-adapted prion strain RML, subsequent observation, and termination were described in detail in (34).
Table 1. List of mouse models assessed in this study.
| Mouse line | Brief description |
| C57BL/6 | WT mouse line |
| A10 cKO | Cre-driven conditional ADAM10 knockout in forebrain neurons (under control of the CamKIIα promotor) (34) |
| Prnp 0/0 | PrP-knockout mouse line (99) |
| tga20 | PrP-overexpressing mouse line (100) |
| 5xFAD | Mouse model for AD-associated amyloid formation |
| 5xFAD/Prnp0/0 | As above but depleted for PrP (by crossing with Prnp0/0 mice) |
Antibodies
The list of primary antibodies used in this study is shown in Table 2.
Table 2. List of primary antibodies used herein (including application).
WB, Western blot; EM, electron microscopy; T, treatment.
| Name | Description | Used for… | Company/source |
| sPrPG228 | Rb polyclonal specific for sPrP | WB, IHC, IF | UKE Hamburg (26) |
| POM1 | Ms monoclonal anti-PrP (C-terminal half) | T, WB, SPT-QD | A.A., Zürich |
| scPOM1 | Single-chain Fv of above | T | A.A., Zürich |
| POM2 | Ms monoclonal anti-PrP (N-terminal half) | T, WB, SPT-QD | A.A., Zürich |
| scPOM2 | Single-chain Fv of above | T | A.A., Zürich |
| scPOM-bi | Joined complementarity-determining regions of POM1 and POM2 | T | L.V., Bellinzona (41) |
| POM3 | Ms monoclonal anti-PrP (central part) | SPT-QD | A.A., Zürich |
| POM11 | Ms monoclonal anti-PrP (N-terminal half) | SPT-QD | A.A., Zürich |
| POM19 | Ms monoclonal anti-PrP (C-terminal half) | SPT-QD | A.A., Zürich |
| 3F4 | Ms monoclonal anti-human PrP (central) | T | Millipore |
| 6D11 | Ms monoclonal anti-PrP (central) | T, EM | BioLegend |
| SAF84 | Ms monoclonal anti-PrP/anti-PrPSc | IHC | Cayman Chemical |
| D18 | Monoclonal anti-PrP | WB | D. Burton, Scripps, La Jolla |
| β-Actin | Ms monoclonal anti–β-actin, clone C4 | WB | Millipore |
| NeuN | Anti-NeuN, Alexa Fluor 488 conj. | IF | Millipore |
| NeuN | Rb polyclonal anti-NeuN (ABN78) | WB | Millipore |
| 1D4B | Rat monoclonal anti-LAMP1 | IF | DSHB Univ. Iowa |
| 6E10 | Ms monoclonal anti-human β amyloid | WB, IHC, IF | BioLegend |
| APP/sAPPα | Rb polyclonal anti-mouse/rat β amyloid | WB | BioLegend |
| Flotillin-1 | Ms monoclonal anti–flotillin-1 | WB | BD Biosciences |
| ADAM10 | Rb monoclonal anti-ADAM10 (EPR5622) | WB | Abcam |
Murine COCS
COCS (350 μm thick) were prepared from 9- to 12-day-old Prnp0/0, C57BL/6, and tga20 pups according to a previously published protocol (85). Six to nine COCS were plated per polytetrafluoroethylene (PTFE)–coated cell culture insert (Millipore) in organotypic slice culture medium (85). Pharmacological treatment was initiated 10 to 14 days after dissection of COCS and was re-added with every medium change [treatment duration: 8 days with three medium changes for Fig. 3 (A to C); Fig. 3D: IgG and 6D11, 14 days; STS, 3 days]. The final concentration of antibodies (6D11, 3F4, and pooled murine IgG) was 67 nM diluted in organotypic slice culture medium. Protein analysis and IHC analysis of COCS were described extensively before (41).
Quantitative reverse transcription PCR
RNA was isolated from N2a cells upon antibody treatment for 18 hours using the RNeasy Mini Kit (Qiagen) following the manufacturer’s instructions. Deoxyribonuclease-treated RNA (2 μg) was used for complementary DNA (cDNA) synthesis using the RevertAid cDNA Synthesis Kit (Thermo Fisher Scientific). The gene expression analysis for Prnp and Gapdh was performed using the respective TaqMan probes (Thermo Scientific Fisher). PrPC expression levels were depicted as percentage of GAPDH (glyceraldehyde-3-phosphate dehydrogenase) expression using the ΔCT method for calculation.
Preparation of mouse brain homogenates
Pieces of fresh or frozen frontal brain were processed as 10% (w/v) homogenates in radioimmunoprecipitation assay (RIPA) buffer [50 mM tris-HCl (pH 8), 150 mM NaCl, 1% NP-40, 0.5% Na-deoxycholate, and 0.1% SDS] with freshly added cOmplete protease inhibitor cocktail (PI; Roche). Tissue was ground by 30 strokes on ice using a Dounce homogenizer and subsequently incubated on ice for 15 min before resuspending by pipetting up and down. Homogenates were then spun down at 11,000g for 10 min at 4°C. Resulting supernatants were collected, aliquoted, and stored at −80°C. For further use, 30 μl was mixed with 120 μl of ddH2O and 50 μl of 4× loading buffer [250 mM tris-HCl, 8% SDS, 40% glycerol, 20% β-mercaptoethanol, 0.008% bromophenol blue (pH 6.8)] and boiled for denaturation for 8 min at 96°C.
Prion-infected mice and samples were handled in respective biosafety facilities/laboratories at the UKE Hamburg. For detection of PrPSc in prion-infected mouse brains, 20% (w/v) homogenates of frontal brain were prepared using phosphate-buffered saline (PBS) lacking protease inhibitors. Homogenates were prepared as above using a Dounce homogenizer and then centrifuged at 1500g for just 2 min to pellet crude cellular debris. The supernatant (4 μl) was incubated with PK (20 μg/ml; Roche) in a total volume of 22 μl of RIPA buffer for 1 hour at 37°C to digest all proteins except for PK-resistant PrPSc. Digestion was terminated by the addition of 2.5 μl of 10× loading buffer and boiling for 8 min at 96°C.
Cell culture, treatments, lysis, harvesting of medium, and sample preparation
Preparations and treatments
Murine neuroblastoma cells [N2a; ACC148, Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) Germany] and the embryonic mouse hippocampal cell line mHippoE-14 (CLU198; CELLutions Biosystems Inc.) were cultured at 37°C in an atmosphere of 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM; Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific). Generation of PrP-depleted N2a cells (Prnp KO) was described elsewhere (86). For overexpression of murine WT (PrP-WT) or 3F4-tagged PrP (PrP-3F4), these PrP knockout cells were transiently transfected with the respective constructs (26) using Lipofectamine 2000 (Thermo Fisher Scientific) following the manufacturer’s instructions.
One day before treatments, 250,000 cells were seeded per well in six-well plates. The next day, the medium was freshly exchanged to prewarmed Opti-MEM (Gibco) and treatments were started by adding antibodies and/or compounds in the stated concentration to the medium. Treatments were carried out overnight for 18 hours in a total of 1 ml of Opti-MEM.
Lysis of N2a and mHippo cells and processing of conditioned medium
Following treatments, conditioned medium and corresponding adherent cells were quickly yet carefully harvested in parallel working on ice or at 4°C. The medium was aspirated, transferred to a 1.5-ml Eppendorf tube already containing 50 μl of a predissolved and 20× concentrated PI (in PBS), and gently inverted for mixing. Cells were washed twice with cold PBS and incubated for 10 min with 150 μl of RIPA buffer (with freshly added PI), scraped off from the plate, transferred to a 1.5-μl tube, shortly vortexed, and incubated on ice for additional 15 min. After a centrifugation step at 12,000g and 4°C for 12 min, the supernatant (=cleared lysate) was transferred to a fresh tube and either stored at −80°C or mixed (50 μl of lysate + 50 μl of 4× sample buffer + 100 μl of ddH2O), boiled (10 min at 96°C), and loaded for SDS–polyacrylamide gel electrophoresis (SDS-PAGE).
Harvested medium supernatants were centrifuged at 500g and then 5000g (both for 5 min at 4°C), and at each step, 50 μl was left at the bottom (as a pellet of cellular debris was mostly invisible). The remaining 900 μl was subjected to protein precipitation as follows: 1:100 volume (i.e., 9 μl) of a 2% sodium deoxycholate solution was added to the sample and mixed by vortexing followed by 30 min of incubation on ice. Samples were then mixed with a 1:10 volume (i.e., 90 μl) of 100% (6.1 N) trichloroacetic acid (Sigma-Aldrich) and incubated for another 30 min on ice. After centrifugation at 15,000g for 15 min at 4°C, the supernatant was aspirated and the pellet was air-dried for 5 min. The pellet was completely resuspended (by intensively pipetting up and down and mixing at 800 rpm at 50°C for 15 min) in 100 μl of 1× loading buffer. As the blue color changed to yellow (indicating low pH), 1.5 μl of 2 M NaOH was mixed into the sample and blue color reappeared. Samples were boiled for 10 min at 96°C and stored at −80°C or directly used for SDS-PAGE.
