Abstract
Background
Despite clinical success with anti-spike vaccines, the effectiveness of neutralizing antibodies and vaccines has been compromised by rapidly spreading SARS-CoV-2 variants. Viruses can hijack the glycosylation machinery of host cells to shield themselves from the host's immune response and attenuate antibody efficiency. However, it remains unclear if targeting glycosylation on viral spike protein can impair infectivity of SARS-CoV-2 and its variants.
Methods
We adopted flow cytometry, ELISA, and BioLayer interferometry approaches to assess binding of glycosylated or deglycosylated spike with ACE2. Viral entry was determined by luciferase, immunoblotting, and immunofluorescence assays. Genome-wide association study (GWAS) revealed a significant relationship between STT3A and COVID-19 severity. NF-κB/STT3A-regulated N-glycosylation was investigated by gene knockdown, chromatin immunoprecipitation, and promoter assay. We developed an antibody-drug conjugate (ADC) that couples non-neutralization anti-spike antibody with NGI-1 (4G10-ADC) to specifically target SARS-CoV-2-infected cells.
Findings
The receptor binding domain and three distinct SARS-CoV-2 surface N-glycosylation sites among 57,311 spike proteins retrieved from the NCBI-Virus-database are highly evolutionarily conserved (99.67%) and are involved in ACE2 interaction. STT3A is a key glycosyltransferase catalyzing spike glycosylation and is positively correlated with COVID-19 severity. We found that inhibiting STT3A using N-linked glycosylation inhibitor-1 (NGI-1) impaired SARS-CoV-2 infectivity and that of its variants [Alpha (B.1.1.7) and Beta (B.1.351)]. Most importantly, 4G10-ADC enters SARS-CoV-2-infected cells and NGI-1 is subsequently released to deglycosylate spike protein, thereby reinforcing the neutralizing abilities of antibodies, vaccines, or convalescent sera and reducing SARS-CoV-2 variant infectivity.
Interpretation
Our results indicate that targeting evolutionarily-conserved STT3A-mediated glycosylation via an ADC can exert profound impacts on SARS-CoV-2 variant infectivity. Thus, we have identified a novel deglycosylation method suitable for eradicating SARS-CoV-2 variant infection in vitro.
Funding
A full list of funding bodies that contributed to this study can be found in the Acknowledgements section
Keywords: SARS-CoV-2 variant, STT3A, NGI-1, ADC, Deglycosylation
Abbreviations: SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; ADC, Antibody-drug conjugate; STT3A/B, STT3 oligosaccharyltransferase complex catalytic subunit A/B; ACE2, angiotensin-converting enzyme 2; OST, oligosaccharyltransferase; eQTL, expression quantitative trait locus; GWAS, genome-wide association study; RBD, receptor-binding domain
Graphical Abstract
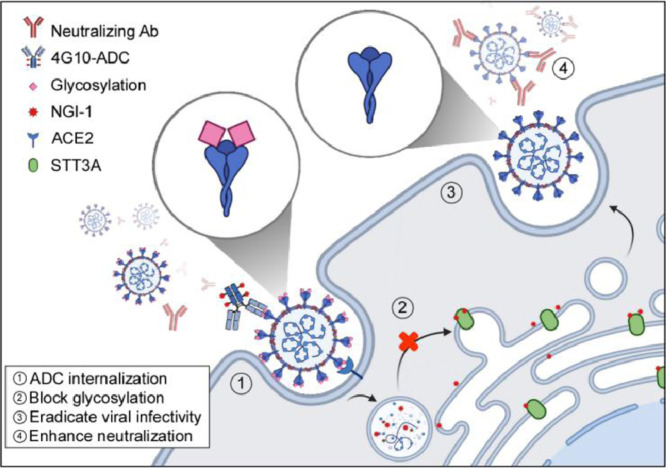
4G10-ADC targets and enters SARS-CoV-2 infected cells. NGI-1 is released from the ADC to block spike glycosylation. Virions with non-glycosylated spike display a higher antibody binding affinity and low infectivity.
Research in context.
Evidence before this study
Rapid spread of SARS-CoV-2 variants resistant to first-generation vaccines has become a global challenge. Glycosylation of viral proteins may protect against antibody recognition, enabling viruses to escape immune attack. Accordingly, deglycosylation may represent a potent antiviral mechanism, but an effective deglycosylation strategy has yet to be established.
Added value of this study
Using the NCBI-Virus-database, flow cytometry, ELISA, and BioLayer interferometry, we show that N-linked glycosylation sites on spike are highly conserved among SARS-CoV-2 and its variants and contribute to ACE2 interaction. We reveal STT3A as a key enzyme catalyzing spike glycosylation. NGI-1-mediated inhibition of STT3A reduced SARS-CoV-2 variant infectivity in vitro. We developed an antibody-drug conjugate (ADC) comprising a non-neutralizing anti-spike antibody coupled to NGI-1 (4G10-ADC).
Implications of all the available evidence
Our results show that removing the N-glycosylation of spike proteins enhances the binding affinity of neutralizing antibodies and reduces viral infectivity. We have developed an antibody-drug-conjugate to inhibit SARS-CoV-2 infection in vitro. Further testing in preclinical models is needed to determine in vivo efficacy against SARS-CoV-2 and its variants.
Alt-text: Unlabelled box
1. Introduction
The emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is posing a considerable threat to human health worldwide. SARS-CoV-2 is a positive-sense, single-stranded RNA virus that encodes four structural proteins [spike (S), envelope (E), nucleocapsid (N), and membrane protein (M)], as well as auxiliary proteins used for virus replication [1]. Binding of viral S protein to its host cell receptor, angiotensin-converting enzyme 2 (ACE2), initiates SARS-CoV and SARS-CoV-2 entry into host cells [2,3]. Several vaccines, such as mRNA-1273 and BNT162b2, have been developed to induce antibodies that neutralize S protein of the original Wuhan-HU-1 strain of SARS-CoV-2 (denoted WT hereafter) [4,5]. However, the neutralization ability of those vaccines is diminished or lost against the new SARS-CoV-2 variants Alpha (B.1.117) and Beta (B.1.351) that emerged in the United Kingdom and Republic of South Africa, respectively [5,6]. These two variants are more transmissible, cause more severe illness, and can evade vaccination-induced immune responses [7,8]. Effective strategies for impairing SARS-CoV-2 variant spread remain limited.
Protein glycosylation is integral to life, with diverse classes of glycans carrying out critical roles in determining protein structure, function, and stability [9,10]. N-glycans are a common [11], clinically relevant [12], and structurally diverse [13] set of molecules attached to secretory and membrane proteins. In mammals, N-linked glycosylation occurs in the lumen of the endoplasmic reticulum (ER) by a membrane-associated enzyme complex called oligosaccharyltransferase (OST). Two catalytic isoforms [STT3 oligosaccharyltransferase complex catalytic subunit A (STT3A) or STT3B] are assembled into two different OST isoforms. When the STT3A isoform enters the lumen of the ER, it is mainly responsible for co-translational N-linked glycosylation of nascent polypeptides. The STT3B isoform is required for effective co-translational and post-translocational glycosylation of acceptor sites adjacent to the secreted protein N-terminal signal sequence [14). Viruses can hijack the glycosylation machinery in the ER-Golgi system of host cells, forming offspring virions that display the characteristic glycosylation pattern of the host cell [15]. Importantly, glycosylation may enhance the ability of a virus to evade host immune surveillance [15] by masking them from antibody recognition [16]. Ablation of N-glycosylation by the OST inhibitor NGI-1 has been shown to disrupt virion formation and effectively reduced Lassa, herpes simplex, and SARS-CoV-2 virus infectivity [17], [18], [19].
Antibody-drug conjugates (ADCs) are an emerging class of cancer therapeutics that can deliver highly effective drugs specifically to malignant cells. ADCs target specific antigens, thereby lowering off-target toxicity. To date, nine ADCs have been approved by the U.S. Food and Drug Administration (FDA), and more than 80 others are currently undergoing clinical studies [20]. In this study, we demonstrate that N-linked glycosylation of viral S protein is highly conserved among SARS-CoV-2 variants. We identify STT3A as a key enzyme triggering glycosylation of S protein, thereby enhancing virus infectivity. Deglycosylation of S protein by means of an STT3A inhibitor exerted potent antiviral activity that limited SARS-CoV-2 variant infectivity. Our study unveils an effective strategy for eradicating Alpha and Beta variant SARS-CoV-2 infection and could potentially be adapted to treat other aggressive variants.
2. Methods
2.1. Plasmids
The gene encoding amino acids 1-1273 of the SARS-CoV-2 spike glycoprotein (Gene ID: MN908947) was cloned into the pMD.G vector (Lentivirus package plasmid, National RNAi Core Facility, Academia Sinica, Taipei, Taiwan) to generate pMD.G-spike construct for transient transfection. The gene encoding human angiotensin-converting enzyme 2 (ACE2, Gene ID: NM_021804) was cloned into the pLAS2w-pPuro lentiviral vector (National RNAi Core Facility) to generate pLAS-ACE2-Flag construct for establishing ACE2 expression cell lines. The human STT3A (−513/−1) promoter was amplified by PCR and cloned into the pGL3-basic vector (E1751, Promega, Madison, WI, USA). The deleted NF-κB binding motif construct had a deletion at bases −363CGTAGTTTCC−353 in the STT3A promoter. pBabe-Puro-IκBalpha-mut (RRID:Addgene_15291; Addgene plasmid #15291, Watertown, MA, USA) [21] is a dominant-negative mutant NF-κB inhibitor (IκBαSR) and inhibits NF-κB activity. The coding region of IκBαSR was cloned into the pcDNA3.1 vector (v79020, Invitrogen, Waltham, Massachusetts, USA). pRL-TK vector (E2241, Promega).
2.2. Antibodies, convalescent sera, and chemicals
The following antibodies were used: STT3A antibody (RRID:AB_2877818; 12034-1-AP), STT3B antibody (RRID:AB_2198046; 15323-1-AP), ACE2 antibody (RRID:AB_2882052; 66699-1-Ig), β-Tubulin antibody (RRID:AB_2881629; 66240-1-Ig), and GAPDH antibody (60004-1-Ig), were purchased from Proteintech (Chicago, IL, USA). 2019-nCoV Spike antibody (RRID:AB_2887482; GTX135356) was purchased from GeneTex (Irvine, CA, USA), N protein antibody (a gift from Dr. An-Suei Yang's lab, Genomics Research Center, Academia Sinica, Taiwan), NF-κB antibody (RRID:AB_2793231; 39369) was purchased from Thermo Fisher Scientific Inc. (Waltham, MA, USA), IκBα antibody (RRID:AB_733068; ab32518), HRP-conjugated Goat anti-Mouse IgG (RRID:AB_955439; ab6789), and HRP-conjugated Goat anti-Rabbit IgG (RRID:AB_955447; ab6721) were purchased from Abcam (Cambridge, MA, USA), Alexa Fluor® 488 AffiniPure Goat Anti-Human IgG (RRID:AB_2337831; 109-545-003) was purchased from Jackson ImmunoResearch (West Grove, PA, USA). Spike monoclonal antibody 4G10 (MAB22854-M13, Abnova, Taipei, Taiwan). APC-conjugated anti-His antibody (RRID:AB_2715818; 362605) and PE-conjugated mouse anti-His Abs (RRID:AB_2563634; 362603) were purchased from Biolegend (San Diego, CA, USA). Serum specimens from COVID-19 convalescent patients were isolated from COVID-19 patient in Taiwan (#1, #2, and #3; gifts from Dr. Yi-Ling Lin's lab, Institute of Biomedical Sciences, Academia Sinica, Taipei, Taiwan).
The following chemicals and inhibitors were used: Tunicamycin (TM; T7765), Benzyl 2-acetamido-2-deoxy-α-D-galactopyranoside (BADG; B4894), NGI-1 (SML1620), Thiamet G (SML0244), 2-Deoxy-D-glucose (2-DG; D8375), and Concanavalin A (ConA; L6397) were purchased from Sigma (St. Louis, MO, USA). Swainsonine (SW; 16860), PUGNAc (17151), and Castanospermine (CSA; 11313) were purchased from Cayman Chemical Company (Ann Arbor, MI, USA); PNGase F (P0704S) and Endo H (P0702S) were purchased from New England Biolabs (Ipswich, MA, USA). 7-amino-actinomycin D (7-AAD; 420404) was purchased from Biolegend.
