Abstract
Background:
Although several candidate-gene association studies have been conducted to investigate noise-induced hearing loss (NIHL) in humans, most are underpowered, unreplicated and account for only a fraction of the genetic risk. Mouse genome-wide association studies (GWASs) have revolutionized the field of genetics and have led to the discovery of hundreds of genes involved in complex traits. The Hybrid Mouse Diversity Panel (HMDP) is a collection of classic inbred and recombinant inbred strains whose genomes have been either genotyped at high resolution or sequenced. To further investigate the genetics of NIHL, we report the first GWAS based on distortion product otoacoustic emission (DPOAE) measurements and the HMDP.
Methods:
A total of 102 strains (n = 635) from the HMDP were evaluated based on DPOAE suprathreshold amplitudes before and after noise exposure. DPOAE amplitude variation was set at 60 dB and 70 dB SPL of the primary tones for each frequency separately (8, 11.3, 16, 22.6, and 32 kHz). These values provided an indirect assessment of outer hair cell integrity. Six-week-old mice were exposed for 2 hours to 10 kHz octave-band noise at 108 dB SPL. To perform local expression quantitative trait locus (eQTL) analysis, gene expression microarray profiles were generated using cochlear RNA from 64 hybrid mouse strains (n = 3 arrays per strain).
Results:
Several new loci were identified and positional candidate genes associated with NIHL were prioritized, especially after noise exposure (1 locus at baseline and 5 loci after exposure). A total of 35 candidate genes in these 6 loci were identified with at least 1 probe whose expression was regulated by a significant cis-eQTL in the cochlea. After careful analysis of the candidate genes based on cochlear gene expression, 2 candidate genes were prioritized: Eya1 (baseline) and Efr3a (post-exposure).
Discussion and Conclusion:
For the first time, an association analysis with correction for population structure was used to map several loci for hearing traits in inbred strains of mice based on DPOAE suprathreshold amplitudes before and after noise exposure. Our results identified a number of novel loci and candidate genes for susceptibility to NIHL, especially the Eya1 and Efr3a genes. Our findings validate the power of the HMDP for detecting NIHL susceptibility genes.
Keywords: Animal models, Distortion product otoacoustic emission, Genome wide association study, Noise induced hearing loss, Noise
Introduction
According to the Centers for Disease Control, up to 25% of the U.S. adult population may develop noise-induced hearing loss (NIHL) [Kujawa & Liberman, 2009; Liberman, 2017]. In addition to human injury, NIHL generates a considerable economic impact. An estimated $1.2 billion are spent annually in the United States on disability compensation for NIHL [Yankaskas, 2013].
There is evidence for a genetic contribution to NIHL. A study evaluating the genetic component of NIHL in twins showed an approximately 36% contribution of inheritance to this condition [Heinonen-Guzejev et al., 2004]. Elucidating specific genetic effects is critical to understanding complex diseases such as NIHL. Although some studies of gene-environment interactions in human hearing loss have been conducted, only a few of them have been replicated to date [Lavinsky et al., 2015].
A major difficulty in human studies is the inability to control or quantify environmental exposures consistently, which results in a lack of well-characterized and well-monitored populations for study. This is notably the case of NIHL, for which only a few ill-designed and poorly characterized studies have been published [Lavinsky et al., 2015]. For this reason, many researchers are turning to animal model organisms.
The study of hearing loss in mouse models appears to be a valid method, since the mouse inner ear is structurally similar to the human inner ear and their genetic material is homologous to a considerable extent. This can create an identical environment that avoids confounding factors and allows analysis through a genome-wide association study (GWAS) using inbred mouse lines from the Hybrid Mouse Diversity Panel (HMDP).
Studies evaluating NIHL in mice have characterized different susceptibility genes for NIHL using different methods [Sliwinska-Kowalska & Pawelczyk, 2013]. An example is the identification that some mouse strains (C57BL/6J) are more susceptible to noise than others [Erway et al., 1996]. Knockout mice tested by auditory brainstem response (ABR) with targeted mutation of SOD1−/− [Ohlemiller et al., 1999], GPX1−/− [Ohlemiller et al., 2000], and PMCA2−/− [Kozel et al., 2002] were more sensitive to noise than their wild-type littermates. CDH23+/− mice were also characterized as more susceptible to noise [Holme & Steel, 2004], with their auditory function tested by measuring compound action potential (CAP) thresholds.
Lavinsky et al. (2015), in a GWAS with correction for population structure, described for the first time the mapping of several susceptibility loci for NIHL in inbred mouse strains using ABR. The analysis revealed a genome-wide significant peak on chromosome 17 within a haplotype block containing NADPH oxidase-3 (Nox3), providing evidence to support its role in susceptibility to NIHL. In addition, frequency-specific genetic susceptibility to hearing loss after noise exposure was demonstrated within the mouse cochlea [Lavinsky et al., 2015].
Based on what has already been discovered, this study aimed to investigate other auditory phenotypes using a different method of auditory assessment, the distortion product otoacoustic emission (DPOAE) suprathreshold amplitude. This method indirectly assesses the outer hair cells, which are affected both before and after noise exposure.
