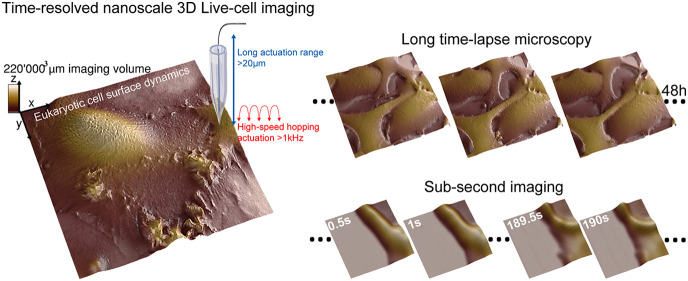
Abstract
Nanocharacterization plays a vital role in understanding the complex nanoscale organization of cells and organelles. Understanding cellular function requires high-resolution information about how the cellular structures evolve over time. A number of techniques exist to resolve static nanoscale structure of cells in great detail (super-resolution optical microscopy, EM, AFM). However, time-resolved imaging techniques tend to either have a lower resolution, are limited to small areas, or cause damage to the cells, thereby preventing long-term time-lapse studies. Scanning probe microscopy methods such as atomic force microscopy (AFM) combine high-resolution imaging with the ability to image living cells in physiological conditions. The mechanical contact between the tip and the sample, however, deforms the cell surface, disturbs the native state, and prohibits long-term time-lapse imaging. Here, we develop a scanning ion conductance microscope (SICM) for high-speed and long-term nanoscale imaging of eukaryotic cells. By utilizing advances in nanopositioning, nanopore fabrication, microelectronics, and controls engineering, we developed a microscopy method that can resolve spatiotemporally diverse three-dimensional (3D) processes on the cell membrane at sub-5-nm axial resolution. We tracked dynamic changes in live cell morphology with nanometer details and temporal ranges of subsecond to days, imaging diverse processes ranging from endocytosis, micropinocytosis, and mitosis to bacterial infection and cell differentiation in cancer cells. This technique enables a detailed look at membrane events and may offer insights into cell–cell interactions for infection, immunology, and cancer research.
Keywords: long-term time-lapse microscopy, scanning ion conductance microscopy (SICM), live-cell 3D imaging, subsecond imaging, nanoscale cell surface dynamics, cancer cells, bacterial infection
Visualizing dynamic structural changes in live eukaryotic cells at the nanoscale is essential to understand the mechanisms by which cellular components fulfill their function in key cellular processes. The time frame of those processes ranges from seconds/minutes for events such as endocytosis to hours/days for cell differentiation. To study long-term biological mechanisms, light-based time-lapse microscopy (TLM) methods have been implemented, capturing a sequence of images at regular intervals.1,2 Among the plethora of TLM techniques on eukaryotic cells, phase-contrast, differential interference contrast, and fluorescence microscopy are the most relevant. Visualizing morphological changes in the cell membrane, however, is challenging as they involve dynamic nanostructures with dimensions below the diffraction limit of light.3,4 Super-resolution microscopy methods have become invaluable in studying live cells.5 Yet, due to the axial resolution, photobleaching, and phototoxic effects, it remains challenging to resolve the membrane surface of a living cell using light-based TLM techniques. In particular, determining the exact boundary of the cell membrane is often impossible at the nanometer scale.6 Atomic force microscopy offers excellent axial resolution and can be used to image live cells. However, the forces applied by the cantilever tip deform the cell, thereby revealing the cytoskeleton rather than the native shape of the cell membrane.7 In addition, the tip–sample forces deform soft structures such as microvilli, making them difficult to resolve.8 Maintaining long imaging times with AFM on mammalian cells is technologically very difficult due to the complexity and time variance of the tip–sample force interaction. This drastically decreases the viability of cells, in particular those of mammalian origin. High-speed AFM imaging is limited to relatively small imaging ranges, and the need for small AFM cantilevers limits the height of the samples that can be imaged. The ultimate method for single-cell biology would combine the high-resolution 3D information on scanning probe microscopy, with the imaging range, the long-time time-lapse capability, and the high temporal resolution of optical microscopy.
Scanning ion conductance microscopy (SICM)9 has been shown to be particularly well-suited for imaging soft samples.8,10,11 By measuring the distance-dependent current through a nanopipette, SICM yields high-resolution images of fragile membrane features, without touching the cell. Together with the high axial resolution from scanning probes, the label-free characteristic of SICM is a crucial benefit over other optical methods such as super-resolution microscopies. Possibly the most notable evolution in SICM was the development of backstep/hopping mode by Happel12 and the seminal work by Novak and Korchev,11 making SICM an ideal starting point for long time-lapse imaging of mammalian cells. However, hopping mode SICM is traditionally a rather slow technique with acquisition times in the order of tens of minutes per image, which prevents the study of fast dynamic events.
Here, we develop a high-performance SICM system for high-speed and long time-lapse imaging by implementing recent advances in nanopositioning,13 nanopore fabrication,14 microelectronics,15 and controls engineering.16 In order to demonstrate the capabilities of our system, we selected a diverse set of three-dimensional biological processes as test cases.
Results and Discussion
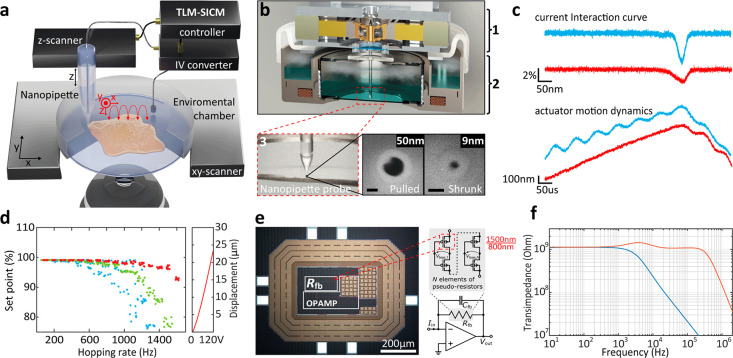
The imaging speed in SICM is largely determined by the hopping rate11 and pipet velocity at which the pipet approaches the surface and retracts when the drop in ion current reaches a certain set point threshold (Figure 1a in red). Usually, a set point below 2% of the baseline current is used to avoid pipet/sample crashing. The hopping rate is limited by the speed at which the piezo scanner can retract the pipet without leading to excessive overshoot. Once the current drop reaches the set point, the SICM controller tries to retract the pipet as fast as possible. This motion creates an inertia that excites the resonance frequency of the piezo actuator. The larger the pipet velocity is at the time the current reaches the set point, the stronger this parasitic excitation of the scanner resonances is. The resonances are also excited stronger the closer the hopping rate is to the resonance frequency of the piezo actuator. To overcome the second issue, previous efforts have been made in designing SICM scanners with high resonance frequencies, which unfortunately goes at the expense of the Z-range.17,18 Imaging of eukaryotic cells, however, requires piezo actuators with a long range (>10–20 μm). This traditionally leads to a trade-off between resonance frequency and the range of the actuator. In addition to using a custom high-bandwidth, large-range actuator (Figure 1b–d and Figures S1 and S2), we overcame this trade-off in our instrument one way by adaptively slowing down the pipet velocity already before it reaches the trigger set point.19 We used an adaptive gain applied to the piezo motion as a function of the current interaction curve (Figure S2a,b). In addition, the motion dynamics of the actuator are reshaped by a data-driven controller,16 implemented in a 16th-order discrete-time filter that compensates for the resonances of the electromechanical actuators (Figure S2c–e and Supporting Information Note 1). The adaptive hopping mode and the data-driven controller increased the achievable hopping rate by a factor of 5 (Figure 1c,d).
