Graphical abstract
Keywords: Evolutionary mutations, Allosteric modulators, Homodimeric protein, Natural compounds, Dynamic residue network analysis, MD-TASK, MDM-TASK-web, Betweenness centrality, Closeness centrality, Degree centrality, Eigencentrality, Katz centrality
Abstract
The rational search for allosteric modulators and the allosteric mechanisms of these modulators in the presence of mutations is a relatively unexplored field. Here, we established novel in silico approaches and applied them to SARS-CoV-2 main protease (Mpro) as a case study. First, we identified six potential allosteric modulators. Then, we focused on understanding the allosteric effects of these modulators on each of its protomers. We introduced a new combinatorial approach and dynamic residue network (DRN) analysis algorithms to examine patterns of change and conservation of critical nodes, according to five independent criteria of network centrality. We observed highly conserved network hubs for each averaged DRN metric on the basis of their existence in both protomers in the absence and presence of all ligands (persistent hubs). We also detected ligand specific signal changes. Using eigencentrality (EC) persistent hubs and ligand introduced hubs we identified a residue communication path connecting the allosteric binding site to the catalytic site. Finally, we examined the effects of the mutations on the behavior of the protein in the presence of selected potential allosteric modulators and investigated the ligand stability. One crucial outcome was to show that EC centrality hubs form an allosteric communication path between the allosteric ligand binding site to the active site going through the interface residues of domains I and II; and this path was either weakened or lost in the presence of some of the mutations. Overall, the results revealed crucial aspects that need to be considered in rational computational drug discovery.
1. Introduction
With the advent of COVID-19, researchers, world-wide reacted quickly to design multiple potential inhibitors to abrogate viral protein activity using rational drug design approaches and wet lab experiments. This concept primarily involves targeting critical viral life-cycle proteins [1], [2], [3], [4]. The SARS-CoV-2 main protease (Mpro) protein plays a crucial role in the viral maturation cycle by lysing itself (autocatalysis) and other viral polyproteins [5]. This presents SARS-CoV-2 Mpro as a key drug target for designing wide-spectrum [6], [7] anti-COVID-19 inhibitors or allosteric modulators that terminate the viral replication cycle [8]. Among the multitude of studied COVID-19 related proteins, the active site of SARS-CoV-2 Mpro has been extensively targeted by virtual screening of both natural and non-natural compounds [9], [10], [11]. In contrast, the rational search for allosteric modulators of the protein is still relatively unexplored [12], [13]. Additionally, allosteric mechanisms in the presence of mutations are rarely considered in drug screening. In our previous study, a potential dual allosteric pocket of SARS-CoV-2 Mpro was identified through multiple in silico tools in the presence of 50 early pandemic mutations [14]. These two pockets are mirrored across the dimer interface and are individually composed of residues from each protomer. Continuing our previous SARS-CoV-2 Mpro work [14], we now set up alternative innovative therapeutic concepts to identify allosteric modulators in the presence of early evolutionary mutations of the virus. These concepts are explained under three subsequent sections:
PART I: Here, we identified potential allosteric modulators for the dimeric SARS-CoV-2 Mpro protein, at a protonation state corresponding to pH 7.0, by screening it against 625 South African natural compounds [15], [16]. Parallel to this, we also docked the natural compounds against the Mpro protein of one of the seven human coronaviruses, HCoV-OC43. Previously, HCoV-OC43 was suggested as a model to study SARS-HCoV without the need for Biosafety Level 3 facilities [17]. This strain is, indeed, used as a laboratory strain. Both strains are under the genus Betacoronavirus, and HCoV-OC43 belongs to the subgenus Embecovirus, while SARS-CoV-2 is Sarbecovirus [18]. Thus, using in silico techniques we wanted to see if similar results would be obtained from the Mpro protein in each strain. This analysis sheds light on potential considerations to factor in when transferring findings of whole virus particle experiments from HCoV-OC43 to SARS-CoV-2.
PART II: Next, our focus was to understand the allosteric effects of the selected hit compounds (PART I) on each protomer of the reference Mpro protein (wild type, WT). In our previous study, we encountered the problem of protein symmetry, where we observed that protomer dynamics could be switched between identical copies of a protomer in a homodimer. Symmetry correction had been performed then by aligning single equilibrium conformations. In this study, we investigated the phenomenon in greater detail using a combinatorial approach to examine patterns of change and conservation of critical nodes, according to five independent criteria of network centrality (betweenness centrality (BC), closeness centrality (CC), degree centrality (DC), eigencentrality (EC) and katz centrality (KC)), with each being used as averages. While doing so, we investigated the relationships and effectiveness of each metric in characterizing allosteric behavior. We hypothesized that allosteric change might be expressed through complex routes involving intraprotomeric and interprotomeric combinations of critical residues. By monitoring the centrality patterns of these residues across the homodimer under the influence of intrinsic (e.g. protein mutations and ligand binding) and extrinsic (simulation parameters) factors during molecular dynamics (MD) simulations, we aimed to extract further details from the homodimer state of the protease. To our knowledge this phenomenon is not commonly addressed in the case of homodimeric protein complexes, even though some other examples of asymmetric behavior of proteins have been reported, such as Hsp90 [19] and KatG [20]. While the same phenomenon exists at the homomultimeric level [21], a less complex case involving allosterically bound dimeric Mpro is investigated herein, with a combinatorial approach as indicated in Table 1 which is only applicable to dimeric proteins.
Table 1.
Hub combination possibilities for any given residue between two dimers. A tick symbol (✓) denotes the presence of a hub from a given protomer, while a cross (x) denotes absence of that same hub from a chain. Apo - A: Apo protein, protomer A; Apo - B: Apo protein, protomer B; Complex A: Protomer A of protein–ligand complex; Complex B: Protomer B of protein–ligand complex.
| Apo - A | Apo - B | Complex A | Complex B | Score | Interpretation | |
|---|---|---|---|---|---|---|
| 1 | ✓ | x | x | x | 1 | Potential ligand effect inferred by asymmetry |
| 2 | x | ✓ | x | x | 1 | Potential ligand effect inferred by asymmetry |
| 3 | x | x | ✓ | x | 1 | Potential ligand effect inferred by asymmetry |
| 4 | x | x | x | ✓ | 1 | Potential ligand effect inferred by asymmetry |
| 5 | ✓ | ✓ | x | x | 2 | Complete hub loss: ligand effect |
| 6 | x | ✓ | ✓ | x | 2 | Inconclusive effect |
| 7 | x | x | ✓ | ✓ | 2 | Hub gain on ligand presence |
| 8 | ✓ | x | ✓ | x | 2 | Inconclusive effect |
| 9 | ✓ | x | x | ✓ | 2 | Inconclusive effect |
| 10 | x | ✓ | x | ✓ | 2 | Inconclusive effect |
| 11 | ✓ | ✓ | ✓ | x | 3 | Potential ligand effect inferred by asymmetry |
| 12 | x | ✓ | ✓ | ✓ | 3 | Potential ligand effect inferred by asymmetry |
| 13 | ✓ | x | ✓ | ✓ | 3 | Potential ligand effect inferred by asymmetry |
| 14 | ✓ | ✓ | x | ✓ | 3 | Potential ligand effect inferred by asymmetry |
| 15 | ✓ | ✓ | ✓ | ✓ | 4 | No ligand effect from symmetry |
| 16 | x | x | x | x | 0 | Not applicable |
Further, we, for the first time, introduced the concept of analyzing globally central nodes (i.e. the 5% most central nodes measured across all samples) for each of the five metrics of dynamic residue networks (DRNs). The metrics comprised averaged versions of BC, CC, DC, EC and KC. Even though some of these metrics were previously used for protein structure analysis [22], [23], [24], to our knowledge this is the second study that gathers five metrics information together in protein analysis and applies over molecular dynamics (MD) simulations [25]. Additionally, the hub data was itself reformulated as a set of network graphs, which were queried in order to decipher the complex patterns of hub conservation and transition (according to each DRN metric) from the apo state to one that is allosterically occupied.
PART III: Here, we examined the effects of mutations on allosteric behavior of the protein in the presence of selected potential allosteric modulators and investigated ligand stability. Structure-based drug discovery approaches have been successfully used for the design of many orthosteric drugs [26] (and to some extent allosteric modulators [27]) for the treatment of communicable and non-communicable diseases. A good example is that of HIV protease inhibitors [28]. However, the impact of evolutionary mutations of pathogens, including those linked to drug resistance, is mostly undetermined in rational drug design. Depending on their position and physicochemical properties, mutations can modulate protein behavior by altering their stability and/or affinity to other interacting biological molecules [29], [30], [31], [32]. A more complex, yet subtle phenomenon may be observed at the level of entropic effects of mutations, whereby differences may be seen at the level of the rate of visiting certain states, and not by the mere presence or absence of a defined state (or set thereof) [33], [34], [35]. A classic case is the distance effect of pathogenic mutations that maintain protein function while gaining resistance [30], [36]; hence our purpose is to understand the effect of evolutionary mutations in COVID-19 rational drug design. We believe the information gleaned here may help develop drugs that could potentially minimize the risks of having premature drug inactivation; and may reduce potential drug resistance effects to provide a longer-lasting treatment option.
For that purpose, mutant protein-allosteric modulator complexes were subjected to 20 ns all-atom MD simulations at a fixed pH, and the results were then evaluated in the same manner as introduced in the second part of the article. The potential effectiveness of the allosteric modulators was identified in the presence of some of the early pandemic mutations of the protein. Even though no solid evidence of the effect of these mutations has been reported, involving them in drug development might help further our understanding of the enzyme’s mechanics and pre-empt the most worrying feature of mutations: drug resistance.
Overall, the results of this study revealed crucial aspects that need to be considered in structure-based drug discovery, such as the way in which the allosteric modulators should be identified; and how the stability of these modulators should be considered in the presence of mutations. We further argue that the potential consideration of asymmetric behavior in homodimeric proteins; of novel DRN approaches and data analysis that are presented here would be applicable and useful in any computational drug discovery research.
2. Materials and methods
2.1. Preparation of the reference and mutant SARS-CoV-2 Mpro and HCoV-OC43 Mpro structures
The three-dimensional (3D) structure of the SARS-CoV-2 Mpro was retrieved from the Protein Data Bank (PDB) [37] (PDB ID: 5RFV [38]), and its dimeric unit was assembled as described in our previous study [14]. In this study, we also utilized a set of 50 SARS-CoV-2 Mpro mutant proteins that were prepared in our previous study [14]. The list of mutations that were acquired from the Global Initiative on Sharing All Influenza Data (GISAID) [39] as described in our previous work is presented in Table S1 [53].
5RFV was further used as a template to model the 3D structure of the human coronavirus strain (HCoV-OC43) Mpro via MODELLER, using the automodel function parameterized with a slow refinement and a deviation of 2.5 Å [40]. This protein is a homolog of the SARS-CoV-2 Mpro, and the strain is generally used in inhibition assays in the laboratory. Prior to homology modelling, the HcoV-OC43 protein sequence was retrieved from the replicase polyprotein 1a record available from UniProt (Entry ID: P0C6U7; position 3247–3549), and was aligned against the sequence and structure of 5RFV using PROMALS3D [41]. The model with the lowest z-DOPE score was selected from a parallel run of 50 models. The PROPKA tool under the PDB2PQR algorithm [42] was then utilized to assign protonation states of all the proteins at a pH of 7. The calculations were done with the AMBER force field [43].
Based on the assembled and protonated SARS-CoV-2 Mpro dimeric structure, all 50 mutations were inserted using BIOVA Discovery Studio Visualizer [44]. This approach was utilized to minimize structural variations across the proteins. All mutated structures were subsequently protonated using the same procedure as for the reference structure.
2.2. High-throughput virtual screening of SANCDB compounds against Mpro proteins
A total of 623 compounds were first obtained from the South African natural compound database (SANCDB) [15], [16]. Partial charges were assigned to compounds and the protonated proteins using the Gasteiger-Hückel protocol in AutoDockTools (ADT) [45]. The AutoDock/Vina plugin from PyMOL was used to place the docking grid around the dimeric SARS-CoV-2 Mpro reference protein. A docking box size of 65 × 71 × 80 Å with a grid spacing of 1 Å was centered at coordinates (0.00, 0.65 and 0.00). An exhaustiveness of 1000 was used, and the maximum number of docking poses was increased to 20. Blind docking (BD) simulations were performed in parallel, with 12 cores per job at the Center for High Performance Computing (CHPC) using the QuickVina-W program [46]. After having docked the SANCDB compounds, the ligand PDBQT files were split into their separate poses before being converted to the PDB format. Preliminary filtering was then applied using an in-house C++ script to every file to retain ligand poses that had a centroid distance of less than 10 Å to any of the allosteric pockets irrespective of binding energy. The pre-filtered poses were then manually curated in PyMOL (version 2.4) [47] to remove those that did not localize to the allosteric pocket. For each of the filtered ligands, the number of poses was tallied and ranked in ascending order of binding energy [48], [49]. The top six compounds from the SARS-CoV-2 Mpro were then short-listed based on the residue interactions of their respective lowest energy poses. HCoV-OC43 Mpro underwent the same steps, to be used as a comparator.
2.3. Molecular dynamics simulations protocol of Mpro and mutant systems
100 ns all-atom molecular dynamics (MD) simulations were conducted using GROMACS (version 2019) [50] for the SARS-CoV-2 Mpro reference protein and the HCoV-OC43 strain homolog protein both in the absence and presence of six hit compounds bound at the previously identified allosteric site. In order to investigate the effect of mutations on ligand stability, 50 ligand-bound SARS-CoV-2 Mpro mutants were similarly taken into 20 ns MD runs for each of the six compounds. GROMACS-compatible structure and ligand topology input files were derived using the AMBER03 force field [43] and the ACPYPE tool [51] respectively. A total of 314 systems [(reference protein × 6) + (homolog protein × 6) + Apo-reference protein + Apo-homolog protein + (50 mutant × 6 compounds)] were solvated using the TIP3P water model [52] in a cubic box, with a minimum distance of 1 nm between the box edge and the protein. All systems were subsequently neutralized with 0.15 M NaCl. Solvated systems were first minimized for 5000 steps using the steepest descent algorithm until the relaxed systems converged to a maximum force of 1000 kJ/mol/nm. Following minimization, systems were equilibrated assuming a constant number of particles, volume and temperature (NVT) (300 K) using the modified Berendsen thermostat algorithm [53], followed by an NPT (constant number of particles, pressure and temperature) equilibration step parametrised at 1 bar pressure using the Parrinello–Rahman barostat algorithm [54]. An integration time step of 2 fs was used in all cases. All bonds were constrained under the LINCS holonomic constraints algorithm [55], whereas the Particle-mesh Ewald (PME) algorithm [56] was set to include the contribution of long-range electrostatic interactions. The overall MD protocol was carried out on the Center for High Performance Computing (CHPC), Cape Town, South Africa using 384 cores for a total of ∼ 2,921,472 CPU hours. Structure coordinates were written after every 10 ps, and periodic boundary conditions (PBC) were removed prior to analysis.
2.4. Calculation of dynamic residue network metrics
To study the effect of ligand binding on the active site, as well as on inter-and intra-domain residue dynamics over the course of MD simulations, dynamic residue network analysis (DRN) was done using MDM-TASK-web scripts [57]. DRN [58] was applied on the last 10 ns trajectories of the apo and ligand-bound Mpro systems, after post-processing the MD trajectories to remove previously introduced water molecules, and sodium chloride ions. Residue network analysis uses graph theory concepts and represents residues in a protein structure as nodes (Cβ and Gly Cα atoms), with inter-connected residues (Cβ - Cβ, Gly Cα - Cβ and Gly Cα- Gly Cαatoms) are depicted as edges based on a specified cut-off distance (6.7 Å) [58]. DRNs were analysed based on five metrics: averaged betweenness centrality (BC), averaged closeness centrality (CC), averaged degree centrality (DC), averaged eigencentrality (EC) and averaged katz centrality (KC) via the cal_network.py script incorporated in the web server, MDM-TASK-web [57]. Each of the metrics is a time-averaged summary of the network metrics obtained during MD simulations.