Materials and Methods
Hybrid Mouse Diversity Panel (HMDP)
A detailed description of the HMDP (strain selection, statistical power, and mapping resolution) is provided by Bennett et al. (2010). Four 4-week-old female mice of each HMDP strain were purchased from the Jackson Laboratory (Bar Harbor, ME). Only female mice were tested to avoid potential confounding effects of sex. The mice were allowed to acclimate to a common environment for 1 week (until 5 weeks of age). We used 5-week-old mice to minimize potential effects of age-related hearing loss that could contribute to our phenotype. All mice were fed a standard chow diet until sacrifice.
Genotyping
Classic inbred and recombinant inbred strains were previously genotyped by the Broad Institute (www.mousehapmap.org). Of the 140,000 single-nucleotide polymorphisms (SNPs) available, 108,064 were informative and used in the association analysis.
Noise Exposure Protocol
Using a method adapted from Kujawa and Liberman (2009), at 6 weeks of age, mice were exposed for 2 hours to 10 kHz octave-band noise (OBN) at 108 dB SPL. The noise exposure protocol was previously described by White et al. (2009). The cage was arranged in a soundproof chamber (MAC-1) created by Industrial Acoustics (IAC, Bronx, NY). The sound chamber was lined with soundproofing acoustic foam. Noise recordings were performed with a speaker (Fostex FT17H Tweeter) built on the top of the sound chamber. Harmful noise was calibrated with a B&K sound level meter, with a variation of 1.5 dB across the cage. The mice were placed in a circular exposure cage with 4 compartments and, for 2 hours, they were allowed to move around within the compartment. Testing involved the right ear only.
Determination of DPOAE Suprathreshold Amplitudes
DPOAEs were analyzed as input/output functions at 2f1-f2 (primary measure). The primary tones were set at a ratio of f2/f1 = 1.2, with f2 ranging from 8 to 32 kHz (f2 level adjusted at 10 dB lower than f1 level) and L2 ranging from 60 to 70 dB. The noise level was calculated as the average of 6 spectral points (above and below 2f1-f2). Both waveform and spectral mean DPOAEs were then extracted. The L2 level required to produce 0 dB SPL with signal-to-noise ratio (SNR) was 3 dB. DPOAE amplitude variation was set at 60 dB and 70 dB SPL of the primary tones for each frequency separately. These values provided an indirect assessment of outer hair cell integrity.
Results
Genome-Wide Association Analysis of NIHL Reveals Frequency-Specific Genetic Susceptibility
The efficient mixed-model association (EMMA) algorithm was applied to each phenotype separately to identify genetic associations for the 6 tone-burst stimuli [Kang et al., 2008]. Adjusted association p-values were calculated for the 108,064 SNPs with minor allele frequency > 5% (p < 0.05 genome-wide equivalent for genome-wide association using EMMA in the HMDP is p = 4.1 × 10−6, −log10p = 5.39). At this threshold, genome-wide significant associations were identified on chromosome 1 at baseline and on chromosomes 2, 3, 15, and 16 after noise exposure.
GWAS and DPOAE Suprathreshold Amplitudes at Each Frequency Tested
An association analysis was performed for each phenotype separately in order to identify genetic associations. A GWAS was performed both at baseline (Figure 1A) and after noise exposure (Figure 1B, 1C, 1D, and 1E) to identify NIHL-associated loci at the various frequencies tested.
