Summary:
In most tetrapod vertebrates, limb skeletal progenitors condense with postaxial dominance. Posterior elements (such as ulna and fibula) appear prior to their anterior counterparts (radius and tibia), followed by digit appearance order with continuing postaxial polarity. The only exceptions are urodele amphibians (salamanders), whose limb elements develop with preaxial polarity and who are also notable for their unique ability to regenerate complete limbs as adults. The mechanistic basis for this preaxial dominance has remained an enigma and has even been proposed to relate to the acquisition of novel genes involved in regeneration. However, recent fossil evidence suggests that preaxial polarity represents an ancestral rather than derived state. Here we report that 5’Hoxd (Hoxd11-d13) gene deletion in mouse is atavistic and uncovers an underlying preaxial polarity in mammalian limb formation. We demonstrate this shift from postaxial to preaxial dominance in mouse results from excess Gli3 repressor (Gli3R) activity due to the loss of 5’Hoxd-Gli3 antagonism and is associated with cell cycle changes promoting precocious cell cycle exit in the anterior limb bud. We further show that Gli3 knock-down in axolotl results in a shift to postaxial dominant limb skeleton formation, as well as expanded paddle-shaped limb bud morphology and ensuing polydactyly. Evolutionary changes in Gli3R activity level, which also played a key role in the fin-to-limb transition, appear to be fundamental to the shift from preaxial to postaxial polarity in formation of the tetrapod limb skeleton.
Graphical Abstract
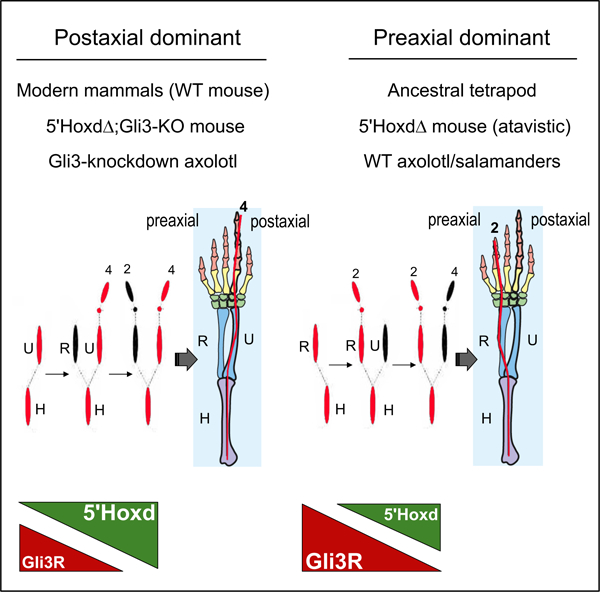
eTOC:
Uniquely in salamanders, the tetrapod primary limb axis forms with ancestral preaxial dominance but the underlying basis is unknown. Here, Trofka et al. show Gli3 has a central role; mice with elevated Gli3 repressor activity revert to preaxial dominance. Conversely, Gli3 knockdown shifts the axolotl limb axis from preaxial to postaxial polarity.
Introduction:
Tetrapod limb anterior-posterior (A-P) polarization and patterning is highly conserved and governed by posterior Sonic hedgehog (Shh) activity. Gli3 repressor form (Gli3R) acts as the major Shh effector in the limb; Shh signaling prevents Gli3 cleavage to form Gli3R, creating an A-P gradient of Shh-target repression activity[1]. Among Shh-regulated targets, the 5’Hoxd genes, together with Hoxa genes, play key roles in specifying the different proximo-distal components of the limb skeleton, including Hox9/10 paralogs in stylopod (upper arm, leg)[2], Hox11 in zeugopod (forearm, foreleg bones)[2, 3], and 5’Hoxd (Hoxd11–12-13) and Hoxa13 in autopod (handplate, footplate)[4–6]. 5’Hoxd expression occurs in two phases: early, posteriorly nested domains colinear with their genomic order (phase one), and later domains extending across the A-P distal limb bud in reverse colinear order during digit formation (phase two)[7]. Additionally, 5’Hoxd proteins also play a role both in initiating and maintaining Shh expression[8], and antagonizing Gli3R[9–12] by directly binding and altering Gli3R function[9, 13], both of which contribute to maintaining limb polarization. In distinction to all other living tetrapods, in urodele amphibians (salamanders), the limb axis develops with dominant preaxial rather than postaxial polarity[14–17], with anterior skeletal mesenchymal condensations forming prior to, rather than after, their posterior counterparts (Figure 1A). Consequently, the urodele primary limb axis extends through the anterior zeugopod element (radius or tibia) and digit 2 distally, rather than the posterior elements as occurs in other vertebrates (ulna or fibula in the zeugopod, and digit 4 in distal autopod). Despite this reversed appearance order, many features of limb A-P patterning appear to be largely preserved, and early spatial expression domains of most regulators of A-P limb polarity, including Shh, Hox and Gli3[18–20], are not reversed, but appear similar to taxa with dominant postaxial polarity. However, altered 5’Hoxd expression has been noted during the second phase, with curtailed distal A-P expansion, and was proposed to play a role in regulating limb axis polarity[18, 20]. But arguably, this altered expression may be rather late to impact limb axis polarity and as yet no functional studies have examined a potential 5’Hoxd role in determining axis polarity. Consequently, the mechanistic basis underlying determination of primary limb axis polarity remains a puzzle.
Figure 1. 5’Hoxd genes (d11-d13) act in concert to regulate polarity of limb condensation order in mouse.
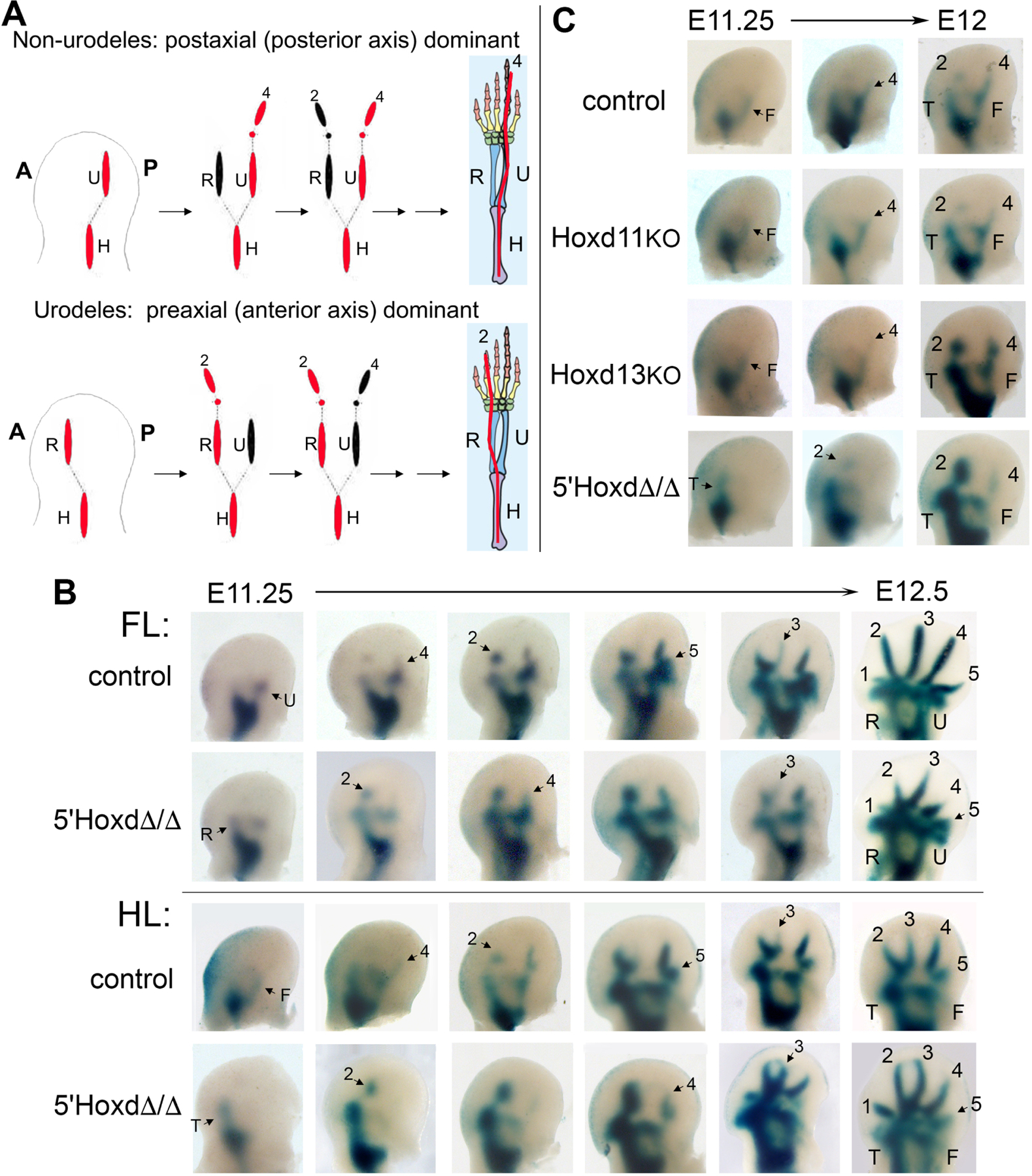
(A) Schematic of limb condensation appearance order in wildtype non-urodele tetrapods (such as mouse), with postaxial dominance (posterior elements form first) compared to wildtype urodeles, with preaxial dominance (anterior elements form first). Forelimb example shown; final adult skeletal pattern shows idealized 5-digit phalangeal formula for tetrapod ground state. Red indicates axial dominance; A, anterior; P, posterior limb bud. Modified from Shubin et al.[17] and from Xu et al.[36].
(B) NogginLacZ staining of forming limb condensations ranging from stage E11.25 – E12.5 in control (WT or 5’HoxdΔ/+) and 5’HoxdΔ/Δ forelimb (FL) and hindlimb (HL) buds. Posterior zeugopod elements form first in WT followed by alternating digit condensations beginning with digit 4 (4–2-5–3 order), whereas anterior elements form first in 5’HoxdΔ/Δ and digit condensations begin with digit 2 (2–4-3–5 order).
(C) NogginLacZ staining of forming limb condensations in single Hoxd11−/− (KO) and Hoxd13−/− (KO) mutant limb buds compared with control and 5’HoxdΔ/Δ. Posterior elements condense first in the single mutants, similar to control, in contrast to 5’HoxdΔ/Δ. (See also Figure S1). In all figure panels of NogginLacZ staining (in Figures 1, 2, 3C–D), limb buds are shown with anterior to left – posterior to right and distal at top of panel, and with U=ulna, R=radius, T=tibia, F=fibula, digits 1–5 = d1 - d5. All skeletal elements are annotated in the rightmost panel, latest stage limb bud shown. In other panels, only newly appearing elements are annotated with arrows. See also Figure S1.