2.3. Western blotting
Place the cell culture dish on ice and wash the cells with cold PBS. The proteins of cells are lysed with RIPA lysis buffer (50 mM Tris-HCl, pH 7.6, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate) with protease (04693132001, Merck KGaA, Darmstadt, Germany). Proteins of interest were studied by hybridizing with corresponding antibodies such as GAPDH antibody, 2019-nCoV Spike antibody, N protein antibody, NF-κB antibody, IκBα antibody, STT3A antibody, STT3B antibody. The protein expression was quantified by the band density using the ImageJ software (https://imagej.nih.gov/ij/).
2.4. Cell lines and generation of stable cells using lentiviral infection
HEK293T (RRID:CVCL_YA60) and A549 cells (RRID:CVCL_0023) were obtained from American Type Culture Collection (ATCC), and Vero E6 cell (RRID:CVCL_0574) was a gift from Dr. Yi-Ling Lin's Lab. All cells were confirmed by STR genotyping. These cells were grown in DMEM/F12 with 10% fetal bovine serum (FBS). To generate stably-expressed ACE2 cell lines, HEK293T and A549 cells were transfected with pLAS2w-ACE2, dR8.2 (RRID:Addgene_8455), and pCMV-VSV-G (RRID:Addgene_8454) plasmids via calcium phosphate transfection method to produce lentivirus containing ACE2 transgene. After 48 h post-transfection, the medium containing ACE2 lentivirus was collected and filtered through 0.45-μm filters. The pLKO.1-based lentivirus shRNA vectors for STT3A intRNAi Core Facility in Academia Sinica. The culture medium of A549 cells was then replaced by the medium with ACE2 lentivirus and 10 μg/mL polybrene (H9268, Sigma) and then centrifuged at 400 x g for 2 h. After 16 h of viral transduction, the medium was refreshed, and the transduced cells were selected with 2 μg/mL puromycin (ant-pr-5, InvivoGen, Occitanie, France) for a week.
2.5. Fluorescence-activated Cell Sorting (FACS) binding assay
For measuring the binding capacity of wild-type or deglycosylated SARS-CoV-2 S protein with ACE2, HEK293T cells were transfected with pMD.G-spike using lipofectamine 2000 (Life Technologies, Carlsbad, CA, USA) and treated with 20 mM 2-DG, 10 μM NGI-1, 10 μM Thiamet G or remained non-treated for 2 days. Cells were then dissociated from the culture plate and incubated with 2 μg of ACE2 (615)-His protein (Dr. Mi-Hua Tao's Lab, Institute of Biomedical Sciences, Academia Sinica, Taipei, Taiwan) in staining buffer (1% FBS in DPBS) on ice for 1 h. After washing out the non-binding ACE2 (615)-His protein, cells were then incubated with 0.5 μg APC-conjugated anti-His antibody in staining buffer on ice for 30 min. 7-amino-actinomycin D (7-AAD) was used to exclude the non-viable cells. The stained cells were analyzed using LSR II cytometer (RRID:SCR_002159; BD Biosciences, San Jose, CA, USA), and data were processed using FlowJo V10 software (RRID:SCR_008520).
2.6. Enzyme-linked immunosorbent assay (ELISA)
SARS-CoV-2 spike-ECD protein (Z03481, GenScript, Piscataway, NJ, USA) was coated on ELISA plate at 4°C overnight and then treated with PNGase F at 37°C overnight. After ECD was blocked with 3% BSA in phosphate-buffered saline (PBS) at room temperature (RT) for 2 h, serially diluted ACE2 (a gift from Dr. Tao's lab, Institute of Biomedical Sciences, Academia Sinica, Taipei, Taiwan) was added at RT for 2 h and followed by three times washing with PBS containing 0.05% Tween 20 (PBST). ACE2 was captured by antibody at RT for 2 h, and then incubated with HRP-conjugated anti-mouse IgG (1:5000) at RT for 1 h. After three times washing, the reaction was colored by substrate 3,3’,5,5’-Tetramethylbenzidine (TMB) (T0440, Sigma), and stopped by 2 N HCl. Finally, the absorbance at 450 nm was detected by an ELISA reader, VERSAmax (Molecular Devices, San Jose, CA, USA).
2.7. Glycoprotein staining and Coomassie blue staining
After SARS-CoV-2 spike protein treated with PNGase F or Endo H for 3 h, the proteins were analyzed by SDS-PAGE with the positive control (Horseradish Peroxidase), and negative control (Soybean Trypsin Inhibitor) in Pierce™ Glycoprotein Staining Kit (24562, Thermo Fisher Scientific Inc.). Then gel was oxidized, stained with 25 mL glycoprotein stain solution, and reduced following the manufacture's protocol; Bio-Safe™ Coomassie Stain (1610786, Bio-Rad, Hercules, CA, USA) was used to stain total protein, all gel images were captured by the scanner.
2.8. Amino acid sequence alignment
Thirteen full-length virus S protein sequences relating to SARS-CoV-2 were obtained from the NCBI Genbank database. In Homo sapiens, SARS-CoV-2 (accession: QIA98606 Taiwan, QHO62877.1 USA, YP 009724390 China, EPI_ISL_741243 Alpha variant (lineage B.1.1.7), and EPI_ISL_736967 Beta variant (lineage B.1.351) and SARS-CoV (accession: accession: AAP13441.1 Urbani, AAP13567.1 CUHK-W1, AAP41037.1 Tor2, and AAS00003.1 GZ02). The sequence alignment, including 13 NXT motifs in SARS-CoV-2 were conducted by QIAGEN CLC main workbench 20.0.4 (https://digitalinsights.qiagen.com).
2.9. Structure modeling
The structure of interaction between SARS-CoV-2 and ACE2 was downloaded from Protein Data Bank (PDB). The PDB IDs were 7A95. The structure figures were modified using the PyMOL Molecular Graphics System, Version 2.3.4 (Schrödinger, LLC, https://pymol.org/2/).
2.10. Identification of N-glycopeptide
Purified proteins were reduced with 10 mM dithiothreitol at 60°C for 1 h, alkylated with 50 mM iodoacetamide in 25 mM ammonium bicarbonate buffer for 45 minutes in the dark at room temperature. Trypsin enzyme was used to digest substrate at an enzyme-to-substrate ratio of 1:50 at 37°C overnight. The digested products were then diluted with formic acid to a final concentration with 0.1% and further cleaned up by ZipTip C18 (Millipore) before LC-MS/MS analysis. The peptide mixture was analyzed by nanospray LC-MS/MS on a Tribrid Mass Spectrometer (RRID:SCR_020562; Orbitrap Fusion Lumos, Thermo Fisher Scientific Inc.) coupled to an Easy-nLC 1200 System (Thermo Fisher Scientific Inc.). Peptide mixtures were loaded onto an Acclaim PepMap RSLC 25 cm × 75 μm i.d. column (Dionex) and separated at a flow rate of 300 nL/min using a gradient of 6% to 50% solvent B (100% acetonitrile with 0.1% formic acid) in 90 min. Solvent A was 0.1% formic acid in water. An HCD-pd-EThcD workflow was employed to additionally trigger EThcD events upon detecting the HCD productions at m/z 204.0867, 138.0545, and 366.1396. The parameters used for spectra acquisition were: top speed mode with 3s cycle time; FTMS: scan range (m/z) = 400-1800; resolution = 120K; HCD collision energy (%) = 28; resolution = 30 K. For EThcD, the calibrated charge-dependent ETD parameter was used and supplemental activation was allowed with a 15% SA collision energy. For glycopeptide identification, the MS raw data were searched using Byonic algorithm (v2.16.11) with the following search parameters: peptide tolerance = 20 ppm; fragment tolerance = 0.05 Da; missed cleavages = 2; modifications: carbamidomethyl cysteine (fixed), methionine oxidation (common 2), deamidation at NQ (common 1). In addition, the MS data were further searched using pGlyco algorithm (v2.2) to retrieve glycopeptides information using the parameters the same as Byonic search. The mass spectrometry proteomics data have been deposited to MassIVE database and the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier MSV000087686 and PXD026852 [22,23].
2.11. Chromatin immunoprecipitation (ChIP) assays
The chromatin samples were prepared from A549-ACE2 cells with a ChIP-IT Express kit (Active Motif, Carlsbad, CA). The chromatin samples (50 μL) were incubated with NF-κB (39369, Active Motif) (3 μg) for 12 h at 4 °C. Primers used to detect the STT3A promoter regions as follows:
STT3A forward: 5’-TCAGCGTAGTTTCCGCTTCT-3’. STT3A reverse: 5’-GAGGAGTG ACGTGTCGGTTT-3’
2.11. Promoter assay
X-tremeGENE HP DNA Transfection Reagent (XTGHP-RO, Roche, Mannheim, Germany) was used to co-transfect an expression vector (pcDNA3.1) encoding IκBαSR, a vector with the STT3 promoter construct, and pRL-TK vector that served as an internal control in A549-ACE2 cells for 1d. Luciferase activity was performed by utilizing the Dual-Luciferase Reporter Assay System kit (E1980, Promega), and detected by GloMax® 96 Microplate Luminometer (RRID:SCR_018614; Promega).
2.12. Pseudovirus neutralization assay
Neutralization assay for mAbs were performed using a SARS-CoV-2, Alpha and Beta variants spike-pseudotyped lentivirus expressing luciferase purchased from National RNAi Core Facility. In brief, mAbs were four-fold serially diluted in culture medium starting at 20 μg/mL, and 50 μL was mixed with 10 μL (1000 TCID50) SARS-CoV-2 pseudovirus for 30 min. The mixture was then added to HEK293T-ACE2 cells and incubated for 48 h, after which the cells were collected and lysed. Upon pseudovirus infection for 48 h, luciferase activity was determined according to the manufacturer's instruction (E1500, Promega). The percentage of infectivity was calculated as a ratio of luciferase readout in the presence of mAbs normalized to luciferase readout in the absence of mAb. The half-maximal inhibitory concentrations (IC50) were determined using 4-parameter logistic regression (GraphPad Prism version 8; RRID:SCR_002798). Cell viability was assessed by the MTT method.
2.13. Virus isolation and infection
The SARS-CoV-2 strain used in this research was isolated from a COVID-19 patient in Taiwan (TCDC#4), and passaged on Vero E6 cells, which were grown in MEM supplemented with 2% FBS and incubated at 37 °C with 5 % CO2. For the in vitro infection study, target cells were infected at a MOI of 0.1 (2000 pfu/well) with SARS-CoV-2 for 24 – 48 h. The cells were then fixed with 10% formaldehyde and permeabilized with 0.5% Triton X-100. All procedures followed the laboratory biosafety guidelines of the Taiwan CDC and were conducted in a Biosafety Level-3 (BSL-3) facility in the Institute of Biomedical Sciences, Academia Sinica, Taipei, Taiwan.
2.14. Real-time quantitative PCR (RT-qPCR)
Total RNA of A549 and A549-ACE2 cells were isolated using Quick-RNA Miniprep Kit (R1055, ZYMO Research, Irvine, CA, USA). The cDNA was prepared using ToolsQuant II Fast RT Kit (KRT-BA06-2, Biotools, Taipei, Taiwan) according to the manufacture's protocol with 1 μg of total RNA. All RT-qPCR reactions were performed in a 10 μL mixture containing 1X iQ™ SYBR® Green supermix (1708880, Bio-Rad), 0.5 μmol/L of each primer, and 100 ng of cDNA template. Primers used are as follows:
STT3A forward: 5’-TACCCTTTGGGACGAATCATTGG-3’.
STT3A reverse: 5’-GGTAAGGTGGTACGTGACGATGG-3’. The RT-qPCR result was acquired by CFX Connect Real-Time PCR Detection System (RRID:SCR_018064; Bio-Rad).
