Abstract
Wolbachia, a maternally inherited intracellular bacterial species, can manipulate host insect reproduction by cytoplasmic incompatibility (CI), which results in embryo lethality in crosses between infected males and uninfected females. CI is encoded by two prophage genes, cifA and cifB. Wolbachia, coupled with the sterile insect technique, has been used in field trials to control populations of the dengue vector Aedes albopictus, but CI-inducing strains are not known to infect the malaria vector Anopheles gambiae. Here we show that cifA and cifB can induce conditional sterility in the malaria vector An. gambiae. We used transgenic expression of these Wolbachia-derived genes in the An. gambiae germline to show that cifB is sufficient to cause embryonic lethality and that cifB-induced sterility is rescued by cifA expression in females. When we co-expressed cifA and cifB in male mosquitoes, the CI phenotype was attenuated. In female mosquitoes, cifB impaired fertility, which was overcome by co-expression of cifA. Our findings pave the way towards using CI to control malaria mosquito vectors.
Subject terms: Microbial genetics, Molecular biology
Wolbachia cifB expression in Anopheles gambiae males is sufficient to induce cytoplasmic incompatibility that is sensitive to the expression level of the factors involved.
Main
Wolbachia endosymbionts are successful insect colonizers. Some strains of these bacteria induce cytoplasmic incompatibility (CI) in host insects. CI is the failure of Wolbachia-infected males to produce viable progeny when mated with uninfected females1. Fertility is rescued in females colonized by Wolbachia, providing the endosymbionts with a reproductive advantage that, when paired with maternal transmission, favours invasion of insect populations when infection frequencies reach a certain threshold2–4. Two genes (cifA and cifB) present in WO prophage regions in the Wolbachia genome (with homologues in all known CI-inducing Wolbachia strains) were shown to encode factors that mediate CI in Drosophila melanogaster5,6. While it is known that cifA expression in females rescues fertility7, either one (cifB) or both of these factors are necessary for inducing CI8–12.
Wolbachia biology has attracted considerable interest because of the potential for exploiting CI in the control of vector-borne diseases. Control programmes based on the release of Wolbachia-infected mosquitoes to reduce transmission of dengue and other arboviruses by Aedes mosquitoes have advanced to field trials13–15. Additionally, a strategy known as Incompatible Insect Technique (IIT), which uses the infertility induced by Wolbachia-infected males mating with uninfected females to achieve suppression of insect populations, has been successfully applied in field trials of Aedes mosquitoes16,17.
The implementation of similar Wolbachia-based strategies to tackle malaria-transmitting Anopheles mosquitoes holds appeal because widespread insecticide resistance threatens strategies for vector control18,19. However, Wolbachia does not appear to form stable endosymbiosis with Anopheles species. Although there is evidence that Wolbachia can limit the ability of Plasmodium to infect Anopheles mosquitoes20–25, only one artificial Wolbachia infection has been achieved in the germline of an anopheline species. Moreover, upon endosymbiosis of An. stephensi with a wAlbB strain (a strong CI-inducing strain from Aedes albopictus), only partial rescue of CI and limited capacity for population invasion were observed26. With the exception of one report of high-density Wolbachia infection in An. moucheti and An. demeilloni27, only few low-titre natural Wolbachia infections have been reported in field populations of Anopheles23,28–32, and these findings have been questioned by some researchers33.
We hypothesized that cifA and cifB genes alone might be capable of inducing CI in An. gambiae, the most important malaria vector in Africa, and report our findings here.
Results
cifA and cifB expression in An. gambiae induces embryonic lethality
We chose to use the Type I cif genes, cifA and cifB, from the Wolbachia strain wPip, which induces strong CI in its natural mosquito host, Culex pipiens. Two different nomenclature systems exist for CI in Wolbachia, and the wPip Type I cif genes are also known as CI-inducing deubiquitinases (cidA and cidB)6,34. After codon optimization, we separately cloned each wPip gene under the control of the zero population growth (zpg) promoter35, which drives germline-limited expression in both male and female germ cells of An. gambiae36 (Fig. 1a). Co-injection of zpg-cifA and zpg-cifB constructs yielded F1 transgenics expressing either cifA alone, or both cifA and cifB (zpg-cifA;B), but none that expressed cifB only.
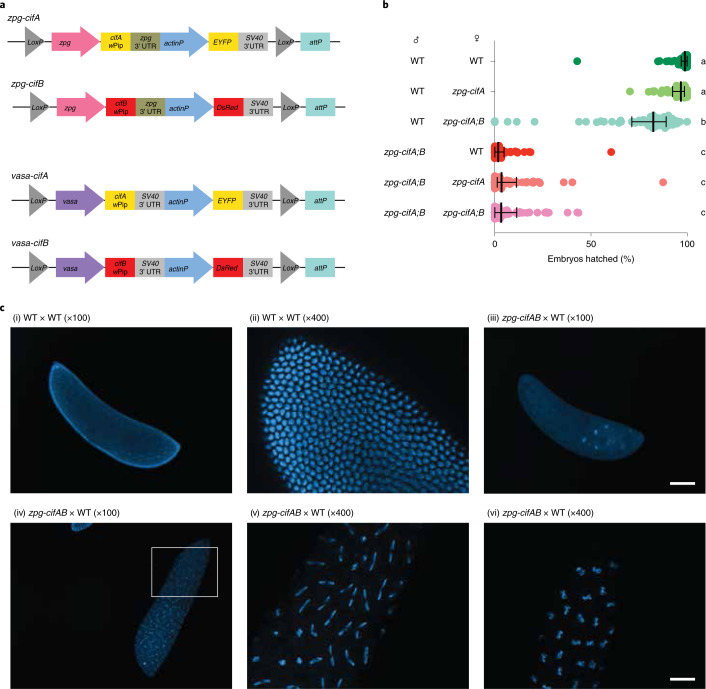
Fig. 1. Co-expression of cifA and cifB in male An. gambiae causes embryonic lethality in progeny.
