Abstract
Ubiquitin-specific processing proteases (UBPs) presently form the largest enzyme family in the ubiquitin system, characterized by a core region containing conserved motifs surrounded by divergent sequences, most commonly at the N-terminal end. The functions of these divergent sequences remain unclear. We identified two isoforms of a novel testis-specific UBP, UBP-t1 and UBP-t2, which contain identical core regions but distinct N termini, thereby permitting dissection of the functions of these two regions. Both isoforms were germ cell specific and developmentally regulated. Immunocytochemistry revealed that UBP-t1 was induced in step 16 to 19 spermatids while UBP-t2 was expressed in step 18 to 19 spermatids. Immunoelectron microscopy showed that UBP-t1 was found in the nucleus while UBP-t2 was extranuclear and was found in residual bodies. For the first time, we show that the differential subcellular localization was due to the distinct N-terminal sequences. When transfected into COS-7 cells, the core region was expressed throughout the cell but the UBP-t1 and UBP-t2 isoforms were concentrated in the nucleus and the perinuclear region, respectively. Fusions of each N-terminal end with green fluorescent protein yielded the same subcellular localization as the native proteins, indicating that the N-terminal ends were sufficient for determining differential localization. Interestingly, UBP-t2 colocalized with anti-γ-tubulin immunoreactivity, indicating that like several other components of the ubiquitin system, a deubiquitinating enzyme is associated with the centrosome. Regulated expression and alternative N termini can confer specificity of UBP function by restricting its temporal and spatial loci of action.
The ubiquitin system is a major cytosolic and nuclear pathway of proteolysis in all eukaryotic cells (reviewed in references 17 and 19). In this proteolytic pathway, ubiquitin, a 76-amino-acid peptide, is ligated covalently to specific intracellular proteins by a series of enzymes. The first step is catalyzed by ubiquitin-activating enzyme (14). The activated ubiquitin is then transferred from ubiquitin-activating enzyme to a specific cysteine residue of one of a family of ubiquitin-conjugating enzymes (13). Some ubiquitin-conjugating enzymes transfer ubiquitin to substrates directly, but most support ubiquitin conjugation to substrates by interaction with one of the ubiquitin protein ligases (39, 44, 46). The covalent attachment of ubiquitin to proteins commonly targets them for degradation by a multisubunit complex, the 26S proteasome (reviewed in reference 5), but may also serve other signaling functions such as endocytosis (18).
Interestingly, besides these enzymes involved in linking ubiquitin to proteins, there are a large number of deubiquitinating enzymes, which remove ubiquitin from covalent attachments to itself or other proteins (reviewed in reference 51). In fact, sequence analysis of the Saccharomyces cerevisiae genome indicates that there are more deubiquitinating enzymes than ubiquitin-conjugating enzymes (6, 51).
Analyses of these enzymes to date indicate that deubiquitinating enzymes can have a number of possible functions. First, these enzymes process the products of ubiquitin genes. Ubiquitin is encoded by two distinct classes of genes, neither of which encodes the monomer form of ubiquitin. One is a polyubiquitin gene encoding a linear polymer of ubiquitins that are linked through peptide bonds between the C-terminal Gly residue and the N-terminal Met residue of contiguous ubiquitin molecules (36). The other encodes a fusion protein consisting of ubiquitin and a ribosome subunit (10, 43). Thus, the generation of free monomeric ubiquitin from the linear polyubiquitin and from the ubiquitin fusion protein needs deubiquitinating enzymes which have peptidase activity (3, 9, 28). Second, deubiquitinating enzymes can also remove esters and amides from ubiquitin to produce free monomeric ubiquitin in the cell (40). Third, ubiquitin-dependent protein degradation requires attachment of at least one ubiquitin to a target protein via an isopeptide bond between the carboxy-terminal glycine of ubiquitin and the ɛ amino group of the side chain of a lysine residue on the target protein. In many cases, additional ubiquitin molecules can be successively conjugated via isopeptide bonds to form a polyubiquitin chain (17, 19). The polyubiquitin chain must be disassembled by deubiquitinating enzymes with isopeptidase activity during or directly after proteolysis, regenerating free monomeric ubiquitin (1, 15, 38). In this way, deubiquitinating enzymes can potentially stimulate protein degradation. Fourth, deubiquitinating enzymes can possibly counteract the effects of ubiquitin-conjugating enzyme- and ubiquitin protein ligase-mediated conjugation by competitively removing the polyubiquitin chain from the conjugated protein (20, 26). This might represent a means of preventing degradation by the proteasome or a means of negatively regulating ubiquitination-dependent functions other than protein degradation.
Deubiquitinating enzymes can be divided broadly on the basis of sequence homology into two classes, the ubiquitin-specific processing protease (UBP, also known as USP or type 2 UCH) and the ubiquitin C-terminal hydrolase (UCH, also known as type 1 UCH) (reviewed in references 6 and 51). It has been shown that UBPs are capable of cleaving the linear ubiquitin gene products (3) and disassembling branched polyubiquitin chains (9). They contain two very highly conserved motifs, the CYS and HIS boxes which presumably play important roles in catalysis (51). In contrast, UCH enzymes hydrolyze primarily carboxy-terminal esters and amides of ubiquitin (40) but may also cleave ubiquitin gene products (27). The active site of these UCH enzymes contains a catalytic triad consisting of cysteine, histidine, and aspartate and utilizes a chemical mechanism similar to that of papain (22, 23).
A very large number of deubiquitinating enzymes exist. Possibly, these deubiquitinating enzymes recognize distinct substrates and are therefore involved in specific cellular processes. For example, BAP1, which belongs to the UCH class, binds to the breast cancer tumor suppressor protein BRCA1, augmenting the growth-suppressive effects of BRCA1 (21). In addition, the yeast UBPs DOT4 and UBP3 interact with SIR4 and mediate an inhibition of silencing (24, 31). Although there is evidence of specificity of these deubiquitinating enzymes, their structure-function relationships remain poorly studied.
We have been studying the role of ubiquitin-dependent protein degradation during male germ cell development (42, 52, 53). Spermatogenesis is a complex developmental process in which undifferentiated stem cells become committed to the spermatid lineage, pass through the spermatocyte stages during which DNA replication and genetic recombination occur, and undergo the two divisions of meiosis to generate haploid round spermatids, which then differentiate into mature elongated spermatids (reviewed in reference 4). This last process involves condensation and removal of much of the cytoplasm as well as reorganization of the chromatin into a tighter structure. Some of this reorganization comes from condensation of cytoplasm into a residual body, which is then phagocytosed by Sertoli cells (45). However, ubiquitination appears also to be involved in this developmental process. We have previously demonstrated the activation of ubiquitin conjugation during spermatogenesis (42). Since deubiquitinating enzymes can play a role in modulating ubiquitination, we have begun to examine the role of these enzymes in this developmental process. To this end, we attempted to find UBP enzymes which were regulated during spermatogenesis. In this paper we report the identification of one UBP enzyme which is expressed in the testis as two isoforms with the same core region but distinct amino termini, allowing precise analysis of the function of the divergent termini. Intriguingly, we observed that the two isoforms were expressed at different stages of spermatid development and that the amino termini played a determining role in the subcellular distribution of the enzymes.
MATERIALS AND METHODS
Cloning of cDNAs encoding UBP-t1 and UBP-t2.
3′ rapid amplification of cDNA ends (RACE) (12) was used to identify a rat cDNA fragment with sequence similarity to the conserved CYS box identified for the UBP enzyme superfamily (55). cDNA was synthesized from rat testis RNA (2 μg) in a 20-μl reaction mixture using reverse transcriptase (Gibco-BRL Superscript Preamp kit), as specified by the supplier, and the oligonucleotide (20 pmol) 5′-GACTCGAGTCGACATCGAT17-3′ as a primer. The 3′-tailed cDNA (2 μl) was used as a template in a PCR in which the oligonucleotides 5′-GGIAA(T/C)ACITG(T/C)T(T/A)(T/C)(C/A/T)TGAA-3′, derived from CYS box residues of the UBP sequence (see Fig. 1) and 5′-GACTCGAGTCGACATCGA-3′ were used. Annealing was carried out at 45°C for 1 min, and extension was carried out at 72°C for 3 min, and 35 cycles were performed. A 1.5-kb DNA fragment was amplified, subcloned, and sequenced. Since the predicted protein sequence from the 1.5-kb fragment indicated marked similarity to other UBP sequences, the PCR-amplified DNA fragment was labeled with 32P and used as a probe to screen a rat testis cDNA library in the λzapII vector (Stratagene). An aliquot containing 106 recombinants was screened by transfer of plaques to nitrocellulose membranes and hybridization with the probe. Purified positive phage were grown and the pBluescript plasmid containing the insert was excised from the phage as specified by the manufacturer and sequenced.