Treatment of HEK293 cells with PrP-directed chemical compounds
HEK293 cells were stably transfected with a pcDNA3.1 plasmid carrying a hygromycin resistance cassette and encoding for mouse WT PrP under the control of the cytomegalovirus promoter. On day 1, cells were seeded at ~70% confluency in 24-well plates [medium containing DMEM, 10% FBS, non-essential amino acids, penicillin-streptomycin, l-glutamine, and hygromycin (150 μg/ml)]. On day 2, medium was replaced with fresh medium containing each chemical compound at 0.1, 0.3, 1, or 3 μM (but lacking hygromycin); on day 3, conditioned medium was collected and diluted 2:1 in 4× Laemmli sample buffer [2% SDS, 10% glycerol, 100 mM tris-HCl (pH 6.8), 0.002% bromophenol blue, and 100 mM dithiothreitol (DTT)], while the corresponding adherent cells were directly lysed with 2× Laemmli sample buffer.
Cell surface protein biotinylation assay
After 18 hours of treatment with antibodies, N2a cells were washed twice with cold PBS and incubated for 30 min with EZ-Link Sulfo-NHS-SS-Biotin (0.5 mg/ml; Thermo Fisher Scientific) in PBS shaking at 4°C. Cells were washed three times for 5 min at 4°C with 0.1% bovine serum albumin and lysed with 500 μl of RIPA buffer. After complete lysis, the supernatant was diluted 1:1 with Triton dilution buffer [100 mM triethanolamine (TEA), 100 mM NaCl, 5 mM EDTA, 0.02% NaN3, 2.5% Triton X-100 (pH 8.6), + PI] and incubated for 1 hour with 200 μl of NeutrAvidin agarose beads (Thermo Fisher Scientific) at 4°C. Beads were washed with wash buffer three times [20 mM TEA, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 0.2% SDS, 0.02% NaN3 (pH 8.6), + PI] and centrifuged at 1000g. Two additional washing steps were performed with the final wash buffer [20 mM TEA, 150 mM NaCl, 5 mM EDTA (pH 8.6), + PI]. Last, 50 μl of 4× sample buffer (including DTT) was added, and the samples were boiled for 6 min at 96°C to release and denature biotinylated proteins from the beads. The supernatants were loaded for SDS-PAGE for further biochemical analysis.
SDS-PAGE and Western blot analyses
SDS-PAGE and Western blotting
Denatured samples (tissue homogenates, cell lysates, or precipitated medium) were loaded on precast Nu-PAGE 4 to 12% bis-tris protein gels (Thermo Fisher Scientific), self-cast 10% or 12% SDS-gels, or Any kD Mini-PROTEAN TGX Precast Protein Gels (Bio-Rad). After electrophoretic separation, wet blotting (at 200 mA per gel for 1 hour) was applied to transfer proteins onto nitrocellulose membranes (Bio-Rad). Membranes were then blocked for 45 min with either 1× RotiBlock (Carl Roth) in Tris-buffered saline with Tween 20 (TBS-T) or 5% skimmed dry milk (in TBS-T) under gentle agitation at room temperature. Membranes were incubated overnight with the respective primary antibodies (see list above) diluted 1:1000 (1:2500 for POM antibodies; 1:5000 for β-actin) in the respective blocking reagents at 4°C on a shaking platform. The next day, membranes were washed four times for 5 min with TBS-T and incubated for 45 min at room temperature with horseradish peroxidase–conjugated secondary antibodies. After extensive washes (at least six times for 5 min) in TBS-T, signal detection was performed [after incubating blots for 5 min with either Pierce ECL Pico or SuperSignal West Femto substrate (Thermo Fisher Scientific)] with a ChemiDoc imaging station (Bio-Rad). Densitometric quantification was done using the Quantity One software (Bio-Rad) followed by further analysis in Excel (Microsoft).
SDS-PAGE and Western blotting (of compound-treated HEK293 cells)
A 30-μl aliquot of either medium or lysate for each sample was heated at 95°C for 10 min and loaded on two separate SDS-PAGE gels (Any kD Mini-PROTEAN TGX Stain-Free Protein Gels; Bio-Rad). Proteins were electrophoretically transferred to polyvinylidene fluoride membranes, which were then blocked for 20 min in 5% (w/v) nonfat dry milk in tris-buffered saline (TBS) containing 0.05% Tween 20. After incubation with appropriate primary [D18 (1:5000) and sPrPG228 (1:3000)] and secondary antibodies, signals were revealed using enhanced chemiluminescence (Luminata, Bio-Rad) and visualized by a Bio-Rad XRS ChemiDoc image scanner (Bio-Rad). Values were obtained by densitometric quantification of PrP bands using the Image Lab 5.2.1 software (ChemiDoc, Bio-Rad). Each PrP signal (D18 or sPrPG228) was normalized to the signal of total proteins in cell lysates (directly acquired by detecting the fluorescence of proteins in stain-free gels) and expressed as percentage of the dimethyl sulfoxide (DMSO)–treated control.
Annexin V cell toxicity assay
N2a cells were grown on six-well plates until 80% confluency, treated overnight with antibodies, washed with PBS, and detached by incubating for 5 min with Accutase (Thermo Fisher Scientific) at 37°C. Next, cells were washed with PBS and resuspended in 1× binding buffer (provided by the manufacturer) at 1 × 106 to 5 × 106 cells/ml. Phycoerythrin (PE)–conjugated annexin V (5 μl; Thermo Fisher Scientific) was added to 100 μl of the cell suspension and incubated at room temperature in the dark. After 15 min of incubation, cells were again washed with 1× binding buffer and subsequently analyzed by flow cytometry. PE-positive, apoptotic cells were counted by using flow cytometry in FACSCanto II (BD Biosciences, Franklin Lakes, NJ, USA). As a positive control, cells were treated with different concentrations of STS, a known inducer of apoptotic cell death.
Immunohistochemistry
Mouse brains were dissected and immediately fixed overnight in an excess volume of 4% buffered formalin. Prion-infected samples were inactivated for 1 hour in 98 to 100% formic acid before transfer into clean tubes and export from the biosafety facility. The latter samples were then again incubated for another night in 4% buffered formalin at 4°C. Formalin-fixed samples then underwent dehydration and were embedded in low–melting point paraffin blocks following standard histological protocols. Sections of 3 to 4 μm were prepared with a microtome submitted to immunostaining following standard IHC procedures using a Ventana BenchMark XT machine (Roche Diagnostics). Briefly, sections were deparaffinated and then boiled for 30 to 60 min in 10 mM citrate buffer (pH 6.0) for antigen retrieval. All applied solutions were purchased from Ventana. Afterward, sections were incubated with primary antibodies diluted in 5% goat serum (Dianova, Hamburg, Germany), 45% TBS (pH 7.6), 0.1% Triton X-100 in antibody diluent solution (Zytomed, Berlin, Germany) for 1 hour. Primary antibodies used for IHC analyses were as follows (for further information, also refer to list above): sPrPG228 (1:30, for detection of sPrP), SAF84 (1:100, for PrPC and PrPSc), and 6E10 (1:100, for human APP and Aβ deposits). Secondary antibody treatment was performed using anti-rabbit Histofine Simple Stain MAX PO Universal immunoperoxidase polymer or Mouse Stain Kit (for detection of mouse antibodies on mouse sections), all purchased from Nichirei Biosciences (Tokyo, Japan). Final visualization of antibodies was achieved with the Ultra View Universal DAB Detection Kit (brownish signals) or Ultra View Universal Alkaline Phosphatase Red Detection Kit (yielding pink signals) from Ventana using standard machine settings. Notably, whenever possible, experimental groups were stained in one run, thus ensuring identical conditions and good comparability. Light blue counterstaining was likewise done by the machine following standard procedures. Secondary antibody–only controls were always run when establishing a given staining yet never revealed a relevant signal. For PrPSc detection using SAF84 antibody, sections of 4 μm were pretreated with 98% formic acid for 5 min. Moreover, sections were boiled in 1.1 mM sodium citrate buffer [2.1 mM tris-HCl and 1.3 mM EDTA (pH 7.8)] at 95°C for 30 min, then incubated with PK for 16 min, in Superblock for 10 min, and finally incubated with primary antibody and further processed as above.
Stained sections were assessed, and representative pictures were taken in TIF format on a digital microimaging device (DMD108, Leica). Pictures were further processed by performing a white balance against a negative, nonstained area within the same field, and finally assembled (using fixed/scaled aspect ratio) for comparison of experimental groups using Photoshop (Adobe).
IF stainings and (live) microscopy
IF stainings of free-floating murine brain sections and microscopic assessment
Mice were deeply anesthetized with Rompun/ketamine followed by transcardial perfusion with 0.1 M phosphate buffer (PB; pH 7.4) and, consecutively, 4% paraformaldehyde (PFA). Brains were removed and postfixed by immersion. After 4 hours of postfixation, PFA was removed and replaced by 30% sucrose (w/v) in 0.1 M PB. After incubation overnight, brains were sectioned sagittally at 35-μm thickness as free-floating sections with a Leica 9000s sliding microtome (Leica, Wetzlar, Germany). For IF staining, the sections were blocked in blocking solutions [0.5% Triton X-100 and 4% normal goat serum in 0.1 M PB (pH 7.4)] and incubated in blocking solution containing the primary antibodies at 4°C overnight. After three washes with wash solution [0.1 M PB (pH 7.4) containing 0.25% Triton X-100], sections were incubated for 90 min with secondary antibody in solution, washed again three times in wash solution containing 4′,6-diamidino-2-phenylindole (DAPI), and finally brought on glass slides and embedded in Mowiol/DABCO (1,4-diazabicyclo-[2.2.2]-octane). Images were acquired with the Olympus FV1000D Laser Scanning Confocal Microscope (model: FV10-292-115) with a 60× lens (UPLSAPO) and processed with the FV10-ASW 4.2 Viewer Software (Olympus, Germany).
