Abstract
Type I interferon (IFN) is essential for viral clearance but also contributes to the pathogenesis of autoimmune diseases, such as systemic lupus erythematosus (SLE), via aberrant nucleic acid sensing pathways leading to autoantibody production. Type III IFN (IFN-λ) is now appreciated to have a non-redundant role in viral infection, but few studies have addressed the effects of IFN-λ on immune cells given the more restricted expression of its receptor primarily to the epithelium. Here, we demonstrate that B cells display a prominent IFN gene expression profile in lupus patients. Serum levels of IFN-λ are elevated in SLE and positively correlate with B cell subsets associated with autoimmune plasma cell (PC) development, including CD11c+ T-bet+ CD21- B cells. Although B cell subsets express all IFN receptors, IFNLR1 strongly correlates with the CD11c+ CD21- B cell expansion, suggesting that IFN-λ may be an unappreciated driver of the SLE IFN signature and B cell abnormalities. We show that IFN-λ potentiates gene transcription in human B cells typically attributed to type I IFN as well as expansion of T-bet expressing B cells after BCR and TLR7/8 stimulation. Further, IFN-λ promotes TLR7/8-mediated plasmablast differentiation and increased IgM production. CD11c+ B cells demonstrate IFN-λ hyper-responsive signaling compared to other B cell subsets suggesting that IFN-λ accelerates PC differentiation through this putative extra-follicular pathway. In summary, our data support type III IFN-λ as a cytokine promoting the antibody-secreting cell pool in human viral and autoimmune disease.
Introduction
Production of a wide variety of autoantibodies is a hallmark of systemic lupus erythematosus (SLE). Thus, therapies that directly target B cells have long been sought for the treatment of lupus, with belimumab FDA approved (1) and recent promising results in a phase II clinical trial of obinutuzumab (anti-CD20) (2). An alternative approach is to target factors that lead to B cell dysfunction in lupus. There is strong evidence for a role of interferons (IFN) in the disease pathogenesis and the B cell dysfunction characteristic of the disease. However, most work has focused on type I IFN (reviewed in (3, 4)).
The effects of type I IFN on B cells in lupus are particularly important (5–9). Type I IFN sensitizes naïve B cells to RNA-associated antigens through the upregulation of RNA-binding TLR7 (5). Type I IFN also upregulates co-stimulatory molecules such as CD69, CD86 and MHC class II thus enhancing B cell ability to present antigen, receive T cell help and generate germinal centers (reviewed in (10)). Moreover, Type I IFN promotes the differentiation of B cells to plasma cells (PC) and the production of antibodies (5, 8, 9). In addition, SLE is associated with perturbations in B cell homeostasis with increases in B cell subsets normally seen during viral infection (11–13) including IgD- CD27- double negative (DN), memory, and plasmablast populations. These B cell abnormalities increase during disease flare (14, 15).
Recent studies have also highlighted the potential role of type II IFN (IFN-γ) in SLE. IFN-γ promotes ABC (age or autoimmunity-enriched B cells), a B cell subset first described in aged mice (16–18) and subsequently in SLE (17, 19, 20). Phenotypic markers for ABCs have varied between studies but include CD11c+, CD21-, CXCR5-, and the transcription factor T-bet (17, 21, 22). Studies in murine and human lupus indicate that ABCs are poised for PC differentiation and enriched in auto-specificities (18). CD11c+ T-bet+ B cells in human SLE patients are overrepresented in the IgD- CD27- (DN) compartment, where they are designated DN2 (22). In lupus, DN2/ABC cells are expanded in both the blood and tissues (22) where they are speculated to contribute to local production of autoantibodies that result in end-organ damage. Thus, ABC are of interest as they may be directly pathogenic in lupus patients given that autoantibody producing PC can be derived from DN2/ABC, DN2/ABC cells are found in lupus patients with high disease activity (22), and certain autoantibodies are directly responsible for disease manifestations (reviewed in (23)).
While type I and type II IFN attracted considerable attention as drivers of B cell aberrations in SLE and in infection models, type III IFN remains poorly studied. For many years, the ability to respond to type III IFN (IFN-λ, previously called IL-28 and IL-29) was thought to be restricted to primarily epithelial-derived, plasmacytoid dendritic, and monocyte-derived dendritic cells (24, 25). In responsive cell types, a gene signature that was very similar, if not identical, as that of type I IFN is triggered by IFN-λ (26). More recently, reports suggest human B lymphocytes also respond to IFN-λ (22, 24, 27–31). This is in contrast to murine B cells that cannot be stimulated by type III IFN (28). We hypothesized that the so-called type I IFN gene signature, which is a hallmark of SLE-derived cells, might in part be derived from type III IFN. This has important clinical implications given that type I IFN receptor neutralizing agents under development for the treatment of SLE such as anifrolumab have no effect on type III IFN signaling as type III IFN utilize a different receptor (32).
Here, we examine the impact of IFN-λ1 on human B cells and its ability to accelerate PC differentiation. We demonstrate that DN B cells in SLE patients have an IFN gene signature. Further, B cells transcribe IFN regulated transcripts historically associated with type I IFN in response to type III IFN. Notably, IFN-λ1 was elevated in SLE serum and correlates with the frequency of DN2/ABC cells in peripheral blood. We show that CD11c+ B cells robustly phosphorylate STAT1 signaling proteins in response to IFN- λ1 and that IFN-λ1 augments T-bet-expressing B cell and plasmablast formation in the setting of TLR7/8 human B cell activation. Thus, our results demonstrate that IFN-λ1 can have effects on human B cells with critical implications for both viral infection and autoimmune disease.
Materials and methods
Patients and samples
Samples were collected from SLE and healthy donors via venipuncture at the University of Rochester Medical Center or affiliated outpatient clinics after informed consent in accordance with approved Institutional Review Board protocol. For RNA-seq analysis, blood was obtained from 8 SLE patients and 5 healthy donors (Supplemental Table 1). For ex vivo B cell subset flow cytometry phenotyping studies, blood was obtained from 26 SLE and 6 healthy donors (Table 1). All patients fulfilled the ACR SLE classification criteria (33). Disease activity was assessed by the Systemic Lupus Erythematosus Disease Activity Index (SLEDAI) (34). All patients were on pharmacologic therapy for lupus (Table 1). All patients had a positive ANA (indirect immunofluorescence with Hep-2 cells) of at least 1:80. Anti-dsDNA titers were determined by the clinical laboratory. Anticoagulated venous blood from SLE patients and healthy donors was subjected to density gradient centrifugation to obtain peripheral blood mononuclear cells (PBMC). RNA-seq, cultures and PCR experiments utilized fresh cells without freezing. Samples for ex vivo phenotyping and phosphorylation flow cytometry were stored in liquid nitrogen and rapidly thawed in batches for analysis. Serum was stored at −80°C.
Table 1: Demographic Data for Flow Cytometry Cohort.
B cell phenotyping was performed on a cohort of 26 SLE and 6 healthy donors. * Of those on systemic steroids, mean±SD prednisone-equivalent dose was 10.2±11.8mg (range 2.5–25mg). Patients on B cell depletion therapy (rituximab) were excluded.
| SLE (n = 26) | Healthy (n = 6) | |
|---|---|---|
| Age (mean±SD, range) | 42.8±13.8 (19 to 69) | 51.7±7.7 (38 to 58) |
| Sex | 88.6% Female | 83.3% Female |
| Race | 38.5% African American 53.9% Caucasian 7.6% Other |
83.3% Caucasian 16.7% African American |
| Ethnicity | 11.5% Hispanic 88.5% Non-Hispanic |
100% Non-Hispanic |
| SLEDAI (mean±SD, range) | 3.2±3.0 (0–10) | -- |
| % Active (SLEDAI≥6) | 23% | -- |
| ANA (mean±SD, range) | 1:809 (1:80–1:2560) | -- |
| dsDNA (mean±SD, range) | 706±2443 (0–9523) | -- |
| Therapy | 92.3% Hydroxychloroquine 42.3% Systemic Steroids* 19.2% Belimumab 19.2% Azathioprine 19.2% Mycophenolate Mofetil 7.7% Tacrolimus 3.8% Quinacrine 3.8% Methotrexate |
-- |
Bulk RNA-seq
Fresh PBMC were FACS sorted into Qiagen RLT lysis buffer. Total RNA was isolated using the RNeasy Plus Micro Kit (Qiagen, Valencia, CA) per manufacturer’s recommendations. RNA concentration was determined with the NanopDrop 1000 spectrophotometer (NanoDrop, Wilmington, DE) and RNA quality assessed with the Agilent Bioanalyzer 2100 (Agilent, Santa Clara, CA). Total RNA (1ng) was pre-amplified with the SMARTer Ultra Low Input kit v4 (Clontech, Mountain View, CA) per manufacturer’s recommendations. The quantity and quality of the subsequent cDNA was determined using the Qubit Fluorimeter (Life Technnologies, Carlsbad, CA) and the Agilent Bioanalyzer 2100 (Agilent, Santa Clara, CA). Amongst the DN B cell samples, one sample for the SLE group and one from the healthy controls were excluded based upon poor RNA quality. cDNA (150pg) was used to generate Illumina compatible sequencing libraries with the NexteraXT library preparation kit (Illumina, San Diego, CA) per manufacturer’s protocols. All samples were sequenced as one batch to eliminate batch effects. The amplified libraries were hybridized to the Illumina single end flow cell and amplified using the cBot (Illumina, San Diego, CA). Single end reads of 100nt were generated by the HiSeq2500 (Illumina, San Diego, CA). Raw reads generated from the Illumina basecalls were demultiplexed using bcl2fastq version 2.19.1. Quality filtering and adapter removal are performed using FastP version 0.20.0 with the following parameters: “--length_required 35 --cut_front_window_size 1 --cut_front_mean_quality 13 --cut_front --cut_tail_window_size 1 --cut_tail_mean_quality 13 --cut_tail -y -r”. Processed/cleaned reads were then mapped to the Homo sapiens reference genome (GRCh38 + Gencode-28 Annotation) using STAR_2.7.0f with the following parameters: “—twopass Mode Basic --runMode alignReads --outSAMtype BAM SortedByCoordinate – outSAMstrandField intronMotif --outFilterIntronMotifs RemoveNoncanonical –outReads UnmappedFastx” (35, 36). Genelevel read quantification was derived using the subread-1.6.4 package (featureCounts) with a GTF annotation file (Gencode-28) and the following parameters: “-s 0 -t exon -g gene_name” (37). Differential expression analysis was performed using DESeq2–1.22.1 with a P-value threshold of 0.05 at a false discovery rate of 0.05 within R version 3.5.1 (https://www.R-project.org/) (38). Counts were normalized using DESeq2 which utilizes a median of ratios methods where counts are divided by sample-specific size factors determined by median ratio of gene counts relative to the geometric mean per gene. Heatmaps were generated using the ComplexHeatmap package with vst variance-stabilized expression values. Gene ontology analyses were performed using the EnrichR and clusterProfiler packages (39). Data were deposited in the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE175913.