Here, we examined whether deletion of 5’Hoxd genes could alter limb axis polarity in mice, which normally develop with postaxial dominance. We reasoned that if altered 5’Hoxd gene expression levels play a role in determining limb axis polarity in salamanders, then removing their mammalian function may uncover an atavistic shift to preaxial polarity, if postaxial dominance indeed represents a derived state[15]. We find that loss of 5’Hoxd function results in preaxial dominant limb formation. Furthermore, 5’Hoxd proteins and Gli3R antagonize each other functionally during limb development[9–13] and we show that genetic Gli3R elevation results in preaxial dominance in wildtype mice, whereas Gli3R removal restores postaxial dominance in the 5’Hoxd mutant. Gli3 acts to promote mesenchymal condensation of limb mesenchyme and to promote cell cycle exit[21], both of which are prerequisite for chondrogenic differentiation of limb skeletal anlage to proceed. We find that the shift to preaxial dominance is associated with cell cycle changes fostering cell cycle exit in anterior limb bud, but is not driven by changes in mesenchymal condensation rate per se. Like the 5’Hoxd mutant mouse, relative Gli3R activity appears to be high and the direct Gli3 cell cycle target Cdk6 is reduced in the preaxial dominant axolotl limb bud, but not in postaxial dominant Xenopus. Moreover CRISPR/Cas9 knockdown of axolotl Gli3 shifts limb skeletal formation to postaxial dominance. Together, these results suggest that the 5’Hoxd mouse mutant is atavistic and that Gli3 modulation played a central role in the evolution of modern tetrapods with a postaxial dominant primary limb axis.
Results and Discussion:
5’Hoxd genes jointly regulate zeugopod postaxial dominance and alternating digit appearance order in mouse.
To determine if altered 5’Hoxd gene expression levels play a role in regulating polarity of limb axis formation, we examined the appearance of skeletal condensations in 5’Hoxd deleted mouse limb buds (5’HoxdΔ/Δ, deletion of Hoxd11-d13)[6, 7], using NogginLacZ as a sensitive reporter to visualize forming condensations early[22, 23] and the 5’HoxdΔ(RXII) allele, which lacks a LacZ transgene present in the conventional 5’HoxdΔ/+ mutant[6, 7]. In contrast to controls, in 5’HoxdΔ/Δ embryos polarity of axis formation is clearly shifted with anterior zeugopod appearing first in both fore and hindlimb buds (Figure 1B), and digit 2 extending from the anterior zeugopod as the first forming digit condensation. We previously showed that digit condensations appear in an alternating A-P order[23] with the sequence 4–2-5–3, and digit 4 extending earliest from the posterior zeugopod in mouse. The basis for this alternating digit order was unknown. In 5’HoxdΔ/Δ limb buds this order appears to be reversed, with a sequence of 2–4-3–5, suggesting that digit appearance order is a consequence of, and linked to, the polarity of primary limb axis formation.
We also examined single 5’Hoxd gene mutants and found that removal of either Hoxd11 or Hoxd13 function alone was insufficient to alter limb axis polarity, indicating that the shift to preaxial dominance requires functional reduction of all three 5’Hoxd genes (Figures 1C, S1).
The balance of antagonistic 5’Hoxd and Gli3 levels determines the dominance of limb axis polarity.
The 5’Hoxd genes have been shown to antagonize Gli3 function in several contexts during early and late phases of limb development[9–13]. We previously found that each of the three 5’Hoxd members can interact directly with Gli3[9], and co-expression studies indicate that the stoichiometric balance of this protein interaction dictates the net effect on 5’Hoxd- and Gli3R- regulated target gene expression[9, 13], rather than strictly by transcriptional regulation (see Figure S3 in Huang et al.[10], and see Bastida et al.[24]). The functional level of Gli3R controls limb A-P polarity and we hypothesized that 5’Hoxd proteins may act in concert in different limb domains to modulate Gli3 activity levels and regulate limb axis polarity. We first determined the sequence of condensation appearance in Gli3−/− embryos. Consistent with prior reports[21], the initial appearance of condensations retains normal postaxial polarity and a 4–2-5 digit order, but the anterior handplate domain expands to accommodate the late formation of additional digit condensations between digits 2 and 3, which is more pronounced in fore than in hindlimb buds, correlating with the more severe forelimb polydactyly in Gli3−/− (Gli3 KO, Figures 2A, S2).
Figure 2. Modulating Gli3 repressor (Gli3R) level determines polarity of limb axis condensation in wildtype (WT) and 5’HoxdΔ/Δ limb buds.
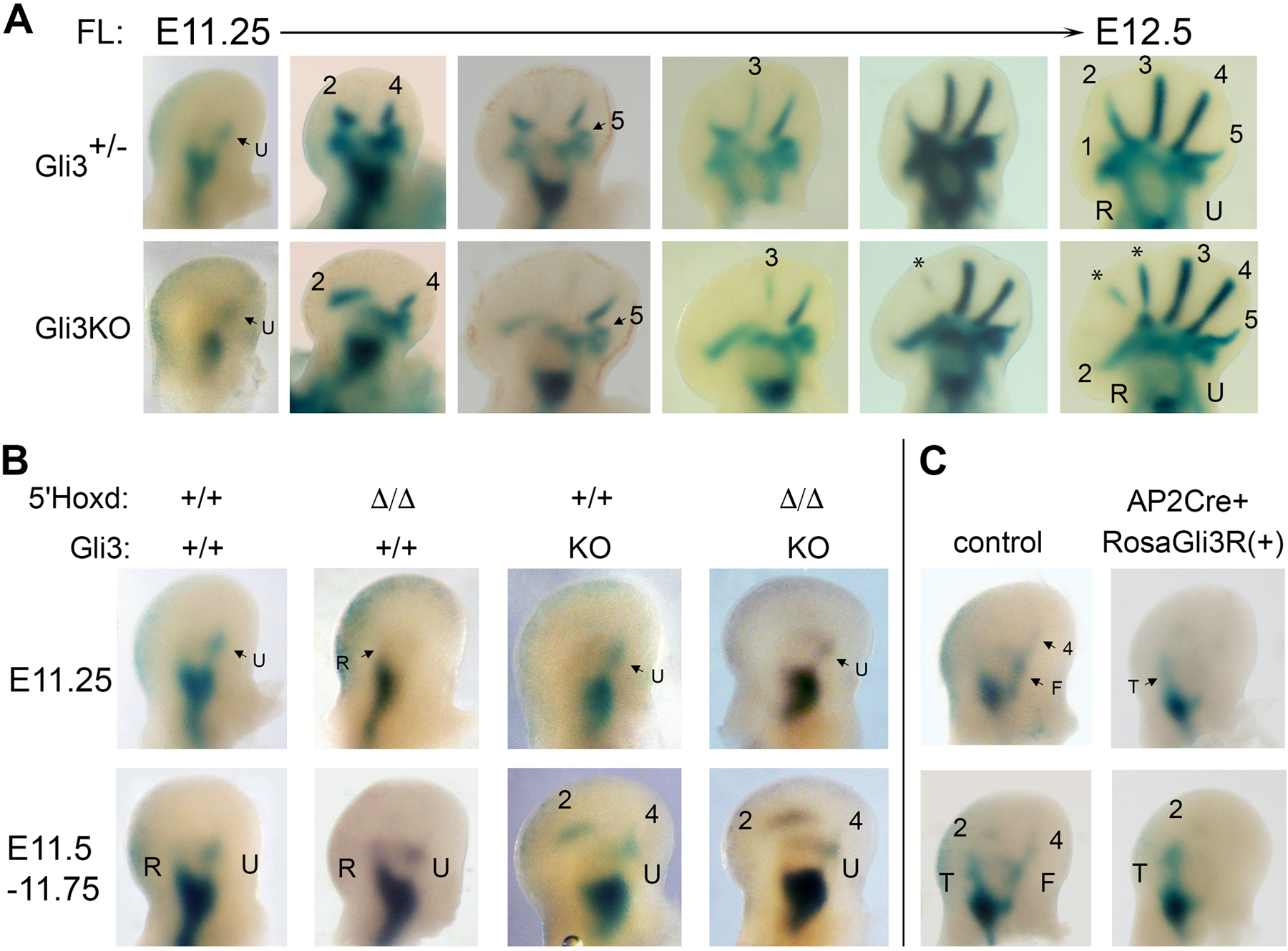
(A) NogginLacZ staining of forming limb condensations in Gli3−/− (KO) forelimb buds compared to Gli3+/- controls ranging from time E11.25-E12.5. Postaxial polarity in appearance order is preserved in zeugopod and digits, but with late appearance of additional anterior digit condensations between d2 and d3 (denoted by *). See also Figure S2.
(B) NogginLacZ staining of forming limb condensations in 5’HoxdΔ/Δ with intact Gli3R function (+/+), and with Gli3 function removed (KO). Gli3 deletion restores postaxial polarity of condensation appearance in the 5’HoxdΔ/Δ mutant, similar to wildtype (+/+) and Gli3KO controls.
(C) NogginLacZ staining of forming limb condensations in wildtype (WT) hindlimb buds from a line carrying a conditional Cre-dependent RosaGli3R transgene allele. Embryos also expressing AP2Cre show a shift to preaxial dominance of condensation appearance, similar to that seen in 5’HoxdΔ/Δ limb buds.
Related to Figure S2.
To check whether preaxial polarity in 5’HoxdΔ/Δ results from excess Gli3R protein activity, we generated Gli3−/−;5’HoxdΔ/Δ compound mutants. Indeed, Gli3 removal restores normal postaxial dominance in 5’HoxdΔ/Δ limb buds (Figure 2B). Conversely, expression of a Gli3-repressor transgene in wild-type embryos is able to shift limb axis polarity from the normal postaxial to preaxial dominance (Figure 2C). These results demonstrate that the relative level of Gli3R activity determines the dominance of primary limb axis formation, and establishes a role for the 5’Hoxd genes as important modulators of Gli3R in specifying primary limb axis polarity.
Previous analysis of Gli3 function in limb showed that Gli3 regulates and restrains autopod expansion by dual mechanisms that could also act to regulate limb axis polarity. Gli3R down-regulates cell cycle gene expression to modulate the G1-S transition and promote cell cycle exit[21], and Gli3R increases Bmp signaling to promote formation/differentiation of skeletal mesenchymal condensations[10, 21, 25, 26].
Mesenchymal aggregation/condensation rates are unaltered ex vivo by loss of 5’Hoxd function.
Ex vivo formation of condensations by limb mesenchyme aggregation in high density culture (HDC) recapitulates the features of this process in vivo[27, 28]. To assess if relative Gli3R activity excess in 5’HoxdΔ/Δ limb buds acts by promoting precocious mesenchymal condensation, we examined the rate at which anterior and posterior limb mesenchyme cells condense in the 5’HoxdΔ/Δ mutant compared to wild type (WT) in mixed HDCs. Limb buds from WT and from 5’HoxdΔ/Δ embryos also expressing a fluorescent protein (EYFP or TdTomato) were dissociated for live imaging to compare WT and mutant cell aggregation behavior in mixed, short-term, HDCs (Figure 3A). We used limb bud stages slightly younger than those in which the zeugopod condensations first begin to form and become clearly detectable (see Figure S3), because the polarity of skeletal condensation is strictly anterior or posterior. In contrast, in the autopod (handplate), anterior and posterior digit condensations appear in an alternating order (albeit in reversed sequence), confounding interpretation of aggregation rates when autopod condensations form. HDC cultures were imaged for up to 18–24 hrs; however visible prechondrogenic condensations were already consistently present in similar distributions for both WT and 5’HoxdΔ/Δ cells in mixed cultures by 12 hrs, and aggregate sizes based on cell fluorescence tracking were plotted from time 0 to 10 hrs to assess relative rates. Cell sorting[27, 29] was not apparent at this stage during the timeframe examined, except in mixed A-P cultures of either WT or 5’HoxdΔ/Δ cells after longer culture times. To compare independent HDC data, aggregate ratios at each time were calculated and normalized to a ratio of 1.0 for time zero (equal plating; see STAR Methods and Figure S4 for details). With no differences between aggregation rates in mixed HDC, the ratio should remain 1 over time, yielding a slope of zero, and a deviation from zero slope indicates rate differences.
