Abstract
The bare lymphocyte syndrome, a severe combined immunodeficiency due to loss of major histocompatibility complex (MHC) class II gene expression, is caused by inherited mutations in the genes encoding the heterotrimeric transcription factor RFX (RFX-B, RFX5, and RFXAP) and the class II transactivator CIITA. Mutagenesis of the RFX genes was performed, and the properties of the proteins were analyzed with regard to transactivation, DNA binding, and protein-protein interactions. The results identified specific domains within each of the three RFX subunits that were necessary for RFX complex formation, including the ankyrin repeats of RFX-B. DNA binding was dependent on RFX complex formation, and transactivation was dependent on a region of RFX5. RFX5 was found to interact with CIITA, and this interaction was dependent on a proline-rich domain within RFX5. Thus, these studies have defined the protein domains required for the functional regulation of MHC class II genes.
Type II bare lymphocyte syndrome (BLS), an inherited severe combined immunodeficiency in humans, is caused by the inability to transcribe major histocompatibility complex (MHC) class II genes (9, 15, 32). MHC class II genes encode heterodimeric glycoproteins that present antigens to CD4+ T cells to initiate the acquired arm of the immune response. They are also crucial for determining the repertoire of CD4+ T cells during positive and negative selection in the thymus. Patients with BLS typically present in the first year of life with recurrent infections and have reduced levels of CD4+ T cells (9, 11). Their humoral immune response is severely impaired as well, and most patients die before reaching puberty. Patient and experimentally derived cell lines were used to separate the BLS phenotype into four complementation groups: BLS groups A, B, C, and D (3, 46, 54). The genes responsible for each of these groups have been identified and found to encode proteins required for MHC class II gene transcription.
MHC class II genes are expressed on the surface of B cells, dendritic cells, macrophages, thymic epithelia, and activated T cells. Additionally, non-antigen-presenting cells can be induced to express MHC class II by exposure to the cytokine gamma interferon (IFN-γ) (8). Aberrant expression of MHC class II genes is associated with autoimmunity, tumor growth, and failure to mount an immune response. The three MHC class II isotypes, HLA-DR, HLA-DP, and HLA-DQ, contain conserved cis-acting elements in their promoters (the W, X1, X2, and Y boxes) that allow their coordinate regulation (reviewed in references 4 and 31). Homologous sequence elements are also found in the HLA-DM, invariant chain, and MHC class I genes. These elements allow the coordinate expression of the different isotypes in antigen-presenting cells and the induction of these genes by IFN-γ. Regulatory factor X (RFX) and the X2 box-binding protein (X2BP), which was identified as the cyclic AMP response element-binding protein (CREB) (34), bind to the X1 and X2 boxes, respectively. The Y box, an inverted CAAT box, is bound by the heterotrimeric nuclear factor Y (NF-Y) (4). The W box has not been extensively studied, but it was suggested to bind the X1 box factor, RFX (22). While all of these promoter-bound factors are required for MHC class II expression, they are not sufficient. The class II transactivator, CIITA, is also required. CIITA does not bind DNA and is believed to interact with factors on the MHC class II promoter, as well as the general transcriptional machinery, to activate transcription through its acidic activation domain (44, 49, 55). CIITA expression correlates directly with MHC class II expression and is regulated by IFN-γ (6, 7, 50). Thus, the presence of CIITA functions as a molecular switch for MHC class II gene regulation.
In vivo genomic footprinting of MHC class II promoters from BLS cell lines defined two distinct patterns (24, 25). Cell lines from complementation group A, which have mutations in the CIITA gene, showed fully occupied X1, X2, and Y boxes at MHC class II promoters. In contrast, cell lines from BLS groups B, C, and D, which are defective in RFX binding (41, 51), displayed no occupancy at the X1, X2, or Y box sites (24). This finding led to the hypothesis that not only was RFX binding critical for the binding of X2BP and NF-Y but also RFX itself could be a multisubunit complex with groups B, C, and D representing mutations in each subunit (36). Immunoprecipitation of the RFX complex and the cloning of the genes for RFX-B/RFXANK (BLS group B), RFX5 (BLS group C), and RFXAP (BLS group D) confirmed that RFX was a heterotrimeric complex (10, 33, 36, 38, 48).
It is known that all three subunits are required for RFX DNA-binding activity in vivo (33, 48), but nothing is known about how the subunits interact with each other to form the RFX complex, how this complex binds DNA, or how it activates transcription. Additionally, while one report showed weak interactions between CIITA and RFX5 by the yeast two-hybrid system (45), no additional information or confirmation of that finding has been reported. To further understand the nature and function of the RFX complex, a mutational study was undertaken to define domains in each subunit that are responsible for transactivation of an MHC class II promoter, DNA binding, subunit association, and the ability of the subunits to interact with CIITA. The results of this analysis identified regions of the RFX subunits responsible for these activities. Notably, the ankyrin repeats of RFX-B were responsible for interactions with both RFX5 and RFXAP. The C-terminal 93 amino acids of RFXAP, which include a glutamine-rich region, were sufficient for all its activity. A region in RFX-B was found to be important for DNA binding of the RFX complex. RFX5 was required for transactivation and could be shown to interact with CIITA both in vitro and in vivo. Thus, these studies define the interactions between the BLS proteins CIITA, RFX-B, RFX5, and RFXAP and define their functional role in the regulation of MHC class II gene expression.
MATERIALS AND METHODS
Construction of expression plasmids.
A linker containing a Kozak consensus sequence (27), a hexahistidine tag, and either an XbaI or EcoRI restriction site was cloned into the eukaryotic and T7 polymerase expression vector pcDNA3.1(−) (Invitrogen, Inc.) to create plasmids pXbaHis6 and pEcoHis6, respectively. PCR primers carrying the appropriate restriction site and the gene sequences indicated were synthesized and used to generate a series of 5′ or 3′ deletions in the three RFX subunit genes. All mutants were generated by PCR using Pfu polymerase (Stratagene, Inc.). Deletion mutations for RFXAP and RFX-B and the ankyrin repeat mutations in RFX-B were cloned into pEcoHis6. Primers used for the PCR of these deletions contained a 5′ EcoRI restriction site and a 3′ HindIII site and encompassed the following amino acids: RFX-BFL, 1 to 260; RFX-BΔ1, 1 to 221; RFX-BΔ2, 69 to 260; RFX-BΔ3, 123 to 260; RFXAPFL, 1 to 272; RFXAPΔ1, 122 to 272; RFXAPΔ2, 179 to 272; RFXAPΔ3, 1 to 245. The overlap-PCR primers used to introduce the alanine substitutions into the ankyrin repeats were as follows: ANK15′, GGAGAGGCTGAGACCGTTCGCGCCGCGGCGGAGTGGGGTGCCG; ANK13′, CGGCACCCCACTCCGCCGCGGCGCGAACGGTCTCAGCCTCTCC; ANK25′, GGCTACACAGACGCTGTGGGGGCGGCGGCGGAGCGTGACGTGG; ANK23′, CCACGTCACGCTCCGCCGCCGCCCCCACAGCGTCTGTGTAGCC; ANK3A5′, GGAGGGACGCCAGCGGCGTACGCTGTGCGC; ANK3A3′, GCGCACAGCGTACGCCGCTGGCGTCCCTCC; ANK3B5′, TGCGTTGAGGCCGCGGCGGCCCGAGGCGC; and ANK3B3′, GCGCCTCGGGCCGCCGCGGCCTCAACGCA. Deletion mutations for RFX5 were generated in the same manner and cloned into pXbaHis6. The primers used for PCR of these deletions contained a 5′ XbaI site and a 3′ NotI site and encompassed the following amino acids: RFX5FL, 1 to 616; RFX5Δ1, 201 to 616; RFX5Δ2, 261 to 616; RFX5Δ3, 410 to 616; RFX5Δ4, 1 to 92; RFX5Δ5, 1 to 170; and RFX5Δ6, 1 to 409. The full-length RFX-B gene was cloned into pGEX-5X-3 (Pharmacia, Inc.) to generate GST-RFX-B. RFX-BSV, the RFX-BΔ5 splice variant, has been previously described (38). The sequences of all clones were verified by automated DNA sequencing using the Emory University DNA sequencing core facility. HA-CIITA contains the hemagglutinin epitope tag placed at the N terminus of the CIITA gene (37).
Cell lines and transfections.
The cell lines Ramia, SJO, and 6.1.6, representing BLS groups B, C, and D, respectively, were described previously (2, 14, 29). Ramia and SJO cells were cultured in F12-Dulbecco's modified Eagle's medium supplemented with 20% fetal bovine serum, 2 mM glutamine, penicillin (100 U/ml), and streptomycin (100 μg/ml). 6.1.6 cells were grown in Iscove's modified Dulbecco's medium supplemented with 5% fetal bovine serum, 5% bovine serum, and the above supplements. Transient-transfection assays were preformed as described previously (43). To determine if the RFX mutants could rescue MHC class II surface expression, the appropriate BLS cell line was cotransfected with 40 μg of the indicated RFX construct and 10 μg of the green fluorescence protein (GFP) expression vector pd2EGFP-control (Clontech, Inc.). At 72 h after transfection, the cells were stained with phycoerythrin-conjugated HLA-DR antibody (Becton Dickinson, Inc.) and analyzed on a FACSCalibur. GFP-positive cells were selected and analyzed for MHC class II expression on the FL2 channel.
Transfection mixtures for HLA-DRA reporter gene transient transfections analyzing the RFX-B, RFX5, and RFXAP deletion mutants contained 20 μg of the MHC class II-dependent chloramphenicol acetyltransferase (CAT) reporter construction, pDRWXY (16), 5 μg of the indicated expression plasmid, and 2 μg of pGL3 (Promega Inc., Madison, Wis.), which carries the firefly luciferase gene. Mixtures for transient transfections for the ankyrin repeat mutants contained 10 μg of the pDRWXY reporter, 10 μg of the indicated expression plasmid, and 0.5 μg of pGL3. Cells were harvested 72 h posttransfection, and 3% of the cell lysate was analyzed for expression of the control luciferase product using the luciferase assay system (Promega Inc.). The remaining sample was analyzed for CAT protein using an enzyme-linked immunosorbent assay (Boehringer Mannheim Inc., Indianapolis, Ind.) as specified by the manufacturer. The data were normalized to the expression of the luciferase reporter. The average of three experiments is presented with the standard error of the mean.