Figure 1.
Time-resolved SICM principle and implementation. (a) Illustration of the SICM principle. (b) Schematic rendering of a cross-section of the time-resolved SICM system based on a high-bandwidth large-range SICM actuator (1), integrated into a miniature incubator (2). Nanopipettes are fabricated through laser pulling (50 nm radius) and are successively shrunk through scanning electron microscopy (SEM) radiation to a sub-10-nm pore radius, shown in SEM images (3). Scale bars, 50 nm. (c) SICM interaction curve (top) and actuator motion dynamics (bottom), with the conventional hopping mode in blue and with the time-resolved SICM implementation (adaptive hopping mode and data-driven controller) in red. One kilohertz hopping rate, 1 μm hopping height, and 98% set point. (d) Actuator hopping rate performance with the conventional hopping mode (blue), adaptive hopping mode (green), and adaptive hopping mode together with data-driven controller (red). One micrometer hopping height and 99% set point. (e) Die micrograph of custom TIA composed of a low-noise operational amplifier and a pseudoresistor in its feedback. The pseudoresistor consists of N series-connected pMOS transistor pairs with specific biasing to achieve a large and precise resistance value. (f) Transimpedance measurements of the 1st stage TIA (blue) and the overall transimpedance using the subsequent amplifier stage for bandwidth extension (red).
Another essential aspect for a fast and accurate feedback response is the bandwidth of the low noise current to voltage conversion. Rosenstein et al. demonstrated that an increased performance in nanopore sensing platforms is obtainable by integrating custom tailored complementary metal oxide semiconductor (CMOS) amplifiers.20 We developed a custom, bandwidth-extended transimpedance amplifier (BE-TIA), which was manufactured in a 180 nm CMOS silicon-on-insulator technology for decreased parasitic well capacitances and leakage currents. It has a subsequent discrete amplifier stage to further increase the overall bandwidth.15 The TIA’s feedback resistor is set to a very large value of 1 GΩ to achieve low noise and is composed of a multielement pseudoresistor (Figure 1e), i.e., a large number of series-connected small-sized p-channel MOS transistors of W/L = 1500/800 nm with inherent linearization and a specific biasing circuit that facilitates a precise and tunable high-value resistance (Figure 1e, Figure S4, and Supporting Information Note 2). The subsequent amplifier increases the overall bandwidth from 10 to 600 kHz (Figure 1f). The BE-TIA multiplied the achievable hopping rate in our time-resolved SICM by a factor of 8 (Figure S5).
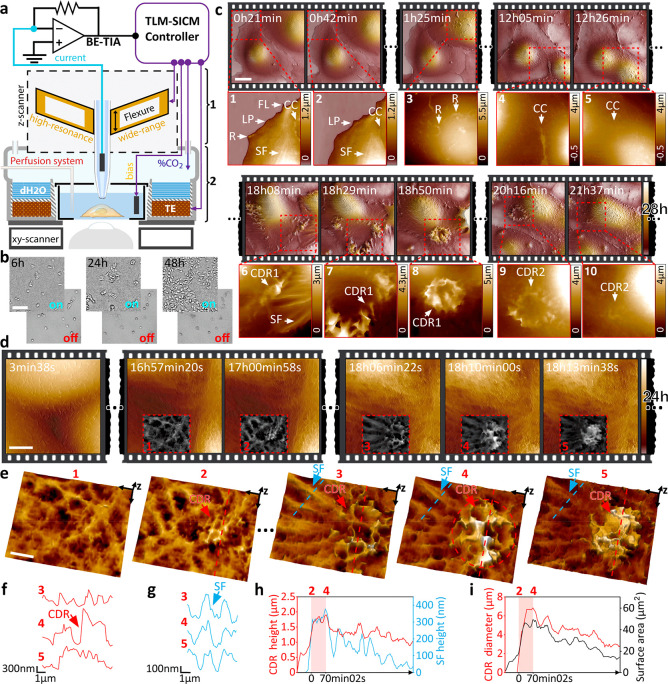
Maintaining cell viability during long-term time-lapse imaging requires accurate control of temperature, humidity, and CO2 levels in the sample area, without adversely affecting the SICM performance. We integrated our time-resolved SICM with a custom miniature incubator (Figure 2a and Figure S6), which ensured cell viability for well over 48 h (Figure 2b). Figure 2c and Movie S1 show a 28 h continuous time-lapse imaging sequence of kidney cells revealing highly dynamic protrusions on the apical cell surface (Figure 2c) such as lamellipodia (“LP” in Figure 2c-1,2) and ruffles (“R” in Figure 2c-1,3), which play a major role in cell motility (Figure S7). Furthermore, we observed filopodia, which are finger-like protrusions that cells use for probing the environment (“FL” in Figure 2c-1 and Figure S7). We also identified stress fibers that have an essential role in cell adhesion, migration, and mechanotransduction (“SF” in Figure 2c-1,3). Additionally, we observed cell-to-cell contacts that play a fundamental role in cell-communication and the development of multicellular organisms (“CC” in Figure 2c-1,2,4,5 and Figure S7). Figure 2d-4,5 and Figure S8 reveal how the interfaces of two cells fuse in syncytium. Of particular interest is the appearance and disappearance of circular dorsal ruffles (“CDR1” and “CDR2” in Figure 2c-5–1, and Figures S9 and S10). Being able to observe the dynamics of these diverse structures within one long experiment lets us correlate the seemingly independent structures and investigate possible interdependencies.
Figure 2.
Time-resolved allows long-term 3D visualization of the eukaryotic cell membrane and the tracking of dynamic structures at nanometer resolution. (a) High-performance pipet z-actuator (1) is integrated into a miniature incubator (2). (b) Demonstration of cell viability and proliferation over 48 h in our time-resolved SICM system, with controlled atmosphere ON vs OFF. Scale bar, 50 μm.(c) Twenty-eight hour time-lapse scanning of live kidney cells (COS-7) in DMEM-Hi glucose medium. Scale bar, 20 μm. Several membrane structures can be visualized: ruffles (R), lamellipodia (LP), filopodia (FL), cell–cell contacts (CC), stress fibers (SF), and CDR. (d) Twenty-four hour time-lapse scanning of live kidney cells tracking the formation of a CDR structure. Scale bar, 5 μm. (e) 3D view of the CDR with the initiation spot in panels 1 and 2 and maximum expansion in panels 3–5. Scale bar, 2 μm. (f) Height profile of the CDR ring (red arrow in f). (g) Height profile of a nearby SF protruding the cell surface (blue arrow in g). (h) Plot of CDR and SF max-height, showing colocalization during the expansion period (2–4); (i) Plot of CDR ring diameter and surface area; over 130 frames.
