Abstract

According to WHO, cervical cancer is considered as one of the most frequently diagnosed cancers and the fourth main source of cancer death in women in 2020 worldwide. Hence, there is a need for development of cervical cancer screening with new rapid and cost-effective methods. Although there are few methods available for HPV identification, these techniques are less sensitive, time-consuming, and costly. An ultra-sensitive, selective, and label-free DNA-based impedimetric electrochemical genosensor is developed in this study to detect HPV-18 for cervical cancer. Electrochemical analysis was performed for the characterization of the sensing platform and for the detection of analyte. A single-stranded 25mer oligonucleotide DNA probe was immobilized onto a nitrogen-doped carbon nanodot-modified ITO electrode. Furthermore, the hybridization event was measured by testing the complementary single stranded DNA sequence in the samples. The sensor could distinguish between complementary as well as non-complementary sequences. Herein, impedance quantification demonstrated a limit of detection of 0.405 fM. The developed genosensor showed high selectivity toward HPV-18 in the clinical samples. This sensing platform can be considered as a rapid and selective method for the screening of HPV-18.
1. Introduction
Globally, among women between the age group of 20 to 39 years,1 cervical cancer in 2020 has estimated around 604,000 new and 342,000 death cases.2 Therefore, its diagnosis at an early stage is a worldwide significant health challenge.3 The vital human causative agent of cervical cancer is human papillomavirus (HPV). HPV is a non-enveloped double-stranded circular DNA genome and the most common sexually transmitted virus. It is protected by an icosahedral capsid, which forms a 55 nm molecule.4−7 Epidemiologic research has shown that 93 to 100% of invasive cervical cases are linked to HPV-16 and HPV-18.6 The oncogenic regions E6 and E7 present in HPV are highly infectious. These regions in HPV-16 or 18 strains promote malignant transformation and tumor growth.4 HPV-18 is the second most carcinogenic viral strain affecting the cervix after HPV-16. Out of all HPV-18 infections, 37% of symptoms belong to cervical adenocarcinomas (ADC) compared to 12% of squamous cell carcinomas (SCC). The vast majority of the HPV-18 are asymptomatic and have the potential to evade clearance by the immune system. Therefore, for the prevention of cancer at an early stage, methods for HPV-18 DNA detection have become a necessity.8
Approachable diagnostic techniques for early detection of cervical cancer are the Papanicolaou test (Pap Smear), dot blot, Hybrid Capture 2 (HC2) assays, and specific nucleic acid amplification methods. Moreover, these techniques have certain limitations such as poor specificity and inability to identify the specific oncogenes. In addition, they require more time in generating reliable and accurate data,9 limited to laboratory testing due to complex workflows, lack of skilled professionals, expensiveness,10 and limited clinical settings.11 Therefore, these techniques are not suitable for the countries with inadequate resources as well as personnel.12
Thus, DNA biosensors can be a better alternative to the traditional methods because of the demand for miniaturized, affordable,13 simple, specific, and rapid detection of pathogens.14,15 They serve as a point of care testing device with high selectivity, sensitivity, and real-time response.16 These diagnostic devices are fabricated to detect target DNA (TDNA) from a million-fold surplus of non-targeted species. DNA probes (PDNA) are immobilized onto the surface of the electrode to detect the hybridized event with complementary DNA.14,15 Electrochemical DNA biosensors offer high sensitivity with rapid detection through signal amplification17 and a low detection limit.18 To enhance the performance of the biosensor, the electrode surface is modified with distinct nanomaterials.16 These nanostructured platforms act as significant interfaces between biomolecules and the electrode surface, providing stable binding for the particular molecule without any biological activity loss. Numerous materials such as nanoparticles, nanowires, nanorods, nanotubes, and quantum dots have been widely used in biosensing.6,19
In this research work, we have reported a label-free impedimetric genosensor that is potentially suitable for the specific identification of HPV-18 DNA sequences. The electrodeposition of N-CDs was done onto the ITO glass electrode for immobilization of PDNA specific for HPV-18. Hybridization of the TDNA and PDNA was performed in the sample solution (10 mL of 0.1 M electrolyte solution (K3[Fe(CN)6])) and measured after the incubation time. Various parameters have been optimized, and electrochemical evaluation has been done. The present label-free genosensor shows high selectivity and wide linear range with a low limit of detection.
2. Results and Discussion
Herein, a novel HPV-18 genosensor with a probe sequence that matches with the sequence retrieved from the CDD (Conserved Domain Database)20 was designed. The probe sequence is selected from the conserved E6 region (105–581 genome seq) of the HPV-18 E6 region. This E6 sequence is cross-checked against the CD-search database.21 It was found that it matched well with other protein clusters found in Prk (PRotein K(c)lusters) database correlating to the non-specific hits of E6 proteins: PHA02779 (50–422 nt) and PHA02775 (40–422 nt) provisional under the cl27673: E6 Superfamily. The alignment showed a significant conserved region between 40 and 422 nt. The best way to detect HPV-18 causing cervical cancer via electrochemical detection is to trace a common HPV sequence conserved in most HPV strains. Specific regions (protein sequence) in a virus that remains unchanged after multiple generations of mutations and strain formations can help us to identify the particular virus family easily. These regions are called conserved sequences, which resulted due to the recurring gene sequence that are translating them.22 A novel way to make the diagnostic kit far more efficient is by detecting a large group of similar viruses (multi-strains) and making it effective against future mutants, as this concept is still followed in recently designed biosensors for SARS-COV-2.23
Furthermore, electrochemical studies were performed to evaluate the step-by-step fabrication of the sensing platform. The electrode fabrication process includes nanodot deposition on ITO for genosensing of HPV DNA. First, nanodots (N-CDs) are deposited onto the substrate surface, i.e., electrode, through the electrodeposition method (as mentioned in Section 2.6) to develop the nanodot-modified ITO electrode. Second, immobilization onto the modified electrode was performed. In this step, the physical method was performed for PDNA immobilization. Physical adsorption is the simplest method for immobilization and does not denature the immobilized enzyme. This adsorption method is based on the combined action of van der Waals force and hydrophobic, ionic, and hydrogen interactions, due to which the bioreceptor attaches to the surface of the sensor. This is a simple-to-use and cost-effective process.24
In the last step, immobilized PDNA was allowed to hybridize with a complementary TDNA present in the sample. This step is followed by the conversion of DNA hybridization events into a measurable current signal (changes in current with and without hybridization have been measured).