COS-7 cells seeded at 106 cells/100-mm culture dish were transfected with Fugene-6 (Boehringer Mannheim, Inc.) as described by the manufacturer, using 3 μg of pRFX5FL or RFX5Δ6 and 6 μg of pHA-CIITA DNA. In the indicated transfections, 1 μg of each pRFX-BFL and pRFXAPFL were included. Cells were harvested after 48 h, lysed in a solution of 50 mM Tris (pH 8.0)–150 mM NaCl–1% NP-40, and used for coimmunoprecipitation analysis as described below.
Recombinant proteins and EMSAs.
For the native RFX complex, partially purified RFX from Raji B cells (38) was used. DNA-binding reactions were carried out as in our previous studies (19, 30, 35). An X2 box DNA competitor was added to the native RFX-binding reaction mixtures to prevent RFX-X2BP-DNA complexes from forming (30). To generate recombinant RFX subunits, in vitro transcription and translation reactions were carried out using the TNT quick coupled transcription translation system (Promega, Inc.) as specified by the manufacturer. Electrophoretic mobility shift assays (EMSAs) with in vitro-transcribed and -translated (IVT) proteins contained a total of 5 μl of the reticulocyte lysate. Before its addition to the DNA-binding reaction mixture, 1.7 μl of each subunit was mixed and incubated at 30°C for 1 h. For IVT RFX proteins, the same DNA-binding reaction (30) was carried out except that 1 μg of poly(dI-dC)-poly(dI-dC), 0.05 μg of salmon sperm DNA, and 0.025% NP-40 were used. DNA competition assay mixtures contained 100 ng of the specified competitor. DNA competitors X1m, X2m, and X1X2m contain mutations in the X1 box, X2 box, or both boxes that have been found to disrupt the binding of RFX, X2BP, and both proteins, respectively (16, 17, 30). The binding-reaction mixtures were incubated on ice for 15 min upon addition of protein and were incubated on ice for 30 min after the addition of 50,000 cpm of an X-box probe, DRAX (17). The binding-reaction mixtures were loaded on a 5% glycerol-tolerant gel containing 0.5 mM EDTA, 89 mM Tris, and 28.5 mM taurine and run for 2 h at 200 V and 4°C.
Purification of GST–RFX-B and GST-binding assays.
Glutathione S-transferase (GST)–RFX-B was expressed in E. coli BL21(DE3) cells. The cells were induced with isopropyl-β-d-thiogalactopyranoside (IPTG) (1 mM) for 2 h, harvested, and lysed in phosphate buffer (50 mM sodium phosphate [pH 7.4])–5% glycerol–1 mM EDTA using a French press. GST–RFX-B was bound to glutathione-Sepharose 4 beads (Pharmacia, Inc.) as specified by the manufacturer and washed three times with buffer containing 150 mM NaCl, 50 mM Tris (pH 8.0), and 1% NP-40. The washed beads corresponding to 2 μg of GST–RFX-B were incubated with 10 μl of each of in vitro-translated RFX5 and RFXAP at 30°C for 1 h. The beads were again washed with the same wash buffer six times. A corresponding amount of GST-containing beads was used as a control. After the washes, the beads were boiled in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) buffer containing 100 mM dithiothreitol and the samples were analyzed by SDS-PAGE.
Coimmunoprecipitations.
Affinity-purified polyclonal anti-RFX5c antibody was obtained as described earlier (38). The antibody was bound to anti-rabbit Dynabead M-280 magnetic beads (Dynal, Inc.) as specified by the manufacturer. For coimmunoprecipitation studies, IVT RFX5, RFXAP, and RFX-B (8 μl each of RFX5 and RFXAP and 4 μl of RFX-B) were incubated together at 30°C for 30 min. Depending on the reaction, one or more of the protein products were labeled with either [35S]methionine or [35S]cysteine (Amersham, Inc.). Anti-RFX5 antibody-saturated magnetic beads (5 μl) were added to this reaction mixture, which was then rotated overnight at 4°C. The beads were washed four times with buffer containing 300 mM NaCl, 50 mM Tris (pH 8.0), and 1% NP-40 and then boiled in SDS-PAGE buffer as above and loaded on SDS-PAGE gels. Autoradiography was carried out on the dried gel. In some cases, a PhosphorImager (Molecular Dynamics, Inc.) was used to quantify the coimmunoprecipitated products. Anti-CIITA polyclonal antibodies (5) were purified on an N-hydroxy-succinimide column (Pharmacia, Inc.) linked to Escherichia coli-generated maltose binding protein-CIITA fusion protein (5). The antibody was bound to anti-rabbit Dynabead M-280 magnetic beads as specified by the manufacturer. For CIITA-RFX coimmunoprecipitation, 5 μl each of IVT RFX5, RFXAP, and RFX-B were incubated together at 30°C for 30 min. To the complex was added 15 μl of IVT CIITA, and the mixture was incubated again for 30 min at 30°C. CIITA and associated proteins were then immunoprecipitated overnight using anti-CIITA antibodies attached to magnetic beads. The precipitated complexes were washed four times using a buffer containing 1% NP-40, 150 mM NaCl, and 50 mM Tris (pH 8.0).
Lysates of transiently transfected COS-7 cells described above were sonicated, and immunoprecipitation was carried out using 25 μl of anti-His or anti-HA antibodies (Santa Cruz, Inc.) bound to either rabbit or murine immunoglobulin G magnetic beads. Immunoprecipitates were washed once in lysis buffer, once in lysis buffer containing 300 mM NaCl and 0.1% NP-40, and once in lysis buffer containing no NaCl and 0.1% NP-40. All the immunoprecipitates were analyzed by SDS-PAGE and Western blotting as indicated in the figure legends.
RESULTS
Coimmunoprecipitation studies using an antiserum generated against RFX5 peptides first showed that the RFX complex consists of three proteins (36). The three proteins or subunits, RFX-B, RFX5, and RFXAP, complement the MHC class II deficiency in cell lines representing BLS groups B, C, and D, respectively. The cloning of the genes for these subunits did not provide information about the interactions among these proteins, their transactivation potential, or, for RFX-B and RFXAP, a DNA-binding motif. To investigate the mechanism of RFX function, we coupled mutagenesis of the different RFX subunits with in vitro protein-protein interaction assays, DNA-binding assays, and transient-transfection assays for MHC class II expression. To accomplish this goal, full-length cDNAs for each subunit gene were subcloned into a modified version of the T7 polymerase and mammalian expression vector pcDNA3.1 containing an N-terminal His6 tag. Recombinant RFX subunits generated in vitro using a coupled T7 transcription-reticulocyte translation kit (IVT) were tested for their ability to associate. Immunoprecipitation of metabolically labeled IVT reaction products using an anti-RFX5 antibody showed that efficient association occurred when individual subunit reaction mixtures were combined and incubated for 30 to 60 min at 30°C (Fig. 1A). Additionally, it was found that under these conditions RFX5 could interact directly with RFXAP, albeit to a lesser extent than when all three proteins were present. However, RFX5 did not interact directly with RFX-B.
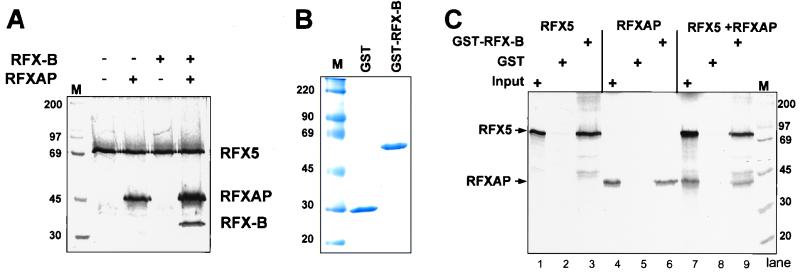
FIG. 1.
Recombinant generated RFX complexes assemble in vitro. (A) IVT RFX subunits were metabolically labeled and incubated in the reactions indicated for 30 min at 30°C. Anti-RFX5-specific antibodies were used to immunoprecipitate RFX5-containing complexes, which were then analyzed by SDS-PAGE and autoradiography. (B) Coomassie blue-stained SDS-PAGE gel containing purified E. coli-generated GST and GST–RFX-B. (C) GST- or GST–RFX-B-loaded glutathione-Sepharose beads were incubated with IVT-produced RFX5 and RFXAP as above. The beads were then pelleted, washed, and analyzed by SDS-PAGE and autoradiography. One-tenth of the input reaction is shown. M, molecular mass standards (in kilobases).
Because the above evaluation of the complex is dependent on the RFX5 antiserum, it was important to verify the interactions from another point of view. Because high-affinity antisera to RFX-B and RFXAP are not available, a chimeric RFX-B protein containing an N-terminal GST tag was generated. Recombinant GST–RFX-B and control GST proteins were produced in E. coli and purified (Fig. 1B). When GST–RFX-B was incubated with IVT-produced RFX5 and RFXAP, association of the three proteins could be detected and purified using glutathione-Sepharose beads (Fig. 1C). When analyzed separately, GST–RFX-B interacted independently with both RFXAP and RFX5 (Fig. 1C), suggesting that multiple protein-protein interactions occur between the RFX subunit. GST alone did not interact with either RFX5 or RFXAP.
All three subunits are required for X-box-specific DNA-binding activity.
