Summary
Zinc (Zn) is essential for normal plant growth and development. The ZIP (ZRT, IRT-like Protein) family members are involved in Zn transport and cellular Zn homeostasis throughout the domains of life. In this study, we have characterized four ZIP transporters from Arabidopsis thaliana (IRT3, ZIP4, ZIP6 and ZIP9) to better understand their functional roles. The four ZIP proteins can restore the growth defect of a yeast Zn-uptake mutant and are up-regulated under Zn deficiency. Single and double mutants show no phenotypes under Zn-sufficient or Zn-limited growth conditions. In contrast, triple and quadruple mutants show impaired growth irrespective of external Zn supply due to reduced Zn translocation from root to shoot. All four ZIP genes are highly expressed during seed development, and siliques from all single and higher order mutants exhibited an increased number of abnormal seeds, and decreased Zn levels in mature seeds relative to wild type. The seed phenotypes could be reversed by supplementing the soil with Zn. Our data demonstrate that IRT3, ZIP4, ZIP6 and ZIP9 function redundantly in maintaining Zn homeostasis and seed development in A. thaliana.
Keywords: ZIP transporter, Arabidopsis, zinc, seeds, high order mutants, redundancy, Zinc homeostasis, abiotic stress
Introduction
Zinc is an essential element in all living organisms and acts as a structural or catalytic cofactor in a large number of enzymes and regulatory proteins, including RNA polymerase, superoxide dismutase, alcohol dehydrogenase, carbonic anhydrase, and the Zn finger family of transcription factors (Palmer and Guerinot, 2009; Sinclair et al., 2012). Bioinformatic approaches estimate that the zinc-binding proteome ranges from 5% to 6% in prokaryotes and about 10% in eukaryotes (Andreini et al., 2009).
Zinc-deficient soils are widespread globally and Zn deficiency is estimated to affect about one-third of the world’s human population who live on plant-based diets. Under Zn deficiency, plants have decreased growth, are more susceptible to stress and have decreased chlorophyll synthesis, leading to a severe reduction in crop production (Sinclair et al., 2012). The damage caused by Zn deficiency at the cellular level is presumably due to reactive oxygen species (ROS) (Cakmak, 2000). Excess Zn can also cause serious harm, due to unregulated high affinity binding to inappropriate intracellular ligands and uncontrolled displacement of essential cofactor metal cations (Palmgren et al., 2008). Therefore, plants must maintain physiologically adequate intracellular Zn levels using a tightly controlled metal homeostasis network capable of responding to fluctuations in Zn availability. This is largely achieved through Zn dependent changes in the expression of genes involved in the uptake, transport and storage of Zn (Lilay et al., 2021).
Zn transporters mediate the cellular influx and efflux of Zn. The ZIP (zinc-regulated transporter (ZRT)/iron-regulated transporter (IRT)-like proteins) family members have been reported to transport Zn as well as Fe and Mn into the cytoplasm from either outside of cells or from intracellular organelles/vesicles (Hu, 2021). The ZIP family was first identified in Arabidopsis thaliana (Eide et al., 1996) and we now know that members of this family are found throughout all domains of life (Hu, 2021). Many members of the A. thaliana ZIP gene family have been shown to be expressed throughout the plant, are up-regulated under Zn deficiency, and many can transport Zn when expressed in yeast (Grotz and Guerinot, 2006; Lin et al., 2009; Milner et al., 2013). However up until now we did not have strong in planta experimental evidence as to which A. thaliana ZIP family members are important for zinc uptake from the soil, and for maintaining Zn homeostasis. Overexpression of IRT3 results in increased accumulation of Zn in the shoot and Fe in the root of transgenic lines, suggesting that IRT3 functions as a Zn and Fe uptake transporter in A. thaliana but this study did not include characterization of a irt3 mutant (Lin et al., 2009). A number of loss of function mutants (zip1, zip2, zip5, zip6, zip9, zip12) have been examined, including A. halleri zip6, but none have a strong Zn-related phenotype (Wu et al., 2009; Milner et al., 2013; Inaba et al., 2015; Spielmann et al., 2020). This certainly suggests that many of the ZIP family members play redundant roles in Zn homeostasis. Indeed, recent work in rice has shown that two tandem duplicated genes, OsZIP5 and OsZIP9, function redundantly in Zn uptake (Tan et al., 2020). OsZIP5 and OsZIP9 are both expressed in the root epidermis and respond to the local Zn status in the root; OsZIP9 is also regulated by systemic signals of Zn status from the shoot. oszip9 mutants show impaired Zn uptake and growth retardation (Tan et al., 2020; Huang et al., 2020; Yang et al., 2020). oszip5 mutants also show decreased Zn uptake, although their phenotype is less severe compared to the oszip9 mutant (Tan et al., 2020). The double mutant oszip5oszip9 showed an enhanced Zn deficiency phenotype compared with the single mutants (Tan et al., 2020). These are the first Zn transporters unequivocally shown to be responsible for uptake from the soil and the first example of the functional overlap among ZIP family members (Tan et al., 2020).
In order to understand the role of the A. thaliana ZIP family members in Zn transport, we examined the function of four putative zinc transporters, IRT3, ZIP4, ZIP6 and ZIP9. We were particularly interested in the role of these transporters in seed development as previous loss of function studies in A. thaliana did not examine seed phenotypes. Zn plays a vital role in seed development with reproductive organs having a high requirement for Zn (Krämer and Clemens, 2005). Using single mutants for each gene, we generated double, triple and quadruple knockout mutants to investigate their functional redundancy. Triple and quadruple mutants showed decreased growth irrespective of Zn supply due to reduced Zn translocation from root to shoot. Furthermore, all single and higher order mutants exhibited abnormal seed development, with higher order mutants having more severe defects, suggesting redundant roles for both vegetative growth and reproductive organ development.
Results
Single and double mutants of IRT3, ZIP4, ZIP6 and ZIP9 do not affect seedling growth or Zn distribution.
In order to begin to understand the function of various understudied members of the ZIP family in Arabidopsis and to determine whether any of these members act redundantly, we decided to create higher order mutants of three ZIP genes (IRT3, ZIP4, ZIP9) that encode proteins with high amino acid similarity and form a subgroup based on phylogenetic analysis (Evens et al., 2017). We also decided to investigate ZIP6 which does not cluster with any other ZIP family members (Evens et al., 2017) and, as such, might be expected to have a unique role in Zn homeostasis.
Functional complementation in yeast showed that all four ZIP proteins can transport Zn (Figure 1a) but not Fe or Mn (Figure S1). We note here that Milner et al. (2013) had previously reported that ZIP6 and ZIP9 cannot transport Zn but can transport Mn, and Lin et al. (2009) had reported that IRT3 could transport Fe as well as Zn. The differences may be explained by the use of different cDNA clones and expression systems. We confirmed that the expression of all four genes is induced in roots when plants are grown under Zn deficient conditions (Figure 1b). Expression of IRT3 and ZIP6 was also increased in Zn deficient shoots whereas ZIP4 and ZIP9 expression levels were not affected. Under Fe deficient growth conditions, expression of ZIP9 was upregulated both in shoot and roots (Figure 1b). We also examined the temporal and spatial expression of the four ZIP genes, using transgenic plants harboring 1 kb of promoter sequence fused to a GUS reporter gene. When plants were grown under Zn deficient growth conditions, expression in roots was enhanced in all transgenic plants (Figure 2a), consistent with our qPCR analysis (Figure 1b). The GUS staining indicated that the four ZIP genes were mainly expressed in the root stele when plants were grown under Zn limitation. In Zn-limited shoots, expression of IRT3 and ZIP6 was increased relative to Zn sufficient conditions, whereas ZIP4 and ZIP9 expression was low regardless of growth conditions (Figure 2a), again in agreement with our qPCR data. In older leaves, we could detect GUS expression mainly in the vascular tissues for all ZIP proteins except ZIP9 (Figure 2b–d). During the reproductive stage, GUS expression driven by the IRT3, ZIP4 and ZIP6 promoters was seen in the flowers and developing seeds (Figure 2b–d). Examination of publicly available expression profiles of A. thaliana ZIP genes revealed that IRT3, ZIP4, ZIP6 are all highly expressed during flower and seed development (Figure S2), agreeing with the results from the promoter-GUS lines.
Figure 1.
