Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has killed millions of people and continues to cause massive global upheaval. Coronaviruses are positive-strand RNA viruses with an unusually large genome of ~30 kb. They express an RNA-dependent RNA polymerase and a cohort of other replication enzymes and supporting factors to transcribe and replicate their genomes. The proteins performing these essential processes are prime antiviral drug targets, but drug discovery is hindered by our incomplete understanding of coronavirus RNA synthesis and processing. In infected cells, the RNA-dependent RNA polymerase must coordinate with other viral and host factors to produce both viral mRNAs and new genomes. Recent research aiming to decipher and contextualize the structures, functions and interplay of the subunits of the SARS-CoV-2 replication and transcription complex proteins has burgeoned. In this Review, we discuss recent advancements in our understanding of the molecular basis and complexity of the coronavirus RNA-synthesizing machinery. Specifically, we outline the mechanisms and regulation of RNA translation, replication and transcription. We also discuss the composition of the replication and transcription complexes and their suitability as targets for antiviral therapy.
Subject terms: SARS-CoV-2, DNA replication, Transcription
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and other coronaviruses rely on a cohort of specialized viral proteins to transcribe and replicate their RNA genomes. Recent studies have improved our understanding of coronavirus RNA translation, replication and transcription, and offer new therapeutic targets.
Introduction
The devastation caused by the COVID-19 pandemic has led to an extraordinary expansion of research focused on the causative agent, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). This virus has been classified within the species Severe acute respiratory syndrome-related coronavirus, which belongs to the genus Betacoronavirus of the family Coronaviridae1. The coronavirus family belongs to the order Nidovirales, which includes a rapidly expanding and diverse group of enveloped viruses with a single-stranded RNA genome of positive polarity. Different nidoviruses use similar strategies to organize, express and replicate their genomes. They constitute a monophyletic virus cluster that is characterized by the universal conservation of seven domains in their large replicase gene, which encodes the functions required for viral RNA replication and transcription in the infected cell2. Much of the foundational research on this replicase addressed viruses in related nidovirus families such as the Arteriviridae and, more relevantly, other members of the family Coronaviridae. The features unique and common to viruses within and outside the order Nidovirales have been described in several comprehensive reviews3–5. Using the first available genome sequences, early studies unravelled the genome organization and expression strategy used by coronaviruses and other nidoviruses. These studies were followed by pioneering bioinformatics, biochemical and genetic studies that established or confirmed the essential functions of many of the replicase proteins, thereby laying a road map for the currently ongoing SARS-CoV-2 research.
Betacoronaviruses have caused previous deadly epidemics, including the 2003 SARS outbreak and the ongoing Middle East respiratory syndrome (MERS) epidemic, which was first detected in 2012 (ref.6). These zoonotic events inspired earlier efforts to develop coronavirus-specific antiviral drugs6–9. The onset of the COVID-19 pandemic spurred scientists around the globe to apply their respective expertise to address how SARS-CoV-2 infects humans, avoids or delays host immune responses, copies its genome and expresses its proteins to make new virions. As successful replication directly depends on efficient synthesis of viral RNA, the replicase proteins responsible for this process are obvious antiviral drug targets.
Coronaviruses use an unusually large collection of RNA-synthesizing and RNA-processing enzymes to express and replicate a genome that is two to three times larger than that of most other RNA viruses. The central enzyme of transcription and replication is the RNA-dependent RNA polymerase (RdRp), which synthesizes all viral RNA and is a proven target for antiviral drugs. Successful replication and transcription also entails the use of specific RNA recognition signals to initiate RNA synthesis, sustaining RdRp processivity and fidelity, viral mRNA capping to ensure translation by host ribosomes, and the spatial and temporal regulation of the viral cycle within the infected cell. The non-structural proteins that assist the RdRp to perform these functions (Table 1) constitute additional targets for antiviral drug development. Like the RdRp, some of these non-structural proteins are conserved across most RNA viruses, whereas others are unique to coronaviruses3–5.
Table 1.
Coronavirus replicase proteins and their functions
| Open reading frame | Non-structural protein | Function | SARS-CoV-2 sizea |
|---|---|---|---|
| ORF1a | nsp1 | Host mRNA degradation, mediates host translation shut-off by targeting the ribosome23–25,143 | 180 |
| nsp2 | Unknown | 638 | |
| nsp3 | Amino-terminal Ubl domain binds RNA144 and the nucleocapsid protein77, mono-ADP-ribosylhydrolase22,145,146, PLpro (ref.147), replication organelle formation148,149, main component of putative RNA transport pore in RNA-synthesizing double-membrane vesicles150 | 1,945 | |
| nsp4 | Replication organelle formation148,149 | 500 | |
| nsp5 | Picornavirus 3C-like protease or Mpro (refs151,152) | 306 | |
| nsp6 | Replication organelle formation149 | 290 | |
| nsp7 | Subunit of the RdRp holoenzyme83,94 | 83 | |
| nsp8 | Subunit of the RdRp holoenzyme83,94, putative primase153, putative 3′-terminal adenylyltransferase154 | 198 | |
| nsp9 | Putative RNA-binding protein155, NiRAN UMP-transferase substrate92 | 113 | |
| nsp10 | Stimulatory subunit of the nsp14 exonuclease and nsp16 methyltransferase activities141,156,157, regulation of ribosomal frameshifting40 | 139 | |
| nsp11 | Unknown | 13 | |
| ORF1b | nsp12b | NiRAN domain: NMPylase92 and (proposed) RNA-capping enzyme93, RdRp3,83,94,151 | 932 |
| nsp13 | Zinc-binding domain-containing RNA helicase111,122,151, RNA 5′-phosphatase115 | 601 | |
| nsp14 | Proofreading 3′-5′ exonuclease22,101,103,105, N7-methyltransferase158,159 | 527 | |
| nsp15 | Uridylate-specific endoribonuclease22,43 | 346 | |
| nsp16 | 2′-O-Methyltransferase22,160 | 298 |
Mpro, main protease; NiRAN, nidovirus RNA-dependent RNA polymerase-associated nucleotidyltransferase; NMP, nucleoside monophosphate; PLpro, papain-like protease; RdRp, RNA-dependent RNA polymerase; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; Ubl, ubiquitin-like; UMP, uridine monophosphate. aNumber of amino acids for SARS-CoV-2 isolate Wuhan-Hu-1 (GenBank entry MN908947.3). bThe amino-terminal 13 amino acids of nsp12 are encoded in the 3′-terminal part of ORF1a.
In this Review, we discuss recent advances in deciphering the molecular mechanisms of coronavirus gene expression and RNA replication, with special focus on SARS-CoV-2. We aim to contextualize the more recent studies with the foundational work, to provide a coherent view of our current understanding of the successive steps in SARS-CoV-2 replication and transcription. We focus on the processes required for viral gene expression and replication: translation, replication organelle formation, and production and capping of genomic RNA (gRNA) and subgenomic RNAs (sgRNAs). We also describe the known and proposed functions of the viral nucleic acid-metabolizing proteins, as revealed by new biochemical, structural and virological studies. We then discuss two well-studied examples of antiviral nucleoside analogues that target the RdRp; owing to space constraints, we do not address therapeutics designed to target viral proteins other than the RdRp. Likewise, we do not extensively cover the mechanisms of viral protein synthesis, or the (proposed) involvement of a substantial number of host factors in coronavirus replication and transcription. We conclude the Review by describing the remaining gaps in our knowledge to help guide new research.
The SARS-CoV-2 infection cycle
To infect a cell, coronaviruses use multiple host factors, whose expression patterns therefore co-determine viral tropism. Delivery into the cell and translation of the large RNA genome launches a cytoplasmic replication cycle that integrates a remarkable variety of strategies to fine-tune viral gene expression on both the translational level and the transcriptional level. The successive steps that ultimately lead to the release of viral progeny are coordinated temporally and spatially, and rely extensively on the infrastructure and metabolism of the host cell. In this section, we outline the key steps of the SARS-CoV-2 infection cycle.
Entry into the host cell
SARS-CoV-2 entry into the cell depends on several host attachment and entry factors, chiefly among them on the receptor angiotensin-converting enzyme 2 (ACE2), to which the SARS-CoV-2 spike (S) glycoprotein binds (reviewed in refs10,11) (Fig. 1). The fusogenic spike protein consists of two parts, S1 and S2, which mediate attachment to and fusion with the cell membrane, respectively. Cellular proteases such as transmembrane serine protease 2 cleave the spike protein, a step required to prime its membrane fusion activity12,13.
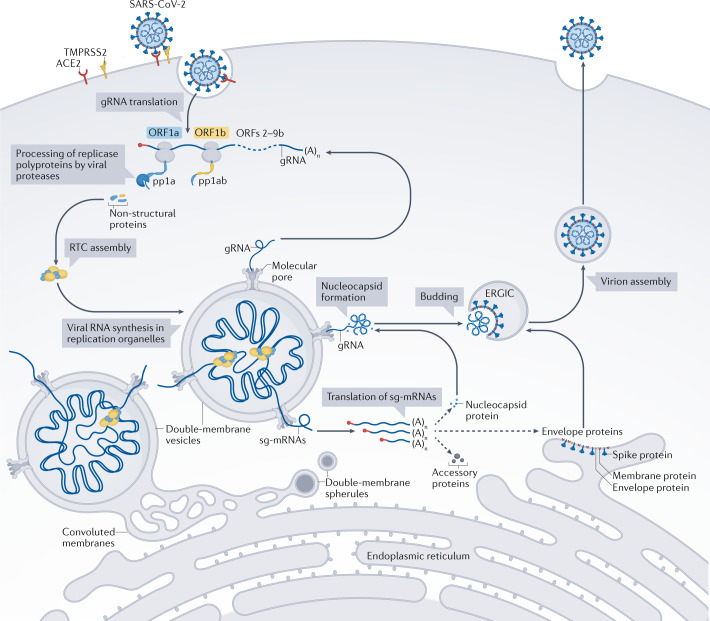
Fig. 1. The SARS-CoV-2 infection cycle.
To enter a host cell, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein interacts with the cellular surface protein angiotensin-converting enzyme 2 (ACE2), while being cleaved by cellular proteases such as transmembrane serine protease 2 (TMPRSS2) to activate its membrane-fusion capacity. The genomic RNA (gRNA), which is capped on its 5′ end (red circle) and polyadenylated ((A)n) on its 3′ end, is released from the viral particle and — after recruiting host-cell ribosomes — translated into two replicase polyproteins, pp1a and pp1ab. Proteases embedded in viral non-structural protein 3 (nsp3) and nsp5 cleave pp1a and pp1ab into 16 non-structural proteins that assemble into replication–transcription complexes (RTCs). Viral RNA synthesis occurs within double-membrane vesicles that are part of virus-induced membranous replication organelles (Box 1). The RTCs produce new gRNAs and a set of subgenomic mRNAs (sg-mRNAs) that include open reading frames (ORFs) 2–9b, which encode the structural spike, membrane, envelope and nucleocapsid proteins, and also a number of accessory proteins. Newly made gRNAs can be translated to yield additional non-structural proteins, serve as a template for further RNA synthesis or be packaged into new virions. SARS-CoV-2 assembly starts with the coating of gRNAs with nucleocapsid proteins, generating nucleocapsid structures that bud into the endoplasmic reticulum–Golgi intermediate compartment (ERGIC), thereby acquiring a lipid bilayer containing the viral spike, membrane and envelope proteins. Adapted from ref.161, CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/).
Following membrane fusion, the SARS-CoV-2 gRNA is released into the cytosol. The genome is a 5′-capped, single-stranded RNA of 29,870 bases with a 3′ poly(A) tail of variable length14,15. It encodes at least 13 recognized open reading frames (ORFs), organized largely linearly from the 5′ end to the 3′ end (Fig. 2a). The coding part of the gRNA is flanked by a 5′ untranslated region (UTR) and a 3′ UTR of 265 and 337 nucleotides (excluding the poly(A) tail), respectively. The gRNA possesses a number of regulatory sequences and higher-order RNA structures (discussed later) that are involved in its translation, replication and transcription16–19.
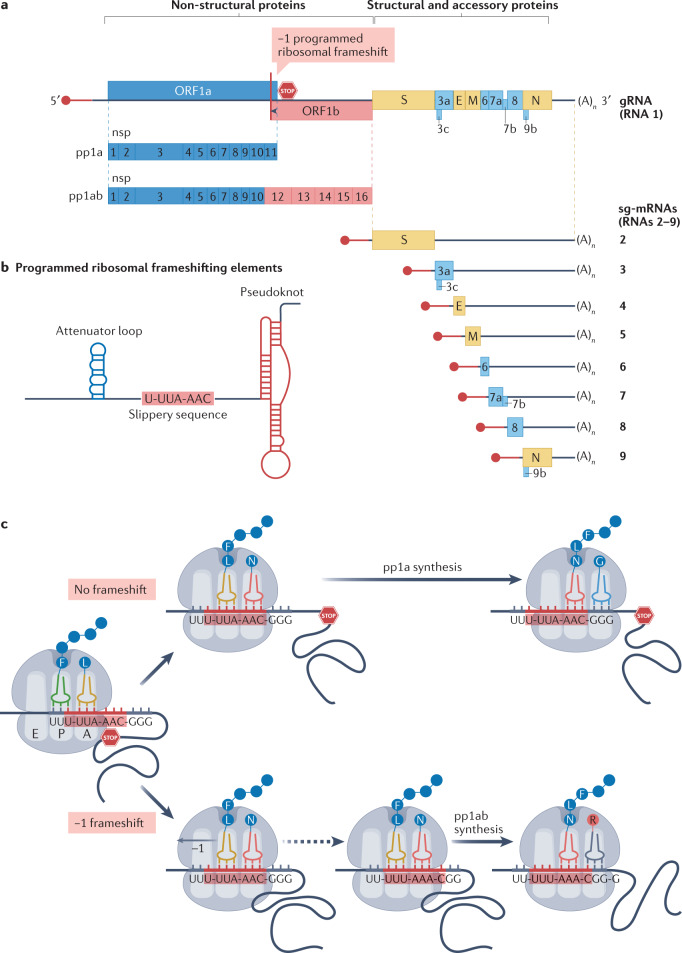
Fig. 2. Regulation of SARS-CoV-2 gene expression on the translational level.
a | Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) genome organization. Open reading frames (ORFs) are not drawn exactly to scale. The SARS-CoV-2 genomic RNA (gRNA) has a 5′ cap (red circle) followed by a leader sequence (red line) that is shared with all subgenomic mRNAs (sg-mRNAs), and a 3′ poly(A) tail. The 5′-proximal three quarters of the genome encode the replicase polyproteins pp1a and pp1ab, which are cleaved to yield 16 non-structural proteins (nsp1–nsp16; blue from ORF1a and red from ORF1b). The 3′-proximal one quarter of the genome encodes the structural (brown) and accessory (azure) proteins. Structural and accessory proteins are expressed from a nested set of sg-mRNAs, with ORF3c, ORF7b and ORF9b being expressed via ribosomal ‘leaky scanning’. b | RNA motif and structures that promote a frameshift from ORF1a to ORF1b, thereby controlling the synthesis of pp1ab. The key programmed ribosomal frameshifting (PRF)-stimulating RNA structure — a pseudoknot — interacts with the ribosome and induces its pausing, which generates tension in the gRNA template. As a result, ribosomes can slip one nucleotide backwards on the ‘slippery sequence’ (−1 PRF). An attenuating RNA loop located upstream of the slippery sequence also contributes to modulating PRF frequency. c | Model of −1 PRF at the ORF1a–ORF1b junction, showing the regulatory RNA elements inducing a simultaneous −1 shift of the tRNAs bound to the A and P sites of the ribosome, which can then translate ORF1b. The one-letter code for amino acids (circles) is used. A stop sign represents the ORF1a stop codon. E, envelope protein; M, membrane protein; N, nucleocapsid protein; S, spike protein.