FIG. 1.
UBP-t1 and UBP-t2 sequences. Translation of cDNA clones encoding this testis-deubiquitinating enzyme revealed the presence of two isoforms, UBP-t1 and UBP-t2. UBP-t1-N indicates the amino-terminal sequence of the short isoform, and UBP-t2-N indicates the amino-terminal sequence of the long isoform. UBP-Core indicates the common sequence of both isoforms. The asterisk indicates an active-site cysteine; bold residues indicate the Cys and His motifs; other regions conserved in UBP family are underlined; residues in the box indicate the peptide sequence encoded by the degenerate oligonucleotide used in the 3′ RACE reaction to identify this cDNA.
In vitro assay of recombinant UBP-t1 and UBP-t2 enzyme activities.
DNA fragments encompassing the coding region of UBP-t1 and UBP-t2 were amplified by PCR and subcloned into the pET-11d vector (Novagen) and sequenced. The pET-11d plasmids containing UBP-t1 and UBP-t2 were transformed individually into Escherichia coli BL21(DE3). Following induction with isopropyl-β-d-thiogalactopyranoside (IPTG) for 2 h at 30°C, the cells were harvested, sonicated, and centrifuged at 10,000 × g at 4°C. The supernatants containing the individual UBP-t1 and UBP-t2 enzymes were used in the assays. For the in vitro assays, 125I-labeled linear Nα-diubiquitin or 125I-labeled branched-chain Nɛ-triubiquitin was used as substrate. Aliquots (8 μl) of lysates containing UBP-t1 or UBP-t2 enzymes were incubated in a total volume of 25 μl containing 150 nM 125I-Nα-diubiquitin or 125I-Nɛ-triubiquitin, 50 mM Tris-HCl (pH 7.8), 1 mM EDTA, and 1 mM dithiothreitol at 37°C for 1 h. The reactions were stopped with Laemmli sample buffer plus 2-mercaptoethanol, and the products were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 20% acrylamide gels and detected by autoradiography.
Northern hybridization.
RNA was prepared from different rat tissues by the guanidium thiocyanate CsCl method. RNA blotting was performed by resolving 10 μg of RNA on 1% agarose gels containing formaldehyde, transferring the products to nylon membranes, and cross-linking them with UV light. The membranes were hybridized with 32P-labeled cDNA probes, washed, and then subjected to autoradiography.
Preparation of antibodies.
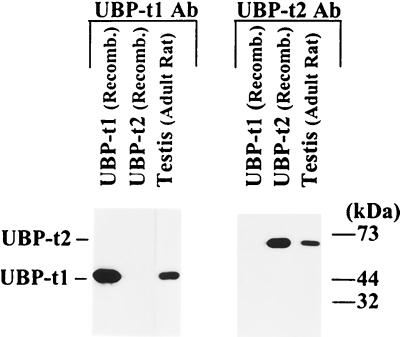
Antibodies specific for either UBP-t1 or UBP-t2 were prepared by immunizing rabbits with Freund's adjuvant mixed with proteins consisting of specific amino-terminal sequences of the isoforms (UBP-t1 residues 1 to 47, UBP-t2 residues 127 to 270) fused distal to maltose binding protein or a His6 tag, respectively. Antibodies were affinity purified by passing crude antiserum over Affi-Gel 10 columns coupled separately to glutathione S-transferase (GST) fusions of the same amino-terminal sequences. Use of these purified antibodies on immunoblots of samples of testis extract or bacterial extracts expressing UBP-t1 or UBP-t2 confirmed the isoform specificities of the antibodies.
Tissue fixation and immunohistochemical staining.
Adult Sprague-Dawley rats were anesthetized with sodium pentobarbital. The testes were fixed with Bouin's fixative by perfusion through the abdominal aorta. The testes were removed, dehydrated in graded ethanol, and embedded in paraffin. Paraffin sections (5 μm thick) were processed for immunostaining, incubated with anti-UBP-t1 or UBP-t2 N-terminus-specific antibody, and reacted with a peroxidase-conjugated anti-rabbit immunoglobulin G (IgG). As negative controls, the antibodies were preincubated with excess purified GST protein fused to the N termini of UCH-t1 or UCH-t2 before being immunostained.
Electron microscopic immunocytochemistry.
Rat testes were fixed by a 10-min perfusion through the abdominal aorta with 0.5% glutaraldehyde and 4% paraformaldehyde in 0.1 M phosphate buffer. Tissue was then embedded in Lowicryl K4M as previously described (35). Ultrathin sections were mounted on 300-mesh Formvar-coated nickel grids. Each grid containing numerous sections was floated on a drop of 20 mM Tris-buffered saline (TBS) (pH 7.4) containing 10% goat serum and then incubated for 1 h with anti-UBP-t1- or anti-UBP-t2-specific antibody in TBS. The grids were washed three times for 5 min each in TBS containing 0.5% Tween 20 and then incubated for 1 h with colloidal gold (diameter, 10 nm)-conjugated goat anti-rabbit antibody (diluted 1:20 in TBS). The sections were washed three times for 5 min each in TBS containing 0.05% Tween 20 and once in distilled water. They were counterstained with uranyl acetate in 30% ethanol for 2 min followed by lead citrate for 30 s. As a negative control, the anti-UBP-t1 or anti-UBP-t2 antibody was preincubated with excess purified GST–UBP-t1 or GST–UBP-t2 protein, respectively, for 2 h at 37°C before being used on the sections. Electron micrographs were obtained on a Philips 400 electron microscope.
Expression of green fluorescent protein (GFP) fusion in COS-7 cells.
Sequences encoding the UBP-t1 N-terminal extension (residues 1 to 49) or UBP-t2 N-terminal extension (residues 1 to 271) were amplified by PCR, subcloned separately into the pEGFP-N1 vector (Clontech), and sequenced. COS-7 cells on several round coverslips (12 mm in diameter) per 100-mm petri dish were seeded into Dulbecco's modified Eagle's medium (DMEM) containing 10% Nud serum. Either pEGFP-UBP-t1 N terminus, pEGFP-UBP-t2 N terminus, or pEGFP plasmids (5 μg) were transfected by the DEAE-dextran method. At 40 h later, cells on coverslips were washed with cold 1× phosphate-buffered saline (PBS), fixed with 3.8% paraformaldehyde, and mounted in Mowiol. The cells were examined and images were analyzed by confocal microscopy.
Expression of UBP-t1–His6-myc, UBP-t2–His6-myc, and UBP-core–His6-myc in COS-7 cells and immunostaining.
UBP-t1, UBP-t2 full-length sequences, or UBP-core sequence were amplified by PCR separately and subcloned into the pcDNA3.1 vector (Invitrogen) to express the protein with a His6-myc tag. These plasmids were transfected into COS-7 cells as described above for the pEGFP plasmids. At 40 h following transfection, the cells were fixed using 3.8% paraformaldehyde in PBS for 30 min at room temperature and washed with PBS. The cells were permeabilized with 0.5% Triton X-100 in PBS for 30 min at room temperature and then blocked with 10% goat serum in PBS for 1 h at room temperature. After being washed with PBS, the cells were incubated with anti-myc monoclonal antibody (1:200 dilution) (Sigma) at 4°C overnight. Following three washes with 0.05% Tween 20 in PBS, the cells were incubated with a 1:100 dilution of fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse secondary antibody. After three washes with 0.5% Tween 20 in PBS and one wash with water, coverslips were mounted on microscope slides using Mowiol and visualized with a confocal microscope (Zeiss LSM41c).
