Abstract
Background and purpose
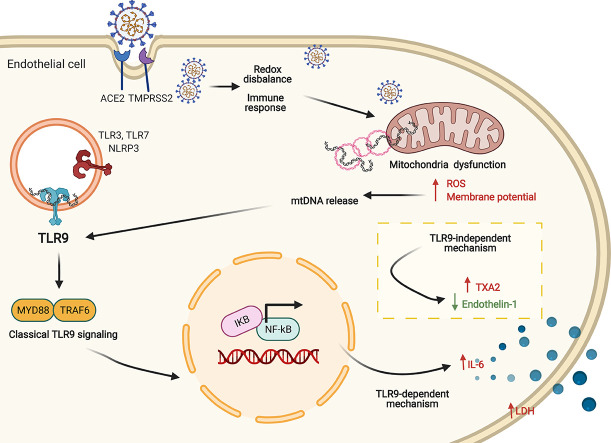
Mitochondria play a central role in the host response to viral infection and immunity, being key to antiviral signaling and exacerbating inflammatory processes. Mitochondria and Toll-like receptor (TLR) have been suggested as potential targets in SARS-CoV-2 infection. However, the involvement of TLR9 in SARS-Cov-2-induced endothelial dysfunction and potential contribution to cardiovascular complications in COVID-19 have not been demonstrated. This study determined whether infection of endothelial cells by SARS-CoV-2 affects mitochondrial function and induces mitochondrial DNA (mtDNA) release. We also questioned whether TLR9 signaling mediates the inflammatory responses induced by SARS-CoV-2 in endothelial cells.
Experimental approach
Human umbilical vein endothelial cells (HUVECs) were infected by SARS-CoV-2 and immunofluorescence was used to confirm the infection. Mitochondrial function was analyzed by specific probes and mtDNA levels by real-time polymerase chain reaction (RT-PCR). Inflammatory markers were measured by ELISA, protein expression by western blot, intracellular calcium (Ca2+) by FLUOR-4, and vascular reactivity with a myography.
Key results
SARS-CoV-2 infected HUVECs, which express ACE2 and TMPRSS2 proteins, and promoted mitochondrial dysfunction, i.e. it increased mitochondria-derived superoxide anion, mitochondrial membrane potential, and mtDNA release, leading to activation of TLR9 and NF-kB, and release of cytokines. SARS-CoV-2 also decreased nitric oxide synthase (eNOS) expression and inhibited Ca2+ responses in endothelial cells. TLR9 blockade reduced SARS-CoV-2-induced IL-6 release and prevented decreased eNOS expression. mtDNA increased vascular reactivity to endothelin-1 (ET-1) in arteries from wild type, but not TLR9 knockout mice. These events were recapitulated in serum samples from COVID-19 patients, that exhibited increased levels of mtDNA compared to sex- and age-matched healthy subjects and patients with comorbidities.
Conclusion and applications
SARS-CoV-2 infection impairs mitochondrial function and activates TLR9 signaling in endothelial cells. TLR9 triggers inflammatory responses that lead to endothelial cell dysfunction, potentially contributing to the severity of symptoms in COVID-19. Targeting mitochondrial metabolic pathways may help to define novel therapeutic strategies for COVID-19.
Keywords: SARS-CoV-2, Mitochondria, Endothelial dysfunction, Toll like receptor 9
Graphical abstract
1. Introduction
The recently human-pathogenic β-coronavirus SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) is responsible for the global pandemic coronavirus disease 2019 (COVID-19) [1] and understanding the wide range of cardiovascular manifestations due to COVID-19 is crucial particularly because patients with cardiovascular risk increased mortality.
Among the different clinical forms, patients with severe and critical COVID-19 exhibit exacerbated pulmonary inflammation, characterized by cytokine storm, predominant influx of myeloid cells, influx of lymphocytes, significant capillary congestion, and microthrombi of alveolar capillaries [2]. SARS-CoV-2 infects and induces the death of alveolar epithelial cells [3], induces the formation of hyaline membrane [4], exudative and proliferative diffuse alveolar damage [2] and promote Acute Respiratory Distress Syndrome (ARDS).
Infection of extrapulmonary cells by SARS-CoV-2, also contributes to the strength of the inflammatory response [5] . SARS-CoV-2 RNA was also detected in heart, kidneys, liver, spleen, lymph nodes, muscle, and intestine of patients with COVID-19 [6].
Extra-respiratory clinical manifestations, with cardiac, renal, neurological, and hematological symptoms as well as multiple organ failure, have been consistently reported in COVID-19 patients [7]. In the cardiovascular system, SARS-CoV-2 infection promotes venous thromboembolic events [8], cardiac injury and dysrhythmias, angiogenesis, and glycocalyx shedding [9,10]. Of importance, microvascular thrombi formation, with complete stagnated capillaries and thromboembolic occlusion, is not restricted to the lungs and has been reported in the systemic circulation [11]. However, the infection of SARS-CoV-2 in endothelial cells of vascular system not well characterized.
Mitochondria play a central role in host immune responses upon viral infection. Mitochondria are key to antiviral signaling [mitochondria serves as a signaling platform for mitochondrial anti-viral signal (MAVS)], but also contribute to exacerbated inflammatory processes [12]. Mitochondrial dysfunction increases reactive oxygen species (ROS) production, alters calcium (Ca2+) homeostasis, deregulates lipid metabolism, and leads to release of mitochondrial DNA (mtDNA) [13,14]. Circulating mtDNA, whose primary source is cell injury and necrosis, is also found in patients affected by multiple systemic conditions, as observed in ARDS [15] and arterial hypertension [16,17]. ROS and Ca2+ are important to vascular homeostasis, and redox disbalance, abnormal Ca2+ handling, and chronic inflammation are all key hallmarks in cardiovascular pathological processes.
mtDNA, released by damaged cells and tissues, is recognized by endosomal/extracellular TLR9 triggering inflammation. TLR9 is activated by cytosine-phosphate-guanine (CpG)-rich unmethylated DNA motifs, which are primarily found in bacterial, mitochondrial, and fetal DNA but rarely in vertebrate adult DNA (genomic DNA) [18] . TLR9 responds to herpes DNA virus [19] as well as to dengue RNA virus [20]. However, whether SARS-CoV-2 infection activates TLR9 is unknown. TLR9, expressed in endothelial cells (ECs) and vascular smooth muscle cells (VSMCs), contributes to endothelial and vascular dysfunction. Prolonged activation of TLR9 causes vascular hypertrophy, fibrosis, and abnormal reactivity by mechanisms that involve ROS generation, decreased NO bioavailability, augmented expression of pro-inflammatory cytokines, and activation of autophagic and pro-contractile pathways [16,17,21].
Considering that SARS-CoV-2 infection produces signs of mitochondrial dysfunction [22], e.g. increased ROS generation in the lungs [23] and low levels of Ca2+ in the blood [24], in the present study we tested the hypothesis that SARS-CoV-2 infection of endothelial cells promotes mitochondrial dysfunction, increases mtDNA levels and activates TLR9 signaling, leading to endothelial dysfunction. Therefore, the aims of this study were: 1) to determine mtDNA levels in control and SARS-CoV-2-infected endothelial cells as well as in serum of COVID-19 patients; 2) to evaluate the impact of SARS-CoV-2-infection in mitochondrial function of endothelial cells, and 3) to investigate the molecular mechanisms involved in SARS-CoV-2-induced endothelial cell dysfunction.
2. Material and methods
2.1. Human samples
The Brazilian National Committee for Ethics in Research (CONEP) approved all procedures performed in the study (CONEP CAAE: 30248420.9.0000.5440 and 30,816,620.0.0000.5440) and the investigation conforms with the principles outlined in the Declaration of Helsinki. In addition, written informed consent was obtained from all recruited patients before entering study. We included male and female patients older than 18 years-old, with confirmed moderate or severe forms of COVID-19 (n = 19) (Table 1 ). The diagnosis was based on results from RT-PCR in nasopharyngeal swab specimens. For clinical classification of COVID-19 severity, we used the World Health Organization Clinical Progression Scale [25]. Patients that required Intensive Care Unit admission and mechanical ventilation or oxygen therapy by non-invasive ventilation or high-flow nasal cannula were classified as severe disease. Moderate COVID-19 was considered for patients who needed hospitalization and oxygen therapy by nasal prongs, with an oxygen saturation in room air bellow 90%, and without clinical evidence respiratory failure. Health age- and sex-matched individuals (n = 7) and age- and sex- matched patients with comorbidities usually observed in COVID-19 (n = 8) were also included in the study for comparison.
Table 1.
Baseline characteristics of SARS-CoV-2 positive patients and health and comorbid age- and sex- matched individuals SARS-CoV-2 negative.
| Characteristics | SARS-CoV-2 positive patients (n = 19) |
SARS-CoV-2 negative patients (comorbidities) (n = 8) |
SARS-CoV-2 negative patients (healthy) (n = 7) |
p value |
|---|---|---|---|---|
| Demographic | ||||
| Age (years), median IQR | 57 (50–70) | 58 (52–63.25) | 45 (36–70) | 0.175 |
| Male gender (%) | 50% | 37,5% | 43% | 0.828 |
| Comorbidities | ||||
| Hypertension (%) | 65% | 100% | ||
| Heart disease | 50% | 12,5% | ||
| Diabetes (%) | 45% | 50% | ||
| Obesity (%) | 50% | 25% | ||
| BMI, median, IQR | 28 (25–35) | 27.9 (26.9–31.7) | 0.975 | |
| Clinical characteristics of COVID-19 patients | ||||
| Days of symptoms, mean, ± sd | 13 ± 5 | – | – | |
| Hospital stays (days), mean, ± sd | 18 ± 11 | – | – | |
| Mechanical ventilation (%) | 50% | – | – | |
| Acute Kidney Injury (%) | 35% | – | – | |
| Mortality rate (%) | 10% | – | – | |
| Laboratory tests | ||||
| Creatinine, mg/dL, mean (IQR) | 1.1 ± 0.9 | – | – | |
| Lymphocyte; mm3, median (IQR) | 1100 (600–1650) | – | – | |
| Platelets; x 103/l (IQR) | 246 (217–338) | – | – | |
| RCP; mg/dL, median (IQR) | 13 (9–20) | – | – | |
| D dimer; mg/dl, median (IQR) | 1.19 (0.93–1.98) | – | – | |
| Lactate; mg/dL, median (IQR) | 2.2 (1.7–2.6) | – | – | |
| Lactate dehydrogenase, U/L (IQR) | 361 (302–434) | – | – | |
| Ferritin, ng/mL (IQR) | 608 (345–1076) | – | – | |
| Fibrinogen (mg/dLL L) | 686 (619–815 | – | – | |
| Severity | ||||
| PaO2/FiO2 ratio, median (IQR) | 205 (139–315) | – | – | |
| SOFA score | 2 (1–3) | – | – | |
|
Simplified acute physiology score 3 SAPS-3 |
47 (38–50) |
- - |
- - |
IQR interquartile range, sd standard deviation, RCP reactive C protein, SOFA sequential organ failure assessment,
All male and female healthy individuals and patients with comorbidities included were submitted to IgG and IgM antibodies test. Blood samples from COVID-19 patients were collected between 1 and 5 days after hospital admission to the State Hospital of Serrana, a unit of the Ribeirao Preto Medical School Health Complex. Blood samples of all subjects were collected in tubes containing ethylenediaminetetraacetic acid (EDTA) and in tubes without anticoagulant. The blood was centrifuged at 3500 ×g for 15 min at 18 °C, obtaining plasma and serum, respectively. Samples were stored at −80 °C for later analysis. Table 1 summarizes age, sex, clinical information, biochemical parameters, and therapeutic treatments for the subjects.