Figure 3. Aggregation rates of limb mesenchyme ex vivo are similar for wildtype and 5’HoxdΔ/Δ, and preaxial polarity of condensation order in vivo is unaltered by reducing Bmp activity in 5’HoxdΔ/Δ.
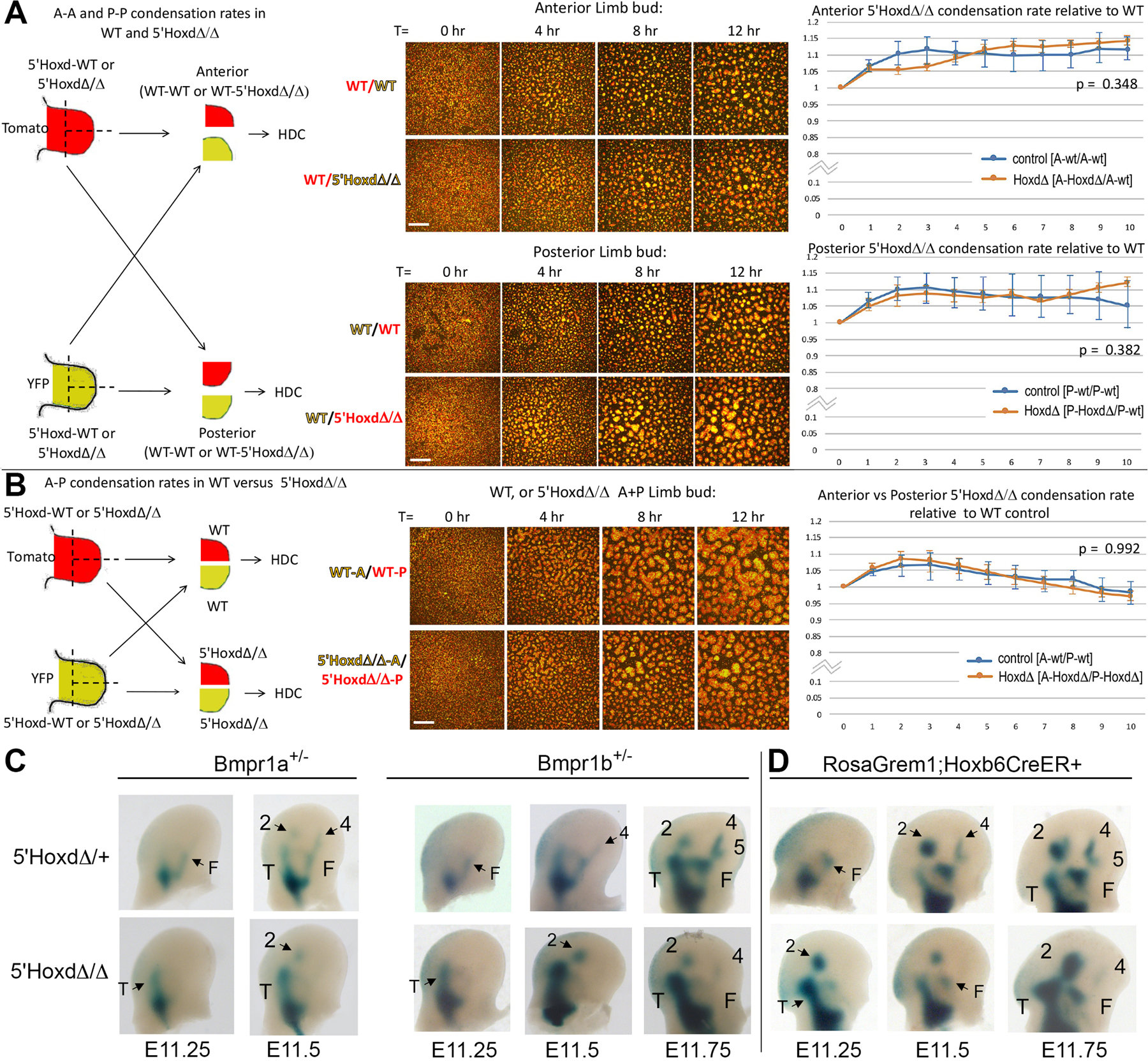
(A,B) Aggregation rates in high density cultures (HDC) measured by live cell fluorescence imaging in genetically tagged (EYFP, TdTomato) wildtype control (WT) and 5’HoxdΔ/Δ cells from anterior or posterior hindlimb bud halves in mixed HDCs. Left diagrams summarize experimental strategies. Middle panels show representative static images of mixed control vs mutant mesenchyme in HDC at different times post-plating. Scale bar for each set of panels is 200 μm. Right graphs show normalized data and p values comparing cell aggregation rates for control and mutant 5’HoxdΔ/Δ in mixed HDC as indicated in diagrams. (N=4 independent experiments for each type of mixed HDC imaged). See also Figures S3, S4.
(A) Aggregation rates in HDC comparing mixed anterior (upper panels) or mixed posterior (lower panels) hindlimb bud halves from either WT/WT or WT/5’HoxdΔ/Δ limb mesenchyme.
(B) Aggregation rates in HDC comparing mixed anterior and posterior hindlimb bud halves from either WT/WT or 5’HoxdΔ/Δ/5’HoxdΔ/Δ limb mesenchyme.
(C) NogginLacZ staining of forming limb condensations in control and 5’HoxdΔ/Δ hindlimb buds from stages E11.25 - E11.75, from embryos also carrying a Bmpr1a or Bmpr1b mutant allele to reduce Bmp receptor gene dosage (Bmpr1a+/-; Bmpr1b+/-).
(D) NogginLacZ staining of forming limb condensations in control and 5’HoxdΔ/Δ hindlimb buds from stages E11.25 - E11.75, from embryos also carrying conditional, Cre-dependent RosaGrem1 transgene and Hoxb6CreER alleles, and tamoxifen treated at E10.5 to activate Grem1 expression and reduce net Bmp activity.
Related to Figures S3, S4.
We did not detect any significant differences in aggregation rates for either anterior or for posterior 5’HoxdΔ/Δ compared to WT limb bud cells in HDC (Figure 3A). We also compared aggregation rates for anterior vs. posterior 5’HoxdΔ/Δ limb cells in mixed culture to determine if relative A-P aggregation rates differed from WT anterior vs. posterior HDC at matched stages. Although we observed a slight trend for anterior cells to aggregate more slowly than posterior in mixed HDC (slope < 0), this did not reach statistical significance and was similarly observed in both the 5’HoxdΔ/Δ and WT cultures (Figure 3B).
Bmp pathway modulation in vivo does not alter condensation appearance order or polarity of primary limb axis formation.
Endogenous signals could arguably be disturbed during ex vivo culture of limb cells. Bmp activity, induced by Gli3R, drives mesenchymal compaction, the earliest step in skeletal condensation[21, 26, 27]. We therefore also modulated Bmp activity levels in vivo to assess whether lowering effective Bmp activity genetically could restore postaxial polarity in 5’HoxdΔ/Δ limb buds. However, reducing the dosage of either of the major limb Bmp receptors (Bmpr1a, Bmpr1b) did not alter the preaxial polarity of limb axis formation in the 5’HoxdΔ/Δ mutant (Figure 3C). We also introduced a RosaGrem1 transgene into the 5’HoxdΔ/Δ mutant to more strongly inhibit net Bmp activity. Mis-expression of this conditional transgenic Bmp antagonist in wildtype embryos causes highly penetrant phenotypes comparable to Bmp4 gene deletion[30]. However, RosaGrem1 activation in 5’HoxdΔ/Δ limb buds also failed to alter the preaxial polarity of condensation appearance (Figure 3D), indicating that Bmp activity levels are not rate-determining in driving the anterior shift in 5’HoxdΔ/Δ limb axis polarity.
Removal of 5’Hoxd function causes precocious cell cycle exit/G1 phase lengthening in anterior limb bud.
Previously, Gli3 was found to repress expression of cell cycle regulators Cdk6 and CyclinD1, and to promote G1 phase lengthening and cell cycle exit[21]. We compared cell cycle progression by flow cytometry in dissected anterior and posterior limb buds from control and 5’HoxdΔ/Δ embryos at several stages bracketing the time at which zeugopod condensations begin to form (see Figure S3), because, as mentioned above, the later stage condensation order in the autopod is more complex (alternating A-P order). Unexpectedly, cell cycle changes were restricted to the anterior limb bud in 5’HoxdΔ/Δ; cell cycle parameters in posterior limb bud were similar for 5’HoxdΔ/Δ and controls at all 3 stages analyzed. At an early stage (E10.75 hindlimb), prior to overt appearance of any limb condensations, the proportion of cells in G1 phase was higher in 5’HoxdΔ/Δ anterior cells than in wildtype and at mid-stage, when the stylopod has formed and onset of zeugopod formation is just beginning (E11.25 hindlimb), this difference became more pronounced and was highly significant (Figure 4A). Conversely, the proportion of cells in S phase was reduced (Figure 4B). After zeugopod elements have condensed and autopod condensations are appearing (late stage, E11.75 hindlimb), anterior limb bud cell cycle parameters for 5’HoxdΔ/Δ were similar to controls. The selective lengthening of G1/S transit seen in anterior 5’HoxdΔ/Δ is consistent with the known effects of Gli3R on proliferation by restricting G1-S regulators and constraining S phase entry selectively in the anterior limb bud[21], and with the observed elevation in Gli3R activity in the 5’HoxdΔ/Δ mutant limb[10, 24] (see also Figures 5A, S5), leading to accelerated cell cycle exit and thereby enabling prechondrogenic condensation to commence.
Figure 4. Precocious cell cycle G1 arrest in 5’HoxdΔ/Δ anterior limb bud relative to controls.