To check whether UBP-t2 colocalizes with γ-tubulin, cells transfected with pcDNA3.1 vector expressing full-length UBP-t2 were incubated with both anti-γ-tubulin monoclonal antibody (Sigma, 1:100 dilution) and anti-UBP-t2-amino-terminus polyclonal antibody (1:50 dilution) at 4°C overnight. Following three washes with 0.05% Tween-20 in PBS, the cells were incubated with trimethylrhodamine-5-isothiocyanate (TRITC)-conjugated goat anti-mouse secondary antibody (1:100 dilution) and FITC-conjugated goat anti-rabbit secondary antibody (1:100 dilution).
Nucleotide sequence accession numbers.
The sequences for UBP-t1 and UBP-t2 have been deposited in the GenBank database under accession numbers AF202453 and AF202454, respectively.
RESULTS
Identification of cDNAs encoding UBP-t1 and UBP-t2.
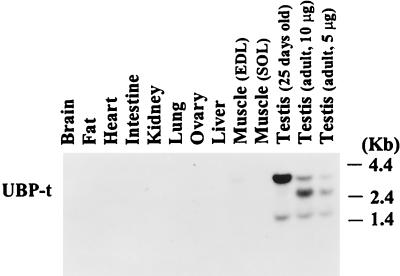
To identify ubiquitin-specific processing proteases expressed in rat testis, 3′ RACE was used in a PCR with a degenerate oligonucleotide encoding the sequence conserved in the CYS box of UBP enzymes (55) and testis RNA as the template. A 1.5-kb DNA fragment was amplified and sequenced. Comparison of the deduced protein sequence with entries in the GenBank database revealed the presence of a HIS box as well as other conserved motifs found in UBP enzymes (51) (Fig. 1). To check which tissues express this deubiquitinating enzyme, RNA from various rat tissues were analyzed by Northern blotting using the DNA fragment as a probe. High expression of this UBP mRNA was seen in the testis (Fig. 2). Two bands (3.8 and 1.8 kb) were seen in testes from 25-day-old rats, and three bands (3.8, 2.4, and 1.8 kb) were seen in testes from adult rats. To obtain full-length clones, a cDNA library derived from this tissue was screened. Two types of clones were observed. Both of them possessed the sequence at the 3′ end, identified by 3′ RACE. However, two different 5′-end sequences were found. One encoded 271 amino acids forming a long N-terminal extension, and the other encoded 49 amino acids forming a short N-terminal extension (Fig. 1). Several pieces of evidence confirmed that these two isoforms were indeed expressed in the testis. First, 5′ RACE (12) analysis confirmed the presence of these two types of RNA transcripts. In this method, cDNA was synthesized using an oligonucleotide primer from the common core region and then tagged at the 5′ end. The cDNA was then used as template in a PCR using an oligonucleotide primer which would recognize the sequence tag at the 5′ end and a primer also from the common core region. Two products were obtained which contained the same core region sequence but different 5′ ends corresponding to the two distinct cDNA clones (data not shown). Second, reverse transcription-PCR using primers from the 5′ ends of the two different forms separately with a primer from the 3′ end of the common core region yielded the two expected products. Finally, as described below, each of the N-terminal ends could be assigned to specific mRNA transcripts. Since the core region contains the CYS and HIS boxes as well as other motifs which are conserved in ubiquitin-specific processing proteases (51) and since this deubiquitinating enzyme is expressed predominantly in the testis, this enzyme was named ubiquitin-specific processing protease testis (UBP-t). We refer to the short isoform as UBP-t1 and the long isoform as UBP-t2.
FIG. 2.
Expression of UBP mRNA in rat tissues. RNA samples (10 μg) from the indicated tissues of adult (unless otherwise indicated) rats were resolved by electrophoresis on 1% agarose gels and transferred to a nylon membrane. Following hybridization with a 32P-labeled fragment from the UBP-t core region cDNA, the membrane was washed and exposed to film. EDL, extensor digitorum longus; SOL, soleus.
UBP-t1 and UBP-t2 have deubiquitinating enzyme activities.
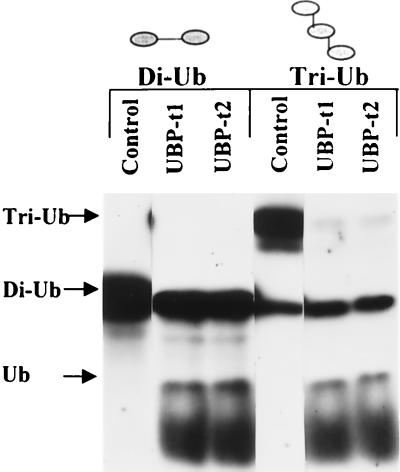
To determine whether UBP-t1 and UBP-t2 possess ubiquitin-specific peptidase or isopeptidase activities, UBP-t1 and UBP-t2 were individually expressed in E. coli. The bacterial lysates were tested against diubiquitin joined together as a peptide bond between the C terminus of one ubiquitin molecule and the N terminus of another ubiquitin molecule. They were also tested against a branched triubiquitin chain linked via isopeptide bonds between the ɛ-amino group of lysine 48 in one ubiquitin molecule and the C terminus of another ubiquitin molecule (Fig. 3). Both UBP-t1 and UBP-t2 can digest diubiquitin to produce monomeric ubiquitin and can digest triubiquitin to produce monomeric ubiquitin and a branched diubiquitin. The cleavages were not due to bacterial proteases, since lysates from uninduced bacteria had no such activities (Fig. 3). Furthermore, the cleavages were blocked by ubiquitin aldehyde, a specific inhibitor of many deubiquitinating enzymes (data not shown). Thus, UBP-t1 and UBP-t2 can hydrolyze both types of bonds.
FIG. 3.
UBP-t1 and UBP-t2 have deubiquitinating-enzyme activities. Enzymatic activities toward 125I-diubiquitin (Di-Ub) joined in a peptide bond and 125I-triubiquitin polymer (Tri-Ub) linked via lysine 48 isopeptide bonds were measured in lysates prepared from E. coli cells expressing either UBP-t1 or UBP-t2. Reaction products were separated by SDS-PAGE and detected by autoradiography. An equivalent volume of bacterial lysate not expressing UBP-t1 or UBP-t2 was used as the negative control.
Induction of UBP-t1 and UBP-t2 mRNA during postnatal development of the testis.
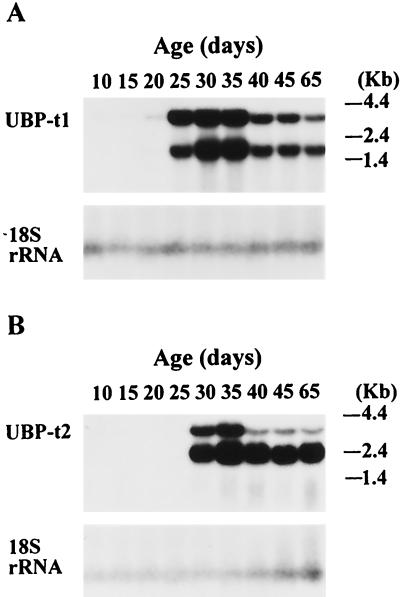
Since the testis undergoes important postnatal development during which progressively more mature germ cells appear, we examined whether UBP-t1 and UBP-t2 are also developmentally regulated. RNA from testes of rats of different ages were analyzed by Northern blotting with probes specific to each of the distinct 5′ regions of UBP-t1 and UBP-t2 cDNAs (Fig. 4). Two transcripts of UBP-t1, 1.8 and 3.8 kb long, were observed. UBP-t1 mRNA was undetectable in testis samples from 20-day-old rats but was markedly induced in samples from 25-day-old rats (Fig. 4A). Two transcripts of UBP-t2, 3.8 kb and 2.4 kb long, were found. UBP-t2 mRNA was undetectable in testis until 30 days of age, when its expression was dramatically increased (Fig. 4B). The later induction of UBP-t2 explains the earlier observations that an extra transcript was detected by Northern blotting in adult samples compared to samples from 25-day-old rats (Fig. 2).
FIG. 4.
