Abstract
BACKGROUND
Schizophrenia afflicts 1% of the world population. Clinical studies suggest that schizophrenia patients may have an imbalance of mitochondrial energy metabolism via inhibition of mitochondrial complex I activity. Moreover, recent studies have shown that ERVWE1 is also a risk factor for schizophrenia. Nevertheless, there is no available literature concerning the relationship between complex I deficits and ERVWE1 in schizophrenia. Identifying risk factors and blood-based biomarkers for schizophrenia may provide new guidelines for early interventions and prevention programs.
AIM
To address novel potential risk factors and the underlying mechanisms of mitochondrial complex I deficiency caused by ERVWE1 in schizophrenia.
METHODS
Quantitative polymerase chain reaction (qPCR) and enzyme-linked immunosorbent assay were used to detect differentially expressed risk factors in blood samples. Clinical statistical analyses were performed by median analyses and Mann-Whitney U analyses. Spearman’s rank correlation was applied to examine the correlation between different risk factors in blood samples. qPCR, western blot analysis, and luciferase assay were performed to confirm the relationship among ERVWE1, cytoplasmic polyadenylation element-binding protein 1 (CPEB1), NADH dehydrogenase ubiquinone flavoprotein 2 (NDUFV2), and NDUFV2 pseudogene (NDUFV2P1). The complex I enzyme activity microplate assay was carried out to evaluate the complex I activity induced by ERVWE1.
RESULTS
Herein, we reported decreasing levels of CPEB1 and NDUFV2 in schizophrenia patients. Further studies showed that ERVWE1 was negatively correlated with CPEB1 and NDUFV2 in schizophrenia. Moreover, NDUFV2P1 was increased and demonstrated a significant positive correlation with ERVWE1 and a negative correlation with NDUFV2 in schizophrenia. In vitro experiments disclosed that ERVWE1 suppressed NDUFV2 expression and promoter activity by increasing NDUFV2P1 level. The luciferase assay revealed that ERVWE1 could enhance the promoter activity of NDUFV2P1. Additionally, ERVWE1 downregulated the expression of CPEB1 by suppressing the promoter activity, and the 400 base pair sequence at the 3′ terminus of the promoter was the minimum sequence required. Advanced studies showed that CPEB1 participated in regulating the NDUFV2P1/NDUFV2 axis mediated by ERVWE1. Finally, we found that ERVWE1 inhibited complex I activity in SH-SY5Y cells via the CPEB1/NDUFV2P1/NDUFV2 signaling pathway.
CONCLUSION
In conclusion, CPEB1 and NDUFV2 might be novel potential blood-based biomarkers and pathogenic factors in schizophrenia. Our findings also reveal a novel mechanism of ERVWE1 in the etiology of schizophrenia.
Keywords: ERVWE1, CPEB1, NADH dehydrogenase ubiquinone flavoprotein 2, complex I, Pseudogene
Core Tip: Schizophrenia is a devastating psychiatric disorder. Clinical studies suggest complex I deficits and abnormal expression of ERVWE1 in schizophrenia. We found that CPEB1 might be a novel blood-based biomarker for schizophrenia. Analyses indicated that ERVWE1 was negatively correlated with CPEB1 and NADH dehydrogenase ubiquinone flavoprotein 2 (NDUFV2) in schizophrenia patients. CPEB1 functioned as a mediator of the NDUFV2 pseudogene (NDUFV2P1)/NDUFV2 pathway induced by ERVWE1. Moreover, ERVWE1 inhibited complex I activity via the CPEB1/NDUFV2P1/NDUFV2 pathway. Thus, CPEB1 and NDUFV2 may be independent risk factors for schizophrenia. Our findings also reveal the role of ERVWE1 in modulating mitochondrial energy metabolism in schizophrenia.
INTRODUCTION
Schizophrenia is a chronic brain disorder with a worldwide prevalence of approximately 1%[1]. It causes many symptoms including hallucinations, delusions, breaking from reality, and a lack of motivation[2]. Schizophrenia imposes a financial burden on society because of high indirect costs[3]. There are numerous hypotheses about schizophrenia. One of the currently prevailing hypothesis proposes that mitochondrial metabolic dysfunction[4-6] is strongly involved in the pathophysiology of schizophrenia. Mitochondrial complex I deficiency is the most frequent cause of mitochondrial metabolic dysfunction implicated in the development of schizophrenia[7-9]. The mitochondrial oxidative phosphorylation (OXPHOS) system comprises five multimeric protein complexes, and plays a central role in cellular energy metabolism. Complex I is the first entry point of electrons into the respiratory chain and is suggested as the rate-limiting step in OXPHOS. NADH dehydrogenase ubiquinone flavoprotein 2 (NDUFV2) assembles in the catalytic component of complex I. Ample evidence has revealed that NDUFV2 is a candidate gene for schizophrenia[10-12]. NDUFV2 pseudogene (NDUFV2P1), an inactive locus of the NDUFV2 subunit of the complex I[13], has increased expression in schizophrenia-derived cells[14]. Knockout of CPEB1, a sequence-specific RNA-binding protein[15], reduces NDUFV2 protein expression and inhibits complex I activity in mice[16]. Nonetheless, there is no definitive hypothesis on the pathogenesis of schizophrenia.
Human endogenous retroviruses (HERVs), which comprise about 8% of the human genome, have emerged as novel risk factors for schizophrenia[17-19]. As remnants of ancient retroviral infections, most HERVs have acquired inactivating mutations during evolution[20]. In some instances, some HERVs sequences remain dormant functional copies encoding retroviral proteins[21] and play an active role in human placental development and the innate immune system. HERVs possess a similar genome structure as typical exogenous retroviruses and are composed of gag, pol, and env regions sandwiched between two long terminal repeats[21]. Conventionally, based on sequence similarity with their exogenous retrovirus, HERVs are divided into three classes: class I (gamma-retroviruses), class II (beta-retroviruses), and class III (spuma-retroviruses)[22].
The HERV W family (HERV-W), the oldest group of HERVs, belongs to class I transposable elements. ERVWE1, HERV-W-derived envelope proteins at chromosome 7, is also known as HERV-W env or Syncytin-1[23]. ERVWE1 is a cell-cell fusion glycoprotein that is usually expressed on the surface of cytotrophoblasts and syncytiotrophoblast. It mediates trophoblast fusion[24] and blocks maternal immune[25], which are essential for normal placental development. ERVWE1 is aberrantly expressed in schizophrenia[18,19,26]. Results from our previous study showed that ERVWE1 is significantly increased in the plasma of recent-onset schizophrenia patients[18,19]. Perron et al[26] also reported antigenemia of ERVWE1 was elevated in the serum of schizophrenic patients.
Both complex I deficiency and ERVWE1 are risk factors for developing schizophrenia. However, no detailed study has been performed on the cytoplasmic polyadenylation element-binding protein 1 (CPEB1) expression profile in schizophrenia. Furthermore, the relationships among these factors in schizophrenia remain unknown. In this study, we found that CPEB1 protein was decreased and negatively correlated with ERVWE1 in recent-onset schizophrenia patients. Quantitative polymerase chain reaction (qPCR) and enzyme-linked immunosorbent assay (ELISA) showed a significant reduction of NDUFV2 in schizophrenia. We further demonstrated the increased levels and positive correlation between ERVWE1 and NDUFV2P1 in schizophrenia patients. Moreover, the mRNA level of ERVWE1 and NDUFV2P1 was negatively correlated with NDUFV2 protein expression in schizophrenia patients. A series of cell and molecular biology experiments were used to reveal the relationship among CPEB1, NDUFV2P1/NDUFV2, ERVWE1, and complex I deficiency. The results suggested that ERVWE1 could decrease the mRNA and protein expression of NDUFV2 and suppress NDUFV2 promoter activation by increasing NDUFV2P1 transcript levels. The luciferase assay showed that overexpression of ERVWE1 enhanced NDUFV2P1 promoter activity. ERVWE1 also inhibited CPEB1 mRNA and protein expression by regulating its promoter. Moreover, CPEB1 was essential in the NDUFV2P1/NDUFV2 axis triggered by ERVWE1 in SH-SY5Y cells. Finally, ERVWE1 induced complex I inactivation via the CPEB1/NDUFV2P1/NDUFV2 signaling pathway in SH-SY5Y cells.
In summary, CPEB1 and NDUFV2 might be novel schizophrenia risk factors. To the best of our knowledge, no similar observations have been published to date. Moreover, our results indicated that ERVWE1 abolished complex I activity by regulating parental- and pseudo-NDUFV2 genes via suppressing CPEB1 in schizophrenia. Our findings might provide new insights into how ERVWE1 impairs mitochondrial energy metabolism and contributes to the pathogenesis of schizophrenia.
MATERIALS AND METHODS
Blood samples
Blood samples from patients and normal individuals, including whole peripheral blood for RNA isolation and serum for ELISA analyses, were collected from the Renmin Hospital, Wuhan University (Wuhan, China). All patients with recent-onset schizophrenia met the Diagnostic and Statistical Manual of Mental Disorders, 4th Edition criteria. None of the patients had a history of psychiatric illness. Informed written consent was obtained from all subjects, and the study was approved by the Institutional Review Board of Wuhan University, School of Basic Medical Sciences. All samples were preserved at -80°C before utilization. There were no significant differences in median age, education, body mass index, smoking status, and sex between control and schizophrenia patients. Demographics are presented in Supplementary Tables 1 and 2.
Plasmid construction
The pCMV-ERVWE1 plasmid was constructed as previously described[18]. Human CPEB1 (NM_030594.5) and NDUFV2P1 (NC_000019.10:c53224879-53223909) genes were amplified separately according to sequences of these genes in National Center for Biotechnology Information (Bethesda, MD, United States). The promoters of human CPEB1 (-800 to 0, -600 to 0, -400 to 0, and -200 to 0), NDUFV2 (-1100 to +100), and NDUFV2P1 (-900 to 0) were cloned into the luciferase reporter vector pGL3-basic, which contains a firefly luciferase gene. All primers were designed using Primer premier 5.0 and described in Supplementary Table 3.