IF analyses and (live) microscopy of antibody-treated rat primary neurons
Primary hippocampal cultures were prepared as described previously (87) from E18 Wistar rats Unilever HsdCpb:WU (Envigo) and plated at a density of 60,000 cells per coverslip in a 12-well plate. For live imaging experiments, neurons were transfected with enhanced GFP (eGFP)–tagged PrP (for cloning information, see below; 1 μg of DNA per well) at day in vitro (div) 14 using Lipofectamine 2000 according to the manufacturer’s instructions. At div15, live imaging of transfected neurons was performed with a Nikon Eclipse Ti microscope Ti-E controlled by VisiView software. Illumination was achieved by a 488-nm excitation laser, coupled to a CSU-X1 spinning disk unit via a single-mode fiber. Emission was collected through a quad band filter (Chroma, ZET 405/488/561/647m) on an Orca-Flash4.0 LT complementary metal-oxide semiconductor camera (Hamamatsu). Imaging in confocal mode was done with a step size of 0.5 μm and pixel size of 108.3 nm [using the 60× 1.4 numerical aperture objective (Nikon, CFI Plan Apo Lambda Oil)]. Neurons of similar PrP-GFP expression rate (judged by signal intensity) were imaged in 500 μl of medium at basal conditions for 5 min and upon administration of 1 μg of antibody (POM2, 6D11, and 3F4) for 10 min with 1 image per minute, up to 1 hour with fewer imaging time points. For image representation, maximum projections were created and brightness/contrast was adjusted per cell. To visualize changes in PrP-GFP localization in an individual image, time points 5 and 0 min were subjected to a 1-pixel Gaussian blur and then subtracted in Fiji using the image calculator. The resulting images are shown in a pseudocolor scale.
For staining of endogenous PrP, hippocampal cultures at div19 were treated with 1 μg of POM2 antibody in 500 μl of culturing medium for 30 min and, immediately afterward, fixed in 4% PFA/4% sucrose for 10 min at room temperature. Immunocytochemistry was performed as in (87) using POM2 antibody (1:250) followed by a secondary anti-mouse Alexa Fluor 488 antibody. Actin staining was obtained by using phalloidin-647N (1:100 dilution) in PBS and incubation for 4 hours at room temperature. Images were acquired at a Leica TCS SP5 microscope (Leica Microsystems), controlled by Leica Application Suite Advanced Fluorescence software. Excitation of fluorophores was done with an argon 488-nm laser and a HeNe 633-nm laser, and signals were detected on HyD detectors. Neurons were imaged using a 63× objective (1.4 HCX PL APO CS) at 400 Hz and frame averages of 2 and z-steps of 0.5 μm (overview) or 0.3 μm (dendrite). Picture format was set to 8 bit, and for the overview image to 1024 pixels, using no zoom (pixel size: 240 nm), and for dendrites to 5× zoom at 512 pixels (pixel size: 96 nm). For image representation, average projections were created and brightness/contrast was adjusted per cell.
eGFP-tagged PrP
The moPrP-eGFP_39/40 fusion construct carrying eGFP within the flexible N-terminal tail between amino acids 39 and 40 of mouse PrP was created using NEB Golden Gate Assembly (New England Biolabs). Briefly, three fragments (PrP1-39, eGFP lacking initial MET and terminal STOP, PrP40-254) were amplified via PCR using oligonucleotides as primers in respective combinations (sequence provided as 5′-3′): PrP1-39, SP_PrP-fwd (TTTTAAGCTTATCAGTCATCATGGCGAACCTTGGCTAC) + Bsa I–PrP39-rev (TGGTCTCTTCACGGGATACCGGCTTC); eGFP, Bsa I–GFP-fwd (AGGTCTCTGTGAGCAAGGGCGAGGAG) + Bsa I–GFP-rev (TGGTCTCTCTTGTACAGCTCGTCCATGC); and PrP40-254, Bsa I–PrP40-fwd (AGGTCTCTCAAGGGGCAGGGAAGCC) + pcDNA3.1-F1-rev (AGGGAAGAAAGCGAAAGG). PCR products were fused to a full-length construct using a standard Golden Gate Assembly protocol (1× T4 Ligase Buffer, 500 U of T4 DNA Ligase, 15 U of Bsa I–HFv2, and respective fragments; 30 cycles of two-step incubation in thermocycler: 37°C for 5 min followed by additional 5 min at 16°C). The resulting construct was cloned into an existing pcDNA3.1-moPrP plasmid using the moPrP endogenous restriction sites Age I and Bst EII and T4 DNA ligase. Cloning success was verified by sequencing.
Single-particle tracking using quantum dots
SPT-QD studies were performed on hippocampal neurons prepared from Sprague-Dawley rat embryos (Janvier Labs, France). The SPT-QD protocol and analysis methods have been used and described in several previous publications (88, 89). POM-x (POM1, POM2, POM3, POM11, or POM19) exposure was performed on live neurons in the culture medium maintained in an incubator at 37°C and 5% CO2. MEM recording medium (phenol red–free MEM, 33 mM glucose, 20 mM Hepes, 2 mM glutamine, 1 mM sodium pyruvate, and 1× B27) was used for QD labeling and imaging. Neurons were labeled using POM-x antibody precoupled with QD-605 nm. To precouple antibodies with QDs, the following protocol was applied: Mix 1 μl of POM-x antibody + 1 μl of Fab′-QD-605 + 7 μl of 1× PBS and gently shake for 30 min followed by addition of 1 μl of 1× casein solution for additional 15 min (88). Synapses were labeled and identified using FM4-64 labeling. SPT-QD experiments were performed on neurons aged 20 to 21 div. Tracking and analysis was performed using SPTrack_v4, homemade software in MATLAB (MathWorks) (88). First, the center of the QD fluorescence spot was determined by Gaussian fit with a spatial resolution of 10 to 20 nm. Trajectories were generated by associating spots in a given frame with spots in the previous/next frame based on a maximum likely approach. Trajectories with a minimum length of 15 consecutive frames were used, and trajectories below 15 points were excluded. The mean square displacement (MSD) was calculated using MSD(ndt) = (N − n)−1 Σi=1 N−n [(xi+n − xi)2 + (yi+n − yi)2], where xi and yi are the coordinates of an object on frame I, N is the total number of steps in the trajectory, dt is the time between two successive frames, and ndt is the time interval over which displacement is averaged (62, 90). MSD plot was generated and used to compute diffusion coefficient D by fitting the first two to five points of the MSD plot versus time with the equation MSD(t) = 4D2−5t + 4σx2, with σx representing the spot localization accuracy in one direction.
SAXS—Sample preparation, data acquisition, and analysis
The recombinant mouse PrP 23–230 (recPrP) was obtained following a previously reported protocol (91). After lyophilization, recPrP was dissolved in a buffer containing 150 mM NaCl and 10 mM Hepes (pH 7.5). The recPrP solution was subjected to centrifugation at 10,000g at 4°C, and the supernatant was used for downstream preparation after the concentration determination. The 6D11 antibody (in PBS) was measured in a concentration series (0.75, 1, 1.5, and 2 mg/ml) in the same buffer. RecPrP and 6D11 IgG were mixed in a 2:1 molar ratio to obtain the concentration series for the recPrP/6D11-antibody complex (i.e., 0.3, 0.5, 1.5, and 1.8 mg/ml). After mixing recPrP and 6D11, the mixtures were incubated for 1 hour on ice to achieve maximum recPrP-to-6D11 binding.
Synchrotron SAXS data were acquired on the EMBL beamline P12 (92) at the Petra III storage ring of DESY (Hamburg, Germany). The data were obtained using an automatic sample changer. To avoid radiation damage, the samples were exposed under a continuous flow in a quartz capillary (0.9 or 1.7 mm diameter). The parameters of data collection are shown in table S1. The scattered intensity was calibrated to absolute units using the scattering of water at 293 K.
Initial data processing and analysis was carried out using PRIMUS (93), and overall parameters were calculated using ATSAS suite (94). A previously described SAXS dataset and corresponding CORAL model of the murine recPrP data were used from Small Angle Scattering Biological Data Bank (SASBDB; www.sasbdb.org; accession code: SASDHV9). The modeling for the 6D11 antibody was performed with CORAL, a program allowing the representation of models of partially disordered proteins as quasi-random loops (93).
The recPrP/6D11-antibody complex was interactively modeled in SASpy (95) by optimizing the conformation of a 2:1 complex between a representative CORAL model of the antibody and an extended conformation of recPrP. The antibody Fab domains were placed to maintain contact with the epitope on the extended N-terminal part of recPrP (residues 94 to 109; as reported by the manufacturer). UCSF Chimera (96) was used for model depiction. Additional information is provided in association with table S1.