Induction of UBP-t1 and UBP-t2 mRNAs during postnatal development of the testis. RNA (10 μg) from rat testes of different ages were electrophoresed on a 1% agarose gel and transferred to a nylon membrane. After hybridization with cDNA probes derived from the 5′ end of UBP-t1 (A) or UBP-t2 (B), the blots were subjected to autoradiography. Following removal of the probe, the membranes were rehybridized with a probe based on the 18S rRNA to evaluate loading and transfer of the samples to the membrane.
The UBP-t1 and UBP-t2 proteins are expressed in different elongated spermatids.
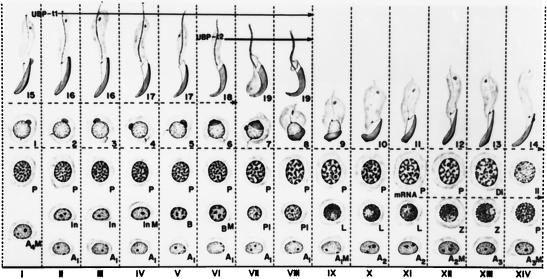
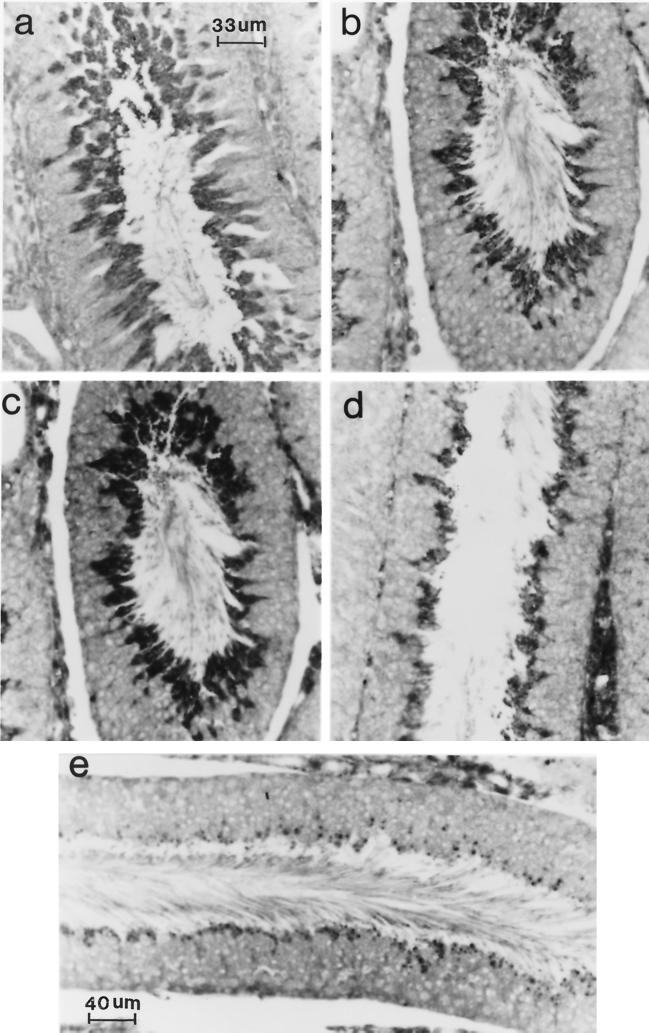
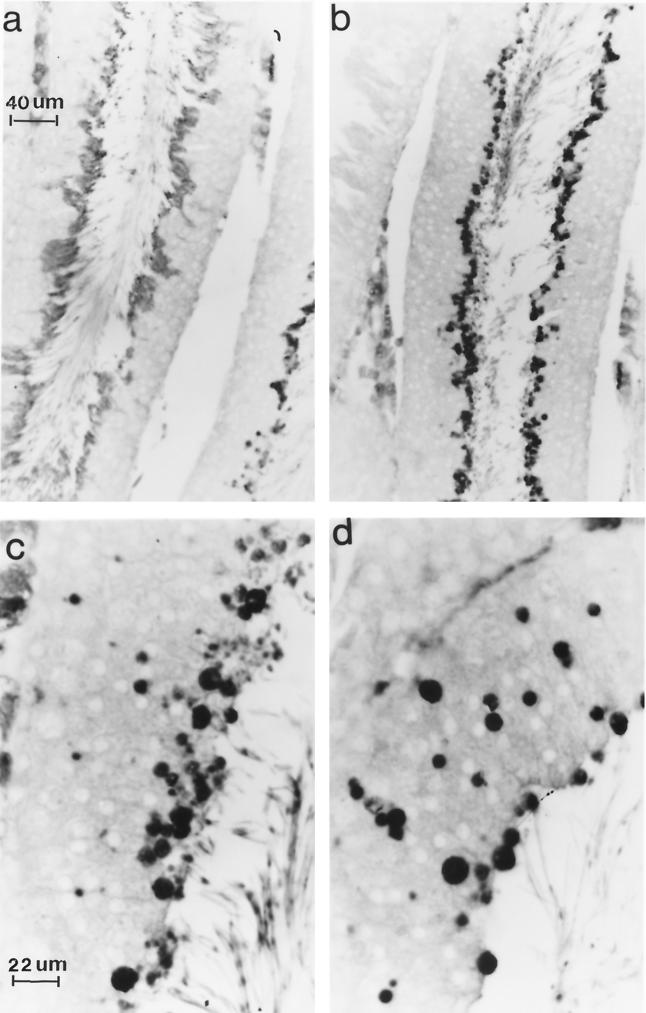
Since the expression of UBP-t appeared during puberty, this suggested that UBP-t1 and UBP-t2 might be expressed in the developing germ cells of the testis. Spermatid development in the rat testis has been well delineated morphologically into steps. In the rat, 14 stages of the cycle of the seminiferous epithelium have been characterized (Fig. 5) (29), each formed by several generations of germ cells, where a generation is defined as a group of cells at the same step of development. As a consequence, any stage will be contributed by one generation of spermatogonia resting directly on the basal lamina adjacent to the somatic Sertoli cell, one or two generations of spermatocytes usually located above the layer of spermatogonia, and one or two generations of spermatids found near the lumen. These specific cellular associations have constant duration. To identify the cells expressing UBP-t1 and UBP-t2 proteins, affinity-purified antibodies specific to each N terminus were produced, confirmed to be specific to each isoform (Fig. 6), and then used in immunocytochemical staining of testis sections. UBP-t1 protein was expressed earlier than UBP-t2 protein during spermatogenesis (Fig. 7 and 8). UBP-t1 was first detected in step 16 spermatids in stage III and was maintained at a high level until step 18 spermatids in stage VI. A decreased level of staining was observed in step 19 spermatids in stage VII, and the staining was even more suppressed in step 19 spermatids in stage VIII. UBP-t2 was first detected in step 18 spermatids in stage VI. Interestingly, it was also highly distributed in residual bodies. As negative controls, the antibodies were preincubated with excess purified GST protein fused to the N termini of UCH-t1 or UCH-t2 before immunostaining. These control immunostainings did not yield any reaction (data not shown).
FIG. 5.
The 14 cellular associations or stages observed in the seminiferous epithelium of the rat. Each vertical column, depicted by a Roman numeral, represents a cellular association and shows the various cell types present at that stage. The stage of the cycle is identified by means of 14 of the 19 steps of spermiogenesis (numbers 1 to 19). These steps are defined by the changes observed in the nucleus and acrosomal system in semithin sections (0.5 μm thick) stained with toluidine blue. The cellular associations or stages of the cycle succeed one another in time in any given area of the seminiferous epithelium according to the sequence indicated from left to right in the figure. Following stage XIV, stage I reappears, so that the sequence starts over again. The succession of the 14 stages makes up the cycle of the seminiferous epithelium. The mitotic divisions of the spermatogonia are indicated by the letter M. The germ cells present are A1, A2, A3, and A4 (type A spermatogonia); In (intermediate-type spermatogonia); B (type B spermatogonia); P1 (preleptotene primary spermatocytes); L (leptotene primary spermatocytes); Z (zygotene primary spermatocytes); P (pachytene primary spermatocytes); Di (diplotene primary spermatocytes); II (secondary spermatocytes); and 1 to 19 (steps of spermiogenesis). The dotted line indicates the cell types which express UBP-t mRNA, as determined by in situ hybridization (data not shown). Solid lines indicate cell types expressing UBP-t1 and UBP-t2 proteins. Modified from reference 7 and reproduced with permission of the publisher.