One short hairpin RNA (shRNA) targeting NDUFV2P1 (shNDUFV2P1, 5′-CCAAGGAGTGGACGCTTCT-3′) and the control shRNA (sh-Control, 5′-CCAAGTGGGCACGTAGTCT-3′) were cloned into the pSilencer 2.1-U6 neo shRNA expression vector (AM5764; Ambion Inc., Austin, TX, United States). All of the constructs were confirmed by sequencing (Sangon Biotech, Shanghai, China).
Cell culture and transfection
SH-SY5Y (CRL-2266) human neuroblastoma cells were obtained from American Type Culture Collection. SH-SY5Y cells were cultured in a 1:1 mixture of minimum essential medium (2225320; Gibco, Gaithersburg, MD, United States) and F-12 nutrient mixture (2209586; Gibco). All media were supplemented with 10% fetal bovine serum (2001003, Biological Industries, Beit HaEmek, Israel), 1% penicillin-streptomycin (2211093; Gibco) and 1% sodium pyruvate (2185865; Gibco) under 5% CO2 at 37 °C. Transfection was performed with Lipofectamine 2000 transfection reagent (11668-019; Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s instructions. Cells were incubated for further analyses after 24, 36, and 48 h post-transfection.
RNA extraction and reverse transcription
Total blood RNA was extracted from whole blood according to the standard protocols for TRIzol LS reagent (10296028; Invitrogen). Total cellular RNA was isolated with TRIzol reagent (15596018; Invitrogen), followed by chloroform separation and isopropanol precipitation. Then, 1 μg total RNA samples were reverse-transcribed into cDNA using the ReverTra Ace qPCR RT master mix with gDNA remover (FSQ-301; Toyobo, Osaka, Japan) according to the manufacturer’s recommendations.
Quantitative PCR
The mRNA levels were measured by quantitative PCR (qPCR) utilizing the 2x Sybr Green qPCR mix (2992239AX; Aidlab Biotechnologies Co. Ltd., Beijing, China) using a mini opticon detector (Bio-Rad, Hercules, CA, United States). GAPDH was used as an internal control, and the relative mRNA expression was calculated using the 2-ΔΔCt method. The primers were designed according to gene sequences in NCBI using primer premier 5.0, and are listed in Supplementary Table 4.
Protein extraction and western blot analysis
Cultured cells were homogenized and lysed in M-PER TM mammalian protein extraction reagent (UC282138; Thermo Fisher Scientific, Waltham, MA, United States) containing protease and phosphatase inhibitors (Roche Applied Science, Indianapolis, IN, United States). Protein concentrations were quantified using the Pierce TM BCA Protein Assay (UD281372; Thermo Fisher Scientific). Protein samples were loaded onto a 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and was then electrotransferred to a PVDF membrane (IPVH00010; Amersham Biosciences, Piscataway, NJ, United States). The following antibodies purchased from ABclonal Technology (Woburn, MA, United States) were used in this study: rabbit anti-ERVWE1 antibody (dilution 1:1000, A16522), rabbit anti-CPEB1 antibody (dilution 1:1000, A5913), rabbit anti-NDUFV2 antibody (dilution 1:1500, A7442), mouse anti-GAPDH antibody (dilution 1:50000, AC002), goat anti-mouse IgG-horseradish peroxidase (HRP) (dilution 1:5000, AS003), and goat anti-rabbit IgG-HRP (dilution 1:5000, AS014). The bands were visualized using the Millipore Immobilon Western Chemiluminescent HRP Substrate (WBKLS0500; Millipore, Burlington, MA, United States). Images were captured using the Tanon 5200 chemiluminescence imaging system (Tanon, Shanghai, China). Target protein expression levels were normalized to GAPDH.
Luciferase assay
The luciferase activity was carried out using the Dual-Luciferase Assay System (E1960; Promega, Madison, WI, United States) in accordance with the manufacturer’s protocol. Firefly luciferase activity was normalized to pRL-CMV Renilla luciferase activity. The promoter activity is expressed as the mean ± standard deviation (SD) of at least triplicate replicates.
ELISA
The expression levels of human CPEB1 (072020; Huaer Biotech Co. Ltd, Wuhan, China) and NDUFV2 (202103; Meiman Industrial Co. Ltd, Jiangsu, China) in serum were measured by an ELISA kit according to the manufacturer’s recommendations. Absorbance was measured at 450 nm using a spectrophotometer (Thermo Fisher Scientific). The concentrations of CPEB1 and NDUFV2 were determined by comparing the optical density of the samples to the standard curve.
Analyses of mitochondrial OXPHOS complex I activity
Mitochondrial OXPHOS complex I activity was detected by the Immunocapture ELISA Kit following the manufacturer’s instructions (AB109721; Abcam, Cambridge, United Kingdom). After cells were lysed with detergent solution, protein concentration was adjusted to 1 mg/mL in incubation solution. The diluted 200 μL sample and control were loaded onto the wells of a microplate pre-coated with complex I capture antibody for 3 h at room temperature. Complex I activity was determined by measuring the oxidation of NADH to NAD+ and simultaneous reduction of a dye, which leads to increased absorbance at 450 nm. The activity was expressed as the change in absorbance per min using this equation: complex I activity = [Abs 1 – Abs 2] / [Time 1 – Time 2].
Statistical analyses
SPSS and GraphPad Prism were used for the statistical analyses. For the blood sample results, the median analyses and Mann-Whitney U analyses were performed to compare the different expressions of ERVWE1, CPEB1, NDUFV2P1, and NDUFV2 between the schizophrenia and control groups. Correlation analyses were performed using the Spearman’s rank correlation. All data were obtained from at least three independent experiments and presented as the mean ± SD. Statistical and data analyses were performed by one-way analysis of variance and Student’s t-tests. Significance was considered at P < 0.05.
RESULTS
Degradation of CPEB1 and NDUFV2 proteins and the correlation among ERVWE1, NDUFV2, and NDUFV2P1 in schizophrenia
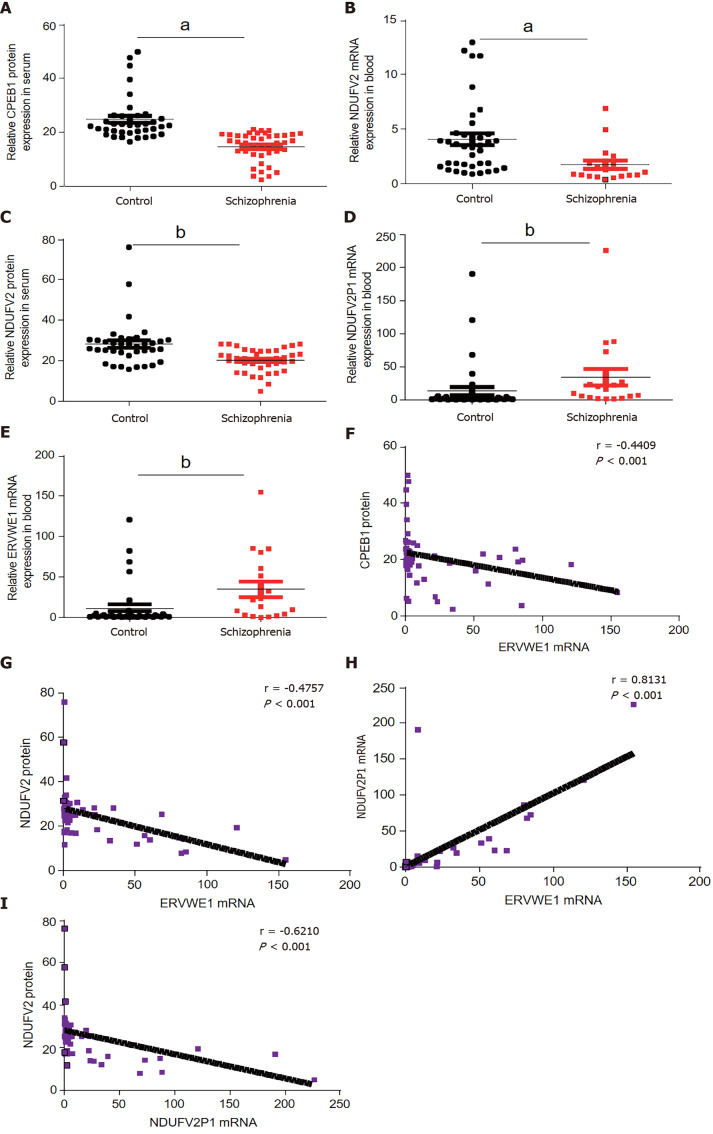
Many biomarkers in serum have been studied to determine whether they can help diagnose schizophrenia. A genome-wide association study (GWAS) identified CPEB1 as a new susceptibility locus for schizophrenia[27]. Nevertheless, there has been no report on the expression of CPEB1 in patients with schizophrenia. Here, we showed that CPEB1 was significantly downregulated in schizophrenia compared to healthy people in blood samples using ELISA (P < 0.05; Figure 1A and Table 1). We further found decreased mRNA and protein levels of NDUFV2 in schizophrenia patients (P < 0.05, Figure 1B; P < 0.01, Figure 1C and Table 2). NDUFV2P1 (NDUFV2 pseudogene) was significantly upregulated in schizophrenia patients (P < 0.01; Figure 1D). We also found increased ERVWE1 mRNA expression in schizophrenia patients (P < 0.01; Figure 1E). Correlation analyses through linear regression revealed that ERVWE1 mRNA was negatively correlated with CPEB1 (P < 0.001; Figure 1F) and NDUFV2 protein (P < 0.001; Figure 1G), and positively correlated with NDUFV2P1 transcripts (P < 0.001; Figure 1H) in schizophrenia. Moreover, high levels of NDUFV2P1 mRNA were correlated with decreased NDUFV2 protein in schizophrenia (P < 0.001; Figure 1I). Univariate and multivariate analyses identified CPEB1 and NDUFV2 as two possible independent risk factors for schizophrenia (Supplementary Table 5).
Figure 1.