2.15. Immunofluorescence assay
Vero E6 cells were fixed with 4% buffered formalin for 30 min at RT. After washing with PBST, cells or spheroids were blocked with 2% donkey serum in PBS for 30 min at RT. Cells or spheroids were then incubated with N protein antibody (1:1000) at 4°C overnight, and followed by Alexa Fluor® 488 AffiniPure Goat Anti-Human IgG secondary antibody (1:1000) for 1 h at RT. Nuclei were stained with 4’,6-diamidino-2-phenylindole (DAPI) for 5 min in the dark. Images were taken by a Zeiss confocal microscope (RRID:SCR_017377; LSM 700 Stage, Zeiss, Oberkochen, Germany).
2.16. Mice and immunohistochemical staining
AAV6-mock or AAV6/CB-hACE2 was produced by AAV core facility in Academia Sinica. 8-10 weeks old C57BL/6J mice (RRID:IMSR_JAX:000664) were anesthetized by intraperitoneal injection of a mixture of Atropine (0.4 mg/ml)/Ketamine (20 mg/ml)/Xylazine (0.4%). Mice were then intratracheally injected with 3 × 1011 vg of AAV6-mock or AAV6/hACE2 in 100 μL saline as previously described [24,25]. Lung tissue sections were gifts from Dr. Tao's lab, Institute of Biomedical Sciences, Academia Sinica, Taipei, Taiwan and were deparaffinized with xylene, rehydrated in graded ethanol, and rinsed in distilled water. Antigen retrieval was performed with 1X Citrate Buffer (C9999, Sigma) at 95°C for 30 min. Prior to incubation with primary antibodies, sections were incubated with endogenous peroxidase for 30 min and blocked with 2% donkey serum for 30 min at RT. Sections were then incubated with anti-STT3A or anti-STT3B primary antibodies (1:100) at 4°C overnight, and ImmPRESS® horse anti-rabbit IgG polymer kit ((RRID:AB_2336536; MP-5401, Vector Laboratories, Burlingame, CA, USA) for 30 min at RT. Sections were then detected with ImmPACT® Vector® Red (RRID:AB_2336524; SK-5105, Vector Laboratories). Following staining, slides were counterstained in hematoxylin, dehydrated in graded ethanol, cleared in xylene, and coverslipped. Images were taken by Olympus BX51 microscope (RRID:SCR_018949; Olympus, Shinjuku, Tokyo, Japan).
2.17. Generation of anti-SARS-CoV-2 monoclonal antibodies (mAbs)
C57BL/6 mice were sequentially immunized in 2 weeks intervals with purified SARS-CoV-2 S1 and S2 proteins. Antigens were injected at 30 μg/mouse using Alum Adjuvant (Thermo Fisher Scientific Inc.) freshly prepared according to the manufacturer's instruction for first and boosting injection. Fourteen days after the last injection, spleen and lymph nodes are harvested, and hybridomas are made by a standard method using a myeloma cell line (RRID:CVCL_2199; ATCC CRL-1581) as a fusion partner. Hybridomas were screened in antigen-specific ELISA, and those selected for further development were subcloned and produced on a small scale (100 mL of medium). For this purpose, hybridomas are cultured in serum- and protein-free medium for hybridoma culturing (PFHM-II, Thermo Fisher Scientific Inc.) with addition of non-essential amino acids (BE13-114E, Biowhittaker Lonza, Basel, Switzerland). Monoclonal antibodies were purified from hybridoma culture supernatants using Protein-G affinity chromatography (16-266, Merck KGaA). Purified antibodies were stored at -80°C until use.
2.18. Mice vaccinations and vaccine formulation
Ten micrograms of purified RBD-His protein were adding to aluminum hydroxide (77161, Thermo Fisher Scientific Inc.) and continued mixing for 1 h before mouse immunization. Female BLAB/c (6- to 8-week-old; RRID:MGI:5568760) were immunized with aluminum adjuvanted RBD vaccine subcutaneously at weeks 0, 2, and 4. Sera were collected 10 weeks post first immunization for neutralizing assay.
2.19. GTEx and GWAS analysis
TPM-normalized RNA-sequence expression values, including 32 types of tissues, were retrieved from GTEx_V8 and plotted using GraphPad Prism version 8 (GraphPad). For eQTL analysis, the result of rs201008304 and rs140479100 genotypes associated with STT3A expression in cells-cultured fibroblasts was retrieved from GTEx website, while the plot of regional analysis for STT3A cis-eQTL was obtained from LocusCompare (locuscompare.com) using cultured_fibroblasts_GTEx_V7. Summarized statistics of SNPs in different COVID19 GWAS, including A2, B2, C2, and D1 analyses, were sequentially downloaded from the COVID19-HGI website on October 16th 2020 (20200930 version) [26].
2.20. Sequence retrieval
As of February 19th 2021, the NCBI-Virus-database has 57,311 Spike glycoprotein sequences of SARS-CoV-2. All these (57,311) sequences were aligned with BioEdit and analyzed the amino acid identity of glycosylation sites in N17, N149, N331, N343, and N657 and mutated sites including E484, N501, and D614 in Alpha, and Beta variants.
2.21. Chemistry synthesis of NGI-1 MC-Val-Cit-PAB-OH
Commercially available 6-maleimidohexanoic acid (600 mg; 2.84 mmol; 1.0 eq) was dissolved in dry CH2Cl2 (27 mL) under argon atmosphere, and the solution was cooled to 0°C. EDC•HCl (600 mg; 3.13 mmol; 1.1 eq), DIPEA (380 μL; 2.178 mmol; 2.3 eq) and H-Val-OtBu•HCl (654 mg; 3.13 mmol; 1.1 eq) were added subsequently. The reaction was stirred at room temperature for 8 h. The mixture was diluted with AcOEt (100 mL), washed with NaHCO3 (saturated aqueous solution; 2 × 30 mL), and brine (1 × 20 mL). The organic phase was dried and concentrated, affording compound as a pale, yellow oil (782 mg; 75% yield). tert-butyl ester (750 mg) was dissolved in dry CH2Cl2 (8 mL) under an argon atmosphere. TFA (5 mL) was added, and the mixture was stirred at room temperature for 2 h. The solution was concentrated and precipitated with hexane, affording carboxylic acid as a yellow solid (650 mg; 90% yield). S9 Fmoc-Ala-OH (312 mg; 1.00 mmol; 1.0 eq) was dissolved in dry DMF (4 mL) under argon atmosphere, and the solution was cooled to 0°C. HATU (420 mg; 1.11 mmol; 1.1 eq), HOAt (151 mg; 1.11 mmol; 1.1 eq) and DIPEA (520 μL; 2.98 mmol; 3.0 eq) were added subsequently. The mixture was stirred at the same temperature for 15 min. 4-aminobenzyl alcohol was added as a solution in dry DMF (2 mL). The solution was allowed to reach room temperature and stirred over-night. The mixture was diluted with AcOEt (100 mL) and, NaHCO3 (saturated aqueous solution; 3 × 20 mL) and brine (3 × 20 mL). The organic phase was dried and concentrated under a vacuum. The crude mixture was purified via flash column (gradient from 99:1 to 95:5 CH2Cl2/MeOH) to afford compound (264 mg; 0.64 mmol; 64% yield). Compound (274 mg; 0.66 mmol) was dissolved in dry DMF (30 mL) under an argon atmosphere and cooled to 0°C. Piperidine (325 μL; 3.29 mmol) was added, and the mixture was stirred at room temperature for 1 h. The solution was concentrated under a high vacuum, dissolved in AcOEt (100 mL), washed with NaHCO3 (saturated aqueous solution; 2 × 20 mL), and brine (2 × 20 mL). The organic phase was dried and concentrated. The crude material was dissolved in AcOEt: Hexane (1:1; 30 mL) and purified over a pad of silica (gradient from 1:1 AcOEt/Hexane to 9:1 CH2Cl2/MeOH with 0.5% TEA), to afford NH2 -Ala-PABOH as a brown oil (60 mg; 0.31 mmol). Carboxylic acid (105 mg; 0.34 mmol; 1.1 eq) was dissolved in dry DMF (4 mL) under an argon atmosphere, and the solution was cooled to 0°C. HATU (129 mg; 0.34 mmol; 1.1 eq), NH2-Ala-PABOH (60 mg; 0.31 mmol; 1.0 eq) and DIPEA (80 μL; 0.46 mmol; 1.5 eq) were added subsequently. The reaction was allowed to slowly reach room temperature and stirred overnight. DMF was removed under vacuum. The crude was dissolved in AcOEt (80 mL) and washed with KH (1 M aqueous solution; 2 × 20 mL) and brine (1 × 20 mL). The organic phase was dried and concentrated under a vacuum. The crude was purified via flash column to afford compound. MC-Val-Cit-PAB-OH, (8.68 g, 15.2 mmol, 572.66 g/mol) was suspended in DMF (43.4 mL) and heated to 80°C to dissolve all solids. After cooling to 0°C, thionyl chloride (1.22 mL; 16.7 mmol) was added dropwise. Following the addition, the reaction was held at 0°C for 30 min and then treated slowly with water (130 mL) to precipitate a yellow solid, which was collected by filtration. The solid was washed sequentially with water, CH3CN, and MTBE. After drying under vacuum, 6.90 g (77% yield) of yellow solid was recovered. MC-Val-Cit-PAB-NGI:In a small vial, the NGI-1 (1 eq) and MC-Val-Cit-PAB-Cl (1.1 eq) were combined in DMF (0.25 M) at room temperature. To the solution was added 0.2-0.5eq of tetrabutylammonium iodide followed by N,N-diisopropylethylamine (2.5 eq), and the mixture stirred until all starting material amine was consumed or until decomposition was observed by LC-MS. The mixture was diluted with 1.5 mL of DMF and injected directly on HPLC for purification eluting with 20-60% acetonitrile: 0.1% formic acid in water to afford the quaternary salt product.
2.22. Antibody-drug conjugate (ADC)
Conjugation of antibodies with NGI-1 was performed by using a MC-Val.Cit-PAB-OH linker. For conjugate preparation, the spike mAb (> 5 mg/mL) in PBS containing 50 mM borate was treated with dithiothreitol (final concentration 10 mM). After gel filtration (G-25, PBS containing 1 mM DTPA), the number of SH groups per mAb was determined using 5,5’-dithiobis (2-nitrobenzoic acid). The reduced mAb was then mixed with the maleimide drug derivatives at 4°C (1.1 equiv./SH group) in cold CH3CN (20% v/v) for 1 h. The ADC was concentrated by centrifugal ultrafiltration, gel filtered (G-25, PBS), and sterile filtered. Protein and drug concentrations were determined by spectral analysis, residual thiol group assays, and amino acid analysis. Size-exclusion HPLC was established to ensure all conjugates used in this study > 98% monomeric, and C18 RP-HPLC was established to ensure < 0.5% unconjugated cysteine quenched drug exists.
2.23. Flow cytometry-based receptor-binding inhibition assay
Monoclonal antibody interference of spike extracellular domain (ECD) protein with His tag binding to human ACE2 receptor on the cell surface was measured by flow cytometry. HEK-293T cells were seeded at a density of 2.5 × 105 cells per mL in a 15 cm dish. After reaching 70-80% confluency, cells were transformed with lentiviral vectors encoding human ACE2. Cells were treated G418 for two weeks. ECD protein (0.03 μg) was pre-incubated with mAb (50 μL supernatant) for 30 min at room temperature. ECD protein and mAbs mixtures were added into 24 well plates containing 1 × 105 HEK293T-ACE2 cells for 1 h at room temperature and subjected to flow cytometry. Single-cell suspensions in FACS buffer were centrifuged at 400 × g for 10 min. Cells were subsequently incubated with PE-conjugated mouse anti-His Abs (362603, BioLegend) for 30 min on ice. The stained cells were analyzed using an LSR II cytometer, and data were processed using FlowJo V10 software.