Gearing up for RNA synthesis
Similarly to all positive-strand RNA viruses of eukaryotes, the replication of SARS-CoV-2 occurs entirely in the cytoplasm. The SARS-CoV-2 gRNA first recruits host ribosomes and serves as mRNA for translation of the two large replicase ORFs ORF1a and ORF1b, which constitute about three quarters of the genome15 (Fig. 2a). The resulting, amino-terminally (N-terminally) collinear replicase polyproteins pp1a and pp1ab are 4,405 and 7,096 amino acids long, respectively. Production of pp1ab depends on the occurrence of a −1 programmed ribosomal frameshift (PRF) just upstream of the ORF1a termination codon, thus extending pp1a with the ORF1b-encoded polyprotein. The estimated frameshifting efficiency is 45–70%, resulting in a 1.5–2-fold overexpression of ORF1a-encoded proteins relative to ORF1b-encoded proteins20. Sixteen mature non-structural proteins are released from pp1a and pp1ab following 15 proteolytic cleavages performed by the virus-encoded papain-like protease (PLpro) in non-structural protein 3 (nsp3) and chymotrypsin-like or main protease (Mpro) in nsp5 (Table 1). In this manner, pp1a yields nsp1 to nsp11, whereas pp1ab is cleaved into nsp1 to nsp10 and nsp12 to nsp16 (refs21,22) (Fig. 2a).
The rapidly released nsp1 mediates the shutdown of the translation of host mRNAs23–25, while the other non-structural proteins form protein complexes, yet to be definitely determined, that engage in viral RNA synthesis and are referred to as the replication–transcription complexes (RTCs). Replication and transcription are driven primarily by the enzymes contained in nsp12, nsp13, nsp14 and nsp16 (Table 1). Nsp12, the subunit containing the RdRp domain, catalyses RNA synthesis with the assistance of nsp7 and nsp8, together forming the holoenzyme RdRp (holo-RdRp). Other RTC subunits have supporting roles in the RTC, modulate the host’s innate immune responses or remodel cell membranes into peculiar double-membrane structures known as ‘replication organelles’, which accommodate viral RNA synthesis26,27. The formation of replication organelles typically precedes the exponential phase of viral RNA synthesis and is discussed in Box 1.
Box 1 Coronavirus replication organelles.
Coronavirus infection induces the extensive remodelling of endoplasmic reticulum (ER) membranes26,27,163–165. This yields a complex vesiculotubular network of convoluted membranes (CMs) and unusual double-membrane vesicles (DMVs), in part interconnected through their outer membranes. Collectively these structures are referred to as ‘replication organelles’, which presumably help to organize viral RNA synthesis in time and space. Moreover, replication organelles may hinder detection of the negative-strand RNA and double-stranded RNA intermediates of viral RNA synthesis by innate immunity receptors. The formation of double-membrane structures can be induced by co-expression of coronavirus transmembrane non-structural protein 3 (nsp3), nsp4 and nsp6 (refs148,149). Coronavirus replication organelles can be visualized by (cryo) electron tomography, as shown for Middle East respiratory syndrome coronavirus-infected cells in the figure, part a (tomogram slice) and part b (surface-rendered model)27. These images show large numbers of DMVs, commonly 200–300 nm in diameter; double-membrane spherules (DMS; arrowheads) of unknown function and having a diameter of ~80 nm; CMs; and ER. Metabolic labelling (with 3H-uridine) of newly synthesized coronavirus RNA in combination with autoradiography and electron microscopy identified DMVs as the main platform for coronavirus RNA synthesis27. This notion was further supported by the abundant immunolabeling of DMVs for double-stranded RNA164 and the presence of ‘fibrillar material’ in their interior150,164,166. However, for many years, coronavirus DMVs were observed as fully sealed compartments27,164,165, lacking a connection to the cytosol that could be used to export genomic RNA and subgenomic mRNAs, thus maintaining controversy about the exact site of coronavirus RNA synthesis.
Recently, cryotomography of coronavirus-infected cells was used to study replication organelles as closely as possible to their native state150,166. This approach revealed that murine hepatitis virus DMVs contain membrane-spanning structures (see the figure, part c). Subsequent subtomogram averaging revealed a hexameric, crown-shaped pore complex (total mass ~3 MDa) surrounding a central channel that would be wide enough to allow RNA transport150 (see the figure, part d). Use of a recombinant murine hepatitis virus expressing GFP-tagged nsp3 revealed that this protein, which is the largest replicase subunit (~2,000 residues), is a major component of the DMV pore complex150. On the cytosolic side of the pore, protein assemblies strongly resembling coronavirus nucleocapsid structures were visualized (see the figure, part e), suggesting that newly made RNA is encapsidated following its export from DMVs150 (see the figure, part f). The localization of the coronavirus replication–transcription complex within DMVs and its potential association with the DMV pore complex remain to be investigated in more detail. Size bars are 250 nm (part a) and 50 nm (parts c and e). Parts a and b adapted from ref.27, CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/); parts c–f adapted from ref.150, CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/).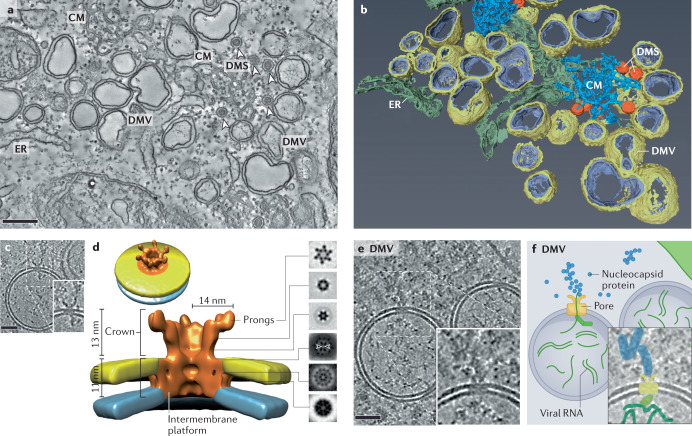
RNA synthesis and virion assembly
Using the gRNA template, RNA synthesis by the RTC starts with producing both a full-length genome complement (the anti-genome) and a set of minus-strand sgRNAs, which are derived from the gRNA region downstream of ORF1a and ORF1b (the replicase gene). Whereas the anti-genome serves as a template to produce new gRNA, the minus-strand sgRNAs direct the synthesis of a nested set of subgenomic mRNAs (sg-mRNAs) (discussed later). Although transcription is defined principally as the synthesis of RNA from a DNA template, in this Review we use the term to describe the synthesis of sg-mRNAs from RNA templates, to conform with the terminology used in the coronavirus literature.
The sg-mRNAs are crucial for the production of the four coronavirus structural proteins, which are required for virion assembly and egress. New virions were recently reported to leave the cell via lysosomal trafficking rather than the biosynthetic secretory pathway used by many other enveloped viruses28. A number of the sg-mRNAs are used to express so-called accessory proteins, many of which have been implicated in modulating cellular innate immune responses20,29,30 (Fig. 1).
Regulation of SARS-CoV-2 translation
Expression of the SARS-CoV-2 proteins in infected cells depends primarily on the translation of gRNA and the eight ‘canonical’ sg-mRNAs20,29,30. The viral genes fall into three groups (Fig. 2a): replicase ORF1a and ORF1b, which are translated from gRNA with ORF1b expression depending on −1 PRF; ORFs encoding the four ‘universal’ coronavirus structural proteins (the spike, membrane, envelope and nucleocapsid proteins) (Fig. 1), which are translated from sg-mRNAs; and ORFs encoding accessory proteins, which are translated from the remaining sg-mRNAs, but differ widely between various coronavirus lineages31 (Fig. 2a). In addition, small (putative) ORFs that overlap with several of the ORFs outlined above were identified by theoretical and experimental approaches. These small ORFs were also the subject of nomenclature confusion32, and their expression and biological relevance continue to be investigated32–35. In this light, we have included only ORF3c and ORF9b of the small ORFs in the current map of the SARS-CoV-2 genome (Fig. 2a).
Like many RNA viruses, coronaviruses use non-canonical translation mechanisms to expand their coding capacity and fine-tune the expression levels of particular viral proteins36. Specifically, PRF (for ORF1b) and ‘leaky ribosomal scanning’ (for some ORFs in sg-mRNAs) co-regulate SARS-CoV-2 genome expression. In leaky ribosomal scanning, ribosomes load onto the 5′ end of the viral sg-mRNAs, but initiate translation from a more downstream, internal start codon. The use of leaky scanning has been suggested or demonstrated for SARS-CoV-2 ORF3c33,34, and for ORF7b20,22,37 and ORF9b38 of both SARS-CoV and SARS-CoV-2. In these cases, leaky scanning and expression of the more downstream ORF appear to be promoted by the suboptimal nature of upstream translation initiation signals.
Expression of ORF1b from gRNA depends on −1 PRF occurring just upstream of the ORF1a stop codon39, a highly conserved feature among coronaviruses and other nidoviruses. Termination of SARS-CoV-2 ORF1a translation yields the 4,405-residue-long pp1a, and −1 PRF results in extension to yield the 7,096-residue-long pp1ab. The ORF1a–ORF1b PRF mechanism directs the expression of the key RNA metabolism enzymes of the RTC and regulates the relative expression levels of the proteins encoded by ORF1a and ORF1b. Frameshifting occurs on a specific ‘slippery sequence’ (5′-U UUA AAC-3′ (nucleotides 13,462–13,468 in GenBank genome entry MN908947.3; all genome reference numbers in this Review refer to this sequence)), followed by GGG in the case of SARS-CoV-2) — ribosomes translating the UUA and AAC codons of ORF1a can shift one nucleotide backwards, and translation then continues with a CGG codon in ORF1b40,41 (Fig. 2b,c). The importance of maintaining the optimal ratio between ORF1a expression and ORF1b expression was demonstrated experimentally with use of SARS-CoV mutants with altered PRF levels, which were found to be dramatically crippled41.
Regulation of PRF efficiency is achieved by the formation of several RNA structures and through interactions of the nascent protein chain with the ribosome. The key PRF-stimulating element is a three-stemmed RNA pseudoknot structure located downstream of the slippery sequence39–42 (Fig. 2b). This element interacts with the ribosome at the entry of the mRNA channel of the 40S ribosomal subunit and induces translational pausing before −1 PRF; complete unfolding of this tertiary RNA structure is slow and thought to promote ribosomal frameshifting on the viral mRNA40. The position of the ORF1a stop codon, five codons downstream of the frameshift site, may also regulate PRF levels by allowing the pseudoknot to refold, thus preventing trailing ribosomes from continuing along the viral RNA that was unfolded by the leading ribosome40. PRF frequency is further modulated by a translation-attenuating RNA loop upstream of the slippery sequence40, which may either directly inhibit frameshifting42 or force elongating ribosomes to dissociate before reaching the PRF site41. Finally, interactions between specific residues in the nascent viral polyproteins and the ribosome exit tunnel are thought to co-determine PRF efficiency40.
RNA replication and transcription
As described earlier, ORF1a and ORF1b are translated into the pp1a and pp1ab precursors that give rise to 16 non-structural proteins1–4. Fourteen of these replicase subunits have been ascribed some type of function in coronavirus replication, several with manifold activities, as listed in Table 1. These non-structural proteins either have been directly implicated in nucleic acid metabolism or enable or promote the activity of the catalytic non-structural proteins, to stimulate RNA synthesis and processing or participate in the formation of replication organelles. In this Review, we discuss in detail the molecular and biochemical features of these proteins, focusing mostly on nsp7, nsp8, nsp9, nsp10, nsp11, nsp12, nsp13, nsp14 and nsp16, but do not delve into the details of the role of nsp15, which includes a unique uridylate-specific endoribonuclease43 that is conserved in most vertebrate nidoviruses. Nsp15 has been implicated in innate immunity evasion44,45, possibly by shortening the poly(U) stretches that are present at the 5′ end of viral minus-strand RNAs45. In coronaviruses, uridylate-specific endoribonuclease activity is required for efficient replication, but that requirement can be bypassed in host cells with depressed type I interferon sensing or production44,45. In the following subsections we summarize our current understanding of the mechanisms of coronavirus gRNA and sg-mRNA synthesis, and the involvement of specific viral proteins in controlling these processes. We note that some non-structural proteins have other functions, which we do not mention because they are outside the scope of this Review. For a more in-depth summary of the literature on the functions of each non-structural protein, see Table 1 and references therein.