Circular dorsal ruffles (CDR) are elusive dynamic structures, likely related to cell migration, macropinocytosis, and internalization of membrane receptors.21 These important roles are receiving increasing attention in the scientific community due to their involvement in cancer progression and the facilitation of the pathogen infection.22 While most studies report that CDRs appear in cells stimulated with mitogenic and/or motogenic factors, our long time-lapse measurements on kidney cells show that these transient structures can appear in optimal culture conditions without external stimulation (Figure 2c-6–10 and Figure 2d). With time-resolved SICM, we tracked a CDR that appeared at 17 h 00 min of scanning with the maximum expansion of the wave reached at 18 h 10 min (Figure 2d,e and Movie S2). Recently, Bernitt et al. modeled the appearance of CDRs and associated them with actin polymerization23 using confocal microscopy. Our time-resolved SICM data corroborate their findings, adding three-dimensional details of the process at sub-100-nm lateral and sub-10-nm axial resolution. Figure 2e clearly shows the proximity of the ring-shaped actin mediated wave (CDR) (Figure 2f) to stress fibers (SF) (Figure 2g). The polymerization of the stress fibers extrudes the cell membrane and correlates with the formation of the CDR ring (Figure 2h,i). After expansion, these fibers gradually fade from the surface topography while CDR decreases in height and diameter, suggesting their interdependence.
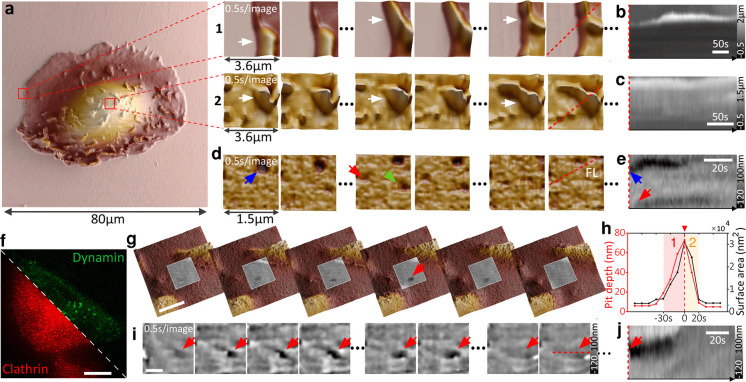
Many of the processes on the level of whole cells occur at a time scale of minutes or hours and can therefore be tracked easily with time-lapse SICM. Subcellular events such as endocytosis or infection, however, often occur much faster. Increasing the imaging speed of SICM has thus far been at the expense of imaging volume to only 7000 μm3.15 This makes studying natural processes on live cells impractical. Our technique combines the ability to address large imaging volumes up to 220 000 μm3 at moderately fast speeds with high-speed SICM imaging of small details on live cells (Figure 3a, Figures S11–S13, and Movie S3). The small-scale, high-speed data can thus be put in the context of the overall cell morphology and growth patterns. Figure 3a shows a large area 80 × 80 μm2 overview scan of a kidney cell. Subsequent zooming-in increased the temporal resolution to frame periods of 0.5 s/image at hopping rates of 2.5 kHz. We tracked protrusions at the cell periphery and on top of the cell membrane (Figure 3a–c and Movie S4). Parts d and e of Figure 2 show endocytic pit formation on live kidney cells (Movie S4). We also observed endocytosis on modified melanoma cells (SKMEL) that coexpress clathrin-RFP and dynamin-GFP (Figure 3f). Figure 3g depicts larger area scans (3 μm field of view) with a higher resolution (100 × 100 pixels, Movie S5) on the cell membrane to identify endocytic pits and to extract dynamic key parameters such as pit depth and pit area and plot them as a function of time (Figure 3h). Another endocytic event was imaged at 0.5 s/image, but a smaller size scale and lower number of pixels are shown in Figure 3i,j. The large dynamic range of the measurements in terms of scan size (from 500 × 500 nm2 to 100 × 100 μm2), imaging speed (from 0.5 s/image to 20 min/image), number of pixels per image (from 1 Kp to 1 Mp), and a depth of view (22 μm with axial resolution below 10 nm) greatly enhances the type of biological questions that can be studied with time-resolved SICM.
Figure 3.
Time-resolved SICM allows for a large dynamic scan range essential for long-term monitoring of cells and high-speed performance to track transient biological events at the nanoscale. (a) Large area scanning of a single kidney cell (80 μm: 512 × 512 pixels). Fast image acquisition at 0.5 s/image (2.5 kHz hopping rate) on the cell periphery (1) and on top of the cell (2). Arrows point to dynamic ruffles. (b and c) Kymogram showing the dynamics of ruffles over time (red dashed lines in a at 1 and 2). (d) Fast image acquisition of 0.5 s/image of the kidney cell membrane with arrows pointing to several endocytic events; with the (e) respective kymogram. (f) Fluorescence image of a transformed melanoma cell that coexpresses clathrin-RFP and dynamin-GFP. Scale bar, 20 μm. (g) Fast image acquisition at 10 s/image (1 kHz hopping rate) detecting the formation of an endocytic pit (red arrow) in a large area. Scale bar, 1 μm. (h) Plot of endocytic pit depth and surface area: 1, growing; and 2, closing. (i) Fast image acquisition at 0.5 s/image of an endocytic pit (red arrow) within 50 s and 100 data points with a 15 nm radius pipet, with the (j) respective kymogram. Scale bar, 500 nm.
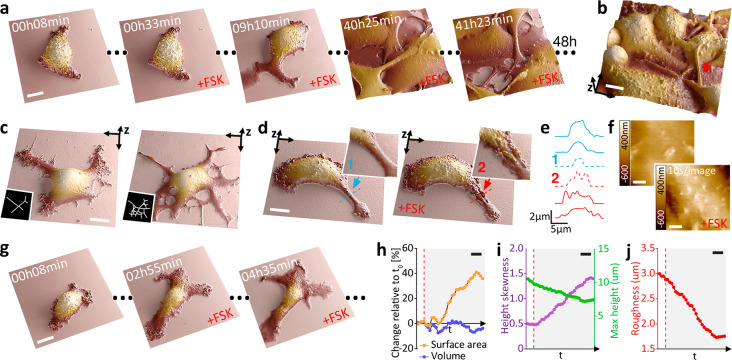
Time-resolved SICM is particularly well-suited for biological questions that relate either to a change in 3D cell morphology or where membrane trafficking plays a major role. Morphological changes of transformed cells, for example, are essential in cancer diagnosis and treatment decision-making24 but are difficult to track over long durations. Knowledge about associated events on cell membranes and their dynamics and roles in tumor evolution is limited by a lack of tools for long-term imaging. In cultured melanoma cells (B16–F1), important morphological changes linked to cell differentiation and motility are induced by 3′-5′-cyclic adenosine monophosphate (cAMP) signaling, which is implicated in mediating resistance to BRAF inhibitor therapy25 and can be activated in vitro by treatment with the adenylate cyclase (ADCY) agonist forskolin (FSK)26 (Figure S14). To evaluate the spatiotemporal resolution of time-resolved SICM to reveal cAMP-induced changes, we monitored melanoma cell morphology during 48 h of sustained FSK treatment27 (Figure 4a and Movie S6). Within this time frame, FSK treatment drastically increased the outgrowth and branching of interwoven cell dendrites that were frequently suspended >5 μm above the substrate (Figure 4b,c).