2.24. Binding affinity assay
An Octet RED96e BioLayer Interferometry system (FortéBio, Menlo Park, USA) was used to measure the binding affinities of non-glycosylated spike (ng-spike) extracellular domain (ECD) to ACE2 and a spike monoclonal mouse antibody. Briefly, Anti-Mouse IgG Fc Capture (AMC) biosensors (FortéBio) were pre-equilibrated in PBS-TB binding buffer (1x PBS, pH 7.4, 0.2% BSA, 0.05% Tween-20) for 10 min prior to loading. 200 nM ACE2-Fc and 40 nM spike antibodies were loaded onto the sensor for 240 s and washed by binding buffer for 120 s to reach a stable baseline. The sensors were then subjected to association phase immersion for 180 s in wells containing both 100 nM g-spike and ng-spike ECD-His diluted in binding buffer. After association, the sensors were placed back into the binding buffer to allow the dissociation phase to occur for 600 to 1,800 s. The kinetic constants were calculated via the Data Analysis 10.0 (FortéBio) software and were fitted by using 1:1 and 2:1 binding models for ACE2 and antibodies, respectively. The final equilibrium dissociation constant (KD) was calculated as the dissociation constant divided by the association constant (KD = Kd/Ka).
2.25. Ethics statement
All animal procedures approved by Institutional Animal Care and Use Committee (IACUC) Academia Sinica. Serum specimens from COVID-19 convalescent patients were gifts from Dr. Yi-Ling Lin's lab, through approval from the Institutional Review Board on Biomedical Science Research, Academia Sinica.
2.26. Statistical analysis
The data from three individual experiments are assessed by one-way or two-way ANOVA with Tukey's post hoc test for multiple comparisons (GraphPad Prism Software Inc, San Diego, CA, USA) and presented as mean ± SD (standard deviation). A p-value < 0.05 was considered statistically significant. The relationships between NF-κB with STT3A and STT3B in the TCGA lung cancer dataset retrieved from TCGA database were analyzed by Pearson correlation analysis.
2.27. Role of funding source
Funders of this study had no role in study design, data collection, data analyses, interpretation, or writing of the report.
3. Results
3.1. Highly evolutionarily-conserved N-linked glycosylation sites on spike protein are essential for ACE2 binding
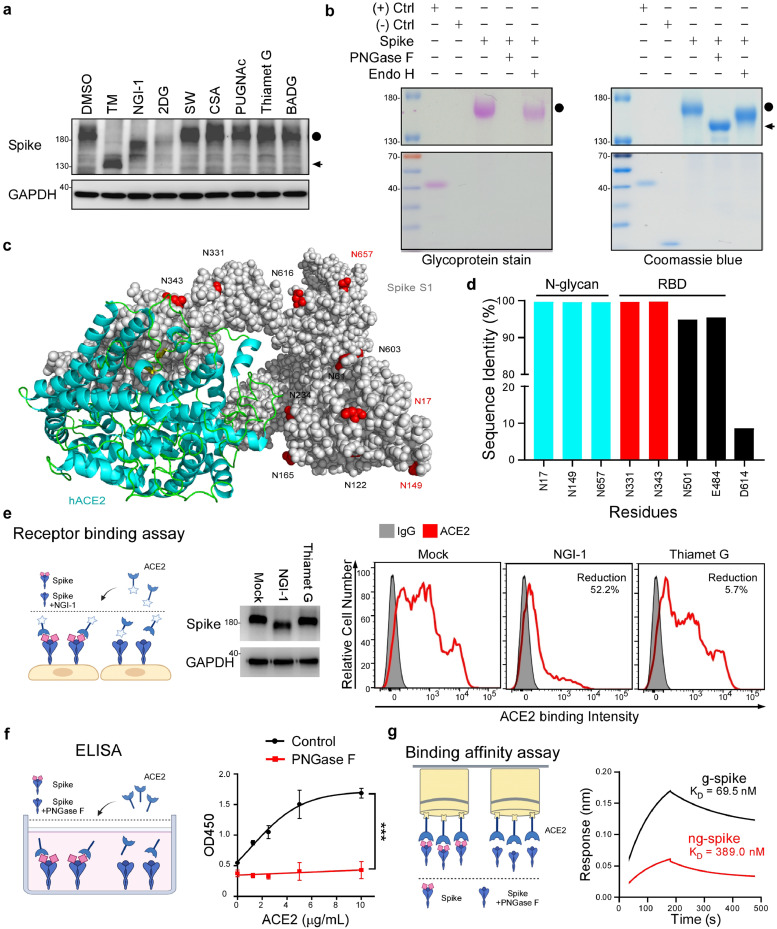
To investigate post-translational modifications of SARS-CoV-2 S protein, we stably transfected S protein into HEK293T cells and inspected expression patterns upon administering various glycosyltransferase inhibitors. We noticed that some treatments shifted the molecular weight of S protein significantly upward (from 140 kDa to 180 kDa) (Figure 1a), indicating that S protein is modified post-translationally. Interestingly, we observed that the molecular weight of S protein (Figure 1a, lanes 2-4) was reduced after treating with N-linked glycosyltransferase inhibitors, including tunicamycin, NGI-1, and 2-DG. However, O-GlcNAc-β-N-acetylglucosaminidase, O-GlcNAcase, and O-linked glycosyltransferase inhibitors did not affect the molecular weight of S protein (Figure 1a, lanes 7-9). Moreover, treatment of S protein with PNGase F (to remove both complex and hybrid N-linked glycans) but not Endo H (to remove hybrid N-linked glycans only) impaired glycostaining intensity, indicating that S protein is extensively modified by complex N-linked glycosylation (Figure 1b).
Figure 1.
N-glycosylation in spike is highly conserved in SARS-CoV-2 variants and essential for ACE2 binding. (a) Western blot analysis of S protein expression in HEK293T-spike cells that transfected with spike for 24h, followed by treatment with the indicated inhibitor for an additional 24 h. N-linked inhibitors [2.5 μg/mL Tunicamycin (TM), 10 μM NGI-1, 20 mg/mL 2-DG, 10 μM Swainsonine (SW), and 50 μM Castanospermine (CSA)]; 10 μM O-GlcNAc-β-N-acetylglucosaminidase inhibitor (PUGNAc); 10 μM O-GlcNAcase inhibitor (Thiamet G), and 10 μM O-linked glycosyltransferase inhibitor [Benzyl 2-acetamido-2-deoxy-α-D-galactopyranoside (BADG)]. The non-glycosylated form in lane 2 represents a spike with TM treatment. Circle, glycosylated spike (g-spike); arrow, non-glycosylated spike (ng-spike). (b) The glycosylation pattern of the S protein. S protein was treated with PNGase F or Endo H, and analyzed by glycoprotein staining (left panel) and Coomassie blue staining (right panel). Black circle, g-spike; arrowhead, ng-spike. Negative control (-), Trypsin inhibitor soybean (20 kDa); positive control (+), horseradish peroxidase (44 kDa). (c) 3D structure diagrams of the ACE2 and spike in SARS-CoV-2 (7A95) structure rendered as a surface with NXT/S motif glycans resolved in the cryo-EM map rendered as red spheres. (d) Sequence identity analysis of three SARS-CoV-2 distinct surface residues (N17, N149, N657), RBD residues (N331, N343), and the susceptible residues (E484, N501, and D614) of the variants in 57311 S proteins retrieved from NCBI-Virus-database. (e) Flow cytometry analysis of ACE2 binding to HEK293T-spike cells with NGI-1 and Thiamet G treatment (right panel). Glycosylated and de-glycosylated S protein molecular weight were assessed by Western blot (left panel). (f) ELISA detection of the indicated concentration of ACE2 binding to precoated SARS-CoV-2 ECD after DMSO or PNGase F treatment. Data shown are means ± SD from representative triplicates from three independent experiments. ***p < 0.001 (Two-way ANOVA). (g) Binding affinity of g-spike or ng-spike with immobilized ACE2 measured by BioLayer interferometry.
Glycosylation is an enzyme-directed site-specific process and N-glycosylation predominantly occurs at the consensus NXT/S motif [27]. To better understand differences among the NXT/S motifs of S proteins from SARS-CoV and SARS-CoV-2, we analyzed twenty-two sequences obtained from the National Center for Biotechnology Information (NCBI) (Supaplementary Material, Figure S1a). Most NXT/S motifs were highly conserved between the two viruses. Notably, SARS-CoV-2 and its variants possessed four additional NXT/S motifs of S protein—N17XT, N74GT, N149KS, and N657NS—not present in SARS-CoV S protein (Supplementary Material, Figure S1a). We further elucidated glycan structure via liquid chromatography-tandem mass spectrometry (LC-MS/MS). Glycopeptides carrying N-glycans, including the complex type, were detected at all four of those SARS-CoV-2-specifc N-glycosylation sites (Supplementary Material, Table S1a), consistent with our glycostaining analysis (Figure 1b). Three-dimensional protein structure analysis showed that three of these four NXT/S motifs in SARS-CoV-2 S protein were on the protein surface, i.e., N17XT, N149KS, and N657NS (PDB: 7A95) (Figure 1c). Thus, N-glycosylation of SARS-CoV-2 S protein may positively mediate viral attachment to ACE2 [28].
To investigate if the N-linked glycosites of SARS-CoV-2 S protein are conserved, we retrieved 57,311 S protein sequences from the NCBI-Virus-database (https://www.ncbi.nlm.nih.gov/labs/virus, accessed on 19 February 2021). Sequence analysis revealed that glycosylation sites in the receptor binding domain (RBD; N331 and N343) and the three surface glycosylation sites (N17, N149, and N657) were all highly conserved (>99.67% identity). However, sequence identity was lower for some mutated sites of the Alpha and Beta variants [D614 (8.68%), N501 (94.96%), and E484 (95.54%)] (Figure 1d), indicating that N-glycosylation sites are highly conserved among SARS-CoV-2 variants.
Next, to determine if N-linked glycosylation of S protein is necessary for ACE2 binding, we conducted flow cytometry, ELISA, and receptor binding assays. We transfected S protein into HEK293T cells for 24 h and then treated the cells with NGI-1 to prevent N-linked glycosylation. NGI-1 treatment still retains surface spike protein expression (Supplementary Material, Figure S1b). Importantly, N-linked glycosyltransferase inhibitor treatment reduced the binding ability of S protein for ACE2 (Figure 1e). Furthermore, we treated S protein with PNGase F and coated the non-glycosylated extracellular domain (ECD) of S protein (ng-spike) on ELISA plates. ELISA revealed that soluble ACE2 specifically bound to non-treated glycosylated S protein (g-spike) but not to ng-spike (Figure 1f). Consistently, the ACE2 binding affinity for g-spike (KD = 69.5 nM) was 5.6-fold stronger than that for ng-spike (KD = 389.0 nM), as determined by BioLayer interferometry (Figure 1g). Together, these results indicate that N-linked glycosylation sites in S protein of SARS-CoV-2 and its variants sustain ACE2 interaction.
