Abstract
Cell membrane-coated (CMC) mimics are micro/nanosystems that combine an isolated cell membrane and a template of choice to mimic the functions of a cell. The design exploits its physicochemical and biological properties for therapeutic applications. The mimics demonstrate excellent biological compatibility, enhanced biointerfacing capabilities, physical, chemical, and biological tunability, ability to retain cellular properties, immune escape, prolonged circulation time, and protect the encapsulated drug from degradation and active targeting. These properties and the ease of adapting them for personalized clinical medicine have generated a significant research interest over the past decade. This review presents a detailed overview of the recent advances in the development of cell membrane-coated (CMC) mimics. The primary focus is to collate and discuss components, fabrication methodologies, and the significance of physiochemical and biological characterization techniques for validating a CMC mimic. We present a critical analysis of the two main components of CMC mimics: the template and the cell membrane and mapped their use in therapeutic scenarios. In addition, we have emphasized on the challenges associated with CMC mimics in their clinical translation. Overall, this review is an up to date toolbox that researchers can benefit from while designing and characterizing CMC mimics.
Keywords: cell membrane, template, biomimetic, biointerfacing, drug delivery, personalized medicine, tumor microenvironment, good manufacturing practice, detoxification, therapeutic applications
Pharmacological/drug-based therapies are the most common and foremost recourse prescribed for treating diseases and disorders in the human body. In practice, for many years, these therapies have improved health and extended lives without the need for aggressive interventions.1−6 However, the advent of nanomedicine has revolutionized this traditional approach for disease diagnosis and treatment. Nanomedicine combines the principles of nanotechnology, immunology, and biomaterials to create delivery systems with significantly improved safety and efficacy.7−9
Delivery systems have two main functions: to execute a specific application that they are designed for and to interact favorably with the complex physiological environment surrounding them to support and enhance their function. Loading a drug of interest or modulating its physiochemical properties can improve these functions partially. However, it is vital to ensure that they have biointerfacing capabilities to avoid roadblocks during clinical translation.10−12 Biointerfacing capabilities include improving stimuli responsiveness, reducing nonspecific interactions, increasing circulation times, and evading uptake or clearance by the reticuloendothelial system.13−15 While PEGylation offered some respite by introducing stealth properties, minimizing nonspecific interactions and prolonging circulation, yet negative immunogenic response and allergic reactions were unavoidable.16,17 An alternative approach is incorporating ligands (antibodies,18,19 aptamers,20,21 peptides,22,23 and small molecules24,25) to improve target efficacy, but this rendered the system overly complicated for scale-up. These strategies were only partial remedies and not universally applicable or sufficient for clinical translation.
Vital clues to improve the biointerfacing capabilities of synthetic delivery systems can be obtained by understanding the structure, function, and homeostasis of cells in the complex physiological environment surrounding them. Incorporating cell properties like shape, flexibility,26,27 compartmentalization,28−30 lipid bilayer structure,31,32 autonomous and specific functionality,33−35 and protecting cargo36,37 can be advantageous in delivery systems. In this regard, researchers have attempted to use liposomes,38,39 polymeric micelles,40 or naturally occurring extracellular vesicles41 as delivery systems. For example, Doxil and GenexoltPM are the first FDA-approved liposomal and polymeric micelle formulations, respectively, translated into the clinic and many more under different phase trials.42,43 However, liposome and polymeric micelle’s long-term stability issues, degradation during sterilization, and complex surface modifications for active targeting still remain a challenge for large scale-up.42−46 For avoiding surface modification complexity, extracellular vesicles are viable alternatives as a delivery system. These are lipid bilayer vesicles, naturally secreted by the cells that display the same proteins, ligands, and targeting moieties like a cell on its surface.47 Unfortunately, the existing isolation and purification methods for vesicle production cause functional heterogenicity and low yield.48−50 Besides, low drug loading efficiency also limits their use for a wide range of applications.51−53 The cell membrane is a major structural component of a cell and extracellular vesicles and replicates their surface functionality. If done correctly, the cell membrane conserves this functionality post-isolation, and its coating improves biointerfacing capabilities. Referred to as cell membrane-coated (CMC) mimics henceforth, these intelligently engineered delivery systems combine the biomimetic features of the cell membrane and the functional versatility of a template. The template (spherical or nonspherical) acts as the central scaffold that carries a payload of interest and provides a structural basis. The cell membrane offers surface functionality that mimics a natural cell to improve accumulation and efficacy at the target site.54 Their assembly process utilizes noncovalent interactions and physical and soft techniques and eliminates a need for complex chemical processing and traditional synthetic modifications.55−57 Compared to the conventional delivery systems, these CMC mimics demonstrate excellent biological compatibility, stealth properties, and retain cellular properties for active targeting using receptor–ligand interactions.58−66
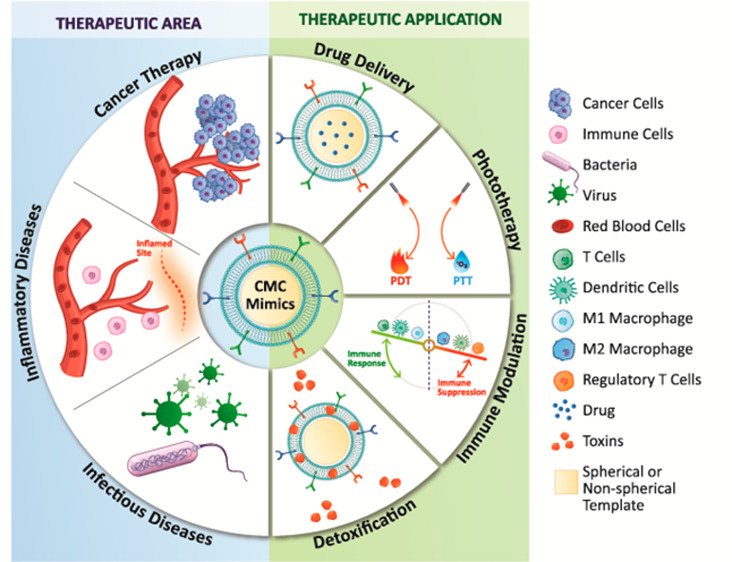
In this review, we focus on providing a detailed insight into the various aspects of designing CMC mimics. We begin with an overview of different cell types, their inherent biological properties, and suitability for specific therapeutic applications including cancer, inflammatory diseases, infectious diseases, and their potential use in personalized medicine. In the next section, we present protocols for isolating cell membranes from both nucleus-free and nucleus-containing cells with minimal nuclear and mitochondrial contamination. It is vital to follow protocols that conserve their surface functionality and mechanical stability during the isolation process. Next, we present an overview of templates and their properties available for cell membrane coating. Selecting the right template allows for the chemical and genetic tunability of the mimics and improves bioimaging,67,68 drug delivery,55,69,70 diagnostic,71−74 biosensing,75 detoxification,76,77 and phototherapy performance.56,78,79 We then highlight the processes used for CMC assembly and the challenges for large-scale production, followed by physiochemical and biological characterization techniques that validate their structural integrity and functionality. The last part of the review presents examples of CMC mimics designed for therapeutic applications and in vitro and in vivo models that evaluate their efficacy. Finally, we conclude with an overview of current challenges en route to clinical translation.
Biological Properties of Different Cell Membranes in CMC Mimics
The cell membrane is the outermost protective layer of a cell with a thickness of around 5–10 nm, mainly composed of lipids, proteins, and carbohydrates, and it interacts and performs complex biological functions with the surrounding environment for survival and proliferation.80,81 Bilayer assembly of lipids incorporates structural rigidity and fluidity,82 while carbohydrates are responsible for cellular recognition,83,84 and proteins play a vital part in signaling and adhesion, briefly.85 The composition and properties of these three components of the cell membrane differentiate them. The possibility of benefiting from native functionalities originating from cell membranes has resulted in significant research interest in CMC mimics.86−89
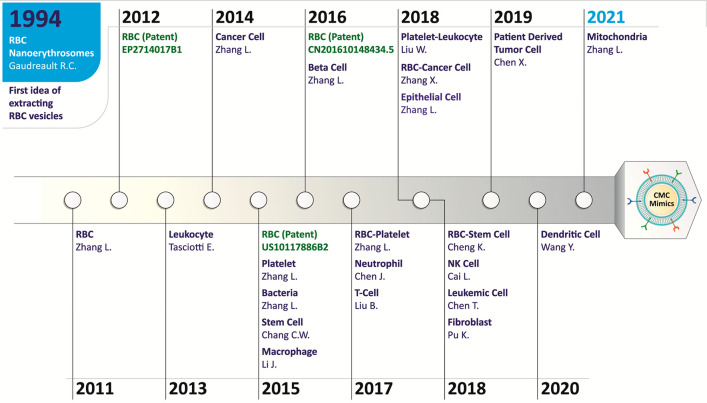
Figure 1 provides a timeline of different cell sources utilized in the CMC mimics fabrication. The idea of isolating RBC vesicles was explored in 199490 and gained significant research interest in utilizing cell membrane vesicles for coating onto a template to design CMC mimics in 2011.62 Until 2020, the natural cell membrane has widely been used from different cell types, but recently, the outer intracellular membrane from the mitochondria has also been explored to enhance biointerfacing capabilities.91 This section describes the specific biological function of the cell membrane of various cell types and the intracellular organelle that they offer to a CMC mimic.
Figure 1.
Timeline of different cell sources utilized in CMC mimics fabrication. The idea of isolating the RBC vesicles was reported in 199490 and gained significant research interest in utilizing cell membrane vesicles for coating onto a template to design CMC mimics in 2011.62 For designing these mimicking systems, the cell membrane from a wide variety of cells source (leukocyte,63 cancer cell,92 platelet,93 bacteria,94 stem cell,95 macrophage,69 β-cell,96 RBC-platelet hybrid,97 neutrophil,55 T-cell,98 platelet-leukocyte hybrid,99 RBC-cancer cell hybrid,100 epithelial cell,101 RBC-stem cell hybrid,86 natural killer (NK) cell,102 leukemic cell,103 fibroblast,104 patient-derived tumor cell,105 dendritic cell106) has been explored depending upon the importance of cells for a specific application. Recently, intracellular organelle membrane coating was investigated using mitochondria as a model organelle. These CMC mimics have shown great potential for use in personalized medicine.92 Some patents granted on these CMC mimics using the RBC membrane are highlighted in green in this figure.
Red Blood Cell Membrane
Red blood cells (RBCs) are the most abundant cell type of the human body, with the longest circulation time of approximately 120 days.107 RBCs transmembrane express protein cluster of differentiation 47 (CD47), also known as the ‘do not eat me’ marker,108 selectively binds to signal-regulatory protein alpha (SIRPα) glycoprotein expressed by macrophages to prevent its uptake.109,110 RBCs are also responsible for oxygen transport to various tissues and organs in the body111 and are involved in pathogen removal by oxycytosis.112 Their membrane is rich in glycophorins that attract pathogens to their surface to release oxygen for killing them.113 Thus, coating the template with an RBC membrane improves long-term circulation,62 pathogens removal,64,114 and toxins absorption115,77 for detoxification applications. These specific advantages have popularized the use of RBC membrane-coated CMC mimics.
Platelet Cell Membrane
Platelets, also known as thrombocytes, inhibit bleeding by forming clots and help in tissue repair.116 Platelets membrane like RBCs express CD47 receptor proteins on their surface that help in evading macrophages. Additional membrane proteins on platelets: integrin like αIIbβ3, α6β1, and P-selectin help in targeting tumor cells,117 glycoprotein Ib (GPIb/IX/V) complex in binding to exposed subendothelial collagen at the injury site in blood vessels by interacting with von Willebrand factor (VWF),117 clusters of differentiation 55 (CD55), and clusters of differentiation 59 (CD59) for immune modulation,118 toll-like receptors for pathogen removal.119 Platelets are involved in a cross-talk with inflamed endothelium cells and bind with immune cells to redirect them to the injury site.120 Thus, coating the template with a platelet membrane offers an escape from macrophage detection, selective adhesion to tumor tissues or injured vessels,121,70 targeting of vascular disorders,93,122,123 and binding ability to circulatory tumor cells87 and pathogen removal.93
Macrophage Cell Membrane
Macrophages are part of the innate immune system, known for removing unwanted or foreign materials/bacteria/viruses from the human body by engulfing them (phagocytosis) using recognition receptors such as scavenger receptors, mannose receptors, and toll-like receptors ((TLR)-2, -4, -5).124,125 Derived from circulatory monocytes, they are present in all the tissues. During infections or tissue damage, cytokines actively recruit monocytes where they differentiate into macrophages.126 Chemokine receptors on the macrophage membrane like C–C chemokine receptors type 2 (CCR2), C–X–C chemokine receptor type 1 (CXCR1), C–C chemokine receptor type 7 (CCR7), etc., facilitate their recruitment at the inflammation site.127 Along with other leukocytes, macrophage membranes also express adhesions molecules like P-selectin glycoprotein ligand-1 (PSGL-1), L-selectin, lymphocyte function-associated antigen 1 (LFA-1), L-selectin, and very late antigen-4 (VLA-4) that assist in their recruitment and cell adhesion.128,129 Thus, coating the template with the macrophage membrane has the potential to bind pathogens and can also easily escape from macrophage detection to provide active targeting at inflammatory sites130 and tumors.69,131,132
Neutrophil Cell Membrane
Neutrophils belong to the innate immune system and constitute around 40–60% of the white cell population in a healthy human body.133 In response to inflammation, their production rate in bone marrow increases by at least 10-fold.134 After leaving the bone marrow, their targeting abilities depend on their phenotypic changes and surface. Neutrophils are usually resting when circulating in healthy body receptors.135,136 They become activated by cytokines or chemokines like tumor necrosis factor-alpha (TNF-α), granulocyte-macrophage colony-stimulating factor (GM-CSF), interleukin 8 (IL-8), and interferon gamma (IFN-γ) which mobilize them to the infection or inflammation site.137 Conformational changes in integrin adhesion receptors like very late antigen-4 (VLA-4), lymphocyte function-associated antigen 1 (LFA-1), macrophage-1 antigen (Mac-1), P-selectin glycoprotein ligand-1 (PSGL-1), and L-selectin also facilitate neutrophil migration through extravasation from blood vessels.133,138 Thus, coating the template with the activated neutrophil membrane actively targets the tumors139,55 and inflammatory sites.66
Natural Killer Cell Membrane
Natural killer (NK) cells are part of the innate immune system and the first line of defense against tumor and virally infected cells that do not require any prior activation like other immune cells (T cells, B cells).140 In human peripheral blood, the NK cells comprise 10–15% of the total lymphocyte population. These cells contain many activating and inhibitory receptors on their surface that selectively target tumor/virally infected cells without affecting healthy cells.141 Some of the important activating receptors are NK group 2D (NKG2D), DNAX accessory molecule-1 (DNAM-1), natural cytotoxicity receptor (NKp30), etc.;142 integrin adhesion receptors are LFA-1, VLA-4, Mac-1, PSGL-1, and L-selectin (along with other leukocytes), etc., that help in extravasation from blood vessels.142,143 These cells also activate other immune cells like T cells by releasing cytokines and chemokines.144 NK cell lines, for example, KHYG-1 and NK-92 membranes, also contain activating and adhesion receptors like primary NK cell membrane, facilitating their use in clinical trials.145−147 These cell lines are also easy to culture and expand in vitro. Therefore, utilizing NK cell line membrane in CMC mimics could also be a potential alternative. Recently, chimeric antigen receptor (CAR)-NK and CAR-NK-92 technologies began undergoing clinical trials for immunotherapy.148,149 Thus, coating the template with the NK cell membrane has the potential to actively target inflammation, infection, and tumor sites without prior activation.102,150
T-Cell Membrane
T cells are part of the adaptive immune system that can recognize antigens using T-cell receptors (TCR).151 TCRs cannot bind to antigens directly and require peptides fragments of antigens for binding. These fragments are presented to them by major histocompatibility complex molecules (MHC I or II) present on antigen-presenting cells (dendritic cells or macrophages).152 Naive T cells recognize these specific fragments and differentiate into subsets like cytotoxic, helper, or regulatory T cells. Cytotoxic T cells express cluster of differentiation 8 (CD8) coreceptor (CD8+ T cell) that recognizes antigens on MHC-I molecules and can kill the infected cells (virus/bacteria/cancer cells) by releasing cytotoxic granules or Fas/FasL interaction.153 Helper T cells express cluster of differentiation 4 (CD4) coreceptor (CD4+ T cells) recognize antigens on MHC-II molecules and regulate immune response that indirectly affects the infected cells.154 According to literature reports, helper T cells play an important role in treating HIV due to its high-affinity receptor (CD4+ T).155 CAR-T cell therapy is an FDA-approved therapy for multiple myeloma (ABECMA) and is under evaluation for treating other cancer types and avoid unwanted side effects.156 Therefore, utilizing the T-cell membrane in CMC mimics could be a potential strategy for treating cancer and infectious diseases.98,157−159
Dendritic Cell Membrane
Dendritic cells (DCs) are central players of the immune system that link innate and adaptive immune systems. These cells are also known as “professional” antigen-presenting cells (APCs).160 DCs are the first immune cells to become activated in the human body post a pathogenic attack (bacteria, virus, or cancer cells).161,162 Even in their resting immature state, iDCs are involved in phagocytosis. They encapsulate pathogens and process them, degrade them into fragments, and present them on the MHC molecules on their surface.163 During this activation process, iDCs mature and migrate to adaptive immune cells (T cells and B cells) and present antigens for their activation. During antigen presentation, DCs upregulate the expression of co-stimulatory receptors molecules CD86, CD83, CD80, and CD40 on their cell membrane.164 These molecules effectively bind to their corresponding receptors on T cells and trigger the release of cytokines (interleukin, IL-12 or IL-10) from DCs that differentiate T cells into their pro-inflammatory or anti-inflammatory subsets. According to experimental reports, one mature DC can stimulate up to 100–3000 T cells.165,166 Thus, CMC mimics fabricated with mature dendritic cell membrane can generate sufficient immune response to activating T cells, required to treat several tumors and infectious diseases.106,167,168
Cancer Cell Membrane
Cancer cells can escape the immune system and are known for their rapid and infinite proliferation. Because of their robust nature, it is easy to culture and expands them in vitro. Different types of cancer cell membranes express numerous tumor-specific antigens and adhesion molecules on their surface. Some of them include cadherins, integrins, galectin-3, lymphocyte-homing receptors (like clusters of differentiation 44 (CD44)), epithelial adhesion molecules, and mucoprotein-1 that play a vital role in cell-to-cell and cell-to-matrix interactions.169−171 Mainly, cancer cell membranes have self-targeting abilities to adhere to their homologous cells.65,172 Thus, coating a template with the cancer cell membrane allows it to escape from macrophage detection and for homotypic tumor targeting173−175 and helps in the development of personalized medicine for cancer.105
Stem Cell Membrane
Stem cells are known for their ability to replicate indefinitely and differentiate into specialized cell types in the body. Among other stem cells, mesenchymal stem cell (MSC)-based therapies have shown immense potential as regenerative medicine176 and have entered many clinical trials.177,178 These cells can specifically target different cancerous and metastatic diseases because of their intrinsic tumor tropic properties,179−181 they are readily isolated, are stable through multiple in vitro passages, and are produced under good manufacturing practice (GMP) conditions.182,183 Various chemokines and cytokine receptors like CCR1, CCR2, CXCR1, CXCR2, etc., help the MSCs to migrate to the inflammatory or injured site.184 Like leukocytes, stem cells also undergo rolling, adhesion, and an extravasation process. Thus, coating the template with a stem cell membrane provides actively targeting abilities toward tumor95,185,186 and degenerative diseases.187,188
Bacterial Cell Membrane
Bacteria have an additional peptidoglycan cell wall, unlike other mammalian cell types. Gram-positive bacteria have a thick peptidoglycan cell wall and no outer membrane, while Gram-negative bacteria have thin cell walls as well as lipopolysaccharide outer membranes.189 Both the Gram-positive and Gram-negative bacteria secrete membrane vesicles. Gram-positive bacteria secrete extracellular vesicles (EVs), whereas Gram-negative bacteria secrete outer membrane vesicles (OMVs).190 These membrane vesicles express several immunogenic antigens with adjuvant properties and pathogens-associated patterns that help immune modulation.94,191 Thus, coating the template with the bacterial membrane vesicles (Escherichia coli (E. coli); Staphylococcus aureus (S. aureus); Klebsiella pneumonia (K. pneumonia)) provides an antibacterial immune response,94 vaccination against bacterial infection,94,192,193 and tumor targeting abilities.194,195
Hybrid Cell Membrane
The hybrid cell membrane coating strategy fuses cell membranes from multiple cell types to incorporate multiple cell-specific functional properties in a single mimic.168,196,197 For example, CMC mimics designed using RBC and B16-F10 melanoma cancer cell membrane express both CD47 transmembrane protein from RBCs and self-recognition markers (glycoprotein, gp100) from the cancer cell membrane.100,198 Overall, these RBC-cancer hybrid membranes provide several features like long-term circulation, immune evasion, and homotypic targeting abilities in the CMC mimics.196 Depending on the specific target application, the relative amount of each membrane can be varied for designing CMC mimics. Thus, hybrid membrane coating by coupling different cell types (refer to previous sections) provides the possibility of designing CMC mimics with multiple desired functionalities, thus offering several advantages in various therapeutic applications.97,99,199−201
Intracellular Cell Membrane (Organelle)
Intracellular membranes from organelles of eukaryotes display the same fundamental structure as the plasma membrane, with the phospholipid bilayer responsible for specific functions.202 Targeting intracellular membrane functions can be an intelligent strategy for treating several diseases. For example, the delivery of biomolecules across nuclear membranes is considered safe and effective gene therapy.203,204 For drug-resistant bacterial or viral infections, it is preferable to block the alteration of intracellular membranes with pathogens and inhibit their intracellular replication.205 Inducing permeability in the mitochondrial, nucleus, and lysosomal membranes is a well-established strategy to overcome drug resistance during cancer treatment.206 Recently, CMC mimics fabricated using intracellular membranes were explored to targeted detoxification and molecular detection in ABT-263-induced thrombocytopenia.91 Therefore, coating templates with the intracellular membranes can be an innovative approach to probe the complex intracellular machinery for several therapeutic applications.
Protocols for Cell Membrane Extraction
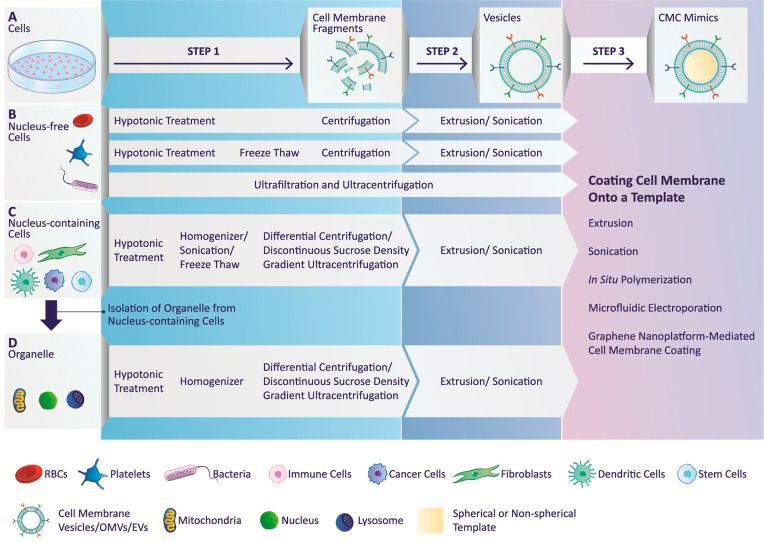
There are two categories of cells: nucleus-free or nucleus-containing cells. There are several reports on cell membrane isolation from various cell types. An attempt has been made to simplify the procedure and discuss the main steps involved during the isolation (Figure 2).
Figure 2.
A schematic illustration of isolating and preparing membrane vesicles from nucleus-free, nucleus-containing cells, and organelle before coating: (A) The two-step process involves extracting cell membrane fragments (step 1) and preparing cell membrane vesicles (step 2). Depending on the type of cell used, cell membrane extraction and vesicle formation require a combination of techniques: (B) nucleus-free cells and (C) nucleus-containing cells. (D) Organelles: Step 3 is the final step of coating cell membrane onto a template (spherical or nonspherical) using suitable technique mentioned. Abbreviations: RBCs, red blood cells; OMVs, outer membrane vesicles; EVs, extracellular vesicles.
Cell membranes isolation protocols aim to separate the cell membrane from the cell with minimal or no nuclear/mitochondrial/cytosol contamination depending on the cell type. Using a pure cell membrane helps in assembling CMC mimics by enhancing an efficient and homogeneous surface coating with maximal functional replication on the template surface. The extraction buffers (pH 7–7.4) are supplemented with protease/phosphatase inhibitor cocktails in ice-cold conditions to protect the membrane proteins from degradation.55,106,188,207,208 Prior to isolation, cells are washed multiple times with 1× phosphate-buffered saline (PBS) buffer to remove remnants from cell culture media. Post-isolation, the cell membrane is lyophilized and usually stored at −80 °C to maintain the long term stability and function of membrane proteins.175,187,209,100,173
The cell membrane isolation mainly involves two steps depending on the cell type (Figure 2):
-
(1)
Gentle rupturing of cells using detergent-free hypotonic treatment (osmotic imbalance) or a combination of hypotonic treatment and physical disruption technique
-
(2)
Separation and purification of the cell membrane from intracellular components using multiple centrifugation steps, differential centrifugation, or discontinuous sucrose density gradient centrifugation.
In this section, we have discussed the membrane isolation methodology from nucleus-free, nucleus-containing cells and the recently explored intracellular organelle (mitochondria) in designing CMC mimics. All the different conditions (hypotonic buffers, physical disruption techniques, and centrifugation speeds) used in cell membrane isolation are summarized in Tables 1 and 2.
Table 1. Cell Membrane Isolation from Nucleus-Free Cellsa.
| cell type | hypotonic buffer (ice cold) | disruption technique (ice cold) | method of isolation (4 °C) | ref |
|---|---|---|---|---|
| RBCs | 0.25× PBS | hypotonic treatment; centrifuge 3–4 times to get pink pellet | 800g, 5 min | (62) |
| 0.25× PBS, 0.2 mM EDTAK2 | 10,000 rpm, 5 min | (211) | ||
| deionized water, EDTA | 4000 rpm, 10 min | (210) | ||
| 4800 rpm, 20 min | ||||
| platelets | 1× PBS, 1 mM EDTA, protease inhibitor cocktail | repeated freeze thaw cycles | 4000g, 3 min | (93) |
| – | 7 freeze–thaw cycles and sonication | discontinuous sucrose gradient ultracentrifugation | (213) | |
| 5%, 40%, 55% | ||||
| cell membrane: interface between 5% and 40% | ||||
| deionized water | 3 freeze–thaw cycles | 21,000g, 7 min | (212) |
| bacteria | ultrafiltration | method of OMVs or EVs isolation | ref |
|---|---|---|---|
| E. coli | 4000g for 10 min, filtration through a 0.45 μm vacuum filter, concentration of filtrate using ultrafiltration with a 100 kDa amicon centrifugal filter | 150,000g, 2 h | (94) |
| S. aureus | 10,000g for 20 min, filtration through a 0.45 μm vacuum filter, concentration of filtrate using ultrafiltration with a 100 kDa amicon centrifugal filter | 150,000g, 3 h | (192) |
| K. pneumoniae | 3220g for 15 min, filtration through a 0.22 μm vacuum filter, concentration of filtrate using a 100 kDa ultrafiltration tube | 150,000g, 1 h | (193) |
Abbreviations: RBC, red blood cells; PBS, phosphate-buffered saline; EDTA, ethylenediaminetetraacetic acid; EDTAK2, dipotassium EDTA; E. coli, Escherichia coli; S. aureus, Staphylococcus aureus; K. pneumonia, Klebsiella pneumonia; g, relative centrifugal force; rpm, revolutions per minute; min, minutes, h, hour; OMVs, outer membrane vesicles; EVs, extracellular vesicles.
Table 2. Cell Membrane Isolation from Nucleus-Containing Cells and Intracellular Organellea.
| cell type | hypotonic buffer (ice cold) | disruption technique (ice cold) | method of isolation (4 °C) | ref |
|---|---|---|---|---|
| Intracellular Organelle: Mitochondria | ||||
| isolation of mitochondria from mouse liver | 210 mM mannitol, 70 mM sucrose, 5 mM Tris-HCl,1 mM EDTA | Kinematic Polytron PT-2000 homogenizer (power setting 7 for 15 strokes) | 2000g, 10 min | (91) |
| 7000g, 10 min | ||||
| followed by sucrose density gradient ultrafiltration | ||||
| top: 15 mL of 1.0 M sucrose solution | ||||
| bottom: 15 mL of 1.5 M sucrose solution | ||||
| 60,000g, 20 min | ||||
| mitochondria- interface between the two sucrose layers | ||||
| isolation of membrane from mitochondria | ultapure water (20 min) followed by addition of 1.4 M sucrose solution (5 min) | Kinematic Polytron PT-2000 homogenizer (power setting 7 and 30 strokes) | differential centrifugation | |
| 12,000g, 10 min | ||||
| 100,000g, 30 min | ||||
| Immune Cells | ||||
| neutrophils | 0.5% (w/v) BSA, 75 mM sucrose, 225 mM mannitol, 0.5 mM EDTA, 30 mM Tris-HCl, protease inhibitor cocktail | Dounce homogenizer, tight-fitting pestle (50–100 passes) | differential centrifugation | (55) |
| 800g, 10 min | ||||
| 10,000g, 20 min | ||||
| 100,000g, 60 min | ||||
| 75 mM sucrose, 225 mM d- mannitol, 0.5 mM EGTA, 30 mM Tris-HCl, protease inhibitor cocktail | Dounce homogenizer, tight-fitting pestle (20 passes) | differential centrifugation | (66) | |
| 20,000g, 25 min | ||||
| 100,000g, 35 min | ||||
| NK-92 | 10 mM Tris-HCl, 10 mM MgCl2, 1 mM KCl, 25 mM sucrose, 2 mM PMSF, 200 μg/mL trypsin chymotrypsin inhibitor, 10 μg/mL DNase, 10 μg/mL RNase | homogenized, 5 min (20 s pulse, 30 s in between pulses) | discontinuous sucrose gradient ultracentrifugation | (67, 102) |
| 30%, 40%, and 55% sucrose in 0.85% saline | ||||
| cell membrane: interface between 30% and 40% | ||||
| cell lysis buffer, protease inhibitor cocktail | water bath sonication, 20–30 min | differential centrifugation | (150) | |
| 3500g, 10 min | ||||
| 20,000g, 25 min | ||||
| 100,000g, 50 min | ||||
| mouse natural killer cells | cell lysis buffer, protease inhibitor cocktail | water bath sonication, 20–30 min | differential centrifugation | (150) |
| 3500 g, 10 min | ||||
| 20,000g, 25 min | ||||
| 100,000g, 50 min | ||||
| human cytotoxic T-lymphocyte cells | 10 mM Tris-HCl, 1 mM KCl, 1 mM MgCl2, phosphatase inhibitor cocktail | Dounce homogenizer, tight-fitting pestle (20 passes) | discontinuous sucrose gradient ultracentrifugation | (98) |
| 30%, 40%, and 55% | ||||
| cell membrane: interface between 30% and 40% | ||||
| RAW264.7 | 10 mM Tris-HCl, 1 mM MgCl2 | Mini-extruder without a polycarbonate membrane (20 times) | homogenate mixed with 1 M sucrose to a final concentration of 0.25 M sucrose | (129) |
| centrifugations | ||||
| 2000g, 10 min | ||||
| 3000g, 30 min | ||||
| 20 mM Tris-HCl, 2 mM MgCl2, 10 mM KCl, protease inhibitor cocktail | Dounce homogenizer, tight-fitting pestle (20 passes) | differential centrifugation | ||
| 3200g, 5 min | ||||
| 20,000g, 30 min | ||||
| 80,000g, 1.5 h | ||||
| 1 mmol/L NaHCO3, 0.2 mmol/L EDTA, 1 mmol/L PMSF | repeatedly grinding (20 times) | differential centrifugation | (69) | |
| 3200g, 5 min | ||||
| 100,000g, 30 min | ||||
| THP-1 | 10 mM Tris-HCl, 10 mM MgCl2, 1 mM KCl, 25 mM sucrose, 2 mM PMSF, 200 μg/mL trypsin chymotrypsin inhibitor, 10 μg/mL DNase, 10 μg/mL RNase | Dounce homogenizer, tight-fitting pestle (20–30 passes) | discontinuous sucrose gradient ultracentrifugation | (63) |
| 30%, 40%, and 55% sucrose in 0.85% saline | ||||
| cell membrane: interface between 30% and 40% | ||||
| J774 | 10 mM Tris-HCl, 10 mM MgCl2, 1 mM KCl, 25 mM sucrose, 2 mM PMSF, 200 μg/mL trypsin chymotrypsin inhibitor, 10 μg/mL DNase, 10 μg/mL RNase | Dounce homogenizer, tight-fitting pestle (20–30 passes) | discontinuous sucrose density gradient ultracentrifugation | (63) |
| 30%, 40%, and 55% sucrose in 0.85% saline | ||||
| cell membrane: interface between 30% and 40% | ||||
| 75 mM sucrose, 20 mM Tris-HCl, 2 mM MgCl2, 10 mM KCl, protease inhibitor cocktail | Dounce homogenizer, tight-fitting pestle (20 passes) | differential centrifugation | (130) | |
| 3200g, 5 min | ||||
| 20,000g, 25 min | ||||
| 100,000g, 35 min | ||||
| dendritic cells | 25 mM sucrose, 10 mM Tris-HCl, 1 mM MgCl2, 1 mM KCl, 2 mM PMSF, protease and phosphatase inhibitor cocktail | Dounce homogenizer, tight-fitting pestle (25 passes) | differential centrifugation | (106) |
| 800g, 5 min | ||||
| 21,000g, 10 min | ||||
| Cancer Cells | ||||
| HeLa | membrane and cytosol protein extraction kit and PMSF | 3 freeze–thaw cycles | differential centrifugation | (219) |
| 700g, 10 min | ||||
| 14,000g, 30 min | ||||
| MCF-7 | HEPES B buffer: 2.38 g/L Hepes, 0.476 g/L MgCl2, 0.292 g/L EDTA, 0.154 g/L DTT, 0.746 g/L KCl, proteinase inhibitor cocktail | IKA T10 basic homogenizer (three times, four-speed position) | discontinuous sucrose density gradient ultracentrifugation | (198) |
| 30%, 40%, and 55% | ||||
| cell membrane: interface between 30% and 40% | ||||
| 0.5% (w/v) BSA, 75 mM sucrose, 225 mM mannitol, 0.5 mM EDTA, 30 mM Tris-HCl, protease inhibitor cocktail | sonication | differential centrifugation | (175) | |
| 800g, 10 min | ||||
| 10,000g, 20 min | ||||
| 100,000g, 60 min | ||||
| 4T1 | 10 mM Tris, 10 mM MgCl2, protease inhibitor cocktail | homogenizer (22,000 rpm for 1 min) | differential centrifugation | (220, 221, and 72) |
| 500g, 10 min | ||||
| 10,000g, 20 min | ||||
| 100,000g, 60 min | ||||
| B16-F10 | 30 mM Tris-HCl, 75.9 mM sucrose, 225 mM d-mannitol, phosphatase inhibitor, protease inhibitor cocktail | Kinematica Polytron PT 10/35 probe homogenizer (70% power, 15 passes) | differential centrifugation | (222) |
| 10,000g, 25 min | ||||
| 150,000g, 35 min | ||||
| 20 mM Tris HCl, 10 mM KCl, 2 mM MgCl2, protease inhibitor cocktail | pipetting thoroughly | differential centrifugation | (214) | |
| 3200g, 5 min | ||||
| 21,000g, 25 min | ||||
| 45,000g, 5 min | ||||
| 20 mM Tris HCl, 10 mM KCl, 2 mM MgCl2, protease inhibitor cocktail | Dounce homogenizer, tight-fitting pestle (20 passes) | differential centrifugation | (92) | |
| 3200g, 5 min | ||||
| 20,000g, 25 min | ||||
| 100,000g, 35 min | ||||
| LL/2, CMT64.OVA, MB49, A549, SKOV-3 | 20 mM Tris HCl, 10 mM KCl, 2 mM MgCl2, protease inhibitor cocktail | pipetting thoroughly | differential centrifugation | (214) |
| 3200g, 5 min | ||||
| 21,000g, 25 min | ||||
| 45,000g, 5 min | ||||
| K562 | 20 mM Tris HCl, 10 mM KCl, 2 mM MgCl2, protease inhibitor cocktail | Dounce homogenizer, tight-fitting pestle (20 passes) | differential centrifugation | (92, 103) |
| 3200g, 5 min | ||||
| 20,000g, 25 min | ||||
| 100,000g, 35 min | ||||
| MDA-MB 435 | 20 mM Tris HCl, 10 mM KCl, 2 mM MgCl2, protease inhibitor cocktail | Dounce homogenizer, tight-fitting pestle (20 passes) | differential centrifugation | (92, 223) |
| 3200g, 5 min | ||||
| 20,000g, 25 min | ||||
| 100,000g, 35 min–1.5 h | ||||
| patient-derived tumor cell | 20 mM Tris HCl, 10 mM KCl, 2 mM MgCl2, protease inhibitor cocktail | Dounce homogenizer, tight-fitting pestle (20 passes) | differential centrifugation | (105) |
| 3200g, 5 min | ||||
| 20,000g, 25 min | ||||
| 80,000g, 1.5 h | ||||
| DU145, CAL27, HCT116 | 20 mM Tris HCl, 10 mM KCl, 2 mM MgCl2, protease inhibitor cocktail | Dounce homogenizer, tight-fitting pestle (20 passes) | differential centrifugation | (223) |
| 3200g, 5 min | ||||
| 20,000g, 25 min | ||||
| 80,000g, 1.5 h | ||||
| U87 | 225 mM mannitol, 75 mM sucrose, mM EDTA, 30 mM Tris-HCl, protease inhibitor cocktail | motor-driven homogenizer with an overhead stirrer (2000 rpm); tissue grinder set with a smooth PTFE pestle (Wheaton Potter-Elvehjem) | differential centrifugation | (224) |
| 1000g, 5 min | ||||
| 7000g, 10 min | ||||
| 100,000g, 30 min | ||||
| followed with discontinuous sucrose gradient ultracentrifugation | ||||
| 2 M, 1.6 M, 1.2 M sucrose buffer | ||||
| cell membrane: 1 M density | ||||
| U-251 MG | distilled water | high-pressure homogenizer (6000 psi/412.5 bar, 10 passages) | differential centrifugation | (67) |
| 10,000g, 10 min | ||||
| 60,000g, 90 min | ||||
| LNCaP-AI | Mem-PERPlus membrane protein extraction kit | stirring for 10 min | 700g, 5 min | (225) |
| 16,000g, 15 min | ||||
| H22 | 18 mM Tris-HCl, 9 mM KCl, 1.5 mM MgCl2, protease inhibitor cocktail | Freeze–thawing, bath ultrasonic cleaner (42 kHz, 100 W) | differential centrifugation | (226) |
| 3000g, 6 min | ||||
| 20,000g, 25 min | ||||
| 120,000g, 1 h | ||||
| Stem Cells | ||||
| neural stem cells | 10 mM Tris, 10 mM MgCl2, protease inhibitor cocktail | freeze–thawing | differential centrifugation | (188) |
| 7000g, 10 min | ||||
| 13,000g, 60 min | ||||
| human adipose-derived stem cells | 10 mM Tris, 10 mM MgCl2, protease inhibitor cocktail | IKA T10 basic homogenizer (22,000 rpm for 1 min) | 6000 g, 15 min | (227) |
| Miscellaneous | ||||
| MIN6 | 20 mM Tris-HCl, 10 mM KCl, 2 mM MgCl2, protease inhibitor cocktail | Dounce homogenizer, tight-fitting pestle (20 passes) | differential centrifugation | (96) |
| 20,000g, 20 min | ||||
| 100,000g, 45 min | ||||
| C6/36 | 20 mM Tris HCl, 10 mM KCl, 2 mM MgCl2, protease inhibitor cocktail | Dounce homogenizer, tight-fitting pestle (20 passes) | differential centrifugation | (228) |
| 3200g, 5 min | ||||
| 20,000g, 25 min | ||||
| 100,000g, 35 min | ||||
Abbreviations/cell lines: min, minutes, h, hour; BSA, bovine serum albumin; EDTA, ethylenediaminetetraacetic acid; EGTA, ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid; Tris-HCl, tris(hydroxymethyl) aminomethane (THAM) hydrochloride; MgCl2, magnesium chloride; KCl, potassium chloride; PMSF, phenylmethylsulfonyl fluoride; HEPES, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid; DTT, dithiothreitol; DNase, deoxyribonuclease; RNase, ribonuclease; NK-92, human natural killer cell line; RAW 264.7, murine macrophage cell line; J774, murine macrophage cell line; THP-1, human acute monocytic leukemia cell line; HeLa, human cervical cancer cell line; MCF-7, human breast cancer cell line; 4T1, human breast cancer cell line; B16-F10, murine melanoma cell line; LL/2, murine lewis lung carcinoma cell line; CMT64.OVA, lung carcinoma cell line expressed ovalbumin; MB49, murine bladder carcinoma cell line; A549, human nonsmall cell lung cancer cell line; SKOV-3, ovarian cancer cell line; CAL27, human squamous cancer cell line; H22, murine hepatocarcinoma cell line; K562, human myelogenous leukemia cell line; MDA-MB-435, human breast carcinoma cell line; DU145, human prostate cancer cell line; CAL27, human squamous cancer cell line; U-251 MG, glioblastoma multiforme cell line; HCT116, human colorectal cancer cell line; U87, human primary glioblastoma cell line; MIN6, pancreatic β-cell line; C6/36, Aedes albopictus mosquito medium host cell line; LNCaP-AI, prostate cancer cell line.