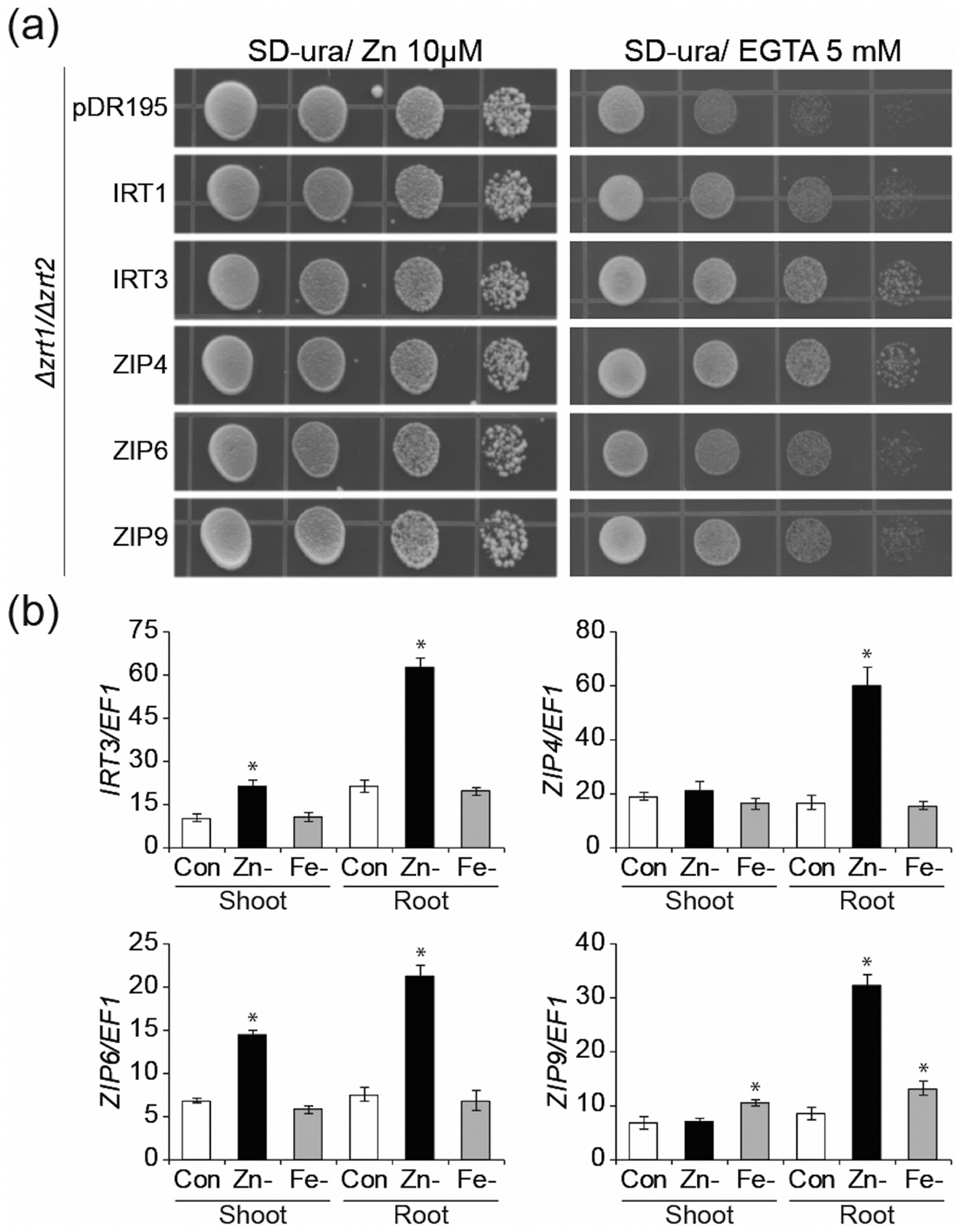
Functional complementation of a yeast Zn uptake mutant and expression analysis of A. thaliana ZIP genes under micronutrient deficiencies. (a) Complementation test of the Zn-uptake defective yeast mutant zrt1 zrt2. Yeast cells were transformed with empty vector pDR195 or a construct containing a ZIP cDNA. IRT1 was used as a positive control. After growth overnight, cell concentration was adjusted to OD600 = 1.0 and then serially diluted 10-, 100-, and 1000-fold. For the assay, 5 μl of each dilution were plated out onto the control or Zn-limited medium and grown for 3 days at 30 °C. (b) Quantitative real time PCR analysis of four ZIP genes in shoot and root of A. thaliana seedlings grown on control or Zn− or Fe-deficient medium. Expression is shown relative to EF1α. Error bars represent the standard error of 3 replicates. Asterisks above the bars indicate significant differences from the control growth condition by Student’s t-test (p< 0.05).
Figure 2.
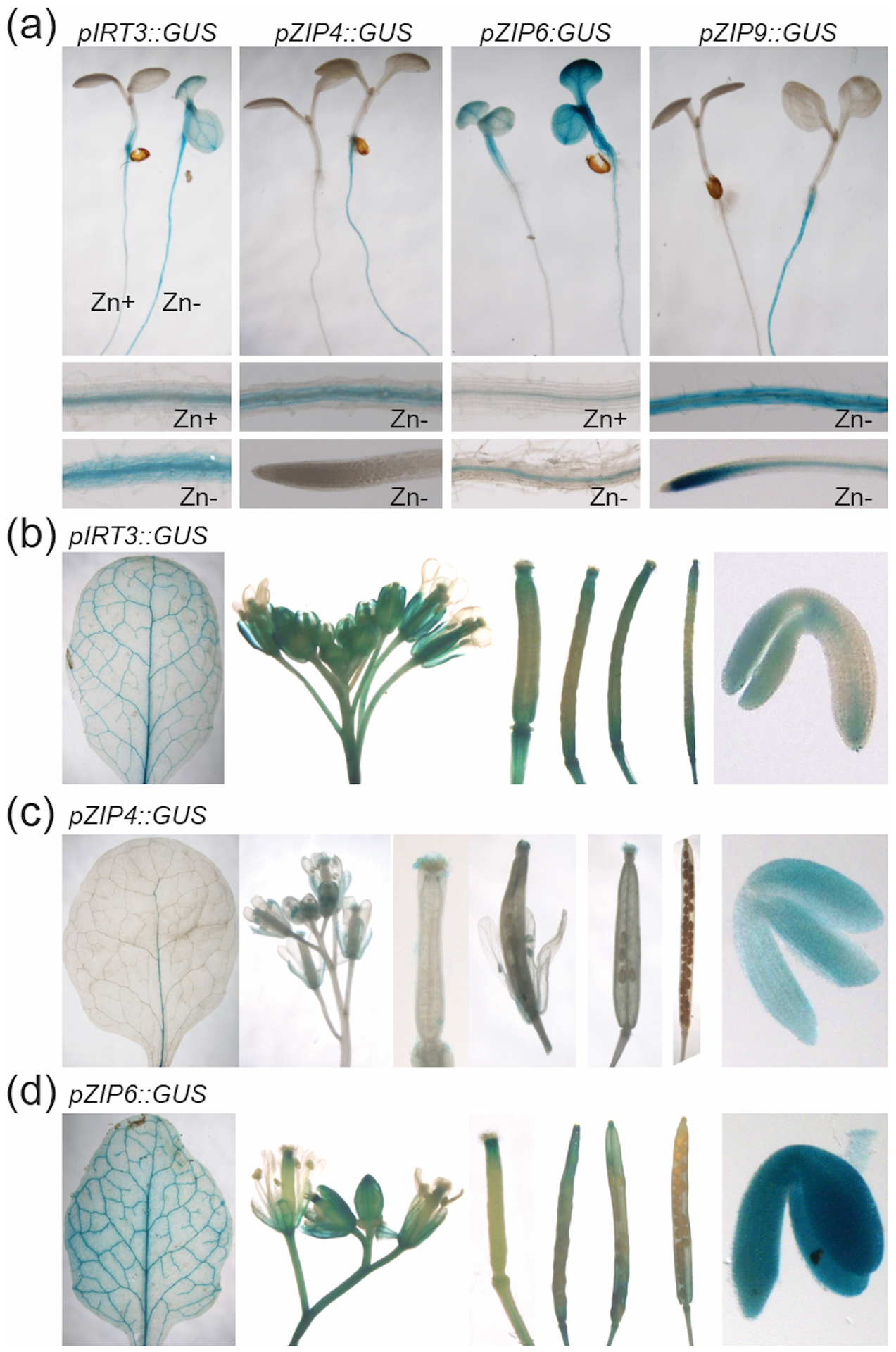
Spatial and temporal expression of A. thaliana ZIP genes in promoter-GUS transgenic plants. (a) four-day old seedlings grown under Zn deficiency (Zn−) or Zn sufficiency (Zn+). Whole seedlings shown in top row with sections of root shown in the middle and bottom rows (a); GUS expression patterns of 7-week old leaf, flowers, siliques and developing seeds from pIRT3::GUS (b), pZIP4::GUS (c) and pZIP6::GUS (d).