2.1. Characterization of N-CDs
To determine the size and morphology of carbon dots, transmission electron microscopy (TEM) was performed. The TEM image of N-CDs, as shown in Figure 1a, depicts that nanodots are spherical and dispersed uniformly (inset shows the zoom image of N-CDs). The average diameter was measured to be ∼15.47 nm.25
Figure 1.

(a) TEM of N-CDs; the inset shows the N-CDs of 20 nm. (b) FTIR spectroscopic analysis of N-CDs. (c) UV–visible spectra of N-CDs.
FT-IR (Fourier-transform infrared spectroscopy) analysis was performed for the nanomaterial to identify their chemical composition26 and surface functional groups present on N-CDs. The broad peak at 3331 cm–1 was due to the stretching vibrations of O–H and N–H. A band was assigned to C–H at 2928 cm–1. The peak at 1639 cm–1 was assigned to the C=O and C=C bond stretching vibrations, while the C–O–C characteristic stretching band was observed at 1381 cm–1. Moreover, the band at 1131 cm–1 was attributed to C–N and C–S bonds.27,28 The FT-IR analysis is shown in Figure 1b. At around 365 nm, a typical absorption band displays carbon dots (Figure 1c) synthesis by hydrothermal carbonization containing amide function and extending to the visible region.29−31 This wide range of 300–400 nm was due to the transition of electrons, and a peak at 365 nm could attribute to the appearance of aromatic π orbitals of carbon dots. Upon the illumination of 365 nm UV light, a blue-green color was generated immediately by the carbon dots solution (Figure 1c inset).32
2.2. Electrochemical Detection of the Sensing Surface
The design and fabrication of the HPV genosensor were shown schematically (Scheme 1). The N-CD-modified electrode is considered to be an excellent platform for further immobilization process. N-dopants may help to improve the positive charge density with the help of neighboring carbon atoms.33 N-CDs are positively charged enabling electrostatic interactions between PDNA and the carbon nanodots.34,35 Thus, enhancing the electrochemical signals significantly.
Scheme 1. The Present HPV-18 DNA Biosensor Design Based on the Immobilization of the Probe DNA and Hybridization of Target DNA onto the N-CD-Modified ITO Glass Electrode.

For the detection of DNA in label-free biosensing, EIS detection is considered to be the most optimistic technique. In this, the dielectric parameters of biological complexes are studied in broad frequency. The functional and fundamental information between the electrolyte and electrode at the interface is provided by the EIS technique.36 Also, the Nyquist diagram demonstrates two regions on the electrode surface during the reactions. The semicircle was obtained by high-frequency modulation, which represents transfer of electron processes, and low-frequency modulation, which represents the mass transport of the redox type. Charge transfer resistance (Rct) being the important parameter in EIS is used for sequence immobilization and hybridization.37Figure 2a presents the Nyquist plots at bare ITO, N-CDs/ITO, PDNA/N-CDs/ITO, and TDNA/PDNA/N-CDs/ITO; the noticeable semicircle of bare ITO (light green semi-circle), PDNA/N-CD-modified ITO (light blue semi-circle), is smaller than that of the bare ITO. After immobilization, the semicircle was increased (light violet semi-circle) in the modified PDNA/N-CDs/ITO and with the hybridization of target DNA (Light Orange semi-circle). The bare ITO showed a semicircle with Rct of about 32.54 Ω. N-CDs/ITO-modified electrode displayed a much lower resistance of 18.55 Ω, which indicated N-CDs as an excellent electric conducting material and accelerated the transfer of electrons. The Rct increased perceptibly to 45.65 Ω when PDNA was immobilized onto the modified electrode surface, formed an auxiliary barrier, and prevented the reaction onto the electrode surface. Further enhancement in Rct (60.42 Ω) is elevated after the hybridization of TDNA. This was because of the increase in the negative charge owing to the formation of double-stranded DNA.15
Figure 2.