Nucleus-Free Cells
RBCs and platelets do not contain nuclei, making their membrane extraction process relatively simple. These cells are isolated first from whole blood using appropriate methodologies. For RBCs, hypotonic treatment easily ruptures the cells followed by centrifugation to collect a pink RBC membrane/ghost pallet. Multiple cycles of centrifugation remove hemoglobin impurities from the pallet.62,210,211 For platelets, it is common to use multiple freeze–thaw cycles to damage their cell membrane by breakage of ice crystals to remove the cytosol followed by centrifugations to obtain the cell membrane.93,212 According to one report, the obtained platelet vesicles were subjected to a discontinuous sucrose gradient (5%, 40%, 55%) step to remove any free proteins, intact platelets, and high-density granules to collect pure platelet vesicles from the interface of 5% and 40% sucrose gradient.213
Bacteria are interesting exceptions in this nucleus-free cell category. Besides containing peptidoglycans in addition to the cell membrane, their cell membrane extraction process can be laborious.189,190 Therefore, they undergo ultrafiltration to separate their membrane as OMV without a cell lysis step. The reported protocols for isolating OMVs and EVs from Gram-negative (E. coli, K. pneumonia) and Gram-positive (S. aureus) bacteria, respectively, are quite similar. In the first step, the bacterial cultures were centrifuged, and the supernatant was collected. The supernatant was further vacuum filtered through a micron filter and concentrated using ultrafiltration. Finally, the obtained filtrate was subjected to ultracentrifugation to get OMV pellets or EV pellets.94,192,193
Some groups reported further purification of these OMVs or EVs with some modifications. For example, after the first ultrafiltration step, the concentrate was reprecipitated overnight using ammonium sulfate (4 °C) and ultracentrifuged to get E. coli OMVs.194,195 OMVs resuspended in PBS were further purified using sucrose gradient (1 mL each of 2.5, 1.6, and 0.6 M sucrose), separated by ultracentrifugation. In another report, after ultrafiltration and ultracentrifugation steps, the obtained S. aureus EVs pellet was resuspended in 50% Optiprep/HEPES (2.2 mL).190 The suspension was applied to the bottom of a step-density gradient (2.0 mL of 40% and 0.8 mL of 10% Optiprep in 10 mM HEPES, supplemented with 150 mM NaCl, pH 7.0), and obtained the pure S. aureus EVs floating at 1.16–1.20 g/mL.
Nucleus-Containing Cells
For nucleus-containing cells, cell membrane isolation and purification are slightly more tedious than that with nucleus-free cells. Examples include immune cells (macrophages/monocytes, neutrophils, NK cells, T cells), cancer stem cells, fibroblasts, and β-cells. These cells can either be obtained from established cell lines (like human breast cancer cell lines (MCF-7, 4T1), mouse macrophage cell line (J447), human NK cell line (NK-92), etc.), or isolated from tissues or blood (neutrophils, cancer cells, T cells, NK cells, stem cells).
On average, 200–300 million cells are required for cell membrane isolation to assemble a CMC mimic.63,67 These cells are ruptured using the hypotonic treatment and physical disruption techniques, resulting in a mixture containing pure cell membrane, intact cells, and high-density granules. Differential centrifugation or discontinuous sucrose gradient ultrafiltration of the mixture finally isolates the cell membrane. These methods are described in detail below.
Differential centrifugation method: This method is the one most commonly used for isolating cell membranes.55,130,150 It works by a stepwise increase in the centrifugation speed. The lower g at the beginning of the process removes heavy particles like a nucleus. A gradual increase in g removes other particles like mitochondria. Finally, very high g is used to pellet down the cell membrane, as it is lighter in weight. For example, the commonly reported centrifugation speeds for isolating cell membrane are 800 g (4 °C, 10 min), followed by 10,000 g (4 °C, 30 min), and finally 100,000 g (4 °C, 60 min) to isolate pure cell membrane.175,214
Discontinuous sucrose gradient ultracentrifugation method: In this method, sucrose concentration increases discretely from top to bottom, aiding density-based separation of particles in the solution. The particles move across the density gradient stopping in a region where their density matches that of the medium. For example, this method was used to demonstrate the isolation of leukocyte cell membrane using 55%, 40%, and 30% (w/v) sucrose gradients in a physiological saline solution.63 The cell membrane was collected from a 30/40% interface with minimal/no nuclear and mitochondrial contamination. A similar approach has been preferred to isolate several cell membranes using this method.63,98,102
During the membrane isolation there can be a loss of functional components like transmembrane proteins/receptors or structural components like cholesterol from the membrane. Cholesterol is mainly responsible for maintaining the rigidity of the cell membrane.215,216 Such loss may result in a decrease in the mechanical stability of the membrane. Therefore, to reduce protein loss and maintain membrane stability, hypotonic buffers with divalent ions (like MgCl2) or the addition of cholesterol can be useful. These stabilize the membrane skeleton by specifically binding to the junction complex and other membrane proteins like tropomyosin, etc.217,218 Additionally, mild lysis buffers, gentle rupturing techniques, the right pH, and ice-cold conditions must be used for membrane isolation to prevent the degradation of the transmembrane proteins and receptors.
Intracellular Organelle
Cell membrane isolation of intracellular organelle requires additional steps, unlike nucleus-containing cells. Before cell membrane isolation, it is essential to first isolate the desired organelle from nucleus-containing cells in their pure form. Isolating organelles from cells is a three-step process: hypotonic treatment, physical disruption, and ultracentrifugation. The final step is carried out in a sucrose density gradient to get the purified organelle in a specific sucrose band. The process is repeated with the purified organelle to extract the pure cell membrane. The mitochondrial outer membrane was isolated from the mouse liver using a similar protocol91 (Table 2).
Choice of Template Based on Its Properties
The template is a central component of a CMC mimic that provides a structural basis during its assembly. Inherent properties of templates extend the application of CMC mimics for diagnosis, drug delivery, and disease suppression/treatment.
There are two major categories of templates (spherical or nonspherical): organic and inorganic. Poly(lactic-co-glycolic acid) (PLGA), gelatin, and liposomes are examples of organic templates. Mesoporous silica, gold and iron oxide (Fe3O4), upconversion nanoparticles (UCNPs), persistent luminescent nanoparticles (PLNPs), and metal–organic frameworks (MOFs) are examples of inorganic templates. Organic templates offer features like biocompatibility, biodegradability, and nontoxicity and are often straightforward choices.229 In comparison, inorganic templates display additional features like magnetic, optical, and electrical properties that determine their selection in a CMC mimic.230
In this section, we have provided the general overview of the templates categorized based on their properties in the context of CMC mimics like Food and Drug Administration (FDA) approval, biocompatibility, biodegradability, and low toxicity to understand the clinical translation perspective, phototherapy for cancer suppression/treatment, bioimaging for disease diagnosis, and detoxification for enhancing the removal/absorption of toxins (summarized in Figure 3).
Figure 3.
The schematic highlights specific physiochemical and biological properties ingrained in a CMC mimics by selecting an appropriate template. The template is one of the components that provide additional value in CMC mimics. The schematic provides a summary of all the possible properties that help in the choice of the reported template while designing CMC mimics. Abbreviations: FDA, Food and Drug Administration; O2, oxygen; Fe3O4, iron oxide; CTC, circulatory tumor cells; MR, magnetic resonance; NIR, near-infrared; PLGA, polylactic-co-glycolic acid; IM, immunomagnetic; ZIF, zeolitic imidazolate; MOFs, metal–organic frameworks; GdTPP, gadolinium porphyrin; ZnTPP, zinc porphyrin; PCN-224, porphyrin (TCPP)-based Zr6 cluster; PFCs, perfluorocarbons; NPs, nanoparticles; MPs, microparticles; NDs, nanodroplets; SP, semiconducting polymer; UC, upconversion; PL, persistent luminescent; UUNCs, ultrasmall unimolecular nanoclusters; PEGDA: poly(ethylene glycol) diacrylate.
FDA-Approved, Biocompatible, Biodegradable, Low Toxicity Templates
CMC mimics are biocompatible, as the cell membrane protects every template from the external microenvironment. For clinical translation, it is also vital to consider template biodegradability and biocompatibility. Byproducts after the biodegradation and their interaction with the human body also determine their toxicity.231 Renal clearance helps to evade undesirable side effects.231 FDA-approved templates are considered the safest, nontoxic, or nonhazardous to the human body in every aspect. These properties help to protect the healthy cells in the body and avoid any unwanted immune response.
Most organic templates are generally thought to be safer than inorganic templates and were, therefore, entered easily into clinical trials.232,233 Examples of organic templates used in CMC mimics are PLGA, gelatin, and liposomes that are FDA approved, biocompatible, and biodegradable in nature. In 2011, the possibility of designing these mimicking systems was demonstrated using a PLGA nanoparticle as a template.62 PLGA is a versatile synthetic polymer that molds into both nano- and micro-sized particles. It is the most common template used for several cell membrane coatings like RBC,62,114,211,234 platelets,87,93,121,123 cancer cells,65,92,235 neutrophils,55,66 cardiac stem cells,187 macrophages,130 NK-92 cells,67 dendritic cells,106 and so on. Gelatin is a natural polypeptide used in cosmetics, pharmaceuticals, the food industry, and in the assembly of CMC mimics.236 For designing CMC mimics, several cell membranes used for coating on gelatin templates are RBC,64 stem cell,186 T cell,237 mosquito medium host Aedes albopictus (C6/36) cell,228 and patient-derived tumor cells.105 Liposomes are spherical vesicles having at least one lipid bilayer. Liposomes have been used for coating macrophages,129 RBCs,238 and cancer cell membranes.175 As reported in the literature, liposomes can also easily fuse with cell membrane vesicles like RBC239 and NK-92 cells102 for designing CMC mimics. Perfluorocarbons (PFCs) are another example of a regulatory-approved template. In 1989, PFCs (Fuosol-DA) were approved in the US, Japan, and Europe for clinical use but were taken off from the market after 5 years due to difficulties in their storage-related issues.240 Nevertheless, PFCs are biocompatible and biodegradable and have a high oxygen-carrying capacity. Many PFCs have a capacity for oxygen dissolution that is nearly 20 times that of water. They can, moreover, be easily fabricated at the nanoscale for oxygen delivery even to the smallest capillaries.241 Therefore, several CMC mimics reported using PFC can supply oxygen at the tumor sites to relieve hypoxic conditions.242,243
Most of the inorganic templates are biocompatible, but toxicity depends on the metal used for their synthesis and its degradation in the cell. Among inorganic templates, mesoporous silica is considered the safest (approved by FDA) and is biocompatible and biodegradable.244 It degrades into nontoxic silicic acid (water-soluble).245 It has been a popular template for many years in research due to its high porosity, large surface area, and high drug/photosensitizer loading capacity.246 CMC mimics reported with spherical silica nanoparticles used several cell membranes from RBCs,61 cancer cells,247 and macrophages.247 Other templates like liposome-PEG,175 UCNPs,185 and PLNPs68 were used in combination with silica to increase their drug/photosensitizer loading capacity. Mesoporous silica nanoparticles are tunable to different sizes and shapes.248,249 According to the reports, rod-shaped silica nanoparticles can enhance antimicrobial properties250 and regulate the endogenous reactive oxygen species for oxidative therapy.251 These tunable properties coupled with CMC mimics could offer potential therapeutic benefits if explored further. Silica templates can also have several desired surface functionalities post-chemical modifications.252 For example, positively charged 3-aminopropyl triethoxysilane (APTES) was used to modify the surface charge of silica microparticles to coat a negatively charged leukocyte membrane115 and platelet membrane for CTC detections.213 For Fe3O4 nanoparticles, iron ions are its biodegradation byproducts and are mostly nontoxic.231 Several CMC mimics reported using Fe3O4 templates used cell membranes like macrophage,131 MSCs,95 and HeLa cells.173 Similarly, MOFs are well-defined 3D architectures formed by the complexation between organic ligands and inorganic metal ions.253 These are biocompatible, and their toxicity depends on the nature of the metal and organic linker used. For example, zinc-based MOFs (zeolitic imidazolate (ZIF-8)) release Zn2+ ions post-degradation, an endogenous element that causes a less harmful effect on the human body if present in a low amount.254 MOFs of porphyrin (TPP)-based Gd/Zn nanocomposites release gadolinium (Gd3+) and zinc (Zn2+) ions post-degradation. Gd3+ can cause a toxic effect in the abnormal functioning of kidneys and cross the blood–brain barrier to accumulate in the brain.255 Several CMC mimics reported using cancer cell membrane-coated MOFs for homologous targeting.219,256−258 MOFs also have high porosity, large surface area, and high photosensitizer loading capacity256−258 due to their structural arrangement. Gold particles are another commonly used biocompatible inorganic template because of their inert nature. However, they are not biodegradable and may be cause for concern.259 To overcome these issues, the use of nano or ultrasmall templates to facilitate rapid renal clearance is preferred.231,260 Gold particles are tunable to different shapes: nanoparticles, nanocages, nanorods, and nanoshells, and all are used as templates for fabricating CMC mimics. Examples of these are gold nanocages with RBC membrane coating56 and H22 liver cancer cell membrane coating,226 nanorods with RBC membrane261 and platelets membrane coating,262 and nanoshells with macrophage membrane coating139 for a specific application.
Phototherapy
Phototherapy is a noninvasive and effective cancer treatment. It includes photothermal therapy (PTT) and photodynamic therapy (PDT).263 With the right choice of template, these photothermal or photodynamic properties can be explored with CMC mimics. PTT involves the photo absorbing agents to generate heat under near-infrared region (NIR) laser irradiation to kill cancer cells thermally and is less harmful to other cells or tissues.264 Gold templates have a large NIR absorption cross-section and tunable localized surface plasmon resonance (LSPR) band in the NIR region.226 This makes them most suitable to incorporate in CMC mimics for PTT.221,226 Similarly, magnetic templates like Fe3O4 are also good alternatives for their use in CMC mimics for PTT. Fe3O4 templates are efficient in photothermal conversion and are outstanding options for hyperthermia treatment.265 Fe3O4 nanoclusters showed a significant increase in NIR absorption,265 in contrast to their nanoparticles.
PDT involves reactive oxygen species (ROS) generation with photosensitizers under the light of a specific wavelength for oxidation and killing cancer cells. Mainly ROS are like singlet oxygen (1O2), superoxide anion radical (O2-•), or hydroxyl radical (•OH).266 Some combinations of photosensitizers and templates used together in CMC mimics are chlorin e6 (Ce6) in hollow mesoporous silica,267 merocyanine 540 (MC540) in UCNPs,268 zinc phthalocyanine (ZnPC) and MC540 in mesoporous silica encapsulated UCNPs,185 5,10,15,20-tetraphenylchlorin (TPC) in (ROS)-responsive paclitaxel (PTX) dimer (PTX2-TK),79 and silicon phthalocyanine in PLPNs.72 PFCs used in combination with photosensitizers provide an adequate oxygen supply to accelerate the generation of reactive singlet oxygen (1O2) and enhance PDT therapy.269 In porphyrin-based MOFs,168,219,257 porphyrin acts as a photosensitizer due to its ability to readily absorb visible light and improve overall ROS generation efficiency.270 The template used in CMC mimics for both PDT and PTT is a semiconducting polymer (SP) nanoparticles-poly(cyclopentadithiophenealt-benzothiadiazole) (PCPDTBT)). SP nanoparticles are known for their excellent optical properties and high NIR absorbing capacity and can generate signet oxygen and heat.104 Verteporfin is a photodynamic agent approved by the US FDA for eliminating abnormal blood vessels in the eyes.271 Recently, platelet membrane-coated verteporfin loaded PLGA nanoparticles reduced skin damage in PDT in combination with solar radiation.121 Indocyanine green (ICG) is an FDA-approved photosensitizer and photothermal agent for template encapsulation.174,209,269,272
Bioimaging
Bioimaging technology has significantly enhanced the ability to diagnose, treat, and prevent diseases by enabling early detection. It helps in imaging inside the animal and human body. Bioimaging includes magnetic resonance imaging (MRI), near-infrared (NIR) imaging, and fluorescence (FL) imaging.
Fe3O4 nanoparticles are the most commonly used as negative (T2) contrast agents for MRI in CMC mimics.57,265,273 Currently, standard probes used in MRI scans are gadolinium (Gd3+)-based compounds. These are positive (T1) contrast agents in MRI and also have been a preferred choice in the clinic for their better image resolution and easy detection, tunable magnetic properties, and higher colloidal stability.274 But these agents also limit their use in patients with renal imparitment and have been reported to cross blood–brain barriers to accumulate in the brain.255 Some examples of Gd3+-based templates used in CMC mimics are PLGA-Gd-lipid67,71 and MOFs like porphyrin (TPP)-based Gd/Zn nanocomposites.219 Manganese (Mn2+) ions can also be a potential alternative to gadolinium as positive (T1) MRI contrast agents.275 Since Mn2+ is one of the essential elements in the human body, its intake in small amounts does not produce toxic effects.274 Recently, CMC mimics designed using porphyrin (TCPP)-based Zr4+ clusters MOFs-coated with MnO2 nanosheets converted MnO2 into Mn2+ because of the generation of H2O2 in the system used for MRI.256 Porphyrin-based MOFs can absorb the energy produced by the excitation of light and generates fluorescence for imaging.270 Gold nanoparticles, PLNPs, UCNPs, and semiconducting polymer (SP) nanoparticles are examples of templates in CMC mimics used for NIR imaging. Gold templates have a large NIR absorption cross-section and a tunable LSPR band in the NIR region, providing greater penetration depth in the imaging.226,276 PLNPs have a long-lasting near-infrared afterglow and avoid tissue autofluorescence from in situ excitation.68,72 SP nanoparticles have a high NIR absorption capacity.104 UCNPs have significant light penetration depth, narrow emission peaks, no background fluorescence, and exceptional photostability.132,277 Indocyanine green (ICG) is best known for NIR fluorescence imaging65 along with phototherapy.
Detoxification
Detoxification removes infections caused by pathogens. The RBC membrane alone278 or in combination with platelet membrane59 has toxin absorbing capabilities. There are also some templates/devices used in the CMC mimics to enhance the detoxification process differently. These include olive oil nanodroplets, Janus micromotors, redox-responsive hydrogels, and a 3D bioprinted nanoparticle-hydrogel hybrid device.
RBC membrane wrapped olive oil nanodroplets were used to form biomimetic oil nanosponges.77 In these nanosponges, the olive oil core soaked nonspecific toxicants through the physical partition, and RBC absorbed and neutralized toxicants through biological binding. They also found greater detoxification than that obtained with PLGA-RBC nanosponges. RBC membrane-coated antibiotic-loaded redox-responsive hydrogels (RBC-nanogels) were reported to absorb and neutralize the pore-forming toxins in the extracellular environment.114 This facilitated their intracellular uptake into the bacteria. Once entered within the bacteria, the cross-linked hydrogel cleaved to release the antibiotics to inhibit the bacterial growth. These redox-responsive hydrogels were more effective in inhibiting bacterial growth than the free antibiotics and nonresponsive hydrogels. Further, RBC membrane-coated Janus micromotors were used to improve the speed of absorption and neutralization of both nerve agent stimulants and biological protein toxins.76 The water-driven mimicking systems were designed by integrating RBC membranes, gold nanoparticles, and alginate (ALG) onto the exposed surface areas of magnesium (Mg) microparticles partially embedded in parafilm. This partial embedding leads to a small hole in the Mg particles. Hydrogen bubbles produced by the spontaneous redox reaction between Mg and water provided the guided propulsion without any external fuel. The 3D bioprinted nanoparticle-hydrogel hybrid device was designed with multiple inner channels for encapsulating many RBC nanoparticles.201 Many RBC nanoparticles in one device enhanced the detoxification process while at the same time absorbing various nonspecific toxins flowing through the channel.
Assembly of Cell Membrane-Coated Mimics
The most crucial step in designing a CMC mimic is the assembly of extracted cell membranes from the cell of interest with the template of choice that can incorporate its physiochemical properties to the CMC mimics. The isolated cell membrane may either be in the form of fragments or of vesicles. Before coating, it may be necessary to include an additional extrusion56,69,92 or sonication55,93,279,280 steps to form cell membrane vesicles (Figure 2). This section describes commonly employed CMC assembly techniques. We have also highlighted other less explored assembly techniques such as microfluidic electroporation, in situ polymerization, and graphene nanoplatform-mediated cell membrane coating. Additionally, we will emphasize the scope and challenges of assembly processes, manufacturing difficulties (reproducibility and scale-up), and limitations for clinical translation.
Extrusion
Producing uniformly sized particles by pushing material through a porous membrane of the desired cross-section is called extrusion.281 The formation of a wide range of nanomaterials like nanoparticles, liposomes, nanotubes, nanofibers, and emulsions use of extrusion technique is preferred. The commonly used membrane extrusion strategies are vesicle extrusion (for liposomes),282 membrane emulsification (for emulsions),283,284 precipitation extrusion (for nanofibers and nanoparticles),285 and biological membrane extrusion (for CMC mimics).62,286
For fabricating CMC mimics, a solution of the cell membrane vesicles and the template repeatedly passes through a porous polycarbonate membrane in a mini extruder. The mechanical force applied during the process disrupts the membrane structure and helps it to wrap around the template. In 2011, extrusion was reported for uniform coating of an RBC cell membrane onto a PLGA nanoparticle template through 400 and 100 nm polycarbonate porous membranes.62 Since then, several groups have reported this technique for assembling CMC mimics using different pore sizes of polycarbonate membranes, cells, and template types.56,69,130,132,186,235 After repeated extrusion, centrifugation separates the left/unbound cell membrane vesicles from the mixture. The main limitation of this technique is the loss of sample due to the accumulation of the material on the porous membrane, leading to difficulty in large-scale production.
Sonication
Sonication is the process of applying sound energy to disperse the particles in the liquid using an ultrasonic bath or probe sonicator. In this technique, both the cell membrane and the template are co-incubated, followed by sonication in ice-cold conditions for few minutes to fabricate CMC mimics. Sonication disrupts the cell membrane layer, and the noncovalent interactions between the template and the cell membrane facilitate their assembly. Several groups have reported this technique for CMC mimic assembly using different cell and template types, for example, RBC membrane coating onto cross-linked 2-hydroxyethyl acrylate (HEA) hydrogel microparticles;287 cardiac stem cell membrane onto PLGA microparticles;187 and stem cell,227 platelet,93 neutrophil membranes55 onto PLGA nanoparticles; and a hybrid of RBC and platelet membrane onto gold nanowires.196 After the sonication, centrifugation of the mixture separates the left/unbound cell membrane vesicles. Sonication, unlike the extrusion technique, avoids the loss of material during the coating process. It requires optimization of parameters like inputs of power, frequency, and time to avoid sample damage or denaturation of protein due to heat energy. However, the resulting particles may vary in size and uniformity of coating.187,227 This technique might also not be appropriate for soft/some templates, as it might affect their size and stability.288,289
In Situ Polymerization
In situ polymerization is a technique of preparing nanocomposites. It consists of polymeric molecules bound to nanoparticles290 (like carbon nanotubes, graphene oxide, etc.) or to biomolecules291 (like DNA, RNA, or proteins, etc.) in a reaction polymerization mixture to form linear conjugates or nanocapsules. The reaction mixture consists of a monomer, initiator, and a cross-linker, exposed to a source of heat or radiation to initiate the polymerization mechanism.
In 2015, this technique was reported using RBC membrane-derived vesicles as a nanoreactor to synthesize polymeric cores via in situ polymerization to prepare cell membrane-coated hydrogel nanoparticles.114,292 Membrane vesicles were prepared by extruding a mixture containing RBC ghosts, monomer (acrylamide), cross-linker (N,N′-methylene bisacrylamide), and an initiator (lithium phenyl-2,4,6-trimethylbenzolyphosphinate) through a polycarbonate membrane filter. A PEG-modified 2,2,6,6-tetramethylpiperidin-1-yl)oxyl (TEMPO) inhibitor was added to this solution to prevent cross-linking of monomers on the outside of the cell membrane vesicles. This inhibitor selectively promotes in situ cross-linking, protects outer cell proteins from denaturing, and inhibits nonspecific interactions and leaking of inner monomers across the cell membrane. Upon UV exposure for 5 min, monomers inside the cell membrane selectively polymerized to form a stable template at room temperature. This process is opposite to the traditional coating methods. It has the potential to be extended for other cross-linking mechanisms and materials and for templates-cell membrane combinations that are not currently feasible due to their unfavorable surface properties. However, preparing cell membrane vesicles using extrusion technique can lead to sample loss during large-scale production.
Microfluidic Electroporation
Electroporation is a high-throughput technique of incorporating nanoparticles within cells.293,294 In this technique, cells subjected to rapid high-voltage electric field pulses create temporary hydrophilic pores within the cell membrane.
In 2017, the microfluidic-assisted fabrication of CMC mimics was demonstrated using electroporation.57 An electroporation setup was integrated with a microfluidic chip with an S-shaped channel to facilitate efficient mixing of RBC vesicles and nanoparticles, fed through a Y-shaped polydimethylsiloxane microchannel. During electroporation, the pores formed with the cell membrane allow passive transport of nanoparticles within the RBC vesicles and fabrication of uniformly coated RBC-Fe3O4 nanoparticles with improved colloidal stability, uniform size, and in vivo efficacy. The advantages of this technique are in the autologous extraction of RBCs, allowing for personalized diagnosis and therapy. Scalability and storing capacity of this technique promote its feasibility for industry translation.
Graphene Nanoplatform-Mediated Cell Membrane Coating
In 2019, a single-step methodology for extraction and assembly of the leukocyte cell membrane was reported.295 The design aimed to increase the antileukocyte targeting ability of CMC mimics using a leukocyte cell membrane. The selective ability of graphene nanosheets to extract phospholipids from the cells vigorously was the innovative aspect of this CMC mimic. Initially, negatively charged Fe3O4 magnetic nanoparticles were modified with graphene and prepared by the layer-by-layer technique. A positively charged polyethylenimine (PEI) facilitated the immobilization of negatively charged graphene nanosheets onto Fe3O4 nanoparticles. The CMC mimics were assembled in a quick single-step process by co-incubating graphene-modified nanoparticles with leukocytes in serum-free media. High phospholipid content on the surface of CMC mimics helped to immobilize lipids for antibody conjugation to target epithelial cell adhesion molecule (EpCAM)-positive CTCs, for example, MCF-7 (the human breast cancer cell line) and HepG2 (human hepatocellular cancer cell line). They also demonstrated CMC mimics’ selectivity with a very high antileukocyte targeting efficacy when tested in synthetic samples (blood mixed with green fluorescent protein (GFP)-MCF-7 cells). The advantage of this protocol is the selective extraction and immobilization of phospholipids from different cell types296 and the efficient separation and proliferation of the captured CTCs for several passages. Further, it is possible to use these CTCs to design a biomimetic system with homotypic target abilities.
Characterization of CMC Mimics
Physiochemical Characterization
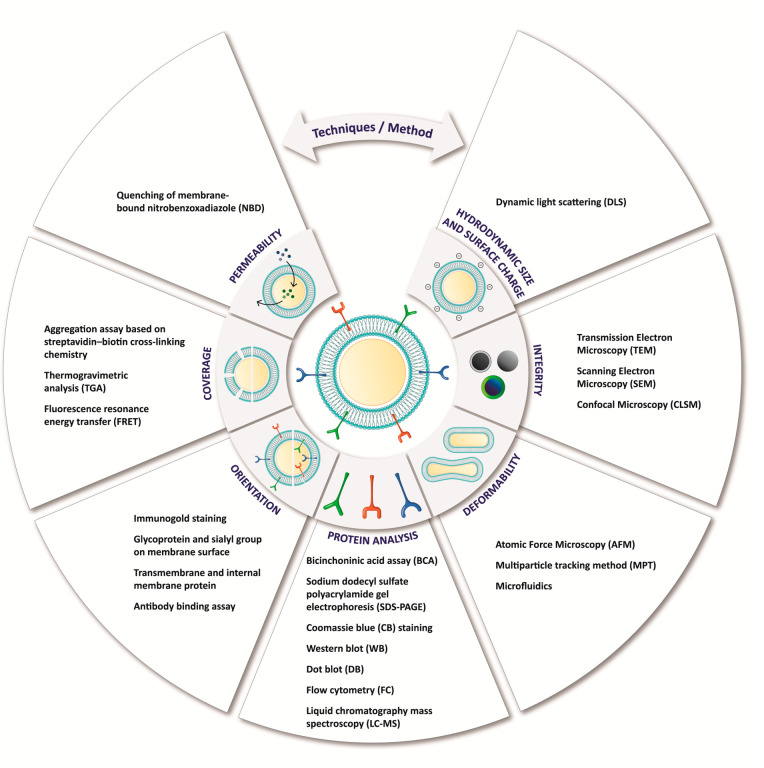
After the fabrication of a CMC mimic, it is essential to analyze its structural features involving the cell membrane and template interface to enhance its colloidal stability. The incomplete/unstable membrane may lead to template exposure and impair the effectiveness of the CMC mimics. Therefore, it is critical to perform qualitative and quantitative evaluations of their structural integrity. The quantitative determination of the number of templates coated with the cell membrane remains unexplored even though this is an important parameter for its clinical translation. In this section, we collate all reported physiological techniques to quantify and visualize thickness, uniformity, stability of the cell membranes, and deformable and permeability properties of CMC mimics post-assembly (Figure 4).
Figure 4.
The schematic illustrates the qualitative and quantitative physicochemical and biological properties of CMC mimics that validate their formation. Some essential parameters need to be considered while designing CMC mimics like surface charge, the thickness of cell membrane-coated onto a template, elasticity, protein quantification and identification of right orientation, amount and area of cell membrane covered onto a template, and permeability of the mimics for the diffusion process. This helps in confirmation and visualization of the cell membrane with right-side-out in CMC mimics. The schematics also list the methods and instruments used for characterizing specific physicochemical and biological properties of CMC mimics.
Size and Surface Charge
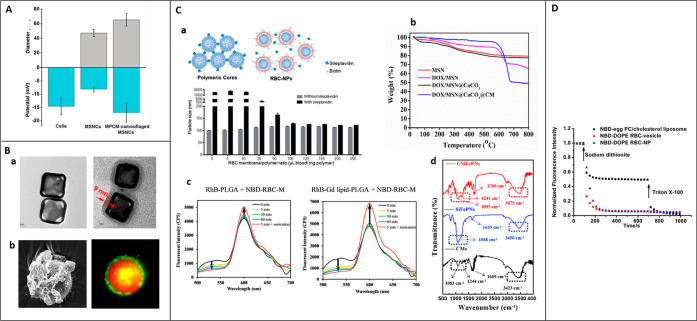
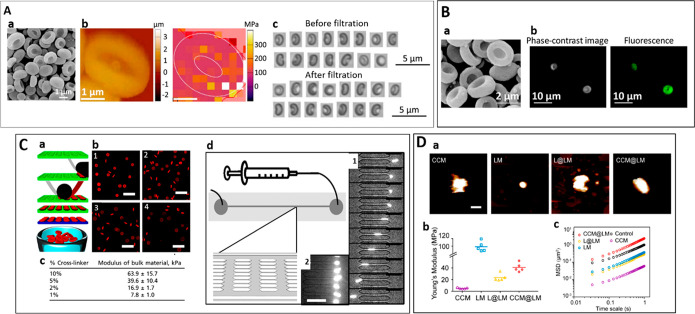
Size and surface charge are two parameters monitored in real-time during the assembly of a CMC mimic. Size (hydrodynamic radius) and surface charge of CMC mimics can be measured using a dynamic light scattering (DLS) analyzer and zeta sizer (Figure 5A).
Figure 5.
Surface charge, hydrodynamic diameter, coverage, and permeability. (A) Size and surface charge of mesoporous silica nanocapsules (MSNCs) and macrophage cell membrane (MPCM)-coated MSNCs. Reprinted with permission from ref (69). Copyright 2015 John Wiley and Sons. (B) Structural integrity: (a) TEM image of gold nanocages (left) and H22 liver cancer cell membrane-coated gold nanocages (right), scale bar = 20 nm. Reprinted with permission under a Creative Commons CC BY-NC License from ref (226). Copyright 2018 Ivyspring International Publisher. (b) Cardiac stem cell membrane fragments-coated onto PLGA microparticles, SEM (left), confocal microscopic images (right) microparticles (texas red), cell membrane (DiO, green), scale bar = 20 μm. Reprinted with permission under a Creative Commons CC-BY License from ref (187). Copyright 2017 Springer Nature. (C) Coverage: (a) Aggregation assay based on streptavidin-biotin cross-linking chemistry, size change of RBC-NPs (RBC-coated PLGA nanoparticles) due to streptavidin-biotin cross-linking at various RBC membrane-to-polymer ratios. Reprinted with permission under a Creative Commons CC-BY License from ref (300). Copyright 2014 Royal Society of Chemistry. (b) TGA curves of MSN, doxorubicin-loaded MSN (DOX/MSN), DOX loaded MSN with calcium carbonate interlayer (DOX/MSN@CaCO3), prostate cancer cell membrane-coated DOX/MSN@CaCO3 (DOX/MSN@CaCO3@CM). Reprinted with permission from ref (225). Copyright 2019 Elsevier. (c) FRET study to demonstrate the insertion of RBC-M onto the gadolinium inserted poly(lactic-co-glycolic acid) nanoparticles (Gd-PLGA) and bare PLGA. Reprinted with permission from ref (71). Copyright 2020 Royal Society of Chemistry. (d) FT-IR spectra of CMs (MDA-MB-231 cell membranes), SiFePNs (MSNs containing superparamagnetic ferroferric oxide), and CSiFePN (cell membrane coated mimics). Reprinted with permission under a Creative Commons CC-BY License from ref (247). Copyright 2019 Springer Nature. (D) NBD fluorescence quenching to study and compare the permeability of RBC vesicles, RBC-NPs (RBC coated PLGA nanoparticle), liposome (egg PC/cholesterol). Reprinted with permission from ref (302). Copyright 2014 Royal Society of Chemistry.