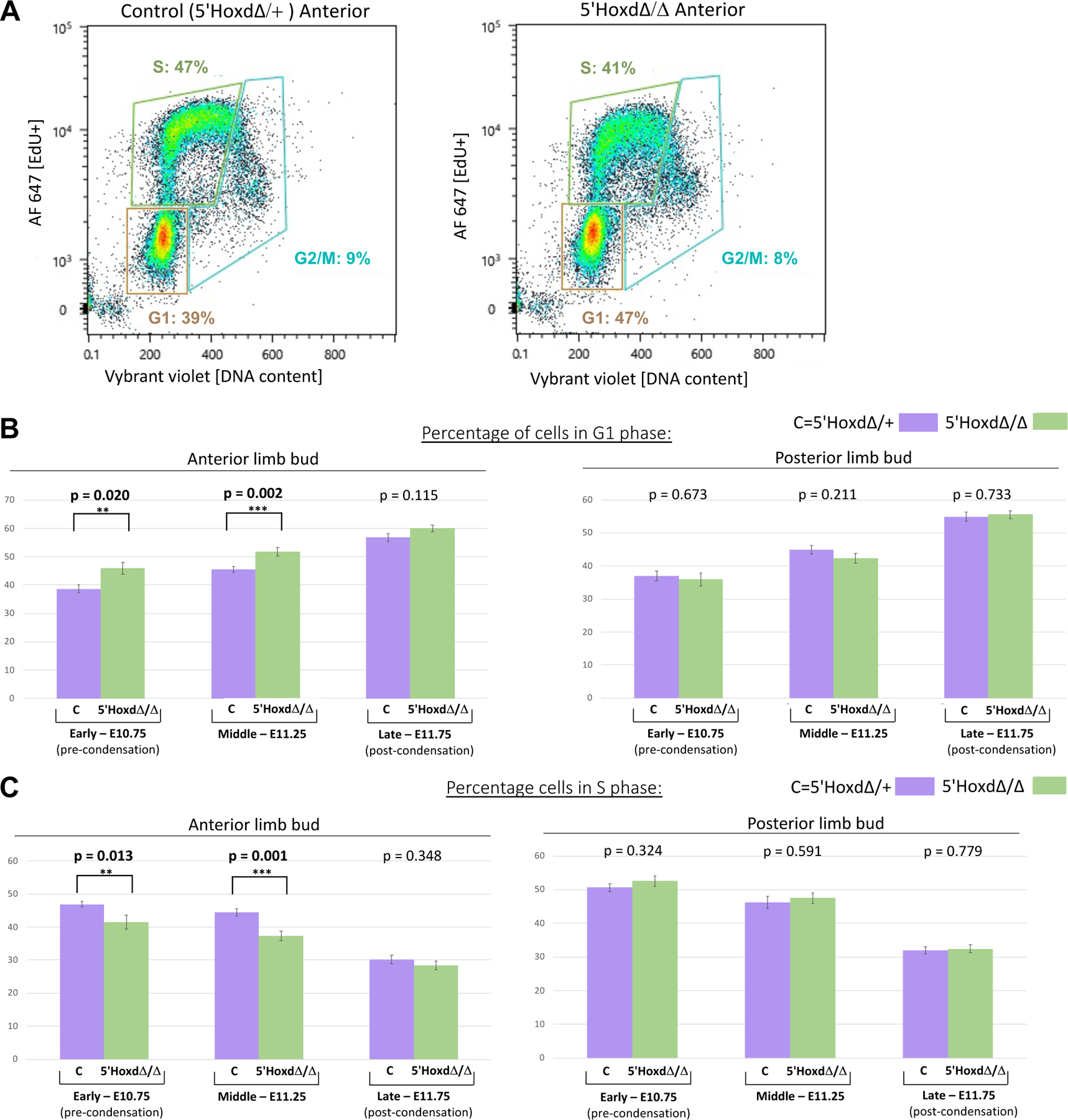
(A) Representative scatter plots of Flow cytometry data showing EdU incorporation and cell DNA content from control 5’HoxdΔ/+ and from 5’HoxdΔ/Δ dissociated anterior limb bud cells. Boxed areas indicate G1, S, and G2/M phase cells and percentages of total population.
(B,C) Bar graphs summarizing cell percentages in G1 (B) and in S (C) phase from flow data at three different stages (E10.75, E11.25, E11.75) spanning condensation appearance time (see also Figure S3) in control (5’HoxdΔ/+, lilac) and in 5’HoxdΔ/Δ (green) for anterior (left panel graphs) and posterior (right panel graphs) limb bud halves. P-values comparing control and mutant for each stage is indicated over each pair of bar graphs. Numbers of independent limb buds analyzed by flow cytometry: for control anterior and posterior, n=11 for early, n=9 for middle, and n=7 for late stage; for 5’HoxdΔ/Δ anterior and posterior, n=8 for early, n=8 for middle, and n=10 for late stage. **, significant at p<0.05; ***, significant at p<0.01. error bars indicate SD.
Related to Figure S3.
Figure 5. Relative Gli3R activity levels in mouse 5’HoxdΔ/Δ mutant compared to WT.
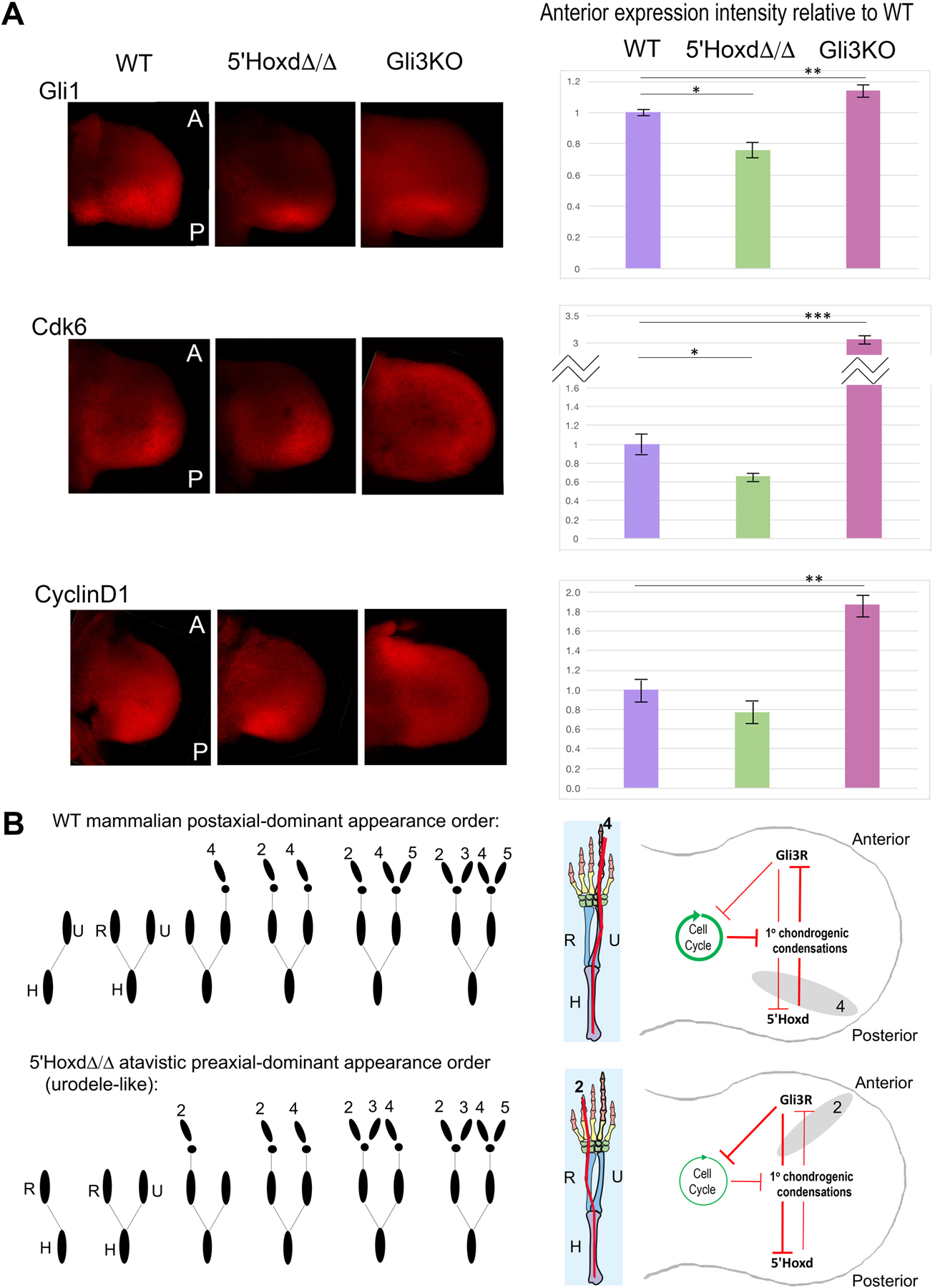
(A) Whole mount in situ transcripts in wildtype (WT), 5’Hoxd mutant (5’HoxdΔ/Δ) and Gli3−/− (Gli3KO) E10.75 forelimb buds were detected using HCR and quantitated with confocal microscopy. All limb buds oriented with anterior (A) at top, posterior (P) at bottom, and distal at right side of panel. 5’Hoxd mutants show reduced anterior Gli1 and Cdk6 expression compared to WT, in contrast to Gli3 KO in which levels of all three transcripts are elevated, as previously shown [21]. CyclinD1 changes did not reach significance between WT and 5’HoxdΔ/Δ. For each probe and each genotype, a minimum of 3 embryos were hybridized at the same time under uniform conditions and evaluated. Total fluorescence in equal-sized anterior domains across all confocal sections was calculated and intensities normalized relative to the average WT level. Graphs to right show mean fluorescence and error bars indicate SEM. Significance determined by standard 2-tailed T test. *, p<0.05; **, p<0.01; ***, p<0.001.
(B) Diagrammatic summary of model: The net Gli3R activity level, governed by Gli3–5’Hoxd mutual antagonism, regulates G1-S phase transit and onset of cell cycle arrest in the anterior limb bud. Cell cycle arrest enables response to differentiation cues leading to chondrogenic condensation (this paper; Lopez-Rios et al.[21]). Shifting the balance in favor of high Gli3R activity promotes earlier anterior cell cycle exit and leads to preaxial polarity in condensation appearance order, as seen normally in urodeles and in 5’Hoxd mutant mice; low Gli3R activity leads to postaxial dominance seen in most tetrapod vertebrates.
Related to Figures S5, S6.
Our results show a clear effect of Gli3R-5’Hoxd modulation on G1 lengthening and cell cycle exit, but not on condensation rates per se in the anterior limb bud, both ex vivo and in vivo (by manipulating Bmp activity levels). Apparently Bmp activity, even when down-modulated, is in excess during limb skeletal element formation and suffices for mesenchymal condensation following cell cycle exit, but the timing of cell cycle slowing in anterior relative to posterior limb bud (Figure 5B) determines when cells become permissive to Bmp response and sets the polarity of limb axis formation. Notably, even direct-developing salamanders with a “paddle” handplate stage, have a comparatively narrow shaped limb bud[31], suggesting a lower degree of proliferative expansion. Bmps have recently been implicated in directly regulating cell cycle in the limb [32], but our results indicate a more direct effect of Gli3 on cell cycle gene expression. The identification of multiple direct Gli3 response elements within the Cdk6 genomic locus is in keeping with such direct regulation[33]; Hoxd13 binding elements have likewise been identified[34], raising the possibility that both factors regulate Cdk6 directly and antagonize each other’s effects.
Gli3R activity and target expression in 5’Hoxd mutant mice and amphibian species with preaxial or postaxial dominance.
5’Hoxd proteins antagonize Gli3R functionally via direct protein interaction[9, 13] and previous analysis in the context of late (phase two) 5’Hoxd expression indicates that neither Gli3 transcript nor protein level is altered in 5’HoxdΔ/Δ limb buds[10, 24], and we confirmed this is also the case in early stage (phase one) 5’Hoxd mutant limb buds (Figure S5). As an indicator of relative Gli3R “functional activity” level, we examined Gli1 expression in the anterior limb bud since elevated Gli3R function would be expected to modulate the extent of Shh pathway activity. Indeed, Gli1 expression was reduced in the anterior 5’HoxdΔ/Δ limb bud compared to wildtype (Figure 5A). Furthermore, expression of the direct Gli3 target Cdk6 was also reduced in the 5’HoxdΔ/Δ anterior limb bud consistent with the observed cell cycle changes. CyclinD1, which is also elevated in the Gli3 KO[21] and may be an indirect target, was also modestly reduced in 5’HoxdΔ/Δ anterior limb bud, but didn’t reach statistical significance (Figure 5A). To further assess if Gli3R activity correlates with polarity of limb axis formation, we compared Gli1, Cdk6 and CyclinD1 expression in two amphibia with postaxial (Xenopus) or preaxial (axolotl) polarity. Since transcript levels per se are difficult to compare between different species, the posterior-to-anterior (P/A) limb bud expression ratios were determined within a given limb bud for each gene in both amphibian species and compared to mouse (Figure S6), which revealed clear A-P polarization of Gli1 and Cdk6 in Xenopus indicative of robust Shh pathway activity, as in mouse. In contrast, Gli1 and Cdk6 expression in axolotl were strikingly uniform across the A-P limb bud axis. Unlike mouse or Xenopus, a lower distal than proximal expression level of all three genes in axolotl also suggests that the distal expression levels are comparatively low, consistent with a weaker A-P limb bud polarization in axolotl.
