Abstract
BACKGROUND & AIMS:
We recently showed that alcoholic hepatitis (AH) is characterized by dedifferentiation of hepatocytes and loss of mature functions. Glucose metabolism is tightly regulated in healthy hepatocytes. We hypothesize that AH may lead to metabolic reprogramming of the liver, including dysregulation of glucose metabolism.
METHODS:
We performed integrated metabolomic and transcriptomic analyses of liver tissue from patients with AH or alcoholic cirrhosis or normal liver tissue from hepatic resection. Focused analyses of chromatin immunoprecipitation coupled to DNA sequencing was performed. Functional in vitro studies were performed in primary rat and human hepatocytes and HepG2 cells.
RESULTS:
Patients with AH exhibited specific changes in the levels of intermediates of glycolysis/gluconeogenesis, the tricarboxylic acid cycle, and monosaccharide and disaccharide metabolism. Integrated analysis of the transcriptome and metabolome showed the used of alternate energetic pathways, metabolite sinks and bottlenecks, and dysregulated glucose storage in patients with AH. Among genes involved in glucose metabolism, hexokinase domain containing 1 (HKDC1) was identified as the most up-regulated kinase in patients with AH. Histone active promoter and enhancer markers were increased in the HKDC1 genomic region. High HKDC1 levels were associated with the development of acute kidney injury and decreased survival. Increased HKDC1 activity contributed to the accumulation of glucose-6-P and glycogen in primary rat hepatocytes.
CONCLUSIONS:
Altered metabolite levels and messenger RNA expression of metabolic enzymes suggest the existence of extensive reprogramming of glucose metabolism in AH. Increased HKDC1 expression may contribute to dysregulated glucose metabolism and represents a novel biomarker and therapeutic target for AH.
Keywords: Alcoholic Liver Disease, Metabolomics, Therapeutic Targets
Alcohol consumption is a leading cause of global morbidity and mortality, with much of the burden resulting from alcohol-related liver disease.1 Alcoholic hepatitis (AH) is a particularly severe form of acute-on-chronic liver disease that has a 20%–50% mortality rate at 3 months because of profound liver insufficiency and a marked systemic inflammatory response that leads to acute kidney injury (AKI) and multiorgan failure.2 Despite the significant impact of alcohol-related liver disease on public health, the disease pathogenesis remains poorly understood, and medical treatment for alcoholic liver disease (ALD) has not changed significantly in 40 years. Indeed, corticosteroids, the current first-line therapy for AH, are not useful in many patients.3 Identification of new targets for therapy for patients with AH represents one of the most urgent needs in clinical hepatology.
Glucose metabolism is an essential cellular physiologic process for controlling systemic energy homeostasis, and its dysregulation has been implicated in the development of diseases including cancer,4 Alzheimer disease,5 and liver diseases.6 Hepatocytes are important regulators of energy homeostasis via control of the storage, breakdown, and use of carbohydrates, lipids, and proteins. It is well known that acute alcohol consumption can contribute to dysregulation of hepatocyte metabolism via multiple mechanisms including depletion of enzyme cofactor levels, increase of endoplasmic reticulum stress, transcriptomic regulation, and alteration of metabolic enzyme activity. In contrast, the effects of long-term alcohol consumption on hepatic metabolism, particularly in the setting of acute-on-chronic liver diseases such as AH, remain unclear.
We recently reported that AH is associated with a loss of hepatocyte differentiation and liver regeneration.7-9 We hypothesize that the dedifferentiation of hepatocytes that occurs during AH may lead to metabolic reprogramming of the liver, including dysregulation of glucose metabolism. To address this question, we performed an integrative analysis of the hepatic metabolome and transcriptome in patients with AH compared to alcoholic cirrhosis (AC) or unaffected liver tissue. One of the limitations for analysis of the hepatic metabolome is the small quantity of liver tissue available from transjugular biopsies. To ensure a sufficient amount of liver tissue sample, we used a unique cohort of patients with AH (or AC) who were undergoing early liver transplantation.10 Furthermore, to our knowledge, this is the first integrated analysis of the intrahepatic metabolome and transcriptome from individual patients. Our findings show that AH is associated with a unique metabolic signature, which includes dysregulation of key metabolic pathways including glucose metabolism. Additionally, we identified the up-regulated expression and altered epigenetic environment of a recently described hexokinase (HKDC1) in patients with AH. HKDC1 has intermediate affinity for glucose, and its expression may be induced to meet increased metabolic demands during pregnancy11 and in cancer, including hepatocellular carcinoma,12 but its role in alcohol-related liver disease is unclear. Here, we report that increased HKDC1 expression is associated with poor patient outcomes, representing a novel metabolic target for AH.
Materials and Methods
Patient Samples and Clinical Data
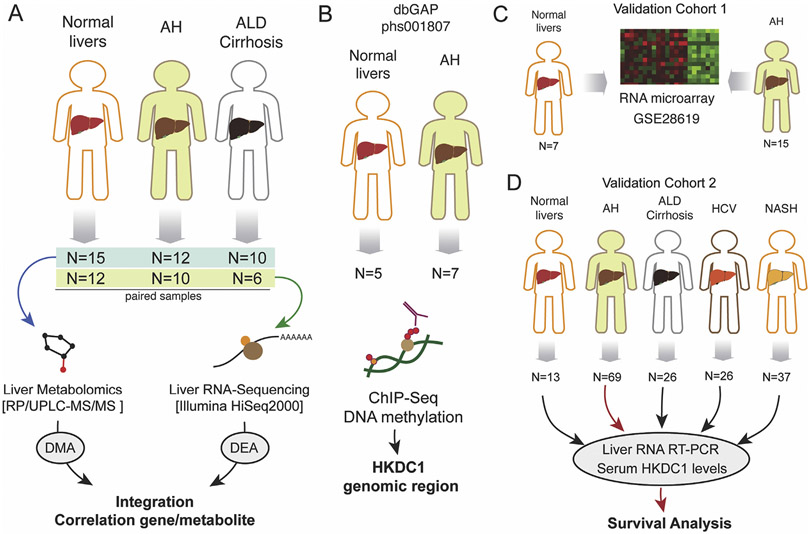
Sections of liver tissue explants from AH were collected at the University Hospital in Lille, France (n = 9) or Johns Hopkins Medical Center (n = 3). Sections of liver tissue explants from AC were collected at the University Hospital in Lille, France (n = 10). All samples were identified by histologic analysis. Control liver tissues were collected from patients undergoing resection for hepatic metastases by the Tissue Procurement Facility at the University of North Carolina at Chapel Hill in Chapel Hill, NC (n = 10) and at the University Hospital in Lille, France (n = 5). We could perform metabolomics analyses from the complete cohort (normal, n = 15; AH, n = 12; and AC, n = 10), whereas for RNA sequencing (RNA-seq), because of limited sample availability, we used lower sample size (normal, n = 12; AH, n = 10; and AC, n = 6) (Figure 1A). All liver tissue samples were snap frozen immediately after collection and stored at −80°C until RNA-seq and metabolomics analysis. To analyze histone chromatin immunoprecipitation coupled to DNA sequencing (ChIP-seq) in the HKDC1 genomic region, we performed in silico analysis from our data previously generated from AH (n= 7) and normal livers (n = 5)9 (Figure 1B). As a validation cohort for our RNA expression study, we used microarray data obtained from patients with clinical, analytical, and histologic features of AH (n = 15) and control individuals (n = 7), as described previously13 (Figure 1C). HKDC1 tissue expression was evaluated in healthy control individuals and in patients with AH (n = 5), ALD cirrhosis (n = 5), and non-ALD cirrhosis (n = 3). Samples were collected at Hospital Santa Creu i Sant Pau, Spain. HKDC1 messenger RNA (mRNA) and serum protein levels were also analyzed in an additional cohort that included the following phenotypes: (1) normal liver tissue (n = 13), (2) patients with AH (n = 69), (3) patients with compensated alcohol-related cirrhosis (n = 26), (4) patients with genotype 1 hepatitis C virus (HCV) chronic infection (n = 26), and (5) patients with morbid obesity and associated nonalcoholic steatohepatitis (NASH) (n = 37) (Figure 1D). Serum HKDC1 was also evaluated in heavy drinkers (n = 10). Samples were collected at Veterans Affairs Long Beach Healthcare System. Figure 1 contains a schematic flowchart of human samples used in this work.