To investigate the subcellular localization of the four ZIP proteins, we generated transgenic Arabidopsis plants expressing GFP translational fusions for the four ZIP proteins, driven by the CaMV 35S promoter. Green fluorescence signals were detected in the plasma membrane (PM) (Fig. S3), and propidium iodide (PI) was used to stain the plasma membrane. Co-localization of GFP and PI signals strongly suggests that the four ZIP proteins all localize to the plasma membrane. Taken together, our results suggest that these four ZIPs act as PM-localized Zn transporters.
In order to characterize the functional role of each ZIP gene in regulating Zn homeostasis, we isolated T-DNA mutants for each of the four ZIP genes (Figure S4a). The T-DNA insertion in irt3-1 is located in the promoter (Figure S4a), leading to reduced expression relative to WT when this mutant is grown under Zn sufficient conditions and no induction of IRT3 expression under Zn deficiency (Figure S4b). zip4-2, zip4-3, zip6-1, zip6-2 and zip9 are loss of function mutants, with no expression detected (Figure S4c–e). When the T-DNA lines were grown on medium containing Zn, no obvious phenotypes were observed compared to WT (Figure S5a). Their fresh weight and root length were similar to those of WT (Figure S5b, c). Under Zn deficiency, the growth of the T-DNA mutants was similar to that of WT, as shown by the similar biomass and root length (Figure S5a–c). To determine whether disruption of any of the four ZIP genes affected Zn distribution, we measured the Zn concentrations in shoots and roots of plants grown under Zn-sufficient and Zn deficient conditions. Compared with WT, the mutants accumulated similar levels of Zn in shoots and roots, reflecting the lack of phenotypic differences (Figure S5d, e).
We next generated double mutants and grew the plants under Zn sufficient or Zn deficient growth conditions. The double mutants did not show any obvious morphological phenotypes, as compared to WT regardless of growth conditions (Figure S6a). Quantification of fresh weight and root length also indicated that there were no significant differences between WT and double mutants (Figure S6b, c). When we compared the Zn concentrations in shoots and roots, double mutants showed levels similar to those of WT (Figure S6d, e).
Quadruple and triple mutants have impaired seedling growth and altered Zn distribution
As single and double T-DNA mutants did not show any obvious phenotypes at the seedling stage, we generated triple and quadruple mutants. Two triple mutants (tko-1: irt3-1/zip4-2/zip6-1; tko-2: irt3-1/zip4-2/zip9-1) and one quadruple (qko: irt3-1/zip4-2/zip6-1/zip9-1) mutant were isolated. Quadruple and triple mutants were smaller than WT, irrespective of Zn supply (Figure 3a, b). Under Zn sufficiency, the fresh weights of qko, tko-1 and tko-2 mutants were 76.6%, 84.8% and 88.4% compared to WT; without Zn, fresh weights were 83.2%, 87.9% and 88.5% compared to WT fresh weight (Figure 3c). Root lengths of qko, tko-1 and tko-2 were 72.7%, 82.8% and 86.7% compared to WT under Zn supply, and 65.8%, 80.7% and 82.1% compared to WT under Zn deficiency, relative to WT (Figure 3d).
Figure 3.
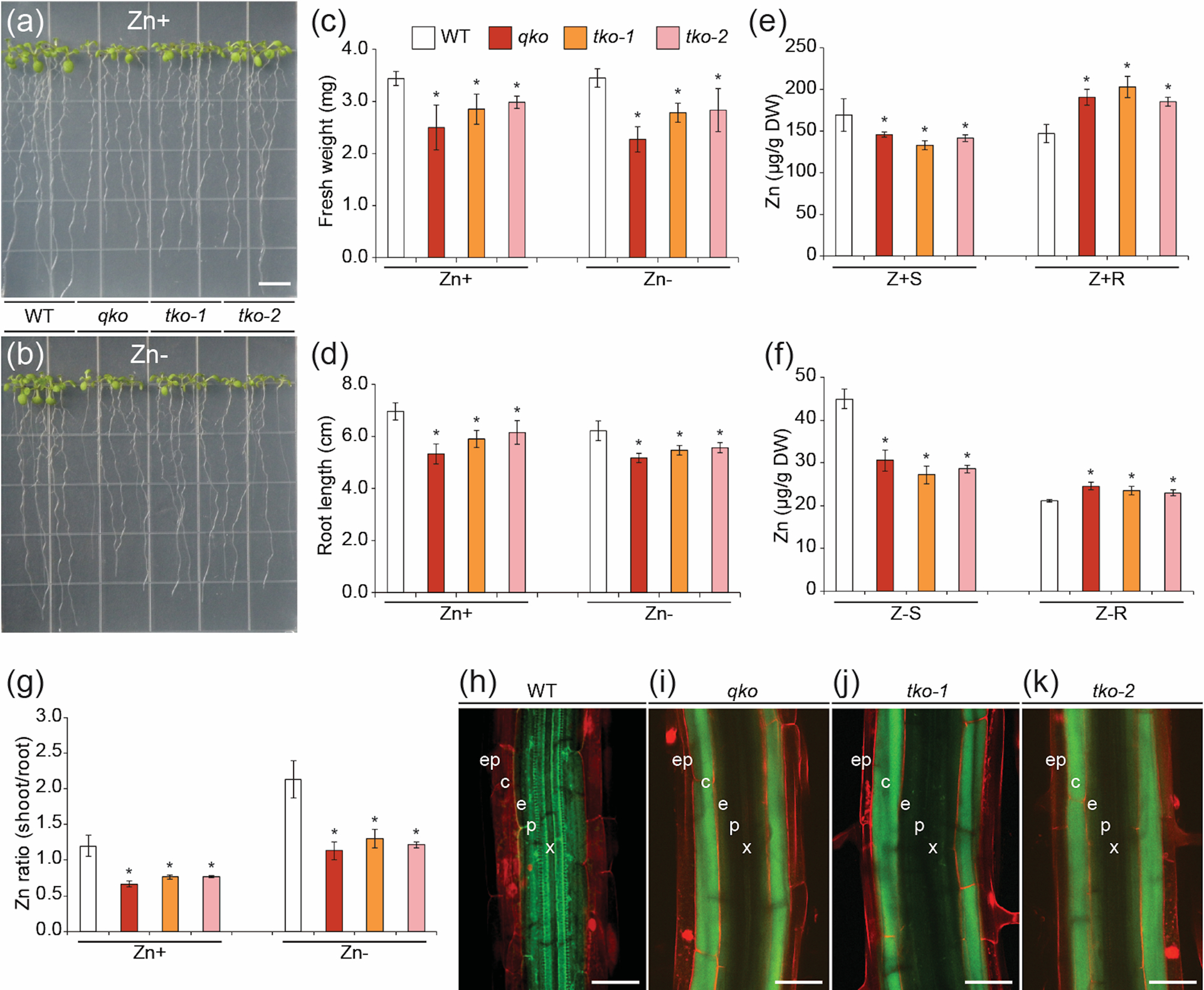
Seedling growth of triple and quadruple mutants. Growth of WT and mutant seedlings on Zn-sufficient (Zn+) (a) and Zn-deficient (Zn−) (b) medium. Photographs were taken 8 d after germination. Bars = 1 cm. Fresh weight (c) and root length (d) of fifteen seedlings of each line grown on Zn+ or Zn− medium. Zn concentrations in shoot and roots from WT, quadruple and triple mutants grown under Zn+ (e) or Zn− (f) for eight days. Tissues were harvested, digested and analyzed by ICP-MS. (g) Zn shoot:root ratios of Zn concentrations were calculated from the data shown in panels e and f. qko indicates irt3-1/zip4-2/zip6-1/zip9-1; tko-1, irt3-1/zip4-2/zip6-1; tko-2, irt3-1/zip4-2/zip9-1. Z+S denotes shoots from Zn sufficient medium; Z+R, roots from Zn sufficiency; Z-S, shoots from Zn deficiency; Z-R, roots from Zn deficiency. Mean values and standard errors were obtained from four independent experiments. *, P < 0.05 for significant difference from WT. (h-k) Visualization of Zn localization in roots by Zinpyr-1. Confocal laser-scanning microscope images of roots of WT (h), qko (i), tko-1 (j) and tko-2 (k) stained with Zinpyr-1 and propidium iodide to visualize Zn (green) and cell walls (red), respectively. x, xylem; p, pericycle; e, endodermis; c, cortex; ep, epidermis. Bars = 50 μm.