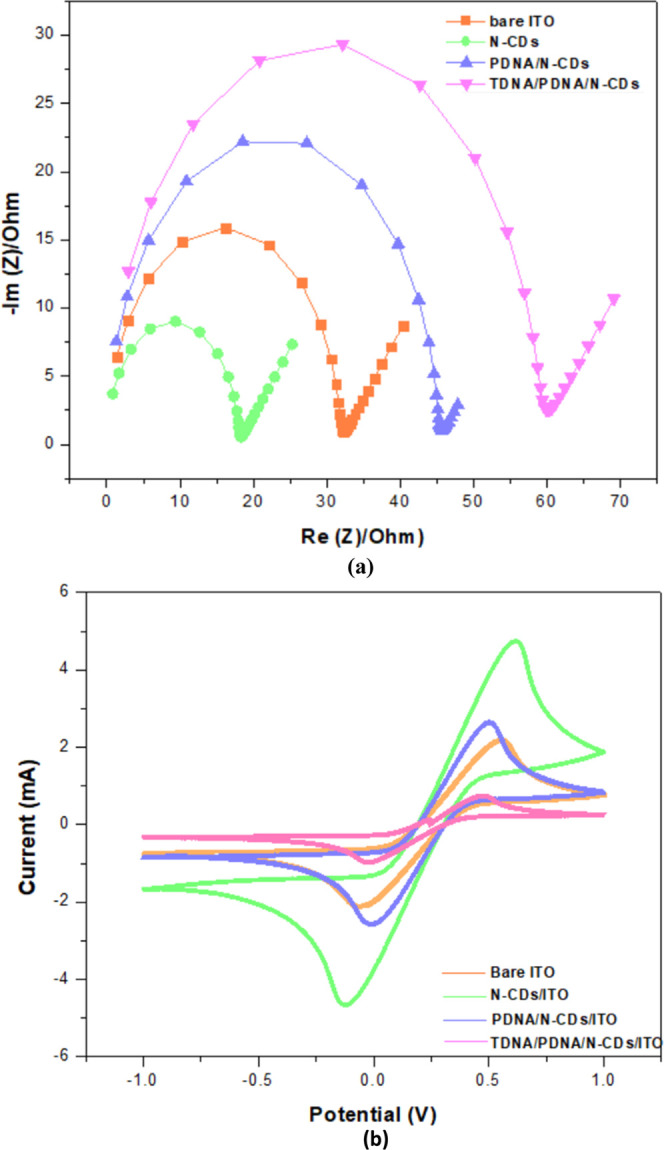
(a) Nyquist plots of bare ITO, N-CDs/ITO, immobilization of probe DNA onto N-CDs/ITO, and hybridization with complementary target DNA. (b) CV analysis of bare ITO, N-CDs/ITO, after the immobilization of probe DNA, and after hybridization electrode containing 0.1 M K3[Fe(CN)6] at 50 mV s–1.
As Figure 2b depicts, the current peak at Ipa = 2.25 mA and Ipc = −2.23 mA shows bare electrode with the smallest current when compared to the N-CD-modified electrode with Ipa = 4.79 mA and Ipc = −4.71 mA. This increase in current was because of the conductive nature of N-CDs, which eases the transfer of electrons. Furthermore, probe DNA was immobilized onto the modified ITO and a decrease in current signal was seen of Ipa = 2.71 mA and Ipc = −2.61 mA. The decreased sensing signal response illustrates the successful and effective immobilization onto the nanodot-modified working electrode. Furthermore, the current decreased to Ipa = 0.81 mA and Ipc = −1.02 mA, when target DNA was hybridized. The difference in the peak currents was based on response analysis of the HPV genosensor. Due to the hybridization and non-conductive layer formation of the probe and target DNA over the ITO, there was a notable decrease in the current.
The above results showed uniformity in the CV and EIS graph confirming a desirable electrode modification.
2.3. Optimization of Target DNA at Different Concentrations
The proposed electrochemical biosensor exhibited a dynamic detection range of 0.1 fM to 100 pM and a sensitivity of 0.266 Ω/fM. This can be accredited to the advance and implicit properties of N-CDs as well as the electrochemical biosensing-associated advantages. The limit of detection (LOD) and limit of quantification (LQD) for the fabricated biosensor were 0.405 and 1.228 fM, respectively. These were calculated using the 3.3σ (LOD) and the 10σ (LQD) method, where σ is the standard deviation. It can be observed from Figure 3a that the current response decreased upon changing the concentration of TDNA from 0.1 fM to 100 pM on the surface of TDNA/ PDNA/N-CDs/ITO. It is thus noticed that by increasing the concentration of TDNA, the hybridization with PDNA was increased. A calibration curve of the Rct values with various concentrations of TDNA was plotted as represented in Figure 3b. As shown in this Figure, a linear dynamic range exhibited with a correlation coefficient of 0.96.
Figure 3.

(a) Optimization of target DNA concentration with the help of Nyquist plots at the TDNA/PDNA/N-CDs/ITO glass electrode containing 0.1 M K3[Fe(CN)6] at 50 mV s–1. (b) dependence of log of Rct on log concentration (fM).
2.4. Evaluation Parameters of HPV Genosensor
The modified sensing platform was evaluated with the help of the EIS study. The Rct value was obtained at PDNA/N-CDs/ITO after target DNA hybridization was compared with hybridization of Mis-Match target-1 (MMT-1) and Mis-Match target-2 (MMT-2) sequences as illustrated in Figure 4. It shows the reduced response of Rct similar to PDNA as seen in the figure. Hence, no significant change in Rct response was noticed; therefore, the PDNA immobilized selectively to TDNA and demonstrated high selectivity of the present genosensor.
Figure 4.

Nyquist plots of the TDNA/PDNA/N-CDs/ITO electrode and hybridization with target complementary DNA along with response toward the non-complimentary DNAs (MMT-1 and MMT-2).
The designed sensing platform for long-term stability was tested after a week. It was observed that when stored at 4 °C, it keeps the original activity by 80% for 5 weeks.