Post-assembly of CMC mimics, it is typical to note a negative surface charge close to that of the cell membrane and a few nanometers increase in their size, confirming the coating.64,94,100,198,226,257 Measuring size pre and post-CMC assembly by using DLS helps to determine the thickness of the cell membrane. However, the thickness of outer membrane coating can vary depending on the number of layers and their extent of fusion with the template.63 In this section, we have mentioned a few variations of the template and cell membrane thickness in different CMC mimics.
For example, a T cell membrane-coated PLGA system was reported with an observed size change from 88.3 ± 1.3 nm to 105.4 ± 4.4 nm (thickness ∼17.1 nm) and a surface charge of −29.5 ± 1.2 mV similar to that of the cell membrane.159 In T1 cell membrane-coated cerium oxide dotted CS, there was an increase in 20 nm size from 131.7 ± 5.2 nm to 152.8 ± 3.9 nm post-assembly with −26.1 ± 0.9 mV ζ potential after cell membrane coating.297 In monocyte membrane (U837)-coated PLGA systems, there was an increase of size by ∼20–40 nm with −16.5 mV ζ potential (PLGA: −8.3 mV; U837: −13.6 mV).298 In MDA-MB-231 cell membrane-coated mesoporous silica loaded with ferric oxide, an increase in average size from 164 to 220 nm (thickness ∼56 nm) with surface charge −20.88 ± 0.4 mV post-assembly was observed.247 In fact, in MCF-7 cell membrane-coated mesoporous silica PEG-liposomes, the size change from 74.07 ± 0.7 nm to 188.5 ± 3.3 nm (thickness ∼114 nm) with −23.8 ± 1.1 mV surface charge closer to that of the cell membrane was observed.175 In addition to using a DLS analyzer, the thickness and coverage of the cell membrane on each template can be visualized using microscopic techniques discussed in the next section.
Structure Integrity
Different microscopic techniques help to visualize the structural integrity of CMC mimics. The three microscopic techniques most often used to gain insight into the structural integrity and uniformity of assembled CMC mimics are cryo-transmission electron microscopy (cryo-TEM) or TEM, field-emission scanning electron microscopy (FESEM) or SEM, and confocal laser scanning microscopy (CLSM) (Figure 5B).
CMC mimics have a characteristic core–shell structure. This consists of a dense inner core of a template and a thin outer coating of a cell membrane. Due to differences in composition, there is a difference in electron density between these two layers. However, TEM imaging visualizes the structure containing a dark core and a light outer coating. The variation in the thickness of the cell membrane in different CMC mimics was also visualized in TEM analysis, same as observed in the above section. For example, in the leukocyte membrane-coated silica-APTES system, the outer layer thickness could reach 500 nm by increasing the membrane:particle ratio.63 For leukocyte cell membrane-coated Fe3O4-PEI-graphene-modified nanoparticles, variable thickness of the membrane around 11.78 and 16.94 nm was observed.295 In case of hybrid RBC and MCF-7 breast cancer cell membrane-coated melanin nanoparticles, an ∼9.1 nm-thick membrane was reported.198 Similarly, for 4T1 cancer cell membrane-coated MOFs257 and RBC membrane-coated PFCs nanoparticles,269 an ∼10 and 20 nm-think membrane was observed, respectively. Overall, TEM can provide a qualitative estimation of the membrane homogeneity around the template post-coating, mostly in the case of nanoscale CMC mimics.
SEM is another qualitative technique used to visualize the change in surface morphology/texture after the cell membrane coating and complete/incomplete coverage of cell membrane, predominantly in the case of microscale CMC mimics. For example, using SEM, the complete coverage of leukocyte membrane onto APTES-silica microparticles63 and incomplete coverage of cardia stem fragments onto PLGA microparticles were observed.187 In case of motor sponge designed using RBC membrane and gold nanowires of 400 nm diameter and 3 μm in length, no change in the concave end of the nanowire was observed after complete RBC membrane coating.299 Uniform RBC membrane coating onto Mg Janus motors was observed with spherical geometry of size 20 μm and a small circular opening of 2 μm.76
CLSM is a qualitative technique that provides an insight into the efficiency of the whole process of assembly by determining an estimation of the number of templates that are well coated. For CLSM, the cell membrane and the inner core are fluorescently tagged with different dyes to aid visualization. 3,3′-Dioctadecyloxacarbocyanine perchlorate (DiO),187,211,219,235 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine perchlorate (DiD),75,175,185 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (DiI),239 fluorescein,55 rhodamine-DMPE (1,2-bis(dimethylphosphino)ethane),62 wheat germ agglutinin,63 rhodamine B (RhB),299 CellVue,287 and Cy5.560 are examples of dyes exclusively used to label cell membranes.
Coverage
The coverage of cell membrane onto the template can be validated using an aggregation assay based on streptavidin-biotin cross-linking ability, Förster resonance energy transfer (FRET) analysis, thermogravimetric analysis (TGA), and Fourier transform infrared (FT-IR) spectroscopy (Figure 5C).
In 2014, the completeness of RBC membrane was demonstrated onto PLGA nanoparticles using the streptavidin-biotin cross-linking chemistry (aggregation assay).300 The RBC membrane-coated biotinylated PLGA nanoparticles were mixed with the streptavidin and monitored for a change in particle size. When the membrane coverage is low, exposed biotin on the PLGA surface binds to streptavidin and induces significant aggregation and size change. Therefore, the suitable membrane to polymeric ratio can be optimized when the aggregation or size changes upon the addition of streptavidin impede. Such an aggregation assay helps in determining the efficiency of membrane coating in CMC mimics.
TGA technique measures the percentage loss in weight of samples when heated.301 The TGA profiles can differ from material to material (template, cell membrane, and the final CMC mimic). The difference in percentage weight loss between the template and the CMC mimic determines the amount of cell membrane-coated onto a template. It also helps to study the stability of the membrane onto a template. The temperature range in TGA depends on the thermal degradation of the samples. For example, the amount and stability of leukocyte membrane coating onto APTES-silica microparticles were demonstrated in the range from 30 to 150 °C. About 15 wt % membrane and 8.0 wt % membrane were observed after 1 and 24 h incubation of CMC mimics in PBS, respectively.63 In case of U-251 MG glioblastoma cell membrane-coated magnetic nanocubes (a hybrid of Fe3O4 and MnO2), particles were heated from 100 to 600 °C and observed 12% weight of cell membrane absorbed on nanocubes.273 Similarly, for LNCaP-AI prostate cancer cell membrane-coated CaCO3 capped mesoporous silica nanoparticles (MSN@CaCO3), around 30% weight of cell membrane was absorbed onto MSN@CaCO3 after heating samples from 25 to 800 °C.225
FT-IR spectroscopy is another versatile technique to qualitatively characterize cell membrane coating by comparing spectra before and after CMC assembly. Only a few reports mentioned this technique to confirm CMC mimics assembly are discussed in this section. The sharp IR characteristics peaks of the template weaken after the cell membrane coating confirms the immobilization of the membranes on the template surface. Along with this, additional strong stretching vibration peaks of C–H/C–N/C=O/N–H/O–H of cell membrane phospholipids, proteins, and carbohydrates are observed if there are no overlaps with the template characteristic peaks itself.
For example, the U-251 MG glioblastoma membrane coating was confirmed onto magnetic nanocubes by observing an additional peak in the range of 1500–3000 cm–1.273 The additional broad peak at approximately 1750 cm–1 was due to the C–N and C=O vibrations of the cell membranes. In the case of MDA-MB-231 breast cancer cell membrane-coated mesoporous silica nanoparticles (SiFePNs), an additional peak was also observed around 2950 cm–1 by the C–H band stretching vibration of the methyl group of cell membrane phospholipids and weakening of the sharp IR peaks of SiFePNs after coating.247 The characteristic peak of C=O and −NH2 in the spectra was reported to confirm the RBC membrane coating onto persistent luminescence nanocarriers.68 On occasion, additional peaks can also be attributed due to chemical interaction between the cell membrane and the template surface. For example, in the leukocyte membrane-coated APTES-silica microparticles, two strong peaks were observed at 1652 and 1544 cm–1 that correspond to the amide I and II modes of the proteins of the membrane.63 The C=O stretching vibrations of the peptide bonds lead to the amide I band, while the C–N stretch coupled with N–H bending mode leads to the amide II band. This amide linkage arose from the peptide bonds between protein residues and the covalent bond between the carboxylic moieties of protein and the primary amines of the APTES molecule in the microparticles. The spectra of CMC mimics also exhibited a weak peak for the Si–O moieties and a strong peak for the C–H stretching compared to that of the APTES-silica particles spectra. This leads to the immobilization of the membranes on the particle’s surface, shielding the silicon surface while exposing the long C–H and C–C chains of phospholipids and proteins.
FRET is another assay used to characterize CMC mimics as they assemble using one or two different cell membranes. In this study, each cell membrane labels with a donor or acceptor fluorescent dye from FRET pairs. 7-Nitrobenz-2-oxa-1,3-diazol-4-yl (NBD) and RhB (rhodamine), DiI, and DiD are the commonly used FRET pairs. This study identifies the molecular distance between the fluorophores using the energy transfer mechanism. Donor energy minimizes when energy transfer occurs from the donor to the acceptor when they are nearby.
For example, fusion of NK cell membrane and liposome to create CMC mimics was validated using a FRET study.102 The two sets of liposomes were tagged with fluorescence donor (PE-NBD) and fluorescent acceptor (PE-RhB) and mixed them with NK cell membrane. As the coating progressed, FRET was masked by the NK cell membrane, resulting in a decrease in the fluorescent intensity of the acceptor and an increase in the fluorescent intensity of the donor. Similarly, FRET study has been used to confirm fusion of various cell membranes, for example, the fusion of platelet and RBC membrane using DOPE-RhB/C6-NBD-doped platelet membrane,97 fusion of the B16-F10 cancer cell membrane and RBC membrane using DiD/DiO-doped B16-F10 cancer cell membrane,100 fusion of liposomes and RBC membrane using DHPE-RhB/C6-NBD-doped liposomes,239 and the fusion of RBC and MCF-7 cancer cell membrane using DOPE-RhB/C6-NBD-doped MCF-7 cancer cell membrane.198
FRET also helps to validate the fusion of the cell membrane onto a template. In this case, the cell membrane and template are both doped with a FRET pair. For example, the fusion of RBC membrane onto PLGA was confirmed by doping the RBC membrane with NBD-PE and PLGA with RhB-egg phosphatidylcholine (PC).71 As the distance between the NBD-labeled RBC membrane and RhB-labeled polymeric core becomes shorter, NBD-energy donor fluorophore becomes more efficient in donating energy to RhB-energy acceptor fluorophore, therefore increasing the fluorescent intensity of the acceptor and decreasing the fluorescent intensity of the donor.
Membrane Permeability
The permeability of the cell membrane in the CMC mimics has been little explored. The cell membrane in the live cells is permeable in nature and controls the in and out of the ions using their ion pumps. It helps to understand the better control of the drug encapsulation and release in CMC mimics. The permeability of RBC-PLGA systems was investigated by using the cell membrane permeable molecular probe.302 The permeable molecular probe succinimidyl 6-(N-(7-nitrobenz-2-oxa-1,3-diazoyl-4-yl)- amino)hexanoate (NBD-NHS) was used to label both the sides of the RBC membrane and dithionite (S2O42–) ion for quenching the NBD fluorescence. It was observed that the RBC-PLGA has a higher permeability to the dithionite ions than to living RBC cells and egg-PC/cholesterol liposomes (Figure 5D).
Deformability
Deformability is a vital design parameter that affects the behavior of particles on both the micro and nanoscales.303,304 It is mostly dependent on the shape and average particle elasticity. Atomic force microscopy (AFM) and compression testing machine/universal testing machines are some of the techniques used for measuring the mechanical properties of CMC mimics. The multiparticle tracking (MPT) method and microfluidic technique help to validate and visualize the deformability of CMC mimics.
There are ongoing efforts to incorporate the elastic properties (Figure 6) in the designed particle system for enhancing mobility and biodistribution in animal studies. For example, RBC-shaped microparticles (RBC-MPs) were prepared using an electrospinning-based technique and showed intraparticle elasticity (IED), which was measured by AFM.305 The Young’s modulus (E) of the dent in the RBC-MPs was <100 MPa, whereas that of the thick rim was 100–300 MPa. The difference in the E value of the dent and the rim in RBC-MPs shaped the particles. This shape helped these particles to deform and retain their original shape after passing through a membrane filter with 1 μm pores. These RBC-MPs also showed less accumulation in the lungs and the spleen. The same group used these RBC-MPs for RBC membrane coating to mimic the shape and the surface structure of RBC for increasing its circulation time in blood.287
Figure 6.
Validation of elastic properties in CMC mimics. (A) RBC-MPs using electrospinning-based technique: (a) SEM, scale bar = 1 μm; (b) IED of RBC-MPs, scale bar = 1 μm; (c) images of RBC-MPs before and after filtration, scale bar = 5 μm. Reprinted with permission from ref (305). Copyright 2018 Elsevier. (B) RBC membrane-coated RBC-MPs mentioned above (a) SEM; (b) phase contrast and CellVue labeled cell membrane-coated RBC-MPs. Reprinted with permission from ref (287). Copyright 2018 American Chemical Society. (C) Disc shaped RBC microparticles (RBCM) replication in: (a) nonwetting templates (PRINT) technique, cross-linked by PEGDA, (b) fluorescence imaging of RBCM varying percent cross-linker: (1) 10% cross-linked, (2) 5% cross-linked, (3) 2% cross-linked, and (4) 1% cross-linked, scale bar = 20 μm; (c) bulk modulus of different cross-linked RBCM; (d) microfluidic evaluation of deformability: (1) 1% cross-linked RBCM and (2) 10% cross-linked RBCM, scale bar = 30 μm. Reprinted with the permission from ref (26). Copyright 2011 United States National Academy of Sciences. (D) MCF-7 breast cancer cell membrane-coated silica nanoparticles-supported PEGylated liposomes: (a) AFM deformation images of CCM, LM (silica nanoparticles-supported PEGylated liposomes), L@LM (1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC)-coated LM, CM@LM (cancer cell membrane-coated LM), scale bar = 100 nm; (b) Young’s modulus of nanoparticles; (c) MSD values of nanoparticles in a simulated tumor ECM. Reprinted with permission from ref (175). Copyright 2020 American Chemical Society.
In 2011, the Prof. DeSimone group also prepared tunable elastic RBC-shaped hydrogel microparticles (RBCMs) in the nonwetting templates (PRINT) technique.26 They tested their mechanical properties (bulk modulus) with a Universal Testing Machine (Instron) with a strain rate of 5 mm/min. They achieved tunable modulus of the hydrated polymer from 63.9 to 7.8 KPa by varying the cross-linker from 10% to 1%, respectively, which overlapped with the reported modulus of RBCs (26.7 KPa). They successfully demonstrated the deformability of RBCMs under the flow condition using microfluidic models of vascular constriction. Microfluidics has also widely been used to study the deformability of cells like RBCs,306 leukocytes,307etc. Therefore, microfluidics can also be explored to analyze and visualize the flexibility of CMC mimics.
Recently, the mechanical properties of the yolk–shell structured MCF-7 cancer-cell-membrane-coated mesoporous silica nanoparticles supported liposome (CCM@LM) was validated using AFM and demonstrated using MPT method.175 These yolk–shell structures showed moderate rigidity with young’s modulus around 40 MPa. During filtration, these CMC mimics could also transform into an ellipsoidal shape. These properties facilitated its penetration through spheroids in vitro. They evaluated its ECM diffusion capability using the MPT method in which the MPT medium was collagen (I) hydrogels. The MSD value of CCM@LM was approximately 7.1- and 2.6-fold higher than that of LM and PLGA nanoparticles, respectively. Incorporating the elastic properties in CMC mimics can enhance its mobility and penetration in the tumors. This property in CMC mimics needs to be explored more in-depth.
Biological Characterization of CMC Mimics
The cell membrane provides surface functionality to the CMC mimics that allows communication with other cells and helps to escape from macrophages and circulate more in the bloodstream. For example, the CD47 receptor on the RBC membrane selectively binds to signal-regulatory protein alpha (SIRPα) glycoprotein expressed by macrophages to prevent its uptake.109,110 Therefore, for CMC mimics to function efficiently, the cell membrane must maintain the right orientation post-coating and contain the maximum amount of translocated protein on its surface. The isolated membrane should have minimal nuclear, mitochondrial, and cytosol contamination, and the transmembrane proteins must face outward for active targeting and accumulation at the intended site. Improper membrane orientation (integral protein exposed onto the other surface of the mimics) will affect cell-to-cell communication, overall function, and risk of macrophage detection and causes unwanted side effects. Therefore, proper qualitative and quantitative evaluation of intact membrane proteins, purity, and orientation is required in CMC mimics to improve their functional efficiency and reproducibility for clinical translation. All the properties are also summarized in Figure 4.
Protein Analysis
The protein composition and expression on the isolated cell membrane and CMC mimics can be analyzed, identified, and quantified using several techniques. These include bicinchoninic acid assay (BCA)/Bradford assay, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), Coomassie brilliant blue staining, Western blot/dot blot, flow cytometry, and liquid chromatography-mass spectroscopy (LC-MS) (Figure 7).
Figure 7.
Protein analysis. (A) BCA assay. Quantification of protein concentration of gold nanoparticles before and after bacterial cell membrane coating. Reprinted with permission from ref (94). Copyright 2015 American Chemical Society. (B) Protein profile and identification using (a) SDS PAGE and (b) Western blot on 4T1 breast cancer cell membrane-coated iron oxide nanoparticles. Reprinted with permission from ref (73). Copyright 2016 American Chemical Society. (C) Protein identification using (a) dot blot (CD47, CD41a) in discontinuous sucrose gradient solution and (b) flow cytometry (CD42b, CD47, CD41, and CD61) on platelet membrane-coated APTES-Si (3-aminopropyl triethoxysilane-silica) microparticles. Reprinted with permission from ref (213). Copyright 2016 Elsevier. (D) LC-MS, analysis of human NK cell membrane in NK-NPs (NK cell membrane-coated nanoparticles): (a) the total number of protein and plasma-membrane associated protein detected on NK-NPs, (b) a pie chart of the proteins identified on NK-NPs according to UniProt and GO information and the literature, and (c) flow cytometry analysis to validate the presence of specific proteins on NK-NPs. Reprinted with permission from ref (150). Copyright 2018 American Chemical Society.
Before the protein analysis, the proteins need to be extracted from the isolated cell membrane and CMC mimics using lysis buffer (for example, radioimmune precipitation (RIPA) buffer). The lysis buffer should be supplemented with protease and phosphatase inhibitor cocktail and phenylmethylsulfonyl fluoride (PMSF) to prevent degradation of proteins and stored at −80 °C for protein analysis. BCA assay and Bradford assay are most commonly used for colorimetric detection and quantification of total protein concentration from various cell membrane and CMC mimics.65,67,100,150,195,308,309 Although most of the studies have discussed the use of these assays, there are only a few reports that have mentioned the amount of total protein translocated onto the isolated cell membrane or CMC mimics. In addition, there are no standards reported on total protein content that should be present on CMC mimics for their therapeutic effects. For example, around 300 mg of protein content was found in NK cell (NK-92) membranes extracted from 1 × 107 cells using the Bradford assay.102 A 7.9 ± 2.0 wt % protein loading yield of the bacterial cell membrane onto the gold nanoparticles,94 2.8 ± 0.5 wt % protein loading yield of the MIN6 cell membrane onto fibers,96 and 18.6 ± 5.7 wt % protein loading yield of the RBC membrane coating onto mesoporous silica nanoparticles61 were found using BCA assay. In fact, determination of the protein content before and after the cell membrane coating can also provide an insight into validate the membrane coating onto a template.
The protein profile in the cells, isolated cell membrane, and CMC mimics can be visualized, analyzed, and compared qualitatively by loading and running the same amount of protein in a specific % of SDS-PAGE gel that allows the separation of protein-based on mass.99,150,207,277 After the separation of proteins, these gels can be stained with irreversible Coomassie brilliant blue dye. This binds nonspecifically to proteins because of ionic interactions between sulfonic acid groups and positive protein amine groups through van der Waals attractions and appears as blue protein bands.63,87,106,175,235,310 The intensity of the blue protein bands helps to compare the total protein profile translocated from a natural cell to the isolated cell membrane and CMC mimics. This is the basic analysis that is reported in almost every literature to validate the successful coating of cell membrane onto a template.
Western blot is the most widely used technique to identify and compare the expression of specific proteins from among a mixture of proteins on the cell lysate, isolated cell membrane, and CMC mimics. For example, using Western blot, comparable presence of CD47 receptor on natural RBC lysate, RBC membrane and RBC membrane-coated Fe3O4 nanoparticles was observed.211 Similarly, comparable DNAM-1, NKG2D, and CD56 (neural cell adhesion molecule receptors) on the NK cell lysate, NK cell membrane, and its membrane-coated PLGA nanoparticle67 and mPEG-PLGA nanoparticle150 were found, respectively. In the case of hybrid RBC and MCF-7 cancer cell membrane-coated melanin nanoparticles,198 comparable RBC-specific membrane proteins (band 3, GPA, CD55, and CD47) and MCF-7 specific membrane proteins (EpCAM, N-cadherin, galectin-3) on hybrid RBC-MCF-7 vesicles and its membrane-coated mimics were also observed. The enrichment of protein-like clusters of differentiation receptors (CD11c, CD86, CD40) expression was found on the dendritic cell membrane and its membrane-coated PLGA nanoparticles than dendritic cell lysate.106 Similarly, the significant protein enrichment for LPS binding proteins (CD14, TLR4), cytokine binding receptors (CD126 and CD130 for IL-6, CD120a, and CD120b for TNF, and CD119 for IFN-γ) was observed on macrophage membrane and membrane-coated PLGA nanoparticles than macrophage cell lysate.130 Similarly, the enrichment of surface protein like TNF alpha R, IL-1R, LFA-1 receptors was also observed on the neutrophil membrane and its membrane-coated PLGA as compared to the neutrophil lysate.66
Dot blot is another blotting technique that requires only a few microliter samples directly onto the PVDF or nitrocellulose membrane followed by blotting procedures. Dot blot is a quick and fast technique used to identify the position of isolated membrane fraction in the sucrose gradient using specific protein markers. For example, leukocyte cell membrane was isolated using 55–40–30% sucrose (w/v) gradient.63 The gradient was divided into 10 different fractions, analyzed for the specific markers using dot blot, and found the lipid ring between 40 and 30% sucrose interface (fractions 5 and 6) enriched with plasma membranes. Similarly, a 44–40–5% sucrose gradient was performed and divided the gradient into eight fractions for the dot blot analysis213 and found that the majority of platelet membranes were present in the lipid ring between 5 and 40% sucrose interfaces. There are very few reports that have used dot blot to identify the expression of a specific protein on the isolated cell membrane and CMC mimics because it does not provide information on the actual size of the target protein like a Western blot. For example, the presence of CD47 and glycophorin A (GPA) on the RBC membrane and its coated PLGA-Gd nanoparticles was reported using both dot blot and Western blot.71 Western blot or dot blot also helps to determine the purity of isolated cell membrane by using nuclear or mitochondrial or cytosol specific antibodies that help to assist in detecting its content in the membranes. For example, histone H3209,257 or nucleoporin p6263 antibodies as a nuclear marker, cytochrome c-oxidase (COX IV)63,209,257 or ATP5a224 antibody as a mitochondrial marker, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH)209,224,257 as cytosol marker and P-cadherin92 or Na+/K+-ATPase73,92,150 as plasma membrane markers have been used.
Flow cytometry is a powerful qualitative and quantitative technique, useful to identify and quantify specific protein on the CMC mimics by measuring the fluorescent intensity. Only a few reports explore this technique for some protein analysis. For example, the fluorescence intensity of CD42b, CD47, CD41, and CD61 on platelet membrane-coated APTES modified silica nanoparticles213 and the fluorescence intensity of CXCR4 on U87 cancer cell membrane-coated PLGA nanoparticles224 was identified. An equal amount of LFA-1 receptor on a neutrophil membrane-coated PLGA nanoparticles and neutrophil cells66 was observed. Similarly, the comparable fluorescence intensity of MHC-II protein on dendritic cell membrane-coated mimics and natural dendritic cells with an equal amount of MHC-II surface protein was observed.106 Therefore, this technique has a lot of potential to analyze and compare the amount of specific protein on CMC mimics with the natural cell in a given batch that may overcome the reproducibility issues.
LC-MS/proteomics analysis is a quantitative large-scale protein study. This study involves fractionation of complex peptide or protein mixtures, acquiring the data necessary to identify individual proteins using mass spectroscopy, and finally, analyzing and organizing the mass spectroscopy data using bioinformatics.311 The total number of proteins identified can be further characterized based on their cellular function (integral or peripheral plasma proteins, cytoskeletal or junctional protein), biological process (transport, immunity, cell–cell adhesion, developmental process, proteolysis, lipid metabolism, etc.), and molecular function (GTP (guanosine triphosphate) binding, protein binding, GTPase activity, GDP (guanosine diphosphate) binding, actin-binding, etc.).150,239,312 For example, the shotgun proteomics method identified 868 distinct proteins on human NK cell membrane-coated mPEG-PLGA nanoparticles.150 They also analyzed specific proteins on CMC mimics like immunity-related GTPase family M protein 1 (IRGM1), cannabinoid type 1 receptor (CB1), ras-related protein encoded with RAB10 gene (RAB 10), and receptor activator of nuclear factor κ-Β ligand (RANKL) involved in the polarization of M1-macrophages and NKG2D, DNAM-1 in targeting tumor cells. 2215 common proteins were identified on U-251 MG cell membranes and their coated magnetic nanocubes.273 Those identified proteins were clusters of differentiation 59 (CD59), epidermal growth factor receptor (EGFR), CD44, tight junction protein 1 (TJP1), myosin light-chain kinase (MYLK), and others. A label-free quantification proteomics method identified 474 membrane proteins on RBC membrane-coated PLGA nanoparticles313 and 148 common proteins in erythroliposomes (a hybrid of RBC vesicles and liposomes) and RBC vesicles.313 All the research papers reported above have provided a long list of proteins identified on the cell membrane and their CMC mimics.
The Orientation of Cell Membranes (Right-Side-Out)
The orientation of the cell membrane in the CMC mimics helps in determining the direction of the receptors while coating onto a template. The extracellular proteins must be directing outward, whereas the intracellular proteins should be directing inward, to maintain the functionality of the CMC mimics. Among the techniques/method for studying the orientation are immunogold staining, antibody binding assays, quantification of glycoproteins or sialic acid content, and quantification of transmembrane or internal membrane proteins (Figure 8).
Figure 8.
Protein orientation. (A) Immunogold staining: (a) TEM images to identify CD235a (red arrows, large gold) and CD61 (orange arrows, small gold) on RBCNP (RBC membrane-coated PLGA), [RBC-P] NP (hybrid RBC and platelet membrane coated PLGA), and PNP (platelet membrane-coated PLGA). Scale bar = 50 nm. Reprinted with permission from ref (97). Copyright 2017 John Wiley and Sons. (b) TEM images to identify CD47 (red arrow, large gold nanoparticle) and CD340 (blue arrow, small gold nanoparticle) on melanin nanoparticles, melanin@RBC, melanin@MCF-7, melanin@RBC-MCF-7 (hybrid). Scale bar = 50 nm. Reprinted with permission from ref (198). Copyright 2019 Elsevier. (c) TEM image of Si/PNPs@Hela (siRNA/PTX coloaded PLGA nanoparticle) stained with extracellular-domain specific AuNPs-AS1411. Scale bar = 100 nm Reprinted with permission under a Creative Commons CC-BY License from the ref (235). Copyright 2020 Ivyspring International Publisher. (B) Antibody binding assay. TEM images of RBC-AuNPs (RBC membrane-coated gold nanoparticles) with (a) anti-CD47 (exoplasmic) antibody-modified polystyrene microspheres, (b) anti-CD47 (cytoplasmic) antibody-modified polystyrene microspheres, control groups, and AuNPs alone, (c) anti-CD47 (exoplasmic) antibody-modified polystyrene microspheres, and (d) CD47 (cytoplasmic) antibody-modified polystyrene microspheres. Scale bar = 1 μm. Reprinted with permission from ref (286). Copyright 2013 John Wiley and Sons. (C) Sialyl acid and glycoprotein content on the membrane. (a) Comparison of relative glycoprotein content and sialyl acid recovered from RBC ghost, RBC-NPs (RBC-coated PLGA nanoparticles), bare NPs (PLGA). Reprinted with permission under a Creative Commons CC-BY License from ref (300). Copyright 2014 Royal Society of Chemistry. (b) Glycoprotein content on RBC vesicles, motor sponges (RBC membrane-coated gold nanowires), and bare motors (gold nanowires). Reprinted with permission from ref (299). Copyright 2015 John Wiley and Sons. (D) Transmembrane and internal membrane proteins identification. (a) Transmembrane fluorescence intensity (PE-labeled antimouse MHC II antibody) of bone marrow-derived dendritic cells (BMDC) and mini DC (mature dendritic cell membrane-coated PLGA). Reprinted with permission under a Creative Commons CC-BY License from ref (106). Copyright 2020 Wiley-VCH. (b) Transmembrane fluorescence intensity comparison measured in neutrophils and neutrophils-NPs (membrane-coated PLGA nanoparticles) stained with allophycocyanin antimouse LFA antibodies. Reprinted with permission from ref (66). Copyright 2018 Springer Nature. (c) Fluorescence intensity ratios after immunofluorescence staining of intracellular (IgG-FITC) and extracellular domains (IgG-Cy3) of CXCR4 receptor on 4T1 cancer cell membrane (CCCM) and cell membrane-coated on magnetic iron oxide nanoparticles loaded with doxorubicin (MNP@DOX@CCCM). Reprinted with permission from ref (73). Copyright 2016 American Chemical Society.
Immunogold staining or labeling is a qualitative technique used in electron microscopy for the identification and distribution of a specific protein of interest located on the interior or the exterior of the cell membrane of the CMC mimics. It utilizes gold nanoparticles conjugated with the secondary antibody of interest and, in turn, attaches to the primary antibody designed to bind a specific protein on the CMC mimic. The gold nanoparticles are visible as a black spot under electron microscopy and help visualize the arrangement of a specific protein onto CMC mimics. Some of the reported specific biomarkers on RBC membrane are CD47198 and CD235a,97 platelet membranes are CD6197 and CD47,93 and MCF-7 breast cancer cell membrane is CD340198 that have been used in the immunogold staining and analyzed under TEM. The orientation of the HeLa membrane onto PLGA nanoparticles was also demonstrated using immunogold staining, but tagged the gold nanoparticles with AS1411, a nucleic acid aptamer that targets the extracellular region of nucleolin in the cell membrane and is analyzed under a transmission electron microscope.235
Antibody binding assay was reported to examine the coating and sidedness of RBC membrane onto gold nanoparticles.286 Two distinct anti-CD47 antibodies (exoplasmic and cytoplasmic) conjugated polystyrene microspheres were used for the interaction with the membrane and further analyzed under TEM.
Glycans and sialic acid groups are asymmetrically distributed on the extracellular side of the cell membrane. Hence, their quantification has also been reported for validating the sidedness of the RBC membrane in a CMC mimic.211,299,300 The glycoprotein content and terminal sialic groups were first enzymatically removed from CMC mimics using trypsin and sialidase, respectively. Further, the glycoprotein content was quantified using a periodate-based glycoprotein detection assay or mouse glycoprotein ELISA kit and sialic acid content using a sialic acid quantification kit.
Transmembrane protein and/or internal membrane protein in a CMC mimic can also be identified, quantified, and compared with the cell source using flow cytometry or immunoblotting or fluorescence microscopy to validate the orientation of the cell membrane. The localization of CD3z (intracellular) and LFA-1 (extracellular) proteins on leukocyte membrane-coated mimics was determined using both flow cytometry and immunoblotting. The fluorescence intensity of LFA-1 on CMC mimics was observed to be three times higher than that of the leukocyte vesicles but detected CD3z signals only after permeabilization.63 The comparable intensity of major histocompatibility complex II (MHC-II) protein (extracellular) on dendritic cell membrane-coated mimics and dendritic cells was confirmed using flow cytometry.106 Similarly, the comparable intensity of LFA-1 (extracellular) protein on neutrophils membrane coated mimics and neutrophils cell66 and comparable intensity of CD47 antibody (extracellular) on RBC membrane-coated mimics and RBC ghosts was observed using flow cytometry.278 In the case of platelet membrane-coated mimics, two anti-CD41 (glycoprotein (Gp) IIb/IIIa integrin) primary antibodies were used for binding to the N-terminal and C-terminal of CD41 present on the extracellular and intracellular regions, respectively.213 Extracellular CD41 intensity was observed four times higher than that in its intracellular domain on the outer surface of CMC mimics using flow cytometry. Similarly, for UM-SCC-7 membrane (human squamous carcinoma cell line) and its mimics, two anti-CXCR4 antibodies were used to bind the extracellular and intracellular region.73 A comparable fluorescence intensity of CXCR4 on CMC mimics and cells using a fluorometer was observed. Epifluorescence microscopy has also been reported to analyze the immunostained CD3 receptor (extracellular) on T cell membrane-coated mimics.280 Fluorescence intensity per particle using ImageJ software was quantified and observed that 40% of the CMC mimics displayed some or all the CD3 receptors in the correct orientation.
Role of CMC Mimics in Various Therapeutic Applications
CMC mimics have gained attention in several therapeutic applications like cancer, inflammatory diseases, and infectious diseases. The purpose of designing these mimics is to achieve target efficacy and accumulation at the target site. Figure 9 presents an overview of the different cell membrane and template combinations used for applications, along with a summary of in vitro and in vivo models used to validate their efficacy in Table 3.
Figure 9.
Schematic overview of the types of CMC mimics used for a wide variety of applications. Designs of CMC mimics have utilized combinations of a template and cell membrane that endows the final system with specific therapeutic properties. We have grouped and color-coded them based on their type and modifications. Abbreviations/cell line: CM, cell membrane; RBC, red blood cell; WBCs, white blood cells; NK, natural killer; CSCs, cardiac stem cells; NSCs, neural stem cells; MDSCs, myeloid-derived suppressor cells; TRAIL, TNF-related apoptosis-inducing ligand; RGD, arginylglycylaspartic acid; SHp, stroke homing peptide; APTES, 3-aminopropyl triethoxysilane; PLGA, poly(lactic-co-glycolic acid); Fe3O4, iron oxide; MnO2, manganese oxide; Au, gold; GdTPP, gadolinium porphyrin; ZnTPP, zinc porphyrin; PCN-224, porphyrin (TCPP)-based Zr6 cluster; PFCs, perfluorocarbons; UCNPs, upconversion nanoparticles; PLNPs, persistent luminescent nanoparticles; MOFs: metal–organic frameworks; ZIF-8, zeolitic imidazolate framework; UUNCs, ultrasmall unimolecular nanocluster; NPs: nanoparticles; NDs, nanodroplets; SPNPS: semiconducting polymer nanoparticles; IM, immunomagnetic; PCL, polycaprolactone; CDA, cystine dimethacrylate; PEG, poly(ethylene glycol); mPEG, methoxy polyethylene glycol; NK-92, human natural killer cell line; RAW 264.7, murine macrophage cell line; J774, murine macrophage cell line; U-251 MG, glioblastoma multiforme cell line; MCF-7, human breast cancer cell line; 4T1, human breast cancer cell line; H22, murine hepatocarcinoma cell line; HeLa, human cervical cancer cell line; B16-F10, murine melanoma cell line; B16.OVA, B16F10 cells expressed ovalbumin; 143B, human osteosarcoma cell line; LNCaP-AI, prostate cancer cell line; S. aureus, Staphylococcus aureus; E. coli:, Escherichia coli; C6/36, mosquito medium host cell line (Aedes albopictus).