3.2. S protein deglycosylation alleviates viral infection
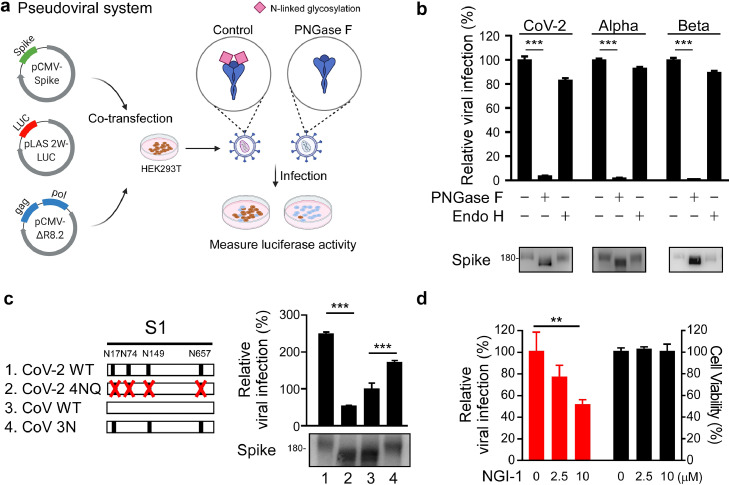
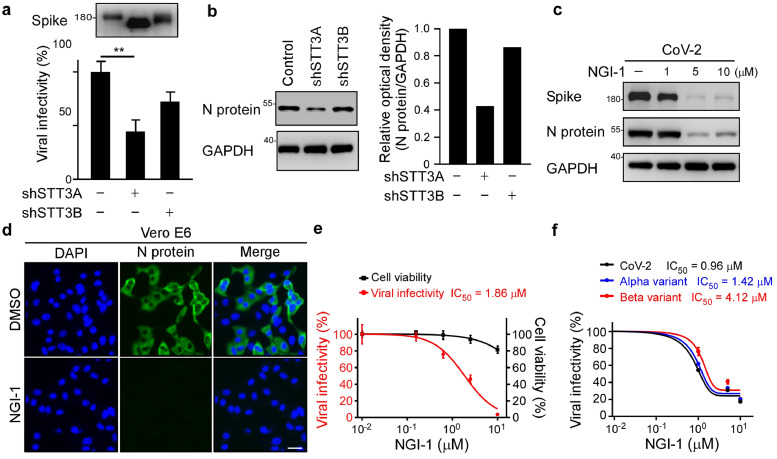
To examine if N-linked glycosylation of S protein is necessary for viral spread, we subjected SARS-CoV-2 pseudoviruses to PNGase F or Endo H treatments (Figure 2a). Removal of hybrid-type N-linked glycosylation by Endo H treatment only slightly reduced pseudovirus infectivity, whereas removal of both complex and hybrid N-linked glycosylation by PNGase F dramatically reduced infectivity (Figure 2b), indicating that glycosylation contributed significantly to pseudovirus infectivity. Relative to SARS-CoV, SARS-CoV-2 harbors four unique surface NXT/S motifs (N17LT, N74GT, N149KS, and N657NS) in its S protein (Figure 2c). We substituted the four asparagines (N) of those NXT/S glycosylation sites with glutamine (Q) (4NQ), with the molecular weight of S protein from SARS-CoV-2 4NQ being lower than that of WT (Figure 2c). We added three of the surface-exposed NXT/S motifs to SARS-CoV (N17LT, N149KS, and N657NS; 3N), which increased the molecular weight of recombinant S protein from SARS-CoV 3N relative to SARS-CoV WT. Next, we infected HEK293T-ACE2 cells with SARS-CoV-2 4NQ or SARS-CoV 3N pseudovirus to elucidate if the NXT/S motifs contribute to viral infectivity. We found that while viral activity of SARS-CoV-2 4NQ was weaker than that of SARS-CoV-2 WT (Figure 2c), SARS-CoV 3N displayed significantly greater infectivity than SARS-CoV WT (Figure 2c), supporting that the three NXT/S motifs are important mediators of viral infectivity. Moreover, SARS-CoV-2 WT pseudovirus displayed lower infectivity in HEK293T-ACE2 cells treated with NGI-1 than non-treated control cells (Figure 2d). Together, these results indicate that deglycosylation of S protein reduces SARS-CoV-2 viral infectivity.
Figure 2.
N-glycosylation in spike is critical to viral infection. (a) Diagram of the spike pseudovirus package. (b) Effect of the deglycosylated spike on viral infection. Luciferase activity was measured to evaluate pseudotype viral entry ability in HEK293T-ACE2 cells challenged with SARS-CoV-2, Alpha, or Beta variant spike pseudovirus for 2 days. Before infection, pseudovirus were pretreated with PNGase F, or Endo H by a non-denaturing reaction protocol (https://international.neb.com/protocols/2014/07/31/pngase-f-protocol). The control represents pseudovirus treated with glycosidase-free glycobuffer. Glycosylated and de-glycosylated S protein molecular weight were assessed by Western blot. n=3. Statistical method: two-way ANOVA, Tukey post hoc tests, ***p < 0.001. (c) Diagram of the NXT motifs in spike S1 of CoV-2, CoV-2 4NQ, CoV, and CoV 3N (left panel). Luciferase activity was measured in HEK293T-ACE2 cells infected with distinct pseudovirus generated from wild-type and mutant spike SARS-CoV or SARS-CoV-2 (right panel). Western blotting was performed to examine S protein molecular weight in CoV-2, CoV-2 4NQ, CoV, and CoV 3N. ***p < 0.001 (One-way ANOVA, Tukey post hoc tests). (d) The infectivity of pseudoviruses generated from DMSO or NGI-1-treated HEK293T cells was evaluated by luciferase assay (red). Cell viability was determined by MTT assay (black bar). n = 3 with mean ± SD shown. ***p < 0.001 (One-way ANOVA, Tukey post hoc tests).
3.3. SARS-CoV-2 hijacks the host NF-κB/STT3A axis to upregulate N-glycosylation of S protein
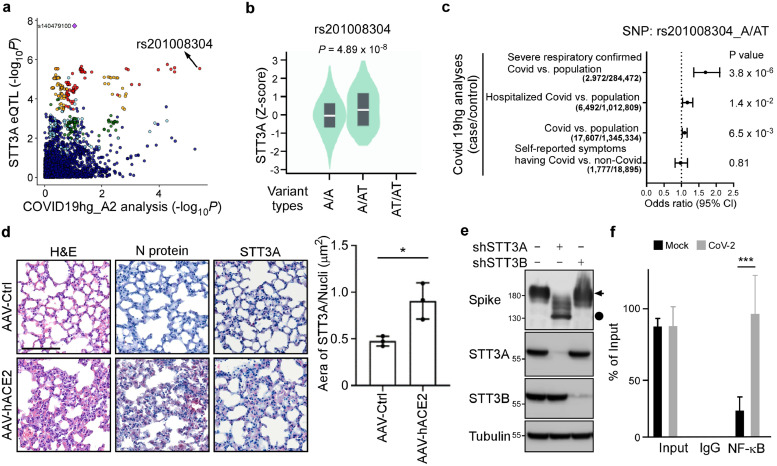
STT3A is responsible for cotranslational N-glycosylation [14] and its ablation induces viral dysfunction [17]. We utilized expression quantitative trait locus (eQTL) mapping to assess differential STT3A expression and explore its association with the COVID-19 phenotype. Rs140479100 represents the most significant cis-eQTL (p = 1.96 × 10−8) for STT3A (Supplementary Material, Figure S2a-b) in a dataset of cell-cultured fibroblasts from the GTEx portal (https://gtexportal.org/). We examined rs140479100 as part of an ongoing COVID-19 genome-wide association study (GWAS) [26], but it was undetectable. As an alternative, we investigated cis-eQTL rs201008304, which displays locus linkage to rs140479100 of r2 > 0.8, and observed a strong correlation with GWAS data (Figure 3a). Thus, people with the rs201008304 variant (i.e. a T insertion) are prone to higher STT3A expression in their cells (Figure 3b) and are at greater risk of developing severe COVID-19 (Figure 3c; Supplementary Material, Figure S2c), implying a link between STT3A expression and severe COVID-19. To mimic human conditions of COVID-19 disease, we used AAV-hACE2-transduced mice susceptible to SARS-CoV-2 and found that STT3A expression was elevated in lung tissue of SARS-CoV-2-infected mice (Figure 3d). Next, to determine if STT3 isoforms play a role in glycosylation, we knocked down the STT3A or STT3B isoforms from Vero E6 cells and then assayed S protein expression. We found that both STT3A and STT3B mediate S protein glycosylation (Figure 3e). Importantly, knocking down STT3A dramatically reduces S protein glycosylation supporting the notion that greater STT3A-mediated N-glycosylation of S protein promotes SARS-CoV-2 infection and leads to severe COVID-19 disease.
Figure 3.
SARS-CoV-2 induces N-glycosylation through NF-κB/STT3A axis. (a) Locus compare plot for the comparison between the STT3A eQTLs and the GWAS result, the COVID-19 A2 analysis for patients with severe respiratory symptoms via COVID-19 Host Genetics Initiative (COVID-19-HGI) project. The points are colored based on linkage disequilibrium (LD) bins. rs201008304, the indel variant in strong LD with the lead STT3A eQTL, is the top and most significant SNP in COVID-19 A2 analysis. (b) The eQTL plot for the association between STT3A variant types [A/A (n = 437), A/AT (n = 46), and AT/AT (n = 0)] expression and rs201008304 in cells-cultured fibroblasts was queried from the GTEx. (c) Forest plot shows the risk of rs201008304 in different COVID-19 phenotypes, originally from the summarized statistic results. (d) Representative data of immunohistochemistry staining of STT3A (red cells) and N protein in lung tissues of AAV-Ctrl and AAV-hACE2 mice infected by SARS-CoV2 at 5 dpi. Images were captured at 40x objective lens magnification. Scale bar, 100 μm. Three randomly selected STT3A/nuclei expression fields in lung tissues were exported from CaseViewer software (3DHistech) and quantified by the HistoQuant plugin in QuantCenter (3DHistech). Statistical method: t test, *p < 0.05, (n = 3). (e) Western blotting of S protein in Vero E6 cells after knockdown of STT3A or STT3B. Circle, g-spike; arrow, ng-spike. (f) ChIP analysis of enrichment of NF-κB in STT3A promoters after SARS-CoV-2 infection for 1 d. The enrichment levels were analyzed by RT-qPCR and shown as the percentage of input. Statistical method: two-way ANOVA, Tukey post hoc tests, ***p < 0.001 (n = 3 with mean ± SD shown).
Several studies have indicated that SARS-CoV-2 induces the NF-κB pathway, which may serve as a potential target for treating COVID-19 patients [29,30]. Based on the lung cancer dataset of The Cancer Genome Atlas (TCGA) (n = 1,018), we observed that NF-κB is significantly and positively correlated with STT3A (p < 0.0001, R = 0.3088) but not STT3B (p = 0.3292, R = -0.0306) (Supplementary Material, Figure S3a). Hence, we used the Encyclopedia of DNA Elements (ENCODE) database to identify that NF-κB binds transcriptional regulatory regions of STT3A (Supplementary Material, Figure S3b). Consistently, our luciferase reporter assay demonstrated that NF-κB activates STT3A promoters containing wild-type but not mutant NF-κB binding motifs (Supplementary Material, Figure S3c). Moreover, a dominant-negative IκBαSR mutant diminished the ability of NF-κB to activate the STT3A promoter (Supplementary Material, Figure S3c). Similar regulation by NF-κB and IκBαSR on endogenous STT3A was observed at both mRNA and protein levels (Supplementary Material, Figure S3d), indicating that NF-κB induces the expression of STT3 isoform. We also conducted chromatin immunoprecipitation (ChIP) assays to demonstrate that SARS-CoV-2 induced NF-κB binding to the promoter region of STT3A in A549-ACE2 cells (Figure 3f). Our previous study showed that STT3 is involved in N-linked glycosylation [31], and this mechanistic studies have indicated that SARS-CoV-2 infection may trigger NF-κb-mediated STT3 activation, which could result in an accumulation of N-linked glycosylation on S protein. Previously we showed that spike glycosylation increases ACE2 binding (Figures 1e-g). This effect may be reinforced by SARS-CoV-2 infection, which triggers NF-κB mediated STT3A expression to increase S protein N-linked glycosylation
3.4. Inhibition of STT3A reduces SARS-CoV-2 infectivity
Given that N-glycosylation of S protein is necessary for viral infection, inhibition of STT3A may represent an alternative therapeutic strategy for impairing SARS-CoV-2 infectivity and that of its variants. Accordingly, we isolated pseudotyped and SARS-CoV-2 virus from STT3 isoform-knockdown cells and then transfected them into ACE2-expressing cells. Compared to the pseudovirus produced from shSTT3B-HEK293T cells, we observed that the pseudovirus produced from shSTT3A-HEK293T had significantly lower infectivity (Figure 4a). Consistently, SARS-CoV-2 produced from shSTT3A-Vero E6 cells showed lower infectivity to A549-ACE2 cells (Figure 4b). Next, we found that administration of an STT3-specific inhibitor, NGI-1, impaired SARS-CoV-2 infection in Vero E6 cells, as demonstrated by reduced viral protein expression according to Western blotting and immunofluorescence assay (IFA) (Figure 4c-d). NGI-1 resulted in a half-maximal inhibitory concentration (IC50) of 1.863 μM in Vero E6 cells, without concomitant cytotoxicity (Figure 4e), indicating that blocking N-glycosylation via NGI-1 treatment effectively impeded SARS-CoV-2 WT infection. Importantly, NGI-1 also impaired infection by the Alpha and Beta variants in a dose-dependent manner (Figure 4f), indicating that the impact of deglycosylation in terms of preventing viral entry is consistent. Together, these results indicate that inhibition of STT3A significantly reduces SARS-CoV-2 WT and variant infectivity.
Figure 4.