The averaged BC metric is defined as how often a residue is traversed along the shortest paths connecting every other residue pairs [59]. This metric was calculated based on the equation:
| (1) |
where δ(s,t|v) symbolises the number of shortest paths bridged between a residue v and other nodes s and t. δ(s,t) denotes the averaged shortest paths existing between residues s and t where s and t are part of the set V, which comprises the set of all nodes, while m indicates the overall number of frames. n denotes the total number of residues.
Averaged closeness centrality (CC) of a residue is calculated as the reciprocal of the average number of the shortest paths linking a residue v and all other residues in the network.
| (2) |
where d (v, u) is the total distance between residue v and all other residues u.
Additionally, metric degree centrality (DC) defines the number of neighboring nodes (the local connectivity) around a given node. It is normalized by both the number of nodes in the network and the number of MD frames. The equation for computing the averaged DC is as follows:
| (3) |
where n indicates the number of residues, m denotes the number of frames; Aijk indicates adjacency at time frame i, being 1 if residues with indices j and k are adjacent and 0 otherwise.
Eigencentrality (EC) measures the high centrality given to high degree residue, or to a residue that is connected to other high degree residues. The procedure for calculating EC is summarized here, and further details are in literature [60]. The formula to compute EC for a single residue i for the kth frame is as follows:
| (4) |
The weighted multiplication operation between the adjacency matrix A is repeated against the vector EC until convergence. Aij is an adjacency, k is a frame, ECik is the jth component of the EC vector for the kth frame, and n is the number of nodes. The averaged EC for the ith node is then computed from the matrix of EC likewise using MDM-TASK-web as follows:
| (5) |
Lastly, Katz centrality (KC) measures the relative degree of influence of a residue i within connected residues in a network. The procedure for calculating KC is summarized here, and further details are in literature [60]. The KC of node i is
| (6) |
| (7) |
where A represents the adjacency matrix and KC is the eigenvector computed by NetworkX in MDM-TASK-web. α and β denote the attenuation factor and weight assigned to each node. The same metric is computed for each frame before averaging the value across frames for each residue.
2.5. Identification of top 5% global high network centrality residues
DRN metrics were computed for the reference and the mutant SARS-CoV-2 Mpro samples using MDM-TASK-web for both the apo and the six ligand-bound complexes. In order to estimate residue hubs, all related samples that were to be compared were combined in order to have a common scale. Therefore, for each individual DRN metric, the data points of samples (apo, mutant and ligand-bound) belonging to that metric were concatenated into a single vector, which was sorted in descending order to focus on nodes of highest overall centrality. The top 5% of these values were extracted [304 residues × 2 chains × (1 apo reference + 6 bound reference) systems × 0.05 = 212 elements]. The value at this index was used as a threshold for the selection of entries from the original data set. Then, each of the original matrices was searched for any component greater than or equal to that minimum number. To accomplish that, a binary matrix was built that contained the number “one” for any cell that satisfied the condition, and from which the row sums were then computed, in order to select any row with a non-zero row sum. This generated a set of row indices that were used to subset the original matrix of centrality values. In this manner, the globally high network centrality values were obtained in the presence of their counterpart values in other samples, thus showing how the hubs perform sample-wide. This approach was performed separately for each of the 5 metrics.
2.6. Application of a binary logic to investigate protomer hub combinations from DRN analysis
For each DRN metric, a global network was built using as nodes the detected globally central hubs for all of the reference protein states (ApoA/B, SANC00302A/B, SANC00303A/B, SANC00467A/B, SANC00468A/B, SANC00469A/B), which have as labels the protein state and the protomer to which each hub residue belongs. These labels were inserted as nodes, and undirected edges were created from them by linking their respective hub nodes to them. As this global network was too dense to analyze, a sub-network was extracted for each individual complex and was merged to the apo protomers. In this way, one could identify whether a hub was shared, gained or lost from the apo state upon ligand binding. This representation was applied and analyzed in a systematic manner (according to Table 1) to investigate whether ligand binding had any effect, as we posited that the effects of a ligand's presence in the allosteric site may manifest itself not only in the bound protomer, but also in the unbound one. In this way it was possible to track patterns of hub conservation and divergence.
3. Results and discussion
3.1. Revisiting the structure of Mpro and mutants
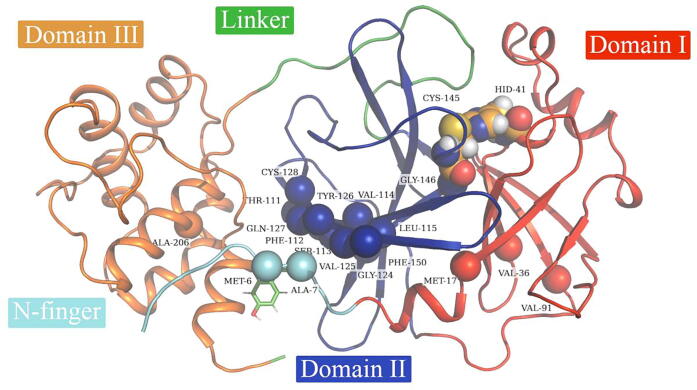
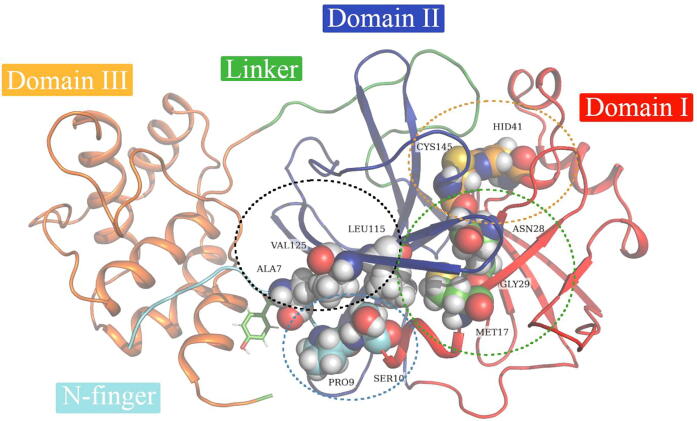
The SARS-CoV-2 Mpro protein comprises 306 residues [2] and is active in its dimeric state at a pH of 7.0 [6], [61]. The dimeric functional state regulates catalytic turnover using the subunit flip-flop mechanism where the two monomers are used alternately in acylation and deacylation steps [62], [63]. Each monomer (designated protomer A and B) harbors three distinct domains (I-III) [2], [10] and contains a His-Cys catalytic dyad signature (HIS41 and CYS145) located within a well-defined hydrophobic substrate-binding site formed between domains I and II (Fig. 1). The catalytic dyad residues are key for hydrolysis in which HIS41 functions as a general base [6], [64]. SARS-CoV-2 Mpro domains I (residue 10–99) and II (100–183) consist of an antiparallel β-barrel structure [2] that form the catalytic domains of the protein as the active site is located between domain I and II. Domain III (198–303) is predominantly compsed of antiparallel α-helices [61], [65] and is connected to the catalytic domains by a long loop region (184–197). This domain is involved in the regulation of enzymatic activity of the virus [66]. The interaction interface, which is crucial for dimerization and enzymatic activity, is formed between domain II of protomer A and the N-finger region (1–9) of protomer B and vice versa [64], [67]. These two N-finger signatures interact with Glu166 to maintain the correct orientation of the substrate-binding site. The N-finger feature is similar to that of previously reported Mpro from other coronaviruses [8], [61], [68], [69]. Each protomer has subsites (S1 – S5) located in the active site cavity, which comprises the following residues: THR25 [70], LEU27, HIS41 [2], [6], [71], CYS44 [70], THR45, SER46 [70], MET49 [2], [6], [70], [71], ILE54 [2], [70], PHE140 [2], [6], [70], LEU141 [2], [70], SER144 [6], [71], CYS145 [2], [6], [71], HIS163 [2], [6], [70], [71], HIS164 [2], [70], MET165 [2], [70], [71], LEU166 [2], [6], [70], [71], LEU167 [2], [70], PRO168 [2], [6], [70], [71], HIS172 [2], [70], [71], ASP187 [2], [70], ARG188 [70], GLN189 [2], [6], [70], [71], THR190 [2], [70], ALA191 [2], [70], GLN192 [2], [70].
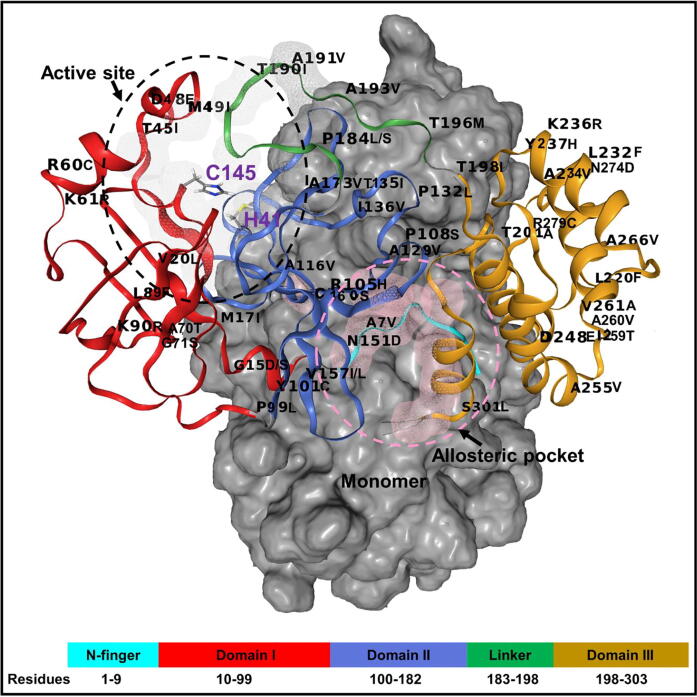
Fig. 1.
A structural representation of the homo-dimeric nature of SARS-CoV-2 Mpro. The structural domains (I-III) are shown in red, royal blue and orange cartoons, respectively. The N-finger region (residue 1–9) and the long loop connecting domain II to III (linker) are colored cyan and green, respectively. The substrate-binding pocket and allosteric pocket on a monomer are illustrated in grey and pink wireframe and dotted lines, respectively. The distribution of SARS-CoV-2 Mpro mutations identified from the GISAID database [72] is labeled on the structure. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
In our previous study, we identified dual allosteric pockets located at the interface of protomer A and B (Fig. 1), that concur with key residues for functional dimerization and enzymatic activities [53]. The residues of this allosteric pocket of SARS-CoV-2 Mpro are ALA116, TYR118, SER123, GLY124, SER139, and LEU141 on protomer A and residues LYS5, MET6, ALA7, PHE8, THR111, GLN127, PHE291, ASP295, ARG298, GLN299, GLY302, and VAL303 on protomer B; or vice versa. We also demonstrated that there is a correlation in compaction between the substrate binding site and the predicted allosteric sites, and that this correlation varied in the presence of some of the studied 50 mutations which spanned several secondary structures in Mpro domains as well as the N-finger and linker regions (Fig. 1).
Part I:
3.2. Identification of allosteric modulators against dimeric SARS-CoV-2 Mpro protein
From our calculations, SARS-CoV-2 and HCoV-OC43 Mpro share a sequence identity of 48.5%, and two structures have an RMSD value of 0.46 Å. We identified six compounds in SARS-CoV-2 and 15 compounds in HCoV-OC43 by blind docking and preliminary filtering of the 625 SANCDB compounds against the dimeric Mpro proteins (Fig. 2A). The high degree of search exhaustiveness increased the likelihood of finding certain binding poses more than once, despite having less favorable binding energy scores. This approach draws from the idea of the use of pose clustering in AutoDock [73], as we have noticed that non-equilibrium binding energy scores tend to be affected by the length of the ligand. The poses corresponding to either copy of the allosteric site were tallied for each compound to be compared across all hit compounds in both coronavirus strains. As seen in Fig. 2A, the lowest energy hits for the mirrored allosteric site occur in HCoV-OC43 but are not the most abundant hit compounds. Of notable interest are compounds SANC00209, SANC00210 and SANC00211, which are halogenated monoaromatic terpenoids produced from the marine alga Plocamium corallorhiza, with anti-proliferative properties. The four most abundant hits for the SARS-CoV-2 allosteric site (SANC00467, SANC00468, SANC00469 and SANC00630) occurred in both coronavirus strains, despite showing less favorable energy scores. While SANC00467, SANC00468 and SANC00469 all come from Drimia robusta [74], [75], SANC00630 is from Senecio oxyodontus [76]. All are monophenolic compounds. The binding of this allosteric site by various small compounds agrees with our previous hypothesis suggesting the pocket’s accessibility to such compounds [14]. Their aromaticity does not necessarily designate the exclusiveness of the pocket to such compounds, but is a result of the properties of the screening library obtained from SANCDB. Nevertheless, this indicates that the pocket is accessible to small aromatic moieties, which may be evaluated as scaffolds for designing small molecules targeting this site. Compounds SANC00302 and SANC00303 did not fare as well as the other compounds, both in terms of energy scoring and in the number of poses in SARS-CoV-2 Mpro; however, we carried them forward for MD analysis to cross-check their stability. The latter two compounds are halogenated indoles from Distaplia skoogi that have shown moderate cytotoxicity against cancerous cells [77]. Interestingly, from the literature, El-Baba et al., [12] also identified a compound (x1187), via mass spectroscopy based assay, binding to this region, slowing the rate of substrate processing of the enzyme. This compound has very low MSC Tanimoto similarity scores to our SANCDB compounds; ranging from 0.12 to 0.25 [78].
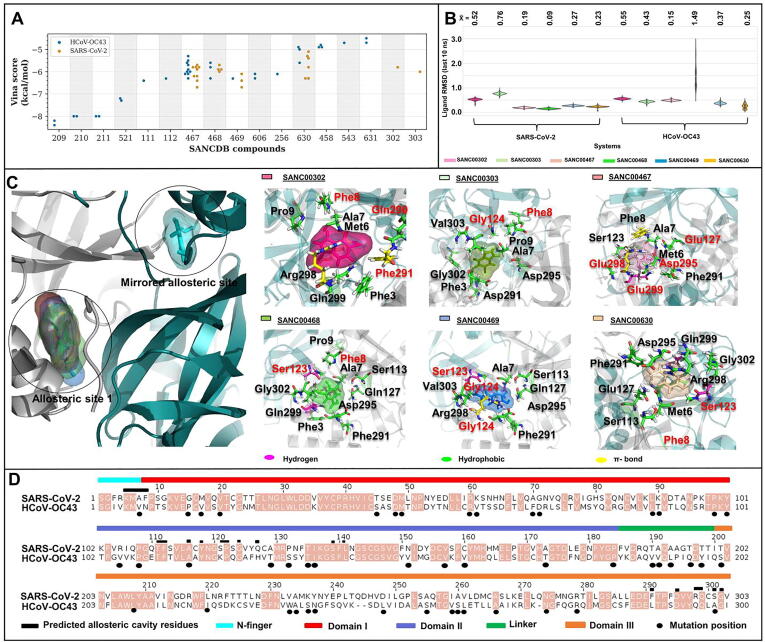
Fig. 2.
Ligand binding and characteristics of the predicted Mpro allosteric site in SARS-CoV-2 and HCoV-OC43. (A) Scatter plot of selected allosteric site ligands and their respective binding energies in SARS-CoV-2 (orange) and HCoV-OC43 (blue). (B) Kernel density plots of the ligand RMSD values for the last 10 ns of the 100 ns MD simulations. (C) Protein-ligand interactions for the six compounds in SARS-CoV-2. Residue contributions from protomer A and B are labelled in black and red respectively. (D) Sequence alignment of Mpro from the two strains, showing residue conservation (highlighted in light brown) with additional functional annotations. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Ligand RMSD graphs of the last 10 ns of the 100 ns MD simulations (Fig. 2B) showed that these six compounds behaved slightly differently in the Mpro protein of SARS-CoV-2 compared to that of HCoV-OC43. Overall, the RMSD distributions spanned a range of under 1 Å, with the exception of SANC00408 in HCoV-OC43, which produced a significantly wider range. Unimodal distributions indicated the presence of single dominant ligand conformations for each of the compounds, with mode shifts possibly linked to minor variations resulting from mutations and/or the part stochasticity of the in silico modeling algorithms. In HCoV-OC43, both SANC00468 and SANC00630 displayed multimodal distributions, suggesting their reduced stability, being clear in the case of SANC00468. Conversely, compounds SANC00467, SANC00468 and SANC00469 demonstrated the most stable conformations in the SARS-CoV-2 protein. Ligand RMSDs of the 100 ns simulations are presented in Fig. S1. The different behavior of the compounds can be attributed to the compound-protein residue interaction differences obtained from the docking stage (Fig. 2C, Table S2), as well as the residue differences between the two homologous proteins at the allosteric sites (Fig. 2D). Residues ALA7, PHE8, GLN127, PHE291 and ARG298 of SARS-CoV-2 Mpro are respectively replaced by VAL7, ASN8, HIS127, LEU291 and GLN298 in the Mpro in the HCoV-OC43 lab strain (Fig. 2D). In SARS-CoV-2, residues ALA7 and PHE8 form part of the N-finger - a region crucial for dimer stabilization [79]. GLN127, PHE291 and ARG298 have also been reported to play important roles in the dimerization and the enzymatic activity of SARS-CoV Mpro [80].