Figure 4.
Time-resolved SICM enables a large dynamic scan range of cells and suspended structures, allowing for long-term visualization of differentiation and morphological changes in melanoma cells. (a) Forty-eight hour time-lapse scanning of melanoma cell (B16–F1) differentiation during prolonged treatment with 20 μM forskolin (FSK). Scale bar, 10 μm. (b) Long actuation range allows for the nanoscale visualization of dendrites (branched cytoplasmic protrusions), suspended 5 μm above the substrate (red arrow). Scale bar, 10 μm. (c) FSK-induced dendrite outgrowth. Scale bar, 10 μm. (d) Tracking a single dendrite before (blue arrow) and after adding FSK (red arrow). Scale bar, 10 μm. (e) Height profile of the dendrite over a time sequence. Profile 1 and 2 locations are shown in panel d. (f) Visualization of the real-time effect of FSK on the cell membrane with fast image acquisition at 10 s/image (1 kHz hopping rate). Scale bar, 1 μm. (g) Visualization of long-term morphological changes associated with FSK-induced melanoma cells differentiation. Scale bar, 20 μm. (h) Surface area and volume percentage change relative to the first frame. (i) Maximal height and height skewness. (j) Membrane roughness. Scale bar, 40 min. The dashed line in red represents the addition of FSK.
The high spatial resolution afforded by time-resolved SICM imaging revealed that FSK treatment rapidly increased the width of dendrites (Figure 4d,e and Figures S15 and S16) and revealed the presence of membrane protrusions already during the earliest time points examined (Figure 4f, Figure S17, and Movie S7). Prolonged FSK treatment resulted in a gradual increase in cell surface area with no significant change in cell volume (Figure 4g,h, Figure S18a,b, and Movie S8). Curiously, these changes were accompanied by a rise in cell height skewness (Figure S18c), decreased cell height (Figure 4i and Figure S18d), and a long-term decrease in membrane roughness (Figure 4j and Figure S18e). The roughness is associated with the presence of membrane protrusions, a feature related to focal adhesions that are mediated by rearrangement in actin cytoskeleton28 and increased invasiveness in cancer.29
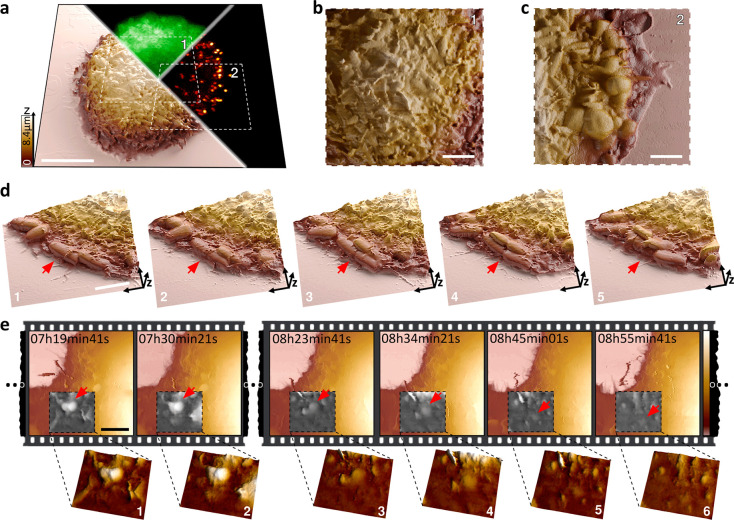
We also applied time-resolved SICM to investigate the mechanisms of bacterial adhesion of mammalian host cells, a critical step toward infection by many pathogens. We monitored the binding of E. coli to epithelial cells through the display of synthetic adhesins (on bacteria) that mimic a pathogenic context. These bacterial cells display anti-GFP nanobodies (VHH) and bind to GFP anchored to the plasma membrane of HeLa cells. Interestingly, we observed an increase in the GFP signal around bacteria, suggesting a local accumulation of the VHH–GFP bonds that represents an increase in the overall affinity of the bacterium to the host cell membrane over time (Figure 5a–c and Figure S19). In addition, time-resolved SICM revealed bacteria dividing directly on the host cell membrane, proving that the attachment to the mammalian cell does not prevent bacterial proliferation (Figure 5d, Figure S20, and Movie S9). Occasionally, the binding of the bacterium tightened to the point it triggered its internalization by the mammalian cell (Figure 5e and Figure S21), as confirmed also by confocal microscopy (Figure S22).
Figure 5.
Long-term visualization of infection mechanisms in bacterial adhesion and entry into host cells using time-resolved SICM. (a) Correlated three-dimensional representation of the live cell membrane surface with SICM. HeLa cells displaying a synthetic GFP receptor enabling its direct visualization (GFP, green channel) sequestered by its ligand; VHH expressed by E. coli (red channel). Scale bar, 10 μm. (b) E. coli can be resolved on top of the host cell body (area 1 marked in a) and (c) on the host cell edge (area 2 marked in a). Scale bar, 4 μm. (d) 3D visualization of bacteria infecting a mammalian cell. The red arrow points to a bacterium adhered to the cell, elongating (1 and 2) and dividing (3–5) on the mammalian cell membrane. Scale bar, 4 μm. (e) Live monitoring of bacteria engulfment and internalization; the red arrow points to a bacterium adhered to the host (1 and 2) and being internalized (3–6). Scale bar, 4 μm.
Complementary to fluorescence microscopy, time-resolved SICM reveals the precise topography and position of the bacteria relative to the membrane, as well as the pathogen–membrane interactions. SICM is label-free and does not suffer from phototoxicity and photobleaching that could prevent the long-term visualization of these processes with fluorescence microscopy. In contrast to AFM, SICM is a truly noncontact measurement that prevents detachment of the settled bacteria or artificially perturbs their weak interactions with the host cell body that govern infection. SICM imaging did not prevent adhesion and invasion and was able to monitor the infection process over long imaging times. Due to the noncontact nature and minimized perturbations, we will be able to investigate a broad range of infection mechanisms and internalization processes in the future.
Conclusions and Prospects
We have demonstrated in this work some of the benefits of time-resolved SICM for studying nanoscale processes in cell biology. However, some technological hurdles still need to be overcome for time-resolved SICM to become a routine tool in cell biology. One key requirement for good quality SICM imaging is a high-quality nanopipette with an appropriate aperture diameter for the specific application. While high-speed imaging generally benefits from a lower resistance pipet (i.e., a larger opening), high-resolution requires a small opening.18 The resolution in SICM is determined by the pipet pore dimensions,30 with a lateral resolution of approximately three times the pore radius.31 In terms of axial resolution, pipettes with pore dimensions of ∼15 nm radius have been used to resolve features ∼5 nm height.32 Obtaining reproducible and robust capillaries at small and precise diameters is still a challenge for many groups starting to use SICM. For SICM to reach its full potential, a reliable off-the-shelf source of SICM probes would be beneficial (similar to what is available for AFM cantilevers).