2.2. Virus
SARS-CoV-2 stock was obtained by serial propagation of the strain Brazil/SPBR-02/2020, starting from the passage 1 obtained in Vero-E6 from a COVID-19 patient in São Paulo. This sample was kindly provided by Professor Edison Luiz Durigon (Institute of Biological Science, University of São Paulo).
2.3. IgM-IgG combined antibody test
Analysis of anti-SARS-CoV-2 IgG and IgM antibodies was performed in serum from COVID-19 negative subjects (control) and COVID-19 patients using the rapid test Asan Easy Test® COVID-19 IgG/IgM from Asan Pharmaceutical (Gyeonggi-do, Korea). This test is based in an immunochromatographic assay for the rapid qualitative detection of IgG and IgM antibodies through combination of particles coated with SARS-CoV-2 antigen. The National Agency of Sanitary Vigilance (ANVISA, Brazil) licensed the Asan Easy Test COVID-19 IgG/IgM in May of 2020 (https://consultas.anvisa.gov.br/#/saude/q/?numeroRegistro=80198110005). The control group comprises subjects whose samples were used in a previous study [10].
2.4. Immunofluorescence
Considering that microvascular thrombi formation, with complete stagnated capillaries and thromboembolic occlusion, is not restricted to the lungs and has been reported in the systemic circulation [11], protocols with endothelial cells were performed in human umbilical vein endothelial cells (HUVECs, ATCC® PCS-100-013 ™; sex unknown) and Vero-E6 (lineage of African green monkey kidney cells - ATCC® RL1586 ™; sex unknown). HUVECs and Vero-E6 were plated on glass coverslips pretreated with 0.4% gelatin. When cells reached confluence, they were infected by SARS-CoV-2. Cells were fixed with 4% v/v paraformaldehyde and permeabilized with 0.1% v/v Triton in Hanks' Balanced Salt Solution (HBSS). Cells were incubated with an anti-SARS-CoV-2 rabbit polyclonal antibody (CABT-CS024, Creative Diagnostics) (1:1000 in permeabilizing/blocking solution of PBS (phosphate buffer saline) with 0.01% v/v Triton and 1% w/v BSA (bovine serum albumin) overnight at 4 °C. Slides were washed three times with PBS and incubated with the SuperBlock® blocking buffer (Pierce, Rockford, USA) and 5% v/v goat serum for 20 min (min) at room temperature. Slides were incubated with FITC-labelled goat anti-rabbit IgG antibody Apl24F (Millipore, Temecula, CA, USA) diluted 1:700 for 30 min, washed three times with PBS, and then incubated for 5 min at room temperature with DAPI (1 μg/mL) for nuclei staining. Slides were then mounted in mounting media (Dako) and analyzed in a fluorescence microscope (Zeiss).
2.5. Immunohistochemistry
Tissue samples obtained from autopsy of COVID-19 fatal cases were tested by immunohistochemistry using anti-SARS-CoV-2 mouse polyclonal antibody (1:100 in PBS containing 1% w/v BSA and 0.1% v/v saponin). Briefly, sections were submitted to antigen retrieval in 10 mM citrate buffer, pH 6 (Sigma, St Louis, MO, USA), with heating in a microwave oven at maximum power for 5 min, and then at a power equivalent to 10% of the maximum for 15 min. To block endogenous peroxidase activity, sections were incubated with 4% v/v H2O2 (Synth, Diadema, SP) for 30 min and washed in PBS (Gibco, Grand Island, NY, USA). Sections were then blocked for 20 min in the SuperBlock® blocking buffer (Pierce, Rockford, USA) with 5% v/v goat serum, and then incubated with the anti-SARS-CoV-2 overnight at 4 °C. Detection of the primary antibody, and signal development was achieved using a SensiTek horseradish peroxidase (HRP) anti-polyvalent (AEC) ready-to-use kit (AEN080-IFU, Scytek Laboratories, Utah, USA). Counterstaining was performed with Harris hematoxylin and slides were mounted with coverslips with aqueous mounting medium. Slides were scanned using a VS120 ScanScope (Olympus) at 400× magnification.
2.6. Real-Time PCR
SARS-CoV-2 genome quantification was done according Lu et al. 2020 [26]. Briefly, RNA was extracted by Trizol® (Invitrogen, CA, EUA), and amplification was done by one-step real-time RT-PCR using a Step-One Plus real-time PCR thermocycler (Applied Biosystems, Foster City, CA, USA). 100 ng of RNA was used for genome amplification, adding specifics primers (20 μM), probe (5 μM), and qPCRBIO Probe 1-Step Go Master Mix (PCRbiosystems, London, UK), with the following cycling parameters: 45 °C for 10 min, 95 °C for 2 min, followed by 40 cycles of 95 °C for 5 s, and 60 °C for 30s. To assess the viral load of SARS-CoV-2, a standard curve was constructed using plasmid contains the N gene insert (944 bp amplicon).
2.7. Endothelial cell culture
HUVECs were chosen considering we sought to determine effects of SARS-CoV-2 on endothelial cells of the systemic circulation and not only effects of SARS-Cov-2 restricted to the pulmonary endothelial cells. HUVECs were cultured in 6- or 96- well plates, using Dulbecco's Modified Eagle Medium (DMEM), supplemented with sodium bicarbonate, penicillin (100 U/mL), streptomycin (100 μg/mL), amphotericin B, 10% v/v FBS, and maintained in an incubator at 37 °C and 5% CO2. Confluent cells between passage 4 and 6 were used in these experiments. After confluence, cells were exposed for 24 h or 48 h to either endothelial cell culture medium (control), virus-free supernatant of Vero-E6 cells mock medium (MOCK), SARS-CoV-2 [multiplicity of infection (MOI) = 0.5, 1, 2 and 5), or ultraviolet light (UV)-inactivated SARS-CoV-2, in the presence of vehicle or ODNA 2088 (1.7 μg/mL, InvivoGen, San Diego, CA, USA), a TLR9 inhibitor. After stimulation, cells were washed with ice-cold PBS, harvested in lysis buffer, and kept frozen until further use. All procedures were performed in a biosafety level 3 (BSL3) laboratory in the Department of Biochemistry and Immunology at the Ribeirao Preto Medical School.
2.8. Cytotoxicity assay
Supernatants of non-infected/non-treated HUVECs (Control), HUVECs infected with SARS-CoV-2 and HUVECs infected with SARS-CoV-2 and treated with TLR9 inhibitor were used to measure lactate dehydrogenase (LDH), a stable cytosolic enzyme that is released upon cell lysis or damage, using the CytoTox 96® Non-Radioactive Cytotoxicity colorimetric assay, according to the manufacturer's instructions (G1780, Promega Corporation, Madison, WI, USA). The results are expressed as the percentage of LDH release in relation to the positive LDH control (100%) provided by the manufacturer.
2.9. Western Blot
Control and infected HUVECs were homogenized in RIPA lysis buffer, consisting of (in mM) 50 Tris·HCl (pH 7.4), 5 EGTA, 2 EDTA, 0.1 PMSF (phenylmethylsulphonyl fluoride), 1 pepstatin A, 1 leupeptin, and 1 aprotinin. Thirty (30) μg of proteins from HUVECs extracts were separated by electrophoresis on 8% or 10% polyacrylamide gels, transferred to nitrocellulose membranes (0.22 μm), and blocked using 3% w/v BSA in TBS (Tris-buffered saline) and Tween 20 (0.1% v/v) for 1 h. Primary antibodies were incubated overnight at 4 °C as follows: ACE2 (#abcam 105282), TMPRSS2 (#sc515727); TLR9 (#ab12121, 1:1000, lot GR153977–2), total and phosphor (at Serine536)-NF-κB (#8242S and #3033S, 1:1000, lot 9 12/2017 and lot 16 12/2017, respectively), total (#9572S, 1:1000, lot 3 03/2019 and lot 2 08/2014, respectively), VDAC channel (#4866S, 1:2000, lot 3 10/2011), Complex 1 of mitochondria (#ab110245, 1:2000, lot GR83667–4), and anti-β-actin (#4967 L, 1:5000, lot 7 03/2915). Respective secondary antibodies were incubated at room temperature for 1 h. Bands were detected using chemiluminescence reaction (Luminata Forte, WBLUF0100, Merck-Millipore, UK). The bands were detected by a chemiluminescent system (ImageQuant LAS 400, GE Life Science, Chicago, IL, USA) and quantified using the ImageJ Software (NIH Image). All western results were normalized by endogenous expression of β-actin. ACE-2 expression in HUVECs was confirmed by using protein extracts from Hep G2 cells, wildtype and submitted to ACE2 deletion by CRISPR-Cas9 (abcam #275495).
2.10. Mitochondrial superoxide generation, membrane potential and mass in endothelial cells
To assess mitochondria-specific superoxide generation, mitochondrial membrane potential and mitochondrial mass in HUVECs infected by SARS-CoV-2 and respective control cells, the fluorescent probes MitoSOX™ Red (#M36008 - 5 μM), MitoTracker® Red CMXRos (#M7512 10 μM) and MitoTracker® Green FM (#M7514–10 μM) were used according to the manufacturer's instructions. Then, cells were protected from light, incubated with the probe at 37 °C for 45 min and washed three times with warm PBS. For the MitoTracker® Red CMXRos assay, cells were fixed in paraformaldehyde 4% v/v for 15 min. DAPI (10−7 M, 15 min) or acridine orange (0.5 μg/mL, 10 min) was used for nuclear staining. Rotenone and FCCP were used as positive controls in MitoSOX™ Red and MitoTracker® Red CMXRos assays, respectively. Fluorescence was quantified in a FlexStation® fluorimeter (Molecular Devices, Sunnyvale, CA, United States) and representative images were obtained in a Zeiss fluorescence microscope after the FlexStation® analysis. Data are expressed by FlexStation® fluorimeter relative fluorescence/total fluorescence of cell campus. After this analysis, the values were corrected by % of the control.