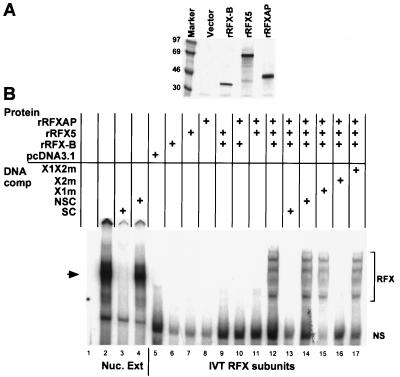
To test the ability of the recombinant proteins to bind DNA in an X1-box-specific manner, a series of EMSAs was performed with a probe containing the X-box region (X1 and X2) of the HLA-DRA promoter. As above, the subunits were synthesized separately using IVT, mixed, and incubated before addition to the DNA-binding reaction mixture. During synthesis, a sample of IVT reaction mixture was removed and metabolically labeled to ascertain the quality of the reactions (Fig. 2A). The presence of all three subunits was required for DNA binding (Fig. 2B, lanes 12 to 17), since individual proteins or all combinations of two of the subunits did not result in a gel shift (lanes 6 to 11). The pattern generated contains four bands and was similar to that described by Masternak et al. (33). For comparison, the native RFX complex using a nuclear extract prepared from the wild-type B-cell line Raji was generated (lanes 2 to 4). The native RFX complex forms a single band (16, 17, 30), which comigrates with the third band from the IVT reactions. The pattern was not affected by cosynthesis of the subunits, changing the order of addition, varying the concentration of one subunit over the others, altering incubation times, or adding IVT-generated CIITA (data not shown). Both the IVT complexes and the native complex were specifically competed by excess cold X-box region DNA but not by a nonspecific competitor. To further test the specificity of DNA binding, competitors with mutations in the X1 box that prevent binding of the native RFX complex were used (30). As shown, the X1 mutant competitors, X1m and X1X2m, did not compete for recombinant RFX binding, but the DNA mutant with a mutation in the X2 box did (lanes 15 to 17). We do not know why a single complex is not formed in this recombinant system. There are several possible explanations. First, all RFX subunits are modified by phosphorylation in their native state (U. M. Nagarajan and J. M. Boss, unpublished data). This and other modifications may contribute to a uniform subunit association and binding conformation that is lacking in the IVT system. Alternatively, the IVT reactions have low levels of partially synthesized protein products, which may influence the conformation of the RFX-DNA complex and lead to complexes with different mobilities. It is also possible that under these conditions, nonequimolar amounts of the subunits bind to the RFX complex. One such possibility may be that RFX-B, which can associate with itself, is causing the multiple complexes. Nonetheless, as stated above, the binding of the IVT complexes is competed by the appropriate RFX specific competitor DNAs and not by X2BP/CREB-specific DNAs, suggesting that RFX DNA-binding specificity is being measured.
FIG. 2.
All three recombinant RFX subunits are required for DNA binding. (A) A portion of the IVT reactions of each subunit was metabolically labeled and analyzed by SDS-PAGE and autoradiography. (B) Upper half of an autoradiograph of an EMSA analyzing the binding of native and recombinant RFX complexes to the X box of HLA-DRA. Lane 1 contains probe with no protein. Lanes 2 to 4 contain native RFX, partially purified from B cell nuclear extracts shown without DNA competitor or with cold X-box DNA (SC) or nonspecific competitor DNA (NSC) added to the reaction. Nuc. Ext, nuclear extract. Lane 5 contains the products of an IVT reaction with vector alone. Lanes 6 to 17 contain the indicated recombinant RFX complexes generated by IVT as described in Materials and Methods. Specific (SC) and nonspecific (NSC) competitor DNA and X-box mutant competitors are indicated. X1m contains a mutated X1 box and wild-type X2 box. X2m contains a mutated X2 box and a wild-type X1 box. X1X2m has mutations in both the X1 and X2 boxes. The major native RFX complex is indicated by the arrow. Specific recombinant RFX-DNA complexes generated by IVT-produced proteins are indicated by the bracket. The bottom band is a nonspecific band (NS) that is derived from the reticulocyte translation mix. The free probe was removed from the picture.
Analysis of RFX-B.
The gene encoding RFX-B (38), also called RFXANK (33), complements BLS-group B cell lines. The amino-terminal portion of RFX-B has homology to a PEST domain (Fig. 3A). PEST domains are found in proteins with a short half-life; however, RFX-B is missing conserved amino acids at the end of the PEST domain, which are crucial for rapid protein turnover, possibly explaining why RFX-B is not seen in diminished amounts compared to the other RFX subunits. The C-terminal portion of RFX-B contains three ankyrin repeats. Ankyrin repeats are typically involved in protein-protein interactions. A role for the RFX-B ankyrin repeats had not been determined. Additionally, previous work showed that RFX-B could be photo-cross-linked to specific base pairs within the 3′ half of the X1 box, suggesting that RFX-B contains a region important for DNA binding of the RFX complex (52).
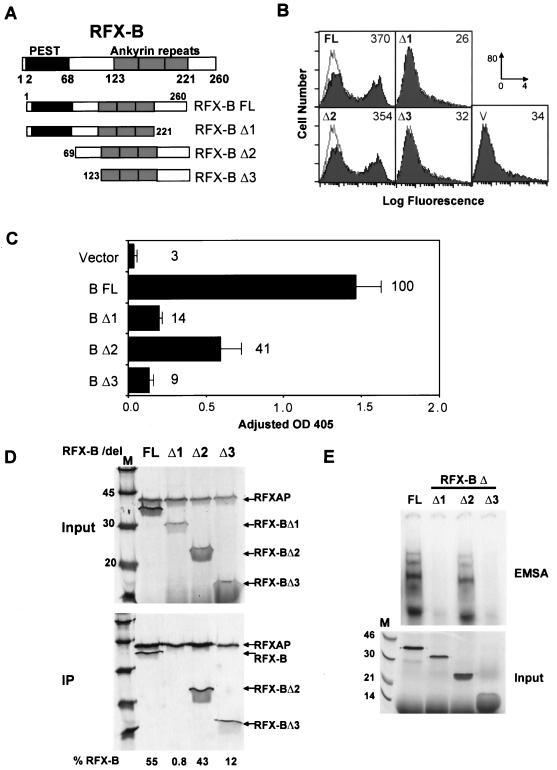
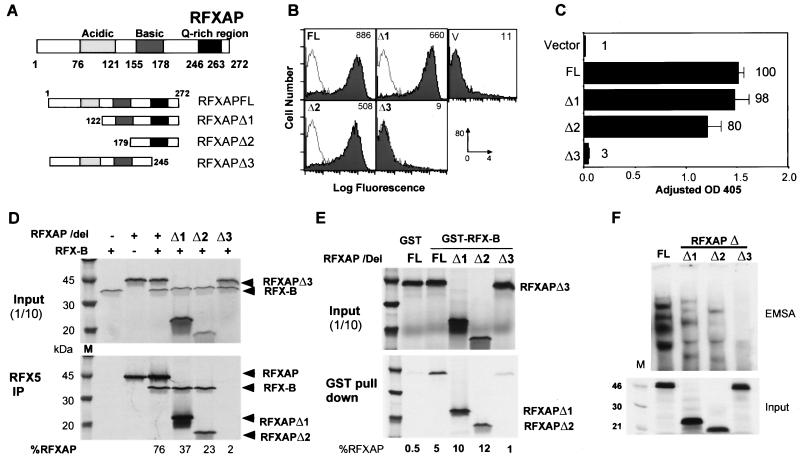
FIG. 3.
Analysis of RFX-B. (A) Schematics of wild-type (FL) and mutant (Δ1 to Δ3) RFX-B constructions shown. RFX-B is 260 amino acids in length, and sequences with homology to PEST domains and ankyrin repeats are indicated. Amino acid boundaries of the domains and the mutant constructions are indicated. (B) BLS group B (Ramia) cells were transiently cotransfected with the indicated RFX-B construction and a GFP expression vector. Cells were stained for surface HLA-DR and analyzed by flow cytometry, gating on the GFP-positive pool. The values in the upper right of each graph indicate the mean fluorescence intensity of the HLA-DR-positive fraction of cells. Panel V shows the vector control transfection that is reiterated as an open histogram on the other panels. The x-axis scale of fluorescence intensity was 100 to 104, and the y-axis scale was 0 to 80 cells. (C) Ramia cells were transiently transfected with the indicated RFX-B construction, a simian virus 40-driven luciferase control vector, and a CAT reporter vector driven by the WXY conserved sequences of the HLA-DRA gene (pDRWXY). CAT assays were normalized to the luciferase values to control for transfection efficiency. The average of three experiments is shown, with the standard error of the mean indicated. The percentage of wild-type RFX-B (B FL) expression is indicated in the graph. OD 405, optical density at 405 nm. (D) Anti-RFX5 antibodies bound to magnetic beads were used to coimmunoprecipitate RFXAP and wild-type or mutant RFX-B proteins. RFXAP and the RFX-B proteins were labeled metabolically. Ten percent of the input and the entire immunoprecipitation (IP) reactions are shown. The percentage of RFX-B in the immunoprecipitation was determined by PhosphorImager analysis of the gel shown. (E) The upper portion of an EMSA using an HLA-DRA X-box probe performed with IVT-generated RFX5, RFXAP, and full-length or mutant RFX-B proteins as indicated is shown. The input panel contains 10 to 20% of each IVT reaction mixture labeled with [35S]methionine and analyzed by SDS-PAGE (12% polyacrylamide) and autoradiography.
To determine the regions of RFX-B responsible for its function, a small series of deletion mutants was constructed (Fig. 3A) and tested for their ability to restore MHC class II expression in transiently transfected cells by using two assays. Transient transfections were carried out in the BLS group B-derived cell line Ramia. Ramia cells are homozygous for a splice site mutation in RFX-B (39). This mutation leads to an unstable mRNA and a frameshifted, truncated protein that is effectively devoid of activity. RFX5, RFXAP, and CIITA are wild type in this cell line. Transfection with full-length RFX-B was previously shown to complement the defect and activate endogenous class II expression (39). In the first assay, the RFX-B wild-type and mutant series were cotransfected with a constitutively expressing GFP expression vector into Ramia cells. The GFP-positive cells were analyzed for HLA-DR surface expression. As shown in Fig. 3B, cells transfected with wild-type RFX-B or RFX-BΔ2 restored endogenous HLA-DR expression. RFX-BΔ1 and RFX-BΔ3 transfections did not restore HLA-DR expression, producing flow cytometry profiles identical to that of the vector control. In the second assay, expression from an X-box-dependent HLA-DRA promoter reporter gene was determined. This assay had similar results to the flow cytometry assay and was able to distinguish between the ability of the wild type and RFX-BΔ2 to complement the defect. RFX-BΔ2 generated 41% of the wild-type RFX-B signal, whereas deletion of the C-terminal 39 amino acids in RFX-BΔ1 or deletion of the N-terminal 122 amino acids in RFX-BΔ3 resulted in only 14 and 9% of the wild-type signal, respectively. These results suggest that deletion of the C-terminal domain or the sequences just N-terminal to the ankyrin repeats but not the PEST-like domain is essential for full RFX-B function.