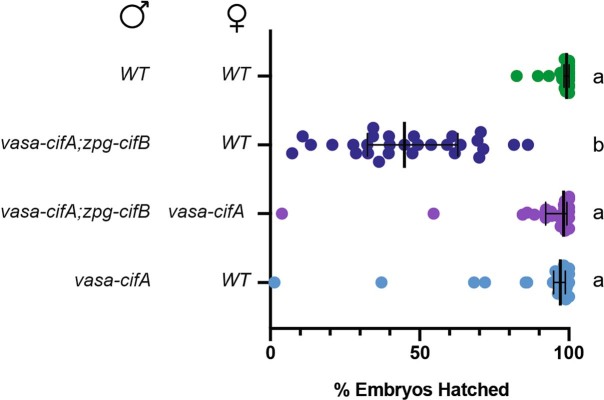
a, Construct design of zpg-cifA, zpg-cifB, vasa-cifA and vasa-cifB. b, Males that express zpg-cifA;B produced largely inviable progeny, regardless of whether their female mate expresses zpg-cifA. Expression of zpg-cifA;B in females caused a decrease in female fertility compared with WT females, but expression of cifA alone did not (Dunn’s multiple comparisons test (two-sided), P ≤ 0.0071 for groups a vs b, P < 0.0001 for groups a vs c and groups b vs c). Median and interquartile ranges are shown. For each group (top to bottom), the n (number of broods) is as follows: 58, 52, 59, 51, 53, 62. Kruskal–Wallis test: H = 265, P < 0.0001, d.f. = 5. c, Embryos from zpg-cifA;B males crossed with WT females (or WT crosses, as controls in (i) and (ii)) were fixed and imaged with DAPI 3–4 h post oviposition, showing developmental arrest of most CI embryos during early nuclear divisions (iii), while some embryos completed multiple rounds of nuclear division but showed mitotic defects, such as chromatin bridging ((iv), with a close-up in (v)), and other chromosomal abnormalities resulting in delayed or arrested development (vi). Scale bars, 100 μm for ×100 images and 25 μm for ×400 images.
We set up crosses between zpg-cifA;B males and different female lines (zpg-cifA;B, zpg-cifA and wild type (WT) females), using WT males as control. In all crosses, females mated to zpg-cifA;B males showed a striking degree of infertility (only 2–4% viable progeny) compared with controls (Fig. 1b). Most infertile embryos were arrested early in development, but a minority initiated development then failed to hatch (Extended Data Fig. 1). Embryo cytology revealed the hallmarks of CI5,6,37, with most embryos showing early developmental arrest, while others showed fewer nuclear divisions or were arrested later in the blastoderm stage due to mitotic failures (Fig. 1c). We did not observe any substantive rescue of infertility when females expressed either cifA alone, or both cifA and cifB (Fig. 1b). We also observed a minor (17%) decrease in fertility of zpg-cifA;B females compared with their WT and zpg-cifA counterparts when mated with WT males (Fig. 1b).
Extended Data Fig. 1. Embryos from zpg-cifA;B males show either early or late arrest.
Brightfield images of eggs 5 days after oviposition from crosses between WT mosquitoes (left) or zpg-cifA;B males and WT females (right). While WT embryos show full development and the standard opening of the hatching cap following larval hatching, embryos from zpg-cifA;B males are inviable and arrested either during early development (EA) with a pale brown color, or late development (LA), which show stemmata, but also present severe abnormalities and do not hatch. Scale bar represents 400μm.
High expression levels of cifA in females rescues CI in An. gambiae
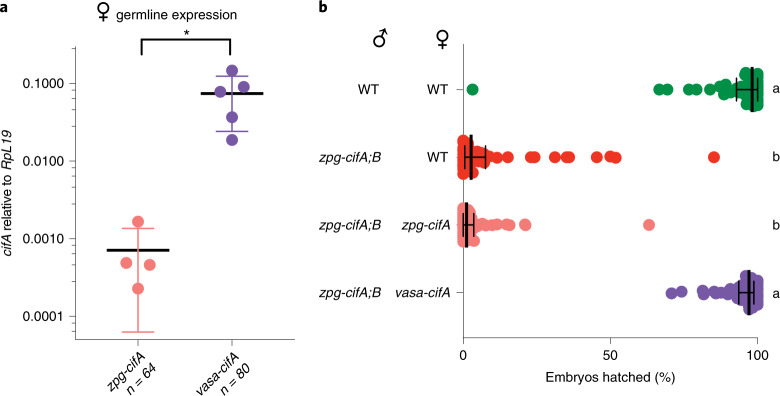
We speculated that the lack of fertility rescue by zpg-cifA could be due to insufficient expression of cifA in females, as the rescue effect has been shown to be promoter-dependent7. To test this possibility, we engineered transgenic expression of cifA from the vasa promoter38 (Fig. 1a) because the vasa promoter has considerably higher expression levels in the female germline than the zpg promoter (Fig. 2a). In D. melanogaster, in addition to different expression levels, vasa has a different expression pattern compared with zpg as it is also expressed in somatic gonadal precursors39, although it is not known whether this is true in Anopheles38. When mated to zpg-cifA;B males, high levels of infertility were observed in both zpg-cifA and WT females as above, but in this case fertility was fully restored in crosses with vasa-cifA females, demonstrating effective rescue by this transgene (Fig. 2b). Combined, these results reveal that CI can be recapitulated in An. gambiae mosquitoes by transgenic expression of cifA and cifB. We also attempted co-injections of vasa-cifA and vasa-cifB constructs (Fig. 1a), but failed to isolate any cifB-expressing progeny.
Fig. 2. High expression of female cifA rescues cifA;B-induced CI in An. gambiae.
a, Transcript abundance of cifA was higher in vasa-cifA females compared with zpg-cifA females relative to RpL19 (unpaired t-test (two-tailed), P = 0.0232, mean and s.d. are shown). b, The expression of vasa-cifA in females rescued infertility caused by zpg-cifA;B expression in males to WT levels, while expression of zpg-cifA in females did not (Dunn’s multiple comparisons test (two-sided), P < 0.0001 for differences between all statistical groups). Median and interquartile ranges are shown. For each group (top to bottom), the n is as follows: 51, 50, 52, 52. Kruskal–Wallis test: H = 153.1, P < 0.0001, d.f. = 3.