To assess whether disruption of three or four ZIP genes affect Zn distribution, we measured the Zn concentrations in shoots and roots at the seedling stage grown on Zn-sufficient or Zn-limited media (Figure 3e, f). Zn concentrations in shoots from qko, tko-1 and tko-2 were lower than that of WT, regardless of the Zn supply (Figure 3e, f). On the contrary, roots from quadruple and triple mutants accumulated more Zn, as compared to WT (Figure 3e, f). Therefore, we observed that the shoot:root ratios of Zn concentrations of WT were higher than those of qko, tko-1 and tko-2, suggesting less efficient root-to-shoot translocation of Zn in mutants than in WT (Figure 3g). Because root Fe uptake can interact and interfere with Zn uptake (Milner et al., 2013), we assessed the role of four ZIPs on the distribution of Fe at the seedling stage under Zn-sufficient or Zn deficient growth conditions. The results indicated that there were no differences in Fe concentration among genotypes and therefore, the shoot:root ratios of Fe concentrations were not different (Figure S7).
Next, we examined whether the triple or quadruple mutants changed the distribution of Zn within the roots, using Zinpyr-1 (Figure 3h–k). Zinpyr-1 is Zn fluorophore that has been previously used to examine the localization of Zn in roots in A. thaliana (Sinclair et al., 2007). In WT, green fluorescence was abundant in the stele where the four ZIP genes are normally expressed (Figure 3h). By contrast, the strongest fluorescence signal was observed in the cortex and endodermis in the quadruple and triple mutants (Figure 3i–k).
Because quadruple and triple mutants modified Zn distribution and translocation between shoot and root, we monitored the sensitivity of seedlings to Zn toxicity. When quadruple and triple mutants were exposed to excess Zn at the seedling stage, they showed better growth and had longer roots than WT (Figure S8).
Altered expression of Zn homeostasis related genes in seedlings of the quadruple and triple mutants
As quadruple and triple mutants exhibited defective seedling growth, we investigated the transcript levels of several genes related to Zn homeostasis in the WT, quadruple and triple mutants under Zn sufficient or Zn deficient growth conditions (Figure S8). First, we analyzed the expression of four A. thaliana nicotianamine synthetase (NAS) genes. NAS catalyzes the formation of nicotianamine (NA) which is involved in the long-distance distribution of metals in plants and is important for cellular metal homeostasis in all tissues (Schuler et al., 2012). In WT, expression of NAS2, NAS3 and NAS4 was higher when plants were grown under Zn deficient than under Zn sufficient growth conditions, while NAS1 was not changed by different external Zn supply (Figure S9a–d). In the quadruple and triple mutants, Zn deficiency enhanced the expression of NAS1 and NAS3 (Figure S9a, c), and transcript levels of NAS2 were increased in shoot and root irrespective of Zn status (Figure S9b). The expression of NAS4 was increased only in Zn-deficient shoots (Figure S9d).
Next, we analyzed the expressions of HMA2 and HMA4, which are located in the plasma membrane and involved in the long distance translocation of Zn (Hussain et al., 2004). In the mutants, expression of HMA2 was decreased in roots relative to WT regardless of Zn concentration (Figure S9e), and HMA4 transcripts were decreased in both roots and shoots of mutants both under Zn sufficient and Zn deficient growth conditions (Figure S9f). We also analyzed the transcript levels of MTP1 and MTP3, which are located at the vacuole membrane and contribute to basal Zn tolerance (Arrivault et al., 2006; Kawachi et al., 2009; Sinclair et al., 2018). Zn deficiency did not affect the mRNA levels of MTP1 in mutants as compared to WT (Figure S9g). In WT, expression levels of MTP3 were higher in roots than in shoots, whereas MTP3 expression in roots from mutant plants showed increased mRNA levels relative to WT irrespective of external Zn status (Figure S9h).
All mutants have altered seed development
We examined characteristics of seeds from WT and mutants to further elucidate the relationship between seed development and ZIP genes. When grown in soil, quadruple and triple mutants show no obvious visible phenotypes, as compared to WT (Figure S10a). Next, we dissected individual siliques and examined seeds from the quadruple and triple mutants to compare to WT at different developmental stages (Figure 4a). As compared to the young and green WT siliques which had embryos of similar size and shape, siliques from quadruple and triple mutants exhibited several defects. Mutant siliques contained more aborted seeds which appeared brown and shrunken (Figure 4a). Quantification of seed abortion revealed that 20.8%, 17.2% and 16.6% of the seeds from qko, tko-1 and tko-2 mutants aborted, while 3.2 % of seeds were aborted in WT siliques (Figure 4b). The number of seeds per silique was also lower than that of WT (Figure 4c). At the late dry maturation stages, mutants contained aborted dark brown and small seeds, while WT mature siliques consisted of uniform brown seeds (Figure 4a). Seed length of mutants was also decreased (Figure 4d), and therefore the 100-seed mass of quadruple and triple mutants was significantly lower than WT (Figure 4e). We tested whether Zn supplementation could rescue the seed phenotypes of the mutants. WT, quadruple and triple mutants were grown side by side in soil and watered with 1 mM ZnSO4 every week. Although Zn supplementation resulted in reduced growth compared to normal soil (Figure S10b), siliques from the WT and mutants were indistinguishable, suggesting that the Zn supplement rescued the seed morphology of triple and quadruple mutants (Figure S10c).
Figure 4.
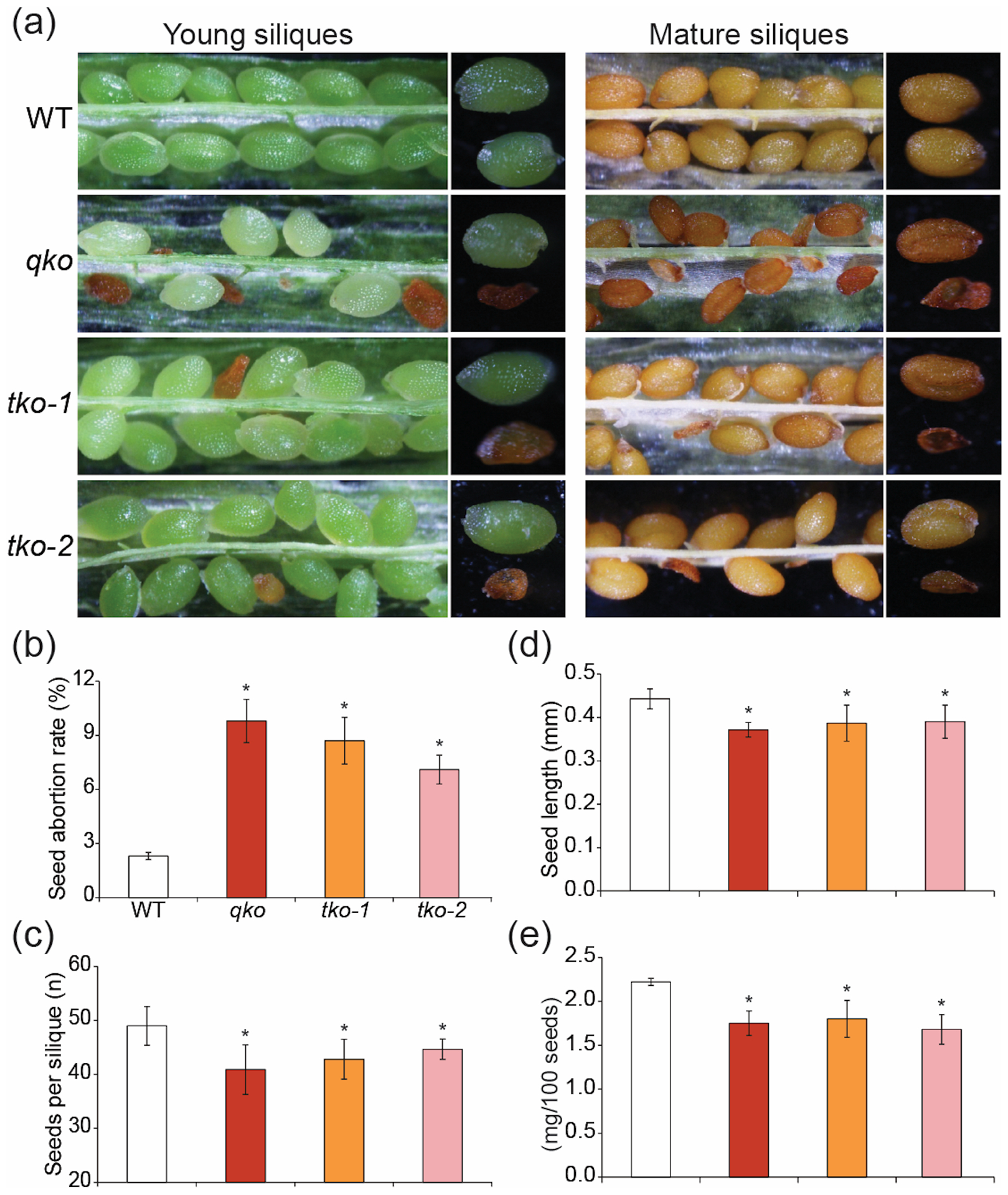
Altered seed development in triple and quadruple mutants. (a) Seed characteristics in the siliques from WT, the quadruple mutant and two triple mutants Quantification of seed abortion in WT and mutants (b) and the number of seeds per silique (c). Values are mean ± SE of 15 to 20 siliques from four independent plants. (d) Average seed length was measured after scanning the seeds via ImageJ software (https://imagej.nih.gov/ij/). Data represent mean ± SE of a minimum of 30 seeds. (e) Seed weight obtained by measuring 100 dry seeds (n = 5, mean ± SE). Asterisks above the bars indicate significant differences from WT (Student’s t-test, P < 0.05).
