Abstract
Background
Multidrug-resistant (MDR) and extensively drug-resistant (XDR) strains of Pseudomonas aeruginosa are the leading cause of healthcare-associated infections worldwide.
Objective
The aim was to identify the resistant phenotypes among P. aeruginosa and to characterize different aminoglycosides and carbapenem resistance genes as major mechanisms of resistance in these isolates, in Theodor Bilharz Research Institute (TBRI), a tertiary care hospital in Cairo, Egypt.
Methods
During a period of 11 months, 42 P. aeruginosa clinical isolates were collected from the microbiology laboratory by routine culture. Antimicrobial sensitivity testing to the aminoglycosides gentamicin and amikacin, and other classes of antibiotics, was performed by a disk diffusion method. Isolates were tested for aminoglycoside resistance genes, aac(6ʹ)-lb, aac-(3)-lla, rmtB, rmtC, armA, rmtD, and rmtF, and carbapenemase resistance genes blaNDM, blaVIM, and blaIMP, using conventional PCR.
Results
Thirty-three (78.5%) of the clinical P. aeruginosa isolates showed MDR and XDR phenotypes at 42.4% and 57.65%, respectively, and these were included in the study. Aminoglycoside resistance was found in 97%, whereas carbapenem resistance was found in 81% of the isolates phenotypically. Only 59.4% (19/26) of the aminoglycoside-resistant isolates harbored resistance genes; none of the amikacin-susceptible isolates harbored any of the tested aminoglycoside resistance genes. Aminoglycoside resistance genes rmtB, armA, aac(6ʹ)-lb, and rmtF were found at rates of 17/33 (51.5%), 3/33 (9%), 2/33 (6%), and 2/33 (6%), respectively, whereas rmtD, acc(3)-II, and rmtC were not detected. Only 40.7% (11/27) of the carbapenem-resistant isolates harbored resistance genes. Carbapenem resistance genes, blaNDM andblaVIM, were found at rates of 7/33 (21.2%) and 6/33 (18.1%), respectively, and blaIMP was not detected.
Conclusion
Rates of MDR and XDR P. aeruginosa and resistance to aminoglycosides and carbapenems in our setting are high. Methyltransferases and metallo-beta-lactamases are the main mechanisms of resistance to aminoglycosides and carbapenems, respectively. The presence of blaNDM and rmtF in the strains confirms their rapid dissemination in the Egyptian environment.
Keywords: aminoglycosides, carbapenems, antibiotic resistance, Pseudomonas aeruginosa, MDR, XDR, rmtB, rmtF, NDM
Introduction
Pseudomonas aeruginosa is a Gram-negative, rod-shaped, strictly aerobic bacterium. This bacterium is an opportunistic pathogen; it causes serious acute and persistent infections that often occur during existing diseases or conditions, including cystic fibrosis and traumatic burns. It is considered a cornerstone pathogen among the important resistant ESKAPE bacteria (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, P. aeruginosa, and Enterobacter species), which are the main pathogens for community and hospital drug resistance.1 It is also placed at the top of the WHO’s ranking list of critical pathogens for which there is a need to develop and discover novel therapeutic modalities.2 Pseudomonas aeruginosa is the cause of about 10% of hospital-acquired infections worldwide, leading to outbreaks in adult, pediatric, and neonatal intensive care units (ICUs) due to the spread of multidrug-resistant (MDR) or extensively drug-resistant (XDR) strains. Thus, it increases morbidity, mortality, length of hospitalization, and treatment costs. Antibiotics against P. aeruginosa include beta-lactam/beta-lactamase inhibitors, such as piperacillin–tazobactam, third generation cephalosporin (ceftazidime), fourth-generation cephalosporin (cefepime), carbapenems (imipenem and meropenem), aminoglycosides (amikacin, gentamicin, and tobramycin), fluoroquinolones (ciprofloxacin, ofloxacin, and levofloxacin), monobactam (aztreonam), and colistin.2
Among the eight categories of anti-pseudomonal agents, aminoglycosides are widely used to treat acute and chronic infections.3 During the past few years, increased bacterial resistance to aminoglycosides with anti-pseudomonal activities, including gentamicin, tobramycin, and amikacin, has been documented among healthcare-associated bacterial pathogens in almost all parts of the world.4 The identification of aminoglycoside resistance mechanisms is thus needed to avoid therapeutic failures, and is a prerequisite for the development of novel and innovative inhibitory molecules.5
There are four main mechanisms for aminoglycoside resistance: aminoglycoside-modifying enzyme (AME) production, reduced cell permeability and uptake, and modification of the ribosomal binding site.6 However, enzymatic modification of hydroxyl and amino groups is considered the most common mechanism.7 Modifying enzyme genes result in reduced binding of the aminoglycoside molecule to the ribosome. There are three families of aminoglycoside-modifying enzymes, namely, aminoglycoside acetyltransferases (AACs), aminoglycoside phosphotransferases (APHs), and aminoglycoside nucleotidyltransferases (ANTs).7 AACs constitute the main type of AME in P. aeruginosa.8
Carbapenems are among the primary treatment options for serious P. aeruginosa infection. The evolution of carbapenem-resistant Pseudomonas aeruginosa (CRPA) strains, mediated by acquiring genes encoding class B enzymes, is a global concern. CRPA occurs mainly through carbapenem-hydrolyzing enzymes. The metallo-beta-lactamases (MβLs) (eg, Verona integron-encoded metallo-beta-lactamases [VIMs], imipenemases [IMPs], and New Delhi metallo-beta-lactamases [NDMs]) are considered to be the most important enzymes in P. aeruginosa. The bla genes encoding these enzymes are either plasmid or chromosome mediated, usually located as horizontally transferable cassettes that classically cluster with other drug resistance determinants. So, the spread of P. aeruginosa with MβL activity could result in a pan-resistant phenotype, thus resulting in further limitation of therapeutic treatment options for these isolates.9
In this study, we aimed to evaluate the predominance of aminoglycoside resistance among MDR and XDR P. aeruginosa isolates, and to detect the main mechanisms of resistance and the most prevalent aminoglycoside resistance genes. Since carbapenem resistance represents a crucial problem that further limits therapeutic options in these isolates, blaMβLs were also investigated.