Impairment of STT3A compromises SARS-CoV-2 infectivity. (a) Luciferase assay was used to monitor the infection rate of pseudovirions generated from shSTT3A or shSTT3B HEK293T cells in HEK293T-ACE2 cells. Expression of spike in pseudovirus produced from shSTT3A and shSTT3B. Statistical method: one-way ANOVA, Tukey post hoc tests. **p < 0.01 (n = 3 with mean ± SD shown). (b) Western blot analysis of N protein expression in A549-ACE2 cells infected with SARS-CoV-2 generated from control, shSTT3A or shSTT3B Vero E6 cells (left panel). N protein intensity was quantified by ImageJ (right panel). (c) Western blot analysis of spike and N protein expression in DMSO and NGI-1 treated SARS-CoV-2 infected Vero E6 cells. (d) Representative immunofluorescence images of N protein expression in DMSO and NGI-1 treated Vero E6 cell after SARS-CoV-2 infection. Scale bar, 100 μm. (e) SARS-CoV-2 neutralization IC50 of NGI-1 in Vero E6. Vero E6 cells was pretreated with NGI-1 1 h ahead at the indicated doses prior to SARS-CoV-2 infection (red). Cell viability of Vero E6 cells following NGI-1 treatment determined by MTT assay (black). Data shown are means ± SD from representative triplicates. (f) Luciferase activity was measured at 48 h to determine SARS-CoV-2, Alpha, and Beta variants pseudoviral infectivity in DMSO and NGI-1 treated HEK293T-ACE2; Data shown are means ± SD from representative triplicates.
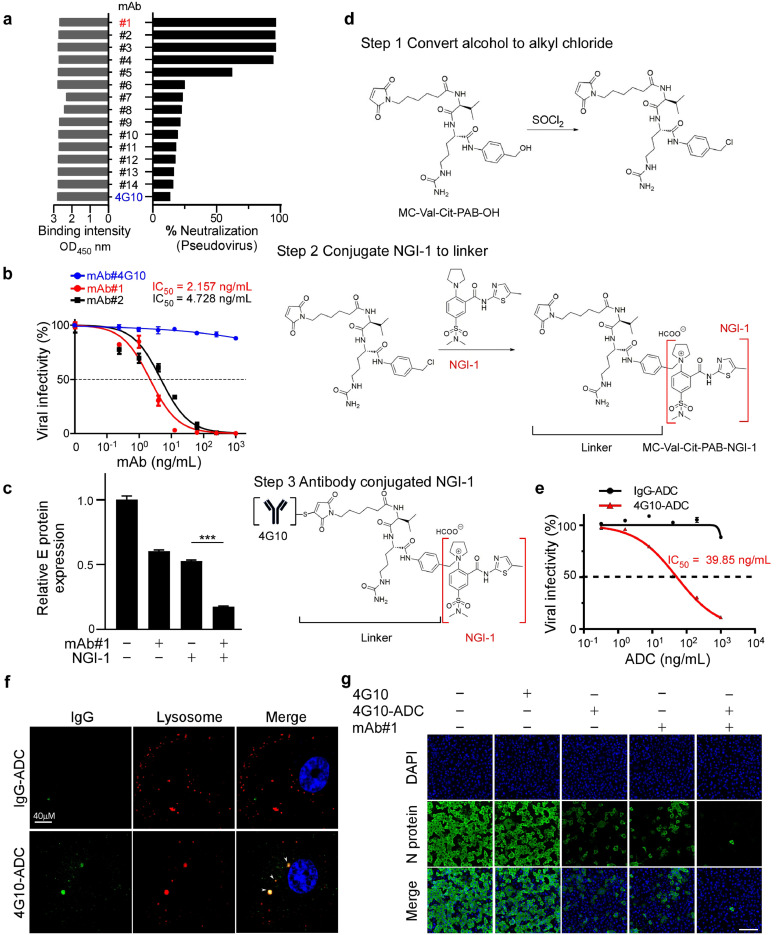
3.5. An antibody-drug conjugate (ADC) targeting SARS-CoV-2-infected cells
To specifically target SARS-CoV-2-infected cells, we generated full-length anti-spike antibodies from hybridoma culture supernatant (Supplementary Material, Figure S4a-b). Fifteen SARS-CoV-2 antibodies were identified as displaying high ELISA reactivity to full-length S protein, as well as its RBD (residues 319–541) (Figure 5a; Supplementary Material, Figure S4a-b). These monoclonal antibodies were further evaluated by receptor binding inhibition assay. Our results show that mAb#1 and mAb#2 antibodies, but not mAb#4G10, neutralized S protein and efficiently prevented it from binding to HEK293T-ACE2 cells (Figure 5a; Supplementary Material, Figure S4c). The neutralizing antibodies mAb#1 and mAb#2 potently inhibited SARS-CoV-2 pseudovirus infection of HEK293T-ACE2 cells in a dose-dependent manner (Supplementary Material, Figure S4c). Moreover, consistently, anti-spike mAb#1 and mAb#2 prevented SARS-CoV-2 infection, with IC50 values of 2.157 ng/mL and 4.728 ng/mL, respectively (Figure 5b).
Figure 5.
Antibody-drug conjugates (ADCs) internalize SARS-CoV-2 infected cells and impair viral infection. (a) ELISA binding assays and binding neutralization assays were performed to demonstrate the binding and neutralization ability of antibodies to RBD. (b) SARS-CoV-2 neutralization IC50 of the indicated concentration of anti-spike antibody. Vero E6 cells was pretreated with mAb#4G10, mAb#1, or mAb#2 1 h ahead at the indicated doses prior to SARS-CoV-2 infection. Data shown are means ± SD from representative triplicates. (c) Detection of SARS-CoV-2 spike pseudovirions after administering 1 ng/mL anti-spike mAb#1 and 1 μM NGI-1 on HEK293T-ACE2 cells. Statistical method: one-way ANOVA, Tukey post hoc tests, ***p < 0.001. Data shown are means ± SD from representative triplicates. (d) Chemistry synthesis of 4G10-ADC. Process in the conjugation of non-neutralizing antibody 4G10 and NGI-1 with the MC-Val.Cit-PAB-OH linker. (e) SARS-CoV-2 pseudoviral neutralization IC50 of the indicated concentration of anti-spike antibody. HEK293T-ACE2 cells were pretreated with IgG-ADC and 4G10-ADC 1 h ahead at the indicated doses prior to pseudoviral infection. Data shown are means ± SD from representative triplicates. (f) Internalization of 4G10-ADC and its co-localization in the lysosome. The antibodies conjugated NGI-1 were labeled with Alexa Fluro 488-Goat anti-mouse IgG and then added to A549-ACE2 cells with LysoTracker red. Arrow indicates localization of 4G10-ADC at lysosome. Scale bar, 10 μm. (g) Representative immunofluorescence images of the 4G10-ADC (40ng/mL) and mAb#1 (1ng/mL) effects on the expression of viral N protein after SARS-CoV-2 infection in Vero E6 cells. Scale bar, 200 μm.
To optimize NGI-1 and anti-S antibody inhibition ability in cell models, we administered NGI-1 and anti-spike neutralizing antibodies at an IC50 concentration. We found that a combination of NGI-1 with mAb#1 had a more substantial neutralizing effect than NGI-1 or mAb#1 alone in HEK293T-ACE2 cells (Figure 5c).
ADCs represent an emerging type of highly effective biopharmaceutical therapeutic composed of antibodies and active drugs. They can target specific antigens, thereby lowering off-target toxicity. To date, nine ADCs have been approved by the FDA for various diseases, and more than 80 others are currently undergoing clinical studies [20]. S protein has been shown to internalize into susceptible cells via ACE2 [32]. To avoid off-target effects of NGI-1 and increase specificity towards SARS-CoV-2-infected cells, we developed an ADC consisting of non-neutralizing mAb#4G10 conjugated to NGI-1 (4G10-ADC; Figure 5d) that binds to S protein (Figure 5a) and prevents SARS-CoV-2 pseudovirus infection, with an IC50 value of 39.85 ng/mL (Figure 5e). We employed a cleavable linker (MC-Val-Cit-PAB-OH) to crosslink NGI-1 to the anti-spike antibody (Supplementary Material, Figure S5a-b). The cleavable linker relies on lysosomal proteolytic degradation to release the conjugated drug [33]. IFA revealed that 4G10-ADC (green), but not IgG-ADC or mAb#1-ADC, was internalized into lysosomes (red) in susceptible cells and released NGI-1 (Figure 5f; Supplementary Material, Figure S5c). In addition, we also performed Western blotting to examine how our 4G10-ADC modifies the molecular mass of SARS-CoV-2 S protein, which revealed that it deglycosylated and destabilized the viral protein (Supplementary Material, Figure S5d).
We further combined mAb#1 with 4G10-ADC to evaluate their impact on SARS-CoV-2 viral infectivity and found that this combination significantly reduced viral infection in Vero E6 cells (Figure 5g). Thus, we have developed an ADC (4G10-ADC) that explicitly targets SARS-CoV-2-infected cells.
3.6. Deglycosylation by 4G10-ADC enhances anti-spike antibody recognition and limits SARS-CoV-2 variant infectivity
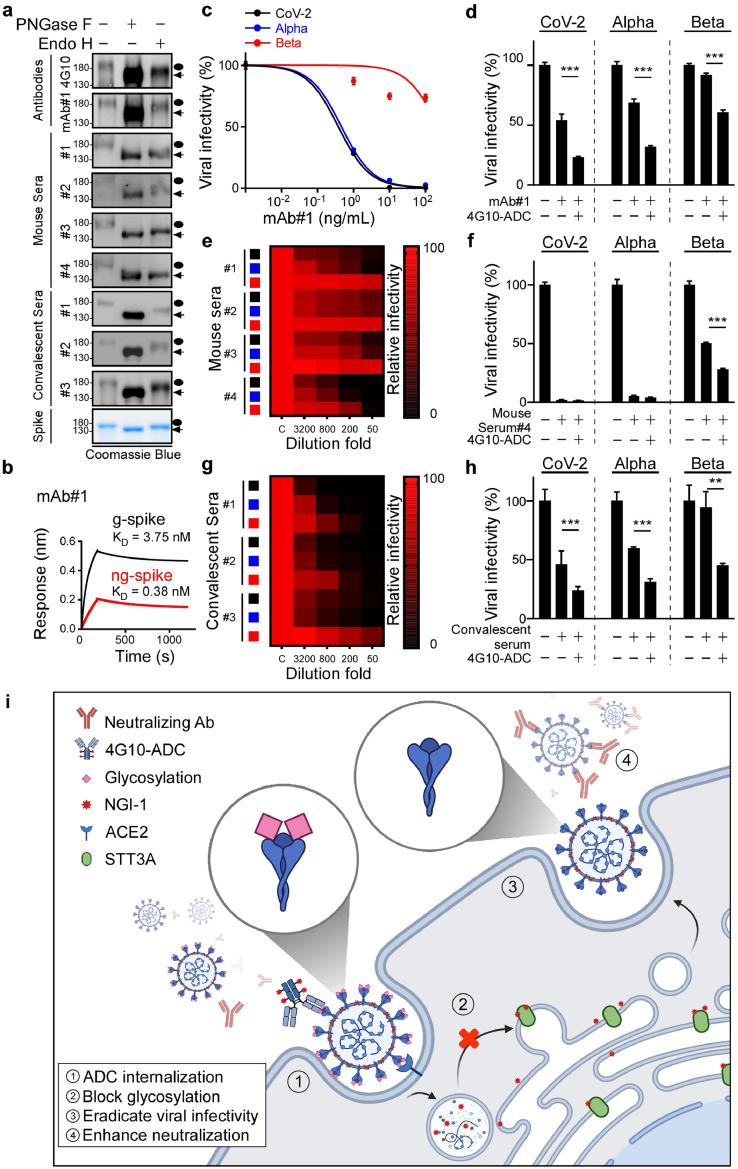
Glycosylation of viral proteins enables them to escape from both epitope-specific monoclonal antibodies and autologous antibodies [34]. As shown in Figure 6a, the binding ability of ng-spike to monoclonal antibodies, vaccinated mouse sera, and COVID-19 convalescent sera was significantly increased upon PNGase F treatment. Additionally, the binding affinity of anti-spike mAb#1 antibody for ng-spike (KD = 0.38 nM) was 9.8-fold stronger than for g-spike (KD = 3.75 nM), as revealed by BioLayer interferometry (Figure 6b). Consistent with a previous study showing that removing glycan moieties from cell surface antigens via enzymatic digestion enhances antibody binding affinity and signal intensity [35], our results imply that N-glycosylation plays a pivotal role in shielding immunogenic epitopes on viral S protein in cell models.