Table 3. Summary of In Vitro and In Vivo Models Used for Validating the Efficacy of CMC Mimics with/without Modification in Various Treatmentsa.
| cell membrane source/modification | template | encapsulation | in vitro models | in vivo models | mode of administration | treatment | ref |
|---|---|---|---|---|---|---|---|
| platelets | PLGA nanoparticles | vancomycin | MRSA252, THP-1, HUVECs | male CD-1 mice (25g), MRSA252 | intravenous | MRSA bacterial infection | (93) |
| PLGA nanoparticles | docetaxel | – | male Sprague–Dawley rats (300–350 g), angioplasty-induced arterial denudation | intravenous | restenosis | (93) | |
| PLGA nanoparticles | – | antiplatelets antibodies | male CD-1 mice (6 week old, 20–24 g) | intraperitoneal | immune thrombocytopenia purpura | (212) | |
| PAMAM-PVL ultrasmall unimolecular nanoparticles | endothelium-protective epigenetic inhibitor (JQ1), or Rapamycin | – | Male Sprague–Dawley rats (∼400 g), carotid artery balloon angioplasty | intravenous | restenosis | (122) | |
| PLGA nanoparticles | – | J774, HUVECs | ApoE knockout mice | intravenous | Atherosclerosis | (123) | |
| PLGA nanoparticles | doxorubicin, Indocyanine green | MCF-7, MDA-MB-231, CAMA-1, HeLa | female nude mice, MDA-MB-231 (footpad) | intratumoral | circulating tumor cells of breast cancer Lung metastasis of breast cancer | (87) | |
| female nude mice, MDA-MB-231 (back) | intravenous | ||||||
| female nude mice, MDA-MB-231-luc (orthotropic) | intravenous | ||||||
| PLGA nanoparticles | verteporfin | 4T1, NIH-3T3 | BALB/c mice (3–5 week old), 4T1 | intravenous | breast cancer | (121) | |
| platelets/RGD | melanin nanoparticles | doxorubicin | RAW264.7, HUVECs, MDA-MB-231, MDA-MB-231/ADR | Female BALB/c nude mice (18–20 g), MDA-MB-231/ADR | intravenous | lung metastasis of breast cancer | (408) |
| platelets/TRAIL | acrylamide nanogels | doxorubicin | MDA-MB-231, RAW264.7 | Sprague–Dawley (SD) rats (200 ± 20 g), MDA-MB-231 | intravenous | circulating tumor cells of breast cancer Lung metastasis of breast cancer | (339) |
| platelets/TRAIL | silica microparticles-APTES | – | MDA-MB-231, PC3, THP-1 | C57BL/6 mice (6–8 week old), MDA-MB-231,COLO 205, B16-F10, MC38 | intravenous | circulating tumor cells of breast cancer | (213) |
| microrenathane microtubes coated human fibrinogen (thrombosis model) | |||||||
| platelets + WBCs | immunomagnetic beads | – | MCF-7, HCT116, HeLa | – | – | circulating tumor cells of breast cancer | (99) |
| platelets + RBC | gold nanowire | – | MRSA USA300, α-toxins | – | – | S. aureus bacterial infection | (196) |
| platelets + RBC | PLGA nanoparticles | – | MDA-MB-231, HFF-1 foreskin fibroblasts, THP-1 | ApoE knockout mice | intravenous | atherosclerosis | (97) |
| RBC | PLGA nanoparticles | vancomycin | MRSA USA300, THP-1, α-toxins | – | – | MRSA bacterial infection | (114) |
| PLGA nanoparticles | rapamycin | RAW264.7 | ApoE knockout mice (8 week old, 25–30 g) | intravenous | atherosclerosis | (211) | |
| olive oil nanodroplets | – | organophosphates (paraoxon, diisopropyl fluorophosphate, dichlorvos) | ICR mice (6 week old) | intravenous, intraperitoneal | organophosphate poisoning | (77) | |
| PLGA nanoparticles | – | HUVECs, α-toxins, streptolysin-O, melittin | female nu/nu nude mice (6 week-old), α-toxins | subcutaneous | pore forming toxins | (115) | |
| PLGA nanoparticles | mesenchymal stem cell, growth factors | THLE-2, HSC-T6, mouse lung cells, human primary M1 macrophages | C57BL/6 mice (18–25 g) | intravenous | acute liver failure | (86) | |
| Human serum albumin nanoparticles | Perfluorotributylamine, indocyanine green | HeLa, CT-26, RAW264.7 | male BALB/C mice (5–6 week), CT26 | intravenous | colon cancer | (269) | |
| Fe3O4 nanoclusters | – | MCF-7, RAW264.7 | female nude mice (4 week old), MCF-7 | intravenous | Breast cancer | (265) | |
| E. coli | – | primary macrophages | female BALB/C mice (6–8 week old), CT26 and 4T1 | intravenous | colon cancer, breast cancer | (59) | |
| PLNPs (Zn1.25Ga1.5Ge0.25O4:Cr3+,Yb3+,Er3+) coated with SiO2 | – | Balb/3T3, HeLa, MCF-7, 4T1, RAW264.7 | BALB/c mice, 4T1 | intravenous | breast cancer | (68) | |
| gelatin nanoparticles | vancomycin | RAW 264.7, S. aureus, E. coli, S. epidermidis, P. vulgaris, S. marcescens, P. aeruginosa | – | – | Gram positive bacterial infection | (64) | |
| gold nanorods | – | Capan-2 | male Balb/c nude mice (6–8 week old), Capan-2 | intravenous | pancreatic ductal adenocarcinomas | (261) | |
| PLGA nanoparticles | – | HUVEC, mouse RBCs | Male CD-1 mice (8 week old) | intravenous | MRSA bacterial infection | (278) | |
| PLGA nanoparticles | – | anti-RBC polyclonal IgG | CD-1 mice, anti-RBCs | intravenous | autoimmune anemia | (409) | |
| PLGA nanoparticles | Fe3O4 nanoparticles | influenza virus, Mardin Darby canine kidney cells | – | – | influenza virus | (74) | |
| carbon nanotube-based field effect transistor | – | MRSA USA300, Pore forming toxins (melittin, streptolysin O, Hlα) | – | – | MRSA bacterial infection | (75) | |
| RBC/cRGD peptide | docetaxel nanocrystal | – | HUVECs, U87 | male nude mice (20–22 g), U87, antisubcutaneous tumor- and antiorthotropic glioma | intravenous | glioblastoma | (333) |
| BBTB model (HUVEC/U87 coculture) | |||||||
| RBC/T7, NGR peptide | solid lipid nanoparticles | – | HUVECs, bEnd.3, C6 | Tg (fli1:egfp) strain of zebrafish, ICR mice (22–24 g), C6 | intracranial | glioblastoma | (335) |
| BBB model(upper: bEnd.3) | |||||||
| BBTB model (upper: HUVECs, lower: C6) | |||||||
| RBC/anti-EGFR-iRGD | – | paclitaxel | MKN45 | male BALB/c nude mice weighing 18–20 g (5–6 week old), MKN45 | intravenous | gastric cancer | (332) |
| RBC/iRGD | polycaprolactone nanoparticles | paclitaxel | 4T1, RAW264.7 | Balb/c mice (18–20 g), orthotopic 4T1 | Intravenous | Lung metastasis of breast cancer | (309) |
| RBC/folate receptor | UCNPs (β-NaYF4:Er3+,Yb3+) | – | MCF-7 | BALB/c nude mice, MCF-7 | intravenous | breast cancer | (277) |
| RBC/ DCDX peptide | PLGA nanoparticles | doxorubicin | primary brain capillary endothelial cells | nude mice, U87 | intravenous | glioblastoma | (234) |
| BBB model (Wistar rat primary brain capillary endothelial cells) | |||||||
| RBC/stroke homing peptide (SHp) | boronic acid conjugated dextran nanoparticles | NR2B9C | PC-12, BBB model (rat brain capillary endothelial cell line) | male Sprague–Dawley (SD) rats (5–6 week, 200 ± 20 g) | intravenous | ischemic stroke | (410) |
| RBC+ MCF-7 | Melanin nanoparticles | - | MCF-7, 4T1, MCF-10A, RAW264.7 | female athymic Balb/c nude mice (20–22 g), MCF-7 | intravenous | breast cancer | (198) |
| NK-92 | poly(ethylene glycol) methyl ether-block-PLGA nanoparticles | 4,4′,4″,4‴-(porphine-5,10,15,20-tetrayl) tetrakis (benzoic acid) (TCPP) | 4T1, MCF-7, MCF10A | female BALB/c mice (6 week old, 18–22 g) | intravenous | breast cancer | (150) |
| liposomes | doxorubicin | MCF-7, NHost | female nu/nu nude mice (6 week old), MCF-7 | intravenous | breast cancer | (102) | |
| neutrophils | PLGA nanoparticles | carfilzomib | HUVECs, 4T1, 4T1 (GFP+ or luc+) | female Balb/c nude mice (20 ± 2 g), GFP+4T1, luc+4T1 | intravenous | lung metastasis of breast cancer | (55) |
| PLGA nanoparticles | – | HUVECs, primary human chondrocytes | human TNF-α male transgenic mice (5 week old) DBA/1J mice in collagen-induced arthritis (6 week old, 18–22 g) | intra-articular | inflammatory arthritis | (66) | |
| natural macrophage cells | gold nanoshells | – | 4T1 | male Balb/c nude mice, 4T1 | intravenous | breast cancer | (139) |
| silica nanocapsules | doxorubicin | RAW 264.7, NIH/3T3, 4T1 | male BALB/c nude mice, 4T1 | intravenous | breast cancer | (69) | |
| RAW 264.7 | liposomes | emtansine | RAW 264.7, 4T1, 4T1-luc | female BALB/c nude mice (18–22 g), 4T1-luc | intravenous | lung metastasis of breast cancer | (129) |
| Fe3O4 nanoparticles | – | RAW 264.7, MCF-7 | Female BALB/c nude mice (6–8 week old), MCF-7 | intravenous | breast cancer | (131) | |
| UCNPs (β-NaYF4:Er3+,Yb3+ rare earth) | – | RAW 264.7, MCF-7 | female BALB/c nude mice (6–8 week old), MCF-7 male ICR mice (6–8 week old) | intravenous | breast cancer | (132) | |
| bismuth selenide nanoparticles | quercetin | RAW 264.7, 4T1, HUVECs | BALB/c mice, 4T1 | intravenous | lung metastasis of breast cancer | (88) | |
| transwell cell invasion (upper: 4T1) | |||||||
| wound healing (4T1) | |||||||
| dual-chamber transwell (5 μm, 0.4-μm sized microporous membranes) | |||||||
| J774 | PLGA nanoparticles | – | J774, HEK-blue mTLR4, HUVECs | male BALB/c mice (6 week old), LPS (endotoxemia model) | intravenous | sepsis | (130) |
| U937 | PLGA nanoparticles | – | MCF-7 | – | – | breast cancer | (298) |
| human cytotoxic T-lymphocyte | PLGA nanoparticles | paclitaxel | MKN-45, differentiated THP-1 | male Balb/c nude athymic mice (4–6 week old), MKN-45 | intravenous | gastric cancer | (98) |
| CD4+ T cells | PLGA nanoparticles | – | HIV stains (X4 and R5) | – | – | HIV | (159) |
| dendritic Cells | PLGA nanoparticles | IL-2 | primary CD8+ T, T cells, NIH-3T3, 293T, ID8 | C57BL/6 mice (6–8 week old), ID8 | subcutaneous | ovarian cancer | (106) |
| bone marrow-derived dendritic cells +4T1 | PCN-224 MOFs | – | 4T1, CT26, 3T3 | Balb/c mice, 4T1 | intravenous | breast cancer | (168) |
| Human adiposed- derived stem cells | PLGA nanoparticles | – | J774, THP-1 | female C57BL/6 mice (10 week old, 22–24 g) | intravenous | hindlimb ischemia | (227) |
| transwell: HUVECs | |||||||
| adipose-derived mesenchymal stem cells | Fe3O4 nanoparticles | – | TRAMP-C1, RAW246.7 | – | – | prostate cancer | (95) |
| cardiac stem cell | PLGA microparticles | cardiac stem cells growth factors | cardiomyocytes | male SCID Beige mice | intramyocardial | myocardial infarction | (187) |
| bone marrow derived mesenchymal stem cell membrane | gelatin nanogels | doxorubicin | HeLa, L02 | female BALB/c nude mice (∼4 week old), HeLa | intravenous | cervical cancer | (186) |
| UCPNs (β-NaYF4:Er3+,Yb3+ rare earth) | – | HeLa, L02 | female BALB/c nude mice (∼4 week old), HeLa | intravenous | cervical cancer | (185) | |
| neural stem cell/CXCR4 | PLGA nanoparticles | – | – | male C57BL/6 mice (≈20 g) | intravenous | ischemic stroke | (188) |
| B16-OVA/mannose | PLGA nanoparticles | imiquimod, R837 (toll-like receptor 7 (TLR-7) agonist | bone marrow-derived dendritic cells | female BALB/c mice (6–8 week old), B16-OVA | Intradermal | melanoma | (343) |
| U87/CXCR4 | PLGA nanoparticles | – | U87, U87-CXCR4, MDA-MB-231, BT-474, HMFs | female athymic Balb/c (nu/nu) mice, Female immunocompetent Balb/c mice(6–8 week old) | intravenous | lung metastasis of breast cancer | (224) |
| transwell model: HMFs | |||||||
| LNCaP-AI | CaCO3 capped mesoporous silica nanoparticles | doxorubicin | LNCaP-AI, QSG-7701, MCF-7 | BALB/c nude mice (6 week old), LNCaP-AI | intravenous | prostate cancer | (225) |
| HeLa | Fe3O4 nanoparticles | doxorubicin | HeLa, UM-SCC-7, COS7, RAW 264.7 | female BALB/c nude mice (4–5 week old), UM-SCC-7 | intravenous | cervical cancer | (173) |
| PLGA nanoparticles | paclitaxel, small interfering RNA (siRNA-E7) | HeLa, Ect1, LO2, RAW264.7 | female BALB/c nude mice (4–5 week old, 18–22 g), HeLa | intravenous | cervical cancer | (235) | |
| GdTPP and ZnTPP nanocomposites | – | HeLa | nude mice, HeLa | intravenous | cervical cancer | (219) | |
| ZIF-8 MOFs | catalase, Al(III) phthalocyanine chloride tetrasulfonic acid | 4T1, HeLa, SCC-7, COS7, RAW264.7 | BALB/c-nu mice (4–5 week old), HeLa | intravenous | cervical cancer | (342) | |
| 143B | silica nanoparticles | indocyanine green | 143B | female Balb/c nude mice (6–8 week old), 143B | intravenous | bone cancer | (174) |
| MCF-7 | PLGA nanoparticles | indocyanine green | MCF-7, 293T, MCF-10A, HepG2, A549, MDA-MB-231 | female BALB/c mice (4–6 week old), MCF-7 | intravenous | breast cancer | (65) |
| mesoporous silica nanoparticles supported with PEGylated liposomes | – | SK-hep-1, Caco-2, HeLa, Bxpc-3, Huh-7, MCF-7 | BALB/c nude mice (20g), MCF-7 | intravenous | breast cancer | (175) | |
| 3D- spheroids of MCF-7 | |||||||
| MDA-MB-435, B16-F10 | PLGA nanoparticles | – | bone marrow-derived DCs, MDA-MB-435, B16-F10, HFFs | female C56BL/6 mice (6 week old), MDA-MB-435 Female C56BL/6 mice (6 week old), B16-F10 | intradermal | melanoma | (92) |
| 4T1 | poly(caprolactone) nanoparticles | paclitaxel | RAW264.7, WML2, 4T1, MDA-MB-435, A549, BEL-7402 | female BALB/c nude mice, 4T1, 4T1-luc, female BALB/c mice (4–5 week old, 18–22 g), 4T1 | intravenous | lung metastasis of breast cancer | (220) |
| gold nanocages | doxorubicin | 4T1 | BALB/c nude mice (18–22 g) orthotopic, 4T1 | intravenous | lung metastasis of breast cancer | (221) | |
| PLNPs | – | 4T1 | female BALB/c nude mice (4 week old, 18–22 g), 4T1 | intravenous | lung metastasis of breast cancer | (72) | |
| carbon sphere dotted with cerium oxide nanoparticles | – | COS7, 293T, 4T1 | female BALB/c mice (6 week old), 4T1 | intravenous | breast cancer | (297) | |
| SGC7901 | silica nanoparticles | chlorins e6 | SGC7901 | female BALB/c nude mice (6–8 week old), SGC7901 | intravenous | gastric cancer | (174) |
| U-251 MG | Fe3O4 nanoparticles | – | U-251 MG, SH-SY5Y, C8-D1A, bEnd.3 | – | – | glioblastoma | (273) |
| U-251 MG 3D spheroids | |||||||
| BBB model (upper chamber: bEnd.3; lower chamber: U-251 MG) | |||||||
| HNSCC | gelatin nanoparticles | cisplatin | patient-derived tumor cell | BALB/c nude mice (male, 6–8 week old), patient derived tumor cells | intravenous | head and neck squamous cell carcinoma | (105) |
| K562 | Fe3O4 nanoparticles | doxorubicin, indocyanine green | MG-63, A549, SW380, A375, HepG2, MCF-7, MGC-803, L02, RAW 264.7 | BALB/c nude mice, MG-63 | intravenous | osteosarcoma | (103) |
| MDA-MB-435, DU145, CAL27, HCT116 | UCNPs (β-NaYF4:Er3+,Yb3+) | – | RAW264.7, MDA-MB-435, DU145, CAL27, HCT116 | female BALB/c nude mice (6–8 week old), MDA-MB-435 | intravenous | breast cancer | (223) |
| B16.OVA, B16.F10, LL/2, CMT64.OVA, MB49, A549, SKOV-3 | adenovirus serotype 5 particles | – | A549, SKOV-3 | female C57BL/6 mice (4–6 week old) B16.OVA, B16.F10, LL/2, | intratumoral | melanoma lung cancer | (214) |
| H22 | gold nanocages | doxorubicin | RAW 264.7, H22 | Sprague–Dawley (SD) male rats (6–8 week old) male BALB/c mice (6 week old, 20–22 g), H22 | intratumoral; intravenous | liver cancer | (226) |
| AF | semiconducting polymer (poly(cyclopentadithiophenealt-benzothiadiazole) nanoparticles | – | AF, fibroblasts, chondrocytes, 4T1, HeLa, SKOV3 | female NCr nude mice (4–6 week old), 4T1 | intravenous | breast cancer-associated fibroblasts | (104) |
| MIN6 | polycaprolactone nanofibers | – | MIN6 | – | – | β-cell proliferation and function | (96) |
| E. coli DH5α + B16-F10 | polydopamine nanoparticles | – | B16-F10, NHDF, MCF-7, A549 | female C57BL/6 mice (6–8 week old), B16-F10 | intravenous | melanoma | (200) |
| E. coli K12/ Rhizobium etli tyrosinase | – | melanin | 4T1 | female athymic Fox-N-1 nude mice (6 week old), 4T1; female C57BL/6 mice (6 week old) | intravenous, intratumoral | breast cancer | (194) |
| E. coli | Gold nanoparticles | – | – | Male CD-1 mice (6 week old), E. coli | subcutaneous | E. coli bacterial infection | (94) |
| S. aureus | PLGA nanoparticles | Vancomycin or Rifampicin | Ana-1, NIH-3T3 | Balb/c mice ((5–6 week old), S. aureus | intravenous | S. aureus bacterial infection | (192) |
| CRKP | BSA nanoparticles | – | RAW 264.7, DC2.4 | female C57BL/6 mice (6–8 week old) | subcutaneous | CRKP bacterial infection | (193) |
| C6/36 | gelatin nanoparticles | – | Vero, HeLa, HEK 293T | type I interferon (IFN-α/β) receptor deficient (A129) mice | intravenous | Zika viral infection | (228) |
| 293T/ACE2 + THP-1 | – | – | Vero-E6, Caco-2 | ICR mice (20 to 25 g, 4- 6 week old) | intratracheal | SARS-CoV-2 infection | (397) |
| NL-20, THP-1 | PLGA nanoparticles | – | Vero E6 | C57BL/6NHsd mice | intratracheal | SARS-CoV-2 infection | (400) |
| mitochondria (mouse liver) | PLGA nanoparticles, carbon nanotube-based field-effect transistors (FET) | – | ABT-263, HL-60 | female C57BL/6 mice (6 week old) | oral gavage | ABT-263-induced thrombocytopenia | (91) |
Abbreviations/cell lines: APTES, 3-aminopropyl triethoxysilane; PAMAM-PVL, poly(amidoamine)-polyvalerolactone; PBMC, peripheral blood mononuclear cells; SHp, stroke homing peptide; BSA, bovine serum albumin; UCNPs, upconversion nanoparticles; PLNPs, persistent luminescent nanoparticles; MOFs, metal–organic frameworks; ZIF-8, zeolitic imidazolate framework; PCN, porous coordination network; MSCs, mesenchymal stem cells; HMFs, human mammary fibroblasts; HFFs, human foreskin fibroblasts; HNSCC, head and neck squamous cell carcinoma; AF, activated fibroblast; DCs, dendritic cells; CXCR4, C–X–C chemokine receptor type 4; MRSA252, methicillin-resistant S. aureus; THP-1, human acute monocytic leukemia cell line; HUVECs, human umbilical vein endothelial cell line; RAW 264.7, murine macrophage cell line; J774, murine macrophage cell line; MCF-7, human breast cancer cell line; 4T1, human breast cancer cell line; 4T1-Luc, 4T1 cells expressing luciferase; HeLa, human cervical cancer cell line; MDA-MB-231, breast cancer cell line; MDA-MB-231/ADR, DOX-resistant breast cancer cell line; 3T3, murine normal fibroblast cell line; CT26, murine colon cancer cell line; NIH-3T3, fibroblast cell line; ID8, murine ovarian surface epithelial cell line; MKN-45, human gastric cancer cell line; CAMA-1, human breast cancer cell line; CAL 27, human squamous carcinoma cell line; RD, human rhabdomyoma cell line; MKN45, gastric cell line; Capan-2, human pancreatic cancer cell line; C6, glioma cell line; 143B, human osteosarcoma cell line; SCC-7, murine squamous cell carcinoma cell line; Ect1, endocervical cell line; L02, normal hepatic cell line; LNCaP-AI, prostate cancer cell line; QSG-7701, human normal liver cell line; UM-SCC-7, head and neck squamous cell carcinoma cell line; HCT116, human colorectal cancer cell line; TRAMP-C1, mouse prostate cancer cell line; COS7, african green monkey kidney cell line; 293T, human embryonic kidney cell line; WML2, murine lung fibroblast cell line; BEL-7402, human hepatoma cell line; SK-hep-1, human hepatic adenocarcinoma cell line; Caco-2, human colorectal adenocarcinoma cell line; Bxpc-3, human pancreatic cancer cell line; Huh-7, human liver cell line; MCF-10A, nontumorigenic epithelial cell line; U-251 MG, glioblastoma multiforme cell line; SH-SY5Y, neuroblastoma cell line; C8-D1A, astrocytes type 1 clone; bEnd.3, mouse brain endothelial cell line; MDA-MB-435, human breast carcinoma cell line; DU145, human prostate cancer cell line; HCT116, human colorectal cancer cell line; CAL27, human squamous cancer cell line; A375, human melanoma cell line; HepG2, human hepatocellular carcinoma cell line; K562, human myelogenous leukemia cell line; MGC-803, human gastric carcinoma cell line; MG-63, human osteosarcoma cell line; L02, human hepatic cell line; SW380, human colorectal adenocarcinoma cells; B16-F10, murine melanoma cell line; B16.OVA, B16F10 cells expressed ovalbumin; LL/2, murine lewis lung carcinoma cell line; CMT64.OVA, lung carcinoma cell line expressed ovalbumin; MB49, murine bladder carcinoma cell line; A549, human nonsmall cell lung cancer cell line; SKOV-3, ovarian cancer cell line; CAL27, human squamous cancer cell line; H22, murine hepatocarcinoma cell line; MIN6, pancreatic β-cell line; C6/36, A. albopictus (mosquito medium host cell line; E. coli, Escherichia coli; E. coliK12, Escherichia coli derived from W3110; CRKP, carbapenem-resistant Klebsiella pneumonia; S. aureus, Staphylococcus aureus; P. aeruginosa, Pseudomonas aeruginosa; S. marcescens, Serratia marcescens; P. vulgaris, Proteus vulgaris; S. epidermidis, Staphylococcus epidermidis; Ana-1, murine macrophage cell line; DC2.4, murine dendritic cell line; NL-20, human lung epithelial cells; HL-60, human leukemia cell line; ABT-263, B-cell lymphoma 2 (Bcl-2) inhibitor.
Cancer Therapy
Cancer causes abnormal and uncontrolled growth of cells in the human body. The primary tumor is the initial region from where the cancer cells begin to spread. These tumor cells secrete various chemokines to redirect the platelets, immune cells (neutrophils, macrophages, T cells, NK cells) to facilitate their growth and progression in different parts of the body.314,315 Circulating tumor cells (CTCs) rapidly spread to the blood and lymph nodes and cause life-threatening metastasis.316,317 Therefore, during chemotherapy, delivering the drug at the metastatic site and neutralizing the CTCs in the blood and lymph nodes is crucial. Chimeric antigen receptor T cell immunotherapy (CAR-T),318,319 adoptive immunotherapy,320,321 immune checkpoint blockade therapy,322,323 vaccines, treatment with oncolytic viruses,324,325 monoclonal antibodies,326 cytokines,327 and immunomodulatory treatment328 are immunotherapies currently under consideration. However, tumor heterogeneity, immune cell dysfunction, acquired resistance to immunotherapy, and immunotoxicity complicate their clinical translation.329−331 Therefore, there is a pressing need to discover and deliver tumor neoantigens to activate the patient’s immune system efficiently.
Incorporating a cancer cell membrane within a CMC mimic provides the required neoantigens, particularly in the case of highly mutagenic tumors. Current treatment options include modifying nano/microparticle surfaces, delivering immunomodulators, and chemo drugs. To resolve the complexity of modification, several CMC mimics with natural biocompatible characteristics have been designed with various combinations of the cell membrane and templates to effectively target primary, metastatic, and CTCs. If required, the cell membrane can be modified further with desired active targeting moieties using lipid insertion71,234,332,333 or membrane fusion97,100,196,198 to increase its target efficacy toward tumor cells in various organs (brain, breasts, lungs, cervical, colon, pancreas, etc.). This section discusses CMC mimics used for cancer treatment, as summarized in Table 3.
RBC membrane-coated PLGA nanoparticles were designed in Zhang’s lab to enhance the low circulation half-life of nanoparticle drug delivery systems by utilizing CD47 receptors on the RBC membrane.62 Compared to the PEGylated systems, the half-life of these mimics improved by at least 2-fold, and they remained in circulation for up to 72 h post-injection. Following this work, RBC cell membrane coating of gold nanocages, Fe3O4 nanoparticles, and PFCs improved their circulation times. Additionally, this enhanced their suitability for bioimaging and phototherapy applications.265,269,59,68 Due to the lack of tumors targeting proteins on the RBC membrane, many researchers have attempted modifying with peptides or fusing it with another cell membrane to enhance its targeting efficacy toward specific primary or metastatic tumors. For example, arginyl glycyl aspartic acid (RGD)-modified RBC membrane-coated paclitaxel loaded polycaprolactone (PCL) nanoparticles inhibited the growth of the primary tumor in breast cancer and lung metastasis significantly.309 Similarly, brain-targeted peptide (DCDX,234 T7,334,335 and NGR335−337)-modified RBC membrane facilitated CMC mimic’s crossing of the blood–brain barrier and improved their ability to target glioma.
Platelet cell membranes gained interest in designing CMC mimics for targeting CTCs due to the effective interaction between P-selectin and CD44 receptors on platelets and tumor cells.338 According to these reports, platelet membrane-coated mimics captured and killed CTCs in blood and lymph nodes and inhibited breast cancer metastasis effectively.87,119 Further, TNF-related apoptosis-inducing ligand (TRAIL) modified platelet membrane-coated templates also eliminated CTCs effectively. TRAIL additionally activates apoptosis in tumor cells by binding to the death receptors (DR4, DR5) on the cell surface.213,339,340 Additionally, coating a hybrid membrane of platelets and leukocytes on commercially available immunomagnetic beads was very effective in isolating pure CTCs from clinical blood samples collected from breast cancer patients, demonstrating the possibility of extending these CTCs for in vitro applications and their potential for use in personalized medicine.99
The cancer cell membrane is known for its homologous targeting abilities attributed to the presence of different adhesion molecules on their surface. These adhesion molecules play an important role in the development of invasive and distant metastasis.341 Designing mimics using cancer cell membranes could be a potential strategy to develop personalized tumor-specific therapies or vaccines. In this context, CMC mimics of cisplatin-loaded gelatin nanoparticles coated with the patient-derived tumor cell membrane (head and neck squamous cell carcinoma) (Figure 10) were fabricated and tested for efficacy in a patient-derived xenograft model.105 These autologous cell membrane-coated mimics were able to ablate the tumor completely and inhibit tumor recurrence. However, the mismatch of membrane donors and hosts resulted in weaker targeting. Numerous cancer cell membrane-coated mimics using different templates are developed for such homotypic targeting (Table 3).235,173,219,342,223,174,103,342,175,220,221
Figure 10.
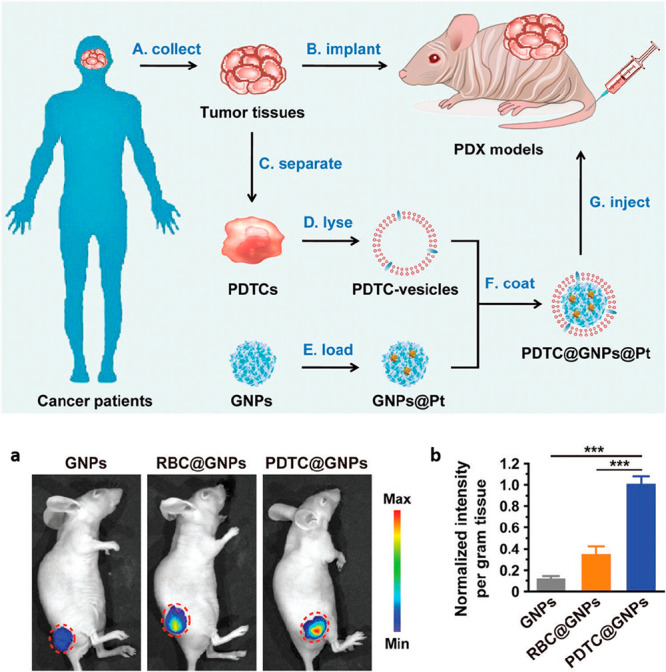
Design of CMC mimics using patient derived tumor cell membrane-coated nanocarriers for personalized cancer therapy. (a) In vivo fluorescence images of nude mice bearing PDX at 24 h after intravenous injection of gelatin nanoparticles (GNPs), RBC-coated gelatin nanoparticles (RBC@GNPs), patient-derived tumor cells-coated gelatin nanoparticles (PDTC@GNPs). (b) Normalized ex vivo intensity per gram of the PDX tissues in the mice injected with GNPs, RBC@GNPs, and PDTC@GNPs. Reprinted with permission from ref (105). Copyright 2019 John Wiley and Sons.
Recently, CMC mimics were redesigned as nanovaccines for cancer. These vaccines combine the cancer cell membrane with an adjuvant that facilitates delivery of tumor-associated antigens to dendritic cells and stimulates the tumor antigen specific-T cells. For example, B16-F10 (murine melanoma cell line) membrane-coated PLGA mimics were designed as nanovaccines for cancer. They incorporated an adjuvant, monophosphoryl lipid A, MPL (FDA-approved LPS derivative) that binds specifically to toll-like receptor 4 to boost the immune response. Additionally, CMC mimics of cancer cell (B16-OVA) membrane and PLGA nanoparticles were formulated as nanovaccines to specifically targeted antigen-presenting cells (APCs) and enhance their uptake to trigger cell maturation. Imiquimod R837, an adjuvant and agonist for toll-like receptor 7 (TLR-7), was preloaded into PLGA particles and coated with a mannose-modified cell membrane.343 These mimics effectively inhibited the growth of melanoma tumors when combined with antiprogrammed cell death protein 1 (PD-1) checkpoint blockade therapy. Viruses also have natural adjuvant properties that initiate an immune response. For example, the oncolytic virus replicates inside tumor cells by releasing tumor antigens and causes tumor lysis without affecting healthy cells.344 Antigen-presenting cells (APCs) engulf these antigens and redirect dendritic and T cells toward the infected site. Therefore, coating adenovirus serotype-5 virus particles with the melanoma (B16.F10) and lung cancer (LL/2) cell membranes can help explore their properties and treat aggressive melanoma and lung tumor.214 Despite coating the virus with different cancer cell membranes, binding efficacy was maximized using homologous and tumor-matched cell membranes.
Macrophages are the most abundant cells in the tumor microenvironment of solid tumors. Interactions between α4 and β1 integrins present on the macrophage surface with the vascular cell adhesion molecule-1 (VCAM-1) present on cancer cells are responsible for progression and metastasis of tumor.345−348 Using these interactions, CMC mimics of macrophages (RAW 264.7) and emtansine-loaded liposomes targeted and inhibited lung metastasis in breast cancer models.129 In another report, using CCL2/CCR2 chemokines interactions, macrophages were recruited. Quercetin-loaded bismuth selenide nanosystems acted as the templates for mimic assembly.88 Post-targeting, nanosystems inhibited primary cancer and lung metastasis by photothermal therapy. Quercetin released inhibited thermoresistant tumors by damaging their heat shock protein 70 (HSP70). Subsequently, several groups have reported the fabrication of CMC mimics with macrophage membranes on various templates that target breast cancer for photothermal therapy.131,132,139
Inflammatory neutrophils are activated and directed by the granulocyte-colony stimulating factor (G-CSF) and C–X–C chemokines (CXCL1, CXCL2, CXCL5) toward early premetastatic niche formation.349,350 Inspired by this mechanism, neutrophil membrane-coated PLGA nanoparticles loaded with carfilzomib were designed.55 These mimics neutralized CTCs, prevented early lung metastasis, and inhibited the progression of already-formed lung metastasis in breast cancer.
Among other immune cells, NK cells can detect and target tumor cells without preactivation. These cells also regulate immune response and T cell activation to kill tumor cells.144 The NK cell membrane from murine NK cells and NK-92 cell line-coated on poly(ethylene glycol) methyl ether-block-poly(lactide-co-glycolide) (mPEG-PLGA) nanoparticles loaded with 4,4′,4″,4‴-(porphine-5,10,15,20-tetrayl) tetrakis (benzoic acid) (TCPP)150 were able to polarize M1 macrophages, kill primary tumors, and inhibit the growth of distant tumors. Further, the fusion of the NK cell membrane from NK-92 with liposomes to create NKsomes loaded with doxorubicin demonstrated an excellent tumor homing potential of NKsomes against breast cancer cells.102
MSCs have inherent tumor-targeting properties and exhibit immunomodulatory activities.351,352 However, biosafety concerns, stability, and reproducibility issues limit their use in clinical applications.353 Taking advantage of MSCs mechanism, adipose-derived MSCs membrane-coated Fe3O4 nanoparticles were constructed as a proof-of-concept to inhibit prostate tumor cells via hyperthermia mechanism. Furthermore, bone marrow-derived MSC membranes were used to coat doxorubicin-loaded gelatin nanogels186 and UCPNs185 to enhance the target efficacy toward cervical tumor cells. The presence of tumor recognition receptors and adhesion molecules on T-cells surfaces has prompted their use for fabricating CMC mimics.354
Taking advantage of T-cell receptors, human cytotoxic T-lymphocyte cell membrane-coated paclitaxel-loaded PLGA nanoparticles were designed in combination with low-dose irradiation (LDI) to target gastric tumor cells.98 These mimics inhibited gastric tumor growth significantly when used in combination with LDI than mimics alone. Further, an azide-modified T-cell membrane was modified with azide and assembled with a PLGA template for biorthogonal targeting of bicyclo[6.1.0]nonyne (BCN)-modified tumor cells.158 Modified cell membrane-coated mimics showed a 1.5-fold higher accumulation around Raji tumor cells.
Dendritic cells (DCs) are the initiators of the primary immune response and capable of activating naïve T cells.355 DC-based cancer vaccines have drawn attention in immunotherapy for treating prostate cancer with one of their variants approved by the US FDA.356 There is also evidence that immunotherapy could benefit patients with ovarian cancer.357−359 Recently, in a clinical phase 1 study for ovarian cancer patients, DC vaccines initiated T-cell responses in only half of the total patients. Their clinical efficacy was affected by low immunogenicity of tumor-associated antigens (TAAs), immunosuppressive tumor-associated microenvironment, restricted migration due to physiological barriers, and downregulation of major histocompatibility complex (MHC).360−362 To overcome these limitations and utilize interesting functions of DCs, cell membranes from mature DCs (primed by ovarian cancer cell lysate) were coated on interleukin-2 (IL-2)-loaded PLGA nanoparticles to fabricate mini DCs.106 These mini DCs enhanced the activation of the T-cell immune response and effectively inhibited the progression and metastasis of ovarian cancer.
There are reports of CMC mimics designed from the cell membrane of AF to target fibroblast-associated cancer cells. This approach enabled crossing the protective physical barriers built around tumor cells by cancer-associated fibroblasts for delivering anticancer drugs.363 Chemically modified nanoparticles are known for their ability to target and kill cancer-associated fibroblasts, prevent biological interactions between tumor and stroma, and enhance chemotherapy.364 Based on these reports, CMC mimics were designed using activated fibroblast cell membranes and semiconducting polymeric nanoparticles and compared their efficacy to that of 4T1 cancer cell membrane-coated mimics in breast cancer models.104 The AF and 4T1-coated mimics showed superior targeting efficacy for cancer-associated fibroblast and 4T1 cells, respectively, due to their homologous targeting capabilities.
OMVs have the potential to induce the production of antitumor cytokines and trigger the antitumor immune response.365−367 Utilizing their mechanism of action, cell membranes of OMVs, and cancer cells were fused to induce both an immune response and increase the homotypic ability. The hybrid cell membrane of E. coli DH5α membrane vesicles and B16-F10 cell membrane coating on hollow polydopamine nanoparticles significantly inhibited melanoma growth and stimulated DC maturation in lymph nodes.200
Inflammation and Immune Diseases
Inflammation is a physiological process that protects the body from harmful stimuli using immune cells, blood vessels, and molecular mediators and promotes tissue repair.368 Chronic or uncontrolled inflammation causes diseases like atherosclerosis, ischemic diseases (myocardial infarction, ischemic stroke, hindlimb ischemia), rheumatoid arthritis, and acute liver failure. Modulation of inflammatory responses to balance the immune homeostasis helps to overcome the disease progression.369 Some of the inflammation-related cells that play a vital role in shaping its microenvironment are neutrophils, NK cells, macrophages, lymphocytes, platelets, and stem cells. These cells are in their resting state during circulation but are activated by cytokines or chemokines during inflammation and migrate to the infected site.370,371 Therefore, designing CMC mimics using these cells has considerable potential to treat inflammatory diseases.