Materials and Methods
Clinical Specimens and Culture Conditions
From January 2020 to November 2020, 42 non-duplicate P. aeruginosa isolates were collected from different clinical specimens (urine, sputum, pus, and blood) obtained from inpatients and outpatients attending the microbiology laboratory at Theodor Bilharz Research Institute (TBRI), a tertiary care hospital in Cairo, Egypt. All specimens included in the study were archived and codes were used instead of patients’ names. The patients’ informed consent was waived as all patient data were anonymized. The protocol of the study was approved by TBRI institutional review board under Federal Wide Assurance (FWA00010609) and the work was carried out in accordance with the Code of Ethics of the World Medical Association (Declaration of Helsinki) for Experiments in Humans and its later amendments (GCP guidelines) or comparable ethical standards.
Identification of P. aeruginosa was carried out using standard techniques, and biochemical tests were conducted according to the procedures and protocols for the identification of non-fermenting Gram-negative bacilli. Pseudomonas aeruginosa identification was based on Gram staining, colony morphology, motility, pigment production, and oxidase reaction (HiMedia, India). Identification was confirmed using API 20 NE (Biomerieux, France). Pseudomonas aeruginosa ATCC 27853 was used as a positive control for bacterial identification. The P. aeruginosa isolates were stored in the form of glycerol stocks in sterile Eppendorf tubes and kept at −70°C until processed.10
Phenotypic Detection and Susceptibility Testing
Susceptibility testing for P. aeruginosa strains was carried out by the Kirby–Bauer disk diffusion method on Mueller–Hinton agar plates, against gentamicin (GM) (10 µg), amikacin (AK) (30 µg), ofloxacin (OFX) (5 µg), ciprofloxacin (CIP) (5 µg), levofloxacin (LEV) (5 µg), norfloxacin (NOR) (10 µg), ceftazidime (CAZ) (30 µg), cefepime (FEP) (30 µg), piperacillin/tazobactam (TZP) (100/10 µg), aztreonam (ATM) (30 µg), meropenem (MEM) (10 µg), and imipenem (IPM) (10 µg). The zone of inhibition was measured after 24-hour incubation at 35–37°C and interpreted according to the Clinical and Laboratory Standards Institute (CLSI) guidelines.11 Resistance phenotypes were defined as MDR P. aeruginosa for an isolate that is non-susceptible to at least one agent in three or more antimicrobial categories; an XDR isolate is non-susceptible to at least one agent in all but two or fewer categories; and pan-drug resistant means that an isolate is non-susceptible to all antimicrobial agents.12
DNA Extraction from P. aeruginosa Isolates
Genomic DNA was extracted from pure colonies of isolated P. aeruginosa isolates using the boiling method. Bacterial colonies were suspended in 1 mL TE buffer (1 mM EDTA, 10 mM Tris, pH 8) and boiled for 10 minutes. Subsequently, the samples were kept at −70°C until processed.13
PCR Analysis of Aminoglycoside and Carbapenem Resistance Genes
Detection of aac(6ʹ)-lb, aac-(3)-lla, rmtB, rmtC, armA, rmtD, andrmtF genes, using a conventional PCR assay (Bio Rad T100 thermal cycler, USA), was performed in a volume of 20 μL, with approximately 2 µL of extracted DNA, using 1 μL (1 pmol/1 μL) of each primer (Invitrogen Co, USA), both forward and reverse primers (Table 1), nuclease-free water, and ready-made PCR master mix solution (Thermoscientific, USA). An initial denaturation at 95°C for 5 minutes was followed by 35 cycles of denaturation at 95ºC for 30 seconds, and then annealing at 59°C for aac(6ʹ)-lb gene, 61°C for aac-(3)-lla gene, 59°C for rmtB gene, 55°C for rmtC gene, 53°C for armA gene, 58°C for rmtD gene, 60°C for rmtF gene, 60°C for blaNDM gene, 52°C for blaVIM gene, and 52°C for blaIMP gene, for 60 seconds. This was followed by extension at 72°C for 50 seconds. Final extension at 72°C for 10 minutes was the last step. The PCR for carbapenemase production genes, blaNDM, blaVIM, and blaIMP, was performed as described before.14 Each PCR product was separated on 2% agarose gel with ethidium bromide, and a 100-bp ladder was used as a DNA molecular weight standard. The sizes of the expected amplicons and sequences of primers are shown in Table 1.
Table 1.