Continuous and discontinuous RNA synthesis in replication and gene expression
Following gRNA translation and proteolytic maturation of the replicase polyproteins, a relatively complex programme of SARS-CoV-2 RNA synthesis and gene expression is initiated, which depends on the interplay between viral RNA and non-structural proteins on the one hand (Fig. 3), and host-cell proteins and membranes on the other hand (Box 1). A variety of RNA sequences and structural elements in the terminal regions of the coronavirus genome have been implicated in the specific recognition of RNA templates by the coronavirus RTC16–18 (Fig. 3a). Long-range RNA–RNA interactions may be important for replication and transcription, although in many cases direct experimental support for their biological relevance remains to be obtained.
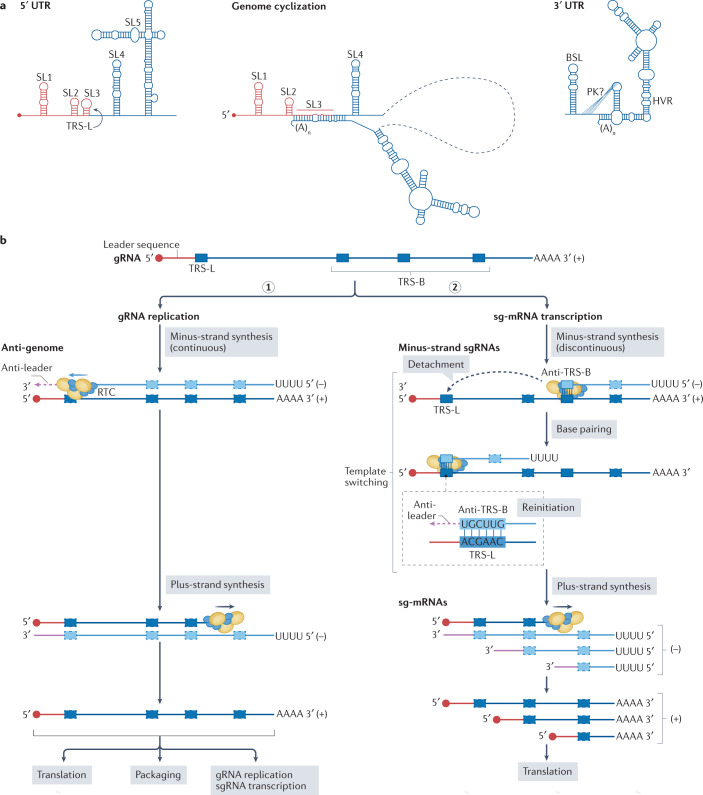
Fig. 3. SARS-CoV-2 RNA replication and transcription.
a | The genomic RNA (gRNA) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has cis-acting structures at its 5′ untranslated region (UTR) and 3′ UTR, and engages in long-distance intramolecular RNA–RNA interactions; these structures and interactions are thought to be involved in the regulation of replication and transcription. The 5′ UTR contains five conserved stem–loop (SL) structures, and the 3′ UTR contains a bulged stem–loop (BSL), a (predicted) pseudoknot (PK) and a stem–loop structure with a hypervariable region (HVR). SARS-CoV-2 gRNA cyclization results in complete opening of SL3, where the leader transcription regulatory sequence (TRS-L) resides. b | The gRNA serves as a template for gRNA replication (step 1) and for subgenomic mRNA (sg-mRNA) transcription (step 2); each process requires dedicated minus-strand templates: the anti-genome and a set of minus-strand sgRNAs, respectively. Synthesis of the latter involves a discontinuous step in which the replication–transcription complex (RTC) pauses RNA synthesis after copying one of the body transcription regulatory sequences (TRS-B), and detaches from the template. Subsequently, the RTC relocates to a position near the 5′ end of the gRNA template, where the complement of the TRS-B (anti-TRS-B) in the nascent minus-strand sgRNA engages in base pairing with the TRS-L. This template switch leads to the addition of the complement of the gRNA leader sequence (anti-leader) to the 3′ end of each of minus-strand sgRNA, which are used as templates for sg-mRNA production, thereby ensuring that all coronavirus sg-mRNAs include a 5′-terminal leader sequence of ~75 nucleotides that is identical to the 5′-terminal sequence of the gRNA. Positions of the TRSs and anti-TRSs are schematic and not drawn exactly to scale. Part b adapted from ref.161, CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/).
As outlined earlier, SARS-CoV-2 RNA synthesis can be divided into genome replication and sg-mRNA transcription (Fig. 3b). Replication yields full-length viral plus-strand gRNA, which can be translated into additional replicase polyproteins, serve as a template for additional minus-strand RNA synthesis or be packaged into progeny virions. Transcription produces the nested set of sg-mRNAs used to express the structural and accessory proteins. Replication and transcription both require dedicated minus-strand RNA templates; full-length minus strands serve as a template for gRNA replication, whereas a nested set of minus-strand sgRNAs serve as templates for transcription, as first proposed about a quarter of a century ago46. The sg-mRNAs have the same 3′-terminal sequence, and carry a common 5′ leader sequence that is identical to the 5′-terminal 75 nucleotides of the gRNA. The leader derives from a discontinuous step (that is, from template switching47 during minus-strand sgRNA synthesis), which occurs when the RTC stalls at the 3′-proximal quarter of the gRNA template (Fig. 3b). This interruption mediates RTC detachment and relocation to a position near the 5′ end of the gRNA template (discussed later), where minus-strand synthesis resumes. This process yields a set of nested minus-strand sgRNAs with common 5′-terminal and 3′-terminal sequences, which serve as templates for the synthesis of a complementary set of sg-mRNAs47,48.
The presence of the common 5′ leader sequence in coronavirus mRNAs may offer several advantages. Its complement (the anti-leader sequence) offers a conserved starting point for plus-strand RNA synthesis, which can be used to initiate the synthesis of both gRNA and all sg-mRNAs. Although not studied in detail thus far, the common 5′ leader sequence may also serve as a recognition signal for the viral mRNA capping machinery (Box 2). Furthermore, the nsp1 proteins of SARS-CoV, MERS-CoV and SARS-CoV-2 all mediate a translation shut-off in the infected cell49, which is based on their ability to block the ribosomal mRNA entry channel24,25 and induce endonucleolytic cleavage of host mRNAs23. The common 5′ leader sequence present in all coronavirus mRNAs allows escape from translation shut-off50, by yet unknown mechanisms, resulting in simultaneous viral mRNA translation and impairment of host-cell gene expression, including of genes mediating the early responses to virus infection.
Box 2 Capping of the SARS-CoV-2 RNA.
RNA capping is required for efficient translation of eukaryotic mRNAs and is also thought to protect the viral RNA from host immune response and nucleases167,168. However, although synthesis of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA occurs in the cytoplasm of the infected cell, endogenous capping enzymes are mostly sequestered in the nucleus167. To address this complication, the coronavirus replicase acquired enzymatic domains to perform the four activities required for mRNA capping in the cytoplasm. Following transcription, the postulated sequence of capping steps, verified in vitro93, is as follows: the γ-phosphate of the 5′-triphosphate of the RNA is hydrolysed and released (see the figure). This RNA triphosphatase (TPase) activity is mediated by the helicase non-structural protein 13 (nsp13) using the same ATPase site as required for its translocation and unwinding activity115,169. The identity of the capping enzyme, defined as the protein that transfers the cap — guanosine monophosphate (GMP) — to the RNA eluded researchers for several decades, until it was discovered that the nidovirus RNA-dependent RNA polymerase (RdRp)-associated nucleotidyltransferase (NiRAN) domain of the RdRp of the arterivirus equine arteritis virus, a distant coronavirus relative, possesses nucleotidyltransferase activity and can be covalently modified with GMP and UDP upon the addition of GTP and UTP, respectively82. This finding led to the proposal that the NiRAN domain is a guanylyltransferase (GMPylase) and transfers the covalently linked GMP to the now exposed 5′ β-phosphate of RNA. The GMPylase activity of the SARS-CoV-2 nsp12 NiRAN towards an RNA substrate was recently verified biochemically 93. Following the capping reaction, N7 of the guanine base is methylated (CH3). The bifunctional nsp14 performs the N7-methyltransferase (N7-MTase) activity using S-adenosylmethionine (SAM) as the methyl donor and releasing the reaction by-product S-adenosylhomocysteine (SAH)158,159. The resulting intermediate cap structure is called ‘cap 0’ or ‘m7GpppN-RNA’, and is presumed to protect viral RNAs from 5′-3′ exonucleases and to secure ribosomal recognition for translation167,168. Nsp10, which binds and stimulates the exonuclease activity of nsp14, does not affect nsp14’s MTase activity158. The crystal structures of the SARS-CoV nsp10–nsp14 complex revealed that nsp10 is bound to the exonuclease domain but not to the MTase domain of nsp14 and that the two domains are linked by a flexible hinge109,157. Notably, the structures revealed that the active site represents a new fold for MTases. This finding is helpful in developing inhibitors specifically targeting the nsp14 MTase activity without collateral inhibition of host methylases. The crystal structure of nsp10–nsp14 revealed the basis of the MTase reaction by capturing the SAM donor and a capped base (GpppN) acceptor in the active site157, potentially facilitating drug development. The last step of the capping pathway is the methylation of the ribose 2′-O of the first nucleotide of the mRNA. This ‘cap 1’ product or m7GpppN1m is formed by a separate MTase, nsp16. The stimulatory subunit, nsp10, is crucial for the activity of this 2′-O-methyltransferase (2′-O-MTase), and, as for the nsp14 N7-MTase, SAM serves as the methyl donor for this final step158,160. Nsp10–nsp16 will only methylate m7GpppA-RNA substrates158. 2′-O-Methylation is crucial for the escape of the virus from sensing by innate immunity receptors that can activate the type I interferon signalling pathway, and allows the virus to ‘camouflage’ its mRNA as host mRNAs170. The NiRAN nucleotidyltransferase activity82 and nsp14 MTase activity159 are essential for viral replication. Therefore, the enzymes in the capping pathway are attractive targets for antiviral drugs (reviewed in ref.168). It is currently unclear how each protein coordinates its individual activity towards the capping pathway or whether a larger capping complex exists to facilitate this process.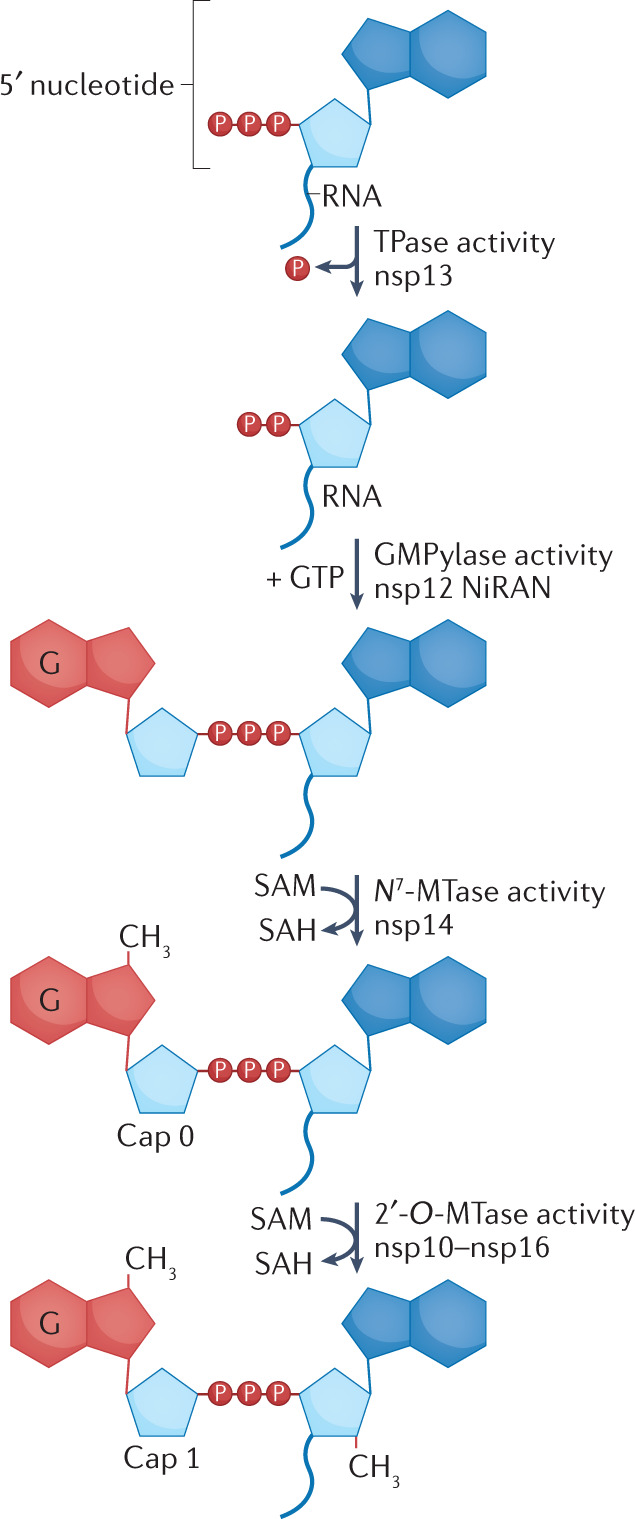
Regulation of template switching
The template switching required to extend the ‘body’ of the nascent minus-strand sgRNA with the anti-leader is primarily guided by the body transcription regulatory sequence (TRS-B) elements. These short sequences are found just upstream of the ORFs that encode structural and accessory proteins (except for those expressed through leaky scanning). After copying of a TRS-B sequence, minus-strand RNA synthesis stalls and the 3′ end of the nascent RNA strand is translocated to reinitiate RNA synthesis at the leader TRS (TRS-L) near the 5′ end of the gRNA template. This step is strongly facilitated by a base pairing interaction between the TRS-B complement at the 3′ end of the nascent minus strand (anti-TRS-B) and the TRS-L sequence in the gRNA template, as demonstrated previously in related viruses by site-directed mutagenesis studies51–54 (Fig. 3b). Coronavirus TRSs comprise a conserved core sequence (5′-ACGAAC-3′ in the case of SARS-CoV and SARS-CoV-2) that is flanked by sequences of variable length that may also contribute to the base pairing interaction with the TRS-L region20,29,30. In addition to the strength of the RNA duplex that is formed with the TRS-L region, other factors may co-determine the relative activity of a TRS-B element, and consequently the level at which the corresponding sg-mRNA is produced. These factors include the relative position of a TRS-B with respect to the 3′ end of the gRNA template, flanking RNA sequences51 and the local or overall RNA structure of the gRNA template16. Genome cyclization driven by long-distance RNA–RNA interactions (Fig. 3a), was recently proposed to expose the TRS-L for base pairing during discontinuous minus-strand synthesis16, similarly to what was previously postulated for arteriviruses55.