Cell Culture
For cell culture experiments, up to 120cc of peripheral blood was obtained from healthy donors per IRB protocol. After density centrifugation with Lymphocyte Separation Media, mononuclear cells were positively sorted by CD19 positive magnetic microbead selection using LS columns (Miltenyi Biotec). All cell cultures were performed in 96-well round bottom plates with freshly isolated cells in 200ul of RPMI 1640 with 10% fetal calf serum supplemented with penicillin/streptomycin at 250,000 cells per well. Stimulated cultures contained combinations of BAFF (100ng/ml), R848 (1μg/ml), IL-21 (10ng/ml), IFN-γ (20ng/ml), anti-IgG/A/M F(ab)2 fragments (10μg/ml), IFN-λ1 (500U/ml), or IFN-α2 (500U/ml). As no CD27hiCD38+ cells were detectable when BCR stimulation was included for the duration of the 7 day culture in agreement with previous studies (20), cultures to generate plasma cells did not contain anti-IgG/A/M.
ELISA
After thaw, patient serum was centrifuged and supernatant ran on ELISA per manufacturer’s instructions. IFN-α level was measured by an all alpha subtypes ELISA. IFN-β was measured by using a high sensitivity ELISA kit. Both kits were labeled for use in serum (PBL Assay Science, Piscataway, NJ, USA). To measure IFN-λ1, the IL-29 ELISA kit was utilized (Invitrogen, Carlsbad, CA, USA). To measure IgM or IgG, B cells cultures were spun down and supernatants removed and frozen at −20C until time of ELISA (Bethyl Laboratories).
Flow Cytometry
Cell viability was assessed by incubation of cells with LIVE/DEAD™ Fixable Aqua Dead Cell Stain Kit (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol. Samples were incubated with antibodies for 30 minutes at 4°C. Intracellular staining was performed using eBioscience FoxP3/Transcription Factor Staining Buffer Set (Invitrogen) following the manufacturer’s recommendations. Cells were fixed for 20 minutes in 1% methanol-free formaldehyde then diluted in 1% BSA in PBS in a 2:1 ratio. Cells were run on LSR II Fortessa flow cytometer (BD). Data analysis was performed using FlowJo software (USA). The following monoclonal anti-human antibodies were used for B cell phenotyping: CD19-APC-CY7 (clone SJ25C1, BD 557791), IGD-FITC (clone IA62, BD 555778), CD11c-BUV395 (clone B-ly6, BD 563787), CD27-BV605 (clone O323, Biolegend 302830), CD24-PE610 (clone SN3, Lifetech MHCD2422), CD38-PerCP-5.5 (clone HIT2, BD 551400), CD95-APC (clone DX2, BD 558814), CD20-ALX700 (clone 2H7, Biolegend 302322), CXCR3-PE (clone 1C6, BD 550633), CD21-PE-CY5 (clone B-LY4, BD 551064), CD3-BV421 (clone UCHT1, BD 562426), and T-Bet-PECY7 (clone 4B10, Biolegend 644824). Gating shown in each figure. B cell subsets were defined as: naïve = IgD+ CD27-, DN = IgD- CD27-, unswitched memory (USM) = IgD+ CD27+, switched memory (SWM) = IgD- CD27+, PC = IgD- CD27hi CD38+, pre-plasmablast = CD27- CD38+, DN2 = IgD- CD27- CD21- CD24-.
RNA Extraction, cDNA synthesis, and qPCR
B cell pellets were suspended in RLT plus lysis buffer (Qiagen) with 10μl/ml β-mercaptoethanol and frozen at −80C until RNA extraction. RNA was purified using Quick-RNA prep kits (Zymo Research). RNA was transcribed to cDNA using qScript Supermix (Quanta Bio). Quantitative real-time PCR was performed on QuantStudio™ 3 System (Applied Biosystems) using ROX as a reference dye in PerfeCTa Fastmix II (Quanta Bio) and TaqMan specific probes (Hs02758991-g1 GAPDH, Hs01911452_s1 IFIT1, Hs01921425_s1 ISG15, Hs00271467_m1 IFI27, and Hs00276441_m1 USP18). GAPDH was used as an endogenous control. Fold change was calculated using the 2–∆∆Ct method.
Phosphorylation by Flow Cytometry
Frozen healthy PBMC were thawed and rested at 37°C for 1.5hrs in RPMI 1640 with 10% fetal calf serum and 1x penicillin/streptomycin (Gibco) media. Experiments were performed to establish kinetics and concentration of maximal STAT1 phosphorylation for IFN-α2 and IFN-λ1. Cells were washed then treated with media, media with 500U/ml of IFN-α2 or 2000U/ml of IFN-λ1 for 25 minutes at 37°C. For interferon receptor neutralization conditions, anti-IFNAR2 (Millipore Sigma MAB1155, 50μg/ml) or anti-IFNLR1 (PBL Assay Science, 21885–1, 1μg/ml) were incubated with cells 1 hour prior to stimulation. After stimulation, cells were fixed with 1% formalin for 10 minutes at room temperature, surfaced stained with antibodies for 30 min at 4°C followed by LIVE/DEAD™ Fixable Aqua Dead Cell Stain Kit (Invitrogen) for dead cell exclusion. Surface stain panel included, CD3-PE-Cy5 (clone UCHT1, BD 555334), CD11c- BUV395 (clone B-ly6, BD563787), CD14-ALX700 (clone M5E2, BD 557923), CD19-BV605 (clone SJ25C1, Biolegend 363024), CD20-APC-Cy7 (clone 2H7, Biolegend 302314), CD27-BUV737 (clone L128, Biolegend 612829), CD38-BB700 (clone HIT2, BD 566445), IgD-BV421 (clone IA6–2, Biolegend 348226) antibodies. Cells were then fixed using BD CYTOFIX for 10 minutes at room temperature followed by 90% BD Phosflow perm buffer III for 20 minutes at 4°C. Two washes were performed prior to intracellular fluorescent antibody staining of STAT1-PE (clone 1/STAT1, BD 558537) and pSTAT1-Alexa 488 (clone pY701, BD 612596) for 45 minutes at 4°C. Cells were then washed and fixed in 1% formalin for 15 minutes at room temperature. An additional 2:1 of 1%BSA/PBS was added and samples stored at 4°C overnight until flow cytometry. Fold change was calculated by dividing median fluorescent intensity of pSTAT1 for the treated sample by that of the unstimulated sample.
Statistical Analysis
Data were plotted with GraphPad Prism or R Studio with ggplot2 (40) and ggpubr (41) packages. Statistical tests were performed as described in the figure legends. Statistical significance are indicated with * p <0.05, ** p <0.01, *** p< .001.
Results
Naïve and Double Negative B Cells in Lupus Display an Interferon Gene Signature
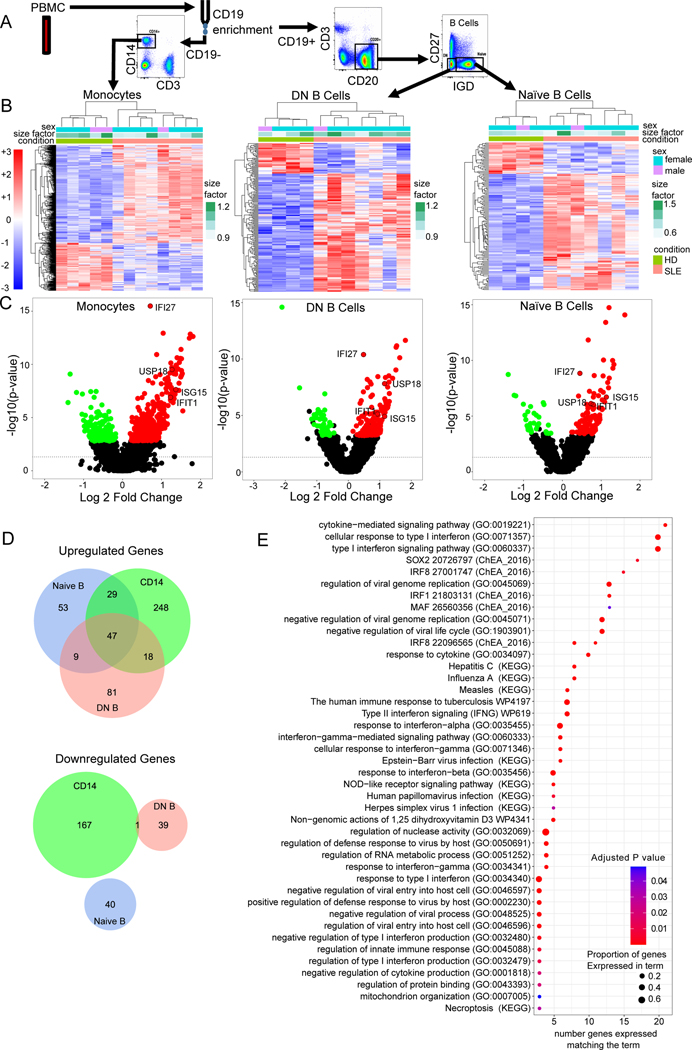
In order to identify key signals mediating B cell activation in lupus, we performed transcriptomic analysis on sorted naïve and DN B cells. Human peripheral blood from healthy donors (n=5) or SLE (n=8) patients meeting ACR criteria were flow sorted into CD14+ (monocytes), IgD-CD27-CD19+ (DN B cells), or IgD+CD27-CD19+ (naïve/transitional B cells) populations and RNA isolated for bulk RNA-seq transcriptomic analysis (Fig. 1A). SLEDAI on the lupus samples ranged from 0–12 (Supplemental Table 1). Hierarchical clustering analysis demonstrated that lupus samples clustered separately from controls for each cell type (Fig. 1B). Differential gene expression analysis between lupus and healthy showed 179 differentially expressed genes for naïve B cells, 196 for DN, and 511 for monocytes (p adj <0.05, Supplemental Table 2). Notably, interferon stimulated genes (ISGs) were prevalent amongst the top 30 upregulated genes for each cell type (volcano plots, Fig. 1C). Naïve and DN B cells had no differentially downregulated genes in common (Fig. 1D). Amongst the upregulated genes, 56 were shared between naïve and DN B cells and 47 shared amongst all three cell types (Fig. 1D). These shared upregulated genes included ISGs such as IFI27, USP18, RSAD2, IFI44, IFI44L, ISG15, IFIT1, IFIT3, STAT1, MX1, MX2, and OAS2 (Table 2). The pathways with the greatest number of genes upregulated in SLE that were shared amongst all three cell types (Fig. 1E) were the cytokine mediated signaling pathway (GO:0019221), cellular response to type I interferon (GO:0071357), and type I interferon signaling pathway (GO:00600337). Gene set enrichment analysis independently performed for the DN cells identified type I interferon signaling pathway (GO:0060337, GO:0071357), regulation of viral genome replication (GO:0045069, GO:0045071, GO:1903901), and double-stranded RNA binding genes as among the top statistically significant upregulated pathways in SLE (Fig. 2A and Supplemental Fig. 1B). These pathways were also upregulated in the SLE naïve and monocytes (Supplemental Fig. 1A–C). Since type I and type III interferon receptors share many components of their signaling pathways these data cannot distinguish which interferon is inducing these ISGs.