We also analyzed the seed phenotype of the single and double mutant plants. When grown in soil, the seed number per silique of single and double mutants was similar to that of WT (Figure S11a). In contrast, the seed abortion rate of mutants was higher than that of WT (Figure S11b). Seed length was similar to that of WT (Figure S11c), while the 100 seed mass of mutants was significantly lower than the WT (Figure S11d).
All mutants accumulate less metal ions in seeds
Because quadruple and triple mutants showed abnormal seed phenotypes, we next investigated metal accumulation and distribution in seeds. The Zn concentrations in seeds from the single and double mutant lines were significantly lower (83.3% ~91.2%) than that of WT (Figure S11e). Furthermore, the Zn concentrations of qko, tko-1and tko-2 mutants were 69.1%, 72.5% and 69.2% compared to WT (Figure 5a). Interestingly, Fe, Cu and Mn concentrations were also decreased in seeds from the single, double, triple and quadruple mutants (Figure 5b–d and Figure S11f–h). At present we cannot easily explain why the concentrations of these cations are also decreased in the mutants but, as noted in a recent review by Kumar et al. (2021), more work is needed to understand how nutrient pathways interact and influence each other.
Figure 5.
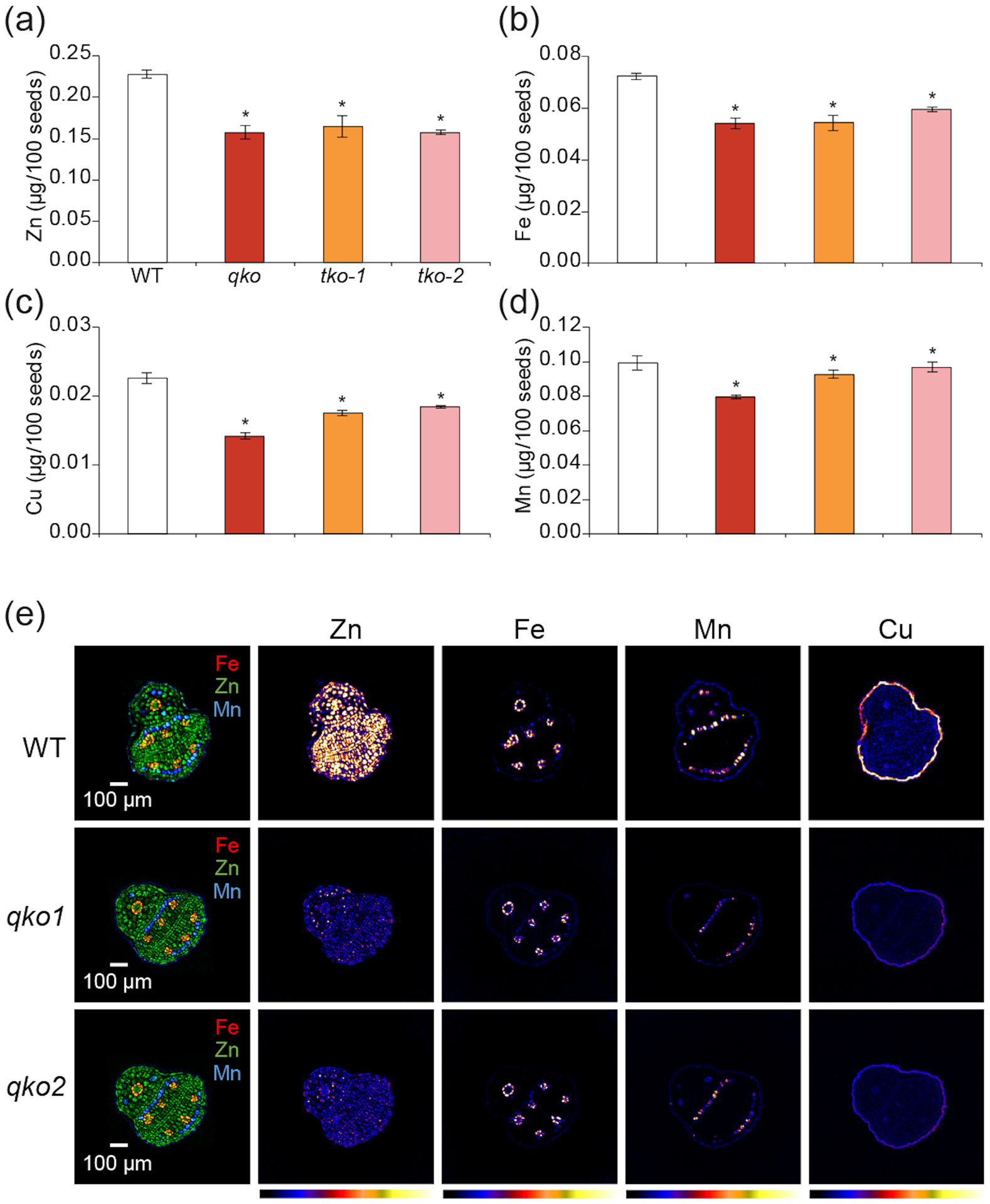
Metal content in mutant seeds measured by ICP-MS and Synchrotron X-ray fluorescence (SXRF) tomograms of mutant seeds. Quantification of Zn (a), Fe (b), Mn (c) and Cu (d) from WT, quadruple mutant and two triple mutants. Values are mean ± SE of 100 seeds from four independent plants. Asterisks above the bars indicate significant differences between mutant lines and WT (P < 0.05). (e) SXRF microtomography of Arabidopsis seeds. (Left row) SXRF composite images of Fe, Zn, and Mn internal distributions in WT (top) and qko (middle, bottom) seeds. (Right) Intensity-normalized SXRF tomographic slices of Zn Kα, Fe Kα, Mn Kα and Cu Kα fluorescence for WT and qko seeds. Tomograms for each element are scaled to the highest maximum pixel abundance. On the color bar, cooler colors indicate lower abundance and warmer colors higher abundance. qko1 and qko2 are two different seeds.
Next, we used synchrotron X-ray fluorescence (SXRF) elemental mapping to visualize the metal distribution in WT, quadruple and triple mutant seeds. High-resolution SXRF microtomography can be used to provide spatial localization and quantification of elements in plant tissues and has been successfully used to document unique localization patterns of micronutrients in seeds (Kim et al., 2006; Ricachenevsky et al., 2018). Zn was clearly less abundant in seeds from the quadruple mutant compared to WT, but there were no changes in distribution (Figure 5e). 2D imaging also showed less Zn in quadruple mutant seed, as compared to WT seeds (Figure S12a). Concentration of other metals such as Fe, Cu and Mn also decreased as measured by ICP-MS, but localization remained unchanged (Figure 5b–e). Seeds from triple mutants also had less Zn, consistent with ICP-MS analysis (Figure S12b).