Figure 1. Manhattan Plots: DPOAE GWAS.
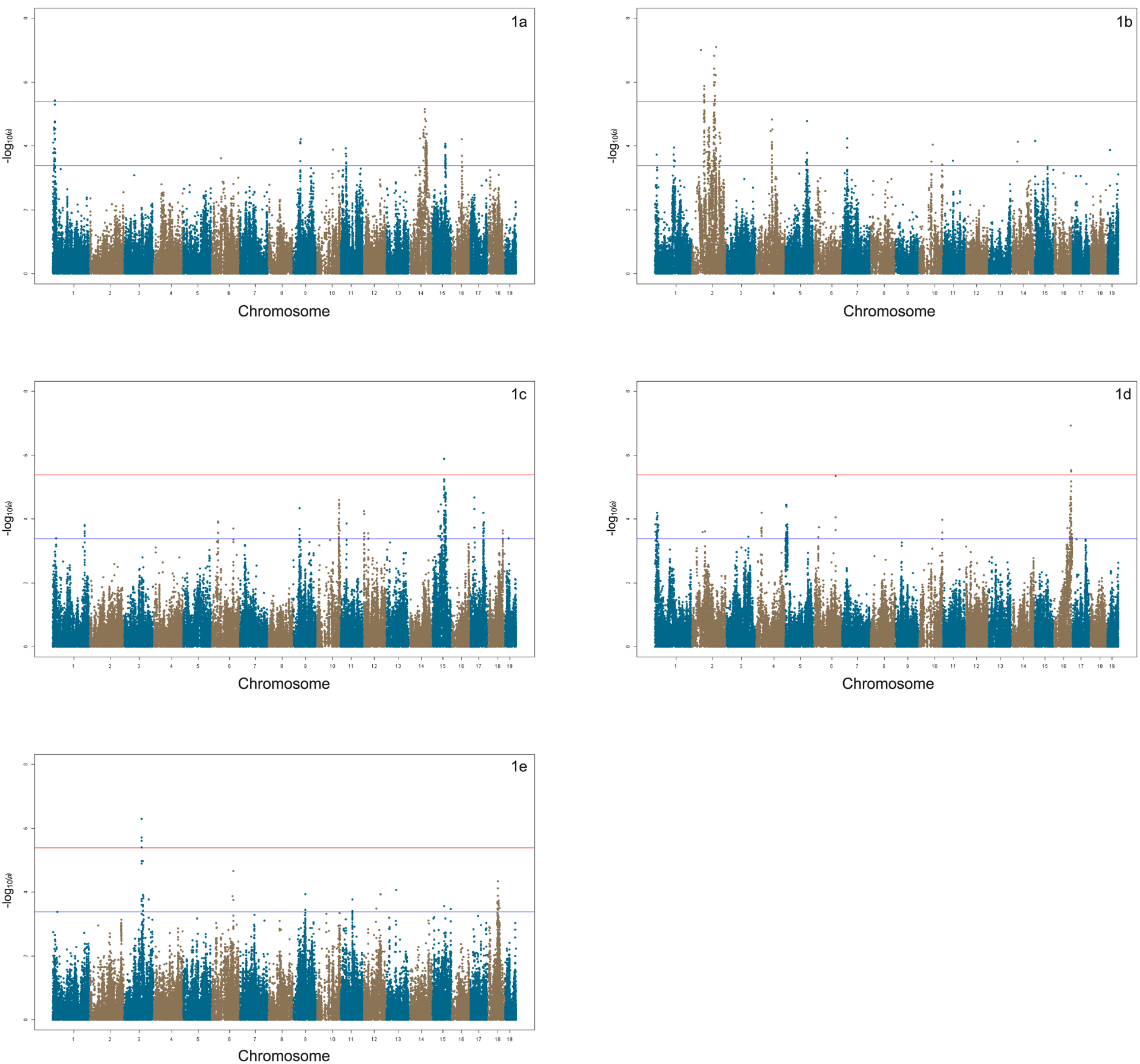
Association peaks (−log10) of p-values (−log10p) for 32 kHz - 70 dB (1a) before noise exposure and 8 kHz - 70 dB (1b), 11.3 kHz - 70 dB (1c), 16 kHz - 60 dB (1d), and 32 kHz - 70 dB (1e) after noise exposure (HMDP inbred strains). Analysis was performed using approximately 200,000 SNPs with minor allele frequency > 5%. Each chromosome is plotted on the x-axis in alternating brown and blue colors.
Association Analysis and Regional Plots
At baseline, a DPOAE association exceeding the genome-wide significance threshold was identified on chromosome 1 (Figure 2A). After exposure, the associations were statistically significant on chromosome 2 (Figure 2B and 2C), chromosome 15 (Figure 2D), chromosome 16 (Figure 2E), and chromosome 3 (Figure 2F).
Figure 2.
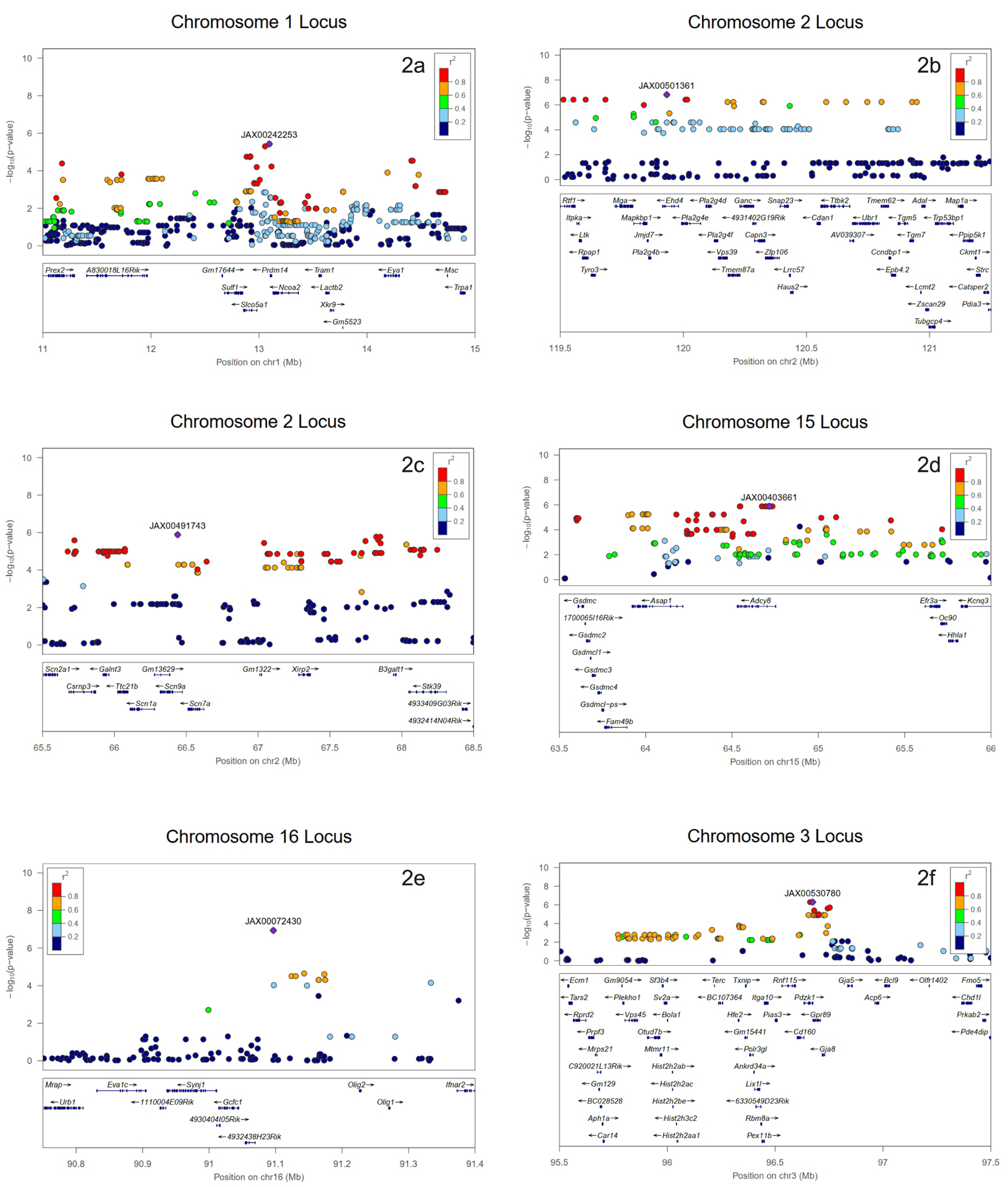
Association peak centered on the lead SNP for 32 kHz - 70 dB (2a) before noise exposure and 8 kHz - 70 dB (2b and 2c), 11.3 kHz - 70 dB (2d), 16 kHz - 60 dB (2e), and 23 kHz - 70 dB (2f) after noise exposure (HMDP inbred strains). The blue diamond represents the most significant SNP. Other SNPs are colored based on their linkage disequilibrium (LD), as follows: red SNPs in LD at r2 > 0.8, orange SNPs in LD at r2 > 0.6, and green SNPs in LD at r2 > 0.4. The positions of all RefSeq genes are plotted using genomic sites (NCBI Build 37).
Prioritizing Our Candidate Genes
Our cochlear expression data allowed us to analyze all 144 candidate genes in each association interval. Genes were identified within each of the intervals regulated by a local expression quantitative trait locus (eQTL). In order to perform eQTL analysis, gene expression microarray profiles were generated using cochlear RNA from 64 hybrid mouse strains (n = 3 arrays per strain). Subsequently, EMMA was used to perform an association analysis between all SNPs and array probes mapping within each region. After excluding the probes that overlapped SNPs present among the classic inbred strains used in the HMDP, as described in the methods, 18,138 genes were represented by at least 1 probe.
Loci in which peak SNPs mapped within 2 Mb of the gene whose expression was regulated were considered “local” or cis-acting eQTLs, while SNPs mapped elsewhere were considered “distal” and presumably trans-acting eQTLs. The significant p-value cutoff (p = 1 × 10−6) was calculated for local and distal associations. These genes were prioritized based on whether they were regulated by a local eQTL.
A total of 35 genes in the 6 identified loci had at least 1 probe whose expression was regulated by a significant cis-eQTL in the cochlea (Table 1). The functions of candidate genes and associated diseases are shown in Table 2.
Table 1.
Association peaks of candidate genes within cis-regulated eQTLs in the cochlea.
| Gene | RefSeq | Chr | txStart (bp)a | txEnd (bp)b | Local eQTL Pc |
|---|---|---|---|---|---|
| Xkr9 | ILMN_3159558 | 1 | 13668771 | 13701723 | 1.79E-01 |
| Eya1 | ILMN_2638487 | 1 | 14168954 | 14310200 | 6.26E-03 |
| Msc | ILMN_2769777 | 1 | 14753346 | 14755966 | 2.50E-04 |
| Mapkbp1 | ILMN_2873793 | 2 | 119972699 | 120027408 | 3.77E-02 |
| Pla2g4d | ILMN_3156124 | 2 | 120265595 | 120289197 | 4.79E-02 |
| Ltk | ILMN_1219860 | 2 | 119751320 | 119760431 | 6.37E-02 |
| Rpap1 | ILMN_2602151 | 2 | 119763304 | 119787537 | 1.65E-02 |