Fig. 1. Peripheral Blood B Lymphocytes Demonstrate an Interferon Gene Signature In Lupus.
A) Flow sorting strategy. Human peripheral blood from healthy donors (n=5) or SLE (n=8) patients meeting ACR criteria were stained for CD3, CD11c, CD19, CD14, CD27 and IgD then flow sorted into CD14+ (monocytes), CD27- IgD- CD19+ (DN B cells), or CD27-IgD+ CD19+ (naïve/transitional B cells) populations and their RNA isolated for bulk RNA-Seq transcriptomic analysis. B) Hierarchical clustering of all significantly differentially expressed genes in each cell type (monocytes, DN B cells, and naïve B cells). Sex, size factor, and disease status shown in bars above heat map. For each cell type, healthy donor cells (olive green) clustered together. Scale for normalized expression for all plots shown in red/blue on the left. C) Volcano plots for each cell type demonstrating healthy versus SLE differential gene expression. Select (prototypical ISG) differentially expressed genes are labeled. D) Venn diagram demonstrating number of upregulated genes (top) in SLE for monocytes (green), double negative B cells (salmon) and naïve B cells (blue) compared to HD. Venn diagram demonstrating numbers of downregulated genes (bottom) in SLE differential expression gene analysis for naïve B cells (blue), double negative B cells (salmon) and monocytes (green). E) Gene set enrichment analysis for differentially expressed genes upregulated in SLE common between monocytes, naïve and DN B cells. Size of dot corresponds to the percentage of genes in the pathway identified as upregulated. Color of dot corresponds to adjusted P-value.
Table 2: Shared Upregulated Differentially Expressed Genes.
Overlapping Upregulated Differentially Expressed Genes: Differentially expressed genes upregulated in SLE shared amongst monocytes, naïve B cells and DN B cells.
| IFI27 EPSTI1 PARP9 HERC6 SAMD9L MX2 EIF2AK2 DDX60 USP18 OAS2 USP41 IFI44L |
IFI44 RSAD2 DDX60L IFI6 CHMP5 CMPK2 DTX3L OAS3 OAS1 STAT1 ISG15 RP11- 273G15.2 |
SPATS2L IFIT3 LY6E XXbac- B33L19.12 NRIR HERC5 IFIT1 MX1 OASL XAF1 IFIT2 IFITM1 |
UBE2L6 IFITM2 IFITM3 ADAR LGALS3BP TRIM14 GBP1 IFI35 HLA-V PPM1K WDFY1 |
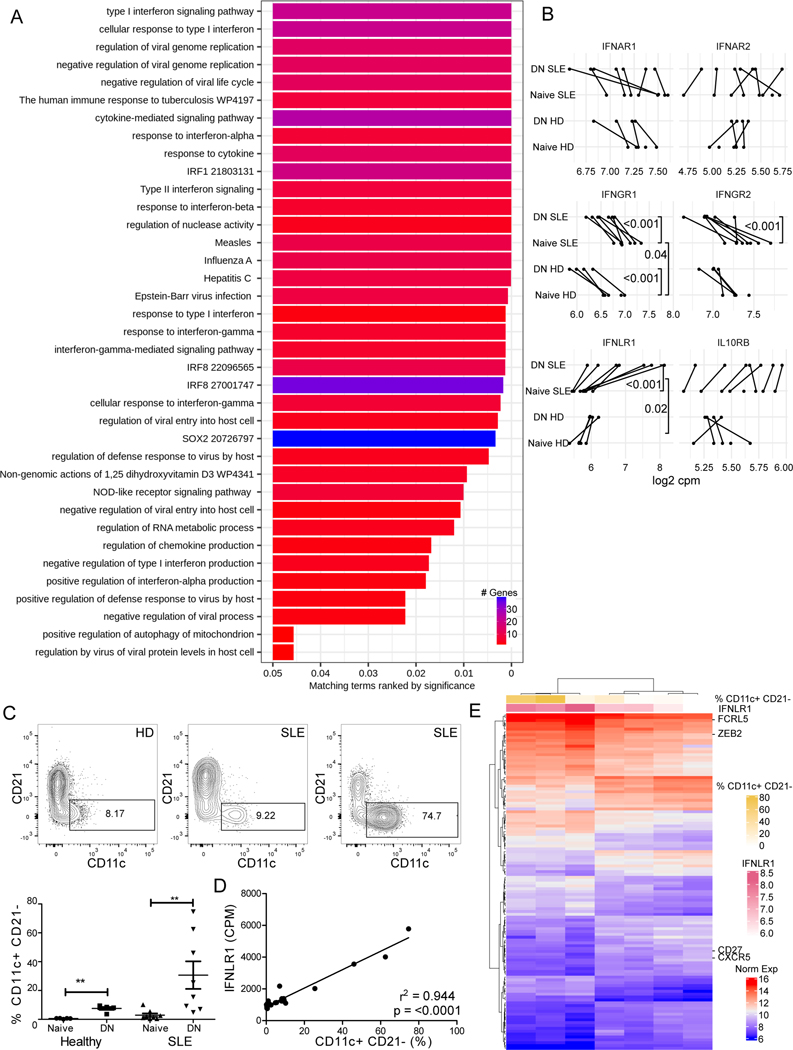
Fig. 2. Type III IFN receptor expression in DN B cells.
A) Gene set enrichment analysis for genes upregulated in SLE DN B cells from bulk transcriptomic analysis. Pathways ranked by significance. Color of bar indicates number of genes identified in the pathway. B) Log2 Counts-per-million for interferon receptor gene transcripts in SLE and healthy controls from bulk transcriptomic analysis. Negative binomial regressions with subject-level random effect and fixed effects for disease, B cell subset and their interaction were fit for each gene. Genes with Bayes’ factors favoring the aforementioned model and individual comparisons with Bayesian p-values < 0.05 are reported. C) Representative dot plots of DN B cells from healthy donor, SLE with low, and SLE with high CD11c+ CD21- cells as measured by flow cytometry in samples used for bulk RNA-seq transcriptomic analysis (top). CD11c+ CD21- Frequency amongst Naïve and DN B cells from healthy and SLE samples (bottom). Statistics calculated by Mann-Whitney U Test. ** P<0.01. D) Linear regression of expression of type III IFN receptor transcript vs frequency of CD11c+ CD21- (% cells) in naive and DN B cells subsets from healthy and SLE donors. Goodness of fit (r2) and p-value (F test) shown. E) Heat map of genes correlated with IFNLR1 transcript (pink) in SLE DN bulk transcriptomic analysis. Percentage of CD11c+ CD21- cells in each sample shown in gold. Increasing IFNLR1 transcript was associated with an increase in frequency of CD11c+ CD21- cells in the sample as measured by flow cytometry and increase in DN2-associated mRNA transcripts FCRL5 and ZEB2 and decrease transcript for negative markers for DN2 cells (such as CD27 and CXCR5) as measured by bulk RNA-seq
Type III interferon receptor mRNA is increased in DN B cells
Conflicting reports exist within the literature regarding whether B cells express functional type III IFN receptor (29, 42). To determine which IFNs could contribute to the ISG signature in lupus B cells, we compared IFN receptor RNA transcript expression in the healthy versus SLE naïve and DN B cells. For the type I IFN receptor, there was no difference in the IFNAR2 normalized transcript counts between SLE and healthy donors for either naïve or DN B cells (Fig. 2B). For the type II receptor, more IFNGR1 transcript was detected amongst the naive cells compared to the DN for both HD and SLE (Fig. 2B). DN B cells expressed more transcript for the unique IFNLR1 subunit of the type III IFN receptor than did naïve for both healthy and SLE donors (Fig. 2B). For IFNLR1, a significant interaction (p < 0.02) was found between B cell subset and disease status. Amongst the SLE DN B cells, there was more heterogeneity in IFNLR1 expression compared to the naïve B cell subset (Fig. 2B and supplemental Fig. 2A).
In order to examine which IFNs contribute to the DN B cell transcriptomic heterogeneity, we evaluated the relationship between CD11c+ CD21- B cell abundance as determined by flow cytometry and the expression of each of the IFN receptor transcripts (Fig. 2D for IFNLR1 and Supplemental Fig. 2B). IFNLR1 was highly and positively correlated (Pearson r = 0.97) with percentage of CD11c+ CD21- B cells (Fig. 2D). High numbers of IFNLR1 transcripts in DN from SLE patients positively associated with expression of known DN2/ABC markers, including FCRL3, FCRL5, and ZEB2, and negatively associated with CXCR5 and CD27 transcript (Fig. 2E). This association of the IFNLR1 transcript with DN2/ABC cells suggests that IFN-λ could influence the extra-follicular B cell differentiation pathways in SLE.