DISCUSSION
Understanding the molecular basis of Zn uptake and transport in plants is critical for improving both the Zn content of plants and the ability of plants to grow in low Zn soils. Our study demonstrates the importance of members of the ZIP family in root to shoot translocation of Zn as well as in proper seed development. Single or double mutants of IRT3, ZIP4 and ZIP9, three closely related ZIP genes, exhibited no apparent phenotypes as seedlings whereas lines carrying mutations in all three of these ZIP genes had altered Zn distribution between shoots and roots, leading to impaired growth irrespective of external Zn status. This is a classic example of genetic redundancy where each of these three ZIP genes can compensate for loss of the other two genes. zip6 single or double mutants also did not show any phenotypes as seedlings but the triple mutant of irt3 zip4 zip6 had a very similar phenotype to that of the irt3 zip4 zip9 triple mutant, showing altered distribution of Zn between the root at the shoot. This suggests overlapping functions among these four genes and in the absence of ZIP6, ZIP9 can no longer compensate for the simultaneous loss of IRT3 and ZIP4, even though ZIP6 does not cluster phylogenetically with the other three ZIP genes. We propose that these four genes play a role in xylem loading based on 1) all are plasma-membrane localized Zn transporters; 2) all four proteins are expressed in the central cylinder of the root and 3) Zn does not localize to the stele in the roots of triple and quadruple mutants. Our GUS expression data agrees well with previously published IRT3-GUS data (Lin et al., 2009), ZIP4-GUS and ZIP4-GFP data (Lin et al., 2011) and ZIP6-GUS data (Speilmann et al., 2020). In addition, IRT3 had previously been shown to localize to the plasma membrane in agreement with our localization data.
The decreased ability to load Zn into the xylem then explains the lower levels of Zn observed in shoot tissues of the triple and quadruple mutants. This reduction of Zn translocation from root to shoot is similar to that reported for the A. thaliana hma2 hma4 double mutant. In roots of the hma2 hma4 double mutant, Zn was predominantly accumulated in the pericycle and endodermal cell layers, where HMA2 and HMA4 are normally expressed (Sinclair et al., 2007). Their lack of expression leads to a failure to export Zn into the xylem whereas a lack of expression of ZIP genes leads to failure to load the xylem parenchyma cells with Zn. The net result is the same: less Zn for transport to the shoots.
Although single and double mutants showed seed phenotypes with higher seed abortion rates and decreased seed weight compared to WT, quadruple and triple mutants had more pronounced phenotypes, with fewer seeds per siliques, higher rates of seed abortion as well as a reduction in seed weight compared to WT (Figure 4). Overall, the quadruple mutant had the most drastic seed phenotypes, likely owing to the additive effect of the disruption of four ZIP genes. The Zn concentration in the mature seed of all mutants was decreased relative to WT, but the quadruple mutant had the lowest seed Zn concentration, suggesting functional redundancy among these ZIP genes.
We can offer two possibilities for how disruption of ZIP genes leads to the observed seed phenotypes. One is that these four ZIP proteins directly affect seed development by loading Zn into seeds during the development of the endosperm and the embryo. The other is that the reduction in Zn translocation from root to shoot means that there is less Zn available to supply reproductive organs as they develop. And, of course, given that these ZIP genes are expressed in both the roots and in developing seeds, it is most likely a combination of both. Such a dual role has been shown for HMA2 and HMA4 (Olsen et al., 2016). By grafting mutant shoots to WT roots to ensure that the HMA pumps were delivering zinc to the shoot, Olsen et al. (2016) could then see that the pumps were also needed in the developing seed.
In rice, two members of the ZIP family, OsZIP3 and OsZIP7, have been implicated in xylem loading and Zn distribution to developing grain. OsZIP7 is expressed in parenchyma cells of vascular bundles in roots and in nodes. It mediates Zn influx from the cortex to pericycle in roots and is involved in intervascular transfer of Zn in nodes (Tan et al., 2019). OsZIP3 is localized to xylem transfer cells in enlarged vascular bundles (EVBs) of nodes and is involved in unloading of Zn from the xylem of EVBs (Sasaki et al., 2015).
The use of higher order mutants has allowed us to show that four of the Zn-deficiency inducible Zn transporters IRT3, ZIP4, ZIP6 and ZIP9 in A. thaliana have overlapping functions. Future studies using cell type specific promoters to drive expression of the different ZIP genes may allow further dissection of how various ZIP family members contribute to Zn homeostasis.
EXPERIMENTAL PROCEDURES
Plant materials and growth conditions
Seeds were surface sterilized and plated on half-strength MS medium supplemented with 0.5 g/L MES, 1.5% sucrose, 0.8% agar, pH 5.8. Plates were kept at 4°C in the dark for 4 days before being incubated under constant light at 21°C for 7 days. Seedlings then transferred to control, or Zn-deficient, or Fe-deficient media. For Zn or Fe deficiency, the Zn chelator TPEN (20 μM) (N,N,N′,N′-tetrakis (2-pyridinylmethyl)-1,2-ethanediamine) was supplemented at 20 μM, or the Fe chelator ferrozine (300 μM) (3-(2-pyridyl)-5, 6-diphenyl-1, 2, 4-triazine sulfonate) was added to the agar medium, respectively. The seedlings were grown for an additional three days and shoot and root samples from seedlings were frozen with liquid nitrogen for RNA isolation.
For the analysis of seedling phenotypes, sterilized seeds were sown on strength MS medium containing 15 μM ZnSO4 (control; Zn sufficient media), no added Zn (Zn−; Zn-deficient media) with 20 μM TPEN, or excess Zn (120 and 125 μM ZnSO4). Plants were grown for 8 days. Growth conditions for both plate-grown and soil-grown plants were 16/8-hour light-dark at 22 °C.
Yeast Growth Conditions
Yeast cells were transformed with empty vector pDR195 or constructs containing ZIP cDNAs. Three yeast strains were used in these studies: the Zn uptake-defective mutant zrt1/zrt2Δ (MATα ade6 can1 his3 leu2 trp1 ura3 zrt1::LEU2 zrt2::HIS3), the Fe uptake-defective mutant fet3/fet4Δ (MATa can1 his3 leu2 trp1 ura3 fet3::HIS3 fet4::LEU2), and the Mn uptake-defective mutant smf1Δ (SLY8; MATa ura3 lys2 ade2 trp1 his3 leu2 smf1::HIS3). Yeast cells were transformed by the Li-acetate method (Gietz et al., 2007). Transformants were grown in an YPD medium (1% yeast extract, 2% peptone, and 2% glucose) and densities were adjusted to an OD600 of 1, 0.1, 0.01, or 0.001 by serial dilution. Afterward, 5 μL aliquots were spotted onto a synthetic agar medium (SD) supplemented with 20 g/L of glucose and the necessary auxotrophic supplements. Primers used to design expression constructs are listed in Table S1.
Quantitative RT-PCR
RNA was prepared from each tissue. cDNA was reverse transcribed from 1 μg RNA with M-MLV reverse transcriptase (New England Biolabs). Quantitative PCR (qPCR) analysis performed to determine gene expression levels (ABI Model 7700, USA) using a SYBR premix ExTaq kit (Takara Bio, Shiga, Japan). Primers used are listed in Table S1. Samples were run in triplicate, normalized to EF1α, and arbitrary transcriptional units calculated. Values represent the average of three biological replicates.
Plasmid construction, Histochemical GUS Assay, and Confocal Microscopy Analysis of GFP
For GUS constructs, promoter regions (~1 kb) of IRT3, ZIP4, ZIP6 or ZIP9 were amplified from WT genomic DNA with two specific primers (Table S1), and the fragment was subcloned into the pGEM T-easy plasmid (Promega) and sequenced. It was then excised by digestion and ligated into pCAMBIA1381xA (CAMBIA). For the GFP constructs, the cDNAs spanning the entire ORF of IRT3, ZIP4, ZIP6 and ZIP9 were amplified with two specific primers listed in Table S1. Amplicons were introduced into the pGEM T-easy plasmid (Promega) for sequencing. After digestion with NcoI and BglII, the fragments were inserted into pCAMBIA 1302 (CAMBIA). All constructs were moved into Agrobacterium tumefaciens strain GV3101 and transformants were used to transform plants using the floral dip method (Clough and Bent, 1998). Transformants were isolated by selection on agar medium containing hygromycin (25 mg/mL).