Figure 6.
4G10-ADC impairs SARS-CoV-2 variants infectivity. (a) Western blot analysis of PNGase F or Endo H treated S protein immunoblotted by purified antibodies, mice sera, or COVID-19 convalescent sera. Black circle, g-spike; arrowhead, ng-spike. (b) Binding affinity of g-spike or ng-spike with immobilized anti-spike mAb#1 measured by BioLayer Interferometry. (c-h) Luciferase activity was measured at 48 h to determine antibody neutralizing activity or viral infectivity in HEK293T-ACE2 cells; n=3 for all experiments. Statistically significant by one-way ANOVA, Tukey post hoc tests. **p < 0.01, ***p < 0.001 (n = 3 with mean ± SD shown). (c,e,g) Heat maps of the relative neutralizing activity of indicated concentration of antibodies from purified mAb#1 (c), mice sera (e), COVID-19 convalescent sera (g) against indicated SARS-CoV-2 (black), Alpha variant (blue), and Beta variant (red) pseudovirus. (d,f,h) Assessment of SARS-CoV-2, Alpha, and Beta variants infectivity in HEK293T-ACE2 cells after administering 40ng/mL 4G10-ADC with 1ng/mL anti-spike mAb#1 (d) 200 folds diluted mouse serum#4 (f) or 3200 folds diluted COVID-19 convalescent serum#3 (h). (i) Proposed mechanism of 4G10-ADC action on SARS-CoV-2 infected cells.
Variants of SARS-CoV-2 have displayed reduced neutralization sensitivity to purified monoclonal antibodies, vaccines, and convalescent sera [36], [37], [38]. Accordingly, we evaluated if 4G10-ADC could reinforce neutralizing ability by targeting N-glycosylation. Although mAb#1 only exhibited low neutralizing ability against Beta variant pseudovirus (Figure 6c), a combination of 4G10-ADC and mAb#1 had a strong effect in reducing Beta variant infectivity (Figure 6d). Moreover, we developed four vaccinated mouse sera to determine their neutralizing ability against WT, Alpha and Beta variant pseudoviruses (Figure 6e). Most of the vaccinated mouse sera exhibited potent neutralizing ability against WT and Alpha SARS-CoV-2, but not the Beta variant. Only vaccinated mouse serum#4 showed moderate neutralizing ability against the Beta variant (Figure 6e). Addition of 4G10-ADC enhanced the neutralizing ability of vaccinated mouse serum#4 against the Beta variant (Figure 6f). The Beta variant was also more resistant to neutralization by some convalescent sera than WT and Alpha variant pseudoviruses (Figure 6g). Consistent with Figure 6f, addition of 4G10-ADC also enhanced the neutralizing ability of COVID-19 convalescent sera against the Beta variant (Figure 6h). In summary, ADCs comprising non-neutralizing anti-spike antibodies can target and internalize into SARS-CoV-2-infected cells. The ADC employed in this study contains a cleavable dipeptide (Valine-Citrulline) linker that can be cleaved by lysosomal enzymes, such as Cathepsin B [39,40]. Therefore, upon infected cell entry, NGI-1 is released from 4G10-ADC in lysosomes to block glycosylation of S protein. Importantly, virus with non-glycosylated S protein (Figure 2b) displays a higher antibody binding affinity (Figure 6a) and poor viral infectivity (Figure 6i). Taken together, deployment of an ADC with monoclonal antibodies may serve as an alternative strategy for eradicating SARS-CoV-2 variant infectivity.
4. Discussion
Although the outlook from the global SARS-CoV-2 vaccination program is promising, rapidly spreading variants have emerged as a new challenge. Effectiveness of the mRNA-1273 COVID-19 vaccine against the Beta variant is 12.4-fold lower than against SARS-CoV-2 WT, and it is 10.3-fold lower for the BNT162b2 vaccine [36]. Notably, the binding affinity of ACE2 for SARS-CoV-2 S protein is 20-fold higher than that for the SARS-CoV S protein [41]. Moreover, the Alpha and Beta variants bind ACE2 with two-fold and five-fold greater affinity than SARS-CoV-2 WT [42]. This stable interaction between SARS-CoV-2 S protein and ACE2 might explain the highly contagious nature of the virus and its variants [43]. Glycan shielding has been widely reported for HIV-1 envelope protein and influenza hemagglutinin, enabling those viruses to evade the host humoral immune system and causing steric barriers that prevent the acquisition of neutralizing antibodies and weaken antibody-antigen recognition [16,[44], [45], [46], [47]]. Targeting glycosylation of viral envelopes to prevent viral spread has attracted increasing attention [48,49]. Our study demonstrates that deglycosylation of S protein reduced its binding affinity for ACE2, thereby reducing viral infectivity (Figure 1e-g, Figure 2b). Moreover, neutralizing antibodies displayed a stronger affinity for non-glycosylated S protein. Accordingly, targeting glycosylation exerts a dual impact in terms of impairing SARS-CoV-2 infection.
In this study, we mainly focused on spike glycosylation rather than ACE2. However, our results in Figure 4c indicate that NGI-1 pretreatment could have multiple effects on SARS-CoV-2 infected cells, resulting in low virus infectivity and generation. We cannot exclude that NGI-1 may interfere with ACE2 receptor function. However, we found that the deglycosylated pseudovirus weakened viral infectivity (Figure 2b), and this effect may come from ACE2 binding (Figures 1e-g). We also applied genetic analysis to reinforce our laboratory-based findings and support the positive association between STT3A expression and COVID-19 severity via the T insertion of the rs201008304 eQTL. According to the 1,000 Genomes Project published in the Ensembl genome browser (http://www.ensembl.org/Homo_sapiens/Info/Index), 4% of the global human population carries this T insertion (minor allele) variant, but proportions vary across different ethnic groups. For example, approximately 7% of Europeans and 9% of South Asians carry this variant, but it is absent from African and East Asian populations. It is noteworthy that both African and East Asian reported fewer COVID-19 cases than the rest of the world. It may be due to different public health measures, population densities, or weather. Further investigation of this coincidence will be favorable in uncovering the interaction between genetics and COVID-19 susceptibility and severity.
Glycosylation of viruses and target cells affects viral infection at several levels, including binding of viral S protein to the host cell ACE2 receptor and anti-spike antibody recognition [50]. The binding interaction between S protein and ACE2 depends on glycan-protein, glycan-glycan, and protein-protein interactions [51]. Antibody-antigen recognition depends on protein-protein interactions [52], but glycans on S protein mask the antibody recognition site to prevent antibody-mediated neutralization [53,54]. It has been reported that removing N-linked glycans from Programmed death ligand 1 (PD-L1) can enhance PD-L1 antibody recognition [35]. Therefore, due to impaired glycan-glycan or glycan-protein interactions, deglycosylation of S protein may reduce its binding affinity for ACE2. In addition, the stronger binding affinity between the antibody and deglycosylated S protein may be due to greater exposure of a recognition site.
NGI-1 disrupts oligosaccharyltransferase function by blocking the transfer of lipid-linked oligosaccharides to recipient glycoproteins, thereby inhibiting N-linked glycosylation [55]. Blocking N-linked but not O-linked glycosylation of S proteins by means of glycosylation inhibitors (such as NGI-1 and kifunensine) impedes viral entry [19,56]. Mutation of N-linked glycosylation sites in the RDB of S protein (N331 and N343) dramatically reduced viral infectivity [57], indicating that N-glycosylation of S protein is essential for viral infectivity. While Nunes Santos et al. observed that 1mM CST induces spike protein up shift by inhibiting glucosidases [58], we herein used 50uM of CST to remove O-linked glycosylation and found no significant change in spike molecular weight.
The Spike protein of SARS-CoV-2 is extensively glycosylated and glycosylation is required for effective infection. Similar to this notion, Rajasekharan et al. and Clarke et al. described that iminosugars, glycosyltransferase inhibitors, can be used to reduce SARS-CoV-2 infection [59,60]. To reduce the off-target effect, we herein used a novel antibody-drug conjugate (ADC) to selectively target virus-infected cells and then remove glycosyaltion by conjugating NGI-1 with anti-spike. Deglycosylation virus-infected cells may also induce ER stress and UPR to trap virus spike protein from virion assembly. ADCs exploit their specific binding properties to deliver drugs to targeted cells while sparing normal cells [61]. Enveloped viruses such as SARS-CoV-2 enter host cells by fusing to the host plasma membrane or endosomal membranes, with the former enabling S protein to be displayed on the plasma membrane [62], [63], [64]. We have developed 4G10-ADC to target S protein displayed on the surface of SARS-CoV-2-infected cells and to enhance antibody recognition by hindering N-linked glycosylation, thereby reducing viral infectivity. Our study shows that 4G10-ADC treatment enhanced the neutralizing abilities of purified antibody and of various sera-derived antibodies. The possible reason why neutralizing mAb#1-ADC was not internalized because it binds to RBD region of spike, therefore prevent spike bind to ACE2. Although it has previously been reported that NGI-1 nanoparticles had no toxic or significant weight loss effects in an animal model [65], the efficacy and safety of ADC combination treatments needs to be explored further. Nevertheless, our 4G10-ADC strategy represents a new and promising therapeutic approach for treating SARS-CoV-2 and its variants.
This study only examined SARS-CoV-2, Alpha, and Beta variants. Other variants such as Delta or Epsilon have not been included. In addition, all experiments are performed using a cell-based system. Further analysis using preclinical models is needed to determine the efficacy of ADC in vivo.
Contributors
H.-C.H. and Y.-J.L. designed and performed the experiments, analyzed data, and wrote the manuscript; C.-C.L., K.-B.H., I.-J.L., W.-F. Y., C.-P.S., C.-T.K., J.W., T.-C.H., P.-J.Y., and T.-A.L. performed experiments and analyzed data; W.H., W.-C.C., L.-H.W., J.-M.H., and F.-A.L. analyzed data; I.-J.L., and. S.-H.W. provided scientific input and wrote the manuscript; C.-Y.S., Y.-L.L., and M.-H.T. provided scientific input. C.-W.L. supervised the entire project, designed the experiments, analyzed data, and wrote the manuscript.
Data Sharing
All mass spectrometry proteomics relevant data is available online at ProteomeXchange database with the dataset identifier PXD026852.
Declaration of Competing Interest
H.-C. H., Y.-J. L., and C.-W.L. declared inventorship with US/ 63/262,926 patent listed on 2021, ANTIBODY-DRUG CONJUGATE FOR REDUCING MEMBRANE RECEPTOR GLYCOSYLATION. The remaining authors declare no conflicts of interest.
Acknowledgments
This work was funded in part by the following: Ministry of Science and Technology (MOST 109-2314-B-001-002 and MOST 109-2314-B-001-008 to C.-W.Li). Academia Sinica (AS-SUMMIT-109, AS-KPQ-109-BioMed) and Ministry of Science and Technology (MOST-108-3114-Y-001-002) (to M.-H.T.). Additionally, we thank Taiwan CDC for providing SARS-CoV-2 TCDC#4 (hCoV-19/Taiwan/4/2020) and funding support from Academia Sinica for IBMS P3 facility (AS-CFII-108-102) and the Ministry of Science and Technology, Taiwan for COVID-19 study (MOST 109-3114-Y-001-001). We thank National RNAi Core Facility of Academia Sinica for production of plasmid and shRNA. We also thank the Proteomic Core Facility, DNA Sequencing Core Facility (AS-CFII-108-115), Flow Cytometry Core Facility, and Light Microscopy Core Facility at the Institute of Biomedical Sciences of Academia Sinica for assistance in N-glycopeptide identification, DNA sequencing, analysis of flow cytometry, and morphological studies.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103712.