Primers Used for the Detection of aac(6ʹ)-lb, aac(3)-lla, rmtB, rmtC, armA, rmtD, blaNDM, blaVIM, and blaIMP Genes, and Expected Product Size
| Gene Name | Sequence (5ʹ–3ʹ) | Amplicon Size | Reference |
|---|---|---|---|
| aac(6ʹ)-lb | F: TTGCGATGCTCTATGAGTGGCTA R: CTCGAATGCCTGGCGTGTTT |
482 bp | [15] |
| aac(3)-lla | F: GGCAATAACGGAGGCGCTTCAAAA R: TTCCAGGCATCGGCATCTCATACG |
563 bp | [15] |
| rmtB | F- GCTTTCTGCGGGCGATGTAA R- ATGCAATGCCGCGCTCGTAT |
173 bp | [16] |
| rmtC | F- GCTGCCCTTTGTATTGTC R-AGATGTTGGGTTAAGTCCC |
711 bp | [16] |
| armA | F- ATTCTGCCTATCCTAATTGG R- ACCTATACTTTATCGTCGTC |
315 bp | [16] |
| rmtD | F- CGGCACGCGATTGGGAAGC R- CGGAAACGATGCGACGAT |
401 bp | [16] |
| rmtF | F- GCGATACAGAAAACCGAAGG R-ACCAGTCGGCATAGTGCTTT |
589 bp | [14] |
| blaNDM | F-CCATGCGGGCCGTATGAGTGATTG R-TCGCGAAGCTGAGCACCGCATTAG |
700 bp | [14] |
| blaVIM | F- GATGGTGTTTGGTCGCATA R- CGAATGCGCAGCACCAG |
390 bp | [14] |
| blaIMP | F- GGAATAGAGTGGCTTAAYTCTC R- GGTTTAAYAAAACAACCACC |
232 bp | [14] |
Statistical Analysis
Statistical analysis was carried out using SPSS software (version 21.0) for Windows (#x1D712;2-test). A p value of ≥0.05 was considered significant.
Results
Bacterial Isolates and Susceptibility Testing
Forty-two clinical P. aeruginosa isolates were collected from the microbiology laboratory at TBRI; 33 of these isolates showed resistance phenotypes, as follows: 14 (42.4%) were MDR, 19 (57.6%) were XDR, and they were all included in the current study. The resistance profiles of studied P. aeruginosa isolates showed that 97% (32/33) were aminoglycoside resistant, whether resistant to both gentamicin and amikacin, 78.8% (26/33), or resistant to gentamicin only, 18.1% (6/33), by the disk diffusion method. Isolates were recovered from different wards, including urology (10/33, 30.3%), outpatients (10/33, 30.3%), ICU (7/33, 21.2%), tropical (4/32, 12.1%), and nephrology (2/33, 6%). They were collected from different clinical specimens, including urine (22/33, 66.7%), sputum (4/33, 12.1%), pus (5/33, 15.1%), and blood (2/33, 6%).
The resistance of P. aeruginosa isolates against other tested antibiotics was as follows: OFX (31/33, 94%), CIP (31/33, 94%), LEV (30/33, 91%), CAZ (23/33, 69.7%), FEP (29/33, 88%), and TZP (21/32, 63.6%), whereas only 7/33 (21.2%) were resistant to ATM. Carbapenem resistance was found in 81.8% (27/33) of the MDR P. aeruginosa isolates that were resistant to both imipenem and meropenem.
Molecular Detection of Aminoglycoside and Carbapenem Resistance Genes
Only 59.4% (19/26) of the aminoglycoside-resistant isolates harbored resistance genes; none of the amikacin-susceptible isolates harbored the tested aminoglycoside resistance genes.
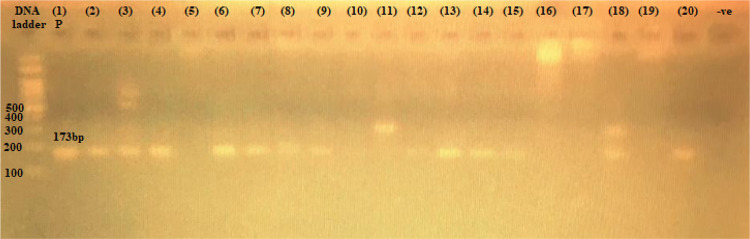
Aminoglycoside resistance genes rmtB, armA, aac(6ʹ)-lb, and rmtF were found at the following rates: 17/33 (51.5%), 3/33 (9%), 2/33 (6%), and 2/33 (6%), respectively, whereas rmtD, acc(3)-II, and rmtC were not detected (Table 2). The gene products of rmtB, armA, and aac(3)-lb genes are shown in Figures 1–3.
Table 2.
Association Between Aminoglycoside Resistance and Carbapenem Resistance Genotypes in 19 Positive Aminoglycoside Resistance Genes in P. aeruginosa Clinical Isolates
| Aminoglycoside Resistance Genotypes (n= 19)a | Carbapenem Resistance Genotypes (n=8)b | Type of Sample | Source of Specimens | |||
|---|---|---|---|---|---|---|
| Urine (n=14) | Blood (n=1) | Pus (n=1) | Sputum (n=3) | |||
| rmtB 14 (73.6%) |
- blaNDM + blaVIM, 1 (12.5%) - blaVIM, 2 (25%) - blaNDM, 1 (12.5%) |
11 (78.5%) | 0 (0.0%) | 0 (0.0%) | 3 (21.4%) | ICU, 4 (28.6%) Urology, 3 (21.4%) Nephrology, 1 (7.1) Tropical, 2 (14.2%) Outpatient, 4 (28.6%) |
| rmtB + armA 1 (7%) | ——– | 1 (100%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | Outpatient, 1 (100%) |
| rmtB + aac(6ʹ)lb 1 (7%) | blaNDM, 1 (12.5%) | 0 (0.0%) | 1 (100%) | 0 (0.0%) | 0 (0.0%) | ICU, 1 (100%) |
| rmtB + armA + aac(6ʹ)-lb 1 (7%) | — | 1 (100%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | Outpatient, 1 (100%) |
| rmtF+ armA 1 (7%) | blaNDM, 1 (12.5%) | 1 (100%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | Outpatient, 1 (100%) |
|
rmtF 1 (7%) |
blaNDM, 1 (12.5%) | 0 (0.0%) | 0 (0.0%) | 1 (100%) | 0 (0.0%) | Urology, 1 (100%) |
Notes: All parameters are represented as F (%) [frequency and percent]; data were analyzed by the χ2 test. aIndicates the presence of aminoglycoside resistance genes, including the detected methyltransferases (rmtB, armA, and rmtF), either alone or in combination with the only detected aminoglycoside-modifying enzyme, aac(6ʹ)lb. bThe detected metallo-beta-lactamases responsible for carbapenem resistance, including blaNDM and blaVIM, either alone or combined.