Figure 1.
Schematic work flowchart of human samples analyses. (A) Liver metabolomics and RNA-seq were performed by using a cohort of normal liver fragments from resection for liver metastasis (n = 15), livers from patients with AH (n = 12), and livers from patients with AC (n = 10). (B) Liver ChIP-seq data were retrieved from a previous study (Database of Genotypes and Phenotypes phs001807). Data from a validation cohort of patients with AH (n = 15) and control individuals (n = 7) were retrieved from previous study (GSE28619). (D) Real-time PCR validation and serum HKDC1 measurements were performed in a cohort of normal liver tissue (n = 13) and from patients with AH (n = 69), AC (n = 26), HCV-infected patients (n = 26), and patients with NASH (n = 37). dbGAP, Database of Genotypes and Phenotypes; RP/UPLC-MS/MS, reversed phase/ultraperformance liquid chromatography coupled to tandem mass spectrometry.
Experiments were performed with the approval of the respective institutional review boards and in accordance with the relevant guidelines established at the institution of collection. All patients included in the study gave written informed consent.
Metabolomics of Human Liver Tissues
Sample preparation for metabolomics was performed at Metabolon, Inc, where samples were prepared by using the automated MicroLab STAR system (Hamilton Company). After protein removal, sample extracts were divided into fractions for analysis by either reversed-phase/ultraperformance liquid chromatography coupled to tandem mass spectrometry with positive ion mode electrospray ionization (ESI), reversed-phase/ultraperformance liquid chromatography coupled to tandem mass spectrometry with negative ion mode ESI, or hydrophilic interaction/ultraperformance liquid chromatography coupled to tandem mass spectrometry with negative ion mode ESI. Raw data were peak-identified by using Metabolon’s library of authenticated standards or recurrent unknown entities and quality-control processed by using Metabolon’s hardware and software. Peaks were quantified by using area under the curve, normalized to raw area counts, and rescaled to set the median equal to 1. Statistical significance was determined with 1-way analysis of variance. The false discovery rate was estimated by using q values to account for multiple comparisons. Heatmap visualization was created using the pheatmap package in R (R Foundation for Statistical Computing).
RNA Sequencing of Human Liver Tissues
A subset of liver tissue samples from AH patients (n = 9), AC patients (n = 6), and control individuals (n = 10) were used for total RNA-seq. Sequencing was performed by the University of North Carolina’s High-Throughput Sequencing Facility. Total RNA was extracted from human liver tissues by using the Qiagen AllPrep DNA/RNA/Protein Mini kit, following manufacturer’s instructions. Extracted RNA was analyzed with the Agilent 2100 Bioanalyzer system (Agilent Biotechnologies). High-quality RNA was used for library construction by using the Illumina TruSeq Stranded Total RNA Ribo-Zero Gold kit. Multiplexed samples were sequenced by using the Illumina HiSeq2000 platform using a read length of 2 × 50 bases. Short read alignment was performed using the STAR alignment algorithm with default parameters.14 Differential gene expression was determined by using the DESeq2 package, and data were visualized by using DESeq2, pheatmap, or ggplot2 packages in R. Changes in disease-associated pathways were identified by using Ingenuity Pathway Analysis (IPA) (Qiagen) and visualized by using the ggplot2 package in R.
Integrative Analysis of the Hepatic Metabolome and Transcriptome
Both RNA-seq and metabolomic data sets were filtered for expression level (sum of expression over all samples, >0) and sample variability (standard deviation over all samples, >0). Raw RNA-seq count data and median-scaled metabolomic data were then log2-transformed before all analyses. Pairwise Spearman rank correlation tests were performed between each gene and metabolite by using the base R function cor.test to generate both Spearman rho and a P value estimate; P < .05 was used to determine significance.
Chromatin Immunoprecipitation Coupled to DNA Sequencing of Histone Marks
ChIP-seq was performed by the Mayo Epigenomics Development Laboratory (EDL) using recently described procedures.9 ChIP-seq of liver tissue from 5 control and 7 severe AH explants (provided by the University of Lille, France) were analyzed for 4 histone modifications, using antibodies against histone H3 lysine 27 acetylation (H3K27ac, Cell Signaling no. 8173), histone H3 lysine 27 trimethylation (H3K27me3, Cell Signaling no. 9733), histone H3 lysine 4 monomethylation (H3K4me1, EDL, Mayo Clinic, lot no. 1), and histone H3 lysine 4 trimethylation (H3K4me3, EDL, Mayo Clinic, lot no. 1). Briefly, reads were aligned to the hg19 genome assembly using BWA. The Integrative Genomics Viewer was then used to visualize the H3K4me1, H3K4me3, H3K27ac, and H3K27me3 peaks in the context of the HKDC1 genomic region.
Adenovirus
Human HKDC1 complementary DNA was purchased from the OpenBiosystems clone library. The complementary DNA encoding for HKDC1 was amplified by polymerase chain reaction (PCR) using sequence-specific primers (forward: 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTACCATGTTTGCGGTCC ACTTGATGGC-3′; reverse: 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTAGTTCTCCTTCTGTGCCTGC-3′). The resulting DNA sequence was cloned into the pAd/CMV/V5-DEST Gateway vector (Invitrogen) using the Gateway cloning system (Invitrogen). The resulting adenoviral vector was linearized with the PacI restriction enzyme and transfected into HEK293A cells. After 7 days, the medium and cells were collected, freeze-thawed 3 times, and centrifuged. The supernatant containing the adenovirus encoding HKDC1 was collected for further adenoviral amplification.
Transcript Profiling
Liver RNA-seq data are available in the Gene Expression Omnibus (GSE142530). Liver microarray data are available in the Gene Expression Omnibus (GSE28619). Liver ChIP-seq peak analysis is available in the Database of Genotypes and Phenotypes (phs001807.v1.p1). Liver raw and processed metabolomic data are provided in the Supplementary Materials.
Results
Patients With Alcoholic Hepatitis Exhibit a Unique Hepatic Metabolome
Metabolomics and transcriptomics were performed on liver tissue from normal control individuals, patients with AH, and patients with AC. Half of the patients with AH were male, with a mean age of 56 years, similar to the unaffected control (66% male; mean age, 60 years) and AC (60% male; mean age, 51 years) cohorts. Patients with AH had a significantly higher Model for End-Stage Liver Disease score (32) and serum aspartate aminotransferase levels (123 U/L) than those with AC (Model for End-Stage Liver Disease, 25; aspartate aminotransferase level, 71 U/L). Clinical characteristics of the control, AC, and AH cohorts used for metabolomic and transcriptomic analysis are shown in Supplementary Table 1.
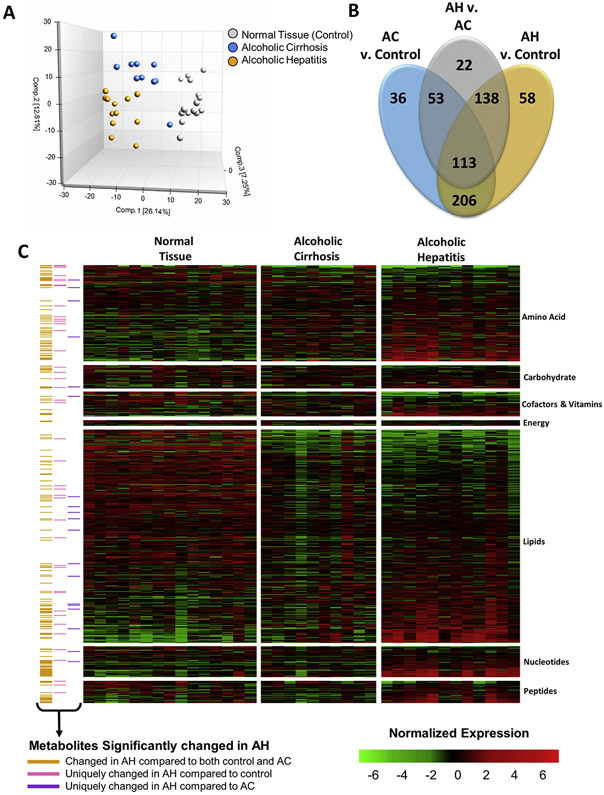
Metabolomic analysis of liver tissue from patients with AH or AC or normal liver tissue detected a total of 869 metabolites across all samples, including 176 intermediates of amino acid metabolism, 43 intermediates of carbohydrate metabolism, 45 cofactors and vitamins, 11 intermediates of energy metabolism, 390 intermediates of lipid metabolism, 57 intermediates of nucleotide metabolism, 41 intermediates of peptide metabolism, and 87 xenobiotics. Principal component analysis showed that the 3 patient cohorts were distinguishable by distinct metabolic signatures (Figure 2A). Indeed, 515 metabolites (254 increased, 261 decreased) were significantly changed in the AH cohort compared to the control cohort, and 326 (225 increased, 101 decreased) were significantly changed in the AH cohort compared to the AC cohort. There were 206 metabolites that differentiated the AH and AC cohorts from the control cohort (Figure 2B), consistent with the similar etiologies of these alcohol-induced chronic liver diseases. Importantly, 58 metabolites (24 increased, 34 decreased) were uniquely changed in patients with AH compared to control individuals (Supplementary Table 2), and 22 metabolites were uniquely changed between the AH and AC cohorts (Supplementary Table 3). The metabolites significantly altered in AH were involved in amino acid, carbohydrate, vitamin, energy, and nucleotide metabolism (Figure 2C, yellow annotation). These changes in hepatic metabolite levels defined an AH-specific metabolite profile.