The series of ‘stop-or-go decisions’ at the consecutive TRS-B elements encountered by the minus strand-transcribing RTC is thought to fine-tune the relative abundances of the various sg-mRNAs, which remain largely similar throughout the course of infection56. The TRS-L is effectively ‘merged’ with one of the TRS-B elements in each of the sg mRNAs, thus positioning the ORF downstream of that TRS-B at the 5′-proximal position in the sg-mRNA and allowing it to be accessed by host ribosomes. Thus, coronavirus sg-mRNAs are nested and, except for the smallest species, polycistronic. However, they are presumed to be functionally monocistronic, with translation being restricted to the ORF most proximal to the 5′ end of the RNA, except in the case of sg-mRNAs on which leaky ribosomal scanning occurs to access a second ORF.
The canonical 5′-ACGAAC-3′ TRS core sequence occurs only nine times in the SARS-CoV-2 genome (in TRS-L and eight TRS-Bs), coordinating the production of eight sg-mRNA species (RNAs 2–9)20,29,30,57 (Fig. 2a). The smallest of these (mRNA 9) encodes the nucleocapsid protein and is by far the most abundant transcript30,56. For most sg-mRNAs, transcript abundance strongly correlates with ribosome footprint densities, indicating that they are translated with similar efficiencies, in line with the fact that their 5′ UTRs starting with the 75-nucleotide common leader sequence are largely identical (Fig. 2a). The detection of a separate sg-mRNA to express ORF7b was reported, derived from template switching at a TRS-B-like sequence (5′-AAGAAC-3′) located just upstream of ORF7b. Although the effective contribution of this TRS-B-like sequence to ORF7b expression may be limited20, this exemplifies how the (low-frequency) use of TRS-B-resembling sequences may yield additional subgenomic transcripts.
SARS-CoV-2 in-depth transcriptomics
Recently, the use of different highly sensitive techniques to study the SARS-CoV-2 transcriptome has identified numerous ‘non-canonical’ subgenomic transcripts20,29,30,57. These derive from TRS-L-dependent transcription, with the TRS-L, for example, being fused to downstream TRS-B-like sequences located in the middle of known ORFs; from large or local deletions generated without the apparent involvement of TRSs; or from the generation of (possibly) defective RNAs that may interfere with replication of the full-length genome by competing for the viral RdRp and other crucial replication factors, as described in several other coronaviruses58. These RNA species may in part derive from RNA recombination, which occurs at high frequency in coronaviruses59–61. The most accepted model for recombination in RNA viruses, similarity-assisted copy-choice RNA recombination, bears strong resemblance to the mechanism of coronavirus discontinuous minus-strand sgRNA synthesis47. Recombination involving host RNAs has also been invoked to explain gene acquisition during the evolution of coronaviruses and other nidoviruses3. It was hypothesized that TRS-B elements serve as recombination hotspots60,61, and that RNA secondary structures promote template switching in a TRS-independent manner61. Together, in-depth transcriptomics and ribosome-profiling experiments have revealed a complex landscape of SARS-CoV-2 RNAs and (potential) proteins, which extends well beyond the ‘canonical’ gene expression programme based on translation of the gRNA and canonical sg-mRNA20,29,30,57. Similar observations were made in other coronaviruses62,63. The additional transcripts may serve to express previously unknown small ORFs, truncated proteins or fused (partial) gene products, but their potential roles in SARS-CoV-2 replication and pathogenesis remain to be thoroughly investigated.
Balancing replication and transcription
It is unknown whether the composition of RTCs engaging in synthesis of minus-strand gRNA versus minus-strand sgRNAs is identical. Interactions with specific protein factors may govern the balance between replication and transcription. Two examples of transcription-specific protein functions have been documented in arteriviruses, which are distant coronavirus relatives in the order Nidovirales that also use discontinuous RNA synthesis to generate a nested set of sg-mRNAs64. The N-terminal subunit of the arterivirus replicase, nsp1, controls the accumulation of gRNA and sg-mRNAs by determining the levels at which their respective minus-strand templates are produced65. Specifically, mutagenesis of the N-terminal zinc-finger domain of nsp1 fully abrogated sg-mRNA synthesis, whereas gRNA production by such mutants increased 2.5–3-fold66. A serendipitous mutation just downstream of the zinc-binding domain of the helicase subunit also decreased arterivirus transcription and increased replication67,68, a finding that may be relevant to a recent hypothesis69,70 postulating that helicase-induced RTC backtracking contributes to the interruption of minus-strand RNA synthesis and/or to template switching (discussed later).
The role of the nucleocapsid protein in RNA synthesis
Although the primary role of the coronavirus nucleocapsid protein is gRNA encapsidation, it has also been implicated in a variety of other functions and interactions, including in regulating or modulating viral replication and transcription, although the interpretation of the supporting evidence is often complicated by the generally strong affinity of the nucleocapsid protein for RNA71–73. Both nonspecific RNA binding and binding to specific RNA sequences, including the TRS, have been reported, but often based on in vitro assays using purified nucleocapsid protein (reviewed in ref.71). A human coronavirus 229E RNA replicon lacking the nucleocapsid-encoding gene (and all other structural protein genes) retained the capability to replicate itself and synthesize sg-mRNAs74. This finding, and the fact that nucleocapsid-protein expression promotes viral replication75,76, suggests that the nucleocapsid protein has a modulatory rather than an essential role in coronavirus RNA synthesis. In line with this notion, the launching of coronavirus replication from in vitro-generated gRNA can be enhanced by co-expression of the nucleocapsid protein71. The nucleocapsid proteins of different coronaviruses interact with numerous other proteins, including with coronavirus replicase subunits such as nsp3 (ref.77) and host cell factors such as the RNA helicase DDX1 (ref.78). In the latter case, a complex formed between DDX1 and phosphorylated nucleocapsid protein was proposed to control the balance between replication and transcription by modulating the level of template switching at the successive TRS-B elements encountered by the RTC. The SARS-CoV-2 nucleocapsid protein was also shown to promote the cooperative association of the nsp7–nsp8–nsp12 complex with poly(U) RNA in vitro, thereby possibly facilitating initiation and/or elongation of viral RNA synthesis79.
The replication–transcription complex
The nsp7–nsp8–nsp12 holo-RdRp is the central component of the coronavirus RTC, and investigating the molecular basis of its RNA-synthesizing activity will facilitate rational drug design. Akin to other polynucleotide polymerases, the RdRp catalyses the incorporation of ribose nucleoside triphosphates (NTPs) into a nascent ‘product’ RNA using the information provided by the template RNA. However, maintaining the integrity of the template poses significant challenges to coronaviruses as their genomes are approximately 30 kb long, which is a burden considering the so-called error threshold that demarcates the genome size above which the long-term survival of an RNA virus species would be tenuous80,81. Thus, to preserve the integrity of their genetic information, coronaviruses have evolved mechanisms to mitigate the impact of nucleotide misincorporations during RNA synthesis3. This section highlights recent biochemical and structural studies of how coronaviruses orchestrate their replication and transcription, with the added aim of enhancing our understanding of the druggable SARS-CoV-2 proteome.
Molecular mechanism of RNA synthesis
Structural and bioinformatics work classified the coronavirus polymerase subunit (nsp12) into three domains: an N-terminal nidovirus RdRp-associated nucleotidyltransferase (NiRAN) domain (residues 1–250), an interface region between the NiRAN domain and the RdRp domain (residues 251–398) and the core RdRp domain (residues 399–932)82,83 (Fig. 4). The core RdRp domain assumes an architecture analogous to a cupped right hand composed of three subdomains: the fingers, palm and thumb84 (Fig. 4b). Within the core RdRp domain, the active site is further subdivided into seven functional features, known as motifs A–G, which are highly conserved across positive-stranded RNA viruses84 (Fig. 4d).
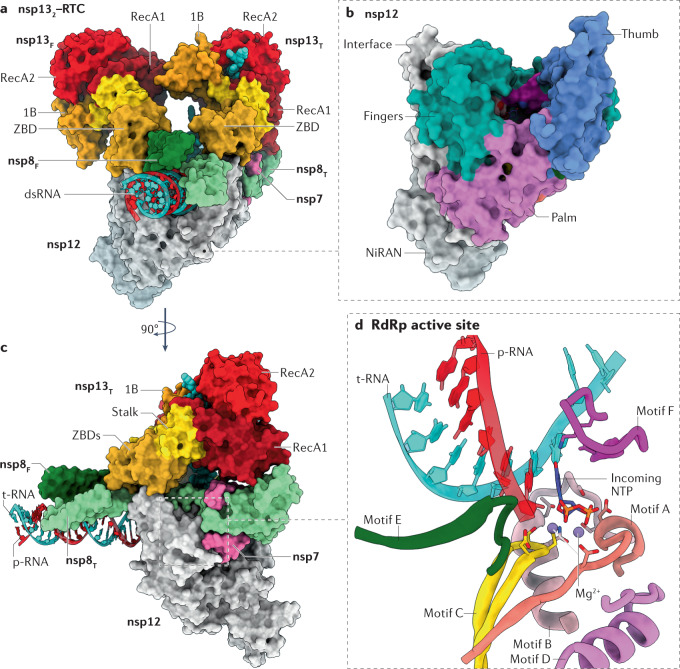
Fig. 4. Architecture of SARS-CoV-2 RNA replication and transcription complexes.
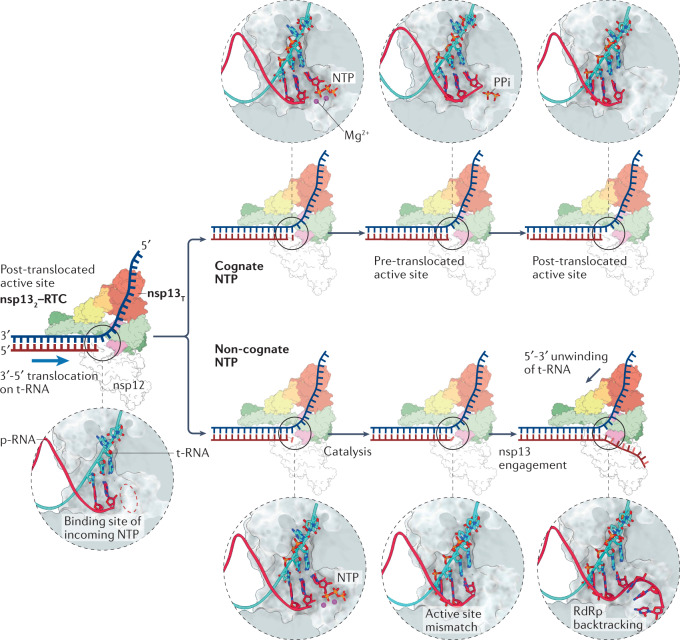
a | Surface-rendered representation illustrating two molecules of non-structural protein 13 (nsp13) bound to the replication–transcription complex (nsp132–RTC) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Depicted are the nsp7–nsp8–nsp12 RNA-dependent RNA polymerase (RdRp) holoenzyme bound to a double-stranded RNA (dsRNA) scaffold and to two molecules of nsp13: nsp13 thumb (nsp13T) and nsp13 fingers (nsp13F). The 1B, RecA1 and RecA2 domains of nsp13 are highlighted. b | The nsp12 RdRp domain adopts the canonical conformation of a ‘cupped right hand’ and its structural motifs are named accordingly. The nidovirus RdRp-associated nucleotidyltransferase (NiRAN) domain constitutes the amino-terminal 250 residues of nsp12. c | View of the nsp132–RTC complex rotated by 90° compared with part a. d | Zoomed-in view of the RdRp active site and its conserved structural motifs, except for motif G, which was excluded from the illustration for clarity. The active site is depicted with an incoming nucleotide, modelled using the pre-incorporation structures of the hepatitis C virus RdRp (Protein Data Bank entry 4WTL162). Residues involved in both chelating the catalytic Mg2+ ions and orientating the incoming nucleotide are shown in a stick representation. Protein Data Bank entry 6XEZ69 was used in preparation of this figure. NTP, nucleoside triphosphate; p-RNA, product RNA; t-RNA, template RNA; ZBD, zinc-binding domain.
Studies of smaller RdRps, amenable to X-ray crystallography, have provided a wealth of information on the role of these conserved structural motifs during the nucleotide addition cycle84,85. Initial nucleotide recognition is mediated by nonspecific charge–charge interactions of the nucleotide substrate with a series of positively charged Lys and Arg residues in motifs D and F of the nsp12 RdRp domain86. Molecular dynamics simulations of the hepatitis C virus RdRp indicate that the nucleotide diffuses into the central cleft through the NTP entry channel, until it reaches the active site located in the main channel86. The nucleotide ribose and base moieties subsequently flip into the active site through stabilizing hydrogen-bonding interactions with residues in motifs A and B and base-specific interactions with residues in motif F to form a Watson–Crick base pair with the template nucleotide86–88 (Fig. 4d). Correct positioning of the incoming nucleotide stabilizes the two catalytic Mg2+ ions that are necessary for the condensation reaction via interactions with the α and β phosphates of the incoming nucleotide, the product RNA 3′-hydroxy group and the catalytic Asp residues of motif C87,89. Closure of the RdRp active site via the stabilizing interactions of motifs A and B with the base enables the acid–base chemistry that drives the attack of the deprotonated product RNA 3′-hydroxy on the α-phosphorus atom of the incoming NTP, resulting in nucleotide addition and the release of pyrophosphate87,89.
The conformational state immediately after catalysis, in which the product RNA 3′ base occupies the incoming-nucleotide site, is referred to as the ‘pre-translocated state’ (Fig. 5). The conversion into the ‘post-translocated’ state mandates the release of pyrophosphate via opening of the active site through a subtle rotation of motif A84. Entry into the post-translocated state resets the active site for the next nucleotide addition cycle. A generalized kinetic scheme indicates that forward translocation is driven by the high nucleotide concentrations in the cellular milieu, which saturate the incoming-nucleotide site (Fig. 5). Failure to translocate would arrest the RTC and lead to the termination of RNA synthesis unless the translocation impediment is cleared. As we discuss later, one mechanism of action of remdesivir, an RdRp inhibitor used for COVID-19 treatment, can be summarized as hindering RdRp translocation through steric effects of its base in the active site.