Expression of cifB alone in males induces CI
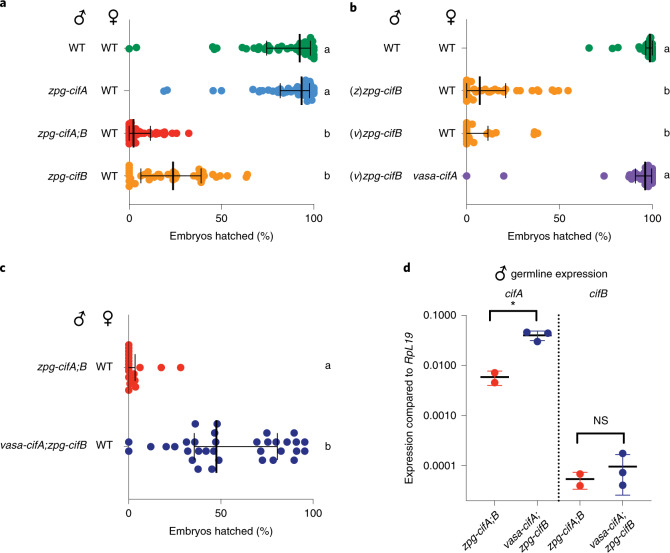
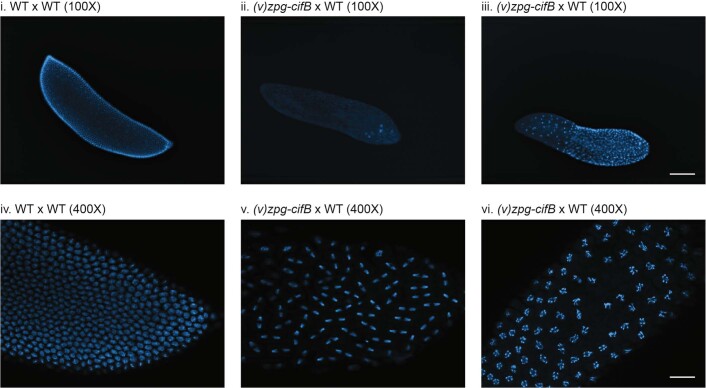
Next, we investigated whether cifB alone can induce CI. Although we could not maintain a zpg-cifB colony in the absence of cifA, we were able to isolate, by fluorescent screening, a limited number of F1 zpg-cifB males from natural colony matings between heterozygous mosquitoes. We found that zpg-cifB males induced high infertility when mated to WT females, at a rate that was statistically indistinguishable from the infertility levels induced by zpg-cifA;B males (Fig. 3a). In contrast, we found that progeny sired by zpg-cifA males were fully fertile (Fig. 3a). CI induction did not differ whether zpg-cifB males were isolated from zpg-cifA;B or vasa-cifA;zpg-cifB colonies (denoted (z)zpg-cifB or (v)zpg-cifB, respectively) (Fig. 3b). We also showed that vasa-cifA expression in females was sufficient to completely rescue sterility caused by zpg-cifB males, ruling out CI-independent effects (Fig. 3b). Cytology of 69 embryos confirmed the results obtained with zpg-cifA;B males, revealing the canonical features of CI (Extended Data Fig. 2). These findings show that conditional sterility can be induced by cifB alone in mosquitos.
Fig. 3. Male cifB expression is sufficient to cause CI, while male cifA attenuates it.
a, zpg-cifB males caused infertility in WT females, while zpg-cifA males did not (Dunn’s multiple comparisons test (two-sided), P ≤ 0.0001 for differences between all statistical groups). Median and interquartile ranges are shown. For each group (top to bottom), the n is as follows: 55, 55, 44, 39. Kruskal–Wallis test: H = 133.8, P < 0.0001, d.f. = 3. b, The expression of vasa-cifA in females rescued infertility caused by (v)zpg-cifB expression in males, which induced CI to the same extent as (z)zpg-cifB males (Dunn’s multiple comparisons test (two-sided), P ≤ 0.0001 for differences between all statistical groups). Median and interquartile ranges are shown. For each group (top to bottom), the n is as follows: 36, 39, 24, 32. Kruskal–Wallis test: H = 95.08, P < 0.0001, d.f. = 3. c, Expression of vasa-cifA;zpg-cifB in males caused only partial induction of CI (Mann–Whitney test (two-tailed), P < 0.0001). Median and interquartile ranges are shown. For each group (top to bottom), the n is as follows: 18, 34. d, Expression of cifA in the male germline was higher in vasa-cifA than in zpg-cifA (unpaired t-test (two-tailed), *P = 0.0135, mean and s.d. are shown), while the expression of cifB was similar (unpaired t-test (two-tailed), P = 0.4882, mean and s.d. are shown). For each group (left to right), the total n is as follows: 32, 48, 32, 48.
Extended Data Fig. 2. Cytology of the progeny of cifB males shows hallmarks of CI.
F1 embryos of crosses between either WT or (v)zpg-cifB males with WT females were stained for DAPI and imaged 3-4 hours after oviposition. At 100X, while WT controls (i. and iv.) show normal development, (v)zpg-cifB embryos show various hallmarks of CI, including (ii.) early arrest, (iii.) regional mitotic failure, and (v., vi.) chromatin bridging or chromosomal abnormalities and delayed or arrested nuclear division. Scale bars represent 100μm for 100X images and 25μm for 400X images.
cifA expression at high levels in males attenuates CI
Given that vasa-cifA rescues inviability caused by cifB in the embryo while zpg-cifA does not, we next asked whether expressing cifA under the vasa promoter in males may impact the strength of CI. To this end, we compared fertility of crosses between males expressing either zpg-cifA;B or vasa-cifA;zpg-cifB and WT females (Fig. 3c). Intriguingly, vasa-cifA;zpg-cifB males were considerably more fertile (median of 48% hatched embryos) compared with zpg-cifA;B males (median of 0% hatched embryos) (Fig. 3c). Consistent with female data, cifA expression was higher in vasa-cifA;zpg-cifB males compared with zpg-cifA;zpg-cifB males, while cifB expression levels were similar (Fig. 3d). Also in this case, the intermediate sterility effects caused by vasa-cifA;zpg-cifB males were rescued when females expressed vasa-cifA (Extended Data Fig. 3). Further, vasa-cifA expression in males did not induce embryonic lethality, supporting the idea that vasa-cifA does not contribute to CI in males and acts solely as a rescue factor (Extended Data Fig. 3). Higher expression of cifA in males (and/or a difference in localization or timing of expression compared with those achieved by the zpg promoter) thus reduces CI penetrance rather than favoring it, possibly either by limiting CifB activity within the male germline, or by rescuing CifB toxicity in the embryo following transfer of CifA in sperm40.