In SARS-CoV-2 Mpro, several ligand interactions (such as hydrogen bonds, hydrophobic and pi interactions) with allosteric site residues were observed (Table S2). Compounds SANC00467, SANC00468, SANC00469 and SANC00630 formed at least two hydrogen bonding interactions with some polar residue side chains (MET6, SER123, GLN299 and VAL303) that may affect ligand stabilization and retention within the pocket. The replacement of valine by a longer side chain in isoleucine at position 303 in HCoV-OC43, suggests that the site in HCoV-OC43 may not behave in the same way as that of SARS-CoV-2. At least seven hydrophobic interactions were observed across all modulators, indicating the enrichment of hydrophobic interactions at allosteric sites. The substitution of the non-polar PHE8 by the polar ASN8; the uncharged SER121 by the positively charged LYS121; and of the polar SER301 by the non-polar ALA301 in HCoV-OC43 may be responsible for the altered pocket topology and charges that together result in different ligand-binding patterns.
Our results indicate that the use of this strain for experimentation on allosteric modulation in SARS-CoV-2 Mpro may have some limitations.
Part II:
3.3. Identification of hub residues while considering symmetry in homodimers
Depending on the level of resolution desired for the analysis of homodimers, comparing MD-simulated pairs of a homodimeric protein can introduce conceptual challenges. For instance, one cannot easily know with certainty whether protomer A (or sections thereof) in one dimer behaves the same as its homologous position in protomer A in the second dimer. While a simpler protomer assignment approach based on permuted structural alignments was used in our earlier work [14] for single conformations, our attempt here investigates this issue in more depth, firstly by isolating potential hubs, and secondly by producing a representation of all the possible hub node combinations (Table 1) in order to obtain a scheme by which hub node importance can be assessed. While a hub is generally accepted as a high connectivity (degree) node, it has also been used to mean high BC [81], but can also be understood as any node that may cause non-negligible topological alterations to a network when removed [82]. In this analysis the term is used in its more general sense to mean any node that forms part of the set of highest centrality nodes, here arbitrarily specified as the top 5% centrality nodes measured across all related samples, independently measured for each of the averaged centrality metrics. This procedure differs from the identification of 1 to 2 standard deviations from the mean or top 5% residues in individual samples that we generally used in our previous studies [83], [84], [85], in that it considers the strongest actors from each protein sample and shows how other non-hub residues behave at homologous positions. We assume that investigating hub transitions in this manner is more likely to detect the most significant shifts in residue importance when exposed to a particular environment. We also used this approach to be able to handle the large amount of centrality data present in the current analysis.
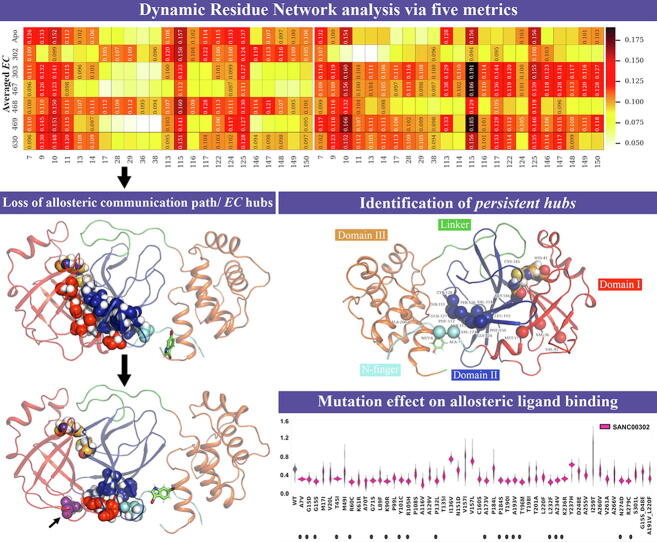
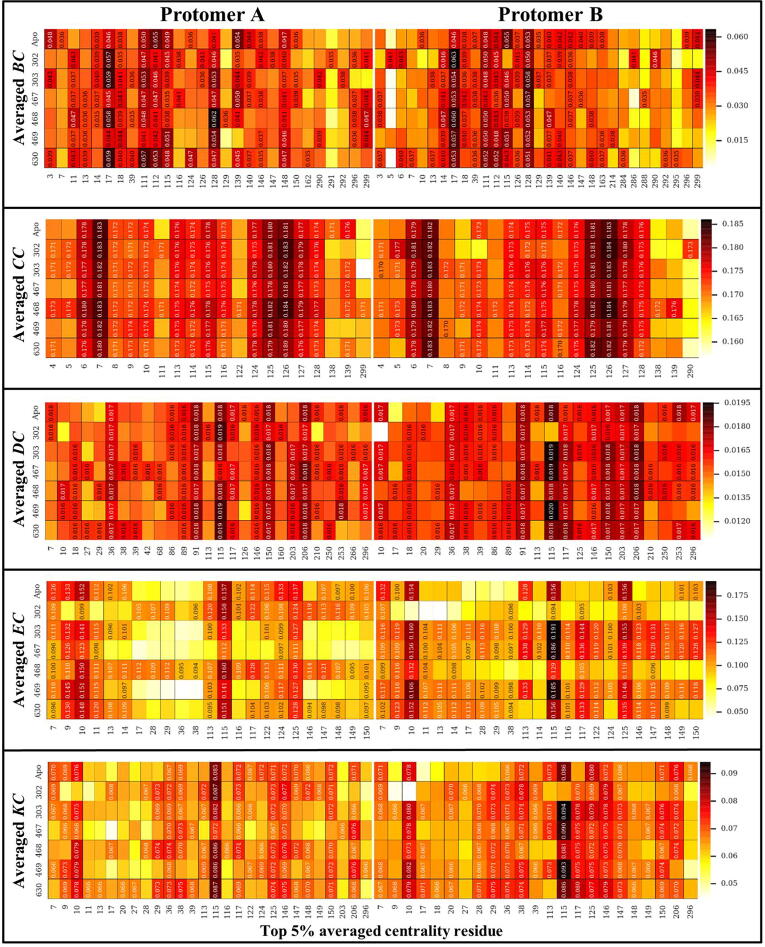
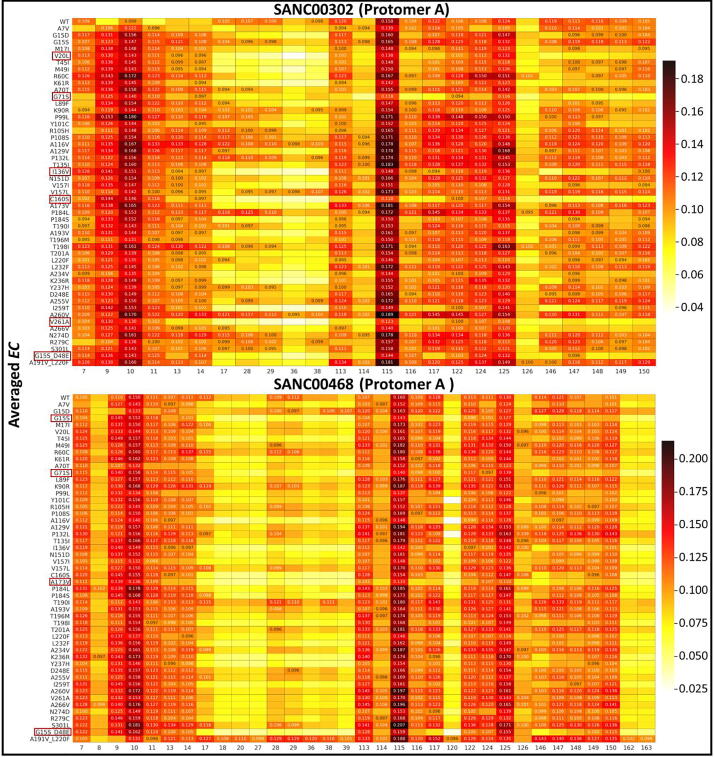
Fig. 3 shows the heat maps of the five DRN metrics for the reference protease in the absence and presence of ligand binding; with the designated ligand-bound allosteric pocket of the dimer always being referenced as protomer A. Specifically, the ligand was assigned to protomer A based on its proximity to a terminal alpha helix in the same chain. In the case of SANC00467, where the allosteric compound had bound protomer B, the protomer label was swapped.
Fig. 3.
Heat maps for the potential hubs according to the global top 5% for each of the five DRN metrics, for the reference protein in the apo and the six allosterically bound states. Detected hubs are annotated with their centrality values, while their homologous residues in alternate samples are not, but are only shown for the sake of comparison. For each metric, low to high centrality values are colored white, through yellow, orange and red to black. Measurements for the ligand-bound protomer (chain A) have been systematically presented on the left side, while those of the unbound protomer are on the right – this does not apply to the apo state. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
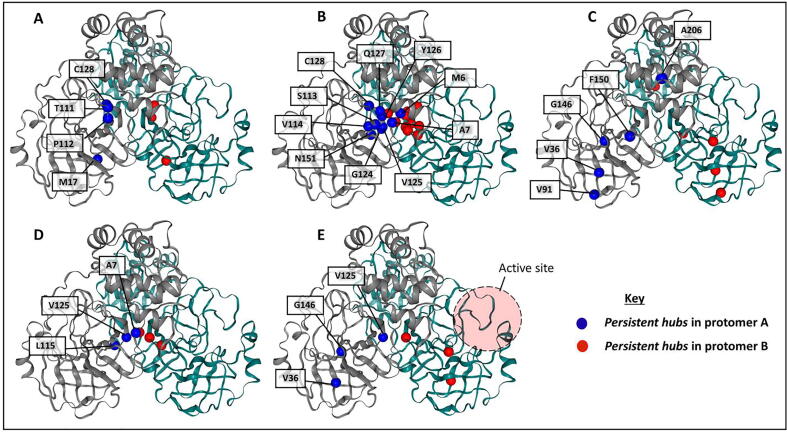
Preliminary examination of Fig. 3 showed that there are some residues that preserve their hub statuses. We, here, introduce the following terms: (1) Constitutive hub: If a hub is present in both protomers of the reference protein and remains as a hub irrespective of the apo or a ligand-bound state, it will be called a constitutive hub (see Table 1; score 4); (2) Persistent hub: If a hub remains across all systems compared, then the hub will be called persistent; in Part II, across all systems would be apo protomers and all ligand-bound dimers of reference protein, and in Part III it would be both protomers of the reference and mutant proteins with a specific ligand. (3) Super-persistent hub: In Part III, we will use the concept of a “Super-persistent hub”, meaning that the hub is persistent across all the ligands considered in both reference and mutant proteins. Most of the constitutive and persistent hubs are metric-specific giving a different perspective to the network. As the five averaged centrality metrics refer to different measures of importance within a network, these terms will be used with respect to a given centrality metric and will not be shared between them.
3.4. Metric based investigation of persistent hubs
3.4.1. Betweenness centrality
According to Fig. 3, MET17, THR111, PHE112 and CYS128 hub residues were found to be unaltered from the reference protein apo state, or upon any selected ligand binding irrespective of protomer for the averaged BC. At individual ligand level, each of these hubs is constitutive and indicates that there is no ligand effect due to preserved symmetry (Table 1, score 4).
For the entire system (apo + 6 ligand systems), these hubs are persistent hubs indicating that the allosteric modulators did not change the information path for these key residues; and any loss to these hubs may disrupt the communication.
Residues MET17 and CYS128 had been previously picked up from multiple simulations, but were not examined in depth in our previous work involving several Mpro mutants in the apo state [14]. The current analysis of the networks derived from the MD simulations further showed that all conserved averaged BC hubs occurred as intrachain or interchain hinges within the dimer (Fig. 4A), both in the absence and presence of different allosterically bound compounds. As averaged BC quantifies the extent by which a node positions itsef along the shortest communication path across all other node pairs over time, and because its hubs were enriched at different types of protein interface, such nodes were designated as hinges. Residue MET17 establishes intraprotomer contacts within the domain I/II interface by interacting with several residues of the beta hairpin. More specifically, it forms alkyl interactions with LEU115, PRO122 and CYS117. Of notable interest is the alpha helix that supports the N-finger. Being also part of the high BC hubs, it is possible that LEU115 and MET17 form an important bridge that relays interdomain information, potentially influencing N-finger stability, and by extension impacting the activity of the alternate protomer. THR111 (from domain II) also plays a role in maintaining intraprotomeric interdomain stability by forming periodic H-bonds with ASP295 (from domain III), and at the same time mediating information flow. THR111 is also firmly bound to the other hub residue CYS128 via multiple hydrogen bonds and carbon H-bonding. CYS128 is firmly seated on a beta-strand, forming non-bonded interactions with TYR126, VAL114 and PHE112.
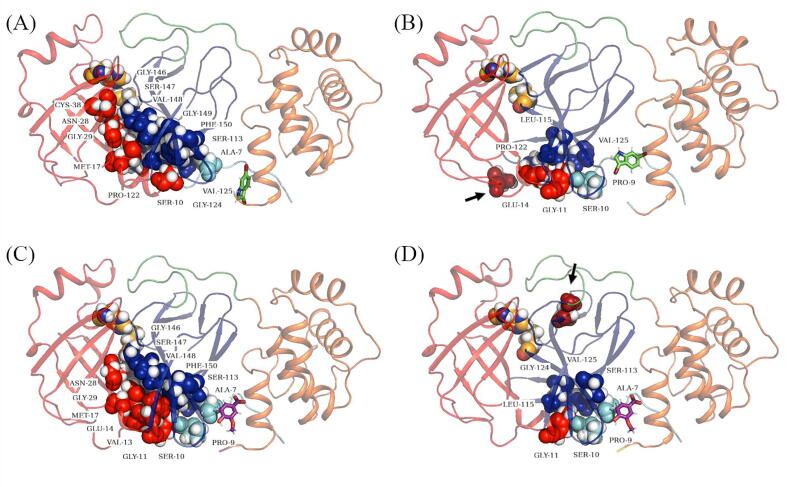
Fig. 4.
Cartoon representation of SARS-CoV-2 Mpro dimeric structure with the distribution of the persistent hubs as per five metrics of DRNs. (A) Averaged BC, (B) Averaged CC, (C) Averaged DC, (D) Averaged EC, (E) Averaged KC. Protomers A and B are shown as cartoon in teal and grey respectively. Protomer A persistent hubs are depicted in red spheres and protomer B ones are in blue. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.4.2. Closeness centrality
CC is calculated as the inverse of the average of the shortest path length from the node to every other node; hence identifies the central nodes which are closer to most of the nodes. Previously we showed that residues with low averaged shortest paths are correlated with the low mobility (increased rigidity) of the protein [59]. Thus, high CC values are most likely to occur within the protein core. Previously, CC metric calculations over single static structures were used to identify active site residues with the support of other approaches, e.g. conservation, solvent accessibility [86], [87] to distinguish them from residues located in the core. Here, our persistent averaged CC hubs are MET6, ALA7, SER113, VAL114, LEU115, GLY124, VAL125, TYR126, GLN127 and CYS128 (Fig. 3). Visual inspection of the residue mappings showed that they are all located at the vicinity of the dimeric center of mass found within the very stable domain II (Fig. 4B), as reported in our earlier work [14]. In the same work, ALA7 (part of the N-finger) was reported to be very rigid, and probably is the reason for the similar behavior in its immediate neighbor residue 6 within the same chain. SER113, VAL114, LEU115 and TYR126 are juxtaposed within the same beta sheet, supported by networks of H-bonding interactions, and are next to ALA7, which forms intraprotomeric alkyl interactions with VAL125 and interprotomeric H-bonds with VAL125 from the alternate protomer. More generally, these residues are mainly in direct contact with the center of the opposing protomer, and the reason for their high averaged closeness may be related to the way in which the protomers were reported to slide over each other, remaining at the same pivot point, centered at domain II (with the N-finger ALA7 also sandwiched in-between). Residue 7 appears crucial for maintaining the bulk of the averaged CC hubs.