| Pla2g4f | ILMN_2727722 | 2 | 120299957 | 120314165 | 1.03E-03 |
| Vps39 | ILMN_3129752 | 2 | 120316461 | 120353137 | 1.52E-02 |
| Tmem87a | ILMN_2969172 | 2 | 120355312 | 120404113 | 1.57E-02 |
| Ganc | ILMN_2791355 | 2 | 120403896 | 120461700 | 1.85E-01 |
| Zfp106 | ILMN_2482427 | 2 | 120506820 | 120563843 | 2.30E-01 |
| Tgm5 | ILMN_2690147 | 2 | 121046111 | 121085841 | 3.14E-01 |
| Ccndbp1 | ILMN_1240178 | 2 | 121008403 | 121016904 | 3.18E-01 |
| Adal | ILMN_1242772 | 2 | 121140428 | 121156680 | 3.35E-01 |
| Trp53bp1 | ILMN_1243753 | 2 | 121193281 | 121271407 | 3.80E-02 |
| Tubgcp4 | ILMN_1257622 | 2 | 121170654 | 121198770 | 3.82E-01 |
| Adcy8 | ILMN_2607127 | 15 | 64699042 | 64922296 | 3.79E-01 |
| Olig1 | ILMN_2760105 | 16 | 91269769 | 91271939 | 2.14E-06 |
| Ifnar2 | ILMN_2598703 | 16 | 91485238 | 91507441 | 6.66E-07 |
| Gpr89 | ILMN_2953098 | 3 | 96868281 | 96905346 | 3.07E-02 |
| Cd160 | ILMN_2707181 | 3 | 96798763 | 96829351 | 1.49E-02 |
| Fmo5 | ILMN_1230890 | 3 | 97628804 | 97655282 | 3.52E-01 |
| Chd1l | ILMN_2986172 | 3 | 97560742 | 97610203 | 1.57E-02 |
| Pde4dip | ILMN_3064283 | 3 | 97689824 | 97888707 | 4.00E-01 |
| Ankrd34a | ILMN_3020599 | 3 | 96596636 | 96599775 | 6.43E-02 |
| Rbm8a | ILMN_1252850 | 3 | 96629928 | 96633779 | 6.80E-02 |
| Pex11b | ILMN_1258372 | 3 | 96635376 | 96653101 | 1.64E-01 |
| Sv2a | ILMN_1213495 | 3 | 96181151 | 96195521 | 3.01E-01 |
| Mtmr11 | ILMN_2676662 | 3 | 96162004 | 96171718 | 3.77E-01 |
| Hist2h2ac | ILMN_1218380 | 3 | 96220361 | 96220880 | 4.10E-01 |
| Hist2h2be | ILMN_1231066 | 3 | 96221121 | 96223738 | 9.24E-03 |
| Prpf3 | ILMN_2941430 | 3 | 95830124 | 95855885 | 5.17E-02 |
| BC028528 | ILMN_2656422 | 3 | 95883954 | 95892005 | 2.21E-01 |
| Aph1a | ILMN_1232821 | 3 | 95893969 | 95898288 | 3.09E-02 |
txStart, location of transcription (NCBI Build 37 genome assembly) start.
txEnd, location of transcription (NCBI Build 37 genome assembly) end.
Statistically significant p-value ≤ E-06 (Bonferroni corrected for the number of probes tested).
Table 2.
Biological processes and phenotypes associated with cis-eQTL candidate genes. Retrieved from ensembl.org. Copyright 2020.
| Gene | Chr | Biological process | Phenotype |
|---|---|---|---|
| Xkr9 | 1 | Apoptosis | Prostatic neoplasms |
| Eya1 | 1 | Kidney, eye, and ear development | Branchio-oto-renal syndrome |
| Msc | 1 | Regulation of transcription by RNA polymerase II | Adolescent idiopathic scoliosis; Alzheimer disease |
| Mapkbp1 | 2 | Negative regulation of interleukin-8 production | Nephronophthisis |
| Pla2g4d | 2 | Glycerophospholipid catabolic process | - |
| Ltk | 2 | Signal transduction | Type 2 diabetes |
| Rpap1 | 2 | Transcription by RNA polymerase II | - |
| Pla2g4f | 2 | Phospholipid metabolic process | Stroke |
| Vps39 | 2 | Late endosome to lysosome transport | Breast neoplasms; Schizophrenia |
| Tmem87a | 2 | - | Stroke |
| Ganc | 2 | Carbohydrate metabolic process | - |
| Zfp106 | 2 | - | Amyotrophic lateral sclerosis |
| Tgm5 | 2 | Peptide cross-linking | Peeling skin syndrome |
| Ccndbp1 | 2 | Regulation of cell cycle | - |
| Adal | 2 | Nucleotide metabolic process | Thrombocytopenia |
| Trp53bp1 | 2 | DNA damage checkpoint | Thymus hypoplasia |
| Tubgcp4 | 2 | Cytoplasmic microtubule organization | Autosomal recessive chorioretinopathy-microcephaly syndrome |