2.11. Mitochondrial DNA expression
mtDNA expression was analyzed in supernatant media fromHUVECs incubated with culture media (control), virus-free vector MOCK medium, UV-inactivated SARS-CoV-2 and active SARS-CoV-2. mtDNA expression was also determined in serum samples from SARS-CoV-2 positive and negative patients. One mL of cell supernatant or serum were centrifuged at 12,000 g for 15 min at 4 °C, this step was taken to exclude intact mitochondria from samples. DNA-free serum was isolated using QIAamp DNA Blood Mini kit (51104, Qiagen, Hilden, Germany) according to the manufacturer's instructions. Isolated DNA was amplified and quantified by real-time polymerase chain reaction (RT-PCR) with 5 ng of DNA from each sample [22]. mtDNA was determined using primers for NADH dehydrogenase (forward 5′-ATTAGACCCTCAAGTCTCCG-3′ and reverse 5′-TTGACTGGTTGTCTAGGGTT-3′) and cytochrome B (forward 5′-TCCACTTCATCCTCCCATTC-3′ and reverse 5′- CTGCGTCGGAGTTTAATCCT-3′). To confirm the absence of bacterial contamination variation in samples, the bacterial 16S rRNA (forward 5′-CGTCAGCTCGTGTTGTGAAA-3′ and reverse 5′-GGCAGTCTCCTTGAGTTCC-3′) was also used as a negative control. Samples were normalized to those from virus-free vector MOCK group, where the values were considered 1. Results were calculated as mean ± SEM of the levels of infected subjects in comparison with their respective control group and expressed as mtDNA/total DNA.
2.12. Ca2+ influx in SARS-CoV-2-infected endothelial cells
Cytosolic-free Ca2+ was measured using the cell permeant Fluo-4 acetoxymethyl ester probe (Invitrogen, UK). HUVECs and EA. hy926 (ATCC® CRL2922) cells were cultured in black-walled, clear-bottomed 96-well polystyrene plates (Corning, NY, United States) at a density of 80,000 cells/well in DMEM with 10% FBS and incubated for 24 h at 37 °C and 5% CO2. On the following day, cells were incubated with virus-free MOCK, UV-inactivated SARS-CoVs-2 or active SARS-CoV-2 for 48 h. The medium was replaced with 50 μl of DMEM without serum plus 50 μl of the dye solution (FLUO-4 AM, probenecid, BSA and pluronic F127) and plates were incubated for 1 h at 37 °C protected from light. Then, each well was washed three times with salt-balanced Hanks' solution (37 °C) and after 15 min of equilibration, the real-time fluorescence was acquired (at 494/525 nm; excitation and emission) using the FlexStation® fluorimeter coupled to SoftMax® Pro software (Molecular Devices, Sunnyvale, CA, United States). Ca2+ mobilization was induced with adenosine 5′-triphosphate (ATP, 10−4 M), ionomycin (10−5 M) and A23187 (10−6 M). Responses were determined as the area under the curve of the peak of fluorescence.
2.13. ELISA assay
Cell supernatant and serum samples were used to evaluate circulating cytokine levels, thromboxane A2 (TXA2) and endothelin-1 (ET-1) levels by enzyme-linked immunosorbent assay (ELISA) kits, as follows: IL-6 (R&D Systems, Minneapolis, MN, USA), Thromboxane (Cayman #501020), and ET-1 (Enzo Life Science #ADI-900-020A). The assay was performed according to the manufacturer’ instructions and the results were expressed in the range of detection (pg/mL).
2.14. Vascular reactivity
The protocol was approved by the Institutional Animal Ethics Committee of the Ribeirao Preto Medical School, University of Sao Paulo (Protocol 183/2013 and 144/2020) and experimental procedures followed the Guide for the Care and Use of Laboratory Animals published by the US National Institute of Health (NIH Publication No. 85–23, revised 1996). Endothelium-intact segments (2 mm) of resistance mesenteric arteries of male C57BL/6 J wild-type (n = 3) and TLR9 knockout (TLR9KO) (n = 4) mice were carefully dissected to remove excess of fat and connective tissue in ice-cold Krebs solution (in mM): 118 NaCl, 4.7 KCl, 25 NaHCO3, 2.5 CaCl2-2H2O, 1.2 KH2PO4, 1.2 MgSO4-7H2O, 5.5 glucose and 0.01 EDTA), and gassed with 95% O2 and 5% CO2. Resistance mesenteric arteries were submitted to a normalization procedure. After a 60-min equilibration period, vessels were initially exposed to 120 mM KCl to determine the functional integrity of smooth muscle cells. Endothelial integrity was assessed by testing the relaxant effect of ACh (10−6 M) in vessels contracted with phenylephrine (~ 80% maximum response – 10−6 M). Concentration-response curves to ET-1 (10−10 to 3 × 10−7 M) were performed in mesenteric resistance arteries from wild type and TLR9KO mice. The area under curve (AUC) was calculated from each individual concentration-response curve and expressed as arbitrary units.
2.15. Statistical analysis
The results are expressed as the mean ± standard error of the mean (SEM) and interquartile interval (IQR), depending on distribution tested by Shapiro Wilk test. In the experiments where HUVECs were used, n indicates the number of independent experiments. The statistical significance was determined by unpaired Student's t-test and one-way ANOVA multiple comparations with Tukey post-hoc test, with a single pooled variance. In the vascular reactivity analysis, maximum response and pEC50 (negative logarithm of the EC50) values were determined from the concentration-response curves. The statistical analysis was performed with the Prism GraphPad 8.0 software and differences statistically significant were considered when p < 0.05.
3. Results
3.1. SARS-CoV-2 infects endothelial cells and induces mitochondrial and endothelial cell dysfunction
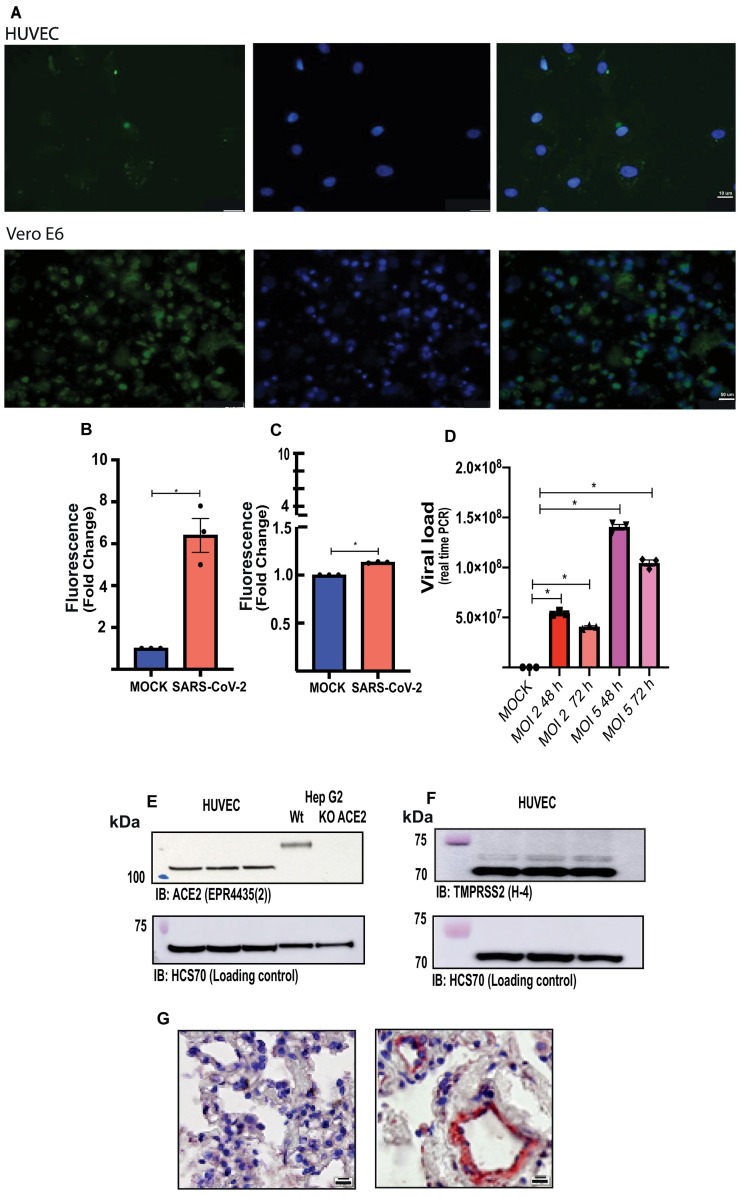
Immunofluorescence analysis, using an anti-SARS-CoV-2 rabbit polyclonal antibody and the FITC-labelled goat anti-rabbit IgG antibody Apl24F, demonstrates that HUVECs are infected with SARS-CoV-2. Fluorescence intensity in SARS-CoV-2-infected HUVECs was 13% higher compared with fluorescence in MOCK-treated cells. Vero-E6 cells infected with SARS-CoV-2, used as a positive control, presented a more significant infection than HUVECs (Fig. 1A, B and C).
Fig. 1.
Endothelial cells are infected by SARS-CoV-2. (A) Immunofluorescence in HUVECs (top panels) and Vero-E6 cells (positive control, bottom panels) infected with SARS-CoV-2 (GFP signals were detected at 48 h post-infection). (B and C) fluorescence intensity (FI) in the region of interest (ROI) was determined using the Image J software in HUVECs and Vero-E6 cells, respectively. (D) Viral load in endothelial cells 48 and 72 h post-infection (MOI 2 and 5). (E) Representative immunoblot image of ACE2 protein in HUVECs and Hep G2 cells, wildtype and submitted to ACE2 deletion by CRISPR-Cas9. (F) Representative immunoblot image of TMPRSS2 protein in HUVECs. HCS70 was used as the loading control. (G) Immunochemistry for SARS-CoV-2 in lung samples of COVID-19 positive (right) and COVID-19 negative (left) patients (Scale bars, 10 μm.). The results are expressed as the mean ± SEM. Statistical significance was determined by unpaired test t-test (n = 3) or one-way ANOVA multiple comparations using the Prism GraphPad 8.0 software. Statistically significant differences were considered when p < 0.05. MOCK, MOI, multiplicity of infection.
Although the viral load increased according to the MOI, viral replication in HUVECs was not observed either after 48 or 72 h of infection (Fig. 1D), suggesting that infection per se but not viral replication impact endothelial cell function. HUVECs express ACE2 and TMPRSS2, two doorway proteins used by SARS-CoV-2 to enter cells, as determined using Hep G2 cell extracts - control Hep G2 and Hep G2 that underwent ACE2 deletion by CRISPR-Cas9 (Fig. 1E and F, respectively).
Lastly, autopsy analysis, using an anti-SARS-CoV-2 antibody, revealed positive immunolabeling through out endothelial cells in lungs of COVID-19 patients, highlighting the endothelial tropism of the new coronavirus during disease (Fig. 1G).