Figure 2.
The hepatic metabolome is altered in patients with AH. Metabolomics was performed at Metabolon, Inc, as described in the Materials and Methods section. (A) Plot of the Principal Component Analysis of the hepatic metabolome of normal liver tissue (control), AC, and AH. (B) Venn diagram representing the metabolites found to be significantly changed by multiple comparison testing between AH and control, between AC and control, and between AH and AC. (C) Heatmap showing the normalized expression of all metabolites detected by metabolomics analysis of the liver tissue. Annotation indicates which metabolites were significantly changed in patients with AH compared to either control individuals or patients with AC (yellow), only significantly changed in patients with AH compared to control individuals (gray), and only significantly changed in patients with AH compared to patients with AC (blue).
Profound Carbohydrate Metabolism Dysregulation in Alcoholic Hepatitis
To understand the effect of chronic alcohol consumption on hepatic metabolism in the setting of AH, we performed a focused analysis of the metabolites involved in glycolysis/gluconeogenesis, the tricarboxylic acid (TCA) cycle, and monosaccharide and disaccharide metabolism (Table 1). Liver tissue from patients with AH showed a unique profile of carbohydrate metabolism intermediates compared to either the control or AC groups. Hepatic glucose, 3-phosphoglycerate, and phosphoenolpyruvate (PEP) levels were all significantly decreased in AH compared to the control or AC cohorts. In contrast, the levels of glucose-6-phosphate (G6P) were increased by more than 200% in the AH compared to the control or AC cohorts. Citrate and aconitate, the products of the first 2 steps of the TCA cycle, were also increased in AH by 170% and 380%, respectively, compared to the control cohort. The TCA cycle intermediates succinyl-carnitine (a surrogate marker for succinyl-coenzyme A [CoA]), fumarate, and malate, as well as the monosaccharide mannose, were all significantly decreased in AH compared to control or AC samples. Cofactors are important for the allosteric regulation of glucose and energy metabolism; thus, changes in cofactor levels or ratios can affect enzyme activity. Liver tissue from patients with AH exhibited an 80% depletion of oxidized nicotinamide adenine dinucleotide (NAD+) stores compared to control samples, which contributed to a significant decrease in the NAD+/reduced nicotinamide adenine dinucleotide (NADH) ratio. Collectively, these results support our hypothesis that AH is characterized by profound reprogramming of glucose metabolism.
Table 1.
Hepatic Metabolite Levels in Fragments of Normal Liver Tissue (Control), AC, or AH
| Biochemical | Control (n = 14) | AC (n = 6) | AH (n = lO) |
|---|---|---|---|
| Glycolysis/gluconeogenesis | |||
| Glucose | 1.22 ± 0.14 | 1.11 ± 0.15 | 0.61 ± 0.lQa,b |
| G6P | 0.84 ± 0.12 | 0.73 ± 0.24a | l.73 ± 0.27a,b |
| Fructose 6-phosphate | 1.07 ± 0.20 | 0.86 ± 0.14 | 1.35 ± 0.34 |
| Fructose 1,6-diphosphate/glucose 1,6-diphosphate | 0.66 ± 0.14 | l.35 ± 0.25a | l.15 ± 0.53a |
| Dihydroxyacetone phosphate (DHAP) | 1.05 ± 0.16 | 1.09 ± 0.19 | 0.98 ± 0.12 |
| 3-Phosphoglycerate | 1.71 ± 0.24 | 1.34 ± 0.19 | 0.79 ± 0.05a,b |
| Glycerate | 1.36 ± 0.16 | 1.01 ± 0.17 | 1.34 ± 0.15 |
| PEP | 1.53 ± 0.18 | 1.43 ± 0.17 | 0.82 ± 0.06a,b |
| Pyruvate | 0.90 ± 0.14 | 1.25 ± 0.39 | l.91 ± 0.32a |
| Lactate | 0.90 ± 0.05 | 0.99 ± 0.03 | l.06 ± 0.04a |
| 1,5-Anhydroglucito | 1.42 ± 0.19 | 0.98 ± 0.18 | 0.89 ± 0.16a |
| Glycogenesis | |||
| UDP-glucose | 2.78 ± 0.92 | 0.92 ± 0.23 | 0.58 ± 0.lP |
| TCA cycle | |||
| Citrate | 0.70 ± 0.07 | l.62 ± 0.35a | 1.21 ± 0.15a |
| Aconitate (cis or trans) | 0.32 ± 0.03 | 0.76 ± 0.11a | l.22 ± 0.23a |
| Alpha-ketoglutarate | 1.36 ± 0.34 | 1.44 ± 0.56 | 1.81 ± 0.29 |
| Succinylcarnitine (C4-DC) | 3.34 ± 0.54 | l.48±0.6P | 0.59 ± 0.18a,b |
| Succinate | 5.04 ± 1.78 | 2.63 ± 1.97a | l.41 ± 0.37a |
| Fumarate | 1.14 ± 0.06 | 1.11 ± 0.11a | 0.82 ± 0.06a,b |
| Malate | 1.10 ± 0.05 | 1.02 ± 0.10a | 0.84 ± 0.05a,b |
| Tricarballylate | 3.95 ± 1.01 | 0.60 ± 0.08a | l.01 ± 0.32a |
| 2-Methylcitrate/homocitrate | 1.17 ± 0.09 | 0.52 ± 0.19a | l.30 ± 0.27b |
| Monosaccharide and disaccharide | |||
| Mannitol/sorbitol | 0.84 ± 0.14 | l.33 ± 0.24a | l.19 ± 0.15a |
| Fructose | 0.99 ± 0.15 | 1.01 ± 0.13 | 1.06 ± 0.06 |
| Galactose 1-phosphate | 0.35 ± 0.07 | 0.87 ± 0.21a | l.45 ± 0.20a |
| Galactonate | 1.29 ± 0.20 | 1.19 ± 0.22 | 1.11 ± 0.22 |
| Mannose | 1.42 ± 0.18 | 1.15 ± 0.17 | 0.79 ± 0.08a,b |
| 2-ketoguIonate | 1.03 ± 0.26 | ND | ND |
| Cofactors | |||
| NAD+ | 2.17 ± 0.43 | 0.89 ± 0.15a | 0.37 ± 0.07a,b |
| NADH | 1.64 ± 0.76 | 0.43 ± 0.02a | 0.41 ± 0.0la |
| NAD+/NADH | 2.29 ± 0.27 | 2.06 ± 0.36 | 0.91 ± 0.18a,b |
| Adenosine 5′-monophosphate | 1.95 ± 0.29 | 0.95 ± 0.18a | 0.61 ± 0.11a |
| Adenosine 5′-diphosphate | 0.71 ± 0.09 | 0.96 ± 0.24 | 0.74 ± 0.13 |
NOTE. Data are shown as mean ± standard error of the mean.
P < .05 compared to control.
P < .05 compared to AC.