While time-resolved SICM can obtain a high-resolution characterization of the cell shape and surface characteristics, these aspects often need to be correlated with biochemical information and changes to the internal organization of the cells. The combination of SICM with high-performance optical microscopy is a very promising area.33 The SICM is integrated with an inverted optical microscope (Figure S23) and can thus be used with many of the recently developed super-resolution microscopy techniques.34
The SICM field has developed rapidly in recent years, with many additional measurement capabilities being added, such as sample stiffness,35 surface charge,32 and local pH.36 Combining these developments with time-resolved SICM or high-speed SICM will provide insights into eukaryotic membrane processes with three-dimensional nanometer detail.
Methods
Cell Lines and Cell Culture
In this study, we used monkey kidney fibroblast-like cells COS-7, SKMEL human melanoma cells, B16–F1 mouse melanoma cells (ATCC), and HeLa cervical cancer cells. SKMEL cells (modified to coexpress dynamin-GFP and clathrin-RFP)37 and COS-7, were cultured in DMEM high glucose without phenol red medium (Gibco, Thermo Fisher Scientific), containing 10% of fetal bovine serum (Gibco, Thermo Fisher Scientific), 1% penicillin–streptomycin (Gibco, Thermo Fisher Scientific), and 4 mM l-glutamine (Gibco, Thermo Fisher Scientific). B16–F1 mouse melanoma cells were maintained in DMEM (Sigma) supplemented with 10% fetal bovine serum, 50 μg/mL gentamicin (Gibco, Thermo Fisher Scientific), and 1% GlutaMAX (Gibco, Thermo Fisher Scientific). Cells were regularly tested negative for Mycoplasma (SouthernBiotech 13100-01). HeLa cells were engineered to express a doxycycline-inducible GFP fused to a mouse CD80 transmembrane domain using standard second-generation lentivector production protocols and the plasmids pMD2G (Addgene 12259), pCMVR8.74 (Addgene 22036), and pXP34038 [eGFP(N105Y, E185 V, Y206F)_mCD80_C-terminal into pRRLSIN.cPPT.GFP.WPRE (Addgene 12252)]. Before coculture experiments, HeLa cells were seeded at 50 000 cells/ml on 35 mm glass dishes (Ibidi) in 400 μL of fluorobrite supplemented with 10% FBS and 1% Glutamax (Life Technologies). After 4–6 h of attachment, the medium was renewed and supplemented with 500 ng/mL doxycycline (HiMedia) for the induction of GFP display and with 3 μL of Deglycosylation Mix II (New England Biolab) to digest the glycocalyx and ease the access of bacteria to the cell membrane. E. coli K12 (BW25113), a flagellated strain (deltaFliCDST) with stable expression mScarlet, were retransformed with pDSG339 for tetracycline-inducible VHH anti-GFP display.39 Stationary precultures of bacteria were diluted 1:3000 and induced overnight with 250 ng/mL tetracycline (Sigma) in LB.
Time-Resolved SICM Instrumentation
The time-resolved SICM setup consists of a custom-built pipet Z-actuator integrated into a controlled-atmosphere device, critical for cell viability during imaging (Figure S6e). The temperature is regulated with a temperature controller from THORLABS (TC200), and the percentage and flow rate of CO2 gas infusion are regulated with a gas mixer from Life Imaging Services (The Brick). While keeping optimal cell culture conditions of 37 °C and 5% CO2 for several days, the high-performance Z-actuator configuration achieves a wide mechanical displacement amplification of 22 μm scanning range on the cell surface and preserved high-speed SICM performance with the first resonance at above 13 kHz. The Z-actuator is driven by a custom-made low-phase lag piezo controller13 and integrated with a stepper-motor stage for sample approach. The XY-scanner used was a Piezo-Nanopositioning stage with a 100 μm x–y travel range on the sample (P-733 Piezo NanoPositioner, Physik Instrumente), driven by a low-voltage piezo amplifier (E-500 Piezo Controller System, Physik Instrumente). At higher scan speeds, the dynamics of the XY scanner can distort the image. We rounded the sawtooth drive signal to avoid excessive resonances. For even higher line rates, applying more advanced controls techniques (like the data driven controller we use in the Z-direction) would be beneficial. The XY-scanner is assembled in a custom-built micro translation stage, mounted atop an inverted Olympus IX71 microscope body. Therefore, the system offers correlative SICM capabilities with fluorescence microscopy. For fluorescence excitation, we used a four-color (405, 488, 561, and 647 nm) pigtailed Monolithic Laser Combiner (400B, Agilent Technologies), controlled by a custom-written LabVIEW software. Images were acquired with a sCMOS camera (Photometrics, Prime 95B) and Micromanager software. The nanopipettes used as a probe in SICM were made of borosilicate (Sutter Instrument) and quartz (Hilgenberg GmbH). They were fabricated with a CO2 laser puller (Model P-2000, Sutter Instruments) with a radius from 60 to 20 nm sizes. Afterward, quartz capillaries can be shrunk to a sub-10-nm radius using electron beam irradiation.14,40 The pipet current was amplified by a transimpedance amplifier NF-SA-605F2 (100MΩ gain, NF corporation) and a 1GΩ custom-made TIA (Supporting Information Note 2) for high-speed current to voltage conversion15 (Figure 3a,g). A low-pass filter setting between 10 and 100 kHz was implemented after the preamplifier (Stanford SR560). A custom SICM controller (Supporting Information Note 1) consisting of a data-driven scan engine16,41 and adaptive hopping mode was implemented in LabVIEW on a NI USB-7856R OEM R Series (National Instruments, Austin, TX) in the framework of a home-built extensible high-performance scanning probe controller. We have the policy to make our instrument developments available as open hardware projects.42 The designs and assembly procedures can be requested directly from the corresponding author.