FIG. 6.
UBP-t1 and UBP-t2 antibodies are specific. Bacterial lysates containing the full length of UBP-t1 or UBP-t2 or the homogenates of 65-day-old rat testes were resolved by SDS-PAGE on 10% polyacrylamide gels and transferred to nitrocellulose membranes. The membranes were probed with anti-UBP-t1 N-terminal extension or UBP-t2 N-terminal extension antibody separately and then incubated with horseradish peroxidase-conjugated protein A and subjected to chemiluminescent detection.
FIG. 7.
UBP-t1 protein is expressed in step 16 to 19 spermatids. A cross section of rat seminiferous epithelium was immunostained with anti-UBP-t1 N-terminus-specific antibody and reacted with a peroxidase-conjugated anti-rabbit IgG. The reaction was visualized under a light microscope. (a) A cross section of seminiferous tubule at stage III of the cycle of the seminiferous epithelium demonstrates that the reaction is over step 16 spermatids. (b) A cross section of seminiferous tubule at stage V of the cycle of the seminiferous epithelium shows that the reaction is over step 17 spermatids. (c) A cross section of seminiferous tubule at stage VI of the cycle of the seminiferous epithelium shows that the reaction is over step 18 spermatids. (d) A cross section of seminiferous tubule at stage VII of the cycle of the seminiferous epithelium demonstrates that the reaction is over step 19 spermatids. (e) A cross section of seminiferous tubule at stage VIII of the cycle of the seminiferous epithelium shows that the reaction is over step 19 spermatids. Residual bodies were not stained but appeared dark because a blue filter used during photography did not eliminate the methylene blue counterstain. (Preincubation of antibody with GST fused to the UBP-t1 N terminus before hybridization with the sections resulted in the absence of any staining [data not shown]). Magnification in panels b, c, and d is the same as in panel e.
FIG. 8.
UBP-t2 protein is expressed in step 18 to 19 spermatids. A cross section of rat seminiferous epithelium was immunostained with anti-UBP-t2 N-terminus-specific antibody and reacted with a peroxidase-conjugated anti-rabbit IgG. The reaction was visualized under a light microscope. (a) A cross section of the seminiferous tubule at stage VI of the cycle of the seminiferous epithelium demonstrates that the reaction is over step 18 spermatids. (b) A cross section of the seminiferous tubule at stage VIII of the cycle of the seminiferous epithelium shows that the reaction is over step 19 spermatids. (c) A cross section of the seminiferous tubule at stage VIII of the cycle of the seminiferous epithelium shows that the reaction is over step 19 spermatids and residual bodies. (d) A cross section of the seminiferous tubule at stage VIII of the cycle of the seminiferous epithelium shows that the reaction is over residual bodies. (Preincubation of antibody with GST fused to the UBP-t2 N terminus before hybridization with the sections resulted in the absence of any staining [data not shown]). Magnification in panel b is the same as in panel a; magnification of panel d is the same as in panel c.
As described above, UBP-t1 and UBP-t2 mRNAs first appear in the testis on days 25 and 30, respectively. The most mature cells present in the testis at these ages are steps 3 and 8 round spermatids, respectively. This suggests that the mRNAs are synthesized in early round spermatids and stored and are used as templates to translate into protein only when the cells have been transformed into late elongated spermatids (Fig. 5). Indeed, in situ hybridization studies confirmed that UBP-t mRNAs were first found in late pachytene spermatocytes and were highly expressed in round spermatids (data not shown).
The N-terminal extensions of UBP-t1 and UBP-t2 regulate their subcellular distribution.
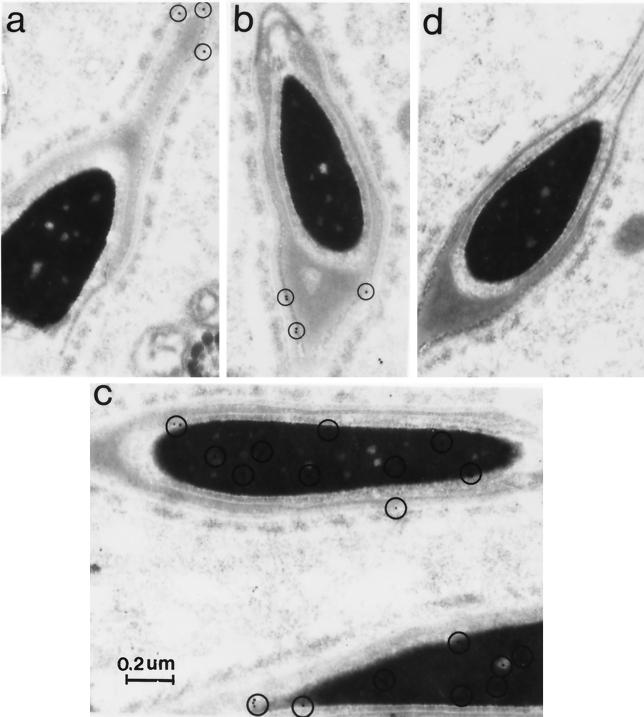
To examine the subcellular distribution of UBP-t1 and UBP-t2 in spermatids, anti-UBP-t1 and anti-UBP-t2 specific antibodies were used in electron microscopic immunocytochemical staining of rat testis tissue (Fig. 9). UBP-t1 staining was found mainly in the nucleus. In contrast, UBP-t2 staining was found in residual bodies (data not shown), as was seen with immunocytochemical staining under the light microscope (Fig. 8). However, within the late-elongating spermatids, UBP-t2 was found in the spermatid head but was absent from the nucleus and acrosome. Therefore, it is located perinuclearly in the thin area between the outer acrosomal membrane and the plasma membrane. Since UBP-t1 and UBP-t2 differ only in their amino-terminal extensions, these sequences must be involved in the differential distribution of these two isoforms in the spermatids.
FIG. 9.
UBP-t1 and UBP-t2 are differentially distributed in spermatids. Rat testis ultrathin sections were mounted on Formvar-coated nickel grids, stained with anti-UBP-t1 or anti-UBP-t2 specific antibody, and incubated with colloidal gold-conjugated goat anti-rabbit antibody. Sections were counterstained with uranyl acetate followed by lead citrate. As a negative control, the anti-UBP-t1 or anti-UBP-t2 antibody was preincubated with excess GST–UBP-t1 or GST–UBP-t2 protein for 2 h at 37°C prior to use on the sections. Electron micrographs were taken on a Philips 400 electron microscope. The circled small black dots represent positive staining. (a and b) Anti-UBP-t2-specific antibody staining. (c) Anti-UBP-t1-specific antibody staining. (d) Negative control.
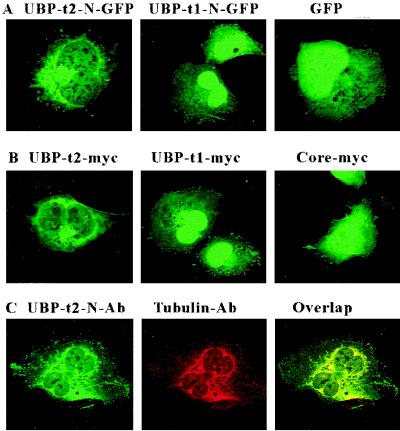
To test whether the amino-terminal extensions of UBP-t1 and UBP-t2 are sufficient for determining this distribution, they were fused separately to the amino-terminal ends of GFP and expressed in COS-7 cells (Fig. 10A). The N-terminal extension of UBP-t1 fused to GFP was located predominantly in the nucleus. However, the N-terminal extension of UBP-t2 fused to GFP protein was found mainly in a perinuclear location. Thus, these distribution patterns were consistent with the distribution of UBP-t1 and UBP-t2 in the spermatids. To exclude the possibility that GFP sequences were involved in determining this localization in COS-7 cells, the full-length UBP-t1 and UBP-t2 forms as well as the common UBP-core region were expressed in the cells separately as myc-tagged proteins. UBP-core was found in both cytoplasmic and nuclear compartments. However, the distribution patterns of UBP-t1 and UBP-t2 were exactly the same as the distribution patterns of the N-terminal extensions of UBP-t1 and UBP-t2 fused to the GFP protein (Fig. 10B). Thus, the N-terminal extensions of UBP determine the subcellular localization of the UBP.