Deregulated Hepatic Messenger RNA Expression of Enzymes Involved in Carbohydrate Metabolism in Alcoholic Hepatitis
To explore changes in the hepatic expression of genes encoding enzymes involved in glucose metabolism in the context of AH, total RNA-seq of control liver tissue or tissue from patients with AC or AH was performed. Principal component analysis showed observable grouping of the 3 disease types into separate clusters (Supplementary Figure 1A). Differential gene expression analysis showed that nearly 7500 genes were differentially expressed in AH compared to control samples, with more than 5000 of those genes being uniquely altered in AH (Supplementary Figure 1B). Consistent with previous findings, KRT23 and extracellular matrix–associated genes (eg, MMP2, MMP14, SPP1, LAMA2) were significantly increased in patients with ALD (Supplementary Figure 1C-E). IPA was performed to identify disease-associated pathways up-regulated and down-regulated in AH compared to normal tissue (Supplementary Figure 2). Consistent with chronic liver disease, IPA identified liver inflammation as the third most up-regulated disease-associated pathway along with “increased response of liver” and “inflammation of organ.” IPA analysis predicted changes in several metabolic pathways, including down-regulation of d-glucose concentration, quantity of carbohydrates, and metabolism of carbohydrates. Thus, these results suggest attenuated intermediate metabolism in AH, which is consistent with decreased hepatic glucose levels and altered levels of intermediates of carbohydrate metabolism identified by metabolomics.
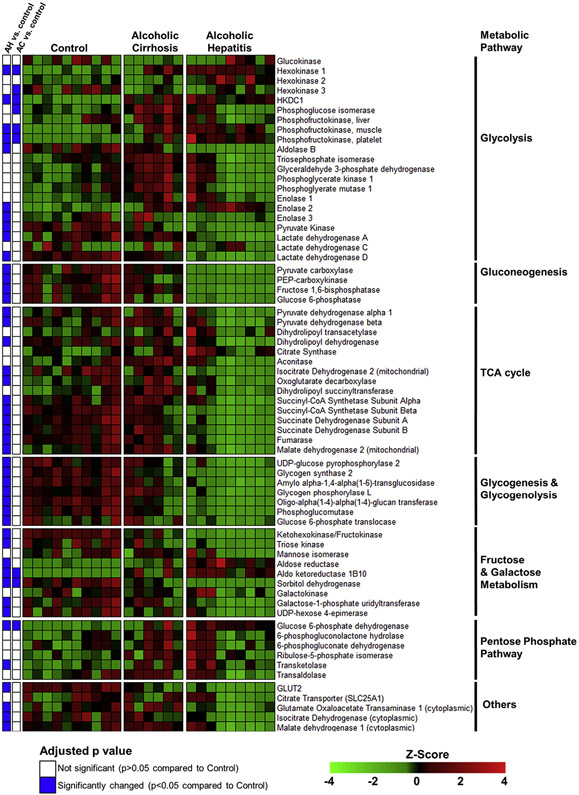
To further explore changes in the gene expression of enzymes involved in intermediate metabolism, we performed a focused analysis on genes encoding the enzymes of glycolysis, gluconeogenesis, the TCA cycle, glycogen metabolism, and monosaccharide and disaccharide metabolism (Figure 3). The mRNA expression of glucokinase and the liver-enriched phosphofructokinase (PFKL), which govern 2 of the irreversible steps of glycolysis, were not significantly changed in patients with AH. However, the phosphofructokinase isozymes PFKM and PFKP, which have appreciable hepatic expression levels, were increased by 180% and 460%, respectively, in AH compared to control samples. HKDC1, a novel hexokinase that converts glucose into G6P, was increased by 600% in patients with AH compared to control individuals. The mRNA expression levels of enolase 3, pyruvate kinase, and lactate dehydrogenase enzymes A and D were significantly decreased in AH, whereas enolase 2 mRNA expression was significantly increased. Pyruvate kinase, which produces pyruvate via the third irreversible glycolytic reaction, was decreased to 35% of control levels in AH. The mRNA expression levels of enzymes that regulate reactions unique to gluconeogenesis were all decreased in AH compared to control samples, including pyruvate carboxylase, malate dehydrogenase 1 and 2, PEP-carboxykinase (PCK1), fructose 1,6-bisphosphatase, and glucose 6-phosphatase. Similarly, enzymes of the TCA cycle tended to be significantly decreased in AH, including the pyruvate dehydrogenase E1 beta subunit (PDHB), pyruvate dehydrogenase complex component X (PDHX) and dihydrolipoamide dehydrogenase enzymes of pyruvate dehydrogenase complex, isocitrate dehydrogenase (NADP+) 2 (IDH2), succinate-CoA ligase (SUCLG) 1 and SUCLG2, succinate dehydrogenase complex (SDH) A and SDHB, fumarase, and malate dehydrogenase 2 (MDH2). The mRNA expression levels of all identified enzymes involved in glycogen metabolism were significantly decreased, including glycogen synthase, which catalyzes the rate-limiting step of glycogen synthesis, and glycogen phosphorylase, which catalyzes the rate-limiting step of glycogen breakdown. Importantly, mRNA expression levels of only 8 of these enzymes were significantly altered in the AC compared to the control group, suggesting that dysregulation of intermediate metabolism gene expression is unique to AH.
Figure 3.
Heatmap showing normalized mRNA expression levels of enzymes involved in intermediate metabolism. RNA-seq of human liver tissue was performed as described in the Materials and Methods section. A subset of genes involved in intermediate metabolism pathways is shown. The color scale represents sample Z-score. Statistical significance was determined by using the DESeq2 package and was adjusted for multiple comparisons. Statistical significance is reported as adjusted P values compared to control.
Glucose Metabolism Reprogramming in Alcoholic Hepatitis Shown by Integrated Analysis of Hepatic Metabolomics and Transcriptomics
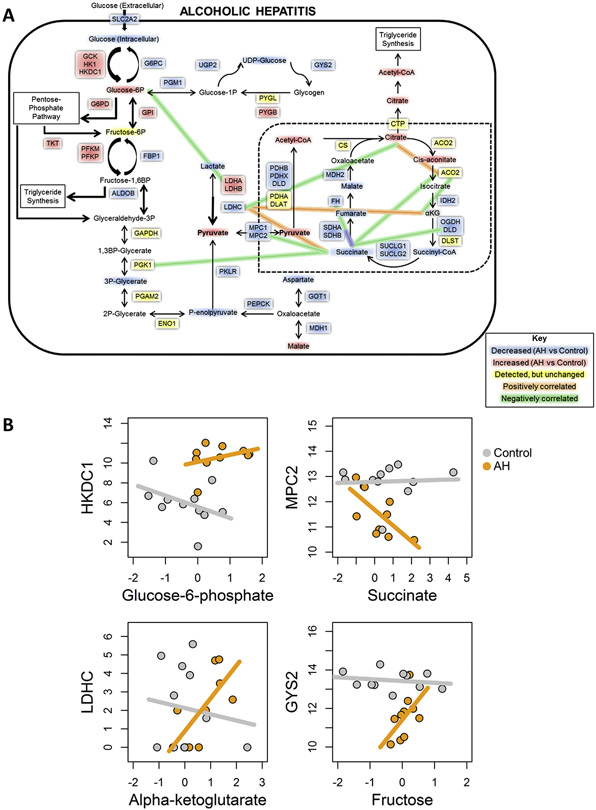
We performed a systems biology integrative analysis examining changes in expression and correlation between genes and metabolites related to glycolysis, gluconeogenesis, and the TCA cycle. Using a network-based approach, we identified several possible metabolite sinks, including G6P, acetyl-CoA, and citrate (Figure 4A). These metabolites showed increased abundance in AH versus control samples, along with a decrease of downstream intermediates, suggesting that there are multiple points of localized metabolite buildup. The TCA cycle in particular shows multiple metabolites with significantly decreased levels, suggesting that normal liver energetic processes are disrupted by AH in favor of alternate pathways, such as triglyceride synthesis and the pentose-phosphate pathway (Figure 4A). These changes in metabolites are also accompanied by altered expression levels of related genes, such as HKDC1, glucose-6-phosphate dehydrogenase (G6PD), and PDHB, which may contribute to these metabolite sinks or to altered glucose metabolism, for example, by promoting use of the pentose-phosphate shunt. Glycogen synthesis also appears to be affected by AH, with several key enzymes showing significant down-regulation; at the same time, glycogenolysis appears to be slightly up-regulated (Figure 3). This suggests that AH liver is incapable of proper glucose use and storage, which may contribute to the lack of liver regeneration and, ultimately, to the hepatocellular failure seen in these patients.