To determine the nature of the loss of transactivation potential, subunit association and DNA binding of the mutant RFX-B proteins were examined. The three RFX subunits were synthesized by IVT, and coimmunoprecipitations with the anti-RFX5 antibody were performed to probe interactions with the RFX-B deletions (Fig. 3D). In these reactions, only RFXAP and RFX-B were metabolically labeled. As above, full-length RFX-B formed an efficient complex with RFX5 and RFXAP; 55% of the input material was immunoprecipitated. RFX-BΔ2 and, to a lesser extent, RFX-BΔ3 (43 and 12% of input, respectively) associated with the RFX complex. These mutations remove the N-terminal region that contains the PEST homology domain and the sequences linking it to the ankyrin repeats, respectively. In contrast, RFX-BΔ1 did not associate (0.8% of input), even though this deletion retained the three ankyrin repeats originally described (33, 38). The C terminus of RFX-B shows weak homology to a fourth ankyrin repeat (28). RFX-BΔ2 and RFX-BΔ3 both contain this region and were able to associate with the other subunits, suggesting that this region is acting as an association domain essential for RFX-B function.
The failure to associate with the other subunits explains the reduced activity of RFX-BΔ1 but does not fully explain the reason why RFX-BΔ3 has less than 10% of wild-type RFX-B activity. To determine if the RFX-BΔ3 RFX complex could bind X1 box DNA, EMSAs were carried out using IVT-generated RFX subunits. The DNA-binding ability of the RFX-B deletions correlated with their ability to transactivate (Fig. 3E). RFX-BΔ2 showed a strong shift equal to that of full-length RFX-B, even though it showed reduced transactivation. As expected, RFX-BΔ1 did not lead to a DNA-binding complex. Importantly, RFX-BΔ3 did not produce a complex that could bind DNA either. Thus, the deletion in RFX-BΔ3 results in a loss of RFX DNA-binding activity, suggesting that the region between the PEST homology domain and the first ankyrin repeat (amino acids 69 to 123) is required for DNA binding. Using this sequence, protein homology searches (Blocks + [18], Pfam [47], ProDom [1], and PROSITE [20]) failed to identify homologous sequences. Thus, this region of RFX-B may contain a novel DNA-binding domain. Alternatively, this region may stabilize the RFX complex to allow DNA binding by the other subunits of the complex.
Analysis of RFX5.
RFX5 was the first subunit of the RFX complex to be identified. It was cloned by complementation of the BLS group C cell line SJO (48). RFX5 (Fig. 4A) has homology to the DNA-binding motif in the RFX family of proteins and is the only RFX subunit that contains a defined DNA-binding domain. Members of the RFX family also share a conserved dimerization domain, but RFX5 lacks this feature. The region of RFX5 responsible for interactions with the other RFX subunits is not known. The C-terminal portion of RFX5 contains a proline-rich region that is found in some transcriptional activators (53), but the ability of this region to transactivate is also not known.
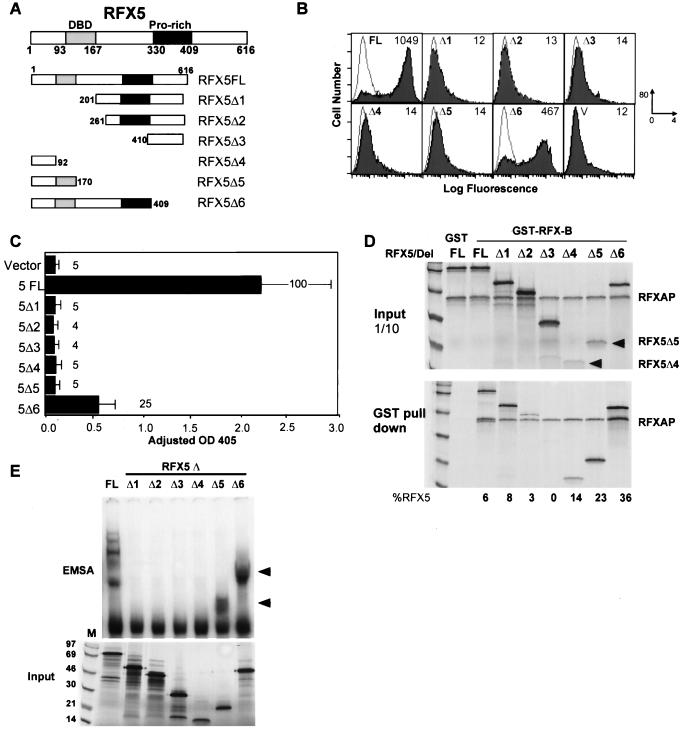
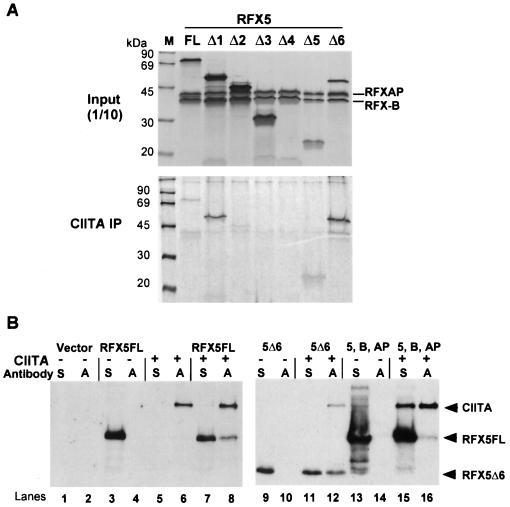
FIG. 4.
Analysis of RFX5. (A) Schematics of wild-type (FL) and mutant RFX5 (Δ1 to Δ6) constructions are shown. RFX5 is 616 amino acids in length. The known DNA-binding domain and proline-rich region of RFX5 are indicated. Domain borders and construction borders are indicated. (B) Flow-cytometric analysis of transient transfections in BLS-group C (SJO) cells is shown. GFP and RFX5 construction cotransfections were performed and analyzed as in Fig. 3. (C) CAT reporter gene assays using wild-type and the indicated mutant RFX5 construction were performed and analyzed as in Fig. 3. OD 405, optical density at 405 nm. (D) GST–RFX-B bound to glutathione-Sepharose was used to isolate RFX5-containing RFX complexes. GST alone did not interact with any of the proteins. In these reactions, RFXAP and RFX5 proteins were metabolically labeled during IVT. Ten percent of the input and the entire pull-down are shown. (E) EMSAs with RFX complexes containing RFX-B, RFXAP, and full-length or mutant RFX5 proteins were performed as in Fig. 3. The arrowheads indicate the positions of DNA complexes containing RFX5Δ5 and RFX5Δ6.
To examine the functionality of RFX5, a series of 5′ and 3′ deletions were introduced into the RFX5 gene (Fig. 4A). The ability of these mutants to transactivate a class II promoter was investigated by performing transient transfections in the SJO cell line with the two assay systems described for the RFX-B mutants. Both assays showed that RFX5 is highly sensitive to mutagenesis. Flow cytometry of the cotransfected SJO cells showed that the wild-type RFX5 restores high levels of HLA-DR surface expression (Fig. 4B). With the exception of RFX5Δ6, all other mutant RFX5 constructs failed to restore surface expression greater than that of the vector control. In the HLA-DRA reporter gene assay, RFX5Δ1 to RFX5Δ5 displayed less than 5% of the wild-type activity (Fig. 4C). RFX5Δ6, which contains a 207-amino-acid deletion in the C terminus, displayed one-fourth the wild-type transactivation activity. The absence of the DNA-binding domain in RFX5Δ1 to RFX5Δ4 is the most probable explanation for their lack of function. The partial activity of RFX5Δ6 compared to RFX5Δ5 indicates that the sequences included in RFX5Δ6 must be important for either subunit association or transactivation.
To assay the ability of the RFX5 mutants to associate with the other RFX subunits, recombinant GST–RFX-B was incubated with in vitro-translated RFXAP and the various RFX5 mutant proteins. Both RFXAP and RFX5 proteins were metabolically labeled. The complexes associating with GST–RFX-B were analyzed following purification on glutathione-Sepharose beads. In contrast to the transactivation data, most of the RFX5 mutants were able to associate with the other subunits (Fig. 4D). RFX5Δ2 showed a 50% reduction in binding, and RFX5Δ3 failed to associate. In most cases, the amount of RFXAP associating with RFX-B remained constant, suggesting some independence in their association. The fact that RFX5Δ1, RFX5Δ2, and RFX5Δ4 were able to associate indicated that there were two regions of RFX5 that were important for interacting with the other subunits: one in the N-terminal 92 amino acids and one between amino acids 201 and 410. The N-terminal domain appeared to be the stronger of the two. It is interesting that all three C-terminal truncation mutants displayed a higher degree of subunit association than did the full-length protein or the N-terminal deletion mutant.