Abbreviation: ICU, intensive care unit.
Figure 1.
The 173-bp PCR amplification of rmtB gene among P. aeruginosa isolates. First lane, DNA ladder, 100-bp DNA size marker; (1) P, positive control, from 2 to 20 clinical isolates; −ve, negative control.
Figure 2.
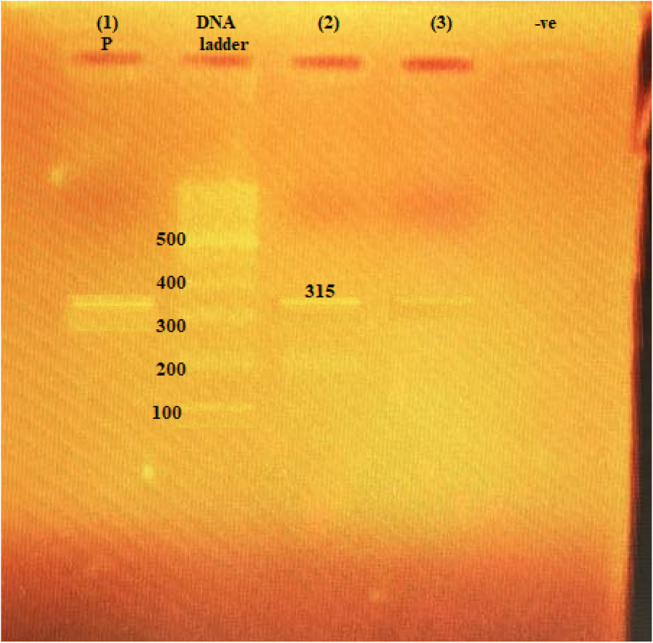
The 315-bp PCR amplification of armA gene among P. aeruginosa isolates. (1) P, positive control; second lane, DNA ladder, 100-bp DNA size marker; (2) and (3), clinical isolates with positive results; −ve, negative control.
Figure 3.
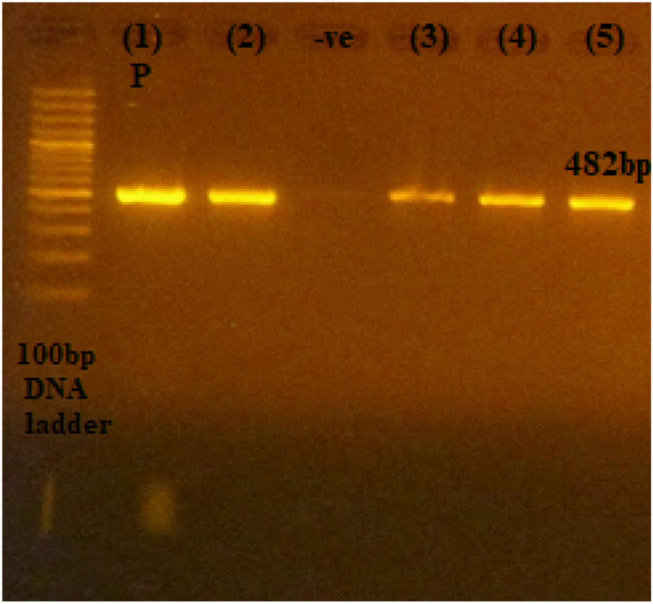
The 482-bp PCR amplification of aac(6ʹ)-lb gene among P. aeruginosa isolates. First lane, DNA ladder, 100-bp DNA size marker; (1) P, positive control; (2), (3), (4), and (5), clinical isolates; −ve, negative control.
Genes coding for methyltransferases (MTs) were the main resistance genes in aminoglycoside-resistant P. aeruginosa isolates. rmtB was the main gene and it was found alone in 14 isolates; they were collected from the ICU (4/14, 28.6%), outpatients (4/14, 28.6%), urology (3/14, 21.4%), nephrology (2/14, 14.2%), and tropical medicine departments (1/14, 7.1%); they were isolated from urine (11/14, 78.5%) and sputum (3/14, 21.4%). One isolate harbored rmtB + armA and another harbored rmtF + armA; both were collected from outpatient urine samples, whereas one isolate harbored only rmtF, from a pus specimen from the urology department. Aminoglycoside-modifying genes were not found alone in any of aminoglycoside-resistant isolates; one isolate harbored rmtB + aac(6ʹ)lb and was collected from the ICU department from a blood sample. One isolate harbored rmtB + armA + aac(6ʹ)lb and was collected from an outpatient urine sample. Only 40.7% (11/27) of carbapenem-resistant isolates harbored resistance genes. Carbapenem resistance genes, blaNDM and blaVIM, were found at rates of 7/33 (21.2%) and 6/33 (18.1%), respectively, while blaIMP was not detected (Figures 4 and 5). Only one carbapenem-susceptible isolate harbored blaVIM. Eight P. aeruginosa isolates harbored genes for both aminoglycoside and carbapenem resistance (Table 2).
Figure 4.