Figure 4.
Integrated analysis of metabolomics and RNA-seq. (A) Metabolic map showing significant changes in gene expresses (boxes) and intermediates (text) of glucose metabolism in patients with AH compared to normal control samples. (B) Representative plots showing correlations between genes and metabolites for control (gray) and AH (orange) samples.
We also performed correlation analysis between gene expression and metabolite changes. We found no significant correlations in AH or control samples (Figure 4B, panel 1). However, there were significant inverse correlations in AH only (Figure 4B, panel 2), opposite correlations in AH and control livers (Figure 4B, panel 3), and significant direct correlations in AH only (Figure 4B, panel 4). The differences in enzyme expression/metabolite correlations between control individuals and patients with AH were mostly related to the TCA cycle (Figure 4A and Supplementary Figure 3). Succinate in particular appears strongly inversely correlated with several TCA cycle-relevant genes, suggesting the presence of a bottleneck in the mitochondrial Krebs cycle. Taken together, our results suggest that dysregulation of glucose metabolism, through altered storage/release mechanisms, use of alternate energetic pathways, and metabolite sinks and bottlenecks, could contribute to abnormal liver function in AH.
Relationship Between Hepatic HKDC1 Expression and Clinical Outcomes in Patients With Alcoholic Hepatitis
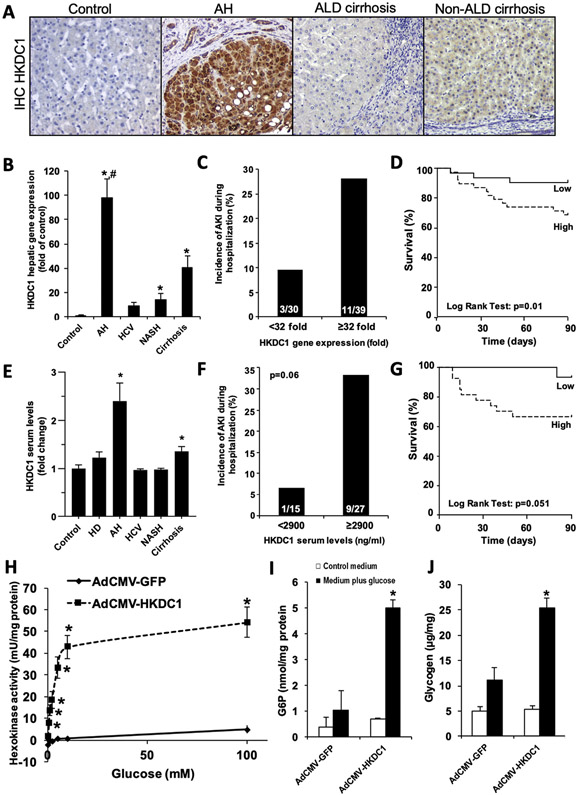
RNA-seq analysis showed that HKDC1 is increased more than 6-fold in AH compared to control samples. HKDC1 is a recently discovered hexokinase that shares nearly 70% nucleotide and amino acid sequence homology with hexokinase 1 (HK1)15 and contributes to cellular hexokinase activity in vitro.16,17 Analysis of microarray data18 confirmed increased HKDC1 as the most up-regulated kinase in AH (Supplementary Figure 4). Therefore, we performed a focused analysis on the role of HKDC1 in AH. HKDC1 expression was found to be robustly induced specifically in hepatocytes in AH, as determined by immuno-histochemical analysis (Figure 5A and Supplementary Figure 5). Increased hepatic HKDC1 mRNA was validated by real-time PCR in cohorts of patients with biopsy-proven AH, other etiologies of liver disease, or control individuals. Expression of HKDC1 was increased by nearly 100-fold in livers from patients with AH than in healthy control individuals (Figure 5B). HKDC1 mRNA was also overexpressed in livers from patients with NASH and ACs. The incidence of AKI was significantly increased in patients with AH with high HKDC1 mRNA expression (>32-fold expression) compared to patients with AH with low HKDC1 mRNA expression (Figure 5C). High HKDC1 mRNA expression was also a good predictor of short-term mortality, as determined by Kaplan-Meier analysis (area under the receiver operating characteristic curve, 0.79; 95% confidence interval, 0.62–0.91) with a cutoff value of 32-fold (Figure 5D). Confirmatory analysis of HKDC1 in liver and plasma was performed in a separate cohort of patients with AH and control individuals. Circulating HKDC1 levels were higher in AH and compensated patients with AC compared to patients with other liver diseases and heavy drinkers (Figure 5E). Additionally, there was a trend (P = .06) toward increased incidence of AKI during hospitalization in patients with high serum HKDC1 levels (>2900 UI/dL) compared to patients with low serum HKDC1 levels (Figure 5F) as well as a trend toward decreased short-term survival (area under the receiver operating characteristic curve; 0.69; 95% confidence interval, 0.50–0.86) in patients with high serum HKDC1 levels, as determined by Kaplan-Meier analysis. These results suggest that increased hepatic expression of HKDC1 is accompanied by a raise in its serum levels and that HKDC1 could be useful as a noninvasive biomarker to predict short-term mortality in patients with AH.
Figure 5.
Increased hepatic HKDC1 mRNA expression and serum HKDC1 levels in patients with AH are associated with higher incidence of AKI and decreased short-term survival. (A) Representative pictures of HKDC1 immunostaining (original magnification, ×200) in control livers and in livers from patients with AH, ALD cirrhosis, and non-ALD cirrhosis. (B) Gene expression of HKDC1 was analyzed in control (n = 13), AH (n = 69), HCV (n = 26), NASH (n = 37), and compensated AC liver tissue (n = 26). Data are shown as mean ± standard error of the mean. *P < .05 compared to control samples, #P < .05 compared to the other groups. (C) High HKDC1 mRNA expression (≥32-fold of control) is associated with the development of AKI during hospitalization in patients with AH. (D) Kaplan-Meier curve showing the 90-day survival in patients with AH with high (>32-fold of control) or low (<32-fold of control) hepatic HKDC1 expression. (E) Fasting serum levels of HKDC1 were evaluated in samples from healthy control individuals (n = 14), heavy drinkers (n = 10), and in patients with AH (n = 46), HCV (n = 17), NASH (n = 12), and compensated AC (n = 22). Data are shown as mean ± standard error of the mean; *P < .05 compared to all other groups. (F) High HKDC1 serum level (>2900 ng/mL) is associated with the development of AKI during hospitalization in patients with AH. (G) Kaplan-Meier curve showing the 90-day survival in patients with AH with high (>2900 ng/mL) or low (<2900 ng/mL) serum HKDC1 level. (H–J) Rat primary hepatocytes were treated with either AdCMV-HKDC1 or AdCMV-GFP as described in the Materials and Methods section. Cells were serum- and glucose-starved overnight followed by incubation with 30 mmol/L glucose for 4 hours. Hexokinase activity (H), G6P levels (I), and glycogen content (J) were determined as described in the Materials and Methods section. Data represent the mean for 3 independent experiments ± standard error; *P < .05 compared to the AdCMV-GFP–treated cells in medium plus glucose.
Demonstration of Glucose-Phosphorylating Activity by HKDC1
Since immunohistochemistry showed a marked increased HKDC1 expression in hepatocytes (Figure 5A), the effect of ethanol exposure on HKDC1 expression in HepG2 cells was determined. Ethanol significantly increased HKDC1 mRNA expression by 50% in HepG2 cells (Supplementary Figure 6). Previous studies have shown that the overexpression of HKDC1 leads to increased hexokinase activity in HepG2 cells, by using plasmid transfection.17 HKDC1 is very active in tumoral cell lines, but its capacity to overcome or replace the strong HK1, HK2, HK3, and glucokinase (GCK) activities in primary hepatocytes has not been fully explored. By using adenovirus-mediated HKDC1 overexpression, we tested the contribution of HKDC1 to cellular hexokinase activity in primary rat hepatocytes. HKDC1 overexpression significantly increased glucose phosphorylating activity in primary rat hepatocytes compared to Ad-GFP–infected control (Figure 5H). G6P production was also increased by more 5-fold (Figure 5I) in Ad-HKDC1–transduced cells. Because G6P is an allosteric activator of glycogen synthase, the rate-limiting enzyme in glycogen synthesis, we also determined the effect of HKDC1 overexpression on hepatocyte glycogen levels. Primary rat hepatocytes overexpressing HKDC1 produced significantly more glycogen compared to GFP-expressing controls (Figure 5J). These results confirm both that ethanol can directly induce hepatocyte HKDC1 expression and that HKDC1 overexpression promotes hepatocyte glucose phosphorylation and increase in glycogen synthesis.