Fig. 5. The nucleotide addition cycle.
Model of the interactions between non-structural protein 13 thumb (nsp13T) and nsp12 during RNA synthesis. Zoomed-in views of the active site during the nucleotide addition cycle are presented above or below each panel. In the presence of a natural, cognate nucleoside triphosphate (NTP), the incoming NTP forms a phosphodiester bond with the product RNA (p-RNA) (top). Nsp13T is not engaged with the template RNA (t-RNA), and forward translocation of the nsp12 RNA-dependent RNA polymerase (RdRp) on the t-RNA strand is unimpeded. In the event of incorporation of a non-natural or non-cognate nucleotide, the active site is perturbed and activates the helicase nsp13 to induce RdRp backtracking, as shown in the last panel (bottom). nsp132, nsp13T–nsp13F; PPi, pyrophosphate; RTC, replication–transcription complex.
The enigmatic NiRAN domain of coronavirus nsp12
The NiRAN domain (Fig. 4b) attracted considerable interest following revelations that it is an essential enzymatic domain that is absent in RNA viruses outside the order Nidovirales82. Early investigations revealed that the NiRAN domain of the RdRp of the arterivirus equine arteritis virus possesses a self-nucleotidylating activity (NMPylation), which may prime the domain to transfer a nucleoside monophosphate (NMP) to another protein or nucleic acid substrate82. This activity is essential for nidovirus replication, as mutations that catalytically inactivate the NiRAN domains of equine arteritis virus and SARS-CoV were lethal82. Recent studies of the coronavirus NiRAN domain suggest that it might function as an RNA ligase, serve as a guanylyltransferase that catalyses the transfer of guanine monophosphate (GMP) during mRNA capping (Box 2) or transfer an NMP to another viral protein to serve in protein-primed initiation of RNA synthesis82. It is possible that the NiRAN domain performs multiple activities or each of these activities, depending on the substate and context. The possible role of the NiRAN in the capping mechanism is discussed in Box 2, and the RNA ligase activity of NiRAN has not been demonstrated so far.
The NiRAN domain’s possible role in protein-primed RNA synthesis warrants further exploration. Members of another clade of positive-strand RNA viruses, the order Picornavirales, initiate RNA synthesis using the protein primer viral protein genome-linked (VPg). A dinucleotide (UpU) that is covalently linked to a hydroxy group of a tyrosine, serine or threonine residue in VPg serves to prime RNA synthesis on the poly(A)-tailed template90. Consistent with the earlier work on equine arteritis virus82, the NiRAN domains of SARS-CoV, human coronavirus 229E and SARS-CoV-2 display higher specificity in vitro for UTP over GTP (UMPylation activity over GMPylation activity)91,92. Given this UTP specificity, it was posited that NiRAN could facilitate the UMPylation of a priming protein to initiate minus-strand RNA synthesis at the 3′ poly(A) tail of the gRNA template92. Recent in vitro evidence indicates that nsp9, a single-stranded RNA-binding protein, is a substrate for UMPylation at or near its N terminus92. Consistent with this finding, mutagenesis of the N-terminal residues of nsp9 severely affected viral replication. These results, combined with a structure of nsp9 bound to the NiRAN domain in an inhibited state93, implicate nsp9 in the initiation of RNA synthesis. Given observations that other RTC components and purification contaminants (such as proteins from bacterial expression systems) can be NMPylated in vitro91,92, further studies are needed to define the biologically relevant repertoire of substrates targeted by NiRAN’s NMPylation activity.
The expanded replication machinery
Replication and transcription are assumed to be executed by various subcomplexes, which include the holo-RdRp associated with other non-structural protein enzymes and accessory subunits. These viral enzymes are required to promote the fidelity of RNA synthesis, equip viral mRNAs with a 5′ cap structure and orchestrate the template switching needed for sgRNA synthesis. The interplay between these subunits and their interactions with regulatory viral RNA elements coordinate the timely replication and expression of the coronavirus genome, and provide a platform for continuous coronavirus evolution.
The holo-RdRp
To faithfully replicate the coronavirus genome, an arsenal of factors is needed to enhance the processivity of the RTC and repair errors in RNA synthesis4. Early biochemical experiments revealed that the primer-extension processivity of SARS-CoV RdRp (nsp12) is greatly increased in the presence of nsp7 and nsp8, cementing their role as essential subunits of the holo-RdRp complex94. Advancements in cryo-electron microscopy led to the seminal structure of the SARS-CoV apo-holo-RdRp complex83 and more recently to several structures of the SARS-CoV-2 RTC95–98. In the presence of an RNA duplex, the N termini of two nsp8 subunits form ordered helices that have nonspecific ionic interactions with the RNA backbone, illuminating the importance of nsp8 for enhanced RdRp processivity95 (Fig. 4a,c). In addition to enhancing processivity, nsp8 interacts in vitro with various other non-structural proteins thought to assist the RTC99,100. Thus, nsp8 is posited to be important for forming higher-order RTCs that couple replication and transcription with template unwinding (nsp13), proofreading (nsp10, nsp12, nsp13 and nsp14) and RNA capping (nsp10, nsp13, nsp14 and nsp16).
The exoribonuclease activity of nsp14
Coronaviruses encode a unique proofreading activity that is not found in other RNA viruses, including in nidoviruses with small genomes such as arteriviruses3,22,101. This activity is encoded in the N-terminal exoribonuclease (ExoN) domain of nsp14, which together with nsp10 forms an RNA proofreading complex102–104 that is presumed to promote faithful replication of large nidovirus genomes22. In some coronaviruses, such as murine hepatitis virus (MHV) and SARS-CoV, nsp14-ExoN knockout yielded crippled but viable viruses exhibiting a mutator phenotype105–107. Remarkably, equivalent ExoN-inactivating substitutions completely abolish MERS-CoV and SARS-CoV-2 replication, suggesting a function for ExoN in primary RNA synthesis108. Because of its role in enhancing the fidelity of genome replication, nsp14 ExoN can promote antiviral drug resistance. For example, in an in vitro assay, its 3′-5′ exoribonuclease activity can efficiently cleave ribavirin, an antiviral nucleoside analogue, from the 3′ end of an RNA substrate, indicating that the enzyme may promote high-level resistance to certain nucleoside analogue antiviral drugs109. Recent cryo-electron microscopy structures have revealed the molecular basis for how the ExoN domain of the nsp10–nsp14 complex interacts with double-stranded RNA containing a 5′ overhang and a one-nucleotide mismatch at the 3′ end110. The mismatched base enters the shallow ExoN active site and interacts with conserved catalytic residues via its 3′-hydroxy and 2′-hydroxy groups. In addition, the double-stranded portion of the RNA interacts with both the nsp10 N terminus and nsp14-ExoN residues outside the catalytic site. These structures provide direct visualization of recognition by ExoN of its preferred mismatched RNA substrate110.
Colliding motors: how the helicase induces RdRp backtracking
Nsp13 is an SF1B-family RNA helicase that is essential for coronavirus replication111–115. Biochemical assays revealed that its 5′-3′ nucleic acid unwinding activity is enhanced twofold in the presence of nsp12 (ref.116). Nsp13 makes stable interactions with the SARS-CoV-2 RTC, which enabled the structural determination of nsp13 bound to the RTC (nsp132–RTC)69,117. Single-particle image classification revealed that the major particle class consists of two molecules of nsp13 bound to the RTC. Both nsp13 molecules (nsp13F (fingers) and nsp13T (thumb)) have extensive interactions with the holo-RdRp, whereas only one copy (nsp13T) is bound to the RNA scaffold at the 5′ end of the template RNA (Fig. 4a,c). Nsp13T, bound to the template strand downstream of the RdRp active site, is positioned to translocate in the opposite direction relative to the RdRp69 (Fig. 5). The opposing directionalities of nsp13 and the RdRp are hypothesized to trigger a translocation conflict. Forward translocation of nsp13T on the template strand was proposed to lead to the reverse threading of the RdRp on the product RNA strand69. The reverse movement of polymerases, relative to their nucleic acid substrate, is well known for all cellular RNA polymerases and is termed ‘backtracking’118 (Fig. 5). The role of the second nsp13, nsp13F, is poorly understood but it has been proposed to regulate the unwinding activity of the substrate-bound nsp13T (ref.117).
A perspective on the role of backtracking in proofreading by the RdRp
Similarly to its role in cellular RNA polymerases, backtracking may be essential for excision of misincorporated nucleotides in coronavirus RNA synthesis69,70,119,120. Indeed, behaviours consistent with backtracking have been observed in single-molecule magnetic tweezer experiments for the SARS-CoV-2 RdRp and RdRps from the Φ6 bacteriophage and poliovirus, illuminating the potentially widespread nature of backtracking in the viral realm119–121. Furthermore, recent evidence indicates that the NTP entry channel can accommodate a single-stranded product RNA 3′ overhang, which mirrors the backtracking product70. Molecular dynamics simulations further showed that entry into the backtracking state occurs when a misincorporated RNA base flips from the pre-translocated state towards the mouth of the NTP entry channel70. Subsequently, the engagement of nsp13 with the template RNA would enhance the backtracking activity and offer a means to control entry into a long-lived backtracked state (Fig. 5).
Backtracking may grant nsp14 ExoN access to any misincorporated nucleotide at the 3′ end of nascent product RNAs, thereby coupling proofreading with RNA synthesis69,70. Alternatively, as suggested by the recent cryo-electron microscopy structures of nsp10–nsp14 bound to a double-stranded RNA substrate with a 5′ overhang and a 3′ nucleotide mismatch110, misincorporation events may lead the RdRp to release the mismatch-containing RNA duplex, thereby granting ExoN access for proofreading. Additionally, it is envisaged that backtracking could have a role in discontinuous RNA synthesis by exposing the anti-TRS-B at the 3′ end of the nascent minus-strand sgRNAs and mediating template switching69,70 (Fig. 3b). This hypothesis is supported by a mutation in the arterivirus helicase that does not affect genome replication but abolishes all sgRNA transcription68,122.
Coupling of backtracking with nsp14-ExoN activity could also explain how coronaviruses excise non-natural nucleotides from their nascent product RNA. Genetic loss of function experiments indicated that nsp14 ExoN mitigates the effect of nucleoside and base analogues such as ribavirin and fluorouracil, respectively, in the betacoronavirus MHV102,105,106. These inhibitors are ineffective for treating SARS-CoV, MERS-CoV and SARS-CoV-2 infections, highlighting that the protection conferred by nsp14 ExoN is of clinical concern in the hunt for promising nucleoside analogues106,123. Combination therapy approaches may yield fruitful outcomes given the likely synergy between nsp14-ExoN and nsp12 (RdRp) inhibition. Therefore, it is pertinent to better understand how nsp14 is recruited for excision and repair124. In addition, shedding light on why some nucleoside analogues, such as remdesivir and molnupiravir (discussed in the next section), are effective could provide valuable insight into the design of novel antiviral nucleoside analogues for monotherapy.
A rational design of RdRp inhibitors
Although vaccines against SARS-CoV-2 have shown remarkable efficacy, COVID-19 continues to spread and affect communities globally. The reasons for this are multiple, and include vaccine shortages, public vaccine hesitancy, reduced vaccine effectiveness in immunosuppressed people and the emergence of new virus variants. It is therefore anticipated that SARS-CoV-2 will become endemic125, potentially evolving in the human host and leading to gradual or more sudden reductions of vaccine efficacy. Given such concerns, the search for drugs against SARS-CoV-2 and related viruses remains a priority in the research community. In this section, we discuss the mechanisms of action of two RdRp inhibitors, remdesivir and molnupiravir, that show clinical benefit in treating COVID-19.
Mechanisms of action of remdesivir and molnupiravir
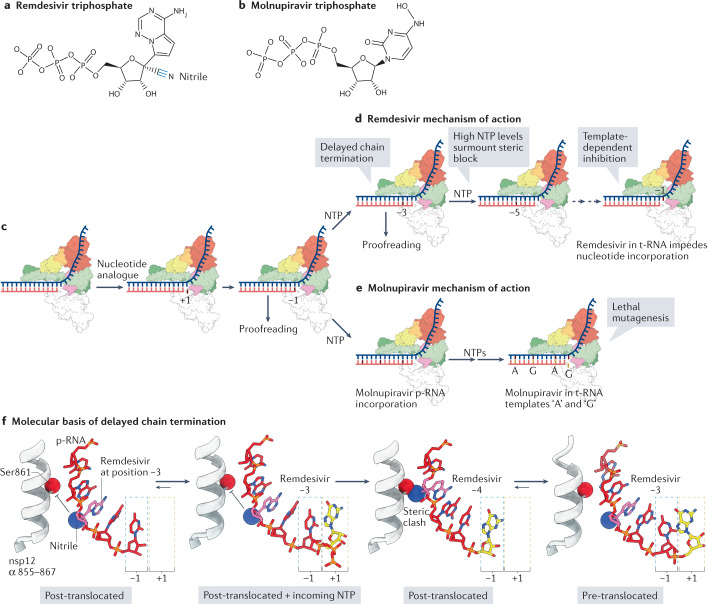
Nucleoside analogues, which can target the RdRp, are common antiviral therapeutics125. Currently remdesivir and molnupiravir are two antiviral drugs used to treat COVID-19 (ref.126). Studies indicate that treatment with remdesivir decreases the duration of the infection in hospitalized patients127. Biochemical evidence demonstrates that the SARS-CoV-2 RdRp preferably incorporates remdesivir (Fig. 6a) rather than its natural analogue adenosine and can incorporate molnupiravir (Fig. 6b) rather than its natural analogue cytidine126,128–131. Once incorporated, neither inhibitor induces immediate pausing of RNA synthesis, in contrast to classical chain terminators126,129,131 (Fig. 6c–e). Initial studies had suggested that remdesivir inhibits RNA synthesis via a delayed chain termination mechanism126,130,132,133. Delayed chain termination occurs when remdesivir impedes RdRp translocation following a steric clash between its nitrile group and nsp12 Ser861, which occurs when remdesivir reaches the fourth position from the 3′ end of the product RNA126,132,133 (Fig. 6f). This steric inhibition is surmounted in vitro in the presence of subphysiological concentrations of NTPs, indicating it is unlikely the major inhibitory hurdle for viral replication in living cells. Instead, recent data suggest that remdesivir may impair replication when incorporated into the template strand following an initial round of viral RNA synthesis134. In the template strand, remdesivir hinders the incorporation of the incoming nucleotide. This mode of activity has been termed ‘template-dependent inhibition’134 (Fig. 6d). Following the eventual incorporation of this incoming nucleotide, a second potential checkpoint was proposed, in which remdesivir would bias the RdRp towards the pre-translocated state, although direct evidence for this is lacking134.