Extended Data Fig. 3. vasa-cifA does not cause CI, and likely inhibits its induction when co-expressed in males.
vasa-cifA expression alone in males does not cause CI, and its expression in females is sufficient to rescue the intermediate CI phenotype caused by expression of vasa-cifA;zpg-cifB in males. (Dunn’s multiple comparisons (two-sided), p≤0.0001 for differences between all statistical groups). Median and interquartile ranges are shown. For each group (top to bottom) the n is as follows: 30, 29, 28, 28. Kruskal-Wallis results: H = 62.87, p < 0.0001, df=3.
cifB expression in females disrupts fertility and fecundity
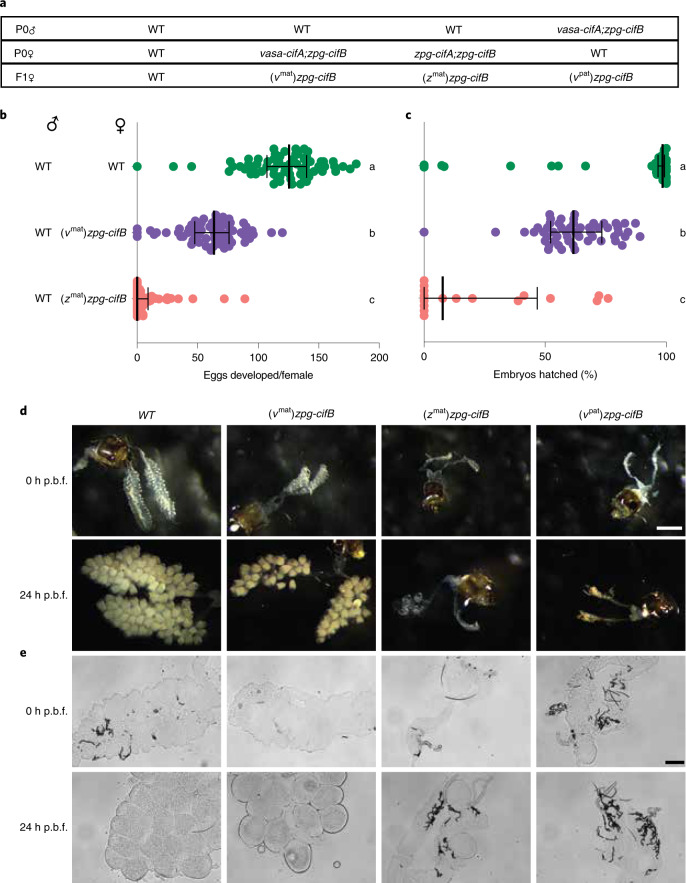
Our finding that cifB expression in males is sufficient to induce significant sterility prompted us to investigate toxicity of this factor in females. We designed crosses between WT males and either zpg-cifA;B or vasa-cifA;zpg-cifB females (Fig. 4a) and then characterized egg development and fertility of the zpg-cifB F1 female progeny after mating to WT males. We noticed that, in contrast to males (Fig. 3b), cifB-mediated effects in females were dependent on the colony of origin. When derived from zpg-cifA;B mothers, most F1 zpg-cifB females (called (zmat)zpg-cifB) failed to develop eggs following a blood meal, and only a few females yielded fertile progeny (Fig. 4b,c). Additionally, morphological analysis of the ovaries before and after ingestion of a blood meal showed that follicles were largely absent, suggestive of defects in germline development (Fig. 4d,e). When derived from vasa-cifA;zpg-cifB mothers, F1 females ((vmat)zpg-cifB) showed intermediate phenotypes with substantial follicle development, although both fecundity and fertility were reduced compared with WT females (Fig. 4b–e). However, when the cifB transgene was inherited from vasa-cifA;zpg-cifB fathers (Fig. 4a), most F1 females ((vpat)zpg-cifB) had ovaries similar to those of (zmat)zpg-cifB females, showing remarkably reduced follicle development (Fig. 4d,e). As the zpg-cifB insertion site and promoter is the same in all these groups, these results reveal rescue effects possibly caused by maternal deposition of cifA (as either mRNA of protein) from vasa-cifA-expressing mothers, although we cannot rule out a difference in other host factors. This is consistent with data showing that transgenes expressed under the vasa promoter, but not the zpg promoter, are maternally deposited38,41. cifB expression is therefore highly deleterious in the female germline when unchecked by the presence of cifA, and it seems to function during the early stages in germline development based on the capacity for maternally derived cifA to rescue these defects.
Fig. 4. cifB expression in females causes severely impaired follicle development in the absence of cifA.
a, Crosses were set up to isolate zpg-cifB females, F1 progeny derived from either mothers that also expressed vasa-cifA ((vmat)zpg-cifB) or zpg-cifA ((zmat)zpg-cifB), or fathers also expressing vasa-cifA ((vpat)zpg-cifB). b, Egg development was nearly abolished in (zmat)zpg-cifB-expressing females, while nearly all (vmat)zpg-cifB females showed egg development, although with decreased numbers of eggs compared with WT females (Dunn’s multiple comparisons test (two-sided), P < 0.0001 for differences between all statistical groups). Medians and interquartile ranges are shown. For each group (top to bottom), the n is as follows: 68, 66, 68. Kruskal–Wallis test: H = 153.9, P < 0.0001, d.f. = 2. c, (zmat)zpg-cifB and (vmat)zpg-cifB females showed impaired fertility compared with WT females (Dunn’s multiple comparisons test (two-sided), P < 0.0257 between groups b and c, P < 0.0001 for other comparisons). Median and interquartile ranges are shown. For each group (top to bottom), the n is as follows: 60, 54, 17. Kruskal–Wallis test: H = 64,64, P < 0.0001, d.f. = 2. d,e, Ovaries from cifB females showed severely impaired follicle development unless derived from a vasa-cifA-expressing mother, when imaged at either 0 h or 24 h post blood feeding (p.b.f.) before fixing under brightfield microscopy (d) (scale bar, 800 μm) or after fixing using differential interference contrast microscopy (e) (scale bar, 100 μm).
Discussion
Using cif genes from wPip in An. gambiae, we show that cifB expression is sufficient to induce embryonic lethality via CI. We show that it is possible to induce and rescue CI in An. gambiae, suggesting that it may be feasible to apply Wolbachia or Wolbachia-derived genes for anopheline vector control13. However, the reproductive toxicity observed in both sexes upon cifB expression may partially explain why infections using CI-inducing Wolbachia strains have been difficult to establish in laboratory colonies of these mosquitoes. Previous efforts to generate cifBwPip-expressing D. melanogaster were unsuccessful6, consistent with our own difficulties in isolating cifB-expressing individuals using two different promoters unless cifA was also co-expressed, and with our results demonstrating cifB toxicity.