| Adcy8 | 15 | Adenylate cyclase-activating G protein-coupled receptor signaling pathway | Chronic kidney disease; myocardial infarction |
| Olig1 | 16 | Regulation of transcription, DNA-templated | Albuminuria |
| Ifnar2 | 16 | Cell surface receptor signaling pathway | Immunodeficiency |
| Gpr89 | 3 | Ion transmembrane transport | Autism spectrum disorder; Schizophrenia |
| Cd160 | 3 | Regulation of adaptive immune response | Schizophrenia |
| Fmo5 | 3 | Oxidation-reduction process | Autism spectrum disorder; chromosome 1q21.1 deletion syndrome |
| Chd1l | 3 | Nucleotide-excision repair, preincision complex assembly | Congenital anomalies of kidney and urinary tract; type 2 diabetes |
| Pde4dip | 3 | Cellular protein-containing complex assembly; regulation of Golgi organization | Breast, central nervous system, gastrointestinal, and skin tumors |
| Ankrd34a | 3 | - | - |
| Rbm8a | 3 | RNA processing | Thrombocytopenia-absent radius syndrome |
| Pex11b | 3 | Peroxisome proliferation | Peroxisome biogenesis disorder; neonatal adrenoleukodystrophy; infantile refsum disease |
| Sv2a | 3 | Chemical synaptic transmission; regulation of gamma-aminobutyric acid secretion; cellular calcium ion homeostasis | Seizures |
| Mtmr11 | 3 | Phosphatidylinositol dephosphorylation | Adolescent idiopathic scoliosis |
| Hist2h2ac | 3 | DNA binding | - |
| Hist2h2be | 3 | Antibacterial humoral response; negative regulation of tumor necrosis factor-mediated signaling pathway | - |
| Prpf3 | 3 | RNA binding | Retinitis pigmentosa |
| BC028528 | 3 | - | - |
| Aph1a | 3 | Notch receptor processing; beta-amyloid precursor protein | Bipolar disorder; schizophrenia |
Discussion
This study evaluated pre- and post-noise exposure auditory variation in mice by means of genome-wide association analysis using the HMDP and DPOAE suprathreshold amplitudes. The HMDP has been previously used successfully in the genomic mapping of blood cell traits in mice [Davis et al., 2013], in genomic association and systems genetics in mice by identifying Asxl2 as a regulator of bone mineral density and osteoclastogenesis [Farber et al., 2011], in the genetic regulation of atherosclerotic plaque size and morphology in the innominate artery of hyperlipidemic mice [Bennett et al., 2009], in the identification of gene networks associated with conditional fear in mice using a systems genetics approach [Park et al., 2011], in the genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice [Parks et al., 2013], in the identification of Nox3 as a critical gene for susceptibility to NIHL [Lavinsky et al., 2015], and in diabetes [Parks et al., 2013] and heart failure [Rau et al., 2015]. In many of these studies, including the present GWAS, the use of modified mouse models allowed the genes in the identified loci to be validated as the causal genes, and several of them matched loci identified in human GWASs.
The present GWAS generated significant associations in 1 region at baseline and in 5 separate regions after exposure to noise, resulting in 144 candidate genes of which 35 in these 6 loci were identified with at least 1 probe whose expression was regulated by a significant cis-eQTL in the cochlea. After careful analysis of the candidate genes based on cochlear gene expression and after a review of the current literature, 2 main candidate genes were prioritized: Eya1 (baseline) and Efr3a (post-exposure).