Fig. 6. Mechanisms of inhibition of coronavirus RNA synthesis by remdesivir and molnupiravir.
a | Chemical structure of remdesivir triphosphate, the activated form of remdesivir (Veklury). The ribose 1′-nitrile group is highlighted. b | Chemical structure of molnupiravir triphosphate, the activated form of molnupiravir (EIDD-2801/MK-4482). c | Schematic showing initial recognition and incorporation of a nucleotide analogue. ‘+1’ refers to the position of the nucleotide analogue before incorporation, whereas ‘−1’ refers to the position of the nucleotide analogue following catalysis and movement into the post-translocated register. d | Remdesivir exerts its inhibitory effect through ‘delayed chain termination’ and ‘template-dependent inhibition’126,134. Delayed chain termination impedes RNA-dependent RNA polymerase (RdRp) translocation through a steric clash that occurs when remdesivir reaches the fourth position (see part f) from the 3′ end of the product RNA (p-RNA) strand (red strand). Delayed chain termination is alleviated by high nucleoside trisphosphate (NTP) concentrations, leading to the proposal of template-dependent inhibition, which occurs when remdesivir is positioned at the RdRp active site in the template RNA (t-RNA) strand. e | Molnupiravir perturbs replication through ‘lethal mutagenesis’, by enabling the indiscriminate incorporation of either ATP (A) or GTP (G) when it is positioned in the t-RNA strand129,131. f | Structural analysis of the delayed chain termination mechanism. Following incorporation of remdesivir and its translocation to the −3 active site position, the ribose 1′-nitrile group is positioned to collide with the side chain of non-structural protein 12 (nsp12) Ser861 (expected clash shown by inhibitory arrows). Addition of the next nucleotide triggers the steric clash of the Ser861 side chain and the remdesivir nitrile group, forcing the growing RNA chain to populate the ‘pre-translocated’ state133. High NTP concentrations may alleviate the energetic cost imposed by the translocation barrier, by occupying the +1 site and thereby driving the forward movement of the RdRp. The structural models are based on Protein Data Bank entries 7B3B, 7B3C and 7B3D133.
Like remdesivir, molnupiravir is a prodrug that is converted in cells into its triphosphate form, thereby serving as a nucleotide analogue. Molnupiravir inhibits replication through lethal mutagenesis of the genomes of multiple viruses, including SARS-CoV-2 (refs135–138). Molnupiravir treatment presents a high barrier to resistance in cell culture assays135,136. Importantly, like remdesivir, molnupiravir escapes from the coronavirus nsp14-ExoN proofreading activity136. Unlike remdesivir, molnupiravir is delivered orally, which, combined with its high barrier to resistance and potent antiviral activity, led to its pursuit as an alternative therapeutic for COVID-19 (ref.139). Molnupiravir triphosphate is a cytidine analogue that exerts its effect by indiscriminately serving as a template for the incorporation of either adenine or guanine, thus explaining the observation of the transition mutations G>A and C>U in coronaviruses exposed to molnupiravir129 (Fig. 6e). Two recently resolved cryo-electron microscopy structures of molnupiravir base-paired with adenine or guanine revealed the structural basis of molnupiravir-mediated lethal mutagenesis131.
The RdRp possesses high selectivity for remdesivir and molnupiravir due to their excellent mimicry of natural nucleotides. Therefore, these compounds do not significantly affect the initial round of RNA synthesis after incorporation, a feature which likely reduces their recognition and excision by nsp14 ExoN surveying the fidelity of RTCs.
Overcoming the proofreading barrier in antiviral drug design
Designing nucleoside analogues that escape the proofreading activity of nsp14 ExoN is a trial-by-error endeavour since it is challenging to pinpoint chemical properties that would lead to nucleotide mimicry. A more rational approach could entail targeting the enzymatic activity of ExoN or its interfaces with nsp10 or the RTC. The ExoN activity is essential for SARS-CoV-2 and MERS-CoV replication, but it is not vital for viral propagation across the betacoronavirus clade108. nsp14-ExoN inactivation in MHV and SARS-CoV, although not lethal, enhances the susceptibility of the virus to nucleoside analogues, highlighting the benefits of dual-inhibition strategies in coronaviruses105. One concern is the potential for off-target effects when the ExoN active site is being targeted with a small-molecule inhibitor, due to structural similarities to other cellular DEDD-family exonucleases. Designing inhibitors against the interface of nsp10 with nsp14 ExoN and nsp16 has attracted interest given that viral replication is abrogated in interface mutants140,141.
A long-standing research interest is the characterization of how the proofreading complex, nsp10–nsp14, interacts with the RTC. Pull-down experiments using a series of truncated proteins indicated that nsp12 interacts with nsp14 and its subdomains109. Recent structural work showed that SARS-CoV-2 nsp10–nsp14 can be recruited to the RTC by forming a covalent link with nsp9, which is bound to the nsp12 NiRAN domain124. Prior observations in MHV inspired the rationale for using as nsp9–nsp10 fusion protein in that study, as ablation of the protease cleavage site between nsp9 and nsp10 in MHV maintains a viable phenotype. This nsp9–nsp10 protease cleavage mutant, however, experienced a pronounced overall defect in RNA synthesis, and the propagation of this mutant was severely compromised142. Given the crippled phenotype of the MHV nsp9–nsp10 cleavage-site mutant142, it remains to be shown whether the same interaction with nsp10–nsp14 can occur when nsp9 and nsp10 are separated. The recent structural analysis of the SARS-CoV-2 nsp12–nsp9–nsp10–nsp14 complex did not reveal any features that would suggest that the incorporation of an nsp9–nsp10 fusion protein affects RTC assembly124. Probing the role of nsp10–nsp14 in greater detail will benefit from single-molecule experiments using reconstituted RTC components. This approach could also test whether nsp14 ExoN alleviates backtracking as proposed69,70. Furthermore, both the engagement of proteins with RTCs and the proposed proofreading activity of nsp14 ExoN will have to be demonstrated in vivo.
Future perspectives
Although the surge of new coronavirus research has expanded our understanding of the molecular mechanisms of SARS-CoV-2 replication and gene expression, the foundation of our knowledge of these processes is built primarily on previous research of other coronaviruses and the distantly related arteriviruses. Corroborating our understanding will likely reveal properties and processes shared between viral species, which are of potential value for the design of pan-coronavirus inhibitors. More specifically, it is pertinent to unravel poorly understood intricacies of spatiotemporal regulation of RNA synthesis in coronaviruses. Unknown to us is the complete repertoire of host-cell factors involved in assisting the coronavirus infection cycle and how these factors may, for example, be subverted to assist in the formation of replication organelles or contribute to the formation of RTCs. The spatial segregation of coronavirus replication in virus-induced membranous organelles appears to be a requisite for successful virus propagation — a feature shared among positive-strand RNA viruses, although it remains to be elucidated where in the cell RNA synthesis occurs during the earliest phase of infection.
Downstream of replication organelle formation, key questions include how RNA synthesis is primed on its templates and how regulation of the two major synthesis pathways is orchestrated. Such regulation must be achieved by the concerted action of the replicase proteins and may be further assisted by host factors, whose role in these pathways is relatively unexplored. Regulating RNA synthesis necessitates the faithful maintenance of the encoded genetic information, as unwanted mutations can alter the RNA elements required for processes such as template switching and that lead to the production of nonsense transcripts. Yet to be worked out in detail is how the coronavirus proofreading complex coordinates its activity with the polymerase, leading to the excision of misincorporated RNA nucleotides and nucleotide analogues. Understanding how mutations accumulate during replication and how they are corrected can inform us on the evolution of drug resistance mutations and aid the design of inhibitors that directly target the replicase complex. We hope that such considerations may guide research that will shape our response to future deadly outbreaks of coronaviruses, a consequence of the encroaching footprint of humanity on the natural world.
Acknowledgements
The authors apologize to colleagues whose work could not be cited due to scope and space limits of the manuscript. Figures 1–3 and the figure in Box 2 were created with BioRender. N.U. was supported by grant LSHM20047 from the Foundation Life Sciences Health-TKI. E.A.C. is grateful for support from NIH grants 2-R01 GM114450 and 1-R01 AI161278. E.J.S. was supported by the #wakeuptocorona crowdfunding initiative of the Leiden University Fund and Leiden University Medical Center Bontius Foundation.
Glossary
- RNA genome of positive polarity
An RNA genome that has mRNA polarity and, when released from viral particles, can be used directly by host ribosomes to produce viral proteins.
- Replication and transcription
Process of amplification of the viral genome through a full-length minus-strand intermediate serving as template and synthesis of subgenomic mRNAs through a set of subgenomic minus-strand templates.
- −1 programmed ribosomal frameshift
(−1 PRF). A regulated switch of the translating ribosome to an alternative open reading frame by shifting one nucleotide backwards on the mRNA.
- Nucleocapsid
Complex of genomic RNA and nucleocapsid proteins that forms the core of a coronavirus particle.
- Pseudoknot
A structural motif that arises as a result of base paring between the loop of an RNA hairpin and a complementary single-stranded (unpaired) region within RNA.
- Similarity-assisted copy-choice RNA recombination
A process of template switching during replication of an RNA virus genome that is guided by local sequence complementarity between the nascent strand and an alternative template, resulting in progeny genomes of mixed ancestry.
- RNA replicon
A self-replicating RNA molecule, derived from a viral genome, that contains the replicase gene but lacks at least one essential structural protein gene and thus is unable to produce infectious progeny.
- Error threshold
The size limit of a viral genome, above which too many mutations accumulate to sustain long-term viability of the virus.
Author contributions
The authors contributed equally to all aspects of the article.
Competing interests
E.A.C has received funding from Gilead Sciences to fund research on remdesivir’s incorporation into the RNA-dependent RNA polymerase. The other authors declare no competing interests.
Footnotes
Peer review information
Nature Reviews Molecular Cell Biology thanks Hauke Hillen, Kenneth Johnson and Yi Shi for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
GenBank entry MN908947.3: https://www.ncbi.nlm.nih.gov/genome/?term=MN908947.3
These authors contributed equally: Brandon Malone, Nadya Urakova.
Contributor Information
Eric J. Snijder, Email: e.j.snijder@lumc.nl
Elizabeth A. Campbell, Email: elizabeth.campbell@rockefeller.edu
References
- 1.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gulyaeva AA, Gorbalenya AE. A nidovirus perspective on SARS-CoV-2. Biochem. Biophys. Res. Commun. 2021;538:24–34. doi: 10.1016/j.bbrc.2020.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gorbalenya AE, Enjuanes L, Ziebuhr J, Snijder EJ. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117:17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Snijder, E. J., Decroly, E. & Ziebuhr, J. in Advances in Virus Research Vol. 96 (ed. Ziebuhr, J.) 59–126 (Elsevier, 2016). [DOI] [PMC free article] [PubMed]
- 5.Saberi A, Gulyaeva AA, Brubacher JL, Newmark PA, Gorbalenya AE. A planarian nidovirus expands the limits of RNA genome size. PLOS Pathog. 2018;14:e1007314. doi: 10.1371/journal.ppat.1007314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hilgenfeld R, Peiris M. From SARS to MERS: 10 years of research on highly pathogenic human coronaviruses. Antivir. Res. 2013;100:286–295. doi: 10.1016/j.antiviral.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Subissi L, et al. SARS-CoV ORF1b-encoded nonstructural proteins 12-16: replicative enzymes as antiviral targets. Antivir. Res. 2014;101:122–130. doi: 10.1016/j.antiviral.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zumla A, Chan JFW, Azhar EI, Hui DSC, Yuen K-Y. Coronaviruses - drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pruijssers AJ, Denison MR. Nucleoside analogues for the treatment of coronavirus infections. Curr. Opin. Virol. 2019;35:57–62. doi: 10.1016/j.coviro.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.V’kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whittaker GR, Daniel S, Millet JK. Coronavirus entry: how we arrived at SARS-CoV-2. Curr. Opin. Virol. 2021;47:113–120. doi: 10.1016/j.coviro.2021.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shang J, et al. Cell entry mechanisms of SARS-CoV-2. Proc. Natl Acad. Sci. USA. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ou X, et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu N, et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu F, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ziv O, et al. The short- and long-range RNA-RNA interactome of SARS-CoV-2. Mol. Cell. 2020;80:1067–1077.e5. doi: 10.1016/j.molcel.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manfredonia I, et al. Genome-wide mapping of SARS-CoV-2 RNA structures identifies therapeutically-relevant elements. Nucleic Acids Res. 2020;48:12436–12452. doi: 10.1093/nar/gkaa1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rangan R, et al. De novo 3D models of SARS-CoV-2 RNA elements from consensus experimental secondary structures. Nucleic Acids Res. 2021;49:3092–3108. doi: 10.1093/nar/gkab119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huston NC, et al. Comprehensive in vivo secondary structure of the SARS-CoV-2 genome reveals novel regulatory motifs and mechanisms. Mol. Cell. 2021;81:584–598.e5. doi: 10.1016/j.molcel.2020.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finkel Y, et al. The coding capacity of SARS-CoV-2. Nature. 2021;589:125–130. doi: 10.1038/s41586-020-2739-1. [DOI] [PubMed] [Google Scholar]
- 21.Ziebuhr J, Snijder EJ, Gorbalenya AE. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 2000;81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- 22.Snijder EJ, et al. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 Lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lokugamage KG, Narayanan K, Huang C, Makino S. Severe acute respiratory syndrome coronavirus protein nsp1 is a novel eukaryotic translation inhibitor that represses multiple steps of translation initiation. J. Virol. 2012;86:13598–13608. doi: 10.1128/JVI.01958-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thoms M, et al. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science. 2020;369:1249. doi: 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schubert K, et al. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat. Struct. Mol. Biol. 2020;27:959–966. doi: 10.1038/s41594-020-0511-8. [DOI] [PubMed] [Google Scholar]
- 26.Cortese M, et al. Integrative imaging reveals SARS-CoV-2-induced reshaping of subcellular morphologies. Cell Host Microbe. 2020;28:853–866.e5. doi: 10.1016/j.chom.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Snijder EJ, et al. A unifying structural and functional model of the coronavirus replication organelle: tracking down RNA synthesis. PLoS Biol. 2020;18:e3000715. doi: 10.1371/journal.pbio.3000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghosh S, et al. β-Coronaviruses use lysosomes for egress instead of the biosynthetic secretory pathway. Cell. 2020;183:1520–1535.e14. doi: 10.1016/j.cell.2020.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang D, et al. The SARS-CoV-2 subgenome landscape and its novel regulatory features. Mol. Cell. 2021;81:2135–2147.e5. doi: 10.1016/j.molcel.2021.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim D, et al. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181:914–921.e10. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masters, P. S. & Perlman, S. in Fields Virology (eds Knipe, D. M. & Howley, P. M.) 825–858 (Lippincott, Williams & Wilkins, 2013).