Atherosclerosis is a condition caused by the accumulation of lipids, cholesterol, and fibrous elements in the artery wall that restricts blood flow.372 The primary challenge of this disease is it is asymptomatic until the very late stages. Surgically stenting the artery is often the preferred intervention route. However, this can lead to potential side effects such as restenosis and stent thrombosis, eventually triggering neointimal hyperplasia.373,374 Using a noninvasive strategy to image and monitor plaque development would be the preferred mode of treatment, as platelets are responsible for hemostasis in the body and are involved in atherogenesis.375−378 CMC mimics assembled with their cell membranes may provide a viable alternative to the existing line of treatment. The platelet membrane-coated PLGA nanoparticles loaded with docetaxel were reported for restenosis therapy.93 Their results concluded that CMC mimics localized better than the drug alone at the plaque site and inhibited neointima growth. Further, the MRI-based platelet membrane-coated PLGA nanoparticles localize better at the plaque-forming and atherosclerosis areas than the PLGA or RBC membrane-coated PLGA nanoparticles, providing crucial information for managing atherosclerosis.123 The nonthrombogenic and stent-free restenotic therapy was also treated using platelet membrane-coated nanoclusters of poly(amidoamine) and polyvalerolactone (PAMAM-PVL).122 These dendritic, unimolecular nanoclusters were preloaded with an endothelium-protective epigenetic inhibitor (JQ1) before coating. Comparing the efficacy of the JQ1 and rapamycin (endothelium-toxic status quo drug) showed up to a 60% reduction of neointimal hyperplasia. However, rapamycin impairs the endothelial recoverage, while JQ1 protects the endothelial coverage in the inner artery wall. Therefore, these noninvasive platelet membrane-coated mimics loaded with MRI contrast agents or endothelium-protective inhibitors or drugs have the potential for live imaging to assess, prevent, and manage the development of atherosclerosis at an early stage.
Restricted blood flow in blood vessels resulting in tissue damage or dysfunction is a characteristic of ischemic disease.379 Mesenchymal stem cells are a promising candidate for their treatment as they can interact effectively with the stromal-derived factor (SDF) overexpressed in ischemic tissue through their CXCR4.380,381 Researchers have tried to overexpress the CXCR4 receptor on stem cells to increase its efficiency. For example, they bioengineered human adipose-derived stem cell (hASCs) membranes to overexpress CXCR4 receptors for coating PLGA nanoparticles loaded with vascular endothelial growth factor (VEGF). These particles showed a higher accumulation at the ischemic site than the hASC-coated mimics.227 Likewise, a neural stem cell membrane overexpressing CXCR4 was coated on PLGA nanoparticles loaded with glyburide for testing in ischemia stroke models.188 Even in the injured brain, these CMC mimics could effectively cross the blood–brain barrier for drug delivery. PLGA microparticles loaded with secretomes were coated with cardiac stem cell membrane for myocardial infarction.187 Their functional efficacy was comparable to cardiac stem cell therapy and allowed for surgical transfer intramyocardially.
Acute liver failure causes deterioration of liver function and requires liver transplantation to cure the patient.382 Stem cell therapy can be a promising treatment for liver failure, as MSCs secret anti-inflammatory factors reducing inflammation and promote healing.383 Cultured MSCs with 20 μm in diameter are larger than the width of the microcapillaries of the lung;384 therefore, intravenously infused MSCs are short-lived and easily filtered by the lungs and do not reach the liver.385 Thus, using RBC membrane-coated PLGA nanoparticles loaded with MSCs regenerative factors of 200 nm in size resolved this size issue.86 The small size of the mimics helped them pass through the lungs and reach the liver, and additionally, the RBC membrane coating prolonged their circulation time.
Rheumatoid arthritis (RA) is another autoimmune disorder that leads to joint damage and disability. Current treatment focuses on targeting the inflammatory responses such as inhibiting the interleukin (IL)-1 and tumor necrosis factor-alpha (TNF-α).386,387 Various chemoattractant are known to promote neutrophil migration into the joints during RA.388 Microvesicles produced by neutrophils can readily enter cartilage and protect joints.389 Therefore, neutrophil membrane-coated PLGA nanoparticles were designed.66 These mimics neutralized pro-inflammatory cytokines (IL-1β and TNF-α), suppressed synovial inflammation, targeted cartilage matrix, and protected chondrocytes against damage. Apart from neutrophils, other cell membrane-like T cells, dendritic cells, macrophages, and monocytes are present in the primary stage of RA, working together for its progression. Therefore, CMC mimics using the membrane of these cells have the potential to show stealth properties for the treatment of RA.390,391
Infectious Diseases
Pathogens, viruses, and bacteria cause infectious diseases by interacting and penetrating the host cell membrane.392,393 A lack of effective drugs, specific treatment options, and increasing drug-resistant strains caused by overuse of antibiotics are the main challenges to their treatment. CMC mimics can target viruses, bacteria, and multidrug-resistance bacteria and absorb toxins.
Viral Infection
Infections caused by viruses are the most difficult to treat, as viruses do not follow a regular cell division process for their growth. They replicate by binding to the receptors on the host cell membrane and infusing genetic material inside them. CMC mimics can be potential alternatives to neutralizing virus-caused infections. Many cells are rich in virus binding receptors, and fabricating CMC mimics from the cell membrane of these cells diverts viruses from host cells. This mechanism is employed for treating infections caused by the influenza virus, zika virus, and human immunodeficiency virus (HIV), and recently by COVID-19 (Figure 11).
Figure 11.
Potential of CMC mimics in treating COVID-19 infection. (A) (a) Preparation of nanodecoys using fused cell membrane vesicles derived from genetically edited 293T/ACE2 and THP-1 cells. (b) Immunofluorescence images of SARS-CoV-2-infected Vero-E6 cells after treatment with nanodecoys, scale bar = 100 μm, cell nuclei: DAPI (blue), N protein of SARS-CoV-2: Alexa 488 (green). (c) Inhibitory activity of nanodecoys against SARS-CoV-2 on Vero-E6 cells. Reprinted with permission under a Creative Commons CC-BY License from ref (397). Copyright 2020 United States National Academy of Sciences. (B) (a) Preparation of nanosponges using epithelial cell membrane and macrophage cell membrane-coated PLGA nanoparticles. (b) Neutralization of SARS-CoV-2 infection by epithelial nanosponges, macrophage (MΦ) nanosponges, RBC nanosponges (control), using SARS-CoV-2 on Vero-E6 cells. Reprinted with permission from ref (400). Copyright 2020 American Chemical Society.
The surface of the influenza virus is rich in hemagglutinin, a glycoprotein that has a high affinity toward sialic acid residues present in cells.394 RBCs membrane is rich in sialic acid and glycoproteins. Designing CMC mimics using their cell membrane can favorably interact with the influenza virus shown by RBC membrane-coated PLGA nanoparticles.74 These mimics bind efficiently with the influenza virus and form clusters that can be readily isolated in vitro by magnetic extraction.
COVID-19 is a viral infection caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The spike protein (S) of SARS-CoV-2 consists of S1 and S2 subunits. The S1 subunit engages human angiotensin-converting enzyme II (ACE2) for binding with the host cells, and the S2 subunit facilitates the entry and fusion of the virus within the host cells. Preclinical and clinical studies report that monoclonal antibodies targeting interleukin 6 (IL-6) and granulocyte-macrophage colony-stimulating factor (GM-CSF) can prevent the infection caused by SARS-CoV-2.395,396 Based on these reports, the human embryonic kidney 293T cell membrane genetically engineered to express ACE2 receptor and monocyte THP-1 cell membrane with abundant cytokine binding receptors were combined to form vesicles.397 These fused vesicles effectively bind and neutralize IL-6 and GM-CSF, suppressing the immune disorders and lung injury in the acute pneumonia mouse model. The ACE2 receptor on SARS-CoV-2 also binds to CD147 expressed on the host cells, human alveolar epithelial type II cells, and human macrophages.398,399 PLGA nanoparticles coated with their cell membrane effectively target SARS-CoV-2.400 Overall epithelial and macrophage-based CMC mimics are preferable for inhibition and neutralization of SARS-CoV-2 (Figure 11).
Zika virus is a mosquito-borne flavivirus, transmitted by Aedes mosquitos (Aedes albopictus, Aedes aegypti).401,402 They can easily pass physiological barriers like the brain–blood barrier and placental–blood barrier, causing fetal microcephaly and other neurological complications.403,404 In general, the nanoparticles cannot enter the immune-privileged sites.405 Coating gelatin-nanoparticles with the Aedes albopictus (C6/36) cell membrane circumvents this limitation.228 These CMC mimics divert the Zika virus away from the fetal brain, suppress fetal microcephaly in pregnant mice, negate virus-induced degenerative changes, prevent replication, and improve overall survival rate.
HIV infects leukocytes via interaction between glycoproteins on its surface (gp120) and cluster of differentiation 4 (CD4) receptor, C–C chemokine receptor type 5 (CCR5), or CXCR4 coreceptors on CD4+ T cells.406 CD4+ T-cell membrane-coated PLGA nanoparticles effectively treated two distinct HIV strains: X4 and R5.159 These mimics neutralized the strains and prevented the HIV-1 from binding to and entering the healthy CD4+ T cells.
Bacterial Infection
Bacteria show both positive and negative impacts on the human body. While probiotic bacteria aid the digestive process, other bacterial strains (Gram-positive: S. aureus and Gram-negative: E. coli, K. pneumonia) cause mild to severe infections and host cell death. Additionally, excessive use of antibiotics results in drug-resistant bacterial strains (e.g., methicillin-resistant S. aureus (MRSA), carbapenem-resistant K. pneumonia (CRKP)) in the human body, posing additional therapeutic challenges. This section highlights antimicrobial activity and toxin neutralization using bacterial- and cell membrane-coated mimics.
The α-toxins are a class of pore-forming toxins secreted by bacteria (E. coli, S. aureus, MRSA, etc.) that create pores in the host cell membrane, causing cell lysis.407 RBCs and platelets express several surface markers (e.g., glycophorin A in RBCs,112 toll-like receptors in platelets119) that readily interact with such pathogens. Using these receptors, RBC membrane-coated PLGA nanoparticles sequestered α-toxins115 and platelet membrane-coated PLGA nanoparticles93 delivered vancomycin to MRSA252. Platelet membrane-coated particles loaded with vancomycin (PNP-Vanc) showed superior efficacy, specificity, and retention due to the presence of platelet-specific serine-rich (SraP) adhesion sites on MRSA252.395 Similarly, RBC membrane-coated vancomycin-loaded redox responsive hydrogels absorbed α-toxins and killed MRSA.114 The intracellular reducing environment of the bacteria triggered vancomycin release from the hydrogels. Other examples include acoustic nanorobots of gold nanorods coated with the hybrid membrane (platelet and RBC). These fuel-free nanorobots accelerated toxin neutralization and removal of bacteria (MRSA USA300).196 RBC membrane coating on carbon nanotubes-based field transistors resulted in rapid detection of several pore-forming toxins. These biomimetic nanosensors could quantitatively detect live pathogens without involving traditional colony-counting methods.75
Homotypic targeting utilizes the membrane of the targeted pathogen for specificity. For example, bovine serum albumin nanoparticles coated with CRKP bacterial membranes enhanced the immune response by secreting cytokines from macrophages and activating dendritic cells.193 Similarly, S. aureus membrane-coated PLGA nanoparticles showed superior targeting efficacy toward S. aureus-infected macrophages.192 These mimics actively targeted all organs except the liver and showed improved efficacy in kidneys and lungs prone to a higher risk of S. aureus infections. Similarly, E. coli membrane-coated gold nanoparticles coated with E. coli membrane removed E. coli bacterial infection.94 They also activated dendritic cells in lymph nodes and increased the production of IFN-γ and IL-17, but not IL-4 generating type 1 T-helper cell (Th1) and type 17 T-helper cell (Th17)-based cell responses against bacterial infections.
Current Challenges
Ingraining complex biological functionalities in delivery systems is a significant outcome of coating with cell membranes that differentiates them from synthetic mimics. Throughout this review, we have emphasized why these CMC mimics utilizing surface functional properties of cells are better suited for therapeutic applications over their synthetic counterparts. Thus far, fabrication and in vitro and in vivo evaluation studies on CMC mimics are limited to the lab setting. However, the actual therapeutic dose of materials required and conditions for clinical studies are higher and stringent. In this context, it is vital to have standardized protocols and well-established characterization tools for scale-up and GMP production of these mimics to maintain quality and reduce batch-to-batch variability. Herein, we discuss some of the challenges associated with their clinical translation.
-
(1)
Large-scale expansion: Cell membrane isolation requires at least a 100 million cells that maintain their phenotype, purity, and quality while passaging. A standardized and well-established cell culture protocol specific to each cell type is essential for large-scale production. In this regard, one can benefit from the existing well-established biomanufacturing platforms using 3D bioreactors (like stirred-tanked bioreactor, WAVE bioreactor, etc.) for the ultralarge scale-up of stem cells, T cells, and dendritic cells.411−414
-
(2)
Cell membrane yield: Lab-scale procedures for cell membrane isolation are multistep and specific to each cell type and may result in loss of sample, functional receptors, and nuclear/mitochondrial/cytosol contamination. Therefore, there is a requirement for an established protocol with minimal manual steps for cell membrane isolation with high yield and purity for various cells, especially for nucleus-containing cells.
-
(3)
Assembly of CMC mimics: In CMC assembly, it is vital to control the cell membrane layers coated onto each template while achieving a homogeneous coating. The physiological effects of differences in membrane layers on CMC mimics remain unexplored. Using automated technology to improve the coating efficiency and avoid uneven coating can be a viable alternative. For example, assembly of RBC-coated CMC mimics using the microfluidic electroporation technique resulted in uniform-sized mimics.57
-
(4)
Long-term storage: Optimizing long-term storage conditions and membrane stability of CMC mimics are critical to improve their shelf life. Post-lyophilization isolated cell membranes can be stored in cold conditions and resuspended in buffers before use. However, shelf life studies to determine the stability and functional efficacy of isolated cell membranes remain unexplored.
-
(5)
Quantitative evaluation of CMC mimics: An in-depth quantitative characterization of CMC mimics is essential to avoid batch-to-batch variability in their biological efficacy. These include the number of templates coated evenly with and without the cell membrane and the amount of transmembrane protein translocated on the mimics in the correct orientation.
-
(6)
Quality control: A standard quality control criteria must be defined to ensure that the cell membranes are free of contamination like viruses, bacteria, or pyrogens. Additionally, removal of denatured proteins from the CMC mimics avoids potential immune responses to endogenous antigens. Also, every assembly step (isolation of cell membrane, synthesis of template, fabrication of CMC mimics) should be carried out under sterile conditions to avoid chemical and biological contamination and maintain GMP requirements.
-
(7)
Unwanted proteins on the CMC mimics: There are several proteins present on the cell membrane. Some are accountable for effective targeting and evading immune responses, and others are involved in interacting with the host environment affecting biodistribution, immune response, and toxicity profile. Optimizing protocols to selectively retain proteins of interest and remove unwanted proteins from the cell membrane can enhance the CMC performance and remains to explore.
-
(8)
Surface modification of cell membrane: Numerous membrane modification strategies are known, but not all of them offer proper orientation, linkage strength, and conserve membrane protein functionality. For example, noncovalent modifications protect the membrane protein functionality, but the interactions are weaker in linkage strength.415 Conversely, covalent bonding with a template is robust, but there is a risk of altering the natural membrane functionality and compromising the protein profiles. Changes in the ζ potential only primary measure the membrane modification,333 creating a necessitous gap in qualitative and quantitative evaluation techniques. It is also complicated to observe small-molecule conjugation on the cell membrane and evaluate the overall impairment.
-
(9)
Autologous cells: For designing CMC mimics, most studies have utilized immortal cell lines. However, certain cell types (like leukocytes) can be heterogeneous and induce hemolysis during a blood transfusion. In such cases, autologous cells are the most suitable option, but would require screening of the donors to prevent the use of allogeneic cells as membrane sources.
Acknowledgments
This publication has emanated from research conducted in part by a grant from Science Foundation Ireland (SFI) and the European Regional Development Fund (ERDF) under grant number 13/RC/2073_P2. K.V.K. has received funding from Science Foundation of Ireland (SFI) Industry Fellowship under grant number 19/IFA/7447. The authors would also like to acknowledge the editorial assistance of Dr. Raghvendra Bohara and Mr. Anthony Sloan and the graphic design support of Mr Maciek Dozkyk.
Glossary
Vocabulary
- Biointerface
point of interaction between nano/microparticles and the surrounding cells
- personalized medicine
modifying disease treatment to suit the patient’s genetic profile
- autologous therapy
utilizing patient-derived cells for disease treatment
- tumor microenvironment
cellular environment surrounding tumors consisting of immune cells, blood vessels, and extracellular matrix that facilitates their growth
- receptor
proteins present on the cell surface that binds selectively to a ligand to transmit signals.
Author Contributions
† These authors contributed equally.
The authors declare no competing financial interest.
References
- Latourte A.; Kloppenburg M.; Richette P. Emerging Pharmaceutical Therapies for Osteoarthritis. Nat. Rev. Rheumatol. 2020, 16, 673–688. 10.1038/s41584-020-00518-6. [DOI] [PubMed] [Google Scholar]
- Sanders J. M.; Monogue M. L.; Jodlowski T. Z.; Cutrell J. B. Pharmacologic Treatments for Coronavirus Disease 2019 (COVID-19): A Review. J. Am. Med. Assoc. 2020, 323, 1824–1836. 10.1001/jama.2020.6019. [DOI] [PubMed] [Google Scholar]
- Sahin U.; Kariko K.; Tureci O. mRNA-Based Therapeutics-Developing a New Class of Drugs. Nat. Rev. Drug Discovery 2014, 13, 759–780. 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- Van de Glind E. M.; Van Enst W. A.; Van Munster B. C.; Olde Rikkert M. G.; Scheltens P.; Scholten R. J.; Hooft L. Pharmacological Treatment of Dementia: A Scoping Review of Systematic Reviews. Dementia Geriatr. Cognit. Disord. 2013, 36, 211–228. 10.1159/000353892. [DOI] [PubMed] [Google Scholar]
- Talsky A.; Pacione L. R.; Shaw T.; Wasserman L.; Lenny A. M.; Verma A.; Hurwitz G.; Waxman R.; Morgan A.; Bhalerao S. Pharmacological Interventions for Traumatic Brain Injury. B. C. Med. J. 2011, 53, 26–31. [Google Scholar]
- Sartori S. B.; Singewald N. Novel Pharmacological Targets in Drug Development for the Treatment of Anxiety and Anxiety-Related Disorders. Pharmacol. Ther. 2019, 204, 107402–107434. 10.1016/j.pharmthera.2019.107402. [DOI] [PubMed] [Google Scholar]
- Chidambaram M.; Manavalan R.; Kathiresan K. Nanotherapeutics to Overcome Conventional Cancer Chemotherapy Limitations. J. Pharm. Pharm. Sci. 2011, 14, 67–77. 10.18433/J30C7D. [DOI] [PubMed] [Google Scholar]
- Veiseh O.; Tang B. C.; Whitehead K. A.; Anderson D. G.; Langer R. Managing Diabetes with Nanomedicine: Challenges and Opportunities. Nat. Rev. Drug Discovery 2015, 14, 45–57. 10.1038/nrd4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nel A. E. Transformational Impact of Nanomedicine: Reconciling Outcome with Promise. Nano Lett. 2020, 20, 5601–5603. 10.1021/acs.nanolett.0c02738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjidemetriou M.; Kostarelos K. Nanomedicine: Evolution of the Nanoparticle Corona. Nat. Nanotechnol. 2017, 12, 288–290. 10.1038/nnano.2017.61. [DOI] [PubMed] [Google Scholar]
- Cai P.; Zhang X.; Wang M.; Wu Y. L.; Chen X. Combinatorial Nano-Bio Interfaces. ACS Nano 2018, 12, 5078–5084. 10.1021/acsnano.8b03285. [DOI] [PubMed] [Google Scholar]
- Nienhaus K.; Wang H.; Nienhaus G. U. Nanoparticles for Biomedical Applications: Exploring and Exploiting Molecular Interactions at the Nano-Bio Interface. Mater. Today Adv. 2020, 5, 100036–100055. 10.1016/j.mtadv.2019.100036. [DOI] [Google Scholar]
- Mura S.; Nicolas J.; Couvreur P. Stimuli-Responsive Nanocarriers for Drug Delivery. Nat. Mater. 2013, 12, 991–1003. 10.1038/nmat3776. [DOI] [PubMed] [Google Scholar]
- Karimi M.; Ghasemi A.; Sahandi Zangabad P.; Rahighi R.; Moosavi Basri S. M.; Mirshekari H.; Amiri M.; Shafaei Pishabad Z.; Aslani A.; Bozorgomid M.; Ghosh D.; Beyzavi A.; Vaseghi A.; Aref A. R.; Haghani L.; Bahrami S.; Hamblin M. R. Smart Micro/Nanoparticles in Stimulus-Responsive Drug/Gene Delivery Systems. Chem. Soc. Rev. 2016, 45, 1457–1501. 10.1039/C5CS00798D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parodi A.; Molinaro R.; Sushnitha M.; Evangelopoulos M.; Martinez J. O.; Arrighetti N.; Corbo C.; Tasciotti E. Bio-Inspired Engineering of Cell and Virus-Like Nanoparticles for Drug Delivery. Biomaterials 2017, 147, 155–168. 10.1016/j.biomaterials.2017.09.020. [DOI] [PubMed] [Google Scholar]
- Moncalvo F.; Martinez Espinoza M. I.; Cellesi F. Nanosized Delivery Systems for Therapeutic Proteins: Clinically Validated Technologies and Advanced Development Strategies. Front. Bioeng. Biotechnol. 2020, 8, 89. 10.3389/fbioe.2020.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fam S. Y.; Chee C. F.; Yong C. Y.; Ho K. L.; Mariatulqabtiah A. R.; Tan W. S. Stealth Coating of Nanoparticles in Drug-Delivery Systems. Nanomaterials 2020, 10, 787–804. 10.3390/nano10040787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slastnikova T. A.; Ulasov A. V.; Rosenkranz A. A.; Sobolev A. S. Targeted Intracellular Delivery of Antibodies: The State of the Art. Front. Pharmacol. 2018, 9, 1208–1228. 10.3389/fphar.2018.01208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firer M. A.; Gellerman G. Targeted Drug Delivery for Cancer Therapy: The Other Side of Antibodies. J. Hematol. Oncol. 2012, 5, 70. 10.1186/1756-8722-5-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z.; Xiang J. Aptamer-Functionalized Nanoparticles in Targeted Delivery and Cancer Therapy. Int. J. Mol. Sci. 2020, 21, 9123–9161. 10.3390/ijms21239123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan B.; Zhang X. Aptamers As Versatile Ligands for Biomedical and Pharmaceutical Applications. Int. J. Nanomed. 2020, 15, 1059–1071. 10.2147/IJN.S237544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian Z.; Ji T. Functional Peptide-Based Drug Delivery Systems. J. Mater. Chem. B 2020, 8, 6517–6529. 10.1039/D0TB00713G. [DOI] [PubMed] [Google Scholar]
- Hoppenz P.; Els-Heindl S.; Beck-Sickinger A. G. Peptide-Drug Conjugates and Their Targets in Advanced Cancer Therapies. Front. Chem. 2020, 8, 571–594. 10.3389/fchem.2020.00571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang C.; Guan X.; Ma H.; Cong H.; Zhang W.; Miao Z. Small Molecule-Drug Conjugates: A Novel Strategy for Cancer-Targeted Treatment. Eur. J. Med. Chem. 2019, 163, 883–895. 10.1016/j.ejmech.2018.12.035. [DOI] [PubMed] [Google Scholar]
- Ekladious I.; Colson Y. L.; Grinstaff M. W. Polymer-Drug Conjugate Therapeutics: Advances, Insights and Prospects. Nat. Rev. Drug Discovery 2019, 18, 273–294. 10.1038/s41573-018-0005-0. [DOI] [PubMed] [Google Scholar]
- Merkel T. J.; Jones S. W.; Herlihy K. P.; Kersey F. R.; Shields A. R.; Napier M.; Luft J. C.; Wu H.; Zamboni W. C.; Wang A. Z.; Bear J. E.; DeSimone J. M. Using Mechanobiological Mimicry of Red Blood Cells to Extend Circulation Times of Hydrogel Microparticles. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 586–591. 10.1073/pnas.1010013108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palange A. L.; Palomba R.; Rizzuti I. F.; Ferreira M.; Decuzzi P. Deformable Discoidal Polymeric Nanoconstructs for the Precise Delivery of Therapeutic and Imaging Agents. Mol. Ther. 2017, 25, 1514–1521. 10.1016/j.ymthe.2017.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoonen L.; van Hest J. C. Compartmentalization Approaches in Soft Matter Science: From Nanoreactor Development to Organelle Mimics. Adv. Mater. 2016, 28, 1109–1128. 10.1002/adma.201502389. [DOI] [PubMed] [Google Scholar]
- Mason A. F.; Yewdall N. A.; Welzen P. L. W.; Shao J.; van Stevendaal M.; van Hest J. C. M.; Williams D. S.; Abdelmohsen L. Mimicking Cellular Compartmentalization in a Hierarchical Protocell through Spontaneous Spatial Organization. ACS Cent. Sci. 2019, 5, 1360–1365. 10.1021/acscentsci.9b00345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian V.; Poillucci A.; Correia A.; Zhang H.; Celia C.; Santos H. A. Cell Membrane-Based Nanoreactor to Mimic the Bio-Compartmentalization Strategy of a Cell. ACS Biomater. Sci. Eng. 2018, 4, 1471–1478. 10.1021/acsbiomaterials.7b00944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Tanner P.; Graff A.; Palivan C. G.; Meier W. Mimicking the Cell Membrane with Block Copolymer Membranes. J. Polym. Sci., Part A: Polym. Chem. 2012, 50, 2293–2318. 10.1002/pola.26000. [DOI] [Google Scholar]
- Peetla C.; Stine A.; Labhasetwar V. Biophysical Interactions with Model Lipid Membranes: Applications in Drug Discovery and Drug Delivery. Mol. Pharmaceutics 2009, 6, 1264–1276. 10.1021/mp9000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo C.; Bhardwaj K.; Marculescu R. Towards Cell-Based Therapeutics: A Bio-Inspired Autonomous Drug Delivery System. Nano Commun. Netw 2017, 12, 25–33. 10.1016/j.nancom.2017.01.006. [DOI] [Google Scholar]
- Petrenko V. A. Autonomous Self-Navigating Drug-Delivery Vehicles: From Science Fiction to Reality. Ther. Delivery 2017, 8, 1063–1107. 10.4155/tde-2017-0086. [DOI] [PubMed] [Google Scholar]
- Large D. E.; Soucy J. R.; Hebert J.; Auguste D. T. Advances in Receptor-Mediated, Tumor-Targeted Drug Delivery. Adv. Therap. 2019, 2, 1800091–1800117. 10.1002/adtp.201800091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M.; Sahandi Zangabad P.; Baghaee-Ravari S.; Ghazadeh M.; Mirshekari H.; Hamblin M. R. Smart Nanostructures for Cargo Delivery: Uncaging and Activating by Light. J. Am. Chem. Soc. 2017, 139, 4584–4610. 10.1021/jacs.6b08313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury A.; Kunjiappan S.; Panneerselvam T.; Somasundaram B.; Bhattacharjee C. Nanotechnology and Nanocarrier-Based Approaches on Treatment of Degenerative Diseases. Int. Nano Lett. 2017, 7, 91–122. 10.1007/s40089-017-0208-0. [DOI] [Google Scholar]
- Deng W.; Chen W.; Clement S.; Guller A.; Zhao Z.; Engel A.; Goldys E. M. Controlled Gene and Drug Release from a Liposomal Delivery Platform Triggered by X-Ray Radiation. Nat. Commun. 2018, 9, 2713–2723. 10.1038/s41467-018-05118-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimov N.; Kastner E.; Hussain M.; Perrie Y.; Szita N. Formation and Purification of Tailored Liposomes for Drug Delivery Using a Module-Based Micro Continuous-Flow System. Sci. Rep. 2017, 7, 12045–12057. 10.1038/s41598-017-11533-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gothwal A.; Khan I.; Gupta U. Polymeric Micelles: Recent Advancements in the Delivery of Anticancer Drugs. Pharm. Res. 2016, 33, 18–39. 10.1007/s11095-015-1784-1. [DOI] [PubMed] [Google Scholar]
- Moller A.; Lobb R. J. The Evolving Translational Potential of Small Extracellular Vesicles in Cancer. Nat. Rev. Cancer 2020, 20, 697–709. 10.1038/s41568-020-00299-w. [DOI] [PubMed] [Google Scholar]
- Bulbake U.; Doppalapudi S.; Kommineni N.; Khan W. Liposomal Formulations in Clinical Use: An Updated Review. Pharmaceutics 2017, 9, 12–44. 10.3390/pharmaceutics9020012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela-Moreira A.; Shi Y.; Fens M. H. A. M.; Lammers T.; Hennink W. E.; Schiffelers R. M. Clinical Application of Polymeric Micelles for the Treatment of Cancer. Mater. Chem. Front. 2017, 1, 1485–1501. 10.1039/C6QM00289G. [DOI] [Google Scholar]
- Toh M.-R.; Chiu G. N. C. Liposomes As Sterile Preparations and Limitations of Sterilisation Techniques in Liposomal Manufacturing. Asian J. Pharm. Sci. 2013, 8, 88–95. 10.1016/j.ajps.2013.07.011. [DOI] [Google Scholar]
- Harrell C. R.; Djonov V.; Fellabaum C.; Volarevic V. Risks of Using Sterilization by Gamma Radiation: The Other Side of the Coin. Int. J. Med. Sci. 2018, 15, 274–279. 10.7150/ijms.22644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia-Guerrero Y. S.; Del Prado-Audelo M. L.; Borbolla-Jimenez F. V.; Giraldo Gomez D. M.; Garcia-Aguirre I.; Colin-Castro C. A.; Morales-Gonzalez J. A.; Leyva-Gomez G.; Magana J. J. Effect of UV and Gamma Irradiation Sterilization Processes in the Properties of Different Polymeric Nanoparticles for Biomedical Applications. Materials 2020, 13, 1090–1108. 10.3390/ma13051090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R.; LeBleu V. S. The Biology, Function, and Biomedical Applications of Exosomes. Science 2020, 367, eaau6977 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klyachko N. L.; Arzt C. J.; Li S. M.; Gololobova O. A.; Batrakova E. V. Extracellular Vesicle-Based Therapeutics: Preclinical and Clinical Investigations. Pharmaceutics 2020, 12, 1171–1196. 10.3390/pharmaceutics12121171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa F.; Quarto R.; Tasso R. Extracellular Vesicles As Natural, Safe and Efficient Drug Delivery Systems. Pharmaceutics 2019, 11, 557–562. 10.3390/pharmaceutics11110557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgio S.; Noori L.; Marino Gammazza A.; Campanella C.; Logozzi M.; Fais S.; Bucchieri F.; Cappello F.; Caruso Bavisotto C. Extracellular Vesicles-Based Drug Delivery Systems: A New Challenge and the Exemplum of Malignant Pleural Mesothelioma. Int. J. Mol. Sci. 2020, 21, 5432–5460. 10.3390/ijms21155432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitford W.; Guterstam P. Exosome Manufacturing Status. Future Med. Chem. 2019, 11, 1225–1236. 10.4155/fmc-2018-0417. [DOI] [PubMed] [Google Scholar]