Solubility/precipitation assay
Quantification of soluble mouse PrP and antibodies (POM1 and POM2-IgG) was performed using SDS-PAGE following a published protocol (41). The mPrP:antibody complexes were formed in a 2:1 ratio at a concentration of 10 and 5 μM in 20 mM Hepes (pH 7.2) and 150 mM NaCl. After complex formation, samples were centrifuged for 2 min at 20,000g at 4°C and supernatants were collected and loaded on polyacrylamide gels (4% stacking ± 12% running). Proteins on gels were then acquired using Fusion FX (Vilber) according to standard procedures, and quantification of soluble proteins was then performed using ImageJ, normalizing all samples to mPrP or antibodies alone. Three different time points after complex formation were analyzed: t0, t10′ (after 10 min), and t60′ (after 60 min).
Atomic force microscopy
Equal volumes of 10 μM recPrP (in 100 mM Hepes and 0.5% Triton X-100) and 5 μM anti-PrP antibodies POM1 or POM2 (each in 100 mM Hepes and 0.5% Triton X-100) were separately incubated at room temperature for 1 hour, with gentle shaking. Freshly cleaved muscovite mica (001) substrates were coated with abovementioned dissolution buffer only, recPrP-POM1 preparation, or recPrP-POM2 preparation at a surface density of 25 μl/cm2. The coated mica substrates were then allowed to dry under gentle vacuum and were used for AFM. A Nanoscope IIIa MultiMode AFM (Bruker Cooperation, Billerica, MA, USA) was used for surface characterization of PrP-POM aggregates. Data evaluation image representation was performed using WSxM 5.0 software (97). The scans were made over the scan ranges of 5 μm by 5 μm and 1 μm by 1 μm. Tapping-mode NSL-20 probes were obtained from NanoWorld Holdings AG (Schaffhausen, Switzerland). The probes had an average force constant of 20 N m−1 and were driven to oscillate at 200 ± 5 kHz for AFM imaging.
Immunogold labeling and electron microscopy
N2a cells were treated with POM2 or POM1 (4 μg/ml) for 5 min at 37°C. Then, they were prepared, sectioned, and labeled according to (98). Briefly, cells were fixed in a mixture of 4% PFA and 0.1% glutaraldehyde and covered with 1% (w/v) gelatin in PBS after several washing steps. They were scraped and transferred into Eppendorf tubes and spun down. The pellet was resuspended in 12% (w/v) gelatin in PBS and solidified on ice. Small pieces of the gelatin-embedded cells were cut, and blocks were left in cold 2.3 M sucrose overnight. Cryoprotected pieces were mounted on specimen holders immersed in liquid nitrogen, and ultrathin sections (70 nm) were cut and collected on Formvar/carbon-coated grids (Science Services GmbH, Germany). Sections were labeled with either POM2 or 6D11 antibody (dilution 1:25) and recognized with 10-nm large protein A–coupled gold (purchased from G. Posthuma, University Medical Center Utrecht). Ultrathin sections were examined in an EM902 (Zeiss, Germany). Pictures were taken with a TRS 2K digital camera (A. Tröndle, Moorenweis, Germany).
Statistical analyses
Applied statistical tests and consideration of significance were as indicated in figure captions.
Acknowledgments
We thank the Mouse Pathology Core Facility (K. Hartmann), the FACS Sorting Core Unit, and the Microscopy Imaging Facility (UMIF) (all at UKE Hamburg) for excellent technical support; C. Giudici and M. Willem (LMU Munich) for providing N2a PrP−/− cells; and G. Spagnolli (University of Trento) for the design of fig. S8A. Molecular graphics and analyses were performed with UCSF Chimera, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from NIH P41-GM103311.
Funding: The authors are thankful for financial research support by the CJD Foundation Inc., the Alzheimer Forschung Initiative e.V. (AFI), the Werner Otto Stiftung (all to H.C.A.), the Deutsche Forschungsgemeinschaft [DFG; Collaborative Research Centre 877 (SFB877) projects A12 (to M.G.), B12 (to M.M.), and A3 (to P.S.); Emmy Noether programme MI1923/1-2 (to M.M.); Germany’s Excellence Strategy—EXC-2033–390677874—RESOLV, TA 167/6-3 and TA 167/11-1 (to J.T.), EXC-2049–390688087 (to M.M.)], Hertie Network of Excellence in Clinical Neuroscience and Excellence Strategy Program (to M.M.), the U.S. NIH (grant R01 NS065244 to D.A.H.), and the Italian Telethon Foundation (TCP14009 to E.B.). L.V. gratefully acknowledges support from SNF (Swiss National Science Foundation) grant 31003A_182270. D.S., S.D.V., M.Sh., and M.G. gratefully acknowledge funding from the Joachim Herz Stiftung (Hamburg, Germany).
Author contributions: Performed experiments/investigation and/or analyzed datasets: H.C.A., L.L., B.M., M.Sh., K.F., J.B., A.N.S., M.D., F.S., A.S., S.D.V., T.M., S.J., A.C., M.Schm., M.Schw., L.A., F.M., and S.T. Provided critical research tools, infrastructure, and ideas and supervised individual experiments and scientific discussion: L.L., B.M., B.P., S.H., I.Z., J.T., E.B., P.S., D.S., L.V., S.G., H.S., D.A.H., A.T., M.M., A.A., H.C.A., and M.G. Visualization/final figure assembly: H.C.A. Conceived and designed the study, project administration, funding, supervision, and writing: H.C.A. and M.G. All authors contributed to, commented on, and/or edited the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The SAXS models and the corresponding data have been submitted to the SASBDB database (www.sasbdb.org), with accession codes SASDLT2 (6D11 IgG monoclonal antibody) and SASDLU2 (1:2 ratio complex between 6D11 IgG and recombinant murine prion protein).
Supplementary Materials
This PDF file includes:
Figs. S1 to S12
Tables S1 and S2
Legend for movie S1
References
Other Supplementary Material for this manuscript includes the following:
Movie S1
REFERENCES AND NOTES
- 1.Aguzzi A., Haass C., Games played by rogue proteins in prion disorders and Alzheimer’s disease. Science 302, 814–818 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Vaquer-Alicea J., Diamond M. I., Propagation of protein aggregation in neurodegenerative diseases. Annu. Rev. Biochem. 88, 785–810 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Checler F., Vincent B., Alzheimer’s and prion diseases: Distinct pathologies, common proteolytic denominators. Trends Neurosci. 25, 616–620 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Benilova I., De Strooper B., Prion protein in Alzheimer’s pathogenesis: A hot and controversial issue. EMBO Mol. Med. 2, 289–290 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lauren J., Gimbel D. A., Nygaard H. B., Gilbert J. W., Strittmatter S. M., Cellular prion protein mediates impairment of synaptic plasticity by amyloid-β oligomers. Nature 457, 1128–1132 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prusiner S. B., Novel proteinaceous infectious particles cause scrapie. Science 216, 136–144 (1982). [DOI] [PubMed] [Google Scholar]
- 7.Weissmann C., Molecular biology of prion diseases. Trends Cell Biol. 4, 10–14 (1994). [DOI] [PubMed] [Google Scholar]
- 8.Resenberger U. K., Harmeier A., Woerner A. C., Goodman J. L., Müller V., Krishnan R., Vabulas R. M., Kretzschmar H. A., Lindquist S., Hartl F. U., Multhaup G., Winklhofer K. F., Tatzelt J., The cellular prion protein mediates neurotoxic signalling of β-sheet-rich conformers independent of prion replication. EMBO J. 30, 2057–2070 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferreira D. G., Temido-Ferreira M., Miranda H. V., Batalha V. L., Coelho J. E., Szegö É. M., Marques-Morgado I., Vaz S. H., Rhee J. S., Schmitz M., Zerr I., Lopes L. V., Outeiro T. F., α-synuclein interacts with PrPC to induce cognitive impairment through mGluR5 and NMDAR2B. Nat. Neurosci. 20, 1569–1579 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Ondrejcak T., Klyubin I., Corbett G. T., Fraser G., Hong W., Mably A. J., Gardener M., Hammersley J., Perkinton M. S., Billinton A., Walsh D. M., Rowan M. J., Cellular prion protein mediates the disruption of hippocampal synaptic plasticity by soluble tau in vivo. J. Neurosci. 38, 10595–10606 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corbett G. T., Wang Z., Hong W., Colom-Cadena M., Rose J., Liao M., Asfaw A., Hall T. C., Ding L., DeSousa A., Frosch M. P., Collinge J., Harris D. A., Perkinton M. S., Spires-Jones T. L., Young-Pearse T. L., Billinton A., Walsh D. M., PrP is a central player in toxicity mediated by soluble aggregates of neurodegeneration-causing proteins. Acta Neuropathol. 139, 503–526 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith L. M., Kostylev M. A., Lee S., Strittmatter S. M., Systematic and standardized comparison of reported amyloid-β receptors for sufficiency, affinity, and Alzheimer’s disease relevance. J. Biol. Chem. 294, 6042–6053 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beland M., Roucou X., The prion protein unstructured N-terminal region is a broad-spectrum molecular sensor with diverse and contrasting potential functions. J. Neurochem. 