Chromatin Organization on the HKDC1 Promoter Region
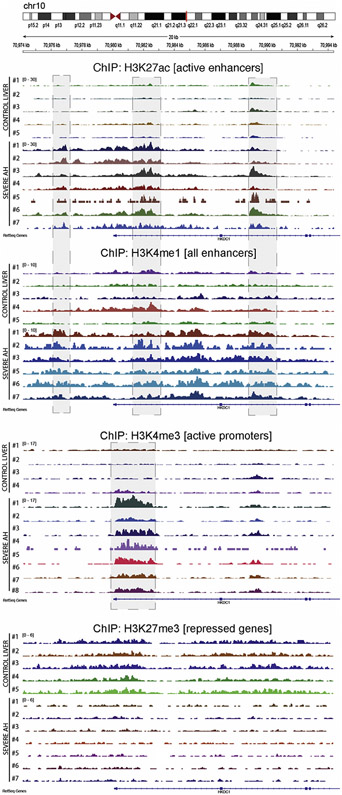
We recently described that the livers of patients with alcoholic hepatitis undergo significant epigenetic modification.9 Compared to normal livers, AH livers present a severe down-regulation of metabolically essential genes such as hepatocyte nuclear factor 4 (HNF4A) targets. The chromatin landscape of their regulatory regions show an increase in silencing (H3K27me3) and a decrease in activating histone marks (H3K4me1, H3K4me3, H3K27ac), which correlates with the hepatocellular failure in these patients.9 We thus explored chromatin states of HKDC1 region. In agreement with the significant increase in HKDC1 expression, patients with AH displayed an altered chromatin pattern in HKDC1 genomic region, showing an increase in density and signal of H3K27ac peaks in enhancers 4–5 kilo base pairs upstream of the transcription start site and within the first and second introns (Figure 6A and B). Interestingly, the first 2 sites are expression-quantitative trait loci for the HKDC1 gene and have been related to gestational diabetes.17 Further studies with larger AH populations might evaluate if single-nucleotide polymorphisms in these expression-quantitative trait loci could explain AH severity or survival. Trimethylation of H3K4 was increased in HKDC1 promoter in patients with AH, alongside increased H3K27ac (Figure 6C). Contrary to normal livers, the trimethylation of H3K27 was absent in livers of patients with AH, which could partially explain the activation of HKDC1 transcription (Figure 6D).
Figure 6.
Genomic view of histone ChIP-seq data showing increased activity in HKDC1 regulatory regions. Data from ChIP-seq in livers from patients with AH (n = 7) and normal livers (n = 5) is presented in the genomic context of the HKDC1 gene. H3K27ac and H3K4me1 were used to mark the activity and location of enhancers, respectively. H3K4me3 was used to verify the activity of the promoter. The H3K27me3 mark indicates the presence of gene silencing. Shadow squares indicate the detected peaks by MACS2.
Discussion
The specific treatment of patients with AH has not evolved over the 4 decades since prednisolone, the only effective drug, was proposed in 1971.19 In the absence of suitable animal models of true AH, there is an urgent need for translational studies using human samples to identify molecular drivers. Previous studies investigating mechanisms of AH have focused on the activation of hepatic inflammatory pathways.20 As a consequence, current efforts to develop novel therapies are focused on targeting inflammatory mediators such as the inflammasome or lipopolysaccharide. However, because of the profound hepatocellular dysfunction in AH and the central role of hepatocytes in intermediate metabolism, it is plausible that glucose metabolism is altered in these patients and could represent a potential avenue for biomarker and drug discovery. Despite the fact that alcohol exposure is known to affect hepatocyte metabolism both in vivo and in vitro,21-23 the hepatic metabolome has not been studied in patients with AH. The current study was designed to fill this knowledge gap.
Previous studies from our group have shown that AH is characterized by hepatocyte dedifferentiation and loss of hepatocyte regenerative capacity.7-9 We hypothesize that hepatocyte dedifferentiation is associated with dysregulation of glucose homeostatic function of hepatocytes, leading to metabolic reprogramming. To address this question, we performed metabolomic and transcriptomic analyses of human liver tissue. Because the limited size of biopsy tissue samples precludes their use for discovery-based metabolite analysis, we used explant liver tissue collected from a unique set of AH patients who underwent early liver transplantation.10 We also analyzed explant tissues from patients with AC as well as fragments of normal liver tissue from patients undergoing hepatic resection for liver metastases. Using an integrative omics approach, the current study showed that the hepatic metabolome is significantly altered in AH and is characterized by profound glucose metabolism reprogramming. Indeed, we found that more than 450 metabolites were significantly changed in AH compared to normal liver tissue, 200 of which were not altered in AC, suggesting that these metabolites are uniquely altered during AH. As expected, based on the natural history of alcohol-induced liver disease and previous serum studies,24 we found that AH was associated with dramatic changes in hepatic lipid metabolism, including increased monoacylglycerols, polyunsaturated fatty acids, and free fatty acid levels compared to both normal control and AC. We further report significant changes in hepatic levels of intermediates of glucose metabolism and concordant changes in the mRNA expression of key metabolic enzymes (eg, HKDC1, G6PC, glycogen synthase 2 [GYS2]) that correlate with changes in hepatic metabolite levels. In addition, network analysis suggested that alternate energetic pathways, such as the pentose-phosphate shunt and triglyceride synthesis, were the way glucose is used instead of feeding the TCA cycle.
Although previous studies investigating metabolic changes during AH have identified changes in serum levels of glucose metabolism intermediates,24 to our knowledge, we are the first to describe global changes in hepatic glucose metabolism in the context of AH. Our analysis showed significant increases in early intermediates of glycolysis including G6P and fructose 1,6-diphosphate/glucose 1,6-diphosphate despite lower levels of intracellular glucose in patients with AH. Our transcriptomic data suggest that in the context of AH, increased expression of hexokinases, including HKDC1, and a concomitant decrease in G6PC expression may contribute to accumulation of G6P. Previous studies have shown that G6PC and phosphoenolpyruvate carboxykinase (PEPCK) can be down-regulated via the interleukin 6–signal transducer and activator of transcription 3 pathway, which is known to be activated in AH.25 Furthermore, decreased mRNA expression of phosphoglucomutase-1 (PGM1), which diverts G6P into glycogenesis, may prevent storage of excess G6P into glycogen. Decreased use of G6P for glycogenesis is further supported by decreased mRNA expression of UGP2 and glycogen synthase 1 (GYS1) as well as reduced levels of uridine diphosphate–glucose, a glycogen precursor. These findings are consistent with previous studies showing decreased glycogen stores and reduced mRNA expression of glycogen synthase and phosphorylase in cirrhotic liver.6 Significant changes in fructose 1,6-bisphosphate, 3-phosphoglycerate, PEP, and pyruvate further suggest dysregulation of hepatic glucose metabolism in the context of AH. Interestingly, AH was also associated with a robust decrease in PEPCK, an irreversible reaction of gluconeogenesis. In mice, the loss of PEPCK has been shown to contribute to the development of hepatic steatosis, consistent with increases in lipid intermediates that were found in AH.25 Further studies should investigate the functional effects of these changes on alcohol-induced liver injury in true mouse models of AH.
Our integrative analysis also identified changes in the intermediates of the TCA cycle. The TCA cycle is compartmentalized to mitochondria, where it contributes to adenosine triphosphate production through the oxidation of acetyl-CoA derived from carbohydrates (eg, glucose), fats, and proteins. We found that AH was associated with a 2-fold increase in pyruvate, a glycolytic product that can be converted into acetyl-CoA. Citrate and aconitate, the products of the first 2 steps of the TCA cycle, were also increased in AH. In contrast, AH was associated with lower levels of the intermediates (eg, succinyl CoA, succinate, fumarate) and enzyme gene expression (eg, SUCLG, SDH) involved in the final steps of the TCA cycle. The mechanism of ethanol hepatotoxicity includes the depletion of glutathione (GSH) and the consequent production of NADH, reducing the NAD+/NADH ratio. Thus, it is possible that the lack of NAD+ undermines the sequence flow of the TCA cycle. Metabolomic analysis did not allow discrimination between mitochondrial and cytosolic levels of intermediates; however, cellular localization of pyruvate, acetyl-CoA, and citrate play an important role in how they are used by the cells. Therefore, despite increases in acetyl-CoA and citrate, decreased levels of succinylcarnitine and succinate, along with concomitant increases in lipid intermediates, could indicate decreased TCA cycle activity. In this case, increased acetyl-CoA and citrate levels may favor fatty acid synthesis, contributing to lipogenesis and fat accumulation associated with ALD.26,27 As one of the first studies to investigate the effects of long-term alcohol consumption on intrahepatic metabolism, our results suggest that chronic alcohol consumption can disrupt the hepatic redox status, similar to acute intoxication.