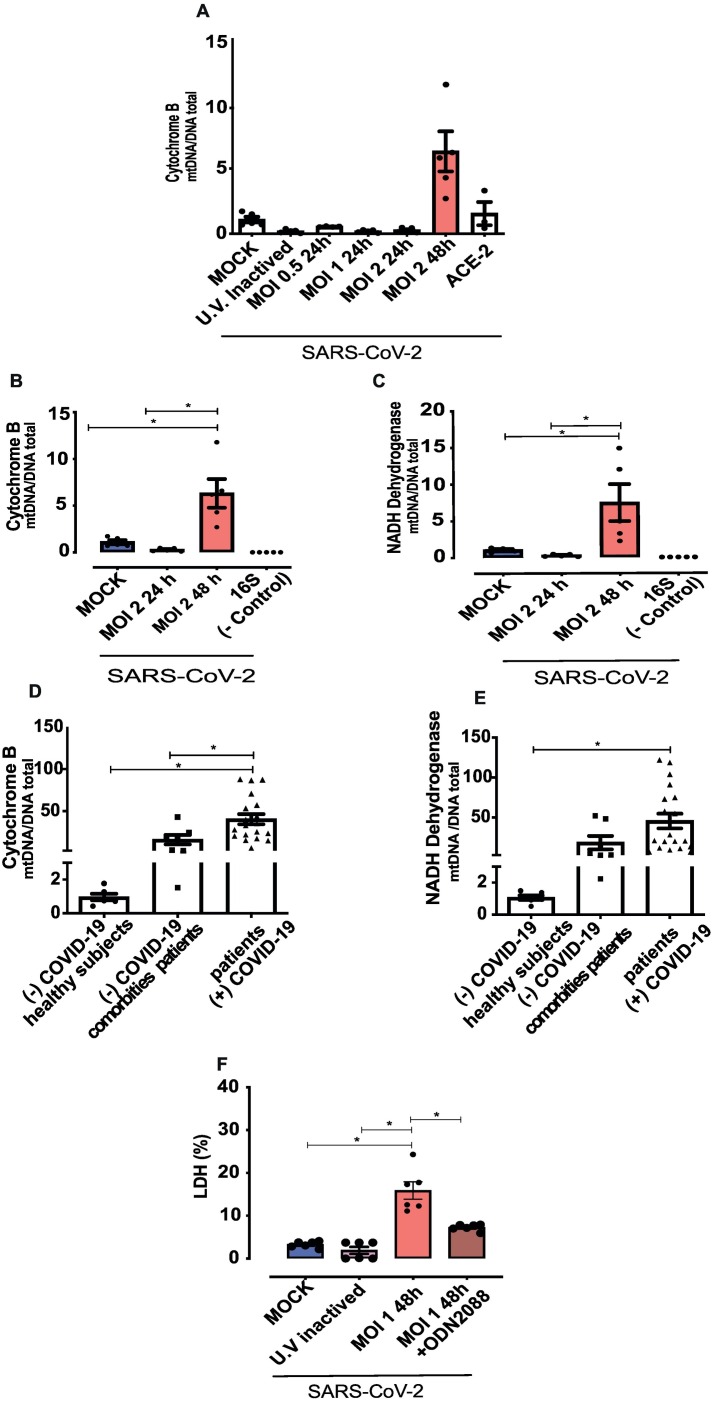
Mitochondrial DNA is a specific marker of mitochondrial dysfunction. In vitro time course analysis showed that SARS-CoV-2 (MOI 2) infection increased levels of cytochrome B after 48 h of infection (Figs. 2A and Fig. 2B). ACE2 antibody inhibited SARS-CoV-2-induced mtDNA release after 48 h of infection (Fig. 2A). In addition, SARS-CoV-2 infection increased levels of NAD dehydrogenase (Fig. 2C) compared to cells in MOCK medium and cells exposed to UV-inactivated SARS-CoV-2. Therefore, subsequent experimental protocols were performed 48 h post-infection.
Fig. 2.
SARS-CoV-2 induces endothelial cell damage and mitochondrial DNA release.
(A) Time and MOI-dependent infection of endothelial cells bySARS-CoV-2. (B and C), cytochrome B and NADH dehydrogenase expression in the supernatant of SARS-CoV-2-infected HUVECs. (D and E) Cytochrome B and NADH dehydrogenase analysis in the serum of positive and negative COVID-19 patients. (F) Lactate dehydrogenase (LDH) assay to identify cell damage. Results are expressed as % of control. NADH dehydrogenase and cytochrome B values are expressed as mtDNA/total DNA). The results are expressed as the mean ± SEM. Statistical significance was determined by one-way ANOVA followed by the multiple comparations Tukey post-hoc test (cells n = 3–5; patients n = 7–19), using the Prism GraphPad 8.0 software. Statistically significant differences were considered when p < 0.05.
Serum of COVID-19 patients presented very high levels of cytochrome B and NAD dehydrogenase, supporting significant mtDNA release in patients with COVID-19 (Fig. 2D and E). Serum of negative COVID-19 patients with comorbidities normally found in COVID-19 patients (hypertension and diabetes) also presented increased levels of cytochrome B compared to healthy subjects (Fig. 2D and E).
Plasma membrane integrity upon in vitro infection was determined by LDH release. SARS-CoV-2 triggered LDH release in HUVECs and this was prevented by ODNA 2088, a TLR9 inhibitor (Fig. 2F). Caspase-3 markers were not altered in SARS-CoV-2-infected cells (date no shown).
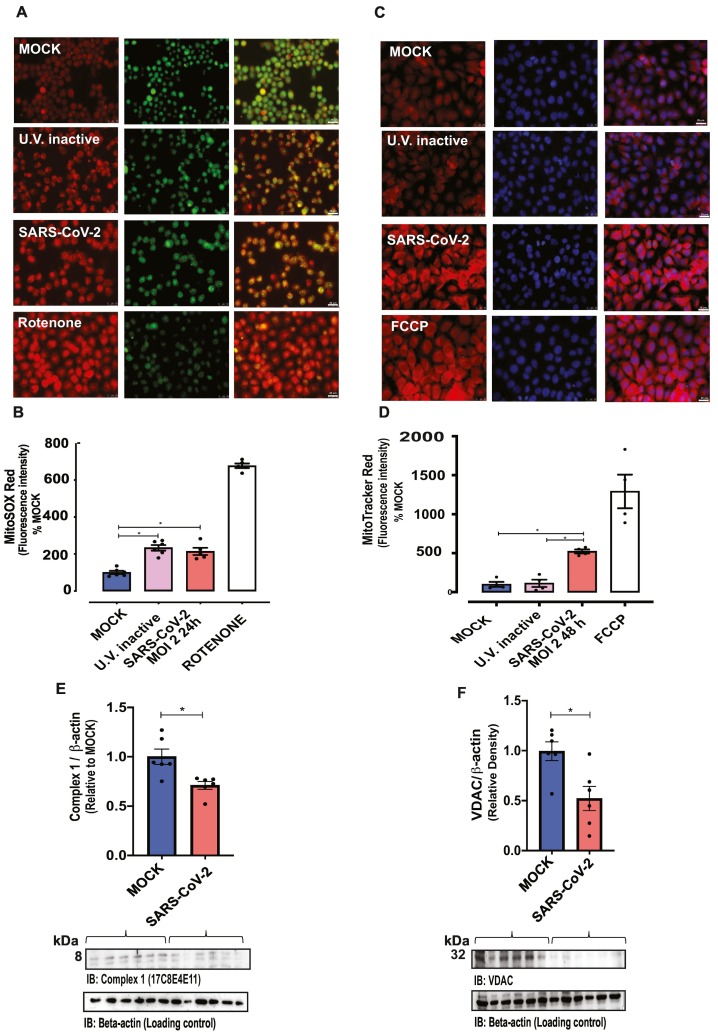
Mitochondria-derived superoxide anion, determined by MitoSOX™ Red fluorescence intensity, did not differ among control HUVECs and HUVECs in virus-free MOCK medium (Supplementary Fig. 1A). MitoSOX™ fluorescence intensity increased in HUVECs exposed to UV-inactivated SARS-CoV-2 and SARS-COV-2-infected HUVECs, compared with the virus-free MOCK medium group (Fig. 3A and B). Rotenone, which inhibits mitochondrial complex 1 and increases superoxide anion production, was used as a positive control.
Fig. 3.
SARS-CoV-2 increases mitochondrial dysfunction.
Fluorescence intensity depicting (A and B) mitochondrial ROS (MitoSOX Red®) and (C and D) mitochondrial potential (MitoTracker® Red CMXRos, FlexStation-3) in control (MOCK and UV-inactivated SARS-CoV-2-exposed HUVECs) and SARS-CoV-2-infected (MOI 2–24 or 48 h) HUVECs. Rotenone and FCCP were used as positive controls. Acridine orange (green) and DAPI (blue) were utilized for the nucleus staining. Immunoblot representative imagens (bottom) and densitometric analysis (top) of protein levels of (E) mitochondrial complex I and (F) VDAC (voltage-dependent ion channel) normalized by β-actin in MOCK and SARS-CoV-2-infected HUVECs. The results are expressed as the mean ± SEM. Statistical significance was determined by one-way ANOVA multiple comparations with Tukey post-hoc test or t-test when appropriated (n = 5–6), using the Prism GraphPad 8.0 software. Statistically significant differences were considered when p < 0.05. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Mitochondrial membrane potential, determined by MitoTracker® Red CMXRos, did not differ among control cells (cells in virus-free MOCK medium, and cells exposed to UV-inactivated SARS-CoV-2 (Supplementary Fig. 1B), but was increased in HUVECs infected by SARS-CoV-2 (Fig. 3C and D). FCCP, a potent uncoupler of mitochondrial oxidative phosphorylation that depolarizes mitochondrial membrane potential, was used as a positive control. Mitochondrial mass, determined by MitoTracker® Green FM, was similar among the groups (Supplementary Fig. 1C ).
SARS-COV-2 infection decreased mitochondrial complex-1 protein levels, compared with virus-free MOCK medium (Fig. 3E). In addition, VDAC (voltage-dependent anion channel) protein levels were decreased in endothelial cells infected with SARS-CoV-2 compared with the virus-free MOCK medium group (Fig. 3F). Complex-1 and VDAC protein levels did not differ between HUVECs exposed to MOCK or UV-inactivated SARS-CoV-2 (Supplementary Fig.1D ).