Cytoplasmic polyadenylation element-binding protein 1 (CPEB1) and NADH dehydrogenase ubiquinone flavoprotein 2 (NDUFV2) deficiency and the correlation among ERVWE1, CPEB1, NDUFV2, and NDUFV2 pseudogene (NDUFV2P1) in schizophrenia patients. A and C: Respectively represent the protein expression of cytoplasmic polyadenylation element-binding protein 1 (CPEB1) and NADH dehydrogenase ubiquinone flavoprotein 2 (NDUFV2) in the schizophrenia patients (n = 41) and in the control groups (n = 38) using enzyme-linked immunosorbent; B, D, and E: Respectively represent the mRNA levels of NDUFV2, NDUFV2 pseudogene (NDUFV2P1), and ERVWE1 in schizophrenia patients (n = 19) and the control groups (n = 38) using quantitative polymerase chain reaction; F: Correlation between ERVWE1 mRNA levels and CPEB1 protein levels in patients with schizophrenia and control groups (n = 57), where Y is the protein expression for CPEB1 and X is ERVWE1 mRNA value for each sample; G: Correlation between ERVWE1 mRNA levels and NDUFV2 protein levels in patients with schizophrenia and control groups (n = 57), where Y is the protein expression for NDUFV2 and X is ERVWE1 mRNA value for each sample; H: Correlation between ERVWE1 and NDUFV2P1 mRNA levels in patients with schizophrenia and control groups (n = 57), where Y is the mRNA expression for NDUFV2P1 and X is ERVWE1 mRNA value for each sample; I: Correlation between NDUFV2P1 mRNA levels and NDUFV2 protein levels in patients with schizophrenia and control groups (n = 57), where Y is the protein expression for NDUFV2 and X is NDUFV2P1 mRNA value for each sample. aP < 0.05; bP < 0.01.
Table 1.
The concentration of CPEB1 in the blood of control and schizophrenia patients
|
Control
|
n
|
38
|
Schizophrenia
|
n
|
41
|
| Mean | 25.76 | Mean | 14.63 | ||
| Median | 22.47 | Median | 16.19 | ||
| SD | 11.70 | SD | 5.39 | ||
| Skewness | 2.89 | Skewness | -0.93 | ||
| SE of Skewness | 0.38 | SE of Skewness | 0.37 | ||
| Range | 53.26 | Range | 18.63 | ||
| Minimum | 16.46 | Minimum | 2.14 | ||
| Maximum | 69.70 | Maximum | 23.67 |
SD: Standard deviation; SE: Standard error.
Table 2.
The concentration of NADH dehydrogenase ubiquinone flavoprotein 2 in the blood of control and schizophrenia patients
|
Control
|
n
|
38
|
Schizophrenia
|
n
|
41
|
| Mean | 28.16 | Mean | 20.14 | ||
| Median | 26.87 | Median | 20.51 | ||
| SD | 11.02 | SD | 5.47 | ||
| Skewness | 2.691 | Skewness | -0.67 | ||
| SE of Skewness | 0.38 | SE of Skewness | 0.37 | ||
| Range | 60.09 | Range | 23.23 | ||
| Minimum | 15.76 | Minimum | 4.91 | ||
| Maximum | 75.85 | Maximum | 28.14 |
SD: Standard deviation; SE: Standard error.
In summary, CPEB1 and NDUFV2 might be novel potential blood-based biomarkers and possible pathogenic factors for schizophrenia, and are negatively correlated with ERVWE1 in schizophrenia.
ERVWE1 reduces the expression of NDUFV2 by upregulating NDUFV2P1.
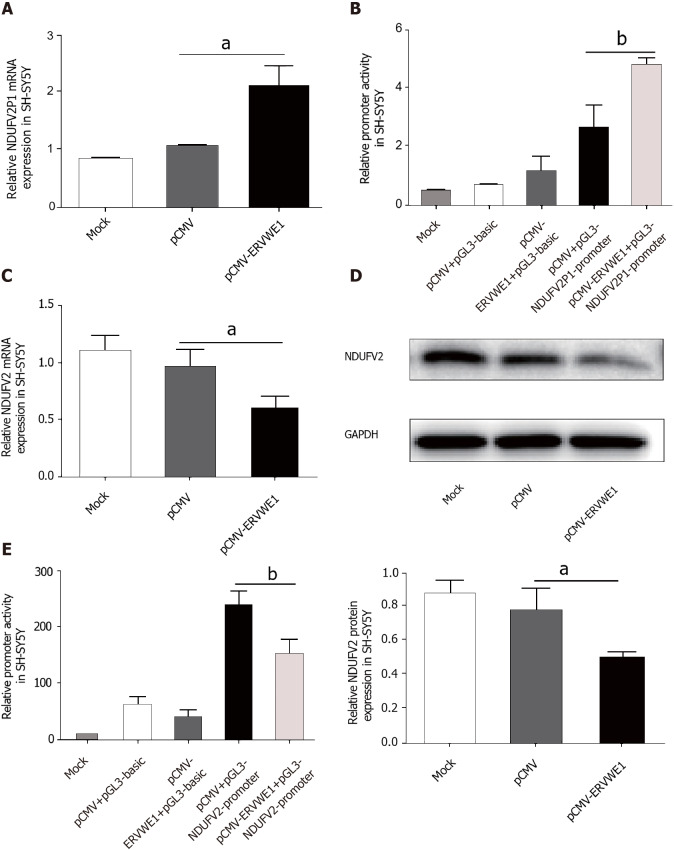
The clinical data showed a significant correlation among ERVWE1, NDUFV2P1, and NDUFV2 gene expression in schizophrenia. Thus, to evaluate the causal relationship among ERVWE1, NDUFV2P1, and NDUFV2, experiments were performed in human neuroblastoma cells. ERVWE1 expression in transfected cells was detected by qPCR and western blotting (Supplementary Figure 1A and B). The mRNA level of NDUFV2P1 was significantly increased in SH-SY5Y cells after overexpression of ERVWE1 (P < 0.05; Figure 2A). Luciferase assays showed that overexpression of ERVWE1 greatly enhanced NDUFV2P1 promoter activity in SH-SY5Y cells (P < 0.01; Figure 2B). NDUFV2 is the parental gene of NDUFV2P1[13]. We found that ERVWE1 decreased both the mRNA (P < 0.05; Figure 2C) and protein levels of NDUFV2 in SH-SY5Y cells (P < 0.05; Figure 2D). Moreover, ERVWE1 reduced the expression of NDUFV2 by suppressing promoter activity in SH-SY5Y cells (P < 0.01; Figure 2E). These findings suggest that ERVWE1 downregulates the expression of NDUFV2 while upregulating its pseudogene, NDUFV2P1, in SH-SY5Y cells.
Figure 2.
Overexpression of ERVWE1 elevated NADH dehydrogenase ubiquinone flavoprotein 2 (NDUFV2) pseudogene (NDUFV2P1) and reduced NDUFV2 levels. A and C: Respectively represent the mRNA levels of NADH dehydrogenase ubiquinone flavoprotein 2 (NDUFV2) pseudogene (NDUFV2P1) and NDUFV2 in ERVWE1-transfected SH-SY5Y cells using quantitative polymerase chain reaction; B: Luciferase assays of pGL3-NDUFV2P1 promoter co-transfected with pCMV-ERVWE1 or control vector in SH-SY5Y cells; D: Western blots of NDUFV2 and GAPDH in SH-SY5Y cells; E: Luciferase assays of pGL3-NDUFV2 promoter co-transfected with pCMV-ERVWE1 or control vector in SH-SY5Y cells. Each bar represents the mean ± SD of three independent experiments. aP < 0.05; bP < 0.01.
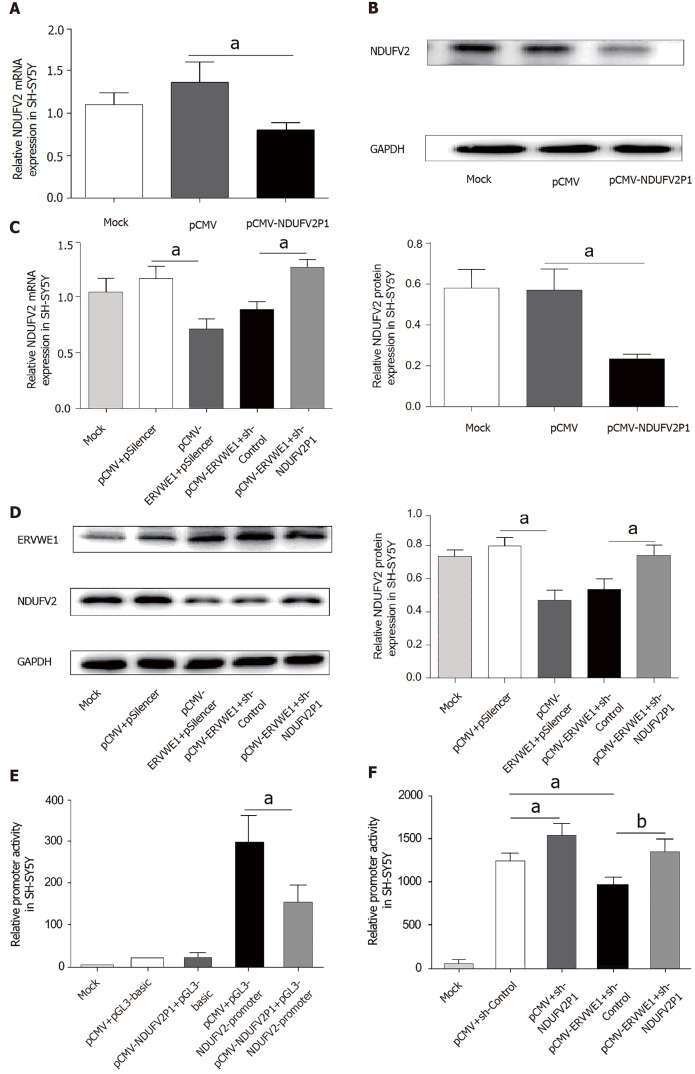
Pseudogene transcripts usually regulate the expression of their parental genes[28]. As shown in Figure 3A, the mRNA levels of NDUFV2 were significantly decreased in the NDUFV2P1 (+) group compared with the control (P < 0.05). Consistently, western blot analysis also confirmed that NDUFV2P1 inhibited the expression levels of NDUFV2 in SH-SY5Y cells (P < 0.05; Figure 3B). NDUFV2P1 expression in transfected cells was detected by qPCR (Supplementary Figure 2).
Figure 3.