A striking finding in our study is the demonstration that HKDC1, a recently discovered hexokinase,16,17 is the most up-regulated kinase in the whole transcriptome of patients with AH. HKDC1 was identified as the most up-regulated kinase in our study and was confirmed to be up-regulated in additional AH cohorts compared to control cohorts. Previous studies have shown that HKDC1 shares significant homology with the canonical HK1 hexokinase and that HKDC1 contributes to glycogen phosphorylation.17,28,29 In our study, we found that increased hepatic HKDC1 expression in AH was associated with incidence of AKI, an early complication associated with poor survival in patients with AH.2 In addition, high HKDC1 expression was associated with decreased 90-day survival AH. In vitro studies using primary rat hepatocytes supported a functional role of HKDC1 in glucose metabolism in the liver and in hepatocytes specifically. Thus, increased HKDC1 serum levels and concomitant increases in hepatocyte expression in AH suggest that HKDC1 may serve as a novel biomarker of AH and a potential therapeutic target. The mechanisms by which the expression of HKDC1 is associated with poorer outcome in AH are unclear and deserve further study. The hexokinase isoforms expressed in the healthy liver have a low avidity to phosphorylated glucose, so it is mostly stored as glycogen for further use by organs such as the brain and heart. The fact that hepatocytes in AH de novo express a hexokinase with intermediate avidity to use glucose for energy generation represents a sign of profound metabolic reprogramming. It is unclear, however, whether HKDC1 overexpression is an epiphenomenon reflecting profound epigenome reprogramming in hepatocytes or is actually a molecular driver leading to hepatocyte dysfunction and loss of mature synthetic functions. Further gain- and loss-of-function studies are needed to assess if HKDC1 mediates hepatocyte failure in AH.
Our integrated analysis of human liver tissues was faced with certain limitations. First, our discovery-based metabolomics approach did not capture the complete hepatic metabolome and was unable to discriminate important intermediates of carbohydrate metabolism, including fructose 6-phosphate and glycogen. Additionally, metabolism is dynamically regulated at multiple levels, including transcriptional regulation, protein phosphorylation, allosteric activation, and inhibition, and via complex protein-protein interactions. Because of limitations of tissue availability, we were restricted to transcriptomic and metabolomic analysis for this study and were unable to analyze changes in protein expression or enzyme activity in these patient cohorts. Therefore, we can only infer changes in enzyme activity based on metabolite level. As early liver transplants become more common for patients with AH, the field should focus on strengthening translational research opportunities that make explanted liver tissue available for more focused metabolic studies in this understudied patient population.
In conclusion, to our knowledge, we report the first integrative analysis of the intrahepatic metabolome and transcriptome. Our integrated correlative analysis suggests that glucose is not effectively being stored as glycogen, used to produce energy via the TCA cycle or to produce glucose. Instead, glucose appears to be accumulating as G6P, preventing meaningful energy use or storage. This study expands on previous reports from our group and others, which have reported abnormal gene expression involved in multiple pathways but not in intermediate metabolism. Importantly, we performed a focused analysis that identified HKDC1 as a potential biomarker of AH. In vitro studies support the hypothesis that increased HKDC1 expression in the liver of patients with AH can contribute to altered glucose metabolism.
Supplementary Material
WHAT YOU NEED TO KNOW.
BACKGROUND AND CONTEXT
Alcoholic hepatitis (AH) is characterized by profound changes in hepatic gene expression.
HKDC1 is a recently described hexokinase which is associated with increased metabolic demand of cancer tissues, including HCC.
NEW FINDINGS
AH is associated with increased levels of hepatic G6P and pyruvate, and alterations of intermediates of the TCA cycle, glycogenesis, and mono- and disaccharide metabolism.
Liver tissue from AH exhibits a massive up-regulation of HKDC1 in hepatocytes, which is associated with worse clinical outcomes.
LIMITATIONS
Human liver tissue availability imposes limitations in the variety of experiments we can perform.
IMPACT
Dysregulated hepatic glucose metabolism represents a novel pathway for drug development in AH. HKDC1 appears to be a promising prognostic biomarker and a potential molecular target for AH.
Acknowledgments
The authors acknowledge the Lineberger Bioinformatics Core of the University of North Carolina at Chapel Hill for their support.
Funding
Jose Altamirano was partially supported by the Mexican National Council of Science and Technology (Mexico City, Mexico) for his predoctoral stay in Barcelona, Spain. Paul Sancho-Bru was supported by grants from Fondo de Investigación Sanitaria Carlos III, cofinanced by Fondo Europeo de Desarrollo Regional, Unión Europea, “Una manera de hacer Europa” PI17/00673 and PI20/00765, and Miguel Servet, CPII16/0004. Andrew Cameron was supported by National Institutes of Health (NIH)/National Institute on Alcohol Abuse and Alcoholism (NIAAA) (grant P50AA027054). Zhaoli Sun was supported by the NIH/NIAAA (grant R24 AA025017). Joan Guinovart was supported by grants from the Spanish MINECO (BFU2017-84345-P) and the CIBER de Diabetes y Enfermedades Metabólicas. Ramon Bataller is supported by the NIH/NIAAA (grants AA026972, AA026978, and AA026264) and National Institute of Diabetes and Digestive and Kidney Diseases (grant P30DK120531).