Because most of the RFX5 mutants were able to associate with RFX-B and RFXAP, it was of interest to determine if the associated RFX complex could bind DNA even if the RFX5 DNA-binding domain was deleted. Thus, an X-box EMSA was performed using IVT-generated RFX proteins (Fig. 4E). The results showed that only RFX complexes with RFX5 proteins containing the DNA-binding domain were able to interact with the X-box DNA, despite their ability to form RFX protein complexes. None of the individual RFX5 deletions were able to bind DNA on their own (data not shown). These data suggest that DNA binding of the RFX complex was dependent on both RFX protein complex formation and the DNA-binding domain of RFX5. Due to changes in the sizes of the RFX5 deletions, the banding pattern migrated faster in the gel. The RFX5Δ5 and RFX5Δ6 mutants were either inactive or less active transcriptionally, respectively, than was the wild type, suggesting that the sequences in RFX5Δ6 not included in RFX5Δ5 may be required for transactivation. This analysis therefore suggests that RFX-B and RFXAP cannot contribute to transcriptional activation of this system in the absence of these sequences in RFX5.
Analysis of RFXAP.
RFXAP, the subunit mutated in BLS complementation group D (10), bears no homology to the RFX family of proteins and contains regions rich in acidic and basic amino acids and glutamine (Fig. 5A). Whereas RFX5 and RFX-B made discrete base pair-specific contacts that were detected by photo-cross-linking studies, RFXAP appeared to interact with most of the base pairs across the X1 box (52), suggesting that it may play different role. To further analyze the role of RFXAP in the RFX complex, two N-terminal mutants and one C-terminal mutant were created and analyzed as above (Fig. 5A). In contrast to the above transcriptional activation assays for RFX-B and RFX5, RFXAP required only a small portion of the protein for activity (Fig. 5B and C). Mutations removing increasing amounts of the N-terminal region of the protein (RFXAPΔ1 and RFXAPΔ2) retained near-wild-type levels of activity in both the flow cytometry surface expression and reporter gene assays, while RFXAPΔ3, which lacked the C-terminal portion of the protein, including the glutamine-rich region, had no activity. Based on these results, the glutamine-rich region and the C terminus of the protein were required for activity.
FIG. 5.
Analysis of RFXAP. (A) Schematics of wild-type (FL) and mutant (Δ1 to Δ3) RFXAP constructions are shown. RFXAP is 272 amino acids in length. No homology to other proteins has been identified, although several regions can be found that are rich in acidic, basic, or glutamine residues as indicated. (B and C) Transient cotransfections were carried out as in Fig. 3, except that BLS group D (6.1.6) cells and the wild-type RFXAP and deletion series were used as indicated. OD 405, optical density at 405 nm. (D and E) Anti-RFX5 antibodies bound to magnetic beads (D) or GST-RFX-B bound to glutathione-Sepharose (E) were used to detect RFX complexes containing wild-type or truncated RFXAP subunits. (D) RFXAP and RFX-B were labeled. (E) Only RFXAP was labeled. Ten percent of the input and the entire immunoprecipitation (IP) or pull-down are shown. (F) EMSAs analyzing RFX complexes containing either full-length or mutant RFXAP proteins were performed as in Fig. 3.
The glutamine region was also required for association with RFX5 and RFX-B. Coimmunoprecipitation of IVT RFX-B and RFX5 showed that while RFXAPΔ1 and RFXAPΔ2 had a diminished association with RFX5, RFXAPΔ3 displayed greatly reduced levels of association (Fig. 5D). In this assay, coimmunoprecipitation of RFX5 with RFX-B was dependent on the presence of the glutamine-rich region of RFXAP as well, suggesting that the major interaction between RFX-B and the RFX complex is probably mediated by RFXAP. When investigated by using GST–RFX-B, associations between RFXAPΔ1 and RFXAPΔ2 were increased over those in the wild type (Fig. 5E). However, virtually all the interactions were dependent on the glutamine-rich region and C terminus of RFXAP. When RFX-B was omitted from the reactions in Fig. 5D or RFX5 was omitted from the reactions in Fig. 5E, the results were similar qualitatively (data not shown). However, the strength of binding was decreased compared with the experiments shown. Thus, RFXAP can interact independently with RFX5 and RFX-B. These data are therefore consistent with the hypothesis that RFXAP functions to bridge RFX5 and RFX-B or to stabilize the complex. The DNA-binding activity of the RFXAP deletion series was analyzed by EMSA (Fig. 5F). The results showed that while RFXAPΔ1 and RFXAPΔ2 associate better with the other RFX subunits, they do not bind as tightly to X1-box DNA. Due to changes in the sizes of the RFXAP subunits, the banding pattern migrates slightly faster in the gel. RFXAPΔ3 did not bind DNA, which was most probably due to its poor complex association characteristics.
Ankyrin repeats are required for RFX-B function.
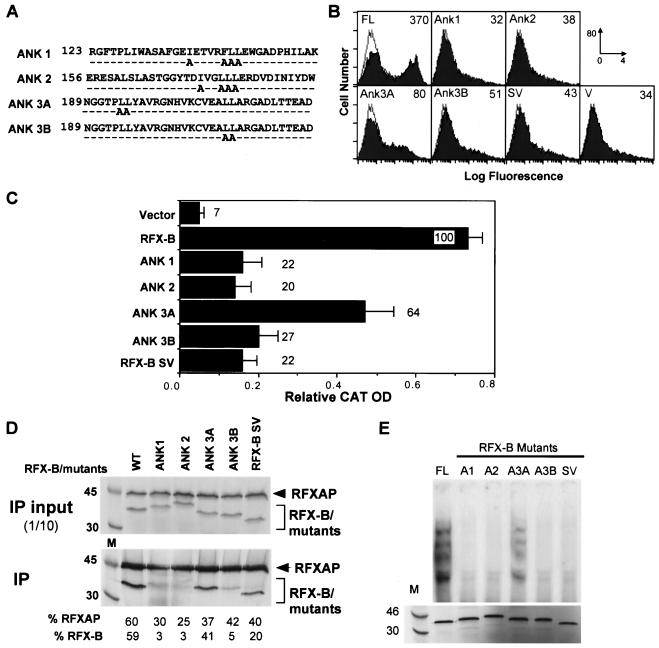
The ankyrin repeats of RFX-B were hypothesized to be important in the association of the RFX complex (33, 38). To test this hypothesis, alanine substitution mutations were introduced into the three ankyrin repeats, with two separate mutations being made in the third ankyrin repeat. The sites chosen are conserved among most ankyrin repeats and are in a hydrophobic region of the repeat (12). In addition to the ankyrin mutations, the naturally occurring splice variant of RFX-B, RFX-BSV (previously termed RFX-BΔ5 [38]), was investigated. RFX-BSV has an in-frame deletion that removes exon 5 but retains the ankyrin repeat region. The transactivation potential of these mutants was tested in BLS group B Ramia cells. Flow cytometry of wild-type RFX-B transfectants revealed reversion of HLA-DR expression in about 50% of the cells (Fig. 6B). A 4.6- to 11.6-fold reduction in activity was seen with all of the mutants in this assay, with the ANK3A mutant displaying the highest level of HLA-DR expression. Similarly, mutants ANK1, ANK2, ANK3B, and RFX-BSV showed approximately a fivefold reduction in the reporter gene assay, while ANK3A displayed 64% of the wild-type level of activity (Fig. 6C). This indicates that the ankyrin repeats are important to RFX-B activity, with the leucines in ANK3B being more crucial than those in ANK3A. Because ankyrin repeats are involved in protein-protein interactions, the effect of the ankyrin mutations on complex association was examined (Fig. 6D). Mutations in ANK1, ANK2, and ANK3B abolished complex association, indicating that the ankyrin repeats were critical for subunit association. This was further enforced by the fact that RFX-BSV, with intact ankyrin repeats, was able to associate. The ability of the ankyrin mutants to bind DNA followed the transactivation results; the only mutant that was able to bind DNA was ANK3A, albeit slightly more weakly than the wild type did (Fig. 6E). Interestingly, RFX-BSV did not bind DNA (Fig. 6E). This splice variant lacks the domain suggested above to be important for DNA binding.
FIG. 6.
Ankyrin repeats in RFX-B are essential for subunit association and function. (A) The alanine substitution mutations introduced into each of the three ankyrin repeats of RFX-B are shown. (B and C) Wild-type RFX-B (FL) or the mutant constructions were analyzed for their ability to restore surface HLA-DR expression (B) or to drive the expression of a HLA-DRA CAT reporter gene (C) following transient transfection of the indicated constructions into Ramia cells as described in Fig. 3. The naturally occurring RFX-B splice variant (RFX-BSV) was also included. (D) Using an anti-RFX5 specific antibody, complex association was assayed by coimmunoprecipitation (IP) of RFX complexes containing wild-type or the indicated mutant RFX-B proteins. RFXAP and RFX-B were labeled during their synthesis. Ten percent of the input is shown. (E) RFX complexes containing IVT-generated RFX-B mutants in panel A were analyzed by EMSA for their ability to bind X-box DNA as in Fig. 3.
CIITA interacts with the RFX complex, principally through RFX5.
CIITA, the gene responsible for the defect in BLS group A cells (49), is responsible for transactivation of this system (44, 49). CIITA does not interact directly with DNA but has been shown to contain a potent transcriptional activation domain in its N terminus (44, 49, 55). It has been proposed by several groups in the field that CIITA interacts with the MHC class II-bound factors and that these interactions lead to activation of gene expression. However, proof of this model and direct interactions have been difficult to obtain. Yeast two-hybrid analysis using RFX5 and CIITA suggested that these two proteins could interact (45). However, the interactions were weak, and it was not clear at the time if the entire RFX complex was required for that interaction, since only RFX5 was tested. The data presented in Fig. 4 (see above) suggest that the proline-rich and C-terminal domains of RFX5 are responsible for transactivation of this system. This leads to the question whether CIITA interacts with RFX5 in a manner dependent on the proline-rich region and C-terminal domain. To begin to answer this question, we developed an antiserum to recombinant CIITA. The antiserum was found to be specific to CIITA by Western blot analysis (5). To determine if CIITA interacted directly with the RFX subunits, CIITA, RFX-B, RFX5, and RFXAP were all synthesized by IVT. CIITA was incubated with each of the subunits separately, and then the CIITA-specific antibodies were used to isolate CIITA and any other coimmunoprecipitating proteins. Only RFX5 was able to associate independently with CIITA, albeit weakly (data not shown). However, when CIITA was incubated with a preformed wild-type RFX complex, CIITA-specific antibodies could reproducibly coimmunoprecipitate some of the input RFX complex (Fig. 7A). Because the initial CIITA-RFX5 observation used a truncated RFX5, the RFX5 mutants shown in Fig. 4 were tested for their ability to interact with CIITA (Fig. 7A). RFX complexes containing RFX5Δ1 and RFX5Δ6 displayed the strongest associations with CIITA (Fig. 7A). In each of these immunoprecipitations, weak interactions with RFXAP could be detected, but only with full-length RFX5 or RFX5Δ6 could RFX-B be detected as well. This is consistent with the data showing the strong associations of RFX5Δ6 with the other subunits. Additionally, RFX5Δ1 and RFX5Δ6 could be coimmunoprecipitated with CIITA without the other subunits, although this interaction was weaker than when RFX-B and RFXAP were present. These RFX5 mutant proteins share the proline-rich domain, which is absent in the other RFX5 mutants except RFX5Δ2. While RFX5Δ2 also shares this domain, it does not associate efficiently with the RFX complex, and this may be the reason why it was not detected. The addition of X-box DNA did not increase reactivity (data not shown). Thus, while these interactions are relatively weak compared to the RFX subunit association reactions, the data provide direct evidence that RFX and CIITA interact and that the interaction is principally through RFX5.