- 32.Jungreis I, et al. Conflicting and ambiguous names of overlapping ORFs in the SARS-CoV-2 genome: a homology-based resolution. Virology. 2021;558:145–151. doi: 10.1016/j.virol.2021.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jungreis I, Sealfon R, Kellis M. SARS-CoV-2 gene content and COVID-19 mutation impact by comparing 44 Sarbecovirus genomes. Nat. Commun. 2021;12:2642. doi: 10.1038/s41467-021-22905-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Firth AE. A putative new SARS-CoV protein, 3c, encoded in an ORF overlapping ORF3a. J. Gen. Virol. 2020;101:1085–1089. doi: 10.1099/jgv.0.001469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bojkova D, et al. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature. 2020;583:469–472. doi: 10.1038/s41586-020-2332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Firth AE, Brierley I. Non-canonical translation in RNA viruses. J. Gen. Virol. 2012;93:1385–1409. doi: 10.1099/vir.0.042499-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schaecher SR, Mackenzie JM, Pekosz A. The ORF7b protein of severe acute respiratory syndrome coronavirus (SARS-CoV) is expressed in virus-infected cells and incorporated into SARS-CoV particles. J. Virol. 2007;81:718–731. doi: 10.1128/JVI.01691-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu K, et al. Severe acute respiratory syndrome coronavirus accessory protein 9b is a virion-associated protein. Virology. 2009;388:279–285. doi: 10.1016/j.virol.2009.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brierley I, Digard P, Inglis SC. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bhatt PR, et al. Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome. Science. 2021;372:1306–1313. doi: 10.1126/science.abf3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Plant EP, Rakauskaite R, Taylor DR, Dinman JD. Achieving a golden mean: mechanisms by which coronaviruses ensure synthesis of the correct stoichiometric ratios of viral proteins. J. Virol. 2010;84:4330–4340. doi: 10.1128/JVI.02480-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Su M-C, Chang C-T, Chu C-H, Tsai C-H, Chang K-Y. An atypical RNA pseudoknot stimulator and an upstream attenuation signal for -1 ribosomal frameshifting of SARS coronavirus. Nucleic Acids Res. 2005;33:4265–4275. doi: 10.1093/nar/gki731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ivanov KA, et al. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc. Natl Acad. Sci. USA. 2004;101:12694–12699. doi: 10.1073/pnas.0403127101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kindler E, et al. Early endonuclease-mediated evasion of RNA sensing ensures efficient coronavirus replication. PLoS Pathog. 2017;13:e1006195. doi: 10.1371/journal.ppat.1006195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hackbart M, Deng X, Baker SC. Coronavirus endoribonuclease targets viral polyuridine sequences to evade activating host sensors. Proc. Natl Acad. Sci. USA. 2020;117:8094–8103. doi: 10.1073/pnas.1921485117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sawicki SG, Sawicki DL. Coronaviruses use discontinuous extension for synthesis of subgenome-length negative strands. Adv. Exp. Med. Biol. 1995;380:499–506. doi: 10.1007/978-1-4615-1899-0_79. [DOI] [PubMed] [Google Scholar]
- 47.Sawicki SG, Sawicki DL, Siddell SG. A contemporary view of coronavirus transcription. J. Virol. 2007;81:20–29. doi: 10.1128/JVI.01358-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pasternak AO, Spaan WJM, Snijder EJ. Nidovirus transcription: how to make sense…? J. Gen. Virol. 2006;87:1403–1421. doi: 10.1099/vir.0.81611-0. [DOI] [PubMed] [Google Scholar]
- 49.Nakagawa K, Makino S. Mechanisms of coronavirus nsp1-mediated control of host and viral gene expression. Cells. 2021;10:300. doi: 10.3390/cells10020300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Banerjee AK, et al. SARS-CoV-2 disrupts splicing, translation, and protein trafficking to suppress host defenses. Cell. 2020;183:1325–1339.e21. doi: 10.1016/j.cell.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sola I, Moreno JL, Zúñiga S, Alonso S, Enjuanes L. Role of nucleotides immediately flanking the transcription-regulating sequence core in coronavirus subgenomic mRNA synthesis. J. Virol. 2005;79:2506–2516. doi: 10.1128/JVI.79.4.2506-2516.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yount B, Roberts RS, Lindesmith L, Baric RS. Rewiring the severe acute respiratory syndrome coronavirus (SARS-CoV) transcription circuit: engineering a recombination-resistant genome. Proc. Natl Acad. Sci. USA. 2006;103:12546–12551. doi: 10.1073/pnas.0605438103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sola I, Almazán F, Zúñiga S, Enjuanes L. Continuous and discontinuous RNA synthesis in coronaviruses. Annu. Rev. Virol. 2015;2:265–288. doi: 10.1146/annurev-virology-100114-055218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pasternak AO, van den Born E, Spaan WJ, Snijder EJ. Sequence requirements for RNA strand transfer during nidovirus discontinuous subgenomic RNA synthesis. EMBO J. 2001;20:7220–7228. doi: 10.1093/emboj/20.24.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van den Born E, Posthuma CC, Gultyaev AP, Snijder EJ. Discontinuous subgenomic RNA synthesis in arteriviruses is guided by an RNA hairpin structure located in the genomic leader region. J. Virol. 2005;79:6312–6324. doi: 10.1128/JVI.79.10.6312-6324.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ogando NS, et al. SARS-coronavirus-2 replication in Vero E6 cells: replication kinetics, rapid adaptation and cytopathology. J. Gen. Virol. 2020;101:925–940. doi: 10.1099/jgv.0.001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davidson AD, et al. Characterisation of the transcriptome and proteome of SARS-CoV-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein. Genome Med. 2020;12:68. doi: 10.1186/s13073-020-00763-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brian DA, Baric RS. Coronavirus genome structure and replication. Curr. Top. Microbiol. Immunol. 2005;287:1–30. doi: 10.1007/3-540-26765-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Masters PS, Rottier PJM. Coronavirus reverse genetics by targeted RNA recombination. Curr. Top. Microbiol. Immunol. 2005;287:133–159. doi: 10.1007/3-540-26765-4_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang Y, Yan W, Hall AB, Jiang X. Characterizing transcriptional regulatory sequences in coronaviruses and their role in recombination. Mol. Biol. Evol. 2021;38:1241–1248. doi: 10.1093/molbev/msaa281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chrisman BS, et al. Indels in SARS-CoV-2 occur at template-switching hotspots. BioData Min. 2021;14:20. doi: 10.1186/s13040-021-00251-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Irigoyen N, et al. High-resolution analysis of coronavirus gene expression by RNA sequencing and ribosome profiling. PLoS Pathog. 2016;12:e1005473. doi: 10.1371/journal.ppat.1005473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Viehweger A, et al. Direct RNA nanopore sequencing of full-length coronavirus genomes provides novel insights into structural variants and enables modification analysis. Genome Res. 2019;29:1545–1554. doi: 10.1101/gr.247064.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Snijder EJ, Kikkert M, Fang Y. Arterivirus molecular biology and pathogenesis. J. Gen. Virol. 2013;94:2141–2163. doi: 10.1099/vir.0.056341-0. [DOI] [PubMed] [Google Scholar]
- 65.Nedialkova DD, Gorbalenya AE, Snijder EJ. Arterivirus Nsp1 modulates the accumulation of minus-strand templates to control the relative abundance of viral mRNAs. PLoS Pathog. 2010;6:e1000772. doi: 10.1371/journal.ppat.1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tijms MA, Nedialkova DD, Zevenhoven-Dobbe JC, Gorbalenya AE, Snijder EJ. Arterivirus subgenomic mRNA synthesis and virion biogenesis depend on the multifunctional nsp1 autoprotease. J. Virol. 2007;81:10496–10505. doi: 10.1128/JVI.00683-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van Dinten LC, den Boon JA, Wassenaar AL, Spaan WJ, Snijder EJ. An infectious arterivirus cDNA clone: identification of a replicase point mutation that abolishes discontinuous mRNA transcription. Proc. Natl Acad. Sci. USA. 1997;94:991–996. doi: 10.1073/pnas.94.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Marle G, van Dinten LC, Spaan WJM, Luytjes W, Snijder EJ. Characterization of an equine arteritis virus replicase mutant defective in subgenomic mRNA synthesis. J. Virol. 1999;73:5274. doi: 10.1128/JVI.73.7.5274-5281.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen J, et al. Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex. Cell. 2020;182:1560–1573.e13. doi: 10.1016/j.cell.2020.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Malone B, et al. Structural basis for backtracking by the SARS-CoV-2 replication–transcription complex. Proc. Natl Acad. Sci. USA. 2021;118:e2102516118. doi: 10.1073/pnas.2102516118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Masters PS. The molecular biology of coronaviruses. Adv. Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chang C, Hou M-H, Chang C-F, Hsiao C-D, Huang T. The SARS coronavirus nucleocapsid protein — forms and functions. Antivir. Res. 2014;103:39–50. doi: 10.1016/j.antiviral.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bai Z, Cao Y, Liu W, Li J. The SARS-CoV-2 nucleocapsid protein and its role in viral structure, biological functions, and a potential target for drug or vaccine mitigation. Viruses. 2021;13:1115. doi: 10.3390/v13061115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thiel V, Herold J, Schelle B, Siddell SG. Viral replicase gene products suffice for coronavirus discontinuous transcription. J. Virol. 2001;75:6676–6681. doi: 10.1128/JVI.75.14.6676-6681.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schelle B, Karl N, Ludewig B, Siddell SG, Thiel V. Selective replication of coronavirus genomes that express nucleocapsid protein. J. Virol. 2005;79:6620–6630. doi: 10.1128/JVI.79.11.6620-6630.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zúñiga S, et al. Coronavirus nucleocapsid protein facilitates template switching and is required for efficient transcription. J. Virol. 2010;84:2169–2175. doi: 10.1128/JVI.02011-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cong Y, et al. Nucleocapsid protein recruitment to replication-transcription complexes plays a crucial role in coronaviral life cycle. J. Virol. 2020;94:e01925–19. doi: 10.1128/JVI.01925-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu C-H, Chen P-J, Yeh S-H. Nucleocapsid phosphorylation and RNA helicase DDX1 recruitment enables coronavirus transition from discontinuous to continuous transcription. Cell Host Microbe. 2014;16:462–472. doi: 10.1016/j.chom.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Savastano A, Ibáñez de Opakua A, Rankovic M, Zweckstetter M. Nucleocapsid protein of SARS-CoV-2 phase separates into RNA-rich polymerase-containing condensates. Nat. Commun. 2020;11:6041. doi: 10.1038/s41467-020-19843-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Biebricher CK, Eigen M. What is a quasispecies? Curr. Top. Microbiol. Immunol. 2006;299:1–31. doi: 10.1007/3-540-26397-7_1. [DOI] [PubMed] [Google Scholar]
- 81.Domingo E, Sheldon J, Perales C. Viral quasispecies evolution. Microbiol. Mol. Biol. Rev. 2012;76:159–216. doi: 10.1128/MMBR.05023-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lehmann KC, et al. Discovery of an essential nucleotidylating activity associated with a newly delineated conserved domain in the RNA polymerase-containing protein of all nidoviruses. Nucleic Acids Res. 2015;43:8416–8434. doi: 10.1093/nar/gkv838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kirchdoerfer RN, Ward AB. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Peersen OB. Picornaviral polymerase structure, function, and fidelity modulation. Virus Res. 2017;234:4–20. doi: 10.1016/j.virusres.2017.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jia H, Gong P. A Structure-function diversity survey of the RNA-dependent RNA polymerases from the positive-strand RNA viruses. Front. Microbiol. 2019;10:1945. doi: 10.3389/fmicb.2019.01945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ben Ouirane K, Boulard Y, Bressanelli S. The hepatitis C virus RNA-dependent RNA polymerase directs incoming nucleotides to its active site through magnesium-dependent dynamics within its F motif. J. Biol. Chem. 2019;294:7573–7587. doi: 10.1074/jbc.RA118.005209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shu B, Gong P. Structural basis of viral RNA-dependent RNA polymerase catalysis and translocation. Proc. Natl Acad. Sci. USA. 2016;113:E4005–E4014. doi: 10.1073/pnas.1602591113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gong P, Peersen OB. Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase. Proc. Natl Acad. Sci. USA. 2010;107:22505–22510. doi: 10.1073/pnas.1007626107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Steitz TA. A mechanism for all polymerases. Nature. 1998;391:231–232. doi: 10.1038/34542. [DOI] [PubMed] [Google Scholar]
- 90.Paul AV, Wimmer E. Initiation of protein-primed picornavirus RNA synthesis. Virus Res. 2015;206:12–26. doi: 10.1016/j.virusres.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Conti BJ, Leicht AS, Kirchdoerfer RN, Sussman MR. Mass spectrometric based detection of protein nucleotidylation in the RNA polymerase of SARS-CoV-2. Commun. Chem. 2021;4:41. doi: 10.1038/s42004-021-00476-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Slanina H, et al. Coronavirus replication–transcription complex: Vital and selective NMPylation of a conserved site in nsp9 by the NiRAN-RdRp subunit. Proc. Natl Acad. Sci. USA. 2021;118:e2022310118. doi: 10.1073/pnas.2022310118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yan L, et al. Cryo-EM structure of an extended SARS-CoV-2 replication and transcription complex reveals an intermediate state in cap synthesis. Cell. 2021;184:184–193.e10. doi: 10.1016/j.cell.2020.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Subissi L, et al. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl Acad. Sci. USA. 2014;111:E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hillen HS, et al. Structure of replicating SARS-CoV-2 polymerase. Nature. 2020;584:154–156. doi: 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- 96.Yin W, et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gao Y, et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang Q, et al. Structural basis for RNA replication by the SARS-CoV-2 polymerase. Cell. 2020;182:417–428.e13. doi: 10.1016/j.cell.2020.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.von Brunn A, et al. Analysis of intraviral protein-protein interactions of the SARS coronavirus ORFeome. PLoS ONE. 2007;2:e459. doi: 10.1371/journal.pone.0000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Imbert I, et al. The SARS-Coronavirus PLnc domain of nsp3 as a replication/transcription scaffolding protein. Virus Res. 2008;133:136–148. doi: 10.1016/j.virusres.2007.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Minskaia E, et al. Discovery of an RNA virus 3′→5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl Acad. Sci. USA. 2006;103:5108. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Denison MR, Graham RL, Donaldson EF, Eckerle LD, Baric RS. Coronaviruses: an RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 2011;8:270–279. doi: 10.4161/rna.8.2.15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bouvet M, et al. RNA 3′-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proc. Natl Acad. Sci. USA. 2012;109:9372–9377. doi: 10.1073/pnas.1201130109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Smith EC, Sexton NR, Denison MR. Thinking outside the triangle: replication fidelity of the largest RNA viruses. Annu. Rev. Virol. 2014;1:111–132. doi: 10.1146/annurev-virology-031413-085507. [DOI] [PubMed] [Google Scholar]