Time-Resolved SICM Measurements
The time-resolved SICM measurements were performed in the optimal culture conditions mentioned above at 37 °C and 5% CO2; COS-7 and SKMEL cells were seeded at 30 000 cells/mL in 35 mm glass dishes (Ibidi) for the long time-lapse experiments and 50 000 cells/mL for the high-speed measurements. For transfected SKMEL cells, the surface was coated with poly-d-lysine (Gibco, Thermo Fisher Scientific). Coverslips were washed with PBS before seeding the cells in 2 mL of DMEM medium. Prior to the experiments, the cells were grown overnight (12–16 h) at 37 °C and 5% CO2. In experiments using B16–F1 mouse melanoma cells, 50 000 cells/mL were seeded in 35 mm glass dishes (Ibidi) in a 2 mL volume of completed culture medium. After overnight culture (13–16 h), cells were used for scanning. Forskolin (10-2073 Focus Biomolecules) was added to the cells through a perfusion system integrated with the environmental mini-chamber to a final concentration in the cell medium of 20 μM FSK. In the cell infection coculture measurements, HeLa cell supernatant was renewed to 2 mL of medium and 200 μL of stationary bacterial culture was added for 30 min (Figure 5a–c) and 15 min (Figure 5d) before coculture. The samples were intensively washed five times with PBS. Two milliliters of DMEM high glucose without phenol red medium (Gibco, Thermo Fisher Scientific) containing 10% fetal bovine serum (Gibco, Thermo Fisher Scientific) was added prior to imaging. In the long time-lapse experiments (Figure 5e), stationary bacteria (100 μL) were added to the medium through a perfusion system integrated into the environmental mini-chamber after 1 h of scanning. A DC voltage bias in a range of +150 to +200 mV was applied to the pipet. The current set point used in the hopping actuation was 99% of the normalized current recorded (98% for high-speed imaging). The SICM image sequence in Figure 2c was acquired at a 200 Hz hopping rate and a 6 μm hopping height at 512 × 256 pixels. Image sequence in Figure 2d was acquired at a 200 Hz hopping rate and a 3 μm hopping height at 256 × 128 pixels. Image sequences in Figure 3a,d,e were acquired at a 2.5 kHz hopping rate and a 200 nm hopping height at 40 × 25 pixels. The image sequence in Figure 3g was acquired at a 1 kHz hopping rate and a 500 nm hopping height at 100 × 100 pixels. Image sequences in Figure 4a,b,g were acquired at a 125 Hz hopping rate and a 6 μm hopping height at 300 × 200 pixels. Figure 4c was acquired at a 200 Hz hopping rate and a 5 μm hopping height at 256 × 256 pixels. The image sequence in Figure 5 was acquired at a 125 Hz hopping rate and a 5 μm hopping height at 512 × 512 pixels (Figure 5a–c) and 256 × 256 pixels (Figure 5d,e).
Data Processing and Analysis
SICM images were further processed using Gwyddion.43 Images were corrected for scanline mismatch with a median of difference row alignments, and line artifacts characteristic of scanning prope microscopy were corrected. Afterward, a 2-pixel conservative denoise filter was applied. Images were exported with uniform pixels aspect ratios in 16 bit Portable Network Graphics format. For better visibility, the topography channel is merged with the slope channel to highlight the surface structures in all the data sets shown in panels with a video frame. In SICM images with a grayscale color map, the background was flattened with a subtraction of a median level. To generate the three-dimensional shape of cells, we used the advanced open-source 3D rendering tool Blender 3D for data visualization. Normalized topographical SICM data was imported as a height map and scaled in the axial direction. The corresponding SICM colormap was used as a color, projected on the topography. Videos were generated in ffmpeg format in Fiji.44
Acknowledgments
The authors would like to thank Navid Asmari and Santiago Andany for assistance. The authors would also like to thank Mustafa Kangül, Matthias Neuenschwander, Mélanie Hannebelle, Benjamin Rothe, Pierpaolo Ginefra, and Patrick Frederix for their valuable discussion. The authors would like to thank Prof. Aurélien Roux, Prof. Hilal Lashuel, Anne-Laure Mellier, and Veronika Cencen for providing biological samples. The authors would like to thank ATPR (EPFL) for mechanical fabrication.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsnano.1c05202.
Time-resolved SICM imaging over 28 hours time-lapse scanning of live kidney cells in optimal culture conditions; 80 μm area (MP4)
Time-resolved SICM imaging over 24 hours time-lapse scanning of live kidney cells in optimal culture conditions; 20 μm area (MP4)
Figures of 3D renderings, finite element simulation, mechanical displacement range, actuator frequency response curve, wiring diagram, working principle of the time-resolved SICM controller, schematics of the time-resolved SICM setup implementation, schematic of the implemented multi-element pseudo-resistor, custom monolithic bandwidth-extended transimpedance amplifier for highspeed current-to-voltage conversion, brightfield microscopy, time-resolved SICM setup, topography images, effect of FSK on melanoma cells, time-resolved SICM imaging sequence, topography profiles, changes in surface area, volume change, max height change, height skewness change, and roughness change, fluorescence images, and three-dimensional representation and discussions of time-resolved SICM controller implementation and MEPR implementation (PDF)
Time-resolved SICM with high dynamic range of XYZ-actuation and high-speed performance capabilities on live kidney cells (MP4)
Time-resolved SICM imaging at 0.5 s/image time-lapse scanning of live kidney cells (MP4)
Time-resolved SICM tracking biologic processes on the cell membrane-endocytic pits, on human melanoma cells (MP4)
Time-resolved SICM imaging over 48 hours time-lapse scanning of melanoma cell differentiation during prolonged treatment with forskolin (FSK) (MP4)
Time-resolved SICM tracking FSK-induced dendrite outgrowth in melanoma cells (MP4)
Time-resolved SICM tracking morphological of melanoma cell differentiation during prolonged treatment with forskolin (FSK) (MP4)
Time-resolved SICM tracking bacteria infecting a host mammalian cell (MP4)
Author Contributions
S.L. conceived the idea, designed and built the instrument, designed experiments, prepared samples, performed measurements, analyzed data, created the figures, and wrote the final manuscript. B.D. conceived the idea, designed and built the instrument, designed experiments and performed measurements, and analyzed data. K.P. designed experiments, prepared samples, analyzed data, and together with D.B.C. gave valuable input on the application in melanoma. X.P. designed experiments, prepared samples and, together with A.P., gave valuable input on the application in infection mechanisms of bacteria. A.P.N. and C.B. developed controller hardware and software. D.D. and J.A. designed and built the TIA chip for high-speed current to voltage conversion. V.N. processed data and together with A.R. gave valuable input on fluorescence correlative data. G.E.F. conceived the idea, designed the instrument, supervised the project, and wrote the final manuscript. All authors edited the manuscript.
This research was funded by multiple funding sources. G.E.F. gratefully acknowledges the support of the Swiss Commission for Technology and Innovation under the grant CTI-18330.1 and the European Research Council under grant number ERC-2017-CoG; InCell. D.C. and K.P. gratefully acknowledge the support of the swiss national science foundation under grant number 31003A_179330. A.P. gratefully acknowledges the Gebert Rüf Foundation under project number GRS-057/16. V.N. and A.R. gratefully acknowledge the support of the Max Planck-EPFL Center for Molecular Nanoscience and Technology and Swiss National Science Foundation through the National Centre of Competence in Research Bio-Inspired Materials. D.D. and J.A. gratefully acknowledge support from the Zeiss foundation and the DFG under contract no. AN 984/12.
A previous version of this manuscript was submitted to the preprint server bioRxiv. Samuel M. Leitao, Barney Drake, Katarina Pinjusic, Xavier Pierrat, Vytautas Navikas, Adrian P. Nievergelt, Charlène Brillard, Denis Djekic, Aleksandra Radenovic, Alexandre Persat, Daniel B. Constam, Jens Anders and Georg E. Fantner. Time-Resolved Scanning Ion Conductance Microscopy for Three-Dimensional Tracking of Nanoscale Cell Surface Dynamics. BioRxiv. 2021.10.1101/2021.05.13.444009 (accessed May 15, 2021).
The authors declare no competing financial interest.