120, 853–868 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Trevitt C. R., Hosszu L. L. P., Batchelor M., Panico S., Terry C., Nicoll A. J., Risse E., Taylor W. A., Sandberg M. K., al-Doujaily H., Linehan J. M., Saibil H. R., Scott D. J., Collinge J., Waltho J. P., Clarke A. R., N-terminal domain of prion protein directs its oligomeric association. J. Biol. Chem. 289, 25497–25508 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rösener N. S., Gremer L., Wördehoff M. M., Kupreichyk T., Etzkorn M., Neudecker P., Hoyer W., Clustering of human prion protein and α-synuclein oligomers requires the prion protein N-terminus. Commun. Biol. 3, 365 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Um J. W., Kaufman A. C., Kostylev M., Heiss J. K., Stagi M., Takahashi H., Kerrisk M. E., Vortmeyer A., Wisniewski T., Koleske A. J., Gunther E. C., Nygaard H. B., Strittmatter S. M., Metabotropic glutamate receptor 5 is a coreceptor for Alzheimer Aβ oligomer bound to cellular prion protein. Neuron 79, 887–902 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Linden R., The biological function of the prion protein: A cell surface scaffold of signaling modules. Front. Mol. Neurosci. 10, 77 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Büeler H., Raeber A., Sailer A., Fischer M., Aguzzi A., Weissmann C., High prion and PrPSc levels but delayed onset of disease in scrapie-inoculated mice heterozygous for a disrupted PrP gene. Mol. Med. 1, 19–30 (1994). [PMC free article] [PubMed] [Google Scholar]
- 19.Sandberg M. K., Al-Doujaily H., Sharps B., Clarke A. R., Collinge J., Prion propagation and toxicity in vivo occur in two distinct mechanistic phases. Nature 470, 540–542 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Sailer A., Büeler H., Fischer M., Aguzzi A., Weissmann C., No propagation of prions in mice devoid of PrP. Cell 77, 967–968 (1994). [DOI] [PubMed] [Google Scholar]
- 21.Mallucci G., Dickinson A., Linehan J., Klöhn P. C., Brandner S., Collinge J., Depleting neuronal PrP in prion infection prevents disease and reverses spongiosis. Science 302, 871–874 (2003). [DOI] [PubMed] [Google Scholar]
- 22.White M. D., Farmer M., Mirabile I., Brandner S., Collinge J., Mallucci G. R., Single treatment with RNAi against prion protein rescues early neuronal dysfunction and prolongs survival in mice with prion disease. Proc. Natl. Acad. Sci. U.S.A. 105, 10238–10243 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferreira N. C., Ascari L. M., Hughson A. G., Cavalheiro G. R., Góes C. F., Fernandes P. N., Hollister J. R., da Conceição R. A., Silva D. S., Souza A. M. T., Barbosa M. L. C., Lara F. A., Martins R. A. P., Caughey B., Cordeiro Y., A promising antiprion trimethoxychalcone binds to the globular domain of the cellular prion protein and changes its cellular location. Antimicrob. Agents Chemother. 62, e01441-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raymond G. J., Zhao H. T., Race B., Raymond L. D., Williams K., Swayze E. E., Graffam S., le J., Caron T., Stathopoulos J., O’Keefe R., Lubke L. L., Reidenbach A. G., Kraus A., Schreiber S. L., Mazur C., Cabin D. E., Carroll J. B., Minikel E. V., Kordasiewicz H., Caughey B., Vallabh S. M., Antisense oligonucleotides extend survival of prion-infected mice. JCI Insight 5, e131175 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jarosz-Griffiths H. H., Corbett N. J., Rowland H. A., Fisher K., Jones A. C., Baron J., Howell G. J., Cowley S. A., Chintawar S., Cader M. Z., Kellett K. A. B., Hooper N. M., Proteolytic shedding of the prion protein via activation of metallopeptidase ADAM10 reduces cellular binding and toxicity of amyloid-β oligomers. J. Biol. Chem. 294, 7085–7097 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Linsenmeier L., Mohammadi B., Wetzel S., Puig B., Jackson W. S., Hartmann A., Uchiyama K., Sakaguchi S., Endres K., Tatzelt J., Saftig P., Glatzel M., Altmeppen H. C., Structural and mechanistic aspects influencing the ADAM10-mediated shedding of the prion protein. Mol. Neurodegener. 13, 18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borchelt D. R., Rogers M., Stahl N., Telling G., Prusiner S. B., Release of the cellular prion protein from cultured cells after loss of its glycoinositol phospholipid anchor. Glycobiology 3, 319–329 (1993). [DOI] [PubMed] [Google Scholar]
- 28.Taylor D. R., Parkin E. T., Cocklin S. L., Ault J. R., Ashcroft A. E., Turner A. J., Hooper N. M., Role of ADAMs in the ectodomain shedding and conformational conversion of the prion protein. J. Biol. Chem. 284, 22590–22600 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altmeppen H. C., Prox J., Puig B., Kluth M. A., Bernreuther C., Thurm D., Jorissen E., Petrowitz B., Bartsch U., de Strooper B., Saftig P., Glatzel M., Lack of a-disintegrin-and-metalloproteinase ADAM10 leads to intracellular accumulation and loss of shedding of the cellular prion protein in vivo. Mol. Neurodegener. 6, 36 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lammich S., Kojro E., Postina R., Gilbert S., Pfeiffer R., Jasionowski M., Haass C., Fahrenholz F., Constitutive and regulated alpha-secretase cleavage of Alzheimer’s amyloid precursor protein by a disintegrin metalloprotease. Proc. Natl. Acad. Sci. U.S.A. 96, 3922–3927 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hartmann D., de Strooper B., Serneels L., Craessaerts K., Herreman A., Annaert W., Umans L., Lübke T., Lena Illert A., von Figura K., Saftig P., The disintegrin/metalloprotease ADAM 10 is essential for Notch signalling but not for alpha-secretase activity in fibroblasts. Hum. Mol. Genet. 11, 2615–2624 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Endres K., Fahrenholz F., Lotz J., Hiemke C., Teipel S., Lieb K., Tuscher O., Fellgiebel A., Increased CSF APPs-α levels in patients with Alzheimer disease treated with acitretin. Neurology 83, 1930–1935 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Endres K., Mitteregger G., Kojro E., Kretzschmar H., Fahrenholz F., Influence of ADAM10 on prion protein processing and scrapie infectiosity in vivo. Neurobiol. Dis. 36, 233–241 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Altmeppen H. C., Prox J., Krasemann S., Puig B., Kruszewski K., Dohler F., Bernreuther C., Hoxha A., Linsenmeier L., Sikorska B., Liberski P. P., Bartsch U., Saftig P., Glatzel M., The sheddase ADAM10 is a potent modulator of prion disease. eLife 4, e04260 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saftig P., Lichtenthaler S. F., The alpha secretase ADAM10: A metalloprotease with multiple functions in the brain. Prog. Neurobiol. 135, 1–20 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Shi M., Dennis K., Peschon J. J., Chandrasekaran R., Mikecz K., Antibody-induced shedding of CD44 from adherent cells is linked to the assembly of the cytoskeleton. J. Immunol. 167, 123–131 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Schelter F., Kobuch J., Moss M. L., Becherer J. D., Comoglio P. M., Boccaccio C., Krüger A., A disintegrin and metalloproteinase-10 (ADAM-10) mediates DN30 antibody-induced shedding of the met surface receptor. J. Biol. Chem. 285, 26335–26340 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hartmann M., Parra L. M., Ruschel A., Lindner C., Morrison H., Herrlich A., Herrlich P., Inside-out regulation of ectodomain cleavage of cluster-of-differentiation-44 (CD44) and of neuregulin-1 requires substrate dimerization. J. Biol. Chem. 290, 17041–17054 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beland M., Bedard M., Tremblay G., Lavigne P., Roucou X., Aβ induces its own prion protein N-terminal fragment (PrPN1)-mediated neutralization in amorphous aggregates. Neurobiol. Aging 35, 1537–1548 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Polymenidou M., Moos R., Scott M., Sigurdson C., Shi Y. Z., Yajima B., Hafner-Bratkovič I., Jerala R., Hornemann S., Wuthrich K., Bellon A., Vey M., Garen G., James M. N. G., Kav N., Aguzzi A., The POM monoclonals: A comprehensive set of antibodies to non-overlapping prion protein epitopes. PLOS ONE 3, e3872 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bardelli M., Frontzek K., Simonelli L., Hornemann S., Pedotti M., Mazzola F., Carta M., Eckhardt V., D’Antuono R., Virgilio T., González S. F., Aguzzi A., Varani L., A bispecific immunotweezer prevents soluble PrP oligomers and abolishes prion toxicity. PLOS Pathog. 14, e1007335 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Falsig J., Sonati T., Herrmann U. S., Saban D., Li B., Arroyo K., Ballmer B., Liberski P. P., Aguzzi A., Prion pathogenesis is faithfully reproduced in cerebellar organotypic slice cultures. PLOS Pathog. 