Abbreviations used in this paper:
- AC
alcoholic cirrhosis
- AH
alcoholic hepatitis
- AKI
acute kidney injury
- ALD
alcoholic liver disease
- ChIP-seq
chromatin immunoprecipitation coupled to DNA sequencing
- CoA
coenzyme A
- EDL
Epigenomics Development Laboratory
- ESI
electrospray ionization
- G6P
glucose-6-phosphate
- G6PC
glucose-6-phosphatase, catalytic subunit
- HCV
hepatitis C virus
- HK
hexokinase
- IPA
Ingenuity Pathway Analysis
- mRNA
messenger RNA
- NAD+
oxidized nicotinamide adenine dinucleotide
- NASH
nonalcoholic steatohepatitis
- PCR
polymerase chain reaction
- PEP
phosphoenolpyruvate
- PEPCK
phosphoenolpyruvate carboxykinase
- RNA-seq
RNA sequencing
- TCA
tricarboxylic acid cycle
Footnotes
CRediT Authorship Contributions
Veronica Massey, PhD (Data curation: Equal; Formal analysis: Equal; Investigation: Equal; Methodology: Equal; Validation: Equal; Visualization: Equal; Writing – original draft: Equal; Writing – review & editing: Equal); Austin Parrish, PhD (Data curation: Equal; Formal analysis: Equal; Investigation: Equal; Methodology: Equal; Visualization: Equal; Writing – original draft: Equal; Writing – review & editing: Equal); Josepmaria Argemi, MD, PhD (Data curation: Equal; Formal analysis: Equal; Investigation: Equal; Methodology: Equal; Visualization: Equal; Writing – original draft: Equal; Writing – review & editing: Equal); Montserrat Moreno, PhD (Data curation: Supporting; Formal analysis: Supporting; Investigation: Supporting; Methodology: Supporting); Aline Mello, PhD (Data curation: Supporting; Formal analysis: Supporting; Writing – review & editing: Supporting); Mar Garcia-Rocha, PhD (Conceptualization: Supporting; Data curation: Supporting; Formal analysis: Supporting); Jose Altamirano, MD (Data curation: Supporting; Formal analysis: Supporting; Investigation: Supporting); Gemma Odena, PhD (Data curation: Supporting; Formal analysis: Supporting); Laurent Dubuquoy, PhD (Data curation: Supporting); Alexandre Louvet, MD (Data curation: Supporting); Carlos Martinez-Pons, PhD (Data curation: Supporting; Formal analysis: Supporting); Anna Adrover, BS (Data curation: Supporting; Formal analysis: Supporting); Silvia Affo, PhD (Data curation: Supporting; Formal analysis: Supporting); Oriol Morales-Ibañez, PhD (Data curation: Supporting; Formal analysis: Supporting); Pau Sancho-Bru, PhD (Data curation: Supporting; Formal analysis: Supporting; Writing – review & editing: Supporting); Cristina Millán, PhD (Data curation: Supporting; Formal analysis: Supporting); Edilmar Alvarado-Tapias, MD, PhD (Resources: Supporting; patient selection, clinical data collection: Supporting); Dalia Morales-Arraez, MD (Data curation: Supporting; Formal analysis: Supporting); Juan Caballeria, MD (Data curation: Supporting; Formal analysis: Supporting; Writing – review & editing: Supporting); Jelena Mann, PhD (Formal analysis: Supporting; Software: Supporting; Writing – review & editing: Supporting); Sheng Cao, PhD (Data curation: Supporting; Formal analysis: Supporting;
Methodology: Supporting); Zhaoli Sun, PhD (Data curation: Supporting; Formal analysis: Supporting); Vijay H. Shah, MD (Data curation: Supporting; Formal analysis: Supporting; Writing – review & editing: Supporting); Andrew Cameron, MD (Data curation: Supporting; Formal analysis: Supporting; Writing – review & editing: Supporting); Phillipe Mathurin, MD (Data curation: Supporting; Formal analysis: Supporting; Writing – review & editing: Supporting); Natasha Snider, PhD (Data curation: Supporting; Formal analysis: Supporting; Writing – review & editing: Supporting); Càndid Villanueva, MD, PhD (Resources: Supporting); Timothy R. Morgan, MD (Resources: Supporting); Joan Guinovart, PhD (Data curation: Supporting; Formal analysis: Supporting; Writing – review & editing: Supporting); Rajanikanth Vadigepalli, PhD (Conceptualization: Supporting; Data curation: Supporting; Formal analysis: Supporting; Investigation: Supporting; Methodology: Supporting; Software: Supporting; Supervision: Supporting; Visualization: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting); Ramon Bataller, MD (Conceptualization: Lead; Funding acquisition: Lead; Project administration: Lead; Resources: Lead; Supervision: Lead; Writing – original draft: Lead; Writing – review & editing: Lead).
Conflicts of interest
The authors disclose no conflicts.
Supplementary Material
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at https://doi.org/10.1053/j.gastro.2020.12.008.
References
- 1.Mandrekar P, Bataller R, Tsukamoto H, et al. Alcoholic hepatitis: translational approaches to develop targeted therapies. Hepatology 2016;64:1343–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altamirano J, Fagundes C, Dominguez M, et al. Acute kidney injury is an early predictor of mortality for patients with alcoholic hepatitis. Clin Gastroenterol Hepatol 2012;10:65–71. [DOI] [PubMed] [Google Scholar]
- 3.Mathurin P, Louvet A, Duhamel A, et al. Prednisolone with vs without pentoxifylline and survival of patients with severe alcoholic hepatitis: a randomized clinical trial. JAMA 2013;310:1033–1041. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646–674. [DOI] [PubMed] [Google Scholar]
- 5.Zhu Y, Shan X, Yuzwa SA, et al. The emerging link between O-GlcNAc and Alzheimer disease. J Biol Chem 2014;289:34472–34481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krahenbuhl L, Lang C, Ludes S, et al. Reduced hepatic glycogen stores in patients with liver cirrhosis. Liver Int 2003;23:101–109. [DOI] [PubMed] [Google Scholar]
- 7.Dubuquoy L, Louvet A, Lassailly G, et al. Progenitor cell expansion and impaired hepatocyte regeneration in explanted livers from alcoholic hepatitis. Gut 2015;64:1949–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sancho-Bru P, Altamirano J, Rodrigo-Torres D, et al. Liver progenitor cell markers correlate with liver damage and predict short-term mortality in patients with alcoholic hepatitis. Hepatology 2012;55:1931–1941. [DOI] [PubMed] [Google Scholar]
- 9.Argemi J, Latasa MU, Atkinson SR, et al. Defective HNF4alpha-dependent gene expression as a driver of hepatocellular failure in alcoholic hepatitis. Nat Commun 2019;10(1):3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mathurin P, Moreno C, Samuel D, et al. Early liver transplantation for severe alcoholic hepatitis. N Engl J Med 2011;365:1790–1800. [DOI] [PubMed] [Google Scholar]
- 11.Hayes MG, Urbanek M, Hivert MF, et al. Identification of HKDC1 and BACE2 as genes influencing glycemic traits during pregnancy through genome-wide association studies. Diabetes 2013;62:3282–3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Z, Huang S, Wang H, et al. High expression of hexokinase domain containing 1 is associated with poor prognosis and aggressive phenotype in hep-atocarcinoma. Biochem Biophys Res Commun 2016;474:673–679. [DOI] [PubMed] [Google Scholar]
- 13.Dominguez M, Rincon D, Abraldes JG, et al. A new scoring system for prognostic stratification of patients with alcoholic hepatitis. Am J Gastroenterol 2008;103:2747–2756. [DOI] [PubMed] [Google Scholar]
- 14.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Irwin DM, Tan H. Molecular evolution of the vertebrate hexokinase gene family: identification of a conserved fifth vertebrate hexokinase gene. Comp Biochem Physiol Part D Genomics Proteomics 2008;3:96–107. [DOI] [PubMed] [Google Scholar]
- 16.Kuwajima M, Newgard CB, Foster DW, et al. The glucose-phosphorylating capacity of liver as measured by three independent assays. Implications for the mechanism of hepatic glycogen synthesis. J Biol Chem 1986;261:8849–8853. [PubMed] [Google Scholar]
- 17.Guo C, Ludvik AE, Arlotto ME, et al. Coordinated regulatory variation associated with gestational hyperglycaemia regulates expression of the novel hexokinase HKDC1. Nat Commun 2015;6:6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Affo S, Dominguez M, Lozano JJ, et al. Transcriptome analysis identifies TNF superfamily receptors as potential therapeutic targets in alcoholic hepatitis. Gut 2013;62:452–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thursz MR, Richardson P, Allison M, et al. Prednisolone or pentoxifylline for alcoholic hepatitis. N Engl J Med 2015;372:1619–1628. [DOI] [PubMed] [Google Scholar]
- 20.Singal AK, Shah VH. Therapeutic strategies for the treatment of alcoholic hepatitis. Semin Liver Dis 2016;36:56–68. [DOI] [PubMed] [Google Scholar]
- 21.Lieber CS. Hepatic and metabolic effects of alcohol (1966 to 1973). Gastroenterology 1973;65:821–846. [PubMed] [Google Scholar]
- 22.Cederbaum AI, Lieber CS, Rubin E. Effect of acetaldehyde on fatty acid oxidation and ketogenesis by hepatic mitochondria. Arch Biochem Biophys 1975;169:29–41. [DOI] [PubMed] [Google Scholar]
- 23.Ontko JA. Effects of ethanol on the metabolism of free fatty acids in isolated liver cells. J Lipid Res 1973;14:78–86. [PubMed] [Google Scholar]
- 24.Rachakonda V, Gabbert C, Raina A, et al. Serum metabolomic profiling in acute alcoholic hepatitis identifies multiple dysregulated pathways. PLoS One 2014;9(12):e113860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.She P, Shiota M, Shelton KD, et al. Phosphoenolpyruvate carboxykinase is necessary for the integration of hepatic energy metabolism. Mol Cell Biol 2000;20:6508–6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Satapati S, Sunny NE, Kucejova B, et al. Elevated TCA cycle function in the pathology of diet-induced hepatic insulin resistance and fatty liver. J Lipid Res 2012;53:1080–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rui L Energy metabolism in the liver. Compr Physiol 2014;4:177–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ronimus RS, Morgan HW. Cloning and biochemical characterization of a novel mouse ADP-dependent glucokinase. Biochem Biophys Res Commun 2004;315:652–658. [DOI] [PubMed] [Google Scholar]
- 29.Ludvik AE, Pusec CM, Priyadarshini M, et al. HKDC1 is a novel hexokinase involved in whole-body glucose use. Endocrinology 2016;157:3452–3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.