- 105.Eckerle LD, Lu X, Sperry SM, Choi L, Denison MR. High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J. Virol. 2007;81:12135–12144. doi: 10.1128/JVI.01296-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Smith EC, Blanc H, Vignuzzi M, Denison MR. Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: evidence for proofreading and potential therapeutics. PLoS Pathog. 2013;9:e1003565. doi: 10.1371/journal.ppat.1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Eckerle LD, et al. Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 2010;6:e1000896. doi: 10.1371/journal.ppat.1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ogando NS, et al. The enzymatic activity of the nsp14 exoribonuclease is critical for replication of MERS-CoV and SARS-CoV-2. J. Virol. 2020;94:e01246–20. doi: 10.1128/JVI.01246-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ferron F, et al. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc. Natl Acad. Sci. USA. 2018;115:E162. doi: 10.1073/pnas.1718806115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Liu C, et al. Structural basis of mismatch recognition by a SARS-CoV-2 proofreading enzyme. Science. 2021;373:1142–1146. doi: 10.1126/science.abi9310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Seybert A, Hegyi A, Siddell SG, Ziebuhr J. The human coronavirus 229E superfamily 1 helicase has RNA and DNA duplex-unwinding activities with 5′-to-3′ polarity. RNA. 2000;6:1056–1068. doi: 10.1017/S1355838200000728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Seybert A, et al. A complex zinc finger controls the enzymatic activities of nidovirus helicases. J. Virol. 2005;79:696. doi: 10.1128/JVI.79.2.696-704.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tanner JA, et al. The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5′ to 3′ viral helicases. J. Biol. Chem. 2003;278:39578–39582. doi: 10.1074/jbc.C300328200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lee N-R, et al. Cooperative translocation enhances the unwinding of duplex DNA by SARS coronavirus helicase nsP13. Nucleic Acids Res. 2010;38:7626–7636. doi: 10.1093/nar/gkq647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ivanov KA, et al. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 2004;78:5619–5632. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Adedeji AO, et al. Mechanism of nucleic acid unwinding by SARS-CoV helicase. PLoS ONE. 2012;7:e36521. doi: 10.1371/journal.pone.0036521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yan L, et al. Architecture of a SARS-CoV-2 mini replication and transcription complex. Nat. Commun. 2020;11:5874. doi: 10.1038/s41467-020-19770-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nudler E. RNA polymerase backtracking in gene regulation and genome instability. Cell. 2012;149:1438–1445. doi: 10.1016/j.cell.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dulin D, et al. Signatures of nucleotide analog incorporation by an RNA-dependent RNA polymerase revealed using high-throughput magnetic tweezers. Cell Rep. 2017;21:1063–1076. doi: 10.1016/j.celrep.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dulin D, et al. Backtracking behavior in viral RNA-dependent RNA polymerase provides the basis for a second initiation site. Nucleic Acids Res. 2015;43:10421–10429. doi: 10.1093/nar/gkv1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dulin D, et al. Elongation-competent pauses govern the fidelity of a viral RNA-dependent RNA polymerase. Cell Rep. 2015;10:983–992. doi: 10.1016/j.celrep.2015.01.031. [DOI] [PubMed] [Google Scholar]
- 122.van Dinten LC, Wassenaar AL, Gorbalenya AE, Spaan WJ, Snijder EJ. Processing of the equine arteritis virus replicase ORF1b protein: identification of cleavage products containing the putative viral polymerase and helicase domains. J. Virol. 1996;70:6625. doi: 10.1128/jvi.70.10.6625-6633.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Agostini ML, et al. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) Is mediated by the viral polymerase and the proofreading exoribonuclease. mBio. 2018;9:e00221-18. doi: 10.1128/mBio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yan L, et al. Coupling of N7-methyltransferase and 3′-5′ exoribonuclease with SARS-CoV-2 polymerase reveals mechanisms for capping and proofreading. Cell. 2021;184:3474–3485.e11. doi: 10.1016/j.cell.2021.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hall MD, et al. Report of the national institutes of health SARS-CoV-2 antiviral therapeutics summit. J. Infect. Dis. 2021;224:S1–S21. doi: 10.1093/infdis/jiab305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gordon CJ, et al. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020;295:6785–6797. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Beigel JH, et al. Remdesivir for the treatment of Covid-19 — final report. N. Engl. J. Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gordon CJ, Tchesnokov EP, Feng JY, Porter DP, Götte M. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J. Biol. Chem. 2020;295:4773–4779. doi: 10.1074/jbc.AC120.013056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gordon CJ, Tchesnokov EP, Schinazi RF, Götte M. Molnupiravir promotes SARS-CoV-2 mutagenesis via the RNA template. J. Biol. Chem. 2021;297:100770. doi: 10.1016/j.jbc.2021.100770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Dangerfield TL, Huang NZ, Johnson KA. Remdesivir is effective in combating COVID-19 because it is a better substrate than ATP for the viral RNA-dependent RNA polymerase. iScience. 2020;23:101849. doi: 10.1016/j.isci.2020.101849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kabinger F, et al. Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis. Nat. Struct. Mol. Biol. 2021;28:740–746. doi: 10.1038/s41594-021-00651-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bravo JPK, Dangerfield TL, Taylor DW, Johnson KA. Remdesivir is a delayed translocation inhibitor of SARS-CoV-2 replication. Mol. Cell. 2021;81:1548–1552.e4. doi: 10.1016/j.molcel.2021.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kokic G, et al. Mechanism of SARS-CoV-2 polymerase stalling by remdesivir. Nat. Commun. 2021;12:279. doi: 10.1038/s41467-020-20542-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Tchesnokov EP, et al. Template-dependent inhibition of coronavirus RNA-dependent RNA polymerase by remdesivir reveals a second mechanism of action. J. Biol. Chem. 2020;295:16156–16165. doi: 10.1074/jbc.AC120.015720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yoon J-J, et al. Orally efficacious broad-spectrum ribonucleoside analog inhibitor of influenza and respiratory syncytial viruses. Antimicrob. Agents Chemother. 2018;62:e00766-18. doi: 10.1128/AAC.00766-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Agostini ML, et al. Small-molecule antiviral β-d-N4-hydroxycytidine inhibits a proofreading-intact coronavirus with a high genetic barrier to resistance. J. Virol. 2019;93:e01348-19. doi: 10.1128/JVI.01348-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sheahan TP, et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl Med. 2020;12:eabb5883. doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Urakova N, et al. β-d-N4-Hydroxycytidine is a potent anti-alphavirus compound that induces a high level of mutations in the viral genome. J. Virol. 2018;92:e01965-17. doi: 10.1128/JVI.01965-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Businesswire. Interim results from phase 2/3 studies of molnupiravir, an investigational oral antiviral therapeutic for mild to moderate COVID-19, presented at the European Congress of Clinical Microbiology & Infectious Diseases (ECCMID). businesswirehttps://www.businesswire.com/news/home/20210712005251/en/ (2021).
- 140.Saramago M, et al. New targets for drug design: importance of nsp14/nsp10 complex formation for the 3′-5′ exoribonucleolytic activity on SARS-CoV-2. FEBS J. 2021;288:5130–5147. doi: 10.1111/febs.15815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bouvet M, et al. Coronavirus Nsp10, a critical co-factor for activation of multiple replicative enzymes. J. Biol. Chem. 2014;289:25783–25796. doi: 10.1074/jbc.M114.577353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Deming DJ, Graham RL, Denison MR, Baric RS. Processing of open reading frame 1a replicase proteins nsp7 to nsp10 in murine hepatitis virus strain A59 replication. J. Virol. 2007;81:10280–10291. doi: 10.1128/JVI.00017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Huang C, et al. SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: viral mRNAs are resistant to nsp1-induced RNA cleavage. PLoS Pathog. 2011;7:e1002433. doi: 10.1371/journal.ppat.1002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Serrano P, et al. Nuclear magnetic resonance structure of the N-terminal domain of nonstructural protein 3 from the severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:12049. doi: 10.1128/JVI.00969-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Putics Á, Filipowicz W, Hall J, Gorbalenya AE, Ziebuhr J. ADP-ribose-1″-monophosphatase: a conserved coronavirus enzyme that is dispensable for viral replication in tissue culture. J. Virol. 2005;79:12721. doi: 10.1128/JVI.79.20.12721-12731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Alhammad YMO, et al. The SARS-CoV-2 conserved macrodomain is a mono-ADP-ribosylhydrolase. J. Virol. 2021;95:e01969-20. doi: 10.1128/JVI.01969-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lee HJ, et al. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology. 1991;180:567–582. doi: 10.1016/0042-6822(91)90071-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Oudshoorn D, et al. Expression and cleavage of middle east respiratory syndrome coronavirus nsp3-4 polyprotein induce the formation of double-membrane vesicles that mimic those associated with coronaviral RNA replication. mBio. 2017;8:e01658-17. doi: 10.1128/mBio.01658-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Angelini MM, Akhlaghpour M, Neuman BW, Buchmeier MJ. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. mBio. 2013;4:e00524-13. doi: 10.1128/mBio.00524-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wolff G, et al. A molecular pore spans the double membrane of the coronavirus replication organelle. Science. 2020;369:1395. doi: 10.1126/science.abd3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Gorbalenya AE, Koonin EV, Donchenko AP, Blinov VM. Coronavirus genome: prediction of putative functional domains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res. 1989;17:4847–4861. doi: 10.1093/nar/17.12.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Anand K, Ziebuhr J, Wadhwani P, Mesters JR, Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 153.Imbert I, et al. A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO J. 2006;25:4933–4942. doi: 10.1038/sj.emboj.7601368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Tvarogová J, et al. Identification and characterization of a human coronavirus 229E nonstructural protein 8-associated RNA 3′-terminal adenylyltransferase activity. J. Virol. 2019;93:e00291-19. doi: 10.1128/JVI.00291-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Egloff M-P, et al. The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world. Proc. Natl Acad. Sci. USA. 2004;101:3792–3796. doi: 10.1073/pnas.0307877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Decroly E, et al. Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2′-O-methyltransferase nsp10/nsp16 Complex. PLoS Pathog. 2011;7:e1002059. doi: 10.1371/journal.ppat.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ma Y, et al. Structural basis and functional analysis of the SARS coronavirus nsp14–nsp10 complex. Proc. Natl Acad. Sci. USA. 2015;112:9436–9441. doi: 10.1073/pnas.1508686112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bouvet M, et al. In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathog. 2010;6:e1000863. doi: 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Chen Y, et al. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc. Natl Acad. Sci. USA. 2009;106:3484–3489. doi: 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Decroly E, et al. Coronavirus nonstructural protein 16 is a cap-0 binding enzyme possessing (nucleoside-2′O)-methyltransferase activity. J. Virol. 2008;82:8071. doi: 10.1128/JVI.00407-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hartenian E, et al. The molecular virology of coronaviruses. J. Biol. Chem. 2020;295:12910–12934. doi: 10.1074/jbc.REV120.013930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Appleby TC, et al. Viral replication. Structural basis for RNA replication by the hepatitis C virus polymerase. Science. 2015;347:771–775. doi: 10.1126/science.1259210. [DOI] [PubMed] [Google Scholar]
- 163.Gosert R, Kanjanahaluethai A, Egger D, Bienz K, Baker SC. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 2002;76:3697–3708. doi: 10.1128/JVI.76.8.3697-3708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Knoops K, et al. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Maier HJ, et al. Infectious bronchitis virus generates spherules from zippered endoplasmic reticulum membranes. mBio. 2013;4:e00801–e00813. doi: 10.1128/mBio.00801-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Klein S, et al. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat. Commun. 2020;11:5885. doi: 10.1038/s41467-020-19619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ramanathan A, Robb GB, Chan S-H. mRNA capping: biological functions and applications. Nucleic Acids Res. 2016;44:7511–7526. doi: 10.1093/nar/gkw551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ferron F, Decroly E, Selisko B, Canard B. The viral RNA capping machinery as a target for antiviral drugs. Antivir. Res. 2012;96:21–31. doi: 10.1016/j.antiviral.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ivanov KA, Ziebuhr J. Human coronavirus 229E nonstructural protein 13: characterization of duplex-unwinding, nucleoside triphosphatase, and RNA 5′-triphosphatase activities. J. Virol. 2004;78:7833–7838. doi: 10.1128/JVI.78.14.7833-7838.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Züst R, et al. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 2011;12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]