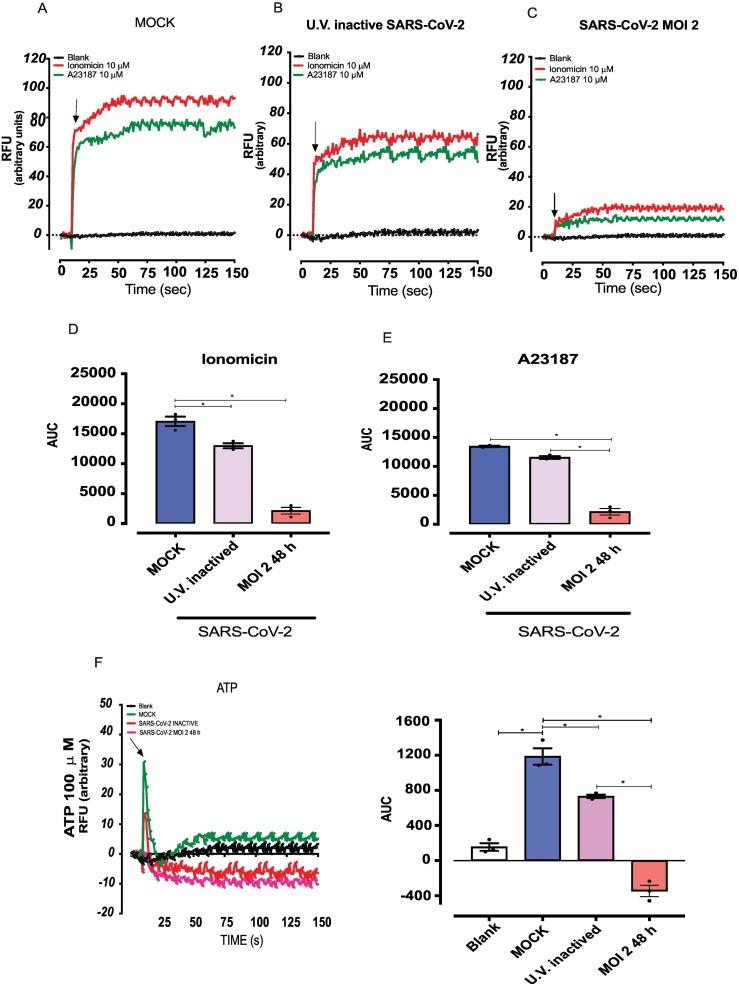
Host cell dysfunction following viral infection is accompanied by abnormal intracellular Ca2+ concentrations [27] and changes in cytosolic Ca2+ levels is an important marker of endothelial cell and mitochondrial dysfunction [28]. To eliminate cell line bias, e.g., that HUVECs differentially respond to Ca2+ mobilization stimuli, responses to Ca2+ ionophores and ATP were determined in 2 endothelial cell lineages, HUVECs and immortal human lineages Ea. hy926. Both cell lines displayed similar responses to these agents (Supplementary Fig. 2A). Responses to ionomycin and A23187 were similar in control and virus-free MOCK medium HUVECs (Suplemmentary Fig. 2B and C). However, SARS-CoV-2-infected cells exhibited decreased responses to both Ca2+ ionophores when compared with cells exposed to UV-inactivated SARS-CoV-2, as determined by the AUC (Figs. 4A-4E). ATP-induced Ca2+ responses were also abolished in HUVECs infect by SARS-CoV-2 compared with cells in virus-free MOCK medium or exposed to UV-inactivated SARS-CoV-2 (Fig. 4F and G). These data suggest that SARS-CoV-2 induces mitochondrial dysfunction in HUVECs, an effect associated with loss of ability to respond to Ca2++-mobilizing agents.
Fig. 4.
SARS-CoV-2 decreases calcium responses in endothelial cells.
Calcium mobilization in response to ionomycin (10−5 M) and A23187 (10−6 M) in HUVECs cells in (A) free MOCK medium, (B) exposed to UV-inactivated SARS-CoV-2, (C) SARS-CoV-2 (MOI 2) for 48 h. (D) and (E) represent the AUC. (F and G) Calcium mobilization in response to ATP. The results are expressed as the mean ± SEM. Statistical significance was determined by one-way ANOVA multiple comparations with Tukey post-hoc test (n = 3), using the Prism GraphPad 8.0 software. Statistically significant differences were considered when p < 0.05.
3.2. Mitochondrial dysfunction activates TLR9 and pro-inflammatory signaling, events linked to mitochondrial and vascular dysfunction
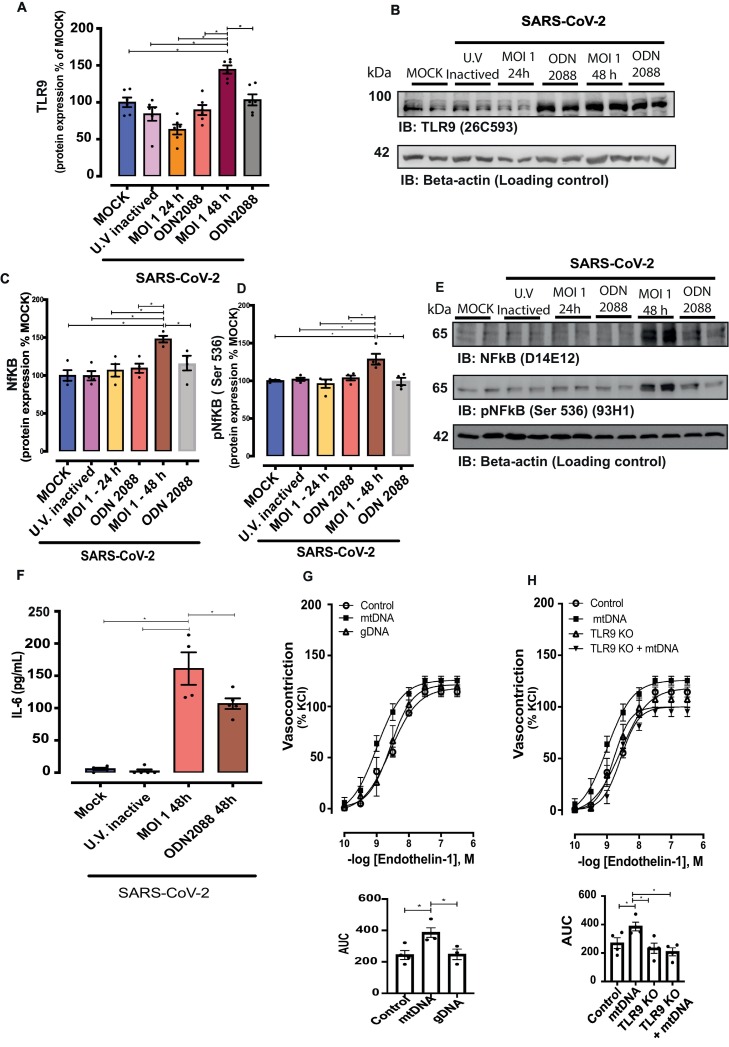
Since mitochondrial dysfunction and mtDNA release have been shown to activate TLR9 responses, TLR9 signaling was investigated. TLR9 protein levels increased in SARS-Cov-2-infected HUVECs (48 h post-infection) compared with cells in virus-free MOCK medium or HUVECs exposed to UV-inactivated virus. TLR9 expression at 24 h was not altered, suggesting that the effect is time-dependent (Fig. 5A). Myd88 expression did not differ among the groups (data not shown). SARS-CoV-2 infection activated inflammatory signaling, indicated by protein levels of total and phosphorylated (at Serine536) (Fig. 5C, D and E).
Fig. 5.
SARS-CoV-2 increases protein levels of TLR9 and NF-kB signaling. Representative immunoblot images and densitometric analysis of protein levels of (A and B) TLR9, (C, D and E) total and phosphorylated (Serine536) NF-kB in HUVECs infected with SARS-CoV-2 for 24 and 48 h treated with vehicle or ODN 2088, TLR9 antagonist. Expression of the protein of interest was normalized by β-actin expression. Results are expressed as a percentage of MOCK control and represent the mean ± SEM from 4 experiments. The results are expressed as the mean ± SEM. Statistical significance was determined by one-way ANOVA multiple comparations with Tukey post-hoc test (n = 4), using the Prism GraphPad 8.0 software. Statistically significant differences were considered when p < 0.05. (F) IL-6 determined by Elisa assay in the supernatant of HUVECs infected by SARS-CoV-2; values represent pg/mL. Cumulative concentration-response curves to endothelin-1 in resistance mesenteric arteries of C57BL/J6 (G) and TLR9KO (H) mice, in the presence of mtDNA (mitochondrial DNA) or gDNA (genomic DNA). Results are expressed as a percentage of KCl-induced vasoconstriction and represent the mean ± SEM from 3 to 4 different experiments. Statistical significance was determined by one-way ANOVA multiple comparations with Tukey post-hoc test (n = 3–6), using the Prism GraphPad 8.0 software. Statistically significant differences were considered when p < 0.05.
Since activation of TLR9 contributes to endothelial dysfunction, levels of ET-1, TXA2, and eNOS, key molecules in endothelial cell function and important in the balance of vascular tone, platelet aggregation, membrane permeability, and Ca2+ homeostasis, were determined. ET-1 levels decreased in SARS-CoV-2-infected cells, but this effect was not dependent on TLR9 activation (Supplementary Fig. S3). COVID-19 negative patients with comorbidities presented high serum levels of ET-1 compared with healthy COVID-19-negative subjects (Supplementary Fig. S3). Although critical patients with COVID-19 presented increased levels of ET-1 compared with moderate COVID-19 patients, ET-1 levels were lower in COVID-19 patients compared to COVID-19 negative patients with comorbidities (Supplementary Fig. S3).
HUVECs infected with SARS-COV-2 presented high levels of IL-6 compared with cells in virus-free MOCK medium or exposed to UV-inactivated virus. ODN 2088 decreased IL-6 levels in SARS-CoV-2-infected HUVECs (Fig. 5F), in agreement with ODN 2088-induced effects on total and phosphorylated NF-kB. Levels of TXA2 increased in SARS-CoV-2-infected cells, but this effect was not dependent on TLR9 activation (Supplementary Fig. 3D).
Considering that SARS-CoV-2 induces endothelial cell dysfunction linked to mtDNA leakage and TLR9 activation, we investigated whether mtDNA, a TLR9 agonist, affects vascular reactivity. mtDNA, but not genomic DNA (gDNA) increased vasoconstriction to ET-1 (Fig. 5G). mtDNA effects on ET-1 vascular reactivity were not observed in arteries from TLR9KO mice (Fig. 5H). These results indicate that, in addition to promoting endothelial cell dysfunction, SARS-CoV-2 infection negatively impacts vascular function since endothelium-derived key vasoactive factors as well as mtDNA and TLR9 activation impair vascular responses.
4. Discussion
The present study shows that SARS-CoV-2 infection of endothelial cells induces cell damage by promoting mitochondrial dysfunction, mtDNA release, TLR9 activation, leading to impaired Ca2+ responses, increased IL-6 levels. Of importance, mtDNA is increased in serum from COVID-19 patients (Graphical Abstract).
The link between coronavirus infection and cardiovascular diseases was initially suggested by studies showing that patients with cardiovascular diseases are prone to COVID-19 complications and a more severe clinical presentation. Infection of endothelial cells by SARS-CoV-2 raised the hypothesis that COVID-19, particularly in patients with complicated outcomes, represents a vascular endothelial disease [29]. Although ACE-2 expression was identified in endothelial cells, we did not observe viral replication in these cells partially supporting a previous report [30]. Although recent studies have shown that other viral receptors and cellular proteases are important in facilitating viral entry and transmission of SARS-CoV-2 in target cells [31,32], this was not investigated in the present study.
It is not clear whether SARS-CoV-2 directly or indirectly activates TLR9. Potential interactions between SARS-CoV-2 spike glycoprotein, ACE-2 receptor homologs, and human TLRs have been proposed. Binding interfaces between TLRs and the spike protein consist of hydrogen and hydrophobic interactions. An in-silico study that examined possible interactions between surface TLRs in human cells and the spike protein of SARS-CoV-2 indicated that TLR4, followed by TLR6 and TLR1, but not TLR9, has a strong binding affinity to spike protein [33].
The vascular endothelium provides a crucial interface between the blood compartment and tissues, modulating vascular tone, angiogenesis, and coagulation processes. During viral infection, endothelial cells mediate important inflammatory processes, contributing to increased blood flow, leakage of plasma proteins, and neutrophil, monocyte, and lymphocyte recruitment [34]. Endothelial dysfunction in COVID-19 patients is linked to thrombotic events [35], anticoagulation imbalance, systemic endotheliitis [36], glycocalyx shedding [10], elevated D-dimer levels [37], increased ROS production [38] and NO deficiency [39].