- Yang M.; Wu S. Y. The Advances and Challenges in Utilizing Exosomes for Delivering Cancer Therapeutics. Front. Pharmacol. 2018, 9, 735–739. 10.3389/fphar.2018.00735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng W.; He C.; Hao Y.; Wang L.; Li L.; Zhu G. Prospects and Challenges of Extracellular Vesicle-Based Drug Delivery System: Considering Cell Source. Drug Delivery 2020, 27, 585–598. 10.1080/10717544.2020.1748758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Luo J.; Chen X.; Liu W.; Chen T. Cell Membrane Coating Technology: A Promising Strategy for Biomedical Applications. Nano-Micro Lett. 2019, 11, 1–46. 10.1007/s40820-019-0330-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang T.; Zhu Q.; Wei D.; Feng J.; Yao J.; Jiang T.; Song Q.; Wei X.; Chen H.; Gao X.; Chen J. Nanoparticles Coated with Neutrophil Membranes can Effectively Treat Cancer Metastasis. ACS Nano 2017, 11, 1397–1411. 10.1021/acsnano.6b06477. [DOI] [PubMed] [Google Scholar]
- Piao J.-G.; Wang L.; Gao F.; You Y.-Z.; Xiong Y.; Yang L. Erythrocyte Membrane is an Alternative Coating to Polyethylene Glycol for Prolonging the Circulation Lifetime of Gold Nanocages for Photothermal Therapy. ACS Nano 2014, 8, 10414–10425. 10.1021/nn503779d. [DOI] [PubMed] [Google Scholar]
- Rao L.; Cai B.; Bu L. L.; Liao Q. Q.; Guo S. S.; Zhao X. Z.; Dong W. F.; Liu W. Microfluidic Electroporation-Facilitated Synthesis of Erythrocyte Membrane-Coated Magnetic Nanoparticles for Enhanced Imaging-Guided Cancer Therapy. ACS Nano 2017, 11, 3496–3505. 10.1021/acsnano.7b00133. [DOI] [PubMed] [Google Scholar]
- Shao J.; Pijpers I. A. B.; Cao S.; Williams D. S.; Yan X.; Li J.; Abdelmohsen L.; van Hest J. C. M. Biomorphic Engineering of Multifunctional Polylactide Stomatocytes toward Therapeutic Nano-Red Blood Cells. Adv. Sci. 2019, 6, 1801678–1801685. 10.1002/advs.201801678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z.; Cheng S.; Wang X.; Pang Y.; Liu J. Camouflaging Bacteria by Wrapping with Cell Membranes. Nat. Commun. 2019, 10, 3452–3461. 10.1038/s41467-019-11390-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X.; Ye X.; Wang C.; Xing C.; Miao Q.; Xie Z.; Chen X.; Zhang X.; Zhang H.; Mei L. Photothermal Cancer Immunotherapy by Erythrocyte Membrane-Coated Black Phosphorus Formulation. J. Controlled Release 2019, 296, 150–161. 10.1016/j.jconrel.2019.01.027. [DOI] [PubMed] [Google Scholar]
- Xuan M.; Shao J.; Zhao J.; Li Q.; Dai L.; Li J. Magnetic Mesoporous Silica Nanoparticles Cloaked by Red Blood Cell Membranes: Applications in Cancer Therapy. Angew. Chem., Int. Ed. 2018, 57, 6049–6053. 10.1002/anie.201712996. [DOI] [PubMed] [Google Scholar]
- Hu C. M.; Zhang L.; Aryal S.; Cheung C.; Fang R. H.; Zhang L. Erythrocyte Membrane-Camouflaged Polymeric Nanoparticles As a Biomimetic Delivery Platform. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 10980–10985. 10.1073/pnas.1106634108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parodi A.; Quattrocchi N.; van de Ven A. L.; Chiappini C.; Evangelopoulos M.; Martinez J. O.; Brown B. S.; Khaled S. Z.; Yazdi I. K.; Enzo M. V.; Isenhart L.; Ferrari M.; Tasciotti E. Synthetic Nanoparticles Functionalized with Biomimetic Leukocyte Membranes Possess Cell-Like Functions. Nat. Nanotechnol. 2013, 8, 61–68. 10.1038/nnano.2012.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L. L.; Xu J. H.; Qi G. B.; Zhao X.; Yu F.; Wang H. Core-Shell Supramolecular Gelatin Nanoparticles for Adaptive and “On-Demand” Antibiotic Delivery. ACS Nano 2014, 8, 4975–4983. 10.1021/nn501040h. [DOI] [PubMed] [Google Scholar]
- Chen Z.; Zhao P.; Luo Z.; Zheng M.; Tian H.; Gong P.; Gao G.; Pan H.; Liu L.; Ma A.; Cui H.; Ma Y.; Cai L. Cancer Cell Membrane-Biomimetic Nanoparticles for Homologous-Targeting Dual-Modal Imaging and Photothermal Therapy. ACS Nano 2016, 10, 10049–10057. 10.1021/acsnano.6b04695. [DOI] [PubMed] [Google Scholar]
- Zhang Q.; Dehaini D.; Zhang Y.; Zhou J.; Chen X.; Zhang L.; Fang R. H.; Gao W.; Zhang L. Neutrophil Membrane-Coated Nanoparticles Inhibit Synovial Inflammation and Alleviate Joint Damage in Inflammatory Arthritis. Nat. Nanotechnol. 2018, 13, 1182–1190. 10.1038/s41565-018-0254-4. [DOI] [PubMed] [Google Scholar]
- Pitchaimani A.; Nguyen T. D. T.; Marasini R.; Eliyapura A.; Azizi T.; Jaberi-Douraki M.; Aryal S. Biomimetic Natural Killer Membrane Camouflaged Polymeric Nanoparticle for Targeted Bioimaging. Adv. Funct. Mater. 2019, 29, 1806817–1806827. 10.1002/adfm.201806817. [DOI] [Google Scholar]
- Liu J. M.; Zhang D. D.; Fang G. Z.; Wang S. Erythrocyte Membrane Bioinspired Near-Infrared Persistent Luminescence Nanocarriers for in Vivo Long-Circulating Bioimaging and Drug Delivery. Biomaterials 2018, 165, 39–47. 10.1016/j.biomaterials.2018.02.042. [DOI] [PubMed] [Google Scholar]
- Xuan M.; Shao J.; Dai L.; He Q.; Li J. Macrophage Cell Membrane Camouflaged Mesoporous Silica Nanocapsules for in Vivo Cancer Therapy. Adv. Healthcare Mater. 2015, 4, 1645–1652. 10.1002/adhm.201500129. [DOI] [PubMed] [Google Scholar]
- Zuo H.; Tao J.; Shi H.; He J.; Zhou Z.; Zhang C. Platelet-Mimicking Nanoparticles Co-Loaded with W18O49 and Metformin Alleviate Tumor Hypoxia for Enhanced Photodynamic Therapy and Photothermal Therapy. Acta Biomater. 2018, 80, 296–307. 10.1016/j.actbio.2018.09.017. [DOI] [PubMed] [Google Scholar]
- Nguyen T. D. T.; Marasini R.; Rayamajhi S.; Aparicio C.; Biller D.; Aryal S. Erythrocyte Membrane Concealed Paramagnetic Polymeric Nanoparticle for Contrast-Enhanced Magnetic Resonance Imaging. Nanoscale 2020, 12, 4137–4149. 10.1039/D0NR00039F. [DOI] [PubMed] [Google Scholar]
- Li Y. J.; Yang C. X.; Yan X. P. Biomimetic Persistent Luminescent Nanoplatform for Autofluorescence-Free Metastasis Tracking and Chemophotodynamic Therapy. Anal. Chem. 2018, 90, 4188–4195. 10.1021/acs.analchem.8b00311. [DOI] [PubMed] [Google Scholar]
- Zhu J. Y.; Zheng D. W.; Zhang M. K.; Yu W. Y.; Qiu W. X.; Hu J. J.; Feng J.; Zhang X. Z. Preferential Cancer Cell Self-Recognition and Tumor Self-Targeting by Coating Nanoparticles with Homotypic Cancer Cell Membranes. Nano Lett. 2016, 16, 5895–5901. 10.1021/acs.nanolett.6b02786. [DOI] [PubMed] [Google Scholar]
- Chen H. W.; Fang Z. S.; Chen Y. T.; Chen Y. I.; Yao B. Y.; Cheng J. Y.; Chien C. Y.; Chang Y. C.; Hu C. J. Targeting and Enrichment of Viral Pathogen by Cell Membrane Cloaked Magnetic Nanoparticles for Enhanced Detection. ACS Appl. Mater. Interfaces 2017, 9, 39953–39961. 10.1021/acsami.7b09931. [DOI] [PubMed] [Google Scholar]
- Gong H.; Chen F.; Huang Z.; Gu Y.; Zhang Q.; Chen Y.; Zhang Y.; Zhuang J.; Cho Y. K.; Fang R. H.; Gao W.; Xu S.; Zhang L. Biomembrane-Modified Field Effect Transistors for Sensitive and Quantitative Detection of Biological Toxins and Pathogens. ACS Nano 2019, 13, 3714–3722. 10.1021/acsnano.9b00911. [DOI] [PubMed] [Google Scholar]
- Wu Z.; Li J.; de Ávila B. E.-F.; Li T.; Gao W.; He Q.; Zhang L.; Wang J. Water-Powered Cell-Mimicking Janus Micromotor. Adv. Funct. Mater. 2015, 25, 7497–7501. 10.1002/adfm.201503441. [DOI] [Google Scholar]
- Chen Y.; Zhang Y.; Zhuang J.; Lee J. H.; Wang L.; Fang R. H.; Gao W.; Zhang L. Cell-Membrane-Cloaked Oil Nanosponges Enable Dual-Modal Detoxification. ACS Nano 2019, 13, 7209–7215. 10.1021/acsnano.9b02773. [DOI] [PubMed] [Google Scholar]
- Ouyang J.; Wang L.; Chen W.; Zeng K.; Han Y.; Xu Y.; Xu Q.; Deng L.; Liu Y. N. Biomimetic Nanothylakoids for Efficient Imaging-Guided Photodynamic Therapy for Cancer. Chem. Commun. (Cambridge, U. K.) 2018, 54, 3468–3471. 10.1039/C8CC00674A. [DOI] [PubMed] [Google Scholar]
- Pei Q.; Hu X.; Zheng X.; Liu S.; Li Y.; Jing X.; Xie Z. Light-Activatable Red Blood Cell Membrane-Camouflaged Dimeric Prodrug Nanoparticles for Synergistic Photodynamic/Chemotherapy. ACS Nano 2018, 12, 1630–1641. 10.1021/acsnano.7b08219. [DOI] [PubMed] [Google Scholar]
- Goni F. M. The Basic Structure and Dynamics of Cell Membranes: An Update of the Singer-Nicolson Model. Biochim. Biophys. Acta, Biomembr. 2014, 1838, 1467–1476. 10.1016/j.bbamem.2014.01.006. [DOI] [PubMed] [Google Scholar]
- Bernardino de la Serna J.; Schutz G. J.; Eggeling C.; Cebecauer M. There is no Simple Model of the Plasma Membrane Organization. Front. Cell Dev. Biol. 2016, 4, 106–122. 10.3389/fcell.2016.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casares D.; Escriba P. V.; Rossello C. A. Membrane Lipid Composition: Effect on Membrane and Organelle Structure, Function and Compartmentalization and Therapeutic Avenues. Int. J. Mol. Sci. 2019, 20, 2167–2196. 10.3390/ijms20092167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucior I.; Scheuring S.; Engel A.; Burger M. M. Carbohydrate-Carbohydrate Interaction Provides Adhesion Force and Specificity for Cellular Recognition. J. Cell Biol. 2004, 165, 529–537. 10.1083/jcb.200309005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucior I.; Burger M. M. Carbohydrate-Carbohydrate Interactions in Cell Recognition. Curr. Opin. Struct. Biol. 2004, 14, 631–637. 10.1016/j.sbi.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Simons K.; Vaz W. L. Model Systems, Lipid Rafts, and Cell Membranes. Annu. Rev. Biophys. Biomol. Struct. 2004, 33, 269–295. 10.1146/annurev.biophys.32.110601.141803. [DOI] [PubMed] [Google Scholar]
- Liang H.; Huang K.; Su T.; Li Z.; Hu S.; Dinh P. U.; Wrona E. A.; Shao C.; Qiao L.; Vandergriff A. C.; Hensley M. T.; Cores J.; Allen T.; Zhang H.; Zeng Q.; Xing J.; Freytes D. O.; Shen D.; Yu Z.; Cheng K. Mesenchymal Stem Cell/Red Blood Cell-Inspired Nanoparticle Therapy in Mice with Carbon Tetrachloride-Induced Acute Liver Failure. ACS Nano 2018, 12, 6536–6544. 10.1021/acsnano.8b00553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H.; Wang K.; Wang M.; Liu R.; Song H.; Li N.; Lu Q.; Zhang W.; Du Y.; Yang W.; Zhong L.; Wang Y.; Yu B.; Wang H.; Kan Q.; Zhang H.; Wang Y.; He Z.; Sun J. Bioinspired Nanoplatelets for Chemo-Photothermal Therapy of Breast Cancer Metastasis Inhibition. Biomaterials 2019, 206, 1–12. 10.1016/j.biomaterials.2019.03.024. [DOI] [PubMed] [Google Scholar]
- Zhao H.; Li L.; Zhang J.; Zheng C.; Ding K.; Xiao H.; Wang L.; Zhang Z. CCL2 Recruits Macrophage-Membrane-Camouflaged Hollow Bismuth Selenide Nanoparticles To Facilitate Photothermal Sensitivity and Inhibit Lung Metastasis of Breast Cancer. ACS Appl. Mater. Interfaces 2018, 10, 31124–31135. 10.1021/acsami.8b11645. [DOI] [PubMed] [Google Scholar]
- Zou S.; Wang B.; Wang C.; Wang C.; Zhang L. Cell Membrane-Coated Nanoparticles: Research Advances. Nanomedicine 2020, 15, 625–641. 10.2217/nnm-2019-0388. [DOI] [PubMed] [Google Scholar]
- Lejeune A.; Moorjani M.; Gicquaud C.; Lacroix J.; Poyet P.; Gaudreault R. Nanoerythrosome, a New Derivative of Erythrocyte Ghost: Preparation and Antineoplastic Potential As Drug Carrier for Daunorubicin. Anticancer Res. 1994, 14, 915–920. [PubMed] [Google Scholar]
- Gong H.; Zhang Q.; Komarla A.; Wang S.; Duan Y.; Zhou Z.; Chen F.; Fang R. H.; Xu S.; Gao W.; Zhang L. Nanomaterial Biointerfacing via Mitochondrial Membrane Coating for Targeted Detoxification and Molecular Detection. Nano Lett. 2021, 21, 2603–2609. 10.1021/acs.nanolett.1c00238. [DOI] [PubMed] [Google Scholar]
- Fang R. H.; Hu C. M.; Luk B. T.; Gao W.; Copp J. A.; Tai Y.; O’Connor D. E.; Zhang L. Cancer Cell Membrane-Coated Nanoparticles for Anticancer Vaccination and Drug Delivery. Nano Lett. 2014, 14, 2181–2188. 10.1021/nl500618u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C. M.; Fang R. H.; Wang K. C.; Luk B. T.; Thamphiwatana S.; Dehaini D.; Nguyen P.; Angsantikul P.; Wen C. H.; Kroll A. V.; Carpenter C.; Ramesh M.; Qu V.; Patel S. H.; Zhu J.; Shi W.; Hofman F. M.; Chen T. C.; Gao W.; Zhang K.; Chien S.; Zhang L. Nanoparticle Biointerfacing by Platelet Membrane Cloaking. Nature 2015, 526, 118–121. 10.1038/nature15373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W.; Fang R. H.; Thamphiwatana S.; Luk B. T.; Li J.; Angsantikul P.; Zhang Q.; Hu C. M.; Zhang L. Modulating Antibacterial Immunity via Bacterial Membrane-Coated Nanoparticles. Nano Lett. 2015, 15, 1403–1409. 10.1021/nl504798g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai P.-Y.; Huang R.-Y.; Lin S.-Y.; Lin Y.-H.; Chang C.-W. Biomimetic Stem Cell Membrane-Camouflaged Iron Oxide Nanoparticles for Theranostic Applications. RSC Adv. 2015, 5, 98222–98230. 10.1039/C5RA17447C. [DOI] [Google Scholar]
- Chen W.; Zhang Q.; Luk B. T.; Fang R. H.; Liu Y.; Gao W.; Zhang L. Coating Nanofiber Scaffolds with Beta Cell Membrane to Promote Cell Proliferation and Function. Nanoscale 2016, 8, 10364–10370. 10.1039/C6NR00535G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehaini D.; Wei X.; Fang R. H.; Masson S.; Angsantikul P.; Luk B. T.; Zhang Y.; Ying M.; Jiang Y.; Kroll A. V.; Gao W.; Zhang L. Erythrocyte-Platelet Hybrid Membrane Coating for Enhanced Nanoparticle Functionalization. Adv. Mater. 2017, 29, 1606209–1606216. 10.1002/adma.201606209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.; Li R.; Chen H.; Wei J.; Qian H.; Su S.; Shao J.; Wang L.; Qian X.; Liu B. Human Cytotoxic T-Lymphocyte Membrane-Camouflaged Nanoparticles Combined with Low-Dose Irradiation: A New Approach to Enhance Drug Targeting in Gastric Cancer. Int. J. Nanomed. 2017, 12, 2129–2142. 10.2147/IJN.S126016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao L.; Meng Q.-F.; Huang Q.; Wang Z.; Yu G.-T.; Li A.; Ma W.; Zhang N.; Guo S.-S.; Zhao X.-Z.; Liu K.; Yuan Y.; Liu W. Platelet-Leukocyte Hybrid Membrane-Coated Immunomagnetic Beads for Highly Efficient and Highly Specific Isolation of Circulating Tumor Cells. Adv. Funct. Mater. 2018, 28, 1803531–1803539. 10.1002/adfm.201803531. [DOI] [Google Scholar]
- Wang D.; Dong H.; Li M.; Cao Y.; Yang F.; Zhang K.; Dai W.; Wang C.; Zhang X. Erythrocyte-Cancer Hybrid Membrane Camouflaged Hollow Copper Sulfide Nanoparticles for Prolonged Circulation Life and Homotypic-Targeting Photothermal/Chemotherapy of Melanoma. ACS Nano 2018, 12, 5241–5252. 10.1021/acsnano.7b08355. [DOI] [PubMed] [Google Scholar]
- Angsantikul P.; Thamphiwatana S.; Zhang Q.; Spiekermann K.; Zhuang J.; Fang R. H.; Gao W.; Obonyo M.; Zhang L. Coating Nanoparticles with Gastric Epithelial Cell Membrane for Targeted Antibiotic Delivery against Helicobacter Pylori Infection. Adv. Therap. 2018, 1, 1800016–1800024. 10.1002/adtp.201800016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitchaimani A.; Nguyen T. D. T.; Aryal S. Natural Killer Cell Membrane Infused Biomimetic Liposomes for Targeted Tumor Therapy. Biomaterials 2018, 160, 124–137. 10.1016/j.biomaterials.2018.01.018. [DOI] [PubMed] [Google Scholar]
- Huang Y.; Mei C.; Tian Y.; Nie T.; Liu Z.; Chen T. Bioinspired Tumor-Homing Nanosystem for Precise Cancer Therapy via Reprogramming of Tumor-Associated Macrophages. NPG Asia Mater. 2018, 10, 1002–1015. 10.1038/s41427-018-0091-9. [DOI] [Google Scholar]
- Li J.; Zhen X.; Lyu Y.; Jiang Y.; Huang J.; Pu K. Cell Membrane Coated Semiconducting Polymer Nanoparticles for Enhanced Multimodal Cancer Phototheranostics. ACS Nano 2018, 12, 8520–8530. 10.1021/acsnano.8b04066. [DOI] [PubMed] [Google Scholar]
- Rao L.; Yu G. T.; Meng Q. F.; Bu L. L.; Tian R.; Lin L. S.; Deng H.; Yang W.; Zan M.; Ding J.; Li A.; Xiao H.; Sun Z. J.; Liu W.; Chen X. Cancer Cell Membrane-Coated Nanoparticles for Personalized Therapy in Patient-Derived Xenograft Models. Adv. Funct. Mater. 2019, 29, 1905671–1905680. 10.1002/adfm.201905671. [DOI] [Google Scholar]
- Cheng S.; Xu C.; Jin Y.; Li Y.; Zhong C.; Ma J.; Yang J.; Zhang N.; Li Y.; Wang C.; Yang Z.; Wang Y. Artificial Mini Dendritic Cells Boost T Cell–Based Immunotherapy for Ovarian Cancer. Adv. Sci. 2020, 7, 1903301–1903314. 10.1002/advs.201903301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosman G. J. Survival of Red Blood Cells After Transfusion: Processes and Consequences. Front. Physiol. 2013, 4, 376–383. 10.3389/fphys.2013.00376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Q.; Zhang Y.; Li Z.; Hou X.; Feng N. Red Blood Cell Membrane-Camouflaged Nanoparticles: A Novel Drug Delivery System for Antitumor Application. Acta Pharm. Sin. B 2019, 9, 675–689. 10.1016/j.apsb.2019.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa C. H.; Anselmo A. C.; Mitragotri S.; Muzykantov V. Red Blood Cells: Supercarriers for Drugs, Biologicals, and Nanoparticles and Inspiration for Advanced Delivery Systems. Adv. Drug Delivery Rev. 2016, 106, 88–103. 10.1016/j.addr.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.; Sun Y.; Zhou X.; Chen C.; Jiao L.; Li W.; Gou S.; Li Y.; Du J.; Chen G.; Zhai W.; Wu Y.; Qi Y.; Gao Y. CD47/SIRP Alpha Blocking Peptide Identification and Synergistic Effect with Irradiation for Cancer Immunotherapy. J. Immunother. Cancer 2020, 8, e000905 10.1136/jitc-2020-000905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedik P. S.; Hamlin S. K. The Physiologic Role of Erythrocytes in Oxygen Delivery and Implications for Blood Storage. Crit. Care Nurs. Clin. North Am. 2014, 26, 325–335. 10.1016/j.ccell.2014.04.002. [DOI] [PubMed] [Google Scholar]
- Minasyan H. Phagocytosis and Oxycytosis: Two Arms of Human Innate Immunity. Immunol. Res. 2018, 66, 271–280. 10.1007/s12026-018-8988-5. [DOI] [PubMed] [Google Scholar]
- Pretini V.; Koenen M. H.; Kaestner L.; Fens M.; Schiffelers R. M.; Bartels M.; Van Wijk R. Red Blood Cells: Chasing Interactions. Front. Physiol. 2019, 10, 945–951. 10.3389/fphys.2019.00945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Zhang J.; Chen W.; Angsantikul P.; Spiekermann K. A.; Fang R. H.; Gao W.; Zhang L. Erythrocyte Membrane-Coated Nanogel for Combinatorial Antivirulence and Responsive Antimicrobial Delivery against Staphylococcus Aureus Infection. J. Controlled Release 2017, 263, 185–191. 10.1016/j.jconrel.2017.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C. M.; Fang R. H.; Copp J.; Luk B. T.; Zhang L. A Biomimetic Nanosponge That Absorbs Pore-Forming Toxins. Nat. Nanotechnol. 2013, 8, 336–340. 10.1038/nnano.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y.; Carrim N.; Wang Y.; Gallant R. C.; Marshall A.; Ni H. Platelets in Hemostasis and Thrombosis: Novel Mechanisms of Fibrinogen-Independent Platelet Aggregation and Fibronectin-Mediated Protein Wave of Hemostasis. J. Biomed. Res. 2015, 29, 437–444. 10.7555/JBR.29.20150121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavergne M.; Janus-Bell E.; Schaff M.; Gachet C.; Mangin P. H. Platelet Integrins in Tumor Metastasis: Do they Represent a Therapeutic Target?. Cancers 2017, 9, 133–149. 10.3390/cancers9100133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews R. K.; López J.; Berndt M. C. Molecular Mechanisms of Platelet Adhesion and Activation. Int. J. Biochem. Cell Biol. 1997, 29, 91–105. 10.1016/S1357-2725(96)00122-7. [DOI] [PubMed] [Google Scholar]
- Portier I.; Campbell R. A. Role of Platelets in Detection and Regulation of Infection. Arterioscler., Thromb., Vasc. Biol. 2020, 41, 70–78. 10.1161/ATVBAHA.120.314645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews R. K.; Lopez J. A.; Berndt M. C. Molecular Mechanisms of Platelet Adhesion and Activation. Int. J. Biochem. Cell Biol. 1997, 29, 91–105. 10.1016/S1357-2725(96)00122-7. [DOI] [PubMed] [Google Scholar]
- Xu L.; Gao F.; Fan F.; Yang L. Platelet Membrane Coating Coupled with Solar Irradiation Endows a Photodynamic Nanosystem with Both Improved Antitumor Efficacy and Undetectable Skin Damage. Biomaterials 2018, 159, 59–67. 10.1016/j.biomaterials.2017.12.028. [DOI] [PubMed] [Google Scholar]
- Wang B.; Chen G.; Urabe G.; Xie R.; Wang Y.; Shi X.; Guo L. W.; Gong S.; Kent K. C. A Paradigm of Endothelium-Protective and Stent-Free Anti-Restenotic Therapy Using Biomimetic Nanoclusters. Biomaterials 2018, 178, 293–301. 10.1016/j.biomaterials.2018.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X.; Ying M.; Dehaini D.; Su Y.; Kroll A. V.; Zhou J.; Gao W.; Fang R. H.; Chien S.; Zhang L. Nanoparticle Functionalization with Platelet Membrane Enables Multifactored Biological Targeting and Detection of Atherosclerosis. ACS Nano 2018, 12, 109–116. 10.1021/acsnano.7b07720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama D.; Iida T.; Nakase H. The Phagocytic Function of Macrophage-Enforcing Innate Immunity and Tissue Homeostasis. Int. J. Mol. Sci. 2018, 19, 92–105. 10.3390/ijms19010092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh Z.; Brooks P. J.; Barzilay O.; Fine N.; Glogauer M. Macrophages, Foreign Body Giant Cells and Their Response to Implantable Biomaterials. Materials 2015, 8, 5671–5701. 10.3390/ma8095269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C.; Pamer E. G. Monocyte Recruitment during Infection and Inflammation. Nat. Rev. Immunol. 2011, 11, 762–574. 10.1038/nri3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor P. R.; Martinez-Pomares L.; Stacey M.; Lin H. H.; Brown G. D.; Gordon S. Macrophage Receptors and Immune Recognition. Annu. Rev. Immunol. 2005, 23, 901–944. 10.1146/annurev.immunol.23.021704.115816. [DOI] [PubMed] [Google Scholar]
- Si J.; Shao S.; Shen Y.; Wang K. Macrophages As Active Nanocarriers for Targeted Early and Adjuvant Cancer Chemotherapy. Small 2016, 12, 5108–5119. 10.1002/smll.201601282. [DOI] [PubMed] [Google Scholar]
- Cao H.; Dan Z.; He X.; Zhang Z.; Yu H.; Yin Q.; Li Y. Liposomes Coated with Isolated Macrophage Membrane can Target Lung Metastasis of Breast Cancer. ACS Nano 2016, 10, 7738–7748. 10.1021/acsnano.6b03148. [DOI] [PubMed] [Google Scholar]
- Thamphiwatana S.; Angsantikul P.; Escajadillo T.; Zhang Q.; Olson J.; Luk B. T.; Zhang S.; Fang R. H.; Gao W.; Nizet V.; Zhang L. Macrophage-Like Nanoparticles Concurrently Absorbing Endotoxins and Proinflammatory Cytokines for Sepsis Management. Proc. Natl. Acad. Sci. U. S. A. 2017, 114, 11488–11493. 10.1073/pnas.1714267114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Q. F.; Rao L.; Zan M.; Chen M.; Yu G. T.; Wei X.; Wu Z.; Sun Y.; Guo S. S.; Zhao X. Z.; Wang F. B.; Liu W. Macrophage Membrane-Coated Iron Oxide Nanoparticles for Enhanced Photothermal Tumor Therapy. Nanotechnology 2018, 29, 134004. 10.1088/1361-6528/aaa7c7. [DOI] [PubMed] [Google Scholar]
- Rao L.; He Z.; Meng Q. F.; Zhou Z.; Bu L. L.; Guo S. S.; Liu W.; Zhao X. Z. Effective Cancer Targeting and Imaging Using Macrophage Membrane-Camouflaged Upconversion Nanoparticles. J. Biomed. Mater. Res., Part A 2017, 105, 521–530. 10.1002/jbm.a.35927. [DOI] [PubMed] [Google Scholar]
- Wright H. L.; Moots R. J.; Bucknall R. C.; Edwards S. W. Neutrophil Function in Inflammation and Inflammatory Diseases. Rheumatology 2010, 49, 1618–1631. 10.1093/rheumatology/keq045. [DOI] [PubMed] [Google Scholar]
- Selders G. S.; Fetz A. E.; Radic M. Z.; Bowlin G. L. An Overview of the Role of Neutrophils in Innate Immunity, Inflammation and Host-Biomaterial Integration. Regen. Biomater 2017, 4, 55–68. 10.1093/rb/rbw041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortaz E.; Alipoor S. D.; Adcock I. M.; Mumby S.; Koenderman L. Update on Neutrophil Function in Severe Inflammation. Front. Immunol. 2018, 9, 2171–2184. 10.3389/fimmu.2018.02171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira S.; Rosowski E. E.; Huttenlocher A. Neutrophil Migration in Infection and Wound Repair: Going Forward in Reverse. Nat. Rev. Immunol. 2016, 16, 378–391. 10.1038/nri.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosales C. Neutrophil: A Cell with Many Roles in Inflammation or Several Cell Types?. Front. Physiol. 2018, 9, 113–129. 10.3389/fphys.2018.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikis V. A.; Simon S. I. Neutrophil Mechanosignaling Promotes Integrin Engagement with Endothelial Cells and Motility within Inflamed Vessels. Front. Immunol. 2018, 9, 2774–2787. 10.3389/fimmu.2018.02774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan M.; Shao J.; Dai L.; Li J.; He Q. Macrophage Cell Membrane Camouflaged Au Nanoshells for in Vivo Prolonged Circulation Life and Enhanced Cancer Photothermal Therapy. ACS Appl. Mater. Interfaces 2016, 8, 9610–9618. 10.1021/acsami.6b00853. [DOI] [PubMed] [Google Scholar]
- Abel A. M.; Yang C.; Thakar M. S.; Malarkannan S. Natural Killer Cells: Development, Maturation, and Clinical Utilization. Front. Immunol. 2018, 9, 1869–1872. 10.3389/fimmu.2018.01869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S. Y.; Fu T.; Jiang Y. Z.; Shao Z. M. Natural Killer Cells in Cancer Biology and Therapy. Mol. Cancer 2020, 19, 1–26. 10.1186/s12943-020-01238-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chester C.; Fritsch K.; Kohrt H. E. Natural Killer Cell Immunomodulation: Targeting Activating, Inhibitory, and Co-stimulatory Receptor Signaling for Cancer Immunotherapy. Front. Immunol. 2015, 6, 601–609. 10.3389/fimmu.2015.00601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo-Herrero S.; Lopez-Soto A.; Sordo-Bahamonde C.; Gonzalez-Rodriguez A. P.; Vitale M.; Gonzalez S. NK Cell-Based Immunotherapy in Cancer Metastasis. Cancers 2019, 11, 29. 10.3390/cancers11010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivier E.; Ugolini S.; Blaise D.; Chabannon C.; Brossay L. Targeting Natural Killer Cells and Natural Killer T Cells in Cancer. Nat. Rev. Immunol. 2012, 12, 239–252. 10.1038/nri3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M.; Zhang J.; Jiang W.; Chen Y.; Tian Z. Natural Killer Cell lines in Tumor Immunotherapy. Front. Med. 2012, 6, 56–66. 10.1007/s11684-012-0177-7. [DOI] [PubMed] [Google Scholar]
- Suck G.; Branch D. R.; Smyth M. J.; Miller R. G.; Vergidis J.; Fahim S.; Keating A. KHYG-1, A Model for the Study of Enhanced Natural Killer Cell Cytotoxicity. Exp. Hematol. 2005, 33, 1160–1171. 10.1016/j.exphem.2005.06.024. [DOI] [PubMed] [Google Scholar]
- Zhang J.; Zheng H.; Diao Y. Natural Killer Cells and Current Applications of Chimeric Antigen Receptor-Modified NK-92 Cells in Tumor Immunotherapy. Int. J. Mol. Sci. 2019, 20, 317–336. 10.3390/ijms20020317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T.; Nakazawa T.; Natsume A.; Nishimura F.; Nakamura M.; Matsuda R.; Omoto K.; Tanaka Y.; Shida Y.; Park Y. S.; Motoyama Y.; Nakagawa I.; Yamada S.; Tamura K.; Takeshima Y.; Takamura Y.; Wakabayashi T.; Nakase H. Novel Human NK Cell Line Carrying CAR Targeting EGFRvIII Induces Antitumor Effects in Glioblastoma Cells. Anticancer Res. 2018, 38, 5049–5056. 10.21873/anticanres.12824. [DOI] [PubMed] [Google Scholar]
- Xie G.; Dong H.; Liang Y.; Ham J. D.; Rizwan R.; Chen J. CAR-NK Cells: A Promising Cellular Immunotherapy for Cancer. EBioMedicine 2020, 59, 102975–102984. 10.1016/j.ebiom.2020.102975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng G.; Sun Z.; Li S.; Peng X.; Li W.; Zhou L.; Ma Y.; Gong P.; Cai L. Cell-Membrane Immunotherapy Based on Natural Killer Cell Membrane Coated Nanoparticles for the Effective Inhibition of Primary and Abscopal Tumor Growth. ACS Nano 2018, 12, 12096–12108. 10.1021/acsnano.8b05292. [DOI] [PubMed] [Google Scholar]
- Kumar B. V.; Connors T. J.; Farber D. L. Human T Cell Development, Localization, and Function throughout Life. Immunity 2018, 48, 202–213. 10.1016/j.immuni.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudino S. J.; Kumar P. Cross-Talk between Antigen Presenting Cells and T Cells Impacts Intestinal Homeostasis, Bacterial Infections, and Tumorigenesis. Front. Immunol. 2019, 10, 360–373. 10.3389/fimmu.2019.00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzhachenko R. V.; Shanker A. CD8(+) T Lymphocyte and NK Cell Network: Circuitry in the Cytotoxic Domain of Immunity. Front. Immunol. 2019, 10, 1906–1912. 10.3389/fimmu.2019.01906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain S. L.; McKinstry K. K.; Strutt T. M. Expanding Roles for CD4(+) T Cells in Immunity to Viruses. Nat. Rev. Immunol. 2012, 12, 136–148. 10.1038/nri3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okoye A. A.; Picker L. J. CD4(+) T-cell Depletion in HIV Infection: Mechanisms of Immunological Failure. Immunol. Rev. 2013, 254, 54–64. 10.1111/imr.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner R. C.; Sterner R. M. CAR-T Cell Therapy: Current Limitations and Potential Strategies. Blood Cancer J. 2021, 11, 69–79. 10.1038/s41408-021-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W.; Zhu D.; Li J.; Chen X.; Xie W.; Jiang X.; Wu L.; Wang G.; Xiao Y.; Liu Z.; Wang F.; Li A.; Shao D.; Dong W.; Liu W.; Yuan Y. Coating Biomimetic Nanoparticles with Chimeric Antigen Receptor T Cell-Membrane provides High Specificity for Hepatocellular Carcinoma Photothermal Therapy Treatment. Theranostics 2020, 10, 1281–1295. 10.7150/thno.40291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y.; Pan H.; Li W.; Chen Z.; Ma A.; Yin T.; Liang R.; Chen F.; Ma Y.; Jin Y.; Zheng M.; Li B.; Cai L. T Cell Membrane Mimicking Nanoparticles with Bioorthogonal Targeting and Immune Recognition for Enhanced Photothermal Therapy. Adv. Sci. 2019, 6, 1900251–1900259. 10.1002/advs.201900251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X.; Zhang G.; Ran D.; Krishnan N.; Fang R. H.; Gao W.; Spector S. A.; Zhang L. T-Cell-Mimicking Nanoparticles can Neutralize HIV Infectivity. Adv. Mater. 2018, 30, 1802233–1802241. 10.1002/adma.201802233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patente T. A.; Pinho M. P.; Oliveira A. A.; Evangelista G. C. M.; Bergami-Santos P. C.; Barbuto J. A. M. Human Dendritic Cells: Their Heterogeneity and Clinical Application Potential in Cancer Immunotherapy. Front. Immunol. 2019, 9, 3176–3193. 10.3389/fimmu.2018.03176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Xiang Y.; Xin V. W.; Wang X. W.; Peng X. C.; Liu X. Q.; Wang D.; Li N.; Cheng J. T.; Lyv Y. N.; Cui S. Z.; Ma Z.; Zhang Q.; Xin H. W. Dendritic Cell Biology and its Role in Tumor Immunotherapy. J. Hematol. Oncol. 2020, 13, 107–114. 10.1186/s13045-020-00939-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geginat J.; Nizzoli G.; Paroni M.; Maglie S.; Larghi P.; Pascolo S.; Abrignani S. Immunity to Pathogens Taught by Specialized Human Dendritic Cell Subsets. Front. Immunol. 2015, 6, 527–539. 10.3389/fimmu.2015.00527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt C. D.; Ma J. K.; Chalouni C.; Ebersold M.; Bou-Reslan H.; Carano R. A.; Mellman I.; Delamarre L. Mature Dendritic Cells Use Endocytic Receptors to Capture and Present Antigens. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 4287–4292. 10.1073/pnas.0910609107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbongue J. C.; Nieves H. A.; Torrez T. W.; Langridge W. H. The Role of Dendritic Cell Maturation in the Induction of Insulin-Dependent Diabetes Mellitus. Front. Immunol. 2017, 8, 327–335. 10.3389/fimmu.2017.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiko G. E.; Horvat J. C.; Beagley K. W.; Hansbro P. M. Immunological Decision-Making: How Does the Immune System Decide to Mount a Helper T-Cell Response?. Immunology 2008, 123, 326–338. 10.1111/j.1365-2567.2007.02719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anne Gowda V. M.; Smitha T. The Dendritic Cell Tool for Oral Cancer Treatment. J. Oral Maxillofac. Pathol. 2019, 23, 326–329. 10.4103/jomfp.JOMFP_325_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Gayo E.; Yu X. G. Role of Dendritic Cells in Natural Immune Control of HIV-1 Infection. Front. Immunol. 2019, 10, 1306–1314. 10.3389/fimmu.2019.01306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W. L.; Zou M. Z.; Liu T.; Zeng J. Y.; Li X.; Yu W. Y.; Li C. X.; Ye J. J.; Song W.; Feng J.; Zhang X. Z. Expandable Immunotherapeutic Nanoplatforms Engineered from Cytomembranes of Hybrid Cells Derived from Cancer and Dendritic Cells. Adv. Mater. 2019, 31, 1900499–1900508. 10.1002/adma.201900499. [DOI] [PubMed] [Google Scholar]
- Nath S.; Mukherjee P. MUC1: A Multifaceted Oncoprotein with a Key Role in Cancer Progression. Trends Mol. Med. 2014, 20, 332–342. 10.1016/j.molmed.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokeland G.; Schumacher U. The Functional Role of Integrins during Intra- and Extravasation within the Metastatic Cascade. Mol. Cancer 2019, 18, 12–30. 10.1186/s12943-018-0937-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon-Alonso M.; Hirsch T.; Wildmann C.; van der Bruggen P. Galectin-3 Captures Interferon-Gamma in the Tumor Matrix Reducing Chemokine Gradient Production and T-cell Tumor Infiltration. Nat. Commun. 2017, 8, 793–805. 10.1038/s41467-017-00925-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose R. J.; Paulmurugan R.; Moon J.; Lee S. H.; Park H. Cell Membrane-Coated Nanocarriers: The Emerging Targeted Delivery System for Cancer Theranostics. Drug Discovery Today 2018, 23, 891–899. 10.1016/j.drudis.2018.02.001. [DOI] [PubMed] [Google Scholar]