Peripheral Blood Human DN2/ABC Cells Correlate with IFN-λ Serum Levels
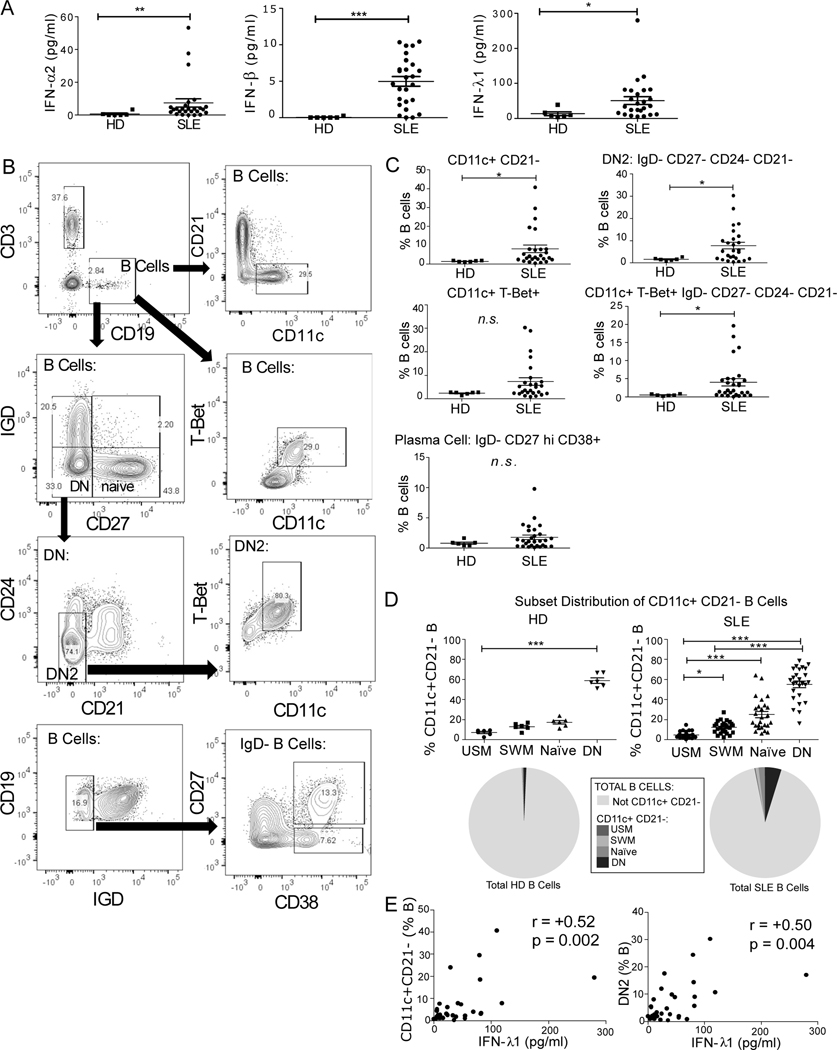
Given that transcriptomic gene set enrichment analysis identified a strong IFN gene signature and both type I and type III IFN receptors in SLE B cells, we next sought to determine which IFNs are present in SLE serum. We obtained serum and peripheral blood cells from SLE (n=26) and healthy (n=6) donors (Table 1) and measured both type I IFN (all subtype IFN-α as well as IFN-β) and type III IFN (IFN-λ1). All three IFN were detectable at increased levels in the SLE patients compared to healthy controls (Fig. 3A). B cell phenotype from these donors was determined by flow cytometry (Fig. 3B). A statistically significant expansion in phenotypes associated with extra-follicular pathway PC precursors including CD19+ CD11c+ CD21-, CD19+ IgD- CD27- CD21- CD24- (DN2), and CD19+ T-bet+ CD11c+ DN2 B cell subsets was found in SLE patients compared to healthy controls (Fig. 3C). The highest percentage of CD11c+ CD21- B cells were found in the DN compartment (Fig. 3D, HD 58.9±2.7%, SLE 55.1±3.2% of CD11c+ CD21- B cells, mean±SEM). In SLE there is an expansion of the CD11c+ CD21- DN cells (Fig. 3D, mean for HD or SLE shown as pie graph) amongst total B cells. CD11c+ CD21- B cells showed a positive correlation with serum levels of IFN-λ1 (Fig. 3D). IFN-α (all subtypes) and IFN-β showed weaker positive correlation with these subsets and did not achieve statistical significance (Table 3). These data raised the possibility that IFN-λ could be playing a role in induction of atypical B cells in lupus.
Fig. 3. IFN-λ1 Positively Correlates with DN2 B cells.
A) Type I and type III IFN serum levels are increased in SLE. Type I and type III interferon was measured in the serum of healthy (n = 6) and SLE (n = 26) by ELISA. IFN-α, IFN-β, and IFN-λ1 were all detectable in subsets of the lupus patients. Statistical significance calculated by Mann-Whitney U Test. B) Flow cytometry gating strategy for ex vivo peripheral blood mononuclear cells from HD and SLE donors. C) Expansion of B cell subsets associated with plasma cell development in SLE patients. Statistical significance calculated by Mann-Whitney U Test * P <0.05, ** P<0.01. D) CD11c+ CD21- B Cell distribution in each B cell subset in HD and SLE (expressed as percentage of total CD11c+ CD21- cells (top) and as pie graph of total B cells (bottom)). Statistics calculated as a Friedman Test with Dunn’s post-test. E) Correlation between IFN-λ1 serum levels and CD11c+ CD21- or DN2 (IgD- CD27- CD24- CD21-) B cells in the cohort of SLE and healthy patients. Spearman correlation coefficient (r) shown.
Table 3: Correlation of interferon serum levels correlate with ABC-associated B cell phenotypes.
Type I (all subtype IFN-α and IFN-β) and type III IFN (IFN-λ1) serum levels were measured in healthy (n = 6) and SLE (n = 26) by ELISA. Spearman correlation coefficient and p-value shown between interferon serum level and peripheral blood CD19+ B cell subsets.
| B cell Subset: Interferon: | CD11c+ CD21- | DN (IgD-, CD27-) | DN2 (IgD- CD27- CD24- CD21-) | DN2 T-Bet+ CD11c+ | T-Bet+ CD11c+ CD21- CD24- | T-Bet+ | T-Bet+ CD11c+ |
|---|---|---|---|---|---|---|---|
| IFN-α | p = 0.08 r= +0.31 |
p = 0.26 r = +0.20 |
p = 0.19 r = +0.24 |
p = 0.41 r = +0.15 |
p = 0.09 r = +0.31 |
p = 0.67 r = +0.07 |
p = 0.32 r = +0.18 |
| IFN-β | p = 0.10 r = +0.29 |
p = 0.31 r = +0.19 |
p = 0.04
r = +0.37 |
p= 0.09 r= + 0.31 |
p = 0.06 r = +0.34 |
p = 0.51 r= +0.12 |
p = 0.49 r = +0.13 |
| IFN-λ1 |
p = 0.002
r = +0.52 |
p = 0.05
r = +0.35 |
p = 0.004
r = +0.50 |
p = 0.03
r = +0.38 |
p = 0.003
r = +0.52 |
p = 0.45 r = +0.14 |
p = 0.03
r = +0.38 |
Human B Cells Are Responsive to IFN-λ
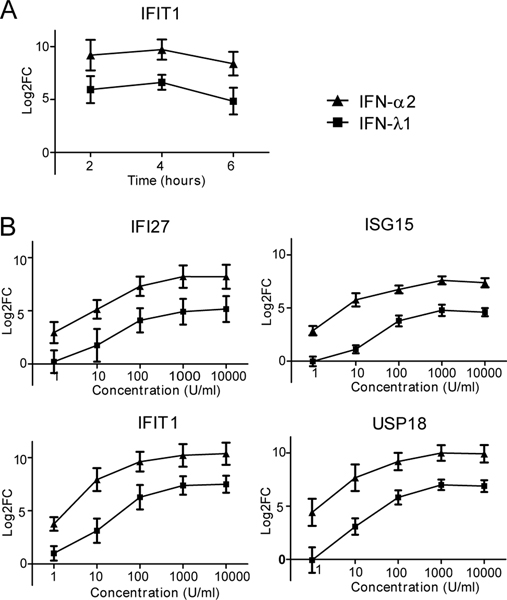
As a first step to understand the influence of IFN-λ on B cells, we asked if B cells directly respond to type III IFN stimulation. Time course experiments demonstrated expression of IFIT1 at 2, 4 and 6 hours for both IFN-λ1 and IFN-α2 treatment (Fig. 4A, n=4 donors, independent experiments, log2fold change mean±SEM). We measured select ISGs identified in our SLE B cell RNA-seq transcriptomic analysis. Relative expression of IFIT1, IFI27, ISG15 and USP18 was quantitated for IFN-λ1 and IFN-α2 stimulation at concentrations from 1 to 10,000U/ml (Fig. 4B, n=4 donors, independent experiments, log2fold change mean±SEM). Both IFN-λ1 and IFN-α2 induced expression of these ISGs, but higher concentration of IFN-λ1 were required, and the magnitude of mRNA induction was greater with IFN-α2. IFN-λ1 and IFN-α2 treatment did not induce B cell expression of IFNA2, IFNB1, or IFNL1, which were undetectable by quantitative RT-PCR (data not shown). Thus, we confirmed based on induction of ISG mRNA that human B cells are directly responsive to type III IFN, but also found IFN-α2 had a more potent effect.
Fig. 4. Human B cells are responsive to IFN-λ1.
Interferon stimulated gene expression in HD CD19+ B lymphocytes as measured by RT-qPCR log2 fold change (mean±SEM) relative to GAPDH in response to IFN-α2 or IFN-λ1. A) Time course of IFIT1 mRNA expression (n=4 independent experiments with n=4 donors, 500U/ml of IFN). B) Dose response. Gene expression for IFIT1, IFI27, ISG15 and USP18 in response to IFN-α2 (left) and IFN-λ1 (right) at concentrations from 1 to 10,000U/ml at 4 hours (n = 4 independent experiments with n=4 donors).