Mitochondria play a central role in host immune responses to viral infection. Mitochondrial dysfunction leads to the release of mtDNA that exacerbates viral pathogenesis [40]. mtDNA is an important DAMP that activates immune response and inflammation, and mitochondrial DAMPs are effective inducers of inflammation and cytokine production [41].
SARS-CoV-1 infection targets host cell mitochondria and disables the mitochondrial antiviral signaling protein (MAVS) adaptor, thus manipulating mitochondrial function [42]. Computational analysis suggests that SARS-CoV-2 RNA genome and subgenomic RNA colocalize with the host mitochondrial matrix [43]. Although there are no biological data showing colocalization of viral SARS-CoV-2 RNA and mitochondrial proteins, it has been hypothesized that SARS-CoV-2 RNA deposition in mitochondria may impair mitochondrial function, contributing to changes in membrane potential and anion channels, reducing energy and increasing ROS generation [44,45]. ROS generation by mitochondria is an important index of mitochondrial dysfunction [46]. Complex I, is the largest multimeric enzyme belonging to the group of five enzyme complexes that act in the mitochondrial respiratory chain, is key for oxidative phosphorylation . Complex I is also a major source of ROS and its dysfunction is associated with mitochondrial damage [47]. Mitochondrial dysfunction has also been reported in several viral infections, including Influenza A, herpes viruses [48,49], human immunodeficiency virus-1 (HIV-1) infection [50], and Pneumoviridae family, caused by respiratory syncytial virus [51].
Mitochondrial membrane potential was also determined to further verify mitochondrial dysfunction. Similarly, the respiratory syncytial virus promotes loss of mitochondrial membrane potential, impairs mitochondrial respiration, and increases production of mitochondrial ROS in pulmonary epithelial cells [51]. Zika virus from the Flaviviridae family also changes mitochondrial transmembrane potential, associated with a mitochondrial morphodynamical perturbation in retinal pigment epithelial cells [52]. Changes in mitochondrial membrane potential disrupts ATP synthesis, induces membrane depolarization, mitochondrial swelling, and increases mitochondrial ROS [53]. Increased mitochondrial ROS, in turn, disturbs mtDNA [54]. Cytochrome B and NADH dehydrogenase are components of the respiratory chain complex and participate in the electron transport chain, therefore contributing to ATP synthesis [55]. The fact that gene expression of cytochrome B and NADH dehydrogenase was increased in HUVECs infected by SARS-CoV-2, and that levels of LDH, an intracellular enzyme released in injury conditions, were also increased in the supernatant of SARS-CoV-2-infected cells, support extravasation of mtDNA in SARS-CoV-2-infected cells. Of importance, increased levels of cytochrome B and NADH dehydrogenase were found in plasma from COVID-19 patients, reinforcing mitochondrial dysfunction with consequent cell damage in COVID-19. Mitochondrial dysfunction and metabolic alterations (i.e. increased glycolysis) were reported in peripheral blood mononuclear cells from patients with COVID-19. It was speculated that SARS-CoV-2 uses glucose as source of energy and for cell survival, exacerbating inflammatory responses that contribute to the severity of symptoms in COVID-19 [22].
The voltage-dependent anion channel (VDAC) is permeable to small solutes such as Ca2+, and regulates mitochondrial Ca2+ flux [28]. Ca2+ is one of most genuine second messenger in cellular physiology and of its homeostasis is impaired in endothelial and smooth muscle cells in pathological conditions. Whereas Ca2+ induces smooth muscle cells contraction, in endothelial cells Ca2+ turns on the outflow of several mediators such as NO and prostacyclin (PGI2), often related to endothelial cell health. A reduction of Ca2+ in endothelial cells abrogates the release of vasodilator mediators and induces endothelial cell dysfunction [56]. In HeLa cells and skeletal myotubes, overexpression of VDCA favors Ca2+ mobilization through the mitochondrial matrix by facilitating the release of Ca2+ from the endoplasmic reticulum [57]. TLR9 inhibits the sarcoplasmic reticulum/ER Ca2+-ATPase 2 (SERCA2) activity and impairs Ca2+ uptake into the ER, leading to decreased mitochondrial Ca2+ concentrations and less mitochondrial ATP generation in cardiomyocytes and neurons [58,59], but not in vascular smooth muscle cells [16]. SARS-CoV-2-infected endothelial cells exhibited reduced expression of VDAC and decreased Ca2+ entry induced by ionomycin, A23187 and ATP, confirming that VDAC expression controls Ca2+ transport. We have not carried out experiments to assess the specific role of Ca2+ uptake by the mitochondria, but our results clearly suggest that mitochondrial dysfunction is associated with impaired Ca2+ mobilization in HUVECs.
In general, mtDNA activates inflammatory responses by TLR9-dependent mechanisms. TLR9 recognizes unmethylated CpG dinucleotides present in high frequency in bacterial and viral DNA. mtDNA is structurally like bacterial DNA and share unmethylated CpG DNA repeats [41]. SARS-CoV-2 has a high number of CpG-motifs in the E-ORF and ORF10 nucleotide sequences, suggesting TLR9 activation in many cells types during COVID-19 [60]. mtDNA is a well-known DAMP that triggers inflammatory responses directly via TLR9 during injury and/or infection [17]. Since SARS-CoV-2 exhibits unmethylated CpG motifs in its genetic material, it is possible that SARS-CoV-2 directly activates TLR9 in endothelial cells, contributing to COVID-19 complications [61].
Since mtDNA activates TLR9 and TLR9 activation itself contributes to cell death, a positive feedback loop between mitochondrial dysfunction and TLR9 activation is suggested. The TLR9 blockade with ODN2088 significantly decreases LDH release, cytochrome B as well and NADH levels in the supernatant of HUVECs, 48 h after infection with SARS-CoV-2, indicating that TLR9 blockade decreases the release of mtDNA.
TLR9 activation by SARS-CoV-2 induced inflammatory process in COVID-19. Although in vivo data suggest that TLR9 activation by mtDNA regulates vasoconstriction, there is a lack of in vitro study addressing the direct interaction of endothelial TLR9 and mtDNA. Activation of TLR9 may be a silent, but driving force that explains hyperinflammation and thrombotic complications caused by SARS-CoV-2 [61].
Corroborating the data reported in HUVECs and translating these data to a clinical model, plasma of patients with COVID-19 exhibit increased mtDNA when compared to non-infected patients. Plasma mtDNA may be linked to apoptosis and necrosis of many cell types, including endothelial cells, as shown in the present study. Increased mtDNA in HUVECs (48 h post-infection) increased TLR9 protein levels, increased MyD88-independent intracellular and NF-kB phosphorylation, and increased IL-6.
TXA2 and ET-1, vasoactive and endothelium-derived factors, may lead to vascular dysfunction. COVID-19 patients present high levels of TXB2, a stable product of TXA2 [62], but increased ET-1 levels have not been consistently found in COVID-19 patients [63]. In fact, we found lower ET-1 levels in the plasma of mild-to-severe COVID-19 patients. Although increased only in severely patients, ET-1 levels were lower than those observed in patients with comorbidities. Changes in TXA2 and ET-1 levels in SARS-CoV-2-infected endothelial cells occurred independently of TLR9 activation.
These data clearly show that SARS-CoV-2 activates inflammatory processes in HUVECs via TLR9. SARS-CoV-2 leads to dysfunction and increased inflammatory process in endothelial cells, which may contribute to the cytokine storm and thrombotic complications in patients with severe coronavirus infection. Pro-inflammatory cytokines, such as IL-6, impact vascular tone via endothelium-dependent mechanisms, such as inhibition of eNOS activity and protein kinase C activation [64]. Increased levels of pro-inflammatory cytokines (e.g., IL-6, TNF-α) were reported in patients diagnosed with severe COVID-19 [65]. These cytokines play a central role in the loss of normal antithrombotic and anti-inflammatory functions of endothelial cells.
In summary, our data show that SARS-CoV-2 infection of endothelial cells promotes mitochondrial dysfunction, increases mtDNA release and activates TLR9 signaling. TLR9 contributes to inflammatory responses, via NF-kB signaling and IL-6 production, and compromises endothelial cell function, by decreasing eNOS expression. Our study contributes to the understanding of how SARS-CoV-2 induces endothelial cells dysfunction and opens new perspectives for better therapeutic approaches.
Financial support
This study was supported by The São Paulo State Research Foundation (FAPESP; Grant 2013/08216–2 – Research Center in Inflammatory Diseases), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES).
Tiago J. Costa is a FAPESP post-doctoral fellow (2017/25116–2 and 2019/26376–0). Simone R. Potje received a FAPESP post-doctoral fellowship (2016/21239–0).
Author contributions
TJC and RT designed the research. MNB, CEA and MAM supervised the clinical study. TJC, SRP, TFCFS, JASN, PRB, DR, MS, RBM, RASE, KSGS, IAC, MCP, and LS performed experiments. TJC, SRP, JASN, PRB, DR, GFB and RT analyzed and interpreted data. TJC, SRP, CB, VLDB, EA, PLJ, RDRO, DSZ, NEZ, MAM, FRCG, and RT discussed data and manuscript. TJC, SRP, GFB and RT wrote the manuscript.
All authors approved the manuscript.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgments
The authors thanks Carla P. Manzato for excellent technical assistance and Prof Dr. Emer Suavinho Ferro for laboratory support (FAPESP grant 16/04000-3).
Tiago J. Costa is grateful to João Victor Cabral-Costa (University of Sao Paulo) and Dr. Paula Reventun (Johns Hopkins University) for excellent support and discussion on protocols involving mitochondrial probes and ACE-2 expression in HUVECs, respectively.
The graphical abstract was created using BioRender.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.vph.2021.106946.
Appendix A. Supplementary data
Fig. S1 SARS-CoV-2 induced mitochondrial dysfunction
(A and B) Ffluorescence intensity of mitochondrial ROS (MitoSOX Red®) and mitochondrial potential (MitoTracker® Red CMXRos, FlexStation-3) in control and MOCK HUVECs. C MitoTracker® green representative imagens and D mitochondrial complex I and VDAC (voltage-dependent ion channel) in MOCK, U.V. inactived SARS-CoV-2, and SARS-CoV-2. The western blot was normalized by β-actin. The results are expressed as the mean ± SEM. Statistical significance was determined by one-way ANOVA multiple comparations with Tukey post-hoc test or t-test when appropriate (n = 5–6), using the Prism GraphPad 8.0 software. Statistically significant differences were considered when p < 0.05.
Fig. S2 Calcium responses in HUVECs and EA. hy296 cells.
Calcium mobilization in response to ionomycin (10−5 M) and A23187 (10−6 M) in (A) HUVECs and EA.hy296 cells. HUVECs under (B) control and (C) free MOCK medium conditions. The results are expressed as the mean ± SEM. Statistical significance was determined by one-way ANOVA multiple comparations with Tukey post-hoc test (n = 5–6), using the Prism GraphPad 8.0 software. Statistically significant differences were considered when p < 0.05.