- Zhu J.; Zhang M.; Zheng D.; Hong S.; Feng J.; Zhang X. Z. A Universal Approach to Render Nanomedicine with Biological Identity Derived from Cell Membranes. Biomacromolecules 2018, 19, 2043–2052. 10.1021/acs.biomac.8b00242. [DOI] [PubMed] [Google Scholar]
- Zhang J.; Miao Y.; Ni W.; Xiao H.; Zhang J. Cancer Cell Membrane-Coated Silica Nanoparticles Loaded with ICG for Tumour Specific Photothermal Therapy of Osteosarcoma. Artif. Cells, Nanomed., Biotechnol. 2019, 47, 2298–2305. 10.1080/21691401.2019.1622554. [DOI] [PubMed] [Google Scholar]
- Nie D.; Dai Z.; Li J.; Yang Y.; Xi Z.; Wang J.; Zhang W.; Qian K.; Guo S.; Zhu C.; Wang R.; Li Y.; Yu M.; Zhang X.; Shi X.; Gan Y. Cancer-Cell-Membrane-Coated Nanoparticles with a Yolk–Shell Structure Augment Cancer Chemotherapy. Nano Lett. 2020, 20, 936–946. 10.1021/acs.nanolett.9b03817. [DOI] [PubMed] [Google Scholar]
- Han Y.; Li X.; Zhang Y.; Han Y.; Chang F.; Ding J. Mesenchymal Stem Cells for Regenerative Medicine. Cells 2019, 8, 886–917. 10.3390/cells8080886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Fuentes D. E.; Fernandez-Garza L. E.; Samia-Meza J. A.; Barrera-Barrera S. A.; Caplan A. I.; Barrera-Saldana H. A. Mesenchymal Stem Cells Current Clinical Applications: A Systematic Review. Arch. Med. Res. 2021, 52, 93–101. 10.1016/j.arcmed.2020.08.006. [DOI] [PubMed] [Google Scholar]
- Kabat M.; Bobkov I.; Kumar S.; Grumet M. Trends in Mesenchymal Stem Cell Clinical Trials 2004–2018: Is Efficacy Optimal in a Narrow Dose Range?. Stem Cells Transl. Med. 2020, 9, 17–27. 10.1002/sctm.19-0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuckey D. W.; Shah K. Stem Cell-Based Therapies for Cancer treatment: Separating Hope from Hype. Nat. Rev. Cancer 2014, 14, 683–691. 10.1038/nrc3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. H.; Zhou Y.; Tabata Y.; Gao J. Q. Mesenchymal Stem Cell-Based Drug Delivery Strategy: From Cells to Biomimetic. J. Controlled Release 2019, 294, 102–113. 10.1016/j.jconrel.2018.12.019. [DOI] [PubMed] [Google Scholar]
- Hmadcha A.; Martin-Montalvo A.; Gauthier B. R.; Soria B.; Capilla-Gonzalez V. Therapeutic Potential of Mesenchymal Stem Cells for Cancer Therapy. Front. Bioeng. Biotechnol. 2020, 8, 43–54. 10.3389/fbioe.2020.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Sousa P.A.; Downie J.M.; Tye B.J.; Bruce K.; Dand P.; Dhanjal S.; Serhal P.; Harper J.; Turner M.; Bateman M. Development and Production of Good Manufacturing Practice Grade Human Embryonic Stem Cell Lines As Source Material for Clinical Application. Stem Cell Res. 2016, 17, 379–390. 10.1016/j.scr.2016.08.011. [DOI] [PubMed] [Google Scholar]
- Vitillo L.; Durance C.; Hewitt Z.; Moore H.; Smith A.; Vallier L. GMP-Grade Neural Progenitor Derivation and Differentiation from Clinical-Grade Human Embryonic Stem Cells. Stem Cell Res. Ther. 2020, 11, 406–415. 10.1186/s13287-020-01915-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heirani-Tabasi A.; Toosi S.; Mirahmadi M.; Mishan M. A.; Bidkhori H. R.; Bahrami A. R.; Behravan J.; Naderi-Meshkin H. Chemokine Receptors Expression in MSCs: Comparative Analysis in Different Sources and Passages. Tissue Eng. Regener. Med. 2017, 14, 605–615. 10.1007/s13770-017-0069-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C.; Lin Z.; Wu Z.; Lin X.; He Q. Stem-Cell-Membrane Camouflaging on Near-Infrared Photoactivated Upconversion Nanoarchitectures for in Vivo Remote-Controlled Photodynamic Therapy. ACS Appl. Mater. Interfaces 2016, 8, 34252–34260. 10.1021/acsami.6b12865. [DOI] [PubMed] [Google Scholar]
- Gao C.; Lin Z.; Jurado-Sanchez B.; Lin X.; Wu Z.; He Q. Stem Cell Membrane-Coated Nanogels for Highly Efficient in Vivo Tumor Targeted Drug Delivery. Small 2016, 12, 4056–4062. 10.1002/smll.201600624. [DOI] [PubMed] [Google Scholar]
- Tang J.; Shen D.; Caranasos T. G.; Wang Z.; Vandergriff A. C.; Allen T. A.; Hensley M. T.; Dinh P. U.; Cores J.; Li T. S.; Zhang J.; Kan Q.; Cheng K. Therapeutic Microparticles Functionalized with Biomimetic Cardiac Stem Cell Membranes and Secretome. Nat. Commun. 2017, 8, 13724–13732. 10.1038/ncomms13724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J.; Zhang S.; Liu J.; Liu F.; Du F.; Li M.; Chen A. T.; Bao Y.; Suh H. W.; Avery J.; Deng G.; Zhou Y.; Wu P.; Sheth K.; Wang H.; Zhou J. Targeted Drug Delivery to Stroke via Chemotactic Recruitment of Nanoparticles Coated with Membrane of Engineered Neural Stem Cells. Small 2019, 15, 1902011–1902018. 10.1002/smll.201902011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhavy T. J.; Kahne D.; Walker S. The Bacterial Cell Envelope. Cold Spring Harbor Perspect. Biol. 2010, 2, a000414. 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E. Y.; Choi D. Y.; Kim D. K.; Kim J. W.; Park J. O.; Kim S.; Kim S. H.; Desiderio D. M.; Kim Y. K.; Kim K. P.; Gho Y. S. Gram-Positive Bacteria Produce Membrane Vesicles: Proteomics-Based Characterization of Staphylococcus Aureus-Derived Membrane Vesicles. Proteomics 2009, 9, 5425–5436. 10.1002/pmic.200900338. [DOI] [PubMed] [Google Scholar]
- Kaparakis-Liaskos M.; Ferrero R. L. Immune Modulation by Bacterial Outer Membrane Vesicles. Nat. Rev. Immunol. 2015, 15, 375–387. 10.1038/nri3837. [DOI] [PubMed] [Google Scholar]
- Gao F.; Xu L.; Yang B.; Fan F.; Yang L. Kill the Real with the Fake: Eliminate Intracellular Staphylococcus Aureus Using Nanoparticle Coated with its Extracellular Vesicle Membrane As Active-Targeting Drug Carrier. ACS Infect. Dis. 2019, 5, 218–227. 10.1021/acsinfecdis.8b00212. [DOI] [PubMed] [Google Scholar]
- Wu G.; Ji H.; Guo X.; Li Y.; Ren T.; Dong H.; Liu J.; Liu Y.; Shi X.; He B. Nanoparticle Reinforced Bacterial Outer-Membrane Vesicles Effectively Prevent Fatal Infection of Carbapenem-Resistant Klebsiella Pneumonia. Nanomedicine 2020, 24, 102148–102158. 10.1016/j.nano.2019.102148. [DOI] [PubMed] [Google Scholar]
- Gujrati V.; Prakash J.; Malekzadeh-Najafabadi J.; Stiel A.; Klemm U.; Mettenleiter G.; Aichler M.; Walch A.; Ntziachristos V. Bioengineered Bacterial Vesicles As Biological Nano-Heaters for Optoacoustic Imaging. Nat. Commun. 2019, 10, 1114–1123. 10.1038/s41467-019-09034-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gujrati V.; Kim S.; Kim S. H.; Min J. J.; Choy H. E.; Kim S. C.; Jon S. Bioengineered Bacterial Outer Membrane Vesicles As Cell-Specific Drug-Delivery Vehicles for Cancer Therapy. ACS Nano 2014, 8, 1525–1537. 10.1021/nn405724x. [DOI] [PubMed] [Google Scholar]
- Esteban-Fernandez de Avila B.; Angsantikul P.; Ramirez-Herrera D. E.; Soto F.; Teymourian H.; Dehaini D.; Chen Y.; Zhang L.; Wang J. Hybrid Biomembrane-Functionalized Nanorobots for Concurrent Removal of Pathogenic Bacteria and Toxins. Sci. Rob. 2018, 3, eaat0485 10.1126/scirobotics.aat0485. [DOI] [PubMed] [Google Scholar]
- Chen H. Y.; Deng J.; Wang Y.; Wu C. Q.; Li X.; Dai H. W. Hybrid Cell Membrane-Coated Nanoparticles: A Multifunctional Biomimetic Platform for Cancer Diagnosis and Therapy. Acta Biomater. 2020, 112, 1–13. 10.1016/j.actbio.2020.05.028. [DOI] [PubMed] [Google Scholar]
- Jiang Q.; Liu Y.; Guo R.; Yao X.; Sung S.; Pang Z.; Yang W. Erythrocyte-Cancer Hybrid Membrane-Camouflaged Melanin Nanoparticles for Enhancing Photothermal Therapy Efficacy in Tumors. Biomaterials 2019, 192, 292–308. 10.1016/j.biomaterials.2018.11.021. [DOI] [PubMed] [Google Scholar]
- Gong C.; Yu X.; You B.; Wu Y.; Wang R.; Han L.; Wang Y.; Gao S.; Yuan Y. Macrophage-Cancer Hybrid Membrane-Coated Nanoparticles for Targeting Lung Metastasis in Breast Cancer Therapy. J. Nanobiotechnol. 2020, 18, 1–17. 10.1186/s12951-020-00649-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D.; Liu C.; You S.; Zhang K.; Li M.; Cao Y.; Wang C.; Dong H.; Zhang X. Bacterial Vesicle-Cancer Cell Hybrid Membrane-Coated Nanoparticles for Tumor Specific Immune Activation and Photothermal Therapy. ACS Appl. Mater. Interfaces 2020, 12, 41138–41147. 10.1021/acsami.0c13169. [DOI] [PubMed] [Google Scholar]
- Chen M. S.; Zhang Y.; Zhang L. Fabrication and Characterization of a 3D Bioprinted Nanoparticle-Hydrogel Hybrid Device for Biomimetic Detoxification. Nanoscale 2017, 9, 14506–14511. 10.1039/C7NR05322C. [DOI] [PubMed] [Google Scholar]
- Klymchenko A. S.; Kreder R. Fluorescent Probes for Lipid Rafts: From Model Membranes to Living Cells. Chem. Biol. 2014, 21, 97–113. 10.1016/j.chembiol.2013.11.009. [DOI] [PubMed] [Google Scholar]
- Yao J.; Fan Y.; Li Y.; Huang L. Strategies on the Nuclear-Targeted Delivery of Genes. J. Drug Target 2013, 21, 926–939. 10.3109/1061186X.2013.830310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandinetti G.; Smith A. E.; Reineke T. M. Membrane and Nuclear Permeabilization by Polymeric pDNA Vehicles: Efficient Method for Gene Delivery or Mechanism of Cytotoxicity?. Mol. Pharmaceutics 2012, 9, 523–538. 10.1021/mp200368p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S.; Krijnse-Locker J. Modification of Intracellular Membrane Structures for Virus Replication. Nat. Rev. Microbiol. 2008, 6, 363–374. 10.1038/nrmicro1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurunnabi M.; Khatun Z.; Badruddoza A. Z. M.; McCarthy J. R.; Lee Y.-k.; Huh K. M. Biomaterials and Bioengineering Approaches for Mitochondria and Nuclear Targeting Drug Delivery. ACS Biomater. Sci. Eng. 2019, 5, 1645–1660. 10.1021/acsbiomaterials.8b01615. [DOI] [Google Scholar]
- Rao L.; Bu L.-L.; Meng Q.-F.; Cai B.; Deng W.-W.; Li A.; Li K.; Guo S.-S.; Zhang W.-F.; Liu W.; Sun Z.-J.; Zhao X.-Z. Antitumor Platelet-Mimicking Magnetic Nanoparticles. Adv. Funct. Mater. 2017, 27, 1604774–1604783. 10.1002/adfm.201604774. [DOI] [Google Scholar]
- Suski J. M.; Lebiedzinska M.; Wojtala A.; Duszynski J.; Giorgi C.; Pinton P.; Wieckowski M. R. Isolation of Plasma Membrane-Associated Membranes from Rat Liver. Nat. Protoc. 2014, 9, 312–322. 10.1038/nprot.2014.016. [DOI] [PubMed] [Google Scholar]
- Zhang N.; Li M.; Sun X.; Jia H.; Liu W. NIR-Responsive Cancer Cytomembrane-Cloaked Carrier-Free Nanosystems for Highly Efficient and Self-Targeted Tumor Drug Delivery. Biomaterials 2018, 159, 25–36. 10.1016/j.biomaterials.2018.01.007. [DOI] [PubMed] [Google Scholar]
- Han X.; Shen S.; Fan Q.; Chen G.; Archibong E.; Dotti G.; Liu Z.; Gu Z.; Wang C. Red Blood Cell-Derived Nanoerythrosome for Antigen Delivery with Enhanced Cancer Immunotherapy. Sci. Adv. 2019, 5, eaaw6870. 10.1126/sciadv.aaw6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Zhang K.; Qin X.; Li T.; Qiu J.; Yin T.; Huang J.; McGinty S.; Pontrelli G.; Ren J.; Wang Q.; Wu W.; Wang G. Biomimetic Nanotherapies: Red Blood Cell Based Core-Shell Structured Nanocomplexes for Atherosclerosis Management. Adv. Sci. 2019, 6, 1900172–1900184. 10.1002/advs.201900172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X.; Gao J.; Fang R. H.; Luk B. T.; Kroll A. V.; Dehaini D.; Zhou J.; Kim H. W.; Gao W.; Lu W.; Zhang L. Nanoparticles Camouflaged in Platelet Membrane Coating As an Antibody Decoy for the Treatment of Immune Thrombocytopenia. Biomaterials 2016, 111, 116–123. 10.1016/j.biomaterials.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.; Ai Y.; Wang L.; Bu P.; Sharkey C. C.; Wu Q.; Wun B.; Roy S.; Shen X.; King M. R. Targeted Drug Delivery to Circulating Tumor Cells via Platelet Membrane-Functionalized Particles. Biomaterials 2016, 76, 52–65. 10.1016/j.biomaterials.2015.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusciello M.; Fontana F.; Tahtinen S.; Capasso C.; Feola S.; Martins B.; Chiaro J.; Peltonen K.; Ylosmaki L.; Ylosmaki E.; Hamdan F.; Kari O. K.; Ndika J.; Alenius H.; Urtti A.; Hirvonen J. T.; Santos H. A.; Cerullo V. Artificially Cloaked Viral Nanovaccine for Cancer Immunotherapy. Nat. Commun. 2019, 10, 5747–5759. 10.1038/s41467-019-13744-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grouleff J.; Irudayam S. J.; Skeby K. K.; Schiott B. The Influence of Cholesterol on Membrane Protein Structure, Function, and Dynamics Studied by Molecular Dynamics Simulations. Biochim. Biophys. Acta, Biomembr. 2015, 1848, 1783–1795. 10.1016/j.bbamem.2015.03.029. [DOI] [PubMed] [Google Scholar]
- Chakraborty S.; Doktorova M.; Molugu T. R.; Heberle F. A.; Scott H. L.; Dzikovski B.; Nagao M.; Stingaciu L. R.; Standaert R. F.; Barrera F. N.; Katsaras J.; Khelashvili G.; Brown M. F.; Ashkar R. How Cholesterol Stiffens Unsaturated Lipid Membranes. Proc. Natl. Acad. Sci. U. S. A. 2020, 117, 21896–21905. 10.1073/pnas.2004807117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An X.; Salomao M.; Guo X.; Gratzer W.; Mohandas N. Tropomyosin Modulates Erythrocyte Membrane Stability. Blood 2007, 109, 1284–1288. 10.1182/blood-2006-07-036954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong P. A Basis of the Crenation of Erythrocyte Ghosts by Electrolytes. Open Biol. J. 2013, 6, 14–19. 10.2174/1874196701306010014. [DOI] [Google Scholar]
- Wang J.; Wang Z.; Zhong Y.; Zou Y.; Wang C.; Wu H.; Lee A.; Yang W.; Wang X.; Liu Y.; Zhang D.; Yan J.; Hao M.; Zheng M.; Chung R.; Bai F.; Shi B. Central Metal-Derived Co-Assembly of Biomimetic Gdtpp/Zntpp Porphyrin Nanocomposites for Enhanced Dual-Modal Imaging-Guided Photodynamic Therapy. Biomaterials 2020, 229, 119576–119588. 10.1016/j.biomaterials.2019.119576. [DOI] [PubMed] [Google Scholar]
- Sun H.; Su J.; Meng Q.; Yin Q.; Chen L.; Gu W.; Zhang P.; Zhang Z.; Yu H.; Wang S.; Li Y. Cancer-Cell-Biomimetic Nanoparticles for Targeted Therapy of Homotypic Tumors. Adv. Mater. 2016, 28, 9581–9588. 10.1002/adma.201602173. [DOI] [PubMed] [Google Scholar]
- Sun H.; Su J.; Meng Q.; Yin Q.; Chen L.; Gu W.; Zhang Z.; Yu H.; Zhang P.; Wang S.; Li Y. Cancer Cell Membrane-Coated Gold Nanocages with Hyperthermia-Triggered Drug Release and Homotypic Target Inhibit Growth and Metastasis of Breast Cancer. Adv. Funct. Mater. 2017, 27, 1604300–1604308. 10.1002/adfm.201604300. [DOI] [Google Scholar]
- Kroll A. V.; Fang R. H.; Jiang Y.; Zhou J.; Wei X.; Yu C. L.; Gao J.; Luk B. T.; Dehaini D.; Gao W.; Zhang L. Nanoparticulate Delivery of Cancer Cell Membrane Elicits Multiantigenic Antitumor Immunity. Adv. Mater. 2017, 29, 1703969–1703977. 10.1002/adma.201703969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao L.; Bu L.-L.; Cai B.; Xu J.-H.; Li A.; Zhang W.-F.; Sun Z.-J.; Guo S.-S.; Liu W.; Wang T.-H.; Zhao X.-Z. Cancer Cell Membrane-Coated Upconversion Nanoprobes for Highly Specific Tumor Imaging. Adv. Mater. 2016, 28, 3460–3466. 10.1002/adma.201506086. [DOI] [PubMed] [Google Scholar]
- Jin J.; Krishnamachary B.; Barnett J. D.; Chatterjee S.; Chang D.; Mironchik Y.; Wildes F.; Jaffee E. M.; Nimmagadda S.; Bhujwalla Z. M. Human Cancer Cell Membrane-Coated Biomimetic Nanoparticles Reduce Fibroblast-Mediated Invasion and Metastasis and Induce T-Cells. ACS Appl. Mater. Interfaces 2019, 11, 7850–7861. 10.1021/acsami.8b22309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. M.; Chen G. B.; Chen H. H.; Zhang J. B.; Li H. Z.; Sheng M. X.; Weng W. B.; Guo S. M. Cancer Cell Membrane-Cloaked Mesoporous Silica Nanoparticles with a pH-Sensitive Gatekeeper for Cancer Treatment. Colloids Surf., B 2019, 175, 477–486. 10.1016/j.colsurfb.2018.12.038. [DOI] [PubMed] [Google Scholar]
- Xu Q.; Wan J.; Bie N.; Song X.; Yang X.; Yong T.; Zhao Y.; Yang X.; Gan L. A Biomimetic Gold Nanocages-Based Nanoplatform for Efficient Tumor Ablation and Reduced Inflammation. Theranostics 2018, 8, 5362–5378. 10.7150/thno.27631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose R. J.; Kim B. J.; Arai Y.; Han I. B.; Moon J. J.; Paulmurugan R.; Park H.; Lee S. H. Bioengineered Stem Cell Membrane Functionalized Nanocarriers for Therapeutic Targeting of Severe Hindlimb Ischemia. Biomaterials 2018, 185, 360–370. 10.1016/j.biomaterials.2018.08.018. [DOI] [PubMed] [Google Scholar]
- Rao L.; Wang W.; Meng Q. F.; Tian M.; Cai B.; Wang Y.; Li A.; Zan M.; Xiao F.; Bu L. L.; Li G.; Li A.; Liu Y.; Guo S. S.; Zhao X. Z.; Wang T. H.; Liu W.; Wu J. A Biomimetic Nanodecoy Traps Zika Virus to Prevent Viral Infection and Fetal Microcephaly Development. Nano Lett. 2019, 19, 2215–2222. 10.1021/acs.nanolett.8b03913. [DOI] [PubMed] [Google Scholar]
- Virlan M. J.; Miricescu D.; Radulescu R.; Sabliov C. M.; Totan A.; Calenic B.; Greabu M. Organic Nanomaterials and Their Applications in the Treatment of Oral Diseases. Molecules 2016, 21, 207–229. 10.3390/molecules21020207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anselmo A. C.; Mitragotri S. A Review of Clinical Translation of Inorganic Nanoparticles. AAPS J. 2015, 17, 1041–1054. 10.1208/s12248-015-9780-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlerding E. B.; Chen F.; Cai W. Biodegradable and Renal Clearable Inorganic Nanoparticles. Adv. Sci. 2016, 3, 1500223–1500230. 10.1002/advs.201500223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A. Z.; Langer R.; Farokhzad O. C. Nanoparticle Delivery of Cancer Drugs. Annu. Rev. Med. 2012, 63, 185–198. 10.1146/annurev-med-040210-162544. [DOI] [PubMed] [Google Scholar]
- Su S.; Kang P. M. Systemic Review of Biodegradable Nanomaterials in Nanomedicine. Nanomaterials 2020, 10, 656–676. 10.3390/nano10040656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Z.; Hu X.; Wei X.; Zhan C.; Lu L.; Jiang K.; Su B.; Ruan H.; Ran D.; Fang R. H.; Zhang L.; Lu W. A Facile Approach to Functionalizing Cell Membrane-Coated Nanoparticles with Neurotoxin-Derived Peptide for Brain-Targeted Drug Delivery. J. Controlled Release 2017, 264, 102–111. 10.1016/j.jconrel.2017.08.027. [DOI] [PubMed] [Google Scholar]
- Xu C.; Liu W.; Hu Y.; Li W.; Di W. Bioinspired Tumor-Homing Nanoplatform for Co-Delivery of Paclitaxel and siRNA-E7 to HPV-Related Cervical Malignancies for Synergistic Therapy. Theranostics 2020, 10, 3325–3339. 10.7150/thno.41228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Nimry S.; Dayah A. A.; Hasan I.; Daghmash R. Cosmetic, Biomedical and Pharmaceutical Applications of Fish Gelatin/Hydrolysates. Mar. Drugs 2021, 19, 145–167. 10.3390/md19030145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y.; Ran W.; Su J.; Lang T.; Meng J.; Wang G.; Zhang P.; Li Y. Traceable Bioinspired Nanoparticle for the Treatment of Metastatic Breast Cancer via NIR-Trigged Intracellular Delivery of Methylene Blue and Cisplatin. Adv. Mater. 2018, 30, 1802378–1802387. 10.1002/adma.201802378. [DOI] [PubMed] [Google Scholar]
- Xie J.; Shen Q.; Huang K.; Zheng T.; Cheng L.; Zhang Z.; Yu Y.; Liao G.; Wang X.; Li C. Oriented Assembly of Cell-Mimicking Nanoparticles via a Molecular Affinity Strategy for Targeted Drug Delivery. ACS Nano 2019, 13, 5268–5277. 10.1021/acsnano.8b09681. [DOI] [PubMed] [Google Scholar]
- He Y.; Li R.; Li H.; Zhang S.; Dai W.; Wu Q.; Jiang L.; Zheng Z.; Shen S.; Chen X.; Zhu Y.; Wang J.; Pang Z. Erythroliposomes: Integrated Hybrid Nanovesicles Composed of Erythrocyte Membranes and Artificial Lipid Membranes for Pore-Forming Toxin Clearance. ACS Nano 2019, 13, 4148–4159. 10.1021/acsnano.8b08964. [DOI] [PubMed] [Google Scholar]
- Castro C. I.; Briceno J. C. Perfluorocarbon-Based Oxygen Carriers: Review of Products and Trials. Artif. Organs 2010, 34, 622–634. 10.1111/j.1525-1594.2009.00944.x. [DOI] [PubMed] [Google Scholar]
- Lowe K. C. Blood Substitutes: From Chemistry to Clinic. J. Mater. Chem. 2006, 16, 4189–4196. 10.1039/b604923k. [DOI] [Google Scholar]
- Gao M.; Liang C.; Song X.; Chen Q.; Jin Q.; Wang C.; Liu Z. Erythrocyte-Membrane-Enveloped Perfluorocarbon As Nanoscale Artificial Red Blood Cells to Relieve Tumor Hypoxia and Enhance Cancer Radiotherapy. Adv. Mater. 2017, 29, 1701429–1701435. 10.1002/adma.201701429. [DOI] [PubMed] [Google Scholar]
- Zou M. Z.; Liu W. L.; Gao F.; Bai X. F.; Chen H. S.; Zeng X.; Zhang X. Z. Artificial Natural Killer Cells for Specific Tumor Inhibition and Renegade Macrophage Re-Education. Adv. Mater. 2019, 31, 1904495–1904504. 10.1002/adma.201904495. [DOI] [PubMed] [Google Scholar]
- Watermann A.; Brieger J. Mesoporous Silica Nanoparticles As Drug Delivery Vehicles in Cancer. Nanomaterials 2017, 7, 189–195. 10.3390/nano7070189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho G. C.; Sabio R. M.; de Cassia Ribeiro T.; Monteiro A. S.; Pereira D. V.; Ribeiro S. J. L.; Chorilli M. Highlights in Mesoporous Silica Nanoparticles As a Multifunctional Controlled Drug Delivery Nanoplatform for Infectious Diseases Treatment. Pharm. Res. 2020, 37, 191–220. 10.1007/s11095-020-02917-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafari S.; Derakhshankhah H.; Alaei L.; Fattahi A.; Varnamkhasti B. S.; Saboury A. A. Mesoporous Silica Nanoparticles for Therapeutic/Diagnostic Applications. Biomed. Pharmacother. 2019, 109, 1100–1111. 10.1016/j.biopha.2018.10.167. [DOI] [PubMed] [Google Scholar]
- Cai D.; Liu L.; Han C.; Ma X.; Qian J.; Zhou J.; Zhu W. Cancer Cell Membrane-Coated Mesoporous Silica Loaded with Superparamagnetic Ferroferric Oxide and Paclitaxel for the Combination of Chemo/Magnetocaloric Therapy on MDA-MB-231 Cells. Sci. Rep. 2019, 9, 14475–14484. 10.1038/s41598-019-51029-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastegari E.; Hsiao Y. J.; Lai W. Y.; Lai Y. H.; Yang T. C.; Chen S. J.; Huang P. I.; Chiou S. H.; Mou C. Y.; Chien Y. An Update on Mesoporous Silica Nanoparticle Applications in Nanomedicine. Pharmaceutics 2021, 13, 1067–1072. 10.3390/pharmaceutics13071067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersting M.; Olejnik M.; Rosenkranz N.; Loza K.; Breisch M.; Rostek A.; Westphal G.; Bunger J.; Ziegler N.; Ludwig A.; Koller M.; Sengstock C.; Epple M. Subtoxic Cell Responses to Silica Particles with Different Size and Shape. Sci. Rep. 2020, 10, 21591–21607. 10.1038/s41598-020-78550-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen Karaman D.; Sarwar S.; Desai D.; Bjork E. M.; Oden M.; Chakrabarti P.; Rosenholm J. M.; Chakraborti S. Shape Engineering Boosts Antibacterial Activity of Chitosan Coated Mesoporous Silica Nanoparticle Doped with Silver: A Mechanistic Investigation. J. Mater. Chem. B 2016, 4, 3292–3304. 10.1039/C5TB02526E. [DOI] [PubMed] [Google Scholar]
- Hao N.; Yang H.; Li L.; Li L.; Tang F. The Shape Effect of Mesoporous Silica Nanoparticles on Intracellular Reactive Oxygen Species in A375 Cells. New J. Chem. 2014, 38, 4258–4266. 10.1039/C4NJ00736K. [DOI] [Google Scholar]
- Natarajan S. K.; Selvaraj S. Mesoporous Silica Nanoparticles: Importance of Surface Modifications and its Role in Drug Delivery. RSC Adv. 2014, 4, 14328. 10.1039/c4ra00781f. [DOI] [Google Scholar]
- Cook T. R.; Zheng Y. R.; Stang P. J. Metal-Organic Frameworks and Self-Assembled Supramolecular Coordination Complexes: Comparing and Contrasting the Design, Synthesis, and Functionality of Metal-Organic Materials. Chem. Rev. 2013, 113, 734–777. 10.1021/cr3002824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoop M.; Walde C. F.; Riccò R.; Mushtaq F.; Terzopoulou A.; Chen X.-Z.; deMello A. J.; Doonan C. J.; Falcaro P.; Nelson B. J.; Puigmartí-Luis J.; Pané S. Biocompatibility Characteristics of the Metal Organic Framework ZIF-8 for Therapeutical Applications. Appl. Mater. Today 2018, 11, 13–21. 10.1016/j.apmt.2017.12.014. [DOI] [Google Scholar]
- Guo B. J.; Yang Z. L.; Zhang L. J. Gadolinium Deposition in Brain: Current Scientific Evidence and Future Perspectives. Front. Mol. Neurosci. 2018, 11, 335–346. 10.3389/fnmol.2018.00335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D.; Ye Z.; Wei L.; Luo H.; Xiao L. Cell Membrane-Coated Porphyrin Metal-Organic Frameworks for Cancer Cell Targeting and O2-Evolving Photodynamic Therapy. ACS Appl. Mater. Interfaces 2019, 11, 39594–39602. 10.1021/acsami.9b14084. [DOI] [PubMed] [Google Scholar]
- Li S. Y.; Cheng H.; Xie B. R.; Qiu W. X.; Zeng J. Y.; Li C. X.; Wan S. S.; Zhang L.; Liu W. L.; Zhang X. Z. Cancer Cell Membrane Camouflaged Cascade Bioreactor for Cancer Targeted Starvation and Photodynamic Therapy. ACS Nano 2017, 11, 7006–7018. 10.1021/acsnano.7b02533. [DOI] [PubMed] [Google Scholar]
- Li S. Y.; Cheng H.; Qiu W. X.; Zhang L.; Wan S. S.; Zeng J. Y.; Zhang X. Z. Cancer Cell Membrane-Coated Biomimetic Platform for Tumor Targeted Photodynamic Therapy and Hypoxia-Amplified Bioreductive Therapy. Biomaterials 2017, 142, 149–161. 10.1016/j.biomaterials.2017.07.026. [DOI] [PubMed] [Google Scholar]
- Carnovale C.; Bryant G.; Shukla R.; Bansal V. Identifying Trends in Gold Nanoparticle Toxicity and Uptake: Size, Shape, Capping Ligand, and Biological Corona. ACS Omega 2019, 4, 242–256. 10.1021/acsomega.8b03227. [DOI] [Google Scholar]
- Zhou C.; Long M.; Qin Y.; Sun X.; Zheng J. Luminescent Gold Nanoparticles with Efficient Renal Clearance. Angew. Chem., Int. Ed. 2011, 50, 3168–3172. 10.1002/anie.201007321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T.; Zhang B.; Shen S.; Tuo Y.; Luo Z.; Hu Y.; Pang Z.; Jiang X. Tumor Microenvironment Modulation by Cyclopamine Improved Photothermal Therapy of Biomimetic Gold Nanorods for Pancreatic Ductal Adenocarcinomas. ACS Appl. Mater. Interfaces 2017, 9, 31497–31508. 10.1021/acsami.7b09458. [DOI] [PubMed] [Google Scholar]
- Rao L.; Bu L. L.; Ma L.; Wang W.; Liu H.; Wan D.; Liu J. F.; Li A.; Guo S. S.; Zhang L.; Zhang W. F.; Zhao X. Z.; Sun Z. J.; Liu W. Platelet-Facilitated Photothermal Therapy of Head and Neck Squamous Cell Carcinoma. Angew. Chem., Int. Ed. 2018, 57, 986–991. 10.1002/anie.201709457. [DOI] [PubMed] [Google Scholar]
- Li X.; Lovell J. F.; Yoon J.; Chen X. Clinical Development and Potential of Photothermal and Photodynamic Therapies for Cancer. Nat. Rev. Clin. Oncol. 2020, 17, 657–674. 10.1038/s41571-020-0410-2. [DOI] [PubMed] [Google Scholar]
- Zou L.; Wang H.; He B.; Zeng L.; Tan T.; Cao H.; He X.; Zhang Z.; Guo S.; Li Y. Current Approaches of Photothermal Therapy in Treating Cancer Metastasis with Nanotherapeutics. Theranostics 2016, 6, 762–772. 10.7150/thno.14988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X.; Zheng R.; Fang X.; Wang X.; Zhang X.; Yang W.; Sha X. Red Blood Cell Membrane Camouflaged Magnetic Nanoclusters for Imaging-Guided Photothermal Therapy. Biomaterials 2016, 92, 13–24. 10.1016/j.biomaterials.2016.03.026. [DOI] [PubMed] [Google Scholar]
- Zhou Z.; Song J.; Nie L.; Chen X. Reactive Oxygen Species Generating Systems Meeting Challenges of Photodynamic Cancer Therapy. Chem. Soc. Rev. 2016, 45, 6597–6626. 10.1039/C6CS00271D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z.; Zhou P.; Pan W.; Li N.; Tang B. A Biomimetic Nanoreactor for Synergistic Chemiexcited Photodynamic Therapy and Starvation Therapy against Tumor Metastasis. Nat. Commun. 2018, 9, 5044–5052. 10.1038/s41467-018-07197-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H.; Lv Y.; Ni D.; Wang J.; Tian Z.; Wei W.; Ma G. Erythrocyte Membrane-Coated NIR-Triggered Biomimetic Nanovectors with Programmed Delivery for Photodynamic Therapy of Cancer. Nanoscale 2015, 7, 9806–9815. 10.1039/C5NR02470F. [DOI] [PubMed] [Google Scholar]
- Ren H.; Liu J.; Li Y.; Wang H.; Ge S.; Yuan A.; Hu Y.; Wu J. Oxygen Self-Enriched Nanoparticles Functionalized with Erythrocyte Membranes for Long Circulation and Enhanced Phototherapy. Acta Biomater. 2017, 59, 269–282. 10.1016/j.actbio.2017.06.035. [DOI] [PubMed] [Google Scholar]
- Chen J.; Zhu Y.; Kaskel S. Porphyrin-Based Metal-Organic Frameworks for Biomedical Applications. Angew. Chem., Int. Ed. 2021, 60, 5010–5035. 10.1002/anie.201909880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressler N. M. Verteporfin Therapy of Subfoveal Choroidal Neovascularization in Age-Related Macular Degeneration: Two-Year Results of a Randomized Clinical Trial including Lesions with Occult with No Classic Choroidal Neovascularization—Verteporfin in Photodynamic Therapy Report 2. Am. J. Ophthalmol. 2001, 131, 541–560. 10.1016/s0002-9394(01)00967-9. [DOI] [PubMed] [Google Scholar]
- Liu W.; Ruan M.; Wang Y.; Song R.; Ji X.; Xu J.; Dai J.; Xue W. Light-Triggered Biomimetic Nanoerythrocyte for Tumor-Targeted Lung Metastatic Combination Therapy of Malignant Melanoma. Small 2018, 14, 1801754–1801768. 10.1002/smll.201801754. [DOI] [PubMed] [Google Scholar]
- Tapeinos C.; Tomatis F.; Battaglini M.; Larranaga A.; Marino A.; Telleria I. A.; Angelakeris M.; Debellis D.; Drago F.; Brero F.; Arosio P.; Lascialfari A.; Petretto A.; Sinibaldi E.; Ciofani G. Cell Membrane-Coated Magnetic Nanocubes with a Homotypic Targeting Ability Increase Intracellular Temperature due to ROS Scavenging and Act As a Versatile Theranostic System for Glioblastoma Multiforme. Adv. Healthcare Mater. 2019, 8, 1900612. 10.1002/adhm.201900612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspani S.; Magalhaes R.; Araujo J. P.; Sousa C. T. Magnetic Nanomaterials As Contrast Agents for MRI. Materials 2020, 13, 2586–2604. 10.3390/ma13112586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D.; Schmieder A. H.; Wickline S. A.; Lanza G. M. Manganese-Based MRI Contrast Agents: Past, Present and Future. Tetrahedron 2011, 67, 8431–8444. 10.1016/j.tet.2011.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y.; Qiang S.; Wang L. Gold Nanomaterials for Imaging-Guided Near-Infrared in Vivo Cancer Therapy. Front. Bioeng. Biotechnol. 2019, 7, 398–406. 10.3389/fbioe.2019.00398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao L.; Meng Q. F.; Bu L. L.; Cai B.; Huang Q.; Sun Z. J.; Zhang W. F.; Li A.; Guo S. S.; Liu W.; Wang T. H.; Zhao X. Z. Erythrocyte Membrane-Coated Upconversion Nanoparticles with Minimal Protein Adsorption for Enhanced Tumor Imaging. ACS Appl. Mater. Interfaces 2017, 9, 2159–2168. 10.1021/acsami.6b14450. [DOI] [PubMed] [Google Scholar]
- Chen Y.; Zhang Y.; Chen M.; Zhuang J.; Fang R. H.; Gao W.; Zhang L. Biomimetic Nanosponges Suppress in Vivo Lethality Induced by the Whole Secreted Proteins of Pathogenic Bacteria. Small 2019, 15, 1804994–1805002. 10.1002/smll.201804994. [DOI] [PubMed] [Google Scholar]
- Zhang Z.; Qian H.; Yang M.; Li R.; Hu J.; Li L.; Yu L.; Liu B.; Qian X. Gambogic Acid-Loaded Biomimetic Nanoparticles in Colorectal Cancer Treatment. Int. J. Nanomed. 2017, 12, 1593–1605. 10.2147/IJN.S127256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y.; Gao Y.; Yu Y. ″Waltz″ of Cell Membrane-Coated Nanoparticles on Lipid Bilayers: Tracking Single Particle Rotation in Ligand-Receptor Binding. ACS Nano 2018, 12, 11871–11880. 10.1021/acsnano.8b04880. [DOI] [PubMed] [Google Scholar]
- Guo P.; Huang J.; Zhao Y.; Martin C. R.; Zare R. N.; Moses M. A. Nanomaterial Preparation by Extrusion through Nanoporous Membranes. Small 2018, 14, 1703493 10.1002/smll.201703493. [DOI] [PubMed] [Google Scholar]
- Ong S. G.; Chitneni M.; Lee K. S.; Ming L. C.; Yuen K. H. Evaluation of Extrusion Technique for Nanosizing Liposomes. Pharmaceutics 2016, 8, 36–47. 10.3390/pharmaceutics8040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charcosset C.; Limayem I.; Fessi H. The Membrane Emulsification Process—A Review. J. Chem. Technol. Biotechnol. 2004, 79, 209–218. 10.1002/jctb.969. [DOI] [Google Scholar]
- Vladisavljevic G. T.; Williams R. A. Recent Developments in Manufacturing Emulsions and Particulate Products Using Membranes. Adv. Colloid Interface Sci. 2005, 113, 1–20. 10.1016/j.cis.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Maas M.; Guo P.; Keeney M.; Yang F.; Hsu T. M.; Fuller G. G.; Martin C. R.; Zare R. N. Preparation of Mineralized Nanofibers: Collagen Fibrils Containing Calcium Phosphate. Nano Lett. 2011, 11, 1383–1388. 10.1021/nl200116d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W.; Hu C.-M. J.; Fang R. H.; Luk B. T.; Su J.; Zhang L. Surface Functionalization of Gold Nanoparticles with Red Blood Cell Membranes. Adv. Mater. 2013, 25, 3549–3553. 10.1002/adma.201300638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K.; Yamada S.; Sakamoto W.; Usugi E.; Watanabe M.; Yogo T. Red Blood Cell-Shaped Microparticles with a Red Blood Cell Membrane Demonstrate Prolonged Circulation Time in Blood. ACS Biomater. Sci. Eng. 2018, 4, 2729–2732. 10.1021/acsbiomaterials.8b00197. [DOI] [PubMed] [Google Scholar]
- Arrigo R.; Teresi R.; Gambarotti C.; Parisi F.; Lazzara G.; Dintcheva N. T. Sonication-Induced Modification of Carbon Nanotubes: Effect on the Rheological and Thermo-Oxidative Behaviour of Polymer-Based Nanocomposites. Materials 2018, 11, 383–396. 10.3390/ma11030383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennhofer H.; Zanghellini B. Dispersion State and Damage of Carbon Nanotubes and Carbon Nanofibers by Ultrasonic Dispersion: A Review. Nanomaterials 2021, 11, 1469–1495. 10.3390/nano11061469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H.; Yang B. In Situ Preparation of Nanoparticles/Polymer Composites. Sci. China, Ser. E: Technol. Sci. 2008, 51, 1886–1901. 10.1007/s11431-008-0109-6. [DOI] [Google Scholar]