Our findings are in contrast with results reported for CI in D. melanogaster where both cifA and cifB from wMel were required to induce CI, and where a cifB transgenic line was isolated in the absence of cifA5. Further, cifBwMel females showed no defects in fertility, contrary to our results5. Many possible reasons could explain these discrepancies, ranging from different promoters and transgene insertion sites to specific differences in CifB function in its natural host (such as in the case of cifB from wMel in D. melanogaster) compared to a novel host (cifB from wPip in An. gambiae). Additionally, the observation that wMel causes weak CI in its natural host D. melanogaster42,43 (though it induces strong CI in Drosophila simulans and Ae. aegypti44,45), while wPip causes strong CI in its natural host C. pipiens46, highlights the possibility for host-dependent and strain-dependent differences. In future studies, it will be interesting to determine whether wMel cifB can induce embryonic lethality in An. gambiae.
Other studies have shown infertility induced by cifB alone in D. melanogaster, induced by wPip’s Type IV cifB homologue (also called cinB) and by the Type I cifB homologue from wRec, a CI-inducing strain found in Drosophila recens47,48. Neither study was able to demonstrate rescue of these effects and thus could not conclude that they were CI related47,48. However, when cifA was expressed alongside cifBwRec in males, very little embryonic lethality was observed, reminiscent of our results showing that high cifA expression in males can attenuate CI47.
Combined with the data we present indicating that high levels of cifA can rescue CI in females but attenuate cifB activity in males, it is possible that Wolbachia may need to fine-tune the relative expression of cifA and cifB in males and females to induce CI in Anopheles mosquitoes without causing lethal toxicity. Such a balancing act might make maintenance of CI-inducing Wolbachia strains in anopheline insects difficult and one outcome could be silencing of the toxic cifB gene by mutation. Interestingly, cifB nonsense mutations were identified by sequencing wAnM and wAnD strains recently discovered in some Anopheles species, although one cifB homologue in wAnD seems to be intact and it will be interesting to learn if this strain can induce CI27. Of note, cifB pseudogenization is not uncommon and is consistent with early evolutionary models that predict male incompatibility not to be selected for within a host lineage49,50.
It may be possible to enable stable colonization of Anopheles by Wolbachia by limiting cifB toxicity using germline expression of cifA. This would create a route to screening for Wolbachia strains that can block transmission of Plasmodium parasites and pave the way to using Wolbachia endosymbiosis in population replacement strategies for malaria control. Plus, the remarkable sterility induced by cifB or cifA;B co-expression could be utilized for sterile male releases to suppress Anopheles populations even in the absence of Wolbachia infection, similar to the IIT programmes implemented in Aedes mosquito control16,17. Due to the difficulty in rearing both cifB and cifA;B mosquitoes, conditional expression of these genes may be required if mass releases were to occur. At a time when novel malaria control strategies are urgently needed, our data presents a step towards utilizing Wolbachia, or Wolbachia-derived genes, in control programmes targeting Anopheles mosquitoes.
Methods
Generation of constructs
The amino acid sequences for cifA (wPa_0282) and cifB (wPa_0283) coding regions from the published wPipI Pel strain of wPip from C. pipiens51 were codon-optimized for expression in An. gambiae using published codon bias information52. Gene blocks were ordered from Integrated DNA Technologies, using custom gene synthesis to create the desired DNA fragments (accession codes OK352257 (cifA) and OK352258 (cifB)). Transgenesis constructs were engineered to express the wPip CI genes cifA and cifB under the control of the germline-specific promoters zpg (zpg, AGAP006241)35 and vasa (vasa2, AGAP008578)38,53. The constructs also express a fluorescent marker under control of the ubiquitous actin promoter to enable selection of transgenic mosquitoes. Integration into the mosquito genome was mediated by piggyBac transposition and rearing lines to homozygosity was accomplished through pupae sorting via fluorescence intensity.
Embryonic micro-injection
PiggyBac transgenic construct pairs corresponding to each germline promoter (zpg-cifA-EYFP and zpg-cifB-DsRed; or vasa-cifA-EYFP and vasa-cifB-DsRed) were co-injected into the posterior of freshly laid embryos from An. gambiae (vasa n = 1,434, zpg n = 512) at a concentration of 250 ng µl−1. Pupae that survived injection were separated according to sex, reared to adulthood, and backcrossed to wild-type G3 to identify and isolate transgenics. A total of 17 EYFP/DsRed double positive F1 transgenics were recovered from the zpg promoter-driven CI constructs injections. In contrast, only vasa-cifA-EYFP positive transgenics were recovered from the vasa-cifA/cifB co-injections. Irrespective of germline promoter, no cifB transgenic mosquitoes were identified post-injection.
Mosquito lines and rearing
An. gambiae mosquitoes from the G3 strain and transgenic derivatives of the G3 strain were maintained in a 27 °C insectary environment with 70–80% humidity and a 12 h light:12 h dark cycle. Adults were given 10% glucose and water ad libitum and fed on human blood (Research Blood Components). Larvae were fed a mixture of Tetramin fish flakes and pellets.
Separate colonies containing the following transgenes were maintained: Colony 1, zpg-cifA (Chromosome 3R insertion); Colony 2, zpg-cifA (3R); zpg-cifB (Chromosome X insertion); Colony 3, vasa-cifA (putative Chromosome 2L insertion (2L*)); Colony 4, vasa-cifA (2L*); zpg-cifB (X); and Colony 5, zpg-cifA (unknown insertion); zpg-cifB (X). To establish Colony 4, zpg-cifB males isolated from Colony 2 or 5 were crossed with vasa-cifA females from Colony 3. All colonies were maintained as heterozygotes and screened for fluorescent markers as pupae to select for the presence of transgenes. For all experiments using zpg-cifA mosquitoes, Colony 1 was used. For all experiments using zpg-cifA;B mosquitoes, Colony 2 was used. For all experiments using vasa-cifA mosquitoes, Colony 4 was used. Experiments using (z)zpg-cifB mosquitoes used mosquitoes isolated from either Colony 2 or 5. Experiments using (v)zpg-cifB males used mosquitoes isolated from Colony 4.