FIG. 10.
N-terminal sequences of UBP-t1 and UBP-t2 determine subcellular localization. (A) COS-7 cells were transfected with plasmids expressing either the UBP-t1 N-terminal extension (UBP-t1-N-GFP) or the UBP-t2 N-terminal extension (UBP-t2-N-GFP) fused to GFP or the GFP alone. Cells were fixed and examined using a confocal fluorescence microscope. (B) COS-7 cells were transfected with plasmids expressing myc epitope tags of either UBP-t1, UBP-t2, or the core region alone. Following fixation, the cells were stained sequentially with anti-myc antibody and FITC-conjugated goat anti-mouse IgG antibody and examined with a confocal fluorescence microscope. (C) COS-7 cells were transfected with a plasmid expressing myc-tagged UBP-t2. Following fixation, the cells were stained with both anti-amino-terminal extension of UBP-t2 polyclonal antibody and anti-γ-tubulin monoclonal antibody followed by both FITC-conjugated goat anti-rabbit IgG antibody (green) and TRITC-conjugated goat anti-mouse IgG antibody (red). Confocal microscopy was used to detect anti-UBP-t2 fluorescence (UBP-t2-N-Ab) and anti-γ-tubulin fluorescence (Tubulin-Ab) and to analyze for colocalizing signals (Overlap, yellow).
Since UBP-t2, although extranuclear, was not evenly distributed in the cytoplasm, we suspected that it may be located to a specific organelle or structure. Therefore, we performed confocal microscopy using cells transfected with UBP-t2 and stained with anti-UBP-t2 N-terminal antibody and a panel of organelle-specific antibodies. Confocal microscopic analysis following staining with antibodies specific to the endoplasmic reticulum, Golgi, and actin did not yield significant overlap with the anti-UBP-t2 signal (data not shown). However, anti-γ-tubulin immunofluorescence colocalized tightly with that of the anti-UBP-t2 antibody (Fig. 10C), indicating that UBP-t2 may be associated with the centrosome and other γ-tubulin-containing structures (33, 47). Similar analysis using cells transfected with UBP-t1 and staining with anti-UBP-t1 N-terminal antibody did not produce any colocalization, confirming the dependence of the centrosomal localization on the N-terminal sequence of UBP-t2 (data not shown).
DISCUSSION
Members of the UBP family of deubiquitinating enzymes possess a core region containing six highly conserved sequence motifs including the presumptive active-site cysteine and histidine residues (51). Surrounding the core region are extensions, most commonly amino terminal, which contain divergent sequences. The functions of these divergent sequences remain unclear. We have reported here the characterization of a novel UBP. This enzyme is unique among UBPs described to date in having two isoforms characterized by identical core regions containing the conserved elements in the UBP family but divergent amino terminal sequences (Fig. 1). This provided a unique opportunity to critically examine the function of these divergent amino-terminal sequences and resulted in a number of intriguing findings. Although these findings are derived from studies of testis-specific enzymes, the derived concepts are probably broadly applicable to the UBP family.
We have shown, for the first time, that one of the functions of these divergent sequences is to determine the subcellular localization of the enzymes to specific organelles (Fig. 9 and 10). Within spermatids, the UBP-t1 isoform was localized primarily to the nucleus while the UBP-t2 isoform was found extranuclearly and in residual bodies extruded from maturing spermatids. When expressed in cultured cells without the amino-terminal extensions, the core region alone was distributed in both the nucleus and cytoplasm. In contrast, intact UBP-t1 was localized in the nucleus while UBP-t2 was localized primarily in a discrete perinuclear region of the cell. Finally, the two amino-terminal sequences fused to a heterologous protein were able to target the reporter protein to subcellular distributions identical to those of the native UBP-t isoforms. Together, these findings demonstrate that these amino-terminal sequences were both necessary and sufficient to mediate the differential localization of the isoforms.
Interestingly, the localization of UBP-t1 to the nucleus was determined by only a 49-residue extension to the core region. No apparent similarities to known nuclear localization signals were detectable in this sequence, suggesting the presence of either a novel signal or the transport of this protein into the nucleus bound to another protein bearing such a signal. Intriguingly, the perinuclear distribution of UBP-t2 matched perfectly the location of γ-tubulin. γ-Tubulin is found predominantly in the microtubular organizing center or centrosome but also occurs in other cytoplasmic structures, although the distribution may vary depending on the stage of the cell cycle (25, 32, 33). Centrosomes are present in spermatids but appear to have somewhat different functions. In elongated spermatids, the centrosome is reduced and segregated away from the centrioles and serves mainly as attachment points for the flagellar apparatus. The centrosomal apparatus is larger in less developed cells, but as the centrosome becomes reduced, it becomes sequestered in the residual body, as shown by localization of γ-tubulin to these structures (30). Thus, our localization of UBP-t2 to the residual body would also be consistent with association of this isoform with the centrosome or other γ-tubulin-containing structures during spermatogenesis.
Interestingly, a number of other components of the ubiquitin system have been recently localized to the centrosome. These include some components of the Skp1-Cullin-F box and anaphase-promoting complex families of ubiquitin-protein ligases, the 20S proteasome and its modulatory complexes, the 19S regulatory particle, and the PA700 activator and certain heat shock proteins (2, 11, 49, 50). Several of these components show a perinuclear distribution strikingly similar to what we have observed with UBP-t2 (50). Recently, centrosomes were reported to contain deubiquitinating activity (8), but no specific enzymes had been identified at this site. Two deubiquitinating enzymes are physically associated with the proteasome (26, 37) and so may indirectly be centrosome associated and responsible for the deubiquitinating activity. Recently, the amino terminus of one of them, DOA4, was shown to be able to confer binding to the proteasome, indicating that amino-terminal extensions can modulate protein-protein interactions (37). From these data, it has been proposed that the centrosome may be an intracellular locus of the ubiquitin-dependent proteolytic pathway. In such a scenario, centrosomal UBPs may be involved in the disassembly of branched multiubiquitin chains during or after proteasome-mediated destruction of the substrate. Whether UBP-t2 plays such a role in the centrosomes of the testis is unclear due to the atypical nature of this structure in the testis. Interestingly, part of the centrosome is segregated to the residual body, where UBP-t2 protein is also partly localized. Residual bodies are inclusions containing cytoplasmic contents that become phagocytosed by the adjacent Sertoli cells. Although believed to be ultimately hydrolyzed in lysosomes of the Sertoli cells, it is possible that the cytoplasmic contents include other enzymes of the ubiquitin/proteasome system. Therefore, UBP-t2 could be involved in supporting ubiquitin-mediated proteolysis in the residual bodies by regenerating free ubiquitin.
Our biochemical characterization (Fig. 3) to date is consistent with such a function, since the enzyme was capable of hydrolyzing branched polyubiquitin chains. Interestingly, both isoforms were capable of this and also of processing linear ubiquitin chains and ubiquitin fused to other peptides. Thus, UBP-t could also be involved in processing the products of translation of the polyubiquitin genes or ubiquitin-protein fusion genes. These data would suggest that these amino-terminal ends do not influence the ability of the enzyme to discriminate between these two distinct classes of substrates. In fact, the core domain itself is sufficient to carry out such cleavages (data not shown). However, our recent studies suggest that the amino-terminal sequences can cause subtle differences in preferences among these classes of substrates (unpublished data). It has also been hypothesized that the divergent sequences may be involved in substrate selectivity, but at the level of discrimination of the protein to which the ubiquitin is attached. This is a possibility that remains to be explored.