The Eya1 gene, which was present with a significant association peak at baseline at 32 kHz - 70 dB intensity, is a gene-encoding protein that acts on other genes, thus being considered a transcription factor. It is interesting to note that the so-far identified functions of Eya1 correlate with certain forms of genetic hearing loss in humans and normal cochlear development [Xu et al., 2017]. The Eya1 protein interacts with several other proteins by activating and inactivating genes that are important for normal development. Key roles include the development of the second branchial arch, which gives rise to the tissues of the neck, eyes, ears, and kidneys [Kochhar et al., 2007].
The Eya1 gene has grown in prominence with the discovery of its expression in mammalian cochlear stem cells. A population of Eya1-expressing progenitor cells capable of differentiating into hair cells, supporting cells, and ganglion cells after hair cell ablation up to 1 week of age was identified in mice. The identification of cochlear regeneration after birth in mammals and the characterization of this cell population (Eya1-positive) provided a new cellular source with potential for cochlear repair and regeneration [Xu et al., 2017].
Furthermore, Eya1 gene mutations are the most common cause of the branchio-oto-renal (BOR) syndrome. More than 80 pathogenic mutations were identified in Eya1 in different populations [Zhang et al., 2004]. BOR syndrome is a rare autosomal dominant disorder characterized by sensorineural, mixed, or conductive hearing loss, branchial fistulas or cysts, preauricular pits, ear malformations, and renal anomalies.
Another important correlation was the confirmation that the inactivation of Eya1 and Six1 in mice results in a significant alteration in ear development owing to an early interruption of development at the otocyst stage [Zheng et al., 2003]. Another robust evidence was the discovery of the involvement of Eya1/Six1 in hair cell development and their coexpression in mouse cochlear explants. Expression of these genes was sufficient to induce hair cell fate in the nonsensory epithelium expressing low-level Sox2 by activating not only Atoh1-dependent but also Atoh1-independent pathways, and both pathways induce Pou4f3 to promote the differentiation of hair cells [Ahmed, 2012].
Efr3a is our main candidate gene, which was found at the frequency of 11.3 kHz at 70 dB after noise exposure. It encodes a plasma membrane protein whose insufficiency has been initially reported to increase the survival of hippocampal neurons in adult mice. This is the first correlation of the gene with NIHL, which has been further investigated in age-related hearing loss and ototoxicity. These findings are in agreement with previous studies investigating Efr3a expression in the auditory neural pathway, which is upregulated soon after hair cell destruction. The impact of Efr3a insufficiency has also been evaluated by the functional and morphological comparison of the cochlea between Efr3a-deficient mice and their wild-type littermates. The present study suggests that Efr3a deficiency is associated with less severe hearing loss after ototoxic exposure and confirms greater morphological damage to cochlear spiral ganglion neurons. These beneficial effects of Efr3a deficiency can be attributed to increased expression of some neurotrophic factors and their receptors, thus suppressing neurodegeneration [Hu et al., 2017].