The 700-bp PCR amplification of blaNDM gene among P. aeruginosa isolates. First lane, DNA ladder, 100-bp DNA size marker; −ve, negative control.
Figure 5.
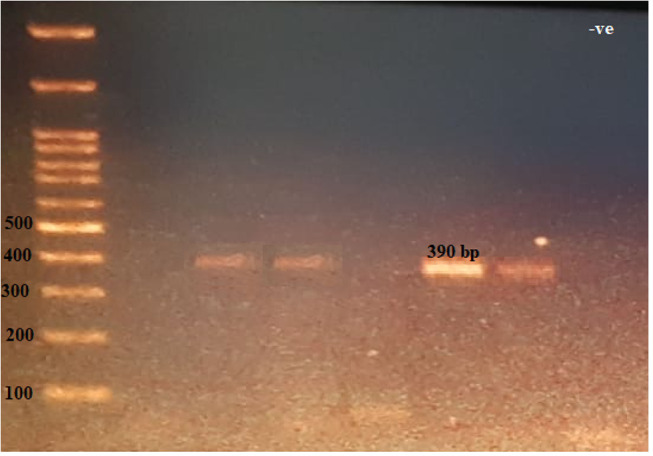
The 390-bp PCR amplification of blaVIM gene among P. aeruginosa isolates. First lane, DNA ladder, 100-bp DNA size marker; −ve, negative control.
Discussion
Infection with MDR P. aeruginosa presents a major health challenge as it is a common cause of healthcare-associated infections that are usually life-threatening, such as ventilator-associated pneumonia, bacteremia, and urinary tract infections, as well as wound and soft-tissue infections, which lead to significant morbidity and mortality.2
Treatment of P. aeruginosa is hampered by its ability to develop resistance to multiple classes of antibacterial agents, even during the course of treatment.17 Pseudomonas aeruginosa normally has low intrinsic antibiotic susceptibility, mediated through different mechanisms of resistance, such as inducible AmpC cephalosporinase, MexAB-OprM efflux pumps, and inducible MexXY efflux pump, together with its low outer membrane permeability and OXA-type oxacillinase. This is in addition to its tendency to acquire resistance genes and its inherently low cell membrane permeability.2,18
Aminoglycosides are among the most frequently prescribed antimicrobial agents against Gram-negative bacterial infections, including P. aeruginosa. The emergence of resistance to aminoglycosides in pathogenic Gram-negative bacteria is a growing concern. The aim of this study was to identify the resistance phenotypes in P. aeruginosa and the prevalence of genes encoding resistance to aminoglycosides, as well as carbapenem resistance genes associated in the isolates, as a major mechanism of resistance in these isolates, from TBRI, a tertiary care hospital in Cairo, Egypt.
Thirty-three (78.6%) of MDR and XDR P. aeruginosa isolates were collected from different clinical samples received at the microbiology laboratory at TBRI during a period of 11 months (January to November 2020). This rate was comparable to that found in previous studies, where all P. aeruginosa isolates were found to be MDR in one study,19 and 72.5% were MDR in Mansoura governorate in Egypt,20 but much higher than the rates reported previously from Menofia governorate (19%)21 and Upper Egypt (22.5%).22 Research from Iran estimated the rates of MDR and XDR P. aeruginosa as 16.5% and 15.53%, respectively.23 Another study showed that 88.9% of the isolates from Greece were MDR and XDR, whereas fewer isolates from Spain (33.3%) and Italy (43.5%) showed antibiotic resistance.24 Isolates showed the highest resistance rates to fluoroquinolones (91–94%), while 69.7% were resistant to ceftazidime, 88% to cefepime, 63.3% to tazobactam–piperacillin, 21% to aztreonam, and 81.8% to carbapenems. Similar results were reported from neighboring countries, such as Bahrain, where resistance of 72–100% was found to third-generation cephalosporins, carbapenems, aminoglycosides, fluoroquinolones, and piperacillin–tazobactam. In a previous Egyptian study, aztreonam was the most effective antibiotic.9 Another study from Europe showed that 64% of isolates were resistant to carbapenems and 54.7% of isolates were resistant to fluoroquinolones.24 It is suggested that the rise in resistance to anti-pseudomonal antibiotics, especially carbapenems, aminoglycosides, and fluoroquinolones has contributed to the emergence of P. aeruginosa MDR/XDR strains, posing a serious and critical therapeutic situation.20
Urine was the main type of specimen from which isolates were collected, representing almost 67%, as in previous studies from Egypt and Iraq (both 100%), and from Saudi Arabia (88.9%), thus showing the difficulty in managing urinary tract infections secondary to MDR P. aeruginosa. Different results were found in other areas, such as in Lebanon, with a lower rate of MDR P. aeruginosa (30%).25
Resistance to aminoglycosides was found in 97% of our isolates; it was manifested either as resistance to both gentamicin and amikacin, in 79%, or as resistance to gentamicin only, in 18%. However, aminoglycoside resistance genes were detected only in 59% of amikacin and gentamicin-resistant isolates, indicating the possibility of the presence of another mechanism that confers resistance to different types of aminoglycosides, such as activated efflux pumps. This figure is comparable to another study from Egypt, where resistance to gentamicin and amikacin was 70% and 66%, respectively.9 None of the susceptible isolates harbored aminoglycosides resistance genes, as in previous studies. A study from Saudi Arabia mentioned that the resistance profiles of P. aeruginosa isolates showed that 46.1% were resistant to one or more aminoglycoside antibiotics and that only 43.3% of the aminoglycoside-resistant isolates harbored resistance genes; none of the susceptible isolates harbored the tested resistance genes.16
Enzymatic modification of the aminoglycoside molecular structure with APH, AAC, or ANT AMEs represents the predominant mechanism of resistance in P. aeruginosa worldwide.26 However, methyltransferases were the main aminoglycoside resistance genes identified in this study, with a predominance of rmtB (51.5%), followed by armA(9%) and then rmtF (6%). However, aac(6ʹ)-lb, although known to be the commonest resistance gene among P. aeruginosa isolates, was detected only in two isolates (6%).8 That rate of rmtB was much higher than those reported from Brazil (44.4%)27 and from Saudi Arabia, where 43% of MDR P. aeruginosa isolates were resistant to amikacin and 18.5% to gentamicin, with rmtB also being the main resistance gene (7.6%), followed by aac(6ʹ)-Ib (6.1%), rmtC (4.6%), and armA (1.5%).16 Our results confirm findings from the USA, showing that AMEs play a relatively minor role in aminoglycoside resistance.28
In the current study, a single aminoglycoside resistance gene was detected in 15 (45%) of the isolates, while a combination of two genes was found in three (9%), and the combination of three genes was detected in only one isolate (3%) of the 33 studied MDR P. aeruginosa strains (Table 2).