Crosses and fertility assays
To perform crosses between different transgenic lines, individuals were isolated as pupae from these colonies and their transgenes were identified by their respective fluorescent markers. We did not verify whether individuals were homozygous or heterozygous for their transgenes. Pupae were separated by sex under a dissecting microscope, placed in cages with a male to female ratio between 1:1 and 2:1, and allowed to eclose in small BugDorm cages. Natural mating proceeded and mosquitoes were given ad libitum access to 10% glucose solution and water for 5–7 d before blood-feeding females and allowing oviposition in individual cups lined with filter paper. Once laid, eggs were stimulated daily by spraying water and allowed to hatch for a minimum of 4 d. We then assessed fertility of females by counting and scoring eggs under a Leica M80 dissecting microscope, and additionally noting the presence or absence of hatched larvae. For any female that showed no fertile embryos, mating status was verified by checking microscopically for the presence of sperm in the spermatheca. For egg development experiments, egg counts for all females were included regardless of whether they had mated or oviposited, while only those that were mated and oviposited were included in fertility experiments.
RNA extraction and quantitative reverse transcription PCR (RT–qPCR)
Male or female reproductive tracts were dissected in pools of 16, collected in TRI reagent (Thermo Fisher Scientific), and stored at −80 °C. RNA was extracted according to the manufacturer’s instructions, with an additional three ethanol washes of pelleted RNA. Following resuspension, RNA was treated with Turbo DNAse (Thermo Fisher Scientific), quantified with a Nanodrop 2000C (Thermo Fisher Scientific), and then 0.75–2 µg were used in a 100 µl complementary DNA synthesis reaction, following standard protocols. We designed primers for RT–qPCR (QuantStudio 6 pro, Thermo Fisher Scientific) using NCBI PrimerBLAST54 and after evaluating four different primer sets for cifB, we used the following primers for cifA and cifB at the following concentrations: cifAF, 5ʹ tcgccgagctgatcgtgaa 3ʹ (300 nM); cifAR, 5ʹ atcatgtccaggatctccttcttctc 3ʹ (300 nM); cifBF, 5ʹ AGAAGGACCGCCTGATCG 3ʹ (900 nM); cifBR, 5ʹ AGGCTATCGGCGTAGTAGCC 3ʹ (900 nM); RpL19F, 5ʹ CCAACTCGCGACAAAACATTC 3ʹ (300 nM); and RpL19R, 5ʹ ACCGGCTTCTTGATGATCAGA 3ʹ (900 nM). Relative quantification was determined using the 2−(ΔCt (cycle threshold)) equation, using RpL19 as the standard. For female cifA expression (Fig. 2a), transcript levels were not found to be different between samples from cifA only or cifA and cifB co-expressing individuals, so these data were pooled.
Microscopy and tissue staining
Embryo cytology
Embryos were collected from 10–12 WT females after natural matings with zpg-cifA;B males. Four hours after oviposition, embryos were bleached, washed and dechorionated according to methods by Goltsev et al.55, and the endochorion was peeled according to methods by Juhn and James56. Embryos were then fixed and stained with 4ʹ,6-diaminidino-2-phenylindole and imaged on a Zeiss Inverted Observer Z1 at ×100 or ×400 magnification.
Brightfield microscopy of embryos
A sample of oviposited embryos were imaged on filter paper at either ×5 or ×7.5 on a Leica M80 dissecting scope.
Brightfield and differential interference contrast imaging of ovaries
Ovaries of 4–7-day-old females were dissected in PBS at either 0 h or 24 h post-blood-meal and imaged with a Leica M80 dissecting scope at ×7.5 magnification for general morphology. After initial imaging, ovaries were fixed in 4% paraformaldehyde and then mounted in Vectashield mounting medium with 4ʹ,6-diaminidino-2-phenylindole counterstain (Vector Laboratories). Ovaries were then imaged using differential interference contrast on a Zeiss Inverted Observer Z1 at ×100 magnification.
Statistical methods
In all comparisons of fertility or egg development, Anderson–Darling normality tests showed that all data were not normally distributed, so non-parametric Kruskal–Wallis tests with Dunn’s multiple comparisons were used. Distinct samples were used for comparisons. Tests were performed in GraphPad Prism 8. For all fertility or egg development experiments, 2–4 replicates were performed for all groups. We conducted power analysis in G*Power 3.1 to detect a 50% reduction in fertility, yielding n = 5, non-centrality parameter = 21.3, critical chi-squared = 11.07 and total sample size = 10 to estimate the sample size required for detecting differences. On average, we used far greater sample sizes than this power analysis suggested, as we planned to use more stringent tests for non-parametric data (which cannot be estimated by power analysis). Only one replicate was performed for embryo cytology experiments. For Fig. 4d,e, where representative images were selected, one replicate of dissection and imaging of 5–10 individuals from each group was performed; however, these phenotypes were confirmed by dissections performed in experiments for Fig. 4b,c. For all RT–qPCR data, 2–5 technical replicates with 16 individuals each were performed for each group, with exact n given in figure legends.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We thank W. R. Shaw for careful reading of the manuscript and advice, and A. Smidler for help with construct design. This study was funded by a joint Howard Hughes Medical Institute (HHMI) and Bill and Melinda Gates Foundation (BMGF) Faculty Scholars Award to F.C. (Grant ID: OPP1158190), a BMGF research grant (Grant ID: OPP1174120) and by a fellowship of the National Sciences and Engineering and Research Council of Canada (NSERC) to K.A. The findings and conclusions in this publication are those of the authors and do not necessarily reflect positions or policies of the HHMI, the BMGF or the NSERC.
Extended data
Source data
Statistical source data and exact P values.
Statistical source data and exact P values.
Statistical source data and exact P values.
Statistical source data and exact P values.
Statistical source data and exact P values.
Author contributions
K.L.A. contributed to literature searches, study design, data collection, data analysis, data interpretation, figure creation and writing; D.G.A. contributed to literature searches, study design, data collection, data analysis, data interpretation and writing; B.C.W. contributed to literature searches, study design, data analysis and data collection; E.K.S. contributed to data analysis and data collection; M.A.I. contributed to data collection; F.C. supervised the project and contributed to study design, data analysis, data interpretation, figure creation and writing. The corresponding authors had full access to all data in the study and final responsibility in the decision to submit for publication.
Data availability
Source data are provided with this paper. Sequence information can be found in GenBank with the accession numbers OK352257 (codon-optimized cifA) and OK352258 (codon-optimized cifB).
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Nature Microbiology thanks Francis Jiggins and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Kelsey L. Adams, Daniel G. Abernathy.
Change history
3/7/2022
A Correction to this paper has been published: 10.1038/s41564-022-01098-9
Extended data
is available for this paper at 10.1038/s41564-021-00998-6.