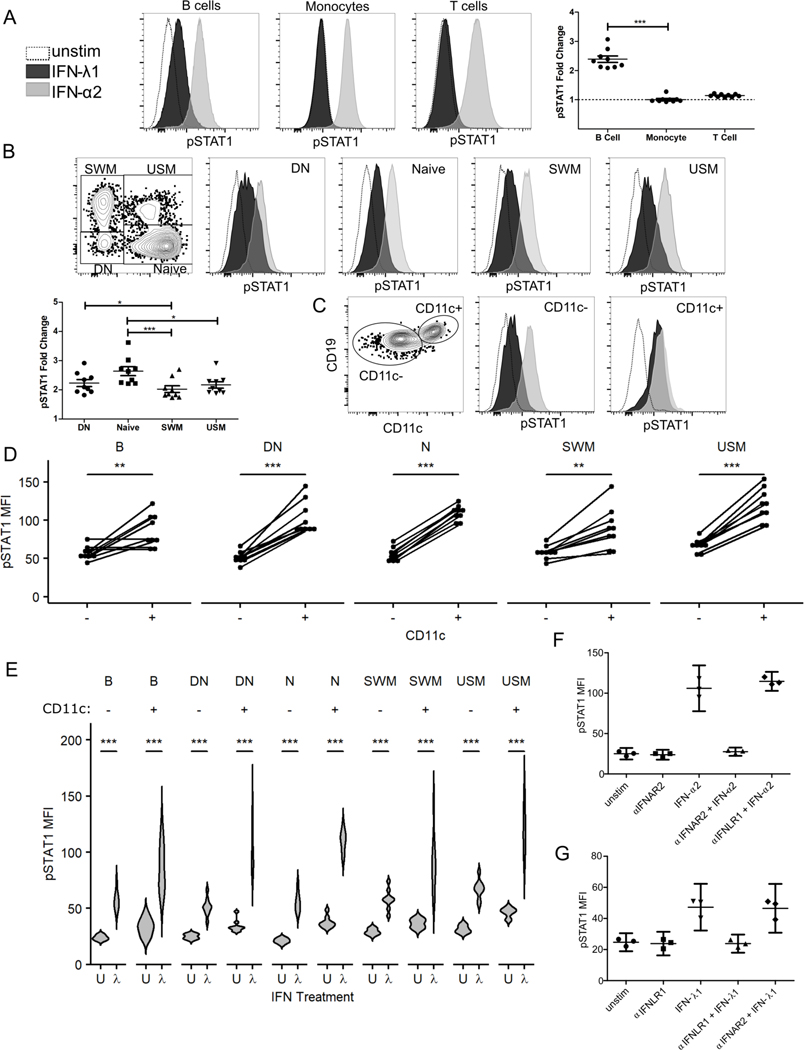
IFN-λ Promotes STAT1 Phosphorylation especially in CD11c+ B Cells
In order further define the B cell subsets targeted by IFN-λ, peripheral blood B cells were stimulated with IFN-λ1 or IFN-α2 and phosphorylated STAT1 measured by flow cytometric analysis. STAT1 is a protein which becomes phosphorylated during signal transduction for both IFN-α2 and IFN-λ1. A time course experiment was performed that determined B cells phosphorylate STAT1 faster after IFN-α2 stimulation compared to IFN-λ1 (Supplemental Fig. 2C). A dose response experiment demonstrated maximal STAT1 phosphorylation with 500U/ml of IFN-α2 and 2000U/ml for IFN-λ1 (data not shown). These concentrations were used for subsequent STAT1 phosphorylation experiments at a time of 25 minutes. Total CD3+ T lymphocytes and CD14+ monocytes demonstrated an increased median fluorescent intensity for phosphorylated STAT1 protein after IFN-α2 treatment, but not for IFN-λ1 stimulation (Fig. 5A) in agreement with recent reports (30). Phosphorylated STAT1 protein increased after IFN-λ1 stimulation in total human B cells, though to a lesser degree than with IFN-α2 (Fig. 5B). Further phenotyping of B cells showed that naïve (IgD+ CD27-), double negative (IgD- CD27-), switched memory (IgD- CD27+) and unswitched memory (IgD+ CD27+) B cells all increased phosphorylation of STAT1 with IFN-λ1 and IFN-α2 stimulation (Fig. 5B). Of these, naïve cells had the highest fold change. In each population, IFN-α2 had a larger effect than IFN-λ1. For donors with a clearly defined CD11c+ DN population, a robust phosphorylation of STAT1 was evident in response to IFN-λ1 that approached that of IFN-α2 (Fig. 5C). CD11c+ B cells had a higher baseline pSTAT1 compared to their CD11c- counterparts and higher levels of pSTAT1 in response to IFN-λ1 in all B cell subsets (Fig. 5D, E). CD11c+ DN B cells (mean±SD, 3.03±0.43, n=9) and CD11c+ naïve B cells (mean±SD, 2.97±0.41, n=9) had the highest fold changes with IFN-λ1 stimulation. These results are in agreement with previous reports that DN2, which are CD11c+, are hyper-responsive to IFN-λ (22). Interferon receptor neutralization experiments using blocking antibody to IFNAR2 (type I IFN) or IFNLR1 (type III IFN) confirmed the specificity of the effect for both type I and type III IFN (Fig. 5F and 5G respectively, n=3 HD, each point indicates a donor, each run as independent experiments, mean±95% confidence interval shown for median pSTAT1 for CD19+ B cells). As expected, these results demonstrate a type I IFN (IFN-α2) broadly stimulates multiple types of peripheral human blood cells including monocytes, T cells, and B cells. In contrast, type III IFN (IFN-λ1) is specific for B cells and is particularly effective for CD11c+ expressing B cells, such as the DN2/ABC subset.
Fig. 5. CD11c+ B cells are hyper-responsive to IFN-λ1 stimulation.
Peripheral blood mononuclear cells were stimulated with IFN-α2 (500U/ml) or IFN-λ1 (2000U/ml) for 25 minutes for detection of phosphorylated STAT1 protein by flow cytometry. Histograms shown pSTAT1 for unstimulated (dashed), IFN-λ1 (black), and IFN-α2 (gray) stimulated cells. A) Fold change of IFN-λ1 stimulated/unstimulated pSTAT1 median fluorescent intensity expression shown for CD14+ monocyte, CD3+ T or CD19+ CD20+ B cells (n=9 HD, mean±SEM). Friedman test with p-values shown from Dunn’s multiple comparison post-test. B) B cell subpopulation gating strategy (left top). Histograms and fold change (bottom left) of IFN-λ1 stimulated/unstimulated for pSTAT1 for each B cell subset (n=9 HD, mean±SEM). Statistics calculated by Friedman test with p-values shown from Dunn’s multiple comparison post-test. C) CD11c+ expression in DN cells from donor with well-defined DN2 population. CD11c+ and CD11c- DN gating strategy and pSTAT1 histograms shown. D) pSTAT1 MFI after IFN-λ1 stimulation for CD11c- and CD11c+ paired data for each B cell subset. Lines represent individual donors with p-values from Wilcoxon Signed Rank Test. E) Violin plots of pSTAT1 MFI for unstimulated (U) and after IFN-λ1 (λ) stimulation for CD11c- and CD11c+ B cell subsets. Statistics calculated as paired, non-parametric two-tailed Wilcoxon Signed Rank Test. * P <0.05, ** P<0.01, *** P<0.001. F & G) IFN receptor neutralization specifically and significantly reduced pSTAT1 phosphorylation induced by IFN stimulation of CD19+ HD B cells. Antibody blockade of type I IFN receptor (IFNAR2) reduced pSTAT1 MFI after IFN-α2 stimulation which was unaffected by type III IFN receptor (IFNLR1) blockade (n=3, F). Similarly, IFN-λ1 induced STAT1 phosphorylation was inhibited with anti-IFNLR1 but not by type I IFN receptor blockade with anti-IFNAR2 (n=3, G). Mean± 95% confidence interval shown (F & G).
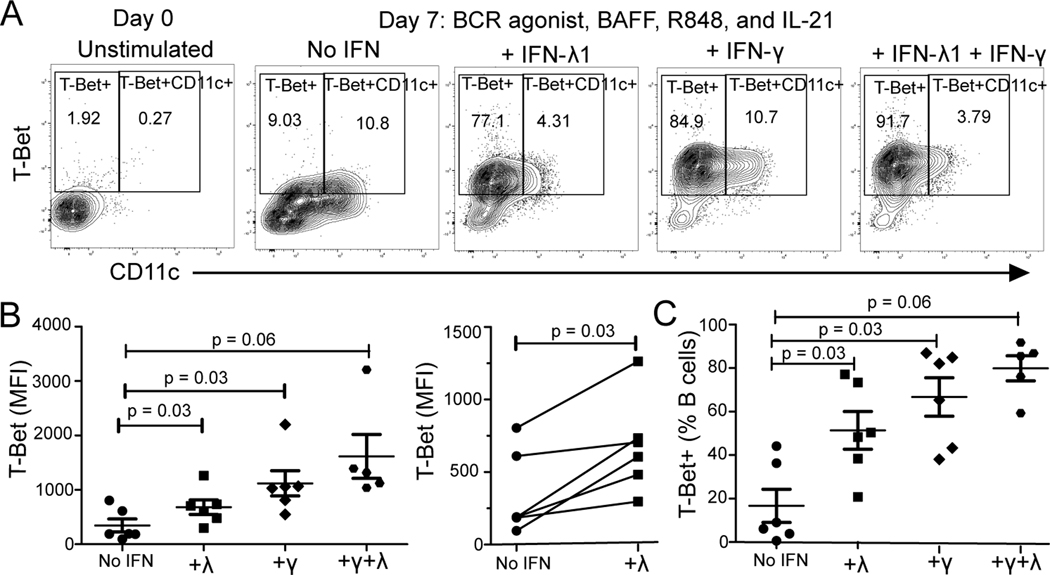
IFN-λ can induce T-bet+ B cells
Antibody-secreting cell precursors DN2 or ABCs have been reported to be generated from human B cultures containing anti-immunoglobulin B cell receptor (BCR) stimulation, TLR7 stimulation, IL-21, BAFF and IFN-γ (type II IFN) (20, 43). As our data demonstrate that ABC phenotype and serum IFN-λ1 (type III IFN) correlate in SLE patients in vivo (Fig. 3D), we assessed whether type III IFN-λ can induce DN2, similar to IFN-γ in vitro. Human CD19+ B cells from healthy donors (n=6) were BCR and TLR7/8 stimulated with or without IFN-λ1 or IFN-γ in the presence of BAFF and IL-21 for 7 days. T-bet+ CD11c+ B cells developed under these culture conditions (Fig. 6A). BCR and TLR7/8 stimulation upregulated CD11c expression in the TLR7/8 activated cultures regardless of whether IFN was included but the percentage of T-bet+ B cells increased only in conditions with either IFN (Fig. 6A, n=6). Increased frequencies of T-bet+ B cells were found amongst B cells differentiated with BCR and TLR7/8 stimulation in cultures containing IFN-λ1 compared to cultures without IFN (Fig. 6C) and as expected, cultures containing IFN-γ generated T-bet+ B cells at high frequency (Fig. 6C, Wilcoxon signed rank test). Additionally, T-bet protein levels measured by geometric mean fluorescent intensity (MFI) increased in the IFN-λ1 containing conditions compared to those without IFN, but to a lesser degree than in IFN-γ-containing cultures (Fig. 6B).
Fig. 6. IFN-γ and IFN-λ Effects on T-bet+ B cell Differentiation via TLR7/8 (R848) and BCR Agonist Activation.
Human CD19+ magnetic bead selection was used to isolate total B cells from healthy donors (n=6) for 7 day culture with TLR7/8 agonist R848 + BCR stimulation (anti-Ig(A+M+G)), with or without IFN-λ1 or IFN-γ in the presence of BAFF and IL-21. A) Flow cytometry plots of CD11c versus T-bet gated on B cells ex vivo and on day 7 of stimulation. B) Geometric mean fluorescent intensity (MFI) of T-bet in B cells in TLR7/8 and BCR activated cultures with no IFN, IFN-λ1, IFN-γ or both (λ+γ). Points connected by line represent individual donors after IFN-λ1 treatment. C) Frequency of T-bet+ B cells. Data are represented at mean±SEM. P-values shown from paired, non-parametric Wilcoxon Signed Rank Test. * indicates p’ <0.05 after Bonferroni correction for multiple testing applied for three comparisons.