2.5. Genosensor Performances at Optimum Scan Rate, pH, Temperature and Time
Figure 5a shows the voltammogram analysis of the TDNA/PDNA/N-CDs/ITO-modified electrode. Cyclic voltammogram (CV) analysis was recorded at different scan rates from 20 to 180 mVs–1. The figure illustrates the increase in peak current with the increase in the scan rate. With increasing scan rates, no peak shift was seen, thus confirming the stability of the sensor. Figure 5b depicts that the peak current quantitative increases with the increase in the square root function of the scan rate, demonstrating moderate electron-transfer kinetics on the CV time scale and dispersion-controlled conduct by plotting log current (I) versus log potential (V). All ensuring experiments were conducted at 50 mVs–1. The peak current dependency on scan rate was expressed as follows:
Figure 5.
(a) CV obtained at TDNA/PDNA/N-CDs/ITO for scan rates of 20–180 mV s–1 TDNA/PDNA/N-CDs/ITO. (b) Dependency of peak currents on the square root of potential sweep rates in a wide range of 20–180 mV s–1.
The pH of the solution being highly acidic or basic affects the solubility and stability of DNA. Various pH values ranging from 4.5 to 8.5 in 0.1 M solution of electrolyte was exposed to the TDNA/PDNA/N-CDs/ITO electrode, and their electrochemical response was recorded. From Figure 6a, it is apparent that the Rct values of pH from 4.5 to 7.0 decrease and then increases. Therefore, the sensing signal was optimum at pH 7.0. Furthermore, TDNA was incubated at different temperatures ranging from 10 to 45 °C, and 35 °C was considered as the optimum temperature for hybridization as shown in Figure 6b. The response time was optimized from 0 to 40 s and a 90% response time was obtained at 40 s as illustrated in Figure 6c.
Figure 6.
Bar representation of the CV data TDNA/PDNA/N-CDs/ITO for different (a) pH values (4.5–8.5), (b) temperatures (10–45 °C), and (c) times (0–40 s) containing 0.1 M K3[Fe(CN)6] at 50 mV s–1.
2.6. Performance of Genosensor in Real Sample
The performance of the present HPV-18 genosensor was investigated in the spiked biological samples (as mentioned in 4.8) within the calibrated concentration range. In Table 1, the relative standard deviation (RSD) was calculated from 95.38 to 99.65%, showing good accuracy and precision of the genosensor for HPV 18 detection in the cervical samples.
Table 1. HPV-18 Recoveries in the Biological Samples Using Nano-Based Genosensor.
| spiked samples (fM) | measured samples (fM) | recovery (%) |
|---|---|---|
| 40.9 | 41.04 | 99.65 |
| 32.3 | 33.22 | 97.23 |
| 20.25 | 21.15 | 95.74 |
| 14.63 | 15.39 | 95.38 |
The proposed sensor showed an improved linearity response with high sensitivity, specificity, and repeatability compared to previous works. In addition, the lower limit of detection was obtained. Thus, the TDNA/PDNA/N-CDs/ITO modified electrode was considered to be an efficient platform for the detection of HPV-18. It is due to the large surface area of the N-CDs that improved the immobilization of PDNA. Even though a lot of research on electrochemical detection of HPV-18 causing cervical cancer has been done, none of them has elaborated the use of a conserved HPV sequence. A detailed comparison of the electrochemical HPV-18 DNA-based biosensor is shown in Table 2.38−46
Table 2. Comparison of the Analytical Performance of the Proposed DNA Sensor Detection with Other Sensors for the Detection of HPV18a.
| sensor mechanism platform | types of biosensor | technique | LOD | linear range | sensitivity | stability | reference |
|---|---|---|---|---|---|---|---|
| gold interdigitated electrode 3-aminopropyl triethoxysilane (APTES) | voltammetric | (I–V) electrical characterization | NR | NR | NR | NR | (38) |
| carbon-based electrode chips | genomagnetic LAMP-based electrochemical | amperometry | 0.1 ng | 0.1 ng and 250 ng | ∼1 ng | 21 days | (39) |
| T–Hg(II) onglassy carbon electrode | electrochemical | SWV | several fM (1.2 × 10–5 nmol/L) | 1 × 10–15 up to 1 × 10–6 M | NR | 30 days | (40) |
| gold nanoparticle IDE | electrochemical | EIS | 1 pM | NR | NR | NR | (41) |
| CdTe quantum dots | optical (QD-sFRET) | fluorescence spectra of QD-FRET sandwich assay | 0.2 nM | 1.0 to 50.0 nM | NR | 3 months | (42) |
| Au/Ag core–shell nanoparticles on glass substrate | optical | photodiode | 50 nM | 0.05 to 0.5 pmol/μL | NR | NR | (43) |
| gold nanoparticle | optical | photodiode | 30 pM | 1.2 nM to 30 pM | NR | NR | (44) |
| gold working electrode | electrochemical | CV | 170 pM | 0.1–1 nM | 1.02 μA nM–1 | 1 month | (45) |
| screen-printed carbon electrode and an anthraquinone-labeled signaling probe | electrochemical | DPV | 153 pM | 0.5–100 nM | NR | NR | (46) |
| N-CDs/ITO | electrochemical | CV, EIS | 0.405 fM | 0.1 fM to 100 pM | 0.266 Ω/fM | 5 weeks | present |
NR* = not reported, SWV = square wave voltammetry, RSD = relative standard deviation, EIS = electrochemical impedance spectroscopy, FRET = fluorescence resonance energy transfer, DPV = differential pulse voltammetry.