Supplementary Material
References
- Baker M. Taking a Long, Hard Look. Nature 2010, 466 (7310), 1137–1140. 10.1038/4661137a. [DOI] [PubMed] [Google Scholar]
- Noctor S. C.; Martínez-Cerdeño V.; Ivic L.; Kriegstein A. R. Cortical Neurons Arise in Symmetric and Asymmetric Division Zones and Migrate through Specific Phases. Nat. Neurosci. 2004, 7 (2), 136–144. 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- Sauvanet C.; Wayt J.; Pelaseyed T.; Bretscher A. Structure, Regulation, and Functional Diversity of Microvilli on the Apical Domain of Epithelial Cells. Annu. Rev. Cell Dev. Biol. 2015, 31, 593–621. 10.1146/annurev-cellbio-100814-125234. [DOI] [PubMed] [Google Scholar]
- Innocenti M. New Insights into the Formation and the Function of Lamellipodia and Ruffles in Mesenchymal Cell Migration. Cell Adhes. Migr. 2018, 12 (5), 401–416. 10.1080/19336918.2018.1448352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S. A.; Shim S. H.; He J.; Zhuang X. Fast, Three-Dimensional Super-Resolution Imaging of Live Cells. Nat. Methods 2011, 8 (6), 499–505. 10.1038/nmeth.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T.; Wang Y.; Wang H.; Zhu Z.; Xiao Z. Visualizing of the Cellular Uptake and Intracellular Trafficking of Exosomes by Live-Cell Microscopy. J. Cell. Biochem. 2010, 111 (2), 488–496. 10.1002/jcb.22733. [DOI] [PubMed] [Google Scholar]
- Eghiaian F.; Rigato A.; Scheuring S. Structural, Mechanical, and Dynamical Variability of the Actin Cortex in Living Cells. Biophys. J. 2015, 108 (6), 1330–1340. 10.1016/j.bpj.2015.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert J.; Rheinlaender J.; Novak P.; Korchev Y. E.; Schäffer T. E. Comparison of Atomic Force Microscopy and Scanning Ion Conductance Microscopy for Live Cell Imaging. Langmuir 2015, 31 (24), 6807–6813. 10.1021/acs.langmuir.5b01124. [DOI] [PubMed] [Google Scholar]
- Hansma P. K.; Drake B.; Marti O.; Gould S. A. C.; Prater C. B. The Scanning Ion-Conductance Microscope. Science (Washington, DC, U. S.) 1989, 243 (4891), 641–643. 10.1126/science.2464851. [DOI] [PubMed] [Google Scholar]
- Korchev Y. E.; Bashford C. L.; Milovanovic M.; Vodyanoy I.; Lab M. J. Scanning Ion Conductance Microscopy of Living Cells. Biophys. J. 1997, 73 (2), 653–658. 10.1016/S0006-3495(97)78100-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak P.; Li C.; Shevchuk A. I.; Stepanyan R.; Caldwell M.; Hughes S.; Smart T. G.; Gorelik J.; Ostanin V. P.; Lab M. J.; Moss G. W. J.; Frolenkov G. I.; Klenerman D.; Korchev Y. E. Nanoscale Live-Cell Imaging Using Hopping Probe Ion Conductance Microscopy. Nat. Methods 2009, 6 (4), 279–281. 10.1038/nmeth.1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Happel P.; Dietzel I. D. Backstep Scanning Ion Conductance Microscopy as a Tool for Long Term Investigation of Single Living Cells. J. Nanobiotechnol. 2009, 7 (1), 7. 10.1186/1477-3155-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nievergelt A. P.; Andany S. H.; Adams J. D.; Hannebelle M. T.; Fantner G. E. Components for High-Speed Atomic Force Microscopy Optimized for Low Phase-Lag. IEEE/ASME Int. Conf. Adv. Intell. Mechatronics, AIM 2017, 731–736. 10.1109/AIM.2017.8014104. [DOI] [Google Scholar]
- Steinbock L. J.; Steinbock J. F.; Radenovic A. Controllable Shrinking and Shaping of Glass Nanocapillaries under Electron Irradiation. Nano Lett. 2013, 13 (4), 1717–1723. 10.1021/nl400304y. [DOI] [PubMed] [Google Scholar]
- Djekic D.; Fantner G.; Lips K.; Ortmanns M.; Anders J. A 0.1% THD, 1-MΩ to 1-GΩ Tunable, Temperature-Compensated Transimpedance Amplifier Using a Multi-Element Pseudo-Resistor. IEEE J. Solid-State Circuits 2018, 53 (7), 1913–1923. 10.1109/JSSC.2018.2820701. [DOI] [Google Scholar]
- Kammer C.; Nievergelt A. P.; Fantner G. E.; Karimi A. Data-Driven Controller Design for Atomic-Force Microscopy. IFAC-PapersOnLine 2017, 50 (1), 10437–10442. 10.1016/j.ifacol.2017.08.1972. [DOI] [Google Scholar]
- Simeonov S.; Schäffer T. E. High-Speed Scanning Ion Conductance Microscopy for Sub-Second Topography Imaging of Live Cells. Nanoscale 2019, 11 (17), 8579–8587. 10.1039/C8NR10162K. [DOI] [PubMed] [Google Scholar]
- Watanabe S.; Kitazawa S.; Sun L.; Kodera N.; Ando T. Development of High-Speed Ion Conductance Microscopy. Rev. Sci. Instrum. 2019, 90 (12), 123704. 10.1063/1.5118360. [DOI] [PubMed] [Google Scholar]
- Jung G.-E.; Noh H.; Shin Y. K.; Kahng S.-J.; Baik K. Y.; Kim H.-B.; Cho N.-J.; Cho S.-J. Closed-Loop ARS Mode for Scanning Ion Conductance Microscopy with Improved Speed and Stability for Live Cell Imaging Applications Closed-Loop ARS Mode for Scanning Ion Conductance Microscopy with Improved Speed and Stability for Live Cell Imaging Applications. Nanoscale 2015, 7 (25), 10989–10997. 10.1039/C5NR01577D. [DOI] [PubMed] [Google Scholar]
- Rosenstein J. K.; Wanunu M.; Merchant C. A.; Drndic M.; Shepard K. L. Integrated Nanopore Sensing Platform with Sub-Microsecond Temporal Resolution. Nat. Methods 2012, 9 (5), 487–492. 10.1038/nmeth.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivia A.; Goicoechea S. M.; Awadia S.; Zinn A.; Garcia-Mata R. Regulation of Circular Dorsal Ruffles, Macropinocytosis, and Cell Migration by RhoG and Its Exchange Factor, Trio. Mol. Biol. Cell 2017, 28 (13), 1768–1781. 10.1091/mbc.e16-06-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoon J.-L.; Wong W.-K.; Koh C.-G. Functions and Regulation of Circular Dorsal Ruffles. Mol. Cell. Biol. 2012, 32 (21), 4246–4257. 10.1128/MCB.00551-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernitt E.; Döbereiner H. G.; Gov N. S.; Yochelis A. Fronts and Waves of Actin Polymerization in a Bistability-Based Mechanism of Circular Dorsal Ruffles. Nat. Commun. 2017, 8 (1), 1–8. 10.1038/ncomms15863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irshad H.; Veillard A.; Roux L.; Racoceanu D. Methods for Nuclei Detection, Segmentation, and Classification in Digital Histopathology: A Review-Current Status and Future Potential. IEEE Rev. Biomed. Eng. 2014, 7, 97–114. 10.1109/RBME.2013.2295804. [DOI] [PubMed] [Google Scholar]