FIG. 7.
CIITA interacts with RFX5. (A) Recombinant CIITA, RFXAP, RFX-B, and each of the indicated RFX5 mutants were synthesized by IVT. With the exception of CIITA, all subunits were labeled metabolically. RFX complexes were assembled first, and CIITA was added later. Anti-CIITA antibodies were used to immunoprecipitate CIITA and any of the associated proteins. Autoradiographs of the SDS-PAGE analysis of the precipitates or 10% of the input proteins are shown. (B) Transient cotransfections of COS-7 cells were performed using HA-tagged CIITA and the indicated His-tagged RFX vectors. Lysates from transfected cells were subjected to immunoprecipitation with anti-HA (A) or anti-His (S) antibodies. The immunoprecipitates were analyzed by Western blotting using biotinylated anti-RFX5 and anti-CIITA antibodies. The arrowheads point to the RFX5- and CIITA-specific bands.
CIITA is expressed at very low levels in cells and is difficult to reproducibly coimmunoprecipitate with the RFX complex. Thus, to show that RFX5 and CIITA interact in cells, both CIITA and RFX5 were overexpressed in COS-7 cells by using an HA-tagged version of CIITA, which has fully activity (data not shown). Following transient transfection, immunoprecipitations were carried out using antibodies specific for the HA tag on CIITA or the His tag on RFX5. The immunoprecipitates were assayed by Western blotting using both RFX5 and CIITA antisera (Fig. 7B). In the anti-His (RFX5) coimmunoprecipitation, a barely detectable CIITA-specific band was observed. However, the anti-HA (CIITA) coimmunoprecipitation showed clear coimmunoprecipitation of RFX5 (Fig. 7B, lane 8), indicating that these proteins interact in cells. Because RFX5Δ6 interacted more strongly in the in vitro assay, it was also analyzed for its ability to interact with CIITA in cells. The results showed an identical pattern to that of the wild-type RFX5 (lanes 11 and 12), supporting the in vitro data. Moreover, the inclusion of RFX-B and RFXAP in the transfections led to the detection of both CIITA and RFX5 irrespective of the antibody used for the coimmunoprecipitation (lanes 15 and 16). Thus, like the IVT interaction experiment, CIITA association with the RFX subunits is enhanced when all three are present. Control transfections and immunoprecipitations showed no background interactions (Fig. 7B).
DISCUSSION
The appropriate regulation of MHC class II gene expression is an important aspect of acquired immune responses. The lack of proper transcriptional control is highlighted in patients with BLS. The discovery of the four genes that are deficient in BLS patients has allowed the present analysis of the interaction and function of their products, CIITA, RFX-B, RFX5, and RFXAP. Using a limited mutagenesis scheme, regions of the RFX subunits responsible for subunit association, DNA binding, and transactivation were defined. The results showed that all four gene products responsible for BLS interact directly with each other and that these interactions are required for MHC class II expression. Our data suggest that RFX complex association is a required first step, followed by DNA binding and subsequent transactivation by RFX interactions with CIITA. A schematic diagram of the interaction domains is presented in Fig. 8A.
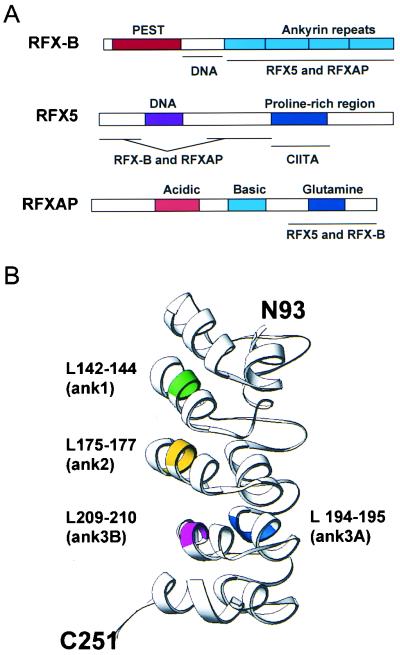
FIG. 8.
Functional domains of the RFX proteins. (A) Schematic representation of the RFX subunits are indicated, with the functionally important regions shown. Interacting domains are listed below each sketch. (B) A model of the RFX-B ankyrin repeat region was generated by the SWISS-MODEL program (http://www.expasy.ch/swissmod/). Colored regions indicate the positions of the alanine substitutions used in the analysis of the ankyrin repeats in Fig. 6.
RFX subunit association.
While it has been known for several years that RFX is a multiprotein complex (36), little was known about how the subunits associate. The ankyrin repeats found during the cloning of RFX-B (33, 38) suggested a series of interacting domains that may have been important for RFX association. The experiments presented here show that these domains are indeed important for interactions with both RFX5 and RFXAP. Alanine substitutions of conserved amino acids within each of the three repeats showed that all three are required for interactions with the RFX complex. The X-ray structures of several ankyrin repeat-containing proteins are known, allowing the computer-generated modeling of the ankyrin repeats of RFX-B. Using the SWISS-MODEL program (40), a hypothetical structure was generated from amino acids 93 to 251 (Fig. 8B). The positions of the alanine substitutions are highlighted in this model. The three mutations that displayed the greatest effect on expression and association all lie in similar positions of the three ankyrin repeats. Interestingly, ANK3A, which retained 64% of the wild-type activity, lies on a different face of the modeled ankyrin repeat. This suggests that the interaction surface in the RFX-B ankyrin repeats is on the left side of the model, which is consistent with other ankyrin-containing proteins (21, 23). Computer modeling of the four mutants showed similar structures to the wild-type structure. Thus, the reduced activity of the mutant protein may be due to reduced hydrophobicity caused by the combined Leu-to-Ala substitutions.
In addition to the three ankyrin repeats reported originally (33, 38), a fourth contiguous ankyrin repeat, displaying weak homology, was identified using computer modeling programs (40). This fourth repeat was also noted by Lin et al., who also cloned RFX-B in a two-hybrid search for Raf-interacting factors (28). Lin et al. suggested that RFX-B could interact with itself. Indeed, GST–RFX-B was able to bind IVT-generated RFX-B (data not shown), suggesting that RFX-B may have other cellular functions, such as interactions with Raf or Raf-like proteins. The loss of this repeat results in the inability of RFX-B to associate with the complex.
RFX5 associated with the complex through two distinct domains that surround its DNA-binding domain. The data suggest that the 92-amino-acid N-terminal domain is most important for association, since it alone can associate with the complex. The second domain is likely to be between the proline-rich region and the DNA-binding domain, since the proline-rich region appears to function in transactivation. There also appears to be an inhibitory region located at the C terminus of RFX5, at amino acids 410 to 616, which, when removed, allows greater subunit association. Its removal in RFX5Δ6 allowed the stronger interactions with CIITA to be detected. While the detection of such a region may be an artifact of designing a minimal-analysis system, it is possible that this region is necessary for interactions with the other class II promoter DNA-binding factors NF-Y or X2BP/CREB, which were not present in our system.
RFXAP was found to associate independently with both RFX5 and RFX-B. However, RFXAP interactions with either protein were increased when all three proteins were present, suggesting stabilization of the complex. The C-terminal Glu-rich domain (amino acids 246 to 272) was required for interactions with both RFX5 and RFX-B. It is also likely that the acidic and basic regions may play a role in subunit association, since the loss of the acidic region resulted in a decrease in association with RFX5. Interactions with RFX-B required only the Glu-rich region, since N-terminal RFXAP mutants retaining this domain still interact with RFX-B. Using a photo-cross-linking system to determine the orientation of the RFX subunits with respect to the X1 box, it was found that RFX5 bound the 5′ half and RFX-B bound the 3′ half (52). Intriguingly, RFXAP was cross-linked with most of the site specific X1 box probes, suggesting that it may have made contacts with the phosphate backbone. Our results are consistent with the idea that RFXAP acts to bridge RFX-B and RFX5 and, in doing so, may place the protein in direct contact with the entire length of the X1 box.
DNA binding of the RFX complex.
For many years, detection of the DNA-binding activity of RFX was controversial (16, 19, 26, 41). Native RFX does not bind DNA with high affinity, having a half-life of <3 min (42). In vivo and in vitro RFX binding to the X1 box is aided by cooperative binding of X2BP/CREB and NF-Y (30, 35, 42). In vitro, these proteins form a very stable protein-DNA complex with a half-life of >4 h (30). Of the three RFX subunits, only RFX5 contains a known DNA-binding motif. This motif is homologous to the motif found in the RFX family of proteins. Recently, the structure of the RFX1 DNA-binding motif was solved and found to belong to the winged-helix subfamily of helix-turn-helix DNA-binding motifs (13). Unlike the other family members, full-length RFX5 is unable to bind DNA independently. A previously described C-terminal truncation of RFX5 is able to bind DNA, leading to the hypothesis that RFX5 must be a member of a multisubunit complex (48). In contrast, RFX complexes containing RFX5Δ5 and RFX5Δ6, the two C-terminal deletions that retain the DNA-binding domain and functioned better than wild-type RFX5 in EMSAs, did not bind DNA independently of the other subunits or as a dimer in combination with either RFX-B or RFXAP (data not shown). This suggests that the original mutant that did bind DNA may have had unusual properties.