3.4.3. Degree centrality
DC defines the number of neighboring nodes around a given node, hence provides information on the local connectivity, but not on how central it is in the entire network. Persistent averaged DC hubs in Mpro are comprised of residues VAL36, VAL91, GLY146, PHE150 and ALA206 (Fig. 3). 3D visualization of this metric showed that averaged DC tends to be concentrated at the confluence of secondary structural elements, irrespective of inter- or intraprotomeric locations (Fig. 4C). VAL36 occurs within a beta sheet and establishes several types of non-bonded interactions with multiple residues within domain I, namely LYS88, LEU89, VAL68 and VAL18. Residue 91 occurs on another strand of the same beta sheet, next to VAL36, and establishes several types of non-bonded interactions with residues ASP34, LEU75, ARG76 and VAL77. Residue GLY146 is another persistent DC hub of potential significance, found next to the catalytic residue CYS145. It was found that GLY146 established durable intraprotomeric contacts with 10 residues, namely LEU27, ASN28, GLY29, CYS38, PRO39, CYS145, SER147, VAL148, MET162 and HIS163. This involved both intradomain (domain I) and interdomain interactions, and occurred in each protomer and both in the presence and absence of allosteric binding. The high averaged DC of GLY146 may be related to the fact that this area has to be kept relatively stable for the proper positioning of the catalytic residues CYS145 and HIS41, from domains II and I respectively. Given the presence of such a residue at the interface of domains I and II, this suggests that it may have a high BC as well, which is generally observed in both protomers A and B (Fig. 3). PHE150 similarly interacted with several residues in each protomer across all samples, and we observed high contact frequencies for 10 residues, namely VAL13, PHE112, SER113, VAL148, GLY149, ASN151, VAL157, SER158, PHE159 and CYS160, once more involving residues from domains I and II. ALA206 was similarly surrounded by 10 durable intraprotomeric contacts within domain III, being composed of residues VAL202, ASN203, LEU205, TRP207, TYR209, ALA210, PHE291, THR292, PRO293 and VAL296, in all cases. More generally, the shared high DC hubs seem to occur in each domain of the protein, probably due to their independent roles in maintaining the organization and integrity of the individual domains.
3.4.4. Eigencentrality
EC measures both the number of connections of a given node and its relevance in terms of information flow. It is based on a recursive allocation of centrality on the basis of nodes that draw importance from that of their successive connections, given that initial centrality is based on DC. Based on this calculation, one would expect high EC values to also have high DC values, or be in spatial proximity to high connectivity residues. However, we found that many of the high DC residues did not show up among the EC hubs, suggesting that EC is mostly gained via proximities to high DC residues, and do not necessarily have high connectivities themselves.
Persistent hubs of averaged EC for the Mpro reference protein comprised residues ALA7, LEU115 and VAL125 (Fig. 3). LEU115 is the only residue that maintained its importance according to averaged DC and EC measurements. Weighted residue contact analysis of this residue showed that LEU115 maintained high contact frequencies (>0.60) with residues CYS117, PRO122, VAL125 and SER147, irrespective of ligand binding. SER10 and VAL13 also showed high frequencies, except in the presence of SANC00302 where notable contact asymmetry was experienced; a similar pattern was observed for residues VAL148 and GLY149 in the presence of SANC00467. 3D visualization of the EC residues shows that it is concentrated around the interface of protomers A and B (Fig. 4D). The main message here is that high DC residues are sharing centrality to their immediate neighbors, and that the vicinity of the dimer interface seems to be the most residue-crowded area within the dimer. One should also bear in mind that centrality may also be coming from further degrees of separation. Other residues picked up as hubs in DC may be surrounded by fewer residues of high centrality, thus giving them less importance.
3.4.5. Katz centrality
KC measures the relative degree of influence of a residue i within connected residues in a network. Irrespective of chain and ligand binding, nodes VAL36, VAL125 and GLY146 remained as hub nodes according to the averaged KC metric (Fig. 3).
Visualization of the averaged KC metric (Fig. 4E) showed that it is an intermediate between averaged EC and averaged DC, with the former being more conservative than the latter when assigning relative node importance. Persistent averaged KC hub 125 was also central according to averaged EC; and VAL36, GLY146 were also persistent hubs according to averaged DC. The default attenuation coefficient (alpha = 0.1) appears to minimize the effect of more distant nodes in the network, such that it assigns centrality patterns intermediate to DC and EC.
In order to give an estimate of the hub similarities between those of KC and those from DC and EC, the Jaccard similarity coefficient (J) of hubs from protomer permutations was calculated, using as a rough estimate from the union of hubs across all states (ligand-bound and unbound) of the reference Mpro, for each of the protomers. The similarities were evaluated likewise: [J(KCA, KC’A), J(KCA, KC’B), J(KCB, KC’B) and J(KCB, KC’A)], where the subscript denotes protomer label and KC’ denotes the complement of KC, in this case DC or EC. The hub similarities J(KC, DC) had a range of [0.375, 0.53] and those from J(KC, EC) had a range of [0.53, 0.76]. The observed ranges suggest that KC is more similar to EC than to DC. For comparison, J(KC, BC) and J(KC, CC) had ranges of [0.2, 0.3] and [0.16, 0.23], respectively denoting they tended not to share many hubs.
The reasons for the high centrality values for residues SER10, LEU115 and VAL125 are as explained in averaged EC, with the main difference being that the effect of distal nodes was reduced due to the dampening coefficient. In this manner, averaged KC appears to improve the resolving power of averaged EC.
Overall, the heat map representation of the identified hubs according to the global top 5% for each of the five DRN metrics (Fig. 3) allowed us to identify persistent hubs according to each of the centrality measurements, suggesting that they are exposing different key aspects of mechanical signal transduction within the protease regardless of apo and allosteric ligand bound forms of both monomers (Fig. 4A-E). Collectively, these persistent hubs are spreading out from the allosteric site along the protein interface as well as along the antiparallel beta strands (Fig. 5). Even though previously it is not reported, we believe that the antiparallel beta strands, and especially the first two nearest to the dimer interface, are functionally highly important.
Fig. 5.
Cartoon representation of SARS-CoV-2 Mpro protomer A with the collective presentation of persistent hubs in spheres colored according to their domains. Catalytic residues are HIS41 and CYS145 are also depicted as spheres.
We also identified a number of changes to hub existence in the presence of potential allosteric modulators and these changes were investigated in the next section.
3.5. Establishing subnetworks for further investigation of hub changes upon allosteric binding within the reference homodimer
We also observed another layer of information within the homodimer, which exists due to the symmetry of the protomers, despite the adjustment made to present the ligand-bound protomer as the one left-hand side (protomer A) in Fig. 3. We hypothesize that it is possible for a homodimer to switch states, because of their sequence identity. This is likely true for the apo state, but may also apply to the asymmetrically occupied allosteric sites, depending on how effective the allosteric pocket occupation is. This approach may also reveal if allosteric activity is manifested as a change of hub symmetry in the protein dynamics - for instance one or more hubs consistently appearing in one or even both of the protomers, when the allosteric site is occupied by a ligand. For this reason, the same data set (Fig. 3) is further analyzed using another concept that we demonstrate in this section.
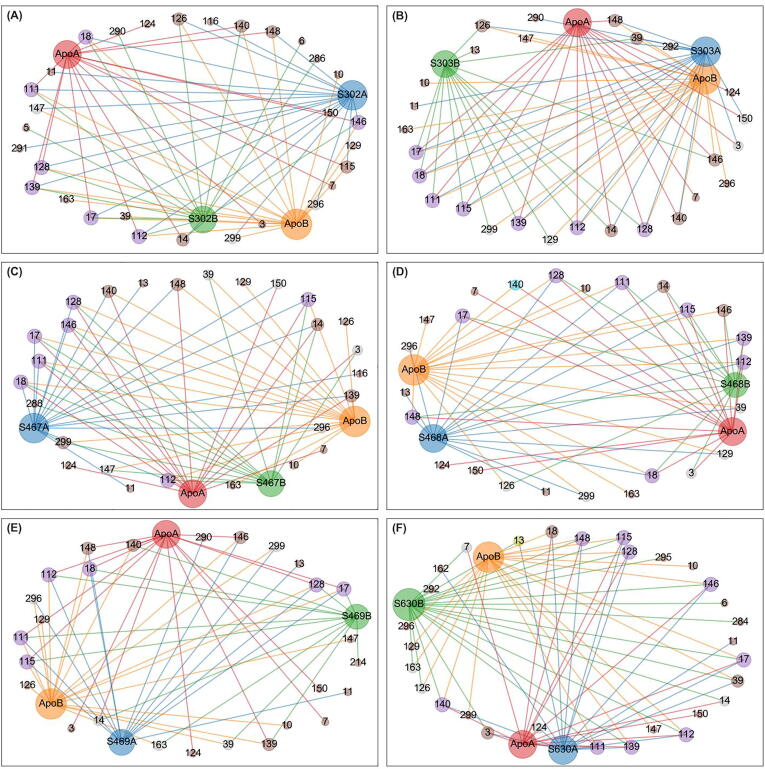
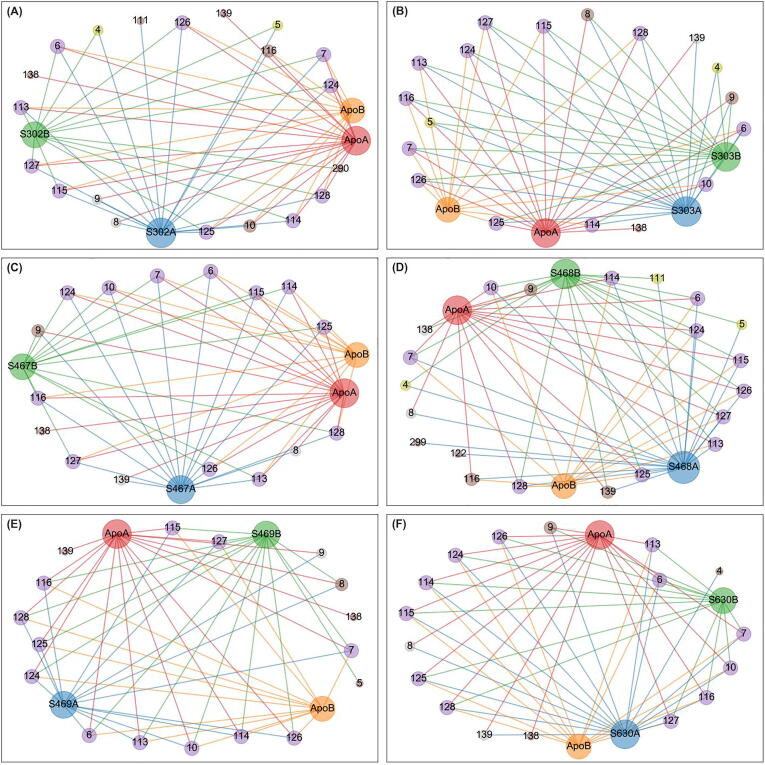
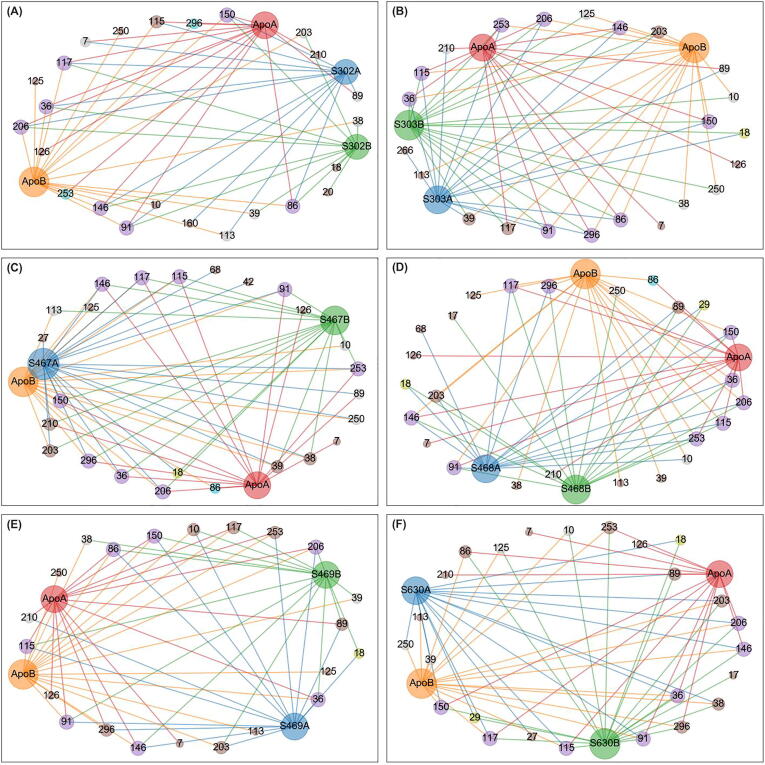
For each allosterically bound ligand, a network was built using the detected centrality hubs as nodes, and the chains to which they belong. A subnetwork was then prepared by combining the edges from the apo protomers A and B, and those from the ligand-exposed dimer, while also tracking the protomer labels, given the ligands had settled at one chain of the blindly docked dimer (Fig. 6, Fig. 7, Fig. 8, Fig. 9, Fig. 10). As indicated before the ligand was assigned to protomer A. In the case of SANC00467 where the compound bound to chain B, the chain label was swapped. The systematic hub representation was done to further investigate whether ligand binding had an effect, keeping a record of the chain labels, as we hypothesized that a ligand's presence in the allosteric site may manifest its effects not only in the bound protomer, but also in the unbound protomer, within the same dimer.
Fig. 6.
Averaged betweenness centrality hubs represented as a networks for both the apo state and the six allosterically-bound complexes. Each of the sub-figures represents the subnetwork obtained for each of the allosterically bound ligands, namely (A) SANC00302, (B) SANC00303, (C) SANC00467, (D) SANC00468, (E) SANC00469 and (F) SANC00630, when merged with the apo protein, in each case. Red, orange, blue and green nodes (and edges) depict the protomers (apo chains A and B, and complex chains A and B, respectively) to which a hub belongs. Each node is also scaled by its score – i.e. the number of edges it holds. Hubs that are present in all 4 protomers are in purple. Score 2 loss and gains from the reference are colored yellow and cyan, respectively. Score 1 losses and gains are colored brown. Inconclusive hubs are in grey. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 7.
Averaged closeness centrality hubs represented as a networks for both the apo state and the six allosterically-bound complexes for the reference dimer. Each of the sub-figures represents the subnetwork obtained for each of the allosterically bound ligands, namely (A) SANC00302, (B) SANC00303, (C) SANC00467, (D) SANC00468, (E) SANC00469 and (F) SANC00630, when merged with the apo protein, in each case. Red, orange, blue and green nodes (and edges) depict the protomers (apo chains A and B, and complex chains A and B, respectively) to which a hub belongs. Each node is also sized by its score – i.e. the number of edges it holds. Hubs that are present in all 4 protomers are in purple. Score 2 loss and gains from the reference are colored yellow and cyan, respectively. Score 1 losses and gains are colored brown. Inconclusive hubs are in grey. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 8.
Averaged degree centrality hubs represented as a networks for both the apo state and the six allosterically-bound complexes. Each of the sub-figures represents the subnetwork obtained for each of the allosterically bound ligands, namely (A) SANC00302, (B) SANC00303, (C) SANC00467, (D) SANC00468, (E) SANC00469 and (F) SANC00630, when merged with the apo protein, in each case. Red, orange, blue and green nodes (and edges) depict the protomers (apo chains A and B, and complex chains A and B, respectively) to which a hub belongs. Each node is also sized by its score – i.e. the number of edges it holds. Hubs that are present in all 4 protomers are in purple. Score 2 loss and gains from the reference are colored yellow and cyan, respectively. Score 1 losses and gains are colored brown. Inconclusive hubs are in grey. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 9.