3. Conclusions
In this study, an ultra-sensitive HPV label-free genosensor consisting of a conserved sequence of HPV-18 E6 serotype was developed with a label-free approach. HPV-18 is considered to be one of the most hazardous types of HPVs. This proposed sensor overcomes the limitations of the available conventional methods. The use of N-CDs further provides a better sensing platform due to their positive charge and enabling significant interactions between PDNA and carbon nanodots. This genosensor showed a detection limit of 0.405 fM, using a calibration curve that evaluated the concentration range of 0.1 fM to 100 pM. The biosensor exhibited good sensitivity and differentiated between the complementary and non-complementary sequences for HPV-18. Also, specific response and stability of the sensor were investigated and showed significant results. Furthermore, it can be exploited for the development of point-of-care lab-on-chip detection platforms by employing the N-CDs. Thus, the fabricated genosensor can be used as a successful tool for the easy, rapid, sensitive, and early detection of HPV-18 causing cervical cancer.
4. Methods
4.1. Materials and Reagents
The anhydrous citric acid (C6H8O7), uric acid (C5H4N4O3), potassium ferricyanide (K3[Fe(CN)6]), and potassium ferrocyanide (K4[Fe(CN)6]) were purchased from SRL, Mumbai, India. All reagents were of analytical grade. All aqueous solutions were prepared using double distilled water (resistivity = ∼18.2 MΩ cm). TE buffer (10 mM) was prepared by mixing the stock solutions of 10 mM Tris and 0.10 mM EDTA.
4.2. Apparatus
The synthesized nanodots were characterized using a UV–vis spectrophotometer (Shimadzu 2600), FT-IR (Nicolet iS5, Thermo Scientific), and TEM (FEI-Tecnai).
All electrochemical studies were performed with CV, EIS, and electrodeposition of the nanomaterial using Potentiostat (Biologics; SP-150 with EC-Lab software). The electrochemical-sensing platform was a three-electrode system approach consisting of auxiliary, reference, and working electrodes. Here, the ITO glass electrode was used as the working electrode, silver/silver chloride (Ag/AgCl) as the reference electrode, and platinum (Pt) as the auxiliary electrode.
4.3. DNA Sequences
Probe and target DNA were selected from the gene encoding sequences of human papillomavirus type 18, the complete genome from the NCBI server (GenBank: GQ180792.1). The E6 region sequence was selected having around 470 base pairs (105–581). Probe sequences were generated using the IDT PrimerQuest online Tool setting qPCR mode in the parameters.47,48 For confirmation, the probe sequence was matched with the BLASTN tool and BLAST results confirmed the alignment with E6 sequences. These generated sequences were purchased from Integrated DNA Technologies (IDT), India, in lyophilized form and are as follows:
4.4. Synthesis of Nanomaterial
For N-CD synthesis, the hydrothermal method was used. In brief, citric acid (2 mM) and urea (6 mM) were dispersed in double-distilled water. The mixture was stirred until a transparent homogenous solution was obtained. Furthermore, this solution was shifted to a Teflon-lined autoclave chamber and heated for 5 h at a temperature of 200 °C for further reactions. Then, the solution was cooled at room temperature (RT) and centrifuged to obtain the final product.49,50
4.5. Sensing Platform Fabrication
The ITO electrode was cleaned with 10 mL of acetone and distilled water kept in an ultrasonication bath. Later, it was wiped with a tissue. The size of the working electrode was about 4.8 cm2. N-CDs were electrodeposited on the working electrode using the CV technique with a potential range of −0.4 to 0.8 V for 30 cycles at a scan rate of 50 mV/s.
4.6. DNA Oligonucleotide Immobilization and Hybridization
The PDNA was immobilized onto the nanomaterial modified electrode by dropping 1.0 μL (1 μM concentration) probe DNA suspension. For immobilization, the electrode was incubated for 12 h at RT. The ITO electrode was then washed with double distilled water to remove the unbounded strands. The modified electrode was further used for electrochemical techniques. Next, hybridization was performed using various concentrations from 100 pM to 0.1 fM of TDNA onto the PDNA/N-CD-modified electrode using [Fe(CN)6]3–/4–. Furthermore, the electrode was optimized at different parameters (scan rates, pH, temperature, and time). In addition, to confirm the selectivity of the biosensor, a thorough evaluation was performed with two non-complementary sequences MMT-1 and MMT-2.
4.7. Electrochemical Analysis
All the experiments were performed at RT using [Fe(CN)6]3–/4– as a redox coupler of 0.1 M concentration. The TDNA modified electrode was denatured for 10 min at a temperature of 100 °C using water bath sonication for further optimization.
4.8. Procedure for Sensing HPV-18 in Biological Samples
To determine the clinical parameters, 10 endocervical samples were collected from Shri Krishna Hospital and Fertility Centre, Rajasthan. The DNA was extracted from the small piece of the dried specimens using a sterile new scalpel blade. It was then transferred to microcentrifuge tubes consisting of 50 μL distilled water and boiled in a microwave oven for 5 min. Polymerase chain reaction (PCR) and gel electrophoresis were then performed to obtain the amplified products.51 The samples were then spiked with known concentrations of the HPV-18 probe and injected into the electrolyte in the electrochemical cell for the measurement.
Acknowledgments
S.P. is grateful to Indian Council Medical Research (ICMR), New Delhi, India, for providing Senior Research Fellow (SRF) (project no. 45/55/2019/-NAN/BMS). The authors are also grateful to the Science & Engineering Research Board, DST, India (SERB file no. EMR/2016/007564) for providing financial supports to carry out this research work.
Author Contributions
S.P. did the conceptualization, experimentation, data curation, and writing. V.R. did the bioinformatics investigation. U.J. did the data curation and writing/editing. M.B. did the data curation and writing/editing. N.C. did the supervision, conceptualization, investigation, data curation, and writing/editing.
The authors declare no competing financial interest.