A comparison between the mutations in RFX-BΔ2 and RFX-BΔ3 suggests that the region between the PEST homology domain and the first ankyrin repeat of RFX-B (amino acids 69 to 123) is required for DNA binding of the RFX complex. This conclusion is based on the fact that RFX-BΔ2 functions fully but RFX-BΔ3, which retains complex association, lacks DNA-binding activity. Additional evidence for this is derived from the analysis of the naturally occurring splice variant, RFX-BSV (38). The in-frame deletion of this transcript removes exon 5 (amino acids 91 to 112) but contains an intact ankyrin repeat region. RFX-BSV is able to associate with RFX5 and RFXAP in a manner similar to that of wild-type RFX-B, but the complex does not bind DNA. There are several ways in which this region may contribute to DNA binding of the RFX complex. The first is that this region may encode a DNA-binding domain. Computer searches for homologous DNA-binding domains using this region failed to detect any known motif, suggesting that if it does encode such a domain, this domain has a novel structure. Close interactions between RFX-B and the 3′ end of the X1 box were observed by site-specific cross-linking experiments (52), supporting the argument that this region may contact DNA directly. Second, this domain may contribute to the stability of RFX5 interactions with DNA. This may occur by the domain altering the conformation of the RFX complex in such a manner as to improve the DNA-binding activity of RFX5. The analysis of RFXAP did not reveal any DNA-binding regions, although it is likely that the charged regions of RFXAP will interact with DNA. As alluded to above, it is likely that if these interactions occur, they will be nonspecific.
Transactivation and association with CIITA.
Only the RFX5 mutants distinguished between proteins that were able to associate, bind DNA, and transactivate, allowing the identification of a transactivation domain. RFX5Δ6, which lacked the region C-terminal to the proline-rich domain, displayed weak transactivation, suggesting that both the Pro-rich domain and the C-terminal domain were important for transactivation. RFX5Δ5, a mutant that formed RFX complexes with DNA-binding activity and that lacked these sequences, was unable to transactivate. Thus, the N-terminal region of RFX5 does not contribute to transactivation. It should be noted that this analysis does not rule out the other subunits from contributing to transactivation. How does this transactivation domain function? CIITA, the class II transactivator, is required for transcriptional activation in a manner that is dependent on the X box and on the presence of the X-box DNA-binding proteins (44, 55). As mentioned above, Scholl et al. (45) found weak interactions between a truncated RFX5 and CIITA in a two-hybrid analysis, but physical interactions were not shown. Using the recombinant system here, weak interactions were detected between RFX5 and CIITA. Interactions were also detected with some of the RFX5 deletions. These interactions were strengthened by the presence of RFXAP and RFX-B. In each case, the Pro-rich region of RFX5 and at least one complex association domain was present. Immunoprecipitations did not reveal interactions of CIITA with RFX-B or RFXAP. In vivo interactions between CIITA and RFX5 were also found; however, these interactions were substantially weaker than was the association of the RFX subunits with each other. One interpretation of this result is that CIITA associates only transiently with RFX on the MHC class II promoter. This would allow more precise regulation of the system and allow the system to be sensitive to changes in CIITA concentrations and regulation. Additionally, the ability to immunoprecipitate CIITA complexes in cellular lysates may be reduced because the other class II-specific DNA-bound transcription factors X2BP/CREB and NF-Y may not be present. If either of these two hypotheses are correct, the interactions that were detected may be expected. Chromatin immunoprecipitation analysis (34) for CIITA has been able to demonstrate CIITA association at the class II promoter in a manner consistent with RFX binding (G. Beresford and J. M. Boss, unpublished data). Hence, it is most likely that CIITA interacts at the MHC class II promoter. However, it is not clear whether X2BP/CREB or NF-Y function to stabilize the interactions of CIITA at the MHC class II promoter.
These studies have identified regions of each of the RFX proteins that are important for their ultimate function: activating MHC class II gene expression. The interplay between the subunits, DNA binding, and transactivation potential through CIITA present numerous surfaces for intervention with small molecules or peptides, which could be used to modulate MHC class II expression in future clinical and experimental settings.
ACKNOWLEDGMENTS
A. DeSandro and U. M. Nagarajan contributed equally to this work.
This work was supported by NIH grant AI34000.
We thank G. Beresford for discussions and review of the manuscript.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baxter-Lowe L A, Hunter J B, Casper J T, Gorski J. HLA gene amplification and hybridization analysis of polymorphism. HLA matching for bone marrow transplantation of a patient with HLA-deficient severe combined immunodeficiency syndrome. J Clin Investig. 1989;84:613–618. doi: 10.1172/JCI114206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benichou B, Strominger J L. Class II-antigen-negative patient and mutant B-cell lines represent at least three, and probably four, distinct genetic defects defined by complementation analysis. Proc Natl Acad Sci USA. 1991;88:4285–4288. doi: 10.1073/pnas.88.10.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boss J M. Regulation of transcription of MHC class II genes. Curr Opin Immunol. 1997;9:107–113. doi: 10.1016/s0952-7915(97)80166-5. [DOI] [PubMed] [Google Scholar]
- 5.Brown J A, Rogers E M, Boss J M. Mutational analysis of the MHC class II transactivator (CIITA) indicates a functional requirement for conserved LCD motifs and for interactions with the conserved W-box promoter element. Nucleic Acids Res. 1998;26:4128–4136. doi: 10.1093/nar/26.18.4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang C-H, Fontes J D, Peterlin B M, Flavell R A. Class II transactivator (CIITA) is sufficient for the inducible expression of major histocompatibility complex class II genes. J Exp Med. 1994;180:1367–1374. doi: 10.1084/jem.180.4.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chin K-C, Mao C, Skinner C, Riley J L, Wright K L, Moreno C S, Stark G R, Boss J M, Ting J P-Y. Molecular analysis of G1B and G3A IFN-γ mutants reveals that defects in CIITA or RFX result in defective class II MHC and Ii gene induction. Immunity. 1994;1:687–697. doi: 10.1016/1074-7613(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 8.Collins T, Korman A J, Wake C T, Boss J M, Kappes D J, Fiers W, Ault K A, Gimbrone M A, Jr, Strominger J L, Pober J S. Immune interferon activates multiple class II major histocompatibility complex genes and the associated invariant chain gene in human endothelial cells and dermal fibroblasts. Proc Natl Acad Sci USA. 1984;81:4917–4921. doi: 10.1073/pnas.81.15.4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeSandro A, Nagarajan U M, Boss J M. The bare lymphocyte syndrome: molecular clues to the transcriptional regulation of major histocompatibility complex class II genes. Am J Hum Genet. 1999;65:279–286. doi: 10.1086/302519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Durand B, Sperisen P, Emery P, Barras E, Zufferey M, Mach B, Reith W. RFXAP, a novel subunit of the RFX DNA binding complex, is mutated in MHC class II deficiency. EMBO J. 1997;16:1045–1055. doi: 10.1093/emboj/16.5.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elhasid R, Etzioni A. Major histocompatibility complex class II deficiency: a clinical review. Blood Rev. 1996;10:242–248. doi: 10.1016/s0268-960x(96)90008-9. [DOI] [PubMed] [Google Scholar]
- 12.Ewaskow S P, Sidorova J M, Hendle J, Emery J C, Lycan D E, Zhang K Y, Breeden L L. Mutation and modeling analysis of Saccharomyces cerevisiae Swi6 ankyrin repeats. Biochemistry. 1998;37:4437–4450. doi: 10.1021/bi972652e. [DOI] [PubMed] [Google Scholar]
- 13.Gajiwala K S, Chen H, Cornille F, Roquest B P, Reith W, Mach B, Burley S K. Structure of the winged-helix protein hRFX1 reveals a new mode of DNA binding. Nature. 2000;403:916–921. doi: 10.1038/35002634. [DOI] [PubMed] [Google Scholar]
- 14.Gladstone P, Pious D. Stable variants affecting B cell alloantigens in human lymphoid cells. Nature. 1978;271:459–461. doi: 10.1038/271459a0. [DOI] [PubMed] [Google Scholar]
- 15.Griscelli C, Lisowska-Grospierre B, Mach B. Combined immunodeficiency with defective expression in MHC class II genes. Immunodefic Rev. 1989;1:135–153. [PubMed] [Google Scholar]
- 16.Hasegawa S L, Riley J L, Sloan J H, Boss J M. Protease treatment of nuclear extracts distinguishes between class II major histocompatibility complex X1 box DNA-binding proteins in wild type and class II deficient B cells. J Immunol. 1993;150:1781–1793. [PubMed] [Google Scholar]
- 17.Hasegawa S L, Sloan J H, Reith W, Mach B, Boss J M. Regulatory factor-X binding to mutant HLA-DRA promoter sequences. Nucleic Acids Res. 1991;19:1243–1249. doi: 10.1093/nar/19.6.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henikoff S, Henikoff J G. Protein family classification based on searching a database of blocks. Genomics. 1994;19:97–107. doi: 10.1006/geno.1994.1018. [DOI] [PubMed] [Google Scholar]