The other intriguing characteristic of these deubiquitinating enzymes was their highly precise regulation. Among all tissues examined, the enzyme was expressed predominantly in the testis (Fig. 2). Within the testis, expression appeared germ cell specific (Fig. 7 and 8). Furthermore, within the germ cell lineage, each isoform was induced at different stages of development (Fig. 4), with the shorter UBP-t1 isoform appearing earlier than the longer UBP-t2 isoform. Interestingly, UBP-t1 and UBP-t2 mRNAs were detected at days 25 and 30, respectively, coincident with the appearance of round spermatids. These distinct mRNAs probably arise from alternate splicing, and genomic cloning is under way to evaluate this. Although UBP-t mRNAs were definitely present in round spermatids, UBP-t1 and UBP-t2 proteins were detectable only in elongating spermatids (Fig. 7 and 8). The differential expression of the two isoforms was also preserved at the protein level in these cells, since UBP-t1 protein appeared in step 16 spermatids whereas UBP-t2 was first clearly expressed only in step 18 spermatids. This argues that the UBP-t mRNAs were probably sequestered following transcription and were released only for translation at a later stage. Such sequestration is well described during spermatid development (16, 34, 48), particularly with respect to the expression of protamines whose genes are transcribed in round spermatids (54) but translated only in elongating spermatids (41). Finally, as described above, within the elongating spermatids, the two isoforms were differentially localized in the cell (Fig. 9).
The UBP enzymes currently form the largest defined family in the ubiquitin system. To date, the rationale for such a large number of isoforms is unknown. Taken together, our data indicate that numerous isoforms can permit specific regulation of expression and subcellular localization of individual deubiquitinating enzymes. Even in the absence of substrate specificity intrinsic to the enzymes, such temporal and physical specificity of their expression can result in specificity of function by catalyzing the removal of ubiquitin from proteins located at specific sites under particular developmental, physiological, or pathological conditions. Since UBP-t1 and UBP-t2 appear to be expressed only in elongating spermatids for which there are no cell culture models, further characterization of function of these particular enzymes will require genetic manipulation in transgenic models.
ACKNOWLEDGMENTS
We are grateful to Keith Wilkinson and Cecile Pickart for supplying us with diubiquitin and triubiquitin, respectively.
This work was supported by grants from the Medical Research Council of Canada to S.S.W. and C.R.M. H.L. was the recipient of a studentship from the Royal Victoria Hospital Research Institute and Department of Medicine. P.H. and L.C. received fellowships from the Medical Research Council of Canada and the Canadian Diabetes Association, respectively. S.S.W. held a Medical Research Council of Canada Clinician Scientist Award.
REFERENCES
- 1.Amerik A, Swaminathan S, Krantz B A, Wilkinson K D, Hochstrasser M. In vivo disassembly of free polyubiquitin chains by yeast Ubp14 modulates rates of protein degradation by the proteasome. EMBO J. 1997;16:4826–4838. doi: 10.1093/emboj/16.16.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anton L C, Schubert U, Bacik I, Princiotta M F, Wearsch P A, Gibbs J, Day P M, Realini C, Rechsteiner M C, Bennink J R, Yewdell J W. Intracellular localization of proteasomal degradation of a viral antigen. J Cell Biol. 1999;146:113–124. doi: 10.1083/jcb.146.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker R T, Tobias J W, Varshavsky A. Ubiquitin-specific proteases of Saccharomyces cerevisiae. Cloning of UBP2 and UBP3, and functional analysis of the UBP gene family. J Biol Chem. 1992;267:23364–23375. [PubMed] [Google Scholar]
- 4.Clermont Y, Oko R, Hermo L. Cell biology of mammalian spermiogenesis. In: Desjardins C, editor. Cell and molecular biology of the testis. New York, N.Y: Oxford University Press; 1993. pp. 332–375. [Google Scholar]
- 5.Coux O, Tanaka K, Goldberg A L. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 6.D'Andrea A, Pellman D. Deubiquitinating enzymes: a new class of biological regulators. Crit Rev Biochem Mol Biol. 1998;33:337–352. doi: 10.1080/10409239891204251. [DOI] [PubMed] [Google Scholar]
- 7.Dym M, Clermont Y. Role of spermatogonia in the repair of the seminiferous epithelium following X-irradiation of the rat testis. Am J Anat. 1970;128:265–282. doi: 10.1002/aja.1001280302. [DOI] [PubMed] [Google Scholar]
- 8.Fabunmi R P, Wigley W C, Thomas P J, DeMartino G N. Activity and regulation of the centrosome-associated proteasome. J Biol Chem. 2000;275:409–413. doi: 10.1074/jbc.275.1.409. [DOI] [PubMed] [Google Scholar]
- 9.Falquet L, Paquet N, Frutiger S, Hughes G J, Hoang-Van K, Jaton J C. A human de-ubiquitinating enzyme with both isopeptidase and peptidase activities in vitro. FEBS Lett. 1995;359:73–77. doi: 10.1016/0014-5793(94)01451-6. [DOI] [PubMed] [Google Scholar]
- 10.Finley D, Bartel B, Varshavsky A. The tails of ubiquitin precursors are ribosomal proteins whose fusion to ubiquitin facilitates ribosome biogenesis. Nature. 1989;338:394–401. doi: 10.1038/338394a0. [DOI] [PubMed] [Google Scholar]
- 11.Freed E, Lacey K R, Huie P, Lyapina S A, Deshaies R J, Stearns T, Jackson P K. Components of an SCF ubiquitin ligase localize to the centrosome and regulate the centrosome duplication cycle. Genes Dev. 1999;13:2242–2257. doi: 10.1101/gad.13.17.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frohman M A, Dush M K, Martin G R. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haas A L, Siepmann T J. Pathways of ubiquitin conjugation. FASEB J. 1997;11:1257–1268. doi: 10.1096/fasebj.11.14.9409544. [DOI] [PubMed] [Google Scholar]
- 14.Haas A L, Warms J V, Hershko A, Rose I A. Ubiquitin-activating enzyme. Mechanism and role in protein-ubiquitin conjugation. J Biol Chem. 1982;257:2543–2548. [PubMed] [Google Scholar]
- 15.Hadari T, Warms J V, Rose I A, Hershko A. A ubiquitin C-terminal isopeptidase that acts on polyubiquitin chains. Role in protein degradation. J Biol Chem. 1992;267:719–727. [PubMed] [Google Scholar]
- 16.Hecht N B. Post-meiotic gene expression during spermatogenesis. Prog Clin Biol Res. 1988;267:291–313. [PubMed] [Google Scholar]
- 17.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 18.Hicke L. Ubiquitin-dependent internalization and down-regulation of plasma membrane proteins. FASEB J. 1997;11:1215–1226. doi: 10.1096/fasebj.11.14.9409540. [DOI] [PubMed] [Google Scholar]
- 19.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 20.Huang Y, Baker R T, Fischer-Vize J A. Control of cell fate by a deubiquitinating enzyme encoded by the fat facets gene. Science. 1995;270:1828–1831. doi: 10.1126/science.270.5243.1828. [DOI] [PubMed] [Google Scholar]
- 21.Jensen D E, Proctor M, Marquis S T, Gardner H P, Ha S I, Chodosh L A, Ishov A M, Tommerup N, Vissing H, Sekido Y, Minna J, Borodovsky A, Schultz D C, Wilkinson K D, Maul G G, Barlev N, Berger S L, Prendergast G C, Rauscher F J., III BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene. 1998;16:1097–1112. doi: 10.1038/sj.onc.1201861. [DOI] [PubMed] [Google Scholar]