Among all studied methyltransferases tested in this study, rmtF represents the most recently discovered and the least reported globally29,30. Thus, rmtF has rarely been reported in P. aeruginosa, and, to our knowledge, this is the first time that it has been reported in Egypt in P. aeruginosa isolates. It has only been previously associated with NDM-1 production in Enterobacteriaceae isolates in Egypt.14,30 In the current study, it was also found in two isolates that co-produced NDM enzyme. Aminoglycoside resistance is most frequently driven by aminoglycoside-modifying enzymes in P. aeruginosa, including the AACs and ANT. However, the prevalence of 16S rRNA MTs, which confer resistance to all aminoglycosides on the market, varies geographically, with the highest rates in Asia.3,31 The predominance of MTs as the main aminoglycoside resistance genes in the Egyptian isolates is an essential finding, as it is known that these enzymes confer resistance to all aminoglycosides, including the novel ones such as plazomicin.32
The high resistance rate to carbapenems (81.8%), associated with relatively low resistance to aztreonam (21%), indicated that the production of MβLs is the main mechanism of resistance to carbapenems. Carbapenemase production genes, mainly blaNDM (21%), followed by blaVIM (18%), were detected. Our findings are comparable to a previous report from Egypt, where blaNDM-1 was the main MβL (90.9%), followed by blaVIM-1 (18.1%),9 whereas blaIMP was not detected, as in another previous study which reported its low rate,33 while other studies did not find it at all.9,20,34 This is in contrast to other parts of the world, such as Iran and China, where blaIMP is the prevailing MβL in P. aeruginosa.35,36 The occurrence of blaVIM in a carbapenem-sensitive isolate was also previously documented from our healthcare setting by another study.37 However, carbapenem resistance MβL genes were found in only 40.7% of these isolates, with blaNDM as a predominant carbapenemase-producing gene (21%), suggesting the possible presence of other mechanisms of resistance to carbapenems other than the production of carbapenemases. Carbapenem-resistant isolates harbored aminoglycoside resistance genes together with their carbapenem-resistant genes in seven isolates. The co-production of MTs and IMP- or VIM-type MβL is not common globally; however, such an association has been reported from Korea, Greece, and Sweden,31 and from India.30 In the absence of MβL enzymes, carbapenem resistance may be attributed to increasing production of AmpC chromosome-encoded cephalosporinase, increasing the expression of efflux pump permeability and loss of porins.20
Middle Eastern countries are recognized as crossroads, with a high volume of travel and varied nationalities and populations. They are also recognized for their high antibiotic consumption, despite endeavors to implement antimicrobial stewardship programs.2 In the past few years, reports from Egypt have called again for the judicious use of conventional antimicrobials in an attempt to retain antibiotic effectiveness and avoid the spread of resistance genes.19
Conclusion
Our study confirms the prevalence of MTs as the main mechanisms of resistance to aminoglycosides in P. aeruginosa isolates, a finding that suggests the uselessness of all aminoglycosides in these isolates. This is in addition to the emergence of blaNDM in P. aeruginosa, which is worrying as it further limits the choice of suitable antibiotics. These findings raise concern over the dynamic state of antimicrobial resistance genes and confirm our urgent demand for the rational use of antibiotics through effective antimicrobial stewardship programs and the need to follow strict infection control measures to avoid the spread of these resistant determinants. This was a single-institution study; in the future we intend to enlarge the sample size by collaboration with other health settings in Egypt, to determine the current situation at the country level.
Funding Statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Abbreviations
ESKAPE bacteria, Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species; ICU, intensive care unit; MDR, multidrug-resistant; XDR, extensively drug-resistant; AME, aminoglycoside-modifying enzyme; AAC, aminoglycoside acetyltransferase; APH, aminoglycoside phosphotransferase; ANT, aminoglycoside nucleotidyl transferase; CRPA, carbapenem-resistant Pseudomonas aeruginosa; MβL, metallo-beta-lactamase; VIM, Verona integron-encoded metallo-beta-lactamase; IMP, imipenemase; NDM, New Delhi metallo-beta-lactamase; TBRI, Theodor Bilharz Research Institute; CLSI, Clinical and Laboratory Standards Institute; PCR, polymerase chain reaction; MT, methyltransferase.
Data Sharing Statement
Data and material are available from the corresponding author upon reasonable request.