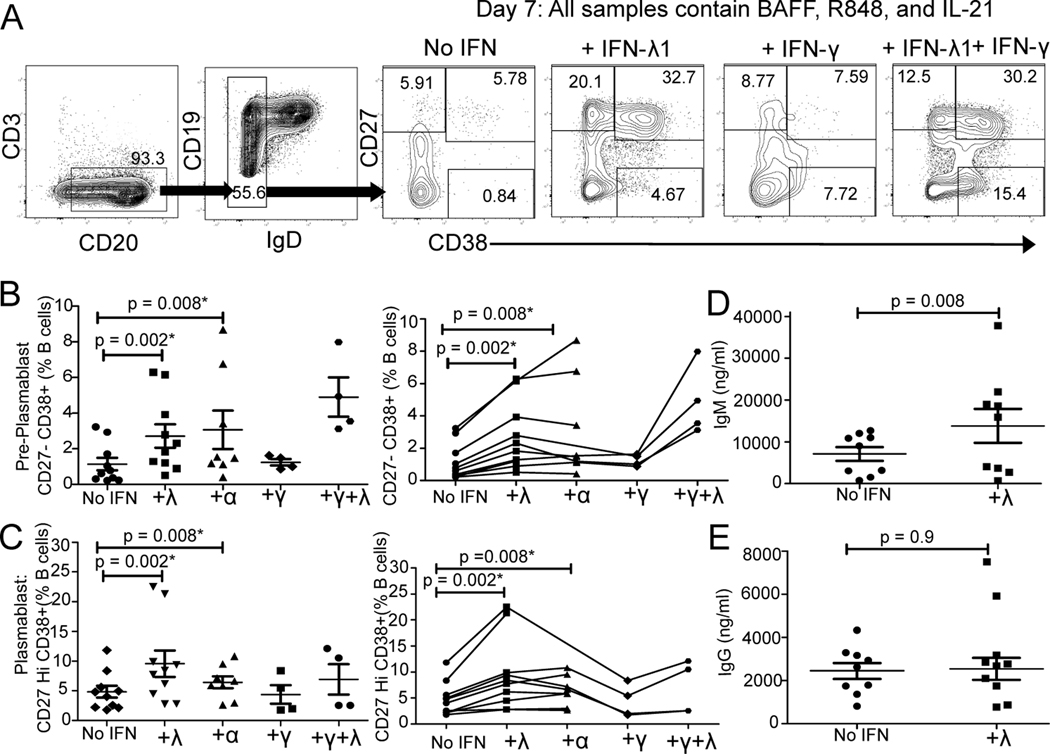
IFN-λ Accelerates Plasmablast Differentiation
Given T-bet expression increased in our TLR7/8-activated cultures exposed to IFN-λ1 and T-bet promotes antibody-secreting cell differentiation in mice (44), we sought to determine the effects of IFN-λ stimulation on the differentiation of plasmablasts. Human B cells from healthy donors were cultured for 7 days with the TLR7/8 agonist +/− IFN-λ1 and plasmablasts enumerated based on high CD27 and CD38 expression. We found a statistically significant enhancement in CD27negCD38+ pre-plasmablasts and CD27hi CD38+ plasmablasts with the addition of IFN-λ1 (Fig. 7A–C, No IFN versus +α or +λ). We also directly compared the effects of IFN-λ1 to type I IFN-α2. The addition of either type I (IFN-α2, n=9) or type III (IFN- λ1, n=10) to the cultures caused a similar statistically significant increase in CD27negCD38+ pre-plasmablasts and CD27hi CD38+ plasmablasts (Fig. 7B–C, No IFN versus +α or +λ).
Fig. 7. TLR7/8 (R848) and IFN-λ Accelerates Plasmablast Differentiation.
Human CD19+ positively selected total B cells from healthy donors were cultured for 7 days with the TLR7/8 agonist R848 with or without interferon in the presence of BAFF and IL-21. A) Flow cytometry plots of CD38 versus CD27 gated on IgD- B cells. B-E) Effects of addition of IFN-λ1 (λ, type III, n=10), IFN-α2 (α, type I, n=8) IFN-γ (γ, type II, n=4) or IFN-γ plus IFN-λ1 (n=4) are compared. Points connected by lines represent an individual donor. B) Percentage of IgD- CD27- CD38+ pre-plasmablasts and C) Percentage of IgD- CD27 Hi CD38+ plasmablasts quantitated by flow cytometry. D) IgM and E) IgG quantitation in the culture supernatants as measured by ELISA. Data are represented at mean±SEM. P values shown from paired, non-parametric Wilcoxon Signed Rank Test. * indicates p’ <0.05 after Bonferroni correction for multiple testing applied for four comparisons.
To further define the effects of IFN-λ1 on PC differentiation, we measured IgG and IgM secretion in the cultures. R848, BAFF and IL-21 stimulation resulted in IgG and IgM secretion (Fig. 7D–E, No IFN). Inclusion of IFN-λ1 in the R848, BAFF and IL-21 stimulated cultures significantly enhanced IgM, but not IgG production (Fig. 7D, +λ). IFN-λ enhances B cell differentiation into plasmablasts and increases secretion of IgM.
Discussion
Type I IFN has long been associated with SLE. However, the type I IFN gene transcription profile is indistinguishable from that produced by the type III IFN (IFN-λ) in cells that express IFN-λ receptor (IFNLR1). Here, we have found that type III IFN (IFN-λ1), in addition to type I IFN (IFN-α and IFN-β), is increased in SLE serum. IFN-λ1 serum levels, but not IFN-α and IFN-β levels, correlate with B cell phenotypes associated with DN2/ABC (increased CD11c expression and down-regulation of CD21). We show that human B cells express transcripts for the receptors of all three types of IFN and that the level of IFNLR1 mRNA transcripts in DN B cells correlates with the frequency of DN2/ABC. By measuring STAT1 phosphorylation we were able to show that the IFN-λ receptor is functional in all major B cell subsets and that levels of STAT1 phosphorylation after IFN-λ stimulation are greatest in B cells with cell surface markers for DN2/ABC. Moreover, in all major B cell subsets CD11c+ B cells responded better to IFN-λ than CD11c- B cells. In marked contrast, peripheral blood T cells and monocytes are not responsive to IFN-λ. Functionality of the IFN-λ receptor on B cells was further demonstrated by induction of ISG transcripts upon stimulation with IFN-λ1. We also demonstrate that IFN-λ1 promotes TLR7/8-mediated differentiation of human B cells to antibody-secreting plasma cells. As CD11c+ B cells are associated with plasma cell differentiation (45), we also establish that IFN-λ1 increases the numbers of T-bet+ B cells and plasmablasts amongst TLR7-activated B cell cultures. Thus, our data confirms recent reports that IFN-λ stimulates human B cells (22, 24, 27–31) and is likely a driver of the prominent IFN signature and B cell dysregulation in SLE.
Even though type I IFN induces a similar transcriptional profile on a broader distribution of cell types, non-redundant roles for IFN-λ are described. In a murine TLR7-driven model of lupus, IFNLR1−/− mice had reduced skin and renal inflammation compared to wild type mice (28). In this setting, IFN-λ effects were mediated via chemokine expression in tissue leading to recruitment of inflammatory cells (28). However, murine models cannot completely recapitulate human SLE given murine B cells are not directly responsive to IFN-λ (28). In epithelial cell viral infections, IFN-λ1 is produced at greater concentrations and with more prolonged duration of action than IFN-α (46, 47). If similar IFN-λ1 kinetics are also found in SLE, we speculate there may be points in the disease course where IFN-λ becomes more dominant in its effects on human B cells.
During lupus flare, there is an increase in the IgD- CD27- B cell compartment due to expansion of CD11c+ CD21- DN2/ABC cells (48, 49). Our data show a correlation between DN2/ABC frequency and IFN-λ1 serum levels. This subset is of significant interest as it is poised for PC differentiation and enriched in auto-specificities (18). In SLE, these DN2/ABC cells share transcriptomic and epigenetic features with the naïve compartment (20, 50, 51). Trajectory analysis supports a resting naïve to activated naïve (CD11c+T-bet+) to DN2 to antibody-secreting cell differentiation pathway (20, 50, 51). IFN-λ3 augmentation of IgM plasmablast differentiation from naïve B cells using a BCR-activated system was recently described where effects were mediated via the mTORC1 pathway (30). Our data also shows that IFN-λ1 augments production of IgM in connection with increased T-bet+ B cell and plasma cell differentiation using TLR7 for activation. Our experiments do not directly address the effects of IFN-λ on class switching and IgG plasma cell differentiation in part because of the lack of CD40 stimulation in the cultures.
TLR7 stimulation generates DN2/ABCs via an extrafollicular pathway in viral and autoimmune disease (22). In infection, the extrafollicular response provides IgM-producing plasmablasts early in the infection course (52–55). In lupus, DN2/ABC cells are expanded in both the blood and tissues (51), such as the renal microenvironment, where they are speculated to contribute to local production of autoantibodies that result in end-organ damage. Plasmacytoid dendritic cells, activated T cells and cells of epithelial origin are capable of type III IFN production. Thus, RNA-containing immune complexes can stimulate plasmacytoid dendritic cells to produce IFN-λ1 (56). Immunohistochemistry staining of renal lupus nephritis biopsies identified IFN-λ protein (57, 58). We previously reported that primary renal tubule epithelial produce IFN-λ in vitro in response to TLR3 stimulation (59). Thus, within the renal microenvironment, we posit that renal tubule epithelial cells could promote T-bet-expressing DN2/ABC cells and subsequent differentiation towards plasmablasts. In other autoimmune diseases, similar networks might contribute to the generation of autoimmunity in other epithelial tissues. The relative contribution of type III versus other classes of IFN to autoimmunity remains to be parsed apart.
These findings are of critical clinical importance given type I IFN remains a promising SLE therapeutic target of interest culminating in clinical trials for type I IFN receptor blockade with the monoclonal antibody anifrolumab (60). However, type I IFN monoclonal antibody blockade would not neutralize IFN-λ given type I and type III IFN signal through unique receptors. As type III IFN induces similar genes as type I IFN in cell types capable of expressing the receptor, type III IFN might also drive disease pathogenesis. Alternatively, escape of IFN-λ from mechanisms targeting type I IFN might serve as a safety net to prevent severe immunosuppression that might occur if all IFNs were universally blocked therapeutically or via viral proteins. In contrast to IFN blockade, pegylated IFN-λ was developed as antiviral therapeutic for hepatitis C (61) and is in trial for use in the treatment of SARS-CoV-2 and may be less inflammatory than type I IFN treatment (62). Our phosphorylation data suggests this is likely true given minimal responsiveness of monocytes and resting T cells to IFN-λ, but we would anticipate this therapy to still have effects on human B cells.