Averaged eigencentrality hubs represented as a networks for both the apo state and the six allosterically-bound complexes. Each of the sub-figures represents the subnetwork obtained for each of the allosterically bound ligands, namely (A) SANC00302, (B) SANC00303, (C) SANC00467, (D) SANC00468, (E) SANC00469 and (F) SANC00630, when merged with the apo protein, in each case. Red, orange, blue and green nodes (and edges) depict the protomers (apo chains A and B, and complex chains A and B, respectively) to which a hub belongs. Each node is also sized by its score – i.e. the number of edges it holds. Hubs that are present in all 4 protomers are in purple. Score 2 loss and gains from the reference are colored yellow and cyan, respectively. Score 1 losses and gains are colored brown. Inconclusive hubs are in grey. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 10.
The path traced by averaged EC hubs, starting from the allosteric ligand towards the catalytic residue. The protease is depicted by a cartoon representation onto which the averaged EC hub residues are overlaid as sphere representations, together with the non-hub catalytic residues HIS41 and CYS145 (circled in orange). EC persistent hub residues are circled in black; the alternate path is circled in blue; and the one triggered by the binding of all ligands is circled in green. One of the compounds is also shown in stick figure representation, as an example. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The hub data set was analyzed based on the logic described in Table 1. To simplify the terminology used to describe the presence of hubs within any combination of protomers, the term “score” is used to specify the number of protomers where the hub is present. In other words, if a hub is present in both protomers of the apo protein, the hub (irrespective of the DRN metric) will have a score of 2. Similarly, if a hub is present in each protomer of the apo dimer, and in each protomer of the ligand-bound dimer, this hub will have a score of 4 (constitutive hub). Further, higher confidence was assumed on the basis of complete loss or complete gain of any hub in each constitutive protomer from either the apo protein or the ligand-exposed enzyme. Lower confidence was assumed when a single hub was gained (i.e. score 0 to 1, or 1 to 2) or lost (i.e. score 1 to 0, or 2 to 1) within a single protomer, out of all four protomers (i.e. the set of protomers: apo chains A/B and complex chains A/B), to account for the part stochasticity of MD simulations. Higher confidence was given to these weaker signals when they were conserved across several ligand-bound states. Cases where asymmetric hub distributions occurred (i.e. a hub was found in only one protomer from each of the apo and the ligand-bound dimers) were ambiguous, given the fact that the apo dimer already expressed both hub states.
Here we will mainly focus on cases where we observed score of 2 and score of 1 gains or losses from each ligand-bound dimer with respect to the apo protein, in order to extract high likelihood ligand-induced changes.
3.5.1. Betweenness centrality
While PHE140 was a constitutive hub in the presence of SANC00630, SANC00468 completely lost node 140 upon ligand binding (score of 2 loss). SANC00630 gained hub 13 (score of 2 gain) with respect to apo structure (Fig. 6). The loss of BC from this “chameleon” switch residue (PHE140) suggests contact loss in its vicinity. VAL13, on the other hand, is found close to the N-finger – in a region where we previously reported lengthening and shortening of the alpha helix and suggested its possible involvement in dimer stability [14].
While there are many score 1 hub gains and losses with each ligand-bound dimer (with respect to the dimeric apo protein), we report the ones which have the highest conservation among all these lower confidence cases, independent of the bound ligand. Hub residue GLY11 systematically changed from a score of 0 in the apo dimer to a score of 1 in the dimeric complexes, suggesting an increased use or stabilization around this residue upon ligand binding. Coincidentally, hub residue SER10 systematically transited from a score of 1 in the apo state to a score of 0 upon ligand binding. The fact that these two residues are next to each other, and in fact interact with their interprotomeric counterpart suggests a possible rerouting of information flow in their vicinity upon introduction of a ligand. Score 2 to 1 (i.e. from the apo to the ligand-bound dimer) changes appeared not as consistent, but showed some agreement on hub nodes being lost from one of the “chameleon switches” in subsite S1, similar to what was seen more strongly in the presence of SANC00468, where both nodes were lost. This was observed for residues SER139 and/or PHE140 when exposed to SANC00302, SANC00303, SANC00467 and SANC00469. Score 1 to 0 changes were not observed when shifting the reference protein from an apo to a ligand-bound state, for any of the centrality metrics.
3.5.2. Closeness centrality
SER10 was a constitutive hub to five ligand-bound states, except SANC00302. Score 2 gains of high CC hubs were observed for residue 4 and 5 in the presence of SANC00302, SANC00303 and SANC00468 – residue 4 also experienced a score 1 gain in the presence of SANC00630, while residue 5 experienced a similar gain in the presence of SANC00469 (Fig. 7). THR111 was also gained as a score of 2 hub, only in the presence of SANC00468. GLY138, which is part of the S1 subsite, manifested itself as a hub in only one monomer of the apo protein, transitioning to a score of 0 upon ligand binding in five out of the six bound states. Upon visual inspection, we find that this residue is next to residue ARG4 on the alternate protomer, even though they do not appear to interact via non-bonded interactions. By measuring the change in their C-alpha interprotomer distances [i.e. the distance between residue pair (4A, 138B) minus that between residues (4B, 138A)] in each of the apo and the ligand-bound Mpro showed us that one of the residue pairs from the apo form had a visibly larger variance in equilibrium distance compared to those all the ligand-bound proteins, as it had an interprotomer distance interquartile range (IQR) of 0.12 nm, compared to a maximum IQR of 0.07 nm across all six the ligand-bound states. The maximum upper quartile additionally informs us of the higher extent of dimeric asymmetry for the residue pair for the apo protein (0.22 nm), compared to that observed in the ligand-bound proteins, which displayed a maximum value of 0.13 nm overall. While counterintuitive, it would seem that asymmetry favors a higher centrality at one of the GLY138-ARG4 (the N-finger from one protomer and domain II from the other) interfaces at the expense of the other in the apo state, while ligand occupation of only one of the detected allosteric sites, has a general stabilizing effect, which dilutes the centrality more evenly.
3.5.3. Degree centrality
LEU115 was a constitutive hub to five ligand-bound states, except for SANC00302 (Fig. 8). Scaffold-related conservation patterns were not apparent using this metric, however some differences did occur. Residue LEU115, which occurs in proximity to the persistent hub PHE150, was highly crowded and formed several durable contacts with its neighbors, namely VAL114, ALA116, CYS117, PRO122 and VAL125. LEU115 had a high frequency contact with PRO9 in only one chain in the presence of SANC00469 and a low frequency contact with VAL13 in only one chain in the presence of SANC00302.
A score 2 hub gain was experienced by VAL18 when exposed to SANC00303, SANC00467, SANC00468, SANC00469 and SANC00630. The same residue incurred a score 1 gain in the presence of SANC00302. Upon contact visualization, we found the systematic significant increase in contact frequency between VAL18 and GLN69 in each protomer upon ligand binding. While their C-alpha distances were relatively similar throughout the apo and ligand-bound Mpro (averaging 0.59 nm), the C-beta distances were significantly larger in the apo (average of 0.69 and 0.70 nm in the apo protomers) compared to those of the ligand-bound states (averages ranging from 0.63 to 0.65 nm), which suggests a rotational decrease of the C-beta distance upon ligand binding. A score 2 gain was also experienced by residue GLY29 when exposed to SANC00468 and SANC00630. 3D visualization shows that GLY29 is H-bonded to VAL18, and together with GLN69 they form a geodesic path travelling directly across antiparallel beta strands. The proximity and arrangement of these three residues may suggest they may act in a concerted manner.
Score 2 losses were observed for VAL86 when exposed to SANC00467 and SANC00468; and for residues LEU253 and VAL296 in the presence of SANC00302, indicating that the connectivity around these areas was reduced. Conserved score 1 to 0 changes were observed for residue TYR126 in the presence of ligand binding, suggesting a possible increase in local compaction in that area in the presence of any of the ligands.
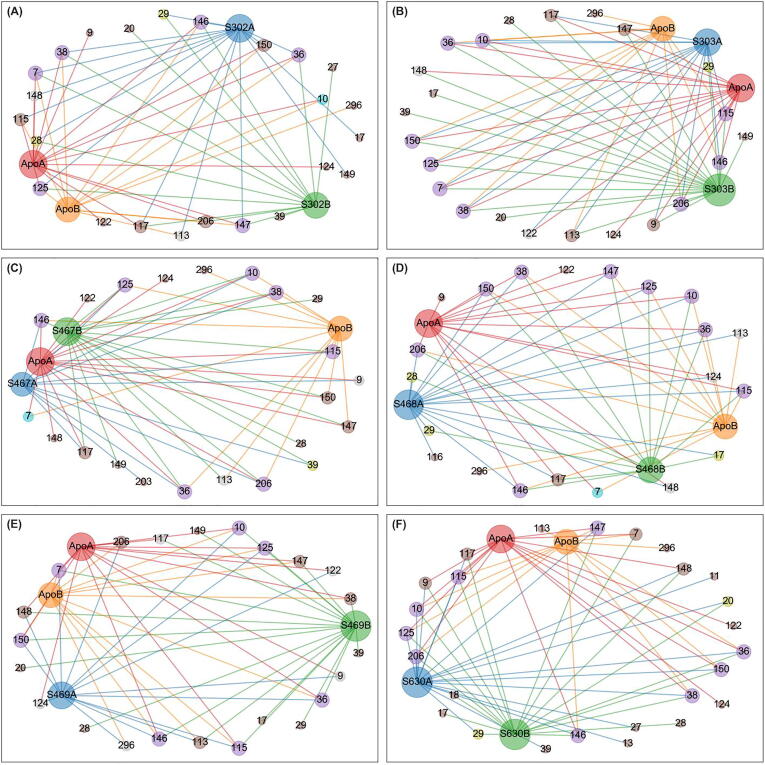
3.5.4. Eigencentrality
PRO9 and SER10 were constitutive hubs to five ligand-bound states, except for SANC00302 (Fig. 9). GLY146 experienced a score 2 gain in the presence of SANC00302 and SANC00630. The same was observed for CYS38 in the presence of SANC00302. While score 1 gains from 1 to 2 were not completely conserved, hub score changes from 0 to 1 were conserved, comprising residues MET17, ASN28 and GLY29 in the presence of ligand binding, suggesting an increase in centrality in the vicinity of these residues. Visual inspection shows that MET17 is proximal to ASN28, which is next to GLY29 on a beta strand. The high averaged EC for MET17 is likely due to its high degree centrality combined to that of VAL18. It is possible that ligand binding further stabilizes its residue neighborhood, compared to the absence of occupation of the allosteric pocket. Hub residues ASN28 and GLY29 appear to draw centrality from the higher degree centrality residues VAL36 and VAL18. Together these domains I residues line the interface of domain II, indicating a possible stabilization around this area in the presence of an occupied allosteric pocket.
A very interesting communication path emerges when combining the persistent averaged EC hubs (ALA7, LEU115 and VAL125) and ones collectively gained by ligand binding (MET17, ASN28 and GLY29) (Fig. 10). We thus describe the path MET7-VAL125-LEU115-MET17-GLY29-ASN28-HIS145, which originates from the N-finger to converge towards the catalytic HIS145, which is itself proximal to second catalytic residue CYS41. From these observations, it is possible that the intradomain MET17-LEU115 contact may be a crucial information path for the SARS-CoV-2 Mpro, as it plays a pivotal role in relaying information from the allosteric pocket. This complements our previous observation of the bridging function of SER17 in the apo Mpro using averaged BC. Extending on the finding of a common path shared by ligand binding, we also describe an alternate path SER9-PRO10-LEU115-MET17-GLY29-ASN28-HIS145 being specifically used in the apo state and the ligand-bound states, with the exception of SANC00302. It is possible that this difference stems from the lack of stability of this compound in the pocket. The complete communication paths are further analyzed in Part III.
A score 2 loss was observed by GLY149 in the presence of SANC00630. The same residue experienced a score 2 to 1 change in the presence of the other compounds. This residue is found at the bifurcation of two beta hairpins that do not completely line up all the way into a beta sheet, close to a main contributor of degree centrality, residue LEU115. The generic decrease in averaged EC in at least one protomer points to a possible loss of contact in this hub's vicinity that occurs upon ligand exposure. Residue 9 was also lost as a score 2 hub, only in the presence of SANC00302. Score 1 to 0 changes were inconsistent.
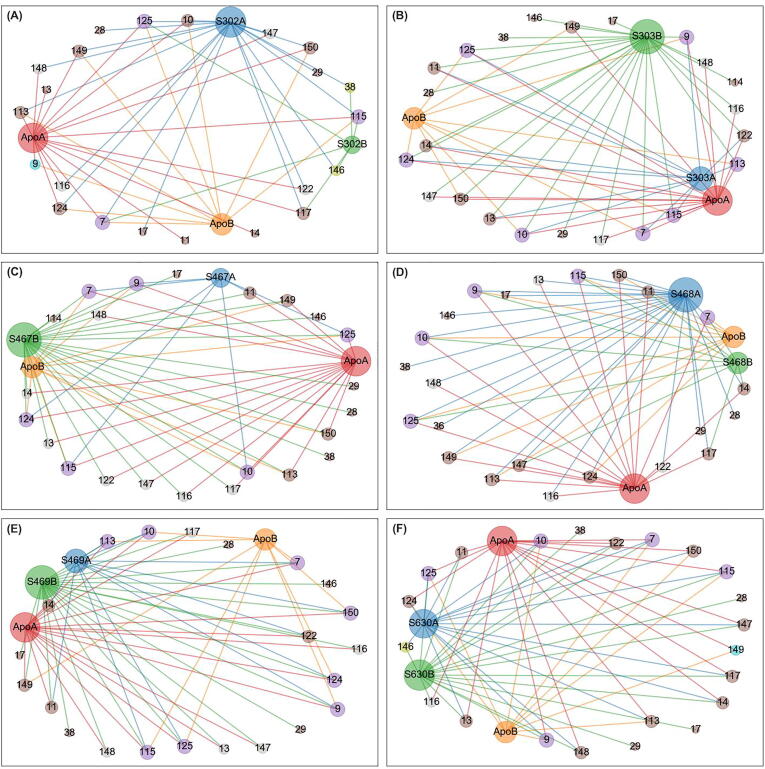
3.5.5. Katz centrality
SER10 and LEU115 were constitutive hubs to five ligand-bound states, except for SANC00302 (Fig. 11). SER10 was also a constitutive hub and LEU115 was a persistent hub in EC. KC hubs residues VAL36, GLY146 and LEU115 were also central according to averaged DC.
Fig. 11.
Averaged Katz centrality hubs represented as a networks for both the apo state and the six allosterically-bound complexes. Each of the sub-figures represents the subnetwork obtained for each of the allosterically bound ligands, namely (A) SANC00302, (B) SANC00303, (C) SANC00467, (D) SANC00468, (E) SANC00469 and (F) SANC00630, when merged with the apo protein, in each case. Red, orange, blue and green nodes (and edges) depict the protomers (apo chains A and B, and complex chains A and B, respectively) to which a hub belongs. Each node is also sized by its score – i.e. the number of edges it holds. Hubs that are present in all 4 protomers are in purple. Score 2 loss and gains from the reference are colored yellow and cyan, respectively. Score 1 losses and gains are colored brown. Inconclusive hubs are in grey. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Hub node 29 gained a score of 2 in the presence of SANC00302, SANC00303, SANC00468 and SANC00630, with respect to the apo protein, and the same residue incurred a score 0 to 1 gain in the presence of SANC00467 and SANC00469, indicating that this residue gains in KC in at least one protomer upon ligand exposure. Score 2 gains are also observed for residues 28 (in the presence of SANC00302 and SANC00468), 39 (in the presence of SANC00467), 17 (in the presence of SANC00468) and 20 (in the presence of SANC00630). Score 2 losses comprised residue 7 in the presence of SANC00467 and SANC00468; and residue 10 in the presence of SANC00302. The similarities in hub response patterns in the presence of SANC00467 and SANC00468 may suggest that these changes may be related to their common scaffold, or to a similar conformational sampling.