Role of NADH dehydrogenase ubiquinone flavoprotein 2 (NDUFV2) pseudogene (NDUFV2P1) in ERVWE1-induced downregulation of NDUFV2. A: The mRNA levels of NADH dehydrogenase ubiquinone flavoprotein 2 (NDUFV2) in NDUFV2 pseudogene (NDUFV2P1)-overexpressing SH-SY5Y cells using quantitative polymerase chain reaction (qPCR); B: Western blots of NDUFV2 and GAPDH in SH-SY5Y cells; C: The mRNA levels of NDUFV2 in SH-SY5Y cells after co-transfection with ERVWE1 and shNDUFV2P1 using qPCR; D: Western blots of ERVWE1, NDUFV2, and GAPDH in SH-SY5Y cells; E: Luciferase assays of pGL3-NDUFV2 promoter co-transfected with pCMV-NDUFV2P1 or control vector in SH-SY5Y cells; F: Luciferase assays of pGL3-NDUFV2 promoter co-transfected with pCMV-ERVWE1 + shNDUFV2P1 or control in SH-SY5Y cells. Each bar represents the mean ± SD of three independent experiments. aP < 0.05; bP < 0.01.
To further explore whether NDUFV2P1 mediated the regulatory effect of ERVWE1 on the expression of NDUFV2, the shNDUFV2P1 plasmid was constructed. Knockdown of NDUFV2P1 substantially elevated the ERVWE1-induced mRNA expressions of NDUFV2 in SH-SY5Y cells (P < 0.05; Figure 3C). Moreover, western blotting showed similar results of increased NDUFV2 protein expression after NDUFV2P1 knockdown (P < 0.05; Figure 3D). Hence, these data demonstrated that ERVWE1 could mediate the downregulation of NDUFV2 by upregulating NDUFV2P1 in SH-SY5Y cells.
We further examined whether NDUFV2P1 had an effect on NDUFV2 promoter activity. Luciferase assays indicated that NDUFV2P1 inhibited NDUFV2 promoter activity (P < 0.05; Figure 3E) in SH-SY5Y cells. To explore the role of NDUFV2P1 in the ERVWE1-induced promoter activity of NDUFV2, shNDUFV2P1 was used. The reduced promoter activity was significantly recovered after knockdown of NDUFV2P1 (P < 0.01; Figure 3F), suggesting that ERVWE1 suppressed NDUFV2 promoter by elevating NDUFV2P1 transcript.
ERVWE1 mediates NDUFV2P1/NDUFV2 signaling pathway through the regulation of CPEB1
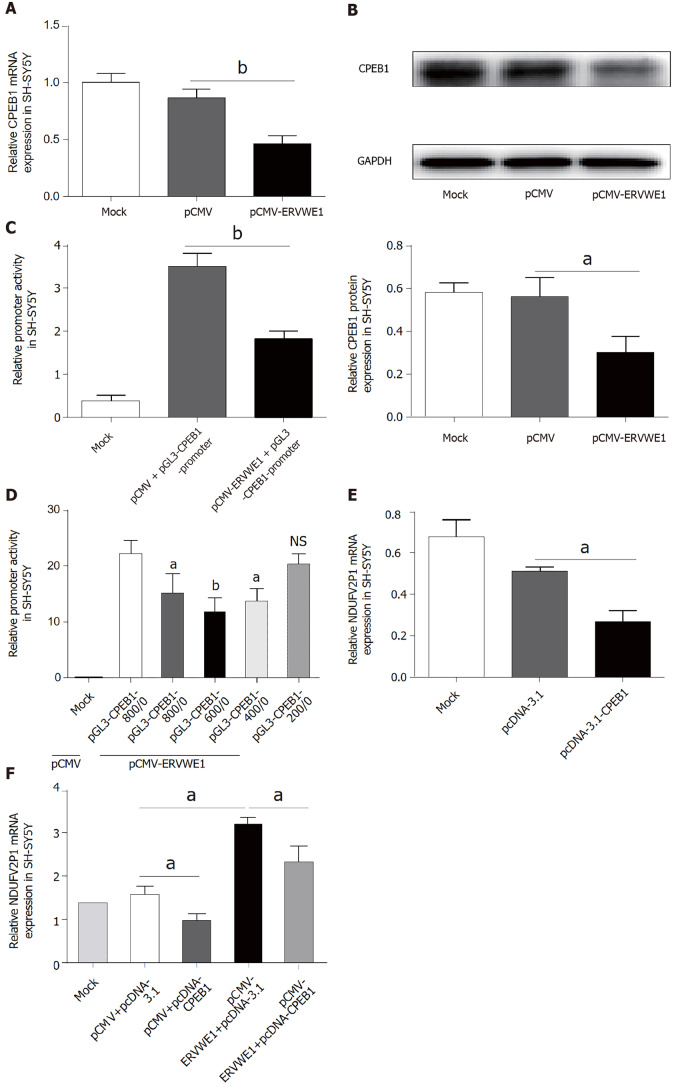
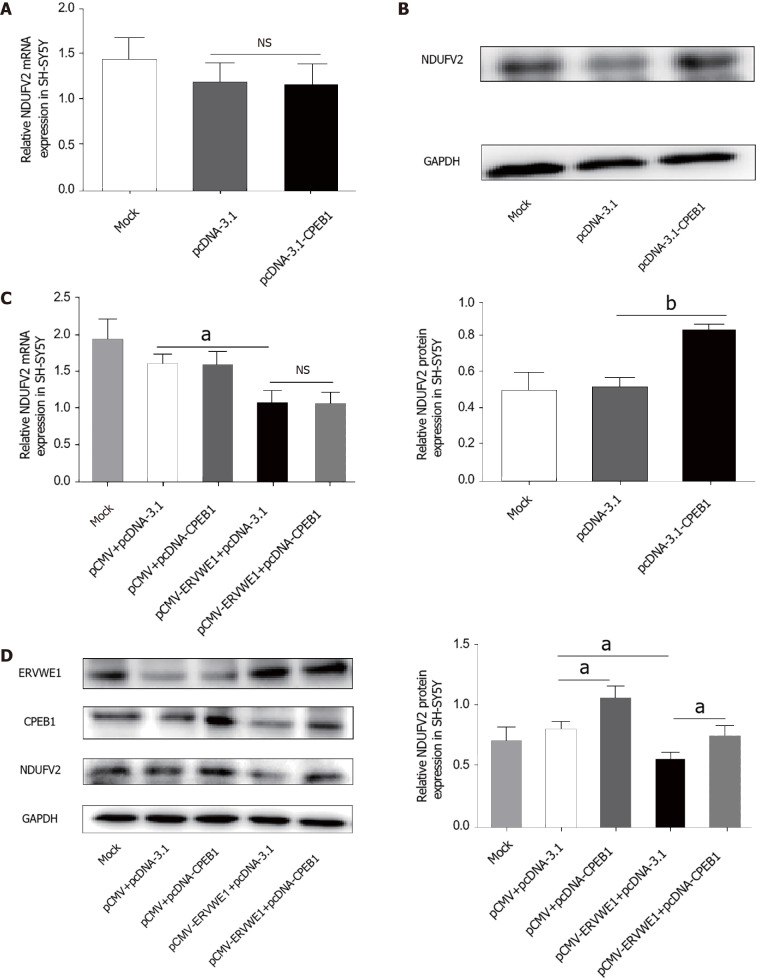
Our clinical data revealed CPEB1 was downregulated, and demonstrated a negative correlation between CPEB1 and ERVWE1 in schizophrenia patients. It would be intriguing to assess the effect of ERVWE1 on CPEB1. We found that both mRNA (P < 0.01; Figure 4A) and protein (P < 0.05; Figure 4B) expression of CPEB1 was significantly lower after overexpression of ERVWE1 in SH-SY5Y cells than in control cells. Moreover, the luciferase assay demonstrated that ERVWE1 suppressed the promoter activity of CPEB1 (P < 0.01; Figure 4C). In order to define the minimum sequence essential for the transcriptional control of CPEB1 gene, four sequentially truncated promoters were examined in transfection assay. The luciferase reporter assay demonstrated that the promoter ranged from -400 to 0 was the minimum sequence required for biological activity (P < 0.05; Figure 4D). Furthermore, we found that CPEB1 could downregulate NDUFV2P1 mRNA levels in SH-SY5Y cells (P < 0.05; Figure 4E). qPCR and western blot analysis confirmed CPEB1 transfection (Supplementary Figure 3A and B). Additionally, overexpression of CPEB1 substantially impaired the ERVWE1-induced mRNA expression of NDUFV2P1 in SH-SY5Y cells, suggesting that ERVWE1-induced NDUFV2P1 was mediated by CPEB1 (P < 0.05; Figure 4F).
Figure 4.
ERVWE1 elevated NADH dehydrogenase ubiquinone flavoprotein 2 pseudogene (NDUFV2P1) expression by suppressing cytoplasmic polyadenylation element-binding protein 1 (CPEB1). A: The mRNA levels of cytoplasmic polyadenylation element-binding protein 1 (CPEB1) after overexpression of ERVWE1 in SH-SY5Y cells using quantitative polymerase chain reaction (qPCR); B: Western blots of CPEB1 and GAPDH in SH-SY5Y cells; C: Luciferase assays of pGL3-CPEB1 promoter co-transfected with pCMV-ERVWE1 or control vector in SH-SY5Y cells; D: Luciferase assays of different lengths of pGL3-CPEB1 promoter co-transfected with pCMV-ERVWE1 or control vector in SH-SY5Y cells; E: The mRNA levels of NADH dehydrogenase ubiquinone flavoprotein 2 pseudogene (NDUFV2P1) after overexpression of CPEB1 in SH-SY5Y cells using qPCR; F: Represent the mRNA levels of NDUFV2P1 in SH-SY5Y cells after co-transfection with ERVWE1 and CPEB1 using qPCR. Each bar represents the mean ± SD of three independent experiments. aP < 0.05; bP < 0.01; NS: Not significant.