For GUS histochemical staining, plants were grown on half strength MS medium with or without Zn for 4 days. Whole seedlings were incubated with the substrate 5-bromo-4-chloro-3-indolyl b-D-glucuronide as described (Jefferson et al., 1998). Laser scanning confocal microscopy (Leica TCS SP) was used to observe the GFP florescence from roots of transgenic plants stained with 10 mM propidium iodide.
Isolation of mutant lines
irt3-1 (WiscDsLox429D04) and zip 6-2 (WiscDsLox233237_03O) were obtained from the WiscDsLox T-DNA collection (Woody et al., 2007). zip4-2 (SALK_043236), zip4-3 (SALK_145371), zip6-1 (SALK_116013), zip9-1 (SALK_074682) were obtained from the SALK collection (Alonso et al., 2003). Homozygous lines were identified by PCR (Figure S4) using the left border T-DNA-specific and the gene-specific primers (Table S1). Double-, triple- and quadruple-mutant lines were obtained by crossing confirmed single mutants, and genotyping performed on plants from the third generation after crossing.
Tissue elemental analysis
Samples were dried at 60 °C for 3 days. Elemental analysis was done by ICP-MS as described (Lahner et al., 2003).
Zinpyr-1 staining
The zinc distribution in roots of WT and mutants was examined using Zinpyr-1, and propidium iodide was used to stain cell walls. Seven‐ to 8‐d‐old seedlings were washed three times in deionized water and immersed in a working solution of 20 μm Zinpyr‐1 and incubated at room temperature in darkness for 3 h. Samples were rinsed in deionized water, immersed in 75 μm propidium iodide and rinsed again. Preparation and staining process of Zinpyr-1 and propidium iodide were performed as described previously (Sinclair et al., 2007). Images were obtained using a confocal microscope (LSM700; Carl Zeiss) with 488-nm excitation.
Synchrotron-based X-ray Fluorescence Computed Microtomography (F-CMT) at XFM
Internal distributions of zinc and other micronutrient elements in WT and mutant-line Arabidopsis seeds were measured in vivo by synchrotron-based X-ray Fluorescence Computed Microtomography (F-CMT) at the X-ray Fluorescence Microprobe (XFM) beamline at the National Synchrotron Light Source II (NSLS-II) in Upton, NY. XFM beamline is designed for monochromatic operation (2.3 to 23 keV) and optimized for spatially-resolved X-ray absorption spectroscopy in conjunction with element-specific imaging and microdiffraction. XFM beamline can also be operated in a pink beam “imaging” mode that delivers up to 1000X more flux than the XFM monochromatic beam. XFM filtered pink beam (12 – 20 keV broadband) was focused by rhodium-coated, silicon Kirkpatrick-Baez (KB) mirrors to a 1 μm spot for F-CMT measurements of seeds. Seeds were mounted to a quartz post that was centered on a rotation stage attached to a fast-scanning translation stage. Seeds were translated horizontally through the focused X-ray beam in step sizes ranging from 2 to 4 μm and then rotated at intervals of between 0.5° and 1.5° angular steps, repeating the translation through a total of 360°. X-ray fluorescence spectra were recorded with a 7-element Canberra Mirion SXD-7 silicon drift detector coupled to Quantum Detectors Xspress3 electronics. Thin-film standard reference materials (SRM 1832 & 1833) were measured as part of the data set to establish elemental sensitivities (counts per second per μg cm−2). Two-dimensional sinograms were computationally reconstructed using filtered back projection method available in LARCH package (Newville, 2013) to provide images of the cross-sectional internal element distribution.
Synchrotron-based X-ray Imaging at NSLS and SSRL
Intact seeds were analyzed for metal distribution using SXRF at now decommissioned beamline X26A of the National Synchrotron Light Source, Brookhaven National Laboratory as described in detail previously (Kim et al., 2006; Ricachenevsky et al., 2018), and at BL2–3 at Stanford Synchrotron Radiation Lightsource, as described previously (Ricachenevsky et al., 2021).
Supplementary Material
Figure S1. Complementation of yeast metal uptake mutants.
Figure S2. In silico expression analysis of ZIP genes during development.
Figure S3. ZIP-GFP translational fusion proteins localize to the plasma membrane.
Figure S4. T-DNA mutants.
Figure S5. Seedling phenotypes of single T-DNA mutants grown under Zn-sufficient or Zn-deficient conditions.
Figure S6. Seedling phenotypes of double mutants grown under Zn-sufficient or Zn-deficient conditions.
Figure S7. Measurement of Fe levels of WT, triple and quadruple mutants.
Figure S8. Increased Zn resistance of triple and quadruple mutants.
Figure S9. Quantitative RT-PCR analysis of Zn homeostasis related genes at the seedling stage.
Figure S10. Growth morphology of WT, triple and quadruple mutants.
Figure S11. Characterization of seed phenotypes and metal contents in single and double mutants.
Figure S12. Synchrotron X-ray fluorescence (SXRF) elemental imaging of triple and quadruple mutants.
Table S1. List of primers used in this study.
Significance.
Deficiencies of micronutrients commonly limit plant growth and crop yields. Furthermore, as most people rely on plants as their dietary source of micronutrients, plants that serve as better sources of essential nutrients would improve human health. The use of higher order mutants has allowed us to show that four of the Arabidopsis thaliana ZIP transporters, IRT3, ZIP4, ZIP6 and ZIP9, function redundantly in maintaining Zn homeostasis and in seed development.
ACKNOWLEDGEMENTS
This work was supported by DOE grant DE-FG02-06ER15809 to MLG, NSF grant DBI 0701119 to MLG and DES, NIH grant R01GM078536 to MLG and DES and NIH grant P42ES007373 to MLG and TP. FKR was awarded a fellowship from CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico). Use of NSLS facility was supported by the Department of Energy under Contract DE-AC02-98CH10886. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. Parts of this research used the XFM Beamline of the National Synchrotron Light Source II, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Brookhaven National Laboratory under Contract No. DE-SC0012704.
Footnotes
CONFLICT OF INTEREST
The authors have declared no conflict of interest.
DATA STATEMENT
All relevant data can be found within the manuscript and its supporting materials.
REFERENCES
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Aguilar-Henonin L, Schmid M, Weigel D, Carter DE, Marchand T, Risseeuw E, Brogden D, Zeko A, Crosby WL, Berry CC and Ecker JR (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science, 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Andreini C, Bertini I and Rosato A (2009) Metalloproteomes: a bioinformatic approach. Acc. Chem. Res 42, 1471–1479. [DOI] [PubMed] [Google Scholar]
- Arrivault S, Senger T and Krämer U (2006) The Arabidopsis metal tolerance protein AtMTP3 maintains metal homeostasis by mediating Zn exclusion from the shoot under Fe deficiency and Zn oversupply. Plant J. 46, 861–879. [DOI] [PubMed] [Google Scholar]
- Cakmak I (2000) Possible roles of zinc in protecting plant cells from damage by reactive oxygen species. New Phytol. 146, 185–205. [DOI] [PubMed] [Google Scholar]
- Clough SJ and Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Eide D, Broderius M, Fett J and Guerinot ML (1996) A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc. Natl. Acad. Sci. USA, 93, 5624–5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evens NP, Buchner P, Williams LE and Hawkesford MJ (2017) The role of ZIP transporters and group F bZIP transcription factors in the Zn-deficiency response of wheat (Triticum aestivum). Plant J. 92, 291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD and Schiestl RH (2007) Quick and easy yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc 2, 35–37. [DOI] [PubMed] [Google Scholar]
- Grotz N and Guerinot ML (2006) Molecular aspects of Cu, Fe and Zn homeostasis in plants. Biochim. Biophys. Acta, 1763, 595–608. [DOI] [PubMed] [Google Scholar]
- Hu J (2021) Towards unzipping the ZIP transporters: structure, evolution and implications on drug discovery against cancer. FEBS J 10.1111/febs.15658. [DOI] [PubMed] [Google Scholar]