- 22.Johnston S C, Larsen C N, Cook W J, Wilkinson K D, Hill C P. Crystal structure of a deubiquitinating enzyme (human UCH-L3) at 1.8 A resolution. EMBO J. 1997;16:3787–3796. doi: 10.1093/emboj/16.13.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnston S C, Riddle S M, Cohen R E, Hill C P. Structural basis for the specificity of ubiquitin C-terminal hydrolases. EMBO J. 1999;18:3877–3887. doi: 10.1093/emboj/18.14.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kahana A, Gottschling D E. DOT4 links silencing and cell growth in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:6608–6620. doi: 10.1128/mcb.19.10.6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khodjakov A, Rieder C L. The sudden recruitment of gamma-tubulin to the centrosome at the onset of mitosis and its dynamic exchange throughout the cell cycle, do not require microtubules. J Cell Biol. 1999;146:585–596. doi: 10.1083/jcb.146.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lam Y A, Xu W, DeMartino G N, Cohen R E. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature. 1997;385:737–740. doi: 10.1038/385737a0. [DOI] [PubMed] [Google Scholar]
- 27.Larsen C N, Krantz B A, Wilkinson K D. Substrate specificity of deubiquitinating enzymes: ubiquitin C-terminal hydrolases. Biochemistry. 1998;37:3358–3368. doi: 10.1021/bi972274d. [DOI] [PubMed] [Google Scholar]
- 28.Larsen C N, Price J S, Wilkinson K D. Substrate binding and catalysis by ubiquitin C-terminal hydrolases: identification of two active site residues. Biochemistry. 1996;35:6735–6744. doi: 10.1021/bi960099f. [DOI] [PubMed] [Google Scholar]
- 29.Leblond C P, Clermont Y. Definition of the stages of the cycle of the seminiferous epithelium in the rat. Ann N Y Acad Sci. 1952;55:548–573. doi: 10.1111/j.1749-6632.1952.tb26576.x. [DOI] [PubMed] [Google Scholar]
- 30.Manandhar G, Sutovsky P, Joshi H C, Stearns T, Schatten G. Centrosome reduction during mouse spermiogenesis. Dev Biol. 1998;203:424–434. doi: 10.1006/dbio.1998.8947. [DOI] [PubMed] [Google Scholar]
- 31.Moazed D, Johnson D. A deubiquitinating enzyme interacts with SIR4 and regulates silencing in S. cerevisiae. Cell. 1996;86:667–677. doi: 10.1016/s0092-8674(00)80139-7. [DOI] [PubMed] [Google Scholar]
- 32.Moudjou M, Bordes N, Paintrand M, Bornens M. gamma-Tubulin in mammalian cells: the centrosomal and the cytosolic forms. J Cell Sci. 1996;109:875–887. doi: 10.1242/jcs.109.4.875. [DOI] [PubMed] [Google Scholar]
- 33.Murphy S M, Urbani L, Stearns T. The mammalian gamma-tubulin complex contains homologues of the yeast spindle pole body components spc97p and spc98p. J Cell Biol. 1998;141:663–674. doi: 10.1083/jcb.141.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nayernia K, Adham I, Kremling H, Reim K, Schlicker M, Schluter G, Engel W. Stage and developmental specific gene expression during mammalian spermatogenesis. Int J Dev Biol. 1996;40:379–383. [PubMed] [Google Scholar]
- 35.Oko R, Hermo L, Chan P T, Fazel A, Bergeron J J. The cytoplasmic droplet of rat epididymal spermatozoa contains saccular elements with Golgi characteristics. J Cell Biol. 1993;123:809–821. doi: 10.1083/jcb.123.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ozkaynak E, Finley D, Varshavsky A. The yeast ubiquitin gene: head-to-tail repeats encoding a polyubiquitin precursor protein. Nature. 1984;312:663–666. doi: 10.1038/312663a0. [DOI] [PubMed] [Google Scholar]
- 37.Papa F R, Amerik A Y, Hochstrasser M. Interaction of the Doa4 deubiquitinating enzyme with the yeast 26S proteasome. Mol Biol Cell. 1999;10:741–756. doi: 10.1091/mbc.10.3.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Papa F R, Hochstrasser M. The yeast DOA4 gene encodes a deubiquitinating enzyme related to a product of the human tre-2 oncogene. Nature. 1993;366:313–319. doi: 10.1038/366313a0. [DOI] [PubMed] [Google Scholar]
- 39.Peters J M. SCF and APC: the Yin and Yang of cell cycle regulated proteolysis. Curr Opin Cell Biol. 1998;10:759–768. doi: 10.1016/s0955-0674(98)80119-1. [DOI] [PubMed] [Google Scholar]
- 40.Pickart C M, Rose I A. Ubiquitin carboxyl-terminal hydrolase acts on ubiquitin carboxyl-terminal amides. J Biol Chem. 1985;260:7903–7910. [PubMed] [Google Scholar]
- 41.Prigent Y, Muller S, Dadoune J P. Immunoelectron microscopical distribution of histones H2B and H3 and protamines during human spermiogenesis. Mol Hum Reprod. 1996;2:929–935. doi: 10.1093/molehr/2.12.929. [DOI] [PubMed] [Google Scholar]
- 42.Rajapurohitam V, Morales C R, El-Alfy M, Lefrancois S, Bedard N, Wing S S. Activation of a UBC4-dependent pathway of ubiquitin conjugation during postnatal development of the rat testis. Dev Biol. 1999;212:217–228. doi: 10.1006/dbio.1999.9342. [DOI] [PubMed] [Google Scholar]
- 43.Redman K L, Rechsteiner M. Identification of the long ubiquitin extension as ribosomal protein S27a. Nature. 1989;338:438–440. doi: 10.1038/338438a0. [DOI] [PubMed] [Google Scholar]
- 44.Reiss Y, Heller H, Hershko A. Binding sites of ubiquitin-protein ligase. Binding of ubiquitin-protein conjugates and of ubiquitin-carrier protein. J Biol Chem. 1989;264:10378–10383. [PubMed] [Google Scholar]
- 45.Russell L, Clermont Y. Anchoring device between Sertoli cells and late spermatids in rat seminiferous tubules. Anat Rec. 1976;185:259–278. doi: 10.1002/ar.1091850302. [DOI] [PubMed] [Google Scholar]
- 46.Scheffner M, Nuber U, Huibregtse J M. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature. 1995;373:81–83. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- 47.Stearns T, Evans L, Kirschner M. Gamma-tubulin is a highly conserved component of the centrosome. Cell. 1991;65:825–836. doi: 10.1016/0092-8674(91)90390-k. [DOI] [PubMed] [Google Scholar]
- 48.Steger K. Transcriptional and translational regulation of gene expression in haploid spermatids. Anat Embryol. 1999;199:471–487. doi: 10.1007/s004290050245. [DOI] [PubMed] [Google Scholar]
- 49.Tugendreich S, Tomkiel J, Earnshaw W, Hieter P. CDC27Hs colocalizes with CDC16Hs to the centrosome and mitotic spindle and is essential for the metaphase to anaphase transition. Cell. 1995;81:261–268. doi: 10.1016/0092-8674(95)90336-4. [DOI] [PubMed] [Google Scholar]
- 50.Wigley W C, Fabunmi R P, Lee M G, Marino C R, Muallem S, DeMartino G N, Thomas P J. Dynamic association of proteasomal machinery with the centrosome. J Cell Biol. 1999;145:481–490. doi: 10.1083/jcb.145.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilkinson K D. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J. 1997;11:1245–1256. doi: 10.1096/fasebj.11.14.9409543. [DOI] [PubMed] [Google Scholar]
- 52.Wing S S, Bedard N, Morales C, Hingamp P, Trasler J. A novel rat homolog of the Saccharomyces cerevisiae ubiquitin-conjugating enzymes UBC4 and UBC5 with distinct biochemical features is induced during spermatogenesis. Mol Cell Biol. 1996;16:4064–4072. doi: 10.1128/mcb.16.8.4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wing S S, Jain P. Molecular cloning, expression and characterization of a ubiquitin conjugation enzyme (E2(17)kB) highly expressed in rat testis. Biochem J. 1995;305:125–132. doi: 10.1042/bj3050125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wykes S M, Nelson J E, Visscher D W, Djakiew D, Krawetz S A. Coordinate expression of the PRM1, PRM2, and TNP2 multigene locus in human testis. DNA Cell Biol. 1995;14:155–161. doi: 10.1089/dna.1995.14.155. [DOI] [PubMed] [Google Scholar]
- 55.Zhu Y, Carroll M, Papa F R, Hochstrasser M, D'Andrea A D. DUB-1, a deubiquitinating enzyme with growth-suppressing activity. Proc Natl Acad Sci USA. 1996;93:3275–3279. doi: 10.1073/pnas.93.8.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]