Several studies have reported that CPEB1 regulates NDUFV2 expression by a post-transcriptional mechanism[16]. We also found no change in the mRNA expression of NDUFV2 after transfection with CPEB1 (Figure 5A). Western blotting showed that CPEB1 elevated the protein levels of NDUFV2 (P < 0.01; Figure 5B). We further assessed the possible role of CPEB1 in ERVWE1-mediated NDUFV2 expression. Our results revealed that NDUFV2 mRNA was not altered, regardless of whether CPEB1 was overexpressed compared with control cells (Figure 5C). Regardless of whether CPEB1 was highly expressed, western blot analyses revealed that CPEB1 could relieve the inhibitory effect of ERVWE1 on NDUFV2 (P < 0.05; Figure 5D). Together, these results suggest that ERVWE1 regulates the NDUFV2P1/NDUFV2 signaling pathway through the suppression of CPEB1.
Figure 5.
ERVWE1 reduced NADH dehydrogenase ubiquinone flavoprotein 2 (NDUFV2) expression by suppressing cytoplasmic polyadenylation element-binding protein 1 (CPEB1). A: The mRNA levels of NADH dehydrogenase ubiquinone flavoprotein 2 (NDUFV2) after overexpression of cytoplasmic polyadenylation element-binding protein 1 (CPEB1) in SH-SY5Y cells using quantitative polymerase chain reaction (qPCR); B: Western blots of NDUFV2 and GAPDH in SH-SY5Y cells; C: Represent the mRNA levels of NDUFV2 in SH-SY5Y cells after co-transfection with ERVWE1 and CPEB1 using qPCR; D: Western blots of ERVWE1, CPEB1, NDUFV2, and GAPDH in SH-SY5Y cells. Each bar represents the mean ± SD of three independent experiments. aP < 0.05; bP < 0.01; NS: Not significant.
ERVWE1 impairs electron transport chain complex I activity via the CPEB1/NDUFV2P1/NDUFV2 signaling pathway
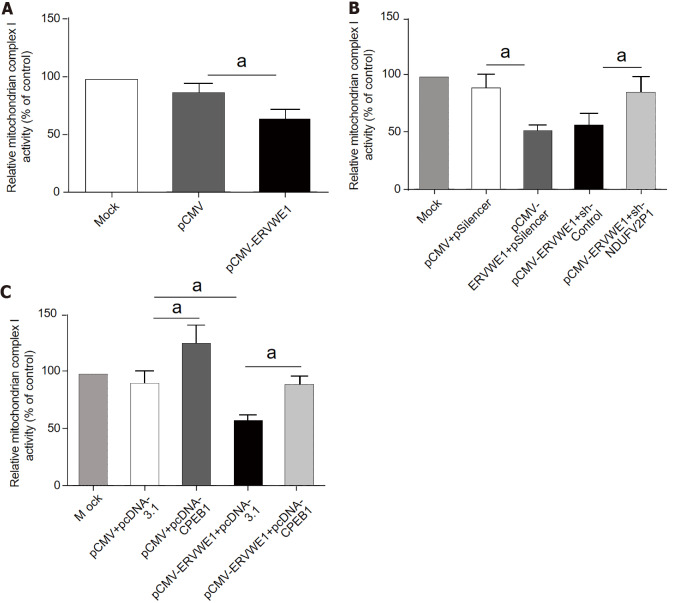
NDUFV2 is a subunit of mitochondrial complex I[11]. Our above results demonstrated that ERVWE1 significantly reduced the mRNA and protein expressions of NDUFV2. Thus, we further analyzed the impact of ERVWE1 on complex I activity. Successful transfection was verified by western blot analyses in the transfected cells (Supplementary Figure 4). The complex I activity was decreased in ERVWE1-overexpressing cells compared to that in control cells (P < 0.05; Figure 6A).
Figure 6.
ERVWE1 contributed to complex I deficits through the cytoplasmic polyadenylation element-binding protein 1 (CPEB1)/ NADH dehydrogenase ubiquinone flavoprotein 2 (NDUFV2) pseudogene (NDUFV2P1)/NDUFV2 signaling pathway. A: Complex I activity in SH-SY5Y cells transfected with pCMV-ERVWE1 or control; B: Complex I activity in SH-SY5Y cells co-transfected with pCMV-ERVWE1 + shNDUFV2P1 or control; C: Complex I activity in SH-SY5Y cells co-transfected with pCMV-ERVWE1 + pcDNA3.1-CPEB1 or control. Each bar represents the mean ± SD of three independent experiments. aP < 0.05.
We measured complex I activity after the knockdown of NDUFV2P1 in SH-SY5Y cells. Downregulating NDUFV2P1 could mitigate ERVWE1-induced inhibition of mitochondrial complex I in SH-SY5Y cells (P < 0.05; Figure 6B), indicating that ERVWE1 inhibited complex I activity by increasing NDUFV2P1.
Another experiment showed that CPEB1 could reverse the complex I deficiency induced by ERVWE1 in SH-SY5Y cells (P < 0.05; Figure 6C), suggesting that ERVWE1 could regulate complex I activity via suppressing CPEB1 expression.
In summary, our data indicate that ERVWE1 modulates mitochondrial energy metabolism, primarily of complex I, via the CPEB1/NDUFV2P1/NDUFV2 signaling pathway in schizophrenia.
DISCUSSION
Schizophrenia is a multifactorial mental disorder with abnormal cognitive functions and behavior. It is characterized by continuous or relapsing episodes of psychosis. Potential blood-based biomarkers have the potential to increase the precision of the diagnostic and prognostic process for schizophrenia. The field of blood-biomarker discovery for schizophrenia is coming to fruition as patient blood is an easily accessible biological sample[29]. An increasing number of molecular detection techniques with high specificity and sensitivity are currently applied in the search for serum markers or genetic markers in peripheral blood. Previous studies have addressed potential blood-based biomarkers for schizophrenia, such as microRNA[30,31], insulin-related peptides[32,33], and inflammatory cytokines[34]. However, no biomarkers have been successfully used in the clinical diagnosis of schizophrenia. GWAS has implicated CPEB1, which possesses a highly conserved RNA-binding domain and controls the cytoplasmic polyadenylation of mRNA by combining with the specific sequences, as a novel schizophrenia susceptibility gene[27,35]. However, there is no experimental evidence of an association between CPEB1 and schizophrenia. Clinical data of this study do not follow a normal distribution, so Spearman’s rank correlation was used for correlation analyses. In this paper, clinical data showed that CPEB1 was decreased and negatively correlated with ERVWE1 in schizophrenia. Univariate and multivariate analyses suggested that CPEB1 might be a novel potential blood-based biomarker and risk factor for schizophrenia.
The mouse model has shown that knockout of CPEB1 downregulates NDUFV2 protein expression[16]. The abnormal expression of NDUFV2 has been found in schizophrenia[7,36]. Our data displayed decreased NDUFV2 transcription in schizophrenia, in contrast to the data reported by Haghighatfard et al[36]. The reason for this might be the primer used by Haghighatfard’s group can detect not only NDUFV2 but also NDUFV2P1. Herein, our primers could successfully detect the transcript levels of NDUFV2 and NDUFV2P1. The NDUFV2P1 transcript was upregulated in schizophrenia and was negatively regulated NDUFV2. Additionally, our previous study showed false positives using nonspecific primers to detect both NDUFV2 and NDUFV2P1 (Supplementary Figure 5). We also found decreased NDUFV2 protein expression, consistent with the report by Bergman et al[14]. However, Bergman et al[14] showed no changes in NDUFV2 mRNA. The primary factor driving the difference between our and Bergman's findings is that we focused on recent-onset schizophrenia patients. In our sample, all subjects with recent-onset schizophrenia were free from antipsychotics, whereas in the schizophrenia samples of Bergman's group, all but one participant were using antipsychotic medications. A previous study demonstrated that exposure to antipsychotic medication can increase NDUFV2 mRNA expression in Sprague-Dawley rats[37]. Hence, antipsychotic medication treatment might reverse the downward trend of NDUFV2 and result in no change at the mRNA level. NDUFV2P1 is a non-transcribed pseudogene of NDUFV2. There are few studies on NDUFV2P1. Only one study has reported that schizophrenia-derived cells have increased expression of NDUFV2P1[14]. Similarly, we found that NDUFV2P1 was significantly increased and negatively correlated with NDUFV2 in schizophrenia patients. ERVWE1 is associated with schizophrenia[18]. Further analyses indicated that ERVWE1 was negatively correlated with NDUFV2 and positively correlated with NDUFV2P1 in schizophrenia patients.
The etiology of schizophrenia remains unclear. Scientists believe that both environmental and genetic factors contribute to the risk of schizophrenia. Several studies have indicated that ERVWE1, a new pathogenic factor in schizophrenia, acts as a bridge and trigger between environmental and genetic factors. A series of environmental factors may activate the ERVWE1, a protein displaying inflammation and neurotoxicity[38]. Several studies have shown that ERVWE1 mediates neuroinflammation via the Toll-like receptor pathway in schizophrenia[19,38,39] or multiple sclerosis (MS)[40]. Moreover, ERVWE1 can activate inducible nitric oxide synthase activity and induce cytotoxic T lymphocyte responses[38]. Further studies on the delineation of a role for retroviruses in schizophrenia will bring new diagnosis and treatment of the devastating schizophrenia.
NDUFV2P1 is a potential predictive marker in schizophrenia by negatively regulating its parent gene, NDUFV2[14]. Based on the findings that ERVWE1 can regulate some schizophrenia-related genes[18,38], we conducted additional studies to examine the potential relationship among ERVWE1, NDUFV2P1, and NDUFV2 in schizophrenia. Previous research suggests that ERVWE1 is overexpressed in brain tissues[41]. To study the possible role of ERVWE1, we selected the SH-SY5Y cell line, a human-derived neuroblastoma cell with neuronal function and differentiation and widely used as in vitro model for neuronal studies. Our results indicated that ERVWE1 induced upregulation of NDUFV2P1 and downregulation of NDUFV2. Pseudogenes exhibit essential roles in gene expression and gene regulation. They are capable of regulating parental genes or act as a competitive endogenous RNA (ceRNA)[28] or small interfering RNA[42]. Our results showed that NDUFV2P1 acted as a ceRNA in NDUFV2 expression induced by ERVWE1.