Fig. S3 SARS-CoV-2 impact on ET-1 and TXA2, key molecules in endothelial cell function. (A) ET-1 levels in HUVECs exposed to MOCK, UV-inactivated SARS-CoV-2 and SARS-CoV-2 . (B and C) ET-1 levels in plasma from COVID-19 negative patients (healthy subjects and patients with comorbidities) and COVID-19 positive patients (moderate an critically ill patients with COVID-19). (D) Thombroxane (TXA2) levels in HUVECs exposed to MOCK, UV-inactived SARS-CoV-2 and SARS-CoV-2. ODN2088, TLR9 inhibitor. The results are expressed as the mean ± SEM. Statistical significance was determined by one-way ANOVA multiple comparations with Tukey post-hoc test (n = 5–6), using the Prism GraphPad 8.0 software. Statistically significant differences were considered when p < 0.05.
References
- 1.CSSE-JHU, C. F. S. S. A. E. A. J. H. U COVID-19 Dashboard n.d. 2020. https://coronavirus.jhu.edu/map.html
- 2.Menter T., et al. Postmortem examination of COVID-19 patients reveals diffuse alveolar damage with severe capillary congestion and variegated findings in lungs and other organs suggesting vascular dysfunction. Histopathology. Aug 2020;77(2):198–209. doi: 10.1111/his.14134. https://www.ncbi.nlm.nih.gov/pubmed/32364264 ISSN 1365–2559. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li S., et al. SARS-CoV-2 triggers inflammatory responses and cell death through caspase-8 activation. Signal Transduct Target Ther. 2020;5(1):235. doi: 10.1038/s41392-020-00334-0. https://www.ncbi.nlm.nih.gov/pubmed/33037188 ISSN 2059–3635. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dorward D.A., et al. Tissue-Specific Immunopathology in Fatal COVID-19. Am. J. Respir. Crit. Care Med. 2021;203(2):192–201. doi: 10.1164/rccm.202008-3265OC. https://www.ncbi.nlm.nih.gov/pubmed/33217246 ISSN 1535–4970. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodrigues T.S., et al. Inflammasomes are activated in response to SARS-CoV-2 infection and are associated with COVID-19 severity in patients. J. Exp. Med. 2021;218(3) doi: 10.1084/jem.20201707. https://www.ncbi.nlm.nih.gov/pubmed/33231615 ISSN 1540–9538. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Puelles V.G., et al. Multiorgan and renal tropism of SARS-CoV-2. N. Engl. J. Med. 2020;383(6):590–592. doi: 10.1056/NEJMc2011400. https://www.ncbi.nlm.nih.gov/pubmed/32402155 ISSN 1533–4406. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta A., et al. Extrapulmonary manifestations of COVID-19. Nat. Med. 2020;26(7):1017–1032. doi: 10.1038/s41591-020-0968-3. https://www.ncbi.nlm.nih.gov/pubmed/32651579 ISSN 1546-170X. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chi G., et al. Venous thromboembolism among hospitalized patients with COVID-19 undergoing thromboprophylaxis: a systematic review and meta-analysis. J. Clin. Med. Aug 2020;9(8) doi: 10.3390/jcm9082489. https://www.ncbi.nlm.nih.gov/pubmed/32756383 ISSN 2077–0383. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stahl K., et al. Injury to the Endothelial Glycocalyx in Critically Ill Patients with COVID-19. Am. J. Respir. Crit. Care Med. 2020;202(8):1178–1181. doi: 10.1164/rccm.202007-2676LE. https://www.ncbi.nlm.nih.gov/pubmed/32833500 ISSN 1535–4970. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Potje S.R., et al. Heparin prevents in vitro glycocalyx shedding induced by plasma from COVID-19 patients. Life Sci. Mar 2021:119376. doi: 10.1016/j.lfs.2021.119376. https://www.ncbi.nlm.nih.gov/pubmed/33781826 ISSN 1879–0631. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Do Espírito Santo D.A., Lemos A.C.B., Miranda C.H. In vivo demonstration of microvascular thrombosis in severe COVID-19. J. Thromb. Thrombolysis. Nov 2020;50(4):790–794. doi: 10.1007/s11239-020-02245-x. https://www.ncbi.nlm.nih.gov/pubmed/32789730 ISSN 1573-742X. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seth R.B., et al. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. Sep 2005;122(5):669–682. doi: 10.1016/j.cell.2005.08.012. https://www.ncbi.nlm.nih.gov/pubmed/16125763 ISSN 0092–8674. Disponível em. [DOI] [PubMed] [Google Scholar]
- 13.Gorman G.S., et al. Mitochondrial diseases. Nat Rev Dis Primers. 2016;2:16080. doi: 10.1038/nrdp.2016.80. https://www.ncbi.nlm.nih.gov/pubmed/27775730 ISSN 2056-676X. Disponível em. [DOI] [PubMed] [Google Scholar]
- 14.Pereira O.R., et al. Changes in mitochondrial morphology modulate LPS-induced loss of calcium homeostasis in BV-2 microglial cells. J. Bioenerg. Biomembr. Apr 2021;53(2):109–118. doi: 10.1007/s10863-021-09878-4. https://www.ncbi.nlm.nih.gov/pubmed/33585958 ISSN 1573–6881. Disponível em. [DOI] [PubMed] [Google Scholar]
- 15.Simmons J.D., et al. Potential contribution of mitochondrial DNA damage associated molecular patterns in transfusion products to the development of acute respiratory distress syndrome after multiple transfusions. J. Trauma Acute Care Surg. 2017;82(6):1023–1029. doi: 10.1097/TA.0000000000001421. https://www.ncbi.nlm.nih.gov/pubmed/28301393 ISSN 2163–0763. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mccarthy C.G., et al. Circulating mitochondrial DNA and Toll-like receptor 9 are associated with vascular dysfunction in spontaneously hypertensive rats. Cardiovasc. Res. Jul 2015;107(1):119–130. doi: 10.1093/cvr/cvv137. https://www.ncbi.nlm.nih.gov/pubmed/25910936 ISSN 1755–3245. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Echem C., et al. Mitochondrial DNA: A new driver for sex differences in spontaneous hypertension. Pharmacol. Res. 2019;144:142–150. doi: 10.1016/j.phrs.2019.04.008. https://www.ncbi.nlm.nih.gov/pubmed/30965087 ISSN 1096–1186. Disponível em. [DOI] [PubMed] [Google Scholar]
- 18.Hemmi H., et al. A Toll-like receptor recognizes bacterial DNA. Nature. Dec 2000;408(6813):5–740. doi: 10.1038/35047123. https://www.ncbi.nlm.nih.gov/pubmed/11130078 ISSN 0028–0836. Disponível em. [DOI] [PubMed] [Google Scholar]
- 19.Lund J., et al. Toll-like receptor 9-mediated recognition of Herpes simplex virus-2 by plasmacytoid dendritic cells. J. Exp. Med. Aug 2003;198(3):513–520. doi: 10.1084/jem.20030162. https://www.ncbi.nlm.nih.gov/pubmed/12900525 ISSN 0022–1007. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai J.H., et al. Infection with the dengue RNA virus activates TLR9 signaling in human dendritic cells. EMBO Rep. 2018;19(8) doi: 10.15252/embr.201846182. https://www.ncbi.nlm.nih.gov/pubmed/29880709 ISSN 1469–3178. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim J.S., et al. The role of TLR9 in stress-dependent autophagy formation. Biochem. Biophys. Res. Commun. Dec 2016;481(3–4):219–226. doi: 10.1016/j.bbrc.2016.10.105. https://www.ncbi.nlm.nih.gov/pubmed/27793667 ISSN 1090–2104. Disponível em. [DOI] [PubMed] [Google Scholar]
- 22.Ajaz S., et al. Mitochondrial metabolic manipulation by SARS-CoV-2 in peripheral blood mononuclear cells of patients with COVID-19. Am. J. Phys. Cell Phys. 01 2021;320(1):C57–C65. doi: 10.1152/ajpcell.00426.2020. https://www.ncbi.nlm.nih.gov/pubmed/33151090 ISSN 1522–1563. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abouhashem A.S., et al. Is low alveolar Type II cell. Antioxid. Redox Signal. 2020;33(2):59–65. doi: 10.1089/ars.2020.8111. https://www.ncbi.nlm.nih.gov/pubmed/32323565 ISSN 1557–7716. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou X., et al. Low serum calcium: a new, important indicator of COVID-19 patients from mild/moderate to severe/critical. Biosci. Rep. Nov 2020;40(12) doi: 10.1042/BSR20202690. https://www.ncbi.nlm.nih.gov/pubmed/33252122 BSR20202690. ISSN 1557–7716. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.WHO Working Group on the Clinical Characterisation and Management of COVID-19 infection A minimal common outcome measure set for COVID-19 clinical research. Lancet Infect. Dis. 08 2020;20(8):e192–e197. doi: 10.1016/S1473-3099(20)30483-7. https://www.ncbi.nlm.nih.gov/pubmed/32539990 ISSN 1474–4457. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu X., et al. US CDC real-time reverse transcription PCR panel for detection of severe acute respiratory syndrome coronavirus 2. Emerg. Infect. Dis. Aug 2020;26(8) doi: 10.3201/eid2608.201246. https://www.ncbi.nlm.nih.gov/pubmed/32396505 ISSN 1080–6059. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Y., Frey T.K., Yang J.J. Viral calciomics: interplays between Ca2+ and virus. Cell Calcium. Jul 2009;46(1):1–17. doi: 10.1016/j.ceca.2009.05.005. https://www.ncbi.nlm.nih.gov/pubmed/19535138 ISSN 1532–1991. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosencrans W.M., et al. VDAC regulation of mitochondrial calcium flux: From channel biophysics to disease. Cell Calcium. 2021;94:102356. doi: 10.1016/j.ceca.2021.102356. https://www.ncbi.nlm.nih.gov/pubmed/33529977 ISSN 1532–1991. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Libby P., Lüscher T. COVID-19 is, in the end, an endothelial disease. Eur. Heart J. 2020;41(32):3038–3044. doi: 10.1093/eurheartj/ehaa623. https://www.ncbi.nlm.nih.gov/pubmed/32882706 ISSN 1522–9645. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mccracken I.R., et al. Lack of evidence of angiotensin-converting enzyme 2 expression and replicative infection by SARS-CoV-2 in human endothelial cells. Circulation. Feb 2021;143(8):865–868. doi: 10.1161/CIRCULATIONAHA.120.052824. https://www.ncbi.nlm.nih.gov/pubmed/33405941 ISSN 1524–4539. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masre S.F., et al. Classical and alternative receptors for SARS-CoV-2 therapeutic strategy. Rev. Med. Virol. Dec 2020:e2207. doi: 10.1002/rmv.2207. https://www.ncbi.nlm.nih.gov/pubmed/33368788 ISSN 1099–1654. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amraie R., et al. CD209L/L-SIGN and CD209/DC-SIGN Act as Receptors for SARS-CoV-2. ACS Cent Sci. Jul 28 2021;7(7):1156–1165. doi: 10.1021/acscentsci.0c01537. https://www.ncbi.nlm.nih.gov/pubmed/34341769 ISSN 2374-7943, Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choudhury A., Mukherjee S. In silico studies on the comparative characterization of the interactions of SARS-CoV-2 spike glycoprotein with ACE-2 receptor homologs and human TLRs. J. Med. Virol. 2020;92(10):2105–2113. doi: 10.1002/jmv.25987. https://www.ncbi.nlm.nih.gov/pubmed/32383269 ISSN 1096–9071. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pober J.S., Sessa W.C. Evolving functions of endothelial cells in inflammation. Nat. Rev. Immunol. Oct 2007;7(10):803–815. doi: 10.1038/nri2171. https://www.ncbi.nlm.nih.gov/pubmed/17893694 . ISSN 1474–1741. Disponível em. [DOI] [PubMed] [Google Scholar]