Collectively, the analysis of hub transitions induced by allosteric binding in the homodimer via subnetworks showed that the DRN metrics shared similarities, but also emphasized their importance in different ways. By focusing on sample-wide centrality at the expense of protein-specific importance, and possibly narrowing the information content, we extracted key features in the modulation of the mirrored allosteric pocket of Mpro. Changes in averaged BC hub patterns hinted at a possible rerouting of information flow from residue 10 to 11, induced by ligand stabilization. CC hub transitions lead to the observation of a ligand-induced stabilization, verified by the decreased interprotomer asymmetry of the distances between residues 4 and 138, compared to the apo protein. DC hubs detected a consistent decrease in compaction around TYR126, in the presence of any ligand; and further analysis showed an associated intraprotomeric side-chain rotation involving residues VAL18 and GLN89, upon ligand binding. The aggregation of persistent and gained averaged EC hubs revealed a common path connecting the allosteric occupation by ligands, to the active site, involving the interaction between MET17 and LEU115 (Fig. 10). It is possible that due to its proximity to one of the EC hubs (VAL125) that TYR126 may have a role in the path traced via averaged EC. Averaged KC mainly showed similarities with averaged EC and DC.
PART III:
3.6. The stability of allosteric modulators in the presence of evolutionary mutations
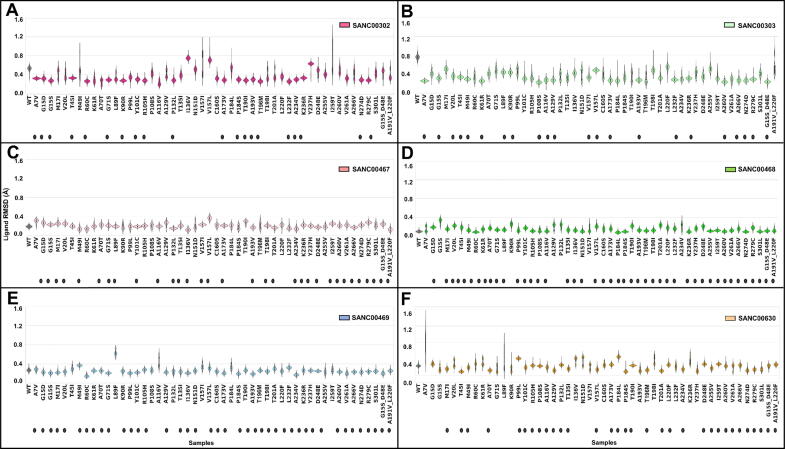
Coronaviruses, including SARS-COV-2, depend on RNA-dependent RNA polymerase (RdRp) for RNA synthesis [8], [88]. Due to the error-prone nature of RdRp, they can accumulate high rates of mutations, some of which may alter their virulence and antigenicity. As there is no FDA-approved drug for COVID-19 at the time of writing, we do not know which of these mutations could affect drug efficacy, or cause drug resistance. Hence it is important to understand the potential effects of a range of mutations in hit identification studies. To date, only a few studies have considered the impact of SARS-COV-2 Mpro variations in apo protein [4], [14], [89]. However, to our knowledge, there is no systematic research incorporating hit identification with the analysis of structural and functional effects of variations. Here, we further analyzed the behavior of six potential allosteric modulators that we identified in the reference protein in the presence of early evolutionary mutations. In order to quickly examine the stability of ligands in a total of 300 mutant systems (6 ligands × 50 mutant proteins), the ligand poses via ligand RMSDs were calculated, from which ligand kernel plots were produced from the last 10 ns of both 20 ns simulations of the mutant-ligand complexes, and 100 ns MD simulations of reference protein–ligand complexes (Fig. 12, Fig. S1). Overall, all ligands were well anchored in the allosteric pocket of mutant proteins, as seen from the ligand RMSD median values below 2.0 Å. Regarding the variations in ligand motion, a more stable conformation (unimodal distribution) was observed across compounds SANC00468, SANC00467 and SANC00469 when bound to mutant proteins, followed by SANC00630, as compared to the conformational stability of SANC00302 and SANC00303 (Fig. 12). This observation was in agreement with docking results where the first four compounds exhibited high stability through various hydrogen bond interactions with key allosteric site residues (Fig. 2C, Table S2). A closer view of each ligand revealed the subtle movement of the bromide group from SANC00302 and SANC00303, and the hydroxyl group of SANC00630 in some mutant proteins as seen from the bimodal distributions (Fig. 12).
Fig. 12.
Kernel density distribution plot of ligand RMSD values in ligand-bound wildtype (WT) and mutant systems extracted from the last 10 ns simulation. Each panel A to F is for the ligand indicated in the figure legend.
Fig. 12 was further evaluated to calculate a consensus score across six ligands within each mutant system. For that, a table (Table S3) was prepared in which the y-axis contains individual mutant proteins and the x-axis for the six ligands. For each ligand, kernel plots were checked and the ligands with a unimodal distribution in each mutant protein system received a tick (✓) in the table; the selected ones are also indicated in Fig. 12 with black oval shape along the x-axis. Surprisingly, out of 50 mutant proteins, only three of them (A173V, N274D and R279C) received a consensus score of six (Table S3), meaning all ligands in these mutant proteins stayed stable over the MD simulation. Over all the systems, the best performing ligands were SANC00468 and SANC00469, which gave highly stable motions for 43 and 41 mutant samples, respectively (Table S3).
The concept of allosteric effects of mutations and their role in the modulation of protein activity was previously discussed in literature [29], [90]. Our results, here, demonstrated the importance of incorporating mutation information in hit identification, as mutations might have distal-allosteric effects (allosteric mutations) to the ligand-binding site. As a next step, we further calculated the five DRN metrics (BC, CC, DC, EC, KC) for these 50 mutant systems complexed with each allosteric modulator and compared them to the reference system, as detailed in the next section.
3.7. Persistent and super persistent hubs of the averaged DRN metrics in the presence of mutations
In Part II, we identified the persistent hubs for each averaged DRN metric on the basis of their existence in both protomers in the absence and presence of all ligands. This gave us MET17, THR111, PHE112 and CYS128 for averaged BC; MET6, ALA7, SER113, VAL114, LEU115, GLY124, VAL125, TYR126, GLN127 and CYS128 for averaged CC; VAL36, VAL91, GLY146, PHE150 and ALA206 for averaged DC; ALA7, LEU115 and VAL125 for averaged EC; and VAL36, VAL125 and GLY146 for averaged KC (Table 2; reference rows).
Table 2.
Persistent hubs (in grey) as observed in the reference protein (apo and all ligand-bound states) and their comparison to 51 protein systems (reference protein + mutants) in the presence of each allosteric modulator. Super-persistent hubs are highlighted in orange. The hubs that are lost across all ligand systems are in pale blue.
 |
Here, to analyze the residue-residue communications, in the presence of potential allosteric modulators in mutant protein systems, we calculated the global top 5% averaged BC, CC, DC, EC, KC metrics for 51 protein systems (50 mutant protein systems and reference protein) (Figs. S2-S6); and extracted the persistent hubs on the basis of their conservation in both reference protein and mutants bound to a specific ligand (Table 2). If a persistent hub is retained across all the ligand-bound systems (in both protomers), then we called it a super-persistent hub.
In the case of averaged BC (Table 2, Fig. S2), we did not observe any super-persistent hub; however, MET17 was retained as the main persistent hub in all protein–ligand systems except in the presence of SANC00630 in which the protomer B of the mutant M49I protein lost the hub node. Hub 111 was retained as persistent hub in the presence of SANC00468, SANC00469 and SANC00630; and hub CYS128 was persistent in the presence of SANC00302 and SANC00467. PHE112 remained as a persistent hub in all 51 protein systems complexed with SANC00630.
Super-persistent hubs of averaged CC for 51 protein systems of all allosteric modulators were observed for residues MET6, ALA7, SER113, LEU115, VAL125, TYR126 and GLN127 (Fig. S3). In the presence of SANC00302, mutant protein A7V and in the presence of SANC00468, the mutant protein G15S lost hub 124 in their protomer B; hence GLY124 stayed as a persistent hub in only SANC00303, SANC00467, SANC00469 and SANC00630-bound systems. Additionally, hub 128 stayed persistent in all ligand systems except for SANC00468 in which it was lost in protomer B of K61R, A193V, I259T and N274D mutant protein systems in the presence of SANC00468. We also observed a new persistent hub for residue 10 in the presence of SANC00303.
In the case of averaged DC (Table 2, Fig. S4), we did not observe any super-persistent hub over all the ligand systems. The key persistent averaged DC hubs in the presence of most allosteric ligands were comprised of residues 150 and 206. PHE150 was a persistent hub to all ligand-bound systems, but was missed as a hub in protomer B of P184L and A116V mutant proteins in the presence of SANC00467. ALA206 was again a persistent hub to all ligand-bound systems, except for SANC00468 in which the hub was missing in protomer A of the L220F mutant protein. Interestingly persistent hub GLY146 was lost in the presence of all allosteric modulators.
Again, we did not observe any super-persistent hub using the EC metric either (Table 2, Fig. S5). But a new persistent hub (residue 10) in the presence of SANC00303 and SANC00469 was obtained. LEU115 was retained as persistent hub in all ligand-bound systems, except in the presence of SANC00468 (the hub node was lost in protomer B of the double mutant protein (A191V, L220F)); and in the presence of SANC00630 (the hub node was lost in protomer B of two mutant proteins M49I and A193V). Interestingly, the persistent hub, ALA7, was lost in the presence of all allosteric modulators according to the averaged EC metric.
LEU115 was also the key persistent hub according to the KC metric, and it was only lost in the presence of SANC00302 due to absence of the hub node in protomer B of the reference protein (Table 2, Fig. S6). Two new persistent hubs (residues 10 and 150) were introduced in the presence of SANC00469. Interestingly, the persistent hub VAL36 was lost across all allosteric modulators according to the averaged KC metric.
In general, by tracking the conservation of the persistent hubs as defined in PART II, we observed that the presence of the mutations affected the highly conserved communication hubs. This was evident by some of them being completely absent, i.e. hub 146 (DC), hub 7 (EC), hub 36 (KC). Some of the persistent hubs were lost in the presence of some ligand-bound systems. We also observed newly introduced persistent hubs in some of the metrics, i.e. hub 10 (KC; SANC00499 and EC; SANC00303 and 469). The super-persistent hubs were only observed from the CC metric and this is probably because CC identifies short communication paths (the central nodes which are closer to most of the nodes).
3.8. Mutation cold spots via analysis of five DRN metrics
There are only limited studies about identification of mutation cold spots with varying definitions of what it means [70], [91], [92], [93]. The techniques that have been used include in silico saturation mutagenesis, meaning mutating every residue to all the other 19 residues and predicting the change in stability [91]; or simply identifying regions where the mutations have not yet occurred in an organism [70]. Here we propose to use DRN metric analysis and define the cold spots as the regions that are the least affected or not affected at all, by mutations. In the previous sections, we introduced persistent hubs and super-persistent hubs, and we will consider the cold spots as being those hubs that are super-persistent, or almost so. The super-persistent hubs of the CC metric are all located mainly at the interface of the dimer as well as in the first two antiparallel beta strands. We believe that these regions should be strongly considered in structure based drug discovery.
3.9. Identification of ligand specific allosteric communication paths and changes in the presence of mutations
In this section we zoomed into the global top 5% averaged metric calculations for the reference and the 50 mutant protein systems in the presence of allosteric modulators (Figs. S2-S6). We picked up two ligands as specific examples: SANC00302 being the least stable compound and SANC00468 being the most stable within all mutant systems. We specifically focused on the protomer A EC results (Fig. 13) as a follow up on the allosteric communication path defined in Section 3.5.4, in which the path from allosteric site to the active site was defined via averaged EC persistent hubs (ALA7, LEU115 and VAL125) and hub score changes from 0 to 1 (MET17, ASN28 and GLY29) in the presence of ligand binding.
Fig. 13.
Heat map for the potential hubs according to the global top 5% for the averaged EC metric for the reference and 50 mutant proteins in allosterically bound state to SANC00302 and SANC00468. Detected hubs are annotated with their centrality values, while their homologous residues in alternate samples are not, but are only shown for the sake of comparison. Low to high centrality values are colored white, through yellow, orange and red to black. Mutants demonstrating highly different centrality hub profiles are marked with red boxes. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Protomer A of the Mpro-SANC00302 reference protein – ligand complex has 18 centrality hubs for EC (residues 7, 10, 17, 28, 29, 38, 113, 115, 116, 117, 122, 124, 125, 146, 147, 148, 149, 150), including the residue path identified in Section 3.5.4 (Fig. 13). When we collectively mapped these centrality hubs to the protein–ligand system, we had another very interesting observation: These centrality hubs form a communication path between the allosteric ligand binding site to the active site going through the interface residues of Domain I and II (Fig. 14A). In the case of Protomer A of Mpro- SANC00468 reference protein – ligand complex, some new centrality hubs are gained (9, 11, 13, 14), and some lost (38, 149) compared to that of Mpro-SANC00302 system; totaling to 20 EC hub residues (7, 9, 10, 11, 13, 14, 17, 28, 29, 113, 115, 116, 117, 122, 124, 125, 146, 147, 148, 150) (Fig. 13).
Fig. 14.
Differentiation of the communication paths traced by the averaged EC hubs (labeled), starting from the allosteric ligand towards the catalytic residue. The protease is represented as a cartoon, onto which the averaged EC hub residues are overlaid as sphere representations, together with the non-hub catalytic residues HIS41 and CYS145 (in orange spheres). The top panels show the hubs obtained in (A) the SANC00302-bound reference Mpro, and (B) the SANC00302-bound G71S mutant. The bottom panels show the hubs obtained in (C) the SANC00468-bound reference Mpro, and (D) the SANC00468-bound A173V mutant. Compounds SANC00302 and SANC00468, the positions of which are as observed after MD simulation, are depicted as green and purple stick figure representations respectively. The mutation loci are represented as purple spheres and are each indicated by an arrow. Additional hubs in the two ligand-bound reference proteins include residues LEU115, ALA116 and CYS117 in the case of panel (A), and residues LEU115, ALA116, CYS117, PRO122, GLY124 and VAL125 in panel (C), but are not shown as labels, for improved visibility. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Next, we looked at how these averaged EC hubs of protomer A change in the presence of mutations. In general, we observed that, there are more mutant cases where a large number of the centrality hubs is lost in the presence of SANC00302 compared to SANC00468. Some examples of these cases from SANC00302 are the V20L, G71S, I136V, C160S and V261A mutant proteins. We further observed that the decreased number of EC hubs leads to diminished communication paths that are either weakened or totally lost. The G71S-SANC00302 mutant system for instance, could only maintain 7 EC hubs out of 18 (residues 9, 10, 11, 14, 115, 122, 125) (Fig. 14B). Extreme examples showing the loss of the communication path in the presence of SANC00468 include G15S, G71S and A173V mutants. The A173V-SANC00468 complex with 8 EC hubs (7, 9, 10, 11, 113, 115, 124, 125) is presented in Fig. 14D.
4. Conclusion
In this study we have provided important new insights towards computational drug discovery, and applied them to the SARS-CoV-2 Mpro protein. Here, we will list the novel aspects and link them to our findings for Mpro protein.
We previously proposed a post-hoc analysis approach of MD simulations using DRN analysis to consider the dynamic nature of functional proteins and protein-drug complexes and to probe the impact of mutations and their allosteric effects. We also established a tool for DRN [57]. We and others, in a number of publications, showed the effectiveness of our DRN approach [14], [19], [32], [59], [85], [94], [95], [96], [97], [98], [99], [100], [101], [102]. In this study, for the first time, we investigated the relationships and effectiveness of five DRN metrics (BC, CC, DC, EC and KC) in characterizing key communication residues of the reference Mpro protein and in its allosteric behavior in the presence of potential allosteric modulators and evolutionary mutations. Further, we introduced the concept of analyzing globally central nodes (i.e. the 5% most central nodes measured across all samples) and developed an algorithm to pinpoint key hub residues, meaning any node that forms part of the set of highest centrality nodes for any given averaged centrality metric.