The NDUFV2 promoter plays essential roles in its expression[11]. ERVWE1 has been proposed to regulate the activation of promoters and control the expression of some schizophrenia-related genes[18,19]. We found that NDUFV2P1 participated in the process of ERVWE1 suppressing the NDUFV2 promoter. It has been suggested that NDUFV2P1 possibly competes with NDUFV2 by interfering with its mRNA[43]. Retrocopies or processed pseudogenes, the majority of the human pseudogenes, are gene copies originating from mRNA retrotransposition. Our observation might reveal another way of regulating parental genes by their retrocopies. From these, we could conclude that ERVWE1 inhibited the NDUFV2 promoter by raising the NDUFV2P1 transcript.
Schizophrenia is linked to mitochondrial energy metabolism dysfunction. CPEB1 controls polyadenylation-induced translation of NDUFV2 mRNA and results in impaired brain-specific mitochondrial respiration[16]. In this work, we found that ERVWE1 dramatically lowered CPEB1 levels via inhibiting its promoter and that the range of -400 to 0 was necessary. We found a new regulatory mechanism that is different from the conserved mechanism of CPEB1. Interestingly, overexpression of CPEB1 led to increased protein levels of NDUFV2 with no change at the transcript level. The data are in line with another study[16]. Furthermore, CPEB1 reduced the NDUFV2P1 transcript. To date, there have been no published data on the mechanisms that regulate NDUFV2P1. Our finding might be the first report of NDUFV2P1 regulation in schizophrenia. Further studies are needed to determine precisely how CPEB1 affects pseudogene transcripts. Moreover, overexpression of CPEB1 decreased the NDUFV2P1 levels induced by ERVWE1. Simultaneously, overexpression of CPEB1 substantially elevated the expression of NDUFV2 inhibited by ERVWE1. Our results suggested that ERVWE1 regulated NDUFV2P1/NDUFV2 signaling pathway through the suppression of CPEB1.
Ample evidence indicates that impaired mitochondrial energy metabolism may be compromised in schizophrenia patients[44,45]. Complex I deficiency is among the most encountered defects of mitochondrial energy metabolism[46]. Complex I, also names NADH:ubiquinone oxidoreductase, is the largest and the most elaborate component of the mitochondrial respiratory chain. It catalyzes the oxidation of NADH, together with the transfers of two electrons to the ubiquinone. Complex I deficiency is associated with abnormalities in calcium signaling[46,47]. The possible role of calcium signaling in schizophrenia is initially presented by Jimerson et al[48]. Our previous report demonstrated that ERVWE1 can induce calcium influx in human neuroblastoma cells[49]. Therefore, we speculated that ERVWE1 might cause the decrease of complex I activity in schizophrenia. Herein, we found overexpression of ERVWE1 suppressed complex I activity. Knockdown of NDUFV2P1 significantly restored the complex I activity suppressed by ERVWE1. This observation suggested that NDUFV2P1 was required in the ERVWE1-induced complex I activity deficiency. CPEB1 is essential to promote mitochondrial energy production in neurons[15]. CPEB1 knockdown cells have reduced complex I activity and mitochondria number[16,50]. Our results showed that CPEB1 overexpression could rescue complex I activity, which was inhibited by ERVWE1. As a core subunit of this complex, NDUFV2 is a crucial regulator of complex I activity, depletion and phosphorylation of this protein causes a decrease in complex I activity[51,52].
Schizophrenia mostly sets in after the age of 20. Mitochondrial energy metabolism dysfunction has been implicated in the etiology of schizophrenia in early adulthood from age 20-30[53]. Research from our and others suggests that environmental factors, including viral infection, drug stimulation, and genetic variation, can cause abnormal expression of ERVWE1. In this study, we demonstrated that increased ERVWE1 induces mitochondrial metabolism deficits, which ultimately leads to the pathogenesis of schizophrenia. In summary, ERVWE1 overexpression contributes to complex I deficits in schizophrenia via the CPEB1/NDUFV2P1/NDUFV2 signaling pathway.
CONCLUSION
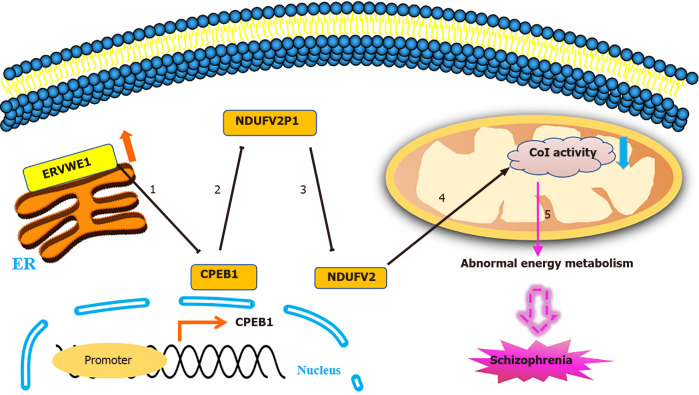
In conclusion, the purpose of this study was to discover novel potential blood-based biomarkers and reveal the potential mechanisms underlying ERVWE1-mediated mitochondrial energy metabolism defects in schizophrenia. We reported that the expression of CPEB1 was lower and negatively correlated with ERVWE1 in schizophrenia blood samples. This study is to identify CPEB1 as a novel potential blood-based biomarker for schizophrenia. We also found that NDUFV2 exerted an inverse expression with NDUFV2P1 and ERVWE1 in schizophrenia. Further analyses showed a positive correlation and marked consistency between expressions of NDUFV2P1 and ERVWE1. In vitro experiments demonstrated that ERVWE1 mediated the downregulation of NDUFV2 by upregulating its pseudogene, NDUFV2P1. Moreover, CPEB1 was decreased after ERVWE1 overexpression and worked as a mediator of the NDUFV2P1/NDUFV2 signaling pathway induced by ERVWE1. Further studies indicated that ERVWE1 impaired mitochondrial complex I activity, which might lead to mitochondrial metabolic dysregulation through the CPEB1/NDUFV21P1/NDUFV2 signaling pathway (Figure 7). As a novel potential blood-based biomarker and risk factor, further studies and clinical research are needed better to understand the role of decreased CPEB1 and NDUFV2 in schizophrenia to develop this paradigm into an effective treatment strategy.
Figure 7.
The proposed signaling pathways for ERVWE1 contributes to mitochondrial energy metabolism deficiency in schizophrenia. 1: ERVWE1 suppressed cytoplasmic polyadenylation element-binding protein 1 (CPEB1) mRNA and protein expression by regulating its promoter activity; 2: ERVWE1 upregulated the expression of NADH dehydrogenase ubiquinone flavoprotein 2 (NDUFV2) pseudogene (NDUFV2P1) through enhancing its promoter activity. CPEB1 downregulated NDUFV2P1 expression and functioned as a mediator of NDUFV2P1 transcript induced by ERVWE1; 3: NDUFV2P1 reduced NDUFV2 expression through inhibiting its promoter activation. Additionally, ERVWE1 decreased the mRNA and protein expression of NDUFV2 and suppressed NDUFV2 promoter through increasing NDUFV2P1 transcript. Moreover, CPEB1 was involved in regulating NDUFV2P1/NDUFV2 signaling pathway triggered by ERVWE1; 4: ERVWE1 reduced complex I activity via regulating NDUFV2 and its retrocopies expression. Decreased CPEB1 was involved in the complex I activity deficiency induced by ERVWE1; 5: Deficient complex I activity might result in mitochondrial metabolic dysregulation and contributed to the development of schizophrenia.
ARTICLE HIGHLIGHTS
Research background
Schizophrenia is a devastating psychiatric disorder that impairs mental and social functioning. There are several hypotheses regarding the pathogenesis of schizophrenia, including mitochondrial dysfunction. Both genetic and environmental factors contribute to the development of schizophrenia. Our previous studies showed that ERVWE1 acts as a bridge between genetic and environmental factors and plays an important role in the occurrence of schizophrenia.
Research motivation
Schizophrenia is a chronic disorder requiring long-term treatment and having a significant impact on patients and their families. Biomarkers and pathogenic mechanisms have become the most important undertaking for schizophrenia. In this manuscript, we focused on the relationship between mitochondrial energy metabolic deficits and ERVWE1 in schizophrenia.
Research objectives
This study aimed to identify novel potential blood-based biomarkers and risk factors for schizophrenia and to explore the underlying mechanisms ERVWE1-mediated mitochondrial metabolic deficits in schizophrenia.
Research methods
Quantitative polymerase chain reaction (qPCR) and enzyme-linked immunosorbent assay were used to detect differentially risk genes expression in blood samples. Statistical analyses of clinical data were performed by median analysis, Mann-Whitney U analysis, and Spearman’s rank correlation analysis. qPCR, western blotting, luciferase assay, and complex I enzyme activity microplate assay were performed to study the relationship between ERVWE1, CPEB1, NADH dehydrogenase ubiquinone flavoprotein 2 (NDUFV2), and NDUFV2 pseudogene (NDUFV2P1).
Research results
CPEB1 and NDUFV2 were deceased and negatively correlated with ERVWE1 in schizophrenia. Moreover, Increased NDUFV2P1 demonstrated a significant positive correlation with ERVWE1 and negative correlation with NDUFV2 in schizophrenia. Cytological experiments demonstrated that ERVWE1 suppressed CPEB1 by suppressing its promoter activity and downregulated NDUFV2 expression through increasing NDUFV2P1 transcript. Further studies showed that CPEB1 participated in regulating the NDUFV2P1/NDUFV2 axis mediated by ERVWE1 in SH-SY5Y cells. Finally, we found that ERVWE1 regulated complex I activity through the CPEB1/NDUFV2P1/NDUFV2 signaling pathway in SH-SY5Y cells.
Research conclusions
CPEB1 and NDUFV2 were identified as novel risk factors in schizophrenia. We also proposed a novel mechanism that ERVWE1 mediated mitochondria metabolic deficits through the CPEB1/NDUFV2P1/NDUFV2 signaling pathway in schizophrenia.
Research perspectives
Further study is needed to verify if CPEB1 and NDUFV2 can be used as blood-based biomarker of schizophrenia. As a risk factor, further efforts need to be focused on better understand the role of ERVWE1, CPEB1 and NDUFV2 in the pathogenesis of schizophrenia. Although our study suggests that ERVWE1 may be a potential therapeutic target for schizophrenia, whether ERVWE1 can really benefit clinical treatment requires further research in animal experiments and clinical trials to confirm these data.