The use of clinically relevant human and SLE patient samples is a strength of our study. Previous studies relying upon Western blot for the measurement of STAT1 phosphorylation events suggested B cells were poorly responsive to IFN-λ1 (42, 63). The use of single cell measurements of phosphorylation events via flow cytometry allowed phenotyping of responsive B cells in our study. Weaknesses of our study include the use of bulk transcriptomic analysis as opposed to single cell transcriptomic analysis and a reliance on in vitro cultures given the limitations associated with working with human samples.
In conclusion, our studies highlight that IFN-λ directly targets human B cells to promote differentiation to plasmablasts. IFN-λ preferentially targets CD11c+ B cells, the majority of which are found in the DN compartment and expanded in SLE. Therefore, IFN-λ may contribute to autoantibody formation via the TLR7-stimulated extra-follicular B cell activation pathway from naïve to DN2/ABC to antibody-secreting plasma cell. In conclusion, IFN-λ is likely an underappreciated player in promoting antibody responses for autoimmune diseases like SLE and may have implications in viral infection.
Supplementary Material
Key points:
A type I IFN transcriptional profile is upregulated in SLE B cells.
Human B cells express type I IFN genes in response to IFN-λ (type III IFN).
IFN-λ may enhance TLR7-mediated B cell activation and PC differentiation in SLE.
Acknowledgements
The authors thank Tyler Cavin, Lin Gao, PhD; Amanda Howell, Neha Nandedkar-Kulkarni, PhD; Mary O’Connell, and Javier Rangel-Moreno, PhD for their technical assistance. We thank the staff of the University of Rochester Genomics Core for performing RNA sequencing for this manuscript.
This work was supported by the Rheumatology Research Foundation (Scientist Development Award 060631-02 to J.L.B), National Institute of Arthritis and Musculoskeletal and Skin Diseases (Accelerated Medicines Partnership Grant 1UH2-Ar-067690 to J.H.A.) and the Bertha and Louis Weinstein Research Fund (to J.H.A.).
References
- 1.Furie R, Rovin BH, Houssiau F, Malvar A, Teng YKO, Contreras G, Amoura Z, Yu X, Mok CC, Santiago MB, Saxena A, Green Y, Ji B, Kleoudis C, Burriss SW, Barnett C, and Roth DA 2020. Two-Year, Randomized, Controlled Trial of Belimumab in Lupus Nephritis. The New England journal of medicine 383: 1117–1128. [DOI] [PubMed] [Google Scholar]
- 2.Furie R, Cascino MD, Garg JP, Aroca G, Alvarez A, Fragoso-Loyo H, Santillán EZ, Malvar A, Brunetta PG, Schindler T, Looney CMD, Hassan I, and Rovin BH 2020. O35 B-cell depletion and response in a randomized, controlled trial of obinutuzumab for proliferative lupus nephritis. Lupus Science & Medicine 7: A27–A28. [Google Scholar]
- 3.Postal M, Vivaldo JF, Fernandez-Ruiz R, Paredes JL, Appenzeller S, and Niewold TB 2020. Type I interferon in the pathogenesis of systemic lupus erythematosus. Current opinion in immunology 67: 87–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang J, Zhao M, Chang C, Wu H, and Lu Q. 2020. Type I Interferons in the Pathogenesis and Treatment of Autoimmune Diseases. Clinical reviews in allergy & immunology 59: 248–272. [DOI] [PubMed] [Google Scholar]
- 5.Bekeredjian-Ding IB, Wagner M, Hornung V, Giese T, Schnurr M, Endres S, and Hartmann G. 2005. Plasmacytoid dendritic cells control TLR7 sensitivity of naive B cells via type I IFN. Journal of immunology (Baltimore, Md. : 1950) 174: 4043–4050. [DOI] [PubMed] [Google Scholar]
- 6.Braun D, Caramalho I, and Demengeot J. 2002. IFN-α/β enhances BCR-dependent B cell responses. International Immunology 14: 411–419. [DOI] [PubMed] [Google Scholar]
- 7.Liu M, Guo Q, Wu C, Sterlin D, Goswami S, Zhang Y, Li T, Bao C, Shen N, Fu Q, and Zhang X. 2019. Type I interferons promote the survival and proinflammatory properties of transitional B cells in systemic lupus erythematosus patients. Cellular & Molecular Immunology 16: 367–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruuth K, Carlsson L, Hallberg B, and Lundgren E. 2001. Interferon-alpha promotes survival of human primary B-lymphocytes via phosphatidylinositol 3-kinase. Biochem Biophys Res Commun 284: 583–586. [DOI] [PubMed] [Google Scholar]
- 9.Jego G, Palucka AK, Blanck JP, Chalouni C, Pascual V, and Banchereau J. 2003. Plasmacytoid dendritic cells induce plasma cell differentiation through type I interferon and interleukin 6. Immunity 19: 225–234. [DOI] [PubMed] [Google Scholar]
- 10.Lam JH, Smith FL, and Baumgarth N. 2020. B Cell Activation and Response Regulation During Viral Infections. Viral Immunology 33: 294–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kratzer B, Trapin D, Ettel P, Kormoczi U, Rottal A, Tuppy F, Feichter M, Gattinger P, Borochova K, Dorofeeva Y, Tulaeva I, Weber M, Grabmeier-Pfistershammer K, Tauber PA, Gerdov M, Muhl B, Perkmann T, Fae I, Wenda S, Fuhrer H, Henning R, Valenta R, and Pickl WF 2020. Immunological imprint of COVID-19 on human peripheral blood leukocyte populations. Allergy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sosa-Hernández VA, Torres-Ruíz J, Cervantes-Díaz R, Romero-Ramírez S, Páez-Franco JC, Meza-Sánchez DE, Juárez-Vega G, Pérez-Fragoso A, Ortiz-Navarrete V, Ponce-de-León A, Llorente L, Berrón-Ruiz L, Mejía-Domínguez NR, Gómez-Martín D, and Maravillas-Montero JL 2020. B Cell Subsets as Severity-Associated Signatures in COVID-19 Patients. Frontiers in Immunology 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woodruff MC, Ramonell RP, Nguyen DC, Cashman KS, Saini AS, Haddad NS, Ley AM, Kyu S, Howell JC, Ozturk T, Lee S, Suryadevara N, Case JB, Bugrovsky R, Chen W, Estrada J, Morrison-Porter A, Derrico A, Anam FA, Sharma M, Wu HM, Le SN, Jenks SA, Tipton CM, Staitieh B, Daiss JL, Ghosn E, Diamond MS, Carnahan RH, Crowe JE Jr., Hu WT, Lee FE, and Sanz I. 2020. Extrafollicular B cell responses correlate with neutralizing antibodies and morbidity in COVID-19. Nature immunology 21: 1506–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Odendahl M, Jacobi A, Hansen A, Feist E, Hiepe F, Burmester GR, Lipsky PE, Radbruch A, and Dorner T. 2000. Disturbed peripheral B lymphocyte homeostasis in systemic lupus erythematosus. Journal of immunology (Baltimore, Md. : 1950) 165: 5970–5979. [DOI] [PubMed] [Google Scholar]
- 15.Grammer AC, and Lipsky PE 2003. B cell abnormalities in systemic lupus erythematosus. Arthritis research & therapy 5 Suppl 4: S22–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hao Y, O’Neill P, Naradikian MS, Scholz JL, and Cancro MP 2011. A B-cell subset uniquely responsive to innate stimuli accumulates in aged mice. Blood 118: 1294–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rubtsov AV, Rubtsova K, Fischer A, Meehan RT, Gillis JZ, Kappler JW, and Marrack P. 2011. Toll-like receptor 7 (TLR7)-driven accumulation of a novel CD11c(+) B-cell population is important for the development of autoimmunity. Blood 118: 1305–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rubtsov AV, Rubtsova K, Kappler JW, and Marrack P. 2013. TLR7 drives accumulation of ABCs and autoantibody production in autoimmune-prone mice. Immunol Res 55: 210–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang S, Wang J, Kumar V, Karnell JL, Naiman B, Gross PS, Rahman S, Zerrouki K, Hanna R, Morehouse C, Holoweckyj N, Liu H, Manna Z, Goldbach-Mansky R, Hasni S, Siegel R, Sanjuan M, Streicher K, Cancro MP, Kolbeck R, and Ettinger R. 2018. IL-21 drives expansion and plasma cell differentiation of autoreactive CD11c(hi)T-bet(+) B cells in SLE. Nature communications 9: 1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zumaquero E, Stone SL, Scharer CD, Jenks SA, Nellore A, Mousseau B, Rosal-Vela A, Botta D, Bradley JE, Wojciechowski W, Ptacek T, Danila MI, Edberg JC, Bridges SL Jr., Kimberly RP, Chatham WW, Schoeb TR, Rosenberg AF, Boss JM, Sanz I, and Lund FE 2019. IFNgamma induces epigenetic programming of human T-bet(hi) B cells and promotes TLR7/8 and IL-21 induced differentiation. eLife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rubtsova K, Rubtsov AV, Thurman JM, Mennona JM, Kappler JW, and Marrack P. 2017. B cells expressing the transcription factor T-bet drive lupus-like autoimmunity. The Journal of clinical investigation 127: 1392–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jenks SA, Cashman KS, Zumaquero E, Marigorta UM, Patel AV, Wang X, Tomar D, Woodruff MC, Simon Z, Bugrovsky R, Blalock EL, Scharer CD, Tipton CM, Wei C, Lim SS, Petri M, Niewold TB, Anolik JH, Gibson G, Lee FE, Boss JM, Lund FE, and Sanz I. 2018. Distinct Effector B Cells Induced by Unregulated Toll-like Receptor 7 Contribute to Pathogenic Responses in Systemic Lupus Erythematosus. Immunity 49: 725–739.e726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gatto M, Iaccarino L, Ghirardello A, Punzi L, and Doria A. 2016. Clinical and pathologic considerations of the qualitative and quantitative aspects of lupus nephritogenic autoantibodies: A comprehensive review. Journal of autoimmunity 69: 1–11. [DOI] [PubMed] [Google Scholar]
- 24.Kelly A, Robinson MW, Roche G, Biron CA, O’Farrelly C, and Ryan EJ 2016. Immune Cell Profiling of IFN-lambda Response Shows pDCs Express Highest Level of IFN-lambdaR1 and Are Directly Responsive via the JAK-STAT Pathway. Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research 36: 671–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hemann EA, Gale M Jr., and Savan R. 2017. Interferon Lambda Genetics and Biology in Regulation of Viral Control. Front Immunol 8: 1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou Z, Hamming OJ, Ank N, Paludan SR, Nielsen AL, and Hartmann R. 2007. Type III interferon (IFN) induces a type I IFN-like response in a restricted subset of cells through signaling pathways involving both the Jak-STAT pathway and the mitogen-activated protein kinases. Journal of virology 81: 7749–7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Groen RA, Groothuismink ZM, Liu BS, and Boonstra A. 2015. IFN-lambda is able to augment TLR-mediated activation and subsequent function of primary human B cells. Journal of leukocyte biology 98: 623–630. [DOI] [PubMed] [Google Scholar]