CRISPR/Cas9 directed knock-down of axolotl Gli3 results in primary limb axis formation with postaxial polarity and polydactyly.
To ascertain directly if a high relative Gli3R activity level dictates preaxial polarity in axolotl, we used CRISPR/Cas9 editing to knockdown Gli3 expression with sgRNAs targeting Gli3 coding sequences upstream of the DNA-binding Zn finger domain (amino acid residues 176–181, and 218–223; see Figure 6A). Sequencing of the targeted genomic DNA in Gli3 knockdown larvae showed perturbation of sequencing peaks, which indicated several nucleotide deletions near the PAM sequences (Figure 6B). At late skeletal stages, when all limb elements have completely formed, a high proportion of the CRISPR-knockdown (crispant) larvae (7/11) displayed striking polydactyly highly reminiscent of the mouse Gli3−/− phenotype compared to control larvae; notably the most prominent, elongated digit was postaxial (*, Figure 6D), in contrast to controls. At earlier stages (st47–50) when the zeugopod elements are just forming and differentiating, 64% (14/22) of the Gli3 knockdown larval limbs displayed postaxial dominance (Figure 6C, ulna appearing first) compared to controls. Furthermore, most of the postaxial dominant crispants (13/14, 93%) had a clearly paddle-shaped limb bud as well, which was not observed in any of the knockdown limb buds (8/8) that retained preaxial dominance. These results clearly demonstrate that modulating Gli3 in axolotl alters primary limb axis polarity and, together with genetic evidence in mouse, implicates Gli3 as a key regulator governing the polarity of limb axis formation.
Figure 6. CRISPR-mediated Gli3 knockdown in Axolotl causes limb skeletal condensation shift to postaxial polarity and polydactyly.
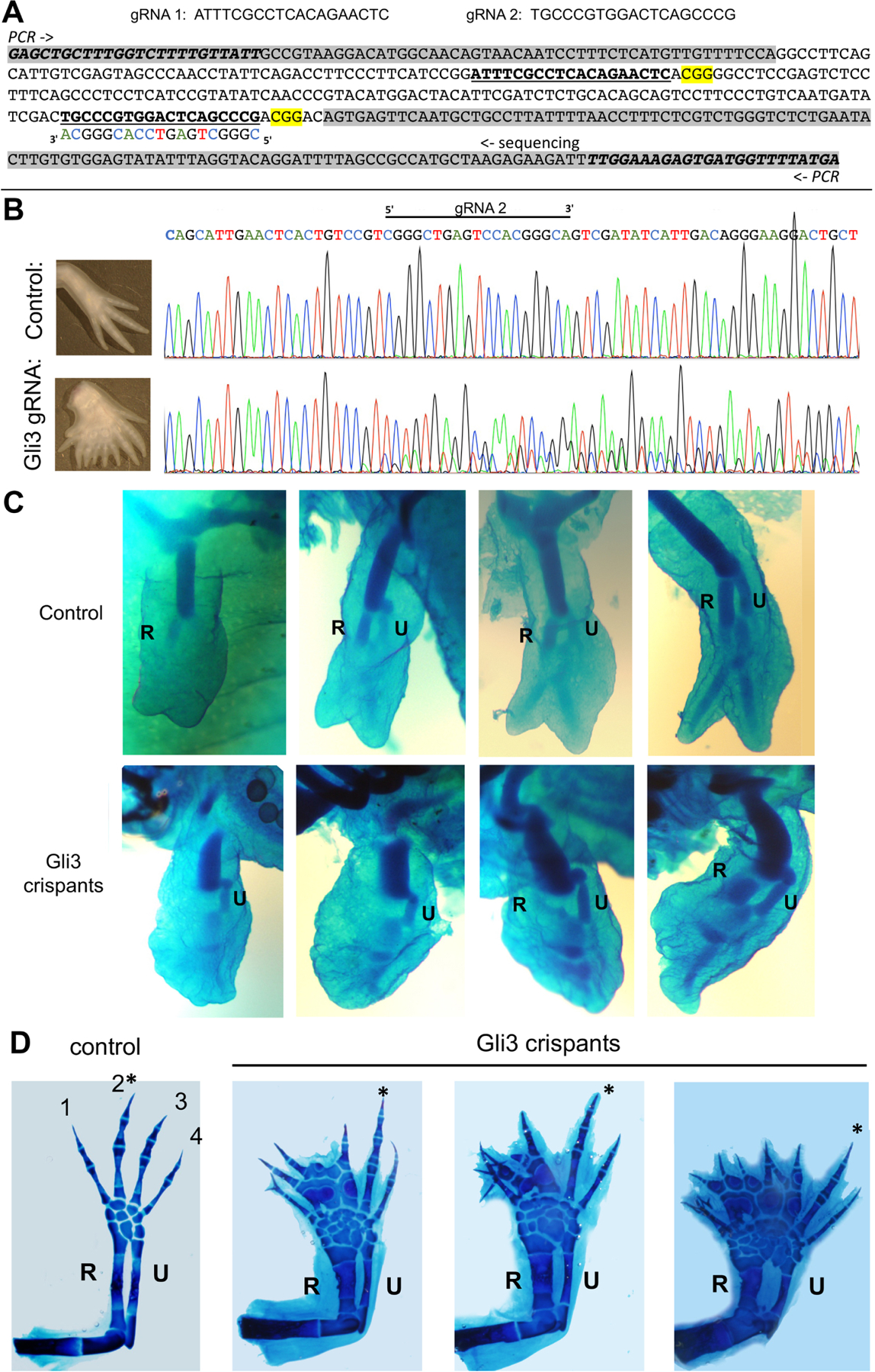
(A, B) Axolotl Gli3 guide RNAs used and sequence analysis of targeted region in crispant larvae.
(A) Top - sequences of co-injected guide RNAs targeting Axolotl Gli3 coding regions (crRNAs) together with tracrRNA and Cas9. Bottom - DNA sequence of axolotl Gli3 genomic region PCR-amplified for sequencing analyses. Intronic sequences are shaded in grey. PCR primers italicized, guides shown as bold underlined sequences, PAM sequences highlighted yellow, and complementary strand guide sequence shown as indicated in sequencing shown in B.
(B) Representative sequence trace of control compared to Gli3 crispant to verify mutagenesis (typical associated limb phenotype shown in panel to left of sequence). Direct sequencing of the PCR amplicon shows multiple nucleotide changes and sequence degeneracy following the guide/PAM region in injected founder larvae, indicative of multiple mutations.
(C) Appearance order of primary zeugopod condensations in Gli3 crispants injected with gRNAs targeting Gli3 coding regions between amino acids 176–223 (shown in Figure 6A) compared to uninjected controls ranging from approximately stage 47–50. In contrast to controls in which the anterior zeugopod (R, radius) invariably appears first (upper panels), 64% of Gli3 crispants (n=14/22, shown in lower panels) displayed postaxial dominance with posterior zeugopod (U, ulna) forming first.
(D) Mature skeletal phenotypes in Gli3 crispants compared to controls. Representative examples of preaxial polydactyly observed in 64% of Gli3 crispants (n=7/11) compared to controls (6cM, ~stage 55–57). Note that the longest digit (*) is invariably postaxial compared to controls, in which digit 2 is always largest. In addition to uninjected controls, injections of guide RNAs designed against other unrelated loci (n=500) have not resulted in any abnormal limb phenotypes. R, radius; U, ulna.
Although Shh expression and early A-P signaling are overall preserved in urodeles, A-P polarization may be relatively less well developed in these taxa, as suggested by the weak or absent polarity in Gli1, Cdk6 or CyclinD1 expression. 5’Hoxd proteins down-modulate Gli3R activity both by direct protein interaction[9] and by upregulating Shh expression level[8]. Consequently, a reduced Shh posterior spatial domain[19] and activity (inferred from low Gli1 and Ptch1 direct target response (see Figure 4 in Bickelmann et al.[18]), and the failure in late phase 5’Hoxd expansion in urodeles[18, 20] may both reflect an earlier excess in Gli3R activity level relative to other tetrapods, which governs the urodele preaxial dominance in limb skeleton formation.
These results reveal the genetic mechanism underlying the evolutionary switch from preaxial to postaxial polarity in limb axis formation as a consequence of changes in Gli3R activity. In light of recent evidence that preaxial dominance in urodeles represents the ancestral state[15], our results demonstrate that a modern mammal can revert to an atavistic state with preaxial dominance in the context of mutations (such as 5’HoxdΔ/Δ) that alter limb A-P polarization via excess net Gli3R activity. Notably, such changes in A-P polarization do not compromise limb functionality drastically, probably because Hoxa13 function is retained and partly compensates in the 5’Hoxd mutant[24]. 5’HoxdΔ/Δ mutant mice are viable and locomote normally, despite brachydactyly (see Figure S1F). Gli3 activity changes may also underlie key events in the emergence of early tetrapods during the fin-to-limb transition[35], as the A-P extent of Gli3 expression declined with the acquisition of strong posterior Shh activity. These features may have co-evolved, with modern urodeles retaining some features of the basal state in the form of preaxial dominance, while other vertebrate taxa with postaxial dominance diverged.
Conclusions:
Using mouse and axolotl, we demonstrate that Gli3R plays a central role in determining primary limb axis polarity and therefore Gli3/Hedgehog pathway changes likely underlie the evolutionary transition from the ancestral preaxial to postaxial polarity present in most modern tetrapods excepting urodele amphibians (salamanders). Accordingly, genetic manipulations in mice, such as 5’Hoxd (Hoxd11-d13) gene deletion, that elevate Gli3R activity uncover an atavistic shift to preaxial polarity. Postaxial polarity can be restored in these mutants by Gli3 removal. Gli3 has been shown to promote both mesenchymal condensation and cell cycle exit in limb mesenchyme[21]. We find that preaxial dominance is associated with precocious cell cycle exit in the anterior limb bud, presumably fostering premature condensation, but is not driven by changes in mesenchymal condensation rates per se. In both mouse and amphibians with either preaxial (axolotl) or postaxial (xenopus) polarity, Gli3/Hedgehog activity levels correlate with limb axis dominance. We further show that CRISPR/Cas9 knock-down of axolotl Gli3 shifts limb axis formation from preaxial to postaxial dominant, and results in an expanded paddle-shaped limb bud and ensuing polydactyly. Together these results suggest that the evolutionary increase in Shh activity and A-P polarization, with consequent Gli3R reduction, played a fundamental role in the shift from preaxial to postaxial polarity in tetrapod limb axis formation.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Susan Mackem (mackems@mail.nih.gov)
Materials availability
The mouse lines (activated fluorescent protein reporter alleles; Hoxd13DsRed2) generated in this study are available from the lead contact (Susan Mackem) or directly from H. Scott Stadler (for Hoxd13DsRed2) on request, but may require a completed Materials Transfer Agreement if there is potential for commercial application. No other new unique reagents were generated.