- 19.Herrero-Sanchez C, Reith W, Silacci P, Mach B. The DNA-binding defect observed in major histocompatibility complex class II regulatory mutants concerns only one member of a family of complexes binding to the X boxes of class II promoters. Mol Cell Biol. 1992;12:4076–4083. doi: 10.1128/mcb.12.9.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hofmann K, Bucher P, Falquet L, Bairoch A. The PROSITE database, its status in 1999. Nucleic Acids Res. 1999;27:215–219. doi: 10.1093/nar/27.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huxford T, Huang D-B, Malek S, Ghosh G. The crystal structure of the I Kappa B alpha/NF-Kappa B complex reveals mechanisms of NF-Kappa B inactivation. Cell. 1998;95:759–770. doi: 10.1016/s0092-8674(00)81699-2. [DOI] [PubMed] [Google Scholar]
- 22.Jabrane-Ferrat N, Fontes J D, Boss J M, Peterlin B M. Complex architecture of major histocompatibility complex class II promoters: reiterated motifs and conserved protein-protein interactions. Mol Cell Biol. 1996;16:4683–4690. doi: 10.1128/mcb.16.9.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacobs M D, Harrison S C. Structure of an I kappa B alpha/NF-kappa B complex. Cell. 1998;95:749–758. doi: 10.1016/s0092-8674(00)81698-0. [DOI] [PubMed] [Google Scholar]
- 24.Kara C J, Glimcher L H. In vivo footprinting of MHC class II genes: bare promoters in the bare lymphocyte syndrome. Science. 1991;252:709–712. doi: 10.1126/science.1902592. [DOI] [PubMed] [Google Scholar]
- 25.Kara C J, Glimcher L H. Developmental and cytokine-mediated regulation of MHC class II gene promoter occupancy in vivo. J Immunol. 1993;150:4934–4942. [PubMed] [Google Scholar]
- 26.Kouskoff V, Mantovani R M, Candeias S M, Dorn A, Staub A, Lisowska Grospierre B, Griscelli C, Benoist C O, Mathis D J. NF-X, a transcription factor implicated in MHC class II gene regulation. J Immunol. 1991;146:3197–3204. [PubMed] [Google Scholar]
- 27.Kozak M. The scanning model for translation: an update. J Cell Biol. 1999;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin J-H, Makris A, McHahon C, Bear S E, Patriotis C, Prasad V R, Brent R, Golemis E A, Tsichlis P N. The Ankyrin repeat-containing adaptor protein Tvl-1 is a novel substrate and regulator of Raf-1. J Biol Chem. 1999;274:14706–14715. doi: 10.1074/jbc.274.21.14706. [DOI] [PubMed] [Google Scholar]
- 29.Lisowska-Grospierre B, Charron D J, de Preval C, Durandy A, Griscelli C, Mach B. A defect in the regulation of major histocompatibility complex class II gene expression in human HLA-DR negative lymphocytes from patients with combined immunodeficiency syndrome. J Clin Investig. 1985;76:381–385. doi: 10.1172/JCI111974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Louis-Plence P, Moreno C S, Boss J M. Formation of a regulatory factor X/X2 box-binding protein/nuclear factor-Y multiprotein complex on the conserved regulatory regions of HLA class II genes. J Immunol. 1997;159:3899–3909. [PubMed] [Google Scholar]
- 31.Mach B, Steimle V, Martinez-Soria E, Reith W. Regulation of MHC class II genes: lessons from a disease. Annu Rev Immunol. 1996;14:301–331. doi: 10.1146/annurev.immunol.14.1.301. [DOI] [PubMed] [Google Scholar]
- 32.Mach B, Steimle V, Reith W. MHC class II-deficient combined immunodeficiency: a disease of gene regulation. Immunol Rev. 1994;138:207–221. doi: 10.1111/j.1600-065x.1994.tb00853.x. [DOI] [PubMed] [Google Scholar]
- 33.Masternak K, Barras E, Zufferey M, Conrad B, Corthals G, Aebersold R, Sanchez J-C, Hochstrasser D F, Mach B, Reith W. A gene encoding a novel RFX-associated transactivator is mutated in the majority of MHC class II deficiency patients. Nat Genet. 1998;20:273–277. doi: 10.1038/3081. [DOI] [PubMed] [Google Scholar]
- 34.Moreno C S, Beresford G, Louis-Plence P, Morris A C, Boss J M. CREB regulates MHC class II expression in a CIITA-dependent manner. Immunity. 1999;10:143–151. doi: 10.1016/s1074-7613(00)80015-1. [DOI] [PubMed] [Google Scholar]
- 35.Moreno C S, Emery P, West J E, Durand B, Reith W, Mach B, Boss J M. Purified X2BP cooperatively binds the class II MHC X box region in the presence of purified RFX, the X box factor deficient in the bare lymphocyte syndrome. J Immunol. 1995;155:4313–4321. [PubMed] [Google Scholar]
- 36.Moreno C S, Rogers E M, Brown J A, Boss J M. RFX, a bare lymphocyte syndrome transcription factor, is a multimeric phosphoprotein complex. J Immunol. 1997;158:5841–5848. [PubMed] [Google Scholar]
- 37.Morris A C, Riley J L, Fleming W H, Boss J M. MHC class II gene silencing in trophoblast cells is caused by inhibition of CIITA expression. Am J Reprod Immunol. 1998;40:385–394. doi: 10.1111/j.1600-0897.1998.tb00423.x. [DOI] [PubMed] [Google Scholar]
- 38.Nagarajan U M, Louis-Plence P, DeSandro A, Nilsen R, Bushey A, Boss J M. RFX-B is the gene responsible for the most common cause of the bare lymphocyte syndrome, a MHC class II immunodeficiency. Immunity. 1999;10:153–162. doi: 10.1016/s1074-7613(00)80016-3. [DOI] [PubMed] [Google Scholar]
- 39.Nagarajan U M, Peijnenburg A, Gobin S J P, Boss J M, van den Elsen P J. Novel mutations within the RFX-B gene and partial rescue of MHC and related genes through exogenous class II transactivator in RFX-B-deficient cells. J Immunol. 2000;164:3666–3674. doi: 10.4049/jimmunol.164.7.3666. [DOI] [PubMed] [Google Scholar]
- 40.Peitsch N, Guex N. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modelling. Electrophoresis. 1997;18:2714. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 41.Reith W, Satola S, Herreo-Sanchez C, Amaldi I, Lisowska-Grospierre B, Griscelli C, Hadam M R, Mach B. Congenital immunodeficiency with a regulatory defect in MHC class II gene expression lacks a specific HLA-DR promoter binding protein, RF-X. Cell. 1988;53:897–906. doi: 10.1016/s0092-8674(88)90389-3. [DOI] [PubMed] [Google Scholar]
- 42.Reith W, Siegrist C A, Durand B, Barras E, Mach B. Function of major histocompatibility complex class II promoters requires cooperative binding between factors RFX and NF-Y. Proc Natl Acad Sci USA. 1994;91:554–558. doi: 10.1073/pnas.91.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Riley J L, Boss J M. Class II MHC transcriptional mutants are defective in higher order complex formation. J Immunol. 1993;151:6942–6953. [PubMed] [Google Scholar]
- 44.Riley J L, Westerheide S D, Price J A, Brown J A, Boss J M. Activation of class II MHC genes requires both the X box region and the class II transactivator (CIITA) Immunity. 1995;2:533–543. doi: 10.1016/1074-7613(95)90033-0. [DOI] [PubMed] [Google Scholar]
- 45.Scholl T, Mahanta S K, Strominger J L. Specific complex formation between the type II bare lymphocyte syndrome-associated transactivators CIITA and RFX5. Proc Natl Acad Sci USA. 1997;94:6330–6334. doi: 10.1073/pnas.94.12.6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seidl C, Saraiya C, Osterweil Z, Fu Y P, Lee J S. Genetic complexity of regulatory mutants defective for HLA class II expression. J Immunol. 1992;148:1576–1584. [PubMed] [Google Scholar]
- 47.Sonnhammer E L, Eddy S R, Birney E, Bateman A, Durbin R. Pfam: multiple sequence alignments and HMM-profiles of protein domains. Nucleic Acids Res. 1998;26:320–322. doi: 10.1093/nar/26.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steimle V, Durand B, Emmanuele B, Zufferey M, Hadam M R, Mach B, Reith W. A novel DNA-binding regulatory factor is mutated in primary MHC class II deficiency (bare lymphocyte syndrome) Genes Dev. 1995;9:1021–1032. doi: 10.1101/gad.9.9.1021. [DOI] [PubMed] [Google Scholar]
- 49.Steimle V, Otten L A, Zufferey M, Mach B. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency (or bare lymphocyte syndrome) Cell. 1993;75:135–146. [PubMed] [Google Scholar]
- 50.Steimle V, Siegrist C-A, Mottet A, Lisowska-Grospierre B, Mach B. Regulation of MHC class II expression by interferon-gamma mediated by the transactivator gene CIITA. Science. 1994;265:106–108. doi: 10.1126/science.8016643. [DOI] [PubMed] [Google Scholar]
- 51.Stimac E, Urieli-Shoval S, Kempin S, Pious D. Defective HLA DRA X box binding in the class II transactive transcription factor mutant 6.1.6 and in cell lines from class II immunodeficient patients. J Immunol. 1991;146:4398–4405. [PubMed] [Google Scholar]
- 52.Westerheide S D, Boss J M. Orientation and positional mapping of the subunits of the multicomponent transcription factors RFX and X2BP to the major histocompatibility complex class II transcriptional enhancer. Nucleic Acids Res. 1999;27:1635–1641. doi: 10.1093/nar/27.7.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiao H, Lis J T, Greenblatt J, Friesen J D. The upstream activator CTF/NF1 and RNA polymerase II share a common element involved in transcriptional activation. Nucleic Acids Res. 1994;22:1966–1973. doi: 10.1093/nar/22.11.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang Z, Accolla R S, Pious D, Zegers B J, Strominger J L, Latron F, Jotterand Bellomo M, Maffei A, Scarpellino L, Bernard M. Two distinct genetic loci regulating class II gene expression are defective in human mutant and patient cell lines. Proc Natl Acad Sci USA. 1988;85:2229–2233. [Google Scholar]
- 55.Zhou H, Glimcher L H. Human MHC class II gene transcription directed by the carboxyl terminus of CIITA, one of the defective genes in type II MHC combined immune deficiency. Immunity. 1995;2:545–553. doi: 10.1016/1074-7613(95)90034-9. [DOI] [PubMed] [Google Scholar]