- 28.Goel RR, Wang X, O’Neil LJ, Nakabo S, Hasneen K, Gupta S, Wigerblad G, Blanco LP, Kopp JB, Morasso MI, Kotenko SV, Yu ZX, Carmona-Rivera C, and Kaplan MJ 2020. Interferon lambda promotes immune dysregulation and tissue inflammation in TLR7-induced lupus. Proc Natl Acad Sci U S A 117: 5409–5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santer DM, Minty GES, Golec DP, Lu J, May J, Namdar A, Shah J, Elahi S, Proud D, Joyce M, Tyrrell DL, and Houghton M. 2020. Differential expression of interferon-lambda receptor 1 splice variants determines the magnitude of the antiviral response induced by interferon-lambda 3 in human immune cells. PLoS pathogens 16: e1008515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Syedbasha M, Bonfiglio F, Linnik J, Stuehler C, Wuthrich D, and Egli A. 2020. Interferon-lambda Enhances the Differentiation of Naive B Cells into Plasmablasts via the mTORC1 Pathway. Cell Rep 33: 108211. [DOI] [PubMed] [Google Scholar]
- 31.Coto-Llerena M, Lepore M, Spagnuolo J, Di Blasi D, Calabrese D, Suslov A, Bantug G, Duong FH, Terracciano LM, De Libero G, and Heim MH 2021. Interferon lambda 4 can directly activate human CD19(+) B cells and CD8(+) T cells. Life Sci Alliance 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, Langer JA, Sheikh F, Dickensheets H, and Donnelly RP 2003. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nature immunology 4: 69–77. [DOI] [PubMed] [Google Scholar]
- 33.Hochberg MC 1997. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis and rheumatism 40: 1725. [DOI] [PubMed] [Google Scholar]
- 34.Bombardier C, Gladman DD, Urowitz MB, Caron D, and Chang CH 1992. Derivation of the SLEDAI. A disease activity index for lupus patients. The Committee on Prognosis Studies in SLE. Arthritis and rheumatism 35: 630–640. [DOI] [PubMed] [Google Scholar]
- 35.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England) 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen S, Zhou Y, Chen Y, and Gu J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics (Oxford, England) 34: i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liao Y, Smyth GK, and Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics (Oxford, England) 30: 923–930. [DOI] [PubMed] [Google Scholar]
- 38.Love MI, Huber W, and Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu G, Wang L-G, Han Y, and He Q-Y. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16: 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ginestet C. 2011. ggplot2: Elegant Graphics for Data Analysis. Journal of the Royal Statistical Society: Series A (Statistics in Society) 174: 245–246. [Google Scholar]
- 41.Kassambara A. 2018. ggpubr: ‘ggplot2’ based publication ready plots. [Google Scholar]
- 42.Witte K, Gruetz G, Volk HD, Looman AC, Asadullah K, Sterry W, Sabat R, and Wolk K. 2009. Despite IFN-lambda receptor expression, blood immune cells, but not keratinocytes or melanocytes, have an impaired response to type III interferons: implications for therapeutic applications of these cytokines. Genes and immunity 10: 702–714. [DOI] [PubMed] [Google Scholar]
- 43.Rubtsova K, Rubtsov AV, van Dyk LF, Kappler JW, and Marrack P. 2013. T-box transcription factor T-bet, a key player in a unique type of B-cell activation essential for effective viral clearance. Proc Natl Acad Sci U S A 110: E3216–3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stone SL, Peel JN, Scharer CD, Risley CA, Chisolm DA, Schultz MD, Yu B, Ballesteros-Tato A, Wojciechowski W, Mousseau B, Misra RS, Hanidu A, Jiang H, Qi Z, Boss JM, Randall TD, Brodeur SR, Goldrath AW, Weinmann AS, Rosenberg AF, and Lund FE 2019. T-bet Transcription Factor Promotes Antibody-Secreting Cell Differentiation by Limiting the Inflammatory Effects of IFN-gamma on B Cells. Immunity 50: 1172–1187 e1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Golinski M-L, Demeules M, Derambure C, Riou G, Maho-Vaillant M, Boyer O, Joly P, and Calbo S. 2020. CD11c+ B Cells Are Mainly Memory Cells, Precursors of Antibody Secreting Cells in Healthy Donors. Frontiers in Immunology 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jewell NA, Cline T, Mertz SE, Smirnov SV, Flaño E, Schindler C, Grieves JL, Durbin RK, Kotenko SV, and Durbin JE 2010. Lambda Interferon Is the Predominant Interferon Induced by Influenza A Virus Infection In Vivo. Journal of virology 84: 11515–11522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marcello T, Grakoui A, Barba-Spaeth G, Machlin ES, Kotenko SV, MacDonald MR, and Rice CM 2006. Interferons alpha and lambda inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology 131: 1887–1898. [DOI] [PubMed] [Google Scholar]
- 48.Jenks SA, Cashman KS, Woodruff MC, Lee FE, and Sanz I. 2019. Extrafollicular responses in humans and SLE. Immunol Rev 288: 136–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nehar-Belaid D, Hong S, Marches R, Chen G, Bolisetty M, Baisch J, Walters L, Punaro M, Rossi RJ, Chung CH, Huynh RP, Singh P, Flynn WF, Tabanor-Gayle JA, Kuchipudi N, Mejias A, Collet MA, Lucido AL, Palucka K, Robson P, Lakshminarayanan S, Ramilo O, Wright T, Pascual V, and Banchereau JF 2020. Mapping systemic lupus erythematosus heterogeneity at the single-cell level. Nature immunology 21: 1094–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scharer CD, Blalock EL, Mi T, Barwick BG, Jenks SA, Deguchi T, Cashman KS, Neary BE, Patterson DG, Hicks SL, Khosroshahi A, Eun-Hyung Lee F, Wei C, Sanz I, and Boss JM 2019. Epigenetic programming underpins B cell dysfunction in human SLE. Nature immunology 20: 1071–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arazi A, Rao DA, Berthier CC, Davidson A, Liu Y, Hoover PJ, Chicoine A, Eisenhaure TM, Jonsson AH, Li S, Lieb DJ, Zhang F, Slowikowski K, Browne EP, Noma A, Sutherby D, Steelman S, Smilek DE, Tosta P, Apruzzese W, Massarotti E, Dall’Era M, Park M, Kamen DL, Furie RA, Payan-Schober F, Pendergraft WF 3rd, McInnis EA, Buyon JP, Petri MA, Putterman C, Kalunian KC, Woodle ES, Lederer JA, Hildeman DA, Nusbaum C, Raychaudhuri S, Kretzler M, Anolik JH, Brenner MB, Wofsy D, Hacohen N, Diamond B, and S. L. E. n. Accelerating Medicines Partnership in. 2019. The immune cell landscape in kidneys of patients with lupus nephritis. Nature immunology 20: 902–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luther SA, Gulbranson-Judge A, Acha-Orbea H, and MacLennan IC 1997. Viral superantigen drives extrafollicular and follicular B cell differentiation leading to virus-specific antibody production. The Journal of experimental medicine 185: 551–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.2020. B Cell Activation and Response Regulation During Viral Infections. Viral Immunology 33: 294–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hastey CJ, Elsner RA, Barthold SW, and Baumgarth N. 2012. Delays and diversions mark the development of B cell responses to Borrelia burgdorferi infection. Journal of immunology (Baltimore, Md. : 1950) 188: 5612–5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Racine R, Jones DD, Chatterjee M, McLaughlin M, Macnamara KC, and Winslow GM 2010. Impaired germinal center responses and suppression of local IgG production during intracellular bacterial infection. Journal of immunology (Baltimore, Md. : 1950) 184: 5085–5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hjorton K, Hagberg N, Pucholt P, Eloranta ML, and Ronnblom L. 2020. The regulation and pharmacological modulation of immune complex induced type III IFN production by plasmacytoid dendritic cells. Arthritis research & therapy 22: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oke V, Brauner S, Larsson A, Gustafsson J, Zickert A, Gunnarsson I, and Svenungsson E. 2017. IFN-lambda1 with Th17 axis cytokines and IFN-alpha define different subsets in systemic lupus erythematosus (SLE). Arthritis research & therapy 19: 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen JY, Wang CM, Chen TD, Jan Wu YJ, Lin JC, Lu LY, and Wu J. 2018. Interferon-lambda3/4 genetic variants and interferon-lambda3 serum levels are biomarkers of lupus nephritis and disease activity in Taiwanese. Arthritis research & therapy 20: 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barnas JL, Albrecht J, Meednu N, Nandedkar-Kulkarni ND, O’Connell M, Looney RJ, and Anolik JH 2020. Type III interferon networks in systemic lupus erythematosus. The Journal of Immunology 204: 224.216–224.216. [Google Scholar]
- 60.Morand EF, Furie R, Tanaka Y, Bruce IN, Askanase AD, Richez C, Bae SC, Brohawn PZ, Pineda L, Berglind A, Tummala R, and Investigators T-T. 2020. Trial of Anifrolumab in Active Systemic Lupus Erythematosus. The New England journal of medicine 382: 211–221. [DOI] [PubMed] [Google Scholar]
- 61.Muir AJ, Shiffman ML, Zaman A, Yoffe B, de la Torre A, Flamm S, Gordon SC, Marotta P, Vierling JM, Lopez-Talavera JC, Byrnes-Blake K, Fontana D, Freeman J, Gray T, Hausman D, Hunder NN, and Lawitz E. 2010. Phase 1b study of pegylated interferon lambda 1 with or without ribavirin in patients with chronic genotype 1 hepatitis C virus infection. Hepatology 52: 822–832. [DOI] [PubMed] [Google Scholar]
- 62.Feld JJ, Kandel C, Biondi MJ, Kozak RA, Zahoor MA, Lemieux C, Borgia SM, Boggild AK, Powis J, McCready J, Tan DHS, Chan T, Coburn B, Kumar D, Humar A, Chan A, O’Neil B, Noureldin S, Booth J, Hong R, Smookler D, Aleyadeh W, Patel A, Barber B, Casey J, Hiebert R, Mistry H, Choong I, Hislop C, Santer DM, Lorne Tyrrell D, Glenn JS, Gehring AJ, Janssen HLA, and Hansen BE 2021. Peginterferon lambda for the treatment of outpatients with COVID-19: a phase 2, placebo-controlled randomised trial. Lancet Respir Med. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dickensheets H, Sheikh F, Park O, Gao B, and Donnelly RP 2013. Interferon-lambda (IFN-lambda) induces signal transduction and gene expression in human hepatocytes, but not in lymphocytes or monocytes. Journal of leukocyte biology 93: 377–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.