References
- Tomita L. Y.; Horta B. L.; da Silva L. L. S.; Malta M. B.; Franco E. L.; Cardoso M. A. Fruits and vegetables and cervical cancer, a systematic review and meta-analysis. Nutr. Cancer 2021, 73, 62–74. 10.1080/01635581.2020.1737151. [DOI] [PubMed] [Google Scholar]
- Sung H.; Ferlay J.; Siegel R. L.; Laversanne M.; Soerjomataram I.; Jemal A.; Bray F. Global cancer statistics 2020, GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca-Cancer J. Clin. 2021, 71, 209–249. 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- Cohen P. A.; Jhingran A.; Oaknin A.; Denny L. Cervical cancer. Lancet. 2019, 393, 169–182. 10.1016/S0140-6736(18)32470-X. [DOI] [PubMed] [Google Scholar]
- Peng X.; Zhang Y.; Lu D.; Guo Y.; Guo S. Ultrathin Ti3C2 nanosheets based “off-on” fluorescent nanoprobe for rapid and sensitive detection of HPV infection. Sens. Actuators, B 2019, 286, 222–229. 10.1016/j.snb.2019.01.158. [DOI] [Google Scholar]
- Farzin L.; Sadjadi S.; Shamsipur M.; Sheibani S. Electrochemical genosensor based on carbon nanotube/amine-ionic liquid functionalized reduced graphene oxide nanoplatform for detection of human papillomavirus (HPV16)-related head and neck cancer. J. Pharm. Biomed. Anal. 2020, 179, 112989. 10.1016/j.jpba.2019.112989. [DOI] [PubMed] [Google Scholar]
- Avelino K. Y. P. S.; Oliveira L. S.; Lucena-Silva N.; de Melo C. P.; Andrade C. A. S.; Oliveira M. D. L. Metal-polymer hybrid nanomaterial for impedimetric detection of human papillomavirus in cervical specimens. J. Pharm. Biomed. Anal. 2020, 113249. 10.1016/j.jpba.2020.113249. [DOI] [PubMed] [Google Scholar]
- Parmin N. A.; Hashim U.; Gopinath S. C. B.; Nadzirah S.; Rejali Z.; Afzan A.; Uda M. N. A. Human Papillomavirus E6 biosensing, Current progression on early detection strategies for cervical Cancer. Int. J. Biol. Macromol. 2019, 126, 877–890. 10.1016/j.ijbiomac.2018.12.235. [DOI] [PubMed] [Google Scholar]
- Parmin N. A.; Hashim U.; Gopinath S. C. B.; Rejali Z.; Afzan A.; Uda M. N. A Sensitive DNA Biosensor using Screen Printed Gold Electrode Interdigitated Electrode (IDE) Pattern based for Identification of Human Papillomavirus Type 18 Variants. J. Futuristic Biosci.and Biomed. Eng. 2019, 1, 1–8. [Google Scholar]
- Ramesh T.; Foo K. L.; Haarindrasad R.; Sam A. J.; Solayappan M. Gold-Hybridized Zinc oxide nanorods as Real-time Low-cost nanoBiosensors for detection of virulent DNA signature of HPV-16 in cervical carcinoma. Sci. Rep. 2019, 9, 1–17. 10.1038/s41598-019-53476-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z.; Lyu W.; Yu M.; Wang Q.; Qu H.; Ismagilov R. F.; Han X.; Lai D.; Shen F. Self-partitioning Slip Chip for slip-induced droplet formation and human papillomavirus viral load quantification with digital LAMP. Biosens. Bioelectron. 2020, 155, 112107. 10.1016/j.bios.2020.112107. [DOI] [PubMed] [Google Scholar]
- Mukama O.; Yuan T.; He Z.; Li Z.; de Dieu Habimana J.; Hussain M.; Li W.; Yi Z.; Liang Q.; Zeng L. A high fidelity CRISPR/Cas12a based lateral flow biosensor for the detection of HPV16 and HPV18. Sens. Actuators, B 2020, 316, 128119. 10.1016/j.snb.2020.128119. [DOI] [Google Scholar]
- Jampasa S.; Wonsawat W.; Rodthongkum N.; Siangproh W.; Yanatatsaneejit P.; Vilaivan T.; Chailapakul O. Electrochemical detection of human papillomavirus DNA type 16 using a pyrrolidinyl peptide nucleic acid probe immobilized on screen-printed carbon electrodes. Biosens. Bioelectron. 2014, 54, 428–434. 10.1016/j.bios.2013.11.023. [DOI] [PubMed] [Google Scholar]
- Campos-Ferreira D. S.; Nascimento G. A.; Souza E. V. M.; Souto-Maior M. A.; Arruda M. S.; Zanforlin D. M. L.; Ekert M. H. F. E.; Bruneska D.; Lima-Filho J. L. Electrochemical DNA biosensor for human papillomavirus 16 detection in real samples. Anal. Chim. Acta 2013, 804, 258–263. 10.1016/j.aca.2013.10.038. [DOI] [PubMed] [Google Scholar]
- Mahmoodi P.; Rezayi M.; Rasouli E.; Avan A.; Gholami M.; Mobarhan M. G.; Karimi E.; Alias Y. Early-stage cervical cancer diagnosis based on an ultra-sensitive electrochemical DNA nanobiosensor for HPV-18 detection in real samples. J. Nanobiotechnol. 2020, 18, 1–12. 10.1186/s12951-020-0577-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H.; Bai W.; Dong C.; Guo R.; Liu Z. An ultrasensitive electrochemical DNA biosensor based on graphene/Au nanorod/polythionine for human papillomavirus DNA detection. Biosens. Bioelectron. 2015, 68, 442–446. 10.1016/j.bios.2015.01.039. [DOI] [PubMed] [Google Scholar]