ACKNOWLEDGEMENTS
We thank Wang Y (Master of Science in Epidemiology and Biostatistics) for assistance with the biostatistical analyses.
Footnotes
Institutional review board statement: The protocol of this study was approved by the Institutional Review Board of Wuhan University, School of Basic Medical Sciences, and the study was carried out in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki) for experiments involving humans. Informed consent was obtained for experimentation with human subjects and their privacy rights were always observed.
Conflict-of-interest statement: No potential conflicts of interest.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review started: April 25, 2021
First decision: July 14, 2021
Article in press: August 25, 2021
Specialty type: Psychiatry
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Pethe P S-Editor: Wu YXJ L-Editor: Filipodia P-Editor: Guo X
Contributor Information
Ya-Ru Xia, State Key Laboratory of Virology and Hubei Province Key Laboratory of Allergy & Immunology, Department of Medical Microbiology, School of Medicine, Wuhan University, Wuhan 430071, Hubei Province, China.
Xiao-Cui Wei, State Key Laboratory of Virology and Hubei Province Key Laboratory of Allergy & Immunology, Department of Medical Microbiology, School of Medicine, Wuhan University, Wuhan 430071, Hubei Province, China.
Wen-Shi Li, State Key Laboratory of Virology and Hubei Province Key Laboratory of Allergy & Immunology, Department of Medical Microbiology, School of Medicine, Wuhan University, Wuhan 430071, Hubei Province, China.
Qiu-Jin Yan, State Key Laboratory of Virology and Hubei Province Key Laboratory of Allergy & Immunology, Department of Medical Microbiology, School of Medicine, Wuhan University, Wuhan 430071, Hubei Province, China.
Xiu-Lin Wu, State Key Laboratory of Virology and Hubei Province Key Laboratory of Allergy & Immunology, Department of Medical Microbiology, School of Medicine, Wuhan University, Wuhan 430071, Hubei Province, China.
Wei Yao, State Key Laboratory of Virology and Hubei Province Key Laboratory of Allergy & Immunology, Department of Medical Microbiology, School of Medicine, Wuhan University, Wuhan 430071, Hubei Province, China.
Xu-Hang Li, State Key Laboratory of Virology and Hubei Province Key Laboratory of Allergy & Immunology, Department of Medical Microbiology, School of Medicine, Wuhan University, Wuhan 430071, Hubei Province, China.
Fan Zhu, State Key Laboratory of Virology and Hubei Province Key Laboratory of Allergy & Immunology, Department of Medical Microbiology, School of Medicine, Wuhan University, Wuhan 430071, Hubei Province, China. fanzhu@whu.edu.cn.
Data sharing statement
The data used and analyzed during the current study are available from the corresponding author upon request.
References
- 1.GBD 2016 Disease and Injury Incidence and Prevalence Collaborators. Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet. 2017;390:1211–1259. doi: 10.1016/S0140-6736(17)32154-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fletcher PC, Frith CD. Perceiving is believing: a Bayesian approach to explaining the positive symptoms of schizophrenia. Nat Rev Neurosci. 2009;10:48–58. doi: 10.1038/nrn2536. [DOI] [PubMed] [Google Scholar]
- 3.Chong HY, Teoh SL, Wu DB, Kotirum S, Chiou CF, Chaiyakunapruk N. Global economic burden of schizophrenia: a systematic review. Neuropsychiatr Dis Treat. 2016;12:357–373. doi: 10.2147/NDT.S96649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prabakaran S, Swatton JE, Ryan MM, Huffaker SJ, Huang JT, Griffin JL, Wayland M, Freeman T, Dudbridge F, Lilley KS, Karp NA, Hester S, Tkachev D, Mimmack ML, Yolken RH, Webster MJ, Torrey EF, Bahn S. Mitochondrial dysfunction in schizophrenia: evidence for compromised brain metabolism and oxidative stress. Mol Psychiatry. 2004;9:684–697, 643. doi: 10.1038/sj.mp.4001511. [DOI] [PubMed] [Google Scholar]
- 5.Rajasekaran A, Venkatasubramanian G, Berk M, Debnath M. Mitochondrial dysfunction in schizophrenia: pathways, mechanisms and implications. Neurosci Biobehav Rev. 2015;48:10–21. doi: 10.1016/j.neubiorev.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 6.Kim Y, Vadodaria KC, Lenkei Z, Kato T, Gage FH, Marchetto MC, Santos R. Mitochondria, Metabolism, and Redox Mechanisms in Psychiatric Disorders. Antioxid Redox Signal. 2019;31:275–317. doi: 10.1089/ars.2018.7606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karry R, Klein E, Ben Shachar D. Mitochondrial complex I subunits expression is altered in schizophrenia: a postmortem study. Biol Psychiatry. 2004;55:676–684. doi: 10.1016/j.biopsych.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 8.Dror N, Klein E, Karry R, Sheinkman A, Kirsh Z, Mazor M, Tzukerman M, Ben-Shachar D. State-dependent alterations in mitochondrial complex I activity in platelets: a potential peripheral marker for schizophrenia. Mol Psychiatry. 2002;7:995–1001. doi: 10.1038/sj.mp.4001116. [DOI] [PubMed] [Google Scholar]
- 9.Holper L, Ben-Shachar D, Mann JJ. Multivariate meta-analyses of mitochondrial complex I and IV in major depressive disorder, bipolar disorder, schizophrenia, Alzheimer disease, and Parkinson disease. Neuropsychopharmacology. 2019;44:837–849. doi: 10.1038/s41386-018-0090-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ayalew M, Le-Niculescu H, Levey DF, Jain N, Changala B, Patel SD, Winiger E, Breier A, Shekhar A, Amdur R, Koller D, Nurnberger JI, Corvin A, Geyer M, Tsuang MT, Salomon D, Schork NJ, Fanous AH, O'Donovan MC, Niculescu AB. Convergent functional genomics of schizophrenia: from comprehensive understanding to genetic risk prediction. Mol Psychiatry. 2012;17:887–905. doi: 10.1038/mp.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ben-Shachar D, Karry R. Sp1 expression is disrupted in schizophrenia; a possible mechanism for the abnormal expression of mitochondrial complex I genes, NDUFV1 and NDUFV2. PLoS One. 2007;2:e817. doi: 10.1371/journal.pone.0000817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Washizuka S, Kametani M, Sasaki T, Tochigi M, Umekage T, Kohda K, Kato T. Association of mitochondrial complex I subunit gene NDUFV2 at 18p11 with schizophrenia in the Japanese population. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:301–304. doi: 10.1002/ajmg.b.30285. [DOI] [PubMed] [Google Scholar]
- 13.de Coo R, Buddiger P, Smeets H, Geurts van Kessel A, Morgan-Hughes J, Weghuis DO, Overhauser J, van Oost B. Molecular cloning and characterization of the active human mitochondrial NADH:ubiquinone oxidoreductase 24-kDa gene (NDUFV2) and its pseudogene. Genomics. 1995;26:461–466. doi: 10.1016/0888-7543(95)80163-g. [DOI] [PubMed] [Google Scholar]
- 14.Bergman O, Karry R, Milhem J, Ben-Shachar D. NDUFV2 pseudogene (NDUFV2P1) contributes to mitochondrial complex I deficits in schizophrenia. Mol Psychiatry. 2020;25:805–820. doi: 10.1038/s41380-018-0309-9. [DOI] [PubMed] [Google Scholar]
- 15.Ivshina M, Lasko P, Richter JD. Cytoplasmic polyadenylation element binding proteins in development, health, and disease. Annu Rev Cell Dev Biol. 2014;30:393–415. doi: 10.1146/annurev-cellbio-101011-155831. [DOI] [PubMed] [Google Scholar]
- 16.Oruganty-Das A, Ng T, Udagawa T, Goh EL, Richter JD. Translational control of mitochondrial energy production mediates neuron morphogenesis. Cell Metab. 2012;16:789–800. doi: 10.1016/j.cmet.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karlsson H, Bachmann S, Schröder J, McArthur J, Torrey EF, Yolken RH. Retroviral RNA identified in the cerebrospinal fluids and brains of individuals with schizophrenia. Proc Natl Acad Sci U S A. 2001;98:4634–4639. doi: 10.1073/pnas.061021998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang W, Li S, Hu Y, Yu H, Luo F, Zhang Q, Zhu F. Implication of the env gene of the human endogenous retrovirus W family in the expression of BDNF and DRD3 and development of recent-onset schizophrenia. Schizophr Bull. 2011;37:988–1000. doi: 10.1093/schbul/sbp166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, Liu Z, Wang P, Li S, Zeng J, Tu X, Yan Q, Xiao Z, Pan M, Zhu F. Syncytin-1, an endogenous retroviral protein, triggers the activation of CRP via TLR3 signal cascade in glial cells. Brain Behav Immun. 2018;67:324–334. doi: 10.1016/j.bbi.2017.09.009. [DOI] [PubMed] [Google Scholar]