Data and Code Availability
No standardized data sets were generated in this study.
This study does not report any original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
All animals were housed and maintained and all animal studies were carried out according to the ethical guidelines of the Institutional Animal Care and Use Committee (IACUC) at NCI-Frederick under protocol #ASP-20–405. All mice were maintained in a specific pathogen-free facility with a 12-hr light cycle (6 am-6 pm) on a standard chow diet (LabDiet mouse breeder diet O/HS). Both male and female mice with mutant alleles (or late-stage embryos for non-viable Gli3−/− mutants) displayed no sex differences in phenotypes, excepting infertility in homozygous 5’HoxdΔ(d11–d13), Hoxd13 and Hoxd11 null males, which were maintained as heterozygotes. Embryos of both genders ranging from E10.5–17.5 were included in all analyses, but individual gender determination was not carried out. The AP2Cre[37], BmpR1a[38], Bmpr1b[39], Gli3(XtJ)[40], Hoxb6CreER[41], 5’HoxdΔ(d11–d13)[6], 5’HoxdΔ(d11–d13)(RXII) allele that lacks LacZ insertion[7], Hoxd11[42], NogginLacZ[22], PrrxCre[43], RosaGrem1[30], and RosaGli3R[33] lines used have all been reported previously. All single and compound mouse lines were maintained on an outbred, mixed strain background (predominantly FVB/n and C57BL/6). The 5’HoxdΔ(RXII) allele[7] was used in all experiments where early skeletal condensations were visualized with NogginLacZ, and the phenotypically equivalent 5’HoxdΔ(d11–d13) containing a Hoxd11-LacZ transgene[6] was used in other experiments (HDC, cell cycle analysis). For timed matings, noon on the day of post-coital plug was considered to be E0.5. For inducible Hoxb6CreER, a single dose of 3mg tamoxifen was injected intraperitoneally at E10.5[41].
Axolotl
The colonies of Mexican axolotls (Ambystoma mexicanum) were bred and maintained at the Ambystoma Genetic Stock Center (AGSC) and analyses carried out according to the ethical guidelines of the Institutional Animal Care and Use Committee (IACUC) at the College of Medicine at the University of Kentucky (protocol # 2017–2580). Wild-type and white A. mexicanum larvae from the Ambystoma Genetic Stock Center (RRID:SCR_006372) were staged as described[44].
Founder CRISPR/Cas9 larvae, generated as described under Method Details, and untreated larvae were maintained in 100% v/v axolotl rearing water (ARW: 1.75 g NaCl, 100 mg MgSO4, 50 mg CaCl2, and 25 mg KCl per liter, with NaHCO3 to achieve a pH in the range of 7.1–7.6.) and fed brine shrimp until reaching 3cM, and then California blackworms subsequently until euthanasia at developmental stages (as indicated in text) ranging from stage 45–57 for HCR in situ hybridization, genomic sequencing and skeletal analyses. Larvae were housed at 16–18°C with a 12-hr light cycle. Both male and female embryos/larvae were included in all analyses with no indications of sex-dependent patterns of variation, but individual gender determination was not carried out.
Xenopus
X. tropicalis albino tadpoles were obtained from NASCO (Fort Atkinson, WI) and staged according to Nieuwkoop and Faber[45]. Tadpoles were then housed in a circulating flow-through system with at least 100 ml water/tadpole and 12-hr light/dark cycle. The tadpoles used in this study were euthanized at stage 50–51 (approximately 2–3 weeks of age) for analysis, and were of indeterminate gender since sexual maturation occurs at later stages of development and is not solely determined by genetic chromosome composition. All animal procedures were performed as approved by the Animal Use and Care Committee of the Eunice Kennedy Shriver National Institute of Child Health and Human Development (ASP 20.061).
METHOD DETAILS
Generation of Mouse Lines
Activated Fluorescent protein alleles:
Mutant and control mice with activated fluorescent protein alleles were generated by crossing Rosa-tdTomato[46] and RosaEYFP[47] males with Prrx1Cre females[43] to produce germ-line recombination and offspring were crossed with the HoxdΔ/+ line to generate both control and mutant lines with either ubiquitous TdTomato or EYFP expression for high density cultures (HDC, detailed below).
Hoxd13 null allele:
The Hoxd13DsRed2 null allele (Hoxd13+/-) was produced by gene targeting using a Ds2Red-Stop-LoxP Neo LoxP cassette introduced into a BamHI restriction site present in Exon 1 of murine Hoxd13 (Figure S1A,B). Homologous recombination of the Hoxd13DsRed2 targeting vector was confirmed by southern blot analysis of DNA isolated following positive/negative selection of the cultured ES cells electroporated with the Hoxd13DsRed2 targeting vector as described[48]. Germline transmission of the targeted allele was confirmed by southern blot (Figure S1C). In vivo removal of the neomycin resistance cassette was accomplished by breeding heterozygous mice with a CMV-Cre mouse. Removal of the neomycin resistance marker was confirmed by PCR. Pups identified as positive for DsRed2 and lacking the neomycin resistance marker were used in all subsequent analyses. Intercrosses of Hoxd13DsRed2 heterozygous adult mice produced homozygous mutant offspring at the expected Mendelian Ratios. Analysis of homozygous mutant Hoxd13DsRed2 embryos by confocal microscopy revealed no detectable signal for DsRed2 fluorescence. The presence of the DsRed2 cassette in the Hoxd13 locus was confirmed by in situ hybridization using an antisense riboprobe specific for DsRed2 which was detected in the autopod Hoxd13 expression domain (Figure S1E). Phenotype analysis of the Hoxd13DsRed2 mutant limbs revealed reduction in the number of phalangeal joints, digit brachydactyly, and malformation of digit 1 (Figure S1F), phenotypes previously reported for Hoxd13 loss of function alleles[4, 5]. Primer sequences for Hoxd13DsRed2 genotyping depicted in Figure S1B,D are D13-F: 5-’GCA AGA GCC AAG GAA GTG-3’, D13-R: 5’-ATG GAG ATG TAT GCC TCG TG-3’, and DSRED2R: 5-GAG CCG TAC TGG AAC TGG-3’. Skeletal staining with Alcian Blue/Alizarin Red in acid-alcohol followed by clearing in 1% w/v KOH was performed on E17.5 limbs as described[10].
LacZ staining
For β-galactosidase (LacZ) staining, mouse embryos were fixed in 2% w/v paraformaldehyde (PFA) and 0.2% v/v glutaraldehyde for 1–2 h, washed in PBS with 0.2% v/v Tween (PBST) and stained with XGal (1mg/ml) in PBST and 2mM MgCl2, 5mM Ferro-CN, 5mM Ferri-CN, at 37°C for several hours to overnight.
High density culture (HDC) of limb mesenchyme
Cell dissociation and imaging:
E11.25 hind limbs were dissected into anterior and posterior halves and pooled based on genotype and EYFP or tdTomato fluorescent marker allele expression (for A-A, P-P, or A-P comparisons, as indicated in Figure 3). Pooled limb bud halves were rinsed in PBS, dissociated in 0.25% w/v trypsin at 37°C and digestion terminated with DMEM + 20% v/v FBS. For each mixed culture, cell suspensions were filtered through 40μm cell strainers (Falcon 352340), yellow and red fluorescent cells counted using Countess II FL Automated Cell Counter (ThermoFisher AMQF1000), and only mixed HDC pools with approximately equal YFP and tdTomato cell counts were analyzed (less than 30% difference). HDC were cultured under conditions previously described[27]. Cells were resuspended in DMEM without phenol + 10% v/v FBS + Primocin (InvivoGen amt-pm-2) at a final concentration of 5 x 103 cells μl-1. 10μl cell suspension was spotted on uncoated Ibidi µ-Dish 35 mm, high glass bottom dish (81158) and allowed to attach for 1 hour at 37°C, after which the Ibidi dish was flooded with 2ml of the same media, and the live culture was imaged every 10min using a Nikon Ti-Eclipse microscope fitted with a 4X objective, Andor Zyla camera, and a heated stage insert (humidified 5% CO2 chamber at 37°C) for up to 24hrs.
Condensation rate analysis:
For analysis of cell aggregate sizes over time relative to time zero (single cells), images of the entire culture were analyzed using Imaris v9.1 (Bitplane) software by generating surfaces (representing cell aggregates) with a smooth surface area detail level of 1.0μm and a thresholding background subtraction (local contrast) with the diameter of the largest sphere which fits into the surface object set at 30.0μm (example shown in Figure S4). The manual threshold intensity was then adjusted to select all images just above a level where a background signal becomes detected. The surfaces over 10 μm in area were used for the analysis. Once parameters were defined, average surface (aggregate) sizes and numbers were calculated for EYFP+ and for tdTomato+ cells for each time point in a given HDC imaging set. Calculations were performed for every hour between 0–12 hours; after 12 hours of culture, most cells for both anterior and posterior limb bud of either genotype had aggregated into visible clusters. To compare aggregation over time for independent cultures and to control for variation in cell plating numbers and inherent differences in EYFP and tdTomato fluorescence intensity, we determined the ratios of EYFP to tdTomato values for each time point within each mixed culture and normalized these values to a ratio of 1.0 for time zero (single cells); only cultures in which the difference in EYFP and tdTomato cell numbers plated was less than 30% were included in analysis. Normalized data of independent experiments were graphed and standard deviations determined, and slopes were compared using Graphpad Prism 8 software to calculate statistical significance of slope differences. Where there are no differences between two cell types in mixed HDC, the aggregate size ratios should be 1 over different time points, yielding a slope of zero. If a slope deviates significantly from zero over time, aggregation rates between the two cell types compared are different.
Flow cytometry cell cycle analysis
Pregnant females were injected with 2mg EdU at 1 hour prior to embryo collection. Hind limb buds were dissected into anterior and posterior halves, rinsed in PBS, dissociated in 100μl 0.25% w/v trypsin at 37°C and the reaction stopped with 100μl DMEM + 10% v/v FBS. Dissociated cells were fixed in 800μl 4% w/v PFA for 15min at room temperature, PFA was quenched with 100μl 1M Tris, and cells were washed in PBS + 1% w/v BSA, and finally resuspended in 100μl 1% w/v saponin buffer. To detect EdU incorporation, a Click-iT kit was used (ThermoFisher C10634) and 250μl reaction cocktail plus Alexa Fluor 647 Picolyl Azide added, and incubated 30min at room temperature, followed by 2 washes in 1ml of 1% w/v saponin buffer, and final suspension in 200μl 1% w/v saponin buffer containing 1:4000 Vybrant DyeCycle Violet Stain (ThermoFisher V35003). After an additional 30min incubation at 37°C, cells were analyzed by flow cytometry on a Sony Biotechnology LE-SH800 Cell Sorter using 100μm sorting chip (LE-C3110). For each stage evaluated, at least 7 independent samples (N= 7–11 as indicated in Figure 4 legend) were collected and analyzed for cell cycle parameters. For statistical analysis of flow data, standard 2-sided T-test was used.