Our finding is also consistent with another recent finding from research into age-related hearing loss that Efr3a deficiency results in better hearing function in senescent mice (at 10 and 12 months of age). It was demonstrated that Efr3a-deficient senescent mice had less severe degeneration of cochlear spiral ganglion neurons in the basal turn; when degeneration occurred, it occurred later [Hu et al., 2016].
Moreover, ototoxic injury in mice showed to rapidly induce hair cell loss, resulting in progressive degeneration of the spiral ganglion 5 days after drug administration. The number of spiral ganglion cells began to decrease on day 15. Efr3a expression was notably increased in the spiral ganglion on day 5, decreasing to near normal levels by day 10. This suggests that the change in Efr3a expression in the spiral ganglion coincides with the time of spiral ganglion degeneration, which implies that this high Efr3a expression may be important to the onset of spiral ganglion degeneration after hair cell loss [Nie et al., 2015].
Conclusion
For the first time, an association analysis with correction for population structure was used to map several loci for hearing traits in inbred strains of mice based on DPOAE suprathreshold amplitudes before and after noise exposure. Our results identified a number of novel loci for susceptibility to NIHL. Also, our findings validate the power of the HMDP for detecting NIHL susceptibility candidate genes.
Funding Sources
This work was supported by National Institutes of Health grant NIDCD R01DC010856-01 to RAF. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Statement of Ethics
This study was carried out in strict accordance with the recommendations of the American Association for Laboratory Animal Sciences (AALAS). The protocol was approved by the University of Southern California Institutional Animal Care and Use Committee (Permit Number: 12033) and the Department of Animal Resources.
Animals were housed with ambient noise not exceeding that of normal air conditioning. All techniques were performed on mice under intraperitoneal anesthesia (ketamine 80mg/kg body weight and xylazine 16mg/kg body weight) and all efforts were made to minimize suffering.
Disclosure Statement
The authors have no conflicts of interest to declare.
References
- 1.Ahmed M, Wong EY, Sol J, Xu J, Wang F, Xu PX. Eya1-Six1 interaction is sufficient to induce hair cell fate in the cochlea by activating Atoh1 expression in cooperation with Sox2. Dev Cell. 2012; 22 (2): 377–390. DOI: 10.1016/j.devcel.2011.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett BJ1, Wang SS, Wang X, Wu X, Lusis AJ. Genetic regulation of atherosclerotic plaque size and morphology in the innominate artery of hyperlipidemic mice. Arterioscler Thromb Vasc Biol. 2009. March;29(3):348–55. DOI: 10.1161/ATVBAHA.108.176685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett BJ, Farber CR, Orozco L, Kang HM, Ghazalpour A, et al. (2010) A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Res 20: 281–290. DOI: 10.1101/gr.099234.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis RC, van Nas A, Bennett B, Orozco L, Pan C, Rau CD, Eskin E, Lusis AJ. Genome-wide association mapping of blood cell traits in mice. Mamm Genome. 2013. April;24(3–4):105–118. DOI: 10.1007/s00335-013-9448-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Erway LC, Shiau YW, Davis RR, Krieg EF (1996) Genetics of age-related hearing loss in mice. III. Susceptibility of inbred and F1 hybrid strains to noise-induced hearing loss. Hear Res 93: 181–187. DOI: 10.1016/0378-5955(95)00226-x [DOI] [PubMed] [Google Scholar]
- 6.Farber CR, Bennett BJ, Orozco L, et al. Mouse genome-wide association and systems genetics identify Asxl2 as a regulator of bone mineral density and osteoclastogenesis. PLoS Genet. 2011;7(4):e1002038. DOI: 10.1371/journal.pgen.1002038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holme RH, Steel KP. Progressive hearing loss and increased susceptibility to noise-induced hearing loss in mice carrying a Cdh23 but not a Myo7a mutation. J Assoc Res Otolaryngol. 2004;5(1):66–79. DOI: 10.1007/s10162-003-4021-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu H, Ma Y, Ye B, et al. The role of Efr3a in age-related hearing loss. Neuroscience. 2016. November 17;341:1–8. DOI: 10.1016/j.neuroscience.2016.11.013 [DOI] [PubMed] [Google Scholar]
- 9.Hu H, Ye B, Zhang L, et al. Efr3a Insufficiency Attenuates the Degeneration of Spiral Ganglion Neurons after Hair Cell Loss. Front Mol Neurosci. 2017;10:86. Published 2017 Mar 29.DOI: 10.3389/fnmol.2017.00086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang HM, Zaitlen NA, Wade CM, Kirby A, Heckerman D, et al. (2008) Efficient control of population structure in model organism association mapping. Genetics 178: 1709–1723.DOI: 10.1534/genetics.107.080101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kochhar A, Fischer SM, Kimberling WJ, Smith RJ. Branchio-oto-renal syndrome. Am J Med Genet 2007;143:1671–8. DOI: 10.1002/ajmg.a.31561 [DOI] [PubMed] [Google Scholar]