Supplementary information
The online version contains supplementary material available at 10.1038/s41564-021-00998-6.
References
- 1.Laven H. Crossing experiments with Culex strains. Evolution. 1951;5:370–375. [Google Scholar]
- 2.Werren JH, Baldo L, Clark ME. Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol. 2008;6:741–751. doi: 10.1038/nrmicro1969. [DOI] [PubMed] [Google Scholar]
- 3.Turelli M, Hoffmann AA. Rapid spread of an inherited incompatibility factor in California Drosophila. Nature. 1991;353:440–442. doi: 10.1038/353440a0. [DOI] [PubMed] [Google Scholar]
- 4.Turelli M. Cytoplasmic incompatibility in populations with overlapping generations. Evolution. 2010;64:232–241. doi: 10.1111/j.1558-5646.2009.00822.x. [DOI] [PubMed] [Google Scholar]
- 5.LePage DP, et al. Prophage WO genes recapitulate and enhance Wolbachia-induced cytoplasmic incompatibility. Nature. 2017;543:243–247. doi: 10.1038/nature21391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beckmann JF, Ronau JA, Hochstrasser M. A Wolbachia deubiquitylating enzyme induces cytoplasmic incompatibility. Nat. Microbiol. 2017;2:17007. doi: 10.1038/nmicrobiol.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shropshire JD, On J, Layton EM, Zhou H, Bordenstein SR. One prophage WO gene rescues cytoplasmic incompatibility in Drosophila melanogaster. Proc. Natl Acad. Sci. USA. 2018;115:4987–4991. doi: 10.1073/pnas.1800650115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beckmann JF, et al. The toxin–antidote model of cytoplasmic incompatibility: genetics and evolutionary implications. Trends Genet. 2019;35:175–185. doi: 10.1016/j.tig.2018.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shropshire JD, et al. Models and nomenclature for cytoplasmic incompatibility: caution over premature conclusions – a response to Beckmann et al. Trends Genet. 2019;35:397–399. doi: 10.1016/j.tig.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 10.Shropshire JD, Bordenstein SR. Two-by-one model of cytoplasmic incompatibility: synthetic recapitulation by transgenic expression of cifA and cifB in Drosophila. PLoS Genet. 2019;15:e1008221–e1008221. doi: 10.1371/journal.pgen.1008221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beckmann JF, et al. Caution does not preclude predictive and testable models of cytoplasmic incompatibility: a reply to Shropshire et al. Trends Genet. 2019;35:399–400. doi: 10.1016/j.tig.2019.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shropshire JD, Leigh B, Bordenstein SR. Symbiont-mediated cytoplasmic incompatibility: what have we learned in 50 years? eLife. 2020;9:e61989. doi: 10.7554/eLife.61989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoffmann AA, et al. Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature. 2011;476:454–457. doi: 10.1038/nature10356. [DOI] [PubMed] [Google Scholar]
- 14.Hoffmann AA, et al. Stability of the wMel Wolbachia infection following invasion into Aedes aegypti populations. PLoS Negl. Trop. Dis. 2014;8:e3115. doi: 10.1371/journal.pntd.0003115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Neill SL. The use of Wolbachia by the World Mosquito Program to interrupt transmission of Aedes aegypti transmitted viruses. Adv. Exp. Med. Biol. 2018;1062:355–360. doi: 10.1007/978-981-10-8727-1_24. [DOI] [PubMed] [Google Scholar]
- 16.Crawford JE, et al. Efficient production of male Wolbachia-infected Aedes aegypti mosquitoes enables large-scale suppression of wild populations. Nat. Biotechnol. 2020;38:482–492. doi: 10.1038/s41587-020-0471-x. [DOI] [PubMed] [Google Scholar]
- 17.Zheng X, et al. Incompatible and sterile insect techniques combined eliminate mosquitoes. Nature. 2019;572:56–61. doi: 10.1038/s41586-019-1407-9. [DOI] [PubMed] [Google Scholar]
- 18.World Malaria Report 2020: 20 Years of Global Progress and Challenges (WHO, 2020).
- 19.Ranson H, Lissenden N. Insecticide resistance in African Anopheles mosquitoes: a worsening situation that needs urgent action to maintain malaria control. Trends Parasitol. 2016;32:187–196. doi: 10.1016/j.pt.2015.11.010. [DOI] [PubMed] [Google Scholar]
- 20.Hughes GL, Koga R, Xue P, Fukatsu T, Rasgon JL. Wolbachia infections are virulent and inhibit the human malaria parasite Plasmodium falciparum in Anopheles gambiae. PLoS Pathog. 2011;7:e1002043. doi: 10.1371/journal.ppat.1002043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kambris Z, et al. Wolbachia stimulates immune gene expression and inhibits Plasmodium development in Anopheles gambiae. PLoS Pathog. 2010;6:e1001143. doi: 10.1371/journal.ppat.1001143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shaw WR, et al. Wolbachia infections in natural Anopheles populations affect egg laying and negatively correlate with Plasmodium development. Nat. Commun. 2016;7:11772. doi: 10.1038/ncomms11772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gomes FM, et al. Effect of naturally occurring Wolbachia in Anopheles gambiae s.l. mosquitoes from Mali on Plasmodium falciparum malaria transmission. Proc. Natl Acad. Sci. USA. 2017;114:12566–12571. doi: 10.1073/pnas.1716181114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dutra HLC, et al. Wolbachia blocks currently circulating Zika virus isolates in Brazilian Aedes aegypti mosquitoes. Cell Host Microbe. 2016;19:771–774. doi: 10.1016/j.chom.2016.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moreira LA, et al. A Wolbachia symbiont in Aedes aegypti limits infection with dengue, Chikungunya, and Plasmodium. Cell. 2009;139:1268–1278. doi: 10.1016/j.cell.2009.11.042. [DOI] [PubMed] [Google Scholar]
- 26.Bian G, et al. Wolbachia invades Anopheles stephensi populations and induces refractoriness to Plasmodium infection. Science. 2013;340:748–751. doi: 10.1126/science.1236192. [DOI] [PubMed] [Google Scholar]
- 27.Walker, T. et al. Stable high-density and maternally inherited Wolbachia infections in Anopheles moucheti and Anopheles demeilloni mosquitoes. Curr. Biol.10.1016/j.cub.2021.03.056 (2021). [DOI] [PMC free article] [PubMed]