8, e1002985 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beland M., Motard J., Barbarin A., Roucou X., PrP(C) homodimerization stimulates the production of PrPC cleaved fragments PrPN1 and PrPC1. J. Neurosci. 32, 13255–13263 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Engelke A. D., Gonsberg A., Thapa S., Jung S., Ulbrich S., Seidel R., Basu S., Multhaup G., Baier M., Engelhard M., Schätzl H. M., Winklhofer K. F., Tatzelt J., Dimerization of the cellular prion protein inhibits propagation of scrapie prions. J. Biol. Chem. 293, 8020–8031 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carter L., Kim S. J., Schneidman-Duhovny D., Stöhr J., Poncet-Montange G., Weiss T. M., Tsuruta H., Prusiner S. B., Sali A., Prion protein-antibody complexes characterized by chromatography-coupled small-angle x-ray scattering. Biophys. J. 109, 793–805 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sonati T., Reimann R. R., Falsig J., Baral P. K., O’Connor T., Hornemann S., Yaganoglu S., Li B., Herrmann U. S., Wieland B., Swayampakula M., Rahman M. H., Das D., Kav N., Riek R., Liberski P. P., James M. N. G., Aguzzi A., The toxicity of antiprion antibodies is mediated by the flexible tail of the prion protein. Nature 501, 102–106 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Le Brun A. P., Haigh C. L., Drew S. C., James M., Boland M. P., Collin S. J., Neutron reflectometry studies define prion protein N-terminal peptide membrane binding. Biophys. J. 107, 2313–2324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Herrmann U. S., Sonati T., Falsig J., Reimann R. R., Dametto P., O’Connor T., Li B., Lau A., Hornemann S., Sorce S., Wagner U., Sanoudou D., Aguzzi A., Prion infections and anti-PrP antibodies trigger converging neurotoxic pathways. PLOS Pathog. 11, e1004662 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu B., McDonald A. J., Markham K., Rich C. B., McHugh K. P., Tatzelt J., Colby D. W., Millhauser G. L., Harris D. A., The N-terminus of the prion protein is a toxic effector regulated by the C-terminus. eLife 6, e23473 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shyng S. L., Huber M. T., Harris D. A., A prion protein cycles between the cell surface and an endocytic compartment in cultured neuroblastoma cells. J. Biol. Chem. 268, 15922–15928 (1993). [PubMed] [Google Scholar]
- 51.Barreca M. L., Iraci N., Biggi S., Cecchetti V., Biasini E., Pharmacological agents targeting the cellular prion protein. Pathogens 7, 27 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kamatari Y. O., Hayano Y., Yamaguchi K., Hosokawa-Muto J., Kuwata K., Characterizing antiprion compounds based on their binding properties to prion proteins: Implications as medical chaperones. Protein Sci. 22, 22–34 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baral P. K., Swayampakula M., Rout M. K., Kav N. N. V., Spyracopoulos L., Aguzzi A., James M. N. G., Structural basis of prion inhibition by phenothiazine compounds. Structure 22, 291–303 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Stincardini C., Massignan T., Biggi S., Elezgarai S. R., Sangiovanni V., Vanni I., Pancher M., Adami V., Moreno J., Stravalaci M., Maietta G., Gobbi M., Negro A., Requena J. R., Castilla J., Nonno R., Biasini E., An antipsychotic drug exerts anti-prion effects by altering the localization of the cellular prion protein. PLOS ONE 12, e0182589 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Priola S. A., Raines A., Caughey W. S., Porphyrin and phthalocyanine antiscrapie compounds. Science 287, 1503–1506 (2000). [DOI] [PubMed] [Google Scholar]
- 56.Massignan T., Cimini S., Stincardini C., Cerovic M., Vanni I., Elezgarai S. R., Moreno J., Stravalaci M., Negro A., Sangiovanni V., Restelli E., Riccardi G., Gobbi M., Castilla J., Borsello T., Nonno R., Biasini E., A cationic tetrapyrrole inhibits toxic activities of the cellular prion protein. Sci. Rep. 6, 23180 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Korth C., May B. C. H., Cohen F. E., Prusiner S. B., Acridine and phenothiazine derivatives as pharmacotherapeutics for prion disease. Proc. Natl. Acad. Sci. U.S.A. 98, 9836–9841 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vogtherr M., Grimme S., Elshorst B., Jacobs D. M., Fiebig K., Griesinger C., Zahn R., Antimalarial drug quinacrine binds to C-terminal helix of cellular prion protein. J. Med. Chem. 46, 3563–3564 (2003). [DOI] [PubMed] [Google Scholar]
- 59.Biggi S., Pancher M., Stincardini C., Luotti S., Massignan T., Dalle Vedove A., Astolfi A., Gatto P., Lolli G., Barreca M. L., Bonetto V., Adami V., Biasini E., Identification of compounds inhibiting prion replication and toxicity by removing PrPC from the cell surface. J. Neurochem. 152, 136–150 (2020). [DOI] [PubMed] [Google Scholar]
- 60.Vallabh S. M., Nobuhara C. K., Llorens F., Zerr I., Parchi P., Capellari S., Kuhn E., Klickstein J., Safar J. G., Nery F. C., Swoboda K. J., Geschwind M. D., Zetterberg H., Arnold S. E., Minikel E. V., Schreiber S. L., Prion protein quantification in human cerebrospinal fluid as a tool for prion disease drug development. Proc. Natl. Acad. Sci. U.S.A. 116, 7793–7798 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Minikel E. V., Zhao H. T., le J., O’Moore J., Pitstick R., Graffam S., Carlson G. A., Kavanaugh M. P., Kriz J., Kim J. B., Ma J., Wille H., Aiken J., McKenzie D., Doh-ura K., Beck M., O’Keefe R., Stathopoulos J., Caron T., Schreiber S. L., Carroll J. B., Kordasiewicz H. B., Cabin D. E., Vallabh S. M., Prion protein lowering is a disease-modifying therapy across prion disease stages, strains and endpoints. Nucleic Acids Res. 48, 10615–10631 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Triller A., Choquet D., New concepts in synaptic biology derived from single-molecule imaging. Neuron 59, 359–374 (2008). [DOI] [PubMed] [Google Scholar]
- 63.Aguilar-Calvo P., Sevillano A. M., Bapat J., Soldau K., Sandoval D. R., Altmeppen H. C., Linsenmeier L., Pizzo D. P., Geschwind M. D., Sanchez H., Appleby B. S., Cohen M. L., Safar J. G., Edland S. D., Glatzel M., Nilsson K. P. R., Esko J. D., Sigurdson C. J., Shortening heparan sulfate chains prolongs survival and reduces parenchymal plaques in prion disease caused by mobile, ADAM10-cleaved prions. Acta Neuropathol. 139, 527–546 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kanaani J., Prusiner S. B., Diacovo J., Baekkeskov S., Legname G., Recombinant prion protein induces rapid polarization and development of synapses in embryonic rat hippocampal neurons in vitro. J. Neurochem. 95, 1373–1386 (2005). [DOI] [PubMed] [Google Scholar]
- 65.Amin L., Nguyen X. T., Rolle I. G., D’Este E., Giachin G., Tran T. H., Šerbec V. Č., Cojoc D., Legname G., Characterization of prion protein function by focal neurite stimulation. J. Cell Sci. 129, 3878–3891 (2016). [DOI] [PubMed] [Google Scholar]
- 66.Megra B. W., Eugenin E. A., Berman J. W., The role of shed PrPc in the neuropathogenesis of HIV infection. J. Immunol. 199, 224–232 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martellucci S., Santacroce C., Santilli F., Piccoli L., Delle Monache S., Angelucci A., Misasi R., Sorice M., Mattei V., Cellular and molecular mechanisms mediated by recPrPC involved in the neuronal differentiation process of mesenchymal stem cells. Int. J. Mol. Sci. 20, 345 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mantuano E., Azmoon P., Banki M. A., Lam M. S., Sigurdson C. J., Gonias S. L., A soluble derivative of PrPC activates cell-signaling and regulates cell physiology through LRP1 and the NMDA receptor. J. Biol. Chem. 295, 14178–14188 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Küffer A., Lakkaraju A. K. K., Mogha A., Petersen S. C., Airich K., Doucerain C., Marpakwar R., Bakirci P., Senatore A., Monnard A., Schiavi C., Nuvolone M., Grosshans B., Hornemann S., Bassilana F., Monk K. R., Aguzzi A., The prion protein is an agonistic ligand of the G protein-coupled receptor Adgrg6. Nature 536, 464–468 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Haas L. T., Salazar S. V., Kostylev M. A., Um J. W., Kaufman A. C., Strittmatter S. M., Metabotropic glutamate receptor 5 couples cellular prion protein to intracellular signalling in Alzheimer’s disease. Brain 139, 526–546 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jarosz-Griffiths H. H., Noble E., Rushworth J. V., Hooper N. M., Amyloid-β receptors: The good, the bad and the prion protein. J. Biol. Chem. 291, 3174–3183 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hu N. W., Nicoll A. J., Zhang D., Mably A. J., O’Malley T., Purro S. A., Terry C., Collinge J., Walsh D. M., Rowan M. J., mGlu5 receptors and cellular prion protein mediate amyloid-β-facilitated synaptic long-term depression in vivo. Nat. Commun. 5, 3374 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Calella A. M., Farinelli M., Nuvolone M., Mirante O., Moos R., Falsig J., Mansuy I. M., Aguzzi A., Prion protein and Aβ-related synaptic toxicity impairment. EMBO Mol. Med. 2, 306–314 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Foley A. R., Roseman G. P., Chan K., Smart A., Finn T. S., Yang K., Lokey R. S., Millhauser G. L., Raskatov J. A., Evidence for aggregation-independent, PrPC-mediated Aβ cellular internalization. Proc. Natl. Acad. Sci. U.S.A. 117, 28625–28631 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Solforosi L., Criado J. R., McGavern D. B., Wirz S., Sánchez-Alavez M., Sugama S., DeGiorgio L. A., Volpe B. T., Wiseman E., Abalos G., Masliah E., Gilden D., Oldstone M. B., Conti B., Williamson R. A., Cross-linking cellular prion protein triggers neuronal apoptosis in vivo. Science 303, 1514–1516 (2004). [DOI] [PubMed] [Google Scholar]