Mouse limb bud protein immunoblot analysis
Comparison of Gli3 full-length and repressor protein levels in WT and 5’HoxdΔ/Δ mutant mouse embryo limb buds was performed as previously described[10, 24]. Briefly, single pairs of E10.75 hindlimb buds from embryos of indicated genotypes were isolated (no pooling), lysed and sonicated in 1% w/v SDS in PBS containing protease inhibitors. After boiling, the supernatant of lysates of one pair of limb buds were loaded into 1 well and and electrophoresed in 3–8% w/v acrylamide Tris-Acetate gels and transferred to nitrocellulose blots. Blots were probed with anti-Gli3 (1:1,000, R&D #AF3690) and with anti-Vinculin (1:1,000, Sigma #V4139) to control for differences in amounts loaded and visualized with secondary donkey anti-goat and donkey anti-rabbit antibodies (LI-COR #926–32214 and 926–68073). Band intensities were quantitated with the Odyssey LI-COR system v5.2 to quantify fluorescence signals and 8 independent samples were analyzed for each genotype. Significance of differences was determined using the standard 2-sided T-test.
Hybridization chain reaction (HCR) in situ analysis
Whole mouse embryos at E10.5–10.75, albino Xenopus tadpole hindlimb buds (stage 50–51) with attached torso segment for orientation, or white axolotl larval forelimb buds (stage 45–46) with attached torso segment for orientation, were each fixed in 4% w/v paraformaldehyde in PBS overnight at 4°C, washed, bleached in 5:1 methanol/30% w/v hydrogen peroxide for 15 min. at 25°C and stored in methanol at −20°C until hybridization. Whole mount in situ hybridization of mouse, Xenopus, and axolotl limbs at stages indicated was performed using recommended conditions and solutions for third generation hybridization chain reaction (HCR) probes[49] designed by Molecular Instruments (Los Angeles, CA) for each species and analyzed by confocal microscopy. Quantitation of total fluorescence intensity in confocal image stacks of representative equivalent anterior and posterior domains were performed using Image J software, and at least 3 or more independent hybridized limb buds per probe for each stage and genotype examined were analyzed.
Axolotl Gli3 CRISPR-Cas9 knock-down analysis
Guide RNAs were designed against N-terminal region axolotl Gli3 exon sequences using CRISPRscan algorithm, and two target-specific Alt-R® crRNAs (Figure 6A) and common Alt-R® tracrRNA were synthesized by Integrated DNA Technologies (Amex_PQ.v4 genome assembly AMEXTC_0340000049779_GLI3)[50]. Alt-R® – Cas9 Ribonucleoprotein complexes were prepared[51] and injected into 1-cell stage axolotl embryos (RRID:AGSC-100E) from the Ambystoma Genetic Stock Center. For injection, embryos were placed in 100% axolotl rearing water (ARW: 1.75 g NaCl, 100 mg MgSO4, 50 mg CaCl2, and 25 mg KCl per liter, with NaHCO3 to achieve a pH in the range of 7.1–7.6.) with 1x Penicillin/Streptomycin. A Parker Picospritzer III (20 psi) was used to deliver 4 nL of 5μM crRNA:tracrRNA:Cas9 RNP per embryo. At 24 hours post-injection, embryos were transferred to 50% v/v AWR with 1x Penicillin/Streptomycin, and thereafter, water was changed daily. Under University of Kentucky IACUC protocol #2017–2580, larvae were maintained in 100% v/v ARW and fed brine shrimp until reaching 3cM, and then California blackworms. Injected founder larvae were euthanized in 0.02% w/v benzocaine at developmental stage 46–49, head and tail tissue was flash frozen for DNA isolation, and torsos were processed for limb skeletal analysis: fixed in ethanol, stained with alcian blue in acid-alcohol overnight (0.1% w/v alcian blue, 1% v/v HCl, 70% v/v ethanol), rinsed several times in acid-alcohol, dehydrated in 100% ethanol, and cleared and stored in methyl salicylate. Some larvae were reared to 6cM, anesthetized in 0.02% w/v benzocaine and limbs were processed for skeletal staining from injected and from age-matched, non-injected larvae. Axolotl genomic DNA was isolated using the Monarch genomic DNA purification kit (Cat. # T3010S; New England Biolabs). Genomic DNA spanning the guide RNA region was PCR amplified (Figure 6A). PCR products were treated with ExoSap-IT (Cat. #78205; Applied Biosystems) and the resulting templates were sequenced by Eurofins Genomics (Louisville, KY) to verify mutagenesis (Figure 6B).
QUANTIFICATION AND STATISTICAL ANALYSIS
For HDC aggregation rate analyses, to compare aggregation rates for independent cultures and to control for variation in cell plating numbers and inherent differences in EYFP and tdTomato fluorescence intensity, we determined the ratios of EYFP to tdTomato values for each time point within each mixed culture and normalized these values to a ratio of 1.0 for time zero (single cells); only cultures in which the difference in EYFP and tdTomato cell numbers plated was less than 30% were included in analyses. Normalized data of independent experiments were graphed and standard deviations determined, and slopes were compared using GraphPad Prism 8 software to calculate statistical significance of slope differences. Where there are no differences between two cell types in mixed HDC, the aggregate size ratios should be 1 over different time points, yielding a slope of zero. If a slope deviates significantly from zero over time, aggregation rates between the two cell types compared are different. Quantitation of total fluorescence intensity in HCR images were performed using Image J v2.1.0 software[52]. For immunoblots, the Odyssey LI-COR system v5.2 was used to quantify band fluorescence signals. For all experiments, statistical significance of differences was determined using the standard 2-sided T-test. Numbers of biological replicates, means, SEM or SD (as indicated), and p values for all experiments are reported in the main text and/or figure legends for the relevant data. The level of alpha used in all t-tests was 0.05.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat anti-Gli3 antibody | R&D Systems | CAT#AF3690; RRID: AB_2232499 |
| Rabbit anti-Vinculin antibody | Sigma-Aldrich | CAT#V4139; RRID: AB_262053 |
| IRDye 800CW donkey anti-goat secondary antibody | LI-COR | CAT#926–32214; RRID: AB_621846 |
| IRDye 680RD donkey anti-rabbit secondary antibody | LI-COR | CAT#926–68073; RRID: AB_10954442 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Tamoxifen | Sigma-Aldrich | CAT#T-5648 |
| Alcian Blue 8 GX | Sigma-Aldrich | CAT#A-5268 |
| Alizarin Red S | Sigma-Aldrich | CAT#A5533 |
| Paraformaldehyde | Sigma-Aldrich | CAT#441244 |
| Glutaraldehyde | Polyscience | CAT#00216 |
| 0.25% Trypsin | ThermoFisher Scientific | CAT#15050065 |
| Primocin | InvivoGen | CAT#ant-pm-2 |
| β-galactosidase | GoldBio | CAT#X4281C |
| Critical Commercial Assays | ||
| Click-iT™ Plus EdU Alexa Fluor™ 647 Flow Cytometry Assay Kit | ThermoFisher Scientific | CAT#C10634 |
| Vybrant™ DyeCycle™ Violet Stain | ThermoFisher Scientific | CAT#V35003 |
| in situ Hybridization Chain Reaction v3.0 | Molecular Instruments49 | N/A |
| Alt-R CRISPR-Cas9 System | Integrated DNA Technologies | Alt-R CRISPR-Cas9 |
| Experimental Models: Organisms/Strains | ||
| Mouse: AP2-Cre tg | 37 | N/A |
| Mouse: BmpR1a+/- | 38 | N/A |
| Mouse: BmpR1b+/- | 39 | N/A |
| Mouse: Gli3XtJ/+ | 40 | N/A |
| Mouse: 5’HoxdΔ/+ | 6 | N/A |
| Mouse: 5’HoxdΔ/+ (RXII) (lacking LacZ transgene) | 7 | N/A |
| Mouse: Hoxd11+/- | 42 | N/A |
| Mouse: Prrx-Cre tg | 43 | CAT#005584 |
| Mouse: Rosa-Grem1 tg | 30 | N/A |
| Mouse: Rosa-Gli3R tg | 33 | N/A |
| Mouse: NogginLacZ/+ | 22 | CAT#003383 |
| Mouse: Hoxb6-CreER tg | 41 | N/A |
| Mouse: Rosa-tdTomato tg | 46 | CAT#007914 |
| Mouse: Rosa-tdTomato tg-Activated | This study | N/A |
| Mouse: Rosa-EYFP tg | 47 | CAT#006148 |
| Mouse: Rosa-EYFP tg-Activated | This study | N/A |
| Mouse: Hoxd13+/- (Hoxd13DsRed2) | This study | N/A |
| X. tropicalis, Wild Type and albino | NASCO | N/A |
| A. mexicanum, Wild Type and white | Ambystoma Genetic Stock Center | RRID:SCR_006372 |
| Oligonucleotides | ||
| Hoxd13 (WT) | This study | D13-F: 5’-GCAAGAGCCAAGGAAGTG-3’ D13-R: 5’-ATGGAGATGTATGCCTCGTG-3’ |
| Hoxd13DsRed2 | This study | DSRED2-R: 5’-GAGCCGTACTGGAACTGG-3’ |
| A. mexicanum Gli3 gRNA | This study | gRNA 1: ATTTCGCCTCACAGAACTC gRNA 2: TGCCCGTGGACTCAGCCCG |
| A. mexicanum Gli3 sequencing primers | This study | Axo Gli3-F: 5’-GAGCTGCTTTGGTCTTTTGTTATT-3’ Axo Gli3-R: 5’-TCATAAAACCATCACTCTTTCCAA-3’ |
| Recombinant DNA | ||
| Hoxd13DsRed2 targeting vector | This study | N/A |
| Software and Algorithms | ||
| LI-COR Odyssey imaging system v5.2 | LI-COR | RRID: SCR_014579 |
| ImageJ v2.1.0 | 52 | RRID: SCR_003070 |
| Imaris v9.1 | Bitplane/Oxford Instruments | RRID: SCR_007370 |
| Prism 8 v8.4.3 | GraphPad | RRID: SCR_002798 |
Gli3 repressor (Gli3R) activity level governs tetrapod limb axis formation polarity
5’Hoxd-Gli3 balance modulates cell cycle exit to determine mouse limb axis polarity
Alternating A-P digit appearance in mammals is linked to primary limb axis polarity
Axolotl Gli3 knockdown shifts the ancestral preaxial dominance to postaxial
Acknowledgements
We thank Denis Duboule for generously providing the 5’HoxdDel(RXII) (d11-d13 deleted) mutant line, Deneen Wellik for the Hoxd11 mutant line, Steve Vokes for both RosaGrem1 and RosaGli3R transgenic lines, Trevor Williams for the AP2Cre transgenic line, Alan Godwin for help in generating the DSRed2 cassette used to create the Hoxd13 targeting construct, Matt Anderson for advice on HCR protocols, Rich Hwang for advice on tadpole analysis, and members of the CDBL for critical discussion.
This research was supported by the Center for Cancer Research (SM; intramural Research Program) and the Frederick National Laboratory (WFH, VM; OMAL Contract No. 75N91019D00024), NCI, NIH, by the Office of Infrastructure Programs, NIH (SRV and MK, P40-OD019794), and by grants from OHSU Medical Research Foundation and Shriners Hospitals for Children (HSS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
Authors declare no competing interests.
