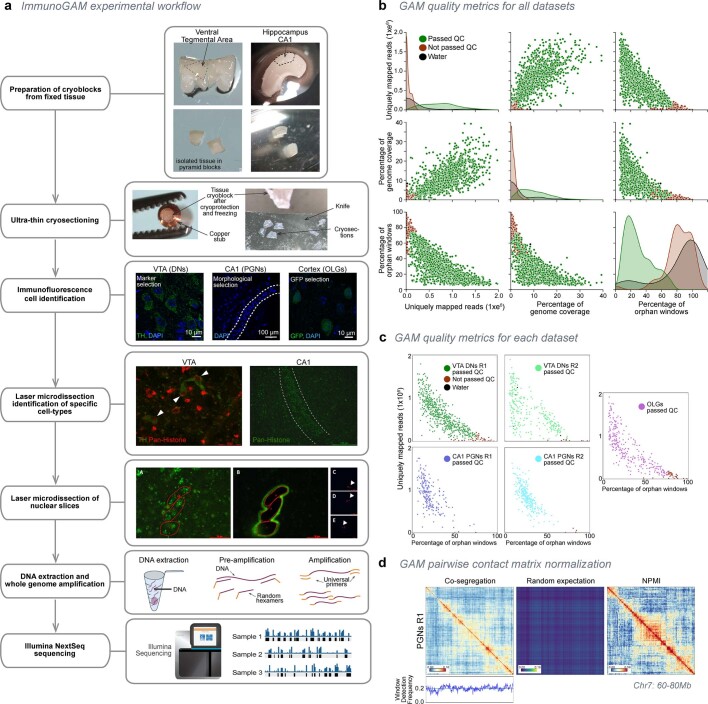
Extended Data Fig. 1. ImmunoGAM experimental pipeline and GAM data quality control.
a, ImmunoGAM experimental pipeline. VTA and CA1 dissections and cryoblock preparations are shown as examples. After fixation, brain tissue is dissected and cryopreserved in sucrose/PBS solution, before sectioning on an ultracryomicrotome (~220nm thick tissue slices; −100 °C). For confocal imaging, DAPI staining labels nuclear slices and helps to morphologically identify the CA1 PGN layer in the hippocampus, or was combined with TH immunolabelling to identify DNs in the midbrain, or with GFP immunolabelling to identify OLG lineage cells in the cortex (scale bars = 10 μm for OLGs and DNs, 100 μm for PGNs). For laser microdissection, nuclei were identified by indirect immunofluorescence using anti-pan-histone antibodies to morphologically select PGNs of the pyramidal neuron layer, or were combined with immunofluorescence detection of TH for DNs or GFP for OLGs. Laser microdissection images are shown as examples (scale bars = 30 μm for DNs, 200 μm for PGNs). Three nuclear slices were selected and laser microdissected from the tissue to fall into the same PCR lid, as described for multiplex-GAM9 (scale bars = 30 μm for panels a and b, 400 μm for panels c-e). Genomic DNA content was extracted from each sample and amplified using whole-genome amplification, followed by Illumina NextSeq sequencing. b, Quality control parameters (uniquely mapped reads, genome coverage of positive windows, and percentage of orphan windows; see Methods) for all combined GAM samples collected from brain cell types. Each data point represents a GAM sample. Samples passing QC are shown in green, samples not passing QC in red. c, Percentages of uniquely mapped reads and orphan windows per GAM sample, shown separately for each dataset produced in this study. Samples not passing QC are shown in red, water control samples (laser-microdissected material not containing a nuclear profile) are shown in black. d, Normalized point-wise mutual information (NPMI) normalization corrects for differences in the co-segregation matrix caused by changes in the window detection frequency (WDF; see Methods). Example shown for PGNs replicate 1 (R1; chr7:60,000,000-80,000,000).