- 76.Parra L. M., Hartmann M., Schubach S., Li Y., Herrlich P., Herrlich A., Distinct intracellular domain substrate modifications selectively regulate ectodomain cleavage of NRG1 or CD44. Mol. Cell. Biol. 35, 3381–3395 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Meier P., Genoud N., Prinz M., Maissen M., Rülicke T., Zurbriggen A., Raeber A. J., Aguzzi A., Soluble dimeric prion protein binds PrP(Sc) in vivo and antagonizes prion disease. Cell 113, 49–60 (2003). [DOI] [PubMed] [Google Scholar]
- 78.Chesebro B., Trifilo M., Race R., Meade-White K., Teng C., LaCasse R., Raymond L., Favara C., Baron G., Priola S., Caughey B., Masliah E., Oldstone M., Anchorless prion protein results in infectious amyloid disease without clinical scrapie. Science 308, 1435–1439 (2005). [DOI] [PubMed] [Google Scholar]
- 79.DeMarco M. L., Daggett V., Characterization of cell-surface prion protein relative to its recombinant analogue: Insights from molecular dynamics simulations of diglycosylated, membrane-bound human prion protein. J. Neurochem. 109, 60–73 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cheng C. J., Koldso H., Van der Kamp M. W., Schiott B., Daggett V., Simulations of membrane-bound diglycosylated human prion protein reveal potential protective mechanisms against misfolding. J. Neurochem. 142, 171–182 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Camacho M. V., Telling G., Kong Q., Gambetti P., Notari S., Role of prion protein glycosylation in replication of human prions by protein misfolding cyclic amplification. Lab. Invest. 99, 1741–1748 (2019). [DOI] [PubMed] [Google Scholar]
- 82.Kang H. E., Bian J., Kane S. J., Kim S., Selwyn V., Crowell J., Bartz J. C., Telling G. C., Incomplete glycosylation during prion infection unmasks a prion protein epitope that facilitates prion detection and strain discrimination. J. Biol. Chem. 295, 10420–10433 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Féraudet C., Morel N., Simon S., Volland H., Frobert Y., Créminon C., Vilette D., Lehmann S., Grassi J., Screening of 145 anti-PrP monoclonal antibodies for their capacity to inhibit PrPSc replication in infected cells. J. Biol. Chem. 280, 11247–11258 (2005). [DOI] [PubMed] [Google Scholar]
- 84.Reidenbach A. G., Mesleh M. F., Casalena D., Vallabh S. M., Dahlin J. L., Leed A. J., Chan A. I., Usanov D. L., Yehl J. B., Lemke C. T., Campbell A. J., Shah R. N., Shrestha O. K., Sacher J. R., Rangel V. L., Moroco J. A., Sathappa M., Nonato M. C., Nguyen K. T., Wright S. K., Liu D. R., Wagner F. F., Kaushik V. K., Auld D. S., Schreiber S. L., Minikel E. V., Multimodal small-molecule screening for human prion protein binders. J. Biol. Chem. 295, 13516–13531 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Falsig J., Aguzzi A., The prion organotypic slice culture assay—POSCA. Nat. Protoc. 3, 555–562 (2008). [DOI] [PubMed] [Google Scholar]
- 86.Mohammadi B., Linsenmeier L., Shafiq M., Puig B., Galliciotti G., Giudici C., Willem M., Eden T., Koch-Nolte F., Lin Y. H., Tatzelt J., Glatzel M., Altmeppen H. C., Transgenic overexpression of the disordered prion protein N1 fragment in mice does not protect against neurodegenerative diseases due to impaired ER translocation. Mol. Neurobiol. 57, 2812–2829 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.van Bommel B., Konietzny A., Kobler O., Bär J., Mikhaylova M., F-actin patches associated with glutamatergic synapses control positioning of dendritic lysosomes. EMBO J. 38, e101183 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Renner M., Lacor P. N., Velasco P. T., Xu J., Contractor A., Klein W. L., Triller A., Deleterious effects of amyloid β oligomers acting as an extracellular scaffold for mGluR5. Neuron 66, 739–754 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shrivastava A. N., Redeker V., Pieri L., Bousset L., Renner M., Madiona K., Mailhes-Hamon C., Coens A., Buée L., Hantraye P., Triller A., Melki R., Clustering of Tau fibrils impairs the synaptic composition of α3-Na+/K+-ATPase and AMPA receptors. EMBO J. 38, e99871 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Saxton M. J., Jacobson K., Single-particle tracking: Applications to membrane dynamics. Annu. Rev. Biophys. Biomol. Struct. 26, 373–399 (1997). [DOI] [PubMed] [Google Scholar]
- 91.Studier F. W., Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 (2005). [DOI] [PubMed] [Google Scholar]
- 92.Blanchet C. E., Spilotros A., Schwemmer F., Graewert M. A., Kikhney A., Jeffries C. M., Franke D., Mark D., Zengerle R., Cipriani F., Fiedler S., Roessle M., Svergun D. I., Versatile sample environments and automation for biological solution x-ray scattering experiments at the P12 beamline (PETRA III, DESY). J. Appl. Cryst. 48, 431–443 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Petoukhov M. V., Franke D., Shkumatov A. V., Tria G., Kikhney A. G., Gajda M., Gorba C., Mertens H. D. T., Konarev P. V., Svergun D. I., New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Cryst. 45, 342–350 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Franke D., Petoukhov M. V., Konarev P. V., Panjkovich A., Tuukkanen A., Mertens H. D. T., Kikhney A. G., Hajizadeh N. R., Franklin J. M., Jeffries C. M., Svergun D. I., ATSAS 2.8: A comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J. Appl. Cryst. 50, 1212–1225 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Panjkovich A., Svergun D. I., SASpy: A PyMOL plugin for manipulation and refinement of hybrid models against small angle x-ray scattering data. Bioinformatics 32, 2062–2064 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 97.Horcas I., Fernández R., Gómez-Rodríguez J. M., Colchero J., Gómez-Herrero J., Baro A. M., WSXM: A software for scanning probe microscopy and a tool for nanotechnology. Rev. Sci. Instrum. 78, 013705 (2007). [DOI] [PubMed] [Google Scholar]
- 98.Slot J. W., Geuze H. J., Cryosectioning and immunolabeling. Nat. Protoc. 2, 2480–2491 (2007). [DOI] [PubMed] [Google Scholar]
- 99.Büeler H., Fischer M., Lang Y., Bluethmann H., Lipp H.-P., De Armond S. J., Prusiner S. B., Aguet M., Weissmann C., Normal development and behaviour of mice lacking the neuronal cell-surface PrP protein. Nature 356, 577–582 (1992). [DOI] [PubMed] [Google Scholar]
- 100.Fischer M., Rülicke T., Raeber A., Sailer A., Moser M., Oesch B., Brandner S., Aguzzi A., Weissmann C., Prion protein (PrP) with amino-proximal deletions restoring susceptibility of PrP knockout mice to scrapie. EMBO J. 15, 1255–1264 (1996). [PMC free article] [PubMed] [Google Scholar]
- 101.Marella M., Lehmann S., Grassi J., Chabry J., Filipin prevents pathological prion protein accumulation by reducing endocytosis and inducing cellular PrP release. J. Biol. Chem. 277, 25457–25464 (2002). [DOI] [PubMed] [Google Scholar]