- Jia X.; Wang L.; Du J. In Situ Polymerization on Biomacromolecules for Nanomedicines. Nano Res. 2018, 11, 5028–5048. 10.1007/s12274-018-2080-2. [DOI] [Google Scholar]
- Zhang J.; Gao W.; Fang R. H.; Dong A.; Zhang L. Synthesis of Nanogels via Cell Membrane-Templated Polymerization. Small 2015, 11, 4309–4313. 10.1002/smll.201500987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.; Lee W. G. Electroporation for Nanomedicine: A Review. J. Mater. Chem. B 2017, 5, 2726–2738. 10.1039/C7TB00038C. [DOI] [PubMed] [Google Scholar]
- Behzadi S.; Serpooshan V.; Tao W.; Hamaly M. A.; Alkawareek M. Y.; Dreaden E. C.; Brown D.; Alkilany A. M.; Farokhzad O. C.; Mahmoudi M. Cellular Uptake of Nanoparticles: Journey Inside the Cell. Chem. Soc. Rev. 2017, 46, 4218–4244. 10.1039/C6CS00636A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X.; Luo B.; Kang K.; Zhang Y.; Jiang P.; Lan F.; Yi Q.; Wu Y. Leukocyte-Repelling Biomimetic Immunomagnetic Nanoplatform for High-Performance Circulating Tumor Cells Isolation. Small 2019, 15, 1900558–1900565. 10.1002/smll.201900558. [DOI] [PubMed] [Google Scholar]
- Tu Y.; Lv M.; Xiu P.; Huynh T.; Zhang M.; Castelli M.; Liu Z.; Huang Q.; Fan C.; Fang H.; Zhou R. Destructive Extraction of Phospholipids from Escherichia Coli Membranes by Graphene Nanosheets. Nat. Nanotechnol. 2013, 8, 594–601. 10.1038/nnano.2013.125. [DOI] [PubMed] [Google Scholar]
- Zhang C.; Liu W. L.; Bai X. F.; Cheng S. X.; Zhong Z. L.; Zhang X. Z. A Hybrid Nanomaterial with NIR-Induced Heat and Associated Hydroxyl Radical Generation for Synergistic Tumor Therapy. Biomaterials 2019, 199, 1–9. 10.1016/j.biomaterials.2019.01.041. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy S.; Gnanasammandhan M. K.; Xie C.; Huang K.; Cui M. Y.; Chan J. M. Monocyte Cell Membrane-Derived Nanoghosts for Targeted Cancer Therapy. Nanoscale 2016, 8, 6981–6895. 10.1039/C5NR07588B. [DOI] [PubMed] [Google Scholar]
- Wu Z.; Li T.; Gao W.; Xu T.; Jurado-Sánchez B.; Li J.; Gao W.; He Q.; Zhang L.; Wang J. Cell-Membrane-Coated Synthetic Nanomotors for Effective Biodetoxification. Adv. Funct. Mater. 2015, 25, 3881–3887. 10.1002/adfm.201501050. [DOI] [Google Scholar]
- Luk B. T.; Hu C. M.; Fang R. H.; Dehaini D.; Carpenter C.; Gao W.; Zhang L. Interfacial Interactions between Natural RBC Membranes and Synthetic Polymeric Nanoparticles. Nanoscale 2014, 6, 2730–2737. 10.1039/C3NR06371B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saadatkhah N.; Carillo Garcia A.; Ackermann S.; Leclerc P.; Latifi M.; Samih S.; Patience G. S.; Chaouki J. Experimental Methods in Chemical Engineering: Thermogravimetric Analysis—TGA. Can. J. Chem. Eng. 2020, 98, 34–43. 10.1002/cjce.23673. [DOI] [Google Scholar]
- Fan Z.; Zhou H.; Li P. Y.; Speer J. E.; Cheng H. Structural Elucidation of Cell Membrane-Derived Nanoparticles Using Molecular Probes. J. Mater. Chem. B 2014, 2, 8231–8238. 10.1039/C4TB00980K. [DOI] [PubMed] [Google Scholar]
- Guo D.; Xie G.; Luo J. Mechanical Properties of Nanoparticles: Basics and Applications. J. Phys. D: Appl. Phys. 2014, 47, 013001–013025. 10.1088/0022-3727/47/1/013001. [DOI] [Google Scholar]
- Ye H.; Shen Z.; Yu L.; Wei M.; Li Y. Manipulating Nanoparticle Transport within Blood Flow through External Forces: An Exemplar of Mechanics in Nanomedicine. Proc. R. Soc. London, Ser. A 2018, 474, 20170845–20170869. 10.1098/rspa.2017.0845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K.; Yamada S.; Hayashi H.; Sakamoto W.; Yogo T. Red Blood Cell-Like Particles with the Ability to Avoid Lung and Spleen Accumulation for the Treatment of Liver Fibrosis. Biomaterials 2018, 156, 45–55. 10.1016/j.biomaterials.2017.11.031. [DOI] [PubMed] [Google Scholar]
- Kang Y. J.; Ha Y.-R.; Lee S.-J. Deformability Measurement of Red Blood Cells Using a Microfluidic Channel Array and an Air Cavity in a Driving Syringe with High Throughput and Precise Detection of Subpopulations. Analyst 2016, 141, 319–330. 10.1039/C5AN01988E. [DOI] [PubMed] [Google Scholar]
- Mohd Noor A.; Masuda T.; Arai F. Microfluidic Device for Rapid Investigation of the Deformability of Leukocytes in Whole Blood Samples. ROBOMECH J. 2020, 7, 5–15. 10.1186/s40648-019-0153-y. [DOI] [Google Scholar]
- Yang N.; Ding Y.; Zhang Y.; Wang B.; Zhao X.; Cheng K.; Huang Y.; Taleb M.; Zhao J.; Dong W. F.; Zhang L.; Nie G. Surface Functionalization of Polymeric Nanoparticles with Umbilical Cord-Derived Mesenchymal Stem Cell Membrane for Tumor-Targeted Therapy. ACS Appl. Mater. Interfaces 2018, 10, 22963–22973. 10.1021/acsami.8b05363. [DOI] [PubMed] [Google Scholar]
- Su J.; Sun H.; Meng Q.; Yin Q.; Tang S.; Zhang P.; Chen Y.; Zhang Z.; Yu H.; Li Y. Long Circulation Red-Blood-Cell-Mimetic Nanoparticles with Peptide-Enhanced Tumor Penetration for Simultaneously Inhibiting Growth and Lung Metastasis of Breast Cancer. Adv. Funct. Mater. 2016, 26, 1243–1252. 10.1002/adfm.201504780. [DOI] [Google Scholar]
- Song Y.; Huang Z.; Liu X.; Pang Z.; Chen J.; Yang H.; Zhang N.; Cao Z.; Liu M.; Cao J.; Li C.; Yang X.; Gong H.; Qian J.; Ge J. Platelet Membrane-Coated Nanoparticle-Mediated Targeting Delivery of Rapamycin Blocks Atherosclerotic Plaque Development and Stabilizes Plaque in Apolipoprotein E-Deficient (ApoE(−/−)) Mice. Nanomedicine 2019, 15, 13–24. 10.1016/j.nano.2018.08.002. [DOI] [PubMed] [Google Scholar]
- Aslam B.; Basit M.; Nisar M. A.; Khurshid M.; Rasool M. H. Proteomics: Technologies and Their Applications. J. Chromatogr. Sci. 2017, 55, 182–196. 10.1093/chromsci/bmw167. [DOI] [PubMed] [Google Scholar]
- Li R.; He Y.; Zhu Y.; Jiang L.; Zhang S.; Qin J.; Wu Q.; Dai W.; Shen S.; Pang Z.; Wang J. Route to Rheumatoid Arthritis by Macrophage-Derived Microvesicle-Coated Nanoparticles. Nano Lett. 2019, 19, 124–134. 10.1021/acs.nanolett.8b03439. [DOI] [PubMed] [Google Scholar]
- Li H.; Jin K.; Luo M.; Wang X.; Zhu X.; Liu X.; Jiang T.; Zhang Q.; Wang S.; Pang Z. Size Dependency of Circulation and Biodistribution of Biomimetic Nanoparticles: Red Blood Cell Membrane-Coated Nanoparticles. Cells 2019, 8, 881–896. 10.3390/cells8080881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen L. M. E.; Ramsay E. E.; Logsdon C. D.; Overwijk W. W. The Immune System in Cancer Metastasis: Friend or Foe?. J. Immunother. Cancer 2017, 5, 79–92. 10.1186/s40425-017-0283-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shihab I.; Khalil B. A.; Elemam N. M.; Hachim I. Y.; Hachim M. Y.; Hamoudi R. A.; Maghazachi A. A. Understanding the Role of Innate Immune Cells and Identifying Genes in Breast Cancer Microenvironment. Cancers 2020, 12, 2226–2241. 10.3390/cancers12082226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo A. S.; Follain G.; Patthabhiraman S.; Harlepp S.; Goetz J. G. Metastasis of Circulating Tumor Cells: Favorable Soil or Suitable Biomechanics, or Both?. Cell Adh Migr 2015, 9, 345–356. 10.1080/19336918.2015.1059563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micalizzi D. S.; Maheswaran S.; Haber D. A. A Conduit to Metastasis: Circulating Tumor Cell Biology. Genes Dev. 2017, 31, 1827–1840. 10.1101/gad.305805.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigor E. J. M.; Fergusson D.; Kekre N.; Montroy J.; Atkins H.; Seftel M. D.; Daugaard M.; Presseau J.; Thavorn K.; Hutton B.; Holt R. A.; Lalu M. M. Risks and Benefits of Chimeric Antigen Receptor T-Cell (CAR-T) Therapy in Cancer: A Systematic Review and Meta-Analysis. Transfus. Med. Rev. 2019, 33, 98–110. 10.1016/j.tmrv.2019.01.005. [DOI] [PubMed] [Google Scholar]
- Feins S.; Kong W.; Williams E. F.; Milone M. C.; Fraietta J. A. An Introduction to Chimeric Antigen Receptor (CAR) T-Cell Immunotherapy for Human Cancer. Am. J. Hematol. 2019, 94, S3–S9. 10.1002/ajh.25418. [DOI] [PubMed] [Google Scholar]
- Restifo N. P.; Dudley M. E.; Rosenberg S. A. Adoptive Immunotherapy for Cancer: Harnessing the T Cell Response. Nat. Rev. Immunol. 2012, 12, 269–281. 10.1038/nri3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maus M. V.; Fraietta J. A.; Levine B. L.; Kalos M.; Zhao Y.; June C. H. Adoptive Immunotherapy for Cancer or Viruses. Annu. Rev. Immunol. 2014, 32, 189–225. 10.1146/annurev-immunol-032713-120136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert C. A Decade of Immune-Checkpoint Inhibitors in Cancer Therapy. Nat. Commun. 2020, 11, 3801–3803. 10.1038/s41467-020-17670-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargadon K. M.; Johnson C. E.; Williams C. J. Immune Checkpoint Blockade Therapy for Cancer: An Overview of FDA-Approved Immune Checkpoint Inhibitors. Int. Immunopharmacol. 2018, 62, 29–39. 10.1016/j.intimp.2018.06.001. [DOI] [PubMed] [Google Scholar]
- Schirrmacher V. Cancer Vaccines and Oncolytic Viruses Exert Profoundly Lower Side Effects in Cancer Patients Than Other Systemic Therapies: A Comparative Analysis. Biomedicines 2020, 8, 61–80. 10.3390/biomedicines8030061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi T.; Song X.; Wang Y.; Liu F.; Wei J. Combining Oncolytic Viruses with Cancer Immunotherapy: Establishing a New Generation of Cancer Treatment. Front. Immunol. 2020, 11, 683–695. 10.3389/fimmu.2020.00683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahavi D.; Weiner L. Monoclonal Antibodies in Cancer Therapy. Antibodies 2020, 9, 34–53. 10.3390/antib9030034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berraondo P.; Sanmamed M. F.; Ochoa M. C.; Etxeberria I.; Aznar M. A.; Perez-Gracia J. L.; Rodriguez-Ruiz M. E.; Ponz-Sarvise M.; Castanon E.; Melero I. Cytokines in Clinical Cancer Immunotherapy. Br. J. Cancer 2019, 120, 6–15. 10.1038/s41416-018-0328-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita M.; Kawaguchi M. Immunomodulatory Effects of Drugs for Effective Cancer Immunotherapy. J. Oncol. 2018, 2018, 8653489–8653495. 10.1155/2018/8653489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restifo N. P.; Smyth M. J.; Snyder A. Acquired Resistance to Immunotherapy and Future Challenges. Nat. Rev. Cancer 2016, 16, 121–126. 10.1038/nrc.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell D. R.; Swanton C. The Role of Tumour Heterogeneity and Clonal Cooperativity in Metastasis, Immune Evasion and Clinical Outcome. BMC Med. 2017, 15, 133–141. 10.1186/s12916-017-0900-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambi M.; Bagheri L.; Szewczuk M. R. Current Challenges in Cancer Immunotherapy: Multimodal Approaches to Improve Efficacy and Patient Response Rates. J. Oncol. 2019, 2019, 4508794–4508805. 10.1155/2019/4508794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.; Sha H.; Zhang L.; Qian H.; Chen F.; Ding N.; Ji L.; Zhu A.; Xu Q.; Meng F.; Yu L.; Zhou Y.; Liu B. Lipid Insertion Enables Targeted Functionalization of Paclitaxel-Loaded Erythrocyte Membrane Nanosystem by Tumor-Penetrating Bispecific Recombinant Protein. Int. J. Nanomed. 2018, 13, 5347–5359. 10.2147/IJN.S165109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Z.; Ran D.; Lu L.; Zhan C.; Ruan H.; Hu X.; Xie C.; Jiang K.; Li J.; Zhou J.; Wang J.; Zhang Y.; Fang R. H.; Zhang L.; Lu W. Ligand-Modified Cell Membrane Enables the Targeted Delivery of Drug Nanocrystals to Glioma. ACS Nano 2019, 13, 5591–5601. 10.1021/acsnano.9b00661. [DOI] [PubMed] [Google Scholar]
- Kang T.; Jiang M.; Jiang D.; Feng X.; Yao J.; Song Q.; Chen H.; Gao X.; Chen J. Enhancing Glioblastoma-Specific Penetration by Functionalization of Nanoparticles with an Iron-Mimic Peptide Targeting Transferrin/Transferrin Receptor Complex. Mol. Pharmaceutics 2015, 12, 2947–2961. 10.1021/acs.molpharmaceut.5b00222. [DOI] [PubMed] [Google Scholar]
- Fu S.; Liang M.; Wang Y.; Cui L.; Gao C.; Chu X.; Liu Q.; Feng Y.; Gong W.; Yang M.; Li Z.; Yang C.; Xie X.; Yang Y.; Gao C. Dual-Modified Novel Biomimetic Nanocarriers Improve Targeting and Therapeutic Efficacy in Glioma. ACS Appl. Mater. Interfaces 2019, 11, 1841–1854. 10.1021/acsami.8b18664. [DOI] [PubMed] [Google Scholar]
- Huang N.; Cheng S.; Zhang X.; Tian Q.; Pi J.; Tang J.; Huang Q.; Wang F.; Chen J.; Xie Z.; Xu Z.; Chen W.; Zheng H.; Cheng Y. Efficacy of NGR Peptide-Modified PEGylated Quantum Dots for Crossing the Blood-Brain Barrier and Targeted Fluorescence Imaging of Glioma and Tumor Vasculature. Nanomedicine 2017, 13, 83–93. 10.1016/j.nano.2016.08.029. [DOI] [PubMed] [Google Scholar]
- Dunne M.; Zheng J.; Rosenblat J.; Jaffray D. A.; Allen C. APN/CD13-Targeting As a Strategy to Alter the Tumor Accumulation of Liposomes. J. Controlled Release 2011, 154, 298–305. 10.1016/j.jconrel.2011.05.022. [DOI] [PubMed] [Google Scholar]
- Ding D.; Yao Y.; Zhang S.; Su C.; Zhang Y. C-Type Lectins Facilitate Tumor Metastasis. Oncol. Lett. 2017, 13, 13–21. 10.3892/ol.2016.5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q.; Sun W.; Qian C.; Wang C.; Bomba H. N.; Gu Z. Anticancer Platelet-Mimicking Nanovehicles. Adv. Mater. 2015, 27, 7043–7050. 10.1002/adma.201503323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X.; Gajan A.; Chu Q.; Xiong H.; Wu K.; Wu G. S. Developing TRAIL/TRAIL Death Receptor-Based Cancer Therapies. Cancer Metastasis Rev. 2018, 37, 733–748. 10.1007/s10555-018-9728-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J.; Bhujwalla Z. M. Biomimetic Nanoparticles Camouflaged in Cancer Cell Membranes and Their Applications in Cancer Theranostics. Front. Oncol. 2020, 9, 1560–1570. 10.3389/fonc.2019.01560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H.; Zhu J.-Y.; Li S.-Y.; Zeng J.-Y.; Lei Q.; Chen K.-W.; Zhang C.; Zhang X.-Z. An O2 Self-Sufficient Biomimetic Nanoplatform for Highly Specific and Efficient Photodynamic Therapy. Adv. Funct. Mater. 2016, 26, 7847–7860. 10.1002/adfm.201603212. [DOI] [Google Scholar]
- Yang R.; Xu J.; Xu L.; Sun X.; Chen Q.; Zhao Y.; Peng R.; Liu Z. Cancer Cell Membrane-Coated Adjuvant Nanoparticles with Mannose Modification for Effective Anticancer Vaccination. ACS Nano 2018, 12, 5121–5129. 10.1021/acsnano.7b09041. [DOI] [PubMed] [Google Scholar]
- Howells A.; Marelli G.; Lemoine N. R.; Wang Y. Oncolytic Viruses-Interaction of Virus and Tumor Cells in the Battle to Eliminate Cancer. Front. Oncol. 2017, 7, 195–209. 10.3389/fonc.2017.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noy R.; Pollard J. W. Tumor-Associated Macrophages: From Mechanisms to Therapy. Immunity 2014, 41, 49–61. 10.1016/j.immuni.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian B. Z.; Pollard J. W. Macrophage Diversity Enhances Tumor Progression and Metastasis. Cell 2010, 141, 39–51. 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L.; Pantel K.; Kang Y. Tumor Metastasis: Moving New Biological Insights into the Clinic. Nat. Med. 2013, 19, 1450–1464. 10.1038/nm.3391. [DOI] [PubMed] [Google Scholar]
- Chen Q.; Zhang X. H.; Massague J. Macrophage Binding to Receptor VCAM-1 Transmits Survival Signals in Breast Cancer Cells That Invade the Lungs. Cancer Cell 2011, 20, 538–549. 10.1016/j.ccr.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonska J.; Lang S.; Sionov R. V.; Granot Z. The Regulation of Pre-Metastatic Niche Formation by Neutrophils. Oncotarget 2017, 8, 112132–112144. 10.18632/oncotarget.22792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M.; Ma M.; Tan Z.; Zheng H.; Liu X. Neutrophil: A New Player in Metastatic Cancers. Front. Immunol. 2020, 11, 565165–565178. 10.3389/fimmu.2020.565165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Miguel M. P.; Julián S. F.; Blázquez-Martínez A.; Pascual C. Y.; Aller M. A.; Arias J.; Arnalich-Montiel F. Immunosuppressive Properties of Mesenchymal Stem Cells: Advances and Applications. Curr. Mol. Med. 2012, 12, 574–591. 10.2174/156652412800619950. [DOI] [PubMed] [Google Scholar]
- Li L.; Guan Y.; Liu H.; Hao N.; Liu T.; Meng X.; Fu C.; Li Y.; Qu Q.; Zhang Y.; Ji S.; Chen L.; Chen D.; Tang F. Silica Nanorattle-Doxorubicin Anchored Mesenchymal Stem Cells for Tumor-Tropic Therapy. ACS Nano 2011, 5, 7462–7470. 10.1021/nn202399w. [DOI] [PubMed] [Google Scholar]
- Wei X.; Yang X.; Han Z. P.; Qu F. F.; Shao L.; Shi Y. F. Mesenchymal Stem Cells: A New Trend for Cell Therapy. Acta Pharmacol. Sin. 2013, 34, 747–754. 10.1038/aps.2013.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apetoh L.; Ladoire S.; Coukos G.; Ghiringhelli F. Combining Immunotherapy and Anticancer Agents: The Right Path to Achieve Cancer Cure?. Ann. Oncol. 2015, 26, 1813–1823. 10.1093/annonc/mdv209. [DOI] [PubMed] [Google Scholar]
- Howard C. J.; Charleston B.; Stephens S. A.; Sopp P.; Hope J. C. The Role of Dendritic Cells in Shaping the Immune Response. Anim. Health Res. Rev. 2004, 5, 191–195. 10.1079/AHR200468. [DOI] [PubMed] [Google Scholar]
- Kantoff P. W.; Higano C. S.; Shore N. D.; Berger E. R.; Small E. J.; Penson D. F.; Redfern C. H.; Ferrari A. C.; Dreicer R.; Sims R. B.; Xu Y.; Frohlich M. W.; Schellhammer P. F. Sipuleucel-T Immunotherapy for Castration-Resistant Prostate Cancer. N. Engl. J. Med. 2010, 363, 411–422. 10.1056/NEJMoa1001294. [DOI] [PubMed] [Google Scholar]
- Zhang X.; He T.; Li Y.; Chen L.; Liu H.; Wu Y.; Guo H. Dendritic Cell Vaccines in Ovarian Cancer. Front. Immunol. 2021, 11, 613773–613786. 10.3389/fimmu.2020.613773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisoni E.; Imbimbo M.; Zimmermann S.; Valabrega G. Ovarian Cancer Immunotherapy: Turning Up the Heat. Int. J. Mol. Sci. 2019, 20, 2927–2932. 10.3390/ijms20122927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q.; Yang Q.; Li J.; Liu G.; Nikoulin I.; Jia S. Advanced Clinical Trials of Dendritic Cell Vaccines in Ovarian Cancer. J. Invest. Med. 2020, 68, 1223–1227. 10.1136/jim-2020-001355. [DOI] [PubMed] [Google Scholar]
- Cubillos-Ruiz J. R.; Silberman P. C.; Rutkowski M. R.; Chopra S.; Perales-Puchalt A.; Song M.; Zhang S.; Bettigole S. E.; Gupta D.; Holcomb K.; Ellenson L. H.; Caputo T.; Lee A. H.; Conejo-Garcia J. R.; Glimcher L. H. ER Stress Sensor XBP1 Controls Anti-Tumor Immunity by Disrupting Dendritic Cell Homeostasis. Cell 2015, 161, 1527–1538. 10.1016/j.cell.2015.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cintolo J. A.; Datta J.; Mathew S. J.; Czerniecki B. J. Dendritic Cell-Based Vaccines: Barriers and Opportunities. Future Oncol. 2012, 8, 1273–1299. 10.2217/fon.12.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill L. A.; Pearce E. J. Immunometabolism Governs Dendritic Cell and Macrophage Function. J. Exp. Med. 2016, 213, 15–23. 10.1084/jem.20151570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B.; Dai W.; Mei D.; Liu T.; Li S.; He B.; He B.; Yuan L.; Zhang H.; Wang X.; Zhang Q. Comprehensively Priming the Tumor Microenvironment by Cancer-Associated Fibroblast-Targeted Liposomes for Combined Therapy with Cancer Cell-Targeted Chemotherapeutic Drug Delivery System. J. Controlled Release 2016, 241, 68–80. 10.1016/j.jconrel.2016.09.014. [DOI] [PubMed] [Google Scholar]
- Ji T.; Ding Y.; Zhao Y.; Wang J.; Qin H.; Liu X.; Lang J.; Zhao R.; Zhang Y.; Shi J.; Tao N.; Qin Z.; Nie G. Peptide Assembly Integration of Fibroblast-Targeting and Cell-Penetration Features for Enhanced Antitumor Drug Delivery. Adv. Mater. 2015, 27, 1865–1873. 10.1002/adma.201404715. [DOI] [PubMed] [Google Scholar]
- Kim O. Y.; Park H. T.; Dinh N. T. H.; Choi S. J.; Lee J.; Kim J. H.; Lee S. W.; Gho Y. S. Bacterial Outer Membrane Vesicles Suppress Tumor by Interferon-Gamma-Mediated Antitumor Response. Nat. Commun. 2017, 8, 626–634. 10.1038/s41467-017-00729-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J. X.; Li Z. H.; Liu X. H.; Zheng D. W.; Chen Y.; Zhang X. Z. Bacteria-Mediated Tumor Therapy Utilizing Photothermally-Controlled TNF-alpha Expression via Oral Administration. Nano Lett. 2018, 18, 2373–2380. 10.1021/acs.nanolett.7b05323. [DOI] [PubMed] [Google Scholar]
- Lee J.; Yoon Y. J.; Kim J. H.; Dinh N. T. H.; Go G.; Tae S.; Park K. S.; Park H. T.; Lee C.; Roh T. Y.; Di Vizio D.; Gho Y. S. Outer Membrane Vesicles Derived from Escherichia coli Regulate Neutrophil Migration by Induction of Endothelial IL-8. Front. Microbiol. 2018, 9, 2268–2279. 10.3389/fmicb.2018.02268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M.; Clevers H. Reparative Inflammation Takes Charge of Tissue Regeneration. Nature 2016, 529, 307–315. 10.1038/nature17039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama M.; Rhee C.; Utsunomiya T.; Zhang N.; Ueno M.; Yao Z.; Goodman S. B. Modulation of the Inflammatory Response and Bone Healing. Front. Endocrinol. (Lausanne, Switz.) 2020, 11, 386–399. 10.3389/fendo.2020.00386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y.; Gao J.; Kaur P.; Wang Z. Neutrophils and Macrophages As Targets for Development of Nanotherapeutics in Inflammatory Diseases. Pharmaceutics 2020, 12, 1222–1245. 10.3390/pharmaceutics12121222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luster A. D.; Alon R.; von Andrian U. H. Immune Cell Migration in Inflammation: Present and Future Therapeutic Targets. Nat. Immunol. 2005, 6, 1182–1190. 10.1038/ni1275. [DOI] [PubMed] [Google Scholar]
- Goldberg I. J.; Sharma G.; Fisher E. A. Atherosclerosis: Making a U Turn. Annu. Rev. Med. 2020, 71, 191–201. 10.1146/annurev-med-042418-011108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne R. A.; Joner M.; Kastrati A. Stent Thrombosis and Restenosis: What Have We Learned and Where are We Going? The Andreas Gruntzig Lecture ESC 2014. Eur. Heart J. 2015, 36, 3320–3331. 10.1093/eurheartj/ehv511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borhani S.; Hassanajili S.; Ahmadi Tafti S. H.; Rabbani S. Cardiovascular Stents: Overview, Evolution, and Next generation. Prog. Biomater 2018, 7, 175–205. 10.1007/s40204-018-0097-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nording H.; Baron L.; Langer H. F. Platelets As Therapeutic Targets to Prevent Atherosclerosis. Atherosclerosis 2020, 307, 97–108. 10.1016/j.atherosclerosis.2020.05.018. [DOI] [PubMed] [Google Scholar]
- von Hundelshausen P.; Schmitt M. M. Platelets and Their Chemokines in Atherosclerosis-Clinical Applications. Front. Physiol. 2014, 5, 294–310. 10.3389/fphys.2014.00294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M. D.; Atkinson T. M.; Lindner J. R. Platelets and Von Willebrand Factor in Atherogenesis. Blood 2017, 129, 1415–1419. 10.1182/blood-2016-07-692673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer H.; Gawaz M. Platelet-Vessel Wall Interactions in Atherosclerotic Disease. Thromb. Haemostasis 2008, 99, 480–486. 10.1160/TH07-11-0685. [DOI] [PubMed] [Google Scholar]
- Albakri A. Ischemic Heart Failure: A Review of Clinical Status and Meta-analysis of Diagnosis and Clinical Management Methods. Clin Med. Investig 2018, 3, 1–15. 10.15761/CMI.1000171. [DOI] [Google Scholar]
- Marquez-Curtis L. A.; Janowska-Wieczorek A. Enhancing the Migration Ability of Mesenchymal Stromal Cells by Targeting the SDF-1/CXCR4 Axis. BioMed Res. Int. 2013, 2013, 561098–561112. 10.1155/2013/561098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H.; Yim H. W.; Park H. J.; Cho Y.; Hong H.; Kim N. J.; Oh I. H. Mesenchymal Stem Cell Therapy for Ischemic Heart Disease: Systematic Review and Meta-analysis. Int. J. Stem Cells 2018, 11, 1–12. 10.15283/ijsc17061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arshad M. A.; Murphy N.; Bangash M. N. Acute Liver Failure. Clin. Med. 2020, 20, 505–508. 10.7861/clinmed.2020-0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas C.; Wang Y.; Luebke-Wheeler J.; Nyberg S. L. Stem Cell Therapies for Treatment of Liver Disease. Biomedicines 2016, 4, 907–910. 10.3390/biomedicines4010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crop M. J.; Baan C. C.; Korevaar S. S.; Ijzermans J. N.; Pescatori M.; Stubbs A. P.; van Ijcken W. F.; Dahlke M. H.; Eggenhofer E.; Weimar W.; Hoogduijn M. J. Inflammatory Conditions Affect Gene Expression and Function of Human Adipose Tissue-derived Mesenchymal Stem Cells. Clin. Exp. Immunol. 2010, 162, 474–486. 10.1111/j.1365-2249.2010.04256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggenhofer E.; Benseler V.; Kroemer A.; Popp F. C.; Geissler E. K.; Schlitt H. J.; Baan C. C.; Dahlke M. H.; Hoogduijn M. J. Mesenchymal Stem Cells are Short-Lived and Donot Migrate Beyond the Lungs After Intravenous Infusion. Front. Immunol. 2012, 3, 297–304. 10.3389/fimmu.2012.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. J.; Anzaghe M.; Schulke S. Update on the Pathomechanism, Diagnosis, and Treatment Options for Rheumatoid Arthritis. Cells 2020, 9, 880–922. 10.3390/cells9040880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmester G. R.; Feist E.; Dorner T. Emerging Cell and Cytokine Targets in Rheumatoid Arthritis. Nat. Rev. Rheumatol. 2014, 10, 77–88. 10.1038/nrrheum.2013.168. [DOI] [PubMed] [Google Scholar]
- Wipke B. T.; Allen P. M. Essential Role of Neutrophils in the Initiation and Progression of a Murine Model of Rheumatoid Arthritis. J. Immunol. 2001, 167, 1601–1608. 10.4049/jimmunol.167.3.1601. [DOI] [PubMed] [Google Scholar]
- Headland S. E.; Jones H. R.; Norling L. V.; Kim A.; Souza P. R.; Corsiero E.; Gil C. D.; Nerviani A.; Dell’Accio F.; Pitzalis C.; Oliani S. M.; Jan L. Y.; Perretti M. Neutrophil-Derived Microvesicles Enter Cartilage and Protect the Joint in Inflammatory Arthritis. Sci. Transl. Med. 2015, 7, 315ra190. 10.1126/scitranslmed.aac5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer J. M.; Westhovens R.; Leon M.; Di Giorgio E.; Alten R.; Steinfeld S.; Russell A.; Dougados M.; Emery P.; Nuamah I. F.; Williams G. R.; Becker J.-C.; Hagerty D. T.; Moreland L. W. Treatment of Rheumatoid Arthritis by Selective Inhibition of T-Cell Activation with Fusion Protein CTLA4Ig. N. Engl. J. Med. 2003, 349, 1907–1915. 10.1056/NEJMoa035075. [DOI] [PubMed] [Google Scholar]
- McInnes I. B.; Schett G. Cytokines in the Pathogenesis of Rheumatoid Arthritis. Nat. Rev. Immunol. 2007, 7, 429–442. 10.1038/nri2094. [DOI] [PubMed] [Google Scholar]
- Kirtane A. R.; Verma M.; Karandikar P.; Furin J.; Langer R.; Traverso G. Nanotechnology Approaches for Global Infectious Diseases. Nat. Nanotechnol. 2021, 16, 369–384. 10.1038/s41565-021-00866-8. [DOI] [PubMed] [Google Scholar]
- Van der Meer J. W. The Infectious Disease Challenges of our Time. Front. Public Health 2013, 1, 1–2. 10.3389/fpubh.2013.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamblin S. J.; Skehel J. J. Influenza Hemagglutinin and Neuraminidase Membrane Glycoproteins. J. Biol. Chem. 2010, 285, 28403–28409. 10.1074/jbc.R110.129809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siboo I. R.; Chambers H. F.; Sullam P. M. Role of SraP, a Serine-Rich Surface Protein of Staphylococcus Aureus, in Binding to Human Platelets. Infect. Immun. 2005, 73, 2273–2280. 10.1128/IAI.73.4.2273-2280.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X.; Han M.; Li T.; Sun W.; Wang D.; Fu B.; Zhou Y.; Zheng X.; Yang Y.; Li X.; Zhang X.; Pan A.; Wei H. Effective Treatment of Severe COVID-19 Patients with Tocilizumab. Proc. Natl. Acad. Sci. U. S. A. 2020, 117, 10970–10975. 10.1073/pnas.2005615117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao L.; Xia S.; Xu W.; Tian R.; Yu G.; Gu C.; Pan P.; Meng Q. F.; Cai X.; Qu D.; Lu L.; Xie Y.; Jiang S.; Chen X. Decoy Nanoparticles Protect against COVID-19 by Concurrently Adsorbing Viruses and Inflammatory Cytokines. Proc. Natl. Acad. Sci. U. S. A. 2020, 117, 27141–27147. 10.1073/pnas.2014352117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R.; Zhang Y.; Li Y.; Xia L.; Guo Y.; Zhou Q. Structural Basis for the Recognition of the SARS-CoV-2 by Full-Length Human ACE2. Science 2020, 367, 1444–1448. 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi F.; Qian S.; Zhang S.; Zhang Z. Single Cell RNA Sequencing of 13 Human Tissues Identify Cell Types and Receptors of Human Coronaviruses. Biochem. Biophys. Res. Commun. 2020, 526, 135–140. 10.1016/j.bbrc.2020.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q.; Honko A.; Zhou J.; Gong H.; Downs S. N.; Vasquez J. H.; Fang R. H.; Gao W.; Griffiths A.; Zhang L. Cellular Nanosponges Inhibit SARS-CoV-2 Infectivity. Nano Lett. 2020, 20, 5570–5574. 10.1021/acs.nanolett.0c02278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Liu J.; Du S.; Shan C.; Nie K.; Zhang R.; Li X. F.; Zhang R.; Wang T.; Qin C. F.; Wang P.; Shi P. Y.; Cheng G. Evolutionary Enhancement of Zika Virus Infectivity in Aedes Aegypti Mosquitoes. Nature 2017, 545, 482–486. 10.1038/nature22365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.; Valderramos S. G.; Wu A.; Ouyang S.; Li C.; Brasil P.; Bonaldo M.; Coates T.; Nielsen-Saines K.; Jiang T.; Aliyari R.; Cheng G. From Mosquitos to Humans: Genetic Evolution of Zika Virus. Cell Host Microbe 2016, 19, 561–565. 10.1016/j.chom.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt D. J.; Smith A. M.; Arora N.; Diamond M. S.; Coyne C. B.; Miner J. J. Zika Virus-Related Neurotropic Flaviviruses Infect Human Placental Explants and Cause Fetal Demise in Mice. Sci. Transl. Med. 2018, 10, eaao7090 10.1126/scitranslmed.aao7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M.; Luan R. Research Progress of the Causal Link between Zika Virus and Microcephaly. Glob. Health J. 2018, 2, 11–18. 10.1016/S2414-6447(19)30178-2. [DOI] [Google Scholar]
- Kreuter J. Nanoparticulate Systems for Brain Delivery of Drugs. Adv. Drug Delivery Rev. 2001, 47, 65–81. 10.1016/S0169-409X(00)00122-8. [DOI] [PubMed] [Google Scholar]
- Campbell E. M.; Hope T. J. HIV-1 Capsid: The Multifaceted Key Player in HIV-1 Infection. Nat. Rev. Microbiol. 2015, 13, 471–483. 10.1038/nrmicro3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Los F. C.; Randis T. M.; Aroian R. V.; Ratner A. J. Role of Pore-Forming Toxins in Bacterial Infectious Diseases. Microbiol. Mol. Biol. Rev. 2013, 77, 173–207. 10.1128/MMBR.00052-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing L.; Qu H.; Wu D.; Zhu C.; Yang Y.; Jin X.; Zheng J.; Shi X.; Yan X.; Wang Y. Platelet-Camouflaged Nanococktail: Simultaneous Inhibition of Drug-Resistant Tumor Growth and Metastasis via a Cancer Cells and Tumor Vasculature Dual-Targeting Strategy. Theranostics 2018, 8, 2683–2695. 10.7150/thno.23654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copp J. A.; Fang R. H.; Luk B. T.; Hu C. M.; Gao W.; Zhang K.; Zhang L. Clearance of Pathological Antibodies Using Biomimetic Nanoparticles. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, 13481–13486. 10.1073/pnas.1412420111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv W.; Xu J.; Wang X.; Li X.; Xu Q.; Xin H. Bioengineered Boronic Ester Modified Dextran Polymer Nanoparticles As Reactive Oxygen Species Responsive Nanocarrier for Ischemic Stroke Treatment. ACS Nano 2018, 12, 5417–5426. 10.1021/acsnano.8b00477. [DOI] [PubMed] [Google Scholar]
- Hassan M.; Yazid M. D.; Yunus M. H. M.; Chowdhury S. R.; Lokanathan Y.; Idrus R. B. H.; Ng A. M. H.; Law J. X. Large-Scale Expansion of Human Mesenchymal Stem Cells. Stem Cells Int. 2020, 2020, 9529465–9529281. 10.1155/2020/9529465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y.; Sun J.; Hu T.; Ma Y.; Du T.; Kong C.; Zhang G.; Yu T.; Piao H. Rapid Expansion in the WAVE Bioreactor of Clinical Scale Cells for Tumor Immunotherapy. Hum. Vaccines Immunother. 2018, 14, 2516–2526. 10.1080/21645515.2018.1480241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coeshott C.; Vang B.; Jones M.; Nankervis B. Large-Scale Expansion and Characterization of CD3 (+) T-cells in the Quantum® Cell Expansion System. J. Transl. Med. 2019, 17, 258–270. 10.1186/s12967-019-2001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson M.; Grayson W. Recent Advances in Bioreactors for Cell-Based Therapies. F1000Research 2018, 7, 517–525. 10.12688/f1000research.12533.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjij I.; Khan S.; Ramharak P.; Cherqaoui D.; Soliman M. E. S. Distinguishing the Optimal Binding Mechanism of an E3 Ubiquitin Ligase: Covalent Versus Non Covalent Inhibition. J. Cell. Biochem. 2019, 120, 12859–12869. 10.1002/jcb.28556. [DOI] [PubMed] [Google Scholar]