- 28.Baldini F, et al. Evidence of natural Wolbachia infections in field populations of Anopheles gambiae. Nat. Commun. 2014;5:3985. doi: 10.1038/ncomms4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baldini F, et al. First report of natural Wolbachia infection in the malaria mosquito Anopheles arabiensis in Tanzania. Parasit. Vectors. 2018;11:635. doi: 10.1186/s13071-018-3249-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeffries CL, et al. Novel Wolbachia strains in Anopheles malaria vectors from Sub-Saharan Africa. Wellcome Open Res. 2018;3:113. doi: 10.12688/wellcomeopenres.14765.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niang EHA, et al. First report of natural Wolbachia infection in wild Anopheles funestus population in Senegal. Malar. J. 2018;17:408. doi: 10.1186/s12936-018-2559-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ayala D, et al. Natural Wolbachia infections are common in the major malaria vectors in Central Africa. Evol. Appl. 2019;12:1583–1594. doi: 10.1111/eva.12804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chrostek E, Gerth M. Is Anopheles gambiae a natural host of Wolbachia? mBio. 2019;10:e00784–19. doi: 10.1128/mBio.00784-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beckmann JF, Sharma GD, Mendez L, Chen H, Hochstrasser M. The Wolbachia cytoplasmic incompatibility enzyme CidB targets nuclear import and protamine-histone exchange factors. eLife. 2019;8:e50026. doi: 10.7554/eLife.50026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kyrou K, et al. A CRISPR–Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes. Nat. Biotechnol. 2018;36:1062–1066. doi: 10.1038/nbt.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tazuke SI, et al. A germline-specific gap junction protein required for survival of differentiating early germ cells. Development. 2002;129:2529–2539. doi: 10.1242/dev.129.10.2529. [DOI] [PubMed] [Google Scholar]
- 37.Landmann F, Orsi GA, Loppin B, Sullivan W. Wolbachia-mediated cytoplasmic incompatibility is associated with impaired histone deposition in the male pronucleus. PLoS Pathog. 2009;5:e1000343. doi: 10.1371/journal.ppat.1000343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Papathanos PA, Windbichler N, Menichelli M, Burt A, Crisanti A. The vasa regulatory region mediates germline expression and maternal transmission of proteins in the malaria mosquito Anopheles gambiae: a versatile tool for genetic control strategies. BMC Mol. Biol. 2009;10:65. doi: 10.1186/1471-2199-10-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Renault AD. vasa is expressed in somatic cells of the embryonic gonad in a sex-specific manner in Drosophila melanogaster. Biol. Open. 2012;1:1043–1048. doi: 10.1242/bio.20121909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beckmann JF, Fallon AM. Detection of the Wolbachia protein WPIP0282 in mosquito spermathecae: implications for cytoplasmic incompatibility. Insect Biochem. Mol. Biol. 2013;43:867–878. doi: 10.1016/j.ibmb.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hammond A, et al. Regulating the expression of gene drives is key to increasing their invasive potential and the mitigation of resistance. PLoS Genet. 2021;17:e1009321. doi: 10.1371/journal.pgen.1009321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bourtzis K, Nirgianaki A, Markakis G, Savakis C. Wolbachia infection and cytoplasmic incompatibility in Drosophila species. Genetics. 1996;144:1063–1073. doi: 10.1093/genetics/144.3.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reynolds KT, Hoffmann AA. Male age, host effects and the weak expression or non-expression of cytoplasmic incompatibility in Drosophila strains infected by maternally transmitted Wolbachia. Genet. Res. 2002;80:79–87. doi: 10.1017/s0016672302005827. [DOI] [PubMed] [Google Scholar]
- 44.Poinsot D, Bourtzis K, Markakis G, Savakis C, Merçot H. Wolbachia transfer from Drosophilamelanogaster into D. simulans: host effect and cytoplasmic incompatibility relationships. Genetics. 1998;150:227–237. doi: 10.1093/genetics/150.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walker T, et al. The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature. 2011;476:450–453. doi: 10.1038/nature10355. [DOI] [PubMed] [Google Scholar]
- 46.Duron O, Weill M. Wolbachia infection influences the development of Culex pipiens embryo in incompatible crosses. Heredity. 2006;96:493–500. doi: 10.1038/sj.hdy.6800831. [DOI] [PubMed] [Google Scholar]
- 47.Shropshire JD, Rosenberg R, Bordenstein SR. The impacts of cytoplasmic incompatibility factor (cifA and cifB) genetic variation on phenotypes. Genetics. 2020;217:iyaa007. doi: 10.1093/genetics/iyaa007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen H, Ronau JA, Beckmann JF, Hochstrasser M. A Wolbachia nuclease and its binding partner provide a distinct mechanism for cytoplasmic incompatibility. Proc. Natl Acad. Sci. USA. 2019;116:22314–22321. doi: 10.1073/pnas.1914571116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meany MK, et al. Loss of cytoplasmic incompatibility and minimal fecundity effects explain relatively low Wolbachia frequencies in Drosophila mauritiana. Evolution. 2019;73:1278–1295. doi: 10.1111/evo.13745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Turelli M. Evolution of incompatibility-inducing microbes and their hosts. Evolution. 1994;48:1500–1513. doi: 10.1111/j.1558-5646.1994.tb02192.x. [DOI] [PubMed] [Google Scholar]
- 51.Klasson L, et al. Genome evolution of Wolbachia strain wPip from the Culex pipiens group. Mol. Biol. Evol. 2008;25:1877–1887. doi: 10.1093/molbev/msn133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Volohonsky G, et al. Tools for Anopheles gambiae transgenesis. G3. 2015;5:1151–1163. doi: 10.1534/g3.115.016808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Werling K, et al. Steroid hormone function controls non-competitive Plasmodium development in Anopheles. Cell. 2019;177:315–325.e314. doi: 10.1016/j.cell.2019.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ye J, et al. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goltsev Y, Hsiong W, Lanzaro G, Levine M. Different combinations of gap repressors for common stripes in Anopheles and Drosophila embryos. Dev. Biol. 2004;275:435–446. doi: 10.1016/j.ydbio.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 56.Juhn, J. & James, A. A. Hybridization in situ of salivary glands, ovaries, and embryos of vector mosquitoes. J. Vis. Exp.10.3791/3709 (2012). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data are provided with this paper. Sequence information can be found in GenBank with the accession numbers OK352257 (codon-optimized cifA) and OK352258 (codon-optimized cifB).