- Kowalczyk A. Trends and perspectives in DNA biosensors as diagnostic devices. Curr. Opin. Electrochem. 2020, 23, 36–41. 10.1016/j.coelec.2020.03.003. [DOI] [Google Scholar]
- Rasouli E.; Shahnavaz Z.; Basirun W. J.; Rezayi M.; Avan A.; Ghayour-Mobarhan M.; Khandanlou R.; Johan M. R. Advancements in electrochemical DNA sensor for detection of human papilloma virus-A review. Anal. Biochem. 2018, 556, 136–144. 10.1016/j.ab.2018.07.002. [DOI] [PubMed] [Google Scholar]
- Bartosik M.; Jirakova L. Electrochemical analysis of nucleic acids as potential cancer biomarkers. Curr. Opin. Electrochem. 2019, 14, 96–103. 10.1016/j.coelec.2019.01.002. [DOI] [Google Scholar]
- Lv Q.; Wang Y.; Su C.; Lakshmipriya T.; Gopinath S. C. B.; Pandian K.; Perumal V.; Liu Y. Human papilloma virus DNA-biomarker analysis for cervical cancer, signal enhancement by gold nanoparticle-coupled tetravalent streptavidin-biotin strategy. Int. J. Biol. Macromol. 2019, 134, 354–360. 10.1016/j.ijbiomac.2019.05.044. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A.; et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, 222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A.; et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2015, 43, D225–D229. 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulop L.; Barrett A. D. T.; Phillpotts R.; Martin K.; Leslie D.; Titball R. W. Rapid identification of flaviviruses based on conserved NS5 gene sequences. J. Virol. Methods 1993, 44, 179–188. 10.1016/0166-0934(93)90053-T. [DOI] [PubMed] [Google Scholar]
- Fani M.; Zandi M.; Soltani S.; Abbasi S. Future developments in biosensors for field-ready SARS-CoV-2 virus diagnostics. Biotechnol. Appl. Biochem. 2021, 68, 695. 10.1002/bab.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inshyna N. M; Chorna I. V; Primova L. O; Hrebenyk L. I; Khyzhnia Y.V.. Biosensors, Design; Classification and Application. 2020.
- Duan J.; Yu J.; Feng S.; Su L. A rapid microwave synthesis of nitrogen–sulfur co-doped carbon nanodots as highly sensitive and selective fluorescence probes for ascorbic acid. Talanta 2016, 153, 332–339. 10.1016/j.talanta.2016.03.035. [DOI] [PubMed] [Google Scholar]
- Zahoor A.; Christy M.; Hwang Y. J.; Lim Y. R.; Kim P.; Nahm K. S. Improved electrocatalytic activity of carbon materials by nitrogen doping. Appl. Catal., B 2014, 147, 633–641. 10.1016/j.apcatb.2013.09.043. [DOI] [Google Scholar]
- Xu Y.; Li D.; Liu M.; Niu F.; Liu J.; Wang E. Enhanced-quantum yield sulfur/nitrogen co-doped fluorescent carbon nanodots produced from biomass Enteromorpha prolifera, synthesis; posttreatment; applications and mechanism study. Sci. Rep. 2017, 7, 4499. 10.1038/s41598-017-04754-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C.; Zhang Y.; Wang P.; Yang Y.; Wang Y.; Xu J.; Wang Y.; William W. Y. Synthesis of nitrogen and sulfur co-doped carbon dots from garlic for selective detection of Fe3+. Nanoscale Res. Lett. 2016, 11, 1–9. 10.1186/s11671-016-1326-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H.; Zhou F.; Gu J.; Shu C.; Xi K.; Jia X. Nitrogen-doped carbon dots as a new substrate for sensitive glucose determination. Sensors 2016, 16, 630. 10.3390/s16050630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.; Xu M.; Liu Y.; He F.; Gao F.; Su Y.; Wei H.; Zhang Y. Nitrogen-doped; carbon-rich; highly photoluminescent carbon dots from ammonium citrate. Nanoscale 2014, 6, 1890–1895. 10.1039/C3NR05380F. [DOI] [PubMed] [Google Scholar]
- Wang H.; Sun P.; Cong S.; Wu J.; Gao L.; Wang Y.; Dai X.; Yi Q.; Zou G. Nitrogen-doped carbon dots for “green” quantum dot solar cells. Nanoscale Res. Lett. 2016, 11, 27. 10.1186/s11671-016-1231-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.; Zhou H. S. Green synthesis of luminescent nitrogen-doped carbon dots from milk and its imaging application. Anal. Chem. 2014, 86, 8902–8905. 10.1021/ac502646x. [DOI] [PubMed] [Google Scholar]
- Miao Y.-E; Liu T.. Electrospun nanofiber electrodes, A promising platform for supercapacitor applications. InElectrospinning, Nanofabrication and Applications; 2019, (pp. 641–669).