- 12.Kozel PJ, Davis RR, Krieg EF, Shull GE, Erway LC (2002) Deficiency in plasma membrane calcium ATPase isoform 2 increases susceptibility to noise-induced hearing loss in mice. Hear Res 164: 231–239.DOI: 10.1016/s0378-5955(01)00420-8 [DOI] [PubMed] [Google Scholar]
- 13.Kujawa SG, Liberman MC. Adding insult to injury: cochlear nerve degeneration after “temporary” noise-induced hearing loss. J Neurosci. 2009;29(45):14077–85. DOI: 10.1523/JNEUROSCI.2845-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lavinsky J, Crow AL, Pan C, et al. Genome-wide association study identifies nox3 as a critical gene for susceptibility to noise-induced hearing loss [published correction appears in PLoS Genet. 2015 Jun;11(6):e1005293]. PLoS Genet. 2015;11(4):e1005094. Published 2015 Apr 16. DOI: 10.1371/journal.pgen.1005094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liberman MC. Noise-induced and age-related hearing loss: new perspectives and potential therapies. F1000Res. 2017;6:927. Published 2017 Jun 16. DOI: 10.12688/f1000research.11310.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heinonen-Guzejev M, Vuorinen HS, Mussalo-Rauhamaa H, Heikkilä K, Koskenvuo M, Kaprio J. Somatic and psychological characteristics of noise-sensitive adults in Finland. Arch Environ Health. 2004;59(8):410–417. DOI: 10.3200/AEOH.59.8.410-417. [DOI] [PubMed] [Google Scholar]
- 17.Sliwinska-Kowalska M, Pawelczyk M. Contribution of genetic factors to noise-induced hearing loss: a human studies review Mutat. Res, 752 (2013), pp. 61–65. DOI: 10.1016/j.mrrev.2012.11.001 [DOI] [PubMed] [Google Scholar]
- 18.Nie C, Hu H, Shen C, Ye B, Wu H, Xiang M (2015) Expression of EFR3A in the Mouse Cochlea during Degeneration of Spiral Ganglion following Hair Cell Loss. PLoS ONE 10(1): e0117345. Published 2015 Jan 26. DOI: 10.1371/journal.pone.0117345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohlemiller KK, McFadden SL, Ding DL, et al. Targeted deletion of the cytosolic Cu/Zn-superoxide dismutase gene (Sod1) increases susceptibility to noise-induced hearing loss. Audiol Neurootol. 1999;4(5):237–246. DOI: 10.1159/000013847 [DOI] [PubMed] [Google Scholar]
- 20.Ohlemiller KK, McFadden SL, Ding DL, Lear PM, Ho YS. Targeted mutation of the gene for cellular glutathione peroxidase (Gpx1) increases noise-induced hearing loss in mice. Assoc Res Otolaryngol. 2000;1(3):243–254. DOI: 10.1007/s101620010043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park CC, Gale GD, de Jong S, et al. Gene networks associated with conditional fear in mice identified using a systems genetics approach. BMC Syst Biol. 2011;5:43. Published 2011 Mar 16. DOI: 10.1186/1752-0509-5-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parks BW, Nam E, Org E, et al. Genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice. Cell Metab. 2013;17(1):141–152. DOI: 10.1016/j.cmet.2012.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rau CD, Parks B, Wang Y, et al. High-Density Genotypes of Inbred Mouse Strains: Improved Power and Precision of Association Mapping. G3 (Bethesda). 2015;5(10):2021–2026. Published 2015 Jul 28. DOI: 10.1534/g3.115.020784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stelzer G, Rosen R, Plaschkes I, et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analysis, Current Protocols in Bioinformatics(2016), 54:1.30.1–1.30.33.DOI: 10.1002/cpbi.5. [DOI] [PubMed] [Google Scholar]
- 25.White CH, Ohmen JD, Sheth S, et al. Genome-wide screening for genetic loci associated with noise-induced hearing loss. Mamm Genome. 2009;20(4):207–213. DOI: 10.1007/s00335-009-9178-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu J, Ueno H, Xu CY, Chen B, Weissman IL, Xu PX. Identification of mouse cochlear progenitors that develop hair and supporting cells in the organ of Corti. Nat Commun. 2017;8:15046. Published 2017 May 11. DOI: 10.1038/ncomms15046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yankaskas K Prelude: noise-induced tinnitus and hearing loss in the military. Hear Res. 2013January;295:38. DOI: 10.1016/j.heares.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, Knosp BM, Maconochie M, Friedman RA, Smith RJ. A comparative study of Eya1 and Eya4 protein function and its implication in branchio-oto-renal syndrome and DFNA10. J Assoc Res Otolaryngol. 2004;5(3):295–304. DOI: 10.1007/s10162-004-4044-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng W, Huang L, Wei ZB, Silvius D, Tang B, Xu PX. The role of Six1 in mammalian auditory system development. Development. 2003;130(17):3989–4000. DOI: 10.1242/dev.00628 [DOI] [PMC free article] [PubMed] [Google Scholar]