We investigated hub transition when exposed to a particular environment (i.e. ligand binding) by considering these strongest actors (hubs) across samples and showed how other non-hub residues behave at homologous positions. The key reason for using DRN analysis in Mpro protein was to tackle the problem of protein symmetry that we identified in our previous study [14], where we observed that protomer dynamics could be switched between identical copies of a protomer in a homodimer. In this study, we investigated the phenomenon in greater detail using a combinatorial approach to examine patterns of change and conservation of critical nodes, according to five independent criteria of network centrality. Asymmetric behavior of multimeric proteins, in general, is not considered in computational analysis. To our knowledge, this is the first study of this problem using five DRN metrics, and emphasizing the importance of this aspect while analyzing a protein’s allosteric behavior in the presence of ligands and mutations.
Applications of our approaches pinpointed a number of important aspects in SARS-CoV-2 Mpro protein: (1) we identified hubs that stayed the same in the apo state and upon a ligand binding (constitutive hubs), indicating that there is no ligand effect from symmetry; (2) we captured different persistent hubs from each metric, and collectively they gave us highly crucial functional residues which were spreading out from the allosteric site to the interface and antiparallel beta strands. We believe that the antiparallel beta strands, especially the first two near to the dimer interface, are crucial in the mechanical signal transduction; (3) we also looked at the symmetry problem and analyzed hub losses and gains in the presence of allosteric modulators. The identified residues that informed us about communication changes due to the presence of ligands and mutations. A few examples of hub gains and losses that we observed in functional residues are VAL13 (next to the N-finger), GLY 138 (part of S1 subsite) and PHE140 (chameleon switch). We also observed a number of hub transitions in antiparallel beta strands; (4) very interestingly, we showed that EC centrality hubs form ligand specific communication paths between the allosteric ligand binding site to the active site going through the interface residues of domains I and II.
In general, structure based drug discovery approaches have been used successfully for the design of many orthosteric drugs and to some extent of allosteric modulators. However, the impact of evolutionary mutations of pathogens is mostly undetermined in rational drug design; even though the information obtained may help to develop drugs that could circumvent or reduce potential drug resistance issues. Here, we applied this concept to identify potential allosteric modulators in the presence of 50 early evolutionary mutations of the SARS-CoV-2. We made several observations: (1) stability of the ligands drastically changed in the presence of some of the mutations. The R60C, N151D, V157I, C160S and A255V mutant proteins could only hold two compounds out of six stably. SANC00302 was the least stable compound (in 20 mutant systems) and SANC00468 was the most stable (in 43 mutant systems); (2) the persistent hubs, residues 7 (EC), 36 (KC) and 146 (DC), lost their importance in network communication in the presence of mutations; (3) in the presence of mutants some new persistent hubs (residues 10 (EC), 115 (DC and KC) and 150 (KC)) were gained; (4) Further, we defined super-persistent hubs, and we considered cold spots as being those hubs that are super-persistent, or almost so. These regions could be considered in structure-based drug discovery; (5) in the presence of some of the mutations, the network communication within each protomer drastically differed from each other, emphasizing the asymmetric behavior of the dimer protein; (6) most importantly, the allosteric communication path, that was identified via EC hubs, between the allosteric ligand binding site and the active site was lost in some of the mutant protein-ligand systems.
Collectively, our approaches offer routes for novel rational drug discovery methods and provide computationally feasible platforms (1) to determine globally central nodes that form part of the set of highest centrality nodes (hubs) for any given averaged centrality metric; (2) to identify key functional residues implicated in allosteric signaling in the presence of allosteric modulators; (3) to understand the potential asymmetric behavior of dimeric proteins under internal and external forces and to distinguish those introduced by ligand binding or by evolutionary mutations; (4) to utilize five DRN metrics to pinpoint cold spot residues that can potentially be chosen for structure guided drug discovery.
Finally, experimental verification of the predicted Mpro inhibitors, and thus of the algorithms presented here, is highly desirable; and we hope that this study will inspire wet-lab investigation.
Funding
O.S.A. is funded as a postdoctoral fellow by H3ABioNet, which is supported by the National Human Genome Research Institute of the National Institutes of Health under Award Number U24HG006941. R.A.B. is funded by DELTAS Africa Initiative under Wellcome Trust (DELGEME grant number 107740/Z/15/Z) for a Ph.D. fellowship. V.B is funded by the Grand Challenges Africa programme [GCA/DD/rnd3/03] African Academy of Sciences (AAS). This work is further supported by Funding for COVID-19 Research and Development Goals for Africa Programme (Grant number: SARSCov2-2-20-002) African Academy of Sciences (AAS). Both programmes of the AAS are implemented through the Alliance for Accelerating Excellence in Science in Africa (AESA) platform, an initiative of the AAS and the African Union Development Agency (AUDA-NEPAD). Grand Challenges Africa is supported by the Bill & Melinda Gates Foundation (BMGF), Swedish International Development Cooperation Agency (SIDA), German Federal Ministry of Education and Research (BMBF), Medicines for Malaria Venture (MMV), and Drug Discovery and Development Centre of University of Cape Town (H3D). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The content of this publication is solely the responsibility of the authors and does not necessarily represent the official views of the funders.
Notes
The authors declare no competing financial interest.
Data and software availability
All data reported in this article are presented in the article and the Supporting Information section. Dynamic residue network analysis metric scripts are implemented in the MDM-TASK-web platform (https://mdmtaskweb.rubi.ru.ac.za/) and are available at https://github.com/RUBi-ZA/MD-TASK/tree/mdm-task-web. MD simulations will be made available upon request.
CRediT authorship contribution statement
Olivier Sheik Amamuddy: Formal analysis, Methodology, Software, Supervision, Visualization, Writing – original draft, Writing – review & editing. Rita Afriyie Boateng: Formal analysis, Methodology, Visualization, Writing – original draft. Victor Barozi: Methodology, Visualization. Dorothy Wavinya Nyamai: Methodology. Özlem Tastan Bishop: Conceptualization, Formal analysis, Funding acquisition, Methodology, Project administration, Resources, Supervision, Visualization, Writing – original draft, Writing – review & editing.
Acknowledgment
Authors acknowledge the use of the Centre for High Performance Computing (CHPC), Cape Town, South Africa for the simulations. Authors thank Dr Thommas M. Musyoka for the Tanimoto coefficient score calculations.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.11.016.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Xiu S., Dick A., Ju H., Mirzaie S., Abdi F., Cocklin S., et al. Inhibitors of SARS-CoV-2 entry: current and future opportunities. J. Med. Chem. 2020;63:2256–12274. doi: 10.1021/acs.jmedchem.0c00502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., et al. Structure of Mpro from SARS-CoV-2 and Discovery of Its Inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 3.Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., et al. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeng L., Li D., Tong W., Shi T., Ning B. Biochemical features and mutations of key proteins in SARS-CoV-2 and their impacts on RNA therapeutics. Biochem. Pharmacol. 2021;189 doi: 10.1016/j.bcp.2021.114424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu C., Zhou Q., Li Y., Garner L.V., Watkins S.P., Carter L.J., et al. Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent. Sci. 2020;6:315–331. doi: 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science:abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sisay M. 3CLpro inhibitors as a potential therapeutic option for COVID-19: available evidence and ongoing clinical trials. Pharmacol. Res. 2020;156 doi: 10.1016/j.phrs.2020.104779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang F., Chen C., Tan W., Yang K., Yang H. Structure of main protease from human coronavirus NL63: insights for wide spectrum anti-coronavirus drug design. Sci. Rep. 2016;6:22677. doi: 10.1038/srep22677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Majumder R., Mandal M. Screening of plant-based natural compounds as a potential COVID-19 main protease inhibitor: an in silico docking and molecular dynamics simulation approach. J. Biomol. Struct. Dyn. 2020;1–16 doi: 10.1080/07391102.2020.1817787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dai W., Zhang B., Jiang X.-M., Su H., Li J., Zhao Y., et al. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368:1331–1335. doi: 10.1126/science:abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J. Fast identification of possible drug treatment of coronavirus disease-19 (COVID-19) through computational drug repurposing study. J. Chem. Inf. Model. 2020;60:3277–3286. doi: 10.1021/acs.jcim.0c00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El‐Baba T.J., Lutomski C.A., Kantsadi A.L., Malla T.R., John T., Mikhailov V., et al. Allosteric inhibition of the SARS-CoV-2 main protease – insights from mass spectrometry-based assays. Angew. Chemie. 2020;59:23544–23548. doi: 10.1002/anie.202010316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Günther S., Reinke P.Y.A., Fernández-García Y., Lieske J., Lane T.J., Ginn H.M., et al. X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease. Science. 2021;372:642–646. doi: 10.1126/science:abf7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheik Amamuddy O., Verkhivker G.M., Tastan Bishop Ö. Impact of early pandemic stage mutations on molecular dynamics of SARS-CoV-2 M Pro. J. Chem. Inf. Model. 2020;60:5080–5102. doi: 10.1021/acs.jcim.0c00634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hatherley R., Brown D.K., Musyoka T.M., Penkler D.L., Faya N., Lobb K.A., et al. SANCDB: A South African natural compound database. J. Cheminform. 2015;7:29. doi: 10.1186/s13321-015-0080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diallo B.N., Glenister M., Musyoka T.M., Lobb K., Tastan Bishop Ö. SANCDB: An update on South African natural compounds and their readily available analogs. J. Cheminform. 2021;13:37. doi: 10.1186/s13321-021-00514-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.St-Jean, J. R.; Jacomy, H.; Desforges, M.; Vabret, A.; Freymuth, F.; Talbot, P. J. Human Respiratory Coronavirus OC43: Genetic Stability and Neuroinvasion. J. Virol. 2004, 78, 8824-34, 10.1128/jvi.78.16.8824-8834.2004. [DOI] [PMC free article] [PubMed]
- 18.Liu, D. X.; Liang, J. Q.; Fung, T. S. Human Coronavirus-229E, -OC43, -NL63, and -HKU1 (Coronaviridae). In Encyclopedia of Virology, 2021, pp 428-440, Elsevier, 10.1016/b978-0-12-809633-8.21501-x.
- 19.Penkler D.L., Atilgan C., Tastan Bishop Ö. Allosteric modulation of human Hsp90α conformational dynamics. J. Chem. Inf. Model. 2018;58:383–404. doi: 10.1021/acs.jcim.7b00630. [DOI] [PubMed] [Google Scholar]
- 20.Munir A., Wilson M.T., Hardwick S.W., Chirgadze D.Y., Worrall J.A.R., Blundell T.L., et al. Using Cryo-EM to understand antimycobacterial resistance in the catalase-peroxidase (KatG) from Mycobacterium tuberculosis. Structure. 2021;29:899–912.e4. doi: 10.1016/j.str.2020.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brennecke J.T., De Groot B.L. Quantifying asymmetry of multimeric proteins. J. Phys. Chem. A. 2018;122:7924–7930. doi: 10.1021/acs.jpca.8b06843. [DOI] [PubMed] [Google Scholar]
- 22.Chea E., Livesay D.R. How accurate and statistically robust are catalytic site predictions based on closeness centrality? BMC Bioinform. 2007;8:153. doi: 10.1186/1471-2105-8-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.del Sol A., Fujihashi H., Amoros D., Nussinov R. Residue centrality, functionally important residues, and active site shape: analysis of enzyme and non-enzyme families. Protein Sci. 2006;15:2120–2128. doi: 10.1110/ps.062249106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Negre C.F.A., Morzan U.N., Hendrickson H.P., Pal R., Lisi G.P., Loria J.P., et al. Eigenvector centrality for characterization of protein allosteric pathways. Proc. Natl. Acad. Sci. 2018;115:E12201–E12208. doi: 10.1073/pnas.1810452115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okeke C.J., Musyoka T.M., Sheik Amamuddy O., Barozi V., Tastan Bishop Ö. Allosteric pockets and dynamic residue network hubs of falcipain 2 in mutations including those linked to artemisinin resistance. Comput. Struct. Biotechnol. J. 2021;19:5647–5666. doi: 10.1016/j.csbj.2021.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Westbrook J.D., Burley S.K. How structural biologists and the protein data bank contributed to recent FDA new drug approvals. Structure. 2019;27:211–217. doi: 10.1016/j.str.2018.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheik Amamuddy O., Veldman W., Manyumwa C., Khairallah A., Agajanian S., Oluyemi O., et al. Integrated computational approaches and tools for allosteric drug discovery. Int. J. Mol. Sci. 2020;21:847. doi: 10.3390/ijms21030847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drag M., Salvesen G.S. Emerging principles in protease-based drug discovery. Nat. Rev. Drug Discov. 2010;9:690–701. doi: 10.1038/nrd3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guarnera E., Berezovsky I.N. Allosteric drugs and mutations: chances, challenges, and necessity. Curr. Opin. Struct. Biol. 2020;62:149–157. doi: 10.1016/j.sbi.2020.01.010. [DOI] [PubMed] [Google Scholar]
- 30.Sheik Amamuddy O., Musyoka T.M., Boateng R.A., Zabo S., Tastan Bishop Ö. Determining the unbinding events and conserved motions associated with the pyrazinamide release due to resistance mutations of Mycobacterium tuberculosis pyrazinamidase. Comput. Struct. Biotechnol. J. 2020;18:1103–1120. doi: 10.1016/j.csbj.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ricatti J., Acquasaliente L., Ribaudo G., De Filippis V., Bellini M., Llovera R.E., et al. Effects of point mutations in the binding pocket of the mouse major urinary protein MUP20 on ligand affinity and specificity. Sci. Rep. Sci. Rep. 2019;9:300. doi: 10.1038/s41598-018-36391-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown D.K., Sheik Amamuddy O., Tastan Bishop Ö. Structure-based analysis of single nucleotide variants in the renin-angiotensinogen complex. Glob. Heart. 2017;12:121–132. doi: 10.1016/j.gheart.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nussinov R., Tsai C.J. Allostery without a conformational change? Revisiting the paradigm. Curr. Opin. Struct. Biol. 2015;30:17–24. doi: 10.1016/j.sbi.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 34.Sheik Amamuddy O. Rhodes University, Makhanda; South Africa: 2019. Application of machine learning, molecular modelling and structural data mining against antiretroviral drug resistance in HIV-1. Ph.D Thesis. [Google Scholar]
- 35.Guo J., Zhou H.-X. Protein allostery and conformational dynamics. Chem. Rev. 2016;116:6503–6515. doi: 10.1021/acs.chemrev.5b00590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sheik Amamuddy O., Bishop N.T., Tastan Bishop Ö. Characterizing early drug resistance-related events using geometric ensembles from HIV protease dynamics. Sci. Rep. 2018;8:1–11. doi: 10.1038/s41598-018-36041-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burley, S. K.; Berman, H. M.; Kleywegt, G. J.; Markley, J. L.; Nakamura, H.; Velankar, S. Protein Data Bank (PDB): The Single Global Macromolecular Structure Archive; Wlodawer, A., Dauter, Z., Jaskolski, M., Eds.; Springer New York: New York, NY, 2017; pp 627–641, 10.1007/978-1-4939-7000-1_26. [DOI] [PMC free article] [PubMed]
- 38.Fearon D., Owen C.D., Douangamath A., Lukacik P., Powell A.J., Strain-Damerell C.M., et al. PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen. Nat. Commun. 2020;11:5047. doi: 10.1038/s41467-020-18709-w. https://www.ebi.ac.uk/pdbe/entry/pdb/5rfv [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elbe, S.; Buckland-Merrett, G. Data, Disease and Diplomacy: GISAID’s Innovative Contribution to Global Health. Glob. Challenges 2017, 1, 33–46, /10.1002/gch2.1018. [DOI] [PMC free article] [PubMed]
- 40.Fiser, A.; Šali, A. Modeller: Generation and Refinement of Homology-Based Protein Structure Models. In Methods in Enzymology; Department of Biochemistry and Seaver Foundation Center for Bioinformatics, Albert Einstein College of Medicine, Bronz, New York 10461, USA., 2003; Vol. 374, pp 461–491, 10.1016/S0076-6879(03)74020-8. [DOI] [PubMed]
- 41.Pei J., Kim B.H., Grishin N.V. PROMALS3D: A tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36:2295–2300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dolinsky T.J., Nielsen J.E., McCammon J.A., Baker N.A. PDB2PQR: An automated pipeline for the setup of poisson-boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32:W665–7. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pearlman D.A., Case D.A., Caldwell J.W., Ross W.S., Cheatham T.E., DeBolt S., et al. AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comput. Phys. Commun. 1995;1:1–41. doi: 10.1016/0010-4655(95)00041-D. [DOI] [Google Scholar]
- 44.San Diego: Accelrys Software Inc. Discovery Studio Modeling Environment, 2012, Release 3.5, San Diego, CA.