References:
- 1.Lopez-Rios J (2016). The many lives of SHH in limb development and evolution. Semin Cell Dev Biol 49, 116–124. [DOI] [PubMed] [Google Scholar]
- 2.Wellik DM, and Capecchi MR (2003). Hox10 and Hox11 genes are required to globally pattern the mammalian skeleton. Science 301, 363–367. [DOI] [PubMed] [Google Scholar]
- 3.Davis AP, Witte DP, Hsieh-Li HM, Potter SS, and Capecchi MR (1995). Absence of radius and ulna in mice lacking hoxa-11 and hoxd-11. Nature 375, 791–795. [DOI] [PubMed] [Google Scholar]
- 4.Davis AP, and Capecchi MR (1996). A mutational analysis of the 5’ HoxD genes: Dissection of genetic interactions during limb development in the mouse. Development 122, 1175–1185. [DOI] [PubMed] [Google Scholar]
- 5.Fromental-Ramain C, Warot X, Messadecq N, LeMeur M, Dolle P, and Chambon P (1996). Hoxa-13 and Hoxd-13 play a crucial role in the patterning of the limb autopod. Development 122, 2997–3011. [DOI] [PubMed] [Google Scholar]
- 6.Zakany J, and Duboule D (1996). Synpolydactyly in mice with a targeted deficiency in the HoxD complex. Nature 384, 69–71. [DOI] [PubMed] [Google Scholar]
- 7.Tarchini B, and Duboule D (2006). Control of Hoxd genes’ collinearity during early limb development. Dev Cell 10, 93–103. [DOI] [PubMed] [Google Scholar]
- 8.Lettice LA, Devenney P, De Angelis C, and Hill RE (2017). The Conserved Sonic Hedgehog Limb Enhancer Consists of Discrete Functional Elements that Regulate Precise Spatial Expression. Cell Rep 20, 1396–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Y, Knezevic V, Ervin V, Hutson R, Ward Y, and Mackem S (2004). Direct interaction with Hoxd proteins reverses Gli3-repressor function to promote digit formation downstream of Shh. Development 131, 2339–2347. [DOI] [PubMed] [Google Scholar]
- 10.Huang BL, Trofka A, Furusawa A, Norrie JL, Rabinowitz AH, Vokes SA, Mark Taketo M, Zakany J, and Mackem S (2016). An interdigit signalling centre instructs coordinate phalanx-joint formation governed by 5’Hoxd-Gli3 antagonism. Nat Commun 7, 12903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sheth R, Bastida MF, and Ros M (2007). Hoxd and Gli3 interactions modulate digit number in the amniote limb. Developmental Biology 310, 430–441. [DOI] [PubMed] [Google Scholar]
- 12.Zakany J, Zacchetti G, and Duboule D (2007). Interactions between HOXD and Gli3 genes control the limb apical ectodermal ridge via Fgf10. Developmental Biology 306, 883. [DOI] [PubMed] [Google Scholar]
- 13.Galli A, Robay D, Osterwalder M, Bao X, Benazet JD, Tariq M, Paro R, Mackem S, and Zeller R (2010). Distinct roles of Hand2 in initiating polarity and posterior Shh expression during the onset of mouse limb bud development. PLoS Genet 6, e1000901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brockes JP, and Gates PB (2014). Mechanisms underlying vertebrate limb regeneration: lessons from the salamander. Biochem Soc Trans 42, 625–630. [DOI] [PubMed] [Google Scholar]
- 15.Frobisch NB, Bickelmann C, Olori JC, and Witzmann F (2015). Deep-time evolution of regeneration and preaxial polarity in tetrapod limb development. Nature 527, 231–234. [DOI] [PubMed] [Google Scholar]
- 16.Frobisch NB, and Shubin NH (2011). Salamander limb development: integrating genes, morphology, and fossils. Dev Dyn 240, 1087–1099. [DOI] [PubMed] [Google Scholar]
- 17.Shubin NH, and Alberch P (1986). A Morphogenetic Approach to the Origin and Basic Organization of the Tetrapod Limb, (Evolutionary Biology: Springer, Boston, MA: ). [Google Scholar]
- 18.Bickelmann C, Frota-Lima GN, Triepel SK, Kawaguchi A, Schneider I, and Frobisch NB (2018). Noncanonical Hox, Etv4, and Gli3 gene activities give insight into unique limb patterning in salamanders. J Exp Zool B Mol Dev Evol 330, 138–147. [DOI] [PubMed] [Google Scholar]
- 19.Torok MA, Gardiner DM, Izpisua-Belmonte JC, and Bryant SV (1999). Sonic hedgehog (shh) expression in developing and regenerating axolotl limbs. J Exp Zool 284, 197–206. [PubMed] [Google Scholar]
- 20.Torok MA, Gardiner DM, Shubin NH, and Bryant SV (1998). Expression of HoxD genes in developing and regenerating axolotl limbs. Dev Biol 200, 225–233. [DOI] [PubMed] [Google Scholar]
- 21.Lopez-Rios J, Speziale D, Robay D, Scotti M, Osterwalder M, Nusspaumer G, Galli A, Hollander GA, Kmita M, and Zeller R (2012). GLI3 constrains digit number by controlling both progenitor proliferation and BMP-dependent exit to chondrogenesis. Dev Cell 22, 837–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brunet LJ, McMahon JA, McMahon AP, and Harland RM (1998). Noggin, cartilage morphogenesis, and joint formation in the mammalian skeleton. Science 280, 1455–1457. [DOI] [PubMed] [Google Scholar]
- 23.Zhu J, Nakamura E, Nguyen MT, Bao X, Akiyama H, and Mackem S (2008). Uncoupling Sonic hedgehog control of pattern and expansion of the developing limb bud. Dev Cell 14, 624–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bastida MF, Perez-Gomez R, Trofka A, Zhu J, Rada-Iglesias A, Sheth R, Stadler HS, Mackem S, and Ros MA (2020). The formation of the thumb requires direct modulation of Gli3 transcription by Hoxa13. Proc Natl Acad Sci U S A 117, 1090–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bastida MF, Delgado MD, Wang B, Fallon JF, Fernandez-Teran M, and Ros MA (2004). Levels of Gli3 repressor correlate with Bmp4 expression and apoptosis during limb development. Dev Dyn 231, 148–160. [DOI] [PubMed] [Google Scholar]
- 26.Pizette S, and Niswander L (2000). BMPs are required at two steps of limb chondrogenesis: formation of prechondrogenic condensations and their differentiation into chondrocytes. Dev Biol 219, 237–249. [DOI] [PubMed] [Google Scholar]
- 27.Barna M, and Niswander L (2007). Visualization of cartilage formation: insight into cellular properties of skeletal progenitors and chondrodysplasia syndromes. Dev Cell 12, 931–941. [DOI] [PubMed] [Google Scholar]
- 28.Owens EM, and Solursh M (1981). In vitro histogenic capacities of limb mesenchyme from various stage mouse embryos. Dev Biol 88, 297–311. [DOI] [PubMed] [Google Scholar]
- 29.Ide H, Wada N, and Uchiyama K (1994). Sorting out of cells from different parts and stages of the chick limb bud. Dev Biol 162, 71–76. [DOI] [PubMed] [Google Scholar]
- 30.Norrie JL, Lewandowski JP, Bouldin CM, Amarnath S, Li Q, Vokes MS, Ehrlich LIR, Harfe BD, and Vokes SA (2014). Dynamics of BMP signaling in limb bud mesenchyme and polydactyly. Dev Biol 393, 270–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kerney RR, Hanken J, and Blackburn DC (2018). Early limb patterning in the direct-developing salamander Plethodon cinereus revealed by sox9 and col2a1. Evol Dev 20, 100–107. [DOI] [PubMed] [Google Scholar]
- 32.Pickering J, Chinnaiya K, and Towers M (2019). An autoregulatory cell cycle timer integrates growth and specification in chick wing digit development. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vokes SA, Ji H, Wong WH, and McMahon AP (2008). A genome-scale analysis of the cis-regulatory circuitry underlying sonic hedgehog-mediated patterning of the mammalian limb. Genes Dev 22, 2651–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheth R, Barozzi I, Langlais D, Osterwalder M, Nemec S, Carlson HL, Stadler HS, Visel A, Drouin J, and Kmita M (2016). Distal Limb Patterning Requires Modulation of cis-Regulatory Activities by HOX13. Cell Rep 17, 2913–2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Onimaru K, Kuraku S, Takagi W, Hyodo S, Sharpe J, and Tanaka M (2015). A shift in anterior-posterior positional information underlies the fin-to-limb evolution. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu X, and Mackem S (2013). Tracing the evolution of avian wing digits. Current biology: CB 23, R538–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Creaser PC, D’Argenio DA, and Williams T (1996). Comparative and functional analysis of the AP2 promoter indicates that conserved octamer and initiator elements are critical for activity. Nucleic Acids Res 24, 2597–2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mishina Y, Suzuki A, Ueno N, and Behringer RR (1995). Bmpr encodes a type I bone morphogenetic protein receptor that is essential for gastrulation during mouse embryogenesis. Genes Dev 9, 3027–3037. [DOI] [PubMed] [Google Scholar]
- 39.Yi SE, Daluiski A, Pederson R, Rosen V, and Lyons KM (2000). The type I BMP receptor BMPRIB is required for chondrogenesis in the mouse limb. Development 127, 621–630. [DOI] [PubMed] [Google Scholar]
- 40.Buscher D, Grotewold L, and Ruther U (1998). The XtJ allele generates a Gli3 fusion transcript. Mamm Genome 9, 676–678. [DOI] [PubMed] [Google Scholar]
- 41.Nguyen MT, Zhu J, Nakamura E, Bao X, and Mackem S (2009). Tamoxifen-dependent, inducible Hoxb6CreERT recombinase function in lateral plate and limb mesoderm, CNS isthmic organizer, posterior trunk neural crest, hindgut, and tailbud. Dev Dyn 238, 467–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davis AP, and Capecchi MR (1994). Axial homeosis and appendicular skeleton defects in mice with a targeted disruption of hoxd-11. Development 120, 2187–2198. [DOI] [PubMed] [Google Scholar]
- 43.Logan M, Martin JF, Nagy A, Lobe C, Olson EN, and Tabin CJ (2002). Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis 33, 77–80. [DOI] [PubMed] [Google Scholar]
- 44.Nye HL, Cameron JA, Chernoff EA, and Stocum DL (2003). Extending the table of stages of normal development of the axolotl: limb development. Dev Dyn 226, 555–560. [DOI] [PubMed] [Google Scholar]
- 45.Nieuwkoop PD, and Faber J (1994). Normal table of Xenopus laevis (Daudin) : a systematical and chronological survey of the development from the fertilized egg till the end of metamorphosis, (New York: Garland Pub.). [Google Scholar]
- 46.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13, 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, and Costantini F (2001). Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol 1, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mansour SL, Thomas KR, and Capecchi MR (1988). Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes. Nature 336, 348–352. [DOI] [PubMed] [Google Scholar]
- 49.Choi HMT, Schwarzkopf M, Fornace ME, Acharya A, Artavanis G, Stegmaier J, Cunha A, and Pierce NA (2018). Third-generation in situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith JJ, Timoshevskaya N, Timoshevskiy VA, Keinath MC, Hardy D, and Voss SR (2019). A chromosome-scale assembly of the axolotl genome. Genome research 29, 317–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoshijima K, Jurynec MJ, Klatt Shaw D, Jacobi AM, Behlke MA, and Grunwald DJ (2019). Highly Efficient CRISPR-Cas9-Based Methods for Generating Deletion Mutations and F0 Embryos that Lack Gene Function in Zebrafish. Dev Cell 51, 645–657 e644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No standardized data sets were generated in this study.
This study does not report any original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