- 35.Bilaloglu S., et al. Thrombosis in Hospitalized Patients With COVID-19 in a New York City Health System. JAMA. 2020;324(8):799–801. doi: 10.1001/jama.2020.13372. https://www.ncbi.nlm.nih.gov/pubmed/32702090 ISSN 1538–3598. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Varga Z. Endotheliitis in COVID-19. Pathologe. Dec 2020;41(Suppl. 2):99–102. doi: 10.1007/s00292-020-00875-9. https://www.ncbi.nlm.nih.gov/pubmed/33306138 ISSN 1432–1963. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang L., et al. D-dimer levels on admission to predict in-hospital mortality in patients with Covid-19. J. Thromb. Haemost. 2020;18(6):1324–1329. doi: 10.1111/jth.14859. https://www.ncbi.nlm.nih.gov/pubmed/32306492 ISSN 1538–7836. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goud P.T., Bai D., Abu-Soud H.M. A multiple-hit hypothesis involving reactive oxygen species and myeloperoxidase explains clinical deterioration and fatality in COVID-19. Int. J. Biol. Sci. 2021;17(1):62–72. doi: 10.7150/ijbs.51811. https://www.ncbi.nlm.nih.gov/pubmed/33390833 ISSN 1449–2288. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Green S.J. Covid-19 accelerates endothelial dysfunction and nitric oxide deficiency. Microbes Infect. 2020 May - Jun 2020;22(4–5):149–150. doi: 10.1016/j.micinf.2020.05.006. https://www.ncbi.nlm.nih.gov/pubmed/32425647 ISSN 1769-714X. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Riley J.S., Tait S.W. Mitochondrial DNA in inflammation and immunity. EMBO Rep. 2020;21(4):e49799. doi: 10.15252/embr.201949799. https://www.ncbi.nlm.nih.gov/pubmed/32202065 ISSN 1469–3178. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Q., et al. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. Mar 2010;464(7285):104–107. doi: 10.1038/nature08780. https://www.ncbi.nlm.nih.gov/pubmed/20203610 ISSN 1476–4687. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi C.S., et al. SARS-coronavirus open reading frame-9b suppresses innate immunity by targeting mitochondria and the MAVS/TRAF3/TRAF6 signalosome. J. Immunol. Sep 2014;193(6):3080–3089. doi: 10.4049/jimmunol.1303196. https://www.ncbi.nlm.nih.gov/pubmed/25135833 ISSN 1550–6606. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu K.E., et al. RNA-GPS Predicts SARS-CoV-2 RNA Residency to Host Mitochondria and Nucleolus. Cell Syst. 2020;11(1):102–108. doi: 10.1016/j.cels.2020.06.008. https://www.ncbi.nlm.nih.gov/pubmed/32673562 ISSN 2405–4720. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burtscher J., et al. Mitochondria: In the cross fire of SARS-CoV-2 and immunity. iScience. Oct 2020;23(10):101631. doi: 10.1016/j.isci.2020.101631. https://www.ncbi.nlm.nih.gov/pubmed/33015593 ISSN 2589–0042. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh K.K., et al. Decoding SARS-CoV-2 hijacking of host mitochondria in COVID-19 pathogenesis. Am. J. Phys. Cell Phys. 2020;319(2):C258–C267. doi: 10.1152/ajpcell.00224.2020. https://www.ncbi.nlm.nih.gov/pubmed/32510973 ISSN 1522–1563. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kowaltowski A.J., Vercesi A.E. Mitochondrial damage induced by conditions of oxidative stress. Free Radic. Biol. Med. Feb 1999;26(3–4):463–471. doi: 10.1016/s0891-5849(98)00216-0. https://www.ncbi.nlm.nih.gov/pubmed/9895239 ISSN 0891–5849. Disponível em. [DOI] [PubMed] [Google Scholar]
- 47.Stroud D.A., et al. Accessory subunits are integral for assembly and function of human mitochondrial complex I. Nature. Oct 2016;538(7623):123–126. doi: 10.1038/nature19754. https://www.ncbi.nlm.nih.gov/pubmed/27626371 ISSN 1476–4687. Disponível em. [DOI] [PubMed] [Google Scholar]
- 48.Ackermann W.W., Klernschmidt E. Concerning the relation of the Krebs cycle to virus propagation. J. Biol. Chem. Mar 1951;189(1):421–428. https://www.ncbi.nlm.nih.gov/pubmed/14832255 ISSN 0021–9258. Disponível em. [PubMed] [Google Scholar]
- 49.Ackermann W.W., Johnson R.B. Some energy relations in a host-virus system. J. Exp. Med. Mar 1953;97(3):315–322. doi: 10.1084/jem.97.3.315. https://www.ncbi.nlm.nih.gov/pubmed/13052802 ISSN 0022–1007. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ladha J.S., Tripathy M.K., Mitra D. Mitochondrial complex I activity is impaired during HIV-1-induced T-cell apoptosis. Cell Death Differ. Nov 2005;12(11):1417–1428. doi: 10.1038/sj.cdd.4401668. https://www.ncbi.nlm.nih.gov/pubmed/15905875 ISSN 1350–9047. Disponível em. [DOI] [PubMed] [Google Scholar]
- 51.Hu M., et al. Respiratory syncytial virus co-opts host mitochondrial function to favour infectious virus production. Elife. 2019;8 doi: 10.7554/eLife.42448. https://www.ncbi.nlm.nih.gov/pubmed/31246170 ISSN 2050-084X. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.García C.C., et al. Cellular organelles reorganization during zika virus infection of human cells. Front. Microbiol. 2020;11:1558. doi: 10.3389/fmicb.2020.01558. https://www.ncbi.nlm.nih.gov/pubmed/32774331 ISSN 1664-302X. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Di Lisa F., Bernardi P. A CaPful of mechanisms regulating the mitochondrial permeability transition. J. Mol. Cell. Cardiol. Jun 2009;46(6):775–780. doi: 10.1016/j.yjmcc.2009.03.006. https://www.ncbi.nlm.nih.gov/pubmed/19303419 ISSN 1095–8584. Disponível em. [DOI] [PubMed] [Google Scholar]
- 54.Nissanka N., Moraes C.T. Mitochondrial DNA damage and reactive oxygen species in neurodegenerative disease. FEBS Lett. 2018;592(5):728–742. doi: 10.1002/1873-3468.12956. https://www.ncbi.nlm.nih.gov/pubmed/29281123 ISSN 1873–3468. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sharma L.K., Lu J., Bai Y. Mitochondrial respiratory complex I: structure, function and implication in human diseases. Curr. Med. Chem. 2009;16(10):1266–1277. doi: 10.2174/092986709787846578. https://www.ncbi.nlm.nih.gov/pubmed/19355884 ISSN 0929–8673. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tran Q.K., Ohashi K., Watanabe H. Calcium signalling in endothelial cells. Cardiovasc. Res. Oct 2000;48(1):13–22. doi: 10.1016/s0008-6363(00)00172-3. https://www.ncbi.nlm.nih.gov/pubmed/11033104 ISSN 0008–6363. Disponível em. [DOI] [PubMed] [Google Scholar]
- 57.Rapizzi E., et al. Recombinant expression of the voltage-dependent anion channel enhances the transfer of Ca2+ microdomains to mitochondria. J. Cell Biol. Nov 2002;159(4):613–624. doi: 10.1083/jcb.200205091. https://www.ncbi.nlm.nih.gov/pubmed/12438411 ISSN 0021–9525. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shintani Y., et al. Toll-like receptor 9 protects non-immune cells from stress by modulating mitochondrial ATP synthesis through the inhibition of SERCA2. EMBO Rep. Apr 2014;15(4):438–445. doi: 10.1002/embr.201337945. https://www.ncbi.nlm.nih.gov/pubmed/24610369 ISSN 1469–3178. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shintani Y. TLR9 mediates cellular protection by modulating energy metabolism in cardiomyocytes and neurons. Proc. Natl. Acad. Sci. U. S. A. Mar 2013;110(13):5109–5114. doi: 10.1073/pnas.1219243110. https://www.ncbi.nlm.nih.gov/pubmed/23479602 ISSN 1091–6490. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Digard P., et al. Intra-genome variability in the dinucleotide composition of SARS-CoV-2. Virus Evol. Jul 2020;6(2):veaa057. doi: 10.1093/ve/veaa057. https://www.ncbi.nlm.nih.gov/pubmed/33029383 ISSN 2057–1577. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bezemer G.F.G., Garssen J. TLR9 and COVID-19: a multidisciplinary theory of a multifaceted therapeutic target. Front. Pharmacol. 2020;11:601685. doi: 10.3389/fphar.2020.601685. https://www.ncbi.nlm.nih.gov/pubmed/33519463 ISSN 1663–9812. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Conti P., et al. IL-1 induces throboxane-A2 (TxA2) in COVID-19 causing inflammation and micro-thrombi: inhibitory effect of the IL-1 receptor antagonist (IL-1Ra) J. Biol. Regul. Homeost. Agents. Sep-Oct, 2020;34(5):1623–1627. doi: 10.23812/20-34-4EDIT-65. https://www.ncbi.nlm.nih.gov/pubmed/32744052 ISSN 0393-974X. Disponível em. [DOI] [PubMed] [Google Scholar]
- 63.Sanghavi D.K., et al. Endotheliitis, endothelin, and endothelin receptor blockers in COVID-19. Med. Hypotheses. May 2021;150:110564. doi: 10.1016/j.mehy.2021.110564. https://www.ncbi.nlm.nih.gov/pubmed/33823371 ISSN 1532–2777. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Orshal J.M., Khalil R.A. Interleukin-6 impairs endothelium-dependent NO-cGMP-mediated relaxation and enhances contraction in systemic vessels of pregnant rats. Am. J. Phys. Regul. Integr. Comp. Phys. Jun 2004;286(6) doi: 10.1152/ajpregu.00729.2003. https://www.ncbi.nlm.nih.gov/pubmed/15142856 R1013–23. ISSN 0363–6119. Disponível em. [DOI] [PubMed] [Google Scholar]
- 65.Chen G., et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 05 2020;130(5):2620–2629. doi: 10.1172/JCI137244. https://www.ncbi.nlm.nih.gov/pubmed/32217835 ISSN 1558–8238. Disponível em. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 SARS-CoV-2 induced mitochondrial dysfunction