- 20.Grandi N, Tramontano E. Human Endogenous Retroviruses Are Ancient Acquired Elements Still Shaping Innate Immune Responses. Front Immunol. 2018;9:2039. doi: 10.3389/fimmu.2018.02039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Küry P, Nath A, Créange A, Dolei A, Marche P, Gold J, Giovannoni G, Hartung HP, Perron H. Human Endogenous Retroviruses in Neurological Diseases. Trends Mol Med. 2018;24:379–394. doi: 10.1016/j.molmed.2018.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vargiu L, Rodriguez-Tomé P, Sperber GO, Cadeddu M, Grandi N, Blikstad V, Tramontano E, Blomberg J. Classification and characterization of human endogenous retroviruses; mosaic forms are common. Retrovirology. 2016;13:7. doi: 10.1186/s12977-015-0232-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mallet F, Bouton O, Prudhomme S, Cheynet V, Oriol G, Bonnaud B, Lucotte G, Duret L, Mandrand B. The endogenous retroviral locus ERVWE1 is a bona fide gene involved in hominoid placental physiology. Proc Natl Acad Sci U S A. 2004;101:1731–1736. doi: 10.1073/pnas.0305763101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gal H, Lysenko M, Stroganov S, Vadai E, Youssef SA, Tzadikevitch-Geffen K, Rotkopf R, Biron-Shental T, de Bruin A, Neeman M, Krizhanovsky V. Molecular pathways of senescence regulate placental structure and function. EMBO J. 2019;38:e100849. doi: 10.15252/embj.2018100849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tolosa JM, Schjenken JE, Clifton VL, Vargas A, Barbeau B, Lowry P, Maiti K, Smith R. The endogenous retroviral envelope protein syncytin-1 inhibits LPS/PHA-stimulated cytokine responses in human blood and is sorted into placental exosomes. Placenta. 2012;33:933–941. doi: 10.1016/j.placenta.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 26.Perron H, Mekaoui L, Bernard C, Veas F, Stefas I, Leboyer M. Endogenous retrovirus type W GAG and envelope protein antigenemia in serum of schizophrenic patients. Biol Psychiatry. 2008;64:1019–1023. doi: 10.1016/j.biopsych.2008.06.028. [DOI] [PubMed] [Google Scholar]
- 27.Li Z, Chen J, Yu H, He L, Xu Y, Zhang D, Yi Q, Li C, Li X, Shen J, Song Z, Ji W, Wang M, Zhou J, Chen B, Liu Y, Wang J, Wang P, Yang P, Wang Q, Feng G, Liu B, Sun W, Li B, He G, Li W, Wan C, Xu Q, Wen Z, Liu K, Huang F, Ji J, Ripke S, Yue W, Sullivan PF, O'Donovan MC, Shi Y. Genome-wide association analysis identifies 30 new susceptibility loci for schizophrenia. Nat Genet. 2017;49:1576–1583. doi: 10.1038/ng.3973. [DOI] [PubMed] [Google Scholar]
- 28.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Comes AL, Papiol S, Mueller T, Geyer PE, Mann M, Schulze TG. Proteomics for blood biomarker exploration of severe mental illness: pitfalls of the past and potential for the future. Transl Psychiatry. 2018;8:160. doi: 10.1038/s41398-018-0219-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He K, Guo C, He L, Shi Y. MiRNAs of peripheral blood as the biomarker of schizophrenia. Hereditas. 2018;155:9. doi: 10.1186/s41065-017-0044-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma J, Shang S, Wang J, Zhang T, Nie F, Song X, Heping Zhao, Zhu C, Zhang R, Hao D. Identification of miR-22-3p, miR-92a-3p, and miR-137 in peripheral blood as biomarker for schizophrenia. Psychiatry Res. 2018;265:70–76. doi: 10.1016/j.psychres.2018.03.080. [DOI] [PubMed] [Google Scholar]
- 32.Cheng TM, Lu YE, Guest PC, Rahmoune H, Harris LW, Wang L, Ma D, Stelzhammer V, Umrania Y, Wayland MT, Lió P, Bahn S. Identification of targeted analyte clusters for studies of schizophrenia. Mol Cell Proteomics. 2010;9:510–522. doi: 10.1074/mcp.M900372-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guest PC, Schwarz E, Krishnamurthy D, Harris LW, Leweke FM, Rothermundt M, van Beveren NJ, Spain M, Barnes A, Steiner J, Rahmoune H, Bahn S. Altered levels of circulating insulin and other neuroendocrine hormones associated with the onset of schizophrenia. Psychoneuroendocrinology. 2011;36:1092–1096. doi: 10.1016/j.psyneuen.2010.12.018. [DOI] [PubMed] [Google Scholar]
- 34.Schwarz E, Guest PC, Rahmoune H, Harris LW, Wang L, Leweke FM, Rothermundt M, Bogerts B, Koethe D, Kranaster L, Ohrmann P, Suslow T, McAllister G, Spain M, Barnes A, van Beveren NJ, Baron-Cohen S, Steiner J, Torrey FE, Yolken RH, Bahn S. Identification of a biological signature for schizophrenia in serum. Mol Psychiatry. 2012;17:494–502. doi: 10.1038/mp.2011.42. [DOI] [PubMed] [Google Scholar]
- 35.Schmidt-Kastner R, Guloksuz S, Kietzmann T, van Os J, Rutten BPF. Analysis of GWAS-Derived Schizophrenia Genes for Links to Ischemia-Hypoxia Response of the Brain. Front Psychiatry. 2020;11:393. doi: 10.3389/fpsyt.2020.00393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haghighatfard A, Andalib S, Amini Faskhodi M, Sadeghi S, Ghaderi AH, Moradkhani S, Rostampour J, Tabrizi Z, Mahmoodi A, Karimi T, Ghadimi Z. Gene expression study of mitochondrial complex I in schizophrenia and paranoid personality disorder. World J Biol Psychiatry. 2018;19:S133–S146. doi: 10.1080/15622975.2017.1282171. [DOI] [PubMed] [Google Scholar]
- 37.Ji B, La Y, Gao L, Zhu H, Tian N, Zhang M, Yang Y, Zhao X, Tang R, Ma G, Zhou J, Meng J, Ma J, Zhang Z, Li H, Feng G, Wang Y, He L, Wan C. A comparative proteomics analysis of rat mitochondria from the cerebral cortex and hippocampus in response to antipsychotic medications. J Proteome Res. 2009;8:3633–3641. doi: 10.1021/pr800876z. [DOI] [PubMed] [Google Scholar]
- 38.Wang X, Huang J, Zhu F. Human Endogenous Retroviral Envelope Protein Syncytin-1 and Inflammatory Abnormalities in Neuropsychological Diseases. Front Psychiatry. 2018;9:422. doi: 10.3389/fpsyt.2018.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X, Wu X, Huang J, Li H, Yan Q, Zhu F. Human endogenous retrovirus W family envelope protein (HERV-W env) facilitates the production of TNF-α and IL-10 by inhibiting MyD88s in glial cells. Arch Virol. 2021;166:1035–1045. doi: 10.1007/s00705-020-04933-8. [DOI] [PubMed] [Google Scholar]
- 40.Perron H, Germi R, Bernard C, Garcia-Montojo M, Deluen C, Farinelli L, Faucard R, Veas F, Stefas I, Fabriek BO, Van-Horssen J, Van-der-Valk P, Gerdil C, Mancuso R, Saresella M, Clerici M, Marcel S, Creange A, Cavaretta R, Caputo D, Arru G, Morand P, Lang AB, Sotgiu S, Ruprecht K, Rieckmann P, Villoslada P, Chofflon M, Boucraut J, Pelletier J, Hartung HP. Human endogenous retrovirus type W envelope expression in blood and brain cells provides new insights into multiple sclerosis disease. Mult Scler. 2012;18:1721–1736. doi: 10.1177/1352458512441381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sotgiu S, Mameli G, Serra C, Zarbo IR, Arru G, Dolei A. Multiple sclerosis-associated retrovirus and progressive disability of multiple sclerosis. Mult Scler. 2010;16:1248–1251. doi: 10.1177/1352458510376956. [DOI] [PubMed] [Google Scholar]
- 42.Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, Hannon GJ. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kubiak MR, Szcześniak MW, Makałowska I. Complex Analysis of Retroposed Genes' Contribution to Human Genome, Proteome and Transcriptome. Genes (Basel) 2020;11 doi: 10.3390/genes11050542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ni P, Chung S. Mitochondrial Dysfunction in Schizophrenia. Bioessays. 2020;42:e1900202. doi: 10.1002/bies.201900202. [DOI] [PubMed] [Google Scholar]
- 45.Roberts RC. Mitochondrial dysfunction in schizophrenia: With a focus on postmortem studies. Mitochondrion. 2021;56:91–101. doi: 10.1016/j.mito.2020.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bergman O, Ben-Shachar D. Mitochondrial Oxidative Phosphorylation System (OXPHOS) Deficits in Schizophrenia: Possible Interactions with Cellular Processes. Can J Psychiatry. 2016;61:457–469. doi: 10.1177/0706743716648290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Willems PH, Valsecchi F, Distelmaier F, Verkaart S, Visch HJ, Smeitink JA, Koopman WJ. Mitochondrial Ca2+ homeostasis in human NADH:ubiquinone oxidoreductase deficiency. Cell Calcium. 2008;44:123–133. doi: 10.1016/j.ceca.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 48.Jimerson DC, Post RM, Carman JS, van Kammen DP, Wood JH, Goodwin FK, Bunney WE Jr. CSF calcium: clinical correlates in affective illness and schizophrenia. Biol Psychiatry. 1979;14:37–51. [PubMed] [Google Scholar]
- 49.Chen Y, Yan Q, Zhou P, Li S, Zhu F. HERV-W env regulates calcium influx via activating TRPC3 channel together with depressing DISC1 in human neuroblastoma cells. J Neurovirol. 2019;25:101–113. doi: 10.1007/s13365-018-0692-7. [DOI] [PubMed] [Google Scholar]
- 50.Burns DM, Richter JD. CPEB regulation of human cellular senescence, energy metabolism, and p53 mRNA translation. Genes Dev. 2008;22:3449–3460. doi: 10.1101/gad.1697808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thu VT, Kim HK, Long le T, Nyamaa B, Song IS, Thuy TT, Huy NQ, Marquez J, Kim SH, Kim N, Ko KS, Rhee BD, Han J. NecroX-5 protects mitochondrial oxidative phosphorylation capacity and preserves PGC1α expression levels during hypoxia/reoxygenation injury. Korean J Physiol Pharmacol. 2016;20:201–211. doi: 10.4196/kjpp.2016.20.2.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu J, Bian X, Liu Y, Hong L, Teng T, Sun Y, Xu Z. Adenosine A2 receptor activation ameliorates mitochondrial oxidative stress upon reperfusion through the posttranslational modification of NDUFV2 subunit of complex I in the heart. Free Radic Biol Med. 2017;106:208–218. doi: 10.1016/j.freeradbiomed.2017.02.036. [DOI] [PubMed] [Google Scholar]
- 53.E Silva LFS, Brito MD, Yuzawa JMC, Rosenstock TR. Mitochondrial Dysfunction and Changes in High-Energy Compounds in Different Cellular Models Associated to Hypoxia: Implication to Schizophrenia. Sci Rep. 2019;9:18049. doi: 10.1038/s41598-019-53605-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used and analyzed during the current study are available from the corresponding author upon request.