- Huang S, Sasaki A, Yamaji N, Okada H, Mitani-Ueno N and Ma JF (2020) The ZIP transporter family member OsZIP9 contributes to root zinc uptake in rice under zinc-limited conditions. Plant Physiol. 183, 1224–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain D, Haydon MJ, Wang Y, Wong E, Sherson SM, Young J, Camakaris J, Harper JF and Cobbett CS (2004) P-type ATPase heavy metal transporters with roles in essential zinc homeostasis in Arabidopsis. Plant Cell, 16, 1327–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba S, Kurata R, Kobayashi M, Yamagishi Y, Mori I, Ogata Y and Fukao Y (2015) Identification of putative target genes of bZIP19, a transcription factor essential for Arabidopsis adaptation to Zn deficiency in roots. Plant J. 84, 323–334. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawachi M, Kobae Y, Mori H, Tomioka R, Lee Y and Maeshima M (2009) A mutant strain Arabidopsis thaliana that lacks vacuolar membrane zinc transporter MTP1 revealed the latent tolerance to excessive zinc. Plant Cell Physiol. 50, 1156–1170. [DOI] [PubMed] [Google Scholar]
- Kim SA, Punshon T, Lanzirotti A, Li L, Alonso JM, Ecker JR, Kaplan J and Guerinot ML (2006) Localization of iron in Arabidopsis seed requires the vacuolar membrane transporter VIT1. Science, 314, 1295–1298. [DOI] [PubMed] [Google Scholar]
- Krämer U, Clemens S (2005) Functions and homeostasis of zinc, copper and nickel in plants. In Topics in Current Genetics 14 (Tamás M and Martinoia E, eds). Heidelberg; Springer, pp. 215–271. [Google Scholar]
- Kumar S, Kumar S and Mohapatra T (2021) Interaction between macro- and micro-nutrients in plants. Front. Plant Sci doi: 10.3389/fpls.2021.665583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahner B, Gong J, Mahmoudian M, Smith EL, Abid KB, Rogers EE, Guerinot ML, Harper JF, Ward JM, McIntyre L, Schroeder JI and Salt DE (2003) Genomic scale profiling of nutrient and trace elements in Arabidopsis thaliana. Nat. Biotechnol 21, 1215–1221. [DOI] [PubMed] [Google Scholar]
- Lilay GH, Persson DP, Castro PH, Liao F, Alexander RD, Aarts MGM, Assunção AGL (2021) Arabidopsis bZIP19 and bZIP23 act as zinc sensors to control plant zinc status. Nat. Plants, 7, 137–143. [DOI] [PubMed] [Google Scholar]
- Lin YF, Hassan Z, Talukdar S, Schat H and Aarts MG (2016) Expression of the ZNT1 Zinc Transporter from the Metal Hyperaccumulator Noccaea caerulescens Confers Enhanced Zinc and Cadmium Tolerance and Accumulation to Arabidopsis thaliana. PLoS One 11, e0149750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YF, Liang HM, Yang SY, Boch A, Clemens S, Chen CC, Wu JF, Huang JL and Yeh KC (2009) Arabidopsis IRT3 is a zinc-regulated and plasma membrane localized zinc/iron transporter. New Phytol. 182, 392–404. [DOI] [PubMed] [Google Scholar]
- Milner MJ, Seamon J, Craft E and Kochian LV (2013) Transport properties of members of the ZIP family in plants and their role in Zn and Mn homeostasis. J. Exp. Bot 64, 369–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newville M (2013) Larch: An analysis package for XAFS and related spectroscopies, J. Phys. Conf. Ser 430, 012007 [Google Scholar]
- Olsen LI, Hansen TH, Larue C, Østerberg JT, Hoffmann RD, Liesche J, Krämer U, Surblé S, Cadarsi S, Samson VA, Grolimund D, Husted S and Palmgren M (2016) Mother-plant-mediated pumping of zinc into the developing seed. Nat. Plants, 2, 16036. [DOI] [PubMed] [Google Scholar]
- Palmer CM and Guerinot ML (2009) A Question of Balance: Facing the challenges of Cu, Fe and Zn Homeostasis. Nat. Chem. Biol 5, 333–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmgren MG, Clemens S, Williams LE, Krämer U, Borg S, Schjørring JK and Sanders D (2008) Zinc biofortification of cereals: problems and solutions. Trends Plant Sci. 13, 464–473. [DOI] [PubMed] [Google Scholar]
- Ricachenevsky FK, Punshon T, Salt DE, Fett JP, Guerinot ML (2021). Arabidopsis thaliana zinc accumulation in leaf trichomes is correlated with zinc concentration in leaves. Sci. Rep, 11, 5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricachenevsky FK, Punshon T, Lee S, Oliveira B, Trenz TS, Maraschin F, Hindt MN, Danku J, Salt DE, Fett JP and Guerinot ML (2018) Elemental profiling of rice FOX lines leads to characterization of a new Zn plasma membrane transporter, OsZIP7. Front. Plant Sci 9, 865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A, Yamaji N, Mitani-Ueno N, Kashino M and Ma JF (2015) A node-localized transporter OsZIP3 is responsible for the preferential distribution of Zn to developing tissues in rice. Plant J. 84, 374–384. [DOI] [PubMed] [Google Scholar]
- Schuler M, Rellán-Álvarez R, Fink-Straube C, Abadía J and Bauer P (2012) Nicotianamine functions in the Phloem-based transport of iron to sink organs, in pollen development and pollen tube growth in Arabidopsis. Plant Cell. 24, 2380–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair SA and Krämer U (2012) The zinc homeostasis network of land plants. Biochim. Biophys. Acta, 1823, 1553–1567. [DOI] [PubMed] [Google Scholar]
- Sinclair SA, Senger T, Talke IN, Cobbett CS, Haydon MJ and Krämer U (2018) Systemic upregulation of MTP2‐ and HMA2‐mediated Zn partitioning to the shoot supplements local Zn deficiency responses. Plant Cell, 30, 2463–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair SA, Sherson SM, Jarvis R, Camakaris J and Cobbett CS (2007) The use of the zinc-fluorophore, Zinpyr-1, in the study of zinc homeostasis in Arabidopsis roots. New Phytol. 174, 39–45. [DOI] [PubMed] [Google Scholar]
- Spielmann J, Ahmadi H, Scheepers M, Weber M, Nitsche S, Carnol M, Bosman B, Kroymann J, Motte P, Clemens S and Hanikenne M (2020) The two copies of the zinc and cadmium ZIP6 transporter of Arabidopsis halleri have distinct effects on cadmium tolerance. Plant Cell Environ. 43, 2143–2157. [DOI] [PubMed] [Google Scholar]
- Tan L, Qu M, Zhu Y, Peng C, Wang J, Gao D and Chen C (2020) ZINC TRANSPORTER5 and ZINC TRANSPORTER9 function synergistically in zinc/cadmium uptake. Plant Physiol. 183, 1235–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Zhu Y, Fan T, Peng C, Wang J, Sun L and Chen C (2019) OsZIP7 functions in xylem loading in roots and inter-vascular transfer in nodes to deliver Zn/Cd to grain in rice. Biochem. Biophys. Res. Commun 512, 112–118. [DOI] [PubMed] [Google Scholar]
- Woody ST, Austin-Phillips S, Amasino RM and Krysan PJ (2007). The WiscDsLox T-DNA collection: an arabidopsis community resource generated by using an improved high-throughput T-DNA sequencing pipeline. J. Plant Res 120, 157–165. [DOI] [PubMed] [Google Scholar]
- Wu J, Zhao FJ, Ghandilyan A, Logoteta B, Guzman MO, Schat H, Wang X, Aarts MGM (2009) Identification and functional analysis of two ZIP metal transporters of the hyperaccumulator Thlaspi caerulescens. Plant Soil, 325, 79–95. [Google Scholar]
- Yang M, Li Y, Liu Z, Tian J, Liang L, Qiu Y, Wang G, Du Q, Cheng D, Cai H, Shi L, Xu F, Lian X (2020). A high activity zinc transporter OsZIP9 mediates zinc uptake in rice. Plant J. 103, 1695–1709. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Complementation of yeast metal uptake mutants.
Figure S2. In silico expression analysis of ZIP genes during development.
Figure S3. ZIP-GFP translational fusion proteins localize to the plasma membrane.
Figure S4. T-DNA mutants.
Figure S5. Seedling phenotypes of single T-DNA mutants grown under Zn-sufficient or Zn-deficient conditions.
Figure S6. Seedling phenotypes of double mutants grown under Zn-sufficient or Zn-deficient conditions.
Figure S7. Measurement of Fe levels of WT, triple and quadruple mutants.
Figure S8. Increased Zn resistance of triple and quadruple mutants.
Figure S9. Quantitative RT-PCR analysis of Zn homeostasis related genes at the seedling stage.
Figure S10. Growth morphology of WT, triple and quadruple mutants.
Figure S11. Characterization of seed phenotypes and metal contents in single and double mutants.
Figure S12. Synchrotron X-ray fluorescence (SXRF) elemental imaging of triple and quadruple mutants.
Table S1. List of primers used in this study.
Data Availability Statement
All relevant data can be found within the manuscript and its supporting materials.