- Li F.; Cai Q.; Hao X.; Zhao C.; Huang Z.; Zheng Y.; Lin X.; Weng S. Insight into the DNA adsorption on nitrogen-doped positive carbon dots. RSC Adv. 2019, 9, 12462–12469. 10.1039/C9RA00881K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.; Song Z.; Gu J.; Li S.; Wu Y.; Han H. Nitrogen-doped carbon quantum dots for preventing biofilm formation and eradicating drug-resistant bacteria infection. ACS Biomater. Sci. Eng. 2019, 5, 4739–4749. 10.1021/acsbiomaterials.9b00583. [DOI] [PubMed] [Google Scholar]
- Shariati M.; Ghorbani M.; Sasanpour P.; Karimizefreh A. An ultrasensitive label free human papilloma virus DNA biosensor using gold nanotubes based on nanoporous polycarbonate in electrical alignment. Anal. Chim. Acta 2019, 1048, 31–41. 10.1016/j.aca.2018.09.062. [DOI] [PubMed] [Google Scholar]
- Faria H. A. M.; Zucolotto V. Label-free electrochemical DNA biosensor for zika virus identification. Biosens. Bioelectron. 2019, 131, 149–155. 10.1016/j.bios.2019.02.018. [DOI] [PubMed] [Google Scholar]
- Akhir M. M; Parmin N. A; Hashim U; Gopinath S. C. B.; Rejali Z; Afzan A; Uda M. N. A.; Uda M. N. A.; Hong V.C.. Voltammetric DNA Biosensor for Human Papillomavirus (HPV) Strain 18 Detection. In IOP Conference Series, Materials Science and Engineering ;2020. (Vol. 864, p. 012166).
- Bartosik M.; Jirakova L.; Anton M.; Vojtesek B.; Hrstka R. Genomagnetic LAMP-based electrochemical test for determination of high-risk HPV16 and HPV18 in clinical samples. Anal. Chim. Acta 2018, 1042, 37–43. 10.1016/j.aca.2018.08.020. [DOI] [PubMed] [Google Scholar]
- Kowalczyk A.; Nowicka A. M. Application of mercury–mediated thymine–base pairs for successful voltammetric detection of HPV 18. Sens. Actuators, B 2016, 237, 810–816. 10.1016/j.snb.2016.07.004. [DOI] [Google Scholar]
- Kim N. S. K. S.; Parmin N. A; Hashim U; Gopinath S. C. B.; Rejali Z; Afzan A; Uda M. N. A.; Uda M. N. A.; Hong V.C.. Electrochemical DNA Biosensor based on 30 nM Gold Nanoparticle Modified Electrode by Electro Less Deposition for Human Papillomavirus (HPV) 18 E6 Region. InIOP Conference Series, Materials Science and Engineering; 2020, (Vol. 864, p. 012167).
- Shamsipur M.; Nasirian V.; Mansouri K.; Barati A.; Veisi-Raygani A.; Kashanian S. A highly sensitive quantum dots-DNA nanobiosensor based on fluorescence resonance energy transfer for rapid detection of nanomolar amounts of human papillomavirus 18. J. Pharm. Biomed. Anal. 2017, 136, 140–147. 10.1016/j.jpba.2017.01.002. [DOI] [PubMed] [Google Scholar]
- Li X. Z.; Kim S.; Cho W.; Lee S. Y. Optical detection of nanoparticle-enhanced human papillomavirus genotyping microarrays. Biomed. Opt. Express 2013, 4, 187–192. 10.1364/BOE.4.000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek T. J.; Park P. Y.; Han K. N.; Kwon H. T.; Seong G. H. Development of a photodiode array biochip using a bipolar semiconductor and its application to detection of human papilloma virus. Anal. Bioanal. Chem. 2008, 390, 1373–1378. 10.1007/s00216-007-1814-x. [DOI] [PubMed] [Google Scholar]
- Civit L.; Fragoso A.; Hölters S.; Dürst M.; O’Sullivan C. K. Electrochemical genosensor array for the simultaneous detection of multiple high-risk human papillomavirus sequences in clinical samples. Anal. Chim. Acta 2012, 715, 93–98. 10.1016/j.aca.2011.12.009. [DOI] [PubMed] [Google Scholar]
- Jampasa S.; Siangproh W.; Laocharoensuk R.; Yanatatsaneejit P.; Vilaivan T.; Chailapakul O. A new DNA sensor design for the simultaneous detection of HPV type 16 and 18 DNA. Sens. Actuators, B 2018, 265, 514–521. 10.1016/j.snb.2018.03.045. [DOI] [Google Scholar]
- Salihah N. T.; Hossain M. M.; Ahmed M. U.. A fast and sensitive real-time PCR assay to detect Legionella pneumophila with the ZENTM double-quenched probe. Scientia Bruneiana. 2018, 17, 10.46537/scibru.v17i1.69. [DOI] [Google Scholar]
- Agarwal M.; Tomar R. S.; Jyoti A. Designing and In-silico validation of duplex molecular beacon probes for quantification of Salmonellae. J. Chem. Pharm. Sci. 2017, 10, 1101–1104. [Google Scholar]
- Wang L.; Jana J.; Chung J. S.; Hur S. H. Glutathione modified N-doped carbon dots for sensitive and selective dopamine detection. Dyes Pigm. 2021, 186, 109028. 10.1016/j.dyepig.2020.109028. [DOI] [Google Scholar]
- Zhu W.; Zhou Y.; Tao M.; Yan X.; Liu Y.; Zhou X. An electrochemical and fluorescence dual-signal assay based on Fe3O4@ MnO2 and N-doped carbon dots for determination of hydrogen peroxide. Microchim. Acta 2020, 187, 1–10. 10.1007/s00604-020-4163-5. [DOI] [PubMed] [Google Scholar]
- Kailash U.; Hedau S.; Gopalkrishna V.; Katiyar S.; Das B. C. A simple ‘paper smear’ method for dry collection, transport and storage of cervical cytological specimens for rapid screening of HPV infection by PCR. J. Med. Microbiol. 2002, 51, 606–683. 10.1099/0022-1317-51-7-606. [DOI] [PubMed] [Google Scholar]