Ethical Considerations
All specimens included in the study were archived and codes were used instead of patients’ names. The patients’ informed consent was waived. The protocol of the study was approved by TBRI institutional review board under Federal Wide Assurance (FWA00010609) and the work has been carried out in accordance with the Code of Ethics of the World Medical Association (Declaration of Helsinki) for Experiments in Humans and its later amendments (GCP guidelines) or comparable ethical standards.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, analysis and interpretation of data; took part in drafting the article and revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest for this work.
References
- 1.El-Far A, Samir S, El-Gebaly E, et al. Assessment of eugenol inhibitory effect on biofilm formation and biofilm gene expression in methicillin resistant Staphylococcus aureus clinical isolates in Egypt. Infect, Genet Evol. 2021;89:104722. doi: 10.1016/j.meegid.2021.104722 [DOI] [PubMed] [Google Scholar]
- 2.Al-Orphaly M, Hadi HA, Eltayeb FK, et al. Epidemiology of multidrug-resistant Pseudomonas aeruginosa in the Middle East and North Africa Region. mSphere. 2021;6(3). doi: 10.1128/msphere.00202-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krause KM, Serio AW, Kane TR, Connolly LE. Aminoglycosides: an overview. Cold Spring Harbor Perspect Med. 2016;6:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horcajada JP, Montero M, Oliver A, et al. Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clin Microbiol Rev. 2019;32(4):1–52. doi: 10.1128/CMR.00031-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kashfi M, Hashemi A, Eslami G, Amin MS, Tarashi S, Taki E. The prevalence of aminoglycoside-modifying enzyme genes among Pseudomonas aeruginosa strains isolated from burn patients. Arch Clin Infect Dis. 2017;12(1):1–5. doi: 10.5812/archcid.40896 [DOI] [Google Scholar]
- 6.Garneau-Tsodikova S, Labby KJ. Mechanisms of resistance to aminoglycoside antibiotics: overview and perspectives. MedChemComm. 2016;7(1):11–27. doi: 10.1039/c5md00344j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith CA, Bhattacharya M, Toth M, Stewart NK, Vakulenko SB. Aminoglycoside resistance profile and structural architecture of the aminoglycoside acetyltransferase AAC(6ʹ)-Im. Microbial Cell. 2017;4(12):402–410. doi: 10.15698/mic2017.12.602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costello SE, Deshpande LM, Andrew PD, Rodrigo E, Mendes MC. Aminoglycoside-modifying enzyme and 16S ribosomal RNA methyltransferase genes among a global collection of Gram-negative isolates. J Global Antimicrob Resist. 2019;16:278–285. doi: 10.1016/j.jgar.2018.10.020 [DOI] [PubMed] [Google Scholar]
- 9.Basha AM, El-Sherbiny GM, Mabrouk MI. Phenotypic characterization of the Egyptian isolates “extensively drug-resistant Pseudomonas aeruginosa” and detection of their metallo-β-lactamases encoding genes. Bull National Res Centre. 2020;44(1). doi: 10.1186/s42269-020-00350-8 [DOI] [Google Scholar]
- 10.Sambrook J, Russell DW. Molecular cloning: a laboratory manual. In: Molecular Cloning: A Laboratory a Manual. 3rd ed. 2001. [Google Scholar]
- 11.Weinstein MP, Lewis JS, Kraft CS. The clinical and laboratory standards institute subcommittee on Antimicrobial susceptibility testing: background, organization, functions, and processes. J Clin Microbiol. 2020;58(3):1–16. doi: 10.1128/JCM.01864-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- 13.Sadeghi HMM, Najafabadi AJ, Abedi D, Dehkordi AJ. Identification of an isolate of pseudomonas aeroginosa deposited in PTCC as a PHA producer strain: comparison of three different bacterial genomic DNA extraction methods. J Biol Sci. 2008;8:826–830. doi: 10.3923/jbs.2008.826.830 [DOI] [Google Scholar]
- 14.Gamal D, Fernández-Martínez M, Salem D, et al. Carbapenem-resistant Klebsiella pneumoniae isolates from Egypt containing blaNDM-1 on IncR plasmids and its association with rmtF. Int J Infect Dis. 2016;43:17–20. doi: 10.1016/j.ijid.2015.12.003 [DOI] [PubMed] [Google Scholar]
- 15.Miró E, Grünbaum F, Gómez L, et al. Characterization of aminoglycoside-modifying enzymes in enterobacteriaceae clinical strains and characterization of the plasmids implicated in their diffusion. Microb Drug Resist. 2013;19(2):94–99. doi: 10.1089/mdr.2012.0125 [DOI] [PubMed] [Google Scholar]
- 16.Asghar AH, Ahmed OB. Prevalence of aminoglycoside resistance genes in Pseudomonas aeruginosa isolated from a tertiary care hospital in Makkah, KSA. Clin Pract. 2018;15(2):541–547. doi: 10.4172/clinical-practice.1000391 [DOI] [Google Scholar]
- 17.Bassetti M, Vena A, Croxatto A, Righi E, Guery B. How to manage Pseudomonas aeruginosa infections. Drugs Context. 2018;7:1–18. doi: 10.7573/dic.212527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valderrama-Carmona P, Cuartas JH, Carolina CD, Corredor M. The role of Pseudomonas aeruginosa RNA methyltransferases in antibiotic resistance. IntechOpen. 2012;13:297. [Google Scholar]