Appendix. Supplementary materials
References
- 1.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang H., Penninger J.M., Li Y., Zhong N., Slutsky A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46:586–590. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walls A.C., et al. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baden L.R., et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. New England Journal of Medicine. 2020;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Polack F.P., et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. New England Journal of Medicine. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galloway S.E., et al. Emergence of SARS-CoV-2 B.1.1.7 Lineage - United States, December 29, 2020-January 12, 2021. MMWR Morb Mortal Wkly Rep. 2021;70:95–99. doi: 10.15585/mmwr.mm7003e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walensky R.P., Walke H.T., Fauci A.S. SARS-CoV-2 Variants of Concern in the United States-Challenges and Opportunities. Jama. 2021;325:1037–1038. doi: 10.1001/jama.2021.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou D., et al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell. 2021 doi: 10.1016/j.cell.2021.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casalino L., et al. Beyond Shielding: The Roles of Glycans in the SARS-CoV-2 Spike Protein. ACS Cent Sci. 2020;6:1722–1734. doi: 10.1021/acscentsci.0c01056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hernández E.Ramírez, et al. The Role of the SARS-CoV-2 S-Protein Glycosylation in the Interaction of SARS-CoV-2/ACE2 and Immunological Responses. Viral Immunol. 2021;34:165–173. doi: 10.1089/vim.2020.0174. [DOI] [PubMed] [Google Scholar]
- 11.Fan Y., et al. Loss and gain of N-linked glycosylation sequons due to single-nucleotide variation in cancer. Scientific reports. 2018;8:4322. doi: 10.1038/s41598-018-22345-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dennis J.W., Nabi I.R., Demetriou M. Metabolism, cell surface organization, and disease. Cell. 2009;139:1229–1241. doi: 10.1016/j.cell.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cummings R.D. The repertoire of glycan determinants in the human glycome. Molecular bioSystems. 2009;5:1087–1104. doi: 10.1039/b907931a. [DOI] [PubMed] [Google Scholar]
- 14.Ruiz-Canada C., Kelleher D.J., Gilmore R. Cotranslational and posttranslational N-glycosylation of polypeptides by distinct mammalian OST isoforms. Cell. 2009;136:272–283. doi: 10.1016/j.cell.2008.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watanabe Y., Bowden T.A., Wilson I.A., Crispin M. Exploitation of glycosylation in enveloped virus pathobiology. Biochim Biophys Acta Gen Subj. 2019;1863:1480–1497. doi: 10.1016/j.bbagen.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watanabe Y., et al. Vulnerabilities in coronavirus glycan shields despite extensive glycosylation. Nature Communications. 2020;11:2688. doi: 10.1038/s41467-020-16567-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu H., Cherepanova N.A., Gilmore R., Contessa J.N., Lehrman M.A. Targeting STT3A-oligosaccharyltransferase with NGI-1 causes herpes simplex virus 1 dysfunction. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2019;33:6801–6812. doi: 10.1096/fj.201802044RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu S., et al. Comprehensive Interactome Analysis Reveals that STT3B Is Required for N-Glycosylation of Lassa Virus Glycoprotein. Journal of virology. 2019;93 doi: 10.1128/JVI.01443-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.A. Casas-Sanchez et al., Protein <em>N</em>-glycosylation is essential for SARS-CoV-2 infection. bioRxiv 10.1101/2021.02.05.429940, 2021.2002.2005.429940 (2021).
- 20.Joubert N., Beck A., Dumontet C., Denevault-Sabourin C. Antibody-Drug Conjugates: The Last Decade. Pharmaceuticals (Basel) 2020:13. doi: 10.3390/ph13090245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boehm J.S., et al. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell. 2007;129:1065–1079. doi: 10.1016/j.cell.2007.03.052. [DOI] [PubMed] [Google Scholar]
- 22.Perez-Riverol Y., et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47 doi: 10.1093/nar/gky1106. D442-d450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deutsch E.W., et al. The ProteomeXchange consortium in 2020: enabling 'big data' approaches in proteomics. Nucleic Acids Res. 2020;48 doi: 10.1093/nar/gkz984. D1145-d1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santry L.A., et al. AAV vector distribution in the mouse respiratory tract following four different methods of administration. BMC Biotechnol. 2017;17:43. doi: 10.1186/s12896-017-0365-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun C.P., et al. Rapid generation of mouse model for emerging infectious disease with the case of severe COVID-19. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Initiative C.-H.G. The COVID-19 Host Genetics Initiative, a global initiative to elucidate the role of host genetic factors in susceptibility and severity of the SARS-CoV-2 virus pandemic. Eur J Hum Genet. 2020;28:715–718. doi: 10.1038/s41431-020-0636-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Breitling J., Aebi M. N-linked protein glycosylation in the endoplasmic reticulum. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a013359. a013359-a013359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Londrigan S.L., et al. N-linked glycosylation facilitates sialic acid-independent attachment and entry of influenza A viruses into cells expressing DC-SIGN or L-SIGN. J Virol. 2011;85:2990–3000. doi: 10.1128/JVI.01705-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hariharan A., Hakeem A.R., Radhakrishnan S., Reddy M.S., Rela M. The Role and Therapeutic Potential of NF-kappa-B Pathway in Severe COVID-19 Patients. Inflammopharmacology. 2020:1–10. doi: 10.1007/s10787-020-00773-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kircheis R., et al. NF-κB Pathway as a Potential Target for Treatment of Critical Stage COVID-19 Patients. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.598444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsu J.M., et al. STT3-dependent PD-L1 accumulation on cancer stem cells promotes immune evasion. Nat Commun. 1908;9(2018) doi: 10.1038/s41467-018-04313-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang S., et al. Endocytosis of the receptor-binding domain of SARS-CoV spike protein together with virus receptor ACE2. Virus Res. 2008;136:8–15. doi: 10.1016/j.virusres.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao P., et al. Recent advances of antibody drug conjugates for clinical applications. Acta Pharm Sin B. 2020;10:1589–1600. doi: 10.1016/j.apsb.2020.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei X., et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 35.Lee H.H., et al. Removal of N-Linked Glycosylation Enhances PD-L1 Detection and Predicts Anti-PD-1/PD-L1 Therapeutic Efficacy. Cancer Cell. 2019;36:168–178. doi: 10.1016/j.ccell.2019.06.008. e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang P., et al. Antibody Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7. Nature. 2021 doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 37.Prévost J., Finzi A. The great escape? SARS-CoV-2 variants evading neutralizing responses. Cell Host Microbe. 2021;29:322–324. doi: 10.1016/j.chom.2021.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Supasa P., et al. Reduced neutralization of SARS-CoV-2 B.1.1.7 variant by convalescent and vaccine sera. Cell. 2021 doi: 10.1016/j.cell.2021.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thevanayagam L., et al. Novel detection of DNA-alkylated adducts of antibody-drug conjugates with potentially unique preclinical and biomarker applications. Bioanalysis. 2013;5:1073–1081. doi: 10.4155/bio.13.57. [DOI] [PubMed] [Google Scholar]
- 40.Senter P.D., Sievers E.L. The discovery and development of brentuximab vedotin for use in relapsed Hodgkin lymphoma and systemic anaplastic large cell lymphoma. Nature biotechnology. 2012;30:631–637. doi: 10.1038/nbt.2289. [DOI] [PubMed] [Google Scholar]
- 41.Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369:330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramanathan M., Ferguson I.D., Miao W., Khavari P.A. SARS-CoV-2 B.1.1.7 and B.1.351 spike variants bind human ACE2 with increased affinity. Lancet Infect Dis. 2021 doi: 10.1016/s1473-3099(21)00262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wrapp D., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grant O.C., Montgomery D., Ito K., Woods R.J. Analysis of the SARS-CoV-2 spike protein glycan shield reveals implications for immune recognition. Scientific reports. 2020;10:14991. doi: 10.1038/s41598-020-71748-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolfert M.A., Boons G.J. Adaptive immune activation: glycosylation does matter. Nature chemical biology. 2013;9:776–784. doi: 10.1038/nchembio.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doe B., Steimer K.S., Walker C.M. Induction of HIV-1 envelope (gp120)-specific cytotoxic T lymphocyte responses in mice by recombinant CHO cell-derived gp120 is enhanced by enzymatic removal of N-linked glycans. Eur J Immunol. 1994;24:2369–2376. doi: 10.1002/eji.1830241017. [DOI] [PubMed] [Google Scholar]
- 47.Li H., et al. Proximal glycans outside of the epitopes regulate the presentation of HIV-1 envelope gp120 helper epitopes. J Immunol. 2009;182:6369–6378. doi: 10.4049/jimmunol.0804287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Helle F., et al. Role of N-linked glycans in the functions of hepatitis C virus envelope proteins incorporated into infectious virions. J Virol. 2010;84:11905–11915. doi: 10.1128/JVI.01548-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sterjovski J., et al. Asn 362 in gp120 contributes to enhanced fusogenicity by CCR5-restricted HIV-1 envelope glycoprotein variants from patients with AIDS. Retrovirology. 2007;4:89. doi: 10.1186/1742-4690-4-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reis C.A., Tauber R., Blanchard V. Glycosylation is a key in SARS-CoV-2 infection. Journal of molecular medicine (Berlin, Germany) 2021:1–9. doi: 10.1007/s00109-021-02092-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao P., et al. Virus-Receptor Interactions of Glycosylated SARS-CoV-2 Spike and Human ACE2 Receptor. Cell host & microbe. 2020;28:586–601. doi: 10.1016/j.chom.2020.08.004. e586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davies D.R., Cohen G.H. Interactions of protein antigens with antibodies. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:7–12. doi: 10.1073/pnas.93.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Galili U. Amplifying immunogenicity of prospective Covid-19 vaccines by glycoengineering the coronavirus glycan-shield to present α-gal epitopes. Vaccine. 2020;38:6487–6499. doi: 10.1016/j.vaccine.2020.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grant O.C., Montgomery D., Ito K., Woods R.J. Analysis of the SARS-CoV-2 spike protein glycan shield reveals implications for immune recognition. Scientific reports. 2020;10:14991. doi: 10.1038/s41598-020-71748-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lopez-Sambrooks C., et al. Oligosaccharyltransferase inhibition induces senescence in RTK-driven tumor cells. Nature chemical biology. 2016;12:1023–1030. doi: 10.1038/nchembio.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang Q., et al. Inhibition of SARS-CoV-2 viral entry upon blocking N- and O-glycan elaboration. Elife. 2020;9 doi: 10.7554/eLife.61552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Q., et al. The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity. Cell. 2020;182:1284–1294. doi: 10.1016/j.cell.2020.07.012. e1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nunes-Santos C.J., Kuehn H.S., Rosenzweig S.D. N-Glycan Modification in Covid-19 Pathophysiology: In vitro Structural Changes with Limited Functional Effects. J Clin Immunol. 2021;41:335–344. doi: 10.1007/s10875-020-00905-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rajasekharan S., et al. Inhibitors of Protein Glycosylation Are Active against the Coronavirus Severe Acute Respiratory Syndrome Coronavirus SARS-CoV-2. Viruses. 2021;13 doi: 10.3390/v13050808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Clarke E.C., Nofchissey R.A., Ye C., Bradfute S.B. The iminosugars celgosivir, castanospermine and UV-4 inhibit SARS-CoV-2 replication. Glycobiology. 2021;31:378–384. doi: 10.1093/glycob/cwaa091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sievers E.L., Senter P.D. Antibody-drug conjugates in cancer therapy. Annu Rev Med. 2013;64:15–29. doi: 10.1146/annurev-med-050311-201823. [DOI] [PubMed] [Google Scholar]
- 62.Zhu X., Liu Q., Du L., Lu L., Jiang S. Receptor-binding domain as a target for developing SARS vaccines. Journal of thoracic disease. 2013;5(Suppl 2):S142–S148. doi: 10.3978/j.issn.2072-1439.2013.06.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hoffmann M., et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tang T., Bidon M., Jaimes J.A., Whittaker G.R., Daniel S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antiviral research. 2020;178 doi: 10.1016/j.antiviral.2020.104792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sambrooks C.Lopez, et al. Oligosaccharyltransferase Inhibition Overcomes Therapeutic Resistance to EGFR Tyrosine Kinase Inhibitors. Cancer Res. 2018;78:5094–5106. doi: 10.1158/0008-5472.CAN-18-0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.