- 45.Gasteiger J., Marsili M. Iterative partial equalization of orbital electronegativity-a rapid access to atomic charges. Tetrahedron. 1980;36:3219–3228. doi: 10.1016/0040-4020(80)80168-2. [DOI] [Google Scholar]
- 46.Trott, O.; Olson, A. AutoDock Vina: Inproving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization and Multithreading. J. Comput. Chem. 2010, 31 (2), J. Comput. Chem. 2010, 31, 455–461, 10.1002/jcc.21334.AutoDock. [DOI] [PMC free article] [PubMed]
- 47.Schrödinger, LLC: New York 2015. The PyMOL Molecular Graphics System, Version 2.4, https://pymol.org/2/support.html?.
- 48.McKinney, W. Pandas: Powerful Python Data Analysis Toolkit — Pandas 0.19.0+128.G43c24e6.Dirty Documentation, 2016.
- 49.Hunter J.D. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 2007;9:90–95. doi: 10.1109/MCSE.2007.55. [DOI] [Google Scholar]
- 50.Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1-2:19–25. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- 51.Sousa da Silva A.W., Vranken W.F. ACPYPE - AnteChamber PYthon Parser InterfacE. BMC Res Notes. 2012;5:367. doi: 10.1186/1756-0500-5-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mark P., Nilsson L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J. Phys. Chem. A. 2001;105:9954–9960. doi: 10.1021/jp003020w. [DOI] [Google Scholar]
- 53.Lemak A.S., Balabaev N.K. On the Berendsen Thermostat. Mol. Simul. 1994;13:177–187. doi: 10.1080/08927029408021981. [DOI] [Google Scholar]
- 54.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. doi: 10.1063/1.328693. [DOI] [Google Scholar]
- 55.Hess B., Bekker H., Berendsen H.J.C., Fraaije J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. doi: 10.1002/(SICI)1096-987X(199709)18:12<1463::AID-JCC4>3.0.CO;2-H. [DOI] [Google Scholar]
- 56.Petersen H.G. Accuracy and efficiency of the particle mesh ewald method. J. Chem. Phys. 1995;103:3668. doi: 10.1063/1.470043. [DOI] [Google Scholar]
- 57.Sheik Amamuddy O., Glenister M., Bishop Ö.T. MDM-TASK-Web: MD-TASK and MODE-TASK Web Server for Analyzing Protein Dynamics. Comput. Struct. Biotechnol. J. 2021;19:5059–5071. doi: 10.1016/j.csbj.2021.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brown, D. K.; Penkler, D. L.; Sheik Amamuddy, O.; Ross, C.; Atilgan, A. R.; Atilgan, C.; Bishop, Ö. T.; Sheik Amamuddy, O.; Ross, C.; Atilgan, A. R.; Atilgan, C.; Tastan Bishop, Ö. MD-TASK: A Software Suite for Analyzing Molecular Dynamics Trajectories. Bioinformatics 2017, 33, 2768–2771, 0.1093/bioinformatics/btx349. [DOI] [PMC free article] [PubMed]
- 59.Penkler D.L., Tastan Bishop Ö. Modulation of human Hsp90α conformational dynamics by allosteric ligand interaction at the C-Terminal domain. Sci. Rep. 2019;9:1600. doi: 10.1038/s41598-018-35835-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hagberg, A.; Swart, P.; S Chult, D. Exploring Network Structure, Dynamics, and Function Using NetworkX. In 7th Python in Science Conference (SciPy 2008); Varoquaux, E., Vaught, T., Millman, J., Eds.; Los Alamos National Lab. (LANL), Los Alamos, NM (United States): Pasadena, CA USA, 2008; pp 1–15.
- 61.Anand K., Palm G.J., Mesters J.R., Siddell S.G., Ziebuhr J., Hilgenfeld R. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra α-helical domain. EMBO J. 2002;21:3213–3224. doi: 10.1093/emboj/cdf327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chang, G. G. Quaternary Structure of the SARS Coronavirus Main Protease. Molecular Biology of the SARS-Coronavirus 2010, 115–128, 10.1007/978-3-642-03683-5_8.
- 63.Ramos-Guzmán C.A., Ruiz-Pernía J.J., Tuñón I. Unraveling the SARS-CoV-2 main protease mechanism using multiscale methods. ACS Catal. 2020;10:12544–12554. doi: 10.1021/acscatal.0c03420. [DOI] [PubMed] [Google Scholar]
- 64.Chen S., Hu T., Zhang J., Chen J., Chen K., Ding J., et al. Mutation of Gly-11 on the dimer interface results in the complete crystallographic dimer dissociation of severe acute respiratory syndrome coronavirus 3C-like protease: crystal structure with molecular dynamics simulations. J. Biol. Chem. 2008;283:554–564. doi: 10.1074/jbc.M705240200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shi J., Wei Z., Song J. Dissection study on the severe acute respiratory syndrome 3C-like protease reveals the critical role of the extra domain in dimerization of the enzyme. J. Biol. Chem. 2004;279:24765–24773. doi: 10.1074/jbc.M311744200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shi J., Song J. The catalysis of the SARS 3C-like protease is under extensive regulation by its extra domain. FEBS J. 2006;273:1035–1045. doi: 10.1111/j.1742-4658.2006.05130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhong N., Zhang S., Zou P., Chen J., Kang X., Li Z., et al. Without its N-finger, the main protease of severe acute respiratory syndrome coronavirus can form a novel dimer through Its C-terminal domain. J. Virol. 2008;82:4227–4234. doi: 10.1128/JVI.02612-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang H., Yang M., Ding Y., Liu Y., Lou Z., Zhou Z., et al. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. 2003;100:13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang H., Xie W., Xue X., Yang K., Ma J., Liang W., et al. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005;3 doi: 10.1371/journal.pbio.0030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Krishnamoorthy N., Fakhro K. Identification of mutation resistance coldspots for targeting the SARS-CoV2 main protease. IUBMB Life. 2021;73:670–675. doi: 10.1002/iub.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Douangamath A., Fearon D., Gehrtz P., Krojer T., Lukacik P., Owen C.D., et al. Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease. Nat. Commun. 2020;11 doi: 10.1038/s41467-020-18709-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shu Y., McCauley J. GISAID: global initiative on sharing all influenza data – from vision to reality. Eurosurveillance. 2017;22:30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Forli W., Halliday S., Belew R., Olson A. AutoDock Version 4.2. Citeseer. 2012:1–66. [Google Scholar]
- 74.Koorbanally N.A., Koorbanally C., Harilal A., Mulholland D.A., Crouch N.R. Bufadienolides from Drimia Robusta and Urginea Epigea (Hyacinthaceae) Chem. Inform. 2005;36:1. doi: 10.1002/chin.200516167. [DOI] [PubMed] [Google Scholar]
- 75.Koorbanally C., Mulholland D.A., Crouch N.R. A Novel Homoisoflavonoid from Drimia Delagoensis (Urgineoideae: Hyacinthaceae) Biochem. Syst. Ecol. 2005;33:743–748. doi: 10.1016/j.bse.2004.11.009. [DOI] [Google Scholar]
- 76.Bohlmann F., Zdero C. New sesquiterpenes from senecio oxyodontus. Phytochemistry. 1978;17:1591–1593. doi: 10.1016/S0031-9422(00)94649-1. [DOI] [Google Scholar]
- 77.Bromley C.L., Parker-Nance S., De La Mare J.A., Edkins A.L., Beukes D.R., Davies-Colemanf M.T. Halogenated Oxindole and indoles from the South African marine ascidian Distaplia Skoogi. South African J. Chem. 2013;66:64–68. http://www.scielo.org.za/pdf/sajc/v66/15.pdf [Google Scholar]
- 78.Backman T.W.H., Cao Y., Girke T. ChemMine tools: an online service for analyzing and clustering small molecules. Nucleic Acids Res. 2011;39:W486–W491. doi: 10.1093/nar/gkr320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kneller D.W., Phillips G., Weiss K.L., Pant S., Zhang Q., O’Neill H.M., et al. Unusual zwitterionic catalytic site of SARS-CoV-2 main protease revealed by neutron crystallography. J. Biol. Chem. 2020;295:P17365–17373. doi: 10.1074/jbc.AC120.016154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shi J., Sivaraman J., Song J. Mechanism for controlling the dimer-monomer switch and coupling dimerization to catalysis of the severe acute respiratory syndrome coronavirus 3C-like protease. J. Virol. 2008;82:9. doi: 10.1128/jvi.02680-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Krukow P., Jonak K., Karpiński R., Karakuła-Juchnowicz H. Abnormalities in hubs location and nodes centrality predict cognitive slowing and increased performance variability in first-episode schizophrenia patients. Sci. Rep. 2019;9:9594. doi: 10.1038/s41598-019-46111-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fornito, A., Zalesky, A., Bullmore, E. T. B. T.-F. of B. N. A. Chapter 5 - Centrality and Hubs, Academic Press: San Diego, 2016; pp 137–161, 10.1016/B978-0-12-407908-3.00005-4.
- 83.Manyumwa, C. V.; Bishop, Ö. T. In Silico Investigation of Potential Applications of Gamma Carbonic Anhydrases as Catalysts of Co2 Biomineralization Processes: A Visit to the Thermophilic Bacteria Persephonella Hydrogeniphila, Persephonella Marina, Thermosulfidibacter Takaii, and Thermus Thermophilus. Int. J. Mol. Sci. 2021, 22, 10.3390/ijms22062861. [DOI] [PMC free article] [PubMed]
- 84.Amusengeri, A.; Tastan Bishop, Ö. Discorhabdin N, a South African natural compound, for Hsp72 and Hsc70 allosteric modulation: combined study of molecular modeling and dynamic residue network analysis. Molecules 2019, 24, 188, 10.3390/molecules24010188. [DOI] [PMC free article] [PubMed]
- 85.Allan Sanyanga T., Nizami B., Bishop Ö.T. Mechanism of action of non-synonymous single nucleotide variations associated with α-carbonic anhydrase II deficiency. Molecules. 2019;24:3987. doi: 10.3390/molecules24213987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Amitai G., Shemesh A., Sitbon E., Shklar M., Netanely D., Venger I., et al. Network analysis of protein structures identifies functional residues. J. Mol. Biol. 2004;344:1135–1146. doi: 10.1016/j.jmb.2004.10.055. [DOI] [PubMed] [Google Scholar]
- 87.Thibert B., Bredesen D.E., del Rio G. Improved prediction of critical residues for protein function based on network and phylogenetic analyses. BMC Bioinform. 2005;6:213. doi: 10.1186/1471-2105-6-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Snijder E.J., Decroly E., Ziebuhr J. The nonstructural proteins directing coronavirus RNA synthesis and processing. Adv. Virus Res. 2016;96:59–126. doi: 10.1016/bs.aivir.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cross T.J., Takahashi G.R., Diessner E.M., Crosby M.G., Farahmand V., Zhuang S., et al. Sequence characterization and molecular modeling of clinically relevant variants of the SARS-CoV-2 main protease. Biochemistry. 2020;59:3741–3756. doi: 10.1021/acs.biochem.0c00462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tee W.-V., Guarnera E., Berezovsky I.N. On the allosteric effect of NsSNPs and the emerging importance of allosteric polymorphism. J. Mol. Biol. 2019;431:3933–3942. doi: 10.1016/j.jmb.2019.07.012. [DOI] [PubMed] [Google Scholar]
- 91.Vedithi S.C., Rodrigues C.H.M., Portelli S., Skwark M.J., Das M., Ascher D.B., et al. Computational saturation mutagenesis to predict structural consequences of systematic mutations in the beta subunit of RNA polymerase in Mycobacterium leprae. Comput. Struct. Biotechnol. J. 2020;18:271–286. doi: 10.1016/j.csbj.2020.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shirian J., Sharabi O., Shifman J.M. Cold spots in protein binding. Trends Biochem. Sci. 2016;41:739–745. doi: 10.1016/j.tibs.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 93.Naftaly S., Cohen I., Shahar A., Hockla A., Radisky E.S., Papo N. Mapping protein selectivity landscapes using multi-target selective screening and next-generation sequencing of combinatorial libraries. Nat. Commun. 2018;9:3935. doi: 10.1038/s41467-018-06403-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fischer A., Häuptli F., Lill M.A., Smieško M. Computational assessment of combination therapy of androgen receptor-targeting compounds. J. Chem. Inf. Model. 2021;61:1001–1009. doi: 10.1021/acs.jcim.0c01194. [DOI] [PubMed] [Google Scholar]
- 95.Wang S., Xu Y., Yu X.W. A Phenylalanine dynamic switch controls the interfacial activation of rhizopus Chinensis lipase. Int. J. Biol. Macromol. 2021;173:1–12. doi: 10.1016/j.ijbiomac.2021.01.086. [DOI] [PubMed] [Google Scholar]
- 96.Ma S., Li H., Yang J., Yu K. Molecular simulation studies of the interactions between the Human/Pangolin/Cat/Bat ACE2 and the receptor binding domain of the SARS-CoV-2 spike protein. Biochimie. 2021;187:1–13. doi: 10.1016/j.biochi.2021.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chebon-Bore L., Sanyanga T.A., Manyumwa C.V., Khairallah A., Bishop Ö.T. Decoding the molecular effects of atovaquone linked resistant mutations on Plasmodium falciparum Cytb-Isp complex in the phospholipid bilayer membrane. Int. J. Mol. Sci. 2021;22:2138. doi: 10.3390/ijms22042138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Amusengeri A., Tata R.B., Tastan Bishop Ö. Understanding the pyrimethamine drug resistance mechanism via combined molecular dynamics and dynamic residue network analysis. Molecules. 2020;25:904. doi: 10.3390/molecules25040904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Arifuzzaman M., Mitra S., Das R., Hamza A., Absar N., Dash R. In Silico analysis of nonsynonymous single-nucleotide polymorphisms (NsSNPs) of the SMPX gene. Ann. Hum. Genet. 2020;84:54–71. doi: 10.1111/ahg.12350. [DOI] [PubMed] [Google Scholar]
- 100.Xiao F., Song X., Tian P., Gan M., Verkhivker G.M., Hu G. Comparative dynamics and functional mechanisms of the CYP17A1 tunnels regulated by ligand binding. J. Chem. Inf. Model. 2020;60:3632–3647. doi: 10.1021/acs.jcim.0c00447. [DOI] [PubMed] [Google Scholar]
- 101.Dehury B., Tang N., Mehra R., Blundell T.L., Kepp K.P. Side-by-side comparison of notch- And C83 binding to γ-secretase in a complete membrane model at physiological temperature. RSC Adv. 2020;10:31215–31232. doi: 10.1039/d0ra04683c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Keretsu S., Ghosh S., Cho S.J. Molecular modeling study of C-Kit/Pdgfrα dual inhibitors for the treatment of gastrointestinal stromal tumors. Int. J. Mol. Sci. 2020;21:8232. doi: 10.3390/ijms21218232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this article are presented in the article and the Supporting Information section. Dynamic residue network analysis metric scripts are implemented in the MDM-TASK-web platform (https://mdmtaskweb.rubi.ru.ac.za/) and are available at https://github.com/RUBi-ZA/MD-TASK/tree/mdm-task-web. MD simulations will be made available upon request.
CRediT authorship contribution statement
Olivier Sheik Amamuddy: Formal analysis, Methodology, Software, Supervision, Visualization, Writing – original draft, Writing – review & editing. Rita Afriyie Boateng: Formal analysis, Methodology, Visualization, Writing – original draft. Victor Barozi: Methodology, Visualization. Dorothy Wavinya Nyamai: Methodology. Özlem Tastan Bishop: Conceptualization, Formal analysis, Funding acquisition, Methodology, Project administration, Resources, Supervision, Visualization, Writing – original draft, Writing – review & editing.