- Johannessen C. M.; Johnson L. A.; Piccioni F.; Townes A.; Frederick D. T.; Donahue M. K.; Narayan R.; Flaherty K. T.; Wargo J. A.; Root D. E.; Garraway L. A. A Melanocyte Lineage Program Confers Resistance to MAP Kinase Pathway Inhibition. Nature 2013, 504 (7478), 138–142. 10.1038/nature12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englaro W.; Bertolotto C.; Buscà R.; Brunet A.; Pagès G.; Ortonne J. P.; Ballotti R. Inhibition of the Mitogen-Activated Protein Kinase Pathway Triggers B16 Melanoma Cell Differentiation. J. Biol. Chem. 1998, 273 (16), 9966–9970. 10.1074/jbc.273.16.9966. [DOI] [PubMed] [Google Scholar]
- Cho D. H.; Bae C. D.; Juhnn Y. S. Multi-Facet Expressions of Adenylate Cyclase Isoforms in B16-F10 Melanoma Cells Differentiated by Forskolin Treatment. Exp. Mol. Med. 2000, 32, 235–242. 10.1038/emm.2000.39. [DOI] [PubMed] [Google Scholar]
- Chang C. H.; Lee H. H.; Lee C. H. Substrate Properties Modulate Cell Membrane Roughness by Way of Actin Filaments. Sci. Rep. 2017, 7 (1), 1–11. 10.1038/s41598-017-09618-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Xu C.; Jiang N.; Zheng L.; Zeng J.; Qiu C.; Yang H.; Xie S. Quantitative Analysis of the Cell-Surface Roughness and Viscoelasticity for Breast Cancer Cells Discrimination Using Atomic Force Microscopy. Scanning 2016, 38 (6), 558–563. 10.1002/sca.21300. [DOI] [PubMed] [Google Scholar]
- Proksch R.; Lal R.; Hansma P. K.; Morse D.; Stucky G. Imaging the Internal and External Pore Structure of Membranes in Fluid: TappingMode Scanning Ion Conductance Microscopy. Biophys. J. 1996, 71 (4), 2155. 10.1016/S0006-3495(96)79416-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheinlaender J.; Schäffer T. E. Lateral Resolution and Image Formation in Scanning Ion Conductance Microscopy. Anal. Chem. 2015, 87 (14), 7117–7124. 10.1021/acs.analchem.5b00900. [DOI] [PubMed] [Google Scholar]
- Klausen L. H.; Fuhs T.; Dong M. Mapping Surface Charge Density of Lipid Bilayers by Quantitative Surface Conductivity Microscopy. Nat. Commun. 2016, 7 (1), 1–10. 10.1038/ncomms12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann P.; Gesper A.; Happel P. Correlative Stimulated Emission Depletion and Scanning Ion Conductance Microscopy. ACS Nano 2018, 12, 5807–5815. 10.1021/acsnano.8b01731. [DOI] [PubMed] [Google Scholar]
- Navikas V.; Leitao S. M.; Grussmayer K. S.; Descloux A.; Drake B.; Yserentant K.; Werther P.; Herten D.-P.; Wombacher R.; Radenovic A.; Fantner G. E. Correlative 3D Microscopy of Single Cells Using Super-Resolution and Scanning Ion-Conductance Microscopy. Nat. Commun. 2021, 12 (1), 1–9. 10.1038/s41467-021-24901-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke R. W.; Novak P.; Zhukov A.; Tyler E. J.; Cano-Jaimez M.; Drews A.; Richards O.; Volynski K.; Bishop C.; Klenerman D. Low Stress Ion Conductance Microscopy of Sub-Cellular Stiffness. Soft Matter 2016, 12 (38), 7953–7958. 10.1039/C6SM01106C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Takahashi Y.; Hong S. P.; Liu F.; Bednarska J.; Goff P. S.; Novak P.; Shevchuk A.; Gopal S.; Barozzi I.; Magnani L.; Sakai H.; Suguru Y.; Fujii T.; Erofeev A.; Gorelkin P.; Majouga A.; Weiss D. J.; Edwards C.; Ivanov A. P.; et al. High-Resolution Label-Free 3D Mapping of Extracellular PH of Single Living Cells. Nat. Commun. 2019, 10 (1), 1–9. 10.1038/s41467-019-13535-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon J. B.; Zeitler B.; Cheng J.; Cheng A. T.; Cherone J. M.; Santiago Y.; Lee A. H.; Vo T. D.; Doyon Y.; Miller J. C.; Paschon D. E.; Zhang L.; Rebar E. J.; Gregory P. D.; Urnov F. D.; Drubin D. G. Rapid and Efficient Clathrin-Mediated Endocytosis Revealed in Genome-Edited Mammalian Cells. Nat. Cell Biol. 2011, 13 (3), 331–337. 10.1038/ncb2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierrat X.; Wong J. P. H.; Al-Mayyah Z.; Persat A. The Mammalian Membrane Microenvironment Regulates the Sequential Attachment of Bacteria to Host Cells. mBio 2021, 12 (4), 1. 10.1128/mBio.01392-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass D. S.; Riedel-Kruse I. H. A Synthetic Bacterial Cell-Cell Adhesion Toolbox for Programming Multicellular Morphologies and Patterns. Cell 2018, 174 (3), 649–658. 10.1016/j.cell.2018.06.041. [DOI] [PubMed] [Google Scholar]
- Navikas V.; Leitao S. M.; Marion S.; Davis S. J.; Drake B.; Fantner G. E.; Radenovic A. High-Throughput Nanocapillary Filling Enabled by Microwave Radiation for Scanning Ion Conductance Microscopy Imaging. ACS Appl. Nano Mater. 2020, 3 (8), 7829–7834. 10.1021/acsanm.0c01345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nievergelt A. P.; Kammer C.; Brillard C.; Kurisinkal E.; Bastings M. M. C.; Karimi A.; Fantner G. E. Large-Range HS-AFM Imaging of DNA Self-Assembly through in Situ Data-Driven Control. Small Methods 2019, 3 (7), 1900031. 10.1002/smtd.201900031. [DOI] [Google Scholar]
- Fantner G. E.; Oates A. C. Instruments of Change for Academic Tool Development. Nat. Phys. 2021, 17 (4), 421–424. 10.1038/s41567-021-01221-3. [DOI] [Google Scholar]
- Nečas D.; Klapetek P. Gwyddion: An Open-Source Software for SPM Data Analysis. Open Phys. 2012, 10 (1), 181–188. 10.2478/s11534-011-0096-2. [DOI] [Google Scholar]
- Schindelin J.; Arganda-Carreras I.; Frise E.; Kaynig V.; Longair M.; Pietzsch T.; Preibisch S.; Rueden C.; Saalfeld S.; Schmid B.; Tinevez J.-Y.; White D. J.; Hartenstein V.; Eliceiri K.; Tomancak P.; Cardona A. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9 (7), 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.