- 102.Kim J. I., Surewicz K., Gambetti P., Surewicz W. K., The role of glycophosphatidylinositol anchor in the amplification of the scrapie isoform of prion protein in vitro. FEBS Lett. 583, 3671–3675 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Freir D. B., Nicoll A. J., Klyubin I., Panico S., Mc Donald J. M., Risse E., Asante E. A., Farrow M. A., Sessions R. B., Saibil H. R., Clarke A. R., Rowan M. J., Walsh D. M., Collinge J., Interaction between prion protein and toxic amyloid β assemblies can be therapeutically targeted at multiple sites. Nat. Commun. 2, 336 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Guillot-Sestier M. V., Sunyach C., Ferreira S. T., Marzolo M.-P., Bauer C., Thevenet A., Checler F., α-Secretase-derived fragment of cellular prion, N1, protects against monomeric and oligomeric amyloid β (Aβ)-associated cell death. J. Biol. Chem. 287, 5021–5032 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nieznanski K., Choi J.-K., Chen S., Surewicz K., Surewicz W. K., Soluble prion protein inhibits amyloid-β (Aβ) fibrillization and toxicity. J. Biol. Chem. 287, 33104–33108 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fluharty B. R., Biasini E., Stravalaci M., Sclip A., Diomede L., Balducci C., la Vitola P., Messa M., Colombo L., Forloni G., Borsello T., Gobbi M., Harris D. A., An N-terminal fragment of the prion protein binds to amyloid-β oligomers and inhibits their neurotoxicity in vivo. J. Biol. Chem. 288, 7857–7866 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yuan J., Zhan Y. A., Abskharon R., Xiao X., Martinez M. C., Zhou X., Kneale G., Mikol J., Lehmann S., Surewicz W. K., Castilla J., Steyaert J., Zhang S., Kong Q., Petersen R. B., Wohlkonig A., Zou W. Q., Recombinant human prion protein inhibits prion propagation in vitro. Sci. Rep. 3, 2911 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Scott-McKean J. J., Surewicz K., Choi J. K., Ruffin V. A., Salameh A. I., Nieznanski K., Costa A. C. S., Surewicz W. K., Soluble prion protein and its N-terminal fragment prevent impairment of synaptic plasticity by Aβ oligomers: Implications for novel therapeutic strategy in Alzheimer’s disease. Neurobiol. Dis. 91, 124–131 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bove-Fenderson E., Urano R., Straub J. E., Harris D. A., Cellular prion protein targets amyloid-β fibril ends via its C-terminal domain to prevent elongation. J. Biol. Chem. 292, 16858–16871 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Corda E., du X., Shim S. Y., Klein A. N., Siltberg-Liberles J., Gilch S., Interaction of peptide aptamers with prion protein central domain promotes α-cleavage of PrPC. Mol. Neurobiol. 55, 7758–7774 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Heppner F. L., Musahl C., Arrighi I., Klein M. A., Rülicke T., Oesch B., Zinkernagel R. M., Kalinke U., Aguzzi A., Prevention of scrapie pathogenesis by transgenic expression of anti-prion protein antibodies. Science 294, 178–182 (2001). [DOI] [PubMed] [Google Scholar]
- 112.Enari M., Flechsig E., Weissmann C., Scrapie prion protein accumulation by scrapie-infected neuroblastoma cells abrogated by exposure to a prion protein antibody. Proc. Natl. Acad. Sci. U.S.A. 98, 9295–9299 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Peretz D., Williamson R. A., Kaneko K., Vergara J., Leclerc E., Schmitt-Ulms G., Mehlhorn I. R., Legname G., Wormald M. R., Rudd P. M., Dwek R. A., Burton D. R., Prusiner S. B., Antibodies inhibit prion propagation and clear cell cultures of prion infectivity. Nature 412, 739–743 (2001). [DOI] [PubMed] [Google Scholar]
- 114.White A. R., Enever P., Tayebi M., Mushens R., Linehan J., Brandner S., Anstee D., Collinge J., Hawke S., Monoclonal antibodies inhibit prion replication and delay the development of prion disease. Nature 422, 80–83 (2003). [DOI] [PubMed] [Google Scholar]
- 115.Gilch S., Wopfner F., Renner-Müller I., Kremmer E., Bauer C., Wolf E., Brem G., Groschup M. H., Schätzl H. M., Polyclonal anti-PrP auto-antibodies induced with dimeric PrP interfere efficiently with PrPSc propagation in prion-infected cells. J. Biol. Chem. 278, 18524–18531 (2003). [DOI] [PubMed] [Google Scholar]
- 116.Perrier V., Solassol J., Crozet C., Frobert Y., Mourton-Gilles C., Grassi J., Lehmann S., Anti-PrP antibodies block PrPSc replication in prion-infected cell cultures by accelerating PrPC degradation. J. Neurochem. 89, 454–463 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Donofrio G., Heppner F. L., Polymenidou M., Musahl C., Aguzzi A., Paracrine inhibition of prion propagation by anti-PrP single-chain Fv miniantibodies. J. Virol. 79, 8330–8338 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pankiewicz J., Prelli F., Sy M. S., Kascsak R. J., Kascsak R. B., Spinner D. S., Carp R. I., Meeker H. C., Sadowski M., Wisniewski T., Clearance and prevention of prion infection in cell culture by anti-PrP antibodies. Eur. J. Neurosci. 23, 2635–2647 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Song C. H., Furuoka H., Kim C. L., Ogino M., Suzuki A., Hasebe R., Horiuchi M., Effect of intraventricular infusion of anti-prion protein monoclonal antibodies on disease progression in prion-infected mice. J. Gen. Virol. 89, 1533–1544 (2008). [DOI] [PubMed] [Google Scholar]
- 120.Sadowski M. J., Pankiewicz J., Prelli F., Scholtzova H., Spinner D. S., Kascsak R. B., Kascsak R. J., Wisniewski T., Anti-PrP Mab 6D11 suppresses PrP(Sc) replication in prion infected myeloid precursor line FDC-P1/22L and in the lymphoreticular system in vivo. Neurobiol. Dis. 34, 267–278 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Müller-Schiffmann A., Petsch B., Leliveld S. R., Muyrers J., Salwierz A., Mangels C., Schwarzinger S., Riesner D., Stitz L., Korth C., Complementarity determining regions of an anti-prion protein scFv fragment orchestrate conformation specificity and antiprion activity. Mol. Immunol. 46, 532–540 (2009). [DOI] [PubMed] [Google Scholar]
- 122.Chung E., Ji Y., Sun Y., Kascsak R. J., Kascsak R. B., Mehta P. D., Strittmatter S. M., Wisniewski T., Anti-PrPC monoclonal antibody infusion as a novel treatment for cognitive deficits in an Alzheimer’s disease model mouse. BMC Neurosci. 11, 130 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Barry A. E., Klyubin I., Mc Donald J. M., Mably A. J., Farrell M. A., Scott M., Walsh D. M., Rowan M. J., Alzheimer’s disease brain-derived amyloid-β-mediated inhibition of LTP in vivo is prevented by immunotargeting cellular prion protein. J. Neurosci. 31, 7259–7263 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ohsawa N., Song C. H., Suzuki A., Furuoka H., Hasebe R., Horiuchi M., Therapeutic effect of peripheral administration of an anti-prion protein antibody on mice infected with prions. Microbiol. Immunol. 57, 288–297 (2013). [DOI] [PubMed] [Google Scholar]
- 125.Klyubin I., Nicoll A. J., Khalili-Shirazi A., Farmer M., Canning S., Mably A., Linehan J., Brown A., Wakeling M., Brandner S., Walsh D. M., Rowan M. J., Collinge J., Peripheral administration of a humanized anti-PrP antibody blocks Alzheimer’s disease Aβ synaptotoxicity. J. Neurosci. 34, 6140–6145 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pankiewicz J. E., Sanchez S., Kirshenbaum K., Kascsak R. B., Kascsak R. J., Sadowski M. J., Anti-prion protein antibody 6D11 restores cellular proteostasis of prion protein through disrupting recycling propagation of PrPSc and targeting PrPSc for lysosomal degradation. Mol. Neurobiol. 56, 2073–2091 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cox T. O., Gunther E. C., Brody A. H., Chiasseu M. T., Stoner A., Smith L. M., Haas L. T., Hammersley J., Rees G., Dosanjh B., Groves M., Gardener M., Dobson C., Vaughan T., Chessell I., Billinton A., Strittmatter S. M., Anti-PrPC antibody rescues cognition and synapses in transgenic Alzheimer mice. Ann. Clin. Transl. Neurol. 6, 554–574 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dyer C., British man with CJD gets experimental treatment in world first. BMJ 363, k4608 (2018). [DOI] [PubMed] [Google Scholar]
- 129.Ferrer I., Blanco R., Carmona M., Puig B., Ribera R., Rey M. J., Ribalta T., Prion protein expression in senile plaques in Alzheimer’s disease. Acta Neuropathol. 101, 49–56 (2001). [DOI] [PubMed] [Google Scholar]
- 130.Schwarze-Eicker K., Keyvani K., Görtz N., Westaway D., Sachser N., Paulus W., Prion protein (PrPc) promotes β-amyloid plaque formation. Neurobiol. Aging 26, 1177–1182 (2005). [DOI] [PubMed] [Google Scholar]
- 131.Falker C., Hartmann A., Guett I., Dohler F., Altmeppen H., Betzel C., Schubert R., Thurm D., Wegwitz F., Joshi P., Verderio C., Krasemann S., Glatzel M., Exosomal cellular prion protein drives fibrillization of amyloid beta and counteracts amyloid beta-mediated neurotoxicity. J. Neurochem. 137, 88–100 (2016). [DOI] [PubMed] [Google Scholar]
- 132.Boon B. D. C., Bulk M., Jonker A. J., Morrema T. H. J., van den Berg E., Popovic M., Walter J., Kumar S., van der Lee S. J., Holstege H., Zhu X., van Nostrand W. E., Natté R., van der Weerd L., Bouwman F. H., van de Berg W. D. J., Rozemuller A. J. M., Hoozemans J. J. M., The coarse-grained plaque: A divergent Aβ plaque-type in early-onset Alzheimer’s disease. Acta Neuropathol. 140, 811–830 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Takahashi R. H., Yokotsuka M., Tobiume M., Sato Y., Hasegawa H., Nagao T., Gouras G. K., Accumulation of cellular prion protein within β-amyloid oligomer plaques in aged human brains. Brain Pathol. 31, e12941 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Harris L. J., Larson S. B., Hasel K. W., McPherson A., Refined structure of an intact IgG2a monoclonal antibody. Biochemistry 36, 1581–1597 (1997). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S12
Tables S1 and S2
Legend for movie S1
References
Movie S1