- 19.Khalifa HO, Soliman AM, Ahmed AM, et al. High prevalence of antimicrobial resistance in gram-negative bacteria isolated from clinical settings in egypt: recalling for judicious use of conventional antimicrobials in developing nations. Microb Drug Resist. 2019;25(3):371–385. doi: 10.1089/mdr.2018.0380 [DOI] [PubMed] [Google Scholar]
- 20.El-Mahdy R, El-Kannishy G. Virulence factors of carbapenem-resistant pseudomonas aeruginosa in hospital-acquired infections in Mansoura, Egypt. Infect Drug Resist. 2019;12:3455–3461. doi: 10.2147/IDR.S222329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahmoud A, Zahran W, Hindawi G, Labib A, Galal R. Prevalence of multidrug-resistant pseudomonas aeruginosa in patients with nosocomial infections at a University Hospital in Egypt, with special reference to typing methods. J Virol Microbiol. 2013;2013:1–13. doi: 10.5171/2013.290047 [DOI] [Google Scholar]
- 22.Hassuna NA, Mandour SA, Mohamed ES. Virulence constitution of multi-drug-resistant pseudomonas aeruginosa in upper Egypt. Infect Drug Resist. 2020;13:587–595. doi: 10.2147/IDR.S233694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mirzaei B, Bazgir ZN, Goli HR, Iranpour F, Mohammadi F, Babaei R. Prevalence of multi-drug resistant (MDR) and extensively drug-resistant (XDR) phenotypes of Pseudomonas aeruginosa and Acinetobacter baumannii isolated in clinical samples from Northeast of Iran. BMC Res Notes. 2020;13(1):4–9. doi: 10.1186/s13104-020-05224-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pérez A, Gato E, Pérez-Llarena J, et al. High incidence of MDR and XDR Pseudomonas aeruginosa isolates obtained from patients with ventilator-associated pneumonia in Greece, Italy and Spain as part of the MagicBullet clinical trial. J Antimicrob Chemother. 2019;74(5):1244–1252. doi: 10.1093/jac/dkz030 [DOI] [PubMed] [Google Scholar]
- 25.Al Wutayd O, Al Nafeesah A, Adam I, Babikir IH. The antibiotic susceptibility patterns of uropathogens isolated in Qassim, Saudi Arabia. J Infect Dev Ctries. 2018;12(11):946–952. doi: 10.3855/jidc.10553 [DOI] [PubMed] [Google Scholar]
- 26.Pang Z, Raudonis R, Glick BR, Lin TJ, Cheng Z. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnol Adv. 2019;37(1):177–192. doi: 10.1016/j.biotechadv.2018.11.013 [DOI] [PubMed] [Google Scholar]
- 27.Rodrigues YC, Furlaneto IP, Pinto Maciel AHP, et al. High prevalence of atypical virulotype and genetically diverse background among Pseudomonas aeruginosa isolates from a referral hospital in the Brazilian Amazon. PLoS One. 2020;15(9):1–21. doi: 10.1371/journal.pone.0238741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Atassi G, Scheetz M, Nozick S, et al. Genomics of aminoglycoside resistance in pseudomonas aeruginosa bloodstream infections at a United States Academic Hospital. medRxiv. 2021. doi: 10.1101/2021.01.15.21249897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hidalgo L, Hopkins KL, Gutierrez B, et al. Association of the novel aminoglycoside resistance determinant RmtF with NDM carbapenemase in enterobacteriaceae isolated in India and the UK. J Antimicrob Chemother. 2013;68(7):1543–1550. doi: 10.1093/jac/dkt078 [DOI] [PubMed] [Google Scholar]
- 30.Rahman M, Prasad KN, Pathak A, et al. RmtC and RmtF 16S rRNA methyltransferase in NDM-1–producing Pseudomonas aeruginosa. Emerg Infect Dis. 2015;21(11):2059–2062. doi: 10.3201/eid2111.150271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jun-ichiWachinoa J-I, Arakawa Y. Exogenously acquired 16S rRNA methyltransferases found in aminoglycoside-resistant pathogenic Gram-negative bacteria: an update. Drug Resist Updates. 2012;15(3):133–148. doi: 10.1016/j.drup.2012.05.001 [DOI] [PubMed] [Google Scholar]
- 32.Golkar T, Bassenden AV, Maiti K, Arya DP, Schmeing TM, Berghuis AM. Structural basis for plazomicin antibiotic action and resistance. Commun Biol. 2021;4(1). doi: 10.1038/s42003-021-02261-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hashem H, Hanora A, Abdalla S, Shaeky A, Saad A. Dissemination of metallo-β-lactamase in Pseudomonas aeruginosa isolates in Egypt: mutation in blaVIM-4. Apmis. 2017;125(5):499–505. doi: 10.1111/apm.12669 [DOI] [PubMed] [Google Scholar]
- 34.El MN, Said M, Emad R, Salama M, Hashish A. Detection of IMP and VIM genes in Pseudomonas aeruginosa isolated from Egyptian patients. Arch Med Sci Civilization Dis. 2019;4(1):58–63. doi: 10.5114/amscd.2019.86742 [DOI] [Google Scholar]
- 35.Dogonchi AA, Ghaemi EA, Ardebili A, Yazdansetad S, Pournajaf A. Metallo‑β‑lactamase‑mediated resistance among clinical carbapenem‑resistant. Tzu Chi Medical j. 2018;30(2):90–96. doi: 10.4103/tcmj.tcmj [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang W, Wang X. Prevalence of metallo-β-lactamase genes among Pseudomonas aeruginosa isolated from various clinical samples in China. J Lab Med. 2020;44(4):197–203. doi: 10.1515/labmed-2019-0162 [DOI] [Google Scholar]
- 37.Diab M, Fam N, El-said M, EL-Defrawy EE. Occurrence of VIM-2 Metallo– lactamases in imipenem resistant and susceptible Pseudomonas aeruginosa clinical isolates from Egypt. Afr J Microbiol Res. 2013;7(35):4465–4472. doi: 10.5897/AJMR2013.6181 [DOI] [Google Scholar]